Summary
Glioblastoma (GBM) is the most common form of malignant primary brain tumor with a dismal prognosis. Currently, the standard treatments for GBM rarely achieve satisfactory results, which means that current treatments are not individualized and precise enough. In this study, a multiomics-based GBM classification was established and three subclasses (GPA, GPB, and GPC) were identified, which have different molecular features both in bulk samples and at single-cell resolution. A robust GBM poor prognostic signature (GPS) score model was then developed using machine learning method, manifesting an excellent ability to predict the survival of GBM. NVP−BEZ235, GDC−0980, dasatinib and XL765 were ultimately identified to have subclass-specific efficacy targeting patients with a high risk of poor prognosis. Furthermore, the GBM classification and GPS score model could be considered as potential biomarkers for immunotherapy response. In summary, an integrative genomic analysis was conducted to advance individual-based therapies in GBM.
Subject areas: Cancer systems biology, Cancer, Omics, Artificial intelligence
Graphical abstract

Highlights
-
•
A multiomics-based classification of GBM was established
-
•
Single-cell transcriptomic profiling of GBM subclasses was revealed using Scissor
-
•
A robust prognostic risk model was developed for GBM by machine learning method
-
•
Prediction of potential agents based on molecular and prognostic risk stratification
Cancer systems biology; Cancer; Omics; Artificial intelligence
Introduction
Glioblastoma (GBM) is the most lethal and common form of brain tumor. The standard treatment for GBM includes surgical resection, concurrent radiotherapy and chemotherapy, and further adjuvant chemotherapy, but the median survival of approximately 15 months remains unsatisfactory (Tan et al., 2020; Wang et al., 2021). Given the unfavorable overall prognosis for GBM, it is indispensable to establish advanced directives at the early stage of disease. Despite all the strides made in this field, there are currently no standardized prognostic biomarkers for GBM, and reliable biomarkers are still needed. Moreover, temozolomide (TMZ) represents the first and thus far the only agent shown to be effective against GBM (Yang et al., 2022). Therefore, there is also an urgent need to find more available agents to improve the survival rate with GBM. However, little headway has been made in any new attempts to treat GBM.
The era of precision oncology heralds significant prospects for exploring more tailored therapies to fight against aggressive disease. Thus, exploring targeted agents for individuals with specific characteristics may be expected to solve this clinical dilemma. The current multiomics-based GBM classification includes three subtypes: classical, mesenchymal, and proneural (Brennan et al., 2013). Recently, Alhalabi revealed that dasatinib showed distinct therapy responsiveness in different molecular subtypes (Alhalabi et al., 2021). Indeed, increasing evidence suggests that the lack of individual-based therapeutic strategies results in the failure of novel treatment in GBM (Omuro and DeAngelis, 2013). Moreover, high heterogeneity, one of the major hallmarks of GBM, makes it scarcely possible to identify versatile strategies suitable for all GBM patients. However, current GBM classification ignores the internal relationship between GBM clinical classification and therapeutic response, and only a few studies have focused on this issue. Accordingly, it is imperative to generate a novel classification available to explore personalized treatment strategies. Moreover, a number of pharmacogenomic databases, such as Genomics of Drug Sensitivity in Cancer (GDSC) (Yang et al., 2013), the Cancer Therapeutics Response Portal (CTRP)(Rees et al., 2016), PRISM Repurposing (Corsello et al., 2020), the DrugBank (Wishart et al., 2018), and the Drug Repurposing Hub (Corsello et al., 2017), have served as precise and reliable clinical data sources for the exploration of tailored treatment strategies in recent years. GDSC, CTRP, and PRISM could provide drug sensitivity response data for numerous cell lines, which have proven reliable for predicting drug response in clinical samples (Yang et al., 2021a). The Drug Repurposing Hub and DrugBank allow for drug screening against identified targets. However, pharmacogenomic databases are often underutilized during the exploration of tailored treatment strategies in GBM.
To compensate for current shortcomings, in this study, we establish a novel GBM molecular classification that provides a preclinical platform for exploring subtype-specific agents for GBM to improve precision medicine. Moreover, we develop a robust prognostic risk signature for the improvement of clinical risk stratification. Precision treatment strategies are finally generated through a comprehensive in silico analysis leveraging ample pharmacogenomic databases. Collectively, we aim to change the state of the art of population-based therapies in GBM and allow new insights into precision oncology.
Results
CancerSubtypes identifies three clusters in GBM
The detailed analysis procedures of this study are shown in Figure 1. In all, 1333 GBM patients were included in our analysis, and they were divided into five cohorts and assigned to three sets: the training set (TCGA training cohort, n = 323), the internal validation set (TCGA testing cohort, n = 195), and the external validation set (CGGA cohort, n = 374; GSE13041 cohort, n = 267; GSE108474 cohort, n = 174). Detailed information on each cohort is illustrated in Table S1. Figures S1A and S1B present the results of principal component analysis (PCA) before and after batch effect correction of the CGGA and GSE13041 cohorts.
Figure 1.
Overview of the study design
Schematic depicting included datasets, discovery of novel classification, construction of prognostic model and corresponding precision treatment strategies in glioblastoma.
Driver mutations, a small part of all gene aberrations in cancer, are able to confer selective advantages on cancer cells and could cause abnormal and uncontrolled cellular growth (Martinez-Jimenez et al., 2020). In this study, 42 potential diver mutations were identified in the training set by the DriverNet and MutSigCV algorithms (Figure S2) (Table S2). Nine of the 10 most altered genes have been proven to be well-known genetic aberrations associated with GBM by a previous study, including PTEN, TP53, EGFR, NF1, SPTA1, RB1, ATRX, PIK3CA, and PIK3R1 (Reifenberger et al., 2017). The CancerSubtypes package was subsequently used to identify GBM subclusters using multiomics data (the top 1000 genes, copy number variation (CNV) with maximum variations, and 42 mutated genes) derived from the training set. The maximum value of ASW was found at k = 3, which would be indicative of the optimal number of clusters (Figure S3). At this point, 323 GBM patients from the training set were allocated into three clusters, which were correspondingly named Cluster 1 (C1, n = 144), Cluster 2 (C2, n = 77), and Cluster 3 (C3, n = 102).
Significant differences in CNV, mRNA, and mutation profiles were observed among these three clusters (Figure S4A). For survival analysis, the results reflected significant prognostic differences among the three clusters in the training set, with a longer median survival time (MST) for C2 than C1 and C3, for progression-free interval (PFI) (P = 2e-04) and overall survival (OS) (p = 0.0088) as an endpoint (Figures S4B and S4C). Following differential expression analysis, 136 differentially expressed genes (DEGs) were identified among these three clusters (log2FC > 1, adjusted p< 0.05) (Table S3). According to the gene set enrichment analysis (GSEA) results based on the 136 DEGs, E2F targets for C1, G2/M checkpoint for C2, and epithelial mesenchymal transition for C3 represented three pathways that showed the most enrichment significance in corresponding clusters (Figures S4D–S4F). Moreover, the ESTIMATE algorithm was leveraged to calculate the immune scores and stromal scores of the three clusters. We then found that the results of the two scores showed good consistency, and the scores of C3 were the highest (p < 0.0001) (Figures S4G and S4H). We also uncovered the correlation between clusters and immune infiltration in the training set. As shown in Figure S4I, there was a remarkable difference in immune infiltration among the three clusters, with a higher absolute abundance of four immune and one stromal cell populations (fibroblasts, monocytic lineage, myeloid dendritic cells, neutrophils, and T cells) for C3 than for C1 and C2 (p < 0.001) and one immune cell population (cytotoxic lymphocytes) for C2 than for C1 and C3 (p < 0.001).
Establishment of a novel GBM molecular classification based on 136 DEGs
To expand the application of our multiomics GBM classification model to other cohorts, a gene expression-based surrogate was needed. Based on the expression data of 136 DEGs in the training set and all validation sets, a novel GBM molecular classification was established, which comprised three subclasses (i.e., GPA, GPB, and GPC). As shown in Figure S5A, PCA revealed that patients in different subclasses were primarily distributed in three different directions.
Figure 2A depicts the main gene expression data, somatic mutation variation (SNV), CNV and the phenotype features of GBM subclasses. As shown in the heatmap, the expression levels of 136 DEGs were markedly different in the three subclasses. PTEN, the main negative regulatory factor of the PI3K/AKT/mTOR pathway, has been proven to be commonly inactivated or deleted in GBM (Brennan et al., 2013; Verhaak et al., 2010). Consistent with this view, PTEN was identified as the most frequent somatic mutation and CNV of all common alterations in our study. The Genomic Identification of Significant Targets in Cancer (GISTIC) scores of the three subclasses are depicted in Figures 2B and 2C. Figure S6 suggests that GPC was correlated with a high O(6)-methylguanine-DNA methyltransferase (MGMT) promoter methylation level (p < 0.05), the presence of isocitrate dehydrogenase enzyme 1 (IDH1) mutation (p < 0.0001) and the glioma CpG island methylator phenotype (G-CIMP) (p < 0.0001). Moreover, in terms of the correlation between our GBM subclasses and the existing GBM classification, GPA was linked to the classical subtype, GPB was associated with the mesenchymal subtype, and GPC was related to the proneural subtype (p < 0.0001).
Figure 2.
The molecular, mutational and prognostic characteristics of novel classification
(A) An overview of clinical phenotype, somatic mutation and CNV characteristics across three subclasses. Composite GISTIC amplification (B) and deletion (C) profiles for GPA, GPB and GPC with annotations highlighting differences. The survival analysis among three subclasses in the TCGA training cohort (PFI and OS) (D), the TCGA testing cohort (OS) and the CGGA cohort (OS) (E). CNV, Copy Number Variation; PFI, Progression-free interval; OS, Overall survival; GISTIC, Genomic Identification of Significant Targets in Cancer; MGMT: O (6)-methylguanine-DNA methyltransferase; G-CIMP, Glioma CpG island methylator phenotype; IDH1, Isocitrate dehydrogenase enzyme 1.
According to the survival analysis, GPC was associated with better PFS (p = 0.0028) and OS (p = 0.0024) than GPA and GPB in the training set (Figure 2D). The survival analysis of the three subclasses by PFI or OS was also conducted in the validation sets (Figures 2E and S5B–S5D). Consistent with the results of the training set, GPC also had a significantly better prognosis than the other two subclasses in almost all cohorts from the validation sets except the GSE13041 cohort. Although no significant difference was observed in the GSE13041 cohort (p = 0.06), it remained noticeable that GPC was related to a tendency toward better survival.
The subclass-related biological processes were identified by GSEA based on the DEGs (Table S4), and the visual analysis results are presented in Figure S7. Moreover, GSEA of hallmark gene sets revealed that these three subclasses were significantly differentially enriched in metabolism-associated processes (bile acid metabolism, glycolysis, fatty acid metabolism, and cholesterol homeostasis), cell cycle-associated pathways (E2F targets and G2/M checkpoint), immune activation-related processes (interferon alpha response and inflammatory response), and other processes (Figure S8A).
Based on the expression matrix from Ivy Glioblastoma Atlas Project (IGAP) (Figure S8B), we conducted enrichment analysis of anatomic structures in the three subclasses (Puchalski et al., 2018). IGAP researchers have performed RNA-seq on 5 major GBM anatomical structures, including cellular tumor (CT), pseudopalisading cells around necrosis (CTpan), microvascular proliferation (CTmvp), leading edge (LE), and infiltrating tumor (IT), identified from hematoxylin and eosin (H&E) staining. Enrichment analysis showed that GPA was highly enriched in CT samples. GPB was highly derived from CTmvp, and GPC was mainly localized in regions of LE (Figure S8C).
