Abstract
With the increasing availability of high-resolution mass spectrometers, suspect screening and non-targeted analysis are becoming popular compound identification tools for environmental researchers. Samples of interest often contain a large (unknown) number of chemicals spanning the detectable mass range of the instrument. In an effort to separate these chemicals prior to injection into the mass spectrometer, a chromatography method is often utilized. There are numerous types of gas and liquid chromatographs that can be coupled to commercially available mass spectrometers. Depending on the type of instrument used for analysis, the researcher is likely to observe a different subset of compounds based on the amenability of those chemicals to the selected experimental techniques and equipment. It would be advantageous if this subset of chemicals could be predicted prior to conducting the experiment, in order to minimize potential false-positive and false-negative identifications. In this work, we utilize experimental datasets to predict the amenability of chemical compounds to detection with liquid chromatography-electrospray ionization-mass spectrometry (LC–ESI–MS). The assembled dataset totals 5517 unique chemicals either explicitly detected or not detected with LC–ESI–MS. The resulting detected/not-detected matrix has been modeled using specific molecular descriptors to predict which chemicals are amenable to LC–ESI–MS, and to which form(s) of ionization. Random forest models, including a measure of the applicability domain of the model for both positive and negative modes of the electrospray ionization source, were successfully developed. The outcome of this work will help to inform future suspect screening and non-targeted analyses of chemicals by better defining the potential LC–ESI–MS detectable chemical landscape of interest.
Keywords: Non-targeted analysis, Suspect screening analysis, Mass spectrometry, Machine learning, Random forest, Predictive modeling
Introduction
Previous studies have shown that humans are exposed to thousands of chemicals each day, either through near-field or far-field sources [1-4]. The totality of these exogenous chemical exposures comprises a large portion of the human exposome, the sum of all exposures (external and internal) experienced by an individual throughout their lifetime [5]. Comprehensive exposome analyses benefit from a combination of bottom-up and top-down approaches using various biological (top-down) and environmental/consumer (bottom-up) samples. As the field of exposomics continues to evolve, so does the ability to thoroughly examine the chemical complexity of these samples.
Whereas high-throughput screening assays for bioactivity have generated data on thousands of biological endpoints for thousands of chemicals (see, e.g., ToxCast [6] and Tox21 [7]), these assays are typically based on chemical-independent probes (for example, luminescence from green fluorescent protein) [8]. Other chemical properties key to understanding chemical risk posed to public health [9]. including physicochemical and toxicokinetic (absorption, distribution, metabolism, excretion), require the development of targeted (that is, chemical-specific) analysis methods [10, 11]. Nicolas et al. examined higher-throughput methods for measuring physicochemical properties (for example, hydrophobicity, water solubility) based on high-performance liquid chromatography—out of 200 ToxCast chemicals, methods could not be developed ~ 15% of the time [11]. In Wetmore et al., developing analytical methods for measuring chemical fraction unbound in plasma failed for 38% of the chemicals [12]. In both cases, resources were expended attempting to develop methods for those chemicals.
Whereas targeted analytical methods can be used to determine the presence and concentration of small numbers of chemicals (on the order of 10s to 100s) in a given sample, this approach is not feasible for comprehensive chemical analysis. Two alternative techniques, known as non-targeted analysis (NTA) and suspect screening analysis (SSA), provide a means to address such a need. NTA uses high-resolution mass spectrometry (HRMS) to deduce the identity of unknown/understudied compounds without the use of chemical standards or chemical suspect lists. Similarly, SSA uses HRMS to tentatively identify chemicals in samples of interest using lists of chemical suspects and, in many cases, supporting data (e.g., reference spectra).
Non-targeted and suspect screening analyses are commonly performed using a chromatograph in tandem with a mass spectrometer. Both gas and liquid chromatography have been successfully used to aid in the characterization of large numbers of small molecules in various media [13-19]. However, neither approach on its own is capable of determining the entire chemical composition of a sample as some chemicals are not amenable to specific methods, ionization techniques, etc. For example, liquid chromatography-electrospray ionization-mass spectrometry (LC–ESI–MS) has proven valuable for the analysis of chemicals with low volatility, such as some perfluoroalkyl substances (PFAS), e.g., perfluorocarboxylic acids and perfluorosulfonic acids. Likewise, many volatile chemicals, including fluorotelomer alcohols commonly found in aqueous film-forming foams, are much more amenable to analysis via gas chromatography-mass spectrometry (GC–MS) [20]. In a recent evaluation of NTA method performance (part of the Environmental Protection Agency’s, EPA’s Non-Targeted Analysis Collaborative Trial [ENTACT]), 1269 diverse chemical substances were analyzed using multiple LC–ESI–MS methods, with up to 40% noted as being unamenable to detection and/or identification [13]. Considering this result, a clear benefit would exist to having model(s) that can accurately predict the amenability of compounds in LC–ESI–MS experiments to aid in the interpretation of positive (compound reported as present) and negative (compound not reported as present) findings. Having this predictive capability could also reduce the costs of time and resources associated with analyzing unamenable compounds.
Herein, we investigate the application of quantitative structure-activity relationship (QSAR) modeling, where “activity” is defined in this case as “amenability to detection with LC–ESI–MS.” Random forest models were used to predict a compound’s amenability to detection with LC–ESI–MS. Specifically, we collected a large (6342 representatives) dataset of chemicals with known LC–ESI–MS amenability, represented our chemicals using PaDEL molecular descriptors, and built random forest models to predict the LC–ESI–MS amenability of compounds for detection using both positive and negative modes of an electrospray ionization source. Model predictivity is evaluated using statistics from Y-randomization, fivefold cross validation (CV), and external validation sets. An applicability domain is defined using the class probability estimates from each of the random forest models. These models provide a new technique to add weight-of-evidence when selecting and eliminating tentative chemical identities in NTA and/or SSA experiments. Whereas no model will likely ever predict the amenability of all (> 1060) organic molecules [21], the models presented here attempt to predict within the subspace of compounds commonly identified in environmental analysis.
Methods
Dataset assembly
Experimental spectra and associated metadata were assembled from the MassBank of North America (MoNA) database [22]. Assembled spectra were for compounds observed via LC–ESI–MS/MS analysis with electrospray ionization in either positive or negative modes. Spectra were restricted to those acquired via tandem mass spectrometry to increase the chance of correct chemical identification. All data were downloaded as an SD file [23] and parsed using the ChemmineR package [24] in the R programming language [25]. All forms of LC-based methods were considered acceptable, and no attempt was made to filter any data based on the specific use of columns, mobile phases, or method conditions. For the purposes of this analysis, a chemical was considered detected if spectra existed for that record in MoNA. Chemicals were identified by InChIKey [26], if available, or by chemical name. Note that the InChIKey first block (the first section of an InChIKey) represents the molecular skeleton and is generally the extent of structural representation possible for a chemical identified using mass spectrometry. The second block of the InChIKey encodes stereochemistry, charge, and isotopic labeling. Quality issues associated with incorrect representations of stereochemistry and charge can be common in online databases, including MoNA [22]. Thus, for each chemical, only the first block of the InChIKey identifier (or the chemical name) was searched using the US EPA’s CompTox Chemicals Dashboard [27] (referred to hereafter as the “Dashboard”). The batch search feature of the Dashboard was used to download structural identifiers for each searched compound, thus enabling additional curation of chemical names and structures retrieved from MoNA [28]. Any chemical that could not be identified by InChIKey or name using the Dashboard search was discarded without any attempt to further curate the data. Ultimately, we assembled 3007 unique chemicals as being detected via ESI negative mode, 4103 as being detected in ESI positive mode, and 1542 of the 7110 chemicals detected in both modes.
