Summary
Increasing evidence indicates that gut microbiota may play a key role in vaccination immunity. Here, we investigate whether the human gut microbiota and metabolic function correlate with the BBIBP-CorV vaccine response. A total of 207 participants who received the BBIBP-CorV vaccine are enrolled. The gut microbiome and metabolic functions are investigated using metagenomic sequencing and metabolomic assays. We find that BBIBP-CorV vaccination is accompanied by altered microbiome composition and functional pathways, and the gut microbiome and its functional profiles correlate with the vaccine response. The levels of short-chain fatty acids (SCFAs) are much higher in the high antibody response group compared to the low response group, and several SCFAs display a positive correlation with the antibody response. Our study highlights that the gut microbiome and its function is associated with the BBIBP-CorV vaccine response, providing evidence for further exploration of microbiome modulation to improve COVID-19 vaccine efficacy.
Keywords: COVID-19 vaccine, gut microbiota, metabolic functions, SCFAs
Graphical abstract
Highlights
-
•
BBIBP-CorV vaccination is accompanied by altered microbiome profile
-
•
Gut microbiome and functional profiles could predict BBIBP-CorV vaccine response
-
•
Fecal and serum SCFA levels positively correlate with BBIBP-CorV vaccine response
Tang et al. report that gut microbiome and functional profiles are altered following BBIBP-CorV vaccination, and microbiome-related SCFA production positively correlated with the immune response of this inactivated COVID-19 vaccine. The findings give insights into microbiota regulation in improving the performance of COVID-19 vaccines.
Introduction
Coronavirus disease 2019 (COVID-19) is a respiratory infectious disease caused by severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), which has spread widely across the world1,2 and as of June 10, 2022 has caused approximately 5.5 million deaths worldwide (https://covid19.who.int/). Thus, safe and effective vaccine development is very important and necessary.3 Several types of vaccines have been developed using different approaches, such as recombinant viral vectors, viral recombinant protein, viral mRNA, or inactivated whole virus.4 There are currently >194 candidate vaccines in preclinical evaluation, and 139 vaccines were in clinical development as of January 14, 2022, according to a World Health Organization (WHO) report (https://www.who.int/publications/m/item/draft-landscape-of-covid-19-andidate-vaccines). Several candidate vaccines have been approved for use in the population and have shown efficacy,5, 6, 7, 8, 9, 10, 11 and ongoing studies continue to provide long-term safety and effectiveness.
Host environmental factors, such as gut microbiota, dietary nutrition, gender, age, genetics, health status, and regional factors have been reported to be involved in vaccine efficacy.12,13 Although the specific factors contributing to the differences in vaccine response are not fully understood, nutrition, intestinal microbiota, genetics, and regional factors have been identified as important determinants.12 Previous studies have indicated that there are important effects of gut microbiota in various physiological procedures such as metabolism, central nervous system function, inflammatory disorders, autoimmune diseases, and immunotherapy.12,14 Given the increasingly established link between gut microbiota and the immune system, the impact of microbiota on vaccine immunity remains poorly elucidated.12,15 Previous studies demonstrated that antibiotic-driven perturbation of the intestinal microbiome influences vaccine immunogenicity in healthy adults,14,16 providing increasing evidence that gut microbiota may be a major determinant of immunity to vaccination.
SARS-CoV-2 infection induces the host immune response to eliminate the virus, and disease severity is attributed to viral infection and host immune responses.15 Previous studies have shown that COVID-19 patients have prolonged gut microbiome dysbiosis, and the gut microbiota composition is associated with serum cytokines, inflammation markers, and disease severity of COVID-19.17,18 Moreover, gut microbiota dysbiosis is related to a wide spectrum of diseases.19 Increasing evidence has revealed that vaccination could drive the microbiota changes, and the composition and function of gut microbiota are crucial factors modulating immune responses to vaccination.15
The present study aimed to investigate whether the human gut microbiota and metabolic function correlate with the inactivated SARS-CoV-2 vaccine (BBIBP-CorV, CNBG) response. We recruited 207 participants to receive the BBIBP-CorV vaccine and performed metagenomic sequencing and targeted metabolomics to examine the role of the gut microbiome and the related metabolites in the BBIBP-CorV vaccination response.
Results
Study participants and clinical characteristics
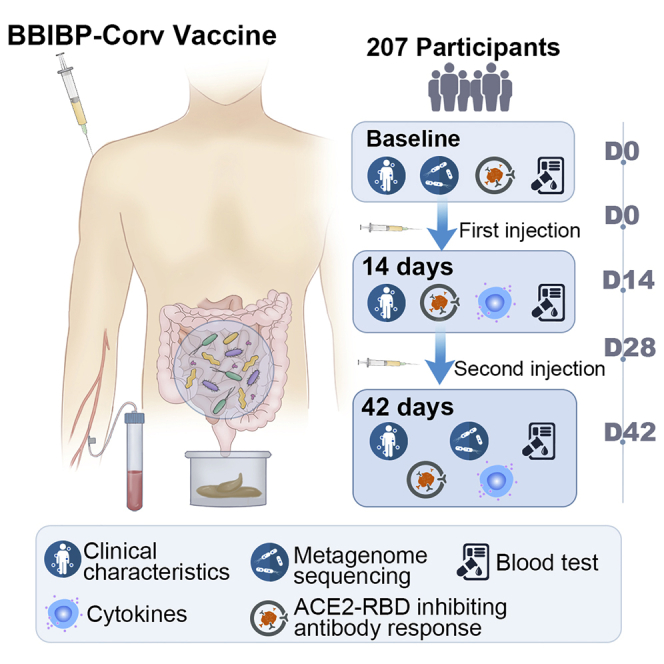
A total of 207 participants were enrolled in this study. All of the participants completed the two-dose BBIBP-CorV vaccination (day 0 and day 28). The demographics, clinical characteristics, and stool and serum collection schedules are shown in Figure 1A. All of the participants provided blood and fecal samples within the stipulated time. The baseline characteristics of the participants are shown in Table 1. The median (interquartile range [IQR]) age was 34 (29–46) years, and there were 136 females and 71 males in this cohort.
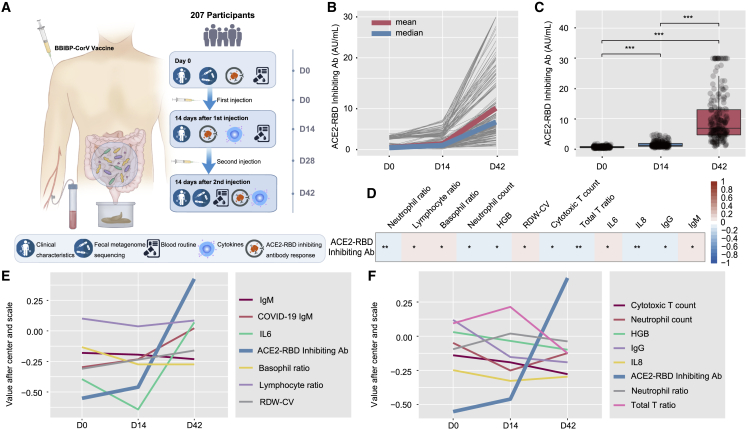
Figure 1.
Clinical characteristics and antibody response
(A) Schematic diagram of the experimental design.
(B) Levels of the ACE2-RBD inhibiting antibody of each participant at baseline (day 0), after the first injection (day 14), and after the second injection (day 42) (n = 207).
(C) Comparison of ACE2-RBD inhibiting antibody levels between days 0, 14, and 42 (n = 207). p values were determined by Welch ANOVA Games-Howell’s multiple comparisons test.
(D) Correlation of ACE2-RBD inhibiting antibody levels and clinical features by calculating the Pearson correlation coefficient between ACE2-RBD inhibiting antibodies (Abs) at days 0, 14, and 42 and clinical features at days 0, 14, and 42 (n = 207). Red represents the positive correlation. Blue represents the negative correlation. p values were determined by Pearson correlation analysis with FDR corrected.
(E) The variation trend of clinical parameters that were positively correlated with ACE2-RBD inhibiting antibody levels at days 0, 14, and 42. p values were determined by Pearson correlation analysis with FDR corrected (n = 207).
(F) The variation trend of clinical parameters that were negatively correlated with ACE2-RBD inhibiting antibody levels at days 0, 14, and 42. p values were determined by Pearson correlation analysis with FDR corrected. n = 207. ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001.
Table 1.
