Abstract
Drought stress is major abiotic stress that affects soybean production. Therefore, it is widely desirable that soybean becomes more tolerant to stress. To provide insights into regulatory mechanisms of the stress response, we compared the global gene expression profiles from leaves of two soybean genotypes that display different responses to water-deficit (BR 16 and Embrapa 48, drought-sensitive and drought-tolerant, respectively). After the RNA-seq analysis, a total of 5335 down-regulated and 3170 up-regulated genes were identified in the BR16. On the other hand, the number of genes differentially expressed was markedly lower in the Embrapa 48, 355 up-regulated and 471 down-regulated genes. However, induction and expression of protein kinases and transcription factors indicated signaling cascades involved in the drought tolerance. Overall, the results suggest that the metabolism of pectin is differently modulated in response to drought stress and may play a role in the soybean defense mechanism against drought. This occurs via an increase of the cell wall plasticity and crosslink, which contributed to a higher hydraulic conductance (Kf) and relative water content (RWC%). The drought-tolerance mechanism of the Embrapa 48 genotype involves remodeling of the cell wall and increase of the hydraulic conductance to the maintenance of cell turgor and metabolic processes, resulting in the highest leaf RWC, photosynthetic rate (A), transpiration (E) and carboxylation (A/Ci). Thus, we concluded that the cell wall adjustment under drought is important for a more efficient water use which promoted a more active photosynthetic metabolism, maintaining higher plant growth under drought stress.
Supplementary Information
The online version contains supplementary material available at 10.1007/s42994-021-00043-4.
Keywords: Gene expression, RNA-seq, Molecular physiology, Water-use efficiency
Introduction
Drought is the main environmental factor that negatively influences both plant growth and development, thus restricting productivity and agricultural expansion. Projections of climate change indicate that drought will become more intense in some areas of the world and therefore the development of tolerant plants is necessary to maintain production (Passioura 2007; Stanke et al. 2013; Spinoni et al. 2017). On the other hand, plants have evolved to create several strategies to cope with drought, including a short life cycle or phenotypical plasticity, enhance in water uptake and reduction water loss, as well as osmotic adjustment, antioxidant capacity, and drought tolerance (Fang and Xiong 2015). The evaluation of tolerance mechanisms and drought-responsive genes from soybean is essential for genetic breeding programs (Rodrigues et al. 2012; Brown 2017). Thus, a transcriptome analysis of contrasting genotypes may contribute to the understanding of the molecular and physiological responses.
The main difficulty in selecting genes as targets for plant breeding that is aimed at drought tolerance is the complexity of the physiological responses to drought stress. Plant survival strategies under drought involve transient responses, such as reduced transpiration, changes in the root system, reduction of leaf area and adjusted osmotic status leading to a minimal water loss and improving water uptake (Hu and Xiong 2014). Transient response and developmental changes require a substantial rebuilding of plant metabolism and changes in the expression of a high number of genes. Global transcriptome analysis has been used to provide a deeper insight into the complexity of plant response to drought stress on the molecular level.
External drought stimuli are perceived by sensors on the membrane, and then the signals are delivered through multiple signaling pathways, resulting in the expression of responsive genes so as to confer drought tolerance in the plants (Zhu 2002; Hirayama and Shinozaki 2010). In general, gene expression studies of various plant species have been performed to classify several groups of genes, which are regulated in response to drought. Among them are those encoding calcium-dependent protein kinases, calmodulin and calmodulin-related calcium sensor proteins and protein phosphatases class 2C (PP2C) (Molina et al. 2008; Guo et al. 2009; Ranjan and Sawant 2015), along with a number of transcription factors (TFs) (Sahoo et al. 2013; Janiak et al. 2018). These signaling proteins are usually classified as ABA-dependent and ABA-independent stress response pathways (Shinozaki and Yamaguchi-Shinozaki 2007). Genes involved in biosynthesis and signaling pathways of other plant hormones, such as auxin, ethylene, jasmonic or salicylic acid, were also identified as differentially expressed under drought (Jakoby et al. 2002; Aimar et al. 2011). Moreover, genes related to anti-oxidation processes, osmo-protectant synthesis and various factors from late embryogenesis abundant (LEA) family were also reported as differentially expressed in response to drought (Shinozaki and Yamaguchi-Shinozaki 2007; Talame et al. 2007).
The main challenge when performing gene expression studies is to identify which genes are not only responsive, but also confer a differential physiological behavior when compared to a sensitive genotype. To achieve this goal, it is necessary to use parental plant genotypes contrasting in drought-tolerance. The physiological response of the genotypes BR 16 (drought-sensitive) and Embrapa 48 (drought-tolerant) under drought conditions was studied by Oya et al. (2004), Carvalho et al. (2015) and our research group (Mesquita et al. 2020). They found that, in the vegetative stage under drought conditions in the field, the drought-tolerant genotype had the highest number of pods. The studies of the proteome, phosphoproteome and metabolomic profile were also performed by Lima et al. (2019) to detect the metabolic pathways which are affected by drought stress. An integrative overview showed that tolerant plants maintain cell homeostasis and photosynthetic metabolism under stress conditions, as indicated by an abundance of protein and regulation by phosphorylation. Drought-stress marker in roots was also evaluated to understand the mechanism of tolerance in these genotypes. The GmaxRD20A-like and GmaxRD22-like genes, homologs of Arabidopsis genes of the ABA-independent pathway, are highly induced by water-deficit, being these potential drought marker genes in these genotypes (Neves-Borges et al. 2012).
Knowledge about drought-tolerance mechanisms in soybean has been reported in recent decades. However, determinate the main mechanism operating in a given plant genotype is a challenge that requires multiple analytical approaches, involving physiological and gene expression analyzes. The physiological responses revealed that the tolerant genotype Embrapa 48 postpones the leaf dehydration by a mechanism involving a more efficient use and translocation of water from root to the shoot to maintain unchanged the cell homeostasis and the photosynthetic metabolism under the stress (Mesquita et al. 2020). Furthermore, an integrative overview involving proteomic, phosphoproteomic and metabolomic profiles also showed that the Embrapa 48 tolerant plants maintain cell metabolism unchanged under the stress condition in contrast to BR16 sensitive genotype that showed several dysregulated pathways (Lima et al. 2019). Only small deviations of the metabolic pathways were observed for drought-tolerant plants in comparison to the sensitive genotype. These findings indicated that osmoprotection and/or oxidative protection does not appear to be the major mechanisms for tolerance, as indicated by the accumulation of the metabolite, enzymes assays and the phytohormone profiles from tolerant and sensitive soybean plants (Lima et al. 2019; Mesquita et al. 2020). Here, we analyzed physiological traits essential for drought-tolerance elucidation, such as assimilation rate of CO2, stomatal conductance (gs), transpiration (E), water use efficiency (WUEi), carboxylation efficiency (A/Ci), relative water content (RWC), hydraulic conductance (Kf), correlating with data from gene-expression responses to dehydration through RNA-Seq analysis from soybean parental genotypes (Embrapa 48 and BR 16). The metabolic/regulatory pathways and biological processes were explored via Gene Ontology (GO) enrichment and indicated differences in the gene expression reprogramming in the drought-tolerant genotype that correlated with the physiological mechanisms of the tolerance. The results revealed that in response to stress, the tolerant genotype Embrapa 48 has less gene reprogramming, however expressed specifically protein kinases and transcription factors in response to drought tolerance. Expression of the genes relating to pectin remodeling in the leaves may be involved in a mechanism that contributes to the maintenance of leaf turgor and metabolism of the Embrapa 48 genotype under drought stress.
Materials and methods
Plant material, growth and drought-stress treatments
Seeds of soybean genotypes BR 16 and Embrapa 48 were obtained from the Empresa Brasileira de Pesquisa Agropecuária (EMBRAPA SOJA, Londrina, Paraná). Seedlings were grown in plastic trays containing Plantmax® commercial substrate, where they remained for 10 days. After germination, seedlings were transplanted to pots containing 10 L of a mixture of soil, sand and manure (2/1/1) each. Plants were grown under natural sunlight in a greenhouse with average daytime temperature 15–35 °C and relative humidity 65–85%.
The plants were grown under normal water conditions until reaching the development stage V3 (fully expanded third trifoliate). The control plants were watered daily with approximately 30 mL water per plant. The plants were exposed to a slow drying soil treatment, which consisted of a reduction in irrigation to 40% of the daily normal until the plant reached the hydraulic potential of Ψw = − 1 MPa (Valente et al. 2009). The hydric regimes were assigned as irrigated (IR) and non-irrigated (NI). The leaf water potential (Ψw) was measured in the third emerging trifoliate at dawn using a Scholander pump (Scholander et al. 1965) during the stress period. Samples were collected in liquid nitrogen when the plants had a water potential − 1 MPa and then stored at − 80 °C until use. For each treatment, 3 pots were used and each pot contained three plants. A trifoliate leaf from each plant from one pot was collected together (3 plants by replicate). Thus, the analyses were performed using 3 distinct pools, resulting in 3 biological replicates. These procedures were performed on both plant cultivars.
RNA extraction, library preparation, and sequencing
Total RNA was extracted from leaves using a Trizol reagent (Invitrogen) according to the manufacturer’s instructions. Five micrograms of total RNA was used to prepare paired-end 100 bp libraries using the BIOO NEXT flex Rapid Directional RNA-Seq Kit (Bioo-Scientific, Austin, TX). Library qualities were analyzed with the Bioanalyzer 2100 (Agilent, Santa Clara, CA) and the barcoded libraries were quantified by fluorometry using a Qubit instrument (LifeTechnologies, Carlsbad, CA). The libraries were then pooled in equimolar ratios, quantified by qPCR with a Kapa Library Quant kit (Kapa, Cape Town, South Africa). Three biological replicates of each treatment were sequenced using the Illumina Hi-Seq 2500 (Illumina, San Diego, CA) from NuBioMol (Center for Biomolecules Analysis—UFV, Brazil).
Raw reads were subsequently subjected to trimming using Trimmometic software (Bolger et al. 2014) with a Phred quality threshold of 20. Reads were aligned to the Glycine max genotype Williams 82 primary transcriptome (Wm82.a2.v1) (Schmutz et al. 2010) using the Kallisto aligner (Bray et al. 2016).