In addition to the spectra acquired from MoNA, amenability data were also compiled from analyses of selected ToxCast Screening Library substances. Spectra were acquired on individual standards via LC–ESI–MS/MS in both ESI+ and ESI− modes and manually reviewed for quality. Only [M+H]+ and [M-H]− adducts were considered. Spectra were reviewed by evaluating base peak height, chemical noise, and mass accuracy of precursor and fragment ions, and consisted of a final visual review. For these purposes, spectra were excluded (considered not detected) when base peak heights were below 1000 counts, when significant impurities were present in the LC–ESI–MS/MS chromatogram, or when a confirmatory spectral match score was less than 90 (out of 100). Low match scores resulted from excessive chemical noise, impurities in MS/MS spectra, or poor mass accuracy of fragment ions relative to the theoretical accurate mass. Only those spectra meeting quality criteria were considered as “detected” for the purposes of modeling. The resulting dataset included 849 positively detected compounds (393 in ESI+ and 456 in ESI−) and 858 compounds that were undetected or considered unamenable due to not meeting the established quality criteria (456 in ESI+ and 402 in ESI−). The undetected compounds were thus assumed to be true-negative results for the purposes of this analysis, but it is acknowledged that there may be certain LC–ESI–MS/MS conditions where these compounds are amenable. In the event that an undetected compound was detected in the MoNA dataset, it was assumed as detected in the final dataset. In total, the following data compilations were produced: 4613 with ESI+ data (4226 amenable and 387 unamenable); 3490 with ESI− data (3130 amenable and 360 unamenable); and 1761 detected in both methods as either amenable (1604 in ESI+ and 1583 in ESI−) or unamenable (202 in ESI+ and 178 in ESI−). It is important to note the large difference in amenable and unamenable compounds in this dataset. Methods for dealing with this imbalance will be discussed in a later section.
Finally, an external dataset consisting of 1767 chemicals that were tested in phase II of the ToxCast screening program, and for which LC–ESI–MS data were collected, was used as an external validation set to evaluate the predictive capability of the models derived in this study. A description of the phase II library contents can be found in Richard et al. [29]. The LC–ESI–MS data in this case were generated under EPA contract with Evotec BioCT (Branford, CT) on test sample plates run in high-throughput mode, i.e., in ESI− and ESI+ detection modes with a single retention time scan window. The results were available only as a summary call in which a positive parent mass ID in either or both modes yielded a summary assessment of “amenable” and a negative parent mass ID in both modes yielded a summary assessment of “unamenable.”
Molecular descriptor calculation and reduction
Using the Dashboard’s batch search feature, QSAR-ready SMILES [30] were obtained for all chemicals identified in both the MoNA and ToxCast datasets. Briefly, QSAR-ready SMILES are representations of desalted, de-isotoped, stereoneutral forms of chemical structures which are appropriate for QSAR modeling (typically excluding inorganics). The QSAR-ready SMILES were used as the basis to obtain both 1D and 2D molecular descriptors using the PaDEL-descriptor software [31]. These descriptors capture aspects of the chemical composition and topology of molecules and have been used in a number of previous QSAR studies [32-35]. A total of 1444 molecular descriptors were calculated. To increase the efficiency and interpretability of the models, constant and highly correlated descriptors were eliminated, reducing the number of descriptors down to 451. Correlation coefficients for descriptor pairs were calculated using the Spearman method. Descriptors were considered highly correlated if the computed correlation coefficient was greater than 0.96. If the correlation threshold was exceeded, then the descriptor with the largest mean absolute correlation across all possible pairwise correlations was eliminated and the other retained. Descriptions of the 1444 PaDEL descriptors (with the 451 used descriptors highlighted in red) are availble in the supplemental file “PaDEL_descriptions.xlsx.”
Publicly available ToxPrint fingerprints (https://toxprints.org) were downloaded for all chemicals using the Dashboard batch search feature. ToxPrint chemotypes are designed to capture salient features of environmental and regulated chemical inventories, including reactive groups, bonding patterns, and scaffolds relevant to safety assessment workflows within regulatory agencies, such as EPA and the Food and Drug Administration [36]. ToxPrint chemotypes are used herein to examine model results and compare inventories with intuitive and visualizable chemical substructure features.
Model learning approach
The random forest (RF) classification algorithm was chosen as the modeling method for this study. A random forest is constructed using a user-specified number of decision trees, denoted here as ntree. These decision trees rely on splitting each decision node using a random number of descriptors (sampled with replacement), denoted here as mtry and also specified by the user, to classify an endpoint. The majority of the predicted classifications from this ensemble of decision trees is used to predict the classification of novel compounds. This sampling technique coupled with a suitably large number of decision trees leads to an unbiased prediction model resistant to overfitting. Random forest was implemented using both caret and randomForest packages in the R programming language [37]. A grid search procedure was used to optimize the hyperparameter mtry for each model using the highest receiver operating characteristic (ROC) metric value. The ROC metric was chosen to find the best balance between sensitivity and specificity (discussed in detail later). The grid search for mtry spanned the set [7, 11, 22, 44, 66], whose values were chosen as multiples of the default mtry value for classification, . The ntree parameter was set to 1000, as no performance increase was observed at higher values.
Validation datasets
Training and test datasets were constructed using the PaDEL descriptors and the ESI+ and ESI− endpoint values discussed previously. For both endpoints, 75% of the compounds were randomly selected for the training set and the remaining 25% were withheld from the model as a test set. Care was taken to stratify the random selection of compounds to ensure a similar ratio of amenable and unamenable compounds in both the training and test sets. Because of the large imbalance of amenable versus unamenable compounds, the application of sampling techniques was necessary to impose a balance on the training set. Two approaches were taken to either upsample the minority class (unamenable compounds) or downsample the majority class (amenable compounds). Upsampling is defined here as resampling the minority class with replacement (meaning the same compound/amenability classification pair can appear multiple times in a new sample) and setting the number of samples to match the majority class. Downsampling is defined here as the random removal of observations in the majority class to match the number of observations in the minority class. Neither sampling method is applied to the test sets. These training and test sets are provided in the supplemental file “Supplemental_train_test.xlsx.”
The test sets are used to validate the predictive power of the constructed model. Two additional validation techniques, fivefold cross validation (CV) and Y-randomization, were utilized. Fivefold cross validation essentially builds a model using 80% of the training set and 20% as a test, which is then repeated five times. Y-randomization randomly permutes the values of the endpoint in the training set, then attempts to model this mislabeled data. This model is then used to make predictions for the test set. Ideally, the Y-randomization model suffers poor performance compared to other models built on non-random data.
Model performance
The performance of the models was evaluated using three metrics: sensitivity, specificity, and balanced accuracy. Sensitivity (Sn) is the rate at which true positives are correctly identified, specificity (Sp) is the rate at which true negatives are correctly identified, and balanced accuracy is the average of the two other metrics. These metrics are calculated from the confusion matrix [38]. This matrix is a cross-tabulation of experimental observations and model-predicted classes and classifies each observation as a true positive (TP), false positive (FP), true negative (TN), or false negative (FN). Balanced accuracy (BA) is expressed as where and . Here, we consider a compound correctly predicted as amenable as a TP and a compound incorrectly predicted as amenable as a FP. Similarly, a compound correctly predicted as unamenable is considered a TN and a compound incorrectly predicted as unamenable as a FN.
