Abstract
Targeted gene manipulation is highly desirable for fundamental plant research, plant synthetic biology, and molecular breeding. The clustered regularly interspaced short palindromic repeats-associated (Cas) nuclease is a revolutionary tool for genome editing, and has received snowballing popularity for gene knockout applications in diverse organisms including plants. Recently, the nuclease-dead Cas (dCas) proteins have been repurposed as programmable transcriptional regulators through translational fusion with portable transcriptional repression or activation domains, which has paved new ways for flexible and multiplex control over the activities of target genes of interest without the need to generate DNA lesions. Here, we review the most important breakthroughs of dCas transcriptional regulators in non-plant organisms and recent accomplishments of this growing field in plants. We also provide perspectives on future development directions of dCas transcriptional regulators in plant research in hope to stimulate their quick evolution and broad applications.
Keywords: Nuclease-Dead Cas9 (dCas9), sgRNA, Transcriptional repressor, Transcriptional activator, Target promoter
Introduction
Targeted gene manipulation is of paramount importance for interrogating gene functions and rewiring cellular activities in basic plant research and for intensifying beneficial agronomic traits in molecular breeding. Although the gene activity can be manipulated at either the DNA or mRNA level, the former is apparently easier because only a few copies of target DNA rather than a large quantity of target mRNA need to be dealt with in individual cells. As such, the past two decades have witnessed slow advances in RNA interference (RNAi) technologies but a quick evolution of genome editing technologies from the zinc finger nuclease (ZFN) to transcription activation-like effector nuclease (TALEN) to clustered regularly interspaced short palindromic repeats (CRISPR)-associated (Cas) nuclease (Wang et al. 2016; Zhang et al. 2016; Adli 2018; Chen et al. 2019). These site-specific nucleases (SSNs) can be programmed to bind a specific DNA sequence in a target gene and, subsequently, cleave the two DNA strands to create a double-strand break (DSB), which can induce frame-shift indels during DNA repair to abolish the gene function. However, DNA recognition by ZFNs and TALENs is based on protein–DNA interactions so that ZFNs or TALENs with different DNA-binding domains have to be re-designed and re-constructed for individual new target sequences, which requires plenty of time, labor, and expertise. In contrast, a single-guide RNA (sgRNA) can be easily customized to direct the same Cas nuclease to a new target site through RNA–DNA base pairing, thus exhibiting unparalleled ease-of-use, incredible multiplexability, and tolerance to DNA methylation. Therefore, the CRISPR-Cas technology has gained explosive popularity since its birth and has been applied to gene loss-of-function (LOF) studies in a broad range of organisms from animals to plants. Detailed introduction about the CRISPR-Cas technology can be found in several excellent reviews published recently (Adli 2018; Chen et al. 2019). However, gene dysfunctions induced by the CRISPR-Cas system are permanent and irreversible, and can cause lethality when essential genes for the organismic survival have been disrupted. Also, the CRISPR-Cas users in mammalian research have been warned of complicated DSB-induced chromosomal rearrangements and truncations in gene LOF studies (Kosicki et al. 2018; Cullot et al. 2019). Therefore, other DNA-based gene LOF technologies with inducibility and reversibility are also wanted.
The opposite direction of gene manipulation is gene gain-of-function (GOF), which is as important and useful as gene LOF because: (1) gene GOF can obtain novel insights on gene functions when LOF studies are hindered by mutant lethality or undetectable phenotypic changes due to functional redundancy (Abe and Ichikawa 2016). (2) Attractive agronomic traits can also be quantitatively enhanced by gene GOF in molecular breeding (Petolino and Davies 2013). (3) Metabolic engineering to promote the production of invaluable plant metabolites often requires coordinated activation of multiple dormant genes controlling a branched metabolic pathway (Zorrilla-López et al. 2013). Nowadays, the mainstream gene GOF strategy in plants is the complementary DNA (cDNA) overexpression using a constitutive strong promoter, such as the cauliflower mosaic virus 35S promoter (Abe and Ichikawa 2016). However, this strategy is less powerful for multigene overexpression or genome-wide GOF screens due to the limited vector capacity and increased labor for cloning. Also, cDNA overexpression cannot fully mimic gene activation under the native cellular context due to the lack of transcript-stabilizing elements (e.g., UTRs or introns) or alternative mRNA splicing (Karve et al. 2011), which may lead to failure of gene overexpression or misinterpretation of gene functions. Therefore, gene GOF technologies that enable multiplex gene activation from the endogenous chromosomal loci are urgently in demand.
Recently, the CRISPR/Cas system has been repurposed as synthetic transcriptional regulators to meet the needs of gene LOF and GOF. In the CRISPR-derived transcriptional regulators, the nuclease-dead Cas (dCas) proteins that lose the key catalytic residues for cleaving DNA double strands serve as sgRNA-directed promoter-binding domains with programmable binding specificity. These dCas proteins are then fused with autonomous transcriptional repression or activation domains (TRDs or TADs) to confer multiplexable transcriptional regulation of genes of interest from their endogenous loci in the chromosome, therefore opening up new exciting possibilities for gene LOF and GOF applications. This review will introduce up-to-date progress in developing dCas transcriptional regulators in plants. Since the plant research in this field is still in its infancy when compared with that in mammals, we will also summarize landmark innovations of dCas-TRDs and dCas-TADs in non-plant model organisms, and discuss future perspectives of these tools in plant research to stimulate plant biologists to explore their full potentials.
