Abstract
The current viral pandemic, coronavirus disease 2019 (COVID-19), caused by severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), creates health, mental, economic, and other serious challenges that are better to say global crisis. Despite the existence of successful vaccines, the possible mutations which can lead to the born of novel and possibly more dangerous variants of the virus as well as the absence of definitive treatment for this potentially fatal multiple-organ infection in critically ill patients make us keep searching. Theoretically targeting human and viral receptors and enzymes via molecular docking and dynamics simulations can be considered a wise, rational, and efficient way to develop therapeutic agents against COVID-19. In this way, The RNA-dependent RNA polymerase (RdRP), main protease, and spike glycoprotein of SARS-CoV-2 as well as the human angiotensin-converting enzyme 2 receptor and transmembrane serine protease 2 are the most discussed and studied targets that play essential roles in the viral life and infection cycle. In the current in silico investigation, the guanidine functionality containing drugs and medicinal substances such as metformin, famotidine, neuraminidase inhibitors, antimalarial medications, anticancer drug imatinib, CGP compounds, and human serine protease inhibitor camostat were studied against the above-mentioned therapeutic targets and most of them (especially imatinib) have revealed an incredible spectrum of free docking scores and MD results. The current in silico investigation that its novel perspective of view is corroborated by the different experimental and clinical evaluations, confirms that the guanidine moiety can be considered as a missing promising pharmacophore in drug design and development approaches against SARS-CoV-2. Considering the chemical potency of this polyamine group in chemical interaction creation, the observed outcomes in this virtual screening were not surprising. On the other hand, the guanidine functional group has unique physico-chemical properties such as basicity that can make the target cells intracellular pH undesirable for the virus entry, uncoating, and cytosolic lifecycle. According to the obtained results in the current study that are interestingly confirmed by the previously reported efficacy of some the guanidine carrying drugs in COVID-19, guanidine as a potential multi-target anti-SARS-CoV-2 functional scaffold deserves further comprehensive investigations.
Graphical abstract
Supplementary Information
The online version contains supplementary material available at 10.1007/s11696-022-02528-y.
Keywords: Guanidine, SARS-CoV-2, Imatinib, TMPRSS2, Camostat, Intracellular pH
Introduction
COVID-19 caused by the enveloped RNA virus SARS-CoV-2 leads to potentially lethal pneumonia and sometimes multi-organ failures (Zanin et al. 2016 Feb 10). The genome of this newly appeared coronavirus encodes different kinds of structural and functional proteins such as the spike glycoproteins, RNA-dependent RNA polymerase (RdRp), and main protease (Mpro) which as well as some of the host cells receptors and enzymes have shown critical roles in the viral life cycle, entry, replication, and the spread of infection in an infected person’s body or in the community (Krumm et al. 2021 Dec). Therefore, targeting these viral and human cellular fragments can prevent these walking dead germs from pathogenicity.
The spike receptors that can be considered as the most unoccupied viral antigen in coronaviruses (CoVs) are responsible for host receptor binding and virus entry. These highly mutable surface glycoproteins initiate the internalization of SARS-CoV-2 via binding to the host cellular receptor angiotensin-converting enzyme 2 (ACE2) (Maurya et al. 2020 Jun; Baig et al. 2020 Sep). Apart from the importance of spike and ACE2, the transmembrane serine protease 2 (TMPRSS2) has also revealed a crucial role in SARS-CoV-2 cell entry spike receptor priming that makes a TMPRSS2 inhibitor a viral spread and pathogenesis suppressor in the infected host (Hoffmann et al. 2020; Gunst et al. 2021 May). Furthermore, SARS-CoV-2 RdRp and Mpro are the main responsible enzymes for the viral genome replication, and maturation of the functional proteins, respectively (Byléhn et al. 2021 Jan 6; Liang 2006 Feb 1). Inhibition of the viral RdRp and Mpro leads to effective management of COVID-19.
The critical roles of the viral spike receptors, RdRp, and Mpro, as well as the human ACE2, and TMPRSS2 make them promising therapeutic targets to combat SARS-CoV-2 and develop antiviral agents against COVID-19. SARS-CoV-2 RdRp has been shown to get suppressed by non-specific antiviral drugs such as remdesivir, ribavirin, and favipiravir that have revealed highly stable bonding interactions in the numerous theoretical investigations (Byléhn et al. 2021 Jan 6) as well as the proved experimental and clinical efficacy (Singh et al. 2020 Dec). Carmofur that is an approved antineoplastic drug has also shown effectiveness against COVID-19 by inhibiting the SARS-CoV-2 main protease (Jin et al. 2020 Jun). Furthermore, camostat and its active metabolite (guanidinobenzoyloxy) phenylacetic acid (GBPA) blocked SARS-CoV-2 spread in human lung tissue, which is the primary target organ for this viral infection by the inhibition of human TMPRSS2 (Hoffmann et al. 2021).
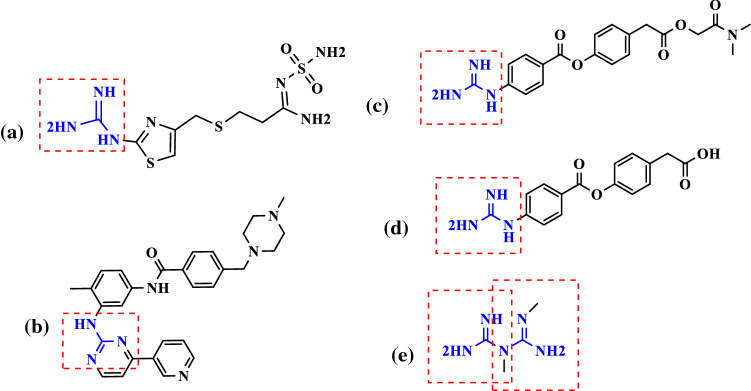
Guanidine-containing natural products or synthetic substances have received lots of attention via their precious and wide variety of biological activities (Xian et al. 2001 Sep 3). In 1962, Loddo and his co-workers published an article in Nature journal about the efficacy of guanidine against polio viruses (Loddo et al. 1962 Jan). They studied the anti-polio activity of some simple guanidine derivatives through an in vitro investigation that confirmed their claim. At the beginning of this article, it has been said that the presence of a guanidine moiety in several antiviral drugs has prompted the authors to examine the antiviral effects of these compounds. As can be seen in Fig. 1, numerous efficient therapeutic agents in COVID-19 patients are carrying this functional chemical group (Xian et al. 2001 Sep 3; Loffredo et al. 2021 Mar 8; Aman et al. 2021 Sep 1; Esam 2020 Sep). Thus, the aim of the current in silico study was to investigate the therapeutic and prophylactic potential of guanidine-containing medicinal substances against SARS-CoV-2.
Fig. 1.
Some of the selected guanidine-containing medications with previously reported efficacy against COVID-19. Famotidine (a), imatinib (b), camostat (c), GBPA (d), metformin (e)
The current interest in the guanidine-containing compounds is not surprising since guanidine as a polyamine moiety with its unique physicochemical properties can play decisive structural and functional roles in the chemical structure of biologically active molecules (Saczewski and Balewski 2009 Oct 1). The potential of guanidine to form different kinds of chemical interactions, especially the strong non-covalent interactions such as hydrogen bonds with biomolecules, and its highly basic nature (pKa ~ 13) while retaining the lipophilic properties by the electron delocalization capability (Saczewski and Balewski 2009 Oct 1) make this scaffold a promising chemical group to get noticed in drug discovery and development.
Although the molecular docking analysis was originally developed to help understanding the possibility and involved mechanisms of intermolecular interactions between small molecules and proteins, there is no doubt that this molecular modeling technique has become one of the best, fast, and cost-effective approaches to predict the efficacy of the newly designed medicinal agents and also achieve the safe and efficient therapeutic candidates among the existing biologically active compounds and already approved drugs (Pinzi and Rastelli 2019 Sep 4; Pourhajibagher and Bahador 2022 Sep 1). Since the beginning of the current serious global health crisis, we have not been successful to find an absolute and target-specific therapeutic agent for treating COVID-19. Considering this fact, drug repurposing strategy such as molecular docking seems one of the main methods to develop novel therapeutic agents against the discovered and introduced viral and human druggable targets (Fadlalla et al. 2022 Apr; Macip et al. 2022 Mar).
Thus, in this perspective in silico investigation, 35 guanidine- and also guanidine-like-containing therapeutic agents (Table 1) with a broad spectrum of biological activities have been studied theoretically against SARS-CoV-2 Mpro, RdRp, and spike receptor in complex with human ACE2, as well as the host TMPRSS2. Furthermore, at the next phase, the pKa and basicity of the best-tested ligands in this study with confirmed therapeutic activity against SARS-CoV-2 were probed in comparison with hydroxy/chloroquine to explore the other possible mechanisms that can be involved in their potential efficacy in COVID-19 and the whole achieved resulted seem impressive.
Table 1.
