Abstract
Background:
The serotonin transporter (SERT) mRNA was previously reported as a quantitative and pathophysiology-based biomarker of heavy drinking in 5HTTLPR:LL genotype-carriers treated with ondansetron. Here, we further validated the potential use of SERT mRNA for quantitative prediction of recent alcohol consumption (in the absence of treatment) and compared with known biomarkers ethyl glucuronide (EtG) and sulfate (EtS).
Methods:
Binge drinking men and women of European ancestry aged 21–65 years were enrolled in a 12-day, in-patient, randomized, double-blind, crossover study, where three beverage doses (placebo, 0.5g/kg (0.4g/kg), and 1g/kg (0.9g/kg) for men (women)) were administered individually in three four-day periods (experiments), separated by minimum of seven-day washout. Diet, sleep, and physical activity were controlled throughout the inpatient experiments. Twenty-nine participants were randomized to receive beverage doses counterbalancing sequence of treatment and gender within subgroups stratified by SERT genotypes 5HTTLPR:LL+rs25531:AA (LALA) vs 5HTTLPR:LS/SS. Peripheral venous blood was collected daily for (1) quantification of SLC6A4 mRNA (primary outcome measure) using qRT-PCR and (2) plasma EtG and EtS levels using tandem mass-spectrometry.
Results:
The association between administered beverage dose and SERT mRNA from completers of at least one four-day experiment (N=18) assessed by a linear mixed model was not statistically significant. Significant positive associations were found with beverage dose and plasma EtG, EtS and EtG/EtS ratio (beta=5.8, SE=1.2, p<0.0001; beta=1.3, SE=0.6, p=0.023; and beta=3.0, SE=0.7, p<0.0001, respectively; C-statistic for discriminating outcomes were 0.97, 0.8, and 0.92, respectively). Additionally, we observed a sequence effect with greater placebo effect on SERT mRNA when administered during the first experiment (p=0.0009), but not on EtG/EtS measures.
Conclusion:
Larger and more innovative studies addressing effects of placebo, race, gender, and response to treatment with serotonergic agents are needed to fully assess SERT, and possibly other mRNA biomarkers of heavy and binge drinking.
Keywords: Serotonin, mRNA, plasma EtG/EtS, biomarker, binge drinking
INTRODUCTION
Excessive alcohol consumption leads to detrimental health and social consequences through dependence, intoxication, and direct toxic effects on almost every organ system in the human body (Rehm, 2011, Tapia-Rojas et al., 2017, https://www.niaaa.nih.gov/alcohols-effects-health/alcohols-effects-body). Despite recent advances in treatments for alcohol use disorder (AUD), excessive alcohol use remains the third leading preventable cause of death in the U.S. since 2001, accounting for approximately 95,000 annual deaths (https://www.niaaa.nih.gov/publications/brochures-and-fact-sheets/alcohol-facts-and-statistics). Epidemiological studies show a significant positive correlation between the amounts and patterns (severity) of alcohol consumption and harmful health outcomes (Dwivedi et al., 2017, Flensborg-Madsen et al., 2011, Choi and Dinitto, 2011, Casswell et al., 2011). Despite the clinical importance of assessing drinking severity, its evaluation still relies heavily on self-report of alcohol use, which is subject to recall errors or underreporting of drinking behaviors due to social stigma and other factors (Arfer et al., 2018, Cherpitel et al., 2018, Mun et al., 2021). These inaccuracies may result in unanticipated cases of alcohol withdrawal or inaccurate assessment of efficacy of novel treatments in clinical trials. Currently available biomarkers have several drawbacks (Seneviratne and Johnson, 2012); for example, commonly used biological markers (biomarkers), ethyl glucuronide (EtG) and ethyl sulfate (EtS), are better predictors of alcohol exposure and reported to be less informative of quantity or patterns of drinking (Mastrovito and Strathmann, 2020, Helander et al., 2009). Some studies report elevated EtG and/or EtS levels following consumption of non-alcoholic beverages such as non-alcoholic beer and wine (Thierauf et al., 2010, Hoiseth et al., 2010) or incidental exposures to alcohol in hand sanitizers or cosmetic hair products (Reisfield et al., 2011, Muller and Iwersen-Bergmann, 2018), questioning their utility for predicting abstinence. Hence, the development of objective biomarkers to supplement self-report promises a more effective strategy to assess acute or recent drinking (Papas et al., 2016, Howlett et al., 2018).
We previously demonstrated that the serotonin transporter (SERT) mRNA might be a sensitive and quantitative biomarker of drinking severity in individuals with AUD who were carriers of LL genotype of the gene that encodes SERT (SLC6A4) (Seneviratne and Johnson, 2012). Serotonin is released acutely following alcohol consumption and involved in mediating its rewarding effects (Lovinger, 1997), dysphoria or depression, and diminutions in neuroadaptive mechanisms during withdrawal (Koob and Le Moal, 2001). The SERTs regulate the potency of serotonergic signaling via re-uptake of synaptic serotonin into pre-synaptic neurons (Mdawar et al., 2020). Numerous human and animal studies show that the availability of SERTs is regulated by the presence or absence of a 44-bp repeat length in the promoter region (5HTTLPR) of SLC6A4 (long (L) or short (S) alleles, respectively) (Lesch et al., 1997). The 5HTTLPR:L allele is associated with increased transcription levels of SERT mRNA compared to the 5HTTLPR:S allele, both in vitro and in healthy living individuals (van der Zwaluw et al., 2010, Wang et al., 2012, Singh et al., 2010). The SERT mRNA levels in carriers of one or two copies of the 5HTTLPR:L allele diminish with chronic alcohol use, but remain unchanged in homozygous 5HTTLPR:S allele carriers (Johnson et al., 2008). Furthermore, in population genetic analyses, both 5-HTTLPR:L and S alleles are associated with alcohol and other substance use disorders, alcohol use severity, and personality traits related to anxiety, depression, aggression, neuroticism, harm avoidance and disagreeableness (Katsuragi et al., 1999, Hariri and Holmes, 2006, Canli and Lesch, 2007, Cao et al., 2018). These findings collectively provide a solid rationale for testing SERT mRNA levels in 5HTTLPR:LL genotype carriers as a biomarker reflective of a pathophysiological response to alcohol use.
In the present study, we sought to further validate the potential use of SERT mRNA levels in 5HTTLPR:LL genotype carriers as a quantitative biomarker of drinking, addressing limitations in our previous study (Seneviratne and Johnson, 2012). Our previous analysis was conducted using samples collected from an outpatient research population whose drinking data were self-reported. To control for these potentially confounding factors, we employed a randomized, double-blind, placebo-controlled crossover, oral alcohol administration paradigm in a controlled human laboratory setting to test the validity of SERT mRNA as a biomarker. Furthermore, we compared SERT mRNA levels with plasma EtG and EtS, widely used biomarkers of recent alcohol exposure (Mastrovito and Strathmann, 2020).
Materials and METHODS
Participants
The study was conducted at the University of Virginia CARE center (UVA CARE) from August 2013 to March 2014 and at the General Clinical Research Center (GCRC) of the University of Maryland School of Medicine (UMSOM) from October 2015 to August 2019. The study was approved by the institutional review boards at each institution and monitored by a three-member data and safety monitoring board (DSMB). All participants gave written informed consent prior to starting study procedures.
Binge drinking, healthy volunteers were recruited through local outreach efforts. The study was restricted to one racial group to limit potential confounding from genetic differences between ancestral populations particularly in a small sample (Yang et al., 2021, Zhong et al., 2019, Cao et al., 2013). We included Hispanic and non-Hispanic individuals of European ancestry as it was the predominant racial group in the area where the study was started. Binge drinking healthy volunteers, rather than individuals with AUD were enrolled to explore the broader application of SERT mRNA as a biomarker of binge alcohol use in the absence of molecular pathology underlying AUD. Inclusion of both populations would have been ideal for direct comparisons; however, it was outside the scope of this relatively small project. Individuals who responded to advertisements were briefly screened by telephone after obtaining oral consent. Those who qualified were invited for a 4–8-hour in-person screening visit where they provided written informed consent and further screening to establish eligibility for study participation. Inclusion criteria included men and women aged 21 to 65 years; experienced at least one binge drinking episode (≥ 5 (men) or ≥ 4 (women) standard drinks in one sitting (Robbins et al., 2020)) in the past 30 days; Alcohol Use Disorders Identification Test (AUDIT) (Bohn et al., 1995) score of >8 and <15 (men) or >7 and <13 (women) (i.e., meeting criteria for heavy drinking but not for DSM-IV alcohol dependence nor for DSM-5 AUD); good physical health (determined by medical history and physical examination, 12-lead ECG, standard clinical laboratory tests [chemistry panel, CBC, urine analysis]); and negative urine pregnancy test and willingness to use adequate contraception throughout the study (for women with reproductive potential). Exclusion criteria included current or past DSM-IV diagnosis of alcohol and/or other substance dependence (including tobacco/nicotine dependence), any psychiatric disorder as assessed with the Mini International Neuropsychiatric Interview (MINI) version 6.0(Sheehan et al., 1998), current use of prescription or over-the-counter medications that could not be discontinued while in the study, and blood donation within the past eight weeks.
