Abstract
Tobacco smoking is positively correlated with nonalcoholic fatty liver disease (NAFLD)1–5, but the underlying mechanism for this association is unclear. Here, we report that nicotine accumulates in the intestine during tobacco smoking and activates intestinal AMPKα. Notably, we identify the gut bacterium Bacteroides xylanisolvens as an effective nicotine degrader. B. xylanisolvens colonization reduces intestinal nicotine concentrations in nicotine-exposed mice, and it improves nicotine-exacerbated NAFLD progression. Mechanistically, AMPKα promotes the phosphorylation of sphingomyelin phosphodiesterase 3 (SMPD3), stabilizing the latter and thus increasing intestinal ceramide formation, which contributes to NAFLD progression. Our results establish a role for intestinal nicotine accumulation in NAFLD progression and reveal an endogenous bacterium in the human intestine capable of metabolizing nicotine. These findings suggest a possible avenue to reduce tobacco smoking-exacerbated NAFLD progression.
As one of the most common unhealthy behaviors among humans, the World Health Organization lists tobacco smoking as the leading cause of preventable death in the world6,7. Smoking cessation is the most effective way to extend life expectancy, but it is not easily achieved due to the powerful addictive property and withdrawal symptoms of nicotine8. Nicotine degradation has been recently considered as a novel strategy for blocking nicotine-induced pathology9,10. NAFLD encompasses a broad spectrum of pathologies, that usually progress from simple steatosis (defined as NAFL) to non-alcoholic steatohepatitis (NASH), and then in some cases to cirrhosis and hepatocellular carcinoma. As metabolites produced from the intestine play a critical role in the progression of NAFLD via the gut-liver axis11, establishing a relationship between intestinal nicotine accumulation, intestine-derived metabolites, and NAFLD progression could shed fresh light on tobacco smoking-related liver diseases. Here, we report that nicotine accumulates in the intestine during tobacco smoking and accelerates the progression of NAFLD, but that it can be degraded effectively by the human symbiont B. xylanisolven. These findings address the pathological effects of intestinal nicotine accumulation, and identify an endogenous nicotine-degrading gut bacterium, which is of valuable potential for the treatment of NAFLD in smokers.
Gut bacteria degrade ileal nicotine
High levels of nicotine can be detected in human saliva and gastric juice during tobacco smoking or nicotine patch application, indicating its accumulation in the digestive tract12, where it may act pathologically. To confirm whether nicotine accumulates in the intestine during tobacco smoking, we collected terminal ileum mucosa biopsies, serum, and stool samples from 30 nonsmokers and 30 smokers. High levels of nicotine were detected in the ileal mucosa tissue of smokers, suggesting nicotine accumulates in the intestine during tobacco smoking (Fig. 1a). We further assessed nicotine levels in lung, ileal contents, ileal tissue, brain, liver, epididymal white adipose tissue (eWAT) and serum in specific pathogen free (SPF) mice given nicotine via three previously described administration methods (i.e., smoke exposure, oral and subcutaneous)13–16. Ileal nicotine accumulation was consistently observed in the above mouse models (Fig. 1b, c, Extended Data Fig. 1a). These results indicate that nicotine levels during smoking accumulate in the intestine to a relatively large degree, which may be of pathophysiological significance.
Fig. 1. Identification of intestinal nicotine accumulation and gut bacterial-derived intestinal nicotine degradation.
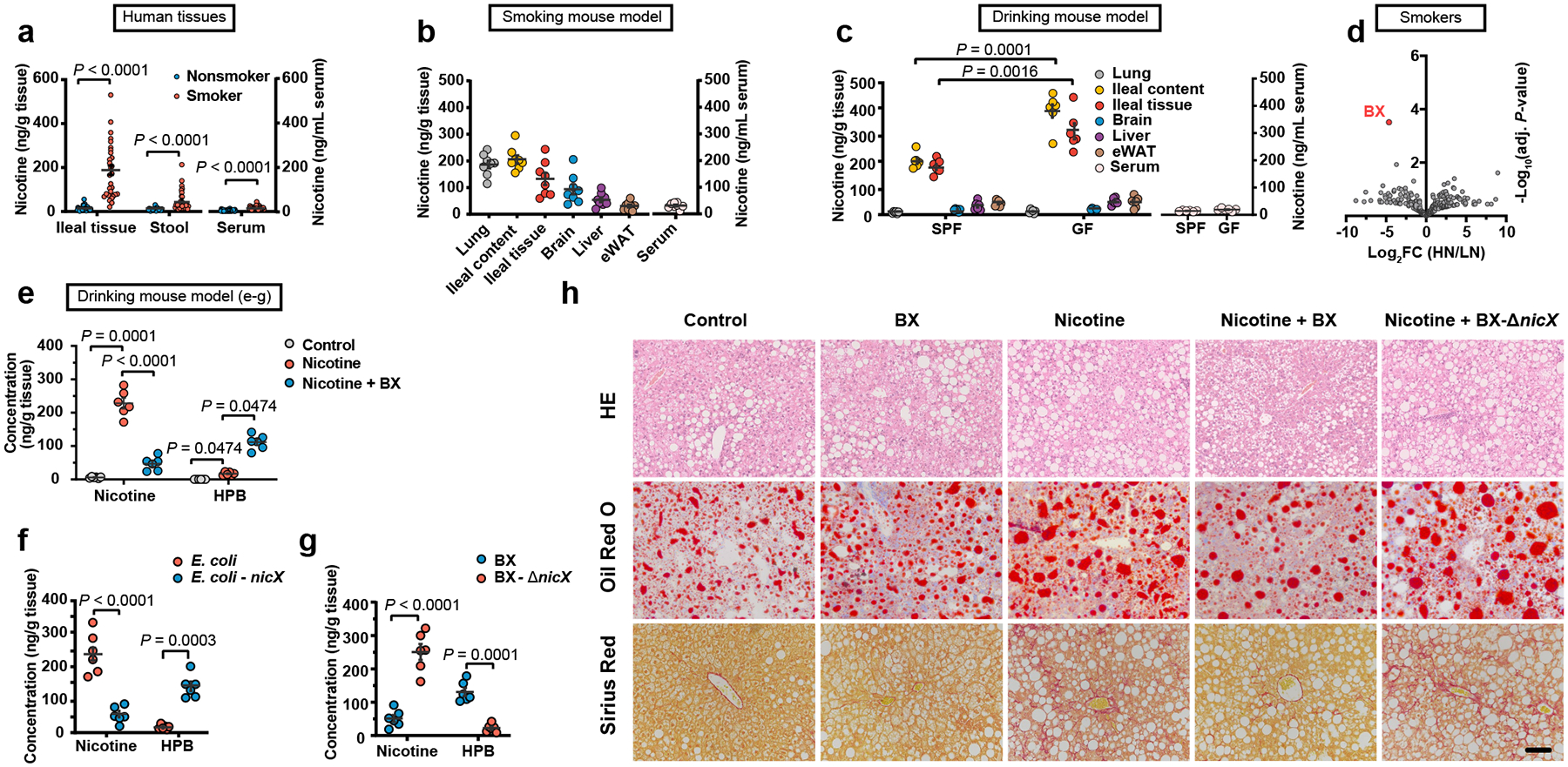
a, Quantification of the nicotine concentrations in terminal ileum mucosa biopsies, serum and stool samples collected from smokers (n = 30) and nonsmokers (n = 30). For the assessment of ileal nicotine accumulation, the following mouse models were treated for two weeks (b, c, e-g). b, Nicotine concentrations in lung, ileum content, ileum tissue, brain, liver, eWAT and serum samples in smoking SPF mouse model (n = 8 mice/group). c, Tissue nicotine concentrations of SPF and GF mice in nicotine drinking model (n = 6 mice/group). d, Volcano plot of human stool metagenomic sequencing data in HN and LN groups. The adjusted P value was calculated using the moderated Student’s t-test followed by the Benjamini-Hochberg procedure via the false discovery rate (FDR). FC: fold change. e, Nicotine and HPB concentration in ileal tissue of control, nicotine and nicotine + B. xylanisolvens treated SPF mice, separately (n = 6 mice/group). f, g, Nicotine and HPB concentration in ileal tissue of E. coli or nicX knock-in E. coli treated SPF mice (f) and B. xylanisolvens or nicX knock-out B. xylanisolvens treated SPF mice (g). (n = 6 mice/group). HFHCD-fed SPF mice were treated with PBS, B. xylanisolvens colonization, nicotine water, nicotine water + B. xylanisolvens colonization, and nicotine water plus nicX knock-out B. xylanisolvens colonization for 20 weeks (n = 6 mice/group). h, Representative H&E staining (top), Oil Red O staining (middle) and Sirius Red staining (bottom) of liver sections (n = 3 mice/group, 3 images/mouse). Scale bar, 100 μm. Data are the means ± s.e.m. a, Two-tailed Mann-Whitney U test. c, f, g, Two-tailed Student’s t-test. e, for Nicotine, one-way ANOVA with Dunnett’s T3 post hoc test; for HPB, Kruskal-Wallis test with Dunn’s test.
In the nicotine drinking water-delivery model, the nicotine concentrations in both ileal content and ileal tissue in germ-free (GF) mice were higher than those in SPF mice, suggesting that endogenous gut bacteria contribute to nicotine degradation (Fig. 1c). To identify the specific endogenous nicotine-degrading gut microbiota in humans, we screened for potential strains carrying known nicotine catabolic genes from public metagenome databases. A homologous sequence of nicA, one of the known nicotine degrading enzymes17,18, was found in several strains, suggesting such strains have the potential to degrade nicotine (Extended Data Fig. 1b). Stool samples from 30 smokers were subjected to a metagenomics study to identify the most correlative species with reduced ileal nicotine concentrations. The participants were defined as either low-nicotine (LN) or high-nicotine (HN), according to the ileal nicotine levels (Extended Data Fig. 1c). Notably, there was no significant difference between the LN and HN groups regarding smoking history, including years of smoking and number of cigarettes per day (Supplementary Table. 1, 2). No differences in the α- and β-diversities were found between two groups, however, several strains showed different abundances between the LN and HN individuals (Fig. 1d, Extended Data Fig. 1d–g). Based on metagenome databases and metagenomics, we found that B. xylanisolvens potentially harboured a nicotine catabolic gene and was identified as the main driver associated with the separation between the LN and HN groups (Fig. 1d, Extended Data Fig. 1h, i).
The supplementation of nicotine in the culture medium had no influence on the growth rate of B. xylanisolvens (Extended Data Fig. 2a); however, B. xylanisolvens efficiently degraded the nicotine added in the medium (Extended Data Fig. 2b). To explore the degraded metabolites of nicotine in B. xylanisolvens, we performed fermentation of B. xylanisolvens and isolated the nicotine metabolite through chromatography. The metabolite was further characterized by nuclear magnetic resonance (NMR), and identified as 4-hydroxy-1-(3-pyridyl)-1-butanone (HPB; Extended Data Fig. 2c), which could be detected in the culture medium of B. xylanisolvens (Extended Data Fig. 2d). To verify the nicotine-degrading properties of B. xylanisolvens in vivo, we colonized this bacterium to SPF mice administrated nicotine by the above three delivery strategies and found that B. xylanisolvens could degrade nicotine in vivo, accompanied by an increase of HPB (Fig. 1e, Extended Data Fig. 2e, f).
Next, we performed whole-genome sequencing of B. xylanisolvens and explored the biosynthetic gene responsible for the catabolism of nicotine in B. xylanisolvens. Among the potential nicotine catabolic proteins in B. xylanisolvens encoded by these sequenced genes, NicX, which is encoded by nicX has a similar predicted core structure with the computational model of a known nicotine-degrading enzyme NicA (Extended Data Fig. 2g). NicX degraded nicotine with a Vmax value of 3.83 ± 0.13 [μmol/ (min*μg protein)] in vitro (Extended Data Fig. 2h). To further verify the nicotine-degrading capacity of NicX, nicX was introduced into E. coli or knocked out in B. xylanisolvens (Extended Data Fig. 2i). Compared with wild-type (WT) E. coli, nicX knock-in E. coli gained the capacity to degrade nicotine and produce HPB in vitro and in vivo (Extended Data Fig. 2j, Fig. 1f). Moreover, nicX deficiency in B. xylanisolvens resulted in the loss of its nicotine degradation capability Extended Data Fig. 2k, Fig. 1g). No significant change was found in the growth curves of the WT and nicX knock-out B. xylanisolvens (Extended Data Fig. 2l). In summary, the results show that nicotine accumulates in the intestine during various routes of nicotine administration, and B. xylanisolvens is capable of degrading nicotine in the presence of NicX.
Nicotine degradation delays NAFLD progression
Oral nicotine administration via drinking water is a widely used model to study nicotine-associated chronic metabolic diseases19,20. To determine the role of intestinal nicotine accumulation and the effect of its degradation by B. xylanisolvens in NAFLD progression, PBS (Control)-, B. xylanisolvens (BX)-, nicotine water (Nicotine)-, B. xylanisolvens plus nicotine water (Nicotine + BX)-, or nicX knock-out B. xylanisolvens plus nicotine water (Nicotine + BX-ΔnicX)-treated SPF mice were administrated a high-fructose and high-cholesterol diet (HFHCD) for 20 weeks. WT and nicX knock-out B. xylanisolvens were both successfully colonized in HFHCD-fed SPF mice (Extended Data Fig. 3a). However, the nicotine degradation is only observed in the WT B. xylanisolvens-colonized group (Extended Data Fig. 3b). No obvious change of body weight was observed among groups (Extended Data Fig. 3c, d). The liver weights, serum levels of ALT and AST, and hepatic triglyceride (TG) levels of nicotine-treated mice were higher than those of control-treated SPF mice (Extended Data Fig. 3e–i). No significant differences in the serum TG, the hepatic and serum cholesterol ester (CE) and serum non-esterified fatty acid (NEFA) levels were found between the two groups (Extended Data Fig. 3j–m). By H&E, Oil Red O, and Sirius Red staining, nicotine supplementation in the water accelerated NAFLD progression under HFHCD feeding conditions, and this acceleration was accompanied by more severe hepatic steatosis, inflammation, and fibrosis (Fig. 1h, Extended Data Fig. 3n–r). Furthermore, increases in the relative expression of mRNAs involved in hepatic lipid metabolism, proinflammatory cytokine production, and collagen synthesis were induced by nicotine-supplemented water treatment (Extended Data Fig. 3s–u), whereas these NAFLD indicators were ameliorated by WT B. xylanisolvens colonization, but not nicX-deficient B. xylanisolvens colonization (Fig. 1h, Extended Data Fig. 3c–u). Together, these data suggest that the colonization of nicotine-degrading bacteria could reduce nicotine-induced NAFLD progression and this effect is dependent on the expression of NicX.
Nicotine activates intestinal AMPKα1
Consistent with previous studies21, we found nicotine treatment dose-dependently induced the phosphorylation of AMPKα at T172 in ileal organoids (Extended Data Fig. 4a, b). Furthermore, ileal AMPKα was activated by tobacco smoking in the terminal ileum, and the strength of the effect was positively related to the level of nicotine in the tissue of smokers (Fig. 2a). In addition, p-AMPKα in the ileum was upregulated in all three models of nicotine-exposed SPF mice (Extended Data Fig. 4c–e).
Fig. 2. Nicotine-induced activation of intestinal AMPKα-SMPD3 axis in NAFLD progression.
