Abstract
Fusion of transmitter-containing vesicles with plasma membranes at the synaptic and neuromuscular junctions mediates neurotransmission and muscle contractions, respectively, thereby underlying all thoughts and actions. The fusion process is driven by the coupled folding and assembly of three synaptic SNARE proteins - syntaxin-1 and SNAP-25 on the target plasma membrane (t-SNAREs) and VAMP2 on the vesicular membrane (v-SNARE) into a four-helix bundle. Their assembly is chaperoned by Munc18–1 and many other proteins to achieve the speed and accuracy required for neurotransmission. However, the physiological pathway of SNARE assembly and its coupling to membrane fusion remain unclear. Here, we review recent progress in understanding SNARE assembly and membrane fusion, with a focus on results obtained by single-molecule manipulation approaches and electric recordings of single fusion pores. We describe two pathways of synaptic SNARE assembly, their associated intermediates, energetics, and kinetics. Assembly of the three SNAREs in vitro begins with the formation of a t-SNARE binary complex, on which VAMP2 folds in a stepwise zipper-like fashion. Munc18–1 significantly alters the SNARE assembly pathway: syntaxin-1 and VAMP2 first bind on the surface of Munc18–1 to form a template complex, with which SNAP-25 associates to conclude SNARE assembly and displace Munc18–1. During membrane fusion, multiple trans-SNARE complexes cooperate to open a dynamic fusion pore in a manner dependent upon their copy number and zippering states. Together, these results demonstrate that stepwise and cooperative SNARE assembly drive stagewise membrane fusion.
Keywords: SNAREs, Munc18–1, membrane fusion, fusion pores, template complexes, protein folding, optical tweezers, electric recording
Introduction
When arriving at a neuron ending, an action potential activates voltage-gated calcium channels, eliciting a Ca2+ influx (Sabatini and Regehr 1996). The resultant fast rise of local Ca2+ concentration triggers further folding of a partially assembled complex containing three soluble N-ethylmaleimide-sensitive factor attachment protein receptor (SNARE) proteins. The Ca2+-triggered SNARE assembly drives fusion of neurotransmitter-containing vesicles to the presynaptic plasma membrane, causing release of the neurotransmitters into the synaptic junction (Rothman 2014; Sudhof 2014). The neurotransmitters then bind to the receptors in the post-synaptic neurons to induce action potentials, completing the neurotransmission. Similar SNARE-dependent and calcium-triggered fusion machinery mediates numerous other vesicle fusion events, including muscle contraction, insulin release, glucose uptake, and antibody secretion (Sudhof and Rothman 2009; Rorsman and Ashcroft 2018). Consequently, dysfunction of the SNARE machinery has widely been linked to human diseases (zur Stadt et al. 2009; Stamberger et al. 2016; Rebane et al. 2018; Cali et al. 2021). However, many aspects of the SNARE fusion machinery are not well understood at a molecular level, especially with respect to SNARE assembly (Brose et al. 2019).
Synaptic SNAREs consist of VAMP2 (also called synaptobrevin) anchored on vesicular membranes (v-SNARE) and syntaxin-1 and SNAP-25 attached to the target plasma membrane (t-SNAREs) (Sollner et al. 1993; Sudhof 2014). These SNAREs share characteristic SNARE motifs ~60 amino acids in length, one in syntaxin-1 and VAMP2 and two in SNAP-25 (Figure 1(A)) (Fasshauer et al. 1998). Individual SNARE motifs are disordered in solution (Fasshauer et al. 2002), but four of them associate into an extraordinarily stable four-helix bundle (Figure 1(B)) (Sollner et al. 1993; Sutton et al. 1998; Stein et al. 2009). However, the SNARE assembly typically takes minutes to hours in vitro (Fasshauer and Margittai 2004). In contrast, synaptic vesicle fusion occurs within 200 microseconds upon calcium triggering, which is essential for fast neurotransmission in the brain (Sabatini and Regehr 1996). The apparent slow assembly in vitro is partly caused by SNARE misfolding or misassembly into many stable fusion-incompetent products, including syntaxin oligomers, 2:1 syntaxin-1:SNAP-25 t-SNARE complexes, and various anti-parallel helical bundles (Brunger 2005). These misassembled products serve as kinetic traps for SNARE folding, thereby greatly slowing down proper SNARE assembly. Thus, efficient SNARE assembly must occur via specific pathway(s) assisted by other proteins (Shen et al. 2007; Wickner 2010; Brunger et al. 2019; Koike and Jahn 2022; Rizo 2022).
Figure 1.
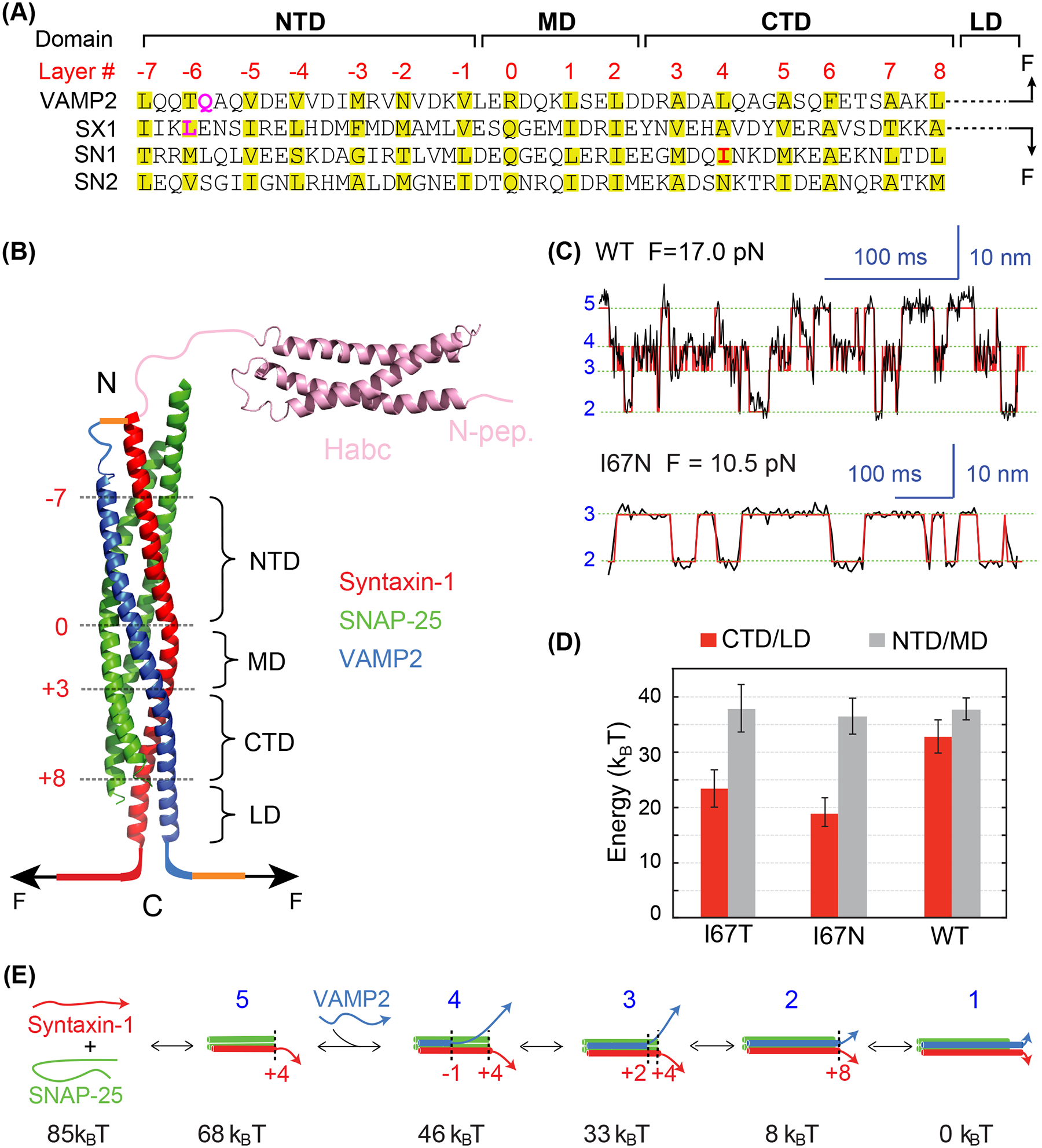
Energetics of stepwise SNARE zippering revealed by optical tweezers. (A) Amino acid sequences of SNARE motifs of synaptic SNAREs VAMP2, syntaxin-1A (SX1), and SNAP-25B (SN1 and SN2), and their associated folding domains and layer numbers. The folding domains include the N-terminal domain (NTD), the middle domain (MD), the C-terminal domain (CTD), and the linker domain (LD). The residues in VAMP2 Q36 and syntaxin-1 L205 in the red rectangle are mutated to cysteine and crosslinked to facilitate detection of reversible SNARE assembly in some single-molecule manipulation experiments (see results in C-E). (B) Crystal structure of the synaptic SNARE complex and its associated folding domains and layer numbers (red). The yellow lines indicate the disulfide bonds used to crosslink syntaxin and VAMP2 at their N-termini (top) and VAMP2 to the DNA handle (bottom, as in Figure 3(A)). The N-terminal SNARE crosslinking site varies in different experiments. (C) Time-dependent extension of the wild-type (top) or mutant (bottom) SNARE complex showing their reversible and stepwise zippering at constant force. The mutant contains a disease mutation at SNAP-25B I67N (highlighted red in A). The red lines represent idealized transitions derived from hidden-Markov modelling (Zhang et al. 2016). The green lines indicate average positions of the states with blue numbers shown in E. (D) Folding energy of different domains of the wild-type and mutant SNARE complexes. The disease mutations SNAP-25B I67N and I67T reduce the total folding energy of CTD and LD (CTD/LD), but not that of the NTD and MD (NTD/MD). (E) Diagrams of the different SNARE folding/assembly states and their energy relative to the fully assembled state.
The physiological pathway of synaptic SNARE assembly has been a matter of debate (Ma et al. 2013; Jakhanwal et al. 2017; Jiao et al. 2018; Zhang and Hughson 2021). A key feature of the assembly pathway is the order of the three synaptic SNAREs joining into a ternary SNARE complex (Figure 2). Within three binary combinations of synaptic SNAREs in the absence of regulatory proteins, only two t-SNAREs associate into a stable 1:1 syntaxin:SNAP-25 t-SNARE complex, as well as the 2:1 t-SNARE complex (Fasshauer et al. 1997). The former efficiently binds VAMP2 to form the four-helix bundle, while the latter is a misassembled intermediate that does not bind VAMP2 (Brunger 2005). However, recent results from biophysical, structural, and single-molecule studies have suggested an alternative Munc18–1-dependent pathway (Ma et al. 2013; Zhang and Hughson 2021). In this pathway, syntaxin-1 and VAMP2 first partially pair with each other on the surface of Munc18–1 to form a ternary template complex (Baker et al. 2015; Jiao et al. 2018). Then SNAP-25 binds to the templated SNAREs to form the four-helix bundle and displace the Munc18–1 molecule. Munc18–1 is the founding member of the highly conserved Sec1/Munc18 (SM) - family proteins that are essential for all SNARE-mediated membrane fusion (Sudhof and Rothman 2009; Zhang and Hughson 2021). Thus, this new SNARE assembly pathway is chaperoned by SM proteins and probably physiologically relevant.
Figure 2.
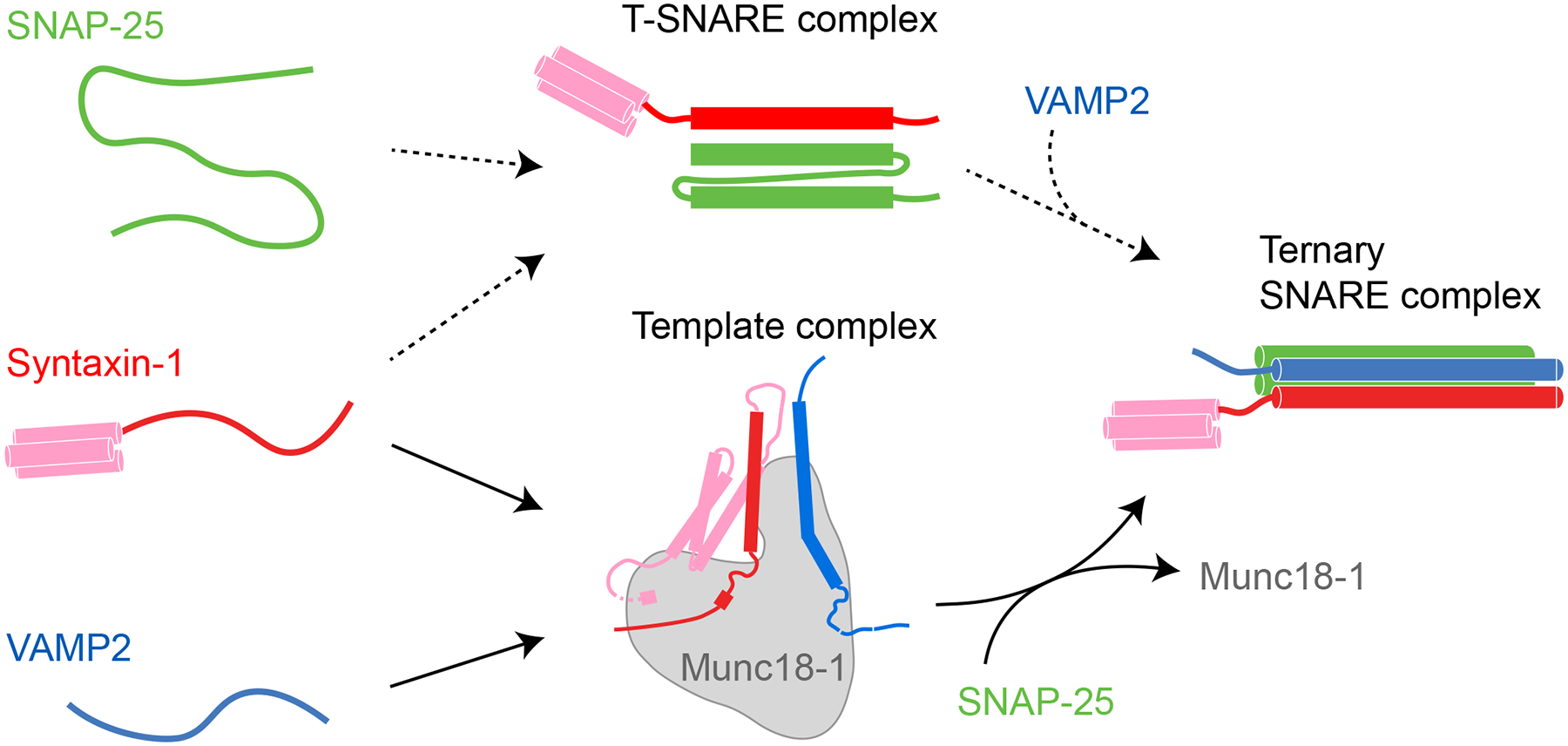
Schematics of two SNARE assembly pathways and their associated characteristic intermediates, the 1:1 syntaxin-1:SNAP-25 t-SNARE complex and the Munc18–1:syntaxin-1:VAMP2 template complex. Additional intermediates and their structural models and transition kinetics are shown in Figures 1(E) and 4(A).