Study Design
This was a randomized, placebo-controlled, double-blind human laboratory trial with a mixed design: parallel groups for genotype and within participant crossover for alcohol dose. Enrolled participants were randomized to receive placebo and two alcohol doses in counterbalanced order, balancing sequence of treatment (i.e., beverage dose) and gender within subgroups stratified by 5HTTLPR genotypes (1) 5HTTLPR:LL + rs25531:AA (LALA), and (2) 5HTTLPR:LS/SS regardless of rs25531 genotypes (S-carriers). The participants, all investigators (except for the statistician and a designated team member), and the staff members who participated in direct participant care (both at UVA CARE and GCRC) were blinded to the beverage dose; participants and staff members who participated in direct participant care were also blinded to the genotype.
Genotyping was carried out at the in-person screen visit using genomic DNA extracted from peripheral venous whole blood (WB) samples according to established procedures used in our previous studies (Seneviratne and Johnson, 2012, Seneviratne et al., 2009). Briefly, both 5-HTTLPR and rs25531 were genotyped by using restriction fragment-length polymorphism method in duplicates. Discrepancies were verified with an additional genotyping assay. Genotype results for eligible participants were then sent to the study statistician, along with information on gender, height, and weight, to generate randomization codes. Participants within each genotype group received three alcohol beverage doses, each dose given in separate, but otherwise identical four-day long (three full-days and two half-days) human laboratory experiments. The three doses were: (1) placebo, (2) 0.5g/kg (men) or 0.4g/kg (women) alcohol (medium-dose that was half the concentration of high-dose), and (3) 1g/kg (men) or 0.9g/kg (women) alcohol (high-dose) that corresponded to binge drinking conditions. The three experiments were counterbalanced using a single 3 × 3 Latin square and the sequence effects was balanced by adding a second diagram-balanced sequence in another group of three participants.
Each experiment consisted of four days: one day prior to beverage administration followed by three consecutive days with daily administration of that experiment’s beverage. As the exact half-life (t1/2) of SERT mRNA was not clear and the t1/2 of median human cell mRNA was reported to be about 10h (Yang et al., 2003), it was possible that daily alcohol consumption (<5 t1/2) could have cumulative effects on SERT mRNA expression levels. Therefore, to better understand exposure–response relationship of SERT mRNA biomarker, we administered three identical repeated doses rather than independent measures of the three tested doses. The beverage free starting day of each experiment will be referred to as “baseline”, and a beverage administration day within an experiment as a “session”. Thus, each experiment consisted of a baseline and three sessions. Participants received the same beverage dose on all three sessions within an experiment. Beverage dose differed between experiments.
Experiments were separated by a minimum of seven days living in the community, during which there were no restrictions on alcohol or other substance use (except for the 72-hour period before the next experiment). This duration allowed for washout between experiments (more than five t1/2 of median human cell mRNA (t1/2=10h) (Yang et al., 2003) and alcohol (t1/2= 4–5h) (Miller, 2013). Each participant had an initial familiarization procedure 72 hours prior to their first experiment. They were met in-person or contacted over the phone 72 hours before subsequent experiments to encourage abstinence from alcohol and other drugs, over-the-counter medications, vitamins and supplements until admission to minimize external effects on baseline SERT mRNA expression levels. Participants who had a positive urine drug screen (UDS) or above zero breath alcohol concentration (BrAC) reading upon admission were either rescheduled or withdrawn from the study. The study procedures during experiments are listed in Supplementary Figure 1.
We used commercially available 5% beer (Budweiser, Anheuser-Busch, Saint Louis, MO) for the high-dose alcohol beverage and non-alcoholic beer O’Douls (0.2% wv alcohol content), with a similar taste, aroma, and texture, for the placebo alcohol beverage (Anheuser-Busch, Saint Louis, MO)(https://www.bloomberg.com/profile/company/0812545D:US, February 5, 2017.). The medium-dose beverage was prepared by mixing O’Douls with the 5% beer in volumes adjusted for a participant’s height, weight, and gender. On each session, participants were served a cold beverage (allocated for that experiment) every 15 minutes for two hours in cups with identical appearance. The total volume consumed in each session was the same for all three doses within a participant but differed between participants based on their height, weight, and gender. At admission to a participants’ first experiment, they were asked to choose a daily meal plan (from a list of few choices to reduce variability), which was kept unchanged throughout the study period. A total of 24 mL of WB per day was drawn into three sealed glass collection tubes containing acid citrate dextrose (ACD) buffer (Vacutainer®, Becton-Dickinson, Franklin Lakes, NJ) from each participant at baseline and 17.5 to 18h after the end of each drinking session to account for late-onset expression of genes that reportedly occurs after about 16h (Bhattacharya et al., 2018, Garret L. Yount, 1994, Saban et al., 2001). Blood samples were promptly stored at 4°C and transported to the genomics lab within two to three hours. At the genomics lab, blood samples were centrifuged promptly to separate plasma and white blood cell (WBC) components from WB as follows: WB samples were centrifuged using an Allegra-6R centrifuge from Beckman Coulter (Beckman Coulter Inc., California, United States), for 10 minutes at 912.3 RCF. The plasma layer was then transferred carefully to cryovials (multiple aliquots each containing up to 2 mL each) without agitating the buffy coat and the underlying WBC layer and preserved at −80°C until use. The remaining red blood cell (RBC) and WBC layers were transferred to tubes containing RBC lysis solution for further isolation of WBC as we reported previously (Seneviratne and Johnson, 2012).
At discharge (end of an experiment), participants were educated about the possibility of developing withdrawal symptoms upon sudden cessation of high-dose alcohol consumption.
Allocation concealment:
Upon receipt of de-identified participant information, the study statistician generated the randomization codes and emailed it only to the designated study team member (Research assistant; RA) involved in beverage dose preparation, prior to a participant’s admission for the first experiment. The randomization code was kept in an envelope in a locked drawer, accessible only to the RA. The volumes of beverages (regular or non-alcoholic beer) required for all three experiments for an individual participant were also calculated by the statistician and included in the email sent to the RA with randomization codes. Both regular and non-alcoholic beer needed for all three experiments for a given participant were purchased on the same day in equal quantities from a local liquor store after scheduling a participant to begin the first experiment. Beer cases were stored in a locked cabinet at the GCRC with access limited to the RA. Access to the room was restricted to others on session days when beverage doses were prepared by the RA. Once prepared (30 minutes prior to dosing), the pitchers with study beverage were stored in the refrigerator in the procedure room that was accessible to staff members providing care.
Total RNA extractions and qRT-PCR assays
Total RNA was isolated from WBC using Macherey-Nagel’s NucleoSpin® miRNA kit and RNA/DNA Buffer kit (Takara Bio USA, Inc. Doral, Fl, USA) according to manufacturer’s recommendations. All total RNA samples from an individual participant were processed simultaneously to synthesize cDNA and subsequently in quantitative reverse transcription polymerase chain reaction (qRT-PCR) to minimize inter-assay variability. Inter-plate controls and two housekeeping genes (GAPDH and MPP1) were used to normalize comparative threshold (Ct) values from qRT-PCR measurements. Conditions used for cDNA synthesis and qRT-PCR reactions were as previously described (Seneviratne and Johnson, 2012).
