Abstract
β cells are defined by the ability to produce and secret insulin. Recent studies have evaluated that human pancreatic β cells are heterogeneous and demonstrated the transcript alterations of β cell subpopulation in diabetes. Single-cell RNA sequence (scRNA-seq) analysis helps us to refine the cell types signatures and understand the role of the β cells during metabolic challenges and diseases. Here, we construct the pseudotime trajectory of β cells from publicly available scRNA-seq data in health and type 2 diabetes (T2D) based on highly dispersed and highly expressed genes using Monocle2. We identified three major states including 1) Normal branch, 2) Obesity-like branch and 3) T2D-like branch based on biomarker genes and genes that give rise to bifurcation in the trajectory. β cell function-maintain-related genes, insulin expression-related genes, and T2D-related genes enriched in three branches, respectively. Continuous pseudotime spectrum might suggest that β cells transition among different states. The application of pseudotime analysis is conducted to clarify the different cell states, providing novel insights into the pathology of β cells in T2D.
Supplementary Information
The online version contains supplementary material is available at 10.1007/s43657-021-00024-z.
Keywords: Single-cell RNA seq, Islets, β cell, Pseudotime, T2D
Introduction
The pancreas is a vital metabolic organ consisting largely of exocrine ductal and acinar cells which secret digestive enzymes. As part of the pancreas, the islets of Langerhans are essential for blood glucose homeostasis. The endocrine islets comprise of at least five kinds of endocrine cells: α cells (30–45%), β cells (50–60%), γ/PP cells (less than 10%), δ cells (less than 10%), and ε cells (less than 1%), which secrete glucagon (GCG), insulin (INS), pancreatic polypeptide (PPY), somatostatin (SST) and ghrelin (GHRL), respectively; however, the cell-type composition is variable among individuals (Cabrera et al. 2006). Accumulating evidence suggests islet β cells play an important role in glucose level maintained by stimulating the glucose uptake of peripheral organs (Ashcroft and Rorsman 2012). However, it has been known that β cells are not functionally identical to each other, including the rate of insulin synthesis and secretion.
Type 2 diabetes (T2D) is characterized by increasing insulin resistance in peripheral tissues and decreasing insulin secretion due to the loss of functional β cells (Kahn 2003). According to our understanding, the correlation between T2D and the changes in the transcriptome of pancreatic cells is obtained from whole-islets transcriptome data (Fadista et al. 2014; Taneera et al. 2012), which makes it difficult to clarify cell-type changes. Single-cell transcriptomic analysis of the pancreas has undergone dramatic advances over the past 5 years (Segerstolpe et al. 2016; Xin et al. 2016), of which it is now able to identify the cell-type-specific transcriptomic changes in the development of T2D (Wang and Kaestner 2019). Previous studies have demonstrated the great potential of single-cell RNA sequence (scRNA-seq) in islet biology and confirmed the heterogeneity of human β cells. However, the single-cell analysis represents the dynamic states of β cells rather than stable and distinct disease-related subpopulations (Fang et al. 2019; Xin et al. 2018), which means more efforts are needed for evaluating the transcript alteration of β cell subpopulations in diseases.
In this study, we traced pancreatic cells from publicly available scRNA-Seq data in healthy and T2D status. The β cells were ordered by pseudotime and projected onto a constructed trajectory. The gene expression of ordered cells was used to study dynamic state change. This provides a higher resolution view of the gene expression landscape of β cells in health and T2D. The gene signatures, together with the enriched pathways for each state and inter-state are described.
Materials and Methods
Single Cell RNA-seq Data Acquisition
In this study, we analyzed scRNA-seq data of the human pancreas obtained from the NCBI GEO DataSets (https://www.ncbi.nlm.nih.gov/gds/) with accession id: GSE101207 (Fang et al. 2019). It contains 39,905 pancreatic islet single cells isolated from six healthy and three T2D donors. The donor information is provided in Table 1. The data analysis was conducted by R (R Core Team 2020).
Table 1.
Donor information of GSE101207
| Donor | Gender | Age | BMI | HbA1c (%) | # of cells | # of β cells |
|---|---|---|---|---|---|---|
| Normal1 | Male | 27 | 20.6 | 5.4 | 1455 | 279 |
| Normal2 | Male | 21 | 22.6 | 5.2 | 1206 | 218 |
| Normal3 | Female | 38 | 34.4 | 5.0 | 9409 | 2744 |
| Normal4 | Male | 52 | 22.0 | 5.6 | 6058 | 2358 |
| Normal5 | Male | 28 | 30.8 | 4.9 | 5405 | 1213 |
| Normal6 | Male | 44 | 34.6 | 5.4 | 6969 | 2030 |
| T2D1 | Male | 58 | 39.3 | 8.9 | 1203 | 291 |
| T2D2 | Male | 61 | 28.1 | 5.2 | 1823 | 444 |
| T2D3 | Male | 51 | 35.6 | 7.1 | 6377 | 1715 |
Data Preprocessing and Cluster Identification
The gene expression matrix was converted to Seurat objects using the Seurat R package (Stuart et al. 2019). Cells were removed when the number of detected genes was less than 100. The top 2000 variable genes were used to correlate and integrate data from different individuals. A total of 500 variable genes were used for the principal component analysis. The first 20 principal components were chosen to reduce dimensions. The first two uniform manifold approximation and projection (UMAP) dimensions were used to visualize cell clusters. Cell clusters were identified using FindClusters (10 principal components and 0.15 resolution), and the cell clusters express the same marker genes will be merged. The β cell cluster was used for next-step pseudotime analysis.
Constructing Single-Cell Trajectory in Pseudotime
Pseudo-time analysis was performed using Monocle 2 (Qiu et al. 2017), which utilized reverse graph embedding based on a user-defined gene list to generate a pseudotime plot that can account for both branched and linear differentiation processes. For pseudo-time analysis of the β cells, the raw count data were normalized by estimating the size factors for the trajectory inference. The highly dispersed and highly expressed genes (empirical dispersion/dispersion fit ≥ 1 and mean expression ≥ 0.01) were used to construct pseudotime trajectory (Karmaus et al. 2019). Default values were chosen for parameters of the DDRTree algorithm. To further analyze these branching events, we used Branched Expression Analysis Modeling (BEAM) implemented in Monocle 2. It helps to identify all genes that show significant branch-dependent expression (Qiu et al. 2017). Visualization of the branch-dependent expression patterns as a heatmap was performed using Monocle 2.
