Abstract
Although first described in the context of disease, cross-β (amyloid) fibrils have also been found as functional entities in all kingdoms of life. However, what are the specific properties of the cross-β fibril motif that convey biological function, make them especially suited for their particular purpose, and distinguish them from other fibrils found in biology? This review approaches these questions by arguing that cross-β fibrils are highly periodic, stable, and self-templating structures whose formation is accompanied by substantial conformational change that leads to a multimerization of their core and framing sequences. A discussion of each of these properties is followed by selected examples of functional cross-β fibrils that show how function is usually achieved by leveraging many of these properties.
Keywords: functional amyloid, cross-β motif, protein fibrils, structure-function relationship, protein aggregation
Overview
How can cross-β fibrils, best known for being hallmarks of disease, carry positive function? In this review, I am going to address this question by discussing the specific properties of cross-β fibrils and showing how these are utilized in selected examples of functional amyloids.
Because the definition of the term ‘amyloid’ has undergone several changes, is used differently depending on the field (Benson et al. 2018), and is somehow a misnomer meaning starch-like (from the Latin amylum or Greek amylon) (Sipe and Cohen 2000), I will be using “cross-β motif” and “cross-β fibril” in this review.
Cross-β (amyloid) fibrils are unbranched protein fibrils with a diameter of 5-10 nm. They were originally identified via several experimental parameters including fibrillar appearance in electron micrographs (Cohen and Calkins 1959; Gras et al. 2011), their ability to induce birefringence when stained with Congo Red or fluorescence when stained with Thioflavin T (Tht) (Vassar and Culling 1959; Puchtler et al. 1962), and their specific X-ray fiber diffraction pattern (Sunde et al. 1997).
Cross-β fibril deposits were first described in the context of diseases. In each of these diseases, specific proteins misfold from their typically soluble state into cross-β fibrils that are hallmarks of the disease. A disproportionate amount of these diseases are neurodegenerative in nature, as for example Alzheimer’s disease (aggregation of Aβ, and tau) (Soria Lopez et al. 2019), Parkinson’s disease (aggregation of α-synuclein) (Baba et al. 1998), Huntington’s disease (aggregation of huntingtin) (DiFiglia et al. 1997), and several others (Eisenberg and Jucker 2012; Knowles et al. 2014). Other examples of diseases with cross-β formation are type II diabetes (human IAPP) (Höppener et al. 2000) and systemic amyloidosis that can be caused by several proteins and can affect multiple organs (Wechalekar et al. 2016).
Later, cross-β fibrils that were unrelated to diseases but carried specific functions (functional amyloids), were discovered (Geddes et al. 1968; Huff et al. 2003). The functional advantage and practical use of cross-β fibrils was consequently recognized and explored in biotechnological applications of the cross-β motif (Wei et al. 2017; Li et al. 2018). Due to the backbone driven nature of cross-β fibrils, most proteins are potential amyloid formers and can be artificially forced into this structure (Chiti et al. 1999; Goldschmidt et al. 2010).
Because cross-β fibrils are intrinsically insoluble, obtaining high-resolution structures had been a challenge for a long time. However, crystal structures of fibrils formed by small peptides (Nelson et al. 2005; Eisenberg and Sawaya 2017), solid-state NMR (Petkova and Tycko 2002; Jaroniec et al. 2004; Wasmer et al. 2008; Loquet et al. 2018), and especially recent advances in cryo electron microscopy (cryo EM) (Ragonis-Bachar and Landau 2021) have provided important structural insights. We now have a comprehensive understanding of the complexity and variety of cross-β fibril structures, both from disease and functional contexts (Sawaya et al. 2021).
These structures confirmed the basic cross-β motif for these fibrils i.e. a β-sheet rich structure in which β-strands and the β-sheet normal are perpendicular to the fibril axis. The majority of these structures also fall in the category of in-register parallel β-sheet structures in which the same residues of individual monomers within the fibril form backbone-backbone hydrogen bonds (see Fig. 1). However, the arrangement of the individual sheets, kinks, and loops results in a surprising diversity of folds. This structural diversity not only occurs among fibrils formed by different proteins but also between fibrils formed by the same protein. A prominent example of this are fibrils formed by α-synuclein (Guerrero-Ferreira et al. 2019; Watson and Lee 2019), which can form a variety of different cross-β structures known as fibril strains or polymorphs (Gallardo et al. 2020; Sawaya et al. 2021). Comparisons between functional and disease associated cross-β fibrils indicate that functional cross-β fibrils (i) have generally one polymorph in contrast to cross-β fibrils in disease, (ii) are often less stable or have conditions under which they disassemble, and (iii) have different amino acid composition (Ragonis-Bachar and Landau 2021; Sawaya et al. 2021).
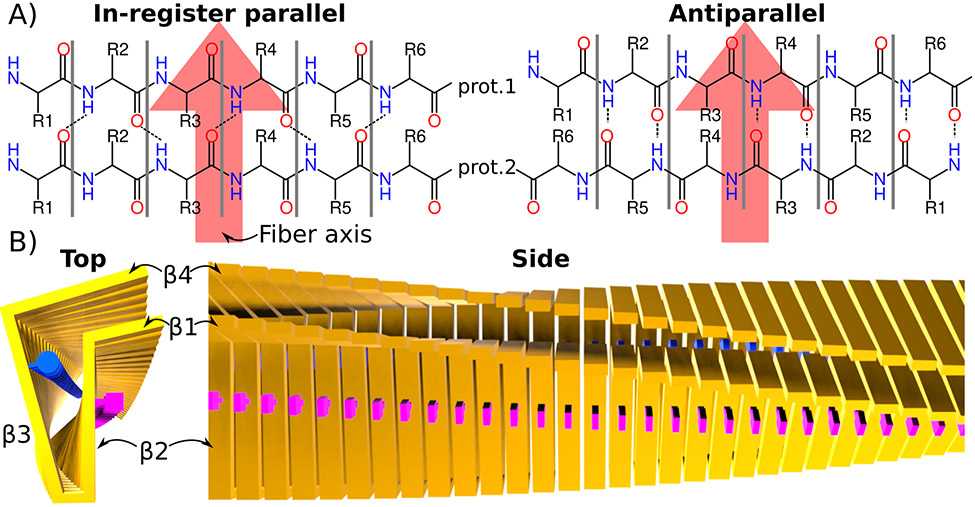
Figure 1: Core structure of cross-β fibrils.
A) Schematics of in-register cross-β (found in most cross-β fibrils) and antiparallel cross-β structures. Two layers formed by two monomers are shown. The fibril axis is indicated with a large red arrow. In the parallel-in register motif, the same residues from each monomer are right on top of each other, which is not the case in the antiparallel (or out of register parallel) case. B) Idealized in-register, parallel cross-β fibril. The protein backbone of each monomer is represented by yellow bars. Two examples of side chains are illustrated as blue circles and pink squares. 4 β-strands connected via kinks and turns can be identified in the top view. The side view shows the large continuous β-sheets formed by strands β2 and β4 from each monomer.
Despite the tremendous progress in understanding cross-β fibril structures over the last few years, the exact role that amyloid fibrils play in disease remains elusive. There are ongoing debates and active research about a) whether or not fibrils are a source of toxicity or rather a symptom of the diseases, b) which fibril species or intermediates are the most toxic, and c) the mechanism of toxicity (Marshall et al. 2014; Chun Ke et al. 2017; Iadanza et al. 2018).
Because many functional cross-β fibrils have well-defined functions that can be tested experimentally, we are in a better position to understand the role of cross-β formation in functional contexts. In the following, I am going to review our current understanding of functional cross-β fibrils with a focus on the functional advantages of the cross-β motif. I will first review the properties of the cross-β motif, then use selected examples to discuss how these features are used and combined to achieve the specific function, and end with concluding remarks. For related questions of the regulation of functional cross-β formation and the role of cross-β motifs in liquid-liquid phase separation, I refer to the following reviews (McGlinchey and Lee 2017; Chuang et al. 2018; Alberti and Hyman 2021).
Properties of cross-β fibrils and consequences of their formation
The functional advantage of cross-β fibrils is either associated with the intrinsic properties of the cross-β motif itself or a consequence of the structural change induced when the soluble non-fibrillar state of a protein transitions into the cross-β form. In the following, I list the major properties of cross-β fibrils and consequences of their formation, indicate the potential functional implication of each of these, and give one example of a functional cross-β fibril that uses this property.
Periodic
The cross-β structure is highly periodic. The cross-β motif itself results in a periodicity along the fibril axis. In its simplest form, i.e. when each strand in an in-register parallel β-sheet structure comes from a different monomer, the periodicity is 4.8 Å. For antiparallel β-sheets or when one monomer makes up several layers of a β-sheet (e.g. in fibrils formed by HET-s(218-289) (Wasmer et al. 2008)) the periodicity becomes a multiple of 4.8 Å (see Fig. 2A). This periodicity creates a continuous array of equally spaced side chains that is unlike anything in the cell. In biotechnological applications of cross-β fibrils, the periodic arrangement of side chains has, for example, been used to coordinate zinc and allow catalysis (Lee et al. 2017).