SNARE assembly outputs the energy to counteract the energy barrier of membrane fusion (Jahn and Scheller 2006). The energy output from a single SNARE complex likely dictates the copy number of SNARE complexes required for membrane fusion, with reported values ranging from one to more than ten (Mohrmann et al. 2010; van den Bogaart et al. 2010; Shi et al. 2012; Mostafavi et al. 2017; Rothman et al. 2017; Bao et al. 2018; Manca et al. 2019; Radhakrishnan et al. 2021). A major difficulty in determining the copy number is the lack of effective methods to control and count the number of SNARE complexes associated with a fusion event. To overcome this difficulty, Shi et al. pioneered the use of nanodiscs to accommodate a specific average number of SNARE complexes and detected the fusion of nanodiscs with liposomes as a function of the copy number of SNARE complexes (Shi et al. 2012). In addition, the small size of nanodiscs (typically 10–20 nm) can be used to modulate the dynamics of fusion pores. Recently, Bao and co-workers improved the approach to detect the copy number-dependent dynamics of a single fusion pore formed between the VAMP2-anchored nanodisc and the suspended planar bilayer with microsecond resolution using electrical recording (Bao et al. 2018; Das et al. 2020). These experiments revealed that one SNARE complex is enough to open small fusion pores, but more SNAREs are required to dilate the fusion pores. Thus, multiple SNARE complexes cooperate to drive efficient membrane fusion.
In this review, we first introduce the single-molecule manipulation approach to protein folding using a single two-stranded coiled coil. Then we summarize the results of SNARE folding pathways, energetics, and kinetics mainly obtained from this approach. Finally, we relate SNARE assembly to fusion pore dynamics. This review focuses primarily on the core fusion machinery for synaptic fusion, i.e., the three synaptic SNAREs and Munc18–1. Other key proteins that regulate SNARE assembly, including NSF, SNAP, Munc13–1, synaptotagmin, and complexin, have recently been reviewed elsewhere (Baker and Hughson 2016; Brunger et al. 2018; Brunger et al. 2018; Rizo 2018; Brose et al. 2019; Brunger et al. 2019; Zhang and Hughson 2021; Koike and Jahn 2022; Rizo 2022).
Single-molecule manipulation approach to protein folding
Among many approaches applied to study SNARE folding and assembly, single-molecule methods are advantageous in dissecting the multiple assembly and misassembly pathways and their associated intermediates (Zhang et al. 2018; Zhang and Hughson 2021; Ha et al. 2022). For example, single-molecule fluorescence studies have shown different orientations of SNARE motifs in the misassembled SNARE complexes and SNARE disassembly by the AAA+ ATPase NSF (Weninger et al. 2008; Ryu et al. 2015; Lai et al. 2017). Single-molecule force spectroscopy, including optical tweezers (Gao et al. 2012; Zhang et al. 2013), magnetic tweezers (Min et al. 2013; Shon et al. 2018; Kim et al. 2021), and atomic force microscopy (Oelkers et al. 2016), utilize mechanical force to induce unfolding of SNARE complexes and detect their folding often with subnanometer and submillisecond resolution. This approach can detect SNARE folding and unfolding under thermal equilibrium conditions that are essential for measurements of SNARE folding energy and kinetics (Rebane et al. 2016; Zhang 2017; Shon et al. 2018). In addition, the mechanical force mimics the membrane repulsive force applied to SNAREs during membrane fusion, which can modulate SNARE assembly pathway and kinetics (Li et al. 2007; Zorman et al. 2014). For instance, while membrane-anchored SNAREs assemble directionally from the membrane distal N-terminal end to the membrane proximal C-terminal end, isolated SNAREs tend to zipper in the opposite direction (Li et al. 2016). Given the importance of single-molecule force spectroscopy in studies of SNARE assembly, we first introduce the methodology using optical tweezers and a simple two-stranded coiled coil as a model system (Figure 3(A)).
Figure 3.
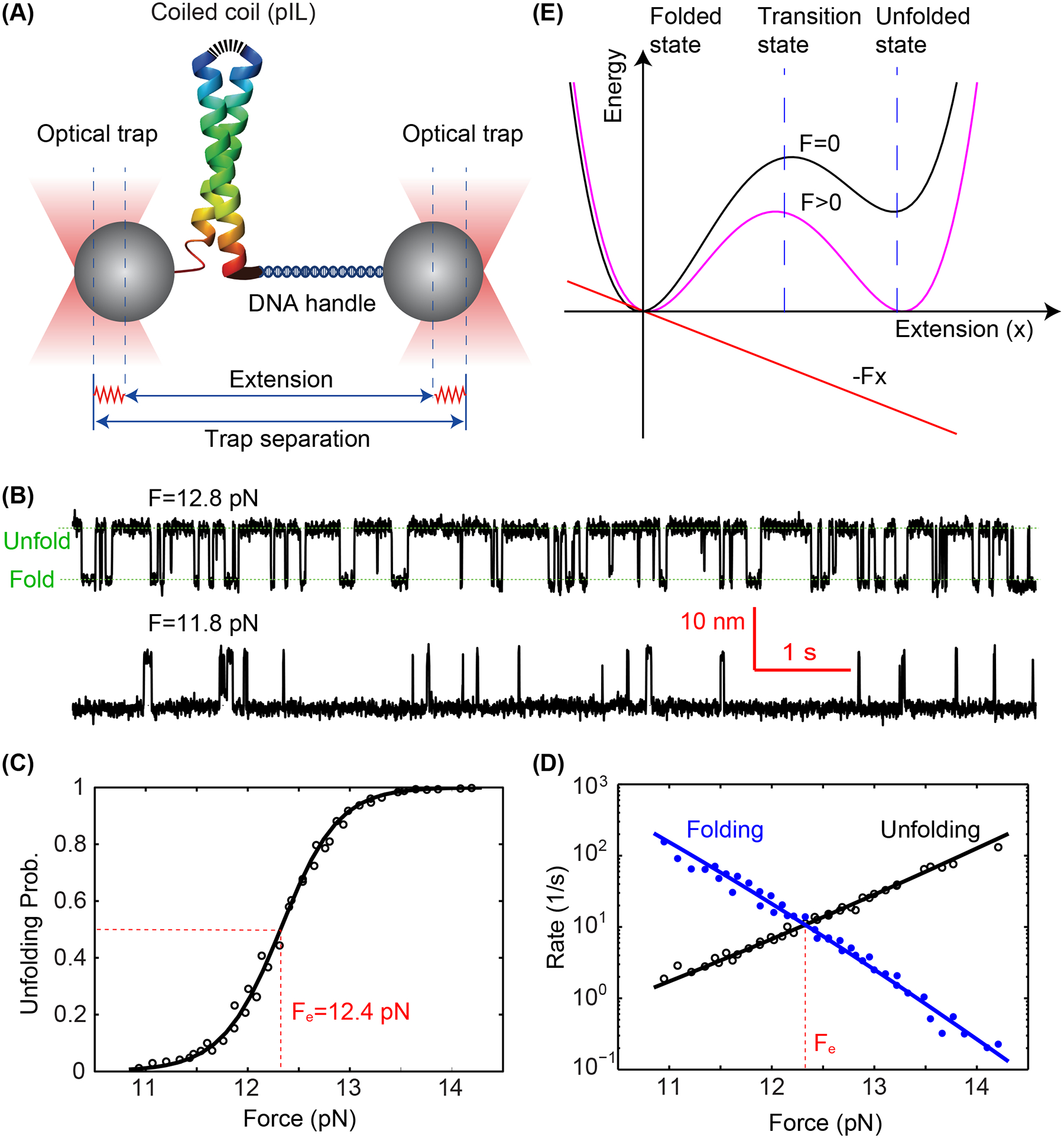
Folding energy and kinetics of a strong coiled coil revealed by optical tweezers. (A) Schematic diagram of the experimental setup. The homodimeric GCN4 variant is crosslinked at its N-terminus (top) and pulled from its C-terminus via a DNA handle. The force applied to the protein is controlled by the separation between two optical traps and the extension of the protein-DNA tether is measured to probe protein folding and unfolding transitions with subnanometer and submillisecond resolution. (B) Time-dependent extensions due to reversible unfolding and refolding of the coiled coil at two constant forces (F). (C) Unfolding probability of the coiled coil as a function of force, with the measured values and theoretical predictions shown in symbols and the line, respectively. The reversible folding transition has an equilibrium force of Fe=12.4 pN as indicated. (D) Measured folding and unfolding rate (symbols) and their theoretical predicts (lines) as a function of force. (E) A constant external force (F) applied to the protein tilts its folding and unfolding energy landscape to facilitate protein unfolding. Specifically, the force lowers the energy of the unfolded state and unfolding energy barrier in their extension-dependent manner.
Optical tweezers have widely been applied to study the folding of proteins and nucleic acids (Liphardt et al. 2001; Cecconi et al. 2005; Bustamante et al. 2020). They utilize focused laser light to form harmonic potentials to trap micron-sized polystyrene or silica beads as force and displacement sensors (Figure 3(A)) (Ashkin et al. 1986; Sirinakis et al. 2012; Zhang et al. 2013). Specifically, the bead displacements are detected by laser interferometry. To appreciate the stability of the synaptic SNARE complex, Xi et al. studied one of the strongest coiled coil pIL derived from the GCN4 transcription factor, which remains folded even in boiling water (Gao et al. 2011; Xi et al. 2012). A single pIL is tethered between two trapped beads via a 2,260 bp DNA handle (Cecconi et al. 2005). The force applied to the protein is controlled by the separation between the two optical traps. The extension response of the protein-DNA tether is detected to derive protein conformational transitions. At constant trap separations with a mean force range of 11 – 14 pN, the coiled coil reversible folds and unfolds between a fully folded state (low extension) and a completely unfolded state (high extension) in a force-dependent manner, as indicated by the corresponding extension changes (Figure 3(B)). Note that the mechanical properties of DNA molecules have been well characterized, which are inert under force below 60 pN (Smith et al. 1996). The force-dependent unfolding probability and transition rates are derived from these extension trajectories, leading to an equilibrium force of 12.4 pN with half unfolding probability (Figures 3(C,D)). Force tilts the energy landscape of protein folding and unfolding such that the free energy of the unfolded state and the transition state relative to that of the folded state linearly decreases as force increases (Figure 3E), leading to the characteristic sigmoid unfolding probability and approximately exponential rates as a function of force (Figures (C,D)). Consequently, the folding energy and folding and unfolding rates can be derived as parameters to fit these curves (Rebane et al. 2016; Jiao et al. 2017). Particularly, the unfolding energy can be estimated by the mechanical work to unfold the protein, which is the equilibrium force multiplied by the corresponding extension change, minus the small entropic energy to stretch the unfolded polypeptide to the equilibrium force. Accordingly, a high unfolding energy of 24 kBT is measured for the coiled coil (Xi et al. 2012), corroborating its extraordinary stability.
Pathways, energetics, and kinetics of SNARE assembly in vitro
Syntaxin-1 consists of a disordered N-terminus called N-peptide (residues 1–26), an Habc domain folded into a three-helix bundle (residues 27–146) (Fernandez et al. 1998), a disordered linker region (residues 147–187), the SNARE motif (residues 188–255), a juxtamembrane linker domain (LD, residues 256–265), and the transmembrane domain (TMD, residues 266–288) (Figures 1(A,B)). Similarly, VAMP2 contains a disordered N-terminal region (residues 1–30) and a juxtamembrane linker domain (residues 86–96). SNAP-25 bears two SNARE motifs connected by a disordered linker. Structural studies show that the fully assembled SNARE complex contains a parallel four-helix bundle formed by the SNARE motifs and an extended coiled coil formed by the linker domains and the transmembrane domains of syntaxin-1 and VAMP2 (Sutton et al. 1998; Stein et al. 2009). The core of the four-helix bundle consists of 15 layers of hydrophobic residues and one middle layer of hydrophilic residues, which are numbered from −7 at the N-terminus to +8 at the C-terminus with the middle zero layer.
