Summary
Mitochondrial diseases are a heterogeneous group of genetic disorders caused by pathogenic variants in genes encoding gene products that regulate mitochondrial function. These genes are located either in the mitochondrial or in the nuclear genome. The TOMM7 gene encodes a regulatory subunit of the translocase of outer mitochondrial membrane (TOM) complex that plays an essential role in translocation of nuclear-encoded mitochondrial proteins into mitochondria. We report an individual with a homozygous variant in TOMM7 (c.73T>C, p.Trp25Arg) that presented with a syndromic short stature, skeletal abnormalities, muscle hypotonia, microvesicular liver steatosis, and developmental delay. Analysis of mouse models strongly suggested that the identified variant is hypomorphic because mice homozygous for this variant showed a milder phenotype than those with homozygous Tomm7 deletion. These Tomm7 mutant mice show pathological changes consistent with mitochondrial dysfunction, including growth defects, severe lipoatrophy, and lipid accumulation in the liver. These mice die prematurely following a rapidly progressive weight loss during the last week of their lives. Tomm7 deficiency causes a unique alteration in mitochondrial function; despite the bioenergetic deficiency, mutant cells show increased oxygen consumption with normal responses to electron transport chain (ETC) inhibitors, suggesting that Tomm7 deficiency leads to an uncoupling between oxidation and ATP synthesis without impairing the function of the tricarboxylic cycle metabolism or ETC. This study presents evidence that a hypomorphic variant in one of the genes encoding a subunit of the TOM complex causes mitochondrial disease.
Keywords: mitochondria, TOM, translocase, TOMM7, skeletal dysplasia, lipoatrophy, mouse model, growth plate, developmental delay, fatty liver
Young et al. reports the first case of syndromic short stature associated with a missense variant in the TOMM7 gene, encoding a regulatory subunit of the translocase of outer mitochondrial membrane complex. Mouse models showed that this variant caused a bioenergetic defect and a phenotype of mitochondrial diseases.
Main text
Mitochondria are double-membrane-bound organelles that are essential for diverse cellular processes, including energy production through the process called oxidative phosphorylation (OXPHOS) and supplying a variety of bioactive metabolites.1 Dysfunctions of mitochondria are associated with various human conditions such as metabolic,2 neurogenerative,3 kidney,4 cardiovascular,5 and neuromuscular diseases.6 Mitochondrial diseases can be caused by pathogenic variants in genes located in the mitochondrial genome DNA (mtDNA) or in the nuclear genome that encode mitochondrial proteins.7,8
The mitochondrion contains about 1,500 proteins in humans. Since mtDNA encodes only 13 mitochondrial proteins that are subunits of the respiratory chain complexes, the majority of mitochondrial proteins are encoded in the nuclear DNA.9 These proteins are synthesized in the cytoplasm and imported into mitochondria. These nuclear DNA-encoded mitochondrial proteins localized inside of the mitochondria need to be transported through the outer mitochondrial membrane (OMM) into the intermembrane space (IMS). This process is mediated by the translocase of the outer membrane (TOM) complex.10 Adequate control of both quantity and quality of mitochondrial proteins via their synthesis, degradation, and transportation is essential for healthy cellular function, and aberration in mitochondrial proteostasis leads to mitochondrial dysfunction and diseases.11
The TOM complex consists of multiple subunits including the core proteins TOMM40, which forms a barrel structure through which mitochondrial proteins translocate from cytosolic space into IMS, and TOMM20 and TOMM22, which provide a binding interface for mitochondrial proteins by associating with their N-terminal cleavable extension, called a “pre-sequence,” that is present in a majority of nuclear-encoded mitochondrial proteins.12 Three evolutionarily conserved regulatory subunits, TOMM5, -6, and -7, are directly associated with TOMM40.13,14 These regulatory subunits have been suggested to modulate the assembly and stability of the TOM complex and thereby control its function.15, 16, 17 In addition to the role of TOM complex assembly, TOMM7 has been implicated in regulation of mitophagy through stabilization of PINK1, a crucial regulator of parkin (PARK2) that initiates mitochondrial protein degradation upon mitochondrial damage.18,19 Thus far, no human diseases have been reported to be associated with variants in genes encoding TOM complex subunits. Here, we present a boy with a short stature and growth failure associated with a homozygous missense variant in TOMM7 (MIM: 607980) and experimental evidence for its pathogenic role.
