Summary
Effective drugs for atrial fibrillation (AF) are lacking, resulting in significant morbidity and mortality. This study demonstrates that network proximity analysis of differentially expressed genes from atrial tissue to drug targets can help prioritize repurposed drugs for AF. Using enrichment analysis of drug-gene signatures and functional testing in human inducible pluripotent stem cell (iPSC)-derived atrial-like cardiomyocytes, we identify metformin as a top repurposed drug candidate for AF. Using the active compactor, a new design analysis of large-scale longitudinal electronic health record (EHR) data, we determine that metformin use is significantly associated with a reduced risk of AF (odds ratio = 0.48, 95%, confidence interval [CI] 0.36–0.64, p < 0.001) compared with standard treatments for diabetes. This study utilizes network medicine methodologies to identify repurposed drugs for AF treatment and identifies metformin as a candidate drug.
Keywords: network medicine, atrial fibrillation, human inducible pluripotent stem cells, drug repurposing, pharmacoepidemiology, systems biology, electronic health record, EHR
Graphical abstract
Highlights
-
•
Network proximity analysis predicts metformin as repurposed drug for AF
-
•
Metformin induces directional changes in AF disease module genes
-
•
Large-scale health record data support metformin use for reduced AF risk
Lal et al. identify metformin as a repurposed drug candidate for atrial fibrillation using human atrial tissue transcriptomics-based network proximity analysis and drug-gene signature-based enrichment analysis. In vitro testing from iPSC-derived atrial-like cardiomyocyte models and large-scale EHR data-based pharmacoepidemiologic analysis validates metformin’s effect on reduced risk of AF.
Introduction
Atrial fibrillation (AF), the most prevalent cardiac arrhythmia in the western world, affects 1%–2% of the general population.1, 2, 3 The incidence in the United States, currently estimated at 5–6 million individuals, is expected to increase to 12 million by 2030.4 Because AF is silent in 5%–35% of diagnosed patients, its overall prevalence is likely higher. Although the early stages of AF are considered benign,5 persistent and long-standing forms of AF are associated with a substantial increase in mortality, with a 1.5- and 1.9-fold increase in the odds ratio in men and women, respectively.6 AF is also associated with a higher risk of stroke, heart failure, and dementia.7,8 Patients with AF are 30% more likely to be hospitalized at least once annually, resulting in a significant financial burden to them and the healthcare system.9
Clinical management of AF requires improvement. The primary goals of AF clinical management are controlling heart rate, restoring and maintaining sinus rhythm, and preventing thromboembolism. Drugs that control the ventricular rate include β-adrenergic blockers, calcium channel blockers, and cardiac glycosides. Rhythm control of AF is limited by side effects of anti-arrhythmic drugs, which can include risk of life-threatening proarrhythmia.10, 11, 12 AF ablation strategies include pulmonary vein isolation and/or atrial substrate ablation. These are invasive procedures with limited success, significant recurrence rates, and risk of major complications.13,14 Anticoagulants and/or left atrial appendage closure prevent thromboembolism but carry bleeding or procedure risks.8
Genome-wide association studies (GWASs) have identified ∼100 AF susceptibility loci.15 Despite this progress in understanding the genetic risk of AF, predisposition to AF involves a complex, polygenic, and pleiotropic genetic architecture. Network medicine exploits the network paradigm that considers the functional and topological organization of gene products as neighborhoods (called “disease modules”) in the human protein-protein interactome.16,17 Recent advances in network medicine methodologies that shed light on the relationship between drugs (drug targets) and diseases (molecular [protein] determinants in disease modules) can serve as useful tools for efficient screening of potentially new indications for approved drugs in multiple complex diseases,18, 19, 20, 21 including cardiovascular disease. We therefore posited that network medicine approaches may enable identification of candidate drugs with well-established pharmacokinetics/pharmacodynamics, safety, and tolerability profiles for patients with AF by using drug repurposing strategies.19,22, 23, 24, 25, 26, 27
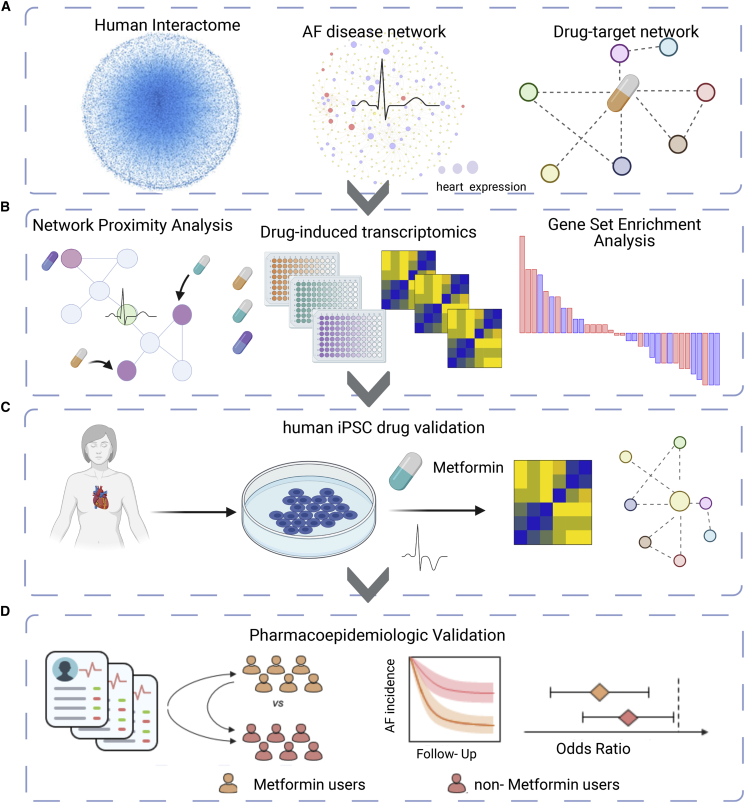
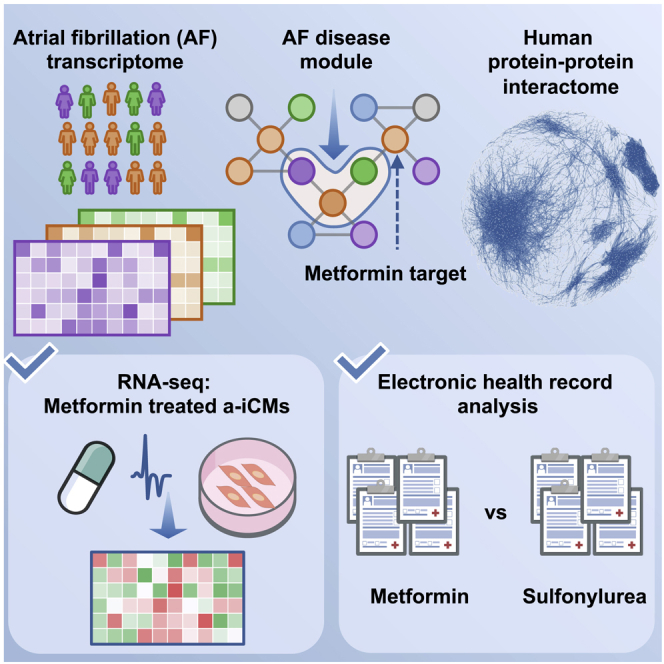
Pathways for AF treatment can include multiple targets that share biological networks. Known drug targets can be mapped to the human proteome and, subsequently, to disease modules. This network medicine approach allows discovery of repurposable drug targets and minimizes the risk of toxicity by considering all drug-protein interactions within a protein-protein network.28,29 Integrative analyses of genomics, transcriptomics, and interactomics (protein-protein interactions [PPIs]) have not been fully exploited for AF drug repurposing. Here we used network-based approaches to prioritize drug repurposing for AF by integration of transcriptomics data from human left atrium (LA) tissue and drug-induced gene signatures from human inducible pluripotent stem cell-derived atrial-like cardiomyocytes under the protein-protein interactome (Figure 1). We also validate metformin use for reduced AF risk using large-scale longitudinal pharmacoepidemiologics analyses.
Figure 1.
Study overview
(A) We utilized a systems pharmacology-based network medicine platform to quantify the proximity of interactions between the atrial fibrillation (AF) interactome nodes and drug targets in the PPI network.
(B) GSEA of known targets are used to validate the in silico approach and nominate candidate drugs.
(C and D) Using human induced pluripotent stem cell-derived atrial-like cardiomyocytes (a-iCMs; C) as well as large-scale pharmacoepidemiologic analysis (D), we are able to validate metformin as a repurposed drug for AF. Created with BioRender.com.
Results
Network-based drug repurposing framework for AF
To understand the drug-disease relationships and framework for nominating putative repurposed drugs for AF, we performed the following: (1) construction of the AF disease module from LA tissue transcriptomics in AF and non-AF controls and reconstruction of the drug-target network (Figure 1A); (2) calculation of network proximity of drug targets to the AF disease module finding and subsequent calculation of the gene set enrichment scores of screened drugs (Figure 1B); (3) in vitro validation of nominated drug candidates and performance of mechanistic studies in human induced pluripotent stem cell (hiPSC)-derived atrial-like cardiomyocytes (a-iCMs) (Figure 1C); and (4) large-scale population-based validation to test candidate drug outcomes with AF (Figure 1D). Our in silico drug repurposing approach applies additional quality control steps for nominating drug candidates. We posited that, if a drug significantly reverses dysregulated gene expression in cell lines, then such a drug has the potential of reversing gene neighborhoods in AF disease modules derived from LA tissue transcriptomics profiles. We harnessed multiple lines of complementary evidence from a-iCM models and large-scale epidemiology data to ensure the plausibility of metformin as a repurposed drug for AF.
AF disease network
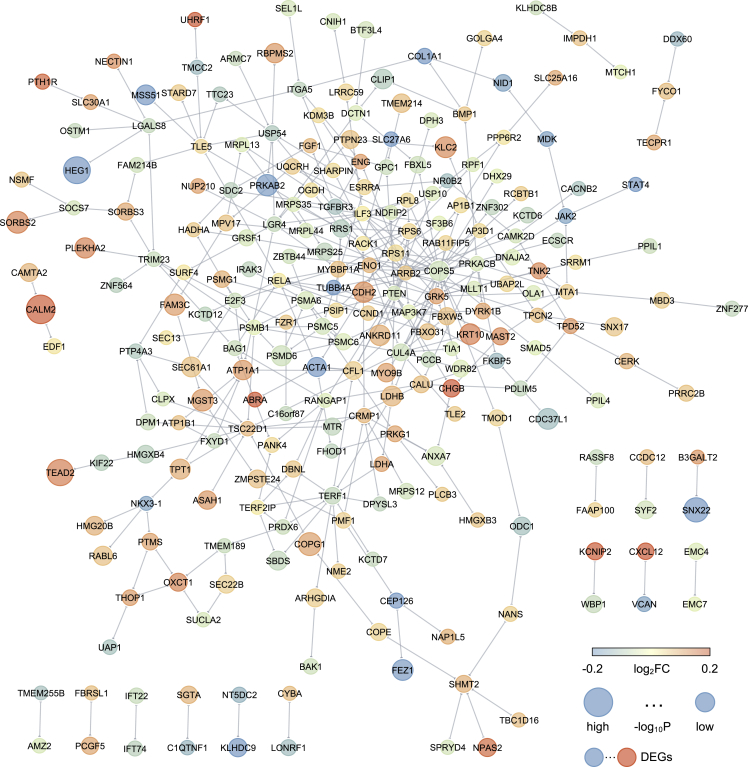
We generated an integrative network-based approach to identify molecular networks of AF. We hypothesized that this network medicine approach would enhance candidate drug prioritization for treatment of AF. In prior work, we described a human protein-protein interactome generated from five types of PPIs.20,24,25,30,31 To depict the human interactome network, we gathered associated proteins from five levels of evidence: (1) binary PPIs tested by high-throughput yeast two-hybrid (Y2H) systems; (2) binary, physical PPIs from protein 3D structures; (3) kinase-substrate interactions; (4) signaling networks; and (5) literature-curated PPIs identified by affinity purification followed by low-throughput mass spectrometry, Y2H, and other experimental approaches. To identify testable AF disease modules under the human PPI network model, we gathered RNA sequencing (RNA-seq) data from LA collected from 251 patients who underwent elective cardiac surgery to treat AF, valve disease, or other cardiac disorders and 14 nonfailing donor hearts not used for transplantation (Table S1). We identified 491 DEGs in AF cases (i.e., hearts in AF at the time of acquisition) compared with sinus rhythm cases (adjusted p < 0.05). The AF disease module (defined by the largest connected component in the human interactome) shown in Figure 2 includes 245 unique proteins (nodes) and 350 PPIs (edges). We identified several AF-specific hub proteins related to cardiac integrity and metabolic fitness, including LDHB, CDH2, UQCRH, PDLIM5, COPS5, and OXCT1. Gene Ontology (GO) enrichment analysis identified the following biological processes significant in AF differentially expressed genes (DEGs): the endoplasmic reticulum unfolded protein response, calcium and potassium ion transport, succinyl-coenzyme A (CoA) metabolism, collagen biosynthesis, membrane repolarization, and cardiac muscle relaxation (Figure S1A). Similarly, Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment analysis identified propanoate metabolism, proteasome, regulated calcium reabsorption, tricarboxylic acid (TCA) cycle, and cardiac muscle contraction as pathways significantly enriched in AF (Figure S1B).
Figure 2.
