Abstract
The ubiquitin-proteasome system (UPS) is critical for protein quality control and regulating protein lifespans. Following ubiquitination, UPS substrates bind multi-domain receptors that, in addition to ubiquitin-binding sites, contain functional domains that bind to deubiquitinating enzymes (DUBs) or the E3 ligase E6AP/UBE3A. We provide an overview of the proteasome, focusing on its receptors and DUBs. We highlight the key role of dynamics and importance of the substrate receptors having domains for both binding and processing ubiquitin chains. The UPS is rich with therapeutic opportunities, with proteasome inhibitors used clinically and ongoing development of small molecule proteolysis targeting chimeras (PROTACs) for the degradation of disease-associated proteins. We discuss the therapeutic potential of proteasome receptors, including hRpn13, for which PROTACs have been developed.
Keywords: Proteasome, ubiquitin, substrate receptors, deubiquitinating enzymes
The cellular investment into protein quality control and degradation
Cellular investment into protein degradation is substantial – with two prominent degradation systems that are sophisticated and highly regulated: autophagy and the ubiquitin-proteasome system (UPS). Central to both degradation systems is ubiquitin, which frequently serves as a kiss of death to the proteins decorated with it, although ubiquitination can also signal for nonproteolytic cellular events [1]. Multiple rounds of ubiquitination yields ubiquitin chains, which can be highly variable (Box 1). This review focuses on the UPS.
Box 1: The chains of ubiquitin.
Ubiquitination begins with ATP-dependent charging of the C-terminal end of the 76-amino acid protein ubiquitin by a ubiquitin E1 activating enzyme, resulting in a ubiquitin thioester attached to the E1 active cysteine. Ubiquitin is then transferred to a ubiquitin E2 conjugating enzyme to form an E2-ubiquitin thioester. E2s bind to ubiquitin E3 ligase enzymes, which can serve as scaffolds to facilitate transfer of ubiquitin from the E2 to a substrate directly or as ubiquitin carriers, accepting ubiquitin at an active site cysteine before its transfer to a substrate. In the final transfer step, the ubiquitin thioester forms an isopeptide bond with a side chain lysine or N-terminal end of a protein substrate or another ubiquitin. Ubiquitin chains are formed by isopeptide linkages involving the N-terminal M1 or ε-amino group of one of the seven lysines (K6, K11, K27, K29, K33, K48, K63). Chains can be formed by using a single linkage type (homotypic), multiple linkage types (heterotypic), or a single ubiquitin can have multiple ubiquitins added to it to form a branched chain. Protein substrates can also be ubiquitinated at multiple amino acids.
The proteasome is central to the UPS and, like ubiquitin, can be diversified into various forms by altering subunit constituency and interactors [2]. Degradation pathways are likely important to the very nature of evolution and adaptation of cells to environmental stress as they provide mechanisms to neutralize harmful mutations. Failure in or hijacking of the UPS is associated with myriad diseases and this degradation system is replete with therapeutic opportunities. We provide a brief overview of the proteasome and its ubiquitin-binding substrate receptors, highlighting functional mechanisms and opportunities for therapeutic targeting. We apologize in advance for our many omissions, as the vastness and complexity of the proteasome prevents us from including all valuable contributions to this field.
Proteasome nuts and bolts
At the core of the proteasome is the aptly named 20S core particle (CP), which by itself can degrade intrinsically disordered proteins, including cells cycle regulators, tumor suppressors, and proteins that are unfolded by damage or mutations. The peptide products of the CP are determined by the catalytic β-subunits, which are altered to form specialized proteasomes, namely, the immunoproteasome (iP) and thymoproteasomes (tP). Inhibition of the CP, in combination with other therapeutics including lenalidomide, dexamethasone, or doxorubicin, is standard treatment of care for hematological cancers. Continued development of inhibitors against the CP, iP, and tP, including alterations that allow for greater specificity by taking advantage of their subtle differences, is of great therapeutic interest. The first clinically approved inhibitors, including bortezomib and carfilzomib, target both the CP and iP; recently, the specific iP inhibitor KZR-616 [3] was approved to treat autoimmune inflammatory myopathies.
The narrow gate opening of the CP coupled with an inability to catalyze protein unfolding renders it ineffective against structured proteins. Degradation of structured proteins by the CP requires the 19S regulatory particle (RP), with the CP-RP complex forming the 26S proteasome (Figure 1A). A heterohexameric ATPase ring of Regulatory particle triphosphatase (Rpt) subunits (Rpt1-Rpt6) in the RP docks against the CP and translocates substrates into its proteolytic interior (Figure 1B, light purple). In the cytosol, AKIRIN2 similarly binds to the CP α-ring and by also binding to the nuclear import factor Importin-9 (IPO9), drives nuclear import of intact proteasomes (CP and CP-RP) [4]. AKIRIN2 is degraded by the proteasome once in the nucleus, perhaps in response to release of IPO9, generating an uncapped end or ends of the CP. This mechanism of generating an uncapped CP surface may enable its binding by the regulator 11Sγ [5], which is abundant in the nucleus where it degrades cell cycle regulators and transcription factors [6].
Figure 1. Architecture of the human 26S proteasome.
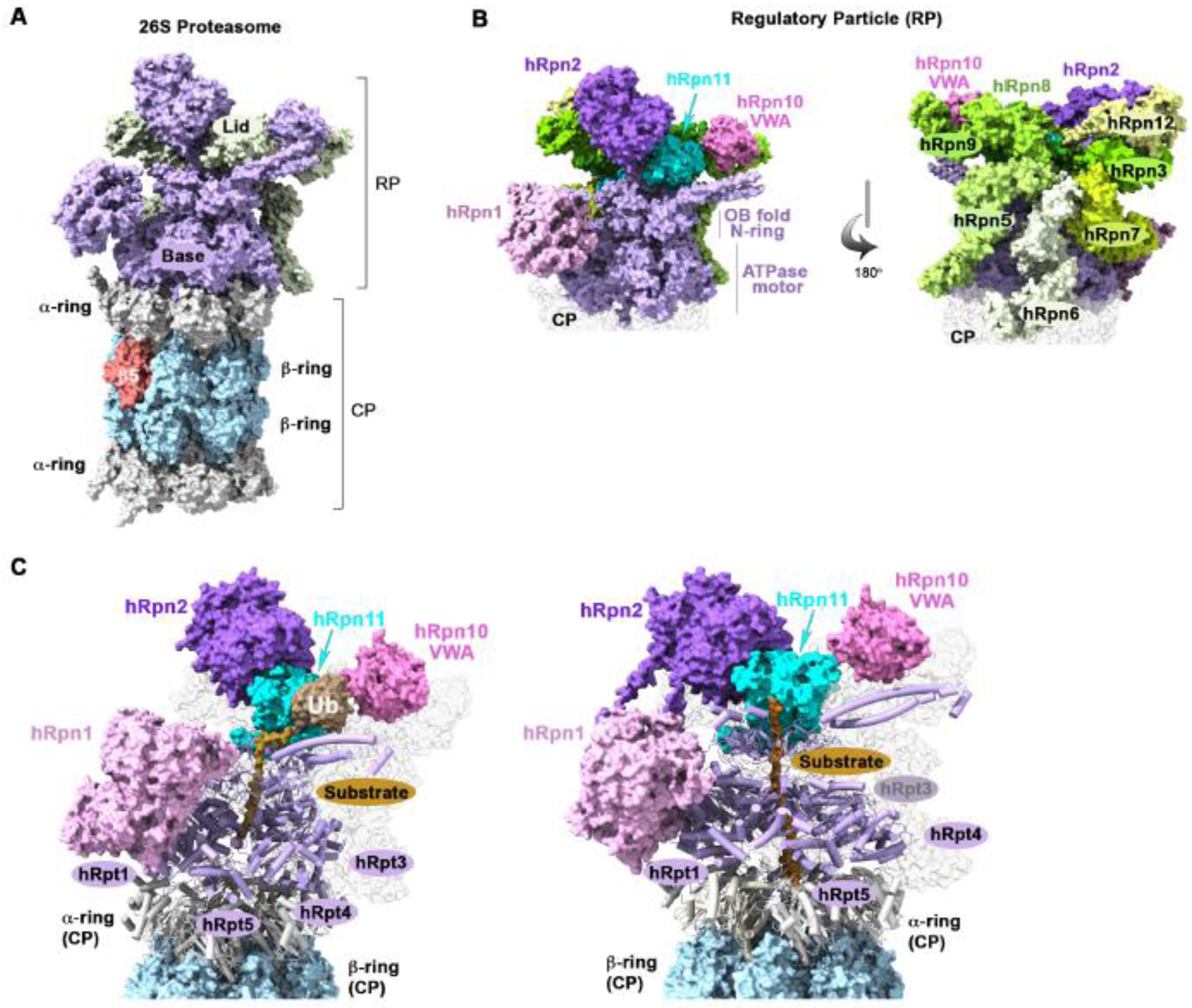
(A) Surface view of the human 26S proteasome (PDB: 5GJQ), highlighting the CP α-rings (grey), β-rings (blue), and subunit β5 (red) and the base (purple) and lid (light green) subcomplexes of the RP. (B) Expanded view of the RP from panel (A) to highlight the base (left) and lid (right) subunits. Base subunits hRpn1 (light pink), hRpn2 (purple), and the AAA-ATPase ring (hRpt1-hRpt6; light purple) are displayed and all lid subunits are in shades of light green except for hRpn11, which is cyan. hRpn10 (pink) is located at the base-lid interface (center). (C) Structure of the human 26S proteasome (6MSE and 6MSJ) with a bound ubiquitinated model substrate (gold) extending through the substrate entry channel of the ATPase ring. The α-ring (grey) and ATPase (purple) subunits are shown in cartoon representation. This figure was generated in ChimeraX.
Structures of the 26S proteasome have been solved by cryo-electron microscopy (cryo-EM) with the RP in different conformations [7–18], including with ubiquitinated substrates [19–22]. Substrate engagement causes alignment of the ATPase ring over the CP and generates an extended substrate entry channel (Figure 1C). To initiate degradation, a flexible unstructured sequence in the substrate extends into this channel to engage the ATPase ring [23]. When not present natively, an unstructured initiation sequence can be generated by the ubiquitin-binding ATPase Cdc48/p97 [24,25] and ubiquitin conjugation to the substrate itself can destabilize certain protein folds [26]. Greater mechanistic detail on the actions of the ATPase ring and its interaction with substrates during processing can be found in a recent review [27].
