Abstract
Splicing factor B subunit 4 (SF3B4), a component of the U2-pre-mRNA spliceosomal complex, contributes to tumorigenesis in several types of tumors. However, the oncogenic potential of SF3B4 in lung cancer has not yet been determined. The in vivo expression profiles of SF3B4 in non-small cell lung cancer (NSCLC) from publicly available data revealed a significant increase in SF3B4 expression in tumor tissues compared to that in normal tissues. The impact of SF3B4 deletion on the growth of NSCLC cells was determined using a siRNA strategy in A549 lung adenocarcinoma cells. SF3B4 silencing resulted in marked retardation of the A549 cell proliferation, accompanied by the accumulation of cells at the G0/G1 phase and increased expression of p27, p21, and p53. Double knockdown of SF3B4 and p53 resulted in the restoration of p21 expression and partial recovery of cell proliferation, indicating that the p53/p21 axis is involved, at least in part, in the SF3B4-mediated regulation of A549 cell proliferation. We also provided ubiquitination factor E4B (UBE4B) is essential for p53 accumulation after SF3B4 depletion based on followings. First, co-immunoprecipitation showed that SF3B4 interacts with UBE4B. Furthermore, UBE4B levels were decreased by SF3B4 depletion. UBE4B depletion, in turn, reproduced the outcome of SF3B4 depletion, including reduction of polyubiquitinated p53 levels, subsequent induction of p53/p21 and p27, and proliferation retardation. Collectively, our findings indicate the important role of SF3B4 in the regulation of A549 cell proliferation through the UBE4B/p53/p21 axis and p27, implicating the therapeutic strategies for NSCLC targeting SF3B4 and UBE4B.
Keywords: non-small cell lung cancer, p21, p27, p53, SF3B4, UBE4B
INTRODUCTION
Splicing factor B subunit 4 (SF3B4) is a component of the U2-pre-mRNA spliceosomal complex (Champion-Arnaud and Reed, 1994). Through interaction with another SF3b subunit, spliceosome-associated protein 145 (SAP145), SF3B4 mediates the tethering of the U2 complex to the branch site of pre-mRNA splicing (Gozani et al., 1996). The haploinsufficiency or mutation of SF3B4 is a major genetic cause of the Nager syndrome, characterized by a defective craniofacial formation and preaxial upper limb defect (Bernier et al., 2012; Cassina et al., 2017; Castori et al., 2014; Drozniewska et al., 2020; Hayata et al., 2019; Zhao and Yang, 2020). Even though the molecular basis by which SF3B4 mutation leads to the phenotypes of Nager syndrome is yet to be identified, a recent study indicates that SF3B4 is involved in the translational control of secretory proteins, such as collagen 1 or fibronectin, as cofactors for p180 in the endoplasmic reticulum (Ueno et al., 2019). Thus, the impaired synthesis of collagen by alteration of SF3B4 activity or expression might be a possible mechanism for the development of Nager syndrome.
The dysregulation of alternative splicing might contribute to tumorigenesis through alteration in the expression of key molecules involved in the cell cycle, apoptosis, migration, and invasion (Anczukow and Krainer, 2016). In addition to a mutation in splicing regulatory cis-elements in specific genes involved in survival or proliferation, the mutation or aberrant expression of splicing regulatory factors also frequently contributes to carcinogenesis. Several studies have demonstrated the oncogenic role of SF3B4 in hepatocellular carcinoma (HCC). SF3B4 expression is increased in HCC tissues compared to that in normal tissues and is frequently accompanied by an increased abundance of SF3B4 genes, which are associated with intrahepatic metastasis and poor prognosis (Iguchi et al., 2016; Wang et al., 2020; Xu et al., 2015). Recently, Shen et al. (2018) found that SF3B4 is one of the diagnostic factors for early-stage HCC. They also showed that the depletion or deletion of SF3B4 suppressed the proliferation and metastasis of HCC cells both in vitro and in vivo, probably through the regulation of p27 expression followed by alternative splicing of an upstream transcription factor, Kruppel-like factor 4 (KLF4). Moreover, the miRNA-133b and Ser/Arg (SR)-rich splicing factor (SRSF) 3 are known to regulate SF3B4 levels (Lee et al., 2020; Liu et al., 2018). Clinicopathological and transcriptome data also demonstrated an oncogenic role of SF3B4 in esophageal squamous cell carcinoma and adrenocortical carcinoma (Ding et al., 2021; Kidogami et al., 2020; Lv et al., 2020). In contrast, SF3B4 expression is lower in pancreatic cancer than in adjacent normal tissues. Furthermore, overexpression of SF3B4 suppresses, while its downregulation promotes, the proliferation of pancreatic cancer cells in vitro, suggesting a tumor-suppressive activity of SF3B4 in pancreatic cancers (Zhou et al., 2017). Thus, SF3B4 exhibits diverse regulatory activity of tumorigenesis in different cancers.
Ubiquitination factor E4B (UBE4B) is a member of the E4 ubiquitination factor class, a novel class of ubiquitination enzymes that catalyzes ubiquitin chain assembly, resulting in the elongation of a polyubiquitin chain (Koegl et al., 1999). Subsequent studies indicated that UBE4B can also function as an E3 ubiquitin ligase and that UBE4B mediates degradation of several selected targets including ataxin-2, Tau, and Tax (Matsumoto et al., 2004; Mohanty et al., 2020; Subramanian et al., 2021). Notably, while murine double minute 2 (MDM2) alone only catalyzes mono-ubiquitination of p53 (Lai et al., 2001), UBE4B promotes the poly-ubiquitination of p53, rendering p53 perceptible by the proteasome (Wu et al., 2011). Thus, UBE4B and MDM2 interdependently promote p53 degradation and inhibit p53-dependent transactivation (Du et al., 2016; Zhang et al., 2014). The negative regulatory role for p53 implicates the oncogenic feature of UBE4B, which was supported by aberrant expression of UBE4B in various types of cancers (Antoniou et al., 2019). However, the molecular mechanisms leading to the alterations in the UBE4B gene or in protein levels have not been clarified yet.
Lung cancer is one of the leading causes of cancer-associated death worldwide (Meza et al., 2015). The predominant histological subtype of lung cancer is the non-small cell lung cancer (NSCLC), accounting for 85% of all lung cancers. With regard to the relevance of the aberrant expression of splicing factors in lung cancer, overexpression or mutation of several splicing factors has been reported, including SRSF1, SRSF5, heterogeneous nuclear ribonucleoparticule A1, SRC associated with mitosis of 68 kDa (Sam68), and RNA binding motif protein 10 (de Miguel et al., 2014; Guo et al., 2013; Sumithra et al., 2020; Yan et al., 2019; Zhang et al., 2020). However, whether SF3B4 has oncogenic potential in NSCLC has not been studied yet. Hence, in the present study, we aimed to investigate the effects of SF3B4 silencing on the proliferation of A549 NSCLC cells and its underlying mechanisms. Our results demonstrated that SF3B4 depletion repressed the proliferation of A549 NSCLC cells, accompanied by the accumulation of p53, p21, and p27. We also provided evidence of the involvement of UBE4B in the SF3B4-mediated regulation of p53 stability.