Gene LOF by dCas-TRDs in non-plant organisms
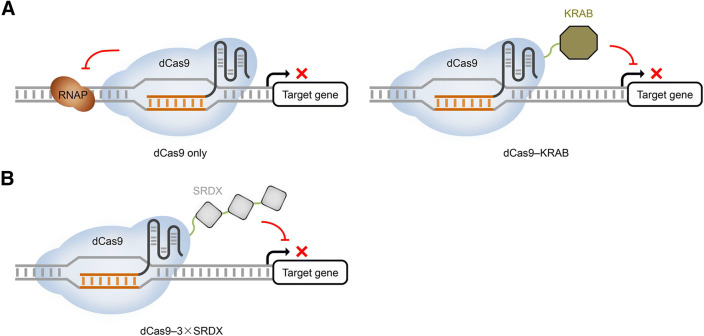
Shortly after the emergence of the CRISPR-Cas technology, it was found in both Escherichia coli and mammalian cells that the nuclease-dead Streptococcus pyogenes Cas9 (hereafter referred to as dCas9) is able to physically inhibit the transcriptional initiation and elongation of a target gene by binding to its core promoter (i.e., from the TATA box to the transcription starting site or TSS) or coding sequence (Qi et al. 2013) (Fig. 1A). This so-called CRISPR interference (CRISPRi) offers a powerful gene silencing strategy for prokaryotes since most prokaryotic organisms lack the RNAi machinery (Peters et al. 2016). The CRISPRi strategy was soon upgraded in mammalian and yeast cells by supplementing dCas9 with specialized TRDs to strengthen the transcriptional repression activities. These TRDs include the chromo shadow (CS) domain of HP1α (Gilbert et al. 2013), Trp-Arg-Pro-Trp (WRPW) motif of Hes1 (Gilbert et al. 2013), krüppel-associated box (KRAB) domain of Kox1 (Gilbert et al. 2014), and Max-interacting protein 1 (Mxi1) domain (Lawhorn et al. 2014). Among these dCas9-TRDs, the dCas9-KRAB protein exhibits the most consistent and robust effects on transcriptional repression (Xu and Qi 2019) (Fig. 1A). Of note, dCas9-TRD-mediated gene LOF, as a gene knockdown rather than gene knockout strategy, will not provoke genetic compensation response in a given organism to confound gene functional studies (Rossi et al. 2015).
Fig. 1.
Representative dCas9-based transcriptional repressors. (A) CRISPR interference (CRISPRi) platforms for transcriptional repression in non-plant organisms. The dCas9 protein coupled with the sgRNA can serve as an effective transcriptional repressor by interfering with the assembly of RNA polymerase (RNAP) and sterically blocking transcriptional initiation or elongation. KRAB, a transcriptional repression domain, can be tethered to dCas9 to further strengthen target gene repression. (B) Potent dCas9-based transcriptional repressor in plants. Three tandem repeats of SRDX, a transcriptional repression motif, can be fused to dCas9 for strong target gene repression. Black arrows represent the initiation of transcription, whereas red lines indicate the effects of transcriptional repression
Gene LOF by dCas-TRDs in plants
The CRISPRi strategy using dCas9 alone has been later proven effective for the targeted transcriptional repression of reporter genes or endogenous protein-coding genes in Nicotiana benthamiana (Piatek et al. 2015; Vazquez-Vilar et al. 2016). Two Arabidopsis endogenous TRDs, namely the SRDX and BRD motifs, have been, respectively, fused to dCas9 with an intention to enhance its transcriptional repression effect in plants (Piatek et al. 2015; Vazquez-Vilar et al. 2016). However, both dCas9-SRDX and dCas9-BRD have exhibited similar transcriptional repression effects on a pNOS::LUC reporter like dCas9 alone (Piatek et al. 2015; Vazquez-Vilar et al. 2016). Notably, it seems that arming dCas9 with three tandem repeats of SRDX (3 × SRDX) could cause stronger transcriptional repression (Lowder et al. 2015) (Fig. 1B). In addition, another orthogonal dCas protein, namely dCas12a (also known as dCpf1) (Zetsche et al. 2015), could be used to substitute dCas9 to create more potent synthetic transcriptional repressors in plants (Tang et al. 2017). Both the dCas9-3 × SRDX and dCas12a-SRDX have been applied to the repression of protein-coding or non-coding genes (e.g., microRNAs) in transgenic Arabidopsis plants (Lowder et al. 2015; Tang et al. 2017). The invention of dCas12a-TRDs can increase genome-wide targetable sites within the promoters, particularly in the T-rich regions, due to different protospacer adjacent motif (PAM) requirements by Cas9 and Cas12a. Moreover, compared to dCas9-based gene repressors, dCas12a gene repressors may enable simplified multiplex transcriptional repression, since dCas12a has the capability to self-process a single transcript of tandem crRNA arrays to generate multiple crRNAs (Fonfara et al. 2016; Zetsche et al. 2017). Table 1 of this review summarizes the applications of dCas-based gene repression in plants.
Table 1.
Applications of dCas-based gene repressors in plant cells
| Repressor | Target gene (sgRNA numbersa) | Repression levelb (%) | Plant species | Assay | References |
|---|---|---|---|---|---|
| dCas9 | PDS (3) | 20 | N. benthamiana | Agroinfiltration | Piatek et al. (2015) |
| pNOS::LUC reporter (3) | 80 | N. benthamiana | Agroinfiltration | Vazquez-Vilar et al. (2016) | |
| dCas9-SRDX | PDS (3) | 33 | N. benthamiana | Agroinfiltration | Piatek et al. (2015) |
| pNOS::LUC reporter (3) | 50 | N. benthamiana | Agroinfiltration | Vazquez-Vilar et al. (2016) | |
| dCas9-BRD | pNOS::LUC reporter (3) | 60 | N. benthamiana | Agroinfiltration | Vazquez-Vilar et al. (2016) |
| dCas9-3 × SRDX | CSTF64 (3) | 60 | Arabidopsis | Transgenic assay | Lowder et al. (2015) |
| miR159a (1) | 80 | Arabidopsis | Transgenic assay | Lowder et al. (2015) | |
| miR159b (2) | 70 | Arabidopsis | Transgenic assay | Lowder et al. (2015) | |
| dLbCpf1-SRDX | miR159b (1) | 90 | Arabidopsis | Transgenic assay | Tang et al. (2017) |
| dAsCpf1-SRDX | miR159b (1) | 90 | Arabidopsis | Transgenic assay | Tang et al. (2017) |
pNOS nopaline synthase promoter, LUC the luciferase gene
aOnly the number of sgRNAs for maximal gene repression is shown
bMaximal percentage of gene expression decline obtained using the indicated number of sgRNAs
It is worthy to mention that, even using the same dCas-TRD gene repressor, different sgRNAs could induce the transcriptional repression of the same promoter with distinct efficiencies (Piatek et al. 2015; Vazquez-Vilar et al. 2016). However, it seems that the sgRNAs targeting dCas-TRD to the proximal regions flanking the TSS could generally be more effective than those targeting the distal promoter (Lowder et al. 2015; Vazquez-Vilar et al. 2016). Also, multiple sgRNAs targeting the same promoter could lead to more efficient transcriptional repression than a single sgRNA (Piatek et al. 2015; Vazquez-Vilar et al. 2016).
