Abstract
Background
Existing risk scores appear insufficient to assess the individual survival risk of patients with advanced pancreatic ductal adenocarcinoma (PDAC) and do not take advantage of the variety of parameters that are collected during clinical care.
Methods
In this retrospective study, we built a random survival forest model from clinical data of 203 patients with advanced PDAC. The parameters were assessed before initiation of systemic treatment and included age, CA19-9, C-reactive protein, metastatic status, neutrophil-to-lymphocyte ratio and total serum protein level. Separate models including imaging and molecular parameters were built for subgroups.
Results
Over the entire cohort, a model based on clinical parameters achieved a c-index of 0.71. Our approach outperformed the American Joint Committee on Cancer (AJCC) staging system and the modified Glasgow Prognostic Score (mGPS) in the identification of high- and low-risk subgroups. Inclusion of the KRAS p.G12D mutational status could further improve the prediction, whereas radiomics data of the primary tumor only showed little benefit. In an external validation cohort of PDAC patients with liver metastases, our model achieved a c-index of 0.67 (mGPS: 0.59).
Conclusions
The combination of multimodal data and machine-learning algorithms holds potential for personalized prognostication in advanced PDAC already at diagnosis.
Key words: pancreatic cancer, machine learning, genetics, computed tomography, prognosis, survival analysis
Highlights
-
•
We developed a machine-learning-based prediction model that outperforms the AJCC staging system and mGPS.
-
•
Applying our model to an external validation cohort demonstrates generalizability.
-
•
Explainable machine learning enables to understand the decision making of our model and identifies relevant parameters.
-
•
Combining clinical, imaging and genetic data holds potential for personalized prognostication in advanced PDAC.
Introduction
Pancreatic ductal adenocarcinoma (PDAC) is a disease with a very poor prognosis. Due to few symptoms of early-stage tumors, >80% of the patients are diagnosed in an unresectable stage including ∼50% diagnosed with metastatic disease.1 With 90%, PDAC represents the most common subtype of pancreatic cancer.2 While PDAC is projected to be soon the third leading cause of cancer death in the European Union, only little progress has been made in developing individual treatment strategies. The current standard of care involves combination chemotherapy regimens that were able to significantly prolong survival in some patients with the downside of potentially severe side effects.3,4 To allow for personalized treatment and identify patients who will likely benefit from enrollment into clinical trials, it is essential to assess the individual prognosis of patients at the time of diagnosis. Until now, the most widely used tool is the eighth edition of the American Joint Committee on Cancer (AJCC) Staging Manual.5 Given that most of the patients are classified as stage III or IV at initial diagnosis, this system does not allow for an appropriately granular risk stratification. The Glasgow Prognostic Score (GPS) and a modified variant (mGPS) are validated scores to predict the patient survival risk based on the inflammatory and nutritional status in unresectable pancreatic cancer and additional cancer entities.6,7 Nevertheless, it seems likely that the two included parameters, C-reactive protein (CRP) and albumin, are not sufficient to reflect the individual disease biology. Several nomograms have been proposed to predict the survival of patients with advanced pancreatic cancer to address this issue.8, 9, 10, 11, 12, 13, 14, 15 However, these studies often only included limited clinical information, whereas the integration of multimodal data such as clinical and imaging data was shown to improve the survival prediction in resectable pancreatic cancer as well as in unresectable patients undergoing radiotherapy.16, 17, 18, 19, 20, 21, 22 This is becoming increasingly relevant as novel methods in automatic image segmentation provide the opportunity to exploit in-depth information from standard-of-care radiological imaging.23 Also, existing Cox proportional hazard models assume a linear combination between covariables. As machine-learning algorithms become more accessible, methods like random survival forest are proposed to model more complex interactions in survival risk prediction.24,25
Due to the absence of suitable tools which include sufficient clinical parameters, the decision on further treatment currently is largely based on the experience of the treating physician. Consequently, this study aims to develop a model for survival risk prediction in patients with unresectable PDAC at the time of diagnosis by combining multimodal clinical data for training random survival forests.
Methods
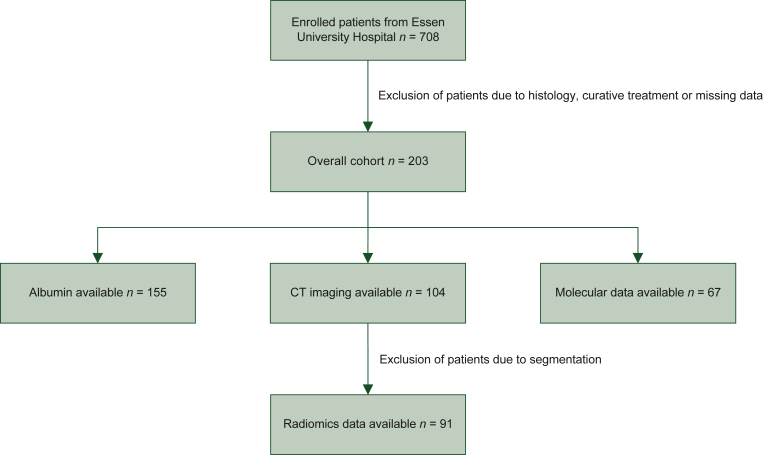
In this retrospective study, 708 patients with pancreatic cancer treated at University Hospital Essen were enrolled. After exclusion of patients due to tumor histology, curative treatment or missing data, 203 patients were included in the final analysis (Figure 1). All patients were treated between 2011 and 2021, had a histologically confirmed diagnosis of PDAC and were not eligible for curatively intended tumor resection. The external validation cohort consisted of 22 PDAC patients with liver metastases who were palliatively treated between 2014 and 2021 at the University Hospital Hamburg-Eppendorf, Germany. All data were collected with a maximum interval of 2 months to the date of initial diagnosis, and before the start of systemic treatment. When necessary, missing values were substituted with data from the next available record before treatment. We excluded patients for whom no suitable data were available. The overall survival was defined as the period from the documented date of