Abstract
Caveolins are an unusual family of membrane proteins whose primary biological function is to build small invaginated membrane structures at the surface of cells known as caveolae. Caveolins and caveolae regulate numerous signaling pathways, lipid homeostasis, intracellular transport, cell adhesion, and cell migration. They also serve as sensors and protect the plasma membrane from mechanical stress. Despite their many important functions, the molecular basis for how these 50–100 nm “little caves” are assembled and regulate cell physiology has perplexed researchers for 70 years. One major impediment to progress has been the lack of information about the structure of caveolin complexes that serve as building blocks for the assembly of caveolae. Excitingly, recent advances have finally begun to shed light on this long-standing question. In this review, we highlight new developments in our understanding of the structure of caveolin oligomers, including the landmark discovery of the molecular architecture of caveolin-1 complexes using cryo-electron microscopy.
Keywords: caveolin, caveolae, cryo-electron microscopy, membrane nanodomains, membrane protein structure
Introduction
Almost 70 years ago, numerous 65-nm diameter vesicles now known as caveolae were observed closely nestled to the plasma membrane of endothelial cells in capillaries of multiple tissues (Palade 1953). The integral membrane protein caveolin was identified as a major protein component of caveolae some 40 years later (Glenney and Soppet 1992, Rothberg et al. 1992, Kurzchalia et al. 1994). It was also among one of the earliest proteins to be identified in detergent resistant membranes (Lisanti et al. 1993, Sargiacomo et al. 1993). The initial discovery of caveolin was rapidly followed by the discovery of two additional family members, caveolin-2 (Cav2) and caveolin-3 (Cav3), leading to re-naming of caveolin as caveolin-1 (Cav1) (Way and Parton 1995, Scherer et al. 1996, Tang et al. 1996). Expression of Cav1 is required to drive biogenesis of caveolae in non-muscle cells (Fra et al. 1995, Drab et al. 2001, Razani et al. 2001, Zhao et al. 2002), whereas Cav3 is essential for caveolae in muscle (Galbiati et al. 2001).
Caveolins and caveolae are found in diverse cell types, including endothelial cells, smooth muscle cells, fibroblasts, myoblasts, and adipocytes (Williams and Lisanti 2004, Bastiani and Parton 2010, Chidlow and Sessa 2010, Ariotti and Parton 2013, Hansen et al. 2013, Parton and del Pozo 2013, Parton 2018). Mice lacking caveolins exhibit defects in multiple organ systems such as the vasculature, lungs, heart, muscle, and adipose tissue, suggesting these proteins play critical regulatory roles in multiple pathways, cell types, and tissues (Razani and Lisanti 2001, Mercier et al. 2009). Consistently, in humans, dysregulation of caveolae and caveolins are linked to pulmonary arterial hypertension, cardiovascular disease, lipodystrophies and other systemic diseases (Mercier et al. 2009, Lamaze and Torrino 2015, Patni and Garg 2015, Ma and Chung 2017, Shu et al. 2017, Parton 2018, Pradhan and Proszynski 2020, Mathew 2021). Caveolae are also multi-functional at the cellular level. They regulate signaling, intracellular trafficking, lipid homeostasis, and cell migration, and serve as mechano-sensors and mechano-protectors of the plasma membrane (Parton and del Pozo 2013, Lamaze et al. 2017, Parton 2018, Parton et al. 2020, Parton et al. 2020, Del Pozo et al. 2021). Several models have been advanced to explain the role of caveolae in these events at a mechanistic level. For example, caveolae are now thought to function as plasma membrane reservoirs that buffer the plasma membrane in response to stress by reversibly flattening (Sinha et al. 2011, Nassoy and Lamaze 2012, Cheng et al. 2015, Lamaze et al. 2017, Torrino et al. 2018, Andrade et al. 2022). Flattening of caveolae also serves a mechanotransduction role, leading to the release of accessory proteins that go on to perform biological activities elsewhere in the cell (Torrino et al. 2018, McMahon et al. 2021, Qifti et al. 2022).
Cav1 is a relatively small protein (~21 kDa) and consists of several distinct domains [reviewed in (Williams and Lisanti 2004, Parton et al. 2006, Root et al. 2019)]. The N-terminal region of the protein contains a predicted disordered domain (residues 1–48), a highly conserved signature motif (residues 68–75), a scaffolding domain (residues 82–101), and an oligomerization domain (residues 61–101). The central region of the protein consists of a highly hydrophobic region of the protein known as intramembrane domain (residues 102–134). The C-terminal region of the protein (residues 135–178) contains three cysteines which serve as palmitoylation sites. Multiple regions of Cav1 control its trafficking and specific residues required for caveolae biogenesis have also been identified (Machleidt et al. 2000, Ren et al. 2004, Kirkham et al. 2008, Ariotti et al. 2015). On a mechanistic level, however, our understanding of how caveolins facilitate the formation and function of caveolae been limited by lack of information about the structure of caveolins. In particular, most structural studies have focused on truncated forms of the proteins or used experimental conditions where caveolins are monomeric [reviewed in (Root et al. 2015, Root et al. 2019)]. Until recently, much less was known about the three-dimensional structure of caveolins organized in biologically relevant complexes that function as the fundamental building blocks of caveolae (Parton and Collins 2022). Excitingly, the molecular organization of oligomeric Cav1 complexes has finally been uncovered. Here, we review these new discoveries and the foundational studies that made them possible.
8S complexes function as the major structural unit of caveolin
To support caveolae assembly, Cav1 must first undergo a series of oligomerization and trafficking events (Monier et al. 1995, Scheiffele et al. 1998, Hayer et al. 2010, Busija et al. 2017). The process begins early in the secretory pathway where newly synthesized Cav1 self-assembles to form highly stable oligomeric complexes. These complexes were originally estimated to contain 7–16 Cav1 protomers, and were subsequently dubbed 8S complexes based on their sedimentation in sucrose density gradients (Monier et al. 1995, Sargiacomo et al. 1995, Schlegel et al. 1999, Hayer et al. 2010). Although the exact site of the assembly of 8S complexes in the secretory pathway has recently been questioned (Morales-Paytuvi et al. 2022), it is clear that 8S complexes eventually organize into higher-order 70S complexes and are subsequently trafficked to the plasma membrane where they recruit specific lipids and accessory proteins that assist in sculpting caveolar membranes and regulating caveolae dynamics (Hill et al. 2008, Hansen and Nichols 2010, Hansen et al. 2011, Moren et al. 2012, Stoeber et al. 2012, Ariotti and Parton 2013, Ludwig et al. 2013, Kovtun et al. 2014, Kovtun et al. 2015). Mutant forms of Cav1 that are unable to form oligomers correctly fail to be delivered to the plasma membrane and cannot drive caveolae assembly (Galbiati et al. 1999, Sotgia et al. 2003, Ren et al. 2004, Pol et al. 2005, Hayer et al. 2010). The formation of 8S complexes of Cav1 is thus essential for caveolae assembly.
Early insights into the structure of caveolin complexes
Several potential clues as to the architecture of caveolin complexes emerged from analysis of isolated complexes using electron microscopy (EM). A very early study reported detergent-solubilized homo-oligomers appear as spherical particles, 4–6 nm in diameter, when visualized by EM using platinum shadowing (Sargiacomo et al. 1995). In contrast, oligomers generated by a truncated version of the protein (residues 1–101) were shown to assemble into ring-like complexes 11 nm in diameter containing 7 copies of Cav1, which themselves were capable of forming higher-order filamentous structures (Fernandez et al. 2002). More recent studies of 8S complexes purified from mammalian cells showed full length Cav1 organizes into flat discs with an average diameter of 15 to 17 nm and estimated thickness of 4–5 nm (Stoeber et al. 2016). However, no other structural features were readily obvious (Stoeber et al. 2016).