The atrial fibrillation interactome
The network highlights the atrial fibrillation (AF) interactome that connects 245 AF enriched genes. Node size is proportional to −log10 p value, and color corresponds to log2 fold change (log2FC) in AF compared with sinus rhythm (STAR Methods). The AF disease module (defined by the largest connected component in the human interactome) shown includes 245 unique proteins (nodes) and 350 PPIs (edges). ∗p < 0.05. See also Figures S1, S5, and S7 and Tables S1 and S8.
Network-based drug repurposing for AF
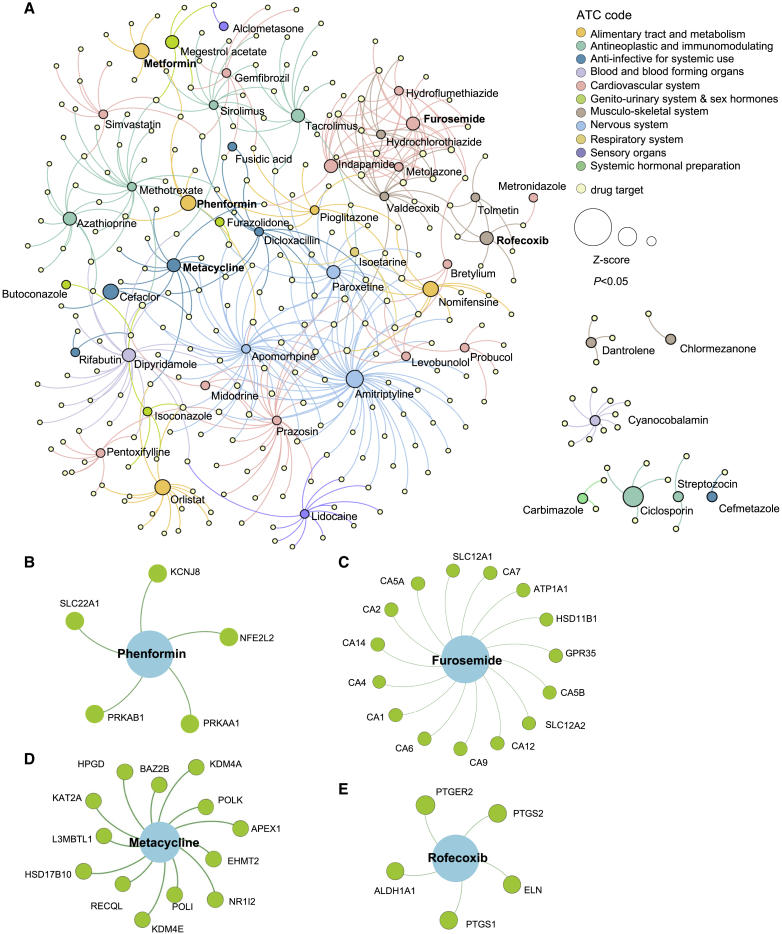
We harnessed drug-target interactions from 2,891 US Food and Drug Administration (FDA)-approved or clinically investigational drugs obtained from the Drugbank and Therapeutic Target (TTD) databases to assemble the drug-target network (Figure 1A).32 The network proximity of drug targets to the AF disease module was calculated using the Jaccard index and overlap coefficient (STAR Methods). To correct the literature bias of well-studied proteins in the human interactome, we normalized calculated values to Z score values and performed 1,000 permutation tests as described in previous work.20,31 A higher network proximity (quantified by a lower Z score) represents a strong network relationship. Using a Z score cutoff value of Z < −1, we focused on 54 drug candidates (Figure 3A). To further narrow our repurposed drug list, we used gene expression data of drug-treated human cell lines from the Connectivity Map database to prioritize drug candidates.33 Gene set enrichment analysis (GSEA) was performed, and enrichment scores (ESs) were calculated to nominate drug candidates using a cutoff of ES > 0 and p < 0.05. Here we identified 9 potential candidates: phenformin (Z = −2.475), metformin (Z = −1.97), furosemide (Z = −1.912), indapamide (Z = −1.691), metacycline (Z = −1.674), rofecoxib (Z = −1.67), dantrolene (Z = −1.193), alclometasone (Z = −1.094), and streptozocin (Z = −1.048). We found enrichment in the following pharmacological categories: metabolism, ion transport drugs, ion transport, and anti-inflammatory drugs. To compare our approach with the traditional disease-associated gene-based approach, we also tested our drug repurposing pipeline using 34 AF disease-associated genes collected from the Human Gene Mutation Database (HGMD).34 Using the same performance cutoff, Z < −1 and p < 0.05, we identified 282 candidates of repurposed drugs. Using this approach, neither metformin (Z = 0.037, p = 0.419) nor phenformin (Z = −1.36, p = 0.116) passed the significance threshold for drug repurposing from 34 AF-associated genes. The top drug candidate using the disease genes is propafenone, a class 1C antiarrhythmic agent. We performed a Pearson correlation coefficient (PCC) analysis between the drug Z scores derived from the network proximity analysis using the HGMD gene list versus DEGs from LA samples. We found modest correlation (PCC = 0.375, p < 1.0 × 10−8, coefficient of determination [R2] = 0.14) (Figure S2). The data suggest high reproducibility of transcriptomics-derived disease module proximity and traditional gene-based approaches.24,25,35,36 We next discuss several candidate drug classes.
Figure 3.
Network medicine applied to AF drug repurposing
(A) A subnetwork is shown to highlight the 54 candidate drugs associated with AF DEGs and their associated targets. Node size is proportional to Z score. Drugs are colored by their first-level anatomical therapeutic chemical (ATC) classification.
(B–E) Four candidate drugs with gene set enrichment analysis (GSEA) ES > 0 and p < 0.05 and their target genes using our drug-target network analysis.
See also Figures S2 and S5 and Table S7.
Anti-hypertension and heart failure drugs
AF and heart failure (HF) share risk factors and may represent additive risks for morbidity and mortality. Consequently, both are frequently treated simultaneously. A recent GWAS of HF showed overlapping genetics with AF.37 The top GWAS loci for AF and HF are on chromosome 4 near PITX2, a gene implicated in formation of the pulmonary veins. Also, AF and HF are associated with loci near SYNPO2L, which encodes a protein that binds to actin, and KLHL3, which encodes a protein involved in ubiquitination of proteins; abnormal proteostasis has been implicated in AF.37 Our AF network proximity analysis shows a significant association with indapamide (Z = −1.69, p = 0.038, ES = 0.38) and furosemide (Z = −1.91, p = 0.031, ES = 0.38) (Figures 3A and 3C). Furosemide is an FDA-approved diuretic for treating edema secondary to clinical conditions like HF by targeting sodium, potassium, and chloride transporters (SLC12A2 and SLC12A1). We found that furosemide targets 13 additional AF module genes (GPR35, HSD11B1, CA1, CA2, CA4, CA5A, CA5B, CA6, CA7, CA9, CA12, CA14, and ATP1A1) (Figure 3C). Previous studies have shown that GPR35 contributes to angiotensin II-induced hypertension and cardiotoxicity, suggesting potential use of anti-hypertensive drugs for treatment of AF.38, 39, 40
Anti-inflammatory drugs
Prior evidence suggests that inflammation plays a role in the etiology of AF disease.41, 42, 43 Comorbidities associated with AF include those associated with low-grade inflammation and metabolic dysfunction, like obesity, hypertension, and coronary artery disease. Structural and electrical remodeling after AF onset can propagate inflammation. Here we identified the anti-inflammatory agents rofecoxib (Z = −1.67, p = 0.004, ES = 0.22) and alclometasone (Z = −1.05, p < 0.001, ES = 0.32) as potential AF repurposable drug candidates. Rofecoxib, a COX2 inhibitor that is FDA approved to treat osteoarthritis and rheumatoid arthritis, has been shown to target five AF module genes (ALDH1A1, ELN, PTGER2, PTGS1, and PTGS2) (Figure 3E).
Cardiac metabolism drugs
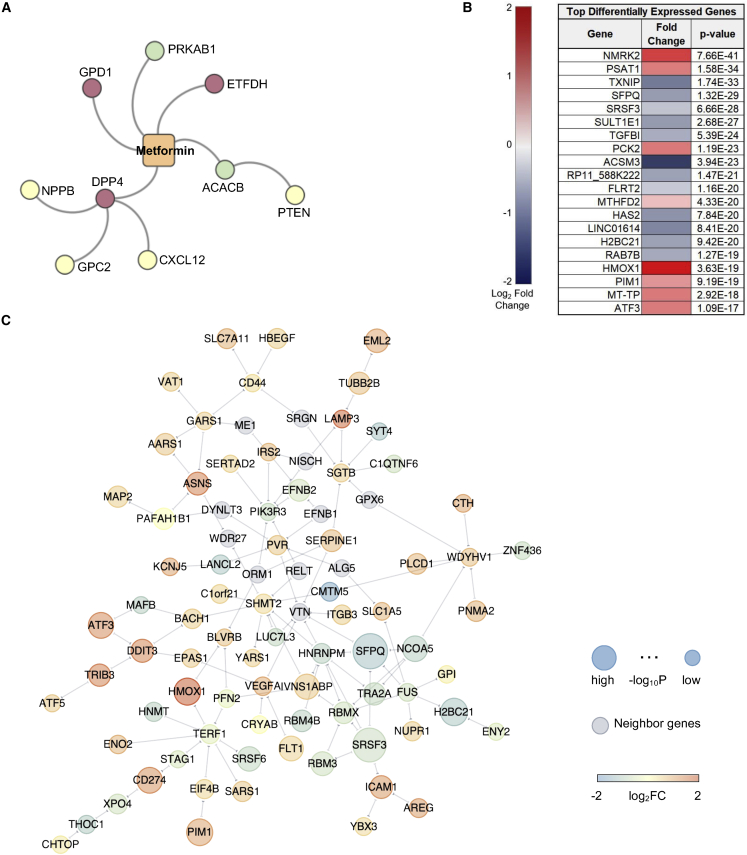
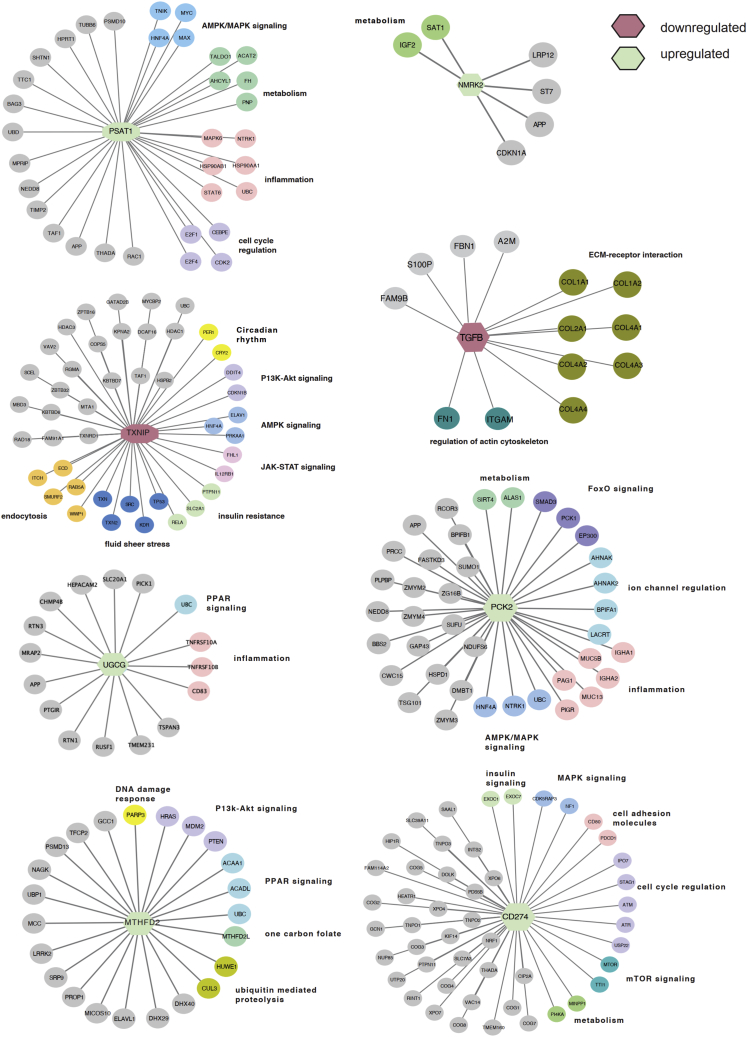
Dysregulation of cardiac metabolism has been well described in AF.44 AMP-activated protein kinase (AMPK) agonists, including the drug family members phenformin and metformin, rank at the top of our AF drug repurposable candidates (Z = −2.475, p = 0.002, ES = 0.255, and Z = −1.97, p = 0.015, ES = 0, respectively) (Figures 3B and 4A). An association between type 2 diabetes and AF was established over two decades ago.45, 46, 47 A prospective case-control analysis of individuals treated with metformin demonstrated a decrease of 19% in the incidence of new-onset AF.48 AMPK is considered a master regulator of energy status in the heart. Activation of AMPK stimulates pathways that utilize glucose and fatty acids to generate ATP.49 In addition to targeting the AMPK subunits PRKAB1 and PRKAA1, phenformin also targets a key regulator of oxidative stress, NFE2L2 (NRF-2), as well as the ATP-sensitive potassium channel pore subunit KCNJ8, a metabolically sensitive ion channel and regulator of cardiac repolarization (Figure 3B). In addition to PRKAB1, metformin also targets ACACB, GPD1, ETFDH, and DPP4, genes that play a role in metabolic pathways. Metformin drug target neighbors that were significant and DEGs in the AF network module (Figure 4C) were PTEN, NPPB, GPC2, and CXCL12 (Figure 4A).