Gene GOF by dCas-TADs in animals
Pilot efforts for the targeted gene activation in mammalian cells have frequently involved the dCas9-VP64 gene activator that contains a tetramer of the TAD from the Herpes simplex viral protein 16 (Mali et al. 2013; Maeder et al. 2013; Perez-Pinera et al. 2013; Gilbert et al. 2013). However, dCas9-VP64 could only weakly activate the target gene expression in mammalian cells. Although the strategy of tiling multiple sgRNAs along the proximal promoter has been employed to reinforce the dCas9-VP64-mediated transcriptional activation (Mali et al. 2013; Maeder et al. 2013; Perez-Pinera et al. 2013; Gilbert et al. 2013), it limits the scalability of the system (Konermann et al. 2015) and increases the risk of off-target transcriptional perturbations (Cheng et al. 2013; Farzadfard et al. 2013; Braun et al. 2016).
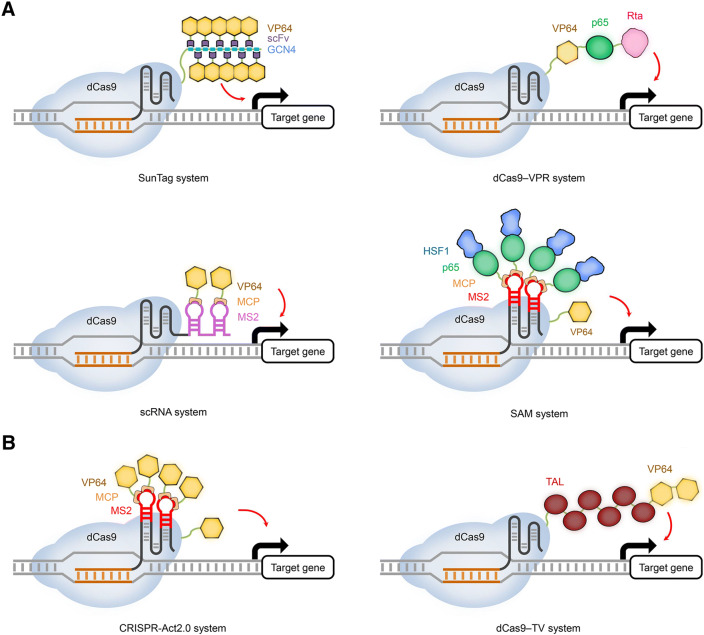
Several innovative strategies have been subsequently developed to improve the transcriptional activation using only a single sgRNA. The first strategy is known as the SunTag system, where dCas9 is fused to tandem GCN4 peptide repeats and each repeat can recruit VP64 through an anti-GCN4 antibody termed scFv (Tanenbaum et al. 2014; Gilbert et al. 2014) (Fig. 2A). In a second strategy, dubbed the dCas9-VPR system, dCas9 is fused to a tripartite transcriptional activation module consisting of VP64 and two additional TADs, namely p65 from the nuclear factor kappa B and Rta from the Epstein–Barr virus R transactivator (Chavez et al. 2015) (Fig. 2A). In a third strategy, dubbed the scaffold RNA (scRNA) strategy, the sgRNA is appended with up to two MS2 RNA hairpins at the 3′ end and each hairpin can recruit VP64 via a MS2-binding protein (MCP) (Mali et al. 2013; Zalatan et al. 2015) (Fig. 2A). The fourth strategy known as the synergistic activation mediator (SAM) system is more like an upgraded scRNA strategy, where dCas9-VP64, rather than dCas9, is used in combination with a modified sgRNA containing two internal MS2 RNA hairpins that can recruit tandem TADs of p65 and human heat shock factor 1 (HSF1) via MCPs (Konermann et al. 2015) (Fig. 2A). The SunTag, VPR, and SAM systems have all been demonstrated to trigger potent targeted transcriptional activation in a variety of animal cells (Chavez et al. 2016). A key concept shared by these three improved dCas9-TAD systems is to engage multiple copies of the same or different TADs through dCas9 or/and the sgRNA to induce additive or synergistic effects on gene activation. By following this concept, although attempts to further increase the transcriptional activation efficiency through combining dCas9-VPR with the SAM strategy were not very successful (Chavez et al. 2016), a new system referred to as SunTag-p65-HSF1 (SPH), which utilized the chimeric p65-HSF1 TADs to replace VP64 in the SunTag system, could induce more potent transcriptional activation in transgenic mice than the VPR, SunTag or SAM system (Zhou et al. 2018).
Fig. 2.
Representative dCas9-based transcriptional activators. (A) Innovative systems for potent transcriptional activation in animal cells. The SunTag system consists of dCas9 fused to tandem repeats of GCN4 peptide, which can recruit VP64 via the antibody scFv that can bind to GCN4. The VPR system contains three transcriptional activation domains (VP64, p65, and Rta), which are fused to dCas9 in tandem. In the scRNA system, a sgRNA bearing two MS2 hairpins at its 3′ end can recruit VP64 via the fusion partner MCP. In the SAM system, a dCas9-VP64 fusion is utilized in combination with a modified sgRNA harboring two MS2 hairpins that can recruit tandem p65-HSF1 transcriptional activation domains via MCP. (B) Improved dCas9-based transcriptional activation systems in plant cells. In the CRISPR-Act2.0 system, a dCas9-VP64 fusion is utilized in combination with a modified sgRNA harboring two MS2 hairpins that can recruit additional VP64 via MCP. The dCas9-TV gene activator contains six tandem repeats of the TAL transcriptional activation domain from Xanthamonas and two copies of VP64. Red arrows indicate the effects of transcriptional activation, while black arrows represent the initiation of transcription
To further expand genome-wide targetable sites within the promoters, dCas12a has also been explored for the targeted transcriptional activation. A fusion of the catalytically inactive Lachnospiraceae bacterium Cas12a (dLbCas12a) to the VPR could efficiently activate target genes in human cells with comparable performance to dCas9-VPR (Tak et al. 2017), while the nuclease-dead Acidaminococcus sp. BV3L6 Cas12a (dAsCas12a)-based VPR gene activator could be even more potent than dCas9-VPR (Liu et al. 2017). It has been demonstrated in another study that the dLbCas12a-based SunTag system can also enable robust targeted gene activation (Zhang et al. 2018b). Very recently, researchers have engineered an enhanced variant of AsCas12a termed enAsCas12a, which has a substantially expanded target range and improved editing efficiency (Kleinstiver et al. 2019). A fusion of the catalytically inactive enAsCas12a to the VPR could outperform dAsCas12a-, dLbCas12a-, and dCas9-based VPR gene activators (Kleinstiver et al. 2019). Compared to dCas9-based gene activators, dCas12a gene activators may be more powerful for multiplex gene activation (Tak et al. 2017; Zhang et al. 2018b; Kleinstiver et al. 2019), as dCas12a is able to self-process a single transcript of tandem crRNA arrays to generate multiple crRNAs (Fonfara et al. 2016; Zetsche et al. 2017).
