Summary
Programmed cell death 1 (PD1) and cytotoxic T lymphocyte-associated protein 4 (CTLA4) suppress CD4+ T cell activation and may promote latent HIV infection. By performing leukapheresis (n = 21) and lymph node biopsies (n = 8) in people with HIV on antiretroviral therapy (ART) and sorting memory CD4+ T cells into subsets based on PD1/CTLA4 expression, we investigate the role of PD1 and CTLA 4 in HIV persistence. We show that double-positive (PD1+CTLA4+) cells in blood contain more HIV DNA compared with double-negative (PD1−CTLA4−) cells but still have a lower proportion of cells producing multiply spliced HIV RNA after stimulation as well as reduced upregulation of T cell activation and proliferation markers. Transcriptomics analyses identify differential expression of key genes regulating T cell activation and proliferation with MAF, KLRB1, and TIGIT being upregulated in double-positive compared with double-negative cells, whereas FOS is downregulated. We conclude that, in addition to being enriched for HIV DNA, double-positive cells are characterized by negative signaling and a reduced capacity to respond to stimulation, favoring HIV latency.
Keywords: HIV, HIV latency, HIV persistence, HIV cure, HIV reservoirs, PD1, CTLA4, immune checkpoints, T cell exhaustion
Graphical abstract

Highlights
-
•
CD4+ T cells co-expressing PD1 and CTLA4 (double positive [DP]) are enriched for HIV DNA
-
•
DP cells contain virus that is more resistant to stimulation
-
•
DP cells display differential expression of genes regulating T cell activation
-
•
These features favor persistence of HIV latency in cells co-expressing PD1 and CTLA4
Memory CD4+ T cells co-expressing PD1 and CTLA4 are enriched for HIV DNA but have a reduced response to stimulation, including limited induction of latent HIV. This may be explained by differential expression of key genes regulating T cell activation and proliferation in CD4+ T cells double positive for PD1 and CTLA4.
Introduction
Combination antiretroviral therapy (ART) for people with HIV (PWH) has provided major benefits by suppressing HIV replication, restoring immune function, and reducing HIV-related morbidity and mortality, but lifelong treatment is required to maintain virus suppression.1 This is due to the long-term persistence of latent HIV in long-lived and proliferating CD4+ T cells from which HIV rapidly rebounds when ART is stopped. Understanding where and how latent HIV infection persists on suppressive ART is fundamentally important for developing curative strategies. Because latently infected cells constitute the main barrier to a cure, identifying specific cellular subsets that preferentially favor latent infection is of great importance and may reveal novel therapeutic targets.
Although previous studies have shown enrichment of HIV within central memory (Tcm), transitional memory (Ttm), and stem cell memory (Tscm) CD4+ T cells,2, 3, 4 other studies have demonstrated the role of immune checkpoint proteins for establishment of latent infection.5 Immune checkpoints constitute a functional network of co-stimulatory and co-inhibitory receptors that play a central role in maintaining a balance between T cell activation and autoimmunity.6 The co-inhibitory receptors programmed cell death 1 (PD1) and cytotoxic T-lymphocyte associated protein 4 (CTLA4) are key receptors of this network. Both are expressed at higher levels on CD4+ and CD8+ T cells during chronic HIV infection and mediate negative T cell signaling that drives HIV-specific T cell functional exhaustion.7, 8, 9, 10
In addition to their role in HIV-associated immune exhaustion,7,8 it has been shown that CD4+ T cells expressing certain immune checkpoints, in particular PD1, are enriched for HIV in PWH on ART. Tcm and Ttm cells that express PD1 have been shown initially to contain higher levels of HIV DNA than their PD1− counterparts.2 It has also been demonstrated that increased expression of PD1 and other immune checkpoints—lymphocyte activation gene 3 (LAG3) and T cell immunoglobulin and ITIM domain (TIGIT)—on CD4+ T cells correlates with a higher frequency of integrated HIV DNA and that CD4+ T cells co-expressing these markers are highly enriched for HIV.11,12 The role of CTLA4 or PD1/CTLA4 co-expression in HIV persistence in PWH on ART has not been investigated previously.
Understanding the contribution of immune checkpoint proteins to HIV persistence in cellular subsets in lymph node (LN) tissue is critically important because most T cells reside in lymphoid tissue and are functionally distinct from those in blood.13 B cell follicles in LNs may provide a sanctuary for HIV-infected cells,14 and not all antiretroviral drugs achieve high concentrations in LNs.15 PD1+ and CTLA4+ CD4+ T cells are highly frequent in LN tissue. In LN tissue from PWH on suppressive ART, CD4+ T cells expressing PD1, the majority of which are follicular helper T cells (Tfh), constituted the major source of replication-competent HIV, but the contribution of these cells to the pool of persistent HIV decreased with time on ART.16 In ART-treated simian immunodeficiency virus (SIV)-infected rhesus macaques, a subset of CTLA4-expressing memory CD4+ T cells in LNs significantly contributed to viral persistence,17 but whether this also occurs in PWH remains unclear. Finally, PD1 and CTLA4 can be targeted therapeutically by monoclonal antibodies, which are now widely used in cancer treatment.18
To investigate the role of PD1 and CTLA4 in HIV persistence on ART, we enrolled PWH on suppressive ART and performed leukapheresis and LN biopsies to enable sorting of memory CD4+ T cells into distinct subsets based on expression of PD1 and/or CTLA4. We show that double-positive (PD1+CTLA4+) compared with double-negative (PD1−CTA4−) cells in blood had a higher frequency of HIV DNA. In addition to being enriched for HIV infection, double-positive cells displayed a reduced capacity to respond to stimulation, including reduced induction of latent HIV, favoring latent infection. Single-cell transcriptomics analyses identified differential expression of key genes regulating T cell activation and proliferation, which may explain why HIV latency is favored in double-positive cells.
Results
Study participants and clinical details
We enrolled 21 PWH older than 18 years who had been on suppressive ART for at least 3 years at clinical trial sites in San Francisco, California, USA and Melbourne, Victoria, Australia. Eleven participants enrolled in Melbourne consented to leukapheresis and excisional biopsy of an inguinal LN under general anesthesia, but one participant withdrew consent prior to the procedures. 11 participants recruited at the University of California, San Francisco (UCSF), consented to leukapheresis. Four of ten participants (40%) who underwent excisional LN biopsy developed seromas at the surgical site 7–17 days after the procedure, which all resolved without further intervention after 4–8 weeks. One participant developed superficial cellulitis of the biopsy incision site 1 day after LN biopsy. The infection fully resolved after 1 week of treatment with oral flucloxacillin, and localized seromas had resolved after 7 weeks. In two participants, the excised LN tissue did not contain any CD3+ T cells and could not be used for the study. We therefore analyzed LN and leukapheresis samples from 8 individuals and leukapheresis samples alone from 13 individuals. All study participants were male and had plasma HIV RNA below the detection limit of the utilized assay (<20 or <40 copies/mL) at study entry. The mean duration of suppressed plasma HIV RNA prior to study entry was 9.4 years (95% confidence interval [CI] 8.1–10.8), and the mean nadir CD4+ T cell count was 222 cells/μL (95% CI 138–305). Additional clinical characteristics are shown in Table S1.
Distribution of PD1+ and CTLA4+ subsets in blood and LNs
The number of peripheral blood mononuclear cells (PBMCs) collected by leukapheresis ranged from 3.2–17.2 billion cells, and the number of LN mononuclear cells (LNMCs) acquired from LN tissue ranged from 6.1–137.0 million cells. We isolated CD45RA− memory CD4+ T cells using negative selection and sorted memory CD4+ T cells into four populations based on their expression of PD1 and CTLA4: PD1−CTLA4− (double-negative), PD1+CTLA4−, PD1−CTLA4+, and PD1+CTLA4+ (double-positive) memory CD4+ T cell subsets (Figure S1). We used flow cytometry to assess the distribution of these four subsets in non-sorted memory CD4+ T cells. This showed that, in blood and LNs, PD1+CTLA4− cells followed by double-negative cells were far more abundant than any of the CTLA4+ subsets with the exception that there was no difference in relative frequency of double-negative and double-positive cells in LNs (Figures 1A and 1B). Frequencies of double-negative cells were higher among blood memory CD4+ T cells compared with LN memory CD4+ T cells, whereas double-positive cells were more frequent in LNs compared with blood (Figure 1C).
Figure 1.
Distribution of memory CD4+ T cells expressing PD1 and/or CTLA4
(A–C) The frequency of PD1/CTLA4 subsets within memory CD4+ T cells in blood (A) (n = 21) and LNs (B) (n = 8), including pairwise comparison of the frequency of each subset in blood versus LNs cells (C). Horizontal bars indicate median levels. Statistical comparisons were done using paired t test with or without log transformation as required to achieve normal distribution.
CTLA4 is commonly expressed on regulatory T (Treg), cells, which have been shown previously to be enriched for HIV in ART-treated PWH19,20 and SIV-infected rhesus macaques.17 To determine the relationship of CTLA4+ and PD1 to markers of Treg cells, we assessed the expression of CD25 and FoxP3 in addition to PD1 and CTLA4 in three uninfected donors and three PWH on suppressive ART. As expected, Treg cells constituted a considerable proportion (15%–20%) of PD1−CTLA4+ cells, whereas Treg cells contributed less to double-positive cells and were almost absent in CTLA4− subsets (Figure S2A). Conversely, around 80% of Treg cells express CTLA4 (Figure S2B).
Memory CD4+ T cells that co-express PD1 and CTLA4 are enriched for HIV-infected cells in blood but not in LNs
To address whether HIV was enriched within any of the PD1/CTLA4 memory CD4+ T cell subsets, we quantified the frequency of cells containing total HIV DNA and cell-associated unspliced HIV RNA (CA-US HIV RNA). CA-US HIV RNA measures initiation of HIV transcription on ART,21,22 whereas cell-associated multiply spliced HIV RNA (CA-MS HIV RNA) measures spliced RNA products a later step in virus transcription.23 CA-US HIV RNA is detected in nearly all people on suppressive ART. It was therefore chosen as the primary measure for the level of cell-associated HIV RNA. In blood memory CD4+ T cells, we found modest but statistically significant differences in the content of HIV DNA across the four subsets, as determined by a Friedman test (p = 0.016; Figure 2A). Using Wilcoxon matched-pairs signed rank tests, we found that total HIV DNA levels were higher in double-positive compared with double-negative memory CD4+ T cells in blood (p = 0.018; Figure 2A). The direct comparison between these two subsets revealed that, in 17 of 21 participants, the level of total HIV DNA was higher in double-positive compared with double-negative blood memory CD4+ T cells (Figure 2B), corresponding to a median 1.8-fold (interquartile range [IQR] 1.1–2.5) higher level of total HIV DNA in double-positive compared with double-negative cells (Figure 2C). The level of total HIV DNA in PD1−CTLA4+ cells was similar to that seen in double-positive cells (Figure 2A), with total HIV DNA being a median 1.2-fold (IQR 0.7–1.9) higher in PD1−CTLA4+ compared with double-negative cells, but this difference was not statistically significant (Figure 2C). In contrast to HIV DNA, we did not find any difference in levels of CA-US HIV RNA across memory CD4+ T cell PD1/CTLA4 subsets in blood (Figure 2D), nor was there any difference in the ratio of CA-US HIV RNA to total HIV DNA (Figure 2E).
Figure 2.