Analysis of detergent-solubilized Cav3 oligomers provided a more detailed picture of the architecture of caveolin complexes (Whiteley et al. 2012). Here, negatively stained complexes were reported to be toroidal, with a diameter of ~16.5 nm, and height of ~ 5.5 nm (Whiteley et al. 2012). In 3D reconstructions, the complex consists of an outer ring of protein connected by spoke-like densities to a central domain that forms a cone on one surface. Although a distinct symmetry was not obvious in the individual images of particles, 2D averages, or the 3D structure, the discs were proposed to consist of nine copies of Cav3, with the C-terminus of the protein facing the interior portion of the disc and the N-terminus on the outside of the disc. The resolution of these structures was low (>17 Å), most likely due to the heterogeneity of the disc combined with the inherent resolution limits of negative stain. Nevertheless, until recently this represented the highest resolution structural data available for any caveolin oligomeric complex.
EM finally reveals the molecular architecture of the human Cav1 8S complex
Insights into the structure and organization of 8S Cav1 complexes have advanced rapidly over the last two years through studies from our groups of human Cav1 8S-like complexes expressed and purified from E. coli. These advances were enabled by the discovery that heterologously expressed Cav1 oligomerizes and induces membrane curvature in E. coli, generating caveolae-like structures known as heterologous caveolae or h-caveolae (Walser et al. 2012, Ariotti et al. 2015, Shin et al. 2015). These features make E. coli an attractive model for structural studies of Cav1.
In an initial series of studies, we used negative stain single particle EM analysis to define the overall architecture of human 8S Cav1 complexes and identify key determinants of their structure and stoichiometry (Han et al. 2020). We found that the Cav1 complexes form ~15 nm diameter disc-shaped structures composed of an inner and outer ring that are flat on one surface and contain a central protrusion generated from the C-terminal most residues of Cav1 on the other (Figure 1). Since the N- and C-termini of Cav1 are both known to face the cytoplasm, this allowed us to assigned the protrusion-containing surface as the cytoplasmic face of the complex and the flat surface as membrane facing. These complexes could also be directly visualized in purified h-caveolae (Figure 2), reinforcing the idea that they form the fundamental building blocks of these membranous structures (Han et al. 2020).
Figure 1. Overall organization of human CAV1 8S complexes as revealed by single particle EM analysis.
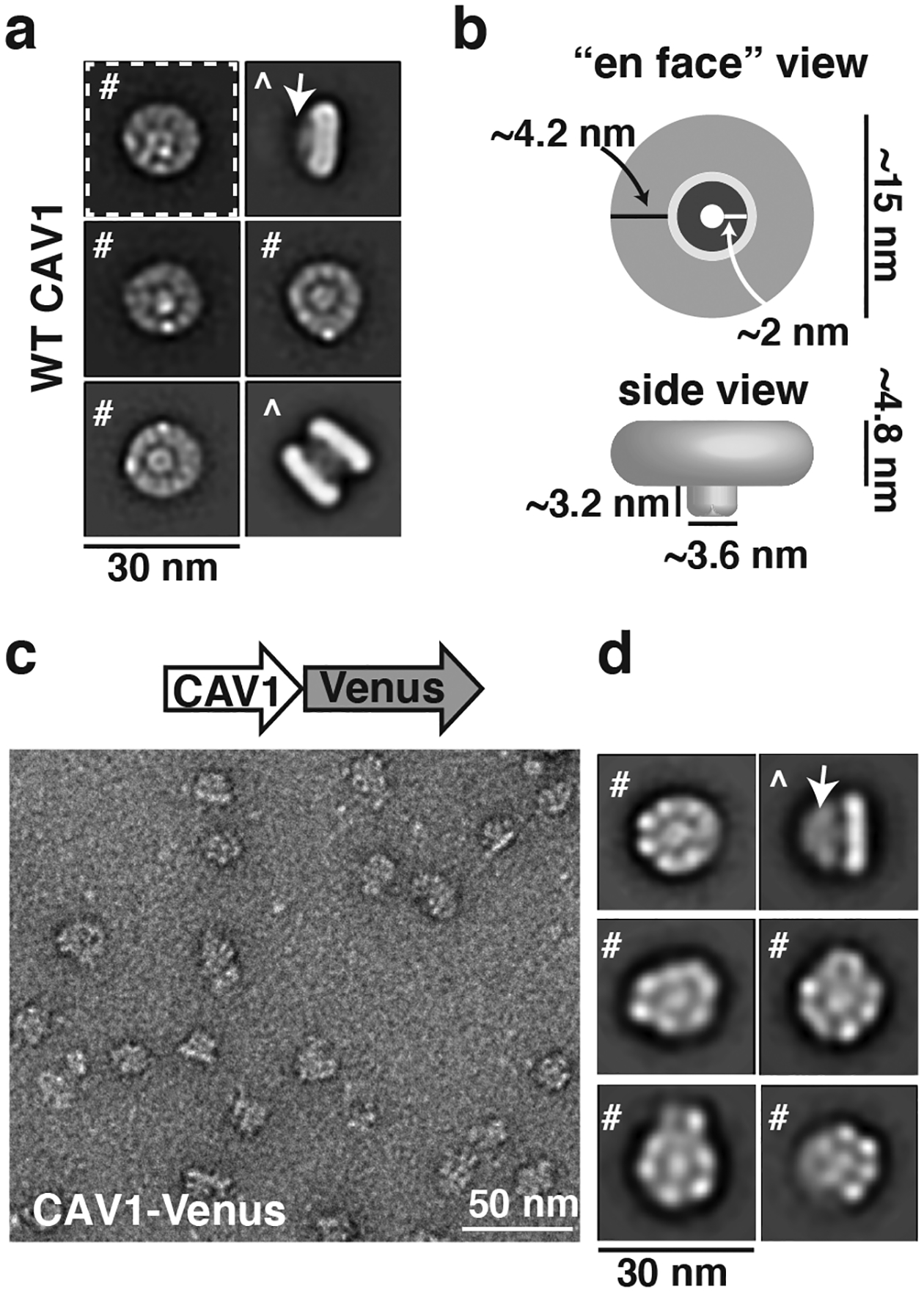
(a) Representative 2D class averages of negatively stained Cav1 8S complexes are disc-like structures with a protruding central stalk (arrow). Examples of both en face views (#) and side views (^) are shown. Scale bar, 30 nm. (b) Dimensions and major features of the Cav1 8S complexes. (c) Representative image of negatively stained Cav1-Venus 8S complexes. Venus serves as a fiducial marker of the C-terminus. Scale bar, 50 nm. (d) Representative 2D class averages of Cav1-Venus 8S complexes. The overall shape and size of the complexes are similar to those shown in panel a, with the exception of the extra fan-like density corresponding to the Venus tag emerging from the central protrusion (arrow). This suggests that the C-terminus of Cav1 is localized in the center of the complex. Scale bar, 30 nm. Adapted from Han, B, JC Porta, JL Hanks, Y Peskova, E Binshtein, K Dryden, DP Claxton, HS McHaourab, E Karakas, MD Ohi and AK Kenworthy (2020). Structure and assembly of CAV1 8S complexes revealed by single particle electron microscopy. Sci Adv 6(49): eabc6185
Figure 2. 8S complexes consisting of Cav1-Venus 8S can be visualized on h-caveolae isolated from E. coli.
