Abstract
Lipid transfer proteins mediate non-vesicular transport of lipids at membrane contact sites to regulate the lipid composition of organelle membranes. Recently, a new type of rod-like lipid transfer protein has emerged; these proteins contain a long hydrophobic groove and can mediate bulk transport of lipids between organelles. Here we review recent insights into the structure of these proteins and identify a repeating modular unit that we propose to name the repeating β-groove (RBG) domain. This new structural understanding conceptually unifies all the RBG domain-containing lipid transfer proteins as members of a RBG protein superfamily. We also examine the biological functions of these lipid transporters in normal physiology and disease and speculate on the evolutionary origins of RBG proteins in bacteria.
Keywords: lipids, lipid transfer proteins, membrane contact sites, Vps13, autophagy, AlphaFold
Non-vesicular lipid transfer occurs at membrane contact sites
The lipid composition of each organelle membrane is unique and plays an essential role in maintaining organelle identity and function. Lipids can be moved between organelles via vesicular or non-vesicular trafficking. Seminal work in the 1980s suggested that phospholipids and sterols are primarily transported via non-vesicular routes [1,2]; since then, evidence supporting the primacy and importance of nonvesicular lipid transport has continued to grow. Non-vesicular lipid trafficking is carried out by lipid transfer proteins (LTPs), cytoplasmic proteins that lower the energy barrier for lipids to move between membranes across aqueous spaces [3].
Most known LTPs fold to form a box-like shape with a hydrophobic lining capable of holding a single lipid [4,5]. These box-like LTPs transfer lipids via a shuttling mechanism, selecting one lipid molecule at a time based on headgroup and transferring it from donor to acceptor membranes [4]. Often this occurs at membrane contact sites, locations where two organelles are in close enough proximity that a single protein can bridge the gap [6].
In the last four years, a new class of LTP has emerged. These LTPs are large proteins that fold to form long bridge-like structures that span the entire distance between membranes at membrane contact sites [7–13]. The hydrophobic lining of these LTPs enables them to function like lipid superhighways connecting organelle membranes [7,9,12,13]. Five members of this LTP family have been identified: VPS13, ATG2, the Hob proteins, Tweek/Csf1/KIAA1109 and SHIP164 [7–9,12–18]. In this review, we examine recent advances in understanding the structure of this novel superfamily of bridge-like LTPs, highlighting a shared structural feature composed of a repeating series of β-sheets that we call the repeating β-groove (RBG) domain. We also review recent developments in characterizing the molecular, cellular and physiological functions of these proteins and speculate on their evolutionary origins.
A new family of eukaryotic LTPs with long hydrophobic grooves
VPS13 and ATG2 are large (3000–4000 and ~2000 amino acids, respectively) proteins that are highly conserved among eukaryotes. Structural studies in the last four years showed that both proteins fold to form a rod with an internal hydrophobic cavity along its length. A crystal structure of the N-terminal 345 residues of fungal VPS13 revealed a β-sheet curved into a U-shape; crucially, the interior of the “U” was hydrophobic, while the exterior was hydrophilic [7]. This work was followed by single particle cryo-electron microscopy (EM) studies of a much longer fragment (N-terminal 1400 amino acids), which showed that the curved β-sheet structure continued down the length of the protein to form a long hydrophobic groove [9] (Fig. 1). A crystal structure of an N-terminal fragment of ATG2 [13] and cryo-EM analysis of full-length human ATG2A [12] also found a similar long hydrophobic groove (Fig. 1). VPS13 and ATG2 were shown to transfer lipids in vitro [7,11–13,19]. Thus, these two proteins were the founding members of a new superfamily of LTPs with long hydrophobic grooves.
Figure 1. Long hydrophobic groove lipid transfer proteins share structural similarities.
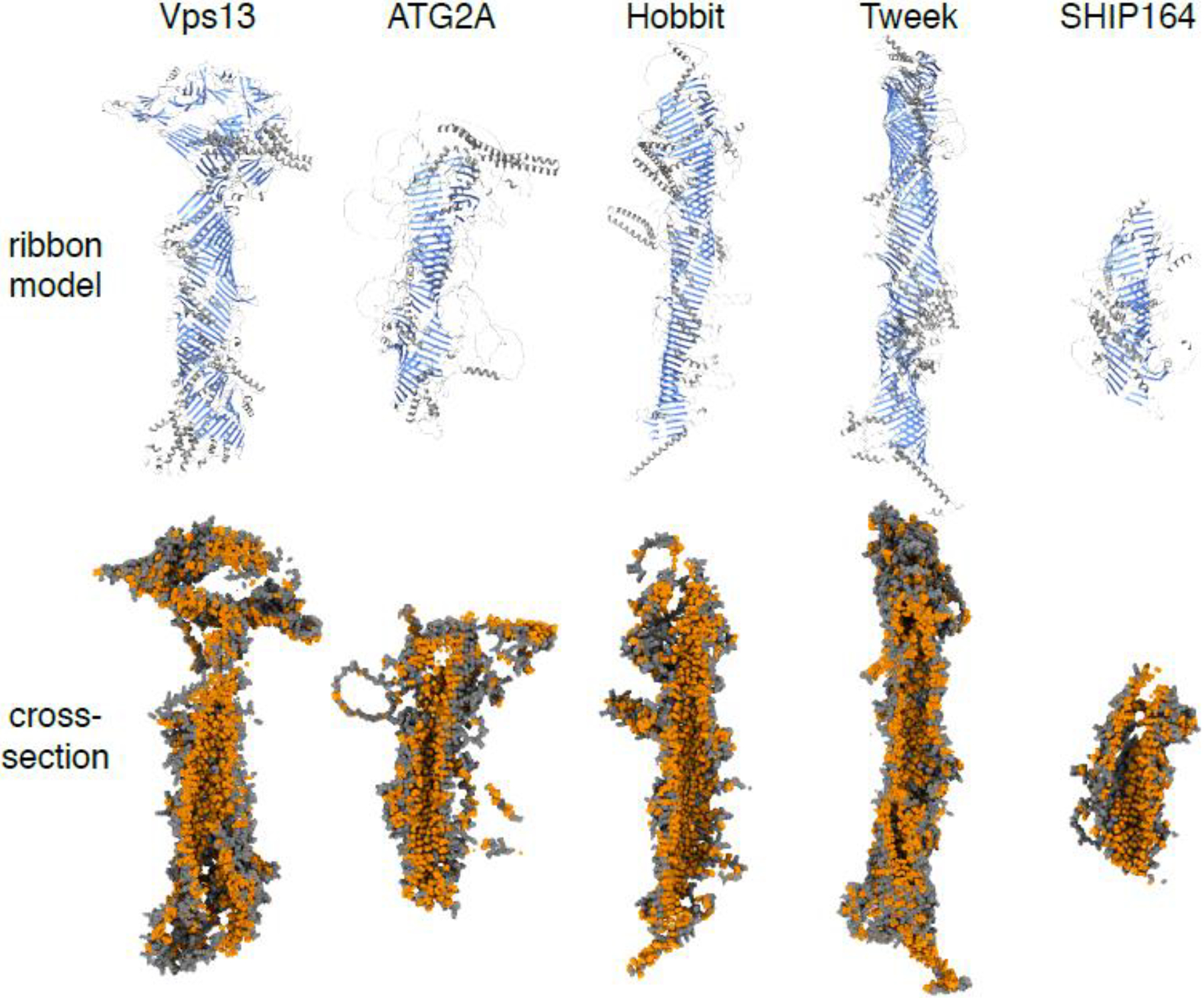
Ribbon models (top) and cross-sections of sphere models (bottom) showing representative members of five protein families that consist of long bridge-like structures comprised primarily of β-sheets (labeled blue in ribbon models), which form a groove lined with hydrophobic residues (glycine, alanine, valine, leucine, isoleucine, proline, phenylalanine, methionine and tryptophan; labeled orange in sphere models). The origins of the models are S. cerevisiae Vps13, H. sapiens ATG2A, H. sapiens Hobbit (KIAA0100), S. cerevisiae Tweek (Csf1) and H. sapiens SHIP164. The SHIP164 rendering omits two long unstructured loops (residues 873–1170 and 1360–1464). PDB files for Vps13 and Csf1 were obtained from Jerry Yang and William Prinz, who used trRosetta to predict regions of ~1000 amino acids, then assembled a complete model from overlapping segments [16]. PDB files for ATG2A, Hob and SHIP164 were obtained from the AlphaFold database [21]. All renderings were generated in ChimeraX [78]. Note that the VPS13 model contains a disconnect in the hydrophobic groove coincident with a large insert called the VAB domain ([23]; further discussed in Fig. 3).
