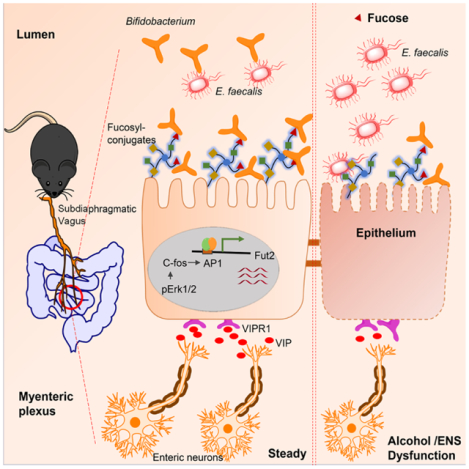
Summary
Interactions between the enteric nervous system (ENS) and intestinal epithelium are thought to play a vital role in intestinal homeostasis. How the ENS monitors the frontier with commensal and pathogenic microbes while maintaining epithelium function remains unclear. Here, by combining subdiaphragmatic vagotomy with transcriptomics, chemogenetic strategy and co-culture of enteric neuron-intestinal organoid, we show that enteric neuron expressing VIP shapes the α1,2-fucosylation of intestinal epithelial cells (IECs). Mechanistically, neuropeptide VIP activates fut2 expression via the Erk1/2-C-fos pathway through the VIPR1 receptor on IECs. We further demonstrate that perturbation of enteric neurons leads to gut dysbiosis through α1,2-fucosylation in the steady state and results in increased susceptibility to alcohol-associated liver disease (ALD). This was attributed to an imbalance between beneficial Bifidobacterium and opportunistic pathogenic Enterococcus faecalis in ALD. In addition, Bifidobacterial α1,2-fucosidase may promote Bifidobacterium adhesion to the mucosal surface, which restricts Enterococcus faecalis overgrowth and prevents ALD progression.
Keywords: VIPR1, fut2, Bifidobacterium, Enterococcus faecalis, ALD, fucosidase, enteric nervous, vagus nerve
Graphical Abstract
eTOC
Lei et al. show that the enteric neuron expressing VIP directs a program of gut mucosal fucosylation via the Erk1/2-C-fos pathway. The reduced abundance of enteric VIPergic neurons leads to the imbalance of abundance between beneficial Bifidobacterium and pathogenic E. faecalis, and enhances the susceptibility of ALD.
Introduction
Proper intestinal function results from interactions between multiple components including the enteric nervous system (ENS)(Walsh and Zemper, 2019; Wang et al., 2021), which permeates the entirety of the intestinal tissue. The intestinal mucosal barrier facilitates a non-inflammatory, symbiotic relationship with commensal microflora, while also providing essential protection from pathogenic microbial invasion(Hooper and Macpherson, 2010). Although it is well known that gut immune cells and ENS communication arbitrates this balance, how epithelial and ENS components interact to influence this critical axis during homeostasis or upon bacterial infection is under-explored.
An emerging paradigm suggests that gut glycosylation is a key force in maintaining a homeostatic relationship between the gut epithelium and microbiota(Moran et al., 2011; Ohtsubo and Marth, 2006), in particular gut epithelium fucosylation (Kashyap et al., 2013). α1,2-fucosylation of IECs is induced by increased Fut2 (fucosyltransferase 2) expression, stimulated when Toll-like receptor ligands interact with the host immune system(Goto et al., 2014; Pham et al., 2014; Pickard et al., 2014). Fut2-mediated intestinal α1,2-fucosylation enables expression of α1,2-fucosylated carbohydrates on IECs and in luminal contents, which can serve as substrates for metabolites, energy source, and adhesion receptors for many symbiotic beneficial bacteria(Goto et al., 2016; Kashyap et al., 2013). Absence of proper gut fucosylation, strengthens pathogenic bacteria, which leads to the breakdown of the epithelial barrier and gut inflammation(Terahara et al., 2011). For example, fucosylation deficiency in mice leads to expansion and invasion of a pathobiont, Enterococcus faecalis (E. faecalis), from the inflamed gut to systemic tissues(Pham et al., 2014). Many intestinal Bifidobacteria use α1,2-L-fucosidases (AfcA) to ferment the glycans of host glycoconjugates and human milk oligosaccharides (HMOs) to produce fucose(Kononova et al., 2021). Fucose could bolster the beneficial members of the gut community, like the Bifidobacterial species, thus maintaining their beneficial functions, such as colonization(Kononova et al., 2021). Therefore, membrane-bound fucosylated glycoproteins on mucosal ECs function as important communication tools between the host and luminal microbes. Nevertheless, it is unclear whether and how host ENS machinery contributes to fucosylation-associated intestinal barrier function.
Alterations in the intestinal epithelial barrier are increasingly recognized as major pathogenic factors in the development and progression of alcohol-associated liver disease (ALD) (Bajaj, 2019; Mutlu et al., 2012). Dysbiosis of the gut microbiota has been reported to accelerate progression of ALDs via the gut-liver axis(Gao and Bataller, 2011; Szabo, 2015). Recent studies indicated that people carrying toxin cytolysin-producing strains of E. faecalis had more severe alcoholic hepatitis and greater risk of death(Duan et al., 2019; Llorente et al., 2017). Interestingly, fucosylation deficiency is linked to cytolytic E. faecalis overgrowth(Zhou et al., 2020), which causes more severe alcoholic hepatitis(Duan et al., 2019). Chronic alcohol administration leads to neurodegeneration in the central nervous system(Rose et al., 2010) but, whether alcohol consumption induces quantitative, functional and neurochemical changes to the ENS is not well investigated. Furthermore, whether changes in ENS activity correlates with mucus dysregulation and microbial dysbiosis in ALD is currently unknown.
In this study, we showed that a vasoactive intestinal polypeptide (VIP)-positive enteric neuron is critical in maintaining IEC α1,2-fucosylation. We identified α1,2-fucosylated carbohydrate as the essential adhesion receptor of Bifidobacterium that restricts cytolytic E. faecalis overgrowth and prevents ALD progression. This study demonstrates that the ENS is a critical arm of mucosal barrier homeostasis and is essential for coordinating not only beneficial bacterial colonization but also combatting opportunistic pathogen infections.
Results
Denervation via vagotomy regulates fucosylation of intestinal epithelium
Co-staining neuronal marker (βIII-Tubulin) with epithelial cells (EpCAM) or Goblet cells (Mucin 2) markers revealed that neuronal fibers were in close proximity to intestinal epithelium (Figures S1A–S1B). Many, but not all, extrinsic nerves enter the intestine through the vagus nerve. Bilateral sub-diaphragmatic vagotomy, a surgical technique removing vagal fibers has uncovered a role for the vagus nerve in regulating intestinal epithelial cellular proliferation(Hakanson et al., 1984; Zhao et al., 2014), colon tumor development(Liu et al., 2015) and colitis(Di Giovangiulio et al., 2016; Teratani et al., 2020). To examine the functions of the vagal extrinsic input to the ENS on intestinal epithelial gene expression, we performed subdiaphragmatic truncal vagotomy (VGx) in wild-type C57BL/6 (B6) mice (Figure S1C). The global transcriptomes of IECs isolated from vagotomized and sham-operated mice were evaluated by RNA sequencing. Gene ontology enrichment analysis showed that factors involved in glycoprotein metabolic processes, inflammatory responses, antimicrobial processes, and defense responses to bacteria were significantly regulated in VGx mice (Figure 1A). VGx profoundly altered the gene expression profile of IECs in the ileum including the genes involved in glycosylation (Figure S1D). Notably, we observed an enrichment of protein glycosylation genes Fut2, B3gnt7, B3gnt8, Ugt1a6a and St6galnac4 (Figure 1B). The IECs from VGx mice exhibited lower level of Fut2 mRNA (Figure 1B) than those from Sham mice. We confirmed this by real-time PCR Of of Fut2 in IECs from small intestine, particularly ileum. (Figures 1C and S1E). Immunofluorescence whole-mount imaging and section staining on segments of mouse ileum and colon showed that fucosylated IECs, detected by the α1,2-fucose-recognizing lectin Ulex europaeus agglutinin-1(UEA-1), were significantly lower in the ileum of VGx mice compared to Sham mice (Figures 1D–1E). Moreover, western blotting (Figure 1F) and FACS (Figures 1G and S1F) analysis showed a significant reduction in the α1,2-fucosylation of IECs in the small intestine of the vagotomized mice compared with sham-operated mice. We further confirmed the reduction of α1,2-fucosylation in goblet cells after vagotomy (Figure S1G). However, no differences were found in the fut2 expression and α1,2-fucosylation in colonic IECs between vagotomized mice compared with sham-operated mice (Figures S1H–S1K). These data suggest that disrupting the vagal extrinsic input to the ENS affects the epithelial fucosylation in the small intestine.
Figure1. Denervation suppresses fucosylation of intestinal epithelium.
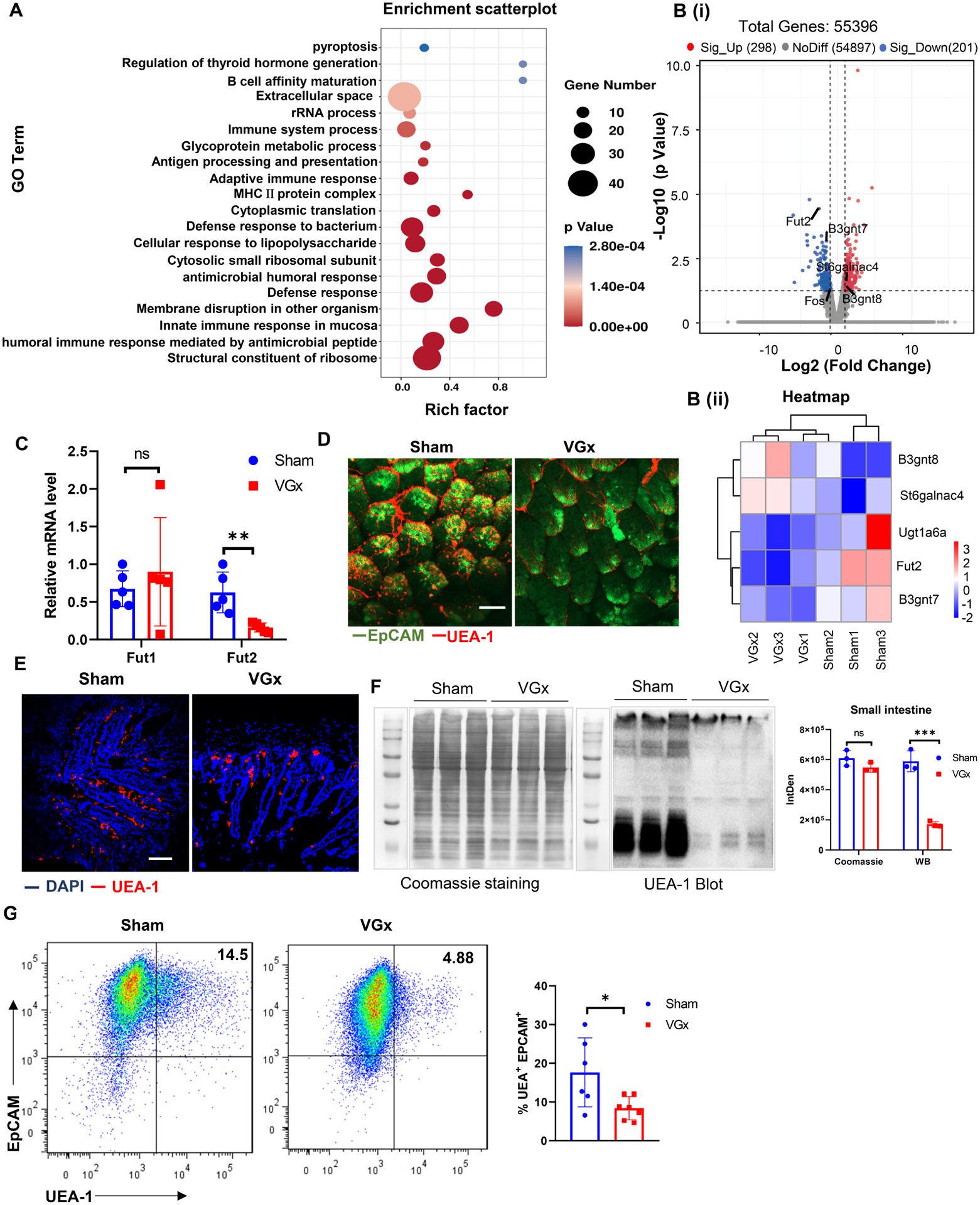
(A) GO enrichment analysis of genes defining the 20 most significant terms is represented in the accompanying bubble plot. Bubble colors represent the corrected p-value. Bubble sizes indicate the number of genes.
(B) Volcano plots (i) showing fold change (FC) and p-value for the comparison of IECs from VGx mice versus IECs from Sham mice on the basis of RNA-seq data (n=3). Genes up- or downregulated in IECs (at FC>2 and p < 0.05) are highlighted (red and blue). Shown on B (ii) is a heatmap representation of changes (log2) versus Sham controls, in all replicates.
(C) Real-time PCR analysis of Fut1 and Fut2 mRNA in ileal IECs from Sham and VGx mice.
(D-E) Whole mount microscopy (D) and immunofluorescence section (E) of UEA-1 staining of ileum in Sham mice or VGx mice. Scale bar, 50 μm.
(F) Pattern of fucosylation modification in the IECs of small intestine (SI) from Sham mice or VGx mice. Coomassie blue staining (left, total protein input), UEA-1 staining blot (middle) and quantification (right) for showing fucosylated-protein.
(G) Representative plots of FACS and summary graphs of the percentage of UEA-1 stained IECs of ileum from Sham and VGx mice.
Data are from (C) 2, (D-F) 2–3, or (G) 3 independent experiments performed. Each dot represents one mouse. Here and thereafter, Error bars represent mean ± SEM. *p < 0.05, **p < 0.01. ns, not significant from unpaired Student’s t test unless otherwise specified. See also Figure S1.
Enteric neuron derived VIP regulates IECs fucosylation
Since cutting the input from central nervous system (CNS) led to the changes in epithelium, we wanted to explore which specific ENS changes are responsible for conveying signals to IECs. To this end, the expression level of neurotransmitter receptors and neuropeptide receptors on IECs were analyzed from RNA-seq data of VGx and Sham mice fed chow diet. The IECs exhibited highest levels of VIP receptor type 1 (VIPR1; also known as VPAC1) compared to the receptors for other neurotransmitters, regardless of vagotomy (Figure 2A). Interestingly, VIP-producing neurons and VIP production was significantly inhibited in the myenteric and submucosal plexus of ileum, but not colon upon vagotomy, as evidenced by whole-mount imaging (Figure 2B), section imaging (Figure S2A), ELISA detection (Figure 2C and S2B) real-time PCR analysis (Figure S2C). These data suggest that VIP-producing neurons, that we refer to as VIPergic neurons, may regulate IECs function via VIPR1. To determine whether VIP directly affects the fucosylation of IECs, we treated mice with VIPR1 ligand VIP or its competitive antagonist VIPx. Treatment with VIP significantly induced IECs-associated fucosylation (Figures 2D–2E and S2D–S2E) and Fut2 mRNA expression (Figures 2F and S2F). In contrast, mice treated with VIPx exhibited lower IEC fucosylation and Fut2 mRNA expression when compared with PBS-treated mice (Figures 2D–2F and S2D–S2F). Next, knockout of VIPR1 decreased the IEC fucosylation and Fut2 expression in the colon (Figures S2G–S2H) and reversed the effect of VIP on IECs fucosylation (Figures 2G and S2I). To explore more directly the functional crosstalk between enteric neurons and IECs, the myenteric plexus (MP) was dissociated with the surrounding muscle layers (Zhang and Hu, 2013), and cells were dissociated and cultured for 6–8 days in “Neurobasal-A medium” that supports enteric neurons that produce VIP in vitro (Figure S2J). We performed co-culture experiments using an intestinal organoid and enteric neuron cells (Figures S2K–S2L), in which differentiated enteric neuron cells are included or excluded. Enteric neurons increased the levels of fucosylation in intestinal organoids derived from WT mice (Figure 2H). Gene expression analysis of organoids following enteric neuron co-culture revealed significantly increased Fut2 expression (Figure 2I), but not other markers involved in intestinal fucosylation (Figure S2M). Notably, addition of VIP to organoids without enteric neurons can directly induce Fut2 expression (Figure 2I). Furthermore, VIPx treatment inhibited fucosylation induction (Figure 2H) and Fut2 expression (Figure 2I) in enteric neurons co-culture experiments. Importantly, co-culture of enteric neurons or VIP treatment had no effect on the fucosylation (Figure 2H) and expression of Fut2 (Figure 2J) and other fucosylation-related markers (Figure S2M) in the intestinal organoids derived from VIPR1−/− mice. Taken together, these data suggest that the enteric neuron is able to drive intestinal fucosylation via VIP-VIPR1 signaling.
Figure 2. Enteric VIPergic neuron regulates epithelial fucosylation.