Gene GOF by dCas-TADs in plants
Considering the functional conservation of transcriptional machineries between animals and plants, it is not surprising to see that some of the dCas-TADs used in mammalian cells have been successfully transferred to plant cells for the targeted gene activation. Four plant research groups independently evaluated the transcriptional activation activity of dCas9-VP64 using either transient or transgenic gene activation assays in Arabidopsis and N. benthamiana (Piatek et al. 2015; Lowder et al. 2015; Vazquez-Vilar et al. 2016; Li et al. 2017). The dCas9-VP64 only weakly activated target gene expression when a single sgRNA was used (Piatek et al. 2015; Vazquez-Vilar et al. 2016; Li et al. 2017). However, it could lead to significantly higher gene activation when multiple sgRNAs were used to target the same promoter (Piatek et al. 2015; Vazquez-Vilar et al. 2016; Lowder et al. 2015). These trends were reminiscent of what has been observed in mammalian cells (Mali et al. 2013; Maeder et al. 2013; Perez-Pinera et al. 2013; Gilbert et al. 2013). Of note, when the SAM or SunTag strategy was introduced into transgenic Arabidopsis plants, they could potently stimulate transcriptional activation of endogenous target genes (Park et al. 2017; Papikian et al. 2019). Interestingly, Papikian et al. found that the SunTag-mediated gene activation in Arabidopsis was coupled with the reduction of promoter methylation at the target region. Encouraged by these observations, they could further employ the SunTag system to reactivate transposable elements in plants (Papikian et al. 2019). Consistently, the Arabidopsis FIS2 gene, which has been assumed to be shut down by DNA methylation, could be activated by 400-fold using dCas9-VP64 in combination with 3 sgRNAs targeting the methylated CpG island within its promoter (Lowder et al. 2015). Similar to the design of the SAM strategy, a new strategy called CRISPR-Act2.0 has been developed recently in plants, where dCas9-VP64 was used in combination with a modified sgRNA containing two internal MS2 RNA hairpins that can recruit additional VP64 via MCPs (Lowder et al. 2018) (Fig. 2B). When using the same sgRNAs, the CRISPR-Act2.0 system could significantly outperform dCas9-VP64 for activating both protein coding and non-coding genes in Arabidopsis and rice cells (Lowder et al. 2018).
The EDLL motif, a plant-derived TAD from the AP2 transcription factor, and the TAD from the bacterial TALE protein (hereafter referred to as TAL), have also been evaluated for building dCas9-based gene activators in plants (Piatek et al. 2015; Vazquez-Vilar et al. 2016; Lowder et al. 2018; Li et al. 2017). However, both dCas9-EDLL and dCas9-TAL only exhibited modest transcriptional activation activities in combination with a single sgRNA in plant cells (Piatek et al. 2015; Vazquez-Vilar et al. 2016). Similar to the design of the VPR strategy, the EDLL motif has also been linked with VP64 for boosting the transcriptional activation efficiency but failed to work efficiently in plant cells (Lowder et al. 2018). Recently, Li et al. devised ten dCas9 gene activators using VP64, TAL, EDLL, and ERF2m, another plant-derived TAD from the ERF2 transcription factor, as basic TAD building blocks, which were used in different combinations and copy numbers (Li et al. 2017). Among these dCas9 gene activators, the one containing six copies of TAL and two copies of VP64 was screened out as the most potent gene activator and was dubbed dCas9-TV (Li et al. 2017) (Fig. 2B). Addition of more TALs to dCas9-TV could trigger severe protein degradation presumably due to high sequence repetition (Li et al. 2017), reminiscent of the protein instability issue in the SunTag strategy where the bulky dCas9 is fused to multiple GCN4 repeats (Tanenbaum et al. 2014). Nevertheless, using only a single sgRNA, the dCas9-TV gene activator has already enabled robust transcriptional activation of target genes in Arabidopsis and rice cells in a multiplex manner. Surprisingly, dAsCas12a-TV could only marginally activate target gene expression for unknown reason (Li et al. 2017). Both dCas9-TV and the SunTag systems have been demonstrated to be highly specific for gene activation in plant cells based on genome-wide RNA-sequencing analyses (Li et al. 2017; Papikian et al. 2019). Table 2 of this review summarizes applications of dCas-based gene activation in plants.
Table 2.
