Summary
Harnessing innate immunity is emerging as a promising therapeutic approach in cancer. We report here the design of tetraspecific molecules engaging natural killer (NK) cell-activating receptors NKp46 and CD16a, the β-chain of the interleukin-2 receptor (IL-2R), and a tumor-associated antigen (TAA). In vitro, these tetraspecific antibody-based natural killer cell engager therapeutics (ANKETs) induce a preferential activation and proliferation of NK cells, and the binding to the targeted TAA triggers NK cell cytotoxicity and cytokine and chemokine production. In vivo, tetraspecific ANKETs induce NK cell proliferation and their accumulation at the tumor bed, as well as the control of local and disseminated tumors. Treatment of non-human primates with CD20-directed tetraspecific ANKET leads to CD20+ circulating B cell depletion, with minimal systemic cytokine release and no sign of toxicity. Tetraspecific ANKETs, thus, constitute a technological platform for harnessing NK cells as next-generation cancer immunotherapies.
Keywords: Cancer immunotherapy, Natural Killer cells, multispecific antibodies, cytokine
Graphical abstract

Highlights
-
•
Tetraspecific ANKETs constitute a technological platform to harness NK cells in cancer
-
•
Tetraspecific ANKETs target NKp46, CD16a, IL-2Rβ, and a tumor antigen
-
•
Tetraspecific ANKETs stimulate NK cell proliferation, activation, and antitumor functions
-
•
In vivo, tetraspecific ANKETs promote NK cell tumor accumulation and antitumor activity
Harnessing NK cells is emerging as a promising therapeutic approach in cancer treatment that may overcome resistance to current immunotherapies targeting T cells. Here, Demaria et al. report the design of tetraspecific ANKETs, which constitute a technological platform to enhance and harness NK cell antitumor functions as next-generation immunotherapies.
Introduction
Natural killer (NK) cells are cytotoxic innate lymphoid cells (ILCs) that can kill tumor cells and secrete an array of cytokines and chemokines that shape adaptive immunity.1,2 The recent clinical success of chimeric antigen receptor (CAR) NK cells for treating malignant hemopathies, such as non-Hodgkin’s lymphoma and chronic lymphocytic leukemia, has illustrated the potential of NK cells as a potent cellular player for next-generation cancer immunotherapies.3 Cytotoxic monoclonal antibodies (mAbs), such as rituximab, trastuzumab, and cetuximab, can induce NK cell activation and cytotoxicity against malignant cells expressing the targeted antigen, via CD16a, their receptor for the Fc fragment of immunoglobulin G (IgG) antibodies. Despite the clinical successes reported for these therapeutic antibodies, many patients have refractory tumors or experience rapid relapse, highlighting the need to develop innovative therapeutics for improving NK cell effector functions.
Activating receptors other than CD16a, such as NKG2D and the natural cytotoxicity receptors NKp46 and NKp30, can be targeted with agonistic antibodies to promote NK cell effector functions. CD16a and NKp46 induce signaling by associating with the ITAM-bearing molecules CD3ζ and FcRγ, which transduce kinase-dependent signals and are responsible for the induction of effector cell functions.4,5 The co-engagement of different activating receptors is important to achieve strong NK cell activation.6,7 We previously reported the design of NK cell engagers co-engaging NKp46 and CD16a,8 as a first generation of trispecific antibody-based NK cell engager therapeutics showing more potent antitumor activity than antibody-dependent cell-mediated cytoxicity (ADCC)-inducing antibodies.
Interleukin-2 (IL-2) is a cytokine with pleiotropic functions in innate and adaptive immune responses.9 High-dose IL-2 was among the first immunotherapies to be approved for cancer but its use has been hindered by its high toxicity and its efficacy restricted to a limited subset of patients, partly due to the ability of this cytokine to activate immunosuppressive regulatory T cells (Tregs).10 IL-2 can bind a dimeric receptor composed of IL-2Rβ (CD122) and the common gamma chain cytokine receptor (γc; CD132), expressed on NK cells, with intermediate affinity or a trimeric receptor including IL-2Rα (CD25)11 with high affinity. The CD122-CD132 dimeric receptor is found mostly on memory CD8+ T cells and NK cells, whereas the CD122-CD132-CD25 trimeric receptor is found predominantly on Tregs at steady state and is transiently upregulated on recently activated T and NK cells. IL-2 triggers the JAK/STAT signaling pathway and induces NK cell activation and proliferation.12,13
Molecular engineering is providing tremendous opportunities for generating molecules capable of inducing synthetic immunity. IL-2 variants (IL-2vs) have been designed with point mutations that abolish binding to CD25, with the goal of limiting interaction with Tregs but retaining the ability to interact with CD122/CD132 and to promote NK and CD8 T cell activation and proliferation.14 With the aim of further exploiting synthetic biology in cancer therapies, we used the versatile NK cell engager platform,8 which we refer to as “ANKETs,” to generate tetraspecific CD20-ANKETs (IL-2v/aNKp46/Fc/aCD20), which consist of an IL-2v peptide stimulating IL-2R without CD25 involvement, an antibody fragment targeting NKp46 (aNKp46-1),8 a Fc domain of human IgG1 mediating interaction with Fcγ receptors such as CD16a, and an antibody fragment targeting CD20 (aCD20) as a model tumor-associated antigen (TAA). Assessments of CD20-ANKETs activity revealed a high efficacy for inducing proliferation, chemokine and cytokine secretion, and cytotoxicity in human primary NK cells and for controlling invasive and subcutaneous tumors in mice. In non-human primates, CD20-ANKETs induced B cell depletion with no sign of toxicity. These data support the development of tetrafunctional ANKETs directed against various tumor antigens as promising and innovative immunotherapies for multiple cancer indications.
Results
Induction of human NK cell activation by tetraspecific ANKETs
NK cells express NKp46, CD16a, and the CD122-CD132 dimeric IL-2 receptor at steady state in human blood (Figure S1A). CD16a and other NK cell-activating receptors, such as NKp30, are not stably expressed at the surface of NK cells infiltrating tumors, but NKp46 expression remains stable in the tumor microenvironment (Figure S1B). These data provided a strong rationale for generating tetraspecific ANKETs engaging NKp46, CD16a, and CD122 to induce the optimal activation of NK cells. We therefore assessed the ability of an IL-2 peptide linked to an ANKET to induce IL-2R signaling (i.e., STAT5 tyrosine phosphorylation) relative to recombinant IL-2. Wild-type IL-2 fused to a mutated ANKET (IL-2 wild type [WT]/IC/Fc-null/aCD20) unable to bind NKp46 (IC, isotype control) and FcγR (Fc-null) induced a dose-dependent increase in STAT5 phosphorylation of a magnitude similar to that for free recombinant human IL-2, in both NK and T lymphocytes, demonstrating that the incorporation of IL-2 into an ANKET format had no effect on its signaling function (Figure S1C). Surface plasmon resonance analysis confirmed that the IL-2v included in ANKETs was unable to bind CD25 (Figure S1D) but had a binding affinity similar to that of wild-type IL-2 for binding to the intermediate affinity receptor, interacting with CD122 with a KD of 2.4 μM, but not with CD132, as expected (Figure S1E). Consistently, the IL-2v included in the IL-2v/IC/Fc-null/aCD20 molecule had a much weaker IL-2R signaling capacity on Tregs than the IL-2WT control molecule (Figure S1C). We then investigated whether the targeting of NKp46 and CD16a on NK cells by ANKET-IL-2v led to preferential NK cell activation. The tetraspecific ANKET preferentially promoted IL-2R signaling on NK cells and induced a decrease of about two orders of magnitude in the half-maximal effective concentration (EC50) for STAT5 phosphorylation relative to the IL-2v/IC/Fc null/aCD20 molecule, which did not target NK cells (Figure 1A). By contrast, NK-targeting and non-NK-targeting molecules activated IL-2R signaling similarly on Tregs, CD4+ T cells, and CD8+ T cells. Our findings confirm that the CD16a and NKp46 binding elements on ANKET-IL-2v were essential for maximal IL-2R signaling on NK cells, as mutated ANKET-IL-2v, which cannot interact with NKp46 (IL-2v/IC/Fc/aCD20) or CD16a (IL-2v/aNKp46/Fc-null/aCD20), was a less potent inducer of STAT5 phosphorylation (Figure 1B). Thus, unlike recombinant IL-2, the tetraspecific ANKET limited Treg activation and redirected IL-2v activity, preferentially promoting IL-2R signaling on NK cells.
Figure 1.
In vitro characterization of ANKET-IL-2v efficacy
(A) pSTAT5 monitoring on NK cells (CD3−CD56+), Tregs (CD3+CD4+CD25hiFoxP3+), CD4+ T cells (CD3+CD4+CD25lowFOXP3−), and CD8+ T cells (CD3+CD8+) gated on peripheral blood mononuclear cells (PBMCs) activated by the IL-2v/aNKp46/Fc/aCD20 molecule or the IL-2v/IC/Fc-null/aCD20 molecule. The picture is representative of at least 3 independent experiments.
(B) pSTAT5 monitoring on NK cells (CD3−CD56+) gated on PBMCs activated with IL-2v/aNKp46/Fc/aCD20, IL-2v/aNKp46/Fc-null/aCD20, IL-2v/IC/Fc/aCD20, and IL-2v/IC/Fc-null/aCD20 molecules. The dose response of a representative donor is presented in the left panel, and a boxplot, with whiskers showing minimal and maximal value, depicting the EC50 obtained for three to seven donors is shown in the right panel.
(C) Purified NK cell proliferation induced by IL-2v/aNKp46/Fc/aCD20, IL-2v/IC/Fc-null/IC, and aNKp46/Fc/aCD20 molecules in the absence of the target tumor cells. Left, the percentage of NK cells displaying proliferation induced by a dose response to the indicated molecules. Right, CellTrace Violet (CTV) dilution profile induced by the indicated molecules at a concentration of 3.7 nM. The data shown are representative of results obtained for five donors.
(D) IFN-γ and MIP-1β production by purified NK cells in the presence of Raji tumor cells induced by IL-2v/aNKp46/Fc/aCD20, IL-2v/aNKp46/Fc-null/aCD20, IL-2v/IC/Fc/aCD20, and IL-2v/aNKp46/Fc/IC molecules. The data shown are the mean ± SD for three donors.
(E) NK cell cytotoxicity toward Raji tumor cells induced by IL-2v/aNKp46/Fc/aCD20, IL-2v/aNKp46/Fc-null/aCD20, IL-2v/IC/Fc/aCD20, and IL-2v/aNKp46/Fc/IC molecules. Left, NK cell-mediated specific lysis from a representative donor analyzed with experimental duplicate ± SD. Right, boxplot, with whiskers showing minimal and maximal value, depicting the EC50 of specific lysis obtained for five donors. The statistical analysis is described in the STAR methods, ∗p < 0.05; ∗∗p < 0.01.
We then evaluated the functional effects of the tetraspecific ANKET on NK cells. Contrasting strongly with the trispecific aNKp46/Fc/aCD20 molecule and consistent with its ability to induce STAT5 phosphorylation in NK cells, the tetraspecific ANKET induced robust NK cell proliferation and stimulated NK cell expansion at lower doses than a non-NK-targeted IL-2v-bearing molecule (Figure 1C). In the presence of the CD20+ Raji B cell lymphoma line as a tumor target, NK cells activated by the tetraspecific ANKETs were also able to produce interferon (IFN)-γ and MIP-1β (Figure 1D). The level of cytokine induction was dependent on CD16a and NKp46 binding on NK cells, as ANKET control molecules lacking the CD16a- or NKp46-binding moieties had a lower cytokine induction capacity (Figure 1D). Targeting of both the tumor antigen and NK cell receptors was essential for the induction of cytokine production, as molecules with no tumor antigen-binding or NK cell-binding elements (NKp46 and CD16a) produced little or no cytokine (Figures 1D and S1F). The tetraspecific ANKET induced NK cell cytokine production more strongly than the trispecific aNKp46/Fc/aCD20 molecule without IL-2v (Figure S1F), supporting that a cooperation between the ITAM and JAK/STAT pathways underlies the enhancement of the functional activity of NK cells.
Using the CD20+ Raji B cell line as a tumor target in a killing assay, we observed that the co-engagement of CD16a and NKp46 increased the potency of the tetraspecific ANKET, with no off-target effect (Figures 1E and S1G), as previously described for the trispecific NKp46/Fc/CD20 molecule.8 Co-engagement of the tumor antigen and the activating NK cell receptors was essential for cytotoxicity, as IL-2v/aNKp46/Fc/IC and IL-2v/IC/Fc-null/aCD20 molecules did not promote NK cell killing activity (Figures 1E and S1G). Importantly, the cytotoxicity mediated by the tetraspecific ANKET was maximal at low concentrations, at which neither the effector NK cell nor the targeted tumor cell binding sites were saturated, indicating that very partial ANKET binding to NK cells and target cells was sufficient to promote the maximal killing activity of tetraspecific ANKET (Figure S1H). The tetraspecific ANKET, thus, forces interactions between NK and tumor cells, inducing strong tumor cell killing even at low receptor occupancy of the TAA.