Figure 4.
Validation of the AF repurposed drug candidate metformin using a-iCMs
(A) A subnetwork of metformin, drug targets, and PPI neighbors. The node color corresponds to the expected effect of metformin on target expression; green indicates agonist, and red indicates antagonist.
(B) DE genes of a-iCMs treated with metformin (n = 3) or water (n = 3) for 30 h. Data are expressed as FC, with red designating increased expression and blue decreased expression. All experimental combinations had 3 replicates.
(C) A subnetwork representing the significant differentially expressed genes (DEGs) and their PPI neighbors (gray). The node size is proportional to −log10 p value, and color corresponds to log2FC in metformin-treated a-iCMs versus the control after 1-Hz pacing stimulation.
See also Figures S3, S4, and S5 and Tables S2 and S9.
a-iCM validation of metformin
To validate our in silico repurposed drug findings, we generated a-iCMs for downstream analysis. 30 days from differentiation onset, the a-iCMs exhibited spontaneous beating and expressed high troponin levels. Beating cells were pretreated with metformin or vehicle for 6 h and then paced for 24 h, stimulating cell stress (Figure 1C).
Bulk RNA-seq was used to broadly assess the DEGs associated with metformin exposure (Figure 4B) using a similar design as the quantitative real-time PCR study. Following multiple testing correction (Bonferroni method with 16,895 expressed genes and family-wise error rate of 0.05), 251 genes exhibited significant differential expression (marginal DEG p < 2.96 × 10−6) for metformin versus vehicle water exposure in the context of 1-Hz pacing stimulation. Several key cardiovascular or drug-related markers were significantly upregulated with metformin, such as NMRK2, PSAT1, PCK2, and HMOX1 (Figure 4B). RT-PCR was performed to validate the expression of AF PPI neighbors. Other known markers associated with low expression in AF (CACNA1C, CACNA1D, HSF1, HSF2, SCN5A, and HSPB1 [HSP27]) increased after metformin treatment (Figure S3; Table S2). Network analysis was performed using the DEGs to identify gene neighborhoods associated with metformin exposure and showed several interactions associated with a decrease in P1K3R3 and an increase in the mitochondrially expressed gene SHMT2 (Figures 4C and S4B). Pathway enrichment analysis suggests that the top upregulated genes, including PSAT1, NMRK2, and PCK2, had neighbors enriched in the AMPK/mitogen-activated protein kinase (MAPK) signaling pathways, metabolism, inflammation, and mitochondrial function (Figures 5 and S4B). TGFB1 and TXNIP were significantly downregulated, indicating enhanced control of redox state and cellular homeostasis. Specifically, we found a subset of known PPI neighbors of thioredoxin interacting protein (TXNIP) corresponding to insulin resistance (RELA, SLC2A1, and PTPN11) (Figure 5). ACSM3, a rate-limiting enzyme involved in fatty acid metabolism, was significantly downregulated. However, ACSF2, HADHB, and ZADH2, genes that also play a role in lipid metabolism, were upregulated (Figure S4B). CS, a marker of mitochondrial content, and the electron transport chain genes SDHA2 and COX6A2 were also upregulated, suggesting potential upregulation of metabolic pathways, which, in the context of AF, could better enable the atria to meet the energy demand. We observed decreased expression of an AF network hub, OXCT1, with metformin treatment, implying a shift in energy utilization. These functional observations in a-iCMs mechanistically support the hypothesis that network-predicted metformin is a potential treatment for AF.
Figure 5.
Subnetworks of upregulated and downregulated genes after metformin treatment in a-iCMs
The central node color corresponds to upregulated (green) or downregulated (red) genes after metformin treatment in pacing a-iCMS. Outer node colors correspond to KEGG pathway classification, and class is listed next to each pathway cluster. See also Table S9.
Metformin induced changes of DEGs in AF
We next looked at the overlap between significant genes (adjusted p < 0.05) in the AF cohort (n = 960) and the metformin treated a-iCMs (n = 491). We found 30 DEGs significantly overlapped between the two datasets (p = 2.2 × 10−16) (Figure S5A). Furthermore, we observed that metformin induced directional changes of six genes upregulated in AF (COL21A1, KRT10, OXCT1, SGCD, SORBS2, and TMEM214) and two genes downregulated in AF (ITGA5 and PPTC7) (Figure S5B). Specifically, we observed that OXCT1 had several connections within the AF module. Succinyl-CoA-3-oxaloacid CoA transferase (SCOT) plays a central role in mitochondrially mediated metabolism. We observed upregulation of PPTC7, which is responsible for regulating coenzyme Q10. Together, the data indicate a shift from starvation to pro-oxidant conditions. Furthermore, the decrease in expression of COL21A1, SGCD, and SORBS2 indicates improved structural integrity of the cardiomyocytes.
Pharmacoepidemiologic analysis suggests metformin use with reduced AF risk
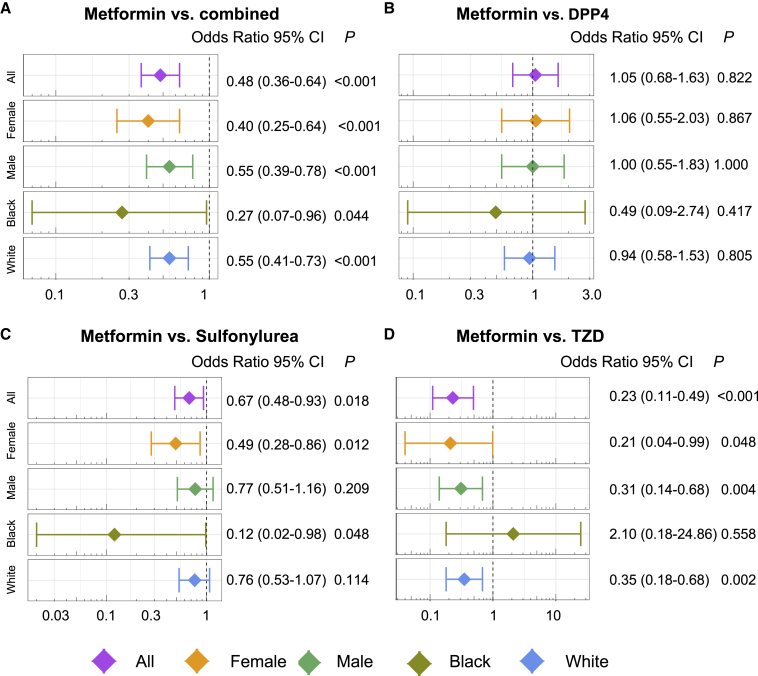
We evaluated the relationship in patients on metformin and AF onset using large-scale patient record data extracted from the Northwestern Medicine Enterprise Data Warehouse (NMEDW) between 2011 and 2021. We conducted five drug cohort comparison analyses: (1) metformin versus dipeptidyl-peptidase 4 sulfonylurea (DPP4) (n = 1,244), (2) metformin versus thiazolidinedione (TZD, n = 288), (3) metformin versus sulfonylurea (n = 2,352), (4) metformin versus glucagon-like peptide 1 receptor agonists (GLP1RAS; n = 258), and (5) metformin versus combination of all four drug cohorts (DPP4, TZD, sulfonylurea, and GLP1RAs, n = 3,578). Tables 1 and S5 summarizes the data for each group in our dataset. The comparator drugs did not pass the threshold for significance in our network proximity pipeline (Table S6). We conducted propensity score (PS) matching for age, gender, race, and comorbidities (including renal disease and other AF-related comorbidities; Table 1). We found that metformin was significantly associated with a 52% reduced likelihood of AF compared with the combined drug cohort (odds ratio [OR] = 0.48, 95% confidence interval [CI] 0.36–0.64, p < 0.001; Figure 6A). Metformin usage was significantly associated with reduced risk of AF compared with sulfonylurea (OR = 0.67, 95% CI 0.48–0.93, p = 0.018; Figure 6C) and TZD (OR = 0.23, 95% CI 0.11–0.49, p < 0.001; Figure 6D). However, metformin use is not significantly associated with AF risk compared with GLP1RA (OR = 1.25, 95% CI 0.33–4.73, p = 0.737; Figure S6) and DPP4 (OR = 1.05, 95% CI 0.68–1.63, p = 0.822; Figure 6B).
Table 1.
Patient demographics for pharmacoepidemiologics analysis
| Metformin | DPP4 | GLP1RA | Sulfonylurea | TZD | Combination | |
|---|---|---|---|---|---|---|
| N | 13522 | 3,477 | 678 | 7,014 | 919 | 10,214 |
| Age (mean [SD]) | 62.41 (15.59) | 70.20 (13.84) | 62.35 (12.61) | 69.34 (14.53) | 69.03 (13.44) | 68.95 (14.41) |
| Gender (male %) | 6,923 (51.2) | 1,870 (53.8) | 333 (49.1) | 3,942 (56.2) | 563 (61.3) | 5,613 (55.0) |
| Race | ||||||
| White (%) | 10,293 (76.1) | 2,819 (81.1) | 519 (76.5) | 5,658 (80.7) | 777 (84.5) | 8,217 (80.4) |
| Black (%) | 1,452 (10.7) | 261 (7.5) | 103 (15.2) | 543 (7.7) | 57 (6.2) | 841 (8.2) |
| Other (%) | 1,559 (11.5) | 358 (10.3) | 41 (6.0) | 737 (10.5) | 73 (7.9) | 1,031 (10.1) |
| Comorbidity | ||||||
| AMI (%) | 540 (4.0) | 169 (4.9) | 33 (4.9) | 193 (2.8) | 33 (3.6) | 359 (3.5) |
| CHF (%) | 1,375 (10.2) | 625 (18.0) | 143 (21.1) | 778 (11.1) | 83 (9.0) | 1,352 (13.2) |
| PVD (%) | 1,118 (8.3) | 412 (11.8) | 102 (15.0) | 338 (4.8) | 92 (10.0) | 789 (7.7) |
| CEVD (%) | 1,552 (11.5) | 471 (13.5) | 76 (11.2) | 599 (8.5) | 107 (11.6) | 1,049 (10.3) |
| Dementia (%) | 321 (2.4) | 114 (3.3) | 5 (0.7) | 128 (1.8) | 29 (3.2) | 227 (2.2) |
| COPD (%) | 1,946 (14.4) | 577 (16.6) | 134 (19.8) | 605 (8.6) | 127 (13.8) | 1,205 (11.8) |
| RHEUMD (%) | 306 (2.3) | 99 (2.8) | 27 (4.0) | 67 (1.0) | 15 (1.6) | 176 (1.7) |
| PUD (%) | 118 (0.9) | 46 (1.3) | 10 (1.5) | 26 (0.4) | 14 (1.5) | 71 (0.7) |
| MLD (%) | 594 (4.4) | 197 (5.7) | 59 (8.7) | 133 (1.9) | 48 (5.2) | 375 (3.7) |
| DIAB (%) | 9,041 (66.9) | 2,577 (74.1) | 534 (78.8) | 3,555 (50.7) | 614 (66.8) | 6,019 (58.9) |
| DIABWC (%) | 1,621 (12.0) | 683 (19.6) | 197 (29.1) | 586 (8.4) | 156 (17.0) | 1,320 (12.9) |
| HP (%) | 245 (1.8) | 49 (1.4) | 17 (2.5) | 54 (0.8) | 13 (1.4) | 109 (1.1) |
| REND (%) | 784 (5.8) | 676 (19.4) | 150 (22.1) | 657 (9.4) | 116 (12.6) | 1,307 (12.8) |
| CANC (%) | 1,458 (10.8) | 440 (12.7) | 74 (10.9) | 463 (6.6) | 110 (12.0) | 893 (8.7) |
| MSLD (%) | 70 (0.5) | 40 (1.2) | 4 (0.6) | 24 (0.3) | 4 (0.4) | 65 (0.6) |
| METACANC (%) | 366 (2.7) | 110 (3.2) | 17 (2.5) | 94 (1.3) | 31 (3.4) | 204 (2.0) |
| AIDS (%) | 32 (0.2) | 7 (0.2) | 2 (0.3) | 7 (0.1) | 3 (0.3) | 16 (0.2) |
Age is shown as mean (SD). All other characteristics are shown as number of and percentage of cases. Race “other” does not include patients with unknown races. AMI, acute myocardial infarction; CHF, congestive heart failure; PVD, peripheral vascular disease; CEVD, cerebrovascular disease; COPD, chronic obstructive pulmonary disease; RHEUMD, rheumatoid disease; PUD, peptic ulcer disease; MLD, mild liver disease; DIAB, diabetes without complications; DIABWC, diabetes with complications; HP, hemiplegia or paraplegia; REND, renal disease; CANC, cancer (any malignancy); MSLD, moderate or severe liver disease; METACANC, metastatic solid tumor; AIDS, AIDS/HIV.
Figure 6.