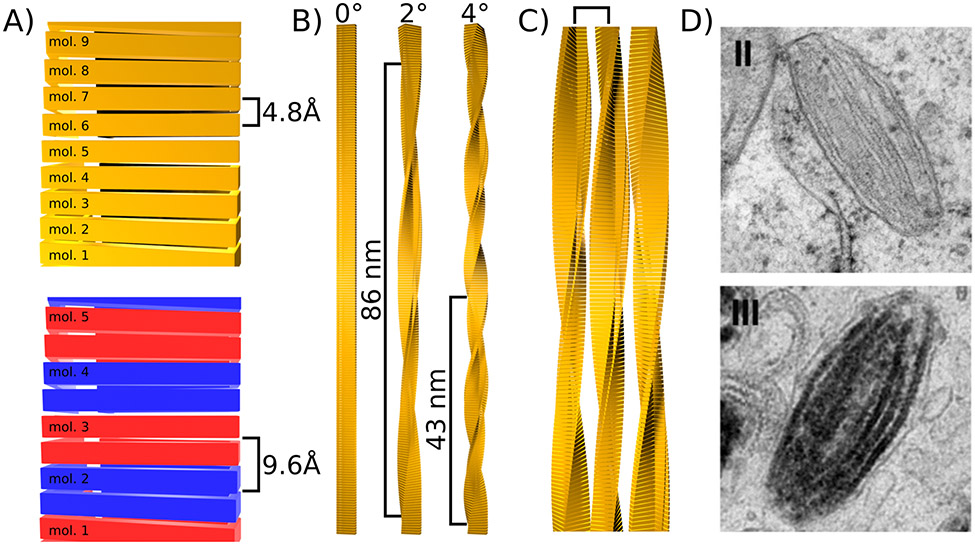
Figure 2: Levels of periodicity in cross-β fibrils.
A) Periodicity along the fibril axis caused by the repetition of monomers. In the simplest case when every strand of a β-sheet is formed by a different monomer, the periodicity is 4.8 Å (top). Otherwise the this periodicity is multiple of 4.8 Å. For example when one monomer provides two strands of the same β-sheet as illustrated with alternating blue and red monomers (bottom). B) Periodicity along the fibril axis caused by a slight rotation of each monomer around the fibril axis (yaw). Three examples with rotation angles of 0°, 2°, and 4° are given and the resulting periodicity is indicated. C) Periodicity induced by lateral association of fibrils. A square bracket indicates the resulting periodicity. D) Example of lateral association of Pmel17 fibrils during stage II of melanosome formation (top), to which melanin binds during stage III (bottom). Images from Hurbain and co-workers (2008) Copyright 2008 National Academy of Sciences.
Another origin of periodicity is the slight rotation around the fibril axis (yaw) between β-strands of consecutive layers typically in the range of zero to a few degrees. This yaw results in an overall twist of the fibril and, depending on the size of the yaw, to periodicities in the high nm to low μm range (see Fig. 2B).
Finally, cross-β fibrils can gain another degree of periodicity via lateral association. Here the periodicity is determined by the thickness of the individual filament and the resulting structure is an array of laterally assembled fibrils (see Fig. 2C). One example of this kind of periodicity is found in the cross-β fibrils formed by Pmel17, which functions as a scaffold for pigment deposition in human skin and eyes (Pfefferkorn et al. 2010; McGlinchey and Lee 2018). Pmel17 fibrils form in melanosomes, acidic organelles that synthesize and store melanin, after proteolytic cleavage. During stage I and II of melanosome development, Pmel17 fibrils form and laterally associate to form a sheet-like fibrillar matrix, which promotes the ellipsoid shape of melanosomes. In stage III and IV these fibril matrices are coated with newly synthesized melanin which provides a template for melanin to aggregate into a large and continuous structure (see Fig. 2D) (Hurbain et al. 2008). The advantage of the highly periodic structure of the Pmel17 sheet-like fibril matrix is that it allows a very high density of melanin to be arranged in a highly ordered manner that would be difficult to produce otherwise.
Multimeric
Related to the periodicity discussed above, is the fact that cross-β fibrils are highly multimeric structures. Where the soluble state of the corresponding protein is often mono- or oligomeric, aggregation into cross-β fibrils dramatically increases the local protein concentration both of the core domain as well as the framing sequences. This multimerization is motif-specific, meaning that only proteins that share a common, compatible β-strand motif are incorporated into the fibril. Usually these are multiple copies of the same protein. However, fibrils formed by Orb2 are made of different isoforms of the same protein (Keleman et al. 2007; Hervas et al. 2020) and fibrils formed by RIP1/RIP3 are even composed of two distinct but complementary proteins that share a RHIM motif, which alternate in the core of the resulting fibril (Li et al. 2012; Mompeán et al. 2018). This multimerization can have important functional consequences. A high protein density is of advantage if the function is protein storage. For example, some peptide hormones are stored in the form of cross-β fibrils (Maji et al. 2009).
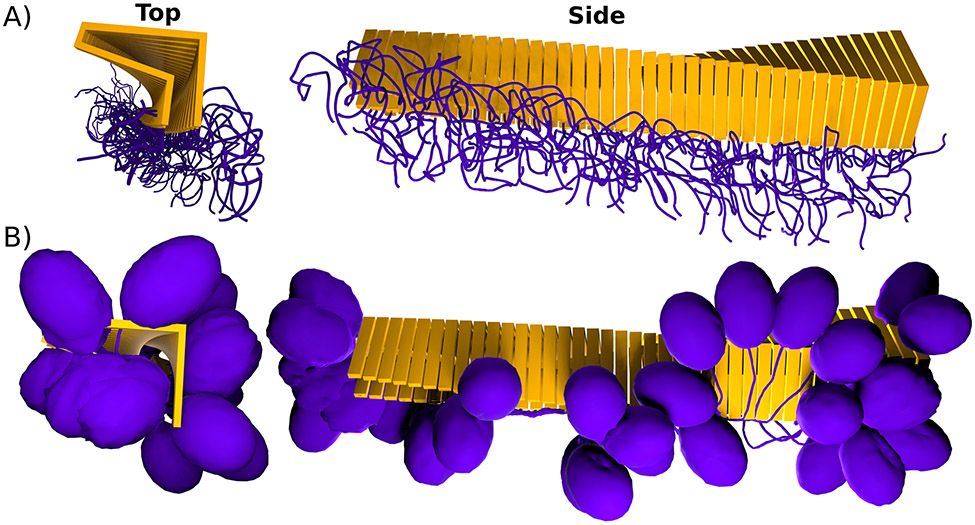
It is important to note that this multimerization not only affects the cross-β fibril core itself, but also its framing sequences i.e. those regions of the protein that are not part of the fibril core but are still part of the fibril. These regions become similarly periodic along the fibril axis or via lateral association and can include well-ordered domains (see Fig. 3A) as for example the RNA recognition motifs (RRMs) in Orb2 (Keleman et al. 2007; Majumdar et al. 2012; Hervas et al. 2020), or intrinsically disordered polymeric brushes (see Fig. 3B) as e.g. in the case of huntingtin exon-1 (Isas et al. 2015; Caulkins et al. 2018; Falk et al. 2020). The high local density of both the surface residues of the cross-β core and the framing sequences potentially increases the binding of ligands with low binding affinities or multimeric ligands which have several binding domains.
Figure 3: Cross-β formation causes multimerization of core and framing sequences.
A) Idealized cross-β fibril with intrinsically disordered framing sequences (purple) that form an entropic brush at the surface of the fibril. B) Idealized cross-β fibril with globular domain (purple) connected to the cross-β core (yellow) via a linker.
Important examples of a cross-β mediated multimerization are adhesins such as the Als5p protein from Candida albicans. Als5p belongs to a class of multidomain cell-surface proteins which are important for cell-cell and cell-ligand interaction. Als5p is C-terminally anchored to the membrane and has globular ligand binding domains at its N-terminus. In between these domains is a Thr-rich domain containing a 7-residue sequence that can cause multimerization via shear stress induced cross-β formation (Ramsook et al. 2010; Chan and Lipke 2014). This cross-β driven nanodomain formation increases the strength of adherence because spontaneous dissociation of a ligand is more likely followed by rebinding to a nearby adhesin (Garcia et al. 2011). Als5p is therefore a good example of cross-β induced multimerization of the same protein, which activates the function of its framing sequences via clustering.
Stable
The cross-β motif is stabilized via dense hydrogen bond networks inherent to the β-sheets that give this structure its name. In addition, side chain hydrogen bond networks known as Gln and Asn ladders (Yoder et al. 1993), tight steric complementarity of the side chains of adjacent β-sheets known as steric zippers (Nelson et al. 2005; Sawaya et al. 2007), and hydrophobic cores and salt bridges known from globular proteins add to the stability of these fibrils (Sawaya et al. 2021). As a consequence, cross-β fibrils are often sodium dodecyl sulfate and protease resistant and can withstand many environmental stresses. In fact, original descriptions of prions (i.e. fibrils formed by the PrP protein) as infective agents in scrapie were based on their resistance to different treatments including heat and UV-radiation (Prusiner 1982). Mechanically, cross-β fibrils can vary considerably depending on their hydrogen bond density. They can be as strong as steel while maintaining a flexibility comparable to that of spider silk (Lamour et al. 2017). An example of functional cross-β fibrils for which stability is of key importance are chorion proteins, which cover the eggs of many fish and insects as protective layers. Dense chorion cross-β fibril networks protect the egg against environmental stress such as dehydration (Iconomidou et al. 2000; Podrabsky et al. 2001).