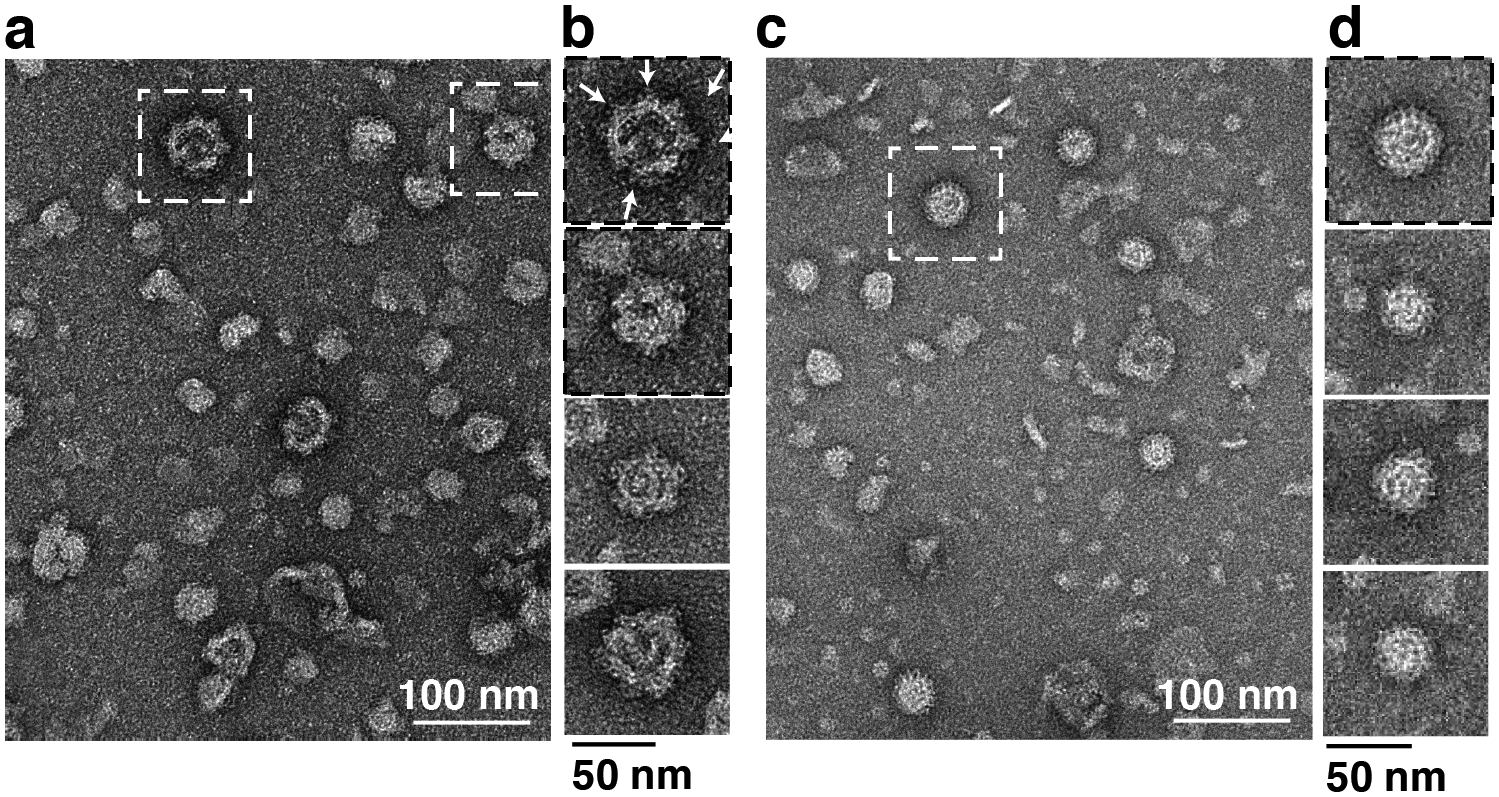
(a) Representative negative stain image of isolated h-caveolae generated by expression of Cav1-Venus in E. coli. The fan shape of the Venus tag (arrows) serves as a fiducial for recognizing the 8S complex. White dashed boxes mark h-caveolae shown in panel b. Scale bar, 100 nm. (b) Close up view of examples of individual h-caveolae isolated from E. coli expressing Cav1-Venus. Note the knob-like structures on the surface of each. Scale bar, 50 nm. (c, d) Representative negative stain image (c) and close up views (d) of isolated h-caveolae generated by expression of WT Cav1 in E. coli. Scale bars are 100 nm in c and 50 nm in d. Adapted from Han, B, JC Porta, JL Hanks, Y Peskova, E Binshtein, K Dryden, DP Claxton, HS McHaourab, E Karakas, MD Ohi and AK Kenworthy (2020). Structure and assembly of CAV1 8S complexes revealed by single particle electron microscopy. Sci Adv 6(49): eabc6185.
The organization of Cav1 protomers within the 8S complex has now been fully uncovered in our 3.5 Å resolution cryo-EM structure of human Cav1 8S complex (Porta et al. 2022) (Figures 3 and 4). The disc-like Cav1 8S complex is composed of eleven closely packed protomers with a diameter of 14 nm (Figure 3). Side views show a centrally located β-barrel protruding from the cytoplasmic face of the complex (Fig. 000). The opposing membrane facing surface is essentially flat and is coated by a detergent micelle, confirming this surface of the complex interacts with membrane (Porta et al. 2022). When viewed en face, the complex consists of an outer rim, curved spokes, and an inner hub region (Fig. 000) (Porta et al. 2022). Along the outer rim, the N-terminus of each protomer forms a loop that tucks under the adjacent protomers, making extensive protomer-protomer contacts that stabilize the overall organization of the disc. The protomers form curved α-helices organized in a “pinwheel”-like arrangement of spokes as they emerge from the rim region and enter the inner β-barrel “hub”, composed of 11 parallel β-strands formed by the C-terminal most residues of each protomer (Figure 3).
Figure 3. Cryo-EM structure of the human Cav1 8S complex reveals it is a tightly packed disc composed of 11 copies of Cav1.
