Abstract
In the past few years, convolutional neural networks (CNNs) have been proven powerful in extracting image features crucial for medical image registration. However, challenging applications and recent advances in computer vision suggest that CNNs are limited in their ability to understand the spatial correspondence between features, which is at the core of image registration. The issue is further exaggerated when it comes to multi-modal image registration, where the appearances of input images can differ significantly. This paper presents a novel cross-modal attention mechanism for correlating features extracted from the multi-modal input images and mapping such correlation to image registration transformation. To efficiently train the developed network, a contrastive learning-based pre-training method is also proposed to aid the network in extracting high-level features across the input modalities for the following cross-modal attention learning. We validated the proposed method on transrectal ultrasound (TRUS) to magnetic resonance (MR) registration, a clinically important procedure that benefits prostate cancer biopsy. Our experimental results demonstrate that for MR-TRUS registration, a deep neural network embedded with the cross-modal attention block outperforms other advanced CNN-based networks with ten times its size. We also incorporated visualization techniques to improve the interpretability of our network, which helps bring insights into the deep learning based image registration methods. The source code of our work is available at https://github.com/DIAL-RPI/Attention-Reg.
Keywords: Multi-modal Registration, Deep learning, Cross-modal attention, Prostate caner imaging
1. Introduction
Image-guided interventional procedures often require registering multi-modal images to visualize and analyze complementary information. For example, prostate cancer biopsy benefits from fusing transrectal ultrasound (TRUS) imaging with magnetic resonance imaging (MR) to optimize targeted biopsy. However, image registration is a challenging task especially for multi-modal images. Traditional multi-modal image registration relies on maximizing the mutual information between images (Maes et al., 1997; Wells et al., 1996), which performs poorly when the input images have complex textural patterns, such as in the case of MR and ultrasound registration. Feature based methods compute the similarity between images by representing image appearances using features. Mutual information (Maes et al., 1997) and its self-similarity enhanced version (Rivaz et al., 2014) describes the similarity between two images from different modalities. Wein et al. (2013) adapts the Linear Correlation of Linear Combination metric, originally designed for CT-US registration, to MR-US registration. Zimmer et al. (2019) argues that the graph Laplacian captures the intrinsic structure of an image’s modality, and that its commutativity is closely related to structures from multi-modal images. Heinrich et al. (2012) proposes a neighborhood descriptor that also takes self-similarity into account. However, these non-learning-bases feature engineering methods limit the registration performance on images in different contrasts, of complicated features, and/or with strong noise.
In the past several years, deep learning has become a powerful tool for medical image registration (Haskins et al., 2020). However, most of the existing methods remain useful only under single-modality settings (Balakrishnan et al., 2019; de Vos et al., 2017; Mok and Chung, 2020; Heinrich, 2019), as the network supervision relies on intensity or texture based information that is only consistent within a single modality. Limited efforts have been put into multi-modal image registration (Fan et al., 2019; Hu et al., 2018; Haskins et al., 2019). In this paper, we focus on registering two distinct imaging modalities, MR and TRUS. It is one of the most challenging tasks among all multi-modal registration settings, as the difference between these two modalities is much more substantial than, e.g., the difference between T1 and T2 MR or the difference between MR and CT. It remains difficult for both conventional and deep learning-based method to align images from these two modalities. Instead of modeling correspondence between images, most CNN-based deep learning methods map the composite features from input images directly into a spatial transformation to align them. So far, the success comes from two primary sources. One is the ability of automatically learning image representations through training a properly designed network; the other is the capability of mapping complex patterns to an image transformation. The current methods mix these two components together for image registration. However, converting image features to a spatial relationship is extremely challenging and highly data-dependent, which is the bottleneck for further improvements of the registration performance.
We hereby propose a cross-modal attention method to efficiently guide the deep neural networks to learn the correspondence between MR and TRUS volumes. Our hypothesis is that explicitly establishing the spatial correspondence between corresponding image features from two different images can significantly improve the performance of image registration. The proposed method adapts the self-attention (Wang et al., 2018; Vaswani et al., 2017) that originally calculates the long-range correspondence within one single feature map to the specific task of image registration. The cross-modal attention aims to incorporate the correspondence between two volumes into the deep learning features for registering multi-modal images. To better bridge the modality difference between the MR and TRUS volumes in the extracted image features, we also introduce a novel contrastive learning-based pre-training method.
1.1. Related works
Starting from the early works of using neural networks for similarity metric computation to direct transformation estimation (Wu et al., 2013; de Vos et al., 2017), deep learning based image registration methods have evolved significantly over the past several years (Haskins et al., 2020). For example, Haskins et al. (2019) developed a deep learning metric to measure the similarity between MR and TRUS volumes. Simonovsky et al. (2016) also proposed a similar idea and experimented on T1-T2 MRI brain registration. The correspondences between the volumes is established by optimizing the similarity iteratively, which can be computationally intensive. de Vos et al. (2017) proposed an end-to-end unsupervised image registration method to train the Spatial Transformer Network (Jaderberg et al., 2015) by maximizing the normalized cross correlation. Their method can directly estimate an image transformation for registration. Balakrishnan et al. (2019) further explored using mean squared voxel-wise difference, local cross-correlation, and Dice similarity coefficient (DSC) of image segmentation to train a registration network to map image features to a spatial transformation. While the methods of transformation estimation changed drastically, the network supervision techniques also underwent significant improvement for image registration. For instance, Hu et al. (2018) proposed an improved version of the DSC metric that applies Gaussian blur to landmark segmentations for smoother learning process. Ha et al. (2020) trained a segmentation network, and used the prediction to guide image registration. Yan et al. (2018) developed an adversarial registration framework using a discriminator to supervise the registration estimator. Siebert et al. (2022) came up with a contrastive learning method that self-supervises the network training.
Some other works have also touched on the idea of establishing correspondence between images for registration. Heinrich (2019) calculated a 6D dissimilarity tensor to map the spatial correspondence between two images, after which a convolutional neural network (CNN) network converts the dense dissimilarity tensor into a deformation map. The method is confined to uni-modal registration due to the direct usage of similarity metric on feature vectors from the a single network. Hu et al. (2019) incorporated contrastive metric to train the network to learn the correspondence between key-points in both images. However, this method also cannot be applied to multi-modal scenarios because the key-point detection relies on Difference of Gaussians, a method sensitive to texture changes. Similar to our proposed pre-training method, Pielawski et al. (2020) also used InfoNCE loss to establish correspondences between multi-modal image pairs. One key difference between these two methods is the choice of negative pairs for contrastive training. Pielawski et al. (2020) used random images from the same dataset as negative pairs to supervise the training, while we believe misaligned patches within the same pair of images is more important for registration.