Baseline characteristics of participants
| All n = 207 |
Low group (n = 52) |
High group (n = 52) |
P | |
|---|---|---|---|---|
| Gender (%) | 0.273 | |||
| Male | 71 (34.3) | 22 (43.1) | 16 (30.8) | |
| Female | 136 (65.7) | 29 (56.9) | 36 (69.2) | |
| Age, y | 34.0 (29.0–46.0) | 35.0 (29.5–49.0) | 31.0 (27.0–39.5) | 0.071 |
| Height, cm | 162.0 (158.0–169.0) | 165.0 (158.0–171.0) | 162.0 (155.0–170.0) | 0.085 |
| Weight, kg | 59.0 (52.0–67.8) | 63.0 (52.0–70.0) | 56.0 (50.0–68.0) | 0.096 |
| BMI | 22.3 (20.5–24.0) | 22.27 (20.99–23.88) | 22.21 (19.48–24.5) | 0.376 |
| CRP | 5.00 (5.00–5.00) | 5.00 (5.00–5.00) | 5.00 (5.00–5.00) | 0.313 |
| WBC, 109/L | 5.72 (4.87–6.60) | 5.62 (4.95–6.68) | 5.72 (4.93–6.70) | 0.764 |
| Neutrophil ratio, % | 59.1 (54.0–65.3) | 59.3 (55.5–64.5) | 58.3 (53.7–64.9) | 0.470 |
| Lymphocyte ratio, % | 32.1 (26.1–36.8) | 31.8 (27.2–35.5) | 33.3 (26.9–37.7) | 0.380 |
| Monocyte ratio, % | 6.10 (4.90–7.35) | 6.10 (4.95–6.75) | 6.10 (4.55–7.15) | 0.825 |
| Eosinophil ratio, % | 1.50 (0.90–2.50) | 1.40 (0.90–2.35) | 1.35 (0.90–2.28) | 0.784 |
| Basophil ratio, % | 0.44 (0.30–0.60) | 0.40 (0.24–0.50) | 0.40 (0.33–0.60) | 0.078 |
| Neutrophil, 109/L | 3.41 (2.72–4.23) | 3.46 (2.84–4.15) | 3.38 (2.72–4.39) | 0.989 |
| Lymphocyte, 109/L | 1.75 (1.44–2.17) | 1.79 (1.42–2.24) | 1.90 (1.58–2.26) | 0.489 |
| Monocyte, 109/L | 0.34 (0.28–0.42) | 0.34 (0.28–0.40) | 0.36 (0.28–0.46) | 0.611 |
| Eosinophil, 109/L | 0.09 (0.05–0.14) | 0.09 (0.05–0.15) | 0.08 (0.06–0.14) | 0.976 |
| Basophil, 109/L | 0.02 (0.02–0.04) | 0.02 (0.01–0.03) | 0.03 (0.02–0.03) | 0.038 |
| HGB, g/L | 137 (128–147) | 142 (132–148) | 136 (128–144) | 0.130 |
| RBC, 1012/L | 4.53 (4.22–4.88) | 4.66 (4.24–4.94) | 4.50 (4.27–4.85) | 0.470 |
| HCT, % | 40.7 (38.4–42.9) | 42.1 (39.2–43.9) | 40.9 (38.8–42.2) | 0.103 |
| MCV, fL | 90.5 (87.4–93.3) | 89.9 (87.7–92.8) | 91.0 (86.4–93.4) | 0.815 |
| MCH, pg | 30.9 (29.6–31.8) | 30.9 (29.6–31.8) | 30.8 (29.6–31.4) | 0.434 |
| MCHC, g/L | 337 (330–346) | 340 (332–348) | 336 (329–344) | 0.153 |
| RDW-CV, % | 12.6 (12.3–13.1) | 12.6 (12.3–13.1) | 12.5 (12.3–12.9) | 0.424 |
| RDW-SD, fL | 41.7 (39.7–43.7) | 41.9 (39.7–43.9) | 41.0 (39.7–43.3) | 0.278 |
| PLT count, 109/L | 230 (196–268) | 234 (201–276) | 230 (198–257) | 0.810 |
| PCT, % | 0.25 (0.22–0.29) | 0.26 (0.22–0.30) | 0.24 (0.22–0.28) | 0.264 |
| MPV, fL | 10.8 (10.1–11.5) | 11.0 (10.4–11.9) | 10.5 (10.0–11.4) | 0.070 |
| P-LCR, % | 31.2 (26.3–37.8) | 33.2 (27.9–39.9) | 29.4 (25.8–36.9) | 0.076 |
| PDW, % | 13.0 (11.9–14.7) | 13.5 (12.4–15.4) | 12.8 (11.6–14.1) | 0.053 |
| Helper T ratio, % | 35.9 (31.6–41.1) | 35.0 (31.6–38.9) | 34.1 (31.1–41.1) | 0.953 |
| CTL ratio, % | 24.9 (21.0–30.3) | 25.4 (21.0–30.1) | 24.5 (21.2–31.7) | 0.680 |
| ALC per μL | 1,938 (1,553–2,400) | 1,976 (1,576–2417) | 2,034 (1,637–2,559) | 0.371 |
| Total T, per μL | 1,321 (1,074–1,650) | 1,385 (992–1,634) | 1,412 (1,172–1,780) | 0.305 |
| Helper T, per μL | 693 (546–844) | 688 (554–818) | 706 (602–876) | 0.287 |
| CTL count, per μL | 497 (372–642) | 496 (372–644) | 530 (404–728) | 0.384 |
| Total T ratio, % | 69.7 (64.8–74.6) | 68.7 (64.2–74.1) | 70.2 (64.2–74.7) | 0.584 |
| IL-6, pg/mL | 2.40 (2.00–3.35) | 2.20 (2.00–3.00) | 2.70 (2.00–3.42) | 0.077 |
| IL-8, pg/mL | 11.1 (7.32–47.1) | 11.5 (8.89–44.7) | 10.7 (6.43–40.8) | 0.402 |
| TNF-α, pg/mL | 6.30 (5.20–7.70) | 6.60 (5.60–7.55) | 6.10 (4.90–7.43) | 0.174 |
| IgG, g/L | 14.8 (13.0–16.6) | 16.1 (14.6–17.4) | 14.2 (12.0–15.5) | <0.001 |
| IgA, g/L | 2.75 (2.01–3.50) | 2.90 (2.11–3.82) | 2.66 (2.13–3.21) | 0.354 |
| IgM, g/L | 1.29 (0.92–1.71) | 1.15 (0.88–1.48) | 1.42 (1.12–1.88) | 0.008 |
| IgE, IU/mL | 27.0 (12.0–60.0) | 24.0 (14.0–47.0) | 39.0 (14.2–75.2) | 0.148 |
All of these parameters were taken at baseline (day 0). Values are expressed in number (percentage, %) and median (IQR, interquartile range). p values (low group versus high group) were calculated by the Mann-Whitney U test or Fisher exact test as appropriate. p < 0.05 is considered to be statistically significant. BMI, body mass index; CRP, C-reactive protein; WBC, white blood cell; HGB, hemoglobin; RBC, red blood cell; HCT, hematocrit; MCV, mean corpuscular volume; MCH, mean corpuscular hemoglobin; MCHC, mean corpuscular hemoglobin concentration; RDW-CV, red blood cell distribution width (coefficient of variation); RDW-SD, red blood cell distribution width (SD); PLT, platelet; PCT, plateletcrit; MPV, mean platelet volume; P-LCR, platelet large cell ratio; PDW, platelet distribution width; CTL, cytotoxic T cell; ALC, absolute lymphocyte count; IL, interleukin; TNF, tumor necrosis factor; Ig, immunoglobulin.
We evaluated the immunogenicity outcomes of these participants. None of the participants had any significant angiotensin-converting enzyme 2-receptor binding domain (ACE2-RBD) inhibiting antibody response or detectable virus-specific immunoglobulin G (IgG) and IgM at baseline (day 0) (Figures 1B and 1C; Table S1). The dynamic changes in the ACE2-RBD inhibiting antibody responses were further analyzed. Most of the participants started to generate an antibody response 14 days after the first injection (day 14) and reached high levels 14 days after the second injection (day 42) (Figure 1B). The median concentration of ACE2-RBD inhibiting antibody at day 14 after the first injection and at day 14 after the second injections (day 42) was 1.043 and 6.753 AU/mL (AU, arbitrary unit), respectively, and the mean concentrations of ACE2-RBD inhibiting antibody at days 14 and 42 were 1.383 and 10.136 AU/mL, respectively (Figures 1B and 1C; Table S1). For virus-specific IgG and IgM, 18.35% of participants tested positive for IgG after the first injection (day 14), and 81.15% of participants tested positive after the second injection (day 42). Moreover, 23.67% of participants were positive for IgM after the first injection (day 14) and 79.71% of participants tested positive after the second injection (day 42) (Table S1). The local injection site and systemic adverse reactions are shown in Table S2. The most common adverse reaction was pain at the injection site, with an occurrence rate of 21.3% of participants at day 14 and 26.1% of participants at day 42 (Table S2). The most common systemic adverse reactions after each vaccination were mild to moderate nausea and vomiting, which was reported by 5.3% of participants at day 14 and by 4.8% of participants at day 42 (Table S2). All of the reported adverse reactions were mild and transient, and they did not require any further treatment.
We then evaluated the laboratory tests and cytokines at days 0 and 14 after each injection. We found that the red blood cell, platelet, monocyte, eosinophil, and basophil counts showed no abnormalities at days 14 and 42 (Table S1). The blood lymphocyte subset and cytokine analysis showed no significant changes at days 14 and 42 (Table S1). We investigated further the potential correlation of baseline characteristics with the levels of ACE2-RBD inhibiting antibody. We observed that the lymphocyte, eosinophil, total T cell, and helper T cell counts as well as interleukin-8 (IL-8) levels at day 0 were positively associated with the ACE2-RBD inhibiting antibody levels at day 42 (Figure S1A). We further analyzed the correlation of trend changes of the laboratory parameters with the trend changes of ACE2-RBD inhibiting antibody levels during the whole vaccination period (days 0, 14, and 42). We found that the basophil ratio, lymphocyte ratio, red cell distribution width (RDW) ratio, IgM, and IL-6 were positively related to the ACE2-RBD inhibiting antibody response, while the neutrophil ratio, neutrophil count, hemoglobin, cytotoxic T cell count, total T cell ratio, and IL-8 were inversely correlated with the ACE2-RBD inhibiting antibody response (days 0, 14, 42) (Figures 1D–1F).
Profile of the gut microbiota after the BBIBP-CorV vaccination
Stool samples were collected at baseline (day 0) and at day 14 after the second injection (day 42). DNA was isolated from these stool samples, amplified, and sequenced using whole-genome sequencing. The metagenomic dataset, which had an average of 36,416,816 ± 4,505,278 paired-end reads per sample, was obtained by an Illumina NovaSeq 6000. Raw reads were preprocessed using KneadData to eliminate human DNA sequences and filter sequences with poor quality, which removed 3.74% of the reads. After quality control, 35,058,535 ± 4,373,185 reads per sample were obtained. Sequences were analyzed with MetaPhLan3 implemented within the HUMAnN3 pipeline. We selected a total of 1,144 clades, including 3 kingdom (L1) 12 phyla (L2), 26 classes (L3), 46 orders (L4), 96 families (L5), 234 genera (L6), and 727 species (L7).
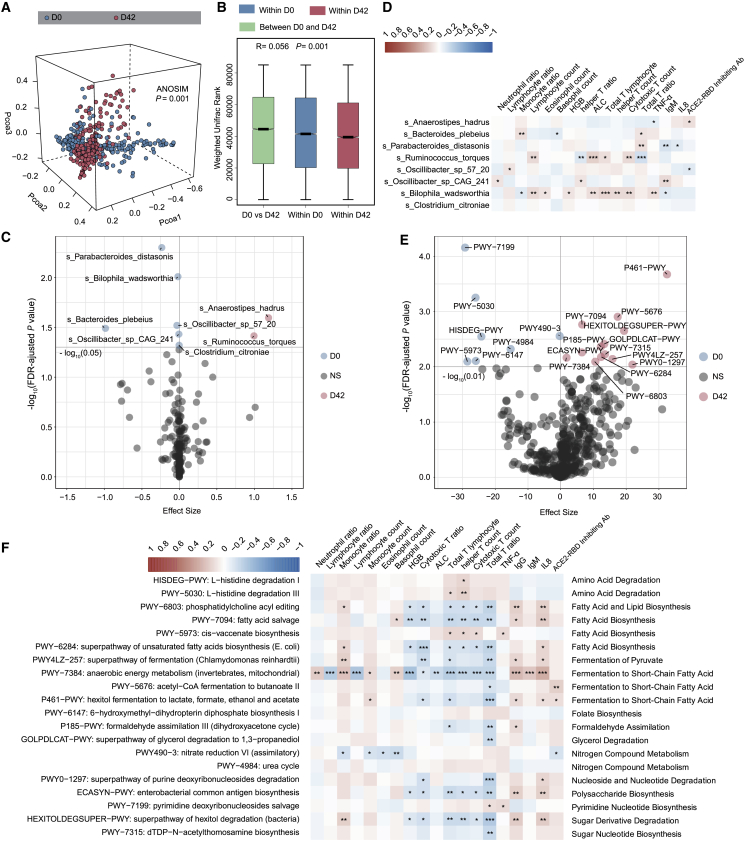
Alpha diversity was calculated to evaluate differences between baseline (day 0) and 14 days after the second injection (day 42). No significant difference at any level between days 0 and 42 was observed (Wilcoxon rank-sum test; Figure S1B; Table S3A). For beta diversity, weighted Unifrac or unweighted Unifrac distances were analyzed and plotted by principal coordinate analysis (PCoA) (analysis of similarities [ANOSIM]; p = 0.001; Figures 2A and S1C; Table S3B). The weighted Unifrac rank or unweighted Unifrac rank between days 0 and 42 was significantly higher than that within days 0 and 42 (ANOSIM; p = 0.001; Figures 2B and S2C; Table S3B). The participants were primarily characterized by a predominance of Parabactoides distasonis, Bilophila wadsworthia, and Bacteroides plebeius at baseline (day 0), whereas Anaerostipes hadrus, Ruminococcus torques, and Oscillibacter were more abundant 14 days after vaccination (day 42) (Figures 2C, S1E, and S1F; Table S3C). We then analyzed the correlation between the differential species at day 0 and the clinical features at day 42. We found that the microbial species at day 0 showed significant correlations with monocytes, total T cells, cytotoxic T cells, helper T cells, IL-8 concentration, and tumor necrosis factor α (TNF-α) concentration at day 42 (Pearson correlation analysis with false discovery rate [FDR] corrected; ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001; Figure 2D). Our data showed that BBIBP-CorV vaccination is accompanied by an altered profile of gut microbiome.