Single-cell transcriptomic context of three GBM subclasses
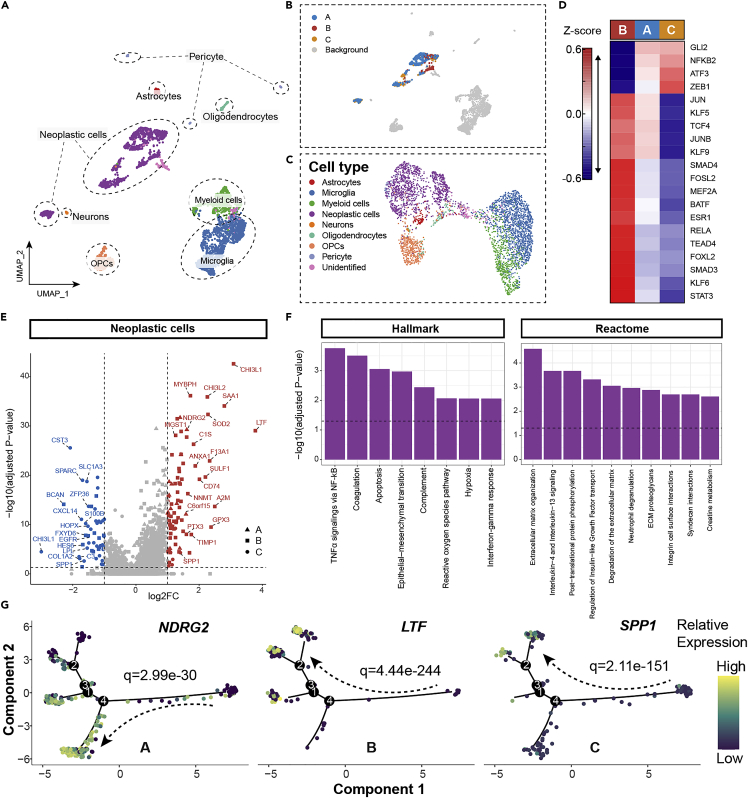
Uniform manifold approximation and projection (UMAP) was adopted to realize cluster visualization of single-cell data (Figure 3A). A total of 3589 cells from GSE84465 were ultimately separated into 8 cell types by manual annotation leveraging canonical gene signature markers of different cell subpopulations (Figure S9A). There was a significant difference between neoplastic cells and non-neoplastic cells through the inferCNV algorithm (Figures S9B and S9C). We then inferred GBM subclass-associated neoplastic cell subpopulations (defined as GPA cells, GPB cells, and GPC cells) using the Scissor algorithm (Figure 3B) (Sun et al., 2022). Scissor can automatically and accurately select highly phenotype-relevant cell subsets from single-cell assays under the guidance of given clinical phenotype information from bulk data, which has been demonstrated to work well in a series of diseases, including lung cancer and melanoma (Sun et al., 2022). To verify the validity of this algorithm in GBM, we used bulk phenotypes, including tumor and normal phenotypes, from the TCGA dataset to guide Scissor analysis. The results showed that 404 Scissor+ cells and 20 Scissor− cells were identified (Figures S9D and S9E). A total of 81.6% of Scissor+ cells were proven to be neoplastic cells, which largely confirmed that Scissor was effective in interpreting GBM datasets (Figure S9E).
Figure 3.
Single-cell transcriptomic characteristics of three GBM subclasses
(A) UMAP plot of scRNA-seq data from four patients.
(B) Cellular maps of subclasses-associated neoplastic cell subpopulations by Scissor displayed by UMAP dimension reduction.
(C) Visualization of single-cell data based on overall TF activities of each cell.
(D) Heatmap of top 20 highly variable TF activities among GPA, GPB and GPC cells. The color of heatmap representing z-scores of TF activities.
(E) Volcano plot of differentially expressed genes among GPA, GPB and GPC cells. The two vertical dashed lines indicate ±1-fold change in gene expression, and the horizontal dashed line demonstrates an FDR cutoff of 0.05. The FDR was the adjusted p value calculated by the two-tailed Wilcoxon rank-sum test.
(F) Enrichment bar plot of differentially expressed genes in GPB cells versus GPC cells based on Reactome and Hallmark pathways. The vertical dashed line denotes an FDR cutoff of 0.05.
(G) Transcriptional changes of NDRG2, LTF and SPP1 along pseudotime. A, GPA cells; B, GPB cells; C, GPC cells; TF, Transcription factor.
In pursuit of upstream regulatory mechanisms, the Dorothea algorithm and the Viper inference tool were applied to infer and score the activities of transcription factor (TF) (Alvarez et al., 2016; Holland et al., 2020). We then revisualized single-cell data based on the overall TF activities of each cell (Figure 3C). We found that neoplastic cells are well circumscribed with non-neoplastic cells. Moreover, TFs with the top 20 highly variant activities were identified (Figure 3D). The activity scores of the 20 TFs in GPA cells were moderate compared with those in GPB cells and GPC cells. We noted higher TF activities for Smad3 in GPB cells, indicative of increased activation of TGF-β/Smad3 signaling, which proved to be a major characteristic of the mesenchymal subtype (related to GPB) (Fan et al., 2021). We also found higher TF activities for STAT3, RELA, TEAD4, and JUN in GPB cells, which are reported to be involved in the regulation of the mesenchymal program in GBM. These results indirectly proved that Scissor can precisely infer most subclass-associated cells to a certain extent (Bhat et al., 2013; Carro et al., 2010; deSouza et al., 2018).
We compared gene expression among GPA, GPB, and GPC cells to further reveal the underlying transcriptional patterns. As a result, 81 upregulated genes (15 genes for GPA cells, 60 genes for GPB cells, 6 genes for GPC cells) and 49 downregulated genes (0 genes for GPA cells, 14 genes for GPB cells, 35 genes for GPC cells) were identified (Figure 3E) (Table S5). Notably, DEGs mainly existed in GPB and GPC cells, which was consistent with the remarkable differences between GPB and GPC cells in terms of TF activities. Of interest, CHI3L1, which was described to figure prominently in modulating tumor microenvironment (TME) of GBM, represented not only the highly overexpressed gene in GPB cells but also the most changed downregulated gene in GPC cells (Chen et al., 2021). The functional enrichment analysis of DEGs in GPB cells over GPC cells ultimately revealed that TNF-α signaling via NF-kB and extracellular matrix organization (part of the TME) were the main enriched pathways and biological processes (Figure 3F). Using the Monocle 2 package, we performed pseudotime analysis of single-cell RNA-seq data to delve into details of DEGs and constructed a cell trajectory of the neoplastic cells in GBM (Figure S9F). NDRG2, LTF, and SPP1, the three most strikingly upregulated genes in corresponding subclass-associated neoplastic cell subpopulations, showed distinct change trends in their expression along pseudotime (Figures 3G and S9G). The composing proportion of three neoplastic cell subpopulations fluctuated dynamically along pseudotime (Figure S9H).
Based on different gene expression patterns, Neftel et al. revealed that human GBM cells corresponded primarily to four states—neural-progenitor-like (NPC-like), mesenchymal-like (MES-like), astrocyte-like (AC-like), and oligodendrocyte-progenitor-like (OPC-like) cellular states at single-cell resolution, which could been considered as classical GBM single cell types (Neftel et al., 2019). According to two dimensional plots and enrichment heatmap (Figures S10A and S10B), AC-like cellular state was linked to GPA cells, MES-like cellular state was related to GPB cells, and NPC- and OPC-like cellular states were associated with GPC cells. In addition, we evaluated GPS score at single-cell resolution. Results showed that neoplastic cells obtained the highest GPS score (Figure S10C), which was in line with this expectation. As for four cellular states, MES-like cells showed the highest GPS score (p < 0.001) (Figure S10D). The finding was also presented at two-dimensional level (Figure S10E).
Construction and prognostic analysis of 36 gene-based GBM poor prognostic signature (GPS) scores
The detailed steps for establishing GPS are described in the STAR methods. Ultimately, a 36-gene list in the WRF model with the minimum out of bag (OOB) error rate was considered GPS and was obtained for further prognostic analysis (Figure S11A; Table S6).
To investigate the prediction ability of GPS in the prognosis of GBM, we performed survival analysis between two groups of patients with high or low GPS scores. Patients from all included cohorts fell into high- and low-GPS groups based on the optimal cutoff points. In the training set, patients in the low-GPS group showed a significantly longer MST than those in the high-GPS group (p < 0.0001) (Figure S12A). This conclusion was further confirmed in validation sets to verify the stability and generalizability of the GPS in different cohorts. As shown in Figure S12A, in agreement with the foregoing conclusion, the results of survival analysis in the validation sets had significant differences in statistics between the two groups, with better survival probability in the low-GPS group than in the high-GPS group (TCGA testing cohort: p = 0.00052; CGGA cohort: p = 0.0026; GSE13041 cohort: p = 0.00043; GSE108474 cohort: p = 0.0019).
To further evaluate the prognostic potential of the GPS, time-dependent receiver operating characteristic (tROC) analysis was conducted to compare the GPS with several previously reported prognostic signatures for GBM, including Hou’s signature (Hou et al., 2019), Lei’s signature (Lei et al., 2021), Zhao’s signature (Zhao et al., 2019c), and Fatai’s signature (Fatai and Gamieldien, 2018) (Table S7). The results of tROC analysis showed that the mean area under the curve values (AUCs) of the GPS ranked first in almost all cohorts (Figures 4A–4C, S11B, and S11C). Actually, in this exceptional case, GPS still exhibited the second highest mean AUCs (Figure S11B). Furthermore, GPS had higher mean AUCs than the others, and the calibration plots suggested that there was a high level of agreement between the GPS model and the ideal model in the training and validation sets (Figures 4D, 4E, S11D, and S11E). Given the above findings, GPS was the most accurate predictor compared with the other four signatures.
Figure 4.
Estimation of prognostic value of 36-gene GPS
(A–C) Time-dependent receiver operating characteristic (tROC) curves comparing GPS and four public signatures in the TCGA training cohort (OS and PFI) (A), the TCGA testing cohort (PFI) (B) and the GSE13041 cohort (OS) (C).
(D) ROC curves of the performance of diverse prognostic signatures in the GSE108474 cohort at 3 and 5 years.
(E) The calibration plots for the comparison of GPS model with ideal model at 3 and 5 years.
(F) Results of the univariate and multivariate Cox regression analysis in the TCGA cohort, the CGGA cohort, the GSE13041 cohort and the GSE108474 cohort. PFI, Progression-free interval; OS, Overall survival.
Univariate and multivariate Cox regression were performed to estimate the independent prognostic value of GPS in GBM patients from TCGA, CGGA, GSE13041, and GSE108474 cohorts (Figure 4F). Variables with statistical significance (p < 0.05) in univariate Cox regression were selected to be used for subsequent multivariate regression. After adjusting for prognosis-related clinical features, variables with p < 0.05 in multivariate Cox regression were regarded as independent predictors. The independent clinical prognostic significance of GPS was verified in TCGA, CGGA, and GSE108474 cohorts by multivariate Cox regression (TCGA: HR = 3.78, 95% CI = 1.80–7.92, p < 0.001; CGGA: HR = 2.33, 95% CI = 1.01–5.42, p = 0.048; GSE108474: HR = 4.10, 95% CI = 1.33–12.57, p = 0.014). The result was not validated in the GSE13041 cohort (HR =1.86, 95% CI =0.86–3.98, p = 0.113), which may be due to its deficiency in GPS-related gene data.
Identification of candidate druggable therapeutic targets for GBM with high GPS scores
Exploration of specific agents for high-risk patients with poor prognosis can facilitate the personalized management of GBM. Thus, 4465 known drug targets from the Drug Repurposing Hub and DrugBank were retained for further two-step analysis to seek potential druggable therapeutic targets for high GPS score GBM. First, by calculating the correlation coefficient between the expression levels of targetable genes and GPS score, we searched for candidate targets (cor > 0.25 and adjusted p < 0.05). Then, identified using correlation analysis between DEMETER/CERES scores and GPS scores, other targets (Spearman’s r < −0.25 and adjusted p < 0.1) were regarded as poor prognosis-related targets. Considering the two results of the above analyses, we ultimately identified 16 potential drug targets (Figure 5A) that may be beneficial to GBM patients with high GPS scores (Table S8).
Figure 5.
Identification of GPS-associated drug targets and therapeutic agents
(A) Volcano plot of Spearman’s correlations and significance between GPS scores and gene expression of drug targets (left). Red dots indicate the significant positive correlations. Volcano plot of Spearman’s correlations and significance between GPS and CERES score of drug targets (mid) as well as between GPS and DEMETER score of drug targets (right) separately. Blue dots indicate the significant positive correlations.
(B) Summary of the most potential therapeutic agents for patients from three subclasses with high risk of poor prognosis according to the evidence from multiple sources. Dose-response curves to 48 h (C) and colony formation assay (D) of NVP-BEZ235, GDC−0980, Dasatinib and XL765 treatment in glioblastoma cell lines of GPA, GPB, and GPC subclasses. Error bars indicate the SEM.
Verifying the availability of drug response databases in clinical samples
This study included three large-scale drug response databases (CTRP, GDSC, and PRISM), from which gene expression and drug sensitivity information on hundreds of cancer cell lines (CCLs) were devoted to estimating the drug sensitivity of clinical samples. Not available agents in 20% of samples, duplicated agents, and hematopoietic and lymphoid tissue-derived cell lines were excluded. There remained a total of 1,689 drugs in these three databases, including 474 CCLs and 1285 compounds in PRISM, 669 CCLs and 402 drugs in CTRP, and 786 CCLs and 320 drugs in GDSC, which were left to be used for additional analysis (Figure S12B).
Before formal analysis, the three databases must be proven to be available and reliable to predict the drug sensitivity of clinical samples by a ridge regression model. TMZ, the only first-line agent for newly diagnosed GBM, was utilized to validate the assumption. MGMT status has been identified as a strong biomarker of response to TMZ by randomized trials (Hegi et al., 2005). Patients with unmethylated MGMT were believed to respond poorly to TMZ. Therefore, we assigned patients in the TCGA cohort to two groups based on MGMT status. The Wilcoxon rank-sum test was carried out to compare the difference in the estimated AUC values of TMZ. As indicated in Figure S12C, patients with unmethylated MGMT exhibited a higher estimated AUC value for TMZ, and the difference was statistically significant (p = 0.036). It should be emphasized that a higher estimated AUC value signified a more insensitive drug response. This result corresponded to the actual clinical response of TMZ, indicating the consistency of the estimated and actual clinical response to a certain extent.