In a typical experiment to study SNARE folding and assembly, a single SNARE complex without transmembrane domains is pulled from the C-termini of syntaxin-1 and VAMP2 (Figure 1(B)) (Gao et al. 2012; Jiao et al. 2017). The N-termini of their SNARE motifs are crosslinked by a disulfide bridge to facilitate reversible SNARE assembly and disassembly. At ~17 pN, a single SNARE complex undergoes reversible and sequential transitions among four distinct states (Figure 1(C)) (Ma et al. 2015). Layer mutations significantly alter the kinetics of the transitions and their associated equilibrium force and energy (Figures 1(C, D)) (Rebane et al. 2018). In combination with results from other experimental approaches, the single-molecule experiments reveal the pathway, energetics, and kinetics of SNARE assembly (Figure 1(E)). SNARE assembly in vitro starts from the formation of the 1:1 t-SNARE complex. The t-SNARE complex contains a three-helix bundle with a frayed C-terminus after the +4 layer, with unfolding energy of ~17 kBT (Zhang et al. 2016). Subsequently, the v-SNARE VAMP2 folds on the t-SNARE complex in four steps, a process termed as SNARE zippering: First, the N-terminal domain of VAMP2 (NTD, from −7 layer to −1 layer) associates with the corresponding domain in the t-SNARE with an association rate constant of 2×106 M−1s−1 (Pobbati et al. 2006; Jiao et al. 2018); Then, the middle domain (MD, from −1 layer to +2 layer) zippers with a small energy barrier of ~5 kBT resulting mainly from pairing of the ionic layer (Ma et al. 2015); Subsequently, the C-terminal domain (CTD, from +3 layer to +8 layer) rapidly folds in a diffusion-limited manner with ~22 kBT energy output and is accompanied by folding of the frayed C-terminal end of the t-SNARE complex; Finally, the linker domain (LD) zippers with an energy output of ~8 kBT and small energy barrier of ~5 kBT. Overall, the assembly of a single synaptic SNARE complex without the transmembrane domain generates total energy of 85 kBT, including 68 kBT from SNARE zippering. SNARE zippering continues to the TMDs of syntaxin and VAMP2 (Stein et al. 2009). However, the corresponding energy contribution remains to be determined. In addition, membranes are expected to strongly modulate LD zippering (Liang et al. 2013; Rathore et al. 2019; Rizo et al. 2022). Compared to the strong coiled coil pIL (Xi et al. 2012), SNARE assembly generates significantly more energy (85 vs 24 kBT) and higher force (~17 pN equilibrium force for the CTD vs ~12 pN for pIL). Interestingly, many SNARE complexes responsible for other membrane traffic pathways zipper in a similar stepwise manner with large energy output and fast CTD zippering (Zorman et al. 2014), consistent with their role as a molecular engine for membrane fusion.
The different SNARE zippering domains likely play distinct roles in synaptic vesicle fusion (Sorensen 2009; Ma et al. 2015). Assembly of the SNARE NTD and CTD are associated with vesicle docking or priming and final fusion, respectively, while MD assembly controls the accuracy of SNARE assembly. The docked vesicles are ready for fusion when triggered by Ca2+, whose number correlates with the strength of neurotransmission, while CTD zippering energy and kinetics control the speed of membrane fusion. The distinct roles of SNARE domains have been confirmed by numerous SNARE mutations whose effects in SNARE assembly, membrane fusion, or neurotransmission have been compared. A prominent example is SNAP-25B mutation I67N and I67T at the +4 layer in the CTD (Figure 1(A)), which are identified from a human patient suffering from intellectual disability and the blind-drunk mouse with the schizophrenia phenotype, respectively (Jeans et al. 2007; Shen et al. 2014). Optical tweezer measurements reveal that both mutations dramatically weaken CTD assembly (Figures 1(C,D)) (Rebane et al. 2018), likely leading to failure in membrane fusion. In contrast, both mutations barely change NTD and MD assembly. Thus, the energy output from SNARE assembly is essential for membrane fusion.
Munc18–1-chaperoned SNARE assembly
Closed and open syntaxin bound by Munc18–1
The SNARE assembly pathway in the cell may be significantly different from the t-SNARE pathway described above. First, there is probably no free syntaxin-1 available for SNARE assembly in the cell. Instead, syntaxin-1 exists in a complex with Munc18–1 as soon as syntaxin-1 is synthesized (Hata et al. 1993; Yin et al. 2018). Munc18–1 serves as a molecular chaperone to help syntaxin-1 traffic from the endoplasmic reticulum membrane to the plasma membrane and protect syntaxin-1 from degradation (Zhou et al. 2013). Knockout of either syntaxin or Munc18–1 leads to a reduced concentration for the other protein. However, Munc18–1 also sequesters syntaxin from binding other SNARE proteins for SNARE assembly, forming closed syntaxin (Burkhardt et al. 2008). Crystal structures reveal that Munc18–1 interacts extensively with the entire cytoplasmic syntaxin (Figure 4(A), state i) (Misura et al. 2000; Colbert et al. 2013). Consequently, a high affinity in the range of 2–10 nM has been reported for the Munc18–1:syntaxin complex (Burkhardt et al. 2008). Thus, it becomes a key puzzle how the closed syntaxin opens to allow SNARE assembly.
Figure 4.
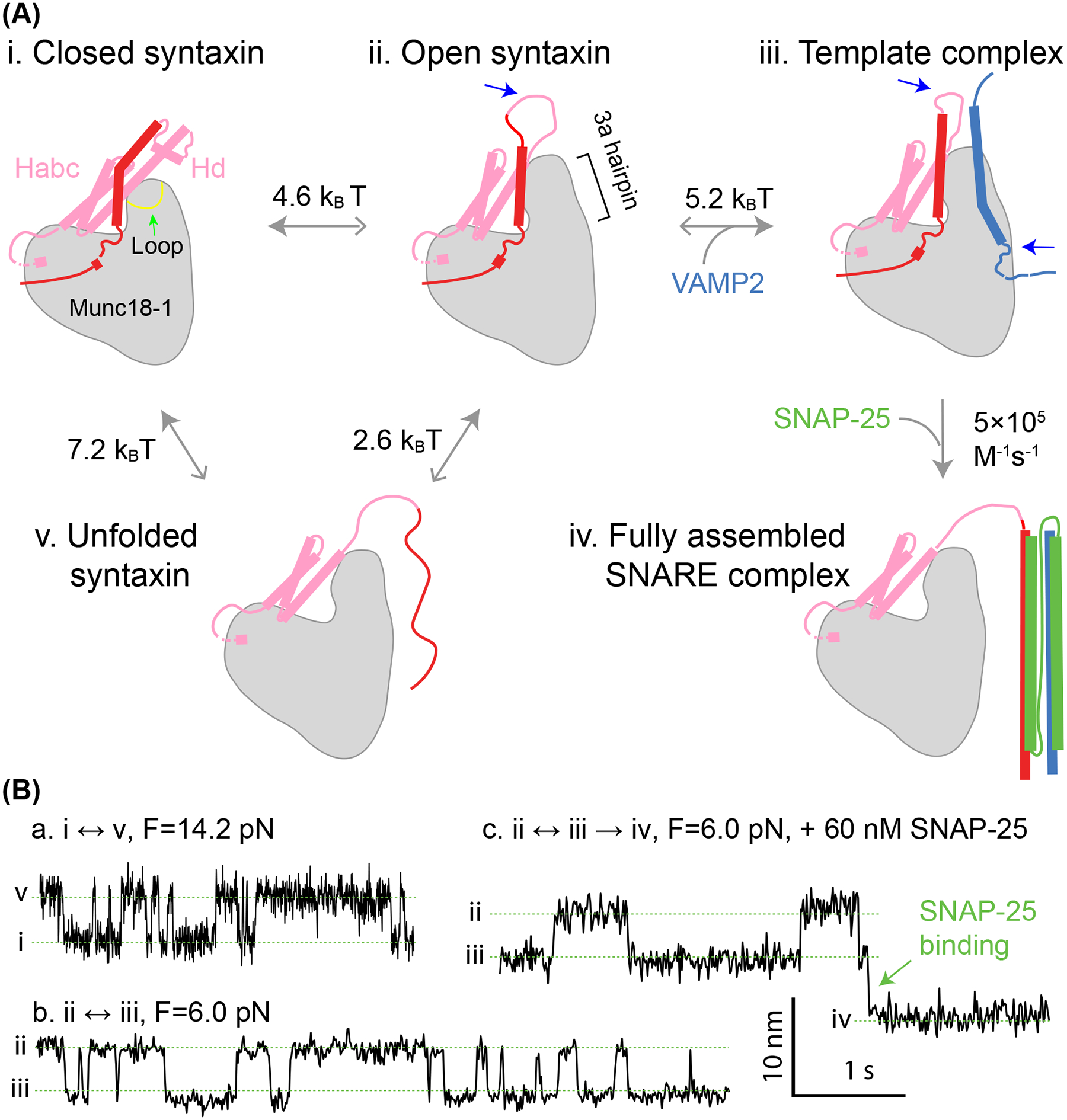
Munc18–1 chaperones SNARE assembly via multiple essential intermediates. (A) The pathway of Munc18–1-chaperoned SNARE assembly. The closed syntaxin (state i) is opened by Munc13–1 (not shown) or mutations to expose the N-terminal region of the SNARE motif in syntaxin (state ii). Syntaxin opening is accompanied by unfurling a loop at the tip of the 3a helical hairpin domain in Munc18–1, as indicated by the green arrow in state i, which extends the 3a domain. Then, the open syntaxin binds to VAMP2 to form the template complex (state iii). The two SNARE binding sites for Munc13–1 (not shown) are indicated by blue arrows. Finally, SNAP-25 associates with the templated SNAREs to form the four-helix bundle (state iv), a process likely accompanied by displacement of Munc18–1 from the SNARE bundle. The free energy to unfold the SNARE motif of syntaxin (state v) has been measured for the closed and open syntaxin. The solid greys arrows point to the states with lower relative free energy. (B) Time-dependent extension trajectories associated with reversible unfolding of the closed syntaxin (a) and the template complex in the absence (b) or presence (c) of SNAP-25 at the indicated force (F).
Evidence suggests that Munc13–1 promotes opening of the closed syntaxin via a series of local conformational changes in both syntaxin and Munc18–1 (Rizo 2022). Munc13–1 is a large multifunctional protein essential for synaptic vesicle fusion (Augustin et al. 1999). With a rod shape of ~19 nm in length, Munc13–1 helps tether synaptic vesicles to the plasma membrane via its terminal C1 and C2 domains (Michelassi et al. 2017; Xu et al. 2017; Quade et al. 2019; Grushin et al. 2022). Through its central MUN domain, Munc13–1 also binds to the linker region of syntaxin-1 (Wang et al. 2017). However, the binding does not dissociate entire syntaxin-1 from Munc18–1. Instead, it likely induces local unfolding of the linker region and the N-terminal region of the SNARE motif to open syntaxin (Shin et al. 2010; Ma et al. 2013; Yang et al. 2015). Consistent with this view, the MUN domain only weakly binds free syntaxin with a dissociation constant greater than 40 μM (Lai et al. 2017; Wang et al. 2017; Magdziarek et al. 2020) and the closed syntaxin can be opened by destabilizing mutations in the linker region such as the LE mutation (L165A/E166A) (Ma et al. 2011). This conformation of open syntaxin is further supported by single-molecule manipulation experiments (Jiao et al. 2018). By pulling single syntaxin molecules at different positions using optical tweezers, Jiao et al. identified a new mutation R198C in the linker region that opens syntaxin for further SNARE assembly. Combining with results from various syntaxin mutations, they found that the parts of the linker region and the N-terminal SNARE motif are either unfolded or dissociated from the Munc18–1, while the C-terminal SNARE motif including +1 to +3 layers remains bound in a cavity in Munc18–1 (Figure 4(A), state ii). Interestingly, Munc18–1 binds the N-peptide and the Habc domain in the open syntaxin similarly to the closed syntaxin. These experiments also reveal the unfolding free energy of the open syntaxin (2.6 kBT) and the closed syntaxin (7.2 kBT) relative to an unfolded syntaxin state in which the SNARE motif, but not the entire syntaxin molecule, is dissociated from Munc18–1 (Figure 4(A), state v; Figure 4(B), a). Thus, syntaxin opening only needs a small energy input of 4.6 kBT, which may be provided by binding of Munc13–1 to syntaxin. In addition, the weak Munc13–1 binding may lower the energy barrier for syntaxin opening. Finally, the derived structural model of open syntaxin is consistent with the crystal structure of a homologous complex Vps45:Tlg2 in which Tlg2 appears to exhibit an intrinsically open conformation (Eisemann et al. 2020).