Applications of dCas-based gene activators in plant cells
| Activator | Target gene (sgRNA numbersa) | Fold changeb | Plant species | Assay | References |
|---|---|---|---|---|---|
| dCas9-EDLL | PDS (3) | 3.5 | N. benthamiana | Agroinfiltration | Piatek et al. (2015) |
| pNOS::LUC reporter (3) | 2.2 | N. benthamiana | Agroinfiltration | Vazquez-Vilar et al. (2016) | |
| dCas9-TAL | PDS (3) | 4 | N. benthamiana | Agroinfiltration | Piatek et al. (2015) |
| dCas9-VP64 | PAP1 (3) | 7 | Arabidopsis | Transgenic assay | Lowder et al. (2015) |
| miR319 (3) | 7.5 | Arabidopsis | Transgenic assay | Lowder et al. (2015) | |
| FIS2 (3) | 400 | Arabidopsis | Transgenic assay | Lowder et al. (2015) | |
| Os03g01240 (2) | 2.1 | Rice | Protoplast assay | Lowder et al. (2018) | |
| Os04g39780 (2) | 1.1 | Rice | Protoplast assay | Lowder et al. (2018) | |
| Os11g35410 (1) | 2.2 | Rice | Protoplast assay | Lowder et al. (2018) | |
| pNOS::LUC reporter (3) | 2.3 | N. benthamiana | Agroinfiltration | Vazquez-Vilar et al. (2016) | |
| WRKY30 (1) | 2.1 | Arabidopsis | Transgenic assay | Li et al. (2017) | |
| RLP23 (1) | 0.9 | Arabidopsis | Transgenic assay | Li et al. (2017) | |
| CDG1 (1) | 4.3 | Arabidopsis | Protoplast assay | Li et al. (2017) | |
| GW7 (1) | 2.7 | Rice | Protoplast assay | Li et al. (2017) | |
| ER1 (1) | 0.3 | Rice | Protoplast assay | Li et al. (2017) | |
| dCas9-VP64 + MS2-p65-HSF1 (SAM) | AVP1 (2) | 5 | Arabidopsis | Transgenic assay | Park et al. (2017) |
| PAP1 (2) | 7 | Arabidopsis | Transgenic assay | Park et al. (2017) | |
| dCas9-VP64-EDLL | PAP1 (3) | 4 | Arabidopsis | Transgenic assay | Lowder et al. (2018) |
| FIS2 (3) | 3 | Arabidopsis | Transgenic assay | Lowder et al. (2018) | |
| dCas9-VP64 + MS2-EDLL | PAP1 (3) | 30 | Arabidopsis | Transgenic assay | Lowder et al. (2018) |
| FIS2 (3) | 30 | Arabidopsis | Transgenic assay | Lowder et al. (2018) | |
| dCas9-VP64 + MS2-VP64 (CRISPR-Act2.0) | PAP1 (3) | 45 | Arabidopsis | Transgenic assay | Lowder et al. (2018) |
| FIS2 (3) | 1500 | Arabidopsis | Transgenic assay | Lowder et al. (2018) | |
| ULC1 (3) | 40 | Arabidopsis | Transgenic assay | Lowder et al. (2018) | |
| miR319 (3) | 6 | Arabidopsis | Transgenic assay | Lowder et al. (2018) | |
| Os03g01240 (2) | 3 | Rice | Protoplast assay | Lowder et al. (2018) | |
| Os04g39780 (2) | 4 | Rice | Protoplast assay | Lowder et al. (2018) | |
| Os11g35410 (1) | 2.8 | Rice | Protoplast assay | Lowder et al. (2018) | |
| dCas9-4 × EE-2 × VP64 | pWRKY30::LUC reporter (1) | 12.6 | Arabidopsis | Protoplast assay | Li et al. (2017) |
| dCas9-6 × TAL-2 × VP64 (dCas9-TV) | WRKY30 (1) | 138.8 | Arabidopsis | Transgenic assay | Li et al. (2017) |
| RLP23 (1) | 32.3 | Arabidopsis | Transgenic assay | Li et al. (2017) | |
| CDG1 (1) | 92.2 | Arabidopsis | Protoplast assay | Li et al. (2017) | |
| GW7 (1) | 78.8 | Rice | Protoplast assay | Li et al. (2017) | |
| ER1 (1) | 62 | Rice | Protoplast assay | Li et al. (2017) | |
| dCpf1-TV | pWRKY30::LUC reporter (1) | 4.7 | Arabidopsis | Protoplast assay | Li et al. (2017) |
| dCas9-2 × GCN4 + scFv-sfGFP-VP64 (SunTag) | FWA (2) | 140 | Arabidopsis | Trangenic assay | Papikian et al. (2019) |
| EVD (2) | 4000 | Arabidopsis | Trangenic assay | Papikian et al. (2019) | |
| AP3 (2) | 350 | Arabidopsis | Trangenic assay | Papikian et al. (2019) | |
| CLV3 (2) | 130 | Arabidopsis | Trangenic assay | Papikian et al. (2019) |
pNOS nopaline synthase promoter, LUC the luciferase gene, EE the bipartite EDLL and ERF2m TADs
aOnly the number of sgRNAs for maximal gene activation is shown
bdCas9 gene activator-mediated expression level versus the basal level
Like the situation of transcriptional repression, different sgRNAs targeting the same promoter in combination with the same dCas9 gene activator could lead to gene activation to different levels (Piatek et al. 2015; Vazquez-Vilar et al. 2016; Li et al. 2017). However, the sgRNAs targeting the proximal promoter (i.e., ~ 200-bp window upstream of the TSS) tended to be more effective for the dCas9-mediated gene activation than those targeting the distal promoter (Piatek et al. 2015; Li et al. 2017; Papikian et al. 2019). Also, multiple sgRNAs targeting the same promoter could significantly enhance transcriptional activation compared to a single sgRNA (Piatek et al. 2015; Vazquez-Vilar et al. 2016; Park et al. 2017; Papikian et al. 2019). Of note, some target genes in plants appeared to be recalcitrant for gene activation even when using potent dCas9 gene activators such as dCas9-TV and the CRISPR-Act2.0 system (Li et al. 2017; Lowder et al. 2018). One possibility behind these observations is that dCas9 gene activators have to compete with endogenous transcriptional activator for binding to the promoter of an actively transcribed target gene. Indeed, genes with lower basal expression are prone to strong transcriptional activation by potent dCas9 gene activators, not only in plant cells but also in mammalian cells (Konermann et al. 2015; Li et al. 2017; Lowder et al. 2018), and weak gene activators (e.g., dCas9-VP64) can cause transcriptional repression rather than activation for the highly expressed genes (Li et al. 2017). Another possibility is that the transcript levels of some plant genes can be under tight regulation, where abnormal transcript upregulation will trigger negative feedback mechanism and post-transcriptional gene silencing to counteract the gene activation effect (Lowder et al. 2018).
