Abstract
The CRISPR/Cas9-mediated base editing technology can efficiently generate point mutations in the genome without introducing a double-strand break (DSB) or supplying a DNA donor template for homology-directed repair (HDR). In this study, adenine base editors (ABEs) were used for rapid generation of precise point mutations in two distinct genes, OsWSL5, and OsZEBRA3 (Z3), in both rice protoplasts and regenerated plants. The precisely engineered point mutations were stably inherited to subsequent generations. These single nucleotide alterations resulted in single amino acid changes and associated wsl5 and z3 phenotypes as evidenced by white stripe leaf and light green/dark green leaf pattern, respectively. Through selfing and genetic segregation, transgene-free, base edited wsl5 and z3 mutants were obtained in a short period of time. We noticed a novel mutation (V540A) in Z3 locus could also mimic the phenotype of Z3 mutation (S542P). Furthermore, we observed unexpected non- A/G or T/C mutations in the ABE editing window in a few of the edited plants. The ABE vectors and the method from this study could be used to simultaneously generate point mutations in multiple target genes in a single transformation and serve as a useful base editing tool for crop improvement as well as basic studies in plant biology.
Electronic supplementary material
The online version of this article (10.1007/s42994-020-00018-x) contains supplementary material, which is available to authorized users.
Keywords: Adenine base editor, CRISPR/Cas9 genome editing, Plant base editing, Precise point mutation, Transgene-free rice
Introduction
Precision genome editing is a powerful tool for accelerating crop improvement. The discovery of the CRISPR/Cas9 system and its repurposing for genome editing revolutionized basic biological research and practical applications in medicine and agriculture (Doudna and Charpentier 2014). In the CRISPR/Cas9-mediated genome editing process, a single guide RNA (sgRNA) can direct Cas9 to create a double strand break (DSB) at a target locus. Higher eukaryotes including plants typically repair the DSB through either non-homologous end joining (NHEJ) or homology-directed repair (HDR) pathways (Molla and Yang 2019a). Higher plants predominantly use the error prone NHEJ which creates random insertion/deletion (indel) causing frameshift mutation and ultimately gene knock out (Huang and Puchta 2019).
NHEJ-mediated gene knock-out has limited application for sophisticated genome engineering, since it cannot create precise indels or specific point mutations. To install precise point mutations and to delete or introduce desired DNA sequences, we highly rely on HDR-mediated precise genome editing. For HDR to occur, one has to supply an exogenous donor template containing the desired changes flanked by homologous sequences to the target locus. Unfortunately, the efficiency of HDR is extremely low in higher plants because of the low innate rate of cellular HDR and difficulties in donor template delivery (Huang and Puchta 2019).
The ability to efficiently generate point mutations in plants has great potential to assist in crop improvement as well as to unravel functions of many natural single nucleotide polymorphisms (Molla and Yang 2019b). CRISPR/Cas-mediated base editing systems have recently been developed to precisely generate point mutations in a genome (Komor et al. 2016; Nishida et al. 2016; Gaudelli et al. 2017). Using cytidine deaminase fused with dCas9 (catalytically dead) or nCas9 (nickase), precise C → T base conversions were achieved in both mammalian and plant systems (Komor et al. 2016; Nishida et al. 2016; Shimatani et al. 2017; Zong et al. 2017; Li et al. 2017). Fusion of a hypothetical DNA adenine deaminase with dCas9/nCas9 would generate an adenine base editor, but all such naturally occurring enzymes deaminate adenine only in RNA substrates (Gaudelli et al. 2017). To develop an A → G (and T → C) base editing system, the E. coli tRNA adenosine deaminase (TadA) has been modified to accept DNA as a substrate through extensive directed evolution (Gaudelli et al. 2017). This laboratory-evolved DNA adenine deaminase, tethered to nCas9, can now deaminate adenine (A) in the non-target strand into inosine (I). Since inosine is read as guanine by cellular replication machinery, the deamination results in a post-replicative conversion of A to G (Alseth et al. 2014; Gaudelli et al. 2017). Among the several adenine base editors (ABEs) developed, ABE7.10 is the most efficient when the target A is within the 4–7 protospacer positions, whereas ABE7.9, ABE7.8, and ABE6.3 perform better than ABE7.10 within the 8-10 protospacer positions (Gaudelli et al. 2017). Although a few of studies have recently been published while preparing this manuscript (Hua et al. 2018; Kang et al. 2018; Li et al. 2018; Yan et al. 2018), relatively little is known about ABE’s wide applicability in the plant system and the heritability of the induced mutation in the subsequent generation in rice plants. Moreover, most of the earlier studies described ABE induced mutation generation without any phenotypic evidence (Hua et al. 2018, 2019a; Yan et al. 2018).
To further demonstrate the application of adenine base editing technology in plants, we attempted to simultaneously edit the white stripe leaf 5 (WSL5) and ZEBRA3 (Z3) loci in rice (Oryza sativa ssp. japonica cv. ‘Kitaake’), a representative cereal crop and monocot model. OsWSL5, a recently characterized rice gene, encodes a novel chloroplast-targeted pentatricopeptide repeat protein, which plays an essential role in rice chloroplast biogenesis (Liu et al. 2018). A single nucleotide polymorphism (T → C) located in the conserved region of exon 1 causes a leucine to proline amino acid substitution in the wsl5 mutant, which can be phenotypically visualized as longitudinal albino leaf striations. OsZ3, another recently characterized rice gene, encodes a citrate transporter (Kim et al. 2018). The mutant plant possesses a single base substitution (T → C) in the third exon, causing a missense mutation (serine to proline at amino acid 542), with mutant z3 plants exhibiting a phenotype of alternating transverse dark-green/light-green stripes in the leaves and growth stunting.