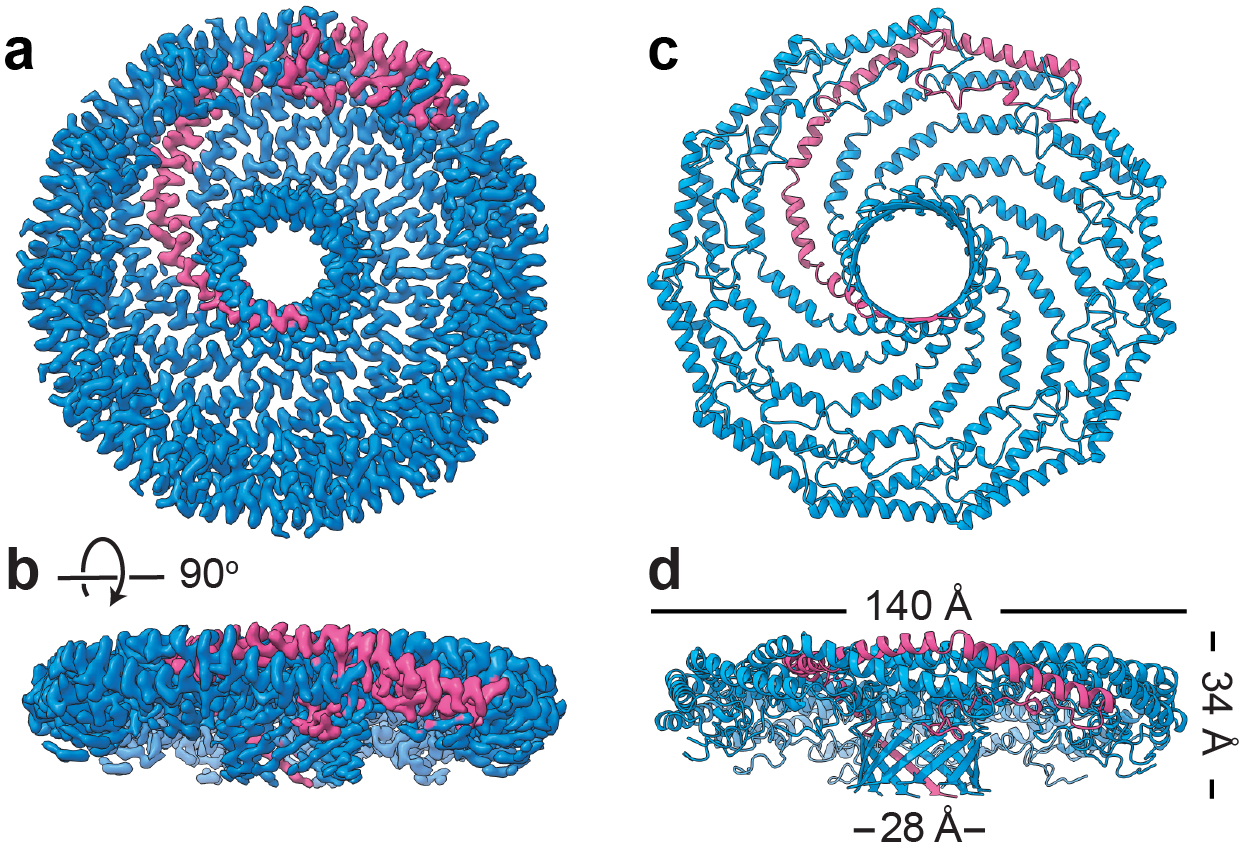
(a) Cryo-EM density map of the Cav1 8S complex at 3.5 Å resolution. A single Cav1 protomer is highlighted in magenta. (b) Side view of the complex. (c) Secondary structure model of the complex shows it is composed of tightly packed α-helices and contains a central β-barrel. (d) Side view of the secondary structure map showing the dimensions of the complex. Adapted from Porta, JC, B Han, A Gulsevin, JM Chung, Y Peskova, S Connolly, HS McHaourab, J Meiler, E Karakas, AK Kenworthy and MD Ohi (2022). Molecular architecture of the human caveolin-1 complex. Sci Adv 8(19): eabn7232.
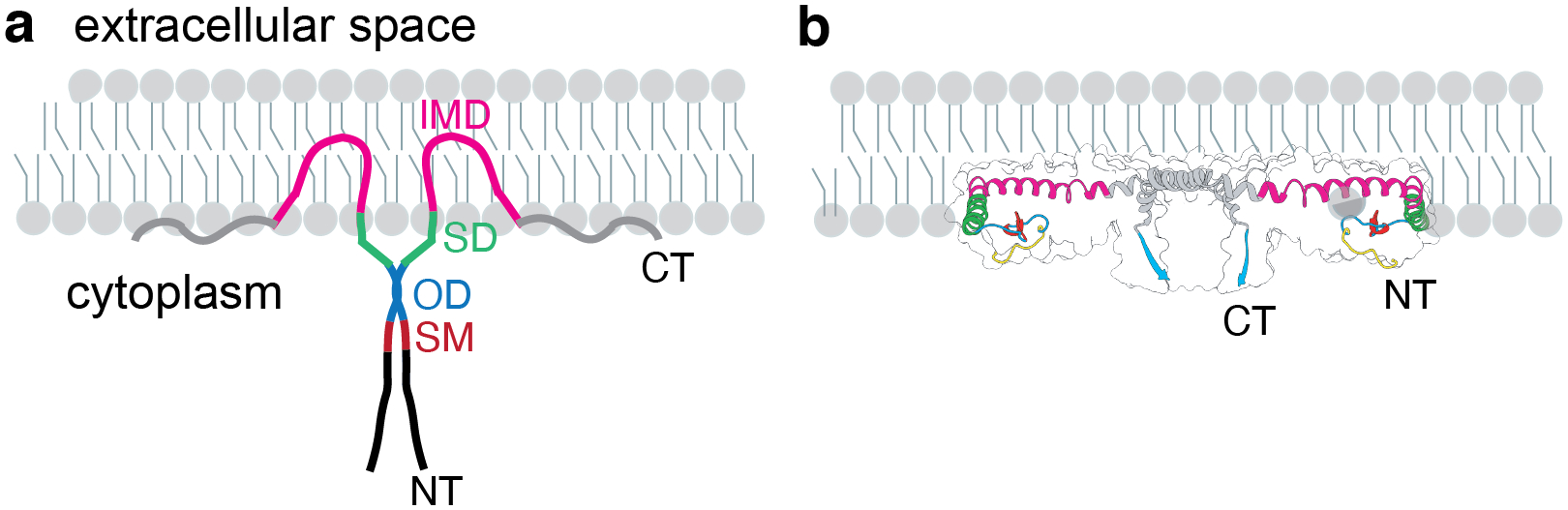
Figure 4. Position of key regions of Cav1 in the atomic model of the human Cav1 8S complex.
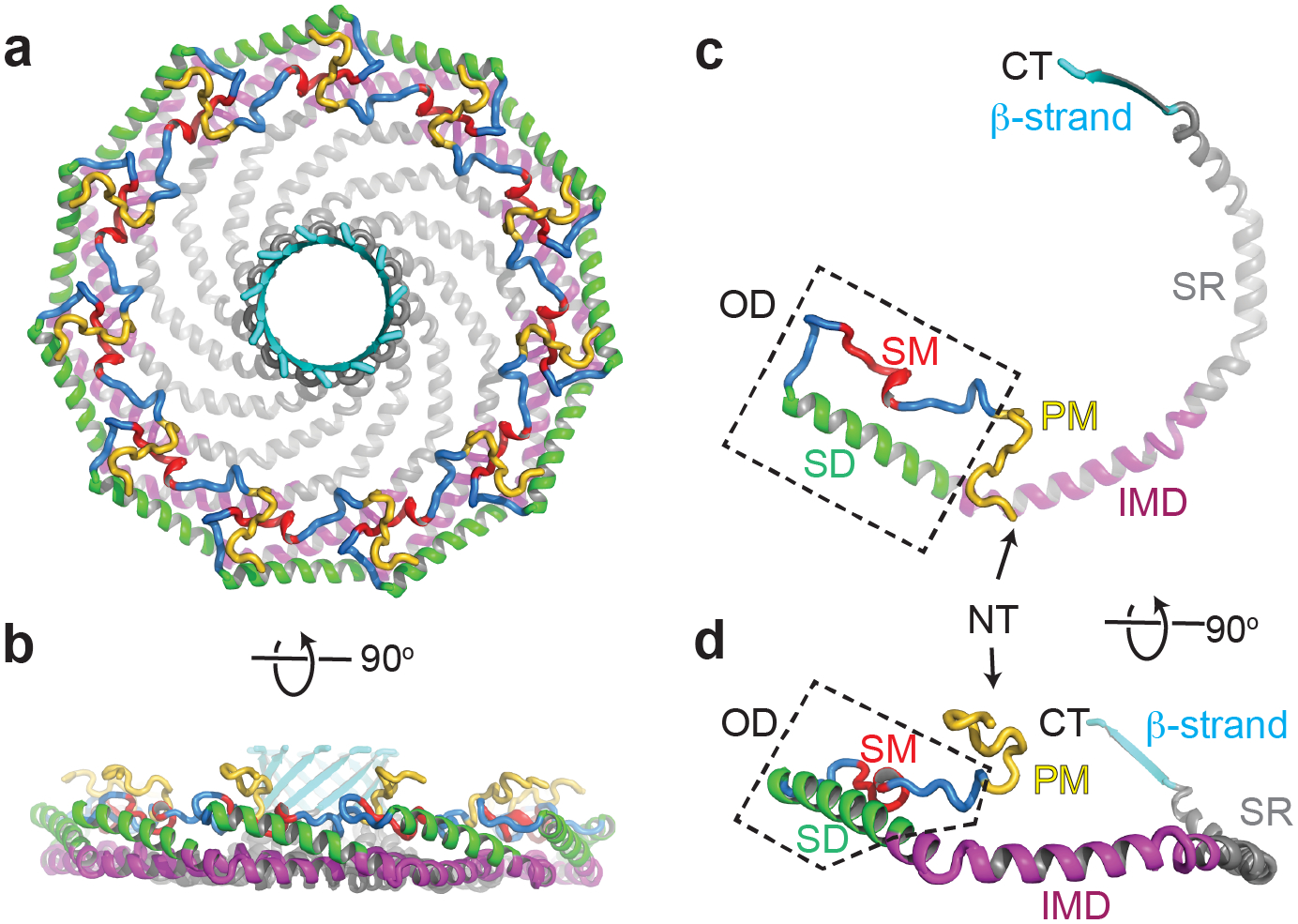
Previously described domains of Cav1 include the signature motif (SM, red), scaffolding domain (SD, green), intramembrane domain (IMD, purple), and oligomerization domain (OD, dashed box). Also shown are the newly described pin motif (PM, yellow), spoke region (SR, gray), and β-strand (cyan). Positions of the N terminus (NT) and C terminus (CT) are marked. Adapted from Porta, JC, B Han, A Gulsevin, JM Chung, Y Peskova, S Connolly, HS McHaourab, J Meiler, E Karakas, AK Kenworthy and MD Ohi (2022). Molecular architecture of the human caveolin-1 complex. Sci Adv 8(19): eabn7232.