Template complex and SNARE assembly
Recent experiments suggest that Munc18–1 can simultaneously bind VAMP2 (Parisotto et al. 2014; Munch et al. 2016; Sitarska et al. 2017; Andre et al. 2020) and Syntaxin-1 to form a Munc18–1:syntaxin-1:VAMP2 ternary template complex (Figure 4(A), state iii) (Baker et al. 2015; Jiao et al. 2018; Wang et al. 2019; Shu et al. 2020). Moreover, this template complex is essential to drive membrane fusion in reconstituted assays using SNARE proteins and accessory proteins at physiologically relevant concentrations (Stepien et al. 2019). To characterize the template complex using optical tweezer at a molecular level, Jiao et al. strategically crosslinked syntaxin residue R198C to VAMP2 residue N29C, both N-terminal to the SNARE motifs (near the extended −8 layer), to create the open syntaxin in the presence of Munc18–1 (Jiao et al. 2018). By pulling the C-termini of syntaxin-1 and VAMP2, they identified the synaptic template complex (Figure 4(A), state iii) and measured its stability (−5.2 kBT) and lifetime (1.4 s) relative to the open syntaxin (Figure 4(B), b). They also examined the stabilities of a battery of mutants and found various domains or regions critical for the template complex. Baker et al. first predicted a homologous template complex in the thermotolerant fungus Chaetomium thermophilum Vps33:Vam3:Nyv1, which are homologs of mammalian Munc18–1, syntaxin-1, and VAMP2, respectively, and derived a structural model based on superposition of two binary crystal structures of Vps33:Vam3 and Vps33:Nyv1 (Baker et al. 2015). In this model, both SNARE motifs of Vam3 and Nyv1 bind on the surface of Vps33 as two broken helices each, with their N-terminal helices aligned on the 3a helical hairpin domain of Munc18–1 while their C-terminal SNARE motifs separated. Consequently, Vps33 interacts extensively with both SNARE motifs, from −7 to +3 layers in syntaxin and from −3 to +6 layers in VAMP2. Particularly, Nyv1 inserts its highly conserved phenylalanine residue at the +6 layer (F77) into a hydrophobic pocket in Vps33. The single-molecules studies demonstrated that the synaptic template complex not only shares some key features of this structural model, including a strong anchor by VAMP2 F77, but also exhibits additional features not predicted by the model (Jiao et al. 2018). The N-terminal hydrophobic layers of VAMP2 from −7 to −4 layers appear to associate with the corresponding layers in syntaxin, as their mutations to alanine destabilize the template complex. Importantly, the N-terminal domains of syntaxin, including the N-peptide and the Habc domain also stabilize the template complex. Particularly, the Habc domain is required for forming the template complex.
The template complex greatly enhances the efficiency of SNARE assembly (Jiao et al. 2018). SNAP-25 quickly binds the template complex to conclude SNARE assembly (Figure 4(A), from state iii to state iv; Figure 4(B), c). After SNARE assembly, Munc18–1 is displaced from the four-helix bundle but may remain bound to the N-terminal domains of syntaxin. The rate constant of SNAP-25 binding to the template complex is estimated to be ~5×105 M−1s−1. This rate is at least 25-fold greater than the rate of spontaneous SNARE assembly in the absence of Munc18–1 under otherwise identical experimental conditions. In this case, the rate of proper SNARE assembly is limited by t-SNARE formation and SNARE misassembly. Note that the single-molecule assay underestimates the rate of SNARE misassembly in bulk, because many known misassembled SNARE complexes, such as syntaxin oligomers and SNARE bundles containing anti-parallel syntaxin and VAMP2, are inhibited in the single-molecule assay. In the presence of Munc18–1, the rate of SNARE misassembly is greatly reduced, and nearly all SNARE assembly is chaperoned by Munc18–1 via the template complex. In addition, Munc18–1 promotes the specificity of proper SNARE assembly. SNARE proteins responsible for different membrane trafficking pathways readily form complexes in vitro in the absence of chaperones, leading to promiscuous SNARE pairing (Yang et al. 1999). The extensive interactions between SNAREs and Munc18–1 in the template complex help proofread SNARE pairing (Shen et al. 2007; Koike and Jahn 2022), such that any noncognate SNAREs that do not associate with Munc18–1 to form the template complex cannot assemble into the ternary SNARE complexes. The essential role of the template complex in SNARE assembly and membrane fusion is supported by extensive mutagenesis studies (Jiao et al. 2018): Mutations in Munc18–1 or SNARE proteins that destabilize the template complex impair the chaperoned SNARE assembly, membrane fusion in vitro, and neurotransmitter release in vivo. In particular, a few Munc18–1 mutations identified from patients with epilepsy are found to destabilize the template complex. In contrast, the mutations that enhance the stability of the template complex promote membrane fusion and neurotransmission. These results suggest that the formation of the template complex is likely a rate-limiting step in membrane fusion. Finally, the template complex and its essential role in chaperoning SNARE assembly are conserved among SNARE-SM fusion machines in different membrane trafficking pathways and species, including Vps33:Vam3:Nyv1 in yeast for vacuole formation (Wickner 2010; Baker et al. 2015; Orr et al. 2017) and Munc18–3:syntaxin-4:VAMP2 in mammals for insulin secretion and GLUT4 translocation (Jiao et al. 2018). In conclusion, the template complex is a conserved intermediate of SM protein chaperoned SNARE assembly that significantly enhances the speed and accuracy of SNARE assembly.
The synaptic template complex is regulated by many proteins that control synaptic vesicle fusion (Zhang and Hughson 2021). Munc13–1 associates with the template complex to enhance its stability, but without interfering with SNAP-25 binding and subsequent SNARE assembly (Shu et al. 2020). The association requires binding of Munc13–1 to the linker region of syntaxin and likely VAMP2 at distant sites on the MUN domain (Wang et al. 2019). Munc13–1 also binds SNAP-25 at its linker region connecting the two SNARE motifs to help recruit SNAP-25 for its association with the template complex (Sundaram et al. 2021). Therefore, Munc13–1 cooperates with Munc18–1 to form a tetrameric complex and helps recruit SNAP-25 for efficient SNARE assembly, in addition to Munc13–1’s role in opening syntaxin. Note that Munc13–1 alone contributes to proper SNARE assembly by promoting parallel pairing between syntaxin and VAMP2 (Lai et al. 2017), likely because Munc13–1 binding to opposite ends of the SNARE motifs of syntaxin-1 and VAMP2 at the two distal ends of the rod-like MUN domain only allows their parallel pairing in the presence of SNAP-25. Moreover, Munc18–1 is phosphorylated at multiple sites by many kinases to regulate the stability of the template, thereby modulating SNARE assembly and neurotransmitter release (Jiao et al. 2018). For example, Munc18–1 phosphorylation at S306 or S313 stabilizes the template complex and promotes neurotransmission (Genc et al. 2014), whereas phosphorylation at Y473 abolishes the template complex and neurotransmission (Meijer et al. 2012). Finally, Munc18–1 prevents the ubiquitous NSF and SNAP proteins in the cell from disassembling the functional intermediates of SNARE assembly but allows NSF/SNAP to specifically disassemble misassembled SNARE complexes (Xu et al. 2010; Prinslow et al. 2019; Stepien et al. 2019). Thus, it is reasonable to hypothesize that the template complex is resistant to NSF/SNAP disassembly, a hypothesis remaining to be tested (Zhang and Hughson 2021). In conclusion, the template complex may serve as a hub to regulate SNARE assembly and synaptic vesicle fusion.
Atomic structure of the template complex
Recently, Stepien et al. solved the first atomic structure of any template complex, which not only confirms many salient features of the structural model described above, but also reveals interesting new features (Figure 5(A)) (Stepien et al. 2022). To stabilize the synaptic template complex, they crosslinked syntaxin-1 L205C and VAMP2 Q36C at a different site near −6 layer as previously tested (Ma et al. 2015; Jiao et al. 2018) (Figures 1(A) and 5(B)). Consequently, they were able to purify the complex in bulk and determine its structure using cryo-electron microscopy (Stepien et al. 2022). Two structures of the complex have been resolved, but with similar architectures. Both structures reveal the aligned helices formed by the NTDs of syntaxin-1 and VAMP2 and their separated CTDs bound on the surface of Munc18–1 (Figure 5(A)). The N-peptide and the Habc domain exhibit similar conformations as in the closed syntaxin. In addition, both structures show an extended loop of the 3a helical hairpin domain (Munc18–1 residues 314–335) expected for the open syntaxin (Hu et al. 2011; Christie et al. 2012; Parisotto et al. 2014; Baker et al. 2015; Sitarska et al. 2017). This loop is furled back in the closed syntaxin to block VAMP2 binding (compare states i and iii in Figure 4(A)) (Misura et al. 2000). All these features are consistent with the results derived from the single-molecule studies and the corresponding structural model (Baker et al. 2015; Jiao et al. 2018; Zhang and Hughson 2021).
Figure 5.
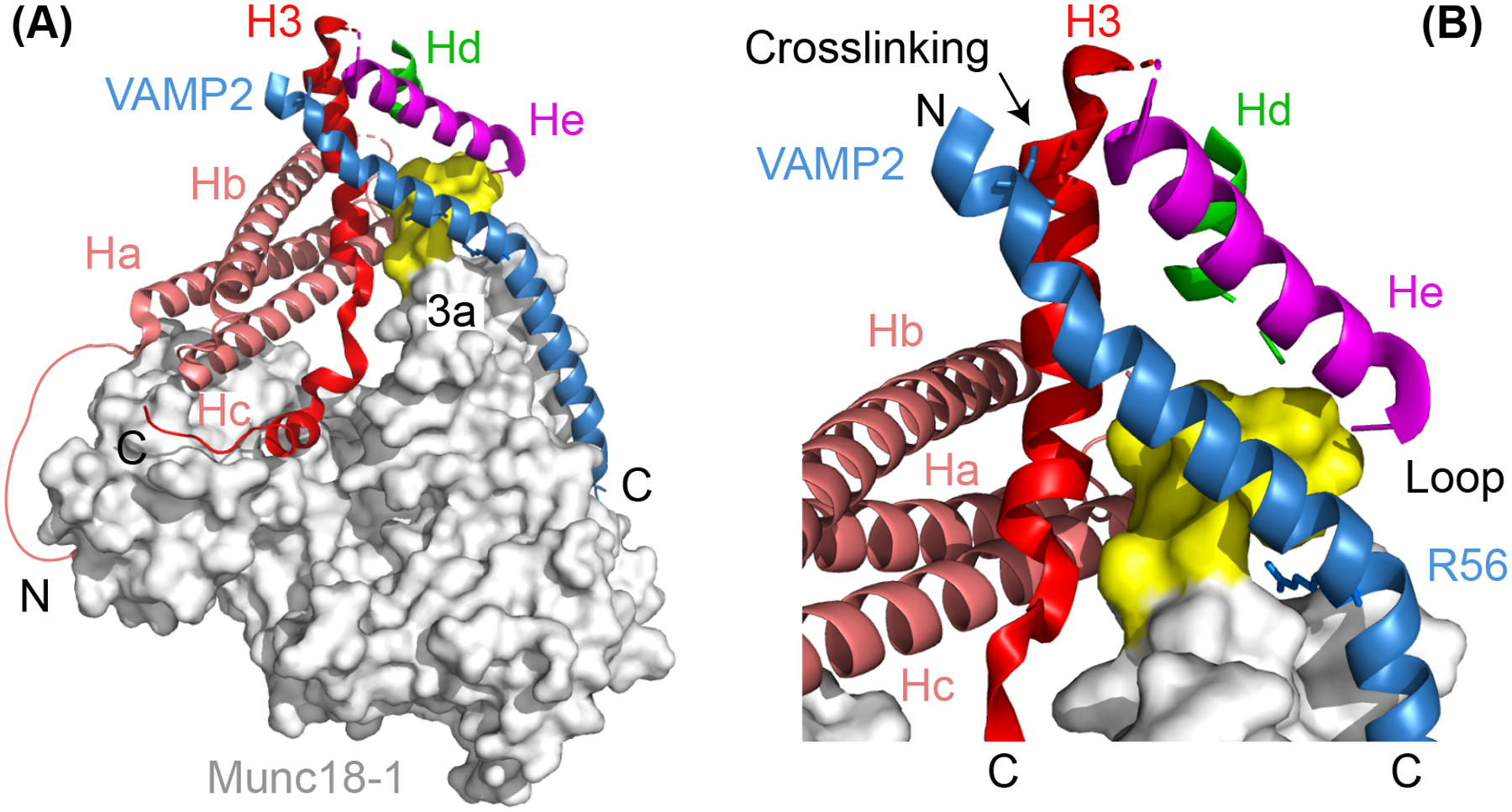
Atomic structure of the template complex (A) and the close-up view of the N-terminal four-helix bundle (B). In (A), the helical N-peptide binds to the back of Munc18–1 (grey) and connects to the Ha helix with a disordered polypeptide. Part of the unfurred loop of the 3a helical hairpin in Munc18–1 (a.a. 327–334) exhibits a helical conformation and is highlighted yellow, while the other part (a.a. 316–326) is not resolved and likely disordered. The amino (‘N’) - and carboxyl (‘C’) -ends of the SNARE polypeptides are labelled. In (B), the zero-layer residue R56 in VAMP2 and the cysteine residues in both VAMP2 and Syntaxin-1 used for crosslinking the two polypeptides are shown in sticks.
