Summary
Simian arteriviruses are endemic in some African primate species and can cause fatal hemorrhagic fevers when they cross into new primate host species. We find that CD163 acts as an intracellular receptor for simian hemorrhagic fever virus (SHFV; a simian arterivirus), a rare mode of virus entry that is shared with other hemorrhagic fever-causing viruses (e.g., Ebola and Lassa viruses). Further, SHFV enters and replicates in human monocytes, indicating full functionality of all of the human cellular proteins required for viral replication. Thus, simian arteriviruses in nature may not require major adaptations to the human host. Given that at least three simian arteriviruses have caused fatal infections in captive macaques after host-switching, and that humans are immunologically naïve to this family of viruses, development of serology tests for human surveillance should be a priority.
Keywords: hemorrhagic fever, arteriviruses, CD163 receptor, disease emergence, virus evolution, primates, virus entry, arms race, positive selection, zoonosis, species tropism
Graphical Abstract
Introduction
Simian hemorrhagic fever is a lethal disease that has been documented in captive Asian macaques (Macaca spp.). Outbreaks have occurred in macaque colonies worldwide (Figure 1A) and have been traced to at least three different simian viruses (Figure 1B in red) classified in subfamily Simarterivirinae (Nidovirales: Arteriviridae) (Kuhn et al., 2016; Lauck et al., 2015). These simian arteriviruses likely entered primate facilities through the import of subclinically infected wild African monkeys. To date, primate facilities remain vigilant about preventing exposure to these viruses (Johnson et al., 2011; Lauck et al., 2015). Human infections at affected primate facilities have not been detected. Further, there is no way to surveille for prior human exposure in Africa, where these viruses are endemic in primates, because no serology tests exist. In the absence of strong biological indications that humans could be at risk of infection with these viruses, there has been little impetus to interrogate the interface between nonhuman primates and humans in Africa to exclude cryptic spillover events or subclinical infection cycles.
Figure 1. Historical outbreaks and natural reservoirs for simian arteriviruses.
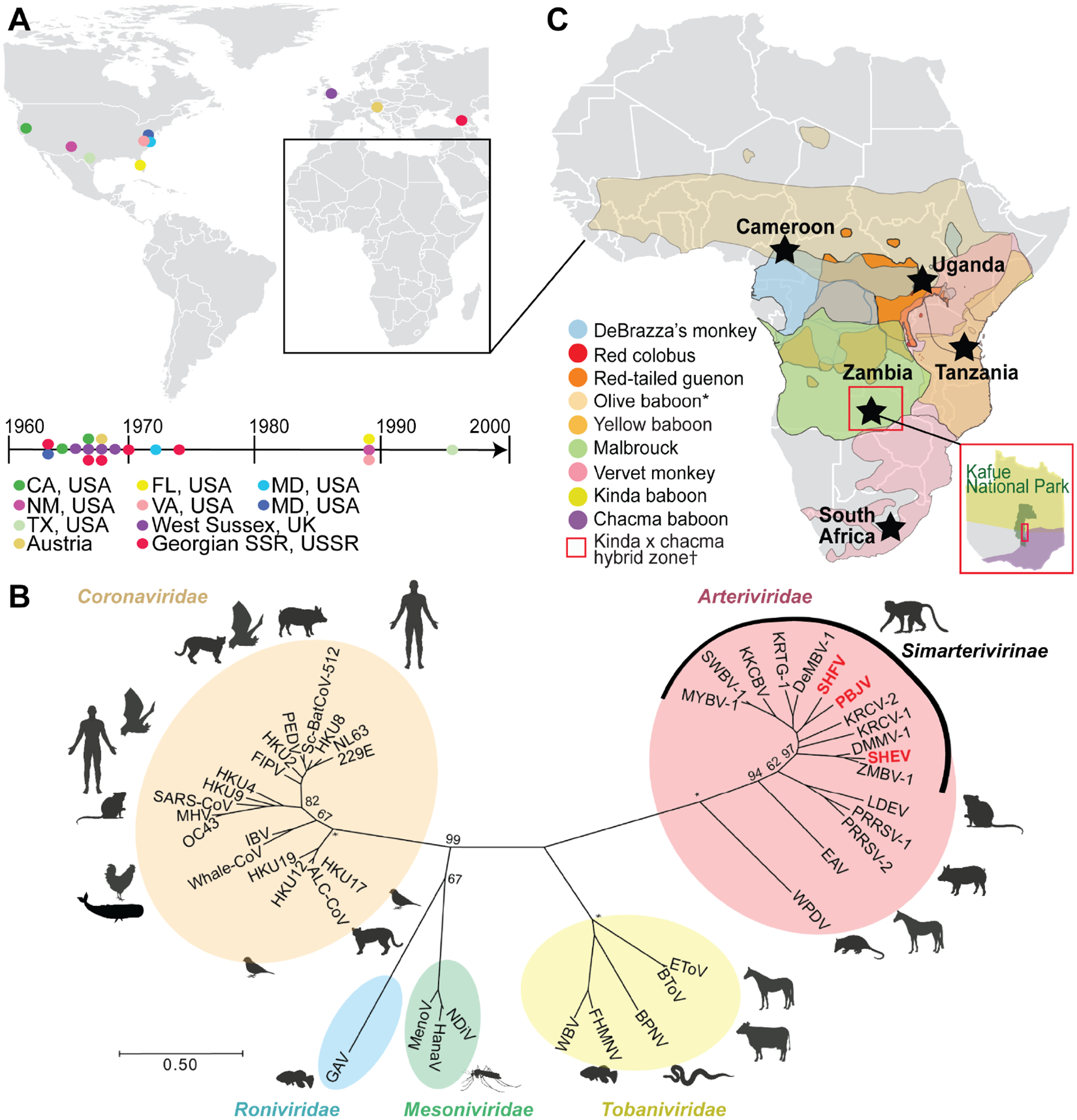
(A) Documented outbreaks of simian arteriviruses in primate facilities. Locations of affected facilities are shown by colored circles on the map and timeline. Several facilities experienced multiple outbreaks. (B) Phylogeny of representative viruses within the order Nidovirales, including all published simian arteriviruses, based on an alignment of concatenated RNA-directed RNA polymerase (RdRp) and helicase genes. Arteriviruses known to have caused outbreaks in primate facilities are written in red. The tree is drawn to scale, with branch lengths measured in the number of substitutions per site and bootstrap values shown for major nodes. Asterisks indicate nodes with 100% support. (C) Geographical ranges of primate species known to be infected with simian arteriviruses. Sampling sites are indicated by black stars. *Only detected in captivity; †Kafue kinda chacma baboon virus (KKCBV) was discovered in kinda x chacma hybrid baboons, within Kafue National Park, as shown as an inset in red (Chiou et al., 2021).
Although the natural primate reservoirs of these viruses remain poorly understood, recent discovery efforts have identified broad arterivirus diversity in apparently healthy African monkeys of numerous species ranging across Africa (Figure 1C) (Bailey et al., 2014b, 2014a, 2016; Lauck et al., 2011, 2013). Infected monkeys can carry high viral loads (Bailey et al., 2016). In some locations, these primate species interact with people in direct and often aggressive ways, leading to high risk of exposure for humans. For instance, red colobus monkeys in western Uganda are infected with high viral loads of two simian arteriviruses (Lauck et al., 2013) and are known to bite and scratch local people (Paige et al., 2014, 2017). Such human–primate antagonism is widespread and increasing across Africa (Hill, 2018; Hockings, 2016).
The recent finding that arteriviruses can replicate in primary ape (chimpanzee and gorilla) cells (Cai et al., 2021) evokes parallels to simian immunodeficiency virus (SIV; Retroviridae). SIV first crossed from African monkeys into apes, a key “stepping-stone” event that ultimately led to the evolution and emergence of the pandemic strain of human immunodeficiency virus (HIV-1 group M) in humans (Sauter and Kirchhoff, 2019; Sharp and Hahn, 2011). Like arteriviruses, SIV can cause spillover infection of captive macaques (Daniel et al., 1985; Mansfield et al., 1995). In addition, simian arteriviruses and SIVs are both endemic in African primates (Bailey et al., 2016; Heuverswyn and Peeters, 2007; Lauck et al., 2013; Sharp et al., 2001), cause long-lasting subclinical infections in their hosts (Bailey et al., 2014b, 2014a; Vatter et al., 2015; Wei et al., 1995), have high mutation rates that produce diverse viral swarms (Bailey et al., 2014b; Coffin, 1995; Kuhn et al., 2016), and primarily induce fatal disease after host switching (Bailey et al., 2015; Sharp and Hahn, 2011). Thus, it is important to determine whether simian arteriviruses have the potential to infect humans (Bailey et al., 2015; Graham and Sullivan, 2018; Warren and Sawyer, 2019). Akin to the example of HIV-1, there is no expectation of resistance in the human population because there are no related human arteriviruses that would give humans cross-protective immunity.
Results
CD163 is necessary and sufficient as the receptor for SHFV infection
Most simian arteriviruses remain uncultured. The few cultured viruses are extremely finicky, replicating in only a handful of nonhuman primate cell lines (Cai et al., 2021; London, 1977; Tauraso et al., 1968; Yú et al., 2016). The best-known simian arterivirus, simian hemorrhagic fever virus (SHFV), was first cultured during a simian hemorrhagic fever outbreak among captive rhesus macaques (Macaca mulatta) at the National Institutes of Health (NIH) Primate Quarantine Unit in Bethesda, Maryland, USA (Palmer et al., 1968; Shelokov et al., 1968; Tauraso et al., 1968). SHFV presumptively entered the macaque population via incidental exposure to fluids from infected African patas monkeys (Erythrocebus patas) cohoused at this facility (Dalgard et al., 1992; Gravell et al., 1986; Lauck et al., 2015; London, 1977). In vitro, SHFV can only be propagated to high titers on the grivet (Chlorocebus aethiops) embryonic kidney MA-104 cell line and its clonal derivatives, such as MARC-145 cells, and a few other cell types (Tauraso et al., 1968; Yú et al., 2016). Grivet cell lines that are routinely used for producing diverse viruses due to their inability to synthesize interferon (Vero cells and subclones) (Desmyter et al., 1968) are not permissive.
Virus entry into mammalian cells is mediated by cellular receptors. For SHFV, we previously found that pre-treating cells with an anti-CD163 antibody impeded infection (Caì et al., 2015; Cai et al., 2021). Based on this clue, we hypothesized that CD163 might be the cellular receptor. Indeed, we found that the relative abundance of CD163 in different grivet cell lines correlates with their relative abilities to produce infectious SHFV (high = MA-104, MARC-145; low = COS-7; none = Vero), potentially corroborating the importance of CD163 expression for SHFV infection (Figure 2A). We next generated CD163 knockout (ΔCD163) single-cell clones of grivet MA-104 cells and confirmed gene deletion (Figure S1). We observed markedly reduced virus production in ΔCD163 cells under singlestep and multi-step growth conditions (Figure 2B, open red circles). Stable re-complementation of these cells with grivet CD163 (gCD163) by retroviral transduction recovered expression and virus production in these cells (Figure 2B, closed black circles). Expression of grivet CD163 in Vero cells also resulted in significant virus production and observable cytopathic effects (Figure S2). Primary skin fibroblasts derived from two different patas monkeys, the presumed natural host of SHFV (Dalgard et al., 1992; Gravell et al., 1986; Lauck et al., 2015; London, 1977), lacked detectable CD163 protein and also did not support virus production. However, stable complementation with patas monkey CD163 (pCD163) rendered cells permissive to infection by SHFV (Figure 2C). Finally, we confirmed that CD163 acts at the step of virus entry into the cell by transfecting ΔCD163 cells with cDNA-launch SHFV rescue plasmids (Cai et al., 2021), effectively bypassing the requirement of CD163 for cellular entry. Transfected ΔCD163 cells produced infectious SHFV but to lower titers, presumably because newly produced virions were incapable of spreading and infecting new cells that lacked CD163 (Figure S3). Therefore, CD163 is necessary and sufficient for infection by SHFV, a finding that should enhance our ability to isolate and study other novel simian arteriviruses that have been historically difficult to propagate in vitro.
Figure 2. CD163 is necessary and sufficient as the receptor for SHFV infection.
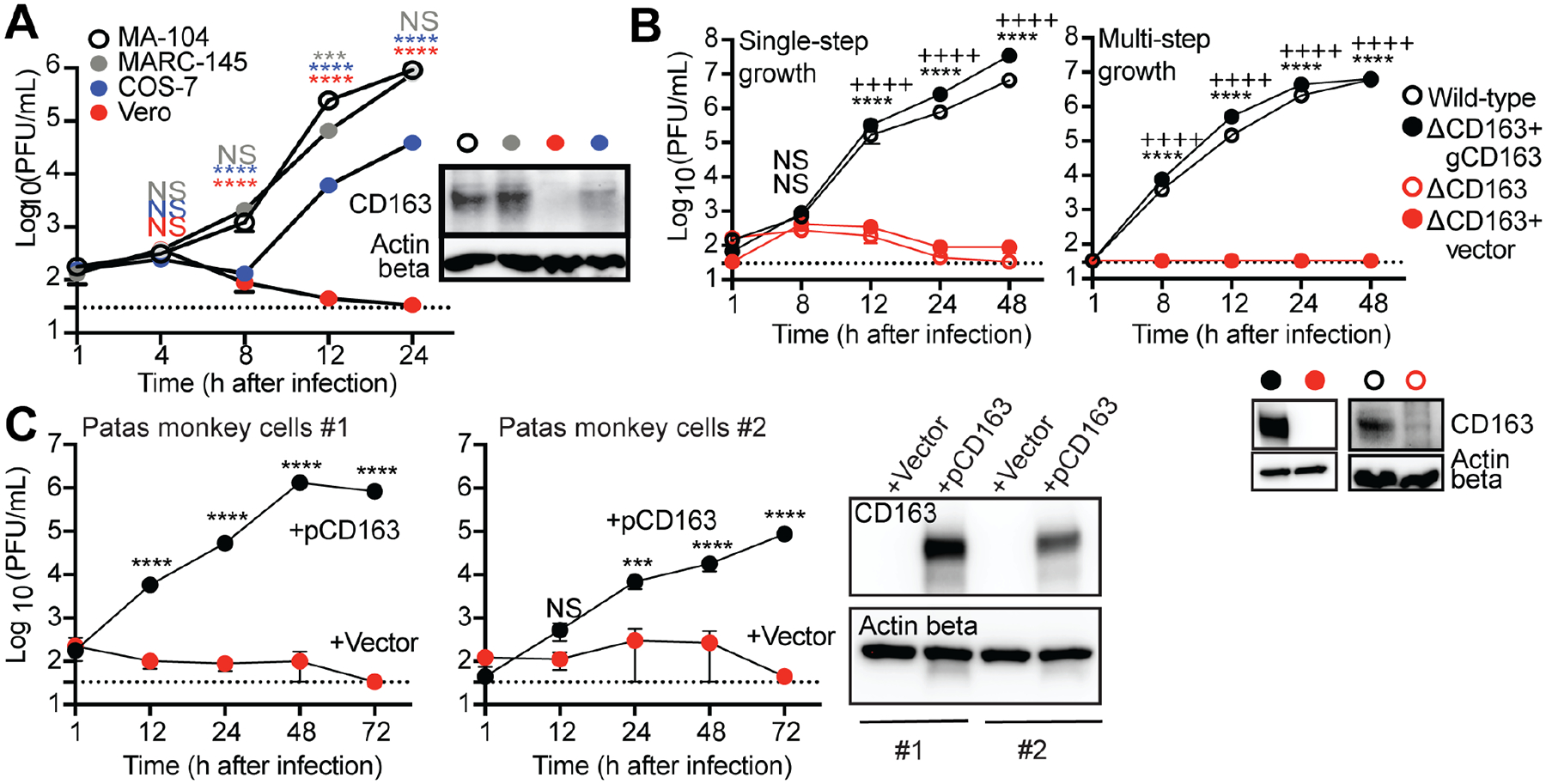
(A) SHFV (at a multiplicity of infection [MOI] of 3) was incubated with cells from four different grivet cell lines (MA-104, MARC-145, COS-7, and Vero). Infectious virion production over a time course was quantified from cell supernatants by plaque assay (PFU/mL) on permissive MA-104 cells. (B) Single-step (MOI = 3) and multi-step (MOI = 0.03) growth curves of SHFV on wild-type MA-104 cells and its derivatives: ΔCD163, ΔCD163 complemented with empty vector (ΔCD163+vector), and ΔCD163 with grivet CD163 (ΔCD163+gCD163). (C) Untransformed primary skin fibroblasts derived from a 6-mo-old (#1) and a 20-yr-old patas monkey (#2) were complemented with empty vector or patas monkey CD163 (pCD163) and exposed to SHFV as described in panel A. (A–C) Lysates from cells were probed for CD163 expression using western blotting. Actin beta served as a loading control. Data show the mean +/− standard error of the mean (SEM) from three independent experiments, with one replicate per experiment. Dotted lines represent the limit of detection for the assay. All plots include error bars; no error bars are shown when the SEM was smaller than the size of the symbols. Statistical tests used: (A) Two-way analysis of variance (ANOVA) with Dunnett’s post-test when compared to MA-104 cells (***P < 0.001, ****P < 0.0001, NS, not significant). (B) Two-way ANOVA with Sidak’s post-test for wild-type compared to ΔCD163 (****P < 0.0001), or for ΔCD163+vector compared to ΔCD163+gCD163 (++++P < 0.0001), and (C) for +pCD163 compared to +vector cells (***P < 0.001, ****P < 0.0001). See also Figures S1 and S2. SHFV, simian hemorrhagic fever virus.
