Abstract
We report a PCR-based assay for the detection of Enterocytozoon bieneusi. We extracted DNA from feces which had been applied to filter paper disks and evaluated four preserving solutions. Infected specimens were identified by electrophoresis of amplicons from concentrated formalin-fixed samples and unconcentrated fresh feces. Our findings demonstrate that this methodology is effective for sample collection, mailing, and diagnosis of this pathogen.
The microsporidian species Enterocytozoon bieneusi and Encephalitozoon intestinalis have been associated with chronic diarrhea and wasting in human immunodeficiency virus-infected patients (2, 12, 40). Diagnosis of gastrointestinal micro sporidiosis depends on direct visualization of spores in stained fecal samples and species differentiation by transmission electron microscopy (6, 40, 41). Identification to the species level is clinically of great value because of the differences in therapy response (1, 13, 26). Most studies involve the use of PCR for detection of E. bieneusi in fresh or formalin-fixed stool specimens (4, 7, 8, 9, 14, 15, 17–20, 28, 29, 33, 37, 38).
Epidemiological investigation of microsporidia has been limited by difficulties in the preservation and transport of feces and diagnostic methods applied to them. Samples collected on a filter paper are a valuable source of DNA for amplification-based methods and may provide a practical solution to many of these problems (3, 21, 30).
The purpose of this study was to describe a PCR assay for E. bieneusi employing stool samples collected in different preserving solutions as impregnated filter paper disks (FPD).
Stool samples and intestinal biopsy specimens were obtained from three human immunodeficiency virus-infected patients with chronic diarrhea and microsporidiosis caused by E. bieneusi. Microsporidial infections were diagnosed using multiple formalinized stool specimens by light-microscopic examination of samples stained according to Weber's modified trichrome method (40) and the quick-hot Gram-chromotrope technique (27). Five biopsy specimens from each patient were obtained from the distal duodenum by flexible fiber-optic endoscopy. Two specimens were fixed in 10% formalin for routine histology and were stained with Giemsa and hematoxylin-eosin. Two other specimens were processed for transmission electron microscopy (11, 31) to confirm E. bieneusi infections. The fifth biopsy specimen was stored in saline solution at −20°C for DNA purification. Stool samples were collected in four solutions: (i) 5% formaldehyde (volume ratio of stool to formalin, 1:3), (ii) 0.05% saline solution, (iii) 2.5% potassium dichromate, and (iv) Merthiolate-formalin (MF) (36) (volume ratio of stool to each of these three solutions, 1:2). FPD (Whatman no. 1) 1 cm in diameter were used to store the fecal specimens. After homogenization, samples were spotted on the FPD in two different forms: as suspensions of homogeneous material and as centrifugation pellets following ethyl ether extraction (34). For pellet spotting, samples were resuspended in the remaining solution and 100 μl was pipetted onto each disk. The FPD were air dried and stored in individual plastic bags at 4°C for at least 6 months.
Each FPD was incubated for 2 h at 56°C in 500 μl of lysis buffer (100 mM Tris-HCl [pH 8.0], 100 mM EDTA [pH 8.0], 2% sodium dodecyl sulfate, 150 mM NaCl, and 200 μg of proteinase K/ml). The DNA was purified by phenol-chloroform extractions and precipitation with absolute ethanol and was resuspended in 10 μl of sterile redistilled water.
The primers Eb.gc (5′-TCAGTTTTGGGTGTGGTATCGG-3′) and Eb.gt (5′-GCTACCCATACACACATCATTC-3′) (38) were used to amplify a 210-bp fragment of the unique rRNA intergenic spacer sequence of E. bieneusi (GenBank accession no. L20290) (43). All PCRs were carried out as previously described (38). Five microliters of DNA purified from each FPD was employed in this assay. The amplification procedure included 30 s of denaturation at 98°C in the first cycle (94°C afterwards) followed by 30 s of annealing at 49°C and 90 s of extension at 72°C for 36 cycles. After the first PCR, 10 μl of amplified material was used to run a second PCR under the same conditions and with the same primers as the first PCR. The amplified material (20 μl) was fractionated by 1.5% agarose gel electrophoresis and visualized by ethidium bromide staining. After the first round of PCR amplification, the analysis was performed on 12.5% polyacrylamide gels (22) followed by silver staining (PlusOne DNA silver stain kit; Pharmacia, Uppsala, Sweden).