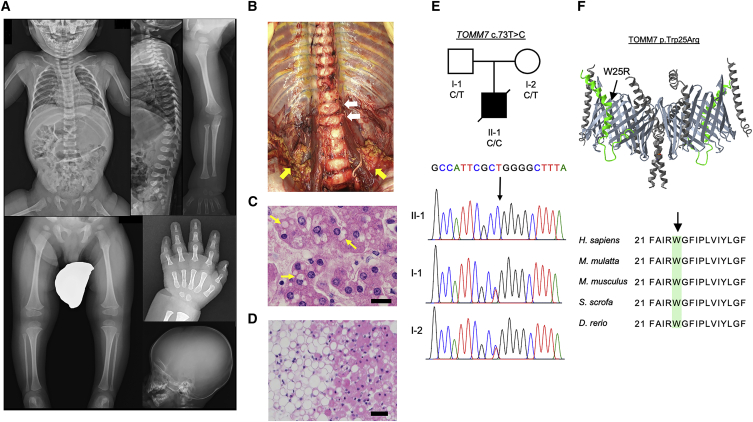
The affected individual and his healthy parents were enrolled in the study of rare congenital skeletal disorders after being diagnosed to have an unknown condition with skeletal abnormalities. The patient, a male infant, born to non-consanguineous Japanese parents, came to medical attention due to progressive growth failure. He was born via Cesarian section due to his mother’s hypertension at the gestational age of 38 weeks and 5 days. His birth weight (3,240 g) and length (50 cm) were normal (Z score +0.3 and +0.06, respectively), and the routine neonatal screening tests for congenital metabolic diseases were negative. He showed progressive postnatal growth retardation. At 1 year and 3 months of age, his body weight and height were 6,905 g (Z score −3.6) and 65.5 cm (Z score −5.4), respectively, and his arm span (61 cm) was shorter than his height (65.2 cm). Other clinical features include narrow thorax, broad hands, small nails, nystagmus, muscular hypotonia with frog-leg posture and poor head control (head raising at 6 months), tachypnea (respiratory rate 32/min), and retractive breathing. The results of his laboratory tests at 12 and 20 months showed increased serum lactate dehydrogenase (LDH) levels and moderately increased white blood cell counts (Table S1). Endocrinological test results showed normal values of thyroid hormone, thyroid-stimulating hormone, and growth hormone (Table S2). Radiology examination revealed relatively large neurocranium, underdeveloped facial bones, open anterior fontanel, mildly flat and rounded outline of the vertebral bodies, and slender diaphyses and relatively wide metaphyses of tubular bones (Figure 1A). The patient died at the age of 2 years and 7 months after developing pneumonia and respiratory failure. Autopsy revealed mild deformity of vertebral bodies and relatively prominent retroperitoneal brown adipose tissues (Figure 1B). Histological analysis showed microvesicular steatosis in the liver (Figure 1C) and relatively small white adipocytes in the retroperitoneal fat tissue (Figure 1D). In addition to diffuse bilateral pneumonia, there were also submucosal fibrosis of the intestine, smooth muscle degeneration of the rectum, and thickening of the tunica intima and edema and proteoglycan deposition in the tunica media of the aorta and coronary arteries, but the clinical significance of these findings was unclear (Figure S1).
Figure 1.
A novel syndromic skeletal dysplasia associated with a TOMM7 variant
(A) Skeletal survey of the affected individual at the age of 1 year and 8 months. Note thin ribs, mild platyspondyly with ovoid vertebral bodies, gracile long tubular bones with relatively large epiphyses in upper and lower extremities, coxa valga, slender metacarpals and proximal phalanges, short middle phalanges with metaphyseal flaring of the proximal ends, and craniofacial disproportion (small facial bones).
(B) Frontal view of the spine and retroperitoneal fat tissues during autopsy. Flat and oval-shaped vertebral bodies (white arrows) and relatively prominent brown adipose tissues (yellow arrows) are shown.
(C) Hematoxylin and eosin-stained liver tissue. Yellow arrows indicate translucent intracellular vesicles in hepatocytes suggesting lipid accumulation. Scale bar, 20 μm.
(D) Hematoxylin and eosin-stained retroperitoneal fat tissue. The size of white adipocytes (left half of the image) is relatively small (15–45 μm in diameter) and is similar to that of brown adipocytes (right half of the image). Scale bar, 50 μm.
(E) The pedigree of the affected individual and Sanger sequencing data for the variant. The parents are heterozygous and the proband is homozygous for TOMM7 c.73T>C variant.
(F) Three-dimensional (3D) structure of the TOM complex dimer. TOMM7 is indicated in green color, TOMM40 barrels are in light gray. Other subunits, TOMM5, TOMM6, and TOM22 are in dark gray. The variant residue (indicated by the arrow) is located next to the arginine residue at the position 24, which is predicted to be important for interaction with TOMM40.20 The image is modified from the data (ID: 195358) of the Molecular Modeling Database.21,22
The initial analysis of whole genome sequencing (WGS) of this affected individual and his parents did not reveal any candidate variants in all known skeletal dysplasia genes (https://panelapp.genomicsengland.co.uk/panels/309/). Extended analysis revealed several possible candidate variants (Table S3) including a single-nucleotide substitution in the TOMM7 gene (chr7:g.22862326T>C (hg19), NM_019059.5 (TOMM7):c.73T>C, p.(Trp25Arg)). This variant was found in the homozygous state in the affected individual and in the heterozygous state in his parents (Figure 1E). The tryptophan at the position 25 of TOMM7, located at a kinked region, is highly conserved across many species (Figure 1F). This variant is expected to replace the tryptophan residue with a positively charged arginine located next to another arginine at the position 24. It is reported that the position 24 arginine is important for the interaction between TOMM7 and -40 barrel.20 Thus, we hypothesized that the Trp25Arg (W25R) variant interfered with the normal interaction between TOMM7 and -40, leading to pathogenic consequences. To test this hypothesis, we generated a knockin mouse model where the identical amino acid variant (W25R) was introduced to the mouse Tomm7 gene. We also generated a mouse line missing a 184 bp genomic region spanning the first exon-intron boundary of the Tomm7 gene (deletion [D]) (Figure 2A). The D allele is expected to be functionally null because the deletion leads to a disruption of normal splicing resulting in premature termination. This deletion also caused a substantial reduction of Tomm7 transcripts, likely due to a nonsense-mediated decay-like mechanism,23 whereas Tomm7(R/R) cells expressed a comparable amount of Tomm7 RNA as wild-type cells (Figure 2B).