CD163 is an intracellular receptor
Several hemorrhagic fever-causing viruses that infect humans, such as Lassa virus, Lujo virus, and Ebola virus, use a unique mode of cell entry that involves intracellular receptors (Carette et al., 2011; Jae and Brummelkamp, 2015; Jae et al., 2014; Raaben et al., 2017). After these viruses enter cells, they interact with a receptor embedded in the membranes of late endosomes/lysosomes. Successful engagement allows viruses to fuse with the endolysosomal membrane and enter the cytoplasm; otherwise the virus gets trapped and degraded. Likewise, we noticed that CD163 was undetectable on the surface of permissive primate cells but could be detected via intracellular staining, and that SHFV particles enter cells (non-productively) even in the absence of CD163 (Figure S4). Thus, we hypothesized that CD163 may act as an intracellular receptor for SHFV. To test this hypothesis, we performed single-molecule RNA fluorescence in situ hybridization (smFISH) (Femino et al., 1998) to detect viral RNA in cells. To perform smFISH, we tiled 48 DNA oligos antisense to the SHFV genome and then conjugated to each a fluorophore. Once hybridized to the viral RNA, the additive signal of these fluorescently labeled probes enabled the visualization of single SHFV genomes either by flow cytometry (Figures S3 and S4) or by microscopy. Using confocal microscopy, we observed smFISH puncta (viral RNA) inside both wild-type and ΔCD163 cells 1 h after exposure to virus (Figures 3A and 3B). The signal was transient in ΔCD163 cells and was absent by 4 h post-exposure (Figure 3B), consistent with flow cytometric analysis of similarly exposed cells (Figure S4). By contrast, in wild-type cells, viral RNA persisted and began forming discreet cellular domains by 6 h post-exposure, likely identifying major sites of viral RNA replication (Figure 3A). SHFV particles can therefore enter cells in the absence of CD163, but viral genomes likely become trapped in intracellular vesicles and become degraded.
Figure 3. CD163 is an intracellular receptor.
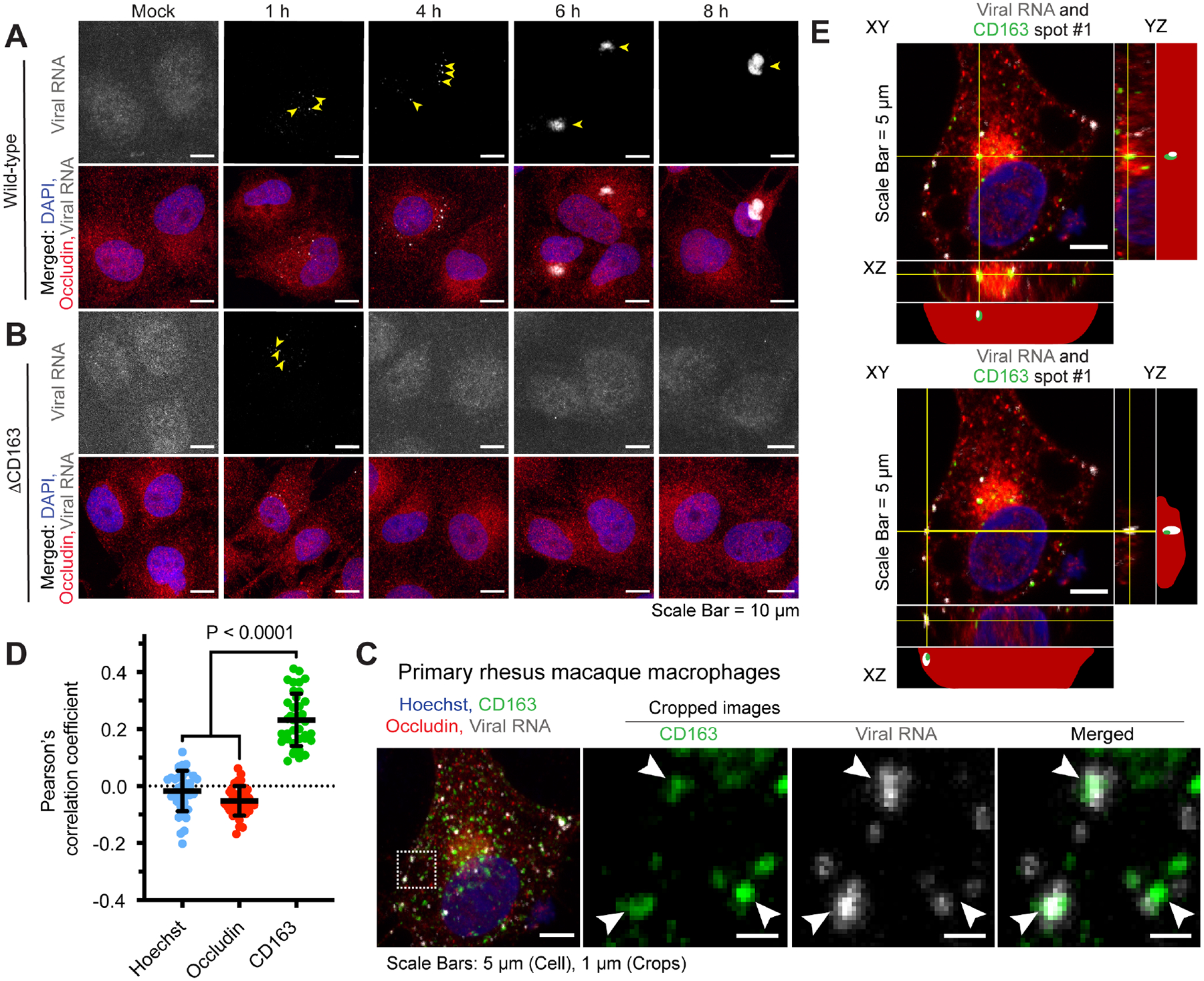
(A) Wild-type MA-104 or (B) ΔCD163 cells were cultured on glass coverslips and mock-exposed or exposed to simian hemorrhagic fever virus (SHFV) at a multiplicity of infection (MOI) of 10 for 1, 4, 6, or 8 h prior to fixation. Cell membranes were immunolabeled (anti-occludin antibody; red) and cells were probed for DNA (4′,6-diamidino-2-phenylindole [DAPI]; blue) and viral RNA (detected by single-molecule RNA fluorescence in situ hybridization [smFISH]; gray). Arrows indicate regions of viral RNA signal. (C) Rhesus macaque monocytes were isolated from whole blood and differentiated into macrophages using macrophage colony-stimulating factor (M-CSF). Exposure to SHFV was performed as described above but for 45 min, representing the earliest timepoint that intracellular viral RNA could reliably be detected inside cells. In addition to Hoechst (DNA, nucleus) and occludin staining, cells were immunolabeled to detect CD163 (green). (A–C) Cells were imaged by laser scanning confocal microscopy (LSCM); a representative maximum intensity projection from each condition is shown. Arrows indicate regions of viral RNA signal. (D) Pearson’s correlation analysis of the LSCM images generated in the experiment in panel C. A Pearson’s correlation coefficient was calculated between viral RNA signal and either Hoechst (blue), occludin (red), or CD163 (green) signal. Average and standard deviations of each correlation are indicated; each data point represents a single cell. (E) Single z-planes are shown for two representative images in which viral RNA (white) and CD163 (green) co-localized and were analyzed in three-dimensional space. The YZ and XZ images are shown for the regions of XY, denoted by the yellow lines and spanning from the top of the imaged cell to the bottom. Actual and simplified illustrations of each orthogonal view are shown, illustrating the relative positions of the viral RNA (white), CD163 (green), nucleus (blue), and cellular boundaries (red dots). In the simplified view, the red profile is the outline of the cell compiled from Z-stack images. See also Figures S3 and S4.
We next confirmed this finding in primary cells. We isolated monocytes from rhesus macaque whole blood and differentiated them into macrophages using macrophage colony stimulating factor (M-CSF). Following a short exposure of macaque macrophages to SHFV, viral RNA colocalized with CD163 (Figure 3C). We calculated the association of each fluorescent channel and smFISH puncta (viral RNA) using Pearson’s correlation coefficient, a metric of colocalization (Dunn et al., 2011). We found a statistically significant correlation of viral RNA with CD163, but not with Hoechst (DNA, nucleus) or occludin (cell surface protein) staining (Figure 3D). Further, orthogonal views of the z-stack images showed smFISH puncta (viral RNA) and CD163 co-localizing within the upper and lower boundaries of the cell, and not at the cell surface, which confirmed the intracellular nature of this interaction (Figure 3E). These findings identify an unusual phenotypic similarity with other zoonotic hemorrhagic fever-causing viruses, i.e., the use of an intracellular receptor.
CD163 is a dynamic barrier to host-switching of SHFV, but the human ortholog is fully functional for SHFV entry
African primates are the long-term reservoir hosts for simian arteriviruses (Bailey et al., 2016; Kuhn et al., 2016). Over evolutionary time, hosts and viruses compete in genetic “arms races” during which each entity experiences natural selection in favor of genetic variants that give them the upper hand (Meyerson and Sawyer, 2011). In the case of receptors, primates are subject to natural selection in favor of mutations that impede virus entry, whereas viruses counter-evolve to regain optimal receptor usage. Indeed, signatures of accelerated evolution can be detected in many virus–receptor pairs (Demogines et al., 2012, 2013; Kaelber et al., 2012; Meyerson et al., 2015; Ng et al., 2015; Warren and Sawyer, 2019; Warren et al., 2019b, 2019a). We found that CD163 has evolved under strong positive natural selection in primates (Figure S5). Nine residue positions in CD163 bear this signature of rapid amino-acid replacement over time. CD163 has nine “scavenger receptor cysteine-rich” (SRCR) domains, a transmembrane region, and a cytoplasmic tail (Law et al., 1993). SRCR domains 5 through 9 contain regions important to arterivirus infection (Gorp et al., 2010). Seven of the nine residues under selection map these domains (Figure 4A).
Figure 4. CD163 is a dynamic barrier to host-switching of SHFV, but the human ortholog is fully functional for virus entry.
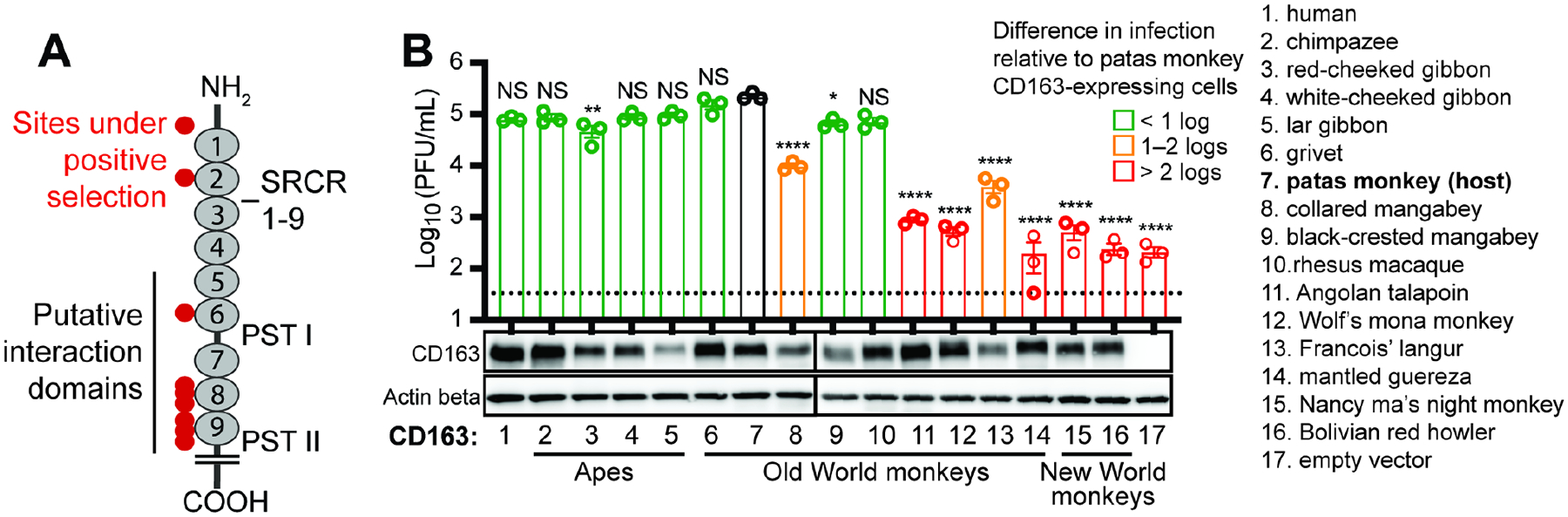
(A) Illustration of the CD163 receptor, showing the extracellular scavenger receptor cysteine-rich (SRCR domains 1–9) and proline domains rich with serine threonine (PST) I and II, and the short cytoplasmic tail. Residues evolving under positive selection are shown with red circles, and the putative arterivirus interaction domain is denoted with a black line. The N-terminus (amine terminus; NH2) and C-terminus (carboxyl terminus; COOH) denote the beginning and end of the protein polypeptide chain, respectively. (B) MA-104 ΔCD163 cells stably expressing the indicated primate CD163 ortholog (X-axis) were exposed to SHFV at a multiplicity of infection (MOI) of 3. Viral titers in cell supernatants 12 h post-exposure were assessed by plaque assay. Bars are color-coded by log-fold differences in SHFV titers compared to cells expressing the receptor of the presumed natural host (patas monkey; #7). The data show the mean +/− standard error of the mean (SEM) from three independent experiments, with one replicate per experiment. One-way analysis of variance (ANOVA) with Dunnett’s post-test compared to control cells (*P < 0.05, **P < 0.01, ****P < 0.0001, NS not significant). The dotted line represents the limit of detection for the assay. CD163 receptor expression was confirmed by western blotting, and actin beta served as a protein loading control. See also Figure S5. SHFV, simian hemorrhagic fever virus.
If primates are involved in a genetic arms race with arteriviruses, it is expected that primates of distinct species would encode receptors with radically different tolerances to any specific arterivirus. This is because each species takes a unique evolutionary trajectory in these battles (Meyerson and Sawyer, 2011). We evaluated this expectation by cloning CD163 orthologs from 15 different primate species and stably integrating them into ΔCD163 cells. These cells were then exposed to SHFV to see if they became infected (Figure 4B). As expected, SHFV entered most efficiently by engaging the CD163 ortholog of its presumed natural host, the patas monkey (black bar in graph). Similarly, based on known outbreaks among captive macaques, rhesus macaque CD163 also efficiently facilitated virus entry. However there were large differences, spanning almost three orders of magnitude, in how well this virus used the CD163 orthologs of other species. The species-specific sequence differences that emerge in virus – receptor arms races have a beneficial side effect to the hosts; they serve as a genetic barrier to virus host switching (Imai and Kawaoka, 2012; Kailasan et al., 2015; Parrish et al., 2008; Warren and Sawyer, 2019). For instance, at least some monkey species are predicted to be resistant to SHFV, based on our finding that the virus does not enter cells efficiently through CD163 of these species (Figure 4B).