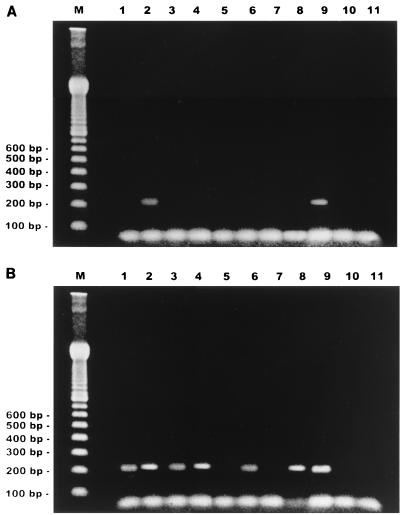
The first round of PCR produced the expected DNA fragments on ethidium bromide- and silver-stained gels when DNA from concentrated fresh feces on FPD was used as a template (Fig. 1A). No bands were detected when DNA from uncentrifuged stool samples or specimens collected in preservative solutions was used. A second round of PCR resulted in efficient amplification of all non-formalin-fixed specimens, independent of the method used to apply feces to the membrane (Fig. 1B, lanes 1 to 4). Higher amounts of product from concentrated samples than from those placed on FPD as suspensions were observed for both fresh feces and potassium dichromate-preserved specimens. Amplicons of 210 bp could also be detected in the second round of PCR amplification using DNA prepared from formalin-fixed samples that had been ethyl ether extracted before FPD spotting (Fig. 1B, lane 6). No PCR products from nonconcentrated fixed specimens could be observed (Fig. 1B, lane 5). The same results were obtained with templates from samples collected in MF solution (Fig. 1B, lanes 7 and 8). Silver staining did not improve the results of the second PCR assay (data not shown). All results were identical for samples obtained from the three studied patients. DNA purified from frozen biopsy specimens of an E. bieneusi-infected patient was used as a positive control. DNA from duodenal biopsy specimens from known E. bieneusi-negative individuals was employed as a negative control in all assays.
FIG. 1.
Agarose gel electrophoresis (ethidium bromide stain) of PCR products. (A) First round of amplification; (B) double PCR amplification. Lanes 1 and 2, fresh sample; lanes 3 and 4, specimen in 2.5% potassium dichromate solution; lanes 5 and 6, formalin-fixed sample; lanes 7 and 8, specimen preserved in MF. Lanes 1, 3, 5, and 7, homogeneous suspension; lanes 2, 4, 6, and 8, concentrated sample. Lanes 9 and 10, positive and negative controls, respectively; lane 11, reaction mixture; lane M, 100-bp ladder.
The epidemiology of E. bieneusi infection has not been completely elucidated, and more information is needed on clinical features in HIV-infected patients and in other hosts, the impact of antiretroviral treatment, and geographical, environmental, and seasonal factors (5, 23, 40). Several PCR protocols for E. bieneusi have used fresh stool samples that were frozen or stored and transported at ambient temperatures for immediate processing (10, 32, 33, 37). Field conditions may limit the handling, transport, and refrigeration of the specimens. Other investigators have employed formalin-fixed fecal specimens, which can be transported and stored, but at present there is no information on the use of other fixatives for fecal specimens to be tested by microsporidial PCR protocols (4, 9, 14, 29).
In our study, examination of products after the first round of PCR showed good amplification with DNA from centrifuged fresh feces spotted on FPD. Although some authors used different procedures to remove PCR inhibitors (15, 25, 36, 42), our results demonstrated that such removal was unnecessary with stool samples eluted from FPD. Negative results observed with preserved specimens showed a low level of DNA recovery. When fecal specimens are preserved in formalin, it is very difficult to amplify the DNA of microsporidial spores because this fixative reacts with the DNA (14). This possibility may be considered for the other fixatives employed (14, 16).