Figure 2.
The gut microbiome is altered in participants receiving the inactivated COVID-19 vaccine (BBIBP-CorV)
(A) Weighted Unifrac 3D PCoA (principal coordinate analysis) plot of samples at baseline (day 0) and after the second injection (day 42) (n = 207). The ANOSIM test was used to calculate the significance of dissimilarity (ANOSIM, p = 0.001).
(B) Comparison of the weighted Unifrac range of samples between baseline (day 0) and after the second injection (day 42) (n = 207). The ANOSIM test was used to calculate the significance of dissimilarity (ANOSIM, p = 0.001).
(C) Volcano plot of differentially enriched bacteria between samples at baseline (day 0) and samples after the second injection (day 42) (n = 207). Pink dots represent bacteria with relatively higher abundance in samples after second injection (day 42). Blue dots represent bacteria with relatively higher abundance in samples at baseline (day 0). Gray dots represent bacteria with no significant difference between the 2 groups. p values were adjusted by the Benjamini-Hochberg (BH) method to control FDR. FDR-adjusted p < 0.05 was shown.
(D) Correlation heatmap of differentially enriched bacteria at day 0 with the clinical characteristics at day 42 (n = 207). Pearson correlation analysis with FDR-adjusted p < 0.05 was shown.
(E) Differentially enriched MetaCyc pathways between samples at days 0 and 42. Pink dots represent MetaCyc pathways with relatively higher counts per million (CPM) in samples at day 42 (n = 207). Blue dots represent MetaCyc pathways with relatively higher CPM in samples at day 0. Gray dots represent bacteria with no significant difference between the 2 groups. p values were adjusted by BH method to control FDR. FDR-adjusted p < 0.01 is shown.
(F) Correlation heatmap of differentially enriched MetaCyc pathways at D0 with the clinical characteristics at day 42. Pearson correlation analysis with FDR-adjusted p < 0.05 is shown. n = 207. ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001.
Altered microbiome functional profile after BBIBP-CorV vaccination
Because there was a difference in microbiome composition between pre- and post-BBIBP-CorV vaccination, we investigated further whether the microbiome compositional differences could translate into functional differences by analyzing the functional pathways. The composition of microbial pathways in participants at baseline (day 0) was significantly different from that of participants at 14 days after the second injection (day 42). Compared to baseline, 7 MetaCyc pathways were decreased and 13 MetaCyc pathways were enriched in participants at day 42 (Maaslin2; p < 0.05; Figures 2E and S1G; Table S4A). The 7 decreased pathways were mainly related to pyrimidine nucleotide biosynthesis, amino acid degradation, fatty acid biosynthesis, folate biosynthesis and the urea cycle, while 13 of the enriched pathways were mainly involved in fermentation to short-chain fatty acids (SCFAs), nucleotide degradation, sugar nucleotide biosynthesis, fermentation to pyruvate, and polysaccharide biosynthesis (Figure S1G).
We then sought to investigate whether these changed pathways correlated with the clinical features to evaluate the potential role of microbial functions in the immune response. We found that several pathways mainly related to fatty acid biosynthesis and fermentation to SCFAs at day 0 showed significant correlations with lymphocytes, the T cell ratio, and IL-8 concentration at day 42 (Figure 2F; Table S4B). More important, these pathways related to fermentation to SCFAs at day 0 were positively correlated with the ACE2-RBD inhibiting antibody response at day 42, while nitrogen compound metabolism at day 0 showed a negative correlation with the ACE2-RBD inhibiting antibody response at day 42 (Figure 2F; Table S4B). We further found that A. hadrus was the primary contributor to fermentation in the SCFA pathway (PWY-5676: acetyl-coenzyme A [CoA] fermentation to butanoate II, and P461-PWY: hexitol fermentation to lactate, formate, ethanol, and acetate) (Figure S1H), suggesting that the microbiome functional profile is altered after BBIBP-CorV vaccination and that the baseline functional pathways may be associated with the vaccine response.
The gut microbiome and its functional profile correlate with the antibody response of the BBIBP-CorV vaccine
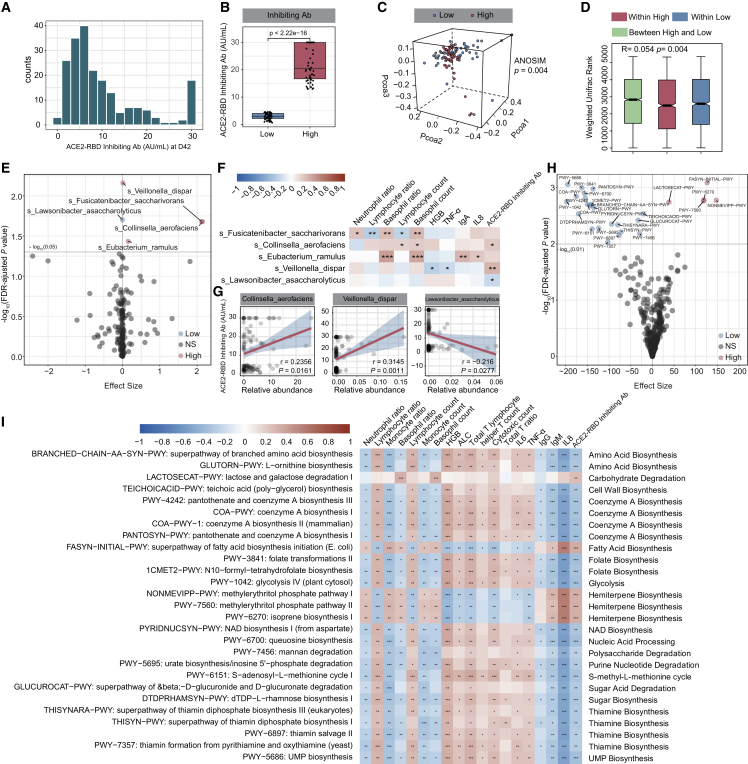
After profiling the signature of the gut microbiota post-BBIBP-CorV vaccination, we explored whether the gut microbiota and functional pathways correlate with the vaccine response. We divided the participants into two groups according to the concentration of ACE2-RBD inhibiting antibody at day 42 as follows: low 25% (low group) and high 25% (high group). The concentration of ACE2-RBD inhibiting antibody at day 42 in the high group was much higher than that in the low group (Mann-Whitney U test; p < 0.001; Figures 3A and 3B). We tested whether the baseline characteristics affect the ACE2-RBD inhibiting antibody response. Interestingly, the baseline characteristics, including age, gender, and BMI, were comparable between the low and high groups (Mann-Whitney U test; Table 1). There was a significant difference in the total IgG and IgM between the low and high groups at day 0 (Table 1). No significant differences in alpha diversity were observed between the low and high groups at day 0 (Mann-Whitney U test; Figure S2A; Table S3A). PCoA analysis showed a notable difference in the gut microbial abundance between the two groups at day 0 (ANOSIM; p = 0.004; Figure 3C; Table S3B). The weighted Unifrac rank between the two groups was significantly higher than that within each group at day 0 (ANOSIM; p = 0.004; Figure 3D; Table S3B). We then performed a comparison of the two groups at day 0 and identified that Collinsella aerofaciens, Fusicatenibacter saccharivorans, Eubacterium ramulus, and Veillonella dispar were significantly increased in the high group, while Lawsonibacter asaccharolyticus was enriched in the low group (Maaslin2; p < 0.05; Figures 3E, S2B, and S2C). We then investigated the correlation between these changed species at day 0 and the clinical parameters at day 42 in the high and low groups. Interestingly, we observed that the abundances of C. aerofaciens and V. dispar at day 0 were positively associated with the concentrations of ACE2-RBD inhibiting antibody at day 42, while L. asaccharolyticus at day 0 was negatively correlated with the ACE2-RBD inhibiting antibody response in the high and low groups at day 42 (Figures 3F and 3G).
Figure 3.
Gut microbiome and microbiome functional profile correlated with BBIBP-CorV vaccine response
Participants were divided into 2 groups (high group versus low group) according to the ACE2-RBD inhibiting antibody levels at day 42 using the second quartile and the third quartile.
(A) Distribution histograph of the ACE2-RBD inhibiting antibody levels at day 42 (n = 207).
(B) Comparison of ACE2-RBD inhibiting antibody levels at day 42 between the high and low groups (low group, n = 52; high group, n = 52). p values were determined by Mann-Whitney U test.
(C) Weighted Unifrac PCoA 3D plot of samples at day 0 between the high and low groups. The ANOSIM test was used to calculate the significance of dissimilarity (ANOSIM, p = 0.004).
(D) Comparison of the weighted Unifrac range of samples at day 0 between the high and low groups (low group, n = 52; high group, n = 52). The ANOSIM test was used to calculate the significance of dissimilarity (ANOSIM, p = 0.004).
(E) Volcano plot of differentially enriched bacteria at day 0 between samples of the high and low groups (low group, n = 52; high group, n = 52). Pink dots represent bacteria with relatively higher abundance in the high group. Blue dots represent bacteria with relatively higher abundance in the low group. Gray dots represent bacteria with no significant difference between the high and low groups. p values were adjusted by the BH method to control FDR. FDR-adjusted p < 0.05 is shown.
(F) Correlation heatmap of differentially enriched bacteria at day 0 with the clinical characteristics at day 42 (low group, n = 52; high group, n = 52). Pearson correlation analysis with FDR-adjusted p < 0.05 is shown.
(G) Correlation of differentially enriched bacteria at day0 with the ACE2-RBD inhibiting antibody levels at day 42. p values were determined by Pearson correlation analysis (low group, n = 52; high group, n = 52).
(H) Volcano plot of differentially enriched MetaCyc pathways at day 0 between samples from the high and low groups (low group, n = 52; high group, n = 52). p values were adjusted by the BH method to control FDR. FDR-adjusted p < 0.01 is shown.
(I) Correlation heatmap of differentially enriched MetaCyc pathways at day 0, with the clinical characteristics at day 42. Pearson correlation analysis with FDR-adjusted p < 0.05 is shown (n = 207). ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001.
See also Figures S2 and S3 and Table S4.