General information of the tested ligands in this study
| Ligand No | Names | Two-dimensional (2D) structures of the selected ligands for the docking studies | Biological activity |
|---|---|---|---|
| 1 | Metformin | 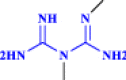 |
Anti-diabetic drug with reported efficacy against SARS-CoV-2 (Esam 2020 Sep) |
| 2 | Phenformin | 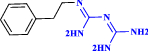 |
Anti-diabetic agent possessing confirmed anti- COVID-19 activity (Lehrer 2020) |
| 3 | Rosuvastatin | 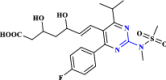 |
An anti-hyperlipidemic drug that demonstrated efficacy in COVID-19 (Rossi et al. 2020 Nov) |
| 4 | Cimetidine | 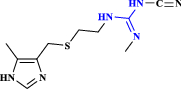 |
A gastric acid secretion inhibitor: H2-receptor antagonist without efficacy against SARS-CoV-2 (Mukherjee et al. 2021) |
| 5 | Famotidine | 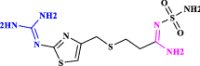 |
A gastric acid secretion inhibitor that has been considered to be a potential treatment for COVID-19 (Loffredo et al. 2021 Mar 8) |
| 6 | Zanamivir | 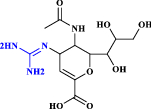 |
Antiviral agent: neuraminidase inhibitor (Wang et al. 2020) |
| 7 | Peramivir | 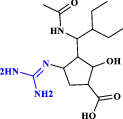 |
Antiviral drug: neuraminidase inhibitor (Wang et al. 2020) |
| 8 | Laninamivir | 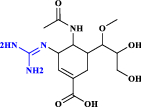 |
Antiviral agent: neuraminidase inhibitor (Wang et al. 2020) |
| 9 | Viramidine* (previously known as Ribamidine) | 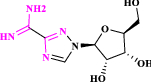 |
Prodrug of ribavirin (Tohme et al. 2012; Liu et al. xxxx) |
| 10 | Taribavirin* | 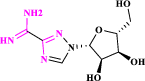 |
Prodrug of ribavirin (Tohme et al. 2012) |
| 11 | Proguanil (chloroguanide) | 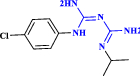 |
An antimalarial agent that revealed a therapeutic effect against SARS-CoV-2 (Carter-Timofte et al. 2021 Oct 18) |
| 12 | Cycloguanil | 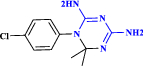 |
The active metabolite of proguanil (Sivaprakasam et al. 2009 Jul 27) |
| 13 | Chlorproguanil | 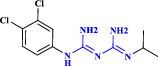 |
Antimalarial drug (Mutabingwa et al. 2001 Oct 13) |
| 14 | Pyrimethamine | 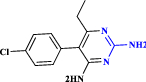 |
Anti-parasitic medication (Waller and Sampson 2018) |
| 15 | Benznidazole | 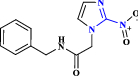 |
Anti-parasitic drug (Zaidel et al. 2020) |
| 16 | Berenil* (Diminazene) | 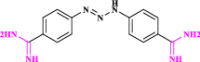 |
Anti-parasitic drug which is ACE2 activator with therapeutic activity in COVID-19 (Pantazi et al. xxxx) |
| 17 | Pentamidine* | Anti-infective agent that can be used against SARS-CoV-2 (Tomar et al. 2021 Mar) | |
| 18 | Hydroxyguanidine | 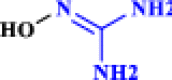 |
Anti-tumor drug with inhibitory effects on coronavirus RNA synthesis (Keck et al. 1989 Sep 1) |
| 19 | Imatinib | 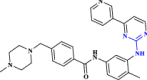 |
Anticancer drug possessing anti-COVID-19 efficacy (Aman et al. 2021 Sep 1) |
| 20 | Mitoguazone | 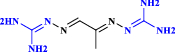 |
Chemotherapeutic agent (Yang et al. 2020 May) |
| 21 | Terbogrel | 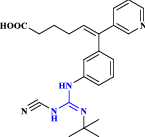 |
Thromboxane A2 receptor and synthase inhibitor (Mulvaney et al. 2020 Dec) |
| 22 | Clonidine | 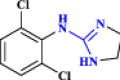 |
α2 adrenoceptor agonist: an anti-hypotensive agent that can manage complications co-occurred with COVID-19 infection (Hyoju et al. 2021 Jan) |
| 23 | Guanadrel | 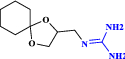 |
Adrenergic antagonist: antihypertensive agent (Palmer and Nugent 1983) |
| 24 | Guanethidine | Antihypertensive drug (Madias 2014 Dec 20) | |
| 25 | Guanoxan | 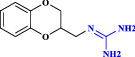 |
Antihypertensive agent (Ruedy and Davies 1967) |
| 26 | Pinacidil | 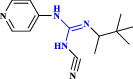 |
ATP-sensitive potassium channel opener: antihypertensive drug (Rajković et al. 2020 Dec) |
| 27 | KR 31,378 | 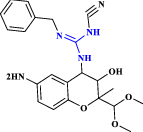 |
ATP-sensitive potassium channel activator (Choi et al. 2009 Jan 1) |
| 28 | CHS-828 | 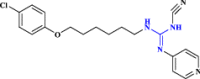 |
Chemotherapeutic agent (Hovstadius et al. 2004 Jan 1) |
| 29 | TP201565 (Analogue of CHS-828) | 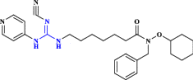 |
Chemotherapeutic agent (Hovstadius et al. 2004 Jan 1) |
| 30 | CGP48664* | 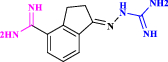 |
S-adenosylmethionine decarboxylase (AdoMetDC) inhibitor: anticancer agent (Thomas et al. 1996 Oct) |
| 31 | CGP35753(Analogue of CGP48664) | 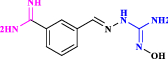 |
Anticancer agent (Tipnis 2000) |
| 32 | CGP40215A (Analogue of CGP35753) | 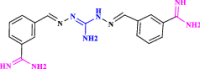 |
Anticancer agent (Tipnis 2000) |
| 33 | CGP39937* | 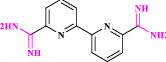 |
AdoMetDC inhibitor: chemotherapeutic agent (Thomas et al. 1996 Oct) |
| 34 | Camostat** | 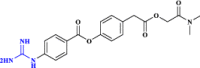 |
Human protease inhibitor (Hoffmann et al. 2021 Mar; Esam et al. 2022 Jan, Esam et al, 2020 May) |
| 35 | 4-(4-guanidinobenzoyloxy)phenylacetic acid (GBPA)** | 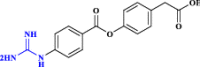 |
The camostat active metabolite (Hoffmann et al. 2021 Mar) |
*Guanidine-like containing molecules are determined by pink colored chemical N–C=N groups
**Reference molecules in this study are ATP, GTP, ribavirin triphosphate (for RdRp), carmofur (for Mpro), camostat, and GBPA (for TMPRSS2). The reference molecules of the first two the above-mentioned targets are investigated in our previously published work (Esam et al. 2022 Jan)
Materials and methods
Preparation of protein structures
The 3D structures of the selected target proteins, SARS-CoV-2 RdRp (PDB ID:6M71-Chain A) (Gao et al. 2020 Jan 1), SARS-CoV-2 Mpro (PDB ID: 6LU7-Chain A) (Jin et al. 2020), also spike receptor in complex with human ACE2 receptor (PDB ID: 6M0J) (Lan et al. 2020 May) and the TMPRSS2 target (PDB ID: 7MEQ) which plays a crucial role in SARS-COV-2 (Fraser et al. 2022 Jun) were retrieved from the protein data bank (www.rcsb.org) (Burley et al. 2021).
Ligand preparation
The 2D structures of all the selected ligands used in the study (Table 1), including the proved reference standard molecules for each target, guanidine-containing drugs, as well as the bioactive substances, were taken from the PubChem (https://pubchem.ncbi.nlm.nih.gov/) (Kim et al. 2016). Some of the guanidine-like groups possessing medicinal agents are also investigated. The constructed structures were energetically minimized using MOPAC (semi-empirical quantum mechanics) and saved as Mol file (∗.mol), and then converted to.pdbqt format by using AutoDock Tools.
Molecular docking
In order to prepare the protein target input files, the water molecules, ligands, and ions were removed from *.pdb files. Polar hydrogen atoms and gasteiger charges were added to each protein, and the atomic potential binding sites were defined using a grid size of (− 19, 12.6, 70) for Mpro and (118, 119, 140) for RdRP. We also considered to create the grid box for ACE2 and Spike with (− 25, 19, 3.1) and (− 32.48, 26.1, 7.92), respectively. Also the grid box size for TMPRSS2 was generated with (32, − 22.14, and − 8). All ligands and proteins were added as *pdbqt format using the AutoDock v4.2 program (Morris et al. 2009 Dec). Molecular docking simulations were carried out through virtual screening using AutoDock Vina (Trott and Olson 2010 Jan 30), and the outcomes of docking results were reported in the form of binding energy (kcal/mol). PLIP webserver was used to find out the involved amino acids with their interactive position in the docked molecule to conduct the hydrogen and hydrophobic bond interaction analysis.
Molecular dynamic simulation
To further evaluate the stability of the predicted complexes between the best-tested ligand with the investigated targets through the molecular docking studies, molecular dynamics (MD) simulations were conducted using the GROMACS version 2019.4 (Abraham et al. 2015 Sep).
All-atom force field on Ubuntu operating system (version 18.04) was used for protonation and minimization steps. SPC water model was chosen to simulate molecular dynamics (MD) of docked complexes in explicit solvation. To neutralize the system, adequate Na+, and Cl– ions were added to the solution. The equilibration was carried out with pressure 1 bar and temperature 300 K by two consecutive 100 ps simulations with canonical NVT ensembles and isobaric NPT ensembles for 1 ns each, respectively. The Particle mesh Ewald approximation was applied for the long-range electrostatic interaction cutoff of 1 nm computing coulomb and the van der Waals interactions (Darden and Pedersen xxxx), after carrying out 100 ns simulation run, the coordinates were saved every 2 fs time frame, and the GROMACS tools were used to analyze trajectories. In addition, a shorter trajectory, consisting of last 200 trajectories, was extracted from the original MD trajectory for the MMPBSA calculations.
Results and discussion
The molecular docking studies and binding mode analysis of the tested ligands against SARS-CoV-2 Mpro in complex with the inhibitor N3 (PDB ID: 6LU7), RdRp (PDB ID: 6M71), spike receptor-binding domain bound with ACE2 (PDB ID: 6M0J) and the host cell transmembrane serine protease 2: TMPRSS2 (PDB ID: 7MEQ)
Molecular docking investigations can be considered as the best primitive step of in silico drug design and discovery approaches. It provides the details of the protein–ligand interactions including the binding affinity as well as the binding modes. Based on our docking protocol, 35 compounds (Table 1) were screened against the critical viral and human targets which possess essential roles in COVID-19 infection. The binding affinities of these compounds against the selected targets are shown in Table 7.
Table 7.