Our findings clearly show the advantage of our double PCR method and confirm that different preserved or fresh specimens can be employed with good diagnostic results. The filter paper specimens facilitate collection, transport, and storage because they have little weight, are cost effective, require minimal storage space, and maintain stable DNA during long periods (3, 24, 39). This study extends the diagnostic possibilities to laboratories employing all the usual collection procedures. Moreover, positive results obtained with unconcentrated fresh samples and the use of FPD as specimen support add great value to this technique because fecal specimens may be collected by untrained persons and mailed to a central diagnostic laboratory for identification.
Acknowledgments
We express our appreciation to colleagues who advised and assisted with the study: José M. Peralta, from the Instituto de Microbiologia, CCS, Universidade Federal de Rio de Janeiro, Rio de Janeiro, Brazil; Raúl Franco, from the Departamento de Bacteriología, Instituto Nacional de Microbiología “Dr. Carlos G. Malbrán,” Buenos Aires, Argentina, and Fernando Sodré, from the Universidade Federal Fluminense, Niteroi, Brazil.
REFERENCES
- 1.Blanshard C, Ellis D S, Tovey D G, Dowell S, Gazzard B G. Treatment of intestinal microsporidiosis with albendazole in patients with AIDS. AIDS. 1992;6:311–313. doi: 10.1097/00002030-199203000-00009. [DOI] [PubMed] [Google Scholar]
- 2.Cali A, Kotler D P, Orenstein J M. Septata intestinalis n.g., n.sp., an intestinal microsporidian associated with chronic diarrhea and dissemination in AIDS patients. J Eukaryot Microbiol. 1993;40:101–112. doi: 10.1111/j.1550-7408.1993.tb04889.x. [DOI] [PubMed] [Google Scholar]
- 3.Carducci C, Ellul L, Antonozzi I, Pontecorvi A. DNA elution and amplification by polymerase chain reaction from dried blood spots. BioTechniques. 1992;13:735–737. [PubMed] [Google Scholar]
- 4.Carville A, Mansfield K, Widmer G, Lackner A, Kotler D, Wiest P, Gumbo T, Sarbah S, Tzipori S. Development and application of genetic probes for detection of Enterocytozoon bieneusi in formalin-fixed stools and in intestinal biopsy specimens from infected patients. Clin Diagn Lab Immunol. 1997;4:405–408. doi: 10.1128/cdli.4.4.405-408.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Conteas C N, Berlin G W, Speck C E, Pandhumas S S, Lariviere M J, Fu C. Modification of the clinical course of intestinal microsporidiosis in acquired immunodeficiency syndrome patients by immune status and anti-human immunodeficiency virus therapy. Am J Trop Med Hyg. 1998;58:555–558. doi: 10.4269/ajtmh.1998.58.555. [DOI] [PubMed] [Google Scholar]
- 6.Conteas C N, Sowerby T, Berlin G W, Speck C E, Pandhumas S S, Lariviere M J, Fu C. Fluorescence techniques for diagnosing intestinal microsporidiosis in stool, enteric fluid, and biopsy specimens from acquired immunodeficiency syndrome patients with chronic diarrhea. Arch Pathol Lab Med. 1996;120:847–853. [PubMed] [Google Scholar]
- 7.Coyle C M, Wittner M, Kotler D P, Noyer C, Orenstein J M, Tanowitz H B, Weiss L M. Prevalence of microsporidiosis due to Enterocytozoon bieneusi and Encephalitozoon (Septata) intestinalis among patients with AIDS-related diarrhea: determination by polymerase chain reaction to the microsporidian small-subunit rRNA gene. Clin Infect Dis. 1996;23:1002–1006. doi: 10.1093/clinids/23.5.1002. [DOI] [PubMed] [Google Scholar]
