Abstract
Background:
HIV is a risk factor for obstructive lung disease (OLD), independent of smoking. We used mass spectrometry (MS) approaches to identify metabolomic biomarkers that inform mechanistic pathogenesis of OLD in persons with HIV (PWH).
Methods:
We obtained bronchoalveolar lavage fluid (BALF) samples from 52 PWH, in case:control (+OLD/−OLD) pairs matched on age, smoking status and antiretroviral treatment. 409 metabolites from eight families were measured on BALF and plasma samples using a MS-based Biocrates platform. After filtering metabolites with a high proportion of missing values and values below the level of detection, we performed univariate testing using paired t-tests followed by false discovery rate corrections. We used distance weighted discrimination (DWD) to test for an overall difference in the metabolite profile between cases and controls.
Results:
After filtering, there were 252 BALF metabolites for analysis from eight metabolite families. DWD testing found that collectively, BALF metabolites differentiated cases from controls, whereas plasma metabolites did not. In BALF samples we identified three metabolites that correlated with OLD at the false discovery rate (FDR) of 10%; all were in the phosphatidylcholine family. We identified additional BALF metabolites when analyzing lung function as a continuous variable, and these included acylcarnitines, triglycerides and a cholesterol ester.
Conclusion:
Collectively, BALF metabolites differentiate PWH with and without OLD. These included several BALF lipid metabolites. These findings were limited to BALF and were not found in plasma from the same individuals. Phosphatidylcholine, the most common lipid component of surfactant, was the predominant lipid metabolite differentially expressed.
Keywords: HIV, OLD, metabolome, bronchoalveolar lavage, biomarker
INTRODUCTION:
Improved survival in persons with HIV (PWH) has led to a higher prevalence of several chronic illnesses including obstructive lung disease (OLD), which affects an estimated 3–23%.1–8 Prior to the common use of effective antiretroviral therapy (ART), airflow obstruction was mostly attributed to pulmonary infections.9 Despite the high prevalence of smoking amongst PWH, observational studies indicate that HIV infection itself is an independent risk factor for accelerated lung function decline and subsequently developing OLD.10 In a simian/human immunodeficiency model OLD develops with Pneumocystis colonization that is not reversed with antibiotic treatment, implying immune activation.11 Pathophysiology underlying at least some cases of HIV-associated OLD may differ from usual smoking-associated COPD. Mechanisms leading to HIV-associated OLD are likely multi-factorial, but may include direct and indirect effects of the HIV on the lung.
We previously identified a plasma metabolite profile in HIV-associated OLD that includes sphingolipids, a class of lipids important in cell signaling and surfactants.12 Abnormal lipid metabolism has been described in chronic lung disease, including COPD.13
In this study, the primary aim was to perform large-scale analysis with mass spectrometry of bronchoalveolar lavage fluid (BALF) to identify lung specific metabolites associated with HIV-associated OLD, including lipids. We found that lipids, particularly phosphatidylcholine, are altered in HIV-associated OLD, similar to COPD in HIV-uninfected individuals.
METHODS:
We performed a cross-sectional, matched case:control study using BALF and plasma samples from two cohort studies.
Study Population:
Cases and controls were PWH selected from the Pittsburgh and Vancouver Lung HIV Cohorts collected from 2011–2018 and 2013–2018 respectively. The Pittsburg cohort were recruited from the Multicenter AIDS Cohort Study (MACS) and the Women’s Inter-agency HIV Study (WIHS) cohorts for the bronchoscopy. The Vancouver cohort consisted of excess BALF from clinically indicated bronchoscopies in PWH. BALF was collected using standard procedures. 14,15 We identified 26 cases with HIV and OLD with available BALF; OLD was defined as the ratio of forced expiratory volume in 1s/forced vital capacity (FEV1/FVC) < lower limit of normal (Table 1). Controls consisted of 26 individuals with HIV and normal lung function (defined as FEV1/FVC > lower limit of normal and FEV1 > 80% of predicted normal) matched on age (+/− 5 years), ART use and smoking status (current vs. nonsmoker). Plasma samples were available for 47 individuals including 21 matched case-control pairs. Participants in the parent cohort studies provided informed consent for BALF collection and storage; the current study was approved by the University of Minnesota Institutional Review Board.
Table 1.