We further observed a notable difference in the composition of MetaCyc pathways at day 0 between the two groups (Figures 3H and S2D; Table S4C). Five microbial pathways, mainly involved in carbohydrate degradation, fatty acid biosynthesis, and hemiterpene biosynthesis, were enriched in the high group, while another 22 pathways (e.g., amino acid biosynthesis, CoA biosynthesis, folate biosynthesis, nucleic acid processing, sugar acid degradation, thiamine biosynthesis) were decreased at day 0 (Figures 3H and S2D; Table S4C). We further found that most of these pathways at day 0 showed a significant correlation with immune cells and cytokines at day 42 (Pearson correlation analysis with FDR corrected; ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001; Figure 3I; Table S4D). Importantly, lactose and galactose degradation I pathways as well as the superpathway of fatty acid biosynthesis initiation mainly involved in fatty acid production at day were positively correlated with the levels of ACE2-RBD inhibiting antibody at day 42 (Figures 3I and S2E; Table S4D). We further investigated which bacterial species contributed most to these functional pathway changes. Interestingly, most of these pathways were dominated by several microbial species. For example, F. saccharivorans and C. aerofaciens were the primary contributing species to the fatty acid production pathways (Figure S3A).
To investigate the potential utility of gut microbial and functional pathways in predicting the ACE2-RBD inhibiting antibody response, we constructed several machine learning classifiers to discriminate the low and high groups by using differentially enriched MetaCyc pathways and differentially enriched species at day 0 as the variables of the classifiers (Figure S3B). Because we did not have an independent test cohort, 10-fold cross-validation was used to test the performance of the differentially enriched species and the differential MetaCyc pathways alone or combined (Figure S3C). The model based on microbial features performed better than MetaCyc pathways with an area under the curve (AUC) of 0.746 (Figure S3C). Moreover, the integration of taxonomic features and MetaCyc pathways improved the performance of classification, with an AUC of 0.769 (Figure S3C).
Fecal and serum levels of SCFAs positively correlated with BBIBP-CorV vaccination response
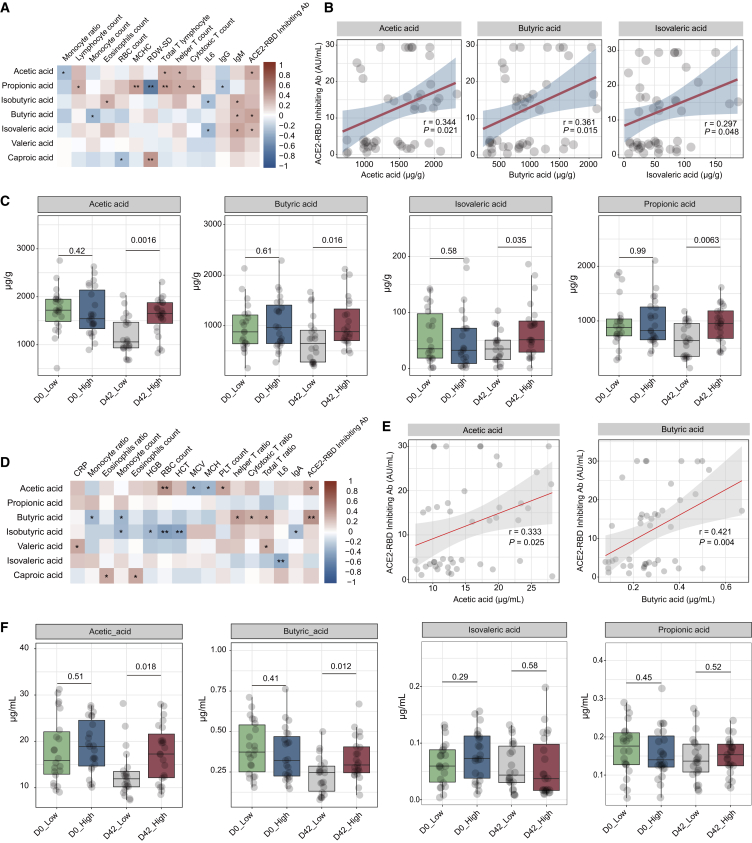
It is well known that the gut microbiota participates in host regulation via the modulation of metabolites and the fecal metabolome. Considering that several pathways involved in carbohydrate degradation and fatty acid biosynthesis showed a positive correlation with the ACE2-RBD inhibiting antibody response, we further used targeted metabolic analysis to verify the changes in fecal and serum metabolites after vaccination. We found that several fecal SCFAs were associated with immune cells at day 42 (Pearson correlation analysis with FDR corrected, ∗p < 0.05, ∗∗p < 0.01; Figure 4A; Table S5). In addition, the levels of acetic acid, butyric acid, and isovaleric acid at day 42 displayed a positive correlation with the ACE2-RBD inhibiting antibody production at day 42 (Figure 4B; Table S5). Fecal levels of SCFAs did not differ significantly between the high and low groups at baseline (day 0), while there was a significant difference in the levels of SCFAs, including acetic acid, butyric acid, isovaleric acid, and propionic acid between high and low groups after vaccination (day 42) (Figure 4C; Figure S4). In addition, we observed that serum levels of acetic acid and butyric acid at day 42 were positively correlated with the ACE2-RBD inhibiting antibody response at day 42 (Figures 4D and 4E), and the serum concentrations of acetic acid and butyric acid were lower in the low group compared to the high group after vaccination (day 42) (Figures 4F and S4). These findings demonstrated that fecal and serum SCFAs levels may be associated with the BBIBP-CorV vaccination response.
Figure 4.
Fecal and serum levels of SCFAs correlated with BBIBP-CorV vaccine response
(A) Correlation heatmap of fecal levels of SCFAs with clinical characteristics at day 42 (n = 45). Pearson correlation analysis with FDR-adjusted p < 0.05 is shown.
(B) Correlation dot plot of fecal levels SCFAs with levels of ACE2-RBD inhibiting antibody at day 42 (n = 45). p values were determined by Pearson correlation analysis.
(C) Boxplots comparing fecal levels SCFAs between the high and low groups at days 0 and 42 (n = 45). Comparisons between subgroups were performed using the Mann-Whitney U test.
(D) Correlation heatmap of serum concentrations of SCFAs with clinical characteristics at day 42 (n = 45). Pearson correlation analysis with FDR-adjusted p < 0.05 is shown.
(E) Correlation dot plot of serum concentrations SCFAs with levels of ACE2-RBD inhibiting antibody at day 42 (n = 45). p values were determined by Pearson correlation analysis.
(F) Boxplots comparing serum concentrations SCFAs between the high and low groups at days 0 and 42. Comparisons between subgroups were performed using the Mann-Whitney U test. D0_Low n = 22, D0_High n = 23, D42_Low n = 22, D42_High n = 23. ∗p < 0.05, ∗∗p < 0.01.
Discussion
The COVID-19 pandemic remains a worldwide medical issue. Currently, there are lack of specific antiviral medicines to treat this infection. Vaccines have become the most effective strategy to prevent the spread of the pandemic. Previous studies reported that good immunogenicity could be observed for all vaccine types (e.g., mRNA vaccine, inactivated vaccine, adenovirus vaccine), and the average efficacies of mRNA-based COVID-19 vaccines, inactivated vaccines, and adenovirus vaccines were 94.6%, 91.3%, and 80.2%, respectively.20,21 The two inactivated SARS-CoV-2 vaccines (BBIBP-CorV and CoronaVac) have been mainly adopted for mass vaccination in China, and showed comparable immune efficacy in the population.22,23 There are many factors that can affect vaccine immunogenicity and efficacy, including genetics, diet, nutrition, infections, and other vaccine-related factors.13,15 Therefore, a sufficient understanding of the factors related to COVID-19 vaccine efficacy is urgently needed. More important, increasing evidence indicates that the gut microbiota is an important factor that modulates the baseline immune status and vaccine response.15 This study investigated the gut microbiota profile changes after the BBIBP-CorV vaccination and evaluated the possible contribution of the gut microbiome and metabolites to this vaccine response. We found that the BBIBP-CorV vaccination is accompanied by an altered gut microbiome composition, and that the specific gut microbiome and related functional profile may be associated with the BBIBP-CorV vaccine response.
In the present study, the BBIBP-CorV vaccine was safe and tolerated in all of the participants, and no serious adverse reactions were observed. The most common adverse reaction was pain at the injection site, and the incidence rates of adverse reactions were comparable to those reported in the previous studies.9,10,22,24 The ACE2-RBD inhibiting antibody response began to increase in most participants 14 days after the first injection (day 14) and reached high levels 14 days after the second injection (day 42), which was consistent with previous reports.6,9 Several recent studies have demonstrated that the SARS-CoV-2 infection could activate innate and adaptive immune responses, causing decreased numbers of circulating CD4+ cells, CD8+ cells, B cells, natural killer (NK) cells, monocytes, eosinophils, and basophils in COVID-19 patients.25, 26, 27, 28 Moreover, increased levels of serum proinflammatory cytokines such as IL-6, IL-1β, IL-8, monocyte chemoattractant protein-1 (MCP-1), chemokine ligand 3 (CCL3), and TNF-α, were observed in most COVID-19 patients.29,30 We did not find any significant changes in the blood lymphocyte subset or cytokine analysis over time after vaccination, suggesting that the vaccination does not affect these cells in the same manner as the natural SARS-CoV-2 virus infection. Moreover, we observed that various immune cells and cytokines correlated with the ACE2-RBD inhibiting antibody response, implying that the baseline immune status may be associated with the vaccine response.
The gut microbiota is well known to be involved in the development of immune cells and maintenance of immune responses,31,32 and intestinal dysbiosis is correlated with immunological disequilibrium.33 In COVID-19 patients, a balanced gut microbiome may be important in maintaining an optimal immune response to limit excessive immune reactions, while the disruption of the gut microbiome may predispose individuals to a severe inflammatory response.17,34 Previous studies have shown that the gut microbiome is disturbed in COVID-19 patients and that the microbiota composition is related to serum cytokines, chemokines, and inflammation markers, indicating that the gut microbiome may modulate the host immune response and affect disease severity.17,18 Here, we observed that BBIBP-CorV vaccination was accompanied by an altered gut microbiome composition. Previous studies demonstrated that the composition of gut microbiota and diverse species of microbes could affect the development and function of the host immune system, regulate the distinct branches of the adaptive immune system, and contribute to the activation, polarization, and function of T cells.35, 36, 37 Emerging evidence reveals a key role for the gut microbiome in controlling the development and function of various immune cells and immune responses to vaccination.12,16 Here, we revealed that several bacterial species were related to altered immune cells and serum cytokines during the BBIBP-CorV vaccination. Among these species, B. wadsworthia has been demonstrated to exacerbate intestinal inflammation and induce systemic inflammation.38,39 R. torques has been reported to regulate the immune response in inflammatory bowel disease.40 In addition, Parabacteroides distasonis may exert protective effects in various inflammatory diseases,41 and A. hadrus is associated with a lower host inflammatory risk.42 However, it remains unknown whether these altered microbiomes in the BBIBP-CorV vaccination play an active role in the BBIBP-CorV vaccine response.
