Abstract
Objective
Metabolomics as an approach to solve biological problems is exponentially increasing in use. Thus, this a pivotal time for the adoption of best practices. It is well known that disrupted tissue oxygen supply rapidly alters cellular energy charge. However, the speed and extent to which delayed mouse tissue freezing after dissection alters the broad metabolome is not well described. Furthermore, how tissue genotype may modulate such metabolomic drift and the degree to which traced 13C-isotopologue distributions may change have not been addressed.
Methods
By combined liquid chromatography (LC)- and gas chromatography (GC)-mass spectrometry (MS), we measured how levels of 255 mouse liver metabolites changed following 30-second, 1-minute, 3-minute, and 10-minute freezing delays. We then performed test-of-concept delay-to-freeze experiments evaluating broad metabolomic drift in mouse heart and skeletal muscle, differential metabolomic change between wildtype (WT) and mitochondrial pyruvate carrier (MPC) knockout mouse livers, and shifts in 13C-isotopologue abundances and enrichments traced from 13C-labled glucose into mouse liver.
Results
Our data demonstrate that delayed mouse tissue freezing after dissection leads to rapid hypoxia-driven remodeling of the broad metabolome, induction of both false-negative and false-positive between-genotype differences, and restructuring of 13C-isotopologue distributions. Notably, we show that increased purine nucleotide degradation products are an especially high dynamic range marker of delayed liver and heart freezing.
Conclusions
Our findings provide a previously absent, systematic illustration of the extensive, multi-domain metabolomic changes occurring within the early minutes of delayed tissue freezing. They also provide a novel, detailed resource of mouse liver ex vivo, hypoxic metabolomic remodeling.
Keywords: Metabolomics, Hypoxia, Quality control, Mitochondrial pyruvate carrier, Stable isotope tracing, Purine degradation
Highlights
-
•
Delayed mouse liver, heart, and muscle tissue freezing leads to broad metabolomic remodeling.
-
•
False negative and false positive between-genotype metabolite level differences are induced.
-
•
Within metabolite 13C-isotopologue abundances and percent enrichments are altered.
-
•
Purine nucleotide degradation is a high dynamic range marker of delayed heart and liver freezing.
1. Introduction
Metabolomics is a powerful investigative approach to solve biological problems that is dramatically increasing in use. The metabolome is the endogenous small molecule pool of reaction substrates and products [1]. It links cellular chemistry, energetics, and environment to impact phenotype, responding nearly instantaneously to homeostatic challenges [2]. Thus, the metabolome is a functional, real-time property of health and disease states. Metabolomics refers to the measurement of the metabolome using NMR or mass spectrometry, with the latter capable of measuring hundreds of metabolites in milligram quantities of sample [3]. As evidence of metabolomics' growing use, a PubMed.gov search for ‘metabolomics or metabolome’ at the time of this writing returned >55,000 original research articles published since the first search-retrievable, mention of “metabolome” in 1998 [4]. Notably, greater than 50% of these manuscripts were published after 2017, with over 90% after 2011.
Because broad metabolomic analysis of animal tissues requires dissection, a major risk to the integrity of metabolomic data is a hypoxia-induced shift of metabolism, between the time of disrupted perfusion and freezing. Indeed, beginning with the evolution of complex life, cellular metabolism became programmed to be potently regulated by oxygen [[5], [6], [7], [8]]. In most eukaryotic cells, mitochondrial electron transport chain flux coupled to oxygen reduction regulates an array of redox-sensitive metabolic reactions and drives bulk ATP production. Furthermore, in addition to supplying nutrients and removing cellular waste, perfusion evolved to provide tissues with oxygen. Consequently, animal tissue metabolism is intricately connected to oxygen availability, and disrupted perfusion rapidly leads to evolutionarily imbedded, coordinated metabolic responses. Thus, to rigorously investigate the in vivo metabolome of animal tissues, it is essential to quickly freeze tissues after dissection to prevent ex vivo, hypoxia-driven metabolomic remodeling.
However, how delayed tissue freezing affects the broad metabolome during the immediate post-dissection period has remained unaddressed. Although it is well known that disrupted oxygen supply rapidly alters cellular energy charge, the speed and extent to which this extends to the larger metabolome has not been defined. Furthermore, most manuscripts reporting tissue metabolomic analysis do not precisely specify the lag time between loss of perfusion and tissue freezing. The absence of time to freeze information makes it difficult to infer how post-dissection metabolism may have impacted the reported metabolomic data. To address this problem, we performed experiments evaluating how the metabolome of three mouse tissues changes with delayed freezing after harvest. We show that tissue harvest-induced hypoxia rapidly alters the in vivo metabolome, between-genotype metabolite level differences, and 13C-tracing enrichments. With metabolomics being increasingly adopted as a mainstream investigative approach and the bloom of multiomics, this is an especially critical time to clarify best practices to obtain reliable metabolomic data. Our findings provide a previously absent, systematic illustration of the extensive and confounding metabolomic changes occurring within the early minutes of delayed tissue freezing. They also provide a novel, detailed resource of mouse liver ex vivo, hypoxia-driven metabolomic remodeling.
2. Materials and methods
2.1. Materials
C57bl6/j mice were obtained from Jackson Laboratories (Stock #000664). Labeled internal standards citric acid (2,2,4,4-D4, 98%; #DLM-3487), succinic acid (2,2,3,3-D4, 98%; #DLM-584), l-valine (2,3,4,4,4,5,5,5-D8; 98%; #DLM-311), l-glutamic acid (13C5, 99%; #CLM-1800), l-glutamine (13C5, 99%, #CLM-1822), l-lysine (13C6, 99%; #CLM-2247), l-methionine (13C5, 99%; #CLM-893), and l-tryptophan (13C11, 99%; #CLM-4290) and tracer glucose (13C6, 99%, #CLM-1396) were obtained from Cambridge Isotope Laboratories, Inc. AAV8-TBG-NULL (#105536-AAV8) and AAV8-TBG-Cre (#107787-AAV8) were purchased from Addgene. Pre-filled bead mill tubes containing 1.4 mm ceramic beads (#15-340-153) were purchased from FisherScientific. Methoxaymine hydrochloride (#226904, MOX) and N-methyl-N-(trimethylsilyl) trifluoroacetamide (#694709, MSTFA) were purchased from Sigma Aldrich. Protease inhibitor cocktail (#786-437) was purchased from G Biosciences. Bio-Rad protein assay dye regent (#5000006) and 0.45 μm nitrocellulose membrane (#1620115) were purchased from Bio-Rad.
2.2. Animal sacrifice
All animal work was performed in accordance with the University of Iowa Animal Use and Care Committee (IACUC) guidelines, protocol #0101249. For all studies, mice were fasted 4 h before sacrifice. 9–11-week-old male mice were used for liver profiling, tibialis anterior muscle (TA) profiling, and U–13C-glucose liver tracing studies. 15-week-old male mice were used for the paired WT versus MPC-LivKO liver and the heart studies. Mice were anesthetized by 3.5% isoflurane inhalation for 1.75 min prior to dissection of tissues (liver, heart, and skeletal muscle). Following dissection, tissue was frozen after about 1 second, which can be reproducibly met for a well-controlled “time zero,” or after the various indicated time points, including up to a 10-minute delay for liver. A liquid-nitrogen cooled Wollenberger-like device referred to here as a freeze-clamp was used to freeze tissue samples instantaneously. Freeze-clamped tissues were stored at −80 °C.
For liver samples: the abdominal space was opened to expose the liver, and care was taken not to disrupt or puncture the diaphragm. The left lateral lobe of the liver was isolated and dissected from anesthetized mice. The freeze-clamp was used to freeze liver samples at the appropriate time points.
For heart samples: the abdominal space was opened to expose the diaphragm. The diaphragm and sternum were quickly cut to open the pericardial space. Forceps were used to quickly expose the heart, and the ventricular portion was excised from the atria (about 2 seconds after opening the pericardial space). Ventricular tissue was frozen at the appropriate time point by freeze clamping. Freeze-clamping the cardiac tissue as described resulted in the expulsion of most ventricular blood.
For skeletal muscle samples: the skin was carefully removed from the leg of the anesthetized mouse. The TA was identified, and the fascia was removed with circulation intact. The fine tip of a micro-fine tweezer was placed under the TA muscle without disrupting the attached tendons. At the time of dissection, the micro-fine tweezer tip was used to disrupt one TA muscle tendon and the second tendon was cut, thereby liberating the TA muscle, which was frozen at the appropriate time point by freeze clamping.
2.3. Generation of Mpc1 liver-specific knockout mice
Mpc1 liver specific knockout (MPC-LivKO) and littermate paired control (WT) mice were generated as previously reported [9]. Briefly, 8-week-old male Mpc1-floxed mice were retroorbitally injected with 1 × 1011 gc/mouse of either AAV8-TBG-NULL or AAV8-TBG-Cre generating WT and MPC-LivKO mice, respectively.
2.4. U–13C-glucose tracing
C57bl/6j mice were i.p. injected with U–13C-glucose at a dosage of 1.4 g/kg body mass. The mice were anesthetized, as described above, and liver tissue was harvested 30 min after injection. Circulating glucose following an i.p. injection peaks at 15–20 min [9]. At 30 min post-injection, glucose clearance includes liver uptake and metabolism, which resulted in >25% 13C enrichment into liver lactate and TCA cycle intermediates.