Pharmacoepidemiologic validation of metformin in reducing AF occurrence
(A–D) OR and 95% confidence interval (CI) for metformin versus (A) combination of the four drug groups (all, dipeptidyl-peptidase 4 sulfonylurea [DPP4], thiazolidinedione [TZD], sulfonylurea, and glucagon-like peptide 1 receptor agonist [GLP1RA]) (n = 3,578), (B) DPP4 (n = 1,244), (C) sulfonylurea (n = 2,352), and (D) TZD (n = 288). For each of the four comparisons, the results for comparisons between subgroups (including female, male, Black, and White) are also shown. Patient groups were matched using PS matching with the variables age, gender, race, and comorbidities (listed in Table 1) for the overall group comparisons. For the subgroup of male and female, the matching variables excluded gender, and for the subgroup Black and White, the matching variables excluded race. Logistic regression models were used for statistical inference of the AF ORs. Subgroup analyses were performed in females (orange), males (green), Black Americans (dark green), and White Americans (blue). ∗p < 0.05. See also Figure S6 and Tables S3, S4, S5, and S6.
We also performed gender- and race-specific subgroup analysis. Several reports indicate that AF is more common in males versus females, but the prevalence of risk factors differs.50, 51, 52, 53 For example, diabetes has been observed as an AF risk factor for women but not men.53 We found that male and female metformin users were significantly associated with reduced likelihood of AF compared with the combined group by assembling all figure drug cohorts (female: OR = 0.40, 95% CI 0.25–0.64, p < 0.001; male: OR = 0.55, 95% CI 0.39–0.78, p < 0.001) and TZD (female: OR = 0.21, 95% CI 0.04–0.99, p = 0.048; male: OR = 0.31, 95% CI 0.14–0.68, p = 0.004) but had a stronger effect size on females (Figures 6A and 6D). Among metformin users versus sulfonylurea users, we observed that females had a reduced AF risk (OR = 0.49, 95% CI 0.28–0.86, p = 0.012) but males did not (OR = 0.77, 95% CI 0.51–1.16, p = 0.209) (Figure 6C).
Despite the greater burden of AF in White individuals of European ancestry, AF risk factors (i.e., diabetes, hypertension, and stroke) are more prevalent in Black Americans. The traditional risk factors only represent a fraction of AF burden in this population.54 The burden of disease explained by genetic risk loci remains unclear for Black Americans.55, 56, 57 We observed a greater reduction in AF risk among Black individuals in metformin users versus the combined group (Black: OR = 0.27, 95% CI 0.07–0.96, p = 0.044; White: OR = 0.55, 95% CI 0.41–0.73, p < 0.001) (Figure 6A). When analyzing specific drug classes, we found that, for Black Americans, metformin users have reduced disease burden compared with sulfonylurea users (OR = 0.12, 95% CI 0.02–0.98, p = 0.048) but White individuals did not (OR = 0.76, 95% CI 0.53–1.07, p = 0.114) (Figure 6C). However, White Americans taking metformin versus TZD had a reduced disease burden (OR = 0.35, 95% CI 0.18–0.68, p = 0.002) but Black Americans did not (OR = 2.10, 95% CI 0.18–24.86, p = 0.558) (Figure 6D). These findings validate our network proximity drug repurposing analysis and indicate that metformin is a candidate medicine that may mitigate AF risk. However, the association between metformin use and decreased incidence of AF will require a randomized controlled trial with an ethnically diverse population to establish causality.
Discussion
Pharmacological management of AF is limited by adverse effects and toxicity rates of anti-arrhythmic drugs and poor long-term efficacy.12 Here we generated an AF disease network module and utilized a network-based approach to prioritize alternative drug options for AF (Figure 1). We showed that our mechanism-based protein-protein interactome can identify drug-AF associations. Specifically, we identified metformin as a high-confidence candidate for drug repurposing for AF using network proximity analysis of drug targets to the AF disease module and validate it using gene expression analysis of drug treatments in human cell lines and large-scale pharmacoepidemiologics analysis.
Recent advances in network medicine have enabled approaches for drug repurposing in cardiovascular diseases.25 We highlighted several improvements of our current study compared with previous network-based approaches.21,58 A strength of our study is the biorepository of LA tissue cohort of patients with AF and control individuals used to generate the AF disease module compared with traditional disease-associated gene approaches in the literature.36,58
We also validate the network proximity-based predictions using drug-gene signature-based enrichments. We posited that, if a drug significantly reverses dysregulated gene expression in human cell lines, then such a drug can potentially reverse gene neighborhood expression in AF disease modules derived from our LA tissues cohort of patients with AF. Finally, we validated a highly promising drug candidate (metformin) using large-scale EHR data.
We tested whether our transcriptomics-based disease module approach outperformed using traditional disease-associated gene-based approaches. We utilized 34 AF risk genes from the HGMD34 and identified 282 candidate drugs (Z < −1.0, p < 0.05). Using this approach, neither metformin (Z = 0.037, p = 0.419) nor phenformin (Z = −1.36, p = 0.116) passed the significance threshold for drug repurposing. The data suggest that human samples as well additional ES analyses can provide a higher-quality list of putative repurposed drug candidates for AF.
Several studies have provided strong evidence for the association of AF with metabolic syndrome diseases. Metformin is a first-line FDA-approved medication for type 2 diabetes mellitus (T2DM). The Framingham study showed a significant risk of developing AF in individuals with diabetes (OR = 1.4 for men and 1.6 for women, respectively) after multivariable adjustment.45 Several follow-up studies have strengthened this observation, showing an increased AF risk with longer disease duration or worse glycemic control.46,47 This evidence suggests that managing metabolic and inflammatory pathways in patients with AF may provide therapeutic benefits.
Metformin targets and activates AMPK, a master regulator of the metabolic stress response that senses AMP/ATP levels. The heterodimeric protein is composed of alpha (catalytic) and beta/gamma (regulatory) subunits encoded by genes such as PRKAA1, PRKAA2, and PRKAB1. PRKAA1 was present in the AF subnetwork.59 AMPK regulates glucose metabolism, fatty acid oxidation, and autophagy via mTORC1.60 AMPK also phosphorylates and inactivates acetyl-CoA carboxylase, a gene (ACACB) our network-based analysis identified as a target of metformin.61 Additionally, metformin suppresses DRP-1-mediated mitochondrial fission via an AMPK mechanism, reducing mitochondrial fragmentation in mice,62 promoting mitophagy to clear dysfunctional mitochondria,63 and reducing endoplasmic reticulum (ER) stress, reactive oxygen species production,64 and protein synthesis, which may reduce proteotoxic stress.65 Isoproterenol suppresses AMPK and can lead to cardiomyocyte apoptosis and ER stress, and metformin protects against this stress.66
Here we found that TGFB1, a significant DEG node in our AF network module, was downregulated following metformin treatment in a-iCMs. In clinical studies of patients with AF compared with patients with sinus rhythm, serum concentrations of tumor necrosis factor alpha (TNF-α) and TGFB1 were increased.67,68 TGFB1 is a well-known pro-fibrotic cytokine that promotes structural and electrical remodeling of the atria.69 Interstitial fibrosis promotes slow heterogeneous electrical conduction between myocytes, contributing to a substrate for reentrant electrical activity. TGFB1 may also affect calcium handling. Reduced L-type calcium channel currents (ICaL) and reduced expression of Cav1.2 (CACNA1C) following TGFB1 exposure have been reported in neonatal rat atrial myocytes but have not yet been reported in human atrial tissues.70 Figure S7 shows an inverse relation of mRNA for TGFB1 with CACNA1C in adult human LA tissues (n = 265, p < 0.001).
Furthermore, we found NPPB (brain natriuretic peptide [BNP]) and CXCL12 to be PPI neighbors of the metformin target DPP4 (Figure 4A). BNP can suppress the activity of the renin-aldosterone-angiotensin system (RAAS).71 Overstimulating the RAAS via angiotensin II has been clinically shown to promote localized oxidative stress by activating nuclear factor κB (NF-κB) and increasing production of interleukin-6 (IL-6) and C-reactive protein (CRP). As noted above, these inflammatory markers are elevated in patients with AF rhythm. The cytokine CXCL12, also a PPI neighbor of DPP4, is responsible for recruiting monocytes and lymphocytes. Systemic levels of macrophages and activated T lymphocytes are elevated in patients with persistent AF.72,73 These findings strengthen the powerful role of inflammation in AF disease etiology.
Still, one might wonder whether metformin’s benefit is due to its effect on metabolic gene expression, the pleiotropic benefits from long-term weight loss, or amelioration of metabolic syndrome.74, 75, 76, 77 It improves insulin resistance and inflammation/oxidative stress response mediated by free fatty acids, leptin, and other adipokines, which may target the pathophysiological link between obesity and AF. Using large-scale EHR data, we associated a significantly reduced risk of AF onset in diabetic patients taking metformin compared with other diabetic drugs, such as sulfonylurea and TZD. However, our pharmacoepidemiologic analysis did not show reduced risk in AF with GLP1RA or DPP4. There are several possible explanations. First, other GLP1RAs are a new-generation anti-T2DM medication with fewer users in our current EHR database compared with sulfonylurea and TZD (Table 1), which may be underpowered during pharmacoepidemiologic analysis. GLP1RA or DPP4 may have strong efficacy in reducing blood glucose levels and weight loss compared with metformin. This suggests that the metabolic effects of metformin may be associated with the utility against AF rather than beneficial outcomes of weight loss. Furthermore, we observed gender- and race-specific AF outcomes. Several reports indicate that AF is more common in males than females, but the prevalence of risk factors differs.50, 51, 52, 53 For example, diabetes has been observed as an AF risk factor for women but not men.53 We found that both male and female metformin users were significantly associated with reduced likelihood of AF, but female metformin users had higher protection. This could be explained by the differences in managing AF risk factors on the effect of AF disease. However, further work would be required to validate this. We also observed a greater reduction in AF risk among Black metformin users versus the combination comparator group. Further work is required to explain whether the reduced risk is environmental or genetic.
We also found that DPP4 is a metformin target responsible for regulating insulin secretion by antagonizing GLP1. Metformin had a more beneficial role than TZD and sulfonylureas, which are also responsible for regulating insulin secretion. This finding suggests that the multitarget effects of metformin are more effective than simply regulating cellular glucose utilization. This analysis was performed using data primarily from patients with diabetes because metformin is less prescribed for non-diabetic patients outside of clinical investigations, although it has been used for pre-diabetes. Activation of AMPK has been shown to improve cardiac function and, in turn, protect from AF.78,79 However, addition of in vitro functional testing on a-iCMs helps to address the potential for more direct effects of metformin in cardiomyocytes beyond potential indirect effects on HF, obesity, or other potential in vivo effects.
In an observational study of 645,710 T2DM subjects over a 13-year follow-up, metformin use was associated with 20% less AF.48 However, no prospective trials have reported metformin for AF in non-diabetics. Metformin has been proposed as upstream therapy in patients scheduled for AF ablation (ClinicalTrials.gov: NCT02931253). An ongoing clinical trial, Upstream Targeting for the Prevention of Atrial Fibrillation: Targeting Risk Interventions and Metformin for Atrial Fibrillation (TRIM-AF), is investigating the benefit of metformin and lifestyle/risk factor modification interventions in patients with AF (ClinicalTrials.gov: NCT03603912). Both interventions have been reported to target AMPK. Two recent studies have utilized healthy insurance claims data to test AF disease burden with metformin treatment. Using the International Business Machines Corporation (IBM) MarketScan Medicare Supplemental Database, Ostropolets et al.80 have reported that patients on metformin monotherapy had significantly reduced risk of atrial arrhythmias compared with monotherapy with DPP4 or TZD medications. In another study, Tseng81 used Taiwan’s National Health Insurance database and reported reduced AF-related hospitalizations in patients with newly diagnosed T2DM who took metformin versus those who did not. However, residual confounding may exist in these studies because insurance claims data are primarily collected for administrative purposes and do not contain detailed clinical information. Our pharmacoepidemiologics analysis, relying on very large patient-level EHR data, has several advantages. First, we use large-scale, longitudinal EHR data which contain various detailed clinical information for adjusting various possible confounding factors, including heart disease, vascular disease, renal disease, and many others (Table 1). We believe that there will be lower residual confounding risk in our EHR data analysis compared with previous health insurance claims databases.80,81
Limitations of the study
We acknowledge several potential limitations of the current study. The incompleteness of the human protein-protein interactome and drug-target networks may influence model performance. Disease heterogeneities of bulk transcriptomics data may influence the AF disease network module analysis. Integration of single-cell transcriptomics may help us to better understand AF disease heterogeneities and identify cell-type-specific drug targets and treatment approaches for AF. In addition, integration of AF genetics findings may help us to identify more likely risk genes and drug targets to accelerate effective therapeutic agent discovery for AF. Treatment of a-iCMs in a monolayer with short-term metformin exposure may not recapitulate long-term treatment in vivo or in the clinical setting, where patients with AF may have multiple comorbidities and exposure to multiple drugs. The TRIM-AF clinical trial (ClinicalTrials.gov: NCT03603912) includes a sub-study that will begin to address clinical response to metformin versus in vitro response in a-iCMs. The association of metformin use and decreased incidence of AF does not establish causality, which will require a randomized controlled trial in the future.