Three additional members of this novel LTP superfamily were recently uncovered. SHIP164 (also called UHRF1BP1L; 1200–1500 residues) was identified via sequence homology and cryo-EM as a VPS13-like protein conserved in all eukaryotes except fungi [14]. Two additional long hydrophobic groove LTPs were identified via remote homology to VPS13/ATG2 using HHpred [20]: Tweek and the Hob proteins [16,17]. Like the other bridge-like LTPs, both are large proteins (3000–5000 and 2300–3000 residues, respectively). Tweek (Csf1 in Saccharomyces cerevisiae, KIAA1109 in humans) is conserved in fungi and metazoans (opisthokonts); the Hob proteins (Hobbit in Drosophila melanogaster, KIAA0100 in humans, SABRE/KIP and APT1 in plants) are conserved throughout eukaryotes. The homology searches were confirmed upon release of the protein structure prediction program AlphaFold [21], which predicts that Tweek and the Hob proteins fold to form rods with internal hydrophobic grooves, structures with marked similarity to VPS13 and ATG2 (Fig. 1). Modeling by trRosetta, another high-performing protein folding prediction program [22], produced similar structures [16]. While there are some caveats that must be considered when working with protein prediction programs, they are generally accurate for folded domains (Box 1). The most conspicuous feature of all five bridge-like LTPs is an extended run of U-shaped β-sheets that together form the hydrophobic groove (Fig. 1). These β-sheets can contain more than 70 β-strands, stretching >30 nm in length (Fig. 2E).
Box 1. Considerations for interpretation of AlphaFold structures.
AlphaFold2, as the first computational tool capable of predicting protein structures with near-experimental accuracy [21], has undoubtedly contributed a striking advance in our understanding of protein folding in general and particularly in understanding the structure of long hydrophobic groove LTPs. However, it is not perfect [59]. When interpreting AlphaFold predictions like those for the proteins under review here, significant issues remain. One is that large proteins are generally not pre-made in the AlphaFold database. In the S. cerevisiae proteome, the ten largest proteins (>2700 amino acids long, including Vps13 and Csf1) are all absent. This problem can be circumvented by submitting overlapping segments (up to 1000 amino acids) to the ColabFold version of AlphaFold2 [60] or to trRosetta [22]. The overlapping regions of the partial models can then be assembled to generate a full-length prediction. This approach was used to generate the models of Vps13 and Tweek shown in this review and in other recent publications [16,17]. A second issue is that AlphaFold provides visualizations of 100% of each protein, even those regions that are predicted with a very low degree of confidence. This particularly applies to unstructured loops. Even short loops are typically missing from experimentally determined structures since they are too mobile to be visualized by crystallography, nuclear magnetic resonance (NMR) imaging or single particle cryo-EM. While having everything predicted is useful for creating complete structures of proteins with multiple short loops like those seen in this family of LTPs, longer loops likely exist in a wide range of positions in vivo. Thus, specification of a single conformation by AlphaFold must be interpreted with caution. A related issue arises in protein regions where there is no strong prediction for secondary structure or any intra-chain interactions. In these situations, AlphaFold tends to predict unstructured loops by default. However, these loops usually do not conform to the stereochemical rules that apply to polypeptide chains, meaning that these they might best be viewed as arbitrary linkers [59]. Although it is critical to keep these caveats in mind when analyzing structural predictions, AlphaFold is a remarkable and powerful tool for folded domains.
Figure 2. The “repeating β-groove” (RBG) domain is the modular building block of RBG lipid transfer proteins.
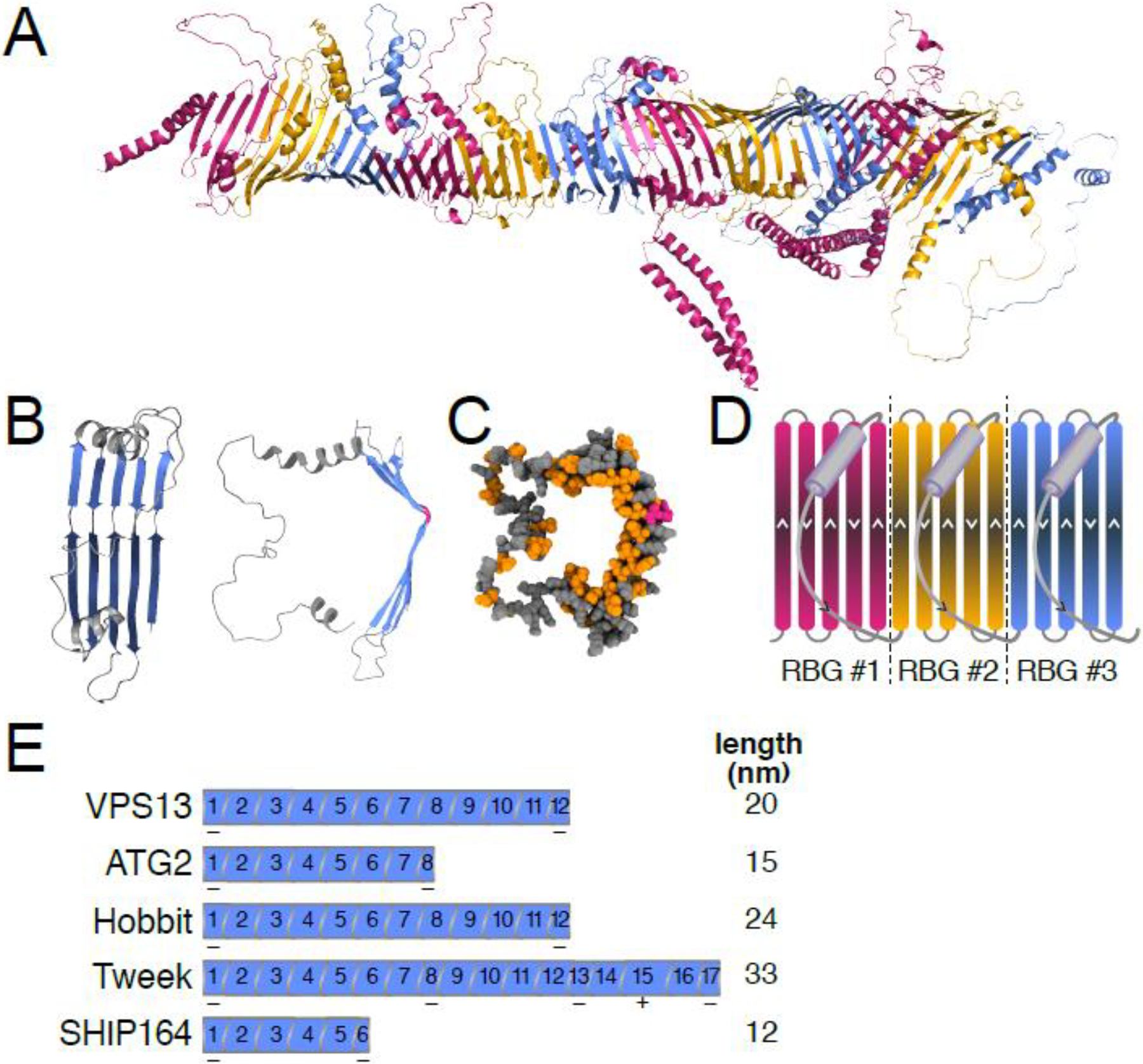
(A) Ribbon model of D. melanogaster Hobbit with RBG domains alternately labeled in pink, yellow and blue. Rendering generated using PyMOL version 2.3.4. PDB file obtained from the AlphaFold database [21]. (B) Model of a single RBG domain from D. melanogaster Hobbit showing the basic RBG domain structure consisting of five antiparallel β-strands (blue) that form a groove, followed by an unstructured loop that crosses back over the β-sheet (gray). Ribbon models: left – front view, middle – side-view (90° rotation). Note the presence of strand-breaking residues (colored pink) near the middle of each strand that generate curvature. (C) Sphere model side-view of the same RBG domain in (B), demonstrating that the concave surface (interior) of the RBG domain’s groove is lined with hydrophobic residues (orange), while the convex surface (exterior) is hydrophilic (gray). Strandbreaking residues shown in pink. Models generated with ChimeraX [78]. (D) Cartoon illustration of three contiguous RBG domains. The direction of each β-strand and loop is indicated by arrowheads. (E) Illustration of the number of RBG domains present in each RBG protein family, as well as the total length of each predicted protein. Note that the initial and final RBG domains are shorter in each protein (indicated by “−”); additionally, Tweek contains two RBG domains with three β-strands and one with seven β-strands (indicated by “+”).