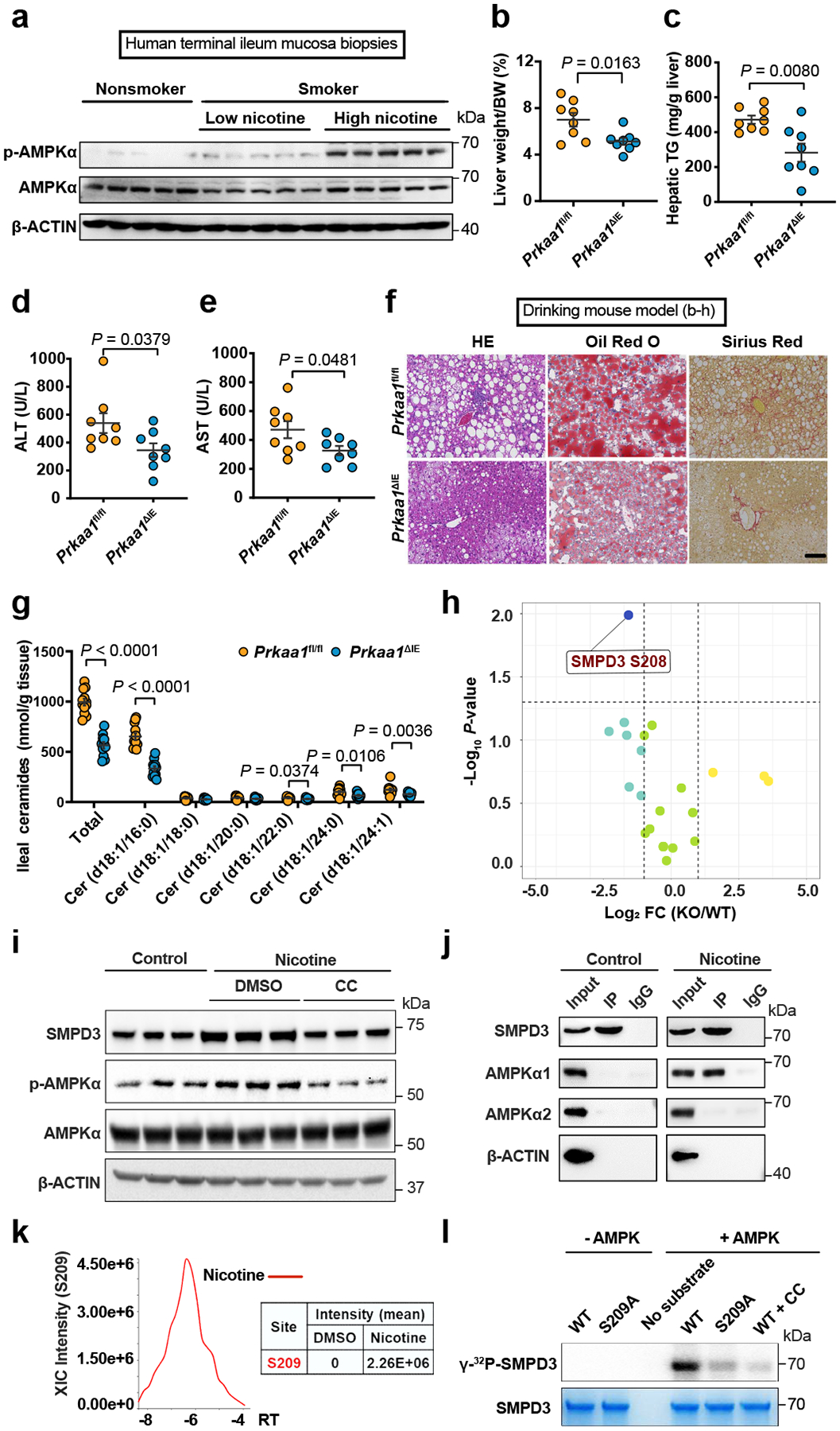
a, Representative western blot of ileal AMPK signaling in nonsmokers (n = 30) and smokers with low nicotine (n = 14) or high nicotine (n = 16) contents in terminal ileum mucosa biopsies. Prkaa1fl/fl and Prkaa1ΔIE mice (SPF) were administrated an HFHCD plus nicotine water for 20 weeks (n = 8 mice/group). b, Liver weight–to–body weight ratios. c, Liver TG content. d, e, Serum ALT (d) and AST (e) levels. f, Representative H&E staining (left), Oil Red O staining (middle) and Sirius Red staining (right) of liver sections (n = 4 mice/group, 3 images/mouse). Scale bar, 100 μm. g, Quantification of ileal ceramide profiles between Prkaa1fl/fl (WT, n = 12) and Prkaa1ΔIE (KO, n = 11) mice administrated an HFHCD plus nicotine water for 20 weeks. h, Volcano map shows all phosphorylated sites of ceramide metabolism-related proteins identified from phosphorylated proteomics analysis of the ileal epithelia of Prkaa1fl/fl (WT) and Prkaa1ΔIE (KO) mice administrated an HFHCD plus nicotine water for 20 weeks (n = 5 mice/group). P values calculated by two-sided Student’s t-test. i, The assessment of AMPK activation and SMPD3 protein levels in ileal organoids treated with control, nicotine or nicotine plus compound C (CC, AMPK inhibitor) for 12 h. j, Co-IP of AMPKα1 with SMPD3. Constructs encoding Flag-tagged SMPD3 were transfected into SW480 cells, and then treated with nicotine for 4 h. k, Mass spectrometry analysis of the phosphorylation intensity at S209 on SMPD3 with or without nicotine treatment for 24 h. l, In vitro phosphorylation of WT or S209A mutant SMPD3 (86–655) by AMPK(α1/β1/γ2) with or without CC treatment. Data are the means ± s.e.m. b, c, e, Two-tailed Student’s t-test. d, g, Two-tailed Mann-Whitney U-test. Experiments in i-l were performed three times independently.
There are two isoforms of AMPKα, including AMPKα1 and AMPKα222. We generated intestinal epithelium specific AMPKα1 (Prkaa1ΔIE) and AMPKα2 (Prkaa2ΔIE) knockout mouse lines (Extended Data Fig. 4f). Nicotine completely lost its capacity to activate AMPKα in the absence of intestinal AMPKα1, whereas the nicotine-induced increase in p-AMPKα still occurred in the absence of intestinal AMPKα2, suggesting that AMPKα1 is the main target of nicotine in the intestine (Extended Data Fig. 4f). B. xylanisolvens colonization inhibited nicotine-induced AMPKα phosphorylation in the ileum (Extended Data Fig. 4g). Similar to B. xylanisolvens, nicX knock-in E. coli had the capability to suppress the nicotine-induced phosphorylation of ileal AMPKα in vivo (Extended Data Fig. 4h). However, the B. xylanisolvens-mediated inhibitory effects were not observed in the absence of NicX (Extended Data Fig. 4i). Hence, B. xylanisolvens mitigates nicotine-induced activation of ileal p-AMPKα in the presence of NicX, which is supported by the observation that the nicotine catabolite, HPB, could not activates ileal p-AMPKα (Extended Data Fig. 4j).
Regarding the role of intestinal AMPKα1 in nicotine-accelerated NAFLD progression, we found that the loss of intestinal epithelium AMPKα1 contribute to improvements in hepatic steatosis, inflammation, and fibrosis in the nicotine-accelerated NAFLD mouse model (Fig. 2b–f, Extended Data Fig. 5a–m).
p-AMPKα phosphorylates SMPD3 (S208/209)
Lipids are considered to play an important role in NAFLD progression, as they are a key mediator of the crosstalk between the intestine and liver in this pathology23. We conducted a targeted lipidomic analysis of the ileum and identified ceramides as the primary metabolites leading to clustering-based differentiation between AMPKα1-knockout (KO) and control (WT) intestinal epithelia with nicotine treatment (Extended Data Fig. 5n, o). Ileal ceramide profiles were lower in the absence of intestinal AMPKα1 in nicotine drinking model (Fig. 2g). Based on the phosphoproteome analysis of ileal epithelium from nicotine drinking model (Extended Data Fig. 5p), S208 of SMPD3, the enzyme that mediates sphingomyelin (SM) metabolism to ceramide, was significantly decreased by intestinal epithelium AMPKα1 loss (Extended Data Fig. 5q, Fig. 2h).
In ileal organoids, nicotine treatment did not alter the Smpd3 mRNA levels (Extended Data Fig. 6a). However, either treatment with an AMPK kinase activity inhibitor or expression of a kinase domain mutant could mitigate nicotine-induced upregulation of SMPD3 protein levels (Fig. 2i, Extended Data Fig. 6b), suggesting the effect of nicotine on SMPD3 is post-translational. Indeed, we found that, under nicotine treatment, SMPD3 bound to AMPKα1 (Fig. 2j). A GPS 2.024 analysis predicted S208 (mouse) as the most likely phosphorylation site by AMPK (Extended Data Fig. 6c). Peptides including S208 (S209 in humans) in SMPD3 satisfied the phosphorylation substrate motif of AMPK and were highly evolutionary conserved (Extended Data Fig. 6d). The stability of SMPD3 is reportedly regulated by the phosphorylation of five conserved serine sites25. Consistently, increased S209 phosphorylation was detected in the nicotine-treated group (Fig. 2k, Extended Data Fig. 6e). SMPD3 (WT) could be directly phosphorylated by AMPKα1, and this phosphorylation was almost abolished when S209 was mutated, or an AMPK inhibitor was administrated (Fig. 2l). To confirm if phosphorylation at S209 influences SMPD3 protein stability, we treated SW480 cells transfected with WT, S209A (lost phosphorylation mutant), and S209D (sustained phosphorylation mutant) SMPD3 with cycloheximide (CHX, a protein translational inhibitor). We found that S209 phosphorylation protected SMPD3 protein from degradation (Fig. 3a). In isolated organoids from nicotine drinking model, faster degradation of SMPD3 was observed that lack intestinal epithelium AMPKα1 (Extended Data Fig. 6f).
Fig. 3. Phosphorylated SMPD3 is more stable due to reduced ubiquitination-mediated degradation.
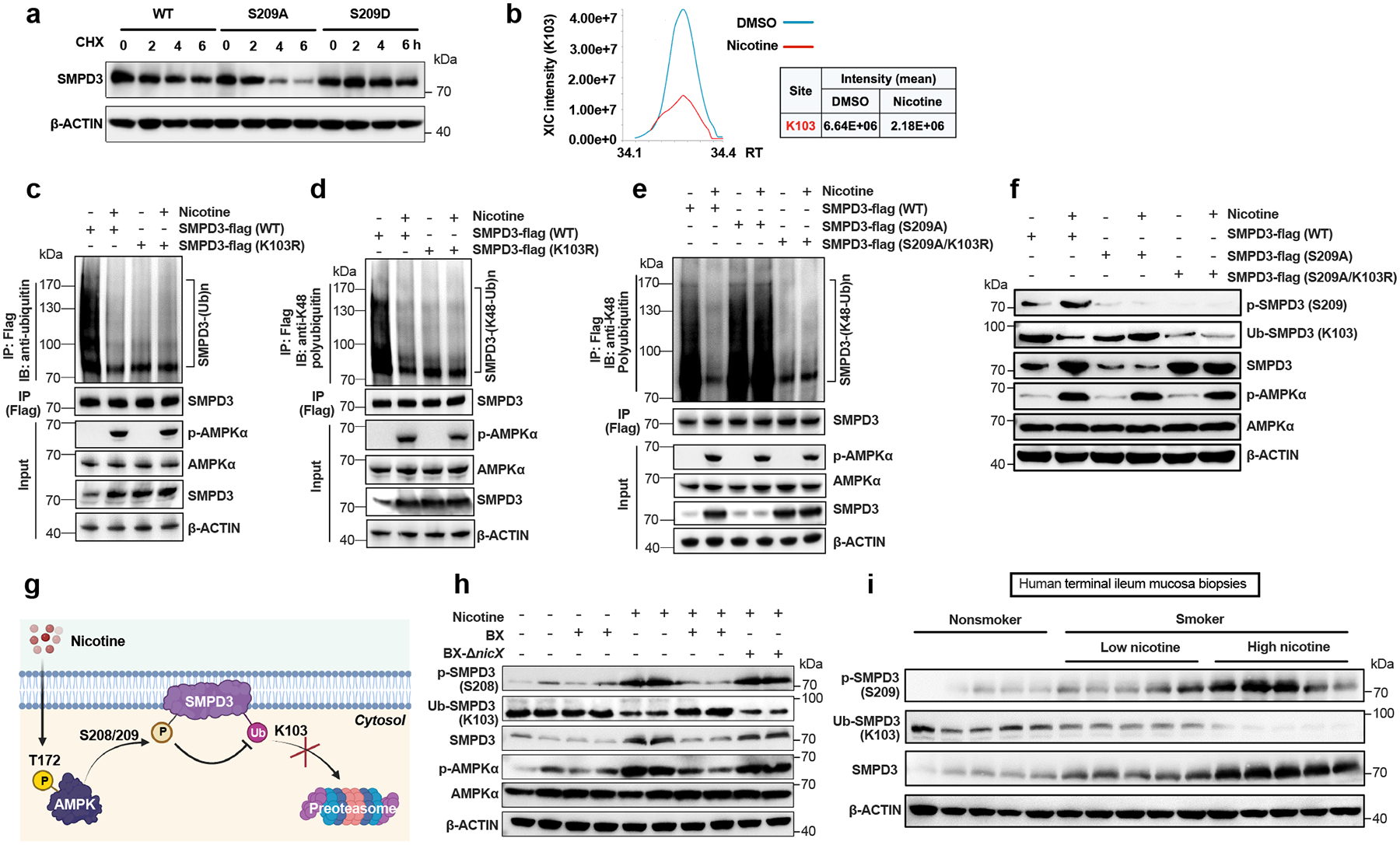
a, Western blot analysis showing the stability of SMPD3 after the administration of cycloheximide (CHX, translation inhibitor) in SW480 cells transfected with WT or mutant SMPD3 (S209A, S209D). b, Mass spectrometry analysis of the ubiquitination intensity at K103 on SMPD3 with or without nicotine treatment. c, Total ubiquitination of SMPD3 in SW480 cells transfected with WT or K103R SMPD3-flag with or without nicotine treatment. d, K48 ubiquitination of SMPD3 in SW480 cells transfected with WT or K103R SMPD3-flag with or without nicotine treatment. e. K48 ubiquitination of SMPD3 in SW480 cells transfected with WT, 209A or 209A plus K103R SMPD3-flag without or with nicotine treatment. f, Western blot analysis showed phosphorylation levels, ubiquitination levels, and protein levels of SMPD3 in SW480 cells transfected with three SMPD3-flag plasmids (WT, S209A, S209A/K103R) and treated with or without nicotine. g, Diagram of the SMPD3 protein phosphorylation and ubiquitination process under normal and nicotine stimulated conditions. Created with BioRender. h, Representative ileal SMPD3 protein phosphorylation and ubiquitination levels in HFHCD-fed mice subjected to PBS, B. xylanisolvens colonization, nicotine water, nicotine water plus B. xylanisolvens colonization, and nicotine water plus nicX knock-out B. xylanisolvens colonization for 20 weeks. (n = 4 mice/group). i, Representative western blot of ileal SMPD3 phosphorylation, ubiquitination and protein levels in nonsmokers and smokers with low nicotine or high nicotine concentration in terminal ileum mucosa biopsies. Experiments were performed with 30 nonsmokers and 30 smokers (14 with low nicotine levels and 16 with high nicotine levels). For b-f, nicotine (1 μg/mL) treatment for 24 h. Experiments in a-f were performed three times independently.
Furthermore, SMPD3 was strikingly more deubiquitinated at K103 in nicotine-treated SW480 cells than in DMSO-treated ones (Fig. 3b, Extended Data Fig. 6g). By immunoprecipitation, we found the ubiquitinated SMPD3 (K103) contained primarily the classical K48-linked Ub chain (K48-Ub) instead of K63-Ub, which was decreased by nicotine treatment (Fig. 3c, d, Extended Data Fig. 6h). S209A mutation reversed the nicotine-induced decrease in K48-Ub of SMPD3, which was further mitigated with K103R mutation (Fig. 3e, Extended Data Fig. 6i). Moreover, we developed anti-p-SPMD3 (S209) and anti-Ub-SMPD3 (K103) antibodies (Extended Data Fig. 6j, k), and confirmed that nicotine-induced increases in total SMPD3 protein resulted from elevated SMPD3 phosphorylation and subsequently reduced ubiquitination (Fig. 3f). These findings suggest that nicotine-activated AMPKα phosphorylates SMPD3 at S209, which in turn inhibits K103 ubiquitination on SMPD3 and the subsequent K48-Ub degradation pathway, thereby increasing SMPD3 protein stability and its protein level (Fig. 3g). We further detected nicotine-induced SMPD3 phosphorylation at S208, accompanied by a decrease of SMPD3 ubiquitination at K103 in the ileal tissues of SPF mice, which was mitigated by B. xylanisolvens colonization in the presence of NicX (Fig. 3h). Phosphorylated and total SMPD3 protein levels were also higher in the ileum of smokers than nonsmokers, which was related to ileal nicotine content; however, ubiquitinated SMPD3 protein levels were decreased sequentially in the non-smokers, LN group and HN group (Fig. 3i).
p-AMPKα-SMPD3-ceramide axis and NAFLD
SMPD3 is considered an accelerator of pathology in metabolic diseases26. GW4869, a SMPD3 inhibitor, was administrated to organoids to suppress the SMPD3 activity (Extended Data Fig. 7a). The ceramide levels in organoids and their supernatants under nicotine treatment were decreased by GW4869 (Extended Data Fig. 7b, c). Lentivirus-expressed SMPD3 was applied to restore the expression of SMPD3 in AMPKα1-knockout ileal organoids from nicotine drinking model (Extended Data Fig. 7d). As expected, SMPD3 overexpression reversed the reduction in ceramide levels resulting from AMPKα1 deficiency in ileal organoids (Extended Data Fig. 7e, f). In nicotine-accelerated NAFLD mouse model, B. xylanisolvens-colonized SPF mice showed a reduction of ileal ceramide in the presence of NicX (Extended Data Fig. 7g), which resulted from the inhibition of intestinal p-AMPKα-SMPD3 axis.
To confirm whether modulation of the ceramide profiles accounts for the NAFLD improvement derived from intestinal epithelium AMPKα1 deficiency in nicotine-accelerated NAFLD mouse model, we administered ceramide (d18:1/16:0) by daily i.p. injection (Extended Data Fig. 8a). The protective effects on hepatic steatosis, inflammation, and fibrosis resulting from the genetic disruption of intestinal epithelium AMPKα1 were reversed by ceramide (d18:1/16:0) administration (Extended Data Fig. 8b–s). Moreover, oral GW4869 delivery decreased the ileal levels of ceramides, and the decrease was accompanied with lower NAFLD severity in nicotine drinking model (Extended Data Fig. 9a–s). In conclusion, nicotine-induced activation of the intestinal AMPKα-SMPD3 axis potentiates NAFLD progression by upregulating intestinal ceramide production and SMPD3 suppression is a novel strategy for relieving hepatic steatosis, inflammation, and fibrosis.