Structural Change
The majority of known proteins follow either Anfisen’s dogma (i.e. their native protein structure is determined by their sequence) (Anfinsen 1973), form quasi-equivalent (i.e. polymorphic) structures, or are intrinsically disordered. Cross-β forming proteins, in contrast, can adopt drastically different conformations: a soluble, monomeric or multimeric conformation and the cross-β fibril. Protein regions that form the cross-β fibril core undergo massive changes in secondary and tertiary structure during fibril formation. Often the cross-β core region is (partially) disordered in the soluble state (Tompa 2009), which removes the necessity of unfolding prior to fibril formation (Goldschmidt et al. 2010). This structural change can be important for function through several mechanisms: it can change the interactome of the protein by expelling ligands from the cross-β core region, it can create new binding epitopes either at the core region or its framing sequences, it can activate catalytic or binding domains located in the framing sequences e.g. via removal of autoinhibitory interactions, etc. A good example of proteins that use this conformational change as a key property are the receptor-interacting protein kinases 1 and 3 (RIPK1 and RIPK3). When RIP1 and RIP3 interact via their RIP homotypic interaction motifs (RHIM) they form a necrosome, which triggers the cell-death pathways known as necrosis (Cho et al. 2009). This RHIM motif interaction is structurally a cross-β fibril formation in which alternating layers are formed by RIP1 and RIP3 (Mompeán et al. 2018). Necrosome formation increases the kinase activity. Furthermore, mutants that prevent cross-β formation also prevent kinase activation, indicating that the structural changes induced by fibril formation increases kinase activity (Li et al. 2012). However, what could the functional advantage of activation via cross-β fibril formation compared to activation via e.g. phosphorylation be? One hypothesis is that multimerization produces a scaffold that recruits other signaling molecules to the necrosome promoting their activation. Another hypothesis is that the slow nucleation process combined with rapid growth via self-templating, once nucleation has occurred, may result in a digital threshold response mechanism (Li et al. 2012).
Self-templating
Cross-β fibrils are self-templating and can grow in the presence of the soluble (monomeric) form without chaperones or the use of energy e.g. in the form of ATP. Once cross-β structures (seeds) are formed, the elongation of fibrils is energetically favorable meaning that the ΔG of the protein in the fibril is lower than in the soluble state. What often prevents spontaneous fibril formation is the low probability of nucleation i.e. the formation of a minimal cross-β motif that makes further growth energetically favorable. Fibril growth can occur via two different nucleation mechanisms. The addition of free monomers to the ends of a fibril is known as primary nucleation. Primary nucleation is inherent to all cross-β fibrils. Nucleation can also occur on the fibril’s surface. This process is known as secondary nucleation (Törnquist et al. 2018). Even if the cross-β state is energetically much more favorable, it always exists in equilibrium with the monomeric state (O’Nuallain et al. 2005), which means that the removal of monomers from the solution will eventually lead to the dissolution of the cross-β fibrils.
The ability of cross-β fibrils to self-template has important functional implications. Self-templating allows the spread of the cross-β structure within the cell, in between cells, and even in between organisms. Through their ability to spread, cross-β fibrils become functionally prions (Prusiner 1982; Prusiner 2013). This ability plays a key role in the infectivity of prions and the spread of neurodegenerative disease through the brain. However, functional cross-β fibrils like those formed by the HET-s prion (Saupe 2011) also take advantage of the ability to self-template. Besides the inherent stability of the cross-β motif (see above), self-templating also contributes to the longevity of the cross-β state. This is because self-templating maintains the cross-β state much longer than the half-life of the corresponding protein in the cell. Even if the core structure and especially the framing sequences are affected by spontaneous or environmental causes of protein degradation, the templating of newly synthesized monomers into the cross-β motif assures the continuous maintenance of the state. Both of these consequences of self-templating are key for functional cross-β fibrils in signaling and result in signals that will last much longer than signals mediated by e.g. hormones. One example of this function is the phenotypic inheritance mediated by cross-β forming prions in yeast (Halfmann et al. 2012). The best studied example of these yeast prions is Sup35. Sup35 is a translation-termination factor that loses its function allowing for stop codon readthrough when it aggregates into a cross-β fibril. Sup35 aggregation can occur spontaneously in yeast and once formed, will be faithfully inherited by daughter cells as the prion termed [PSI+]. Due to the stop codon readthrough, [PSI+] cells will have heritable new traits that can be of advantage in certain environments. Based on its ability to self-template, Sup35 becomes an epigenetic element of inheritance, which makes the [PSI+] prion state persist past the half-life of the individual protein.
Fibrous
Many, if not most, cross-β motifs found in nature manifest themself as fibrillar structures, which can be visualized using electron (Cohen and Calkins 1959) or (super-resolution) fluorescence microscopy (Pinotsi et al. 2014; Rice et al. 2021; Kaur et al. 2022). These fibrils are unbranched and essentially 1D crystals. Some fibrils are made of a single filaments in which single monomers stack on top of each other (see Fig. 4A). An example of such a structure is the fibril formed by HET-s(218-289) (Wasmer et al. 2008). However, many fibrils have multiple protofilaments as their minimal unit, i.e. multiple monomers per fibril layer, bundled into a fibril (see Fig. 4B). Besides this minimal structure, cross-β fibrils often have the tendency to bundle into even larger assemblies. This bundling can occur, depending on the environment, in a relatively uncontrolled fashion as for example by fibrils formed by HTTex1, in which the degree of bundling is inversely correlated with fibril toxicity (Isas et al. 2021). In functional cross-β fibrils this bundling is usually more controlled and can result in arrays of laterally associated fibrils as for example those formed by chorion proteins or Pmel17 discussed above.
Figure 4: Cross-β fibrils can be made of one or multiple protofilaments.
A) View down the fibril axis (Top) and along the fibril axis (Side) of an idealized cross-β core formed by a single (proto)filament. In such a fibril there is only one monomer per layer. B) Idealized cross-β fibril that is made of two protofilaments. The top view shows nicely how this fibil has two identical monomers per layer.
Another potential function of the fibrous nature of the cross-β motif is the ability to form gels and promote biofilm formation. One of the best understood examples are the fibrils formed by curli proteins found in biofilms of E. coli and other Enterobacteriaceae (Barnhart and Chapman 2006; Blanco et al. 2012; Van Gerven et al. 2018). Biofilms are bacterial communities that are held together by an extracellular matrix (ECM), which also protects this community from environmental stresses. Curli fibrils are a major component of this ECM in addition to cellulose and other polysaccharides. Curli fibrils are crucial for the initial surface attachment of the biofilm and are thereby important for host interaction in the case of pathogenic bacteria. Curli also provide a crucial scaffold to maintain the structural integrity of the biofilm which is where fibrous function comes into play (Barnhart and Chapman 2006).
Comparison to other Biological Fibrils
There are many other fibrils in nature besides cross-β fibrils. What makes cross-β fibrils special in comparison? Structurally, cross-β fibrils are by definition unique and quite different from collagen fibrils with their triple helical structure, the coiled-coil structure that stabilizes α-keratin, or the array of globular structures found in actin and tubulins. β-Keratin found in reptiles and birds is structurally more closely related to cross-β fibrils. However, X-ray diffraction analysis indicates that their β-strands run parallel to the fibril axis in contrast to cross-β fibrils (Fraser et al. 1971; Inouye et al. 1993; Fraser and Parry 2008). This is also true for spider silk, which, depending on type, is composed of different fractions of α-helical and β-sheet structure. In contrast to the cross-β fold, β-strands in parallel-β silk run parallel to the fibril axis (van Beek et al. 2002; Kenney et al. 2002). An exception here are the silk stalks on which the green lace-wing fly (Chrysopa flava) lays their eggs, which are one of the first functional cross-β fibrils described (Geddes et al. 1968). In contrast to most other biological fibrils, cross-β fibrils often have substantial framing sequences that stay in globular or disordered conformations unrelated to the fibril structure. This is because the core forming part of many cross-β fibrils is relatively small compared to the size of the overall protein.
Mechanically, cross-β fibrils are stronger than actin and most keratins and can have comparable mechanical properties to dragline spider silk. Generally, cross-β fibrils are more elastic (i.e. have a lower Young’s modulus) compared to other biological fibrils of equal strengths. The softest and weakest cross-β fibrils have unique material properties that are not shared with other biological materials (Lamour et al. 2017).
The assembly and disassembly of cross-β fibrils is another feature that sets them apart from other biological fibrils. Although the initiation or nucleation of functional cross-β fibrils is highly regulated as, for example, in the case of curli fibrils (Barnhart and Chapman 2006), their growth and maintenance occurs via self-templation without requiring energy (e.g. in the form of ATP), assistance from other proteins or chaperones, or the requirement of cross-linking. This is in contrast to actin, which requires ATP, CCT chaperonin, and prefoldin for assembly. Tubulins similarly require GTP and a microtuble-organizing center for their assembly (Alberts et al. 2002). Collagen fibrils self-assemble spontaneously but only after substantial post-translational modifications (PTMs) in the endoplasmic reticulum, and cleavage of collagen propeptides. In addition, collagen fibrils are often crosslinked (Shoulders and Raines 2009). The self-assembly of keratins is similarly regulated via PTMs and cross-linking (Loschke et al. 2015) and soluble silk proteins are thought to fibrilize when leaving the gland through changes in pH and shear forces (Andersson et al. 2016).