Analysis of differentially expressed genes (DEGs)
To identify differentially expressed genes (DEGs), we used the DESeq2 software package, which performs pairwise comparisons (Anders and Huber 2012). DESeq2 analyses were carried out using the Kallisto output. The DEGs were identified using the MA-plot-based method from the package DEGseq version 3.0 (Wang et al. 2009). An absolute fold-change threshold of 2.0 and a false discovery rate (FDR) of ≤ 0.0001 were used to select the DEGs identified by DEGseq.
RT-qPCR
RT-qPCR was performed to validate the differential gene expression data obtained by RNA-Seq analysis. RNA was extracted from leaf tissues of the control and stressed plants using Trizol reagent (Invitrogen) according to the manufacturer’s instructions. RNA quality was analyzed using agarose gel electrophoresis and quantification was performed on a Thermo Scientific NanoDrop 2000c. A total of 4 μg of RNA was used for cDNA synthesis with the SuperScript III kit (Invitrogen) following the manufacturer’s instructions.
The gene expression was assessed using an ABI 7500 Fast Thermocycler (Applied Biosystems, Foster City, CA, USA) and Fast SYBR Green Master Mix (Thermo Fisher Scientific). The cycling conditions were as follows: 15 s at 95 °C, 40 cycles of 95 °C for 3 s; 30 s at 60 °C, and final denaturation at 95 °C for 20 s, followed by a melting curve. Specific primers for RT-qPCR were designed using the Primer-BLAST software (https://www.ncbi.nlm.nih.gov/tools/primer-blast/), with a melting temperature (T) between 59–61 °C, a length of 18–23 bp, an amplicon product size of 120–150 bp, and a GC content of 40–60% (Supplementary Information Table S1). Gene expression was normalized using two soybean housekeeping genes, being them UNK2 and Actin. A total of three biological replicates and three technical replicates were performed for each gene. Relative quantification was calculated according to the 2−ΔΔCt method (Livak and Schmittgen 2001).
Functional classification of the differentially expressed genes
The DEGs were subjected to functional classification using The Arabidopsis Information Resource (http://www.arabidopsis.org). The gene ontology (GO) enrichment analysis of each gene set was performed using the ClueGO version 2.0.7 plugin tool (Bindea et al. 2009) in Cytoscape version 3.2.1 (Shannon et al. 2003) using the GO biological process category. Over-represented biological process categories were identified using an (right-sided) enrichment test based on the hypergeometric distribution. To determine significantly the overrepresented GO terms, the terms with a p value lower than 0.05 were considered as a Kappa significant value. Genes classified as significantly over-represented were validated by the Benjamin test.
Measurement of gas exchange and photosynthetic parameters
The assimilation rate of CO2 (A), stomatal conductance to water vapor (gs), transpiratory rate (E) and internal and external carbon ratio (Ci/Ca ratio) were determined for the fourth leaf from the apical meristem of each plant using an infrared gas analyzer (IRGA, portable model LI-6400XT, LI-COR Biosciences Inc., Lincon, Nebraska, USA) as described by Mesquita et al. (2020).
Measurement of the relative water content (RWC) of the leaves
The fresh weight (FW) of leaf discs was measured immediately after they were removed from the stem. Then, tissues were incubated in distilled water for at least 4 h in the dark, and the turgid weight (TW) was measured. Finally, dry weight (DW) was measured after incubation at 85 °C until the sample reached a constant weight in the oven. The relative water content (RWC) was calculated using the equation: RWC (%) = [(FW − DW)/(TW − DW)] × 100.
Evaluation of leaf hydraulic conductivity (Kf)
Leaf hydraulic conductivity (Kf) determination was performed using the evaporative flow method (EFM), according to the methodology described by Sack et al. (2002) and Brodribb and Holbrook (2003, 2006), with modifications. The water restriction experiment started when the third trefoil was fully expanded. The leaf water potentials (Ψw) were determined before dawn using a Scholander pressure pump and when the leaves reached Ψ of approximately − 1.0 MPa, the gas exchange parameters were determined. The measurement of leaf transpiration rate (E) was carried out between eight and ten in the morning of the same day using an infrared gas analyzer (IRGA, portable model LI-6400xt, LI-COR Biosciences Inc., Lincon, Nebraska, USA). Kf was calculated using the equation: Kf = − E/Ψ.
Results
Analysis of RNA transcripts
The response of the soybean to moderate drought was investigated at the transcriptional level by an RNA-Seq approach. Previous studies performed by our research group showed greater reduction in the growth of the aerial part, simultaneously with a greater induction of the growth of the root system under drought, without changing the leaf area, indicating that the tolerant genotype has a differential mechanism in allocating the carbon for the roots (Mesquita et al. 2020). In the present study, gene expression in leaves of two soybean genotypes, BR 16 and Embrapa 48, drought-sensitive and drought-tolerant, were analyzed by Illumina Hi-Seq 2500 (Illumina, San Diego, CA). The initial sample collected after a water-deficit was designated as “NI”, and control plants “IR” (Fig. 1a). Approximately 45–50 million reads were generated from each sample. Raw reads were subjected to a pre-processing/trimming step to remove short or low-quality sequences and adaptor/primer sequences. The RNA-Seq analysis workflow is shown in Supplementary Information Figure S1 and was used for data analysis.
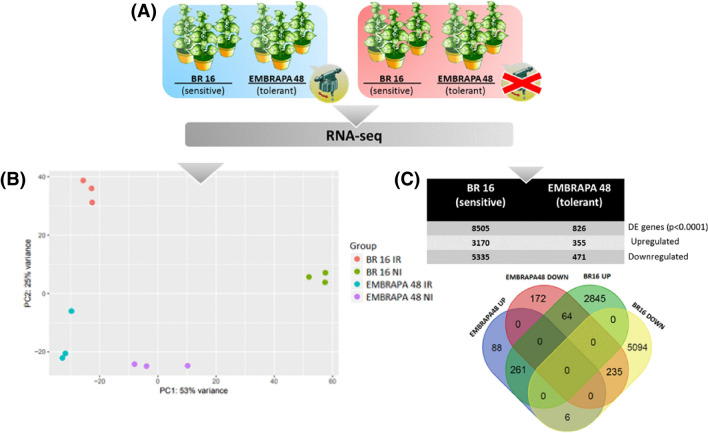
Fig. 1.
Overall gene expression in response to drought stress. In a soybean plants BR 16 and Embrapa 48 exposed to a gradual drought regime to isolate total RNA for transcriptomic analysis. The water potential was measured by Scholander pump. In the blue box irrigated plants and in the red stressed plants. In b transcriptome data used for the principal component analysis, showing distinct clusters of the different soybean genotypes in Irrigated conditions (IR) and not irrigated (NI). In c number of differently expressed genes in drought conditions in both genotypes and Venn diagram showing the comparison of the number of genes differentially expressed. Proportion of significant results (p ≤ 0.0001, log2 fold change ≥ 2 for up-regulated and ≤ − 2 for down-regulated genes)
To understand further about the similarity of the genotypic responses under water-deficit conditions, we used Principal Component Analysis (PCA) (Fig. 1b). The quality of the data obtained can be observed by the analysis of the sample-by-sample Euclidean distance, which is repopulated in the form of a heat map (Supplementary Information Fig. S2). High-throughput RNA-sequencing analysis was performed using a Kallisto pipeline (Bray et al. 2016) comparing the number of genes differentially regulated in response to drought combinations between controls vs. treatments, for each genotype, using DESEq2 (Anders and Huber 2012). When comparing all genes differentially expressed, we identified for sensitive BR 16 8505 genes, including 3170 genes up-regulated and 5335 genes down-regulated under drought conditions. For the tolerant EMBRAPA 48, 826 genes were differentially expressed, including 355 up-regulated and 471 genes down-regulated (Fig. 1c). The global data show that the transcriptional reprogramming was more pronounced in the sensitive plants, considering the highest number of DEGs in BR 16 genotype. In addition, gene-expression data obtained by RNA-seq strategy correlated with RT-qPCR measurements (Fig. 2), confirming the accuracy of our RNA-seq data.
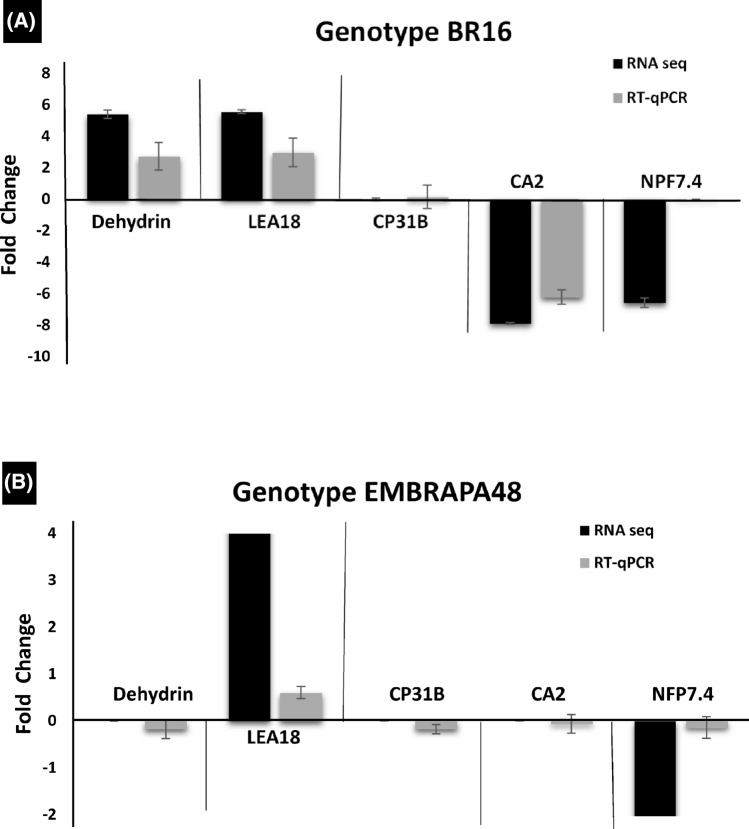
Fig. 2.