HIV RNA and DNA in blood memory CD4+ T cells expressing PD1 and/or CTLA4
(A–E) The frequency of blood memory CD4+ T cells containing total HIV DNA across PD1/CTLA4 subsets (A), pairwise comparison of the level of total HIV DNA in double-positive versus double-negative cells (B), ratio of total HIV DNA in PD1/CTLA4 subsets compared with double-negative cells (C), the level of cell-associated unspliced HIV RNA (CA-US HIV RNA) (D), and the ratio of CA-US HIV RNA to total HIV DNA (E). Samples from 21 participants are analyzed, with each estimate of HIV DNA and CA-US HIV RNA being the average of three and four technical replicates, respectively. Horizontal bars indicate median levels. Statistical comparisons were performed using Friedman’s test and Wilcoxon signed-rank test.
When we assessed LNMCs from 8 participants, we did not detect enrichment of HIV DNA or RNA within any of the PD1/CTLA4 subsets (Figures 3A and 3B), nor were there any differences in the ratio of CA-US HIV RNA to HIV DNA (Figure 3C). We also directly compared the level of HIV DNA and CA-US HIV RNA and the ratio of CA-US HIV RNA with HIV DNA within each subset between LNs and blood but did not detect any differences across these two compartments (Figures 3D–3F).
Figure 3.
HIV RNA and DNA in LN memory CD4+ T cells expressing PD1 and/or CTLA4
(A–C) The frequency of LN memory CD4+ T cells containing total HIV DNA (A), cell-associated unspliced HIV RNA (CA-US HIV RNA) (B), and the ratio of CA-US HIV RNA to total HIV DNA (C) across PD1/CTLA4 subsets.
(D–F) Pairwise comparisons between blood and LN PD1/CTLA4 memory CD4+ T cell subsets in the level of total HIV DNA (D), cell-associated unspliced HIV RNA (CA-US HIV RNA) (E), and the ratio of CA-US HIV RNA to total HIV DNA (F).
Samples from 8 participants are analyzed, with each estimate of HIV DNA and CA-US HIV RNA being the average of three and four technical replicates, respectively. Horizontal bars indicate median levels. Statistical comparisons were performed using paired t test on log-transformed data.
These data demonstrate that blood memory CD4+ T cells that co-express PD1 and CTLA4 are enriched for total HIV DNA, but the same enrichment was not found in LN tissue.
CTLA4− cells are the major contributors to the total pool of HIV DNA in blood and LNs
To determine the relative contribution from each of the PD1/CTLA4 subsets to the total pool of memory CD4+ T cells harboring HIV DNA or CA-US HIV RNA, we adjusted for the frequency of each subset in blood and LN as determined by flow cytometry (Figure 1). This revealed that, in blood, the double-negative and PD1+CTLA4− subsets were the major contributors to the total pool of cells carrying HIV DNA and CA-US HIV RNA (Figures 4A and 4B). A similar pattern was observed in LNs, but because memory CD4+ T cells co-expressing PD1 and CTLA4 were more frequent in LNs compared with blood, these cells provided a larger contribution to the total pool of HIV DNA and CA-US HIV RNA in LNs (Figures 4C and 4D).
Figure 4.
Contribution to the total pool of cell-associated HIV DNA and RNA in blood and LNs
(A–D) The percentage contribution to the total pool of total HIV DNA (A) and cell-associated HIV RNA (B) within peripheral blood memory CD4+ T cells and the percentage contribution to the total pool of total HIV DNA (C) and cell-associated HIV RNA (D) within LN memory CD4+ T cells. Samples from 21 (blood) and 8 (LN) participants are analyzed, with each estimate of HIV DNA and CA-US HIV RNA being the average of three and four technical replicates, respectively. Horizontal bars indicate mean percentage contribution. CA-US HIV RNA, cell-associated unspliced HIV RNA. Statistical comparisons were performed using Wilcoxon signed-rank test.
These data emphasize that the relative abundance of exhausted T cell subsets is a key determinant of the relative contribution to the latent and active HIV reservoir.
Higher CD8+ T cell counts correlate with a higher frequency of HIV DNA but not in cells that co-express PD1 and CTLA4
Previous studies have shown that the frequency of infected PD1+ CD4+ T cells in LN tissue was lower in PWH who had been on ART for a longer duration.16 In contrast, in SIV-infected non-human primates on suppressive ART, PD1−CTLA4+CD4+ infected T cells increased with the duration of viral suppression, consistent with enhanced survival or persistence of this subset.17 To investigate factors associated with the distribution of HIV DNA within blood and LN memory CD4+ T cell subsets, we analyzed the association between clinical characteristics recorded at study entry and the frequencies of HIV DNA and CA-US HIV RNA in each cellular subset.
We found that CD4+ T cell count, CD4+ T cell percentage, CD4+ nadir, and CD4+/CD8+ ratio generally showed negative but non-significant correlations with the frequency of HIV DNA within PD1/CTLA4 subsets (Figure S3A). In contrast, duration of viral load suppression, time since HIV diagnosis, and peak viral load pre-ART trended toward positive correlations but were also mostly non-significant (Figure S3A). The strongest and most consistently significant correlations with infection frequencies in these subsets were seen with CD8+ T cell count and CD8+ T cell percentage (Figure S3A). Higher CD8+ T cell counts correlated with a higher frequency of HIV DNA in blood memory CD4+ T cells but only in double-negative and PD1−CTLA4+ cells, whereas no association was seen in cells that co-expressed PD1 and CTLA4 (Figure S3B). In fact, we found that CD8+ T cell counts were negatively correlated with the ratio of HIV DNA in double-positive versus double-negative cells, indicating that a lower CD8+ T cell count was associated with enriched levels of HIV DNA in double-positive compared with double-negative cells. CD8+ T cell count was also significantly associated with the frequency of HIV DNA in LN memory CD4+ T cell PD1/CTLA4 subsets, but this was driven by a single outlier, and significance was lost when this was removed from analysis (data not shown).
These data identified CD8+ T cell count and percentage as the parameters most strongly associated with the frequency of HIV-infected cells in the blood, and the correlation differed across subsets of exhausted memory CD4+ T cells.
Despite being enriched for HIV, a lower proportion of double-positive memory CD4+ T cells in blood produced CA-MS HIV RNA upon stimulation
Although quantification of total HIV DNA provides a measure of the total frequency of cells that contain HIV, most HIV proviruses are defective and cannot give rise to infectious virus particles.24 To address whether the frequency of cells containing functional virus might differ across PD1/CTLA4 subsets in memory CD4+ T cells, we quantified the expression of CA-MS HIV RNA as well as the proportion of cells that could be induced to produce CA-MS HIV RNA by employing the tat/rev induced limiting dilution assay (TILDA).25 CA-MS transcripts are usually absent in latently infected cells but produced upon viral reactivation.25 Constitutive expression of CA-MS HIV RNA was rarely detected in blood and LN memory CD4+ T cell subsets, although, within double-negative memory CD4+ T cells in blood, CA-MS RNA was detected in 9 of 21 participants at a frequency ranging from 0.25–9.51 copies per million cells (Figures 5A and 5B).
Figure 5.
Multiply spliced HIV RNA in blood and LN memory CD4+ T cells expressing PD1 and/or CTLA4
(A–D) The level of cell-associated, multiply spliced HIV RNA (CA-MS HIV RNA) in PD1/CTLA4 subsets in peripheral blood (A) and LN tissue (B) and the proportion of cells in blood that could be induced to produce CA-MS HIV RNA using the tat/rev limiting dilution assay (TILDA) in double-negative versus double-positive blood memory CD4+ T cells (C) and in double-negative versus PD1−CTLA+ memory CD4+ T cells (D). Samples from 20 (blood) and 5 (LN) participants are analyzed (fewer for some subsets, as indicated in the figures), with each estimate of CA-MS HIV RNA being the average of four technical replicates. Statistical comparisons were performed using Wilcoxon signed-rank test.
We then performed TILDA in blood memory CD4+ T cell subsets where enough cells were available. This could be done in the CTLA4− subsets for 20 participants, in PD1−CTLA4+ cells for 7 participants, and in double-positive cells for 9 participants. Because of the limited number of cells available, TILDA could not be performed in LN PD1/CTLA4 subsets. In paired analyses, we unexpectedly found that the proportion of cells producing CA-MS HIV RNA upon stimulation was higher for double-negative cells than for double-positive cells (p = 0.023; Figure 5C) even though the latter was found to contain total HIV DNA at a higher frequency (Figures 2A–2C). The proportion of PD1−CTLA4+ cells that produced CA-MS HIV RNA upon stimulation tended to be lower than in the double-negative subset, but this comparison did not reach statistical significance (p = 0.063; Figure 5D).
To investigate whether the lower proportion of double-positive cells with inducible CA-MS HIV RNA could be explained by a reduced capacity to respond to stimulation of cells expressing PD1 and CTLA4, we stimulated double-negative, double-positive, and PD1+CTLA4− memory CD4+ T cells from blood with PMA/ionomycin or DMSO for 24 and 72 h and quantified markers of T cell activation (CD38, HLA-DR, and CD69), T cell proliferation (Ki67), and cell death (live/dead stain). We primarily focused on the data from the 72-h stimulation to allow enough time for upregulation of late-phase activation markers and Ki67 expression and to fully appreciate effects on cell death. To avoid too much cell death with this extended stimulation, we used lower concentrations of PMA and ionomycin than what is used in TILDA. T cell activation and proliferation markers were higher in double-positive compared with double-negative cells in the absence of stimulation (Figures S4A–S4E). After stimulation, expression of Ki67 was lower in double-positive compared with double-negative cells (Figures S4A–S4E), whereas rates of cell death were higher in double-positive cells after 72 h in culture with or without stimulation (Figure S4F). The most striking contrast between double-negative and double-positive cells was seen when we compared the fold change in expression of activation and proliferation markers after PMA/ionomycin stimulation relative to DMSO. Although the small sample size (n = 4) in this experiment warrants cautious interpretation, these data indicated a marked reduction in the capacity of double-positive cells to upregulate markers of T cell activation compared with double-negative cells, whereas the rate of cell death was comparable across PD1/CTLA4 subsets (Figure 6).
Figure 6.
Induction of T cell activation, proliferation, and death after stimulation with PMA/ionomycin relative to DMSO
(A–F) Fold change relative to DMSO in the expression of the T cell activation markers CD38 (A), HLA-DR (B), CD69 (C), co-expression of HLA-DR and CD38 (D), the proliferation marker Ki67 (E), and the relative change compared with DMSO conditions in live cells (F). Samples from four participants are analyzed with no technical replicates. Horizontal bars indicate median fold change. No formal statistical comparisons were performed, given the low sample size.
These data indicate that, although blood memory CD4+ T cells co-expressing PD1 and CTLA4 are modestly enriched for total HIV DNA, they produce CA-MS HIV RNA at a lower frequency upon stimulation compared with their double-negative counterparts. This may be explained by lower responsiveness to stimulation, possibly because of the inhibitory signaling mediated by PD1 and CTLA4.