Here we report the development of a plant base editing system based on E. coli TadA-derived adenine base editors, demonstrate its utility for single nucleotide mutations and their germline transmission, and provide evidence for the associated mutant phenotypes. This system allows one to generate non-HDR-based indel-free and multiplexed base change mutations and to readily generate transgene-free, single base edited plants through selfing or backcrossing.
Materials and Methods
Construction of base editing vectors
pENTR11-dual selection vector (Invitrogen, USA) was digested with EcoRI and self-ligated to eliminate the ccdB gene. pCMV-ABE7.10 (Addgene plasmid #102919) and pCMV-ABE7.9 (Addgene Plasmid #102918)(Gaudelli et al. 2017) were digested with NotI/PmeI to release the ABE-7.10 and 7.9 (TadA–Tad*A–nCas9) and cloned in the NotI/EcoRV site of pENTR11 (–ccdB) to generate pENTR11-ABE7.10 and pENTR11-ABE7.9, respectively. The ABE7.10 was digested out from pENTR11-ABE7.10 with BstBI/XbaI and cloned in the same site of pRGE32 (Xie et al. 2015) replacing the SpCas9 to generate pPr-ABE7.10.
Polycistronic tRNA-gRNA (PTG) for targeting both the rice WSL5 and Z3 loci simultaneously was generated following our earlier described method (Xie et al. 2015). Primer sequences used for PTG synthesis are listed in supplementary Table 1. The PTG fragment was digested with FokI and inserted into the BsaI digested pPr-ABE7.10 vector. ABEs were expressed under the control of rice ubiquitin 10 promoter, while the PTG was transcribed under the control of OsU3 promoter. The pPr-ABE7.10 was used for protoplast transfection.
Table 1.
Base editing efficiencies in regenerated rice plants
| Vector | Total no ABE + plants evaluated | WSL5 (sgRNA1) edited | Efficiency (%) | Z3 (sgRNA2) edited | Efficiency (%) |
|---|---|---|---|---|---|
| pKABE7.9 | 149 | 2 | 1.34 | 58 | 38.92 |
| pKABE7.10 | 142 | 4 | 2.81 | 20 | 14 |
For binary vector construction, the pENTR11-ABE7.10 and pENTR11-ABE7.9 was digested with BstBI/XbaI to release ABE7.10 and ABE7.9. The fragments were inserted into the same site of binary vector pRGEB32 (Xie et al. 2015) replacing SpCas9 to generate pK-ABE7.10 and pK-ABE7.9. The PTG containing sgRNA for both WSL5 and Z3 genes was inserted similarly into the BsaI digested pK-ABE7.10 and pK-ABE7.9 vector.
Protoplast transfection
Rice (Oryza sativa ssp. Japonica cv. ‘Kitaake’) protoplasts were isolated and transfected as described previously with few modifications (Xie and Yang 2013). Briefly, rice stem and sheath were cut into 0.5–1 mm strips and immediately transferred into 10 ml of 0.6 M mannitol and incubated for 10 min. Mannitol was replaced with enzyme solution (1.5% Cellulase R10, 0.75% Macerozyme R10, 0.5 M mannitol, 10 mM MES at pH 5.7, 1 mM CaCl2, 5 mM β-marcaptoethanol, and 0.1% BSA) and the rice tissues were digested for 5–8 h in dark with gentle shaking. After adding 10–15 ml W5 solution (2 mM MES at pH 5.7, 154 mM NaCl, 5 mM KCl, 125 mM CaCl2), protoplasts were filtered through 35 µm Nylon mesh. Protoplasts were pelleted down by centrifugation at 250 g for 3 min and re-suspended in 1 ml W5 solution. After incubation for 1 h at room temperature, W5 solution was removed by centrifugation and protoplasts were re-suspended in MMG solution (4 mM MES, 0.6 M Mannitol, 15 mM MgCl2) to a final concentration of 5 × 106 cells ml−1.
PEG-mediated transfection was carried out using 200 μl of protoplasts and 20 μg of DNA (pPr-ABE7.10, and pPr-ABE7.9). A GFP expression cassette containing plasmid was used as the control to determine transformation efficiency. Protoplasts were transfected with ~ 40% efficiency.
The protoplasts and DNA were gently mixed, and 1 ml of freshly prepared PEG solution (0.6 M Mannitol, 100 mM CaCl2, and 40% PEG4000) was added slowly. The mixture was incubated at room temperature for 20 min. Four ml of W5 solution was added to stop the transformation. Protoplasts were collected by centrifugation and resuspended in WI solution (4 mM MES, pH 5.7, 0.6 M Mannitol, 4 mM KCl) and transferred in six well plates. After 48 h of incubation, DNA was extracted from the protoplast for further analysis.
Mutation detection by PCR, CAPS analysis and sequencing
The target regions for both WSL5 and Z3 were amplified by PCR using specific pairs of oligo primers (Supplementary Table 1). DNA was extracted following a previously described method (Molla et al. 2015). PCR products were digested with SacI and SalI for WSL5 and Z3, respectively. Since the target A for both the loci fall within a restriction enzyme recognition site, successful editing destroys the restriction sites. Digestion insensitivity would indicate base editing. After electrophoresis, the undigested DNA fragments were purified from agarose gels for DNA sequencing.