1.2. Contributions
In this paper, we propose a novel cross-modal attention mechanism to explicitly model the spatial correspondence to improve the performance of neural networks for image registration. By extending the non-local attention mechanism (Wang et al., 2018) to an attention operation between two images, we designed a cross-modal attention block that is specifically oriented towards registration tasks. The attention block captures both local features and their global correspondence. To better utilize the novel cross-modal attention block, we also propose a contrastive learning-based pre-training strategy that helps extract discriminative features from the two imaging modalities.
Hu et al. (2018) proposed one of the few methods that addresses the challenging task of MR and TRUS registration. In their work, multiple prostate-related segmentations from both MR and TRUS are incorporated during training. However, accurately segmenting prostate from TRUS images is not easy, especially given that the TRUS images often suffer severe noise. In our work, we will only be using the MR prostate segmentation and manually labeled ground-truth registration, both of which bring no extra burden to the clinicians since these manual labels are already parts of regular clinical protocol. Sun et al. (2014) worked on the same TRUS-MR registration task. However, their method demands a manual step to rigidly align the TRUS and MR images. Only then may a deformable registration be applied. Our work aims at automating the step for which Sun et al. (2014) requires manual labor. In addition, although the deformable registration pipeline from (Hu et al., 2018) encodes higher flexibility, the clinical study in (Venderink et al., 2018) found no significant difference in the clinical effect between rigid and deformable registration methods.
To the best of our knowledge, this is the first work to embed the non-local attention in the deep neural network for image registration, which substantially extends our preliminary work presented at MICCAI 2021 (Song et al., 2021) with the following contributions:
A detailed explanation of the motivation and design of the proposed cross-modal attention module that effectively establishes feature correspondence for image registration is presented.
We have expanded our method from rigid image registration to deformable registration tasks.
A novel contrastive loss-guided pre-training method is proposed to efficiently establish the cross-modal correspondence of image features.
Extensive experiments were performed as presented in Section 3 to demonstrate the significant improvements in performance brought by the proposed methods. An improved visualization of the explainability of the proposed method is also included.
2. Method
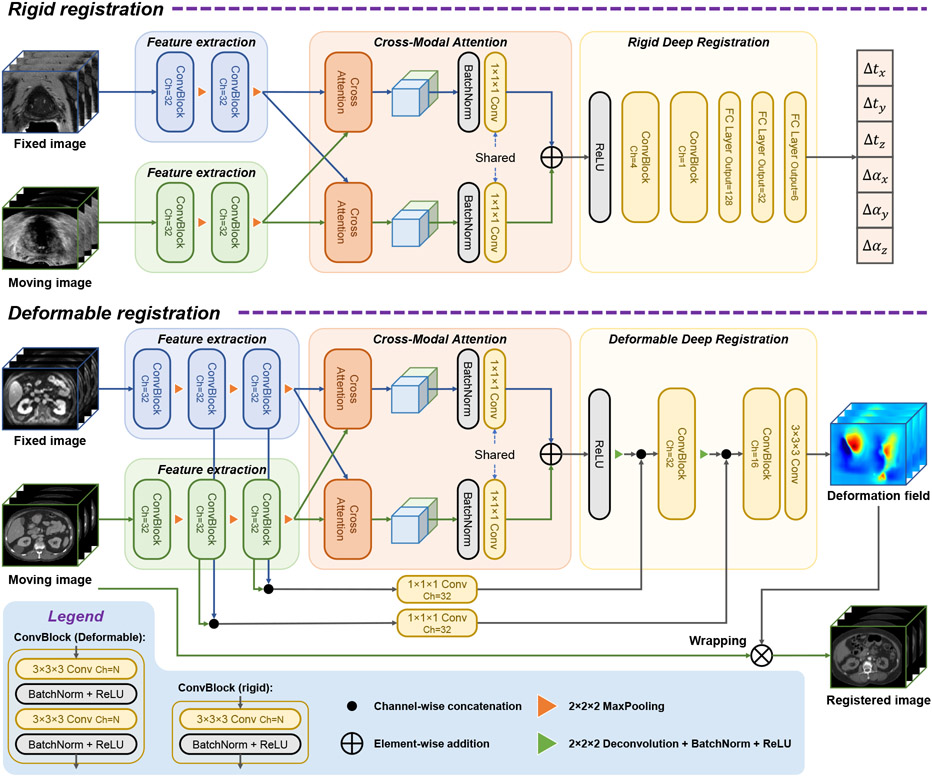
In this multi-modal image registration application, an MRI volume is considered to be the fixed image and a TRUS volume is the moving image. Our registration network consists of three main parts, as shown in Fig. 1. The feature extractor uses convolutional and max pooling layers to capture regional features from the input volumes. Then we use the proposed cross-modal attention block to capture both local features and their global correspondence between modalities. Finally, this information is fed to the deep registrator, which further fuses information from the two modalities and infers the registration parameters. Details of these technical components are presented as follows.
Fig. 1.
Overview of the proposed network structure.
2.1. Feature Extraction
The feature extraction module of the network is designed to extract high-level features that are capable of overcoming the difference between modalities. Due to texture and intensity differences, two different feature extractors are used for each branch of input. For each branch, the input goes through iterations of convolution layer + normalization + ReLU and down-sampling.
2.2. Cross-modal attention
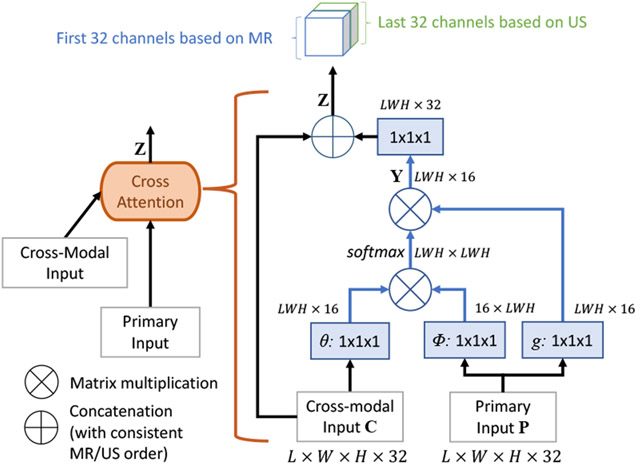
The proposed cross-modal attention block takes as input image features extracted from MR and TRUS volumes by the preceding convolutional layers. Unlike the non-local block (Wang et al., 2018) that computes self-attention on a single image, the proposed cross-modal attention block aims to establish spatial correspondences between features from two images in different modalities. Fig. 2 shows the inner structure of the proposed cross-modal attention block.
