Abstract
DNA nanotechnology has yielded remarkable advances in composite materials with diverse applications in biomedicine. The specificity and predictability of building 3D structures at the nanometer scale make DNA nanotechnology a promising tool for uses in biosensing, drug delivery, cell modulation, and bioimag-ing. However, for successful translation of DNA nanostructures to real-world applications, it is crucial to understand how they interact with living cells, and the consequences of such interactions. In this review, we summarize the current state of knowledge on the interactions of DNA nanostructures with cells. We identify key challenges, from a cell biology perspective, that influence progress towards the clinical translation of DNA nanostructures. We close by providing an outlook on what questions must be addressed to accelerate the clinical translation of DNA nanostructures.
Statement of significance
Self-assembled DNA nanostructures (DNs) offers unique opportunities to overcome persistent challenges in the nanobiotechnology field. However, the interactions between engineered DNs and living cells are still not well defined. Critical systematization of current cellular models and biological responses triggered by DNs is a crucial foundation for the successful clinical translation of DNA nanostructures. Moreover, such an analysis will identify the pitfalls and challenges that are present in the field, and provide a basis for overcoming those challenges.
Keywords: Nanotechnology, DNA nanotechnology, Cytotoxicity, Cellular uptake, Protein corona, Bionano interactions
1. Introduction
Advances in nanotechnology have enabled interesting applications and techniques in various fields, ranging from engineering to pharmacology and medicine [1-5]. The unique physicochemical properties of nanocarriers, in combination with their multifunctional capacity, allows these nanomaterials to be implemented in multiple biomedical applications [2]. In fact, distinct nanoparticles (NPs) were found to be highly useful in drug delivery, diagnosis, and imaging [3,6-8], thanks to improvement of the biodistribution and pharmacokinetics of the active pharmaceutical ingredients [9,10]. Numerous chemically distinct NPs (e.g. gold, metal oxides, silica, polystyrene, etc.) have been synthesized and are now being utilized as drug delivery vehicles, imaging enhancers, biosensing platform components, and other therapeutic and diagnostic uses [3,6-8,11,12]. Successful implementation of nanotechnology in medicine has resulted in clinical approval of 27 nanoparticle-based medicines by the Food and Drug Administration (FDA) and European Medicines Agency (EMA) [13]. Although FDA approval indicates some clinical success of nanomedicines, thus far patients offered these nanomedicines have showed only minor improvement in survival rates [14-16]. Additionally, nanomedicine formulations possess the risk of activating the immune system, which may lead to premature clearance from the body, as well as toxic side effects [17]. Emerging evidence highlights the following major challenges that hamper the clinical success of nanoparticles: difficulties in overcoming various biological barriers, low targeting efficiency, and safety issues [2,5,6,15,16]. As a result, medical applications of NPs are often criticized for their extremely low rate of clinically successful outcomes, despite their long research history and large investments [16,18-21]. Additionally, the lack of detailed understanding of the basic biological foundations of NP-cell interactions have also resulted in poor clinical translation of nanomedicines [6,16,19,21-23].
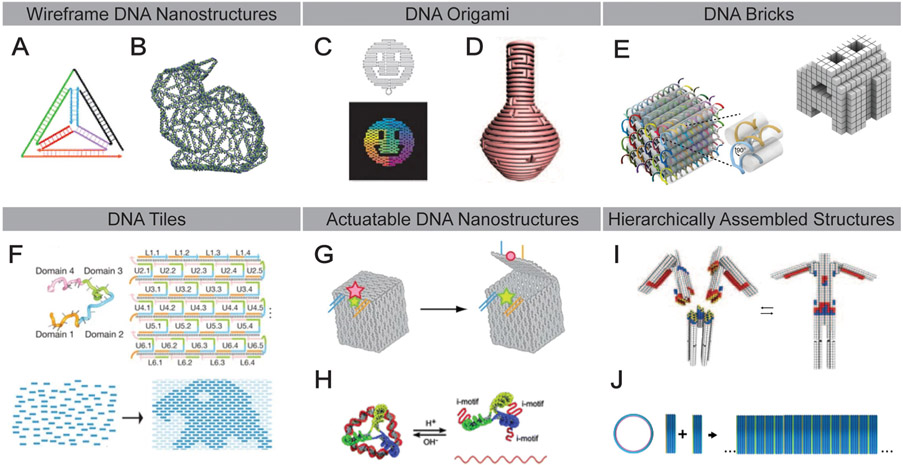
To circumvent translational challenges, it is crucial to reproducibly form nanomaterial complexes, retaining high precision in the nanometer range [2,24]. Indeed, production of complex functionalized NPs on this scale usually lacks a tight control over size, shape, and surface chemistry [24-26]. Self-assembly motifs, which are based on predictable and specific molecular interactions, represent an important direction in nanotechnology, with a promising foundation to overcome the challenges with structural precision [27-29]. Specifically, DNA nanotechnology bears tremendous potential in constructing complex 3D structures with nanometer precision [27,30-32]. DNA nanostructures (DNs), are being extensively investigated and applied in various research fields, such as chemical sensing, nanoelectronics, and biomedicine [27,30-34]. Their architectural diversity is exemplified by the variety of methods that have been developed to assemble these structures, including wireframe DNA structures (Fig. 1A and B) [35-37], DNA origami (Fig. 1C and D) [38-40], DNA brick [41] and tile [42,43] motifs (Fig. 1E and F, respectively). Dynamic, actuatable DNs [44,45] have also been developed (Fig. 1G and H) in addition to larger, hierarchically assembled structures (Fig. 1I and J) [46,47].
Fig. 1.
Key examples of various DNA nanostructure designs. (A) DNA tetrahedron [35]. (B) Three-dimensional wireframe rabbit-shaped DNA structure designed from a polygonal mesh architecture [36]. (C) Two-dimensional DNA origami in the shape of a smiley face [38]. (D) Three-dimensional DNA origami vase structure featuring complex curvature [39]. (E) Modular DNA structures composed of 32-nucleotide “brick” motifs [41]. (F) Single-stranded DNA “tiles” acts as pixels in a two-dimensional array [187]. (G) A DNA box designed to be opened via toehold strand displacement to release a cargo of interest [44]. (H) pH-sensitive DNA i-motifs allow the assembly and disassembly of a DNA tetrahedral structure [45]. (I) Heteromultimeric assembly of complex DNA architectures via shape complementarity [46]. J) Homomultimeric assembly of DNA barrel structures into a hollow DNA tube via sticky end adhesion [47].
Biomedical DN research is progressing impressively quickly, specifically in the direction of diagnostics and therapeutics [2,24,27,32,34,48]. DNs possess several key advantages in biomedical applications over conventional NPs [2,24,27,32,34]. For example, conventional NPs have been shown to induce various adverse reactions [21,49,50]. By contrast, DNs typically exhibit great biocompatibility and thus far lack toxicity in preliminary studies [2,24,27,32,34]. Furthermore, the capacity of DNs for self-assembly allows their construction into well-defined 3D architectures of arbitrary shape and size at the nanoscale. This in turn enables the biological activity of DNs to be finely tuned and modified [2,24,27,32,34,44,51,52]. The surfaces of DNs can be functionalized accurately and precisely using the properties of DNA self-assembly [2,24,27,32,34,44,51,52]. These unique properties of DNs have opened doors to numerous biomedical applications. Thus far, DNs represent great nanomedical potential and are being actively studied as platforms for controlled release of various therapeutic compounds, as imaging modules, and as vehicles for targeted delivery [2,24,27,32,34,44,51,52].
Despite these promising initial results, the translation of DNs to the clinic is still in its infancy. There are only a handful of studies that use DNs under in vivo conditions [34], and the field still lacks a thorough knowledge about the precise molecular determinants that modulate DN-cell interactions [34]. Verifying the key principles of DN-cell interactions is important to understanding the molecular mechanisms underlying therapeutic approaches. Clear knowledge of how certain treatments work is crucial in forthcoming clinical trials, and represents a roadmap for successful implementation of the treatment [53]. Understanding the mode of action at the cellular and molecular level will aid in determining the therapeutic window of a treatment, enabling better dosing, stratifying clinical trials, and eventually helping patients [21,53-57]. Thus, in this review we present an analysis of current knowledge on DN-cell interactions. We discuss challenges currently limiting DNs translation towards real-world applications. Finally, we highlight strategies that may help to overcome these challenges and maximize the biomedical potential of DNs.
2. The protein corona and its impact on DNA nanostructures
Generally, it has been found that contact with physiological fluids results in the formation of a protein corona around many types of nanoparticles [58-60]. Proteins and other biomolecules interact with the surface of the particles, forming a multilayered shell [58-60]. The presence of the corona may shield surface modifications (e.g. chemical moieties, targeting ligands, antibodies, etc.) and affect their function or efficiency [58-60]. Not surprisingly, a protein corona has been shown to form on DNs as well [27,34,61]. Overall, adsorption of proteins onto a particle surface occurs rapidly, approximately one hour after exposure to physiological fluids [58-60]. Accumulating evidence suggests that multiple factors play a role in the composition of the protein corona and biomolecule binding efficacy [58-60,62,63]. NP physicochemical properties (i.e. chemical composition, size, shape, surface functionalization), physiological fluid composition, and exposure time determine the makeup and nature of the protein corona [58-60,62,63]. In turn, protein binding to the NP surface changes the physicochemical properties of the particle itself (e.g. hydrodynamic diameter, zeta potential, solubility), and the protein properties are also altered (e.g. misfolding, aggregation, conformational changes, alteration in enzymatic activity) [58-60,62-65]. These structural and functional changes of proteins upon binding to NP surface may lead to cellular injury [58-60,62,63]; furthermore, the protein corona may greatly hinder the targeting capabilities of NPs by shielding surface functionalization [66]. On the other hand, tuning the surface modification of NPs may affect protein corona composition in a way to improve circulation half-time, mitigate toxic effects, and/or ameliorate targeting issues [60,67].
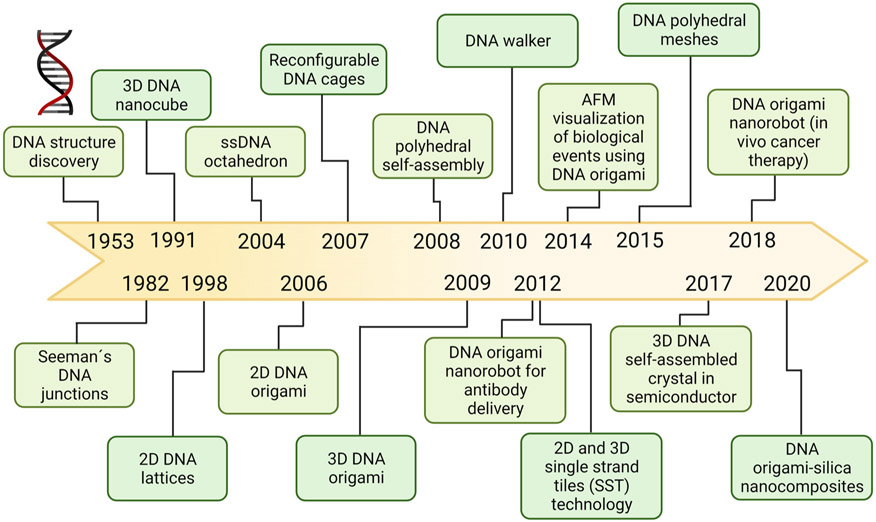
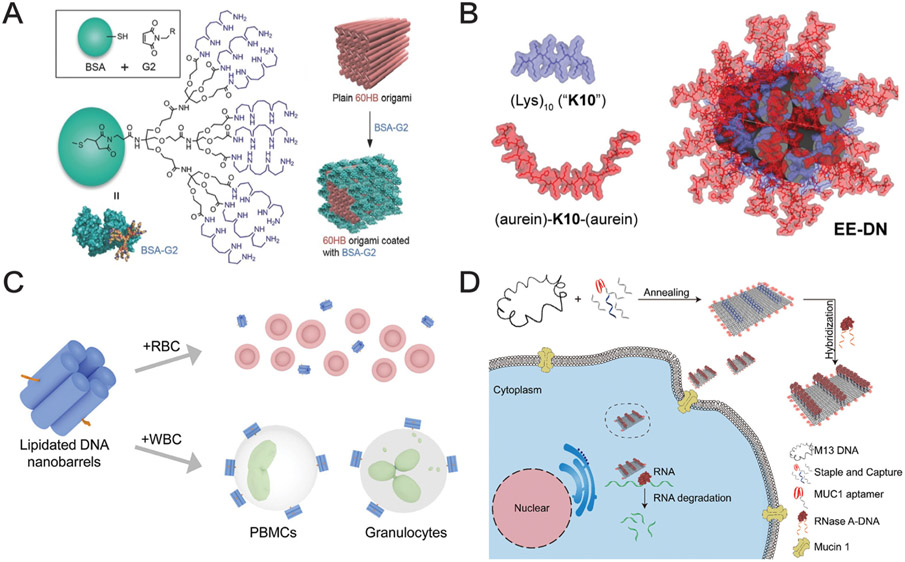
The considerations outlined above demonstrate why it is crucial to study the protein corona formation around DNs in detail. Although DNA nanostructures have now been studied for decades, research on applications in biomedicine (e.g. DNs as tools for imaging and vehicles for gene delivery) and therapeutics (e.g. targeted drug delivery) for DNs are quite recent (Fig. 2). Consequently, little attention has been given thus far to the analysis of DN-protein corona composition and the corona’s functional consequences for DNs [27,34,61]. Recently, more research has been devoted to how protein corona affects DN stability [27,34,61]. Indeed, the limited stability of DNs in physiological fluids represents a challenge for their successful biomedical application [27,34,61]. Nucleases are predominantly responsible for in vivo degradation of DNs [61], and to mitigate this problem, peptides and proteins have been used to create nuclease-protective coatings that give DNs a longer half-life in biological environments [61]. Synthetic protein coronae may also be utilized to create nuclease-resistant DNs [61,68]. For example, bovine serum albumin (BSA)-dendron conjugates attached to DNs protected nanostructures from exposure to 10 U of DNase I (Fig. 3A) [68]. The BSA corona also significantly reduced the immune response against DNs and improved their transfection efficacy [68]. Protein polymers and diblock polypeptides have also been shown to be effective for shielding DNs from enzymatic degradation [69,70]. Another strategy to enhance DN stability is to create stable and enzymatically resistant DNs that simultaneously reduce particle-protein interactions, such as coating the nanostructures with poly(ethylene glycol) (PEG) [2,27,34,61], while ensuring that DN surface functionality is not compromised. One way to achieve PEG passivation is via an electrostatically-adhered oligolysine–PEG coating, which was found not to interfere with the functionality of surface-displayed ligands on DNs [71]. While PEG conjugation is a widely used surface modification for various other types of NPs, some PEG-based nanomaterials have been shown to be immunogenic, resulting in release of anti-drug antibodies [72,73]. PEG itself can trigger anti-PEG IgG and IgM antibody responses [72,73], and high titers of these antibodies may lead to severe allergic reactions such as anaphylaxis [72-74]. Thus, PEG-based coatings of DNs must be designed in a controlled and cautious manner. An additional consideration is that DN coatings (e.g. PEG, the protein corona) may undermine compatibility and functionality of switchable and dynamic DNs [34].