Agrobacterium-mediated rice transformation
The base editing constructs were electroporated into Agrobacterium tumefaciens strain EHA105. The Agrobacterium-mediated transformation of 21–25-day-old calli derived from Kitaake mature seed was performed as described (Mei et al. 2007). Transformed calli were selected on hygromycin (50 µg/ml) containing media for 4–6 weeks and transferred to regeneration media. After regeneration of shoots, they were transferred to rooting media. Transgenic plantlets were transferred to soil and grown in controlled greenhouse for subsequent genotypic and phenotypic analysis.
Analysis of base editing efficiency in the T0 generation
Genomic DNA was extracted from leaves of regenerated T0 plants as described previously (Molla et al. 2015). The target loci were amplified by PCR using specific primers (Supplementary Table 1) and resulting DNA fragments were purified with PCR purification kit (Bio Basic Inc, Canada). The wsl5 and zebra3 PCR product was digested with SacI and SalI, respectively, to determine the editing. The purified PCR products were also subjected to Sanger sequencing.
Segregation analysis in the T1 generation and Phenotyping of mutant lines
Seeds obtained from monoallelic mutation of WSL5 and Z3 were germinated to raise T1 plants. Ten plants from each line were evaluated by RE analysis and Sanger sequencing after targeted amplification. A Chi-square test was performed to ascertain the inheritance pattern.
T0 seeds from mutant plants were germinated and grown at 28 °C/23 °C (day/night) under 12/12 h light and dark cycle. For phenotyping wsl5 mutants, plants were grown at constant temperature of 20 °C.
Off-target analysis
Potential off-targets of the base editing for both the genes were examined. The off-target sites having up to three base mismatches to sgRNA target region were identified using CRISPR-GE (http://skl.scau.edu.cn) software (Xie et al. 2017). Then the corresponding sequence for Kitaake genome was retrieved from Phytozome and primers were designed using primer 3 software (Untergasser et al. 2012). A total of six off-target loci, four for WSL5 and two for Z3, were amplified from the transgenic lines and analyzed with Sanger sequencing to detect any unwanted mutations.
Results
Adenine base-editor vectors for targeted base editing in plants
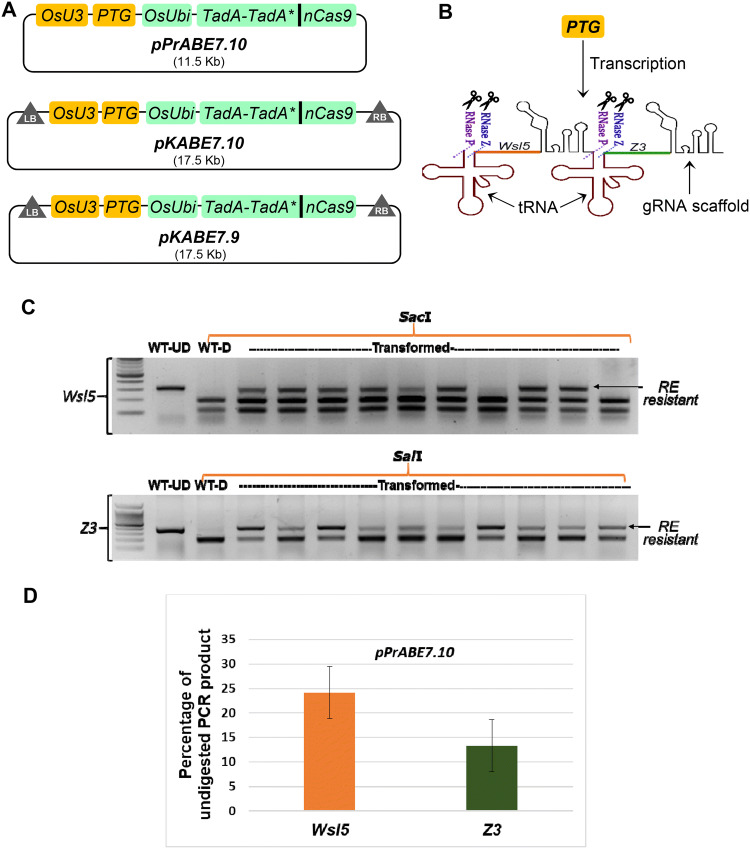
To perform targeted adenine base editing in rice, we have constructed a vector (pPr-ABE7.10) for plant adenine base editing for protoplast transient assays, as well as two binary vectors (pKABE7.10 and pKABE7.9) for Agrobacterium-mediated plant transformation (Fig. 1A). All the vectors have a similar basic structural configuration composed of an engineered adenosine deaminase fused with a Cas9 nickase (nCas9-D10A). To carry out the deamination reactions, a heterodimeric adenosine deaminase composed of a wild type tRNA adenosine deaminase (TadA) and an engineered TadA (TadA*) has been tethered to nCas9 using a linker peptide. The fused TadA–TadA*-nCas9 ORF was constitutively expressed under the control of a rice ubiquitin promoter. The polycistronic tRNA-gRNA (PTG) gene was expressed under the control of a polymerase III promoter, OsU3 (Fig. 1B). Both the binary vectors contained hygromycin resistant gene as a selectable marker.
Fig. 1.