Differential Expression and Downstream Analysis
DESeq 2 was used with default settings for differential expression analysis (Love et al. 2014). Top enriched gene sets selected by adjusting p value < 0.005 were taken for downstream analysis. Differential expression genes and branch-dependent genes were analyzed for pathway enrichment with Gene Ontology (GO), Kyoto Encyclopedia of Genes and Genomes (KEGG), Reactome gene sets, canonical pathways, and CORUM through Metascape (Zhou et al. 2019). The adjusted p value was calculated based on the accumulative hypergeometric distribution. Part resulting data was visualized using ggplot2 (Wickham 2016).
Results
Reidentification of Pancreatic Cell Type and Obtain of β Cell Cluster
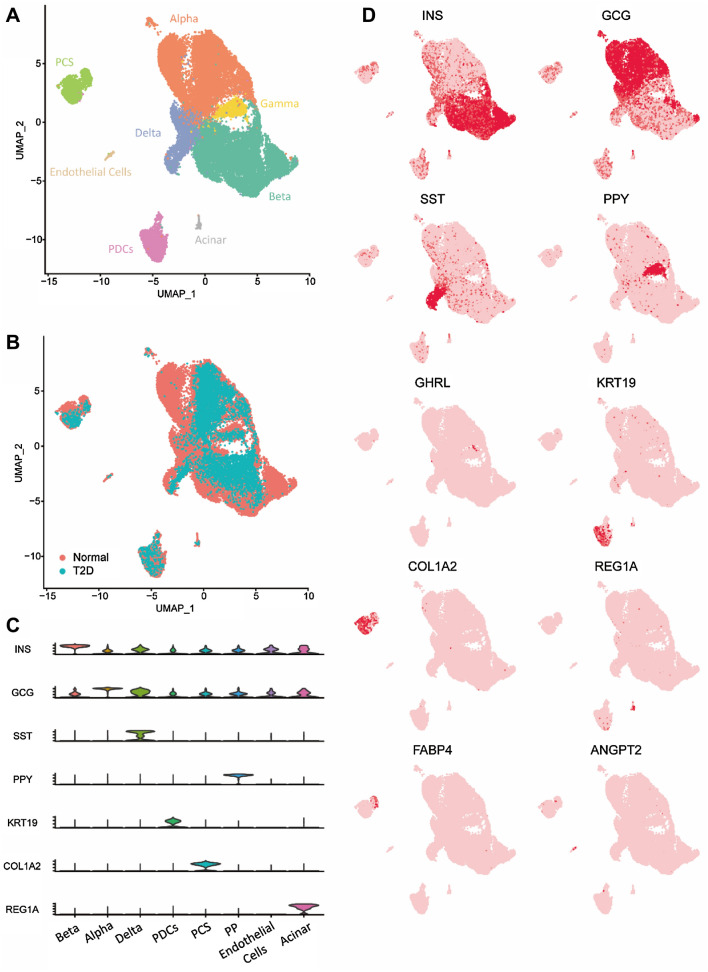
The scRNA-seq data of the islet was obtained from the NCBI (https://www.ncbi.nlm.nih.gov/gds/) with accession id: GSE101207 (Fang et al. 2019). Islets isolated from nine donors in either healthy and T2D status underwent single-cell RNA-seq analysis. We input the read counts matrix into Seurat, after quality control, 30,696 out of 39,905 cells were retained for dimension reduction, as detailed in the Materials and Methods. The cells were divided into eight cell clusters, with different relative unique molecular identifier values per cell (Fig. 1a). When the cells were projected to a two-dimensional uniform manifold approximation and projection (UMAP) plot, the distinction between endocrine cells and non-endocrine cells was observed, which is the same as GSE101207, and homogeneous populations are independent of donor types (Fig. 1b). Using the cell markers in published literatures (Fang et al. 2019; Segerstolpe et al. 2016; Xin et al. 2016), four non-endocrine cells including pancreatic ductal cells (PDCs, n = 1873) marked by several keratin genes (KRTs), pancreatic stellate cells (PSCs, n = 1525) marked by COL1A1, endothelial cells (n = 198) marked by ANGPT2 and acinar cells (n = 143) marked by REG1A were identified (Fig. 1c, d). The differential expression of FABP4, the lipid-processing or adipogenesis gene, shows the heterogeneity of the PSCs cluster. Fang et al. (2019) label the PSCs with high expression of FABP4 as quiescent PSCs, otherwise, as activated PSCs (Fig. 1C). Four endocrine cells were recognized, mainly α cells (n = 11,973), β cells (n = 11,292), γ/PP cells (n = 1121), and δ cells (n = 2571), marked by INS, GCG, PPY, SST, respectively (Fig. 1c, d). Compared with GSE101207, an extra endothelial cell cluster is identified, which might be caused by different criteria of quality control.
Fig. 1.