The repeating β-groove (RBG) domain: a modular building block of long hydrophobic groove LTPs
Analysis of the predicted AlphaFold structures for VPS13, ATG2, Hobbit, Tweek and SHIP164 reveals an intriguing pattern: all the hydrophobic grooves are composed of a simple modular unit that oligomerizes head-to-tail to build a long rod. The module contains five antiparallel β-strands followed by a sixth element consisting of a disordered loop usually starting with a short helix that curves back across the β-sheet (Fig. 2A–D). The β-strands form a U-shape by bending at a 30–60° angle at their mid-points, which are frequently populated by strand-breaking residues (glycine/proline) (Fig. 2B–C). The residues populating the inner face of the U-shaped β-sheet are hydrophobic, while those on the exterior face are hydrophilic (Fig. 2C). Multimerization of these repeating units creates an unbroken chain of structurally identical repeats that together build the hydrophobic groove (Fig. 2A, D). This pattern of repeating units had already been seen in the high resolution cryo-EM structure of VPS13 [9] (Fig. 3A), validating the AlphaFold predictions. The modular structure of these proteins is unique in eukaryotes, since most known LTPs form a box-like shape with a hydrophobic pocket that holds a single lipid (e.g. OSBP and Sec14 family proteins) [4]. We propose to call this repeating module of five antiparallel β-strands followed by a loop the repeating β-groove (RBG) domain and LTPs with long hydrophobic grooves RBG proteins.
Figure 3. Structural features of RBG proteins in eukaryotes and prokaryotes.
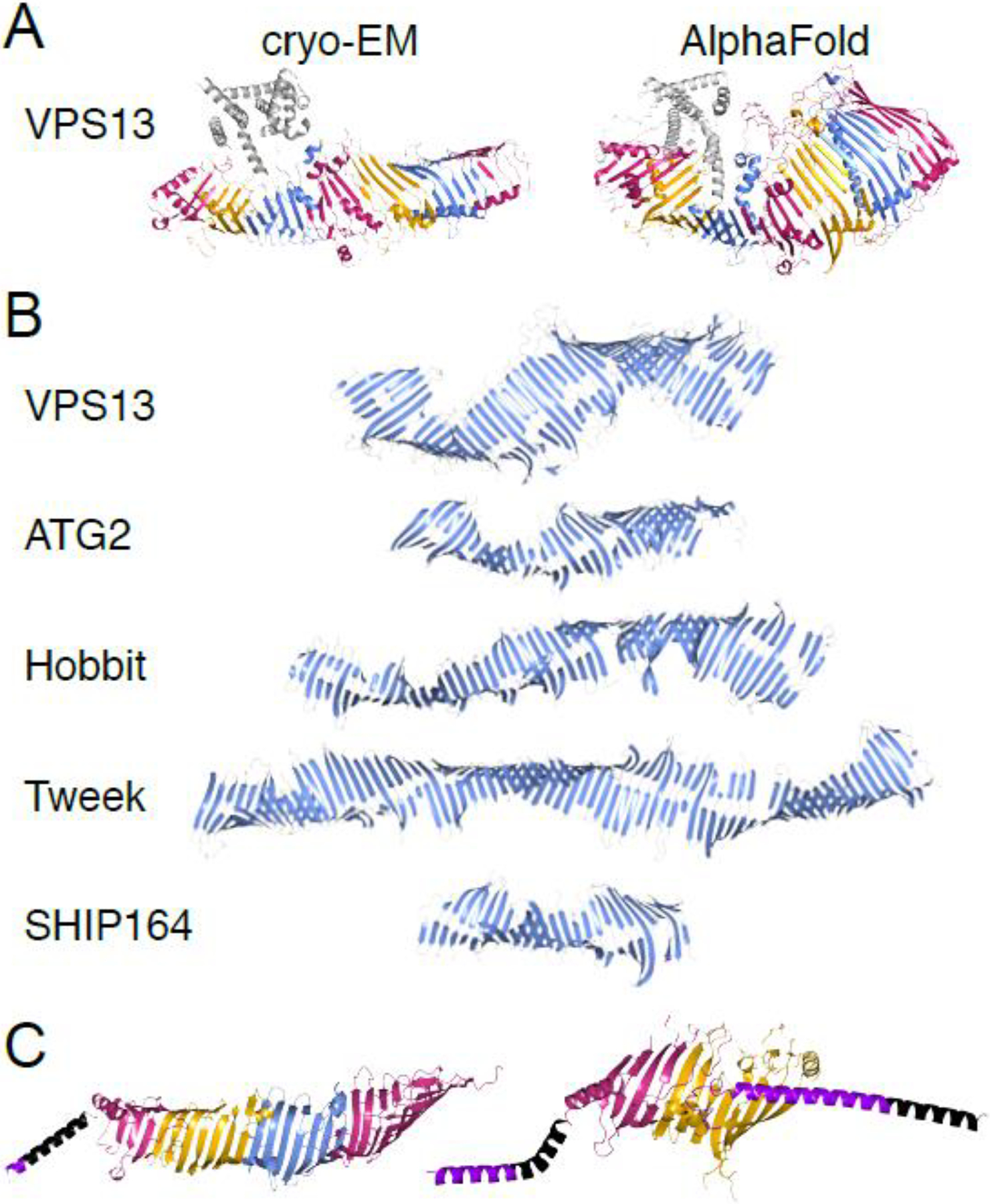
(A) Ribbon models of cryo-EM structure of Chaetomium thermophilum Vps13 (left; residues 6–1381) [9] and AlphaFold prediction of rat VPS13A (right; residues 1–1388) [21] show consistency of RBG domain structure between experimental and predicted models. Differences include diameter of the hydrophobic groove and degree of superhelical rotation. Coloring: RBG domains as Figure 2A; helical “handle” [9] gray. PDB file of Vps13 cryo-EM provided by Karin Reinisch [9]. (B) Ribbon models of the five eukaryotic RBG proteins showing only β-sheets; long loops and all helices omitted. (C) Ribbon models of YicH (E. coli) and Mdm31 (mitochondrial protein) highlight structural similarities with eukaryotic RBG proteins. Coloring: RBG domains as Figure 2A; terminal helices: purple; predicted transmembrane helices: black. Note adaptation of Mdm31 to integrate into both inner and outer mitochondrial membranes. PDB files for ATG2A (human), Hobbit (fly), SHIP164 (human), YicH (E. coli) and Mdm31 (yeast, omitting long unstructured loops) obtained from AlphaFold database [21]. Tweek model is of yeast Csf1, provided by Rosario Valentini [17]. VPS13 model in (B) was generated by submitting residues 1643–2111+2543–2840 from yeast Vps13 (omitting 67% of the VAB domain insert) to AlphaFold [60]. Resulting structure was overlapped with AlphaFold model of rat VPS13A (residues 1–2335). Other published Vps13 models ([16]; Fig. 1) show a disconnect in the hydrophobic groove where VAB domain (>700 residues; [23]) is inserted. Continuity traversing this insert, as shown here, is strongly predicted (probability local distance difference test (pLDDT) >0.9 [21]).