Tetraspecific ANKETs induce an NK cell activation transcriptomic signature
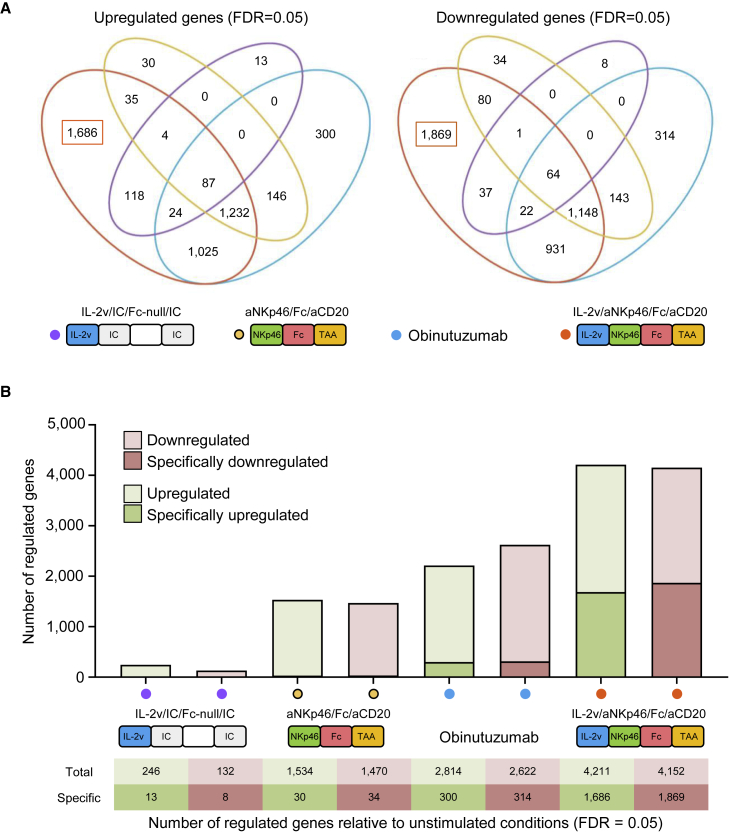
We dissected the mechanisms underlying the impact of tetraspecific ANKETs on NK cells by performing transcriptome analysis on NK cells purified from blood and activated with tetraspecific ANKETs, trispecific aNKp46/Fc/aCD20, the Fc-enhanced anti-CD20 mAb Obinutuzumab, or an IL-2v polypeptide (IL-2v/IC/Fc-null/IC) in the presence of CD20+ tumor target cells. RNA sequencing (RNA-seq) analysis revealed that the tetraspecific ANKET induced a unique gene expression program in NK cells (Figure S2A), with thousands of genes specifically upregulated or downregulated relative to untreated NK cells (Figures 2A and 2B). The genes specifically upregulated by tetraspecific ANKET included genes involved in NK cell activation and functions, such as VAV1, RAC1, and RAC2, shown to regulate cell-mediated killing,15 and structural components, such as ACTB and TUBB, involved in the regulation of immunological synapse formation at the interface with targeted cells16 (Table S1). An overrepresentation analysis (ORA) of the genes specifically upregulated by the tetraspecific ANKET relative to other stimuli revealed an overrepresentation of pathways involved in NK cell effector functions, such as cytokine signaling and IFN-γ responses, membrane trafficking, cell-cycle, oxidative phosphorylation, and mTORC1 pathways (Figure S3A). Remarkably, the gene expression program induced by tetraspecific ANKET differed from that induced by a combination of the trispecific aNKp46/Fc/aCD20 and the IL-2v polypeptide (Figure 3A). Gene set enrichment analysis (GSEA) revealed an enrichment in genes involved in the IFN-γ and cytokine responses, including JAK/STAT signaling pathways, in NK cells stimulated by the tetraspecific ANKET relative to cells stimulated by a combination of the trispecific aNKp46/Fc/aCD20 and the IL-2v polypeptide (Figure 3B). Furthermore, the tetraspecific ANKET specifically drove the upregulation of >1,300 genes for which expression was not induced by the IL-2v polypeptide alone, the trispecific aNKp46/Fc/aCD20 alone, or a combination of the trispecific aNKp46/Fc/aCD20 and the IL-2v polypeptide (Figure 3C). In addition to pathways involved in cytokine and IFN-γ responses, ORA on these genes revealed an overrepresentation of pathways involved in cellular processes linked to cell activation, such as transcriptional regulation, RNA metabolism and processing, posttranslational protein modifications, protein trafficking and secretion, and cell cycle (Figure S3B). Thus, tetraspecific ANKET drives a specific transcriptomic program poising NK cells for more effective antitumor action, with an improved effector and proliferative phenotype. A gene-candidate analysis confirmed that the tetraspecific ANKET induced a specific pattern of gene regulation different from that induced by a combination of the trispecific aNKp46/Fc/aCD20 plus an IL-2v polypeptide or stimulation by single molecules, separately (Figure 3D; Table S2). Tetraspecific ANKET-activated NK cells had higher levels of expression for genes encoding molecules involved in effector functions, such as IFN-γ, MIP-1α (CCL3), and MIP-1β (CCL4), or cytotoxic factors, such as perforin (PRF1), granzyme B (GZMB), and TRAIL (TNFSF10), consistent with their enhanced killing capacities (Figure 3D). Interestingly, enrichment was also observed for genes such as FLT3LG, XCL1, and IL-32, involved in the crosstalk between NK cells and DCs, indicating that ANKET may enable NK cells to stimulate dendritic cells and, hence, to promote antitumor adaptive responses (Figure 3D). Flow cytometry analysis confirmed that synthetic co-stimulation of the NKp46, CD16a, and IL-2R pathways with tetraspecific ANKET was the most efficient way to promote NK cell activation, as monitored by the upregulation of the CD69 activation marker (Figure 3E). These data show that tetraspecific ANKET stimulation resulted in a specific transcriptomic effector program associated with enhanced NK cell responses.
Figure 2.
Transcriptomic landscape of NK cells activated by ANKET-IL-2v
(A and B) Purified human NK cells were activated for 4 h in the presence of the murine huCD20-B16F10 target cells, with 0.1 μg/mL IL-2v/aNKp46/Fc/aCD20, IL-2v/IC/Fc-null/IC, Obinutuzumab, or aNKp46/Fc/aCD20. RNA sequencing was performed, and reads were aligned with the human genome, n = 4 to 6 donors.
(A) Venn diagram representing the number of genes significantly regulated by each of the indicated stimulation conditions relative to unstimulated conditions (false discovery rate [FDR] = 0.05).
(B) Bar plot showing the number of genes regulated (FDR = 0.05) for each stimulation treatment relative to unstimulated conditions.
Figure 3.
A specific transcriptomic effector program associated with enhanced NK cell responses is induced by ANKET-IL-2v stimulation
(A–D and F) Purified human NK cells were activated for 4 h in the presence of the murine huCD20-B16F10 target cells, with 0.1 μg/mL IL-2v/aNKp46/Fc/aCD20, IL-2v/IC/Fc-null/IC, Obinutuzumab, or aNKp46/Fc/aCD20 or a combination of 0.1 μg/mL IL-2v/IC/Fc-null/IC + 0.1 μg/mL aNKp46/Fc/aCD20 molecules. RNA sequencing was performed, and reads were aligned with the human genome, N = 4 to 6 donors.
(A) Volcano plot representing the results of the analysis of differential expression between NK cells activated with IL-2v/aNKp46/Fc/aCD20 and those treated with a mixture of IL-2v/IC/Fc-null/IC + aNKp46/Fc/aCD20 molecules. In green: differentially expressed genes (FDR = 0.05); in gray: genes not differentially expressed.
(B) Gene sets displaying significant enrichment in NK cells stimulated with IL-2v/aNKp46/Fc/aCD20 relative to cells stimulated with a combination of the trispecific aNKp46/Fc/aCD20 and the IL-2v polypeptide, based on GSEA.
(C) Venn diagram representing the number of genes upregulated in NK cells following activation with the indicated molecules, relative to unstimulated conditions (FDR = 0.05).
(D) Heatmap representing the expression of a selection of differentially regulated genes.
(E) CD69 monitored by flow cytometry on purified human NK cells activated for 24 h in the presence of the murine huCD20-B16F10 target cells with 0.1 μg/mL IL-2v/aNKp46/Fc/aCD20, IL-2v/IC/Fc-null/IC, Obinutuzumab, or aNKp46/Fc/aCD20 or a combination of equal doses of IL-2v/IC/Fc-null/IC + aNKp46/Fc/aCD20. Picture is a boxplot obtained with NK cells from four donors. Whiskers show minimal and maximal value. Symbols are conserved for each donor across the conditions of stimulation.
(F) Gene set score for CD56dimCD16a+CD57+ NK1 cells and CIML NK cells regulated by each of the indicated condition of stimulation.
Several NK cell subsets have been reported in humans and mice.17 To analyze whether ANKET stimulation may expand specific NK cell subsets, we built a catalog of NK cell gene sets representative of each NK cell subset described in the literature.17, 18, 19 Gene sets identify the well-documented populations CD56dimCD16a+CD57− or CD57+ (aka NK1) and the CD56brightCD16a− (aka NK2) NK cells, as well as other more recently described subsets of type I IFN-responding NK cells, a small NK cell population with low ribosomal expression, subsets of cytokine-induced memory-like (CIML) phenotype, and adaptive NK cells. Stimulation of purified NK cells with trispecific ANKET (aNKp46/Fc/aCD20) and tetraspecific ANKET (IL-2v/aNKp46/Fc/aCD20) modified the composition of the total NK cell population. ANKET stimulation induced a downregulation of the metagene signature identifying the NK1 CD56dimCD16a+CD57+ NK cell subset and an enrichment in the metagene signature identifying the CIML NK cell subset (Figure 3F). CIML NK cells represent a long-lived NK cell subset exhibiting enhanced NK cell effector functions with a high potential for cancer immunotherapy. Altogether, these data show that tetraspecific ANKET stimulations induce a transcriptomic reprogramming of NK cells driving the amplification of a cytokine-induced memory NK cell subset characterized by increased effector functions.
Induction of tumor immunity by ANKET
We then investigated the ability of ANKET to control tumor growth in vivo. We performed preclinical studies with a tetraspecific ANKET generated with a surrogate antimouse NKp46 antibody fragment, a Fc fragment of human IgG1, and an antibody fragment targeting the human CD20 antigen. The antimouse NKp46 antibody (29A1.4) binds to mouse NKp46 recombinant proteins with similar affinity as the antihuman NKp46 (NKp46-1) for human NKp46 (Figure S4A). Tetraspecific ANKET targeting mouse NKp46 induced potent cytotoxicity by mouse NK cells (Figure S4B). In contrast, the clinically approved anti-CD20 antibody Obinutuzumab showed lower killing capacities (Figure S4B). As already reported for trispecific ANKET,8 an analysis of tetraspecific ANKET levels in plasma after a single intravenous (i.v.) injection demonstrated that the molecule had an IgG1-like pharmacokinetic profile in CB17-SCID mice, providing long-term exposure in vivo (Figure S4C). We tested the tetraspecific ANKET in CD20+ Raji B cell lymphomas engrafted subcutaneously into immunocompromised mice lacking B and T cells but retaining NK cell activity. A single injection of the tetraspecific ANKET induced tumor regression and controlled tumor growth for up to 30 days after treatment, whereas the anti-CD20 mAb Obinutuzumab gave only modest control of tumor growth in this aggressive lymphoma model (Figure 4A). A single injection at the doses of 10 to 70 μg per animals of tetraspecific ANKET efficiently controlled tumor growth in this model (Figure S4D). These doses of tetraspecific ANKET induced a moderate, dose-dependent, and transient weight loss, demonstrating the existence of a safe therapeutic window (Figure S4E).
Figure 4.
In vivo characterization of the antitumor efficacy of ANKET-IL-2v
(A–C and E) Raji B cell lymphoma cells were subcutaneously engrafted in CB17 SCID mice.
(A) Efficacy of 70 μg IL-2v/aNKp46/Fc/aCD20 or 1,500 μg Obinutuzumab injected i.v. once, 9 days after tumor engraftment.
(B) Efficacy of 25 μg IL-2v/aNKp46/Fc/aCD20 or 25 μg IL-2v/aNKp46/Fc/IC injected i.v. once, 9 days after tumor engraftment.
(C) Efficacy of two weekly i.v. injections of 25 μg IL-2v/aNKp46/Fc/aCD20, IL-2v/IC/Fc null/aCD20, or IL-2v/IC/Fc/aCD20 molecules.
(D) Immunostaining for human NKp46 and Gzmb on sections of Raji tumors grown subcutaneously in RAG1ko huNKp46Tg mice, 3 days after treatment with 25 μg IL-2v/aNKp46/Fc/aCD20 or vehicle. The images shown are representative of n = 5 tumors.