Fig. 2.
The proposed cross-modal attention block.
The two input feature maps of the block are denoted as primary input P ∈ and cross-modal input C ∈ , respectively, where LWH indicates the size of each 3D feature channel after flattening. The block computes the cross-modal feature attention as
| (1) |
and the attention weighted primary input as
| (2) |
where ci and pj are features from C and P at location i and j, θ(·), φ(·) and g(·) are all linear embeddings. In Eq. 1, the attention aij is computed as a scalar representing correlations between the features of these two locations, ci and pj. The attention weighted result yi is a normalized summary of features on all locations of P weighted by their correlations with the cross-modal feature on location i as shown in Eq. 2. Thus, the matrix Y composed by yi integrates non-local information from P to every position in C.
Finally, the attention block concatenates Y and C to obtain the output Z to allow efficient back-propagation and prevent potential loss of information. Here we deliberately arrange the ordering of concatenation so that features based on MR are always in the first half channels of Z, and those from TRUS are always in the second half. This maneuver ensures that a common convolution layer can be used for the output of both attention blocks.
2.3. Rigid registration
2.3.1. Contrastive pretraining of the Feature Extraction Module
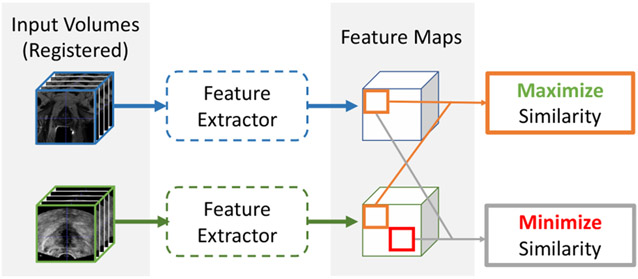
The cross-modal attention module uses the features extracted by the previous feature extraction module to compute the attention. In the early phase of the training, the extracted features may be irrelevant to the image registration task and thus the computed attention may not be correlated with the registration. The overall training can be highly inefficient. To address this issue, we develop a contrastive learning-based pretraining strategy that enforces the feature extractor module of the proposed network to learn similar feature representations from corresponding anatomical regions from two modalities before the end-to-end training of the entire network.
In this task, we are provided with the ground-truth rigid registration between the MR and the TRUS images. When the two images are aligned with the ground-truth transformation, image contents found at the same location in each of the volumes should represent similar anatomical structures. In principle, similar anatomical structures should produce similar feature vectors. Therefore, image contents found at the same location in each volume should produce similar feature vectors. Since aligned image volumes automatically produce aligned feature maps, our contrastive pre-training process aims to maximize the similarity between the feature vectors at the same location in each feature maps, and minimize the similarity between feature vectors at different locations. This process is inspired by (Wang and Liu, 2021) as illustrated in Fig. 3.
Fig. 3.
Illustration of the contrastive pre-training method.
After we obtain the two aligned feature maps, we first normalize all the feature vectors using the L2-norm, and randomly select K pairs of corresponding points from the two feature maps. During the selection process, we avoid feature vectors that are outside of the fan-shaped field-of-view in the original ultrasound image. The selected feature vectors form two K × 32 matrices, one for MR and the other for US, where 32 is the length of each feature vector. We then multiply the two matrices to obtain a K × K cosine similarity map M. Since the feature maps are aligned, our task is to maximize the diagonal of M, and to minimize all other elements. Suppose each row of M represents the similarities between one MR feature vector and all TRUS features, and each column represents similarities between one TRUS feature vector and all MR features. For the MR feature at location i, Eq. 4 will force it to be close to the TRUS feature at the same location i and to be different from the TRUS features at other positions. Similarly, Eq. 5 imposes such a constrain on the TRUS feature at location i.
Iterating this loss across all rows and columns of M could be summarized with Eq. 3
| (3) |
where
| (4) |
| (5) |
In the row-wise and column-wise losses, Lrow(i) and Lcolumn(i), the numerators are the diagonal components {Mi,i, i = [0, K – 1]}. Minimizing the combined contrastive loss will help achieve the effect of maximizing the correlation at the corresponding locations Mi,i and minimizing the correlation Mi,j, (i # j) between patches from misaligned locations.
2.3.2. Rigid Deep Registration module
The deep registration module fuses the concatenated outputs of the two cross-modal attention blocks, and predicts the transformation parameters for registration. Other works have used very deep neural networks to automatically learn the complex features of inputs (Xie et al., 2017). However, since the cross-modal attention blocks help establish the spatial correspondence between the two sets of input volumes, our registration module can afford to be light weighted. Thus, only three convolutional layers are used to fuse the two feature maps. The final fully connected layers convert the learnt spatial information into an estimated transformation. Similar structures for the last few layers are also employed in other works (Guo et al., 2020; Ha et al., 2020).
2.3.3. Rigid registration Implementation Details
In this work, we formulate our work as a rigid transformation task since it is one of the most commonly used registration forms in clinical practice for image-guided prostate intervention (Venderink et al., 2018; Bass et al., 2021). The ground-truth registration labels used in our work are acquired from the clinical procedures of image-fusion guided prostate biopsy. Rigid transformations in our work are performed with 4×4 matrices generated from 6 transformation parameters θ = {Δtx, Δty Δtz, Δax, Δay, Δaz}, which represent translations and rotations along the x, y, and z directions, respectively. We supervise the network training by calculating the Mean Squared Error (MSE) between the prediction and the ground-truth parameters. For the proposed method, we first pre-train the Feature Extraction module of our network as described in Section 2.3.1. We then freeze the pre-trained module to tune the rest of the network. After 300 epochs, we relax the entire network for fine-tuning.