Each RBG protein is composed of a characteristic number of RBG domains (from six in SHIP164 to 17 in Tweek; Fig. 2E). The number of RBG domains in each RBG protein is highly conserved across orthologous proteins in different species. The AlphaFold predictions also suggest that the RBG domains form superhelices in the RBG proteins (Fig. 3A–B). The basic structure of the RBG domain (five β-strands plus a loop) is modified in some modules, often in the length of the loop and uncommonly in the number of β-strands. In all RBG proteins, the N-terminal RBG domain has a helix in place of the first β-strand, while the C-terminal RBG domain is shortened, containing only two β-strands in VPS13, ATG2 and the Hob proteins and one or three β-strands in Tweek and SHIP164, respectively (Fig. 2E). In VPS13, ATG2, SHIP164 and the Hob proteins, all RBG domains in the middle of the protein contain the characteristic five-stranded β-sheet structure. By contrast, in Tweek, two of the central RBG domains contain only three β-strands and a third RBG domain contains seven β-strands (Fig. 2E). The loop element of RBG domains always occurs after an odd number of β-strands, which maintains the symmetry of the structure so that the multimerization that extends the hydrophobic groove is uninterrupted. The final loop element of the RBG domain is most often composed of a single helix and a short, disordered polypeptide chain. But in some cases, the length of the final element varies widely, even encoding an entire domain >700 amino acids long that extends away from the hydrophobic groove. These modifications to the RBG domain are conserved across orthologous RBG proteins, suggesting that they play a role in the function and/or localization of RBG proteins [23].
Although the AlphaFold predictions provide basic insights into the structure of RBG proteins, experimental structural studies will still be required to uncover the precise details of RBG protein folding. A direct comparison of the predicted AlphaFold structure of the N-terminal portion of R. norvegicus VPS13A and the high-resolution cryo-EM structure of fungal Vps13 [9] highlights this fact. AlphaFold predicts that the diameter of the hydrophobic groove remains uniform down the length of the protein (Fig. 3A), while the experimental structure shows that the diameter of the groove narrows towards its C-terminus (Fig. 3A). Additionally, the AlphaFold prediction and the experimental structure indicate different degrees of superhelicity (Fig. 3A–B). The experimental structure also identifies the RBG domains (Fig. 3A), highlighting this unit as the building block of RBG proteins.
Molecular functions of the eukaryotic RBG proteins: Lipid superhighways
The hydrophobic groove of RBG proteins, which seems a priori capable of lipid transport, has had this function confirmed via in vitro analysis of glycerophospholipid transport by VPS13, ATG2 and SHIP164 [7,11–14,19]. Although lipid transport has not yet been assayed for Tweek or the Hob proteins, the marked structural similarity shared among RBG proteins suggests that all are likely to perform the same function. Small box-like LTPs contain a hydrophobic pocket capable of harboring only one lipid [4,5]; in contrast, the hydrophobic cavity formed by the RBG fold is large enough to accommodate tens of glycerophospholipids at a time [9,12]. In ATG2 and VPS13, mutation of a band of hydrophobic residues within the groove to hydrophilic residues inhibits lipid transfer activity [9,12]. Thus, it seems likely that a primary molecular function of RBG proteins is bulk transport of lipids along hydrophobic grooves.
Despite many advances in our understanding of the molecular function of RBG proteins in the last few years, there are still many unknowns about the selection, direction and energetics of lipid transfer. VPS13 and ATG2 take part in reactions that transfer lipids from the N- to the C-terminus [7,12,14], while Tweek/Csf1 has been suggested to transport lipids, including phosphatidylethanolamine (PE), in the opposite direction from the C- to the N-terminus [16]. The mechanisms that determine the directionality of lipid transfer remain largely unknown. ATG2 was recently shown to physically interact with lipid scramblases in both the donor and acceptor compartments for ER to autophagosome lipid transport (TMEM41B/VMP1 and ATG9, respectively) [24]. Scramblases may play a role in dissipating concentration gradients between the inner and outer leaflets of the recipient membranes as lipids arrive. Human VPS13A also interacts with the lipid scramblase XK [25]. Whether Tweek or the Hob proteins interact with scramblases is unknown. Regarding lipid selectivity, it is possible that properties intrinsic to the RBG proteins, in particular the highly conserved sequences at the N- or C-termini (Box 2), play a role in selecting lipid moieties for subsequent transport through the channel; however, this has not yet been tested. Future studies will no doubt examine the molecular mechanisms that govern selectivity and directionality of lipid transport by RBG proteins.
Box 2. Examining domains within RBG proteins.
Domains have traditionally been defined based on conservation of sequence, and most existing domain databases use sequence conservation as their main criterion for domain detection. Domains have also typically been thought of as independent structural modules. However, the low-throughput nature of traditional structural biology techniques has made it impractical to solve the structure of every annotated domain; thus, until the release of a reliable prediction program like AlphaFold, it had not been possible to fully understand the relative importance of conserved sequences versus conserved structures in defining protein domains. The RBG proteins provide an example of this issue. Domain databases annotate an Apt1 domain near the C-terminus of Hobbit; this domain was named for its presence in the C-terminal region of the Z. mays Hob protein APT1 [45]. The remote homology detection program HHpred [20] also detects an Apt1 domain near the C-terminus of VPS13 and ATG2 [32,34,61]. The AlphaFold predictions show that the Apt1 domain folds as an integral part of the hydrophobic groove (the final three RBG domains make up the Apt1 domain; see Fig. 2A). Therefore, it is not an independent structural domain. Yet, this C-terminal region is highly conserved both in terms of sequence and in the potentially significant activity of binding the headgroups of phosphoinositide lipids in vitro (with varying specificity) [18,32,34,61]. Thus, the Apt1 domain may be an independent functional unit, even though there is no recognizable distinction in the predicted folding of the Apt1 domain compared to the rest of the RBG protein. So far, the Apt1 domain is the only region present in multiple RBG proteins that consistently binds to phosphoinositides [7,12,18], suggesting that lipid binding is dictated by conserved protein sequences, not by protein structure. Future work will be required to confirm this hypothesis. Standard domain databases also annotate other domains within the RBG proteins, such as the Chorein_N domain in VPS13, ATG2 and SHIP164 (the first RBG repeat makes up the Chorein_N domain; see Fig. 3A). Like the Apt1 domain, Chorein_N domains fold contiguously with the hydrophobic groove, making it difficult to classify them as distinct structural domains. Chorein_N domains may have a distinct function, though, since specific conserved sequences within this domain are required for ATG2 localization to the ER [62]. Thus, conserved sequences in the context of overall protein structure may be the most critical feature in defining the function of some RBG protein domains.
