Abstract
Allosteric mechanisms are pervasive in nature, but human-designed allosteric perturbagens are rare. The history of KRASG12C inhibitor development suggests that covalent chemistry may be a key to expanding the armamentarium of allosteric inhibitors. In that effort, irreversible targeting of a cysteine converted a non-deal allosteric binding pocket and low affinity ligands into a tractable drugging strategy. Here we examine the feasibility of expanding this approach to other allosteric pockets of RAS and kinase family members, given that both protein families are regulators of vital cellular processes that are often dysregulated in cancer and other human diseases. Moreover, these heavily studied families are the subject of numerous drug development campaigns that have resulted, sometimes serendipitously, in the discovery of allosteric inhibitors. We consequently conducted a comprehensive search for cysteines, a commonly targeted amino acid for covalent drugs, using AlphaFold-generated structures of those families. This new analysis presents potential opportunities for allosteric targeting of validated and understudied drug targets, with an emphasis on cancer therapy.
Keywords: allosteric inhibitor, covalent inhibitor, cysteinome, KRAS inhibitor, kinase inhibitor
Synthetic allosteric inhibitors are rare but desirable
Allosteric regulation by small molecules is pervasive in biology.1 It is therefore tempting to conclude that researchers should also be able to design synthetic small molecules to effectively manipulate biological phenomena using analogous allosteric mechanisms for therapeutic purposes. However, while there are numerous examples of human-designed orthosteric inhibitors,2 far fewer allosteric inhibitors have translated into therapeutic agents. Indeed, a query of drugbank for approved agents yields over 4200 entries, but only 7 in the list are described as allosteric.3-4 Nevertheless, successes are becoming more common, as seen with KRAS,5 SHP26-8 and kinase inhibitors,9 and investment in allosteric drug development is growing.10
The term allosteric comes from the Greek àllos, meaning “other”, and steric, meaning “relating to or involving the relation of atoms in space”.11 Allosteric inhibitors were first described as inhibitors that do not bear a structural resemblance to natural ligands.12 However, the allosteric classification has substantially expanded to include macromolecules that bind outside of the active site, macromolecules that transmit conformational changes from one location on a macromolecule to another, and macromolecules that induce entropic changes, but do not necessarily cause conformational changes.3 Characterization of allosteric changes has extended from consideration of binary states (tense vs. relaxed), to ensembles of conformational states.3,13 Allostery applies to most proteins, which are naturally flexible and undergo conformational changes in response to ligand binding, mutations, covalent modifications or alterations in pH, temperature or ionic strength. Such flexibility is necessary to achieve delicate and predictable regulation of biological processes, in response to environmental changes.
Allosteric drugs have several potential advantages. First, catalytic active sites are often highly conserved across protein families, whereas allosteric pockets are far less conserved. This suggests that achieving inhibitor selectivity may be more straight-forward when targeting allosteric sites. Additionally, allosteric inhibitors have the potential to fine-tune enzymatic activity, whereas orthosteric inhibitors most often work in a binary, on/off fashion.14-15
Allostery in biology
Allostery was first observed by Bohr and colleagues more than a century ago when they observed cooperativity during the binding of oxygen molecules to hemoglobin.16 The structural mechanism was initially thought of as a two-state model between tense (T) and relaxed (R) conformations (Figure 1 (A)).17 However, the model has been extended to include shifting dynamic ensembles of hemoglobin conformations.18 This example illustrates that allosteric ligands often function by altering the binding affinities of other biomolecules.
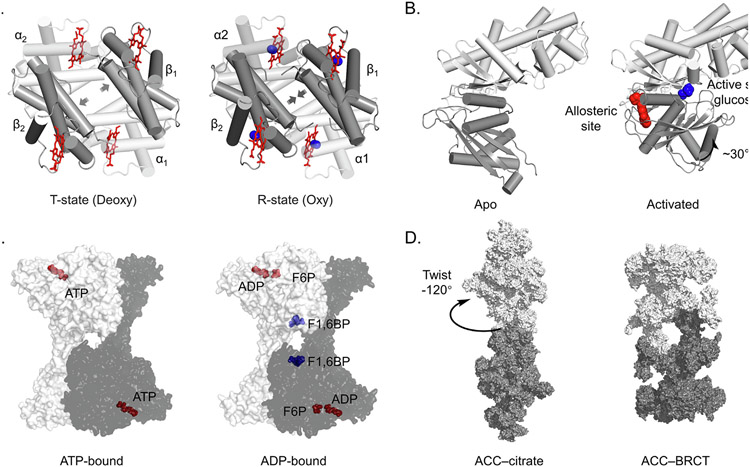
Figure 1.
Allosteric regulation is common in biology and can be targeted with drugs. (A) Hemoglobin in the T (Tense, left) and R (Relaxed, right) conformations. The T conformation is stabilized in the absence of oxygen and has low binding affinity. Binding of oxygen transitions the tetramer to the R state, increasing oxygen binding affinity. PDB: 2HHB, 1HHO. α subunit in white, β in grey, heme in red, oxygen in blue. (B) GK changes conformation to become active when the allosteric site is bound. Glucose in blue, allosteric ligand in red. PDB: 1V4T, 1V4S. (C) Binding of F2,6BP to the allosteric site of the homotetrameric phosphofructokinase-1 (PFK1) complex causes rotation of the protomers relative to each other to stimulate enzymatic activity. ATP, ADP and F6P in red, F1,6BP in blue. PDB: 4XYJ, 4XZ2. (D) Citrate allosterically activates human acetyl-CoA carboxylase (ACC) by stabilizing filament formation. PDB: 6G2D,6G2I.
Other notable early examples include enzymes involved in energy metabolism such as glucokinase (GK), which plays a major role in glucose sensing and homeostasis.19 The mechanism of sensing is allosteric. In addition to the active site, glucose binds GK at a remote site with an affinity corresponding to the ideal physiological serum concentration of glucose, about 5 mM.20 When glucose is elevated and the allosteric site is occupied, GK shifts from an open, less active state, to a closed, highly active form (Figure 1 (B)). This mechanism has been exploited to create GK activators such as dorzagliatin,21 RO-28-1675,22 PB-201,23 and others currently in clinical trials at various stages for type 2 diabetes therapies.19,24-27
Phosphofructokinase-1 (PFK1), the “gatekeeper” of the glycolytic pathway, is another well-studied example. PFK1 converts fructose-6-phosphate (F6P) to fructose 1,6-bisphosphate (F1,6BP), but is allosterically regulated by many other ligands in eukaryotes. ATP, citrate and lactate inhibit PFK1 activity, while ADP, AMP, cAMP, and F2,6BP are activating. The structural mechanism centers on stabilization of PFK1 dimers, tetramers or higher order oligomers with the active site at the interface of these protein–protein interactions.28-29 As an example, structural studies on ATP vs. ADP-bound enzyme showed a rotation of 12 degrees for the ATP vs. ADP-bound structures, leading to opening and deactivation of the catalytic site in the case of ATP binding (Figure 1(C)).30 Evolutionary divergence of allosteric binding pockets between species has enabled development of selective allosteric PFK inhibitors as antibiotics.31
Finally, mammalian Acetyl-CoA Carboxylase (ACC) is a large multifunctional dimeric protein that catalyzes a rate limiting step early in fatty acid synthesis, the carboxylation of acetyl-CoA to form malonyl CoA. In addition to regulation by phosphorylation at multiple sites, ACC activity is allosterically repressed by interactions with malonyl CoA and fatty acid derivatives, but is activated by citrate with an activation constant in the low mM range.32 One mechanism of activation was revealed by electron microscopy, showing that citrate induces large conformational changes that allow ACC to polymerize into unbranched fibers that can extend up to 1 micrometer.33 As fibers, ACC subunits are ‘locked’ into an active conformation (Figure 1(D)). The lipid mimetic TOFA (5-(tetradecyloxy)-2-furancarboxylic acid) was found to allosterically inhibit ACC, presumably in a similar manner to natural fatty acid negative regulators. Derivatives of TOFA have been explored as therapies for various maladies.34-35 More recently, structure-guided design resulted in firsocostat, an allosteric ACC inhibitor currently in human trials for non-alcoholic steatohepatitis (NASH).36
While modulators of GK, PFK1 and ACC show that allosteric mechanisms can be manipulated for therapeutic purposes, major challenges remain for other targets. Allosteric binding pockets are often non-ideal for achieving high affinity drugs. Also, these examples illustrate that targeting naturally-occurring allosteric pockets often means competition with physiological ligands that exist in cells at high concentrations. Nevertheless, recent successes in allosteric drug discovery suggest that utilization of covalent chemistry may be one way to overcome these challenges.
Allosteric RAS inhibitors
RAS was one of the first oncogenes discovered37-39 and its isoforms KRAS, HRAS and NRAS, proved to be some of the most commonly mutated drivers of cancers, particularly bad acting cancers such as pancreatic, lung, colorectal, and skin cancer.40 RAS has long been considered an “undruggable” target but allosteric inhibitors of RAS were recently developed.5 This was accomplished by a combination of good fortune and covalent chemistry.
The function of RAS is contingent on its structure because it transduces signals by transient protein–protein interactions that rely on the conformation of RAS. At baseline those interactions are governed by GTP, which constrains RAS to a compact form, or by GDP, which permits more movement.41 Two key structural elements called the switches form a large portion of the nucleotide binding pocket, and these are the principle mobile elements (Figure 2 (A)). Many disease-associated RAS mutations shift the conformational dynamics of the switches. This can translate into abnormal biochemical phenotypes such as rapid nucleotide exchange42-44 or loss of interactions with GTPase-activating proteins (GAPs) such as NF1 that normally facilitate GTP hydrolysis to inactivate RAS.45 The degree of RAS activation caused by specific mutations can be highly variable, as reflected by the wide range of diseases and syndromes driven by RAS mutations, including cancers of varying aggressiveness and developmental disorders where malignancy does not occur at all.42-43,46-48 However, this phenotypic spectrum is not fully explainable by differences in nucleotide exchange rates or GAP insensitivity. The RAS field is now examining other factors such as tissue-specific contextual variables42 or other biophysical phenomenon such as altering how RAS functions within large signaling complexes.49 Regardless, altered protein dynamics also create opportunities for selective allosteric targeting.
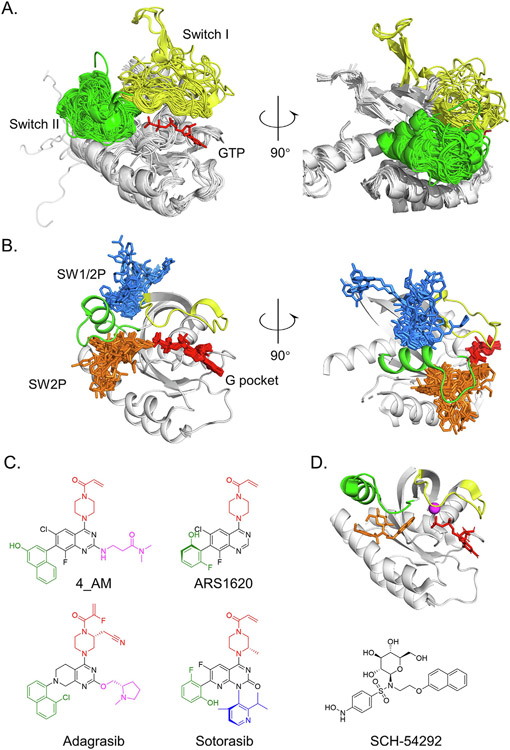
Figure 2.