Importantly, the atomic structures reveal several new features (Stepien et al. 2022). First, the linker region of syntaxin-1 folds into two short helices (Hd and He helices) that associate with the NTD helices of syntaxin-1 and VAMP2 to form an N-terminal four-helix bundle (Figure 5(B)). The Hd helix is also found in the Munc18–1:syntaxin-1 complex to stabilize the closed syntaxin (Figure 4(A), state i), but switches binding partners in the template complex. Two mutations in the He helix (M183A and D184P) that are predicted to destabilize the N-terminal four-helix bundle impair the template complex and membrane fusion in vitro. These observations suggest an important role of the N-terminal four-helix bundle in the chaperoned SNARE assembly. Second, both NTDs of syntaxin-1 and VAMP2, as well as the entire N-terminal four-helix bundle, detach from Munc18–1 (Figure 5(B)). As a result, only the MD/CTD regions directly bind Munc18–1. While the detached syntaxin-1 NTD is a characteristic of open syntaxin (Eisemann et al. 2020; Zhang and Hughson 2021), the hanging VAMP2 NTD is unexpected, because the same region of the homologous Nyv1 binds to the extended 3a helical hairpin domain of Vps33 in the Vps33:Nyv1 binary complex (Baker et al. 2015) and the underlying Munc18–1 binding surface is critical for template complex formation and neurotransmitter release (Munch et al. 2016; Jiao et al. 2018). The two structures of the template complex differ in the separation of the N-terminal four-helical bundle from the underlying Munc18–1 surface (Stepien et al. 2022). The structure with greater separation is the mature template complex (Figure 5(A)), while the other one (not shown) represents an intermediate conformation during the transition from the closed syntaxin to the mature template complex. Finally, VAMP2 exhibits a continuous helix from −7 layer to + 6 layer, but with a kink near the 0 layer in the mature template complex, compared to two broken helices in the previous structural model (Baker et al. 2015). More experiments are required to further test the biological significance of the new structures, especially in the cell.
Fusion pores and SNARE assembly
Membrane fusion often proceeds via distinct stages, including vesicle docking, priming, hemifusion, transient fusion pore opening and closing (pore flickering), and final pore expansion (Fang and Lindau 2014; Zhao et al. 2016; Bao et al. 2018; Sharma and Lindau 2018). Particularly, the fusion pore establishes the first connection between the lumens bounded by the two fusing membranes and is widely observed in vesicle fusion in the cell (Albillos et al. 1997). Fusion pores are generally small and transient with their diameters and lifetimes less than a few nanometers and 10 milliseconds, respectively (Breckenridge and Almers 1987; Albillos et al. 1997; Henkel et al. 2000; Klyachko and Jackson 2002; Jackson and Chapman 2006; Dhara et al. 2020). Then, they either quickly dilate for full fusion or close for ‘kiss-and-run’ fusion (Jackson and Chapman 2008; Alabi and Tsien 2013). Alterations in SNARE proteins significantly change the size and stability of the fusion pores (Han et al. 2004; Kesavan et al. 2007; Fang et al. 2008), suggesting a tight coupling between SNARE assembly and fusion pore dynamics.
It remains an open question how the different steps of SNARE assembly described above underlie the different stages of membrane fusion, especially the fusion pore dynamics. Despite their physiological relevance, the SNARE assembly described above is mainly derived from experiments based on cytosolic SNAREs. In contrast, SNARE assembly during membrane fusion is counteracted by two opposing lipid bilayers, regulated by many accessory proteins, and occurs in concert with multiple SNARE complexes. While the stepwise SNARE assembly is detected in the presence of approximately constant force in the single-molecule manipulation experiments (Gao et al. 2012; Zhang 2017; Jiao et al. 2018; Zhang and Hughson 2021), multiple trans-SNARE complexes cooperatively assemble in the presence of the membrane repulsive force that varies quickly with the distance between the two membranes (Zorman et al. 2014). Therefore, the lifetimes and populations of the derived intermediates may differ in the context of trans-SNARE assembly. In addition, the role of SNARE TMDs in fusion is controversial. SNARE TMDs may simply act as membrane anchors to transduce the SNARE zippering force to membranes (Zhou et al. 2013), provide additional zippering energy required for fusion (Stein et al. 2009), or oligomerize to form proteinaceous fusion pores (Han et al. 2004; Chang et al. 2016). In addition, these challenges called for new experiments to correlate the sequence of events of SNARE assembly with that of membrane fusion.
Membrane fusion has been reconstituted with fixed average numbers of SNARE complexes. Shi et al. first used nanodiscs (NDs) to control the copy number of VAMP2 inserted into the nanodiscs and detected their fusion with small unilamellar vesicles (SUVs) containing excess t-SNARE complexes (Shi et al. 2012). Each ND is a small patch of lipid bilayer enclosed by two molecules of membrane scaffold proteins (MSPs) like a belt (Bayburt et al. 2002). NDs are self-assembled during the gradual removal of detergent from the reconstitution reaction composed of lipids, MSPs, and membrane proteins. Besides their superior solubility and stability, NDs can be made in different sizes ranging from 6 to 100 nm in diameter using a library of MSPs derived from apolipoproteins (Bayburt et al. 2002; Denisov et al. 2004; Hagn et al. 2013; Nasr et al. 2017; Zhang et al. 2021). Consequently, NDs have widely been adopted as model membranes for structural and biophysical characterization of membrane proteins (Autzen et al. 2019; Sligar and Denisov 2021). To control the average copy number of VAMP2 inserted into NDs, Shi et al. varied the molar ratio of VAMP2 to MSP during ND preparation and further purified the NDs containing specific copy numbers of VAMP2 (Shi et al. 2012). An additional benefit of the assay is that the NDs with a typical diameter of ~16 nm prevents dilation of the fusion pores, thereby stabilizing the generally transient fusion pores for characterization of their properties. Using this assay, they found that a single SNARE complex is enough to induce lipid mixing or hemifusion in which only the outer leaflets of the two membranes fuse, while at least three SNARE complexes are required to open the fusion pores with an average diameter of ~3 nm. This observation suggests that the opening of a fusion pore experiences much higher energy barrier than hemifusion, and thus requires energy from cooperative zippering of at least three SNARE complexes. As a potential rate-limiting state for membrane fusion, the fusion pore formation is regulated by many regulatory proteins. For example, in the presence of synaptotagmin-1 (Syt1), the Ca2+ sensor for neurotransmission, two SNARE complexes are sufficient to catalyze glutamate release (Bhalla et al. 2006; Bao et al. 2016; Zhou et al. 2017). Finally, the SNARE TMDs are required for pore opening, as substitution of the TMDs with lipid anchors abolishes glutamate release (Shi et al. 2012; Chang et al. 2016). Thus, SNARE TMDs not only act as a membrane anchor to transduce the SNARE zippering force to membranes, but also play an active role in fusion pore opening (Ngatchou et al. 2010; Bao et al. 2016; Dhara et al. 2016; Sharma and Lindau 2018; Dhara et al. 2020). The latter observation contrasts with other findings that lipid-anchored SNAREs lacking their TMDs support membrane fusion or neurotransmitter release (Jun et al. 2007; Zhou et al. 2013).
The fusion pores stabilized by NDs have been further characterized at a single pore level with sub-millisecond resolution. Wu et al. applied cell-based patch clamp and single-channel recording to characterize single fusion pores between v-SNARE-anchored NDs and cell surfaces ectopically expressing flipped t-SNAREs (Wu et al. 2016; Wu et al. 2017). They observed fast and reversible flickering of fusion pores with an average pore diameter of ~1.2 nm and a lifetime of ~6 s before their irreversible resealing in the presence of 7–9 copies of v-SNAREs in nanodiscs. However, the high background current from the cell surface and the rapid pore closure poses challenges for a more detailed characterization of the fusion pores. Bao et al. significantly improved the electrical recordings of single fusion pores by replacing cell membranes with planar lipid bilayers (Figure 6(A)) (Bao et al. 2018). The high electric impedance of the synthetic planar bilayer leads to high signal-to-noise ratio to detect fusion pores without the need of a patch clamp. In addition, the fusion pore is further stabilized by the lipids in the planar lipid bilayer that lower the membrane bending energy for fusion pore formation. Consequently, individual fusion pores are readily observed with microsecond temporal resolution for over 30 minutes (Figure 6(B)). Consistent with previous studies, the new experiment revealed that the number of SNARE complexes dictates the pore size and lifetime and that the fusion pores are highly dynamic. Moreover, by virture of the improved resolution, the open pores exhibit approximately equal sizes for a given copy number of SNARE complexes, leading to discrete two-state transitions between the open and the closed state (Figure 6(B)) and accurate measurements of pore sizes and lifetimes (Figures 6(C,D)). An average of three VAMP2 molecules in ND3 only forms an unstable 1.1 nm pore with an open lifetime of only ~3 ms (Figure 6 (D)). The addition of two more VAMP2 molecules (ND5) significantly stabilizes the pore with an increase in the pore size to 2.2 nm and the average open lifetime to 0.2 s. An average of 7 VAMP2 molecules in ND7 generates long-lived pores with an average lifetime of ~2 s and a pore size of 3 nm. Interestingly, the shape of the pore lifetime distributions significantly changes as the SNARE copy number increases: For ND3, the distribution is approximately single exponential, indicating the existence of a single energy barrier for pore opening, while for ND5 and ND7, the distributions become unimodal, suggesting multiple energy barriers or at least transient intermediate states for pore opening. Consistent with the latter interpretation, some intermediate states are observed in time-dependent current trajectories (Bao et al. 2018). Finally, pore formation requires the transmembrane domains (TMD) of syntaxin and VAMP2 and is altered by mutations in the TMD. Since the TMDs of three copies of SNARE complexes are too few to form a completely proteinaceous pore, this observation suggests that the fusion pores are formed by both lipids and SNARE TMDs, consistent with the results from molecular dynamic simulations and other studies (Bao et al. 2016; Dhara et al. 2016; Sharma and Lindau 2018; Sharma and Lindau 2018; Dhara et al. 2020).
Figure 6.
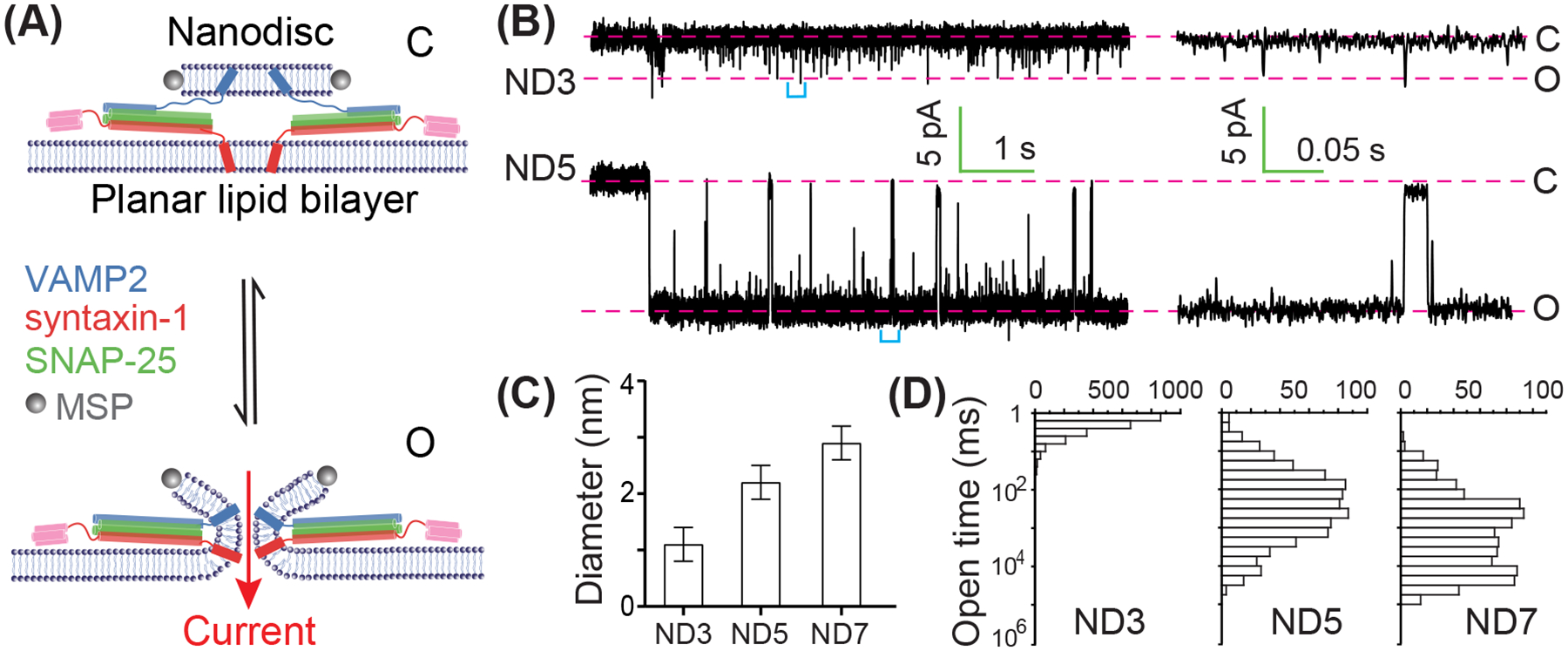
Dynamics of fusion pores as a function of the copy number of SNARE complexes revealed electric recordings. (A) Illustration of closed (“C”) and open (“O”) pores formed between a nanodisc containing two v-SNARE molecules (ND2) and a planar lipid bilayer harbouring t-SNARE complexes. The reversible transition between the two states is detected by the current through the pore under a constant voltage across the two sides of the planar lipid bilayer. (B) Representative time-dependent current showing reversible opening of the pore through a nanodisc containing three (ND3) or five (ND5) v-SNAREs on average. The average copy number of trans-SNARE complexes is approximately half of the total v-SNARE number per nanodisc, as only v-SNAREs anchored on one side of nanodiscs can engage with the t-SNAREs in the planar lipid bilayer. The right panel shows close-up views of the regions in the left panel marked by cyan brackets. (C) The average diameter of the pore as a function of SNARE copy number in the nanodisc. (D) The corresponding distributions of the pore lifetimes.