Exploration of potential subclass-specific agents targeting patients with a high risk of poor prognosis
Figure S12D shows a case of our adopted strategy to explore specific agents for patients with high GPS scores in each subclass. First, we sought all agents corresponding to 16 identified drug targets in the Repurposing Hub and DrugBank. By taking the intersection of the result and drug information from the LINCS Data Portal, 17 drugs were first selected. Next, we performed Spearman correlation analysis between the estimated AUC value and GPS score in our five cohorts. All agents with negative correlation coefficients (Spearman’s r < 0) were collected together for subsequent analysis. Again, we carried out differential analysis of estimated drug response among the three subclasses in all cohorts. Subclass-specific agents were defined as differential-response agents (log2FC < 0 and adjusted p < 0.05) appearing more than twice. Finally, 7 potential agents were acquired through the above three methods, which were deemed to have subclass-specific efficacy according to our classification and show preferential effects for patients with a high risk of poor prognosis. (NVP−BEZ235 for GPA; ponatinib, ZSTK−474, idelalisib, GDC−0980, NVP−BEZ235, dasatinib for GPB; XL765 for GPC). Intriguingly, Alhalabi et al. revealed that the mesenchymal subtype (related to GPB) could benefit more from dasatinib than the others, which partly confirmed our predictions (Alhalabi et al., 2021).
Further screening of the most promising agents based on multiperspective analyses
To identify the most promising agents, we comprehensively analyzed and evaluated the therapeutic significance of these seven potential agents in GBM from multiple perspectives (Figure 5B). First, a complete literature retrieval was conducted in the PubMed dataset to determine the clinical status of these seven agents and find their experimental evidence in a contemporary context. In addition, Log2FC of AUC was presented, and a higher value signified a more obvious subclass-specific efficacy. Finally, the discordance scores of each drug were calculated by the SynergySeq platform, of which the values were deemed to partly exhibit a positive correlation with the ability to combat GBM (Stathias et al., 2018). Considering all these factors, NVP−BEZ235, GDC−0980, dasatinib, and XL765 were treated as the most promising agents in our study.
To go a step further and validate the conclusion that the four currently selected agents had actual therapeutic significance, in vitro experiments on a small scale were performed in three GBM cell lines, including T98G, NP8, and LN229. Based on the results of consensus clustering, the T98G, NP8, and LN229 cell lines were regarded to be equivalent to GPA, GPB, and GPC, respectively. The Cell Counting Kit-8 (CCK8) assay indicated that the four agents reduced the corresponding cell viability in a dose-dependent manner (Figure 5C). Colony formation assays were performed to further confirm their cytotoxic effect on GBM cell viability, where the survival of GBM cells was also inhibited in a dose-dependent manner (Figure 5D). Apoptosis was observed by flow cytometry. The percentage of apoptotic GBM cells increased after using higher concentrations of agents compared to the control groups (Figure S13). Moreover, we compared the IC50 values of four selected agents in different cell lines to demonstrate the differences in drug sensitivity across subclasses. As shown in Table S9, NVP−BEZ235 and dasatinib showed a trend for preferential effect, which was entirely consistent with the prediction results of our bioinformatic analyses. It is interesting that the expected agent in a certain subclass showed a lower IC50 value than the others.
Prediction of immunotherapy response
We first checked the immune landscape of every patient from the training set in terms of immune cells, checkpoint targets, and cytokines. The results were categorized by the patient subclass and GPS score (from high to low) and are presented in a heatmap (Figure 6A). GPB exhibited the highest absolute fraction of immune cells (Figure 6B). Moreover, as a typical example, GPB had a significantly higher abundance of macrophages, which could be found with clarity in Figures 6A and 6B. Tumor-associated macrophages (TAMs) in the GBM microenvironment consist of tissue-resident microglia (MG) and recruited bone-marrow-derived macrophages (BMDMs) (Bowman et al., 2016). As shown in Figure 6C, the enrichment score of BMDM among the three subclasses was significantly different, and GPB showed the highest score, followed by GPA and GPC. Of interest, the enrichment score of MG showed the opposite result. Further analysis at single-cell resolution indicated that MG primarily exhibited the tumoricidal M1 phenotype and that BMDMs were dominated by the tumorigenic M2 phenotype (Figure 6D). GPB was found to be associated with a higher enrichment score of immunosuppressive expression signatures, including PTEN pathway, induced regulatory T cells, and FOXO3 proteins, which was identified by Zhao et al. (Figure 6E) (Zhao et al., 2019b). However, high levels of immunosuppression always restricted the availability of tumor antigens necessary for an effective response to immunotherapy (Lim et al., 2018). Furthermore, microsatellite instability (MSI) was more frequent in GPC patients, suggesting GPC might have meaningful responses to immunotherapy (Figure S14A) (Dudley et al., 2016). Tumor Immune Dysfunction and Exclusion (TIDE) algorithm was utilized to calculate the Dysfunction and Exclusion score among different subclasses (Jiang et al., 2018). As shown in Figure S14C, these three subclasses adopted distinct immune evasion strategies and GPC might response well to immunotherapy because of the low probability of immune evasion. In summary, GPC seemed more likely to be sensitive to immunotherapy and GPB could only benefit from this treatment in a limited way.
Figure 6.
The immune landscape and immunotherapeutic responses among three subclasses
(A) An overview of the GPS score, immune cells, checkpoint targets and cytokines in three subclasses.
(B) The stacked column charts of immune cell fraction in GPA, GPB and GPC.
(C) The boxplots depicting difference of BMDM and MG enrichment score among three subclasses.
(D) The scatterplot of M1 and M2 phenotypes from BMDM and MG based on scRNA-seq data.
(E) Violin plot showing the enrichment scores of immunosuppressive expression signatures, including PTEN pathway, induced regulatory T cells, and FOXO3 proteins. BMDM, Bone marrow-derived macrophage; MG, Microglia; ANOVA was used to compare groups. ∗: p < 0.05, ∗∗: p < 0.01, ∗∗∗: p < 0.001, ∗∗∗∗: p < 0.0001, NS, Not Significant.
A negative correlation was observed between GPS score and MSI events (p < 0.0001) (Figure S14B), indicting low-risk patients were more likely to benefit from immunotherapy (Dudley et al., 2016). Furthermore, TIDE algorithm was also utilized to conduct the sensitivity analysis of the high- and low-GPS groups to immunotherapy and to determine the type of immune evasion in them. As shown in Figure S15A, Dysfunction and Exclusion scores were significantly higher for the high-GPS group than for the low-GPS group. These results suggested that the high- and low-GPS groups utilized distinct immune evasion strategies, with high-GPS group showing a higher level of T cell dysfunction and a lower cytotoxic T lymphocyte (CTL) level. Moreover, the high-GPS group might show an unsatisfactory immunotherapy efficacy due to the high probability of immune evasion. Besides, we further validated the prediction efficacy of the GPS for immunotherapy response in the real immunotherapy cohort (the PRJNA482620 cohort) (Figure S15B) (Zhao et al., 2019b). Although there was no significant statistic difference of GPS scores between responders and non-responders (P = 0.23), it was obvious that patients who did not respond to immunotherapy tended to have a higher GPS score (Figure S15C). The reason that this difference did not reach a statistically significant level may be due to the relatively small size of the dataset and highly heterogeneous among individuals. We also tested the predictive value of GPS on prognosis and immunotherapy efficacy in the PRJNA482620 cohort. The results of tROC analysis showed that GPS has a certain predictive value for the prognosis in the PRJNA482620 cohort (Figure S15D). The AUC value was 0.664, indicating a certain degree of predictive validity of GPS for immunotherapy response (Figure S15E). Collectively, low-GPS group might be sensitive to immunotherapy. However, this conclusion still requires further verification in large-scale immunotherapy cohorts.
Six different immune response–related gene signatures derived from solid tumors (Table S10) were then assessed among the three subclasses and between high- and low-GPS groups. As shown in Figures S16A–S16C, the results showed that these signatures were not suitable for application in predicting immunotherapy response in GBM. This may be attributable to the existence of the blood–brain barrier and the unique immune environment within the brain, which are relatively unique to GBM.
Discussion
The high burden of disease within the context of current medical practices highlights the unmet need for an instructive system for the development of new therapies for GBM. However, new attempts to treat GBM have not made much headway. Indeed, given the high heterogeneity, the notion of precision oncology driven by molecular profiling for the treatment of GBM is attractive and scientifically sound, of which the core concept is the exact matching of the specific drugs and appropriate patients. Although knowledge of the genomic landscape of GBM has advanced tremendously, efforts to translate these findings into better prognosis have not advanced very far. In this study, we created a system that simultaneously covered GBM molecular classification, prognostic risk signature, precision treatment strategies, and their connection. We first established a multiomics-based classification and uncovered the clinical and molecular characteristics of different clusters. Next, to broaden the scope of the classification applications, a gene expression-based surrogate was identified based on 136 DEGs, which successfully stratified GBM patients with different molecular features both in bulk samples and at single-cell resolution. The importance of the novel classification lies in the fact that different subclasses might need different treatment therapies. Then, we developed a prognostic risk signature, 36-gene GPS, to stratify patients with high-risk or low-risk poor prognosis. Finally, four agents (NVP−BEZ235, GDC−0980, dasatinib and XL765) were identified to have subclass-specific efficacy and to target patients with a high risk of dismal prognosis. Compared to previous studies, multiple validation sets in this study ensured stability of the molecular classification and good predictive power of GPS. Targeted agents were predicted based on both the molecular classification and prognostic risk stratification, which ensured the innovation of our research. Moreover, we identified the GBM classification and GPS score model which were considered as potential biomarkers for immunotherapy response, providing more options for the treatment of GBM.
Multiple drug targets identified in our study have been proven to play critical roles in GBM, indicating the feasibility of generating corresponding targeted therapies for GBM with a high GPS score. S100A4 also referred to FSP1/mts-1/metastasin/pEL98. Proteins encoded by S100A4 participate in regulating apoptosis, cell motility, and angiogenesis (Chow et al., 2017). It was reported that high levels of S100A4 expression were related to dismal prognosis in various human tumors (Bresnick et al., 2015). The latest research declared that S100A4 was closely associated with immunosuppression and GBM growth, which confirmed the potential of S100A4 as an immunotherapy target in GBM through single-cell analysis (Abdelfattah et al., 2022). Oncostatin M receptor (OSMR) is a member of the interleukin-6 receptor family, regulating homeostasis, cell growth, and cell differentiation (Heinrich et al., 1998; Hermanns, 2015). The upregulation of OSMR was found in mesenchymal and classical subtypes of GBM, which was strongly associated with poor outcome of GBM (Jahani-Asl et al., 2016; Natesh et al., 2015). Hara revealed that macrophage-derived oncostatin M (OSM), the ligand for OSMR, could induce the mesenchymal-like state of GBM (Hara et al., 2021). Deletion of OSMR contributed to tumor cell death and sensitized the response of GBM to ionizing radiation therapy (Sharanek et al., 2020).
NVP−BEZ235, GDC−0980, and XL765 are dual PI3K/mTOR inhibitors. The PI3K/mTOR signaling pathway is a frequently dysregulated pathway in GBM. The feedback and downstream mechanisms in the PI3K/mTOR pathway remain practicable when using mTOR or PI3K inhibitors alone (Zhao et al., 2017). Therefore, studies have focused more on the dual inhibition of both PI3K and mTOR in GBM. NVP−BEZ235 is the most fully studied of the three agents, with a wide range of applications in preclinical studies on breast cancers, colorectal cancers, lung cancers, and others (Zhao et al., 2017). NVP-BEZ235 has been proven effective in suppressing the growth of GBM cells in vivo and in vitro (Liu et al., 2009). It was shown that NVP-BEZ235 treatment of GBM cell lines induced cell-cycle arrest in G1 phase and evoked endothelial cell apoptosis (Yu et al., 2015). In a clinical trial, NVP-BEZ235 was identified as generally well tolerated in a phase I clinical trial (NCT01343498) (Bendell et al., 2015). Moreover, the treatment effectiveness of the combination of NVP-BEZ235 and MK-3475 (a PD-1 monoclonal antibody) is being evaluated by a phase IIb clinical trial (NCT02430363). Our study suggested that NVP−BEZ23 might obtain better therapeutic effects in GPA and GPB subclasses, thus shedding new light on future clinical trials of NVP−BEZ235. GDC−0980 is a novel dual PI3K/mTOR inhibitor whose treatment effectiveness has been evaluated in phase I/II clinical trials for prostate cancer, pancreatic cancer, and lung cancer cell lines (Omeljaniuk et al., 2021). However, there are limited experimental studies and no clinical trials investigating the treatment effectiveness of GDC−0980 for GBM. Omeljaniuk first revealed that GDC−0980 could significantly inhibit the growth of GBM cell lines by inducing apoptosis (Omeljaniuk et al., 2021). GDC-0980 played an inhibitory role in PERK expression, might thus block its repressive action on protein synthesis, resulting in its intensification, which might contribute to increased apoptosis. CHOP protein, on the other hand, induced protein synthesis and promoted apoptosis (Omeljaniuk et al., 2021). XL765 is a blood brain barrier-permeable PI3K/mTOR inhibitor (Zhao et al., 2019a). XL765 showed potent anti-GBM effects in subcutaneous or intracranial GBM xenografts used as a single agent or in combination with TMZ (Prasad et al., 2011; Yu et al., 2014). Zhao et al. suggested that XL765 executed its inhibitory effects on GBM cell growth by triggering endoplasmic reticulum (ER) stress dependent apoptosis. ER stress led to the activation of CHOP/DR5 pathway, which was thus responsible for inducing the production of apoptosis (Zhao et al., 2019a). A phase I dose-escalation study of XL765 combined with TMZ with or without concurrent radiotherapy indicated a favorable safety performance in high-grade glioma patients (NCT00704080) (Wen et al., 2015). XL765 showed different therapeutic effects in different types of GBM cell lines. Obvious tumor shrinkage was found in the U87-MG model after XL765 treatment, whereas GS-2 was not sensitive to XL765, suggesting that there might be heterogeneity that has not yet been recognized for GBM (Radoul et al., 2016). Our study indicated that XL765 showed a potential preferential effect in the GPC subclass, especially for those with a high risk of poor prognosis. To some extent, our proposed molecular classification could accelerate our understanding of GBM heterogeneity.