MATERIALS AND METHODS
Cell culture and transfection
The human NSCLC cell line A549 was provided by the American Type Culture Collection (USA). Cells were cultured in RPMI 1640 medium with 10% fetal bovine serum (GeneDEPOT Barker, USA), 1% penicillin-streptomycin (Biowest, France), and 0.1% gentamicin sulfate (Biowest) at 37°C in a humidified incubator with 5% CO2. The indicated proteins were knocked down using a small interfering RNA (siRNA) strategy using Lipofectamine 2000 reagent (Thermo Fisher Scientific, USA). Cells were harvested 48 h after transfection for RNA isolation. The sequences of siRNAs used were as follows: SF3B4, 5’- GCAGUACCUCUGUAACCGU-3’; p53, 5’-CACUACAACUACAUGUGUA-3’; p27, 5’-GGAGCAAUGCGCAGGAAUA-3’; UBE4B, 5’-CCCUGUGUGCAAUUUGGUU-3’; control, 5’-CCUACGCCACCAAUUUCGU-3’.
Reverse transcription and quantitative real-time polymerase chain reaction (qRT-PCR)
For gene expression analysis, qRT-PCR was performed as previously described (Yun et al., 2021). Briefly, total RNA was isolated using RNAiso Plus (TaKara Biotechnology, Japan) and cDNA was synthesized using PrimeScript™ RT Master Mix (TaKara Biotechnology) according to the manufacturer’s instructions. RNA expression levels were measured by standard qPCR procedures with SYBR Premix Ex Taq™ (Takara Biotechnology) on an Applied Biosystems 7300 instrument (Applied Biosystems, USA). The relative values for the target mRNAs were calculated using the 2−ΔΔCT method after normalization to the Ct value of β-actin. The change in target mRNA expression level is presented as a fold change compared to the control cells. The primer sequences used are listed in Supplementary Table S1.
Cell proliferation analysis
After treatment with the indicated siRNAs, cells (1 × 104/well) were seeded into 96-well plates. After incubation for the indicated times, cell proliferation was assessed using 100 μl/well of 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT; Sigma-Aldrich, USA) as previously described (Koh et al., 2020). Then, 100 μl 0.4 M hydrochloric acidic isopropanol and deionized water were added to each well. Optical density values at 570 nm were evaluated using an EpochTM microplate spectrophotometer (BioTek, USA). For the colony formation assay, cells were seeded at a density of 2,000 cells per 60-mm dish and incubated for eight days. Plates were then stained with 1 ml of 0.1% crystal violet (Sigma-Aldrich), and colonies were counted using a dissecting microscope.
Cell cycle analysis
The proportion of A549 cells in each phase of the cell cycle was determined by fluorescence-activated cell sorting (FACS) by labeling with bromodeoxyuridine (BrdU) and 7-aminoactinomycin D (7-AAD) using an APC BrDU flow kit (BD Biosciences, USA). Data acquisition and analysis were performed using a flow cytometer (FACS Calibur; BD Biosciences) and CellQuest Pro software (BD Biosciences).
Western blot analysis
The expression levels of specific proteins were determined by western blotting (Yun et al., 2020). The primary antibodies used are as follows: anti-SF3B4 (Cat. No. ab157117; Abcam, UK), anti-UBE4B (Cat. No. sc-100610; Santa Cruz Biotechnology, USA), anti-MDM2 (Cat. No. sc-965; Santa Cruz Biotechnology), anti-p53 (Cat. No. sc-126; Santa Cruz Biotechnology), anti-p27 (Cat. No. 610242; BD Biosciences), anti-p21 (Cat. No. ab109199; Abcam), anti-hemagglutinin (HA)-probe (Cat. No. sc-9133; Santa Cruz Biotechnology), and anti-β-Actin (Cat. No. A5441; Sigma-Aldrich). After incubation with horseradish peroxidase-conjugated anti-rabbit IgG (Cat. No. GTX213110-01; Genetex, USA) or anti-mouse IgG (Cat. No. GTX213111-01; Genetex), the immunoreactive bands were detected with luminol enhancer solution and peroxide solution (Promega, USA) or SuperSignal™ West Femto Maximum Sensitivity Substrate (Sigma-Aldrich). Densitometric analysis of the specific bands was performed using ImageJ version 1.51 (NIH, USA). For the cycloheximide (CHX; Sigma-Aldrich) chase assay, CHX was added for the indicated times before harvesting cell lysates.
Co-immunoprecipitation
Cells were lysed with lysis buffer (Tris-HCl [pH 7.4], 5 M NaCl, 0.2% NP-40, and 0.5 M EDTA) and protease inhibitor cocktail (Roche, Switzerland). One microgram of normal rabbit IgG (Merck KGaA, Germany), p53 antibody (Cat. No. 10442-1-AP; Proteintech Group, USA), SF3B4 antibody (Abcam), or UBE4B antibody (Santa Cruz Biotechnology) were then added to the supernatant and incubated overnight at 4°C. Protein Agarose A/G (Merck KGaA) was added and the samples were mixed by rolling at 4°C for at least 3 h. The beads were washed three times with lysis buffer, and the pellets were dissolved in 2X SDS loading buffer, and then analyzed using western blot. For the polyubiquitination assay, HA-ubiquitin immunoprecipitation was performed. pRK5-HA-Ubiquitin-WT was a gift from Ted Dawson (Addgene plasmid #17608) (Lim et al., 2005). After 24 h of knockdown of p53 or UBE4B, A549 cells were transfected with pRK5-HA-Ubiquitin-WT using Lipofectamine 2000 reagent (Thermo Fisher Scientific). After 24 h, cells were harvested and polyubiquitination was analyzed by immunoprecipitation with anti-p53 antibody (Santa Cruz Biotechnology) and subsequent western blotting with anti-HA antibody (Santa Cruz Biotechnology).
Establishment of UBE4B-overexpressing A549 cells
The coding region of UBE4B was cloned into the BamHI and EcoR1 sites of a retroviral vector (pBabePuro; Cell Biolabs, USA) and transfected into HEK293T cells by the calcium phosphate method as previously described (Yang et al., 2018). After 36 h of transfection, the virus supernatants were collected and infected into A549 cells, which were then exposed to puromycin (Sigma-Aldrich) for 48 h for selection (A549-UBE4B). As a control, A549 cells were infected with retrovirus containing pBabe vector (A549-CON).
Statistics
Data are expressed as the mean ± SD from three independent experiments. Statistical significance between two groups or multiple groups was analyzed by Student’s t-test or one-way ANOVA and Newman–Keuls multiple comparison test, respectively. A P value of ≤ 0.05 was considered statistically significant.
RESULTS
High expression of SF3B4 is related with poor survival in NSCLC patients
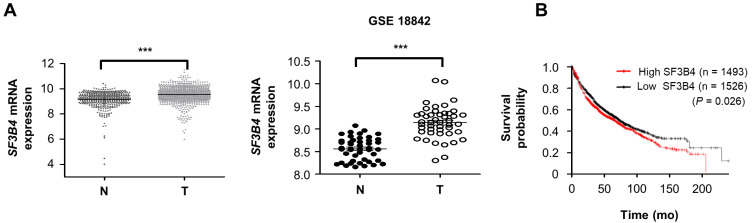
The SF3B4 expression profile in patients with lung cancer or NSCLC was analyzed using GENT2 (http://gent2.appex.kr/gent2/) and the NCBI GEO database (http://www.ncbi.nlm.nih.gov/geo). As shown in Fig. 1A, SF3B4 expression was significantly higher in lung cancer tissues from various types of histology compared to the normal lung tissues, which was also evident in the comparison between paired NSCLC tissues with adjacent normal lung tissue (GSE18842). The clinical significance of the high expression of SF3B4 in NSCLC was evaluated using the Kaplan–Meier survival curve from KMplot.com (https://kmplot.com/analysis). The survival probability of lung cancer patients depending on SF3B4 expression revealed that the high SF3B4 expression group had a relatively poorer overall survival than the low SF3B4 expression group (P = 0.026, Fig. 1B). Hence, SF3B4 may be involved in the progression of NSCLC.