Applicability domain
In the case of random forest, no assumptions are made regarding dataset distribution. Rather, this modeling approach uses the local vicinity of each object to determine the probability of belonging to each end point class [39]. Let N0 represent the indices of k training set objects of the leaf node to which a new object, xnew, is assigned. A decision tree assigns the fraction of objects in class j in N0 as a confidence measure for the class to which xnew belongs. Random forest is an ensemble of decision trees, so the confidence measure is averaged over all B trees:
Here, Bi is the jth decision tree of the ensemble to which the leaf node xnew belongs. The class probability , when j represents the amenable class, determines whether a compound is classified as amenable or unamenable in our model where classifies an object as unamenable and equates to an amenable compound. The value of should be considered when utilizing predictions for the elimination of candidate compounds in a suspect screening analysis. Consider the case where a molecular feature has only two possible tentative chemical identities. The model predicts both compounds as unamenable to LC–ESI–MS with for tentative identity A and for tentative identity B, where j represents the unamenable class. While both compounds may be unamenable in LC–ESI–MS, compound B is more likely to be unamenable based on the model predictions. In this case, compound A would be considered a better candidate for confirmation by chemical standard over compound B.
Results and discussion
Random forest models were constructed using both upsampled and downsampled versions of the ESI+ and ESI− training sets to predict amenability of novel compounds using those ionization modes in LC–ESI–MS. The dataset sizes and model performance metrics are provided in Table 1.
Table 1.
Model performances for fitting, training, and cross-validated sets and for the test and Y-randomized test sets
| Model | Size | Balanced accuracy |
Sensitivity | Specificity | Balanced accuracy | Sensitivity | Specificity |
|---|---|---|---|---|---|---|---|
| Training set | Fivefold CV | ||||||
| ESI+ models (downsampling applied) | 580 | 0.78 | 0.79 | 0.77 | 0.77 | 0.76 | 0.78 |
| ESI+ models (upsampling applied) | 6340 | 0.99 | 1.00 | 0.99 | 0.99 | 0.98 | 1.00 |
| ESI− models (downsampling applied) | 550 | 0.83 | 0.82 | 0.84 | 0.81 | 0.83 | 0.79 |
| ESI− models (upsampling applied) | 4688 Test set |
0.99 | 1.00 | 0.98 | 0.98 Y-randomization |
0.97 | 1.00 |
| ESI+ models (downsampling applied) | 1153 | 0.81 | 0.85 | 0.76 | 0.48 | 0.44 | 0.51 |
| ESI+ models (upsampling applied) | 1153 | 0.58 | 0.98 | 0.19 | 0.55 | 0.48 | 0.63 |
| ESI− models (downsampling applied) | 871 | 0.82 | 0.85 | 0.80 | 0.50 | 0.49 | 0.51 |
| ESI− models (upsampling applied) | 871 | 0.68 | 0.99 | 0.38 | 0.51 | 0.46 | 0.56 |
Comparing the performance metrics for both the model and Y-randomization model predictions for the test sets shows that the specificity value for the ESI+ upsampled model (0.19) is substantially lower than the specificity value for the upsampled Y-randomization model (0.63). This indicates overfitting of the model (as reflected in the overly high-performance metrics in the training set) and a lack of predictive power for unamenable compounds for the upsampled model. Similarly for the ESI− models, there is only an 18% difference between the upsampled model (0.38) and the upsampled Y-randomization mode (0.56), also suggesting an overfitted model with lack of predictive power compared to chance.
Both ESI+ and ESI− downsampled models performed well for both cross-validated and final models with approximately equal performance metrics (Table 1). The application of both downsampled models to the test datasets showed good performance, whereas both performed similarly better for predicting amenable test compounds (higher sensitivity than specificity). Predictions from the Y-randomization models performed worse compared to those from the final models (33% and 32% worse by balanced accuracy for ESI+ and ESI− downsampled models, respectively).
Predictive power and applicability domain
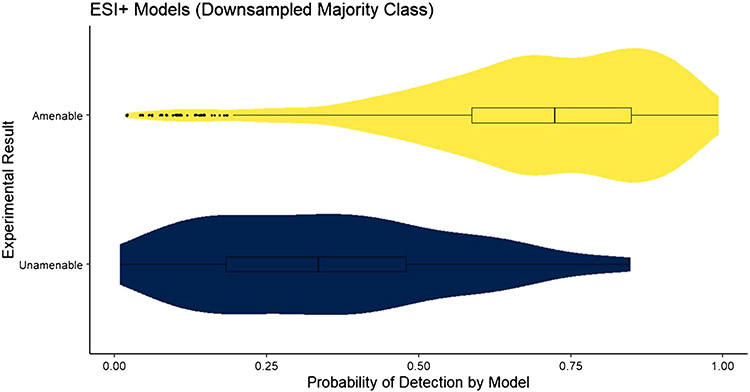
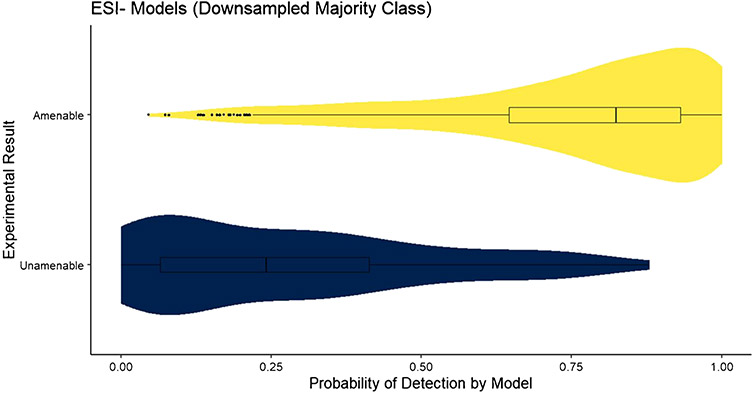
The confidence measure of the applicability domain of the RF model, , was assessed for the test datasets for each model. The distribution of , where j represents the amenable class and xnew is an element of the test set, for the downsampled ESI+ model is shown in Fig. 1. The distribution of predictions for LC–ESI–MS-amenable compounds (upper, yellow) shows a left skew, with the majority of predictions having probabilities > 0.70. The first quartile for amenable compounds is just above 0.5, where the model will predict a compound as unamenable. The distribution of for the downsampled ESI− model is shown in Fig. 2. In this case, the distribution of predictions for LC–ESI–MS-amenable compounds is further skewed toward the left, with a higher density of predictions at high probabilities. The distribution of predictions for the LC–MS ESI-unamenable compounds (lower, blue) is also further skewed right, with the majority of predictions for these unamenable compounds having probabilities strongly in favor of them being unamenable. This suggests that the downsampled ESI− model has better overall performance than the downsampled ESI+ model, which is supported by the results in Table 1.
Fig. 1.
The distribution of for the downsampled ESI+ model applied to the test set. The quartiles of the distribution are provided as a box plot inlayed into the violin plot
Fig. 2.