SRC represents the first discovered human proto-oncogene and the prototype of nonreceptor tyrosine kinases (SRC family kinases, SFKs). Ample evidence supports a pivotal role of SRC and SFKs in GBM. Dasatinib inhibited all SFKs (Kantarjian et al., 2006). A wealth of in vitro and in vivo preclinical results indicated that dasatinib regulated the migration, proliferation, and morphology of GBM through its suppressive effect on SRC kinase (Ahluwalia et al., 2010; Du et al., 2009; Han et al., 2014; Lewis-Tuffin et al., 2015). However, the clinical trials of dasatinib have thus far been disappointing. A phase II clinical trial revealed that the combination of dasatinib and bevacizumab failed to improve the prognosis of GBM patients when compared with monotherapy using bevacizumab (NCT00892177) (Galanis et al., 2019). Another phase II clinical trial revealed that dasatinib did not have enough capacity to combat recurrent GBM (NCT00423735) (Lassman et al., 2015). The difference between the experimental and clinical results was thought-provoking. A review by Cirotti et al. underlined the requirement for studies to improve patient stratification and identify the patient group who could authentically benefit from SRC targeting (Cirotti et al., 2020). Indeed, Alhalabi et al. showed that the SRC phosphorylation status, which is normally used to indicate SRC activation, could not be used to predict the therapeutic response to dasatinib (Alhalabi et al., 2021). Therefore, despite laudable attempts by researchers to select patients most likely to respond well to dasatinib, they might lack the ability to precisely identify a relevant predictive biomarker, which might partly account for the failure of previous dasatinib trials (Schiff and Sarkaria, 2015). In our study, we proposed a new stratification strategy for patient selection in the dasatinib trial. Dasatinib was considered to be effective mostly for patients with high GPS scores in the GPB subclass, which allows new insights into precision medicine for GBM.
The success of immunotherapies, especially immune checkpoint inhibitor (ICI) therapy in several solid tumors, has resulted in clear motivation for their evaluation in GBM. Phase III clinical studies with ICIs in GBM have yielded disappointing results, and ICIs have merely led to responses in isolated case reports to date (Lim et al., 2018; Medikonda et al., 2021; Schalper et al., 2019), which implies that GBM might not be a single entity and that ICIs have a limited effect in unselected patients. Thus, it is necessary to identify the group that benefits from GBM immunotherapy. However, no predictive biomarker has emerged to help identify and select patients most suitable for immunotherapy thus far (McGranahan et al., 2019). In this study, we found that GPB and high-GPS group could only benefit from this treatment in a limited way but GPC and low-GPS group might be sensitive to immunotherapy. Collectively, in view of the robust differences in molecular features and immunity landscape among GBM subclasses in our study, immunotherapies with subtype-specific considerations might lead to highlights for future therapies in GBM.
Another obstacle to effective immunotherapy is the immune-suppressive TME, including TAMs, which predominate the immune infiltrate and have emerged as pivotal players in GBM (Mirzaei et al., 2017; Poon et al., 2017). It has been proven that TAMs have diverse activation states and cellular phenotypes. The high plasticity enables TAMs to switch between tumoricidal M1 and tumorigenic M2 phenotypes (Biswas and Mantovani, 2010). A previous study indicated significant enrichment of TAM-related genes in the mesenchymal subtype, indicating that TAMs could play a subtype-specific role in GBM (Engler et al., 2012). Moreover, the mesenchymal subtype was found to exhibit increased TAM infiltration compared with the proneural subtype in both humans and mice (Chen and Hambardzumyan, 2018). In our study, the mesenchymal subtype was related to GPB, and the proneural subtype was linked to GPC. We not only suggested differences in TAM abundance among the three subclasses but also disclosed the different TAM compositions in subclasses. However, in terms of the number and composition of TAMs, it remains unclear which one is more vital for tumor development. In general, given the abundance of TAMs in the TME and their plasticity, TAM-based immunotherapies might prove beneficial for GBM. Efforts should be made to “re-educate” TAMs and polarize them from M2 toward M1, especially for BMDMs, which proved to be dominated by the tumorigenic M2 phenotype in our study. Moreover, further studies are needed to prove whether blocking TAM-mediated immunosuppression could improve the efficacy of ICIs for GBM.
In summary, we established a novel molecular classification and developed a robust prognostic risk signature, providing references for personalized clinical management of GBM. We then predicted certain specific agents that may be of higher drug sensitivity for patients with high GPS scores in corresponding subclasses, which would translate the molecular classification and prognostic risk stratification into better survival with GBM. In addition, the novel classification and GPS score could serve as an invaluable tool to identify patients who may reap benefits from immunotherapy. In general, this study will allow new insights into precision oncology by combining individualized prognostic prediction with tailored therapeutic strategies.
Limitations of the study
Admittedly, the current research still has some limitations. First, as conclusions in this study were primarily reached by in silico methods, the practical clinical value of our molecular classification and GPS score entailed further validation in future large prospective studies. Second, further experimental validations in the animal model of GBM are essential to provide additional preclinical evidence for these four final selected agents. Finally, the potential benefits of immunotherapy for patients in the GPC and low-GPS group need to be validated prospectively. Furthermore, other forms of immunotherapy, including GBM vaccines, oncolytic viral therapies, and chimeric antigen receptor T-cell therapy, cannot be ignored.
STAR★Methods
Key resources table
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Zhouping Tang (ddjtzp@163.com).
Materials availability
This study did not generate new unique reagents.
Experimental model and subject details
Cell lines
T98G (RRID: CVCL_0556) and LN229 (RRID: CVCL_A8V5) cell lines were purchased from the American Type Culture Collection, and NP8 (RRID: CVCL_4W66) cell line was obtained from Japanese Collection of Research Bioresources Cell Bank. GBM cells were cultured in T25 flasks at 37°C and in incubators containing 5% CO2. MEM medium (Gibco, USA) supplemented with 10% fetal bovine serum (FBS, Gibco, USA) was used for T98G cell line culture. DMEM medium (Gibco, USA) containing 10% FBS was used for NP8, and DMEM with 5% FBS was used for LN229. The cell passages were performed when the cell density reached 80–90% confluency.
Method details
GBM datasets
In this study, a total of four GBM datasets were collected with available survival data, including three microarray datasets (TCGA-GBM, GSE108474, GSE13041) and one RNA-seq dataset (CGGA-GBM). Among the microarray-based datasets, the raw expression files of GSE108474 and GSE13041 were downloaded from the Gene Expression Omnibus (GEO) database (http://www.ncbi.nlm.nih.gov/geo/) and transformed into normalized data by the robust multiarray average (RMA) method located in the R package Affy (Gautier et al., 2004). The normalized TCGA-GBM data were directly obtained from the UCSC Xena website (https://xenabrowser.net/). For the RNA-seq dataset, the Chinese Glioma Genome Atlas (CGGA) database (http://www.cgga.org.cn) provided transcript per million (TPM) values for all samples. The clinical data from TCGA-GBM and GSE13041 were accessed through the supplementary files of corresponding publications (Brennan et al., 2013; Lee et al., 2008), and survival data for GSE108474 and CGGA-GBM were obtained from the GEO and CGGA databases. Furthermore, SNV data and CNV profiles were acquired from The Cancer Genome Atlas (TCGA) database (https://portal.gdc.cancer.gov/repository). Notably, we excluded these non-glioblastoma samples depending on tumor histology information to avoid irrelevant interference and utilized the ComBat method of the SVA package to correct batch effects from technical biases in GSE13041 and CGGA-GBM (Leek et al., 2012).
In addition, we collected a multiregion expression cohort, including 122 RNA-seq samples of 5 anatomic structures in 10 tumors, from the IGAP website (http://glioblastoma.alleninstitute.org/static/download.html).
Driver mutation prediction
We applied two algorithms, DriverNet (Bashashati et al., 2012) and MutSigCV (Lawrence et al., 2013), to detect the potential driver mutation which plays a crucial role in tumorigenesis from thousands of somatic mutations. Specifically, the DriverNet algorithm could predict the candidate genes by integrating genome and expression data of samples on the basis of the influence graph extracted from the Reactome Functional interactions (Wu et al., 2010). In addition, the somatic mutationdata (MAF files) were mapped to hg19 reference genome using MutSigCV with default parameters to infer significantly mutated gene. The genes with adjusted p value <0.05 were kept for the subsequent clustering analysis.
Clustering analysis and subclass prediction
TCGA-GBM dataset was divided into two sets, training set possessing complete multi-omics data (n = 323) and internal validation set with available expression and survival data (n = 195). The remaining three datasets, CGGA-GBM, GSE108474 and GSE13041, were held out as external validation sets. Based on the mutation, copy number variation (CNV) and expression profiles of training set, we performed similarity network fusion-consensus clustering (SNF-CC) using the top 1000 genes, CNVs with maximum variations and 42 mutated genes via CancerSubtypes package (Xu et al., 2017). To obtain the optimal cluster k, we repeated the above steps with the parameters of 500 iterations and cluster k ranging from 2 to 5, and collected the corresponding average silhouette widths (ASW). The value of k that achieved the maximal ASW was reserved as the optimal number of clusters.
To apply this multi-omics classification model to other cohorts, we required a surrogate based on gene expression. Differential analysis among three clusters acquired above was conducted using Limma package (Ritchie et al., 2015), and the genes with an absolute log2 fold change > 1 (adjusted p value < 0.05) were considered as differential expressed genes (DEGs), and kept for the subclass prediction. We repeated the consensus clustering (CC) using the expression data of DEGs on training set and validation sets to predict tumor samples’ subclasses.
CNV analysis
To identify independent significantly altered regions, we conducted Genomic Identification of Significant Targets in Cancer (GISTIC) analysis on the TCGA CNV data using GISTIC 2.0 software (Mermel et al., 2011). We achieved GISTIC value of each common driver mutation and attributed these into two subgroups, including amplification group (GISTIC >1) and deletion group (GISTIC < −1).
Pathway and gene set enrichment analysis (GSEA)
Based on the clusterProfiler package (Wu et al., 2021), we performed GSEA using Gene Ontology Resource to identify pathway differences in biological process (BP), cellular component (CC) and molecular function (MF) levels. Next, EnrichmentMap analysis was conducted on the GSEA results using emapplot function and mutually overlapping gene sets tended to cluster together, making it easy to identify similar functional module. The major biological themes of each cluster were extracted manually from enrichment results and labeled in the figures.
To compare the difference of the certain pathway or signature enrichment, we also conducted gene set variation analysis (GSVA) among three subclasses based on transcriptomic data utilizing the GSVA package (Hänzelmann et al., 2013). Hallmark and other pathway gene sets were downloaded from the Molecular Signatures Database (MSigDB) (Liberzon et al., 2015). Although gene sets representing the bone-marrow-derived macrophage (BMDM) and tissue-resident microglia (MG) activity were obtained from previous publication (Bowman et al., 2016). We calculated the enrichment score of each gene set and conducted differential analysis among three subclasses.