initial diagnosis to the date of death from any cause. Patients for whom no date of death was available were censored at the time of the last follow-up. Seven clinical parameters were selected for a baseline model based on a Cox proportional hazards model and prognostic relevance previously described in the literature: age at diagnosis, metastatic status (no metastases, liver metastases), CRP, neutrophil-to-lymphocyte ratio (NLR), CA19-9 level and total serum protein level. For a total of 91 patients of the Essen cohort, a computed tomography (CT) image was available before the start of systemic treatment and the primary tumor could be automatically segmented using the pretrained nnU-Net architecture.23 Using an internal feature extraction pipeline including PyRadiomics 3.0.1, 1688 image features were extracted.26 From these, three features were selected using permutation-based feature importance and the Boruta package.27,28 For 67 patients of the Essen cohort, data from genomic sequencing of macrodissected tissue samples were available before the start of systemic treatment. Tissues were collected either from the primary cancer site or metastasis. Somatic mutations were confirmed by next-generation sequencing of a focused gene panel. For survival risk prediction, a random survival forest model was built using the clinical parameters either alone or in combination with radiomic or molecular features in the respective subgroups. Our prediction model was compared against the AJCC eighth edition staging system as well as the mGPS. Predictive performance was assessed by calculating the average concordance-index (c-index) from fivefold cross-validation with a nested grid search for hyperparameters. The 10% of patients with the highest and lowest risk scores predicted by our model were defined as high- and low-risk groups, respectively. To compare the survival between subgroups, we created Kaplan–Meier survival curves and calculated the restricted mean survival time difference (ΔRMST). The given ΔRMST is the average of a 10 × 2 cross-validation with nested grid search for hyperparameters. Due to the limited number, stage II patients were excluded from the test set in the calculation of the ΔRMST. To analyze the impact of first-line chemotherapy regimen on overall survival, we built two balanced treatment groups using propensity score matching. We used logistic regression and a nearest neighbor algorithm to match patients receiving 5-fluorouracil (5-FU) and gemcitabine-based combination chemotherapies. Model explainability was achieved using SHapley Additive exPlanations (SHAP).29 Statistical significance between models was assessed using a two-tailed Student's t-test and P values <0.05 were considered statistically significant. All statistical analyses were carried out using Python 3.8 and the packages scikit-survival, lifelines, scipy and psmpy.27,30, 31, 32
Figure 1.
Flowchart depicting the process of enrollment and subgroup formation in the internal cohort.
CT, computed tomography.
Results
Patient characteristics
The internal cohort included 203 patients treated at University Hospital Essen. The patient characteristics are summarized in Table 1. The mean age was 64.2 years. The median survival time (MST) was 6.7 months [95% confidence interval (CI) 5.8-8.6 months; see Supplementary Figure S1, available at https://doi.org/10.1016/j.esmoop.2022.100555]. At the time of analysis, 186 patients had died and 17 (8.4%) patients were censored. At the date of initial diagnosis, 7 (3.4%) patients were in stage II, 28 (13.8%) in stage III and 168 (82.8%) patients were in stage IV. In total, 131 (64.5%) patients presented with synchronous liver metastases. One hundred and eighty patients (88.7%) had received systemic treatment at University Hospital Essen, and 23 (11.3%) patients received best supportive care or palliative treatment at a different facility. The external validation cohort consisted of 22 patients with a mean age of 61.4 years and an MST of 8.4 months. All patients in the external cohort were diagnosed at stage IV with liver metastases and treated at University Hospital Hamburg-Eppendorf.
Table 1.
Characteristics of the internal and external cohorts with unresectable PDAC
| Internal cohort (n = 203) | External validation cohort (n = 22) | |
|---|---|---|
| Age, years | ||
| Mean | 64.2 | 61.4 |
| Range | 30-90 | 38-78 |
| Sex, n (%) | ||
| Male | 111 (54.7) | 14 (63.6) |
| Female | 92 (45.3) | 8 (36.4) |
| AJCC stage, n (%) | ||
| II | 7 (3.4) | 0 (0) |
| III | 28 (13.8) | 0 (0) |
| IV | 168 (82.8) | 22 (100) |
| Liver metastasis, n (%) | ||
| Yes | 131 (64.5) | 22 (100) |
| No | 72 (35.5) | 0 (0) |
| Chemotherapy, n (%) | ||
| FOLFIRINOX | 59 (29.1) | 14 (63.6) |
| FOLFOXIRI | 26 (12.8) | 0 (0) |
| FOLFOX | 14 (6.9) | 1 (4.5) |
| Gemcitabine + nab-paclitaxel | 41 (20.2) | 5 (22.7) |
| Gemcitabine | 20 (9.9) | 1 (4.5) |
| Gemcitabine + oxaliplatin | 13 (6.4) | 0 (0) |
| Other | 7 (3.4) | 1 (4.5) |
| None at our institute | 23 (11.3) | 0 (0) |
| Median survival time, months (95% CI) | 6.7 (5.8-8.6) | 8.4 (3.8-10.2) |
| Censored, n (%) | 17 (8.4) | 0 (0) |
AJCC, American Joint Committee on Cancer; CI, confidence interval; PDAC, pancreatic ductal adenocarcinoma.
Selected clinical parameters for a baseline model
To select parameters for our baseline model, we carried out univariate and multivariate analyses on the Essen cohort using a Cox proportional hazards model (Table 2). The univariate Cox proportional hazards model identified the age at initial diagnosis, CRP, the metastatic status, which is composed of no metastases (M0) and liver metastases [M1 (HEP)], NLR and total serum protein level to be significantly associated with the overall survival. Together with CA19-9, these parameters were used to build a baseline model. A detailed analysis is provided in Supplementary Table S1, available at https://doi.org/10.1016/j.esmoop.2022.100555.