The pore dynamics is intrinsically coupled to SNARE zippering. The use of planar bilayer facilitates the addition of various regulatory proteins or other factors to interrogate the SNARE assembly underlying the pore dynamics. The fusion pore is closed by the addition of the cytosolic fragment of v-SNARE in a concentration-dependent manner (Bao et al. 2018), indicating the presence of at least one partially zippered trans-SNARE complexes in the closed pore state that are available for competitive binding of the cytosolic v-SNARE fragment (Figure 6(A)). Furthermore, mutations that impair CTD zippering of the SNARE complex destabilize the fusion pores. Together, these observations suggest that closed and open pore states correspond to half-zippered and fully zippered states of the SNARE complex, respectively. Regulatory proteins, including Syt1, α-synuclein, and complexin, may promote SNARE zippering or membrane remodelling to facilitate pore opening and membrane fusion (Das et al. 2020; Nellikka et al. 2021; Wu et al. 2021; Courtney et al. 2022). Thus, the electrical recording of fusion pores formed between nanodiscs and planar lipid bilayers provides a powerful platform for interrogating the relationship between SNARE assembly and fusion pore dynamics.
Although fusion pores are observed as a distinct intermediate before their dilation to full fusion in some cells (Breckenridge and Almers 1987; Henkel et al. 2000; Jackson and Chapman 2006), they remain elusive during synaptic vesicle fusion. As a step to detect fusion pores during synaptic vesicle fusion, Bao et al. increased the size of the nanodisc to ~50 nm to allow fusion pore expansion (Bao et al. 2018). They found that flickering fusion pores of ~2 nm diameter frequently appear before they irreversibly expand into full fusion, suggesting the fusion pores as an intermediate in membrane fusion. Importantly, distinct fusion pores are directly observed in SNARE-mediated fusion between small unilamellar vesicles and planar lipid bilayers again with electric recordings (Heo et al. 2021). In this case, α-hemolysin channel proteins are incorporated into the v-SNARE anchored vesicles to facilitate measurements of the current through fusion pores. The experiment revealed small flickering fusion pores with average diameters of 0.4 nm and 0.8 nm mediated by one and two trans-SNARE complexes, respectively. Three or more trans-SNARE complexes lead to irreversible pore expansion without distinct fusion pores, consistent with highly cooperative SNARE assembly among different trans-SNARE complexes. The variable pore sizes observed in the different experiments are likely caused by different membranes used. Thus, the dynamics of fusion pores or membrane fusion in generally can be exquisitely regulated by the size and composition of membranes, SNARE proteins, and various accessory proteins.
Concluding remarks
Experiments in vitro have revealed two pathways of SNARE assembly and their associated intermediate states: the t-SNARE pathway and the template complex pathway. While the t-SNARE pathway has long been exploited in reconstituted membrane fusion assays, the template complex pathway likely represents the physiological pathway of SNARE assembly in vivo. This conclusion is supported by the conserved template complexes observed for multiple SNARE-SM fusion machines and the essential role of the template complex in the chaperoned SNARE assembly and membrane fusion. Note that the total energy of SNARE assembly is independent of the pathways and thus the values measured based on the t-SNARE pathway should be approximately equal to the energy output from the Munc18–1-chaperoned SNARE assembly. In addition, the SNARE assembly along both pathways converges in partially zippered states in which the N-terminal SNARE motifs are zippered while the C-terminal motifs are unzipped (Ma et al. 2015; Jiao et al. 2018; Yu et al. 2018). Other regulatory proteins, including Munc13–1, synaptotagmin, and complexin, may bind to the partially zippered trans-SNARE complexes to form a super-complex primed for Ca2+-triggered membrane fusion (Rothman et al. 2017). Due to the strong coupling between SNARE zippering and pore opening, the stability of the super-complex is also modulated by the interactions of both regulatory proteins and SNAREs with membranes. However, it remains unclear whether and how all these proteins form a primed super-complex with a defined stoichiometry. It will be interesting to reconstitute the primed state in vitro, determine their structures, and further interrogate their functions in Ca2+-triggered membrane fusion. To this end, it will be crucial to combine optical tweezers with single-molecule fluorescence to simultaneously pull trans- or cis-SNARE complexes in membranes and visualize the action of regulatory proteins. In addition, the fusion pore assays need further improvement to examine fusion pores mediated by precise numbers of trans-SNARE complexes in the presence of Munc18–1. Finally, super-resolution microscopy and electron microscopy may be applied to dissect the structure and dynamics of the primed super-complex (Adams et al. 2015; Radhakrishnan et al. 2021). Together, these techniques will help uncover the long-standing puzzle on the key molecular machinery underlying our thinking and action.
Acknowledgements
We thank Jose Rizo and Frederic Pincet for reading our manuscript.
Declaration of interest
This work was supported by NIH grants R35 GM131714 to Y. Z. and DP2 GM140920 to B.H.
Footnotes
Disclosure statement
The authors declare no conflict of interests.
References
- Adams DJ, Arthur CP, Stowell MHB. 2015. Architecture of the synaptophysin/synaptobrevin complex: structural evidence for an entropic clustering function at the synapse. Sci Rep. 5:13659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alabi AA, Tsien RW. 2013. Perspectives on kiss-and-run: role in exocytosis, endocytosis, and neurotransmission. Annu Rev Physiol. 75(75):393–422. [DOI] [PubMed] [Google Scholar]
- Albillos A, Dernick G, Horstmann H, Almers W, Alvarez de Toledo G, Lindau M. 1997. The exocytotic event in chromaffin cells revealed by patch amperometry. Nature. 389(6650):509–512. [DOI] [PubMed] [Google Scholar]
- Andre T, Classen J, Brenner P, Betts MJ, Dorr B, Kreye S, Zuidinga B, Meijer M, Russell RB, Verhage M, Sollner TH. 2020. The interaction of Munc18–1 Helix 11 and 12 with the central region of the VAMP2 SNARE motif is essential for SNARE templating and synaptic transmission. Eneuro. 7(6):ENEURO.0278–20.2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashkin A, Dziedzic JM, Bjorkholm JE, Chu S. 1986. Observation of a single-beam gradient force optical trap for dielectric particles. Opt Lett. 11(5):288–290. [DOI] [PubMed] [Google Scholar]
- Augustin I, Rosenmund C, Sudhof TC, Brose N. 1999. Munc13–1 is essential for fusion competence of glutamatergic synoptic vesicles. Nature. 400(6743):457–461. [DOI] [PubMed] [Google Scholar]
- Autzen HE, Julius D, Cheng YF. 2019. Membrane mimetic systems in CryoEM: keeping membrane proteins in their native environment. Curr Opin Struct Biol. 58(58):259–268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker RW, Hughson FM. 2016. Chaperoning SNARE assembly and disassembly. Nat Rev Mol Cell Biol. 17(8):465–479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker RW, Jeffrey PD, Zick M, Phillips BP, Wickner WT, Hughson FM. 2015. A direct role for the Sec1/Munc18-family protein Vps33 as a template for SNARE assembly. Science. 349(6252):1111–1114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bao H, Das D, Courtney NA, Jiang Y, Briguglio JS, Lou X, Roston D, Cui Q, Chanda B, Chapman ER. 2018. Dynamics and number of trans-SNARE complexes determine nascent fusion pore properties. Nature. 554(7691):260–263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bao H, Goldschen-Ohm M, Jeggle P, Chanda B, Edwardson JM, Chapman ER. 2016. Exocytotic fusion pores are composed of both lipids and proteins. Nat Struct Mol Biol. 23(1):67–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bayburt TH, Grinkova YV, Sligar SG. 2002. Self-assembly of discoidal phospholipid bilayer nanoparticles with membrane scaffold proteins. Nano Lett. 2(8):853–856. [Google Scholar]
- Bhalla A, Chicka MC, Tucker WC, Chapman ER. 2006. Ca2+-synaptotagmin directly regulates t-SNARE function during reconstituted membrane fusion. Nat Struct Mol Biol. 13(4):323–330. [DOI] [PubMed] [Google Scholar]
- Breckenridge LJ, Almers W. 1987. Currents through the fusion pore that forms during exocytosis of a secretory vesicle. Nature. 328(6133):814–817. [DOI] [PubMed] [Google Scholar]
- Brose N, Brunger A, Cafiso D, Chapman ER, Diao J, Hughson FM, Jackson MB, Jahn R, Lindau M, Ma C, Rizo J, Shin YK, Sollner TH, Tamm L, Yoon TY, Zhang YL. 2019. Synaptic vesicle fusion: today and beyond. Nat Struct Mol Biol. 26(8):663–668. [DOI] [PubMed] [Google Scholar]
- Brunger AT. 2005. Structure and function of SNARE and SNARE-interacting proteins. Q Rev Biophys. 38(1):1–47. [DOI] [PubMed] [Google Scholar]
- Brunger AT, Choi UB, Lai Y, Leitz J, White KI, Zhou Q. 2019. The pre-synaptic fusion machinery. Curr Opin Struct Biol. 54(54):179–188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brunger AT, Choi UB, Lai Y, Leitz J, Zhou QJ. 2018. Molecular mechanisms of fast neurotransmitter release. Annual Review of Biophysics. 47:469–497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brunger AT, Leitz J, Zhou QJ, Choi UB, Lai Y. 2018. Ca2+-Triggered Synaptic Vesicle Fusion Initiated by Release of Inhibition. Trends Cell Biol. 28(8):631–645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burkhardt P, Hattendorf DA, Weis WI, Fasshauer D. 2008. Munc18a controls SNARE assembly through its interaction with the syntaxin N-peptide. EMBO J. 27(7):923–933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bustamante C, Alexander L, Maciuba K, Kaiser CM. 2020. Single-molecule studies of protein folding with optical tweezers. Annu Rev Biochem. 89(89):443–470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cali E, Rocca C, Salpietro V, Houlden H. 2021. Epileptic phenotypes associated with SNAREs and related synaptic vesicle exocytosis machinery. Front Neurol. 12(12):806506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cecconi C, Shank EA, Bustamante C, Marqusee S. 2005. Direct observation of the three-state folding of a single protein molecule. Science. 309(5743):2057–2060. [DOI] [PubMed] [Google Scholar]
- Chang CW, Chiang CW, Gaffaney JD, Chapman ER, Jackson MB. 2016. Lipid-anchored synaptobrevin provides little or no support for exocytosis or liposome fusion. J Biol Chem. 291(6):2848–2857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christie MP, Whitten AE, King GJ, Hu SH, Jarrott RJ, Chen KE, Duff AP, Callow P, Collins BM, James DE, Martin JL. 2012. Low-resolution solution structures of Munc18:Syntaxin protein complexes indicate an open binding mode driven by the Syntaxin N-peptide. Proc Natl Acad Sci USA. 109(25):9816–9821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colbert KN, Hattendorf DA, Weiss TM, Burkhardt P, Fasshauer D, Weis WI. 2013. Syntaxin1a variants lacking an N-peptide or bearing the LE mutation bind to Munc18a in a closed conformation. Proc Natl Acad Sci USA. 110(31):12637–12642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Courtney KC, Wu L, Mandal T, Swift M, Zhang Z, Alaghemandi M, Wu Z, Bradberry MM, Deo C, Lavis LD, Volkmann N, Hanein D, Cui Q, Bao H, Chapman ER. 2022. The complexin C-terminal amphipathic helix stabilizes the fusion pore open state by sculpting membranes. Nat Struct Mol Biol. 29(2):97–107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das D, Bao H, Courtney KC, Wu LX, Chapman ER. 2020. Resolving kinetic intermediates during the regulated assembly and disassembly of fusion pores. Nat Commun. 11(1):231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denisov IG, Grinkova YV, Lazarides AA, Sligar SG. 2004. Directed self-assembly of monodisperse phospholipid bilayer nanodiscs with controlled size. J Am Chem Soc. 126(11):3477–3487. [DOI] [PubMed] [Google Scholar]
- Dhara M, Martinez MM, Makke M, Schwarz Y, Mohrmann R, Bruns D. 2020. Synergistic actions of v-SNARE transmembrane domains and membrane-curvature modifying lipids in neurotransmitter release. Elife. 9:e55152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dhara M, Yarzagaray A, Makke M, Schindeldecker B, Schwarz Y, Shaaban A, Sharma S, Bockman RA, Lindau M, Mohrmann R, Bruns D. 2016. v-SNARE transmembrane domains function as catalysts for vesicle fusion. Elife. 5:e17571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisemann TJ, Allen F, Lau K, Shimamura GR, Jeffrey PD, Hughson FM. 2020. The Sec1/Munc18 protein Vps45 holds the Qa-SNARE Tlg2 in an open conformation. Elife. 9:e60724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang QH, Berberian K, Gong LW, Hafez I, Sorensen JB, Lindau M. 2008. The role of the C terminus of the SNARE protein SNAP-25 in fusion pore opening and a model for fusion pore mechanics. Proc Natl Acad Sci USA. 105(40):15388–15392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang QH, Lindau M. 2014. How could snare proteins open a fusion pore? Physiology. 29(4):278–285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fasshauer D, Antonin W, Subramaniam V, Jahn R. 2002. SNARE assembly and disassembly exhibit a pronounced hysteresis. Nat Struct Biol. 9(2):144–151. [DOI] [PubMed] [Google Scholar]