Fig. 1. The expression profile of SF3B4 in patients with lung cancer from public datasets.
(A) The expression level of SF3B4 mRNA between normal (N) and tumor (T) tissues of patients with various types of lung cancer (left, number of samples, N = 466 T = 1,045) and paired samples from patients with NSCLC (GSE18842, N = 45, T = 45). (B) Overall survival analysis of 1493 patients with high SF3B4 expression and 1,526 patients with low SF3B4 expression by the Kaplan–Meier survival curve. ***P < 0.001 between indicated groups.
SF3B4 knockdown repressed the proliferation of A549 cells
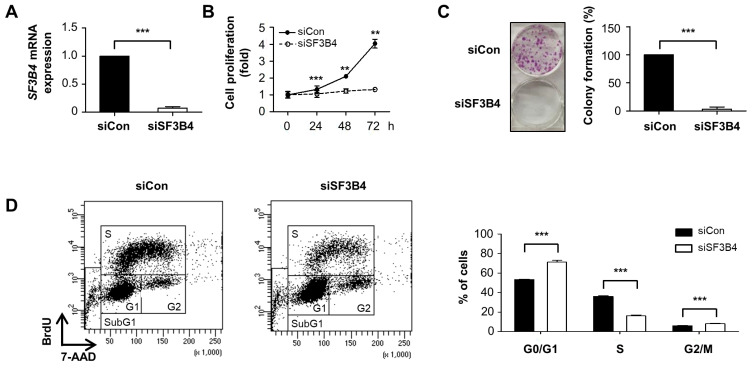
To verify the oncogenic potential of SF3B4 in lung cancer cells, we investigated the effect of SF3B4 knockdown on the proliferation of A549 lung cancer cells in vitro. Suppression of SF3B4 expression was achieved using siRNA, and its efficacy was evaluated by qRT-PCR (Fig. 2A). Using the MTT assay, time-dependent growth was determined in both control and SF3B4-depleted A549 cells. As shown in Fig. 2B, the growth of A549 cells was significantly inhibited by SF3B4 depletion. The absorbance value of control cells increased four-folds 72 h after the start of the experiment, while the value of SF3B4-depleted A549 cells remained almost constant. In addition, SF3B4 depletion markedly suppressed the colony formation ability of A549 cells to 3.2% of the control (Fig. 2C). The levels of cleaved forms of PARP and caspase 3 did not increase significantly, as shown through western blotting (Supplementary Fig. S1A). In addition, propidium iodide (PI) staining revealed that the PI positive population was not increased by SF3B4 depletion (Supplementary Fig. S1B). Hence, the growth retardation induced by SF3B4 depletion was due to neither apoptosis nor necrosis. Subsequently, we analyzed the cell cycle progression following SF3B4 knockdown by FACS analysis after BrdU labeling. Upon SF3B4 depletion, the proportion of cells in the G0/G1 phase increased by almost 20%, from 53.4% to 71.2%, whereas that in the S phase decreased by more than 20%, from 36.2% to 16.1% (Fig. 2D). Thus, cell cycle accumulation in the G1 phase might mainly contribute to cell growth retardation induced by SF3B4 knockdown.
Fig. 2. Proliferation of A549 cells was suppressed by SF3B4 knockdown.
(A) SF3B4 mRNA expression was measured by qRT-PCR 48 h after 100 nM of control siRNA (siCon) or SF3B4 siRNA (siSF3B4) transfection. (B) A549 cells were treated with the indicated siRNA for 0, 24, 48, or 72 h, and the relative viability was determined using MTT assay. Data are presented as mean ± SD of three independent experiments. (C) Colony-forming assay was performed with 2000 cells/well of A549 cells treated with 100 nM of siCon or siSF3B4. After 8 days, the colony number was counted with crystal violet staining (left). The mean values with SD from three independent experiments were provided as a percentage of control (right). (D) Cell cycle analysis was performed after the cells were incubated in siRNA for 48 h by fluorescence-activated cell sorting. The representative staining patterns in control and SF3B4-depleted cells were provided (left). The relative percentages of cells at G0/G1, S and G2/M phase were presented as mean ± SD from three independent experiments. **P < 0.01 and ***P < 0.001 with respect to control values. BrdU, bromodeoxyuridine; 7-AAD, 7-aminoactinomycin D.
p53/p21 axis is involved in SF3B4-mediated regulation of cell growth
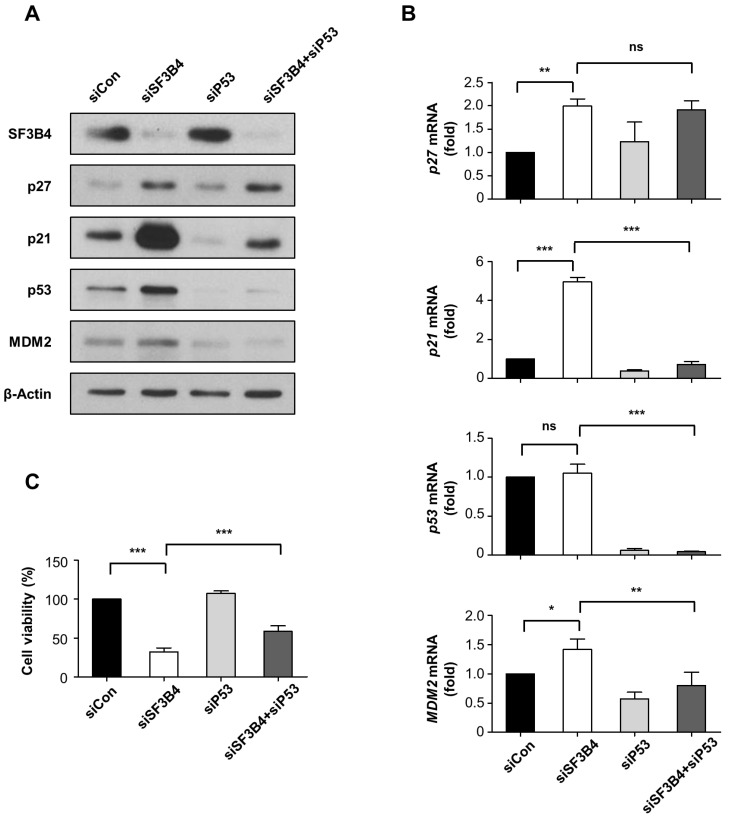
Next, we determined the levels of p27 and p21, two representative cyclin-dependent kinase inhibitors, after SF3B4 depletion. Western blotting assay showed that p27 and p21 expression was notably increased by 3.6- and 5.5-fold, respectively, upon SF3B4 knockdown (Fig. 3A). Subsequent qRT-PCR analysis revealed that p27 and p21 mRNA levels also increased upon SF3B4 depletion, by 2.0- and 4.8-fold, respectively, relative to their levels in the control (Fig. 3B). The mRNA profiles were similar to the protein profiles in western blot, indicating that SF3B4 primarily affects p27 and p21 transcript levels. SF3B4 regulates p27 transcription through exon skipping of KLF4, yielding a nonfunctional product as a transcriptional regulator for p27 in HCC (Shen and Nam, 2018; Shen et al., 2018). Consistent with these previous findings, the depletion of SF3B4 increased functional wild-type KLF4 mRNA while it decreased nonfunctional KLF4 isoform, which seems to contribute to the induction of p27 mRNA in A549 cells (Supplementary Fig. S2). In the present study, since the p21 mRNA level also increased markedly upon SF3B4 depletion, we examined p53 as an upstream regulator of p21 transcription. While SF3B4 depletion induced about two-fold increase in p53 protein levels, the p53 mRNA levels were not altered (Figs. 3A and 3B). MDM2, an E3 ligase to target p53, was also induced by SF3B4 knockdown at the mRNA and protein levels, which appeared to be a p53-dependent gene response (Figs. 3A and 3B). We then performed double knockdown experiments for SF3B4 and p53 to determine whether p53 induction by SF3B4 depletion contributed to the increase in p27 and p21 levels and subsequent cell growth retardation. As shown in Fig. 3A, p53 accumulation induced by SF3B4 depletion was successfully suppressed by p53 siRNA. Upon co-treatment with p53 siRNA, the expression levels of p21 notably decreased at both the protein and mRNA levels, while the expression levels of p27 were not notably affected at both the protein and mRNA levels (Figs. 3A and 3B). The retardation in the A549 cell proliferation, induced by SF3B4 depletion, was partially restored by treatment with p53 siRNA, from 32.2% to 58.8% of that of the control (Fig. 3C). Hence, the p53/p21 axis seems to be involved, at least in part, in the signaling process initiated by SF3B4, leading to cell growth regulation.