2.5. Western blots
Freeze-clamped liver samples were homogenized in 40 volumes:weight of a buffer containing 40 mM HEPES, 120 mM NaCl, 50 mM NaF, 5 mM sodium pyrophosphate decahydrate, 5 mM b-glycerophosphate, 1 mM EDTA, 1 mM EGTA, 10% glycerol (v/v), 1% igepal CA-630 (v/v), with 1× protease inhibitor and 1 μM DTT. Homogenates were rotated at 4 °C for 30 min and centrifuged at 21,000×g. Supernatants were collected, and protein concentrations were determined using the Bio-Rad protein assay reagent. Proteins were separated using TRIS-SDS-PAGE gel electrophoresis, transferred to 0.45 μM nitrocellulose membranes, and blocked with TBST (50 mM Tris, 150 mM NaCl, and 0.05% Tween-20) supplemented with 2.5% BSA. Blocked membranes were incubated with primary antibodies at 4 °C overnight [mouse monoclonal anti-AMPKα (F6) (RRID: AB_915794), rabbit monoclonal anti-pAMPKα (Thr172) (40H9) (RRID: AB_331250), and rabbit monoclonal anti-HSP90 (RRID: AB_2121214) from Cell Signaling Technology]. The following day membranes were washed with TBST and incubated with fluorescently labeled secondary antibodies for 1 h at room temperature [goat anti-Mouse Dylight 800 (RRID: AB_2556756), donkey anti-Rabbit DyLight 680 (RRID: AB_2556622), and donkey anti-Rabbit DyLight 800 (RRID: AB_2556616) from ThermoFisher]. Membranes were visualized using the Li-Cor Odyssey CLx system (Li-Cor Biosciences).
2.6. Metabolite extraction and derivatization
Freeze-clamped liver tissue (40 ± 5 mg) was partitioned and weighed without thawing and lyophilized overnight. Heart and TA muscle tissues were weighed and lyophilized intact. Tissues were extracted in 18 volumes (relative to wet tissue weight) of ice-cold extraction solvent composed of acetonitrile:methanol:water (2:2:1) containing 1 μL/mL of an internal standard mix (0.33 mg of each/mL in water, see “Materials”). Lyophilized tissue was added to pre-filled bead mill tubes containing 1.4 mm ceramic beads. Ice-cold extraction solvent was added, and tissue was homogenized using a BeadRupter bead mill homogenizer (Omni International) for 30 s at 6.45 MHz. Immediately afterwards, homogenization tubes were rotated for 60 min at −20 °C. Samples were centrifuged at 21,000×g for 10 min, after which the supernatant was transferred to a new 1.5 mL microcentrifuge tube and pulse vortexed to ensure uniform mixing of the supernatant. Of this supernatant, 150 μL was transferred to glass autosampler vials for GC–MS analysis and 300 μL was transferred to a 1.7 mL microcentrifuge tube for further processing for LC–MS analysis. A quality control (QC) sample was prepared by pooling equal volumes from each sample; the QC sample was aliquoted into a glass autosampler vial and a 1.7 mL microcentrifuge tube for GC–MS (150 μL) and LC–MS analysis (300 μL), respectively. Samples were dried in a Speedvac Vacuum concentrator (Thermo) for 2 h without heating at a vacuum ramp = 4.
2.7. GC–MS analysis
GC–MS sample derivatization with MOX + MSTFA was accomplished as follows: 30 μL of pyridine containing 11.4 mg/mL of MOX was added to the dried metabolite extracts. Samples were then vortexed for 10 min and heated at 60 °C for 60 min. Next, 20 uL of MSTFA was added to the pyridine/MOX derivatized samples, and they were vortexed for 5 min, and heated at 60 °C for an additional 30 min. GC–MS analysis was performed using a Trace 1300 GC (Thermo) coupled to an ISQ-LT single quadrupole mass spectrometer (Thermo). For each sample, 1 μL of sample was injected into the GC by an autosampler in split mode (split ratio: 20:1; split flow: 24 μL/min, purge flow: 5 mL/min, Carrier mode: Constant Flow, Carrier flow rate: 1.2 mL/min). Separation was accomplished using a standard fused silica TraceGold TG-5SilMS column (Thermo). The temperature gradient was as follows: 80 °C for 3 min, ramped at a rate of 20 °C/min to 280 °C, and held for 8 min. Between sample runs, the injection syringe was washed 3 times with methanol and 3 times with pyridine. The MS was operated from 3.90 to 21.00 min in EI mode (−70eV) using select ion monitoring (SIM). The mass spectrometer was tuned and calibrated daily. The pooled QC sample was analyzed at the beginning and at the end of the GC/MS run, as well as about every eight injections throughout.
2.8. LC–MS analysis
For LC–MS analysis, dried metabolite extracts were resuspended in 30 ul of acetonitrile:water (1:1), vortexed for 10 min, and stored at −20 °C overnight. The following day, resuspended samples were centrifuged at 21,000×g for 10 min, and the resulting supernatant were transferred to autosampler vials for LC–MS analysis. For each prepared sample, 2 μL was separated using a Millipore SeQuant ZIC-pHILIC (2.1 × 150 mm, 5 μm particle size, Millipore Sigma #150460) column with a ZIC-pHILIC guard column (20 × 2.1 mm, Millipore Sigma #150437) attached to a Thermo Vanquish Flex UHPLC. The mobile phase comprised Buffer A [20 mM (NH4)2CO3, 0.1% NH4OH (v/v)] and Buffer B [acetonitrile]. The chromatographic gradient was run at a flow rate of 0.150 mL/min as follows: 0–21 min-linear gradient from 80 to 20% Buffer B; 20–20.5 min-linear gradient from 20 to 80% Buffer B; and 20.5–28 min-hold at 80% Buffer B. Data was acquired using a Thermo Q Exactive MS operated in polarity switching full scan mode with a spray voltage set to 3.0 kV, the heated capillary held at 275 °C, and the HESI probe held at 350 °C. The sheath gas flow was set to 40 units, the auxiliary gas flow was set to 15 units, and the sweep gas flow was set to 1 unit. MS data resolution was set at 70,000, the AGC target at 10e6, and the maximum injection time at 200 ms. The QC sample was analyzed at the beginning and at the end of the LC–MS run, as well as about every eight injections throughout.
2.9. Mass spectrometry data analysis
Acquired GC- and LC–MS data were processed using the Thermo Scientific TraceFinder (4.1 and 5.1) software. Targeted metabolites were identified based on the University of Iowa Metabolomics Core facility standard-confirmed, in-house library defining a target ion and at least 1 confirming ion and retention time (GC) or accurate mass, retention time, and MS/MS fragmentation pattern when present (LC). The NOREVA tool used the QC sample analyzed throughout the instrument run to apply local polynomial fits to metabolite peak areas and correct for instrument drift [10]. NOREVA corrected data were normalized to the D4-succinate signal/sample to control for extraction, derivatization (GC), and/or loading effects. For 13C-tracing analysis, 12C-natural abundance was corrected using previously defined equations [11].
2.10. Quantification and statistical analysis
Post NOREVA corrected, D4-succinate-normalized metabolomic data were log transformed. Following log transformation, the Grubbs' test was employed at an α = 0.01 to detect extreme outliers. This resulted in the exclusion of 130/20388 or 0.64% of values. Log transformed metabolomic data were then analyzed by unpaired Student's t-tests or by one-way ANOVA with the Holm-Sidak post-hoc multiple comparison test compared to time zero. Statistically significance differences were defined as having p-values < 0.05 (∗), p < 0.01 (∗∗) and p < 0.001 (∗∗∗). Results were plotted as mean ± standard error of the means (SEM) relative to time zero. The number of biologically unique samples used in each experiment is specified in the relevant figure legend. Heatmaps were generated using Heatmapper [12].
For calculation of total metabolomic change and t50, normalized metabolite values were processed in R (v4.1.0). For each metabolite, a smooth spline model was fitted for the time course using the npreg (v1.0-7) R package and the maximum likelihood methods for smoothing [13]. Maximum likelihood method was selected due to fewest issues with overfitting of splines after comparison with generalized cross-validation, Bayesian information criterion, and Akaike's information criterion. Time at which 50% of the total change in the metabolite occurred was estimated using the direction and magnitude of the largest change of the metabolite and the fitted smooth spline model. The output of this analysis was plotted using the gpplot2 (3.3.5) R package. Code for t50 calculations and Urchin plot generation is available at https://github.com/ncborcherding/Metabolite.
3. Resource availability
3.1. Lead contact
Additional information and requests for reagents and resources should be directed to and will be fulfilled by the lead contact, Dr. Eric Taylor (eric-taylor@uiowa.edu).
3.2. Materials availability
No new materials were generated during this study.