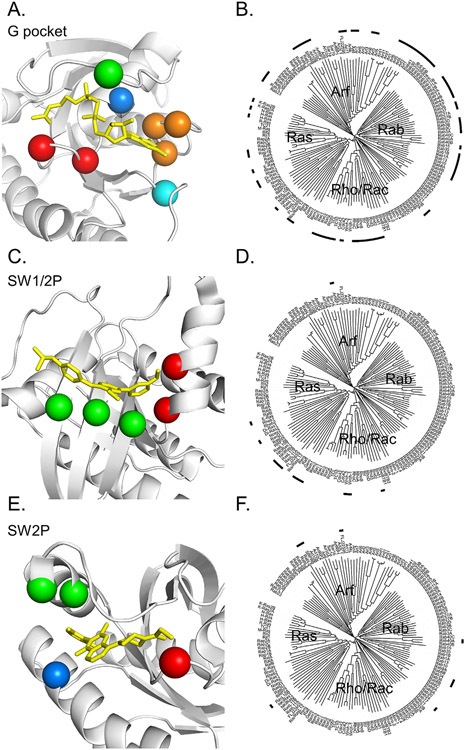
RAS is allosterically targetable because of mobility of the switches. (A) Superposition of 190 experimentally derived KRAS protein structures show the switches are the primary mobile elements. GTP in red, SWI in yellow and SWII in green. (B) Superposition of 91 experimentally derived structures of KRAS proteins bound to inhibitors demonstrates the location of major allosteric pockets. After the alignment, only ligands are displayed relative to a prototypical SW2-bound KRAS structure (PDB: 6OIM). G pocket binders in red, SW2 binders in orange, and SW1/2 binders in blue. (C) KRASG12C inhibitors are structurally related. Structurally similar components are highlighted in color for emphasis. (D) Simulated docking of SCH-54292 (orange) shows binding to the SW2 pocket of KRAS.
Clinical SW2 pocket RAS inhibitors
Targeting RAS for therapeutic purposes was proposed at the time of its discovery, but the conceptually straight-forward strategy of competing for the guanosine nucleotide binding pocket with a small molecule inhibitor was felt infeasible due to the micromolar concentrations of nucleotide in the cell and the picomolar affinity of nucleotides for RAS.50 Therefore, a major effort centered on inhibition of RAS prenylation by farnesyl transferases.51 This was successful in animal models, but ultimately failed in human trials, likely because of compensation by other prenyltransferases and functional redundancy by RAS isoforms.52 Targeting downstream of RAS has not worked because of toxicity or unanticipated signaling feedback.53-54
After many years and challenges, a direct KRAS-targeted therapy, sotorasib, is approved for cancers bearing KRASG12C, a common mutation found in lung cancer.55 Additional approvals for similar drugs, including adagrasib,56 are expected soon and a number of others are currently under evaluation.57-63 Two properties are central to the success of these drug campaigns: all advanced KRASG12C inhibitors, so far, are allosteric and covalent.
Sotorasib and adagrasib bind in a pocket formed by switch 2 that can open when KRAS is GDP-bound (Figure 2(B)). The feasibly of covalently targeting this induced pocket was first demonstrated by Shokat and colleagues by screening with low-affinity disulfide-containing fragments for those that could preferentially covalently label KRASG12C protein.64 The compounds they identified were selective for GDP-bound KRASG12C and were ultimately shown to bind in what is now termed the switch 2 (SW2) pocket. However, it was only later appreciated that KRASG12C is targetable in this way because it has a rapid nucleotide cycling phenotype.65-66 Optimized covalent fragments showed sufficient activity in KRASG12C-driven cancer cells to motivate additional research on the properties of KRASG12C67 and substantial investment in chemistry ensued. Multiple groups converged on a similar chemical scaffold with markedly improved activity (Figure 2 (C)). 4_AM showed important new interactions with KRAS His95, which boosted affinity68 and ARS1620 showed activity in vivo.69 Mirati and Amgen later disclosed structures of their lead clinical candidates, MRTX849 and AMG510, respectively, which showed structural similarities to 4_AM and ARS1620.69-70 The precise mechanism of how these compounds inhibit RAS signaling is not fully resolved at the structural level, but they presumably work by disrupting protein–protein interactions such as RAS-RAS71 or RAS-RAF interactions that72 are important for RAS signaling. These compounds are highly selective for KRASG12C over other RAS forms because non-covalent interactions with the SW2 pocket are relatively weak and therefore require covalent attachment with cysteine 12 for biological activity.68,73-74 These inhibitors demonstrate that covalent chemistry can convert suboptimal allosteric binding pockets with corresponding weak ligands into druggable binding pockets.
Although covalent SW2 pocket binders are relatively new, the idea of targeting the allosteric SW2 pocket of RAS was considered over 15 years before. The first reports came in 1997 from a group at Plough-Shering aiming to develop allosteric RAS-targeted compounds using x-ray crystal structure-guided design.75-77 Binding of their lead compound SCH-54292 to the SW2 pocket (Figure 2(D)) was confirmed by MS mapping and NMR spectroscopy.75-76 These compounds had activity in RAS-dependent cell-based systems in the micromolar range. Several years later, derivatives were made, but gains in affinity were not sufficient to translate into biologically meaningful activity. It is unknown if covalent analogues of SCH-54292 are achievable, but it is intriguing to consider that SCH-54292 was only one conceptual step away from drugging RAS. This idea has motivated us to re-examine other allosteric RAS and kinase binding pockets, for which numerous allosteric compounds have been discovered (Tables 1 and 2), and consider if any of these may be one step away from a breakthrough.
Table 1.
RAS allosteric compounds.
| Year | PDB | Class | Compound Name |
|---|---|---|---|
| 2012 | 4EPY | SW1/2 | 13 |
| 2013 | 4M22 | SW2 | 16 |
| 2013 | 2LWI | SW1/2 | Kobe2601 |
| 2013 | 5ZC6 | SW1/2 | KBFM123 |
| 2014 | 4NMM | G | SML-8-73-1 |
| 2014 | 4PZZ | SW1/2 | 3 |
| 2016 | 5F2E | SW2 | ARS-853 |
| 2017 | 5MLA | G | darpin K55 |
| 2017 | 5V9O | SW2 | 3_AM |
| 2018 | 6F76 | SW1/2 | 3344 |
| 2018 | 6FA4 | SW1/2 | Abd-7 |
| 2018 | 5V9U | SW2 | ARS1620 |
| 2018 | 6N2K | SW2 | 12 |
| 2019 | 6QUV | SW1/2 | 15R |
| 2019 | 6GQY | SW1/2 | CH-3 |
| 2019 | 6V5L | SW1/2 | E22 |
| 2019 | 6GJ8 | SW1/2 | BI 2852 |
| 2019 | 6P8Z | SW2 | 1 |
| 2019 | 6OIM | SW2 | AMG 510* |
| 2020 | 6TAM | SW2 | 3 |
| 2020 | 6UT0 | SW2 | MRTX849 |
Approved.
Table 2.
Kinase allosteric compounds.
| Year | PDB | Target | Class | Compound Name |
|---|---|---|---|---|
| 2004 | 1UKI | JNK1 | III | SP600125 |
| 2005 | 2BFY | AURORA B | III | hesperadin |
| 2009 | 3E8N | MEK1/2 | III | Refametinib |
| 2009 | 3JVS | CHK1 | IV-C1 | Compound 3 |
| 2009 | 3F9N | CHK1 | IV-C1 | Compound 38 |
| 2009 | 3HRF | PDK1 | IV-PIF | PS48 |
| 2010 | 3K5V | ABL1 | IV-Myristic | GNF2 |
| 2011 | 3PYY | ABL1 | IV-Myristic | DPH |
| 2011 | 3PY1 | CDK2 | IV-PIF | SU9516 |
| 2011 | 3O2M | JNK1 | IV-C3 | Compound 3 |
| 2011 | 3NEW | P38A | IV-C2 | CHEMBL1230164 |
| 2012 | 4AW1 | PDK1 | IV-PIF | PS210 |
| 2013 | 4EBV | FAK | III | CHEMBL2333445 |
| 2013 | 4ITH | RIP1 | III | Necrostatin analogue |
| 2014 | 4RQK | PDK1 | IV-PIF | RS1 |
| 2015 | 4TPT | LIMK2 | III | CHEMBL3355498 |
| 2015 | 4U6R | IRE1 | III | CHEMBL3356007 |
| 2016 | 5EHY | MPS1 | III | 1356851-39-2 |
| 2017 | 5MO4 | ABL1 | IV-Myristic | Asciminib |
| 2019 | 6HHF | AKT1 | III | Borussertib |
| 2020 | 7JUY | MEK1/2 | III | Cobimetinib* |
| 2020 | 7JUZ | MEK1/3 | III | Selumetinib* |
| 2021 | 7M0U | MEK1/4 | III | Binimetinib* |
| 2021 | 7M0Y | MEK1/2 | III | Trametinib* |
Approved.
G pocket and switch 1/2 (SW1/2) pocket RAS inhibitors
Concurrent to the discovery of covalent SW2 pocket inhibitors, GTP-competitive (G pocket) covalent inhibitors were also reported. These compounds were GDP derivatives bearing a reactive electrophilic warhead in place of the gamma phosphate of GTP.78 Remarkably these compounds were able to compete with GTP and GDP in biochemical systems, even in the presence of high concentrations of nucleotide and impair important biochemical properties such as RAS-RAF interactions.78-79 These compounds also lock KRASG12C into an inactive conformation.79-80 Although nucleotide mimetics ultimately could not be adapted into compounds that permeate the cell membrane,80 they demonstrate that covalent approaches are sufficient to overcome competition with high levels of high affinity binders. It is also interesting to consider that although these inhibitors bind to the active site of KRASG12C, they function by altering the conformational state of the protein, which is the primary determinant of KRAS function. This raises the question of whether G-pocket inhibitors should also be considered allosteric inhibitors, thus further expanding the definition of an allosteric inhibitor.
Success in targeting the switch 2 and G-pockets in KRASG12C has inspired numerous other RAS targeting efforts and through that process another major allosteric pocket has been identified that sits between switches 1 and 2, termed the SW1/2 pocket. Notable inhibitors include compounds directed at inhibiting SOS-RAS interactions such as compound 1381 and BI2852,82 which function by inducing non-physiological RAS dimers.83 These compounds bind with low affinity, in the micromolar range, and have not been translated into drugs. Nevertheless, compounds in this class have been adapted into useful assay probes that facilitate evaluation of target engagement and protein–protein interactions.73
Allosteric kinase inhibitors
Kinases are distinguished by a rich history that often overlaps with pathways regulated by RAS proteins and is interwoven with targeted drug discovery. Kinases play critical roles in cellular signaling.84-86 Therefore, subtle changes in kinase function, due to aberrant regulation, mutations or other modifications, can lead to large-scale cellular dysfunction that manifests at the tissue or organismal level as disease. Consistent with this idea, kinase dysregulation is a common driver of many diseases.87 Accordingly, perturbation of kinase function by small molecules can generate profound cellular effects, making them prime drug targets. To date, the US FDA has approved 71 small-molecule kinase inhibitors, 35 of which were approved in the last 5 years.2 It is worth noting that many of these were developed in response to the perceived “undruggable” nature of RAS, in an effort to indirectly address aberrant RAS signaling.88 These drugs target an array of malignancies, rheumatologic conditions, connective tissue disorders, glaucoma, chemotherapy-induced myelosuppression and skin conditions.89 However, only a fraction of the kinome, around 70 of the approximate 520 members, has been targeted for therapeutic purposes. Understanding and targeting the “dark kinome” is a current research priority.90
Kinases are druggable in large part because they exhibit remarkable conformational dynamics. The prototypical kinase domain consists of an N-terminal lobe containing sheets and a C-terminal lobe comprised of a series of helices (Figure 3). The lobes are connected by a loop called the hinge domain. ATP is sandwiched between the N and C-lobes and interacts with the hinge. Prominent functional motifs near the active site include dynamic p and activation loops, which are important for substrate binding and kinase activation for many kinases.91-93 Conformational flexibility results in a relatively low affinity (typically in the 50–100 μM range) for ATP.94-95 This affinity makes it possible to design inhibitors that compete for the ATP binding site. Indeed, the first inhibitors of kinases were ATP-competitive. However, conformational flexibility also makes kinases ripe for allosteric targeting. Moreover, while many kinase signals are transmitted by way of their phosphorylation activity, some signals are transduced through protein–protein interactions. Targeting those functions with small molecules may require allosteric approaches.
