Abstract
Amorpha-4,11-diene synthase (ADS) catalyzes the first committed step in the artemisinin biosynthetic pathway, which is the first catalytic reaction enzymatically and genetically characterized in artemisinin biosynthesis. The advent of ADS in Artemisia annua is considered crucial for the emergence of the specialized artemisinin biosynthetic pathway in the species. Microbial production of amorpha-4,11-diene is a breakthrough in metabolic engineering and synthetic biology. Recently, numerous new techniques have been used in ADS engineering; for example, assessing the substrate promiscuity of ADS to chemoenzymatically produce artemisinin. In this review, we discuss the discovery and catalytic mechanism of ADS, its application in metabolic engineering and synthetic biology, as well as the role of sesquiterpene synthases in the evolutionary origin of artemisinin.
Keywords: Amorpha-4,11-diene Synthase; Artemisinin; Catalytic mechanism; Metabolic engineering; Synthetic biology
Introduction
Artemisinin is a well-known antimalarial drug against chloroquine-resistant strains of Plasmodium falciparum (Chen and Xu 2016; Wang et al. 2019). In 2002, the World Health Organization recommended artemisinin-based combinatorial therapies as the first-line treatment for uncomplicated malaria. Artemisia annua L. is the only natural source of artemisinin, which is biosynthesized and accumulated in the glandular trichome cells of the plant. A recent study has proved that artemisinin is also produced in the non-glandular trichome cells (Judd et al. 2019). The low content of artemisinin in A. annua (0.1–1.0% of dry weight) makes its plant-derived production insufficient for global requirements (Ikram and Simonsen 2017). Recent advances in metabolic engineering and synthetic biology have enabled higher yield of artemisinin in microbial or plant heterologous hosts by engineering the artemisinin pathway genes in these hosts. However, complete understanding of artemisinin biosynthesis is still required (Fig. 1).
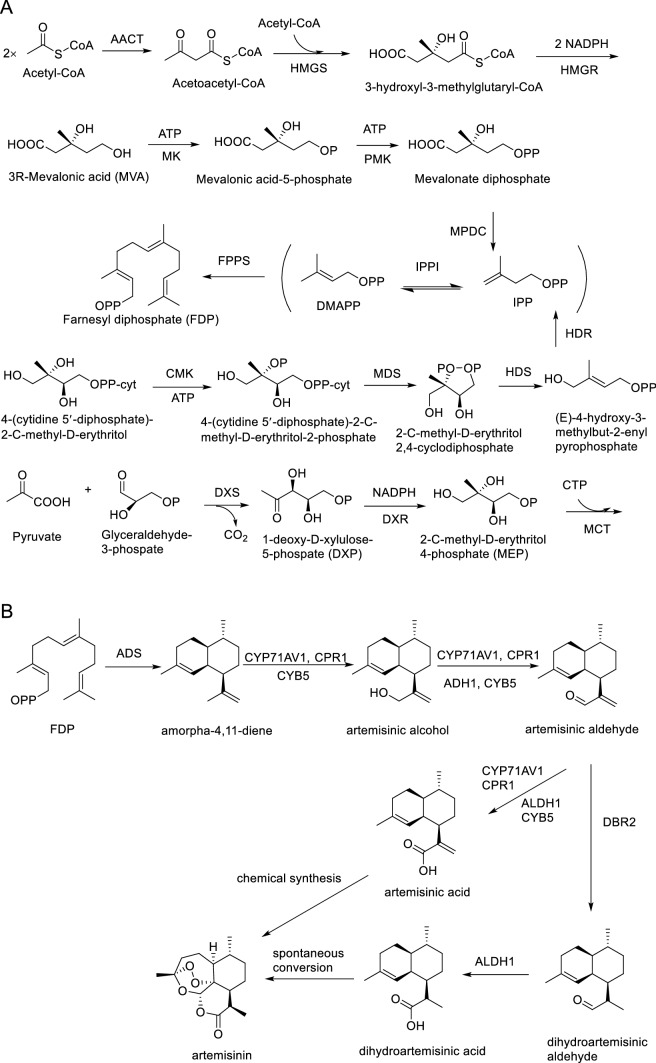
Fig. 1.
Proposed artemisinin biosynthetic pathway in A. annua. A. Carbon flow from MVA (in the cytosol) and MEP (in the chloroplast) pathways to form FDP. AACT, acetyl-coenzyme transferase; HMGS and HMGR, 3-hydroxy-3-methylglutaryl-CoA synthase and reductase; MK, 3R-mevalonic acid kinase; PMK, mevalonic acid-5-phosphate kinase; MPDC, mevalonate 5-pyrophosphate decarboxylase; DXS and DXR, 1-deoxy-D-xylulose 5-phosphate synthase and reductase; MCT, 2-C-methyl-D-erythritol 2,4-cyclodiphosphate synthase; CMK, 4-diphosphocytidyl-2-C-methyl-D-erythritol kinase; MDS, 2-C-methyl-D-erythritol 2,4-cyclodiphosphate synthase; HDS and HDR, 4-hydroxy-3-methylbut-2-enyl pyrophosphate synthase and reductase; IPPI, isopentenyl pyrophosphate isomerase; FPPS, farnesyl pyrophosphate synthase. B. Production of artemisinin in planta and by biological methods. ADS, armorpha-4,11-diene synthase; CYP71AV1, cytochrome P450 monooxygenase; CPR1, cytochrome P450 reductase 1; CYB5, cytochrome b5 monooxygenase; ALDH1, aldehyde dehydrogenase; DBR2, artemisinic aldehyde delta-11(13)-double bond reductase. Enzymes marked in red improved the efficiency of different oxidation steps in yeast (Paddon et al. 2013)
The first step toward the elucidation of the artemisinin biosynthetic pathway began in 1999, when amorpha-4,11-diene synthase (ADS) was purified from A. annua and functionally characterized (Bouwmeester et al. 1999). Since then, studies have been extensively conducted to understand the artemisinin biosynthetic pathway and its evolutionary origin and to develop metabolic engineering and biosynthetic methods for its production. Notably, stable and high-yielding production of artemisinic acid was established in Saccharomyces cerevisiae and chemical conversion of artemisinic acid effectively yielded artemisinin (Paddon and Keasling 2014). Recently, the production of amorpha-4,11-diene has become a touchstone technique because of its important role in metabolic engineering and synthetic biology (Choi et al. 2016; Orsi et al. 2020; Redding-Johanson et al. 2011; Shukal et al. 2019; Wang et al. 2013; Yuan and Ching 2014, 2015a, 2015b, 2016). Furthermore, the substrate promiscuity of ADS was used to develop a chemoenzymatic strategy for artemisinin production (Demiray et al. 2017). The importance of ADS still attracts considerable research attention.
Because advances in the investigation of artemisinin biosynthesis as well as in metabolic engineering or synthetic biology for artemisinin production have been previously reviewed (Farhi et al. 2013; Ikram and Simonsen 2017; Kung et al. 2018; Xie et al. 2016), herein, we mainly focus on the discovery, catalytic mechanism, and engineering of ADS, as well as the impact of the emergence of ADS in the evolutionary origin of artemisinin biosynthetic pathway in this review.
Characterization, catalytic mechanism, and evolutionary origin of ADS
Structurally, artemisinin is an endoperoxide sesquiterpene lactone in which the basic carbon skeleton is constructed from farnesyl diphosphate (FDP) by a sesquiterpene synthase. In 1999, a native ADS protein was purified from A. annua and functionally characterized, suggesting that ADS may catalyze the first rate-determining step in artemisinin biosynthesis (Bouwmeester et al. 1999). Several groups have isolated terpene synthase genes from A. annua and bacterially expressed them in Escherichia coli, resulting in the identification of ADS (Chang et al. 2000; Mercke et al. 2000; Wallaart et al. 2001) and other terpene synthases, such as (3R)-linalool synthase (Jia et al. 1999), 8-epicedrol synthase (Hua and Matsuda 1999; Mercke et al. 1999), and β-caryophyllene synthase (Cai et al. 2002), from A. annua. Overexpression and downregulation of ADS in A. annua plants resulted in increased and reduced artemisinin content in planta, respectively, providing direct genetic evidence of the involvement of ADS in artemisinin biosynthesis (Alam and Abdin 2011; Catania et al. 2018; Han et al. 2016; Ma et al. 2009, 2015). These studies have paved the way for further investigation of artemisinin biosynthesis.