Demographics
| Case (N=26) | Control (N=26) | Total (N=52) | |
|---|---|---|---|
|
| |||
| Sex | |||
| Male | 20 (76.9%) | 18 (69.2%) | 38 (73.1%) |
| Age | |||
| Mean (SD) | 59.6 (8.58) | 53.8 (7.30) | 56.7 (8.41) |
| Median | 58.0 [44.0, 80.0] | 54.0 [42.0, 76.0] | 56.0 [42.0, 80.0] |
| Ethnicity | |||
| Black, Non-Hispanic | 16 (61.5%) | 12 (46.2%) | 28 (53.8%) |
| White, Hispanic/Latino | 10 (38.5%) | 13 (50.0%) | 23 (44.2%) |
| Asian/Pacific Islander | 0 (0%) | 1 (3.8%) | 1 (1.9%) |
| Smoking Status | |||
| Yes | 14 (53.8%) | 14 (53.8%) | 28 (53.8%) |
| Former | 9 (34.6%) | 7 (26.9%) | 16 (30.8%) |
| Never | 3 (11.5%) | 5 (19.2%) | 8 (15.4%) |
| Pack Years | |||
| Mean (SD) | 31.1 (28.3) | 15.2 (13.7) | 23.1 (23.4) |
| Median [Min, Max] | 29.6 [0, 120] | 13.6 [0, 38.0] | 17.2 [0, 120] |
| HIV Status | |||
| ART Therapy | 24 (92.3%) | 24 (92.3%) | 48 (92.3%) |
| Viral Load | |||
| < 50 Copies | 12 (46.2%) | 18 (69.2%) | 30 (57.7%) |
| > 50 Copies | 2 (7.7%) | 3 (11.5%) | 5 (9.6%) |
| Missing | 12 (46.2%) | 5 (19.2%) | 17 (32.7%) |
| FEV1 Percent Predicted | |||
| Mean (SD) | 68.1 (16.0) | 104 (11.3) | 85.8 (22.5) |
| Median [Min, Max] | 68.3 [21.0, 90.4] | 102 [80.6, 128] | 86.5 [21.0, 128] |
| DLCO Percent Predicted | |||
| Mean (SD) | 71.6 (26.3) | 76.6 (23.1) | 74.2 (24.6) |
| Median [Min, Max] | 67.6 [36.3, 139] | 74.5 [14.4, 117] | 74.5 [14.4, 139] |
| Missing | 6 (23.1%) | 5 (19.2%) | 11 (21.2%) |
Sample Processing and Metabolomics:
At study enrollment, BALF was collected as previously described. Blood was drawn into EDTA tubes, and plasma was processed within 4 hours. 14,15 Samples were stored at −80 degrees Celsius prior to processing and underwent one freeze-thaw cycle. BALF samples were vortexed and centrifuged at 5000 × G for 5 minutes at 4°C followed by separation of the pellet and supernatant for the removal of additional debris. To identify metabolites, 10μl of plasma or 200 μl of BALF was manually loaded onto a Biocrates Life Sciences Absolute IDQ p400 HR (Biocrates Life Sciences catalog number 21018) following the manufacturer’s instructions and as previously reported.16 BALF samples were pipetted in four 50 μL increments. The addition of each increment was followed by drying under liquid nitrogen for 30 minutes. Both the supernatant and plasma were pipetted into specific designated wells in a randomized plate layout created in MetIDQ and the plate was sealed with a clean silicon mat. Analysis was performed on a Thermo Scientific, Q Exactive TM, Hybrid Quadrupole-Orbitrap TM, mass spectrometer equipped with a Thermo Scientific Ultimate 3000 UHPLC equipped with an autosampler. The autosampler was set to collect eluent from 0.2 to 1.5 minute retention times. Sample metabolite processing and quantification was performed with the integrated MetIDQ Biocrates software17. The Biocrates platform contains standards for eight families of metabolites for a total of 409 individual metabolites. Internal controls are incorporated for normalization between plates. The limit of detection (LOD) for each metabolite is provided by the Biocrates manufacturer and is calculated by Met/DQ™ and is defined as three times the background noise level. Families (number of metabolites) measured included: acylcarnitines (55), amino acids (21), biogenic amines (21), monosaccharide (1), di- and tri- glycerides (60), phospholipids (lysophosphatidylcholines and phosphatidylcholines) (196), sphingolipids (ceramides and sphingomyelins) (40) and cholesteryl esters (14).
Data Cleaning:
We removed any metabolites that were either missing data or below the limit of detection (LOD) for more than 50% of the cohort. This cleaning procedure left 252 metabolites and 52 individuals (26 matched case-control pairs) in the final processed BALF data and 258 metabolites and 42 individuals (consisting of 21 complete case-control pairs) in the final processed plasma data. In both datasets, remaining missing values after filtering were replaced with 0s (i.e., not present). We applied a log(1+x) transformation to the data and scaled and centered each metabolite to have mean 0 and standard deviation 1. We checked for the presence of outliers or technical artifacts from either fluid using principal components analysis plots.
Statistical Analysis
To evaluate the collective power of the metabolites to differentiate between cases and controls, we used distance-weighted discrimination (DWD)18 to classify individuals based on a weighted combination of their observed metabolite levels for each sample type. To assess the significance and robustness of the results, we used a permutation-testing approach 19 for each fluid where we:
Randomly permuted the case-control labels within matched pairs
Applied cross-validation wherein each case:control pair was held out and the DWD classifier was trained using the remaining pairs
Calculated a one-sided paired t-test comparing the scores assigned to cases and controls for held-out pairs and saved the resulting test statistic (termed ‘permutation test statistic’)
This approach accounted for the matched sampling method. After 500 permutations, we obtained a p-value by calculating the proportion of permutation test statistics that were greater than the paired t-test statistic computed using the data with the true case-control labels.
We used paired t-tests to compare the level of each individual metabolite between matched case-control pairs. In the BALF dataset, we performed 252 paired t-tests for each of the 252 metabolites and 258 paired t-tests for the plasma dataset. We then applied a false discovery rate (FDR) correction to the resulting p-values to account for multiple comparisons. 20 We also evaluated whether families of metabolites cumulatively differed between cases and controls by summing over the measured abundances of each metabolite within a family, for BALF and plasma separately, and then compared these levels between cases and controls.
In addition to considering OLD status as a binary variable, we also considered percent of predicted FEV1 as a continuous outcome measure to account for the wide range in these values (20–90% of predicted normal) in our sample. We used Pearson correlation tests to evaluate correlation between percent predicted FEV1 and levels of each metabolite in lavage and plasma. We then applied an FDR correction to adjust for multiple comparisons. We also used lasso regression 21 within each fluid with percent predicted FEV1 as the outcome and metabolites as the predictors. We followed a similar cross validation approach as described for DWD analysis, where each case:control pair was held out as a test set while the model was trained on the remaining pairs. Within each training iteration, we obtained the optimal L1 penalty term using 10-fold cross validation on the training set. This optimal penalty value was then used in prediction of the outcome on the held-out test pair.
We used Pearson correlation tests with FDR correction for multiple comparisons to assess the association between metabolite levels in BALF and plasma.
Lastly, we considered the association between each metabolite and the diffusing capacity of the lungs for carbon monoxide (DLCO). We used Pearson correlation tests with an FDR adjustment to assess the relationship between the metabolites and DLCO.