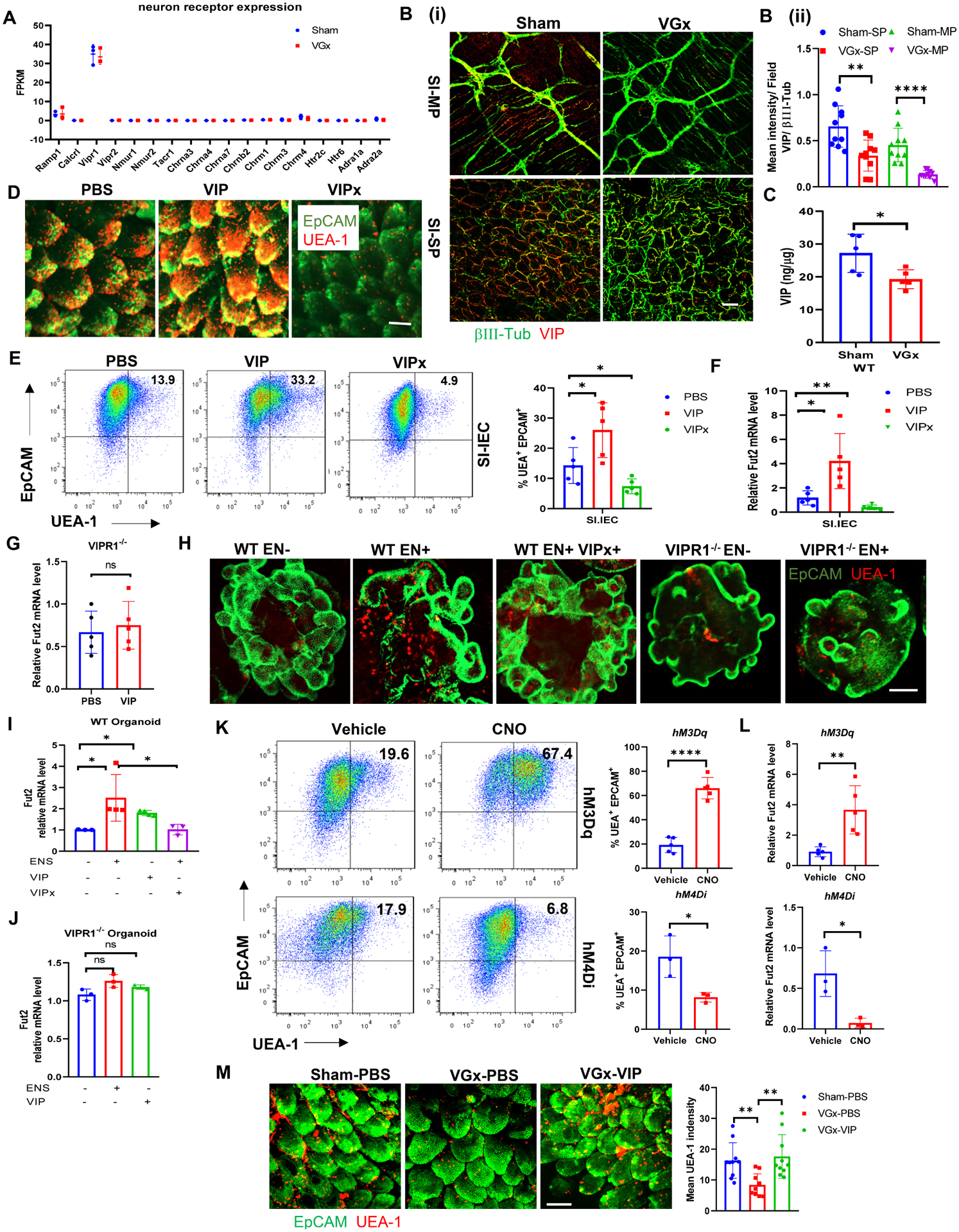
(A) The expression of genes encoding neurotransmitter and neuropeptide receptors in isolated IECs, as determined by RNA-sequencing analysis in Figure1. FPKM, Fragments Per Kilobase of transcript per Million mapped reads.
(B) Whole mount microscopy of βIII-tubulin (green) and VIP (red) staining (i) and quantification (ii) in the myenteric (MP) and submucosal plexus (SP) of ileum in Sham mice or VGx mice. Scale bar, 50 μm. ***p < 0.001.
(C) Concentration of VIP in the ileum of Sham mice or VGx mice by EIA. n = 5, representative of (B-C) 2 independent experiments. Mean ± SEM; *p < 0.05.
(D) Whole mount microscopy of EpCAM and UEA-1 staining of ileum in WT mice 12 h after injection with PBS, VIP (6 μg/mice) or VIPx (100 μg/mice). Data are representative of at least 5 mice per condition. Scale bar, 50 μm.
(E) Representative plots of FACS and summary graphs of the percentage of UEA-1 stained IECs of ileum in WT mice 12 h after injection with PBS, VIP or VIPx.
(F-G) Real-time PCR analysis of Fut2 mRNA expression in IECs of ileum in WT (F) or VIPR1−/− mice (G) 12 h after injection with PBS, VIP or VIPx.
Data (D-G) were shown as Mean ± SEM; n = 5, representative of 3 independent experiments. *p < 0.05. **p < 0.01, ns not significant.
(H-J) UEA-1 staining of intestinal organoid cultures (H), scale bar, 50 μM; and real-time PCR analysis of Fut2 expression in intestinal organoid cultures from WT mice (I) or from VIPR1−/− (J). Representative of 2 or more fields in 3 independent cultures. Mean ± SEM; n = 5, *p < 0.05, ns: not significant.
(K) Representative plots of FACS and summary graphs of the percentage of UEA-1 stained colonic IECs from VipIRES-crehM3Dqfl/+ mice or VipIRES-crehM4Difl/fl after treatment with vehicle or CNO.
(L) Real-time PCR analysis of Fut2 expression in colonic IECs from VipIRES-crehM3Dqfl/+ mice or VipIRES-crehM4Di-fl/fl after treatment with vehicle or CNO. n = 3–5. (L-M) Representative of 2 independent experiments. Mean ± SEM; * p < 0.05, **p < 0.01. ****p < 0.0001, ns: not significant.
(M) Whole mount microscopy of EpCAM and UEA-1 staining of ileum in sham mice or VGx mice after repeated injection with PBS or VIP (6 μg/mice). Scale bar, 50 μm.
See also Figures S2 and S3.
To determine whether direct modulation of VIPergic neurons (inhibition or activation) affects the IECs fucosylation in vivo, we adopted a chemogenetic strategy using mice engineered to express designer receptors exclusively activated by designer drugs (DREADD)(Roth, 2016) in VIP+ cells. Homozygous Vip IRES-cre mice were bred to hemizygotes hM3Dqfl-stop-fl mice (activation of VIPergic neurons, Figure S3A) or homozygous hM4Difl-stop-fl mice (inhibition of VIPergic neurons, Figure S3B). Mice that expressed the activating DREADD hM3Dq (VipIRES-crehM3Dqfl/+ mice) had higher UEA-1+EpCAM+ IECs in the small intestine, but not colon, at 36 h after treatment with the DREADD ligand clozapine-N-oxide (CNO) (Figure 2K and Figure S3C). In parallel, there was increased expression of Fut2 mRNA in ileal, but not colonic, extracts enriched for IECs (Figure 2L and Figure S3D). Conversely, using the inhibitory DREADD hM4Di (VipIRES-crehM4Difl/fl mice), there was a lower frequency and number of UEA-1+EpCAM+ IECs in the small intestine (Figure 2K and Figure S3C), and a lower expression of Fut2 mRNA (Figure 2L and Figure S3D) in ileal extracts enriched for IECs at 36 h after administration of CNO. To provide further evidence to support the link between vagotomy and the VIP, we treated VGx mice with VIP. Injection of VIP rescued the fucosylation of IECs in VGx mice (Figures 2M and S3E). Combined, these results indicate that VIPergic neurons modulate intestinal epithelium homeostasis, acting through VIPR1 on IEC to regulate fucosylation production.
VIPergic neuron regulates IECs fucosylation via Erk-c-fos pathway
Next, we looked for potential mechanisms whereby VIPergic neuron regulates Fut2 expression. RNA-seq analysis of IECs obtained from the VGx mice indicated relatively lower levels of c-fos compared to Sham mice (Figure 3A). C-fos is a neuronal transcription factor controlled in part by gut microbiota and responsible for gut-extrinsic sympathetic activation(Muller et al., 2020). AP-1 transcription complexes are dimers mainly composed of members of the Fos and Jun transcription factors. Sequence analysis of fut2 promoter indicated an AP-1 binding site upstream of the first exon (Figure 3B). To investigate C-fos function on fut2 promoter activity, we co-transfected the doxycycline (DOX) inducible c-fos expressing vector with a luciferase reporter vector in which the luciferase gene was under the control of Fut2 promoter with a naïve or mutant AP-1 binding site. Indeed, doxycycline-induced c-fos promoted the induction of Fut2-luciferase reporter with a naïve AP1 site (Figure 3B), whereas doxycycline failed to activate a Fut2-luciferase reporter with mutant AP-1 binding site, indicating a transactivation mechanism. We next investigated specific signals that might be required for inducing this interaction. C-fos is known to be regulated by several kinases such as Erk1/2, RSK and IKKβ and protein kinase A (PKA)(Nakakuki et al., 2010; Saijo et al., 2011). VIP treatment induced phosphorylation of Erk1/2 in a time-dependent manner in IECs (Figure 3C) and increased expression of c-fos mRNA and in WT organoid in vitro, but not in VIPR1−/− organoid (Figure 3D). In addition, Erk1/2 inhibitor treatment prevented VIP-induced increases of c-fos and fut2 expression in WT organoid (Figure 3D), implicating the activation of c-fos by VIP is dependent of Erk1/2 phosphorylation. Consistent with these observations, VIP injection (Figure 3E) resulted in induction of the levels of p-Erk1/2 and c-Fos in the ileum in vivo. Notably, chemogenetic activation or inhibition of VIPergic neurons resulted in increased or reduced C-Fos and p-Erk levels in EpCAM+ IECs of the ileum, respectively (Figures 3F and 3G). Taken together, VIP neuron regulates IECs fucosylation via Erk1/2-C-fos pathway.
Figure 3. VIP activates Fut2 expression via Erk/c-Fos pathway.
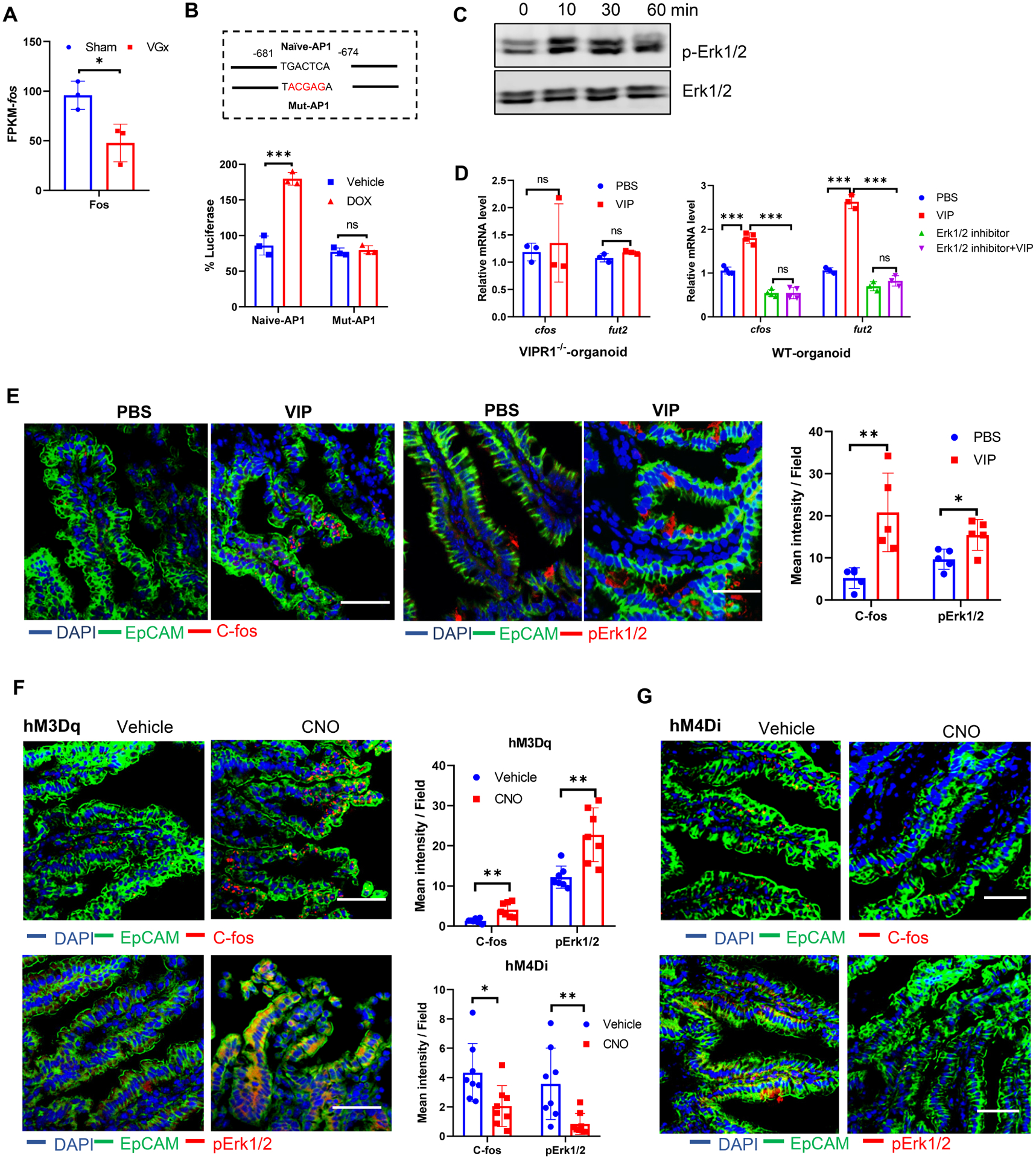
(A) The expression of c-fos in isolated IECs, as determined by RNA-sequencing analysis as Figure 1.
(B) Fut2 promoter activity as demonstrated with luciferase activity. Naïve-AP1 represents fut2 promoter with naïve AP-1 binding site and Mut-AP1 represents fut2 promoter with mutated AP-1 binding site as indicated in the figure. The number indicates the position AP-1 site with the start of first exon set as +1. Data were shown as relative luciferase activity with the ratio of Dox:vehicle. Mean ± SEM, ***p < 0.001, ns not significant.
(C) Representative western blot image of p-Erk1/2 in ileal IECs stimulated with VIP (100 nM) for the indicated time.
(D) C-fos mRNA and Fut2 expression in intestinal organoid cultures from WT mice or from VIPR1−/− stimulated with/without VIP in the presence or absence of Erk1/2 inhibitor (FR180204, 50 μM) for 12 h. (B-D) Data are representative of three independent experiments.
(E) Immunofluorescence section and quantification for C-fos (red) or p-Erk1/2 (red) with EpCAM (green) and DAPI staining of ileum in WT mice 12h after injection with PBS or VIP (6 μg/mice).
(F-G) Immunofluorescence section and quantification for C-fos (red) or p-Erk1/2 (red) with EpCAM (green) and DAPI staining of ileum in VipIRES-crehM3Dqfl/+ mice (F, activating DREADD) or VipIRES-crehM4Difl/fl mice (G, inhibitory DREADD) after treatment with vehicle or CNO. (E-G) Data are representative of at least 5 mice per condition. Scale bar, 50 μm. * p < 0.05, **p < 0.01.
Interaction of VIPergic neurons and IEC fucosylation shapes the gut microbiota composition
Fucosylation is a protective glycosylated mechanism for maintaining host-microbial symbiosis(Pickard and Chervonsky, 2015). To explore whether reduced fucosylation induces the perturbation of the gut microbial community, we performed 16S ribosomal RNA (rRNA) sequencing of feces from VGx and sham-operated mice. Alpha-diversity indexes indicated decreases of bacterial richness in the VGx mice compare to Sham mice (Figure 4A). Beta-diversity represented by Principal Coordinates Analysis (PCoA) revealed clearly separated clusters among the two groups (Figure 4B). Differentiated abundance analysis showed that vagotomy significantly decreases the levels of Bacteroidetes phylum (mean, 65.4% to 34.4%) and Actinobacteria (mean, 0.47% to 0.04%) but increases the level of Firmicutes (mean, 30.9% to 60.8%) (Figures 4C and S4A–4C). Phenotypic prediction based on BugBase (Ward et al., 2017) showed gram-positive bacteria and mobile element containing bacteria enriched; however, the gram-negative bacteria and potentially pathogenic bacteria decreased in the VGx mice (Figure S4D). LEfse analysis showed multiple genera from actinobacterial phylum to be significantly decreased in the VGx-samples compare to the Sham group, including Bifidobacterium, Microbacterium, Enterorhabdus and Rhodococcus, whereas multiple genera of firmicute phylum were significantly increased in the VGx-samples, including Tyzzerella, Angelakisella, Intestinimonas, Ruminococcus, Ruminoclostridium, Clostridiales_unclassified and Lachnospiraceace_unclassified (Figure 4D). Correlation analysis of differentiated taxa showed the abundance of Parasutterella, Microbacterium, Enterorhabdus, Rhodococcus, Dubosiella, Turicibacter, and Brevundimonas are positively correlated to the abundance of Bifidobacterium, whereas the levels of clostridiales sp. and Angelakisella are negatively correlated to the level of Bifidobacterium (Figure S4E). Alpha 1,2-fucosylated carbohydrates on IECs and in luminal contents can serve as substrates for adhesion receptors for many symbiotic beneficial bacteria(Goto et al., 2016; Kashyap et al., 2013) including Bifidobacterium. We thus proposed that the microbiota changes after vagotomy may be due to α1,2-fucosylation. Consistently, the quantity of vagotomy-reduced bacteria including Bifidobacterium were positively correlated with the expression of Fut2 in IECs (Figure 4E). A decrease of Bifidobacterium in VGx mice was confirmed by real-time PCR analysis using primers specific for Bifidobacterium (Figure 4F). Deletion of Fut2 significantly decreased the abundance of those bacteria that are correlated with Bifidobacterium in the gut (Figure 4G), which phenocopies the effect of vagotomy on the changes of those bacteria. Importantly, inhibition of VIPergic neurons reduced the abundance of those bacteria that are correlated with Bifidobacterium in the gut (Figure S4F), which has a similar effect of vagotomy on the changes of those bacteria. Furthermore, injection of VIP increased the level of Bifidobacterium in VGx mice (Figures S4G).
Figure 4. Denervation attenuates Bifidobacterium abundance that correlates with the level of fucosylation.