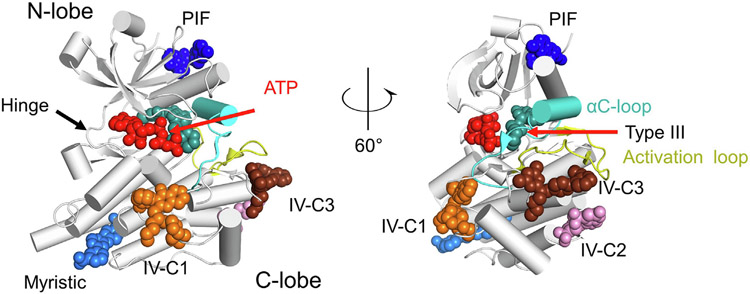
Figure 3.
Kinases are allosterically targetable at multiple sites. Representative binders of allosteric sites are shown relative to a prototypical kinase fold (PDB 2G2F). The conformation of the mobile αC helix is substituted from PDB 7JUY to illustrate the typical conformation seen with type III inhibitors. ATP in red (from PDB 1S9J), Compound 38 in orange (from PDB 3F9N), CHEMBL1230164 in pink (from PDB 3NEW), Compound 3 in chocolate (from PDB 3O2M), RS1 in dark blue (from PDB 4RQK), GNF2 in sky blue (from PDB 3K5V), and cobimetinib in cyan (from PDB 7JUY).
Kinase inhibitor classification
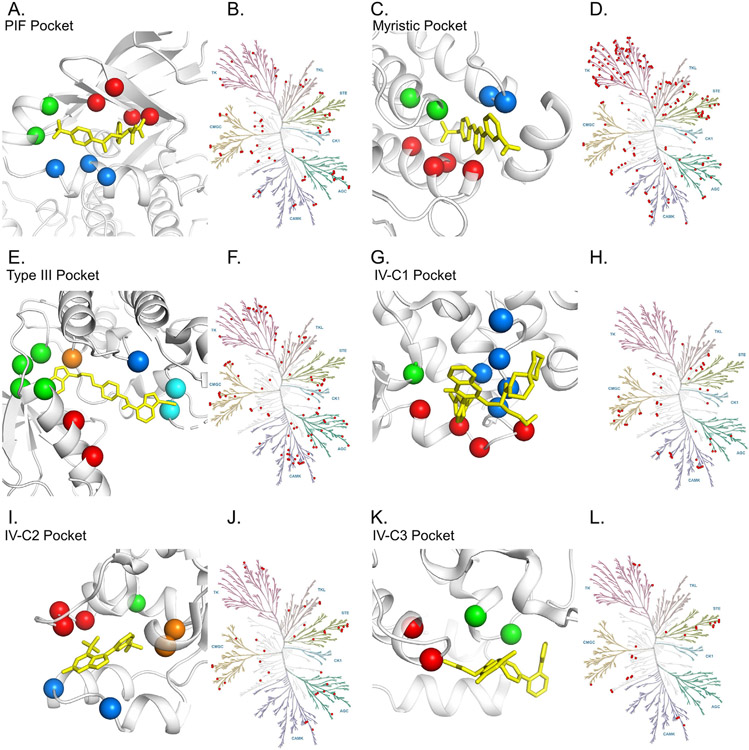
Kinase inhibitors are commonly classified as class I, II, III or IV according to their binding modes.96-97 Type I inhibitors exclusively occupy the ATP-binding pocket, which is highly conserved. Type II inhibitors occupy the ATP site, but also extend into an allosteric pocket that opens when the kinase is inactive. Gefitinib is a prominent early example of a type I inhibitor98 and imatinib is an example of type II inhibitor.99 Multiple examples of covalent type II inhibitors have been reported and are reviewed elsewhere.100 Type III inhibitors bind to a pocket adjacent to the ATP pocket, but are still sandwiched between the N and C lobes. Type IV inhibitors bind to pockets found in the N or C kinase lobes or other domains. Here we briefly review the history and properties of type III and IV inhibitors (Table 2) and explore the potential for covalent targeting of their allosteric pockets.
Type III inhibitors
Interest in the MAPK pathway led to discovery of the first type III inhibitors. Efforts to target MEK resulted in an ATP non-competitive compound PD184352 (also known as CI-1040).101 Determination of the co-crystal x-ray structure of a related compound, PD318088, revealed the type III binding pocket, immediately behind the ATP binding pocket.102 Intense interest in this pocket has resulted in four FDA-approved type III MEK inhibitors: trametinib, cobimetinib, binimetinib, and selumetinib. Type III inhibitors have also been identified for a variety of kinases, including AKT,103 TRKA,104 EGFR,105 IGF-R1,106 LIM-kinase,107 p38a, JNK2,108 cSrc,109 FAK,110 and PAK1.111 The structural mechanism by which type III compounds inhibit target function likely arises from altering the conformational dynamics of their kinase targets which in turn impairs the ability of kinases to bind to substrates or catalyze phosphate transfer.112 There are no examples of covalent type III inhibitors.
Myristic pocket inhibitors
Myristic acid is a known regulator of c-Abl kinase that binds to a hydrophobic pocket located in the C-lobe of its kinase domain.113 Binders of this allosteric pocket were discovered by a high-throughput screen for compounds that produced anti-proliferative activity against ABL-transformed BA/F3 cells. One of the hits, GNF-2, was not competitive with ATP or the ATP-competitive compound imatinib. However, GNF-2 was competitive with myristoylated peptides. Also, mutations to the ABL myristoyl binding pocket conferred resistance to GNF-2 in ABL-dependent cells.114 The x-ray structure of GNF-2 bound to ABL kinase was solved years later, confirming binding to the myristic pocket. Analysis of the impact of GNF-2 binding on protein dynamics showed a large GNF-2 dependent decrease in accessibility of the ATP binding site.115-116 More recently a chemically distinct myristic pocket binder, ABL001 (Asciminib) was reported. It has substantially improved affinity, and works together with orthosteric inhibitors to give more durable responses in CML.117 Asciminib is currently under evaluation in numerous CML-focused clinical trials.118-120 No covalent inhibitors of the myristic pocket have been developed.
PIF pocket inhibitors
The PIF pocket, located in the N-lobe, was originally named for activating interactions between PDK1 and a PDK1-interacting fragment (PIF) of PKA.121 In normal physiology this interaction regulates insulin and growth factor signaling. Manipulation of this pocket with disulfide trapping of small molecule fragments showed the ability to activate or deactivate PDK1.122 Campaigns to target this pocket have yielded probes that interact with PDK1 and CDK2. All PDK1-targeted compounds bind with affinities in the low micro molar range.123 There are no reports of covalent inhibitors of the PIF pocket.
IV-C1 pocket inhibitors
Our search for allosteric inhibitors of the kinase C-lobe identified three distinct pockets, different from the myristic pocket, for which inhibitors have been reported. The first, which we call IV-C1, sits immediately below the ATP binding pocket (Figure 3, orange). Inhibitors of this pocket were discovered in a screening campaign targeting CHK1, motivated by its role in DNA repair and potential applications in cancer therapy. The biochemical screening strategy evaluated for compounds that prevent peptide substrate phosphorylation in a non-ATP competitive manner. The optimized hit, compound 38, bound to CHK1 with an IC50 of 1.5 μM. Crystal structures revealed a shallow pocket in the C-lobe.124
IV-C2 pocket inhibitors
Another allosteric pocket, we term the IV-C2 pocket, sits on the back side of the C-lobe, below the activation loop. Binders were identified from an affinity-based screen using partially phosphorylated p38a as bait. The optimized hit, compound 10, showed an IC50 of 1.2 μM. Structural analysis showed these compounds bind near the “bottom” of the C-lobe, about 30 Å away from the ATP binding site. The mechanism by which these compounds impact enzyme activity is unclear.125
IV-C3 pocket inhibitors
The IV-C3 pocket also sits beneath the activation loop. Binders were identified by an affinity screening approach using JNK1 as bait. Hit compounds bind with an affinity in the low micromolar range to a pocket comprised of the activation loop and MAP insertion loop found in the CMGC family of protein kinases. Mechanistically, these compounds appear to work in JNK1 by stabilizing the activation loop and preventing its phosphorylation.125
Covalent targeting of allosteric pockets in the RAS and kinase families
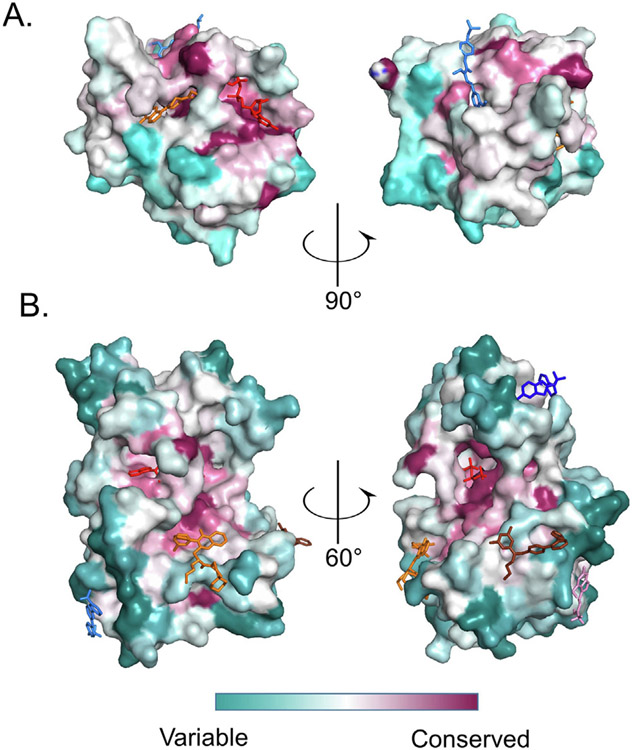
Achieving selectivity has been a major challenge in the kinase inhibitor field and will likely impact RAS inhibitor development strategies for non-G12C targeting. Allosteric inhibitors offer the theoretical advantage of targeting pockets that are less well-conserved than the active sites. This should offer more ways to engineer selectivity. To confirm this is true of allosteric pockets discussed above, we evaluated for primary sequence conservation in the RAS and kinase superfamilies and mapped this onto the three-dimensional structure of prototypical members of these families using methods described previously.79 In general, allosteric pockets were far less conserved than the active site, with the exception of the SW1/2 pocket (Figure 4).
Figure 4.