Consistent with recent findings that SHFV can replicate in primary cells of the common chimpanzee (Pan troglodytes) and western gorilla (Gorilla gorilla) (Cai et al., 2021), we found that SHFV is fully compatible with all tested ape CD163 orthologs. Remarkably, this includes the human CD163 ortholog, which supported robust SHFV replication (Figure 4B). These findings are important because they confirm that SHFV has overcome the first hurdle of successful spillover to humans: SHFV virions are fully capable of entering cells expressing the human version of their receptor. If CD163 is involved in an evolutionary arms race with simian arteriviruses, then the surface glycoproteins of these viruses are counter-evolving as well— being constantly selected for variants with new receptor binding specificities which could enable them to use receptor orthologs encoded by animals typically resistant to infection.
Human blood monocyte-derived macrophages do not support SHFV infection
Beyond the entry receptor, viruses depend on many host proteins inside of cells to support their replication cycles (Han et al., 2018; Li et al., 2020; Ma et al., 2017; Marceau et al., 2016; Park et al., 2016; Savidis et al., 2016). Although little is known about the lifecycle of simian arteriviruses, for other viruses critical human cofactors include proteases, trafficking proteins, etc. For simian arteriviruses to be a threat to humans, they need to be able to use the human orthologs of all the proteins that they require to replicate, and to bypass the interferon response that will be triggered in human cells. CD163 is expressed specifically in myeloid cells, like monocytes and macrophages (Buechler et al., 2000; Fabriek et al., 2005; Maniecki et al., 2006). To test for SHFV replication in primary human cells, monocytes derived from peripheral blood mononuclear cells (PBMCs) from three human donors were differentiated into M0, M1, or M2 macrophages. We found all of these cell types refractory to SHFV, but susceptible to Ebola virus, which is known to infect macrophages (Figures 5 and S6). However, little is known about the precise myeloid-derived cell subtypes that SHFV favors (Vatter and Brinton, 2014). Porcine reproductive and respiratory syndrome virus 1 (PRRSV-1, a suid arterivirus) primarily infects subpopulations of tissue-resident macrophages (Duan et al., 1997a; Teifke et al., 2001) and less efficiently infects circulating (blood) monocytes and monocyte-derived cells (Duan et al., 1997b). Similarly, lactate dehydrogenase-elevating virus 1 (LDV-1, a murine arterivirus) has an extremely narrow specificity for a subpopulation of tissue-resident macrophages and is poorly cultivated in macrophages ex vivo (Onyekaba et al., 1989; Stueckemann et al., 1982). Further studies on the exact cells that replicate simian arteriviruses in vivo are needed, although studies of tissue-embedded immune cell subsets are notoriously difficult (Lee et al., 2018). Based on uncertainties of SHFV tropism in primary cells, we broadened our approach.
Figure 5. Human blood monocyte-derived macrophages do not support SHFV infection.
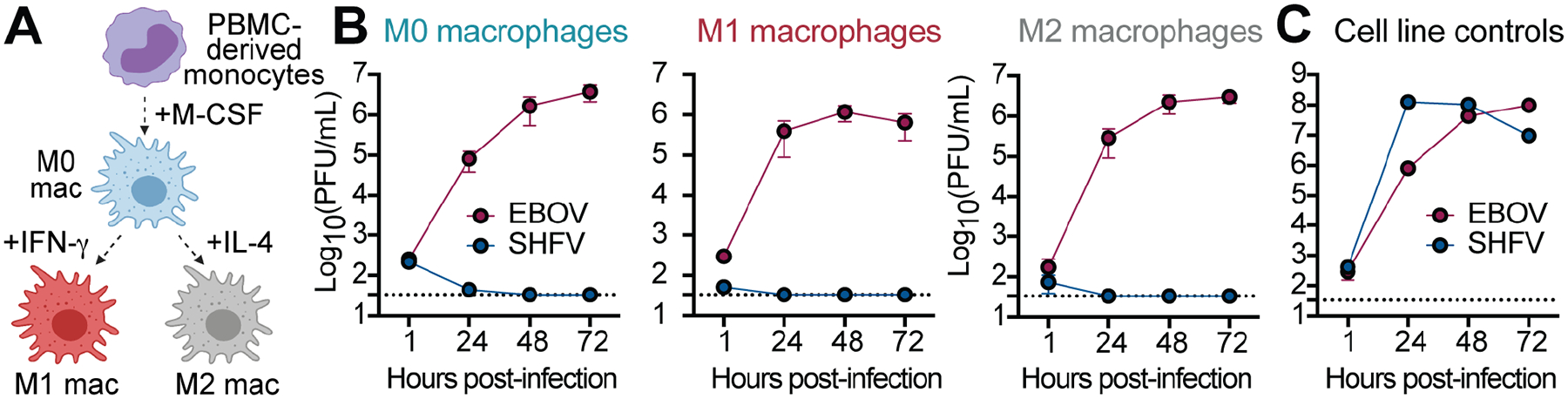
(A) Monocytes were isolated from human peripheral blood mononuclear cells (PBMCs; n = 3 donors) and differentiated into M0 macrophages. M0 macrophages were further polarized into M1 or M2 subsets using IFN-γ or IL-4. (B) Monocyte-derived macrophages were exposed to SHFV at a multiplicity of infection (MOI) of 3 or Ebola virus (MOI = 3), and viral titers were enumerated by plaque assay over time. Exposures were performed in technical triplicates, and the mean viral titers from three donors were plotted. (Error bars represent standard error of the mean [SEM].) (C) In parallel, virus stocks were verified as infectious by exposing them to positive control cell lines (MA-104 for SHFV, Vero E6 for Ebola virus). Data are the mean +/− the standard deviation from technical triplicates. See also Figure S6. SHFV, simian hemorrhagic fever virus; EBOV, Ebola virus.
SHFV is fully competent for replication in human cells
We next added SHFV to several human monocytic cell lines representing the cellular lineage prior to differentiation into macrophages. We included one monocytic cell line that is naturally CD163-positive (SU-DHL-1) and, as controls, two that are CD163-negative (THP-1 and U937) (Buechler et al., 2000). Remarkably, SHFV produced infectious virus in human SU-DHL-1 cells (Figure 6A). SHFV replication in human cells demonstrates that the compatibility of SHFV with humans extends well beyond the viral receptor and includes the human orthologs of all cellular proteins required for viral replication.
Figure 6. SHFV is fully competent for replication in human cells.
(A) Three human monocytic cell lines were exposed to SHFV at a multiplicity of infection (MOI) of 0.3. Infectious virus production over a time course was quantified in cell supernatant by plaque assay (PFU/mL) on SHFV-permissive MA-104 cells. (B) A human cell line was transduced with human CD163 (hCD163) or empty retroviral vectors, and antibiotics were used to select for stable integration. Cells were exposed to recombinant SHFV expressing enhanced green fluorescent protein (rSHFV-eGFP), MOI = 1. Shown are high-content images of mock-exposed or virus-exposed cells counterstained with Hoechst (blue) at 72 h post-exposure. Green fluorescence is indicative of productive viral infection. MA-104 monkey cells served as a positive control. (C) Wild-type MA-104, along with ACHN kidney cells stably transduced with human CD163 (hCD163) or empty vector, were exposed to wild-type SHFV (MOI = 0.3). (A, C) Data shown are representative of two independent experiments, each with three technical replicates. Error bars represent the mean +/− standard deviation. See also Figure S7.
To further verify this highly unexpected result, we aimed to demonstrate replication of SHFV in a second human cell line. Previously it was shown that no adherent human cell line in a large human cell panel (the NCI-60 cancer panel) supported SHFV infection (Cai et al., 2021). However, CD163 expression is highly tissue-restricted to myeloid cells, and myeloid cells are not included in that panel. Indeed, western blotting revealed that none of the human NCI-60 cell lines expressed CD163 (Figure S7). Consequently, we sought to test whether SHFV could replicate in any of these NCI-60 human cell lines if human CD163 was provided. From four randomly chosen cell lines tested, we found a human kidney epithelial cell line (ACHN) that supported replication of SHFV expressing enhanced green fluorescent protein (SHFV-eGFP) after it was stably transduced with human CD163 (hCD163; Figure 6B). In fact, wild-type SHFV exhibited replication kinetics and viral titers on human CD163-expressing ACHN cells that were the same as those on the most permissive known primate cell line (MA-104; Figure 6C). The virus replicated to high titers on these human cells, producing over 107 infectious virions per mL. ACHN cells are documented to be interferon-competent (Shang et al., 2007), leaving us to assume that the virus may escape the human interferon response. Thus, SHFV is able to replicate in human cells, can use the human orthologs of all necessary cofactors, and may bypass interferon mediated pathways of innate immunity. This conclusion is further supported by the fact that SHFV was recently shown to replicate in primary cells of the western gorilla and common chimpanzee (Cai et al., 2021), two apes closely related to humans.
Discussion
We identified CD163 as the intracellular receptor for a primate arterivirus, SHFV. The expression of CD163 is restricted to myeloid cells such as monocytes and macrophages, where it plays in important role in human health (Greaves and Gordon, 2005). When hemoglobin is released into plasma during red blood cell destruction, it is important that it is quickly removed due to the toxicity of its oxidative heme group. Tissue-embedded macrophages take care of this important process, primarily through their CD163 receptor (Kristiansen et al., 2001). Other mammalian arteriviruses have also been associated with CD163 and with tissue-resident monocytes and macrophages (Calvert et al., 2007; Gorp et al., 2010; Huang et al., 2020; Su et al., 2021; Xu et al., 2020; Yu et al., 2020). CD163 is mostly localized intracellularly (Gorp et al., 2009; Nielsen et al., 2006), a common feature shared with many endocytic receptors that traffic between the cell surface and endosomes.
Intracellular processing of the SHFV glycoprotein(s) that interact with CD163 may be required before receptor binding can occur. Neutralization experiments utilizing purified soluble grivet CD163 did not prevent SHFV infection (data not shown). This lack of neutralization suggests that arteriviruses, as they circulate in the bloodstream, cannot bind CD163. Instead, we speculate that the unmasking of CD163 binding sites occurs within intracellular compartments. This may function as a mechanism to evade the humoral immune response, as has also been speculated for Ebola virus (Miller et al., 2012), and would be consistent with the observation of persistent arterivirus infections in monkeys (Bailey et al., 2014b, 2014a). This finding has important consequences that need to be prospectively considered in risk scenarios: severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), which like SHFV is also a nidovirus, in contrast uses a receptor on the cellular surface (ACE2) for cell entry. To do so, SARS-CoV-2 needs to expose its receptor-binding domain prior to adsorbing to cells, thereby leaving it exposed to attack by neutralizing antibodies—the basis for current vaccine approaches. Viruses that use intracellular receptors expose their receptor-binding domains only once inside a cellular compartment. Thus, medical countermeasure development against such viruses is challenging, i.e., strategies that have been successful against SARS-CoV-2 may fail against arteriviruses.
Arteriviruses have been considered relatively obscure since their discovery. However, it should be noted that SIV, and in particular the chimpanzee variant most closely related to the pandemic strain of HIV, was also considered an “obscure” virus prior to its discovery in the early years of the HIV/AIDS pandemic (Keele et al., 2006; Sharp and Hahn, 2011). Given the propensity for RNA viruses such as SHFV and SIV to evolve and emerge, ignoring the threat that arteriviruses pose could be unwise.
Our results raise concerns for global health and pandemic prevention. Not only is the human CD163 ortholog compatible with SHFV entry into human cells, but all other cellular proteins required for SHFV replication are functional in human cells, as evidenced by SHFV titers of up to 107 infectious virions per mL (Figure 6C). Further, we have tested only one of the numerous simian arteriviruses that have been discovered in monkeys (Bailey et al., 2014a, 2016; Lauck et al., 2011, 2013, 2015). They all share the same genetic organization and considerable sequence homology, suggesting that all of them use a similar entry mechanism and that many of them could potentially use human CD163. Further, it appears that interferon-mediated pathways of innate immunity may be ineffective at blocking these viruses, which is concerning given that humans are predicted to be antibody-naïve to arteriviruses. Animal studies are not needed before concern for humans is raised, because at least three different simian arteriviruses have already been shown (on numerous occasions) to cause fatal infection in some nonhuman primates.
It is possible that people in Africa are already subclinically infected with arteriviruses. Similar to HIV-1 infections that spread undetected for decades prior to virus discovery in the 1980s (Korber et al., 2000; Worobey et al., 2008), human arterivirus infections may already cause unmeasured disease. HIV-1-induced AIDS kills by secondary infections and hence was etiologically difficult to pinpoint. Simian arterivirus infection may result in a similar scenario, in subclinical infections, or in acute epidemics of hemorrhagic fever. Development of serology tests and other measures for human surveillance will be required to understand and detect human exposure to these viruses. These tools are needed in African regions where endemically infected nonhuman primates live.
Limitations of the study
Despite its breadth, this study has several limitations. Simian hemorrhagic fever virus (SHFV) is just one simian arteriviruses. However, virus stocks do not exist for other arteriviruses that have caused epizootics in the past (i.e., Pebjah virus [PBJV] and simian hemorrhagic encephalitis virus [SHEV]). This limitation prevented us from performing a more systematic study of simian arterivirus diversity. We speculated based on previous studies that patas monkeys are the natural reservoir host for SHFV (Dalgard et al., 1992; Gravell et al., 1986; Lauck et al., 2015; London, 1977). However, additional viral surveillance of patas monkeys in the wild is required to determine whether or not they are a possible source of SHFV spillover to humans. Although SHFV-exposed primary human blood monocyte-derived macrophages were incapable of producing infectious virus, this does not provide direct evidence that SHFV cannot replicate in more specialized macrophage subsets. It is possible that alternative subpopulations of tissue-resident macrophages may support SHFV infection, like alveolar macrophages (the target for PRRSV-1 arterivirus infection in pigs) or peritoneal macrophages (the target for LDV-1 arterivirus infection in mice).
STAR Methods
RESOURCE AVAILABILITY
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Sara L. Sawyer (ssawyer@colorado.edu).
Materials availability
There are restrictions to the availability of materials. Constructs and reagents in this study will be made available upon request, but a completed Materials Transfer Agreement may be required if there is potential for commercial application.