Fig. 3. The effects of SF3B4 and/or p53 depletion on the expression of cell cycle regulators and relative survival in A549 cells.
(A) After treatment of control siRNA (siCon), SF3B4 siRNA (siSF3B4), p53 siRNA (siP53) or combined treatment with siSF3B4 and siP53, the alterations of SF3B4, p27, p21, p53, and MDM2 expressions were examined by western blotting assay. As a loading control, beta- Actin levels were used. A representative figure from three independent experiments was provided. (B) The mRNA levels of p27, p21, p53, and MDM2 in the indicated groups were analyzed by qRT-PCR and expressed as a fold change compared to control. (C) Proliferation of A549 cells was measured by MTT assay after depletion of SF3B4 and/or p53. Data are presented as mean ± SD from three independent experiments. *P < 0.05; **P < 0.01; and ***P < 0.001 between indicated groups. ns, non-significant.
p53 protein decay was delayed by SF3B4 depletion
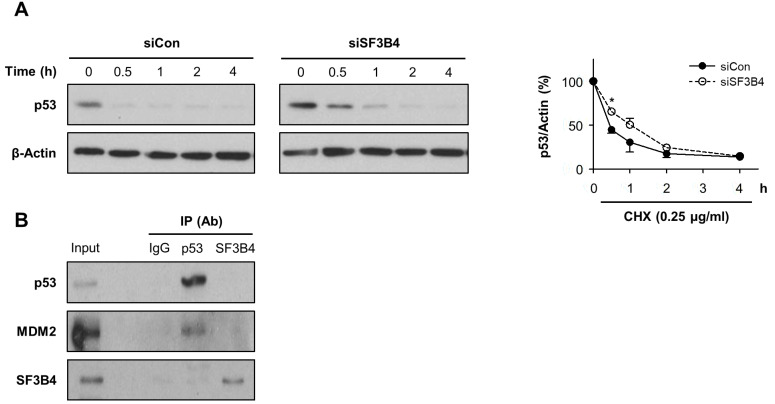
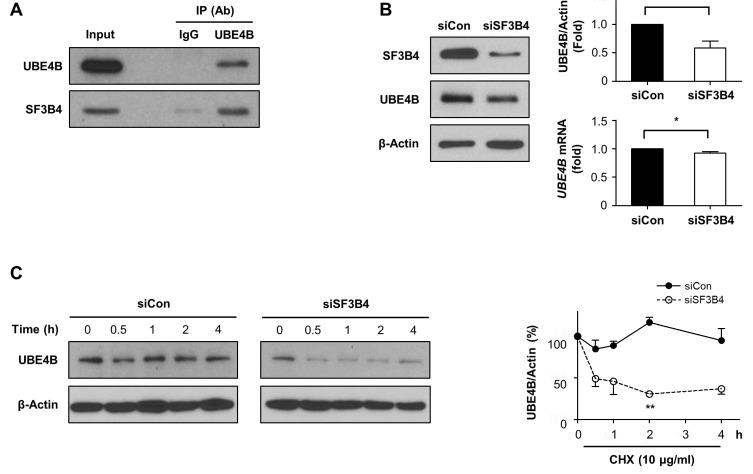
As shown above, p53 induction by SF3B4 depletion was observed at the protein level but not the mRNA level. Hence, we examined whether p53 protein stability was affected by SF3B4 depletion using CHX chase experiments. As expected, the decay of p53 was delayed by SF3B4 depletion; the remaining p53 protein level was 30.6% of its level in the control, while it was 50.6% under SF3B4 depletion 1 h after CHX treatment (Fig. 4A). These findings indicate that, in the absence of SF3B4, p53 degradation was significantly delayed. MDM2 is a critical regulator of p53 protein stability and functions as an E3 ligase (Haupt et al., 1997). However, MDM2 levels increased both at the protein and mRNA levels upon SF3B4 depletion (Figs. 3A and 3B), suggesting that p53 accumulation is not the result of the quantitative reduction in MDM2. We also examined whether SF3B4 interacts with p53 through co-immunoprecipitation. We found that SF3B4 was neither bound to p53 nor MDM2, indicating that p53 stability is not directly affected by protein-protein interaction with SF3B4 (Fig. 4B).
Fig. 4. Degradation of p53 was delayed by SF3B4 depletion.
(A) A549 cells were treated with the control or SF3B4 siRNA for 24 h and subsequently incubated with cycloheximide (CHX, 0.25 μg/ml) for the indicated times. p53 protein levels at the indicated times were determined by western blotting (left) and are provided as mean ± SD from three independent experiments. *P < 0.05 with respect to control values at the same time points. (B) Co-immunoprecipitation was performed by immunoprecipitation (IP) of cell lysates with anti-p53 or anti-SF3B4 and subsequent western blotting with p53, MDM2, or SF3B4. Ab, antibody.