4. Results
4.1. Delayed post-dissection freezing of the liver leads to broad metabolomic drift
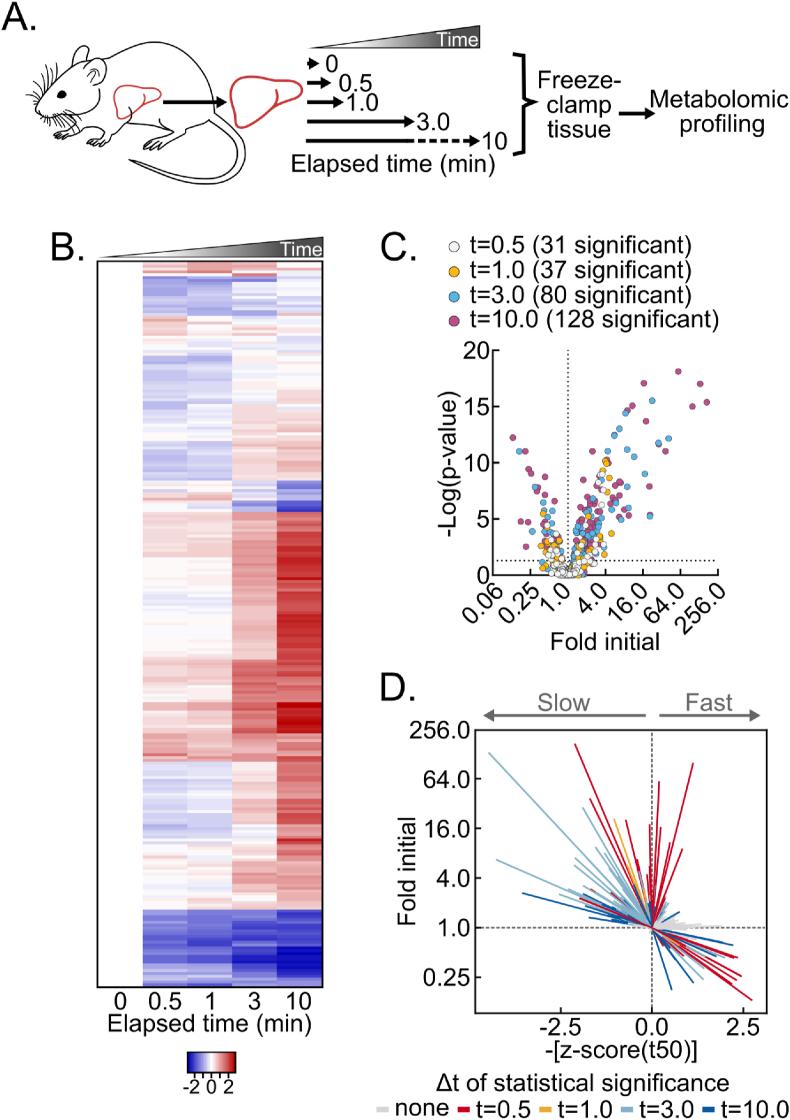
The liver is a metabolically versatile tissue routinely used to investigate basic and translational metabolism. To understand how the mouse liver in vivo metabolome is remodeled after dissection from live, anesthetized mice, we performed time-course delay-to-freeze experiments followed by broad metabolomic analysis. Specifically, the left lateral lobe of the liver was dissected from isoflurane-anesthetized mice and frozen by liquid nitrogen-temperature freeze clamping at several timepoints after dissection: immediately (about 1 s, which can be consistently achieved for a reproducible “time zero”), and after 30 s, 1 min, 3 min, and 10 min (Figure 1A). Compared to immediate freezing, ex vivo metabolomic drift was rapid and progressive (Figure 1B, Table S1). Thirty-one metabolites significantly changed by 30 s after dissection, and 128 did so by 10 min (Figure 1C).
Figure 1.
Delayed post-dissection freezing of the liver leads to broad metabolomic drift. (A) Schematic of experimental procedures. (B) Heatmap depicting the z-scores of metabolite fold changes by delayed freezing time-point compared to time zero. The heatmap is not time-scaled (n = 5–6 biological replicates). (C) Volcano plot showing the interaction between metabolite fold changes and associated p-values by delayed freezing time-point compared to time zero. The -log transformed p-values were calculated by one-way ANOVA with the post-hoc Holm-Sidak multiple comparison test. The number of significant differences for each time-point are listed in the panel (n = 5–6 biological replicates). (D) Urchin plot showing line slopes as an interaction between the t50 kinetics and dynamic ranges of metabolite fold changes with delayed freezing. Metabolites were converted to line vectors displaying dynamic range (10-minute fold change compared to time zero; y-axis) and the negative z-score of the time required for one-half maximum fold change (-[z-score (t50)]; x-axis). Line color indicates the time-point by which statistically significant change was reached calculated by one-way ANOVA with the post-hoc Holm-Sidak multiple comparison test (n = 5–6 biological replicates). Related to Figure S1 and Tables S1 and S2. (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
Given these extensive fold changes across individual metabolites, we aimed to sort and visualize metabolites by both kinetics and dynamic range of change. We expected that binning metabolites by an interaction between kinetics and dynamic range would reveal associations not readily apparent with heat maps, line graphs, or other commonly employed data visualization methods. First, because individual metabolites change with different initial rates relative to their average rate for the full 10-minute time course, we calculated and averaged the time to 50% apparent maximum change for all metabolites. This produced an average time of one-half maximum fold-change (t50) for the broad metabolome of approximately 3.58 ± 0.32 min (Fig. S1A) and a metabolome-wide distribution of per metabolite t50s. Thus, compared to the average metabolite, metabolites with earlier t50s initially change faster by percent of maximum fold-change and vice versa for metabolites with later t50s. Second, we plotted all measured metabolites as individual line vectors, by dynamic range (total fold change), versus negative z-scores of t50s (Figure 1D, Table S2). Time-points by which statistically significant change was reached were then indicated by line color. This vectorized “urchin plot” displays line slopes as an interaction between t50 kinetics and dynamic range (Fig. S1B). For visual comparison between metabolites, a fully labeled web object version of the urchin plot is available online (Supplemental Object1). Here the urchin plot revealed distinctive features of dissection-induced metabolomic drift. Significantly changed metabolites binned to the upper left, upper right, lower right, but not lower left quadrants. This pattern indicates that, on average, metabolite decreases reach time of 50% change more rapidly than metabolite increases. Furthermore, the absence of metabolites in the lower left quadrant is consistent with larger macromolecules not detected by small molecule metabolomic analysis, like glycogen, being slowly depleted while feeding pathways showing metabolite accumulation. Overall, these data show post-tissue dissection metabolomic drift is diverse in kinetics and magnitudes, with both observed metabolomic and likely non-observed macromolecular metabolic interactions.
4.2. The post-dissection liver metabolome is rapidly remodeled by hypoxia
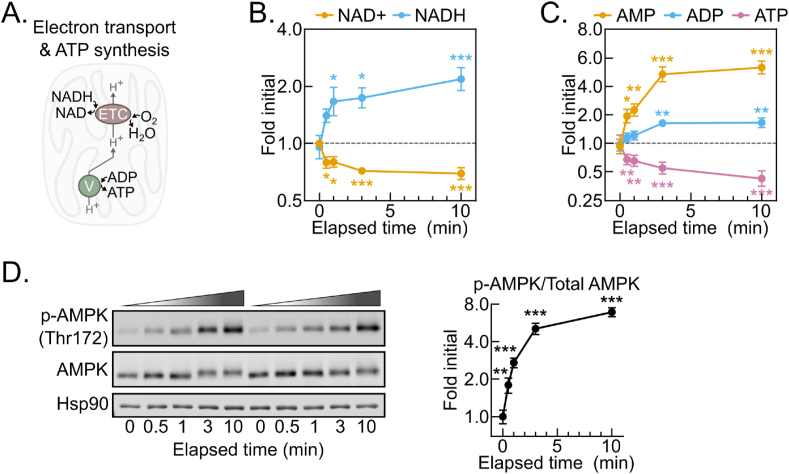
To begin evaluating changes in individual metabolites, we asked whether metabolites known to be influenced by hypoxia changed as expected during delayed tissue freezing. The ratio of the critical redox cofactor couple NAD+ and NADH is highly sensitive to hypoxia. NADH accumulation results from hypoxia because the mitochondrial electron transport chain becomes fully reduced in the absence of a terminal electron acceptor (Figure 2A). NADH levels were strikingly increased by 30 s, reaching a near peak at 2-fold initial levels with minimal further increases by 10 min (Figure 2B). This was accompanied by a concomitant decrease in NAD+ levels and, thus, an increased NADH:NAD+ ratio (Figure 2B, S2A). Without electron transport chain activity, lost mitochondrial inner membrane proton motive force results in decreased mitochondrial ATP synthesis. ATP levels significantly decreased by 30 s of tissue dissection and continued decreasing throughout 10 min (Figure 2C). In parallel, ADP modestly increased after 3 min, while AMP levels doubled by 30 s and accumulated to greater than 500% of initial levels after 10 min. As an additional control, we tested whether phosphorylation of the master energy-sensing kinase AMPK tracked with the AMP:ATP ratio [[14], [15], [16], [17]]. The AMP:ATP ratio increased nearly 3-fold by 30 s after dissection, and it continued to increase through 10 min (Fig. S2B). In parallel, AMPK alpha subunit threonine 172 phosphorylation, which is known to increase when the AMP:ATP ratio increases, rose linearly through 3 min, reaching a nearly 700% increase by 10 min (Figure 2D). Thus, although the tissue ischemia resulting from dissection disrupts aspects of tissue metabolism beyond oxygen supply, the rapid shift to a reduced redox balance and concomitantly decreased energy state are consistent with hypoxia being a major driver of post-dissection metabolomic remodeling.
Figure 2.
The post-dissection liver metabolome is rapidly remodeled by hypoxia. (A) Schematic depicting the dependence of the NADH oxidation and O2 reduction activities of the electron transport chain (ETC) on the ATP synthesis activity of the ATP synthase/Complex V (V). (B–C) Line graphs showing the NAD+ and NADH (B) and AMP, ADP, and ATP (C) metabolite fold changes by delayed freezing time-point compared to time zero. Data are presented as the mean ± SEM. P-values were calculated by one-way ANOVA with the post-hoc Holm-Sidak multiple comparison test (n = 6 biological replicates). (D) Representative Western blot of liver tissue p-AMPK (Thr172), AMPK, and Hsp90 by delayed freezing time-point and a line graph showing p-AMPK/AMPK quantification. Quantified densiometric data are presented as the mean ± SEM. P-values were calculated by one-way ANOVA with the post-hoc Holm-Sidak multiple comparison test (n = 6 biological replicates). ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001; Related to Figure S2 and Table S1.