Cellular functions of the RBG proteins: Membrane biogenesis and lipid homeostasis
Membrane contact sites are often detected by the close apposition (10–30 nm) of two different organelle membranes; these appositions are critical regions for integration of cellular function and physiology and are the primary sites of action for many LTPs [6]. Accordingly, most of the RBG proteins have been detected at one or more membrane contact sites in vivo (Table 1). To date, RBG proteins have been implicated in two types of cellular processes: de novo generation of membranes and lipid homeostasis.
Table 1.
Reported subcellular localization of RBG proteins.
| RBG family | Gene name | Species | Localization | Ref. |
|---|---|---|---|---|
| Vps13 | Vps13 | S. cerevisiae | vacuole to mitochondria (vCLAMP) | [63,64] |
| nuclear ER to vacuole (NVJ) | [23,63] | |||
| endosome to mitochondria | [64] | |||
| pro-spore membrane | [27,64] | |||
| vacuole | [23,63,64] | |||
| endosome | [23,63] | |||
| peroxisome | [65] | |||
| mitochondria | [63–65] | |||
| VPS13A | T. thermophila | phagosome | [66] | |
| Vps13 (~Hs A/C) | D. melanogaster | ER to PM | [50] | |
| Vps13D | D. melanogaster | lysosome | [51] | |
| VPS13A | H. sapiens | ER to mitochondria | [7,67,68] | |
| ER to lipid droplet | [7,67] | |||
| mitochondria to endosome | [68] | |||
| VPS13B | H. sapiens | Golgi | [69] | |
| endosomes | [70] | |||
| VPS13C | H. sapiens | ER to lipid droplet | [7] | |
| ER to endo-lysosome | [7] | |||
| VPS13D | H. sapiens | ER to mitochondria | [8] | |
| peroxisomes | [8] | |||
| Golgi | [8,71] | |||
| Atg2 | Atg2 | S. cerevisiae | pre-autophagosomal structure (PAS) | [72] |
| ATG2A | H. sapiens | ER to autophagosome | [12,73] | |
| ER to mitochondria | [74] | |||
| lipid droplet | [73,75] | |||
| Hobbit | Fmp27 (Hob1) |
S. cerevisiae | ER to PM | [16,18] |
| ER to mitochondria | [16] | |||
| Hob2 (Ypr117w) |
S. cerevisiae | ER to PM | [16,18] | |
| ER to mitochondria | [16] | |||
| Hobbit | D. melanogaster | ER to PM | [18] | |
| SABRE | P. patens | ER (puncta) | [42] | |
| Tweek | Csf1 | S. cerevisiae | ER to PM | [16] |
| ER to mitochondria | [16] | |||
| SHIP164 | SHIP164 | H. sapiens | early endosomes | [14,76] |
| UHRF1BP1 | late endosomes | [14] | ||
| TamB/AsmA family | TamB | E. coli | inner to outer membrane | [54,77] |
| AsmA | inner membrane | [77] | ||
| YicH | [77] | |||
| YhjG | [77] | |||
| YdbH | [77] | |||
| YhdP | outer membrane | [77] | ||
| TamB relatives in endo-symbionts | Mdm31 | S. cerevisiae | inner mitochondrial membrane | [57] |
| TIC236 (At2g25660) |
A. thaliana | inner to outer chloroplast membrane | [58] |
ATG2 and VPS13 function in de novo membrane synthesis. ATG2 is primarily implicated in macroautophagy, an essential cellular process that drives non-selective degradation of cellular detritus, including protein aggregates and defective organelles [26]. Autophagy requires the formation of a new double-membraned organelle, the autophagosome, which encapsulates cellular debris in an isolation membrane that fuses with lysosomes for content degradation [12,26]. Recent studies show that ATG2 localizes to endoplasmic reticulum (ER)-autophagosome membrane contact sites in yeast and human cells, where it drives expansion of the isolation membrane [12,13]. Thus, the specific subcellular localization of ATG2, coupled with its putative bulk lipid transfer capabilities, likely confers the ability to generate new membrane and form autophagosomes largely from scratch. Similarly, one of the functions of VPS13 is pro-spore membrane expansion during S. cerevisiae meiosis [27]. VPS13 is enriched at the pro-spore membrane during sporulation [27] and may form a bridge connecting this membrane directly or indirectly to lipid droplets, which are hypothesized to serve as a source of lipids for pro-spore membrane biogenesis [28]. A significant feature of both the autophagic isolation membrane and the yeast pro-spore membrane is that they are lipid-rich and protein-poor compared to most organelles. For example, autophagosomes have just one integral membrane protein, ATG9, which is the membrane receptor for ATG2 [24]. The mechanism driving bulk lipid traffic to generate protein-poor membranes remained elusive until the discovery of VPS13 and ATG2.
Other RBG proteins appear to function in lipid homeostasis. Defects in the subcellular distribution of PI(4,5)P2 (normally highly enriched on the plasma membrane) are observed in D. melanogaster hobbit mutant cells, with Hobbit (and its yeast orthologs) localizing to ER-plasma membrane contact sites in both organisms [18]. Mutation of hobbit also results in cell-autonomous defects in regulated exocytosis by professional secretory cells; these defects are caused by aberrations in trafficking of membrane fusion proteins, which are absent from secretory granule membranes [29]. tweek mutant cells also exhibit defects in the levels and subcellular distribution of PI(4,5)P2 and in endocytosis of synaptic vesicles [30]. A recent study demonstrated that S. cerevisiae Csf1 (and its human and Caenorhabditis elegans orthologs) plays a role in delivering PE to the ER for synthesis of glycosylphosphatidylinositol (GPI) anchors [16], and other work also suggests a role for yeast Csf1 in lipid homeostasis [31]. SHIP164 appears to function in endocytic/retrograde trafficking of specific cargo proteins [14]. Understanding the connection between lipid trafficking and cellular phenotypes will require further investigation.
The function of RBG proteins is dictated by their localization to specific membrane contact sites. This has stimulated research into mechanisms that regulate the subcellular localization of RBG proteins. Sequence motifs that bind lipids and/or organelle-specific adapter proteins have been identified at the N- and C-termini of the VPS13 and Hob proteins [7,8,15,18,32–34]. However, this work is just beginning, and the full complement of intrinsic and extrinsic factors driving RBG protein localization remains to be uncovered. Additionally, the kinetics and stability of RBG protein localization to membrane contact sites is unknown. Do RBG proteins constitutively localize to their target membrane contact site, or are they dynamically recruited when their function is needed, and what is the regulatory mechanism?
Physiological and developmental functions of the RBG proteins: Mysterious drivers of disease
All experimental evidence to date points to lipid transport as the primary molecular function of RBG proteins, but the significance of this process for organismal development and physiology remains poorly understood. Mutations in several RBG proteins are associated with human diseases; thus, by uncovering the developmental and physiological roles of these proteins, we will gain valuable insights into the etiology of these conditions.
There is a strong connection between mutation of RBG proteins and neurological disease. Mutations in VPS13A are associated with the neurodegenerative disease chorea-acanthocytosis [35], VPS13B with the neurodevelopmental disease Cohen syndrome [36], VPS13C with early-onset Parkinson’s disease [37] and VPS13D with spastic ataxia [38]. Mutations in the human ortholog of tweek (KIAA1109) cause the neurodevelopmental disorder Alkuraya-Kučinskas syndrome [39]. However, the relationship between the putative lipid transfer function of these proteins and disease has not yet been uncovered.