The distribution of for the downsampled ESI− model applied to the test set. The quartiles of the distribution are provided as a box plot inlayed into the violin plot
Descriptor importance
There is a need for a mechanistic explanation of the models presented here to satisfy Organisation for Economic Co-operation and Development (OECD) principle 5 [40], which states that QSAR models should have a mechanistic interpretation, if possible. This is particularly the case when the activity being modeled is biological in nature due to the potential mechanistic complexity in relation to the underlying chemistry. In the present case, the endpoint of interest, i.e., LC–ESI–MS amenability (or unamenability), has more in common with what are typically referred to as quantitative structure–property relationships (QSPR). QSPRs typically model chemical properties, such as log octanol–water partition coefficients, water solubility, vapor pressure, etc., which tend to relate more directly to underlying chemical interactions. In pursuit of some measure of interpretability, however, a descriptor importance metric native to the RF algorithm, the Gini index, can be leveraged to shed light on the underlying chemical factors influencing LC–ESI–MS amenability. Assume an endpoint consists of C total categories and the probability of picking a category i is p(i). The Gini index can be calculated as . The mean decrease of the Gini index for a descriptor in a RF model is a measure of how likely a split on that descriptor will lead to the correct classification of an endpoint. Therefore, the higher the mean decrease in the Gini index, the more important a descriptor is to the success of the model. We should note that removal of underperforming descriptors based on the variable importance discussed here would have no significant effect on the quality of these RF models, due to RF’s inherent ability to consider only useful descriptors. The removal of highly correlated descriptors, as discussed in the “Methods” section, reduces the chance of discussing descriptors that are capturing the same property of the modeled compounds.
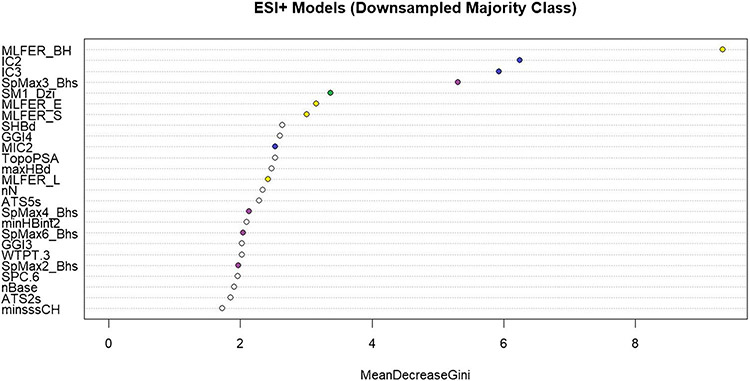
The descriptor importance for the ESI+ downsampled model is shown in Fig. 3. A similar plot of variable importance for the ESI+ upsampled model can be found in Supplemental Figure S1. Of the descriptors shown, the top performers have been selected for discussion. The descriptors plotted as blue points (IC2, IC3, MIC2) are estimations of the information content of a molecule. Information content is an index of the topological information of a molecular graph using the number of neighbors and multiplicity of bonds around individual atoms [41]. Chemical phenomena like isomerism and tautomerism are captured in these descriptors, which affect the retention of molecules on a chromatography column [42].
Fig. 3.
A plot of variable importance based on the mean decrease in the Gini index gain for the downsampled ESI+ model. Descriptors chosen for discussion in the main text are represented as colored points on the graph
The descriptors plotted as yellow points in Fig. 3 (MLFER_BH, MLFER_E, MLFER_S, MLFER_E) represent molecular linear free energy relationship terms. These terms capture specific interaction elements of the solvation property [43]. These interaction elements include polarizability and hydrogen bond basicity and acidity. Each of these properties influences the eluting molecule’s interaction with both mobile and stationary phases of the LC column. Polarizability has been shown to be predictive of LC retention time [44]. The acidity/basicity of a molecule is important to the selectivity of the ionization source of the mass spectrometer [45].
The descriptors plotted as purple points (SpMax4_Bhs, SpMax6_Bhs, SpMax2_Bhs) represent the largest absolute eigenvalue of the Burden modified matrix weighted by intrinsic state [41]. Briefly, the Burden matrix considers atomic number and bond order between atoms in the molecule. The intrinsic state is a ratio of the valence and sigma electrons in an atom [46]. These descriptors capture the electrotopological state of the molecule, which determines the ionization potential of the molecule. Ionization is also represented in the model by the descriptor plotted as a green point in Fig. 3 (SM1_Dzi). This descriptor is the Barysz vertex-distance matrix which represents the heteroatoms of a molecule and is weighted by ionization potential [41]
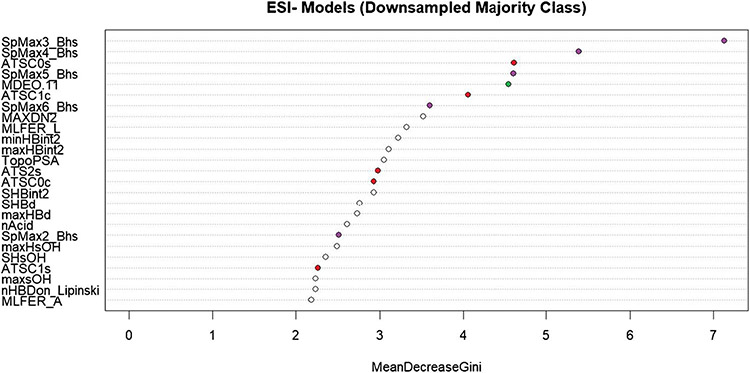
The descriptor importance for the ESI− downsampled model is shown in Fig. 4. A similar plot of variable importance for the ESI− upsampled model can be found in Supplemental Figure S2. Again, the top descriptors are selected for discussion. The descriptors plotted as purple points (SpMax3_Bhs, SpMax4_Bhs, SpMax5_Bhs, SpMax6_Bhs, SpMax2_Bhs) represent the Burden matrix, which, as described earlier, captures the electrotopological state of the molecule, which determines the ionization potential of the molecule.
Fig. 4.
A plot of variable importance based on the mean decrease in Gini-index gain for the downsampled ESI− model. Descriptors chosen for discussion in the main text are represented as colored points on the graph
The descriptors plotted as red points in Fig. 4 (ATSC0s, ATSC1c, ATS2s, ATSC0c, ATSC1s) represent the Centered Broto-Moreau autocorrelation weighted by atomic properties [41]. The calculation of autocorrelation vectors for atomic properties (in this case, atomic charge, intrinsic state, and Sanderson electronegativity were observed) provides a way to see the effect of different atoms in the same position of a molecular skeleton. This will influence both the polarizability of the molecule as it elutes through a LC column and its ionization potential as it enters the ionization source. Lastly, we note the descriptor plotted as a green point in Fig. 4 (MDEO.11), which is a descriptor representing the molecular distance between primary oxygens. Among other chemical phenomena, this can be associated with the molecule’s ability to polarize, which we have already noted influences the retention of the molecule on the column.
Model comparison with simpler models
The models described above can (and should) be described as complex. A more simplistic modeling approach, wherein a small number of hand-picked descriptors are considered using a simpler modeling technique, can be considered as a surrogate for how an analytical chemist would consider the functional aspects of a molecule to determine its amenability. Commonly considered aspects would include the number of acidic or basic functional groups along with its overall tendency to act as a proton donor or acceptor (dependent on the ionization mode). The “nBase” and “MLFER-BH” descriptors were chosen for a simple model of LC–MS ESI+ amenability, and the “nAcid” and “MLFER-A” descriptors were chosen to provide a simple model of LC–MS ESI− amenability. Logistic regression models were then constructed using the ESI+/− downsampled training sets with the two relevant descriptors. The logistic regression models were then applied to the appropriate test set as performed with the earlier random forest models. Table 2 shows the results of these simplistic models. Overall, these models performed significantly better than those built on randomized endpoints (Y-randomized models in Table 1); however, they fall short of the performance of the downsampled random forest models. While acidity/basicity is an important aspect of a molecule’s amenability in LC–ESI–MS, other important factors such as size/shape and polarizability are captured in the more complex random forest models.
Table 2.