4.3. Post-dissection liver hypoxia leads to increased glycolytic and decreased TCA cycle activity
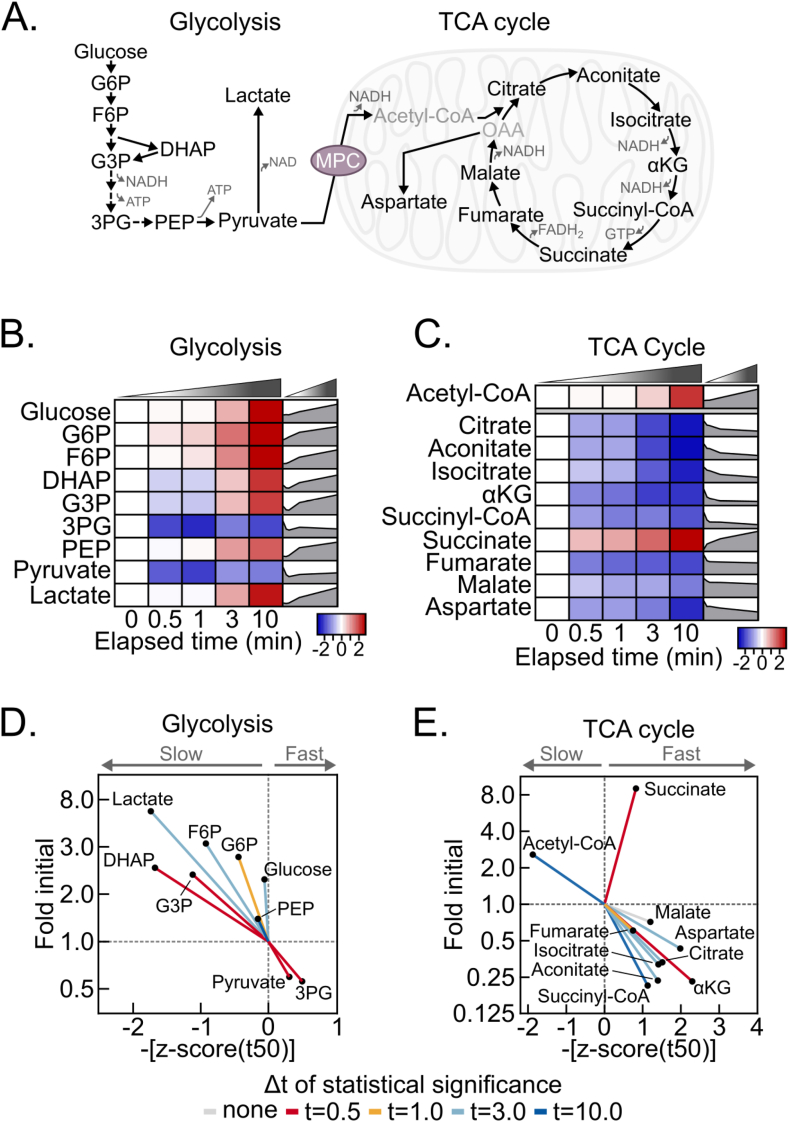
We next considered the rate at which a reduced redox balance and decreased energy state propagate to changes in central carbon metabolism metabolites. We examined metabolites in glycolysis and the TCA cycle, which are regulated by the NADH:NAD+ and AMP:ATP ratios (Figure 3A). Indeed, a hypoxic shift to increased glycolytic and decreased TCA cycle activities throughout the time-course is evident from a final, greater than 20-fold increase in the lactate:citrate ratio (Fig. S3A). In accord, most glycolytic intermediates increased, except for 3-phosphoglycerate and pyruvate, likely because of relatively increased consumption to support ATP production and NAD+ regeneration, respectively (Fig. 3A, 3B, S3B). Conversely, but also consistent with an elevated NADH:NAD+ ratio, TCA cycle intermediates decreased, except for succinate. Notably, succinate is a well-recognized marker of hypoxia and increased 9-fold by 10 min, which further validates our approach (Fig. 3C and S3B).
Figure 3.
Post-dissection liver hypoxia leads to increased glycolytic and decreased TCA cycle activity. (A) Schematic depicting glycolysis and the TCA cycle. G6P, glucose 6-phosphate; F6P, fructose 6-phosphate; DHAP, dihydroxyacetone phosphate; G3P, glyceraldehyde 3-phosphate; 3 PG, 3-phosphoglycerate; PEP, phosphoenolpyruvate; αKG, α-ketoglutarate; OAA, oxaloacetate; MPC, mitochondrial pyruvate carrier. (B–C) Heatmaps depicting the z-scores of glycolytic (B) and TCA cycle (C) metabolite fold changes by delayed freezing time-point compared to time zero. The heatmaps are not time-scaled. Corresponding time-scaled line plots are to the right of each heatmap (n = 5–6 biological replicates). (D–E) Urchin plots for glycolysis (D) and the TCA cycle (E) showing line slopes as an interaction between the t50 kinetics and dynamic ranges of metabolite fold changes with delayed freezing. Metabolites were converted to line vectors displaying dynamic range (10-minute fold change compared to time zero; y-axis) and the negative z-score of the time required for one-half maximum fold change (-[z-score (t50)]; x-axis). Line color indicates the time-point by which statistically significant change was reached calculated by one-way ANOVA with the post-hoc Holm-Sidak multiple comparison test (n = 5–6 biological replicates). Related to Figure S3 and Table S1 and S2. (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
Urchin plots for both glycolysis and the TCA cycle further revealed distinctive features of post-dissection, ex vivo metabolism (Figure 3D and E). For glycolysis, increased versus decreased metabolites binned oppositely to the upper left and lower right quadrants, respectively. Because glucose also increased, this is consistent with a relatively sustained rate of glycogenolysis feeding the overall increases in glycolytic intermediates. In contrast to glycolysis, TCA cycle metabolites strongly binned to the lower right quadrant, with succinate markedly diverging in the upper right (Figure 3E). This suggests that hepatic TCA cycle metabolites are highly sensitive to hypoxia and is consistent with bi-directional flux into succinate as a metabolic dead-end during hypoxia. In addition to succinate, acetyl-CoA binned distinctly, to the upper left, possibly because of cytosolic accumulation. Together, these data demonstrate complex, time-dependent responses of glycolysis and the TCA cycle to dissection-induced hypoxia. This is important to consider for rigorous metabolomic analysis, because it demonstrates that metabolomic changes within and across adjoining pathways during delayed freezing are not equal, and thus they do normalize away during data analysis.
4.4. Post-dissection liver hypoxia leads to enormous increases in purine degradation metabolites
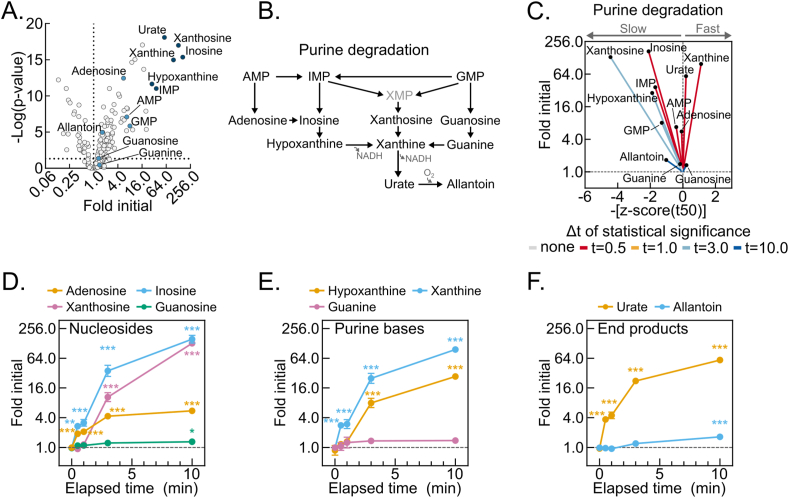
To identify high dynamic-range markers of hepatic post-dissection, hypoxia-induced metabolomic drift, we looked for relationships among the metabolites with the highest fold increases (Figure 4A). Strikingly, the six most increased metabolites were products of the redox- and oxygen-regulated purine degradation pathway (Figure 4B). An urchin plot of the purine degradation pathway shows that its metabolites have distinctive kinetic and magnitude of change features (Figure 4C). In contrast to components of glycolysis and the TCA cycle, all purine nucleotide degradation products increased. Moreover, the line vectors for these metabolites generally centered around the mean line and displayed steep slopes with enormous fold changes. Several purine nucleotide degradation products showed greater than 50-fold increases by 10 min, which for inosine and xanthine exceeded 100-fold (Figure 4D–F). This combination of high-magnitude increases and generally near-mean t50 kinetics suggests that purine nucleotide degradation may serve as a representative, sensitive marker of hepatic post-dissection metabolomic decay.
Figure 4.