RESULTS:
Study participant characteristics:
Cases consisted of 26 PWH with OLD (Table 1). Controls were 26 PWH with normal lung function, matched on age, antiretroviral therapy (ART) use and smoking status. Most of the participants were male in their fifth decade of life. Slightly over half were current smokers, 17% were never smokers and most were on ART. Lung function (FEV1) ranged from 21 – 90% of predicted normal in the OLD cases, compared to 80 – 128% in the controls. Airway obstruction (DLCO) ranged from 36 – 139% in cases and 14.4 – 117% in controls. All cases demonstrated obstruction defined as FEV1/FVC < lower limits of normal.
Metabolite Analysis:
We used DWD to assess the collective power of metabolites in either BALF or plasma to distinguish between cases and controls. DWD is a classification method for high-dimensional data sets with low sample-size that calculates a weighted sum of metabolite expression levels for each sample that distinguishes cases from controls. Following permutation testing, the one-sided permutation testing p-value was 0.014 for BALF metabolites and 0.224 for plasma metabolites. These p-values suggest that BALF metabolite levels collectively distinguish between cases and controls, whereas plasma metabolites do not (Figure 1). Figure 2 shows the metabolite expression across the patient cohort. The metabolites are ordered by the direction and magnitude of their correlation with FEV1pp.
Figure 1:
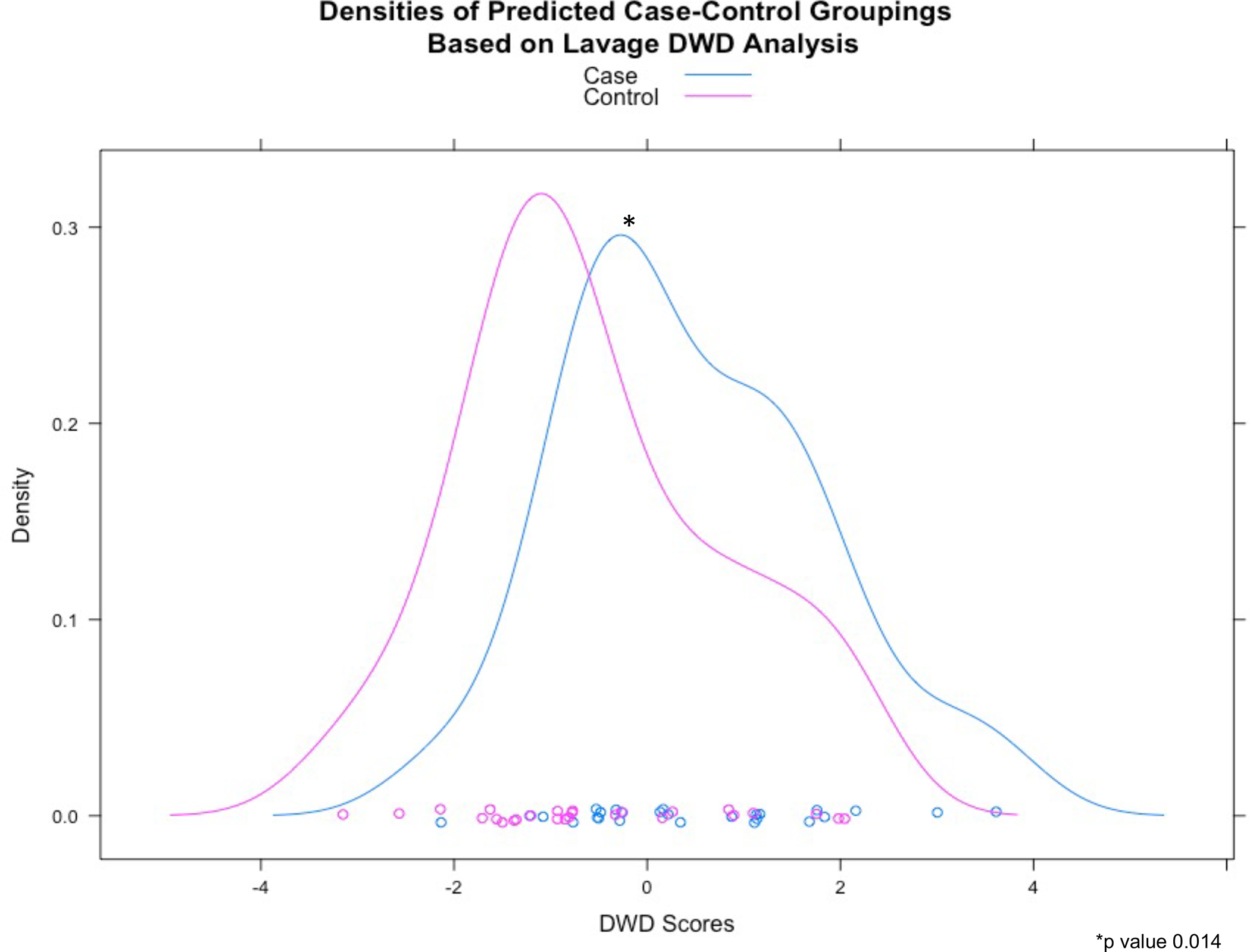
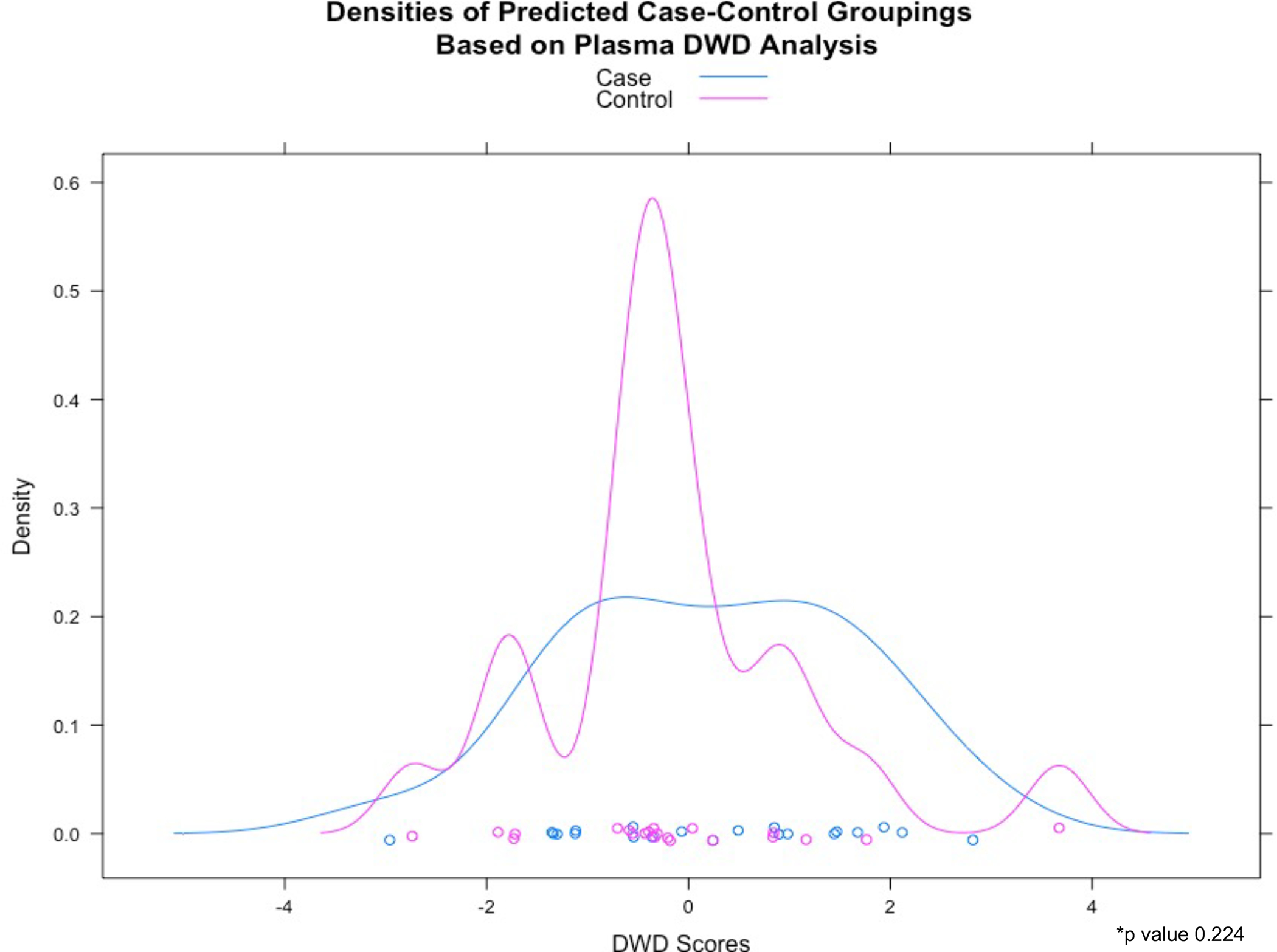
Distribution of DWD scores based on a) bronchoalveolar lavage and b) plasma metabolites
a) BALF (*p value 0.014)
b) Plasma (*p value 0.224)
Figure 2:
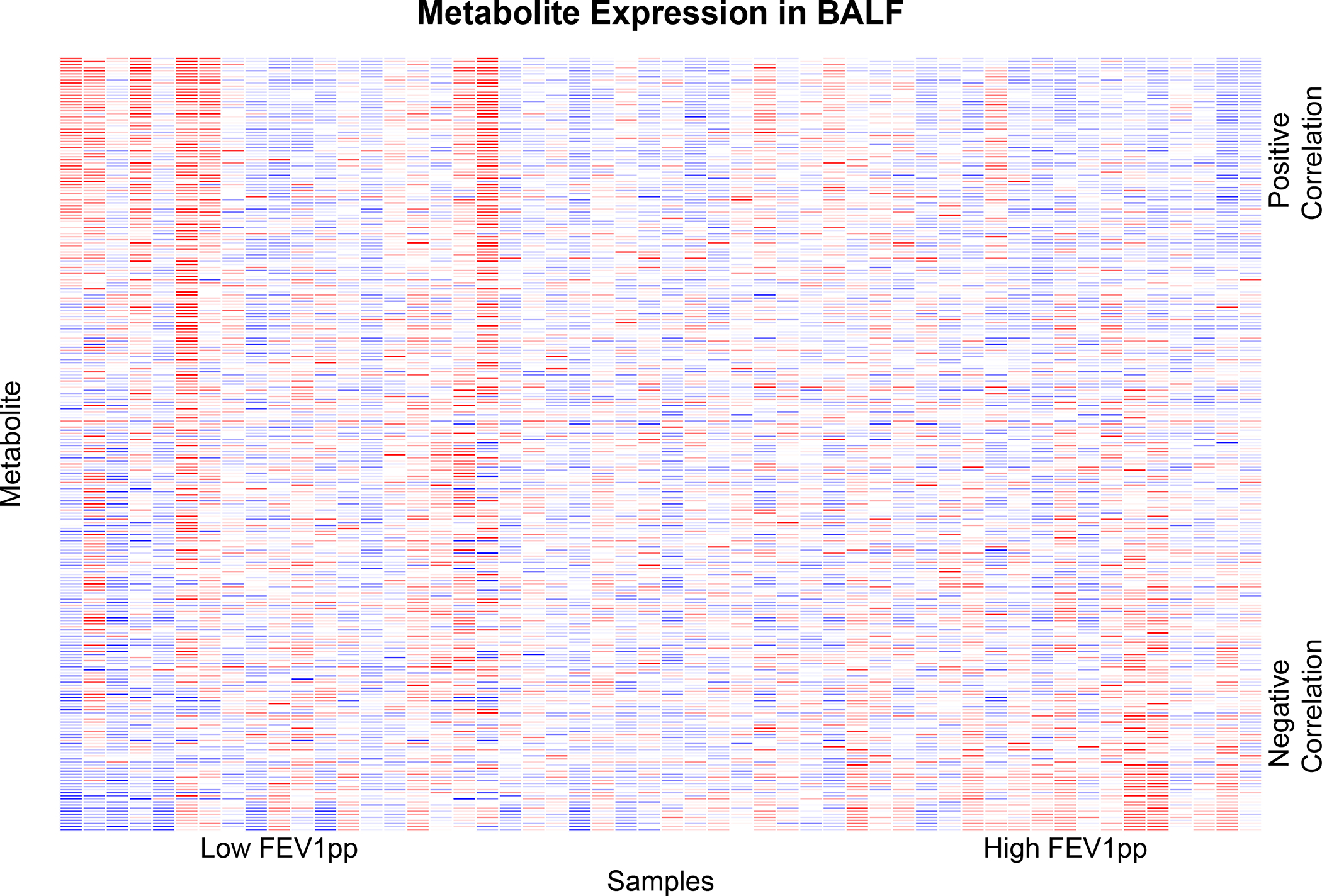
Heatmap showing metabolite expression for the study cohort. Columns reflect samples and rows reflect metabolites. The metabolites are ordered by the direction and magnitude of their correlation with FEV1pp.