Binding free energies of molecular docking studies at a glance
| Entry | Compounds | Docking scores kcal/mol | ||||
|---|---|---|---|---|---|---|
| SARS-CoV-2 Mpro (6LU7)* | Human TMPRSS2 (7MEQ)** | SARS-CoV-2 RdRp (6M71)*** | Human ACE2 receptor (6M0J) | SARS-CoV-2 Spike receptor (6M0J) | ||
| 1 | Metformin | − 4.7 | − 3.7 | − 4.9 | − 4.5 | − 4.2 |
| 2 | Cimetidine | − 5.9 | − 3.5 | − 5.2 | − 5 | − 4.6 |
| 3 | Viramidine | − 6.1 | − 4.1 | − 5.7 | − 5.6 | − 5.5 |
| 4 | Zanamivir | − 7.0 | − 3.9 | − 5.9 | − 5.2 | − 5.6 |
| 5 | Taribavirin | − 6.1 | − 4.0 | − 5.8 | − 6 | − 5.7 |
| 6 | Peramivir | − 6.4 | − 4.3 | − 5.9 | − 4.3 | − 5.4 |
| 7 | Famotidine | − 6.6 | − 4.9 | − 6.2 | − 5.2 | − 5.2 |
| 8 | Chlorproguanil | − 6.3 | − 4.4 | − 6.0 | − 5.2 | − 6 |
| 9 | CHS 828 | − 6.6 | − 4.9 | − 6.4 | − 5.4 | − 6.1 |
| 10 | Clonidine | − 5.3 | − 3.7 | − 5.2 | − 4.8 | − 5.1 |
| 11 | cycloguanil | − 6.1 | − 4.3 | − 6.0 | − 4.7 | − 5.1 |
| 12 | Laninamivir | − 6.8 | − 4.2 | − 5.6 | − 5.2 | − 5.6 |
| 13 | Guanadrel | − 6.0 | − 4.2 | − 5.6 | − 6 | − 5.6 |
| 14 | Guanoxan | − 6.2 | − 4.3 | − 5.9 | − 5.8 | − 5.5 |
| 15 | Hydroxyguanidine | − 4.0 | − 3.1 | − 4.4 | − 3.7 | − 3.1 |
| 16 | KR 31,378 | − 7.5 | − 4.4 | − 7.2 | − 5.1 | − 5.9 |
| 17 | Proguanil | − 6.1 | − 4.4 | − 6.1 | − 5.1 | − 5.9 |
| 18 | Guanethidine | − 5.6 | − 4.0 | − 5.2 | − 5.7 | − 4.9 |
| 19 | Pyrimethamine | − 5.6 | − 4.0 | − 5.3 | − 4.8 | − 5.2 |
| 20 | Rosuvastatin | − 7.8 | − 5.0 | − 6.3 | − 5.9 | − 6.3 |
| 21 | TP201565 | − 6.9 | − 4.0 | − 7.3 | − 5.4 | − 6.4 |
| 22 | Mitoguazone | − 5.4 | − 4.0 | − 5.5 | − 5.9 | − 5.1 |
| 23 | Pinacidil | − 6.4 | − 3.9 | − 5.7 | − 4.9 | − 5.4 |
| 24 | Imatinib | − 8.5 | − 6.2 | − 8.3 | − 7.3 | − 8.2 |
| 25 | Terbogrel | − 8.0 | − 5.0 | − 7.4 | − 5.9 | − 6.7 |
| 26 | Benznidazole | − 6.5 | − 4.8 | − 6.1 | − 5.1 | − 5.6 |
| 27 | Phenformin | − 6.2 | − 4.9 | − 6.2 | − 6.7 | − 6 |
| 28 | CGP 48664A | − 6.6 | − 4.7 | − 6.5 | − 5.4 | − 6.5 |
| 29 | Berenil | − 6.6 | − 4.6 | − 6.4 | − 6 | − 6.6 |
| 30 | CGP 40215A | − 7.4 | − 5.7 | − 8.0 | − 7.7 | − 7.4 |
| 31 | Pentamidine | − 6.6 | − 3.8 | − 5.9 | − 4.6 | − 5.7 |
| 32 | CGP-35753 | − 6.0 | − 4.7 | − 6.6 | − 6.7 | − 5.8 |
| 33 | CGP39937 | − 6.9 | − 5.1 | − 6.6 | − 6.7 | − 6 |
| 34 | GBPA | − 8.0 | − 5.1 | − 7.5 | − 8.2 | − 6.7 |
| 35 | Camostat | − 6.8 | − 5.1 | − 7.7 | − 6.1 | − 6.6 |
*Reference molecule N3 and carmofur for Mpro
**Reference molecules nafamostat, camostat, and GBPA for TMPRSS2
***Reference molecules ATP, GTP, and ribavirin triphosphate for RdRp
The highest binding affinities of − 8.3, − 8.5, − 8.2, − 6.2, and − 7.3 kcal/mol are achieved by imatinib, the magic bullet against SARS-CoV-2 RdRp, Mpro, and spike receptors as well as the host TMPRSS2 and ACE2 receptors, respectively. These resulting data signify that among these compounds, imatinib strongly binds to the investigated targets in this study. These obtained results in this study are confirmed by the numerous reported clinical and pharmacological efficacy of imatinib against SARS-CoV-2 (Aman et al. 2021 Sep 1; Bernal-Bello et al. 2020 Jul; Weston et al. 2020). The free binding energy (kcal/mol) of the best-tested ligands against SARS-CoV-2 RdRp, Mpro, the human ACE2 and viral spike receptors, and the host cell TMPRSS2, as well as the involved amino acids in the active/binding site of the investigated targets, are summarized in Tables 2, 3, 4 and 6, respectively. The docking results of the other evaluated compounds can be found in Tables S1-S5. Considering Figs. 2 and S1-S5, the importance of the guanidine group in the formation of stable ligand–receptor complexes is entirely apparent.
Table 2.
The resulting data (free energies and binding modes) from the docking studies of the best-tested ligands onSARS-CoV-2 RdRp (PDB ID: 6M71)
| Entry | Compounds | Free energy of binding (kcal/mol) | Hydrogen bands | Hydrophobic interactions | Other interactions |
|---|---|---|---|---|---|
| 7 | Famotidine | − 6.2 | ASP452, ARG553, THR556, ARG624, THR680, THR687, ASN691 | – | ASP623 |
| 8 | Chlorproguanil | − 6.0 | ARG553, THR556, ARG624 | TYR455, LYS545, ARG555, ARG624 | ASP452, ASP623 |
| 9 | CHS 828 | − 6.4 | ARG553, THR556, ARG624, SER682 | LYS621, ASP623, ARG624 | ARG624 |
| 11 | cycloguanil | − 6.0 | ARG553, THR556, ARG624 | LYS621,ARG624, TYR455 | ASP452, ASP623 |
| 16 | KR 31,378 | − 7.2 | ARG553, THR556, ARG624 | TYR455, ARG555, ARG624 | ASP452, ASP623 |
| 17 | Proguanil | − 6.1 | ARG553, THR556, ARG624 | TYR455, ARG555, ARG624 | ASP452, ASP623 |
| 20 | Rosuvastatin | − 6.3 | ARG553, ARG555, THR556, LYS621, ASP623, ARG624 | TYR455, LYS621, ARG624 | ARG553, LYS545, ARG555 |
| 21 | TP201565 | − 7.3 | ARG553, THR556, LYS621, ARG624, SER682 | ASP618, PRO620, LYS621, ASP623, LYS798 | ARG624 |
| 24 | Imatinib | − 8.3 | ARG553, LYS621, ASP623, ASN691, SER759 | TYR455, PRO620, ASP623, LYS798 | ARG553, ASP760 |
| 25 | Terbogrel | − 7.4 | GLN408, LEU544, TYR546 | TYR546, LYS545, LYS411, VAL410 | – |
| 26 | Benznidazole | − 6.1 | ARG553, THR556, ASP623 | TYR455, LYS621, ARG624 | ARG553 |
| 27 | Phenformin | − 6.2 | GLY413, TYR546 | LYS411, TYR546 | TYR546, ASP846 |
| 28 | CGP 48664A | − 6.5 | ILE548, ARG555, PHE843, ASP845, ARG858 | ILE548, ARG836, ALA840 | ASP845 |
| 29 | Berenil | − 6.4 | ILE548, ASP845, ARG858 | PHE441, ILE548, ARG836, VAL844 | ARG858 |
| 30 | CGP 40215A | − 8.0 | ASP164, ARG553, THR556, LYS621, ARG624, PHE793 | – | PRO620, LYS798 |
| 32 | CGP-35753 | − 6.6 | ASP452, TYR455, ARG553, THR556, LYS621 | TYR455, LYS621, ARG624 | ASP623 |
| 33 | CGP39937 | − 6.6 | ILE548, ASP845, ARG858 | ILE548, VAL844, ARG858 | ARG858, PHE441 |
| 34 | GBPA | − 7.5 | LYS545, ILE548 | PHE441, ILE548, VAL844 | ARG858 |
| 35 | Camostat | − 7.7 | THR556, TYR619, LYS621, ARG624, ASP760 | ASP618, LYS621, ARG624 | ARG553, LYS621, ARG624, ASP760 |
| Ref1* | ATP | − 7.5 | ASP452, THR556, TYR619, CYS622, ASP623, ASP760 | – | ARG553, ARG555, ARG624 |
| Ref2* | GTP | − 7.9 | ARG553, THR556, TYR619, LYS621, CYS622, ASP623 | – | ARG553, ARG555, ARG55, ASP623, ARG624 |
| Ref3* | Ribavirin triphosphate | − 7.2 | ASP452, ARG553, THR556, ASP623, SER682, ASN691, SER759 | – | ARG553, ARG555, ARG624 |
In addition to the best-investigated ligands, the docking results of the rest tested ligands against 6M71 are summarized in Table S1
*Reference molecules 1, 2, and 3 in docking study against SARS-CoV-2 RdRp
Table 3.