Single islet cell transcriptomes clustering. a Two-dimensional t-SNE plot of distinct islet cell types. b Two-dimensional t-SNE plots with cells colored based on donor T2D condition. c Violin plots confirm that the marker genes of annotated cell types have mutually exclusive expressions. d Expression levels of cell markers are overlaid onto the t-SNE plot in a
Pseudotime Analysis Clarify Three Branches of β Cell
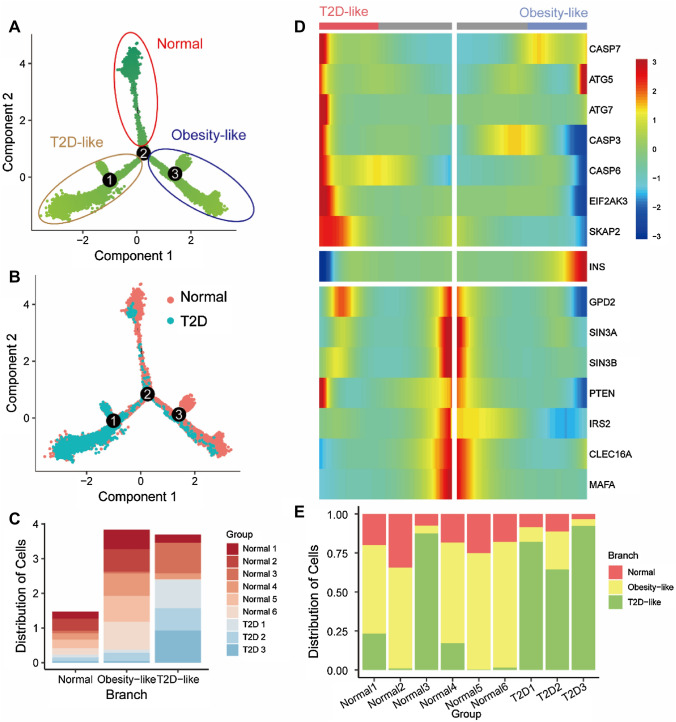
Although cell clustering is helpful to identify cell subtypes, it is difficult to identify the cell subtypes in continuous processes, which can be observed in the two-dimension UMAP plot (Fig. 1a, b), and the asynchrony and heterogeneity of biological samples are not identical with the chronological time of sampling. Therefore, we used a pseudotime analysis to reconstruct the trajectory of β cells to synchronize the process of diabetes. To guide the construction of the trajectory, we used 1138 highly dispersed and expressed genes (mean expression ≥ 0.01 and dispersion empirical ≥ 1 * dispersion fit). Figure 2a shows the trajectory of β cells that bifurcates into two main cell fates on branch point 2. We mapped the cell source label onto the pseudotime trajectory (Fig. 2b). Branch 3 is composed of cells mostly from T2D donors, which means the branch might mostly compose of T2D-like damaged β cells (Fig. 2c). This was confirmed by the expression of T2D-related genes based on the existed researches, and the genes upregulated in the T2D-like branch are linked to autophagy, endoplasmic reticulum stress (ER stress), and apoptosis (Fig. 2d). Autophagy genes (ATGs) encoded multiple proteins to direct the autophagosomes to fuse with lysosomes for cytoplasmic components degradation and recycle. The upregulation of autophagy, particularly the expression levels of Atg5 and Atg7, were reported in ob/ob and High-Fat-Diet (HFD) mice (Yang et al. 2010). Atg7 deficient mice showed impaired glucose tolerance and decreased serum insulin level (Yang et al. 2010). β cell mass and pancreatic insulin content were reduced because of the increased apoptosis and decreased proliferation of β cells (Ebato et al. 2008; Jung et al. 2008). The results identified a unique role of autophagy as an adaptive response of β cells in diabetes. The function of β cells is also influenced by ER stress, in the case of T2D, predominantly mediated PERK–eIF2α (Eukaryotic Translation Initiation Factor 2 Alpha Kinase 3, EIF2AK3) pathways (Eizirik et al. 2020). Collectively, this research highlights the key role of EIF2AK3 in maintaining β cell function. The cysteinyl aspartate protease (CASP) gene family plays a significant role in programmed cell death, and CASP3 and CASP7 from CASP family are also used as the markers for evaluating the apoptosis level in β cells (Good et al. 2019). The upregulation of the CASP gene family means the increase of apoptosis in the T2D-like branch, apoptosis then decreases β cell mass, which increases the stress experienced by the remaining β cells as they try to compensate for the reduced insulin levels and experience increased secretory demand, resulting in more β cell death.
Fig. 2.
Pseudotime analysis identifies three β-cell states. a Pseudotime trajectory was reconstructed in β cells, which contains three states, labeled as normal, obesity-like, and T2D-like branches, respectively. States are circled in different colors. b Pseudotime trajectory with cells colored based on donor T2D condition. c Bar graphs demonstrating the percentage of donors in each branch. d Heat map of differential expressed marker genes between the branches. e Bar graphs demonstrating the percentage of each cell type in each donor
The active autophagy, ER stress, and apoptosis pathway suggest the T2D-like branch, while another two branches composing of cells mostly from normal donors show heterogeneity with each other (Fig. 2d). The branch with high-level INS expression is threefold higher than that in the other one. The high expression of INS means the cells are more specialized in insulin secretion. Upregulation of ATG5 in the high-INS (INShi) branch suggests the high protein burden (Yang et al. 2010). Downregulation in low-INS (INSlo) branch, Swi-independent 3a and 3b (SIN3A and SIN3B) are linked to modulating Ca2+/ion transport, cell survival, vesicle/membrane trafficking, glucose metabolism, and stress responses, and SIN3A and SIN3B deficient reduced islet cell mass at the birth of mice (Yang et al. 2020). The expression of insulin receptor substrate 2 (IRS2) was correlated with β cell mass in HFD-induced insulin resistance (IR) mice (Takamoto et al. 2008), and the Pancreatic β cell-specific IRS2 Knockout (IRS2-KO) mice exhibited a reduction of β cell mass and β cell proliferation rates (Kubota et al. 2004; Lin et al. 2004). These data suggest that IRS2 is critical for regulating β cell mass and function to promote insulin secretion. v-maf musculoaponeurotic fibrosarcoma oncogene homolog A (MAFA, a basic leucine zipper transcription factor) is thought to be critical in the regulation of insulin biosynthesis and secretion (Matsuoka et al. 2004; Wang et al. 2007). Significantly, MAFA was demonstrated to be effectively reprogrammed adult pancreatic acinar cells to islet β-like cells, which not only produced β cell markers but also be similar to endogenous β cells in function and structure (Zhou et al. 2008). The high requirement of insulin processing, maturation, and secretion makes β cells particularly susceptible to mitochondrial dysfunction, which leads to β cell failure in diabetes. Mitophagy is a key component of mitochondrial quality control, which is necessary to maintain mitochondrial function and quality. Correlated to INS expression, C-type lectin domain family 16, member A (CLEC16A) involves the mitochondrial trafficking during mitophagy to maintain mitochondrial function, consequently, maintain insulin secretion of β cell (Sidarala et al. 2020; Soleimanpour et al. 2014). The differential expression of marker genes between two branches shows a high protein synthesis burden of the INShi branch and reminds us of the normal state of the INSlo branch.
Figure 2E shows the distribution of cells from different donors. The different cell composition of female donor Normal 3 (p < 8.252e-5) might suggest the difference of metabolism signature between males and females, which is confirmed in the existed cohort study (Rasouli et al. 2021). The unique cell states of Normal 3 also can be observed in the individual pseudotime analysis (Fig. S1), which might suggest the potential T2D risk of Normal 3.