The gut microbiota modulates immune responses to vaccination, and the roles of the gut microbiome in the immune response to vaccination have been increasingly considered an attractive target.15 A close link between the gut microbiota and vaccine response has been revealed in several observational clinical studies.43,44 Other studies have found that perturbations of the gut microbiota lead to altered responses to vaccination.14,16,45 Moreover, the immune response to vaccines has been closely associated with prevaccination immune status, which may be attributed to the gut microbiota profile before vaccination. In the present study, the baseline characteristics, such as age, BMI, and clinical parameters, did not differ between the high response and low response groups, further emphasizing the possible role of microbiota in the BBIBP-CorV vaccine response. We observed no differences in α diversity between the two groups, suggesting that intestinal microbiota diversity is not related to the antibody response to the BBIBP-CorV vaccine. However, we observed a significant difference in the prevaccination microbiome beta diversity and microbiota composition between the two groups. Microbial species, including C. aerofaciens, F. saccharivorans, E. ramulus, and V. dispar, were significantly enriched in the high response group. In addition, the increased abundance of C. aerofaciens and V. dispar was positively correlated with the high levels of ACE2-RBD inhibiting antibody. Previous studies have reported that C. aerofaciens augments the T cell response and improves anti-programmed cell death ligand-1 (PD-L1) therapy efficacy.46 In addition, V. dispar is dominant in the PD-L1 immunotherapy responder group47 and is associated with influenza-specific H1 and H3 IgA antibody responses.48 Although we used machine learning classifiers to investigate the potential utility of gut microbial in predicting the ACE2-RBD inhibiting antibody response, a lack of an independent test cohort is a limitation, further cause-and-effect study is still needed to investigate the mechanism of how the microbiota modulates the BBIBP-CorV vaccine response.
The gut microbiota produces various metabolites that can modulate the immune response.15 A previous study revealed that antibiotic-driven perturbation of the microbiome affected the bile acid metabolism and immune responses to the influenza vaccination.16 In our study, we did not observe significant changes in the bile acid metabolism pre- and post-vaccination. Antibiotic use would notably disturb the microbiome and metabolome, which may lead to this discrepancy. In this study, we observed that the functional pathways mainly involved in fatty acid production were positively correlated with the levels of ACE2-RBD inhibiting antibody. We revealed that C. aerofaciens was the primary species contributing to the fatty acid production pathways. C. aerofaciens has recently been characterized to be a butyric acid-producing bacterium.49 SCFAs, including acetate, butyrate, propionate, isovaleric acid, caproic acid, and valeric acid, are the main metabolic products of bacterial fermentation.15 A previous study has demonstrated that SCFAs activate B cell metabolism and promote antibody responses in a mouse model of Citrobacter rodentium infection.50 A recent study has observed that microbial-derived SCFAs improve the humoral immune response to seasonal influenza vaccine.51 Fecal concentrations of SCFAs are significantly lower in COVID-19 patients, and SARS-CoV-2 infection may be related to the impaired capacity for SCFA production.52 In the present study, we observed that several SCFAs displayed a slight but nonsignificant decrease after the BBIBP-CorV vaccination. More important, we observed that the fecal and serum levels of SCFAs were much higher in the high response group compared to the low response group after vaccination. Because previous studies demonstrated that SARS-CoV-2 infection might be related to impaired capacity for SCFA production,52 we hypothesize that various factors (e.g., dietary changes, microbiota profiles, vaccination) may contribute to the difference in the SCFAs levels after vaccination. Notably, we observed that the increased SCFAs levels were positively correlated with the levels of ACE2-RBD inhibiting antibody, suggesting that SCFAs may have potential in improving antibody responses during the BBIBP-CorV vaccination.
Our study demonstrated that the BBIBP-CorV vaccination is accompanied by altered gut microbiome composition and functional profiles. The gut microbiome and microbiome-related SCFAs may be associated with the BBIBP-CorV vaccine response. Our study highlighted the potential link between gut microbiome function and the BBIBP-CorV vaccine response, which may help to improve the performance of COVID-19 vaccines.
Limitations of the study
There were several limitations. First, there was a lack of an independent validation cohort to validate the potential utility of gut microbial in predicting antibody response. Second, there was a lack of information for the ACE2-RBD inhibiting antibody decline after the second injection. Long-term follow-up is needed in further research to investigate whether the gut microbiota and its functional metabolites affect the antibody decline. Third, although we adjusted various covariates that may affect the microbial profile, information was lacking for dietary habits or diet consumption, which may affect the microbiota profile and SCFAs production. Fourth, our data revealed the correlations among the gut microbiome, metabolites, and the BBIBP-CorV vaccine response, but we were unable to differentiate whether the identified correlations were the cause of or result of vaccination. Cause-and-effect relationships using a mouse model are needed.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Mouse-anti-human CD3-FITC (clone UCHT1) | Agilent | Cat#F081801; RRID: AB_578681 |
| Mouse-anti-human CD4-APC (clone MT310) | Agilent | Cat#C722601; RRID: AB_578593 |
| Mouse-anti-human CD8-RPE (clone DK25) | Agilent | Cat#R080601; RRID: AB_579542 |
| Mouse-anti-human CD45-APC (clone T29/33) | Agilent | Cat#C723001; RRID: AB_578595 |
| Biological samples | ||
| Serum | This paper | N/A |
| Feces | This paper | N/A |
| Chemicals, peptides, and recombinant proteins | ||
| TIANamp Stool DNA Kit | TIANGEN | Cat#DP328 |
| phosphoric acid | Sigma-Aldrich | Cat#345245 |
| diethyl ether | Sigma-Aldrich | Cat#673811 |
| C-reactive protein kit | Beckman Coulter | Cat#OSR6299 |
| IL-6 detection kit | Siemens | Cat#LK6P1 |
| IL-8 detection kit | Siemens | Cat#LK8P1 |
| TNF-α detection kit | Siemens | Cat#0274 |
| IgG | Beckman Coulter | Cat#OSR61172 |
| IgA | Beckman Coulter | Cat#OSR61171 |
| IgM | Beckman Coulter | Cat#OSR61173 |
| IgE | Beckman Coulter | Cat#IM2122 |
| Critical commercial assays | ||
| Coronavirus (2019-nCoV) IgM antibody diagnostic kit | Bioscience Co. | Cat#20203400182 |
| Coronavirus (2019-nCoV) IgG antibody diagnostic kit | Bioscience Co. | Cat#20203400183 |
| ACE2-RBD inhibiting assay | Bioscience Co. | N/A |
| GC-MS analysis | PANOMIX Tech Co. | N/A |
| Deposited data | ||
| Raw sequencing data | National Genomics Data Center | https://ngdc.cncb.ac.cn/gsa/ |
| Software and algorithms | ||
| GraphPad Prism v.9 | Graphpad | http://graphpad-prism.cn/ |
| R v4.0.2 | R Core Team | https://www.R-project.org/ |
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Dr. Shiming Yang (Yangshiming@tmmu.edu.cn).
Materials availability
This study did not generate new unique reagents.
Experimental model and subject details
Study cohort
This study was approved by Ethics Committee of Xinqiao Hospital, Third Military Medical University, which was registered at Chinese Clinical Trial Registry (ChiCTR2100044539). We undertook the study in accordance with the 2013 Declaration of Helsinki. Written informed consent was signed by all participants before enrollment. Healthy adults aged 18 to 59 years from Xinqiao Hospital were recruited between 1 April 2021 and 1 July 2021. Participants were considered eligible for this research if they met the following criteria1: General good health as established by medical history and physical examination2; seronegative for serum-specific IgM/IgG antibodies against SARS-CoV-23; Women without pregnancy4; Participants are willing to complete the whole research procedure5 Participants have the ability to understand the research procedures, to sign the informed consent voluntarily after explanation, and can comply with the requirements of the research team. Participants who met the inclusion criteria were further evaluated for the following exclusion criteria1: Those with positive antibody tests of the COVID-192; Those with fever, fatigue, nasal obstruction, myalgia, diarrhea, shortness of breath, and dyspnea within 14 days before inoculation3; Those with clinically abnormal parameters from blood biochemical, blood routine, and urine routine before inoculation4; Those who have experienced severe allergic reactions5; Those receiving immune-enhancement or inhibitor treatment (over 14 days within 3 months6; Those receiving live-attenuated vaccines within one month before injection or other vaccines within 14 days before inoculation7; Those receiving antibiotics, probiotics or other study drugs within 1 month before inoculation8; Those under other conditions not suitable for the study.
The recruited participants received inactivated SARS-CoV-2 Vaccine (BBIBP-CorV) which was developed by the Beijing Institute of Biological Products (Beijing, China).53 The inactivated SARS-CoV-2 Vaccine (BBIBP-CorV) was approved for clinical use in China and showed general safety, immunogenicity, and efficacy in previous phase 1/2 clinical trial and phase 3 randomized clinical trial.9,10,22,24 BBIBP-CorV was injected through intramuscular on Days 0 (first injection) and 28 (second injection) (Figure 1A). Before the second injection, the following participants were not allowed to be injected for the second injection: Women with positive urine pregnancy tests; Those with high fever lasting for three days or severe allergic reaction after the first injection; Serious adverse reactions related to the previous injection; If the investigators found that the participant did not meet the inclusion criteria or the participant met the exclusion criteria after the first injection; Other reasons for exclusion evaluated by investigators. The local and systemic reactions were recorded after each injection (Table S2).
The previous literature reported that the bacterial groups were significantly correlated with antibody response of the rotavirus vaccine with a minimum correlation coefficient of 0.2.43 Based on the previous study, a sample size of 194 participants was calculated to be sufficient for a two-tailed significance level of 5% and a power of 80%, assuming a minimum correlation coefficient of 0.2 between bacterial species and BBIBP-CorV vaccine response.54 This was adjusted to 214 participants to allow for 10% dropouts. We totally screened 276 participants during the study period. The participants would be withdrawn if any medications, antibiotics or probiotics were used during the vaccination period (Figure S5). Two participants refused to complete the study after the first injection and were excluded (Figure S5). After exclusion, 207 participants were included in the final analysis (Figure S5).
Sample processing and storage
Blood and fecal samples from all eligible participants were collected on Days 0, 14 and 42. The experienced nurses drew blood from the participants in our hospital, and 5–10 mL of blood in anticoagulant was collected from each participant and transported to clinical laboratory for plasma separation and tests within 2 h. The residual serum was stored in at −80°C until use. Fecal samples were self-collected in DNA preservative tube, transferred to laboratories within 2 h and stored at−80°C until DNA extraction. Routine blood tests, blood lymphocyte subsets, T cell subsets and key cytokines were measured on Days 0, 14 and 42. Specific IgG, specific IgM and neutralizing antibodies were analyzed.
Method details
Fecal genomic DNA extraction
Fecal genomic DNA was extracted from fecal samples using the TIANamp Stool DNA Kit (TIANGEN, Beijing, China) according to the manufacturer’s instructions. The concentration and purity of the extracted DNA were evaluated using a NanoDrop 2000C spectrophotometer (Thermo Fisher Scientific, Waltham, USA).