Table 2.
Univariate and multivariate analysis of the parameters included in the baseline model
| Univariate analysis |
Multivariate analysis |
|||
|---|---|---|---|---|
| HR (95% CI) | P value | HR (95% CI) | P value | |
| Age | 1.03 (1.01-1.04) | <0.005 | 1.03 (1.02-1.05) | <0.005 |
| CA19-9 | 1.00 (1.00-1.00) | 0.065 | 1.00 (1.00-1.00) | 0.201 |
| CRP | 1.08 (1.06-1.11) | <0.005 | 1.08 (1.05-1.11) | <0.005 |
| M0 | 0.53 (0.35-0.81) | <0.005 | 0.56 (0.33-0.95) | 0.030 |
| M1 (HEP) | 1.79 (1.31-2.44) | <0.005 | 1.26 (0.86-1.87) | 0.236 |
| NLR | 1.11 (1.08-1.51) | <0.005 | 1.06 (1.03-1.10) | <0.005 |
| Total protein | 0.80 (0.66-0.97) | 0.024 | 0.78 (0.63-0.97) | 0.024 |
CI, confidence interval; CRP, C-reactive protein; HR, hazard ratio; M0, no metastasis; M1 (HEP), liver metastasis; NLR, neutrophil-to-lymphocyte ratio.
Internal validation of the baseline model
Over the entire Essen cohort (n = 203), a survival model based on the seven selected clinical parameters achieved a c-index of 0.71 (95% CI 0.64-0.79) which was significantly higher than survival risk prediction based on the AJCC system (0.57, 95% CI 0.52-0.62, P < 0.006). In a subgroup of patients for whom albumin levels were available (n = 155), we compared our prediction model with the established mGPS. In this subgroup our proposed model achieved a c-index of 0.72 (95% CI 0.68-0.76) and performed significantly better than the mGPS (0.63, 95% CI 0.59-0.66, P < 0.005).
Identification of high- and low-risk subgroups
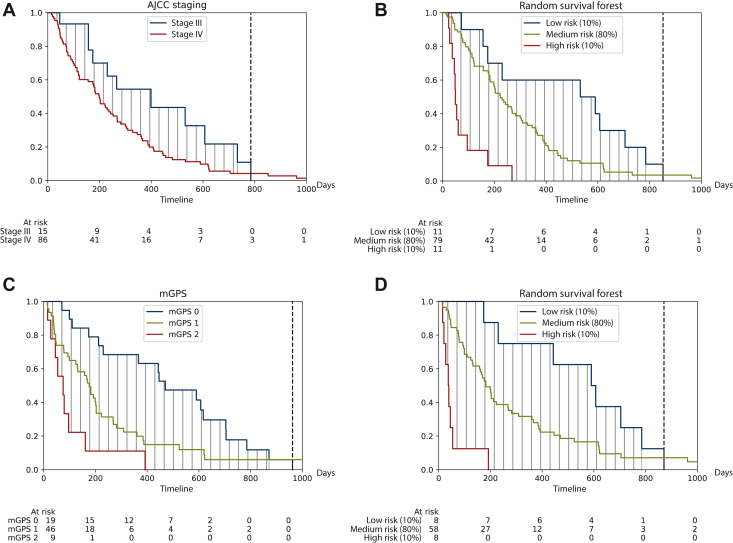
To describe the survival difference between different risk groups, we calculated the ΔRMST which represents the difference between the areas under two Kaplan–Meier survival curves. We compared the high- and low-risk groups identified by our model with subgroups identified by AJCC staging (stage III versus IV, n = 203) or mGPS (score 0 versus 2, n = 155). Calculating the average of a 10 × 2 cross-validation, we could achieve significantly higher survival differences calculated by ΔRMST between the identified subgroups using our model compared to AJCC staging (366, 95% CI 342-390 versus 190, 95% CI 150-230, P < 0.005) or mGPS (434, 95% CI 397-471 versus 241, 95% CI 206-276, P < 0.005). Figure 2 shows representative folds of the identified subgroups.
Figure 2.
Kaplan–Meier survival curves showing: (A) high- and low-risk subgroups identified by the AJCC staging system, our baseline model (B, D), or the mGPS (C). In the overall internal cohort (n = 203), the AJCC staging system (A) was compared to our baseline model (B). In a subgroup where albumin was available (n = 155) the mGPS (C) was compared to our baseline model (D). Experiments show representatives of a 10 × 2 cross-validation.
AJCC, American Joint Committee on Cancer; mGPS, modified Glasgow Prognostic Score.
Model explainability
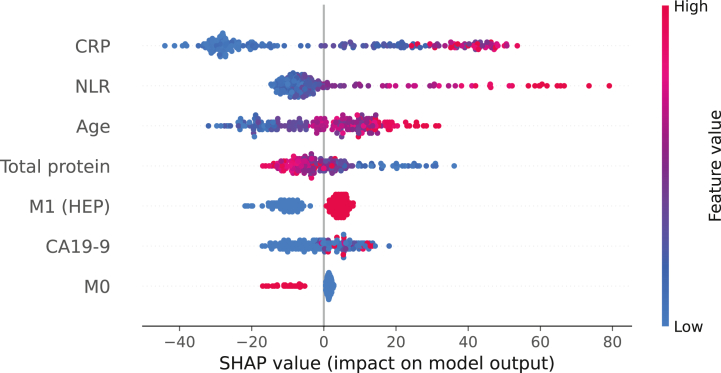
Using SHAP we could investigate the importance of different parameters for the decision making of our model. Figure 3 shows the feature importance of our baseline model in the overall internal cohort (n = 203). It could be shown that a higher value for the features CRP, NLR, age and CA19-9 as well as the presence of liver metastases are associated with higher SHAP values and are therefore predictive of a shorter survival time. A higher value in total serum protein level and an M0 status are associated with lower SHAP values predicting a longer survival. Overall, the inflammatory markers CRP and NLR have the greatest influence on the decision making of our model.