ADS is a class I terpenoid synthase (TPS) belonging to the plant TPS-a subgroup (Salmon et al. 2015). It contains conserved DDXXD (DDTYD) and NSE/DTE (NDLMTHKAE) ion-binding motifs (Chang et al. 2000; Mercke et al. 2000). Accordingly, the effects of divalent metal ions, such as Mg2+, Mn2+, Co2+, Ni2+, Zn2+, and Cu2+, on enzyme activity were tested. No activity was reported when Ni2+, Zn2+, and Cu2+ were used, and the enzyme activity was lower with Mn2+ and Co2+ than with Mg2+ (Picaud et al. 2005). ADS showed 36% and 41% amino acid sequence identity with tobacco 5-epi-aristolochene synthase (TEAS) and cotton ( +)-δ-cadinene synthase, respectively (Chang et al. 2000). Similar to the crystal structure of TEAS, ADS also has an N-terminal glycosyl hydrolase domain and a C-terminal catalytic domain (Mercke et al. 2000).
The catalytic mechanism of ADS has attracted continued interest. Generally, the catalytic mechanism of a class I TPS involves the initial ionization of the substrate diphosphate group, electrophilic cyclization, deprotonation, or capture of a nucleophile, and finally, the release of neutral products (Christianson 2017). Methods used to investigate mechanistic details involve labeled substrates and mutant enzymes and include X-ray crystallography and quantum chemical study (Faraldos et al. 2012). A catalytic model of ADS was suggested by the observation of TEAS, in which FDP binding placed Phe525 next to Trp271 to form an extended aromatic box, and the carbocation intermediates were stabilized by the nucleophilicity of the Trp271 indole ring. The ionization of FDP was facilitated by the positive charges of Arg262 and Arg440 with the help of divalent metal cations coordinated by the DDXXD motif. Other motifs including the Arg10–Pro11 pair and the Asp444–Tyr520–Asp524 triad were also conserved in ADS, but their function was not experimentally investigated (Chang et al. 2000; Mercke et al. 2000).
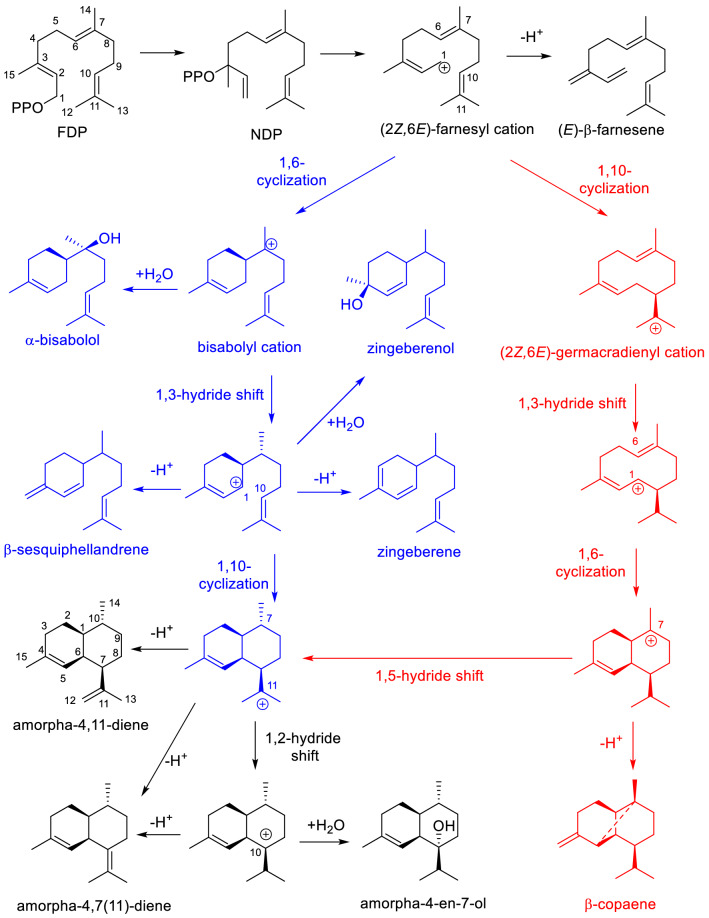
The bicyclic structure of amorpha-4,11-diene is formed by an initial 1,6 or 1,10 cyclization of FDP involving a bisabolyl or (2Z,6E)-germacradienyl cation, respectively (Chang et al. 2000). The recombinant ADS expressed in E. coli produces the by-products β-sesquiphellandrene, α-bisabolol, zingiberene, and zingibernol, supporting the involvement of a bisabolyl cation in the cyclization mechanism (Mercke et al. 2000; Picaud et al. 2005). By using deuterium-labeled FDP (labeled at C-1) as a chemical probe, two study groups independently found that H-1 migrated to the original C-7 of FDP (C-10 of amorpha-4,11-diene). They deduced the occurrence of the initial 1,6 cyclization because the initial 1,10-ring closure led to the shift of H-1 to C-11 (Kim et al. 2006; Picaud et al. 2006). However, a subsequent 1,5-hydride shift could also allow the migrating H-1 to relocate to the original C-7 of FDP (Fig. 2), indicating that these two cyclization mechanisms cannot be determined by using labeled substrates. Indeed, a quantum chemical study concluded that the 1,5-hydride shift is feasible, supporting the occurrence of the initial 1,10 cyclization for the catalytic mechanism of ADS (Hong and Tantillo 2010). Recently, the ADS Q518L variant was reported to generate the initial 1,10 cyclization product β-copaene in addition to amorpha-4,11-diene (Fig. 2), implying that ADS uses both initial 1,6 and 1,10 cyclization mechanisms to produce amorpha-4,11-diene (Huang et al. 2021).
Fig. 2.
Proposed cyclization mechanisms for the formation of amorpha-4,11-diene by ADS. Reactions starting from the initial 1,6-ring closure of FDP and the generation of bisabolene-type by-products are highlighted in blue. Steps proceeding through the initial 1,10-ring closure and corresponding by-products are marked in red
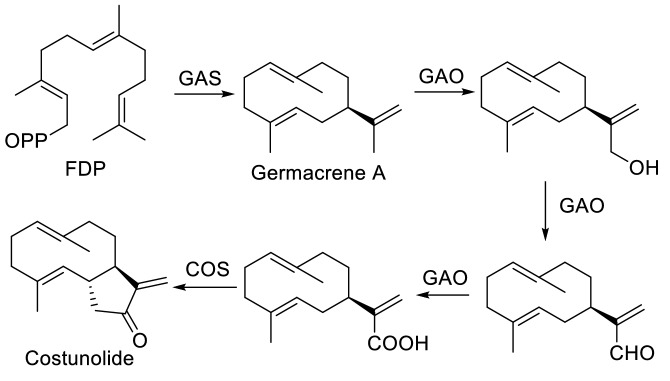
New terpenoid biosynthetic pathways usually initiate from the emergence of functional TPS/CYP gene pairs (Boutanaev et al. 2015), but the sequence of their occurrence is not fixed. Regarding the evolutionary emergence of the artemisinin biosynthetic pathway in A. annua, Nguyen et al. (2010) suggested that the occurrence of ADS is a dominant event mainly shaped by an Y374L mutation in its progenitor (Salmon et al. 2015). They found that in all major subfamilies of Asteraceae, germacrene A oxidase (GAO) is conserved when producing germacrene A acid, the key intermediate of the Asteraceae sesquiterpene lactone biosynthetic pathway (Fig. 3). Remarkably, GAO uses amorpha-4,11-diene as the substrate to produce artemisinic acid, whereas CYP71AV1 or amorpha-4,11-diene oxidase (AMO) is inactive to germacrene A. Thus, they hypothesized that the advent of ADS activity in A. annua removed GAO from the Asteraceae sesquiterpene lactone biosynthetic pathway and eventually replaced it with AMO. In addition, sesquiterpene lactones derived from germacrene A are absent in A. annua but are present in other Artemisia species (Bertea et al. 2006). The promiscuity of GAO and the specificity of AMO were further confirmed by the ability of GAO to oxidize several sesquiterpenes, including germacrene D, 5-epi-aristolochene, valencene, δ-cadinene, α- and δ-guaienes, and valerenadiene to corresponding sesquiterpene acids, whereas AMO showed negligible activities (Nguyen et al. 2019). Similarly, orthologs of CYP71AV1 (94% amino acid identity) from the Artemisia genus (e.g., A. afra and A. absinthium) converted amorpha-4,11-diene to artemisinic alcohol (Komori et al. 2013). However, ADS homologs from other Artemisia species (e.g., A. absinthium, A. kurramensis, and A. maritima) did not produce amorpha-4,11-diene (Muangphrom et al. 2016); a homologous synthase from A. maritima produced amorphen-4,11-ol (Muangphrom et al. 2018). Although further investigation is required, accumulated data is in favor of the hypothesis that artemisinin production in A. annua is attributed to the emergence of ADS.