Conclusion
In summary, we have identified AF-specific dysregulated gene networks enriched in cardiac metabolism, ion transport, and immune pathways. Notably, we utilized multiple lines of complementary approaches, including LA tissue transcriptomics-based network module findings, network proximity, and drug-gene signature-based enrichment analysis to identify nine candidate drugs that are putative repurposed drugs for AF. Utilizing multiple lines of synergistic evidence, including in silico prediction from the AF disease module findings, in vitro testing from a-iCM models, and large-scale EHR data-based pharmacoepidemiologics analysis, we nominated metformin for functional validation and identified key molecular signals that help explain metformin’s mechanism of action. We believe that the network medicine approaches presented here, when broadly applied, would significantly catalyze effective treatment development for AF and other cardiovascular diseases.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Chemicals, peptides, and recombinant proteins | ||
| retinoic acid | Thermo Fisher | AC207341000 |
| lactate (L-lactic acid) | Wako Chemical | 129-02666 |
| Metformin | Sigma-Aldrich (now Millipore Sigma) | PHR1084 |
| Trizol | Invitrogen | 15596026 |
| Critical commercial assays | ||
| RNAeasy Micro Kit kit | Qiagen | 74004 |
| Qiashredder | Qiagen | 79654 |
| TruSeq RNA Sample Prep LT EUC | Illumina | Part # 15015050, Rev A |
| Heat Shock Factor 1 (HSF1) | ThermoFisher-Taqman expression assays | Assay ID Hs01027616_g1, cat# 4331182 |
| Heat Shock Factor 2 (HSF2) | ThermoFisher-Taqman expression assays | Assay ID Hs00988308_g1, cat# 4331182 |
| Heat Shock Protein 27/heat shock protein beta 1 (HSP27/HSBP1) | ThermoFisher-Taqman expression assays | Assay ID Hs00356629_g1, cat# 4331182 |
| Sodium Voltage-Gated Channel Alpha Subunit 5 (SCN5A) | ThermoFisher-Taqman expression assays | Assay ID Hs00165693_m1, cat# 4331182 |
| Calcium Voltage-Gated Channel Subunit Alpha1 C (CACNA1C) | ThermoFisher-Taqman expression assays | Assay ID Hs00167681_m1, cat# 4331182 |
| Calcium Voltage-Gated Channel Subunit Alpha1 D (CACNA1D) | ThermoFisher-Taqman expression assays | Assay ID Hs00167753_m1, cat# 4331182 |
| cardiac troponin T2 (TNNT2) | ThermoFisher-Taqman expression assays | Assay ID Hs00943911_m1, cat# 4448489 |
| TaqMan™ Gene Expression Master Mix | ThermoFisher Scientific/Applied Biosystems | cat# 4369016 |
| Deposited data | ||
| Gene Expression Omnibus database | https://www.ncbi.nlm.nih.gov/geo/ | GSE69890 |
| Experimental models: Cell lines | ||
| ATCC-BXS0116 Human Induced Pluripotent Stem (IPS) Cells | ATCC | ACS-1030 |
| Software and algorithms | ||
| Ingenuity Pathway AnalTOpysis (IPA®) software package | Qiagen | N/A |
| TopHat v2.0.4 | https://tophat.cbcb.umd.edu | N/A |
| Piccard tools | https://broadinstitute.github.io/picard/ | N/A |
| Kallisto | https://patcherlab.github.io/kallisto/ | N/A |
| limma v3.15 | R package (https://bioconductor.org/packages/release/bioc/html/limma.html) | N/A |
| cate v1.1.1 | R package (cran.r-project.org/web/packages/cate/index.html) | N/A |
| enrichR v3.0 | R package (https://cran.r-project.org/web/packages/enrichR/index.html) | N/A |
| sleuth v0.30 | R package (https://www.rdocumentation.org/packages/sleuth/versions/0.30.0) | N/A |
| tximport v1.24 | http://bioconductor.org/packages/release/bioc/html/tximport.html | N/A |
| Gephi v0.9.2 | https://gephi.org/ | N/A |
| Cytoscape v3.8.2 | https://cytoscape.org/ | N/A |
Resource availability
Lead contact
Further information and requests for resources should be directed to and will be fulfilled by the lead contact, Mina Chung (chungm@ccf.org).
Materials availability
This study did not generate new unique reagents.
Experimental model and subject details
Human subjects
Human LA tissues were obtained from 265 patients, including 251 who underwent elective cardiac surgery and 14 from non-failing unused transplant donors. Prior to 2008 verbal consent was obtained and documented in a process approved by the Cleveland Clinic Institutional Review Board. After this time written informed consent was obtained. Consent to use donor hearts was obtained from the family. The patient population, tissue processing, RNA isolation and sequencing have been previously reported.82 Briefly, human LA tissues were obtained from patients undergoing elective surgery to treat AF, valve disease, or other cardiac disorders. Specimens were snap frozen in liquid nitrogen and stored at −80°C until RNA extraction. LA tissue specimens were also obtained from nonfailing donor hearts not used for transplant. These hearts were perfused with cardioplegia before explant and processed in the same manner as hearts used for an organ transplant. As with the surgical specimens, donor tissue samples were snap frozen in liquid nitrogen and kept at −80°C until RNA extraction. Donor information was available for age, race, and gender (see Table S1).
Human induced pluripotent stem cell derived atrial cardiomyocytes
The iPSC line used in this experiment was obtained from the American Type Culture Collection (ATCC, ACS-1030). The iPSCs were reprogrammed from bone marrow derived CD34+ cells obtained from a healthy 31-year-old white female donor using Sendai viral expression of OCT4, SOX2, KLF4, and MYC. iPSCs are routinely used between passage 20–70. Pluripotency to form atrial-like cardiomyocytes (a-iCMs) was confirmed routinely by assessment of pluripotency genes (TRA-160, SSEA-4) and ability to differentiate and create beating cardiomyocytes. iPSCs were grown in E8 media, passaged every 3 days, and incubated at 37°C, 5% CO2, 98% relative humidity.83 Differentiation of a-iCMs was adapted from the protocol of Burridge et al, with the inclusion of retinoic acid (1 μmol/L) in the differentiation media from days 4–8 to enhance atrial-like gene expression.83, 84, 85 A metabolic selection step (no glucose, 4 mM lactate) was used from days 12–14 to limit the abundance of non-myocytes in the cultures. The media was returned to CDM3 from day 16 until day 20, followed by maintenance in CDM3-M media until the cells were used for experiments. Using this method, beating cells were typically evident by day 10, and beating sheets (electrically synchronized cell sheets covering the entire dish) by day 20. One week prior to the stimulation experiment, cells were dissociated, combined, and replated at a density of 150,000 per well in 12 well dishes. Experiments began after differentiation for 30 days and were completed in triplicate.
Method details
RNA isolation and sequencing of LA tissue
Fifty to 100 mg of LAA tissue was used to extract RNA. The tissue, in 1 mL of TRIzol (Invitrogen), was homogenized with sterile Omni Tip Disposable Generator Probes. RNA was isolated from the homogenate after the TRIzol protocol. The RNA pellet was dried and resuspended in 80 μL of RNase-free water, with concentration determined by OD260 nm and RNA stored at −80°C. Library generation for RNAseq was done at the University of Chicago Genomics Facility using standard Illumina protocols (Part No. 15015050, Rev A). Samples were filtered based on RNA quality as ascertained on an Agilent 2100 Bioanalyzer. Unstranded 100-bp paired-end sequencing was performed on the Illumina HiSeq 2000 platform and multiplexed to 6 samples across 2 lanes.
RNA isolation and sequencing of a-iCMs
RNA was isolated from the a-iCMs using the RNeasy Micro Kit (Qiagen). Cells from each treatment group were harvested three wells of a 12 well culture dish by pipetting with appropriate buffer from the kit. A Qiashredder (Qiagen) spin column was used for homogenization. Following the kit protocol, gDNA was removed from the sample with a gDNA eliminator spin column. Samples from each well were transferred to individual RNEasy spin columns and washed as instructed. RNA from each well was eluted from the column with 15 ul of RNase-free water. RNA concentrations were determined by OD260 nm and RNA was stored at −80°C.
cDNA preparation and RNA sequencing from a-iCMs
Libraries were constructed using lllumina oligo-dT based stranded kits following the recommended protocol. Approximately 60 million 100-basepair paired ends reads (30 million clusters) were generated for each library on an Illumina NovaSeq 6000 sequencer with a S1 flowcell. Both library construction and sequencing were done at the University of Chicago Genomics Facility. Tissue processing, RNA isolation and sequencing from the human atrial tissues was described in a prior publication.82
Quantitative RT-PCR
Quantitative reverse transcriptase-polymerase chain reaction (qRT-PCR) was performed using BioRad CRX Thermal Cycler as previously described.86 Briefly, 15 μL of master mix was pipetted into individual wells in a 96-well plate. 10 μL of the diluted cDNA was added to each well. 5 μL of the working mixture was pipetted in triplicate to a 384 well plate. Thermal cycling was performed with a hot start at 95°C for 10 min, followed by 40 cycles of 95°C for 15 s and 60°C for 60 s. Multiplexed Taqman primer/probes were used(Applied Biosystems/Thermofisher), delta C(t) values for: Heat Shock Factor 1 (HSF1), Heat Shock Factor 2 (HSF2), Heat Shock Protein 27/heat shock protein beta 1 (HSP27/HSBP1), Sodium Voltage-Gated Channel Alpha Subunit 5 (SCN5A), Calcium Voltage-Gated Channel Subunit Alpha1 C (CACNA1C) and Calcium Voltage-Gated Channel Subunit Alpha1 D (CACNA1D) expression levels were calculated relative to cardiac troponin T2 (TNNT2) expression. The ΔΔCT method was used to compare expression among samples yielding log2-based expression values, which were converted to fold-change values when indicated by calculating 2−ΔΔCT.86 Each assay was performed on 3 technical replicates from each of three biological replicates per treatment group.
Drug treatment
a-iCMs were pre-treated with 2.5 mM metformin in water for 6 h (n = 3). Cells in the presence of added water (vehicle) were used as controls (n = 3). All experimental combinations had 3 replicates.
Electrical pacing
Functional changes occurring during AF were assessed with our IonOptix C-Pace system for a-iCMs in monolayers grown in 12-well tissue culture plates. All wells were beating prior to and following pacing protocol. A-iCMs were paced at 1 Hz for 24 h in the presence of the drug. All experimental combinations had 3 replicates.
Quantification and statistical analysis
RNAseq analysis from human LA tissue
Samples were demultiplexed and aligned to hg19 using TopHat (v2.0.4)87 with the default options. Reads from exactly matched PCR duplicates were marked using Picard tools (https://broadinstitute.github.io/picard/) and excluded from further analysis. The sequence reads were mapped to the human genome to derive a digital count of the expression of genes, which were defined using the Ensembl (version 71) gene catalog. After preprocessing and quality control, transcript-level quantifications per sample were generated using kallisto with the Gencode version 27 human transcriptome.88 Gene-level quantifications were generated from these transcript-level estimates using the tximport package89 with the length scaled TPM option; after sample and gene expression level filtering, 19,816 genes measured on 265 RNAseq profiles from (distinct patient) LA tissues were used for DEG analyses. A description of the corresponding patient cohort was previously reported.82 Linear regression models per gene with empirical Bayes shrinkage90 as implemented in the limma R package were used to estimate and test the contrast between those whose hearts were in atrial fibrillation versus sinus rhythm at time of heart tissue acquisition (surgery or donor heart harvesting). The per-gene regression models included adjustments for gender, age (modeled as a 2 degree of freedom spline), white race, interactions of gender with age and gender with race, as well as 33 surrogate variables (SV) to help account for unmodeled confounders and heterogeneity. SVs were estimated using a latent high dimensional confounder approach91 as implemented in the cate R package (see Table S8).
Building the human protein-protein interactome
To build a comprehensive list of human PPI, we assembled data from 18 bioinformatics and systems biology databases compromised of five experimental assays: (i) binary PPIs tested by high-throughput yeast-two-hybrid (Y2H) systems; (ii) binary, physical PPIs from protein 3D structures; (iii) kinase-substrate interactions by literature-derived low throughput or high-throughput assays; (iv) signaling network by literature-derived low-throughput experiments; and (v) literature-curated PPIs identified by affinity purification followed by mass spectrometry (AP-MS), Y2H, or by literature-derived low-throughput assays. The genes were mapped to their ENTREZ ID based on the NCBI database92 as well as their official gene symbols on GeneCards (https://www.genecards.org/). The human protein-protein interactome used in this study includes 351,444 unique PPIs (edges or links) connecting 17,706 proteins (nodes).
Building the atrial fibrillation disease network
We used the DEGs from the human LA RNA-seq data (Table S8). We extracted the PPIs for the DEGs from the human interactome.
Pathway enrichment analysis
Gene Ontology (GO) and KEGG pathway enrichment analysis were used to identify pathways enriched in the AF disease module and metformin treated a-iCMs differentially expressed genes using the enrichR R package (version 3.0).93
Building the drug-target network
We collected drug-target interaction information from DrugBank database (accessed April 2021),94 Therapeutic Target Database (TTD).95 Drug-target interactions meeting the following three criteria were used: (i) binding affinities, including Ki, Kd, IC50, or EC50 each ≤10 μM; (ii) the target was marked as “reviewed” in the UniProt database96; and (iii) the human target was represented by a unique UniProt accession number. The final drug-target network contains 22,527 interactions among 2,939 drugs and 2,882 targets.
Network proximity measure
To quantify the relationship of the AF disease network and metformin treated a-iCMs DEGs, we adopted the closest path-based network proximity measure as below.
| (Equation 1) |
where d(c,t) is the shortest distance between gene c and t in the human protein interactome. The network proximity was normalized to Z-score based on permutation tests:
| (Eqaution 2) |
Where and were the mean and standard deviation of the permutation test was repeated 1000 times using two randomly selected gene lists with similar degree distributions to those of C and T. The p-value was calculated based on the permutation test results. Z < −1.0 and p < 0.05 were considered significant proximal AF disease associations. The networks were visualized using Gephi 0.9.2 (https://gephi.org/) or Cytoscape 3.8.2 (https://cytoscape.org/).