UBE4B is involved in SF3B4-mediated regulation of p53 levels
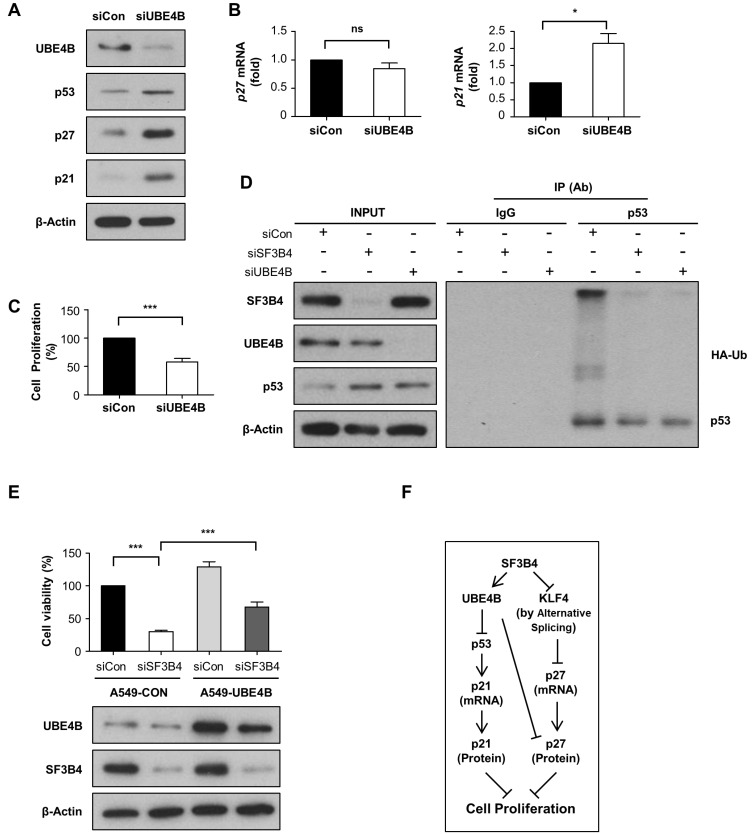
Through a literature search, we recognized that UBE4B, an ubiquitin chain assembly factor, is predicted to be an SF3B4-binding protein based on high throughput complex fractionation and nanoflow liquid chromatography-tandem mass spectroscopy (LC-MS/MS) (Havugimana et al., 2012; Koegl et al., 1999). Furthermore, UBE4B has been previously reported to directly interact with p53 and MDM2, promoting p53 ubiquitination and degradation (Du et al., 2016; Wu et al., 2011; Zhang et al., 2014). Thus, we hypothesized that SF3B4 regulates p53 stability via UBE4B. To test this hypothesis, we investigated the interaction of endogenous SF3B4 with UBE4B in A549 cells by co-immunoprecipitation with UBE4B-specific antibody, followed by western blotting with SF3B4 antibody. As shown in Fig. 5A, SF3B4 interacted physically with UBE4B. We then examined whether UBE4B expression was altered by SF3B4 depletion. UBE4B expression was decreased by SF3B4 knockdown to 58.5% at the protein level but not at the mRNA level relative to their values in control (Fig. 5B). Subsequent CHX chase experiments revealed that, under SF3B4 depletion, UBE4B protein degradation was notably accelerated (Fig. 5C). Thus, SF3B4 affects stability of UBE4B, probably via protein–protein interaction. UBE4B depletion, in turn, increased p53 expression, which concomitantly increased p27 and p21 levels (Fig. 6A). Notably, p21 mRNA levels were upregulated, while p27 mRNA levels were not altered by UBE4B, indicating that UBE4B primarily affects p27 expression at the protein level (Fig. 6B). Furthermore, the growth of A549 cells was significantly suppressed by UBE4B depletion (Fig. 6C). Since UBE4B was shown to polyubiquitinate p53, we determined the polyubiquitination status of p53 after UBE4B or SF3B4 depletion with HA-ubiquitin treatment. As shown in Fig. 6D, multiple bands of ubiquitinated p53 were reduced by the depletion of either protein. Finally, we examined the effects of overexpression of UBE4B on cell viability in the SF3B4 knockdown condition. As shown in Fig. 6E, the viability of A549 cells under SF3B4 depletion were restored by UBE4B overexpression more than two-fold, from 30.2% to 67.8% relative to control cells. Hence, UBE4B might be a critical mediator of SF3B4 signaling, leading to p27- and p53/p21-mediated regulation of proliferation in A549 cells (Fig. 6F).
Fig. 5. UBE4B is involved in p53 stabilization upon SF3B4 depletion in A549 cells.
(A) Interaction between SF3B4 and UBE4B was verified by immunoprecipitation with anti-UBE4B antibody followed by western blotting with anti-SF3B4. IP, immunoprecipitation; Ab, antibody. (B) Effect of SF3B4 knockdown on the expression of UBE4B as determined by western blotting (left, representative result from three independent experiments) and qRT-PCR (right) after exposure to the indicated siRNA for 48 h. (C) Under SF3B4 depletion, the degradation rate of UBE4B was determined by CHX (10 μg/ml) chase assay (left). UBE4B levels are represented as mean ± SD from three independent experiments (right panels in B and C). *P < 0.05 and **P < 0.01 with respect to control values.
Fig. 6. Effect of alteration of UBE4B expression on the cell cycle regulators and cell proliferation in A549 cells.
(A-C) Effect of UBE4B depletion on the expression of cell cycle regulators and cell proliferation by western blotting, qRT-PCR, and MTT assay as in Fig. 3. (D) The ubiquitination status in SF3B4 or UBE4B-depleted A549 cells was measured by transfection with HA-ubiquitin (Ub), IP with anti-p53 or anti-UBE4B antibody, and western blotting with anti-HA antibody. SF3B4 and UBE4B levels in the input were determined by western blotting assay. (E) Effects of overexpression of UBE4B on the cell viability of A549 cells under SF3B4 depletion was assessed by MTT assay and presented as mean ± SD from three independent experiments (upper). The expression levels of UBE4B and SF3B4 were verified by western blotting (lower). The western blot results are representative of three independent experiments. *P < 0.05 and ***P < 0.001 between the indicated groups. ns, non-significant. (F) Proposed model for the molecular pathways by which SF3B4 regulates proliferation of A549 cells.
DISUCUSSION
SF3B4 is a splicing factor whose mutation is a major cause of Nager Syndrome (Liu et al., 2018). SF3B4 has been reported as an oncogenic driver in several tumor types, including HCC and esophageal cancer, while in pancreatic cancer, it acts as a tumor-suppressor protein (Ding et al., 2021; Iguchi et al., 2016; Kidogami et al., 2020; Lee et al., 2020; Liu et al., 2018; Lv et al., 2020; Shen et al., 2018; Wang et al., 2020; Xu et al., 2015; Zhou et al., 2017). In this study, using a siRNA-based knockdown strategy with cell growth and cell cycle analysis, we demonstrated for the first time that SF3B4 exhibits oncogenic properties in A549 NSCLC cells. Our findings were consistent with public data showing a negative association between SF3B4 expression and survival probability in patients with lung cancer.
The molecular mechanisms underlying the oncogenic potential of SF3B4 are yet to be elucidated. Shen and Nam (2018) proposed that SF3B4 regulates p27 transcription level via alternative splicing of tumor suppressor KLF4 in HCC. Overexpression of SF3B4 promotes the generation of nonfunctional KLF4, resulting in the loss of function as a transcriptional regulator for p27 transcription. Likewise, we observed that SF3B4 depletion increased p27 expression in A549 cells with an accumulation of functional KLF4 mRNA, indicating that KLF4-mediated p27 expression is a critical regulatory step in SF3B4 signaling in NSCLC as well.
We also provided evidence for the involvement of the p53/p21 axis in SF3B4-mediated cell cycle regulation. SF3B4 depletion induced the expression of p21 mRNA, as well as the expression of its upstream transcriptional regulator, p53. Furthermore, the double knockdown of SF3B4 and p53 restored the growth of A549 cells concomitantly with the suppression of p21 levels. However, the recovery of cell growth was not perfect since the relative cell survival increased from 32.2% to 58.8% relative to the control, by SF3B4 suppression alone and co-suppression with p53, respectively. This might be because the inhibitory effect of p53 depletion on p27 expression is not as pronounced as that on p21. Thus, the remaining p27 might still contribute to the delayed cell cycle progression under SF3B4 depletion. These findings indicate that both the p53/p21 axis and p27 might contribute to SF3B4-mediated regulation of A549 cells.