Fig. 2.
Historical timeline of the advancements in DNA nanotechnology research [2,24,27,34,61].
Fig. 3.
DNA nanostructures for biological applications. (A) BSA modified with positively charged dendrimers to adhere to a 60-helix bundle (60HB) nanostructure enables enhanced nanostructure stability, uptake, and immunoquiescence [68]. (B) Oligolysine-based peptide coating featuring two functional aurein 1.2 sequences that exhibits endosomal escape of the coated DNA nanostructure (EE-DN) in the absence of serum proteins [76]. (C) Cholesterol-bearing 6-helix bundle DNA nanostructures facilitate targeted uptake in white blood cells compared to red blood cells [188]. (D) A DNA origami sheet bearing MUC1-targeted aptamers capable of targeted intracellular delivery of active RNase A [189].
Importantly, studies that analyze how a protein corona would affect biological and therapeutic properties of DNs are rather rare [75]. However, it is critical to assess not only the stability of DNs in physiological fluids, but rather how those fluids may modify the surface, and impact or hamper the desired function of DNs. We recently showed that the protein corona greatly affects the intracellular function of specifically designed DNs [76]. In absence of serum proteins, DNs coated with aurein 1.2 (a peptide that facilitates endosome escape [77]) showed marked endolysosomal escape in different cell lines (Fig. 3B) [76]. However, upon exposure to serum-containing medium, a protein shell formed around the DNs, significantly hampering the efficiency of endolysosomal escape and leading to accumulation of DNs in lysosomal compartments (which is the usual fate for unmodified nanostructures) [76]. Therefore, protein corona formation over DN particles should be taken into account for successful and clinically relevant design and optimization of DNs.
3. Physical background on the interaction of DNA nanostructure ligands with cell surface proteins
In a light of above discussed, it is crucial to analyze how functionalized DNs physically interact with the surface receptors of cells. Generally, ligand interactions with cell surface proteins predispose subsequent cell entry of exogenous materials and regulate to a large extent the intracellular fate of various materials [78-81]. Interestingly, DNA molecules alone do not cross the plasma membrane of the cell. However, 3D DNA nanostructures are able to efficiently enter the cellular cytosol [82]. Therefore, a study of the physical parameters that modulate cellular interaction and processing of DN represents an important milestone for efficient targeting of cell surface receptors.
Indeed, current progress in understanding nanoparticle-cell interactions revealed several possibilities for the modulation of targeting efficacy and cellular uptake [83]. Those possibilities comprise the orientation, mobility, and surface density of ligands on the nanoparticle [84-88]. Furthermore, accumulating evidence revealed that particle geometry parameters, e.g. size, shape, and aspect ratio, affects largely their uptake and to a larger extend therapeutic efficacy [89-91]. For example, particles having a rod-like geometry showed higher cellular binding efficacy in comparison with spherically-shaped particles [92]. By contrast, spherically-shaped particles showed a higher uptake efficiency compared with rod-shaped ones [93].
However, we have to state that despite this progress, it is still not fully understood how DNs influence the interaction between ligands functionalizing the DN surface and cell surface receptors. Indeed, it was shown that DN functionalization with a protein ligand does not reduce the protein’s ability to bind its receptor [94,95]. Another recent study identified that the affinity of anti-programmed cell death protein 1 antibody (aPD1) incorporated onto DN remains unchanged compared with the free antibody [96]. Interestingly, this study further revealed that the absolute number of bound DNs was significantly lower in comparison with the free antibody, which in turn resulted in lower binding efficiency [96]. In fact, the cell surface composition plays the role of a natural barrier, resulting in limited receptor accessibility for functionalized DNs [96]. As a result, DN orientation and size represent crucial parameters for effective binding to the receptors [96]. In other words, the efficacy of cellular targeting by functionalized DNs is predisposed by an interplay of receptor affinity and accessibility of receptors [96]. Such knowledge is critical in designing programmable DNs for improved applications of nanomedicines towards targeted cell signaling modulation.
DNs offer programmable precision for decorating their surfaces with biomolecule nanopatterns, enabling precise spatial separation between ligands on the nanoscale [36,39,51,96-98]. Furthermore, DNs have been decorated with varying nanopatterns of biomolecules, e.g. ephrin-A5 [94,99], immunogen eOD-GT8 [100], caspase-9 variant [101], antigens of human IgGs and IgMs [102], and Fas ligands [103]. Of note, regulation of the spatial organization of surface receptors at the nanoscale provides a route for controlling cellular responses [104]. A recent study revealed the use of DNs for regulated death receptor 5 (DR5) clustering and subsequent triggering of apoptosis [105]. Furthermore, the study revealed that the required inter-ligand distance for initiation of apoptotic events was less than 10 nm [105]. Interestingly, this approach of DN-mediated clustering of DR5 was effective even against resistant breast cancer cells [105].
Overall, nanometer precision in patterning of various DNs with specific surface ligands offers a significant boost to the potential of DN-based nanomedicines. We see in this technology an opportunity to study also fundamental cell biological questions of receptor function.
4. Analysis of DNA nanostructure cytotoxicity
To bolster the biomedical applicability of DNs, researchers commonly stress that DNA is a natural biological molecule [24,34,61,106], and is therefore readily biodegradable and biocompatible, with minimal toxicity [24,34,61,106]. Therefore, DNs made of DNA molecules are generally assumed to be biocompatible as well as nontoxic [24,34,61,106]. However, this is the so-called “naturalistic fallacy” [107,108]: The “natural” origin does not directly correspond to “safe” or “biocompatible” [107-109], as plenty of “natural” molecules are toxic or immunogenic [107-109]. Specifically, cell-free DNA is known to be present in blood plasma of healthy individuals [110], yet high levels of circulating cell-free DNA are also associated with multiple pathologies, including systemic lupus erythematosus, metastatic cancers, atherosclerosis, primary Sjögren’s syndrome, and rheumatoid arthritis [110-113]. Elevated levels of donor-derived cell-free DNA during transplantation may lead to adverse post transplantation events such as allograft rejection [113-115]. Furthermore, it has been proposed that cell-free DNA may possess cytotoxic properties [110,113,116]. DNA can be released during cellular injury as damage-associated molecular patterns (DAMPs) [117,118]. Injury-issued DAMPs, including extracellular DNA, can result in the activation of innate immunity [117]. For example, circulating cell-free mitochondrial DNA was shown to induce inflammasome-dependent caspase-1 activation and IL-1β and IL-18 release [119]. Therefore, careful assessment of the toxicological and immunogenic potential of DNs is imperative for successful clinical translation of DN-based technologies.
It is worth noting that preliminary studies indicate some DN biocompatibility and potentially favorable clearance kinetics [120]. In light of the aforementioned DNA-related adverse cellular effects, it is important to systematically analyze the available literature regarding DN toxicity, and to our knowledge there is no systematic analysis of their toxic potential [24,27,34,61,75,106]. Thus, we briefly summarize available accounts of in vitro toxicological responses to the DN architectures most studied thus far (Table 1).
Table 1.
A brief summary of in vitro toxicity assessments of different DNs.
| DN type | Specifications | Cell model | Exposure time | Outcome | Ref. |
|---|---|---|---|---|---|
| Deoxyribonucleic acid-nanothread (DNA-NT) | Diameter: 50–150 nm; Length: 300–600 nm; CPT-DNA-NT Attachment of cisplatin | HeLa | 48 h | CPT-DNA-NT reduced cell viability; Signs of apoptosis; DNA-NT No effect on cell viability | [190] |
| DNA nanobarrels (NB) | Six DNA duplexes forming a six-helical bundle (9 × 5 × 5 nm) NB-3C, NB-1C, and NB-0C containing 3, 1, or 0 cholesterol anchors | Red blood cells (RBC); white blood cells (WBC); granulocytes; peripheral blood mononuclear cells (PBMC) | 6 h | No effect on viability | [188] Fig. 3C |
| DNA nanoscaffolds, DNA tetrahedron (Td) rectangle DNA origami | Incorporation of 5-fluoro-2′ - deoxyuridine; (FdUn) oligomers; Attachment of cholesterol | HTB-38; HCC2998 | 24 or 48 h | Reduced proliferation of the HTB-38 cells; Signs of apoptosis higher in HTB-38 cells | [191] |
| DNA duplexes | Attachment of cholesterol; Attachment of alkyl-phosphorothioate (PPT) belt | HeLa; MyrPalm-EGFP HeLa | 5 min | No data on viability | [192] |
| Nanotoroids | 3 types of nanotoroids of different size ρ = 6, ρ = 2, and ρ = 1.5 1 | SMMC-7721; HeLa | 6 h | Slight decrease of cellular viability with higher concentration of nanotoroids | [193] |
| DNA nanopores | NP- EP pore- 6-helix bundle with hydrophobic belt containing ethyl phosphorothioate (EP) groups; 3 negative controls of nanobarels without EP-belt formation | HeLa | 1 and 24 h | Decreased viability of cells after incubation with NP-EP | [194] |
| DNA nanopore | Six DNA duplexes modified with phosphorothioate (PPT) group | MCF-7 | 48 h | No significant effect on cellular viability; Decrease viability after incubation DNA nanopores with doxorubicin | [195] |
| Rectangular DNA origami nanosheets | Binding of RNase A to DNs; With/without decoration with protein MUC1 | MCF-7 | 48 h | Colocalization with lysosomes after 1 h; No effect of bare DNs on cell viability; Increased cell death post incubation with MUC1-modified RNase A loaded DNA origami | [189] Fig. 3D |
| DNA origami nanobox (DON) | Cuboid structure; 36 × 36 × 42 nm; Attachment of AS1411 aptamers and incorporation of doxorubicin (DOX) | HeLa; MCF-7 | 2 h | Decrease viability after incubation DON with doxorubicin; Signs of apoptosis | [196] |
The aspect ratio of the nanotoroid is derived as ρ = R/r, where R - the radius of the nanotoroid and r - the radius of the nanotoroid cavity. A larger ρ value defines a smaller cavity [193].