(E) Efficacy of a single i.v. injection of 25 μg IL-2v/aNKp46/Fc/aCD20 in the context of NK cell depletion with anti-Asialo-GM1. Dotted vertical lines represent the day of treatment. Each curve represents data from a single mouse. The data shown are representative of at least two independent experiments. The statistical analysis is described in the STAR methods, ∗p < 0.05; ∗∗p < 0.01; ∗∗∗p < 0.001; ∗∗∗∗p < 0.0001.
The in vivo efficacy of tetraspecific ANKET required the engagement of NK cells at the tumor site, as an ANKET variant lacking CD20 binding to tumor cells (IL-2v/aNKp46/Fc/IC) was unable to control tumor growth effectively (Figure 4B). Furthermore, the IL-2v/IC/Fc-null/aCD20 molecule, which cannot bind NKp46 and CD16a, and the IL-2v/IC/Fc/aCD20 molecule, which cannot bind NKp46 but retains CD16a-binding capacity, were less effective than the tetraspecific ANKET for controlling tumor growth (Figure 4C). Tumor engagement and binding to NKp46 on NK cells are, therefore, required to induce full antitumor activity.
Consistent with the in vitro data showing that tetraspecific ANKET promoted NK cell proliferation, flow cytometry analysis revealed that tetraspecific ANKET treatment in Raji tumor-bearing mice induced an increase in NK cell number in the spleen and an increase in the percentage of spleen NK cells expressing the CD69 early activation marker and the KI-67 proliferation marker, in contrast to treatments with Obinutuzumab or the trispecific aNKp46/Fc/aCD20 molecule (Figure S5A). Thus, the IL-2v moiety on tetraspecific ANKET was able to expand the population of NK cells outside of the tumor microenvironment, generating a pool of NK cells that could potentially contribute to antitumor activity.
We then investigated whether tetraspecific ANKET treatment increased NK cell infiltration in tumors. We used RAGko huNKp46Tg mice, in which NK cells express both human and mouse NKp46, making it possible to detect NK cells in tissue sections via NKp46 staining and to avoid interference with the antimouse NKp46 mAb fragment present in the therapeutic ANKET. In the Raji B lymphoma model engrafted subcutaneously, the immunostaining of human NKp46 revealed a strong infiltration of NKp46+ cells into the tumor microenvironment, associated with an increase in Gzmb expression after treatment with the tetraspecific ANKET (Figure 4D). Tetraspecific ANKET also induced higher levels of Ncr1 transcripts (encoding NKp46) than Obinutuzumab or the trispecific aNKp46/Fc/aCD20 molecule at the tumor bed (Figure S5B), confirming the infiltration of NK cells into tumors. In parallel, tetraspecific ANKET promoted the upregulation, in tumors, of Ifn-γ and Gzmb transcripts, indicating an enhancement of the effector activity of NK cells at the tumor site (Figure S5B). Ncr1 transcript levels were also increased by an ANKET-IL-2v lacking a CD20-binding element for binding to tumor cells (IL-2v/aNKp46/Fc/IC). Tumor targeting by IL-2v-bearing ANKETs is not, therefore, responsible for the accumulation of NK cells at the tumor site (Figure S5C). However, the absence of tumor engagement abolished the capacity of tetraspecific ANKETs to upregulate Ifn-γ and Gzmb transcript levels (Figure S5C), and the absence of NKp46 engagement limited Gzmb expression at the tumor site (Figure S5D). Thus, IL-2v-bearing ANKETs increase the number of NK cells at the tumor site even in the absence of tumor recognition, but only ANKET molecules engaging a tumor antigen on tumor cells and NKp46 on NK cells induce strong NK cell effector functions. NK cell depletion in IL-2v/aNKp46/Fc/aCD20-treated tumor-bearing mice abolished tumor control, demonstrating the requirement for NK cells to control tumors following treatment with the tetraspecific ANKET (Figure 4E).
Tetraspecific ANKET promoted the expansion and activation of NK cells in the spleen of immunocompetent mice (Figure 5A) and also affected CD8 T cell number, thereby increasing the CD8/CD4 T cell ratio without modulating the Treg compartment (Figure S6A). Tetraspecific ANKET controlled tumors very effectively in melanoma models based on B16F10 tumor cells expressing the human CD20 antigen injected i.v. or engrafted subcutaneously (Figures 5B and 5C), confirming its potent antitumor activity.
Figure 5.
In vivo activity of ANKET-IL-2v in immunocompetent mice
(A) NK cell number and percentage of CD69+ NK cells in spleen of huCD20-B16F10 tumor bearing C57BL6 mice treated with 25 μg tetraspecific ANKET (IL2v/aNKp46/Fc/aCD20) i.v. injection for 4 days. Picture is a boxplot obtained with NK cells from five mice. Whiskers show minimal and maximal value.
(B) Efficacy of a single i.v. injection of 70 μg IL-2v/aNKp46/Fc/aCD20, 70 μg aNKp46/Fc/aCD20, or 600 μg obinutuzumab in the huCD20-B16F10 disseminated melanoma model in C57BL6 wild-type (WT) mice.
(C) Efficacy of i.v. injections performed at days 1, 9, and 16 of 25 μg IL-2v/aNKp46/Fc/aCD20 or 25 μg IL-2v/IC/Fc-null/aCD20 in the model of huCD20-B16F10 melanoma cells engrafted subcutaneously in C57BL6 μM mice. The data shown are representative of at least two independent experiments. The statistical analysis is described in the STAR methods, ∗p < 0.05; ∗∗p < 0.01; ∗∗∗p < 0.001.
Efficacy and safety of ANKET treatment in non-human primates
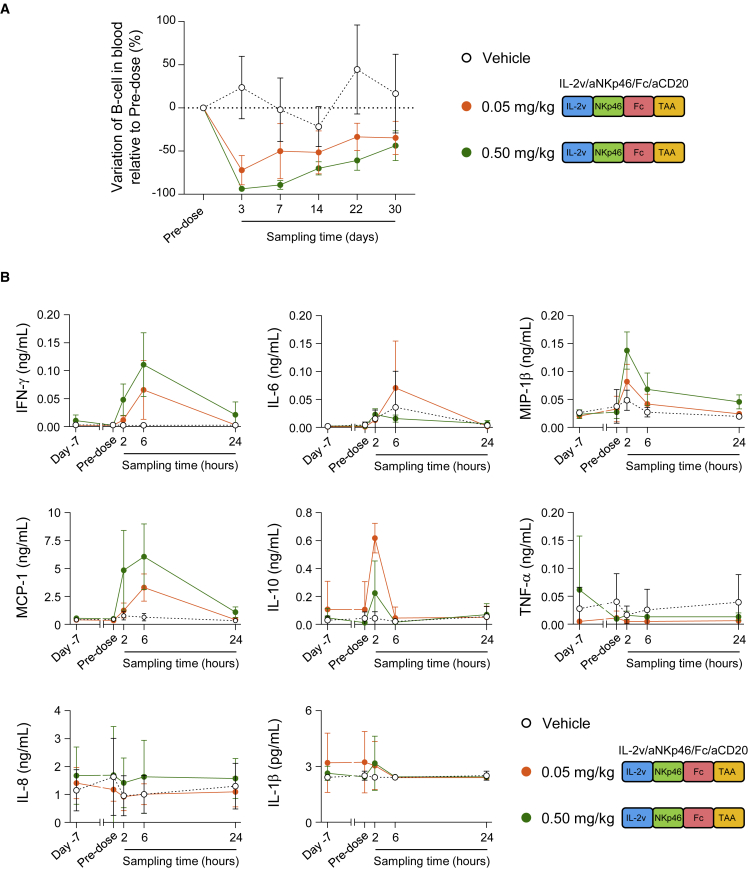
Improved efficacy of cancer treatments may be associated with an increase in toxicity, and IL-2-derived molecules have been reported to lead to adverse events in clinical practice that potentially limit their therapeutic use and benefit.20 We therefore evaluated the efficacy and toxicity of the tetraspecific ANKET in non-human primates. Human and cynomolgus FcγR interact with similar affinities to human IgG1 isotype antibodies.21,22 We performed the experiment with the tetraspecific ANKET generated with the antihuman NKp46 antibody NKp46-1,8 which crossbinds to cynomolgus NKp46 with a similar affinity (KD of about 18 nM) as for human (Figure S4A). Following treatment with a single dose of tetraspecific ANKET, no clinical signs of toxicity were detected in comparisons with control animals treated with vehicle alone (Figure S7A). A single injection of the CD20-targeting tetraspecific ANKET depleted circulating B cells in a dose-dependent manner, demonstrating the efficacy of this molecule for eliminating target cells (Figure 6A). Tetraspecific ANKET injection induced a minimal, rapid, and transient increase in plasma concentrations of various cytokines, including IFN-γ, IL-6, MIP-1β, MCP-1, and IL-10, with peaks at low levels within 2 or 6 h of the injection and a return to baseline values by 24 h (Figure 6B). No production of tumor necrosis factor (TNF), IL-8, or IL-1β was observed (Figure 6B). The pharmacodynamics and the absence of toxicity signals of the tetraspecific ANKET studied here suggest that this molecule is effective and safe at the doses tested, supporting its clinical development for next-generation cancer immunotherapies.
Figure 6.
Efficacy and safety of ANKET-IL-2v in non-human primates
(A) Counts of circulating B cells, expressed as a percentage of the baseline counts in NHPs receiving a single injection of vehicle (n = 6, black) or IL-2v/aNKp46/Fc/aCD20 at a dose of 0.05 (n = 4, orange) or 0.5 mg/kg (n = 4, green).
(B) Cytokine concentrations in the plasma of NHPs receiving a single injection of vehicle (n = 6, black) or IL-2v/aNKp46/Fc/aCD20 at a dose of 0.05 (n = 4, orange) or 0.5 mg/kg (n = 4, green).
Discussion
Antibody engineering is opening up tremendous opportunities to induce synthetic immunity, with the ultimate goal of designing innovative therapeutic agents of greater clinical efficacy. We described here the development of a fit-for-purpose antibody-based tetraspecific platform to harness the antitumor functions of NK cells, boosting their capacity to proliferate, to accumulate at the tumor site and to kill tumor cells. The tetraspecific ANKET was designed to induce synthetic immunity in several different ways: by limiting Treg activation through the addition to the ANKET of an IL-2v devoid of CD25 binding, by redirecting IL-2 activity specifically on NK cells through the cis-engagement of NKp46, CD16a, and IL-2Rβ by a single agonistic molecule, and by creating a specific bridge between NK and tumor cells that leads to targeted killing.
We have shown that the co-engagement of CD16a and NKp46 by trispecific ANKET induces NK cell functions more potently than the engagement of CD16a and NKp46 separately,8 consistent with data showing that optimal NK cell activation requires the triggering of multiple activating receptors on NK cells.6,7 We used IL-2v, which cannot bind CD25, to minimize the action of IL-2 on Tregs and to reduce toxicity,23,24 while inducing NK cell proliferation and enhancing the effector functions of NK cells. By incorporating IL-2v into a tetraspecific ANKET, we were able to retarget this molecule preferentially to NK cells, resulting in higher efficacy at very low doses compared with IL-2 used as a single agent. A dissection of the mechanisms underlying tetraspecific ANKET activity revealed an effect on NK cell responses different from that achieved following the treatment of NK cells with a combination of trispecific ANKETs inducing the ITAM pathway and IL-2v inducing the JAK3-STAT5 pathway. Tetraspecific ANKET stimulation induced a synthetic response poising NK cells for optimal antitumor functions by increasing the expression of molecules with direct cytotoxic activity, such as TNFSF10, PRF1, or GZMB, and by boosting the production of factors stimulating dendritic cells, including XCL1,25 FLT3LG,26 and IL-32,27 involved in the promotion of adaptive immune responses.
CIML NK cells are long-lived memory NK cells with enhanced effector functions.28 Our data showed that ANKET activation promotes CIML NK phenotype, in line with previous data reporting that CD16a engagement prior to IL-2 stimulation enhanced NK cell memory-like phenotype and cell reactivity.29 These key features of CIML NK cells represent a promising approach for immunotherapy, as illustrated by the adoptive transfer of CIML NK cells, which led to favorable clinical responses and remissions in a subset of patients with acute myeloid leukemia (AML).30
T cell harnessing in cancer therapies has been linked to severe toxicity. Such toxicity has been reported for T cell engagers, which target the CD3-T cell antigen receptor complex on T cells and tumor antigens on cancer cells,31,32 for the infusion of allogeneic T cells, which induces graft-versus-host disease, and for the infusion of CAR-T cells, which can lead to neurotoxicity or cytokine release syndrome.33 By contrast, the therapeutic manipulation of NK cells appears to be a safe approach, as none of the toxic effects associated with T cell therapies have been reported for NK cell-based therapies.3,34 Our data confirmed a good tolerability profile for tetraspecific ANKETs, as no off-target cytokine release or cytotoxicity was observed. However, even if the promotion of NK activity appears to be a safe approach, the toxicity linked to IL-2 therapy is a matter of concern. Indeed, high-dose IL-2 therapy is associated with a vascular leak syndrome leading to severe multiorgan toxicity.20 The tetraspecific ANKET was designed with an IL-2v mutated so as to abolish interaction with CD25, thereby preventing Treg activation. The abolition of binding to CD25 may also prevent the risk of toxicity mediated by cytokine release35 and the vascular leak syndrome induced by the direct activation of endothelial cells expressing CD25 and linked to pulmonary edema.23 Consistent with these findings, our study in non-human primates revealed a very good safety profile for tetraspecific ANKETs at the tested doses (i.e., up to 0.5 mg/kg in non-human primates [NHPs]), with no clinical signs of toxicity and a minimal release of inflammatory cytokines36 but with potent cytotoxicity resulting in a profound depletion of circulating B cells. Tetraspecific ANKETs are, therefore a promising tool for inducing the synthetic activation of NK cells by forcing crosstalks between stimulatory pathways and generating potent antitumor activity with a safe profile, supporting their clinical development as the next generation of off-the-shelf cancer immunotherapies.