B. xylanisolvens and clinical NAFLD
To elucidate the correlation between our major findings and human NAFLD progression, we collected data from 83 patients with biopsy-proven NAFLD, which was divided into smokers (n = 41) and nonsmokers (n = 42) (Supplementary Table. 3–6). Among the smokers, there was no significant difference between NAFL, borderline NASH and definite NASH with respect to tobacco smoking history, including years of tobacco smoking and number of cigarettes per day (Supplementary Table. 3, 4). However, we found that B. xylanisolvens levels were negatively correlated with NAFLD severity (ie, from NAFL to borderline NASH to definite NASH), as indicated by NAFLD activity score (NAS) (Fig. 4a, b, Extended Data Fig. 10a–c). As a result, the fecal nicotine levels were step-wise increased with the progression of NAFLD, and the nicotine degradation product HPB in feces was accordingly decreased along with NAFLD progression (Fig. 4c, d). In smokers, B. xylanisolvens abundance was negatively correlated with fecal nicotine content, but positively correlated with HPB levels (Fig. 4e, f).
Fig. 4. B. xylanisolvens-mediated nicotine degradation is negatively correlated with clinical NAFLD progression.
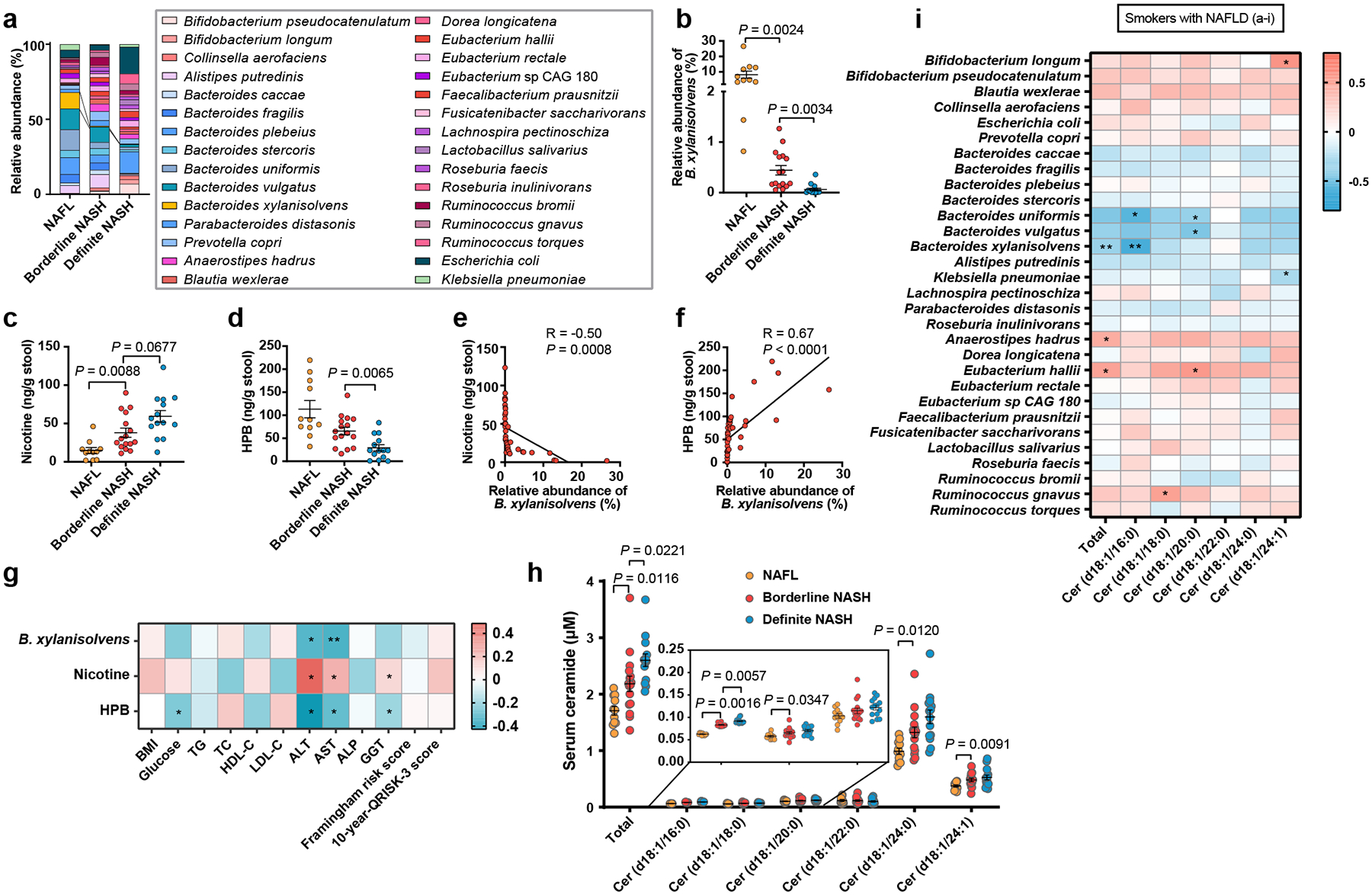
In 41 smokers with NAFLD, including NAFL (n = 11), Borderline NASH (n = 16), Definite NASH (n = 14). a, Bacterial taxonomic profiling of the gut microbiota from different NAFLD stages processed at the species level. b, Relative abundances of B. xylanisolvens associated with different NAFLD stages in smokers with NAFLD. c, d, Concentration of fecal nicotine (c) and HPB (d) with different NAFLD stages in smokers with NAFLD. e, f, Correlative analysis of B. xylanisolvens with fecal nicotine (e) and HPB (f). Correlations between variables were assessed by linear regression analysis. Linear correction index R and P values were calculated. g, Heatmap of the correlation between B. xylanisolvens abundance, fecal nicotine and HPB levels and metabolic indicators. Correlation analysis were determined by Spearman’s rank test. *P < 0.05, **P < 0.01. h, Quantification of serum ceramides in different NAFLD stages. i, Heatmap of the correlation between gut bacteria (top 30) and serum ceramides levels. Correlation analysis were determined by Spearman’s rank test. *P < 0.05, **P < 0.01. The data are presented as the means ± s.e.m. b, c, h, Kruskal-Wallis test with Dunn’s test. d, One-way ANOVA with Dunnett’s T3 post hoc test.
In terms of metabolic indicators, the presence of B. xylanisolvens and HPB in feces was found to be adversely connected to serum ALT and AST levels, while nicotine concentrations were positively related (Fig. 4g). These results suggest that B. xylanisolvens-mediated nicotine degradation potentially safeguards liver function in individuals who smoke. The serum ceramide levels, especially ceramide (d18:1/16:0), which is mainly derived from the intestine, were significantly higher in the individuals with NASH compared to those with NAFL (Fig. 4h), and it was negatively correlated with the abundance of B. xylanisolvens (Fig. 4i).
However, we did not observe the correlation between B. xylanisolvens abundance and NAFLD severity in nonsmokers (Extended Data Fig. 10d–j). In conclusion, these data establish intestinal nicotine accumulation as a risk factor for clinical NAFLD progression in smokers, while also suggesting that B. xylanisolvens, with its nicotine-degrading activities, could exhibited protective effects to improve the progression of NAFLD (Extended Data Fig. 10k).
Discussion
In patients who smoke and in three nicotine-exposed mouse models, high concentrations of intestinal nicotine were detected by an LC/MS-based method. Hence, the distribution of nicotine in the intestine exists irrespective of the route of delivery. As a reminder, intraperitoneal nicotine supplementation in mice induced a nicotine accumulation in the stool16, and previous literature using nicotine skin patches in humans also found an obvious accumulation of nicotine in the saliva and gastric juice27, though the mechanism(s) for this accumulation need to be addressed in future studies.
Although some natural nicotine-degrading bacteria have been discovered28, 29, suggesting a bacteria-based approach to lower nicotine levels, the nicotine-degrading properties of gut microbiota in mammalian have yet to be confirmed. Our founding raised the potential of gut microbial nicotine degradation. B. xylanisolvens has been verified to be safe for in vivo use, and the European Union has authorized the inclusion of heat-treated milk products fermented with B. xylanisolvens DSM 23964 as a novel food30. Moreover, our discovery of HPB, a nicotine-derived catabolic metabolite, could potentially be utilized as a biomarker to assess the degree of gut microbial-mediated nicotine catabolism in patients. Even so, more research is needed to fully understand the gut microbial nicotine-degrading capacity.
The discovery of a novel substrate of AMPK, SMPD3, provides the first link from nicotine activated AMPK to ceramide metabolism, and this pathway may also explain the promotion of ceramide levels by cigarette smoke in other tissues such as serum, lung, liver, and abdominal aorta31, 32. In addition to a high-fat diet and a high-cholesterol diet, we show here that nicotine intake is another important inducer of intestine-derived ceramide production. Based on our findings, this study indicates that interventions with either microbial nicotine degraders or other targets based on the intestinal AMPKα-SMPD3-ceramide axis show translational potential for the treatment of nicotine-associated NAFLD progression.
Methods
Human participants
Three cohorts were included in this study. Cohort 1 was approved by the Ethical Committee of the First Affiliated Hospital, Zhejiang University, School of Medicine [2021(875)]. All participants provided written informed consent. Cohort 2 and cohort 3 were approved by the Ethical Committee of the First Affiliated Hospital of Wenzhou Medical University [2016–246] and informed written consent was obtained from all participants.
For cohort 1, terminal ileum mucosa biopsies, serum, and stool samples from 30 nonsmokers and 30 smokers were collected. The characteristics of the participants are listed in Supplementary Tables 1, 2. All the participants were diagnosed with colon polyps. During the polypectomy under a colonoscopy, we obtained a trace amounts of terminal ileal mucosa tissue with no obvious abnormalities.
For cohort 2, serum and stool samples from 41 smokers diagnosed with biopsy-confirmed NAFLD33. Histological specimens were scored according to the NASH-Clinical Research Network (CRN) scoring system34. Briefly, NAFLD activity score (NAS) was calculated as the sum of three histological components, including steatosis (grades 0–3), ballooning (grades 0–2), and lobular inflammation (grades 0–3). Patients with NAS of ≥ 5 correlated with Definite NASH, patients with NAS at 3 or 4 were correlated with borderline NASH, biopsies with scores of less than 3 were diagnosed as NAFL. The characteristics of the participants are listed in Supplementary Tables 3, 4.
For cohort 3, serum and stool samples from 42 nonsmokers diagnosed with biopsy-confirmed NAFLD, the diagnosis and scoring of NAFLD is described as above. The characteristics of the participants are listed in Supplementary Tables 5, 6.
The exclusion criteria for cohort 1–3 were: diabetic ketoacidosis or hyperglycemic hyperosmolar state; patients with decompensated cirrhosis; stage 3–5 chronic kidney disease; inflammatory bowel disease; cancer; pulmonary tuberculosis and AIDS; alcoholism; diseases judged by clinicians as unsuitable for biopsy; taking antibiotics (including rifaximin), probiotics, prebiotics, proton pump inhibitors, and laxatives during the last 3 months. All clinical information was collected according to standard procedures. The collected metadata covered the participants’ anthropometric features and information related to health status, age, sex, and dietary habits. Peripheral blood samples were centrifuged at 1000×g and 4 °C for 5 min to obtain the serum. The levels of blood biochemical indicators (i.e., TG, TC, AST, and ALT) were measured using an autoanalyzer (BioTek Instruments 800TS). Framingham risk score, 10-year-QRISK-3 score were calculated according previously reports35,36. Feces were collected in collection cups and immediately frozen at −80 °C until analysis. Mucosa biopsies were collected by colonoscopy from terminal ileum with no obvious abnormalities.
Animals
SPF mice of the C57BL/6J strain were randomly divided into different groups, housed 3–5 per cage, and maintained under standard laboratory conditions with free access to rodent feed and water in Department of Experimental Animal Science, Peking University Health Science Center, Beijing, China or National Cancer Institute, National Institutes of Health (NIH), USA. Specifically, the light was on from 8 a.m. to 8 p.m., with temperature kept at 21–24 °C, and humidity at 40–70%. Germ-free C57BL/6J (GemPharmatech, Nanjing, China) were bred within sterile vinyl isolators and maintained at the gnotobiotic mouse facility at Department of Experimental Animal Science, Peking University Health Science Center. All animal experiments complied with the protocols for animal use, treatment, and euthanasia approved by Peking University (permit: LA2020480) and the National Cancer Institute Animal Care and Use Committees of NIH (animal protocols: LM-016 and LM-096).
Prkaa1fl/fl mice were purchased from the Jackson Laboratory (#014141). To achieve intestinal epithelium specific AMPKα1 disruption, Prkaa1fl/fl mice were crossed with Villin-cre mice (gifted by Dr. Deborah L Gumucio, University of Michigan) to obtain Prkaa1ΔIE mice. Villin-cre mice express Cre recombinase in villus and crypt epithelial cells of the small and large intestines. The successful knockout of AMPKα1 was assessed (Extended Data Fig. 4f). Prkaa2fl/fl mice were purchased from the GemPharmatech (#T013159), and crossed with Villin-cre mice to obtain Prkaa2ΔIE mice. The successful knockout of AMPKα2 was assessed (Extended Data Fig. 4f).
Eight-week-old male mice were fed a high-fructose and high-cholesterol diet (HFHCD, Research Diets, #D09100310). In assays of the drinking mouse model, SPF mice or GF mice were treated with nicotine (10 μg/mL, Sigma, #54-11-5) in “sweet water” containing 23.1 g/L D-(−)-fructose (Sigma, #F0127) and 18.6 g/ L D-(+)-glucose (Sigma, #G8270). In assays of the smoking mouse model, the mice were exposed to 60 mg/m3 cigarette smoke (CS) of total suspended particulate matter (TSP) for 2 weeks, with 2 hours per day. In mouse subcutaneous injection model, the mice were received nicotine (0.5 mg/kg) or saline daily for 2 weeks. For GW4869 (Selleck Chemicals, #S7609) treatment, SPF mice were treated with control or 10 mg/kg GW4869 by daily gavage. For ceramide (d18:1/16:0) (Avanti Polar Lipids, #860692) supplementation, littermate Prkaa1fl/fl and Prkaa1ΔIE mice were given vehicle or 10 mg/kg ceramide (d18:1/16:0) by daily i.p. injection.
For bacterial colonization, SPF mice were treated with 2×108 CFUs of B. xylanisolvens or nicX knock-out B. xylanisolvens in 200 μL of sterile anaerobic PBS by gavage every three days37. WT or nicX knock-out B. xylanisolvens was grown anaerobically in BHI medium, the cultures were washed and concentrated in anaerobic PBS to an end concentration of 1×109 cfu/mL under strict anaerobic conditions. The determination of cfu/mL was performed by a dilution-plate method using BHI media containing 1% agarose under anaerobic conditions.
For mice body mass composition analysis, the body fat content and fat ratio were measured and calculated with an MRI body fat measurement instrument (EchoMRI-700, EchoMRI) 1 week before sacrifice. For biochemical analysis, hepatic and serum triglyceride and cholesterol and non-esterified fatty acid levels were determined with assay kits from Wako Diagnostics (Wako Chemicals) following the manufacturer’s instructions. ALT and AST levels were assessed using commercial ALT and AST assay kits (Catachem).
Quantitative mass spectrometry
The concentrations of nicotine and HPB were measured by LC-MS/MS. For tissue analysis, tissue samples of approximately 0.1 g were homogenized after the addition of 10% methanol aqueous solution. Subsequently, 20 μL NaOH (1 M), 1 mL of ethyl acetate and 10 μL of the internal standard (500 ng/mL nicotine-d4 for both nicotine and HPB) were added to 100 μL of the homogenates, and the mixtures were rapidly mixed and centrifuged at 13000 rpm for 10 min. Subsequently, 900 μL of the supernatant was volatilized by nitrogen gas, and the sediment was dissolved in 100 μL of 70% aqueous acetonitrile (v/v). For serum and culture medium of the bacteria, 100 μL samples were prepared as described above. Five microliters of the supernatant were analyzed using a QTRAP® 5500 system (AB SCIEX, Concord, ON, Canada) with a Waters XBridge amide column (100 × 4.6 mm i.d., 3.5 μm). The mobile phase was a mixture of acetonitrile (B) and water containing 0.1% formic acid and 2 mM ammonium formate (A) at a constant flow rate (0.5 mL/min). The proportion of mobile phase B was maintained at 25% by isocratic elution. The analytes [M+H]+ were measured at m/z 163.3 → 130.2 for nicotine, m/z 166.1 → 106.0 for HPB, and m/z 167.3 → 134.2 for nicotine-d4. Lower relative standard deviations (less than 10%) were detected both between and within days. The recoveries of all the analytes were higher than 97%.
Metagenomic Sequencing
Metagenomic sequencing was performed as previously described38. DNA (700 ng/sample) was used for sample preparation. Sequencing libraries were generated with an Ultra DNA Library Prep Kit for Illumina; the manufacturer’s recommendations and index codes were adopted to annotate sequences in each sample. The fragmented DNA ends were repaired, polyA tailed, and ligated with a sequencing adaptor for Illumina sequencing. PCR amplification and DNA purification were performed with an AMPure XP system. The DNA concentration was measured with the Qubit DNA Assay Kit in a Qubit 2.0 fluorometer and diluted to 2 ng/μL. The library insert size was assessed using an Agilent Bioanalyzer 2100 system. Clustering of index-coded samples was performed on a cBot Cluster Generation System using a HiSeq 250 PE Cluster Kit according to the manufacturer’s instructions. After cluster generation, the libraries were sequenced on the Illumina HiSeq 250 platform, and 150-bp paired-end reads were generated.