Fig. 3.
Asteraceae sesquiterpene lactone biosynthetic pathway. GAS, germacrene A synthase; COS, costunolide synthase
Metabolic engineering of ADS
Microbial production of artemisinin is a milestone in the development of synthetic biology (Kung et al. 2018), which initiated from the expression of ADS in engineered E. coli (Martin et al. 2003). To increase amorpha-4,11-diene production, several improvements have been made (Table 1), including the development of a two-phase partitioning bioreactor (Newman et al. 2006); identifying and enhancing the production of rate-limiting enzymes (Fig. 1), such as MK, PMK, HMGS, HMGR, and ADS (Anthony et al. 2009; Ma et al. 2011; Redding-Johanson et al. 2011; Tsuruta et al. 2009); increasing the flux of 1-deoxy-D-xylulose-5-phosphate by engineering the phosphoenolpyruvate-dependent phosphotransferase system (PTS; Zhang et al. 2013, 2015); systematically optimizing transcription and translation in E. coli (Shukal et al. 2019); and constructing multienzyme complexes in E. coli (Wei et al. 2020). Among these improvements, assembling and modulating efflux pumps in E. coli are vital because the accumulation of antimicrobial amorpha-4,11-diene in E. coli inhibited cell growth (Zhang et al. 2016; Wang et al. 2013). Collectively, the highest production of amorpha-4,11-diene was 27.4 g/L (Tsuruta et al. 2009).
Table 1.
Biosynthetic and metabolic engineering approaches to produce amorpha-4,11-diene
| Construction | Organism | Amorphadiene | References |
|---|---|---|---|
| Engineering codon-optimized ADS and mevalonate pathway from S. cerevisiae in E. coli | E. coli | 24 mg/L | Martin et al. (2003) |
| Enhancing production of rate-limiting enzymes MK and ADS | E. coli | 300 mg/L | Anthony et al. (2009) |
| Introducing more active HMG-CoA synthase and HMG-CoA reductase | E. coli | 27.4 g/L | Tsuruta et al. (2009) |
| Enhancing production of rate-limiting enzymes MK and PMK | E. coli | 500 mg/L | Redding-Johanson et al. (2011) |
| Introducing more active HMG-CoA reductase | E. coli | 700 mg/L | Ma et al. (2011) |
| Engineering efflux pumps | E. coli | 363 mg/L | Wang et al. (2013) |
| Deleting PTS | E. coli | 182 mg/L | Zhang et al. (2013) |
| Engineering PTS and GGS | E. coli | 201 mg/L | Zhang et al. (2015) |
| Efflux transporter engineering | E. coli | 150 mg/L | Zhang et al. (2016) |
| Systematically optimizing transcription and translation in E. coli | E. coli | 30 g/L | Shukal et al. (2019) |
| Plasmid integration of ADS into yeast | S. cerevisiae | 0.6 mg/L | Lindahl et al. (2006) |
| Inserting ADS into yeast genome | S. cerevisiae | 0.1 mg/L | Lindahl et al. (2006) |
| Overexpressing tHMGR, ERG20, and upc2-1, and downregulating ERG9 | S. cerevisiae | 153 mg/L | Ro et al. (2006) |
| Increasing copy number of ADS in yeast | S. cerevisiae | 781 mg/L | Ro et al. (2008) |
| Engineering codon-optimized ADS in S. cerevisiae | S. cerevisiae | 123 mg/L | Kong et al. (2009) |
| Integrating HMG1, FDPS, and ADS into yeast mitochondria | S. cerevisiae | 20 mg/L | Farhi et al. (2011a) |
| Overexpressing every enzyme of MVA pathway | S. cerevisiae | 41 g/L | Westfall et al. (2012) |
| Downregulating ERG9 and fusing ADS with FPPS | S. cerevisiae | 25 mg/L | Baadhe et al. (2013) |
| Combinatorial genome integration of MVA pathway genes in yeast | S. cerevisiae | 64 mg/L | Yuan and Ching (2014) |
| Knockout genes outside isoprenoid pathway but improving isoprenoid fluxes | S. cerevisiae | 54.5 mg/L | Sun et al. (2014) |
| Dynamic control of the expression of ERG9 | S. cerevisiae | 350 mg/L | Yuan and Ching (2015a) |
| Assembling MVA pathway genes into yeast chromosomes and reducing ERG9 expression | S. cerevisiae | 500 mg/L | Yuan and Ching (2015b) |
| Integrating MVA pathway genes and ADS into yeast mitochondria | S. cerevisiae | 427 mg/L | Yuan and Ching (2016) |
| Expressing ADS in N. tabacum | N. tabacum | 1.7 ng/g FW | Wallaart et al. (2001) |
| Targeting FPS and ADS in plastids | N. tabacum | 25 μg/g FW | Wu et al. (2006) |
| Introducing tHMGR, FPS, and ADS into N. benthamiana | N. benthamiana | 6.2 μg/g FW | Van Herpen et al. (2010) |
| Targeting FPS and ADS in plastids | N. tabacum | 4 μg/g FW | Zhang et al. (2011) |
| Introducing tHMGR from yeast and ADS, CPR, CYP71AV1, and DBR2 into N. tabacum | N. tabacum | 827 ng/g FW | Farhi et al. (2011b) |
| Introducing whole artemisinin pathway genes into N. tabacum chloroplasts | N. tabacum | Fuentes et al. (2016) | |
| Engineering MVA and artemisinin pathway genes in N. tabacum chloroplasts, nuclei, and mitochondria | N. tabacum | 60 μg/g DW | Malhotra et al. (2016) |
| Engineering ADS in P. patens | Physcomitrella patens | 200 mg/L | Ikram et al. (2017) |
| Engineering dxs, idi, and ADS in B. subtilis | B. subtilis | 20 mg/L | Zhou et al. (2013) |
| Chromosomally integrated GFP-ADS, FPPS, and a plasmid-encoded synthetic operon carrying MEP pathway genes | B. subtilis | 416 mg/L | Pramastya et al. (2020) |
| Engineering MEP pathway genes and ADS in cyanobacteria | Synechococcus elongatus PCC 7942 | 19.8 mg/L | Choi et al. (2016) |
| Engineering codon-optimized ADS in S. avermitilis | S. avermitilis | 30 μg/L | Komatsu et al. (2010) |
| Engineering ADS in R. sphaeroides | R. sphaeroides | 56.4 mg/L | Orsi et al. (2020) |
Although the production of amorpha-4,11-diene in engineered E. coli was successful, several enzymes are necessary to complete the transformation of amorpha-4,11-diene to artemisinin, in which the cytochrome P450 CYP71AV1 catalyzes the first oxidation reaction (Teoh et al. 2006). N-terminal modified CYP71AV1 was heterologously expressed in E. coli (Chang et al. 2007); however, the functional expression of plant P450 in E. coli is extremely challenging because intracellular membrane structures are absent.