Limitations of the study
The mechanisms linked to tetraspecific ANKET stimulation leading to an improved effector and proliferative phenotype in NK cells remain to be dissected further. By forcing crosstalks between the ITAM and JAK/STAT signaling pathways, the single agonistic tetraspecific ANKET may quantitatively enhance both the ITAM and the JAK3-STAT5 pathways and qualitatively modify NK cell responses through the selective impact (induction or silencing) of a set of genes. Cooperative crosstalks between distinct classes of cell-surface receptors have been reported to occur in natural conditions. Indeed, IL-15 plays an important role in priming NK cells for the cytotoxic responses induced by NKG2D,37 and IFN-γR requires FcγR signaling for the induction of a specific subset of antimicrobial functions in macrophages.38
In addition, there are limitations to evaluate the efficacy of molecules with a human IgG1 Fc domain in mice. Even if human CD16a and mouse CD16a bind to human IgG1 Fc within the same range of affinity,39,40 human IgG1 isotype antibodies poorly activate mouse CD16a.41 This feature may be explained in part by an inefficient complex formation between CD16a and the CD3ζ adaptor molecule, which dampens CD16a function and ADCC efficacy.41 In vivo activity in mouse models of human IgG1-based biologics may thus be underestimated. As previously reported in the literature42 and described here (Figure S1B), CD16a is often downmodulated on human NK cells that infiltrate tumors, leading to lower responses to cytotoxic antibodies.42 In contrast, the expression of NKp46 remains stable in patients with cancer (Figure S1B). If there are limits to interpret efficacy data for human IgG1-based molecules in mice, the strong antitumor activity of tetraspecific ANKETs in mouse models supports the advantage of targeting NKp46 in addition of CD16a to induce potent NK cell antitumor functions even in the context of CD16a dysfunction.
The current study demonstrated the strong capacity of tetraspecific ANKETs to induce the antitumor functions of NK cells. However, our transcriptomic analysis of NK cells stimulated by tetraspecific ANKETs revealed an upregulation of the expression of genes usually associated with a dysfunctional state such as KLRC1, HAVCR2, PVR, LAG-3,2 or CISH43 and SOCS family members.44 Further analysis will be required to evaluate whether various schemes of ANKET treatment may impact the expression of these genes and, hence, the efficacy of ANKETs.
Despite these limitations, tetraspecific ANKETs will make a valuable contribution to current efforts to harness NK cell antitumor functions through multispecific NK cell engagers.45 CD16a engagers are currently in clinical development, mostly for the treatment of hematological cancers.45, 46, 47 Bispecific killer cell engagers (BiKEs) engaging CD16a, and trispecific killer cell engagers (TriKEs) engaging CD16a and containing an IL-15 moiety, have also been developed to target antigens expressed on solid tumors.48,49 It will be of considerable interest to determine the potential value of these various NK cell engagers in terms of their manufacturability, safety, and efficacy across multiple cancer conditions.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Anti-human CD3 BUV395 | Becton Dickinson | Cat# 563546, RRID:AB_2744387 |
| Anti-human CD3 PB | Becton Dickinson | Cat# 558117, RRID:AB_397038 |
| Anti-human CD3 FITC | Becton Dickinson | Cat# 555332, RRID:AB_395739 |
| Anti-human CD3 BUV496 | Becton Dickinson | Cat# 612940, RRID:AB_2870222 |
| Anti-human CD4 BUV496 | Becton Dickinson | Cat# 612936, RRID:AB_2870220 |
| Anti-human CD7 FITC | Becton Dickinson | Cat# 555360, RRID:AB_395763 |
| Anti-human CD8 BUV737 | Becton Dickinson | Cat# 612754, RRID:AB_2870085 |
| Anti-human CD14 BUV737 | Becton Dickinson | Cat# 612763, RRID:AB_2870094 |
| Anti-human CD25 PE | Becton Dickinson | Cat# 341011, RRID:AB_2783790 |
| Anti-human CD25 PECy7 | Biolegend | Cat# 356108, RRID:AB_2561975 |
| Anti-human Foxp3 AF647 | Becton Dickinson | Cat# 560045, RRID:AB_1645411 |
| Anti-human pSTAT5 AF488 | Becton Dickinson | Cat# 612598, RRID:AB_399881 |
| Anti-human CD33 PECF594 | Becton Dickinson | Cat# 562492, RRID:AB_2713912 |
| Anti-human CD45 BV711 | Becton Dickinson | Cat# 564357, RRID:AB_2744404 |
| Anti-human CD56 APC | Miltenyi | RRID:AB_2658734 |
| Anti-human CD56 PEVio770 | Miltenyi Biotec | Cat# 130-113-313, RRID:AB_2726091 |
| Anti-human CD56 BV786 | Becton Dickinson | Cat# 564058, RRID:AB_2738569 |
| Anti-human NKp46 APC | Becton Dickinson | Cat# 558051, RRID:AB_398653 |
| Anti-human NKp46 FITC | Biolegend | Cat# 331922, RRID:AB_2561965 |
| Anti-human NKp46 PE | Becton Dickinson | Cat# 557991, RRID:AB_396974 |
| Anti-human CD69 APCCy7 | Becton Dickinson | Cat# 557756, RRID:AB_396862 |
| Anti-human CD69 FITC | Miltenyi Biotec | Cat# 130-113-523, RRID:AB_2733656 |
| Anti-human CD137 PEVio770 | Miltenyi Biotec | Cat# 130-119-885, RRID:AB_2783944 |
| Anti-human CD107a APC | Miltenyi Biotec | Cat# 130-119-869, RRID:AB_2751898 |
| Anti-human CD107b APC | Miltenyi Biotec | Cat# 130-103-993, RRID:AB_2654503 |
| Anti-human CD16 BUV395 | Becton Dickinson | RRID:AB_2744293 |
| Anti-human CD32 APC | Becton Dickinson | Cat# 559769, RRID:AB_398665 |
| Anti-human CD132 BV421 | Becton Dickinson | Cat# 562881, RRID:AB_2737862 |
| Anti-human CD132 PE | Becton Dickinson | Cat# 555900, RRID:AB_396211 |
| Anti-human CD19 BUV395 | Becton Dickinson | Cat# 563549, RRID:AB_2738272 |
| Anti-human CD20 PE | Becton Dickinson | Cat# 555623, RRID:AB_395989 |
| Anti-human CD20 FITC | Becton Dickinson | Cat# 556632, RRID:AB_396501 |
| Cell Trace Violet | Invitrogen | Cat#C34557 |
| Anti-human IFN-γ BV605 | Biolegend | Cat# 502536, RRID:AB_2563881 |
| Anti-human MIP-1β PE | Becton Dickinson | Cat# 550078, RRID:AB_393549 |
| Anti-human NKG2D PE | Beckman Coulter | Cat# A08934, RRID:AB_2801262 |
| Anti-human NKp30 APC | Miltenyi Biotec | Cat# 130-121-995, RRID:AB_2784148 |
| Anti-mouse CD11b V500 | Becton Dickinson | Cat# 562127, RRID:AB_10893815 |
| Anti-mouse CD11b FITC | Beckman Coulter | Cat# IM0530U, RRID:AB_130987 |
| Anti-mouse CD49b PE | Becton Dickinson | Cat# 553858, RRID:AB_395094 |
| Anti-mouse CD49b FITC | Becton Dickinson | Cat# 553857, RRID:AB_395093 |
| Anti-mouse CD25 BV605 | Becton Dickinson | Cat# 563061, RRID:AB_2737982 |
| Anti-mouse CD69 PB | Biolegend | Cat# 104524, RRID:AB_2074979 |
| Anti-mouse CD69 PE | Biolegend | Cat# 104508, RRID:AB_313111 |
| Anti-mouse NKp46 BV605 | Becton Dickinson | Cat# 564069, RRID:AB_2738575 |
| Anti-mouse CD3 BV650 | Biolegend | Cat# 100229, RRID:AB_11204249 |
| Polyclonal anti-asialo-GM1 | Biolegend | Cat#Poly21460 |
| Anti-KI67 BUV395 | Becton Dickinson | Cat# 564071, RRID:AB_2738577 |
| Anti-human CD3 BB700 | Becton Dickinson | Cat# 566517, RRID:AB_2744378 |
| Anti-human CD4 BV786 | Becton Dickinson | Cat# 563877, RRID:AB_2738462 |
| Anti-human CD8 BV510 | Becton Dickinson | Cat# 563256, RRID:AB_2738101 |
| Anti-human CD14 BV650 | Becton Dickinson | Cat# 563419, RRID:AB_2744286 |
| Anti-human CD16 BUV737 | Becton Dickinson | Cat# 612786, RRID:AB_2833077 |
| Anti-human CD19 A700 | Becton Dickinson | Cat#B49212 |
| Anti-human CD45 FITC | Becton Dickinson | Cat# 557803, RRID:AB_396879 |
| Anti-human NKG2A APC | Miltenyi Biotec | Cat# 130-113-563, RRID:AB_2726170 |
| Obinutuzumab | Roche | Gazyva/Gazyvaro |
| RGS·His antibody, BSA-free | Qiagen | Cat# 34670, RRID:AB_2571551 |
| Goat anti-human APC | Jackson Immunoresearch | Cat# 109-136-088, RRID:AB_2337691 |
| Cytokine and chemokine multiplex assay kit | Merck Millipore | Cat#PRCYTOMAG-40K |
| NKp46 antibody (clone 8E5B) | Innate Pharma | N/A |
| Anti-granzyme B (clone D6E9W) | Cell Signaling Technologies | Cat# 46890, RRID:AB_2799313 |
| Goat anti-Human IgG-Fc Fragment | Bethyl Laboratories | Cat# A80-104A, RRID:AB_67061 |
| Goat anti-Human IgG-Fc fragment antibody HRP conjugated | Bethyl Laboratories | Cat# A80-104P, RRID:AB_67064 |
| ANKET-IL-2v | Innate Pharma | In this paper |
| ANKET | Innate Pharma | In this paper |
| Biological samples | ||
| Human whole blood | EFS Marseille | N/A |
| Experimental models: Cell lines | ||
| RAJI cell ine | ATCC | Cat# CCL-86 |
| B16F10 cell line | ATCC | Cat# CRL-6475 |
| EXPI-293F cell line | ThermoFisher Scientific | Cat# A14527 |
| Experimental models: Organisms/strains | ||
| CB-17 SCID mice | Janvier Laboratories | Cat# CB-17/Icr-Prkdcscid/scid/Rj |
| C57Bl/6J mice | Janvier Laboratories | Cat# C57BL/6JRj |
| μMT (C57BL/6J) mice | Jackson Laboratories | Cat# 002288 |
| Cynomolgus macaque | IDMIT | N/A |
| Chemicals, peptides, and recombinant proteins | ||
| His-tagged CD25 | Sino Biological | Cat# 10165-H08H |
| His-tagged CD122 | Sino Biological | Cat# 10696-H08B |
| His-tagged CD132 | Sino Biological | Cat# 10555-H08B |
| RPMI 1640 medium | Gibco | Cat# 31870-025 |
| Fetal bovine serum (FBS) | Gibco | Cat# 10270-106 |
| L-glutamine | Gibco | Cat# 25030-024 |
| Non-essential amino acids solution (NEAA) | Gibco | Cat# 11140-035 |
| Sodium pyruvate | Gibco | Cat# 11360070 |
| HindIII restriction enzyme | Thermo Fisher Scientific | Cat# FD0504 |
| XbaI restriction enzyme | Thermo Fisher Scientific | Cat# FD0684 |
| Matrigel Basement Membrane Matrix | Corning | Cat# 354234 |
| Normal rabbit serum | Abcam | Cat# Ab7487 |
| BD CellFIX | Becton Dickinson | Cat# 340181 |
| BD Cytofix/Cytoperm | Becton Dickinson | Cat# 554722 |
| BD Perm/Wash | Becton Dickinson | Cat# 554723 |
| BD CytoFix | Beckton Dickinson | Cat# 554665 |
| Optilyse C | Beckman Coulter | Cat# A11895 |
| EXPI293 expression medium | Gibco | Cat# A14351-01 |
| PEI max | Polysciences Inc | Cat# 24765 |
| Ficoll Paque Plus | GE healthcare | Cat# 17-1440-03 |
| Hygromycin B | Gibco | Cat# 10687010 |
| Valproic acid sodium salt | Sigma | Cat# P4543-10G |
| D(+) glucose monohydrate | Fluka | Cat# 49161 |
| Tryptone N1 | Organo Technie | Cat# 19553 |
| DNAse1 | Roche | Cat# 11284932001 |
| Collagenase I | Pan-Biotech | Cat# LS0004196 |
| Collagenase IV | Pan-Biotech | Cat# LS0004188 |
| Live/Dead near infrared | Thermo Fisher Scientific | Cat# L34976 |
| Dulbecco's phosphate-buffered saline (DPBS) | Gibco | Cat# 14190-094 |
| Bovine serum albumin (BSA) | Sigma Aldrich | Cat# A9418 |
| Sodium azide | Sigma Aldrich | Cat# 71290 |
| EDTA | Invitrogen | Cat# 15575-038 |
| Versalyse | Beckman Coulter | Cat# A09777 |
| TruCount tubes | Becton Dickinson | Cat# 340334 |
| GolgiStop | Becton Dickinson | Cat# 554724 |
| Calcein | Life Technologies | Cat# C3100MP |
| Triton X-100 | Sigma-Aldrich | Cat# 93443-100ML |