Single-cell RNA-seq analysis
The single-cell RNA-seq dataset, GSE84465, comprising 3,589 cells from four GBM patients, was obtained from the GEO database. The raw count matrix required from Smart-Seq2 was transformed into relative expression values through the CreateSeuratObject function deposited in Seurat package (Butler et al., 2018). Then, all cells passing quality control were clustered leveraging principal component analysis (PCA) with the following parameters: Ngenes = 2000, Npcs = 50, Res = 0.4, N.iters = 10. Referring to these marker genes used in previous research (Darmanis et al., 2017; Lau et al., 2020), all cells were classified to each cell type. For astrocytes, AQP4, GPC5, RYR3. For neurons, GAD2, GRIK1. For microglia, C3, DOCK8, TMEM119, P2RY12, GPR34, OLFML3, SLC2A5, SALL1, ADORA3, CD74. For myeloid cells, CRIP1, S100A8, S100A9, ANXA1, CST3, LYZ, FCGR3B, CSF3R. For OPCs, TNR, IGSF21, NEU4, GPR17. For oligodendrocytes, MBP, PLP1, ST18. For pericyte, DCN, LUM, GSN, FGF7, MME, ACTA2, RGS5, PDGFRB. For neoplastic cells, EGFR. Neoplastic cells were validated using the inferCNV algorithm (https://github.com/broadinstitute/inferCNV). The CNV of each cell was calculated by sorting expressed genes by their chromosomal location and applying a moving average to the relative expression values with a sliding window of 100 genes within each chromosome. Each of the non-neoplastic cells mentioned above was used as normal reference. To discern neoplastic cells from non-neoplastic cells, we scored each cell for two measures, CNV signal and CNV correlation, using the same protocol as in a previous article (Neftel et al., 2019). Specifically, “CNV signal’’ was defined as the mean of the squares of CNV values across the whole genome, representing the overall extent of CNVs. ‘‘CNV correlation’’ reflected the correlation between the CNV profile of each cell and the average CNV profile of all cells except for those classified as non-neoplastic.
Identifying phenotype-associated single cell subpopulations
To explore the difference of neoplastic cell expressions associated with specific phenotypes, Scissor (Sun et al., 2022), a method quantifying the similarity between each single cell and each bulk sample, was applied to identify specific cell clusters with corresponding phenotypes. To further demonstrate whether Scissor could distinguish specific kind of cells from background cells, the expression data of tumor and normal phenotypes were used to guide the identification of cell subpopulations. Afterwards, we integrated three Scissor results to determine the eventual assignment of each neoplastic cell. For example, one cell was regarded by Scissor as GPA associated neoplastic cell subpopulations, not assigned to GPB/GPC associated neoplastic cell subpopulations in other two Scissor results, the cell was finally assigned to GPA associated neoplastic cell subpopulations. Similarly, cells were assigned to background cells only if these assignments were not satisfied with the above conditions. Then, the FindMarkers function in Seurat (Butler et al., 2018) leveraging the Wilcoxon rank sum test was used to identify the DEGs among GPA/GPB/GPC associated neoplastic cell subpopulations. The DEGs which were statistically significant must meet the following criteria: (1) false discovery rate (FDR) less than 0.05. (2) the absolute value of expression fold change among three groups was greater than 1. Similarly, we conducted GSEA using the clusterProfiler based on the Hallmark and Reactome Pathway (Gillespie et al., 2022) gene sets with the default parameters to identify differential biological processes at single cell resolution.
Assignment of GBM single cell subtypes
In this part, we acquired 6 glioma meta-modules, including MES1-like, MES2-like, NPC1-like, NPC2-like, AC-like, and OPC-like subtypes, from previous research (Neftel et al., 2019). Specifically, the MES1 and MES2 groups were assigned into one group of MES-like cells, and similarly the NPC1 and NPC2 into one group of NPC-like cells. Then, we calculated meta-module scores of all neoplastic cells using the “score” function from R package scrabble and achieved the exact position of each cell on two-dimensional plots through scrabble::hierarchy() function. The final plots were visualized through ggplot2 package, where each quadrant corresponds to one cellular state.
Transcription factor (TF) and pseudotime analysis
The DoRothEA (https://saezlab.github.io/dorothea/) resource was applied to decipher the difference of TF activities between single cell subsets within neoplastic cells (Garcia-Alonso et al., 2019). Based on the expression data of specific cells, TF activities were scored with the statistical method VIPER (Alvarez et al., 2016). The DoRothEA’s regulons whose confidence levels ranging from A to C were selected to quantify TF activities which were presented using UMAP with default parameters. To allow comparison of TF activities, we conducted differential analysis on scaled VIPER scores among single cell subsets. TFs were ranked according to the fold change of their corresponding VIPER scores and the top 20 highly variable scores were retained for heatmap visualization. We applied the Monocle 2 package to perform the cell trajectory and pseudotime analysis leveraging the reverse graph embedding algorithm (Qiu et al., 2017). The DifferentialGeneTest function in Monocle 2 was utilized to identify genes differing among GPA/GPB/GPC associated neoplastic cell subpopulations and the trajectory was constructed using differential genes with a q value (adjusted p value) < 0.01.
Construction of prognostic risk signature
As mentioned above, training set was selected to construct a prognostic risk model, whereas the remaining sets were used to evaluate the predictive performance of the prognostic model. Specifically, we first conducted preliminary filtering to exclude those DEGs without significant prognostic association (p value >0.1) using univariate Cox proportional hazards regression (COXPH) analysis. Then weighted random forest (WRF) analysis was performed to further narrow down the retained genes (Chen and Ishwaran, 2012). Based on the training set, the WRF analysis could measure the importance of every gene through integrating 1000 decision trees using RandomForest package. Notably, sliding windows sequential forward feature selection was also applied to find the optimal signatures. These remaining genes were ranked by their importance, gradually included in the WRF model. And at each time this model changed, the out-of-bag (OOB) error was recorded under the current gene combination. The gene list whose OOB error rate reached its minimum was considered as the optimal biomarkers and was used to develop the prognostic prediction model. The GBM poor prognostic signature score (GPS) model was established by these selected genes using multivariate Cox regression as follows:
In addition, four previously published signatures, including Hou’s (Hou et al., 2019), Lei’s (Lei et al., 2021), Zhao’s (Zhao et al., 2019c) and Fatai‘s (Fatai and Gamieldien, 2018), which were used to predict patients’ survival, were achieved from corresponding publications to estimate the effects of GPS in GBM. Based on timeROC package, we calculated areas under the receiver operating characteristic curve (AUC) at 1, 2, 3, 4 and 5 years of each signature on validation sets to demonstrate the superiority of GPS for prognostication. Notably, the exact value of risk scores based on each signature made no sense unless comparisons. For the sake of a better presentation, these scores were scaled into a range from 0 to 1.
Cancer cell line data
The expression profiles and mutationdata of human cancer cell lines (CCLs) were downloaded from the Cancer Cell Line Encyclopedia (CCLE) project (https://portals.broadinstitute.org/ccle/). And drug response data of different compounds against CCLs were obtained from the Cancer Therapeutics Response Portal (CTRP v2.0, released October 2015, https://portals.broadinstitute.org/ctrp), PRISM Repurposing dataset (19Q4, released December 2019, https://depmap.org/portal/prism/) and GDSC 1&2 datasets (Release 8.2, release February 2020, https://www.cancerrxgene.org/downloads/bulk_download), respectively. To explore the targets with potential therapeutic effects, we also collected the genome-wide gene dependency scores, including CERES scores based on clustered regularly interspaced short palindromic repeats (CRISPR) knockout screens (Meyers et al., 2017) and DEMETER scores based on RNA interference (RNAi) screens (Tsherniak et al., 2017), from the Cancer Dependency Map (DepMap) portal (https://depmap.org/portal/download/), which indicated that certain gene possessed lower CERES or DEMETER score was more likely to be essential in cell survival and proliferation of CCLs.
Drug screen analysis
In this study, we conducted Spearman correlation analysis between the targetable gene expressions and GPS in the training set to identify genes with potential therapeutic implications. And those genes with correlation coefficient more than 0.25 (adjusted p value < 0.05) were considered as prognosis-related targets and kept for subsequent process. Then the GPS for each GBM cell line from the CCLE project was calculated based on the corresponding expression data and the Spearman correlation analysis between it and CERES, DEMETER of all GBM cell lines were performed respectively to narrow down gene panel (Yang et al., 2021b). Notably, those targetable genes left should meet two criteria: (1) the mean value of CERES or DEMETER should be less than that for nonessential gene list among all the GBM cell lines; (2) the correlation coefficient should be less than −0.25 and the adjusted p value should be less than 0.1. In addition, we collected the drug-target interactions from the Drug Repurposing Hub database (https://clue.io/repurposing) (Corsello et al., 2017) and DrugBank database (https://go.drugbank.com/) (Wishart et al., 2018), which comprised a total of 11,875 compounds and 4,465 targets after merging two drug databases. Through matching these targets with compounds, we obtained these candidate therapeutic agents. To demonstrate whether these candidate agents could inhibit GBM cells well, we downloaded the Library of Integrated Network-Based Cellular Signatures (LINCS) data from the LINCS Data Portal (http://lincsportal.ccs.miami.edu/signatures/) (Stathias et al., 2020). Next, the disease discordance for each drug was computed which indicated the ability to inhibit GBM tumor based on the platform termed SynergySeq (Stathias et al., 2018) and these drugs with discordance ≥0 were left for drug response analysis.
Predicting drug response in subclass samples
Three large pharmacogenomic datasets, including CTRP, PRISM and GDSC, contained massive drug response data and expression profiles across hundreds of CCLs. And the area under curve values (AUCs) of dose-response acquired from these three datasets were used as evaluation indicators of drug sensitivity, meaning lower AUC value suggests higher response probability to therapy treatment. Compounds with missing AUCs across more than 20% of the CCLs were excluded firstly, and the absent data among the rest of compounds were imputed using the K-nearest neighbors (KNN) method located in the Impute package (Di Lena et al., 2019). In this study, the ridge regression model that exhibited great and precise performance in the previous research was utilized for transcriptome data-based drug response prediction using the pRRophetic package (Geeleher et al., 2017). Through exploiting the expression and drug response data of solid carcinomas from CCLE and GDSC projects (excluding hematopoietic and lymphoid tissue-derived CCLs), this predictive model was trained with the parameter of 10-fold cross-validation and then applied to calculate different drug response across subclass samples.
Immune microenvironment analysis and immunotherapy prediction
Microenvironment cell population-counter (MCPcounter) was applied to evaluate absolute abundance of eight immune and two stromal cell populations among tumor samples based on bulk expression data (Becht et al., 2016). In addition, we utilized ESTIMATE algorithm to calculate immune and stromal scores which indicated the enrichment of immune and stromal cell signatures (Yoshihara et al., 2013). To fully describe the immune landscape of GBM, single-sample GSEA (ssGSEA) measuring the level of immune-infiltration in tumor samples was applied in this study based on 24 different cell gene signatures achieved from previous researches (Bindea et al., 2013; Li et al., 2022). Then, the proportions of immune subgroup cells were inferred by leveraging CIBERSORT algorithm with the parameter of 1000 iterations. The LM22 basis matrix was acquired from the CIBERSORT website (https://cibersort.stanford.edu/) for deconvolution.
To explore the responses to immunotherapy for subclasses, we collected 6 public immunotherapy response–associated signatures, including Chemokine (Messina et al., 2012), Cytolytic activity (CYT) (Rooney et al., 2015), T cell–inflamed GEP (Ayers et al., 2017), IFN-γ (Yonekura and Ueda, 2021), IPRES (Hugo et al., 2016) and MHC score (Lauss et al., 2017). The Tumor Immune Dysfunction and Exclusion (TIDE) algorithm was a computational method used to model two main types of tumor immune evasion mechanisms, including the induction of T cell dysfunction in tumors with high infiltration of cytotoxic T lymphocytes (CTLs) and the prevention of T cell infiltration in tumors with low CTL levels (Jiang et al., 2018). TIDE was utilized to calculate the Dysfunction and Exclusion score of the high- and low-GPS groups to investigate the reasons of immune evasion and predict the clinical response to immunotherapy between groups. In addition, we collected a validation set (PRJNA482620) (Zhao et al., 2019b), which included 17 GBM patients undergoing anti-PD-1 checkpoint inhibition treatment, to measure the prediction ability of the GPS for immunotherapy response in the real immunotherapy cohort.
CCK8 assay
Cell Counting Kit-8 (CCK8, Bimake, China) assay was performed to evaluate cell proliferation. GBM cells were seeded in 96-well plates at the density of 5∗103 cells per well and cultured with various concentrations of drugs. CCK8 was added and incubated for 2 hours and microplate reader was then used to read the absorbance of plates. IC50 values were calculated to evaluate the drug toxicity.
Colony formation assays
GBM cells were seeded in 6-well plates at the density of 800 cells per well and cultured for 12 hours. Drugs were then added to the plates. The medium was replaced every three days. After 7 days, the colonies were fixed with 4% paraformaldehyde and stained with 0.5% crystal violet solution. ImageJ was used to count the surviving colonies.
Flow cytometry analysis of cell apoptosis
GBM cells were harvested using trypsin solution without EDTA (Gibco, USA), washed with cold phosphate buffer saline solution (Biological Industries, Israel) for three times, and resuspended with binding buffer (BD Biosciences, USA). Cells were then stained with Annexin V-fluorescein isothiocyanate (V-FITC) and propidium iodide (PI) for 15 min according to manufacturer’s protocol. Apoptotic glioma cells (annexin V-FITC+/PI-) were detected using flow cytometer (BD Biosciences, USA).