Model performances for logistic regression models using the downsampled training and test sets from the earlier random forest models
| Model | Size | Balanced accuracy |
Sensitivity | Specificity |
|---|---|---|---|---|
| Training set | ||||
| ESI+ logistic model | 580 | 0.72 | 0.61 | 0.82 |
| ESI+ random forest model (downsampling applied) | 580 | 0.78 | 0.79 | 0.77 |
| ESI− logistic model | 550 | 0.75 | 0.67 | 0.84 |
| ESI− random forest model (downsampling applied) | 550 | 0.83 | 0.82 | 0.84 |
| Test set | ||||
| ESI+ logistic model | 1153 | 0.71 | 0.66 | 0.76 |
| ESI+ random forest model (downsampling applied) | 1153 | 0.81 | 0.85 | 0.76 |
| ESI− logistic model | 871 | 0.76 | 0.70 | 0.82 |
| ESI− random forest model (downsampling applied) | 871 | 0.82 | 0.85 | 0.80 |
Model comparison with expert intuition
An analytical chemist with extensive training in LC–ESI–MS will possess chemical intuition capable of hypothesizing the amenability of a molecule based solely on its structure. For instance, the carboxylic acid functional group present on a small molecule like benzoic acid would suggest the compound is amenable to ESI− LC–MS, as the acid group has a high affinity for proton loss, or amenable to ESI+ LC–MS, as the double-bonded oxygen can accept a proton and subsequently be stabilized through resonance. This kind of intuition should align with the results produced by the models presented in this work.
As a simple test for this concordance, we compared the ESI− and ESI+ downsampled model results for a set of chemicals with a common substructure, in this case, the presence of a carboxylic acid group. The subset of chemicals containing this functional group was determined using the ToxPrint “bond:C(= O)O_carboxylicAcid_generic.” A total of 464 chemical compounds with experimentally measured ESI+ data out of 4613 and 773 chemical compounds with experimentally measured ESI− data out of 3490 were found to contain this ToxPrint. Predictions using both ESI+ and ESI− downsampled models were then generated for these compounds. Table 3 shows the confusion matrix and performance metrics for these predictions. As would be expected, a large majority, 92%, of these compounds were detectable using ESI+ LC–MS and 94% using ESI–LC–MS. The ESI+ downsampled model was capable of correctly predicting the amenability of 93% of these compounds, and the ESI− downsampled model was capable of correctly predicting 93%. The ESI− downsampled model had poorer performance predicting unamenable compounds with only 69% correctly predicted; however, the ESI+ downsampled model performed much better, correctly predicting all 10 ESI+ unamenable compounds listed in Table 3. While this is another indicator that more unamenable data would improve modeling, the results are promising as a large majority of these compounds fit with intuition. Future work will strive to highlight the ToxPrints that are commonly used by experts to hypothesize the amenability of compounds in relation to the models presented here.
Table 3.
Confusion matrix and performance metrics for ESI− and ESI+ downsampled model predictions compared to the subset of model datasets containing a carboxylic acid functional group
| Amenable (prediction) |
Unamenable (prediction) |
|
|---|---|---|
| ESI− downsampled model Detected (experiment) | 728 | 4 |
| Not detected (experiment) | 37 | 9 |
| Sensitivity | 0.95 | |
| Specificity | 0.69 | |
| Balanced accuracy | 0.82 | |
| ESI+ downsampled model Detected (experiment) | 573 | 42 |
| Not-detected (experiment) | 0 | 10 |
| Sensitivity | 0.93 | |
| Specificity | 1.00 | |
| Balanced accuracy | 0.97 |
External validation
Although the dataset used to construct the models in this work was split in order to leave out a portion of the data for validation, best modeling practices necessitate an external validation dataset also be considered for predictability. As part of the ToxCast program, a number of chemical standards have been analyzed for quality using multiple analytical methods, including LC–ESI–MS. In total, there are 1768 chemical compounds analyzed for their amenability in LC–ESI–MS that are not found in the training and test sets used in our models. These chemicals were solely reported as detected or not-detected in LC–ESI–MS and were not differentiated between either of the modes of ESI. While we may consider the chemicals detected as amenable to LC–ESI–MS, the not-detected chemicals are not necessarily unamenable to the method as there are considerable method application variables across laboratories and instrumentation. Hence, this exercise attempted to assess the applicability of our models in such cases. Predictions were generated for these chemicals using both ESI+ and ESI− downsampled models and compared to experimental observations in the confusion matrices in Table 4. As there was no differentiation between ESI+/− in this external dataset, model predictions were combined. In the event that a chemical was predicted as amenable by either ESI+ or ESI− downsampled model, the chemical was considered as amenable in the combined model in Table 4. Those predicted as unamenable in both modes were considered unamenable. In this combined model approach, performance metrics are generally favorable compared to those observed for the model test sets in Table 1. A balanced accuracy of 0.76 for predicted amenability for these 1768 chemical compounds suggests our models should be applicable to chemical data generated from LC–ESI–MS setups in labs novel to those included in the model dataset. The dataset and experimental and predicted amenability calls are provided in the supplemental file “Supplemental_ToxCast_PhaseII.xlsx.”
Table 4.
Confusion matrices and performance metrics for ESI+ and ESI− downsampled model predictions compared to external validation data
| Amenable (prediction) |
Unamenable (prediction) |
|
|---|---|---|
| ESI− downsampled model | ||
| Detected (experiment) | 323 | 502 |
| Not-detected (experiment) | 68 | 874 |
| Sensitivity | 0.83 | |
| Specificity | 0.64 | |
| Balanced accuracy | 0.73 | |
| ESI+ downsampled model | ||
| Detected (experiment) | 423 | 402 |
| Not-detected (experiment) | 103 | 839 |
| Sensitivity | 0.80 | |
| Specificity | 0.68 | |
| Balanced accuracy | 0.74 | |
| Combined models | ||
| Detected (experiment) | 505 | 320 |
| Not-detected (experiment) | 129 | 813 |
| Sensitivity | 0.80 | |
| Specificity | 0.72 | |
| Balanced accuracy | 0.76 |
Model application to suspect screening
To show the potential usefulness of the models described herein, we considered a typical suspect screening scenario. As part of the previously mentioned ENTACT study, a set of compounds was identified among multiple laboratories as amenable in LC–MS using electrospray ionization. There were 228 compounds identified in ESI+ (of which only 39 are found in the ESI+ downsampled model training set) and 108 compounds identified in ESI− (of which only 13 are found in the ESI− downsampled model training set), with 37 of the compounds detected in both ESI+ and ESI− modes. To simulate a suspect screening approach, the molecular formula matching each of these compounds was searched on the Dashboard using the batch search feature and each matching structure was downloaded as a candidate. For the 228 compounds identified in ESI+, there were 13,325 candidates, and for the 108 compounds identified in ESI−, there were 7079 candidates. PaDEL descriptors were generated for each candidate and amenability predictions were calculated using both ESI+ and ESI− downsampled models. The resulting dataset is available in the supplemental file “Supplemental_Application.xlsx.”