| Chromium-51 radionuclide | Perkin Elmer | Cat# NEZ030002MC |
| Formalin | BiopSafe | Cat# 3178-200-19 |
| ER1 Epitope Retrieval Solution pH 6.0 | Leica Biosystems | Cat# AR9961 |
| ER2 Epitope Retrieval solution pH 9 | Leica Biosystems | Cat# AR9640 |
| BOND Polymer Refine Detection kit | Leica Biosystems | Cat# DS9800 |
| StartingBlock blocking buffer | Thermo Fisher Scientific | Cat# 37542 |
| RNAprotect Tissue Reagent | Qiagen | Cat# 76106 |
| Lysing matrix A tubes | MP Biomedicals | Cat# 6910050 |
| Taqman universal master mix II | Thermo Fisher Scientific | Cat# 4440040 |
| Critical commercial assays | ||
| NK Cell Isolation Kit, human | MACS Miltenyi Biotec | Cat# 130-092-657 |
| Maxima First-Strand cDNA synthesis kit | Thermo Fisher Scientific | Cat# K1642 |
| RNeasy minikit | Qiagen | Cat# 74106 |
| RNeasy micro kit | Qiagen | Cat# 74004 |
| Qubit RNA HS Assay kit | Thermo Fisher Scientific | Cat# Q32855 |
| KAPA RNA HyperPrep kit | Roche | Cat# 08098115702 |
| Qubit 1X dsDNA HS Assay kit | Thermo Fisher Scientific | Cat# 15860210 |
| Oligonucleotides | ||
| Taqman primers Gapdh | Thermo Fisher Scientific | Mm99999915_g1 |
| Taqman primers Ncr1 | Thermo Fisher Scientific | Mm01337324_g1 |
| Taqman primers Ifn-γ | Thermo Fisher Scientific | Mm01168134_m1 |
| Taqman primers Gzmb | Thermo Fisher Scientific | Mm00442834_m1 |
| Others | ||
| LumaPlate-96 | Perkin Elmer | Cat# 6006633 |
| SuperFrost Plus™ Adhesion glass slides | Thermo Fisher Scientific | Cat# J1800AMNZ |
| Deposited data | ||
| Raw data files for RNAseq | This work | GEO accession number: GSE214378 (https://www.ncbi.nlm.nih.gov/geo/) |
| Software and algorithms | ||
| FlowJo v.10.5.2 | BD Biosciences | https://www.flowjo.com/solutions/flowjo/downloads/previous-versions |
| xPONENT 4.2 | Luminex | https://www.luminexcorp.com/xponent/#overview |
| GraphPad Prism, version 8.1.1 | GraphPad | https://www.graphpad.com/scientific-software/prism/ |
| R v4.0.3 | R Core Team, 2020 | https://www.r-project.org/ |
| R package: DESeq2 v1.30.0 | Love, Huber and Anders, 2014 | https://bioconductor.org/packages/release/bioc/html/DESeq2.html |
| R package: fgsea v1.16.0 | Korotkevich, Sukhov and Sergushichev, 2019 | https://bioconductor.org/packages/release/bioc/html/fgsea.html |
| R package: pheatmap v1.0.12 | Kolde, 2019 | https://cran.r-project.org/web/packages/pheatmap/index.html |
| R package: ggVennDiagram v1.1.0 | Gao, Yu, and Cai, 2021 | https://cran.r-project.org/web/packages/ggVennDiagram/index.html |
| R package: ggplot2 v3.3.5 | Wickham, 2016 | https://cran.r-project.org/web/packages/ggplot2 |
| R package: edgeR v3.32.1 | Robinson, McCarthy and Smyth, 2010 | https://bioconductor.org/packages/release/bioc/html/edgeR.html |
| R package: WebGestaltR v0.4.4 | Liao et al., 2019 | https://cran.r-project.org/web/packages/WebGestaltR/ |
| Picard software suite v2.18.29 | Broad Institute 2019 | https://broadinstitute.github.io/picard/ |
| STAR v2.7.0 | Dobin et al.50 2013 | https://github.com/alexdobin/STAR |
| Subread v1.6.4 | Liao, Smyth and Shi, 2013 | http://subread.sourceforge.net/ |
| FastQC v0.11.8 | Andrews, 2010 | https://www.bioinformatics.babraham.ac.uk/projects/fastqc/ |
| Trimmomatic v0.36 | Bolger, Lohse, Usadel, 2014 | http://www.usadellab.org/cms/?page=trimmomatic |
| Qualimap v2.2.1 | Okonechnikov, Conesa and García-Alcalde, 2015 | http://qualimap.conesalab.org/ |
| SAMtools v1.10 | Li et al., 2009 | http://samtools.sourceforge.net/ |
| MultiQC v1.7.dev0 | Ewels et al., 2016 | https://multiqc.info/ |
Resource availability
Lead contact
Further information and requests for reagents and resources should be directed to and will be fulfilled by the lead contact, Prof Eric Vivier (vivier@ciml.univ-mrs.fr).
Material availability
The molecular organization of the ANKET used in the present study can be found in patent PCT/EP2022/057824. Requests for new materials generated in this paper are to be directed to and will be fulfilled (pending MTA and associated restrictions) by the lead contact (vivier@ciml.univ-mrs.fr).
Experimental model and subject details
Animals
Mice
Female CB17 SCID and C57Bl/6J mice were purchased from Janvier Labs. μMT (C57BL/6J) female mice51 were purchased from the Jackson Laboratory (JAX stock #002288). HuNKp46 tg mice, which have been described elsewhere,52 were backcrossed onto a RAG1−/− background, reared at UMS-CIPHE (Marseille, France) and kept under specific and opportunistic pathogen-free conditions. All mice were used at ages of 7 to 17 weeks. All mouse experiments were performed in accordance with the rules of the Innate Pharma ethics committee and were approved by the Ministère de l’Enseignement Supérieur, de la Recherche et de l’Innovation – France (APAFIS# 19272).
Non-human primates
We used two- to four-year-old Mauritian cynomolgus macaques (Macaca fascicularis). None of the animals used had undergone any previous experimental procedure. The macaques were housed in social groups within the IDMIT animal facility at the CEA, Fontenay-aux-Roses, France. All experimental procedures were performed in accordance with European guidelines for animal care and use for scientific purposes (Directive 63-2010, “Journal Officiel des Communautés Européennes”, L276, September 22, 2010) and CEA institutional guidelines. The study was approved by the local ethics committee under number A18_080 and by the French administration (APAFIS#20525-2019050616506478 v1).
Human primary cells
Buffy coats from healthy donors were obtained from Etablissement Francais du Sang (EFS, Marseille) with the written consent of each volunteer blood donor. PBMCs were isolated from buffy coats by centrifugation on Ficoll (GE Healthcare) density gradients. NK cells were purified from PBMCs by negative magnetic selection, with a human NK cell isolation kit (MACS-Miltenyi Biotec), according to the manufacturer’s instructions.
Human tumor tissues were provided by the Assistance des Hopitaux Publics de Marseille (APHM), via the BIO-003-APHM study. All of the patients provided written informed consent before sampling and for the use of their clinical and biological data.
Cell lines
The Raji human B-cell lymphoma and B16F10 murine melanoma cell lines were obtained from the ATCC. B16F10 cells were transfected to express the human CD20 antigen.
Generation of the human CD20 B16-F10 cell line: The human CD20 sequence (NP_068769.2) was inserted into a SLX expression vector between the HindIII and XbaI restriction sites. After sequencing, the vector was used to transfect the B16-F10 cell line with AMAXA Nucleofector 4D technology. The cells were cultured under selection pressure with 50 μg/mL hygromycin and the positive cells, stained with an anti-CD20 FITC antibody, were sorted.
All the cell lines were cultured in RPMI 1640 medium (Gibco) supplemented with 10% heat-inactivated fetal bovine serum (FBS, Gibco), 2 mM L-glutamine (Gibco), 1% non-essential amino acids (Gibco) and 1 mM sodium pyruvate (Gibco) and maintained at 37°C under an atmosphere containing 5% CO2.
Method details
Multispecific molecule production and purification
The sequences encoding the different fragments of each multispecific molecule were inserted into the pTT-5 vector between the HindIII and BamHI restriction sites. Expression vectors (prepared as endotoxin-free midipreps) were used to cotransfect EXPI-293F cells (Life Technologies) in the presence of PEI (37°C, 5% CO2, 150 rpm). The cells were used to seed culture flasks at a density of 1 x 106 cells per mL and were cultured in EXPI293 medium (Gibco) supplemented with valproic acid (final concentration 0.5 mM), glucose (4 g/L) and tryptone N1 (0.5%). The supernatants were harvested after six days and passed through a Stericup filter with 0.22 μm pores. Multispecific molecules were purified with Protein A beads (250 μL/50 mL SN), eluted with 0.1 M sodium citrate buffer at pH 3 and immediately neutralized with 1 M Tris pH 8. The proteins were then dialyzed overnight against 1 x PBS at 4°C and concentrated to 10 mg/mL before loading on an S200 Increase 10/300 column (AKTA, 500 μL injection loop). The proteins yielding a peak at the expected size were harvested and analyzed by analytical SEC and SDS-PAGE under reducing and non-reducing conditions, with Coomassie brilliant blue staining. Alternatively, ANKET were dialyzed against 25 mM Phosphate buffer pH 6.2 and purified by ion-exchange chromatography on MonoS 4.6/100 PE column (GE Healthcare). All the purified molecules were stored in 1X PBS and analyzed to check for the absence of aggregates and endotoxins.
Surface plasmon resonance (SPR) experiments
SPR measurements were performed on a Biacore T200 apparatus (Biacore GE Healthcare) at 25°C. For IL-2R binding study, the anti-His antibody (Qiagen, 34670) was immobilized by covalent attachment to carboxyl groups on the surface of CM5 Sensor Chips (GE Healthcare). The chip surface was activated by incubation with 1-ethyl-3-(3-dimethylaminopropyl) carbodiimide (EDC)/N-hydroxysuccinimide (NHS) (Biacore GE Healthcare). Anti-His antibody was diluted to 10 μg/mL in coupling buffer (10 mM acetate, pH 5.0) and injected until the appropriate level of immobilization was achieved (i.e. approximately 1000 response units). The remaining activated groups were deactivated with 100 mM ethanolamine (pH 8) (Biacore GE Healthcare). Binding studies were performed by capturing His-tagged antigens (CD25, CD122 or CD132) (Sino Biological, 10165-H08H, 10696-H08B and 10555-H08B, respectively) on the chip surface at 15 μg/mL. ANKET were diluted to 162.5 μg/mL and injected for 120 s at a rate of 10 μL/min. Anti-huCD25-PE/Cy7 (BioLegend, 356108) antibody was diluted to 10 μg/mL and injected for 120 s at a rate of 10 μL/min. Binding studies were performed by capturing CD122-His on the chip surface at 15 μg/mL. NKCE was diluted from 2000 nM to 62.5 nM and injected for 240 s at 40 μL/min. The dissociation time was set to 600 s. The entire data set was fitted with the steady state affinity model and KD calculated from the Scatchard plot. For NKp46 binding study, anti-human NKp46 antibody NKp46-1 and anti-mouse NKp46 antibody 29A1.48,51 were diluted to 5 μg/ml in running buffer and injected onto protein-A chips until the appropriate level of capture was achieved (i.e., 80 to 100 response units). Serial dilutions of soluble human, cynomolgus (7.8 to 250 nM) and mouse NKp46 recombinant proteins (31.25 to 1,000 nM) were injected over the captured antibodies and allowed to dissociate for 10 min before regeneration. The entire data sets were fitted with the kinetic 1:1 binding model after blanc subtraction.