Overall, from Table 1 it is clearly seen that in the majority of studies, DNs that have not been loaded with drugs show low to no cytotoxicity. However, the maximum exposure time used in majority of studies is only 48 h (Table 1). Indeed, designed nuclease-resistant DNs may withstand harsh biological environment for more than 48 h [61]. Thus, longer-term cytotoxic effects have yet to be fully elucidated. Of note, the U.S. Environmental Protection Agency (EPA) explicitly mentions that biodegradability does not guarantee low toxicity of a compound [121]. A number of compounds that showed rapid biodegradability were found to be carcinogenic, mutagenic, or toxic [121-123]. During degradation, decomposition products and/or adducts of initial compounds might be highly reactive and possess significant toxicity [121-123]. A classic example is drug-induced liver injury (DILI) triggered by products of acetaminophen metabolization [124,125]. Indeed, drug-protein adducts, occurring drug metabolism in hepatocytes, may act as neoantigens, triggering an immune response and resulting in cell injury [124,125]. Idiosyncratic (unpredictable) DILI pathology does not require high doses of the drug, and can be profound with relatively low but chronic doses of >50–100 mg per day [124,126]. Specifically, both short oligonucleotides and long DNA pieces show undesired toxic and immunogenic responses [127,128], and it is well known that dsDNA induces a number of autoimmune pathologies [129-131]. An increase in serum DNA concentration is a straightforward marker of systemic inflammatory reaction and sepsis [132,133]. Therefore, it is important to consider not only toxicity of entire DNs but as well, their degradation products and/or adducts forming upon metabolization by cells, and to probe potential side effects of these materials for extended exposure and circulation times.
Extrapolating experience from other nanomaterial studies, some particles may be retained in the human body for weeks before excretion [21,134]. In fact, emerging evidence suggests that many different nanomaterials may possess time-delayed toxicity [135-140], so it is important to carefully and systematically analyze such long-term toxicity.
Another challenge apparent from the analysis in Table 1 is that the toxicological assessments for the majority of studies have been primarily been carried out in only a handful number of standard cell lines. For example, MCF-7 cells are frequently used as a model breast cancer cell line, and HeLa cells are abundantly utilized as a general “cancer” cell model. However, a thorough investigation of different MCF-7 cell line strains revealed substantial genetic heterogeneity among them [141]. When those strains were challenged with 321 anti-cancer compounds, they showed dramatic variability in response. Strikingly, 75% of compounds that induced marked toxicity in some strains were completely ineffective in others tested [141]. Another thorough study demonstrated that different strains of HeLa cells possess great genetic and phenotypic variability, e.g. variations were found in genome-wide copy numbers, mRNAs, proteins, and protein turnover rates [142]. Those studies highlighted an important question regarding the reproducibility of research conducted using MCF-7 and HeLa cells. It is worth noting that cell line authentication is crucial for conducting reproducible and reliable research [143]. Avoiding this authentication can easily lead to unreliable outcomes, resulting in the loss of time, money, and trustworthy publication data [143]. It has been reported that over 20% of cell lines are misidentified or mislabeled, often due to cross-contamination [144]; indeed, HeLa cells are the major contributors to such instances of cross-contaminations [144,145].
Another key challenge in the assessment of DNs toxicology is the scarcity of studies implementing primary cell cultures (Table 1). Although cell lines are very powerful for initial screening, they do not fully recapitulate tissue-specific functions and have limited predictive value towards in vivo applications [146,147]. In this regard, primary cell cultures could mitigate these problems and provide results more closely related to in vivo conditions [146-148]. Even cell lines phenotypically related to primary cells can possess substantial gene expression differences and be functionally distinct by comparison [149-151]. Additionally, highly proliferating tumor-derived cell lines such as HeLa cells tend to redistribute the nanomaterial among daughter cells, resulting in a lower particle load per cell and thus overlooked toxic effects [21,152,153]. By contrast, primary cell cultures with limited proliferative activity may provide more reliable results on nanomaterial toxicity [21,152,153]. Therefore, there is an unmet need to boost research on DN toxic effects utilizing primary cell culture models.
5. Analysis of DNA nanostructures interactions with cells
Aside from toxicological assessments of DNs, a deep analysis of DN-cell interactions and identification of target proteins and/or pathways mediating the cellular effects of DNs is necessary for successful translation of DN technology into any biomedical application. To help meet this end, we summarize the currently most studied DN-induced cellular effects and interactions (Table 2).
Table 2.
A brief summary of DNA nanostructures-induced cellular effects and interactions.
| DN type | Specifications | Cell model | Incubation conditions | Major results | Ref. |
|---|---|---|---|---|---|
| Tetrahedral DNA nanostructure (TDN) | Four 55-base ssDNA strands; each vertex of TDN labeled with cyanine-3 (Cy3) | HeLa; COS-7 | Up to 12 h for uptake; Incubation at 4 °C and 37 °C for 6 h | Time-dependent uptake; Caveolin-dependent endocytosis; Microtubule-dependent transport; Lysosomal internalization after 12 h of incubation | [197] |
| 6-helix bundle (6HB) nanostructure | 7 × 6 nm; attachment of (Lys)10 peptide (K10) and aurein 1.2 | HepG2; Alexander; Huh7 | Up to 24 h at 37 °C in complete medium for uptake and cytotoxicity assays; incubation with/without serum for 6 h 37 °C | Colocalization of DNs with lysosomes; Protein corona formation post incubation in the presence of serum; Reduced endosomal escape of DNs | [76] |
| Framework nucleic acids (FNAs) | 3 different shapes: tetrahedron, triangular prism, and cube labeled with cyanine-5 (Cy5) | HeLa | In FBS-free culture medium at 37 °C for 3 h | Partially clathrin-mediated endocytosis; Scavenger receptor (SR)-mediated endocytosis; Cellular uptake dependent on DN geometry | [198] |
| DNA origami nanostructures (DONs) | 2 different shapes: tetrahedron and rod Cy5 labeled with distinct size: small tetrahedron (ST) 4 × 2 × 11 per edge; small rod (SR) 4 × 4 × 32; large tripod tetrahedron (LT) 7.2 × 12 × 47 per arm; large rod (LR) 8 × 8 × 127 | H1299; DMS53 | Up to 8 h at 37 °C for uptake analysis | Cellular uptake dependent on DN shape and size; larger and rod-shaped structures have higher efficiency of uptake; scavenger receptor-mediated uptake; endolysosomal accumulation of LR after 24 h | [199] |
| DNA origami nanoparticles (DONs) | 11 distinct DNA-origami shapes Cy5-labeled; range of the size: 50–400 nm | HUVEC; HEK293; BMDCs | At 37 °C for 12 h for uptake analysis | Higher uptake of larger DNs with better compactness | [200] |
| DNA-based nanostructure ChloropHore | 61-base pair DNA duplex Cl− reporter domain- Clensor and a pH reporter domain (I-switch) | Primary human dermal fibroblasts | Up to 23 h for stability analysis | Scavenger receptor-mediated endocytic pathway; use for evaluation of pH and Cl− in lysosomes | [201] |
| DNA nanobundles (NB) | 6-duplex nanobundle Hight: 9 nm Width: 6 nm; NB-3C, NB-1C, and NB-0C containing 3,1 or 0 cholesterol anchors | HeLa | For 2 h/24 h in OptiMEM or DMEM+ 10% FCS | Higher cellular uptake of NB-3C, nanobundles containing 3 cholesterol anchors; colocalization of DN with endolysosomal compartments after 24 h | [202] |
| Tube DNA nanostructures | YOYO-1 labelled tube DNs | NIH 3T3 | For 4 h/24 h | Colocalization of DNs with lysosomes | [203] |
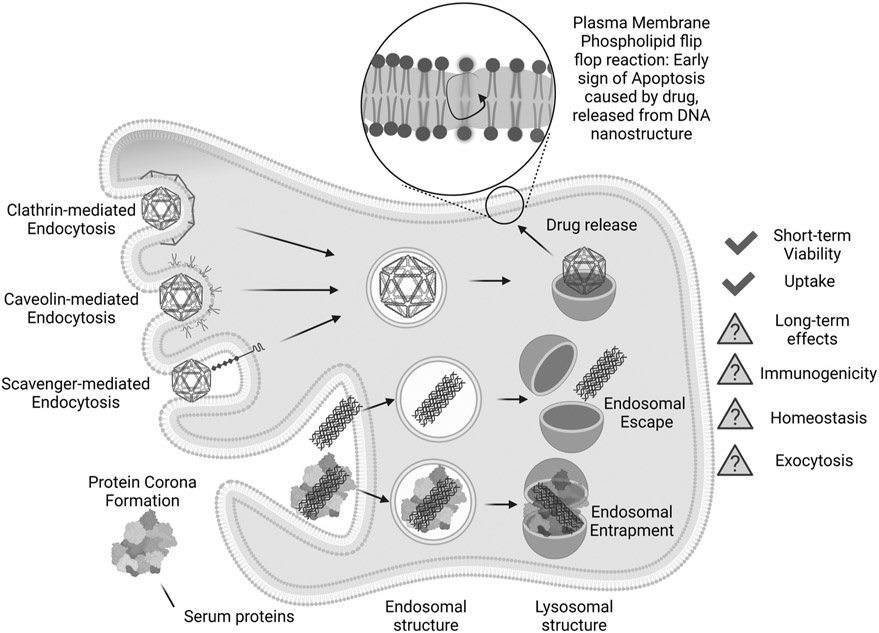
It is evident from Table 2 that a deep analysis of signaling pathways involved in cell-DN interactions is underrepresented in the current literature. Current research has been primarily focused on revealing DN uptake and subcellular localization with minimal attention towards functional changes that DNs may elicit in cells (Table 2). This is likely because research efforts towards biomedical applications for DNs are still relatively new [27,34,61]. Despite this, there has already been substantial progress in understanding how the size and shape of DNs affect cellular uptake and subcellular distribution (Table 2 and Fig. 4). However, it has yet to be seen whether signaling is biased in cells upon DN treatment. We may take lessons from NP studies in which it has been suggested that NPs trigger substantial cellular responses that bias lysosomal function without triggering a cytotoxic response [21,154]. Additionally, current studies on DN-cell interactions suffer from same problems described in the previous section, i.e. lack of primary culture use in research, usage of spurious cell lines, and a lack of studies on their long-term effects.
Fig. 4.
Schematic brief summary of DNA nanostructures interaction with living cells.
We would like to stress that the current developments in the field of DNA nanotechnology are considerable, intriguing, and provide great perspective. Specifically, in biomedically driven studies, DNs have shown promising results in biosensing, drug delivery, cell modulation, and bioimaging [24,27,34,61]. For instance, DN-based biosensors proved advantageous in precise design, specificity, and low-cost synthesis [155,156]. DNs can be designed and functionalized to bear various drug cargos, which opens a route for improved drug delivery applications [24,27,34,61]. Cell behavior and activity can also be altered in a controlled manner using smartly designed DNs [157]. DNs are indispensable for super-resolution DNA-PAINT (DNA-based point accumulation for imaging in nanoscale topography) imaging applications [158]. However, for advanced and successful implementation of these various functional DNs, an understanding the detailed mechanisms of DN-cell interactions and their consequences is vital. Knowledge of the long-term effects, signaling mechanisms, immunogenicity, and excretion of DNs (Table 2) has yet to be fully elucidated.
It is worth noting here, that in addition to nanoparticles, DNA nanotechnology has been applied to tunable hydrogel systems [159,160]. Such systems represent 3-D hydrophilic networks featuring DNA as a part of the system [159-161]. DNA hydrogels are scalable from bulk hydrogels to nanogels [159,160,162]. As DNA hydrogels contain programmable and complementary DNA strands as part of the network, this feature allows resultant hydrogels to be easily manipulated to create different DNA building block with precise geometries, leading to a predictable and controlled resultant DNA networks [159,160,162,163]. Due to the structural programmability of DNA hydrogels, these systems allow to exert various interaction with cells in controlled manner [159,160,162,164]. For example, immunostimulatory CpG DNA hydrogels may be potent in enhancing the antigen-specific antitumor immunity [165]. Additionally, DNA-based hydrogels allow for control of interactions between cells and the extracellular matrix interactions with nanoscale precision [166]. This possibility makes DNA hydrogels a promising platform for programmed tissue engineering [159,160,162]. However, there are still considerable challenges needed to be addressed in the development of DNA-based hydrogels, e.g. cost-effective upscaling, potential for degradation by secreted nucleases, and possible toxic or immunogenic effects [159,160,162].
In fact, DNA assembly into complex customized 3D structures with desired functions has seen great advancements in recent years [24,27,34,61]. It is now possible to produce more stable DNs at a faster rate, while precisely varying the size and shape with higher production yields [24,27,34,61]. However, for successful clinical adaptation of DNA nanotechnology we need to overcome several challenges arising from a biological point of view. We describe and discuss those challenges in the following section.