Figure 2.
Generation and gross phenotype of Tomm7 mutant mice
(A) Edited alleles of the mouse Tomm7 gene in mice. Using CRISPR-mediated genome editing in mouse zygotes, a knockin (R) allele with the Trp25Arg (W25R) substitution and a deletion (D) allele missing a 184 bp sequence including the first exon-intron boundary were created. The locations of primers for qRT-PCR in (B) are indicated by arrows.
(B) qRT-PCR analysis of Tomm7 expression in mutant mice. RNA was isolated from Tomm7(D/D) bone marrow stromal cells and from the liver of Tomm7(R/R) along with their wild-type littermate controls. ± SEM, n = 4, ∗ ∗p < 0.0001, t test.
(C) Growth (body weight) of mice with indicated genotypes and genders. Both wild type and heterozygotes are used as control. Missing values in mutant mouse groups are due to either spontaneous death or euthanasia per IACUC protocol criteria. Left: for (D/D) mice and their control cohorts, both genders are included. n = 4–6 for (D/D), n = 8–12 for control (Ctrl), ∗p = 0.0041, ∗∗p < 0.0001. Mann-Whitney U test. Middle: growth of male (R/R) and compound mutant (R/D) mice. n = 4 for (R/R), n = 4 for (R/D), and n = 5 for Ctrl. ∗p = 0.0286 (R/R) versus (R/D), ∗∗p = 0.0286 Ctrl versus (R/R). Mann-Whitney U test. Right: growth of female (R/R) mice. n = 4 for (R/R), n = 6 for Ctrl. ∗p = 0.0159, ∗∗p = 0.0038. Mann-Whitney U test.
(D) Representative images of lipoatrophy in (R/R), (R/D), and (D/D) mice at indicated ages. The dorsal white adipose tissue covering the interscapular brown adipose tissue of male (D/D) mice and the epididymal adipose tissue of (R/D) and (R/R) mice are indicated by arrows.
(E) Quantification of the fat tissue mass of the epididymal (R/R and R/D) and dorsal (D/D) white adipose tissues. n = 5 for (D/D), (R/R), and their littermate control groups. Each group contains both males and females. The gender ratio between control and experimental groups is matched. n = 4 for (R/D) and their littermate control mice. ± SEM, ∗p = 0.0159, ∗∗p = 0.0579, ∗∗∗p = 0.0079. Mann-Whitney U test.
(F) Frontal views of the lumbar spine of micro-computed tomography X-ray analysis of mice with indicated genotypes and ages. Mild reduction in the height of vertebrae is observed in (R/D) and (D/D) mice. Scale bar, 1 mm.
Mice heterozygous for the D or R allele are grossly normal and indistinguishable from wild-type littermates. Homozygous Tomm7(R/R) mice showed relatively normal growth up to 7–8 weeks, but then they developed rapidly progressing emaciation leading to sudden death at around 9 to 10 weeks of age (Figure 2C). On the other hand, Tomm7(D/D) mice showed a significant growth failure in early postnatal stages, followed by death at 3 to 4 weeks of age after rapid weight loss near the end of their lives (Figure 2C). This phenotype is similar to that of previously reported Tomm7 knockout mice.24 Compound heterozygotes, Tomm7(R/D) mice showed relatively normal growth but again developed rapid weight loss around 5–6 weeks and died by 6–7 weeks (Figure 2C). These observations support the notion that the missense variant causes partial loss of Tomm7 function. The most prominent gross phenotype in these mouse models is lipoatrophy (Figures 2D and 2E). Since OXPHOS defects cause tissue-autonomous lipoatrophy,25,26 these findings suggest that Tomm7 deficiency impairs mitochondrial bioenergetics.