Figure 3.
Explainability of the baseline model with SHAP in the overall internal cohort. Each dot represents one patient. The parameters are ranked from top to bottom according to their impact on the model output. The color gradient represents the feature value while the x-axis shows the respective SHAP values. Positive or negative SHAP values indicate the responsibility of parameters for a longer or shorter predicted overall survival, respectively.
CRP, C-reactive protein; M1 (HEP), liver metastasis; NLR, neutrophil-to-lymphocyte ratio; SHAP, SHapley Additive exPlanations.
Influence of first-line chemotherapy regimen
In our internal cohort we investigated the influence of 5-FU (n = 99) or gemcitabine-based (n = 54) first-line combination therapy and saw no difference in overall survival using Kaplan–Meier survival curves (P = 0.318, see Supplementary Figure S2, available at https://doi.org/10.1016/j.esmoop.2022.100555). Also, when comparing patients with low and high risk according to our model, we found no difference in overall survival depending on chemotherapy regimen (Supplementary Figure S3, available at https://doi.org/10.1016/j.esmoop.2022.100555). We used propensity score matching to create two balanced treatment cohorts (n = 54) and found that age was a significant predictor only in the 5-FU cohort (hazard ratio 1.04, 95% CI 1.00-1.08, P = 0.036 versus hazard ratio 1.02, 95% CI 0.99-1.05, P = 0.215) and total serum protein only in the gemcitabine cohort (0.52, 95% CI 0.32-0.85, P = 0.009 versus 1.02, 95% CI 0.62-1.69, P = 0.92). This relationship was also evident in a SHAP analysis of prediction models trained on each cohort (Supplementary Figure S4, available at https://doi.org/10.1016/j.esmoop.2022.100555).
External validation of the baseline model
We validated the results of our clinical prediction model on an external cohort of 22 palliatively treated PDAC patients with synchronous liver metastases. The patient characteristics are described in Table 1. The baseline model trained on our internal cohort achieved a c-index of 0.67 compared to a c-index of 0.59 by mGPS.
Inclusion of imaging data
In a subgroup of 91 patients of the internal cohort, imaging data were available, and three radiomics features from the primary tumor region were selected (first-order skewness, first-order median, small-area low-gray-level emphasis). Multivariate analysis revealed that tumor skewness and median gray-level intensity were independently associated with overall survival (Supplementary Table S2, available at https://doi.org/10.1016/j.esmoop.2022.100555). To analyze the impact of radiomics features on survival prediction, we built a separate model which was based on the seven clinical variables together with the three selected radiomics features. By the addition of radiomics features, the survival prediction in our internal cohort (0.73, 95% CI 0.61-0.84) could not be significantly improved as compared to the baseline model excluding radiomic features (0.70, 95% CI 0.61-0.78, P = 0.2) in this subgroup.
Inclusion of molecular data
In a subgroup of 67 patients of the internal cohort, genomics data were available. In 53 patients (79.1%), a KRAS mutation was detected. As expected, the KRAS p.G12D mutation was the most frequent subtype (24, 45.3%), followed by p.G12V (20, 37.7%), p.G12R (4, 7.5%) and p.G12C (2, 3.8%). The KRAS p.G12D mutation was an independent predictor of prognosis (hazard ratio 2.79, 95% CI 1.49-5.22, P < 0.005; see Supplementary Table S3, available at https://doi.org/10.1016/j.esmoop.2022.100555). The addition of the KRAS p.G12D mutation status to the clinical parameters could significantly improve survival risk prediction in this subgroup (0.76, 95% CI 0.65-0.86 versus 0.73, 95% CI 0.64-0.83, P < 0.015).
Discussion
To allow for optimal treatment planning in patients with pancreatic ductal adenocarcinoma, it is important to assess the patient’s individual prognosis at the time of diagnosis. We demonstrated that survival risk prediction based on a machine-learning approach incorporating routine clinical parameters achieved a significantly better risk stratification as compared to the AJCC staging system or the mGPS. This enabled us to identify high- and low-risk patients who have substantially different survival outcomes under palliative systemic therapy. We externally validated our risk prediction model in an independent cohort confirming the reproducibility of our findings. In the internal patient cohort, we could further improve risk prediction by the inclusion of the KRAS p.G12D mutational status. Further, we observed a trend toward better risk prediction by the inclusion of imaging data derived from the primary tumors. We suggest our model as an alternative to existing risk stratification tools in care planning of advanced pancreatic cancer patients. In addition, our risk predictor may improve patient stratification in studies of systemic treatment of PDAC.
In contrast to previous prediction tools in unresectable PDAC, our study incorporates a comprehensive set of parameters including clinical, imaging and molecular data and reveals the decision making of our model by using an explainable machine-learning approach.8, 9, 10, 11, 12, 13, 14, 15 The combination of multimodal data provides a more precise characterization of the individual patients and has already shown promising results in resectable pancreatic cancer, although not following a machine-learning approach.16 In contrast to these studies, we used random survival forests, which enables our model to predict non-linear relationships. The parameters used for our baseline model were largely reported in the literature to be associated with survival of patients with solid tumors but, to our knowledge, they have not been used in this combination as risk predictors in advanced pancreatic cancer.11, 12, 13,33,34 The role of inflammation in cancer evolution and progression is well known, and previous studies have described the importance of the inflammatory markers CRP and NLR in advanced pancreatic cancer.35, 36, 37, 38 Through SHAP, we were able to conclude that our model also considers these two parameters to be crucial for the further course of therapy.