The high resolution of the cryo-EM structure allowed us to build an atomic model and locate the position of all of the previously described domains of Cav1, connecting for the first time the structural importance and positions of regions known to be important for biological function (Figure 4). For example, the highly conserved signature motif (SM) (Scherer et al. 1996, Tang et al. 1996) plays an integral role in maintaining close contacts between adjacent protomers in the outer rim region of the complex. The scaffolding domain (SD), long thought to be important for both oligomerization as well as serving as a protein-protein interaction interface (Li et al. 1995, Schlegel et al. 1999), is also localized to the outer rim of the complex where it is both accessible for binding to other proteins and helps define the overall organization and structural integrity of the complex. In contrast, residues 1–48, which are predicted to be disordered (Root et al. 2019), were not resolved.
Several new and surprising features of Cav1 emerged from the structure. First, residues 49–81 at the N-terminal side of the scaffolding domain interact with the neighboring protomer to help lock protomer-protomer interactions in place, leading us to name this region the “pin motif”. The extensive protomer-protomer interactions mediated by this part of the protein helps to explain why this region is required for caveolae formation (Kirkham et al. 2008). A second feature of interest is the intramembrane domain (IMD). Classically, the IMD has been proposed to form a helix-break-helix structure that inserts into the lipid bilayer acting as a wedge (Figure 5A) (Parton et al. 2006, Lee and Glover 2012, Stachowiak et al. 2013, Jarsch et al. 2016, Parton et al. 2021). The structure of the complex shows that instead, this region is tightly packed with neighboring protomers to form a flat membrane-binding surface. This surprising finding has important implications for our understanding of how caveolins shape cell membranes (discussed further below). Immediately following the IMD is an alpha helical region (residues 135–169) which we termed the “spoke region” based on its position in the structure. These helices lie in the same flat plane as the intramembrane domain and are also tightly packed, providing a striking expansion of the hydrophobic surface of the protein. The presence of the 11-stranded parallel β-barrel in the center of the protein, the largest currently known to exist in nature, was yet another surprise. Assembled from the extreme C-terminal residues of each protomer, the barrel is lined on the inside primarily with hydrophobic residues and forms a pore through the center of the complex. Truncation or mutagenesis of residues contributing to the beta barrel causes Cav1 complexes to become structurally heterogenous, suggesting the β-barrel stabilizes the complex (Han et al. 2020). Interestingly, several disease-associated frameshift mutations in the C-terminal domain of Cav1 disrupt the β-barrel (Han et al. 2020), suggesting it may also fulfill as-yet-to-be-determined biological functions in addition to providing structural stability for the complex.
Figure 5. Proposed model for how Cav1 8S complexes associate with membranes.
(a) Classic model of how Cav1 oligomerizes and inserts in membranes. (b) New cryo-EM structure-based model. See text for details. For comparison, positions of the SM (brown), OD (blue), SD (green), and IMD (magenta) are highlighted in each model. Adapted from Porta, JC, B Han, A Gulsevin, JM Chung, Y Peskova, S Connolly, HS McHaourab, J Meiler, E Karakas, AK Kenworthy and MD Ohi (2022). Molecular architecture of the human caveolin-1 complex. Sci Adv 8(19): eabn7232.
The structure of the Cav1 8S complex suggests new models for how it associates with and sculpt membranes
The cryo-EM structure of Cav1 also speaks to how the protein inserts into and bends membranes. Two models for how caveolins induce membrane curvature to generate caveolae in mammalian cells have been proposed. The first model posits that the IMD forms a helix-break-helix motif that adopts a “hairpin” shape and inserts into the cytoplasmic leaflet of the plasma membrane, forming a wedge that induces continuous membrane curvature (Parton et al. 2006, Lee and Glover 2012, Stachowiak et al. 2013, Jarsch et al. 2016, Parton et al. 2021) (Figure 5). Cholesterol has been speculated to control the degree of insertion of the wedge and thus the degree of curvature that Cav1 can generate (Parton et al. 2021). A second model has now arisen based on more recent findings that caveolae and h-caveolae are polyhedral, rather than continuously curved, and contain a regular distribution of repeating polygons (Ariotti et al. 2015, Ludwig et al. 2016, Stoeber et al. 2016, Lamaze et al. 2017, Khater et al. 2019). This model proposes that the 8S complexes comprise the flat surfaces of the polygons (Ariotti et al. 2015, Ludwig et al. 2016, Stoeber et al. 2016, Lamaze et al. 2017). The extensive and flat membrane-facing surface of the disc-shaped 8S complex (Figures 3, 4) supports this latter model.
Cav1 also works together with other caveolar components such as the cavins to dictate the geometry, size, and lipid and protein composition of mammalian caveolae (Ludwig et al. 2016, Busija et al. 2017, Parton et al. 2020, Parton et al. 2021, Tillu et al. 2021, Zhou et al. 2021). Exactly how Cav1 contributes to these events is likely intimately coupled to how the complex inserts into membrane. The cryo-EM structure of Cav1 suggests the entire flat disc region inserts deeply into the membrane, essentially replacing hundreds of lipids on the cytoplasmic face of the membrane (Figure 5B) (Porta et al. 2022). This model has a number of interesting implications, including the possibility that specific lipid species could be enriched in the patch of membrane opposing the complex or around the periphery of the complex. More work will be required to test these intriguing hypotheses.