Determination of EtG and EtS plasma concentrations
We supplemented EtG readings with EtS and calculated EtG/EtS ratios, as bacteria present in urine samples have been shown to generate EtG in vitro but not EtS (Walsham and Sherwood, 2012, Mastrovito and Strathmann, 2020), although it was unlikely that aseptically collected, prepared, and cryopreserved plasma samples would contain bacteria. A liquid chromatography tandem mass spectrometric method was developed for the simultaneous determination of EtS and EtG in plasma (Bakhireva et al., 2019, Stefanak et al., 2020) and used for the present sample analysis. Deuterated internal standards were used for quantification. A Dionex U3000 UHPLC (Dionex, Sunnyvale, CA) coupled to a Thermo TQS Altis Triple Quadrupole Mass Spectrometer ((Thermo Scientific, San Jose, CA) operated in a negative ion mode was used for the analysis with m/z transitions of 124.9 to 96.9 for EtS; m/z 221.0 to 75.1 for EtG; m/z 129.9 to 97.9 for EtS_d5 and m/z 226.1 to 75.0 for EtS_d5. Analyte quantification was achieved in the concentration range of 5–10,000 ng/mL of EtS and EtG in plasma. Lowest limit of quantification (LLOQ) was set at 5 ng/mL for both analytes. Sample extraction was carried out as described in the earlier report (Bakhireva et al., 2019). Briefly, 10 μL of plasma samples were extracted with 200 μL of the internal standard working solution prepared in methanol. The samples were shaken for 2 minutes then centrifuged at 15,000 RPM for 5 minutes at 5°C. Supernatant was collected and evaporated to dryness. Residue was reconstituted in 20003B0043L of 0.1% formic acid and acetonitrile (60:40, v/v) and a 5 μL aliquot was injected. The extracted samples were chromatographed on a Zorbax SB-C18 (4.6 × 100 mm, 3.5 μm; Agilent Technologies, Santa Clara, California) using 0.1% formic acid in water (A) and acetonitrile (B) as mobile phase with a 5 minutes gradient program. The gradient elution started at 40% of B until 1.5 minutes, ramped up to 100% of B in 2.6 minutes, and then held at 100% of B till 3.5 minutes and brought back to 40% of B in 4 minutes. The flow rate was set at 0.4 mL/min. The retention times for EtS and EtG and their corresponding internal standards were 2.235 minutes and 2.05 minutes, respectively.
Power analysis
In our previous pilot study, we estimated a partial correlation coefficient of 0.427 between drinking intensity (drinks per drinking day (DDD)) and SERT mRNA expression levels after controlling for age, gender, time (in weeks), and baseline mRNA expression levels in 5-HTTLPR:LALA genotype carriers who received ondansetron (Seneviratne and Johnson, 2012); We therefore assumed a similar partial correlation when the present study was proposed. Since the proposed study was a cross-over study design, participants were randomly assigned to receive a sequence of three different beverage doses. In our power analysis, we assumed that there was no correlation between three different beverage doses within participants because of at least 7-days washout period between different beverage doses, thus, we would have three ‘independent’ observations for each participant. Based on the approximation normality of Fisher’s Z transformation of the sample correlation coefficient, with a type I error of 0.05, 18 participants in each genotype group (a total of 36 participants for both genotype groups, thus a total of 108 observations (36 participants*3 doses)) was deemed sufficient to achieve greater than 90% statistical power to detect such correlation. Since each level of beverage dose was administered for three days (three repeated measures, thus a total of 324 observations (36 participants*3 doses*3 repeated measures)), we employed a linear mixed-effects regression in our final analysis to account for correlations not only among the observations within the same participant, but also among the repeated measures over three days on a specific beverage dose that was administrated to a participant. Therefore, our initial study design adopted a conservative threshold that likely had even greater statistical power to detect such correlations.
Statistical Analysis
Criterion validity of SERT mRNA:
mRNA expression level ratios between SLC6A4 and GAPDH were calculated for each time point. The mRNA ratios (response variable) were utilized in a linear mixed model. As each measurement point was from the same person, the assumption of independence was violated. Thus, mixed linear model was used where the correlation between the repeated measurements was captured by including participants as a random variable; independent variables included baseline mRNA, age, peak BrAC at each session day as continuous variables; and gender, genotype for SLC6A4, beverage dose (i.e., placebo, high- or medium-dose), session day (1, 2, and 3), experiment (1, 2, and 3) and interaction between session day and experiment as fixed effects. Missing values were handled using listwise deletion.
Criterion validity of EtG and EtS:
We used all EtG and EtS values above LLOQ in three generalized linear mixed-effects models to assess associations between beverage doses and EtG, EtS, and EtG/EtS ratios. The models were adjusted for time, experiments, age, and gender. The Kenward-Roger approximation (Kenward and Roger, 1997) was used for estimating the number of degrees of freedom. A C-index or area under the receiver operative characteristic (ROC) curve (Wu, 2019) was utilized as a measure of overall predictive discrimination, defined as the ability to separate participants with detectable EtG, EtS or EtG/EtS ratios, from those with undetectable values. A C-index of 0.5 indicates no discrimination power, whereas a C-index of 1.0 indicates perfect discrimination (Harrell Jr., 2015). To control type I error rate due to multiple comparisons, Tukey-Kramer adjustment (Tukey, 1949) was used for comparisons of primary interest. The overall significance level was specified at 0.05. All analyses were performed in SAS 9.4 (SAS Institute, Inc., Cary NC).
Criterion validity of BrAC:
The BrAC readings recorded immediately prior to and following each 2h-drinking session were used to calculate mean area under peak BrAC time-curves. Therefore, BrAC data from all drinking sessions (up to 12) per individual were included in the analysis.
RESULTS
Population
Data on participant recruitment, randomization, and study completion are presented in Figure 1. The two genotype subgroups did not differ significantly in any baseline characteristics (Table 1).
Figure 1:
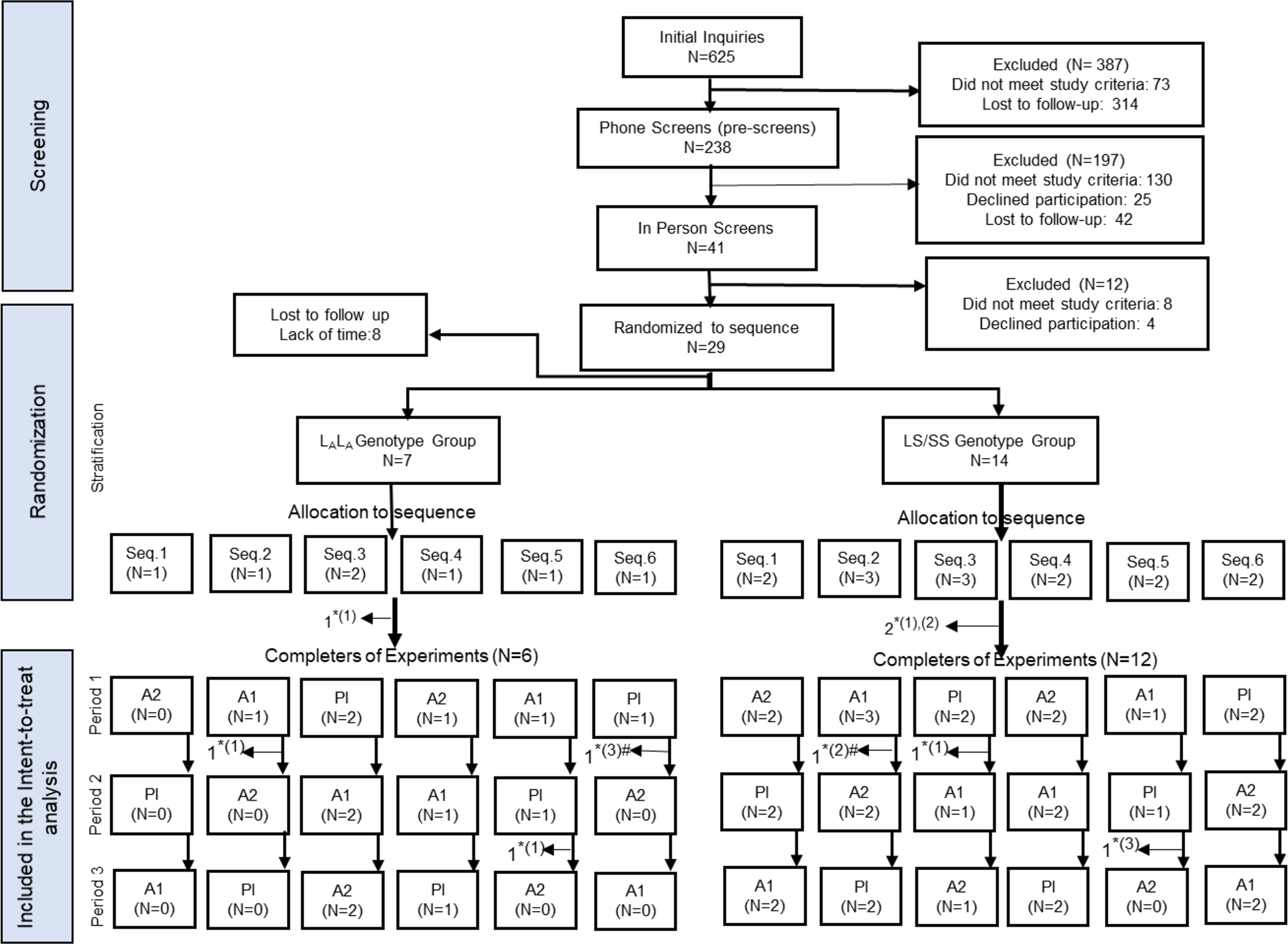
Consort flow diagram of screening, intake, stratification, randomization, and treatment procedures.