Allosteric sites for RAS and kinase families are not conserved. Protein sequences for kinases or RAS families were aligned and relative conservation scores for each amino acid were calculated using the ConSurf server and plotted on the surface of a prototypical family member fold. (A) Conservation scores for the RAS family are plotted on the surface of KRAS PDB 4LDJ. Representative compounds are ARS1620 (orange from PDB 5V9U), XY-02-075 (red from PDB 5KYK) and CH-2 (blue from PDB 6GQX). (B) Conservation scores for 493 kinases are plotted on the 3D structure of Aurora A (PDB 3E5A). Representative compounds: Compound 38 in orange (from PDB 3F9N), GNF2 in cyan (from PDB 3K5V), Compound 3 in chocolate (from PDB 3O2M), RS1 in blue (from PDB 4RQK), CHEMBL1230164 in pink (from PDB 3NEW) and ATP in red (from PDB 1S9J).
We next considered the possibility of covalently targeting these pockets. For this analysis we focused on cysteines, given that cysteines have been successfully targeted in a large number and variety of environments to yield clinically approved inhibitors. It is estimated that between 30–50% of FDA approved covalent drugs target cysteines.126-127 As reviewed elsewhere, cysteines contain nucleophilic sidechains that are highly reactive with electrophiles such as acrylamide or enone groups, but are also stable in vivo.128 Further, cysteine is rare in comparison to other amino acids (2.3%), which limits off-target reactivity.126 The other major amino acid target for approved covalent drugs, serine, is nearly exclusively targeted within the context of enzymatic active sites, and therefore cannot be applied to allosteric pockets. Other theoretically possible amino acids, such as lysines which have been targeted in a handful of examples,129 may be worth considering in future analyses of allosteric pockets after methods for predicting local effects on lysine sidechain pKa’s and compatible chemistries mature.
It is also worth noting the potential limitations of cysteine-directed covalent inhibitors for targeting allosteric pockets. An early concern with covalent inhibitors, in general, was that incorporation of electrophilic warheads into drugs could contribute to off-target toxicity. However, this has not borne out, at least with the chemistries currently used for cysteine-directed compounds.130 Another theoretical concern is that covalent inhibitors may not effectively target proteins that are rapidly turned over or degraded by enzymes. Finally, targeting suboptimal allosteric pockets can mean that high potency inhibitors may not be achievable, so that higher concentrations of drug will be required. Pill fatigue, related to the need for elevated plasma levels of drug, is a known issue with certain KRAS G12C inhibitors.131
Ultimately, our primary rationale for targeting cysteine residues using covalent chemistry is to improve the selectivity and efficacy of inhibitors and to overcome resistance mutations. Our analysis also sought to identify opportunities to convert low or modest affinity ligands into viable drugs, as was done with KRASG12C.
Cysteineome of allosteric RAS pockets
Early in the history of KRASG12C inhibitor discovery we evaluated the RAS family G pocket and SW2 pocket for available cysteines that might be used as handles for covalent drug discovery.79 Here we extend that analysis to include additional structural information, and add the switch 1/2 pocket. To discover RAS family members with cysteine residues that might be approachable we developed a new method by obtaining and aligning AlphaFold132 generated models for 145 members of the RAS superfamily. These models all showed high confidence for modeling the G-domains. Using these aligned models we conducted a 3D search of each pocket to find cysteines within 3 A ofÅ a prototypical, structurally-characterized ligand that binds the pocket. This revealed 96, 38 and 31 family members with potentially accessible cysteines in the SW2, SW1/2 and G-pockets respectively (Figure 5, Table 3, Table S1). The potential disease relevance of many of these members is unclear, but several RAS family members stood out as possible targets based on evaluation of the essentiality of these genes in cancer cell lines133-134 and by review of the literature.
Figure 5.
Potentially accessible cysteines in allosteric RAS pockets. (A, C, E) Spheres highlight αC corresponding to potentially accessible cysteines in indicated allosteric binding sites. Table 2 provides a list of specific family members. RAS protein in white, prototypical compound in yellow. (A, 5KYK; C, 6GQX; and E, 5V9U). (B, D, F) Dendrograms of the RAS family provide a high-level view of which members have targetable cysteines for the indicated pockets. Dendrograms generated using the Interactive Tree of Life (iTOL).
Table 3.
List of RAS superfamily proteins with potentially accessible cysteines in the allosteric pocket.
| Position on KRAS |
Color | RAS superfamily | |
|---|---|---|---|
| G pocket | 12, 14 | red | RabL5 RhoA RhoB RhoC RhoD Rif Rnd1 Rnd2 Rnd3 SRPRB |
| 31 | green | Rab14 Rab39A | |
| 18 | blue | Cdc42 Rab10 Rab13 Rab14 Rab15 Rab1A Rab1B Rab26 Rab2A Rab2B Rab30 Rab33A Rab33B Rab34 Rab37 Rab39A Rab39B Rab41 Rab4A Rab4B Rab8A Rab8B Rac1 Rac2 Rac3 RhoA RhoB RhoC RhoG TC10 TCL |
|
| 145-147 | orange | Arf1 Arf3 Arf4 Arf5 Arf6 Arl10B Arl10C Arl11 Arl4 Arl5 Arl7 Arl8 FLJ22595 Rab15 RabL3 Rerg Ris/RasL12 | |
| 118 | cyan | Di-Ras1 Di-Ras2 E-Ras H-Ras K-Ras2B NKIRas2 N-Ras Rab10 Rab13 Rab1A Rab22A Rab22B Rab32 Rab33A Rab33B Rab38 Rab39A Rab39B Rab3A Rab3B Rab3C Rab3D Rab7L1 Rab8A Rab8B Rap1A Rap1B | |
| SI/SII Pocket | 71, 75 | red | E-Ras Gem NKIRas2 Rad RasL11A RasL11B Rem1 Rnd1 Wrch-1 Wrch-2 |
| 5, 41, 54 | green | ArfRP1 Noey2 | |
| SII-Pocket | 12 | red | RabL5 SRPRB |
| 65, 68 | green | FLJ22595 Rab17 Rab34 Rab36 Rab40A RasL10B | |
| 99 | blue | Rab33A Rab33B RabL5 |
Cell division control protein 42 homolog (CDC42), a member of Rho family, was identified in Saccharomyces cerevisiae as a regulator of reproduction.135 Subsequently, CDC42 has been shown to mediate cellular transformation, division, invasion, migration, and polarity in human cells. Aberrant regulation of CDC42 occurs in several pathogenic processes including cancer, motivating drug discovery efforts.136 Secramine was identified based on its ability to block recruitment of prenylated CDC42 to the cell membrane.137 More recently, ZCL278 was developed as an inhibitor of CDC42-GEF interactions.138 CID2950007 is a non-competitive allosteric inhibitor that induces GTP dissociation.139 Nevertheless, no efforts to date have translated into therapeutic candidates. Our cysteine analysis reveals another potential avenue for CDC42 targeting by way of covalent ligands that interact with cysteines in G and S1/2 pockets, specifically Cys18 or 157 in the G pocket or Cys6 in the SI/SII pocket.
RAC1, also a member of Rho family, has received attention as a potential drug target based on its role in regulating the assembly and disassembly of cytoskeletal elements for a variety of cellular processes.140 Dysregulation of RAC1 leads to various pathological conditions including cancer.141 NSC23766 is a direct RAC1 inhibitor that disrupts RAC1-GEF interactions, thereby preventing RAC1 activation.142 Derivatives of NSC23766 with greater potency, such as EHT 1964 and EHop-016 have been developed141,143 but these have not advanced. Based on our analysis, covalent targeting may be possible for cysteines in the G pocket (Cys18 and 157) and S1/2 pocket (Cys6).
RHOA, another member of Rho family, also regulates cell morphology, cell polarity, and cell migration144 and is dysregulated in a number of cancers.145 RHOA directed inhibitors such as G04, have been identified, but are early stage.146 Our analysis shows potentially accessible cysteines at residues 20 and 159 in the G pocket.
NRAS is frequently mutated in melanoma, leukemia, and thyroid carcinoma and those mutations are associated with poor prognosis.147 No direct NRAS inhibitors have been developed. A cysteine at G pocket residue 118 may provide a foothold.
Cysteineome of allosteric kinase pockets
Analysis of the cysteineome of the ATP-binding pocket of kinases has led to the development of numerous covalent type I and type II inhibitors.148 Different from that analysis, here we examined the kinome to find family members that present with cysteines in allosteric pockets (Figure 6, Table 4, Table S2) using the same method described above for the RAS family. This revealed a number of established cancer targets as well as targets where there is considerable interest in developing targeted therapies. Notable cancer targets for which there are already approved drugs include HER2, HER3, EGFR, FGFR1, CDK4, CDK6, and JAK1. Despite the availability of targeted therapies, development of additional allosteric inhibitors may provide advantages. As an illustration, although potent clinical-grade inhibitors of EGFR such as erlotinib have been available since the early 2000′s, new generations of EGFR inhibitors such as osimertinib have shown significant advantages,149 particularly with respect to acquired resistance mutations.150 Development of allosteric inhibitors may provide additional advantages both for drug resistance and efficacy.
Figure 6.
Potentially accessible cysteines in allosteric kinase pockets. (A, C, E, G, I, K) Similar to Fig 5, spheres highlight α C corresponding to potentially accessible cysteines in indicated allosteric binding sites. Table 4 provides a list of specific family members. Kinase in white, and prototypical compounds in yellow (A, 4AW1; C, 3K5V; E, 3LW0; G, 3F9M; I, 3NEW; and K, 3O2M). (B, D, F, H, J, L) Dendrograms of the kinase family provide a high level view of which members have targetable cysteines for the indicated pockets. Dendrograms generated using KinMap.
Table 4.
List of kinase proteins with potentially accessible cysteines in the allosteric pocket.