The RP can be biochemically separated into two subcomplexes (Figure 1A), a ‘base’ that contains the ATPase ring (Figure 1B, light purple), Regulatory particle non-ATPase (Rpn) 1 (Figure 1B, light pink), Rpn2 (Figure 1B, deep purple), and Rpn13; and a ‘lid’, which resembles a fan-like structure and extends upright from the CP to cover one side of the RP (Figure 1B, shades of green). The lid most likely plays an important role in holding the RP together during substrate-dependent conformational switching and contains the essential deubiquitinating enzymes (DUB), Rpn11/Poh1 (Figure 1B, cyan), which removes ubiquitin from substrates as they translocate into the substrate entry channel of the ATPase ring. Rpn11 contacts the Rpt4 oligonucleotide-binding (OB) domain and following substate engagement, caps the substrate entry channel (Figure 1B, left). Substrate receptor Rpn10 is also in the RP, located at the base-lid interface (Figure 1B, pink). The RP substrate receptors Rpn1, Rpn10, and Rpn13 are multi-modular with binding domains for ubiquitin and ubiquitin-binding shuttle factors and for transient interactions with ubiquitin processing enzymes, as described below.
Proteasome substrate receptors on the move
Proteasome receptors Rpn1, Rpn10, and Rpn13
Ubiquitin chains linked by K48 signal for proteasomal degradation and a commonality of Rpn1, Rpn10, and Rpn13 is their preference for this chain type although their ubiquitin-binding modules differ [28–31]. Each substrate receptor has different affinity for other linkage types, and together, the three receptors allow the proteasome to bind all chain types [28–30,32,33]. Rpn1 has a large toroid structure with a site that includes three outer helices named toroid 1 (T1) that bind to two ubiquitin moieties (Figure 2A), albeit with differential affinity [30]. A study [31] of the isolated scRpn1 protein also revealed ubiquitin binding at a site that is N-terminal to the toroid named N-terminal to Toroid (NT) (Figure 2B). Although future studies are needed to demonstrate binding of the NT site to ubiquitin in the proteasome context, the T1 and NT extend along a common Rpn1 surface (Figure 2B), suggesting that they could potentially bind to a common ubiquitin chain.
Figure 2. The dynamic nature of the proteasome-associated ubiquitin receptors.
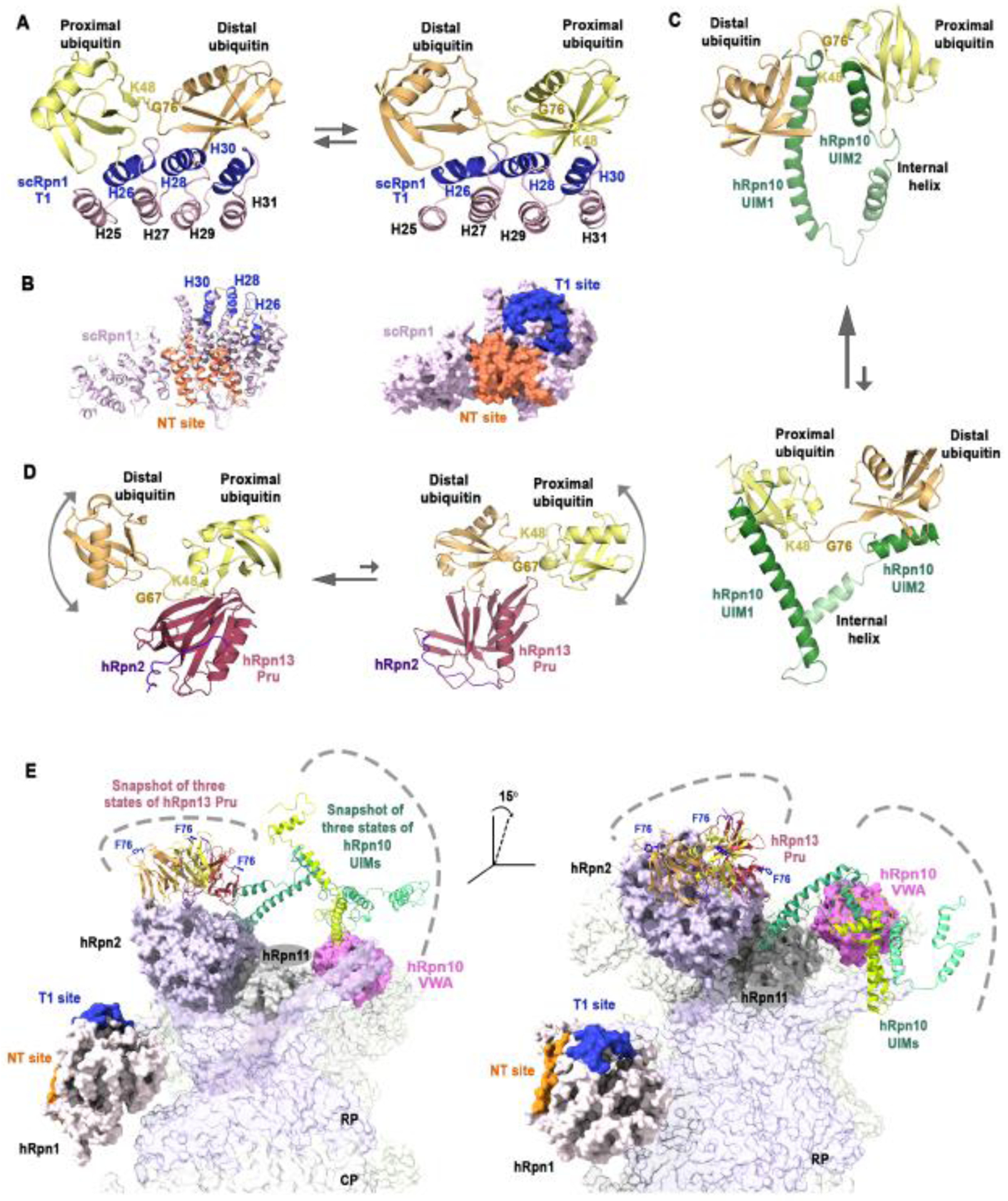
(A) Ribbon diagram representation of the structure of the scRpn1 (light pink) T1 site, formed by three outer α-helices (H26, H28, H30, indigo), bound to K48-linked diubiquitin with the ubiquitin moiety having a free glycine (proximal ubiquitin, yellow) at either H26 (left) or H30 (right); the two states (PDB: 2N3V, left panel and PDB; 2N3W, right panel) are in exchange with equivalent populations in solution (represented by arrows of equal size). The distal ubiquitin (light orange) is linked by its C-terminal G76 (labeled) to K48 (heavy sidechain atoms displayed as sticks and labeled) of the proximal ubiquitin. (B) Ribbon (left) and surface (right) illustration of the scRpn1 structure (light pink; PDB: 5MPC) highlighting the T1 (indigo) and NT (deep orange) ubiquitin-binding sites. (C) Structure of the ubiquitin-binding region of hRpn10 (green) bound to K48 diubiquitin (displayed as in (A)). UIM1 and UIM2 bind to either ubiquitin moiety, with a preference for UIM2 at the proximal ubiquitin (represented by a larger arrow pointing to this state, PDB; 2KDE and PDB: 2KDF). The flexible linker regions between the two UIMs allow for dynamics, as does the exchange of the UIMs between the two ubiquitin moieties. The images presented are a snapshot from a dynamic ensemble. (D) Structure of the hRpn13 Pru (pink) bound to hRpn2 (purple) and to K48 diubiquitin at either the proximal (left; PDB: 6UYI) or distal (right; PDB: 6UYJ) moiety. hRpn13 Pru has higher affinity for the proximal ubiquitin but exchanges between the two moieties (represented by two arrows with the larger pointing to hRpn13 Pru bound to proximal ubiquitin). The unbound ubiquitin is dynamically distributed relative to the hRpn13 Pru-bound ubiquitin (depicted by a curved arrow) and these images represent a snapshot from a dynamic ensemble of conformations. (E) Cartoon depicting the dynamic distribution of the hRpn13 Pru (ribbon, orange, yellow and red) and hRpn10 UIMs (ribbon, shades of green) at the proteasome (surface rendering). Three possible snapshots of hRpn13 Pru (PDB:6CO4) and hRpn10 UIMs (PDB:1YX4) are displayed based on their free structures with hRpn13 F76 in ball and stick rendering (blue). Also displayed are hRpn1 (light pink) NT (orange) and T1 (blue), hRpn2 (purple), hRpn10 VWA (pink), and hRpn11 (gray) (PDB: 5GJQ).
Rpn10 uses single-helix ubiquitin interacting motifs (UIMs) that are connected by a flexible region to bind to ubiquitin chains [34,35]. The number of UIMs in Rpn10 is species dependent with human Rpn10 (hRpn10) having two UIMs, allowing hRpn10 to bind two ubiquitin moieties of a chain, and like the Rpn1 T1, these two sites have differential affinity for ubiquitin [30,34]. The Rpn10 proteasome-binding von Willebrand factor type A (VWA) domain has also been observed to bind to ubiquitin, but this interaction is proposed to promote its monoubiquitination and in turn, Rpn10 expulsion from the proteasome [36].
Rpn13 binds ubiquitin at a single site formed by three loops of a Pleckstrin-like receptor for ubiquitin (Pru) domain [37,38] with interactions at the K48 linker region that establish a preference for this chain type [39]. Nuclear magnetic resonance (NMR) studies have found hRpn13, hRpn10, and Saccharomyces cereviae Rpn1 (scRpn1) to interact dynamically with K48-linked diubiquitin; ubiquitin exchanges at the scRpn1 T1 site with the distal or proximal ubiquitin binding to either the binding site formed by Helix26 (H26) or that at Helix30 (H30) (Figure 2A) and similarly ubiquitin moieties exchange between the two UIMs of hRpn10 (Figure 2C). Although the hRpn13 Pru has only one ubiquitin-binding site, it exchanges dynamically between the ubiquitin moieties of a K48-linked chain [39] (Figure 2D). This dynamic behavior is intrinsic to the low affinity of each site for individual ubiquitin molecules. In larger chains, this low affinity is expected to promote dynamic exchange of the ubiquitin chains attached to a substrate between the multiple receptor sites at the proteasome. As none of the ubiquitin binding sites provide a strong enough affinity to rigidly hold onto ubiquitin moieties within a chain for an extended duration, exchange of ubiquitin moieties between the various substrate binding sites of RP is expected. Such exchange may promote movement of the substrate around the ATPase ring that helps it to find and engage productively with the substrate entry channel [39,40].