A → G base editing efficiencies at two target loci in transient assays using rice protoplasts. A Schematic diagram of plant adenine base editors constructed and used in this study. pPrABE7.10 was used in protoplast transformation; pKABE7.10 and pKABE7.9 were used for Agrobacterium-mediated calli transformation. B Diagrammatic representation of how PTG (polycistronic tRNA-gRNA) gene works in generating more than one single guide RNA (sgRNA). Endogenous RNase P and RNase Z splice out the tRNA releasing sgRNAs for targeting WSL5 (orange protospacer) and Z3 (green protospacer) loci. C Representative image of a 2% agarose gel showing a restriction enzyme (RE) digest of PCR amplicons spanning the target loci. RE resistant band indicates disruption of recognition sites by base editing. SacI and SalI enzymes were used for Wsl5 and Z3, respectively. WT-UD, wild type undigested/untreated; WT-D, wild type digested with RE. D Percentage of undigested PCR product after overnight incubation with RE. Bar diagram represents result for n = 15 ± SE
The original ABE7.10 and ABE7.9 base editors were designed for slightly different activity windows (Gaudelli et al. 2017). The ABE7.10 based vectors are suitable to edit target bases at protospacer position 4–8 (counting the PAM as 21–23), whereas ABE7.9 based vectors are better suited for target bases positioned at 7–10. We have cloned two tandemly arrayed guide-RNAs into our PTG, which target the loci OsWSL5 (OsKitaake04g315500.1) and OsZ3 (OsKitaake03g038700.11). For the OsWSL5 locus, the target A is at the 5th position of the protospacer, whereas the target A for OsZ3 locus is at the 7th position of the protospacer.
Adenine base editing in protoplasts via transient expression assay
To evaluate whether adenine base editors are functional in rice cells, we transfected Kitaake protoplasts with the pPr-ABE7.10 vector, containing a PTG for targeting WSL5 and Z3 loci. After 48 h of incubation, both the target loci were amplified from the transfected protoplast DNA by PCR. To detect mutations, we performed cleaved amplified polymorphic sequence (CAPS) analysis using SacI and SalI restriction endonucleases (RE) for WSL5 and Z3, respectively. Successful conversion of the target A to G abolishes a SacI site (3′-CTCGAG-5′) in the WSL5 protospacer. A 310 bp amplified wild type WSL5 fragment was completely digested by SacI into 197 and 113 bp fragments, whereas the PCR fragment amplified from transfected protoplast DNA showed partial resistance to cleavage (Fig. 1C). Similarly, targeted conversion of A to G in the Z3 locus destroys a SalI restriction site (3′-CAGCTG-5′). Amplified 403 bp wild type Z3 fragment should be digested into 205 and 188 bp upon SalI treatment. Gel profiles of SalI digested PCR product revealed that targeted mutations were induced in a portion of the protoplast population (Fig. 1C). As there were no other A bases in the targetable window, successful mutation of the specific A could be screened by detecting undigested bands (RE resistant bands). Based on the band intensity, we have observed an average of 24.14% and 13.31% RE resistant bands for Wsl5 and Z3 loci, respectively (Fig. 1D). Editing efficiency was determined by the percentage of PCR amplicons insensitive to restriction digestion. Subsequent sequencing of these PCR amplicons further confirmed successful targeted A > G base editing at both the loci in rice protoplasts (Supplementary Fig. 1).
Single base conversion at WSL5 and Z3 loci in regenerated plants
Because we observed low base editing efficiency at the Z3 locus in our protoplast assay (Fig. 1D), we hypothesized that the lower efficiency might be due to the position of the targeted base. The target A for Z3 is more proximal (7th) to the PAM than the target base (5th) for WSL5 (Fig. 2A, B). Therefore, we decided to use both pKABE7.10 and pKABE7.9 binary constructs for stable transformation and regeneration of transgenic plants. Based on the earlier report in the mammalian system, ABE7.9 could be more efficient than ABE7.10 for a target A proximal to PAM (Gaudelli et al. 2017). A total of 160 and 165 hygromycin resistant transgenic plants were regenerated with the pKABE7.10 and pKABE7.9 binary constructs, respectively. To evaluate base editing, genomic DNA was isolated from each of the regenerated plants and initially screened for the presence of base editors by ABE specific PCR primers. We obtained 142 and 149 ABE-positive plants for pKABE7.10 and pKABE7.9, respectively. From these ABE-positive transgenic plants, the target loci (WSL5 and Z3) were amplified by PCR. RE analysis of the PCR amplicons was initially used to calculate the editing efficiency. Amplicons from successfully edited plants were found to be partly insensitive to restriction digestion (data not shown). To verify the result, we performed Sanger sequencing and confirmed the single base conversions that are consistent with the RE assay.