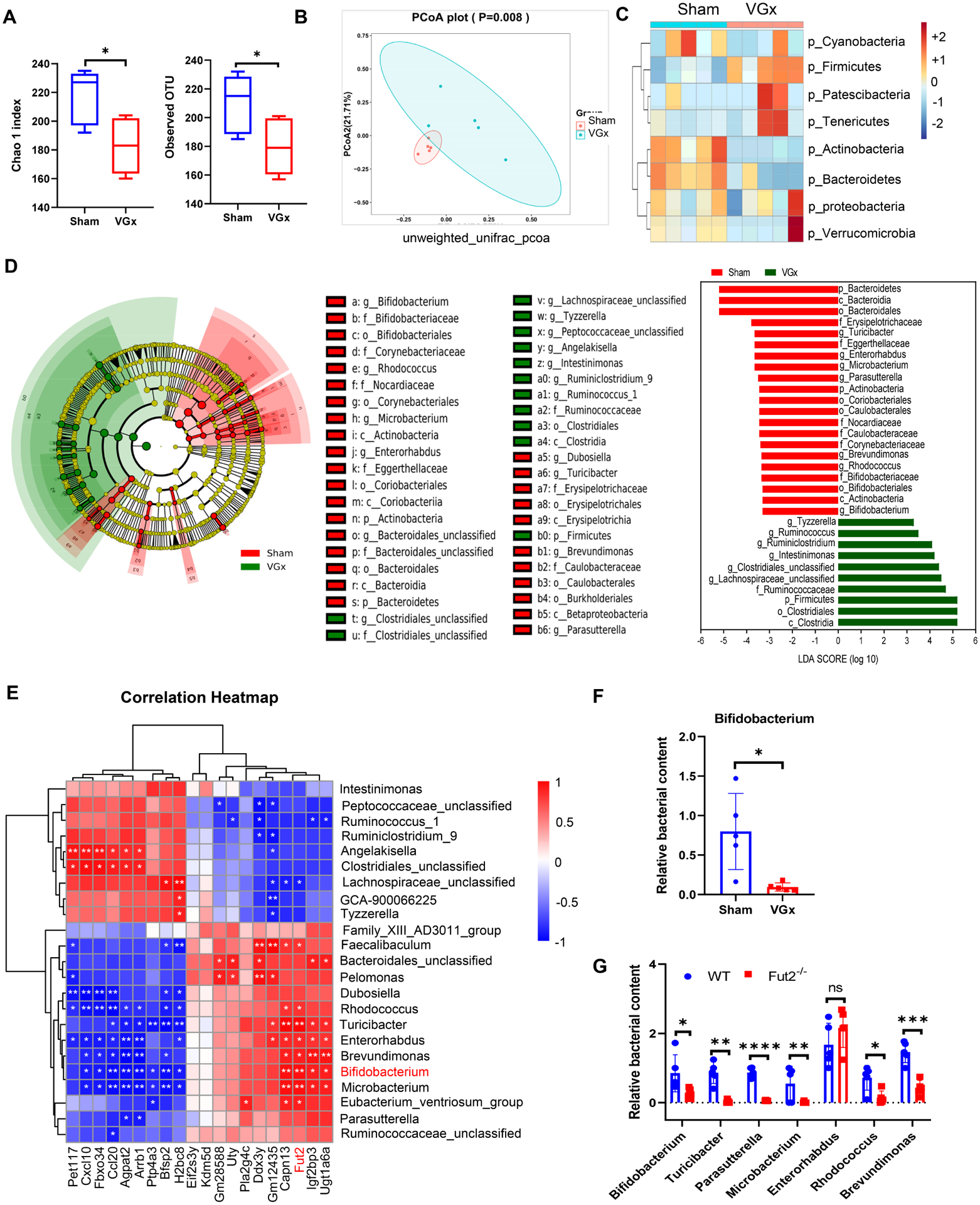
(A) Alpha diversity represented by the Chao1 and observed OTU index.
(B) Principal coordinates analysis (PCoA) of unweighted UniFrac distance based on 16S rRNA sequencing of fecal samples from Sham mice and VGx mice. Ellipse represents 95% confidence of sampling. n = 5, p = 0.008.
(C) Heatmap showing the clustering of the two groups based on log-relative abundances of filtered microbiota at the phylum levels.
(D) Results of a linear discriminant analysis of microbiota composition in VGx mice compared to Sham mice are displayed as a cladogram (left), highlighting differences in the abundance of taxa (red and green highlighting) determined by 16S rRNA gene amplicon sequencing. Taxa are arranged on a circular phylogenetic tree. Green, taxa elevated in VGx mice compared to Sham mice; Red, taxa reduced in VGx mice compared to Sham mice.
LDA scores (right) of the differentially abundant taxa. Taxa enriched in microbiota from Sham mice (red) are indicated with a negative LDA score related to that from VGx mice (green) (Kruskal-Qallis test, p<0.05 and LDA >=±3); n = 5.
(E) Spearman correlation analyses between the abundance of gut bacteria at the genus levels (Top 23 most differentiated) and the level of IEC genes expression (Top 20 most differentiated). R values (value of coefficient) are shown in different colors as indicated in the figure, the red color represents a positive correlation, the blue color represents a negative correlation.
(F) Real-time PCR analysis of relative abundance of Bifidobacterium.
(G) Relative abundance of bacterial genera in fecal samples from WT and Fut2−/− mice determined by real-time PCR, the most correlated genera with fu2 expression levels were chosen for realtime PCR analysis. (F-G) Mean ± SEM; n = 5, *p < 0.05, ** p < 0.01, ***p < 0.001, ****p < 0.0001.
See also Figure S4.
Combined, these results indicate that the regulation of VIPergic neurons might affect intestinal microbiota composition, particularly Bifidobacterium abundance, probably acting through epithelium fucosylation.
α1,2-fucosidase (afcA) promotes B. bifidum adhesion to the mucosal surface
Mucin glycoprotein is a major receptor supporting bacterial colonization of the mucosal surface. α1,2-fucosylation is the terminal glycosyl-conjugation wide spread in the intestine, which can be easily accessible to gut bacteria by providing adhesion sites(Kononova et al., 2021). In vitro adhesion assays showed that Bifidobacterium bifidum (B. bifidum) decreased adhesion to the mucosa of Fut2−/− mice (Figure 5A), which lack mucosal α1,2-fucosylation (Figure S5A), compared to that of WT mice in vitro. The adhesion of B. bifidum to the mucosa was attenuated by pretreatment with UEA-1 (Figure 5B). These data suggest that α1,2-fucosylation is required for the adhesion of Bifidobacterium. A previous report showed cell wall associated sialidase can enhance B. bifidum colonization by binding to sialylation conjugations(Nishiyama et al., 2017). We thus proposed B. bifidum derived α1,2-fucosidase (encoded by afcA) might act as an anchor to bind to host fucosyl-conjugates to enhance bifidobacterial colonization. To determine the contribution of AfcA to bacterial adhesion to the mucosa, we used antisense RNA to knockdown afcA in B. bifidum (Figure S5B). B. bifidum with afcA knockdown showed reduced adhesion to immobilized intestinal mucosa derived WT mice in vitro (Figure 5B). However, no additional reduction was observed in the adhesion to intestinal mucosa pretreated with UEA-1 (Figure 5B). We also expressed afcA in the Bifidobacterium longum 105A (B. longum 105A), which is lacking the afcA gene. Expression of afcA can significantly increase B. longum 105A colonization in the colonic lumen (Figure 5C) and mucosa (Figure 5D) in WT mice compared to B. longum 105A transfected with empty-vector expression (105A). However, such an increase was not observed in Fut2−/− mice (Figures 5C–5D), not even when fed with free 2′-fucosyllactose (2′-FL), one of the most abundant HMOs that can be degraded to produce fucose by the infant bifidobacterial community(Van den Abbeele et al., 2019) (Figure 5C). These results indicate that the binding property of AfcA, but not fucosidase activity, is important for the colonization of B. bifidum through host fucosylation. Protein motif analysis showed the AfcA contains a Fucosidase domain (Fuc) from 577 to 1474 aa, a Bacterial ig2-like domain (Big2) from 1475 to 1728 aa(Katayama et al., 2004) and a N-terminal function unknown fragment without a conserved motif found from 41 to 576 aa (hereafter we named UK) (Figure 5E, top). To further characterize AfcA-mediated B. bifidum adhesion, the recombinant protein for these domains and full-length protein were expressed, purified (Figure 5E bottom and Figure S5C), and examined for the binding to colonic mucosa. ELISA (Figure 5F) and SPR assay (Figure S5D) in vitro showed that full-length AfcA protein can directly bind to mucosal protein from WT mice. The binding ability of AfcA protein was significantly decreased in mucosa protein pretreated by UEA-1 (Figures 5F and S5D). ELISA results revealed that the UK domain is the main domain that is responsible for binding to fucosyl-conjugates (Figure 5F). The Fuc domain exhibits the binding ability (Figure 5F) and is comparable fucosidase activity as full-length afcA (Figure S5E). To confirm the binding of His-AfcA and His-UK to the intestinal fucosylated mucosa, we also carried out binding assays using murine colonic mucosal protein from Fut2−/− mice. His-UK and AfcA showed no binding with colonic mucosal protein from Fut2−/− mice (Figure S5F). Although the UK domain has no fucosidase activity (Figure S5E), B. longum 105A carrying the UK domain, but not the Fuc+Big2 domain, showed comparable in vivo colonization with B. longum 105A carrying full length AfcA (Figure 5G), which further supported the binding ability contributing to the enhanced in vivo colonization of B. longum 105A. As Muc2 is the major O-glycosylated product in mucus of the small intestine and colon and is also reported to be fucosylated(Arike et al., 2017), we thus tested whether AfcA can bind to fucosylated Muc2 using mucosal protein and tissue sections from WT and Fut2−/− mice. Pull down experiments showed that Muc2 is fucosylated (Figure 5H, top) and AfcA protein can directly bind to fucosylated Muc2 (Fuc-Muc2) (Figure 5H, middle), but not nonfucosylated muc2 (Non-muc2, from Fut2−/− mice) (Figure 5H, bottom), and this binding was blocked by pre-incubation with UEA-1 (Figure 5H). We also performed binding assays in tissue sections. Goblet cells were detected by mucin 2 staining (Figure 5I). His-tagged afcA protein was found to specifically react with fucosylated mucin2 in goblet cells in WT mice. His-afcA produces less signal when interacting with mucin2 producing goblet cells of Fut2−/− mice (Figure 5I). These data indicate that AfcA specifically interacts with fucosylated mucus, such as fucosylated Muc2. To test whether the fucosidase-producing bacteria benefit from α1,2-fucosylation of the IEC surface, total α1,2-fucosidase activity was examined in the fecal samples of Fut2−/− mice, VGx mice and VIPR1−/− mice. The total fucosidase activity significantly decreased in Fut2−/−, VGx mice and VIPR1−/− mice compared to control mice (Figure 5J). Real-time PCR analysis also showed decreases of α1,2-fucosidase-producing bacteria in the Fut2−/− mice (Figure 5K). These data suggest α1,2-fucosidase acts as a moonlight protein to enhance bacterial adhesion and colonization to the fucosyl-conjugated mucus in the gut.
Figure 5. α1,2-fucosidase interacts with fucosylated mucosa to enhance adhesion and colonization of Bifidobacterium in the gut.
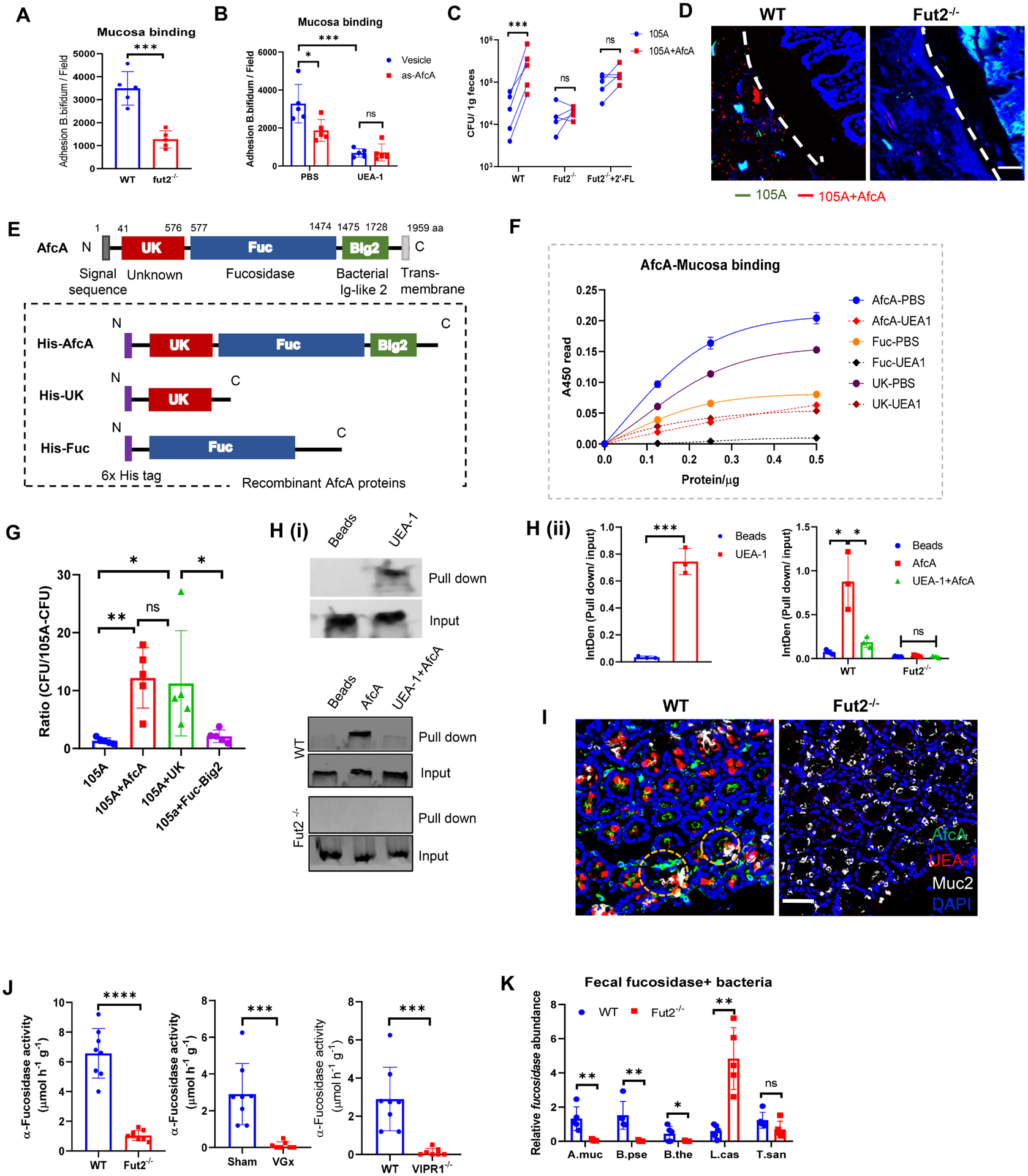
(A) Adhesion of B. bifidum strain to colonic mucosa derived from WT or Fut2−/− mice.
(B) Adhesion of B. bifidum afcA knockdown strain (As-afcA) and control strain to colonic mucosa. UEA-1 was pre-incubated with mucosa for 1h at 37°C before bacteria adding as indicated.
(C) WT mice, Fut2−/− mice or 2’-FL-fed Fut2−/− mice were orally given a 1:1 mixture of B. longum 105A strain (Erythromycin resistant vector-transformed 105A strain (105A)), and afcA-vector-transformed 105A strain (105A+AfcA)). The data show recovery from the feces at 24 h after gavage. (A-C) Data are representative of three independent experiments. Error bars: Mean ± SEM; *p < 0.05; ***p < 0.001.
(D) FISH analysis of B. longum 105-A colonization using a probe for erythromycin (Erm, green) and AfcA (red) within the colonic samples of mice that were gavaged with mixture strains as in C. Scale bar, 50 μm.
(E) (Top) Schematic representation of the primary structure of AfcA from B. bifidum. The grey box at the N terminus indicates a signal peptide. Unknown function domain (UK, 41–576 aa) is depicted as red box. The fucosidase domain (Fuc, 577–1474 aa) responsible for 1,2- fucosidase is depicted as a blue box. The region with Ig-like folds Big2 (1475–1728 aa) is shown by green box; (Bottom) Schematic representation of AfcA-truncated protein expressed as fusions to 6×His tag at the N terminus (purple boxes).
(F) Binding properties of AfcA- full-length or truncated recombinant proteins to mucosa in the absence or presence of UEA-1, as determined by ELISA.
(G) Mice were orally given B. longum 105A strains expressing empty vector (105A), vector with full-length of AfcA (105A+AfcA), the UK domain (105A+UK) or the Fuc+Big2 domain (105A+Fuc−Big2). The data show recovery from the feces at 24 h after infection. Mean ± SEM; n = 5; *p < 0.05; **p < 0.01.
(H) Binding of AfcA recombinant protein with fucosylated Muc2 (i). (Top) Mucosal protein were pulled down by UEA-1 and immunoblotted by anti-Muc2 antibody, a group using streptavidin bead without UEA-1 was set as a blank control (Beads); mucosal protein from WT (Middle) or Fut2−/− mice (Bottom) was pretreated with/without UEA-1 (UEA-1+AfcA or AfcA), pulled down by His-AfcA protein, and then immunoblotted by anti-Muc2 antibody, a group using Ni-NTA bead without his-AfcA was set as blank control (Beads). The qualifications were shown in (ii).
(I) His-AfcA protein binding to murine colonic tissue sections prepared from WT mice or Fut2−/− mice. Total fucosylated conjugates were detected by UEA-1 (red) staining. His-AfcA (green) was detected with anti-His antibody, the goblet cells were indicated by Muc2 (grey) staining. Scale bar, 50 μm.
(J) Measurement of total α-L-fucosidase activity in fecal samples. Fecal supernatant was assayed for cleavage of 4-methylumbelliferyl-fucopyranoside substrate by fluorescence.
(K) Real-time PCR analysis of the relative abundance of α-L-fucosidase producing bacterial strains in the feces of WT mice and Fut2−/− mice. Five representative gut bacteria expressing L-fucosidase were chosen for analysis, including Akkermansia muciniphila (A. muc), Bifidobacterium pseudolongum (B. pse), Bacteroides thetaiotaomicron (B. the), Lactobacillus casei (L. cas) and Turicibacter sanguinis (T. san). Primers target the L-fucosidase genes in the corresponding bacterium were designed for real-time PCR.
Data are from (J-K) at least 2 independent experiments performed. n=5–10, mean ± SEM. *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001, ns not significant. See also Figure S5.
Disturbance of enteric neuron promotes the development of ALD via gut-liver axis
The gut microbiota is a major player in ALD progression via the gut-liver axis(Gao and Bataller, 2011; Szabo, 2015). Fucosylation deficiency changes the composition of gut microbiota(Kashyap et al., 2013) and contributes to the development of ALD(Zhou et al., 2020), probably through regulation of colonization of cytolysin-positive E. faecalis in the intestine. Alcoholic neuropathy is associated with altered neuronal activity and nerve damage(Ammendola et al., 2000; Chopra and Tiwari, 2012). In order to determine the impact of alcohol consumption on the ENS, we quantified enteric neurons along the GI tract in the myenteric plexus. Following alcohol feeding, we observed a 40%−60% reduction in the myenteric plexus day 50 post feeding in the ileum and colon (Figures S6A–S6B). Notably, chronic alcohol consumption reduced the number of VIP neurons in both the ileal and colonic submucosal plexus (Figures S6C–6D). Furthermore, we observed significant differences of VIP production in the ileum, but not in the colon between in Sham and in VGx mice after alcohol consumption (Figure S6E). In in vitro cultured enteric neurons, alcohol treatment significantly inhibited the expression of VIP mRNA, but not of other neuropeptides including CGRP and NPY (Figure S6F). These data indicate that alcohol consumption perturbs the ENS, and may provide an explanation for the observed GI/IEC functional changes in ALD.