The stability of p53 is primarily controlled by MDM2, which targets p53, leading to proteasomal degradation (Haupt et al., 1997). p53 expression was upregulated at the protein but not at the mRNA level upon SF3B4 depletion. The major ubiquitin ligase for p53, MDM2, did not decrease but rather increased upon SF3B4 knockdown, excluding the possibility that the reduction in MDM2 level is the cause of p53 induction under SF3B4 depletion. The increase in MDM2 mRNA levels seems to be due to the autoregulatory feedback loop between p53 and its own inhibitor, MDM2 (Juven-Gershon and Oren, 1999). Furthermore, SF3B4 failed to co-immunoprecipitate with p53 and MDM2, suggesting the presence of additional effector molecules that relay the signal from SF3B4 to p53. Recently, UBE4B, an E3 and E4 ubiquitin ligase, has been shown to interact with MDM2 and p53 as well as to be essential for MDM2-mediated p53 polyubiquitination and degradation (Du et al., 2016; Wu et al., 2011). Moreover, UBE4B is present as a soluble protein complex with SF3B4, as shown through differential biochemical fractionation network and integrative proteomic analysis using cell extracts from HeLa and H293 cells. These co-fractionation results, therefore, increase the interaction probability between two proteins (Havugimana et al., 2012). In this study, we verified the interaction of the endogenous UBE4B and SF3B4 in A549 cells by co-immunoprecipitation. In addition, we showed that the polyubiquitination of p53 was reduced by SF3B4 and UBE4B depletion, indicating that UBE4B is required for polyubiquitination and subsequent degradation of p53 in A549 cells. We also demonstrated that UBE4B expression decreased upon SF3B4 and UBE4B depletion, which, in turn, reproduced SF3B4 depletion-mediated outcomes, including induction of p53, p27, and p21, as well as growth retardation. In addition, overexpression of UBE4B restored the cell viability 30.2% to 67.8% relative to control in the SF3B4 knockdown condition. Notably, UBE4B depletion increased p27 only at the protein level, raising the possibility that UBE4B regulates p27 stabilization. Thus, these results indicate that SF3B4 controls p27 expression via multiple pathways in addition to KLF4, including UBE4B and/or p53, as proposed in Fig. 6F. Collectively, our findings strongly suggest that UBE4B is an essential factor for SF3B4-mediated regulation of p53/p21, p27 expression, and growth retardation in A549 cells. However, the possibility that unknown mechanisms other than cell cycle arrest, such as induction of autophagy or senescence, which may be involved in the control of A549 cell proliferation by SF3B4, cannot be excluded.
As far as the mechanism by which SF3B4 controls UBE4B expression, the splicing regulatory role of SF3B4 does not seem to be involved in this since the molecular weight of UBE4B protein is not altered by SF3B4 depletion. Given that UBE4B expression only decreased at the protein level by SF3B4 depletion and the decay rate of UBE4B was accelerated by SF3B4 depletion, UBE4B seems to be stabilized by interaction with SF3B4. However, in the case of the BMP receptor 1A, SF3B4 decreases its cell surface levels probably by facilitating the internalization of the receptor (Watanabe et al., 2007). Thus, SF3B4, which is known as a splicing factor, could participate in diverse cellular functions through interaction with specific proteins, resulting in the accumulation or degradation of partner proteins depending on the cellular milieu. Recently, UBE4B has been identified as a miR-9 target gene that promotes autophagy-mediated Tau degradation in miRNA libraries in Drosophila (Subramanian et al., 2021). Interestingly, several miRNAs, including miR 9-1, are listed as interactors of SF3B4 (Treiber et al., 2017). Thus, SF3B4 may control UBE4B expression via miR 9-1. However, the effect of the interaction of SF3B4 and miR 9-1 on the stabilization or translation of UBE4B needs to be demonstrated in human cells.
The role of UBE4B as a negative regulator of p53 implicates the oncogenic feature of UBE4B. This is supported by the aberrant expression of UBE4B in diverse cancers frequently associated with poor prognosis (Heuze et al., 2008; Huang et al., 2020; Zage et al., 2013; Zhang et al., 2014; 2016). Furthermore, low UBE4B expression sensitizes neuroblastoma cells to cetuximab or induces apoptosis in nasopharyngeal cancer cells (Memarzadeh et al., 2019; Weng et al., 2019). Thus, targeting either UBE4B, its upstream regulator SF3B4, or both, may have potential therapeutic implications in several types of cancers, including lung cancer.
In summary, our study reinforced the oncogenic properties of SF3B4 in A549 NSCLC cells using a siRNA strategy. We demonstrated for the first time, to our knowledge, that UBE4B is involved in regulating p53/p21 and p27 in SF3B4-mediated regulation of A549 cell growth. Therefore, SF3B4 and UBE4B could serve as molecular targets for the treatment of NSCLC harboring wild-type p53. Further, we discovered a novel role of SF3B4 in stabilizing specific proteins such as UBE4B, beyond its role as a splicing factor, in driving oncogenic processes.
Supplemental Materials
Note: Supplementary information is available on the Molecules and Cells website (www.molcells.org).
ACKNOWLEDGMENTS
This research was funded by grants from the National Research Foundation of Korea (NRF-2019R1A2C1086949).
Footnotes
AUTHOR CONTRIBUTIONS
J.-H.L. conceived and designed the experiments. H.K., J.L., S.-Y.J., and H.H.Y. performed experiments. J.-H.K. analyzed data. All authors have read and agreed to the published version of the manuscript.
CONFLICT OF INTEREST
The authors have no potential conflicts of interest to disclose.