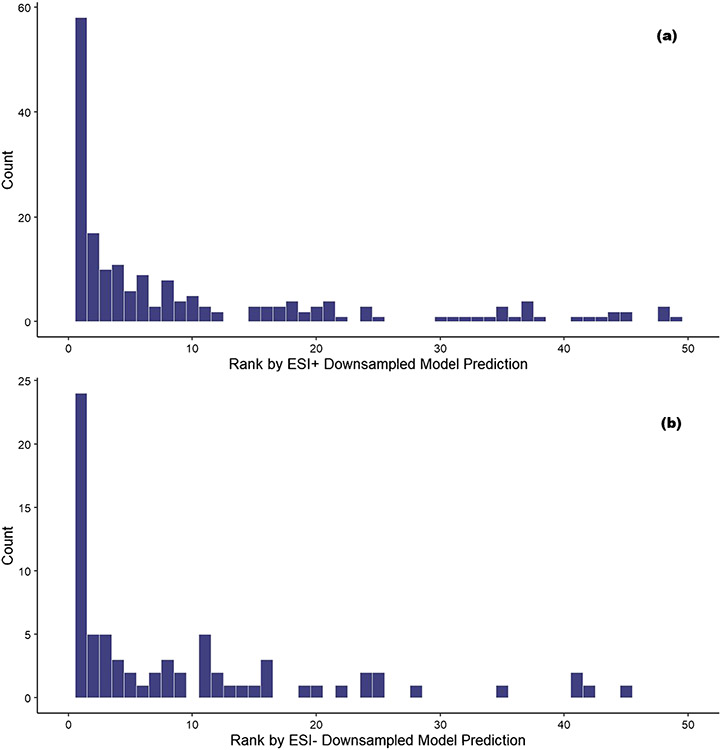
The confidence measure, , where j represents the amenable class, was then used to determine the potential amenability of each candidate compound. For each molecular formula, candidates were ranked based on the value of , with the highest being assigned rank 1, the next highest being assigned rank 2, etc., such that the higher-ranked compounds (those close to rank 1) have the highest probabilities of detection in LCMS and the lower-ranked compounds (those farther away from rank 1) have the lowest probabilities of detection in LCMS. For example, if a formula has two potential candidate matches, with for tentative identity A and for tentative identity B, tentative identity A would be assigned a rank of 2 and tentative identity B would be assigned a rank of 1. These ranks were used in a manner analogous to data source counts [47] to determine the best candidate compounds for each molecular formula. These candidate ranks were then compared to the compound(s) identified in the ENTACT study to test the hypothesis that higher-ranked candidates will match compounds identified in the ENTACT study more often than lower-ranked candidates. The frequency of candidate ranks matching an ENTACT compound are shown in Fig. 5.
Fig. 5.
Frequency counts of candidate compounds found to be a match for an ENTACT compound ordered by prediction rank value (with 1 being the highest confidence rank and 50 the lowest) based on ESI+ LC–MS (a) and ESI− LC–MS (b) amenability predictions. Ranks greater than 50 are omitted due to very low occurrence
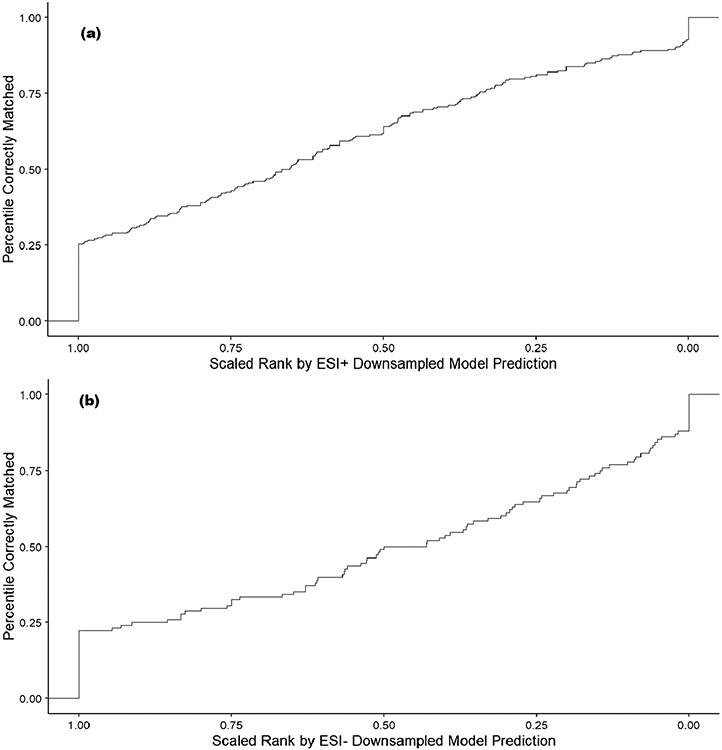
Figure 5 shows that a significant portion of candidate compounds that are matches for ENTACT compounds are assigned to the top ten (1–10) rank values (57.5% in ESI+ and 43.5% in ESI−) based on the amenability prediction value. However, not all molecular formulae have similar numbers of candidate compounds. For example, the molecular formula C6HCl5O matches only one compound in DSSTox, pentachlorophenol, whereas the formula C11H14O3 matches 956 unique compounds. For this reason, each rank was scaled by the number of compounds matching each formula as a percentile between 0 and 1, where 1 is the highest-ranked candidate (rank 1) and 0 is the lowest-ranked candidate. The cumulative amount of correctly matched compounds by scaled rank is shown in Fig. 6. For ESI+, ~ 25% of compounds are correctly matched by the highest-ranked prediction (value at rank 1.00 in the lower left of the figure) and the result for ESI− is similar at ~ 23%. Whereas this initial percentile of candidate compounds correctly matching the scaled rank predictions based on rank 1 predictions is promising, the trend moving to the right of the figure with the lower scaled ranked compounds has an approximately linear relationship with the percentile of correctly matched compounds, indicating only incremental gains in detection of correct matches.
Fig. 6.
A plot of the percentile of correctly matched ENTACT compounds at a given scaled rank value based on ESI+ LC–MS (a) and ESI− LC–MS (b) amenability predictions
Whereas we do not anticipate this kind of modeling replacing other candidate ranking methods (such as data source ranking or molecule fragmentation approaches), it can provide a useful complimentary and/or weight-of-evidence approach. It should be noted that the dataset used in this demonstration is biased toward environmentally relevant chemicals, many of which appear in a large number of chemical lists on the Dashboard (see the DATA_SOURCES column in “Supplemental_Application.xlsx” for both ESI+ and ESI−). In the case of this dataset, the amenability model predictions would underperform relative to ranking candidates based on data source counts alone. However, typical suspect screens of unknown mixtures may not have the advantage of data source–rich candidates, in which case, the present models could be more useful.
Model applicability to environmental datasets
The external validation results presented in Table 4, using the present models to predict LC–ESI–MS amenability of ToxCast chemicals, provided an initial estimate of applicability of the models to environmental datasets. To further anticipate differences between the chemical compounds in the modeling dataset versus those encountered in a typical analysis of an environmental media, we compared ToxPrint chemotype profiles for our dataset versus the larger Tox21 compound library [48]. The Tox21 program was conceived as a cooperative effort of multiple US government agencies to assemble a dataset of 8947 unique chemical compounds spanning a wide range of criteria, particularly those of environmental hazard or exposure concern.