Gene set enrichment analysis
To further prioritize drugs identified by the drug-gene network analysis, we performed gene set enrichment analysis. Differential expressed genes from the AF disease module with p < 0.05 were used, as well as differential gene expression in human cell lines treated with various drugs downloaded from the CMAP database.33 To avoid bias, we queried DEGs of all cell lines (MCF7, PC3, HL60, and SKMEL5) provided in this version of CMAP. We took the average response of four cell lines for each drug. For each drug that was in both the CMAP data set and our drug-target network, we calculated an ES based on previously described methods97 as shown below:
| (Equation 3) |
ESup and ESdown were calculated for up- and down-regulated genes from the AF disease network signature. We first computed aup/down and bup/down using the following equations:
Where j = 1, 2, 3 …., s corresponds to the genes of the AF disease signature in ascending ranked order. V(j) corresponds to the rank of gene j, where , with r being the number of genes (12,849) from the drug profile. ES up/down were defined by:
| (Equation 4) |
Permutation tests were repeated 100 times using a random gene list with the same up- and down- regulated genes as the AF signature dataset used to measure the significance of the ES scores. Drugs were considered to have potential treatment effect if ES > 0 and p < 0.05.
RNAseq analysis from a-iCMs
RNAseq results from 6 wells of a-iCMs in the metformin versus water study were quality accessed using the nf-core/rnaseq pipeline.98 Transcript-level quantifications per sample were generated using kallisto with the Gencode version 31 human transcriptome.88 Transcripts were filtered by expression intensity using the default settings in the sleuth R package (version 0.30). Filtered transcript-level differential expression of Metformin versus Control was performed using Wald tests accounting for transcript uncertainty as implemented in the sleuth R package (sleuth_wt function).99 p-values from these transcript-level tests were aggregated within the corresponding genes using the Lancaster weighted (weighted by transcript relative intensity) combining method to get gene-level differential expression p-values for Metformin versus Control as implemented in the sleuth R package.100 Q-values101 were calculated from the gene-level p-values to obtain false discovery rate adjusted error values per gene (see Table S9).
Ingenuity Pathway Analysis
Upstream regulators and downstream target molecules were identified using the Ingenuity Pathway Analysis (IPA) software package (Qiagen). UniProtKB, p value, and log fold change values for DE-Gs with a p ≤ 0.05. Benjamini-Hochberg (BH) tests were used to determine the p value of association between genes and upstream regulators, where p < 0.05 was considered significant. Genes encoding proteins localized to the mitochondria and associated mitochondrial pathways were identified using MitoCarta 3.0100.
Pharmacoepidemiologic validation
The data for validation is from Northwestern Medicine Enterprise Data Warehouse (NMEDW),100 a data network containing medical and clinical data from 11 hospitals located in Illinois (https://www.nm.org/about-us/northwestern-medicine-newsroom/media-relations/about-our-health-system). We extracted the data from NMEDW between 2011 and 2021 for analysis (see Table 1).
To validate the effect of metformin on AF, besides the target metformin group, we also construct four comparator groups and the combination of the four groups for further comparison. The four comparator groups are sulfonylureas, TZD, DPP4, and GLP1RA. The groups including the metformin group and the 4 comparator groups correspond to drug usages in NMEDW defined by generic names and brand names (Table S3). For each group, we took three steps to get the final cohort, data extraction, filtering, and matching. In data extraction, we extracted all the patients who have the drug administered as recorded in NMEDW. The earliest drug administration date was set as index date. The outcome is the occurrence of AF (Atrial Fibrillation, diagnosis code defined in Table S4) within 180 days after the index date. In the filtering step, we followed the previous studies102 to exclude patients who have been diagnosed with AF or type I diabetes before the index date, who have been diagnosed with gestational diabetes within 1 year before the index date, and who have not been diagnosed with type II diabetes mellitus before the index date (diagnosis code defined in Table S4). For each comparison, to distinguish the effect between the target drug and the comparator drug, we also excluded the patients who have the comparator drug administered before the index date or within 180 days after the index date for the target drug group and made similar exclusion for the comparator drug group. Then, in the matching step, we executed a propensity score matching algorithm to control for potential confounding variables, including age, gender, race, and comorbidities. In propensity score matching, the propensity score is estimated by fitting a logit model where the outcome variable is a binary variable indicating whether the target drug is used, and the predictors are the potential confounding variables those are age, gender, race and comorbidities in our studies. By propensity score matching, each patient in the target group was paired with a patient from the comparator group with similar propensity score if the comparator group has more patients than the target group, and vice versa. Table S5 shows the total number of patients, as well as the number of outcome events in each group in each step. Logistic regression models were used to conduct inference for the odds ratio and the 95% confidence interval of the risk for developing AF when comparing treatment groups.
Acknowledgments
This work was supported by the National Heart, Lung, and Blood Institute of the National Institutes of Health (NIH) under awards R00HL138272 and K99HL138272 and the NIH National Institute on Aging under awards R01AG076448, R01AG066707, and U01AG073323 (to F.C.); NIH R01-HL111314 (to J.B., M.K.C., and D.R.V.W.); and P01HL158502 (to M.K.C., J.B., F.C., D.R.V.W., and J.S.). This work was also supported by the American Heart Association Atrial Fibrillation Strategically Focused Research Network grants 18SFRN34110067, 18SFRN34170013, and 18SFRN34170442 (to M.K.C., D.R.V.W., J.B., and J.S.), the NIH National Center for Research Resources for Case Western Reserve University and Cleveland Clinic Clinical and Translational Science award UL1-RR024989, Cleveland Clinic Department of Cardiovascular Medicine philanthropy research funds, the Tomsich Atrial Fibrillation Research Fund (to M.K.C.), and Howard Hughes Medical Institute Gilliam Fellowship GT14941 (to J.C.L. and F.C.).
Author contributions
F.C., M.K.C., Y.L., and D.R.V.W. conceived the study. J.C.L., C.M., Y.Z., S.R.G.-P., J.D.S., and J.H.R. performed all experiments and data analyses. J.B. contributed to differential expression analysis and helped to interpret the data analysis. J.C.L., C.M., F.C., D.R.V.W., and M.K.C. drafted the manuscript and critically revised the manuscript. All authors critically revised and gave final approval of the manuscript.
Declaration of interests
The authors declare no competing interests.
Inclusion and diversity
One or more of the authors of this paper self-identifies as an underrepresented ethnic minority in science. One or more of the authors of this paper received support from a program designed to increase minority representation in science.
Published: October 11, 2022
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.xcrm.2022.100749.
Contributor Information
Yuan Luo, Email: yuan.luo@northwestern.edu.
Feixiong Cheng, Email: chengf@ccf.org.
Mina K. Chung, Email: chungm@ccf.org.
Supplemental information
Data and code availability
-
•
The DEGs reported here are available as supplemental files (see Tables S8 and S9).
-
•
The codes written for and data used for network proximity analysis to reanalyze the data are publicly available from website. Github:https://github.com/ChengF-Lab/AFnetproximity.
-
•
Any additional information required to reanalyze the data reported in this work is available from the lead author upon request.
References
- 1.Michaud G.F., Stevenson W.G. Atrial fibrillation. N. Engl. J. Med. 2021;384:353–361. doi: 10.1056/NEJMcp2023658. [DOI] [PubMed] [Google Scholar]
- 2.Piccini J.P., Hammill B.G., Sinner M.F., Jensen P.N., Hernandez A.F., Heckbert S.R., Benjamin E.J., Curtis L.H. Incidence and prevalence of atrial fibrillation and associated mortality among Medicare beneficiaries, 1993-2007. Circ. Cardiovasc. Qual. Outcomes. 2012;5:85–93. doi: 10.1161/circoutcomes.111.962688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wilke T., Groth A., Mueller S., Pfannkuche M., Verheyen F., Linder R., Maywald U., Bauersachs R., Breithardt G. Incidence and prevalence of atrial fibrillation: an analysis based on 8.3 million patients. Europace. 2013;15:486–493. doi: 10.1093/europace/eus333. [DOI] [PubMed] [Google Scholar]
- 4.Benjamin E.J., Muntner P., Alonso A., Bittencourt M.S., Callaway C.W., Carson A.P., Chamberlain A.M., Chang A.R., Cheng S., Das S.R., et al. Heart disease and stroke statistics-2019 update: a report from the American heart association. Circulation. 2019;139:e56–e528. doi: 10.1161/cir.0000000000000659. [DOI] [PubMed] [Google Scholar]
- 5.Samol A., Masin M., Gellner R., Otte B., Pavenstädt H.J., Ringelstein E.B., Reinecke H., Waltenberger J., Kirchhof P. Prevalence of unknown atrial fibrillation in patients with risk factors. Europace. 2013;15:657–662. doi: 10.1093/europace/eus366. [DOI] [PubMed] [Google Scholar]
- 6.Benjamin E.J., Wolf P.A., D'Agostino R.B., Silbershatz H., Kannel W.B., Levy D. Impact of atrial fibrillation on the risk of death: the Framingham Heart Study. Circulation. 1998;98:946–952. doi: 10.1161/01.cir.98.10.946. [DOI] [PubMed] [Google Scholar]
- 7.Agarwal M., Apostolakis S., Lane D.A., Lip G.Y.H. The impact of heart failure and left ventricular dysfunction in predicting stroke, thromboembolism, and mortality in atrial fibrillation patients: a systematic Review. Clin. Therapeut. 2014;36:1135–1144. doi: 10.1016/j.clinthera.2014.07.015. [DOI] [PubMed] [Google Scholar]
- 8.Hindricks G., Potpara T., Dagres N., Arbelo E., Bax J.J., Blomström-Lundqvist C., Boriani G., Castella M., Dan G.-A., Dilaveris P.E., et al. 2020 ESC Guidelines for the diagnosis and management of atrial fibrillation developed in collaboration with the European Association for Cardio-Thoracic Surgery (EACTS): the Task Force for the diagnosis and management of atrial fibrillation of the European Society of Cardiology (ESC) Developed with the special contribution of the European Heart Rhythm Association (EHRA) of the ESC. Eur. Heart J. 2020;42:373–498. doi: 10.1093/eurheartj/ehaa612. [DOI] [PubMed] [Google Scholar]
- 9.Kim M.H., Johnston S.S., Chu B.C., Dalal M.R., Schulman K.L. Estimation of total incremental health care costs in patients with atrial fibrillation in the United States. Circ. Cardiovasc. Qual. Outcomes. 2011;4:313–320. doi: 10.1161/circoutcomes.110.958165. [DOI] [PubMed] [Google Scholar]
- 10.Prystowsky E.N., Benson D.W., Fuster V., Hart R.G., Kay G.N., Myerburg R.J., Naccarelli G.V., Wyse D.G. Management of patients with atrial fibrillation. Circulation. 1996;93:1262–1277. doi: 10.1161/01.CIR.93.6.1262. [DOI] [PubMed] [Google Scholar]
- 11.Pollak P. Clinical organ toxicity of antiarrhythmic compounds: ocular and pulmonary manifestations. Am. J. Cardiol. 1999;84:37–45. doi: 10.1016/S0002-9149(99)00700-6. [DOI] [PubMed] [Google Scholar]
- 12.Zimetbaum P. Antiarrhythmic drug therapy for atrial fibrillation. Circulation. 2012;125:381–389. doi: 10.1161/CIRCULATIONAHA.111.019927. [DOI] [PubMed] [Google Scholar]
- 13.Kottkamp H., Bender R., Berg J. Catheter ablation of atrial fibrillation. J. Am. Coll. Cardiol. 2015;65:196–206. doi: 10.1016/j.jacc.2014.10.034. [DOI] [PubMed] [Google Scholar]
- 14.Ouyang F., Antz M., Ernst S., Hachiya H., Mavrakis H., Deger F.T., Schaumann A., Chun J., Falk P., Hennig D., et al. Recovered pulmonary vein conduction as a dominant factor for recurrent atrial tachyarrhythmias after complete circular isolation of the pulmonary veins. Circulation. 2005;111:127–135. doi: 10.1161/01.CIR.0000151289.73085.36. [DOI] [PubMed] [Google Scholar]