Growth failure and mild spondyloepimetaphyseal dysplasia features have been previously described in mitochondrial diseases.27, 28, 29, 30, 31, 32, 33 Variants in several genes encoding proteins regulating mitochondrial protein homeostasis, including TOMM70,34 have been reported to associate with skeletal abnormalities. To characterize the skeletal phenotype of these mouse models, we performed micro-computed tomography (μCT) and histological analyses. The μCT images of the spine revealed a modest shortening of vertebral bodies in Tomm7(R/D) and Tomm7(D/D) mice, reminiscent of the platyspondyly detected in the patient (Figures 2F and S2). The longitudinal growth of vertebral bodies and appendicular bones is driven by the growth plate. To investigate the abnormalities in growth plate cartilage, we analyzed the tibial growth plates that are much larger than those of vertebral bodies. Histological analysis of tibial growth plates of mutant mice showed a shortening of the growth plate with reduced cellular densities and cellular proliferation, assessed by EdU labeling in young mice and with Ki-67 staining in older mice (Figures 3A–3C), suggesting that the reduced chondrocyte proliferation contributes to the skeletal phenotype. Similar changes were also found in Tomm7(R/R) mice that showed only a modest growth defect. Additionally, these results suggest that the normal mitochondrial function plays an important regulatory role in growth plate chondrocytes, which predominantly rely on glycolysis, rather than mitochondrial metabolism, for ATP synthesis.35 Additional histological assessment of Tomm7 mutant tissues showed lipid accumulation in the liver (Figure 4A) and reductions in size of adipocytes (Figure 4B), findings observed also in the patient and consistent with mitochondrial dysfunction.13,15,20
Figure 3.
Hypocellularity and reduced proliferation in the mutant growth plates
(A) Tibial growth plates of P15 Tomm7(D/D) mice and control littermates. Hematoxylin and eosin (H&E)-stained sections (top). The insets are magnified views of the proliferating zone (indicated by yellow bar) of the growth plate (green bar). Quantification of the cellular density and EdU index of the proliferating zone are shown (middle and bottom). ± SEM, n = 3, t test.
(B) Tibial growth plates of P40 Tomm7(R/D) mice and control littermates. H&E-stained sections (top). The insets are magnified views of the proliferating zone (yellow bar) of the growth plate (green bar). Quantification of the cellular density and Ki67 index of the proliferating zone are shown (middle and bottom). ± SEM, n = 4–5, t test.
(C) Tibial growth plates of P61 Tomm7(R/R) mice and control littermates. H&E-stained sections (top). The insets are magnified views of the proliferating zone (yellow bar) of the growth plate (green bar). Quantification of the cellular density and Ki67 index of the proliferating zone are shown (middle and bottom). ± SEM, n = 4, t test. Scale bar, 100 μm.
Figure 4.
Lipid accumulation in the liver and peripheral fat loss in mutant mice
(A) The liver tissues from mice with indicated genotypes and ages. H&E staining of paraffin-processed sections (top) and oil red O staining of frozen sections (bottom). Arrows indicate cytoplasmic vacuoles in hepatocytes, suggesting lipid accumulation. Scale bar, 100 μm.
(B) H&E sections of epididymal (R/R and R/D) and dorsal (D/D) white adipose tissues (top) and quantification of adipocyte diameter. The maximal length of the longitudinal axis of cross sections of adipocytes was counted in 10 randomly selected cells per group. Scale bar, 100 μm. ± SEM, n = 10, ∗ p < 0.0001, t-test
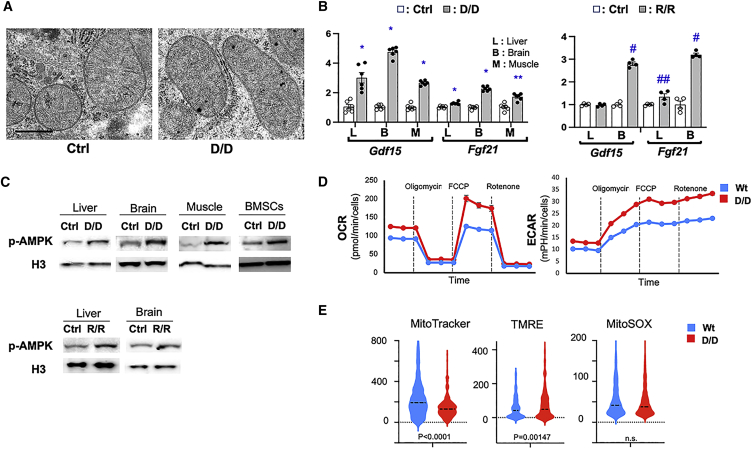
These data provide evidence that the TOMM7 W25R variant found in the affected individual is pathogenic and causes mitochondrial dysfunction at the organismal and tissue levels. This notion is also supported by the fact that this patient presents findings frequently observed in mitochondrial diseases, including hypotonia, nystagmus, elevated LDH levels, and fat accumulation in the liver. At subcellular and molecular levels, Tomm7 deficiency does not appear to significantly affect mitochondrial morphology except that mutant mitochondria in muscle appear generally small in size and that mild structural changes may develop in older mice (Figures 5A and S3–S5), whereas tissue expression of Gdf15 and Fgf21, markers for mitochondrial dysfunction36 were significantly upregulated in both Tomm7(D/D) and Tomm7(R/R) tissues (Figure 5B).
Figure 5.
Altered mitochondrial function in mutant mice
(A) Representative transmission electron microscope images of liver mitochondria in hepatocytes of P15 Tomm7(D/D) mice and control littermates (30,000×). Scale bar, 600 nm.