Cluster Genes with Branch-dependent Differential Expression
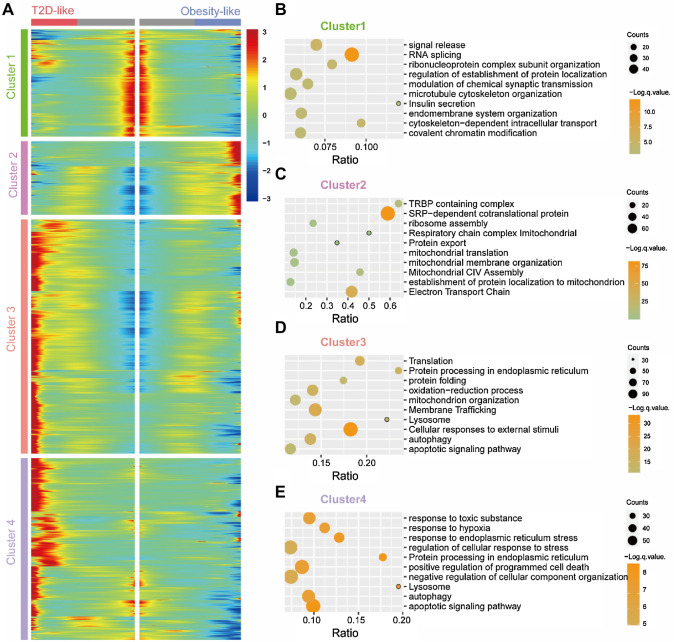
To focus on how fate choices are made among different β cell branches, we performed expression analysis on how genes are differentially regulated after the branch point. In this way, we could identify genes that are specific for β cell fate acquisition. A total of 3094 genes were grouped into four clusters (Fig. 3a). We are interested in the genes differentially expressed in different branches, hence we take them for enrichment analysis. Cluster 1 contains 557 genes upregulated in the INSlo branch that are associated with insulin secretion, endomembrane system, and organelle biogenesis and maintenance (Fig. 3b). Compared to the enriched pathway of genes (n = 381) in cluster 2 upregulated in the T2D-like branch, which comprises responses to external stimuli, autophagy, oxidation stress, lysosome, and apoptosis (Fig. 3c), the enrich pathway of cluster 1 suggests the INSlo branch as a normal-function branch. The cluster 3 genes (n = 1214) upregulated in the INShi branch are linked to protein translation and secretion, mitochondria assembly and electron transport chain (ETC), and ribosome assembly (Fig. 3d). The item ribosome assembly supports the exuberant INS secretion. Mitochondria controls the production of insulin in β cells in an ATP/ADP ratio-dependent manner, and the active mitochondria and ETC increase the ATP/ADP ratio, then insulin is secreted (Rocha et al. 2020). Compared to the expression in the INShi branch, the genes in cluster 4 (n = 942) are highly expressed in the T2D-like branch, and the enrichment analysis reveals the difference in ER stress, apoptosis, glycosylation, and response to hypoxia between the INShi branch and the T2D-like branch (Fig. 3e). Ohtsubo et al. (2011) report the key role of glycosylation in maintaining β cell hemostasis to prevent hyperglycemia, impaired glucose tolerance, and hyperinsulinemia. Hypoxia-inducible factors (HIFs) are a family of transcription factors activated by hypoxia. The deletion of HIF-1α in β cells decreased basal and glucose-stimulated ATP concentrations, and the low ATP generation provides a mechanism for impaired glucose-stimulated insulin secretion (Cheng et al. 2010). Compared to the T2D-like branch, the INShi branch suffers a heavier protein burden, however, the upregulation of mitochondrial and ETC pathway suggests the cells in the INShi branch react to stress normally. The upregulation of INS and molecule enrichment pathways reflect the obesity-like signature of the INShi branch (Alarcon et al. 2016; Kusminski et al. 2020; Sarparanta et al. 2017). Based on the gene expression signatures, we mark the branch with enrichment items related to endomembrane system and insulin secretion as Normal branch, then the one related to mitochondria and ETC as Obesity-like branch, while the one related to apoptotic, ER stress, and autophagy are marked as T2D-like branch (Fig. 2a).
Fig. 3.
Analysis of gene clusters with different expression signatures. a Heatmap showing four-gene clusters with different expression signatures. b–e Pathway enrichment analysis result of differential expressed genes on b cluster 1, c cluster 2, d cluster 3, and e cluster 4
Identification and Comparison of T2D- and Obesity-related Genes
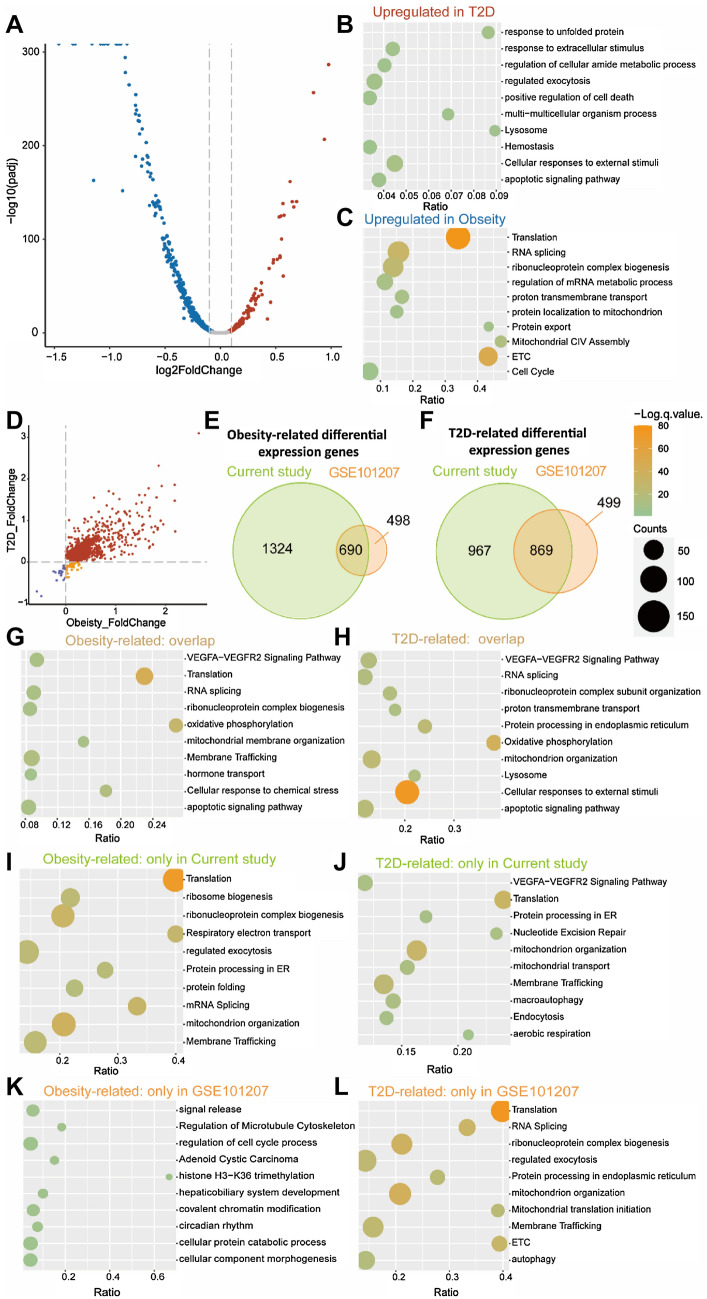
The analysis of differential expression among branches is conducted with R package DEseq2 by input reads count matrix. The differential expression genes between Obesity-like and T2D-like branches indicate the complex transcriptional state of the Obesity-like branch (Fig. 4a). The result of enrichment analysis is related to the genes upregulated in the Obesity-like branch to ETC, mitochondrial membrane potential, and insulin synthesis and secretion (Fig. 4c). Meanwhile, the genes upregulated in T2D-like branch were enriched in Unfolded Protein Response (UPR), ER stress, lysosome, apoptosis, and external stimuli related items (Fig. 4b). The enrichment analysis demonstrated the stress-state of the T2D-like branch, which is consistent with our previous conclusion.