| Position | Color | Kinases | |
|---|---|---|---|
| PIF | Position on PDPK1 148, 150, 155, 157 |
red | BIKE caMLCK CDC7 CDK4 CDK6 CK1d CK1e CLIK1 CLIK1L HIPK1 HIPK2 HIPK3 ILK IRAK1 KIS KSR2 LMR1 MAP2K4 MAP3K1 MAP3K2 MAP3K3 MAPKAPK3 PKCa PKCb PKCe PKCh PKCi PKCz PKN1 PKN2 PKN3 PKR SgK495 TAO1 TAO2 TAO3 TTBK1 TTBK2 TYK2 |
| 115, 118 | green | AXL CLIK1 CLIK1L CTK FAK MPSK1 STLK3 STLK5 STLK6 TYK2 ULK4 YANK1 YANK2 YANK3 | |
| 124, 127, 128 | blue | AAK1 BIKE CDK11 CDK8 DRAK1 DRAK2 EphA7 Erk1 Erk2 HH498 IKKb MAPKAPK2 NEK6 NEK7 PSKH1 PSKH2 SgK071 SgK494 TEC TLK1 TLK2 |
|
| Myristic | Position on ABL1 356, 359, 360, 363 |
red | AAK1 ACTR2 ACTR2B ALK2 BMPR2 BMX BTK CCK4 CHK2 EGFR ErbB2 ErbB4 Erk1 Erk2 GAK GCK HPK1 IRAK2 IRAK4 ITK KHS1 LKB1 LOK MAP3K19 MINK MOS MUSK NRBP1 NRBP2 NRK p70S6K p70S6Kb PAK1 PAK2 PAK3 PAK4 PAK5 PAK6 PDK1 PEK PKACg PKCd PKG2 RIPK3 ROS SLK smMLCK SRPK2 TAK1 TBCK TEC TNIK TXK Wnk1 Wnk2 Wnk3 |
| 448, 452 | green | AAK1 AKT1 AMPKa1 AMPKa2 AurA AurB AurC BIKE CaMK1d CHK2 CYGD FAK GAK HH498 IKKa IKKb LIMK1 LIMK2 MAPKAPK2 MAPKAPK5 MASTL MRCKa MRCKb NEK1 NEK11 NEK2 NEK3 NEK4 NEK5 NIK NRBP1 NRBP2 PIM3 QIK QSK SIK smMLCK SNRK TAO1 TAO2 TAO3 TBCK TESK1 TLK1 TLK2 TSSK1 TSSK2 ULK1 ULK2 VRK1 VRK3 Wnk1 Wnk2 Wnk3 Wnk4 |
|
| 482, 483 | blue | ABL1 ABL2 ACK ARAF AXL BLK BRK CCK4 COT CSK CTK DDR1 DDR2 DYRK2 DYRK3 EGFR EphA1 EphA10 EphA2 EphA3 EphA4 EphA5 EphA7 EphA8 EphB1 EphB2 EphB3 EphB4 EphB6 ErbB2 ErbB3 ErbB4 FAK FER FES FGFR1 FGFR2 FGFR3 FGFR4 FGR FRK FYN HCK IGF1R IRR JAK1 JAK2 JAK3 LCK LIMK1 LKB1 LTK LYN MER MET MLKL MUSK NIK RET RIPK1 RON ROR1 ROR2 ROS RSKL1 RYK SRC SRM SuRTK106 SYK TESK1 TESK2 TIE1 TIE2 TRKA TRKB TRKC TYRO3 YES ZAP70 |
|
| Type III | Position on IGF-1R 1046, 1050 |
red | EphA7 Erk1 Erk2 RYK SgK494 TEC |
| 1054, 1063-1065 | green | BARK1 BARK2 CaMK2a CaMK2b CaMK2d CaMK2g CASK CDC7 EGFR IRAK2 LMR1 LMR3 LTK MAP2K7 MAPKAPK3 MELK MER NIK ROR1 ROR2 RSKL1 smMLCK TBCK TTBK2 TYRO3 VRK3 |
|
| 1134 | blue | DYRK1A DYRK1B DYRK2 DYRK3 DYRK4 LRRK1 PKD1 PKD2 PKD3 ULK4 | |
| 1152 | orange | AAK1 BIKE CDKL1 CDKL2 CDKL3 CDKL4 CDKL5 Erk1 Erk2 Erk7 FLT1 FLT4 Fused GAK KDR KIT KSR1 KSR2 MAP2K1 MAP2K2 MAP2K3 MAP2K4 MAP2K5 MAP2K6 MAP2K7 MAPKAPK5 MNK1 MNK2 NIK PBK PDGFRb PKD1 PKD2 PKD3 PRP4 SuRTK106 TAK1 TGFbR2 ZAK | |
| 1173, 1174 | cyan | AKT1 AurA AurB AurC BUB1 CaMK1g DCAMKL3 MAST1 MAST2 MAST3 MAST4 MASTL MSK1 MSK2 PKCb PKCe PKG1 PLK3 RSK2 STK33 | |
| IV-C1 | Position on CHK1 93, 96, 97, 99 |
red | AurB BMPR2 CaMK2a CaMK2b CaMK2d CaMK2g CASK CDKL3 DRAK1 DRAK2 JNK1 JNK2 JNK3 MAST1 MISR2 MYT1 PEK PFTAIRE1 PKR |
| 133 | green | AurB BMPR2 CaMK2a CaMK2b CaMK2d CaMK2g CASK CDKL3 CHED CRK7 DRAK1 DRAK2 JNK1 JNK2 JNK3 MAST1 MISR2 MYT1 NRBP1 PEK PFTAIRE1 PKR SSTK TSSK1 TSSK2 TSSK3 Wnk1 Wnk2 Wnk3 Wnk4 |
|
| 200, 204-206, 209 | blue | BUB1 CaMKK1 CaMKK2 CASK CDK11 CDK4 CDK8 GCN2 IRAK3 JNK2 MAP3K1 MAST1 MAST2 MAST3 MAST4 NIK PKN3 SBK SgK069 SgK071 SgK110 SSTK Wee1 | |
| IV-C2 | Position on MAPK14 195-197 |
red | CaMK4 CDC7 CDK2 COT HGK LOK MINK MYO3A MYO3B NRK PHKg1 PHKg2 SgK069 SLK TNIK ULK3 |
| 232 | green | BMPR1A BMPR1B FLT1 KDR MAP2K5 PLK2 RIPK1 TRKA TRKC | |
| 249, 252 | blue | Erk1 Erk2 HIPK4 IRE1 PKG2 | |
| 291, 292291-292 | orange | CDK5 DMPK2 HH498 LATS2 MSK2 NDR1 NEK6 PFTAIRE1 PKN3 Wnk2 | |
| IV-C3 | Position on MAPK8 197, 199 |
red | FLT1 JAK3 KDR MAP2K5 NEK6 NEK7 NEK9 |
| 231, 234 | green | CaMK4 CDC7 CDK2 COT HGK KIT LOK MINK MYO3A MYO3B NEK1 NEK5 NEK8 NEK9 NRK PHKg1 PHKg2 SBK SgK069 SgK110 SLK TNIK |
Additionally, we note several emerging drug targets with potentially targetable allosteric pocket cysteines. Tousled-like kinases (TLKs) are relatively understudied kinases that are essential for genome stability and normal embryonic development in a variety of multicellular organisms.151 Human TLK1/2 are often amplified or mutated in human cancers, especially breast and bile duct cancers. Additionally, TLK2 mutations occur in intellectual disability and autism spectrum disorders.152 There are few examples of TLK-directed inhibitor efforts in the literature and no potent and selective TLK inhibitors have been reported. Covalent allosteric inhibitors to the PIF, IV-C1 and IV-C3 pockets may be possible based on our analysis.
Non-receptor tyrosine kinase PTK2 (protein tyrosine kinase 2), also known as FAK (focal adhesion kinase) mediates cell adhesion signal transduction downstream of integrins and growth factor receptors. FAK supports cell survival, migration, and invasion by cancer cells and is commonly overexpressed in ovarian cancers, lung squamous cell neoplasms, esophageal cancers and uveal melanomas. Multiple orthosteric PTK2 inhibitors have been developed, including defactinib, IN10018, VS-4718, GSK2256098, and PF573228, but many show off-target effects that limit their further development as drugs.153 However, FAK is also known to play scaffolding roles, so efforts to develop inhibitors that disrupt PTK2 protein–protein interactions have been developed.154 Covalent allosteric inhibitors of FAK may address both the selectivity problems of current inhibitors and better target the allosteric functions of FAK.
Conclusion
KRASG12C not only showed that KRAS can be drugged, but also that covalent chemistry can convert a suboptimal binding pocket into a druggable pocket. In this analysis we take the first step in extending this concept to other potential drug targets by the identification of members of the RAS and kinase families with cysteines in known allosteric pockets. Whether known inhibitors can be adapted to covalently access these cysteines or if new chemotypes will be required remains to be seen.
Supplementary Material
Acknowledgements
Research in the authors lab is supported by funding from NIH/NCI R01 CA244341, and P30CA142543, Cancer Prevention and Research Institute of Texas RP220145, Welch Research Foundation I1829, and the American Cancer Society RSG-18-039-01.
Footnotes
Competing Interests Statement
Kenneth Westover is a member of the Scientific Advisory Board for Vibliome Therapeutics and on the Advosory Panel for Reactive Biosciences. The Westover lab receives or has received research funding from Astellas and Revolution Medicines. None of these relationships are in conflict with the content of this manuscript.
CRediT authorship contribution statement
Lianbo Li: Conceptualization, Methodology, Software, Formal analysis, Investigation, Writing – original draft, Writing – review & editing, Visualization. Cynthia Meyer: Conceptualization, Writing – original draft, Writing – review & editing. Zhi-Wei Zhou: Writing – review & editing. Ammar Elmezayen: Investigation, Software. Kenneth Westover: Conceptualization, Methodology, Software, Formal analysis, Investigation, Writing – original draft, Writing – review & editing, Visualization, Supervision, Project administration, Funding acquisition.
Appendix A. Supplementary data
Supplementary data to this article can be found online at https://doi.org/10.1016/j.jmb.2022.167626.
References
- 1.Lu S, Shen Q, Zhang J, (2019). Allosteric Methods and Their Applications: Facilitating the Discovery of Allosteric Drugs and the Investigation of Allosteric Mechanisms. Acc. Chem. Res 52, 492–500. [DOI] [PubMed] [Google Scholar]
- 2.Attwood MM, Fabbro D, Sokolov AV, Knapp S, Schioth HB, (2021). Trends in kinase drug discovery: targets, indications and inhibitor design. Nat. Rev. Drug Discov 20, 839–861. [DOI] [PubMed] [Google Scholar]
- 3.Motlagh HN, Wrabl JO, Li J, Hilser VJ, (2014). The ensemble nature of allostery. Nature 508, 331–339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wishart DS, Knox C, Guo AC, Shrivastava S, Hassanali M, Stothard P, et al. , (2006). DrugBank: a comprehensive resource for in silico drug discovery and exploration. Nucleic Acids Res. 34, D668–D672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Moore AR, Rosenberg SC, McCormick F, Malek S, (2020). RAS-targeted therapies: is the undruggable drugged? Nat. Rev. Drug Discov 19, 533–552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fodor M, Price E, Wang P, Lu H, Argintaru A, Chen Z, et al. , (2018). Dual Allosteric Inhibition of SHP2 Phosphatase. ACS Chem. Biol 13, 647–656. [DOI] [PubMed] [Google Scholar]
- 7.Garcia Fortanet J, Chen CH, Chen YN, Chen Z, Deng Z, Firestone B, et al. , (2016). Allosteric Inhibition of SHP2: Identification of a Potent, Selective, and Orally Efficacious Phosphatase Inhibitor. J. Med. Chem 59, 7773–7782. [DOI] [PubMed] [Google Scholar]
- 8.Chen YN, LaMarche MJ, Chan HM, Fekkes P, Garcia-Fortanet J, Acker MG, et al. , (2016). Allosteric inhibition of SHP2 phosphatase inhibits cancers driven by receptor tyrosine kinases. Nature 535, 148–152. [DOI] [PubMed] [Google Scholar]
- 9.Lu X, Smaill JB, Ding K, (2020). New Promise and Opportunities for Allosteric Kinase Inhibitors. Angew. Chem. Int. Ed. Engl 59, 13764–13776. [DOI] [PubMed] [Google Scholar]
- 10.Jarvis LM, (2019). Drug hunters explore allostery’s advantages. Chem. & Eng. News 97 [Google Scholar]
- 11.Merriam-Webster. Merriam-Webster. [Google Scholar]
- 12.Monod J, Jacob F, (1961). General conclusions: teleonomic mechanisms in cellular metabolism, growth, and differentiation. Cold Spring Harbor symposia on quantitative biology: Cold Spring Harbor Laboratory Press, pp. 389–401. [DOI] [PubMed] [Google Scholar]
- 13.Gunasekaran K, Ma B, Nussinov R, (2004). Is allostery an intrinsic property of all dynamic proteins? Proteins: Structure, Function, and Bioinformatics. 57, 433–443. [DOI] [PubMed] [Google Scholar]
- 14.Cheng X, Jiang H, (2019). Allostery in Drug Development. In: Zhang J, Nussinov R (Eds.), Protein Allostery in Drug Discovery. Springer Singapore, Singapore, pp. 1–23. [Google Scholar]
- 15.Nussinov R, Tsai CJ, (2013). Allostery in disease and in drug discovery. Cell 153, 293–305. [DOI] [PubMed] [Google Scholar]
- 16.Ni D, Chai Z, Wang Y, Li M, Yu Z, Liu Y, et al. , (2021). Along the allostery stream: Recent advances in computational methods for allosteric drug discovery. WIREs Comput. Mol. Sci. [Google Scholar]