Post-dissection liver hypoxia leads to enormous increases in purine degradation metabolites. (A) Volcano plot showing the interaction between metabolite fold changes and associated p-values at 10-minutes delayed freezing compared to time zero. Purine degradation pathway metabolite data points are labeled and colored blue, with the six most changed colored dark blue. The -log transformed p-values were calculated by one-way ANOVA with the post-hoc Holm-Sidak multiple comparison test (n = 5–6 biological replicates). (B) Schematic of the purine degradation pathway. (C) Urchin plots for the purine degradation pathway showing line slopes as an interaction between the t50 kinetics and dynamic ranges of metabolite fold changes with delayed freezing. Metabolites were converted to line vectors displaying dynamic range (10-minute fold change compared to time zero; y-axis) and the negative z-score of the time required for one-half maximum fold change (-[z-score (t50)]; x-axis). Line color indicates the time-point by which statistically significant change was reached calculated by one-way ANOVA with the post-hoc Holm-Sidak multiple comparison test (n = 5–6 biological replicates). (D–F) Line graphs showing the purine nucleosides (D), purine bases (E), and purine degradation end-products (F) fold changes by delayed freezing time-point compared to time zero. Data are presented as the mean ± SEM. P-values were calculated by one-way ANOVA with the post-hoc Holm-Sidak multiple comparison test (n = 6 biological replicates). ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001. Related to Tables S1 and S2. (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
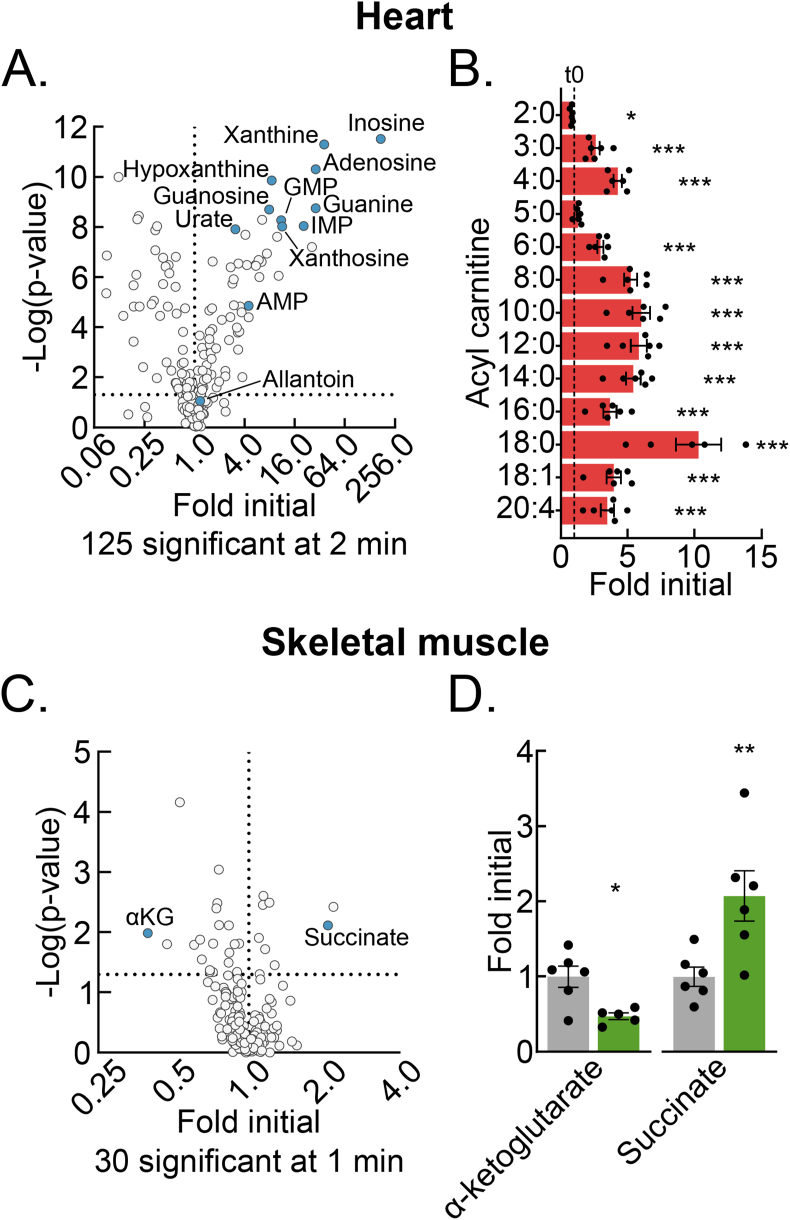
4.5. Delayed post-dissection freezing of heart and skeletal muscle results in a shift to hypoxic metabolism
Compared to the liver, other tissues may have different metabolomic sensitivities to and signatures of delayed post-dissection freezing. To address this question, we performed test of concept experiments with heart and skeletal muscle, two mouse tissues frequently investigated in the translational metabolism literature [[18], [19], [20]]. Heart muscle, like the liver, has a constitutively high metabolic rate but, unlike the liver, its primary function is contractile. Skeletal muscle, like the heart, is striated and has a primary contractile function but, unlike the heart, has a low resting metabolic rate. Thus, comparing metabolomic changes among liver, heart muscle, and skeletal muscle after delayed freezing could inform both generalized properties and tissue-specific functional and metabolic rate correlates of delayed freezing.
Given the high metabolic rate of heart muscle, we chose a 2-minute freezing delay as a period likely to reveal clear signatures of post-dissection, ex vivo metabolism. Metabolomic change with a 2-minute delay to freeze was substantial. Like the liver, this included many-fold increases in several purine nucleotide degradation products and increases in the NADH:NAD+ and AMP:ATP ratios (Figures 5A, S4A–B, Table S3). Acyl-carnitines, which had increased a few-fold in the liver by 10 min delayed freezing, were increased several-fold in the heart at the 2-minute timepoint (Figure 5B, Tables S1 and S3). This trend is consistent with the rapid failure of high baseline β-oxidation. It suggests that the accumulation of acyl-carnitines may be an additional heart-specific signature of hypoxic metabolism.
Figure 5.
Delayed post-dissection freezing of heart and skeletal muscle results in a shift to hypoxic metabolism. (A) Volcano plot showing the interaction between the heart metabolite fold changes and associated p-values at 2-minutes delayed freezing compared to time zero. The purine degradation pathway metabolite data points are labeled and colored blue. The -log transformed p-values were calculated by Student's t-test. The number of significant differences for each time-point are listed in the panel (n = 5–6 biological replicates). (B) Bar graph showing the heart acyl-carnitine fold changes at 2-minutes delayed freezing compared to time zero. Data are presented as the mean ± SEM. P-values were calculated by Student's t-test (n = 5–6 biological replicates). (C) Volcano plot showing the interaction between the skeletal muscle metabolite fold changes and associated p-values at 1-minute delayed freezing compared to time zero. α-Ketoglutarate and succinate data points are labeled and colored blue. The -log transformed p-values were calculated by Student's t-test. The number of significant differences for each time-point are listed in the panel (n = 5–6 biological replicates). (D) Bar graph showing the skeletal muscle α-ketoglutarate and succinate fold changes at 1-minute delayed freezing compared to time zero. Data are presented as the mean ± SEM. P-values were calculated by Student's t-test (n = 5–6 biological replicates). ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001; Related to Figure S4 and Tables S3 and S4. (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
Given the slow metabolic rate of resting skeletal muscle, we used a shorter freezing delay to test for a potential window of metabolomic stability, before the onset of dissection-induced hypoxic metabolism. We utilized the mouse tibialis anterior (TA) muscle, which can be quickly dissected to obtain a representative time zero sample as a control. Thirty metabolites significantly changed with a 1-minute delay to freeze (Figure 5C, Table S4). Notably, the NADH:NAD+ ratio, but not AMP:ATP ratio was significantly changed (Figs. S4C–D). Lack of change in the latter may reflect initial buffering by the large skeletal muscle creatine phosphate pool, which decreased but did not reach statistical significance by 1 min. Like for the liver and heart muscle, the highly oxygen-sensitive TCA cycle metabolite pair, α-ketoglutarate and succinate, which are known to reciprocally regulate several hypoxia-responsive transcriptional control programs, significantly decreased and increased, respectively (Figure 5D). These results from skeletal muscle demonstrate that even metabolically slower tissues must be rapidly frozen after dissection to preserve the in vivo metabolome. Together, data from liver, heart muscle, and skeletal muscle show that although common metabolomic features of hypoxia persist across different tissues, that individual tissues also have unique signatures of post-dissection, hypoxic metabolism.
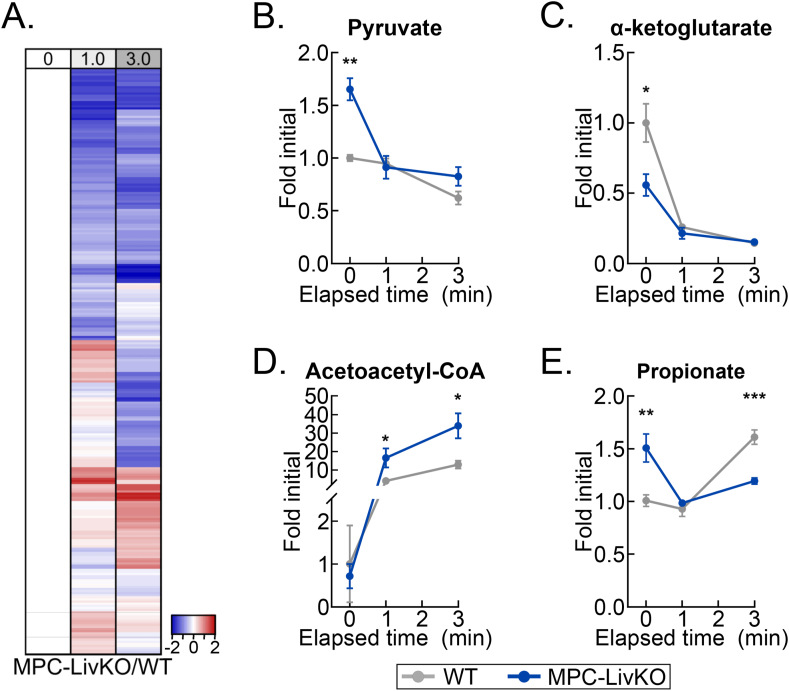
4.6. Delayed liver freezing leads to both false negative and false positive between-genotype metabolite level differences
A critical question for rigorous metabolomic investigation is how individual metabolite levels in control versus experimentally modified tissues change during post-harvest delays to freeze. If such metabolomic drift across different conditions is similar, then equally delaying freezing time could enable accurate comparisons when immediate freezing is not ideal for experimental design. To test this possibility, we returned to our well-vetted liver system and compared dissection-induced, hypoxic metabolomic drift between WT and liver-specific mitochondrial pyruvate carrier (MPC) knockout (MPC-LivKO) mouse livers. Liver tissue was frozen immediately after 0 (about 1 s) and 1- and 3-minute delays. When frozen immediately, as previously reported, MPC-LivKO and WT livers displayed many between-genotype metabolomic differences (Figure 5A, Table S5) [9,21,22]. To directly compare relative rates of change between individual metabolites, MPC-LivKO and WT metabolite-level ratios at all time points were normalized to the time zero ratio. This normalized the time zero ratios to 1, with the per metabolite ratios at 1- and 3-minute freezing delays being greater or less than 1 if they increased more or less in MPC-LivKO mice, respectively (Table S6). Rendering these between-genotype metabolite-level ratios on a heat map and volcano plot revealed myriad genotypic differences in post-harvest metabolomic change (Figs. 6A and S5B). Specific examples of metabolites for which the between-genotype relative abundance changed are: pyruvate (a greater initial level in MPC-LivKO was lost); α-ketoglutarate (a lesser initial level in MPC-LivKO was lost); acetoacetyl-CoA (after a similar initial level, a greater MPC-LivKO level was gained); and propionate (a greater initial level in MPC-LivKO was inverted to a lesser level) (Figure 6B–E). Thus, genotypic differences in ex vivo tissue metabolism cannot be controlled for by equally delaying time to post-harvest freeze, which leads to both false negative and false positive between-genotype differences.