Metagenomic sequencing and data analysis
The libraries were sequenced using the Illumina NovaSeq 6000 platform by Novogene (Tianjin, China). KneadData (v0.7.4) software55,56 was used to process the raw sequencing data. KneadData software was run by calling Bowtie2 (v2.4.2)57 to remove host genome contamination in samples. Trimmomatic (v0.39)58 was used to remove sequencing primers to obtain clean data. Metaphlan3 software55 was run to obtain the taxonomic counts in each sample. Functional annotations and pathways were analyzed using the data files from the HMP Unified Metabolic Analysis Network 3.0 (HUMAnN 3.0).55 HUMAnN is a method for efficiently and accurately profiling the abundance of microbial functional pathways from metagenomic sequencing data. The clean paired-end sequencing data were merged into a single fastq file. The HUMAnN 3.0 toolkit was run by using the “humann --input myseq∗.fq --output humann3/--threads 32 --memory-use maximum -r -v” command, which calls Bowtie257 to compare nucleic acid sequence and calls DIAMOND59 to compare protein sequences to complete gene/protein function annotation and to obtain MetaCyc pathway annotation. HUMAnN 3.0 quantifies genes and pathways in units of RPKs (reads per kilobase). These account for gene length but not sample sequencing depth. Humann3 build-in script humann_renorm_table provides the choice to normalize to relative abundance or copies per million (CPM) units. Both of these represent “total sum scaling (TSS)”-style normalization: in the former case, each sample is constrained to sum to 1, whereas in the latter case (CPMs) samples are constrained to sum to 1 million (HUMAnN 3.0 User Manual: https://github.com/biobakery/humann).
Differences in bacterial abundance and the Metacyc pathway were analyzed using Maaslin2.60 Shannon, Simpson and richness indices were calculated using the R Community Ecology Package vegan.61 Weighted Unifrac distance was calculated using Metaphlan3 R script “Unifrac_distance.r” and root-tree file “mpa_v30_CHOCOPhlAn_201901_species_tree.nwk”. The PCoA results were calculated and visualized using R build-in functions and the plot3D R package.62 The ANOSIM test was used to calculate the significance of dissimilarity using the R Community Ecology Package vegan.61 All machine learning classifiers were built by using the caret R package.63 The ROC-AUC was performed using the pROC R package.56 Pearson correlation and p values were evaluated using the rcorr function in the Hmisc R package.64
Detection of antibody response
Serum samples were collected to measure COVID-19 IgG, IgM and the ACE2-RBD inhibiting antibodies. All serum samples were inactivated at 56°C for 45 min before testing. The levels of IgG and IgM against SARS-CoV-2 were measured using the commercial magnetic chemiluminescence enzyme immunoassay (MCLIA) kits: Coronavirus (2019-nCoV) IgM/G antibody diagnostic kit (plate CLIA) supplied by Bioscience Co. (China National Medical Products Administration, approval numbers 20203400183 [IgG] and 20203400182 [IgM]). According to the manufacturer’s instructions, based on the double-antibody sandwich immunoassay, the recombinant antigens contain the nucleoprotein and a peptide from the spike protein of SARS-CoV-2, which are conjugated with fluorescein isothiocyanate (FITC) and immobilized on anti-FITC antibody-conjugated magnetic particles. Alkaline phosphatase conjugated anti-human IgG/IgM antibody was used as the detection antibody. All tests were performed on an automated magnetic chemiluminescence analyzer (Axceed 260, Bioscience, China). Associated positively with the measured chemiluminescence values, the IgM/IgG titers are presented as chemiluminescence values divided by the cutoff (S/CO). If the S/CO value is > 1, the sample is considered seropositive for IgM or IgG, while if the S/CO value is < 1, the sample is considered seronegative for IgM or IgG.65
The levels of ACE2-RBD inhibiting antibody were measured using ACE2-RBD inhibiting assay by the commercial MCLIA kits (Bioscience, China). Briefly, magnetic beads were coated with Human ACE2-hFc, and the recombinant RBD of SARS-CoV-2 were conjugated with Alkaline phosphatase. The assays are two step immunoassays for the quantitative detection. In the first step, a 20 μL sample and 20 μL of recombinant ACE2-coated magnetic beads were combined. Then, 40 μL of alkaline phosphatase-labeled RBD conjugate was added. Following a wash cycle, the substrate was added to the mixture. Then the tests were performed on an automated magnetic chemiluminescence analyzer (Axceed 260, Bioscience, China). The resulting chemiluminescent reaction was measured as relative light unit (RLU). The calibrators were made from monoclonal antibodies targeting SARS-CoV-2 RBD with designated concentrations and the standard curves were established. The levels of the ACE2-RBD inhibiting antibody in the sample were determined by comparing the RLU of a sample to the RLU determined from standard curves.66
Plasma measurements
Blood samples were collected in anticoagulant tube and analyzed within 2 h. The main parameters in the routine blood test including white blood cell (WBC), neutrophil, lymphocyte, monocyte, eosinophil, basophil, red blood cell and platelet counts as well as hemoglobin levels, neutrophil ratio, lymphocyte ratio, monocyte ratio, eosinophil ratio and basophil ratio were analyzed using a Sysmex XN-9000 (Sysmex Co., Kobe, Japan). The ratio of neutrophil, lymphocyte, monocyte was calculated as (ratio %= (counts of neutrophil or lymphocyte or monocyte/counts of white blood cells)✕100%). C-reactive protein (CRP) was tested by a detection kit using an immunoquantitative analyzer (FIA8600, Getein Biotech, Nanjing, China). The lymphocyte subsets were detected by lymphocyte detection kits (Agilent, Santa Clara, USA) using NovoCyte D2040R flow cytometry (ACEA BIO, Hangzhou, China), and the ratio of T cells was automatically calculated as (ratio %= (counts of total T cells/counts of total lymphocytes) ✕100%). The cytokines were detected by the Immulite 1000 (Siemens, Glyn Rhonwy, United Kingdom). Immunoglobulin was tested by IMMAGE 800 (Beckman Coulter, California, USA) with matched detection kits.
Quantification of fecal and serum SCFAs
Quantification of fecal and serum SCFAs, including acetic acid, propionic acid, butyric acid, isovaleric acid, isobutyric acid, valeric acid and caproic acid, was detected by GC-MS analysis performed by PANOMIX (Suzhou PANOMIX Biomedical Tech Co., LTD, China). Briefly, the fecal samples were self-collected and transferred to laboratories within 2 h and stored at−80° until analysis. Blood samples were obtained and allowed to clot for at least 30 min at room temperature, and then centrifuged at 1300 g for 15 min. The supernatant was immediately aliquoted and stored at −80°C until use.
Fecal or serum samples were thawed. 0.5 g of fecal sample or 50 μL of serum was added to a centrifuge tube with 50 μL phosphoric acid (15%), 100 μL of the internal standard and 400 μL of diethyl ether. The mixture was the centrifuged for 10 min at 12,000 rpm at 4°C. The supernatant was transferred to a vial for further analysis. SCFAs were detected using an Agilent 7890A gas chromatograph coupled with an Agilent 5975C mass spectrometric detector (Agilent Technologies, USA) equipped with an HP-5MS column (30 mm length × 0.25 mm inner diameter × 0.25 μm film thickness). Helium was used as the carrier gas at a flow rate of 1 mL/min. The initial oven temperature was held at 90°C for 1 min, ramped to 250 °C at a rate of 10°C/min and held at 250°C for 5 min. The temperatures of the front inlet, transfer line and electron impact (EI) ion source were set as 280, 250 and 230°C, respectively. Data analysis was performed with an Agilent MSD ChemStation (E.02.00.493, Agilent Technologies, Inc., USA). Authentic standards of acetic acid, propionic acid, butyric acid, isobutyric acid, valeric acid, isovaleric acid and caproic acid were diluted in diethyl ether with concentrations ranging from 0.02 to 500 μg/mL. The standard calibration curves were established using the above standards. Quantification of fecal and serum SCFAs was based on the standard calibration curves. Samples for method validation were extracted using the standard method to evaluate within-day repeatability (eight replicates), inter-day repeatability (three different days and six replicates per day), instrument stability (eight repeat injections of one sample), and accuracy (four replicates).
Quantification and statistical analysis
Statistical analysis
Differential abundance of species and MetaCyc pathway were screened using Microbiome Multivariable Associations with Linear Models (Masslin2). Correlation between continuous variables including species abundance, MetaCyc pathway CPMs, ACE2-RBD inhibiting antibody levels and clinical characteristics were analyzed by using Pearson correlation test. Differences of continuous variables between two groups were analyzed by using the Wilcoxon’s rank-sum test (two-sided) via R build-in function wilcoxon test. Welch ANOVA with Games-Howell’s multiple comparisons test was used to evaluate the statistical significance of differences among three or more groups if equal variances were not assumed. All the analyses and results visualization were performed in R V4.1.2 with R packages: tidyverse, ggplot2, ggsignif, ggsci, readr, dplyr, readxl, plot3D, masslin2, patchwork, caret, pheatmap, Hmisc, magrittr, corrplot, vegan, pROC, factoextra, FactoMineR and RColorBrewer. Generalized linear models (GLM) for binominal outcomes (High group versus Low group) with receiver operating characteristic (ROC) curve was applied to determine the prediction value of the identified biomarkers. Clinical characteristics tables were generated by using R packages compareGroups. Multiple testing was corrected using the Benjamini-Hochberg method to control the false-discovery rate (FDR). p value or FDR-corrected p value < 0.05 was considered statistically significant.
Acknowledgments
The authors would like to thank all of the participants in this study for their willingness to enroll in this study. We would also like to thank the staff at the Laboratory Medicine Center for their excellent technical assistance. We thank all of the nurses and physicians at the Department of Gastroenterology of Xinqiao Hospital for blood draws and medical advice. This work was supported by the Key Project of the National Natural Science Foundation of China (82030020), the National Key R&D Program of China (2021YFC2301300, 2021YFA1301000) and National Natural Science Foundation of China (32025009).
Author contributions
B.T., Z.L., F.Z., and S.Y. designed and supervised the study. W.H., C.H., Y.Z., and S.L. enrolled patients and collected the patient samples and data. B.T., L.T., and K.P. performed the data analyses. Y.L. and S. Liu helped with the data analyses and visualization. X.J., Y.L., Y.Z., S.W., and M.Y. performed laboratory detection and fecal genomic DNA extraction. B.T. and S.Y. wrote the original manuscript. Z.L., F.Z., and S.Y. reviewed and edited the manuscript.
Declaration of interests
The authors declare no competing interests.
Inclusion and diversity
We support inclusive, diverse, and equitable conduct of research.
Published: September 13, 2022
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.xcrm.2022.100752.
Contributor Information
Zhongjun Li, Email: johnneyusc@gmail.com.
Fangqing Zhao, Email: zhfq@biols.ac.cn.
Shiming Yang, Email: yangshiming@tmmu.edu.cn.
Supplemental information
Table S3A is an index of the alpha diversity in all of the samples at baseline (day 0) and samples after the second injection (day 42), related to Figure S2A. Table S3B is an index of principal coordinate analysis of samples at baseline (day 0) and after the second injection (day 42), related to Figure 2A. Table S3C shows differentially enriched bacteria between baseline samples (day 0) and samples after the second injection (day 42), related to Figure 2C.