Conclusions and future directions
Nearly 30 years after the identification of Cav1 as a major structural component of caveolae, a high-resolution structure of the protein in a biologically relevant oligomeric complex has finally been determined. It is now possible to begin to answer a number of long-standing questions in the field. For example, the cryo-EM structure provides a clear explanation for how Cav1 monomers interact with one another to form stable 8S complexes and the role of specific residues and domains of the protein in protomer-protomer interactions. The structure also makes strong predictions about what regions of Cav1 are available to bind other proteins and lipids. These insights will be essential to generate new models for how Cav1 and caveolae regulate cellular functions such as membrane buffering and cell signaling that are essential to ensure proper function of the many tissues and cell types where caveolae are abundant. Other aspects of the structure raise new questions, such as exactly how the complex is positioned in the membrane and how it interacts with specific proteins and lipids to generate curvature. Further investigation into these issues should help resolve the still poorly understood roles of caveolins in the process of caveolae assembly and flattening. Finally, the structure provides a framework for understanding how both common variants and disease-associated mutations of Cav1 impact the structure and function of caveolae. Given the high level of homology between Cav3 and Cav1, it should now become possible to probe the structural basis for the function of this muscle-specific caveolin family member as well. This will be critical moving forward to understand the basis for the shared and isoform-specific features of these two important proteins and could provide insights into diseases linked to defects in Cav3 function.
Data availability statement.
Data sharing is not applicable to this article as no new datasets were generated or analyzed during the current study.
Acknowledgements
We thank all of our colleagues who contributed to our studies of the structure of caveolin-1. This work was supported by National Institutes of Health R01 HL144131 (AKK and MDO). The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Funding:
This work was supported by National Institutes of Health R01 HL144131 (AKK and MDO). The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Footnotes
Disclosures: AKK is a member of the editorial board of Journal of Membrane Biology. The authors have no relevant financial or non-financial interests to disclose.
References
- Andrade V, Bai J, Gupta-Rossi N, Jimenez AJ, Delevoye C, Lamaze C and Echard A (2022). Caveolae promote successful abscission by controlling intercellular bridge tension during cytokinesis. Sci Adv 8(15): eabm5095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ariotti N and Parton RG (2013). SnapShot: caveolae, caveolins, and cavins. Cell 154(3): 704–704 e701. [DOI] [PubMed] [Google Scholar]
- Ariotti N, Rae J, Leneva N, Ferguson C, Loo D, Okano S, Hill MM, Walser P, Collins BM and Parton RG (2015). Molecular characterization of caveolin-induced membrane curvature. J Biol Chem 290(41): 24875–24890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bastiani M and Parton RG (2010). Caveolae at a glance. J Cell Sci 123(Pt 22): 3831–3836. [DOI] [PubMed] [Google Scholar]
- Busija AR, Patel HH and Insel PA (2017). Caveolins and cavins in the trafficking, maturation, and degradation of caveolae: implications for cell physiology. Am J Physiol Cell Physiol 312(4): C459–C477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng JP, Mendoza-Topaz C, Howard G, Chadwick J, Shvets E, Cowburn AS, Dunmore BJ, Crosby A, Morrell NW and Nichols BJ (2015). Caveolae protect endothelial cells from membrane rupture during increased cardiac output. J Cell Biol 211(1): 53–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chidlow JH Jr. and Sessa WC (2010). Caveolae, caveolins, and cavins: complex control of cellular signalling and inflammation. Cardiovasc Res 86(2): 219–225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Del Pozo MA, Lolo FN and Echarri A (2021). Caveolae: Mechanosensing and mechanotransduction devices linking membrane trafficking to mechanoadaptation. Curr Opin Cell Biol 68: 113–123. [DOI] [PubMed] [Google Scholar]
- Drab M, Verkade P, Elger M, Kasper M, Lohn M, Lauterbach B, Menne J, Lindschau C, Mende F, Luft FC, Schedl A, Haller H and Kurzchalia TV (2001). Loss of caveolae, vascular dysfunction, and pulmonary defects in caveolin-1 gene-disrupted mice. Science 293(5539): 2449–2452. [DOI] [PubMed] [Google Scholar]
- Fernandez I, Ying Y, Albanesi J and Anderson RG (2002). Mechanism of caveolin filament assembly. Proc Natl Acad Sci U S A 99(17): 11193–11198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fra AM, Williamson E, Simons K and Parton RG (1995). De novo formation of caveolae in lymphocytes by expression of VIP21-caveolin. Proc. Natl. Acad. Sci. USA 92(19): 8655–8659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galbiati F, Engelman JA, Volonte D, Zhang XL, Minetti C, Li M, Hou H Jr., Kneitz B, Edelmann W and Lisanti MP (2001). Caveolin-3 null mice show a loss of caveolae, changes in the microdomain distribution of the dystrophin-glycoprotein complex, and t-tubule abnormalities. J Biol Chem 276(24): 21425–21433. [DOI] [PubMed] [Google Scholar]
- Galbiati F, Volonte D, Minetti C, Chu JB and Lisanti MP (1999). Phenotypic behavior of caveolin-3 mutations that cause autosomal dominant limb girdle muscular dystrophy (LGMD-1C). Retention of LGMD-1C caveolin-3 mutants within the golgi complex. J Biol Chem 274(36): 25632–25641. [DOI] [PubMed] [Google Scholar]
- Glenney JR Jr. and Soppet D (1992). Sequence and expression of caveolin, a protein component of caveolae plasma membrane domains phosphorylated on tyrosine in Rous sarcoma virus-transformed fibroblasts. Proc Natl Acad Sci U S A 89(21): 10517–10521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han B, Porta JC, Hanks JL, Peskova Y, Binshtein E, Dryden K, Claxton DP, McHaourab HS, Karakas E, Ohi MD and Kenworthy AK (2020). Structure and assembly of CAV1 8S complexes revealed by single particle electron microscopy. Sci Adv 6(49): eabc6185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen CG, Howard G and Nichols BJ (2011). Pacsin 2 is recruited to caveolae and functions in caveolar biogenesis. J Cell Sci 124(Pt 16): 2777–2785. [DOI] [PubMed] [Google Scholar]
- Hansen CG and Nichols BJ (2010). Exploring the caves: cavins, caveolins and caveolae. Trends Cell Biol 20(4): 177–186. [DOI] [PubMed] [Google Scholar]
- Hansen CG, Shvets E, Howard G, Riento K and Nichols BJ (2013). Deletion of cavin genes reveals tissue-specific mechanisms for morphogenesis of endothelial caveolae. Nat Commun 4: 1831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayer A, Stoeber M, Bissig C and Helenius A (2010). Biogenesis of caveolae: stepwise assembly of large caveolin and cavin complexes. Traffic 11(3): 361–382. [DOI] [PubMed] [Google Scholar]