Although mutation of the human Hob protein (KIAA0100) has not yet been associated with disease, the developmental and physiological roles of the Hob proteins have been extensively characterized in D. melanogaster and in plants. In D. melanogaster, hobbit is an essential gene, as loss-of-function hobbit mutant animals exhibit a dramatic reduction in pupal body size (leading to the name) and lethality during metamorphosis [29]. The small body size of hobbit mutant animals is caused by defects in the endocrine signaling axis that drives release of insulin [29], which regulates both carbohydrate metabolism and systemic growth in flies [40]. However, rescue of body size via restoration of this endocrine signaling axis does not rescue the lethality of hobbit mutant animals [29], showing that additional vital functions of hobbit are yet to be uncovered. Mutation of plant orthologs of hobbit also results in stunted growth, including loss of SABRE and KIP in Arabidopsis thaliana and loss of SABRE in Physcomitrium patens [41–44]. Additionally, A. thaliana SABRE/KIP mutants exhibit defects in root hair patterning [44] and abnormal elongation of pollen tubes [43], a phenotype shared in mutants of the Zea mays Hob protein APT1 [45]; P. patens SABRE mutants also have defective cell plate deposition [42]. Root hair growth, pollen tube elongation and cell plate deposition all rely heavily on the secretory pathway [46–48]. Given that fly hobbit function is required for regulated exocytosis in professional secretory cells (as mentioned in the previous section) [29], this reveals a possible conserved requirement for the Hob proteins in secretory trafficking pathways.
Phenotypic analysis of mutant animals has also revealed that D. melanogaster Vps13 and Tweek have essential functions in animal development and physiology. Flies have three Vps13 genes: Vps13 (similar to human VPS13A/C [49]), Vps13B and Vps13D. Vps13 knockouts are viable but exhibit reduced fertility and defects in nurse cell clearance during oogenesis [50]. Vps13D mutants are lethal during larval/pupal stages (indicating that Vps13D is an essential gene) and have defects in mitochondrial morphology [51]. Mutation of tweek usually results in lethality during metamorphosis, but a few flies survive to adulthood and exhibit severe locomotion deficits and seizures [30]. These phenotypes provide a foothold for future studies to connect the molecular and cellular functions of VPS13 and Tweek/KIAA1109 to their role in physiology and disease.
Although the developmental and physiological manifestations of RBG protein mutation are varied, one common theme emerges: all these proteins appear to be required during cellular processes that depend heavily on lipid homeostasis and membrane dynamics. VPS13 and ATG2 appear to drive de novo membrane formation, while the Hob proteins, Tweek and SHIP164 seem to regulate the subcellular distribution of lipids, including phosphoinositides, which are essential for organelle identity. As expected, loss of the Hob proteins, Tweek or SHIP164 results in aberrations in intracellular trafficking. However, some major questions remain. Why are some RBG proteins essential genes (required for survival to adulthood in humans or animal models) [29], while others are not [50]? What specific cellular functions require RBG protein function? Deletion of all five yeast RBG proteins had no effect on growth on rich medium [16], but the complex phenotypes associated with loss of VPS13, ATG2, Hobbit or Tweek in animals suggest that there is little redundancy between RBG proteins. There may also be additional functions for RBG proteins beyond lipid transfer, perhaps in transport of other biomolecules [52]. Future studies that answer these questions will address the developmental and physiological roles of RBG proteins.
Evolutionary origins of RBG proteins: prototypes in prokaryotes
Because four of the RBG proteins (VPS13, ATG2, Hobbit and SHIP164) are conserved across eukaryotes, all of these were likely present in the last eukaryotic common ancestor (LECA). Prokaryotes may provide insights into the evolutionary origins of RBG proteins, since structurally similar proteins are present in the intermembrane space of Gram-negative bacteria [53–56]. The Escherichia coli protein TamB has been shown by crystallography to have a C-terminus forming a β-groove with a hydrophobic interior. Sequence analysis suggests that this structure is shared by the remainder of the protein [52], which forms an elongated rod [55]. This suggests that RBG proteins first arose in prokaryotes. Accordingly, analysis of the AlphaFold predictions for TamB and other related proteins in Gram-negative bacteria reveals groove-like structures resembling RBG proteins, built from a series of modules formed from β-strands and a loop (Fig. 3C). While the number of β-strands per repeat is more variable and generally larger (median of 11) than the eukaryotic RBG domain, a key feature shared with the eukaryotic RBG domain is that the number of β-strands between each loop that crosses the hydrophobic groove is always odd. This results in the bacterial RBG proteins having a similar, though less modular, structure to the eukaryotic RBG proteins.
Further evolutionary links between bacterial and eukaryotic RBG proteins are found in proteins that form elongated hydrophobic grooves in mitochondria and chloroplasts, organelles that arose by endosymbiosis. Mdm31 is a protein present in the mitochondrial inner membrane space in many fungi and some protists. This protein contains an N-terminal transmembrane helix (required for anchoring to the inner mitochondrial membrane [57]) followed by two RBG domains and a second transmembrane helix; the AlphaFold structure suggests this second helix may integrate into the outer mitochondrial membrane (Fig. 3C). TIC236 is a highly conserved protein present in the chloroplast intermembrane space [58]. The predicted AlphaFold structure of TIC236 is strikingly similar to TamB. The RBG domains of Mdm31 and TIC236 display intermediate characteristics between prokaryotic and eukaryotic RBG domains. Like the bacterial proteins, Mdm31 and TIC236 have variable numbers of β-strands per RBG domain, but like the eukaryotic proteins, loops in the RBG domains tend to be longer, adding structural complexity. Together, this structural evidence strongly suggests that RBG proteins first arose in prokaryotes and evolved into the eukaryotic proteins we see today.
Concluding remarks
The RBG proteins are a fascinating new family of bulk lipid transporters. Continued work examining the molecular and cellular functions of these proteins is certain to reveal novel roles for lipid trafficking at membrane contact sites, and many questions remain unanswered (see Outstanding Questions). In the future, it will be particularly important to characterize the developmental and physiological roles of RBG proteins since these studies likely hold the key to understanding how mutation of these proteins causes human disease.
Outstanding Questions.
Which lipid classes can enter hydrophobic grooves? Only glycerophospholipids have been identified so far, but not sphingolipids or sterols. Does this apply generally?
How are lipids are selected? Box-like shuttling LTPs usually select for lipid headgroups, but RBG proteins show little headgroup selectivity in vitro. Instead, do they select lipid cargo by fatty acid composition, chain length, and/or saturation, since they interact mainly with acyl chains?
What are the mechanisms that regulate the dynamics of lipid transfer by RBG proteins? RBG protein function requires multiple concurrent actions: targeting to appropriate membrane contact sites, ensuring no helices (which are likely highly mobile in vivo) block the channel, and crucially, generating a gradient that induces flow in one direction. How is all this regulated?
How do structural properties of RBG proteins, like “springiness,” affect function? The ability to flex side-to-side or to stretch and compact vertically could play a role in RBG protein function.
Is the lipid transfer function of RBG proteins conserved in bacteria? TamB, the best characterized RBG protein in bacteria, directly binds BamA, the insertase for outer membrane porins. This links TamB, and by implication the other bacterial RBG proteins, to export of porin β-strand polypeptides from inner to outer membranes. Could a similar polypeptide transfer function be present in eukaryote RBG proteins?
What are the developmental and physiological functions of RBG proteins, and how does disruption of these functions cause disease? Cell biology experiments strongly suggest that mutation of RBG proteins leads to lipid imbalances. Yet, phenotypes in both unicellular and multicellular organisms are complex and not overtly related to lipids in many cases, implying quite indirect pathways to disease.