Pl = placebo; A1= medium-dose; A2=high alcohol dose; Experiment 1–3 = three human laboratory experiments that were identical in every way except for the administered beverage dose; Seq. 1–6= 1 through 6 sequences of dose allocation; *Lost to follow up due to, (1) lack of time to continue with participation, (2) excluded due to non-compliance, and (3) developed study-unrelated adverse events; #Partially completed the subsequent experiment prior to being excluded. The partial data from incomplete experiments were included in intent-to-treat analyses.
Table 1:
Baseline Participant Characteristics
| Characteristics | Population (N=18) | 5HTTLPR:LALA genotype N=6 (33%) | 5HTTLPR:LS/SS genotype N=12 (67%) | p-value |
|---|---|---|---|---|
|
| ||||
| Age Mean (SD) | 27 (6.9) | 26.7 (6.56) | 27.5 (7.61) | 0.81 |
|
| ||||
| Male (No., %) | 12 (67%) | 4 (67%) | 8 (67%) | |
|
| ||||
| Average BMI (SD) | 24.9 (3.6) | 26.6 (1.85) | 24.1 (4.02) | 0.10 |
|
| ||||
| Educational Level | ||||
| ≥16 Years (No., %) | 14 (78%) | 4 (67%) | 10 (83%) | |
| <16 Years (No., %) | 4 (22%) | 2 (33%) | 2 (17%) | |
|
| ||||
| Average AUDIT Score (SD) | 8.6 (2.3) | 8.7 (1.03) | 8.5 (2.81) | 0.86 |
|
| ||||
| *Past 30 Days Drinking Measures | ||||
| Average DDD (SD) | 4.7 (1.8) | 5.6 (1.2) | 4.3 (1.92) | 0.11 |
| Average Binge Episodes (SD) | 7.3 (4.7) | 8.67 (3.27) | 6.67 (5.25) | 0.34 |
|
| ||||
| **Past 90 Days Other Drug Use | ||||
| Nicotine (No.) | 4 | 2 | 2 | |
| Marijuana (No.) | 7 | 2 | 5 | |
| Cocaine (No.) | 1 | 0 | 1 | |
|
| ||||
| †Average SERT mRNA levels (SD) | 1.1795 (0.08) | 1.2103 (0.07) | 1.1654 (0.08) | 0.06 |
|
| ||||
| EtG Levels: | ||||
| ≥LLOQ (No. of participants, %) | 1 (2%) | 1 (7%) | 0 (0%) | |
| 0-LLOQ (No. of participants, %) | 3 (7%) | 2 (14%) | 1 (3%) | |
| EtS Levels: | ||||
| ≥LLOQ (No. of participants, %) | 5 (11%) | 2 (14%) | 3 (9%) | |
| 0-LLOQ (No. of participants, %) | 1 (2%) | 0 (0%) | 1 (3%) | |
BMI – body mass index
calculated using standard drinks consumed in the 30 days prior to initial in-person screen
Number of participants who used other drugs (excluding alcohol) in the 90 days prior to initial in-person screen
all SERT mRNA levels were normalized by GAPDH mRNA (housekeeping gene) expression levels
SD – standard deviation.
Daily SERT mRNA readings
Table 2 presents results from linear mixed-effects model consisting of age, gender, genotype (HTTLPR:LALA (33%) or 5HTTLPR:LS/SS (67%)), study day (sessions within an experiment), sequence of experiments, and the interaction with alcohol dose on daily changes in SERT mRNA levels. As shown in Table 2 and Figure 2, the sequence of alcohol dose administered to a participant across the three experiments had a significant effect on SERT mRNA expression levels (p=0.003). The alcohol dose and the sequence of its administration interacted significantly to influence SERT mRNA levels (p=0.0002). Placebo effects were greatest when administered on the first of the three experiments (i.e., experiment 1) for each participant, whereas alcohol had greater effects when administered during second or third experiments (p=0.0167, p=0.0156 and p<0.0001, respectively, for placebo, medium- and high-doses). However, SERT mRNA levels did not differ significantly among alcohol dose groups: placebo vs. medium-dose = 0.0189 (SE= 0.0125; t-value = 1.51; p= 0.3084); placebo vs. high-dose = −0.0002 (SE= 0.0115; t-value = −0.02; p= 0.9998); medium- vs. high-dose = −0.0191 (SE= 0.0125; t-value = −1.53; p= 0.2993). Daily SERT mRNA levels are presented in Supplementary Figure 2.
Table 2:
Linear mixed effects model of daily SERT mRNA levels across all dose groups
| Effect | DF | F Value | P-value |
|---|---|---|---|
| Baseline mRNA | 1 | 11.48 | 0.001 |
| Age | 1 | 1.98 | 0.1626 |
| Gender | 1 | 0.23 | 0.6309 |
| Session day | 2 | 0.55 | 0.5776 |
| Alcohol dose | 2 | 0.04 | 0.964 |
| Genotype | 1 | 2.09 | 0.1513 |
| BrAC | 1 | 1.33 | 0.2526 |
| Experiment | 2 | 6.19 | 0.003 |
| Alcohol dose *Experiment | 4 | 6.20 | 0.0002 |
The P-values that remained significant after correction for multiple testing are shown in bold letters. Same beverage dose was administered on all 3 session days within an experiment.
Figure 2:
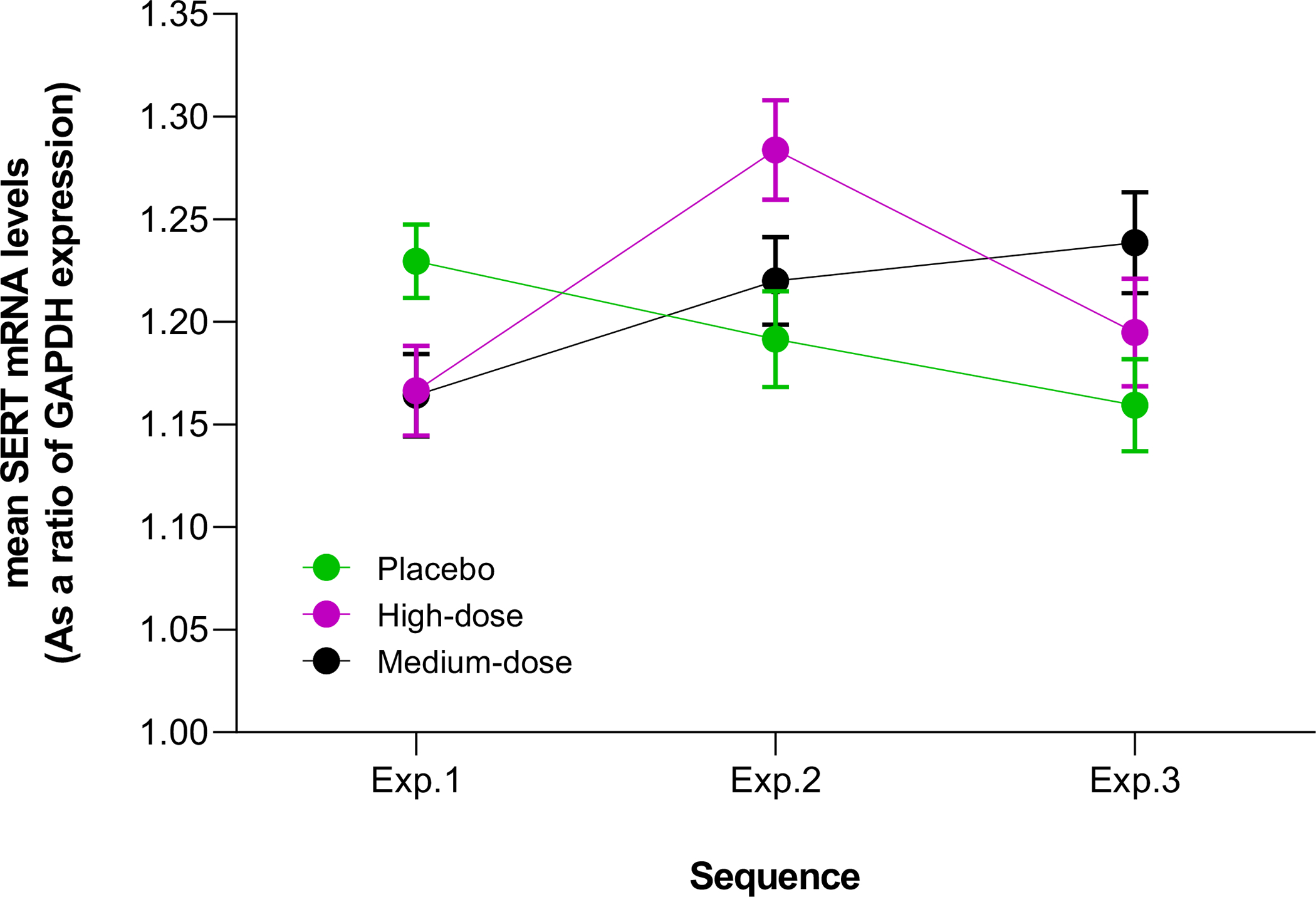
The effect of sequence of beverage dose administration across the three experiments on average daily SERT mRNA levels.