Concluding remarks and future perspectives
Although the dCas-based gene repressors and activators have started to cut a figure in plant research, the overall development of these new tools in plants is still at the stage of proof-of-concept. This means that continuous efforts are still needed to further augment the efficiencies of these tools. We envision that three directions can be explored in the near future. First, considering the functional conservation of transcriptional machineries across eukaryotes, it would be inviting to borrow and test the cutting-edge dCas-TRD and dCas-TAD systems developed in non-plant organisms for applications in plants. Second, current dCas-based gene repressors and activators can be used under new experimental schemes. An interesting example is that a positive feedback circuit can be established by using the target gene promoter to drive the expression of the gene activator (Lowder et al. 2018), thereby leading to self-amplified gene activation. Third, it is worth testing whether combining recently developed dCas-derived epigenetic regulators with transcriptional regulators can synergistically boost the effects of gene repression or activation. For instance, potent dCas-TRDs can be used along with the dCas-based DNA methylation system (Pflueger et al. 2018; Papikian et al. 2019) to maximally shut down the target gene transcription, while dCas-TADs can be orchestrated with the dCas-derived DNA demethylation system (Gallego-Bartolomé et al. 2018) to robustly activate the target gene transcription. In addition to enhancing the potencies of dCas9-TRDs and dCas9-TADs in plants, it is also important to expand the arsenal of dCas gene repressors and activators using new orthogonal CRISPR/Cas systems (Nakade et al. 2017), which can greatly increase genome-wide targetable sites in promoters.
To date, the CRISPR/Cas system is still limited for cell/tissue-specific or conditional manipulation of gene activity (Zhang et al. 2018a). It is readily conceivable that both dCas-TRDs (e.g., dCas-3 × SRDX) and dCas-TADs (e.g., dCas9-TV) can be expressed under a cell/tissue-specific or chemically inducible promoter to confer spatial or temporal control of gene repression and activation, which can perfectly complement the CRISPR/Cas system for flexible gene manipulation in plants. Moreover, given that the potent dCas9 gene repressors or activators have been implemented with rationally designed genome-wide sgRNA libraries for genome-scale gene LOF or GOF screens in mammalian cells (Gilbert et al. 2014; Konermann et al. 2015; Ewen-Campen et al. 2017; Sanson et al. 2018; Gasperini et al. 2019), it will be just a matter of time to see such powerful screens being conducted in plant research.
Acknowledgements
The work in the laboratory of JF Li is supported by the National Natural Science Foundation of China (Grant Nos. 31570276 and 31770295). We apologize to those whose work could not be cited due to the space constraint.
References
- Abe K, Ichikawa H. Gene overexpression resources in cereals for functional genomics and discovery of useful genes. Front Plant Sci. 2016;7:1359. doi: 10.3389/fpls.2016.01359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adli M. The CRISPR tool kit for genome editing and beyond. Nat Commun. 2018;9:1911. doi: 10.1038/s41467-018-04252-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braun CJ, Bruno PM, Horlbeck MA, Gilbert LA, Weissman JS, Hemann MT. Versatile in vivo regulation of tumor phenotypes by dCas9-mediated transcriptional perturbation. Proc Natl Acad Sci USA. 2016;113:E3892–E3900. doi: 10.1073/pnas.1600582113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chavez A, Scheiman J, Vora S, Pruitt BW, Tuttle M, Iyer EPR, Lin S, Kiani S, Guzman CD, Wiegand DJ, Ter-Ovanesyan D, Braff JL, Davidsohn N, Housden BE, Perrimon N, Weiss R, Aach J, Collins JJ, Church GM. Highly efficient Cas9-mediated transcriptional programming. Nat Methods. 2015;12:326–328. doi: 10.1038/nmeth.3312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chavez A, Tuttle M, Pruitt BW, Ewen-Campen B, Chari R, Ter-Ovanesyan D, Haque SJ, Cecchi RJ, Kowal EJK, Buchthal J, Housden BE, Perrimon N, Collins JJ, Church GM. Comparison of Cas9 activators in multiple species. Nat Methods. 2016;13:563–567. doi: 10.1038/nmeth.3871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen K, Wang Y, Zhang R, Zhang H, Gao C. CRISPR/Cas genome editing and precision plant breeding in agriculture. Annu Rev Plant Biol. 2019;70:667–697. doi: 10.1146/annurev-arplant-050718-100049. [DOI] [PubMed] [Google Scholar]
- Cheng AW, Wang H, Yang H, Shi L, Katz Y, Theunissen TW, Rangarajan S, Shivalila CS, Dadon DB, Jaenisch R. Multiplexed activation of endogenous genes by CRISPR-on, an RNA-guided transcriptional activator system. Cell Res. 2013;23:1163–1171. doi: 10.1038/cr.2013.122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cullot G, Boutin J, Toutain J, Prat F, Pennamen P, Rooryck C, Teichmann M, Rousseau E, Lamrissi-Garcia I, Guyonnet-Duperat V, Bibeyran A, Lalanne M, Prouzet-Mauléon V, Turcq B, Ged C, Blouin JM, Richard E, Dabernat S, Moreau-Gaudry F, Bedel A. CRISPR-Cas9 genome editing induces megabase-scale chromosomal truncations. Nat Commun. 2019;10:1136. doi: 10.1038/s41467-019-09006-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ewen-Campen B, Yang-Zhou D, Fernandes VR, González DR, Liu LP, Tao R, Ren X, Sun J, Hu Y, Zirin J, Mohr SE, Ni JQ, Perrimon N. Optimized strategy for in vivo Cas9-activation in Drosophila. Proc Natl Acad Sci USA. 2017;114:9409–9414. doi: 10.1073/pnas.1707635114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farzadfard F, Perli SD, Lu TK. Tunable and multifunctional eukaryotic transcription factors based on CRISPR/Cas. ACS Synth Biol. 2013;2:604–613. doi: 10.1021/sb400081r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fonfara I, Richter H, Bratovič M, Le Rhun A, Charpentier E. The CRISPR-associated DNA-cleaving enzyme Cpf1 also processes precursor CRISPR RNA. Nature. 2016;532:517–521. doi: 10.1038/nature17945. [DOI] [PubMed] [Google Scholar]