(B) mRNA expression of markers for mitochondrial dysfunction, Gdf15 and Fgf21, in indicated tissues. Significant upregulation of these markers was observed in both in Tomm7(D/D) and Tomm7(R/R) tissues. n = 4 - 6, ± SEM, ∗ p = 0.0022, ∗∗ p = 0.0043, # p = 0.0286, ## p=0.0571 vs Ctrl, Mann-Whitney U test.
(C) Upregulation of phospho-AMPK expression in indicated tissues.
(D) Seahorse analysis using primary rib chondrocytes of Tomm7(D/D) and littermate control mice. Oxygen consumption rate (OCR) and extracellular acidification rate (ECAR) were measured at the basal level and upon treatment with the ATP synthase inhibitor, oligomycin, the uncoupler, FCCP, and the ETC inhibitor, rotenone, and antimycin A (Rotenone). Both OCR and ECAR are significantly upregulated. Mean ± SEM. n = 8, p < 0.001 at all data points. t test.
(E) Flow cytometric analysis of primary rib chondrocytes. Cells were pre-incubated with the indicated fluorescent dyes and subjected to flow cytometric analysis. The signal intensity of 500 randomly selected cells per group was evaluated. Dotted lines indicate median values. Mitochondrial membrane potential (TMRE) is slightly upregulated in Tomm7(D/D) despite the decreased mitochondrial mass (MitoTracker).
As expected from the mouse phenotype, phosphorylation of AMP-dependent protein kinase (AMPK), a pivotal energy sensor in the cell,37 was upregulated in multiple tissues of Tomm7 mutants, indicating inefficient ATP synthesis in mutant cells (Figure 5C). Interestingly, multiple types of Tomm7-deficient cells showed increases not only in extracellular acidification rate (ECAR) but also in oxygen consumption rate (OCR) and showed normal responses to inhibitors of the electron transport chain (ETC) and ATP synthase (complex V) (Figures 5D and S6). These results suggest that the function of ETC and the proton transportation of ATP synthase are maintained intact in Tomm7-deficient cells (Figure 5D). Similar to Tomm7(D/D) cells, the upregulation in both OCR and ECAR was also observed in Tomm7(R/R) cells (Figure S7). Flow cytometric analysis showed a slight increase in membrane potential, as assessed by TMRE staining in Tomm7(D/D) chondrocyte samples, whereas the mitochondrial mass, assessed by MitoTracker staining, was modestly reduced (Figure 5E). Unlike in a previous zebrafish study,24 we did not find significant changes in reactive oxygen species (ROS), as assessed by MitoSox staining. These findings may reflect the hyperactive state of the ETC. These changes were generally found also in other cell types of Tomm7(D/D) cells (Figure S8) and were partially found in Tomm7(R/R) cells (Figure S9).
The upregulation both in OCR and in ECAR despite the increase in p-AMPK suggests that there is a dissociation between oxidation and ATP synthesis in Tomm7 mutant mitochondria. Such uncoupling is physiologically observed in brown adipose tissues where uncoupling protein 1 (Ucp1)38 mediates the effect. We analyzed the expression of Ucp1 and its paralogs, Ucp2 and Ucp3,39 in primary bone marrow stromal cells (BMSCs) and in the brain of Tomm7(D/D) mice. We found that these genes were generally downregulated rather than upregulated except for a modest upregulation of Ucp3 in the brain, suggesting that aberrant uncoupling protein expression is an unlikely mechanism for the impaired ATP synthesis in Tomm7 mutant cells (Figure S10). ATP deficiency can be caused by defects in transport of ATP generated in the mitochondrial matrix into the cytoplasm, which is mediated by adenine nucleotide translocase (ANT) family proteins and voltage-dependent anion channels (VDACs).40,41 We examined the expression of Slc25a2, a major ANT, and Vdac1, a major VDAC, in mitochondria-enriched protein fractions of Tomm7(D/D) mice; however, we did not find significant changes in their abundance (Figure S11). The bioenergetic dysfunction and the increased oxygen consumption with normal response to oligomycin, a proton transport inhibitor of complex V, suggest that there is a dissociation between proton transport and catalytic activity of complex V. Complex V consists of two rotary motors, the membrane-bound F0 that mediates proton transport and F1 that is responsible for ATP synthesis. We assessed the expression of several complex V subunits along with ETC subunits, but we did not find overt changes in the abundance of these proteins, suggesting that Tomm7 deficiency may have relatively limited impact on mitochondrial proteostasis (Figure S11). Independently of the mitochondrial bioenergetic dysfunction, it could be possible that Tomm7 deficiency contributes to the phenotype via suppressing PINK1/parkin-mediated mitophagy. We have tested whether Tomm7 deficiency alters PINK1 expression using primary mouse BMSCs. Upon mitochondrial stress, PINK1 expression was induced both in control and in Tomm7-deficient cells to similar extents. This finding suggests that Tomm7’s regulatory role in PINK1 expression in mouse skeletal progenitors is limited (Figure S12).