6. Challenges and future perspectives
DNs as biomolecule-based nanoparticles possess advantages over standard nanomaterials in terms of controllable size, shape, and surface functionality [2,24,27,34,61]. Undoubtedly, those advantages will enable even more therapeutic and diagnostic use of these nanostructures, but as DNs become increasingly complex, more efforts must be taken to overcome hurdles to clinical translation. In this regard, thorough studies of DN-cell interactions are of paramount importance. Below, we summarize current studies of DNs in biological contexts, then identify challenges and pitfalls in their biomedical implementation in order to provide a roadmap for overcoming them.
The first major challenge is the stability of DNs under physiological conditions. Substantial progress in this direction has been already achieved [2,24,27,34,61], but often these approaches are reported as stand-alone studies. It will be critical to integrate these stabilizing approaches with specific applications of nanoparticles for biomedical applications, in conjunction with primary cells or in vivo models. For example, most of the coatings reported were not tested for how they may change bare DN immunogenicity, biodistribution, or pharmacokinetics; probing these factors will aid in more sophisticated, real-world applications of functionalized DNs.
Drug delivery, specifically of cancer therapeutics, is certainly the most investigated and notable DN application [2,24,27,34,61]. Thus, DN-cancer cell interactions must be more thoroughly interrogated to reach the greatest potential of anticancer DN use. For example, both toxicological assessments and analyses of DN interactions with cells are still fragmented and unstandardized (Tables 1 and 2). There are very few studies employing primary cell cultures to investigate these two parameters, and a substantial number of studies still use potentially problematic cell lines, like HeLa and MCF-7 (Tables 1 and 2), that could affect the reproducibility of their results. We also draw attention on the importance of cell line authentication [143]. Apart from these concerns, we propose that a thorough justification of the choice of biological model should be provided, especially when DN research is directed toward therapeutic use. Guidelines for selecting and justifying cancer models already exist [167-169], since human tumor cell lines that are routinely used may possess considerable differences in comparison with primary tumors [168,170]. Misidentification, contamination with mycoplasma, genetic drift, and phenotypic instability in frequently used cell lines are often neglected by many researchers [146]. In order to reliably compare results of DN function within cells, guidelines of cell line selection, authentication, and maintenance must be applied in future studies [146].
Recently, it has been noted that there is substantial variability in the field regarding DN characterization techniques and experimental design [2,24,27,34,61]. Thus, we propose that the biomedical research of DNs adopt a “minimal reporting standard” derived from an already existing one from the field of bionanotechnology [171]. This minimal reporting standard combines guidelines for nanomaterial characterization, biological model justification, and standardized experimental protocols [171]. Implementation of such standards will improve reproducibility and significantly boost quantitative comparisons of results on DN-cell interactions.
Another opportunity to boost biomedical research of DNs lies in the implementation of more sophisticated in vivo models than conventional rodent models. Current research shows that animal model systems currently abundantly used in the biomedical field poorly recapitulate human counterparts, often leading to unreliable results [172-174]. Some researchers suggest that “more complex human conditions” should be used in biomedical research in place of rodent models [172]. For this reason, the U.S. Environmental Protection Agency has plans to dramatically reduce or even eliminate the use of animal models for testing research by 2035 [175]. Organoids—complex multicellular systems that recapitulate in vivo structure and functions of the selected tissue—may help to overcome those challenges and provide systems that are more relevant for use in humans [176-178]. Organoids have already been implemented in biomedical nanoparticle research, showing reliable and progressive results [179,180]. The use of organoids in DN research is still quite fragmented [2,24,27,34,61], so using them for screening toxicity and performing studies on the interactions of DNs with cells would provide important foundation for future clinical translation of DNA nanotechnologies.
A final challenge that must be addressed prior to DN use in the clinic is that of liver sequestration. Cumulative evidence suggests that the liver sequesters up to 99% of intravenously injected nanoparticles [21,23,181,182]. Long-term accumulation in the liver results in adverse effects and greatly limits the clinical efficacy of nanoparticles [21,23,181,182]. Generally, drug-induced liver injury (DILI) is described as a harmful and unexpected impact of drugs on the liver [124]. In fact, DILI represents a serious problem, being one of major causes of acute liver failure in Western countries [124,125]. Moreover, many nanoparticles have been shown to possess hepatotoxicity and display DILI properties that were initially overlooked [21,183-186]. Indeed, studies carefully addressing the hepatotoxic potential of DNs remain unaddressed in current literature. Examining the hepatotoxic properties of DNs will help optimize the design and synthesis of clinically suitable DNs to avoid off-target effects of DNs.
In summary, we would like to emphasize that DNs possess great biomedical potential. We expect to see more diverse applications of DNs capable of translating towards clinical use, but understanding the hurdles and limitations of therapeutic DNA nanotechnology is crucial for its clinical success. Consequently, the critical analysis provided herein will help researchers to establish a roadmap for overcoming these challenges.
Acknowledgments
Figures 2 and 4 were created with BioRender.
Funding
This research was funded by Operational Programme Research, Development and Education financed by European Structural and Investment Funds and the Czech Ministry of Education, Youth and Sports (Project No. SOLID21 - CZ.02.1.01/0.0/0.0/16_019/0000760) and MH CZ - DRO Institute for Clinical and Experimental Medicine – IKEM, IN 00023001. Nicholas Stephanopoulos acknowledges support from the National Science Foundation (DMR-BMAT CAREER award 1,753,387). Research reported in this publication was supported by The National Institute of General Medical Sciences of the National Institutes of Health under grant number DP2GM132931. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.”
Footnotes
Credit author statement
Conception and design: O.L., A.F., B.S., N.S., A.D., and M.L.; Development of Methodology: O.L., A.F., B.S., A.D., M.L., M.U., S.J.W.H., and M.J.; Acquisition of data: O.L., A.F., B.S., A.D., M.L., M.U., and S.J.W.H.; Formal analysis and interpretation of data: O.L., A.F., B.S., A.D., M.L., M.U., S.J.W.H., and M.J.; Drafted the manuscript: O.L., A.F., B.S., N.S., and S.J.W.H.; Review and revision of manuscript: M.L., M.U., M.J., and A.D. Study supervision: O.L., A.D. and N.S.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Data availability
All data needed to support the conclusions are present in the paper and/or the Supplementary Materials. Additional data related to this paper may be requested from the authors.
References
- [1].Dawson KA, Yan Y, Current understanding of biological identity at the nanoscale and future prospects, Nat. Nanotechnol 16 (2021) 229–242. [DOI] [PubMed] [Google Scholar]
- [2].Wang J, Li YY, Nie GJ, Multifunctional biomolecule nanostructures for cancer therapy, Nat. Rev. Mater 6 (2021) 766–783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Lammers T, Ferrari M, The success of nanomedicine, Nano Today 31 (2020) 100853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Seitkalieva MM, Samoylenko DE, Lotsman KA, Rodygin KS, Ananikov VP, Metal nanoparticles in ionic liquids: synthesis and catalytic applications, Coordin. Chem. Rev 445 (2021). [Google Scholar]
- [5].Mitchell MJ, Billingsley MM, Haley RM, Wechsler ME, Peppas NA, Langer R, Engineering precision nanoparticles for drug delivery, Nat. Rev. Drug Discov 20 (2021) 101–124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Shi JJ, Kantoff PW, Wooster R, Farokhzad OC, Cancer nanomedicine: progress, challenges and opportunities, Nat. Rev. Cancer 17 (2017) 20–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Ahrens ET, Bulte JWM, Tracking immune cells in vivo using magnetic resonance imaging, Nat. Rev. Immunol 13 (2013) 755–763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Pelaz B, Alexiou C, Alvarez-Puebla RA, Alves F, Andrews AM, Ashraf S, Balogh LP, Ballerini L, Bestetti A, Brendel C, Bosi S, Carril M, Chan WC, Chen C, Chen X, Chen X, Cheng Z, Cui D, Du J, Dullin C, Escudero A, Feliu N, Gao M, George M, Gogotsi Y, Grunweller A, Gu Z, Halas NJ, Hampp N, Hartmann RK, Hersam MC, Hunziker P, Jian J, Jiang X, Jungebluth P, Kadhiresan P, Kataoka K, Khademhosseini A, Kopecek J, Kotov NA, Krug HF, Lee DS, Lehr CM, Leong KW, Liang XJ, Ling Lim M, Liz–Marzan LM, Ma X, Macchiarini P, Meng H, Mohwald H, Mulvaney P, Nel AE, Nie S, Nordlander P, Okano T, Oliveira J, Park TH, Penner RM, Prato M, Puntes V, Rotello VM, Samarakoon A, Schaak RE, Shen Y, Sjoqvist S, Skirtach AG, Soliman MG, Stevens MM, Sung HW, Tang BZ, Tietze R, Udugama BN, VanEpps JS, Weil T, Weiss PS, Willner I, Wu Y, Yang L, Yue Z, Zhang Q, Zhang Q, Zhang XE, Zhao Y, Zhou X, Parak WJ, Diverse applications of nanomedicine, ACS Nano 11 (2017) 2313–2381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Allen TM, Cullis PR, Drug delivery systems: entering the mainstream, Science 303 (2004) 1818–1822. [DOI] [PubMed] [Google Scholar]
- [10].Blanco E, Shen H, Ferrari M, Principles of nanoparticle design for overcoming biological barriers to drug delivery, Nat. Biotechnol 33 (2015) 941–951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Davis ME, Chen Z, Shin DM, Nanoparticle therapeutics: an emerging treatment modality for cancer, Nat. Rev. Drug Discov 7 (2008) 771–782. [DOI] [PubMed] [Google Scholar]
- [12].Lunov O, Uzhytchak M, Smolkova B, Lunova M, Jirsa M, Dempsey NM, Dias AL, Bonfim M, Hof M, Jurkiewicz P, Petrenko Y, Kubinova S, Dejneka A, Remote actuation of apoptosis in liver cancer cells via magneto-mechanical modulation of iron oxide nanoparticles, Cancers (Basel) 11 (2019) 1873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Anselmo AC, Mitragotri S, Nanoparticles in the clinic: an update, Bioeng. Transl. Med 4 (2019) e10143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Gabizon AA, Patil Y, La-Beck NM, New insights and evolving role of pegylated liposomal doxorubicin in cancer therapy, Drug Resist Update 29 (2016) 90–106. [DOI] [PubMed] [Google Scholar]
- [15].Anchordoquy TJ, Barenholz Y, Boraschi D, Chorny M, Decuzzi P, Dobrovolskaia MA, Farhangrazi ZS, Farrell D, Gabizon A, Ghandehari H, Godin B, La-Beck NM, Ljubimova J, Moghimi SM, Pagliaro L, Park JH, Peer D, Ruoslahti E, Serkova NJ, Simberg D, Mechanisms and barriers in cancer nanomedicine: addressing challenges, looking for solutions, ACS Nano 11 (2017) 12–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Sun DX, Zhou S, Gao W, What went wrong with anticancer nanomedicine design and how to make it right, ACS Nano 14 (2020) 12281–12290. [DOI] [PubMed] [Google Scholar]
- [17].Moghimi SM, Farhangrazi ZS, Nanomedicine and the complement paradigm, Nanomed.-Nanotechnol 9 (2013) 458–460. [DOI] [PubMed] [Google Scholar]