Quantification and statistical analysis
Based on R statistical software (https://cran.r-project.org/), we conducted all the statistical tests and graphical visualization. Student’s t-test or analysis of variance (ANOVA) was applied for comparison of two or more than two groups with normally distributed variables. If not, Wilcoxon rank-sum test or Kruskal–Wallis test was adopted. Similarly, correlation between two continuous variables was measured by either Pearson’s r correlation (measure of linear relationship between two continuous variables) or Spearman’s rank-order correlation (nonparametric measure of statistical dependence between two variables). Contingency table variables were analyzed by Fisher’s exact tests. The Kaplan–Meier method was applied to perform survival analysis and the statistical significance of differences was determined using the log-rank (Mantel-Cox) test. The hazard ratios (HR) were calculated using the univariate and multivariate Cox proportional hazards regression model located in the survival package. The Benjamini-Hochberg method was utilized to adjust p value of multiple testing in those analyses with more than 20 comparisons. A two tailed p value < 0·05 was considered statistically significant for all computational analysis unless otherwise stated.
Additional resources
No additional resource was used in this study.
Acknowledgments
This work was supported by grants from the National Natural Science Foundation of China (92148206, 81901219, 82071330); the Hubei Provincial Department of Science & Technology (2021BCA109) and the Hubei Provincial Natural Science Foundation of China (2021CFB067).
Author contributions
D.Y.C, Z.C.L., and J.X.W. contributed study design, data analysis and paper writing. C.Y., C.P., and Y.X.T. conducted some of the cell culture experiments. P.Z., N.L., G.G.L., Y.L., Z.J.W., F.X., and C.T.Z. participated in editing the manuscript. H.N. and Z.P.T. supervised the study and guided the editing of manuscripts.
Declaration of interests
The authors declare no conflict of interest.
Published: November 18, 2022
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.isci.2022.105276.
Contributor Information
Hao Nie, Email: haonie@tjh.tjmu.edu.cn.
Zhouping Tang, Email: ddjtzp@163.com.
Supplemental information
Data and code availability
-
•
This paper analyzes existing, publicly available data. These accession numbers for the datasets are listed in the key resources table.
-
•
R codes used in this study have been deposited and publicly available at GitHub (https://github.com/Liuzhicheng048/GBM).
-
•
Any additional information required to reanalyze the data reported in this paper is available from the lead contact on request.
References
- Abdelfattah N., Kumar P., Wang C., Leu J.S., Flynn W.F., Gao R., Baskin D.S., Pichumani K., Ijare O.B., Wood S.L., et al. Single-cell analysis of human glioma and immune cells identifies S100A4 as an immunotherapy target. Nat. Commun. 2022;13:767. doi: 10.1038/s41467-022-28372-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahluwalia M.S., de Groot J., Liu W.M., Gladson C.L. Targeting SRC in glioblastoma tumors and brain metastases: rationale and preclinical studies. Cancer Lett. 2010;298:139–149. doi: 10.1016/j.canlet.2010.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alhalabi O.T., Fletcher M.N.C., Hielscher T., Kessler T., Lokumcu T., Baumgartner U., Wittmann E., Schlue S., Göttmann M., Rahman S., et al. A novel patient stratification strategy to enhance the therapeutic efficacy of dasatinib in glioblastoma. Neuro Oncol. 2021;24:39–51. doi: 10.1093/neuonc/noab158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alvarez M.J., Shen Y., Giorgi F.M., Lachmann A., Ding B.B., Ye B.H., Califano A. Functional characterization of somatic mutations in cancer using network-based inference of protein activity. Nat. Genet. 2016;48:838–847. doi: 10.1038/ng.3593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ayers M., Lunceford J., Nebozhyn M., Murphy E., Loboda A., Kaufman D.R., Albright A., Cheng J.D., Kang S.P., Shankaran V., et al. IFN-gamma-related mRNA profile predicts clinical response to PD-1 blockade. J. Clin. Invest. 2017;127:2930–2940. doi: 10.1172/JCI91190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bashashati A., Haffari G., Ding J., Ha G., Lui K., Rosner J., Huntsman D.G., Caldas C., Aparicio S.A., Shah S.P. DriverNet: uncovering the impact of somatic driver mutations on transcriptional networks in cancer. Genome Biol. 2012;13:R124. doi: 10.1186/gb-2012-13-12-r124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becht E., Giraldo N.A., Lacroix L., Buttard B., Elarouci N., Petitprez F., Selves J., Laurent-Puig P., Sautès-Fridman C., Fridman W.H., de Reyniès A. Estimating the population abundance of tissue-infiltrating immune and stromal cell populations using gene expression. Genome Biol. 2016;17:218. doi: 10.1186/s13059-016-1070-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bendell J.C., Kurkjian C., Infante J.R., Bauer T.M., Burris H.A., 3rd, Greco F.A., Shih K.C., Thompson D.S., Lane C.M., Finney L.H., Jones S.F. A phase 1 study of the sachet formulation of the oral dual PI3K/mTOR inhibitor BEZ235 given twice daily (BID) in patients with advanced solid tumors. Invest. New Drugs. 2015;33:463–471. doi: 10.1007/s10637-015-0218-6. [DOI] [PubMed] [Google Scholar]
- Bhat K.P.L., Balasubramaniyan V., Vaillant B., Ezhilarasan R., Hummelink K., Hollingsworth F., Wani K., Heathcock L., James J.D., Goodman L.D., et al. Mesenchymal differentiation mediated by NF-kappaB promotes radiation resistance in glioblastoma. Cancer Cell. 2013;24:331–346. doi: 10.1016/j.ccr.2013.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bindea G., Mlecnik B., Tosolini M., Kirilovsky A., Waldner M., Obenauf A.C., Angell H., Fredriksen T., Lafontaine L., Berger A., et al. Spatiotemporal dynamics of intratumoral immune cells reveal the immune landscape in human cancer. Immunity. 2013;39:782–795. doi: 10.1016/j.immuni.2013.10.003. [DOI] [PubMed] [Google Scholar]
- Biswas S.K., Mantovani A. Macrophage plasticity and interaction with lymphocyte subsets: cancer as a paradigm. Nat. Immunol. 2010;11:889–896. doi: 10.1038/ni.1937. [DOI] [PubMed] [Google Scholar]
- Blanche P., Dartigues J.F., Jacqmin-Gadda H. Estimating and comparing time-dependent areas under receiver operating characteristic curves for censored event times with competing risks. Stat. Med. 2013;32:5381–5397. doi: 10.1002/sim.5958. [DOI] [PubMed] [Google Scholar]
- Bowman R.L., Klemm F., Akkari L., Pyonteck S.M., Sevenich L., Quail D.F., Dhara S., Simpson K., Gardner E.E., Iacobuzio-Donahue C.A., et al. Macrophage ontogeny underlies differences in tumor-specific education in brain malignancies. Cell Rep. 2016;17:2445–2459. doi: 10.1016/j.celrep.2016.10.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brennan C.W., Verhaak R.G.W., McKenna A., Campos B., Noushmehr H., Salama S.R., Zheng S., Chakravarty D., Sanborn J.Z., Berman S.H., et al. The somatic genomic landscape of glioblastoma. Cell. 2013;155:462–477. doi: 10.1016/j.cell.2013.09.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bresnick A.R., Weber D.J., Zimmer D.B. S100 proteins in cancer. Nat. Rev. Cancer. 2015;15:96–109. doi: 10.1038/nrc3893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butler A., Hoffman P., Smibert P., Papalexi E., Satija R. Integrating single-cell transcriptomic data across different conditions, technologies, and species. Nat. Biotechnol. 2018;36:411–420. doi: 10.1038/nbt.4096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carro M.S., Lim W.K., Alvarez M.J., Bollo R.J., Zhao X., Snyder E.Y., Sulman E.P., Anne S.L., Doetsch F., Colman H., et al. The transcriptional network for mesenchymal transformation of brain tumours. Nature. 2010;463:318–325. doi: 10.1038/nature08712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen A., Jiang Y., Li Z., Wu L., Santiago U., Zou H., Cai C., Sharma V., Guan Y., McCarl L.H., et al. Chitinase-3-like 1 protein complexes modulate macrophage-mediated immune suppression in glioblastoma. J. Clin. Invest. 2021;131:147552. doi: 10.1172/JCI147552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X., Ishwaran H. Random forests for genomic data analysis. Genomics. 2012;99:323–329. doi: 10.1016/j.ygeno.2012.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Z., Hambardzumyan D. Immune microenvironment in glioblastoma subtypes. Front. Immunol. 2018;9:1004. doi: 10.3389/fimmu.2018.01004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chow K.H., Park H.J., George J., Yamamoto K., Gallup A.D., Graber J.H., Chen Y., Jiang W., Steindler D.A., Neilson E.G., et al. S100A4 is a biomarker and regulator of glioma stem cells that is critical for mesenchymal transition in glioblastoma. Cancer Res. 2017;77:5360–5373. doi: 10.1158/0008-5472.CAN-17-1294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cirotti C., Contadini C., Barilà D. SRC kinase in glioblastoma news from an old acquaintance. Cancers. 2020;12:E1558. doi: 10.3390/cancers12061558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corsello S.M., Bittker J.A., Liu Z., Gould J., McCarren P., Hirschman J.E., Johnston S.E., Vrcic A., Wong B., Khan M., et al. The Drug Repurposing Hub: a next-generation drug library and information resource. Nat. Med. 2017;23:405–408. doi: 10.1038/nm.4306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corsello S.M., Nagari R.T., Spangler R.D., Rossen J., Kocak M., Bryan J.G., Humeidi R., Peck D., Wu X., Tang A.A., et al. Discovering the anti-cancer potential of non-oncology drugs by systematic viability profiling. Nat. Cancer. 2020;1:235–248. doi: 10.1038/s43018-019-0018-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darmanis S., Sloan S.A., Croote D., Mignardi M., Chernikova S., Samghababi P., Zhang Y., Neff N., Kowarsky M., Caneda C., et al. Single-cell RNA-seq analysis of infiltrating neoplastic cells at the migrating front of human glioblastoma. Cell Rep. 2017;21:1399–1410. doi: 10.1016/j.celrep.2017.10.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Souza C.F., Sabedot T.S., Malta T.M., Stetson L., Morozova O., Sokolov A., Laird P.W., Wiznerowicz M., Iavarone A., Snyder J., et al. A distinct DNA methylation shift in a subset of glioma CpG island methylator phenotypes during tumor recurrence. Cell Rep. 2018;23:637–651. doi: 10.1016/j.celrep.2018.03.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Lena P., Sala C., Prodi A., Nardini C. Missing value estimation methods for DNA methylation data. Bioinformatics. 2019;35:3786–3793. doi: 10.1093/bioinformatics/btz134. [DOI] [PubMed] [Google Scholar]
- Du J., Bernasconi P., Clauser K.R., Mani D.R., Finn S.P., Beroukhim R., Burns M., Julian B., Peng X.P., Hieronymus H., et al. Bead-based profiling of tyrosine kinase phosphorylation identifies SRC as a potential target for glioblastoma therapy. Nat. Biotechnol. 2009;27:77–83. doi: 10.1038/nbt.1513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dudley J.C., Lin M.T., Le D.T., Eshleman J.R. Microsatellite instability as a biomarker for PD-1 blockade. Clin. Cancer Res. 2016;22:813–820. doi: 10.1158/1078-0432.CCR-15-1678. [DOI] [PubMed] [Google Scholar]
- Engler J.R., Robinson A.E., Smirnov I., Hodgson J.G., Berger M.S., Gupta N., James C.D., Molinaro A., Phillips J.J. Increased microglia/macrophage gene expression in a subset of adult and pediatric astrocytomas. PLoS One. 2012;7:e43339. doi: 10.1371/journal.pone.0043339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan X., Fan J., Yang H., Zhao C., Niu W., Fang Z., Chen X. Heterogeneity of subsets in glioblastoma mediated by Smad3 palmitoylation. Oncogenesis. 2021;10:72. doi: 10.1038/s41389-021-00361-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fatai A.A., Gamieldien J. A 35-gene signature discriminates between rapidly- and slowly-progressing glioblastoma multiforme and predicts survival in known subtypes of the cancer. BMC Cancer. 2018;18:377. doi: 10.1186/s12885-018-4103-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galanis E., Anderson S.K., Twohy E.L., Carrero X.W., Dixon J.G., Tran D.D., Jeyapalan S.A., Anderson D.M., Kaufmann T.J., Feathers R.W., et al. A phase 1 and randomized, placebo-controlled phase 2 trial of bevacizumab plus dasatinib in patients with recurrent glioblastoma: alliance/North Central Cancer Treatment Group N0872. Cancer. 2019;125:3790–3800. doi: 10.1002/cncr.32340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Alonso L., Holland C.H., Ibrahim M.M., Turei D., Saez-Rodriguez J. Benchmark and integration of resources for the estimation of human transcription factor activities. Genome Res. 2019;29:1363–1375. doi: 10.1101/gr.240663.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gautier L., Cope L., Bolstad B.M., Irizarry R.A. affy--analysis of Affymetrix GeneChip data at the probe level. Bioinformatics. 2004;20:307–315. doi: 10.1093/bioinformatics/btg405. [DOI] [PubMed] [Google Scholar]