Given that vagotomy and inhibition of VIPergic neurons leads to the similar effects on the IECs fucosylation and the diversity of gut microbiota, we assessed if the disturbance of enteric neuron regulates ALD via the gut-liver axis. VGx mice had significant increases in the alanine transaminase (ALT) and aspartate transaminase (AST) levels compared with those of the sham group after ethanol feeding (Figure 6A). Hepatic steatosis was more severe in the ethanol-fed VGx mice compared to Sham mice, as clearly evidenced by the amount of hepatic triglyceride, and demonstrated with H&E and Oil red-O staining (Figures 6B–6C). FACS analysis also showed more neutrophil infiltration in the liver of ethanol-fed VGx mice (Figure S7A). These histological changes were supported by significantly elevated hepatic expression of the CXC chemokines in the ethanol-fed VGx group, particularly CXCL1 and CCL3, which regulate neutrophil recruitment (Figure S7B). This process resulted in upregulated expression of pro-inflammatory cytokine genes in the liver, such as IL-1β (Figure S7B). Taken together, both the histological and gene expression analyses showed significantly increased hepatic steatosis and inflammation in the ethanol-fed groups after vagotomy. The development of ALD is linked to gut dysbiosis (Wang et al., 2016). We thus analyzed changes in fecal microbiota by 16S rRNA gene sequencing. PCoA showed separate clustering of the microbiota of ethanol-fed Sham mice versus ethanol-fed VGx mice (Figure 6D). Kruskal-Wallis analysis showed that vagotomy significantly reduced alpha diversity with Chao1 and observed OTU in alcohol fed mice (Figure 6E), however, Simpson and Shannon analysis did not differ significantly (Figure S7C). These findings indicate that following chronic ethanol exposure, vagotomy may affect the luminal composition of intestinal microbiota. Since bacteria reside in different niches within the intestine, we also measured the bacterial loads in the mucus layer of gut. Ethanol-fed VGx mice had significantly higher numbers of mucosa-associated bacteria in the mucus layer of the small intestine, but not the that of large intestine, than ethanol-fed Sham mice (Figure 6F). Interestingly, VGx mice, with or without alcohol feeding, were colonized by a higher abundance of Enterococcus spp., especially cytolysin-producing E. faecalis both at the mucus layer and within the lumen, (Figure 6F, 6G and S7D–S7E). Similar to VGx mice in the steady state, ethanol-fed VGx mice also had less Bifidobacterium (Figure 6G). UEA-1 staining of intestinal sections revealed that the mucus layer has been inappropriately fucosylated after chronic alcohol consumption (Figure S7F). Since the presence of cytolysin-positive (cytolytic) E. faecalis correlates with the mortality in patients with alcoholic hepatitis(Duan et al., 2019), we propose that VGx-induced alteration in mucosal carbohydrate availability leads to disruption of the resident microbiota composition including E. faecalis expansion, which may mediate the increased susceptibility of VGx mice to alcohol-induced liver disease. To this end, first, we performed fecal microbiota transplantation (FMT) to germ free (GF) mice. Sham or VGx mice fed with chow diet were used as donors in FMT experiments. The bacteria were transplanted to recipient mice one week before the alcohol diet adaptation phase. The analysis of liver lesions showed that fecal microbiota from VGx promotes steatosis in alcohol-fed mice compared to FMT from Sham mice (Figures 6H–6I). Moreover, transplantation of microbiota from VGx mice resulted in higher levels of serum transaminase (Figure 6J). Second, depletion of the intestinal microbiota led to a similar degree of liver disease in Sham and VGx mice after alcohol feeding (Figures S7G–S7I). Taken together, these data suggest that the vagus nerve might promote VIPnergic-mediated IEC fucosylation and reduce ethanol-induced liver damage in mice via gut-liver axis.
Figure 6. Perturbation of vagal afferents promotes ALD via modulating gut microbiota.
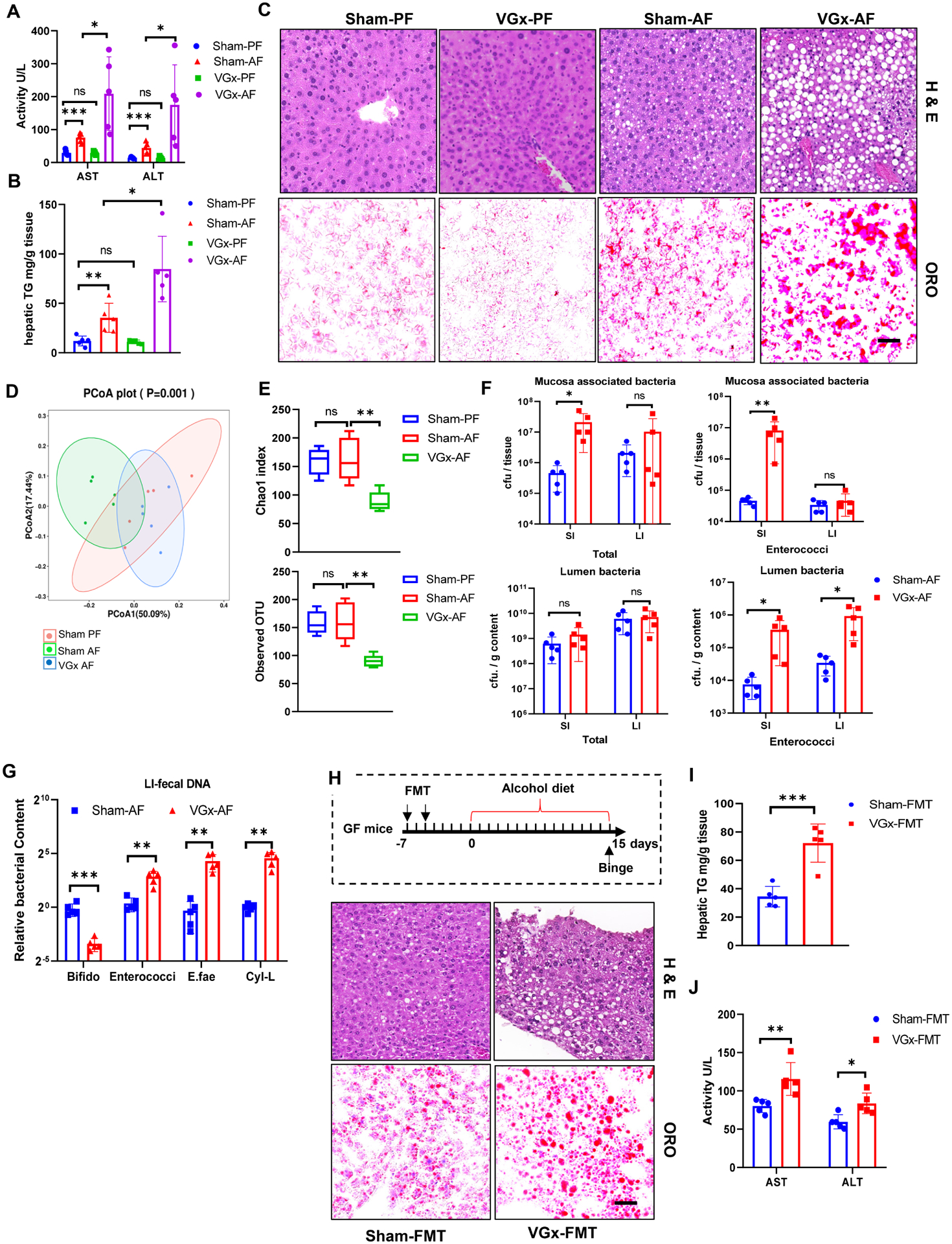
Sham mice or VGx mice were fed control or alcohol diet and sacrificed 15 days later. Pair feeding (PF); Alcohol feeding (AF).
(A) Serum ALT and AST.
(B) Hepatic triglycerides (TG).
(C) H&E staining and Oil red O (ORO) staining of liver, scale bar, 50 μm.
Data are from (A-C) three independent experiments performed. n=5, mean ± SEM. *p < 0.05, **p < 0.01, ***p < 0.001, ns not significant.
(D) PCoA analysis of unweighted UniFrac distance based on 16S rRNA sequencing of fecal samples from Sham mice and VGx mice.
(E) Alpha diversity represented by the Chao1 and observed OTU index.
(F) Total bacteria and enterococci cfu in the mucus layer and lumen assessed by plating.
(G) Relative levels of Bifidobacterium, Enterococci, E. faecalis (E.fae) and cytolysin -producing E. faecalis (Cyl-L) in the lumen in alcohol-fed mice assessed by real-time PCR.
(H-J) FMT from VGx mice increase liver damage in the ALD model. (H) The ethanol diet timeline and H&E and Oil red O staining of liver, scale bar, 50 μm. The FMT group was force-fed (black arrows) fresh fecal bacteria twice a week before alcohol feeding. (I) Hepatic triglycerides. (J) Serum ALT and AST. Sham-FMT, recipient transplanted with fecal microbiota from Sham mice with chow diet; VGx-FMT, recipient transplanted with fecal microbiota from VGx mice with chow diet.
Data are from (F-G) 2, (H-J) 3, independent experiments performed. n=5, mean ± SEM. *p < 0.05, **p < 0.01. ***p < 0.001. See also Figures S6 and S7.
AfcA enables Bifidobacteria to inhibit E. faecalis intestinal colonization in experimental ALD model
Fucosylation has been reported to limit E. faecalis colonization, possibly via the gut microbiota(Pham et al., 2014), while the exact mechanism is still unknown. Based on our above findings, we proposed that fucosidase-producing bacterial strains, like B. bifidum, contributes to the restriction of E.faecalis colonization. Specific pathogen free (SPF) WT littermates or Fut2−/− mice were precolonized with PBS or B. bifidum 2–3 days before colonization with E. faecalis (Figure 7A). Real-time PCR revealed that precolonization with B. bifidum significantly decreased the levels of cytolysin producing E. faecalis (Figure 7B) in WT mice, but not in Fut2−/− mice. Fluorescence in situ hybridization (FISH) analysis of the gut mucosa demonstrated that WT mice appeared to have more B. bifidum, less E. faecalis distributed within the mucosa than Fut2−/− mice with B. bifidum precolonization (Figure 7C), suggesting that AfcA/fucosylation-mediated adhesion is therefore likely to directly prevent the colonization of E. faecalis. To decipher the direct role of B. bifidum on E. faecalis colonization in vivo, we also inoculated germ free C57BL/6J mice with either PBS, B. bifidum or B. bifidum with 2′-FL supplement. One week after colonization, mice were orally inoculated with E. faecalis. Compared to PBS, E. faecalis displayed significantly lower loads after gavage in both the intestinal lumen and mucosa in mice precolonized with B. bifidum (Figure 7D). In addition, the number of E. faecalis in the lumen, but not in mucosa, significantly decreased in B. bifidum-colonized animals with 2′-FL supplementation compared to B. bifidum-colonized animals (Figure 7D), suggesting that Bifidobacterium could utilize free fucosylated HMOs to suppress E. faecalis colonization. Next, to emphasize the role of AfcA in competing with E. faecalis, we pre-colonized SPF WT mice with PBS, B. longum 105A transformed with empty vector (105A) or B. longum 105A transformed with AfcA expression vector (105A+AfcA), with UK expression vector (105A+UK) or with FUC-BIG2 expression vector (105A+FUC-BIG2) 2–3 days before colonization with E. faecalis. Similar to B. bifidum, mice treated with 105A+AfcA or with 105A+UK could significantly reduce the growth of E. faecalis and Enterococcus spp. in the lumen (Figure 7E and S7J–S7K), but precolonization with 105A with 105A+FUC-BIG2 failed to prevent the growth of E. faecalis and Enterococcus spp. (Figure 7E and S7J–S7K). FISH analysis of the gut mucosa demonstrated that E. faecalis appeared to be less distributed within the mucosa of mice with 105A+AfcA than 105A (Figure 7F). Altogether, these experiments demonstrated that AfcA producing Bifidobacterial strains, i.e., B. bifidum, most efficiently promoted E. faecalis clearance from the microbiome of precolonized animals. A previous study showed that experimental expansion of intestinal E. faecalis by gavage during ethanol feeding exacerbates ALD in mice(Llorente et al., 2017). We thus evaluated the role of B. bifidum in the suppression of E. faecalis-mediated ALD. Administration of E. faecalis promotes hepatic steatosis and injury in the ethanol-fed mice by H&E staining, oil red O staining, hepatic triglyceride and transaminase analysis (Figures 7G–7I). In contrast, B. bifidum treatment significantly attenuated the E. faecalis-mediated fat accumulation in the liver (Figures 7G–7I) of mice with ALD. Since VGx mice (Figure 6) had significantly higher level of fecal E. faecalis compared with control mice, we also evaluate the role of B. bifidum in alcohol-induced liver disease in mice with vagotomy. Administration of B. bifidum in VGx mice decreased hepatic steatosis and injury as evidenced by improvement in liver histopathology, lower serum transaminases and hepatic triglyceride levels (Figures 7J–7L). This result demonstrated that ENS enhances resistance to colonization of E. faecalis via the intestinal interaction of fucosidase-producing Bifidobacterium and α1,2-fucosylation.
Figure 7. Fucosidase-producing Bifidobacteria inhibit E. faecalis intestinal colonization and ameliorate ALD.
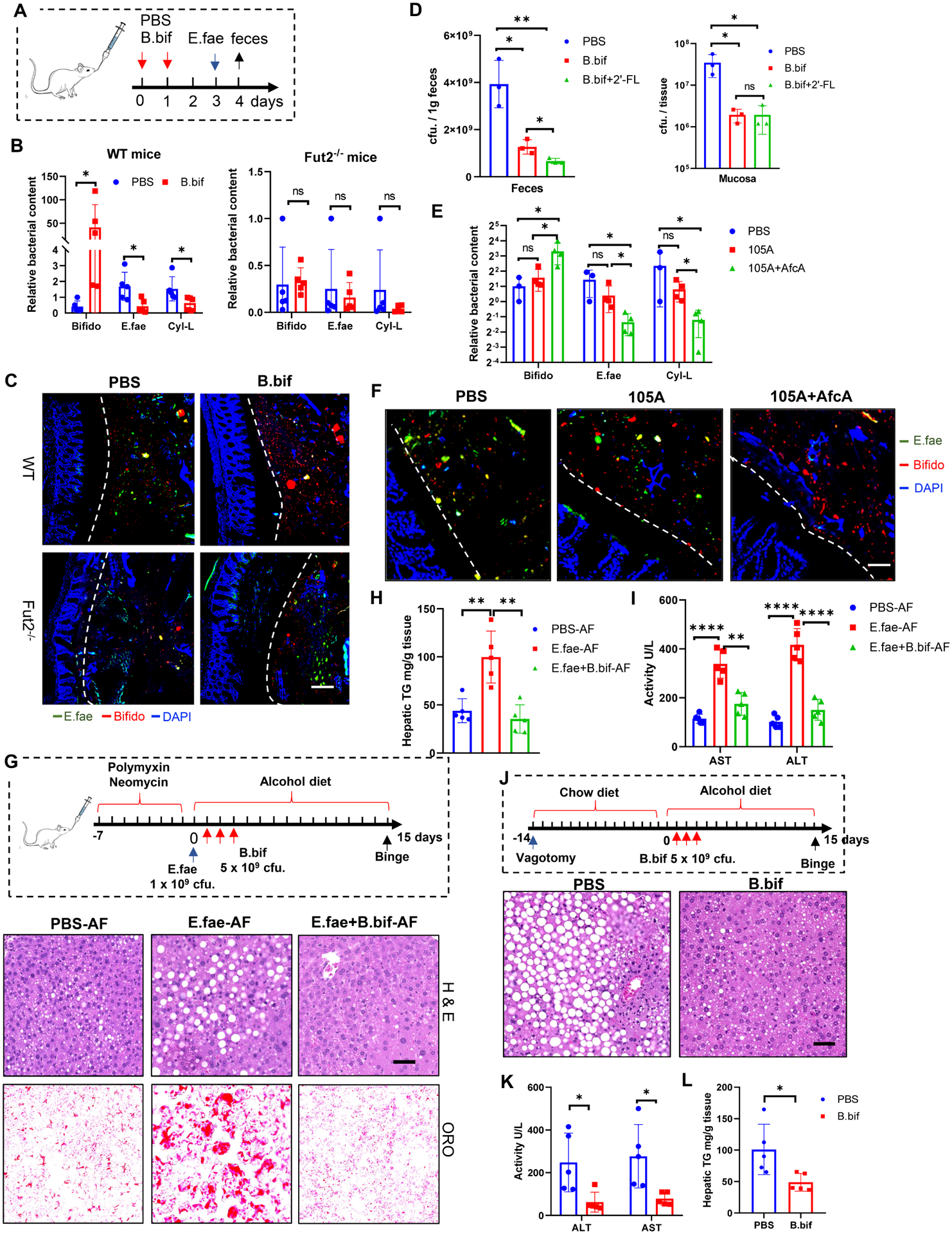
(A-C) Co-colonization of B. bifidum (B. bif) and E. faecalis (E. fae) in both SPF WT and Fut2−/− mice. Schematic diagram of the co-colonization experiment (A), Total Bifidobacterium and E. faecalis in the lumen and mucus layer were assessed by RT-PCR (B) and FISH analysis (C) using a probe targeted to the 16S rRNA of Bifidobacterium (red) and E. faecalis (green). Scale bar, 50 μm.
(D) Co-colonization of B. bifidum (B. bif) and E. faecalis in germ free (GF) mice. E. faecalis CFU in the feces and mucosa was assessed by plating at day 3 of colonization of E. faecalis. 2’-FL (2 mg/kg/day) was given to GF mice by gavage.
(E-F) Co-colonization of B. longum 105A strains and E. faecalis in SPF mice. Total Bifidobacterium (Bifido) and E. faecalis in the feces (E) and mucus layer (F) were assessed by RT-PCR (E) and FISH analysis (F) using a probe targeted to the 16S rRNA of Bifidobacterium (red) and E. faecalis (green). Scale bar, 50 μm.
(G-I) Administration of B. bifidum ameliorated E. faecalis-mediated liver disease in the ALD model. Schematic diagram and H&E staining and Oil red O staining of liver (G); Serum ALT and AST levels (H); liver triglyceride quantification (I). Scale bar, 50 μm.
(J-L) Administration of B. bifidum reverse vagotomy-increased liver damage in the ALD model. H&E staining and Oil red O staining of liver (J); Serum ALT and AST (K); Hepatic triglycerides (L). Data represent (A-C) 3, (D) 2, (E-F) 3, (G-L) 2 independent experiments combined. Scale bar, 50 μm. n=5, mean ± SEM. *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001.
Discussion
In this study, we showed that the enteric neuron, particularly VIPergic innervation, directs mucus fucosylation, which maintains gut microbiota homeostasis. The reduced abundance of enteric VIPergic neurons leads to the imbalance of abundance between beneficial Bifidobacterium and pathogenic E. faecalis, and enhances the susceptibility of ALD.