The resulting data (free energies and binding modes) from the docking studies of the best-tested ligands on SARS-CoV-2 Mpro (PDB ID: 6LU7)
| Entry | Compounds | Free Energy of Binding (kcal/mol) | Hydrogen Bands | Hydrophobic interactions | Other interactions |
|---|---|---|---|---|---|
| 3 | Viramidine | − 6.1 | GLY143, SER144, CYS145, HIS164, GLU166, LEU141 | – | – |
| 4 | Zanamivir | − 7.0 | PHE140, LEU141, ASN142, GLY143, SER144,CYS145, HIS164, GLU166, GLN189 | – | HIS41 |
| 5 | Taribavirin | − 6.1 | LEU141, GLY143, SER144, CYS145, HIS164, GLU166 | – | – |
| 6 | Peramivir | − 6.4 | LEU141, GLY143, SER144, CYS145, GLU166, GLN189 | MET165, GLN189 | – |
| 7 | Famotidine | − 6.6 | MET49, TYR54, LEU141, SER144, CYS145, HIS163, GLU166, GLN189 | – | – |
| 8 | Chlorproguanil | − 6.3 | – | ASP187, GLN189 | HIS41 |
| 9 | CHS 828 | − 6.6 | TYR54, GLU166, ARG188,THR190, GLN192 | THR25, LEU27, GLN189 | HIS41 |
| 11 | cycloguanil | − 6.1 | PHE140, GLY143, SER144, CYS145, GLU166 | ASN142 | GLU166 |
| 12 | Laninamivir | − 6.8 | LEU141, GLY143, SER144, CYS145, HIS164, GLU166, GLN189 | – | HIS41 |
| 13 | Guanadrel | − 6.0 | PHE140 | MET165 | GLU166 |
| 14 | Guanoxan | − 6.2 | LEU141, HIS164 | HIS41, MET165, GLN189 | – |
| 16 | KR 31,378 | − 7.5 | – | THR25, ASP187,GLN189 | HIS41 |
| 17 | Proguanil | − 6.1 | – | THR25, ASP187, GLN189 | HIS41 |
| 20 | Rosuvastatin | − 7.8 | GLY143, GLN189, THR190, GLN192 | HIS41, MET165, GLU166 | GLU166 |
| 21 | TP201565 | − 6.9 | THR45, SER46, GLY143 | THR25, MET165, GLU166, GLN189 | – |
| 23 | Pinacidil | − 6.4 | HIS164, GLU166 | HIS41, ASP187, GLN189 | – |
| 24 | Imatinib | − 8.5 | GLU166, THR190 | HIS41, GLU166, PRO168 | – |
| 25 | Terbogrel | − 8.0 | ARG188, THR190, GLN192 | THR25, LEU27, GLU166 | – |
| 26 | Benznidazole | − 6.5 | GLU166 | HIS41, MET165, GLN189 | – |
| 27 | Phenformin | − 6.2 | ASN142, GLY143, SER144, CYS145, HIS163 | THR25, LEU27 | HIS41, GLU166 |
| 28 | CGP 48664A | − 6.6 | HIS41,HIS164, ARG188, THR190, GLN192 | MET165,GLN189 | HIS41 |
| 29 | Berenil | − 6.6 | TYR54, GLY143, SER144, CYS145, HIS164, GLU166 | MET165, GLU166, GLN189 | – |
| 30 | CGP 40215A | − 7.4 | MET49, TYR54, GLU166, PRO168, GLY170, GLN189, THR190, GLN192 | MET165, PRO168, GLN189 | HIS41 |
| 31 | Pentamidine | − 6.6 | LEU141, SER144, THR190 | MET165 | – |
| 32 | CGP-35753 | − 6.0 | GLY143, SER144, CYS145, LEU141 | MET165 | HIS41 |
| 33 | CGP39937 | − 6.9 | MET49, TYR54, LEU141, ASN142, GLY143, SER144, CYS145, GLN189 | MET165, GLN189 | – |
| 34 | GBPA | − 8.0 | HIS41, TYR54, GLY143, SER144, CYS145 | GLN189 | ASP187 |
| 35 | Camostat | − 6.8 | TYR54, ASP187, GLN189 | ASN142, MET165, GLN189 | HIS41 |
| Ref* | Carmofur | − 6.3 | GLY143, SER144, CYS145 | – | – |
| Ref* | N3 | − 7.7 | ASN142,THR26,GLY143, SER144, CYS145 | ASN142, GLU166 | – |
In addition to the best-investigated ligands, the docking results of the rest tested ligands against 6LU7 are summarized in Table S2
*Reference molecule in docking study against the SARS-CoV-2 main protease
Table 4.
The resulting data (free energies and binding modes) from the docking studies of the best-tested ligands on human ACE2 receptors (PDB ID: 6M0J)
| Entry | Compounds | Free Energy of Binding (kcal/mol) | Hydrogen Bands | Hydrophobic interactions | Other interactions |
|---|---|---|---|---|---|
| 5 | Taribavirin | − 6 | ALA348, ASP350, TYR394, ARG393 | – | – |
| 13 | Guanadrel | − 6 | ALA348, ASP350 | PHE40, PHE390, ARG393 | ASP382 |
| 24 | Imatinib | − 7.3 | LYS353, GLY354, ARG393 | ASN33, GLU37, ALA386, PRO389 | HIS34 |
| 27 | Phenformin | − 6.7 | ALA348, ASP350, TYR385, ARG393 | PHE40, PHE390, ARG393 | ASP350, ASP382 |
| 29 | Berenil | − 6 | ALA384, GLN388, GLY551, ARG559 | ALA387, PHE555 | PHE555 |
| 30 | CGP 40215A | − 7.7 | LYS353, GLY354, ASP382, TYR385, ARG393, ASN394 | PHE40, PHE390, ARG393 | ASP350 |
| 32 | CGP-35753 | − 6.7 | ASP350, PHE390, LEU391, ARG393, ASN394 | ASP350, PHE390 | PHE40 |
| 33 | CGP39937 | − 6.7 | GLU37, ALA348, ASP350, ARG393 | PHE390 | PHE40 |
| 34 | GBPA | − 8.2 | ASP350, ARG393, ASN394 | PHE40, PHE390, ARG393 | – |
| 35 | Camostat | − 6.1 | ASN33, MET383, PHE390 | ALA386 | ARG393 |
In addition to the best tested ligands, the docking results of the rest tested ligands against 6M0J are summarized in Table S3
Table 6.
The resulting data (free energies and binding modes) from the docking studies of the best-tested ligands against TMPRSS2 (PDB ID: 7MEQ)
| Entry | Compounds | Free Energy of Binding (kcal/mol) | Hydrogen Bands | Hydrophobic interactions | Other interactions |
|---|---|---|---|---|---|
| 3 | Viramidine | − 4.1 | ASN177, TYR180, SER215 | – | – |
| 5 | Taribavirin | − 4.0 | TYR180, SER215, LYS224 | – | – |
| 7 | Famotidine | − 4.9 | LYS211, ASN213, THR214, LYS223, TYR226 | – | – |
| 8 | Chlorproguanil | − 4.4 | LYS223 | TYR226 | TYR226 |
| 9 | CHS 828 | − 4.9 | ASN213, LYS223 | TYR226 | TYR226 |
| 11 | cycloguanil | − 4.3 | SER436, SER460 | – | HIS296 |
| 13 | Guanadrel | − 4.2 | LYS211, THR214, TYR226 | – | ASP175 |
| 14 | Guanoxan | − 4.3 | ASN213, LEU225 | – | TYR226 |
| 16 | KR 31,378 | − 4.4 | LYS223 | TYR226 | TYR226 |
| 17 | Proguanil | − 4.4 | LYS223 | TYR226 | TYR226 |
| 18 | Guanethidine | − 4.0 | TYR180, SER215, LYS224 | ASP175, TYR180 | – |
| 19 | Pyrimethamine | − 4.0 | LEU212, THR214, SER215,TYR226 | – | TYR226 |
| 20 | Rosuvastatin | − 5.0 | ASN213, THR214, SER215,TYR226, HIS227 | TYR226 | – |
| 21 | TP201565 | − 4.0 | ASN213, LYS213 | LYS223, LYS224 | – |
| 22 | Mitoguazone | − 4.0 | ASP175, TYR180, SER215, ALA216, LYS224 | THR214 | ASP175 |
| 23 | Pinacidil | − 3.9 | ASN213, LYS223 | TYR226 | – |
| 24 | Imatinib | − 6.2 | LYS223, HIS227, | SER162, TYR222 | TYR226 |
| 25 | Terbogrel | − 5.0 | ASN213, LEU225, HIS227 | – | TYR226 |
| 26 | Benznidazole | − 4.8 | TYR226 | TYR226 | TYR226 |
| 27 | Phenformin | − 4.9 | LYS211, LEU212, THR214, TYR226 | – | TYR226,ASP175 |
| 28 | CGP 48664A | − 4.7 | ASN213, THR214, TYR226, HIS227 | TYR226 | TYR226 |
| 29 | Berenil | − 4.6 | ASP175, LYS211, THR214, TYR226 | – | TYR226 |
| 30 | CGP 40215A | − 5.7 | ASN213, LEU225, HIS227 | TYR226 | TRP461, HIS296 |
| 31 | Pentamidine | − 3.8 | TYR222, LYS223, LEU225 | SER162 | TYR222 |
| 32 | CGP-35753 | − 4.7 | ASN213, LYS223, LEU225 | TYR226 | TYR226 |
| 33 | CGP39937 | − 5.1 | ASN213, SER215, LYS223, LEU225 | – | TYR226 |
| 34 | GBPA | − 5.1 | ASN213, THR214, HIS227 | TYR226 | – |
| 35 | Camostat | − 5 | ASN213, THR214, HIS227 | TYR266 | – |
| Ref* | Nafamostat | − 5.6 | TYR222, LYS223, TYR226 | TYR226 | TYR226 |
In addition to the best-investigated ligands, the docking results of the rest tested ligands against 7MEQ are summarized in Table S5
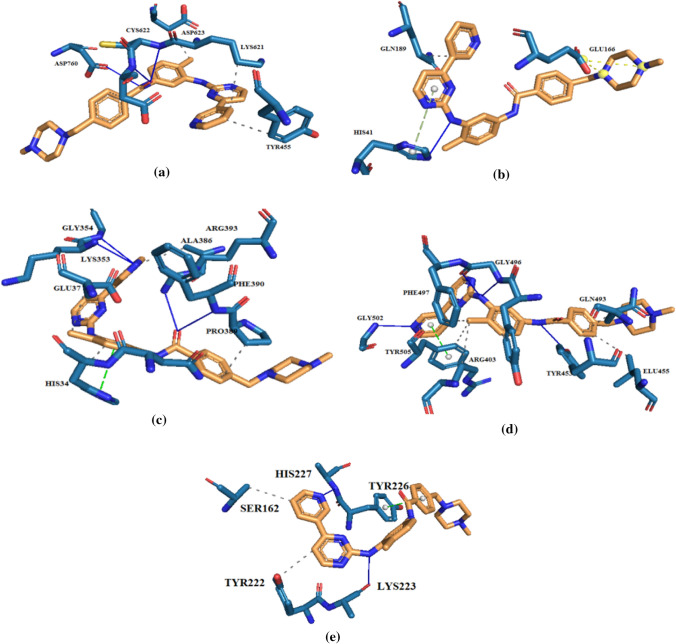
Fig. 2.
Imatinib interactions with binding residues of (a) RdRp (6m71), (b) Mpro (6lu7), (c) ACE2 receptor (6M0J), (d) SARS-CoV-2 spike glycoprotein (6M0J), and (e) TMPRSS2 (7MEQ). The 3D structures of these interactions are shown in Fig. S6
In comparison with ATP, GTP, and ribavirin triphosphate as the endogenous and therapeutic reference standard ligands against SARS-CoV-2 RdRp (with free binding energies of − 7.5, − 7.9, and − 7.2 kcal/mol, respectively) (Esam et al. 2022 Jan), imatinib with the best free energy of binding (− 8.3 kcal/mol) had hydrogen bonds with amino acids ARG553, LYS621, ASP623, ASN691 and SER759, whereas hydrophobic interaction with TYR455, PRO620, ASP623 and LYS798 and one salt bridge with ASP760 and one π-cation interaction with ARG553 of RdRp binding site (Table 2 and Fig. 2a). The importance of the guanidine group in ligand–target complex formation can be found in Figs. 2a and S1.