- 8.da Silva A J, Schwartz D A, Visvesvara G S, de Moura H, Slemenda S B, Pieniazek N J. Sensitive PCR diagnosis of infections by Enterocytozoon bieneusi (microsporidia) using primers based on the region coding for small-subunit rRNA. J Clin Microbiol. 1996;34:986–987. doi: 10.1128/jcm.34.4.986-987.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.da Silva A J, Bornay-Llinares F J, del Aguila de la Puente C, Moura H, Peralta J M, Sobottka I, Schwartz D A, Visvesvara G S, Slemenda S B, Pieniazek N J. Diagnosis of Enterocytozoon bieneusi (microsporidia) infections by polymerase chain reactions in stool samples using primers based on the region coding for small-subunit ribosomal RNA. Arch Pathol Lab Med. 1997;121:874–879. [PubMed] [Google Scholar]
- 10.del Aguila C, Navajas R, Gurbindo D, Ramos J T, Mellado M J, Fenoy S, Muñoz Fernandez M A, Subirats M, Ruiz J, Pieniazek N J. Microsporidiosis in HIV-positive children in Madrid (Spain) J Eukaryot Microbiol. 1997;44:845–855. doi: 10.1111/j.1550-7408.1997.tb05798.x. [DOI] [PubMed] [Google Scholar]
- 11.Desportes I, Hilmarsdottir I, Romaña C, Tanguy S, Datry A, Gentilini M. Characteristics of the microsporidian Enterocytozoon bieneusi: a consequence of its development within short-living enterocytes. J Protozool. 1991;38:111–113. [PubMed] [Google Scholar]
- 12.Desportes I, Le Charpentier Y, Galian A, Bernard F, Cochand-Priollet B, Lavergne A, Ravisse P, Modigliani R. Occurrence of a new microsporidian, Enterocytozoon bieneusi n.g., n.sp., in the enterocytes of a human patient with AIDS. J Protozool. 1985;32:250–254. doi: 10.1111/j.1550-7408.1985.tb03046.x. [DOI] [PubMed] [Google Scholar]
- 13.Dieterich D T, Len E A, Kotler D P, Poles M A, Orenstein J M. Treatment with albendazole for intestinal disease due to Enterocytozoon bieneusi in patients with AIDS. J Infect Dis. 1994;169:178–183. doi: 10.1093/infdis/169.1.178. [DOI] [PubMed] [Google Scholar]
- 14.Dowd S E, Gerba C P, Enriquez F J, Pepper I L. PCR amplification and species determination of microsporidia in formalin-fixed feces after immunomagnetic separation. Appl Environ Microbiol. 1998;64:333–336. doi: 10.1128/aem.64.1.333-336.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fedorko D P, Nelson N A, Cartwright C P. Identification of microsporidia in stool specimens by using PCR and restriction endonucleases. J Clin Microbiol. 1995;33:1739–1741. doi: 10.1128/jcm.33.7.1739-1741.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Johnson D W, Pieniazek N J, Griffin D W, Misener L, Rose J B. Development of a PCR protocol for sensitive detection of Cryptosporidium oocysts in water samples. Appl Environ Microbiol. 1995;61:3849–3855. doi: 10.1128/aem.61.11.3849-3855.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Katzwinkel-Wladarsch S, Lieb M, Heise W, Löscher T, Rinder H. Direct amplification and species determination of microsporidian DNA from stool specimens. Trop Med Int Health. 1996;1:373–378. doi: 10.1046/j.1365-3156.1996.d01-51.x. [DOI] [PubMed] [Google Scholar]
- 18.Katzwinkel-Wladarsch S, Deplazes P, Weber R, Löscher T, Rinder H. Comparison of polymerase chain reaction with light microscopy for detection of microsporidia in clinical specimens. Eur J Clin Microbiol Infect Dis. 1997;16:7–10. doi: 10.1007/BF01575111. [DOI] [PubMed] [Google Scholar]
- 19.Kock N P, Petersen H, Fenner T, Sobottka I, Schmetz C, Deplazes P, Pieniazek N J, Albrecht H, Schottelius J. Species specific identification of microsporidia in stool and intestinal biopsy specimens by the polymerase chain reaction. Eur J Clin Microbiol Infect Dis. 1997;16:369–376. doi: 10.1007/BF01726365. [DOI] [PubMed] [Google Scholar]