Validation of (black filled square) RNA-Seq data by real-time (ash filled square) RT-qPCR. The expression variation of the RNAs analyzed in this study in plants submitted to water-deficit compared with controls. The graph in a shows the expression variation in the genotype BR16, and the graph in b shows the expression variation in the genotype EMBRAPA48. Genes encoding Dehydryn, LEA18, CP31B (chloroplast RNA-binding protein 31B), CA2 (carbonic anhydrase), NPF4.7 (protein NRT1/PTR family 4.7), RGXT2 and RGXT1 were analyzed. The data represent the mean ± SE (n = 3)
The gene expression response to drought has been evaluated extensively in several plants including soybeans (Bhargava and Sawant 2013). Thus, to select genes that may be involved not only in general stress response, but also that conferred drought tolerance in soybean, we analyze the list of genes that were differentially expressed only in EMBRAPA 48 genotype and could be correlated with molecular and physiological data previously generated for both genotypes (Lima et al. 2019; Mesquita et al. 2020). Since the genotype BR16 and EMBRAPA 48 are parental, we believe that the genes differentially expressed only in the genotype EMBRAPA 48 during stress may be responsible for their tolerance. Thus, we first manually inspect the functional categories (ClueGO results) grouped for each genotype to select those enriched only for the tolerant genotype. Followed, the functions of the genes included in these categories were related to molecular mechanisms described for drought tolerance and correlated as physiological traits of both genotypes. This strategy allowed us to select candidates that could play a role in drought adaptation and tolerance, whilst also may explain the lower number of DEGs found in the tolerant genotype. Thus, 260 genes were selected as being differentially expressed exclusively in the tolerant genotype, including 88 up-regulated and 172 down-regulated (Fig. 1c). We identified 20 genes encoding protein kinases (PKs) (Table 1): four genes encoding Serine/threonine-protein kinases (S/T PKs), two were up-regulated (Glyma20G14580.1, Glyma18G18720.1) and two were down-regulated (Glyma09G09800.1, Glyma20G17650.1) under water-deficit; three genes encoding Calcium-dependent protein kinases (CDPKs) all were up-regulated (Glyma16G02240.1, Glyma16G14250.1, Glyma15G14340.1); and fourteen other PKs genes, nine of them were down-regulated (Glyma10G19570.1, Glyma07G21590.1, Glyma13G26610.1, Glyma13G24090.1, Glyma18G12450.1, Glyma13G28310.1, Glyma04G19530.1, Glyma07G21660.1) and five were up-regulated genes (Glyma10G22220.1, Glyma13G22430.18, Glyma13G22430.22, Glyma13G22430.11, Glyma17G14990.2).
Table 1.
Protein Kinases responsive to dehydration only in the drought-tolerant genotype
| Protein kinase | ||
|---|---|---|
| Gene | Log2 ratio | Annotation |
| S/T PKS | ||
| GLYMA09G09800.1 | − 0.709405048 | CBL-interacting serine/threonine-protein kinase |
| GLYMA18G18720.1 | 1.162165 | Serine/threonine-protein kinase wnk with no lysine |
| GLYMA20G14580.1 | 1.035192 | Serine/threonine-protein kinase wnk with no lysine |
| GLYMA20G17650.1 | − 0.624644997 | Serine/threonine-protein kinase |
| CDPKS | ||
| GLYMA16G02240.1 | 1.123958 | Calcium-binding protein |
| GLYMA16G14250.1 | 1.102288 | Calcium-binding protein cml41-related |
| GLYMA15G14340.1 | 0.827979131 | WTF9 |
| PKS | ||
| GLYMA10G19570.1 | − 1.091893292 | Leucine-rich repeat receptor-like protein kinase pepr1-related |
| GLYMA07G21590.1 | − 1.570048333 | Leucine-rich repeat N-terminal domain (LRRNT_2) |
| GLYMA13G26610.1 | − 1.640632312 | Protein tyrosine kinase (Pkinase_Tyr) |
| GLYMA13G24090.1 | − 0.484742282 | AMP-activated protein kinase. gamma regulatory subunit |
| GLYMA18G12450.1 | − 1.188983376 | Protein kinase domain (Pkinase)//Leucine Rich Repeat (LRR_1) |
| GLYMA13G28310.1 | − 0.964734668 | Cysteine-rich receptor-like protein kinase 27-related |
| GLYMA04G19530.1 | − 1.480822749 | SNF1-related protein kinase regulatory subunit gamma-1 |
| GLYMA10G22220.1 | 1.094759 | Cell division protein kinase |
| GLYMA07G21660.1 | − 0.539613982 | 1-Phosphatidylinositol-3-phosphate 5-kinase fyab1c-related |
| GLYMA13G22430.18 | 1.81788 | Protein tyrosine kinase |
| GLYMA13G22430.22 | 1.648667 | Protein tyrosine kinase |
| GLYMA13G22430.11 | 1.506839 | Protein tyrosine kinase |
| GLYMA17G14990.2 | 2.101687 | Protein tyrosine kinase |
In addition, 23 genes were identified encoding transcription factors (TFs) (Table 2). These genes were grouped into major groups. The first group contained one auxin response factor (ARF) gene down-regulated (Glyma18G18450.1) in the drought-tolerant genotype. The second group was composed of zinc-finger protein family genes, containing five members (Glyma07G12680.1, Glyma08G03140.1, Glyma05G22440.1, Glyma01G00500.1, Glyma17G15100.1) which were induced by dehydration, and three (Glyma06G19660.2, Glyma12G17890.1, Glyma04G06660.1) were suppressed. The third group was constituted by five MYB family genes down-regulated (Glyma08G02080.1, Glyma19G22220.1, Glyma11G18340.1, Glyma15G14190.1, Glyma12G18470.1). The fourth group consisted of ring-finger family genes; two members (Glyma20G07670.1, Glyma04G03980.4) were induced by dehydration in leaves, and one (Glyma10G12460.4) was suppressed. The fifth group consisted of heat-shock factors (HSFs); two members were induced (Glyma01G21740.1, Glyma09G19060.1) and two suppressed (Glyma08G02590.1, Glyma08G15850.1) by water-deficit. The remaining TF genes encoded members of families AP2/EREBP (Glyma16G01260.4) and NAC domain protein (Glyma11G18200.1).
Table 2.
Transcription factors responsive to dehydration only in the drought-tolerant genotype
| Transcription factors (TFS) | ||
|---|---|---|
| Gene | Log2 ratio | Annotation |
| AUXIN-RELATED PROTEIN | ||
| GLYMA18G18450.1 | − 0.754799159 | Auxin response factor |
| ZINC FINGER PROTEIN | ||
| GLYMA07G12680.1 | 1.293328 | CCCH Zinc finger protein |
| GLYMA08G03140.1 | 1.437808 | CCCH Zinc finger protein |
| GLYMA06G19660.2 | − 0.967052969 | C3HC4 Zinc finger protein |
| GLYMA12G17890.1 | − 1.279118701 | C2H2 Zinc finger protein |
| GLYMA04G06660.1 | − 2.353948183 | C2H2 zinc finger protein |
| GLYMA05G22440.1 | 1.214217 | CCCH Zinc finger protein |
| GLYMA01G00500.1 | 1.406536 | CCCH Zinc finger protein |
| GLYMA17G15100.1 | 1.06469 | C2C2 Zinc-finger of the FCS-type |
| MYB TRANSCRIPTION FACTOR FAMILY | ||
| GLYMA08G02080.1 | − 3.100653512 | Leucine Rich Repeat (LRR_1)-MYB-LIKE DNA-BINDING PROTEIN MYB // ATMYB103 |
| GLYMA19G22220.1 | − 1.554766923 | MYB transcription factor |
| GLYMA11G18340.1 | − 0.993510023 | MYB transcription fator-MYB 2 |
| GLYMA15G14190.1 | − 0.599749666 | MYB |
| GLYMA12G18470.1 | − 1.655029264 | MYB transcription fator |
| RING-H2 PROTEIN | ||
| GLYMA20G07670.1 | 2.672113 | Ring finger domain |
| GLYMA04G03980.4 | 2.921892 | Ring finger domain |
| GLYMA10G12460.4 | − 2.716919566 | Ring finger domain |
| HEAT SHOCK PROTEIN | ||
| GLYMA08G02590.1 | − 0.507591383 | Heat shock protein 70 kDa |
| GLYMA08G15850.1 | − 0.745310757 | Small heat-shock protein 20 KDa |
| GLYMA01G21740.1 | 1.480779 | Heat stress transcription factor B-2B |
| GLYMA09G19060.1 | 1.187051 | Heat stress transcription factor C-1 |
| AP2/EREBP FAMILY | ||
| GLYMA16G01260.4 | − 0.946836287 | AP2 domain |
| NAC FAMILY | ||
| GLYMA11G18200.1 | 1.480184 | NAC domain protein 61 |
We found that some genes that code for proteins involved in cell wall dynamics were differentially expressed for both genotypes, which appear to be regulated by drought stress (Table 3). The genes related to the metabolism of Rhamnogalacturonan were only expressed in the tolerant genotype (Glyma08G09360.1, Glyma15G07400.1, Glyma05G22440.1). Two expansion protein genes (Glyma05G06580.1, Glyma20G03390.1) and two responsive xyloglucans transferase genes (Glyma10G15100.1, Glyma05G13870.1) were expressed in both genotypes. Two glycosidases responsive (Glyma10G20000.1, Glyma03G10450.1) were only expressed in the tolerant genotype. A synthase-like D3 Cellulose (Glyma01G23250.1) overexposed in the sensitive genotype and the pectinesterase inhibitor (glyma08G14790.1) down-regulated in the tolerant genotype and up-regulated in the susceptible.
Table 3.
Genes coding for proteins involved in cell wall dynamics differentially expressed for both genotypes
| Gene | Protein | BR 16-Log2 | EMBRAPA 48-Log2 |
|---|---|---|---|
| GLYMA10G15100.1 | Xylosyltransferase MGP4 | 3.473208 | 1.975452 |
| GLYMA08G09360.1 | Rhamnogalacturonan xylosyltransferase 1 (RGXT1) | NF | 2.308536 |
| GLYMA15G07400.1 | Rhamnogalacturonan xylosyltransferase 2 (RGXT2) | NF | 2.397853 |
| GLYMA05G22440.1 | Rhamnogalacturonan specific Xylosyltransferase 1 (RGTX3) | NF | 1.214217 |
| GLYMA05G13870.1 | Xyloglucan endotransglucosylase 27 | 3.006961 | 1.068432 |
| GLYMA10G20000.1 | UDP-Glycosyltransferase superfamily protein | NF | 3.68817 |
| GLYMA03G10450.1 | Hydroquinone glucosyltransferase | NF | 1.567052 |
| GLYMA08G14790.1 | Pectinesterase inhibitor 51 | 1.29512 | -2.04434 |
| GLYMA05G06580.1 | Expansin-like B1 | 8.871832 | 3.815932 |
| GLYMA20G03390.1 | Expansin -A14 | 6.47856 | NF |
| GLYMA01G23250.1 | Cellulose Synthase-like D3 | 2.404065 | NF |
Functional classification of differentially expressed genes
We used the enrichment analysis of DEGs based on up- and down-regulated genes, performed by Cytoscape plug-in ClueGO, which identified significantly over-represented enrichment networks present in both genotypes for the drought treatment (Figs. 3, 4). When the lists of functional categories and networks produced by ClueGO for each genotype were verified, we observed similarities, despite the number of genes grouped in the categories to be very small for tolerant Embrapa 48. This lower number was justified by lower number of dysregulated genes in the tolerant genotype, as describe before. However, some categories and genes were highlighted or present only for Embrapa 48.