The free energy of binding between the best tested guanidine-containing molecules and the SARS-CoV-2 main protease are summarized in Table 3. The amino acid interactions between Mpro as target and imatinib as the best-investigated ligand with the lowest free binding energy (− 8.5 kcal/mol) were also identified (Bekhradnia et al. 2015), as shown in Table 3. HIS41, as one of the principal residues of the catalytic dyad (Cys145 and His41) in the main protease (Esam et al. 2022 Jan), was found to have binding interaction with imatinib. To be precise, in comparison with the reference standard ligand carmofur (with the free energy of binding − 6.3 kcal/mol), imatinib was found to have hydrogen binding interaction with GLU166 and THR190 residues in the main protease, while HIS41, GLU166 and PRO168 have hydrophobic interaction with this magic bullet. The crucial role of the guanidine moiety in formation of the highly stable protein–ligand complexes can be found in Figs. 2b and S2.
As can be seen from the results (Table 4), imatinib has also been reported to have the best binding affinity to the host ACE2 receptor (− 7.3 kcal/mol) via hydrogen bond interaction with LYS353, GLY354, PHE390, and ARG393, while the hydrophobic interaction was with ASN33, HIS34, GLU37, ALA386, and PRO389. On the other hand, this magic bullet creates a stable ligand–target complex (− 8.2 kcal/mol) with TYR453, GLN493, GLY496, and GLY502 residues of the virus spike glycoprotein by hydrogen bonds and hydrophobic interaction with ARG403, LEU455, GLN493, TYR495, PHE497,and TYR505 residues (Table 5). Apart from Tables 4 and 5 that contain the docking results of the best-evaluated ligands against the human ACE2 and the viral spike receptors, the obtained data from the molecular docking study of the rest tested ligands in the current investigation are summarized in Tables S3 and S4. The essential role of the guanidine scaffold in the chemical structure of the studied compounds in this research can be seen in Figs. 2c, 2d, S3, and S4.
Table 5.
The resulting data (free energies and binding modes) from the docking studies of the best-tested ligands on virus spike protein (PDB ID: 6M0J)
| Entry | Compounds | Free Energy of Binding (kcal/mol) | Hydrogen Bands | Hydrophobic interactions | Other interactions |
|---|---|---|---|---|---|
| 8 | Chlorproguanil | − 6 | GLY496, GLN498, ASN501 | TYR495 | TYR505 |
| 9 | CHS 828 | − 6.1 | TYR453, GLY496, TYR505 | Tyr449, TYR495, ASN501, TYR505 | TYR505 |
| 20 | Rosuvastatin | − 6.3 | ARG403, TYR453, GLN493, SER494 | ARG403, LEU455, GLN493, TYR495 | TYR505, ANS501, ARG403 |
| 21 | TP201565 | − 6.4 | GLN493, SER494 | TYR449, GLN493,TYR495, ASN501, TYR505 | TYR505 |
| 24 | Imatinib | − 8.2 | TYR453, GLY496, GLY502 | LEU455, GLN493, TYR495, PHE497, TYR505 | TYR505 |
| 25 | Terbogrel | − 6.7 | ARG403, TYR453, GLY496 | TYR505, TYR495, GLN493 | TYR505 |
| 27 | Phenformin | − 6 | TYR453, GLY496, TYR505 | ASN501, TYR505 | TYR505 |
| 28 | CGP 48664A | − 6.5 | TYR453, GLY496, SER494, ASN501 | TYR495, ARG403 | TYR505 |
| 29 | Berenil | − 6.6 | ARG403, TYR453, GLN498, ASN501 | TYR495, ASN501, TYR505 | TYR505, ARG403 |
| 30 | CGP 40215A | − 7.4 | ARG403, ASP405, ARG408, GLN409, TYR453, ASN501 | ARG403, LYS417, TYR495, PHE497 | TYR505, GLU406 |
| 33 | CGP39937 | − 6 | ARG403, GLU406, TYR453, GLY496, ASN501 | ARG403, TYR495, PHE497 | TYR505 |
| 34 | GBPA | − 6.7 | ARG403, GLU406, TYR453, GLY496, ASN501, TYR505 | TYR453, TYR495, TYR505 | – |
| 35 | Camostat | − 6.6 | ARG403, TYR453, GLN498, ASN501 | LYS417, TYR453 | TYR505, ARG403 |
In addition to the best-evaluated ligands, the docking results of the rest tested ligands against 6M0J are summarized in Table S4
Regarding TMPRSS2–imatinib complex, imatinib with the best docking score (6.2 kcal/mol) in comparison with the reference standard ligands camostat (− 5 kcal/mol) and its active metabolite GBPA (− 5.1 kcal/mol), interact with residues LYS223 and HIS227 by hydrogen bonds and hydrophobic interaction with SER162 and TYR222 and π-stacking as with TYR226 summarized in Table 6. The determinative roles of guanidine in the formation of the investigated ligand–target complexes are shown schematically in Figs. 2e and S5.
The calculated docking free energies of all the tested ligands in this study (Table 7) as well as the involved amino acids in RdRp, Mpro, and Tmprss2 that are summarized in Tables 2, 3, 6, S1, S2, and S5 in comparison with the related reference standard molecules have proved that the guanidine-containing molecules can be considered as prophylactic/therapeutic agents or at least the promising lead compounds against SARS-CoV-2. The critical role of the guanidine group in the chemical structure of the best-tested ligands (Tables 2, 3, 4, 5 and 6) and the best investigated ligand imatinib as well as the other investigated molecules (Tables S1-S5), are also demonstrated clearly in Figs. 2 and S1-S5.
Molecular dynamics (MD) simulation
Molecular dynamics simulation was carried out for the apo-form without ligand to validate the MD system. As imatinib showed strong intermolecular interaction with all the investigated target proteins with high binding scores (− 6.2 kcal/mol to − 8.5 kcal/mol) (Table 7), this compound was selected as the potential inhibitor to use for further analysis through 100 ns MD simulation, which reveals the interaction and the stability of inhibitor complexes with protein. The root mean square deviation (RMSD) and root mean square fluctuation (RMSF) were calculated for all frames in the trajectory. The protein RMSD values give insights into its structural conformation and the stability of protein–ligand complexes throughout the simulation. The lower RMSD values indicated the higher stability of the simulation system. The RMSF value that investigates the residual vibration, the structural integrity and the atomic mobility of the complex was also evaluated.
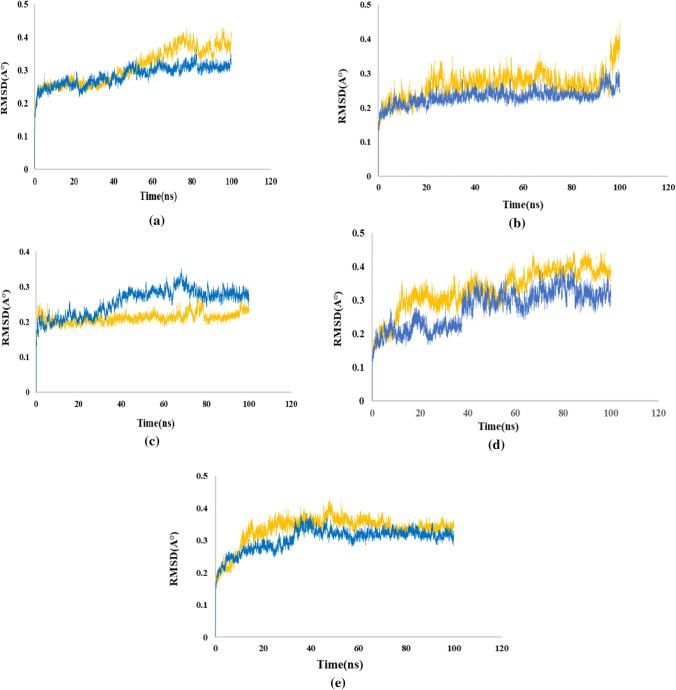
Imatinib with RdRp
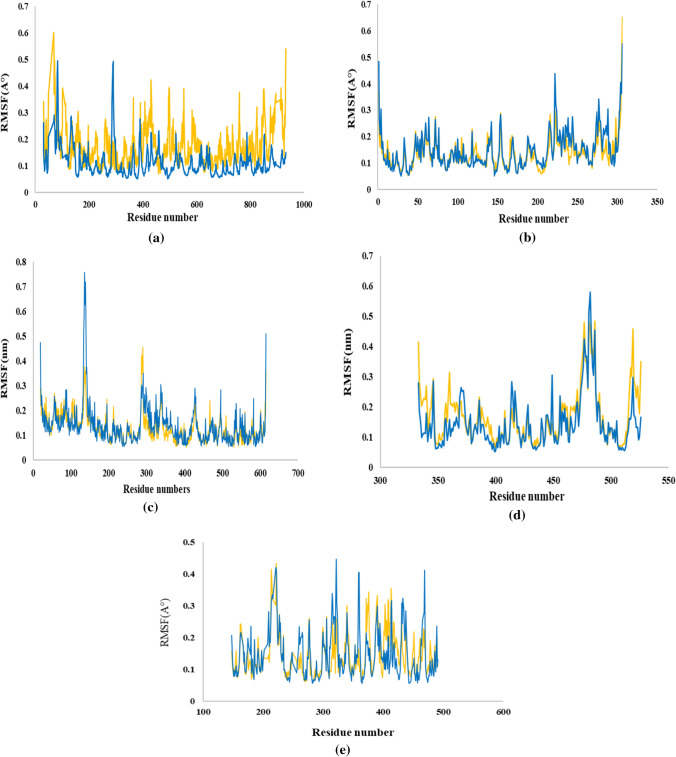
RMSD values for imatinib with RdRp reached at its maximum dynamicity peak 0.428 nm, and the RMSD value of the ligand indicated stabilization after about 80 ns of simulation (Fig. 3a). In this complex, as can be seen in (Fig. 3a), the mean RMSD of protein was 0.3 nm, which was within the acceptable range. On the other hand, the RMSD of apo-RdRp was static over the simulation period with the average of 0.28. Moreover, as (Fig. 4a) shows, the mean RMSF value of the protein-RdRp was 0.6 which shows higher fluctuation for imatinib-RdRp than apo-RdRp. The highest peaks for imatinib belong to TYR69, GLU431, ARG553, SER759, LYS849, which are located outside of the enzyme binding site. The residues with the most interactions in the protein–ligand complexes in molecular docking studies such as ASP623, LYS621, and ASP760 had the least RMSF values in the imatinib–RdRp complexes in MD studies. Thus, RMSF calculations indicated that MD results were following molecular docking outputs.