In summary, this study identifies TOMM7 as a new human disease gene and provides genetic evidence that this variant is pathogenic and creates a hypomorphic allele. This study presents evidence that aberration of mitochondrial protein homeostasis leads to mitochondrial disease with skeletal involvement, as previously suggested by several reports.42 Tomm7 deficiency in mice results in a phenotype consistent with mitochondrial bioenergetic deficiency, yet Tomm7-deficient cells show increased oxygen consumption with normal responses to ETC and ATP synthase inhibitors, suggesting that Tomm7 deficiency does not impair the function of the tricarboxylic cycle, ETC, or proton transport by the complex V. These data suggest a discordance between proton transport (F0) and catalytic function of ATP synthase (F1). However, we did not find overt changes in the stoichiometry of some of complex V subunits; the precise mechanism by which Tomm7 deficiency causes dysfunction in ATP synthesis is currently unclear.
Acknowledgments
We thank the Center for Skeletal Research (P30AR075042) for assistance in histological analysis. We thank Dr. Patrick Ward and Dr. Lawrence Zukerberg for valuable comments on mitochondrial biology and liver pathology. This study was supported by the NIH grant AR056645 (T.K.) and by grants from Stiftelsen Samariten, Stiftelsen Promobilia, and Stiftelsen Frimurare Barnhuset i Stockholm, Stiftelsen Sällsyntafonden, from Region Stockholm (20180131 and 20200500), from Swedish Research Council (2018-03046), and by the Karolinska Institutet (G.G. and D.B).
Declaration of interests
The authors declare no competing interests.
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.xhgg.2022.100148.
Contributor Information
Giedre Grigelioniene, Email: giedre.grigelioniene@ki.se.
Tatsuya Kobayashi, Email: tkobayashi1@mgh.harvard.edu.
Web resources
CADD, https://cadd.gs.washington.edu/.
ClinVar, https://www.ncbi.nlm.nih.gov/clinvar/.
DECIPHER, https://www.deciphergenomics.org/.
Genomics England PannelApp, https://panelapp.genomicsengland.co.uk/#!.
OMIM, https://omim.org/.
Supplemental information
Data availability
The variant has been submitted to the ClinVar database (accession number SCV002505631).
References
- 1.Wallace D.C. Mitochondria and cancer. Nat. Rev. Cancer. 2012;12:685–698. doi: 10.1038/nrc3365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Moos W.H., Faller D.V., Glavas I.P., Harpp D.N., Kamperi N., Kanara I., Kodukula K., Mavrakis A.N., Pernokas J., Pernokas M., et al. Pathogenic mitochondrial dysfunction and metabolic abnormalities. Biochem. Pharmacol. 2021;193:114809. doi: 10.1016/j.bcp.2021.114809. [DOI] [PubMed] [Google Scholar]
- 3.Rey F., Ottolenghi S., Zuccotti G.V., Samaja M., Carelli S. Mitochondrial dysfunctions in neurodegenerative diseases: role in disease pathogenesis, strategies for analysis and therapeutic prospects. Neural Regen. Res. 2022;17:754–758. doi: 10.4103/1673-5374.322430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Forbes J.M., Thorburn D.R. Mitochondrial dysfunction in diabetic kidney disease. Nat. Rev. Nephrol. 2018;14:291–312. doi: 10.1038/nrneph.2018.9. [DOI] [PubMed] [Google Scholar]
- 5.Salnikova D., Orekhova V., Grechko A., Starodubova A., Bezsonov E., Popkova T., Orekhov A. Mitochondrial dysfunction in vascular wall cells and its role in atherosclerosis. Int. J. Mol. Sci. 2021;22:8990. doi: 10.3390/ijms22168990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jankovic M., Novakovic I., Gamil Anwar Dawod P., Gamil Anwar Dawod A., Drinic A., Abdel Motaleb F.I., Ducic S., Nikolic D. Current concepts on genetic aspects of mitochondrial dysfunction in amyotrophic lateral sclerosis. Int. J. Mol. Sci. 2021;22:9832. doi: 10.3390/ijms22189832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.DiMauro S. Mitochondrial diseases. Biochim. Biophys. Acta. 2004;1658:80–88. doi: 10.1016/j.bbabio.2004.03.014. [DOI] [PubMed] [Google Scholar]
- 8.Gorman G.S., Chinnery P.F., DiMauro S., Hirano M., Koga Y., McFarland R., Suomalainen A., Thorburn D.R., Zeviani M., Turnbull D.M. Mitochondrial diseases. Nat. Rev. Dis. Primers. 2016;2:16080. doi: 10.1038/nrdp.2016.80. [DOI] [PubMed] [Google Scholar]