Figure 6.
Delayed liver freezing leads to both false negative and false positive between-genotype metabolite level differences. (A) Heatmap depicting the z-scores of the MPC-LivKO/WT metabolite-level ratios by delayed freezing time-point compared to the MPC-LivKO/WT ratio at time zero. The heatmap is not time-scaled (n = 4–5 biological replicates). (B–E) Line graphs showing MPC-LivKO (blue) and WT (gray) pyruvate (B), α-ketoglutarate (C), acetoacetyl-CoA (D) and propionate (E) fold changes by delayed freezing time-point. Data are presented as the mean ± SEM. P-values between genotypes were calculated using the Student's t-test (n = 4–5 biological replicates). ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001; Related to Figure S5 and Tables S5 and S6. (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
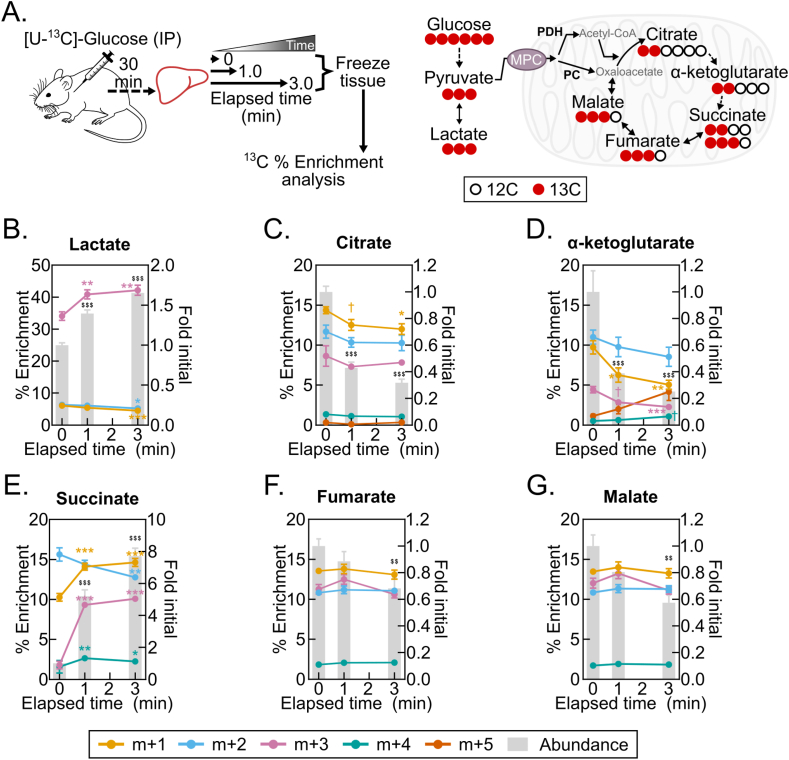
4.7. Post-dissection, ex vivo liver metabolism changes 13C isotopologue abundances and fractional enrichments
Finally, we asked whether liver 13C isotopologue enrichments following [U–13C]-glucose administration are subject to post-harvest change. Stable-isotope tracing is increasingly being employed to investigate substrate-specific metabolism over minutes- to hours-long time intervals [23,24]. This raises the possibility that such a time-integrated signal is more stable than the broad metabolome during post-dissection hypoxia. We administered a single bolus injection of [U–13C]-glucose, harvested livers 30 min later, and then examined changes in liver lactate and TCA cycle intermediate 13C-enrichments following 1- and 3-minute freezing delays (Figure 7A, Table S7). In this pseudo steady-state model, we observed striking post-harvest changes in both metabolite pool sizes and percent 13C enrichments (Figure 7B–G). Notably, the percent enrichment of the succinate m+3 and m+1 isotopologues increased many-fold (Figure 7E), which is consistent with increased flux of labeled pyruvate (m+3) or labeled bicarbonate (m+1) through pyruvate carboxylase and TCA cycle reversal (Figure 7A). Together, these data show that, like metabolite levels, isotopologue abundances and enrichment percentages change with post-dissection freezing delays. This likely represents a shift in the proportional flux from metabolite feeder pools. It demonstrates the requirement to rapidly freeze tissues after dissection to obtain 13C tracing data reflecting the in vivo state.
Figure 7.
Post-dissection, ex vivo liver metabolism changes 13C isotopologue abundances and fractional enrichments. (A) Schematics of the [U–13C]-glucose tracing experiment (left) and the 13C incorporation into TCA cycle intermediates from glycolytically produced pyruvate (right). Pyruvate oxidation by pyruvate dehydrogenase (PDH) results in m+2 TCA cycle isotopologue enrichments, and pyruvate carboxylation by pyruvate carboxylase (PC) results in m+3 TCA cycle isotopologue enrichments. (B–G) Line graphs showing the isotopologue % enrichments (left y-axis) overlaying the corresponding bar graphs depicting the lactate (B), citrate (C), α-ketoglutarate (D), succinate (E), and fumarate (F), and malate (G) fold changes by delayed freezing time-point compared to time zero. Data are presented as the mean ± SEM. P-values were calculated by one-way ANOVA with the post-hoc Holm-Sidak multiple comparison test (n = 8 biological replicates). †p < 0.1, ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001 (% Enrichment); $$p < 0.01, $$$p < 0.001 (Fold change); Related to Table S7.
5. Discussion
Metabolomics is a powerful, mainstreaming research approach that is frequently applied to animal tissues to understand in vivo metabolism. Because broad metabolomic analysis of tissues requires dissection, a major risk to the integrity of metabolomic data is a hypoxia-induced shift of tissue metabolism between the time of disrupted tissue perfusion and freezing. However, most manuscripts reporting metabolomic analysis of mouse or other tissues do not precisely specify the lag time between loss of perfusion and tissue freezing. Thus, it has remained unclear how delayed tissue freezing affects multiple classes of tissue metabolomic data. We addressed this problem by systematically evaluating broad, genotype-specific, and 13C isotopologue metabolomic change resulting from post-dissection, ex vivo mouse tissue metabolism.
Prior work addressing metabolomic change with delayed tissue freezing has been limited in scope or without seconds-to-single minutes coverage of the immediate post-dissection period [[25], [26], [27]]. Notably, the most recent of these carefully demonstrates that delaying mouse liver freezing by as little as 10 seconds alters the lactate:pyruvate ratio and TCA cycle intermediate levels [27]. Although the implications of these findings are critical to the field, how they extend to the larger metabolome has remained unaddressed. Our findings here corroborate this observation and show how it fits within a framework of broad, interdependent post-dissection metabolomic drift. One NMR-based study of intact tumor pieces reports that 16 metabolites in mouse tumor xenografts changed only minimally 30 min after removal but significantly 90 min after removal [28]. However, this publication does not specify the time that elapsed between loss of perfusion and the time zero reference measurement. Thus, substantial metabolomic drift could have occurred that minimized observable changes by the 30-minute time point. Conversely, the observed changes after the very long delay of 90 min might have been chemical versus enzymatic, obscuring the possibility that enzymatic metabolism had ceased much earlier in the time course. In a study of metabolomic and macromolecular change in primary human tumors, only 10% of numerous measured metabolites, lipids, and peptides were found to be affected by a 3-hour delay in freezing, leading to the conclusion that no specific pathway-wide changes occurred. However, the tumors were cut into pieces prior to freezing and, as in the above study, the lag time from loss of perfusion to freezing of the pieces was not stated [29]. Although incomplete technical detail occludes clear interpretation of the data, it is also possible that the findings of lower sensitivity of tumors to hypoxia reflect prior biological adaptation to a lack of perfusion in vivo [[30], [31], [32]]. Future, carefully controlled studies will be required to determine this, which may also apply to poorly vascularized normal tissues like cartilage or other connective tissues. Thus, in comparison to these prior studies, our findings here show that delayed freezing after dissection of liver, heart muscle, and skeletal muscle leads to rapid, extensive metabolomic drift. In addition, the liver dataset provides a resource mapping the time course of broad, ex vivo metabolomic change following dissection. We expect much of this will extend to in vivo ischemia, as we observed for succinate, but this will need to be experimentally tested.