REFERENCES
- Anczukow O., Krainer A.R. Splicing-factor alterations in cancers. RNA. 2016;22:1285–1301. doi: 10.1261/rna.057919.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antoniou N., Lagopati N., Balourdas D.I., Nikolaou M., Papalampros A., Vasileiou P.V.S., Myrianthopoulos V., Kotsinas A., Shiloh Y., Liontos M., et al. The role of E3, E4 ubiquitin ligase (UBE4B) in human pathologies. Cancers (Basel) 2019;12:62. doi: 10.3390/cancers12010062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernier F.P., Caluseriu O., Ng S., Schwartzentruber J., Buckingham K.J., Innes A.M., Jabs E.W., Innis J.W., Schuette J.L., Gorski J.L., et al. Haploinsufficiency of SF3B4, a component of the pre-mRNA spliceosomal complex, causes Nager syndrome. Am. J. Hum. Genet. 2012;90:925–933. doi: 10.1016/j.ajhg.2012.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cassina M., Cerqua C., Rossi S., Salviati L., Martini A., Clementi M., Trevisson E. A synonymous splicing mutation in the SF3B4 gene segregates in a family with highly variable Nager syndrome. Eur. J. Hum. Genet. 2017;25:371–375. doi: 10.1038/ejhg.2016.176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castori M., Bottillo I., D'Angelantonio D., Morlino S., De Bernardo C., Scassellati Sforzolini G., Silvestri E., Grammatico P. A 22-week-old fetus with Nager syndrome and congenital diaphragmatic hernia due to a novel SF3B4 mutation. Mol. Syndromol. 2014;5:241–244. doi: 10.1159/000365769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Champion-Arnaud P., Reed R. The prespliceosome components SAP 49 and SAP 145 interact in a complex implicated in tethering U2 snRNP to the branch site. Genes Dev. 1994;8:1974–1983. doi: 10.1101/gad.8.16.1974. [DOI] [PubMed] [Google Scholar]
- de Miguel F.J., Sharma R.D., Pajares M.J., Montuenga L.M., Rubio A., Pio R. Identification of alternative splicing events regulated by the oncogenic factor SRSF1 in lung cancer. Cancer Res. 2014;74:1105–1115. doi: 10.1158/0008-5472.CAN-13-1481. [DOI] [PubMed] [Google Scholar]
- Ding J., Li C., Cheng Y., Du Z., Wang Q., Tang Z., Song C., Xia Q., Bai W., Lin L., et al. Alterations of RNA splicing patterns in esophagus squamous cell carcinoma. Cell Biosci. 2021;11:36. doi: 10.1186/s13578-021-00546-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drozniewska M., Kilby M.D., Vogt J., Togneri F., Quinlan-Jones E., Reali L., Allen S., McMullan D. Second-trimester prenatal diagnosis of Nager syndrome with a deletion including SF3B4 detected by chromosomal microarray. Clin. Case Rep. 2020;8:508–511. doi: 10.1002/ccr3.2509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du C., Wu H., Leng R.P. UBE4B targets phosphorylated p53 at serines 15 and 392 for degradation. Oncotarget. 2016;7:2823–2836. doi: 10.18632/oncotarget.6555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gozani O., Feld R., Reed R. Evidence that sequence-independent binding of highly conserved U2 snRNP proteins upstream of the branch site is required for assembly of spliceosomal complex A. Genes Dev. 1996;10:233–243. doi: 10.1101/gad.10.2.233. [DOI] [PubMed] [Google Scholar]
- Guo R., Li Y., Ning J., Sun D., Lin L., Liu X. HnRNP A1/A2 and SF2/ASF regulate alternative splicing of interferon regulatory factor-3 and affect immunomodulatory functions in human non-small cell lung cancer cells. PLoS One. 2013;8:e62729. doi: 10.1371/journal.pone.0062729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haupt Y., Maya R., Kazaz A., Oren M. Mdm2 promotes the rapid degradation of p53. Nature. 1997;387:296–299. doi: 10.1038/387296a0. [DOI] [PubMed] [Google Scholar]
- Havugimana P.C., Hart G.T., Nepusz T., Yang H., Turinsky A.L., Li Z., Wang P.I., Boutz D.R., Fong V., Phanse S., et al. A census of human soluble protein complexes. Cell. 2012;150:1068–1081. doi: 10.1016/j.cell.2012.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayata K., Masuyama H., Eto E., Mitsui T., Tamada S., Eguchi T., Maki J., Tani K., Ohira A., Washio Y., et al. A case of Nager syndrome diagnosed before birth. Acta Med. Okayama. 2019;73:273–277. doi: 10.18926/AMO/56872. [DOI] [PubMed] [Google Scholar]
- Heuze M.L., Lamsoul I., Moog-Lutz C., Lutz P.G. Ubiquitin-mediated proteasomal degradation in normal and malignant hematopoiesis. Blood Cells Mol. Dis. 2008;40:200–210. doi: 10.1016/j.bcmd.2007.07.011. [DOI] [PubMed] [Google Scholar]
- Huang X.Q., Hao S., Zhou Z.Q., Huang B., Fang J.Y., Tang Y., Zhang J.H., Xia J.C. The roles of ubiquitination factor E4B (UBE4B) in the postoperative prognosis of patients with renal cell carcinoma and in renal tumor cells growth and metastasis. Onco Targets Ther. 2020;13:185–197. doi: 10.2147/OTT.S229577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iguchi T., Komatsu H., Masuda T., Nambara S., Kidogami S., Ogawa Y., Hu Q., Saito T., Hirata H., Sakimura S., et al. Increased copy number of the gene encoding SF3B4 indicates poor prognosis in hepatocellular carcinoma. Anticancer Res. 2016;36:2139–2144. [PubMed] [Google Scholar]
- Juven-Gershon T., Oren M. Mdm2: the ups and downs. Mol. Med. 1999;5:71–83. doi: 10.1007/BF03402141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kidogami S., Iguchi T., Sato K., Yoshikawa Y., Hu Q., Nambara S., Komatsu H., Ueda M., Kuroda Y., Masuda T., et al. SF3B4 plays an oncogenic role in esophageal squamous cell carcinoma. Anticancer Res. 2020;40:2941–2946. doi: 10.21873/anticanres.14272. [DOI] [PubMed] [Google Scholar]
- Koegl M., Hoppe T., Schlenker S., Ulrich H.D., Mayer T.U., Jentsch S. A novel ubiquitination factor, E4, is involved in multiubiquitin chain assembly. Cell. 1999;96:635–644. doi: 10.1016/S0092-8674(00)80574-7. [DOI] [PubMed] [Google Scholar]
- Koh E.Y., You J.E., Jung S.H., Kim P.H. Biological functions and identification of novel biomarker expressed on the surface of breast cancer-derived cancer stem cells via proteomic analysis. Mol. Cells. 2020;43:384–396. doi: 10.14348/molcells.2020.2230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai Z., Ferry K.V., Diamond M.A., Wee K.E., Kim Y.B., Ma J., Yang T., Benfield P.A., Copeland R.A., Auger K.R. Human mdm2 mediates multiple mono-ubiquitination of p53 by a mechanism requiring enzyme isomerization. J. Biol. Chem. 2001;276:31357–31367. doi: 10.1074/jbc.M011517200. [DOI] [PubMed] [Google Scholar]
- Lee J., Seo G., Hur W., Yoon S.K., Nam S.W., Lee J.H. SRSF3 depletion leads to an increase in SF3B4 expression in SNU-368 HCC cells. Anticancer Res. 2020;40:2033–2042. doi: 10.21873/anticanres.14160. [DOI] [PubMed] [Google Scholar]