Metagenomic Analysis
Sequencing data were converted to fastq files with bcl2fastq. Reads were then QC trimmed using Trimmomatic39 with the following parameters: PE -threads 10 -phred33 -validatePairs ILLUMINACLIP:TruSeq3-PE.fa:2:30:10 LEADING:3 TRAILING:3 MINLEN:50. We used MetaPhlAn240 for taxonomic analysis with the following parameters: –ignore_viruses –ignore_archaea –ignore _eukaryotes. The follow-up analysis was performed with MicrobiomeAnalyst41. The α-diversity was estimated based on the gene profile of each sample according to the ACE, Chao1 and Shannon indices. The LEfSe analysis was performed with Galaxy42.
Microbial strains and gene editing strategies
The B. xylanisolvens strain J1101 was isolated from human feces as previously described43. The strain was identified by comparing the 16S rRNA gene sequence with those in the NCBI reference database (https://www.ncbi.nlm.nih.gov/). The DNA fragments encoding full-length nicX (Supplementary table 7) were cloned into the pET28a vector with a 6×His tag at the N-terminal end using standard molecular cloning procedures. Gene nicX was overexpressed in Escherichia coli Rosseta (DE3), and the cells were grown at 37 °C to an OD600 of 0.6. NicX protein was induced with 200 μM isopropyl-β-d-thiogalactoside (IPTG) overnight at 16 °C. An internal fragment (610 bp) of the nicX gene from B. xylanisolvens was cloned into the pGERM suicide vector containing E. coli (bla) and B. xylanisolvens (ermG) selective markers. The constructed vector was then transformed into the conjugative E. coli S17 strain. The donor E. coli S17 and recipient B. xylanisolvens were co-incubated under aerobic, the bacteria on the filter were transferred onto BHI medium agar plates supplemented (mutant selection) or not supplemented (control) with erythromycin (25 μg/mL) and gentamicin (200 μg/mL). Then, resistant colonies were picked for identification by PCR using primers targeting junction regions between the pGERM and nicX genes. Details of construction processes were according to a previously reported method44. Both B. xylanisolvens and nicX-deficient B. xylanisolvens were grown in BHI medium supplemented with 0.3 g/mL cysteine under anaerobic conditions at 37 °C. E. coli and nicX-overexpressing E. coli were cultured in Luria-Broth (LB) medium at 37 °C for 24 h. To obtain bacteria in PBS for gavage, bacterial culture medium was centrifuged at 8,000×g and 4 °C for 10 min and the pellets were then suspended in PBS (oxygen-free PBS for anaerobic bacteria).
Growth curves of B. xylanisolvens
B. xylanisolvens or nicX-deficient B. xylanisolvens (2×107 CFUs/mL) were inoculated anaerobically into BHI medium at a ratio of 1:50 (v/v). To investigate the effect of nicotine on the growth of B. xylanisolvens, nicotine was added to a medium at the concentration of 100 μg/mL. Two hundred microliters of culture were taken every 2 h over a day to measure the optical density (OD600) using a Spectra Max 190 microplate reader (Molecular Devices Inc.).
Microbial nicotine catabolism in vitro
B. xylanisolvens and nicX-deficient B. xylanisolvens were cultured in BHI medium containing 100 μg/mL nicotine, and then incubated for 48 h at 37 °C anaerobically. The supernatants were sampled for mass spectrometry detection of nicotine and HPB. WT E. coli and nicX-overexpressing E. coli were treated with 200 μM IPTG overnight at 16 °C to induce the expression of NicX protein before incubation. WT E. coli and nicX-overexpressing E. coli were cultured in LB medium and the other steps were the same as those described above.
Kinetic parameters for NicX were determined in assays containing 1 mM flavin mononucleotide (FMN), 25 mM Tris-HCl (pH 7.6), and nicotine. The reaction mixture was incubated for 20 min at 37 °C. Substrate nicotine concentrations varied from 6.16 μM to 789 μM, and a 20 μL sample was pipetted and mixed with an equal volume of acetonitrile to stop the reaction. Product formation was determined using LC-MS/MS, as described above. Reactions were performed in triplicate, and data were fitted to the Michaelis-Menten equation using the least-squares method.
Histological analysis
After sacrifice, liver tissues of mice were preserved in 10% formalin or embedded in OCT compound. Formalin-fixed paraffin-embedded liver tissue slides were subjected to hematoxylin and eosin (H&E) and Sirius red staining to assess lipid accumulation and liver fibrosis degree, respectively. The frozen liver sections were stained with Oil Red O for visualizing lipid droplets. All the processes were according to standard protocols followed by microscopic examination. The histology scores were evaluated according to a reported NAFLD scoring system34.
Extraction, isolation and identification of HPB
The solvents used for extraction and chromatographic separation were of analytical grade. TLC was carried out on silica gel HSGF254, and the spots were visualized by spraying with Bismuth potassium iodide chromogenic agent. Silica gel and Sephadex LH-20 were used for column chromatography. HPLC separation was performed on an Agilent 1200 HPLC system using an ODS column (C18, 250 × 9.4 mm; detector: UV) with a flow rate of 2.0 mL/min. NMR spectral data were obtained with a Bruker Avance-500 spectrometer.
The fermentation broth of B. xylanisolvens in BHI medium containing 100 μg/mL nicotine (1 L) were extracted three times with ethyl acetate (3 × 500 mL), and the organic solvent was evaporated to dryness under vacuum to afford the crude extract (565 mg). The crude extract was subsequently separated on a silica gel column (30.0 g of silica gel) eluted with hexane−ethyl acetate via gradient elution (v/v, 100:0, 100:2, 100:5, 100:10, 100:20, 100:40, each 500 mL) and then with dichloromethane−methanol (v/v, 100:1, 100:2, 100:5, 100:10, 0:100, each 500 mL) to afford 10 fractions (BX-1−BX-10). Compound 1 (55.3 mg, Rt 18.3 min) was obtained from fraction BX-5 by RP-HPLC using 23% methanol in water. The structure of the Compound 1 was identified as the known compound 4-hydroxy-1-(3-pyridyl)-1-butanone (HPB) by comparing the experimental NMR data obtained with literature values45.
Isolation and culture of intestinal primary cells
Intestinal primary cells were prepared from mice on C57BL/6J background as previously described46. Ileum (bottom 1/3 small intestine) was opened longitudinally, rinsed in cold PBS containing 1% Penicillin-Streptomycin (PS) and 2% glucose and the outer muscle layer was removed, then the tissue was chopped into approximately 5 mm pieces in cold PBS and further washed three times. After discarding the supernatant, 10 mL of digestion fluid (DMEM containing 0.1% type 1 neutral protease, 0.2% collagenase and 0.1% Y-27632) was added to the remaining ileum pieces, and the pieces were then incubated at 37 °C for 15 min with intermittent shocks. Stop buffer (DMEM containing 5% fetal bovine serum and 2% sorbitol) was added and filtered through 100 μm and 40 μm cell filters. Single cells were centrifuged at 500 g at 4 °C for 5 minutes, and DMEM containing 10% FBS and 1% PS was used for ileal primary cell culture in an incubator at 37 °C under 5% CO2. Nicotine applied in ileal primary cells treatment was 1 μg/mL.
Organoid isolation, culture and treatment
Isolation and culture of ileal organoids were performed as previously reported26. Briefly, ileum isolated from mice was dissected, and washed with Dulbecco’s PBS 10 times. Then, the ileal fragments were incubated with Gentle Cell Dissociation Reagent (STEMCELL Technologies) to separate the crypts and villi from the intestinal basement membrane. After centrifugation, the crypts were isolated by filtration (70 μm) and resuspended in a 1:1 mixture of Matrigel (Corning) and IntestiCult organoid growth medium (OGM) (STEMCELL Technologies) at a density of 6000 crypts/mL. A droplet of 50 μL containing 300 crypts was placed into the center of each well of a prewarmed 24-well plate, forming a dome. After the domes had solidified, 750 μL of OGM was added to each well. The crypts were cultured at 37 °C under 5% CO2, and the medium was refreshed every 3 days.
For compound C (CC, AMPK inhibitor) treatment, ileal organoids were incubated with control, nicotine (1 μg/mL) or nicotine plus CC (20 μM) for 12 h. For GW4869 treatment, HFHCD-fed WT mice were treated with nicotine water or nicotine water plus 10 mg/kg GW4869 (by daily gavage) for 2 weeks, and then organoids were isolated and cultured. The ileal organoids were cultured for 7 days and treated with GW4869 (10 μM) and nicotine (1 μg/mL) for the last 3 days before being harvested.
Lentiviral transduction and treatment in organoids
HFHCD-fed Prkaa1fl/fl and Prkaa1ΔIE mice were treated with nicotine water for 2 weeks, and then ileal organoids were isolated. To further restore SMPD3 expression, isolated Prkaa1fl/fl and Prkaa1ΔIE organoids were infected with LV (lentivirus)-Ctrl or LV-Smpd3 (1 × 106 PFU/100 crypts), and the organoid-virus mixture was placed in an incubator at 37 °C for 1 h to allow transduction. After transduction, the infected organoids were plated and cultured for 7 days and treated with nicotine (1 μg/mL) for the last 3 days before being harvested.
Targeted lipidomics analysis and ceramide quantification
Targeted lipidomics were performed according previous study with adjustment47. Lipids including phosphatidylcholine (PC), lyso-phosphatidylcholine (LPC), phosphatidylethanolamine (PE), lyso- phosphatidylethanolamine (LPE), sphingomyelin (SM), and ceramide (Cer). 20 mg ileal tissue was added with 250 μL water and homogenized for 1 min, and the protein concentration was determined by BCA. Then 300 μL methanol, 1 mL methyl tert-butyl ether was added and vortexed for 10 s, centrifuged at 4 °C and 12,000 rpm for 10 min. The supernatant (95 μL) and internal standard solution (5 μL) was charged into a 1.5 mL centrifuge tube and vortexed for 2 min. Subsequently, the mixture was transferred into a vial for MS detection. Waters ACQUITY UPLC I-CLASS with a Waters UPLC BEH C8 column (2.1 mm (inner diameter) ×100 mm (length), 1.7 μm (particle dimension)) was used for separation. For the positive ion mode, the mobile phase was consisted of acetonitrile/water (6:4, v/v, phase A) and isopropanol/acetonitrile (9:1, v/v, containing 5 mmol/L ammonium formate and 0.1% formic acid, phase B) with a flow rate of 0.25 mL/min and a column temperature of 55 °C, with the injection volume as 5.0 μL. The MS data under positive ion mode were collected by a Waters XEVO TQ-S Micro system. The parameters were set as follows: ion source voltage 3.0 kV, ion source temperature 150 °C, desolvation temperature 500 °C, desolvation gas flow 1000 L/h, cone voltage 10 V, cone gas flow 10 L/h. For data processing, the peak area was integrated by the TargetLynx software with a retention time tolerance of 15 s. The final concentration was calculated by a single point internal standard method.
For the quantification of ceramides, 25 μL serum, 20 mg ileal tissue or organoids in 12-well plates, was homogenized with chloroform/methanol (v/v, 2:1) containing 1 μM LM6002 (Avanti Polar Lipids) as internal standard. The mixture was oscillated immediately and then centrifuged at 13,000 rpm for 20 min. The lower phase was dried by a SpeedVac. The sediment was dissolved in 100 μL isopropanol/acetonitrile (IPA/CAN, v/v, 1:1) and analyzed by Eksigent LC100 coupled with AB SCIEX Triple TOF 5600 system using a Waters XBridge Peptide BEH C18 column (3.5 μm, 2.1*100 mm). The UPLC and MS parameters were according to previous study26. The lipid metabolites were quantified using MultiQuant 2.1 software (AB SCIEX).
Phosphoproteomics
Phosphoproteomics were performed as previously reported48. Briefly, ileal tissues were added to an appropriate amount of lysis buffer (including 4% SDS, 0.1 M Tris/HCl and 0.1 M DDT, pH 7.6) containing protease inhibitors and phosphatase inhibitors, and vortexed for 30 s. The samples were then ultrasonicated for 60 s (0.2s on and 2s off), with an amplitude of 25%. After standing at room temperature for 30 min, and centrifugation at 15,000 rpm and 4 °C for 10 min, the supernatant was carefully removed and centrifuged again. The protein concentration was measured using BCA protein quantitative assay. Phosphopeptides were enriched using titanium dioxide (TiO2). TiO2 beads in tubes and TiO2 microcolumns packed on centrifugation adaptors (GL Sciences, 5010–21514) were individually equilibrated with 100 μL of loading buffer (70% ACN, 5% TFA and 20% lactic acid (Sigma-Aldrich, L6661)). After centrifugation, supernatants containing TiO2 beads were removed. Peptides from all 6 subfractions in each experiment were diluted in 100 μL of loading buffer, mixed thoroughly with 5 mg of TiO2 beads, and incubated for 30 min. The mixtures of TiO2 beads and peptides were then centrifuged at 800 × g for 5 min, the supernatants were transferred into new clear tubes for a second round of phosphopeptide enrichment, and the enriched TiO2 beads were suspended in 100 μL of loading buffer, transferred into microcolumns and then packed into centrifugation adaptors. Each column with enriched TiO2 beads was washed four times with 120 μL of loading buffer (150 × g, 10 min), washed once with 100 μL of washing buffer 1 (30% ACN and 0.5% TFA) (250 × g, 8 min) and wash twice with washing buffer 2 (0.4% TFA and 80% ACN) (250 × g, 5 min). Phosphopeptides were eluted with 150 μL of elution buffer (15% NH4OH (Wako, 013–23355), 40% ACN), and the first and second phosphopeptides enriched in each subfraction were combined, dried in a SpeedVac and stored at −80 °C. Lumos reversed- phase chromatography fusion was used for protein analysis. The volcano map was performed using the OmicStudio tools at https://www.omicstudio.cn/tool.
Cell culture and transient transfection
SW480 cells were grown in DMEM containing 100 units/mL PS and 10% fetal bovine serum at 37 °C in a humidified incubator with 5% CO2. Lipo3000™ Transfection Reagent was used for the transfection of SW480 cells and the total DNA for each plate was adjusted using the relevant empty vector. 24 hours after transfection, the cells were harvested. All the cell experiments treated with nicotine were under the concentration of 1μg/mL. Cell line was tested for mycoplasma contamination, and no cell lines were detected positive for mycoplasma contamination via a PCR-based method.
In vitro kinase assays.
Bacterially purified SMPD3-His (86–655) proteins (2 μg) were incubated with partially purified rat liver active AMPK heterotrimer (α1/β1/γ2) (Promega, V4012) (1 μg) for 2 h at 30 °C in kinase buffer (25 mM HEPES, pH 7.4, 50 mM NaCl, 5 mM MgCl2, 1 mM CaCl2, 1 mM dithiothreitol, 30 μM ATP and 0.5 mg/mL BSA) containing 1 μCi of [γ−32P] ATP (Perkin Elmer). The reaction mixture was stopped by the addition of SDS sample buffer, and the labeled mixture was resolved by SDS-PAGE. The gels were dried, exposed to phosphor- imager screens for autoradiography, and stained with Coomassie Blue to visualize the proteins. CC was applied at the concentration of 20 μM.
Ubiquitination assay of SMPD3
The mutant proteins of SMPD3 (S209A, S209D, K103R or S209A/K103R) were constructed using a commercial point mutagenesis kit (Invitrogen, #A14604) with seamless cloning and mutagenic primers. S209A: By a codon point mutation, the serine residue (S) at position 209 of SMPD3 is turned into an alanine residue (A), which cannot be phosphorylated, thus mimicking SMPD3 with no phosphorylation at S209. S209D: By a codon point mutation, the serine residue (S) at position 209 of SMPD3 is turned into an aspartate residue (D), which has a side chain similar to phosphorylated serine residue, thus mimicking SMPD3 with sustained phosphorylation at S209. K103R: By a codon point mutation, the lysine residue (K) at position 103 of SMPD3 is turned into an arginine residue (R), which cannot be ubiquitinated, thus mimicking SMPD3 with no ubiquitination at K103.
For SMPD3 ubiquitination assay, SMPD3-flag (WT, K103R, S209A or S209A/K103R) were transfected into SW480 cells with Lipo3000™ Transfection Reagent. Twenty-four hours later, the cells were treated with DMSO or nicotine (1μg/mL) for 24 h. The cells were washed with PBS and lysed in HEPES buffer (20 mM HEPES, pH 7.2, 50 mM NaCl, 1 mM NaF, 0.5% Triton X-100) plus 0.1% SDS and protease-inhibitor cocktail. The lysates were incubated with ANTI-FLAG M2 affinity beads (Sigma-Aldrich, A2220) overnight at 4 °C. After extensive washing, proteins were eluted with 2 × SDS sample buffer and resolved by SDS- PAGE, and blotted with antibodies against Ub, K48-Ub or K63-Ub.
Mass spectrometry analysis
Modified SMPD3-flag samples were prepared by immunoprecipitation from SW480 cells after DMSO or nicotine (1μg/mL) treatment for 24 h. The SMPD3-flag proteins were resolved by SDS-PAGE. The gel was stained with Coomassie Blue reagent and destained and the SMPD3-flag band was excised. The samples were subjected to reduction with 10 mM dithiothreitol (DTT) for 30 min, alkylation with 55 mM iodoacetamide for 45 min, and in-gel digestion with trypsin. The digested samples were analyzed by reversed-phase microcapillary/tandem mass spectrometry (LC/MS/MS) as described previously49.
Real-Time PCR analysis
Liver tissues were flash-frozen in liquid nitrogen and stored at −80 °C. Total RNA from frozen liver tissues or ileal organoids was extracted with the TRIzol reagent (Invitrogen, Carlsbad, CA, USA). cDNA was synthesized from 2 μg of total RNA using 5× All-In-One RT MasterMix. Real-time PCR primer sequences were included in Supplementary table 7. The relative amount of each mRNA was calculated after normalization to the corresponding β-ACTIN gene, and the results are expressed as fold changes relative to the control group.