In our experiments, we included the recent methods of MSReg (Guo et al., 2020) and DVNet (Sun et al., 2018) as benchmarks. We used Adam optimizer (Kingma and Ba, 2014) with maximum of 300 epochs to train all the networks including our proposed Attention-Reg approach. We used a step learning rate scheduler for MSReg training, with initial learning rate 5 × 10−5 which then decays by 0.9 for every 5 epochs, as recommended in (Guo et al., 2020). For DVNet, we used the same scheduler but with initial learning rate adjusted to 1 × 10−3. For all other methods, including our pre-training and tuning phase, we used a starting learning rate of 1 × 10−3 and a step learning rate scheduler with step size of 30 and gamma of 0.5. The models were trained on a NVIDIA DGX-1 deep learning server with batch size of 8. All of the benchmarked methods were implemented in Python using the open source PyTorch library (Paszke et al., 2017). Our implementation of the proposed Attention-Reg is available at: https://github.com/DIAL-RPI/Attention-Reg.
2.4. Deformable registration
2.4.1. Deformable Deep Registration module
The deformable registration module acts similar to the decoder structure of a conventional U-Net. It up-samples the outputs of the two cross-modal attention blocks into a full-size dense deformation field φ The predicted deformation field is then either applied to the moving image for inference, or the segmentation of the moving image for DICE supervision. Feature maps at different resolutions from the feature encoder are passed as residual connections to the registration decoder. Since we used two separate feature extractor/encoders, different from U-Net, the residual connections is twice as large as the up-sampled feature map. To resolve this imbalance, the residual connections are first reduced to the same channel size as the up-sampled decoder feature maps with 1 × 1 × 1 convolution layers.
2.4.2. Deformable registration implementation details
We have formulated our method as a deformable registration network and evaluated the performance on the Learn2Reg 2021 Abdomen CT-MR dataset (Hering et al., 2021). For fair benchmarking, we implement our network within the Voxelmorph framework (Balakrishnan et al., 2019) by replacing the U-Net backbone with the proposed network. Training is guided with DICE similarity loss and encourage smoothness with a diffusion regularizer on the spatial gradients of all displacements in the predicted deformation field, consistent with Balakrishnan et al. (2019). The weights for the two losses are 1.0 for DICE and 0.1 for smoothness for all experiments.
For training, we used the paired images from the training set as well as the auxiliary unpaired images. We conducted data augmentation by performing rotation (±5 degrees around each axis), translation (±10 voxels in each direction), and isotropic scaling (±0.1) to both fixed and moving images. For all experiments on this dataset, we used Adam optimizer with learning rate of 1 × 10−4 for a maximum of 800 epochs. Since only the validation set of the dataset is publicly available, we benchmark our result on the validation set with the model from the last training epoch.
3. Experiments and Results
3.1. TRUS-MR rigid registration
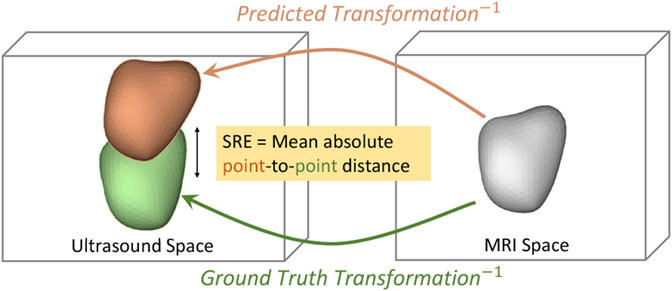
3.1.1. Evaluation Metric: SRE (Surface Registration Error)
The MSE loss used for network training does not directly describe the final registration quality. A clinically meaningful metric should focus on the position and orientation of the relevant organ. Therefore, we use the Surface Registration Error (SRE) to evaluate the registration performance. Let X denote an MR prostate segmentation mesh containing n surface points x. Since we are treating the TRUS as the moving image, both the ground truth Tgt and the estimated transformation Tpred register the TRUS to the MR. Thus, we can use their inverse transformations, and , to map the MR segmentation X to the TRUS space. The SRE is then formally defined as
| (6) |
The SRE describes the Euclidean point-to-point distance between the ground truth registered prostate and the prediction registered prostate, as illustrated in Fig. 4. Such point-to-point distance is not reflected by DICE, which measures the overlap of two volumes regardless of orientation. Nevertheless, we have included DICE as an additional metric for reference.
Fig. 4.
Illustration of the SRE (Surface Registration Error) calculation
3.1.2. Dataset and Preprocessing
In this work, we used 528 cases of MRI-TRUS volume pair for training, 66 cases for validation, and 68 cases for testing. Each case contains a T2-weighted MRI volume and a 3D ultrasound volume. The MRI volumes have 512×512×D voxels, where D ranges from 26 to 32. The ultrasound volume is reconstructed from an electro-magnetic tracked freehand 2D sweep of the prostate. For the purpose of evaluation, all ultrasound volumes are annotated with a mask of whole prostate. 315 volumes are manually annotated by two experienced radiologists. The rest 347 volumes are automatically segmented using a state-of-the-art prostate segmentation method (Xu et al., 2022). Although in an ultrasound guided biopsy workflow the MR should be the moving image, we chose MR as the fixed image in this work due to its much larger physical size and thereby better visualization quality. The initial registration for the training set was generated afresh for every training epoch to boost model robustness. We first perturbed each ground-truth transformation parameter randomly within the range of 5mm of translation or 6 degrees of rotation, and then scale the perturbation to a uniformly sampled SRE within the range of [0,20] mm. We resampled both the MR and the TRUS volumes to 96×96×32 as network input.
The validation set consists of 5 generated initialization matrices for each case, resulting in 330 total samples. For testing, we generated 40 random initialization matrices for each case by using the same random perturbation approach for training, which results in 2,720 test samples. For experiments in Section 3.1.6, we used 10 random initialization matrices per case due to much slower runtime. For experiments in Section 3.1.5, only one non-synthetic clinical initialization matrix is available for each case. Both the validation and test sets are fixed to monitor the epoch-to-epoch performance and evaluate different methods with a consistent basis.