Data and code availability
Primate CD163 sequences have been deposited at GenBank and are publicly available as of the date of publication. Accession numbers are listed in the key resources table. Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Goat polyclonal anti-CD163 | R&D Systems | Cat#AF1607 |
| Mouse monoclonal anti-actin beta (clone 8H10D10) | Cell Signaling Technology | Cat#3700S |
| Donkey anti-goat IgG (H+L) HRP conjugate | Thermo Fisher Scientific | Cat#A16005 |
| Anti-mouse IgG (H+L) HRP conjugate | Promega | Cat#W402B |
| Mouse purified monoclonal anti-CD163 (clone GHI/61) | BioLegend | Cat#333609 |
| Rabbit polyclonal anti-occludin | Thermo Fisher Scientific | Cat#71-150-0 |
| Goat anti-mouse IgG AF488+ | Invitrogen | Cat#A32723 |
| Goat anti-rabbit IgG AF546 | Invitrogen | Cat#A11010 |
| Bacterial and virus strains | ||
| Simian hemorrhagic fever virus (SHFV) isolate LVR42–0/M6941 | American Type Culture Collection (ATCC) | Cat#VR-533 |
| Ebola virus/H.sapiens-wt/GIN/2014/Makona-C05 | Gary Kobinger (Public Health Agency Canada) | N/A |
| Biological samples | ||
| Rhesus macaque peripheral blood mononuclear cells (PBMCs) | G. K. Wilkerson, The University of Texas MD Anderson Cancer Center | N/A |
| Human PBMCs | Lonza | Cat#4W-270A |
| Nancy Ma’s night monkey (Aotus nancymaae) PBMCs | G. K. Wilkerson | N/A |
| Chemicals, peptides, and recombinant proteins | ||
| Human recombinant macrophage colony-stimulating factor (M-CSF) | R&D Systems | Cat#216-MC-010 |
| Interferon-Y (IFN-γ) | R&D Systems | Cat#285-IF |
| Interleukin 4 (IL-4) | MilliporeSigma | Cat#H7291 |
| Polybrene hexadimethrine bromide | MilliporeSigma | 107689–10G |
| 5-propargylamino-2’,3’-dideoxyuridine-5’-triphosphate (ddUTP) ATTO 633 | Jena Bioscience | Cat#NU-1619–633 |
| Cas9 nuclease, S. pyogenes | New England Biolabs (NEB) | Cat#M0386M |
| Eagle’s minimum essential medium (EMEM) | ATCC | 30–2003 |
| 10% fetal bovine serum (FBS) | MilliporeSigma | TMS-013-B |
| 1X penicillin-streptomycin solution (Pen Strep) | Avantor | #45000–652 |
| RPMI-1640 medium | ATCC | #30–2001 |
| Dulbecco’s modified Eagle’s medium (DMEM) | MilliporeSigma | #D6429 |
| L-glutamine | Corning Incorporated | #MT25005CI |
| Hygromycin | Corning Incorporated | #30–240-CR |
| Phosphate-buffered saline (PBS) | Caisson Labs | PBL01–6X500ML |
| ImmunoCult-SF macrophage medium | STEMCELL Technologies | #10961 |
| 1X phenol-free EMEM | VWR | #115-073-101 |
| 1.4% Avicel | FMC Corporation | RC-591 NF |
| 10% neutral buffered formalin | MilliporeSigma | #HT501128–4L |
| 0.2% crystal violet | MilliporeSigma | #C3866–25G |
| Lipofectamine 3000 reagent | Thermo Fisher Scientific | #L3000008 |
| Opti-MEM | Thermo Fisher Scientific | #31985–070 |
| Tragacanth | MilliporeSigma | #G1128 |
| Nonidet P-40 buffer (NP-40) | Promega | #V4221 |
| Tris(hydroxymethyl)aminomethane hydrochloride (HCl) pH 7.4 | Teknova | #T1075 |
| 1% NP-40 | G-Biosciences | #DG001 |
| Dithiothreitol (DTT) | IBI Scientific | #IB21040 |
| Benzonase | MilliporeSigma | #E1014 |
| Protease inhibitor mixture | MilliporeSigma | #11873580001 |
| Ethylenediamine tetraacetic acid (EDTA) | Corning Incorporated | #25–052-CI |
| 10% Tris-glycine eXtended (TGX) stain-free acrylamide premixed gel solution | Bio-Rad | #1610182 |
| 0.1% tris-buffered saline (TBS) | Research Products International (RPI) | #T60075–4000.0 |
| Polysorbate 20 | Amresco | #M147–1L |
| Nestle Carnation 5% nonfat dried milk | Vevey | N/A |
| Enhanced chemiluminescent (ECL) reagent | MilliporeSigma | #GERPN2232 |
| Neon Transfection System Kit | Thermo Fisher Scientific | #MPK10025 |
| T7 endonuclease 1 (E1) assay in the Alt-R Genome Editing Detection Kit | Integrated DNA Technologies (IDT) | #1075931 |
| Phusion high-fidelity polymerase chain reaction (PCR) master mix with buffer | NEB | #M0531L |
| Clontech pLHCX retroviral expression vector | Takara Bio Inc. | #631511 |
| TransIT 293 transfection reagent | Mirus Bio | #MIR2705 |
| 4% PFA | VWR | #100504–858 |
| FACS buffer | MilliporeSigma | #324506–100ML |
| Terminal deoxynucleotidyl transferase (TdT) buffer | Thermo Fisher Scientific | #EP0161 |
| Sodium acetate (NaOAc) | Amresco | #E498–200ML |
| 100% cold ethanol (EtOH) | Thermo Fisher Scientific | # 04-355-223 |
| Nuclease-free water | IBI Scientific | # IB42201 |
| Poly-L-lysine | MilliporeSigma | #P8920 |
| Paraformaldehyde (PFA) | VWR | #100504–85 |
| 0.1% Triton X-100 | VWR | #JTX198–5 |
| Wash Buffer A | Biosearch Technologies | #SMF-WA1–60 |
| Hybridization buffer | Biosearch Technologies | #SMF-HB1–10 |
| Hoechst 33342 dye for DNA and nuclei | Invitrogen | #H3570 |
| Wash Buffer B | Biosearch Technologies | #SMF-WB1–20 |
| Prolong Gold Antifade Mountant with 4’,6-diamidino-2-phenylindole (DAPI) | Thermo Fisher Scientific | #P36941 |
| 10% neutral buffered formalin | VWR | #16004–128 |
| Critical commercial assays | ||
| Bicinchoninic acid (BCA) protein assay | Thermo Fisher Scientific | Cat#23227 |
| Alt-R Genome Editing Detection Kit | IDT | Cat#1075931 |
| TOPO TA cloning kit | Thermo Fisher Scientific | Cat#K457501 |
| Cytofix/cytoperm Fixation/Permeabilization solution kit | BD | Cat#554714 |
| Data and code availability | ||
| CD163 receptor orthologs cloned from nonhuman primate biomaterial | This study | GenBank IDs: MZ695198–MZ695214 |
| Pearson correlation coefficient analysis | Peters and Garcea, 2020 | https://github.com/DouglasPeters/GarceaMatLabAnalyses/blob/52257dea52d610218b7b9b8b7b43818de67f72de/SIM_ColocalizationandCovarianceAnalysis_08152019_CURRENT.m |
| Experimental models: Cell lines | ||
| MA-104 Clone 1 | ATCC | Cat#CRL-2378.1 |
| MA-104 subclone MARC-145 | Kay Faaberg, U.S. Department of Agriculture | N/A |
| COS-7 | ATCC | Cat#CRL-1651 |
| Patas monkey skin fibroblasts | Coriell Institute for Medical Research (CIMR) | Cat#AG06254 and AG06116 |
| Human histiocytic SU-DHL-1 | ATCC | Cat#CRL-2955 |
| Monocytic THP-1 | ATCC | TIB-202 |
| Monocytic U937 | ATCC | Cat#CRL-1593.2 |
| Kidney cell line ACHN | NCI-Frederick Cancer DCTD Tumor/Cell Line Repository | N/A |
| Kidney cell line CAKI-1 | NCI-Frederick Cancer DCTD Tumor/Cell Line Repository | N/A |
| Kidney cell line786–0 | NCI-Frederick Cancer DCTD Tumor/Cell Line Repository | N/A |
| Kidney cell line A498 | NCI-Frederick Cancer DCTD Tumor/Cell Line Repository | N/A |
| HEK293T | ATCC | Cat#11268 |
| Grivet kidney Vero E6 | ATCC | Cat#CRL-1586 |
| MA-104, Vero, patas monkey, human bone marrow | Takara Bio Inc. | #636643 |
| Common chimpanzee (Pan troglodytes) fibroblasts | Emory University | S008886 |
| Red-cheeked gibbon (Nomascus gabriellae) fibroblasts | CIMR | #PR00381 |
| Northern white-cheeked gibbon (Nomascus leucogenys) fibroblasts | CIMR | #PR00712 |
| Lar gibbon (Hylobates lar) fibroblasts | CIMR | #PR01131 |
| Collared mangabey (Cercocebus torquatus) fibroblasts | CIMR | #PR00485 |
| Black crested mangabey (Lophocebus aterrimus) fibroblasts | CIMR | #PR01215 |
| Angolan talapoin (Miopithecus talapoin) fibroblasts | CIMR | #PR00716 |
| Wolf’s mona monkey (Cercopithecus wolfi) fibroblasts | CIMR | #PR01241 |
| Francois’ langur (Trachypithecus francoisi) fibroblasts | CIMR | #PR01099 |
| Mantled guereza (Colobus guereza) fibroblasts | CIMR | #PR00980 |
| Bolivian red howler (Alouatta sara) fibroblasts | CIMR | #PR00708 |
| Oligonucleotides | ||
| Primers for cloning nonhuman primate CD163 receptor orthologs | See Table S1 for primer sequences | N/A |
| CRISPR-Cas9 RNA (crRNA) targeting CD163 exon 3; AATGCGTCCAGAACCTGCAC | This study | N/A |
| Alt-R trans-activating CRISPR-Cas9 RNA (tracrRNA) | IDT | Cat#1072534 |
| Recombinant DNA | ||
| pLHCX retroviral transfer vector encoding CD163 orthologs | This study | N/A |
| Empty pLHCX retroviral transfer vector | Yamashita and Emerman, 2004 | N/A |
| pCS2-mGP encoding MLV gag-pol | Yamashita and Emerman, 2004 | N/A |
| pCMV-VSV-G (encoding vesicular stomatitis Indiana virus glycoprotein “G” | Addgene | Cat#8454 |
| Recombinant SHFV (rSHFV) | Cai et al., 2021 | N/A |
| Enhanced green fluorescent protein-expressing rSHFV | Cai et al., 2021 | N/A |
| Software and algorithms | ||
| Sequencher DNA sequence analysis software | Gene Codes Corporation | N/A |
| Stellaris probe designer | Biosearch Technologies | https://www.biosearchtech.com/support/tools/design-software/stellaris-probe-designer |
| ImageJ | Schneider et al., 2012 | https://imagej.nih.gov/ij/ |
| FlowJo v10.8.0 software | BD Life Sciences | N/A |
| MEGA7 | Kumar et al., 2016 | https://www.megasoftware.net/ |
| MEGAX | Stecher et al., 2020 | https://www.megasoftware.net/ |
| PAL2NAL web server tool | Suyama et al., 2006 | http://www.bork.embl.de/pal2nal/ |
| Nipot v2.3 | Perriere and Gouy, 1996 | http://doua.prabi.fr/software/njplot |
| PAML4.8 | Yang, 2007 | N/A |
| Prism v9.1.0 | GraphPad | N/A |
| QGIS v3.16.4 | QGIS Geographic Information System | www.qgis.org |
| BioRender | Created with Biorender.com | N/A |
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Cells
Nonhuman primate cell lines, including grivet (Chlorocebus aethiops) embryonic kidney MA-104 Clone 1 (American Type Culture Collection [ATCC], Manassas, VA, USA, #CRL-2378.1), MA-104 subclone MARC-145 (originally provided by Dr. Kay Faaberg, U.S. Department of Agriculture [USDA], National Animal Disease Center, Ames, IA, USA), grivet kidney fibroblast COS-7 (ATCC, #CRL-1651), and patas monkey skin fibroblasts (Coriell Institute for Medical Research [CIMR], Camden, NJ, USA, #AG06254 and #AG06116) were cultured in Eagle’s minimum essential medium (EMEM; ATCC, 30–2003) plus 10% fetal bovine serum (FBS; MilliporeSigma, Billerica, MA, USA, TMS-013-B) and 1X penicillin-streptomycin solution (Pen Strep; Avantor, Radnor, PA, USA, #45000–652) (complete EMEM [cEMEM]). Human histiocytic SU-DHL-1 (ATCC, #CRL-2955), monocytic THP-1 (ATCC, #TIB-202), monocytic U937 (ATCC, #CRL-1593.2), and kidney cell lines ACHN, CAKI-1, 786–0, and A498 (obtained from the National Cancer Institute [NCI] Division of Cancer Treatment and Diagnosis [DCTD] Tumor Repository, Frederick, MD, USA) were cultured in RPMI-1640 medium (ATCC, #30-2001) plus 10% FBS and 1X Pen Strep (complete RPMI [cRPMI]). HEK293T cells (ATCC, #11268) were cultured in Dulbecco’s modified Eagle’s medium (DMEM; MilliporeSigma, Billerica, MA, USA, # D6429) plus 10% FBS, 1X Pen Strep, and L-glutamine (Corning Incorporated, Corning, NY, USA, #MT25005CI) (complete DMEM [cDMEM]). Grivet kidney Vero E6 cells (ATCC, #CRL-1586) were maintained in DMEM supplemented with 10% FBS. All cells were cultured at 37°C in a humidified 5% CO2 atmosphere. Hygromycin (Corning, #30–240-CR) selection was maintained throughout culture of the following cell lines, stably transduced with retroviral vectors: MA-104 (300 μg/mL), Vero (200 μg/mL), patas monkey skin fibroblasts (50 μg/mL), 786–0 and CAKI-1 (500 μg/mL), and ACHN and A498 (250 μg/mL). The NCI-60 panel of human cell lines was handled as previously described (Cai et al., 2021).
Rhesus macaque peripheral blood mononuclear cells (PBMCs) were obtained from rhesus macaques housed at the Keeling Center for Comparative Medicine and Research (KCCMR) in Bastrop, TX, USA. All blood collections performed for this study were approved by the MD Anderson Cancer Center Institutional Animal Care and Use Committee (OLAW assurance number A3343–01) and were performed in strict compliance with The Guide for the Care and Use of Laboratory Animals (8th edition). PBMCs were seeded at >5×106 cells per well of a 6-well plate (CELLTREAT Scientific Products, Pepperell, MA, USA, #229105) and cultured overnight in cRPMI. The next day, cells were gently washed three times with phosphate-buffered saline (PBS; Caisson Labs, Smithfield, UT, USA, PBL01–6X500ML) to remove non-adherent lymphocytes. Macrophages were cultured from adherent cells by incubation with cRPMI supplemented with 20 ng/mL human recombinant macrophage colony-stimulating factor (M-CSF; R&D Systems, Minneapolis, MN, USA, #216-MC-010) for 5 d. Human PBMCs (Lonza, Basel, Switzerland, #4W-270A) were maintained in ImmunoCult-SF macrophage medium (STEMCELL Technologies, Vancouver, British Columbia, Canada, #10961), supplemented with 50 ng/mL human recombinant M-CSF (CSF1; R&D Systems, 216-MC) for 6 d to allow M0 macrophage differentiation. For M1 or M2 macrophage differentiation, M0 macrophages were treated with 50 ng/mL interferon-γ (IFN-γ; R&D Systems, 285-IF) or with 10 ng/mL interleukin 4 (IL-4; MilliporeSigma, St. Louis, MO, USA, H7291), respectively, for 2 d.
Viruses
Simian hemorrhagic fever virus (SHFV) isolate LVR42–0/M6941 (ATCC, #VR-533) (Lauck et al., 2014) and cDNA-launch systems for recombinant SHFV (rSHFV) and rSHFV expressing enhanced green fluorescent protein (rSHFV-eGFP) were used as previously described (Cai et al., 2021). Briefly, for passage 0 (P0), a stock of SHFV was propagated in 1.75×107 MA-104 cells plated in 182-cm2 flasks (CELLTREAT Scientific Products, #229381) using a multiplicity of infection (MOI) of 0.001 for 1 h at 37°C (5 mL of EMEM + 2% FBS), with gentle rocking every 15 min. Inocula were then removed and replaced with 15 mL of 2% EMEM. Cell supernatants were harvested 3 d later (>80% cytopathic effect) and clarified by centrifugation at 500 ×g for 10 min at 4°C. This P1 stock was titered by plaque assay. Briefly, MA-104 cells were seeded at 3×105 cells per well in 6-well plates. The following day, cell culture media were removed, virus samples were 10-fold serial-diluted in EMEM, and 300 μL of diluted samples were added to the each well of cells, followed by incubation at 37°C for 1 h, with gentle rocking every 15 min. Cells were then washed three times with PBS, and a volume of 3 mL of overlay medium (final concentration 1X phenol-free EMEM [VWR, #115-073-101] + 5% FBS + Lglutamine + 1.4% Avicel [Avicel RC-591 NF; FMC Corporation, Philadelphia, PA, USA]) was added to each well, followed by undisturbed incubation at 37°C in a humidified 5% CO2 atmosphere for 48 h. Cells were then washed twice with PBS and fixed/stained (final concentration 10% neutral buffered formalin [MilliporeSigma, #HT501128–4L], 0.2% crystal violet [MilliporeSigma, #C3866–25G]) for 15 min. Plaques were visualized following the removal of the fix/stain solution and gentle washing. From the P1 stock, a P2 stock, which was used for all downstream experiments, was generated by exposing MA-104 cells (MOI = 0.01) as described above. rSHFV and rSHFV-eGFP were rescued from plasmids by transfection of MA-104 cells using Lipofectamine 3000 reagent (Thermo Fisher Scientific, Carlsbad, CA, USA, #L3000008). Briefly, cells were seeded at 3×105 cells per well in 6-well plates and, the next day, transfected with 3 μg of plasmid DNA. (Tube A contained 150 mL Opti-MEM [Thermo Fisher Scientific, #31985–070] and 4.5 μL Lipofectamine 3000; Tube B contained 150 μL Opti-MEM, 3 μg plasmid DNA, and 6 μL Lipofectamine p3000. Tube A was added to Tube B, mixed 5 min, and added dropwise to cells.) Viral titers peaked 3 d post-transfection. Ebola virus/H.sapiens-wt/GIN/2014/Makona-C05 (Baize et al., 2014; Hoenen et al., 2014) was provided by Dr. Gary Kobinger (Public Health Agency Canada, Winnipeg, MT, Canada) and propagated in Vero E6 cells as previously described (Logue et al., 2019).