Dynamics in the proteasome substrate receptors is likely to also help with initial substrate capture. The hRpn13 Pru binds to an intrinsically disordered C-terminal tail of hRpn2/PSMD1 [41]. This region snakes along a hydrophobic channel in hRpn13, binding with 12 nM affinity [41–43]. hRpn2 has a large, structured toroid domain that its hRpn13-binding C-terminus seems to pivot about, causing flexibility in the orientation of hRpn13 relative to the toroid and rest of the proteasome (Figure 2E). This model of hRpn13 flexibility is supported by its limited visibility in cryo-EM density maps of the proteasome and to date, it has only been observed at yeast proteasomes and at low resolution [16,17,44,45]. The ubiquitin-binding Rpn10 UIMs are similarly flexible with disordered amino acids separating them from the Rpn10 VWA and each other (Figure 2E). The dynamic nature of the hRpn10 UIMs and hRpn13 Pru likely benefits substrate capture by allowing them to sample a greater spatial volume for binding to ubiquitin chains and in addition, would allow flexibility in ubiquitin chain placement at the proteasome. This latter feature may be important for the large breath of ubiquitin chain architectures and diversity of substrates that are processed by the proteasome.
Extending the proteasome receptors via shuttle factor proteins
Additional ubiquitin-binding sites become available at the proteasome through Rad23, Dsk2, and Ddi1 family members. These so-called ‘shuttle factor’ proteins bind to ubiquitin with one or more ubiquitin-associated (UBA) domain(s) while using ubiquitin-like domains (UBLs) to bind to the ubiquitin-binding sites in Rpn1, Rpn10, or Rpn13 [28–31,37,46–51]. Between the UBL and UBA domains of these proteins are long stretches of intrinsically disordered sequence [50,52]. This disorder further contributes to the flexibility of ubiquitin chain binding at the proteasome, allowing the UBA domains to span the full volume around the RP while the UBL domain is docked. In addition to tethering ubiquitinated substrates to the proteasome, UBL-UBA proteins can promote the formation of liquid-liquid phase separated (LLPS) proteasome-containing foci that actively degrade protein substrates (Box 2).
Box 2: Shuttle factors and proteasome foci.
Studies in human cells revealed cellular stress to cause nuclear proteasomes to localize to LLPS condensates in a RAD23B-dependent [116,117] or p62-dependent [118] manner. These proteasomal foci are proteolytically active and differ by their recruited substrates. Whereas RAD23B-dependent foci promoted degradation of ribosomal or heat shock proteins, p62-mediated proteasome foci contained transcription factors and unassembled proteasomal subunits, perhaps co-localized by moonlighting activities for these proteasomal components in transcription or DNA repair [119]. p62, also known as sequestome-1 (SQSTM-1), is an autophagy regulator [120], that uses its C-terminal UBA domain to mediate formation of LLPS foci that contain ubiquitin conjugation machinery (E1, E2, and E3), DUBs (USP15 and USP26), and either K48-linked or K63-linked ubiquitin chains [118]. By contrast, only K48-linked chains were found in RAD23B-mediated foci, with colocalized E6AP [116]. Following osmotic stress, UCHL5 localizes to proteasome foci and its deletion from cells by gene editing caused increased accumulation of K11/K48 branched ubiquitin chains at proteasome foci [68].
Unique to the hPLIC/UBQLN family of shuttle receptors is a series of stress-inducible heat shock chaperonin-binding (STI1) motifs in the center of their sequence by which they can interact with chaperone or autophagy proteins [121–123]. UBQLN colocalization in inclusion bodies is very common in neurodegenerative disorders, such as amyotrophic lateral sclerosis [124]. Following certain stressors, similar to RAD23B, UBQLN2 colocalizes with LLPS stress granules, mediated by oligomerization of its STI1-II motifs. Binding of ubiquitin to the UBA of UBQLN2 is reported to disrupt its LLPS and speculated to trigger its exit from stress granules to allow it to deliver substrates to the proteasome [125]. This model contrasts the mechanism observed for the RAD23B-mediated and p62-mediated foci with active proteasomes.
Extending the proteasome receptors via DUBs
Ubiquitin binding sites are also present in the proteasome DUBs. A crystal structure was solved of the yeast Rpn11:Rpn8 heterodimer with monoubiquitin, which revealed a three-stranded β-sheet between an Rpn11 β-hairpin and the ubiquitin C-terminus [53]. This interaction is expected to stabilize ubiquitin for isopeptide cleavage by Rpn11 and was also observed in cryo-EM studies of yeast [54] and human [21] proteasome with a trapped model substrate that had conjugated K63-linked ubiquitin chains. As the substrate threaded through the center of the ATPase, ubiquitin was bound to Rpn11 (Figure 1C). Another cryo-EM study revealed Rpn11 binding to an unanchored (without substrate) M1-linked ubiquitin chain at the proteasome [10].
The proteasome has two additional DUBs, Ubp6/USP14 and UCHL5/Uch37, which bind to Rpn1 and Rpn13, respectively. A UBL at the N-terminus of Ubp6 binds the Rpn1 toroid at a location named toroid 2 (T2), which is nearby to its T1 site [30] (Figure 3B); an atomic resolution structure has recently been determined for this interaction at the proteasome [55]. Ubiquitin binding by Ubp6 stabilizes the proteasome in the substrate-engaged state, making the ATPase ring inaccessible to the initiation sequence of a new substrate [45,56,57]. When ubiquitin-bound, the Ubp6 catalytic domain contacts Rpt1 [45,57], thus extending inward to a more central location compared to the peripheral T2 site [55]. Interaction with Rpt1 releases Ubp6 from an autoinhibitory mechanism by releasing steric obstruction of its catalytic region [58], resulting in Ubp6 activation and proteasome inhibition [59]. Thus, Ubp6 hydrolase activity at the proteasome is linked to delayed substrate translocation and proteolysis. This mechanistic coupling may act as a sensor of whether ubiquitin chains have been cleared from the RP well enough to initiate the degradation of a new substrate.
Figure 3. Interactions between proteasome-associated receptors and ubiquitin processing enzymes.
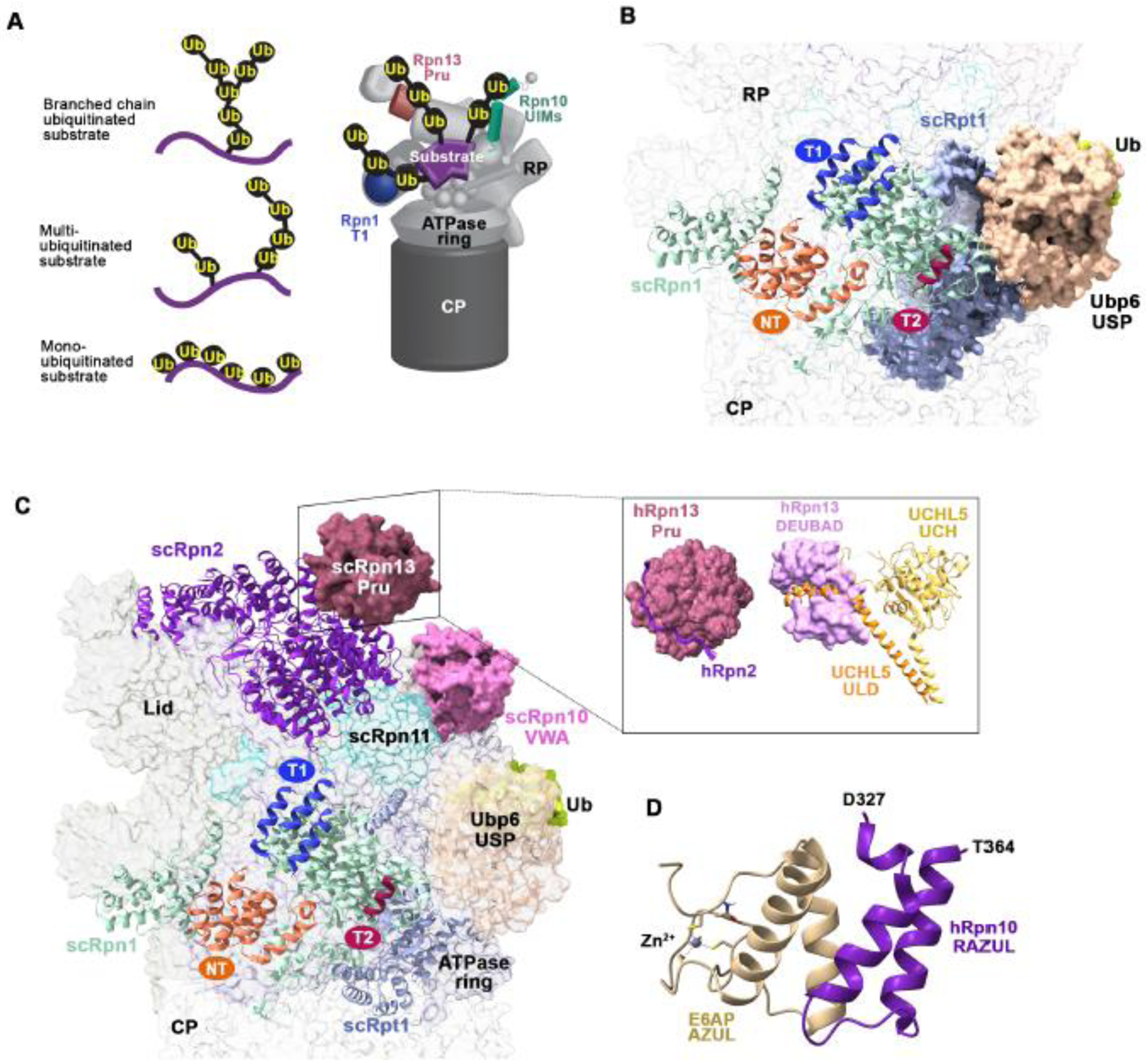
(A) Representation of differentially ubiquitinated substrates including with a single branched ubiquitin chain (top left) or multiubiquitinated substrates with chains (middle left) or monoubiquitin (bottom left) attached. A model of how a multiubiquitinated substrate (purple) may engage the Rpn10 UIMs (green), Rpn13 Pru (pink), and Rpn1 T1 (indigo) is provided (right). (B) View of the Rpn1 and Rpt1 (light blue) regions of the budding yeast 26S proteasome bound to the Ubp6 catalytic domain (USP, light orange) conjugated to ubiquitin aldehyde (green). The Rpn1 (pale green) T1, T2, and NT sites are colored indigo, burgundy, and deep orange, respectively. The Ubp6 N-terminal UBL domain was not visible in this cryo-EM structure (PDB:5A5B). (C) As in (B) but displaying Rpn13 Pru (pink) at Rpn2 (violet) (left; PDB:5A5B). A box insert shows the human domains with interacting partners, including hRpn13 Pru bound to the C-terminus of hRpn2 and the hRpn13 DEUBAD domain (PDB:6CO4) bound to UCHL5 (ULD, orange; catalytic domain (UCH), yellow; PDB:4UEL). (D) Structure of the hRpn10 RAZUL (dark purple) bound to the E6AP AZUL (pale yellow; PDB;6U19). Zinc is displayed as a grey sphere. All structure images were generated with ChimeraX.