- [18].Cheng YH, He CL, Riviere JE, Monteiro-Riviere NA, Lin ZM, Meta-analysis of nanoparticle delivery to tumors using a physiologically based pharmacokinetic modeling and simulation approach, ACS Nano 14 (2020) 3075–3095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Wilhelm S, Tavares AJ, Dai Q, Ohta S, Audet J, Dvorak HF, Chan WCW, Analysis of nanoparticle delivery to tumours, Nat. Rev. Mater 1 (2016) 16014 . [Google Scholar]
- [20].Park K, The beginning of the end of the nanomedicine hype, J. Control Release 305 (2019) 221–222. [DOI] [PubMed] [Google Scholar]
- [21].Frtus A, Smolkova B, Uzhytchak M, Lunova M, Jirsa M, Kubinova S, Dejneka A, Lunov O, Analyzing the mechanisms of iron oxide nanoparticles interactions with cells: a road from failure to success in clinical applications, J. Control Release 328 (2020) 59–77. [DOI] [PubMed] [Google Scholar]
- [22].Gause KT, Wheatley AK, Cui JW, Yan Y, Kent SJ, Caruso F, Immunological principles guiding the rational design of particles for vaccine delivery, ACS Nano 11 (2017) 54–68. [DOI] [PubMed] [Google Scholar]
- [23].Sindhwani S, Syed AM, Ngai J, Kingston BR, Maiorino L, Rothschild J, MacMillan P, Zhang YW, Rajesh NU, Hoang T, Wu JLY, Wilhelm S, Zilman A, Gadde S, Sulaiman A, Ouyang B, Lin Z, Wang LS, Egeblad M, Chan WCW, The entry of nanoparticles into solid tumours, Nat. Mater 19 (2020) 566–575. [DOI] [PubMed] [Google Scholar]
- [24].Dey S, Fan C, Gothelf KV, Li J, Lin C, Liu L, Liu N, Nijenhuis MAD, Sacca B, Simmel FC, Yan H, Zhan P, D.N.A. origami, Nature Rev. Methods Primers 1 (2021) 13. [Google Scholar]
- [25].Abedini A, Bakar AAA, Larki F, Menon PS, Islam MS, Shaari S, Recent advances in shape-controlled synthesis of noble metal nanoparticles by radiolysis route, Nanoscale Res. Lett 11 (2016) 287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].da Silva AGM, Rodrigues TS, Slater TJA, Lewis EA, Alves RS, Fajardo HV, Balzer R, da Silva AHM, de Freitas IC, Oliveira DC, Assaf JM, Probst LFD, Haigh SJ, Camargo PHC, Controlling size, morphology, and surface composition of AgAu nanodendrites in 15s for improved environmental catalysis under low metal loadings, ACS Appl. Mater. Inter 7 (2015) 25624–25632. [DOI] [PubMed] [Google Scholar]
- [27].Jahanban-Esfahlan A, Seidi K, Jaymand M, Schmidt TL, Majdi H, Javaheri T, Jahanban-Esfahlan R, Zare P, Dynamic DNA nanostructures in biomedicine: beauty, utility and limits, J. Control Release 315 (2019) 166–185 . [DOI] [PubMed] [Google Scholar]
- [28].Gangrade A, Stephanopoulos N, Bhatia D, Programmable, self-assembled DNA nanodevices for cellular programming and tissue engineering, Nanoscale 13 (2021) 16834–16846. [DOI] [PubMed] [Google Scholar]
- [29].Stephanopoulos N, Hybrid nanostructures from the self-assembly of proteins and DNA, Chem-Us 6 (2020) 364–405. [Google Scholar]
- [30].Wang WT, Arias DS, Deserno M, Ren X, Taylor RE, Emerging applications at the interface of DNA nanotechnology and cellular membranes: perspectives from biology, engineering, and physics, Apl. Bioeng. 4 (2020) 041507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Ke YG, Castro C, Choi JH, Structural DNA nanotechnology: artificial nanostructures for biomedical research, Annu. Rev. Biomed. Eng 20 (2018) 375–401. [DOI] [PubMed] [Google Scholar]
- [32].Henry SJW, Stephanopoulos N, Functionalizing DNA nanostructures for therapeutic applications, Wires Nanomed. Nanobio 13 (2021) e1729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Hong F, Zhang F, Liu Y, Yan H, origami DNA, Scaffolds for creating higher order structures, Chem. Rev 117 (2017) 12584–12640. [DOI] [PubMed] [Google Scholar]
- [34].Keller A, Linko V, Challenges and perspectives of DNA nanostructures in biomedicine, Angew. Chem. Int. Edit 59 (2020) 15818–15833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Goodman RP, Berry RM, Turberfield AJ, The single-step synthesis of a DNA tetrahedron, Chem. Commun (2004) 1372–1373. [DOI] [PubMed] [Google Scholar]
- [36].Benson E, Mohammed A, Gardell J, Masich S, Czeizler E, Orponen P, Högberg B, DNA rendering of polyhedral meshes at the nanoscale, Nature 523 (2015) 441–444. [DOI] [PubMed] [Google Scholar]
- [37].Wang W, Chen S, An B, Huang K, Bai T, Xu M, Bellot G, Ke Y, Xiang Y, Wei B, Complex wireframe DNA nanostructures from simple building blocks, Nat. Commun 10 (2019) 1067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Rothemund PWK, Folding DNA to create nanoscale shapes and patterns, Nature 440 (2006) 297–302. [DOI] [PubMed] [Google Scholar]
- [39].Han D, Pal S, Nangreave J, Deng Z, Liu Y, Yan H, DNA origami with complex curvatures in three-dimensional space, Science 332 (2011) 342–346. [DOI] [PubMed] [Google Scholar]
- [40].Douglas SM, Dietz H, Liedl T, Högberg B, Graf F, Shih WM, Self-assembly of DNA into nanoscale three-dimensional shapes, Nature 459 (2009) 414–418 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Ke Y, Ong LL, Shih WM, Yin P, Three-dimensional structures self-assembled from DNA bricks, Science 338 (2012) 1177–1183 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Dong YH, Yao C, Zhu Y, Yang L, Luo D, Yang DY, DNA functional materials assembled from branched DNA: design, synthesis, and applications, Chem. Rev 120 (2020) 9420–9481 . [DOI] [PubMed] [Google Scholar]
- [43].Wang X, Chandrasekaran AR, Shen ZY, Ohayon YP, Wang T, Kizer ME, Sha RJ, Mao CD, Yan H, Zhang XP, Liao SP, Ding BQ, Chakraborty B, Jonoska N, Niu D, Gu HZ, Chao J, Gao X, Li YH, Ciengshin T, Seeman NC, Paranemic crossover DNA: there and back again, Chem. Rev 119 (2019) 6273–6289. [DOI] [PubMed] [Google Scholar]
- [44].Andersen ES, Dong M, Nielsen MM, Jahn K, Subramani R, Mamdouh W, Golas MM, Sander B, Stark H, Oliveira CLP, Pedersen JS, Birkedal V, Besenbacher F, Gothelf KV, Kjems J, Self-assembly of a nanoscale DNA box with a controllable lid, Nature 459 (2009) 73–76. [DOI] [PubMed] [Google Scholar]
- [45].Keum J-W, Bermudez H, DNA-based delivery vehicles: pH-controlled disassembly and cargo release, Chem. Commun 48 (2012) 12118–12120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Gerling T, Wagenbauer KF, Neuner AM, Dietz H, Dynamic DNA devices and assemblies formed by shape-complementary, non-base pairing 3D components, Science 347 (2015) 1446–1452. [DOI] [PubMed] [Google Scholar]
- [47].Wickham SFJ, Auer A, Min J, Ponnuswamy N, Woehrstein JB, Schueder F, Strauss MT, Schnitzbauer J, Nathwani B, Zhao Z, Perrault SD, Hahn J, Lee S, Bastings MM, Helmig SW, Kodal AL, Yin P, Jungmann R, Shih WM, Complex multicomponent patterns rendered on a 3D DNA-barrel pegboard, Nat. Commun 11 (2020) 5768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Li F, Lv ZY, Zhang X, Dong YH, Ding XH, Li ZM, Li S, Yao C, Yang DY, Supramolecular self-assembled DNA nanosystem for synergistic chemical and gene regulations on cancer cells, Angew. Chem. Int. Edit 60 (2021) 25557–25566. [DOI] [PubMed] [Google Scholar]
- [49].Wang Y, Santos A, Evdokiou A, Losic D, An overview of nanotoxicity and nanomedicine research: principles, progress and implications for cancer therapy, J. Mater. Chem. B 3 (2015) 7153–7172. [DOI] [PubMed] [Google Scholar]
- [50].Khanna P, Ong C, Bay BH, Baeg GH, Nanotoxicity: an interplay of oxidative stress, inflammation and cell death, Nanomaterials-Basel 5 (2015) 1163–1180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Douglas SM, Dietz H, Liedl T, Hogberg B, Graf F, Shih WM, Self-assembly of DNA into nanoscale three-dimensional shapes, Nature 459 (2009) 414–418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Simmons CR, MacCulloch T, Zhang F, Liu Y, Stephanopoulos N, Yan H, A self-assembled rhombohedral DNA crystal scaffold with tunable cavity sizes and high-resolution structural detail, Angew. Chem. Int. Edit 59 (2020) 18619–18626. [DOI] [PubMed] [Google Scholar]
- [53].Mechanism matters, Nat. Med 16 (2010) 347. [DOI] [PubMed] [Google Scholar]
- [54].Ehrenstein MR, Mauri C, If the treatment works, do we need to know why?: the promise of immunotherapy for experimental medicine, J. Exp. Med 204 (2007) 2249–2252 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Schenone M, Dancik V, Wagner BK, Clemons PA, Target identification and mechanism of action in chemical biology and drug discovery, Nat. Chem. Biol 9 (2013) 232–240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [56].Liebler DC, Guengerich FP, Elucidating mechanisms of drug-induced toxicity, Nat. Rev. Drug Discov 4 (2005) 410–420. [DOI] [PubMed] [Google Scholar]
- [57].Smolkova B, Frtus A, Uzhytchak M, Lunova M, Kubinova S, Dejneka A, Lunov O, Critical analysis of non-thermal plasma-driven modulation of immune cells from clinical perspective, Int. J. Mol. Sci 21 (2020) 6226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [58].Ke PC, Lin S, Parak WJ, Davis TP, Caruso F, A decade of the protein corona, ACS Nano 11 (2017) 11773–11776. [DOI] [PubMed] [Google Scholar]
- [59].Del Pino P, Pelaz B, Zhang Q, Maffre P, Nienhaus GU, Parak WJ, Protein corona formation around nanoparticles - from the past to the future, Mater. Horiz 1 (2014) 301–313. [Google Scholar]
- [60].Cai R, Chen CY, The crown and the scepter: roles of the protein corona in nanomedicine, Adv. Mater 31 (2019) e1805740. [DOI] [PubMed] [Google Scholar]
- [61].Chandrasekaran AR, Nuclease resistance of DNA nanostructures, Nat. Rev. Chem 5 (2021) 225–239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [62].Ge CC, Tian J, Zhao YL, Chen CY, Zhou RH, Chai ZF, Towards understanding of nanoparticle-protein corona, Arch. Toxicol 89 (2015) 519–539. [DOI] [PubMed] [Google Scholar]
- [63].Kharazian B, Hadipour NL, Ejtehadi MR, Understanding the nanoparticle-protein corona complexes using computational and experimental methods, Int. J. Biochem. Cell B 75 (2016) 162–174. [DOI] [PubMed] [Google Scholar]
- [64].Nel AE, Madler L, Velegol D, Xia T, Hoek EMV, Somasundaran P, Klaessig F, Castranova V, Thompson M, Understanding biophysicochemical interactions at the nano-bio interface, Nat. Mater 8 (2009) 543–557. [DOI] [PubMed] [Google Scholar]
- [65].Lunova M, Prokhorov A, Jirsa M, Hof M, Olzynska A, Jurkiewicz P, Kubinova S, Lunov O, Dejneka A, Nanoparticle core stability and surface functionalization drive the mTOR signaling pathway in hepatocellular cell lines, Sci. Rep.-UK 7 (2017) 16049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [66].Salvati A, Pitek AS, Monopoli MP, Prapainop K, Bombelli FB, Hristov DR, Kelly PM, Aberg C, Mahon E, Dawson KA, Transferrin-functionalized nanoparticles lose their targeting capabilities when a biomolecule corona adsorbs on the surface, Nat. Nanotechnol 8 (2013) 137–143. [DOI] [PubMed] [Google Scholar]
- [67].Hamad-Schifferli K, Exploiting the novel properties of protein coronas: emerging applications in nanomedicine, Nanomedicine-UK 10 (2015) 1663–1674. [DOI] [PubMed] [Google Scholar]
- [68].Auvinen H, Zhang HB, Kopilow Nonappa A, Niemela EH, Nummelin S, Correia A, Santos HA, Linko V, Kostiainen MA, Protein coating of DNA nanostructures for enhanced stability and immunocompatibility, Adv. Healthc. Mater 6 (2017) 1700692. [DOI] [PubMed] [Google Scholar]
- [69].Estrich NA, Hernandez-Garcia A, de Vries R, LaBean TH, Engineered diblock polypeptides improve DNA and gold solubility during molecular assembly, ACS Nano 11 (2017) 831–842. [DOI] [PubMed] [Google Scholar]
- [70].Hernandez-Garcia A, Estrich NA, Werten MW, Van Der Maarel JR, LaBean TH, de Wolf FA, Cohen Stuart MA, de Vries R, Precise coating of a wide range of DNA templates by a protein polymer with a DNA binding domain, ACS Nano 11 (2017) 144–152. [DOI] [PubMed] [Google Scholar]