3.1.3. Experiments with different registration methods
We compared our method with other end-to-end rigid registration techniques, including MSReg by Guo et al. (2020) and DVNet by Sun et al. (2018). Table 1 lists the results. The ResNeXt structure adopted by Guo et al. (2020) is one of the more advanced variations of CNN (Xie et al., 2017), adding more weight to this comparison. The 2D CNN network in DVNet (Sun et al., 2018) treats 3D volumes as patches of 2D images, a lighter approach in handling 3D volume registration. To better compare our Attention-Reg with MSReg (Guo et al., 2020), which used two consecutive networks to boost performance, we also trained our network twice on two differently distributed training sets. The model for the 1st stage was trained and tested on a generated dataset with initial SRE uniformly distributed within the range of [0, 20mm], and the range for the 2nd stage was set to be SRE ∈ [0, 8mm]. The trained networks were concatenated together to form a two-stage registration network.
Table 1.
Performance comparison between Attention-Reg, MSReg (Guo et al., 2020), and DVNet (Sun et al., 2018). The initial Dice is 0.619±0.17 and the initial SRE uniformly distributes between 0 and 20 mm.
| Method | Stage 1 | Stage 2 | Registered DICE | #Parameters |
|---|---|---|---|---|
| DVNet (Sun et al., 2018) | 4.80±3.14 | - | 0.794±0.09 | 5,275,832 |
| MSReg (Guo et al., 2020) | 4.79±2.41 | 4.04±2.30 | 0.811±0.06 | 16,106,076 |
| ResNet-50 (He et al., 2016) | 4.64±2.49 | - | 0.799±0.07 | 46,443,622 |
| Attention-Reg | 4.27±2.38 | 3.67±2.00 | 0.818±0.06 | 1,248,905 |
| Ablation Study | ||||
| Attention-Reg w/o attention module | 5.11±2.58 | 0.782±0.07 | 1,244,393 | |
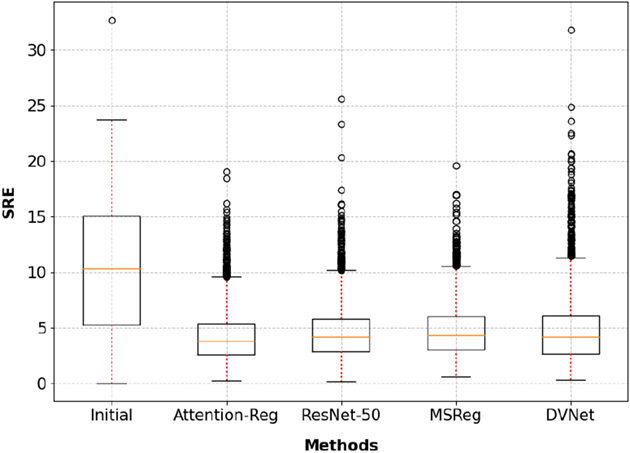
As shown in the top part of Table 1, our cross-modal attention network outperformed MSReg in both registration stages. Furthermore, the significantly better result (p<0.001) was achieved with only 1/10 of number of parameters. The significantly smaller model and simpler calculation demonstrate that the proposed cross-modal attention block can efficiently capture key features of the image registration task. Significant improvement (p<0.001) is also observed when compared to the even larger ResNet-50 (He et al., 2016), which contains 40 × more parameters than the proposed method. Fig 5 plots all the test cases and the registration result from the benchmarked methods. It is evident that the proposed method achieved better performance in every quartile.
Fig. 5.
Boxplot of detailed testing results summarized in Table 1.
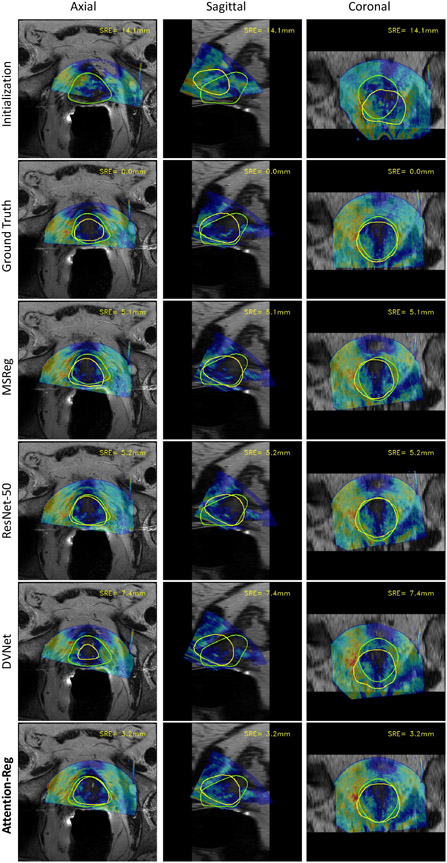
Fig. 6 showcases a sample from our test set to visualize the effect of registration. The SRE of each scenario is shown at the top-right corner of each image. Juxtaposing the ground-truth registration with all the benchmarking methods, we can clearly see how the misalignment intensifies as the corresponding SRE grows.
Fig. 6.
Visualization of registration results from different methods.The contour of MR prostate segmentation is highlighted in green. Ultrasound segmentation is highlighted in yellow.
To demonstrate the contribution of the proposed cross-modal attention block, we trained our Attention-Reg network without the attention block, i.e., directly concatenating the outputs of feature extraction modules and feeding to the deep registration module. The results are shown in the bottom half of Table 1, which shows the importance of the proposed cross-modal attention block. Without the attention module, the registration performance was significantly reduced (p<0.001 with paired t-test) and worse than any other networks of larger size.
It is also worth noting that the residual error achieved with the proposed method (3.67±2.00mm) is approaching the limit of manual rigid alignment. Sun et al. (2014) reports that after initial manual alignment the TRE was reduced to 3.37±1.23mm, only 0.3mm lower than what the proposed method achieved without manual force.
3.1.4. Experiments with different pretraining methods
To examine the effect of the proposed pre-training method, we ran ablation studies to test our network with and without the pretraining steps. The results are summarized in Table 2. When the network is trained from scratch, we observe a significant drop in performance, which demonstrate the effect of the proposed pre-training method. However, even without the pre-training process, the performance of our network is comparable to other benchmark methods in Table 1.
Table 2.