- Lim K.L., Chew K.C., Tan J.M., Wang C., Chung K.K., Zhang Y., Tanaka Y., Smith W., Engelender S., Ross C.A., et al. Parkin mediates nonclassical, proteasomal-independent ubiquitination of synphilin-1: implications for Lewy body formation. J. Neurosci. 2005;25:2002–2009. doi: 10.1523/JNEUROSCI.4474-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Z., Li W., Pang Y., Zhou Z., Liu S., Cheng K., Qin Q., Jia Y., Liu S. SF3B4 is regulated by microRNA-133b and promotes cell proliferation and metastasis in hepatocellular carcinoma. EBioMedicine. 2018;38:57–68. doi: 10.1016/j.ebiom.2018.10.067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lv J., He Y., Li L., Wang Z. Alternative splicing events and splicing factors are prognostic in adrenocortical carcinoma. Front. Genet. 2020;11:918. doi: 10.3389/fgene.2020.00918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsumoto M., Yada M., Hatakeyama S., Ishimoto H., Tanimura T., Tsuji S., Kakizuka A., Kitagawa M., Nakayama K.I. Molecular clearance of ataxin-3 is regulated by a mammalian E4. EMBO J. 2004;23:659–669. doi: 10.1038/sj.emboj.7600081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Memarzadeh K., Savage D.J., Bean A.J. Low UBE4B expression increases sensitivity of chemoresistant neuroblastoma cells to EGFR and STAT5 inhibition. Cancer Biol. Ther. 2019;20:1416–1429. doi: 10.1080/15384047.2019.1647049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meza R., Meernik C., Jeon J., Cote M.L. Lung cancer incidence trends by gender, race and histology in the United States, 1973-2010. PLoS One. 2015;10:e0121323. doi: 10.1371/journal.pone.0121323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohanty S., Han T., Choi Y.B., Lavorgna A., Zhang J., Harhaj E.W. The E3/E4 ubiquitin conjugation factor UBE4B interacts with and ubiquitinates the HTLV-1 Tax oncoprotein to promote NF-kappaB activation. PLoS Pathog. 2020;16:e1008504. doi: 10.1371/journal.ppat.1008504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen Q., Eun J.W., Lee K., Kim H.S., Yang H.D., Kim S.Y., Lee E.K., Kim T., Kang K., Kim S., et al. Barrier to autointegration factor 1, procollagen-lysine, 2-oxoglutarate 5-dioxygenase 3, and splicing factor 3b subunit 4 as early-stage cancer decision markers and drivers of hepatocellular carcinoma. Hepatology. 2018;67:1360–1377. doi: 10.1002/hep.29606. [DOI] [PubMed] [Google Scholar]
- Shen Q., Nam S.W. SF3B4 as an early-stage diagnostic marker and driver of hepatocellular carcinoma. BMB Rep. 2018;51:57–58. doi: 10.5483/BMBRep.2018.51.2.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subramanian M., Hyeon S.J., Das T., Suh Y.S., Kim Y.K., Lee J.S., Song E.J., Ryu H., Yu K. UBE4B, a microRNA-9 target gene, promotes autophagy-mediated Tau degradation. Nat. Commun. 2021;12:3291. doi: 10.1038/s41467-021-23597-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sumithra B., Jayanthi V., Manne H.C., Gunda R., Saxena U., Das A.B. Antibody-based biosensor to detect oncogenic splicing factor Sam68 for the diagnosis of lung cancer. Biotechnol. Lett. 2020;42:2501–2509. doi: 10.1007/s10529-020-02951-9. [DOI] [PubMed] [Google Scholar]
- Treiber T., Treiber N., Plessmann U., Harlander S., Daiss J.L., Eichner N., Lehmann G., Schall K., Urlaub H., Meister G. A compendium of RNA-binding proteins that regulate microRNA biogenesis. Mol. Cell. 2017;66:270–284.e13. doi: 10.1016/j.molcel.2017.03.014. [DOI] [PubMed] [Google Scholar]
- Ueno T., Taga Y., Yoshimoto R., Mayeda A., Hattori S., Ogawa-Goto K. Component of splicing factor SF3b plays a key role in translational control of polyribosomes on the endoplasmic reticulum. Proc. Natl. Acad. Sci. U. S. A. 2019;116:9340–9349. doi: 10.1073/pnas.1901742116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L., Zhang Z., Li Y., Wan Y., Xing B. Integrated bioinformatic analysis of RNA binding proteins in hepatocellular carcinoma. Aging (Albany N.Y.) 2020;13:2480–2505. doi: 10.18632/aging.202281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe H., Shionyu M., Kimura T., Kimata K., Watanabe H. Splicing factor 3b subunit 4 binds BMPR-IA and inhibits osteochondral cell differentiation. J. Biol. Chem. 2007;282:20728–20738. doi: 10.1074/jbc.M703292200. [DOI] [PubMed] [Google Scholar]
- Weng C., Chen Y., Wu Y., Liu X., Mao H., Fang X., Li B., Wang L., Guan M., Liu G., et al. Silencing UBE4B induces nasopharyngeal carcinoma apoptosis through the activation of caspase3 and p53. Onco Targets Ther. 2019;12:2553–2561. doi: 10.2147/OTT.S196132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu H., Pomeroy S.L., Ferreira M., Teider N., Mariani J., Nakayama K.I., Hatakeyama S., Tron V.A., Saltibus L.F., Spyracopoulos L., et al. UBE4B promotes Hdm2-mediated degradation of the tumor suppressor p53. Nat. Med. 2011;17:347–355. doi: 10.1038/nm.2283. [DOI] [PubMed] [Google Scholar]
- Xu W., Huang H., Yu L., Cao L. Meta-analysis of gene expression profiles indicates genes in spliceosome pathway are up-regulated in hepatocellular carcinoma (HCC) Med. Oncol. 2015;32:96. doi: 10.1007/s12032-014-0425-6. [DOI] [PubMed] [Google Scholar]
- Yan J., Zhang D., Han Y., Wang Z., Ma C. Antitumor activity of SR splicing-factor 5 knockdown by downregulating pyruvate kinase M2 in non-small cell lung cancer cells. J. Cell. Biochem. 2019;120:17303–17311. doi: 10.1002/jcb.28992. [DOI] [PubMed] [Google Scholar]
- Yang S., Hwang S., Kim M., Seo S.B., Lee J.H., Jeong S.M. Mitochondrial glutamine metabolism via GOT2 supports pancreatic cancer growth through senescence inhibition. Cell Death Dis. 2018;9:55. doi: 10.1038/s41419-017-0089-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yun H.H., Jung S.Y., Park B.W., Ko J.S., Yoo K., Yeo J., Kim H.L., Park H.J., Youn H.J., Lee J.H. An adult mouse model of dilated cardiomyopathy caused by inducible cardiac-specific Bis deletion. Int. J. Mol. Sci. 2021;22:1343. doi: 10.3390/ijms22031343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yun H.H., Kim S., Kuh H.J., Lee J.H. Downregulation of BIS sensitizes A549 cells for digoxin-mediated inhibition of invasion and migration by the STAT3-dependent pathway. Biochem. Biophys. Res. Commun. 2020;524:643–648. doi: 10.1016/j.bbrc.2020.01.154. [DOI] [PubMed] [Google Scholar]
- Zage P.E., Sirisaengtaksin N., Liu Y., Gireud M., Brown B.S., Palla S., Richards K.N., Hughes D.P., Bean A.J. UBE4B levels are correlated with clinical outcomes in neuroblastoma patients and with altered neuroblastoma cell proliferation and sensitivity to epidermal growth factor receptor inhibitors. Cancer. 2013;119:915–923. doi: 10.1002/cncr.27785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang S., Bao Y., Shen X., Pan Y., Sun Y., Xiao M., Chen K., Wei H., Zuo J., Saffen D., et al. RNA binding motif protein 10 suppresses lung cancer progression by controlling alternative splicing of eukaryotic translation initiation factor 4H. EBioMedicine. 2020;61:103067. doi: 10.1016/j.ebiom.2020.103067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X.F., Pan Q.Z., Pan K., Weng D.S., Wang Q.J., Zhao J.J., He J., Liu Q., Wang D.D., Jiang S.S., et al. Expression and prognostic role of ubiquitination factor E4B in primary hepatocellular carcinoma. Mol. Carcinog. 2016;55:64–76. doi: 10.1002/mc.22259. [DOI] [PubMed] [Google Scholar]
- Zhang Y., Lv Y., Zhang Y., Gao H. Regulation of p53 level by UBE4B in breast cancer. PLoS One. 2014;9:e90154. doi: 10.1371/journal.pone.0090154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao J., Yang L. Broad-spectrum next-generation sequencing-based diagnosis of a case of Nager syndrome. J. Clin. Lab. Anal. 2020;34:e23426. doi: 10.1002/jcla.23426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou W., Ma N., Jiang H., Rong Y., Deng Y., Feng Y., Zhu H., Kuang T., Lou W., Xie D., et al. SF3B4 is decreased in pancreatic cancer and inhibits the growth and migration of cancer cells. Tumour Biol. 2017;39:1010428317695913. doi: 10.1177/1010428317695913. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.