Vagal afferents control epithelial homeostasis and food intake in the intestine (Drokhlyansky et al., 2020; Page, 2021). VIP enteric neurons have previously been implicated in circadian regulation (Harmar et al., 2002), food sensing (Fujimiya and Inui, 2000) and immunoregulation (Seillet et al., 2020; Talbot et al., 2020), and are in close proximity with ileal goblet cells, which suggests a direct ability of the enteric neuron to signal to epithelial cells for mucus secretion (Lelievre et al., 2007), (Schwerdtfeger and Tobet, 2020). Here we found that epithelial expression of Fut2 is directly induced by VIPergic signaling, consistent with a lower level of fucosylated glycans at the ileal mucosa of VGx mice and VipIRES-cre hM4Difl-stop-fl/+ mice. We did observe that there was less requirement of VIPergic signaling for fucosylation of IECs in the colon compared to small intestine. This seemingly incongruent finding could be due to the diversity among VIP-expressing neurons throughout the intestine or indirect neuronal VIP signaling mechanisms may exist to regulate fucosylation production. Indeed, a recent study revealed that VIP+ neurons exclusively innervate the intestinal villi and were evenly distributed across proximal, middle, and distal small intestine(Bai et al., 2019). Our data suggested that VIP ligation of VIPR1 on the epithelium is essential for fucosylation in intestinal organoid cultures and by in vivo injection. Although VIPR1 is more highly expressed in IEC, more in-depth mechanistic approaches including studies in mice with IEC-specific deletion of VIPR1 will be required to fully understand the mechanism of VIP-induced IEC fucosylation. In addition to fucosylation, our IECs RNA-seq data also showed the vagotomy can significantly affect the pathways of bacterial defense and immune system process including innate and adaptive immune responses, indicating the extensive communications between the CNS, ENS, IECs and immune system. Indeed, VIP+ neurons were recently reported to regulate the function of intestinal ILC3s (Talbot et al., 2020). ILC3 cells-related cytokine IL-22 or MyD88-dependent immune signaling were reported to regulate IEC fucosylation in the small intestine(Goto et al., 2014; Pickard et al., 2014). Therefore, although our study highlights that a direct neuronal VIP signaling mechanisms may exist to regulate fucosylation production, there is possibly neuroimmune axis -dependent contribution to IECs fucosylation, and we cannot rule out an additional role of ENS functions in vivo. Since this study did not examine the gut-brain axis to explain the interactions between the vagus nerve, IEC and gut microbiota, we acknowledge that uncovering the specific mechanism through which specific inputs from CNS affects VIPnergic neurons in this context is an area of future studies.
α1,2-Fucosylation plays an important role in influencing interactions between the host microbial populations with mucus(Arike and Hansson, 2016; Herath et al., 2020). We detected a clear impact of vagotomy on the microbiota diversity and composition, characterized by a decreased abundance of some anaerobic commensal populations, particularly bifidobacterial species. A range of mucus-degrading bacteria are present in mucus, including Bifidobacterium bifidium(He et al., 2001) and Bacteroides fragilis(Macfarlane and Gibson, 1991). As an adaptation to residing in a glycan-rich environment, Bifidobacteria produce mucus-degrading enzymes such as fucosidase that cleave the mucus network to enhance the utilization of mucus. In agreement, we also observed a lower colonization of Bifidobacterium and fucosidase activity in the Fut2−/− mice and VGx mice. Moreover, the fucose-utilizing, anaerobic fucosidase-producing bacteria, were remarkably reduced in Fut2−/− mice. These data further support the notion that intestinal fucosylation provides a nutrient foundation for symbiotic commensals(Pickard et al., 2014). Bifidobacteria are thought to employ a variety of mechanisms to facilitate colonization of the host GI tract(Fanning et al., 2012; Gonzalez-Rodriguez et al., 2012; Turroni et al., 2013). Here, our data showed the fucosidase producing bacteria, like B. bifidum, can use α1,2-fucosidase as anchor molecular to facilitate bacteria colonization in the gut. B. bifidum produces a GH95 α-fucosidase (AfcA) with enzymatic activity on α1,2-fucosyl galactose(Katayama et al., 2004). We demonstrated that α-fucosidase (AfcA) not only has enzymatic activity on the assimilation of fucosylated carbohydrates, but also modulates B. bifidum-mucosal surface interactions. Adhesion assays with heterologous AfcA-expressing B. longum 105A strain demonstrated that cell surface-associated AfcA contributes to adhesion to fucosylated mucins in vitro and in vivo. We showed that domain UK, which is lacking fucosidase activity, bound to mucus in a similar degree as full-length AfcA, indicating an unknown fucosyl-binding motif in UK domain. We further demonstrated that the His-AfcA bound to murine colonic tissue, particularly fucosylated Mucin 2, indicating that AfcA probably adheres to the mucosal surface via Mucin 2 produced by goblet cells. Thus, our results provide insights into the molecular mechanisms of nutrient acquisition and adhesion of B. bifidum involving the α-fucosidase AfcA.
Finally, emerging evidence indicates that chronic alcohol consumption decreases neurogenesis and prominently disturbs the ENS leading to gut motility disturbances(Bode and Bode, 2003). Our study revealed that ethanol exposure decreases the cell numbers of enteric VIPergic neurons and inhibits the VIP secretion from the enteric neuron in vivo and in vitro. Additionally, mice with vagotomy showed increased susceptibility to ALD because of overgrowth of cytolysin-positive E. faecalis, which was found to cause more severe alcoholic hepatitis(Duan et al., 2019). We further investigated the effect of commensal Bifidobacterium on E. faecalis colonization. Notably, the AfcA-carrying Bifidobacterium strains significantly reduced E. faecalis colonization in mouse models including SPF mice and gnotobiotic mice, but not in Fut2−/− mice. It is worth noting that the protective effect of Bifidobacterium on inhibiting E. faecalis colonization was enhanced after gnotobiotic mice were fed 2’-FL (Figure 7D). However, B. longum 105A expressed with afcA has less ability to colonize in Fut2−/− mice, even in Fut2−/− mice fed with 2′-FL (Figure 5C). The bacterial α1,2-fucosidase can degrade fucosyl-conjugates or HMOs to produce free L-fucose, which can induce the expression of fut2 in IECs(Bry et al., 1996; Kononova et al., 2021; Pickard et al., 2014). Thus, our data support this elegant regulation network needed to balance the fucosylation level and Bifidobacterial colonization and indicates bidirectional cross-talk between intestinal fucosylation and gut microbiota(Kononova et al., 2021; Pickard et al., 2014). Strikingly, the administration of fucosidase-producing Bifidobacterium also prevents VGx- and E. faecalis-mediated ALD progression. These data strongly suggest Bifidobacterium may restrict the access of E. faecalis or other pathogens to host epithelial cell surfaces following chronic ethanol administration. Based on these observations, it is attractive to speculate about potential applications of probiotic fucosidase-producing Bifidobacterium or VIP for human ALD treatment. Our study elucidates how a high amount of alcohol consumption is associated with disturbances in VIP-producing enteric neuron, reduction in IECs fucosylation and beneficial symbionts, and subsequently overgrowth of cytolysin-positive E. faecalis, all of which contribute to the development of ALD.
STAR ★ Methods
Resource availability
Lead contact
Further information and requests for resources and/or reagents should be directed to and will be fulfilled by Dr. Zhongbin Deng (z0deng01@louisville.edu)
Materials availability
The materials generated in this study are available from the lead contact with a completed Materials Transfer Agreement.
Data and code availability
Next generation 16s rRNA sequencing and poly(A) RNA sequencing data have been deposited at Sequence Read Archive and are publicly available as of the date of publication. Accession numbers are listed in the key resources table. All data reported in this paper will be shared by the lead contact upon request.
This paper does not report any original code
Any additional information required to reanalyze the data reporting in this paper is available from the lead author on request.
KEY RESOURCE TABLE
| Reagent or resource | Source | Identifier |
|---|---|---|
| Antibodies | ||
| Rabbit VIP | Abcam | Cat# ab22736; RRID:AB_447294 |
| β-tubulin3 Alexa Fluor 488 | Thermo Fisher Scientific | Cat# 53-4510-82; RRID:AB_2574421 |
| Hu C/D Alexa Fluor 647 | Abcam | Cat# ab237235 |
| EpCAM-APC | Invitrogen | Cat# 17-5791-82; RRID:AB_2716944 |
| Rabbit Erk1/2 | Cell Signaling Technology | Cat# 4695S |
| Rabbit pErk1/2 | Cell Signaling Technology | Cat# 4370S |
| Rabbit C-fos | Cell Signaling Technology | Cat# 2250S |
| Mouse Muc2 | Abcam | Cat# ab11197; RRID:AB_297837 |
| Rabbit Muc2 | Abcam | Cat# ab272692; RRID:AB_2888616 |
| Mouse Anti-6x His | Thermo Fisher Scientific | Cat# MA1-21315; RRID:AB_557403 |
| Gr-1-PE | Invitrogen | Cat# 12-5931-83; RRID:AB_466046 |
| CD11b-APC | Invitrogen | Cat# 17-0012-83 |
| Goat anti-Rabbit Alexa Fluor 594 | Thermo Fisher Scientific | Cat# A32740; RRID:AB_2762824 |
| Goat anti-Rabbit Alexa Fluor 680 | Thermo Fisher Scientific | Cat# A32734; AB_2633283 |
| Goat anti-Mouse Alexa Fluor 488 | Thermo Fisher Scientific | Cat# A32723; RRID:AB_2633275 |
| IRDye 800CW Goat anti-Mouse | LI-COR Biosciences | Cat# 926-32210; RRID:AB_621842 |
| IRDye 800CW Goat anti-Rabbit | LI-COR Biosciences | Cat# 926-32211; RRID:AB_621843 |
| IRDye® 800CW Streptavidin | LI-COR Biosciences | Cat# 926-32230 |
| UEA-1 Rhodamine | Vector Laboratories | Cat# RL-1062; RRID:AB_2336769 |
| UEA-1 Biotin | Vector Laboratories | Cat# B-1065; RRID:AB_2336766 |
| UEA-1 Unconjugated | Vector Laboratories | Cat# L-1060; RRID:AB_2336768 |
| Bacterial strains | ||
| Bifidobacterium bifidum | ATCC | Cat# 29521 |
| Bifidobacterium longum subsp. longum 105A | (Sakanaka et al., 2019) | N/A |
| Enterococcus faecalis | ATCC | Cat# 29212 |
| E. coli Top10 | Invitrogen | Cat# C404006 |
| E. coli BL21 (DE3) | Invitrogen | Cat# C600003 |
| Chemicals, peptides, and recombinant proteins | ||
| Difco™ Lactobacilli MRS Broth | Becton Dickinson | Cat# 288130 |
| BBL™ Brain Heart Infusion | Becton Dickinson | Cat# 221778 |
| BBL™ Enterococcosel™ Broth | Becton Dickinson | Cat# 212207 |
| LB Broth (Miller) | Sigma | Cat# L3522-250G |
| RNAprotect Bacteria Reagent | Qiagen | Cat#76506 |
| Ni-NTA Agarose | Thermo Fisher Scientific | Cat# R90101 |
| Dynabeads™ M-270 Streptavidin | Invitrogen | Cat# 65305 |
| SuperScript™ IV First-Strand Synthesis System | Invitrogen | Cat# 18091050 |
| RNase-free DNase І | New England Biolabs | Cat# M0303S |
| Kanamycin | Sigma | Cat# K1876 |
| Chloramphenicol | Sigma | Cat# C0378 |
| Erythromycin | Sigma | Cat# E5389 |
| Ampicillin | Sigma | Cat# A9393 |
| 2’-Fucosyllactose (2’-FL) | Sigma | Cat# 41263-94-9 |
| VIP | Tocris Bioscience | Cat# 1911 |
| [D-p-Cl-Phe6,Leu17]-VIP (VIPx) | Tocris Bioscience | Cat# 3054 |
| Clozapine N Oxide (CNO) | Tocris Bioscience | Cat# 4936 |
| 4-methylumbelliferyl-alpha-l-fucopyranoside (4-MUF) | GoldBio | Cat# M-580-50 |
| FR180204 | Cayman Chemical | Cat# 15544 |
| Oil Red O | Sigma | Cat# O0625 |
| Gill™ Hematoxylin | Thermo Fisher Scientific | Cat# 6765007 |
| Shandon™ Eosin-Y | Thermo Fisher Scientific | Cat# 6766007 |
| B-27 Supplement | Gibco | Cat# 17504044 |
| N-2 Supplement | Gibco | Cat# 17502001 |
| Complete™ Protease Inhibitor Cocktail | Sigma | Cat# 11697498001 |
| Phosphatase Inhibitor Cocktail | Sigma | Cat# P2850 |
| Q5® Hot Start HiFi PCR Master Mix | New England Biolabs | Cat# M0543S |
| Critical Commercial Assays | ||
| RNeasy Mini Kit | Qiagen | Cat# 74104 |
| Nunc™ Lab-Tek™ II Chamber Slide™ System | Thermo Scientific | Cat# 154453 |
| RiboPure RNA Purification Kit, bacteria | Invitrogen | Cat# AM1925 |
| PowerUP™ SYBR™ Green Master Mix | Qiagen | Cat# 204143 |
| HiScribe T7 Quick High Yield RNA Synthesis Kit | New England Biolabs | Cat# E2050S |
| QIAamp@ Fast DNA Stool Mini Kit | Qiagen | Cat# 51604 |
| IntestiCult™ Organoid Growth Medium | Stem Cell Biotechnology | Cat# 06005 |
| Luciferase Reporter Gene Detection Kit | Sigma | Cat# LUC1-1KT |
| NEBuilder® HiFi DNA Assembly Master Mix | New England Biolabs | Cat# E2621S |
| Deposited data | ||
| Next generation 16s rRNA sequencing | Sequence Read Archive | PRJNA870429; SRR21103243-SRR21103252 |
| Next generation Poly(A) RNA sequencing | Sequence Read Archive | PRJNA870429; SRR21102741-SRR21102746 |
| Experimental Models: Organisms/Strains Mouse | ||
| C57BL/6 | Jackson Laboratory Stock | Cat# 000664 |
| VIPR1−/− | Jackson Laboratory Stock | Cat# 034744 |
| Fut2−/− | Jackson Laboratory Stock | Cat# 006262 |
| hM3Dq fl-stop-fl | Jackson Laboratory Stock | Cat# 026220 |
| hM4Di fl-stop-fl | Jackson Laboratory Stock | Cat# 026219 |
| Vip IRES-cre | Jackson Laboratory Stock | Cat# 031628 |
| oligos | ||
| Primers see table S1 | Sigma | N/A |
| Recombinant DNA | ||
| pET28b-MBP-TEV | (Currinn et al., 2016) | RRID: Addgene_69929 |
| pET28b-AfcA | This paper | N/A |
| pET28b-UK | This paper | N/A |
| pET28b-FUC | This paper | N/A |
| pBFS38 | (Sakanaka et al., 2014) | N/A |
| pBFS38-AfcA | This paper | N/A |
| pBFS38-Erm | This paper | N/A |
| pBFS38-UK | This paper | N/A |
| p16337 | (Gu et al., 2005) | RRID: Addgene_16337 |
| p16337-fut2p-Naive | This paper | N/A |
| p16337-fut2p-Mutant | This paper | N/A |
| pFUW-tetO-fos | (Pereira et al., 2013) | RRID: Addgene_131593 |
| Software and algorithms | ||
| Prism (GraphPad Software) | GraphPad | https://www.graphpad.com |
| Bugbase | (Ward et al., 2017) | https://bugbase.cs.umn.edu/ |
| Qiime2 | (Hall and Beiko, 2018) | https://qiime2.org/ |
| Bioedit | (Hall, 1999) | https://www.bioedit.com/ |
| SnapGene | SnapGene | https://www.snapgene.com/ |
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Mice
C57BL/6 mice, VIPR1−/− mice (Jax no. 034744), Fut2−/− mice (Jax no. 006262), hM3Dqfl-stop-fl mice (CAG-LSL-Gq-DREADD, Jax no. 026220), hM4Difl-stop-fl mice (CAG-LSL-Gi-DREADD, Jax no. 026219) and VipIRES-cre mice (B6J.Vip-IRES-Cre, Jax no. 031628) were obtained from Jackson Laboratory. For performing chemogenetic activation or inhibition of VIPergic neurons, we bred VipIRES-cre homozygous mice to hemizygotes hM3Dqfl-stop-fl mice (DREADD for activation) or homozygous hM4Difl-stop-fl mice (DREADD for inhibition), generating VipIRES-crehM3Dqfl-stop-fl (activation of VIPergic neurons) and VipIRES-crehM4Difl-stop-fl mice (inhibition of VIPergic neurons). To perform 36-h activation of the DREADDs, mice were treated with CNO (1 mg per kg bodyweight (mg/kg) intraperitoneally, TOCRIS) each 12 h. Age- and sex-matched WT mice and littermates were used in all of experiments. 6–8 weeks old C57BL/6 syngeneic germ-free (GF) mice were maintained in flexible film isolators at the Clean Mouse Facility of the University of Louisville. Animal care was performed following the Institute for Laboratory Animal Research (ILAR) guidelines and all animal experiments were done in accordance with protocols approved by the University of Louisville Institutional Animal Care and Use Committee (IACUC, Louisville, KY). The mice were acclimated for at least 1 week before any experiments were conducted.
Bacteria strains
Bifidobacterium bifidum (B. bifidum) was from ATCC (29521). B. longum subsp. Longum 105-A (B. longum 105-A) was kindly provided by Takane Katayama, Kyoto University and Y. Kano at Kyoto Pharmaceutical University, Japan (currently available as JCM 31944 from RIKEN Bioresource Center)(Sakanaka et al., 2019). All Bifidobacterial strains were grown in de Man, Rogosa, and Sharpe medium (Becton Dickinson, Franklin Lakes, NJ, USA) with 0.05% L-cysteine (MRS-C) at 37°C under an anaerobic atmosphere (10% CO2, 10% H2, and 80% N2). Escherichia coli (E. coli) DH5α was used for genetic manipulations and E. coli BL21(DE3) was used for protein heterologous expression. E. Coli strains were grown at 37°C in Luria-Bertani broth (LB). Enterococcus faecalis (E. faecelis) was originally obtained from the ATCC (29212). E. faecelis was grown in Brain Heart Infusion (BHI) media at 37°C.