Yeast is considered a reliable host for the expression of plant P450. To facilitate the expression of CYP71AV1, two groups independently engineered yeast to produce amorpha-4,11-diene in 2006. Lindahl et al. (2006) expressed ADS in yeast using plasmids and chromosomal integration resulting in 0.6 mg/L and 0.1 mg/L amorpha-4,11-diene production, respectively, whereas Ro et al. (2006) obtained 153 mg/L of amorpha-4,11-diene by introducing ADS into yeast that was simultaneously engineered by the overexpression of truncated HMGR, FPPS (ERG20), and an activated allele of the UPC2 transcription factor (upc2-1) as well as the downregulation of the expression of squalene synthase (ERG9). Since then, other metabolic engineering methods have been used in yeast to increase amorpha-4,11-diene production (Table 1). These methods include mutating the ADS gene to the yeast-conform variant (Kong et al. 2009), using a high-copy plasmid system to express ADS in yeast (Ro et al. 2008), downregulating the expression of ERG9 and fusing ADS with FPPS (Baadhe et al. 2013; Yuan and Ching 2015a), integrating the combinatorial genome of mevalonate (MVA) pathway genes in yeast (Yuan and Ching 2014), using knockout genes outside the isoprenoid pathway but improving isoprenoid fluxes (Sun et al. 2014), assembling MVA pathway genes into yeast chromosomes and reducing ERG9 expression (Yuan and Ching 2015b), and expressing MVA pathway genes and ADS into yeast mitochondria (Farhi et al. 2011b; Yuan and Ching 2016). By overexpressing every enzyme of the MVA pathway, the production of amorpha-4,11-diene reached 41 g/L (Westfall et al. 2012).
By expressing artemisinin pathway genes in microbial hosts, current biosynthetic methods only produced artemisinic acid. However, introducing artemisinin pathway genes in Nicotiana spp. resulted in the heterologous production of artemisinin, suggesting a metabolic engineering application of these plants in the production of artemisinin (Kram and Simonsen 2017). Initially, ADS was expressed in N. tabacum to characterize its function, but it only yielded 1.7 ng/g (fresh weight) of amorpha-4,11-diene (Wallaart et al. 2001). Methods similar to those for the microbial production of amorpha-4,11-diene (Table 1), such as coexpressing MVA pathway genes and targeting ADS into mitochondria, chloroplasts, or plastids, were used to improve the accumulation of amorpha-4,11-diene (Farhi et al. 2011a; Fuentes et al. 2016; Malhotra et al. 2016; van Herpen et al. 2010; Wu et al. 2006; Zhang et al. 2011). Using the moss Physcomitrella patens as a heterologous host avoided the glycosylation of pathway intermediates (Ikram et al. 2017, 2019), and this effect was similar to the expression of artemisinin pathway genes in the chloroplasts, nuclei, and mitochondria of N. tabacum (Fuentes et al. 2016; Malhotra et al. 2016).
The success of microbial production of amorpha-4,11-diene in E. coli and yeast has promoted engineering other organisms for amorpha-4,11-diene production as proof-of-concept studies. For example, Bacillus subtilis was chosen because of its rapid growth rate and safe status (Pramastya et al. 2020; Song et al. 2021; Zhou et al. 2013), and cyanobacteria were engineered as biosolar cell factories for the photosynthetic conversion of CO2 to amorpha-4,11-diene (Choi et al. 2016). The industrial microorganism Streptomyces avermitilis was genetically engineered to produce amorpha-4,11-diene but none of its major endogenous secondary metabolites (Komatsu et al. 2010). Rhodobacter sphaeroides was used to test the growth-independent production of isoprenoids such as amorpha-4,11-diene (Orsi et al. 2020). Another strategy to produce high-value natural products is in vitro metabolic engineering, which has been applied for the production of amorpha-4,11-diene and some inhibitors of ADS such as ATP and pyrophosphate were identified (Chen et al. 2013; 2016).
Protein engineering and chemoenzymatic application of ADS
The above approaches in improving amorpha-4,11-diene production often involve the enhancement of the efficiency and production of rate-limiting enzymes in metabolic flux. However, protein engineering of ADS itself for higher catalytic efficiency has not been attempted. Engineering ADS is important because it catalyzes the first committed step in the formation of the artemisinin carbon skeleton but has poor catalytic activity. The classic metabolic engineering approach of increasing enzyme concentration to increase the production of target molecules is often limited by inherent low enzyme activity, particularly for TPS, which has 30 times lower enzyme activity than the central metabolism enzymes, which is also the case for ADS (Bar-Even et al. 2011). Protein engineering to improve the catalytic efficiency of TPS is a promising solution to this problem, which includes rational and non-rational engineering (Leonard et al. 2010). Non-rational engineering is based on error-prone PCR that introduces random mutations to a target gene, followed by the screening of clones for the desired function. Because of the lack of a high-throughput assay for screening mutant libraries, this method for engineering TPS is difficult (Lauchli et al. 2013). Thus, rational engineering of TPS is an alternative approach, which requires both knowledge of catalytic processes and understanding of the three-dimensional structure of TPS.
In 2013, A. annua α-bisabolol synthase (BOS) was isolated and functionally characterized to understand its crystal structure (Li et al. 2013). It shares 82% amino acid sequence identity and the bisabolyl cation as a common intermediate with ADS, providing a basis to find the active residues involved in ADS catalysis. After partially elucidating ADS catalysis, a T399S ADS variant that showed twofold higher turnover rate (kcat) because of accelerated product release was found (Li et al. 2013). This inspired continuous investigation for rational engineering of ADS. As mentioned earlier, the ADS catalytic mechanism involves sequential 1,6 and 1,10 cyclization (Fig. 2). By using BOS (single 1,6 cyclization) and germacrene A synthase (single 1,10 cyclization) from A. annua as reference (Bertea et al. 2006), A. annua phylogeny-based site-directed substitutions were performed. This led to the identification of seven residues in ADS controlling the whole cyclization process of amorpha-4,11-diene formation and a double mutation T399S/T447S that tripled kcat (Fang et al. 2017). Interestingly, the ADS T296V variant abolished the cyclization to the bisabolyl cation (Fang et al. 2017; Li et al. 2016; Abdallah et al. 2018). Four residues L374, L404, L405, and L439 were collectively responsible for the 1,10 cyclization, and T399 and T447 catalyzed the regioselective deprotonation and product release of ADS (Fang et al. 2017). To further identify active site residues, homology models of ADS based on a BOS variant (Abdallah et al. 2016) and TEAS (Eslami et al. 2017) were constructed. The root-mean-square deviation values between the BOS and TEAS models were 2.35 Å and 0.302 Å, respectively. Guided by the BOS model, extensive mutations were performed, leading to the identification of several residues influencing ADS catalysis. These residues included R262 for binding the PPi group; W271, Y519, and F525 for stabilizing intermediate carbocations; G400, G439, and L515 for the 1,10-ring closure; T399 for regioselective deprotonation; and W271 as an active site catalytic base. A double mutation T399S/H448A that improved kcat by 5 times was also reported (Abdallah et al. 2016, 2018). Similarly, by using the TEAS model, residues identified were involved in FDP binding and determining the fate of the allylic carbocation intermediate. These residues included Y519, D444, W271, N443, T399, R262, V292, G400, and L405, which largely overlapped with those reported by other groups (Eslami et al. 2017). Collectively, these studies have provided insight into the sequence–function relationships of ADS and have impacted the industrial production of artemisinin by microbial fermentation.
In addition to catalytic efficiency, product specificity of ADS is another target of protein engineering. Heterologous expression of ADS in E. coli yielded 89% of amorpha-4,11-diene (Newman et al. 2006), whereas an in vitro enzymatic reaction led to 80% of amorpha-4,11-diene in addition to 15 by-products (Picaud et al. 2005), and one of these by-products, amorpha-4-en-11-ol, was recently found to exist as an epimer of 6(R/S)-amorpha-4-en-11-ol (Huang et al. 2021). In contrast, the recombinant ADS expressed in N. benthamiana produced 97% of amorpha-4,11-diene and 3% of amorpha-4,7(11)-diene in vitro (Kanagarajan et al. 2012), suggesting that CYP71AV1 may not be exposed to the above 14 by-products produced by ADS expressed in E. coli (except for amorpha-4,7(11)-diene) in planta. Recently, it was demonstrated that CYP71AV1 could not use any of the 15 by-products as substrates, including amorpha-4,7(11)-diene and amorpha-4-en-7-ol, which are structurally similar to amorpha-4,11-diene, suggesting an overlooked issue to improve the fidelity of heterologously expressed ADS for more effective production of this artemisinin precursor by fermentation in E. coli (Huang et al. 2021).