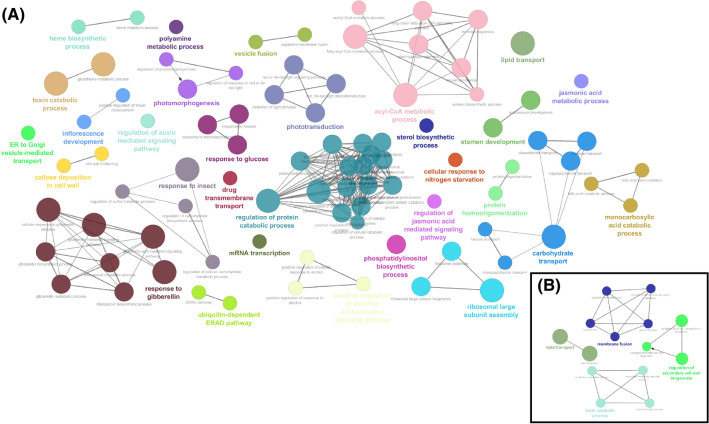
Fig. 3.
Over-representation analysis of down-regulated genes using the Gene Ontology biological process database. In a clusters containing down-regulated genes in the sensitive genotype BR16. In b clusters containing down-regulated genes in the tolerant genotype EMBRAPA 48. The size of the node represents the integration of genes and the thickness of the edge shows a significant Kappa value
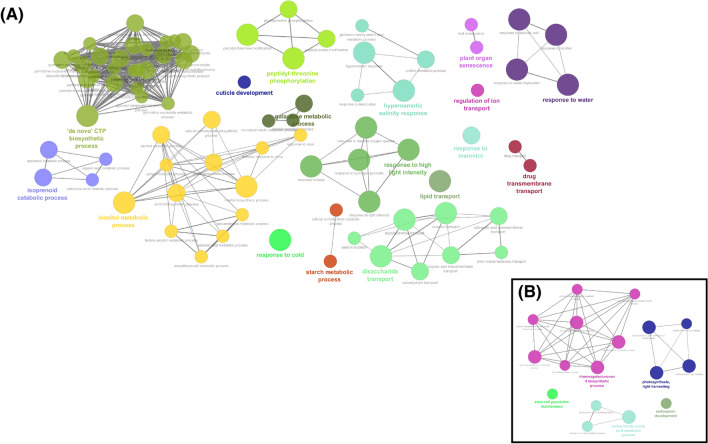
Fig. 4.
Over-representation analysis of up-regulated genes using the Gene Ontology biological process database. In a clusters containing up-regulated genes in the sensitive genotypeBR16. In b clusters containing up-regulated genes in the tolerant genotype EMBRAPA 48. The size of the node represents the integration of genes and the thickness of the edge shows a significant Kappa value
The down-regulated genes from the sensitive genotype BR16 showed clusters relating to biological processes, such as regulation of protein catabolic, gibberellin-responsive, acyl-CoA metabolic process, response to glucose, carbohydrate and lipid transport, proteolysis, response to red or far red light, stamen filament development, among others (Fig. 3a). However, for Embrapa 48 (Fig. 3b), the down-regulated genes showed distinct clusters related to lipid transport, membrane fusion, toxin catabolic process, and regulation of secondary cell wall biogenesis.
Analysis applied for up-regulated genes also showed distinct results for the genotypes. Notably, we observed in the sensitive BR16 under drought the predominance of clusters related to the amino acid catabolic process, alcohol biosynthetic process, response to monosaccharide stimulus, hormone signaling pathway, monocarboxylic acid metabolic process, nucleotide salvage (Fig. 4A). For the tolerant genotype, the up-regulation of the DEGs was mainly corresponded to the pathways rhamnogalacturonan II biosynthetic process, endosperm development, serine family amino acid metabolic process as well as photosynthesis and light-harvesting (Fig. 4b).
Drought-stress assays
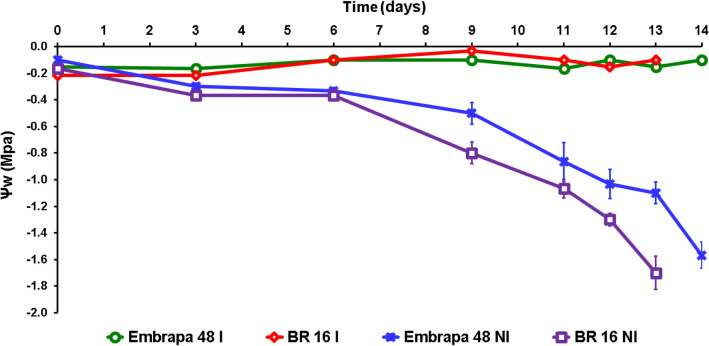
After the irrigation was interrupted, a reduction on ψw was observed. As expected, the decrease on ψw was more noticeable in sensitive BR 16 plants (Fig. 5). The BR 16 plants reached ψw values of − 1.0 MPa on the eleventh day, while the Embrapa 48 plants reached the same ψw levels on the thirteen day after irrigation suspension. These results were consistent with a more efficient water use by cultivar Embrapa 48 and confirmed this cultivar as drought-tolerant in accordance with Lima et al. (2019) and Mesquita et al. (2020).
Fig. 5.
Temporal profile of leaf pre-dawn water potential (ψw) for two soybean cultivars, sensitive (BR 16), and tolerant (Embrapa 48). Each point represents the mean ± standard error (n = 5, where n represents the number of plants), IR irrigated, NI non-irrigated treatments
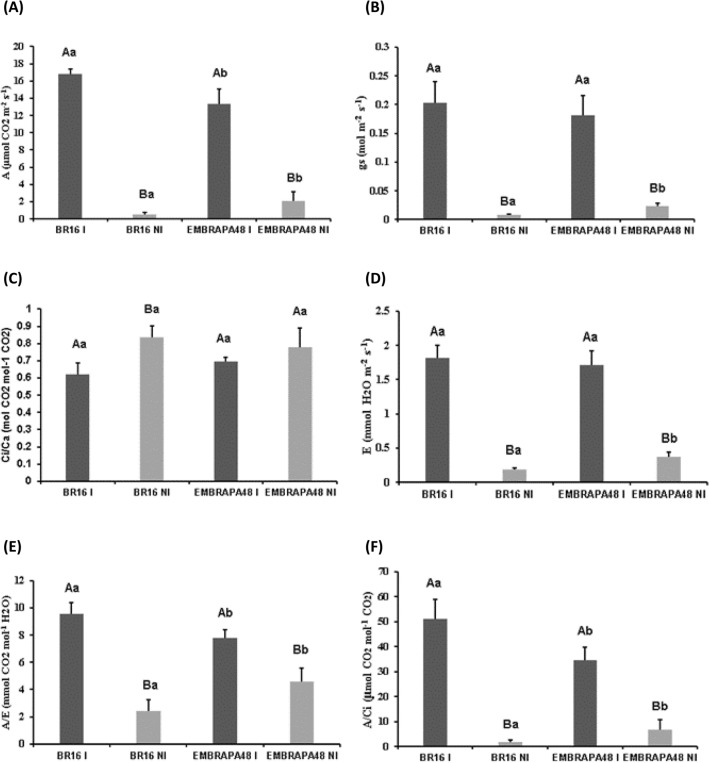
Gas exchange and photosynthetic parameters were evaluated (Fig. 6) and also were in accordance with Mesquita et al. (2020). The net photosynthetic rate (A) decreased for both cultivars under water-deficit; however, the Embrapa 48 maintained higher net photosynthetic rate than BR 16 (Fig. 6a). The stomatal conductance (gs) showed also higher values in Embrapa 48 (Fig. 6b) while the ratio between internal and external CO2 (Ci/Ca) concentrations showed slight differences between genotypes (Fig. 6c). The transpiration rate (E) was reduced in both cultivars when water-deficit was imposed, however was significantly higher in the Embrapa 48 cultivar (Fig. 6d). The proportional decrease in E compared to A was greater in Embrapa 48 plants under irrigation and drought conditions, contributing to a greater instantaneous water use efficiency (A/E) in the tolerant genotype (Fig. 6e). The same behavior was also verified for the A/Ci ratio (carboxylation efficiency) being higher in Embrapa 48 (Fig. 6f) under irrigation and drought conditions. In accordance with Mesquita et al. (2020), these results suggest a greater carboxylation efficiency in the tolerant cultivar, associated with its greater photosynthetic capacity.
Fig. 6.