- 17.Perutz MF, (1970). Stereochemistry of cooperative effects in haemoglobin. Nature 228, 726–739. [DOI] [PubMed] [Google Scholar]
- 18.Yuan Y, Tam MF, Simplaceanu V, Ho C, (2015). New look at hemoglobin allostery. Chem. Rev 115, 1702–1724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kamata K, Mitsuya M, Nishimura T, Eiki J, Nagata Y, (2004). Structural basis for allosteric regulation of the monomeric allosteric enzyme human glucokinase. Structure. 12, 429–438. [DOI] [PubMed] [Google Scholar]
- 20.Matschinsky FM, Wilson DF, (2019). The Central Role of Glucokinase in Glucose Homeostasis: A Perspective 50 Years After Demonstrating the Presence of the Enzyme in Islets of Langerhans. Front. Physiol 10, 148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhu XX, Zhu DL, Li XY, Li YL, Jin XW, Hu TX, et al. , (2018). Dorzagliatin (HMS5552), a novel dual-acting glucokinase activator, improves glycaemic control and pancreatic beta-cell function in patients with type 2 diabetes: A 28-day treatment study using biomarker-guided patient selection. Diabetes Obes. Metab 20, 2113–2120. [DOI] [PubMed] [Google Scholar]
- 22.Li YQ, Zhang YL, Hu SQ, Wang YL, Song HR, Feng ZQ, et al. , (2011). Design, synthesis and biological evaluation of novel glucokinase activators. Chin. Chem. Lett 22, 73–76. [Google Scholar]
- 23.Liu D, Du Y, Yao X, Wei Y, Zhu J, Cui C, et al. , (2021). Safety, tolerability, pharmacokinetics, and pharmacodynamics of the glucokinase activator PB-201 and its effects on the glucose excursion profile in drug-naive Chinese patients with type 2 diabetes: a randomised controlled, crossover, single-centre phase 1 trial. EClinicalMedicine. 42, 101185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Grimsby J, Sarabu R, Corbett WL, Haynes NE, Bizzarro FT, Coffey JW, et al. , (2003). Allosteric activators of glucokinase: potential role in diabetes therapy. Science 301, 370–373. [DOI] [PubMed] [Google Scholar]
- 25.Eli L, Company. A Study of LY2599506 (Oral Agent Medication: Glucokinase Activator 1) in Type 2 Diabetes Mellitus. 2010. [Google Scholar]
- 26.L. PegBio Co, Clinical Trial for the Investigational Drug (PB-201) in Subjects With Type 2 Diabetes Mellitus, 2019. [Google Scholar]
- 27.University of V. Effects of Modulators of Gluconeogenesis, Glycogenolysis and Glucokinase Activity, 2022. [Google Scholar]
- 28.Sola-Penna M, Da Silva D, Coelho WS, Marinho-Carvalho MM, Zancan P, (2010). Regulation of mammalian muscle type 6-phosphofructo-1-kinase and its implication for the control of the metabolism. IUBMB Life 62, 791–796. [DOI] [PubMed] [Google Scholar]
- 29.Kloos M, Bruser A, Kirchberger J, Schoneberg T, Strater N, (2015). Crystal structure of human platelet phosphofructokinase-1 locked in an activated conformation. Biochem. J 469, 421–432. [DOI] [PubMed] [Google Scholar]
- 30.Webb BA, Forouhar F, Szu FE, Seetharaman J, Tong L, Barber DL, (2015). Structures of human phosphofructokinase-1 and atomic basis of cancer-associated mutations. Nature 523, 111–114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.McNae IW, Kinkead J, Malik D, Yen LH, Walker MK, Swain C, et al. , (2021). Fast acting allosteric phosphofructokinase inhibitors block trypanosome glycolysis and cure acute African trypanosomiasis in mice. Nat. Commun 12, 1052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cheng D, Chu CH, Chen L, Feder JN, Mintier GA, Wu Y, et al. , (2007). Expression, purification, and characterization of human and rat acetyl coenzyme A carboxylase (ACC) isozymes. Protein Expr. Purif 51, 11–21. [DOI] [PubMed] [Google Scholar]
- 33.Hunkeler M, Hagmann A, Stuttfeld E, Chami M, Guri Y, Stahlberg H, et al. , (2018). Structural basis for regulation of human acetyl-CoA carboxylase. Nature 558, 470–474. [DOI] [PubMed] [Google Scholar]
- 34.Wang C, Rajput S, Watabe K, Liao D-F, Cao D, (2010). Acetyl-CoA carboxylase-alpha as a novel target for cancer therapy. Front Biosci. 2, 15–26. [DOI] [PubMed] [Google Scholar]
- 35.Chen L, Duan Y, Wei H, Ning H, Bi C, Zhao Y, et al. , (2019). Acetyl-CoA carboxylase (ACC) as a therapeutic target for metabolic syndrome and recent developments in ACC1/2 inhibitors. Expert Opin. Invest. Drugs 28, 917–930. [DOI] [PubMed] [Google Scholar]
- 36.Loomba R, Kayali Z, Noureddin M, Ruane P, Lawitz EJ, Bennett M, et al. , (2018). GS-0976 Reduces Hepatic Steatosis and Fibrosis Markers in Patients With Nonalcoholic Fatty Liver Disease. Gastroenterology 155, (1463–73) e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kirsten WH, Mayer LA, (1967). Morphologic Responses to a Murine Erythroblastosis Virus2. JNCI: J. Natl. Cancer Inst 39, 311–335. [PubMed] [Google Scholar]
- 38.Harvey JJ, (1964). An Unidentified Virus Which Causes the Rapid Production of Tumours in Mice. Nature 204, 1104–1105. [DOI] [PubMed] [Google Scholar]
- 39.Ellis RW, Defeo D, Shih TY, Gonda MA, Young HA, Tsuchida N, et al. , (1981). The p21 src genes of Harvey and Kirsten sarcoma viruses originate from divergent members of a family of normal vertebrate genes. Nature 292, 506–511. [DOI] [PubMed] [Google Scholar]
- 40.Prior IA, Lewis PD, Mattos C, (2012). A comprehensive survey of Ras mutations in cancer. Cancer Res. 72, 2457–2467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Milburn MV, Tong L, deVos AM, Brunger A, Yamaizumi Z, Nishimura S, et al. , (1990). Molecular switch for signal transduction: structural differences between active and inactive forms of protooncogenic ras proteins. Science 247, 939–945. [DOI] [PubMed] [Google Scholar]
- 42.Poulin EJ, Bera AK, Lu J, Lin YJ, Strasser SD, Paulo JA, et al. , (2019). Tissue-Specific Oncogenic Activity of KRAS(A146T). Cancer Discov. 9, 738–755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bera AK, Lu J, Wales TE, Gondi S, Gurbani D, Nelson A, et al. , (2019). Structural basis of the atypical activation mechanism of KRAS(V14I). J. Biol. Chem 294, 13964–13972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lu J, Hunter J, Manandhar A, Gurbani D, Westover KD, (2015). Structural dataset for the fast-exchanging KRAS G13D. Data Brief. 5, 572–578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Scheffzek K, Ahmadian MR, Kabsch W, Wiesmuller L, Lautwein A, Schmitz F, et al. , (1997). The Ras-RasGAP complex: structural basis for GTPase activation and its loss in oncogenic Ras mutants. Science 277, 333–338. [DOI] [PubMed] [Google Scholar]
- 46.Krengel U, Schlichting I, Scherer A, Schumann R, Frech M, John J, et al. , (1990). Three-dimensional structures of H-ras p21 mutants: molecular basis for their inability to function as signal switch molecules. Cell 62, 539–548. [DOI] [PubMed] [Google Scholar]
- 47.Bera AK, Lu J, Lu C, Li L, Gondi S, Yan W, et al. , (2020). GTP hydrolysis is modulated by Arg34 in the RASopathy-associated KRAS(P34R). Birth Defects Res. 112, 708–717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lu J, Bera AK, Gondi S, Westover KD, (2018). KRAS Switch Mutants D33E and A59G Crystallize in the State 1 Conformation. Biochemistry 57, 324–333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Mysore VP, Zhou ZW, Ambrogio C, Li L, Kapp JN, Lu C, et al. , (2021). A structural model of a Ras-Raf signalosome. Nat. Struct. Mol. Biol 28, 847–857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Goody RS, Frech M, Wittinghofer A, (1991). Affinity of guanine nucleotide binding proteins for their ligands: facts and artefacts. Trends Biochem. Sci 16, 327–328. [DOI] [PubMed] [Google Scholar]
- 51.Willumsen BM, Norris K, Papageorge AG, Hubbert NL, Lowy DR, (1984). Harvey murine sarcoma virus p21 ras protein: biological and biochemical significance of the cysteine nearest the carboxy terminus. EMBO J. 3, 2581–2585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Whyte DB, Kirschmeier P, Hockenberry TN, Nunez-Oliva I, James L, Catino JJ, et al. , (1997). K- and N-Ras are geranylgeranylated in cells treated with farnesyl protein transferase inhibitors. J. Biol. Chem 272, 14459–14464. [DOI] [PubMed] [Google Scholar]
- 53.Blumenschein GR Jr., Smit EF, Planchard D, Kim DW, Cadranel J, De Pas T, et al. , (2015). A randomized phase II study of the MEK1/MEK2 inhibitor trametinib (GSK1120212) compared with docetaxel in KRAS-mutant advanced non-small-cell lung cancer (NSCLC)dagger. Ann. Oncol 26, 894–901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Infante JR, Fecher LA, Falchook GS, Nallapareddy S, Gordon MS, Becerra C, et al. , (2012). Safety, pharmacokinetic, pharmacodynamic, and efficacy data for the oral MEK inhibitor trametinib: a phase 1 dose-escalation trial. Lancet Oncol. 13, 773–781. [DOI] [PubMed] [Google Scholar]
- 55.Hong DS, Fakih MG, Strickler JH, Desai J, Durm GA, Shapiro GI, et al. , (2020). KRAS(G12C) Inhibition with Sotorasib in Advanced Solid Tumors. N. Engl. J. Med 383, 1207–1217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Riely GJ, Ou SHI, Rybkin I, Spira A, Papadopoulos K, Sabari JK, et al. , (2021). KRYSTAL-1: Activity and preliminary pharmacodynamic (PD) analysis of adagrasib (MRTX849) in patients (Pts) with advanced non-small cell lung cancer (NSCLC) harboring KRASG12C mutation. J. Thoracic Oncol 16, S751–S752. [Google Scholar]
- 57.Jacobio Pharmaceuticals Co L. FIH Study of JAB-21822 in Adult Patients With Advanced Solid Tumors Harboring KRAS G12C Mutation in China, 2025. [Google Scholar]
- 58.Zhejiang Genfleet Therapeutics Co L, Genfleet Therapeutics I. A Study of GFH925 in Patients With Advanced Solid Tumors With KRAS G12C Mutations, 2024. [Google Scholar]
- 59.P. Novartis, Novartis. Study of JDQ443 in Comparison With Docetaxel in Participants With Locally Advanced or Metastatic KRAS G12C Mutant Non-small Cell Lung Cancer, 2024. [Google Scholar]
- 60.I. Revolution Medicines, Sanofi, Amgen, Combination Study of RMC-4630 and Sotorasib for NSCLC Subjects With KRASG12C Mutation After Failure of Prior Standard Therapies, 2023. [Google Scholar]
- 61.Shanghai YingLi Pharmaceutical Co L. A Phase 1, Study of YL-15293 in Subjects With Advanced Solid Tumors With a KRAS G12C Mutation, 2022. [Google Scholar]