METHOD DETAILS
Viral infection assays
Cells were seeded in individual wells of 6-well plates and, the next day, exposed to SHFV or Ebola virus at the indicated MOIs (see figure legends). Cells were exposed to virus in a minimal volume of media (300 μL) and incubated at 37°C for 1 h, with gentle rocking every 15 min. After incubation, cells were washed three times with PBS and then maintained in cell-type-specific media supplemented with 5% FBS. Cell supernatants were collected at indicated times post-exposure. Viral titers were determined by standard plaque assays using MA-104 cells for SHFV (Caì et al., 2015) and Vero E6 cells for Ebola virus (Logue et al., 2019). Plaque assays used overlays with final concentrations of 0.8% Tragacanth (MilliporeSigma, G1128) or 1.4% Avicel for SHFV and 1.25% Avicel for Ebola virus. At 2 d (SHFV) and 8 d (Ebola virus), overlays were removed, and plaques were counted.
Western blotting
Cells were lysed in Nonidet P-40 buffer (NP-40; 150 mM sodium chloride [NaCl; Promega, Madison, WI, USA, #V4221]), 50 mM tris(hydroxymethyl)aminomethane hydrochloride [HCl] pH 7.4 [Teknova, Hollister, CA, USA, #T1075], 1% NP-40 [G-Biosciences, St. Louis, MO, USA, #DG001], 1 mM dithiothreitol [DTT] [IBI Scientific, Dubuque, IA, USA, #IB21040], 1 μL/mL Benzonase [MilliporeSigma, #E1014]), and protease inhibitor mixture (MilliporeSigma, #11873580001). Briefly, cells were detached from tissue-culture plates by trypsin ethylenediamine tetraacetic acid (EDTA; Corning, Tewksbury, MA, USA, #25–052-CI) treatment, inactivated with FBS containing media, and then pelleted (300 ×g for 5 min) and washed once with PBS. The cells were again pelleted, resuspended in NP-40 buffer, and incubated at 4°C for 30 min on a tube revolver rotator (ThermoScientific, #88881001). Cell lysates were cleared by centrifugation at 16,000 ×g for 15 min at 4°C. Whole-cell extracts were quantified using a bicinchoninic acid (BCA) protein assay (Thermo Fisher Scientific, #23227). Total protein (10 μg) was resolved with polyacrylamide gel electrophoresis using gels prepared with 10% Tris-glycine eXtended (TGX) stain-free acrylamide premixed gel solution (Bio-Rad, Hercules, CA, USA, #1610182) and by applying 180 V until protein-loading dye ran off the gel. Proteins were transferred to Immobilon-P PVDF membranes (MilliporeSigma, #IPVH07850) using a wet transfer apparatus (Bio-Rad Mini-Protean tetra and blotting module, #1660828EDU) set at 100 V for 1 h. Membranes were blocked in 0.1% tris-buffered saline (TBS; Research Products International [RPI], Mount Prospect, IL, USA, #T60075–4000.0) + Polysorbate 20 (TWEEN 20; Amresco, Dallas, TX, USA, #M147–1L) (TBST) + Nestle Carnation 5% nonfat dried milk (Vevey, Switzerland) for 1 h at ambient temperature. Primary antibodies (goat polyclonal anti-CD163 antibody [R&D Systems, #AF1607] and mouse monoclonal actin beta (clone 8H10D10) antibody [mAb; Cell Signaling Technology, Danvers, MA, USA, #3700S]) were diluted in TBST + 5% nonfat dried milk and incubated either overnight at 4°C (anti-CD163 polyclonal antibody) or for 1 h at ambient temperature (actin beta [8H10D10] mouse mAb). After primary incubation, cells were washed three times for 5 min in TBST. Secondary antibodies—donkey anti-goat immunoglobulin G (IgG) (H+L) cross-adsorbed horseradish peroxidase (HRP) conjugate (Thermo Fisher Scientific, #A16005), anti-mouse IgG (H+L) HRP conjugate (Promega, #W402B)—were diluted in TBST + 5% nonfat dried milk and incubated with the membrane for 1 h at ambient temperature. After secondary incubation, membranes were washed three times for 5 min in TBST, developed using enhanced chemiluminescent (ECL) reagent (MilliporeSigma, #GERPN2232), and imaged on a ChemiDoc Imaging System (Bio-Rad).
CRISPR-Cas9 editing of CD163
CRISPR-Cas9 RNA (crRNA) targeting grivet CD163 exon 3 (AATGCGTCCAGAACCTGCAC) was synthesized and mixed with Alt-R trans-activating CRISPR-Cas9 RNA (tracrRNA; Integrated DNA Technologies [IDT], Coralville, IA, USA, #1072534) to a final duplex concentration of 44 μM. RNA duplexes were heated at 95°C for 1 h and cooled to ambient temperature. Cas9 nuclease, S. pyogenes (New England Biolabs [NEB], Ipswich, MA, USA, NEB #M0386M) was diluted with resuspension buffer R from the Neon Transfection System Kit (Thermo Fisher Scientific, #MPK10025) to 36 μM in a 0.5-μL final solution, mixed with 0.5 μL of crRNA:tracrRNA duplex, and incubated at ambient temperature for 20 min. A total of 1×106 MA-104 cells were electroporated with two 30-ms pulses of 1,150 V using the Neon Transfection System (Thermo Fisher Scientific, #MPK5000). Electroporated cells were resuspended in cEMEM and incubated for 4 d in 12-well plates (CELLTREAT Scientific Products, #229111). Genomic editing in the bulk cell populations were confirmed using the T7 endonuclease 1 (E1) assay in the Alt-R Genome Editing Detection Kit (IDT, #1075931). Single-cell clones from the CD163 exon-3-edited bulk cell population were obtained by limited dilution cloning (0.5 cells per well in 96-well plates [CELLTREAT Scientific Products, #229196]). Genomic DNA from single-cell clones was screened by the T7E1 assay to identify edited clones. CD163 knockout was confirmed by western blotting, and clone “F11” was selected for all follow-up studies. Primers flanking the crRNA target site were used to amplify a small fragment of genomic DNA using a Phusion high-fidelity polymerase chain reaction (PCR) master mix with buffer (NEB, #M0531L). A-tailed PCR fragments were then TA-cloned via sequencing with the TOPO TA Cloning Kit (Thermo Fisher Scientific, #K457501). Successful insertion of a PCR product was confirmed by blue-white screening. A total of 15 independent plasmid clones from F11-edited and parental (MA-104) cells were Sanger-sequenced (Quintara Biosciences, Cambridge, MA, USA) and sequences analyzed using Sequencher DNA sequence analysis software (Gene Codes Corporation, Ann Arbor, MI, USA). Bi-allelic deletions in clone F11 (ΔCACTG and ΔCA) resulted in a premature stop codon and ablation of CD163 protein expression (MA-104 ΔCD163) (Figure S1).
CD163 receptor complementation experiments
Total RNA was isolated from the following cells: MA-104, Vero, patas monkey, human bone marrow (Takara Bio Inc., Mountain View, CA, USA, #636643), common chimpanzee (Pan troglodytes) fibroblasts (Emory University, Yerkes National Primate Research Center, Atlanta, GA, USA, S008886), red-cheeked gibbon (Nomascus gabriellae) fibroblasts (CIMR, #PR00381), northern white-cheeked gibbon (Nomascus leucogenys) fibroblasts (CIMR, #PR00712), lar gibbon (Hylobates lar) fibroblasts (CIMR, #PR01131), collared mangabey (Cercocebus torquatus) fibroblasts (CIMR, #PR00485), black crested mangabey (Lophocebus aterrimus) fibroblasts (CIMR, #PR01215), rhesus macaque PBMCs (obtained from G. K. Wilkerson, Michale E. Keeling Center for Comparative Medicine and Research, The University of Texas MD Anderson Cancer Center), Angolan talapoin (Miopithecus talapoin) fibroblasts (CIMR, #PR00716), wolf’s mona monkey (Cercopithecus wolfi) fibroblasts (CIMR, #PR01241), François’ langur (Trachypithecus francoisi) fibroblasts (CIMR, #PR01099), mantled guereza (Colobus guereza) fibroblasts (CIMR, #PR00980), Nancy Ma’s night monkey (Aotus nancymaae) PBMCs (obtained from G. K. Wilkerson), and Bolivian red howler (Alouatta sara) fibroblasts (CIMR, #PR00708). cDNA was generated by reverse transcription with a SuperScript IV First-Strand Synthesis System (Thermo Fisher Scientific, #18091200), and CD163 was amplified using a Phusion high-fidelity PCR master mix with buffer (NEB, #M0531L) with gene-specific primers. CD163 from these host cells was inserted into the Clontech pLHCX retroviral expression vector (Takara Bio Inc., #631511) using a Gibson assembly. Sequences are deposited as Genbank accession numbers MZ695198–MZ695214.
Murine leukemia virus (MLV; Retroviridae: Gammaretrovirus) pseudotypes were generated as described previously (Warren et al., 2019a). Briefly, HEK293T cells were plated in cDMEM without antibiotics (1×106 cells per well in 6-well plates) and, the next day, transfected with 2 μg of pLHCX transfer vector (encoding CD163 receptor orthologs and hygromycin-resistance genes), 1 μg of pCS2-mGP (encoding MLV gag-pol) (Yamashita and Emerman, 2004), and 0.2 μg of pCMV-VSV-G (encoding vesicular stomatitis Indiana virus glycoprotein “G”) (Addgene, Watertown, MA, USA, #8454) using a 3:1 ratio of TransIT 293 transfection reagent (Mirus Bio, Madison, WI, USA, #MIR2705) to DNA. At 48 h post-transfection, cell supernatants containing gammaretrovirus particles were collected, filtered through 0.22-μm cellulose acetate filters (GVS Life Sciences, Sanford, ME, USA, #FJ25BSCCA002AL01), and stored at −80°C. For transduction, target cells were plated at ≈15% confluence and, the next day, exposed to gammaretrovirus-containing supernatant by spinoculation at 1,200 ×g for 75 min in the presence of 3 μg/mL polybrene hexadimethrine bromide (MilliporeSigma, 107689-10G). The following day, media were replaced with fresh media and, 48 h later, cells were placed in media containing hygromycin throughout their time in culture.
CD163 detection by flow cytometry
For each cell line analyzed, 1×105 cells were added to three wells of a 96-well v-bottom plate. Each well was handled as follows: well #1, unstained control; well #2, surface stain only; well #3, intracellular stain only. The cells were pelleted at 250 ×g for 5 mins, washed once with PBS, and then proceeded to surface staining. For surface stains, each well was handled as follows: wells #1 and #3, cells were resuspended in 50 μl fluorescence-activated cell-sorting (FACS) buffer (PBS + 2% FBS + 1 mM EDTA); well #2, cells were resuspended in 50 μL FACS buffer + 1:25 dilution of APC-conjugated CD163 antibody (clone GHI/61, BioLegend, #333609). The cells were incubated at 4°C for 30 mins in the dark and then washed three times with FACS buffer. The cells were then resuspended in 200 μl of fixation/permeabilization solution (BD Cytofix/cytoperm kit, #554714) and then incubated at 4°C for 20 mins in the dark. The cells were then washed two times in 200 μl of perm/wash buffer (BD Cytofix/cytoperm kit) and then proceeded to intracellular staining. For intracellular stains, each well was handled as follows: wells #1 and #2, cells were resuspended in 50 μL of perm/wash buffer; well #3, cells were resuspended in 50 μL of perm/wash buffer + 1:25 dilution of APC-conjugated CD163 antibody (clone GHI/61, BioLegend). The cells were incubated at 4°C for 30 mins in the dark and then washed three times with perm/wash buffer. After the final wash, the cells were resuspended in FACS buffer and analyzed by flow cytometry.
Design and labeling of smFISH probes
A total of 48 probes (20 nucleotides in length, 2 nucleotides minimum spacing) were designed antisense to SHFV ORF1a using the Stellaris probe designer (Biosearch Technologies, Petaluma, CA, USA, https://www.biosearchtech.com/support/tools/design-software/stellaris-probe-designer). Probes were then ordered according to a 96-well plate layout from IDT and directly conjugated with 2’,3’-dideoxyuridine-5’-triphosphate (ddUTP) and labeled with fluorescent dyes by combining the following reagents and incubating overnight: 10 μL 5X terminal deoxynucleotidyl transferase (TdT) buffer, 1 μL TdT (Thermo Fisher Scientific, #EP0161), 5 μL probe mix (10 μL of each DNA oligo were pooled, and then 5 μL of these mixtures were used in the labeling reaction), and 3 μL 5-propargylamino-ddUTP ATTO 633 (Jena Bioscience, Jena, Thuringia, Germany, #NU-1619–633). Probes were incubated for 8 h at 37°C in the dark. Then, an additional volume of 0.75 μL TdT was added and mixed, and incubation was continued overnight (≈24 h total). To precipitate probes, 50 μL of 3 M sodium acetate (NaOAc; Amresco, #E498–200ML) and 400 μL of 100% cold ethanol (EtOH; Thermo Fisher Scientific, # 04-355-223) were added to each reaction, and then the samples were stored at −20°C for 30 min. Oligos were pelleted by centrifugation at 16,000 ×g for 15 min at 4°C, washed two times with 70% cold EtOH, resuspended in nuclease-free water (IBI Scientific, # IB42201), aliquoted into single-use tubes, and stored at −80°C.
Sequential immunofluorescence and smFISH
Wild-type MA-104 and MA-104 ΔCD163 cells were seeded onto acid-etched coverslips in 6-well plates at ≈80% confluency (2.1×105 cells per well). Macaque macrophages (Figures 4C and 4D) were seeded at 1×105 cells per well with 1 μg/mL poly-L-lysine-coated (MilliporeSigma, #P8920) for 1 h at 37°C on glass plates (DOT Scientific, Burton, MI, USA, #MGB096). Cells were exposed to SHFV (MOI = 10) or mock-exposed for the indicated duration. Following exposure, cells were washed three times with PBS, fixed with paraformaldehyde (PFA; VWR, #100504–85) in PBS (4% final concentration) undisturbed for 10 min at ambient temperature. Following two PBS washes, cells were permeabilized using 0.1% Triton X-100 (VWR, # JTX198–5) for 5 min at ambient temperature. Permeabilization solution was removed, and cells were washed once with PBS and then incubated with primary antibodies—mouse purified monoclonal anti-CD163 antibody clone GHI/61 (Biolegend, San Diego, CA, USA, #333602) and/or rabbit polyclonal anti-occludin antibody (Thermo Fisher Scientific, #71-150-0)—for 1 h at ambient temperature. Following three 10-min PBS washes, cells were incubated with Alexa-Fluor-labeled secondary antibodies—goat anti-mouse IgG AF488+ (Invitrogen, Waltham, MA, USA, #A32723) and/or goat anti-rabbit IgG AF546 (Invitrogen, #A11010)—for 1 h at ambient temperature. Cells were then washed three times with PBS for 10 min each and refixed as detailed above.
For smFISH, cells were washed twice with PBS and once with 1X Wash Buffer A (Biosearch Technologies, #SMF-WA1–60) following fixation. Cells were incubated with hybridization buffer (Biosearch Technologies, #SMF-HB1–10), containing smFISH probes (1-μL probe per 100-μL hybridization reaction), at 37°C for 16 h in the dark. Cells were then incubated with Wash Buffer A at 37°C for 30 min. Then, microphages were placed on glass plates, counterstained with Hoechst 33342 dye for DNA and nuclei (Invitrogen, #H3570) at a 1:5,000 dilution in Wash Buffer A for 15 min at 37°C, while cell lines continued processing (on glass coverslips). Staining buffer was removed, the cells were washed once with Wash Buffer B (Biosearch Technologies, #SMF-WB1–20) for 5 min, and then either stored in PBS at 4°C (macrophages) or mounted onto glass slides (cell lines) using Prolong Gold Antifade Mountant with 4′,6-diamidino-2-phenylindole (DAPI; Thermo Fisher Scientific, #P36941). Mounted coverslips were cured for 2 d at ambient temperature prior to imaging.
Microscopy and image processing
Laser scanning confocal microscopy images were acquired on a A1R laser scanning confocal microscope (Nikon Metrology, Brighton, MI, USA) using a 1.45 NA 100x oil objective, 405/488/561/640 laser lines, and photomultiplier tube detectors. Z-stacks were collected at Nyquist resolution through the volume of several cells in each condition, and the acquisition parameters were adjusted to optimize the signal-to-noise ratio for each image and condition. Image processing was performed using ImageJ analysis software (Schneider et al., 2012) (National Institutes of Health [NIH], Bethesda, MD, USA). Maximum-intensity projections are shown for representative images in Figures 3a, 3b, and 3c; single z-planes are shown for orthogonal images in Figure 3e. Brightness and contrast were adjusted, when needed, to display the full dynamic range of each image. Pearson’s correlation coefficient (PCC) analysis (Figure 3D) was performed with a custom MATLAB script that was adapted from (Peters and Garcea, 2020) and based on the image analysis practices outlined in (Aaron et al., 2018). Briefly, a percentile-based threshold was applied to each channel in each image. A PCC value was calculated for all colocalizing pixels in each pair of channels, reflecting the relative co-occurrence of the corresponding fluorescent signals throughout the z-stack image. For imaging rSHFV-eGFP-infected cells, cells were fixed with 10% neutral buffered formalin (VWR, #16004–128) and stained with Hoechst 33342 (Thermo Fisher Scientific, #H3570) 3 d after virus exposure. Images were taken by Operetta CLS (PerkinElmer, Dumfries, VA, USA).