- [71].Ponnuswamy N, Bastings MMC, Nathwani B, Ryu JH, Chou LYT, Vinther M, Li WA, Anastassacos FM, Mooney DJ, Shih WM, Oligolysine-based coating protects DNA nanostructures from low-salt denaturation and nuclease degradation, Nat. Commun 8 (2017) 15654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [72].Kozma GT, Shimizu T, Ishida T, Szebeni J, Anti-PEG antibodies: properties, formation, testing and role in adverse immune reactions to PEGylated nano-biopharmaceuticals, Adv. Drug Deliv. Rev 154 (2020) 163–175. [DOI] [PubMed] [Google Scholar]
- [73].Kozma GT, Meszaros T, Vashegyi I, Fulop T, Orfi E, Dezsi L, Rosivall L, Bavli Y, Urbanics R, Mollnes TE, Barenholz Y, Szebeni J, Pseudo-anaphylaxis to polyethylene glycol (PEG)-coated liposomes: roles of anti-PEG IgM and complement activation in a porcine model of human infusion reactions, ACS Nano 13 (2019) 9315–9324. [DOI] [PubMed] [Google Scholar]
- [74].Shiraishi K, Yokoyama M, Toxicity and immunogenicity concerns related to PEGylated-micelle carrier systems: a review, Sci. Technol. Adv. Mat 20 (2019) 324–336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [75].Wang PF, Meyer TA, Pan V, Dutta PK, Ke YG, The beauty and utility of DNA origami, Chem-Us 2 (2017) 359–382. [Google Scholar]
- [76].Smolkova B, MacCulloch T, Rockwood TF, Liu MH, Henry SJW, Frtus A, Uzhytchak M, Lunova M, Hof M, Jurkiewicz P, Dejneka A, Stephanopoulos N, Lunov O, Protein corona inhibits endosomal escape of functionalized DNA nanostructures in living cells, ACS Appl. Mater. Inter 13 (2021) 46375–46390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [77].Li M, Tao Y, Shu YL, LaRochelle JR, Steinauer A, Thompson D, Schepartz A, Chen ZY, Liu DR, Discovery and characterization of a peptide that enhances endosomal escape of delivered proteins in vitro and in vivo, J. Am. Chem. Soc 137 (2015) 14084–14093. [DOI] [PubMed] [Google Scholar]
- [78].Wymann MP, Schneiter R, Lipid signalling in disease, Nat. Rev. Mol. Cell Bio 9 (2008) 162–176. [DOI] [PubMed] [Google Scholar]
- [79].Kholodenko BN, Cell-signalling dynamics in time and space, Nat. Rev. Mol. Cell Bio 7 (2006) 165–176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [80].Yang NJ, Hinner MJ, Getting across the cell membrane: an overview for small molecules, peptides, and proteins, Methods Mol. Biol 1266 (2015) 29–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [81].Lunov O, Syrovets T, Loos C, Nienhaus GU, Mailander V, Landfester K, Rouis M, Simmet T, Amino-functionalized polystyrene nanoparticles activate the NLRP3 inflammasome in human macrophages, ACS Nano 5 (2011) 9648–9657 . [DOI] [PubMed] [Google Scholar]
- [82].Walsh AS, Yin HF, Erben CM, Wood MJA, Turberfield AJ, DNA cage delivery to mammalian cells, ACS Nano 5 (2011) 5427–5432. [DOI] [PubMed] [Google Scholar]
- [83].Tian XH, Angioletti-Uberti S, Battaglia G, On the design of precision nanomedicines, Sci. Adv 6 (2020) eaat0919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [84].Yong KW, Yuen D, Chen MZ, Porter CJH, Johnston APR, Pointing in the right direction: controlling the orientation of proteins on nanoparticles improves targeting efficiency, Nano Lett. 19 (2019) 1827–1831. [DOI] [PubMed] [Google Scholar]
- [85].Yong KW, Yuen D, Chen MZ, Johnston APR, Engineering the orientation, density, and flexibility of single-domain antibodies on nanoparticles to improve cell targeting, ACS Appl. Mater. Inter 12 (2020) 5593–5600. [DOI] [PubMed] [Google Scholar]
- [86].Johnston APR, Kamphuis MMJ, Such GK, Scott AM, Nice EC, Heath JK, Caruso F, Targeting cancer cells: controlling the binding and internalization of antibody-functionalized capsules, ACS Nano 6 (2012) 6667–6674 . [DOI] [PubMed] [Google Scholar]
- [87].Colombo M, Fiandra L, Alessio G, Mazzucchelli S, Nebuloni M, De Palma C, Kantner K, Pelaz B, Rotem R, Corsi F, Parak WJ, Prosperi D, Tumour homing and therapeutic effect of colloidal nanoparticles depend on the number of attached antibodies, Nat. Commun 7 (2016) 13818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [88].Zhang QY, Reinhard BM, Ligand density and nanoparticle clustering cooperate in the multivalent amplification of epidermal growth factor receptor activation, ACS Nano 12 (2018) 10473–10485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [89].Shimoni O, Yan Y, Wang YJ, Caruso F, Shape-dependent cellular processing of polyelectrolyte capsules, ACS Nano 7 (2013) 522–530. [DOI] [PubMed] [Google Scholar]
- [90].Dasgupta S, Auth T, Gompper G, Shape and orientation matter for the cellular uptake of nonspherical particles, Nano Lett. 14 (2014) 687–693. [DOI] [PubMed] [Google Scholar]
- [91].Shang L, Nienhaus K, Nienhaus GU, Engineered nanoparticles interacting with cells: size matters, J. Nanobiotechnol 12 (2014) 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [92].Barua S, Yoo JW, Kolhar P, Wakankar A, Gokarn YR, Mitragotri S, Particle shape enhances specificity of antibody-displaying nanoparticles, Proc. Natl. Acad. Sci. USA 110 (2013) 3270–3275 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- [93].Chithrani BD, Ghazani AA, Chan WCW, Determining the size and shape dependence of gold nanoparticle uptake into mammalian cells, Nano Lett. 6 (2006) 662–668. [DOI] [PubMed] [Google Scholar]
- [94].Shaw A, Lundin V, Petrova E, Fordos F, Benson E, Al-Amin A, Herland A, Blokzijl A, Hogberg B, Teixeira AI, Spatial control of membrane receptor function using ligand nanocalipers, Nat. Methods 11 (2014) 841–846. [DOI] [PubMed] [Google Scholar]
- [95].Kwon PS, Ren S, Kwon SJ, Kizer ME, Kuo L, Xie M, Zhu D, Zhou F, Zhang FM, Kim D, Fraser K, Kramer LD, Seeman NC, Dordick JS, Linhardt RJ, Chao J, Wang X, Designer DNA architecture offers precise and multivalent spatial pattern-recognition for viral sensing and inhibition, Nat. Chem 12 (2020) 26–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [96].Cremers GAO, Rosier BJHM, Meijs A, Tito NB, van Duijnhoven SMJ, van Eenennaam H, Albertazzi L, de Greef TFA, Determinants of ligand-functionalized DNA nanostructure-cell interactions, J. Am. Chem. Soc 143 (2021) 10131–10142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [97].Castro CE, Kilchherr F, Kim DN, Shiao EL, Wauer T, Wortmann P, Bathe M, Dietz H, A primer to scaffolded DNA origami, Nat. Methods 8 (2011) 221–229. [DOI] [PubMed] [Google Scholar]
- [98].P J, Sobczak J, Martin TG, Gerling T, Dietz H, Rapid folding of DNA into nanoscale shapes at constant temperature, Science 338 (2012) 1458–1461. [DOI] [PubMed] [Google Scholar]
- [99].Verheyen T, Fang T, Lindenhofer D, Wang Y, Akopyan K, Lindqvist A, Hogberg A, Teixeira AI, Spatial organization-dependent EphA2 transcriptional responses revealed by ligand nanocalipers, Nucleic. Acids. Res 48 (2020) 5777–5787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [100].Veneziano R, Moyer TJ, Stone MB, Wamhoff EC, Read BJ, Mukherjee S, Shepherd TR, Das J, Schief WR, Irvine DJ, Bathe M, Role of nanoscale antigen organization on B-cell activation probed using DNA origami, Nat. Nanotechnol 15 (2020) 716–723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [101].Rosier BJHM, Markvoort AJ, Audenis BG, Roodhuizen JAL, den Hamer A, Brunsveld L, de Greef TFA, Proximity-induced caspase-9 activation on a DNA origami-based synthetic apoptosome, Nat. Catal 3 (2020) 295–306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [102].Shaw A, Hoffecker IT, Smyrlaki I, Rosa J, Grevys A, Bratlie D, Sandlie I, Michaelsen TE, Andersen JT, Hogberg B, Binding to nanopatterned antigens is dominated by the spatial tolerance of antibodies, Nat. Nanotechnol 14 (2019) 184–190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [103].Berger RML, Weck JM, Kempe SM, Hill O, Liedl T, Radler JO, Monzel C, Heuer-Jungemann A, Nanoscale FasL organization on DNA origami to decipher apoptosis signal activation in cells, Small 17 (2021) 2101678 . [DOI] [PubMed] [Google Scholar]
- [104].Graves JD, Kordich JJ, Huang TH, Piasecki J, Bush TL, Sullivan T, Foltz IN, Chang W, Douangpanya H, Dang T, O’Neil JW, Mallari R, Zhao XN, Branstetter DG, Rossi JM, Long AM, Huang X, Holland PM, Apo2L/TRAIL and the death receptor 5 agonist antibody AMG 655 cooperate to promote receptor clustering and antitumor activity, Cancer Cell 26 (2014) 177–189. [DOI] [PubMed] [Google Scholar]
- [105].Wang Y, Baars I, Fordos F, Hogberg B, Clustering of death receptor for apoptosis using nanoscale patterns of peptides, ACS Nano 15 (2021) 9614–9626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [106].Xu F, Xia Q, Wang PF, Rationally designed DNA nanostructures for drug delivery, Front. Chem 8 (2020) 751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [107].Harman C, The fallacy of ‘alternative’ medicine, Nat. Rev. Nephrol 5 (2009) 361 . [DOI] [PubMed] [Google Scholar]
- [108].Singh S, Ernst E, Trick Or treatment? : Alternative Medicine On Trial, Transworld, London, 2009. [Google Scholar]
- [109]. https://blogs.bmj.com/bmj/2012/08/15/edzard-ernst-the-natural-equals-safe-fallacy/.
- [110].Volik S, Alcaide M, Morin RD, Collins C, Cell-free DNA (cfDNA): clinical significance and utility in cancer shaped by emerging technologies, Mol. Cancer Res 14 (2016) 898–908 . [DOI] [PubMed] [Google Scholar]
- [111].Bartoloni E, Ludovini V, Alunno A, Pistola L, Bistoni O, Crino L, Gerli R, Increased levels of circulating DNA in patients with systemic autoimmune diseases: a possible marker of disease activity in Sjogren’s syndrome, Lupus 20 (2011) 928–935. [DOI] [PubMed] [Google Scholar]
- [112].Atamaniuk J, Kopecky C, Skoupy S, Saemann MD, Weichhart T, Apoptotic cell-free DNA promotes inflammation in haemodialysis patients, Nephrol. Dial. Transpl 27 (2012) 902–905 . [DOI] [PubMed] [Google Scholar]
- [113].Dholakia S, De Vlaminck I, Khush KK, Adding insult on injury: immunogenic role for donor-derived cell-free DNA? Transplantation 104 (2020) 2266–2271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [114].Verhoeven J, Peeters A, De Jonge E, Von der Thusen J, Baan C, van Schaik R, Hesselink D, Manintveld O, Boer K, Donor-derived cell-free DNA for the detection of heart allograft injury: impact of rejection severity and timing of the liquid biopsy, Transpl. Int 34 (2021) 32–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [115].Oellerich M, Sherwood K, Keown P, Schutz E, Beck J, Stegbauer J, Rump LC, Watson PD, Liquid biopsies: donor-derived cell-free DNA for the detection of kidney allograft injury, Nat. Rev. Nephrol 17 (2021) 591–603. [DOI] [PubMed] [Google Scholar]
- [116].Peneder P, Stuetz AM, Surdez D, Krumbholz M, Semper S, Chicard M, Sheffield NC, Pierron G, Lapouble E, Toetzl M, Erguener B, Barreca D, Rendeiro AF, Agaimy A, Boztug H, Engstler G, Dworzak M, Bernkopf M, Taschner-Mandl S, Ambros IM, Myklebost O, Marec-Berard P, Burchill SA, Brennan B, Strauss SJ, Whelan J, Schleiermacher G, Schaefer C, Dirksen U, Hutter A, Boye K, Ambros PF, Delattre O, Metzler M, Bock C, Tomazou EM, Multimodal analysis of cell-free DNA whole-genome sequencing for pediatric cancers with low mutational burden, Nat. Commun 12 (2021) 3230 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- [117].Zhang Q, Raoof M, Chen Y, Sumi Y, Sursal T, Junger W, Brohi K, Itagaki K, Hauser CJ, Circulating mitochondrial DAMPs cause inflammatory responses to injury, Nature 464 (2010) 104–107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [118].Matzinger P, Tolerance, danger, and the extended family, Annu. Rev. Immunol 12 (1994) 991–1045. [DOI] [PubMed] [Google Scholar]
- [119].Bae JH, Jo S, Kim SJ, Lee JM, Jeong JH, Kang JS, Cho NJ, Kim SS, Lee EY, Moon JS, Circulating cell-free mtDNA contributes to AIM2 inflammasome–mediated chronic inflammation in patients with type 2 diabetes, Cells-Basel 8 (2019) 328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [120].Messaoudi S, Greschner AA, Gauthier MA, Progress toward absorption, distribution, metabolism, elimination, and toxicity of DNA nanostructures, Adv. Ther.-Germany 2 (2019) 1900144. [Google Scholar]
- [121]. https://www.epa.gov/pesticide-labels/criteria-biodegradability-claims-products-registered-under-fifra.