CD4+ T cells that are double positive for PD1 and CTLA4 display distinct differences in their transcriptional program compared with double-negative cells
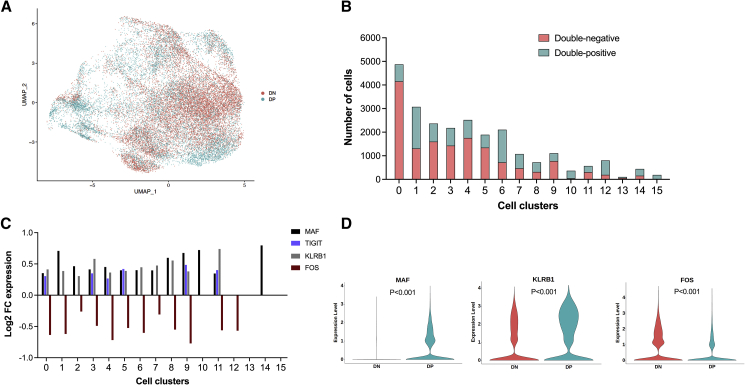
Given the differences between double-negative and double-positive cells in their capacity to respond to stimulation, we performed single-cell RNA sequencing (scRNA-seq) to characterize transcriptomic differences between double-negative and double-positive cells and to identify genes and biological pathways differentially expressed between these two subsets. We chose scRNA-seq over bulk RNA-seq to better characterize the heterogeneity within PD1/CTLA4 subsets and to identify specific clusters/cellular subsets responsible for the differences between double-negative and double-positive cells. We examined 40,000 double-negative and 40,000 double-positive cells from five study participants for whom we had sorted PD1/CTLA4 subsets available. Principal-component analysis followed by dimensionality reduction with uniform manifold approximation and projection (UMAP), and unsupervised clustering on the integrated dataset of double-negative and double-positive cells using Seurat with default settings (resolution 0.8), identified 16 different transcriptomic clusters (Figure S5A) with distinct gene expression profiles (Figures S5B and S5C). Analysis of clustering across different resolutions supported selection of resolution at 0.8 (Figure S5D). As expected, transcripts of PDCD1 and CTLA4 were reduced in double-negative cells, although counts were low in both populations (Figure S6). Expression of other inhibitory receptors, including LAG3, TIM3 (HAVCR2), and TIGIT, showed similar and consistent results with reduced expression in double-negative compared with double-positive cells but with overall low expression in both cell subsets (Figure S6). Although it was not possible to identify a specific cell type for each cluster, grouping clusters into five groups enabled us to assign broad categories for each group that were aligned with available gene profile information. This indicated that clusters 0, 1, 2, 5, and 11 (group 1) resembled resting CD4+ T cells (gene signature SELL, CCR7, and TCF7); clusters 4 and 9 (group 2) were metallothionein-expressing cells (gene signature MT1X, MTA2, MT1G, and MT1F); clusters 3, 8, 10, 13, and 14 (group 3) were cells with a circulating Tfh cell profile (gene signature CXCR5, PDCD1, and ICOS); cluster 7 (group 4) were cells with a Treg cell profile (gene signature FOXP3, interleukin-10 [IL-10], and CD25); and clusters 6, 12, and 15 (group 5) resembled Th1 cells (gene signature CXCR6, GZMB, NKG7, CCL5, and IFNG) (Figures S7A–S7E). It should be noted that, because of similarities between all cells, scoring against gene signatures did not provide consistent annotation of cell subsets, and the above proposed manual annotation should be interpreted with this caveat in mind. UMAP plotting of double-negative and double-positive memory CD4+ T cells revealed an extensive overlap in their clustering but also distinct enrichment of double-negative cells in cluster 0 and of double-positive cells in clusters 6 and 10 (Figures 7A and 7B). This differential abundance was supported by single-cell composition data analysis (scCODA) (Figure S7F). Across all clusters, we identified a total of 241 differentially expressed genes (DEGs) between double-positive and double-negative cells, with 179 significantly upregulated and 62 genes significantly downregulated in double-positive compared with double-negative cells (Table S2). Several genes displayed differential expression in more than one cluster. Of those, MAF, KLRB1, FOS, and TIGIT – genes involved in regulating T cell activation,26 exhaustion,12 proliferation,27,28 apoptosis,29 and production of antiviral cytokines,30 stood out by displaying a highly consistent pattern of differential expression across most T cell clusters (Figure 7C) and when compared across all double-positive versus double-negative cells; i.e., pooling all clusters (Figure 7D). MAF, KLRB1, and TIGIT were consistently upregulated in double-positive compared with double-negative cells, whereas FOS was consistently downregulated (Figures 7C and 7D). FOS protein is known to heterodimerize with JUN to form AP-1, a key transcription factor involved in regulating the remodeling of chromatin required for T cell activation.26 AP-1 can also bind to the HIV long terminal repeat (LTR) to activate transcription31 and is downregulated in latently infected cells.32 We found JUN to be differentially expressed in clusters 4, 6, and 9, and although the difference in JUN expression showed less consistency across all clusters compared with differential expression of FOS, it displayed similar directionality with significantly lower expression in double-positive versus double-negative cells (Table S2).
Figure 7.
Clustering and differential gene expression in double-positive compared to double-negative memory CD4+ T cells
(A–D) Clustering by UMAP of double-positive and double-negative memory CD4+ T cells (A), number of double-negative and double-positive cells (B), gene expression levels of the DEGs MAF, TIGIT, KLRB1, and FOS in double-positive compared with double-negative cells across cell clusters (C), and violin plots of gene expression levels of MAF, KLRB1, and FOS in all double-positive compared with double-negative memory CD4+ T cells (D). The violin plot for TIGIT is not shown because most cells displayed minimal expression. Double-negative cells from four donors and double-positive cells from five donors are analyzed. Differential gene expression analysis was done in Seurat using non-parametric statistics with FDR-adjusted p values shown in (D). DN, double-negative; DP, double-positive.
Using Kyoto Encyclopedia of Genes and Genomes (KEGG) databases for enrichment analysis, we identified multiple biological pathways that were significantly down- or upregulated in double-positive relative to double-negative cells, as determined by a false discovery rate (FDR) adjusted p value of less than 0.05. Although 43 pathways displayed significant upregulation in double-positive compared with double-negative cells, only four pathways were significantly downregulated, and each of those only appeared in a single cell cluster. By sorting identified pathways hierarchically according to the number of cell clusters in which a given pathway was differentially expressed, we identified that the most frequently upregulated pathways in double-positive cells were dominated by key immunological processes regulating Th1 and Th2 cell differentiation, IL-17 signaling, Th17 cell differentiation, Toll-like receptor signaling, T cell receptor signaling, and PD-L1 and PD1 checkpoint pathways (Table S3). Consistent with the reduced response to PMA/ionomycin stimulation in double-positive cells, we also found lower expression of protein kinase C (PKC) and calcium signaling pathways in this subset compared with double-negative cells (Figure S8). In contrast, double-positive cells had increased expression of genes belonging to the apoptosis pathway (Figure S8), which is aligned with the increased expression of MAF in this cellular subset. We found no differential expression in transcription factors NFAT, nuclear factor κB (NF-κB), RUNX, BATF, IRF7, and IRF8 between double-negative and double-positive cells.
In summary, single-cell transcriptomics analyses revealed a considerable overlap between memory CD4+ T cells double positive or double negative for PD1 and CTLA4 but also distinct differences in key genes regulating T cell activation and proliferation as well as differential expression of pathways involved in regulating T cell differentiation processes.
Discussion
In this study, we investigated the role of the immune checkpoints PD1 and CTLA4 in HIV persistence in blood and LNs from PWH on ART. We focused on PD1 and CTLA4 because these are key negative receptors involved in T cell exhaustion, and both have been identified as central pathways involved in viral persistence on ART and can be targeted therapeutically by the clinical administration of monoclonal antibodies to PD1 and/or CTLA4.2,11,16,17,33 We found that blood memory CD4+ T cells co-expressing PD1 and CTLA4 were modestly enriched for HIV DNA compared with their PD1−CTLA4− counterparts, but after stimulation with PMA/ionomycin, double-positive compared with double-negative cells in blood had a lower proportion of cells that produced MS RNA and showed reduced upregulation of markers of T cell activation and reduced proliferation. Single-cell transcriptomics analyses identified differential expression of key genes regulating T cell activation and proliferation. MAF, KLRB1, and TIGIT were consistently upregulated in double-positive compared with double-negative cells, whereas FOS was consistently downregulated, which may explain why double-positive cells respond less to activation stimuli. These data indicate that, in addition to being enriched for HIV DNA, memory CD4+ T cells co-expressing PD1 and CTLA4 are characterized by negative signaling and a reduced capacity to respond to stimulation, favoring maintenance of latent infection.
Based on our data and those of previous studies,5,11 we hypothesize that the reduced capacity to respond to stimulation is an important explanation for the maintenance of HIV in cells that co-express PD1 and CTLA4 because this will limit virus- or immune-mediated lysis of latently infected cells. The in vivo half-life of PD1+CTLA4+ memory CD4+ T cells is not well characterized. It has been shown that, upon PD1 ligation, activated T cells undergo metabolic reprogramming with an increased preference for fatty acid oxidation and lipolysis over glycolysis, which is known to be associated with effector-to-memory conversion and may explain the longevity of PD1+ T cells.34 Irrespective of the half-life of the individual PD1+CTLA4+ memory CD4+ T cell, long-term HIV persistence within these cells may also result from homeostatic or antigen-driven proliferation of infected cells.2,35 The key strengths of this study are the direct quantification of measures of HIV persistence within purified subsets of memory CD4+ T cells defined exclusively by their expression of PD1 and CTLA4 from blood and LN tissue. Previous studies have addressed similar questions but did not perform their investigations on sorted subsets,36 did not include LN tissue,11,37 did not investigate human infection,17 or did not sort for PD1 and CTLA4.12,16 Our study is fundamentally different because we specifically quantified HIV persistence within exhausted memory CD4+ T cells as defined by expression of PD1 and CTLA4.
We unexpectedly found that cells co-expressing PD1 and CTLA4, compared with double-negative cells, contained a lower proportion of cells with inducible CA-MS HIV RNA despite harboring total HIV DNA at a higher frequency. We hypothesized that the reduced inducibility was caused by functional exhaustion of cells expressing PD1 and CTLA4. Supporting this hypothesis, we demonstrated reduced upregulation of T cell activation and proliferation markers in double-positive compared with double-negative cells after PMA/ionomycin stimulation. This was done using peripheral blood cells, and it is possible that double-positive and/or double-negative cells may behave differently in the context of lymphoid tissue. Our findings contrast with previous data, where a higher proportion of memory CD4+ T cells expressing TIGIT and/or LAG-3 and/or PD1 have been shown to produce CA-MS HIV RNA upon stimulation compared with triple-negative cells.11 The divergent results may potentially be explained by the different T cell subsets investigated in the two studies. In the study by Fromentin et al., the cellular subset for this analysis contained a mix of cells expressing one, two, or all three immune checkpoints, with more than 50% being single positive, so it may have included a broader spectrum of functional exhaustion compared with cells double positive for PD1 and CTLA4. It is also possible that inclusion of CTLA4+ cells led to enrichment of cells with more extensive exhaustion. Finally, the CTLA4+ subsets in our study were enriched for Treg cells (Figure S2A), which may also have affected HIV inducibility because Treg cells might suppress HIV expression in other CD4+ T cells during co-culture.38 This could potentially occur through interaction with the surface inhibitory receptors, PD1, CTLA4, and glycoprotein A repetitions predominant (GARP) on Treg cells, as shown previously.38 Also, in double-positive and double-negative CD4+ T cells, HIV infection may differ with respect to inducibility, integration sites, and frequency of defective virus. Further work is needed to understand these differences. We assessed T cell proliferation and activation after stimulation with PMA/ionomycin, the same stimulus used in the TILDA assay. PMA/ionomycin stimulation bypasses the initial steps of T cell receptor (TCR) and co-stimulatory signaling. These experiments (including TILDA) were performed in purified T cell subsets in the absence of antigen-presenting cells, which express PD-L1 (the ligand for PD1) and CD80 and CD86 (the ligands for CTLA4). We believe that our approach is valid for several reasons. First, PD1/CTLA4 inhibitory signaling may affect T cell activation downstream of the TCR, potentially through transcriptomic regulation, as indicated in other studies.39,40 Second, CD4+ T cells themselves can express PD-L1, as demonstrated in other settings.41,42 CD86 and CD80, the ligands for CTLA4, have also been shown to be expressed on CD4+ and CD8+ T cells.43,44
We found differential expression of key genes centrally involved in regulating T cell activation, proliferation, and susceptibility to apoptosis. MAF, KLRB1, and TIGIT were consistently upregulated and FOS consistently downregulated in double-positive compared with double-negative cells. Although MAF is involved in increased CD4+ T cell susceptibility to apoptosis through downregulation of Bcl-2,29 KLRB1 expression has been demonstrated recently to be enriched in peripheral memory CD4+ T cells with high simultaneous production of interferon-γ (IFNγ) and tumor necrosis factor alpha (TNF-α).30 TIGIT is a well-described immune checkpoint protein involved in co-inhibitory signaling on T and natural killer (NK) cells and has been shown previously to be associated with HIV persistence and exhaustion of HIV-specific T cells.11,12 Expression of FOS has been linked to control of cellular responses to stimuli that regulate cell proliferation, cell differentiation, and apoptosis.27,28 More recently, AP-1, the heterodimer of FOS and JUN, was identified as the transcription factor that principally directs the chromatin remodeling required for T cells to respond to activation stimuli.26 In our dataset, JUN also showed significantly lower expression in double-positive cells than in double-negative cells, although less consistently across cell clusters compared with FOS. Collectively, this suggests that downregulation of FOS and, to a lesser extent, JUN and the reduced ability to mediate the downstream chromatin relaxation that is essential for T cell activation45 may help explain why memory CD4+ T cells co-expressing PD1 and CTLA4 respond less to activation stimuli and why, when infected, virus is harder to induce.