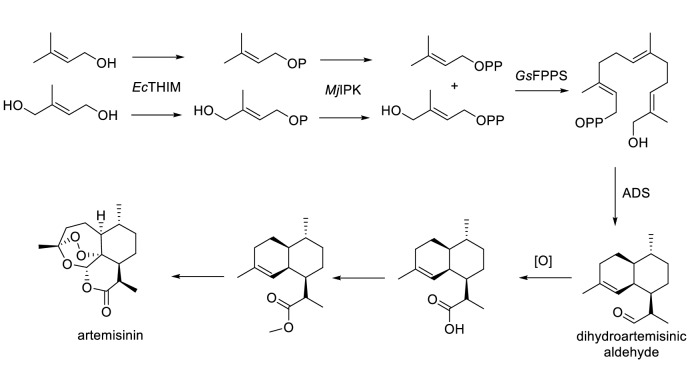
By exploiting the substrate promiscuity of ADS, a chemoenzymatic strategy was recently developed for artemisinin production. Demiray et al. (2017) found that ADS accepted chemically synthesized 12-hydroxy-FDP as the substrate and converted it to dihydroartemisinic aldehyde. When the enzymatic reaction was performed using high-performance counter current chromatography, the yield of dihydroartemisinic aldehyde increased from 20 to 60% and the reaction time reduced about tenfold (Huynh et al. 2020). In a few chemical steps, a high yield of artemisinin was obtained from this intermediate (Tang et al. 2018). Furthermore, by using the substrate promiscuity of kinases and FPPS, 12-hydroxy-FDP was enzymatically synthesized in quantitative yield (Johnson et al. 2020). On reversing the oxidation order, the entire route was complementary to the biosynthetic approach (Fig. 4).
Fig. 4.
Chemoenzymatic approach to synthesize artemisinin. EcTHIM, E. coli hydroxyethylthiazole kinase; MjIPK, Methanocaldococcus jannaschii isopentenyl phosphate kinase; GsFPPS, Geobacillus stearothermophilus farnesyl pyrophosphate synthase
Conclusion and perspectives
ADS catalyzes the first committed step in the artemisinin biosynthetic pathway. Therefore, any approach using synthetic biology and metabolic engineering to synthesize artemisinin heterologously should start with the expression of ADS. Commercial scale production of semi-synthetic artemisinin was developed based on the progress of synthetic biology for artemisinin production. Evolutionarily, the emergence of ADS in A. annua essentially shapes a specialized artemisinin pathway from the costunolide pathway. Collectively, insights from these approaches have improved our knowledge and understanding of secondary metabolism biosynthesis, metabolic engineering, and synthetic biology.
However, some questions still remain unanswered. For example, bacterial systems used to express ADS produce 10% of by-products that cannot be used by downstream CYP71AV1. This reduces the efficiency of metabolic flux for artemisinin production by microbial fermentation and requires further investigation. Regarding the catalytic mechanism of ADS, the current mechanism was proposed based on mutation and labeled substrate experiments but was not supported by quantum chemical studies. Thus, X-ray crystallography data is needed. Besides, the three-dimensional structure of ADS expressed in E. coli and in planta will unravel the molecular basis for product promiscuity of recombinant ADS from E. coli.
More importantly, a question that needs to be answered is whether the conversion from artemisinic acid to artemisinin is dependent or independent of enzymes in planta. Although chemical transformation of these compounds is feasible in plant hosts—artemisinic acid is readily converted to artemisinin, such conversion is not feasible in microbial hosts. This observation suggests a missing enzymatic link between dihydroartemisinic acid and artemisinin in A. annua.
Acknowledgements
This research was supported by the National Natural Science Foundation of China (31872666); the Special Fund for Talent Introduction of Kunming Institute of Botany, CAS; the China Postdoctoral Science Foundation (Grant Nos. 2020M671252 and 2020T130668); the Young Elite Scientists Sponsorship Program by CAST (2019QNRC001); the Open Fund of Shanghai Key Laboratory of Plant Functional Genomics and Resources (PFGR201902). We would also like to thank TopEdit (www.topeditsci.com) for English language editing of this manuscript.
Declarations
Conflicts of interest
All the authors report no conflicts of interest.
Footnotes
The original online version of this article was revised: The spelling of Jin Quan Huang’s name was incorrect.
Change history
9/21/2021
A Correction to this paper has been published: 10.1007/s42994-021-00059-w
References
- Abdallah II, Czepnik M, van Merkerk R, Quax WJ. Insights into the three-dimensional structure of amorpha-4,11-diene synthase and probing of plasticity residues. J Nat Prod. 2016;79:2455–2463. doi: 10.1021/acs.jnatprod.6b00236. [DOI] [PubMed] [Google Scholar]
- Abdallah II, van Merkerk R, Klumpenaar E, Quax WJ. Catalysis of amorpha4,11-diene synthase unraveled and improved by mutability landscape guided engineering. Sci Rep. 2018;8:9961. doi: 10.1038/s41598-018-28177-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alam P, Abdin MZ. Over-expression of HMG-CoA reductase and amorpha-4,11-diene synthase genes in Artemisia annua L. and its influence on artemisinin content. Plant Cell Rep. 2011;30:1919–1928. doi: 10.1007/s00299-011-1099-6. [DOI] [PubMed] [Google Scholar]
- Anthony JR, Anthony LC, Nowroozi F, Kwon G, Newman JD, Keasling JD. Optimization of the mevalonate-based isoprenoid biosynthetic pathway in Escherichia coli for production of the anti-malarial drug precursor amorpha-4,11-diene. Metab Eng. 2009;11:13–19. doi: 10.1016/j.ymben.2008.07.007. [DOI] [PubMed] [Google Scholar]
- Baadhe RR, Mekala NK, Parcha SR, Devi YP. Combination of ERG9 repression and enzyme fusion technology for improved production of amorphadiene in Saccharomyces cerevisiae. J Anal Methods Chem. 2013;2013:140469. doi: 10.1155/2013/140469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bar-Even A, Noor E, Savir Y, Liebermeister W, David D, Tawfi DS, Milo R. The moderately efficient enzyme: evolutionary and physicochemical trends shaping enzyme parameters. Biochemistry. 2011;50:4402–4410. doi: 10.1021/bi2002289. [DOI] [PubMed] [Google Scholar]
- Bertea CM, Voster A, Verstappen FWA, Massimo M, Beekwilder J, Bouwmeester HJ. Isoprenoid biosynthesis in Artemisia annua: cloning and heterologous expression of a germacrene A synthase from a glandular trichome cDNA library. Arch Biochem Biophys. 2006;448:150–155. doi: 10.1016/j.abb.2006.02.026. [DOI] [PubMed] [Google Scholar]
- Boutanaev AM, Moses T, Zi J, Nelson DR, Mugford ST, Peters RJ, Osbourn A. Investigation of terpene diversification across multiple sequenced plant genomes. Proc Natl Acad Sci USA. 2015;112:E81–E88. doi: 10.1073/pnas.1419547112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouwmeester HJ, Wallaart TE, Janssen MHA, et al. Amorpha-4,11-diene synthase catalyses the first probable step in artemisinin biosynthesis. Phytochemistry. 1999;52:843–854. doi: 10.1016/S0031-9422(99)00206-X. [DOI] [PubMed] [Google Scholar]
- Cai Y, Jia JW, Crock J, Lin ZX, Chen XY, Croteau R. A cDNA clone for β-caryophyllene synthase from Artemisia annua. Phytochemistry. 2002;61:523–529. doi: 10.1016/S0031-9422(02)00265-0. [DOI] [PubMed] [Google Scholar]