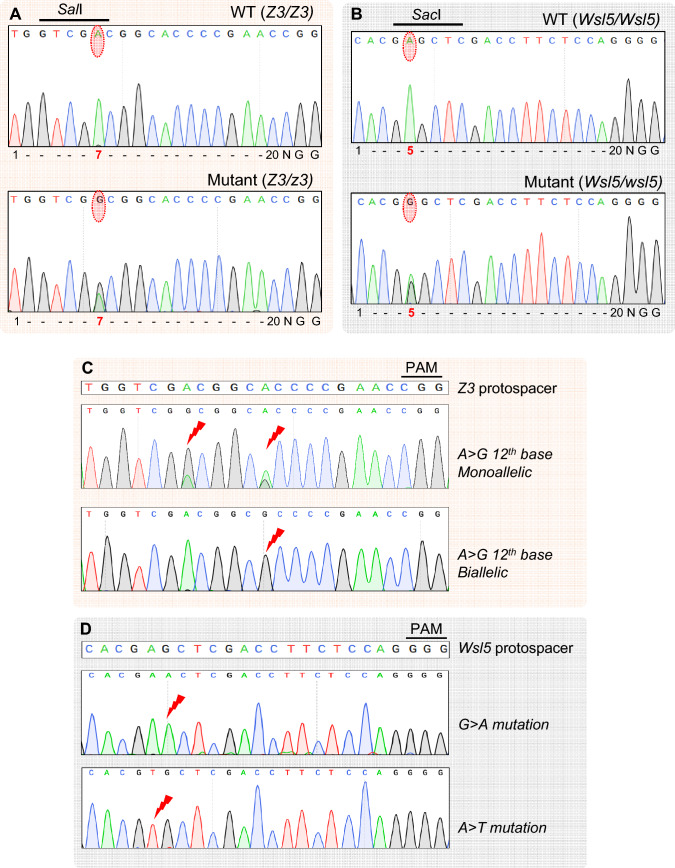
Fig. 2.
Chromatogram showing successful monoallelic targeted A → G editing in T0 and unintended base edition in T1 rice plants. A Editing of 7th A in the Z3 protospacer, B Editing of 5th A in the WSL5 protospacer. Targeted and resultant bases are encircled with red dotted bubble. Upper and lower panels represent chromatogram from wild type (WT) and T0 base edited plants, respectively. C Monoallelic and biallelic editing of 12th A in the Z3 protospacer seq. D G → A and A → T conversion in the WSL5 protospacer
Out of the 142 plants derived from pKABE7.10 transformation, a total of 20 (14.08%) plants were found to be successfully edited for the Z3 locus, whereas only 4 (2.81%) plants were obtained with the edited WSL5 locus (Table 1). On the other hand, pKABE7.9 showed a much higher editing efficiency (38.92%) for the Z3 locus. Again, the target base at WSL5 locus remained recalcitrant to editing, showing success only in 2 (1.34%) out of 149 plants (Table 1). Considering both constructs, for WSL5, a total of six plants with intended A → G conversion were obtained exhibiting 1.7% editing efficiency. Interestingly, all the edited T0 plants obtained were heterozygous in edited loci as evidenced from partial sensitivity to RE digestion and overlapping peaks in the chromatograms (Fig. 2A and B). Analysis of Sanger sequencing data from T0 plants does not reveal any unintended proximal base editing or indel formation. Collectively, these experiments demonstrate the feasibility of simultaneous generation of single base edited mutants for two different genes. While we initially intended to obtain individual plants with both sites mutated, due to the low efficiency of base editing at the WSL5 locus, we were unable to obtain stable transgenic lines with both types of mutations.
Stable inheritance and segregation of targeted mutations
Next, we sought to follow segregation of the targeted mutations in the T1 generation. T1 plants derived from the self-pollination of T0 monoallelic mutant lines were subjected to inheritance pattern analysis. Like the analysis done for the T0 plants, we performed targeted amplification and RE analysis from the T1 plants. Among the four Z3 mutant lines tested, segregation of the mutation in two lines (10–105, and 9–38) did not follow the expected Mendelian ratio (1:2:1) (Supplementary Table 2). The other two lines (10–51, and 9–86) followed the Mendelian law as evidenced from the calculated Chi-square (χ2) value being less than the critical value (p < 0.05).
Similarly, among the WSL5 mutant lines, one line (9–79) exhibited deviation from Mendelian segregation pattern and two lines (10–145, and 9–72) showed segregation according to Mendelian law. On the contrary, when we calculated the segregation pattern of base-editors based on ABE specific PCR, all three lines showed inheritance following Mendelian 3:1 ratio. Taken together, these results suggest that the mutation generated by plant adenine base editors is able to be stably inherited to the next generation.
Unintended proximal base editing in T1
Unintended editing in non-target bases of the protospacer and in the nearby region is termed as proximal base editing. Out of 84 mutant T0 plants, not a single plant exhibited unintended proximal base editing. The observation of non-Mendelian segregation in two lines stimulated us to investigate if there is any new kind of base editing pattern in the T1 plants. We have sequenced the target locus amplified from 14 plants from Z3/10-105 and 12 plants from WSL5/9-79.
Interestingly, we found that the 12th base (A) of the Z3 protospacer sequence has been changed to G in 7 plants out of 14 (Fig. 2C). Out of these seven plants, two were homozygous mutants for the 12th base, and five were heterozygous. The T0 plant, 10–105, had exhibited editing only at the 7th position of the protospacer (targeted). However, the editing of this 12th A indicates the base editor is still active in the T1 generation. pKABE7.10 showed an extended activity window in contrast to what reported in the mammalian cells (Gaudelli et al. 2017).
Surprisingly, in the WSL5 line (9–79), two plants showed unusual base conversion in the activity window. One displayed a single G → A (4th base) conversion, while the other showed a single A → T (5th base) conversion (Fig. 2D), that is an unexpected behavior of adenine base editor. The result prompted us to sequence more T1 plants for unraveling any unintended editing. We did not observe any other types of unwanted mutation in any of the lines. We then assumed if the active base editor could induce mutation in the other targeted locus in the single mutant plants in T1 generation, i.e., checking WSL5 locus in Z3 mutant plant and vice versa. Unfortunately, no single plant was obtained with both the loci edited.