- [122].Zimmermann L, Dombrowski A, Völker C, Wagner M, Are bioplastics and plant-based materials safer than conventional plastics? In vitro toxicity and chemical composition 145 (2020) 106066 Environment International. [DOI] [PubMed] [Google Scholar]
- [123].Levis JW, Barlaz MA, Is biodegradability a desirable attribute for discarded solid waste? Perspectives from a national landfill greenhouse gas inventory model, Environ. Sci. Technol 45 (2011) 5470–5476 . [DOI] [PubMed] [Google Scholar]
- [124].Andrade RJ, Chalasani N, Bjornsson ES, Suzuki A, Kullak-Ublick GA, Watkins PB, Devarbhavi H, Merz M, Lucena MI, Kaplowitz N, Aithal GP, Drug-induced liver injury, Nat. Rev. Dis. Primers 5 (2019) 58. [DOI] [PubMed] [Google Scholar]
- [125].Fernandez-Checa JC, Bagnaninchi P, Ye H, Sancho-Bru P, Falcon-Perez JM, Royo F, Garcia-Ruiz C, Konu O, Miranda J, Lunov O, Dejneka A, Elfick A, McDonald A, Sullivan GJ, Aithal GP, Lucena MI, Andrade RJ, Fromenty B, Kranendonk M, Cubero FJ, Nelson LJ, Advanced preclinical models for evaluation of drug-induced liver injury - consensus statement by the European Drug-Induced Liver Injury Network [PRO-EURO-DILI-NET], J. Hepatol 75 (2021) 935–959. [DOI] [PubMed] [Google Scholar]
- [126].Chen MJ, Borlak J, Tong WD, High lipophilicity and high daily dose of oral medications are associated with significant risk for drug-induced liver injury, Hepatology 58 (2013) 388–396. [DOI] [PubMed] [Google Scholar]
- [127].Roberts TC, Langer R, Wood MJA, Advances in oligonucleotide drug delivery, Nat. Rev. Drug Discov 19 (2020) 673–694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [128].Alyamkina EA, Nikolin VP, Popova NA, Minkevich AM, Kozel AV, Dolgova DV, Efremov YR, Bayborodin SI, Andrushkevich OM, Taranov OS, Omigov VV, Rogachev VA, Proskurina AS, Vereschagin EI, Kiseleva EV, Zhukova MV, Ostanin AA, Chernykh ER, Bogachev SS, Shurdov MA, Combination of cyclophosphamide and double-stranded DNA demonstrates synergistic toxicity against established xenografts, Cancer Cell Int. 15 (2015) 32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [129].Guiducci C, Tripodo C, Gong M, Sangaletti S, Colombo MP, Coffman RL, Barrat DJ, Autoimmune skin inflammation is dependent on plasmacytoid dendritic cell activation by nucleic acids via TLR7 and TLR9, J. Exp. Med 207 (2010) 2931–2942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [130].Pisetsky DS, Ullal AJ, The blood nucleome in the pathogenesis of SLE, Au- toimmun. Rev 10 (2010) 35–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [131].Almqvist N, Winkler TH, Mårtensson I-L, Autoantibodies: focus on anti-DNA antibodies, Self Nonself 2 (2011) 11–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [132].Rhodes A, Wort SJ, Thomas H, Collinson P, Bennett ED, Plasma DNA concentration as a predictor of mortality and sepsis in critically ill patients, Crit. Care 10 (2006) R60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [133].Kung CT, Hsiao SY, Tsai TC, Su CM, Chang WN, Huang CR, Wang HC, Lin WC, Chang HW, Lin YJ, Cheng BC, Su BYJ, Tsai NW, Lu CH, Plasma nuclear and mitochondrial DNA levels as predictors of outcome in severe sepsis patients in the emergency room, J. Transl. Med 10 (2012) 130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [134].Bourrinet P, Bengele HH, Bonnemain B, Dencausse A, Idee JM, Jacobs PM, Lewis JM, Preclinical safety and pharmacokinetic profile of ferumoxtran-10, an ultrasmall superparamagnetic iron oxide magnetic resonance contrast agent, Invest. Radiol 41 (2006) 313–324. [DOI] [PubMed] [Google Scholar]
- [135].Fadeel B, Farcal L, Hardy B, Vazquez-Campos S, Hristozov D, Marcomini A, Lynch H, Valsami-Jones E, Alenius H, Savolainen K, Advanced tools for the safety assessment of nanomaterials, Nat. Nanotechnol 13 (2018) 537–543. [DOI] [PubMed] [Google Scholar]
- [136].Hirai T, Yoshioka Y, Izumi N, Ichihashi K, Handa T, Nishijima N, Uemura E, Sagami K, Takahashi H, Yamaguchi M, Nagano K, Mukai Y, Kamada H, Tsunoda S, Ishii KJ, Higashisaka K, Tsutsumi Y, Metal nanoparticles in the presence of lipopolysaccharides trigger the onset of metal allergy in mice, Nat. Nanotechnol 11 (2016) 808–816. [DOI] [PubMed] [Google Scholar]
- [137].Lombi E, Donner E, Dusinska M, Wickson F, A One Health approach to managing the applications and implications of nanotechnologies in agriculture, Nat. Nanotechnol 14 (2019) 523–531. [DOI] [PubMed] [Google Scholar]
- [138].Levada K, Pshenichnikov S, Omelyanchik A, Rodionova V, Nikitin A, Savchenko A, Schetinin I, Zhukov D, Abakumov M, Majouga A, Lunova M, Jirsa M, Smolkova B, Uzhytchak M, Dejneka A, Lunov O, Progressive lysosomal membrane permeabilization induced by iron oxide nanoparticles drives hepatic cell autophagy and apoptosis, Nano Converg. 7 (2020) 17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [139].Tsang MP, Kikuchi-Uehara E, Sonnemann GW, Aymonier C, Hirao M, Evaluating nanotechnology opportunities and risks through integration of life-cycle and risk assessment, Nat. Nanotechnol 12 (2017) 734–739. [DOI] [PubMed] [Google Scholar]
- [140].Liu YY, Yu NY, Fang WD, Tan QG, Ji R, Yang LY, Wei S, Zhang XW, Miao AJ, Photodegradation of carbon dots cause cytotoxicity, Nat. Commun 12 (2021) 812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [141].Ben-David U, Siranosian B, Ha G, Tang H, Oren Y, Hinohara K, Strathdee CA, Dempster J, Lyons NJ, Burns R, Nag A, Kugener G, Cimini B, Tsvetkov P, Maruvka YE, O’Rourke R, Garrity A, Tubelli AA, Bandopadhayay P, Tsherniak A, Vazquez F, Wong B, Birger C, Ghandi M, Thorner AR, Bittker HA, Meyerson M, Getz G, Beroukhim R, Golub TR, Genetic and transcriptional evolution alters cancer cell line drug response, Nature 560 (2018) 325–330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [142].Liu YS, Mi Y, Mueller T, Kreibich S, Williams EG, Van Drogen A, Borel C, Franks M, Germain PL, Bludau I, Mehnert M, Seifert M, Emmenlauer M, Sorg I, Bezrukov F, Bena FS, Zhou H, Dehio C, Testa G, Saez-Rodriguez J, Antonarakis SE, Hardt WD, Aebersold R, Multi-omic measurements of heterogeneity in HeLa cells across laboratories, Nat. Biotechnol 37 (2019) 314–322 . [DOI] [PubMed] [Google Scholar]
- [143].Marx V, Cell-line authentication demystified, Nat. Methods 11 (2014) 483–488. [DOI] [PubMed] [Google Scholar]
- [144].Masters JR, HeLa cells 50 years on: the good, the bad and the ugly, Nat. Rev. Cancer 2 (2002) 315–319. [DOI] [PubMed] [Google Scholar]
- [145].Horbach SPJM, Halffman W, The ghosts of HeLa: how cell line misidentification contaminates the scientific literature, PLoS One 12 (2017) e0186281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [146].Geraghty RJ, Capes-Davis A, Davis JM, Downward J, Freshney RI, Knezevic I, Lovell-Badge R, Masters JRW, Meredith J, Stacey GN, Thraves P, Vias M, Guidelines for the use of cell lines in biomedical research, Brit. J. Cancer 111 (2014) 1021–1046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [147].Lorsch JR, Collins FS, Lippincott-Schwartz J, Fixing problems with cell lines, Science 346 (2014) 1452–1453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [148].Richter M, Piwocka O, Musielak M, Piotrowski I, Suchorska WM, Trzeciak T, From donor to the lab: a fascinating journey of primary cell lines, Front. Cell Dev. Biol 9 (2021) 711381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [149].Schwende H, Fitzke E, Ambs P, Dieter P, Differences in the state of differentiation of THP-1 cells induced by phorbol ester and 1,25-dihydroxyvitamin d-3, J. Leukocyte Biol 59 (1996) 555–561. [PubMed] [Google Scholar]
- [150].Kohro T, Tanaka T, Murakami T, Wada Y, Aburatani H, Hamakubo T, Kodama T, A comparison of differences in the gene expression profiles of phorbol 12-myristate 13-acetate differentiated THP-1 cells and human monocyte-derived macrophage, J. Atheroscler. Thromb 11 (2004) 88–97. [DOI] [PubMed] [Google Scholar]
- [151].Lunov O, Syrovets T, Loos C, Beil J, Delecher M, Tron K, Nienhaus GU, Musyanovych A, Mailander V, Landfester K, Simmet T, Differential uptake of functionalized polystyrene nanoparticles by human macrophages and a monocytic cell line, ACS Nano 5 (2011) 1657–1669. [DOI] [PubMed] [Google Scholar]
- [152].Lunov O, Syrovets T, Buchele B, Jiang XE, Rocker C, Tron K, Nienhaus GU, Walther P, Mailander V, Landfester K, Simmet T, The effect of carboxydextran-coated superparamagnetic iron oxide nanoparticles on c-Jun N-terminal kinase-mediated apoptosis in human macrophages, Biomaterials 31 (2010) 5063–5071 . [DOI] [PubMed] [Google Scholar]
- [153].Lunov O, Syrovets T, Rocker C, Tron K, Nienhaus GU, Rasche V, Mailander V, Landfester K, Simmet T, Lysosomal degradation of the carboxydextran shell of coated superparamagnetic iron oxide nanoparticles and the fate of professional phagocytes, Biomaterials 31 (2010) 9015–9022. [DOI] [PubMed] [Google Scholar]
- [154].Uzhytchak M, Smolkova B, Lunova M, Jirsa M, Frtus A, Kubinova S, Dejneka A, Lunov O, Iron oxide nanoparticle-induced autophagic flux is regulated by interplay between p53-mTOR axis and Bcl-2 signaling in hepatic cells, Cells-Basel 9 (2020) 1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [155].Chandrasekaran AR, Punnoose JA, Zhou LF, Dey P, Dey BK, Halvorsen K, DNA nanotechnology approaches for microRNA detection and diagnosis, Nucleic. Acids. Res 47 (2019) 10489–10505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [156].Coleridge EL, Dunn KE, Assessing the cost-effectiveness of DNA origami nanostructures for targeted delivery of anti-cancer drugs to tumours, Biomed. Phys. Eng. Expr 6 (2020) 065030. [DOI] [PubMed] [Google Scholar]
- [157].Aldaye FA, Senapedis WT, Silver PA, Way JC, A structurally tunable DNA-based extracellular matrix, J. Am. Chem. Soc 132 (2010) 14727–14729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [158].Jungmann R, Avendano MS, Woehrstein JB, Dai MJ, Shih WM, Yin P, Multiplexed 3D cellular super-resolution imaging with DNA-PAINT and Exchange-PAINT, Nat. Methods 11 (2014) 313–318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [159].Mo FL, Jiang K, Zhao D, Wang YQ, Song J, Tan WH, DNA hydrogel-based gene editing and drug delivery systems, Adv. Drug Deliv. Rev 168 (2021) 79–98 . [DOI] [PubMed] [Google Scholar]
- [160].Gacanin J, Synatschke CV, Weil T, Biomedical applications of DNA-based hydrogels, Adv. Funct. Mater 30 (2020) 1906253. [Google Scholar]