Fig. 3.
RMSD Analysis: (a) RdRp (6m71), (b) Mpro (6lu7), (c) the host ACE2 receptor (6M0J), (d) SARS-CoV-2 spike (6M0J), (e) human TMPRSS2 (7MEQ)
Fig. 4.
RMSF Analysis: (a) RdRp (6m71), (b) Mpro (6lu7), (c) the host ACE2 receptor (6M0J), (d) SARS-CoV-2 spike (6M0J), (e) human TMPRSS2 (7MEQ)
Imatinib with Mpro
The mean RMSD value of imatinib with Mpro was 0.27 nm which is within the acceptable range and confirmed this docked complex was stable during the simulation (Fig. 3b). Moreover, the last 20 ns RMSD of imatinib-Mpro was observed quite similar to the apo-Mpro in Fig. 3b. Regarding Mpro complexes, the RMSF values were carefully investigated (Fig. 4b). The RMSF value of Mpro was 0.14. It was observed that the highest fluctuations belong to TYR154, ASP216, ARG222, and LEU277 that are not found in the binding site. As is illustrated in (Fig. 4b), the amino acids with the most interactions in molecular docking studies of Mpro–imatinib complex, GLU166, THR190, and HIS41 exhibited low values of fluctuation, which verify the outcomes of molecular docking. The result of analysis shows that the fluctuation of residues in imatinib–Mpro complex does not change with respect to apo-Mpro.
Imatinib with ACE2
The obtained RMSD was 0.21 for ACE2, and as is illustrated in (Fig. 3c), the ligand–protein complex reached the equilibrium status during the simulation time. From the above observation with RMSD deviations, it can be concluded that imatinib behaves well within the active site of the ACE2 protein. It can also be observed that apo-form of the ACE2 showed more deviation in the RMSD values in comparison with imatinib–ACE2 complex form. As can be seen from the RMSF plot of the ACE2–protein complex (Fig. 4c), the average RMS fluctuation for ACE2 is 0.12 nm that proved to forma stable complex.
There is a strong fluctuation for amino acids ASN290, GLN139, ASN338, and GLN429, which are far from the protein binding area, while the amino acids on the protein binding region have varying degrees of fluctuation. Moreover, the apo-ACE2 showed similar RMSF values as imatinib–ACE2 complex.
Imatinib with spike
Concerning the spike–imatinib complex, the value of RMSD is found changing between 0.1 and 0.45 nm that reached the equilibrium stage after about 60 ns (Fig. 3d). Minor fluctuations observed at various intervals are because of the conformation changes of ligand at the active sites. As shown in Fig. 3d, the apo form structure showed an RMSD value of 0.27 nm.
Additionally, RMSFs of the Cα atoms belonging to spike protein concerning the mean structure and the apo form structure were measured below about 0.2 nm (Fig. 4d). The RMSF values of the residues with the highest fluctuations are apart from the active site, while the residues with the lowest fluctuations are in the active site of the protein.
Imatinib with TMPRSS2
The RMSD plot (Fig. 3e) shows that the imatinib–TMPRSS2 complex is stable with an average deviation of 0.33 nm in the second 50 ns. Generated trajectories for TMPRSS2 complex during the whole run indicate more stability than apo-TMPRSS2. RMSF was calculated and plotted (Fig. 4e). Compared to apo-protease structure, while bound to imatinib, residues 376 and 413 showed higher mobility, whereas residues 322 and 469 showed higher mobility.
MMPBSA
MMPBSA calculations were carried out for the compound with the best MD results in complex with studied targets by MM-PBSA. The MM-PBSA analysis revealed the contribution of different energies that has a significant impact on the stability of studied complexes. The distribution of MM-PBSA binding free energies is listed in Table 8. As can be seen, imatinib–Mpro complex (− 100.575 ± 6.397 kcal/mol) is the lowest signifying strong binding affinity, while imatinib-SARS-CoV-2 spike has the lowest binding free energy (− 49.124 ± 6.145 kcal/mol). Moreover, imatinib–TMPRSS2 complex displayed stability with − 123.159 ± 13.696 kcal/mol. In addition, the total binding free energy of imatinib with the host ACE2 and SARS-CoV-2 RdRp is (− 65.633 ± 6.657 kcal/mol) and (− 66.663 ± 20.611 kcal/mol), respectively.
Table 8.
Results of the binding free energy calculation for imatinib in complex with the (a) RdRp (6m71), (b) Mpro (6lu7), (c) human ACE2 receptor (6M0J), (d) SARS-CoV-2 spike receptor (6M0J), (e) the host TMPRSS2 (7MEQ)
| Targets | ΔG binding energy | ΔG Vdw | ΔG elec | ΔG polar | SASA energy |
|---|---|---|---|---|---|
| (a) | − 65.633 ± 6.657 | − 56.775 ± 5.667 | − 3.440 ± 5.131 | 3.922 ± 8.621 | − 9.340 ± 1.514 |
| (b) | − 100.575 ± 6.397 | − 70.700 ± 5.096 | − 15.252 ± 3.514 | − 2.463 ± 3.953 | − 12.159 ± 1.271 |
| (c) | − 66.663 ± 20.611 | − 55.409 ± 6.807 | 6.869 ± 13.377 | − 8.473 ± 7.950 | − 9.650 ± 1.499 |
| (d) | − 49.124 ± 6.145 | − 42.884 ± 3.937 | − 0.816 ± 6.919 | 1.738 ± 4.730 | − 7.163 ± 1.081 |
| (e) | − 123.159 ± 13.696 | − 167.144 ± 10.939 | − 12.623 ± 3.025 | 72.632 ± 8.339 | − 16.024 ± 0.946 |
All energies are in kJ/mol
PKa
The PKa values of the guanidine-containing therapeutic agents that have been investigated in the current in silico study have also been checked (Table S6). In comparison with the two well-known efficient anti-SARS-CoV-2 drugs (hydroxy)chloroquine (Altulea et al. 2021 Mar 9; Liu et al. xxxx), most of the investigated compounds in this study especially those possessing proved experimental/clinical activity against SARS-CoV-2 (the bolded name molecules in Table 1 including metformin and phenformin (Langmaier et al. 2016 Sep 22), rosuvastatin, famotidine (Laurence and Lewis 2009), proguanil (Plöger et al. 2018 Jul 1), pentamidine (Paul et al. 1998 Jan 1), benznidazole (Moral Sanchez et al. 2018 Oct), Berenil (Atsriku et al. 2002 Nov 7), imatinib (Manley et al. 2010 Oct 1), camostat and GBPA (Hempel et al. 2021) have shown basic/extremely basic property that considering the presence the guanidine moiety, it is not surprising. Thus, according to the observed results, the guanidine-containing molecules can have a multi mechanism activity against SARS-CoV-2, and this theoretical claim needs further experimental investigations.
Conclusion
The COVID-19 pandemic is still a considerable challenge to many aspects of human life worldwide. The previously reported efficacy of metformin, phenformin, rosuvastatin, famotidine, proguanil, pentamidine, benznidazole, Berenil, imatinib, camostat, etc., in the current potentially fatal viral infection made us to investigate 35 molecules of the guanidine-carrying compounds toward the therapeutic targets against SARS-CoV-2.
The results of the current in silico investigation have revealed that the guanidine-containing medicinal agents can be considered as potential therapeutic as well as prophylactic medications against COVID-19. In this research, the magic bullet, imatinib, revealed the best docking scores and MD results. Furthermore, it seems that aside from the target–ligand concepts, the guanidine functional group with its poly nitrogen structure-derived basicity can make the target cells’ intracellular pH undesirable for the virus entry, uncoating, and cytosolic lifecycle the same as the pH alteration by chloroquine and hydroxychloroquine.
Besides, according to the guanidine antithrombotic activity, preventing effects on the formation of reactive oxygen species (ROS), NO synthase inhibition, and anti-inflammatory effects, as well as its confirmed antiviral properties via inhibition of the viral proteases, neuraminidase, sialidase, chemokine receptors (Saczewski and Balewski 2009 Oct 1), this scaffold and its containing substances can be considered as a particular anti-COVID-19 generation that deserve further comprehensive investigations.
Supplementary Information
Below is the link to the electronic supplementary material.
Funding
Funding was provided by Mazandaran University of Medical Sciences.
Declarations
Conflict of interest
On behalf of all authors, the corresponding author states that there is no conflict of interest.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Change history
3/31/2023
The Original article is revised to update the corresponding author affiliation.