Base conversion at the WSL5 and Z3 loci translate to mutant phenotype
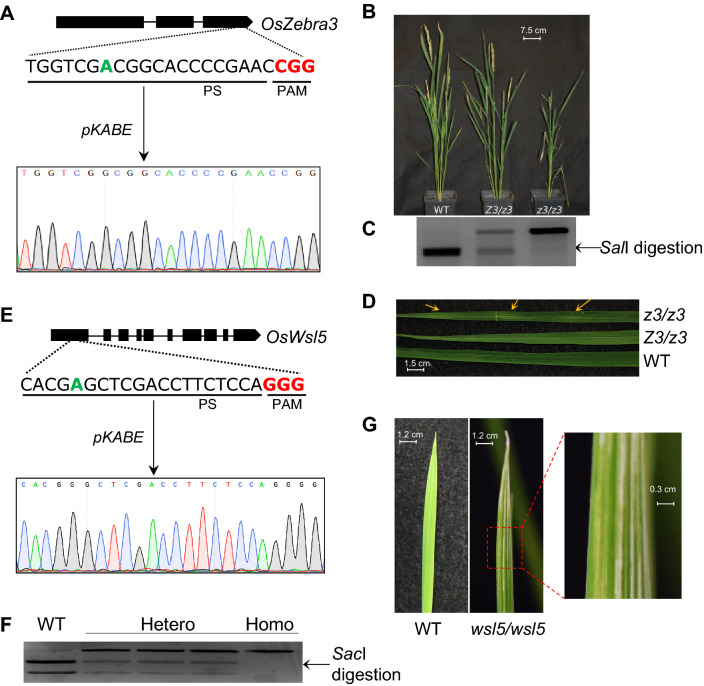
We further looked at whether the successful targeted base editing alters the phenotype of rice plants. A biallelic single nucleotide change (T → C) in the wild type Z3 gene causes the mutant phenotype: transverse dark-green/green sectors in the mature leaves, late flowering and stunted plant growth (Kim et al. 2018). The mutation in exon 3 causes a missense mutation from Serine 542 to Proline 542. Successful creation of the homozygous mutant (z3/z3) line by adenine base-editor displayed all the mutant phenotypes described by Kim et al. (2018) (Fig. 3A–D). As expected, stunted growth phenotype in heterozygous plants was not as severe as in homozygous plants (Fig. 3B). We have noticed one or two of the leaves per mutant plant exhibited leaf variegation phenotype (Fig. 3D), while others were normal. However, delayed flowering was common in all the z3/z3 mutants. We observed 1–2 panicles per plant and fewer number of seeds per panicle in the homozygous mutant, while the panicle numbers in heterozygous (Z3/z3) plants were not significantly different from the wild type plants. We also observed a reduction in seed size in the mutant plants in comparison to the wild type Kitaake plants (Supplementary Fig. 2). Interestingly, the plants with the edited 12th A in the Z3 protospacer showed a similar phenotype as the z3/z3 homozygous mutant (7th A in the protospacer) (Supplementary Fig. 3). The conversion of A → G at the 12th position results in a missense mutation of valine to alanine (V540A) (Fig. 2C).
Fig. 3.
Characteristic phenotypes of z3 and wsl5 mutants generated by adenine base editors. A Schematic representation of the target site in OsZ3 locus and DNA chromatogram of homozygous mutant (z3/z3) plant. Green bold letter is the target base and red bold letters are the protospcaer adjacent motif (PAM). B Phenotypic appearance of wild type (WT), heterozygous (Z3/z3), and homozygous (z3/z3) mutant plants. C Representative restriction analysis from WT, Z3/z3, and z3/z3 plant showing complete, partial and nil digestion, respectively. D Leaf variegation phenotype showing green and dark green sectors in mature leaf. Yellow arrow indicates green sectors. E Schematic representation of the target site in OsWSL5 locus and DNA chromatogram of homozygous mutant (wsl5/wsl5) plant. Green bold letter is the target base and red bold letters are the protospcaer adjacent motif (PAM). F Representative RE analysis for WT, hetero-, and homozygous mutant plants. G White strip leaf phenotype in homozygous (wsl5/wsl5) seedling
The homozygous wsl5 mutant is known to exhibit white-striped leaves in the seedlings (Liu et al. 2018). A single nucleotide polymorphism (T → C) is the causal mutation of wsl5. It alters a CTC codon to CCC resulting in a Leucine151 to Proline151 amino acid substitution in the conserved region of the first exon of the WSL5 gene. We have noticed that the base-editing derived homozygous (wsl5/wsl5) mutant Kitaake seedlings exhibited white stripe leaf phenotype (Fig. 3E–G). Mature mutant plants displayed normal appearance and phenotype as the non-transformed wild type plants or transformed non-edited plants.