Antibiotics and the concentrations used were as follows: chloramphenicol (Cm, 2.5 μg/ml for Bifidobacteria and 10 μg/ml for E. coli), erythromycin (Em, 5 μg/ml for Bifidobacteria), ampicillin (Amp, 100 μg/ml for E. coli) and kanamycin (Kan, 50 μg/ml for E. coli). Growth was monitored by measuring optical density at 600 nm (OD600) unless otherwise indicated.
Mouse enteric neuronal cell (EN) culture
Enteric neuronal cell culture was conducted according to a previous report(Zhang and Hu, 2013). In brief, the small intestine from adult mice (7–12 weeks) was opened lengthwise, and the muscularis myenteric plexus (MMP) was dissected completely under a binocular microscope using watchmaker forceps(Zhang and Hu, 2013). The MMP was cut into pieces of approximately 2 to 5 mm2 and placed in 3 ml pre-warmed collagenase digestion medium (1mg/ml collagenase IV (Worthington), 0.5 mM CaCl2, 10mM HEPES in neutral Hank’s Balanced Salt Solution (HBSS)). After digesting for 15 min at 37°C, the tissue segments were washed with ice-cold HBSS and gently shaken for 10 min in 10 ml HBSS to release smooth muscle cells from myenteric plexuses (MP). The remaining tissue containing MP was further digested with 0.05% Trypsin -EDTA (Thermo Fisher) solution at 37°C for 7 min. After trituration using a P1000 pipette, single cell suspensions were filtered through a 70um filter, cleaned of debris by centrifugation through 1ml fetal bovine serum (FBS), and captured by selective plating on Matrigel (Sigma)-coated cell culture dishes in B27 (Thermo Fisher) and N2 (Thermo Fisher) supplemented Neurobasal-A medium (Thermo Fisher) plus 10 ng/mL fibroblast growth factor-basic (FGF-b, Sigma), 20ng/mL Epidermal growth factor (EGF, Sigma), 50U/mL penicillin and 50ug/mL streptomycin.
Mouse intestinal organoid culture
Intestinal organoids were isolated and cultured in vitro using IntestiCult™ Organoid Growth Medium (Mouse) following the manufacturer’s instructions. Briefly, 20 cm of small intestine proximal to the stomach was harvested and longitudinally opened. The tissue was cut into 2 mm pieces and washed with ice-cold PBS 15 to 20 times with gently shaking until the supernatant was clear. The tissue was then digested with gentle cell dissociation reagent for 15 min at room temperature on a rocking platform at 20 rpm. Tissue pieces were collected and resuspended in PBS with 0.1% BSA. Crypts were isolated by gently shaking and then filtered through a 70-μm strainer. The supernatant containing crypts was collected and pelleted by centrifugation at 200× g for 3 min. Approximately 200–500 crypts were embedded in Matrigel (Sigma) per well of a 24-well plate and submerged in IntestiCult™ Organoid Growth Medium (OGM, Stem Cell Technologies) with included supplements. Organoids were subcultured every 6–7 days at 37°C in a 5% CO2 environment with a 1:4 splitting ratio.
METHOD DETAILS
Chronic exposure to ethanol
We used the binge-on-chronic NIAAA (Gao) model with 8-week-old mice(Chu et al., 2021). Briefly, male or female mice were acclimated to the Lieber-DeCarli liquid control diet (F1259SP; Bio-Serv, Flemington, NJ) or gradually introduced to and increased on the ethanol diet (5% ethanol-w/v; F1258SP; Bio-Serv) for 5 days followed by further feeding with the liquid control (pair feeding, PF) or ethanol diet (alcohol feeding, AF, 5% (v/v)) for an additional 10 days (day 15) or 45 days (day 50). On the last day of feeding, mice were also given ethanol 5 g/kg or maltose dextran 9 g/kg by gavage, and sacrificed 9 hours later. The volume of control diet given to mice was matched to the volume of ethanol diet consumed. Alcohol fed groups were allowed free access to the 5% (vol/vol) ethanol diet. Control mice were fed the isocaloric control diet throughout the entire feeding period. Body weights were measured every other day, and food intake was checked every day.
Subdiaphragmatic vagotomy
Bilateral subdiaphragmatic vagotomy was performed as previously reported(Dezfuli et al., 2018; Teratani et al., 2020) (Figure S1C). Mice were anaesthetized with a combination of Ketamine, Xylazine and butorphanol and a midline incision was made to provide wide exposure of the upper abdominal organs. The bilateral subdiaphragmatic trunks of the vagal nerves along the oesophagus were exposed and cut. In the sham operation group, these vagal trunks were exposed but not cut.
afcA gene cloning and heterologous expression
For heterologous expression in E. coli, the full-length AfcA and UK fragment, Fuc domain fragments were amplified from B. bifidum genomic DNA by primers afcA-Ex-F/R, afcA-Ex-F/UK-Ex-R, Fuc-Ex-F/R, respectively. The expression vector pET28 was linearized by PCR using primer pET28-F/R. The target fragments and linear vector were assembled by using NEBuilder® HiFi DNA Assembly kit following the manufacturer’s instructions. Positive colonies were screened by colony PCR and confirmed by Sanger sequencing. Constructed expression vectors were then transformed into E. coli BL21 (DE3). AfcA proteins were induced by adding 0.1 mM isopropyl-β-d-thiogalactopyranoside (IPTG) when an OD600 = 0.8 was achieved and the bacteria were grown overnight at 18°C. Bacteria were collected by centrifuge at 5,000× g for 10 min and lysed by sonication. AfcA proteins were purified using Ni-NTA agarose according to the manufacturer’s instructions and stored in storage buffer (50 mM Tris-HCl, 150 mM NaCl, 50mM KCl, 1 mM DTT, 10% glycerol, pH 7.6). Purified proteins were confirmed by SDS-PAGE.
For heterologous expression in the B. longum 105-A, the afcA gene was introduced into B. longum 105-A using an E. coli-Bifidobacterium shuttle vector pBFS38 (Sakanaka et al., 2014). The afcA gene was amplified by PCR with B. bifidum genomic DNA as a template using primers afcA-OE-F/R. pBFS38 was linearized by PCR using primers pBFS38-F/R. afcA gene and linearized pBFS38 were assembled by using NEBuilder® HiFi DNA Assembly kit following the manufacture’s instruction. An α1,2-fucosidase negative control vector with erythromycin resistance gene (ErmR) insertion was constructed similarly. The constructed vectors were transformed into the α-L-fucosidase-negative strain B. longum 105-A by electroporation with parameters set as follows: 10 KV/cm, 25 μF, 200 Ω.
Gut bacteria colony-forming unit (cfu) counts
The number of Enterococcus was enumerated on selective agar plates (BBL™ Enterococcosel™ Agar, BD) with appropriate dilutions. The numbers of AfcA-null B.lon 105-A were enumerated on MRS agar plates adding Em and Cm antibiotics with appropriate dilutions. The number of AfcA-expressing B. longum 105-A was calculated from the difference between cfu. on Cm-MRS plates and cfu. on Cm-Em-MRS plates. The number of total bacteria were enumerated on BHIS media (BHI+ 0.5 g/L L-cysteine, 5 mg/L hemin and 0.2 ml vitamin K1 solution) under an anaerobic atmosphere with appropriate dilutions.
Transplantation of gut microbiota to mice with ALD
GF C57BL/6 mice were obtained and maintained in sterile isolators in Functional Microbiomics Core (FMC)-Germ Free facility at University of Louisville. Microbiota transfer was performed by feces gavage using a modified version of a previously described protocol(Gu et al., 2021). Feces were collected from chow diet-fed Sham or VGx mice two weeks after surgery (n=5), and pooled and suspended in 1.5 ml PBS. The suspensions were immediately administered (0.15 ml/mouse) to 8-week-old C57BL/6 GF male mice that were maintained with autoclaved normal chow diet food and water. One week after transferring of microbiota, mice were gradually introduced to the 5% (vol/vol) ethanol diet for 15 (5+10) days. Antibiotics treatment of Sham or VGx mice was started at 1 weeks before liquid diet feeding, and mice were gavaged three times per week until harvesting. The composition of antibiotics mixture was used (polymyxin B (Sigma-Aldrich) 150 mg kg−1 and neomycin (Sigma-Aldrich) 200 mg kg−1 body weight). In some experiments, C57BL/6 mice were treated with antibiotics for 1 weeks and gaveaged E. faecelis (ATCC 29212,1×109 cfu/mouse) once at 0 day of liquid diet feeding. Three separate doses of 1× 109 cfu Bifidobacterium bifidum (B. bifidum, ATCC 29521) were given to C57BL/6 mice by gavage at 1, 2, and 3 days after Enterococcus faecalis. Mice were introduced to the 5% (vol/vol) ethanol diet for 10 days.
Microbiota depletion
8-week-old C57BL/6 male mice were treated with antibiotic cocktail containing ampicillin (1 g/L), vancomycin (0.5g/L), metronidazole (1g/L), neomycin (1g/L) and streptomycin (1g/L) in drink water for 2 weeks to deplete the indigenous gut bacteria. Depletion of gut bacteria were confirmed by plating on BHIS agar plates 10 days and 14 days after antibiotics treatment, respectively.
VIP Rescue experiment
VGx mice were i.p. injected with VIP (6 μg/mouse) or PBS daily for 1 week from day 3 after vagotomy. Mice were sacrificed 12h after last treatment and feces and intestine samples were collected.
Bifidobacterium and E. faecelis co-colonization
For SPF mice colonization, male and female C57BL/6 mice were gavaged with polymyxin B (Sigma-Aldrich) 150 mg kg−1 and neomycin (Sigma-Aldrich) 200 mg kg−1 body weight once daily for 1 week to reduce intestinal bacteria. Following eradication of the commensal microbiota, two separate doses of 1× 1010 cfu Bifidobacterium bifidum (B. bifidum, ATCC 29521) or B. longum 105A (Sakanaka et al., 2019) strains were given to WT or Fut2−/− mice by gavage at 2 and 3 days before Enterococcus faecalis (E. faecelis, ATCC 29212) gavage (1×109 cfu/mouse), fecal samples were collected at 24 h and 72 h after E. faecelis gavage and quantified by RT-PCR. The feces were examined by 16S rRNA PCR. E. faecalis was grown freshly in Bacto Brain-Heart infusion plates (Becton Dickinson) for calculating CFU. For the GF mice colonization, 1× 108 cfu of B. bifidum was given to mice by gavage. Mice were then orally inoculated with 1× 108 cfu of E. faecelis 2 weeks after B. bifidum colonization. 2’-FL (Sigma) was given to mice at a dose of 2 mg/mouse/day starting from B. bifidum colonization until the end of the experiment as indicated. Fecal and mucosal samples were collected at 48 h after E. faecelis gavage and the cfu of E. faecelis was counted as described above.
Bacterial adhesion assay
The bacterial adhesion assay was performed as previously reported with minor modifications(Suzuki et al., 2016). Briefly, diagnostic glass slides with 15mm diameter circles (Fisherbrand) were coated with 100 μL DNase I and RNaseA-treated mucosa protein extracts (1 mg/ml) for 12 h at 4°C. After washing three times with PBS with 0.05% BSA, slides were incubated with 2% BSA in PBS for 1 h at room temperature. A 40-μL volume of bacterial suspension (OD600=0.5) was added to the slides followed by incubation at 25°C for 2 h. Samples were washed three times with 0.05% BSA-PBS to remove unbound bacteria and then stained with methylene blue to visualize bifidobacterial cells. Randomly selected fields were imaged with a microscope and bacteria in five fields were counted.
ELISA for binding of His-AfcA proteins
A binding assay for AfcA to mouse mucosa protein extracts was conducted using an ELISA as described previously(Nishiyama et al., 2017). Briefly, a 96-well microplate was coated with mucosa protein extracts (0.2 mg/well) and blocked with 2% BSA-PBS for 1 h at room temperature. After two washes with 0.05% BSA-PBS, different amounts of his-tagged recombinant AfcA proteins as indicated were added to the wells, followed by incubation for 1 h at room temperature. After washing twice with 0.05% BSA-PBS, proteins were detected with horseradish peroxidase-labeled anti-6×His-tag IgG (Thermo fisher, 1:1,000 dilution). Tetramethylbenzidine (TMB) peroxidase substrate was added until most of wells were blue in color, the reaction was terminated with 2 M H2SO4. Absorbance at 450 nm was measured on a BioTek HT plate reader.
UEA-1 and AfcA pull down assays
For UEA-1 pull down, 10 μg of biotin labeled UEA-1 was incubated with 200 μg of IECs lysate proteins for 30 min at room temperature (RT). The UEA-1-mucosa protein mixture was then incubated with agarose beads conjugated with streptavidin. The bound proteins were released by boiling in 1× SDS loading buffer and then washing the agarose beads 3 times in PBST. The eluted samples were used for western blot analysis. Unlabeled UEA-1 run through the same procedure was used as negative control.
For AfcA pull down, 10 μg of 6×his labeled AfcA protein was incubated with agarose beads conjugated with an anti-his antibody for 2 h at RT with rocking 20 rpm. The beads were incubated with 500 μg of IEC lysate proteins for 2 h at 4°C after which the beads were washed twice with 30 mM imidazole in PBS. The bound proteins were eluted with 300 mM imidazole in PBS after twice wash with 30 mM imidazole. The eluted protein samples were boiled after 1× SDS loading buffer added, and then used for western blot analysis.
Fucosidase activity measurement
The fucosidase activities in the fecal samples were tested according to the previous report(Pickard et al., 2014). Briefly, 1 uL of 50 mM 4-methylumbelliferyl-α-L-fucopyranoside (4-Muf, Carbosynth) was added into 50 ul of fecal supernatant (100 mg feces in 1 ml PBS), and incubated at 37 °C. The reaction was stopped by adding 500 ul of stop solution (0.2 M glycine-NaOH, pH 10.5) at the indicated time and fluorescence measured at 365 nm excitation, 445 nm emission. Fluorescence of a no-substrate control was subtracted from the substrate-containing reaction.
Mucosa associated bacteria isolation and enumeration
Mice were sacrificed and the intestine was dissected in ice-cold PBS. The lumen content in the intestine were flushed out with PBS and rinsed another 3 times with 20 ml PBS. The intestine was longitudinally opened, laid out so the luminal surface was facing up, and then gently rinsed with PBS to remove any attached debris. Mucosa was collected by gently scrapping the surface with a cover slip and homogenized in PBS. The mucosa associated bacteria were then plated on BHIS agar plates with appropriate dilutions, and cultured at 37°C for 48–72 h in an anaerobic chamber.
Construction and assaying of the fut2 promoter reporter system
A high-copy number luciferase containing plasmid was purchased from Addgene (#16337) to construct fut2 promoter reporter. 800 bp sequence at upstream of the fut2 1st exon containing the AP-1 binding site was amplified from mouse genomic DNA using primers fut2-prom-F/R. Plasmid 16337 was linearized by PCR using primer 16337-F/R. The fut2 promoter fragment and linearized 16337 were assembled using the method described above. Positive colonies were screened by colony PCR and confirmed by Sanger sequencing. AP-1 binding site mutation was conducted using a previously reported site-specific mutagenesis method(Kunkel, 1985). The constructed reporter plasmids were co-transfected with a c-fos expressing plasmid pFUW-tetO-fos (Addgene, #131593) (Pereira et al., 2013) into HEK-293T cell. C-fos expression was induced by adding 1 μg/ml of doxycycline (DOX). Luciferase activity was measured by using Luciferase Reporter Gene Detection Kit (Sigma) according to the manufacture’s instruction.
AfcA antisense RNA biosynthesis and gene knockdown
Antisense RNA of afcA was obtained from in-vitro transcription by using a HiScribe T7 Quick High Yield RNA Synthesis Kit following the manufacturer’s instruction. T7 promoter was incorporated into the 5’- terminal of the primer which was used to amplify the template DNA. The RNA was quantified using a NanoDrop 2000 (ThermoFisher scientific). One μg of transcribed asRNA was then encapsulated into lemon exosomal like nanoparticle derived nano vector (LNV) according to our previous report(Lei et al., 2021). asRNA containing LNV was added into B. bifidum cultures when an OD600=0.5 was reached with a ratio of 1:10 (LNV number to B. bifidum number). Three hours after adding asRNA, the bacteria were harvested by centrifugation for 5 min at 4,000× g, and RNA extracted.
Intestinal organoids/neuron co-culture
For intestinal organoids and neuron co-culture, EN were purified and cultured as above. At the day 7 of the EN culture, crypts were isolated and purified from small intestines. Differentiated EN were digested, counted and mixed with crypts with a ratio of 5:1, the EN-Crypt mixtures were embedded in the Maltrigel and coculture in the OGM media with included supplements (1 μM VIPx, for 24h) at 37°C for 3–5 days.
Fluorescence in situ hybridization (FISH) to detect bacteria
Mice were euthanized with CO2 and death was confirmed by cervical dislocation. After euthanasia, intestinal tissues including faecal pellets were cut and fixed in Carnoy’s fixative (600 mL ethanol, 300 mL chloroform, 100 mL CH3COOH) for at least 1 h. The tissues were then washed as follows: 30-min 100% methanol (2X), 20-min 100% ethanol (2X), and 15-min xylene (2X). The tissue was incubated in 55°C paraffin for 12–24 h before embedding. Approximately 5 μm thick histologic sections were hybridized at 50°C for 90 min in the hybridization solution (20 mM Tris-HCl (pH 7.4), 0.9 M NaCl, 0.1% SDS) with 1 μM of following probes: Enf184, probe targeting E.faecelis, Bif228 probe targeting Bifidobacterium, p-AfcA, probe targeting afcA containing bacteria, p-Erm, probes targeting ErmR containing B. longum 105A. Staining was conducted in a humid environment using a glass coverslip to prevent drying of the samples. The sections were washed in buffer (20 mM Tris-HCl (pH 7.4), 0.9 M NaCl) for 20 min at 50°C. The sections were counterstained with DAPI for 5 min at room temperature.
Histology and immunofluorescence
For H&E staining, tissue specimens were fixed in 10% formalin, dehydrated, and then embedded in paraffin. Tissue samples were cut at 5 μm thicknesses and stained with hematoxylin and eosin. Sections were scanned using an PANNORAMIC DESK II DW scanner (3D Histech). Liver steatosis was semi-quantitatively assessed into five grades: none = 0; mild = 1 (1% to 5% of hepatocytes); moderate = 2 (6% to 32%); marked = 3 (33% to 66%); and severe = 4 (67% to 100%). For oil red-O (ORO) staining, liver tissues were embedded in the OCT and were cut at 5 μm thicknesses. The slides were fixed with ice-cold 10% formalin for 10 min, followed by incubation in the 100% propylene glycol for 5 min. The slides were stained with pre-warmed (60°C) ORO solution for 20 min in A 60°C oven. After A 5-min wash in 85% propylene glycol, the slides were counterstained with Hematoxylin for 1 min and mounted with CC/Mount Aqueous Mounting Medium (Sigma).