Change history
3/30/2023
A Correction to this paper has been published: 10.1007/s11696-023-02787-3
References
- Abraham MJ, Murtola T, Schulz R, Páll S, Smith JC, Hess B, Lindahl E. GROMACS: high performance molecular simulations through multi-level parallelism from laptops to supercomputers. SoftwareX. 2015;1(1):19–25. doi: 10.1016/j.softx.2015.06.001. [DOI] [Google Scholar]
- Altulea D, Maassen S, Baranov MV, van den Bogaart G. What makes (hydroxy) chloroquine ineffective against COVID-19: insights from cell biology. J Mol Cell Biol. 2021 doi: 10.1093/jmcb/mjab016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aman J, Duijvelaar E, Botros L, Kianzad A, Schippers JR, Smeele PJ, Azhang S, Bartelink IH, Bayoumy AA, Bet PM, Boersma W. Imatinib in patients with severe COVID-19: a randomised, double-blind, placebo-controlled, clinical trial. Lancet Respir Med. 2021;9(9):957–968. doi: 10.1016/S2213-2600(21)00237-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atsriku C, Watson DG, Tettey JN, Grant MH, Skellern GG. Determination of diminazene aceturate in pharmaceutical formulations by HPLC and identification of related substances by LC/MS. J Pharm Biomed Anal. 2002;30(4):979–986. doi: 10.1016/S0731-7085(02)00450-8. [DOI] [PubMed] [Google Scholar]
- Baig MS, Alagumuthu M, Rajpoot S, Saqib U. Identification of a potential peptide inhibitor of SARS-CoV-2 targeting its entry into the host cells. Drugs R&d. 2020;20(3):161–169. doi: 10.1007/s40268-020-00312-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernal-Bello D, Jaenes-Barrios B, Morales-Ortega A, Ruiz-Giardin JM, García-Bermúdez V, Frutos-Pérez B, Farfán-Sedano AI, de Ancos-Aracil C, Bermejo F, García-Gil M, Zapatero-Gaviria A. Imatinib might constitute a treatment option for lung involvement in COVID-19. Autoimmun Rev. 2020;19(7):102565. doi: 10.1016/j.autrev.2020.102565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bekhradnia A, Norrby P-O. New insights into the mechanism of iron-catalyzed cross-coupling reactions. Dalton Transactions. 2015;44(9):3959–3962. doi: 10.1039/C4DT03491K. [DOI] [PubMed] [Google Scholar]
- Burley SK, Bhikadiya C, Bi C, Bittrich S, Chen L, Crichlow GV, Christie CH, Dalenberg K, di Costanzo L, Duarte JM, Dutta S. Guranovi c, V. Guzenko, D., Hudson, BP, Zhuravleva, M. 2021. 10.1093/nar/gky1004
- Byléhn F, Menéndez CA, Perez-Lemus GR, Alvarado W, De Pablo JJ. Modeling the binding mechanism of remdesivir, favilavir, and ribavirin to SARS-CoV-2 RNA-dependent RNA polymerase. ACS Cent Sci. 2021;7(1):164–174. doi: 10.1021/acscentsci.0c01242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carter-Timofte ME, Arulanandam R, Kurmasheva N, Fu K, Laroche G, Taha Z, van der Horst D, Cassin L, van der Sluis RM, Palermo E, Di Carlo D. Antiviral potential of the antimicrobial drug atovaquone against SARS-CoV-2 and emerging variants of concern. ACS Infect Dis. 2021;7(11):3034–3051. doi: 10.1021/acsinfecdis.1c00278. [DOI] [PubMed] [Google Scholar]
- Choi A, Choi JS, Yoon YJ, Kim KA, Joo CK. KR-31378, a potassium-channel opener, induces the protection of retinal ganglion cells in rat retinal ischemic models. J Pharmacol Sci. 2009;109(4):511–517. doi: 10.1254/jphs.FP0072067. [DOI] [PubMed] [Google Scholar]
- Darden TA, Pedersen LG. Molecular modeling: an experimental tool. 10.1289/ehp.93101410 [DOI] [PMC free article] [PubMed]
- del Moral Sanchez JM, Gonzalez-Alvarez I, Cerda-Revert A, Gonzalez-Alvarez M, Navarro-Ruiz A, Amidon GL, Bermejo M. Biopharmaceutical optimization in neglected diseases for paediatric patients by applying the provisional paediatric biopharmaceutical classification system. Br J Clin Pharmacol. 2018;84(10):2231–2241. doi: 10.1111/bcp.13650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esam Z. A proposed mechanism for the possible therapeutic potential of Metformin in COVID-19. Diabetes Res Clin Pract. 2020;1:167. doi: 10.1016/j.diabres.2020.108282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esam Z, Akhavan M, Bekhradnia A, Mohammadi M, Tourani S. A Novel Magnetic Immobilized Para-Aminobenzoic Acid-Cu(II) Complex: A Green, Efficient and Reusable Catalyst for Aldol Condensation Reactions in Green Media. Catalysis Letters. 2020;150(11):3112–3131. doi: 10.1007/s10562-020-03216-w. [DOI] [Google Scholar]
- Esam Z, Akhavan M, Bekhradnia A. Molecular docking and dynamics studies of Nicotinamide Riboside as a potential multi-target nutraceutical against SARS-CoV-2 entry, replication, and transcription: a new insight. J Mol Struct. 2022;5(1247):131394. doi: 10.1016/j.molstruc.2021.131394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fadlalla M, Ahmed M, Ali M, Elshiekh AA, Yousef BA. Molecular docking as a potential approach in repurposing drugs against COVID-19: a systematic review and novel pharmacophore models. Curr Pharmacol Rep. 2022;1:1–5. doi: 10.1007/s40495-022-00285-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fraser BJ, Beldar S, Seitova A, Hutchinson A, Mannar D, Li Y, Kwon D, Tan R, Wilson RP, Leopold K, Subramaniam S. Structure and activity of human TMPRSS2 protease implicated in SARS-CoV-2 activation. Nat Chem Biol. 2022;8:1–9. doi: 10.1038/s41589-022-01059-7. [DOI] [PubMed] [Google Scholar]
- Gao Y, Yan L, Huang Y, Liu F, Zhao Y, Cao L, Wang T, Sun Q, Ming Z, Zhang L, Ge J. Structure of RNA-dependent RNA polymerase from 2019-nCoV, a major antiviral drug target. BioRxiv. 2020 doi: 10.1126/science.abb7498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gunst JD, Staerke NB, Pahus MH, Kristensen LH, Bodilsen J, Lohse N, Dalgaard LS, Brønnum D, Fröbert O, Hønge B, Johansen IS. Efficacy of the TMPRSS2 inhibitor camostat mesilate in patients hospitalized with Covid-19-a double-blind randomized controlled trial. EClinicalMedicine. 2021;1(35):100849. doi: 10.1016/j.eclinm.2021.100849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamada Y. Novel prodrugs with a spontaneous cleavable guanidine moiety. Bioorganic & Medicinal Chemistry Letters. 2016;26(7):1685–9. doi: 10.1016/j.bmcl.2016.02.060. [DOI] [PubMed] [Google Scholar]
- Hempel T, Raich L, Olsson S, Azouz NP, Klingler AM, Hoffmann M, Pöhlmann S, Rothenberg ME, Noé F. Molecular mechanism of inhibiting the SARS-CoV-2 cell entry facilitator TMPRSS2 with camostat and nafamostat. Chem Sci. 2021;12(3):983–992. doi: 10.1039/d0sc05064d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann M, Kleine-Weber H, Schroeder S, Krüger N, Herrler T, Erichsen S, Schiergens TS, Herrler G, Wu NH, Nitsche A, Müller MA, Drosten C, Pöhlmann S. SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell. 2020;181:271. doi: 10.1016/j.cell.2020.02.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann M, Hofmann-Winkler H, Smith JC, Krüger N, Arora P, Sørensen LK, Søgaard OS, Hasselstrøm JB, Winkler M, Hempel T, Raich L. Camostat mesylate inhibits SARS-CoV-2 activation by TMPRSS2-related proteases and its metabolite GBPA exerts antiviral activity. EBioMedicine. 2021;1(65):103255. doi: 10.1016/j.ebiom.2021.103255,10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann M, Hofmann-Winkler H, Smith JC, Kruger N, Arora P, Sorensen LK, Sogaard OS, Hasselstrom JB, Winkler M, Hempel T, Raich L. Camostat mesylate inhibits SARS-CoV-2 activation by TMPRSS2-related proteases and its metabolite GBPA exerts antiviral activity. EBioMedicine. 2021;65:103255. doi: 10.1016/j.ebiom.2021.103255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hovstadius P, Lindhagen E, Hassan S, Nilsson K, Jernberg-Wiklund H, Nygren P, Binderup L, Larsson R. Cytotoxic effect in vivo and in vitro of CHS 828 on human myeloma cell lines. Anticancer Drugs. 2004;15(1):63–70. doi: 10.1097/00001813-200401000-00010. [DOI] [PubMed] [Google Scholar]
- Hyoju SK, Baral B, Jha PK. Central catecholaminergic blockade with clonidine prevent SARS-CoV-2 complication: a case series. Idcases. 2021;1(25):e01219. doi: 10.1016/j.idcr.2021.e01219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin Z, Zhao Y, Sun Y, Zhang B, Wang H, Wu Y, Zhu Y, Zhu C, Hu T, Du X, Duan Y. Structural basis for the inhibition of SARS-CoV-2 main protease by antineoplastic drug carmofur. Nat Struct Mol Biol. 2020;27(6):529–532. doi: 10.1038/s41594-020-0440-6. [DOI] [PubMed] [Google Scholar]
- Jin Z, Du X, Xu Y, Deng Y, Liu M, Zhao Y, Zhang B. Structure of M. sup. pro from SARS-CoV-2 and discovery of its inhibitors. Nature. 2020;582(7811):289–294. doi: 10.1038/s41586-020-2223-y. [DOI] [PubMed] [Google Scholar]
- Keck JG, Pou-Hsiung W, Lien EJ, Lai MM. Inhibition of murine coronavirus RNA synthesis by hydroxyguanidine derivatives. Virus Res. 1989;14(1):57–63. doi: 10.1016/0168-1702(89)90069-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S, Thiessen PA, Bolton EE, Chen J, Fu G, Gindulyte A, Han L, He J, He S, Shoemaker BA, Wang J. PubChem substance and compound databases. Nucleic Acids Res. 2016;44:1202–1213. doi: 10.1093/nar/gkv951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krumm ZA, Lloyd GM, Francis CP, Nasif LH, Mitchell DA, Golde TE, Giasson BI, Xia Y. Precision therapeutic targets for COVID-19. Virol J. 2021;18(1):1–22. doi: 10.1186/s12985-021-01526-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lan J, Ge J, Yu J, Shan S, Zhou H, Fan S, Zhang Q, Shi X, Wang Q, Zhang L, Wang X. Structure of the SARS-CoV-2 spike receptor-binding domain bound to the ACE2 receptor. Nature. 2020;581(7807):215–220. doi: 10.1038/s41586-020-2180-5. [DOI] [PubMed] [Google Scholar]
- Langmaier J, Pizl M, Samec Z, Zalis S. Extreme basicity of biguanide drugs in aqueous solutions: ion transfer voltammetry and dft calculations. J Phys Chem A. 2016;120(37):7344–7350. doi: 10.1021/acs.jpca.6b04786. [DOI] [PubMed] [Google Scholar]
- Laurence C, Gal JF. Lewis basicity and affinity scales: data and measurement. New York: John Wiley & Sons; 2009. [Google Scholar]