Western blot analysis
Tissue and cell extracts were homogenized in ice-cold RIPA buffer with protease and phosphatase inhibitors and resolved on 8%−10% SDS–polyacrylamide gels and transferred to a PVDF membrane. After blocking with 5% nonfat milk, the PVDF membrane was incubated with the desired primary antibody (diluted in TBST supplemented with 5% BSA) overnight at 4 °C on an orbital shaker with gentle shaking, followed by rinsing with TBST 3 times, 8 min each at room temperature, and then the secondary antibodies were incubated for 2 h at room temperature with gentle shaking. The secondary antibody (diluted in TBST supplemented with 5% BSA) was then removed, and the PVDF membrane was further washed with TBST 3 times, 8 min each at room temperature.
Antibodies
Rabbit polyclonal antibody against phosphor-SMPD3-S209/208 (ABclonal, cat. #AP1312, 1:500 for IB) was raised and validated by Abclonal. Rabbit polyclonal antibody against ubiquitin-SMPD3-K103 (ABclonal, cat. #A21272, 1:500 for IB) was raised and validated by Abclonal. Rabbit anti-phospho-AMPKα-T172 (Cell Signaling Technology, cat. #2535, 1:1000 for IB), anti-AMPKα (Cell Signaling Technology, cat. #2532, 1:1000 for IB), anti-AMPKα1 (Cell Signaling Technology, cat. #2795, 1:1000 for IB) anti-AMPKα2 (Cell Signaling Technology, cat. #2757, 1:500 for IB), anti-ACTB (ABclonal, cat. #ab8227, 1:2000 for IB), anti-SMPD3 (Santa Cruz Biotechnology, cat. #sc-166637, 1:500 for IB), Anti-Ubiquitin (Medical & Biological Laboratories, cat. #MK-11-3, 1:500 for IB), Anti-Ubiquitin (linkage-specific K48) (Cell Signaling Technology, cat. #8081, 1:500 for IB), Anti-Ubiquitin (linkage-specific K63) (Cell Signaling Technology, cat. #5621, 1:500 for IB) were used in this study.
Statistics
Heatmap and all charts were generated using GraphPad Prism software 8.0. Metagenomic sequencing data were collected by lllumina HiSeq PE250. Metabolite content were collected by a QTRAP 5500 system. Phosphoproteome data were collected by EASY-nano-LC1000. Other data were collected with Microsoft Excel 2019. All statistical data were analyzed using SPSS version 26.0. The sample sizes were determined by power analysis using StatMate version 2.0. No data were excluded during the data analysis. All experimental data are reported as the means ± s.e.m. The Shapiro-Wilk normality test was used to determine the normal distribution of samples. Comparisons between two groups were performed by two-tailed unpaired Student’s t-test (normal distribution) or Mann–Whitney U test (non-normal distribution). Comparisons across multiple groups were assessed with one-way ANOVA (normal distribution) followed Tukey’s (with same standard deviation) or Dunnett’s T3 (with different standard deviation) post hoc test, or Kruskal-Wallis test (non-normal distribution), followed Dunn’s test, as indicated in each figure legend.
Extended Data
Extended Data Fig. 1. Gut microbiota composition differences between smokers with high nicotine and low nicotine levels.
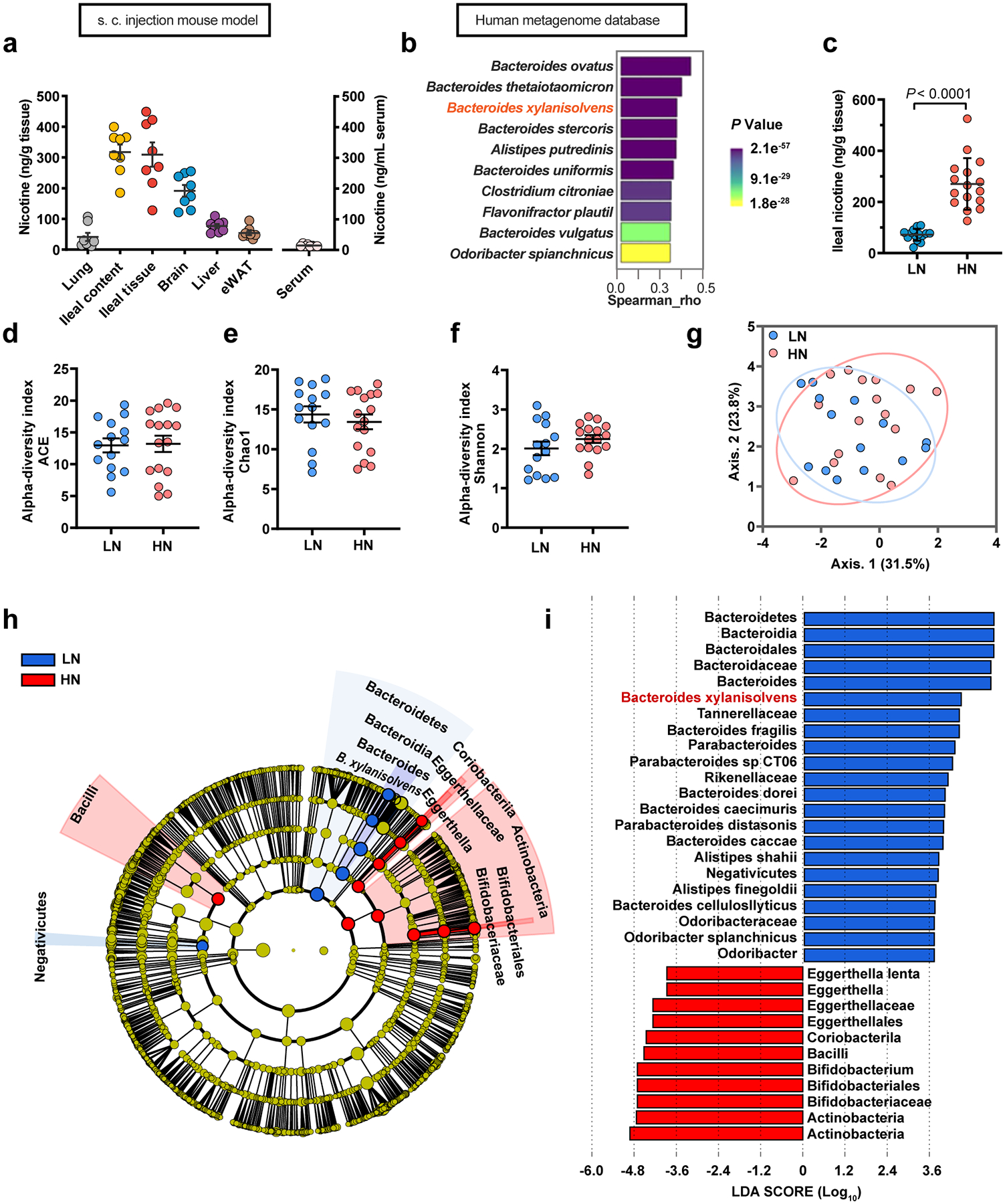
a, Quantification of the nicotine concentrations in lung, ileum content, ileum tissue, brain, liver, eWAT and serum samples obtained from s.c. injection mouse model for two weeks (SPF, n = 8 mice/group). b, Top 10 species of gut bacteria in humans that show high correlation with the known nicotine-degrading enzyme, nicA. These species were identified by MetaQuery. c-i, 30 smokers who were further divided into the HN (high nicotine, n = 16) and LN (low nicotine, n = 14) groups according to their ileal nicotine levels. Ileal nicotine concentrations in HN and LN groups. The data did not obey the normal distribution determined by the Shapiro normality test; thus, the median was used as a break point, and divided into two groups (c). α-diversity of the gut microbiota between the LN and HN individuals, as indicated by the ACE (d), Chao1 (e) and Shannon indices (f). Partial least squares discriminant analysis (PLS-DA) using the Bray-Curtis distance (g). Taxonomic cladogram generated by LefSe of metagenomic analysis data. The blue color indicates enriched taxa in the LN group, and the red color indicates enriched taxa in the HN group. The size of each circle is proportional to the taxon’s abundance (h, i). Data are the means ± s.e.m. b, Correlations were assessed by nonparametric Spearman’s test. c-f, Two-tailed Student’s t-test.
Extended Data Fig. 2. Identification of B. xylanisolvens as a nicotine degrader.
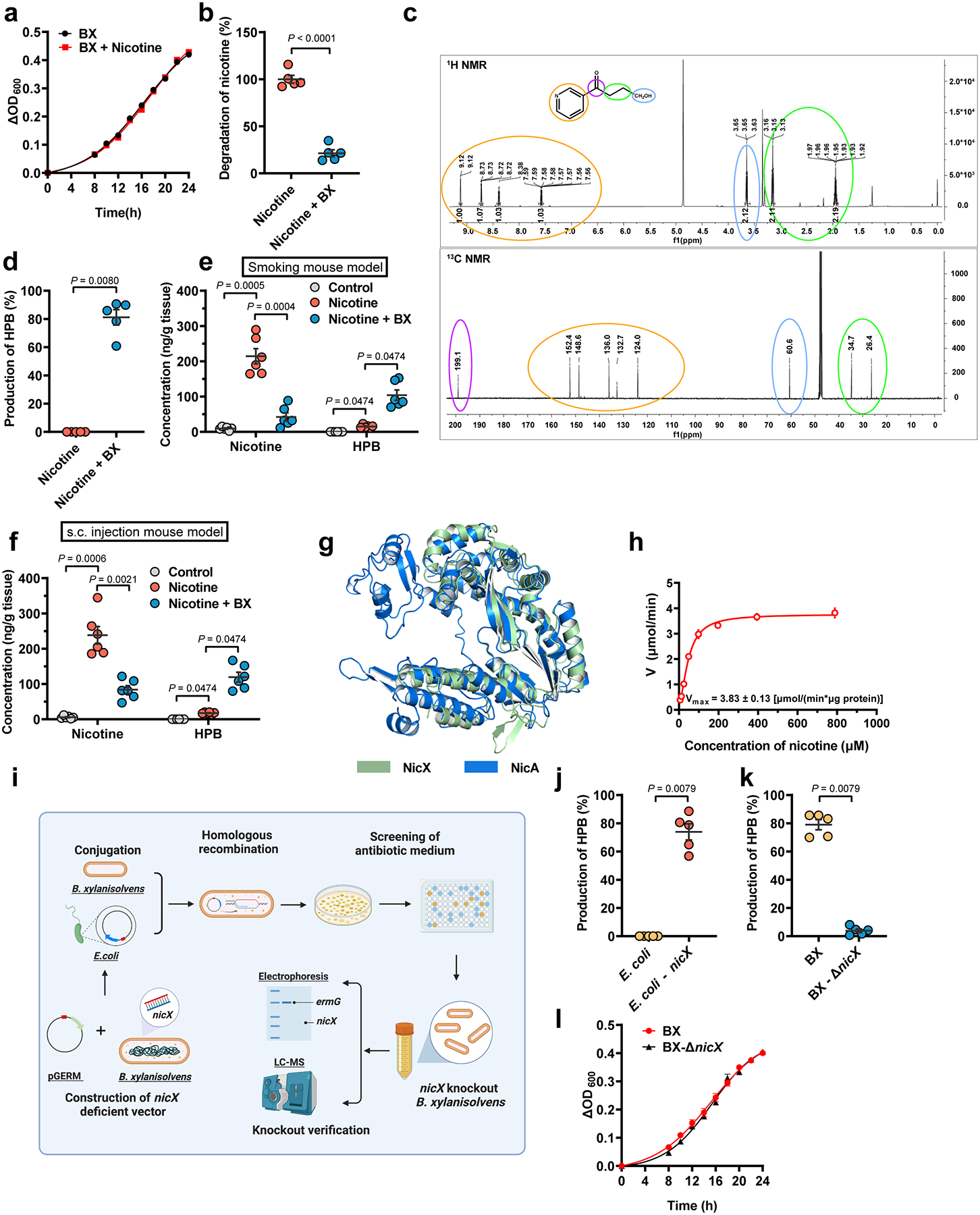
a, Growth curves of B. xylanisolvens with or without nicotine in culture medium (n = 3/group). b, Nicotine concentration in B. xylanisolvens in vitro cultivation compared with control (BHI medium with nicotine supplementation, n = 5/group). c, 1H NMR spectrum (top) and 13C NMR spectrum (bottom) of HPB. d, Production of HPB in B. xylanisolvens in vitro supplementation compared with control (BHI medium with nicotine supplementation, n = 5/group). e, Nicotine and HPB concentration in ileal tissues of smoking exposure mouse model for two weeks (SPF, n = 6 mice/group). f, Nicotine and HPB concentration in ileal tissues of subcutaneous injection mouse model for two weeks (SPF, n = 6 mice/group). g, Structural alignment of the SWISS-MODEL50-predicted B. xylanisolvens NicX and predicted Pseudomonas putida NicA. The Root-Mean-Square-Deviation (RMSD) of 242 aligned residues is 1.323 Å. h, Nonlinear regression for nicotine degradation catalyzed by purified NicX. The reaction mixture contained 1 mM FMN, 25 mM Tris-HCl (pH 7.6), 20 ng NicX, and nicotine in different concentration at 37 °C, n = 3/group. i, Schematic diagram illustrating the workflow for nicX gene deletion in B. xylanisolvens. j, Production of HPB in E. coli and E. coli + nicX in vitro cultivation (LB medium with nicotine supplementation, n = 5/group). k, Production of HPB in B. xylanisolvens and B. xylanisolvens-ΔnicX in vitro cultivation in culture medium (BHI medium with nicotine supplementation, n = 5/group). l, Growth curves of WT and nicX-KO B. xylanisolvens. (n = 3/group). Data are the means ± s.e.m. a, b, l, Two-tailed Student’s t-test. d, j, k, Two-tailed Mann-Whitney U-test. e, f, for Nicotine, one-way ANOVA with Dunnett’s T3 post hoc test; for HPB, Kruskal-Wallis test with Dunn’s test. Experiments in a, b, d, h, j, k, l were performed three times independently.
Extended Data Fig. 3. B. xylanisolvens transplantation alleviates nicotine-accelerated NAFLD progression.
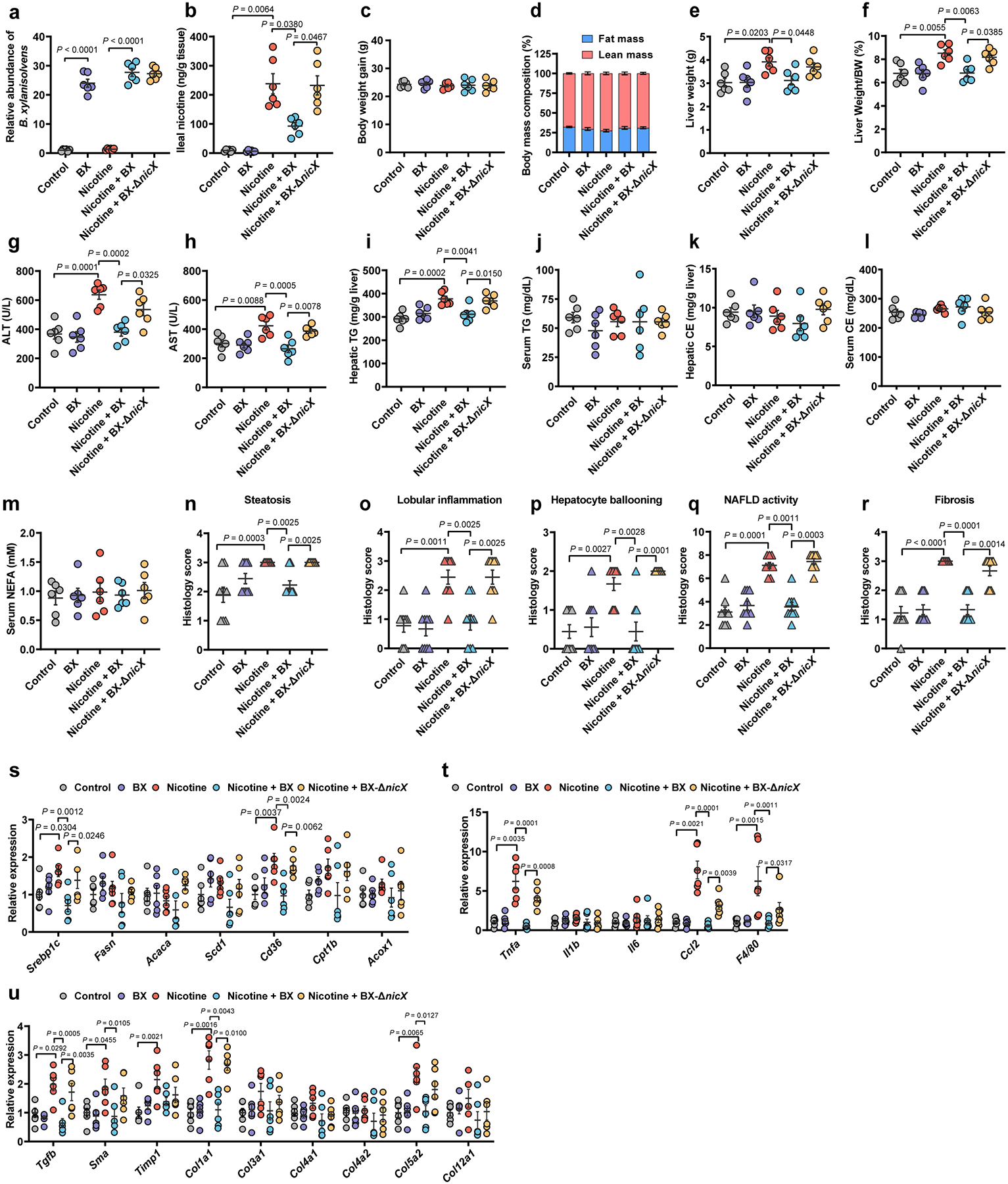
HFHCD-fed SPF mice were treated with PBS, B. xylanisolvens colonization, nicotine water, nicotine water plus B. xylanisolvens colonization, and nicotine water plus nicX knock-out B. xylanisolvens colonization for 20 weeks (n = 6 mice/group). a, Fecal B. xylanisolvens abundance analyses of mice by qPCR. b, Ileal nicotine concentrations. c, Body weight gain. d, Body mass composition. e, Liver weights. f, Liver weight–to–body weight ratios. g, h, Serum ALT (g) and AST (h) levels. i, Hepatic TG content. j, Serum TG content. k, Hepatic CE content. l, Serum CE content. m, Serum NEFA content. n-r, Histology scores of hepatic steatosis (n), lobular inflammation (o), ballooning (p), NAFLD activity (q), and fibrosis stage (r). s-u, Relative mRNA levels of genes related to hepatic lipid metabolism (s), inflammation (t) and fibrosis (u). Data are the means ± s.e.m. c, e-i, k-m, One-way ANOVA with Tukey’s post hoc test. a, b, j, One-way ANOVA with Dunnett’s T3 post hoc test. d, n-u, Kruskal-Wallis test with Dunn’s test.