Our findings suggest that combined targeting of PD1 and CTLA4 may be superior to targeting either immune checkpoint alone with respect to reversing latency and eliminating latently infected cells. This is consistent with recent studies in SIV-infected rhesus macaques46 and PWH and people with cancer,47 which showed a greater increase in latency reversal and reduction in reservoir size when antibodies to PD1 and CTLA4 were co-administered compared with antibodies to PD-1 alone. The frequency of immune-related toxicities associated with concurrent blockade of PD1 and CTLA4 using currently available monoclonal antibodies48 will limit their use as a strategy for an HIV cure. Approaches with fewer adverse events will be needed to interrupt the negative signaling from immune checkpoints.
In contrast to previous studies showing preferential infection of PD1-expressing CD4+ T cells,2,11,49 we did not find a higher level of HIV DNA in PD1− compared with PD1+ cells. This discrepancy may be related to the relatively modest enrichment of HIV within PD1-expressing cells in those studies and, in fact, is only seen within Tcm and Ttm but not effector memory T (Tem) CD4+ T cells.2,11 Because Tem cells express higher levels of PD1 compared with other CD45RA− memory CD4+ T cells,11 the PD1+ subsets in our study were likely enriched for Tem cells and therefore less likely to show enrichment of HIV. Other studies have found an enrichment for HIV, including intact HIV, in Tem cells, but these studies did not directly compare PD1− and PD1+ Tem cells.49,50 Because we sorted cells based on expression of CTLA4 and PD1, we did not directly compare total PD1+ with total PD1− memory CD4+ T cells.
Although previous studies have described that CD8+ T cell activation is associated with a higher frequency of latently infected CD4+ T cells,9,37,51 these studies did not find an association with CD8+ T cell count or percentages. Our observations of a correlation between levels of HIV DNA and CD8+ T cell count are different because they relate specifically to the frequency of HIV DNA within subsets of exhausted CD4+ T cells. We found that a higher CD8+ T cell count was correlated with a higher level of HIV DNA in double-negative cells but not in double-positive cells. One interpretation of this finding is that less negative signaling in double-negative cells leads to more antigens being expressed, which would stimulate HIV-specific CD8+ T cells, but these are a very small proportion of total CD8+ T-cells.52
Limitations of the study
Some limitations of our study require consideration. First, of the 21 participants enrolled, we were only able to acquire LN memory CD4+ T cells from 8 individuals, which limited our ability to detect minor differences in HIV persistence between PD1/CTLA4 subsets in LN tissue and also precluded performing scRNA-seq or additional stimulation experiments in LN cells because not enough cells were available for these studies. However, although analyzing sorted LN subsets in a larger cohort would be favorable, our current findings did not suggest a trend toward enrichment in any of the cellular subsets analyzed here. Second, memory CD4+ T cells expressing CTLA4 were relatively rare. Therefore, there was a limited number of memory CD4+ T cells that could be acquired from a single LN, and, as a result, we were not able to analyze as many CTLA4+ as CTLA4− cells in this tissue. This limitation specifically meant that we could only perform TILDA on memory CD4+ T cells from blood, and we only sorted enough cells across all four subsets in 10 individuals. Third, the cross-sectional study design meant that our correlative analyses of HIV persistence and immunological and clinical parameters cannot infer causality. Studies in non-human primates, where timing of infection, initiation of ART, and extensive tissue sampling can be closely controlled, provide an enhanced opportunity for tracking these dynamics but for a different retroviral infection. Fourth, PMA/ionomycin is a potent T cell activator and has a latency-reversing capacity that is currently not achievable with any clinically available compounds. It is therefore uncertain whether the differences in PMA/ionomycin-induced activation of latent HIV will also be present in vivo using clinically safe latency-reversing agents. Fifth, because the majority of proviral HIV DNA is known to be defective or mutated,24 identifying cells that contain full-length intact HIV DNA53 and addressing whether specific mutations or deletions are enriched in any PD1/CTLA4 subset would be highly desirable. This is the focus of ongoing analyses. Finally, because of the exploratory nature of the study, we did not adjust the significance level for multiple comparisons. Our findings therefore require subsequent confirmation in a separate cohort.
In conclusion, we show that the frequency of infected cells is higher in blood memory CD4+ T cells that co-express PD1 and CTLA4 compared with their double-negative counterparts, but despite being enriched for HIV, double-positive cells displayed a reduced capacity to respond to stimulation, including reduced induction of latent HIV. Single-cell transcriptomics analyses identified differential expression of key genes regulating T cell activation and proliferation, which may explain why double-positive cells respond less to activation stimuli. These observations indicate that, as HIV persistence is established and maintained preferentially in CD4+ T cells co-expressing PD1 and CTLA4, combined targeting of these pathways may be superior in terms of reversing latency and eliminating latently infected cells.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Anti-CD3 BV711 | BD Biosciences | Cat# 563725 Clone UCHT1; RRID:AB_2744392 |
| Anti-CD4 FITC | BD Biosciences | Cat# 561842 Clone RPA-T4; RRID:AB_10892821 |
| Anti-CD45RA PE-Cy7 | BD Bioscineces | Cat# 337167 Clone L48; RRID:AB_647424 |
| Anti-PD1 BV421 | BD Biosciences | Cat# 562516 Clone EH12.1; RRID:AB_11153482 |
| Anti-CTLA4 PE | BD Biosciences | Cat# 557301 Clone BNI3; RRID:AB_396628 |
| MojoSort Human CD4 memory T cell isolation kit | Biolegend | Cat# 480064 |
| Anti-Foxp3 FITC | Biolegend | Cat# 320106 Clone 206D; RRID:AB_439752 |
| Anti-CD45RA PerCP-Cy5.5 | Biolegend | Cat# 304122 Clone HI100; RRID:AB_893357 |
| Anti-PD1 APC | Biolegend | Cat# 329907 Clone EH12; RRID:AB_940473 |
| Anti CD4 BUV 395 | BD Biosciences | Cat# 564550 Clone SK3; RRID:AB_2738273 |
| Anti-CD3 Alexa Fluor 700 | Biolegend | Cat# 561027 Clone UCH1; RRID:AB_10561682 |
| Anti-CCR7 APC-Cy7 | Biolegend | Cat# 353211 Clone G043H7; RRID:AB_10915272 |
| Anti CD27 BUV737 | BD Biosciences | Cat# 564302 Clone L128; RRID:AB_2744350 |
| Anti-CD8 BUV805 | BD Biosciences | Cat#564912 Clone SK1; RRID:AB_2744465 |
| Anti-CD127 BV421 | BD Biosciences | Cat# 562436 Clone HIL-7R-M21; RRID:AB_11151911 |
| Anti-CD25 PE-Cy7 | Biolegend | Cat# 356107 Clone M-A251; RRID:AB_2561974 |
| Anti-CD69 Alexa Fluor 647 | Biolegend | Cat#310918 Clone FN50; RRID:AB_528871 |
| Anti-HLA-DR APC-Fire 750 | Biolegend | Cat# 307658 Clone L243; RRID:AB_2572101 |
| Anti-CD38 V450 | BD Biosciences | Cat# 561378 Clone HIT2; RRID:AB_10689627 |
| Anti-Ki-67 PE | BD Biosciences | Cat# 556027 Clone B56; RRID:AB_2266296 |
| Biological samples | ||
| Fetal Bovine Serum | Bovogen | Cat# SFBSNZ |
| Leukapheresis | Alfred Hospital and University of San Francisco | N/A |
| Lymph node biopsy | The Avenue Hospital | N/A |
| Chemicals, peptides, and recombinant proteins | ||
| RPMI 1640 media | Life Technologies | Cat# 21870092 |
| HEPES buffer | Life Technologies | Cat# 15630080 |
| Penicillin Streptomycin Glutamine | Life Technologies | Cat# 10378016 |
| ACK lysing buffer | Life Technologies | Cat# A1049201 |
| DNase I | Sigma Aldrich | Cat# DN25-100mg |
| Ficoll-Paque PLUS density gradient media | Cytiva | Cat# 17144003 |
| Fixable live/Dead aqua cell viability dye | ThermoFisher | Cat# L34957 |
| True-Nuclear™ Transcription Factor Buffer Set | Biolgend | Cat# 424401 |
| Dimethyl sulfoxide (DMSO) | Sigma Aldrich | Cat# D2650-5X10ML |
| PMA | Sigma Aldrich | Cat# P1585-1MG |
| Ionomycin | Sigma Aldrich | Cat# I3909-1ML |
| BD Cytofix/Cytoperm Buffer | BD Biosciences | Cat# 554714 |
| Qiagen All-prep kit | Qiagen | Cat# 80204 |
| Zidovudine (AZT) | NIH HIV Reagent Program, Division of AIDS, NIAID, NIH: contributed by DAIDS/NIAID | ARP-3485 |
| Efavirenz | Selleckchem | Cat# S4685 |
| Raltegravir | Selleckchem | Cat# S2005 |
| TaqMan Fast Advanced Master Mix | Applied Biosystems | Cat# 4444557 |
| Critical commercial assays | ||
| 18S TaqMan gene expression assay | Applied Biosystems | Cat# 4331182 |
| Chromium Next GEM Single Cell 3′ Reagent Kits v3.1 | 10x Genomics | Cat#PN-1000128, PN-1000127, PN-120262 |
| High Sensitivity D5000 Tapestation Reagents | Agilent | Cat#5067–5592, 5067-5593 |
| Deposited data | ||
| Raw and analyzed scRNAseq data | This paper | ArrayExpress, Accession number E-MTAB-12039 |
| Oligonucleotides | ||
| MH535 (1st Rd US HIV RNA forward) 5-AACTAGGGAACCCACTGCTTAAG-3′ |
Lewin et al. 1999, Pasternak et al. 2013 | N/A |
| SL19 (2ND Rd US HIV RNA forward and total HIV DNA forward) 5' – TCTCTAGCAGTGGCGCCCGAACA |
Lewin et al. 1999, Pasternak et al. 2013 | N/A |
| SL20 (1st and 2ND Rd US HIV RNA Reverse and total HIV reverse) 5'-TCTCCTTCTAGCCTCCGCTAGTC-3′ |
Lewin et al. 1999, Pasternak et al. 2013 | N/A |