For immunofluorescence analysis, OCT (Sakura Finetek)-embedded tissue cryosections (5 μm-thick) were blocked for 1 hour at 22°C with blocking solution (0.5% Triton X-100, 2% normal donkey serum in PBS) and incubated overnight at 4°C with the primary antibodies, antibodies against Beta-Tubulin III (Thermo Fisher, 1:1000), EpCAM (Thermo Fisher, 1:200), and VIP (Abcam 1:500). For lectin staining, the sections were incubated for 30 min at 4°C with Ulex europaeus aggluitinin-1 (UEA-1) conjugated to rhodanmine (Vector Laboratories or Sigma-Aldrich). Tissues were counterstained with DAPI and images were captured on a confocal laser scanning microscopy (Nikon, Melville, NY).
For whole-mount staining, ileums or colons were collected and immersed on ice for 20 min in 1 μM nicardipine hydrochloride (calcium channel blocker) to maximize smooth muscle relaxation, and then flat-opened. The mucosal layer was carefully removed using a glass cover slides, the flat tissue was fixed with 4% paraformaldehyde (PFA) overnight on ice, blocked in the blocking solution overnight at 4°C, and incubated for 2–3 days at 4°C using the following antibodies: Tuj1 (1:500), HuC/D (1:500), VIP (1:250), or EpCAM (1:200) in PBST. Tissue was washed for 24 h at 4 °C and incubated with secondary fluorescently labelled antibodies for 24 h at 4 °C. For lectin staining, the whole-mount tissues were incubated for 1 h on ice with Ulex europaeus aggluitinin-1 (UEA-1) conjugated to Rhodamine (Vector Laboratories, 1:500). Tissue was washed with PBST 3 times for 30–60 min each time. The nucleus was stained with DAPI. Images were acquired using a confocal laser scanning microscopy.
For immunofluorescence staining of organoids, organoids co-culture with or without enteric neuron cells were cultured in the Chamber Slide™ System (Thermo Scientific). The organoids were washed with ice-cold PBS 3 times and then fixed in 4% PFA for 30 min at room temperature (most of Maltrigel dissolved in this step). After washing 3 times with PBST, blocking solution was added to the organoids for 1 h. Samples were then incubated with the following primary antibodies: EpCAM (1:200), Beta-Tubulin III (1:1000), or UEA-1 (1:1000) with gentle shaking at 4°C, overnight. After repeated washing, DAPI (32670, Sigma-Aldrich) was added for 10 min to visualize the nucleus. Images were obtained using a confocal laser scanning microscopy.
Isolation of IECs and hepatic immune cells
The method used for isolation of IECs and LPLs has been reported previously(Deng et al., 2015). In brief, fat tissues and Peyer’s patches (PPs) were removed from intestine tissue. The intestine was open and cut in pieces 1-cm long and incubated in an HBSS solution containing 8 mM EDTA and 10 mM HEPPS for 30 min at 37°C with slow rotation (180 rpm). After passage through a 40-μm mesh filter, IECs were resuspended in Dulbecco’s Modified Eagle Medium (DMEM) containing 10% fetal calf serum (FCS). Livers were perfused with saline solution by way of the portal vein which was followed by enzymatic digestion. Liver immune cells were purified by centrifugation using a Percoll gradient. For flow cytometry analysis, the cells were labelled using standard procedures as described above.
Flow cytometry
Isolated intestinal epithelium cells (IECs) were stained with Ulex Europaeus Agglutinin 1-Rhodamine (UEA-1-Rho, 1:500), anti-CD45-FITC (1:500), anti-EpCAM-APC (1:1000). CD45− EpCAM +UEA-1+ cells were identified as fucosylated IECs (Fuc-IECs). For analysis of surface markers of hepatic immune cells, cells were stained in PBS containing 2% (wt/vol) BSA. The following antibodies were used at a dilution of 1/200–1/600: PE-, FITC- or APC-labeled anti-CD11b (M1/70), PE-, FITC- or APC-labeled anti-CD4 (RM4–5), PE-Cy7-labeled anti-CD3 (145-2C11), PE-anti-Gr-1 (RB6–8C5), PE- or FITC-labeled anti-mouse Ly6G (1A8), APC-conjugated CD45.2 (104). All antibodies were obtained from ThermoFisher unless otherwise noted. Flow cytometry data were acquired on a 5-color FACScan (Becton Dickinson) and analyzed using FlowJo software (Treestar).
Gene expression profiling by RNA-seq
IECs were isolated from the ileum of Sham or VGx mice as described above and total RNA was extracted for RNA-seq. A Poly(A) RNA sequencing library was prepared following Illumina’s TruSeq-stranded-mRNA sample preparation protocol. RNA integrity was checked with Agilent Technologies 2100 Bioanalyzer. Paired-ended sequencing was performed on Illumina’s NovaSeq 6000 sequencing system. Prior to assembly, reads containing sequencing adaptors, reads containing sequencing primers, and sequences with q quality score lower than 20 were removed.
The cleaned sequencing reads were aligned to the reference genome using the HISAT2 package. Multiple alignments were allowed for each read sequence (up to 20 by default), with a maximum of two mismatches allowed. HISAT2 also built a database of potential splice junctions. Aligned reads of individual samples were assembled using StringTie. Transcriptomes from all samples were then merged to reconstruct a comprehensive transcriptome using a proprietary Perl script of LC Sciences (Houston, Texas, U.S.A.). Following transcriptome reconstruction, FPKM reads were evaluated by StringTie, and differential expressed genes were evaluated by edgeR. The differentially expressed mRNAs and genes were selected with log2 (fold change) ≥1 or log2 (fold change) ≤−1, and with p values < 0.05.
Fecal DNA preparation and microbiota 16S-rDNA sequencing
For microbiota analyses of mice fed with the chow diet, fecal samples were collected 14 days after vagotomy or sham surgery was conducted. For microbiota analyses in mice fed the alcoholic diet, fecal samples were collected at the endpoint of the experiments. Total genomic DNA was purified using a QIAamp Stool DNA Extraction Kit following the manufacturer’s instructions. The V3-V4 region was amplified using primer 319F (ACTCCTACGGGAGGCAGCAG) and 806R (GGACTACHVGGGTWTCTAAT) and the amplified product was used to prepare the library for sequencing. The sequencing data were analyzed by qiime2 (Hall and Beiko, 2018) and lianchuan omics.
Enzyme Immunoassay (EIA) for VIP
The quantity of VIP was determined in intestinal tissue using EIA kits according to the manufacturer’s instructions. The sensitivity of the assays was <2.6 ng/ml. (Raybiotech, Cat: EIAM-VIP-1)
RNA extraction and RT-qPCR
For mammalian tissues, organoids and cells, total RNA was isolated using the RNeasy RNA isolation Kit (Qiagen). For bacteria cells, total RNA was extracted using the RiboPure™ RNA Purification Kit (Invitrogen). RNA (1μg) was reverse-transcribed with Superscript IV and random primers (Invitrogen). For quantitation of genes of interest, cDNA samples were amplified in applied biosystems Realtime System using SYBR Green Master Mix (Invitrogen) and specific primers (Supplemental table 1) according to the manufacturer’s instructions. Fold changes in mRNA expression between treatments and controls were determined by the ΔCT method. Results for each sample were normalized to the concentration of β-actin mRNA for intestine and liver tissues, vil1 mRNA for IECs, or rpoB mRNA for bacteria measured in the same samples. All primers were purchased from Eurofins MWG Operon or Sigma.
Gut microbiota quantification by RT-qPCR
Genomic DNA of fecal bacteria, small intestine derived bacteria or mucosa associated bacteria were extracted using a QIAamp Stool DNA Extraction Kit following the manufacturer’s instructions. Purified genomic DNA was dilute to 10 ng/μl using MilliQ water and was used as templates for RT-qPCR analysis according to method described above. Results for each sample were normalized to the concentration of total 16S rRNA by using a universal 16S rRNA primer pair.
Western blot analysis
Intestinal tissue or IECs were disrupted in RIPA lysis buffer with protease and phosphatase inhibitors (Roche) for 30 min on ice. The samples were centrifuged (16,000g, 10 min, 4°C) and the resulting supernatants transferred to fresh tubes. Protein lysates were quantitated using a Bio-Rad protein kit (Bio-Rad) and 50–100 μg of lysates were separated on 10% SDS polyacrylamide gels and transferred to a PVDF membrane (Bio-Rad). Appropriate primary antibodies and IRDye-labeled secondary antibodies were used to detect target proteins. The images were acquired with an Odyssey ® CLx imaging System (LI-COR).
Surface Plasmon Resonance (SPR)
SPR experiments were conducted on an OpenSPR™ (Nicoya, Lifesciences). NTA sensor chips were used for the assay following the manufacture’s instruction (Nicoya, Lifesciences). The sensograms were analyzed using TraceDrawer kinetic analysis software.
Quantification and Statistical Analysis
All statistical analyses were performed using GraphPad Prism 8 software. The data are presented as values with standard deviation (mean ± SEM). The significance was analyzed using t-tests for two-group analyses or ANOVA for multiple-group analyses. The significance is shown as p ≤ 0.05*, p ≤ 0.01**, p ≤ 0.001***, and p ≤ 0.0001****, p > 0.05 was considered not significant (ns).
Supplementary Material
Highlights.
Enteric neuron derived VIP shapes IECs fucosylation and gut microbiota composition
Neuropeptide VIP activates fut2 expression in IECs via the Erk1/2-C-fos pathway
α1,2-fucosidase enhances Bifidobacterial adhesion to fucosyl-conjugated mucus
Bifidobacteria restrict cytolytic E. faecalis growth and prevent ALD progression
Acknowledgments
We thank Dr. Takane Katayama for kindly providing B. longum 105-A strain and Dr. Satoru Fukiya for providing pBFS38 plasmid, and use of anaerobic chambers and germ-free facility in Functional Microbiomics Core in University of Louisville. This work was supported by grants from the NIH R21AA025724, R21AI128206, R21AI159194, R01 DK115406 and R01DK131442 (Z.D.). C.J. M and W.F. are supported by NIH P50 AA024337 and P20 GM113226. W.F. is supported by NIH R01AA023190. Y.T. is supported by NIH R01HL160927. We thank Dr. J. Ainsworth for editorial assistance.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Declaration of Interests
The authors disclose no conflicts of interest.
References
- Ammendola A, Gemini D, Iannaccone S, Argenzio F, Ciccone G, Ammendola E, Serio L, Ugolini G, and Bravaccio F (2000). Gender and peripheral neuropathy in chronic alcoholism: A clinical-electroneurographic study. Alcohol Alcoholism 35, 368–371. [DOI] [PubMed] [Google Scholar]
- Arike L, and Hansson GC (2016). The Densely O-Glycosylated MUC2 Mucin Protects the Intestine and Provides Food for the Commensal Bacteria. J Mol Biol 428, 3221–3229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arike L, Holmen-Larsson J, and Hansson GC (2017). Intestinal Muc2 mucin O-glycosylation is affected by microbiota and regulated by differential expression of glycosyltranferases. Glycobiology 27, 318–328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bai L, Mesgarzadeh S, Ramesh KS, Huey EL, Liu Y, Gray LA, Aitken TJ, Chen YM, Beutler LR, Ahn JS, et al. (2019). Genetic Identification of Vagal Sensory Neurons That Control Feeding. Cell 179, 1129-+. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bajaj JS (2019). Alcohol, liver disease and the gut microbiota. Nat Rev Gastroenterol Hepatol 16, 235–246. [DOI] [PubMed] [Google Scholar]
- Bode C, and Bode JC (2003). Effect of alcohol consumption on the gut. Best Pract Res Clin Gastroenterol 17, 575–592. [DOI] [PubMed] [Google Scholar]
- Bry L, Falk PG, Midtvedt T, and Gordon JI (1996). A model of host-microbial interactions in an open mammalian ecosystem. Science 273, 1380–1383. [DOI] [PubMed] [Google Scholar]
- Chopra K, and Tiwari V (2012). Alcoholic neuropathy: possible mechanisms and future treatment possibilities. Brit J Clin Pharmaco 73, 348–362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu S, Sun R, Gu X, Chen L, Liu M, Guo H, Ju S, Vatsalya V, Feng W, McClain CJ, et al. (2021). Inhibition of Sphingosine-1-Phosphate-Induced Th17 Cells Ameliorates Alcohol-Associated Steatohepatitis in Mice. Hepatology 73, 952–967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Currinn H, Guscott B, Balklava Z, Rothnie A, and Wassmer T (2016). APP controls the formation of PI(3,5)P-2 vesicles through its binding of the PIKfyve complex. Cell Mol Life Sci 73, 393–408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng Z, Mu J, Tseng M, Wattenberg B, Zhuang X, Egilmez NK, Wang Q, Zhang L, Norris J, Guo H, et al. (2015). Enterobacteria-secreted particles induce production of exosome-like S1P-containing particles by intestinal epithelium to drive Th17-mediated tumorigenesis. Nat Commun 6, 6956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dezfuli G, Gillis RA, Tatge JE, Duncan KR, Dretchen KL, Jackson PG, Verbalis JG, and Sahibzada N (2018). Subdiaphragmatic Vagotomy With Pyloroplasty Ameliorates the Obesity Caused by Genetic Deletion of the Melanocortin 4 Receptor in the Mouse. Front Neurosci 12, 104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Giovangiulio M, Bosmans G, Meroni E, Stakenborg N, Florens M, Farro G, Gomez-Pinilla PJ, Matteoli G, and Boeckxstaens GE (2016). Vagotomy affects the development of oral tolerance and increases susceptibility to develop colitis independently of the alpha-7 nicotinic receptor. Mol Med 22, 464–476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drokhlyansky E, Smillie CS, Van Wittenberghe N, Ericsson M, Griffin GK, Eraslan G, Dionne D, Cuoco MS, Goder-Reiser MN, Sharova T, et al. (2020). The Human and Mouse Enteric Nervous System at Single-Cell Resolution. Cell 182, 1606-+. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duan Y, Llorente C, Lang S, Brandl K, Chu HK, Jiang L, White RC, Clarke TH, Nguyen K, Torralba M, et al. (2019). Bacteriophage targeting of gut bacterium attenuates alcoholic liver disease. Nature 575, 505-+. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fanning S, Hall LJ, Cronin M, Zomer A, MacSharry J, Goulding D, Motherway MO, Shanahan F, Nally K, Dougan G, et al. (2012). Bifidobacterial surface-exopolysaccharide facilitates commensal-host interaction through immune modulation and pathogen protection. P Natl Acad Sci USA 109, 2108–2113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujimiya M, and Inui A (2000). Peptidergic regulation of gastrointestinal motility in rodents. Peptides 21, 1565–1582. [DOI] [PubMed] [Google Scholar]
- Gao B, and Bataller R (2011). Alcoholic Liver Disease: Pathogenesis and New Therapeutic Targets. Gastroenterology 141, 1572–1585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez-Rodriguez I, Sanchez B, Ruiz L, Turroni F, Ventura M, Ruas-Madiedo P, Gueimonde M, and Margolles A (2012). Role of Extracellular Transaldolase from Bifidobacterium bifidum in Mucin Adhesion and Aggregation. Appl Environ Microb 78, 3992–3998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goto Y, Obata T, Kunisawa J, Sato S, Ivanov II, Lamichhane A, Takeyama N, Kamioka M, Sakamoto M, Matsuki T, et al. (2014). Innate lymphoid cells regulate intestinal epithelial cell glycosylation. Science 345, 1254009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goto Y, Uematsu S, and Kiyono H (2016). Epithelial glycosylation in gut homeostasis and inflammation. Nat Immunol 17, 1244–1251. [DOI] [PubMed] [Google Scholar]
- Gu PL, Le Menuet D, Chung ACK, Mancini MA, Wheeler DA, and Cooney AJ (2005). Orphan nuclear receptor GCNF is required for the repression of pluripotency genes during retinoic acid-induced embryonic stem cell differentiation (vol 25, pg 8507, 2005). Mol Cell Biol 25, 10204–10204. [DOI] [PMC free article] [PubMed] [Google Scholar] [Research Misconduct Found]
- Gu XM, Sun R, Chen L, Chu SH, Doll MA, Li XH, Feng WK, Siskind L, McClain CJ, and Deng ZB (2021). Neutral Ceramidase Mediates Nonalcoholic Steatohepatitis by Regulating Monounsaturated Fatty Acids and Gut IgA(+) B Cells. Hepatology 73, 901–919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hakanson R, Vallgren S, Ekelund M, Rehfeld JF, and Sundler F (1984). The vagus exerts trophic control of the stomach in the rat. Gastroenterology 86, 28–32. [PubMed] [Google Scholar]
- Hall M, and Beiko RG (2018). 16S rRNA Gene Analysis with QIIME2. Methods Mol Biol 1849, 113–129. [DOI] [PubMed] [Google Scholar]
- Hall TA (1999). BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. In Nucleic acids symposium series ([London: ]: Information Retrieval Ltd., c1979-c2000.), pp. 95–98. [Google Scholar]
- Harmar AJ, Marston HM, Shen SB, Spratt C, West KM, Sheward WJ, Morrison CF, Dorin JR, Piggins HD, Reubi JC, et al. (2002). The VPAC(2) receptor is essential for circadian function in the mouse suprachiasmatic nuclei. Cell 109, 497–508. [DOI] [PubMed] [Google Scholar]
- He F, Ouwehand AC, Hashimoto H, Isolauri E, Benno Y, and Salminen S (2001). Adhesion of Bifidobacterium spp. to human intestinal mucus. Microbiol Immunol 45, 259–262. [DOI] [PubMed] [Google Scholar]