In addition to evaluating the broad metabolome, here we investigated how tissue 13C mass isotopologues change with delayed freezing. In a test of concept experiment, we observed that delayed freezing of the liver leads to altered TCA cycle intermediate 13C isotopologue abundances and fractional enrichments, which were especially prominent for succinate. Numerous prior studies have shown that succinate accumulates with ischemia, but they have not addressed how ex vivo ischemic metabolism impacts 13C labeling [[33], [34], [35]]. Our observation that delayed freezing of liver from mice administered 13C glucose results in not only an increase in the size of the succinate pool but also a many-fold increase in the m+3 and m+1 succinate 13C isotopologues is consistent with reverse TCA cycle flux driven by a high NADH:NAD+ ratio. It is possible that results from a true steady-state would differ from those obtained here with a pseudo steady-state 30 min after i.p. 13C-glucose injection. However, because ex vivo non-steady state hepatic 13C-fluxes are likely negligible compared to the massive changes in metabolism induced by dissection, we expect such differences would also be negligible. Overall, this result suggests that isotopologue distributions of metabolites with differentially redox-regulated feeder pools may be especially sensitive to delays in tissue freezing.
Given that ex vivo tissue metabolism after dissection rapidly alters the broad in vivo and isotope-enriched metabolomes, we highlight best practices for sample collection to obtain accurate in vivo metabolomic data. For most tissues, the need for rapid, controlled freezing can be accommodated by using a liquid nitrogen-cooled Wollenberger-like freeze-clamp device. This instrument maximizes the heat transfer rate and results in nearly instantaneous sample freezing [36]. Therefore, samples destined for metabolomics must be prioritized, and it is likely not possible to preserve the in vivo metabolome of multiple tissues collected from a single animal. Here, the liver and heart showed rapid, high magnitude increases in purine nucleotide degradation products that could serve as a within-study marker of metabolomic quality for these tissues. This is also consistent with the elevated purine degradation products observed in the plasma of hypoxic newborn humans, mouse retinas after extended incubation in culture media, and mouse tissues with protracted ischemia [26,35,37]. Although such changes were not observed in skeletal muscle following a 1-minute delay in freezing, the α-ketoglutarate:succinate ratio decreased several-fold, suggesting this ratio could serve as a skeletal muscle-specific marker of post-dissection metabolism. Thus, when investigating animal tissue metabolomics, tissue- and condition-specific pilot experiments to determine signatures of metabolomic change after loss of perfusion may be useful for informing data quality.
Lastly, we note key limitations of this study, which was not designed to investigate variables other than how delayed mouse tissue freezing after dissection alters the in vivo metabolome. First, we do not account for the effects of anesthesia, which has previously been shown to impact the metabolome [38,39]. For purposes of experimental control, we used isoflurane as a rapidly acting anesthesia with predictable dose- and timing-dependent effects. However, the time zero values that we calculated fold changes from could differ by anesthetic, which could influence fold-change results. Second, compared to humans and larger animals, mice have a much higher metabolic rate. Thus, the rate of post-dissection metabolomic drift in humans and other animals may be slower, which may permit mild freezing delays before the onset of confounding metabolomic drift. Nonetheless this possibility needs to be experimentally tested. Third, we do not attempt to define tolerances for delayed freezing. Based on the substantial metabolomic change observed by 30 s delayed mouse liver freezing, we expect such tolerances to be minimal for moderate to highly metabolically active tissues. In this case, a few seconds window of tolerance would offer minimal practical value and attempting to utilize such a window would increase risk of metabolomic drift. Fourth, our data are presented as fold changes and do not overtly account for differences in absolute metabolite pool sizes. This is important to recognize as small fold differences can equate to large absolute changes when a metabolite has a large pool and vice versa. For example, the dramatic fold changes in purine degradation pathway intermediates accrue from basal pool sizes that are 10-fold less than some TCA cycle intermediates [22,40]. Finally, although we demonstrate that post-dissection metabolomic change is a major risk to research rigor and data integrity, we speculate that such change nonetheless may inform biology. As we observed comparing WT MPC and LivKO liver tissue, delayed freezing led to both false negative and false positive between-genotype differences. Because these differential, genotype-specific rates of metabolomic change reflect differences in metabolic programming, they may inform secondary mechanistic experiments to define the underlying molecular bases. Yet, even in this case, to be most useful, a well-controlled time zero is still required to fully detect such ex vivo metabolomic divergence.
6. Conclusions
In conclusion, in this study we provide a novel illustration of how delayed mouse tissue freezing leads to broad, hypoxia-driven metabolomic remodeling. This is consistent with the fact that, compared to the proteome and transcriptome, the metabolome is highly dynamic, responding nearly instantaneously to metabolic stimuli and other cues. Whereas the transcriptome and base proteome, independent of modification state, are stable for minutes to hours after tissue dissection, we show here that the broad metabolome of mouse liver and likely other metabolically active tissues markedly changes by 30 s after dissection. Because obtaining accurate results requires that the in vivo metabolome is fixed prior to analysis, achieving research rigor in metabolomics brings unique and unavoidable challenges for accurate experimental design. These additional challenges to ensuring data integrity, combined with the rapidly escalating use of metabolomics as an investigative method, make this an important time for the field to embrace best practices. We suggest that all manuscripts reporting metabolomic data clearly and precisely describe experimental procedures and minimize the time elapsing between loss of tissue perfusion and tissue freezing.
Author contributions
EBT conceived the project. AJR and EBT designed experiments. AJR, DJP, ASK, and DAS performed experiments. EBT, AJR, and NB performed data analysis and data visualization. AJR, NB, and EBT wrote and edited the manuscript. All authors read and commented on a draft manuscript.
Acknowledgements
We are grateful to the University of Iowa Fraternal Order of Eagles Diabetes Research Center Metabolomics Core Facility for technical assistance. This work was supported by grants NIH R01 DK104998 (EBT), University of Iowa Healthcare Distinguished Scholars Award (EBT), T32 DK112751 to E. Dale Abel (DAS), ADA 1-18-PDF-060 and AHA CDA851976 (AJR), and the Carver College of Medicine Medical Scientist Training Program (MSTP) grant NIH T32 GM139776 to Steven Lentz (DJP). We are grateful to Dr. Bo Ram Kim, Dr. Alejandro Pezzulo, and Dr. Leonid Zingman, and the Scientific Editing and Research Communication Core (Dr. Christine Blaumueller), at the University of Iowa Carver College of Medicine for critically evaluating a draft manuscript.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.molmet.2022.101596.
CONFLICT OF INTEREST
The authors declare no competing interests.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
Data availability
Data will be made available on request.
References
- 1.Patti G.J., Yanes O., Siuzdak G. Innovation: metabolomics: the apogee of the omics trilogy. Nature Reviews Molecular Cell Biology. 2012;13(4):263–269. doi: 10.1038/nrm3314. Epub 20120322. PubMed PMID: 22436749; PubMed Central PMCID: PMC3682684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wishart D.S. Metabolomics for investigating physiological and pathophysiological processes. Physiological Reviews. 2019;99(4):1819–1875. doi: 10.1152/physrev.00035.2018. PubMed PMID: 31434538. [DOI] [PubMed] [Google Scholar]
- 3.Johnson C.H., Ivanisevic J., Siuzdak G. Metabolomics: beyond biomarkers and towards mechanisms. Nature Reviews Molecular Cell Biology. 2016;17(7):451–459. doi: 10.1038/nrm.2016.25. Epub 20160316. PubMed PMID: 26979502; PubMed Central PMCID: PMC5729912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tweeddale H., Notley-McRobb L., Ferenci T. Effect of slow growth on metabolism of Escherichia coli, as revealed by global metabolite pool (“metabolome”) analysis. Journal of Bacteriology. 1998;180(19):5109–5116. doi: 10.1128/JB.180.19.5109-5116.1998. PubMed PMID: 9748443; PubMed Central PMCID: PMC107546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Falkowski P.G., Isozaki Y. Geology. The story of O2. Science. 2008;322(5901):540–542. doi: 10.1126/science.1162641. PubMed PMID: 18948530. [DOI] [PubMed] [Google Scholar]
- 6.Jablonska J., Tawfik D.S. The evolution of oxygen-utilizing enzymes suggests early biosphere oxygenation. Nature Ecology and Evolution. 2021;5(4):442–448. doi: 10.1038/s41559-020-01386-9. Epub 20210225. PubMed PMID: 33633374. [DOI] [PubMed] [Google Scholar]
- 7.Raymond J., Segre D. The effect of oxygen on biochemical networks and the evolution of complex life. Science. 2006;311(5768):1764–1767. doi: 10.1126/science.1118439. PubMed PMID: 16556842. [DOI] [PubMed] [Google Scholar]