- Hill MM, Bastiani M, Luetterforst R, Kirkham M, Kirkham A, Nixon SJ, Walser P, Abankwa D, Oorschot VM, Martin S, Hancock JF and Parton RG (2008). PTRF-Cavin, a conserved cytoplasmic protein required for caveola formation and function. Cell 132(1): 113–124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jarsch IK, Daste F and Gallop JL (2016). Membrane curvature in cell biology: An integration of molecular mechanisms. J Cell Biol 214(4): 375–387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khater IM, Liu Q, Chou KC, Hamarneh G and Nabi IR (2019). Super-resolution modularity analysis shows polyhedral caveolin-1 oligomers combine to form scaffolds and caveolae. Sci Rep 9(1): 9888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirkham M, Nixon SJ, Howes MT, Abi-Rached L, Wakeham DE, Hanzal-Bayer M, Ferguson C, Hill MM, Fernandez-Rojo M, Brown DA, Hancock JF, Brodsky FM and Parton RG (2008). Evolutionary analysis and molecular dissection of caveola biogenesis. J Cell Sci 121(Pt 12): 2075–2086. [DOI] [PubMed] [Google Scholar]
- Kovtun O, Tillu VA, Ariotti N, Parton RG and Collins BM (2015). Cavin family proteins and the assembly of caveolae. J Cell Sci 128(7): 1269–1278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kovtun O, Tillu VA, Jung W, Leneva N, Ariotti N, Chaudhary N, Mandyam RA, Ferguson C, Morgan GP, Johnston WA, Harrop SJ, Alexandrov K, Parton RG and Collins BM (2014). Structural insights into the organization of the cavin membrane coat complex. Dev Cell 31(4): 405–419. [DOI] [PubMed] [Google Scholar]
- Kurzchalia TV, Dupree P and Monier S (1994). VIP21-Caveolin, a protein of the trans-Golgi network and caveolae. FEBS Lett 346(1): 88–91. [DOI] [PubMed] [Google Scholar]
- Lamaze C, Tardif N, Dewulf M, Vassilopoulos S and Blouin CM (2017). The caveolae dress code: structure and signaling. Curr Opin Cell Biol 47: 117–125. [DOI] [PubMed] [Google Scholar]
- Lamaze C and Torrino S (2015). Caveolae and cancer: A new mechanical perspective. Biomed J 38(5): 367–379. [DOI] [PubMed] [Google Scholar]
- Lee J and Glover KJ (2012). The transmembrane domain of caveolin-1 exhibits a helix-break-helix structure. Biochim Biophys Acta 1818(5): 1158–1164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S, Okamoto T, Chun M, Sargiacomo M, Casanova JE, Hansen SH, Nishimoto I and Lisanti MP (1995). Evidence for a regulated interaction between heterotrimeric G proteins and caveolin. J Biol Chem 270(26): 15693–15701. [DOI] [PubMed] [Google Scholar]
- Lisanti MP, Tang ZL and Sargiacomo M (1993). Caveolin forms a hetero-oligomeric protein complex that interacts with an apical GPI-linked protein: implications for the biogenesis of caveolae. J Cell Biol 123(3): 595–604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ludwig A, Howard G, Mendoza-Topaz C, Deerinck T, Mackey M, Sandin S, Ellisman MH and Nichols BJ (2013). Molecular composition and ultrastructure of the caveolar coat complex. PLoS Biol 11(8): e1001640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ludwig A, Nichols BJ and Sandin S (2016). Architecture of the caveolar coat complex. J Cell Sci 129(16): 3077–3083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma L and Chung WK (2017). The role of genetics in pulmonary arterial hypertension. J Pathol 241(2): 273–280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Machleidt T, Li WP, Liu P and Anderson RG (2000). Multiple domains in caveolin-1 control its intracellular traffic. J. Cell Biol 148(1): 17–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathew R (2021). Critical Role of Caveolin-1 Loss/Dysfunction in Pulmonary Hypertension. Med Sci (Basel) 9(4). [DOI] [PMC free article] [PubMed] [Google Scholar]
- McMahon KA, Stroud DA, Gambin Y, Tillu V, Bastiani M, Sierecki E, Polinkovsky ME, Hall TE, Gomez GA, Wu Y, Parat MO, Martel N, Lo HP, Khanna KK, Alexandrov K, Daly R, Yap A, Ryan MT and Parton RG (2021). Cavin3 released from caveolae interacts with BRCA1 to regulate the cellular stress response. Elife 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mercier I, Jasmin JF, Pavlides S, Minetti C, Flomenberg N, Pestell RG, Frank PG, Sotgia F and Lisanti MP (2009). Clinical and translational implications of the caveolin gene family: lessons from mouse models and human genetic disorders. Lab Invest 89: 614–623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monier S, Parton RG, Vogel F, Behlke J, Henske A and Kurzchalia TV (1995). VIP21-caveolin, a membrane protein constituent of the caveolar coat, oligomerizes in vivo and in vitro. Mol. Biol. Cell 6(7): 911–927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morales-Paytuvi F, Ruiz-Mirapeix C, Fajardo A, Rae J, Bosch M, Enrich C, Collins B, Parton RG and Pol A (2022). Proteostatic regulation of caveolins avoids premature oligomerisation and preserves ER homeostasis. bioRxiv. [Google Scholar]
- Moren B, Shah C, Howes MT, Schieber NL, McMahon HT, Parton RG, Daumke O and Lundmark R (2012). EHD2 regulates caveolar dynamics via ATP-driven targeting and oligomerization. Mol Biol Cell 23(7): 1316–1329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nassoy P and Lamaze C (2012). Stressing caveolae new role in cell mechanics. Trends Cell Biol 22(7): 381–389. [DOI] [PubMed] [Google Scholar]
- Palade GE (1953). Fine structure of blood capillaries. J Appl Phys 24(1): 1424–1436. [Google Scholar]
- Parton RG (2018). Caveolae: structure, function, and relationship to disease. Annu Rev Cell Dev Biol 34: 111–136. [DOI] [PubMed] [Google Scholar]
- Parton RG and Collins BM (2022). The structure of caveolin finally takes shape. Sci Adv 8(19): eabq6985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parton RG and del Pozo MA (2013). Caveolae as plasma membrane sensors, protectors and organizers. Nat Rev Mol Cell Biol 14(2): 98–112. [DOI] [PubMed] [Google Scholar]
- Parton RG, Del Pozo MA, Vassilopoulos S, Nabi IR, Le Lay S, Lundmark R, Kenworthy AK, Camus A, Blouin CM, Sessa WC and Lamaze C (2020). Caveolae: The FAQs. Traffic 21(1): 181–185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parton RG, Hanzal-Bayer M and Hancock JF (2006). Biogenesis of caveolae: a structural model for caveolin-induced domain formation. J Cell Sci 119(Pt 5): 787–796. [DOI] [PubMed] [Google Scholar]
- Parton RG, Kozlov MM and Ariotti N (2020). Caveolae and lipid sorting: Shaping the cellular response to stress. J Cell Biol 219(4). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parton RG, McMahon KA and Wu Y (2020). Caveolae: formation, dynamics, and function. Curr Opin Cell Biol 65: 8–16. [DOI] [PubMed] [Google Scholar]
- Parton RG, Tillu V, McMahon KA and Collins BM (2021). Key phases in the formation of caveolae. Curr Opin Cell Biol 71: 7–14. [DOI] [PubMed] [Google Scholar]
- Patni N and Garg A (2015). Congenital generalized lipodystrophies-new insights into metabolic dysfunction. Nat Rev Endocrinol 11(9): 522–534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pol A, Martin S, Fernandez MA, Ingelmo-Torres M, Ferguson C, Enrich C and Parton RG (2005). Cholesterol and fatty acids regulate dynamic caveolin trafficking through the Golgi complex and between the cell surface and lipid bodies. Mol Biol Cell 16: 2091–2105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porta JC, Han B, Gulsevin A, Chung J, Peskova Y, Connolly S, Mchaourab HS, Meiler J, Karakas E, Kenworthy AK and Ohi MD (2022). Molecular architecture of the human caveolin-1 complex. bioRxiv: 2022.2002.2017.480763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porta JC, Han B, Gulsevin A, Chung JM, Peskova Y, Connolly S, McHaourab HS, Meiler J, Karakas E, Kenworthy AK and Ohi MD (2022). Molecular architecture of the human caveolin-1 complex. Sci Adv 8(19): eabn7232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pradhan BS and Proszynski TJ (2020). A role for caveolin-3 in the pathogenesis of muscular dystrophies. Int J Mol Sci 21(22). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qifti A, Balaji S and Scarlata S (2022). Deformation of caveolae impacts global transcription and translation processes through relocalization of cavin-1. J Biol Chem: 102005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Razani B, Engelman JA, Wang XB, Schubert W, Zhang XL, Marks CB, Macaluso F, Russell RG, Li M, Pestell RG, Di Vizio D, Hou H Jr., Kneitz B, Lagaud G, Christ GJ, Edelmann W and Lisanti MP (2001). Caveolin-1 null mice are viable but show evidence of hyperproliferative and vascular abnormalities. J Biol Chem 276(41): 38121–38138. [DOI] [PubMed] [Google Scholar]