Albumin serves as a ‘negative’ acute-phase protein and indicator of nutrient status of the patient. It could show its potential in survival risk prediction as part of the mGPS.6 More than 80% of PDAC patients are affected by cancer cachexia, which leads to hypoalbuminemia.39 In our study, we have chosen total serum protein level instead of albumin because of the higher availability in our dataset and could show that it was an independent predictor of survival risk. CA19-9 is an established tumor marker in pancreatic cancer and was also proven as an independent risk predictor at baseline in patients with unresectable PDAC.40,41
We also included the metastatic status in our model, but in contrast to other studies we specified the metastatic localization by using the presence of liver metastases as a separate parameter. This allowed our model to include three different metastatic scenarios (M0, M1 with liver metastases, M1 other than liver). Oweira et al. have previously indicated the relevance of metastasis site on overall survival.42 By using an explainable approach, we could infer that our model uses the additional information in a reasonable way and in line with the literature, even though M0 is the least relevant parameter for the model’s decision making.
The reproducibility of classification scores must be a major concern in order to generalize the results to newly presenting patients. While nested cross-validation is a strong method toward this end, most studies have not validated their scores on an external patient cohort. Our results were validated on a cohort of 22 patients with synchronous liver metastases treated at a different hospital, showing that our model generalizes well to patients that are potentially submitted to different treatments. As nearly 50% of PDAC patients are already metastasized at diagnosis and thus classified as stage IV, further risk stratification by the AJCC staging system is not possible in clinical practice. Although the external patient cohort was different from the internal cohort, our model generalized well and outperformed the mGPS.
The addition of the KRAS p.G12D mutation status in our internal cohort further improved the predictive power of our model. KRAS mutations, which are observed in up to 90% of PDAC patients, have been extensively studied, and particularly the KRAS p.G12D mutation was previously identified to be a negative predictor of survival in patients with unresectable PDAC.43, 44, 45, 46 It was shown that this subtype plays a critical role in tumor metabolism and maintenance.47 As the abundance of KRAS mutations in our study is in line with previous studies, this strengthens our results of the KRAS p.G12D mutation being an independent predictor and supports our approach to include cancer genomics in a prediction model.
While several studies could show an association between radiomics features of the primary tumor and overall survival in patients with resectable PDAC,17,18,20, 21, 22,48, 49, 50, 51, 52, 53 only few studies have investigated this relationship for patients with unresectable tumors.19,54, 55, 56 These studies suggested among others tumor skewness as a predictor, which we also included in our model.54,56 Using SHAP we could conclude that while tumor skewness has a particularly strong influence on our model, the small-area low-gray-level emphasis does not provide much benefit (Supplementary Figure S5, available at https://doi.org/10.1016/j.esmoop.2022.100555). Our results suggest a trend toward a benefit from the addition of radiomics features, which, however, did not significantly enhance risk prediction based on our baseline model. In this study, we exclusively investigated the impact of radiomics features from the primary tumor region. Other studies in pancreatic as well as colorectal cancer have shown that imaging information from liver metastases allow survival prediction and improve the prediction based on clinical parameters alone.57,58 The integration of imaging information on liver and other metastases could potentially further improve the survival prediction and should be investigated in further studies.
Patient management, including the selection of first-line therapy, is a significant challenge in the treatment of advanced pancreatic cancer. A prediction model for chemotherapy selection is beyond the scope of this study and requires prospective randomized data collection, which we plan to conduct in the future. Based on the current results, we conclude that overall survival is not markedly affected by treatment choice. Therefore, therapeutic decisions should still be driven initially on the basis of clinical criteria and in consultation with the patient. Nevertheless, our results indicate that younger age is particularly prognostically favorable for 5-FU-based chemotherapies and higher total serum protein for gemcitabine-based therapies. Since our prediction model works independently of the chemotherapy regimen, we see substantial clinical benefit by being able to recommend closer monitoring to patients with poor prognosis and enabling early enrollment in clinical trials of new therapeutic concepts.
Our study has limitations. Firstly, it is a retrospective study and consists only of a medium sample size. We decided not to use data imputation to avoid introducing bias into our data and had to exclude patients with missing data. While automatic image segmentation was carried out to avoid bias due to manual segmentation, we had to exclude patients in whom no primary tumor was visible. To control for overfitting, we carried out cross-validation. While the holdout data were unseen by the model during fitting, it was included in the feature extraction process which leads to potential information leakage. To rule this out, we validated our results on an external dataset which was previously unseen by our model. Since both datasets were obtained from German university hospitals, it will be important in the further course to also validate the model with geographically different datasets as well as datasets from patients treated by community oncologists.
In conclusion, we propose a novel prediction model based on multimodal parameters and a machine-learning approach for the prediction of overall survival risk in patients with advanced PDAC. We could externally validate that our model based on seven clinical parameters outperformed the existing staging system and an established clinical score. We could demonstrate the relevance of cancer genomics data on survival prediction and suggest further studies to investigate the potential of radiomics in pancreatic cancer.
Acknowledgments
Funding
This work was supported by funding from the German Cancer Consortium (DKTK) to JK (no grant number). Work in the lab of JTS is supported by the German Cancer Consortium (DKTK; no grant number), Deutsche Forschungsgemeinschaft (DFG; grant numbers 405344257/SI1549/3-2 and SI1549/4-1); the Deutsche Krebshilfe (German Cancer Aid; grant numbers #70112505/PIPAC, #70113834/PREDICT-PACA); the German Federal Ministry of Education and Research (BMBF; grant number 01KD2206A/SATURN3).