FISH-Flow
Wild-type MA-104 or MA-104 ΔCD163 cells were either (1) exposed to SHFV (Figure S4) or (2) transfected with SHFV-encoding plasmids (Figure S3). At the indicated times following exposure/transfection, cell-culture media was removed, and the cells were washed three times with PBS. Cells were detached with trypsin, inactivated with FBS-containing media, and then transferred to individual wells of a 96-well v-bottom plate (NEST Scientific, Wuxi, China, #701211). Cells were washed once with PBS and then fixed with 4% PFA (VWR, #100504–858) for 10 min at ambient temperature. Cells were pelleted at 500 ×g for 5 min, washed three times with PBS, and then permeabilized with cold 70% EtOH for at least 1 h at 4°C. Cells were pelleted, washed once with Wash Buffer A, and resuspended in 50 μL of hybridization buffer containing probe (one 0.5-μL probe per 50-μL reaction). The plate was incubated in the dark at 37°C for 16 h. After incubation, 150 μL of Wash Buffer A were added. Cells were pelleted, resuspended in 200 μL of Wash Buffer A, and incubated in the dark at 37°C for 30 min. After incubation, cells were pelleted and resuspended in Wash Buffer B. Cells were pelleted once more and resuspended in FACS buffer (MilliporeSigma, #324506–100ML), comprised of PBS + 2% FBS + 1 mM EDTA. Samples were analyzed on an Accuri C6 Cytometer (BD, East Rutherford, NJ, USA). At least 50,000 live cell events were collected following singlet discrimination. Data was analyzed with FlowJo v10.8.0 (Becton, Dickinson and Company, Franklin Lakes, NJ, USA).
Phylogenetic analysis
Sequences of 33 representative nidovirals were downloaded from a previously published analysis (Nga et al., 2011). Basic Local Alignment Search Tool (BLAST) searches using RNA-directed RNA polymerase (RdRp) and helicase domains from SHFV were queried against simian arteriviruses, and multiple sequence alignments (using the Clustal tool in Molecular Evolutionary Genetics Analysis Version 7 [MEGA7] software with default parameters (Kumar et al., 2016)) were generated to identify and parse homologous regions for all simian arteriviruses. Phylogenetic relationships were inferred using the maximum-likelihood method based on the JTT matrix-based model (Jones et al., 1992). The tree with the highest log likelihood (−30036.74) is shown. Initial tree(s) for the heuristic search were obtained automatically by applying Neighbor-Join and BioNJ algorithms using default parameters in MEGA7 (Kumar et al., 2016) to a matrix of pairwise distances estimated using a JTT model and then selecting the topology with superior log likelihood value. The tree is drawn to scale, with branch lengths measured in the number of substitutions per site. The analysis involved 44 amino-acid sequences with a total of 746 positions in the final dataset. Bootstrap analysis was used to test the robustness of the tree topology (100 resamplings).
Evolutionary analysis of positive selection
Gene sequences (Figure S5 were aligned to the longest human isoform in MEGA Version 10 (MEGA X) for MacOS (Stecher et al., 2020) using the ClustalW alignment tool. Multiple sequence alignments were visually inspected, duplicate gene sequences were removed, and the gene isoform from each animal that best aligned to the human reference was retained for further analysis. Terminal stop codons were removed and aligned DNA and protein sequences were exported as fasta files. Codon alignments were generated using the PAL2NAL web server tool (Suyama et al., 2006). Species cladograms for use in the PAML4.8 software package (Yang, 2007) were constructed following the species-rank phylogenetic relatedness of primates (Perelman et al., 2011). Cladograms were generated using Newick-formatted files and viewed with Njplot version 2.3 (Perrière and Gouy, 1996). Codon alignments and unrooted species cladograms were used as input files for analysis of positive selection using PAML4.8. To detect selection, multiple sequence alignments were fit to the NSites models M7 (neutral model, codon values of dN/dS fit to a beta distribution bounded by 0 and 1), M8a (neutral model, similar to M7 but with an extra codon class fixed at a dN/dS value equal to 1), and M8 (positive selection model, similar to M8a but with the extra codon class allowed to have a dN/dS greater than 1). A likelihood ratio test was performed to assess whether the model of positive selection (M8) yielded a significantly better fit to the data compared to null models (model comparisons M7 versus M8 and M8a versus M8). Posterior probabilities (Bayes Empirical Bayes analysis) were assigned to individual codons with dN/dS values greater than 1.
African nonhuman primate range data
Range data of simian arterivirus-infected African nonhuman primates (Figure 1C) were downloaded from the International Union for Conservation of Nature (IUCN) Red List (iucnredlist.org) as shapefiles and visualized in QGIS (v3.16.4) (www.qgis.org). Cartoon schematics were created using Biorender.com.
QUANTIFICATION AND STATISTICAL ANALYSIS
Correlations between viral RNA and cellular markers by microscopy were determined using Pearson’s correlation analysis of the laser scanning confocal microscopy images. Data are presented as mean ± SEM unless otherwise indicated in figure legends. Experimental repeats are indicated in figure legends. Differences in means were considered significantly different at p < 0.05. Significance levels are: * p < 0.05; ** p < 0.01; *** p < 0.001; **** p < 0.0001; n.s., non-significant. Analyses were performed using Prism version 9.1.0 (GraphPad, La Jolla, CA, USA).
Supplementary Material
Table S1. Primers used for cloning CD163 receptor orthologs, related to Figure 4.
SHFV uses an intracellular receptor, CD163, for cellular entry.
CD163 divergence in primates of some species poses a barrier to SHFV entry.
All cellular proteins required for SHFV replication are functional in human cells.
SHFV replication in human cells suggests potential for zoonotic transmission.
Arteriviruses that infect apes and monkeys can cause fatal hemorrhagic fevers. One such virus, SHFV is capable of infecting and replicating in human monocytes by using CD163 as the intracellular receptor. This suggests that SHFV does not require major adaptations for the human host and surveillance as well as therapeutics are needed for this virus family.
Acknowledgments
We are grateful to Kay Faaberg (U.S. Department of Agriculture National Animal Disease Center, Ames, IA, USA) for providing MA-104 subclone MARC-145 and to Gary Kobinger (Public Health Agency Canada, Winnipeg, MT, Canada) for providing Ebola virus/H.sapiens-wt/GIN/2014/Makona-C05. We thank Nicholas Meyerson, Adam Bailey, and Teressa Shaw for their insightful advice on this work and Anya Crane for critically editing the manuscript.
This work was funded in part by grants from the National Institutes of Health (DP1-DA-046108 and R01-AI-137011 to S.L.S.; F32-GM-125442 and K99-AI151256-01A1 to C.J.W.). S.L.S. is a Burroughs Wellcome Fund Investigator in the Pathogenesis of Infectious Disease. The imaging work was performed at the BioFrontiers Institute Advanced Light Microscopy Core (RRID: SCR_018302). Laser scanning confocal microscopy was performed on a Nikon A1R microscope supported by NIST-CU Cooperative Agreement award number 70NANB15H226. R01AI151636 (to D.K.P.). The flow cytometry work was performed at the BioFrontiers Institute Flow Cytometry Core on the BD Accuri C6 Cytometer, supported by NIH Grant S10ODO21601. This work was also supported in part through the Laulima Government Solutions, LLC, prime contract with the NIAID under Contract Number HHSN272201800013C (S.Y., I-C.H., and G.W.); J.H.K. performed this work as an employee of Tunnell Government Services (TGS), a subcontractor of Laulima Government Solutions, LLC, under Contract Number HHSN272201800013C.
The views and conclusions contained in this document are those of the authors and should not be interpreted as necessarily representing the official policies, either expressed or implied, of the U.S. Department of Health and Human Services or of the institutions and companies affiliated with the authors, nor does mention of trade names, commercial products, or organizations imply endorsement by the U.S. Government. In no event shall any of these entities have any responsibility or liability for any use, misuse, inability to use, or reliance upon the information contained herein.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Declaration of Interests
S.L.S. and Q.Y. are founders of, equity holders of, and consultants for Darwin Biosciences. S.L.S is on the scientific advisory board for Darwin Biosciences. S.L.S. serves as a consultant for the MITRE Corporation, and is a member of the Planning Committee for Countering Zoonotic Spillover of High Consequence Pathogens, sponsored by the U.S. National Academies of Sciences, Engineering, and Medicine.
References
- Aaron JS, Taylor AB, and Chew T-L (2018). Image co-localization – co-occurrence versus correlation. J Cell Sci 131, jcs211847. 10.1242/jcs.211847. [DOI] [PubMed] [Google Scholar]
- Bailey AL, Lauck M, Sibley SD, Pecotte J, Rice K, Weny G, Tumukunde A, Hyeroba D, Greene J, Correll M, et al. (2014a). Two novel simian arteriviruses in captive and wild baboons (Papio spp.). J Virol 88, 13231–13239. 10.1128/jvi.02203-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey AL, Lauck M, Weiler A, Sibley SD, Dinis JM, Bergman Z, Nelson CW, Correll M, Gleicher M, Hyeroba D, et al. (2014b). High genetic diversity and adaptive potential of two simian hemorrhagic fever viruses in a wild primate population. PLoS One 9, e90714. 10.1371/journal.pone.0090714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey AL, Lauck M, Sibley SD, Friedrich TC, Kuhn JH, Freimer NB, Jasinska AJ, Phillips-Conroy JE, Jolly CJ, Marx PA, et al. (2015). Zoonotic potential of simian arteriviruses. J Virol 90, 630–635. 10.1128/jvi.01433-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey AL, Lauck M, Ghai RR, Nelson CW, Heimbruch K, Hughes AL, Goldberg TL, Kuhn JH, Jasinska AJ, Freimer NB, et al. (2016). Arteriviruses, pegiviruses, and lentiviruses are common among wild African monkeys. J Virol 90, 6724–6737. 10.1128/jvi.00573-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baize S, Pannetier D, Oestereich L, Rieger T, Koivogui L, Magassouba N, Soropogui B, Sow MS, Keïta S, Clerck HD, et al. (2014). Emergence of Zaire Ebola virus disease in Guinea. New Engl J Medicine 371, 1418–1425. 10.1056/nejmoa1404505. [DOI] [PubMed] [Google Scholar]
- Buechler C, Ritter M, Orsó E, Langmann T, Klucken J, and Schmitz G (2000). Regulation of scavenger receptor CD163 expression in human monocytes and macrophages by pro- and antiinflammatory stimuli. J Leukocyte Biol 67, 97–103. 10.1002/jlb.67.1.97. [DOI] [PubMed] [Google Scholar]
- Caì Y, Postnikova EN, Bernbaum JG, Yú S, Mazur S, Deiuliis NM, Radoshitzky SR, Lackemeyer MG, McCluskey A, Robinson PJ, et al. (2015). Simian hemorrhagic fever virus cell entry is dependent on CD163 and uses a clathrin-mediated endocytosis-like pathway. J Virol 89, 844–856. 10.1128/jvi.02697-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai Y, Yu S, Fang Y, Bollinger L, Li Y, Lauck M, Postnikova EN, Mazur S, Johnson RF, Finch CL, et al. (2021). Development and characterization of a cDNA-launch recombinant simian hemorrhagic fever virus expressing enhanced green fluorescent protein: ORF 2b’ Is not required for in vitro virus replication. Viruses 13, 632. 10.3390/v13040632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calvert JG, Slade DE, Shields SL, Jolie R, Mannan RM, Ankenbauer RG, and Welch S-KW (2007). CD163 expression confers susceptibility to porcine reproductive and respiratory syndrome viruses. J Virol 81, 7371–7379. 10.1128/jvi.00513-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carette JE, Raaben M, Wong AC, Herbert AS, Obernosterer G, Mulherkar N, Kuehne AI, Kranzusch PJ, Griffin AM, Ruthel G, et al. (2011). Ebola virus entry requires the cholesterol transporter Niemann–Pick C1. Nature 477, 340–343. 10.1038/nature10348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiou KL, Bergey CM, Burrell AS, Disotell TR, Rogers J, Jolly CJ, and Phillips-Conroy JE (2021). Genome-wide ancestry and introgression in a Zambian baboon hybrid zone. Mol Ecol 30, 1907–1920. 10.1111/mec.15858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coffin JM (1995). HIV population dynamics in vivo: implications for genetic variation, pathogenesis, and therapy. Science 267, 483–489. 10.1126/science.7824947. [DOI] [PubMed] [Google Scholar]
- Dalgard DW, Hardy RJ, Pearson SL, Pucak GJ, Quander RV, Zack PM, Peters CJ, and Jahrling PB (1992). Combined simian hemorrhagic fever and Ebola virus infection in cynomolgus monkeys. Lab Anim Sci 42, 152–157. [PubMed] [Google Scholar]
- Daniel MD, Letvin NL, King NW, Kannagi M, Sehgal PK, Hunt RD, Kanki PJ, Essex M, and Desrosiers RC (1985). Isolation of T-cell tropic HTLV-III-like retrovirus from macaques. Science 228, 1201–1204. 10.1126/science.3159089. [DOI] [PubMed] [Google Scholar]
- Demogines A, Farzan M, and Sawyer SL (2012). Evidence for ACE2-utilizing coronaviruses (CoVs) related to severe acute respiratory syndrome CoV in bats. J Virol 86, 6350–6353. 10.1128/jvi.00311-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demogines A, Abraham J, Choe H, Farzan M, and Sawyer SL (2013). Dual host-virus arms races shape an essential housekeeping protein. PLoS Biol 11, e1001571. 10.1371/journal.pbio.1001571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desmyter J, Melnick JL, and Rawls WE (1968). Defectiveness of interferon production and of rubella virus interference in a line of African green monkey kidney cells (Vero). J Virol 2, 955–961. 10.1128/jvi.2.10.955-961.1968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duan X, Nauwynck HJ, and Pensaert MB (1997a). Virus quantification and identification of cellular targets in the lungs and lymphoid tissues of pigs at different time intervals after inoculation with porcine reproductive and respiratory syndrome virus (PRRSV). Vet Microbiol 56, 9–19. 10.1016/s0378-1135(96)01347-8. [DOI] [PubMed] [Google Scholar]
- Duan X, Nauwynck HJ, and Pensaert MB (1997b). Effects of origin and state of differentiation and activation of monocytes/macrophages on their susceptibility to porcine reproductive and respiratory syndrome virus (PRRSV). Arch Virol 142, 2483–2497. 10.1007/s007050050256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunn KW, Kamocka MM, and McDonald JH (2011). A practical guide to evaluating colocalization in biological microscopy. Am J Physiol-Cell Ph 300, C723–C742. 10.1152/ajpcell.00462.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fabriek BO, Dijkstra CD, and van den Berg TK (2005). The macrophage scavenger receptor CD163. Immunobiology 210, 153–160. 10.1016/j.imbio.2005.05.010. [DOI] [PubMed] [Google Scholar]
- Femino AM, Fay FS, Fogarty K, and Singer RH (1998). Visualization of single RNA transcripts in situ. Science 280, 585–590. 10.1126/science.280.5363.585. [DOI] [PubMed] [Google Scholar]
- Gorp HV, Breedam WV, Delputte PL, and Nauwynck HJ (2009). The porcine reproductive and respiratory syndrome virus requires trafficking through CD163-positive early endosomes, but not late endosomes, for productive infection. Arch Virol 154, 1939–1943. 10.1007/s00705-009-0527-1. [DOI] [PubMed] [Google Scholar]
- Gorp HV, Breedam WV, Doorsselaere JV, Delputte PL, and Nauwynck HJ (2010). Identification of the CD163 protein domains involved in infection of the porcine reproductive and respiratory syndrome virus. J Virol 84, 3101–3105. 10.1128/jvi.02093-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graham BS, and Sullivan NJ (2018). Emerging viral diseases from a vaccinology perspective: preparing for the next pandemic. Nat Immunol 19, 20–28. 10.1038/s41590-017-0007-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gravell M, London WT, Leon ME, Palmer AE, and Hamilton RS (1986). Differences among isolates of simian hemorrhagic fever (SHF) virus. P Soc Exp Biol Med 181, 112–119. 10.3181/00379727-181-42231. [DOI] [PubMed] [Google Scholar]
- Greaves DR, and Gordon S (2005). Thematic review series: the immune system and atherogenesis. Recent insights into the biology of macrophage scavenger receptors. J Lipid Res 46, 11–20. 10.1194/jlr.r400011-jlr200. [DOI] [PubMed] [Google Scholar]
- Han J, Perez JT, Chen C, Li Y, Benitez A, Kandasamy M, Lee Y, Andrade J, tenOever B, and Manicassamy B (2018). Genome-wide CRISPR/Cas9 screen identifies host factors essential for influenza virus replication. Cell Reports 23, 596–607. 10.1016/j.celrep.2018.03.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heuverswyn FV, and Peeters M (2007). The origins of HIV and implications for the global epidemic. Curr Infect Dis Rep 9, 338–346. 10.1007/s11908-007-0052-x. [DOI] [PubMed] [Google Scholar]
- Hill CM (2018). Human–primate conflict. In The International Encyclopedia of Biological Anthropology. 10.1002/9781118584538.ieba0258. [DOI] [Google Scholar]
- Hockings KJ (2016). Mitigating human–nonhuman primate conflict. In The International Encyclopedia of Primatology. 10.1002/9781119179313.wbprim0053. [DOI] [Google Scholar]