- Fasshauer D, Margittai M. 2004. A transient N-terminal interaction of SNAP-25 and syntaxin nucleates SNARE assembly. J Biol Chem. 279(9):7613–7621. [DOI] [PubMed] [Google Scholar]
- Fasshauer D, Otto H, Eliason WK, Jahn R, Brunger AT. 1997. Structural changes are associated with soluble N-ethylmaleimide-sensitive fusion protein attachment protein receptor complex formation. J Biol Chem. 272(44):28036–41. [DOI] [PubMed] [Google Scholar]
- Fasshauer D, Sutton RB, Brunger AT, Jahn R. 1998. Conserved structural features of the synaptic fusion complex: SNARE proteins reclassified as Q- and R-SNAREs. Proc Natl Acad Sci USA. 95(26):15781–15786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandez I, Ubach J, Dulubova I, Zhang XY, Sudhof TC, Rizo J. 1998. Three-dimensional structure of an evolutionarily conserved N-terminal domain of syntaxin 1A. Cell. 94(6):841–849. [DOI] [PubMed] [Google Scholar]
- Gao Y, Sirinakis G, Zhang YL. 2011. Highly anisotropic stability and folding kinetics of a single coiled coil protein under mechanical tension. J Am Chem Soc. 133(32):12749–12757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao Y, Zorman S, Gundersen G, Xi ZQ, Ma L, Sirinakis G, Rothman JE, Zhang YL. 2012. Single reconstituted neuronal SNARE complexes zipper in three distinct stages. Science. 337(6100):1340–1343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Genc O, Kochubey O, Toonen RF, Verhage M, Schneggenburger R. 2014. Munc18–1 is a dynamically regulated PKC target during short-term enhancement of transmitter release. Elife. 3:e01715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grushin K, Kalyana Sundaram RV, Sindelar CV, Rothman JE. 2022. Munc13 structural transitions and oligomers that may choreograph successive stages in vesicle priming for neurotransmitter release. Proc Natl Acad Sci USA. 119(7):e2121259119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ha T, Kaiser C, Myong S, Wu B, Xiao J. 2022. Next generation single-molecule techniques: Imaging, labeling, and manipulation in vitro and in cellulo. Mol Cell. 82(2):304–314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagn F, Etzkorn M, Raschle T, Wagner G. 2013. Optimized phospholipid bilayer nanodiscs facilitate high-resolution structure determination of membrane proteins. J Am Chem Soc. 135(5):1919–1925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han X, Wang CT, Bai JH, Chapman ER, Jackson MB. 2004. Transmembrane segments of syntaxin line the fusion pore of Ca2+-triggered exocytosis. Science. 304(5668):289–292. [DOI] [PubMed] [Google Scholar]
- Hata Y, Slaughter CA, Sudhof TC. 1993. Synaptic vesicle fusion complex contains Unc-18 homolog bound to syntaxin. Nature. 366(6453):347–351. [DOI] [PubMed] [Google Scholar]
- Henkel AW, Meiri H, Horstmann H, Lindau M, Almers W. 2000. Rhythmic opening and closing of vesicles during constitutive exo- and endocytosis in chromaffin cells. EMBO J. 19(1):84–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heo P, Coleman J, Fleury JB, Rothman JE, Pincet F. 2021. Nascent fusion pore opening monitored at single-SNAREpin resolution. Proc Natl Acad Sci USA. 118(5):e2024922118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu SH, Christie MP, Saez NJ, Latham CF, Jarrott R, Lua LHL, Collins BM, Martin JL. 2011. Possible roles for Munc18–1 domain 3a and Syntaxin1 N-peptide and C-terminal anchor in SNARE complex formation. Proc Natl Acad Sci USA. 108(3):1040–1045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson MB, Chapman ER. 2006. Fusion pores and fusion machines in Ca2+-triggered exocytosis. Annu Rev Biophys Biomol Struct. 35:135–160. [DOI] [PubMed] [Google Scholar]
- Jackson MB, Chapman ER. 2008. The fusion pores of Ca2+-triggered exocytosis. Nat Struct Mol Biol. 15(7):684–689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jahn R, Scheller RH. 2006. SNAREs - engines for membrane fusion. Nat Rev Mol Cell Bio. 7(9):631–643. [DOI] [PubMed] [Google Scholar]
- Jakhanwal S, Lee CT, Urlaub H, Jahn R. 2017. An activated Q-SNARE/SM protein complex as a possible intermediate in SNARE assembly. EMBO J. 36(12):1788–1802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeans AF, Oliver PL, Johnson R, Capogna M, Vikman J, Molnar Z, Babbs A, Partridge CJ, Salehi A, Bengtsson M, Eliasson L, Rorsman P, Davies KE. 2007. A dominant mutation in SNAP25 causes impaired vesicle trafficking, sensorimotor gating, and ataxia in the blind-drunk mouse. Proc Natl Acad Sci USA. 104(7):2431–2436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiao J, He M, Port SA, Baker RW, Xu Y, Qu H, Xiong Y, Wang Y, Jin H, Eisemann TJ, Hughson FM, Zhang Y. 2018. Munc18–1 catalyzes neuronal SNARE assembly by templating SNARE association. Elife. 7:e41771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiao JY, Rebane AA, Ma L, Zhang YL. 2017. Single-molecule protein folding experiments using high-resolution optical tweezers. Methods Mol Biol. 1486:357–390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jun Y, Xu H, Thorngren N, Wickner W. 2007. Sec18p and Vam7p remodel trans-SNARE complexes to permit a lipid-anchored R-SNARE to support yeast vacuole fusion. EMBO J. 26(24):4935–4945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kesavan J, Borisovska M, Bruns D. 2007. v-SNARE actions during Ca2+-triggered exocytosis. Cell. 131(2):351–363. [DOI] [PubMed] [Google Scholar]
- Kim C, Shon MJ, Kim SH, Eun GS, Ryu JK, Hyeon C, Jahn R, Yoon TY. 2021. Extreme parsimony in ATP consumption by 20S complexes in the global disassembly of single SNARE complexes. Nat Commun. 12:3206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klyachko VA, Jackson MB. 2002. Capacitance steps and fusion pores of small and large-dense-core vesicles in nerve terminals. Nature. 418(6893):89–92. [DOI] [PubMed] [Google Scholar]
- Koike S, Jahn R. 2022. SNARE proteins: zip codes in vesicle targeting? Biochem J. 479(3):273–288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai Y, Choi UB, Leitz J, Rhee HJ, Lee C, Altas B, Zhao ML, Pfuetzner RA, Wang AL, Brose N, Rhee J, Brunger AT. 2017. Molecular mechanisms of synaptic vesicle priming by Munc13 and Munc18. Neuron. 95(3):591–607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li F, Pincet F, Perez E, Eng WS, Melia TJ, Rothman JE, Tareste D. 2007. Energetics and dynamics of SNAREpin folding across lipid bilayers. Nat Struct Mol Biol. 14(10):890–896. [DOI] [PubMed] [Google Scholar]
- Li F, Tiwari N, Rothman JE, Pincet F. 2016. Kinetic barriers to SNAREpin assembly in the regulation of membrane docking/priming and fusion. Proc Natl Acad Sci USA. 113(38):10536–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang BY, Kiessling V, Tamm LK. 2013. Prefusion structure of syntaxin-1A suggests pathway for folding into neuronal trans-SNARE complex fusion intermediate. Proc Natl Acad Sci USA. 110(48):19384–19389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liphardt J, Onoa B, Smith SB, Tinoco I, Bustamante C. 2001. Reversible unfolding of single RNA molecules by mechanical force. Science. 292(5517):733–737. [DOI] [PubMed] [Google Scholar]
- Ma C, Li W, Xu Y, Rizo J. 2011. Munc13 mediates the transition from the closed syntaxin-Munc18 complex to the SNARE complex. Nat Struct Mol Biol. 18(5):542–549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma C, Su LJ, Seven AB, Xu YB, Rizo J. 2013. Reconstitution of the vital functions of Munc18 and Munc13 in neurotransmitter release. Science. 339(6118):421–425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma L, Rebane AA, Yang G, Xi Z, Kang Y, Gao Y, Zhang YL. 2015. Munc18–1-regulated stage-wise SNARE assembly underlying synaptic exocytosis. Elife. 4:e09580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magdziarek M, Bolembach AA, Stepien KP, Quade B, Liu XX, Rizo J. 2020. Re-examining how Munc13–1 facilitates opening of syntaxin-1. Protein Sci. 29(6):1440–1458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manca F, Pincet F, Truskinovsky L, Rothman JE, Foret L, Caruel M. 2019. SNARE machinery is optimized for ultrafast fusion. Proc Natl Acad Sci USA. 116(7):2435–2442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meijer M, Burkhardt P, de Wit H, Toonen RF, Fasshauer D, Verhage M. 2012. Munc18–1 mutations that strongly impair SNARE-complex binding support normal synaptic transmission. EMBO J. 31(9):2156–2168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michelassi F, Liu HW, Hu ZT, Dittman JS. 2017. A C1-C2 module in Munc13 inhibits calcium-dependent neurotransmitter release. Neuron. 95(3):577–590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Min D, Kim K, Hyeon C, Cho YH, Shin YK, Yoon TY. 2013. Mechanical unzipping and rezipping of a single SNARE complex reveals hysteresis as a force-generating mechanism. Nat Commun. 4:1705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Misura KMS, Scheller RH, Weis WI. 2000. Three-dimensional structure of the neuronal-Sec1-syntaxin 1a complex. Nature. 404(6776):355–362. [DOI] [PubMed] [Google Scholar]
- Mohrmann R, de Wit H, Verhage M, Neher E, Sorensen JB. 2010. Fast vesicle fusion in living cells requires at least three SNARE complexes. Science. 330(6003):502–505. [DOI] [PubMed] [Google Scholar]
- Mostafavi H, Thiyagarajan S, Stratton BS, Karatekin E, Warner JM, Rothman JE, O’Shaughnessy B. 2017. Entropic forces drive self-organization and membrane fusion by SNARE proteins. Proc Natl Acad Sci USA. 114(21):5455–5460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munch AS, Kedar GH, van Weering JRT, Vazquez-Sanchez S, He EQ, Andre T, Braun T, Sollner TH, Verhage M, Sorensen JB. 2016. Extension of Helix 12 in Munc18–1 induces vesicle priming. J Neurosci. 36(26):6881–6891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nasr ML, Baptista D, Strauss M, Sun ZYJ, Grigoriu S, Huser S, Pluckthun A, Hagn F, Walz T, Hogle JM, Wagner G. 2017. Covalently circularized nanodiscs for studying membrane proteins and viral entry. Nat Methods. 14(1):49–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nellikka RK, Bhaskar BR, Sanghrajka K, Patil SS, Das D. 2021. alpha-Synuclein kinetically regulates the nascent fusion pore dynamics. Proc Natl Acad Sci USA. 118(34):e2021742118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ngatchou AN, Kisler K, Fang QH, Walter AM, Zhao Y, Bruns D, Sorensen JB, Lindau M. 2010. Role of the synaptobrevin C terminus in fusion pore formation. Proc Natl Acad Sci USA. 107(43):18463–18468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oelkers M, Witt H, Halder P, Jahn R, Janshoff A. 2016. SNARE-mediated membrane fusion trajectories derived from force-clamp experiments. Proc Natl Acad Sci USA. 113(46):13051–13056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orr A, Song HK, Rusin SF, Kettenbach AN, Wickner W. 2017. HOPS catalyzes the interdependent assembly of each vacuolar SNARE into a SNARE complex. Mol Biol Cell. 28(7):975–983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parisotto D, Pfau M, Scheutzow A, Wild K, Mayer MP, Malsam J, Sinning I, Sollner TH. 2014. An extended helical conformation in domain 3a of Munc18–1 provides a template for SNARE (soluble N-ethylmaleimidesensitive factor attachment protein receptor) complex assembly. J Biol Chem. 289(14):9639–9650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pobbati AV, Stein A, Fasshauer D. 2006. N- to C-terminal SNARE complex assembly promotes rapid membrane fusion. Science. 313(5787):673–676. [DOI] [PubMed] [Google Scholar]
- Prinslow EA, Stepien KP, Pan YZ, Xu J, Rizo J. 2019. Multiple factors maintain assembled trans-SNARE complexes in the presence of NSF and alphaSNAP. Elife. 8( [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quade B, Camacho M, Zhao XW, Orlando M, Trimbuch T, Xu JJ, Li W, Nicastro D, Rosenmund C, Rizo J. 2019. Membrane bridging by Munc13–1 is crucial for neurotransmitter release. Elife. 8:e42806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radhakrishnan A, Li X, Grushin K, Krishnakumar SS, Liu J, Rothman JE. 2021. Symmetrical arrangement of proteins under release-ready vesicles in presynaptic terminals. Proc Natl Acad Sci USA. 118(5):e2024029118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rathore SS, Liu YH, Yu HJ, Wan C, Lee M, Yin Q, Stowell MHB, Shen JS. 2019. Intracellular vesicle fusion requires a membrane-destabilizing peptide located at the juxtamembrane region of the v-SNARE. Cell Rep. 29(13):4583–4592.e3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rebane AA, Ma L, Zhang YL. 2016. Structure-based derivation of protein folding intermediates and energies from optical tweezers. Biophys J. 110(2):441–454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rebane AA, Wang B, Ma L, Qu H, Coleman J, Krishnakumar SS, Rothman JE, Zhang YL. 2018. Two disease-causing SNAP-25B mutations selectively impair SNARE C-terminal assembly. J Mol Biol. 430(4):479–490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rizo J 2018. Mechanism of neurotransmitter release coming into focus. Protein Sci. 27(8):1364–1391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rizo J 2022. Molecular mechanisms underlying neurotransmitter release. Annu Rev Biophys. 51:377–408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rizo J, Sari L, Qi YE, Im W, Lin MM. 2022. All-atom molecular dynamics simulations of Synaptotagmin-SNARE-complexin complexes bridging a vesicle and a flat lipid bilayer. Elife. 11:e76356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rorsman P, Ashcroft FM. 2018. Pancreatic β-cell electrical activity and insulin secretion: of mice and men. Physiol Rev. 98(1):117–214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rothman JE. 2014. The principle of membrane fusion in the cell (Nobel Lecture). Angew Chem Int Edit. 53(47):12676–12694. [DOI] [PubMed] [Google Scholar]