Disclosure
SK reports honoraria (self) from Merck, Amgen, Roche, Sanofi, Aventis, Servier and Lilly; honoraria (institution) from Merck, Amgen, Roche and Lilly; advisory/consultancy roles with Merck, Amgen, Roche, Sanofi, Aventis, Servier and Lilly; research grants/funding (self) from Merck, Bristol Myers Squibb, Roche and Lilly; research grants/funding (institution) from Merck and Lilly; and travel/accommodation expenses from Merck, Amgen, Roche, Sanofi, Aventis, Servier, Pierre Fabre and Lilly. MW reports honoraria from Amgen, Boehringer Ingelheim, Novartis, Roche and Takeda; and received research funding from Bristol Myers Squibb and Takeda. JTS reports honoraria as consultant or for continuing medical education presentations from AstraZeneca, Bayer, Immunocore, Roche and Servier. His institution receives research funding from Bristol Myers Squibb, Celgene, Roche. He holds ownership and serves on the Board of Directors of Pharma15, all outside the submitted work. MS reports compensated consultancy from Amgen, AstraZeneca, BIOCAD, Boehringer Ingelheim, Bristol Myers Squibb, GlaxoSmithKline, Janssen, Merck Serono, Novartis, Roche, Sanofi and Takeda; honoraria for continuing medical education presentations from Amgen, Boehringer Ingelheim, Bristol Myers Squibb, Janssen and Novartis; and received research funding to his institution from AstraZeneca and Bristol Myers Squibb. All other authors have declared no conflicts of interest.
Ethics approval and consent to participate
This study was carried out in accordance with the Declaration of Helsinki. Anonymized analyses of clinical data are covered by the general treatment contract of the University Hospital Essen, which is signed by the patient. The study was approved by the local Ethics Committee of the Medical Faculty of the University Duisburg-Essen (No. 21-10347).
Data sharing
De-identified datasets analyzed in this study are available from the corresponding author upon reasonable request.
Supplementary data
References
- 1.Siegel R.L., Miller K.D., Fuchs H.E., Jemal A. Cancer statistics, 2022. CA Cancer J Clin. 2022;72:7–33. doi: 10.3322/caac.21708. [DOI] [PubMed] [Google Scholar]
- 2.Park W., Chawla A., O’Reilly E.M. Pancreatic cancer: a review. JAMA. 2021;326:851–862. doi: 10.1001/jama.2021.13027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Von Hoff D.D., Ervin T., Arena F.P., et al. Increased survival in pancreatic cancer with nab-paclitaxel plus gemcitabine. N Engl J Med. 2013;369:1691–1703. doi: 10.1056/NEJMoa1304369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Conroy T., Desseigne F., Ychou M., et al. FOLFIRINOX versus gemcitabine for metastatic pancreatic cancer. N Engl J Med. 2011;364:1817–1825. doi: 10.1056/NEJMoa1011923. [DOI] [PubMed] [Google Scholar]
- 5.Chun Y.S., Pawlik T.M., Vauthey J.N. 8th edition of the AJCC cancer staging manual: pancreas and hepatobiliary cancers. Ann Surg Oncol. 2018;25:845–847. doi: 10.1245/s10434-017-6025-x. [DOI] [PubMed] [Google Scholar]
- 6.Glen P., Jamieson N.B., McMillan D.C., Carter R., Imrie C.W., McKay C.J. Evaluation of an inflammation-based prognostic score in patients with inoperable pancreatic cancer. Pancreatology. 2006;6:450–463. doi: 10.1159/000094562. [DOI] [PubMed] [Google Scholar]
- 7.McMillan D.C. The systemic inflammation-based Glasgow Prognostic Score: a decade of experience in patients with cancer. Cancer Treat Rev. 2013;39:534–540. doi: 10.1016/j.ctrv.2012.08.003. [DOI] [PubMed] [Google Scholar]
- 8.Xue P., Zhu L., Wan Z., et al. A prognostic index model to predict the clinical outcomes for advanced pancreatic cancer patients following palliative chemotherapy. J Cancer Res Clin Oncol. 2015;141:1653–1660. doi: 10.1007/s00432-015-1953-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yi J.H., Lee J., Park S.H., et al. A prognostic model to predict clinical outcomes with first-line gemcitabine-based chemotherapy in advanced pancreatic cancer. Oncology. 2011;80:175–180. doi: 10.1159/000328449. [DOI] [PubMed] [Google Scholar]
- 10.Deng G.C., Lv Y., Yan H., et al. Nomogram to predict survival of patients with advanced and metastatic pancreatic Cancer. BMC Cancer. 2021;21:1227. doi: 10.1186/s12885-021-08943-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hang J., Wu L., Zhu L., et al. Prediction of overall survival for metastatic pancreatic cancer: development and validation of a prognostic nomogram with data from open clinical trial and real-world study. Cancer Med. 2018;7 doi: 10.1002/cam4.1573. 2974-2984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hamada T., Nakai Y., Yasunaga H., et al. Prognostic nomogram for nonresectable pancreatic cancer treated with gemcitabine-based chemotherapy. Br J Cancer. 2014;110:1943–1949. doi: 10.1038/bjc.2014.131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Shibuki T., Mizuta T., Shimokawa M., et al. Prognostic nomogram for patients with unresectable pancreatic cancer treated with gemcitabine plus nab-paclitaxel or FOLFIRINOX: a post-hoc analysis of a multicenter retrospective study in Japan (NAPOLEON study) BMC Cancer. 2022;22:19. doi: 10.1186/s12885-021-09139-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fornaro L., Leone F., Vienot A., et al. Validated nomogram predicting 6-month survival in pancreatic cancer patients receiving first-line 5-fluorouracil, oxaliplatin, and irinotecan. Clin Colorectal Cancer. 2019;18:e394–e401. doi: 10.1016/j.clcc.2019.08.004. [DOI] [PubMed] [Google Scholar]