- Geeleher P., Zhang Z., Wang F., Gruener R.F., Nath A., Morrison G., Bhutra S., Grossman R.L., Huang R.S. Discovering novel pharmacogenomic biomarkers by imputing drug response in cancer patients from large genomics studies. Genome Res. 2017;27:1743–1751. doi: 10.1101/gr.221077.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghandi M., Huang F.W., Jane-Valbuena J., Kryukov G.V., Lo C.C., McDonald E.R., 3rd, Barretina J., Gelfand E.T., Bielski C.M., Li H., et al. Next-generation characterization of the Cancer Cell Line Encyclopedia. Nature. 2019;569:503–508. doi: 10.1038/s41586-019-1186-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gillespie M., Jassal B., Stephan R., Milacic M., Rothfels K., Senff-Ribeiro A., Griss J., Sevilla C., Matthews L., Gong C., et al. The reactome pathway knowledgebase 2022. Nucleic Acids Res. 2022;50:D687–D692. doi: 10.1093/nar/gkab1028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gusev Y., Bhuvaneshwar K., Song L., Zenklusen J.C., Fine H., Madhavan S. The REMBRANDT study, a large collection of genomic data from brain cancer patients. Sci. Data. 2018;5:180158. doi: 10.1038/sdata.2018.158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han X., Zhang W., Yang X., Wheeler C.G., Langford C.P., Wu L., Filippova N., Friedman G.K., Ding Q., Fathallah-Shaykh H.M., et al. The role of Src family kinases in growth and migration of glioma stem cells. Int. J. Oncol. 2014;45:302–310. doi: 10.3892/ijo.2014.2432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hänzelmann S., Castelo R., Guinney J. GSVA: gene set variation analysis for microarray and RNA-seq data. BMC Bioinf. 2013;14:7. doi: 10.1186/1471-2105-14-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hara T., Chanoch-Myers R., Mathewson N.D., Myskiw C., Atta L., Bussema L., Eichhorn S.W., Greenwald A.C., Kinker G.S., Rodman C., et al. Interactions between cancer cells and immune cells drive transitions to mesenchymal-like states in glioblastoma. Cancer Cell. 2021;39:779–792.e11. doi: 10.1016/j.ccell.2021.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hegi M.E., Diserens A.C., Gorlia T., Hamou M.F., de Tribolet N., Weller M., Kros J.M., Hainfellner J.A., Mason W., Mariani L., et al. MGMT gene silencing and benefit from temozolomide in glioblastoma. N. Engl. J. Med. 2005;352:997–1003. doi: 10.1056/NEJMoa043331. [DOI] [PubMed] [Google Scholar]
- Heinrich P.C., Behrmann I., Müller-Newen G., Schaper F., Graeve L. Interleukin-6-type cytokine signalling through the gp130/Jak/STAT pathway. Biochem. J. 1998;334:297–314. doi: 10.1042/bj3340297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hermanns H.M. Oncostatin M and interleukin-31: cytokines, receptors, signal transduction and physiology. Cytokine Growth Factor Rev. 2015;26:545–558. doi: 10.1016/j.cytogfr.2015.07.006. [DOI] [PubMed] [Google Scholar]
- Holland C.H., Tanevski J., Perales-Patón J., Gleixner J., Kumar M.P., Mereu E., Joughin B.A., Stegle O., Lauffenburger D.A., Heyn H., et al. Robustness and applicability of transcription factor and pathway analysis tools on single-cell RNA-seq data. Genome Biol. 2020;21:36. doi: 10.1186/s13059-020-1949-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou Z., Yang J., Wang H., Liu D., Zhang H. A potential prognostic gene signature for predicting survival for glioblastoma patients. Biomed Res. Int. 2019;2019:9506461. doi: 10.1155/2019/9506461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hugo W., Zaretsky J.M., Sun L., Song C., Moreno B.H., Hu-Lieskovan S., Berent-Maoz B., Pang J., Chmielowski B., Cherry G., et al. Genomic and transcriptomic features of response to anti-PD-1 therapy in metastatic melanoma. Cell. 2016;165:35–44. doi: 10.1016/j.cell.2016.02.065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jahani-Asl A., Yin H., Soleimani V.D., Haque T., Luchman H.A., Chang N.C., Sincennes M.C., Puram S.V., Scott A.M., Lorimer I.A.J., et al. Control of glioblastoma tumorigenesis by feed-forward cytokine signaling. Nat. Neurosci. 2016;19:798–806. doi: 10.1038/nn.4295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang P., Gu S., Pan D., Fu J., Sahu A., Hu X., Li Z., Traugh N., Bu X., Li B., et al. Signatures of T cell dysfunction and exclusion predict cancer immunotherapy response. Nat. Med. 2018;24:1550–1558. doi: 10.1038/s41591-018-0136-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kantarjian H., Jabbour E., Grimley J., Kirkpatrick P. Dasatinib. Nat. Rev. Drug Discov. 2006;5:717–718. doi: 10.1038/nrd2135. [DOI] [PubMed] [Google Scholar]
- Lassman A.B., Pugh S.L., Gilbert M.R., Aldape K.D., Geinoz S., Beumer J.H., Christner S.M., Komaki R., DeAngelis L.M., Gaur R., et al. Phase 2 trial of dasatinib in target-selected patients with recurrent glioblastoma (RTOG 0627) Neuro Oncol. 2015;17:992–998. doi: 10.1093/neuonc/nov011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lau S.F., Cao H., Fu A.K.Y., Ip N.Y. Single-nucleus transcriptome analysis reveals dysregulation of angiogenic endothelial cells and neuroprotective glia in Alzheimer's disease. Proc. Natl. Acad. Sci. USA. 2020;117:25800–25809. doi: 10.1073/pnas.2008762117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lauss M., Donia M., Harbst K., Andersen R., Mitra S., Rosengren F., Salim M., Vallon-Christersson J., Törngren T., Kvist A., et al. Mutational and putative neoantigen load predict clinical benefit of adoptive T cell therapy in melanoma. Nat. Commun. 2017;8:1738. doi: 10.1038/s41467-017-01460-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawrence M.S., Stojanov P., Polak P., Kryukov G.V., Cibulskis K., Sivachenko A., Carter S.L., Stewart C., Mermel C.H., Roberts S.A., et al. Mutational heterogeneity in cancer and the search for new cancer-associated genes. Nature. 2013;499:214–218. doi: 10.1038/nature12213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee Y., Scheck A.C., Cloughesy T.F., Lai A., Dong J., Farooqi H.K., Liau L.M., Horvath S., Mischel P.S., Nelson S.F. Gene expression analysis of glioblastomas identifies the major molecular basis for the prognostic benefit of younger age. BMC Med. Genomics. 2008;1:52. doi: 10.1186/1755-8794-1-52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leek J.T., Johnson W.E., Parker H.S., Jaffe A.E., Storey J.D. The sva package for removing batch effects and other unwanted variation in high-throughput experiments. Bioinformatics. 2012;28:882–883. doi: 10.1093/bioinformatics/bts034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leek J.T., Johnson W.E., Parker H.S., Jaffe A.E., Storey J.D. The sva package for removing batch effects and other unwanted variation in high-throughput experiments. Bioinformatics. 2012;28:882–883. doi: 10.1093/bioinformatics/bts034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lei C., Chen W., Wang Y., Zhao B., Liu P., Kong Z., Liu D., Dai C., Wang Y., Wang Y., Ma W. Prognostic prediction model for glioblastoma: a metabolic gene signature and independent external validation. J. Cancer. 2021;12:3796–3808. doi: 10.7150/jca.53827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis-Tuffin L.J., Feathers R., Hari P., Durand N., Li Z., Rodriguez F.J., Bakken K., Carlson B., Schroeder M., Sarkaria J.N., Anastasiadis P.Z. Src family kinases differentially influence glioma growth and motility. Mol. Oncol. 2015;9:1783–1798. doi: 10.1016/j.molonc.2015.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y., Yang C., Liu Z., Du S., Can S., Zhang H., Zhang L., Huang X., Xiao Z., Li X., et al. Integrative analysis of CRISPR screening data uncovers new opportunities for optimizing cancer immunotherapy. Mol. Cancer. 2022;21:2. doi: 10.1186/s12943-021-01462-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liaw A., Wiener M. The R Journal: Classification and regression by randomForest. R News; 2022. https://journal.r-project.org/articles/RN-2002-022/ [Google Scholar]
- Liberzon A., Birger C., Thorvaldsdóttir H., Ghandi M., Mesirov J.P., Tamayo P. The Molecular Signatures Database (MSigDB) hallmark gene set collection. Cell Syst. 2015;1:417–425. doi: 10.1016/j.cels.2015.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim M., Xia Y., Bettegowda C., Weller M. Current state of immunotherapy for glioblastoma. Nat. Rev. Clin. Oncol. 2018;15:422–442. doi: 10.1038/s41571-018-0003-5. [DOI] [PubMed] [Google Scholar]
- Liu T.J., Koul D., LaFortune T., Tiao N., Shen R.J., Maira S.M., Garcia-Echevrria C., Yung W.K.A. NVP-BEZ235, a novel dual phosphatidylinositol 3-kinase/mammalian target of rapamycin inhibitor, elicits multifaceted antitumor activities in human gliomas. Mol. Cancer Ther. 2009;8:2204–2210. doi: 10.1158/1535-7163.MCT-09-0160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez-Jimenez F., Muiños F., Sentís I., Deu-Pons J., Reyes-Salazar I., Arnedo-Pac C., Mularoni L., Pich O., Bonet J., Kranas H., et al. A compendium of mutational cancer driver genes. Nat. Rev. Cancer. 2020;20:555–572. doi: 10.1038/s41568-020-0290-x. [DOI] [PubMed] [Google Scholar]
- McGranahan T., Therkelsen K.E., Ahmad S., Nagpal S. Current state of immunotherapy for treatment of glioblastoma. Curr. Treat. Options Oncol. 2019;20:24. doi: 10.1007/s11864-019-0619-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medikonda R., Dunn G., Rahman M., Fecci P., Lim M. A review of glioblastoma immunotherapy. J. Neuro Oncol. 2021;151:41–53. doi: 10.1007/s11060-020-03448-1. [DOI] [PubMed] [Google Scholar]
- Mermel C.H., Schumacher S.E., Hill B., Meyerson M.L., Beroukhim R., Getz G. GISTIC2.0 facilitates sensitive and confident localization of the targets of focal somatic copy-number alteration in human cancers. Genome Biol. 2011;12:R41. doi: 10.1186/gb-2011-12-4-r41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Messina J.L., Fenstermacher D.A., Eschrich S., Qu X., Berglund A.E., Lloyd M.C., Schell M.J., Sondak V.K., Weber J.S., Mulé J.J. 12-Chemokine gene signature identifies lymph node-like structures in melanoma: potential for patient selection for immunotherapy? Sci. Rep. 2012;2:765. doi: 10.1038/srep00765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyers R.M., Bryan J.G., McFarland J.M., Weir B.A., Sizemore A.E., Xu H., Dharia N.V., Montgomery P.G., Cowley G.S., Pantel S., et al. Computational correction of copy number effect improves specificity of CRISPR-Cas9 essentiality screens in cancer cells. Nat. Genet. 2017;49:1779–1784. doi: 10.1038/ng.3984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mirzaei R., Sarkar S., Yong V.W. T cell exhaustion in glioblastoma: intricacies of immune checkpoints. Trends Immunol. 2017;38:104–115. doi: 10.1016/j.it.2016.11.005. [DOI] [PubMed] [Google Scholar]
- Natesh K., Bhosale D., Desai A., Chandrika G., Pujari R., Jagtap J., Chugh A., Ranade D., Shastry P. Oncostatin-M differentially regulates mesenchymal and proneural signature genes in gliomas via STAT3 signaling. Neoplasia. 2015;17:225–237. doi: 10.1016/j.neo.2015.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neftel C., Laffy J., Filbin M.G., Hara T., Shore M.E., Rahme G.J., Richman A.R., Silverbush D., Shaw M.L., Hebert C.M., et al. An integrative model of cellular states, plasticity, and genetics for glioblastoma. Cell. 2019;178:835–849.e21. doi: 10.1016/j.cell.2019.06.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newman A.M., Steen C.B., Liu C.L., Gentles A.J., Chaudhuri A.A., Scherer F., Khodadoust M.S., Esfahani M.S., Luca B.A., Steiner D., et al. Determining cell type abundance and expression from bulk tissues with digital cytometry. Nat. Biotechnol. 2019;37:773–782. doi: 10.1038/s41587-019-0114-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Omeljaniuk W.J., Krętowski R., Ratajczak-Wrona W., Jabłońska E., Cechowska-Pasko M. Novel dual PI3K/mTOR inhibitor, apitolisib (GDC-0980), inhibits growth and induces apoptosis in human glioblastoma cells. Int. J. Mol. Sci. 2021;22:11511. doi: 10.3390/ijms222111511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Omuro A., DeAngelis L.M. Glioblastoma and other malignant gliomas: a clinical review. JAMA. 2013;310:1842–1850. doi: 10.1001/jama.2013.280319. [DOI] [PubMed] [Google Scholar]