- 15.Roselli C., Chaffin M.D., Weng L.C., Aeschbacher S., Ahlberg G., Albert C.M., Almgren P., Alonso A., Anderson C.D., Aragam K.G., et al. Multi-ethnic genome-wide association study for atrial fibrillation. Nat. Genet. 2018;50:1225–1233. doi: 10.1038/s41588-018-0133-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Menche J., Sharma A., Kitsak M., Ghiassian S.D., Vidal M., Loscalzo J., Barabási A.L. Disease networks. Uncovering disease-disease relationships through the incomplete interactome. Science. 2015;347:1257601. doi: 10.1126/science.1257601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Barabási A.L., Gulbahce N., Loscalzo J. Network medicine: a network-based approach to human disease. Nat. Rev. Genet. 2011;12:56–68. doi: 10.1038/nrg2918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fang J., Zhang P., Zhou Y., Chiang C.-W., Tan J., Hou Y., Stauffer S., Li L., Pieper A.A., Cummings J., Cheng F. Endophenotype-based in silico network medicine discovery combined with insurance record data mining identifies sildenafil as a candidate drug for Alzheimer’s disease. Nat. Aging. 2021;1:1175–1188. doi: 10.1038/s43587-021-00138-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhou Y., Hou Y., Shen J., Mehra R., Kallianpur A., Culver D.A., Gack M.U., Farha S., Zein J., Comhair S., et al. A network medicine approach to investigation and population-based validation of disease manifestations and drug repurposing for COVID-19. PLoS Biol. 2020;18:e3000970. doi: 10.1371/journal.pbio.3000970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhou Y., Hou Y., Shen J., Huang Y., Martin W., Cheng F. Network-based drug repurposing for novel coronavirus 2019-nCoV/SARS-CoV-2. Cell Discov. 2020;6:14. doi: 10.1038/s41421-020-0153-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Guney E., Menche J., Vidal M., Barábasi A.L. Network-based in silico drug efficacy screening. Nat. Commun. 2016;7:10331. doi: 10.1038/ncomms10331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Huang Y., Fang J., Lu W., Wang Z., Wang Q., Hou Y., Jiang X., Reizes O., Lathia J., Nussinov R., et al. A systems pharmacology approach uncovers wogonoside as an angiogenesis inhibitor of triple-negative breast cancer by targeting hedgehog signaling. Cell Chem. Biol. 2019;26:1143–1158.e6. doi: 10.1016/j.chembiol.2019.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fiscon G., Conte F., Amadio S., Volonté C., Paci P. Drug repurposing: a network-based approach to amyotrophic lateral sclerosis. Neurotherapeutics. 2021;18:1678–1691. doi: 10.1007/s13311-021-01064-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cheng F., Lu W., Liu C., Fang J., Hou Y., Handy D.E., Wang R., Zhao Y., Yang Y., Huang J., et al. A genome-wide positioning systems network algorithm for in silico drug repurposing. Nat. Commun. 2019;10:3476. doi: 10.1038/s41467-019-10744-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cheng F., Desai R.J., Handy D.E., Wang R., Schneeweiss S., Barabási A.L., Loscalzo J. Network-based approach to prediction and population-based validation of in silico drug repurposing. Nat. Commun. 2018;9:2691. doi: 10.1038/s41467-018-05116-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Morselli Gysi D., do Valle Í., Zitnik M., Ameli A., Gan X., Varol O., Ghiassian S.D., Patten J.J., Davey R.A., Loscalzo J., Barabási A.L. Network medicine framework for identifying drug-repurposing opportunities for COVID-19. Proc. Natl. Acad. Sci. USA. 2021;118 doi: 10.1073/pnas.2025581118. e2025581118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fiscon G., Conte F., Farina L., Paci P. SAveRUNNER: a network-based algorithm for drug repurposing and its application to COVID-19. PLoS Comput. Biol. 2021;17:e1008686. doi: 10.1371/journal.pcbi.1008686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cheng F., Kovács I.A., Barabási A.L. Network-based prediction of drug combinations. Nat. Commun. 2019;10:1197. doi: 10.1038/s41467-019-09186-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Greene J.A., Loscalzo J. Putting the patient back together - social medicine, network medicine, and the limits of reductionism. N. Engl. J. Med. 2017;377:2493–2499. doi: 10.1056/NEJMms1706744. [DOI] [PubMed] [Google Scholar]
- 30.Xu J., Zhang P., Huang Y., Zhou Y., Hou Y., Bekris L.M., Lathia J., Chiang C.W., Li L., Pieper A.A., et al. Multimodal single-cell/nucleus RNA sequencing data analysis uncovers molecular networks between disease-associated microglia and astrocytes with implications for drug repurposing in Alzheimer's disease. Genome Res. 2021;31:1900–1912. doi: 10.1101/gr.272484.120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cheng F., Zhao J., Wang Y., Lu W., Liu Z., Zhou Y., Martin W.R., Wang R., Huang J., Hao T., et al. Comprehensive characterization of protein–protein interactions perturbed by disease mutations. Nat. Genet. 2021;53:342–353. doi: 10.1038/s41588-020-00774-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wishart D.S., Knox C., Guo A.C., Shrivastava S., Hassanali M., Stothard P., Chang Z., Woolsey J. DrugBank: a comprehensive resource for in silico drug discovery and exploration. Nucleic Acids Res. 2006;34:D668–D672. doi: 10.1093/nar/gkj067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lamb J., Crawford E.D., Peck D., Modell J.W., Blat I.C., Wrobel M.J., Lerner J., Brunet J.P., Subramanian A., Ross K.N., et al. The Connectivity Map: using gene-expression signatures to connect small molecules, genes, and disease. Science. 2006;313:1929–1935. doi: 10.1126/science.1132939. [DOI] [PubMed] [Google Scholar]
- 34.Cooper D.N., Ball E.V., Krawczak M. The human gene mutation database. Nucleic Acids Res. 1998;26:285–287. doi: 10.1093/nar/26.1.285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Song E., Wang R.S., Leopold J.A., Loscalzo J. Network determinants of cardiovascular calcification and repositioned drug treatments. Faseb. J. 2020;34:11087–11100. doi: 10.1096/fj.202001062R. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Paci P., Fiscon G., Conte F., Wang R.S., Handy D.E., Farina L., Loscalzo J. Comprehensive network medicine-based drug repositioning via integration of therapeutic efficacy and side effects. NPJ Syst. Biol. Appl. 2022;8:12. doi: 10.1038/s41540-022-00221-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Shah S., Henry A., Roselli C., Lin H., Sveinbjörnsson G., Fatemifar G., Hedman Å.K., Wilk J.B., Morley M.P., Chaffin M.D., et al. Genome-wide association and Mendelian randomisation analysis provide insights into the pathogenesis of heart failure. Nat. Commun. 2020;11:163. doi: 10.1038/s41467-019-13690-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Diogo D., Tian C., Franklin C.S., Alanne-Kinnunen M., March M., Spencer C.C.A., Vangjeli C., Weale M.E., Mattsson H., Kilpeläinen E., et al. Phenome-wide association studies across large population cohorts support drug target validation. Nat. Commun. 2018;9:4285. doi: 10.1038/s41467-018-06540-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Divorty N., Milligan G., Graham D., Nicklin S.A. The orphan receptor GPR35 contributes to angiotensin II-induced hypertension and cardiac dysfunction in mice. Am. J. Hypertens. 2018;31:1049–1058. doi: 10.1093/ajh/hpy073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yang X., Li G., Yang T., Guan M., An N., Yang F., Dai Q., Zhong C., Luo C., Gao Y., et al. Possible susceptibility genes for intervention against chemotherapy-induced cardiotoxicity. Oxid. Med. Cell. Longev. 2020;2020:4894625. doi: 10.1155/2020/4894625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Shahid F., Lip G.Y.H., Shantsila E. Role of monocytes in heart failure and atrial fibrillation. J. Am. Heart Assoc. 2018;7:e007849. doi: 10.1161/JAHA.117.007849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Harada M., Van Wagoner D.R., Nattel S. Role of inflammation in atrial fibrillation pathophysiology and management. Circ. J. 2015;79:495–502. doi: 10.1253/circj.CJ-15-0138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yao C., Veleva T., Scott L., Jr., Cao S., Li L., Chen G., Jeyabal P., Pan X., Alsina K.M., Abu-Taha I., Dr, et al. Enhanced cardiomyocyte NLRP3 inflammasome signaling promotes atrial fibrillation. Circulation. 2018;138:2227–2242. doi: 10.1161/circulationaha.118.035202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rennison J.H., Li L., Lin C.R., Lovano B.S., Castel L., Wass S.Y., Cantlay C.C., McHale M., Gillinov A.M., Mehra R., et al. Atrial fibrillation rhythm is associated with marked changes in metabolic and myofibrillar protein expression in left atrial appendage. Pflügers Archiv. 2021;473:461–475. doi: 10.1007/s00424-021-02514-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Benjamin E.J., Levy D., Vaziri S.M., D'Agostino R.B., Belanger A.J., Wolf P.A. Independent risk factors for atrial fibrillation in a population-based cohort: the framingham heart study. JAMA. 1994;271:840–844. doi: 10.1001/jama.1994.03510350050036. [DOI] [PubMed] [Google Scholar]
- 46.Xiong Z., Liu T., Tse G., Gong M., Gladding P.A., Smaill B.H., Stiles M.K., Gillis A.M., Zhao J. A machine learning aided systematic Review and meta-analysis of the relative risk of atrial fibrillation in patients with diabetes mellitus. Front. Physiol. 2018;9:835. doi: 10.3389/fphys.2018.00835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Dublin S., Glazer N.L., Smith N.L., Psaty B.M., Lumley T., Wiggins K.L., Page R.L., Heckbert S.R. Diabetes mellitus, glycemic control, and risk of atrial fibrillation. J. Gen. Intern. Med. 2010;25:853–858. doi: 10.1007/s11606-010-1340-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Chang S.-H., Wu L.-S., Chiou M.-J., Liu J.-R., Yu K.-H., Kuo C.-F., Wen M.-S., Chen W.-J., Yeh Y.-H., See L.-C. Association of metformin with lower atrial fibrillation risk among patients with type 2 diabetes mellitus: a population-based dynamic cohort and in vitro studies. Cardiovasc. Diabetol. 2014;13:123. doi: 10.1186/s12933-014-0123-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Dyck J.R.B., Lopaschuk G.D. AMPK alterations in cardiac physiology and pathology: enemy or ally? J. Physiol. 2006;574:95–112. doi: 10.1113/jphysiol.2006.109389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ball J., Løchen M.L., Wilsgaard T., Schirmer H., Hopstock L.A., Morseth B., Mathiesen E.B., Njølstad I., Tiwari S., Sharashova E. Sex differences in the impact of body mass index on the risk of future atrial fibrillation: insights from the longitudinal population-based tromsø study. J. Am. Heart Assoc. 2018;7:e008414. doi: 10.1161/jaha.117.008414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Huxley R.R., Lopez F.L., Folsom A.R., Agarwal S.K., Loehr L.R., Soliman E.Z., Maclehose R., Konety S., Alonso A. Absolute and attributable risks of atrial fibrillation in relation to optimal and borderline risk factors: the Atherosclerosis Risk in Communities (ARIC) study. Circulation. 2011;123:1501–1508. doi: 10.1161/circulationaha.110.009035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Schnabel R.B., Yin X., Gona P., Larson M.G., Beiser A.S., McManus D.D., Newton-Cheh C., Lubitz S.A., Magnani J.W., Ellinor P.T., et al. 50 year trends in atrial fibrillation prevalence, incidence, risk factors, and mortality in the Framingham Heart Study: a cohort study. Lancet. 2015;386:154–162. doi: 10.1016/S0140-6736(14)61774-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Bose A., O'Neal W.T., Wu C., McClure L.A., Judd S.E., Howard V.J., Howard G., Soliman E.Z. Sex differences in risk factors for incident atrial fibrillation (from the reasons for geographic and racial differences in stroke [REGARDS] study) Am. J. Cardiol. 2019;123:1453–1457. doi: 10.1016/j.amjcard.2019.01.056. [DOI] [PubMed] [Google Scholar]
- 54.Lipworth L., Okafor H., Mumma M.T., Edwards T.L., Roden D.M., Blot W.J., Darbar D. Race-specific impact of atrial fibrillation risk factors in blacks and whites in the southern community cohort study. Am. J. Cardiol. 2012;110:1637–1642. doi: 10.1016/j.amjcard.2012.07.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Roberts J.D., Hu D., Heckbert S.R., Alonso A., Dewland T.A., Vittinghoff E., Liu Y., Psaty B.M., Olgin J.E., Magnani J.W., et al. Genetic investigation into the differential risk of atrial fibrillation among black and white individuals. JAMA Cardiol. 2016;1:442–450. doi: 10.1001/jamacardio.2016.1185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Delaney J.T., Jeff J.M., Brown N.J., Pretorius M., Okafor H.E., Darbar D., Roden D.M., Crawford D.C. Characterization of genome-wide association-identified variants for atrial fibrillation in African Americans. PLoS One. 2012;7:e32338. doi: 10.1371/journal.pone.0032338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Huang Y., Wang C., Yao Y., Zuo X., Chen S., Xu C., Zhang H., Lu Q., Chang L., Wang F., et al. Molecular basis of gene-gene interaction: cyclic cross-regulation of gene expression and post-GWAS gene-gene interaction involved in atrial fibrillation. PLoS Genet. 2015;11:e1005393. doi: 10.1371/journal.pgen.1005393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ghiassian S.D., Menche J., Barabási A.L. A DIseAse MOdule detection (DIAMOnD) algorithm derived from a systematic analysis of connectivity patterns of disease proteins in the human interactome. PLoS Comput. Biol. 2015;11:e1004120. doi: 10.1371/journal.pcbi.1004120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Oliveira S.M.J., Ehtisham J., Redwood C.S., Ostman-Smith I., Blair E.M., Watkins H. Mutation analysis of AMP-activated protein kinase subunits in inherited cardiomyopathies: implications for kinase function and disease pathogenesis. J. Mol. Cell. Cardiol. 2003;35:1251–1255. doi: 10.1016/s0022-2828(03)00237-2. [DOI] [PubMed] [Google Scholar]