Cross-β fibrils in disease are very stable and difficult for the protein homeostasis machinery to remove (Chuang et al. 2018; Scior et al. 2018). In contrast, functional cross-β fibrils often have an evolved mechanism of disassembly, similar to other biological fibrils such as tubulin, whose disassembly is regulated via GTP to GDP hydrolysis. For example, the cross-β fibrils formed by the peptide hormone β-endorphin readily form under conditions found in secretory granules but can rapidly dissolve due to a change in pH, ionic strength, glycosaminoglycan concentration (Maji et al. 2009; Nespovitaya et al. 2016).
In summary, the cross-β motif combines a high degree of stability, strength, and elasticity with the ability to self template, and with a compact core structure that leaves other domains open for functional activities.
In the following, I will present a few prominent examples of functional cross-β fibrils and discuss how their function is a result of multiple properties of the cross-β motif. I will also discuss the potential advantages of cross-β formation over alternative solutions to the same biological problem.
Functional Fibrils use Multiple Properties of the Cross-β Motif
Orb2: Stable, self-templating structural change in long-term memory
Orb2 belongs to the cytoplasmic polyadenylation element binding (CPEB) protein family. CPEBs have a relatively well conserved C-terminus containing two RNA-recognition motifs, which bind a CPE element in the 5’UTR of target mRNA. When CPEBs are activated, they initiate mRNA translation by allowing the elongation of the poly(A) tails of bound mRNA. Many CPEBs, as for example CPEB in Xenopus oocytes, are activated via phosphorylation (Richter 2007). In contrast, Orb2 and other CPEBs with roles in synaptic plasticity, are activated by the cross-β fibril formation of their N-terminal low complexity domains (Si, Giustetto, et al. 2003; Si, Lindquist, et al. 2003; Majumdar et al. 2012; Stephan et al. 2015). Orb2 mRNA targets are involved in neuronal growth and synapse formation (Mastushita-Sakai et al. 2010) and Orb2 aggregation into a cross-β fibril is required for long-term memory in Drosophila (Majumdar et al. 2012; Hervas et al. 2020). Functionally, Orb2 activation is a result of the conformational change caused by transition of the glutamine/histidine rich N-terminal domain from a disordered, soluble conformation (Cervantes et al. 2016; Hervás et al. 2016; Oroz et al. 2020) into the cross-β motif. This transition results in a change in protein cofactors binding to Orb2. Monomeric Orb2 interacts with the protein CG13928, which decreases translation of target mRNA. However, Orb2 fibrils do not bind CG13928 but the protein CG4612, which enhances translation (Khan et al. 2015; Hervas et al. 2020).
An important aspect that could explain the functional advantage of activation via fibril formation compared to phosphorylation is the long-lasting nature of the active state. The stability of the cross-β motif makes the protein more resistant to protein degradation compared to the monomeric state, significantly extending its half-life. However, the self-templating nature of the cross-β motif makes the activation fully independent of protein half-life (Raveendra et al. 2013). Li and co-workers illustrated this concept with an elegant experiment (Li et al. 2016). They replaced endogenous Orb2 in Drosophila with Orb2 constructs that had a TEV cleavage site between the cross-β core forming N-terminus and the mRNA binding C-terminus. In addition, the flies expressed neuron-specific TEV protease when fed RU486, which turned Orb2 translationally inactive. As a consequence, flies were not able to acquire long-term memories when fed RU486 while being trained. In addition, flies lost long-term memories when they were put on a RU486-containing diet after training. However, what is most striking is that flies that lost memories due to RU486 induced Orb2 inactivation after training, regained those memories when RU486 was removed from their diet. These data suggest that the self-templating activity of Orb2 fibrils that had the functional mRNA binding domains removed restored function through addition of newly expressed, uncleaved Orb2 monomers (Li et al. 2016). This example shows that protein activation via cross-β formation results in a very stable, self-perpetuating signal that lasts longer than e.g. activation via phosphorylation.
The example of Orb2 also shows that functional cross-β fibrils do not necessarily take advantage of all properties of the cross-β motif. Specifically, the fibrous nature and mechanical stability of the fibrils does not seem to be as important in this case. Orb2 fibrils were calculated to have a rather low per residue stability compared to other cross-β fibrils (Sawaya et al. 2021).
Class I hydrophobins: reducing surface tension using stable, multimeric, and self-templating fibrils
Class I hydrophobins from filamentous fungi such as the protein EAS from Neurospora crassa can self-assemble into amphipathic protein layers at the air-water interface, which allow the fungus to overcome the surface tension and breach the air-water interface. Other class I hydrophobins such as RodA from Aspergillus nidulans can prevent wetting and protect spores and help with host interaction by serving as shields for the underlying fungal cell wall components (Lo et al. 2019). In the following, I will limit the discussion to the functional advantage of cross-β formation of the hydrophobin EAS. Similar to other class I hydrophobins, a large fraction of the monomer surface of EAS is hydrophobic (61%) and the protein itself is stabilized via several disulfide bonds. In the absence of an air-water interface EAS stays monomeric. However, EAS self-assembles into rodlet structures when present at the air water interface. These rodlets are arrays of laterally assembled EAS cross-β fibrils (see Fig. 5A), which lower the surface tension of the solution (Morris et al. 2011; Macindoe et al. 2012). The functional advantage of cross-β formation in class I hydrophobins comes into focus when comparing them to class II hydrophobins, which are structurally similar and also assemble at the air-water interface to lower surface tension (Lo et al. 2019). However, class II hydrophobins do not form rodlets (i.e. laterally assembled cross-β fibrils) compared to class I hydrophobins. This difference in structure results in a striking difference in stability. Where rodlets formed by class I hydrophobins withstand temperatures up to 100°C in detergent and are chemically stable under alkaline (3 M NaOH) and acidic (3 M HCl) conditions, class II hydrophobin layers readily dissolve when treated with alcohols and detergents or by applying mild pressures or temperatures. On the flipside, class II hydrophobins assemble more quickly at the air water interface (Lo et al. 2019). Besides stability as the key functional advantage, other properties of the cross-β motif also play a role in EAS rodlets. The ability to self-template allows the assembly of rodlets at the air-water interface. However, because fibril nucleation occurs spontaneously at the air-water interface, there is no real element of self propagation. Nevertheless, fibril formation is spontaneous without the need of chaperones or additional energy. The fibrous nature clearly adds to the function of EAS by allowing it to form a dense array of laterally assembled cross-β fibrils. It is unclear if the periodicity that is introduced along the fibril axis and via lateral association of fibrils plays a role for function. However, the multimeric nature is important in this example because the cross-β core, which is formed by only a loop of EAS, organizes a dense array of globular, amphiphilic framing sequences (see Fig. 5B) (Kwan et al. 2006; Macindoe et al. 2012).
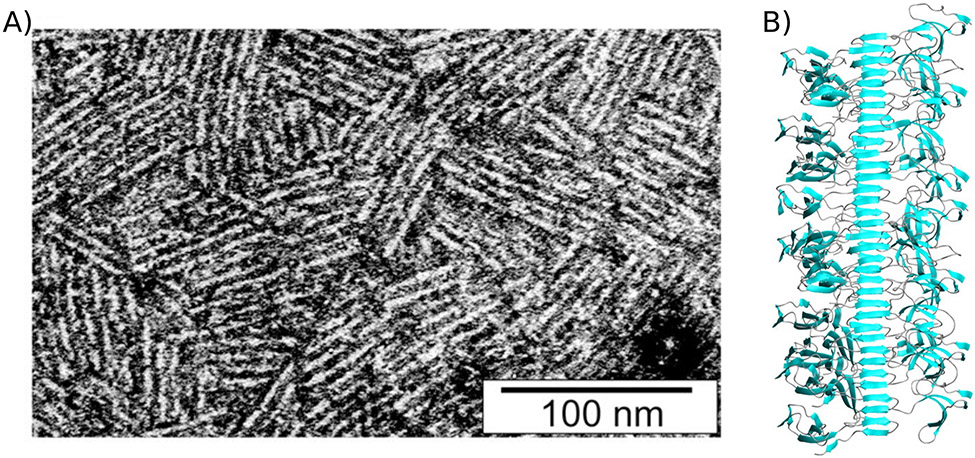
Figure 5: Class I hydrophobin rodlet structures are stabilized by a cross-β core.
A) transmission electron micrograph of negatively stained EASΔ15 rodlet monolayers. Image from Morris et. al. 2011 under a CC BY 4.0 license (https://creativecommons.org/licenses/by/4.0/). B) Model of EASΔ15 fibrils showing the fibril core and globular, amphipathic framing sequences. Image from Macindoe et al. 2012.