| SL28 (1st and 2ND Rd MS HIV RNA forward) 5'-CTTAGGCATCTCCTATGGCAGGAA-3′ |
Zerbato et al., 2021 | N/A |
| SL29 (2ND Rd MS HIV RNA Forward) 5'-TTCCTTCGGGCCTGTCGGGTCCC-3′ |
Zerbato et al., 2021 | N/A |
| TM1 (1st Rd MS HIV RNA Reverse) 5′-TCAAGCGGTGGTAGCTGAAGAGG-3′ |
Zerbato et al., 2021 | N/A |
| LK46(forward CCR5) 5'-GCTGTGTTTGCGTCTCTCCCAGGA-3′ |
Lewin et al., 1999 | N/A |
| LK47 (Reverse CCR5) 5'-CTCACAGCCCTGTGCCTCTTCTTC-3′ |
Lewin et al., 1999 | N/A |
| tat1.4 (TILDA forward) 5′-TGGCAGGAAGAAGCGGAGA-3′ |
Procopio et al., 2015 | N/A |
| Reverse (TILDA Reverse 1st and 2ND Rd) 5′-GGATCTGTCTCTGTCTCTCTCTCCACC-3′ |
Procopio et al., 2015 | N/A |
| tat2 (TILDA Reverse) 5′-ACAGTCAGACTCATCAAGTTTCTCTATCAA AGCA-3′ |
Procopio et al., 2015 | N/A |
| MS-HIV-FAMZEN (TILDA probe) 5’-/56-FAM/TTC CTT CGG/ZEN/GCC TGT CGG GTC CC/3IABkFQ/-3′ |
Procopio et al., 2015 | N/A |
| Software and algorithms | ||
| MxPro qPCR Software, for the Mx3005P qPCR System | Agilent Technologies | https://www.agilent.com/en/product/real-time-pcr-(qpcr)/real-time-pcr-(qpcr)-instruments/mx3000-mx3005p-real-time-pcr-system-software/mxpro-qpcr-software-232751; RRID:SCR_016375 |
| ELDA: Extreme Limiting Dilution Analysis, WEHI Bioinformatics | Hu, Y, and Smyth, GK (2009) | https://bioinf.wehi.edu.au/software/elda/ |
| BD FACSDiva | BD Biosciences | RRID:SCR_001456 |
| FlowJo Version 10 | Tree Star | https://www.flowjo.com/; RRID: SCR_008520 |
| GrahPad Prism Version 8.4.2 | Graphpad | https://www.graphpad.com/; RRID: SCR_002798 |
| Cell Ranger Version 3.1.0 | 10x Genomics | https://support.10xgenomics.com/single-cell-gene-expression/software/downloads/3.1 |
| R | R Development Core Team, 2008 | https://www.r-project.org/ |
| R Studio | N/A | https://www.rstudio.com/ |
| CellSNP | Huang 2021 | https://pypi.org/project/cellSNP/ |
| Vireo | Huang 2019 | https://vireosnp.readthedocs.io/en/latest/index.html |
| R | R Development Core Team, 2008 | https://www.r-project.org/ |
| R Studio | N/A | https://www.rstudio.com/ |
| Seurat Version 3.1.5 | Stuart ∗, Butler∗ et al. 2019 | http://satijalab.org/seurat/ |
| DoubletDecon | DePasquale 2019 | https://github.com/EDePasquale/DoubletDecon/ |
| scVI Version 0.7.0 | Lopez 2018 | https://github.com/scverse/scvi-tools/ |
| Harmony Version 0.1.0 | Korsunsky 2019 | https://github.com/immunogenomics/harmony |
| Other | ||
| Chromium Controller & Next GEM Accessory Kit | 10x Genomics | Cat# PN-1000202 |
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to the lead contact, Sharon Lewin (Sharon.lewin@unimelb.edu.au).
Materials availability
This study did not generate new unique reagents.
Experimental model and subject details
Study participants and sample collection
We enrolled 21 PWH receiving suppressive ART (see clinical characteristics in Table S1) at The Alfred Hospital, Melbourne, Australia (n = 11) and University of California San Francisco (UCSF), California, USA (n = 10). Participants provided written informed consent before inclusion in the study. As per study protocol inclusion criteria, we enrolled individuals with documented HIV infection aged 18 years or older and receiving combination ART with plasma HIV RNA <50 copies/mL for at least 3 years. Due to leukapheresis and LN biopsy procedures, we excluded individuals with skin infection at inguinal area; current lower extremity, gastrointestinal or genitourinary infection; chronic venous stasis or lymphedema of lower extremities; blood coagulation disorder or conditions requiring anticoagulant therapy; liver cirrhosis; weight <50 kg or BMI >35; blood pressure >160/100 mmHg or <100/70 mmHg; pregnant or breast feeding; or any with clinically significant cardiac or cerebrovascular disease. Leukapheresis procedures were performed at The Alfred Hospital (n = 11) or UCSF (n = 10) and excisional LN biopsies were performed at The Avenue Hospital, Melbourne, Australia. The time between the two procedures ranged from 6 to 50 days. LN biopsies were performed by an experienced surgeon using general (n = 9) or local (n = 1) anesthesia.
Study approvals
The study was approved by Human Research Ethics Committees at The Alfred and Avenue Hospitals in Melbourne and the Institutional Review Board at UCSF. The study was also registered at and approved by the University of Melbourne Ethics Committee. The study was conducted in accordance with the principles of the Declaration of Helsinki (1996) and the principles described in the Food and Drug Administration regulations and the Department of Health and Human Services regulations for the protection of human participants. Each participant provided written informed consent.
Method details
Processing of LNs and sorting of LNMCs
To disaggregate LNMCs from LN biopsy tissue, we initially stored LN samples in RPMI 1640 media (Life Technologies, Carlsbad, CA, USA) supplemented with 15% heat-inactivated Fetal Bovine Serum (FBS, Bovogen, Keilor East, VIC, AUS), 10mM HEPES buffer (Life Technologies, Cat no 15630-080), 1X Penicillin-Streptomycin-Glutamine (Life Technologies, Cat no 10378016) on ice in a 4°C cold room overnight for processing the following day. Using sterile disposable scalpels and forceps, LN tissue was torn apart and minced to obtain small fragments. Minced node tissue was transferred to a 70-micron nylon cell strainer (Bio-Strategy Laboratory Products PTY LTD, Tullamarine, VIC, AUS, Cat no 352350) and inserted into a 50 mL collection tube. Remaining tissue within the cell strainer was further grinded to allow cells to pass through and then passed through a second strainer with a 40-micron filter (Bio-Strategy Laboratory Products PTY LTD, Cat no 352340). Cells were then transferred to conical tubes and centrifuged at 400g for 10 min. In case of significant presence of red blood cells, these were lysed with ACK lysing buffer (Life Technologies). Cells were then resuspended in warm media containing DNase (RPMI with 1% FBS and 100 ug/mL DNase I [Sigma Aldrich, St. Louis, MO, USA, Cat no DN25-100mg]) at 10 million cells/mL, centrifuged at 400g for 10 min and resuspended at desired concentration for sorting or immunophenotyping. Following isolation of LNMCs from LN biopsies, unfrozen cells were labeled with the following antibodies and fluorophores all from BD Biosciences (Franklin Lakes, NJ, USA), according to the staining protocol outlined below for blood memory CD4+ T cells: anti-CD3 BV711 (Cat no 563725, clone UCHT1), CD4 FITC (Cat no 561842, clone RPA-T4), CD45RA PE-Cy7 (Cat no 337167, clone L48), PD1 BV421 (Cat no 562516, clone EH12.1) and CTLA4 PE (Cat no 557301, clone BNI3) and sorted on a BD FACS Aria cell sorter. We collected 4 different subsets of LN memory CD4+ T cells defined as double-positive (CD3+CD4+CD45RA-PD1+CTLA4+), PD1 single positive (CD3+CD4+CD45RA-PD1+CTLA4-), CTLA4 single positive (CD3+CD4+CD45RA-PD1-CTLA4+) and double-negative (CD3+CD4+CD45RA-PD1-CTLA4-).
Isolation of CD4+ T cells and cell sorting
Following leukapheresis PBMCs and plasma were isolated using standard Ficoll procedures. To isolate memory CD4+ T cells from fresh PBMCs obtained by leukapheresis, we used a magnetic bead negative selection kit from BioLegend that depleted non-CD4+ memory T cells with a biotin antibody cocktail consisting of anti-CD8a, anti-CD11b, anti-CD14, anti-CD16, anti-CD19, anti-CD20, anti-CD36, anti-CD45RA, anti-CD56, anti-CD123, anti-CD235ab and TC γ/δ (MojoSort Human CD4 memory T cell isolation kit, BioLegend, San Diego, CA, USA, Cat no 480064). Memory CD4+ T cells were first stained with live/dead aqua cell viability dye (ThermoFisher, Waltham, MA, USA, Cat no L34957) followed by labeling with the following antibodies and fluorophores, all from BD Biosciences: anti-CD3 BV711, CD4 FITC, CD45RA PE-Cy7, PD1 BV421 and CTLA4 PE and sorted on a BD FACS Aria Fusion. CTLA4-positive cells are often identified using intracellular staining for CTLA4; however, this was not feasible given the need for live cells in the subsequent TILDA assay. Due to intracellular expression and re-cycling of CTLA4, accurately detecting its surface expression is known to be challenging. In a previous study, 3-h stimulation with PMA/ionomycin was shown to enhance detection of CTLA4-positive cells.17 To avoid activation of cellular subsets or latent HIV, we did not employ T cell activation to enhance CTLA4 detection but instead stained cells with the CTLA4 antibody for 3 h at 37°C before incubation with the remaining antibodies for an additional 30 min at 37°C. This strategy yielded similar surface detection as compared to PMA/ionomycin stimulation and staining at room temperature, and with lower variability compared to PMA/ionomycin stimulation and staining at 37°C (Figure S9). During cell sorting we collected 4 different subsets defined as double-positive (CD3+CD4+CD45RA-PD1+CTLA4+), PD1 single positive (CD3+CD4+CD45RA-PD1+CTLA4-), CTLA4 single positive (CD3+CD4+CD45RA-PD1-CTLA4+) and double-negative (CD3+CD4+CD45RA-PD1-CTLA4-) (gating strategy shown in Figure S1). To reduce cross-contamination for each of the 4 populations sorted, individual gates were generated rather than a quadrant gate with slight gap between the gates to prevent overlap. This meant that a small percentage of each of the 4 populations would have been discarded during the sort. Purity check was performed on a subset of samples with a median purity of 90.1% and an IQR of 76.95–97.3%.