Error bars represent standard errors of mean. The F-values (variance of the group means) for interactions between beverage dose and the experiment in which the dose was administered (i.e., in experiment 1/ experiment 2/ experiment 3) on SERT mRNA levels within each dose group are: placebo = 4.28 (p = 0.0167); middle-dose = 4.35 (p= 0.0156); and high-dose = 10.21 (p<0.0001).
Daily EtG/EtS readings
On all 48 days that placebo was administered across 16 participants, none had EtG levels ≥LLOQ; one participant had EtG levels between zero and LLOQ (0-LLOQ) on a placebo administered day, and another participant had 0-LLOQ at baseline prior to placebo administration. Of the high-dose days across 12 participants, one had a missing data point and 94.3% days had EtG ≥LLOQ; two days had values between 0 and LLOQ and indeterminate EtG levels. The two days with <LLOQ were distributed between two individuals. Two individuals who had ≥LLOQ on all high-dose days also had detectable, but <LLOQ, EtG levels at baseline.
The distribution of EtG levels for the medium-dose alcohol was 14.894% days for ≥LLOQ and 21.273% days for 0-LLOQ. The distribution of EtS levels was ≥LLOQ on 37.14% days with high-dose, 6.38% days with medium-dose, and 6.25% days with placebo. Results from regression models for EtG, EtS and EtG/EtS ratios are presented in Tables 3A and 3B. There were significant associations between the beverage dose and EtG or EtS or EtG/EtS ratio (beta=5.8, se=1.2, p<0.0001, C-statistic=0.97; beta=1.2, se=0.6, p=0.023, C-statistic=0.80; beta=3.0, se=0.7, p<0.0001, C-statistic=0.92; respectively, Table 3A). EtG/EtS ratios were indeterminate at baseline across all dose groups, as EtG and/or EtS values were zero.
Table 3A:
Adjusted association between alcohol dose and EtG or EtS or EtG/EtS ratio
| Analyte (outcome) | Estimated Coefficient | Standard Error | p-Value | C-index for discriminating the outcomes |
|---|---|---|---|---|
| EtG | 5.7792 | 1.1985 | <.0001 | 0.97 |
| EtS | 1.3384 | 0.5794 | 0.0226 | 0.80 |
| EtG/EtS ratio | 3.0457 | 0.6549 | <.0001 | 0.92 |
EtG and EtS measures ≥LLOQ were considered as positive; EtG/EtS ratios were calculated with any detectable EtG and EtS measures and considered as binary measures; All models were adjusted for age, gender, study days, and experiment sequence of beverage dose administration.
Table 3B:
Effects of beverage doses in probability of detecting EtG, EtS and EtG/EtS ratio
| Comparison | Odds Ratio | Tukey-Kramer adjusted 95% CI | Adjusted p-Value |
|---|---|---|---|
| EtG: high- vs. medium-dose treated days | 68.2 | 6.2 to 748.6 | 0.003 |
| EtG: medium- vs. placebo-dose treated days | 15.2 | 2.7 to 87.3 | 0.009 |
| EtG: high- vs. placebo-dose treated days | N/A* | ||
| EtS: high- vs. medium-dose treated days | 10.79 | 1.33 to 87.12 | 0.021 |
| EtS: medium- vs. placebo-dose treated days | 0.79 | 0.06 to 9.65 | 0.972 |
| EtS: high- vs. placebo-dose treated days | 8.50 | 1.07 to 67.65 | 0.042 |
| EtG/EtS ratio: high- vs. medium-dose treated days | 19.0 | 2.0 to 179.0 | 0.019 |
| EtG/EtS ratio: medium- vs. placebo-dose treated days | N/A** | ||
| EtG/EtS ratio: high- vs. placebo-dose treated days | N/A** |
Not available as EtG was not traceable on most placebo treated days.
Not available as either EtG or EtS or both analytes were not traceable on most days to calculate EtG/EtS ratios
BrAC readings
The BrAC readings were zero for all participants immediately prior to alcohol administration sessions. The mean area under peak BrAC time-curves, calculated with BrAC readings taken immediately following the 2-hour -alcohol drinking sessions were as follows: Placebo = 0.0041g/210L (95% CI= −5.813e-006 to 0.0082 g/210L); medium-dose = 0.1365 g/210L (95% CI= 0.0970 to 0.1760 g/210L); high-dose = 0.3017 g/210L (0.2315 to 0.3719 g/210L). The BrAC readings across sessions and experimental sequences are illustrated in Supplementary Figures 3A and 3B.
DISCUSSION
The 5HTTLPR L/S alleles of the SLC6A4 gene have been studied in numerous association analyses examining the role of the serotonergic system in alcohol use, dependence, and treatment response to serotonergic agents (Oo et al., 2016, Hollerbach et al., 2021, Villalba et al., 2015, Johnson et al., 2011, Kenna et al., 2014b). Some studies have demonstrated modulatory effects of 5HTTLPR L/S alleles on SERT mRNA expression levels (Prasansuklab et al., 2011, Philibert et al., 2008, Bradley et al., 2005). The goal of the present study was to assess the possibility of utilizing SERT mRNA as a quantitative biomarker to predict daily alcohol consumption. Our findings from 18 healthy binge drinkers of European ancestry who participated in a crossover, double-blind alcohol administration paradigm in a controlled environment indicated that SERT mRNA levels were not predictive of daily alcohol consumption levels, either within the 5HTTLPR:LALA genotype group or in the combined population. In contrast, widely evaluated biomarkers of plasma EtG, EtS, and EtG/EtS ratios derived from the same samples demonstrated excellent predictive power for discriminating the outcomes (C-statistics ≥ 0.80).
The outpatient pilot study that we previously conducted showed a >50% increase in SERT mRNA levels per unit of standard drink on a drinking day in the ondansetron treated 5HTTLPR:LL genotype carriers compared to the placebo arm (Seneviratne and Johnson, 2012). There are several methodological differences between the pilot and the present study; in the pilot study, (1) drinking levels were self-reported, (2) the mRNA measurements were collected at four-week intervals from baseline for a total of 12 weeks, as opposed to three-day drinking sessions (per dose) in the current study, (3) the participants were heavy drinking alcohol-dependent outpatients whose lifestyle and environmental factors were not controlled, and (4) the duration of alcohol exposure was potentially heavier and continuous, rather than daily two-hour sessions with maximum of five to six standard drinks in the present study. Nonetheless, the absence of significant association observed in the present study is comparable to the lack of effect of drinking intensity on SERT mRNA levels that we observed within the placebo arm of the pilot project. The absence of statistically significant alterations under our stringent conditions suggests that the changes in SERT mRNA observed in the ondansetron treated group in the pilot project may have resulted from ondansetron’s pharmacologic modulation of 5HTTLPR genotype-based SERT mRNA expression levels, rather than biochemical effects of alcohol alone. These observations are consistent with findings from studies that tested SERT mRNA as biomarkers of treatment response to other serotonergic agents under other conditions. SERT mRNA were biomarkers of response to duloxetine and paroxetine in improvement of Hamilton depression rating scale score (Kao et al., 2018), and lamotrigine tested in in vitro human lymphoblastoid cell lines (D’Souza et al., 2013), albeit with the caveat that these studies were not placebo-controlled.
The present study found a significant sequence effect: placebo effects were greatest when administered on the first of the three experiments (experiment 1) for each participant, whereas, alcohol had a greater effect on the two subsequent experiments. These sequence effects on SERT mRNA levels were likely due to biological rather than methodological factors, such as: (1) The t1/2 of alcohol is four to five hours (Miller, 2013), so the minimum washout period of seven days (33 half-lives) between sessions was more than adequate to ensure no carry-over alcohol present at the succeeding baseline. Furthermore, all participants were required to remain abstinent starting 72h prior to each experiment and absence of carry-over effects of alcohol was confirmed by the retrospective analysis of baseline plasma EtG levels that were below the range of quantification (<5 ng/mL) in all participants except for one participant with a marginal EtG level of 5.389 ng/mL at baseline of the middle-dose experiment. The untraceable amounts of alcohol detected by breathalyzer readings measured immediately prior to alcohol administration sessions also support the absence of carry-over effects. The t1/2 of SERT mRNA is not clearly defined (Baudry et al., 2019), so we do not know for certain whether the minimum seven-day washout periods were sufficient to avoid carry-over. However, the baseline SERT mRNA levels during each experiment were not statistically significantly different from each other suggesting an absence of carryover effect; (2) beverage doses were assigned to experiments in blinded, counterbalanced order; (3) all study conditions and procedures, including preparation of beverage doses, were identical across participants and sessions; and (4) We did not detect a sequence effect on the area under BrAC time-curve, EtG or EtS levels across the 3 experiments, and none of the placebo session days yielded EtG levels above LLOQ.