- Catania TM, Branigan CA, Stawniak N, et al. Silencing amorpha-4,11-diene synthase genes in Artemisia annua leads to FPP accumulation. Front Plant Sci. 2018;9:547. doi: 10.3389/fpls.2018.00547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang YJ, Song SH, Park SH, Kim SU. Amorpha-4,11-diene synthase of Artemisia annua: cDNA isolation and bacterial expression of a terpene synthase involved in artemisinin biosynthesis. Arch Biochem Biophys. 2000;383:178–184. doi: 10.1016/j.abb.2000.2061. [DOI] [PubMed] [Google Scholar]
- Chang MCY, Eachus RA, Trieu W, Ro D-K, Keasling JD. Engineering Escherichia coli for production of functionalized terpenoids using plant P450s. Nat Chem Biol. 2007;3:274–277. doi: 10.1038/nchembio875. [DOI] [PubMed] [Google Scholar]
- Chen X-Y, Xu Z. Artemisinin and plant secondary metabolism. Sci Bull. 2016;61:1–2. doi: 10.1007/s11434-015-0987-5. [DOI] [Google Scholar]
- Chen X, Zhang C, Zou R, Zhou K, Stephanopoulos G, Too HP. Statistical experimental design guided optimization of a one-pot biphasic multienzyme total synthesis of amorpha-4,11-diene. PLoS ONE. 2013;8:e79650. doi: 10.1371/journal.pone.0079650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X, Zhang C, Zou R, Stephanopoulos G, Too H-P. In vitro metabolic engineering of amorpha-4,11-diene biosynthesis at enhanced rate and specific yield of production. ACS Synth Biol. 2016;6:1691–1700. doi: 10.1021/acssynbio.6b00377. [DOI] [PubMed] [Google Scholar]
- Choi SY, Lee HJ, Choi J, et al. Photosynthetic conversion of CO2 to farnesyl diphosphate-derived phytochemical (amorpha-4,11-diene and squalene) by engineered cyanobacteria. Biotechnol Biofuels. 2016;9:202. doi: 10.1186/s13068-016-0617-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christianson DW. Structural and chemical biology of terpenoid cyclases. Chem Rev. 2017;117:11570–11648. doi: 10.1021/acs.chemrev.7b00287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demiray M, Tang X, Wirth T, Faraldos JA, Allemann R. An efficient chemoenzymatic synthesis of dihydroartemisinic aldehyde. Angew Chem Int Ed. 2017;56:4347–4350. doi: 10.1002/anie.201609557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eslami H, Mohtashami SK, Basmanj MT, Rahati M, Rahimi H. An in-silico insight into the substrate binding characteristics of the active site of amorpha-4,11-diene synthase, a key enzyme in artemisinin biosynthesis. J Mol Model. 2017;23:202. doi: 10.1007/s00894-017-3374-0. [DOI] [PubMed] [Google Scholar]
- Fang X, Li JX, Huang JQ, Xiao YL, Zhang P, Chen XY. Systematic identification of functional residues of Artemisia annua amorpha-4,11-diene synthase. Biochem J. 2017;474:2191–2202. doi: 10.1042/BCJ20170060. [DOI] [PubMed] [Google Scholar]
- Farhi M, Marhevka E, Ben-Ari J, et al. Generation of the potent anti-malarial drug artemisinin in tobacco. Nat Biotechnol. 2011;29:1072–1074. doi: 10.1038/nbt1251. [DOI] [PubMed] [Google Scholar]
- Farhi M, Marhevka E, Tania M, et al. Harnessing yeast subcellular compartments for the production of plant terpenoids. Metab Eng. 2011;13:474–481. doi: 10.1016/j.ymben.2011.05.001. [DOI] [PubMed] [Google Scholar]
- Farhi M, Kozin M, Duchin S, Vainstein A. Metabolic engineering of plants for artemisinin synthesis. Biotechnol Genet Eng. 2013;29:135–148. doi: 10.1080/02648725.2013.821283. [DOI] [PubMed] [Google Scholar]
- Fuentes P, Zhou F, Erban A, Karcher D, Kopka J, Bock R. A new synthetic biology approach allows transfer of an entire metabolic pathway from a medicinal plant to a biomass crop. eLife. 2016;5:e13664. doi: 10.7554/eLife.13664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han J, Wang H, Kanagarajan S, Hao M, Lundgren A, Brodelius PE. Promoting artemisinin biosynthesis in Artemisia annua plants by substrate channeling. Mol Plant. 2016;9:946–948. doi: 10.1016/j.molp.2016.03.004. [DOI] [PubMed] [Google Scholar]
- Hong YJ, Tantillo DJA. tangled web-interconnecting pathways to amorphadiene and the amorphene sesquiterpenes. Chem Sci. 2010;1:609–614. doi: 10.1039/c0sc00333f. [DOI] [Google Scholar]
- Hua L, Matsuda SPT. The molecular cloning of 8-epicedrol synthase from Artemisia annua. Arch Biochem Biophys. 1999;369:208–212. doi: 10.1016/j.abb.1999.1357. [DOI] [PubMed] [Google Scholar]
- Huang JQ, Li DM, Tian X, Lin JL, Yang L, Xu JJ, Fang X. Side products of recombinant amorpha-4,11-diene synthase and their effect on microbial artemisinin production. J Agric Food Chem. 2021;69:2168–2178. doi: 10.1021/acs.jafc.0c07462. [DOI] [PubMed] [Google Scholar]
- Huynh F, Tailby M, Finniear A, Stephens K, Allemann RK, Wirth T. Accelerating biphasic biocatalysis through new process windows. Angew Chem Int Ed. 2020;59:16490–16495. doi: 10.1002/anie.202005183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikram NKBK, Simonsen HT. A review of biotechnological artemisinin production in plants. Front Plant Sci. 2017;8:1966. doi: 10.3389/fpls.2017.01966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikram NKBK, Kashkooli AB, Peramuna AV, van der Krol AR, Bouwmeester H, Simonsen HT. Stable production of the antimalarial drug artemisinin in the moss Physcomitrella patens. Front Bioeng Biotechnol. 2017;5:47. doi: 10.3389/fbioe.2017.00047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikram NKBK, Kashkooli AB, Peramuna AV, van der Krol AR, Bouwmeester H, Simonsen HT. Insights into heterologous biosynthesis of arteannuin B and artemisinin in Physcomitrella patens. Molecules. 2019;24:3822. doi: 10.3390/molecules24213822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia JW, Crock J, Lu S, Croteau R, Chen XY. (3R)-linalool synthase from Artemisia annua L.: cDNA isolation, characterization, and wound induction. Arch Biochem Biophys. 1999;372:143–149. doi: 10.1016/j.abb.1999.1466. [DOI] [PubMed] [Google Scholar]
- Johnson LA, Dunbabin A, Benton JCR, Mart RJ, Allemann RK. Modular chemoenzymatic synthesis of terpenes and their analogues. Angew Chem Int Ed. 2020;59:8486–8490. doi: 10.1002/anie.202001744. [DOI] [PubMed] [Google Scholar]
- Judd R, Bagley MC, Li M, et al. Artemisinin biosynthesis in non-glandular trichome cells of Artemisia annua. Mol Plant. 2019;12:704–714. doi: 10.1016/j.molp.2019.02.011. [DOI] [PubMed] [Google Scholar]
- Kanagarajan S, Muthusamy S, Gliszczyńska A, Lundgren A, Brodelius PE. Functional expression and characterization of sesquiterpene synthases from Artemisia annua L. using transient expression system in Nicotiana benthamiana. Plant Cell Rep. 2012;31:1309–1319. doi: 10.1007/s00299-012-1250-z. [DOI] [PubMed] [Google Scholar]
- Kim SH, Heo K, Chang YJ, Park SH, Rhee SK, Kim SU. Cyclization mechanism of amorpha-4,11-diene synthase, a key enzyme in artemisinin biosynthesis. J Nat Prod. 2006;69:758–762. doi: 10.1021/np050356u. [DOI] [PubMed] [Google Scholar]
- Komatsu M, Uchiyama T, Omura S, Cane DE, Ikeda H. Genome-minimized Streptomyces host for the heterologous expression of secondary metabolism. Proc Natl Acad Sci USA. 2010;107:2646–2651. doi: 10.1073/pnas.0914833107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komori A, Suzuki M, Seki H, et al. Comparative functional analysis of CYP71AV1 natural variants reveals an important residue for the successive oxidation of amorpha-4,11-diene. FEBS Lett. 2013;587:278–284. doi: 10.1016/j.febslet.2012.11.031. [DOI] [PubMed] [Google Scholar]