UCHL5/Uch37 binds to the proteasome through hRpn13, with its ULD domain enveloped by a C-terminal deubiquitinase adaptor (DEUBAD) domain in Rpn13 [60,61] (Figure 3C). Similar to Ubp6, UCHL5 also has autoinhibitory mechanism; a flexible inhibitory loop in UCHL5 moves away from its catalytic domain upon binding to the hRpn13 DEUBAD, thereby activating its hydrolase activity [60–62]. In the absence of UCHL5 and other binding partners, the hRpn13 Pru and DEUBAD interact with each other [63], perhaps to prevent lower affinity interactions with monoubiquitin, which is used for other signaling processes [1,64]; however, proteasome binding displaces the DEUBAD from the Pru. A similar autoinhibitory mechanism seems to exist in the shuttle factor proteins, as their UBAs interact intra- and inter-molecularly with their UBLs [52,65]. An intrinsically disordered sequence of ~130 amino acids separates the hRpn13 Pru and DEUBAD, perhaps to allow UCHL5 to access a large area for deconjugation of ubiquitin chains [63].
Coordinated binding of ubiquitin chains by the ubiquitin receptors and DUBs is most likely a determinant of degradation efficiency, with each factor making contributions that collectively allow the proteasome to degrade its diverse substrates. Substrates with multiple ubiquitin chains attached most likely engage Ubp6, which removes supernumerary ubiquitin chains from proteasomal substrates [66]. UCHL5 hydrolyzes K48 branched ubiquitin chains, such as K48/K6, K48/K11, and K48/K63 [67,68] due to a K48-specific binding site in UCHL5, which is required for debranching and proteolysis of such chains at the proteasome [69]. Thus, UCHL5 acts on specific chain topologies. The emerging model of Ubp6 removing ‘extra’ chains and UCHL5 removing branched chains suggests that their main activity may be priming ubiquitinated substrates for Rpn11 engagement. The extent to which ubiquitin chains are hydrolyzed into individual ubiquitin molecules at the proteasome is not known and may primarily occur by DUBs that hydrolyze unanchored ubiquitin chain products following their release from the proteasome. Each of the proteasome DUBs have therapeutic promise. A small-molecule inhibitor was developed against Usp14 and found to accelerate the degradation of a subset of proteasome substrates in mammalian cells [59] and several inhibitors against hRpn11 have been developed [70–72].
Other proteasome receptors
Other receptor sites for ubiquitin have been proposed at the proteasome, including ATPase subunit Rpt5 and lid component Sem1/Dss1, as reviewed in [73]. Recently, a cryo-EM study of proteasomes isolated from human cells and mixed with a ubiquitinated model substrate found in small populations, extra density consistent with the size expected for ubiquitin at the Rpn10 VWA as well as a region in Rpn1 distinct from the T1 and NT sites, a location in Rpn2 homologous to Rpn1 T1, and CP subunit α5 [74]. Multiplicity of ubiquitin binding sites throughout the proteasome likely favors a ubiquitination topology that takes advantage of avidity affects by allowing the binding of multiple receptors without compromising substrate proximity to the ATPase ring. These requirements would likely favor substrates with multiple ubiquitin chains of moderate length to enable their distribution to dispersed receptors (Figure 3A). Moreover, interactions with conjugated ubiquitin at various receptor sites may induce additional physical forces on the substrate during translocation that assist unfolding.
It is possible that slowed fluctuations of the proteasome receptor sites caused by ubiquitin/UBL binding promotes activation of the proteasome by extending the lifetime of the substrate-engaged conformational state. Proteasomes were found to be predominantly in the substrate-free state in intact neurons [75] and the cytosol and nucleus of Chlamydomonas reinhardtii [76,77]; however, their substrate engaged state was enriched following the addition of poly-GA containing protein aggregates into neurons [78], perhaps in response to the proteasome substrate receptors binding to the ubiquitinated aggregates. Proteasome binding to ubiquitin or UBLs has been found to induce CP gate opening, proteolysis, and ATP-hydrolysis activities [57,79–82]; however, the mechanisms behind this activation are not yet known. For example, unanchored K48-linked ubiquitin chains stimulated the peptidase activity of the 26S proteasome in an in vitro assay by a mechanism independent of its mode of delivery [81].
Proteasome receptors in health and disease
The many modules of hRpn10
Rpn10 is positioned at a central location of the RP, in contact with Rpn11 and the ATPase ring (Figure 1B–C). Due to the dynamics of Rpn10’s C-terminal half, only its VWA domain is visible in cryo-EM structures of the proteasome. Nonetheless, the Rpn10 UIMs appear to be ideally poised to aid in the recruitment of ubiquitin chains to Rpn11 and to also potentially help drive conformational switching involving the ATPase ring and surrounding regions by their own internal motions.
Efforts to probe the relative contribution of the multiple ubiquitin-binding sites in the proteasome generally invoke deletion of Rpn10, including its VWA domain; however, interpretation of these experiments is complicated by the important structural contributions of the VWA. Positioned at the base-lid interface, the VWA appears important for maintaining the RP structural integrity during conformational switching, as its deletion from yeast [83] and human cell lines [84] causes RP structural instability. Yeast studies demonstrate that the VWA is mono-ubiquitinated and suggest that this modification induces Rpn10 dissociation from the proteasome and in turn, proteasome interaction with Dsk2 [36,85]. The frequency and impact of Rpn10 exchange on and off of the proteasome remains to be elucidated but the resulting proteasome instability from hRpn10 reduction in human cells [84] suggests that the RP base-lid interaction would be disturbed following hRpn10 expulsion.
At its C-terminal end, hRpn10 has an additional functional module that binds to the E3 ligase E6-associated protein (E6AP)/ubiquitin protein ligase E3A (UBE3A), but direct evidence for E6AP ubiquitinating hRpn10 has yet to be found [84,86]. By itself, this region in hRpn10 is intrinsically disordered but upon binding to E6AP, it forms a 4-helix bundle with two helices from the E6AP N-terminal amino-terminal Zn-finger of UBE3A ligase (AZUL) domain (Figure 3D); this domain is thus named Rpn10 AZUL-binding domain (RAZUL) [84]. The RAZUL is the first identified binding site for ubiquitination machinery at the proteasome, although other ligases have been found associated [56,87,88]. E6AP loss at the proteasome reduces the amount of associated ubiquitinated proteins [84], highlighting unique importance for this E3 at the proteasome. The coupling of hRpn10 to E6AP also occurs through correlated protein levels whereby E6AP abundance is reduced in whole cell lysates following loss of hRpn10 [84]. The mechanism of this correlated protein abundance is not known and hRpn10 is not reduced following loss of E6AP [84]. Though non-essential in yeast, deletion of Rpn10 from mice or Drosophila melanogaster causes embryonic lethality [89,90]. Its functional importance in the development of higher eukaryotes may stem from an increased dependency on its role as a proteasome substrate receptor, as Rpn10 has only one UIM in budding yeast and multiple in higher eukaryotes, and/or its activities involving E6AP, as neither the RAZUL domain nor E6AP is present in yeast.
E6AP dysregulation and dysfunction is associated with neurological diseases, including Angelman syndrome [91–93] and autism spectrum disorders [94], as well as cancer. The importance of E6AP to human health may relate to its relationship with hRpn10 and/or the proteasome. E6AP has three isoforms with distinct distribution between the nucleus and cytosol in neurons [95,96], with hRpn10 and the AZUL required for nuclear localization [96]. Loss of nuclear E6AP causes physiological defects and an Angelman syndrome missense mutation in E6AP causes its mislocalization [96]. Moreover, E6AP is hijacked by the high-risk human papilloma virus E6 oncoprotein to induce ubiquitination and degradation of tumor suppressor p53, mechanistically contributing to cervical cancer [97]. Human papilloma virus (HPV) E6 may have evolved to bind E6AP rather than another E3 due to its location at the proteasome. If so, this relationship would suggest that E6AP may be an ideal E3 target for proteolysis targeting chimera (PROTAC) molecules (Figure 4B).
Figure 4. New therapeutic applications involving the proteasome.