HET-s: A functional prion that serves as a template for multimerization and structural change
Podospora anserina, like many other filamentous fungi, can undergo spontaneous cell fusion that leads to cells with multiple nuclei (heterokaryon). These heterokaryons only survive if the cells involved in the fusion are genetically similar. The cell death of the heterokaryon as a non-self recognition process is genetically regulated via differences of the fusion partners at certain loci called het loci. It is speculated that the function of this self, non-self recognition process is to prevent viral infection or parasitism (Saupe and Daskalov 2012). One of these het loci is the gene het-s which has two alternative incompatible alleles, het-s and het-S encoding for the proteins HET-s and HET-S, respectively. The protein HET-s can form a functional cross-β fibril, which can be transmitted between cells and organisms making it a prion termed [HET-s]. Non prion strains which have non-aggregated soluble HET-s protein are designated [HET-s*]. When a heterokaryon forms between a het-s and a het-S strain, it only undergoes cell death when het-s is in the prion state [HET-s] i.e. when HET-s cross-β fibrils are present (Ritter et al. 2005). HET-s has an N-terminal, globular HeLo domain and a C-terminal prion forming domain (PDF), which forms the cross-β core of the fibril (Siemer et al. 2005; Wasmer et al. 2008). HET-S does not form fibrils on its own (Greenwald et al. 2010). HET-s fibrils alone are non-toxic. However, the interaction of HET-S with HET-s fibrils triggers the oligomerization and toxicity of HET-S resulting in cell death. Central to this function is the structural change induced by fibril formation: the interaction with HET-s fibrils induces the multimerization of HET-S, which triggers the release of a hydrophobic transmembrane segment from the HeLo domain of HET-S. Thus activated, HET-S oligomers can interact with membranes and induce membrane leakage and cell death (Mathur et al. 2012; Seuring et al. 2012) reminiscent of the membrane disruption caused by Aβ oligomers (Reiss et al. 2018). Where HET-s’ ability to self-template is not important for the cell death induced by the HET-s/HET-S interaction, its ability to act as a template for HET-S oligomerization very much is. Nevertheless, the self-templating of HET-s allows the maintenance of the functional state of HET-s between cells and Podospora strains. One property of the cross-β motif that does not seem to be important for function is its fibrous nature. Despite the fact that HET-s fibrils were found to be mechanically strong (Lamour et al. 2017), the fibrous nature of these fibrils does not play an obvious role for function.
Concluding remarks
Functional cross-β fibrils (functional amyloids), a well-established concept, are found in all kingdoms of life, and new examples are described regularly. In this review, I wanted to highlight the functional advantages of cross-β fibril formation over other means of solving the same biological problem by focusing on the unique properties of the cross-β motif. These properties are the inherent periodicity, stability, and multimeric and fibrous nature of cross-β fibrils as well as their ability to self-template and the structural change that occurs when proteins aggregate into cross-β fibrils. Each functional cross-β fibril takes advantage of multiple of these properties depending on its specific function. In some cases, the advantages of cross-β fibril formation over other solutions to the same biological problem are quite clear. For example, when properties such as stability and self-templating are used to create long-lasting signals (e.g. Orb2) that can be inherited in a non-Mendelian fashion (e.g. Sup35). For other functional cross-β fibrils the advantage of the cross-β motif is less obvious. For example, what is the advantage of cross-β formation over e.g. phosphorylation for activating the RIP1-RIP3 kinases? Additional research is required to answer this question.
The fact that functional cross-β fibrils are no rare curiosity in nature is supported by mounting evidence that some of these functional motifs are evolutionarily old and well-conserved. A prominent example of this is the similarity between programmed cell death-inducing functional cross-β fibrils: HET-s and HELLP in fungi, and metazoan RHIMs domain proteins such as RIP1/RIP3 (Kajava et al. 2014; Daskalov et al. 2016; Daskalov and Saupe 2021). The long evolutionary history of functional cross-β fibrils should not come as a surprise considering that this fold, which is dominated by backbone hydrogen bonding, can be adopted by virtually all proteins under the right conditions (Chiti et al. 1999). Consequently, Greenwald and co-workers hypothesized that the cross-β motif is among the first protein folds (Greenwald and Riek 2012).
What is the relation between functional cross-β fibrils and cross-β fibrils found in disease? Is there an intrinsic toxicity to cross-β fibrils or oligomeric intermediates formed during their formation? One hypothesis is that functional cross-β fibrils might mitigate oligomer toxicity through controlled and rapid aggregation into mature fibrils which are often found to be less toxic (Hervás et al. 2016). However, if there is an intrinsic toxicity to oligomers or cross-β fibrils, it is surprising that in cross-β fibrils with a function in programmed cell death, the cross-β core itself is not toxic and toxicity is rather caused by conformational change in the framing sequence (RIP1/RIP3) or of binding partners (HET-s/HET-S). These examples of functional cross-β fibrils in programmed cell death, in turn, suggest that cross-β fibrils in disease could potentially induce cell death via conformational change in framing sequences or an influence on their binding partners rather than a direct toxicity of their cross-β core or oligomeric intermediates. This exchange of hypotheses between the field of functional and disease relevant cross-β fibrils is a good example of why studying functional cross-β fibrils not only deepens our understanding of basic biology but it can also result in new ideas and concepts that might help us cure diseases caused by fibril formation.
Acknowledgements
I would like to thank Dr. Maria Soria for fruitful discussions about the initial concept of this review. I would like to thank Sayuri Pacheco and Silvia Cervantes for carefully proofreading and helpful comments on the manuscript.
Declaration of interest statement
The author reports no declarations of interest. This work was supported by the National Institutes of Health under grant numbers R01GM110521, R01AG061865, and R01NS120704.
References
- Alberti S, Hyman AA. 2021. Biomolecular condensates at the nexus of cellular stress, protein aggregation disease and ageing. Nat Rev Mol Cell Biol. 22(3):196–213. doi: 10.1038/s41580-020-00326-6. [DOI] [PubMed] [Google Scholar]
- Alberts B, Johnson A, Lewis J, Raff M, Roberts K, Walter P. 2002. The Self-Assembly and Dynamic Structure of Cytoskeletal Filaments. Mol Biol Cell 4th Ed. https://www.ncbi.nlm.nih.gov/books/NBK26862/. [Google Scholar]
- Andersson M, Johansson J, Rising A. 2016. Silk Spinning in Silkworms and Spiders. Int J Mol Sci. 17(8):1290. doi: 10.3390/ijms17081290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anfinsen CB. 1973. Principles that govern the folding of protein chains. Science. 181(4096):223–230. doi: 10.1126/science.181.4096.223. [DOI] [PubMed] [Google Scholar]
- Baba M, Nakajo S, Tu P-H, Tomita T, Nakaya K, Lee VM-Y, Trojanowski JQ, Iwatsubo T. 1998. Aggregation of α-synuclein in Lewy bodies of sporadic Parkinson’s disease and dementia with Lewy bodies. Am J Pathol. 152(4):879–884. [PMC free article] [PubMed] [Google Scholar]
- Barnhart MM, Chapman MR. 2006. Curli biogenesis and function. Annu Rev Microbiol. 60:131–47. doi: 10.1146/annurev.micro.60.080805.142106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Beek JD, Hess S, Vollrath F, Meier BH. 2002. The molecular structure of spider dragline silk: folding and orientation of the protein backbone. Proc Natl Acad Sci U S A. 99(16):10266–10271. doi: 10.1073/pnas.152162299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benson MD, Buxbaum JN, Eisenberg DS, Merlini G, Saraiva MJM, Sekijima Y, Sipe JD, Westermark P. 2018. Amyloid nomenclature 2018: recommendations by the International Society of Amyloidosis (ISA) nomenclature committee. Amyloid. 25(4):215–219. doi: 10.1080/13506129.2018.1549825. [DOI] [PubMed] [Google Scholar]