Highlights.
VPS13, ATG2, SHIP164, Csf1 and the Hob proteins comprise a novel superfamily of conserved lipid transfer proteins with long hydrophobic grooves.
All these long hydrophobic grooves are built from multiply repeating modules that consist of five β-sheets followed by a loop, for which we propose the name “repeating β-groove” (RBG) domain.
RBG proteins carry out lipid transport at membrane contact sites, with functions in lipid homeostasis and membrane biogenesis. Some of these processes require bulk lipid transfer, which appears to be one of the primary molecular functions of RBG proteins.
Eukaryotic RBG proteins likely evolved from structurally related prokaryotic proteins that transfer lipids between the inner and outer membranes in Gram-negative bacteria.
Acknowledgements
We thank Amy Cavanagh for assistance with illustrations and Rosario Valentini, Maya Schuldiner, Jerry Yang, Karin Reinisch and Will Prinz for sharing original PDB files.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Declaration of Interests
The authors declare no competing interests.
References
- 1.Kaplan MR and Simoni RD (1985) Intracellular transport of phosphatidylcholine to the plasma membrane. J. Cell Biol 101, 441–445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.DeGrella RF and Simoni RD (1982) Intracellular transport of cholesterol to the plasma membrane. J. Biol. Chem 257, 14256–14262 [PubMed] [Google Scholar]
- 3.Wirtz KW and Zilversmit DB (1968) Exchange of phospholipids between liver mitochondria and microsomes in vitro. J. Biol. Chem 243, 3596–3602 [PubMed] [Google Scholar]
- 4.Wong LH et al. (2019) Lipid transfer proteins: the lipid commute via shuttles, bridges and tubes. Nat. Rev. Mol. Cell Biol 20, 85–101 [DOI] [PubMed] [Google Scholar]
- 5.Wong LH et al. (2017) Advances on the Transfer of Lipids by Lipid Transfer Proteins. Trends Biochem. Sci 42, 516–530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Prinz WA et al. (2020) The functional universe of membrane contact sites. Nat. Rev. Mol. Cell Biol 21, 7–24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kumar N et al. (2018) VPS13A and VPS13C are lipid transport proteins differentially localized at ER contact sites. J. Cell Biol 217, 3625–3639 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Guillén-Samander A et al. (2021) VPS13D bridges the ER to mitochondria and peroxisomes via Miro. J. Cell Biol 220, e202010004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Li PQ et al. (2020) Cryo-EM reconstruction of a VPS13 fragment reveals a long groove to channel lipids between membranes. J. Cell Biol 219, e202001161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Otomo T et al. (2018) The Rod-Shaped ATG2A-WIPI4 Complex Tethers Membranes In Vitro. Contact (Thousand Oaks) 2018 Jan-Dec, 10.1177/2515256418819936 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Maeda S et al. (2019) The autophagic membrane tether ATG2A transfers lipids between membranes. Elife 8, e45777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Valverde DP et al. (2019) ATG2 transports lipids to promote autophagosome biogenesis. J. Cell Biol 218, 1787–1798 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Osawa T et al. (2019) Atg2 mediates direct lipid transfer between membranes for autophagosome formation. Nat. Struct. Mol. Biol 26, 281–288 [DOI] [PubMed] [Google Scholar]
- 14.Hanna MG et al. (2021) SHIP164 is a Chorein Motif Containing Lipid Transport Protein that Controls Membrane Dynamics and Traffic at the Endosome-Golgi Interface. bioRxiv DOI: 10.1101/2021.11.04.467353 [DOI] [Google Scholar]
- 15.Neuman SD et al. (2021) The Hob Proteins: Putative, Novel Lipid Transfer Proteins at ER-PM Contact Sites. Contact (Thousand Oaks) 4, 1–3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Toulmay A et al. (2022) Vps13-like proteins provide phosphatidylethanolamine for GPI anchor synthesis in the ER. J. Cell Biol 221, e202111095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Castro IG et al. (2021) Systematic analysis of membrane contact sites in Saccharomyces cerevisiae uncovers modulators of cellular lipid distribution. bioRxiv DOI: 10.1101/2021.10.17.464712 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Neuman SD et al. (2022) The Hob proteins are novel and conserved lipid-binding proteins at ER-PM contact sites. J. Cell Sci 135, jcs259086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Osawa T et al. (2020) Human ATG2B possesses a lipid transfer activity which is accelerated by negatively charged lipids and WIPI4. Genes to Cells 25, 65–70 [DOI] [PubMed] [Google Scholar]
- 20.Gabler F et al. (2020) Protein Sequence Analysis Using the MPI Bioinformatics Toolkit. Curr. Protoc. Bioinforma 72, e108. [DOI] [PubMed] [Google Scholar]
- 21.Jumper J et al. (2021) Highly accurate protein structure prediction with AlphaFold. Nature 596, 583–589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yang J et al. (2020) Improved protein structure prediction using predicted interresidue orientations. Proc. Natl. Acad. Sci. U. S. A 117, 1496–1503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bean BDM et al. (2018) Competitive organelle-specific adaptors recruit Vps13 to membrane contact sites. J. Cell Biol 217, 3593–3607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ghanbarpour A et al. (2021) A model for a partnership of lipid transfer proteins and scramblases in membrane expansion and organelle biogenesis. Proc. Natl. Acad. Sci. U. S. A 118, 2101562118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Park JS and Neiman AM (2020) XK is a partner for VPS13A: a molecular link between Chorea-Acanthocytosis and McLeod Syndrome. Mol. Biol. Cell 31, 2425–2436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yu L et al. (2018) Autophagy pathway: Cellular and molecular mechanisms. Autophagy 14, 207–215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Park JS and Neiman AM (2012) VPS13 regulates membrane morphogenesis during sporulation in Saccharomyces cerevisiae. J. Cell Sci 125, 3004–3011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hsu TH et al. (2017) Lipid droplets are central organelles for meiosis II progression during yeast sporulation. Mol. Biol. Cell 28, 440–451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Neuman SD and Bashirullah A (2018) Hobbit regulates intracellular trafficking to drive insulin-dependent growth during Drosophila development. Development 145, dev161356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Verstreken P et al. (2009) Tweek, an Evolutionarily Conserved Protein, Is Required for Synaptic Vesicle Recycling. Neuron 63, 203–215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.John Peter AT et al. (2022) Rewiring phospholipid biosynthesis reveals resilience to membrane perturbations and uncovers regulators of lipid homeostasis. EMBO J. DOI: 10.15252/embj.2021109998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rzepnikowska W et al. (2017) Amino acid substitution equivalent to human choreaacanthocytosis I2771R in yeast Vps13 protein affects its binding to phosphatidylinositol 3-phosphate. Hum. Mol. Genet 26, 1497–1510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Dziurdzik SK and Conibear E (2021) The vps13 family of lipid transporters and its role at membrane contact sites. Int. J. Mol. Sci 22, 1–17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kolakowski D et al. (2020) The binding of the APT1 domains to phosphoinositides is regulated by metal ions in vitro. Biochim. Biophys. Acta - Biomembr 1862, 183349. [DOI] [PubMed] [Google Scholar]
- 35.Ueno SI et al. (2001) The gene encoding a newly discovered protein, chorein, is mutated in chorea-acanthocytosis. Nat. Genet 28, 121–122 [DOI] [PubMed] [Google Scholar]
- 36.Kolehmainen J et al. (2003) Cohen syndrome is caused by mutations in a novel gene, COH1, encoding a transmembrane protein with a presumed role in vesicle-mediated sorting and intracellular protein transport. Am. J. Hum. Genet 72, 1359–1369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Schormair B et al. (2018) Diagnostic exome sequencing in early-onset Parkinson’s disease confirms VPS13C as a rare cause of autosomal-recessive Parkinson’s disease. Clin. Genet 93, 603–612 [DOI] [PubMed] [Google Scholar]