Effect of water-deficit on the a assimilation rate of CO2 (A), in b stomatal conductance (gs), in c ratio Ci/Ca, in d transpiratory rate E, in e water use efficiency (WUEi) as A/E and in f carboxylation efficiency as A/Ci. IR irrigated, NI non-irrigated treatments. Each bar represents the mean + standard error (n = 5, where n represents the number of plants, t test p < 0.5). Different lower case letters indicate significant differences between averages of the same treatment in different cultivars, and capital letters show significant differences between averages within the same cultivar under different treatments
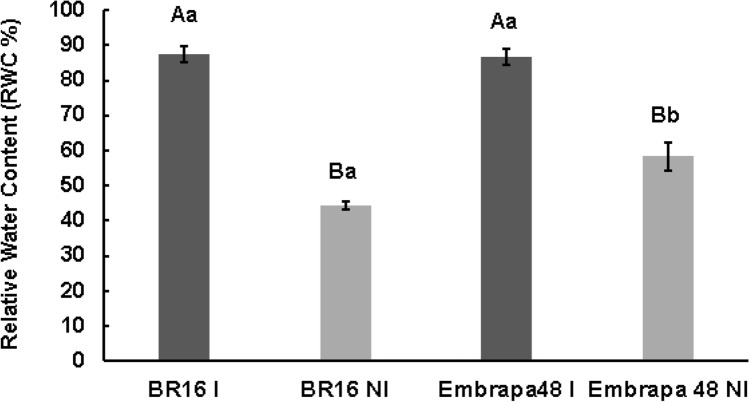
In conditions of stress due to water-deficit, the decrease in stomatal conductance (gs) due to stomata closure leads to a drop in water flow, loss of hydraulic load, resulting from cavitation of the xylem and, ultimately, a drop in hydraulic conductivity. The decrease in water flow through the intercellular spaces and the transpiration through the stomates correlated with the drop in water potential. Thus, we studied the correlation between photosynthetic and hydraulic parameters in the soybean genotypes under conditions of normal irrigation and water-deficit. Leaf hydraulic conductance (Kf) was estimated from the leaf tissues during gas exchanges. As expected, the Kf was abruptly reduced under drought stress. However, the hydraulic conductivities were higher in the Embrapa 48 tolerant genotype under irrigation and drought conditions (Fig. 7a, b). These results are consistent with a better water-absorption efficiency in the Embrapa48 genotype (Lima et al. 2019; Mesquita et al. 2020) and were confirmed by leaf relative water contents (RWC). RWC values decreased with the progression of the stress in both genotypes (Fig. 8). In the leaves under ψw = − 1.0 MPa, the RWC declined in BR16 by 49.14% while for Embrapa genotype, the reduction in the RWC was only of 32.67% (Fig. 8).
Fig. 7.
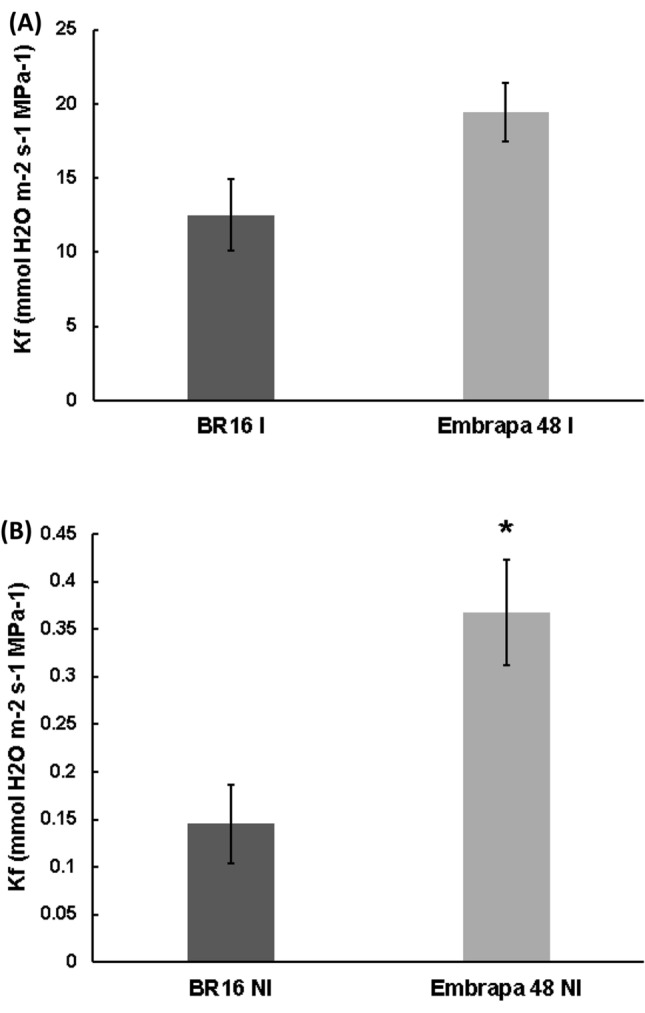
Leaf hydraulic conductance (Kf) of the soybean genotypes. Each bar represents the mean ± standard error (n = 5, where n represents the number of plants, t test p < 0.5). In a IR for irrigated and in b for NI non-irrigated treatments
Fig. 8.
Relative water content RWC (%) of the leaves of BR16 and Embrapa 48 genotypes under different water potential. Bars represent ± standard error (n = 5, where n represents the number of plants, t test p < 0.5). Different lower case letters indicate significant differences between averages of the same treatment in different cultivars, and capital letters show significant differences between averages within the same cultivar under different treatments. IR irrigated, NI non-irrigated treatments
Discussion
The early events of plant responses to drought stress are signal perception and subsequent signal transduction, which lead to the activation of various molecular, biochemical and physiological changes (Rejeb et al. 2014; Joshi et al. 2016). With the availability of genomic sequences from various plant species and the recent advances in sequencing technologies, the genes involved in drought/dehydration responses have been identified in a number of plant species, such as Arabidopsis (De Oliveira et al. 2011; Borkotoky et al. 2013; Shariatipour and Heidari 2018), and crops, such as rice and soybean (Prabha et al. 2011; Nakashima et al. 2014; Zhu et al. 2016; Sahebi et al. 2018). Thus, knowledge on gene expression reprogramming in response to drought stress has been obtained thoroughly. However, identifying the genes that contribute the most to the physiological and molecular adaptation mechanisms is a challenge.
In this study, we focused on two soybean genotypes that share a common ancestor (Davis genotype). The general response of Embrapa 48, when compared to the sensitive BR16, showed a very distinct physiological behavior (Oya et al. 2004; Carvalho et al. 2015; Mesquita et al. 2020) and a molecular response (Lima et al. 2019). The overriding feature observed in the gene expression response of BR16 genotype was the transcriptional induction of a relatively large number of genes. As this genotype is drought-sensitive, this large number of differentially expressed genes is in accordance with the other RNAseq studies in other plants (Yates et al. 2014; Fracasso et al. 2016; Yang et al. 2017), showing that sensitive plants dramatically reprogram gene expression under drought stress. This could be explained by the fact that sensitive species undergo greater changes in physiological and biochemical when mitigating the effects of stress conditions, as also observed by Lima et al. (2019) and Mesquita et al. (2020). On the other hand, the drought-tolerant genotype, Embrapa 48, showed a low alteration of gene expression under drought stress, as indicated by the notably lower number of identified DEGs. This general behavior was also observed in the proteomic and metabolomic data (Lima et al. 2019). Thus, the tolerance may reflect a lower level of stress when compared to BR16 and as a consequence result in a reduced reprogramming of the transcriptome (Fig. 1). These data corroborate with those obtained by Rodrigues et al. (2012), who used the Suppressive Subtractive Hybridization (SSH) technique to investigate differentially expressed genes under water-deficit conditions in these genotypes. This “more subtle” response is probably due to differently expressed genes observed in Embrapa 48, when comparing the genetic background among genotypes (Supplementary Information Fig. S3). Gene expression for Transcriptome study performed by Janiak et al. (2018) reveals that drought tolerance in barley may be attributed to stressed-like expression patterns that exist before the occurrence of stress. Our results suggest that drought stress and pathway activation may vary considerably between the two genotypes and involves genes that are expressed even before the onset of drought treatment in the genotype EMBRAPA48, as differentially expressed genes in genetic background by that participate in the pathways of activation of cellular catabolic process, organic cation transport, dicarboxylic acid biosynthetic process, diterpenoid biosynthetic process and responsive to far red light. Yet, these processes may be responsible for the highest photosynthetic rate, stomatal conductance and carboxylation, observed in this genotype even before stress (Mesquita et al. 2020). In fact, gene expression analyses by RNAseq in the present study and by proteomic profiles (Lima et al. 2019) are in accordance with activities for enzymes involved in antioxidant defenses that were higher in the sensitive genotype BR16 (Mesquita et al. 2020). Moreover, levels of oxidative damage (lipid peroxidation) and activity of antioxidant enzymes under water-deficit were always higher in leaves of BR16 and confirmed by a stronger DAB staining in the leaves BR16 compared with Embrapa48 plants (Mesquita et al. 2020).
The ability to tolerate a water-deficit is a complex trait that could be controlled by many genes (Molina et al. 2008; Ergen and Budak 2009). In this context, plant cells detect stress stimulus through sensors or receptors that activate second messengers and initiate the corresponding signaling pathways to transduce the signals (Bhargava and Sawant 2013). In this study, we focused on the gene expression patterns that were distinct between contrasting genotypes aiming to understand the physiological behavior and identify specific candidates that correlate with drought tolerance.
Genes that stimulate the plant to survive better in drought conditions play a role in the regulatory network of gene expression, including several kinase proteins and transcription factors. The higher levels of phytohormone ABA and proline were observed for the sensitive BR 16 in accordance with a more pronounced perturbation in the metabolic pathways under drought for this genotype (Lima et al. 2019). However, when in stress conditions, the genotype Embrapa 48 showed an less changed metabolism, with higher photosynthetic rate, less oxidative damage in leaves and greater root growth compared to the genotype BR 16, indicating different signaling and regulation of the metabolism of the soybean leaves (Mesquita et al. 2020). Three calcium-dependent protein kinases (CDPKs) were up-regulated, and mainly function in the abscisic acid (ABA) signaling pathway and are plant-specific calcium sensors that play important roles in various aspects of plant physiology (Yang et al. 2011; Huang et al. 2012). Several works report that transcript levels of CDPKs are highly induced by drought, suggesting their important roles during abiotic stress responses in soybean (Hettenhausen et al. 2016), especially in the modulation of ABA signaling to reduce the reactive oxygen species (ROS) (Asano et al. 2012; Neto et al. 2013). Other kinases differentially expressed in the tolerant soybean Embrapa 48 have not been associated to drought response so far.
In recent years, a wide range of TF families holding relevance in drought stress response have been identified, such as AREB, DREB, MYB, WRKY, NAC, ZFP and bZIP (Golldack et al. 2011; Jin et al. 2014; Anbazhagan et al. 2015). Genes that encode C3HC4 and C2H2-type Zinc finger proteins were down-regulated in Embrapa 48 under drought stress (Table 2). Zhang et al. (2016) reported that the family C2H2-type Zinc finger protein negatively regulates the drought response in transgenic Arabidopsis, because the plants, overexpressing these genes, might lose large volumes of water by increasing the width/length and number of completely open stoma, leading to drought stress sensitivity. Most of the family CCCH-type zinc finger proteins have shown up-regulation, which has been associated with RNA metabolism by directly binding to RNA targets and have been involved in abiotic and biotic stresses. Studies have indicated that CCCH zinc finger proteins are associated with senescence delaying effect, and they can interact with ABA and drought response regulators (Jan et al. 2013; Bogamuwa and Jang 2016; Chen et al. 2019). In summary, the higher expression of these CCCH-type zinc finger proteins and CDPKs genes in leaves of the tolerant cultivar may be responsible for the lower production of reactive oxygen species and, consequently, less cell damage, as observed in the ROS in leaf assay conducted by Mesquita et al. (2020).