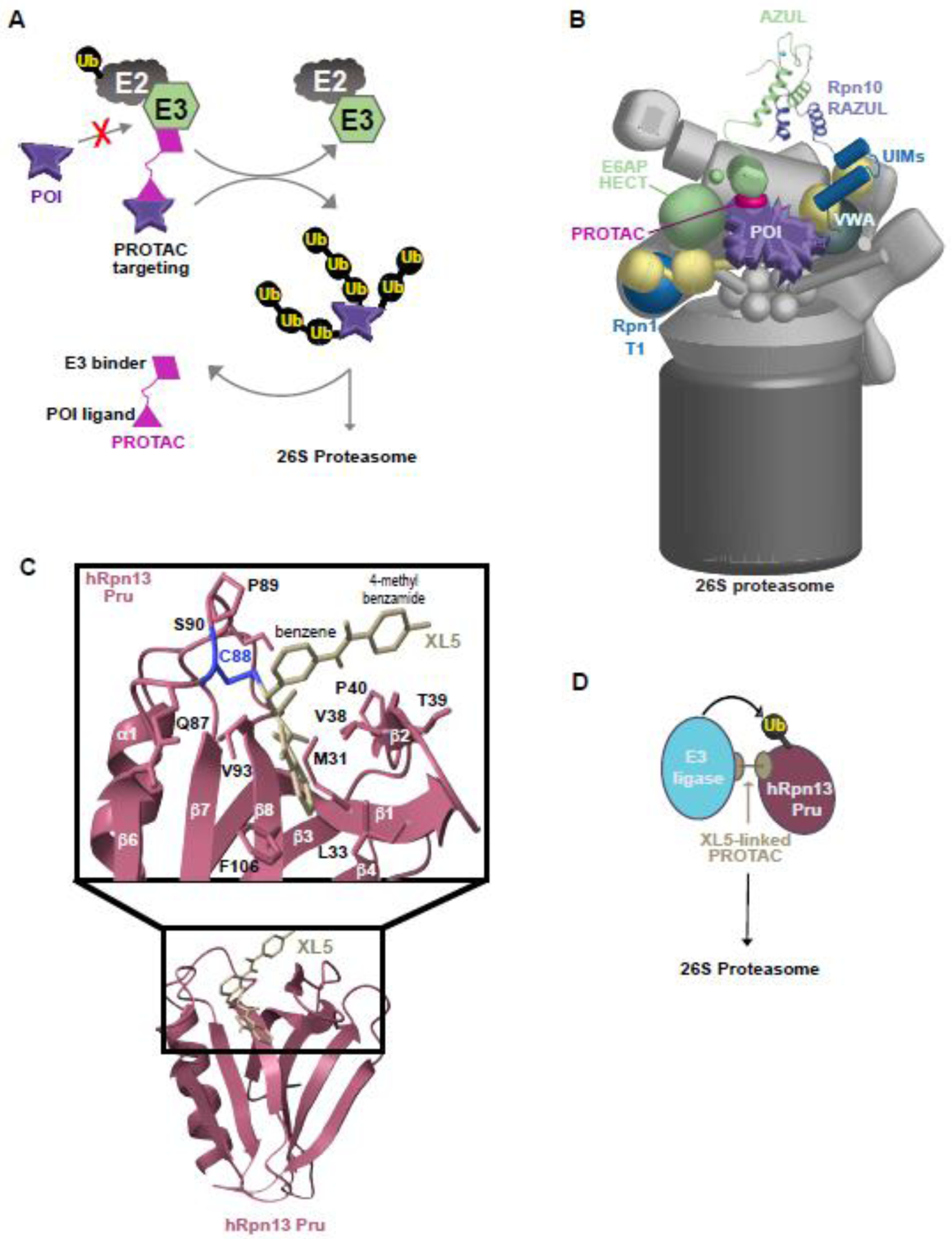
(A) Cartoon depicting a PROTAC (hot pink) linking a non-native substrate (purple) to an E2 (grey) : E3 ligase (green) complex for ubiquitination and degradation by the 26S proteasome. POI, protein of interest. (B) Cartoon representation of the degradation of a protein of interest (POI, purple) by an E6AP (green)-binding PROTAC (hot pink). The AZUL (green) : RAZUL (navy) interaction that binds E6AP to the proteasome is displayed as a ribbon diagram with zinc in light blue. The Rpn1 T1 and Rpn10 VWA and UIMs are colored shades of blue. (C) Structure of XL5 (light yellow) bound to the hRpn13 Pru domain (pink) with C88 (blue) and other interacting side chain heavy atoms highlighted. (D) Schematic of hRpn13 Pru (pink) ubiquitination and degradation by a PROTAC (light yellow) that uses XL5 to bind to hRpn13 Pru while also binding to an E3 ligase (blue).
Similar to the HPV viral oncogene, a PROTAC-like mechanism involving Rpn10 has evolved in insect-vectored plant pathogenic phytoplasmas that results in prolonged host plant lifespans and promotes witches’ broom-like proliferations of leaf and sterile shoots for colonization by phytoplasmas and vectors [98]. This effect is achieved by bifunctional phytoplasma effector SAP05 proteins, which bind to both plant Rpn10 VWA domains as well as transcription factor families SPL and GATA, a scaffolding activity that triggers ubiquitin-independent degradation of SPLs and GATAs. SAP05 does not bind to insect Rpn10 VWA, with specificity defined by only a two amino acid substitution. This discovery highlights the ability of proteasome subunits themselves to serve as inducers of protein degradation, thereby encouraging the development of PROTACs that bypass the requirement for ubiquitination.
The therapeutic promise of hRpn13
The importance of Rpn13 as a ubiquitin receptor at the proteasome has been debated, with Rpn10 proposed to be the essential substrate receptor [99,100]. A study with proteasomes from yeast expressing Rpn1, Rpn10, or Rpn13 with defective ubiquitin binding indicated intact Rpn10 to be sufficient for achieving wild-type single-turnover of a model substrate protein with K48-linked ubiquitin chains and that Rpn1 serves as an effective co-factor with Rpn10 for a substrate protein with K63-linked chains [100]. It is not clear whether these findings can be extended to all substrates and/or whether there are factors in the cellular environment that modulate the need for one proteasome receptor over another. Deletion of UCHL5 or Rpn13 from mice causes embryonic [101] or post-natal [102] death, respectively, suggesting a prominent role for both proteins during development. Loss of hRpn13 causes reduced UCHL5 protein levels [103–105] by an unknown mechanism. Hence, untangling their distinct functional significance is challenging. Human cells with UCHL5 deleted exhibit an increase in bulk ubiquitinated proteins at the proteasome [103], supportive of its unique role in hydrolyzing branched ubiquitin chains [67]. This accumulation is lost, however, in cells deleted of hRpn13 despite greatly reduced levels of UCHL5 [103], suggesting that hRpn13 plays a non-redundant role in the recruitment of these ubiquitin chains to the proteasome or that Rpn13-bound chains particularly require the activity of UCHL5. Deletion of only the hRpn13 Pru domain, maintaining the hRpn13 UCHL5-binding DEUBAD and canonical levels of UCHL5, similarly causes a reduction of proteasome-associated ubiquitinated proteins [103]. It is possible that in the absence of hRpn13 Pru and UCHL5, it is harder for substrates to reach the proteasome. Alternatively, their loss at the proteasome may cause substrates to be processed more quickly. Steady-state analyses of yeast proteasomes found deletion of Rpn13 to accelerate degradation when the proteasome was saturated with substrate [100]. It is also possible that reduced ubiquitinated proteins at the proteasome is caused by their escape due to en bloc hydrolysis by USP14 [106], which otherwise acts on supernumerary ubiquitin chains [66]. The recycled ubiquitin chains may in turn be of a more complex topology than those generated with the benefit of UCHL5 debranching.
The RP has potential as both an alternative therapeutic target to the CP as well as a target synergistic with the CP. hRpn13 has emerged as a promising candidate for therapeutic targeting of the RP, in part because of a cysteine residue (Cys88) available at the periphery of its hRpn2-binding channel that has an accessible sulfur for forming a covalent bond with small molecules [107–109]. The first hRpn13-targeting compounds were bis-benzylidine piperidone derivatives [107,108] which react with exposed cysteines in general [42,107,110]. Nonetheless, these compounds showed hRpn13-dependent accumulation of ubiquitinated proteins and induction of apoptosis in cancer cells [42,103,111], albeit incomplete knockdown of hRpn13 does not show dependency [112]. A next-generation molecule with specificity for hRpn13 (XL5) was developed by using a combined structure-biophysical screening pipeline [109] taking advantage of the hRpn13:hRpn2 structure [42,43]. This compound has a weaker electrophile for Michael addition at Cys88 and forms more stable interactions with the surrounding region, including by interdigitating into a hydrophobic pocket created by lateral movement of a β-hairpin in the Pru domain (Figure 4C). XL5 was linked to an E3 ligase-targeting compound to generate an hRpn13 PROTAC that induced apoptosis in cells expressing hRpn13 with an intact Pru domain [109].
The hRpn13 PROTACs further revealed a plausible mechanism of action for hRpn13 targeting. Whereas full length hRpn13 was not substantially reduced in cells treated with the hRpn13 PROTAC, a fragment lacking the DEUBAD domain (hRpn13Pru) was ubiquitinated and degraded [109]. Intriguingly, hRpn13Pru appears to be generated by the proteasome from full length hRpn13 with cell type dependency, with greater abundance in cells that are sensitive to the hRpn13 PROTAC, such as multiple myeloma cells [109]. This hRpn13 species cannot bind UCHL5 suggesting that it cannot be deubiquitinated by UCHL5 in contrast to the DEUBAD-containing full length protein. hRpn13Pru has also lost any regulatory restrictions imposed by the interdomain DEUBAD:Pru interaction. Ubiquitination of hRpn13 was observed in a previous study that found it to be ubiquitinated by the proteasome E3 ligase Ube3c/Hul5 and following heat-shock, arsenite exposure, or proteasome inhibition [113]. It is possible that the ubiquitinated hRpn13 species observed in this study include hRpn13Pru, as the pattern of ubiquitination appears to match that observed by hRpn13Pru ubiquitination [109].
Altogether, these data suggest that that hRpn13 pharmacological approaches lead to apoptosis in cancer cells by a mechanism of action driven by the targeting of endogenously produced hRpn13Pru, which is up-regulated in certain cancer cell types, including multiple myeloma. The enhanced production of hRpn13Pru in cancers may reflect dysfunction at the proteasome and conversely may impart dysfunction. Hence, this hRpn13 species may be a marker for certain cancers and responsiveness to treatments related to the proteasome.
Concluding remarks
In summary, the proteasome has multiple substrate receptors and DUBs, but each with functional distinctions. A shared physiochemical property of the receptors is the highly dynamic nature of their interactions with ubiquitin chains. The ubiquitin-binding sites are also used to bind ubiquitin shuttle factors, allowing the substrate receptors to also act indirectly and the advantage of additional dynamics imparted by the UBL-UBA proteins. Rpn1, Rpn10 and Rpn13 are multi-modular with distinct elements that appear to couple their ubiquitin binding activities to ubiquitination machinery. This feature is interesting in the context of the proteasome where these elements could have in principle been separated into different subunits and suggests that interaction between the distinct modules, as has been observed by structural studies, provides mechanistic advantages. It is possible for example that binding of substrates to the Rpn1 T1 and/or NT site causes allosteric interactions at the T2 site that impacts interactions with USP14; notably this DUB also has innate flexibility between its UBL and catalytic domain. It is not yet clear whether ubiquitin receptor interactions with the ubiquitin processing enzymes are important for their activity when not assembled into the proteasome, or even what these activities may be. For example, do the DUB binding partners prevent ubiquitin chains from being assembled on hRpn1 or hRpn13? The hRpn13 PROTAC suggests that this might be the case, as only hRpn13Pru is robustly lost following its addition to cells [109].
The hRpn13 DEUBAD domain and hRpn10 RAZUL do not exist in budding yeast, where UCHL5 and E6AP are also absent, suggesting their co-evolution. The importance of UCHL5 in hydrolyzing branched ubiquitin chains suggests that perhaps the E3 ligases that catalyze formation of this chain type are also lacking in yeast. Moreover, UCHL5 and E6AP protein levels correlate with hRpn13 and hRpn10, respectively, suggesting that their functions are tightly coupled. Future studies aimed at fully understanding the mechanistic advantage of the coupling of Rpn1, Rpn10 and Rpn13 to USP14, E6AP, and UCHL5 respectively will no doubt open up new opportunities for therapeutic targeting and reveal advantages of tethering functional activities into a single chain protein that will be applicable to other biomolecular machines (see Outstanding Questions).