- 9.Cotter D., Guda P., Fahy E., Subramaniam S. MitoProteome: mitochondrial protein sequence database and annotation system. Nucleic Acids Res. 2004;32:D463–D467. doi: 10.1093/nar/gkh048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chacinska A., Koehler C.M., Milenkovic D., Lithgow T., Pfanner N. Importing mitochondrial proteins: machineries and mechanisms. Cell. 2009;138:628–644. doi: 10.1016/j.cell.2009.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Moehle E.A., Shen K., Dillin A. Mitochondrial proteostasis in the context of cellular and organismal health and aging. J. Biol. Chem. 2019;294:5396–5407. doi: 10.1074/jbc.TM117.000893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Araiso Y., Imai K., Endo T. Structural snapshot of the mitochondrial protein import gate. FEBS J. 2021;288:5300–5310. doi: 10.1111/febs.15661. [DOI] [PubMed] [Google Scholar]
- 13.Yamano K., Tanaka-Yamano S., Endo T. Tom7 regulates Mdm10-mediated assembly of the mitochondrial import channel protein Tom40. J. Biol. Chem. 2010;285:41222–41231. doi: 10.1074/jbc.M110.163238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bausewein T., Mills D.J., Langer J.D., Nitschke B., Nussberger S., Kühlbrandt W. Cryo-EM structure of the TOM core complex from neurospora crassa. Cell. 2017;170:693–700.e7. doi: 10.1016/j.cell.2017.07.012. [DOI] [PubMed] [Google Scholar]
- 15.Wiedemann N., Kozjak V., Chacinska A., Schönfisch B., Rospert S., Ryan M.T., Pfanner N., Meisinger C. Machinery for protein sorting and assembly in the mitochondrial outer membrane. Nature. 2003;424:565–571. doi: 10.1038/nature01753. [DOI] [PubMed] [Google Scholar]
- 16.Dembowski M., Kunkele K.P., Nargang F.E., Neupert W., Rapaport D. Assembly of Tom6 and Tom7 into the TOM core complex of neurospora crassa. J. Biol. Chem. 2001;276:17679–17685. doi: 10.1074/jbc.M009653200. [DOI] [PubMed] [Google Scholar]
- 17.Pfanner N., Wiedemann N., Meisinger C., Lithgow T. Assembling the mitochondrial outer membrane. Nat. Struct. Mol. Biol. 2004;11:1044–1048. doi: 10.1038/nsmb852. [DOI] [PubMed] [Google Scholar]
- 18.Hasson S.A., Kane L.A., Yamano K., Huang C.H., Sliter D.A., Buehler E., Wang C., Heman-Ackah S.M., Hessa T., Guha R., et al. High-content genome-wide RNAi screens identify regulators of parkin upstream of mitophagy. Nature. 2013;504:291–295. doi: 10.1038/nature12748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sekine S., Wang C., Sideris D.P., Bunker E., Zhang Z., Youle R.J. Reciprocal roles of Tom7 and OMA1 during mitochondrial import and activation of PINK1. Mol. Cell. 2019;73:1028–1043.e5. doi: 10.1016/j.molcel.2019.01.002. [DOI] [PubMed] [Google Scholar]
- 20.Wang W., Chen X., Zhang L., Yi J., Ma Q., Yin J., Zhuo W., Gu J., Yang M. Atomic structure of human TOM core complex. Cell Discov. 2020;6:67. doi: 10.1038/s41421-020-00198-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wang Y., Addess K.J., Chen J., Geer L.Y., He J., He S., Lu S., Madej T., Marchler-Bauer A., Thiessen P.A., et al. MMDB: annotating protein sequences with Entrez's 3D-structure database. Nucleic Acids Res. 2007;35:D298–D300. doi: 10.1093/nar/gkl952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sayers E.W., Bolton E.E., Brister J.R., Canese K., Chan J., Comeau D.C., Connor R., Funk K., Kelly C., Kim S., et al. Database resources of the national center for biotechnology information. Nucleic Acids Res. 2022;50:D20–D26. doi: 10.1093/nar/gkab1112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Brogna S., Wen J. Nonsense-mediated mRNA decay (NMD) mechanisms. Nat. Struct. Mol. Biol. 2009;16:107–113. doi: 10.1038/nsmb.1550. [DOI] [PubMed] [Google Scholar]
- 24.Shi D., Qi M., Zhou L., Li X., Ni L., Li C., Yuan T., Wang Y., Chen Y., Hu C., et al. Endothelial mitochondrial preprotein translocase Tomm7-Rac1 signaling axis dominates cerebrovascular network homeostasis. Arterioscler. Thromb. Vasc. Biol. 2018;38:2665–2677. doi: 10.1161/ATVBAHA.118.311538. [DOI] [PubMed] [Google Scholar]
- 25.Vernochet C., Mourier A., Bezy O., Macotela Y., Boucher J., Rardin M.J., An D., Lee K.Y., Ilkayeva O.R., Zingaretti C.M., et al. Adipose-specific deletion of TFAM increases mitochondrial oxidation and protects mice against obesity and insulin resistance. Cell Metab. 2012;16:765–776. doi: 10.1016/j.cmet.2012.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Vernochet C., Damilano F., Mourier A., Bezy O., Mori M.A., Smyth G., Rosenzweig A., Larsson N.G., Kahn C.R. Adipose tissue mitochondrial dysfunction triggers a lipodystrophic syndrome with insulin resistance, hepatosteatosis, and cardiovascular complications. FASEB J. 2014;28:4408–4419. doi: 10.1096/fj.14-253971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Boal R.L., Ng Y.S., Pickett S.J., Schaefer A.M., Feeney C., Bright A., Taylor R.W., Turnbull D.M., Gorman G.S., Cheetham T., McFarland R. Height as a clinical biomarker of disease burden in adult mitochondrial disease. J. Clin. Endocrinol. Metab. 2019;104:2057–2066. doi: 10.1210/jc.2018-00957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Castro-Gago M., Gómez-Lado C., Pérez-Gay L., Eirís-Puñal J. Abnormal growth in mitochondrial disease. Acta Paediatr. 2010;99:796. doi: 10.1111/j.1651-2227.2010.01738.x. [DOI] [PubMed] [Google Scholar]