To our knowledge, this is the first study to analyze daily EtG and EtS measures in participants in a controlled environment receiving repeated daily alcohol administration at doses mirroring binge drinking patterns. Thus, our results add new information to the existing body of literature on plasma EtG and EtS levels as biomarkers of binge drinking. Previous studies found that EtG and EtS are measurable in plasma from a few minutes up to a few days after ingestion of alcohol (Kwon et al., 2019, Hoiseth et al., 2009, Shukla et al., 2017). Since there is no recommended ideal timeline for plasma/serum sampling, we measured EtG and EtS levels at 17.5–18h to coincide with SERT mRNA measures for direct comparisons of the biomarkers with each other. Our data showed that both EtG and EtS levels in plasma up to 18h after cessation of drinking had excellent ability to discriminate high doses of alcohol, based on the percentage of session days when EtG and EtS were detectable above LLOQ. However, the range of absolute amounts of EtG and EtS overlapped between the middle- and high-doses. Furthermore, the levels of EtG and EtS detected in response to an identical daily dose varied within the same individual, without any observable pattern that suggested carryover effects from consecutive daily dosing. These findings confirm the previously reported findings that EtG and EtS are biomarkers of alcohol exposure, rather than quantitative intake.
This study has several limitations. First, the small sample size of 18 participants rendered the study underpowered to fully assess the interactive effects of alcohol doses and 5HTTLPR genotypes on SERT mRNA levels, even though the highly controlled within-participant, inpatient study design minimized between-participant genetic heterogeneity and environmental confounders. Second, the placebo effects were not analyzed systematically, limiting our ability to interpret the biological underpinnings of response to placebo, particularly related to the sequence effects. As the participants knew that they would receive a placebo in one of the three experiments, although they (and study team) were blinded to the specific details, it is possible that the expectancy of receiving a regular alcohol dose during the third experiment diminished after experiencing alcohol during the first and/or second experiments, based on a participant’s prior experience in consuming alcohol. There are no published studies on how or whether expectancy of receiving alcohol influences SERT mRNA expression levels. As we did not measure expectancy in this study, the underlying mechanisms are only hypothetical. Nonetheless, our hypothesis is supported by the evidence of expectancies playing a significant role in response to treatment of depression with SSRIs (Faria et al., 2017, Rief et al., 2009, Kirsch, 2017) and the lack of sequence effects that we observed on EtG/EtS that are direct biomarkers originating from alcohol metabolism. Third, we did not collect any indices of brain SERT mRNA expression changes to correlate with the changes detected in peripheral WBC. It will be intriguing to know how or if psychological (i.e., placebo) effects can influence SERT mRNA levels in peripheral blood cells. Fifth, a serial quantification of SERT mRNA levels starting immediately after the binge drinking episode would have been more informative than a single time point (17.5–18h post-cessation of drinking) that presumably captured late-onset gene expression that may occur about 16h after exposure. Fifth, our population was entirely of European ancestry and received a single type of beverage (i.e., beer) for a limited period (two hours) each day. Generalizability of these findings to other racial groups with an equal gender distribution particularly as an interaction of 5HTTLPR and DRD4 polymorphisms has been observed in women (Kenna et al., 2014a) and testing of different alcohol beverages for longer durations are all warranted to better understand the utility of SERT mRNA as a biomarker of alcohol consumption.
In summary, our findings are in line with the previous evidence that binge drinking is not associated with significant alterations in peripheral blood cell SERT mRNA levels that would allow their use as a quantitative biomarker of recent alcohol consumption, at the least in healthy binge drinkers at the tested doses. More comprehensive studies addressing the above-mentioned limitations are required to accurately assess the use of SERT mRNA as a biomarker for recent alcohol consumption. Future research should systematically assess whether alcohol may alter SERT mRNA levels differentially in those with AUD under similar conditions, or whether SERT mRNA are simply treatment response biomarkers of pharmacologic agents or behavioral interventions acting on the serotonergic system.
Supplementary Material
ACKNOWLEDGMENTS
Authors thank the staff members at UVA CARE center and the University of Maryland General Clinical Research Center (GCRC), Latrice Linton and John Sivinski for their assistance with participant recruitment, enrollment and conduct of screening, laboratory procedures, and data entry.
FUNDING AND DISCLOSURE
The research was supported by the NIAAA grants K23AA020899 and R01AA026291 (to CS), “micro-grants (vouchers)” to CS through the UMB Clinical Translational Research Initiative (ICTR program) that partially facilitated human laboratory procedures and statistical analyses through CTSA grant 1UL1TR003098, and in part by the University of Maryland Baltimore, School of Pharmacy Mass Spectrometry Center (SOP1841-IQB2014). The funding agencies had no further role in study design; in the collection, analysis, and interpretation of data; in the writing of the report; and in the decision to submit the paper for publication. JC, AC, XQW, JCF, HC, NAD, DAG, NP, MAK and CS have nothing to disclose. BAJ is founder and chief medical officer of Adial Pharmaceuticals.
Footnotes
ClinicalTrials.gov Identifier: NCT02315885
REFERENCES:
- ARFER KB, TOMLINSON M, MAYEKISO A, BANTJES J, VAN HEERDEN A & ROTHERAM-BORUS MJ 2018. Criterion validity of self-reports of alcohol, cannabis, and methamphetamine use among young men in Cape Town, South Africa. Int J Ment Health Addict, 16, 45–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- BAKHIREVA LN, KANE MA, BEARER CF, BAUTISTA A, JONES JW, GARRISON L, BEGAY MG, OZECHOWSKI T & LEWIS J 2019. Prenatal alcohol exposure prevalence as measured by direct ethanol metabolites in meconium in a Native American tribe of the southwest. Birth Defects Res, 111, 53–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- BAUDRY A, PIETRI M, LAUNAY JM, KELLERMANN O & SCHNEIDER B 2019. Multifaceted Regulations of the Serotonin Transporter: Impact on Antidepressant Response. Front Neurosci, 13, 91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- BHATTACHARYA B, CHATTRERJEE S, BERLAND R, PICHUGIN A, GAO Y, CONNOR J, IVANOV A, YAN B-S, KOBZIK L & KRAMNIK I 2018. Increased susceptibility to intracellular bacteria and necrotic inflammation driven by a dysregulated macrophage response to TNF. bioRxiv, 238873. [Google Scholar]
- BOHN MJ, BABOR TF & KRANZLER HR 1995. The Alcohol Use Disorders Identification Test (AUDIT): validation of a screening instrument for use in medical settings. J Stud Alcohol, 56, 423–32. [DOI] [PubMed] [Google Scholar]
- BRADLEY SL, DODELZON K, SANDHU HK & PHILIBERT RA 2005. Relationship of serotonin transporter gene polymorphisms and haplotypes to mRNA transcription. Am J Med Genet B Neuropsychiatr Genet, 136B, 58–61. [DOI] [PubMed] [Google Scholar]