Image registration performance of Attention-Reg under various training strategies. The initial Dice is 0.619±0.17 and the initial SRE uniformly distributes between 0 and 20 mm.
| Training settings | Result SRE | Result Dice | #Parameters |
|---|---|---|---|
| Attention-Reg without pre-training | 4.66±2.49 mm | 0.789±0.07 | 1,248,905 |
| MIND-SSC + Attention-Reg | 5.23±2.76 mm | 0.778±0.08 | 1,187,173 |
| Attention-Reg co-trained with segmentation | 4.98±2.76 mm | 0.783±0.08 | 1,722,995 |
| Constrastive loss pre-trained Attention-Reg | 4.27±2.38 mm | 0.802±0.07 | 1,248,905 |
Similar to feature extracting modules, MIND-SSC (Heinrich et al., 2012) also computes modality independent feature maps from images. Here we replace the feature extracting module with the MIND-SSC algorithm, and directly use the result feature map for attention calculation and the rest of the network.This experiment is listed as MIND-SSC + Attention-Reg in Table 2.
We also experimented the training method proposed by Ha et al. (2020) ), which is to co-train the registration network with an auxiliary segmentation head. For this experiment, we train two U-Nets for the segmentation of MR and TRUS and use the feature maps of these two segmentation networks as input to the registration pipeline.We observed an increase in the registration error compared without pretraining. We speculate that the drop in performance is caused by distractions from segmentation feature maps that aren’t useful for registration. Although the prostate boundary is the key to MR-TRUS registration, prostate boundary isn’t the only focus of segmentation tasks. For region-based segmentation, the network targets at not only the boundary pixels, but also the inner pixels far away from the boundary. Segmentation features from those regions might not match the needs of the registration tasks.
We have experimented with values of 4096, 6144, 8192 for the hyper-parameter K in our pre-training method to test the model sensitivity on K. The corresponding SRE values are 4.27mm, 4.55mm, and 4.29mm, respectively. The performance is not very sensitive to K and a clear trend for an optimal value is not present. Since a larger K leads to larger memory consumption, we decided to use k = 4096, the smallest number that is able to hold sufficient number of positive contrastive samples given any pair of images from the dataset.
3.1.5. Experiments with non-synthetic initialization
Synthetic registration initialization, which have been used in the results from previous sections, might induce bias to the synthetic setting. We have also tested the model that was trained on purely synthetic data on clinically relevant non-synthetic initialization. The results are shown in Table 6 and Figure 3. Although there is a gap between the training distribution of the model and the testing distribution obtained from real-world, the model shows good generalizability in this experiment. In the future we plan to better imitate the real-world initialization during training so that the performance drop may be alleviated.
3.1.6. Experiment with conventional methods
We have also test our methods against conventional registration methods that minimizes a similarity metric. In these experiments, we initialize the registration at different values to test the performance of the benchmarked methods. For each sample from the test set, we generate 10 random initializations at the targeted SRE. The methods and the results are shown in Table 4. From the table we observe that MIND-SSC significantly outperforms mutual information, but the performance degrades a lot when the initialization is far off. This could also be an limitation of the conventional iterative optimizer.
Table 4.
Performance comparison between the proposed method and conventional iterative registration methods with DINO optimizer(Haskins et al., 2019). SRE values are in mm.
| Method | Initialization | Result SRE (mm) |
|---|---|---|
| Mutual Information(Maes et al., 1997) | 8mm | 9.24±3.89 |
| MIND-SSC(Heinrich et al., 2012) | 6.39±2.93 | |
| Attention-Reg | 3.63±1.86 | |
| Mutual Information(Maes et al., 1997) | 16mm | 12.9±5.36 |
| MIND-SSC (Heinrich et al., 2012) | 8.64±3.64 | |
| Attention-Reg | 4.06±2.10 |
3.1.7. Visualization and interpretability
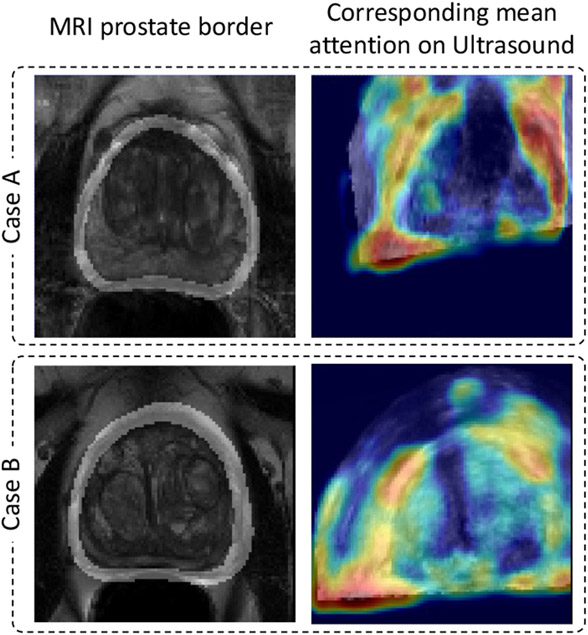
To help understand the function of the proposed cross-modal attention blocks, we visualized prostate boundary from the MR image and their corresponding attention from the US image in Fig. 7. As described in Eq. 1, aij represents the correspondence between feature i from one volume and feature j from the other volume. When the MR feature map serves as the cross-modal input to the proposed attention module, row ai,j∈[1,LWH] represents the correspondence between feature i from MR to every feature vectors from TRUS. We find the average of all ai,j∈[1,LWH] for which ai corresponds to a points on the MR prostate border, and superimpose the reshaped attention heat-map onto the original TRUS volume to create the right column of Fig. 7. The column highlights the corresponding MR prostate boundaries.
Fig. 7.
Visualized attention maps from the multi-modal attention blocks.
Our assumption was that the network should learn to correlate corresponding points from the two volumes, for which this visualization provides strong support. The visualized attention heat-maps showed that the prostate boundary, an important information that guides the registration process, is indeed correlated across modalities as shown in the figure. While registering MR and TRUS volumes can be a abstract task even for humans, overlapping highlighted prostate boundaries is much more straight forward and intuitive.
3.2. CT-MR deformable registration
3.2.1. Dataset and Preprocessing
The Learn2Reg 2021 Abdomen CT-MR dataset (Hering et al., 2021) contains 8 pairs of CT-MR images, 5 for training and 3 for validation. Additionally, 49 CT scans and 40 MR scans can be used as auxiliary training data. All images have been resampled to 192×160×192. For preprocessing, we clip the CT images at HU values from −180 to 400, and MR images from 0 to 95 percentile intensity.