- 29.Jabbour S., Harissi-Dagher M. Recessive mutation in a nuclear-encoded mitochondrial tRNA synthetase associated with infantile cataract, congenital neurotrophic keratitis, and orbital myopathy. Cornea. 2016;35:894–896. doi: 10.1097/ICO.0000000000000847. [DOI] [PubMed] [Google Scholar]
- 30.Mierzewska H., Rydzanicz M., Biegański T., Kosinska J., Mierzewska-Schmidt M., Ługowska A., Pollak A., Stawiński P., Walczak A., Kędra A., et al. Spondyloepimetaphyseal dysplasia with neurodegeneration associated with AIFM1 mutation - a novel phenotype of the mitochondrial disease. Clin. Genet. 2017;91:30–37. doi: 10.1111/cge.12792. [DOI] [PubMed] [Google Scholar]
- 31.Vona B., Maroofian R., Bellacchio E., Najafi M., Thompson K., Alahmad A., He L., Ahangari N., Rad A., Shahrokhzadeh S., et al. Expanding the clinical phenotype of IARS2-related mitochondrial disease. BMC Med. Genet. 2018;19:196. doi: 10.1186/s12881-018-0709-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wolny S., McFarland R., Chinnery P., Cheetham T. Abnormal growth in mitochondrial disease. Acta Paediatr. 2009;98:553–554. doi: 10.1111/j.1651-2227.2008.01148.x. [DOI] [PubMed] [Google Scholar]
- 33.Zhao T., Goedhart C.M., Sam P.N., Sabouny R., Lingrell S., Cornish A.J., Lamont R.E., Bernier F.P., Sinasac D., Parboosingh J.S., et al. PISD is a mitochondrial disease gene causing skeletal dysplasia, cataracts, and white matter changes. Life Sci Alliance. 2019;2 doi: 10.26508/lsa.201900353. e201900353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wei X., Du M., Xie J., Luo T., Zhou Y., Zhang K., Li J., Chen D., Xu P., Jia M., et al. Mutations in TOMM70 lead to multi-OXPHOS deficiencies and cause severe anemia, lactic acidosis, and developmental delay. J. Hum. Genet. 2020;65:231–240. doi: 10.1038/s10038-019-0714-1. [DOI] [PubMed] [Google Scholar]
- 35.Stegen S., Laperre K., Eelen G., Rinaldi G., Fraisl P., Torrekens S., Van Looveren R., Loopmans S., Bultynck G., Vinckier S., et al. HIF-1alpha metabolically controls collagen synthesis and modification in chondrocytes. Nature. 2019;565:511–515. doi: 10.1038/s41586-019-0874-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Maresca A., Del Dotto V., Romagnoli M., La Morgia C., Di Vito L., Capristo M., Valentino M.L., Carelli V., ER-MITO Study Group Expanding and validating the biomarkers for mitochondrial diseases. J. Mol. Med. 2020;98:1467–1478. doi: 10.1007/s00109-020-01967-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Herzig S., Shaw R.J. AMPK: guardian of metabolism and mitochondrial homeostasis. Nat. Rev. Mol. Cell Biol. 2018;19:121–135. doi: 10.1038/nrm.2017.95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kalinovich A.V., de Jong J.M.A., Cannon B., Nedergaard J. UCP1 in adipose tissues: two steps to full browning. Biochimie. 2017;134:127–137. doi: 10.1016/j.biochi.2017.01.007. [DOI] [PubMed] [Google Scholar]
- 39.Nicholls D.G. Mitochondrial proton leaks and uncoupling proteins. Biochim. Biophys. Acta. Bioenerg. 2021;1862:148428. doi: 10.1016/j.bbabio.2021.148428. [DOI] [PubMed] [Google Scholar]
- 40.Ruprecht J.J., King M.S., Zögg T., Aleksandrova A.A., Pardon E., Crichton P.G., Steyaert J., Kunji E.R.S. The molecular mechanism of transport by the mitochondrial ADP/ATP carrier. Cell. 2019;176:435–447.e15. doi: 10.1016/j.cell.2018.11.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Heslop K.A., Milesi V., Maldonado E.N. VDAC modulation of cancer metabolism: advances and therapeutic challenges. Front. Physiol. 2021;12:742839. doi: 10.3389/fphys.2021.742839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhao T., Goedhart C., Pfeffer G., Greenway S.C., Lines M., Khan A., Innes A.M., Shutt T.E. Skeletal phenotypes due to abnormalities in mitochondrial protein homeostasis and import. Int. J. Mol. Sci. 2020;21:E8327. doi: 10.3390/ijms21218327. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The variant has been submitted to the ClinVar database (accession number SCV002505631).