- Kong J-Q, Wang W, Wang L-N, Zheng X-D, Cheng K-D, Zhu P. The improvement of amorpha-4,11-diene production by a yeast-conform variant. J Appl Microbiol. 2009;106:941–951. doi: 10.1111/j.1365-2672.2008.04063.x. [DOI] [PubMed] [Google Scholar]
- Kung SH, Lund S, Murarka A, McPhee D, Paddon CJ. Approaches and recent developments for the commercial production of semi-synthetic artemisinin. Front Plant Sci. 2018;9:87. doi: 10.3389/fpls.2018.00087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lauchli R, Rabe KS, Kalbarczyk KZ, Tata A, Heel T, Kitto RZ, Arnold FH. High-throughput screening for terpene-synthase-cyclization activity and directed evolution of a terpene synthase. Angew Chem Int Ed. 2013;52:5571–5574. doi: 10.1002/anie.201301362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leonard E, Ajikumar PK, Thayer K, et al. Combining metabolic and protein engineering of a terpenoid biosynthetic pathway for overproduction and selectivity control. Proc Natl Acad Sci USA. 2010;113:13654–13659. doi: 10.1073/pnas.1006138107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li JX, Fang X, Zhao Q, et al. Rational engineering of plasticity residues of sesquiterpene synthases from Artemisia annua: product specificity and catalytic efficiency. Biochem J. 2013;451:417–426. doi: 10.1042/BJ20130041. [DOI] [PubMed] [Google Scholar]
- Li Z, Gao R, Hao Q, et al. The T296V mutant of amorpha-4,11-diene synthase is defective in allylic diphosphate isomerization but retains the ability to cyclize the intermediate (3R)-nerolidyl diphosphate to amorpha-4,11-diene. Biochemistry. 2016;36:8332–8339. doi: 10.1021/acs.biochem.6b01004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindahl A-L, Oslsson MK, Mercke P, Tollbom Ö, Schelin J, Brodelius M, Brodelius PE. Production of the artemisinin precursor amorpha-4,11-diene by engineered Saccharomyces cerevisiae. Biotechnol Lett. 2006;28:571–580. doi: 10.1007/s10529-006-0015-6. [DOI] [PubMed] [Google Scholar]
- Ma C, Wang H, Lu X, Wang H, Xu G, Liu B. Terpenoid metabolic profiling analysis of transgenic Artemisia annua L. by comprehensive two-dimensional gas chromatography time-of-flight mass spectrometry. Metabolomics. 2009;5:497–506. doi: 10.1007/s11306-009-0170-6. [DOI] [Google Scholar]
- Ma SM, Garcia DE, Redding-Johanson AM, et al. Optimization of a heterologous mevalonate pathway through the use of variant HMG-CoA reductases. Metab Eng. 2011;13:588–597. doi: 10.1016/j.ymben.2011.07.001. [DOI] [PubMed] [Google Scholar]
- Ma D-M, Wang Z, Wang L, Alejos-Gonzales F, Sun M-A, Xie D-Y. A genome-wide scenario of terpene pathways in self-pollinated Artemisia annua. Mol Plant. 2015;8:1580–1598. doi: 10.1016/j.molp.2015.07.004. [DOI] [PubMed] [Google Scholar]
- Malhotra K, Subramaniyan M, Rawat K, et al. Compartmentalized metabolic engineering for artemisinin biosynthesis and effective malaria treatment by oral delivery of plant cells. Mol Plant. 2016;9:1464–1477. doi: 10.1016/j.molp.2016.09.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin VJJ, Pitera DJ, Withers ST, Newman JD, Keasling JD. Engineering a mevalonate pathway in Escherichia coli for production of terpenoids. Nat Biotechnol. 2003;21:796–802. doi: 10.1038/nbt833. [DOI] [PubMed] [Google Scholar]
- Mercke P, Crock J, Croteau R, Brodelius PE. Cloning, expression, and characterization of epi-cedrol synthase, a sesquiterpene cyclase from Artemisia annua L. Arch Biochem Biophys. 1999;369:213–222. doi: 10.1016/j.abb.1999.1358. [DOI] [PubMed] [Google Scholar]
- Mercke P, Bengtsson M, Bouwmeester HJ, Posthumus MA, Brodelius PE. Molecular cloning, expression, and characterization of amorpha-4,11-diene synthase, a key enzyme of artemisinin biosynthesis in Artemisia annua L. Arch Biochem Biophys. 2000;381:173–180. doi: 10.1016/j.abb.2000.1962. [DOI] [PubMed] [Google Scholar]
- Muangphrom P, Seki H, Suzuki M, et al. Functional analysis of amorpha-4,11-diene synthase (ADS) homologs from non-artemisinin-producing Artemisia species: the discover of novel koidzumiol and (+)-α-bisabolol synthases. Plant Cell Physiol. 2016;57:1678–1688. doi: 10.1093/pcp/pcw094. [DOI] [PubMed] [Google Scholar]
- Muangphrom P, Seki H, Matsumoto S, Nishiwaki M, Fukushima EO, Muranaka T. Identification and characterization of a novel sesquiterpene synthase, 4-amorphen-11-ol synthase, from Artemisia maritima. Plant Biotechnol Nar. 2018;35:113–121. doi: 10.5511/plantbiotechnology.18.0324a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newman JD, Marshall J, Chang M, et al. High-level production of amorpha-4,11-diene in a two-phase partitioning bioreactor of metabolically engineered Escherichia coli. Biotechnol Bioeng. 2006;95:684–691. doi: 10.1002/bit.21017. [DOI] [PubMed] [Google Scholar]
- Nguyen DT, Göpfert JC, Ikezawa N, et al. Biochemical conservation and evolution of germacrene A oxidase in Asteraceae. J Biol Chem. 2010;285:16588–16598. doi: 10.1074/jbc.M110.111757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen T-D, Kwon M, Kim S-U, Fischer C, Ro D-K. Catalytic plasticity of germacrene A oxidase underlies sesquiterpene lactone diversification. Plant Physiol. 2019;181:945–960. doi: 10.1104/pp.19.00629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orsi E, Mougiakos I, Post W, et al. Growth-uncoupled isoprenoid synthesis in Rhodobacter sphaeroides. Biotechnol Biofuels. 2020;13:123. doi: 10.1186/s13068-020-01765-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paddon CJ, Keasling JD. Semi-synthetic artemisinin: a model for the use of synthetic biology in pharmaceutical development. Nat Rev Microbiol. 2014;12:355–357. doi: 10.1038/nrmicro3240. [DOI] [PubMed] [Google Scholar]
- Paddon CJ, Westfall PJ, Pitera DJ, et al. High-level semi-synthetic production of the potent antimalarial artemisinin. Nature. 2013;494:528–532. doi: 10.1038/nature12051. [DOI] [PubMed] [Google Scholar]
- Picaud S, Olofsson L, Brodelius M, Brodelius PE. Expression, purification, and characterization of recombinant amorpha-4,11-diene synthase from Artemisia annua L. Arch Biochem Biophys. 2005;436:215–226. doi: 10.1016/j.abb.2005.02.012. [DOI] [PubMed] [Google Scholar]
- Picaud S, Mercke P, He X, Sterner O, Brodelius M, Cane DE, Brodelius PE. Amorpha-4,11-diene synthase: mechanism and stereochemistry of the enzymatic cyclization of farnesyl diphosphate. Arch Biochem Biophys. 2006;448:150–155. doi: 10.1016/j.abb.2005.07.015. [DOI] [PubMed] [Google Scholar]
- Pramastya H, Xue D, Abdallah II, Setroikromo R, Quax WJ. High level production of amorphadiene using Bacillus subtilis as an optimized terpenoid cell factory. New Biotechnol. 2020;60:159–167. doi: 10.1016/j.nbt.2020.10.007. [DOI] [PubMed] [Google Scholar]