Immunophenotyping
Before sorting cells, we used flow cytometry to assess the distribution of the 4 subsets of memory CD4+ T cells in blood and LN based on their expression of PD1 and CTLA4. For this we used the same antibodies as described for cell sorting and fluorescence minus one (FMO) controls and ran samples on a BD LSR Fortessa flow cytometer. Gates were set using both the FMOs and then adjusted based on expression of PD1 and CTLA4 against CD45RA as this was found to be a more accurate way of determining the populations. Flow Jo version 10 (FlowJo, Ashland, OR, USA) was used for analysing acquired data. Surface and intracellular staining was performed on 1 × 10e6 PBMC of three uninfected donors and three PWH on suppressive ART to determine the relationship of CTLA4+ and PD1 and Tregs using the following reagents and antibodies: anti-Foxp3 FITC (Biolegend, Cat no 320106, clone 206D), anti-CD45RA Percp-Cy5.5 (Biolegend, Cat no 304122, clone HI100), anti-PD1 APC (Biolegend, Cat no 329907, clone EH12), anti-CD3 Alexa Fluor 700 (Biolegend, Cat no 561027, clone UCH1), anti-CCR7 APC-Cy7 (Biolegend, Cat no 353211, clone G043H7), anti-CD4 BUV 395 (BD Biosciences, Cat no 563550, clone SK3), anti-CD27 BUV737 (BD Biosciences, Cat no 564302, clone L128), anti-CD8 BUV805 (BD Biosciences, Cat no 564912, clone SK1), anti-CD127 BV421 (BD Biosciences, Cat no 562436, clone HIL-7R-M21), fixable dead/live stain Aqua, anti-CTLA-4 PE (Biolegend, Cat no 557301, clone BNI3), anti-CD25 PE-Cy7 (Cat no 356107, clone M-A251), True-Nuclear™ Transcription Factor Buffer Set (Biolegend, Cat no 424401).
T cell stimulation
To quantify the level of activation, proliferation and cell death, cryopreserved CD4 subsets remaining from the sort were thawed. Approximately 2 million cells were stimulated with either DMSO (Sigma Aldrich, Cat no D2650-5X10ML) or PMA/ionomycin (final concentration of 10nM PMA and 0.5μM ionomycin) for 72 h. After the stimulation, 1.5 million cells were washed and surface stained for live/dead aqua, CD69 Alexa Fluor 647 (Biolegend, Cat no 310918, clone FN50), HLA-DR APC-Fire750 (Biolegend, Cat no 307658, clone L243), and CD38 V450 (BD biosciences, Cat no 561378, clone HIT2). Cells were then permeabilized using BD Cytofix/Cytoperm buffer (BD Biosciences, Cat no 554714) and stained intracellularly with Ki-67 PE (BD biosciences, Cat no 556027, clone B56). Samples were run within 2 h of staining completion on BD LSR Fortessa flow cytometer and the data was analyzed using FlowJo version 10.
Cell-associated HIV DNA and RNA and TILDA
To quantify levels of cell-associated HIV RNA and DNA, memory CD4+ T cells sorted based on their expression of PD1 and CTLA4 subsets were lysed and lysates stored at −80°C until analysis. We extracted RNA from lysates using the Qiagen All-prep kit (Qiagen, Hilden, Germany, Cat no 80204). CA-US HIV RNA was then quantified in four replicates using a semi-nested real-time qPCR (rt-PCR) as previously described.54 Primers used for first and second round amplified HIV RNA copy numbers, which were standardized to cellular input using the 18S TaqMan gene Expression Assay (Applied Biosystems, Foster City, CA USA). For all samples, a non-RT control was included. If this control was positive, results for this sample were excluded from the analysis. CA-MS HIV RNA was measured using rt-PCR with primers and probes as described previously.54 The number of input cells for quantifying US CA-US HIV RNA and CA-MS HIV RNA were 5 × 104 – 105 for peripheral blood and 104–105 for LN cells. Cell-associated total HIV DNA was analyzed in triplicate with cell lysates using primers and probes as previously described.55 HIV DNA copy numbers were standardized to cellular equivalents through quantification of CCR5. The lower limit of detection of the HIV DNA assay was one copy per well and given the increasing variation in threshold cycles at this low level, we included values down to 0.1 copies. Any value < 0.1 copy per well was included as zero. The number of input cells for quantifying HIV DNA were 105–1.25 × 105 for peripheral blood and 1.7 × 104 – 105 for LN cells. To estimate the percentage contribution of each PD1/CTLA4 subset to the total pool of HIV DNA and CA-US HIV RNA within memory CD4+ T cells, we adjusted for the frequency of each subset in blood and LN as determined by flow cytometry by using the formula A = (A/A + B + C + D) X 100, where A = (% of subset A/100) X (HIV DNA copies/106 cells for subset A); B = (% of subset B/100) X (HIV DNA copies/106 cells for subset B), etc. To measure the frequency of cells with inducible MS HIV RNA, we performed the tat/rev Induced Limiting Dilution Assay (TILDA) adapted from (Procopio et al., 2015). The isolated four memory CD4+ T cell subsets were stimulated with PMA (100 ng/mL) and ionomycin (1 μg/mL) for 12–18 h in the presence of antiretroviral drugs (180nM AZT, 300 nM efavirenz, 200 nM raltegravir), and the frequency of cells producing MS HIV RNA was measured at various dilutions. Limiting dilutions, oligonucleotide and probe sequences, and the pre-amplification PCR reaction was performed as described by (Procopio et al., 2015). 1 μL of the pre-amplified PCR product was used as template in the tat/rev real-time PCR reaction, added to 10 uL TaqMan Fast Advanced Master Mix (Applied Biosystems), 0.4 μL of each primer (tat2 and rev, at 20 μM), 0.4 μL of the probe FamZen (at 5 μM) and 7.8 μL H20. The real-time PCR reaction was carried out on the Stratagene Mx3005P (Agilent Technologies) using the following program: pre-incubation at 95°C for 20 s, and 45 cycles of amplification (95°C 1 s, 60°C 20 s). TILDA was performed in 22 replicates per dilution, and the frequency of cells with inducible HIV MS HIV RNA was calculated with the maximum likelihood method using the online software http://bioinf.wehi.edu.au/software/elda/. For each cellular subset from each participant 18,000, 9,000, 3,000 and 1,000 cells were seeded in a limiting dilution format, where 22 wells were seeded for each of the cell numbers.
Single-cell RNA sequencing
To characterise and compare the transcriptomic program in PD1/CTLA4 double-positive and double-negative memory CD4+ T cells, we used sorted cryopreserved cells from five study participants, where freshly isolated cells had been sorted as described above. For one study participant the double-negative cell population showed poor viability and was excluded, i.e. we used double-negative cells from four donors and double-positive cells from five donors. To analyze an equal number of cells from each subset, we loaded 10,000 double-negative cells per donor but 8,000 double-positive cells per donor. Gel bead-in emulsions were generated using the Chromium controller and 3′ gene expression kits (v3.1, 10x Genomics). We superloaded lanes with 40,000 double-positive and 40,000 double-negative cells originating from five/four different study participants, respectively. cDNA libraries were generated following the manufacturer’s guidelines (10x Genomics) and sequenced using MGISEQ instruments.
Quantification and statistical analysis
Statistical analyses
As robust preliminary data were unavailable for specific power calculations for this type of study, we instead calculated standardised effect sizes and estimated that a sample size of 20 participants would provide 80% statistical power for a standardized effect size of 0.7 (mean difference divided by the SD of the differences). We assessed data distribution through visual inspection of frequency histograms and quantile-quantile plots and also used the Shapiro-Wilk normality test. We explored logarithmic transformation to achieve normal distribution whenever possible. We tested for differences across all 4 subsets using mixed-effect analysis with the Geisser-Greenhouse correction or Friedman test depending on data distribution. We also performed pairwise comparisons across the individual subsets using paired t-test or Wilcoxon matched-pairs signed rank test, depending on data distribution. We investigated associations of HIV persistence measures and baseline characteristics using Spearman or Pearson correlation depending on data distribution. We did not adjust p values or significance level for multiple testing, given the exploratory nature of these studies.56 All statistical analyses were performed in Prism 8 for macOS (version 8.4.2, GraphPad Software, San Diego, CA, USA).
Single-cell RNA sequencing analysis
FASTQ files were processed via Cell Ranger v3.1.0 (10x Genomics) using “cellranger count” pipeline with 10x human genome 3.0.0 release as a reference. Superloaded samples were de-multiplexed into donors by modeling expressed alleles using Vireo v0.3.257 with doublets further removed using DoubletDecon. Seurat v3.1.5 was used for downstream analysis. Quality control of scRNA-seq data included removal of genes expressed in less than 3 cells, removal of cells outside threshold of 200-4000 expressed genes, and removal of cells with more than 15% mitochondrial content. Seurat’s “SCTransform” function was used to normalise, identify highly variable genes, and scale scRNA-seq data. Principal component analysis (PCA) was performed using Seurat’s “RunPCA” function. Top 30 PCs was used as input to generate 2-dimensional Uniform Manifold Approximation Projection (UMAP) projection of the data using Seurat’s “RunUMAP” function. To place PD1/CTLA4 double-positive and double-negative cells from HIV-infected donors in context of similar subsets from uninfected donors, we initially integrated data with those generated in similarly performed experiment using sorted PD1-CTLA4-, PD1+CTLA4-and PD1+CTLA4+ cells from four uninfected controls. Because data from uninfected donors were generated in a separate experiment, we observed substantial batch effects requiring data integration (Figure S10). Seurat’s “IntegrateData” function was used to integrate with scRNA-seq data generated from memory CD4+ T cells of healthy controls to remove any batch effect. Validation of the integration was carried out using scVI v0.7.0 and Harmony v0.1.0. To determine consistency across the three integration approaches (Seurat, Harmony, and scVI), we overlaid the unsupervised clustering of cells from HIV-infected donors onto the UMAPs, as shown in Figure S5A. In all three integration approaches, we noted that clusters 0, 1 and 2 were located close to each other on the UMAP and overlapped substantially with cells from uninfected donors, whereas clusters 6 and 12 were located close to each other but did not overlap with cells from uninfected donors (Figure S11). Overall, we observed similarities and differences between cells from HIV-infected and uninfected donors, but as a detailed comparison between these was beyond the scope of this study, all subsequent analyses were conducted on double-positive cells vs double-negative cells from HIV-infected donors only. Harmony was run using Seurat normalised, scaled and PCA embedded data. The UMAP was generated using the Harmony embeddings as input. scVI was run using the top 3000 genes as input and with latent variables set to default. The model was trained with all parameters kept at default except using 200 epochs and 50 iterations. The UMAP was generated using scVI embeddings as input. Combined dataset was projected onto UMAP and healthy cells were then removed from downstream analysis. Unsupervised clustering was performed using Seurat’s “FindClusters” function. Differential gene expression analysis (DGEA) to identify marker genes for each cell cluster and compare cells from double-positive and double-negative samples were performed using “FindMarkers” function from Seurat.
Acknowledgments
We acknowledge Tina Luke, Angela Hind, Catherine Li, and Lankesha Yapa from the Melbourne Cytometry Platform at the Doherty Institute for Infection and Immunity for assistance with flow cytometry and cell sorting. We also acknowledge Socheata Chea and Surekha Tennakoon for assistance with laboratory procedures. This work was kindly supported by the Harold and Cora Brennen Trust through purchase of the 10X Chromium Controller. The study was funded by a grant from American Foundation for AIDS Research (amfAR; 109226-58-RGRL) and the National Institutes of Health Delaney AIDS Research Enterprise (DARE) Collaboratory (UM1AI126611), The National Health and Medical Research Council (NHMRC; GNT1149990) of Australia. We thank all study participants for donating their time and samples to contribute to the study. A huge thanks goes to Michelle Hagenauer and the clinical research unit at The Department of Infectious Diseases at Alfred Hospital and Marian Kerbleski and the SCOPE team at UCSF for help with identifying study participants and coordinating sample collection and study visits.