Other TFs, such as the MYB and NAC, were also characterized for their role in stomatal movement controlling pore closure or in inhibiting its opening (Cominelli et al. 2005; Baldoni et al. 2015). Soybean MYB genes may contribute to the coordination of both cellulose and lignin biosynthesis in secondary wall formation. Yang et al. (2017) showed that plants engineered to accumulate less lignin or xylan are more tolerant to drought and activate drought responses faster than control plants. This is an important finding because it demonstrates that modification of cell walls must occur in the primary wall, and the analyses showed a low expression of secondary wall biosynthesis genes in the stress-tolerant genotype. In addition, the soybean orthologue coding Cellulose Synthase-like D3 was up-regulated only in the sensitive genotype and not altered in tolerant Embrapa 48. Evidence suggests that C2H2 transcription factors are also involved in the secondary metabolism and cell wall structure (Rao and Dixon 2018).
We found that some genes that code for proteins involved in cell wall dynamics were differentially expressed for both genotypes. Xyloglucan endotransglucosylase/ hydrolase (XTH) and Expansin are cell wall proteins involved in cell wall extension, which appear to be regulated by drought stress as observed for soybean genotypes. Some xyloglucan transferase and glycosidase were also responsive in both genotypes; however, some proteins involved in pectin metabolism were up-regulated in the tolerant genotype Embrapa 48 (Table 3). The major group of polymers in primary dicot cell walls are pectins, a heterogeneous group of homogalacturonic acid, rhamnogalacturonan I (RG-I) and rhamnogalacturonan II (RG-II) (Mohnen 2008). Pectins are often modified in plants exposed to drought, facilitating an increase in cell wall plasticity that can contribute to the maintenance of cell turgor or symplast volume (De Diego et al. 2013; Martínez et al. 2007). This plasticity can be correlated with drought tolerance mainly by increasing side chains of the pectic polymers rhamnogalacturonan I and II (RGI and RGII), possibly because the pectin form hydrated gels, which limit the damage to cells (Leucci et al. 2008). Despite its highly complex structure, RG-II is evolutionarily conserved in the plant kingdom as its present in the primary cell wall of all higher plants (O'Neill et al. 1996; Kobayashi et al. 1996). RG-II biosynthesis is a complex process and it is involved in several glycosyltransferases (GTs). One α3-xylosyltransferase (α3-XylT), named RGXT, was able to transfer a xylose residue onto the fucose of the side chain. The Arabidopsis RGXT family has four members linked to RG-II synthesis (Egelund et al. 2006; Liu et al. 2011). Three soybean orthologous genes RGXT1, RGXT2 and RGXT3 were up-regulated only in the genotype Embrapa 48.
An increase in cell wall elasticity can contribute to the maintenance of cell turgor. These results are an indication that the cell wall molecular modifications on the tolerant genotype could contribute to a more efficient water use observed in Embrapa 48. Only in drought-tolerant wheat genotypes, the side chains of rhamnogalacturonan I and II significantly increased in response to water stress (Leucci et al. 2008). The results confirm the role of the pectic side chains during water stress response. In addition, in this study, we also found a gene encoding for pectinesterase inhibitor differentially expressed between genotypes. Pectin is converted by the pectin methylesterase (PME) in pectate and methanol. PME activity is regulated by inhibitor proteins known as the pectin methylesterase inhibitor (PMEI), which plays a key role in wounding, osmotic stress, senescence and seed development. A gene coding for a Pectinesterase inhibitor 51 was down-regulated in tolerant Embrapa 48 and up-regulated in the sensitive genotype. These results indicate that the metabolism of pectin is differently modulated in response to drought in soybean and may play a role in the plants defense mechanism against water-deficit, through the increase of elasticities and crosslink of the cell wall. Interestingly, the amount of side chains of RGI and/or RGII has been crucial to determine the hydration status of the cell wall matrix (Gall et al. 2015). The comparison with tolerant wheat genotypes indicates an increase in the amount of side chains during water stress, which consequently affects the viscosity status of the cell wall (Piro et al. 2003; Leucci et al. 2008; Gall et al. 2015). In fact, changes in the cell wall in response to abiotic stress, such as drought and cold, have been verified and involve a improve the viscoelastic properties of the primary wall. This is due to increases in the levels of cell wall remodeling and biosynthesis enzymes, as well as by modulating other wall loosening agents, including pectin. Thus contributing to increasing the hydration status of the plant and maintaining turgor pressure for growth (Gall et al. 2015).
The fact that the tolerant genotype leaves were more hydrated under the same water potentials suggests a possible osmotic adjustment; however, the higher levels ABA and proline were observed in the sensitive BR 16 leaves (Lima et al. 2019). Thus, the signal for drought by ABA and proline was more noticeable in the sensitive BR 16. In the same way, amino acids and sugar were more abundant during drought in the sensitive genotype (Lima et al. 2019), which suggests that these compounds were not important for the osmoprotection in the tolerant genotype. Furthermore, we observed also evidences suggesting the participation of a non-stomatal event in the relative drought tolerance of the Embrapa 48 and even though under severe stress, showing lower alteration on the net photosynthesis (Mesquita et al. 2020). Thus, the postponement of water and physiological response suggests that differential hydraulic conductivity may be important to this tolerance. In fact, the relative water content (RWC%) and hydraulic conductance (Kf) were higher in tolerant genotype Embrapa 48. Therefore, the maintenance of higher water content in the leaves in the tolerant cultivar could explain, at least partially, the greater photosynthetic rate of this cultivar.
Conclusion
Drought tolerance in plants is performed by different complex mechanisms, and to evaluate which gene is determinant for this phenotype is a challenge, especially because an extensive gene reprogramming is activated. However, we have used two soybean parental genotypes to investigate the molecular responses under drought stress. Although many genes showed similar gene expression patterns in both genotypes, genes involved in the signal transduction cascades and regulation of gene expression, such as kinase of the family CDPKS and TFs of the family C2H2 and CCCH, were only differentially expressed in the drought-tolerant genotype. The global transcriptomic study also showed that drought tolerance is operating even before the occurrence of stress and makes the plant ready to respond to adverse environmental conditions. This behavior was confirmed by lower genetic reprogramming in the Embrapa 48 genotype when subjected to drought. Physiological traits combined with proteomic and metabolomic profiles are in accordance with gene expression analysis by RNAseq. The drought-tolerance mechanism of the Embrapa 48 genotype involves in an increase in cell wall elasticity and hydraulic conductance contributes to the maintenance of cell turgor, resulting in the highest leaf RWC, photosynthetic rate (A), transpiration (E) and carboxylation (A/Ci) under conditions of water stress (Mesquita et al. 2020). These genetic traits contribute to maintain higher growth of the Embrapa 48 soybean plants under drought conditions (Mesquita et al. 2020).
Gene expression regulation analysis indicated that cell wall metabolism was changed in this genotype and could be correlated with the more efficient water use. Remodeling of the pectin component of the cell wall may be an important mechanism for the drought tolerance in the Embrapa 48 soybean genotype, promoting a differential hydraulic conductivity and a higher relative water content (RWC%).
Supplementary Information
Below is the link to the electronic supplementary material.
Figure S1. Overview of the workflow for analysis of RNA-Seq data. Transcriptomes of three biological replicates of two soybean genotypes, BR 16 and EMBRAPA 48, sensitive and tolerant to drought, respectively, were isolated from leaf tissue for and sequenced on the Illumina Hi-Seq 2500 (Illumina, San Diego, CA). The initial sample collected after water deficit was designated as “NI”, and control plants “IR”. Curated RNA sequence data was quality filtered using Trimmometic tool. High quality raw sequence reads were processed and pseudo aligned to soybean reference transcriptome using Kallisto and output collected was submitted to the DESEq2 for differential expression analysis. Finally, the differential genes expression was submitted to analyzes of prediction and annotation of genes (DOCX 724 kb)
Figure S2. Heatmap generated with DeSeq2 software packages showing the Euclidean distances between the samples. 16IRP- genotype BR 16 irrigated, 16NIP-genotype BR 16 not irrigated, 48IRP- genotype Embrapa 48 irrigated, 48IRP-genotype Embrapa 48 not irrigated. The numbers represent the biological repetitions (DOCX 1014 kb)
Figure S3. The over-representation analysis of up-regulated genes in EMBRAPA 48 genotype observed when studying the genetic background among genotypes by using the Gene Ontology biological process database. The size of the node represents the integration of genes and the thickness of the edge shows a significant Kappa value (DOCX 837 kb)
Acknowledgements
The authors would like to thank the Núcleo de Análise de Biomoléculas (NuBioMol), the Universidade Federal de Viçosa, MG, Brazil, the Instituto de Biotecnologia Aplicada a Agropecuária (BIOAGRO), the Instituto Nacional de Ciência e Tecnologia em Interações Planta-Praga (INC-TIPP) and the Brazilian Soybean Genome Consortium (GENOSOJA).
Funding
This work was supported by the Coordenação de Aperfeiçoamento de Pessoal de Nível Superior (CAPES), Fundação de Amparo à Pesquisa de Minas Gerais (FAPEMIG) and Conselho Nacional de Desenvolvimento Científico e Tecnológico (CNPq).
Declaration
Conflict of interest
On behalf of all authors, the corresponding author states that there is no conflict of interest.