Extended Data Fig. 4. Nicotine-induced activation of intestinal AMPKα.
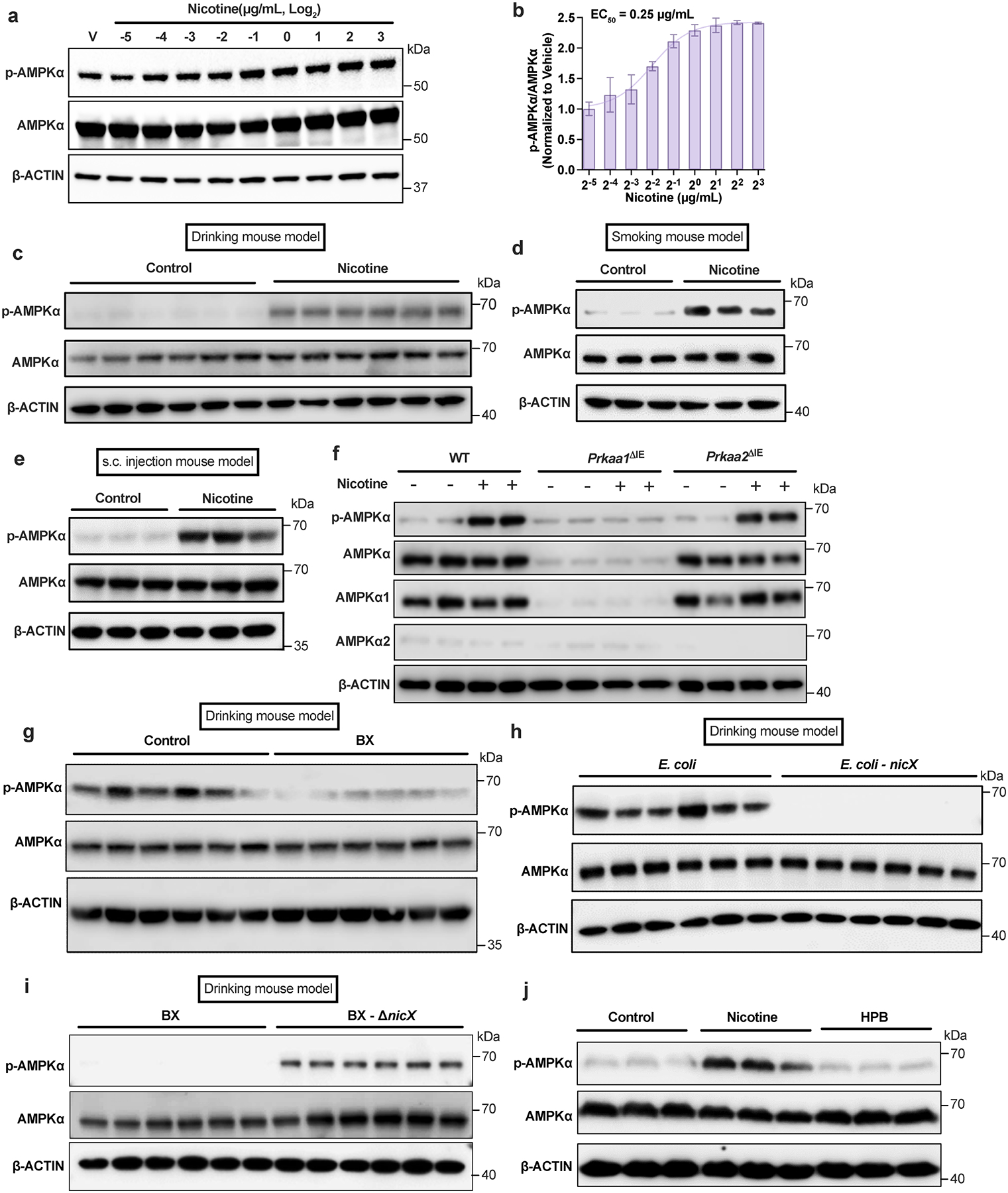
a, b, Activation of the AMPKα in ileal organoids after treatment with nicotine at different concentrations for 4 h (n = 3 independent experiments). c, Western blot analysis indicated that ileal AMPKα was activated in the nicotine drinking mouse model (SPF, n = 6 mice/group). d, e, Western blot analysis indicated that ileal AMPKα was activated in the smoking mouse model (d) and the subcutaneous injection mouse model (e) (SPF, n = 3 mice/group). f, Western blot analysis of ileal primary enterocytes isolated from WT, Prkaa1ΔIE and Prkaa2ΔIE mice (SPF) and then cultured with or without nicotine (1 μg/mL) treatment for 4 h. Experiments were performed with n = 4 mice/group. g, Western blot analysis showing ileal AMPK signaling in the nicotine drinking mouse model transplanted with Control or B. xylanisolvens (SPF, n = 6 mice/group). h, i, Western blot analysis showing ileal AMPK signaling in the nicotine drinking mouse model transplanted with E. coli or nicX knock-in E. coli (h); and WT or nicX knock-out B. xylanisolvens (i) (SPF, n = 6 mice/group). j, Western blot analysis showing AMPK signaling in SW480 cells incubated with nicotine (1 μg/mL) or HPB (1 μg/mL) for 4 h. This result is representative of 3 independent experiments. In c-e, g-i, mice were supplied nicotine plus HFHCD for two weeks. Data are the means ± s.e.m. (b).
Extended Data Fig. 5. Intestinal AMPKα1 deficiency improves NAFLD progression via downregulating ceramides generation.
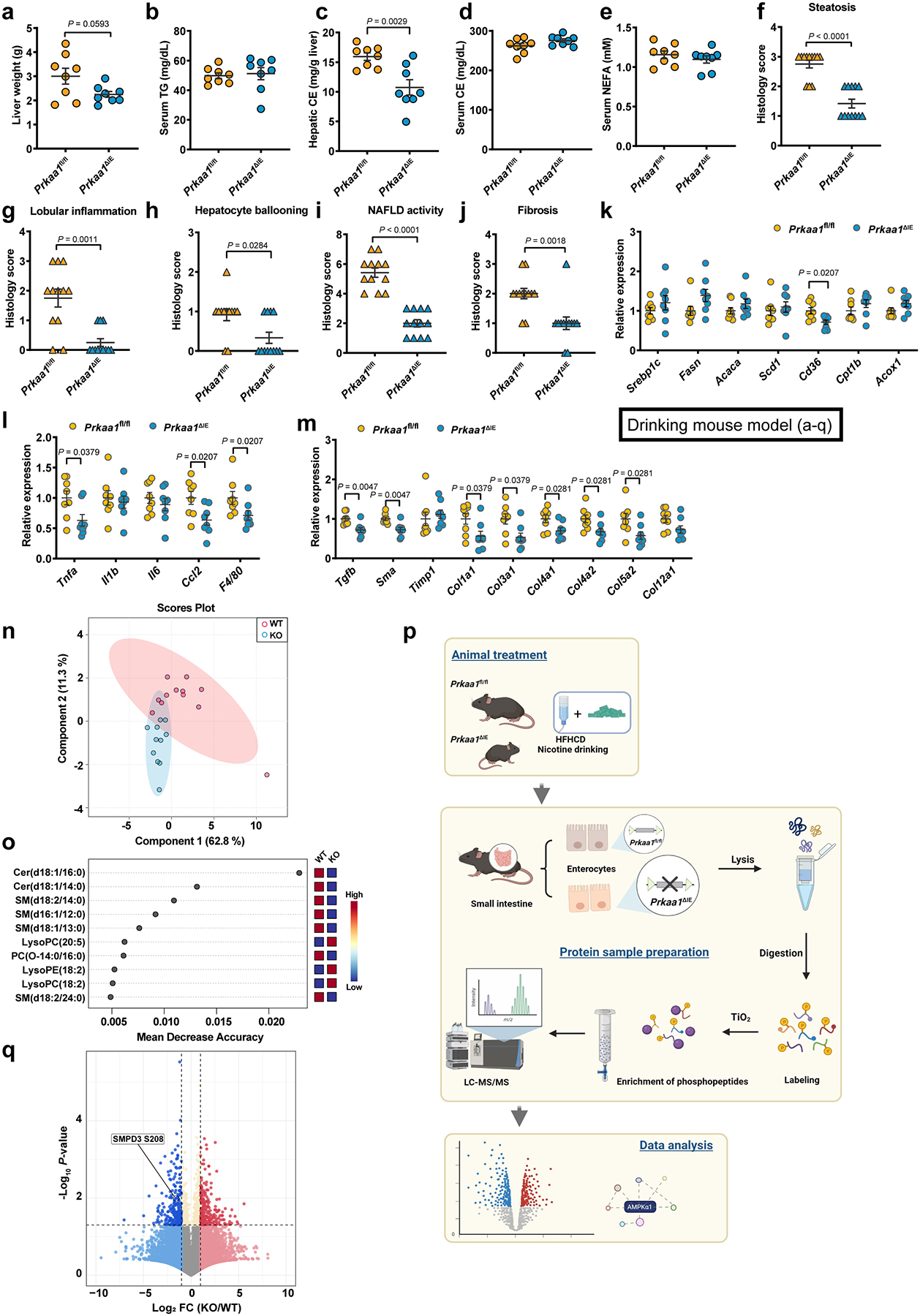
Eight-week-old male Prkaa1fl/fl and Prkaa1ΔIE mice were administered an HFHCD plus nicotine water for 20 weeks (SPF, n = 8 mice/group). a, Liver weight. b-e, Serum TG (b), hepatic CE (c), serum CE (d) and serum NEFA (e) contents. f-j, Histology scores of hepatic steatosis (f), lobular inflammation (g), ballooning (h), NAFLD activity (i), and fibrosis stage (j). k-m, Relative mRNA levels of genes related to hepatic lipid metabolism (k), inflammation (l) and fibrosis (m). n, o, Eight-week-old male Prkaa1fl/fl (WT) and Prkaa1ΔIE (KO) mice were administered an HFHCD plus nicotine water for 20 weeks (SPF, Prkaa1fl/fl, n = 12 mice; Prkaa1ΔIE, n = 11 mice). PLS-DA analysis of lipid metabolites in the ileum (n). Random forest analysis showing the top 10 lipid metabolites that lead to differences in the ileal lipid profiles (o). p, A schematic diagram illustrating the workflow of phosphorylated proteomics. Created with BioRender. q, Volcano map of phosphorylated proteomics analysis from ileal epithelia of Prkaa1fl/fl (WT) and Prkaa1ΔIE (KO) mice (n = 5 mice/group) administrated an HFHCD plus nicotine water for 20 weeks, relative fold change (log2) of phosphorylated sites abundance versus −log10 (P values), analysis from P values calculated by two-tailed t-test. Data are the means ± s.e.m. a-e, Two-tailed Student’s t-test. f-m, Two-tailed Mann- Whitney U-test.
Extended Data Fig. 6. AMPKα phosphorylates SMPD3 protein which became more stable through escaped from ubiquitination degradation.
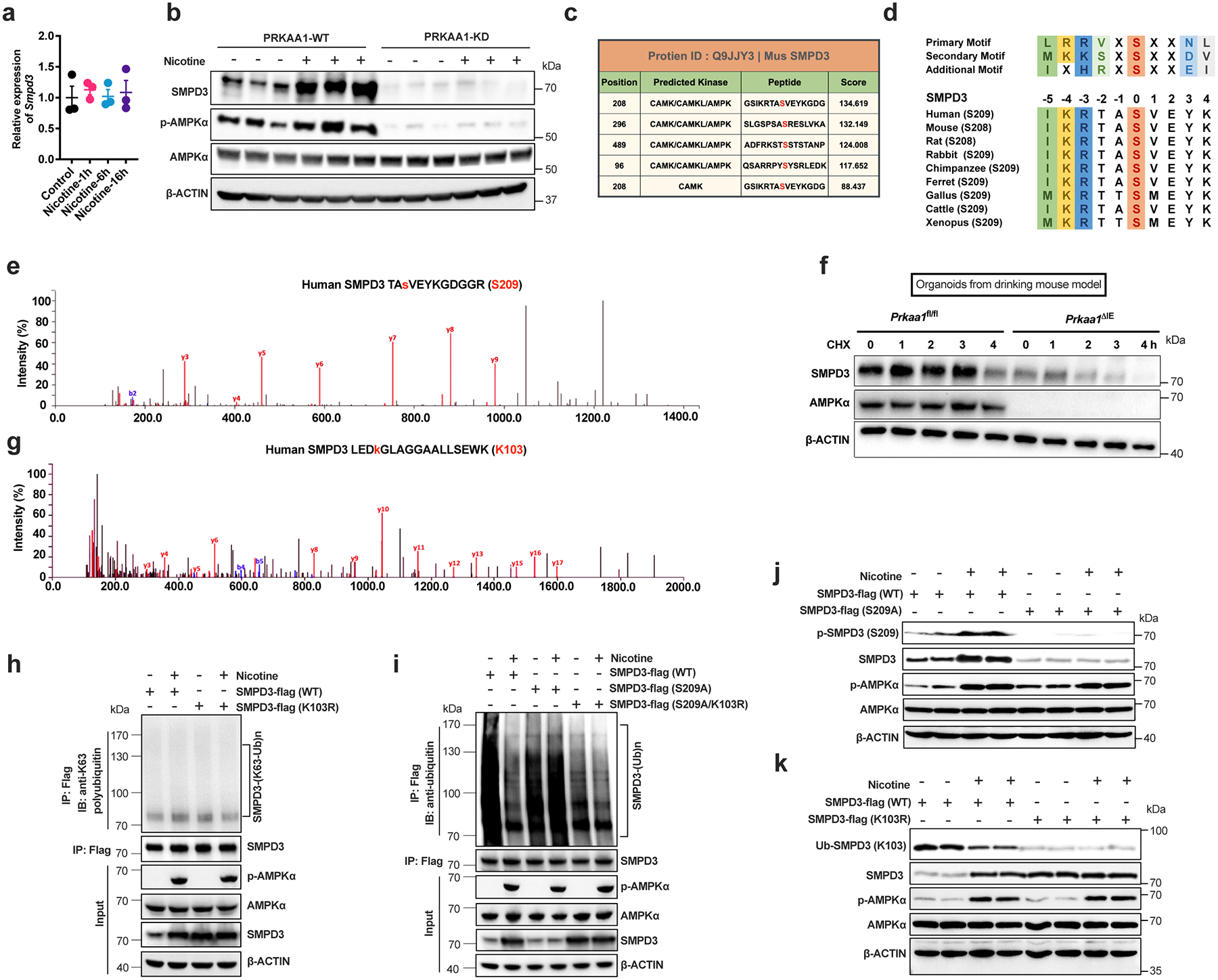
a, Nicotine (1 μg/mL) treatment on Smpd3 mRNA levels in ileal organoids (n = 3/group). b, PRKAA1-WT or PRKAA1-KD (kinase domain mutant) was introduced into SW480 cells, and the cells were then treated with vehicle or nicotine (1 μg/mL) for 12 h. c, GPS2.0 predicts potential kinases and phosphorylation sites for SMPD3. d, The S208/209 peptide of SMPD3 satisfied the AMPK substrate motif and was conserved in different species (data from NCBI database). e, Mass spectrometry analysis of the phosphorylation at S209 on SMPD3. f, HFHCD-fed Prkaa1fl/fl and Prkaa1ΔIE mice (SPF) were treated with nicotine water for 2 weeks, and organoids were then isolated and cultured for 7 days and treated with nicotine (1 μg/mL) for the last 3 days. Western blot analysis showing the stability of SMPD3 after the administration of CHX. g, Mass spectrometry analysis of the ubiquitination at K103 on SMPD3. h, The K63 ubiquitination of SMPD3 in SW480 cells transfected with SMPD3-flag (WT and K103R) with or without nicotine treatment. i, The SMPD3 ubiquitination in SW480 cells transfected with SMPD3-flag (WT, S209A or S209A/K103R) and treated with or without nicotine. j, Phosphorylation level (S209) of SMPD3 was detected by anti-p-SPMD3 (S209) antibody in SW480 cells transfected with SMPD3-flag (WT or S209A) and treated with or without nicotine for 24 h. k, Ubiquitination level (K103) of SMPD3 was detected by anti-ubi-SMPD3 (K103) antibody in SW480 cells transfected with SMPD3-flag (WT or K103R) and treated with or without nicotine for 24 h. For e, g-k, nicotine (1 μg/mL) treatment for 24 h. Data are the means ± s.e.m. Experiments in a, b, e-k were performed three times independently. a, One-way ANOVA with Tukey’s post hoc test.