Table S4A shows differentially enriched MetaCyc pathways between samples at days 0 and 42, related to Figure 2E. Table S4B shows the correlation between MetaCyc pathways at day 0 and the clinical characteristics at day 42, related to Figure 2F. Table S4C shows differentially enriched MetaCyc pathways at day 0 between samples from the high and low groups, related to Figure 3H. Table S4D shows the correlation of differentially enriched MetaCyc pathways between high and low groups at day 0 with the clinical characteristics at day 42, related to Figure 3I.
Data and code availability
The published article includes all datasets generated or analyzed during this study. Data relating to the metagenomic sequencing that support the findings of this study have been uploaded to the National Genomics Data Center (https://ngdc.cncb.ac.cn/gsa/) and are available via the accession number CRA007947.
References
- 1.Chan J.F., Yuan S., Kok K.H., To K.K., Chu H., Yang J., Xing F., Liu J., Yip C.C., Poon R.W., et al. A familial cluster of pneumonia associated with the 2019 novel coronavirus indicating person-to-person transmission: a study of a family cluster. Lancet. 2020;395:514–523. doi: 10.1016/S0140-6736(20)30154-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Dong E., Du H., Gardner L. An interactive web-based dashboard to track COVID-19 in real time. Lancet Infect. Dis. 2020;20:533–534. doi: 10.1016/S1473-3099(20)30120-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Krammer F. SARS-CoV-2 vaccines in development. Nature. 2020;586:516–527. doi: 10.1038/s41586-020-2798-3. [DOI] [PubMed] [Google Scholar]
- 4.Dai L., Gao G.F. Viral targets for vaccines against COVID-19. Nat. Rev. Immunol. 2021;21:73–82. doi: 10.1038/s41577-020-00480-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bhasin S., Gill T.M., Reuben D.B., Latham N.K., Ganz D.A., Greene E.J., Dziura J., Basaria S., Gurwitz J.H., Dykes P.C., et al. A randomized trial of a multifactorial strategy to prevent serious Fall Injuries. N. Engl. J. Med. 2020;383:129–140. doi: 10.1056/NEJMoa2002183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Guo W., Duan K., Zhang Y., Yuan Z., Zhang Y.B., Wang Z., Zhao D., Zhang H., Xie Z., Li X., et al. Safety and immunogenicity of an inactivated SARS-CoV-2 vaccine in healthy adults aged 18 years or older: a randomized, double-blind, placebo-controlled, phase 1/2 trial. EClinicalMedicine. 2021;38:101010. doi: 10.1016/j.eclinm.2021.101010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jackson L.A., Anderson E.J., Rouphael N.G., Roberts P.C., Makhene M., Coler R.N., McCullough M.P., Chappell J.D., Denison M.R., Stevens L.J., et al. An mRNA vaccine against SARS-CoV-2 - Preliminary report. N. Engl. J. Med. 2020;383:1920–1931. doi: 10.1056/NEJMoa2022483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Walsh E.E., Frenck R.W., Jr., Falsey A.R., Kitchin N., Absalon J., Gurtman A., Lockhart S., Neuzil K., Mulligan M.J., Bailey R., et al. Safety and immunogenicity of two RNA-based Covid-19 vaccine candidates. N. Engl. J. Med. 2020;383:2439–2450. doi: 10.1056/NEJMoa2027906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Xia S., Duan K., Zhang Y., Zhao D., Zhang H., Xie Z., Li X., Peng C., Zhang Y., Zhang W., et al. Effect of an inactivated vaccine against SARS-CoV-2 on safety and immunogenicity outcomes: Interim analysis of 2 randomized clinical trials. JAMA. 2020;324:951–960. doi: 10.1001/jama.2020.15543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Xia S., Zhang Y., Wang Y., Wang H., Yang Y., Gao G.F., Tan W., Wu G., Xu M., Lou Z., et al. Safety and immunogenicity of an inactivated SARS-CoV-2 vaccine, BBIBP-CorV: a randomised, double-blind, placebo-controlled, phase 1/2 trial. Lancet Infect. Dis. 2021;21:39–51. doi: 10.1016/S1473-3099(20)30831-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhu F.C., Guan X.H., Li Y.H., Huang J.Y., Jiang T., Hou L.H., Li J.X., Yang B.F., Wang L., Wang W.J., et al. Immunogenicity and safety of a recombinant adenovirus type-5-vectored COVID-19 vaccine in healthy adults aged 18 years or older: a randomised, double-blind, placebo-controlled, phase 2 trial. Lancet. 2020;396:479–488. doi: 10.1016/S0140-6736(20)31605-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.de Jong S.E., Olin A., Pulendran B. The impact of the microbiome on immunity to vaccination in humans. Cell Host Microbe. 2020;28:169–179. doi: 10.1016/j.chom.2020.06.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zimmermann P., Curtis N. Factors that influence the immune response to vaccination. Clin. Microbiol. Rev. 2019;32 doi: 10.1128/CMR.00084-18. e00084-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Harris V.C., Haak B.W., Handley S.A., Jiang B., Velasquez D.E., Hykes B.L., Jr., Droit L., Berbers G.A.M., Kemper E.M., van Leeuwen E.M.M., et al. Effect of antibiotic-Mediated microbiome modulation on rotavirus vaccine immunogenicity: a human, randomized-control Proof-of-Concept trial. Cell Host Microbe. 2018;24:197–207.e4. doi: 10.1016/j.chom.2018.07.005. [DOI] [PubMed] [Google Scholar]
- 15.Lynn D.J., Benson S.C., Lynn M.A., Pulendran B. Modulation of immune responses to vaccination by the microbiota: implications and potential mechanisms. Nat. Rev. Immunol. 2022;22:33–46. doi: 10.1038/s41577-021-00554-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hagan T., Cortese M., Rouphael N., Boudreau C., Linde C., Maddur M.S., Das J., Wang H., Guthmiller J., Zheng N.Y., et al. Antibiotics-driven gut microbiome perturbation Alters immunity to vaccines in humans. Cell. 2019;178:1313–1328.e13. doi: 10.1016/j.cell.2019.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yeoh Y.K., Zuo T., Lui G.C., Zhang F., Liu Q., Li A.Y., Chung A.C., Cheung C.P., Tso E.Y., Fung K.S., et al. Gut microbiota composition reflects disease severity and dysfunctional immune responses in patients with COVID-19. Gut. 2021;70:698–706. doi: 10.1136/gutjnl-2020-323020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zuo T., Zhang F., Lui G.C.Y., Yeoh Y.K., Li A.Y.L., Zhan H., Wan Y., Chung A.C.K., Cheung C.P., Chen N., et al. Alterations in gut microbiota of patients with COVID-19 during time of Hospitalization. Gastroenterology. 2020;159:944–955.e8. doi: 10.1053/j.gastro.2020.05.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hou K., Wu Z.X., Chen X.Y., Wang J.Q., Zhang D., Xiao C., Zhu D., Koya J.B., Wei L., Li J., Chen Z.S. Microbiota in health and diseases. Signal. Transduct. Target. Ther. 2022;7:135. doi: 10.1038/s41392-022-00974-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Du Y., Chen L., Shi Y. Safety, immunogenicity, and efficacy of COVID-19 vaccines in Adolescents, Children, and infants: a Systematic Review and meta-analysis. Front. Public Health. 2022;10:829176. doi: 10.3389/fpubh.2022.829176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pormohammad A., Zarei M., Ghorbani S., Mohammadi M., Razizadeh M.H., Turner D.L., Turner R.J. Efficacy and safety of COVID-19 vaccines: a systematic review and meta-analysis of randomized clinical trials. Vaccines (Basel) 2021;9:467. doi: 10.3390/vaccines9050467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Al Kaabi N., Zhang Y., Xia S., Yang Y., Al Qahtani M.M., Abdulrazzaq N., Al Nusair M., Hassany M., Jawad J.S., Abdalla J., et al. Effect of 2 inactivated SARS-CoV-2 vaccines on Symptomatic COVID-19 infection in adults: a randomized clinical trial. JAMA. 2021;326:35–45. doi: 10.1001/jama.2021.8565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wang G.L., Wang Z.Y., Duan L.J., Meng Q.C., Jiang M.D., Cao J., Yao L., Zhu K.L., Cao W.C., Ma M.J. Susceptibility of circulating SARS-CoV-2 variants to neutralization. N. Engl. J. Med. 2021;384:2354–2356. doi: 10.1056/NEJMc2103022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Xia S., Zhang Y., Wang Y., Wang H., Yang Y., Gao G.F., Tan W., Wu G., Xu M., Lou Z., et al. Safety and immunogenicity of an inactivated COVID-19 vaccine, BBIBP-CorV, in people younger than 18 years: a randomised, double-blind, controlled, phase 1/2 trial. Lancet Infect. Dis. 2022;22:196–208. doi: 10.1016/S1473-3099(21)00462-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Vabret N., Britton G.J., Gruber C., Hegde S., Kim J., Kuksin M., Levantovsky R., Malle L., Moreira A., Park M.D., et al. Immunology of COVID-19: current state of the science. Immunity. 2020;52:910–941. doi: 10.1016/j.immuni.2020.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hue S., Beldi-Ferchiou A., Bendib I., Surenaud M., Fourati S., Frapard T., Rivoal S., Razazi K., Carteaux G., Delfau-Larue M.H., et al. Uncontrolled innate and impaired adaptive immune responses in patients with COVID-19 acute respiratory distress syndrome. Am. J. Respir. Crit. Care Med. 2020;202:1509–1519. doi: 10.1164/rccm.202005-1885OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rodda L.B., Netland J., Shehata L., Pruner K.B., Morawski P.A., Thouvenel C.D., Takehara K.K., Eggenberger J., Hemann E.A., Waterman H.R., et al. Functional SARS-CoV-2-specific immune memory Persists after mild COVID-19. Cell. 2021;184:169–183.e17. doi: 10.1016/j.cell.2020.11.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Brodin P. Immune determinants of COVID-19 disease presentation and severity. Nat. Med. 2021;27:28–33. doi: 10.1038/s41591-020-01202-8. [DOI] [PubMed] [Google Scholar]
- 29.Mehta P., McAuley D.F., Brown M., Sanchez E., Tattersall R.S., Manson J.J. COVID-19: consider cytokine storm syndromes and immunosuppression. Lancet. 2020;395:1033–1034. doi: 10.1016/S0140-6736(20)30628-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Liu Y., Zhang C., Huang F., Yang Y., Wang F., Yuan J., Zhang Z., Qin Y., Li X., Zhao D., et al. Elevated plasma levels of selective cytokines in COVID-19 patients reflect viral load and lung injury. Natl. Sci. Rev. 2020;7:1003–1011. doi: 10.1093/nsr/nwaa037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yang W., Cong Y. Gut microbiota-derived metabolites in the regulation of host immune responses and immune-related inflammatory diseases. Cell. Mol. Immunol. 2021;18:866–877. doi: 10.1038/s41423-021-00661-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pickard J.M., Zeng M.Y., Caruso R., Núñez G. Gut microbiota: role in pathogen colonization, immune responses, and inflammatory disease. Immunol. Rev. 2017;279:70–89. doi: 10.1111/imr.12567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zheng D., Liwinski T., Elinav E. Interaction between microbiota and immunity in health and disease. Cell Res. 2020;30:492–506. doi: 10.1038/s41422-020-0332-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Viana S.D., Nunes S., Reis F. ACE2 imbalance as a key player for the poor outcomes in COVID-19 patients with age-related comorbidities - role of gut microbiota dysbiosis. Ageing Res. Rev. 2020;62:101123. doi: 10.1016/j.arr.2020.101123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Brown E.M., Kenny D.J., Xavier R.J. Gut microbiota regulation of T cells during inflammation and Autoimmunity. Annu. Rev. Immunol. 2019;37:599–624. doi: 10.1146/annurev-immunol-042718-041841. [DOI] [PubMed] [Google Scholar]