Fig. 4.
Differential expression analysis results compared to the previous study by Fang et al. (2019). a Volcano plot of the genes with differential expression (p < 0.05) between Obesity-like and T2D-like branches. b, c Pathway enrichment analysis of the genes upregulated in T2D-like b and obesity-like c branches. d Scatterplot of the genes with differential expression (p < 0.05) of obesity-related and T2D-related genes. Gray dots indicate genes differentially expressed in the obesity-like branch or T2D-like branch. Red dots indicate genes both significantly upregulated in obesity-like and T2D-like branches. Purple dots indicate genes both significantly downregulated in obesity-like and T2D-like branches. Yellow dots indicate genes significantly downregulated in the obesity-like branch while significantly upregulated in the T2D-like branch. e, f Venn diagram displaying the overlap of the sets of obesity-related e and T2D-related f genes between the current study and the previous study by Fang et al. (2019). g, h Pathway enrichment analysis of the overlap of the obesity-related g and T2D-related h genes between the current study and the previous study by Fang et al. (2019). i, j Pathway enrichment analysis of the obesity-related i and T2D-related j genes in the current study. k, l Pathway enrichment analysis of the obesity-related k and T2D-related l genes in the previous study by Fang et al. (2019)
More upregulated obesity-related genes match with the complex transcriptome of the Obesity-like branch. Some genes are upregulated in the T2D-like branch while downregulated in the Obesity-like branch showing the transcriptional heterogeneity of two states (Fig. 4d). To assess the efficiency of our analysis, we compared our results with T2D-related and obesity-related genes list provided by Fang et al. (2019). The overlap between the sets of differentially expressed genes is displayed in the Venn diagram (Fig. 4e,f). We then applied the genes to pathway enrichment analysis. The obesity-related genes were both expressed differentially in two analyses, which were related with protein synthesis and secretion, mitochondrion and ETC, and apoptotic (Fig. 4g). The similarity in enrichment items indicates the functional consistency of the two gene groups. The genes expressed differentially in our analysis are more relevant with the function of β cell than those in Fang et al. (2019), as shown in Fig. 4f. Enriched translation item was shown in our enrichment result, and Ras homolog enriched in brain (RHEB) is demonstrated to activate the mTORC1 pathway to increase β cell mass and the glucose tolerance of mice (Hamada et al. 2009). The expression of INS is related to the ATP/ADP ratio, which is under the control of mitochondrial. In mitochondrial-related pathways, ATF4 acts as a master regulator of cellular stress, and the mitochondrial dysfunction induces FGF21 in ATF4-dependent manner to increase insulin resistance and adipose tissue browning (Kim et al. 2013). In response to the high protein burden, autophagy protects from obesity and IR. Lysosome-associated membrane protein 2 (LAMP2), which is enriched in the autophagy pathway, is essential for lysosomes formation and autophagy, and the LAMP2 depletion causes cell death and lysosome impairment (Sudhakar et al. 2020). The intermittent fast causes cell death in LAMP2 KO mice also demonstrates the key role of autophagy and lysosome pathways in maintaining β cell homeostasis (Liu et al. 2017).
The overlap of differentially expressed genes between T2D-like and Obesity-like branches similarly shows the enriched pathway items (Fig. 4d, g–l). The enrichment result suggests the important role of translation, mitochondrion and autophagy in obesity and T2D. Compared to the T2D-related genes enrichment items in Fang et al. (2019), the translation item missed (Fig. 4j, l) and linked to insulin secretion was found in the enrichment of obesity-related genes. The difference indicates the complex etiology of T2D, individual heterogeneity, and the expression of insulin. Particularly single-cell insulin expression is not completely correlated to individual T2D, and this is demonstrated by the division of Obesity-like and T2D-like branches in pseudotime analysis. The functional difference between Obesity-like and T2D-like branches might interpret how obese people escape from T2D. However, restricted by individual clinical information, the inner heterogeneity in the individual is ignored in Fang et al. (2019). The pseudotime analysis synchronizes the cells from individuals and contributes to clustering the function-similar cell across individuals, which helps us understand the process of T2D at the single-cell level. The genes of the T2D-like branch also provide insight into the etiology of T2D (Fig. 4j). In the item of vesicle-mediated transport, vesicle-associated membrane protein 8 (VAMP8) is confirmed to play a dual role in the regulation of insulin recruitment and negative regulation of β cell proliferation, of which the precise mechanisms are not completely understood. However, VAMP8 KO mice are reported with increased insulin secretion, which improved metabolism homeostasis (Zhu et al. 2012). In the citric acid (TCA) cycle and respiratory electron transport, while syntaxin-1A (STX1A) regulates insulin secretion through binding to β cell ion channels, and STX1A overexpression causes impaired mice insulin secretion (Lam et al. 2005). The enrichment item VEGFA–VEGFR2 signaling is linked to vascularization, on the one hand, the vascular changes contribute to the onset of diabetes (Staels et al. 2019). Nonetheless, diabetes induces vascular complications via VEGFA-related pathway (Sivaskandarajah et al. 2012; Wirostko et al. 2008). The demonstrated T2D related genes suggest that pseudotime can synchronize samples of asynchrony and provide potential insight into obesity and diabetes.