- Blanco LP, Evans ML, Smith DR, Badtke MP, Chapman MR. 2012. Diversity, biogenesis and function of microbial amyloids. Trends Microbiol. 20(2):66–73. doi: 10.1016/j.tim.2011.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caulkins BG, Cervantes SA, Isas JM, Siemer AB. 2018. Dynamics of the Proline-Rich C-Terminus of Huntingtin Exon-1 Fibrils. J Phys Chem B. 122(41):9507–9515. doi: 10.1021/acs.jpcb.8b09213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cervantes SA, Bajakian TH, Soria MA, Falk AS, Service RJ, Langen R, Siemer AB. 2016. Identification and Structural Characterization of the N-terminal Amyloid Core of Orb2 isoform A. Sci Rep. 6(1):38265. doi: 10.1038/srep38265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan CXJ, Lipke PN. 2014. Role of force-sensitive amyloid-like interactions in fungal catch bonding and biofilms. Eukaryot Cell. 13(9):1136–1142. doi: 10.1128/EC.00068-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiti F, Webster P, Taddei N, Clark A, Stefani M. 1999. Designing Conditions for in Vitro Formation of Amyloid Protofilaments and Fibrils. Proc Natl Acad Sci. 96(7):3590–3594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho Y, Challa S, Moquin D, Genga R, Ray TD, Guildford M, Chan FK-M. 2009. Phosphorylation-Driven Assembly of the RIP1-RIP3 Complex Regulates Programmed Necrosis and Virus-Induced Inflammation. Cell. 137(6):1112–1123. doi: 10.1016/j.cell.2009.05.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chuang E, Hori AM, Hesketh CD, Shorter J. 2018. Amyloid assembly and disassembly. J Cell Sci. 131(8):jcs189928. doi: 10.1242/jcs.189928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ke Chun P, Sani M-A, Ding F, Kakinen A, Javed I, Separovic F, Davis TP, Mezzenga R. 2017. Implications of peptide assemblies in amyloid diseases. Chem Soc Rev. 46(21):6492–6531. doi: 10.1039/C7CS00372B. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen AS, Calkins E. 1959. Electron Microscopic Observations on a Fibrous Component in Amyloid of Diverse Origins. Nature. 183(4669):1202–1203. doi: 10.1038/1831202a0. [DOI] [PubMed] [Google Scholar]
- Daskalov A, Habenstein B, Sabaté R, Berbon M, Martinez D, Chaignepain S, Coulary-Salin B, Hofmann K, Loquet A, Saupe SJ. 2016. Identification of a novel cell death-inducing domain reveals that fungal amyloid-controlled programmed cell death is related to necroptosis. Proc Natl Acad Sci. 113(10):2720–2725. doi: 10.1073/pnas.1522361113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daskalov A, Saupe SJ. 2021. The expanding scope of amyloid signalling. Prion. 15(1):21–28. doi: 10.1080/19336896.2021.1874791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DiFiglia M, Sapp E, Chase KO, Davies SW, Bates GP, Vonsattel JP, Aronin N. 1997. Aggregation of huntingtin in neuronal intranuclear inclusions and dystrophic neurites in brain. Science. 277(5334):1990–1993. doi: 10.1126/science.277.5334.1990. [DOI] [PubMed] [Google Scholar]
- Eisenberg D, Jucker M. 2012. The amyloid state of proteins in human diseases. Cell. 148(6):1188–203. doi: 10.1016/j.cell.2012.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisenberg DS, Sawaya MR. 2017. Structural Studies of Amyloid Proteins at the Molecular Level. Annu Rev Biochem. 86:69–95. doi: 10.1146/annurev-biochem-061516-045104. [DOI] [PubMed] [Google Scholar]
- Falk AS, Bravo-Arredondo JM, Varkey J, Pacheco S, Langen R, Siemer AB. 2020. Structural Model of the Proline-Rich Domain of Huntingtin Exon-1 Fibrils. Biophys J 119(10):2019–2028. doi: 10.1016/j.bpj.2020.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fraser RDB, MacRae TP, Parry DAD, Suzuki E. 1971. The structure of feather keratin. Polymer. 12(1):35–56. doi: 10.1016/0032-3861(71)90011-5. [DOI] [Google Scholar]
- Fraser RDB, Parry DAD. 2008. Molecular packing in the feather keratin filament. J Struct Biol. 162(1):1–13. doi: 10.1016/j.jsb.2008.01.011. [DOI] [PubMed] [Google Scholar]
- Gallardo R, Ranson NA, Radford SE. 2020. Amyloid structures: much more than just a cross-β fold. Curr Opin Struct Biol. 60:7–16. doi: 10.1016/j.sbi.2019.09.001. [DOI] [PubMed] [Google Scholar]
- Garcia MC, Lee JT, Ramsook CB, Alsteens D, Dufrêne YF, Lipke PN. 2011. A Role for Amyloid in Cell Aggregation and Biofilm Formation. PLOS ONE. 6(3):e17632. doi: 10.1371/journal.pone.0017632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geddes AJ, Parker KD, Atkins EDT, Beighton E. 1968. “Cross-β” conformation in proteins. J Mol Biol. 32(2):343–358. doi: 10.1016/0022-2836(68)90014-4. [DOI] [PubMed] [Google Scholar]
- Goldschmidt L, Teng PK, Riek R, Eisenberg D. 2010. Identifying the amylome, proteins capable of forming amyloid-like fibrils. Proc Natl Acad Sci U S A. 107(8):3487–92. doi: 10.1073/pnas.0915166107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gras SL, Waddington LJ, Goldie KN. 2011. Transmission Electron Microscopy of Amyloid Fibrils. In: Hill AF, Barnham KJ, Bottomley SP, Cappai R, editors. Protein Folding, Misfolding, and Disease: Methods and Protocols. Totowa, NJ: Humana Press. (Methods in Molecular Biology). p. 197–214. 10.1007/978-1-60327-223-0_13. [DOI] [PubMed] [Google Scholar]
- Greenwald J, Buhtz C, Ritter C, Kwiatkowski W, Choe S, Maddelein M-L, Ness F, Cescau S, Soragni A, Leitz D, et al. 2010. The mechanism of prion inhibition by HET-S. Mol Cell. 38(6):889–899. doi: 10.1016/j.molcel.2010.05.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenwald J, Riek R. 2012. On the Possible Amyloid Origin of Protein Folds. J Mol Biol. 421(4–5):417–426. doi: 10.1016/j.jmb.2012.04.015. [DOI] [PubMed] [Google Scholar]
- Guerrero-Ferreira R, Taylor NM, Arteni A-A, Kumari P, Mona D, Ringler P, Britschgi M, Lauer ME, Makky A, Verasdonck J, et al. 2019. Two new polymorphic structures of human full-length alpha-synuclein fibrils solved by cryo-electron microscopy. eLife. 8. doi: 10.7554/eLife.48907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halfmann R, Jarosz DF, Jones SK, Chang A, Lancaster AK, Lindquist S. 2012. Prions are a common mechanism for phenotypic inheritance in wild yeasts. Nature. 482(7385):363–368. doi: 10.1038/nature10875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hervás R, Li L, Majumdar A, Fernández-Ramírez M del C, Unruh JR, Slaughter BD, Galera-Prat A, Santana E, Suzuki M, Nagai Y, et al. 2016. Molecular Basis of Orb2 Amyloidogenesis and Blockade of Memory Consolidation. PLOS Biol. 14(1):e1002361. doi: 10.1371/journal.pbio.1002361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hervas R, Rau MJ, Park Y, Zhang W, Murzin AG, Fitzpatrick JAJ, Scheres SHW, Si K. 2020. Cryo-EM structure of a neuronal functional amyloid implicated in memory persistence in Drosophila. Science. 367(6483):1230–1234. doi: 10.1126/science.aba3526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Höppener JW, Ahrén B, Lips CJ. 2000. Islet amyloid and type 2 diabetes mellitus. N Engl J Med. 343(6):411–419. doi: 10.1056/NEJM200008103430607. [DOI] [PubMed] [Google Scholar]
- Huff ME, Balch WE, Kelly JW. 2003. Pathological and functional amyloid formation orchestrated by the secretory pathway. Curr Opin Struct Biol. 13(6):674–682. doi: 10.1016/j.sbi.2003.10.010. [DOI] [PubMed] [Google Scholar]
- Hurbain I, Geerts WJC, Boudier T, Marco S, Verkleij AJ, Marks MS, Raposo G. 2008. Electron tomography of early melanosomes: Implications for melanogenesis and the generation of fibrillar amyloid sheets. Proc Natl Acad Sci. 105(50):19726–19731. doi: 10.1073/pnas.0803488105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iadanza MG, Jackson MP, Hewitt EW, Ranson NA, Radford SE. 2018. A new era for understanding amyloid structures and disease. Nat Rev Mol Cell Biol. 19(12):755–773. doi: 10.1038/s41580-018-0060-8. [DOI] [PubMed] [Google Scholar]
- Iconomidou VA, Vriend G, Hamodrakas SJ. 2000. Amyloids protect the silkmoth oocyte and embryo. FEBS Lett. 479(3):141–145. doi: 10.1016/S0014-5793(00)01888-3. [DOI] [PubMed] [Google Scholar]