- 8.Reinhard C.T., Planavsky N.J., Olson S.L., Lyons T.W., Erwin D.H. Earth’s oxygen cycle and the evolution of animal life. Proceedings of the National Academy of Sciences of the United States of America. 2016;113(32):8933–8938. doi: 10.1073/pnas.1521544113. Epub 20160725. PubMed PMID: 27457943; PubMed Central PMCID: PMC4987840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gray L.R., Sultana M.R., Rauckhorst A.J., Oonthonpan L., Tompkins S.C., Sharma A., et al. Hepatic mitochondrial pyruvate carrier 1 is required for efficient regulation of gluconeogenesis and whole-body glucose homeostasis. Cell Metabolism. 2015;22(4):669–681. doi: 10.1016/j.cmet.2015.07.027. Epub 20150903. PubMed PMID: 26344103; PubMed Central PMCID: PMC4754674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Li B., Tang J., Yang Q., Li S., Cui X., Li Y., et al. NOREVA: normalization and evaluation of MS-based metabolomics data. Nucleic Acids Research. 2017;45(W1):W162–W170. doi: 10.1093/nar/gkx449. PubMed PMID: 28525573; PubMed Central PMCID: PMC5570188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yuan J., Bennett B.D., Rabinowitz J.D. Kinetic flux profiling for quantitation of cellular metabolic fluxes. Nature Protocols. 2008;3(8):1328–1340. doi: 10.1038/nprot.2008.131. PubMed PMID: 18714301; PubMed Central PMCID: PMC2710581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Babicki S., Arndt D., Marcu A., Liang Y., Grant J.R., Maciejewski A., et al. Heatmapper: web-enabled heat mapping for all. Nucleic Acids Research. 2016;44(W1):W147–W153. doi: 10.1093/nar/gkw419. Epub 20160517. PubMed PMID: 27190236; PubMed Central PMCID: PMC4987948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Helwig N.E. In: SAGE research methods foundations. Paul Atkinson S.D., Cernat Alexandru., Sakshuag Joseph W., Williams Richard .A., editors. SAGE Publications Ltd.; 2020. Multiple and generalized nonparametric regression. [Google Scholar]
- 14.Winder W.W. Energy-sensing and signaling by AMP-activated protein kinase in skeletal muscle. Journal of Applied Physiology. 2001;91(3):1017–1028. doi: 10.1152/jappl.2001.91.3.1017. PubMed PMID: 11509493. [DOI] [PubMed] [Google Scholar]
- 15.Hardie D.G., Sakamoto K. AMPK: a key sensor of fuel and energy status in skeletal muscle. Physiology. 2006;21:48–60. doi: 10.1152/physiol.00044.2005. PubMed PMID: 16443822. [DOI] [PubMed] [Google Scholar]
- 16.Hardie D.G. AMP-activated protein kinase: an energy sensor that regulates all aspects of cell function. Genes & Development. 2011;25(18):1895–1908. doi: 10.1101/gad.17420111. PubMed PMID: 21937710; PubMed Central PMCID: PMC3185962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Garcia D., Shaw R.J. AMPK: mechanisms of cellular energy sensing and restoration of metabolic balance. Molecular Cell. 2017;66(6):789–800. doi: 10.1016/j.molcel.2017.05.032. PubMed PMID: 28622524; PubMed Central PMCID: PMC5553560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.McGarrah R.W., Crown S.B., Zhang G.F., Shah S.H., Newgard C.B. Cardiovascular metabolomics. Circulation Research. 2018;122(9):1238–1258. doi: 10.1161/CIRCRESAHA.117.311002. PubMed PMID: 29700070; PubMed Central PMCID: PMC6029726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hargreaves M., Spriet L.L. Skeletal muscle energy metabolism during exercise. Nature Metabolism. 2020;2(9):817–828. doi: 10.1038/s42255-020-0251-4. Epub 20200803. PubMed PMID: 32747792. [DOI] [PubMed] [Google Scholar]
- 20.Lopaschuk G.D., Karwi Q.G., Tian R., Wende A.R., Abel E.D. Cardiac energy metabolism in heart failure. Circulation Research. 2021;128(10):1487–1513. doi: 10.1161/CIRCRESAHA.121.318241. Epub 20210513. PubMed PMID: 33983836; PubMed Central PMCID: PMC8136750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.McCommis K.S., Chen Z., Fu X., McDonald W.G., Colca J.R., Kletzien R.F., et al. Loss of mitochondrial pyruvate carrier 2 in the liver leads to defects in gluconeogenesis and compensation via pyruvate-alanine cycling. Cell Metabolism. 2015;22(4):682–694. doi: 10.1016/j.cmet.2015.07.028. Epub 20150903. PubMed PMID: 26344101; PubMed Central PMCID: PMC4598280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rauckhorst A.J., Gray L.R., Sheldon R.D., Fu X., Pewa A.D., Feddersen C.R., et al. The mitochondrial pyruvate carrier mediates high fat diet-induced increases in hepatic TCA cycle capacity. Molecular Metabolism. 2017;6(11):1468–1479. doi: 10.1016/j.molmet.2017.09.002. Epub 20170918. PubMed PMID: 29107293; PubMed Central PMCID: PMC5681281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jang C., Chen L., Rabinowitz J.D. Metabolomics and isotope tracing. Cell. 2018;173(4):822–837. doi: 10.1016/j.cell.2018.03.055. PubMed PMID: 29727671; PubMed Central PMCID: PMC6034115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kim I.Y., Suh S.H., Lee I.K., Wolfe R.R. Applications of stable, nonradioactive isotope tracers in in vivo human metabolic research. Experimental and Molecular Medicine. 2016;48 doi: 10.1038/emm.2015.97. Epub 20160115. PubMed PMID: 26795236; PubMed Central PMCID: PMC4686699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lowry O.H., Passonneau J.V., Hasselberger F.X., Schulz D.W. Effect of ischemia on known substrates and cofactors of the glycolytic pathway in brain. Journal of Biological Chemistry. 1964;239:18–30. PubMed PMID: 14114842. [PubMed] [Google Scholar]
- 26.Zhu S., Yam M., Wang Y., Linton J.D., Grenell A., Hurley J.B., et al. Impact of euthanasia, dissection and postmortem delay on metabolic profile in mouse retina and RPE/choroid. Experimental Eye Research. 2018;174:113–120. doi: 10.1016/j.exer.2018.05.032. Epub 20180601. PubMed PMID: 29864440; PubMed Central PMCID: PMC6110973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fu X., Deja S., Kucejova B., Duarte J.A.G., McDonald J.G., Burgess S.C. Targeted determination of tissue energy status by LC-MS/MS. Analytical Chemistry. 2019;91(9):5881–5887. doi: 10.1021/acs.analchem.9b00217. Epub 20190412. PubMed PMID: 30938977; PubMed Central PMCID: PMC6506803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Haukaas T.H., Moestue S.A., Vettukattil R., Sitter B., Lamichhane S., Segura R., et al. Impact of freezing delay time on tissue samples for metabolomic studies. Frontiers in Oncology. 2016;6:17. doi: 10.3389/fonc.2016.00017. Epub 20160128. PubMed PMID: 26858940; PubMed Central PMCID: PMC4730796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mock A., Rapp C., Warta R., Abdollahi A., Jager D., Sakowitz O., et al. Impact of post-surgical freezing delay on brain tumor metabolomics. Metabolomics. 2019;15(5):78. doi: 10.1007/s11306-019-1541-2. Epub 20190513. PubMed PMID: 31087206. [DOI] [PubMed] [Google Scholar]
- 30.DeBerardinis R.J., Chandel N.S. Fundamentals of cancer metabolism. Science Advances. 2016;2(5) doi: 10.1126/sciadv.1600200. Epub 20160527. PubMed PMID: 27386546; PubMed Central PMCID: PMC4928883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Martinez-Reyes I., Chandel N.S. Cancer metabolism: looking forward. Nature Reviews Cancer. 2021;21(10):669–680. doi: 10.1038/s41568-021-00378-6. Epub 20210716. PubMed PMID: 34272515. [DOI] [PubMed] [Google Scholar]
- 32.Zhong H., De Marzo A.M., Laughner E., Lim M., Hilton D.A., Zagzag D., et al. Overexpression of hypoxia-inducible factor 1alpha in common human cancers and their metastases. Cancer Research. 1999;59(22):5830–5835. PubMed PMID: 10582706. [PubMed] [Google Scholar]
- 33.Taegtmeyer H. Metabolic responses to cardiac hypoxia. Increased production of succinate by rabbit papillary muscles. Circulation Research. 1978;43(5):808–815. doi: 10.1161/01.res.43.5.808. PubMed PMID: 709743. [DOI] [PubMed] [Google Scholar]
- 34.Freminet A., Leclerc L., Poyart C., Huel C., Gentil M. Alanine and succinate accumulation in the perfused rat heart during hypoxia. Journal of Physiology. 1980;76(2):113–117. PubMed PMID: 7400977. [PubMed] [Google Scholar]
- 35.Chouchani E.T., Pell V.R., Gaude E., Aksentijevic D., Sundier S.Y., Robb E.L., et al. Ischaemic accumulation of succinate controls reperfusion injury through mitochondrial ROS. Nature. 2014;515(7527):431–435. doi: 10.1038/nature13909. Epub 20141105. PubMed PMID: 25383517; PubMed Central PMCID: PMC4255242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wollenberger A., Ristau O., Schoffa G. [A simple technic for extremely rapid freezing of large pieces of tissue] Pflugers Archiv für die Gesamte Physiologie des Menschen und der Tiere. 1960;270:399–412. PubMed PMID: 13845757. [PubMed] [Google Scholar]
- 37.Saugstad O.D. Hypoxanthine as a measurement of hypoxia. Pediatric Research. 1975;9(4):158–161. doi: 10.1203/00006450-197504000-00002. PubMed PMID: 1143950. [DOI] [PubMed] [Google Scholar]
- 38.Overmyer K.A., Thonusin C., Qi N.R., Burant C.F., Evans C.R. Impact of anesthesia and euthanasia on metabolomics of mammalian tissues: studies in a C57BL/6J mouse model. PLoS One. 2015;10(2) doi: 10.1371/journal.pone.0117232. Epub 20150206. PubMed PMID: 25658945; PubMed Central PMCID: PMC4319778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Petucci C., Rojas-Betancourt S., Gardell S.J. Comparison of tissue harvest protocols for the quantitation of acylcarnitines in mouse heart and liver by mass spectrometry. Metabolomics. 2012;8(5):784–792. doi: 10.1007/s11306-011-0370-8. [DOI] [Google Scholar]
- 40.Pillwein K., Jayaram H.N., Weber G. Effect of ischemia on nucleosides and bases in rat liver and hepatoma 3924A. Cancer Research. 1987;47(12):3092–3096. PubMed PMID: 3581061. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data will be made available on request.