- Gallego-Bartolomé J, Gardiner J, Liu W, Papikian A, Ghoshal B, Kuo HY, Zhao JM, Segal DJ, Jacobsen SE. Targeted DNA demethylation of the Arabidopsis genome using the human TET1 catalytic domain. Proc Natl Acad Sci USA. 2018;115:E2125–E2134. doi: 10.1073/pnas.1716945115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gasperini M, Hill AJ, McFaline-Figueroa JL, Martin B, Kim S, Zhang MD, Jackson D, Leith A, Schreiber J, Nobel WS, Trapnell C, Ahituv N, Shendure J. A genome-wide framework for mapping gene regulation via cellular genetic screens. Cell. 2019;176:377–390. doi: 10.1016/j.cell.2018.11.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilbert LA, Larson MH, Morsut L, Liu Z, Brar GA, Torres SE, Stern-Ginossar N, Brandman O, Whitehead EH, Doudna JA, Lim WA, Weissman JS, Qi LS. CRISPR-mediated modular RNA-guided regulation of transcription in eukaryotes. Cell. 2013;154:442–451. doi: 10.1016/j.cell.2013.06.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilbert LA, Horlbeck MA, Adamson B, Villalta JE, Chen Y, Whitehead EH, Guimaraes C, Panning B, Ploegh HL, Bassik MC, Qi LS, Kampmann M, Weissman JS. Genome-scale CRISPR-mediated control of gene repression and activation. Cell. 2014;159:647–661. doi: 10.1016/j.cell.2014.09.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karve R, Liu W, Willet SG, Torri KU, Shpak ED. The presence of multiple introns is essential for ERECTA expression in Arabidopsis. RNA. 2011;17:1907–1921. doi: 10.1261/rna.2825811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleinstiver BP, Sousa AA, Walton RT, Tak YE, Hsu JY, Clement K, Welch MM, Horng JE, Malagon-Lopez J, Scarfò I, Maus MV, Pinello L, Aryee MJ, Joung JK. Engineered CRISPR–Cas12a variants with increased activities and improved targeting ranges for gene, epigenetic and base editing. Nat Biotechnol. 2019;37:276–282. doi: 10.1038/s41587-018-0011-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konermann S, Brigham MD, Trevino AE, Joung J, Abudayyeh OO, Barcena C, Hsu PD, Habib N, Gootenberg JS, Nishimasu H, Nureki O, Zhang F. Genome-scale transcriptional activation by an engineered CRISPR-Cas9 complex. Nature. 2015;517:583–588. doi: 10.1038/nature14136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kosicki M, Tomberg K, Bradley A. Repair of double-strand breaks induced by CRISPR-Cas9 leads to large deletions and complex rearrangements. Nat Biotechnol. 2018;36:765–771. doi: 10.1038/nbt.4192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawhorn IE, Ferreira JP, Wang CL. Evaluation of sgRNA target sites for CRISPR-mediated repression of TP53. PLoS One. 2014;9:e113232. doi: 10.1371/journal.pone.0113232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z, Zhang D, Xiong X, Yan B, Xie W, Sheen J, Li JF. A potent Cas9-derived gene activator for plant and mammalian cells. Nat Plants. 2017;3:930–936. doi: 10.1038/s41477-017-0046-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Han J, Chen Z, Wu H, Dong H, Nie G. Engineering cell signaling using tunable CRISPR-Cpf1-based transcription factors. Nat Commun. 2017;8:2095. doi: 10.1038/s41467-017-02265-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowder LG, Zhang D, Baltes NJ, Paul JW, Tang X, Zheng X, Voytas DF, Hsieh TF, Zhang Y, Qi Y. A CRISPR/Cas9 toolbox for multiplexed plant genome editing and transcriptional regulation. Plant Physiol. 2015;169:971–985. doi: 10.1104/pp.15.00636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowder LG, Zhou J, Zhang Y, Malzahn A, Zhong Z, Hsieh TF, Voytas DF, Zhang Y, Qi Y. Robust transcriptional activation in plants using multiplexed CRISPR-Act2.0 and mTALE-Act systems. Mol Plant. 2018;11:245–256. doi: 10.1016/j.molp.2017.11.010. [DOI] [PubMed] [Google Scholar]
- Maeder ML, Linder SJ, Cascio VM, Fu Y, Ho QH, Joung JK. CRISPR RNA-guided activation of endogenous human genes. Nat Methods. 2013;10:977–979. doi: 10.1038/nmeth.2598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mali P, Aach J, Stranges PB, Esvelt KM, Moosburner M, Kosuri S, Yang L, Church GM. CAS9 transcriptional activators for target specificity screening and paired nickases for cooperative genome engineering. Nat Biotechnol. 2013;31:833–838. doi: 10.1038/nbt.2675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakade S, Yamamoto T, Sakuma T. Cas9, Cpf1 and C2c1/2/3-what’s next? Bioengineered. 2017;8:265–273. doi: 10.1080/21655979.2017.1282018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papikian A, Liu W, Gallego-Bartolomé J, Jacobsen SE. Site-specific manipulation of Arabidopsis loci using CRISPR-Cas9 SunTag systems. Nat Commun. 2019;10:729. doi: 10.1038/s41467-019-08736-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park JJ, Dempewolf E, Zhang W, Wang ZY. RNA-guided transcriptional activation via CRISPR/dCas9 mimics overexpression phenotypes in Arabidopsis. PLoS One. 2017;12:e0179410. doi: 10.1371/journal.pone.0179410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perez-Pinera P, Kocak DD, Vockley CM, Adler AF, Kabadi AM, Polstein LR, Thakore PI, Glass KA, Ousterout DG, Leong KW, Guilak F, Crawford GE, Reddy TE, Gersbach CA. RNA-guided gene activation by CRISPR-Cas9-based transcription factors. Nat Methods. 2013;10:973–976. doi: 10.1038/nmeth.2600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peters JM, Colavin A, Shi H, Czarny TL, Larson MH, Wong S, Hawkins JS, Lu CHS, Koo BM, Marta E, Shiver AL, Whitehead EH, Weissman JS, Brown ED, Qi LS, Huang KC, Gross CA. A comprehensive, CRISPR-based functional analysis of essential genes in bacteria. Cell. 2016;165:1493–1506. doi: 10.1016/j.cell.2016.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petolino JF, Davies JP. Designed transcriptional regulators for trait development. Plant Sci. 2013;201:128–136. doi: 10.1016/j.plantsci.2012.12.006. [DOI] [PubMed] [Google Scholar]