Fresh tumor preparation and analysis
Fresh tumors were chopped with tweezers and scissors and incubated with enzymes: collagenase I (Pan-Biotech, LS0004196, 1 mg/mL final concentration), collagenase IV (Pan-Biotech, LS0004188, 1 mg/mL final concentration) and DNAse I (Roche, 11284932001, 20 μg/mL final concentration) on a MACSmix tube rotator (Miltenyi Biotec) for 45 minutes at 37 ± 1°C. Single-cell suspensions were obtained by filtering the cells through a cell strainer with a 70 μm mesh. FcR was then blocked by incubation with 10% human serum diluted in PBS (15 min at +5 ± 3°C), before staining with Live/Dead near infrared (Invitrogen, #L34976, 1/1000 in PBS, 15 min at +5 ± 3°C). Samples were then washed and incubated with the antibody mixture for 30 minutes, treated with Optilyse C (Beckman Coulter) at room temperature to lyse the red blood cells, washed in PBS, incubated in CellFix (Becton Dickinson, 340181, 1/10 in H2O) to fix the cells, before washing and resuspension immediately followed by flow cytometry.
Flow cytometry
Immune cell phenotyping
Cells were incubated with mouse serum to saturate the Fc receptors, and then with the appropriate antibody cocktail diluted in staining buffer: 1XPBS (Gibco), 0.2% BSA (Sigma), 0.02% sodium azide (Prolabo) and 2 mM EDTA (Invitrogen Life Technologies). Red blood cells were lysed in Optilyse C Solution (Beckman Coulter) according to the manufacturer’s instructions. Cells were fixed in Cell Fix solution (BD) according to the manufacturer’s instructions. Data were acquired in an LSRFortessaX20 flow cytometer. The FCS3.0 files obtained were exported from BD FACSDiva software and imported into FlowJo v.10.5.2 (BD Biosciences). Automatic compensation was calculated with FACSDiva software and single-stained compensation beads. This compensation matrix was analyzed in detail in FlowJo, with the N-by-N view feature and by investigating the pairwise expression of all proteins stained in this study. Fluorescence minus one (FMO) experiments were run before this study, to facilitate optimization of the compensation matrix. We then adjusted the compensation matrix where necessary due to over or undercompensation by the automatic algorithm.
STAT5 phosphorylation
PBMCs were used to seed 96-well plates and were treated with test molecules for 20 min at 37°C. After incubation, the cells were fixed immediately with FixPerm Buffer (BD) and incubated for 50 minutes on ice before permeabilization by incubation with Perm buffer III (BD) for 20 minutes at 4°C. Cells were then stained with anti-phospho-STAT5 (BD, 612598) and gating antibodies for 60 minutes at 4°C before flow cytometry analysis.
NHP cell analysis
Whole blood from cynomolgus monkeys was collected into heparinized tubes and stored at +5°C for processing within 24 h. Whole-blood samples were washed in PBS and then stained by incubation for 15 minutes with Live/Dead near IR (Thermo Fisher Scientific, L34976; prepared at 1:250 in PBS). Cells were washed in staining buffer incubated with the antibody mixture for 30 minutes and washed again with staining buffer. Red blood cells were then lysed by incubation with Versalyse (Beckman Coulter, A09777) for 15 minutes, before washing in staining buffer. Samples were fixed by incubation with CytoFix (Beckton Dickinson, 554665) for 15 minutes, before immediate acquisition on a BD LSR Fortessa X20 flow cytometer. Absolute cell counts were performed with TruCount tubes (Becton Dickinson, 340334).
Proliferation assay
NK cells isolated from PBMCs were labeled with Cell Trace Violet (CTV) dye (Invitrogen, C34557) and used to seed a 96-well plate. Cells were then incubated for five days in RPMI medium (10% FBS) supplemented with 10% AB serum (Sigma). Proliferation was analyzed by flow cytometry. Proliferating cells were identified as cells with diluted or negative CTV staining.
Intracellular cytokine assessment
Human NK cells, purified and cultured in RPMI 1640 medium without phenol red (Gibco) supplemented with 1% heat-inactivated fetal bovine serum (FBS, Gibco), 2 mM L-glutamine (Gibco), 1% non-essential amino acids (Gibco) and 1 mM sodium pyruvate (Gibco) and maintained at 37°C under an atmosphere containing 5% CO2, were co-incubated with Raji cells at an effector:target (E:T) ratio of 1:2 for 4 h at 37°C, in the presence of a range of doses of ANKET molecules. GolgiStop (BD) was added to the preparation to block intracellular protein transport. The cells were then washed once in staining buffer and stained with anti-CD3-Pacific Blue (BD) and CD56-Pe-Vio770 (Miltenyi Biotec) antibodies for 30 minutes at 4°C. Cells were washed twice, fixed and permeabilized with Cytofix/Cytoperm (BD). Cells were washed twice in Perm/Wash (BD) and stained by incubation with anti-MIP-1β-PE (BD) and anti-IFN-γ-BV605 (Biolegend) antibodies for 30 min at 4°C. Cells were washed twice and data were acquired on a LSRFortessa X-20 (BD) flow cytometer.
Cytotoxicity assay
Purified human NK cells were plated with Raji tumor cells loaded with Calcein (Life Technologies, C3100MP) at an E:T cell ratio of 10:1 in U-bottomed 96-well plates (BD Falcon, 353077). The cells were incubated for 4 h at 37°C in RPMI 1640 medium without phenol red (Gibco) supplemented with 1% heat-inactivated fetal bovine serum (FBS, Gibco), 2 mM L-glutamine (Gibco), 1% non-essential amino acids (Gibco) and 1 mM sodium pyruvate (Gibco) under an atmosphere containing 5% CO2. We then transferred 100 μL of the culture supernatant to a black flat-bottomed plate (Greinier Bio-one, 655096). Calcein release into the supernatant was measured with Enspire (Perkin Elmer) determinations of fluorescence emission at λ =516 nm after excitation at λ=495 nm. The following formula was used to calculate the percent specific lysis:
Maximal calcein release was determined by adding 2% Triton X-100 (Sigma-Aldrich, 93443-100ML) to the target cells, and spontaneous release was measured in medium alone, without effector cells.
Mouse NK cells were purified from spleen of CB17 SCID or C57BL6 mice and plated with Raji or huCD20-B16F10 tumor cells loaded with chromium-51 (51Cr) (PerkinElmer) at an effector:target (E:T) cell ratio of 10:1 in U-bottom 96-well plates (BD Falcon). Dose ranges of test molecules were added (in triplicate) and plates were incubated for 4 h at 37°C. After incubation, 50 μl of the culture supernatant was transferred to a LumaPlate (Perkin Elmer) coated with solid scintillator, which was then placed in a microplate scintillation counter (TopCount NXT, Perkin Elmer) to measure 51Cr release into the supernatant, which was correlated with target cell lysis. The following formula was used to calculate the percent specific lysis:
Specific lysis (%) = (experimental release – spontaneous release) / (maximal release − spontaneous release) ×100
Maximal calcein release was determined by adding 2% Triton X-100 (Sigma-Aldrich, 93443-100ML) to the target cells, and spontaneous release was measured in medium alone, without effector cells.
Binding assay
Purified human NK cells or Raji cells were incubated for 4 hours at 4°C with various doses of ANKET molecules in RPMI 1640 medium without phenol red (Gibco) supplemented with 1% heat-inactivated fetal bovine serum (FBS, Gibco), 2 mM L-glutamine (Gibco), 1% non-essential amino acids (Gibco) and 1 mM sodium pyruvate (Gibco). Cells were washed once in staining buffer and stained by incubation with a goat anti-human APC antibody (Jackson Immunoresearch, 109-136-088) for 20 minutes at 4°C. Cells were washed once and data were acquired on a LSRFortessa X-20 (BD) flow cytometer.
In vivo mouse tumor models
For the solid tumor models, 5×106 Raji cells in a 1:1 mixture of endotoxin-free PBS and Matrigel (Corning) were injected s.c. into the flank of CB-17 SCID recipient mice. Mice were randomized when tumor volume reached 50 -100 mm3. Treatment was initiated on day 9.
Alternatively, 5×104 huCD20-B16F10 cells in a 1:1 mixture of endotoxin-free PBS and Matrigel (Corning) were injected s.c. into the flank of μMt- recipient mice (C57BL/6 background). Treatment was initiated the day after implantation and repeated on days 9 and 16.
Tumor size was monitored with a digital caliper (Mitutoyo) every three to four days and is expressed as a volume ((length× width2) /2).
For NK cell depletion, 100 μL of polyclonal anti-asialo-GM1 (Biolegend, Poly21460) antibody was injected i.p. into recipient mice two days before treatment and then once a week. Normal rabbit serum was administered as a control.
For the disseminated tumor model, 5×105 huCD20-B16F10 cells were injected intravenously (i.v.) into the tail vein of C57BL/6 mice. The following day, treatments were initiated. Mice were observed daily, to monitor clinical signs. They were killed 13 days after tumor cell inoculation, and the lungs were analyzed for the presence of metastases.
NHP monitoring
Animals were observed daily, by trained staff, throughout the study period. For all procedures (handling, blood collection and treatment), animals were sedated by an intramuscular injection of ketamine (Imalgen 1000, 10 mg/kg) and medetomidine (Domitor 0.03 mg/kg). Antisedan was administrated to reverse anesthesia (atipamezole, 0.03 mg/kg). Body weight and temperature were followed throughout the study. Heart rate and oximetry data were recorded with a tensiometer (Vismo Bedside Monitor PVM-2701/PVM-2703, Nihon Kohden). Behavior scores were determined on the basis of stereotypy, the appearance of the fur, body posture, breath, hydration, interaction with the observer and clinical symptoms (diarrhea, vomiting, mydriasis). Behavior scores ranged from 0 to 27, as follows: 0, healthy animal; 1-7: healthy animal with moderate behavioral alterations; 8-12: animal with marked behavioral alterations; 13-20: animal displaying signs of suffering; 21-27: animal in severe distress.
NHP cytokine and chemokine assessment
Cytokines and chemokines including IFN-γ, IL-1β, IL-10, IL-6, IL-8, MCP-1, MIP-1β and TNF-α were measured in serum of cynomolgus monkeys with multiplex assay kit from MILLIPLEX® (#PRCYTOMAG-40K, Merck Millipore). Detection was performed using a magnetic Luminex instrument (MagPix). The results were analyzed using xPONENT 4.2® software (provided by Luminex with MagPix instrument) using a 5-parameters fitting.
Immunohistochemistry
The freshly harvested Rag-/- huNKp46Tg mouse tumor tissue was placed in vials containing 20 mL 10% formalin (3178-200-19, BiopSafe) and incubated for 24 hours for fixation, before being placed in 70% ethanol before tissue processing. Tissues were placed in standard histological cassettes and dehydrated with a STP120 Spin Tissue Processor (Thermo Fisher Scientific Microm) before being embedded in paraffin with a HistoStar™ Embedding Workstation (Thermo Fisher Scientific). We cut 4 μm-thick serial sections from each formalin-fixed paraffin-embedded (FFPE) block with a microtome (Epredia™ HM 340E, Thermo Fisher Scientific). The sections were deposited onto SuperFrost Plus™ Adhesion glass slides (J1800AMNZ; Thermo Fisher Scientific), and left to dry for 2 hours at 42°C in in a paraffin oven (UN110pa, Memmert).
Immunohistochemical (IHC) staining was performed on a Leica BOND RX automatic research stainer (Leica Biosystems). The sections were deparaffinized and rehydrated, and subjected to heat-induced antigen retrieval (HIER) by incubation in BOND ER1 Epitope Retrieval Solution pH 6.0 (for NKp46 ; AR9961, Leica Biosystems) or ER2 Epitope Retrieval solution pH 9 (for granzyme B; AR9640, Leica Biosystems) at 100°C for 20 minutes. Following peroxidase blockade for 5 minutes, the sections were incubated at room temperature with 1 μg/mL anti-NKp46 antibody (clone 8E5B, Innate Pharma) or 0.12 μg/mL anti-granzyme B antibody (clone D6E9W, Cell Signaling Technologies) diluted in primary antibody diluent (AR9352, Leica Biosystems) for 20 or 45 minutes, respectively. An anti-mouse/rabbit IgG DAB detection kit (BOND Polymer Refine Detection kit, Leica Biosystems) was used for amplification and primary antibody detection. The sections were counterstained with hematoxylin, washed, dehydrated, cleared, and finally mounted in Epredia™ ClearVue™ Mountant XYL (Thermo Fisher Scientific) with a Shandon Clearvue Coverslipper (Thermo Fisher Scientific). Slides were scanned with a NanoZoomer S60 digital slide scanner (Hamamatsu). Positive staining for each marker was assessed visually, taking staining intensity and localization into account. Representative snapshots of regions of interest (ROIs) were taken with NDPView.2 viewing software (Hamamatsu).