- 38.Koh K et al. (2020) VPS13D-related disorders presenting as a pure and complicated form of hereditary spastic paraplegia. Mol. Genet. Genomic Med 8, e1108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kumar K et al. (2020) KIAA1109 gene mutation in surviving patients with Alkuraya-Kučinskas syndrome: A review of literature. BMC Med. Genet 21, 1–11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Texada MJ et al. (2020) Regulation of body size and growth control. Genetics 216, 269–313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pietra S et al. (2013) Arabidopsis SABRE and CLASP interact to stabilize cell division plane orientation and planar polarity. Nat. Commun 4, 1–15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Cheng X and Bezanilla M (2021) SABRE populates ER domains essential for cell plate maturation and cell expansion influencing cell and tissue patterning. Elife 10, e65166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Procissi A et al. (2003) Kinky Pollen encodes a Sabre-like protein required for tip growth in Arabidopsis and conserved among eukaryotes. Plant J. 36, 894–904 [DOI] [PubMed] [Google Scholar]
- 44.Pietra S et al. (2015) SABRE is required for stabilization of root hair patterning in Arabidopsis thaliana. Physiol. Plant 153, 440–453 [DOI] [PubMed] [Google Scholar]
- 45.Xu Z and Dooner HK (2006) The maize aberrant pollen transmission 1 gene is a SABRE/KIP homolog required for pollen tube growth. Genetics 172, 1251–1261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Guan Y et al. (2013) Signaling in pollen tube growth: crosstalk, feedback, and missing links. Mol. Plant 6, 1053–1064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Smertenko A et al. (2017) Plant Cytokinesis: Terminology for Structures and Processes. Trends Cell Biol. 27, 885–894 [DOI] [PubMed] [Google Scholar]
- 48.Hochholdinger F et al. (2018) Genetic Control of Root System Development in Maize. Trends Plant Sci. 23, 79–88 [DOI] [PubMed] [Google Scholar]
- 49.Ugur B et al. (2020) Role of VPS13, a protein with similarity to ATG2, in physiology and disease. Curr. Opin. Genet. Dev 65, 61–68 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Faber AIE et al. (2021) Vps13 is required for timely removal of nurse cell corpses. Dev. 147, dev191759 [DOI] [PubMed] [Google Scholar]
- 51.Anding AL et al. (2018) Vps13D Encodes a Ubiquitin-Binding Protein that Is Required for the Regulation of Mitochondrial Size and Clearance. Curr. Biol 28, 287–295.e6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Josts I et al. (2017) The Structure of a Conserved Domain of TamB Reveals a Hydrophobic β Taco Fold. Structure 25, 1898–1906.e5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Levine TP (2019) Remote homology searches identify bacterial homologues of eukaryotic lipid transfer proteins, including Chorein-N domains in TamB and AsmA and Mdm31p. BMC Mol. Cell Biol 20, 43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Selkrig J et al. (2012) Discovery of an archetypal protein transport system in bacterial outer membranes. Nat. Struct. Mol. Biol 19, 506–510 [DOI] [PubMed] [Google Scholar]
- 55.Shen HH et al. (2014) Reconstitution of a nanomachine driving the assembly of proteins into bacterial outer membranes. Nat. Commun 5, 1–10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Heinz E et al. (2015) Evolution of the translocation and assembly module (TAM). Genome Biol. Evol 7, 1628–1643 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Dimmer KS et al. (2005) Mdm31 and Mdm32 are inner membrane proteins required for maintenance of mitochondrial shape and stability of mitochondrial DNA nucleoids in yeast. J. Cell Biol 168, 103–115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Chen YL et al. (2018) TIC236 links the outer and inner membrane translocons of the chloroplast. Nature 564, 125–129 [DOI] [PubMed] [Google Scholar]
- 59.Jones DT and Thornton JM (2022) The impact of AlphaFold2 one year on. Nat. Methods 19, 15–20 [DOI] [PubMed] [Google Scholar]
- 60.Mirdita M et al. (2021) ColabFold - Making protein folding accessible to all. bioRxiv DOI: 10.1101/2021.08.15.456425 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Kaminska J et al. (2016) Phosphatidylinositol-3-phosphate regulates response of cells to proteotoxic stress. Int. J. Biochem. Cell Biol 79, 494–504 [DOI] [PubMed] [Google Scholar]
- 62.Kotani T et al. (2018) The Atg2-Atg18 complex tethers pre-autophagosomal membranes to the endoplasmic reticulum for autophagosome formation. Proc. Natl. Acad. Sci. U. S. A 115, 10363–10368 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Lang AB et al. (2015) ER-mitochondrial junctions can be bypassed by dominant mutations in the endosomal protein Vps13. J. Cell Biol 210, 883–890 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Park JS et al. (2016) Yeast Vps13 promotes mitochondrial function and is localized at membrane contact sites. Mol. Biol. Cell 27, 2435–49 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.John Peter AT et al. (2017) Vps13-Mcp1 interact at vacuole-mitochondria interfaces and bypass ER-mitochondria contact sites. J. Cell Biol 216, 3219–3229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Samaranayake HS et al. (2011) Vacuolar protein sorting protein 13A, TtVPS13A, localizes to the Tetrahymena thermophila phagosome membrane and is required for efficient phagocytosis. Eukaryot. Cell 10, 1207–1218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Yeshaw WM et al. (2019) Human VPS13A is associated with multiple organelles and influences mitochondrial morphology and lipid droplet motility. Elife 8, e43561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Munõz-Braceras S et al. (2019) VPS13A is closely associated with mitochondria and is required for efficient lysosomal degradation. Dis. Model. Mech 12, dmm036681 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Seifert W et al. (2011) Cohen syndrome-associated protein, COH1, is a novel, giant Golgi matrix protein required for Golgi integrity. J. Biol. Chem 286, 37665–37675 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Koike S and Jahn R (2019) SNAREs define targeting specificity of trafficking vesicles by combinatorial interaction with tethering factors. Nat. Commun 10, 1–16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Baldwin HA et al. (2021) Vps13d promotes peroxisome biogenesis. J. Cell Biol 220, e202001188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Shintani T et al. (2001) Apg2p functions in autophagosome formation on the perivacuolar structure. J. Biol. Chem 276, 30452–30460 [DOI] [PubMed] [Google Scholar]
- 73.Velikkakath AKG et al. (2012) Mammalian Atg2 proteins are essential for autophagosome formation and important for regulation of size and distribution of lipid droplets. Mol. Biol. Cell 23, 896–909 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Tang Z et al. (2019) TOM40 Targets Atg2 to Mitochondria-Associated ER Membranes for Phagophore Expansion. Cell Rep. 28, 1744–1757.e5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Tamura N et al. (2017) Differential requirement for ATG2A domains for localization to autophagic membranes and lipid droplets. FEBS Lett. 591, 3819–3830 [DOI] [PubMed] [Google Scholar]
- 76.Otto GP et al. (2010) A Novel Syntaxin 6-Interacting protein, SHIP164, regulates Syntaxin 6-Dependent sorting from early endosomes. Traffic 11, 688–705 [DOI] [PubMed] [Google Scholar]
- 77.Sueki A et al. (2020) Systematic Localization of Escherichia coli Membrane Proteins. mSystems 5, e00808–19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Pettersen EF et al. (2021) UCSF ChimeraX: Structure visualization for researchers, educators, and developers. Protein Sci. 30, 70–82 [DOI] [PMC free article] [PubMed] [Google Scholar]