Obtainment of transgene-free, base-edited mutants
Developing Cas9 or base editor-free, non-transgenic mutant plants is highly desirable to address the regulatory issues of genome edited crop plants. Segregation of ‘integrated Cas9 transgene at a distant locus from the induced mutation’ would provide the possibility to readily obtain Cas9 free plants with the targeted mutation. While analyzing the segregation pattern of the mutants in T1 generation, we have obtained base-editor (nCas9-adenine deaminase) free z3 and wsl5 mutants. Among all T1 plants derived from both wsl5 and z3 monoallelic T0 mutant lines, we obtained 21% of plants devoid of the base-editor. The absence of the base editor was determined by the negative result in ABE (tadA) specific PCR assays (Supplementary Fig. 4). Both biallelic and monoallelic mutant plants were found to be transgene-free. What emerges from the results reported here is that we can generate precisely base edited plants free from integrated T-DNA within a very short period. Obtaining T1 plants with the desired edit and devoid of any foreign DNA has taken us only a single generation (about 5 months).
Assessment of indels and off-target base editing
To further examine the specificity of the adenine base editor used in our study, we analyzed the base editing percentage at the potential off-target sites. Off-target of base editors might presumably occur at the potential genomic sites that could be targeted by Cas9 through partial homology with the guide RNA. According to CRISPR-GE (skl.scau.edu.cn) designated scores, potential off-target loci were identified. A total of six off-target loci, four for WSL5 and two for Z3 were amplified using site-specific primers (Supplementary Table 1) and sequenced. Genomic loci other than the two selected sites for Z3 did not contain a targetable A in the activity window. Randomly selected 20 plants for each type of mutant was subjected to off-target analysis. No mutations including indels or single base substitutions was detected at any of these putative off-target sites (Table 2). Our data confirmed the high specificity and precision of adenine base editing in rice plants.
Table 2.
Off-target editing efficiency
| sgRNA | Off- target | Sequence | No of mismatches | % of editing |
|---|---|---|---|---|
| sgRNA1 (WSL5) | 1 | CACGAGCTCCAACTTCTCCTTGG | 3 | 0 |
| 2 | CAGGAGCTCCATCTTCTCCATGG | 3 | 0 | |
| 3 | CACGAGCTCCAGCTTCTCCTCGG | 3 | 0 | |
| 4 | CACGAGCTCCAGCTTCTCCTTGG | 3 | 0 | |
| sgRNA2 (Z3) | 1 | TGGTCGGCGGCATCCCGAACCAG | 2 | 0 |
| 2 | TCGTCGACGGCACGCCGAAGCGG | 3 | 0 |
Percent of editing includes both base conversion and indel generation
Mismatches are highlited in bold
Discussion
In this study, we achieved successful and precise A → G base editing at two gene loci in rice using two versions of adenine base editor, ABE7.10 and ABE7.9. The SpCas9 nickase (D10A) (nCas9) fused with deaminase (TadA–Tad*A) via a 32-amino acid {(SGGS)2-XTEN-(SGGS)2} linker was cloned under the control of a rice ubiquitin promoter for both protoplast transient expression and Agrobacterium meditated stable expression (Fig. 1A). The deaminase (TadA–TadA*) was fused at the N terminus of nCas9 as described in the original report (Gaudelli et al. 2017). Fusion of adenosine deaminase at the C terminus has been demonstrated to be ineffective in rice and wheat (Li et al. 2018). We used the polycistronic tRNA–gRNA (PTG) approach to efficiently produce two gRNAs for both the targeted genes (Fig. 1B) (Xie et al. 2015). The expression of PTG was driven by OsU3, a Pol-III promoter. As a tRNA gene contains the internal boxA and boxB which act as promoter elements for the RNA Pol-III, transcription of tRNA-gRNA is enhanced. After transcription as a unit, the endogenous RNase P and RNase Z would splice out the tRNA and as a result, individual sgRNA would be available to form complexes with the base-editor (nCas9-deaminase fusion). Earlier studies in rice reported adenine base editing with single guide RNA (Hua et al. 2018; Li et al. 2018; Yan et al. 2018). This system enables us to clone multiple gRNAs in a single vector to target multiple loci in the genome. To get a visible phenotype resulting from the base editing, we have targeted functional SNPs in two recently identified and cloned rice genes, viz., white stripe leaf 5 (WSL5) and ZEBRA3 (Z3). WSL5 encodes a pentatricopeptide repeat protein and a single nucleotide polymorphism (T → C) in the first exon caused Leucine151 to Proline151 substitution resulting in the wsl5 mutation. The wsl5 mutant displays white-striped leaves at the seedling stage (Liu et al. 2018). Z3 is a putative citrate transporter gene and a single base substitution (T → C) in the third exon gives rise to the z3 mutant plant which exhibits dark-green/green variegation in mature leaves (Kim et al. 2018). To generate T → C mutation in the coding strand, the opposite strand needs to be targeted by ABE. We designed sgRNAs targeting the opposite strand, where target adenines were at protospacer position 5 and 7 for WSL5 and Z3 gene, respectively.
For rapid verification of multiplexed base editing by ABE, we transfected rice protoplasts with the vector pPr-ABE7.10 containing gRNA for both the targets. Target regions of WSL5 and Z3 were amplified by PCR from the genomic DNA harvested from protoplasts at 3–4-day post-transfection. As the target A bases are contained within recognition sites of SacI and SalI in WSL5 and Z3 genes, respectively, successful base editing destroys the restriction sites. As evidenced from CAPS analysis, 24.14% of mutated WSL5 was obtained, whereas the mutation frequency of Z3 locus was about 13.31% (Fig. 1C). A recent study in rice and wheat protoplasts reported A → G conversion frequencies up to 7.5% (Li et al. 2018). Kang et al. (2018) reported an A → G base editing frequencies up to 8.8% and 4.1% in Arabidopsis and rapeseed protoplast, respectively. However, co-transfection of ABE plasmid and mutated GFP plasmid in rice protoplasts exhibited up to 32.8% of GFP fluorescent cells indicative of ABE mediated correction (Li et al. 2018).