Using paired t-tests, we identified three metabolites (PC(31:0), PC(31:3), PC(31:4)) in BALF that were significantly different between cases and controls at the 10% FDR threshold (Table 1S). These metabolites were exclusively in the phosphatidylcholine (PC) metabolite family and are listed using standard lipid nomenclature.22 We did not find any plasma metabolites that were significant at the 10% FDR threshold. When we considered the cumulative expression of each metabolite family, none in BALF were significantly different (p-value>0.05) between cases and controls, though plasma glycerides (p-value=0.01) and glycerophospholipids (p-value=0.03) were both significantly different between cases and controls.
When we considered the correlation between each metabolite and percent predicted FEV1 as a continuous variable, we found 8 metabolites to be significant at an FDR threshold of 0.01 in BALF (Table 2), 34 at a threshold of 5%, and 47 at a threshold of 10%. These metabolites belonged to multiple families, including phosphatidylcholines, triglycerides and acylcarnitines. As indicated by their correlation, phosphatidylcholines were decreased in severe OLD, whereas triglycerides and acylcarnitines were increased. No metabolites in plasma were significantly correlated with percent predicted FEV1 after multiple comparisons adjustment.
Table 2:
Correlation between metabolites and FEV1 percent predicted for metabolites significant at an FDR threshold of 1%. Shown are both p-values and p-values adjusted for FDR (q-values).
| Metabolite | Correlation | P-Value | Q-Value |
|---|---|---|---|
| PC(34:4) | 0.5859 | 0.0000 | 0.0013 |
| AC(10:0) | −0.5047 | 0.0001 | 0.0090 |
| AC(14:1) | −0.4916 | 0.0002 | 0.0090 |
| TG(55:9) | −0.4941 | 0.0002 | 0.0090 |
| PC(33:0) | 0.4968 | 0.0002 | 0.0090 |
| PC(34:3) | 0.5199 | 0.0001 | 0.0090 |
| AC(18:0) | −0.4815 | 0.0003 | 0.0095 |
| PC(32:0) | 0.4855 | 0.0003 | 0.0095 |
Using lasso regression for BALF metabolites with percent predicted FEV1 as the response, the cross-validated predicted outcomes for each individual had a 0.39 correlation with their true observed percent predicted FEV1 values (Figure 1S), suggesting modest predictive accuracy for percent predicted FEV1. In plasma the correlation between predicted and observed FEV1 values was only 0.05, indicating poor prediction accuracy of the model. Table 3 shows the lasso regression results with the top 8 metabolites, their average coefficients across cross validation folds, and the proportion of folds in which their coefficients were non-zero. Most of the metabolites were phosphatidylcholines (PC) along with two acylcarnitines (AC) and one cholesterol ester (CE). Only the single metabolite PC(34:4) overlapped in all three analyses (Figure 3).
Table 3:
Coefficients of metabolites using lasso regression with cross validation. Metabolites shown are those that were selected for more than 50% of cross validation folds. Coefficients were averaged across folds.
| Metabolite | Average Coefficient | Proportion of Folds Selected | |
|---|---|---|---|
| 123 | PC(34:4) | 4.5729 | 1.0000 |
| 22 | AC(10:0) | −1.7843 | 0.9615 |
| 105 | PC(31:0) | 2.9223 | 0.9615 |
| 33 | AC(14:1) | −1.6489 | 0.9231 |
| 163 | PC(41:2) | −1.9888 | 0.9231 |
| 79 | CE(17:0) | −1.1589 | 0.8846 |
| 194 | PC-O(33:6) | −0.7304 | 0.7692 |
| 113 | PC(33:0) | 0.9883 | 0.7308 |
AC = acylcarnitine, PC = phosphatidylcholine, CE = cholesterol ester
Figure 3.