Outstanding Questions.
What is the function of Rpn10 and Rpn13 off the proteasome?
What mechanisms link Rpn10 and Rpn13 protein levels to E6AP and UCHL5 respectively?
Why are Rpn10 and Rpn13 physically coupled to E6AP and UCHL5?
How do ubiquitin/UBL interactions lead to proteasome activation?
What is the conformational status of the Rpn10 UIMs and RAZUL at the proteasome during substrate processing?
Is E6AP required at the proteasome for any cellular activities and is its HPV targeting linked to its presence at the proteasome?
Do other E3 ligases have specific binding sites at the proteasome and if so, where? What induces production of hRpn13Pru in multiple myeloma cells and how do hRpn13-targeting compounds induce apoptosis?
Highlights.
Inhibitors against the proteasome core particle (CP) treat hematological cancers and new therapies that target the proteasome continue to be developed.
At the proteasome, substrate receptors (Rpn1, Rpn10, and Rpn13) have their own intrinsic dynamics and bind to ubiquitin chains dynamically, aiding in substrate capture and processing.
Proteasome substrate receptors are multi-modular, with distinct domains for binding to substrates through ubiquitin chains and to DUBs USP14 and UCHL5 or ubiquitin E3 ligase E6AP.
Inducing proteasomal degradation is done naturally by pathogens that generate neo-substrates through Rpn10 and E6AP and a PROTAC has been developed against Rpn13.
Acknowledgements.
The research of the authors is supported by the Intramural Research Program of the CCR, NCI, NIH (1 ZIA BC011490 and 1 ZIA BC011627). We are grateful to Gwen Buel, Xiang Chen, Xiuxiu Lu, and Hiroshi Matsuo for their critical reading of this manuscript.
Glossary
- 26S proteasome
A 2.5 MDa holoenzyme that degrades proteins into peptides and is formed by a 19S RP capping a 20S CP
- Autophagy
a degradation pathway named from the Greek words for self (auto) and eating (phagy)
- Core particle (CP)
The CP is formed by four stacked heteroheptameric rings and has a hollow interior. Two central β-rings contain the catalytic subunits and outer α-rings restrict entry into the catalytic chamber by steric occlusion as their N-terminal amino acids form a gate that extends across the center of the CP
- Deubiquitinating enzymes (DUBs)
A family of enzymes that deconjugate ubiquitin chains and/or remove them from protein substrates. Humans express ~100 different DUBs
- Immunoproteasome (iP)
An altered form of the proteasome that is generated in cells of hematopoietic origin or following induction by pro-inflammatory cytokines. Peptides generated by the iP are loaded onto major histocompatibility complex (MHC) class I molecules for T-cell presentation at the cell surface
- Proteolysis targeting chimera (PROTAC)
PROTACs bind to an E3 ligase and physically tether it to a protein of interest to induce its ubiquitination and in turn degradation by the proteasome
- Regulatory particle (RP, also known as PA700)
The RP caps either or both ends of the CP and unfolds protein substrates while translocating them into the central interior of the CP for hydrolysis
- Regulatory particle triphosphatase (Rpt)
Regulatory particle triphosphatase (Rpt) subunits (Rpt1-Rpt6) serve as the translocating motor of the RP and their C-termini dock directly into pockets formed at the interface of the CP α-subunits, an interaction that induces opening of the α-ring gate
- Substrate receptor
Ubiquitin-binding subunits of the proteasome, including Rpn1, Rpn10, and Rpn13
- Thymoproteasome (tP)
specialized proteasome of cortical thymic epithelial cells. Peptides from the tP drive positive selection of CD8+ T cells [114,115]
- Ubiquitin-associated domain (UBA)
An ~40 amino acid sequence that folds into a 3-helix structure and typically binds ubiquitin
- Ubiquitin-like domain (UBL)
A protein or region within a protein that is defined by sequence and structural similarity to ubiquitin. UBLs form a β-grasp fold and similar to ubiquitin, some can be covalently conjugated to other proteins
- Ubiquitin-proteasome system (UPS)
a highly regulated degradation pathway that begins with post-translational modification of protein substrates with ubiquitin and culminates in substrate proteolysis by the proteasome. To ubiquitinate substrates, humans have two ubiquitin E1 activating enzymes, ~40 ubiquitin E2 conjugating enzymes, and ~600 ubiquitin E3 ligases
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Liu F and Walters KJ (2010) Multitasking with ubiquitin through multivalent interactions. Trends Biochem Sci 35, 352–360. 10.1016/j.tibs.2010.01.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Yu C et al. (2016) Characterization of Dynamic UbR-Proteasome Subcomplexes by In vivo Cross-linking (X) Assisted Bimolecular Tandem Affinity Purification (XBAP) and Label-free Quantitation. Mol Cell Proteomics 15, 2279–2292. 10.1074/mcp.M116.058271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Johnson HWB et al. (2018) Required Immunoproteasome Subunit Inhibition Profile for Anti-Inflammatory Efficacy and Clinical Candidate KZR-616((2S,3R)-N-((S)-3-(Cyclopent-1-en-1-yl)-1-((R)-2-methyloxiran-2-yl)-1-oxopropan-2-yl)-3-hydroxy-3-(4-methoxyphenyl)-2-((S)-2-(2-morpholinoacetamido)propanamido)propenamide). J Med Chem 61, 11127–11143. 10.1021/acs.jmedchem.8b01201 [DOI] [PubMed] [Google Scholar]
- 4.de Almeida M et al. (2021) AKIRIN2 controls the nuclear import of proteasomes in vertebrates. Nature. 10.1038/s41586-021-04035-8 [DOI] [PubMed] [Google Scholar]
- 5.Chen X and Walters KJ (2022) Nuclear destruction: A suicide mission by AKIRIN2 brings intact proteasomes into the nucleus. Mol Cell 82, 13–14. 10.1016/j.molcel.2021.11.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Li S et al. (2015) Regulation of c-Myc protein stability by proteasome activator REGgamma. Cell Death Differ 22, 1000–1011. 10.1038/cdd.2014.188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lander GC et al. (2012) Complete subunit architecture of the proteasome regulatory particle. Nature 482, 186–191. 10.1038/nature10774 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lasker K et al. (2012) Molecular architecture of the 26S proteasome holocomplex determined by an integrative approach. Proc Natl Acad Sci U S A 109, 1380–1387. 10.1073/pnas.1120559109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Eisele MR et al. (2018) Expanded Coverage of the 26S Proteasome Conformational Landscape Reveals Mechanisms of Peptidase Gating. Cell Rep 24, 1301–1315 e1305. 10.1016/j.celrep.2018.07.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chen X et al. (2020) Cryo-EM Reveals Unanchored M1-Ubiquitin Chain Binding at hRpn11 of the 26S Proteasome. Structure. 10.1016/j.str.2020.07.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chen S et al. (2016) Structural basis for dynamic regulation of the human 26S proteasome. Proc Natl Acad Sci U S A 113, 12991–12996. 10.1073/pnas.1614614113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ding Z et al. (2017) High-resolution cryo-EM structure of the proteasome in complex with ADP-AlFx. Cell Res 27, 373–385. 10.1038/cr.2017.12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Greene ER et al. (2019) Specific lid-base contacts in the 26s proteasome control the conformational switching required for substrate degradation. Elife 8. 10.7554/eLife.49806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Luan B et al. (2016) Structure of an endogenous yeast 26S proteasome reveals two major conformational states. Proc Natl Acad Sci U S A 113, 2642–2647. 10.1073/pnas.1601561113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sledz P et al. (2013) Structure of the 26S proteasome with ATP-gammaS bound provides insights into the mechanism of nucleotide-dependent substrate translocation. Proceedings of the National Academy of Sciences of the United States of America 110, 7264–7269. 10.1073/pnas.1305782110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Unverdorben P et al. (2014) Deep classification of a large cryo-EM dataset defines the conformational landscape of the 26S proteasome. Proceedings of the National Academy of Sciences of the United States of America 111, 5544–5549. 10.1073/pnas.1403409111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wehmer M et al. (2017) Structural insights into the functional cycle of the ATPase module of the 26S proteasome. Proc Natl Acad Sci U S A 114, 1305–1310. 10.1073/pnas.1621129114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhu Y et al. (2018) Structural mechanism for nucleotide-driven remodeling of the AAA-ATPase unfoldase in the activated human 26S proteasome. Nat Commun 9, 1360. 10.1038/s41467-018-03785-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Matyskiela ME et al. (2013) Conformational switching of the 26S proteasome enables substrate degradation. Nat. Struct. Mol. Biol 20, 781–788. 10.1038/nsmb.2616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.de la Pena AH et al. (2018) Substrate-engaged 26S proteasome structures reveal mechanisms for ATP-hydrolysis-driven translocation. Science 362. 10.1126/science.aav0725 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dong Y et al. (2019) Cryo-EM structures and dynamics of substrate-engaged human 26S proteasome. Nature 565, 49–55. 10.1038/s41586-018-0736-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bard JAM et al. (2019) The 26S Proteasome Utilizes a Kinetic Gateway to Prioritize Substrate Degradation. Cell 177, 286–298 e215. 10.1016/j.cell.2019.02.031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Prakash S et al. (2004) An unstructured initiation site is required for efficient proteasome-mediated degradation. Nat Struct Mol Biol 11, 830–837. 10.1038/nsmb814 [DOI] [PubMed] [Google Scholar]