- 20.Liguory O, David F, Sarfati C, Schuitema A R, Hartskeerl R A, Derouin F, Modai J, Molina J M. Diagnosis of infections caused by Enterocytozoon bieneusi and Encephalitozoon intestinalis using polymerase chain reaction in stool specimens. AIDS. 1997;11:723–726. doi: 10.1097/00002030-199706000-00004. [DOI] [PubMed] [Google Scholar]
- 21.Makowsky G S, Aslanzadeh J, Hopfer S M. In situ PCR amplification of Guthrie card DNA to detect cystic fibrosis mutations. Clin Chem. 1995;41:477–479. [PubMed] [Google Scholar]
- 22.Maniatis T, Fritsch E F, Sambrook J. Molecular cloning: a laboratory manual. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory (ed.).; 1982. pp. 173–177. [Google Scholar]
- 23.Mansfield K G, Carville A, Hebert D, Chalifoux L, Shvetz D, Lin K C, Tzipori S, Lackner A A. Localization of persistent Enterocytozoon bieneusi infection in normal rhesus macaques (Macaca mulatta) to the hepatobiliary tree. J Clin Microbiol. 1998;36:2336–2338. doi: 10.1128/jcm.36.8.2336-2338.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.McCabe E R B, Huang S Z, Seltzer W K, Law M L. DNA microextraction from dried blood spots on filter paper blotters: potential applications to newborn screening. Hum Genet. 1987;75:213–216. doi: 10.1007/BF00281061. [DOI] [PubMed] [Google Scholar]
- 25.Michel D, Marre E, Hampl W, Roczkos J, Müller S, Hertenstein B, Kern P, Heymer B, Salzberger B, Arasteh K, Mertens T. Intestinal cytomegalovirus disease in immunocompromised patients may be ruled out by search for cytomegalovirus DNA in stool samples. J Clin Microbiol. 1995;33:3064–3067. doi: 10.1128/jcm.33.11.3064-3067.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Molina J M, Oksenhendler E, Beauvais B, Sarfati C, Jaccard A, Derouin F, Modai J. Disseminated microsporidiosis due to Septata intestinalis in patients with AIDS: clinical features and response to albendazole therapy. J Infect Dis. 1995;171:245–249. doi: 10.1093/infdis/171.1.245. [DOI] [PubMed] [Google Scholar]
- 27.Moura H, Schwartz D A, Bornay-Llinares F, Sodré F C, Wallace S, Visvesvara G S. A new and improved “quick-hot Gram-chromotrope” technique that differentially stains microsporidian spores in clinical samples, including paraffin-embedded tissue sections. Arch Pathol Lab Med. 1997;121:888–893. [PubMed] [Google Scholar]
- 28.Müller A, Stellermann K, Hartmann P, Schrappe M, Fätkenheuer G, Salzberger B, Diehl V, Franzen C. A powerful DNA extraction method and PCR for detection of microsporidia in clinical stool specimens. Clin Diagn Lab Immunol. 1999;6:243–246. doi: 10.1128/cdli.6.2.243-246.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ombrouck C, Ciceron L, Biligui S, Brown S, Marechal P, van Gool T, Datry A, Danis M, Desportes-Livage I. Specific PCR assay for direct detection of intestinal microsporidia Enterocytozoon bieneusi and Encephalitozoon intestinalis in fecal specimens from human immunodeficiency virus-infected patients. J Clin Microbiol. 1997;35:652–655. doi: 10.1128/jcm.35.3.652-655.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Raskin S, Philips III J A, Krishnamani M R S, Vnencak-Jones C, Parker R A, Rozov T, Cardieri J M, Marostica P, Abreu F, Giugliani R, Reis F, Rosario N A, Ludwig N, Pilotto R F. DNA analysis of cystic fibrosis in Brazil by direct PCR amplification from Guthrie cards. Am J Med Genet. 1993;46:665–669. doi: 10.1002/ajmg.1320460612. [DOI] [PubMed] [Google Scholar]