Discussion
Earlier studies were mainly based on the expression of individual genes in different cell types. However, the expression of individual genes does not provide information on the transcriptional programming of cell types. With the multiple methodologies employed in single-cell analysis, several groups identified subpopulations of β, α, ductal and acinar cells based on scRNA-seq data and reported differentially expressed genes in T2D across all kinds of islets cell types (Fang et al. 2019; Lawlor et al. 2017; Segerstolpe et al. 2016; Xin et al. 2016). In this study, we detected heterogeneity among β cells. Pseudotime analysis ordered the β cells into three branches with varying degrees of INS and diabetes-related gene expression. The single-cell transcriptome provides insight into the relationship between individual β cell heterogeneity and T2D while identifying three different β cell branches. According to the expression of marker genes and enrichment analysis of branch differential expression analysis, the three branches are labeled as normal, obesity-like, and T2D-like. Unprecedented insights into the association of β cell transcriptome and disease states were provided through applying pseudotime analysis to connect the progression of gene expression and β cell states. The analysis identified the genes of important role that missed in the analysis result of Fang et al. (2019), which is crucial in the process of diabetes. The genes are enriched in mitochondrial & ETC, protein synthesis and secretion, autophagy, and vascularization. The enrichment pathway and part of enriched genes are demonstrated to be related to obesity and T2D. Further research is needed to prove the role of the rest genes in the etiology of obesity and T2D. We hope the pseudotime analysis will help us understand the change of cell states in the processing of T2D.
Cell branches or states might not be restricted only to pancreatic β cells. Baron et al. (Baron et al. 2016) reported two expression profiles related to different ductal cell states. Segerstolpe et al. (Segerstolpe et al. 2016) reported a subpopulation of α cells and acinar cells. Future studies will focus on the analysis of the whole pancreas cell types to determine the permanent subpopulation or identify the temporary change of cell states and the genes that drive the state change during the disease process.
Concerning the islet cell transcriptome in T2D, the differential expressed genes between the T2D sample and control were reported by several groups, respectively (Lawlor et al. 2017; Segerstolpe et al. 2016; Xin et al. 2016). The differential expressed genes from different groups are largely non-overlapping (Wang and Kaestner 2019). The discrepancy reflects the complex etiology of T2D and asynchrony and heterogeneity of the biological samples. In our study, the distribution of individual cells among branches also suggests the limitation of donor samples (Fig. 2c, e). Applying pseudotime analysis might contribute to synchronizing the inter-individual asynchrony, and expanding the number of islet donors also might be helpful to discover the disease-associated change in gene expression and contribute to the robust analysis.
In summary, we conducted pseudotime analysis to better understand the behavior of β cells in health and disease. We observed the difference between our analysis result and Fang et al. (2019). The difference suggests the development of an analysis method is needed to close the gap between cellular function and transcriptomics. Another limitation of scRNA-seq is “dropout” caused by low-efficiency of mRNA capture and cDNA synthesis, which leads to loss of information and artefactually overestimates the heterogeneity between cell subpopulations. We hope the improvement of methodology can help us better understand the etiology of the disease.
Supplementary Information
Below is the link to the electronic supplementary material.
Acknowledgements
This research was supported by the National Key R&D Program of China (2019YFA0801900, 2018YFA0800300), the National Natural Science Foundation of China (31971074), the Science and Technology Innovation Action Plan of Shanghai Science and Technology Committee (18140901300), the Open Research Fund of the National key laboratory of genetic engineering (SKLGE1803), the Shanghai Municipal Science and Technology Major Project (2017SHZDZX01).
Authors' Contribution
KB contributed to the data analysis and manuscript writing. ZC contributed to scientific discussion and manuscript writing. TL and XK conceived the idea and contributed to the writing of the paper. HW and HX contributed to the final revision of the paper.
Funding
This research was supported by the National Key R&D Program of China (2019YFA0801900, 2018YFA0800300), the National Natural Science Foundation of China (31971074), the Science and Technology Innovation Action Plan of Shanghai Science and Technology Committee (18140901300), the Open Research Fund of the National key laboratory of genetic engineering (SKLGE1803), the Shanghai Municipal Science and Technology Major Project (2017SHZDZX01), and Shanghai Frontiers Science Research Base of Exercise and Metabolic Health.
Data Availability
The datasets generated during the current study are available from the corresponding author on reasonable request.
Code Availability
The codes for performing the Seurat, Monocle2, and DEseq2 analyses are provided in the repository: https://github.com/finchbao/T2D_scRNA_seq/.
Declarations
Conflicts of interest
The authors declare no competing financial interests.
Ethics approval
Not applicable.
Consent to participate
Not applicable.
Consent for publication
All the participants approved to publish.
Footnotes
Kaixuan Bao and Zhicheng Cui contributed equally.
Contributor Information
Xingxing Kong, Email: kongxingxing@fudan.edu.cn.
Tiemin Liu, Email: tiemin_liu@fudan.edu.cn.
References