Author contributions
S.R.L., P.U.C., S.G.D., J.H.M., and T.A.R. conceived and designed the study. S.R.L., T.A.R., and J.H.M. drafted the study protocol. R.H., M.K., S.G.D., S.R.L., J.S.Y.L., and J.M. oversaw all aspects of the clinical trial, including study protocol, ethics submission, and management of study participants. W.B. performed LN biopsies. P.U.C., V.E., R.P., and A.D. performed processing of LN biopsies and cell sorting on biopsy samples. A.R., J.M.Z., and A.D. performed all sample processing on leukaphereses, memory CD4+ T cell isolation, and cell sorting. A.R., J.M.Z. and A.D. performed PCR analyses of CA-US HIV RNA, HIV DNA, CA-MS HIV RNA, and TILDA analyses. V.E., S.R.L., P.U.C., J.M., and T.A.R. coordinated all data generation and data collection and performed data analysis and interpretation. R.F., A.R., J.M.Z., A.D., R.M.V.d.S., J.J.C., and N.C. contributed to cell sorting, flow analyses, data analyses, and interpretation. C.T. performed sample and library preparation for single-cell RNA sequencing. A.H., M.B., and H.J.L. performed analysis of single-cell RNA sequencing data. S.R.L. and T.A.R. performed statistical analyses. The manuscript was drafted by T.A.R. and S.R.L. All authors reviewed and provided input to the manuscript and approved the final version.
Declaration of interests
S.R.L. has received funding from the National Health and Medical Research Council of Australia (NHMRC), National Institutes of Health (NIH), amfAR, Gilead Sciences, Merck, and ViiV outside of the submitted work. S.R.L. is a paid member of advisory boards to Merck, Gilead, Immunocore, Esfam, and Vaxxinity. S.R.L. has consulted for Abbvie and BMS. T.A.R. has received funding from the Danish Research Council, Region Midt Denmark, The Australian Centre for HIV and Hepatitis Research, the Melbourne HIV Cure Consortium, Gilead, and the Novo Nordisk Foundation outside of the submitted work. S.G.D. receives grant support from Gilead, Merck, and ViiV; is a member of the scientific advisory boards for BryoLogyx and Enochian Biosciences; and has consulted for AbbVie, Biotron, and Eli Lilly.
Published: October 4, 2022
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.xcrm.2022.100766.
Supplemental information
Data and code availability
-
•
Single-cell RNA-seq data have been deposited at ArrayExpress (Accession number E-MTAB-12039) and are publicly available as of the date of publication. Accession numbers are listed in the key resources table.
-
•
This paper does not report original code
-
•
Any additional information required to re-analyse the data reported in this paper is available from the lead contact upon request.
References
- 1.Pitman M.C., Lau J.S.Y., McMahon J.H., Lewin S.R. Barriers and strategies to achieve a cure for HIV. Lancet HIV. 2018;5:e317–e328. doi: 10.1016/S2352-3018(18)30039-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chomont N., El-Far M., Ancuta P., Trautmann L., Procopio F.A., Yassine-Diab B., Boucher G., Boulassel M.R., Ghattas G., Brenchley J.M., et al. HIV reservoir size and persistence are driven by T cell survival and homeostatic proliferation. Nat. Med. 2009;15:893–900. doi: 10.1038/nm.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Buzon M.J., Sun H., Li C., Shaw A., Seiss K., Ouyang Z., Martin-Gayo E., Leng J., Henrich T.J., Li J.Z., et al. HIV-1 persistence in CD4(+) T cells with stem cell-like properties. Nat. Med. 2014;20:139–142. doi: 10.1038/nm.3445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Soriano-Sarabia N., Bateson R.E., Dahl N.P., Crooks A.M., Kuruc J.D., Margolis D.M., Archin N.M. Quantitation of replication-competent HIV-1 in populations of resting CD4+ T cells. J. Virol. 2014;88:14070–14077. doi: 10.1128/JVI.01900-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Evans V.A., van der Sluis R.M., Solomon A., Dantanarayana A., McNeil C., Garsia R., Palmer S., Fromentin R., Chomont N., Sékaly R.P., et al. Programmed cell death-1 contributes to the establishment and maintenance of HIV-1 latency. AIDS. 2018;32:1491–1497. doi: 10.1097/QAD.0000000000001849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Attanasio J., Wherry E.J. Costimulatory and coinhibitory receptor pathways in infectious disease. Immunity. 2016;44:1052–1068. doi: 10.1016/j.immuni.2016.04.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Trautmann L., Janbazian L., Chomont N., Said E.A., Gimmig S., Bessette B., Boulassel M.R., Delwart E., Sepulveda H., Balderas R.S., et al. Upregulation of PD-1 expression on HIV-specific CD8+ T cells leads to reversible immune dysfunction. Nat. Med. 2006;12:1198–1202. doi: 10.1038/nm1482. [DOI] [PubMed] [Google Scholar]
- 8.Kaufmann D.E., Kavanagh D.G., Pereyra F., Zaunders J.J., Mackey E.W., Miura T., Palmer S., Brockman M., Rathod A., Piechocka-Trocha A., et al. Upregulation of CTLA-4 by HIV-specific CD4+ T cells correlates with disease progression and defines a reversible immune dysfunction. Nat. Immunol. 2007;8:1246–1254. doi: 10.1038/ni1515. [DOI] [PubMed] [Google Scholar]
- 9.Cockerham L.R., Siliciano J.D., Sinclair E., O'Doherty U., Palmer S., Yukl S.A., Strain M.C., Chomont N., Hecht F.M., Siliciano R.F., et al. CD4+ and CD8+ T cell activation are associated with HIV DNA in resting CD4+ T cells. PLoS One. 2014;9:e110731. doi: 10.1371/journal.pone.0110731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Macatangay B.J.C., Gandhi R.T., Brad Jones R., Mcmahon D.K., Lalama C.M., Bosch R.J., Cyktor J.C., Thomas A.S., Borowski L., Riddler S.A., et al. PD-1HI T cells are associated with lower hiv-specific immune responses despite long-term antiretroviral therapy. AIDS. 2019 doi: 10.1097/QAD.0000000000002406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fromentin R., Bakeman W., Lawani M.B., Khoury G., Hartogensis W., DaFonseca S., Killian M., Epling L., Hoh R., Sinclair E., et al. CD4+ T cells expressing PD-1, TIGIT and LAG-3 contribute to HIV persistence during ART. PLoS Pathog. 2016;12:e1005761. doi: 10.1371/journal.ppat.1005761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chew G.M., Fujita T., Webb G.M., Burwitz B.J., Wu H.L., Reed J.S., Hammond K.B., Clayton K.L., Ishii N., Abdel-Mohsen M., et al. TIGIT marks exhausted T cells, correlates with disease progression, and serves as a target for immune restoration in HIV and SIV infection. PLoS Pathog. 2016;12:e1005349. doi: 10.1371/journal.ppat.1005349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kim C.H., Rott L.S., Clark-Lewis I., Campbell D.J., Wu L., Butcher E.C. Subspecialization of CXCR5+ T cells: B helper activity is focused in a germinal center-localized subset of CXCR5+ T cells. J. Exp. Med. 2001;193:1373–1381. doi: 10.1084/jem.193.12.1373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fukazawa Y., Lum R., Okoye A.A., Park H., Matsuda K., Bae J.Y., Hagen S.I., Shoemaker R., Deleage C., Lucero C., et al. B cell follicle sanctuary permits persistent productive simian immunodeficiency virus infection in elite controllers. Nat. Med. 2015;21:132–139. doi: 10.1038/nm.3781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fletcher C.V., Staskus K., Wietgrefe S.W., Rothenberger M., Reilly C., Chipman J.G., Beilman G.J., Khoruts A., Thorkelson A., Schmidt T.E., et al. Persistent HIV-1 replication is associated with lower antiretroviral drug concentrations in lymphatic tissues. Proc. Natl. Acad. Sci. USA. 2014;111:2307–2312. doi: 10.1073/pnas.1318249111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Banga R., Procopio F.A., Noto A., Pollakis G., Cavassini M., Ohmiti K., Corpataux J.-M., de Leval L., Pantaleo G., Perre M. PD-1 and follicular helper T cells are responsible for persistent HIV-1 transcription in treated aviremic individuals. Nat. Med. 2016 doi: 10.1038/nm.4113. [DOI] [PubMed] [Google Scholar]
- 17.McGary C.S., Deleage C., Harper J., Micci L., Ribeiro S.P., Paganini S., Kuri-Cervantes L., Benne C., Ryan E.S., Balderas R., et al. CTLA-4(+)PD-1(-) memory CD4(+) T cells critically contribute to viral persistence in antiretroviral therapy-suppressed, SIV-infected rhesus macaques. Immunity. 2017;47:776–788.e5. doi: 10.1016/j.immuni.2017.09.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ribas A., Wolchok J.D. Cancer immunotherapy using checkpoint blockade. Science. 2018;359:1350–1355. doi: 10.1126/science.aar4060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jiao Y.M., Liu C.E., Luo L.J., Zhu W.J., Zhang T., Zhang L.G., Su L.S., Li H.J., Wu H. CD4+CD25+CD127 regulatory cells play multiple roles in maintaining HIV-1 p24 production in patients on long-term treatment: HIV-1 p24-producing cells and suppression of anti-HIV immunity. Int. J. Infect. Dis. 2015;37:42–49. doi: 10.1016/j.ijid.2015.06.008. [DOI] [PubMed] [Google Scholar]
- 20.Tran T.A., de Goër de Herve M.G., Hendel-Chavez H., Dembele B., Le Névot E., Abbed K., Pallier C., Goujard C., Gasnault J., Delfraissy J.F., et al. Resting regulatory CD4 T cells: a site of HIV persistence in patients on long-term effective antiretroviral therapy. PLoS One. 2008;3:e3305. doi: 10.1371/journal.pone.0003305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pasternak A.O., Lukashov V.V., Berkhout B. Cell-associated HIV RNA: a dynamic biomarker of viral persistence. Retrovirology. 2013;10:41. doi: 10.1186/1742-4690-10-41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lewin S.R., Vesanen M., Kostrikis L., Hurley A., Duran M., Zhang L., Ho D.D., Markowitz M. Use of real-time PCR and molecular beacons to detect virus replication in human immunodeficiency virus type 1-infected individuals on prolonged effective antiretroviral therapy. J. Virol. 1999;73:6099–6103. doi: 10.1128/jvi.73.7.6099-6103.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zerbato J.M., Khoury G., Zhao W., Gartner M.J., Pascoe R.D., Rhodes A., Dantanarayana A., Gooey M., Anderson J., Bacchetti P., et al. Multiply spliced HIV RNA is a predictive measure of virus production ex vivo and in vivo following reversal of HIV latency. EBioMedicine. 2021;65:103241. doi: 10.1016/j.ebiom.2021.103241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ho Y.C., Shan L., Hosmane N.N., Wang J., Laskey S.B., Rosenbloom D.I.S., Lai J., Blankson J.N., Siliciano J.D., Siliciano R.F. Replication-competent noninduced proviruses in the latent reservoir increase barrier to HIV-1 cure. Cell. 2013;155:540–551. doi: 10.1016/j.cell.2013.09.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Procopio F.A., Fromentin R., Kulpa D.A., Brehm J.H., Bebin A.G., Strain M.C., Richman D.D., O'Doherty U., Palmer S., Hecht F.M., et al. A novel assay to measure the magnitude of the inducible viral reservoir in HIV-infected individuals. EBioMedicine. 2015;2:874–883. doi: 10.1016/j.ebiom.2015.06.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yukawa M., Jagannathan S., Vallabh S., Kartashov A.V., Chen X., Weirauch M.T., Barski A. AP-1 activity induced by co-stimulation is required for chromatin opening during T cell activation. J. Exp. Med. 2020;217:e20182009. doi: 10.1084/jem.20182009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Huen N.Y., Pang A.L.Y., Tucker J.A., Lee T.L., Vergati M., Jochems C., Intrivici C., Cereda V., Chan W.Y., Rennert O.M., et al. Up-regulation of proliferative and migratory genes in regulatory T cells from patients with metastatic castration-resistant prostate cancer. Int. J. Cancer. 2013;133:373–382. doi: 10.1002/ijc.28026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Angel P., Karin M. The role of Jun, Fos and the AP-1 complex in cell-proliferation and transformation. Biochim. Biophys. Acta. 1991;1072:129–157. doi: 10.1016/0304-419x(91)90011-9. [DOI] [PubMed] [Google Scholar]