- CANLI T & LESCH KP 2007. Long story short: the serotonin transporter in emotion regulation and social cognition. Nat Neurosci, 10, 1103–9. [DOI] [PubMed] [Google Scholar]
- CAO H, HARNEIT A, WALTER H, ERK S, BRAUN U, MOESSNANG C, GEIGER LS, ZANG Z, MOHNKE S, HEINZ A, ROMANCZUK-SEIFERTH N, MUHLEISEN T, MATTHEISEN M, WITT SH, CICHON S, NOTHEN MM, RIETSCHEL M, MEYER-LINDENBERG A & TOST H 2018. The 5-HTTLPR Polymorphism Affects Network-Based Functional Connectivity in the Visual-Limbic System in Healthy Adults. Neuropsychopharmacology, 43, 406–414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- CAO J, HUDZIAK JJ & LI D 2013. Multi-cultural association of the serotonin transporter gene (SLC6A4) with substance use disorder. Neuropsychopharmacology, 38, 1737–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- CASSWELL S, YOU RQ & HUCKLE T 2011. Alcohol’s harm to others: reduced wellbeing and health status for those with heavy drinkers in their lives. Addiction, 106, 1087–94. [DOI] [PubMed] [Google Scholar]
- CHERPITEL CJ, YE Y, STOCKWELL T, VALLANCE K & CHOW C 2018. Recall bias across 7 days in self-reported alcohol consumption prior to injury among emergency department patients. Drug Alcohol Rev, 37, 382–388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- CHOI NG & DINITTO DM 2011. Heavy/binge drinking and depressive symptoms in older adults: gender differences. Int J Geriatr Psychiatry, 26, 860–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D’SOUZA UM, POWELL-SMITH G, HADDLEY K, POWELL TR, BUBB VJ, PRICE T, MCGUFFIN P, QUINN JP & FARMER AE 2013. Allele-specific expression of the serotonin transporter and its transcription factors following lamotrigine treatment in vitro. Am J Med Genet B Neuropsychiatr Genet, 162B, 474–83. [DOI] [PubMed] [Google Scholar]
- DWIVEDI AK, CHATTERJEE K & SINGH R 2017. Lifetime alcohol consumption and severity in alcohol dependence syndrome. Ind Psychiatry J, 26, 34–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- FARIA V, GINGNELL M, HOPPE JM, HJORTH O, ALAIE I, FRICK A, HULTBERG S, WAHLSTEDT K, ENGMAN J, MANSSON KNT, CARLBRING P, ANDERSSON G, REIS M, LARSSON EM, FREDRIKSON M & FURMARK T 2017. Do You Believe It? Verbal Suggestions Influence the Clinical and Neural Effects of Escitalopram in Social Anxiety Disorder: A Randomized Trial. EBioMedicine, 24, 179–188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- FLENSBORG-MADSEN T, BECKER U, GRONBAEK M, KNOP J, SHER L & MORTENSEN EL 2011. Alcohol consumption and later risk of hospitalization with psychiatric disorders: prospective cohort study. Psychiatry Res, 187, 214–9. [DOI] [PubMed] [Google Scholar]
- GARRET L. YOUNT PP, WHITE JEFFREYD, 1994. Pentylenetetrazole-induced seizures stimulate transcription of early and late response genes. Molecular Brain Research, 21, 5. [DOI] [PubMed] [Google Scholar]
- HARIRI AR & HOLMES A 2006. Genetics of emotional regulation: the role of the serotonin transporter in neural function. Trends Cogn Sci, 10, 182–91. [DOI] [PubMed] [Google Scholar]
- HARRELL FE JR. 2015. Regression Modeling Strategies, Switzerland: 2015, Springer, Cham. [Google Scholar]
- HELANDER A, BOTTCHER M, FEHR C, DAHMEN N & BECK O 2009. Detection times for urinary ethyl glucuronide and ethyl sulfate in heavy drinkers during alcohol detoxification. Alcohol Alcohol, 44, 55–61. [DOI] [PubMed] [Google Scholar]
- HOISETH G, MORINI L, POLETTINI A, CHRISTOPHERSEN A & MORLAND J 2009. Blood kinetics of ethyl glucuronide and ethyl sulphate in heavy drinkers during alcohol detoxification. Forensic Sci Int, 188, 52–6. [DOI] [PubMed] [Google Scholar]
- HOISETH G, YTTREDAL B, KARINEN R, GJERDE H & CHRISTOPHERSEN A 2010. Levels of ethyl glucuronide and ethyl sulfate in oral fluid, blood, and urine after use of mouthwash and ingestion of nonalcoholic wine. J Anal Toxicol, 34, 84–8. [DOI] [PubMed] [Google Scholar]
- HOLLERBACH P, OLDERBAK S, WILHELM O, MONTAG C, JUNG S, NEUMANN CS, HABERMEYER E & MOKROS A 2021. Associations of the MAOA uVNTR genotype and 5-HTTLPR/rs25531 haplotype with psychopathic traits. Psychoneuroendocrinology, 131, 105275. [DOI] [PubMed] [Google Scholar]
- HOWLETT H, MACKENZIE S, GRAY WK, RANKIN J, NIXON L, RICHARDSON A, STREHLE EM & BROWN NW 2018. Assessing prevalence of alcohol consumption in early pregnancy: Self-report compared to blood biomarker analysis. Eur J Med Genet, 61, 531–538. [DOI] [PubMed] [Google Scholar]
- HTTPS://WWW.BLOOMBERG.COM/PROFILE/COMPANY/0812545D:US February 5, 2017. “Company Overview of Anheuser-Busch Companies, LLC”. Bloomberg Research. Bloomberg.
- HTTPS://WWW.NIAAA.NIH.GOV/ALCOHOLS-EFFECTS-HEALTH/ALCOHOLS-EFFECTS-BODY.
- HTTPS://WWW.NIAAA.NIH.GOV/PUBLICATIONS/BROCHURES-AND-FACT-SHEETS/ALCOHOL-FACTS-AND-STATISTICS.
- JOHNSON BA, AIT-DAOUD N, SENEVIRATNE C, ROACHE JD, JAVORS MA, WANG XQ, LIU L, PENBERTHY JK, DICLEMENTE CC & LI MD 2011. Pharmacogenetic approach at the serotonin transporter gene as a method of reducing the severity of alcohol drinking. Am J Psychiatry, 168, 265–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- JOHNSON BA, JAVORS MA, ROACHE JD, SENEVIRATNE C, BERGESON SE, AIT-DAOUD N, DAWES MA & MA JZ 2008. Can serotonin transporter genotype predict serotonergic function, chronicity, and severity of drinking? Prog Neuropsychopharmacol Biol Psychiatry, 32, 209–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- KAO WT, CHANG CL & LUNG FW 2018. 5-HTT mRNA level as a potential biomarker of treatment response in patients with major depression in a clinical trial. J Affect Disord, 238, 597–608. [DOI] [PubMed] [Google Scholar]
- KATSURAGI S, KUNUGI H, SANO A, TSUTSUMI T, ISOGAWA K, NANKO S & AKIYOSHI J 1999. Association between serotonin transporter gene polymorphism and anxiety-related traits. Biol Psychiatry, 45, 368–70. [DOI] [PubMed] [Google Scholar]
- KENNA GA, ZYWIAK WH, SWIFT RM, MCGEARY JE, CLIFFORD JS, SHOAFF JR, FRICCHIONE S, BRICKLEY M, BEAUCAGE K, HAASS-KOFFLER CL & LEGGIO L 2014a. Ondansetron and sertraline may interact with 5-HTTLPR and DRD4 polymorphisms to reduce drinking in non-treatment seeking alcohol-dependent women: exploratory findings. Alcohol, 48, 515–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- KENNA GA, ZYWIAK WH, SWIFT RM, MCGEARY JE, CLIFFORD JS, SHOAFF JR, VUITTONET C, FRICCHIONE S, BRICKLEY M, BEAUCAGE K, HAASS-KOFFLER CL & LEGGIO L 2014b. Ondansetron reduces naturalistic drinking in nontreatment-seeking alcohol-dependent individuals with the LL 5’-HTTLPR genotype: a laboratory study. Alcohol Clin Exp Res, 38, 1567–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- KENWARD MG & ROGER JH 1997. Small sample inference for fixed effects from restricted maximum likelihood. Biometrics, 53, 983–97. [PubMed] [Google Scholar]
- KIRSCH I 2017. Response Expectancy and the Response to Antidepressant Medication. EBioMedicine, 25, 13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- KOOB GF & LE MOAL M 2001. Drug addiction, dysregulation of reward, and allostasis. Neuropsychopharmacology, 24, 97–129. [DOI] [PubMed] [Google Scholar]
- KWON M, CHOI HJ, JO YH, SON MH, MIN JS, KIM NY & JUNG JE 2019. Analysis of ethyl glucuronide and ethyl sulfate in blood to determine the absorption or elimination phase of alcohol for Korean. Forensic Sci Int, 302, 109857. [DOI] [PubMed] [Google Scholar]
- LESCH KP, BALLING U, SEEMANN M, TEUFEL A, BENGEL D, HEILS A, GODECK P & RIEDERER P 1997. Molecular heterogeneity of neurotransporters: implications for neurodegeneration. J Neural Transm Suppl, 49, 155–67. [DOI] [PubMed] [Google Scholar]