- 24.Olszewski MM et al. (2019) The Cdc48 unfoldase prepares well-folded protein substrates for degradation by the 26S proteasome. Commun Biol 2, 29. 10.1038/s42003-019-0283-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Twomey EC et al. (2019) Substrate processing by the Cdc48 ATPase complex is initiated by ubiquitin unfolding. Science 365. 10.1126/science.aax1033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Carroll EC et al. (2020) Site-specific ubiquitination affects protein energetics and proteasomal degradation. Nat Chem Biol. 10.1038/s41589-020-0556-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chen X et al. (2020) Proteasome interaction with ubiquitinated substrates: from mechanisms to therapies. FEBS J. 10.1111/febs.15638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chen X et al. (2016) Structures of Rpn1 T1:Rad23 and hRpn13:hPLIC2 Reveal Distinct Binding Mechanisms between Substrate Receptors and Shuttle Factors of the Proteasome. Structure 24, 1257–1270. 10.1016/j.str.2016.05.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chen X et al. (2019) Structure of hRpn10 Bound to UBQLN2 UBL Illustrates Basis for Complementarity between Shuttle Factors and Substrates at the Proteasome. J Mol Biol 431, 939–955. 10.1016/j.jmb.2019.01.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Shi Y et al. (2016) Rpn1 provides adjacent receptor sites for substrate binding and deubiquitination by the proteasome. Science 351, aad9421. 10.1126/science.aad9421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Boughton AJ et al. (2021) A novel recognition site for polyubiquitin and ubiquitin-like signals in an unexpected region of proteasomal subunit Rpn1. Journal of Biological Chemistry 297, 101052. 10.1016/j.jbc.2021.101052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Boughton AJ et al. (2020) Branching via K11 and K48 Bestows Ubiquitin Chains with a Unique Interdomain Interface and Enhanced Affinity for Proteasomal Subunit Rpn1. Structure 28, 29–43.e26. 10.1016/j.str.2019.10.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chojnacki M et al. (2017) Polyubiquitin-Photoactivatable Crosslinking Reagents for Mapping Ubiquitin Interactome Identify Rpn1 as a Proteasome Ubiquitin-Associating Subunit. Cell Chem Biol 24, 443–457 e446. 10.1016/j.chembiol.2017.02.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhang N et al. (2009) Structure of the s5a:k48-linked diubiquitin complex and its interactions with rpn13. Mol Cell 35, 280–290. 10.1016/j.molcel.2009.06.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wang Q et al. (2005) Structure of S5a bound to monoubiquitin provides a model for polyubiquitin recognition. J Mol Biol 348, 727–739. 10.1016/j.jmb.2005.03.007 [DOI] [PubMed] [Google Scholar]
- 36.Keren-Kaplan T et al. (2016) Structure of ubiquitylated-Rpn10 provides insight into its autoregulation mechanism. Nat Commun 7, 12960. 10.1038/ncomms12960 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Husnjak K et al. (2008) Proteasome subunit Rpn13 is a novel ubiquitin receptor. Nature 453, 481–488. 10.1038/nature06926 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Schreiner P et al. (2008) Ubiquitin docking at the proteasome through a novel pleckstrin-homology domain interaction. Nature 453, 548–552. 10.1038/nature06924 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lu X et al. (2020) An Extended Conformation for K48 Ubiquitin Chains Revealed by the hRpn2:Rpn13:K48-Diubiquitin Structure. Structure 28, 495–506. 10.1016/j.str.2020.02.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Roelofs J (2020) The Extent of Extended-Ubiquitin Binding to the Proteasome. Structure 28, 489–491. 10.1016/j.str.2020.04.013 [DOI] [PubMed] [Google Scholar]
- 41.Lu X et al. (2015) A High Affinity hRpn2-Derived Peptide That Displaces Human Rpn13 from Proteasome in 293T Cells. PLoS One 10, e0140518. 10.1371/journal.pone.0140518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lu X et al. (2017) Structure of the Rpn13-Rpn2 complex provides insights for Rpn13 and Uch37 as anticancer targets. Nat Commun 8, 15540. 10.1038/ncomms15540 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.VanderLinden RT et al. (2017) Structure and energetics of pairwise interactions between proteasome subunits RPN2, RPN13, and ubiquitin clarify a substrate recruitment mechanism. J Biol Chem 292, 9493–9504. 10.1074/jbc.M117.785287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sakata E et al. (2012) Localization of the proteasomal ubiquitin receptors Rpn10 and Rpn13 by electron cryomicroscopy. Proc Natl Acad Sci U S A 109, 1479–1484. 10.1073/pnas.1119394109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Aufderheide A et al. (2015) Structural characterization of the interaction of Ubp6 with the 26S proteasome. Proc Natl Acad Sci U S A 112, 8626–8631. 10.1073/pnas.1510449112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Elsasser S et al. (2002) Proteasome subunit Rpn1 binds ubiquitin-like protein domains. Nat. Cell Biol 4, 725–730. 10.1038/ncb845 [DOI] [PubMed] [Google Scholar]
- 47.Rosenzweig R et al. (2012) Rpn1 and Rpn2 coordinate ubiquitin processing factors at proteasome. The Journal of biological chemistry 287, 14659–14671. 10.1074/jbc.M111.316323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Walters KJ et al. (2002) Structural studies of the interaction between ubiquitin family proteins and proteasome subunit S5a. Biochemistry 41, 1767–1777. 10.1021/bi011892y [DOI] [PubMed] [Google Scholar]
- 49.Kang Y et al. (2007) Defining how ubiquitin receptors hHR23a and S5a bind polyubiquitin. J Mol Biol 369, 168–176. 10.1016/j.jmb.2007.03.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wang Q et al. (2003) Ubiquitin recognition by the DNA repair protein hHR23a. Biochemistry 42, 13529–13535. 10.1021/bi035391j [DOI] [PubMed] [Google Scholar]
- 51.Mueller TD and Feigon J (2003) Structural determinants for the binding of ubiquitin-like domains to the proteasome. EMBO J 22, 4634–4645. 10.1093/emboj/cdg467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Walters KJ et al. (2003) DNA-repair protein hHR23a alters its protein structure upon binding proteasomal subunit S5a. Proc Natl Acad Sci U S A 100, 12694–12699. 10.1073/pnas.1634989100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Worden EJ et al. (2017) An AAA Motor-Driven Mechanical Switch in Rpn11 Controls Deubiquitination at the 26S Proteasome. Molecular cell 67, 799–811 e798. 10.1016/j.molcel.2017.07.023 [DOI] [PubMed] [Google Scholar]
- 54.de la Peña AH et al. (2018) Substrate-engaged 26S proteasome structures reveal mechanisms for ATP-hydrolysis-driven translocation. Science 362. 10.1126/science.aav0725 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zhang S et al. (2022) USP14-regulated allostery of the human proteasome by time-resolved cryo-EM. Nature. 10.1038/s41586-022-04671-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Leggett DS et al. (2002) Multiple associated proteins regulate proteasome structure and function. Mol. Cell 10, 495–507. 10.1016/S1097-2765(02)00638-X [DOI] [PubMed] [Google Scholar]
- 57.Bashore C et al. (2015) Ubp6 deubiquitinase controls conformational dynamics and substrate degradation of the 26S proteasome. Nat Struct Mol Biol 22, 712–719. 10.1038/nsmb.3075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Hung KYS et al. (2022) Allosteric control of Ubp6 and the proteasome via a bidirectional switch. Nature Communications 13, 838. 10.1038/s41467-022-28186-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Lee B-H et al. (2010) Enhancement of proteasome activity by a small-molecule inhibitor of USP14. Nature 467, 179–184. 10.1038/nature09299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Sahtoe DD et al. (2015) Mechanism of UCH-L5 activation and inhibition by DEUBAD domains in RPN13 and INO80G. Mol Cell 57, 887–900. 10.1016/j.molcel.2014.12.039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Vander Linden RT et al. (2015) Structural basis for the activation and inhibition of the UCH37 deubiquitylase. Mol Cell 57, 901–911. 10.1016/j.molcel.2015.01.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Chen X and Walters KJ (2015) Structural plasticity allows UCH37 to be primed by RPN13 or locked down by INO80G. Mol Cell 57, 767–768. 10.1016/j.molcel.2015.02.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Chen X et al. (2010) Structure of proteasome ubiquitin receptor hRpn13 and its activation by the scaffolding protein hRpn2. Mol Cell 38, 404–415. 10.1016/j.molcel.2010.04.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Randles L and Walters KJ (2012) Ubiquitin and its binding domains. Front Biosci (Landmark Ed) 17, 2140–2157. 10.2741/4042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Kang Y et al. (2007) Ubiquitin receptor proteins hHR23a and hPLIC2 interact. J Mol Biol 365, 1093–1101. 10.1016/j.jmb.2006.10.056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Lee BH et al. (2016) USP14 deubiquitinates proteasome-bound substrates that are ubiquitinated at multiple sites. Nature 532, 398–401. 10.1038/nature17433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Deol KK et al. (2020) Proteasome-Bound UCH37/UCHL5 Debranches Ubiquitin Chains to Promote Degradation. Mol Cell 80, 796–809 e799. 10.1016/j.molcel.2020.10.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Song A et al. (2021) Branched ubiquitin chain binding and deubiquitination by UCH37 facilitate proteasome clearance of stress-induced inclusions. Elife 10. 10.7554/eLife.72798 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Du J et al. (2021) A Cryptic K48 Ubiquitin Chain Binding Site on UCH37 is Required for its Role in Proteasomal Degradation. bioRxiv, 2021.2011.2015.468727. 10.1101/2021.11.15.468727 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Perez C et al. (2017) Discovery of an Inhibitor of the Proteasome Subunit Rpn11. J Med Chem 60, 1343–1361. 10.1021/acs.jmedchem.6b01379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Lauinger L et al. (2017) Thiolutin is a zinc chelator that inhibits the Rpn11 and other JAMM metalloproteases. Nat Chem Biol 13, 709–714. 10.1038/nchembio.2370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Li J et al. (2017) Capzimin is a potent and specific inhibitor of proteasome isopeptidase Rpn11. Nat Chem Biol 13, 486–493. 10.1038/nchembio.2326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Finley D et al. (2016) Gates, Channels, and Switches: Elements of the Proteasome Machine. Trends Biochem Sci 41, 77–93. 10.1016/j.tibs.2015.10.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Wu Z et al. (2020) Deep manifold learning reveals hidden dynamics of proteasome autoregulation. bioRxiv. : 10.1101/2020.12.22.423932 [DOI] [Google Scholar]
- 75.Asano S et al. (2015) Proteasomes. A molecular census of 26S proteasomes in intact neurons. Science 347, 439–442. 10.1126/science.1261197 [DOI] [PubMed] [Google Scholar]