- Rothman JE, Krishnakumar SS, Grushin K, Pincet F. 2017. Hypothesis - buttressed rings assemble, clamp, and release SNAREpins for synaptic transmission. FEBS Lett. 591(21):3459–3480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryu JK, Min D, Rah SH, Kim SJ, Park Y, Kim H, Hyeon C, Kim HM, Jahn R, Yoon TY. 2015. Spring-loaded unraveling of a single SNARE complex by NSF in one round of ATP turnover. Science. 347(6229):1485–1489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sabatini BL, Regehr WG. 1996. Timing of neurotransmission at fast synapses in the mammalian brain. Nature. 384(6605):170–172. [DOI] [PubMed] [Google Scholar]
- Sharma S, Lindau M. 2018. The fusion pore, 60 years after the first cartoon. FEBS Lett. 592(21):3542–3562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma S, Lindau M. 2018. Molecular mechanism of fusion pore formation driven by the neuronal SNARE complex. Proc Natl Acad Sci USA. 115(50):12751–12756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen JS, Tareste DC, Paumet F, Rothman JE, Melia TJ. 2007. Selective activation of cognate SNAREpins by Sec1/Munc18 proteins. Cell. 128(1):183–195. [DOI] [PubMed] [Google Scholar]
- Shen XM, Selcen D, Brengman J, Engel AG. 2014. Mutant SNAP25B causes myasthenia, cortical hyperexcitability, ataxia, and intellectual disability. Neurology. 83(24):2247–2255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi L, Shen QT, Kiel A, Wang J, Wang HW, Melia TJ, Rothman JE, Pincet F. 2012. SNARE proteins: One to fuse and three to keep the nascent fusion pore open. Science. 335(6074):1355–1359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin OH, Lu J, Rhee JS, Tomchick DR, Pang ZPP, Wojcik SM, Camacho-Perez M, Brose N, Machius M, Rizo J, Rosenmund C, Sudhof TC. 2010. Munc13 C2B domain is an activity-dependent Ca2+ regulator of synaptic exocytosis. Nat Struct Mol Biol. 17(3):280–288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shon MJ, Kim H, Yoon TY. 2018. Focused clamping of a single neuronal SNARE complex by complexin under high mechanical tension. Nat Commun. 9(1):3639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shu T, Jin H, Rothman JE, Zhang YL. 2020. Munc13–1 MUN domain and Munc18–1 cooperatively chaperone SNARE assembly through a tetrameric complex. Proc Natl Acad Sci USA. 117(2):1036–1041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sirinakis G, Ren YX, Gao Y, Xi ZQ, Zhang YL. 2012. Combined and versatile high-resolution optical tweezers and single-molecule fluorescence microscopy. Rev Sci Instrum. 83(9):093708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sitarska E, Xu JJ, Park S, Liu XX, Quade B, Stepien K, Sugita K, Brautigam CA, Sugita S, Rizo J. 2017. Autoinhibition of Munc18–1 modulates synaptobrevin binding and helps to enable Munc13-dependent regulation of membrane fusion. Elife. 6:e24278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sligar SG, Denisov IG. 2021. Nanodiscs: A toolkit for membrane protein science. Protein Sci. 30(2):297–315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith CL, Cui Y, Bustamante C. 1996. Overstretching B-DNA: the elastic response of individual double-stranded and single-stranded DNA molecules. Science. 271(5250):795–799. [DOI] [PubMed] [Google Scholar]
- Sollner T, Whiteheart SW, Brunner M, Erdjument-Bromage H, Geromanos S, Tempst P, Rothman JE. 1993. SNAP receptors implicated in vesicle targeting and fusion. Nature. 362(6418):318–324. [DOI] [PubMed] [Google Scholar]
- Sorensen JB. 2009. Conflicting views on the membrane fusion machinery and the fusion pore. Annu Rev Cell Dev Biol. 25:513–537. [DOI] [PubMed] [Google Scholar]
- Stamberger H, Nikanorova M, Willemsen MH, Accorsi P, Angriman M, Baier H, Benkel-Herrenbrueck I, Benoit V, Budetta M, Caliebe A, Cantalupo G, Capovilla G, Casara G, Courage C, Deprez M, Destree A, Dilena R, Erasmus CE, Fannemel M, Fjaer R, Giordano L, Helbig KL, Heyne HO, Klepper J, Kluger GJ, Lederer D, Lodi M, Maier O, Merkenschlager A, Michelberger N, Minetti C, Muhle H, Phalin J, Ramsey K, Romeo A, Schallner J, Schanze I, Shinawi M, Sleegers K, Sterbova K, Syrbe S, Traverso M, Tzschach A, Uldall P, Van Coster R, Verhelst H, Viri M, Winter S, Wolff M, Zenker M, Zoccante L, De Jonghe P, Helbig I, Striano P, Lemke JR, Moller RS, Weckhuysen S. 2016. STXBP1 encephalopathy: A neurodevelopmental disorder including epilepsy. Neurology. 86(10):954–962. [DOI] [PubMed] [Google Scholar]
- Stein A, Weber G, Wahl MC, Jahn R. 2009. Helical extension of the neuronal SNARE complex into the membrane. Nature. 460(7254):525–528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stepien KP, Prinslow EA, Rizo J. 2019. Munc18–1 is crucial to overcome the inhibition of synaptic vesicle fusion by alpha SNAP. Nat Commun. 10:4326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stepien KP, Xu J, Zhang X, Bai XC, Rizo J. 2022. SNARE assembly enlightened by cryo-EM structures of a synaptobrevin-Munc18–1-syntaxin-1 complex. Sci Adv. 8(25):eabo5272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sudhof TC. 2014. The molecular machinery of neurotransmitter release (Nobel lecture). Angew Chem Int Edit. 53(47):12696–12717. [DOI] [PubMed] [Google Scholar]
- Sudhof TC, Rothman JE. 2009. Membrane fusion: grappling with SNARE and SM proteins. Science. 323(5913):474–477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sundaram RV, Jin H, Li F, Shu T, Coleman J, Yang J, Pincet F, Zhang YL, Krishnakumar SS, Rothman JE. 2021. Munc13 binds and recruits SNAP25 to chaperone SNARE complex assembly. FEBS Lett. 595(3):297–309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sutton RB, Fasshauer D, Jahn R, Brunger AT. 1998. Crystal structure of a SNARE complex involved in synaptic exocytosis at 2.4 angstrom resolution. Nature. 395(6700):347–353. [DOI] [PubMed] [Google Scholar]
- van den Bogaart G, Holt MG, Bunt G, Riedel D, Wouters FS, Jahn R. 2010. One SNARE complex is sufficient for membrane fusion. Nat Struct Mol Biol. 17(3):358–364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S, Choi UB, Gong JH, Yang XY, Li Y, Wang AL, Yang XF, Brunger AT, Ma C. 2017. Conformational change of syntaxin linker region induced by Munc13s initiates SNARE complex formation in synaptic exocytosis. EMBO J. 36(6):816–829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S, Li Y, Gong JH, Ye S, Yang X, Zhang R, Ma C. 2019. Munc18 and Munc13 serve as a functional template to orchestrate neuronal SNARE complex assembly. Nat Commun. 10(1):69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weninger K, Bowen ME, Choi UB, Chu S, Brunger AT. 2008. Accessory proteins stabilize the acceptor complex for synaptobrevin, the 1 : 1 syntaxin/SNAP-25 complex. Structure. 16(2):308–320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wickner W 2010. Membrane fusion: Five lipids, four SNAREs, three chaperones, two nucleotides, and a Rab, all dancing in a ring on yeast vacuoles. Annu Rev Cell Dev Biol. 26:115–136. [DOI] [PubMed] [Google Scholar]
- Wu Z, Auclair SM, Bello O, Vennekate W, Dudzinski NR, Krishnakumar SS, Karatekin E. 2016. Nanodisc-cell fusion: control of fusion pore nucleation and lifetimes by SNARE protein transmembrane domains. Sci Rep. 6:27287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Z, Bello OD, Thiyagarajan S, Auclair SM, Vennekate W, Krishnakumar SS, O’Shaughnessy B, Karatekin E. 2017. Dilation of fusion pores by crowding of SNARE proteins. Elife. 6:e22964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Z, Dharan N, McDargh ZA, Thiyagarajan S, O’Shaughnessy B, Karatekin E. 2021. The neuronal calcium sensor Synaptotagmin-1 and SNARE proteins cooperate to dilate fusion pores. Elife. 10:e68215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xi ZQ, Gao Y, Sirinakis G, Guo HL, Zhang YL. 2012. Direct observation of helix staggering, sliding, and coiled coil misfolding. Proc Natl Acad Sci USA. 109(15):5711–5716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu H, Jun Y, Thompson J, Yates J, Wickner W. 2010. HOPS prevents the disassembly of trans-SNARE complexes by Sec17p/Sec18p during membrane fusion. EMBO J. 29(12):1948–1960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu JJ, Camacho M, Xu YB, Esser V, Liu XX, Trimbuch T, Pan YZ, Ma C, Tomchick DR, Rosenmund C, Rizo J. 2017. Mechanistic insights into neurotransmitter release and presynaptic plasticity from the crystal structure of Munc13–1 C1C2BMUN. Elife. 6:e22567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang B, Gonzalez L, Prekeris R, Steegmaier M, Advani RJ, Scheller RH. 1999. SNARE interactions are not selective - Implications for membrane fusion specificity. J Biol Chem. 274(9):5649–5653. [DOI] [PubMed] [Google Scholar]
- Yang XY, Wang S, Sheng Y, Zhang MS, Zou WJ, Wu LJ, Kang LJ, Rizo J, Zhang RG, Xu T, Ma C. 2015. Syntaxin opening by the MUN domain underlies the function of Munc13 in synaptic-vesicle priming. Nat Struct Mol Biol. 22(7):547–754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin P, Gandasi NR, Arora S, Omar-Hmeadi M, Saras J, Barg S. 2018. Syntaxin clusters at secretory granules in a munc18-bound conformation. Mol Biol Cell. 29(22):2700–2708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu H, Shen C, Liu Y, Menasche BL, Ouyang Y, Stowell MHB, Shen J. 2018. SNARE zippering requires activation by SNARE-like peptides in Sec1/Munc18 proteins. Proc Natl Acad Sci USA. 115(36):E8421–E8429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang SW, Ren Q, Novick SJ, Strutzenberg TS, Griffin PR, Bao H. 2021. One-step construction of circularized nanodiscs using SpyCatcher-SpyTag. Nat Commun. 12(1):5451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang XM, Ma L, Zhang YL. 2013. High-resolution optical tweezers for single-molecule manipulation. Yale J Biol Med. 86(3):367–383. [PMC free article] [PubMed] [Google Scholar]
- Zhang XM, Rebane AA, Ma L, Li F, Jiao J, Qu H, Pincet F, Rothman JE, Zhang YL. 2016. Stability, folding dynamics, and long-range conformational transition of the synaptic t-SNARE complex. Proc Natl Acad Sci USA. 113(50):E8031–E8040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang YL. 2017. Energetics, kinetics, and pathway of SNARE folding and assembly revealed by optical tweezers. Protein Sci. 26(7):1252–1265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang YL, Ha T, Marqusee S. 2018. Single-molecule approaches up to difficult challenges in folding and dynamics. J Mol Biol. 430(4):405–408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang YL, Hughson FM. 2021. Chaperoning SNARE folding and assembly. Annu Rev Biochem. 90:581–603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang YL, Jiao J, Rebane AA. 2016. Hidden Markov modeling with detailed balance and its application to single protein folding Biophys J. 111(10):2110–2124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao WD, Hamid E, Shin W, Wen PJ, Krystofiak ES, Villarreal SA, Chiang HC, Kachar B, Wu LG. 2016. Hemi-fused structure mediates and controls fusion and fission in live cells. Nature. 534(7608):548–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou P, Bacaj T, Yang X, Pang ZP, Sudhof TC. 2013. Lipid-anchored snares lacking transmembrane regions fully support membrane fusion during neurotransmitter release. Neuron. 80(2):470–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou P, Pang ZPP, Yang XF, Zhang YS, Rosenmund C, Bacaj T, Sudhof TC. 2013. Syntaxin-1 N-peptide and Habc-domain perform distinct essential functions in synaptic vesicle fusion. EMBO J. 32(1):159–171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Q, Zhou P, Wang AL, Wu D, Zhao M, Sudhof TC, Brunger AT. 2017. The primed SNARE-complexin-synaptotagmin complex for neuronal exocytosis. Nature. 548(7668):420–425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zorman S, Rebane AA, Ma L, Yang GC, Molski MA, Coleman J, Pincet F, Rothman JE, Zhang YL. 2014. Common intermediates and kinetics, but different energetics, in the assembly of SNARE proteins. Elife. 3:e03348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- zur Stadt U, Rohr J, Seifert W, Koch F, Grieve S, Pagel J, Strauss J, Kasper B, Nurnberg G, Becker C, Maul-Pavicic A, Beutel K, Janka G, Griffiths G, Ehl S, Hennies HC. 2009. Familial hemophagocytic lymphohistiocytosis type 5 (FHL-5) is caused by mutations in Munc18–2 and impaired binding to syntaxin 11. Am J Hum Genet. 85(4):482–492. [DOI] [PMC free article] [PubMed] [Google Scholar]