- Hoenen T, Groseth A, Feldmann F, Marzi A, Ebihara H, Kobinger G, Günther S, and Feldmann H (2014). Complete genome sequences of three Ebola virus isolates from the 2014 outbreak in West Africa. Genome Announc 2, e01331–14. 10.1128/genomea.01331-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang C, Bernard D, Zhu J, Dash RC, Chu A, Knupp A, Hakey A, Hadden MK, Garmendia A, and Tang Y (2020). Small molecules block the interaction between porcine reproductive and respiratory syndrome virus and CD163 receptor and the infection of pig cells. Virol J 17, 116. 10.1186/s12985-020-01361-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imai M, and Kawaoka Y (2012). The role of receptor binding specificity in interspecies transmission of influenza viruses. Curr Opin Virol 2, 160–167. 10.1016/j.coviro.2012.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jae LT, and Brummelkamp TR (2015). Emerging intracellular receptors for hemorrhagic fever viruses. Trends Microbiol 23, 392–400. 10.1016/j.tim.2015.04.006. [DOI] [PubMed] [Google Scholar]
- Jae LT, Raaben M, Herbert AS, Kuehne AI, Wirchnianski AS, Soh TK, Stubbs SH, Janssen H, Damme M, Saftig P, et al. (2014). Lassa virus entry requires a trigger-induced receptor switch. Science 344, 1506–1510. 10.1126/science.1252480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson RF, Dodd LE, Yellayi S, Gu W, Cann JA, Jett C, Bernbaum JG, Ragland DR, St. Claire M, Byrum R, et al. (2011). Simian hemorrhagic fever virus infection of rhesus macaques as a model of viral hemorrhagic fever: clinical characterization and risk factors for severe disease. Virology 421, 129–140. 10.1016/j.virol.2011.09.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones DT, Taylor WR, and Thornton JM (1992). The rapid generation of mutation data matrices from protein sequences. Bioinformatics 8, 275–282. 10.1093/bioinformatics/8.3.275. [DOI] [PubMed] [Google Scholar]
- Kaelber JT, Demogines A, Harbison CE, Allison AB, Goodman LB, Ortega AN, Sawyer SL, and Parrish CR (2012). Evolutionary reconstructions of the transferrin receptor of caniforms supports canine parvovirus being a re-emerged and not a novel pathogen in dogs. PLoS Pathog 8, e1002666. 10.1371/journal.ppat.1002666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kailasan S, Agbandje-McKenna M, and Parrish CR (2015). Parvovirus family conundrum: what makes a killer? Ann Rev Virol 2, 1–26. 10.1146/annurev-virology-100114-055150. [DOI] [PubMed] [Google Scholar]
- Keele BF, Heuverswyn FV, Li Y, Bailes E, Takehisa J, Santiago ML, Bibollet-Ruche F, Chen Y, Wain LV, Liegeois F, et al. (2006). Chimpanzee reservoirs of pandemic and nonpandemic HIV-1. Science 313, 523–526. 10.1126/science.1126531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korber B, Muldoon M, Theiler J, Gao F, Gupta R, Lapedes A, Hahn BH, Wolinsky S, and Bhattacharya T (2000). Timing the ancestor of the HIV-1 pandemic strains. Science 288, 1789–1796. 10.1126/science.288.5472.1789. [DOI] [PubMed] [Google Scholar]
- Kristiansen M, Graversen JH, Jacobsen C, Sonne O, Hoffman H-J, Law SKA, and Moestrup SK (2001). Identification of the haemoglobin scavenger receptor. Nature 409, 198–201. 10.1038/35051594. [DOI] [PubMed] [Google Scholar]
- Kuhn JH, Lauck M, Bailey AL, Shchetinin AM, Vishnevskaya TV, Bào Y, Ng TFF, LeBreton M, Schneider BS, Gillis A, et al. (2016). Reorganization and expansion of the nidoviral family Arteriviridae. Arch Virol 161, 755–768. 10.1007/s00705-015-2672-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar S, Stecher G, and Tamura K (2016). MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol 33, 1870–1874. 10.1093/molbev/msw054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lauck M, Hyeroba D, Tumukunde A, Weny G, Lank SM, Chapman CA, O’Connor DH, Friedrich TC, and Goldberg TL (2011). Novel, divergent simian hemorrhagic fever viruses in a wild Ugandan red colobus monkey discovered using direct pyrosequencing. PLoS One 6, e19056. 10.1371/journal.pone.0019056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lauck M, Sibley SD, Hyeroba D, Tumukunde A, Weny G, Chapman CA, Ting N, Switzer WM, Kuhn JH, Friedrich TC, et al. (2013). Exceptional simian hemorrhagic fever virus diversity in a wild African primate community. J Virol 87, 688–691. 10.1128/jvi.02433-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lauck M, Palacios G, Wiley MR, Lǐ Y, Fāng Y, Lackemeyer MG, Caì Y, Bailey AL, Postnikova E, Radoshitzky SR, et al. (2014). Genome sequences of simian hemorrhagic fever virus variant NIH LVR42–0/M6941 isolates (Arteriviridae: Arterivirus). Genome Announc 2, e00978–14. 10.1128/genomea.00978-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lauck M, Alkhovsky SV, Bào Y, Bailey AL, Shevtsova ZV, Shchetinin AM, Vishnevskaya TV, Lackemeyer MG, Postnikova E, Mazur S, et al. (2015). Historical outbreaks of simian hemorrhagic fever in captive macaques were caused by distinct arteriviruses. J Virol 89, 8082–8087. 10.1128/jvi.01046-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Law SKA, Micklem KJ, Shaw JM, Zhang X, Dong Y, Willis AC, and Mason DY (1993). A new macrophage differentiation antigen which is a member of the scavenger receptor superfamily. Eur J Immunol 23, 2320–2325. 10.1002/eji.1830230940. [DOI] [PubMed] [Google Scholar]
- Lee CZW, Kozaki T, and Ginhoux F (2018). Studying tissue macrophages in vitro: are iPSCderived cells the answer? Nat Rev Immunol 18, 716–725. 10.1038/s41577-018-0054-y. [DOI] [PubMed] [Google Scholar]
- Li B, Clohisey SM, Chia BS, Wang B, Cui A, Eisenhaure T, Schweitzer LD, Hoover P, Parkinson NJ, Nachshon A, et al. (2020). Genome-wide CRISPR screen identifies host dependency factors for influenza A virus infection. Nat Commun 11, 164. 10.1038/s41467-019-13965-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Logue J, Licona WV, Cooper TK, Reeder B, Byrum R, Qin J, Murphy ND, Cong Y, Bonilla A, Sword J, et al. (2019). Ebola virus isolation using Huh-7 cells has methodological advantages and similar sensitivity to isolation using other cell types and suckling BALB/c laboratory mice. Viruses 11, 161. 10.3390/v11020161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- London WT (1977). Epizootiology, transmission and approach to prevention of fatal simian haemorrhagic fever in rhesus monkeys. Nature 268, 344–345. 10.1038/268344a0. [DOI] [PubMed] [Google Scholar]
- Ma Y, Walsh MJ, Bernhardt K, Ashbaugh CW, Trudeau SJ, Ashbaugh IY, Jiang S, Jiang C, Zhao B, Root DE, et al. (2017). CRISPR/Cas9 screens reveal Epstein-Barr virus-transformed B cell host dependency factors. Cell Host Microbe 21, 580–591.e7. 10.1016/j.chom.2017.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maniecki MB, Møller HJ, Moestrup SK, and Møller BK (2006). CD163 positive subsets of blood dendritic cells: the scavenging macrophage receptors CD163 and CD91 are coexpressed on human dendritic cells and monocytes. Immunobiology 211, 407–417. 10.1016/j.imbio.2006.05.019. [DOI] [PubMed] [Google Scholar]
- Mansfield KG, Lerche NW, Gardner MB, and Lackner AA (1995). Origins of simian immunodeficiency virus infection in macaques at the New England Regional Primate Research Center. J Med Primatol 24, 116–122. 10.1111/j.1600-0684.1995.tb00156.x. [DOI] [PubMed] [Google Scholar]
- Marceau CD, Puschnik AS, Majzoub K, Ooi YS, Brewer SM, Fuchs G, Swaminathan K, Mata MA, Elias JE, Sarnow P, et al. (2016). Genetic dissection of Flaviviridae host factors through genome-scale CRISPR screens. Nature 535, 159–163. 10.1038/nature18631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyerson NR, and Sawyer SL (2011). Two-stepping through time: mammals and viruses. Trends Microbiol 19, 286–294. 10.1016/j.tim.2011.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyerson NR, Sharma A, Wilkerson GK, Overbaugh J, and Sawyer SL (2015). Identification of owl monkey CD4 receptors broadly compatible with early-stage HIV-1 Isolates. J Virol 89, 8611–8622. 10.1128/jvi.00890-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller EH, Obernosterer G, Raaben M, Herbert AS, Deffieu MS, Krishnan A, Ndungo E, Sandesara RG, Carette JE, Kuehne AI, et al. (2012). Ebola virus entry requires the host-programmed recognition of an intracellular receptor. Embo J 31, 1947–1960. 10.1038/emboj.2012.53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng M, Ndungo E, Kaczmarek ME, Herbert AS, Binger T, Kuehne AI, Jangra RK, Hawkins JA, Gifford RJ, Biswas R, et al. (2015). Filovirus receptor NPC1 contributes to species-specific patterns of ebolavirus susceptibility in bats. ELife 4, e11785. 10.7554/elife.11785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nga PT, del C. Parquet M, Lauber C, Parida M, Nabeshima T, Yu F, Thuy NT, Inoue S, Ito T, Okamoto K, et al. (2011). Discovery of the first insect nidovirus, a missing evolutionary link in the emergence of the largest RNA virus genomes. PLoS Pathog 7, e1002215. 10.1371/journal.ppat.1002215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielsen MJ, Madsen M, Møller HJ, and Moestrup SK (2006). The macrophage scavenger receptor CD163: endocytic properties of cytoplasmic tail variants. J Leukocyte Biol 79, 837–845. 10.1189/jlb.1005602. [DOI] [PubMed] [Google Scholar]
- Onyekaba CO, Harty JT, and Plagemann PGW (1989). Extensive cytocidal replication of lactate dehydrogenase-elevating virus in cultured peritoneal macrophages from 1–2-week-old mice. Virus Res 14, 327–338. 10.1016/0168-1702(89)90025-7. [DOI] [PubMed] [Google Scholar]
- Paige SB, Frost SDW, Gibson MA, Jones JH, Shankar A, Switzer WM, Ting N, and Goldberg TL (2014). Beyond bushmeat: animal contact, injury, and zoonotic disease risk in Western Uganda. Ecohealth 11, 534–543. 10.1007/s10393-014-0942-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paige SB, Bleecker J, Mayer J, and Goldberg T (2017). Spatial overlap between people and non-human primates in a fragmented landscape. Ecohealth 14, 88–99. 10.1007/s10393-016-1194-9. [DOI] [PubMed] [Google Scholar]
- Palmer AE, Allen AM, Tauraso NM, and Shelokov A (1968). Simian hemorrhagic fever. I. Clinical and epizootiologic aspects of an outbreak among quarantined monkeys. Am J Tropical Medicine Hyg 17, 404–412. [PubMed] [Google Scholar]
- Park RJ, Wang T, Koundakjian D, Hultquist JF, Lamothe-Molina P, Monel B, Schumann K, Yu H, Krupzcak KM, Garcia-Beltran W, et al. (2016). A genome-wide CRISPR screen identifies a restricted set of HIV host dependency factors. Nat Genet 49, 193–203. 10.1038/ng.3741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parrish CR, Holmes EC, Morens DM, Park E-C, Burke DS, Calisher CH, Laughlin CA, Saif LJ, and Daszak P (2008). Cross-species virus transmission and the emergence of new epidemic diseases. Microbiol Mol Biol R 72, 457–470. 10.1128/mmbr.00004-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perelman P, Johnson WE, Roos C, Seuánez HN, Horvath JE, Moreira MAM, Kessing B, Pontius J, Roelke M, Rumpler Y, et al. (2011). A molecular phylogeny of living primates. PLoS Genet 7, e1001342. 10.1371/journal.pgen.1001342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perrière G, and Gouy M (1996). WWW-query: An on-line retrieval system for biological sequence banks. Biochimie 78, 364–369. 10.1016/0300-9084(96)84768-7. [DOI] [PubMed] [Google Scholar]
- Peters DK, and Garcea RL (2020). Murine polyomavirus DNA transitions through spatially distinct nuclear replication subdomains during infection. PLoS Pathog 16, e1008403. 10.1371/journal.ppat.1008403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raaben M, Jae LT, Herbert AS, Kuehne AI, Stubbs SH, Chou Y, Blomen VA, Kirchhausen T, Dye JM, Brummelkamp TR, et al. (2017). NRP2 and CD63 are host factors for Lujo virus cell entry. Cell Host Microbe 22, 688–696.e5. 10.1016/j.chom.2017.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sauter D, and Kirchhoff F (2019). Key viral adaptations preceding the AIDS pandemic. Cell Host Microbe 25, 27–38. 10.1016/j.chom.2018.12.002. [DOI] [PubMed] [Google Scholar]
- Savidis G, McDougall WM, Meraner P, Perreira JM, Portmann JM, Trincucci G, John SP, Aker AM, Renzette N, Robbins DR, et al. (2016). Identification of Zika virus and dengue virus dependency factors using functional genomics. Cell Reports 16, 232–246. 10.1016/j.celrep.2016.06.028. [DOI] [PubMed] [Google Scholar]
- Schneider CA, Rasband WS, and Eliceiri KW (2012). NIH Image to ImageJ: 25 years of image analysis. Nat Methods 9, 671–675. 10.1038/nmeth.2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shang D, Liu Y, Ito N, Kamoto T, and Ogawa O (2007). Defective Jak–Stat activation in renal cell carcinoma is associated with interferon-α resistance. Cancer Sci 98, 1259–1264. 10.1111/j.1349-7006.2007.00526.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharp PM, and Hahn BH (2011). Origins of HIV and the AIDS pandemic. CSH Perspect Med 1, a006841. 10.1101/cshperspect.a006841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharp PM, Bailes E, Chaudhuri RR, Rodenburg CM, Santiago MO, and Hahn BH (2001). The origins of acquired immune deficiency syndrome viruses: where and when? Philosophical Transactions Royal Soc Lond Ser B Biological Sci 356, 867–876. 10.1098/rstb.2001.0863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shelokov A, Tauraso NM, Allen AM, and Palmer AE (1968). Simian hemorrhagic fever: II. Studies in pathology. Am J Tropical Medicine Hyg 17, 413–421. 10.4269/ajtmh.1968.17.413. [DOI] [PubMed] [Google Scholar]
- Stecher G, Tamura K, and Kumar S (2020). Molecular evolutionary genetics analysis (MEGA) for macOS. Mol Biol Evol 37, 1237–1239. 10.1093/molbev/msz312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stueckemann JA, Holth M, Swart WJ, Kowalchyk K, Smith MS, Wolstenholme AJ, Cafruny WA, and Plagemann PGW (1982). Replication of lactate dehydrogenase-elevating virus in macrophages. J Gen Virol 59, 263–272. 10.1099/0022-1317-59-2-263. [DOI] [PubMed] [Google Scholar]
- Su C-M, Rowland RRR, and Yoo D (2021). Recent advances in PRRS virus receptors and the targeting of receptor–ligand for control. Nato Adv Sci Inst Se 9, 354. 10.3390/vaccines9040354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suyama M, Torrents D, and Bork P (2006). PAL2NAL: robust conversion of protein sequence alignments into the corresponding codon alignments. Nucleic Acids Res 34, W609–W612. 10.1093/nar/gkl315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tauraso NM, Shelokov A, Palmer AE, and Allen AM (1968). Simian hemorrhagic fever. III. Isolation and characterization of a viral agent. Am J Tropical Medicine Hyg 17, 422–431. [PubMed] [Google Scholar]
- Teifke JP, Dauber M, Fichtner D, Lenk M, Polster U, Weiland E, and Beyer J (2001). Detection of European porcine reproductive and respiratory syndrome virus in porcine alveolar macrophages by two-colour immunofluorescence and in-situ hybridization-immunohistochemistry double labelling. J Comp Pathol 124, 238–245. 10.1053/jcpa.2000.0458. [DOI] [PubMed] [Google Scholar]
- Vatter HA, and Brinton MA (2014). Differential responses of disease-resistant and disease-susceptible primate macrophages and myeloid dendritic cells to simian hemorrhagic fever virus infection. J Virol 88, 2095–2106. 10.1128/jvi.02633-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vatter HA, Donaldson EF, Huynh J, Rawlings S, Manoharan M, Legasse A, Planer S, Dickerson MF, Lewis AD, Colgin LMA, et al. (2015). A simian hemorrhagic fever virus isolate from persistently infected baboons efficiently induces hemorrhagic fever disease in Japanese macaques. Virology 474, 186–198. 10.1016/j.virol.2014.10.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warren CJ, and Sawyer SL (2019). How host genetics dictates successful viral zoonosis. PLoS Biol 17, e3000217. 10.1371/journal.pbio.3000217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warren CJ, Meyerson NR, Dirasantha O, Feldman ER, Wilkerson GK, and Sawyer SL (2019a). Selective use of primate CD4 receptors by HIV-1. PLoS Biol 17, e3000304. 10.1371/journal.pbio.3000304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warren CJ, Meyerson NR, Stabell AC, Fattor WT, Wilkerson GK, and Sawyer SL (2019b). A glycan shield on chimpanzee CD4 protects against infection by primate lentiviruses (HIV/SIV). Proc National Acad Sci 116, 11460–11469. 10.1073/pnas.1813909116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei X, Ghosh SK, Taylor ME, Johnson VA, Emini EA, Deutsch P, Lifson JD, Bonhoeffer S, Nowak MA, Hahn BH, et al. (1995). Viral dynamics in human immunodeficiency virus type 1 infection. Nature 373, 117–122. 10.1038/373117a0. [DOI] [PubMed] [Google Scholar]
- Worobey M, Gemmel M, Teuwen DE, Haselkorn T, Kunstman K, Bunce M, Muyembe J-J, Kabongo J-MM, Kalengayi RM, Marck EV, et al. (2008). Direct evidence of extensive diversity of HIV-1 in Kinshasa by 1960. Nature 455, 661–664. 10.1038/nature07390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu H, Liu Z, Zheng S, Han G, and He F (2020). CD163 antibodies inhibit PRRSV infection via receptor blocking and transcription suppression. Nato Adv Sci Inst Se 8, 592. 10.3390/vaccines8040592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamashita M, and Emerman M (2004). Capsid Is a dominant determinant of retrovirus infectivity in nondividing cells. J Virol 78, 5670–5678. 10.1128/jvi.78.11.5670-5678.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z (2007). PAML 4: phylogenetic analysis by maximum likelihood. Mol Biol Evol 24, 1586–1591. 10.1093/molbev/msm088. [DOI] [PubMed] [Google Scholar]
- Yu P, Wei R, Dong W, Zhu Z, Zhang X, Chen Y, Liu X, and Guo C (2020). CD163ΔSRCR5 MARC-145 cells resist PRRSV-2 infection via inhibiting virus uncoating, which requires the interaction of CD163 with calpain 1. Front Microbiol 10, 3115. 10.3389/fmicb.2019.03115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yú S, Caì Y, Lyons C, Johnson RF, Postnikova E, Mazur S, Johnson JC, Radoshitzky SR, Bailey AL, Lauck M, et al. (2016). Specific detection of two divergent simian arteriviruses using RNAscope in situ hybridization. PLoS One 11, e0151313. 10.1371/journal.pone.0151313. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1. Primers used for cloning CD163 receptor orthologs, related to Figure 4.