- [161].Guo XC, Li F, Liu CX, Zhu Y, Xiao NN, Gu Z, Luo D, Jiang JH, Yang DY, Construction of organelle-like architecture by dynamic DNA assembly in living cells, Angew. Chem. Int. Edit 59 (2020) 20651–20658. [DOI] [PubMed] [Google Scholar]
- [162].Li FY, Lyu DY, Liu S, Guo WW, DNA hydrogels and microgels for biosensing and biomedical applications, Adv. Mater 32 (2020) 1806538. [DOI] [PubMed] [Google Scholar]
- [163].Yao C, Tang H, Wu WJ, Tang JP, Guo WW, Luo D, Yang DY, Double rolling circle amplification generates physically cross-linked DNA network for stem cell fishing, J. Am. Chem. Soc 142 (2020) 3422–3429. [DOI] [PubMed] [Google Scholar]
- [164].Yao C, Zhu CX, Tang JP, Ou JH, Zhang R, Yang DY, T lymphocyte-captured DNA network for localized immunotherapy, J. Am. Chem. Soc 143 (2021) 19330–19340. [DOI] [PubMed] [Google Scholar]
- [165].Umeki Y, Saito M, Kusamori K, Tsujimura M, Nishimura M, Takahashi Y, Takakura Y, Nishikawa M, Combined encapsulation of a tumor antigen and immune cells using a self-assembling immunostimulatory DNA hydrogel to enhance antigen-specific tumor immunity, J. Control Release 288 (2018) 189–198. [DOI] [PubMed] [Google Scholar]
- [166].Shimomura S, Nishimura T, Ogura Y, Tanida J, Photothermal fabrication of microscale patterned DNA hydrogels, R. Soc. Open Sci 5 (2018) 171779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [167].Dawidczyk CM, Russell LM, Searson PC, Recommendations for benchmarking preclinical studies of nanomedicines, Cancer Res. 75 (2015) 4016–4020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [168].Morgan RA, Human tumor xenografts: the good, the bad, and the ugly, Mol. Ther 20 (2012) 882–884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [169].Bjornmalm M, Thurecht KJ, Michael M, Scott AM, Caruso F, Bridging bio–nano science and cancer nanomedicine, ACS Nano 11 (2017) 9594–9613. [DOI] [PubMed] [Google Scholar]
- [170].Lee J, Kotliarova S, Kotliarov Y, Li AG, Su Q, Donin NM, Pastorino S, Purow AW, Christopher N, Zhang W, Park JK, Fine HA, Tumor stem cells derived from glioblastomas cultured in bFGF and EGF more closely mirror the phenotype and genotype of primary tumors than do serum-cultured cell lines, Cancer Cell 9 (2006) 391–403. [DOI] [PubMed] [Google Scholar]
- [171].Faria M, Bjornmalm M, Thurecht KJ, Kent SJ, Parton RG, Kavallaris M, Johnston APR, Gooding JJ, Corrie SR, Boyd BJ, Thordarson P, Whittaker AK, Stevens MM, Prestidge CA, Porter CJH, Parak WJ, Davis TP, Crampin EJ, Caruso F, Minimum information reporting in bio-nano experimental literature, Nat. Nanotechnol 13 (2018) 777–785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [172].Seok J, Warren HS, Cuenca AG, Mindrinos MN, Baker HV, Xu WH, Richards DR, McDonald-Smith GP, Gao H, Hennessy L, Finnerty CC, Lopez CM, Honari S, Moore EE, Minei JP, Cuschieri J, Bankey PE, Johnson JL, Sperry J, Nathens AB, Billiar TR, West MA, Jeschke MG, Klein MB, Gamelli RL, Gibran NS, Brownstein BH, Miller-Graziano C, Calvano SE, Mason PH, Cobb JP, Rahme LG, Lowry SF, Maier RV, Moldawer LL, Herndon DN, Davis RW, Xiao WZ, Tompkins RG, Injury IHR, Genomic responses in mouse models poorly mimic human inflammatory diseases, Proc. Natl. Acad. Sci. USA 110 (2013) 3507–3512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [173].Of men, not mice, Nat. Med 19 (2013) 379 . [DOI] [PubMed] [Google Scholar]
- [174].Mestas J, Hughes CCW, Of mice and not men: differences between mouse and human immunology, J. Immunol 172 (2004) 2731–2738. [DOI] [PubMed] [Google Scholar]
- [175]. https://www.science.org/content/article/us-epa-eliminate-all-mammal-testing-2035.
- [176].Harrison SP, Baumgarten SF, Verma R, Lunov O, Dejneka A, Sullivan GJ, Liver organoids: recent developments, limitations and potential, Front. Med.-Lausanne 8 (2021) 574047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [177].Huch M, Knoblich JA, Lutolf MP, Martinez-Arias A, The hope and the hype of organoid research, Development 144 (2017) 938–941. [DOI] [PubMed] [Google Scholar]
- [178].Xu HX, Jiao Y, Qin S, Zhao WH, Chu Q, Wu KM, Organoid technology in disease modelling, drug development, personalized treatment and regeneration medicine, Exp. Hematol. Oncol 7 (2018) 30 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- [179].Mahapatra C, Lee R, Paul MK, Emerging role and promise of nanomaterials in organoid research, Drug Discov. Today (2021), doi: 10.1016/j.drudis.2021.11.007. [DOI] [PubMed] [Google Scholar]
- [180].Prasad M, Kumar R, Buragohain L, Kumari A, Ghosh M, Organoid technology: a reliable developmental biology tool for organ-specific nanotoxicity evaluation, Front. Cell Dev. Biol 9 (2021) 696668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [181].Zhang YN, Poon W, Tavares AJ, McGilvray ID, Chan WCW, Nanoparticle-liver interactions: cellular uptake and hepatobiliary elimination, J. Control Release 240 (2016) 332–348 . [DOI] [PubMed] [Google Scholar]
- [182].Ouyang B, Poon W, Zhang YN, Lin ZP, Kingston BR, Tavares AJ, Zhang YW, Chen J, Valic MS, Syed AM, MacMillan P, Couture-Senecal J, Zheng D, Chan WCW, The dose threshold for nanoparticle tumour delivery, Nat. Mater 19 (2020) 1362–1371. [DOI] [PubMed] [Google Scholar]
- [183].Easo SL, Mohanan PV, Hepatotoxicity evaluation of dextran stabilized iron oxide nanoparticles in Wistar rats, Int. J. Pharmaceut 509 (2016) 28–34. [DOI] [PubMed] [Google Scholar]
- [184].Mirshafiee V, Sun BB, Chang CH, Liao YP, Jiang W, Jiang JH, Liu XS, Wang X, Xia T, Nel AE, Toxicological profiling of metal oxide nanoparticles in liver context reveals pyroptosis in Kupffer cells and macrophages versus apoptosis in hepatocytes, ACS Nano 12 (2018) 3836–3852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [185].Couto D, Freitas M, Costa VM, Chiste RC, Almeida A, Lopez-Quintela MA, Rivas J, Freitas P, Silva P, Carvalho F, Fernandes E, Biodistribution of polyacrylic acid-coated iron oxide nanoparticles is associated with proinflammatory activation and liver toxicity, J. Appl. Toxicol 36 (2016) 1321–1331. [DOI] [PubMed] [Google Scholar]
- [186].Kazemipour N, Nazifi S, Poor MHH, Esmailnezhad Z, Najafabadi RE, Esmaeili A, Hepatotoxicity and nephrotoxicity of quercetin, iron oxide nanoparticles, and quercetin conjugated with nanoparticles in rats, Comp. Clin. Path 27 (2018) 1621–1628. [Google Scholar]
- [187].Wei B, Dai M, Yin P, Complex shapes self-assembled from single-stranded DNA tiles, Nature 485 (2012) 623–626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [188].Arulkumaran N, Lanphere C, Gaupp C, Burns JR, Singer M, Howorka S, DNA nanodevices with selective immune cell interaction and function, ACS Nano 15 (2021) 4394–4404. [DOI] [PubMed] [Google Scholar]
- [189].Zhao S, Duan FY, Liu SL, Wu TT, Shang YX, Tian R, Liu JB, Wang ZG, Jiang Q, Ding BQ, Efficient intracellular delivery of RNase A using DNA origami carriers, ACS Appl. Mater. Inter 11 (2019) 11112–11118 . [DOI] [PubMed] [Google Scholar]
- [190].Abbas M, Baig MMFA, Zhang YL, Yang YS, Wu SY, Hu YQ, Wang ZC, Zhu DL, A DNA-based nanocarrier for efficient cancer therapy, J. Pharm. Anal 11 (2021) 330–339.34277121 [Google Scholar]
- [191].Jorge AF, Avino A, Pais AACC, Eritja R, Fabrega C, DNA-based nanoscaffolds as vehicles for 5-fluoro-2 ’-deoxyuridine oligomers in colorectal cancer therapy, Nanoscale 10 (2018) 7238–7249. [DOI] [PubMed] [Google Scholar]
- [192].Jones SF, Joshi H, Terry SJ, Burns JR, Aksimentiev A, Eggert US, Howorka S, Hydrophobic interactions between DNA duplexes and synthetic and biological membranes, J. Am. Chem. Soc 143 (2021) 8305–8313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [193].Wu FS, Jin X, Guan Z, Lin JP, Cai CH, Wang LQ, Li YS, Lin SL, Xu PF, Gao L, Membrane nanopores induced by nanotoroids via an insertion and pore-forming pathway, Nano Lett. 21 (2021) 8545–8553. [DOI] [PubMed] [Google Scholar]
- [194].Burns JR, Al-Juffali N, Janes SM, Howorka S, Membrane-spanning DNA nanopores with cytotoxic effect, Angew. Chem. Int. Edit 53 (2014) 12466–12470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [195].Lv C, Gu XY, Li HW, Zhao YM, Yang DL, Yu WF, Han D, Li J, Tan WH, Molecular transport through a biomimetic DNA channel on live cell membranes, ACS Nano 14 (2020) 14616–14626 . [DOI] [PubMed] [Google Scholar]
- [196].Liu K, Xu C, Liu JY, Regulation of cell binding and entry by DNA origami mediated spatial distribution of aptamers, J. Mater. Chem. B 8 (2020) 6802–6809. [DOI] [PubMed] [Google Scholar]
- [197].Liang L, Li J, Li Q, Huang Q, Shi JY, Yan H, Fan CH, Single-particle tracking and modulation of cell entry pathways of a tetrahedral DNA nanostructure in live cells, Angew. Chem. Int. Edit 53 (2014) 7745–7750. [DOI] [PubMed] [Google Scholar]
- [198].Peng XY, Fang SB, Ji B, Li M, Song J, Qiu LP, Tan WH, DNA nanostructure-programmed cell entry via corner angle-mediated molecular interaction with membrane receptors, Nano Lett. 21 (2021) 6946–6951. [DOI] [PubMed] [Google Scholar]
- [199].Wang PF, Rahman MA, Zhao ZX, Weiss K, Zhang C, Chen ZJ, Hurwitz SJ, Chen ZG, Shin DM, Ke YG, Visualization of the cellular uptake and trafficking of DNA origami nanostructures in cancer cells, J. Am. Chem. Soc 140 (2018) 2478–2484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [200].Bastings MMC, Anastassacos FM, Ponnuswamy N, Leifer FG, Cuneo G, Lin AX, Ingber DE, Ryu JH, Shih WM, Modulation of the cellular uptake of DNA origami through control over mass and shape, Nano Lett. 18 (2018) 3557–3564. [DOI] [PubMed] [Google Scholar]
- [201].Leung K, Chakraborty K, Saminathan A, Krishnan Y, A DNA nanomachine chemically resolves lysosomes in live cells, Nat. Nanotechnol 14 (2019) 176–183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [202].Whitehouse WL, Noble JE, Ryadnov MG, Howorka S, Cholesterol anchors enable efficient binding and intracellular uptake of DNA nanostructures, Bioconjugate Chem. 30 (2019) 1836–1844. [DOI] [PubMed] [Google Scholar]
- [203].Fu MF, Dai LR, Jiang Q, Tang YQ, Zhang XM, Ding BQ, Li JB, Observation of intracellular interactions between DNA origami and lysosomes by the fluorescence localization method, Chem. Commun 52 (2016) 9240–9242. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All data needed to support the conclusions are present in the paper and/or the Supplementary Materials. Additional data related to this paper may be requested from the authors.