3.2.2. Experiments with different networks
We compare our method with U-Net-based Voxelmorph. As noted in Section 2.6, the proposed method simply replaces the U-Net in Voxelmorph with a cross-modal attention enhances network. The only variable between methods is network structure. We also include an ablation study to demonstrate the effectiveness of the proposed attention module. The result from this dataset is shown in Table 3. From Table 3, we observe that with a smaller network (fewer parameter count), the proposed method is able to outperform the original U-Net-based Voxelmorph. When we delete the attention module from the proposed network as an ablation study, we observe that the performance drops back to similar values as Voxelmorph.
Table 3.
Performance evaluated on real clinical initialization.
| Metric | Initial Error | Registration Error |
|---|---|---|
| SRE | 7.98±5.01 mm | 5.99±3.52 mm |
| DICE | 0.77±0.14 | 0.82±0.06 |
We also note that the proposed network tends to produce deformation fields with higher standard deviation in terms of log Jacobian determinant, despite the same weight on the smoothness loss during training. The cause of such compromise in smoothness could be two-fold. Firstly, the smoothness loss in the Voxelmorph framework does not directly supervise the standard deviation in terms of log Jacobian determinant, which is used in the Learn2Reg challenge to evaluate smoothness. On the other hand, we also speculate that this is caused by the spatial correspondence established by the proposed attention module. Such spatial correspondence, if not perfectly accurate, could disturb the original spatial alignment.
We emphasize how the proposed method can be used to enhance registration performance when used to improve the CNN neural network while the rest of the registration framework remains the same. Although we have only tested the Voxelmorph framework in this paper, we encourage experimenting it in other registration frameworks in the future.
4. Conclusion
This paper introduced a novel attention architecture for the task of medical image registration, and a contrastive loss-guided pre-training method. Our main motivation is to explicitly model the correspondence between the multi-modal inputs to align these images, because image registration is all about the correspondences. The existing deep learning based image registration methods have achieved promising performance, where they might have implicitly mapped the appearance from image pairs into certain relationship. However, such a black-box nature of those methods has hindered the further development of effective deep learning methods for medical image registration. The cross-modal attention mechanism that we introduced in this paper is an innovative attempt towards building the explicit correspondence in the deep learning framework. It helps enhance the explainability of deep learning based image registration methods. The proposed method also advances the state-of-the-art for multi-modal image registration by improving the registration performance.
By comparing the proposed deep neural network with other methods based on the classical CNNs up to ten times of its size, we demonstrated the effectiveness of the new cross-modal attention block in capturing features relevant to image registration. The proposed pre-training method assists the attention calculation by pre-establishing the correspondences between volumes. These innovations have led to significant improvements in image registration accuracy over other pre-existing rigid registration methods. Through feature map visualization, we also observed that the network indeed extracted meaningful features to guide image registration. We expect that the improvements we made to registration networks will inspire and influence future works in the field.
Table 5.
Performance comparison between the proposed network, Voxelmorph (Unet)(Balakrishnan et al., 2019), and ablation study on Learn2Reg 2021 CT-MR deformable registration dataset. Initial DICE=0.310.
| Method | Registered DICE | SDlogJ | #Parameters |
|---|---|---|---|
| Voxelmorph (Unet) (Balakrishnan et al., 2019) | 0.752 | 0.15 | 880,451 |
| Attention-Reg-Deform w/o attention module | 0.758 | 0.56 | 696,035 |
| Attention-Reg-Deform | 0.804 | 0.71 | 696,611 |
The self-attention mechanism is introduced to map the corresponding features from images for multi-modal image registration.
A novel contrastive learning-guided pretraining process is proposed to capture cross-modality image features that are important for registration.
Performance of a network equipped with both of these methods outperforms conventional CNN-networks up to ten times its size.
Acknowledgments
This work was partially supported by National Institute of Biomedical Imaging and Bioengineering (NIBIB) of the National Institutes of Health (NIH) under awards R21EB028001 and R01EB027898, and through an NIH Bench-to-Bedside award made possible by the National Cancer Institute.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Conflict of interest
The authors declare no conflict of interest.
Declaration of interests
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
References
- Balakrishnan G, Zhao A, Sabuncu MR, Guttag J, Dalca AV, 2019. Voxelmorph: a learning framework for deformable medical image registration. IEEE transactions on medical imaging 38, 1788–1800. [DOI] [PubMed] [Google Scholar]
- Bass EJ, Pantovic A, Connor MJ, Loeb S, Rastinehad AR, Winkler M, Gabe R, Ahmed HU, 2021. Diagnostic accuracy of magnetic resonance imaging targeted biopsy techniques compared to transrectal ultrasound guided biopsy of the prostate: a systematic review and meta-analysis. Prostate Cancer and Prostatic Diseases , 1–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan J, Cao X, Wang Q, Yap PT, Shen D, 2019. Adversarial learning for mono-or multi-modal registration. Medical image analysis 58, 101545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo H, Kruger M, Xu S, Wood BJ, Yan P, 2020. Deep adaptive registration of multi-modal prostate images. Computerized Medical Imaging and Graphics 84, 101769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ha IY, Wilms M, Heinrich M, 2020. Semantically guided large deformation estimation with deep networks. Sensors 20, 1392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haskins G, Kruecker J, Kruger U, Xu S, Pinto PA, Wood BJ, Yan P, 2019. Learning deep similarity metric for 3d mr–trus image registration. International journal of computer assisted radiology and surgery 14, 417–425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haskins G, Kruger U, Yan P, 2020. Deep learning in medical image registration: a survey. Machine Vision and Applications 31, 8. [Google Scholar]