Extended Data Fig. 7. Interaction between p-AMPKα and SMPD3 in intestinal ceramide production.
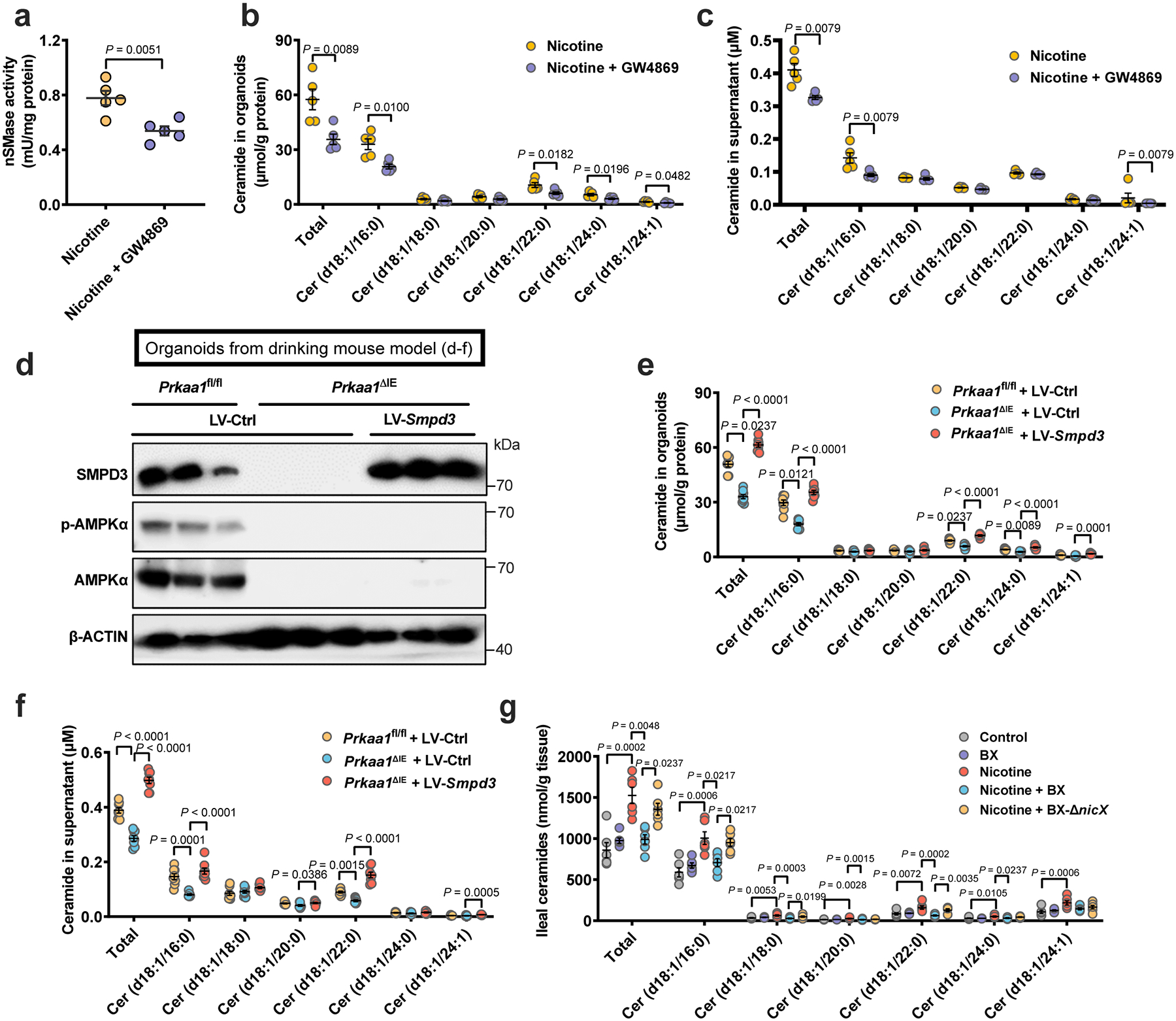
a-c, HFHCD-fed SPF mice were treated with nicotine water or nicotine water plus 10 mg/kg GW4869 (by daily gavage) for 2 weeks, and ileal organoids were then isolated and cultured for 7 days and treated with GW4869 (10 μM) and nicotine (1 μg/mL) for the last 3 days before the detection of ceramide production and secretion (n = 5 mice/group). a, nSMase activity. b, Ceramide profiles in isolated organoids. c, Ceramide profiles in the supernatant of isolated organoids. d-f, HFHCD-fed Prkaa1fl/fl and Prkaa1ΔIE mice (SPF) were treated with nicotine water for 2 weeks, and organoids were then isolated and infected with LV (lentivirus)-Ctrl or LV-Smpd3, the infected organoids were plated and cultured for 7 days and treated with nicotine (1 μg/mL) for the last 3 days before the detection of ceramide production and secretion. Western blot analysis for verifying SMPD3 overexpression. (n = 3 mice/group) (d). Ceramide profiles in isolated organoids. (n = 8 mice/group) (e). Ceramide profiles in the supernatant of isolated organoids. (n = 8 mice/group) (f). g, HFHCD-fed WT mice were transplanted with PBS, B. xylanisolvens colonization, nicotine water, nicotine water plus B. xylanisolvens colonization, and nicotine water plus nicX knock-out B. xylanisolvens colonization for 20 weeks (SPF, n = 6 mice/group), and ileal tissues were collected for the ceramide profiles detection. Data are the means ± s.e.m. a, b, Two-tailed Student’s t-test. c, Two-tailed Mann-Whitney U-test. f, One-way ANOVA with Tukey’s post hoc test. e, g, Kruskal-Wallis test with Dunn’s test.
Extended Data Fig. 8. Ceramide supplementation eliminates the beneficial effects derived from intestinal AMPKα1 deficiency.
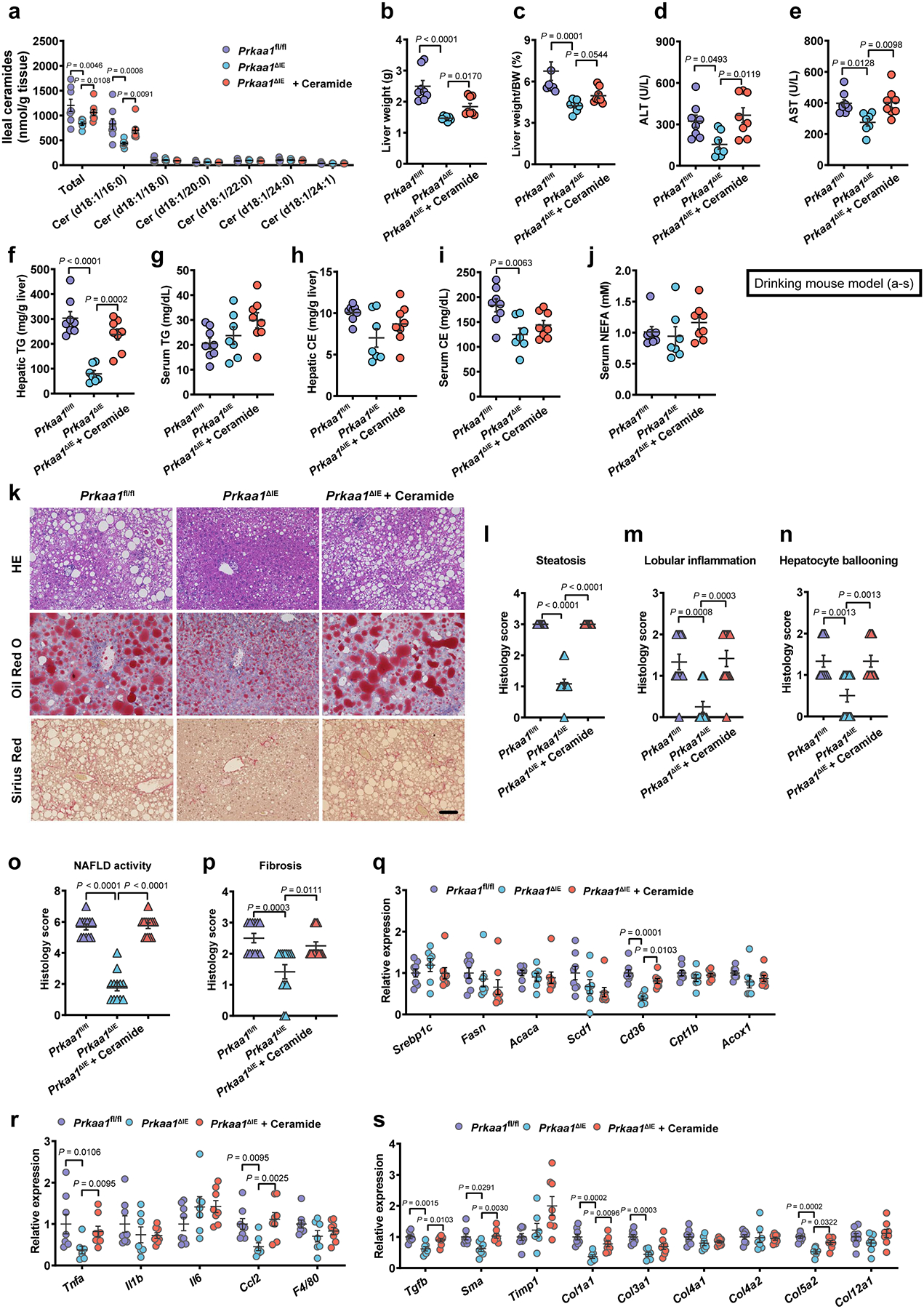
Eight-week-old male Prkaa1fl/fl and Prkaa1ΔIE mice were treated with or without 10 mg/kg ceramide (d18:1/16:0) by daily i.p. injection under HFHCD plus nicotine water treatment for 20 weeks (SPF, Prkaa1fl/fl, n = 8 mice; Prkaa1ΔIE, n = 7 mice; Prkaa1ΔIE + Ceramide, n = 8 mice). a, Ileal ceramide profiles. b, Liver weights. c, Liver weight–to–body weight ratios. d, e, Serum ALT (d) and AST (e) levels. f-j, Hepatic TG (f), serum TG (g), hepatic CE (h), serum CE (i) and serum NEFA (j) contents. k, Representative H&E staining (upper), Oil Red O staining (middle) and Sirius Red staining (lower) of liver sections (n = 4 mice/group, 3 images/mouse). Scale bar, 100 μm. l-p, Histology scores of steatosis (l), lobular inflammation (m), hepatocyte ballooning (n), NAFLD activity (o) and fibrosis stage (p). q-s, Relative mRNA levels of genes related to hepatic lipid metabolism (q), inflammation (r) and fibrosis (s). Data are the means ± s.e.m. d-g, i, One-way ANOVA with Tukey’s post hoc test. h, One-way ANOVA with Dunnett’s T3 post hoc test. a-c, j, l-s, Kruskal-Wallis test with Dunn’s test.
Extended Data Fig. 9. Inhibition of SMPD3 ameliorates nicotine-induced NAFLD progression.
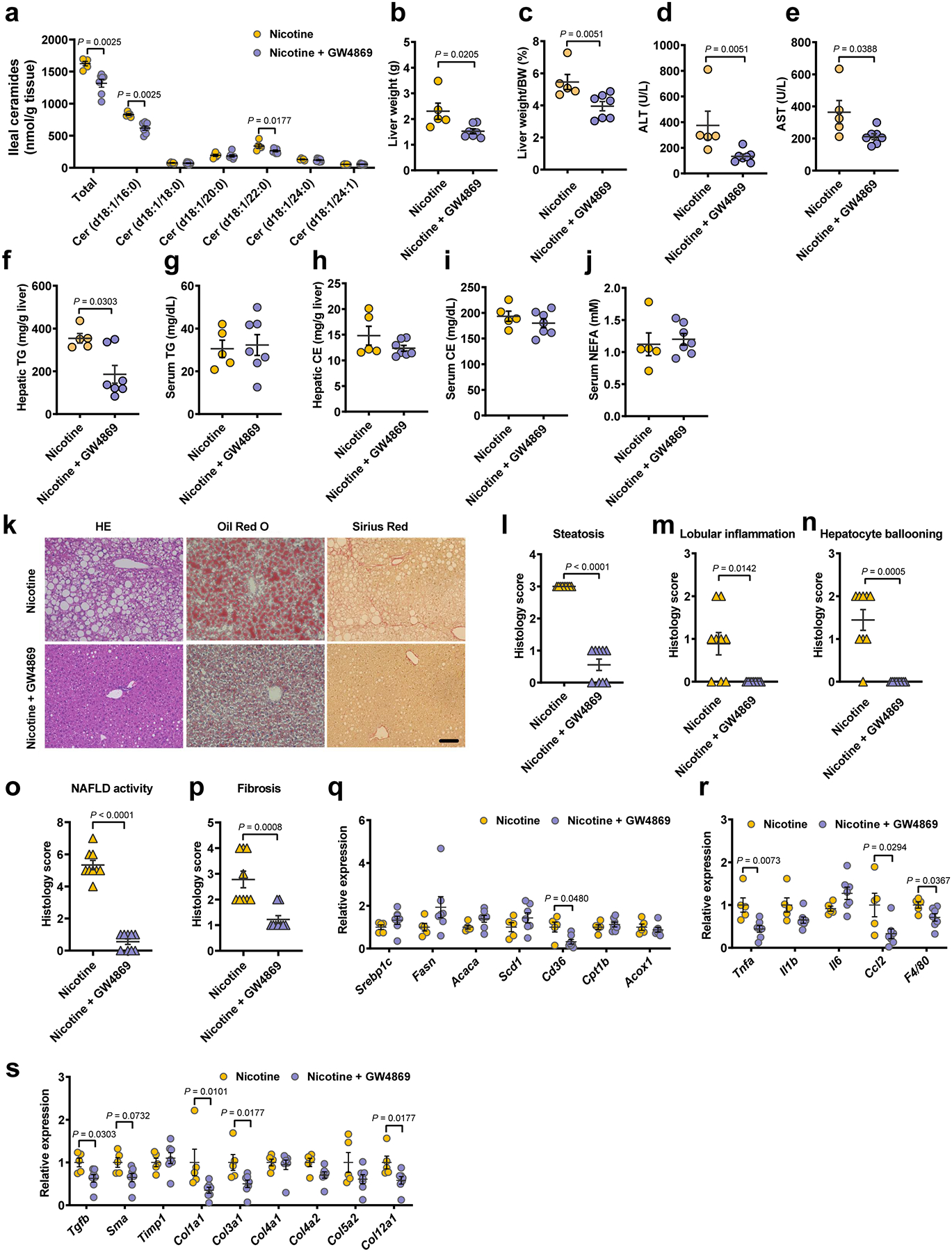
Eight-week-old male SPF mice were randomly grouped and received control or 10 mg/kg GW4869 by daily gavage under HFHCD plus nicotine water treatment for 20 weeks (Nicotine, n = 5 mice; Nicotine + GW4869, n = 7 mice). a, Ileal ceramide profiles. b, Liver weights. c, Liver weight–to–body weight ratios. d, e, Serum ALT (d) and AST (e) levels. f-j, Hepatic TG (f), serum TG (g), hepatic CE (h), serum CE (i) and serum NEFA (j) contents. k, Representative H&E staining (left), Oil Red O staining (middle) and Sirius Red staining (right) of liver sections (n = 3 mice/group, 3 images/mouse). Scale bar, 100 μm. l-p, Histology scores of steatosis (l), lobular inflammation (m), hepatocyte ballooning (n), NAFLD activity (o) and fibrosis stage (p). q-s, Relative mRNA levels of genes related to hepatic lipid metabolism (q), inflammation (r) and fibrosis (s). Data are the means ± s.e.m. b, e, g-j, r, Two-tailed Student’s t-test. a, c, d, f, l-q, s, Two-tailed Mann-Whitney U-test.
Extended Data Fig. 10. B. xylanisolvens-mediated nicotine degradation negatively correlates with clinical NAFLD progression.