- Razani B and Lisanti MP (2001). Caveolin-deficient mice: insights into caveolar function human disease. J Clin Invest 108(11): 1553–1561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren X, Ostermeyer AG, Ramcharan LT, Zeng Y, Lublin DM and Brown DA (2004). Conformational defects slow Golgi exit, block oligomerization, and reduce raft affinity of caveolin-1 mutant proteins. Mol Biol Cell 15(10): 4556–4567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Root KT, Julien JA and Glover KJ (2019). Secondary structure of caveolins: a mini review. Biochem Soc Trans 47(5): 1489–1498. [DOI] [PubMed] [Google Scholar]
- Root KT, Plucinsky SM and Glover KJ (2015). Recent progress in the topology, structure, and oligomerization of caveolin: a building block of caveolae. Curr Top Membr 75: 305–336. [DOI] [PubMed] [Google Scholar]
- Rothberg KG, Heuser JE, Donzell WC, Ying YS, Glenney JR and Anderson RG (1992). Caveolin, a protein component of caveolae membrane coats. Cell 68(4): 673–682. [DOI] [PubMed] [Google Scholar]
- Sargiacomo M, Scherer PE, Tang Z, Kubler E, Song KS, Sanders MC and Lisanti MP (1995). Oligomeric structure of caveolin: implications for caveolae membrane organization. Proc Natl Acad Sci U S A 92(20): 9407–9411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sargiacomo M, Sudol M, Tang Z and Lisanti MP (1993). Signal transducing molecules and glycosyl-phosphatidylinositol-linked proteins form a caveolin-rich insoluble complex in MDCK cells. J. Cell Biol 122(4): 789–807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheiffele P, Verkade P, Fra AM, Virta H, Simons K and Ikonen E (1998). Caveolin-1 and −2 in the exocytic pathway of MDCK cells. J. Cell Biol 140(4): 795–806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scherer PE, Okamoto T, Chun M, Nishimoto I, Lodish HF and Lisanti MP (1996). Identification, sequence, and expression of caveolin-2 defines a caveolin gene family. Proc Natl Acad Sci U S A 93(1): 131–135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlegel A, Schwab RB, Scherer PE and Lisanti MP (1999). A role for the caveolin scaffolding domain in mediating the membrane attachment of caveolin-1. The caveolin scaffolding domain is both necessary and sufficient for membrane binding in vitro. J Biol Chem 274(32): 22660–22667. [DOI] [PubMed] [Google Scholar]
- Shin J, Jung YH, Cho DH, Park M, Lee KE, Yang Y, Jeong C, Sung BH, Sohn JH, Park JB and Kweon DH (2015). Display of membrane proteins on the heterologous caveolae carved by caveolin-1 in the Escherichia coli cytoplasm. Enzyme Microb Technol 79–80: 55–62. [DOI] [PubMed] [Google Scholar]
- Shu L, Chan KHK, Zhang G, Huan T, Kurt Z, Zhao Y, Codoni V, Tregouet DA, Cardiogenics C, Yang J, Wilson JG, Luo X, Levy D, Lusis AJ, Liu S and Yang X (2017). Shared genetic regulatory networks for cardiovascular disease and type 2 diabetes in multiple populations of diverse ethnicities in the United States. PLoS Genet 13(9): e1007040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sinha B, Koster D, Ruez R, Gonnord P, Bastiani M, Abankwa D, Stan RV, Butler-Browne G, Vedie B, Johannes L, Morone N, Parton RG, Raposo G, Sens P, Lamaze C and Nassoy P (2011). Cells respond to mechanical stress by rapid disassembly of caveolae. Cell 144(3): 402–413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sotgia F, Woodman SE, Bonuccelli G, Capozza F, Minetti C, Scherer PE and Lisanti MP (2003). Phenotypic behavior of caveolin-3 R26Q, a mutant associated with hyperCKemia, distal myopathy, and rippling muscle disease. Am J Physiol Cell Physiol 285(5): C1150–1160. [DOI] [PubMed] [Google Scholar]
- Stachowiak JC, Brodsky FM and Miller EA (2013). A cost-benefit analysis of the physical mechanisms of membrane curvature. Nat Cell Biol 15(9): 1019–1027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoeber M, Schellenberger P, Siebert CA, Leyrat C, Helenius A and Grunewald K (2016). Model for the architecture of caveolae based on a flexible, net-like assembly of Cavin1 and Caveolin discs. Proc Natl Acad Sci U S A 113(50): E8069–E8078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoeber M, Stoeck IK, Hanni C, Bleck CK, Balistreri G and Helenius A (2012). Oligomers of the ATPase EHD2 confine caveolae to the plasma membrane through association with actin. EMBO J 31(10): 2350–2364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang Z, Scherer PE, Okamoto T, Song K, Chu C, Kohtz DS, Nishimoto I, Lodish HF and Lisanti MP (1996). Molecular cloning of caveolin-3, a novel member of the caveolin gene family expressed predominantly in muscle. J Biol Chem 271(4): 2255–2261. [DOI] [PubMed] [Google Scholar]
- Tillu VA, Rae J, Gao Y, Ariotti N, Floetenmeyer M, Kovtun O, McMahon KA, Chaudhary N, Parton RG and Collins BM (2021). Cavin1 intrinsically disordered domains are essential for fuzzy electrostatic interactions and caveola formation. Nat Commun 12(1): 931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torrino S, Shen WW, Blouin CM, Mani SK, Viaris de Lesegno C, Bost P, Grassart A, Koster D, Valades-Cruz CA, Chambon V, Johannes L, Pierobon P, Soumelis V, Coirault C, Vassilopoulos S and Lamaze C (2018). EHD2 is a mechanotransducer connecting caveolae dynamics with gene transcription. J Cell Biol 217(12): 4092–4105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walser PJ, Ariotti N, Howes M, Ferguson C, Webb R, Schwudke D, Leneva N, Cho KJ, Cooper L, Rae J, Floetenmeyer M, Oorschot VM, Skoglund U, Simons K, Hancock JF and Parton RG (2012). Constitutive formation of caveolae in a bacterium. Cell 150(4): 752–763. [DOI] [PubMed] [Google Scholar]
- Way M and Parton RG (1995). M-Caveolin, a Muscle-Specific Caveolin-Related Protein. FEBS Letters 376(1–2): 108–112. [DOI] [PubMed] [Google Scholar]
- Whiteley G, Collins RF and Kitmitto A (2012). Characterization of the molecular architecture of human caveolin-3 and interaction with the skeletal muscle ryanodine receptor. J Biol Chem 287(48): 40302–40316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams TM and Lisanti MP (2004). The Caveolin genes: from cell biology to medicine. Ann Med 36(8): 584–595. [DOI] [PubMed] [Google Scholar]
- Williams TM and Lisanti MP (2004). The caveolin proteins. Genome Biol 5(3): 214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao YY, Liu Y, Stan RV, Fan L, Gu Y, Dalton N, Chu PH, Peterson K, Ross J Jr. and Chien KR (2002). Defects in caveolin-1 cause dilated cardiomyopathy and pulmonary hypertension in knockout mice. Proc Natl Acad Sci U S A 99(17): 11375–11380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Y, Ariotti N, Rae J, Liang H, Tillu V, Tee S, Bastiani M, Bademosi AT, Collins BM, Meunier FA, Hancock JF and Parton RG (2021). Caveolin-1 and cavin1 act synergistically to generate a unique lipid environment in caveolae. J Cell Biol 220(3). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Data sharing is not applicable to this article as no new datasets were generated or analyzed during the current study.