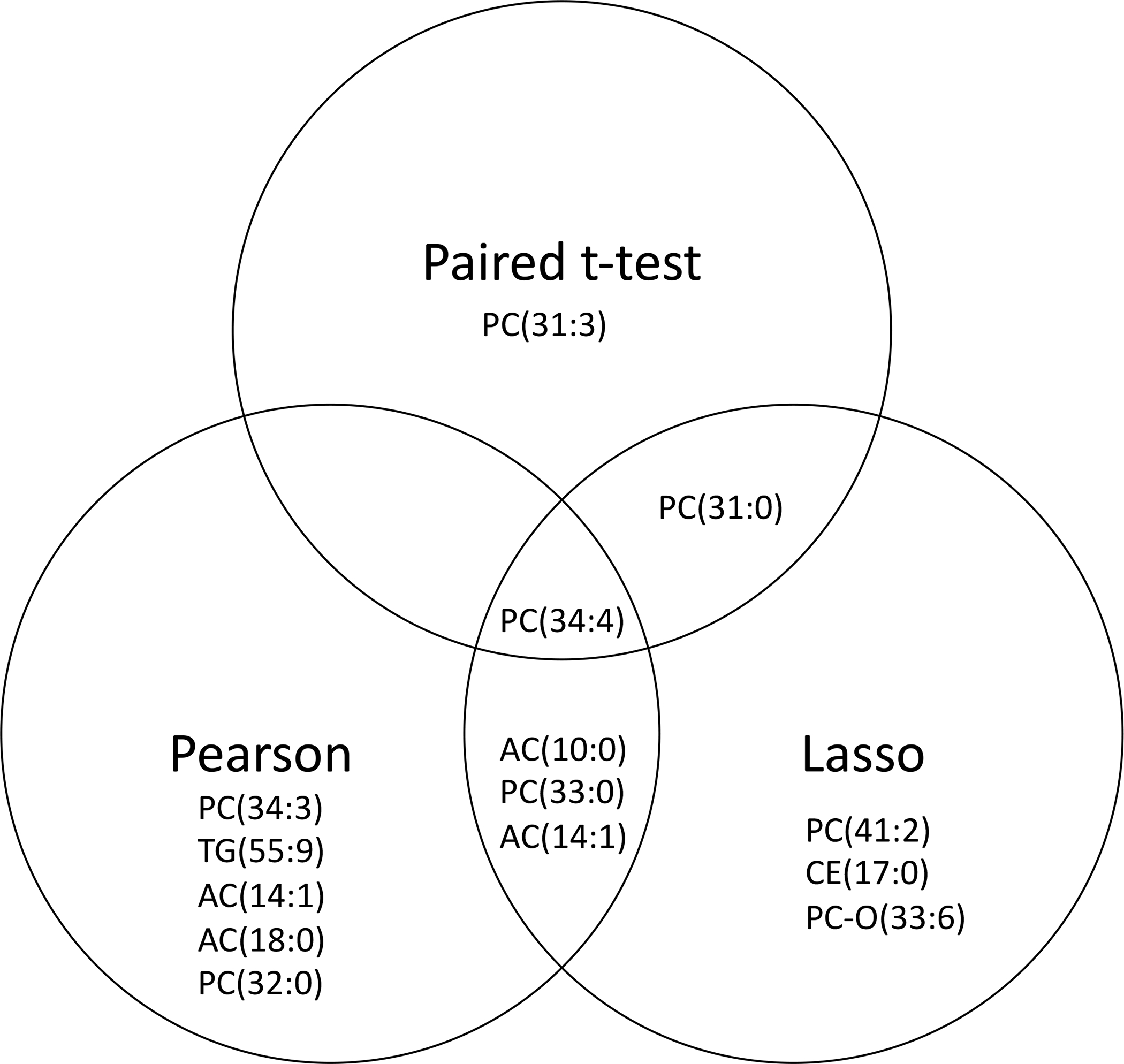
Venn diagram of significant metabolites from separate analyses
For the subjects that had available DLCO, we did not find a correlation with any metabolites with or without FDR correction. In addition, there were 151 metabolites common between BALF and plasma after filtering. When we considered the Pearson correlation between the two fluids for these shared metabolites, we did not find any metabolites that were significantly correlated at the 5%, 10%, or 20% FDR thresholds. This suggests there is little relationship between metabolite levels across the two fluids (Figure 2S).
DISCUSSION:
We found that the BALF metabolome, but not the plasma metabolome, differentiates PWH with and without OLD. The OLD-associated BALF metabolome differed mainly in lipids, particularly phosphatidylcholine species.
This study was designed as a case:control study with controls matched on age, ART and smoking status. Using the case:control status, we identified phosphatidylcholine species as differentially expressed in OLD BALF. Since biomarker expression is influenced by severity of disease, we considered the correlation of metabolites with lung function, i.e. FEV1 percent predicted, which ranged from 21 to 90% predicted. In addition to phosphatidylcholine species, acylcarnitines, triglyceride and a cholesterol ester were associated with the OLD BALF metabolome. The phosphatidylcholine species were decreased in severe OLD whereas acylcarnitines were increased in severe OLD.
Multiple studies have demonstrated abnormal lipid metabolism in COPD.23–26 The lung alveoli participate in lipid metabolism as they contain both type II cells that produce surfactant lipids and alveolar macrophages that participate in surfactant recycling. Pulmonary surfactant is a lipid-protein complex lining the alveolar air-liquid interface that reduces surface tension to prevent alveolar collapse. Pulmonary surfactant is composed of 90% lipid, primarily phospholipids, with phosphatidylcholine being the major phospholipid. Certain surfactants (e.g. surfactant proteins A and D [SP-A and SP-D]), belong to the collectin family and participate in innate immune responses. 27 Surfactant and its major component, phosphatidylcholine, are decreased in smoking-associated COPD, both collectively and individual PC species.28 This decrease in alveolar lipid composition is associated with a change in the alveolar macrophage lipid metabolism transcriptome.29 In addition, metabolomic studies demonstrate altered glycerophospholipid metabolism correlates with worse airflow obstruction. 23 Like these other studies, we found that decreases in phosphatidylcholine correlated with worse OLD severity in PWH, as measured by lower FEV1. Included amongst our metabolites was the commonly measured surfactant phosphatidylcholine species PC(32:0). The decrease in PC with worsening lung function may represent a combination of abnormal lipid synthesis and/or recycling. Loss of lipid and surfactant is also associated with alveoli cell apoptotic death and cellular dropout. Apoptotic death is seen in the emphysema phenotype of COPD. HIV is an independent risk factor for a low DLCO,30 which can be a surrogate for emphysema. Several observational studies have reported an emphysema phenotype associated with HIV,31 however, a recent study from a large Danish study did not find HIV to be independently associated with emphysema. 32 CT lung imaging was not performed in our study and although we found a correlation with metabolites and FEV1 we did not find a correlation between DLCO and metabolites. Therefore, the decrease in PC may represent a functional change as reflected in the FEV1 or airway obstruction that does not correlate with structural changes in the lung, such as emphysema, that would be reflected in the DLCO. It should be noted that control subjects without OLD had marked reduction in some of the DLCO values, that may reflect something other than lung parenchymal disease.