ANKET determination in mouse plasma
The concentration of ANKET in mouse plasma was evaluated with an optimized commercial sandwich ELISA kit (Bethyl Laboratories, A80-104A/A80-104P). Briefly, two polyclonal goat anti-human antibodies specific for the human IgG Fc fragment were used as capture and detection antibodies. The capture antibody was used to coat the plate at a concentration of 5.5 μg/mL and an HRP-conjugated antibody was used at a dilution of 1/50,000 for detection. All sample dilutions and blocking steps were performed in StartingBlock blocking buffer (Thermo Fisher Scientific, 37542). TMB was used as a peroxidase substrate. Once the reaction had been stopped by acidification, the intensity of the absorbance signal was read at 450 nm with Enspire (Perkin Elmer). The linear dynamic range assessed for this method extended from 20 to 5000 ng/mL for pure mouse plasma (mouse C57BL/6j@RJ plasma pool), with a minimum required dilution of 1/10.
Gene expression analysis
Tissues were stored in RNAprotect Tissue Reagent (Qiagen, 76106) at -80°C until processing. They were lysed with a FastPrep 5G (MP Biomedicals) in lysing matrix A tubes (MP Biomedicals, 6910050). mRNA was extracted with the RNeasy minikit (Qiagen, 74106), according to the manufacturer’s protocol, and quantified with Nanodrop spectrometer (Thermo Fisher Scientific). cDNA was synthesized from mRNA with the Maxima First-Strand cDNA synthesis kit for RT-qPCR (Thermo Fisher Scientific, K1642) according to the manufacturer’s protocol. qPCR reactions were run on a CFX Connect Real-Time machine (Bio-Rad) with Taqman universal master mix II (Thermo Fisher Scientific, 4440040) and Taqman primers (Thermo Fisher Scientific: Gapdh, Mm99999915_g1; Ncr1, Mm01337324_g1; Ifn-γ, Mm01168134_m1; Gzmb, Mm00442834_m1). Relative gene expression was quantified and expressed as 2-ΔCt, with Gapdh as an endogenous control.
Ribonucleic acid preparation for RNA sequencing
Total RNA was extracted with the RNeasy micro kit (Qiagen, 74004) according to the manufacturer’s instructions. RNA concentration was assessed with the Qubit RNA HS Assay kit (Thermo Fisher Scientific).
Library preparation and sequencing
RNA-Seq libraries were prepared from 80 ng to 200 ng of total RNA with the KAPA RNA HyperPrep kit (Roche, 08098115702). The preparation was enriched in mRNA with oligo dT magnetic beads and fragmented by incubation for 5.5 minutes at 94°C. Cleaved mRNA fragments were used as templates for double-stranded DNA synthesis followed by the addition of a polyA tail. The 8 bp UMI containing TruSeq Compatible Duplex Y adapters UDI-UMI (unique dual index - unique molecular identifier) from IDT were then ligated on either side of the cDNA. After bead-based purification, libraries were amplified by 10-12 cycles of PCR. Cleaned-up libraries were quantified with the Qubit 1X dsDNA HS Assay kit (Thermo Fisher Scientific, 15860210) and quality was assessed with TapeStation. Pooled libraries were sequenced over four high-output paired-end 71 bp runs on an Illumina NextSeq 500 instrument.
RNAseq analysis
Sequencing data containing unique molecular identifiers (UMIs) were demuliplexed according to IDT guidelines. Briefly, sample barcodes were extracted for every read in every sequencer lane, each lane of bcl files was demultiplexed and per-sample unaligned BAM including UMI data were created, BAM files were converted to FASTQ files, FASTQ reads were mapped and the sorted aligned BAM files were then merged with sorted unaligned BAM files with UMI data. Picard software suite v2.18.29 was used (Broad Institute 2019).
The reads were mapped onto the human genome (GRCh38) and the mouse genome (GRCm38) with STAR v2.7.0.50 PCR replicates were removed with the Picard UmiAwareMarkDuplicatesWithMateCigar program (Broad Institute 2019). Gene expression was assessed with featureCounts in SubReads package v1.6.4,53 using gene annotations from GENCODE (release 32 for the human genome and M23 for the mouse genome).
Differential expression analyses were performed with the R package DESeq2 version 1.30.0. The model included the donor and the treatment (unstimulated, IL-2v polypeptide, obinutuzumab, trispecific aNKp46/Fc/aCD20, combination of trispecific aNKp46/Fc/aCD20 + IL-2v polypeptide and tetraspecific ANKET). Log2 fold-changes were shrunk with the lfcShrink function implemented in DESeq2, by the “apeglm” method. Gene set enrichment analysis (GSEA) was performed with the R package fgsea, version 1.16.0 (fgseaMultilevel function). ORA (overrepresentation analysis) was used to identify the pathways overrepresented in the lists of genes. These enrichment analyses were performed on the Hallmark, Reactome, BioCarta, PID and WikiPathways gene sets (msigdb version 7.4). Redundant gene sets were removed with collapsePathways (fgsea package) and affinityPropagation (WebGestaltR package) for GSEA and ORA, respectively.
NK genesets were built based on the literature17, 18, 19 and analysed by GSEA. Expression score was calculated for each NK geneset and each sample and was visualized by boxplot. Expression score for a geneset was defined as follows: Score = , where n is the number of genes in the geneset, markerPos is a gene upregeulated by the NK subset and markerNeg a gene downregulated by the same NK subset.
Quantification and statistical analysis
Data analysis and statistics
Statistical analyses and plots for in vitro experiments were performed with GraphPad Prism (version 8.1.1).
Four-parameter non-linear regression analysis was used to calculate the EC50 (Figures 1B and 1E). For the comparison of groups (Figures 1B, 1E, S3D, and 3E), non-parametric Kruskall-Wallis tests were performed, with Dunn’s correction for multiple comparisons. Mixed-effect analysis was used, together with Tukey’s multiple comparisons test, for cytokine assessments (Figure 1D). For the multiple comparison of groups in Figure 2G, Friedman tests with Dunn’s correction was performed.
In vivo analysis: For the comparison of treatment groups (Figures 3A, 3B, 3C, and S3C), we used the following linear mixed-effects model: Volume ∼ Treatment∗Time + (1|MouseID) (treatment, time and the interaction being fixed effects and MouseID the random effect). Pairwise differences of least squares means were calculated for the factor “treatment”.
The linear mixed-effects model Volume ∼ Depletion∗Treatment∗Time + (1|MouseID) was also applied and pairwise differences of least squares means for the factor “depletion” were calculated (Figure 3E). Confidence intervals and p-values are based on the t-distribution and degrees of freedom based on Kenward-Roger methods.
Significance is indicated as follows: ∗p < 0.05; ∗∗p<0.01; ∗∗∗p < 0.001, ∗∗∗∗p < 0.0001.
Acknowledgments
The E.V. laboratory at CIML and Assistance-Publique des Hôpitaux de Marseille is supported by funding from the European Research Council (ERC) under the European Union’s Horizon 2020 research and innovation program (TILC, grant agreement no. 694502 and MInfla-TILC, grant agreement no. 875102 - MInfla-Tilc), the Agence Nationale de la Recherche including the PIONEER Project (ANR-17-RHUS-0007), MSDAvenir, Innate Pharma, and institutional grants awarded to the CIML (INSERM, CNRS, and Aix-Marseille University) and Marseille Immunopole.
Author contributions
A.B.A., G.H., W.B., N.B., M.G.-M., C.C., M.C., F.B., O.B., J.L., A.F., J.G., S.T., B.C., F. Carrette, A. Maguer, S.J., A.S., R.L.-D., C.K., N.L., A.D., F. Chanuc, S.B., N.J., C.F., N.G., R.R., and M.L.V. performed all experiments. S.C., R.L.G., C.B., L.V., A.R., B.R., I.P., and Y.M. provided analysis and advice. C.V., L.B., and S.C. performed RNA-seq and statistical analyses. E.L., N.F., S.S., and J.-L.D. provided clinical samples. O.D., L.G., M.V., A. Morel, and E.V. conceptually planned and supervised the study. O.D. and E.V. wrote the manuscript, with contributions from all authors.
Declaration of interests
Innate Pharma has filed patent applications relating to ANKETs and its use of ANKETs in the treatment of tumors (US patent no. 10,113,003, together with other patents and patent applications), as well as applications for use of ANKETs as a trademark. With the exception of E.L., N.F., S.S., J.-L.D., and R.L.G., all the authors are employees of Innate Pharma.
Published: October 18, 2022
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.xcrm.2022.100783.
Contributor Information
Olivier Demaria, Email: olivier.demaria@innate-pharma.fr.
Eric Vivier, Email: vivier@ciml.univ-mrs.fr.
Supplemental information
Data and code availability
-
•
All sequencing data supporting the findings of this study have been deposited into the NCBI -Gene Expression Omnibus database (https://www.ncbi.nlm.nih.gov/geo/). The accession number (GEO: GSE214378) is listed in the key resources table.
-
•
This paper does not report original code. Software packages used in the analysis are also presented in the key resources table.
-
•
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
References
- 1.Vivier E., Artis D., Colonna M., Diefenbach A., Di Santo J.P., Eberl G., Koyasu S., Locksley R.M., McKenzie A.N.J., Mebius R.E., et al. Innate lymphoid cells: 10 years on. Cell. 2018;174:1054–1066. doi: 10.1016/j.cell.2018.07.017. [DOI] [PubMed] [Google Scholar]
- 2.Demaria O., Cornen S., Daëron M., Morel Y., Medzhitov R., Vivier E. Harnessing innate immunity in cancer therapy. Nature. 2019;574:45–56. doi: 10.1038/s41586-019-1593-5. [DOI] [PubMed] [Google Scholar]
- 3.Liu E., Marin D., Banerjee P., Macapinlac H.A., Thompson P., Basar R., Nassif Kerbauy L., Overman B., Thall P., Kaplan M., et al. Use of CAR-transduced natural killer cells in CD19-positive lymphoid tumors. N. Engl. J. Med. 2020;382:545–553. doi: 10.1056/NEJMoa1910607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Vivier E., Nunès J.A., Vély F. Natural killer cell signaling pathways. Science. 2004;306:1517–1519. doi: 10.1126/science.1103478. [DOI] [PubMed] [Google Scholar]
- 5.Lanier L.L. Up on the tightrope: natural killer cell activation and inhibition. Nat. Immunol. 2008;9:495–502. doi: 10.1038/ni1581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bryceson Y.T., March M.E., Ljunggren H.G., Long E.O. Synergy among receptors on resting NK cells for the activation of natural cytotoxicity and cytokine secretion. Blood. 2006;107:159–166. doi: 10.1182/blood-2005-04-1351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bryceson Y.T., Ljunggren H.G., Long E.O. Minimal requirement for induction of natural cytotoxicity and intersection of activation signals by inhibitory receptors. Blood. 2009;114:2657–2666. doi: 10.1182/blood-2009-01-201632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gauthier L., Morel A., Anceriz N., Rossi B., Blanchard-Alvarez A., Grondin G., Trichard S., Cesari C., Sapet M., Bosco F., et al. Multifunctional natural killer cell engagers targeting NKp46 trigger protective tumor immunity. Cell. 2019;177:1701–1713.e16. doi: 10.1016/j.cell.2019.04.041. [DOI] [PubMed] [Google Scholar]
- 9.Boyman O., Sprent J. The role of interleukin-2 during homeostasis and activation of the immune system. Nat. Rev. Immunol. 2012;12:180–190. doi: 10.1038/nri3156. [DOI] [PubMed] [Google Scholar]
- 10.Mullard A. Restoring IL-2 to its cancer immunotherapy glory. Nat. Rev. Drug Discov. 2021;20:163–165. doi: 10.1038/d41573-021-00034-6. [DOI] [PubMed] [Google Scholar]
- 11.Wang X., Rickert M., Garcia K.C. Structure of the quaternary complex of interleukin-2 with its alpha, beta, and gammac receptors. Science. 2005;310:1159–1163. doi: 10.1126/science.1117893. [DOI] [PubMed] [Google Scholar]
- 12.Henney C.S., Kuribayashi K., Kern D.E., Gillis S. Interleukin-2 augments natural killer cell activity. Nature. 1981;291:335–338. doi: 10.1038/291335a0. [DOI] [PubMed] [Google Scholar]