- 62.Janssen R, Development LLC. First-in-Human Study of JNJ-74699157 in Participants With Tumors Harboring the KRAS G12C Mutation, 2020. [Google Scholar]
- 63.L. Eli, Company. A Study of LY3499446 in Participants With Advanced Solid Tumors With KRAS G12C Mutation, 2020. [Google Scholar]
- 64.Ostrem JM, Peters U, Sos ML, Wells JA, Shokat KM, (2013). K-Ras(G12C) inhibitors allosterically control GTP affinity and effector interactions. Nature 503, 548–551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Hunter JC, Manandhar A, Carrasco MA, Gurbani D, Gondi S, Westover KD, (2015). Biochemical and Structural Analysis of Common Cancer-Associated KRAS Mutations. Mol. Cancer Res 13, 1325–1335. [DOI] [PubMed] [Google Scholar]
- 66.Lito P, Solomon M, Li LS, Hansen R, Rosen N, (2016). Allele-specific inhibitors inactivate mutant KRAS G12C by a trapping mechanism. Science 351, 604–608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Patricelli MP, Janes MR, Li LS, Hansen R, Peters U, Kessler LV, et al. , (2016). Selective Inhibition of Oncogenic KRAS Output with Small Molecules Targeting the Inactive State. Cancer Discov. 6, 316–329. [DOI] [PubMed] [Google Scholar]
- 68.Zeng M, Lu J, Li L, Feru F, Quan C, Gero TW, et al. , (2017). Potent and Selective Covalent Quinazoline Inhibitors of KRAS G12C. Cell Chem. Biol 24, (1005–16) e3. [DOI] [PubMed] [Google Scholar]
- 69.Janes MR, Zhang J, Li LS, Hansen R, Peters U, Guo X, et al. , (2018). Targeting KRAS Mutant Cancers with a Covalent G12C-Specific Inhibitor. Cell 172, (578–89) e17. [DOI] [PubMed] [Google Scholar]
- 70.Canon J, Rex K, Saiki AY, Mohr C, Cooke K, Bagal D, et al. , (2019). The clinical KRAS(G12C) inhibitor AMG 510 drives anti-tumour immunity. Nature 575, 217–223. [DOI] [PubMed] [Google Scholar]
- 71.Ambrogio C, Kohler J, Zhou ZW, Wang H, Paranal R, Li J, et al. , (2018). KRAS Dimerization Impacts MEK Inhibitor Sensitivity and Oncogenic Activity of Mutant KRAS. Cell 172, (857–68) e15. [DOI] [PubMed] [Google Scholar]
- 72.Nassar N, Horn G, Herrmann CA, Scherer A, McCormick F, Wittinghofer A, (1995). The 2.2 Å crystal structure of the Ras-binding domain of the serine/threonine kinase c-Raf1 in complex with RaplA and a GTP analogue. Nature 375, 554–560. [DOI] [PubMed] [Google Scholar]
- 73.Vasta JD, Peacock DM, Zheng Q, Walker JA, Zhang Z, Zimprich CA, et al. , (2021). KRAS is vulnerable to reversible switch-II pocket engagement in cells. bioRxiv. 2021.10.15.464544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Hansen R, Peters U, Babbar A, Chen Y, Feng J, Janes MR, et al. , (2018). The reactivity-driven biochemical mechanism of covalent KRAS(G12C) inhibitors. Nat. Struct. Mol. Biol 25, 454–462. [DOI] [PubMed] [Google Scholar]
- 75.Ganguly AK, Pramanik BN, Huang EC, Liberles S, Heimark L, Liu YH, et al. , (1997). Detection and structural characterization of ras oncoprotein-inhibitors complexes by electrospray mass spectrometry. Bioorg. Med. Chem 5, 817–820. [DOI] [PubMed] [Google Scholar]
- 76.Ganguly AK, Wang YS, Pramanik BN, Doll RJ, Snow ME, Taveras AG, et al. , (1998). Interaction of a novel GDP exchange inhibitor with the Ras protein. Biochemistry 37, 15631–15637. [DOI] [PubMed] [Google Scholar]
- 77.Taveras AG, Remiszewski SW, Doll RJ, Cesarz D, Huang EC, Kirschmeier P, et al. , (1997). Ras oncoprotein inhibitors: the discovery of potent, ras nucleotide exchange inhibitors and the structural determination of a drug-protein complex. Bioorg. Med. Chem 5, 125–133. [DOI] [PubMed] [Google Scholar]
- 78.Lim SM, Westover KD, Ficarro SB, Harrison RA, Choi HG, Pacold ME, et al. , (2014). Therapeutic targeting of oncogenic K-Ras by a covalent catalytic site inhibitor. Angew. Chem. Int. Ed. Engl 53, 199–204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Hunter JC, Gurbani D, Ficarro SB, Carrasco MA, Lim SM, Choi HG, et al. , (2014). In situ selectivity profiling and crystal structure of SML-8-73-1, an active site inhibitor of oncogenic K-Ras G12C. Proc. Natl. Acad. Sci. U. S. A 111, 8895–8900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Xiong Y, Lu J, Hunter J, Li L, Scott D, Choi HG, et al. , (2017). Covalent Guanosine Mimetic Inhibitors of G12C KRAS. ACS Med. Chem. Lett 8, 61–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Sun Q, Burke JP, Phan J, Burns MC, Olejniczak ET, Waterson AG, et al. , (2012). Discovery of small molecules that bind to K-Ras and inhibit Sos-mediated activation. Angew. Chem. Int. Ed. Engl 51, 6140–6143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Kessler D, Gmachl M, Mantoulidis A, Martin LJ, Zoephel A, Mayer M, et al. , (2019). Drugging an undruggable pocket on KRAS. Proc. Natl. Acad. Sci. U. S. A 116, 15823–15829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Tran TH, Alexander P, Dharmaiah S, Agamasu C, Nissley DV, McCormick F, et al. , (2020). The small molecule BI-2852 induces a nonfunctional dimer of KRAS. Proc. Natl. Acad. Sci. U. S. A 117, 3363–3364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Ullrich A, Schlessinger J, (1990). Signal transduction by receptors with tyrosine kinase activity. Cell 61, 203–212. [DOI] [PubMed] [Google Scholar]
- 85.Simons K, Toomre D, (2000). Lipid rafts and signal transduction. Nat. Rev. Mol. Cell Biol 1, 31–39. [DOI] [PubMed] [Google Scholar]
- 86.Blume-Jensen P, Hunter T, (2001). Oncogenic kinase signalling. Nature 411, 355–365. [DOI] [PubMed] [Google Scholar]
- 87.Fabbro D, Cowan-Jacob SW, Moebitz H, (2015). Ten things you should know about protein kinases: IUPHAR R eview 14. Br. J. Pharmacol 172, 2675–2700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Stephen AG, Esposito D, Bagni RK, McCormick F, (2014). Dragging ras back in the ring. Cancer Cell 25, 272–281. [DOI] [PubMed] [Google Scholar]
- 89.Roskoski R Jr., (2022). Properties of FDA-approved small molecule protein kinase inhibitors: A 2022 update. Pharmacol. Res 175, 106037. [DOI] [PubMed] [Google Scholar]
- 90.Moret N, Liu C, Gyori BM, Bachman JA, Steppi A, Hug C, et al. , (2021). A resource for exploring the understudied human kinome for research and therapeutic opportunities. bioRxiv. 2020.04.02.022277. [Google Scholar]
- 91.Saladino G, Gervasio FL, (2012). New insights in protein kinase conformational dynamics. Curr. Top. Med. Chem 12, 1889–1895. [DOI] [PubMed] [Google Scholar]
- 92.Huse M, Kuriyan J, (2002). The conformational plasticity of protein kinases. Cell 109, 275–282. [DOI] [PubMed] [Google Scholar]
- 93.Hubbard SR, Till JH, (2000). Protein tyrosine kinase structure and function. Annu. Rev. Biochem 69, 373–398. [DOI] [PubMed] [Google Scholar]
- 94.Manning G, Whyte DB, Martinez R, Hunter T, Sudarsanam S, (2002). The protein kinase complement of the human genome. Science 298, 1912–1934. [DOI] [PubMed] [Google Scholar]
- 95.Becher I, Savitski MM, Savitski MF, Hopf C, Bantscheff M, Drewes G, (2013). Affinity profiling of the cellular kinome for the nucleotide cofactors ATP, ADP, and GTP. ACS Chem. Biol 8, 599–607. [DOI] [PubMed] [Google Scholar]
- 96.Dar AC, Shokat KM, The Evolution of Protein Kinase Inhibitors from Antagonists to Agonists of Cellular Signaling, in: Kornberg RD, Raetz CRH, Rothman JE, Thorner JW (Eds.), Annual Review of Biochemistry, Vol 802011. p. 769–95. [DOI] [PubMed] [Google Scholar]
- 97.Lamba V, Ghosh I, (2012). New directions in targeting protein kinases: focusing upon true allosteric and bivalent inhibitors. Curr. Pharm. Des 18, 2936–2945. [DOI] [PubMed] [Google Scholar]
- 98.Wakeling AE, Guy SP, Woodburn JR, Ashton SE, Curry BJ, Barker AJ, et al. , (2002). ZD1839 (Iressa): an orally active inhibitor of epidermal growth factor signaling with potential for cancer therapy. Cancer Res. 62, 5749–5754. [PubMed] [Google Scholar]
- 99.Deininger MW, Goldman JM, Lydon N, Melo JV, (1997). The tyrosine kinase inhibitor CGP57148B selectively inhibits the growth of BCR-ABL-positive cells. Blood 90, 3691–3698. [PubMed] [Google Scholar]
- 100.Abdeldayem A, Raouf YS, Constantinescu SN, Moriggl R, Gunning PT, (2020). Advances in covalent kinase inhibitors. Chem. Soc. Rev 49, 2617–2687. [DOI] [PubMed] [Google Scholar]
- 101.Bridges AJ, (1998). Preparation of 2-phenylaminobenzoic acids and its amides as MEK inhibitors for treating or preventing septic shock. Warner Lambert Co.. [Google Scholar]
- 102.Ohren JF, Chen H, Pavlovsky A, Whitehead C, Zhang E, Kuffa P, et al. , (2004). Structures of human MAP kinase kinase 1 (MEK1) and MEK2 describe novel noncompetitive kinase inhibition. Nat. Struct. Mol. Biol 11, 1192–1197. [DOI] [PubMed] [Google Scholar]
- 103.Ashwell MA, Lapierre J-M, Brassard C, Bresciano K, Bull C, Cornell-Kennon S, et al. , (2012). Discovery and optimization of a series of 3-(3-Phenyl-3 H-imidazo [4, 5-b] pyridin-2-yl) pyridin-2-amines: orally bioavailable, selective, and potent ATP-independent Akt inhibitors. J. Med. Chem 55, 5291–5310. [DOI] [PubMed] [Google Scholar]
- 104.Bagal SK, Omoto K, Blakemore DC, Bungay PJ, Bilsland JG, Clarke PJ, et al. , (2018). Discovery of allosteric, potent, subtype selective, and peripherally restricted TrkA kinase inhibitors. J. Med. Chem 62, 247–265. [DOI] [PubMed] [Google Scholar]
- 105.Jia Y, Yun C-H, Park E, Ercan D, Manuia M, Juarez J, et al. , (2016). Overcoming EGFR (T790M) and EGFR (C797S) resistance with mutant-selective allosteric inhibitors. Nature 534, 129–132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Heinrich T, Gradler U, Bottcher H, Blaukat A, Shutes A, (2010). Allosteric IGF-1R Inhibitors. ACS Med. Chem. Lett 1, 199–203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Goodwin NC, Cianchetta G, Burgoon HA, Healy J, Mabon R, Strobel ED, et al. , (2015). Discovery of a type III inhibitor of LIM kinase 2 that binds in a DFG-out conformation. ACS Med. Chem. Lett 6, 53–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Klüter S, Grütter C, Naqvi T, Rabiller M, Simard JR, Pawar V, et al. , (2010). Displacement assay for the detection of stabilizers of inactive kinase conformations. J. Med. Chem 53, 357–367. [DOI] [PubMed] [Google Scholar]