- 15.Vernerey D., Huguet F., Vienot A., et al. Prognostic nomogram and score to predict overall survival in locally advanced untreated pancreatic cancer (PROLAP) Br J Cancer. 2016;115:281–289. doi: 10.1038/bjc.2016.212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kaissis G.A., Jungmann F., Ziegelmayer S., et al. Multiparametric modelling of survival in pancreatic ductal adenocarcinoma using clinical, histomorphological, genetic and image-derived parameters. J Clin Med. 2020;9:E1250. doi: 10.3390/jcm9051250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Eilaghi A., Baig S., Zhang Y., et al. CT texture features are associated with overall survival in pancreatic ductal adenocarcinoma-a quantitative analysis. BMC Med Imaging. 2017;17:38. doi: 10.1186/s12880-017-0209-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhang Y., Lobo-Mueller E.M., Karanicolas P., Gallinger S., Haider M.A., Khalvati F. CNN-based survival model for pancreatic ductal adenocarcinoma in medical imaging. BMC Med Imaging. 2020;20:11. doi: 10.1186/s12880-020-0418-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cozzi L., Comito T., Fogliata A., et al. Computed tomography based radiomic signature as predictive of survival and local control after stereotactic body radiation therapy in pancreatic carcinoma. PLoS One. 2019;14 doi: 10.1371/journal.pone.0210758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chakraborty J., Langdon-Embry L., Cunanan K.M., et al. Preliminary study of tumor heterogeneity in imaging predicts two year survival in pancreatic cancer patients. PLoS One. 2017;12 doi: 10.1371/journal.pone.0188022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Parr E., Du Q., Zhang C., et al. Radiomics-based outcome prediction for pancreatic cancer following stereotactic body radiotherapy. Cancers. 2020;12:1051. doi: 10.3390/cancers12041051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Xie T., Wang X., Li M., Tong T., Yu X., Zhou Z. Pancreatic ductal adenocarcinoma: a radiomics nomogram outperforms clinical model and TNM staging for survival estimation after curative resection. Eur Radiol. 2020;30:2513–2524. doi: 10.1007/s00330-019-06600-2. [DOI] [PubMed] [Google Scholar]
- 23.Isensee F., Jaeger P.F., Kohl S.A.A., Petersen J., Maier-Hein K.H. nnU-Net: a self-configuring method for deep learning-based biomedical image segmentation. Nat Methods. 2021;18 doi: 10.1038/s41592-020-01008-z. 203-121. [DOI] [PubMed] [Google Scholar]
- 24.Ishwaran H., Kogalur U.B., Blackstone E.H., Lauer M.S. Random survival forests. Ann Appl Stat. 2008;2 [Google Scholar]
- 25.Kim D.W., Lee S., Kwon S., Nam W., Cha I.-H., Kim H.J. Deep learning-based survival prediction of oral cancer patients. Sci Rep. 2019;9:6994. doi: 10.1038/s41598-019-43372-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.van Griethuysen JJM, Fedorov A., Parmar C., et al. Computational radiomics system to decode the radiographic phenotype. Cancer Res. 2017;77:e104–e107. doi: 10.1158/0008-5472.CAN-17-0339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pölsterl S. Scikit-survival: a library for time-to-event analysis built on top of scikit-learn. J Mach Learn Res. 2020;21:1–6. [Google Scholar]
- 28.Kursa M.B., Rudnicki W.R. Feature selection with the boruta package. J Stat Softw. 2010;36:1–13. [Google Scholar]
- 29.Lundberg S.M., Lee S.-I. Vol. 30. Curran Associates, Inc.; Red Hook, NY: 2017. A unified approach to interpreting model predictions. (Advances in Neural Information Processing Systems). [Google Scholar]
- 30.Davidson-Pilon C. Zenodo; 2021. Lifelines, survival analysis in Python. [DOI] [Google Scholar]
- 31.Virtanen P., Gommers R., Oliphant T.E., et al. SciPy 1.0: fundamental algorithms for scientific computing in Python. Nat Methods. 2020;17:261–272. doi: 10.1038/s41592-019-0686-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kline A., Luo Y. PsmPy: a package for retrospective cohort matching in python. IEEE EMBC. 2022 doi: 10.1109/EMBC48229.2022.9871333. in press. [DOI] [PubMed] [Google Scholar]
- 33.Piciucchi M., Stigliano S., Archibugi L., et al. The neutrophil/lymphocyte ratio at diagnosis is significantly associated with survival in metastatic pancreatic cancer patients. Int J Mol Sci. 2017;18:730. doi: 10.3390/ijms18040730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Fernández A., Salgado M., García A., et al. Prognostic factors for survival with nab-paclitaxel plus gemcitabine in metastatic pancreatic cancer in real-life practice: the ANICE-PaC study. BMC Cancer. 2018;18:1185. doi: 10.1186/s12885-018-5101-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Markus M., Abendroth A., Noureddine R., et al. Combined systemic inflammation score (SIS) correlates with prognosis in patients with advanced pancreatic cancer receiving palliative chemotherapy. J Cancer Res Clin Oncol. 2021;147:579–591. doi: 10.1007/s00432-020-03361-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Stotz M., Gerger A., Eisner F., et al. Increased neutrophil-lymphocyte ratio is a poor prognostic factor in patients with primary operable and inoperable pancreatic cancer. Br J Cancer. 2013;109:416–421. doi: 10.1038/bjc.2013.332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Schlick K., Magnes T., Huemer F., et al. C-Reactive protein and neutrophil/lymphocytes ratio: prognostic indicator for doubling overall survival prediction in pancreatic cancer patients. J Clin Med. 2019;8:1791. doi: 10.3390/jcm8111791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mantovani A., Allavena P., Sica A., Balkwill F. Cancer-related inflammation. Nature. 2008;454:436–444. doi: 10.1038/nature07205. [DOI] [PubMed] [Google Scholar]