- Alarcon C, Boland BB, Uchizono Y, Moore PC, Peterson B, Rajan S, Rhodes OS, Noske AB, Haataja L, Arvan P, et al. Pancreatic β-cell adaptive plasticity in obesity increases insulin production but adversely affects secretory function. Diabetes. 2016;65:438. doi: 10.2337/db15-0792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashcroft FM, Rorsman P. Diabetes mellitus and the β cell: the last ten years. Cell. 2012;148:1160–1171. doi: 10.1016/j.cell.2012.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baron M, Veres A, Wolock SL, Faust AL, Gaujoux R, Vetere A, Ryu JH, Wagner BK, Shen-Orr SS, Klein AM, et al. A single-cell transcriptomic map of the human and mouse pancreas reveals inter- and intra-cell population structure. Cell Syst. 2016;3:346–360.e344. doi: 10.1016/j.cels.2016.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cabrera O, Berman DM, Kenyon NS, Ricordi C, Berggren PO, Caicedo A. The unique cytoarchitecture of human pancreatic islets has implications for islet cell function. Proc Natl Acad Sci USA. 2006;103:2334–2339. doi: 10.1073/pnas.0510790103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng K, Ho K, Stokes R, Scott C, Lau SM, Hawthorne WJ, O'Connell PJ, Loudovaris T, Kay TW, Kulkarni RN, et al. Hypoxia-inducible factor-1alpha regulates beta cell function in mouse and human islets. J Clin Investig. 2010;120:2171–2183. doi: 10.1172/jci35846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ebato C, Uchida T, Arakawa M, Komatsu M, Ueno T, Komiya K, Azuma K, Hirose T, Tanaka K, Kominami E, et al. Autophagy is important in islet homeostasis and compensatory increase of beta cell mass in response to high-fat diet. Cell Metab. 2008;8:325–332. doi: 10.1016/j.cmet.2008.08.009. [DOI] [PubMed] [Google Scholar]
- Eizirik DL, Pasquali L, Cnop M. Pancreatic β-cells in type 1 and type 2 diabetes mellitus: different pathways to failure. Nat Rev Endocrinol. 2020;16:349–362. doi: 10.1038/s41574-020-0355-7. [DOI] [PubMed] [Google Scholar]
- Fadista J, Vikman P, Laakso EO, Mollet IG, Esguerra JL, Taneera J, Storm P, Osmark P, Ladenvall C, Prasad RB, et al. Global genomic and transcriptomic analysis of human pancreatic islets reveals novel genes influencing glucose metabolism. Proc Natl Acad Sci USA. 2014;111:13924–13929. doi: 10.1073/pnas.1402665111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang Z, Weng C, Li H, Tao R, Mai W, Liu X, Lu L, Lai S, Duan Q, Alvarez C, et al. Single-cell heterogeneity analysis and CRISPR screen identify key β-cell-specific disease genes. Cell Rep. 2019;26:3132–3144.e3137. doi: 10.1016/j.celrep.2019.02.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Good AL, Haemmerle MW, Oguh AU, Doliba NM, Stoffers DA. Metabolic stress activates an ERK/hnRNPK/DDX3X pathway in pancreatic β cells. Mol Metab. 2019;26:45–56. doi: 10.1016/j.molmet.2019.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamada S, Hara K, Hamada T, Yasuda H, Moriyama H, Nakayama R, Nagata M, Yokono K. Upregulation of the mammalian target of rapamycin complex 1 pathway by Ras homolog enriched in brain in pancreatic beta-cells leads to increased beta-cell mass and prevention of hyperglycemia. Diabetes. 2009;58:1321–1332. doi: 10.2337/db08-0519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung HS, Chung KW, Won Kim J, Kim J, Komatsu M, Tanaka K, Nguyen YH, Kang TM, Yoon K-H, Kim J-W, et al. Loss of autophagy diminishes pancreatic β cell mass and function with resultant hyperglycemia. Cell Metab. 2008;8:318–324. doi: 10.1016/j.cmet.2008.08.013. [DOI] [PubMed] [Google Scholar]
- Kahn SE. The relative contributions of insulin resistance and beta-cell dysfunction to the pathophysiology of Type 2 diabetes. Diabetologia. 2003;46:3–19. doi: 10.1007/s00125-002-1009-0. [DOI] [PubMed] [Google Scholar]
- Karmaus PWF, Chen X, Lim SA, Herrada AA, Nguyen T-LM, Xu B, Dhungana Y, Rankin S, Chen W, Rosencrance C, et al. Metabolic heterogeneity underlies reciprocal fates of TH17 cell stemness and plasticity. Nature. 2019;565:101–105. doi: 10.1038/s41586-018-0806-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim KH, Jeong YT, Oh H, Kim SH, Cho JM, Kim YN, Kim SS, Kim DH, Hur KY, Kim HK, et al. Autophagy deficiency leads to protection from obesity and insulin resistance by inducing Fgf21 as a mitokine. Nat Med. 2013;19:83–92. doi: 10.1038/nm.3014. [DOI] [PubMed] [Google Scholar]
- Kubota N, Terauchi Y, Tobe K, Yano W, Suzuki R, Ueki K, Takamoto I, Satoh H, Maki T, Kubota T, et al. Insulin receptor substrate 2 plays a crucial role in beta cells and the hypothalamus. J Clin Investig. 2004;114:917–927. doi: 10.1172/jci21484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kusminski CM, Ghaben AL, Morley TS, Samms RJ, Adams AC, An Y, Johnson JA, Joffin N, Onodera T, Crewe C, et al. A novel model of diabetic complications: adipocyte mitochondrial dysfunction triggers massive β-cell hyperplasia. Diabetes. 2020;69:313–330. doi: 10.2337/db19-0327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lam PP, Leung YM, Sheu L, Ellis J, Tsushima RG, Osborne LR, Gaisano HY. Transgenic mouse overexpressing syntaxin-1A as a diabetes model. Diabetes. 2005;54:2744–2754. doi: 10.2337/diabetes.54.9.2744. [DOI] [PubMed] [Google Scholar]
- Lawlor N, George J, Bolisetty M, Kursawe R, Sun L, Sivakamasundari V, Kycia I, Robson P, Stitzel ML. Single-cell transcriptomes identify human islet cell signatures and reveal cell-type-specific expression changes in type 2 diabetes. Genome Res. 2017;27:208–222. doi: 10.1101/gr.212720.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin X, Taguchi A, Park S, Kushner JA, Li F, Li Y, White MF. Dysregulation of insulin receptor substrate 2 in beta cells and brain causes obesity and diabetes. J Clin Investig. 2004;114:908–916. doi: 10.1172/jci22217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H, Javaheri A, Godar RJ, Murphy J, Ma X, Rohatgi N, Mahadevan J, Hyrc K, Saftig P, Marshall C, et al. Intermittent fasting preserves beta-cell mass in obesity-induced diabetes via the autophagy-lysosome pathway. Autophagy. 2017;13:1952–1968. doi: 10.1080/15548627.2017.1368596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15:550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuoka T-A, Artner I, Henderson E, Means A, Sander M, Stein R. The MafA transcription factor appears to be responsible for tissue-specific expression of insulin. Proc Natl Acad Sci USA. 2004;101:2930. doi: 10.1073/pnas.0306233101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohtsubo K, Chen MZ, Olefsky JM, Marth JD. Pathway to diabetes through attenuation of pancreatic beta cell glycosylation and glucose transport. Nat Med. 2011;17:1067–1075. doi: 10.1038/nm.2414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu X, Mao Q, Tang Y, Wang L, Chawla R, Pliner HA, Trapnell C. Reversed graph embedding resolves complex single-cell trajectories. Nat Methods. 2017;14:979–982. doi: 10.1038/nmeth.4402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Core Team . R: a language and environment for statistical computing. Vienna: Austria; 2020. [Google Scholar]