- He K, Zhang X, Ren S, Sun J, 2016. Deep residual learning for image recognition, in: Proceedings of the IEEE conference on computer vision and pattern recognition, pp. 770–778. [Google Scholar]
- Heinrich MP, 2019. Closing the gap between deep and conventional image registration using probabilistic dense displacement networks, in: International Conference on Medical Image Computing and Computer-Assisted Intervention, Springer. pp. 50–58. [Google Scholar]
- Heinrich MP, Jenkinson M, Bhushan M, Matin T, Gleeson FV, Brady SM, Schnabel JA, 2012. MIND: Modality independent neighbourhood descriptor for multi-modal deformable registration. Medical Image Analysis 16, 1423–1435. [DOI] [PubMed] [Google Scholar]
- Hering A, Hansen L,Mok TC, Chung A, Siebert H, Häger S, Lange A, Kuckertz S, Heldmann S, Shao W, et al. , 2021. Learn2reg: comprehensive multi-task medical image registration challenge, dataset and evaluation in the era of deep learning. arXiv preprint arXiv:2112.04489 . [DOI] [PubMed] [Google Scholar]
- Hu J, Sun S, Yang X, Zhou S, Wang X, Fu Y, Zhou J, Yin Y, Cao K, Song Q, et al. , 2019. Towards accurate and robust multi-modal medical image registration using contrastive metric learning. IEEE Access 7, 132816–132827. [Google Scholar]
- Hu Y, Modat M, Gibson E, Li W, Ghavami N, Bonmati E, Wang G, Bandula S, Moore CM, Emberton M, et al. , 2018. Weakly-supervised convolutional neural networks for multimodal image registration. Medical image analysis 49, 1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaderberg M, Simonyan K, Zisserman A, et al. , 2015. Spatial transformer networks. Advances in neural information processing systems 28. [Google Scholar]
- Kingma DP, Ba J, 2014. Adam: A method for stochastic optimization. arXiv preprint arXiv:1412.6980. [Google Scholar]
- Maes F, Collignon A, Vandermeulen D, Marchal G, Suetens P, 1997. Multimodality image registration by maximization of mutual information. IEEE Trans. Medical Imaging 16, 187–198. [DOI] [PubMed] [Google Scholar]
- Mok TC, Chung A, 2020. Large deformation diffeomorphic image registration with laplacian pyramid networks, in: International Conference on Medical Image Computing and Computer-Assisted Intervention, Springer. pp. 211–221. [Google Scholar]
- Paszke A, Gross S, Chintala S, et al. , 2017. Automatic differentiation in pytorch, in: NIPS 2017 Workshop Autodiff, pp. 1–4. [Google Scholar]
- Pielawski N, Wetzer E, Öfverstedt J, Lu J, Wählby C, Lindblad J, Sladoje N, 2020. Comir: Contrastive multimodal image representation for registration. Advances in neural information processing systems 33, 18433–18444. [Google Scholar]
- Rivaz H, Karimaghaloo Z, Collins DL, 2014. Self-similarity weighted mutual information: a new nonrigid image registration metric. Medical image analysis 18, 343–358. [DOI] [PubMed] [Google Scholar]
- Siebert H, Hansen L, Heinrich MP, 2022. Learning a metric for multimodal medical image registration without supervision based on cycle constraints. Sensors 22, 1107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simonovsky M, Gutiérrez-Becker B, Mateus D, Navab N, Komodakis N, 2016. A deep metric for multimodal registration, in: International conference on medical image computing and computer-assisted intervention, Springer. pp. 10–18. [Google Scholar]
- Song X, Guo H, Xu X, Chao H, Xu S, Turkbey B, Wood BJ, Wang G, Yan P, 2021. Cross-modal attention for mri and ultrasound volume registration, in: International Conference on Medical Image Computing and Computer-Assisted Intervention, Springer. pp. 66–75. [Google Scholar]
- Sun Y, Moelker A, Niessen WJ, van Walsum T, 2018. Towards robust ct-ultrasound registration using deep learning methods, in: Understanding and Interpreting Machine Learning in Medical Image Computing Applications. Springer, pp. 43–51. [Google Scholar]
- Sun Y, Yuan J, Qiu W, Rajchl M, Romagnoli C, Fenster A, 2014. Three-dimensional nonrigid mr-trus registration using dual optimization. IEEE transactions on medical imaging 34, 1085–1095. [DOI] [PubMed] [Google Scholar]
- Vaswani A, Shazeer N, Parmar N, Uszkoreit J, Jones L, Gomez AN, Kaiser Ł, Polosukhin I, 2017. Attention is all you need. Advances in neural information processing systems 30. [Google Scholar]
- Venderink W, de Rooij M, Sedelaar JM, Huisman HJ, Fütterer JJ, 2018. Elastic versus rigid image registration in magnetic resonance imaging–transrectal ultrasound fusion prostate biopsy: a systematic review and meta-analysis. European Urology Focus 4, 219–227. [DOI] [PubMed] [Google Scholar]
- de Vos BD, Berendsen FF, Viergever MA, Staring M, Išgum I, 2017. End-to-end unsupervised deformable image registration with a convolutional neural network, in: Deep Learning in Medical Image Analysis and Multimodal Learning for Clinical Decision Support. Springer, pp. 204–212. [Google Scholar]
- Wang F, Liu H, 2021. Understanding the behaviour of contrastive loss, in: Proceedings of the IEEE/CVF conference on computer vision and pattern recognition, pp. 2495–2504. [Google Scholar]
- Wang X, Girshick R, Gupta A, He K, 2018. Non-local neural networks, in: Proceedings of the IEEE conference on computer vision and pattern recognition, pp. 7794–7803. [Google Scholar]
- Wein W, Ladikos A, Fuerst B, Shah A, Sharma K, Navab N, 2013. Global registration of ultrasound to mri using the lc 2 metric for enabling neurosurgical guidance, in: International Conference on Medical Image Computing and Computer-Assisted Intervention, Springer. pp. 34–41. [DOI] [PubMed] [Google Scholar]
- Wells WM, Viola P, Atsumi H, Nakajima S, Kikinis R, 1996. Multi-modal volume registration by maximization of mutual information. Medical image analysis 1, 35–51. [DOI] [PubMed] [Google Scholar]
- Wu G, Kim M, Wang Q, Gao Y, Liao S, Shen D, 2013. Unsupervised deep feature learning for deformable registration of mr brain images, in: International Conference on Medical Image Computing and Computer-Assisted Intervention, Springer. pp. 649–656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie S, Girshick R, Dollár P, Tu Z, He K, 2017. Aggregated residual transformations for deep neural networks, in: Proceedings of the IEEE conference on CVPR, pp. 1492–1500. [Google Scholar]
- Xu X, Sanford T, Turkbey B, Xu S, Wood BJ, Yan P, 2022. Shadow-consistent semi-supervised learning for prostate ultrasound segmentation. IEEE Transactions on Medical Imaging 41, 1331–1345. doi: 10.1109/TMI.2021.3139999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan P, Xu S, Rastinehad AR, Wood BJ, 2018. Adversarial image registration with application for MR and TRUS image fusion, in: International Workshop on Machine Learning in Medical Imaging, Springer. pp. 197–204. [Google Scholar]
- Zimmer VA, Ballester MÁG, Piella G, 2019. Multimodal image registration using laplacian commutators. Information Fusion 49, 130–145. [Google Scholar]