With rapid expansion of robust and sensitive mass spectrometry technology, identification of complex mixtures of species and homologues of surfactant lipids is expanding. This includes a growing availability of mass spectrometry lipid standards for accurate identification and quantification. In this study, we utilized the Biocrates platform that contains 409 metabolite standards including several lipid families and 196 phospholipid species. When comparing across other mass spectrometry-based studies, some species such as PC(32:0) have commonality. However, we and others have identified unique species associated both with OLD and lung function. Further studies are required to determine if these lipid species are generated due to exogenous and/or endogenous influences on metabolism.
There are several limitations to this study. Abnormal lipid metabolites have been seen in HIV-negative COPD, including PC. Although we identified unique PC metabolite species that differentiate OLD in PWH, their individual biological significance cannot be derived from this study. While our controls were matched on age, smoking status and ART, they were not matched on race or sex. There were more Black and male individuals represented in cases. COPD biomarkers are influenced by both race and sex, and specific lipid species, such as acylcarnitines have been found to be differentially expressed in females with COPD. 33 Another limitation is the relatively small sample size, and this study may have been underpowered to determine all significant metabolites differentially expressed in HIV-associated OLD, especially in plasma. We and others have previously identified metabolite biomarkers of OLD in plasma from HIV negative individuals.34–37 However, these were often much larger case studies and lacked concomitant BALF studies.
A major strength of our study is our analysis of BALF. Most of the prior work seeking to identify biomarkers of HIV-associated OLD have been done in plasma. Interestingly, we did not find the BALF metabolite biomarker differences in plasma suggesting these alterations in metabolites are isolated to the lung or only detectable in the lung. Although wide-scale collection of BALF is not feasible in large cohort studies, our data highlight the scientific value of such specimen collection in informing mechanisms of OLD pathogenesis in PWH.
In summary, we found that BALF metabolites differ between PWH with and without OLD. These included several BALF lipid metabolites. These findings were unique to BALF as they were not identified in plasma from the same individuals. Phosphatidylcholine, the most common lipid component of surfactant, was the predominant lipid metabolite differentially expressed.
Future studies would benefit from larger samples sizes and longitudinal study designs to determine if these changes are transient or correlate with outcomes, such as lung function decline or exacerbation rates.
Supplementary Material
Acknowledgements:
The views expressed in this article are those of the authors and do not reflect the views of the United States Government, the Department of Veterans Affairs, the funders, the sponsors, or any of the authors’ affiliated academic institutions.
Support:
Supported by National Institutes of Health grant R01 HL140971-01A1 (all authors)
Footnotes
Summary of Conflict of Interest Statements: All authors report no conflicts.
Disclaimer
The views expressed in this article are those of the authors and do not reflect the views of the United States Government, the Department of Veterans Affairs, the funders, the sponsors, or any of the authors’ affiliated academic institutions.
Prior presentations: This work was previously presented at the 18th European AIDS Conference, October 27–30, 2020, London, UK
References
- 1.Drummond MB, Merlo CA, Astemborski J, et al. The effect of HIV infection on longitudinal lung function decline among injection drug users: a prospective cohort. AIDS. 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.George MP, Kannass M, Huang L, Sciurba FC, Morris A. Respiratory symptoms and airway obstruction in HIV-infected subjects in the HAART era. PloS one. 2009;4(7):e6328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gingo MR, George MP, Kessinger CJ, et al. Pulmonary function abnormalities in HIV-infected patients during the current antiretroviral therapy era. American journal of respiratory and critical care medicine. 2010;182(6):790–796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hirani A, Cavallazzi R, Vasu T, et al. Prevalence of obstructive lung disease in HIV population: a cross sectional study. Respiratory medicine. 2011;105(11):1655–1661. [DOI] [PubMed] [Google Scholar]
- 5.Crothers K, Huang L, Goulet JL, et al. HIV infection and risk for incident pulmonary diseases in the combination antiretroviral therapy era. American journal of respiratory and critical care medicine. 2011;183(3):388–395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cui Q, Carruthers S, McIvor A, Smaill F, Thabane L, Smieja M. Effect of smoking on lung function, respiratory symptoms and respiratory diseases amongst HIV-positive subjects: a cross-sectional study. AIDS research and therapy. 2010;7:6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Madeddu G, Fois AG, Calia GM, et al. Chronic obstructive pulmonary disease: an emerging comorbidity in HIV-infected patients in the HAART era? Infection. 2013;41(2):347–353. [DOI] [PubMed] [Google Scholar]
- 8.Kristoffersen US, Lebech AM, Mortensen J, Gerstoft J, Gutte H, Kjaer A. Changes in lung function of HIV-infected patients: a 4.5-year follow-up study. Clinical physiology and functional imaging. 2012;32(4):288–295. [DOI] [PubMed] [Google Scholar]