- Lehrer S. Inhaled biguanides and mTOR inhibition for influenza and coronavirus (review) World Acad Sci J. 2020;2(3):1. doi: 10.3892/wasj.2020.42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang PH. Characterization and inhibition of SARS-coronavirus main protease. Curr Top Med Chem. 2006;6(4):361–376. doi: 10.2174/156802606776287090. [DOI] [PubMed] [Google Scholar]
- Liu J, Cao R, Xu M. Hydroxychloroquine, a less toxic derivative of chloroquine, is effective in inhibiting SARS-CoV-2 infection in vitro. Cell Discov. 2020 doi: 10.1038/s41421-020-0156-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu C, Zhou Q, Li Y, Garner LV, Watkins SP, Carter LJ, Smoot J, Gregg AC, Daniels AD, Jervey S, Albaiu D. Research and development on therapeutic agents and vaccines for COVID-19 and related human coronavirus diseases. 10.1021/acscentsci.0c00272 [DOI] [PMC free article] [PubMed]
- Loddo B, Ferrari W, Brotzu G, Spanedda A. In vitro inhibition of infectivity of polio viruses by guanidine. Nature. 1962;193(4810):97–98. doi: 10.1038/193097a0. [DOI] [PubMed] [Google Scholar]
- Loffredo M, Lucero H, Chen DY, O’Connell A, Bergqvist S, Munawar A, Bandara A, De Graef S, Weeks SD, Douam F, Saeed M. The in-vitro effect of famotidine on SARS-CoV-2 proteases and virus replication. Sci Rep. 2021;11(1):1–9. doi: 10.1038/s41598-021-84782-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macip G, Garcia-Segura P, Mestres-Truyol J, Saldivar-Espinoza B, Ojeda-Montes MJ, Gimeno A, Cereto-Massagué A, Garcia-Vallvé S, Pujadas G. Haste makes waste: a critical review of docking-based virtual screening in drug repurposing for SARS-CoV-2 main protease (M-pro) inhibition. Med Res Rev. 2022;42(2):744–769. doi: 10.1002/med.21862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madias JE. Reserpine, mecamilamine, guanethidine, atropine for patients with Takotsubo syndrome? Int J Cardiol. 2014;177(3):1078–1079. doi: 10.1016/j.ijcard.2014.10.016. [DOI] [PubMed] [Google Scholar]
- Manley PW, Stiefl N, Cowan-Jacob SW, Kaufman S, Mestan J, Wartmann M, Wiesmann M, Woodman R, Gallagher N. Structural resemblances and comparisons of the relative pharmacological properties of imatinib and nilotinib. Bioorg Med Chem. 2010;18(19):6977–6986. doi: 10.1016/j.bmc.2010.08.026. [DOI] [PubMed] [Google Scholar]
- Maurya VK, Kumar S, Prasad AK, Bhatt ML, Saxena SK. Structure-based drug designing for potential antiviral activity of selected natural products from Ayurveda against SARS-CoV-2 spike glycoprotein and its cellular receptor. Virusdisease. 2020;31(2):179–193. doi: 10.1007/s13337-020-00598-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris GM, Huey R, Lindstrom W, Sanner MF, Belew RK, Goodsell DS, Olson AJ. AutoDock4 and AutoDockTools4: automated docking with selective receptor flexibility. J Comput Chem. 2009;30(16):2785–2791. doi: 10.1002/jcc.21256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukherjee R, Bhattacharya A, Bojkova D, Mehdipour AR, Shin D, Khan KS, Cheung HH, Wong KB, Ng WL, Cinatl J, Geurink PP. Famotidine inhibits toll-like receptor 3-mediated inflammatory signaling in SARS-CoV-2 infection. J Biol Chem. 2021 doi: 10.1016/j.jbc.2021.100925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mulvaney EP, Reid HM, Bialesova L, Bouchard A, Salvail D, Kinsella BT. NTP42, a novel antagonist of the thromboxane receptor, attenuates experimentally induced pulmonary arterial hypertension. BMC Pulm Med. 2020;20(1):1–6. doi: 10.1186/s12890-020-1113-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mutabingwa T, Nzila A, Mberu E, Nduati E, Winstanley P, Hills E, Watkins W. Chlorproguanil-dapsone for treatment of drug-resistant falciparum malaria in Tanzania. Lancet. 2001;358(9289):1218–1223. doi: 10.1016/S0140-6736(01)06344-9. [DOI] [PubMed] [Google Scholar]
- Palmer JD, Nugent CA. Guanadrel sulfate: a postganglionic sympathetic inhibitor for the treatment of mild to moderate hypertension. Pharmacother J Human Pharmacol Drug Ther. 1983;3(4):220–227. doi: 10.1002/j.1875-9114.1983.tb03257.x. [DOI] [PubMed] [Google Scholar]
- Pantazi I, Al-Qahtani AA, Alhamlan FS, Alothaid H, Matou-Nasri S, Sourvinos G, Vergadi E, Tsatsanis C. SARS-CoV-2/ACE2 interaction suppresses IRAK-M expression and promotes pro-inflammatory cytokine production in macrophages. Front Immunol. 2021 doi: 10.3389/fimmu.2021.683800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paul M, Durand R, Boulard Y, Fusai T, Fernandez C, Rivollet D, Deniau M, Astier A. Physicochemical characteristics of pentamidine-loaded polymethacrylate nanoparticles: implication in the intracellular drug release in Leishmania major infected mice. J Drug Target. 1998;5(6):481–490. doi: 10.3109/10611869808997874. [DOI] [PubMed] [Google Scholar]
- Pinzi L, Rastelli G. Molecular docking: shifting paradigms in drug discovery. Int J Mol Sci. 2019;20(18):4331. doi: 10.3390/ijms20184331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plöger GF, Abrahamsson B, Cristofoletti R, Groot DW, Langguth P, Mehta MU, Parr A, Polli JE, Shah VP, Tajiri T, Dressman JB. Biowaiver monographs for immediate release solid oral dosage forms: proguanil hydrochloride. J Pharm Sci. 2018;107(7):1761–1772. doi: 10.1016/j.xphs.2018.03.009. [DOI] [PubMed] [Google Scholar]
- Pourhajibagher M, Bahador A. Molecular docking study of potential antimicrobial photodynamic therapy as a potent inhibitor of SARS-CoV-2 main protease: an in silico insight. Infect Disord Drug Targets. 2022 doi: 10.2174/1871526522666220901164329. [DOI] [PubMed] [Google Scholar]
- Rajković J, Perić M, Novaković R, Djokić V, Gostimirović MZ, Heinle H, Gojković-Bukarica L. Differences in potassium channel-independent effects of pinacidil on the isolated human saphenous veins obtained from diabetic and non-diabetic patients. Atherosclerosis. 2020;1(315):e105. doi: 10.1016/j.atherosclerosis.2020.10.323. [DOI] [Google Scholar]
- Rossi R, Talarico M, Coppi F, Boriani G. Protective role of statins in COVID 19 patients: importance of pharmacokinetic characteristics rather than intensity of action. Intern Emerg Med. 2020;15(8):1573–1576. doi: 10.1007/s11739-020-02504-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruedy J, Davies RO. A comparative clinical trial of guanoxan and guanethidine in essential hypertension. Clin Pharmacol Ther. 1967;8(1part1):38–47. doi: 10.1002/cpt196781part138. [DOI] [PubMed] [Google Scholar]
- Saczewski F, Balewski Ł. Biological activities of guanidine compounds. Expert Opin Ther Pat. 2009;19(10):1417–1448. doi: 10.1517/13543770903216675. [DOI] [PubMed] [Google Scholar]
- Singh TU, Parida S, Lingaraju MC, Kesavan M, Kumar D, Singh RK. Drug repurposing approach to fight COVID-19. Pharmacol Rep. 2020;72(6):1479–1508. doi: 10.1007/s43440-020-00155-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sivaprakasam P, Tosso PN, Doerksen RJ. Structure−activity relationship and comparative docking studies for cycloguanil analogs as PfDHFR-TS inhibitors. J Chem Inf Model. 2009;49(7):1787–1796. doi: 10.1021/ci9000663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas T, Faaland CA, Adhikarakunnathu S, Thomas TJ. Structure-activity relations of S-adenosylmethionine decarboxylase inhibitors on the growth of MCF-7 breast cancer cells. Breast Cancer Res Treat. 1996;39(3):293–306. doi: 10.1007/BF01806157. [DOI] [PubMed] [Google Scholar]
- Tipnis UR. Polymine-traveled pathways: Significance in health and disease. In: Advances in structural biology 2000 Jan 1 (vol. 6, pp. 117–146). JAI. 10.1016/S1064-6000(00)80007-1
- Tohme RA, Holtzman D, Holmberg SD (2012) Hepatitis C Virus. In: Principles and practice of pediatric infectious diseases. Elsevier, pp. 1105–1112. 10.1016/B978-1-4377-2702-9.00222-1
- Tomar PP, Krugliak M, Arkin IT. Blockers of the SARS-CoV-2 3a channel identified by targeted drug repurposing. Viruses. 2021;13(3):532. doi: 10.3390/v13030532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trott O, Olson AJ. AutoDockVina: improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J Comput Chem. 2010;31(2):455–461. doi: 10.1002/jcc.21334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waller DG, Sampson AP. Chemotherapy of infections. Med Pharmacol Ther. 2018;5:581–629. doi: 10.1016/B978-0-7020-7167-6.00051-8. [DOI] [Google Scholar]
- Wang X, Cao R, Zhang H, Liu J, Xu M, Hu H, Li Y, Zhao L, Li W, Sun X, Yang X. The anti-influenza virus drug, arbidol is an efficient inhibitor of SARS-CoV-2 in vitro. Cell Discov. 2020;6:28. doi: 10.1038/s41421-020-0169-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weston S, Coleman CM, Haupt R, Logue J, Matthews K, Li Y, Reyes HM, Weiss SR, Frieman MB. Broad anti-coronavirus activity of food and drug administration-approved drugs against SARS-CoV-2. J Virol. 2020;94:e01218–e1220. doi: 10.1128/JVI.01218-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xian M, Li X, Tang X, Chen X, Zheng Z, Galligan JJ, Kreulen DL, Wang PG. N-Hydroxyl derivatives of guanidine based drugs as enzymatic NO donors. Bioorg Med Chem Lett. 2001;11(17):2377–2380. doi: 10.1016/S0960-894X(01)00456-5. [DOI] [PubMed] [Google Scholar]
- Yang ZW, Zhao YZ, Zang YJ, Wang H, Zhu X, Meng LJ, Yuan XH, Zhang L, Zhang SL. Rapid structure-based screening informs potential agents for coronavirus disease (COVID-19) outbreak. Chin Phys Lett. 2020;37(5):058701. doi: 10.1088/0256-307X/37/5/058701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaidel EJ, Forsyth CJ, Novick G, Marcus R, Ribeiro AL, Pinazo MJ, Morillo CA, Echeverria LE, Shikanai-Yasuda MA, Buekens P, Perel P. COVID-19: implications for people with Chagas disease. Glob Heart. 2020 doi: 10.5334/gh.891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zanin M, Baviskar P, Webster R, Webby R. The interaction between respiratory pathogens and mucus. Cell Host Microbe. 2016;19(2):159–168. doi: 10.1016/j.mehy.2020.109844. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.