- 29.Peng S., Lalani S., Leavenworth J.W., Ho I.C., Pauza M.E. c-Maf interacts with c-Myb to down-regulate Bcl-2 expression and increase apoptosis in peripheral CD4 cells. Eur. J. Immunol. 2007;37:2868–2880. doi: 10.1002/eji.200636979. [DOI] [PubMed] [Google Scholar]
- 30.Truong K.L., Schlickeiser S., Vogt K., Boës D., Stanko K., Appelt C., Streitz M., Grütz G., Stobutzki N., Meisel C., et al. Killer-like receptors and GPR56 progressive expression defines cytokine production of human CD4(+) memory T cells. Nat. Commun. 2019;10:2263. doi: 10.1038/s41467-019-10018-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.van der Sluis R.M., Derking R., Breidel S., Speijer D., Berkhout B., Jeeninga R.E. Interplay between viral Tat protein and c-Jun transcription factor in controlling LTR promoter activity in different human immunodeficiency virus type I subtypes. J. Gen. Virol. 2014;95:968–979. doi: 10.1099/vir.0.059642-0. [DOI] [PubMed] [Google Scholar]
- 32.Jefferys S.R., Burgos S.D., Peterson J.J., Selitsky S.R., Turner A.M.W., James L.I., Tsai Y.H., Coffey A.R., Margolis D.M., Parker J., Browne E.P. Epigenomic characterization of latent HIV infection identifies latency regulating transcription factors. PLoS Pathog. 2021;17:e1009346. doi: 10.1371/journal.ppat.1009346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cook M.R., Kim C. Safety and efficacy of immune checkpoint inhibitor therapy in patients with HIV infection and advanced-stage cancer: a systematic Review. JAMA Oncol. 2019;5:1049–1054. doi: 10.1001/jamaoncol.2018.6737. [DOI] [PubMed] [Google Scholar]
- 34.Patsoukis N., Bardhan K., Chatterjee P., Sari D., Liu B., Bell L.N., Karoly E.D., Freeman G.J., Petkova V., Seth P., et al. PD-1 alters T-cell metabolic reprogramming by inhibiting glycolysis and promoting lipolysis and fatty acid oxidation. Nat. Commun. 2015;6:6692. doi: 10.1038/ncomms7692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Simonetti F.R., Zhang H., Soroosh G.P., Duan J., Rhodehouse K., Hill A.L., Beg S.A., McCormick K., Raymond H.E., Nobles C.L., et al. Antigen-driven clonal selection shapes the persistence of HIV-1-infected CD4+ T cells in vivo. J. Clin. Invest. 2021;131:145254. doi: 10.1172/JCI145254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Khoury G., Fromentin R., Solomon A., Hartogensis W., Killian M., Hoh R., Somsouk M., Hunt P.W., Girling V., Sinclair E., et al. Human immunodeficiency virus persistence and T-cell activation in blood, rectal, and lymph node tissue in human immunodeficiency virus-infected individuals receiving suppressive antiretroviral therapy. J. Infect. Dis. 2017;215:911–919. doi: 10.1093/infdis/jix039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hatano H., Jain V., Hunt P.W., Lee T.H., Sinclair E., Do T.D., Hoh R., Martin J.N., McCune J.M., Hecht F., et al. Cell-based measures of viral persistence are associated with immune activation and programmed cell death protein 1 (PD-1)-expressing CD4+ T cells. J. Infect. Dis. 2013;208:50–56. doi: 10.1093/infdis/jis630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhang M., Robinson T.O., Duverger A., Kutsch O., Heath S.L., Cron R.Q. Regulatory CD4 T cells inhibit HIV-1 expression of other CD4 T cell subsets via interactions with cell surface regulatory proteins. Virology. 2018;516:21–29. doi: 10.1016/j.virol.2017.12.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Shimizu K., Sugiura D., Okazaki I.M., Maruhashi T., Takegami Y., Cheng C., Ozaki S., Okazaki T. PD-1 imposes qualitative control of cellular transcriptomes in response to T cell activation. Mol. Cell. 2020;77:937–950.e6. doi: 10.1016/j.molcel.2019.12.012. [DOI] [PubMed] [Google Scholar]
- 40.Fromentin R., DaFonseca S., Costiniuk C.T., El-Far M., Procopio F.A., Hecht F.M., Hoh R., Deeks S.G., Hazuda D.J., Lewin S.R., et al. PD-1 blockade potentiates HIV latency reversal ex vivo in CD4(+) T cells from ART-suppressed individuals. Nat. Commun. 2019;10:814. doi: 10.1038/s41467-019-08798-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Diskin B., Adam S., Cassini M.F., Sanchez G., Liria M., Aykut B., Buttar C., Li E., Sundberg B., Salas R.D., et al. PD-L1 engagement on T cells promotes self-tolerance and suppression of neighboring macrophages and effector T cells in cancer. Nat. Immunol. 2020;21:442–454. doi: 10.1038/s41590-020-0620-x. [DOI] [PubMed] [Google Scholar]
- 42.Fanelli G., Romano M., Nova-Lamperti E., Werner Sunderland M., Nerviani A., Scottà C., Bombardieri M., Quezada S.A., Sacks S.H., Noelle R.J., et al. PD-L1 signaling on human memory CD4+ T cells induces a regulatory phenotype. PLoS Biol. 2021;19:e3001199. doi: 10.1371/journal.pbio.3001199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Paine A., Kirchner H., Immenschuh S., Oelke M., Blasczyk R., Eiz-Vesper B. IL-2 upregulates CD86 expression on human CD4(+) and CD8(+) T cells. J. Immunol. 2012;188:1620–1629. doi: 10.4049/jimmunol.1100181. [DOI] [PubMed] [Google Scholar]
- 44.Soskic B., Jeffery L.E., Kennedy A., Gardner D.H., Hou T.Z., Halliday N., Williams C., Janman D., Rowshanravan B., Hirschfield G.M., Sansom D.M. CD80 on human T cells is associated with FoxP3 expression and supports treg homeostasis. Front. Immunol. 2020;11:577655. doi: 10.3389/fimmu.2020.577655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lee M.D., Bingham K.N., Mitchell T.Y., Meredith J.L., Rawlings J.S. Calcium mobilization is both required and sufficient for initiating chromatin decondensation during activation of peripheral T-cells. Mol. Immunol. 2015;63:540–549. doi: 10.1016/j.molimm.2014.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Harper J., Gordon S., Chan C.N., Wang H., Lindemuth E., Galardi C., Falcinelli S.D., Raines S.L.M., Read J.L., Nguyen K., et al. CTLA-4 and PD-1 dual blockade induces SIV reactivation without control of rebound after antiretroviral therapy interruption. Nat. Med. 2020;26:519–528. doi: 10.1038/s41591-020-0782-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Rasmussen T.A., Rajdev L., Rhodes A., Dantanarayana A., Tennakoo S., Chea S., Spelman T., Lensing S., Rutishauser R., Bakkour S., et al. Impact of anti-PD-1 and anti-CTLA-4 on the HIV reservoir in people living with HIV with cancer on antiretroviral therapy: the AIDS Malignancy Consortium-095 study. Clin. Infect. Dis. 2021 doi: 10.1093/cid/ciaa1530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Boutros C., Tarhini A., Routier E., Lambotte O., Ladurie F.L., Carbonnel F., Izzeddine H., Marabelle A., Champiat S., Berdelou A., et al. Safety profiles of anti-CTLA-4 and anti-PD-1 antibodies alone and in combination. Nat. Rev. Clin. Oncol. 2016;13:473–486. doi: 10.1038/nrclinonc.2016.58. [DOI] [PubMed] [Google Scholar]
- 49.Neidleman J., Luo X., Frouard J., Xie G., Hsiao F., Ma T., Morcilla V., Lee A., Telwatte S., Thomas R., et al. Phenotypic analysis of the unstimulated in vivo HIV CD4 T cell reservoir. Elife. 2020;9:e60933. doi: 10.7554/eLife.60933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hiener B., Horsburgh B.A., Eden J.S., Barton K., Schlub T.E., Lee E., von Stockenstrom S., Odevall L., Milush J.M., Liegler T., et al. Identification of genetically intact HIV-1 proviruses in specific CD4(+) T cells from effectively treated participants. Cell Rep. 2017;21:813–822. doi: 10.1016/j.celrep.2017.09.081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Martin G.E., Pace M., Shearer F.M., Zilber E., Hurst J., Meyerowitz J., Thornhill J.P., Lwanga J., Brown H., Robinson N., et al. Levels of human immunodeficiency virus DNA are determined before ART initiation and linked to CD8 T-cell activation and memory expansion. J. Infect. Dis. 2020;221:1135–1145. doi: 10.1093/infdis/jiz563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Casazza J.P., Betts M.R., Picker L.J., Koup R.A. Decay kinetics of human immunodeficiency virus-specific CD8+ T cells in peripheral blood after initiation of highly active antiretroviral therapy. J. Virol. 2001;75:6508–6516. doi: 10.1128/JVI.75.14.6508-6516.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Hiener B., Eden J.S., Horsburgh B.A., Palmer S. Amplification of near full-length HIV-1 proviruses for next-generation sequencing. J. Vis. Exp. 2018;140 doi: 10.3791/58016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Van der Sluis R.M., Zerbato J.M., Rhodes J.W., Pascoe R.D., Solomon A., Kumar N.A., Dantanarayana A.I., Tennakoon S., Dufloo J., McMahon J., et al. Diverse effects of interferon alpha on the establishment and reversal of HIV latency. PLoS Pathog. 2020;16:e1008151. doi: 10.1371/journal.ppat.1008151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Rasmussen T.A., McMahon J.H., Chang J.J., Audsley J., Rhodes A., Tennakoon S., Dantanarayana A., Spelman T., Schmidt T., Kent S.J., et al. The effect of antiretroviral intensification with dolutegravir on residual virus replication in HIV-infected individuals: a randomised, placebo-controlled, double-blind trial. Lancet HIV. 2018;5:e221–e230. doi: 10.1016/S2352-3018(18)30040-7. [DOI] [PubMed] [Google Scholar]
- 56.Bacchetti P., Deeks S.G., McCune J.M. Breaking free of sample size dogma to perform innovative translational research. Sci. Transl. Med. 2011;3:87ps24. doi: 10.1126/scitranslmed.3001628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Huang Y., McCarthy D.J., Stegle O. Vireo: bayesian demultiplexing of pooled single-cell RNA-seq data without genotype reference. Genome Biol. 2019;20:273. doi: 10.1186/s13059-019-1865-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
-
•
Single-cell RNA-seq data have been deposited at ArrayExpress (Accession number E-MTAB-12039) and are publicly available as of the date of publication. Accession numbers are listed in the key resources table.
-
•
This paper does not report original code
-
•
Any additional information required to re-analyse the data reported in this paper is available from the lead contact upon request.