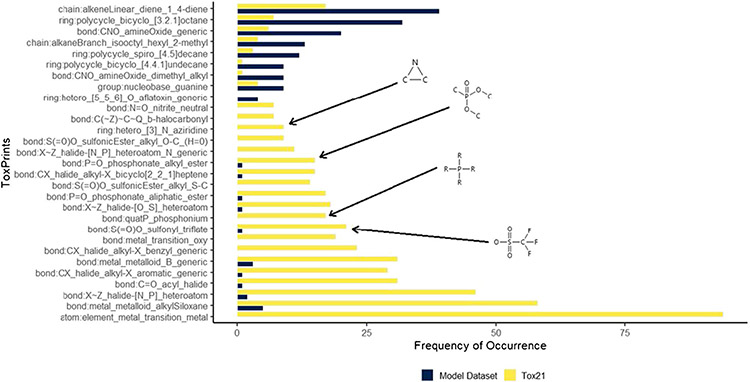
Figure 7 shows the chemotypes with the greatest difference of frequency of occurrence between the two datasets. There are a number of chemotypes not represented in the modeling dataset that are prevalent in the Tox21 dataset. Two conclusions can be drawn based on the chemotypes that are largely absent from the modeling dataset. A number of these chemotypes are metal-containing substructures, which cannot be represented using the current modeling methodology (QSAR-ready structures exclude organometallics and salt components). Capturing compounds containing these substructures would require another modeling approach, beyond the scope of this work. The remaining chemotypes are purely organic (e.g., sample structures illustrated in Fig. 7). Analytical QC data gathered in the course of the Tox21 programs are currently being compiled into datasets for public release and will be represented in a future modeling dataset.
Fig. 7.
A plot of prevalent chemotypes in the Tox21 dataset and the model dataset used in this work, selected based on the absolute difference of frequency of occurrence between datasets
Conclusion
We have constructed models that capture the physicochemical properties of molecules that determine their amenability to detection in different LC–ESI–MS modes of detection. Predictions made using these models can be used to deduce the likelihood of a chemical compound appearing in an LC–ESI–MS analysis and in what polarity of ESI. Furthermore, these predictions can be used to rank a list of potential chemical identities in suspect screens for further evaluation, such as in conjunction with an applicable retention time prediction model, data source ranking scheme, and/or spectral matching in a weight-of-evidence approach. In turn, this should result in the need for fewer chemical standards for confirmation of identity. These amenability models have the potential to save researchers’ time and resources by better anticipating which chemicals are amenable to LC–ESI–MS. Furthermore, additional savings can be found by prioritizing lower-confidence predictions for follow-up analysis using other methods such as GC–MS, rather than attempting to address whether there is a sample issue versus a method issue.
As more analytical data are acquired, these models will continue to be improved. The models herein were trained using chemical compounds detected using a multitude of LC–ESI–MS methods, whereas non-detects were limited to only one method. Future models will incorporate both detects and non-detects from the various instrumentation used in the ENTACT study. This will allow for a comparison of individual methods as well as a consensus model similar to what is presented here. Future models will also attempt to predict compound amenability in other instrumentation, such as GC–MS. Real-time predictions using these models will be available through the predictions page of the Dashboard, https://comptox.epa.gov/dashboard/predictions/index, at a future date.
Supplementary Material
Acknowledgements
The authors would like to acknowledge Ralph Hindle from Vogon Laboratory Services along with Tarun Anumol and Craig Marvin from Agilent Technologies, Inc. for assisting with the spectral collection and curation of the ToxCast Screening Library data used in this study. We would also like to thank Katherine Phillips, Katie Paul-Friedman, and Risa Sayre for preliminary conversations surrounding this study.
Funding
The US EPA Office of Research and Development funded and managed the research described here.
Footnotes
Supplementary Information The online version contains supplementary material available at https://doi.org/10.1007/s00216-021-03713-w.
Consent for publication All authors have consented to publication of this work.
Conflict of interest The authors declare no competing interests.
Disclaimer The information in this document has been funded wholly or in part by the US Environmental Protection Agency. It does not signify that the contents necessarily reflect the views of the agency, nor does mention of trade names or commercial products constitute endorsement or recommendation for use. The paper has been subjected to the agency’s review process and approved for publication. Mention of trade names or commercial products does not constitute endorsement or recommendation for use.
Data availability
All data generated or analyzed during this study are included in this published article and its supplementary information files.
Code availability
The computer code created for the current study is available from the corresponding author on reasonable request.
References
- 1.Wambaugh JF, Setzer RW, Reif DM, Gangwal S, Mitchell-Blackwood J, Arnot JA, et al. High-throughput models for exposure-based chemical prioritization in the ExpoCast Project. Environ Sci Technol. 2013;47(15):8479–88. [DOI] [PubMed] [Google Scholar]
- 2.Csiszar SA, Meyer DE, Dionisio KL, Egeghy P, Isaacs KK, Price PS, et al. Conceptual framework to extend life cycle assessment using near-field human exposure modeling and high-throughput tools for chemicals. Environ Sci Technol. 2016;50(21):11922–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Li L, Westgate JN, Hughes L, Zhang X, Givehchi B, Toose L, et al. A model for risk-based screening and prioritization of human exposure to chemicals from near-field sources. Environ Sci Technol. 2018;52(24):14235–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Isaacs KK, Glen WG, Egeghy P, Goldsmith M-R, Smith L, Vallero D, et al. SHEDS-HT: an integrated probabilistic exposure model for prioritizing exposures to chemicals with near-field and dietary sources. Environ Sci Technol. 2014;48(21):12750–9. [DOI] [PubMed] [Google Scholar]
- 5.Wild CP, Scalbert A, Herceg Z. Measuring the exposome: a powerful basis for evaluating environmental exposures and cancer risk. Environ Mol Mutagen. 2013;54(7):480–99. [DOI] [PubMed] [Google Scholar]
- 6.Dix DJ, Houck KA, Martin MT, Richard AM, Setzer RW, Kavlock RJ. The ToxCast program for prioritizing toxicity testing of environmental chemicals. Toxicol Sci. 2007;95(1):5–12. [DOI] [PubMed] [Google Scholar]
- 7.Tice RR, Austin CP, Kavlock RJ, Bucher JR. Improving the human hazard characterization of chemicals: a Tox21 update. Environ Health Perspect. 2013;121(7):756–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hertzberg RP, Pope AJ. High-throughput screening: new technology for the 21st century. Curr Opin Chem Biol. 2000;4(4):445–51. [DOI] [PubMed] [Google Scholar]
- 9.NRC U. Risk assessment in the federal government: managing the process. National Research Council, Washington DC. 1983;11(3). [PubMed] [Google Scholar]
- 10.Tolonen A, Pelkonen O. Analytical challenges for conducting rapid metabolism characterization for QIVIVE. Toxicology. 2015;332:20–9. [DOI] [PubMed] [Google Scholar]
- 11.Nicolas CI, Mansouri K, Phillips KA, Grulke CM, Richard AM, Williams AJ, et al. Rapid experimental measurements of physicochemical properties to inform models and testing. Sci Total Environ. 2018;636:901–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wetmore BA, Wambaugh JF, Ferguson SS, Sochaski MA, Rotroff DM, Freeman K, et al. Integration of dosimetry, exposure, and high-throughput screening data in chemical toxicity assessment. Toxicol Sci. 2012;125(1):457–74. [DOI] [PubMed] [Google Scholar]