- Redding-Johanson AM, Batth TS, Chan R. Targeted proteomics for metabolic pathway optimization: application to terpene production. Mtab Eng. 2011;13:194–203. doi: 10.1016/j.ymben.2010.12.005. [DOI] [PubMed] [Google Scholar]
- Ro D-K, Paradise EM, Ouellet M, et al. Production of the antimalarial drug precursor artemisinic acid in engineered yeast. Nature. 2006;440:940–943. doi: 10.1038/nature04640. [DOI] [PubMed] [Google Scholar]
- Ro D-K, Ouellet M, Paradise EM, et al. Induction of multiple pleiotropic drug resistance genes in yeast engineered to produce an increased level of anti-malarial drug precursor, artemisinic acid. BMC Biotechnol. 2008;8:83. doi: 10.1186/1472-6750-8-83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salmon M, Laurendon C, Vardakou M, et al. Emergence of terpene cyclization in Artemisia annua. Nat Commun. 2015;6:6143. doi: 10.1038/ncomms7143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shukal S, Chen X, Zhang C. Systematic engineering for high-yield production of viridiflorol and amorphadiene in auxotrophic Escherichia coli. Mtab Eng. 2019;55:170–178. doi: 10.1016/j.ymben.2019.07.007. [DOI] [PubMed] [Google Scholar]
- Song Y, He S, Abdallah II, et al. Engineering of multiple modules to improve amorphadiene production in Bacillus subtilis using CRISPR-Cas9. J Agric Food Chem. 2021;69:4785–4794. doi: 10.1021/acs.jafc.1c00498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Z, Meng H, Li J, Wang J, Li Q, Wang Y, Zhang Y. Identification of novel knockout targets for improving terpenoids biosynthesis in Saccharomyces cerevisiae. PLoS ONE. 2014;9:e112615. doi: 10.1371/journal.pone.0112615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang X, Demiray M, Wirth T, Allemann RK. Concise synthesis of artemisinin from a farnesyl diphosphate analogue. Bioorgan Med Chem. 2018;26:1314–1319. doi: 10.1016/j.bmc.2017.03.068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teoh KH, Polichuk DR, Reed DW, Nowak G, Covello PS. Artemisia annua L. (Asteraceae) trichome-specific cDNAs reveal CYP71AV1, a cytochrome P450 with a key role in the biosynthesis of the antimalarial sesquiterpene lactone artemisinin. FEBS Lett. 2006;580:1411–1416. doi: 10.1016/j.febslet.2006.01.065. [DOI] [PubMed] [Google Scholar]
- Tsuruta H, Paddon CJ, Eng D, et al. High-level production of amorpha-4,11-diene, a precursor of the antimalarial agent artemisinin, in Escherichia coli. PLoS ONE. 2009;4:e4489. doi: 10.1371/journal.pone.0004489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Herpen TWJM, Cankar K, Nogueira M, Bosch D, Bouwmeester HJ, Beekwilder J. Nicotiana benthamiana as a production platform for artemisinin precursors. PLoS ONE. 2010;5:e14222. doi: 10.1371/journal.pone.0014222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wallaart TE, Bouwmeester HJ, Hille J, Poppinga L, Maijers NCA. Amorpha-4,11-diene synthase: cloning and functional expression of a key enzyme in the biosynthetic pathway of the novel antimalarial drug artemisinin. Planta. 2001;212:460–465. doi: 10.1007/s004250000428. [DOI] [PubMed] [Google Scholar]
- Wang J-F, Xiong Z-Q, Li S-Y, Wang Y. Enhancing isoprenoid production through systematically assembling and modulating efflux pumps in Escherichia coli. Appl Microbiol Biotechnol. 2013;97:8057–8067. doi: 10.1007/s00253-013-5062-z. [DOI] [PubMed] [Google Scholar]
- Wang J, Xu C, Wong YK, Li Y, Liao F, Jiang T, Tu Y. Artemisinin, the magic drug discovered from traditional Chinese medicine. Engineering. 2019;5:32–39. doi: 10.1016/j.eng.2018.11.011. [DOI] [Google Scholar]
- Wei Q, He S, Qu J, Xia J. Synthetic multienzyme complexes assembled on virus-like particles for cascade biosynthesis in cellulo. Bioconjugate Chem. 2020;31:2413–2420. doi: 10.1021/acs.bioconjchem.0c00476. [DOI] [PubMed] [Google Scholar]
- Westfall PJ, Pitera DJ, Lenihan JR, et al. Production of amorphadiene in yeast, and its conversion to dihydroartemisinic acid, precursor to the antimalarial agent artemisinin. Proc Natl Acad Sci USA. 2012;109:E111–E118. doi: 10.1073/pnas.1110740109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu S, Schalk M, Clark A, Miles RB, Coates R, Chappell J. Redirection of cytosolic or plastidic isoprenoid precursors elevates terpene production in plants. Nat Biotechnol. 2006;24:1441–1447. doi: 10.1038/nbt1251. [DOI] [PubMed] [Google Scholar]
- Xie DY, Ma DM, Judd R, Jones AL. Artemisinin biosynthesis in Artemisia annua and metabolic engineering: questions, challenges, and perspectives. Phytochem Rev. 2016;15:1093–1114. doi: 10.1007/s11101-016-9480-2. [DOI] [Google Scholar]
- Yuan J, Ching C-B. Combinatorial engineering of mevalonate pathway for improved amorpha-4,11-diene production in budding yeast. Biotechnol Bioeng. 2014;111:608–617. doi: 10.1002/bit.25123. [DOI] [PubMed] [Google Scholar]
- Yuan J, Ching C-B. Dynamic control of ERG9 expression for improved amorpha-4,11-diene production in Saccharomyces cerevisiae. Microb Cell Fact. 2015;14:38. doi: 10.1186/s12934-015-0220-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan J, Ching C-B. Combinatorial assembly of large biochemical pathways into yeast chromosomes for improved production of value-added compounds. ACS Synth Biol. 2015;14:23–31. doi: 10.1021/sb500079f. [DOI] [PubMed] [Google Scholar]
- Yuan J, Ching C-B. Mitochondrial acetyl-CoA utilization pathway for terpenoid productions. Metab Eng. 2016;38:303–309. doi: 10.1016/j.ymben.2016.07.008. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Nowak G, Reed DW, Covello PS. The production of artemisinin precursors in tobacco. Plant Biotechnol J. 2011;9:445–454. doi: 10.1111/j.1467-7652.2010.00556.x. [DOI] [PubMed] [Google Scholar]
- Zhang C, Chen X, Zou R, Stephanopoulos G, Too H-P. Combining genotype improvement and statistical media optimization for isoprenoid production in E. coli. Plos ONE. 2013;8:75164. doi: 10.1371/journal.pone.0075164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang C, Zou R, Chen X, Stephanopoulos G, Too H-R. Experimental design-aided systematic pathway optimization of glucose uptake and deoxyxylulose phosphate pathway for improved amorphadiene production. Appl Microbiol Biotechnol. 2015;99:3825–3837. doi: 10.1007/s00253-015-6463-y. [DOI] [PubMed] [Google Scholar]
- Zhang C, Chen X, Stephanopoulos G, Too H-P. Efflux transporter engineering markedly improved amorphadiene production in Escherichia coli. Biotechnol Bioeng. 2016;113:1755–1763. doi: 10.1002/bit.25943. [DOI] [PubMed] [Google Scholar]
- Zhou K, Zou R, Zhang C, Stephanopoulos G, Too H-P. Optimization of amorphadiene synthesis in Bacillus subtilis via transcriptional, translational, and media modulation. Biotechnol Bioeng. 2013;110:2556–2561. doi: 10.1002/bit.24900. [DOI] [PubMed] [Google Scholar]