- Poon C.C., Sarkar S., Yong V.W., Kelly J.J.P. Glioblastoma-associated microglia and macrophages: targets for therapies to improve prognosis. Brain. 2017;140:1548–1560. doi: 10.1093/brain/aww355. [DOI] [PubMed] [Google Scholar]
- Prasad G., Sottero T., Yang X., Mueller S., James C.D., Weiss W.A., Polley M.Y., Ozawa T., Berger M.S., Aftab D.T., et al. Inhibition of PI3K/mTOR pathways in glioblastoma and implications for combination therapy with temozolomide. Neuro Oncol. 2011;13:384–392. doi: 10.1093/neuonc/noq193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puchalski R.B., Shah N., Miller J., Dalley R., Nomura S.R., Yoon J.G., Smith K.A., Lankerovich M., Bertagnolli D., Bickley K., et al. An anatomic transcriptional atlas of human glioblastoma. Science. 2018;360:660–663. doi: 10.1126/science.aaf2666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu X., Mao Q., Tang Y., Wang L., Chawla R., Pliner H.A., Trapnell C. Reversed graph embedding resolves complex single-cell trajectories. Nat. Methods. 2017;14:979–982. doi: 10.1038/nmeth.4402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radoul M., Chaumeil M.M., Eriksson P., Wang A.S., Phillips J.J., Ronen S.M. MR studies of glioblastoma models treated with dual PI3K/mTOR inhibitor and temozolomide:metabolic changes are associated with enhanced survival. Mol. Cancer Ther. 2016;15:1113–1122. doi: 10.1158/1535-7163.MCT-15-0769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rees M.G., Seashore-Ludlow B., Cheah J.H., Adams D.J., Price E.V., Gill S., Javaid S., Coletti M.E., Jones V.L., Bodycombe N.E., et al. Correlating chemical sensitivity and basal gene expression reveals mechanism of action. Nat. Chem. Biol. 2016;12:109–116. doi: 10.1038/nchembio.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reifenberger G., Wirsching H.G., Knobbe-Thomsen C.B., Weller M. Advances in the molecular genetics of gliomas - implications for classification and therapy. Nat. Rev. Clin. Oncol. 2017;14:434–452. doi: 10.1038/nrclinonc.2016.204. [DOI] [PubMed] [Google Scholar]
- Ritchie M.E., Phipson B., Wu D., Hu Y., Law C.W., Shi W., Smyth G.K. Limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015;43:e47. doi: 10.1093/nar/gkv007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rooney M.S., Shukla S.A., Wu C.J., Getz G., Hacohen N. Molecular and genetic properties of tumors associated with local immune cytolytic activity. Cell. 2015;160:48–61. doi: 10.1016/j.cell.2014.12.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schalper K.A., Rodriguez-Ruiz M.E., Diez-Valle R., López-Janeiro A., Porciuncula A., Idoate M.A., Inogés S., de Andrea C., López-Diaz de Cerio A., Tejada S., et al. Neoadjuvant nivolumab modifies the tumor immune microenvironment in resectable glioblastoma. Nat. Med. 2019;25:470–476. doi: 10.1038/s41591-018-0339-5. [DOI] [PubMed] [Google Scholar]
- Schiff D., Sarkaria J. Dasatinib in recurrent glioblastoma: failure as a teacher. Neuro Oncol. 2015;17:910–911. doi: 10.1093/neuonc/nov086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharanek A., Burban A., Laaper M., Heckel E., Joyal J.S., Soleimani V.D., Jahani-Asl A. OSMR controls glioma stem cell respiration and confers resistance of glioblastoma to ionizing radiation. Nat. Commun. 2020;11:4116. doi: 10.1038/s41467-020-17885-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stathias V., Jermakowicz A.M., Maloof M.E., Forlin M., Walters W., Suter R.K., Durante M.A., Williams S.L., Harbour J.W., Volmar C.H., et al. Drug and disease signature integration identifies synergistic combinations in glioblastoma. Nat. Commun. 2018;9:5315. doi: 10.1038/s41467-018-07659-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stathias V., Turner J., Koleti A., Vidovic D., Cooper D., Fazel-Najafabadi M., Pilarczyk M., Terryn R., Chung C., Umeano A., et al. LINCS Data Portal 2.0: next generation access point for perturbation-response signatures. Nucleic Acids Res. 2020;48:D431–D439. doi: 10.1093/nar/gkz1023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun D., Guan X., Moran A.E., Wu L.Y., Qian D.Z., Schedin P., Dai M.S., Danilov A.V., Alumkal J.J., Adey A.C., et al. Identifying phenotype-associated subpopulations by integrating bulk and single-cell sequencing data. Nat. Biotechnol. 2022;40:527–538. doi: 10.1038/s41587-021-01091-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan A.C., Ashley D.M., López G.Y., Malinzak M., Friedman H.S., Khasraw M. Management of glioblastoma: state of the art and future directions. CA. Cancer J. Clin. 2020;70:299–312. doi: 10.3322/caac.21613. [DOI] [PubMed] [Google Scholar]
- Tsherniak A., Vazquez F., Montgomery P.G., Weir B.A., Kryukov G., Cowley G.S., Gill S., Harrington W.F., Pantel S., Krill-Burger J.M., et al. Defining a cancer dependency map. Cell. 2017;170:564–576.e16. doi: 10.1016/j.cell.2017.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verhaak R.G.W., Hoadley K.A., Purdom E., Wang V., Qi Y., Wilkerson M.D., Miller C.R., Ding L., Golub T., Mesirov J.P., et al. Integrated genomic analysis identifies clinically relevant subtypes of glioblastoma characterized by abnormalities in PDGFRA, IDH1, EGFR, and NF1. Cancer Cell. 2010;17:98–110. doi: 10.1016/j.ccr.2009.12.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z., Dai Z., Zheng L., Xu B., Zhang H., Fan F., Zhang X., Liang X., Liu Z., Yang K., Cheng Q. Ferroptosis activation scoring model assists in chemotherapeutic agents' selection and mediates cross-talk with immunocytes in malignant glioblastoma. Front. Immunol. 2021;12:747408. doi: 10.3389/fimmu.2021.747408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wen P.Y., Omuro A., Ahluwalia M.S., Fathallah-Shaykh H.M., Mohile N., Lager J.J., Laird A.D., Tang J., Jiang J., Egile C., Cloughesy T.F. Phase I dose-escalation study of the PI3K/mTOR inhibitor voxtalisib (SAR245409, XL765) plus temozolomide with or without radiotherapy in patients with high-grade glioma. Neuro Oncol. 2015;17:1275–1283. doi: 10.1093/neuonc/nov083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilkerson M.D., Hayes D.N. ConsensusClusterPlus: a class discovery tool with confidence assessments and item tracking. Bioinformatics. 2010;26:1572–1573. doi: 10.1093/bioinformatics/btq170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wishart D.S., Feunang Y.D., Guo A.C., Lo E.J., Marcu A., Grant J.R., Sajed T., Johnson D., Li C., Sayeeda Z., et al. DrugBank 5.0: a major update to the DrugBank database for 2018. Nucleic Acids Res. 2018;46:D1074–D1082. doi: 10.1093/nar/gkx1037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu G., Feng X., Stein L. A human functional protein interaction network and its application to cancer data analysis. Genome Biol. 2010;11:R53. doi: 10.1186/gb-2010-11-5-r53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu T., Hu E., Xu S., Chen M., Guo P., Dai Z., Feng T., Zhou L., Tang W., Zhan L., et al. clusterProfiler 4.0: a universal enrichment tool for interpreting omics data. Innovation. 2021;2:100141. doi: 10.1016/j.xinn.2021.100141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu T., Le T.D., Liu L., Su N., Wang R., Sun B., Colaprico A., Bontempi G., Li J. CancerSubtypes: an R/Bioconductor package for molecular cancer subtype identification, validation and visualization. Bioinformatics. 2017;33:3131–3133. doi: 10.1093/bioinformatics/btx378. [DOI] [PubMed] [Google Scholar]
- Yang C., Chen J., Li Y., Huang X., Liu Z., Wang J., Jiang H., Qin W., Lv Y., Wang H., Wang C. Exploring subclass-specific therapeutic agents for hepatocellular carcinoma by informatics-guided drug screen. Brief. Bioinform. 2021;22:bbaa295. doi: 10.1093/bib/bbaa295. [DOI] [PubMed] [Google Scholar]
- Yang C., Huang X., Li Y., Chen J., Lv Y., Dai S. Prognosis and personalized treatment prediction in TP53-mutant hepatocellular carcinoma: an in silico strategy towards precision oncology. Brief. Bioinform. 2021;22:bbaa164. doi: 10.1093/bib/bbaa164. [DOI] [PubMed] [Google Scholar]
- Yang K., Wu Z., Zhang H., Zhang N., Wu W., Wang Z., Dai Z., Zhang X., Zhang L., Peng Y., et al. Glioma targeted therapy: insight into future of molecular approaches. Mol. Cancer. 2022;21:39. doi: 10.1186/s12943-022-01513-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang W., Soares J., Greninger P., Edelman E.J., Lightfoot H., Forbes S., Bindal N., Beare D., Smith J.A., Thompson I.R., et al. Genomics of Drug Sensitivity in Cancer (GDSC): a resource for therapeutic biomarker discovery in cancer cells. Nucleic Acids Res. 2013;41:D955–D961. doi: 10.1093/nar/gks1111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yonekura S., Ueda K. EVI2B is a new prognostic biomarker in metastatic melanoma with IFNgamma associated immune infiltration. Cancers. 2021;13:4110. doi: 10.3390/cancers13164110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshihara K., Shahmoradgoli M., Martínez E., Vegesna R., Kim H., Torres-Garcia W., Treviño V., Shen H., Laird P.W., Levine D.A., et al. Inferring tumour purity and stromal and immune cell admixture from expression data. Nat. Commun. 2013;4:2612. doi: 10.1038/ncomms3612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu P., Laird A.D., Du X., Wu J., Won K.A., Yamaguchi K., Hsu P.P., Qian F., Jaeger C.T., Zhang W., et al. Characterization of the activity of the PI3K/mTOR inhibitor XL765 (SAR245409) in tumor models with diverse genetic alterations affecting the PI3K pathway. Mol. Cancer Ther. 2014;13:1078–1091. doi: 10.1158/1535-7163.MCT-13-0709. [DOI] [PubMed] [Google Scholar]
- Yu Z., Xie G., Zhou G., Cheng Y., Zhang G., Yao G., Chen Y., Li Y., Zhao G. NVP-BEZ235, a novel dual PI3K-mTOR inhibitor displays anti-glioma activity and reduces chemoresistance to temozolomide in human glioma cells. Cancer Lett. 2015;367:58–68. doi: 10.1016/j.canlet.2015.07.007. [DOI] [PubMed] [Google Scholar]
- Zhao H., Chen G., Liang H. Dual PI3K/mTOR Inhibitor, XL765, suppresses glioblastoma growth by inducing ER stress-dependent apoptosis. OncoTargets Ther. 2019;12:5415–5424. doi: 10.2147/OTT.S210128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao H.F., Wang J., Shao W., Wu C.P., Chen Z.P., To S.S.T., Li W.P. Recent advances in the use of PI3K inhibitors for glioblastoma multiforme: current preclinical and clinical development. Mol. Cancer. 2017;16:100. doi: 10.1186/s12943-017-0670-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao J., Chen A.X., Gartrell R.D., Silverman A.M., Aparicio L., Chu T., Bordbar D., Shan D., Samanamud J., Mahajan A., et al. Immune and genomic correlates of response to anti-PD-1 immunotherapy in glioblastoma. Nat. Med. 2019;25:462–469. doi: 10.1038/s41591-019-0349-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao J., Wang L., Hu G., Wei B. A 6-gene risk signature predicts survival of glioblastoma multiforme. Biomed Res. Int. 2019;2019:1649423. doi: 10.1155/2019/1649423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Z., Zhang K.N., Wang Q., Li G., Zeng F., Zhang Y., Wu F., Chai R., Wang Z., Zhang C., et al. Chinese Glioma Genome Atlas (CGGA): A Comprehensive Resource with Functional Genomic Data from Chinese Glioma Patients. Genomics Proteomics Bioinformatics. 2021;19:1–12. doi: 10.1016/j.gpb.2020.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
-
•
This paper analyzes existing, publicly available data. These accession numbers for the datasets are listed in the key resources table.
-
•
R codes used in this study have been deposited and publicly available at GitHub (https://github.com/Liuzhicheng048/GBM).
-
•
Any additional information required to reanalyze the data reported in this paper is available from the lead contact on request.