References
- Aimar D, Calafat M, Andrade AM, Carassay L, Abdala GI, Molas ML (2011) Drought tolerance and stress hormones: from model organisms to forage crops. Intech: from model organisms to forage crops
- Anbazhagan K, Bhatnagar-Mathur P, Vadez V, Dumbala SR, Kishor PB, Sharma KK. Dreb1a overexpression in transgenic chickpea alters key traits influencing plant water budget across water regimes. Plant Cell Rep. 2015;34:199–210. doi: 10.1007/s00299-014-1699-z. [DOI] [PubMed] [Google Scholar]
- Anders S, Huber W (2012) Differential expression of RNA-Seq data at the gene level–the DESeq package. Heidelberg, Germany: European Molecular Biology Laboratory (EMBL)
- Asano T, Hayashi N, Kikuchi S, Ohsugi R. CDPK-mediated abiotic stress signaling. Plant Signal Behav. 2012;7:817–821. doi: 10.4161/psb.20351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baldoni E, Genga A, Cominelli E. Plant MYB transcription factors: their role in drought response mechanisms. Int J Mol Sci. 2015;16:15811–15851. doi: 10.3390/ijms160715811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhargava S, Sawant K. Drought stress adaptation: metabolic adjustment and regulation of gene expression. Plant Breed. 2013;132:21–32. [Google Scholar]
- Bindea G, Mlecnik B, Hackl H, Charoentong P, Tosolini M, Kirilovsky A. ClueGO: a Cytoscape plug-in to decipher functionally grouped gene ontology annotation networks. Bioinformatics. 2009;25:1091–1093. doi: 10.1093/bioinformatics/btp101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bogamuwa S, Jang JC. Plant tandem CCCH zinc finger proteins interact with ABA, drought, and stress response regulators in processing-bodies and stress granules. Plos One. 2016;11(3):e0151574. doi: 10.1371/journal.pone.0151574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolger AM, Lohse M, Usadel B. Trimmomatic: a flexible trimmer for Illu-mina sequence data. Bioinformatics. 2014;30:2114–2120. doi: 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borkotoky S, Saravanan V, Jaiswal A, et al. The Arabidopsis stress responsive gene database. Int J Plant Genom. 2013;2013:949564. doi: 10.1155/2013/949564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bray N, Pimentel H, Melsted P, Pachter L. Near-optimal RNA-Seq quantification. Nat Biotechnol. 2016;34:525–527. doi: 10.1038/nbt.3519. [DOI] [PubMed] [Google Scholar]
- Brodribb TJ, Holbrook NM. Stomatal closure during leaf dehydration, correlation with other leaf physiological traits. Plant Physiol. 2003;132:2166–2173. doi: 10.1104/pp.103.023879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brodribb TJ, Holbrook NM. Declining hydraulic efficiency as transpiring leaves desiccate: two types of response. Plant Cell Environ. 2006;29:2205–2215. doi: 10.1111/j.1365-3040.2006.01594.x. [DOI] [PubMed] [Google Scholar]
- Brown HKA. Transcriptional profiling of mechanically and genetically sink-limited soybeans. Plant Cell Environ. 2017;40:2307–2318. doi: 10.1111/pce.13030. [DOI] [PubMed] [Google Scholar]
- Carvalho JFC, Crusiol LGT, Perini LJ, Sibaldelli RNL, Ferreira LC, Guimarães FCM, Nepomuceno AL, Neumaier N, Farias JRB. Phenotyping soybeans for drought responses using remote sensing techniques and non-destructive physiological analysis. Glob Sci Technol. 2015;8:1–16. [Google Scholar]
- Chen L-M, Fang Y-S, Zhang C-J, Hao Q-N, Cao D, Yuan S-L, Zhou X-A. GmSYP24, a putative syntaxin gene, confers osmotic/drought, salt stress tolerances and ABA signal pathway. Sci Rep. 2019;9(1):5990. doi: 10.1038/s41598-019-42332-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cominelli E, Galbiati M, Vavasseur A, Conti L, Sala T, Vuylsteke M, et al. A guard-cellspecific MYB transcription factor regulates stomatal movements and plant drought tolerance. Curr Biol. 2005;15:1196–1200. doi: 10.1016/j.cub.2005.05.048. [DOI] [PubMed] [Google Scholar]
- De Diego N, Sampedro MC, Barrio RJ, Saiz-Fernandez I, Moncalean P, Lacuesta M. Solute accumulation and elastic modulus changes in six radiata pine breeds exposed to drought. Tree Physiol. 2013;33:69–80. doi: 10.1093/treephys/tps125. [DOI] [PubMed] [Google Scholar]
- De Oliveira TM, Cidade LC, Gesteira AS, Coelho Filho MA, Soares Filho WS, Costa MGC. Analysis of the NAC transcription factor gene family in citrus reveals a novel member involved in multiple abiotic stress responses. Tree Genet Genomes. 2011;7(6):1123–1134. [Google Scholar]
- Egelund J, Petersen BL, Motawia MS, Damager I, Faik A, Olsen CE, et al. Arabidopsis thaliana RGXT1 and RGXT2 encode Golgi-localized (1,3)-alpha-d-xylosyltransferases involved in the synthesis of pectic rhamnogalacturonan-II. Plant Cell. 2006;18:2593–2607. doi: 10.1105/tpc.105.036566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ergen NZ, Budak H. Sequencing over 13,000 expressed sequence tags from six subtractive cDNA libraries of wild and modern wheats following slow drought stress. Plant Cell Environ. 2009;32:220–236. doi: 10.1111/j.1365-3040.2008.01915.x. [DOI] [PubMed] [Google Scholar]
- Fang Y, Xiong L. General mechanisms of drought response and their application in drought resistance improvement in plants. Cell Mol Life Sci. 2015;72:673–689. doi: 10.1007/s00018-014-1767-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fracasso A, Trindade LM, Amaducci S. Drought stress tolerance strategies revealed by RNA-Seq in two sorghum genotypes with contrasting WUE. BMC Plant Biol. 2016;16:115. doi: 10.1186/s12870-016-0800-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gall HL, Philippe F, Domon JM, Gillet F, Pelloux J, Rayon C. Cell wall metabolism in response to abiotic stress. Plants (Basel) 2015;4(1):112–166. doi: 10.3390/plants4010112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Golldack D, Lüking I, Yang O. Plant tolerance to drought and salinity: stress regulating transcription factors and their functional significance in the cellular transcriptional network. Plant Cell Rep. 2011;30:1383–1391. doi: 10.1007/s00299-011-1068-0. [DOI] [PubMed] [Google Scholar]
- Guo P, Baum M, Grando S, Ceccarelli S, Bai G, Li R, et al. Differentially expressed genes between drought-tolerant and drought-sensitive barley genotypes in response to drought stress during the reproductive stage. J Exp Bot. 2009;60:3531–3544. doi: 10.1093/jxb/erp194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hettenhausen C, Sun G, He Y, Zhuang H, Sun T, Qi J, Wu J. Genome-wide identification of calcium-dependent protein kinases in soybean and analyses of their transcriptional responses to insect herbivory and drought stress. Sci Rep. 2016;6:18973. doi: 10.1038/srep18973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirayama T, Shinozaki K. Research on plant abiotic stress responses in the post-genome era: past, present and future. Plant J. 2010;61:1041–1052. doi: 10.1111/j.1365-313X.2010.04124.x. [DOI] [PubMed] [Google Scholar]
- Hu H, Xiong L. Genetic engineering and breeding of drought-resistant crops. Annu Rev Plant Biol. 2014;65:715–741. doi: 10.1146/annurev-arplant-050213-040000. [DOI] [PubMed] [Google Scholar]
- Huang GT, Ma SL, Bai LP, Zhang L, Ma H, Jia P. Signal transduction during cold, salt, and drought stresses in plants. Mol Biol Rep. 2012;39:969–987. doi: 10.1007/s11033-011-0823-1. [DOI] [PubMed] [Google Scholar]
- Jakoby M, Weisshaar B, Droge-Laser W, Vicente-Carbajosa J, Tiedemann J, Kroj T, et al. bZIP transcription factors in Arabidopsis. Trends Plant Sci. 2002;7:106–111. doi: 10.1016/s1360-1385(01)02223-3. [DOI] [PubMed] [Google Scholar]
- Jan A, Maruyama K, Todaka D. OsTZF1, a CCCH-tandem zinc finger protein, confers delayed senescence and stress tolerance in rice by regulating stress-related genes. Plant Physiol. 2013;16:1202–1216. doi: 10.1104/pp.112.205385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janiak A, Kwasniewski M, Sowa M, Gajek K, Żmuda K, Kościelniak J, Szarejko I. No time to waste: transcriptome study reveals that drought tolerance in barley may be attributed to stressed-like expression patterns that exist before the occurrence of stress. Front Plant Sci. 2018;8:2212. doi: 10.3389/fpls.2017.02212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin J, Zhang H, Kong L, Gao G, Luo J (2014) PlantTFDB 3.0: a portal for the functional and evolutionary study of plant transcription factors. Nucleic Acids Res 42:1182–1187 [DOI] [PMC free article] [PubMed]
- Joshi R, Wani SH, Singh B, Bohra A, Dar ZA, Lone AA, Pareek A, Singla-Pareek SL. Transcription factors and plants response to drought stress: current understanding and future directions. Front Plant Sci. 2016;7:1029. doi: 10.3389/fpls.2016.01029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi M, Matoh T, Azuma J. Two chains of rhamnogalacturonan II are cross-linked by borate-diol ester bonds in higher plant cell walls. Plant Physiol. 1996;110:1017–1020. doi: 10.1104/pp.110.3.1017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leucci MR, Lenucci MS, Piro G, Dalessandro G. Water stress and cell wall polysaccharide sinthe apical root zone of wheat cultivars varying in drought tolerance. J Plant Physiol. 2008;165:1168–1180. doi: 10.1016/j.jplph.2007.09.006. [DOI] [PubMed] [Google Scholar]
- Lima LL, Balbi BP, Mesquita RO, Silva JCF, Coutinho FS, Carmo FMS, Vital CE, Mehta A, Fontes EPB, Barros EG, Ramos HJO. Proteomic and metabolomic analysis of a drought tolerant soybean genotype from Brazilian Savanna. Crop Breed Genet Genom. 2019;1:e190022. [Google Scholar]
- Liu XL, Liu L, Niu QK, Xia C, Yang KZ, Li R, et al. Male Gametophyte Defective 4 encodes a rhamnogalacturonan II xylosyltransferase and is important for growth of pollen tubes and roots in Arabidopsis. Plant J. 2011;65:647–660. doi: 10.1111/j.1365-313X.2010.04452.x. [DOI] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(−Delta Delta C(T)) Method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- Martínez JP, Silva H, Ledent JF, Pinto M. Effect of drought stress on the osmotic adjustment, cell wall elasticity and cell volume of six genotypes of common beans (Phaseolus vulgaris L.) Eur J Agron. 2007;26:30–38. [Google Scholar]