- Rasouli N, Younes N, Utzschneider KM, Inzucchi SE, Balasubramanyam A, Cherrington AL, Ismail-Beigi F, Cohen RM, Olson DE, DeFronzo RA, et al. Association of baseline characteristics with insulin sensitivity and β-cell function in the glycemia reduction approaches in diabetes: a comparative effectiveness (GRADE) study cohort. Diabetes Care. 2021;44:340–349. doi: 10.2337/dc20-1787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rocha M, Apostolova N, Diaz-Rua R, Muntane J, Victor VM. Mitochondria and T2D: role of autophagy, ER stress, and inflammasome. Trends Endocrinol Metab. 2020;31:725–741. doi: 10.1016/j.tem.2020.03.004. [DOI] [PubMed] [Google Scholar]
- Sarparanta J, García-Macia M, Singh R. Autophagy and mitochondria in obesity and type 2 diabetes. Curr Diabetes Rev. 2017;13:352–369. doi: 10.2174/1573399812666160217122530. [DOI] [PubMed] [Google Scholar]
- Segerstolpe Å, Palasantza A, Eliasson P, Andersson EM, Andréasson AC, Sun X, Picelli S, Sabirsh A, Clausen M, Bjursell MK, et al. Single-cell transcriptome profiling of human pancreatic islets in health and type 2 diabetes. Cell Metab. 2016;24:593–607. doi: 10.1016/j.cmet.2016.08.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sidarala V, Pearson GL, Parekh VS, Thompson B, Christen L, Gingerich MA, Zhu J, Stromer T, Ren J, Reck EC, et al. Mitophagy protects β cells from inflammatory damage in diabetes. JCI Insight. 2020 doi: 10.1172/jci.insight.141138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sivaskandarajah GA, Jeansson M, Maezawa Y, Eremina V, Baelde HJ, Quaggin SE. Vegfa protects the glomerular microvasculature in diabetes. Diabetes. 2012;61:2958–2966. doi: 10.2337/db11-1655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soleimanpour SA, Gupta A, Bakay M, Ferrari AM, Groff DN, Fadista J, Spruce LA, Kushner JA, Groop L, Seeholzer SH, et al. The diabetes susceptibility gene Clec16a regulates mitophagy. Cell. 2014;157:1577–1590. doi: 10.1016/j.cell.2014.05.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staels W, Heremans Y, Heimberg H, De Leu N. VEGF-A and blood vessels: a beta cell perspective. Diabetologia. 2019;62:1961–1968. doi: 10.1007/s00125-019-4969-z. [DOI] [PubMed] [Google Scholar]
- Stuart T, Butler A, Hoffman P, Hafemeister C, Papalexi E, Mauck WM, 3rd, Hao Y, Stoeckius M, Smibert P, Satija R. Comprehensive integration of single-cell data. Cell. 2019;177:1888–1902.e1821. doi: 10.1016/j.cell.2019.05.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sudhakar JN, Lu HH, Chiang HY, Suen CS, Hwang MJ, Wu SY, Shen CN, Chang YM, Li FA, Liu FT, et al. Lumenal Galectin-9-Lamp2 interaction regulates lysosome and autophagy to prevent pathogenesis in the intestine and pancreas. Nat Commun. 2020;11:4286. doi: 10.1038/s41467-020-18102-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takamoto I, Terauchi Y, Kubota N, Ohsugi M, Ueki K, Kadowaki T. Crucial role of insulin receptor substrate-2 in compensatory β-cell hyperplasia in response to high fat diet-induced insulin resistance. Diabetes Obes Metab. 2008;10:147–156. doi: 10.1111/j.1463-1326.2008.00951.x. [DOI] [PubMed] [Google Scholar]
- Taneera J, Lang S, Sharma A, Fadista J, Zhou Y, Ahlqvist E, Jonsson A, Lyssenko V, Vikman P, Hansson O, et al. A systems genetics approach identifies genes and pathways for type 2 diabetes in human islets. Cell Metab. 2012;16:122–134. doi: 10.1016/j.cmet.2012.06.006. [DOI] [PubMed] [Google Scholar]
- Wang YJ, Kaestner KH. Single-cell RNA-Seq of the pancreatic islets—a promise not yet fulfilled? Cell Metab. 2019;29:539–544. doi: 10.1016/j.cmet.2018.11.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, Brun T, Kataoka K, Sharma AJ, Wollheim CB. MAFA controls genes implicated in insulin biosynthesis and secretion. Diabetologia. 2007;50:348–358. doi: 10.1007/s00125-006-0490-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wickham H. ggplot2: elegant graphics for data analysis. New York: Springer; 2016. [Google Scholar]
- Wirostko B, Wong TY, Simó R. Vascular endothelial growth factor and diabetic complications. Prog Retin Eye Res. 2008;27:608–621. doi: 10.1016/j.preteyeres.2008.09.002. [DOI] [PubMed] [Google Scholar]
- Xin Y, Kim J, Okamoto H, Ni M, Wei Y, Adler C, Murphy AJ, Yancopoulos GD, Lin C, Gromada J. RNA sequencing of single human islet cells reveals type 2 diabetes genes. Cell Metab. 2016;24:608–615. doi: 10.1016/j.cmet.2016.08.018. [DOI] [PubMed] [Google Scholar]
- Xin Y, Dominguez Gutierrez G, Okamoto H, Kim J, Lee AH, Adler C, Ni M, Yancopoulos GD, Murphy AJ, Gromada J. Pseudotime ordering of single human β-cells reveals states of insulin production and unfolded protein response. Diabetes. 2018;67:1783–1794. doi: 10.2337/db18-0365. [DOI] [PubMed] [Google Scholar]
- Yang L, Li P, Fu S, Calay ES, Hotamisligil GS. Defective hepatic autophagy in obesity promotes ER stress and causes insulin resistance. Cell Metab. 2010;11:467–478. doi: 10.1016/j.cmet.2010.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang X, Graff SM, Heiser CN, Ho KH, Chen B, Simmons AJ, Southard-Smith AN, David G, Jacobson DA, Kaverina I, et al. Coregulator Sin3a promotes postnatal murine β-cell fitness by regulating genes in Ca(2+) homeostasis, cell survival, vesicle biosynthesis, glucose metabolism, and stress response. Diabetes. 2020;69:1219–1231. doi: 10.2337/db19-0721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Q, Brown J, Kanarek A, Rajagopal J, Melton DA. In vivo reprogramming of adult pancreatic exocrine cells to beta-cells. Nature. 2008;455:627–632. doi: 10.1038/nature07314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Y, Zhou B, Pache L, Chang M, Khodabakhshi AH, Tanaseichuk O, Benner C, Chanda SK. Metascape provides a biologist-oriented resource for the analysis of systems-level datasets. Nat Commun. 2019;10:1523. doi: 10.1038/s41467-019-09234-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu D, Zhang Y, Lam PP, Dolai S, Liu Y, Cai EP, Choi D, Schroer SA, Kang Y, Allister EM, et al. Dual role of VAMP8 in regulating insulin exocytosis and islet β cell growth. Cell Metab. 2012;16:238–249. doi: 10.1016/j.cmet.2012.07.001. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets generated during the current study are available from the corresponding author on reasonable request.
The codes for performing the Seurat, Monocle2, and DEseq2 analyses are provided in the repository: https://github.com/finchbao/T2D_scRNA_seq/.