- 36.Honda K., Littman D.R. The microbiota in adaptive immune homeostasis and disease. Nature. 2016;535:75–84. doi: 10.1038/nature18848. [DOI] [PubMed] [Google Scholar]
- 37.Kayama H., Okumura R., Takeda K. Interaction between the microbiota, Epithelia, and immune cells in the intestine. Annu. Rev. Immunol. 2020;38:23–48. doi: 10.1146/annurev-immunol-070119-115104. [DOI] [PubMed] [Google Scholar]
- 38.Feng Z., Long W., Hao B., Ding D., Ma X., Zhao L., Pang X. A human stool-derived Bilophila wadsworthia strain caused systemic inflammation in specific-pathogen-free mice. Gut Pathog. 2017;9:59. doi: 10.1186/s13099-017-0208-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Natividad J.M., Lamas B., Pham H.P., Michel M.L., Rainteau D., Bridonneau C., da Costa G., van Hylckama Vlieg J., Sovran B., Chamignon C., et al. Bilophila wadsworthia aggravates high fat diet induced metabolic dysfunctions in mice. Nat. Commun. 2018;9:2802. doi: 10.1038/s41467-018-05249-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Matsuoka K., Kanai T. The gut microbiota and inflammatory bowel disease. Semin. Immunopathol. 2015;37:47–55. doi: 10.1007/s00281-014-0454-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ezeji J.C., Sarikonda D.K., Hopperton A., Erkkila H.L., Cohen D.E., Martinez S.P., Cominelli F., Kuwahara T., Dichosa A.E.K., Good C.E., et al. Parabacteroides distasonis: intriguing aerotolerant gut anaerobe with emerging antimicrobial resistance and pathogenic and probiotic roles in human health. Gut Microb. 2021;13:1922241. doi: 10.1080/19490976.2021.1922241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zeevi D., Korem T., Godneva A., Bar N., Kurilshikov A., Lotan-Pompan M., Weinberger A., Fu J., Wijmenga C., Zhernakova A., Segal E. Structural variation in the gut microbiome associates with host health. Nature. 2019;568:43–48. doi: 10.1038/s41586-019-1065-y. [DOI] [PubMed] [Google Scholar]
- 43.Harris V.C., Armah G., Fuentes S., Korpela K.E., Parashar U., Victor J.C., Tate J., de Weerth C., Giaquinto C., Wiersinga W.J., et al. Significant correlation between the infant gut microbiome and rotavirus vaccine response in Rural Ghana. J. Infect. Dis. 2017;215:34–41. doi: 10.1093/infdis/jiw518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Harris V., Ali A., Fuentes S., Korpela K., Kazi M., Tate J., Parashar U., Wiersinga W.J., Giaquinto C., de Weerth C., de Vos W.M. Rotavirus vaccine response correlates with the infant gut microbiota composition in Pakistan. Gut Microb. 2018;9:93–101. doi: 10.1080/19490976.2017.1376162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Grassly N.C., Praharaj I., Babji S., Kaliappan S.P., Giri S., Venugopal S., Parker E.P., Abraham A., Muliyil J., Doss S., et al. The effect of azithromycin on the immunogenicity of oral poliovirus vaccine: a double-blind randomised placebo-controlled trial in seronegative Indian infants. Lancet Infect. Dis. 2016;16:905–914. doi: 10.1016/S1473-3099(16)30023-8. [DOI] [PubMed] [Google Scholar]
- 46.Matson V., Fessler J., Bao R., Chongsuwat T., Zha Y., Alegre M.L., Luke J.J., Gajewski T.F. The commensal microbiome is associated with anti-PD-1 efficacy in metastatic melanoma patients. Science. 2018;359:104–108. doi: 10.1126/science.aao3290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Jang H.J., Choi J.Y., Kim K., Yong S.H., Kim Y.W., Kim S.Y., Kim E.Y., Jung J.Y., Kang Y.A., Park M.S., et al. Relationship of the lung microbiome with PD-L1 expression and immunotherapy response in lung cancer. Respir. Res. 2021;22:322. doi: 10.1186/s12931-021-01919-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Salk H.M., Simon W.L., Lambert N.D., Kennedy R.B., Grill D.E., Kabat B.F., Poland G.A. Taxa of the nasal microbiome are associated with influenza-specific IgA response to live attenuated influenza vaccine. PLoS One. 2016;11:e0162803. doi: 10.1371/journal.pone.0162803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Qin P., Zou Y., Dai Y., Luo G., Zhang X., Xiao L. Characterization a novel butyric acid-producing bacterium Collinsella aerofaciens Subsp. Shenzhenensis Subsp. Nov. Microorganisms. 2019;7:78. doi: 10.3390/microorganisms7030078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kim M., Qie Y., Park J., Kim C.H. Gut microbial metabolites fuel host antibody responses. Cell Host Microbe. 2016;20:202–214. doi: 10.1016/j.chom.2016.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Cait A., Mooney A., Poyntz H., Shortt N., Jones A., Gestin A., Gell K., Grooby A., O'Sullivan D., Tang J.S., et al. Potential association between dietary fibre and humoral response to the seasonal influenza vaccine. Front. Immunol. 2021;12:765528. doi: 10.3389/fimmu.2021.765528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zhang F., Wan Y., Zuo T., Yeoh Y.K., Liu Q., Zhang L., Zhan H., Lu W., Xu W., Lui G.C.Y., et al. Prolonged Impairment of short-chain fatty acid and L-Isoleucine biosynthesis in gut microbiome in patients with COVID-19. Gastroenterology. 2022;162:548–561.e4. doi: 10.1053/j.gastro.2021.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wang H., Zhang Y., Huang B., Deng W., Quan Y., Wang W., Xu W., Zhao Y., Li N., Zhang J., et al. Development of an inactivated vaccine candidate, BBIBP-CorV, with potent protection against SARS-CoV-2. Cell. 2020;182:713–721.e9. doi: 10.1016/j.cell.2020.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Lachin J.M. Introduction to sample size determination and power analysis for clinical trials. Control. Clin. Trials. 1981;2:93–113. doi: 10.1016/0197-2456(81)90001-5. [DOI] [PubMed] [Google Scholar]
- 55.Beghini F., McIver L.J., Blanco-Míguez A., Dubois L., Asnicar F., Maharjan S., Mailyan A., Manghi P., Scholz M., Thomas A.M., et al. Integrating taxonomic, functional, and strain-level profiling of diverse microbial communities with bioBakery 3. Elife. 2021;10:e65088. doi: 10.7554/eLife.65088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Robin X., Turck N., Hainard A., Tiberti N., Lisacek F., Sanchez J.C., Müller M. pROC: an open-source package for R and S+ to analyze and compare ROC curves. BMC Bioinf. 2011;12:77. doi: 10.1186/1471-2105-12-77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Langmead B., Salzberg S.L. Fast gapped-read alignment with Bowtie 2. Nat. Methods. 2012;9:357–359. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Bolger A.M., Lohse M., Usadel B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 2014;30:2114–2120. doi: 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Buchfink B., Xie C., Huson D.H. Fast and sensitive protein alignment using DIAMOND. Nat. Methods. 2015;12:59–60. doi: 10.1038/nmeth.3176. [DOI] [PubMed] [Google Scholar]
- 60.Mallick H., Rahnavard A., McIver L.J., Ma S., Zhang Y., Nguyen L.H., Tickle T.L., Weingart G., Ren B., Schwager E.H., et al. Multivariable association discovery in population-scale meta-omics studies. PLoS Comput. Biol. 2021;17:e1009442. doi: 10.1371/journal.pcbi.1009442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Oksanen J., Blanchet F.G., Friendly M., Kindt R., Wagner H.H. 2020. Vegan Community Ecology Package Version 2.5-7 November 2020.https://github.com/vegandevs/vegan [Google Scholar]
- 62.Soetaert K. 2021. plot3D: Plotting Multi-Dimensional Data.https://CRAN.R-project.org/package=plot3D R package version 1.4. [Google Scholar]
- 63.Wallstrom G., Liebner J., Kass R.E. An Implementation of Bayesian adaptive Regression Splines (BARS) in C with S and R Wrappers. J. Stat. Softw. 2008;26:1–21. [PMC free article] [PubMed] [Google Scholar]
- 64.FEH J. 2021. Hmisc: Harrell Miscellaneous. R Package Version 4.5-0.https://hbiostat.org/R/Hmisc/ [Google Scholar]
- 65.Zhu L., Xu X., Zhu B., Guo X., Xu K., Song C., Fu J., Yu H., Kong X., Peng J., et al. Kinetics of SARS-CoV-2 specific and neutralizing antibodies over seven months after symptom onset in COVID-19 patients. Microbiol. Spectr. 2021;9:e0059021. doi: 10.1128/Spectrum.00590-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Zhang S., Xu K., Li C., Zhou L., Kong X., Peng J., Zhu F., Bao C., Jin H., Gao Q., et al. Long-term Kinetics of SARS-CoV-2 antibodies and impact of inactivated vaccine on SARS-CoV-2 antibodies based on a COVID-19 patients cohort. Front. Immunol. 2022;13:829665. doi: 10.3389/fimmu.2022.829665. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S3A is an index of the alpha diversity in all of the samples at baseline (day 0) and samples after the second injection (day 42), related to Figure S2A. Table S3B is an index of principal coordinate analysis of samples at baseline (day 0) and after the second injection (day 42), related to Figure 2A. Table S3C shows differentially enriched bacteria between baseline samples (day 0) and samples after the second injection (day 42), related to Figure 2C.
Table S4A shows differentially enriched MetaCyc pathways between samples at days 0 and 42, related to Figure 2E. Table S4B shows the correlation between MetaCyc pathways at day 0 and the clinical characteristics at day 42, related to Figure 2F. Table S4C shows differentially enriched MetaCyc pathways at day 0 between samples from the high and low groups, related to Figure 3H. Table S4D shows the correlation of differentially enriched MetaCyc pathways between high and low groups at day 0 with the clinical characteristics at day 42, related to Figure 3I.
Data Availability Statement
The published article includes all datasets generated or analyzed during this study. Data relating to the metagenomic sequencing that support the findings of this study have been uploaded to the National Genomics Data Center (https://ngdc.cncb.ac.cn/gsa/) and are available via the accession number CRA007947.