- Mesquita RO, Coutinho SC, Vital CE, Nepomuceno AL, Williams TCR, Ramos HJO, Loureiro ME. Physiological approaches to decipher the drought tolerance of a soybean genotype from Brazilian Savana. Plant Physiol Biochem. 2020 doi: 10.1016/j.plaphy.2020.03.004. [DOI] [PubMed] [Google Scholar]
- Mohnen D. Pectin structure and biosynthesis. Curr Opin Plant Biol. 2008;11:266–277. doi: 10.1016/j.pbi.2008.03.006. [DOI] [PubMed] [Google Scholar]
- Molina C, Rotter B, Horres R, Udupa SM, Besser B, Bellarmino L, Baum M, Matsumura H, Terauchi R, Kahl G. SuperSAGE: the drought stress-responsive transcriptome of chickpea roots. BMC Genom. 2008;9:553. doi: 10.1186/1471-2164-9-553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakashima K, Yamaguchi-Shinozaki K, Shinozaki K. The transcriptional regulatory network in the drought response and its crosstalk in abiotic stress responses including drought, cold, and heat. Front Plant Sci. 2014;5:170. doi: 10.3389/fpls.2014.00170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neto JRCF, Pandolfi V, Guimaraes FCM, Benko-Iseppon AM, Romero C, Silva RLO, Rodrigues FA, Abdelnoor RF, Nepomuceno AL, Kido EA. Early transcriptional response of soybean contrasting accessions to root dehydration. PLoS ONE. 2013;8:12. doi: 10.1371/journal.pone.0083466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neves-Borges AC, Guimarães-Dias F, Cruz F, Mesquita RO, Nepomuceno AL, Romano E, Loureiro ME, Grossi-De-Sá MF, Alves-Ferreira M. Expression pattern of drought stress marker genes in soybean roots under two water deficit systems. Genet Mol Biol. 2012;35:212–221. doi: 10.1590/S1415-47572012000200003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Neill MA, Warrenfeltz D, Kates K, Pellerin P, Doco T, Darvill AG, Albersheim P. Rhamnogalacturonan II, a pectic polysaccharide in the walls of growing plant cells, forms a dimer that is covalently cross-linked by a borate ester. J Biol Chem. 1996;271:22923–22930. doi: 10.1074/jbc.271.37.22923. [DOI] [PubMed] [Google Scholar]
- Oya T, Nepomuceno AL, Neumaier N, Farias JRB, Tobita S, Ito O. Drought tolerance characteristics of Brazilian soybean genotypes—evaluation and characterization of drought tolerance of various Brazilian soybean genotypes in the field. Plant Prod Sci. 2004;7:129–137. [Google Scholar]
- Passioura J. The drought environment: physical, biological and agricultural perspectives. J Exp Bot. 2007;58:113–117. doi: 10.1093/jxb/erl212. [DOI] [PubMed] [Google Scholar]
- Piro G, Leucci MR, Waldron K, Dalessandro G. Exposure to water stress causes changes in the biosynthesis of cell wall polysaccharides in roots of wheat genotypes varying in drought tolerance. Plant Sci. 2003;165:559–569. [Google Scholar]
- Prabha R, Ghosh I, Singh DP. Plant stress database: a collection of plant genes responding to stress condition. ARPN J Sci Technol. 2011;1:28–31. [Google Scholar]
- Ranjan A, Sawant S. Genome-wide transcriptomic comparison of cotton (Gossypium herbaceum) leaf and root under drought stress. Biotech. 2015;5:585–596. doi: 10.1007/s13205-014-0257-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rao X, Dixon RA. Current models for transcriptional regulation of secondary cell wall biosynthesis in grasses. Front Plant Sci. 2018;9:399. doi: 10.3389/fpls.2018.00399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rejeb IB, Pastor V, Mauch-Mani B. Plant responses to simultaneous biotic and abiotic stress: molecular mechanisms. Plants (Basel) 2014;3(4):458–475. doi: 10.3390/plants3040458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodrigues FA, Gomes JM, Carvalho JFC, Nascimento LC, Neumaier N, Farias JRB, Carazzolle MF, Marcelino FC, Nepomuceno AL. Subtractive libraries for prospecting differentially expressed genes in the soybean under water deficit. Genet Mol Biol. 2012;35:304–314. doi: 10.1590/S1415-47572012000200011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sack L, Melcher PJ, Zwieniecki MA, Holbrook NM. The hydraulic conductance of the angiosperm leaf lamina: a comparison of three measurement methods. J Exp Bot. 2002;53:2177–2184. doi: 10.1093/jxb/erf069. [DOI] [PubMed] [Google Scholar]
- Sahebi M, Hanafi MM, Rafii MY, Mahmud TMM, Azizi P, Osman M, Abiri R, Taheri S, Kalhori N, Shabanimofrad M, Miah G, Atabaki N. Improvement of drought tolerance in rice (Oryza sativa L.): genetics, genomic tools, and the WRKY gene family. BioMed Res Int. 2018;3:3158474. doi: 10.1155/2018/3158474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sahoo KK, Tripathy AK, Pareek A, Singla-Pareek S. Taming drought stress in rice through genetic engineering and transcription factors and protein kinases. Plant Stress. 2013;1:60–72. [Google Scholar]
- Schmutz J, Cannon SB, Schlueter J, Ma J, Mitros T, Nelson W, Hyten DL, Song Q, Thelen JJ, Cheng J, et al. Genome sequence of the palaeopolyploid soybean. Nature. 2010;463:178–183. doi: 10.1038/nature08670. [DOI] [PubMed] [Google Scholar]
- Scholander PE, Hammel HT, Bradstreet ED, Hemmingsen EA. Sap pressure in vascular plants. Science. 1965;148:339–346. doi: 10.1126/science.148.3668.339. [DOI] [PubMed] [Google Scholar]
- Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13:2498–2504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shariatipour N, Heidari B. Investigation of drought and salinity tolerance related genes and their regulatory mechanisms in Arabidopsis (Arabidopsis thaliana) Open Bioinform J. 2018;11:12–28. [Google Scholar]
- Shinozaki K, Yamaguchi-Shinozaki K. Gene networks involved in drought stress response and tolerance. J Exp Bot. 2007;58:221–227. doi: 10.1093/jxb/erl164. [DOI] [PubMed] [Google Scholar]
- Spinoni J, Vogt JV, Naumann G, Barbosa P, Dosio A. Will drought events become more frequent and severe in Europe? Int J Climatol. 2017;38:1718–1736. [Google Scholar]
- Stanke C, Kerac M, Prudhomme C, Medlock J, Murray V. Health effects of drought: a systematic review of the evidence. PLoS Curr. 2013;5:5. doi: 10.1371/currents.dis.7a2cee9e980f91ad7697b570bcc4b004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Talame V, Ozturk NZ, Bohnert HJ, Tuberosa R. Barley transcript profiles under dehydration shock and drought stress treatments: a comparative analysis. J Exp Bot. 2007;58:229–240. doi: 10.1093/jxb/erl163. [DOI] [PubMed] [Google Scholar]
- Valente MAS, Faria JQA, Ramos JRLS, Reis PAB, Pinheiro GL, Piovesan ND, Morais AT, Menezes CC, Cano MAO, Fietto LG, Loureiro ME, Aragao FJL, Fontes EBP. The ER luminal binding protein (BiP) mediates an increase in drought tolerance in soybean and delays drought-induced leaf senescence in soybean and tobacco. J Exp Bot. 2009;60:533–546. doi: 10.1093/jxb/ern296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L, Feng Z, Wang X, Wang X, Zhang X. DEGseq: an R package for identifying differentially expressed genes from RNA-seq data. Bioinformatics. 2009;26:136–148. doi: 10.1093/bioinformatics/btp612. [DOI] [PubMed] [Google Scholar]
- Yang W, Liu XD, Chi XJ, Wu CA, Li YZ, Song LL. Dwarf apple MbDREB1 enhances plant tolerance to low temperature, drought, and salt stress via both ABA-dependent and ABA-independent pathways. Planta. 2011;233:219–229. doi: 10.1007/s00425-010-1279-6. [DOI] [PubMed] [Google Scholar]
- Yang Z, Dai Z, Lu R, Wu B, Tang O, Xu Y, Cheng C, Su J. Transcriptome analysis of two species of jute in response to polyethylene glycol (PEG)-induced drought stress. Sci Rep. 2017;7:16565. doi: 10.1038/s41598-017-16812-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yates SA, et al. De novo assembly of red clover transcriptome based on RNA-Seq data provides insight into drought response, gene discovery and marker identification. BMC Genom. 2014;15:453. doi: 10.1186/1471-2164-15-453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang D, Tong J, Xu Z, Wei P, Xu L, Wan Q, Huang Y, He X, Yang J, Shao H, Ma H. Soybean C2H2-type zinc finger protein GmZFP3 with conserved QALGGH motif negatively regulates drought responses in transgenic Arabidopsis. Front Plant Sci. 2016;7:325. doi: 10.3389/fpls.2016.00325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu JK. Salt and drought stress signal transduction in plants. Annu Rev Plant Biol. 2002;53:247–273. doi: 10.1146/annurev.arplant.53.091401.143329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu K, et al. Evolution of an intron-poor cluster of the CIPK gene family and expression in response to drought stress in soybean. Sci Rep. 2016;6:28225. doi: 10.1038/srep28225. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. Overview of the workflow for analysis of RNA-Seq data. Transcriptomes of three biological replicates of two soybean genotypes, BR 16 and EMBRAPA 48, sensitive and tolerant to drought, respectively, were isolated from leaf tissue for and sequenced on the Illumina Hi-Seq 2500 (Illumina, San Diego, CA). The initial sample collected after water deficit was designated as “NI”, and control plants “IR”. Curated RNA sequence data was quality filtered using Trimmometic tool. High quality raw sequence reads were processed and pseudo aligned to soybean reference transcriptome using Kallisto and output collected was submitted to the DESEq2 for differential expression analysis. Finally, the differential genes expression was submitted to analyzes of prediction and annotation of genes (DOCX 724 kb)
Figure S2. Heatmap generated with DeSeq2 software packages showing the Euclidean distances between the samples. 16IRP- genotype BR 16 irrigated, 16NIP-genotype BR 16 not irrigated, 48IRP- genotype Embrapa 48 irrigated, 48IRP-genotype Embrapa 48 not irrigated. The numbers represent the biological repetitions (DOCX 1014 kb)
Figure S3. The over-representation analysis of up-regulated genes in EMBRAPA 48 genotype observed when studying the genetic background among genotypes by using the Gene Ontology biological process database. The size of the node represents the integration of genes and the thickness of the edge shows a significant Kappa value (DOCX 837 kb)