- Inouye H, Fraser PE, Kirschner DA. 1993. Structure of beta-crystallite assemblies formed by Alzheimer beta-amyloid protein analogues: analysis by x-ray diffraction. Biophys J. 64(2):502–519. doi: 10.1016/S0006-3495(93)81393-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Isas JM, Langen R, Siemer AB. 2015. Solid-State Nuclear Magnetic Resonance on the Static and Dynamic Domains of Huntingtin Exon-1 Fibrils. Biochemistry. 54(25):3942–9. doi: 10.1021/acs.biochem.5b00281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaroniec CP, MacPhee CE, Bajaj VS, McMahon MT, Dobson CM, Griffin RG. 2004. High-resolution molecular structure of a peptide in an amyloid fibril determined by magic angle spinning NMR spectroscopy. Proc Natl Acad Sci U S A. 101(3):711–716. doi: 10.1073/pnas.0304849101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kajava AV, Klopffleisch K, Chen S, Hofmann K. 2014. Evolutionary link between metazoan RHIM motif and prion-forming domain of fungal heterokaryon incompatibility factor HET-s/HET-s. Sci Rep. 4:7436. doi: 10.1038/srep07436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaur A, Adair LD, Ball SR, New EJ, Sunde M. 2022. A Fluorescent Sensor for Quantitative Super-Resolution Imaging of Amyloid Fibril Assembly. Angew Chem Int Ed. 61(10):e202112832. doi: 10.1002/anie.202112832. [DOI] [PubMed] [Google Scholar]
- Keleman K, Krüttner S, Alenius M, Dickson BJ. 2007. Function of the Drosophila CPEB protein Orb2 in long-term courtship memory. Nat Neurosci. 10(12):1587–93. doi: 10.1038/nn1996. [DOI] [PubMed] [Google Scholar]
- Kenney JM, Knight D, Wise MJ, Vollrath F. 2002. Amyloidogenic nature of spider silk. Eur J Biochem. 269(16):4159–4163. doi: 10.1046/j.1432-1033.2002.03112.x. [DOI] [PubMed] [Google Scholar]
- Khan MR, Li L, Pérez-Sánchez C, Saraf A, Florens L, Slaughter BD, Unruh JR, Si K. 2015. Amyloidogenic Oligomerization Transforms Drosophila Orb2 from a Translation Repressor to an Activator. Cell. 163(6):1468–1483. doi: 10.1016/j.cell.2015.11.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knowles TPJ, Vendruscolo M, Dobson CM. 2014. The amyloid state and its association with protein misfolding diseases. Nat Rev Mol Cell Biol. 15(6):384–96. doi: 10.1038/nrm3810. [DOI] [PubMed] [Google Scholar]
- Kwan AHY, Winefield RD, Sunde M, Matthews JM, Haverkamp RG, Templeton MD, Mackay JP. 2006. Structural basis for rodlet assembly in fungal hydrophobins. Proc Natl Acad Sci U S A. 103(10):3621–6. doi:0505704103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamour G, Nassar R, Chan PHW, Bozkurt G, Li J, Bui JM, Yip CK, Mayor T, Li H, Wu H, et al. 2017. Mapping the Broad Structural and Mechanical Properties of Amyloid Fibrils. Biophys J. 112(4):584–594. doi: 10.1016/j.bpj.2016.12.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee M, Wang T, Makhlynets OV, Wu Y, Polizzi NF, Wu H, Gosavi PM, Stöhr J, Korendovych IV, DeGrado WF, et al. 2017. Zinc-binding structure of a catalytic amyloid from solid-state NMR. Proc Natl Acad Sci. 114(24):6191–6196. doi: 10.1073/pnas.1706179114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li C, Qin R, Liu R, Miao S, Yang P. 2018. Functional amyloid materials at surfaces/interfaces. Biomater Sci. 6(3):462–472. doi: 10.1039/C7BM01124E. [DOI] [PubMed] [Google Scholar]
- Li J, McQuade T, Siemer AB, Napetschnig J, Moriwaki K, Hsiao Y-S, Damko E, Moquin D, Walz T, McDermott A, et al. 2012. The RIP1/RIP3 Necrosome Forms a Functional Amyloid Signaling Complex Required for Programmed Necrosis. Cell. 150(2):339–350. doi: 10.1016/j.cell.2012.06.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L, Sanchez CP, Slaughter BD, Zhao Y, Khan MR, Unruh JR, Rubinstein B, Si K. 2016. A Putative Biochemical Engram of Long-Term Memory. Curr Biol. 26(23):3143–3156. doi: 10.1016/j.cub.2016.09.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lo V, I-Chun Lai J, Sunde M. 2019. Fungal Hydrophobins and Their Self-Assembly into Functional Nanomaterials. In: Perrett S, Buell AK, Knowles TPJ, editors. Biological and Bio-inspired Nanomaterials: Properties and Assembly Mechanisms. Singapore: Springer. (Advances in Experimental Medicine and Biology). p. 161–185. 10.1007/978-981-13-9791-2_5. [DOI] [PubMed] [Google Scholar]
- Loquet A, El Mammeri N, Stanek J, Berbon M, Bardiaux B, Pintacuda G, Habenstein B. 2018. 3D Structure Determination of Amyloid Fibrils using Solid-State NMR Spectroscopy. Methods. doi: 10.1016/j.ymeth.2018.03.014. https://www.sciencedirect.com/science/article/pii/S1046202317302517. [DOI] [PubMed] [Google Scholar]
- Loschke F, Seltmann K, Bouameur J-E, Magin TM. 2015. Regulation of keratin network organization. Curr Opin Cell Biol. 32:56–64. doi: 10.1016/j.ceb.2014.12.006. [DOI] [PubMed] [Google Scholar]
- Macindoe I, Kwan AH, Ren Q, Morris VK, Yang W, Mackay JP, Sunde M. 2012. Self-assembly of functional, amphipathic amyloid monolayers by the fungal hydrophobin EAS. Proc Natl Acad Sci U S A. 109(14):E804–11. doi: 10.1073/pnas.1114052109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maji SK, Perrin MH, Sawaya MR, Jessberger S, Vadodaria K, Rissman RA, Singru PS, Nilsson KPR, Simon R, Schubert D, et al. 2009. Functional amyloids as natural storage of peptide hormones in pituitary secretory granules. Science. 325(5938):328–332. doi: 10.1126/science.1173155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Majumdar A, Cesario WC, White-Grindley E, Jiang H, Ren F, Khan MR, Li L, Choi EM-L, Kannan K, Guo F, et al. 2012. Critical Role of Amyloid-like Oligomers of Drosophila Orb2 in the Persistence of Memory. Cell. 148(3):515–29. doi: 10.1016/j.cell.2012.01.004. [DOI] [PubMed] [Google Scholar]
- Isas JM, Pandey NK, Xu H, Teranishi K, Okada AK, Fultz EK, Rawat A, Applebaum A, Meier F, Chen J, et al. 2021. Huntingtin fibrils with different toxicity, structure, and seeding potential can be interconverted. Nat Commun. 12(1):4272. doi: 10.1038/s41467-021-24411-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marshall KE, Marchante R, Xue W-F, Serpell LC. 2014. The relationship between amyloid structure and cytotoxicity. Prion. 8(2):192–196. doi: 10.4161/pri.28860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mastushita-Sakai T, White-Grindley E, Samuelson J, Seidel C, Si K. 2010. Drosophila Orb2 targets genes involved in neuronal growth, synapse formation, and protein turnover. Proc Natl Acad Sci U S A. 107(26):11987–92. doi: 10.1073/pnas.1004433107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathur V, Seuring C, Riek R, Saupe SJ, Liebman SW. 2012. Localization of HET-S to the cell periphery, not to [Het-s] aggregates, is associated with [Het-s]-HET-S toxicity. Mol Cell Biol. 32(1):139–153. doi: 10.1128/MCB.06125-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGlinchey RP, Lee JC. 2017. Reversing the Amyloid Trend: Mechanism of Fibril Assembly and Dissolution of the Repeat Domain from a Human Functional Amyloid. Isr J Chem. 57(7–8):613–621. doi: 10.1002/ijch.201600080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGlinchey RP, Lee JC. 2018. Why Study Functional Amyloids? Lessons from the Repeat Domain of Pmel17. J Mol Biol. 430(20):3696–3706. doi: 10.1016/j.jmb.2018.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mompeán M, Li W, Li J, Laage S, Siemer AB, Bozkurt G, Wu H, McDermott AE. 2018. The Structure of the Necrosome RIPK1-RIPK3 Core, a Human Hetero-Amyloid Signaling Complex. Cell. 173(5):1244–1253.e10. doi: 10.1016/j.cell.2018.03.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris VK, Ren Q, Macindoe I, Kwan AH, Byrne N, Sunde M. 2011. Recruitment of class I hydrophobins to the air:water interface initiates a multi-step process of functional amyloid formation. J Biol Chem. 286(18):15955–15963. doi: 10.1074/jbc.M110.214197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson R, Sawaya MR, Balbirnie M, Madsen AØ, Riekel C, Grothe R, Eisenberg D. 2005. Structure of the cross-beta spine of amyloid-like fibrils. Nature. 435(7043):773–8. doi: 10.1038/nature03680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nespovitaya N, Gath J, Barylyuk K, Seuring C, Meier BH, Riek R. 2016. Dynamic Assembly and Disassembly of Functional β-Endorphin Amyloid Fibrils. J Am Chem Soc. 138(3):846–856. doi: 10.1021/jacs.5b08694. [DOI] [PubMed] [Google Scholar]
- O’Nuallain B, Shivaprasad S, Kheterpal I, Wetzel R. 2005. Thermodynamics of Aβ(1–40) Amyloid Fibril Elongation †. Biochemistry. 44(38):12709–12718. doi: 10.1021/bi050927h. [DOI] [PubMed] [Google Scholar]
- Oroz J, Félix SS, Cabrita EJ, Laurents DV. 2020. Structural transitions in Orb2 prion-like domain relevant for functional aggregation in memory consolidation. J Biol Chem. 295(52):18122–18133. doi: 10.1074/jbc.RA120.015211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petkova AT, Tycko R. 2002. Sensitivity enhancement in structural measurements by solid state NMR through pulsed spin locking. J Magn Reson. 155(2):293–299. doi: 10.1006/jmre.2002.2519. [DOI] [PubMed] [Google Scholar]