- 13.Sobus JR, Grossman JN, Chao A, Singh R, Williams AJ, Grulke CM, et al. Using prepared mixtures of ToxCast chemicals to evaluate non-targeted analysis (NTA) method performance. Anal Bioanal Chem. 2019;411(4):835–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Newton SR, McMahen RL, Sobus JR, Mansouri K, Williams AJ, McEachran AD, et al. Suspect screening and non-targeted analysis of drinking water using point-of-use filters. Environ Pollut. 2018;234:297–306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Schymanski EL, Williams AJ. Open science for identifying “known unknown” chemicals. Environ Sci Technol. 2017;51(10):5357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sobus JR, Wambaugh JF, Isaacs KK, Williams AJ, McEachran AD, Richard AM, et al. Integrating tools for non-targeted analysis research and chemical safety evaluations at the US EPA. J Eposure Sci Environ Epidemiol. 2018;28(5):411–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ulrich EM, Sobus JR, Grulke CM, Richard AM, Newton SR, Strynar MJ, et al. EPA’s non-targeted analysis collaborative trial (ENTACT): genesis, design, and initial findings. Anal Bioanal Chem. 2019;41l(4):853–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.McEachran AD, Chao A, Al-Ghoul H, Lowe C, Grulke C, Sobus JR, et al. Revisiting five years of CASMI contests with EPA identification tools. Metabolites. 2020;10(6):260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Newton SR, Sobus JR, Ulrich EM, Singh RR, Chao A, McCord J, et al. Examining NTA performance and potential using fortified and reference house dust as part of EPA’s Non-Targeted Analysis Collaborative Trial (ENTACT). Anal Bioanal Chem. 2020;412(18):4221–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Favreau P, Poncioni-Rothlisberger C, Place BJ, Bouchex-Bellomie H, Weber A, Tremp J, et al. Multianalyte profiling of per- and polyfluoroalkyl substances (PFASs) in liquid commercial products. Chemosphere. 2017;171:491–501. [DOI] [PubMed] [Google Scholar]
- 21.Reymond J-L, Ruddigkeit L, Blum L, van Deursen R. The enumeration of chemical space. WIREs Comput Mol Sci. 2012;2(5):717–33. [Google Scholar]
- 22.Horai H, Arita M, Kanaya S, Nihei Y, Ikeda T, Suwa K, et al. MassBank: a public repository for sharing mass spectral data for life sciences. J Mass Spectrom. 2010;45(7):703–14. [DOI] [PubMed] [Google Scholar]
- 23.Dalby A, Nourse JG, Hounshell WD, Gushurst AKI, Grier DL, Leland BA, et al. Description of several chemical structure file formats used by computer programs developed at Molecular Design Limited. J Chem Inf Model. 1992;32(3):244–55. [Google Scholar]
- 24.Cao Y, Charisi A, Cheng L-C, Jiang T, Girke T. ChemmineR: a compound mining framework for R. Bioinformatics. 2008;24(15):1733–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.R Core Team (2019). R: A language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria. https://www.R-project.org/. [Google Scholar]
- 26.Heller SR, McNaught A, Pletnev I, Stein S, Tchekhovskoi D. InChI, the IUPAC international chemical identifier. J Cheminform. 2015;7(1):23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Williams AJ, Grulke CM, Edwards J, McEachran AD, Mansouri K, Baker NC, et al. The CompTox Chemistry Dashboard: a community data resource for environmental chemistry. J Cheminform. 2017;9(1):61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lowe CN, Williams AJ. Enabling high-throughput searches for multiple chemical data using the U.S.-EPA CompTox chemicals dashboard. J Chem Inf Model. 2021;61(2):565–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Richard AM, Judson RS, Houck KA, Grulke CM, Volarath P, Thillainadarajah I, et al. ToxCast chemical landscape: paving the road to 21st century toxicology. Chem Res Toxicol. 2016;29(8):1225–51. [DOI] [PubMed] [Google Scholar]
- 30.Mansouri K, Grulke C, Richard A, Judson R, Williams A. An automated curation procedure for addressing chemical errors and inconsistencies in public datasets used in QSAR modelling. SAR QSAR Environ Res. 2016;27(11):911–37. [DOI] [PubMed] [Google Scholar]
- 31.Yap CW. PaDEL-descriptor: an open source software to calculate molecular descriptors and fingerprints. J Comput Chem. 2011;32(7):1466–74. [DOI] [PubMed] [Google Scholar]
- 32.Mansouri K, Grulke CM, Judson RS, Williams AJ. OPERA models for predicting physicochemical properties and environmental fate endpoints. J Cheminform. 2018;10(1):10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mansouri K, Cariello NF, Korotcov A, Tkachenko V, Grulke CM, Sprankle CS, et al. Open-source QSAR models for pKa prediction using multiple machine learning approaches. J Cheminform. 2019;11(1):60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Khan K, Baderna D, Cappelli C, Toma C, Lombardo A, Roy K, et al. Ecotoxicological QSAR modeling of organic compounds against fish: application of fragment based descriptors in feature analysis. Aquat Toxicol. 2019;212:162–74. [DOI] [PubMed] [Google Scholar]
- 35.Gramatica P, Cassani S, Chirico N. QSARINS-chem: Insubria datasets and new QSAR/QSPR models for environmental pollutants in QSARINS. J Comput Chem. 2014;35(13):1036–44. [DOI] [PubMed] [Google Scholar]
- 36.Yang C, Tarkhov A, Marusczyk J, Bienfait B, Gasteiger J, Kleinoeder T, et al. New publicly available chemical query language, CSRML, to support chemotype representations for application to data mining and modeling. J Chem Inf Model. 2015;55(3):510–28. [DOI] [PubMed] [Google Scholar]
- 37.Liaw A, Wiener M. Classification and regression by randomForest. R News. 2002;2(3):18–22. [Google Scholar]
- 38.Altman DG, Bland JM. Diagnostic tests. 1: sensitivity and specificity. BMJ. 1994;308(6943):1552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Klingspohn W, Mathea M, ter Laak A, Heinrich N, Baumann K. Efficiency of different measures for defining the applicability domain of classification models. J Cheminform. 2017;9(1):44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gramatica P Principles of QSAR models validation: internal and external. QSAR Comb Sci. 2007;26(5):694–701. [Google Scholar]
- 41.Todeschini R, Consonni V. Molecular descriptors for chemoinformatics: volume I: alphabetical listing/volume II: appendices, references: John Wiley & Sons; 2009. 10.1002/9783527628766 [DOI] [Google Scholar]
- 42.D’Amboise M, Bertrand MJ. General index of molecular complexity and chromatographic retention data. J Chromatogr A. 1986;361:43–24. [Google Scholar]
- 43.Platts JA, Butina D, Abraham MH, Hersey A. Estimation of molecular linear free energy relation descriptors using a group contribution approach. J Chem Inf Comput Sci. 1999;39(5):835–45. [DOI] [PubMed] [Google Scholar]
- 44.Jinno K, Kawasaki K. The correlation between molecular polarizability of PAHs and their retention data on various stationary phases in reversed-phase HPLC. Chromatographia. 1984;18(2):103–5. [Google Scholar]
- 45.Ehrmann BM, Henriksen T, Cech NB. Relative importance of basicity in the gas phase and in solution for determining selectivity in electrospray ionization mass spectrometry. J Am Soc Mass Spectrom. 2008;19(5):719–28. [DOI] [PubMed] [Google Scholar]
- 46.Hall LH, Mohney B, Kier LB. The electrotopological state: structure information at the atomic level for molecular graphs. J Chem Inf Comput Sci. 1991;31(1):76–82. [Google Scholar]
- 47.McEachran AD, Sobus JR, Williams AJ. Identifying known unknowns using the US EPA’s CompTox Chemistry Dashboard. Anal Bioanal Chem. 2017;409(7):1729–35. [DOI] [PubMed] [Google Scholar]
- 48.Richard AM, Huang R, Waidyanatha S, Shinn P, Collins BJ, Thillainadarajah I, et al. The Tox21 10K Compound Library: collaborative chemistry advancing toxicology. Chem Res Toxicol. 2021;34(2):189–216. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data generated or analyzed during this study are included in this published article and its supplementary information files.
The computer code created for the current study is available from the corresponding author on reasonable request.