- 60.Mihaylova M.M., Shaw R.J. The AMPK signalling pathway coordinates cell growth, autophagy and metabolism. Nat. Cell Biol. 2011;13:1016–1023. doi: 10.1038/ncb2329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Dyck J.R., Kudo N., Barr A.J., Davies S.P., Hardie D.G., Lopaschuk G.D. Phosphorylation control of cardiac acetyl-CoA carboxylase by cAMP-dependent protein kinase and 5'-AMP activated protein kinase. Eur. J. Biochem. 1999;262:184–190. doi: 10.1046/j.1432-1327.1999.00371.x. [DOI] [PubMed] [Google Scholar]
- 62.Wang Q., Zhang M., Torres G., Wu S., Ouyang C., Xie Z., Zou M.-H. Metformin suppresses diabetes-accelerated atherosclerosis via the inhibition of drp1-mediated mitochondrial fission. Diabetes. 2017;66:193–205. doi: 10.2337/db16-0915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Song Y.M., Lee W.K., Lee Y.H., Kang E.S., Cha B.S., Lee B.W. Metformin restores parkin-mediated mitophagy, suppressed by cytosolic p53. Int. J. Mol. Sci. 2016;17:E122. doi: 10.3390/ijms17010122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Li J., Wang Y., Wang Y., Wen X., Ma X.-N., Chen W., Huang F., Kou J., Qi L.-W., Liu B., Liu K. Pharmacological activation of AMPK prevents Drp1-mediated mitochondrial fission and alleviates endoplasmic reticulum stress-associated endothelial dysfunction. J. Mol. Cell. Cardiol. 2015;86:62–74. doi: 10.1016/j.yjmcc.2015.07.010. [DOI] [PubMed] [Google Scholar]
- 65.Dai S., Tang Z., Cao J., Zhou W., Li H., Sampson S., Dai C. Suppression of the HSF1-mediated proteotoxic stress response by the metabolic stress sensor AMPK. EMBO J. 2015;34:275–293. doi: 10.15252/embj.201489062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Zhuo X.Z., Wu Y., Ni Y.J., Liu J.H., Gong M., Wang X.H., Wei F., Wang T.Z., Yuan Z., Ma A.Q., Song P. Isoproterenol instigates cardiomyocyte apoptosis and heart failure via AMPK inactivation-mediated endoplasmic reticulum stress. Apoptosis. 2013;18:800–810. doi: 10.1007/s10495-013-0843-5. [DOI] [PubMed] [Google Scholar]
- 67.Zhao F., Zhang S., Shao Y., Wu Y., Qin J., Chen Y., Chen L., Gu H., Wang X., Huang C., Zhang W. Calreticulin overexpression correlates with integrin-α5 and transforming growth factor-β1 expression in the atria of patients with rheumatic valvular disease and atrial fibrillation. Int. J. Cardiol. 2013;168:2177–2185. doi: 10.1016/j.ijcard.2013.01.239. [DOI] [PubMed] [Google Scholar]
- 68.Wang Y., Hou X., Li Y. Association between transforming growth factor β1 polymorphisms and atrial fibrillation in essential hypertensive subjects. J. Biomed. Sci. 2010;17:23. doi: 10.1186/1423-0127-17-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Liew R., Khairunnisa K., Gu Y., Tee N., Yin N.O., Naylynn T.M., Moe K.T. Role of tumor necrosis factor-α in the pathogenesis of atrial fibrosis and development of an arrhythmogenic substrate. Circ. J. 2013;77:1171–1179. doi: 10.1253/circj.CJ-12-1155. [DOI] [PubMed] [Google Scholar]
- 70.Avila G., Medina I.M., Jiménez E., Elizondo G., Aguilar C.I. Transforming growth factor-beta1 decreases cardiac muscle L-type Ca2+ current and charge movement by acting on the Cav1.2 mRNA. Am. J. Physiol. Heart Circ. Physiol. 2007;292:H622–H631. doi: 10.1152/ajpheart.00781.2006. [DOI] [PubMed] [Google Scholar]
- 71.Cannone V., Ledwidge M., Watson C., McKie P.M., Burnett J.C., Jr., McDonald K. STOP-HF trial: higher endogenous BNP and cardiovascular protection in subjects at risk for heart failure. JACC. Basic Transl. Sci. 2021;6:497–504. doi: 10.1016/j.jacbts.2021.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Hu Y.F., Yeh H.I., Tsao H.M., Tai C.T., Lin Y.J., Chang S.L., Lo L.W., Tuan T.C., Tzeng C.H., Huang S.H., et al. Impact of circulating monocyte CD36 level on atrial fibrillation and subsequent catheter ablation. Heart Rhythm. 2011;8:650–656. doi: 10.1016/j.hrthm.2010.12.036. [DOI] [PubMed] [Google Scholar]
- 73.Liu L., Lee J., Fu G., Liu X., Wang H., Zhang Z., Zheng Q. Activation of peripheral blood CD3(+) T-lymphocytes in patients with atrial fibrillation. Int. Heart J. 2012;53:221–224. doi: 10.1536/ihj.53.221. [DOI] [PubMed] [Google Scholar]
- 74.Diabetes Prevention Program Research Group Long-term safety, tolerability, and weight loss associated with metformin in the Diabetes Prevention Program Outcomes Study. Diabetes Care. 2012;35:731–737. doi: 10.2337/dc11-1299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Cameron A.R., Morrison V.L., Levin D., Mohan M., Forteath C., Beall C., McNeilly A.D., Balfour D.J.K., Savinko T., Wong A.K.F., et al. Anti-inflammatory effects of metformin irrespective of diabetes status. Circ. Res. 2016;119:652–665. doi: 10.1161/circresaha.116.308445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Salpeter S.R., Buckley N.S., Kahn J.A., Salpeter E.E. Meta-analysis: metformin treatment in persons at risk for diabetes mellitus. Am. J. Med. 2008;121:149–157.e2. doi: 10.1016/j.amjmed.2007.09.016. [DOI] [PubMed] [Google Scholar]
- 77.Glueck C.J., Fontaine R.N., Wang P., Subbiah M.T., Weber K., Illig E., Streicher P., Sieve-Smith L., Tracy T.M., Lang J.E., McCullough P. Metformin reduces weight, centripetal obesity, insulin, leptin, and low-density lipoprotein cholesterol in nondiabetic, morbidly obese subjects with body mass index greater than 30. Metabolism. 2001;50:856–861. doi: 10.1053/meta.2001.24192. [DOI] [PubMed] [Google Scholar]
- 78.Gundewar S., Calvert J.W., Jha S., Toedt-Pingel I., Ji S.Y., Nunez D., Ramachandran A., Anaya-Cisneros M., Tian R., Lefer D.J. Activation of AMP-activated protein kinase by metformin improves left ventricular function and survival in heart failure. Circ. Res. 2009;104:403–411. doi: 10.1161/CIRCRESAHA.108.190918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Tong D., Schiattarella G.G., Jiang N., Daou D., Luo Y., Link M.S., Lavandero S., Gillette T.G., Hill J.A. Impaired AMP-activated protein kinase signaling in heart failure with preserved ejection fraction–associated atrial fibrillation. Circulation. 2022;146:73–76. doi: 10.1161/CIRCULATIONAHA.121.058301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Ostropolets A., Elias P.A., Reyes M.V., Wan E.Y., Pajvani U.B., Hripcsak G., Morrow J.P. Metformin is associated with a lower risk of atrial fibrillation and ventricular arrhythmias compared with sulfonylureas. Circ. Arrhythm. Electrophysiol. 2021;14:e009115. doi: 10.1161/CIRCEP.120.009115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Tseng C.-H. Metformin use is associated with a lower incidence of hospitalization for atrial fibrillation in patients with type 2 diabetes mellitus. Front. Med. 2021;7:592901. doi: 10.3389/fmed.2020.592901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Hsu J., Gore-Panter S., Tchou G., Castel L., Lovano B., Moravec C.S., Pettersson G.B., Roselli E.E., Gillinov A.M., McCurry K.R., et al. Genetic control of left atrial gene expression yields insights into the genetic susceptibility for atrial fibrillation. Circ. Genom. Precis. Med. 2018;11:e002107. doi: 10.1161/circgen.118.002107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Burridge P.W., Diecke S., Matsa E., Sharma A., Wu H., Wu J.C. Modeling cardiovascular diseases with patient-specific human pluripotent stem cell-derived cardiomyocytes. Methods Mol. Biol. 2016;1353:119–130. doi: 10.1007/7651_2015_196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Zhang Q., Jiang J., Han P., Yuan Q., Zhang J., Zhang X., Xu Y., Cao H., Meng Q., Chen L., et al. Direct differentiation of atrial and ventricular myocytes from human embryonic stem cells by alternating retinoid signals. Cell Res. 2011;21:579–587. doi: 10.1038/cr.2010.163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Wu S.P., Cheng C.M., Lanz R.B., Wang T., Respress J.L., Ather S., Chen W., Tsai S.J., Wehrens X.H.T., Tsai M.J., Tsai S.Y. Atrial identity is determined by a COUP-TFII regulatory network. Dev. Cell. 2013;25:417–426. doi: 10.1016/j.devcel.2013.04.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Gore-Panter S.R., Hsu J., Barnard J., Moravec C.S., Van Wagoner D.R., Chung M.K., Smith J.D. PANCR, the PITX2 adjacent noncoding RNA, is expressed in human left atria and regulates PITX2c expression. Circ. Arrhythm. Electrophysiol. 2016;9:e003197. doi: 10.1161/circep.115.003197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Trapnell C., Pachter L., Salzberg S.L. TopHat: discovering splice junctions with RNA-Seq. Bioinformatics. 2009;25:1105–1111. doi: 10.1093/bioinformatics/btp120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Bray N.L., Pimentel H., Melsted P., Pachter L. Near-optimal probabilistic RNA-seq quantification. Nat. Biotechnol. 2016;34:525–527. doi: 10.1038/nbt.3519. [DOI] [PubMed] [Google Scholar]
- 89.Soneson C., Love M.I., Robinson M.D. Differential analyses for RNA-seq: transcript-level estimates improve gene-level inferences. F1000Res. 2015;4:1521. doi: 10.12688/f1000research.7563.2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Ritchie M.E., Phipson B., Wu D., Hu Y., Law C.W., Shi W., Smyth G.K. Limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015;43:e47. doi: 10.1093/nar/gkv007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Wang J., Zhao Q., Hastie T., Owen A.B. Confounder adjustment in multiple hypothesis testing. Ann. Stat. 2017;45:1863–1894. doi: 10.1214/16-aos1511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.NCBI Resource Coordinators Database resources of the national center for biotechnology information. Nucleic Acids Res. 2016;44:D7–D19. doi: 10.1093/nar/gkv1290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Kuleshov M.V., Jones M.R., Rouillard A.D., Fernandez N.F., Duan Q., Wang Z., Koplev S., Jenkins S.L., Jagodnik K.M., Lachmann A., et al. Enrichr: a comprehensive gene set enrichment analysis web server 2016 update. Nucleic Acids Res. 2016;44:W90–W97. doi: 10.1093/nar/gkw377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Law V., Knox C., Djoumbou Y., Jewison T., Guo A.C., Liu Y., Maciejewski A., Arndt D., Wilson M., Neveu V., et al. DrugBank 4.0: shedding new light on drug metabolism. Nucleic Acids Res. 2014;42:D1091–D1097. doi: 10.1093/nar/gkt1068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Yang H., Qin C., Li Y.H., Tao L., Zhou J., Yu C.Y., Xu F., Chen Z., Zhu F., Chen Y.Z. Therapeutic target database update 2016: enriched resource for bench to clinical drug target and targeted pathway information. Nucleic Acids Res. 2016;44:D1069–D1074. doi: 10.1093/nar/gkv1230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.The UniProt Consortium UniProt: the universal protein knowledgebase. Nucleic Acids Res. 2017;45 doi: 10.1093/nar/gkw1099. D158–d169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Sirota M., Dudley J.T., Kim J., Chiang A.P., Morgan A.A., Sweet-Cordero A., Sage J., Butte A.J. Discovery and preclinical validation of drug indications using compendia of public gene expression data. Sci. Transl. Med. 2011;3:96ra77. doi: 10.1126/scitranslmed.3001318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Ewels P.A., Peltzer A., Fillinger S., Patel H., Alneberg J., Wilm A., Garcia M.U., Di Tommaso P., Nahnsen S. The nf-core framework for community-curated bioinformatics pipelines. Nat. Biotechnol. 2020;38:276–278. doi: 10.1038/s41587-020-0439-x. [DOI] [PubMed] [Google Scholar]
- 99.Rath S., Sharma R., Gupta R., Ast T., Chan C., Durham T.J., Goodman R.P., Grabarek Z., Haas M.E., Hung W.H.W., et al. MitoCarta3.0: an updated mitochondrial proteome now with sub-organelle localization and pathway annotations. Nucleic Acids Res. 2021;49 doi: 10.1093/nar/gkaa1011. D1541–d1547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Starren J.B., Winter A.Q., Lloyd-Jones D.M. Enabling a learning health system through a unified Enterprise data Warehouse: the experience of the northwestern university clinical and translational sciences (NUCATS) Institute. Clin. Transl. Sci. 2015;8:269–271. doi: 10.1111/cts.12294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Storey J.D., Tibshirani R. Statistical significance for genomewide studies. Proc. Natl. Acad. Sci. USA. 2003;100:9440–9445. doi: 10.1073/pnas.1530509100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Ostropolets A., Elias P.A., Reyes M.V., Wan E.Y., Pajvani U.B., Hripcsak G., Morrow J.P. Metformin is associated with a lower risk of atrial fibrillation and ventricular arrhythmias compared with sulfonylureas: an observational study. Circ. Arrhythm. Electrophysiol. 2021;14:e009115. doi: 10.1161/circep.120.009115. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
-
•
The DEGs reported here are available as supplemental files (see Tables S8 and S9).
-
•
The codes written for and data used for network proximity analysis to reanalyze the data are publicly available from website. Github:https://github.com/ChengF-Lab/AFnetproximity.
-
•
Any additional information required to reanalyze the data reported in this work is available from the lead author upon request.