- Pfefferkorn CM, McGlinchey RP, Lee JC. 2010. Effects of pH on aggregation kinetics of the repeat domain of a functional amyloid, Pmel17. Proc Natl Acad Sci U S A. 107(50):21447–21452. doi: 10.1073/pnas.1006424107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinotsi D, Buell AK, Galvagnion C, Dobson CM, Kaminski Schierle GS, Kaminski CF. 2014. Direct Observation of Heterogeneous Amyloid Fibril Growth Kinetics via Two-Color Super-Resolution Microscopy. Nano Lett. 14(1):339–345. doi: 10.1021/nl4041093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Podrabsky JE, Carpenter JF, Hand SC. 2001. Survival of water stress in annual fish embryos: dehydration avoidance and egg envelope amyloid fibers. Am J Physiol-Regul Integr Comp Physiol. 280(1):R123–R131. doi: 10.1152/ajpregu.2001.280.1.R123. [DOI] [PubMed] [Google Scholar]
- Prusiner SB. 1982. Novel Proteinaceous Infectious Particles Cause Scrapie. Science. 216:136–144. [DOI] [PubMed] [Google Scholar]
- Prusiner SB. 2013. Biology and genetics of prions causing neurodegeneration. Annu Rev Genet. 47:601–23. doi: 10.1146/annurev-genet-110711-155524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puchtler H, Sweat F, Levine M. 1962. On the binding of congo red by amyloid. J Histochem Cytochem. 10(3):355–364. doi: 10.1177/10.3.355. [DOI] [Google Scholar]
- Ragonis-Bachar P, Landau M. 2021. Functional and pathological amyloid structures in the eyes of 2020 cryo-EM. Curr Opin Struct Biol. 68:184–193. doi: 10.1016/j.sbi.2021.01.006. [DOI] [PubMed] [Google Scholar]
- Ramsook CB, Tan C, Garcia MC, Fung R, Soybelman G, Henry R, Litewka A, O’Meally S, Otoo HN, Khalaf RA, et al. 2010. Yeast cell adhesion molecules have functional amyloid-forming sequences. Eukaryot Cell. 9(3):393–404. doi: 10.1128/EC.00068-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raveendra BL, Siemer AB, Puthanveettil SV, Hendrickson W a, Kandel ER, McDermott AE. 2013. Characterization of prion-like conformational changes of the neuronal isoform of Aplysia CPEB. Nat Struct Mol Biol Mol Biol. 20(4):495–501. doi: 10.1038/nsmb.2503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reiss AB, Arain HA, Stecker MM, Siegart NM, Kasselman LJ. 2018. Amyloid toxicity in Alzheimer’s disease. Rev Neurosci. 29(6):613–627. doi: 10.1515/revneuro-2017-0063. [DOI] [PubMed] [Google Scholar]
- Rice LJ, Ecroyd H, van Oijen AM. 2021. Illuminating amyloid fibrils: Fluorescence-based single-molecule approaches. Comput Struct Biotechnol J. 19:4711–4724. doi: 10.1016/j.csbj.2021.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richter JD. 2007. CPEB: a life in translation. Trends Biochem Sci. 32(6):279–285. doi: 10.1016/j.tibs.2007.04.004. [DOI] [PubMed] [Google Scholar]
- Ritter C, Maddelein M-L, Siemer AB, Lührs T, Ernst M, Meier BH, Saupe SJ, Riek R. 2005. Correlation of structural elements and infectivity of the HET-s prion. Nature. 435(7043):844–848. doi: 10.1038/nature03793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saupe SJ. 2011. The [Het-s] prion of Podospora anserina and its role in heterokaryon incompatibility. Semin Cell Dev Biol. 22(5):460–468. doi: 10.1016/j.semcdb.2011.02.019. [DOI] [PubMed] [Google Scholar]
- Saupe SJ, Daskalov A. 2012. The [Het-s] prion, an amyloid fold as a cell death activation trigger. PLoS Pathog. 8(5):e1002687. doi: 10.1371/journal.ppat.1002687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawaya MR, Hughes MP, Rodriguez JA, Riek R, Eisenberg DS. 2021. The expanding amyloid family: Structure, stability, function, and pathogenesis. Cell. 184(19):4857–4873. doi: 10.1016/j.cell.2021.08.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawaya MR, Sambashivan S, Nelson R, Ivanova MI, Sievers SA, Apostol MI, Thompson MJ, Balbirnie M, Wiltzius JJW, McFarlane HT, et al. 2007. Atomic structures of amyloid cross-beta spines reveal varied steric zippers. Nature. 447(7143):453–457. doi: 10.1038/nature05695. [DOI] [PubMed] [Google Scholar]
- Scior A, Buntru A, Arnsburg K, Ast A, Iburg M, Juenemann K, Pigazzini ML, Mlody B, Puchkov D, Priller J, et al. 2018. Complete suppression of Htt fibrilization and disaggregation of Htt fibrils by a trimeric chaperone complex. EMBO J. 37(2):282–299. doi: 10.15252/embj.201797212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seuring C, Greenwald J, Wasmer C, Wepf R, Saupe SJ, Meier BH, Riek R. 2012. The mechanism of toxicity in HET-S/HET-s prion incompatibility. Lindquist SL, editor. PLoS Biol. 10(12):e1001451. doi: 10.1371/journal.pbio.1001451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shoulders MD, Raines RT. 2009. Collagen Structure and Stability. Annu Rev Biochem. 78(1):929–958. doi: 10.1146/annurev.biochem.77.032207.120833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Si K, Giustetto M, Etkin A, Hsu R, Janisiewicz AM, Miniaci MC, Kim J-H, Zhu H, Kandel ER. 2003. A neuronal isoform of CPEB regulates local protein synthesis and stabilizes synapse-specific long-term facilitation in aplysia. Cell. 115(7):893–904. doi: 10.1016/s0092-8674(03)01021-3. [DOI] [PubMed] [Google Scholar]
- Si K, Lindquist S, Kandel ER. 2003. A neuronal isoform of the aplysia CPEB has prion-like properties. Cell. 115(7):879–91. doi: 10.1016/s0092-8674(03)01020-1. [DOI] [PubMed] [Google Scholar]
- Siemer AB, Ritter C, Ernst M, Riek R, Meier BH. 2005. High-Resolution Solid-State NMR Spectroscopy of the Prion Protein HET-s in Its Amyloid Conformation. Angew Chem Int Ed. 44(16):2441–2444. doi:doi.org/ 10.1002/anie.200462952. [DOI] [PubMed] [Google Scholar]
- Sipe JD, Cohen AS. 2000. Review: History of the Amyloid Fibril. J Struct Biol. 130(2):88–98. doi: 10.1006/jsbi.2000.4221. [DOI] [PubMed] [Google Scholar]
- Soria Lopez JA, González HM, Léger GC. 2019. Alzheimer’s disease. Handb Clin Neurol. 167:231–255. doi: 10.1016/B978-0-12-804766-8.00013-3. [DOI] [PubMed] [Google Scholar]
- Stephan JS, Fioriti L, Lamba N, Colnaghi L, Karl K, Derkatch IL, Kandel ER. 2015. The CPEB3 Protein Is a Functional Prion that Interacts with the Actin Cytoskeleton. Cell Rep. 11(11):1772–1785. doi: 10.1016/j.celrep.2015.04.060. [DOI] [PubMed] [Google Scholar]
- Sunde M, Serpell LC, Bartlam M, Fraser PE, Pepys MB, Blake CCF. 1997. Common core structure of amyloid fibrils by synchrotron X-ray diffraction. J Mol Biol. 273(3):729–739. doi: 10.1006/jmbi.1997.1348. [DOI] [PubMed] [Google Scholar]
- Tompa P. 2009. Structural disorder in amyloid fibrils: its implication in dynamic interactions of proteins. FEBS J. 276(19):5406–5415. doi: 10.1111/j.1742-4658.2009.07250.x. [DOI] [PubMed] [Google Scholar]
- Törnquist M, Michaels TCT, Sanagavarapu K, Yang X, Meisl G, Cohen SIA, Knowles TPJ, Linse S. 2018. Secondary nucleation in amyloid formation. Chem Commun. 54(63):8667–8684. doi: 10.1039/C8CC02204F. [DOI] [PubMed] [Google Scholar]
- Van Gerven N, Van der Verren SE, Reiter DM, Remaut H. 2018. The Role of Functional Amyloids in Bacterial Virulence. J Mol Biol. 430(20):3657–3684. doi: 10.1016/j.jmb.2018.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vassar PS, Culling CF. 1959. Fluorescent stains, with special reference to amyloid and connective tissues. Arch Pathol. 68:487–498. [PubMed] [Google Scholar]
- Wasmer C, Lange A, Van Melckebeke H, Siemer AB, Riek R, Meier BH. 2008. Amyloid fibrils of the HET-s(218-289) prion form a beta solenoid with a triangular hydrophobic core. Science. 319(5869):1523–6. doi:319/5869/1523. [DOI] [PubMed] [Google Scholar]
- Watson MD, Lee JC. 2019. N-Terminal Acetylation Affects α-Synuclein Fibril Polymorphism. Biochemistry. 58(35):3630–3633. doi: 10.1021/acs.biochem.9b00629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wechalekar AD, Gillmore JD, Hawkins PN. 2016. Systemic amyloidosis. The Lancet. 387(10038):2641–2654. doi: 10.1016/S0140-6736(15)01274-X. [DOI] [PubMed] [Google Scholar]
- Wei G, Su Z, Reynolds NP, Arosio P, Hamley IW, Gazit E, Mezzenga R. 2017. Self-assembling peptide and protein amyloids: from structure to tailored function in nanotechnology. Chem Soc Rev. 46(15):4661–4708. doi: 10.1039/C6CS00542J. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoder MD, Keen NT, Jurnak F. 1993. New Domain Motif: the Structure of Pectate Lyase C, a Secreted Plant Virulence Factor. Science. 260(5113):1503–1507. doi: 10.1126/science.8502994. [DOI] [PubMed] [Google Scholar]