- LOVINGER DM 1997. Serotonin’s role in alcohol’s effects on the brain. Alcohol Health Res World, 21, 114–20. [PMC free article] [PubMed] [Google Scholar]
- MASTROVITO R & STRATHMANN FG 2020. Distributions of alcohol use biomarkers including ethanol, phosphatidylethanol, ethyl glucuronide and ethyl sulfate in clinical and forensic testing. Clin Biochem, 82, 85–89. [DOI] [PubMed] [Google Scholar]
- MDAWAR B, GHOSSOUB E & KHOURY R 2020. Selective serotonin reuptake inhibitors and Alzheimer’s disease. Neural Regen Res, 15, 41–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MILLER PM 2013. Principles of Addiction: , Academic Press. [Google Scholar]
- MULLER A & IWERSEN-BERGMANN S 2018. Ethyl Glucuronide in Alcoholic Beverages. Alcohol Alcohol, 53, 532–538. [DOI] [PubMed] [Google Scholar]
- MUN EY, LI X, BUSINELLE MS, HEBERT ET, TAN Z, BARNETT NP & WALTERS ST 2021. Ecological Momentary Assessment of Alcohol Consumption and Its Concordance with Transdermal Alcohol Detection and Timeline Follow-Back Self-report Among Adults Experiencing Homelessness. Alcohol Clin Exp Res, 45, 864–876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- OO KZ, AUNG YK, JENKINS MA & WIN AK 2016. Associations of 5HTTLPR polymorphism with major depressive disorder and alcohol dependence: A systematic review and meta-analysis. Aust N Z J Psychiatry, 50, 842–57. [DOI] [PubMed] [Google Scholar]
- PAPAS RK, GAKINYA BN, MWANIKI MM, KETER AK, LEE H, LOXLEY MP, KLEIN DA, SIDLE JE, MARTINO S, BALIDDAWA JB, SCHLAUDT KL & MAISTO SA 2016. Associations Between the Phosphatidylethanol Alcohol Biomarker and Self-Reported Alcohol Use in a Sample of HIV-Infected Outpatient Drinkers in Western Kenya. Alcohol Clin Exp Res, 40, 1779–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- PHILIBERT RA, SANDHU H, HOLLENBECK N, GUNTER T, ADAMS W & MADAN A 2008. The relationship of 5HTT (SLC6A4) methylation and genotype on mRNA expression and liability to major depression and alcohol dependence in subjects from the Iowa Adoption Studies. Am J Med Genet B Neuropsychiatr Genet, 147B, 543–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- PRASANSUKLAB A, POOVORAWAN Y & TENCOMNAO T 2011. Modulation of human serotonin transporter expression by 5-HTTLPR in colon cells. Int J Mol Sci, 12, 6619–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- REHM J 2011. The risks associated with alcohol use and alcoholism. Alcohol Res Health, 34, 135–43. [PMC free article] [PubMed] [Google Scholar]
- REISFIELD GM, GOLDBERGER BA, CREWS BO, PESCE AJ, WILSON GR, TEITELBAUM SA & BERTHOLF RL 2011. Ethyl glucuronide, ethyl sulfate, and ethanol in urine after sustained exposure to an ethanol-based hand sanitizer. J Anal Toxicol, 35, 85–91. [DOI] [PubMed] [Google Scholar]
- RIEF W, NESTORIUC Y, VON LILIENFELD-TOAL A, DOGAN I, SCHREIBER F, HOFMANN SG, BARSKY AJ & AVORN J 2009. Differences in adverse effect reporting in placebo groups in SSRI and tricyclic antidepressant trials: a systematic review and meta-analysis. Drug Saf, 32, 1041–56. [DOI] [PubMed] [Google Scholar]
- ROBBINS T, WEJNERT C, BALAJI AB, HOOTS B, PAZ-BAILEY G, BRADLEY H & GROUP N-YS 2020. Binge Drinking, Non-injection Drug Use, and Sexual Risk Behaviors among Adolescent Sexual Minority Males, 3 US Cities, 2015. J Urban Health, 97, 739–748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- SABAN MR, HELLMICH H, NGUYEN NB, WINSTON J, HAMMOND TG & SABAN R 2001. Time course of LPS-induced gene expression in a mouse model of genitourinary inflammation. Physiol Genomics, 5, 147–60. [DOI] [PubMed] [Google Scholar]
- SENEVIRATNE C, HUANG W, AIT-DAOUD N, LI MD & JOHNSON BA 2009. Characterization of a functional polymorphism in the 3’ UTR of SLC6A4 and its association with drinking intensity. Alcohol Clin Exp Res, 33, 332–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- SENEVIRATNE C & JOHNSON BA 2012. Serotonin transporter genomic biomarker for quantitative assessment of ondansetron treatment response in alcoholics. Front Psychiatry, 3, 23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- SHEEHAN DV, LECRUBIER Y, SHEEHAN KH, AMORIM P, JANAVS J, WEILLER E, HERGUETA T, BAKER R & DUNBAR GC 1998. The Mini-International Neuropsychiatric Interview (M.I.N.I.): the development and validation of a structured diagnostic psychiatric interview for DSM-IV and ICD-10. J Clin Psychiatry, 59 Suppl 20, 22–33;quiz 34–57. [PubMed] [Google Scholar]
- SHUKLA L, SHARMA P, GANESHA S, GHADIGAONKAR D, THOMAS E, KANDASAMY A, MURTHY P & BENEGAL V 2017. Value of Ethyl Glucuronide and Ethyl Sulfate in Serum as Biomarkers of Alcohol Consumption. Indian J Psychol Med, 39, 481–487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- SINGH YS, SAWARYNSKI LE, MICHAEL HM, FERRELL RE, MURPHEY-CORB MA, SWAIN GM, PATEL BA & ANDREWS AM 2010. Boron-Doped Diamond Microelectrodes Reveal Reduced Serotonin Uptake Rates in Lymphocytes from Adult Rhesus Monkeys Carrying the Short Allele of the 5-HTTLPR. ACS Chem Neurosci, 1, 49–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- STEFANAK MP, AL-MUDARES F, EL-METWALLY D, JONES JW, KANE MA & BEARER CF 2020. High concentrations of urinary ethanol metabolites in neonatal intensive care unit infants. Pediatr Res, 88, 865–870. [DOI] [PubMed] [Google Scholar]
- TAPIA-ROJAS C, MIRA RG, TORRES AK, JARA C, PEREZ MJ, VERGARA EH, CERPA W & QUINTANILLA RA 2017. Alcohol consumption during adolescence: A link between mitochondrial damage and ethanol brain intoxication. Birth Defects Res, 109, 1623–1639. [DOI] [PubMed] [Google Scholar]
- THIERAUF A, GNANN H, WOHLFARTH A, AUWARTER V, PERDEKAMP MG, BUTTLER KJ, WURST FM & WEINMANN W 2010. Urine tested positive for ethyl glucuronide and ethyl sulphate after the consumption of “non-alcoholic” beer. Forensic Sci Int, 202, 82–5. [DOI] [PubMed] [Google Scholar]
- TUKEY JW 1949. Comparing Individual Means in the Analysis of Variance. Biometrics, 5, 99–114. [PubMed] [Google Scholar]
- VAN DER ZWALUW CS, ENGELS RC, VERMULST AA, ROSE RJ, VERKES RJ, BUITELAAR J, FRANKE B & SCHOLTE RH 2010. A serotonin transporter polymorphism (5-HTTLPR) predicts the development of adolescent alcohol use. Drug Alcohol Depend, 112, 134–9. [DOI] [PubMed] [Google Scholar]
- VILLALBA K, ATTONITO J, MENDY A, DEVIEUX JG, GASANA J & DORAK TM 2015. A meta-analysis of the associations between the SLC6A4 promoter polymorphism (5HTTLPR) and the risk for alcohol dependence. Psychiatr Genet, 25, 47–58. [DOI] [PubMed] [Google Scholar]
- WALSHAM NE & SHERWOOD RA 2012. Ethyl glucuronide. Ann Clin Biochem, 49, 110–7. [DOI] [PubMed] [Google Scholar]
- WANG YM, CHANG Y, CHANG YY, CHENG J, LI J, WANG T, ZHANG QY, LIANG DC, SUN B & WANG BM 2012. Serotonin transporter gene promoter region polymorphisms and serotonin transporter expression in the colonic mucosa of irritable bowel syndrome patients. Neurogastroenterol Motil, 24, 560–5, e254-5. [DOI] [PubMed] [Google Scholar]
- WU X 3872–2019 Using SAS ® to Validate Prediction Models. 2019.
- YANG E, VAN NIMWEGEN E, ZAVOLAN M, RAJEWSKY N, SCHROEDER M, MAGNASCO M & DARNELL JE JR. 2003. Decay rates of human mRNAs: correlation with functional characteristics and sequence attributes. Genome Res, 13, 1863–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- YANG HC, CHEN CW, LIN YT & CHU SK 2021. Genetic ancestry plays a central role in population pharmacogenomics. Commun Biol, 4, 171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ZHONG Y, PERERA MA & GAMAZON ER 2019. On Using Local Ancestry to Characterize the Genetic Architecture of Human Traits: Genetic Regulation of Gene Expression in Multiethnic or Admixed Populations. Am J Hum Genet, 104, 1097–1115. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