- Pflueger C, Tan D, Swain T, Nguyen T, Pflueger J, Nefzger C, Polo JM, Ford E, Lister R. A modular dCas9-SunTag DNMT3A epigenome editing system overcomes pervasive off-target activity of direct fusion dCas9-DNMT3A constructs. Genome Res. 2018;28:1193–1206. doi: 10.1101/gr.233049.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piatek A, Ali Z, Baazim H, Li L, Abulfaraj A, Al-Shareef S, Aouida M, Mahfouz MM. RNA-guided transcriptional regulation in planta via synthetic dCas9-based transcription factors. Plant Biotechnol J. 2015;13:578–589. doi: 10.1111/pbi.12284. [DOI] [PubMed] [Google Scholar]
- Qi LS, Larson MH, Gilbert LA, Doudna JA, Weissman JS, Arkin AP, Lim WA. Repurposing CRISPR as an RNA-guided platform for sequence-specific control of gene expression. Cell. 2013;152:1173–1183. doi: 10.1016/j.cell.2013.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rossi A, Kontarakis Z, Gerri C, Nolte H, Holer S, Kruger M, Stainier DY. Genetic compensation induced by deleterious mutations but not gene knockdowns. Nature. 2015;524:230–233. doi: 10.1038/nature14580. [DOI] [PubMed] [Google Scholar]
- Sanson KR, Hanna RE, Hegde M, Donovan KF, Strand C, Sullender ME, Vaimberg EW, Goodale A, Root DE, Piccioni F, Doench JG. Optimized libraries for CRIPSR-Cas9 genetic screens with multiple modalities. Nat Commun. 2018;9:5416. doi: 10.1038/s41467-018-07901-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tak YE, Kleinstiver BP, Nuñez JK, Hsu JY, Horng JE, Gong J, Weissman JS, Joung JK. Inducible and multiplex gene regulation using CRISPR-Cpf1-based transcription factors. Nat Methods. 2017;14:1163–1166. doi: 10.1038/nmeth.4483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanenbaum ME, Gilbert LA, Qi LS, Weissman JS, Vale RD. A protein-tagging system for signal amplification in gene expression and fluorescence imaging. Cell. 2014;159:635–646. doi: 10.1016/j.cell.2014.09.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang X, Lowder LG, Zhang T, Malzahn AA, Zheng X, Voytas DF, Zhong Z, Chen Y, Ren Q, Li Q, Kirkland ER, Zhang Y, Qi Y. A CRISPR-Cpf1 system for efficient genome editing and transcriptional repression in plants. Nat Plants. 2017;3:17018. doi: 10.1038/nplants.2017.18. [DOI] [PubMed] [Google Scholar]
- Vazquez-Vilar M, Bernabé-Orts JM, Fernandez-Del-Carmen A, Ziarsolo P, Blanca J, Granell A, Orzaez D. A modular toolbox for gRNA-Cas9 genome engineering in plants based on the GoldenBraid standard. Plant Methods. 2016;12:10. doi: 10.1186/s13007-016-0101-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, La Russa M, Qi LS. CRISPR/Cas9 in genome editing and beyond. Annu Rev Biochem. 2016;85:227–264. doi: 10.1146/annurev-biochem-060815-014607. [DOI] [PubMed] [Google Scholar]
- Xu X, Qi LS. A CRISPR-dCas toolbox for genetic engineering and synthetic biology. J Mol Biol. 2019;431:34–47. doi: 10.1016/j.jmb.2018.06.037. [DOI] [PubMed] [Google Scholar]
- Zalatan JG, Lee ME, Almeida R, Gilbert LA, Whitehead EH, La Russa M, Tsai JC, Weissman JS, Dueber JE, Qi LS, Lim WA. Engineering complex synthetic transcriptional programs with CRISPR RNA scaffolds. Cell. 2015;160:339–350. doi: 10.1016/j.cell.2014.11.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zetsche B, Gootenberg JS, Abudayyeh OO, Slaymaker IM, Makarova KS, Essletzbichler P, Volz SE, Joung J, van der Oost J, Regev A, Koonin EV, Zhang F. Cpf1 is a single RNA-guided endonuclease of a class 2 CRISPR-Cas system. Cell. 2015;163:759–771. doi: 10.1016/j.cell.2015.09.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zetsche B, Heidenreich M, Mohanraju P, Fedorova I, Kneppers J, DeGennaro EM, Winblad N, Choudhury SR, Abudayyeh OO, Gootenberg JS, Wu WY, Scott DA, Severinov K, van der Oost J, Zhang F. Multiplex gene editing by CRISPR-Cpf1 using a single crRNA array. Nat Biotechnol. 2017;35:31–34. doi: 10.1038/nbt.3737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang D, Li Z, Li JF. Targeted gene manipulation in plants using the CRISPR/Cas technology. J Genet Genom. 2016;43:251–262. doi: 10.1016/j.jgg.2016.03.001. [DOI] [PubMed] [Google Scholar]
- Zhang N, Zhang D, Chen SL, Gong BQ, Guo Y, Xu L, Zhang XN, Li JF. Engineering artificial microRNAs for multiplex gene silencing and simplified transgenic screen. Plant Physiol. 2018;178:989–1001. doi: 10.1104/pp.18.00828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Wang W, Shan L, Han L, Ma S, Zhang Y, Hao B, Lin Y, Rong Z. Gene activation in human cells using CRISPR/Cpf1-p300 and CRISPR/Cpf1-SunTag systems. Protein Cell. 2018;9:380–383. doi: 10.1007/s13238-017-0491-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou H, Liu J, Zhou C, Gao N, Rao Z, Li H, Hu X, Li C, Yao X, Shen X, Sun Y, Wei Y, Liu F, Ying W, Zhang J, Tang C, Zhang X, Xu H, Shi L, Cheng L, Huang P, Yang H. In vivo simultaneous transcriptional activation of multiple genes in the brain using CRISPR-dCas9-activator transgenic mice. Nat Neurosci. 2018;21:440–446. doi: 10.1038/s41593-017-0060-6. [DOI] [PubMed] [Google Scholar]
- Zorrilla-López U, Masip G, Arjó G, Bai C, Banakar R, Bassie L, Berman J, Farré G, Miralpeix B, Pérez-Massot E, Sabalza M, Sanahuja G, Vamvaka E, Twyman RM, Christou P, Zhu C, Capell T. Engineering metabolic pathways in plants by multigene transformation. Int J Dev Biol. 2013;57:565–576. doi: 10.1387/ijdb.130162pc. [DOI] [PubMed] [Google Scholar]