- 109.Simard JR, Klüter S, Grütter C, Getlik M, Rabiller M, Rode HB, et al. , (2009). A new screening assay for allosteric inhibitors of cSrc. Nat. Chem. Biol 5, 394–396. [DOI] [PubMed] [Google Scholar]
- 110.Tomita N, Hayashi Y, Suzuki S, Oomori Y, Aramaki Y, Matsushita Y, et al. , (2013). Structure-based discovery of cellular-active allosteric inhibitors of FAK. Bioorg. Med. Chem. Lett 23, 1779–1785. [DOI] [PubMed] [Google Scholar]
- 111.Karpov AS, Amiri P, Bellamacina C, Bellance M-H, Breitenstein W, Daniel D, et al. , (2015). Optimization of a dibenzodiazepine hit to a potent and selective allosteric PAK1 inhibitor. ACS Med. Chem. Lett 6, 776–781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Zhao Z, Xie L, Bourne PE, (2017). Insights into the binding mode of MEK type-III inhibitors. A step towards discovering and designing allosteric kinase inhibitors across the human kinome. PLoS ONE 12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Hantschel O, Nagar B, Guettler S, Kretzschmar J, Dorey K, Kuriyan J, et al. , (2003). A myristoyl/phosphotyrosine switch regulates c-Abl. Cell 112, 845–857. [DOI] [PubMed] [Google Scholar]
- 114.Adrian FJ, Ding Q, Sim T, Velentza A, Sloan C, Liu Y, et al. , (2006). Allosteric inhibitors of Bcr-abl-dependent cell proliferation. Nat. Chem. Biol 2, 95–102. [DOI] [PubMed] [Google Scholar]
- 115.Zhang J, Adrian FJ, Jahnke W, Cowan-Jacob SW, Li AG, Iacob RE, et al. , (2010). Targeting Bcr-Abl by combining allosteric with ATP-binding-site inhibitors. Nature 463, 501–506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Iacob RE, Zhang J, Gray NS, Engen JR, (2011). Allosteric interactions between the myristate- and ATP-site of the Abl kinase. PLoS ONE 6, e15929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Wylie AA, Schoepfer J, Jahnke W, Cowan-Jacob SW, Loo A, Furet P, et al. , (2017). The allosteric inhibitor ABL001 enables dual targeting of BCR-ABL1. Nature 543, 733–737. [DOI] [PubMed] [Google Scholar]
- 118.P. Novartis, Novartis. A Study of Oral Asciminib Versus Other TKIs in Adult Patients With Newly Diagnosed Ph+ CML-CP, 2024. [Google Scholar]
- 119.P. Novartis, Novartis. Study of Efficacy and Safety of CML-CP Patients Treated With Asciminib Versus Best Available Therapy, Previously Treated With 2 or More Tyrosine Kinase Inhibitors, 2023. [Google Scholar]
- 120.U. Augusta, H.J.K.C.C. Consortium, Asciminib as Initial Therapy for Patients With Chronic Myeloid Leukemia in Chronic Phase, 2025. [Google Scholar]
- 121.Biondi RM, Cheung PC, Casamayor A, Deak M, Currie RA, Alessi DR, (2000). Identification of a pocket in the PDK1 kinase domain that interacts with PIF and the C-terminal residues of PKA. EMBO J. 19, 979–988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Sadowsky JD, Burlingame MA, Wolan DW, McClendon CL, Jacobson MP, Wells JA, (2011). Turning a protein kinase on or off from a single allosteric site via disulfide trapping. Proc. Natl. Acad. Sci. U. S. A 108, 6056–6061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Rettenmaier TJ, Sadowsky JD, Thomsen ND, Chen SC, Doak AK, Arkin MR, et al. , (2014). A small-molecule mimic of a peptide docking motif inhibits the protein kinase PDK1. Proc. Natl. Acad. Sci. U. S. A 111, 18590–18595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Converso A, Hartingh T, Garbaccio RM, Tasber E, Rickert K, Fraley ME, et al. , (2009). Development of thioquinazolinones, allosteric Chk1 kinase inhibitors. Bioorg. Med. Chem. Lett 19, 1240–1244. [DOI] [PubMed] [Google Scholar]
- 125.Comess KM, Sun C, Abad-Zapatero C, Goedken ER, Gum RJ, Borhani DW, et al. , (2011). Discovery and characterization of non-ATP site inhibitors of the mitogen activated protein (MAP) kinases. ACS Chem. Biol 6, 234–244. [DOI] [PubMed] [Google Scholar]
- 126.Péczka N, Orgován Z, Ábrányi-Balogh P, Keserű GM, (2022). Electrophilic warheads in covalent drug discovery: an overview. Expert Opin. Drug Discov 17, 413–422. [DOI] [PubMed] [Google Scholar]
- 127.De Vita E, (2021). 10 years into the resurgence of covalent drugs. Future Med. Chem 13, 193–210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Ghosh AK, Samanta I, Mondal A, Liu WR, (2019). Covalent Inhibition in Drug Discovery. ChemMedChem 14, 889–906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Pettinger J, Jones K, Cheeseman MD, (2017). Lysine-Targeting Covalent Inhibitors. Angew. Chem. Int. Ed. Engl 56, 15200–15209. [DOI] [PubMed] [Google Scholar]
- 130.Sutanto F, Konstantinidou M, Dömling A, (2020). Covalent inhibitors: a rational approach to drug discovery. RSC Med. Chem 11, 876–884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Nakajima EC, Drezner N, Li X, Mishra-Kalyani PS, Liu Y, Zhao H, et al. , (2021). FDA Approval Summary: Sotorasib for KRAS G12C-Mutated Metastatic NSCLC. Clin. Cancer Res [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Senior AW, Evans R, Jumper J, Kirkpatrick J, Sifre L, Green T, et al. , (2020). Improved protein structure prediction using potentials from deep learning. Nature 577, 706–710. [DOI] [PubMed] [Google Scholar]
- 133.Meyers RM, Bryan JG, McFarland JM, Weir BA, Sizemore AE, Xu H, et al. , (2017). Computational correction of copy number effect improves specificity of CRISPR-Cas9 essentiality screens in cancer cells. Nat. Genet 49, 1779–1784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Tsherniak A, Vazquez F, Montgomery PG, Weir BA, Kryukov G, Cowley GS, et al. , (2017). Defining a Cancer Dependency Map. Cell 170, (564–76) e16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Johnson DI, Pringle JR, (1990). Molecular characterization of CDC42, a Saccharomyces cerevisiae gene involved in the development of cell polarity. J. Cell Biol 111, 143–152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Arias-Romero LE, Chernoff J, (2013). Targeting Cdc42 in cancer. Expert Opin. Ther. Targets 17, 1263–1273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Pelish HE, Peterson JR, Salvarezza SB, Rodriguez-Boulan E, Chen JL, Stamnes M, et al. , (2006). Secramine inhibits Cdc42-dependent functions in cells and Cdc42 activation in vitro. Nat. Chem. Biol 2, 39–46. [DOI] [PubMed] [Google Scholar]
- 138.Friesland A, Zhao Y, Chen YH, Wang L, Zhou H, Lu Q, (2013). Small molecule targeting Cdc42-intersectin interaction disrupts Golgi organization and suppresses cell motility. Proc. Natl. Acad. Sci. U. S. A 110, 1261–1266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Hong L, Kenney SR, Phillips GK, Simpson D, Schroeder CE, Noth J, et al. , (2013). Characterization of a Cdc42 protein inhibitor and its use as a molecular probe. J. Biol. Chem 288, 8531–8543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Heasman SJ, Ridley AJ, (2008). Mammalian Rho GTPases: new insights into their functions from in vivo studies. Nat. Rev. Mol. Cell Biol 9, 690–701. [DOI] [PubMed] [Google Scholar]
- 141.Bid HK, Roberts RD, Manchanda PK, Houghton PJ, (2013). RAC1: an emerging therapeutic option for targeting cancer angiogenesis and metastasis. Mol. Cancer Ther 12, 1925–1934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Gao Y, Dickerson JB, Guo F, Zheng J, Zheng Y, (2004). Rational design and characterization of a Rac GTPase-specific small molecule inhibitor. Proc. Natl. Acad. Sci. U. S. A 101, 7618–7623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Montalvo-Ortiz BL, Castillo-Pichardo L, Hernandez E, Humphries-Bickley T, De la Mota-Peynado A, Cubano LA, et al. , (2012). Characterization of EHop-016, novel small molecule inhibitor of Rac GTPase. J. Biol. Chem 287, 13228–13238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Mackay DJ, Hall A, (1998). Rho GTPases. J Biol Chem 273, 20685–20688. [DOI] [PubMed] [Google Scholar]
- 145.Karlsson R, Pedersen ED, Wang Z, Brakebusch C, (2009). Rho GTPase function in tumorigenesis. Biochim. Biophys. Acta, Mol. Cell. Biol. Lipids 1796, 91–98. [DOI] [PubMed] [Google Scholar]
- 146.Shang X, Marchioni F, Sipes N, Evelyn CR, Jerabek-Willemsen M, Duhr S, et al. , (2012). Rational design of small molecule inhibitors targeting RhoA subfamily Rho GTPases. Chem. Biol 19, 699–710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Jakob JA, Bassett RL Jr., Ng CS, Curry JL, Joseph RW, Alvarado GC, et al. , (2012). NRAS mutation status is an independent prognostic factor in metastatic melanoma. Cancer-Am Cancer Soc. 118, 4014–4023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Liu Q, Sabnis Y, Zhao Z, Zhang T, Buhrlage SJ, Jones LH, et al. , (2013). Developing irreversible inhibitors of the protein kinase cysteinome. Chem. Biol 20, 146–159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Ramalingam SS, Yang J, Lee CK, Kurata T, Kim D-W, John T, et al. , (2018). Osimertinib as first-line treatment of EGFR mutation-positive advanced non-small-cell lung cancer. J. Clin. Oncol 36, 841–849. [DOI] [PubMed] [Google Scholar]
- 150.Pao W, Miller VA, Politi KA, Riely GJ, Somwar R, Zakowski MF, et al. , (2005). Acquired resistance of lung adenocarcinomas to gefitinib or erlotinib is associated with a second mutation in the EGFR kinase domain. PLoS Med. 2, e73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Mortuza GB, Hermida D, Pedersen AK, Segura-Bayona S, Lopez-Mendez B, Redondo P, et al. , (2018). Molecular basis of Tousled-Like Kinase 2 activation. Nat. Commun 9, 2535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Segura-Bayona S, Stracker TH, (2019). The Tousled-like kinases regulate genome and epigenome stability: implications in development and disease. Cell. Mol. Life Sci 76, 3827–3841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Dawson JC, Serrels A, Stupack DG, Schlaepfer DD, Frame MC, (2021). Targeting FAK in anticancer combination therapies. Nat. Rev. Cancer 21, 313–324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Cance WG, Kurenova E, Marlowe T, Golubovskaya V, (2013). Disrupting the scaffold to improve focal adhesion kinase-targeted cancer therapeutics. Sci. Signal 6 pe10. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.