- 76.Albert S et al. (2017) Proteasomes tether to two distinct sites at the nuclear pore complex. Proc Natl Acad Sci U S A 114, 13726–13731. 10.1073/pnas.1716305114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Albert S et al. (2020) Direct visualization of degradation microcompartments at the ER membrane. Proc Natl Acad Sci U S A 117, 1069–1080. 10.1073/pnas.1905641117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Guo Q et al. (2018) In Situ Structure of Neuronal C9orf72 Poly-GA Aggregates Reveals Proteasome Recruitment. Cell 172, 696–705.e612. 10.1016/j.cell.2017.12.030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Peth A et al. (2009) Ubiquitinated proteins activate the proteasome by binding to Usp14/Ubp6, which causes 20S gate opening. Mol Cell 36, 794–804. 10.1016/j.molcel.2009.11.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Collins GA and Goldberg AL (2020) Proteins containing ubiquitin-like (Ubl) domains not only bind to 26S proteasomes but also induce their activation. Proceedings of the National Academy of Sciences of the United States of America 117, 4664–4674. 10.1073/pnas.1915534117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Li X and Demartino GN (2009) Variably modulated gating of the 26S proteasome by ATP and polyubiquitin. Biochem J 421, 397–404. 10.1042/BJ20090528 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Bech-Otschir D et al. (2009) Polyubiquitin substrates allosterically activate their own degradation by the 26S proteasome. Nat Struct Mol Biol 16, 219–225. 10.1038/nsmb.1547 [DOI] [PubMed] [Google Scholar]
- 83.Glickman MH et al. (1998) A subcomplex of the proteasome regulatory particle required for ubiquitin-conjugate degradation and related to the COP9-signalosome and eIF3. Cell 94, 615–623. 10.1016/s0092-8674(00)81603-7 [DOI] [PubMed] [Google Scholar]
- 84.Buel GR et al. (2020) Structure of E3 ligase E6AP with a proteasome-binding site provided by substrate receptor hRpn10. Nat Commun 11, 1291. 10.1038/s41467-020-15073-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Zuin A et al. (2015) Rpn10 monoubiquitination orchestrates the association of the ubiquilin-type DSK2 receptor with the proteasome. Biochem J 472, 353–365. 10.1042/BJ20150609 [DOI] [PubMed] [Google Scholar]
- 86.Kuhnle S et al. (2018) Angelman syndrome-associated point mutations in the Zn(2+)-binding N-terminal (AZUL) domain of UBE3A ubiquitin ligase inhibit binding to the proteasome. The Journal of biological chemistry 293, 18387–18399. 10.1074/jbc.RA118.004653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Martinez-Noel G et al. (2012) Identification and proteomic analysis of distinct UBE3A/E6AP protein complexes. Mol Cell Biol 32, 3095–3106. 10.1128/MCB.00201-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Chu BW et al. (2013) The E3 ubiquitin ligase UBE3C enhances proteasome processivity by ubiquitinating partially proteolyzed substrates. The Journal of biological chemistry 288, 34575–34587. 10.1074/jbc.M113.499350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Hamazaki J et al. (2007) Rpn10-mediated degradation of ubiquitinated proteins is essential for mouse development. Mol Cell Biol 27, 6629–6638. 10.1128/MCB.00509-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Szlanka T et al. (2003) Deletion of proteasomal subunit S5a/Rpn10/p54 causes lethality, multiple mitotic defects and overexpression of proteasomal genes in Drosophila melanogaster. J Cell Sci 116, 1023–1033. 10.1242/jcs.00332 [DOI] [PubMed] [Google Scholar]
- 91.Kishino T et al. (1997) UBE3A/E6-AP mutations cause Angelman syndrome. Nat Genet 15, 70–73. 10.1038/ng0197-70 [DOI] [PubMed] [Google Scholar]
- 92.Matsuura T et al. (1997) De novo truncating mutations in E6-AP ubiquitin-protein ligase gene (UBE3A) in Angelman syndrome. Nat Genet 15, 74–77. 10.1038/ng0197-74 [DOI] [PubMed] [Google Scholar]
- 93.Cooper EM et al. (2004) Biochemical analysis of Angelman syndrome-associated mutations in the E3 ubiquitin ligase E6-associated protein. J Biol Chem 279, 41208–41217. 10.1074/jbc.M401302200 [DOI] [PubMed] [Google Scholar]
- 94.Samaco RC et al. (2005) Epigenetic overlap in autism-spectrum neurodevelopmental disorders: MECP2 deficiency causes reduced expression of UBE3A and GABRB3. Hum Mol Genet 14, 483–492. 10.1093/hmg/ddi045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Miao S et al. (2013) The Angelman syndrome protein Ube3a is required for polarized dendrite morphogenesis in pyramidal neurons. J Neurosci 33, 327–333. 10.1523/JNEUROSCI.2509-12.2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Avagliano Trezza R et al. (2019) Loss of nuclear UBE3A causes electrophysiological and behavioral deficits in mice and is associated with Angelman syndrome. Nat Neurosci 22, 1235–1247. 10.1038/s41593-019-0425-0 [DOI] [PubMed] [Google Scholar]
- 97.Scheffner M et al. (1993) The HPV-16 E6 and E6-AP complex functions as a ubiquitin-protein ligase in the ubiquitination of p53. Cell 75, 495–505 [DOI] [PubMed] [Google Scholar]
- 98.Huang W et al. (2021) Parasitic modulation of host development by ubiquitin-independent protein degradation. Cell 184, 5201–5214.e5212. 10.1016/j.cell.2021.08.029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Cundiff MD et al. (2019) Ubiquitin receptors are required for substrate-mediated activation of the proteasome’s unfolding ability. Sci Rep 9, 14506. 10.1038/s41598-019-50857-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Martinez-Fonts K et al. (2020) The proteasome 19S cap and its ubiquitin receptors provide a versatile recognition platform for substrates. Nat Commun 11, 477. 10.1038/s41467-019-13906-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Al-Shami A et al. (2010) Regulators of the proteasome pathway, Uch37 and Rpn13, play distinct roles in mouse development. PLoS One 5, e13654. 10.1371/journal.pone.0013654 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Hamazaki J et al. (2015) Redundant Roles of Rpn10 and Rpn13 in Recognition of Ubiquitinated Proteins and Cellular Homeostasis. PLoS Genet 11, e1005401. 10.1371/journal.pgen.1005401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Osei-Amponsa V et al. (2020) Impact of Losing hRpn13 Pru or UCHL5 on Proteasome Clearance of Ubiquitinated Proteins and RA190 Cytotoxicity. Mol Cell Biol 40. 10.1128/MCB.00122-20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Randles L et al. (2016) The Proteasome Ubiquitin Receptor hRpn13 and Its Interacting Deubiquitinating Enzyme Uch37 Are Required for Proper Cell Cycle Progression. J Biol Chem 291, 8773–8783. 10.1074/jbc.M115.694588 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Hamazaki J et al. (2006) A novel proteasome interacting protein recruits the deubiquitinating enzyme UCH37 to 26S proteasomes. EMBO J 25, 4524–4536. 10.1038/sj.emboj.7601338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Yao T and Cohen RE (2002) A cryptic protease couples deubiquitination and degradation by the proteasome. Nature 419, 403–407. 10.1038/nature01071 [DOI] [PubMed] [Google Scholar]
- 107.Anchoori RK et al. (2018) Covalent Rpn13-Binding Inhibitors for the Treatment of Ovarian Cancer. ACS Omega 3, 11917–11929. 10.1021/acsomega.8b01479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Anchoori RK et al. (2013) A bis-benzylidine piperidone targeting proteasome ubiquitin receptor RPN13/ADRM1 as a therapy for cancer. Cancer Cell 24, 791–805. 10.1016/j.ccr.2013.11.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Lu X et al. (2021) Structure-guided bifunctional molecules hit a DEUBAD-lacking hRpn13 species upregulated in multiple myeloma. Nat Commun 12, 7318. 10.1038/s41467-021-27570-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Dickson P et al. (2020) Physical and Functional Analysis of the Putative Rpn13 Inhibitor RA190. Cell Chem Biol. 10.1016/j.chembiol.2020.08.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Song Y et al. (2016) Targeting proteasome ubiquitin receptor Rpn13 in multiple myeloma. Leukemia 30, 1877–1886. 10.1038/leu.2016.97 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Dickson P et al. (2020) Physical and Functional Analysis of the Putative Rpn13 Inhibitor RA190. Cell Chem Biol 27, 1371–1382.e1376. 10.1016/j.chembiol.2020.08.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Besche HC et al. (2014) Autoubiquitination of the 26S proteasome on Rpn13 regulates breakdown of ubiquitin conjugates. EMBO J 33, 1159–1176. 10.1002/embj.201386906 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Ohigashi I et al. (2021) The thymoproteasome hardwires the TCR repertoire of CD8+ T cells in the cortex independent of negative selection. J Exp Med 218. 10.1084/jem.20201904 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Kincaid EZ et al. (2016) Specialized proteasome subunits have an essential role in the thymic selection of CD8(+) T cells. Nat Immunol 17, 938–945. 10.1038/ni.3480 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Yasuda S et al. (2020) Stress- and ubiquitylation-dependent phase separation of the proteasome. Nature 578, 296–300. 10.1038/s41586-020-1982-9 [DOI] [PubMed] [Google Scholar]
- 117.Uriarte M et al. (2021) Starvation-induced proteasome assemblies in the nucleus link amino acid supply to apoptosis. Nat Commun 12, 6984. 10.1038/s41467-021-27306-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Fu A et al. (2021) p62-containing, proteolytically active nuclear condensates, increase the efficiency of the ubiquitin-proteasome system. Proc Natl Acad Sci U S A 118. 10.1073/pnas.2107321118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Kodadek T (2010) No Splicing, no dicing: non-proteolytic roles of the ubiquitin-proteasome system in transcription. J Biol Chem 285, 2221–2226. 10.1074/jbc.R109.077883 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Sun D et al. (2018) Polyubiquitin chain-induced p62 phase separation drives autophagic cargo segregation. Cell Res 28, 405–415. 10.1038/s41422-018-0017-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Hjerpe R et al. (2016) UBQLN2 Mediates Autophagy-Independent Protein Aggregate Clearance by the Proteasome. Cell 166, 935–949. 10.1016/j.cell.2016.07.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Kaye FJ et al. (2000) A family of ubiquitin-like proteins binds the ATPase domain of Hsp70-like Stch. FEBS Lett 467, 348–355. 10.1016/s0014-5793(00)01135-2 [DOI] [PubMed] [Google Scholar]
- 123.Kurlawala Z et al. (2017) The STI and UBA Domains of UBQLN1 Are Critical Determinants of Substrate Interaction and Proteostasis. J Cell Biochem 118, 2261–2270. 10.1002/jcb.25880 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Deng HX et al. (2011) Mutations in UBQLN2 cause dominant X-linked juvenile and adult-onset ALS and ALS/dementia. Nature 477, 211–215. 10.1038/nature10353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Dao TP et al. (2018) Ubiquitin Modulates Liquid-Liquid Phase Separation of UBQLN2 via Disruption of Multivalent Interactions. Mol Cell 69, 965–978.e966. 10.1016/j.molcel.2018.02.004 [DOI] [PMC free article] [PubMed] [Google Scholar]