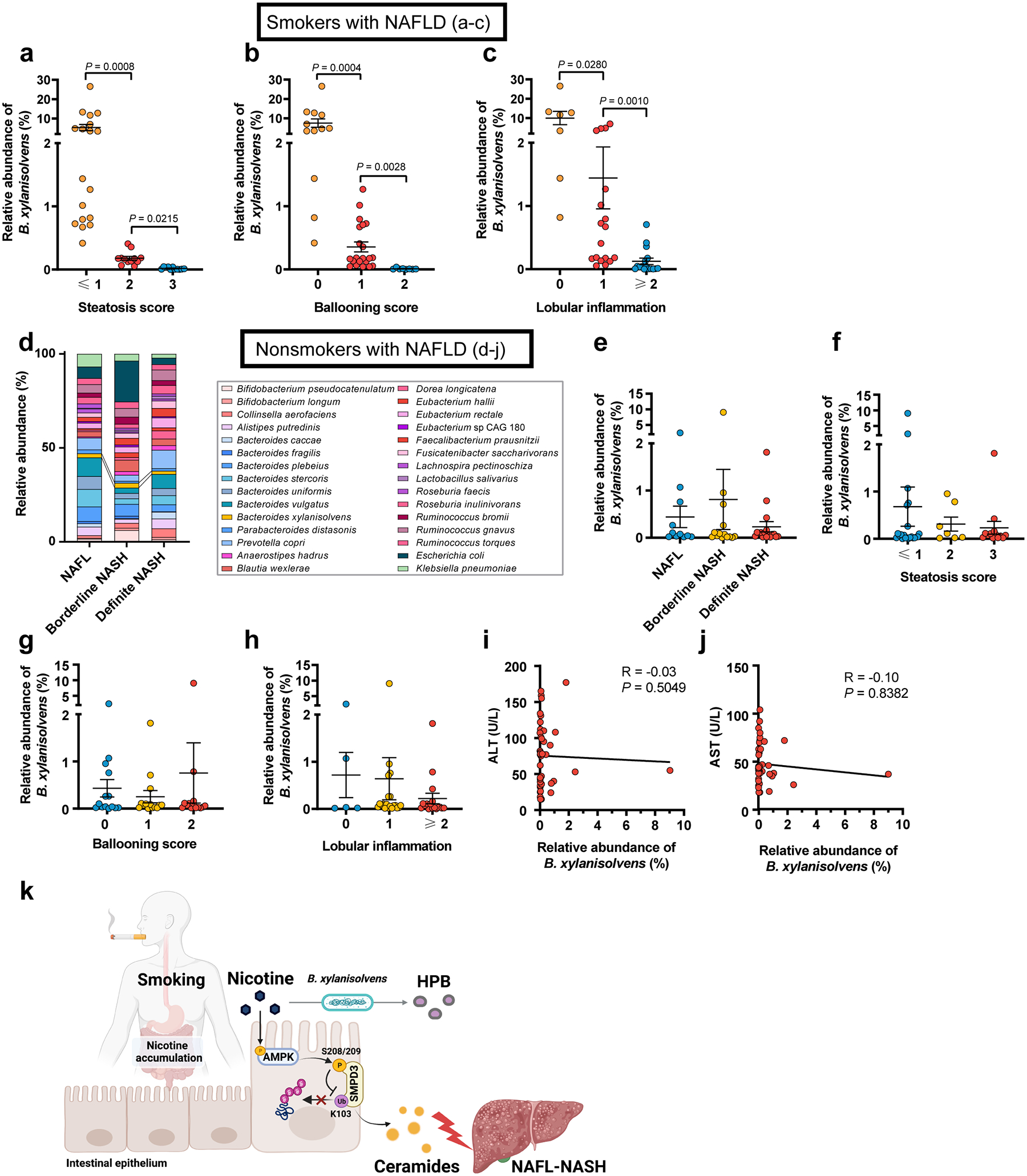
In 41 smokers with NAFLD, NAFL n = 11, borderline NASH n= 16, definite NASH n = 14. a-c, Relative abundances of B. xylanisolvens associated with steatosis score (a), ballooning score (b), and lobular inflammation (c) in smokers with NAFLD. In 42 nonsmokers with NAFLD, including NAFL (n = 11), borderline NASH (n = 14), and definite NASH (n = 17). d, Bacterial taxonomic profiling of the gut microbiota from nonsmokers with different NAFLD stages at the species level. e, Relative abundances of B. xylanisolvens associated with different NAFLD stages in nonsmokers with NAFLD. f-h, Relative abundances of B. xylanisolvens associated with steatosis score (f), ballooning score (g), and lobular inflammation (h) in nonsmokers with NAFLD. i, j, Correlative analysis of B. xylanisolvens with ALT (i) and AST (j). Correlations between variables were assessed by linear regression analysis. Linear correction index R square and P values were calculated. k, Summary diagram illustrating the role of microbial nicotine degradation in ceramide modulation and NAFL-NASH progression. Created with BioRender. Data are the means ± s.e.m. a-c, e-h, Kruskal-Wallis test with Dunn’s test.
Supplementary Material
Acknowledgements.
This work was supported by the National Natural Science Foundation of the P. R. of China (No. 91857115, 31925021), the National Key Research and Development Program of China (No. 2018YFA0800700), the National Natural Science Foundation of the P. R. of China (No. 82130022, 81921001, 92057103, 31872820, and 82070588), the National Cancer Institute Intramural Research Program, the National Natural Science Foundation of the P. R. of China (No. 92057103, and 31872820), the National Key Research and Development Program of China (No. 2022ZD0213000), and the Innovative Research Team of High-Level Local Universities in Shanghai and a Key Laboratory Program of the Education Commission of Shanghai Municipality (ZDSYS14005).
Footnotes
Competing interest declaration. The authors have declared that no conflicts of interest exist.
Supplementary Information is available for this paper.
Data availability
All the data that support the findings of this study are included in the published article and Supplementary Tables. The metagenomic and phosphoproteomic data were uploaded to public database (China National Microbiology Data Center (NMDC), with accession numbers as NMDC10018157, and NMDC10018158, respectively). The following public databases were applied in this study: NCBI reference database, Metaquery database.
References
- 1.Okamoto M et al. Cigarette smoking is a risk factor for the onset of fatty liver disease in nondrinkers: A longitudinal cohort study. PLoS One 13, e0195147, doi: 10.1371/journal.pone.0195147 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ou H, Fu Y, Liao W, Zheng C & Wu X Association between Smoking and Liver Fibrosis among Patients with Nonalcoholic Fatty Liver Disease. Can J Gastroenterol Hepatol 2019, 6028952, doi: 10.1155/2019/6028952 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Jung HS et al. Smoking and the Risk of Non-Alcoholic Fatty Liver Disease: A Cohort Study. Am J Gastroenterol 114, 453–463, doi: 10.1038/s41395-018-0283-5 (2019). [DOI] [PubMed] [Google Scholar]
- 4.Takenaka H et al. Non-Alcoholic Fatty Liver Disease Is Strongly Associated with Smoking Status and Is Improved by Smoking Cessation in Japanese Males: A Retrospective Study. Kobe J Med Sci 66, E102–E112 (2020). [PMC free article] [PubMed] [Google Scholar]
- 5.Yuan S et al. Lifestyle and metabolic factors for nonalcoholic fatty liver disease: Mendelian randomization study. Eur J Epidemiol, doi: 10.1007/s10654-022-00868-3 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.WHO. WHO report on the global tobacco epidemic 2008: the MPOWER oackage, 14 (2008). [Google Scholar]
- 7.WHO. WHO global report on trends in prevalence of tobacco smoking 2000–2025, second edition., 21 (2018). [Google Scholar]
- 8.Holford TR et al. Tobacco control and the reduction in smoking-related premature deaths in the United States, 1964–2012. JAMA 311, 164–171, doi: 10.1001/jama.2013.285112 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Xue S, Schlosburg JE & Janda KD A new strategy for smoking cessation: characterization of a bacterial enzyme for the degradation of nicotine. Journal of the American Chemical Society 137, 10136–10139, doi: 10.1021/jacs.5b06605 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dulchavsky M, Clark CT, Bardwell JCA & Stull F A cytochrome c is the natural electron acceptor for nicotine oxidoreductase. Nature Chemical Biology 17, 344–350, doi: 10.1038/s41589-020-00712-3 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tripathi A et al. The gut-liver axis and the intersection with the microbiome. Nat Rev Gastroenterol Hepatol 15, 397–411, doi: 10.1038/s41575-018-0011-z (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lindell G et al. Acute effects of smoking during modified sham feeding in duodenal ulcer patients. An analysis of nicotine, acid secretion, gastrin, catecholamines, epidermal growth factor, prostaglandin E2, and bile acids. Scand J Gastroenterol 28, 487–494, doi: 10.3109/00365529309098254 (1993). [DOI] [PubMed] [Google Scholar]
- 13.Han XJ et al. Stimulation of alpha 7 Nicotinic Acetylcholine Receptor by Nicotine Suppresses Decidual M1 Macrophage Polarization Against Inflammation in Lipopolysaccharide-Induced Preeclampsia-Like Mouse Model. Frontiers in Immunology 12, doi: 10.3389/fimmu.2021.642071 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sousa MV et al. Smoking accelerates renal cystic disease and worsens cardiac phenotype in Pkd1-deficient mice. Scientific Reports 11, doi: 10.1038/s41598-021-93633-7 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wu XX et al. Nicotine promotes atherosclerosis via ROS-NLRP3-mediated endothelial cell pyroptosis. Cell Death & Disease 9, doi: 10.1038/s41419-017-0257-3 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fluhr L et al. Gut microbiota modulates weight gain in mice after discontinued smoke exposure. Nature 600, 713–+, doi: 10.1038/s41586-021-04194-8 (2021). [DOI] [PubMed] [Google Scholar]
- 17.Wang SN, Liu Z, Tang HZ, Meng J & Xu P Characterization of environmentally friendly nicotine degradation by Pseudomonas putida biotype A strain S16. Microbiology-(UK) 153, 1556–1565, doi: 10.1099/mic.0.2006/005223-0 (2007). [DOI] [PubMed] [Google Scholar]
- 18.Tang HZ et al. A novel gene, encoding 6-hydroxy-3-suceinoylpyridine hydroxylase, involved in nicotine degradation by Pseudomonas putida strain S16. Applied and Environmental Microbiology 74, 1567–1574, doi: 10.1128/aem.02529-07 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wang C et al. Nicotine Accelerates Atherosclerosis in Apolipoprotein E-Deficient Mice by Activating alpha 7 Nicotinic Acetylcholine Receptor on Mast Cells. Arteriosclerosis Thrombosis and Vascular Biology 37, 53–+, doi: 10.1161/atvbaha.116.307264 (2017). [DOI] [PubMed] [Google Scholar]
- 20.Liu R, Kurose T & Matsukura S Oral nicotine administration decreases tumor necrosis factor-alpha expression in fat tissues in obese rats. Metabolism-Clinical and Experimental 50, 79–85, doi: 10.1053/meta.2001.19436 (2001). [DOI] [PubMed] [Google Scholar]
- 21.Wu Y et al. Activation of AMPKα2 in adipocytes is essential for nicotine-induced insulin resistance in vivo. Nat Med 21, 373–382, doi: 10.1038/nm.3826 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Garcia D & Shaw RJ AMPK: Mechanisms of cellular energy sensing and restoration of metabolic balance. Mol Cell 66, 789–800, doi: 10.1016/j.molcel.2017.05.032 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Marra F Lipotoxicity and the gut-liver axis in NASH pathogenesis. Journal of hepatology 68, 280–295, doi: 10.1016/j.jhep.2017.11.014 (2018). [DOI] [PubMed] [Google Scholar]
- 24.Xue Y et al. GPS 2.0, a tool to predict kinase-specific phosphorylation sites in hierarchy. Mol Cell Proteomics 7, 1598–1608, doi: 10.1074/mcp.M700574-MCP200 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Filosto S, Ashfaq M, Chung S, Fry W & Goldkorn T Neutral sphingomyelinase 2 activity and protein stability are modulated by phosphorylation of five conserved serines. J Biol Chem 287, 514–522, doi: 10.1074/jbc.M111.315481 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wu Q et al. Suppressing the intestinal farnesoid X receptor/sphingomyelin phosphodiesterase 3 axis decreases atherosclerosis. J Clin Invest 131, doi: 10.1172/JCI142865 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lindell G, Lunell E & Graffner H Transdermally administered nicotine accumulates in gastric juice. European Journal of Clinical Pharmacology 51, 315–318, doi: 10.1007/s002280050204 (1996). [DOI] [PubMed] [Google Scholar]
- 28.Mu Y et al. Bacterial catabolism of nicotine: catabolic strains, pathways and modules. Environ Res 183, 109258, doi: 10.1016/j.envres.2020.109258 (2020). [DOI] [PubMed] [Google Scholar]
- 29.Gunasekaran M Direct evidence that sunbirds’ gut microbiota degrades floral nectar’s toxic alkaloids. Frontiers in microbiology 12, doi: 10.3389/fmicb.2021.639808 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Agostoni C et al. Scientific Opinion on the safety of ‘heat-treated milk products fermented with Bacteroides xylanisolvens DSM 23964’ as a novel food EFSA Panel on Dietetic Products, Nutrition and Allergies (NDA). Efsa Journal 13, doi: 10.2903/j.efsa.2015.3956 (2015). [DOI] [Google Scholar]
- 31.Lavrynenko O et al. Ceramide ratios are affected by cigarette smoke but not heat-not-burn or e-vapor aerosols across four independent mouse studies. Life Sciences 263, 118753, doi: 10.1016/j.lfs.2020.118753 (2020). [DOI] [PubMed] [Google Scholar]
- 32.Tippetts TS et al. Cigarette smoke increases cardiomyocyte ceramide accumulation and inhibits mitochondrial respiration. BMC Cardiovascular Disorders 14, 165, doi: 10.1186/1471-2261-14-165 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhou YJ et al. Screening for compensated advanced chronic liver disease using refined Baveno VI elastography cutoffs in Asian patients with nonalcoholic fatty liver disease. Alimentary Pharmacology & Therapeutics 54, 470–480, doi: 10.1111/apt.16487 (2021). [DOI] [PubMed] [Google Scholar]
- 34.Kleiner DE et al. Design and validation of a histological scoring system for nonalcoholic fatty liver disease. Hepatology 41, 1313–1321, doi: 10.1002/hep.20701 (2005). [DOI] [PubMed] [Google Scholar]
- 35.Lloyd-Jones DM et al. Framingham risk score and prediction of lifetime risk for coronary heart disease. American Journal of Cardiology 94, 20–24, doi: 10.1016/j.amjcard.2004.03.023 (2004). [DOI] [PubMed] [Google Scholar]
- 36.Hippisley-Cox J, Coupland C & Brindle P Development and validation of QRISK3 risk prediction algorithms to estimate future risk of cardiovascular disease: prospective cohort study. Bmj-British Medical Journal 357, doi: 10.1136/bmj.j2099 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Everard A et al. Cross-talk between Akkermansia muciniphila and intestinal epithelium controls diet-induced obesity. Proceedings of the National Academy of Sciences of the United States of America 110, 9066–9071, doi: 10.1073/pnas.1219451110 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wu Q et al. Intestinal hypoxia-inducible factor 2 alpha regulates lactate levels to shape the gut microbiome and alter thermogenesis. Cell Metabolism 33, 1988–+, doi: 10.1016/j.cmet.2021.07.007 (2021). [DOI] [PubMed] [Google Scholar]
- 39.Bolger AM, Lohse M & Usadel B Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30, 2114–2120, doi: 10.1093/bioinformatics/btu170 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Truong DT et al. MetaPhlAn2 for enhanced metagenomic taxonomic profiling. Nature Methods 12, 902–903, doi: 10.1038/nmeth.3589 (2015). [DOI] [PubMed] [Google Scholar]
- 41.Dhariwal A et al. MicrobiomeAnalyst: a web-based tool for comprehensive statistical, visual and meta-analysis of microbiome data. Nucleic Acids Research 45, W180-W188, doi: 10.1093/nar/gkx295 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Segata N et al. Metagenomic biomarker discovery and explanation. Genome Biology 12, doi: 10.1186/gb-2011-12-6-r60 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Liu C et al. Enlightening the taxonomy darkness of human gut microbiomes with a cultured biobank. Microbiome 9, 119, doi: 10.1186/s40168-021-01064-3 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Despres J et al. Unraveling the pectinolytic function of Bacteroides xylanisolvens using a RNA-seq approach and mutagenesis. BMC Genomics 17, 147, doi: 10.1186/s12864-016-2472-1 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Apsunde TD & Trudell ML Microwave-Assisted Iridium-Catalyzed Synthesis of Nicotine and Anabasine Derivatives. Synthesis-Stuttgart 45, 2120–2124, doi: 10.1055/s-0032-1316859 (2013). [DOI] [Google Scholar]
- 46.Dye FS et al. Characterisation of proguanylin expressing cells in the intestine evidence for constitutive luminal secretion. Scientific Reports 9, doi: 10.1038/s41598-019-52049-0 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Xuan QH et al. Development of a High Coverage Pseudotargeted Lipidomics Method Based on Ultra-High Performance Liquid Chromatography-Mass Spectrometry. Analytical Chemistry 90, 7608–7616, doi: 10.1021/acs.analchem.8b01331 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ren LL et al. TiO2 with Tandem Fractionation (TAFT): An Approach for Rapid, Deep, Reproducible, and High-Throughput Phosphoproteome Analysis. Journal of Proteome Research 17, 710–721, doi: 10.1021/acs.jproteome.7b00520 (2018). [DOI] [PubMed] [Google Scholar]
- 49.Liu P et al. Cell-cycle-regulated activation of Akt kinase by phosphorylation at its carboxyl terminus. Nature 508, 541–545, doi: 10.1038/nature13079 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Waterhouse A et al. SWISS-MODEL: homology modelling of protein structures and complexes. Nucleic Acids Research 46, W296–W303, doi: 10.1093/nar/gky427 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All the data that support the findings of this study are included in the published article and Supplementary Tables. The metagenomic and phosphoproteomic data were uploaded to public database (China National Microbiology Data Center (NMDC), with accession numbers as NMDC10018157, and NMDC10018158, respectively). The following public databases were applied in this study: NCBI reference database, Metaquery database.