- 39.Ronga I., Gallucci F., Riccardi F., Uomo G. Anorexia-cachexia syndrome in pancreatic cancer: recent advances and new pharmacological approach. Adv Med Sci. 2014;59:1–6. doi: 10.1016/j.advms.2013.11.001. [DOI] [PubMed] [Google Scholar]
- 40.Maisey N.R., Norman A.R., Hill A., Massey A., Oates J., Cunningham D. CA19-9 as a prognostic factor in inoperable pancreatic cancer: the implication for clinical trials. Br J Cancer. 2005;93:740–743. doi: 10.1038/sj.bjc.6602760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tas F., Karabulut S., Ciftci R., et al. Serum levels of LDH, CEA, and CA19-9 have prognostic roles on survival in patients with metastatic pancreatic cancer receiving gemcitabine-based chemotherapy. Cancer Chemother Pharmacol. 2014;73:1163–1171. doi: 10.1007/s00280-014-2450-8. [DOI] [PubMed] [Google Scholar]
- 42.Oweira H., Petrausch U., Helbling D., et al. Prognostic value of site-specific metastases in pancreatic adenocarcinoma: a surveillance epidemiology and end results database analysis. World J Gastroenterol. 2017;23:1872–1880. doi: 10.3748/wjg.v23.i10.1872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kawesha A., Ghaneh P., Andrén-Sandberg A., et al. K-ras oncogene subtype mutations are associated with survival but not expression of p53, p16(INK4A), p21(WAF-1), cyclin D1, erbB-2 and erbB-3 in resected pancreatic ductal adenocarcinoma. Int J Cancer. 2000;89:469–474. doi: 10.1002/1097-0215(20001120)89:6<469::aid-ijc1>3.0.co;2-l. [DOI] [PubMed] [Google Scholar]
- 44.Immervoll H., Hoem D., Kugarajh K., Steine S.J., Molven A. Molecular analysis of the EGFR-RAS-RAF pathway in pancreatic ductal adenocarcinomas: lack of mutations in the BRAF and EGFR genes. Virchows Arch. 2006;448:788–796. doi: 10.1007/s00428-006-0191-8. [DOI] [PubMed] [Google Scholar]
- 45.Ogura T., Yamao K., Hara K., et al. Prognostic value of K-ras mutation status and subtypes in endoscopic ultrasound-guided fine-needle aspiration specimens from patients with unresectable pancreatic cancer. J Gastroenterol. 2013;48:640–646. doi: 10.1007/s00535-012-0664-2. [DOI] [PubMed] [Google Scholar]
- 46.Bournet B., Muscari F., Buscail C., et al. KRAS G12D mutation subtype is a prognostic factor for advanced pancreatic adenocarcinoma. Clin Transl Gastroenterol. 2016;7:e157. doi: 10.1038/ctg.2016.18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ying H., Kimmelman A.C., Lyssiotis C.A., et al. Oncogenic Kras maintains pancreatic tumors through regulation of anabolic glucose metabolism. Cell. 2012;149:656–670. doi: 10.1016/j.cell.2012.01.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Salinas-Miranda E., Khalvati F., Namdar K., et al. Validation of prognostic radiomic features from resectable pancreatic ductal adenocarcinoma in patients with advanced disease undergoing chemotherapy. Can Assoc Radiol J. 2021;72:605–613. doi: 10.1177/0846537120968782. [DOI] [PubMed] [Google Scholar]
- 49.Yun G., Kim Y.H., Lee Y.J., Kim B., Hwang J.-H., Choi D.J. Tumor heterogeneity of pancreas head cancer assessed by CT texture analysis: association with survival outcomes after curative resection. Sci Rep. 2018;8:7226. doi: 10.1038/s41598-018-25627-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Attiyeh M.A., Chakraborty J., Doussot A., et al. Survival prediction in pancreatic ductal adenocarcinoma by quantitative computed tomography image analysis. Ann Surg Oncol. 2018;25:1034–1042. doi: 10.1245/s10434-017-6323-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Healy G.M., Salinas-Miranda E., Jain R., et al. Pre-operative radiomics model for prognostication in resectable pancreatic adenocarcinoma with external validation. Eur Radiol. 2021;32:2492–2505. doi: 10.1007/s00330-021-08314-w. [DOI] [PubMed] [Google Scholar]
- 52.Park S., Sham J.G., Kawamoto S., et al. CT radiomics-based preoperative survival prediction in patients with pancreatic ductal adenocarcinoma. AJR Am J Roentgenol. 2021;217:1104–1112. doi: 10.2214/AJR.20.23490. [DOI] [PubMed] [Google Scholar]
- 53.Nasief H., Zheng C., Schott D., et al. A machine learning based delta-radiomics process for early prediction of treatment response of pancreatic cancer. NPJ Precis Oncol. 2019;3:25. doi: 10.1038/s41698-019-0096-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Cheng S.-H., Cheng Y.-J., Jin Z.-Y., Xue H.-D. Unresectable pancreatic ductal adenocarcinoma: role of CT quantitative imaging biomarkers for predicting outcomes of patients treated with chemotherapy. Eur J Radiol. 2019;113:188–197. doi: 10.1016/j.ejrad.2019.02.009. [DOI] [PubMed] [Google Scholar]
- 55.Sandrasegaran K., Lin Y., Asare-Sawiri M., Taiyini T., Tann M. CT texture analysis of pancreatic cancer. Eur Radiol. 2019;29:1067–1073. doi: 10.1007/s00330-018-5662-1. [DOI] [PubMed] [Google Scholar]
- 56.Hang J., Xu K., Yin R., et al. Role of CT texture features for predicting outcome of pancreatic cancer patients with liver metastases. J Cancer. 2021;12:2351–2358. doi: 10.7150/jca.49569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Gebauer L., Moltz J.H., Mühlberg A., et al. Quantitative imaging biomarkers of the whole liver tumor burden improve survival prediction in metastatic pancreatic cancer. Cancers. 2021;13:5732. doi: 10.3390/cancers13225732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Mühlberg A., Holch J.W., Heinemann V., et al. The relevance of CT-based geometric and radiomics analysis of whole liver tumor burden to predict survival of patients with metastatic colorectal cancer. Eur Radiol. 2021;31:834–846. doi: 10.1007/s00330-020-07192-y. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.