- 13.Caligiuri M.A., Zmuidzinas A., Manley T.J., Levine H., Smith K.A., Ritz J. Functional consequences of interleukin 2 receptor expression on resting human lymphocytes. Identification of a novel natural killer cell subset with high affinity receptors. J. Exp. Med. 1990;171:1509–1526. doi: 10.1084/jem.171.5.1509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Klein C., Waldhauer I., Nicolini V.G., Freimoser-Grundschober A., Nayak T., Vugts D.J., Dunn C., Bolijn M., Benz J., Stihle M., et al. Cergutuzumab amunaleukin (CEA-IL2v), a CEA-targeted IL-2 variant-based immunocytokine for combination cancer immunotherapy: overcoming limitations of aldesleukin and conventional IL-2-based immunocytokines. Oncoimmunology. 2017;6:e1277306. doi: 10.1080/2162402X.2016.1277306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Billadeau D.D., Brumbaugh K.M., Dick C.J., Schoon R.A., Bustelo X.R., Leibson P.J. The Vav-Rac1 pathway in cytotoxic lymphocytes regulates the generation of cell-mediated killing. J. Exp. Med. 1998;188:549–559. doi: 10.1084/jem.188.3.549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Orange J.S. Formation and function of the lytic NK-cell immunological synapse. Nat. Rev. Immunol. 2008;8:713–725. doi: 10.1038/nri2381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Crinier A., Milpied P., Escalière B., Piperoglou C., Galluso J., Balsamo A., Spinelli L., Cervera-Marzal I., Ebbo M., Girard-Madoux M., et al. High-dimensional single-cell analysis identifies organ-specific signatures and conserved NK cell subsets in humans and mice. Immunity. 2018;49:971–986.e5. doi: 10.1016/j.immuni.2018.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Smith S.L., Kennedy P.R., Stacey K.B., Worboys J.D., Yarwood A., Seo S., Solloa E.H., Mistretta B., Chatterjee S.S., Gunaratne P., et al. Diversity of peripheral blood human NK cells identified by single-cell RNA sequencing. Blood Adv. 2020;4:1388–1406. doi: 10.1182/bloodadvances.2019000699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Crinier A., Dumas P.Y., Escalière B., Piperoglou C., Gil L., Villacreces A., Vély F., Ivanovic Z., Milpied P., Narni-Mancinelli É., Vivier É. Single-cell profiling reveals the trajectories of natural killer cell differentiation in bone marrow and a stress signature induced by acute myeloid leukemia. Cell. Mol. Immunol. 2021;18:1290–1304. doi: 10.1038/s41423-020-00574-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Schwartz R.N., Stover L., Dutcher J.P. Managing toxicities of high-dose interleukin-2. Oncology (Williston Park) 2002;16:11–20. [PubMed] [Google Scholar]
- 21.Derebe M.G., Nanjunda R.K., Gilliland G.L., Lacy E.R., Chiu M.L. Human IgG subclass cross-species reactivity to mouse and cynomolgus monkey Fcgamma receptors. Immunol. Lett. 2018;197:1–8. doi: 10.1016/j.imlet.2018.02.006. [DOI] [PubMed] [Google Scholar]
- 22.Warncke M., Calzascia T., Coulot M., Balke N., Touil R., Kolbinger F., Heusser C. Different adaptations of IgG effector function in human and nonhuman primates and implications for therapeutic antibody treatment. J. Immunol. 2012;188:4405–4411. doi: 10.4049/jimmunol.1200090. [DOI] [PubMed] [Google Scholar]
- 23.Krieg C., Létourneau S., Pantaleo G., Boyman O. Improved IL-2 immunotherapy by selective stimulation of IL-2 receptors on lymphocytes and endothelial cells. Proc. Natl. Acad. Sci. USA. 2010;107:11906–11911. doi: 10.1073/pnas.1002569107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Li Y., Strick-Marchand H., Lim A.I., Ren J., Masse-Ranson G., Dan L., Jouvion G., Rogge L., Lucas S., Bin L., Di Santo J.P. Regulatory T cells control toxicity in a humanized model of IL-2 therapy. Nat. Commun. 2017;8:1762. doi: 10.1038/s41467-017-01570-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Böttcher J.P., Bonavita E., Chakravarty P., Blees H., Cabeza-Cabrerizo M., Sammicheli S., Rogers N.C., Sahai E., Zelenay S., Reis e Sousa C. NK cells stimulate recruitment of cDC1 into the tumor microenvironment promoting cancer immune control. Cell. 2018;172:1022–1037.e14. doi: 10.1016/j.cell.2018.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Barry K.C., Hsu J., Broz M.L., Cueto F.J., Binnewies M., Combes A.J., Nelson A.E., Loo K., Kumar R., Rosenblum M.D., et al. A natural killer-dendritic cell axis defines checkpoint therapy-responsive tumor microenvironments. Nat. Med. 2018;24:1178–1191. doi: 10.1038/s41591-018-0085-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Schenk M., Krutzik S.R., Sieling P.A., Lee D.J., Teles R.M.B., Ochoa M.T., Komisopoulou E., Sarno E.N., Rea T.H., Graeber T.G., et al. NOD2 triggers an interleukin-32-dependent human dendritic cell program in leprosy. Nat. Med. 2012;18:555–563. doi: 10.1038/nm.2650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cooper M.A., Elliott J.M., Keyel P.A., Yang L., Carrero J.A., Yokoyama W.M. Cytokine-induced memory-like natural killer cells. Proc. Natl. Acad. Sci. USA. 2009;106:1915–1919. doi: 10.1073/pnas.0813192106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pahl J.H.W., Koch J., Götz J.J., Arnold A., Reusch U., Gantke T., Rajkovic E., Treder M., Cerwenka A. CD16A activation of NK cells promotes NK cell proliferation and memory-like cytotoxicity against cancer cells. Cancer Immunol. Res. 2018;6:517–527. doi: 10.1158/2326-6066.CIR-17-0550. [DOI] [PubMed] [Google Scholar]
- 30.Romee R., Rosario M., Berrien-Elliott M.M., Wagner J.A., Jewell B.A., Schappe T., Leong J.W., Abdel-Latif S., Schneider S.E., Willey S., et al. Cytokine-induced memory-like natural killer cells exhibit enhanced responses against myeloid leukemia. Sci. Transl. Med. 2016;8:357ra123. doi: 10.1126/scitranslmed.aaf2341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hutchings M., Morschhauser F., Iacoboni G., Carlo-Stella C., Offner F.C., Sureda A., Salles G., Martínez-Lopez J., Crump M., Thomas D.N., et al. Glofitamab, a novel, bivalent CD20-targeting T-cell-engaging bispecific antibody, induces durable complete remissions in relapsed or refractory B-cell lymphoma: a phase I trial. J. Clin. Oncol. 2021;39:1959–1970. doi: 10.1200/JCO.20.03175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Friberg G., Reese D. Blinatumomab (Blincyto): lessons learned from the bispecific t-cell engager (BiTE) in acute lymphocytic leukemia (ALL) Ann. Oncol. 2017;28:2009–2012. doi: 10.1093/annonc/mdx150. [DOI] [PubMed] [Google Scholar]
- 33.Bonifant C.L., Jackson H.J., Brentjens R.J., Curran K.J. Toxicity and management in CAR T-cell therapy. Mol. Ther. Oncolytics. 2016;3:16011. doi: 10.1038/mto.2016.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Devillier R., Calmels B., Guia S., Taha M., Fauriat C., Mfarrej B., Venton G., Vivier E., Olive D., Chabannon C., et al. Phase I trial of prophylactic donor-derived IL-2-activated NK cell infusion after allogeneic hematopoietic stem cell transplantation from a matched sibling donor. Cancers. 2021;13:2673. doi: 10.3390/cancers13112673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Heaton K.M., Ju G., Grimm E.A. Human interleukin 2 analogues that preferentially bind the intermediate-affinity interleukin 2 receptor lead to reduced secondary cytokine secretion: implications for the use of these interleukin 2 analogues in cancer immunotherapy. Cancer Res. 1993;53:2597–2602. [PubMed] [Google Scholar]
- 36.Teachey D.T., Lacey S.F., Shaw P.A., Melenhorst J.J., Maude S.L., Frey N., Pequignot E., Gonzalez V.E., Chen F., Finklestein J., et al. Identification of predictive biomarkers for cytokine release syndrome after chimeric antigen receptor T-cell therapy for acute lymphoblastic leukemia. Cancer Discov. 2016;6:664–679. doi: 10.1158/2159-8290.CD-16-0040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Horng T., Bezbradica J.S., Medzhitov R. NKG2D signaling is coupled to the interleukin 15 receptor signaling pathway. Nat. Immunol. 2007;8:1345–1352. doi: 10.1038/ni1524. [DOI] [PubMed] [Google Scholar]
- 38.Bezbradica J.S., Rosenstein R.K., DeMarco R.A., Brodsky I., Medzhitov R. A role for the ITAM signaling module in specifying cytokine-receptor functions. Nat. Immunol. 2014;15:333–342. doi: 10.1038/ni.2845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Dekkers G., Bentlage A.E.H., Stegmann T.C., Howie H.L., Lissenberg-Thunnissen S., Zimring J., Rispens T., Vidarsson G. Affinity of human IgG subclasses to mouse Fc gamma receptors. mAbs. 2017;9:767–773. doi: 10.1080/19420862.2017.1323159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bruhns P., Jönsson F. Mouse and human FcR effector functions. Immunol. Rev. 2015;268:25–51. doi: 10.1111/imr.12350. [DOI] [PubMed] [Google Scholar]
- 41.Aguilar O.A., Fong L.K., Ishiyama K., DeGrado W.F., Lanier L.L. The CD3zeta adaptor structure determines functional differences between human and mouse CD16 Fc receptor signaling. J. Exp. Med. 2022;219 doi: 10.1084/jem.20220022. e20220022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Danielou-Lazareth A., Henry G., Geromin D., Khaznadar Z., Briere J., Tamouza R., Cayuela J.M., Thieblemont C., Toubert A., Dulphy N. At diagnosis, diffuse large B-cell lymphoma patients show impaired rituximab-mediated NK-cell cytotoxicity. Eur. J. Immunol. 2013;43:1383–1388. doi: 10.1002/eji.201242733. [DOI] [PubMed] [Google Scholar]
- 43.Delconte R.B., Kolesnik T.B., Dagley L.F., Rautela J., Shi W., Putz E.M., Stannard K., Zhang J.G., Teh C., Firth M., et al. CIS is a potent checkpoint in NK cell-mediated tumor immunity. Nat. Immunol. 2016;17:816–824. doi: 10.1038/ni.3470. [DOI] [PubMed] [Google Scholar]
- 44.Keating N., Nicholson S.E. SOCS-mediated immunomodulation of natural killer cells. Cytokine. 2019;118:64–70. doi: 10.1016/j.cyto.2018.03.033. [DOI] [PubMed] [Google Scholar]
- 45.Demaria O., Gauthier L., Debroas G., Vivier E. Natural killer cell engagers in cancer immunotherapy: next generation of immuno-oncology treatments. Eur. J. Immunol. 2021;51:1934–1942. doi: 10.1002/eji.202048953. [DOI] [PubMed] [Google Scholar]
- 46.Bartlett N.L., Herrera A.F., Domingo-Domenech E., Mehta A., Forero-Torres A., Garcia-Sanz R., Armand P., Devata S., Izquierdo A.R., Lossos I.S., et al. A phase 1b study of AFM13 in combination with pembrolizumab in patients with relapsed or refractory Hodgkin lymphoma. Blood. 2020;136:2401–2409. doi: 10.1182/blood.2019004701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Rothe A., Sasse S., Topp M.S., Eichenauer D.A., Hummel H., Reiners K.S., Dietlein M., Kuhnert G., Kessler J., Buerkle C., et al. A phase 1 study of the bispecific anti-CD30/CD16A antibody construct AFM13 in patients with relapsed or refractory Hodgkin lymphoma. Blood. 2015;125:4024–4031. doi: 10.1182/blood-2014-12-614636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Vallera D.A., Felices M., McElmurry R., McCullar V., Zhou X., Schmohl J.U., Zhang B., Lenvik A.J., Panoskaltsis-Mortari A., Verneris M.R., et al. IL15 trispecific killer engagers (TriKE) make natural killer cells specific to CD33+ targets while also inducing persistence, in vivo expansion, and enhanced function. Clin. Cancer Res. 2016;22:3440–3450. doi: 10.1158/1078-0432.CCR-15-2710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Vallera D.A., Ferrone S., Kodal B., Hinderlie P., Bendzick L., Ettestad B., Hallstrom C., Zorko N.A., Rao A., Fujioka N., et al. NK-Cell-Mediated targeting of various solid tumors using a B7-H3 tri-specific killer engager in vitro and in vivo. Cancers. 2020;12:E2659. doi: 10.3390/cancers12092659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Dobin A., Davis C.A., Schlesinger F., Drenkow J., Zaleski C., Jha S., Batut P., Chaisson M., Gingeras T.R. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29:15–21. doi: 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kitamura D., Roes J., Kühn R., Rajewsky K. A B cell-deficient mouse by targeted disruption of the membrane exon of the immunoglobulin mu chain gene. Nature. 1991;350:423–426. doi: 10.1038/350423a0. [DOI] [PubMed] [Google Scholar]
- 52.Walzer T., Bléry M., Chaix J., Fuseri N., Chasson L., Robbins S.H., Jaeger S., André P., Gauthier L., Daniel L., et al. Identification, activation, and selective in vivo ablation of mouse NK cells via NKp46. Proc. Natl. Acad. Sci. USA. 2007;104:3384–3389. doi: 10.1073/pnas.0609692104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Liao Y., Smyth G.K., Shi W. featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics. 2014;30:923–930. doi: 10.1093/bioinformatics/btt656. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
-
•
All sequencing data supporting the findings of this study have been deposited into the NCBI -Gene Expression Omnibus database (https://www.ncbi.nlm.nih.gov/geo/). The accession number (GEO: GSE214378) is listed in the key resources table.
-
•
This paper does not report original code. Software packages used in the analysis are also presented in the key resources table.
-
•
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.