- 9.Raynaud C, Roche N, Chouaid C. Interactions between HIV infection and chronic obstructive pulmonary disease: Clinical and epidemiological aspects. Respiratory research. 2011;12:117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Verboeket SO, Boyd A, Wit FW, et al. Changes in lung function among treated HIV-positive and HIV-negative individuals: analyses of the prospective AGEhIV cohort study. Lancet Heal Longev 2021;2:e202–e211. [DOI] [PubMed] [Google Scholar]
- 11.Kling HM, Shipley TW, Guyach S, Tarantelli R, Morris A, Norris KA. Trimethoprim-sulfamethoxazole treatment does not reverse obstructive pulmonary changes in pneumocystis-colonized nonhuman primates with SHIV infection. J Acquir Immune Defic Syndr. 2014;65(4):381–389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hodgson S, Griffin TJ, Reilly C, et al. Plasma sphingolipids in HIV-associated chronic obstructive pulmonary disease. BMJ Open Respir Res. 2017;4(1):e000180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Agudelo CW, Samaha G, Garcia-Arcos I. Alveolar lipids in pulmonary disease. A review. Lipids Health Dis. 2020;19(1):122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cribbs SK, Uppal K, Li S, et al. Correlation of the lung microbiota with metabolic profiles in bronchoalveolar lavage fluid in HIV infection. Microbiome. 2016;4:3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Akata K, Leung JM, Yamasaki K, et al. Altered polarization and impaired phagocytic activity of lung macrophages in people with HIV and COPD. J Infect Dis. 2021. [DOI] [PubMed] [Google Scholar]
- 16.Wendt CH, Castro-Pearson S, Proper J, et al. Metabolite profiles associated with disease progression in influenza infection. PloS one. 2021;16(4):e0247493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wenk MR. The emerging field of lipidomics. Nat Rev Drug Discov. 2005;4(7):594–610. [DOI] [PubMed] [Google Scholar]
- 18.Qiao X, Zhang HH, Liu Y, Todd MJ, Marron JS. Weighted Distance Weighted Discrimination and Its Asymptotic Properties. J Am Stat Assoc. 2010;105(489):401–414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wei S, Chihoon L, Wichers L, Marron JS Direction-Projection-Permutation for High-Dimensional Hypothesis Tests. J Comp and Graph Stat. 2016;26(2):549–569. [Google Scholar]
- 20.Benjamini YYH. Controlling the False Discovery Rate: A Practical and Powerful Approach to Multiple Testing. J R Statist Soc. 1995;57(1):289–300. [Google Scholar]
- 21.Tibshirani R Regression Shrinkage and Selection via the Lasso. J R Statist Soc. 1996;58(1):267–288. [Google Scholar]
- 22.Liebisch G, Vizcaino JA, Kofeler H, et al. Shorthand notation for lipid structures derived from mass spectrometry. J Lipid Res. 2013;54(6):1523–1530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cruickshank-Quinn CI, Jacobson S, Hughes G, et al. Metabolomics and transcriptomics pathway approach reveals outcome-specific perturbations in COPD. Sci Rep. 2018;8(1):17132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gillenwater LA, Pratte KA, Hobbs BD, et al. Plasma Metabolomic Signatures of Chronic Obstructive Pulmonary Disease and the Impact of Genetic Variants on Phenotype-Driven Modules. Netw Syst Med. 2020;3(1):159–181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Halper-Stromberg E, Gillenwater L, Cruickshank-Quinn C, et al. Bronchoalveolar Lavage Fluid from COPD Patients Reveals More Compounds Associated with Disease than Matched Plasma. Metabolites. 2019;9(8). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Koike K, Berdyshev EV, Bowler RP, et al. Bioactive Sphingolipids in the Pathogenesis of Chronic Obstructive Pulmonary Disease. Ann Am Thorac Soc. 2018;15(Suppl 4):S249–S252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Casals C, Campanero-Rhodes MA, Garcia-Fojeda B, Solis D. The Role of Collectins and Galectins in Lung Innate Immune Defense. Front Immunol. 2018;9:1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Agudelo CW, Kumley BK, Area-Gomez E, et al. Decreased surfactant lipids correlate with lung function in chronic obstructive pulmonary disease (COPD). PLoS One. 2020;15(2):e0228279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Fujii W, Kapellos TS, Bassler K, et al. Alveolar macrophage transcriptomic profiling in COPD shows major lipid metabolism changes. ERJ Open Res. 2021;7(3). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Crothers K, McGinnis K, Kleerup E, et al. HIV infection is associated with reduced pulmonary diffusing capacity. J Acquir Immune Defic Syndr. 2013;64(3):271–278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Attia EF, Akgun KM, Wongtrakool C, et al. Increased Risk of Radiographic Emphysema in HIV Is Associated With Elevated Soluble CD14 and Nadir CD4. Chest. 2014;146(6):1543–1553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ronit A, Kristensen T, Hoseth VS, et al. Computed tomography quantification of emphysema in people living with HIV and uninfected controls. Eur Respir J. 2018;52(1). [DOI] [PubMed] [Google Scholar]
- 33.Gillenwater LA, Kechris KJ, Pratte KA, et al. Metabolomic Profiling Reveals Sex Specific Associations with Chronic Obstructive Pulmonary Disease and Emphysema. Metabolites. 2021;11(3). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bowler RP, Wendt CH, Fessler MB, et al. New Strategies and Challenges in Lung Proteomics and Metabolomics. An Official American Thoracic Society Workshop Report. Ann Am Thorac Soc. 2017;14(12):1721–1743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ubhi BK, Cheng KK, Dong J, et al. Targeted metabolomics identifies perturbations in amino acid metabolism that sub-classify patients with COPD. Molecular bioSystems. 2012;8(12):3125–3133. [DOI] [PubMed] [Google Scholar]
- 36.Ubhi BK, Riley JH, Shaw PA, et al. Metabolic profiling detects biomarkers of protein degradation in COPD patients. Eur Respir J. 2012;40(2):345–355. [DOI] [PubMed] [Google Scholar]
- 37.Wendt CH, Nelsestuen G, Harvey S, Gulcev M, Stone M, Reilly C. Peptides in Bronchoalveolar Lavage in Chronic Obstructive Pulmonary Disease. PloS one. 2016;11(5):e0155724. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