- Herath M, Hosie S, Bornstein JC, Franks AE, and Hill-Yardin EL (2020). The Role of the Gastrointestinal Mucus System in Intestinal Homeostasis: Implications for Neurological Disorders. Front Cell Infect Mi 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hooper LV, and Macpherson AJ (2010). Immune adaptations that maintain homeostasis with the intestinal microbiota. Nat Rev Immunol 10, 159–169. [DOI] [PubMed] [Google Scholar]
- Kashyap PC, Marcobal A, Ursell LK, Smits SA, Sonnenburg ED, Costello EK, Higginbottom SK, Domino SE, Holmes SP, Relman DA, et al. (2013). Genetically dictated change in host mucus carbohydrate landscape exerts a diet-dependent effect on the gut microbiota. Proc Natl Acad Sci U S A 110, 17059–17064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katayama T, Sakuma A, Kimura T, Makimura Y, Hiratake J, Sakata K, Yamanoi T, Kumagai H, and Yamamoto K (2004). Molecular cloning and characterization of Bifidobacterium bifidum 1,2-alpha-L-fucosidase (AfcA), a novel inverting glycosidase (glycoside hydrolase family 95). J Bacteriol 186, 4885–4893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kononova S, Litvinova E, Vakhitov T, Skalinskaya M, and Sitkin S (2021). Acceptive Immunity: The Role of Fucosylated Glycans in Human Host-Microbiome Interactions. Int J Mol Sci 22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunkel TA (1985). Rapid and Efficient Site-Specific Mutagenesis without Phenotypic Selection. P Natl Acad Sci USA 82, 488–492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lei C, Teng Y, He L, Sayed M, Mu J, Xu F, Zhang X, Kumar A, Sundaram K, Sriwastva MK, et al. (2021). Lemon exosome-like nanoparticles enhance stress survival of gut bacteria by RNase P-mediated specific tRNA decay. iScience 24, 102511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lelievre V, Favrais G, Abad C, Adle-Biassette H, Lu Y, Germano PM, Cheung-Lau G, Pisegna JR, Gressens P, Lawson G, et al. (2007). Gastrointestinal dysfunction in mice with a targeted mutation in the gene encoding vasoactive intestinal polypeptide: a model for the study of intestinal ileus and Hirschsprung’s disease. Peptides 28, 1688–1699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu V, Dietrich A, Kasparek MS, Benhaqi P, Schneider MR, Schemann M, Seeliger H, and Kreis ME (2015). Extrinsic intestinal denervation modulates tumor development in the small intestine of Apc(Min/+) mice. J Exp Clin Cancer Res 34, 39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Llorente C, Jepsen P, Inamine T, Wang LR, Bluemel S, Wang HJ, Loomba R, Bajaj JS, Schubert ML, Sikaroodi M, et al. (2017). Gastric acid suppression promotes alcoholic liver disease by inducing overgrowth of intestinal Enterococcus. Nature Communications 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macfarlane GT, and Gibson GR (1991). Formation of Glycoprotein Degrading Enzymes by Bacteroides-Fragilis. Fems Microbiol Lett 77, 289–293. [DOI] [PubMed] [Google Scholar]
- Moran AP, Gupta A, and Joshi L (2011). Sweet-talk: role of host glycosylation in bacterial pathogenesis of the gastrointestinal tract. Gut 60, 1412–1425. [DOI] [PubMed] [Google Scholar]
- Muller PA, Schneeberger M, Matheis F, Wang P, Kerner Z, Ilanges A, Pellegrino K, Del Marmol J, Castro TBR, Furuichi M, et al. (2020). Microbiota modulate sympathetic neurons via a gut-brain circuit. Nature 583, 441–446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mutlu EA, Gillevet PM, Rangwala H, Sikaroodi M, Naqvi A, Engen PA, Kwasny M, Lau CK, and Keshavarzian A (2012). Colonic microbiome is altered in alcoholism. Am J Physiol-Gastr L 302, G966–G978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakakuki T, Birtwistle MR, Saeki Y, Yumoto N, Ide K, Nagashima T, Brusch L, Ogunnaike BA, Okada-Hatakeyama M, and Kholodenko BN (2010). Ligand-Specific c-Fos Expression Emerges from the Spatiotemporal Control of ErbB Network Dynamics. Cell 141, 884–896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishiyama K, Yamamoto Y, Sugiyama M, Takaki T, Urashima T, Fukiya S, Yokota A, Okada N, and Mukai T (2017). Bifidobacterium bifidum Extracellular Sialidase Enhances Adhesion to the Mucosal Surface and Supports Carbohydrate Assimilation. mBio 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohtsubo K, and Marth JD (2006). Glycosylation in cellular mechanisms of health and disease. Cell 126, 855–867. [DOI] [PubMed] [Google Scholar]
- Page AJ (2021). Gastrointestinal Vagal Afferents and Food Intake: Relevance of Circadian Rhythms. Nutrients 13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pereira CF, Chang B, Qiu JJ, Niu XH, Papatsenko D, Hendry CE, Clark NR, Nomura-Kitabayashi A, Kovacic JC, Ma’ayan A, et al. (2013). Induction of a Hemogenic Program in Mouse Fibroblasts. Cell Stem Cell 13, 205–218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pham TA, Clare S, Goulding D, Arasteh JM, Stares MD, Browne HP, Keane JA, Page AJ, Kumasaka N, Kane L, et al. (2014). Epithelial IL-22RA1-mediated fucosylation promotes intestinal colonization resistance to an opportunistic pathogen. Cell Host Microbe 16, 504–516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pickard JM, and Chervonsky AV (2015). Intestinal Fucose as a Mediator of Host-Microbe Symbiosis. Journal of Immunology 194, 5588–5593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pickard JM, Maurice CF, Kinnebrew MA, Abt MC, Schenten D, Golovkina TV, Bogatyrev SR, Ismagilov RF, Pamer EG, Turnbaugh PJ, et al. (2014). Rapid fucosylation of intestinal epithelium sustains host-commensal symbiosis in sickness. Nature 514, 638–641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rose AK, Shaw SG, Prendergast MA, and Little HJ (2010). The importance of glucocorticoids in alcohol dependence and neurotoxicity. Alcohol Clin Exp Res 34, 2011–2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roth BL (2016). DREADDs for Neuroscientists. Neuron 89, 683–694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saijo K, Collier JG, Li AC, Katzenellenbogen JA, and Glass CK (2011). An ADIOL-ER beta-CtBP Transrepression Pathway Negatively Regulates Microglia-Mediated Inflammation. Cell 145, 584–595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakanaka M, Hansen ME, Gotoh A, Katoh T, Yoshida K, Odamaki T, Yachi H, Sugiyama Y, Kurihara S, Hirose J, et al. (2019). Evolutionary adaptation in fucosyllactose uptake systems supports bifidobacteria-infant symbiosis. Sci Adv 5, eaaw7696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakanaka M, Tamai S, Hirayama Y, Onodera A, Koguchi H, Kano Y, Yokota A, and Fukiya S (2014). Functional analysis of bifidobacterial promoters in Bifidobacterium longum and Escherichia coli using the alpha-galactosidase gene as a reporter. J Biosci Bioeng 118, 489–495. [DOI] [PubMed] [Google Scholar]
- Schwerdtfeger LA, and Tobet SA (2020). Vasoactive intestinal peptide regulates ileal goblet cell production in mice. Physiol Rep 8, e14363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seillet C, Luong K, Tellier J, Jacquelot N, Shen RD, Hickey P, Wimmer VC, Whitehead L, Rogers K, Smyth GK, et al. (2020). The neuropeptide VIP confers anticipatory mucosal immunity by regulating ILC3 activity. Nat Immunol 21, 168–177. [DOI] [PubMed] [Google Scholar]
- Suzuki K, Nishiyama K, Miyajima H, Osawa R, Yamamoto Y, and Mukai T (2016). Adhesion properties of a putative polymorphic fimbrial subunit protein from Bifidobacterium longum subsp. longum. Biosci Microbiota Food Health 35, 19–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szabo G (2015). Gut-Liver Axis in Alcoholic Liver Disease. Gastroenterology 148, 30–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Talbot J, Hahn P, Kroehling L, Nguyen H, Li DY, and Littman DR (2020). Feeding-dependent VIP neuron-ILC3 circuit regulates the intestinal barrier. Nature 579, 575-+. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terahara K, Nochi T, Yoshida M, Takahashi Y, Goto Y, Hatai H, Kurokawa S, Jang MH, Kweon MN, Domino SE, et al. (2011). Distinct fucosylation of M cells and epithelial cells by Fut1 and Fut2, respectively, in response to intestinal environmental stress. Biochem Biophys Res Commun 404, 822–828. [DOI] [PubMed] [Google Scholar]
- Teratani T, Mikami Y, Nakamoto N, Suzuki T, Harada Y, Okabayashi K, Hagihara Y, Taniki N, Kohno K, Shibata S, et al. (2020). The liver-brain-gut neural arc maintains the Treg cell niche in the gut. Nature 585, 591–596. [DOI] [PubMed] [Google Scholar]
- Turroni F, Serafini F, Foroni E, Duranti S, Motherway MO, Taverniti V, Mangifesta M, Milani C, Viappiani A, Roversi T, et al. (2013). Role of sortase-dependent pili of Bifidobacterium bifidum PRL2010 in modulating bacterium-host interactions. P Natl Acad Sci USA 110, 11151–11156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van den Abbeele P, Duysburgh C, Vazquez E, Chow J, Buck R, and Marzorati M (2019). 2 ’-Fucosyllactose alters the composition and activity of gut microbiota from formula-fed infants receiving complementary feeding in a validated intestinal model. J Funct Foods 61. [Google Scholar]
- Walsh KT, and Zemper AE (2019). The Enteric Nervous System for Epithelial Researchers: Basic Anatomy, Techniques, and Interactions With the Epithelium. Cell Mol Gastroenterol Hepatol 8, 369–378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, Foong JPP, Harris NL, and Bornstein JC (2021). Enteric neuroimmune interactions coordinate intestinal responses in health and disease. Mucosal Immunol. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L, Fouts DE, Starkel P, Hartmann P, Chen P, Llorente C, DePew J, Moncera K, Ho SB, Brenner DA, et al. (2016). Intestinal REG3 Lectins Protect against Alcoholic Steatohepatitis by Reducing Mucosa-Associated Microbiota and Preventing Bacterial Translocation. Cell Host Microbe 19, 227–239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ward T, Larson J, Meulemans J, Hillmann B, Lynch J, Sidiropoulos D, Spear JR, Caporaso G, Blekhman R, and Knight RJB (2017). BugBase predicts organism-level microbiome phenotypes. 133462.
- Zhang Y, and Hu W (2013). Mouse enteric neuronal cell culture. Methods Mol Biol 1078, 55–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao CM, Hayakawa Y, Kodama Y, Muthupalani S, Westphalen CB, Andersen GT, Flatberg A, Johannessen H, Friedman RA, Renz BW, et al. (2014). Denervation suppresses gastric tumorigenesis. Sci Transl Med 6, 250ra115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou R, Llorente C, Cao J, Gao B, Duan Y, Jiang L, Wang Y, Kumar V, Starkel P, Bode L, et al. (2020). Deficiency of Intestinal alpha1-2-Fucosylation Exacerbates Ethanol-Induced Liver Disease in Mice. Alcohol Clin Exp Res 44, 1842–1851. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Next generation 16s rRNA sequencing and poly(A) RNA sequencing data have been deposited at Sequence Read Archive and are publicly available as of the date of publication. Accession numbers are listed in the key resources table. All data reported in this paper will be shared by the lead contact upon request.
This paper does not report any original code
Any additional information required to reanalyze the data reporting in this paper is available from the lead author on request.
KEY RESOURCE TABLE
| Reagent or resource | Source | Identifier |
|---|---|---|
| Antibodies | ||
| Rabbit VIP | Abcam | Cat# ab22736; RRID:AB_447294 |
| β-tubulin3 Alexa Fluor 488 | Thermo Fisher Scientific | Cat# 53-4510-82; RRID:AB_2574421 |
| Hu C/D Alexa Fluor 647 | Abcam | Cat# ab237235 |
| EpCAM-APC | Invitrogen | Cat# 17-5791-82; RRID:AB_2716944 |
| Rabbit Erk1/2 | Cell Signaling Technology | Cat# 4695S |
| Rabbit pErk1/2 | Cell Signaling Technology | Cat# 4370S |
| Rabbit C-fos | Cell Signaling Technology | Cat# 2250S |
| Mouse Muc2 | Abcam | Cat# ab11197; RRID:AB_297837 |
| Rabbit Muc2 | Abcam | Cat# ab272692; RRID:AB_2888616 |
| Mouse Anti-6x His | Thermo Fisher Scientific | Cat# MA1-21315; RRID:AB_557403 |
| Gr-1-PE | Invitrogen | Cat# 12-5931-83; RRID:AB_466046 |
| CD11b-APC | Invitrogen | Cat# 17-0012-83 |
| Goat anti-Rabbit Alexa Fluor 594 | Thermo Fisher Scientific | Cat# A32740; RRID:AB_2762824 |
| Goat anti-Rabbit Alexa Fluor 680 | Thermo Fisher Scientific | Cat# A32734; AB_2633283 |
| Goat anti-Mouse Alexa Fluor 488 | Thermo Fisher Scientific | Cat# A32723; RRID:AB_2633275 |
| IRDye 800CW Goat anti-Mouse | LI-COR Biosciences | Cat# 926-32210; RRID:AB_621842 |
| IRDye 800CW Goat anti-Rabbit | LI-COR Biosciences | Cat# 926-32211; RRID:AB_621843 |
| IRDye® 800CW Streptavidin | LI-COR Biosciences | Cat# 926-32230 |
| UEA-1 Rhodamine | Vector Laboratories | Cat# RL-1062; RRID:AB_2336769 |
| UEA-1 Biotin | Vector Laboratories | Cat# B-1065; RRID:AB_2336766 |
| UEA-1 Unconjugated | Vector Laboratories | Cat# L-1060; RRID:AB_2336768 |
| Bacterial strains | ||
| Bifidobacterium bifidum | ATCC | Cat# 29521 |
| Bifidobacterium longum subsp. longum 105A | (Sakanaka et al., 2019) | N/A |
| Enterococcus faecalis | ATCC | Cat# 29212 |
| E. coli Top10 | Invitrogen | Cat# C404006 |
| E. coli BL21 (DE3) | Invitrogen | Cat# C600003 |
| Chemicals, peptides, and recombinant proteins | ||
| Difco™ Lactobacilli MRS Broth | Becton Dickinson | Cat# 288130 |
| BBL™ Brain Heart Infusion | Becton Dickinson | Cat# 221778 |
| BBL™ Enterococcosel™ Broth | Becton Dickinson | Cat# 212207 |
| LB Broth (Miller) | Sigma | Cat# L3522-250G |
| RNAprotect Bacteria Reagent | Qiagen | Cat#76506 |
| Ni-NTA Agarose | Thermo Fisher Scientific | Cat# R90101 |
| Dynabeads™ M-270 Streptavidin | Invitrogen | Cat# 65305 |
| SuperScript™ IV First-Strand Synthesis System | Invitrogen | Cat# 18091050 |
| RNase-free DNase І | New England Biolabs | Cat# M0303S |
| Kanamycin | Sigma | Cat# K1876 |
| Chloramphenicol | Sigma | Cat# C0378 |
| Erythromycin | Sigma | Cat# E5389 |
| Ampicillin | Sigma | Cat# A9393 |
| 2’-Fucosyllactose (2’-FL) | Sigma | Cat# 41263-94-9 |
| VIP | Tocris Bioscience | Cat# 1911 |
| [D-p-Cl-Phe6,Leu17]-VIP (VIPx) | Tocris Bioscience | Cat# 3054 |
| Clozapine N Oxide (CNO) | Tocris Bioscience | Cat# 4936 |
| 4-methylumbelliferyl-alpha-l-fucopyranoside (4-MUF) | GoldBio | Cat# M-580-50 |
| FR180204 | Cayman Chemical | Cat# 15544 |
| Oil Red O | Sigma | Cat# O0625 |
| Gill™ Hematoxylin | Thermo Fisher Scientific | Cat# 6765007 |
| Shandon™ Eosin-Y | Thermo Fisher Scientific | Cat# 6766007 |
| B-27 Supplement | Gibco | Cat# 17504044 |
| N-2 Supplement | Gibco | Cat# 17502001 |
| Complete™ Protease Inhibitor Cocktail | Sigma | Cat# 11697498001 |
| Phosphatase Inhibitor Cocktail | Sigma | Cat# P2850 |
| Q5® Hot Start HiFi PCR Master Mix | New England Biolabs | Cat# M0543S |
| Critical Commercial Assays | ||
| RNeasy Mini Kit | Qiagen | Cat# 74104 |
| Nunc™ Lab-Tek™ II Chamber Slide™ System | Thermo Scientific | Cat# 154453 |
| RiboPure RNA Purification Kit, bacteria | Invitrogen | Cat# AM1925 |
| PowerUP™ SYBR™ Green Master Mix | Qiagen | Cat# 204143 |
| HiScribe T7 Quick High Yield RNA Synthesis Kit | New England Biolabs | Cat# E2050S |
| QIAamp@ Fast DNA Stool Mini Kit | Qiagen | Cat# 51604 |
| IntestiCult™ Organoid Growth Medium | Stem Cell Biotechnology | Cat# 06005 |
| Luciferase Reporter Gene Detection Kit | Sigma | Cat# LUC1-1KT |
| NEBuilder® HiFi DNA Assembly Master Mix | New England Biolabs | Cat# E2621S |
| Deposited data | ||
| Next generation 16s rRNA sequencing | Sequence Read Archive | PRJNA870429; SRR21103243-SRR21103252 |
| Next generation Poly(A) RNA sequencing | Sequence Read Archive | PRJNA870429; SRR21102741-SRR21102746 |
| Experimental Models: Organisms/Strains Mouse | ||
| C57BL/6 | Jackson Laboratory Stock | Cat# 000664 |
| VIPR1−/− | Jackson Laboratory Stock | Cat# 034744 |
| Fut2−/− | Jackson Laboratory Stock | Cat# 006262 |
| hM3Dq fl-stop-fl | Jackson Laboratory Stock | Cat# 026220 |
| hM4Di fl-stop-fl | Jackson Laboratory Stock | Cat# 026219 |
| Vip IRES-cre | Jackson Laboratory Stock | Cat# 031628 |
| oligos | ||
| Primers see table S1 | Sigma | N/A |
| Recombinant DNA | ||
| pET28b-MBP-TEV | (Currinn et al., 2016) | RRID: Addgene_69929 |
| pET28b-AfcA | This paper | N/A |
| pET28b-UK | This paper | N/A |
| pET28b-FUC | This paper | N/A |
| pBFS38 | (Sakanaka et al., 2014) | N/A |
| pBFS38-AfcA | This paper | N/A |
| pBFS38-Erm | This paper | N/A |
| pBFS38-UK | This paper | N/A |
| p16337 | (Gu et al., 2005) | RRID: Addgene_16337 |
| p16337-fut2p-Naive | This paper | N/A |
| p16337-fut2p-Mutant | This paper | N/A |
| pFUW-tetO-fos | (Pereira et al., 2013) | RRID: Addgene_131593 |
| Software and algorithms | ||
| Prism (GraphPad Software) | GraphPad | https://www.graphpad.com |
| Bugbase | (Ward et al., 2017) | https://bugbase.cs.umn.edu/ |
| Qiime2 | (Hall and Beiko, 2018) | https://qiime2.org/ |
| Bioedit | (Hall, 1999) | https://www.bioedit.com/ |
| SnapGene | SnapGene | https://www.snapgene.com/ |