- 31.Rijpstra A C, Canning E U, van Ketel R J, Eeftinck Schattenkerk J K M, Laarman J. Use of light microscopy to diagnose small-intestinal microsporidiosis in patients with AIDS. J Infect Dis. 1988;157:827–831. doi: 10.1093/infdis/157.4.827. [DOI] [PubMed] [Google Scholar]
- 32.Rinder H, Janitschke K, Aspök H, Da Silva A J, Deplazes P, Fedorko D P, Franzen C, Futh U, Hünger F, Lehmacher A, Meyer C G, Molina J-M, Sandfort J, Weber R, Löscher T The Diagnostic Multicenter Study Group on Microsporidia. Blinded, externally controlled multicenter evaluation of light microscopy and PCR for detection of microsporidia in stool specimens. J Clin Microbiol. 1998;36:1814–1818. doi: 10.1128/jcm.36.6.1814-1818.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rinder H, Katzwinkel-Wladarsch S, Lösher T. Evidence for the existence of genetically distinct strains of Enterocytozoon bieneusi. Parasitol Res. 1997;83:670–672. doi: 10.1007/s004360050317. [DOI] [PubMed] [Google Scholar]
- 34.Ritchie L S. An ether sedimentation technique for routine stool examinations. Bull US Army Med Dept. 1948;8:326. [PubMed] [Google Scholar]
- 35.Sapero J J, Lawless D K. The “M.I.F.” stain preservation technique for the identification of intestinal protozoa. Am J Trop Med Hyg. 1953;2:613–619. doi: 10.4269/ajtmh.1953.2.613. [DOI] [PubMed] [Google Scholar]
- 36.Shames B, Fox J G, Dewhirst F, Yan L, Shen Z, Taylor N S. Identification of widespread Helicobacter hepaticus infection in feces in commercial mouse colonies by culture and PCR assay. J Clin Microbiol. 1995;33:2968–2972. doi: 10.1128/jcm.33.11.2968-2972.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Talal A H, Kotler D P, Orenstein J M, Weiss L M. Detection of Enterocytozoon bieneusi in fecal specimens by polymerase chain reaction analysis with primers to the small-subunit rRNA. Clin Infect Dis. 1998;26:673–675. doi: 10.1086/514593. [DOI] [PubMed] [Google Scholar]
- 38.Velásquez J N, Carnevale S, Guarnera E A, Labbé J H, Chertcoff A, Cabrera M G, Rodríguez M I. Detection of the microsporidian parasite Enterocytozoon bieneusi in specimens from patients with AIDS by PCR. J Clin Microbiol. 1996;34:3230–3232. doi: 10.1128/jcm.34.12.3230-3232.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Verlingue C, Mercier B, Lecoq L, Audrézet M P, Laroche D, Travert G, Férec C. Retrospective study of the cystic fibrosis transmembrane conductance regulator (CFTR) gene mutations in Guthrie cards from a large cohort of neonatal screening for cystic fibrosis. Hum Genet. 1994;93:429–434. doi: 10.1007/BF00201669. [DOI] [PubMed] [Google Scholar]
- 40.Weber R, Bryan R T, Schwartz D A, Owen R L. Human microsporidial infections. Clin Microbiol Rev. 1994;7:426–461. doi: 10.1128/cmr.7.4.426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Weber R, Bryan R T, Owen R L, Wilcox S M, Gorelkin L, Visvesvara G S. Improved light-microscopical detection of microsporidia spores in stool and duodenal aspirates. N Engl J Med. 1992;326:161–166. doi: 10.1056/NEJM199201163260304. [DOI] [PubMed] [Google Scholar]
- 42.Wilde J, Eiden J, Yolken R. Removal of inhibitory substances from human fecal specimens for detection of group A rotaviruses by reverse transcriptase and polymerase chain reactions. J Clin Microbiol. 1990;28:1300–1307. doi: 10.1128/jcm.28.6.1300-1307.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhu X, Wittner M, Tanowitz H B, Kotler D, Cali A, Weiss L M. Small subunit rRNA sequence of Enterocytozoon bieneusi and its potential diagnostic role with use of polymerase chain reaction. J Infect Dis. 1993;168:1570–1575. doi: 10.1093/infdis/168.6.1570. [DOI] [PubMed] [Google Scholar]