Data Availability Statement
Primate CD163 sequences have been deposited at GenBank and are publicly available as of the date of publication. Accession numbers are listed in the key resources table. Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Goat polyclonal anti-CD163 | R&D Systems | Cat#AF1607 |
| Mouse monoclonal anti-actin beta (clone 8H10D10) | Cell Signaling Technology | Cat#3700S |
| Donkey anti-goat IgG (H+L) HRP conjugate | Thermo Fisher Scientific | Cat#A16005 |
| Anti-mouse IgG (H+L) HRP conjugate | Promega | Cat#W402B |
| Mouse purified monoclonal anti-CD163 (clone GHI/61) | BioLegend | Cat#333609 |
| Rabbit polyclonal anti-occludin | Thermo Fisher Scientific | Cat#71-150-0 |
| Goat anti-mouse IgG AF488+ | Invitrogen | Cat#A32723 |
| Goat anti-rabbit IgG AF546 | Invitrogen | Cat#A11010 |
| Bacterial and virus strains | ||
| Simian hemorrhagic fever virus (SHFV) isolate LVR42–0/M6941 | American Type Culture Collection (ATCC) | Cat#VR-533 |
| Ebola virus/H.sapiens-wt/GIN/2014/Makona-C05 | Gary Kobinger (Public Health Agency Canada) | N/A |
| Biological samples | ||
| Rhesus macaque peripheral blood mononuclear cells (PBMCs) | G. K. Wilkerson, The University of Texas MD Anderson Cancer Center | N/A |
| Human PBMCs | Lonza | Cat#4W-270A |
| Nancy Ma’s night monkey (Aotus nancymaae) PBMCs | G. K. Wilkerson | N/A |
| Chemicals, peptides, and recombinant proteins | ||
| Human recombinant macrophage colony-stimulating factor (M-CSF) | R&D Systems | Cat#216-MC-010 |
| Interferon-Y (IFN-γ) | R&D Systems | Cat#285-IF |
| Interleukin 4 (IL-4) | MilliporeSigma | Cat#H7291 |
| Polybrene hexadimethrine bromide | MilliporeSigma | 107689–10G |
| 5-propargylamino-2’,3’-dideoxyuridine-5’-triphosphate (ddUTP) ATTO 633 | Jena Bioscience | Cat#NU-1619–633 |
| Cas9 nuclease, S. pyogenes | New England Biolabs (NEB) | Cat#M0386M |
| Eagle’s minimum essential medium (EMEM) | ATCC | 30–2003 |
| 10% fetal bovine serum (FBS) | MilliporeSigma | TMS-013-B |
| 1X penicillin-streptomycin solution (Pen Strep) | Avantor | #45000–652 |
| RPMI-1640 medium | ATCC | #30–2001 |
| Dulbecco’s modified Eagle’s medium (DMEM) | MilliporeSigma | #D6429 |
| L-glutamine | Corning Incorporated | #MT25005CI |
| Hygromycin | Corning Incorporated | #30–240-CR |
| Phosphate-buffered saline (PBS) | Caisson Labs | PBL01–6X500ML |
| ImmunoCult-SF macrophage medium | STEMCELL Technologies | #10961 |
| 1X phenol-free EMEM | VWR | #115-073-101 |
| 1.4% Avicel | FMC Corporation | RC-591 NF |
| 10% neutral buffered formalin | MilliporeSigma | #HT501128–4L |
| 0.2% crystal violet | MilliporeSigma | #C3866–25G |
| Lipofectamine 3000 reagent | Thermo Fisher Scientific | #L3000008 |
| Opti-MEM | Thermo Fisher Scientific | #31985–070 |
| Tragacanth | MilliporeSigma | #G1128 |
| Nonidet P-40 buffer (NP-40) | Promega | #V4221 |
| Tris(hydroxymethyl)aminomethane hydrochloride (HCl) pH 7.4 | Teknova | #T1075 |
| 1% NP-40 | G-Biosciences | #DG001 |
| Dithiothreitol (DTT) | IBI Scientific | #IB21040 |
| Benzonase | MilliporeSigma | #E1014 |
| Protease inhibitor mixture | MilliporeSigma | #11873580001 |
| Ethylenediamine tetraacetic acid (EDTA) | Corning Incorporated | #25–052-CI |
| 10% Tris-glycine eXtended (TGX) stain-free acrylamide premixed gel solution | Bio-Rad | #1610182 |
| 0.1% tris-buffered saline (TBS) | Research Products International (RPI) | #T60075–4000.0 |
| Polysorbate 20 | Amresco | #M147–1L |
| Nestle Carnation 5% nonfat dried milk | Vevey | N/A |
| Enhanced chemiluminescent (ECL) reagent | MilliporeSigma | #GERPN2232 |
| Neon Transfection System Kit | Thermo Fisher Scientific | #MPK10025 |
| T7 endonuclease 1 (E1) assay in the Alt-R Genome Editing Detection Kit | Integrated DNA Technologies (IDT) | #1075931 |
| Phusion high-fidelity polymerase chain reaction (PCR) master mix with buffer | NEB | #M0531L |
| Clontech pLHCX retroviral expression vector | Takara Bio Inc. | #631511 |
| TransIT 293 transfection reagent | Mirus Bio | #MIR2705 |
| 4% PFA | VWR | #100504–858 |
| FACS buffer | MilliporeSigma | #324506–100ML |
| Terminal deoxynucleotidyl transferase (TdT) buffer | Thermo Fisher Scientific | #EP0161 |
| Sodium acetate (NaOAc) | Amresco | #E498–200ML |
| 100% cold ethanol (EtOH) | Thermo Fisher Scientific | # 04-355-223 |
| Nuclease-free water | IBI Scientific | # IB42201 |
| Poly-L-lysine | MilliporeSigma | #P8920 |
| Paraformaldehyde (PFA) | VWR | #100504–85 |
| 0.1% Triton X-100 | VWR | #JTX198–5 |
| Wash Buffer A | Biosearch Technologies | #SMF-WA1–60 |
| Hybridization buffer | Biosearch Technologies | #SMF-HB1–10 |
| Hoechst 33342 dye for DNA and nuclei | Invitrogen | #H3570 |
| Wash Buffer B | Biosearch Technologies | #SMF-WB1–20 |
| Prolong Gold Antifade Mountant with 4’,6-diamidino-2-phenylindole (DAPI) | Thermo Fisher Scientific | #P36941 |
| 10% neutral buffered formalin | VWR | #16004–128 |
| Critical commercial assays | ||
| Bicinchoninic acid (BCA) protein assay | Thermo Fisher Scientific | Cat#23227 |
| Alt-R Genome Editing Detection Kit | IDT | Cat#1075931 |
| TOPO TA cloning kit | Thermo Fisher Scientific | Cat#K457501 |
| Cytofix/cytoperm Fixation/Permeabilization solution kit | BD | Cat#554714 |
| Data and code availability | ||
| CD163 receptor orthologs cloned from nonhuman primate biomaterial | This study | GenBank IDs: MZ695198–MZ695214 |
| Pearson correlation coefficient analysis | Peters and Garcea, 2020 | https://github.com/DouglasPeters/GarceaMatLabAnalyses/blob/52257dea52d610218b7b9b8b7b43818de67f72de/SIM_ColocalizationandCovarianceAnalysis_08152019_CURRENT.m |
| Experimental models: Cell lines | ||
| MA-104 Clone 1 | ATCC | Cat#CRL-2378.1 |
| MA-104 subclone MARC-145 | Kay Faaberg, U.S. Department of Agriculture | N/A |
| COS-7 | ATCC | Cat#CRL-1651 |
| Patas monkey skin fibroblasts | Coriell Institute for Medical Research (CIMR) | Cat#AG06254 and AG06116 |
| Human histiocytic SU-DHL-1 | ATCC | Cat#CRL-2955 |
| Monocytic THP-1 | ATCC | TIB-202 |
| Monocytic U937 | ATCC | Cat#CRL-1593.2 |
| Kidney cell line ACHN | NCI-Frederick Cancer DCTD Tumor/Cell Line Repository | N/A |
| Kidney cell line CAKI-1 | NCI-Frederick Cancer DCTD Tumor/Cell Line Repository | N/A |
| Kidney cell line786–0 | NCI-Frederick Cancer DCTD Tumor/Cell Line Repository | N/A |
| Kidney cell line A498 | NCI-Frederick Cancer DCTD Tumor/Cell Line Repository | N/A |
| HEK293T | ATCC | Cat#11268 |
| Grivet kidney Vero E6 | ATCC | Cat#CRL-1586 |
| MA-104, Vero, patas monkey, human bone marrow | Takara Bio Inc. | #636643 |
| Common chimpanzee (Pan troglodytes) fibroblasts | Emory University | S008886 |
| Red-cheeked gibbon (Nomascus gabriellae) fibroblasts | CIMR | #PR00381 |
| Northern white-cheeked gibbon (Nomascus leucogenys) fibroblasts | CIMR | #PR00712 |
| Lar gibbon (Hylobates lar) fibroblasts | CIMR | #PR01131 |
| Collared mangabey (Cercocebus torquatus) fibroblasts | CIMR | #PR00485 |
| Black crested mangabey (Lophocebus aterrimus) fibroblasts | CIMR | #PR01215 |
| Angolan talapoin (Miopithecus talapoin) fibroblasts | CIMR | #PR00716 |
| Wolf’s mona monkey (Cercopithecus wolfi) fibroblasts | CIMR | #PR01241 |
| Francois’ langur (Trachypithecus francoisi) fibroblasts | CIMR | #PR01099 |
| Mantled guereza (Colobus guereza) fibroblasts | CIMR | #PR00980 |
| Bolivian red howler (Alouatta sara) fibroblasts | CIMR | #PR00708 |
| Oligonucleotides | ||
| Primers for cloning nonhuman primate CD163 receptor orthologs | See Table S1 for primer sequences | N/A |
| CRISPR-Cas9 RNA (crRNA) targeting CD163 exon 3; AATGCGTCCAGAACCTGCAC | This study | N/A |
| Alt-R trans-activating CRISPR-Cas9 RNA (tracrRNA) | IDT | Cat#1072534 |
| Recombinant DNA | ||
| pLHCX retroviral transfer vector encoding CD163 orthologs | This study | N/A |
| Empty pLHCX retroviral transfer vector | Yamashita and Emerman, 2004 | N/A |
| pCS2-mGP encoding MLV gag-pol | Yamashita and Emerman, 2004 | N/A |
| pCMV-VSV-G (encoding vesicular stomatitis Indiana virus glycoprotein “G” | Addgene | Cat#8454 |
| Recombinant SHFV (rSHFV) | Cai et al., 2021 | N/A |
| Enhanced green fluorescent protein-expressing rSHFV | Cai et al., 2021 | N/A |
| Software and algorithms | ||
| Sequencher DNA sequence analysis software | Gene Codes Corporation | N/A |
| Stellaris probe designer | Biosearch Technologies | https://www.biosearchtech.com/support/tools/design-software/stellaris-probe-designer |
| ImageJ | Schneider et al., 2012 | https://imagej.nih.gov/ij/ |
| FlowJo v10.8.0 software | BD Life Sciences | N/A |
| MEGA7 | Kumar et al., 2016 | https://www.megasoftware.net/ |
| MEGAX | Stecher et al., 2020 | https://www.megasoftware.net/ |
| PAL2NAL web server tool | Suyama et al., 2006 | http://www.bork.embl.de/pal2nal/ |
| Nipot v2.3 | Perriere and Gouy, 1996 | http://doua.prabi.fr/software/njplot |
| PAML4.8 | Yang, 2007 | N/A |
| Prism v9.1.0 | GraphPad | N/A |
| QGIS v3.16.4 | QGIS Geographic Information System | www.qgis.org |
| BioRender | Created with Biorender.com | N/A |