When we sequenced the undigested products, we observed monoallelic targeted editing in both loci (Supplementary Fig. 1). This finding, while preliminary, suggests that ABE works efficiently in rice. After this initial indication of successful A > G editing, we sought to generate stable transgenic rice plants harboring the intended mutation.
Since we observed a lower percentage of base editing at the Z3 locus, we assumed the position of target A (7th base of the protospacer) might be one of the influencing factors. Due to this assumption, we prepared two variants of adenine base editors pKABE7.10 and pKABE7.9. Although ABE7.10 was reported as the best one among the variants for the activity window ranging from the 4th to 8th base of the protospacer, ABE7.9 is better suited when the target base is at the 8th–10th position. We reasoned that the window might be different for the plant system and 7th is not favorable for ABE7.10. When we analyzed the regenerated plants from pKABE7.10, in contrast with the protoplast assay result, we obtained only 2.81% edited plants for WSL5 and 14% for Z3.
On the other hand, pKABE7.9 performed far better for Z3 with ~ 39% editing, but poorer for WSL5 with only 1.34%. This very low editing efficiency at WSL5 locus could possibly be attributed to many reasons. When we were planning for the study, we had bioinformatically analyzed RNA fold pattern of sgRNA for both the loci and we noticed that WSL5 sgRNA was far better in its folding pattern with three stem-loop structures than the Z3 sgRNA folding. This indicates the folding pattern is unlikely to have affected the editing outcome for WSL5 and Z3 loci. The efficiency of Cas9 or base editors varies between different sgRNA target genes. For example, rice PMS1 and OMTN1 were reported to be resistant to base editing with the tested sgRNAs (Hua et al. 2019a). Similarly, rice Tms9-1 gene was found to be resistant to ABE (Yan et al. 2018). The low efficiency or nil base editing might be due to the poor accessibility of the locus to the base editor. Association with nucleosomes or other proteins may also reduce or hinder accessibility of the target base to the deaminase. Incidentally, WSL5 protospacer has a stretch of four G bases at its 3′ end, which can act as three consecutive NGG PAMs and one NAG PAM. The presence of additional Gs next to PAM, which could shift the editing window, impede the rate of R-loop resolution or hinder Cas9′s alignment for proper gRNA–DNA interactions (Malina et al. 2015). Proper R-loop resolution is the prerequisite for the generation of accessible single-strand DNA targeted by the base editor. Recent studies expanded the targetability by developing plant base editors with NG PAM compatibility (Hua et al. 2019b; Ren et al. 2019; Zhong et al. 2019). The base editing efficiency at Z3 locus in our study is comparable to those studies (Hua et al. 2019b; Ren et al. 2019).
Although earlier studies showed ABEs applicability in rice base-editing, they have not reported the evidence of germline transmission of the mutation (Hua et al. 2018; Li et al. 2018; Yan et al. 2018). Analysis of segregation of monoallelic mutants in T1 generation revealed two lines from Z3 and one line from WSL5 did not follow the Mendelian pattern (Supplementary Table 2). Most of the T1 plants from both the lines harbor the base editor transgene as evidenced by ABE specific PCR. Generation of new mutations in the T1 generation was likely due to the presence of active base editors. This kind of unpredicted segregation was reported in Cas9 treated Arabidopsis (Fauser et al. 2014) and rice (Xu et al. 2015; Ishizaki 2016) plants. Plants descendent from mutants generated by active Cas9 are prone to further rounds of editing until the PAM and seed region of protospacer are destroyed by editing. However, the situation is quite different for the plants derived from base editing. If the activity window of base-editor contains only one targetable A, creation of biallelic germline mutation of the target in first generation would make the base editor unnoticeable in the following generation. Then the descendent from that mutant plant should all carry the homozygous mutation for that loci. However, if the generated mutation is monoallelic in the first generation, there are chances of activity from the base editor in the next generation. That was likely the case in our study, which could explain why three of the tested lines did not follow Mendelian segregation pattern. The vigilant nature of base editors in the T1 generation was further evidenced by the detection of ‘out of window’ base editing (12th base of the protospacer) by ABE7.10 in Z3 mutant line (10–105) (Fig. 2C). Earlier studies of ABEs in plants reported activity windows similar to that observed in mammalian cells (4–8 base position). However, Hua et al. (2018) reported editing at the 10th base by SpCas9 nickase based-ABE7.10, and at 12th and 14th by SaCas9 nickase based-ABE7.10. It is evident that the activity window of a base editor varies with the target.
Unlike cytosine base editors (CBEs) which are reported to cause unintended C → A or C → G editing, ABEs are not known to generate undesired mutation other than the expected A → G (Reviewed by Molla and Yang 2019b). However, a recent study showed ABE catalyzed cytosine conversion in human cell line (Kim et al. 2019). Strikingly, two T1 plants exhibited unusual base conversion in our study (Fig. 2D). In one plant, the target A was found to be converted to T. Mechanistically, ABE acts by deaminating deoxy-adenosine (dA) to deoxy-inosine (dI) which is read as guanosine by replication machinery, and as a result, it causes a post-replicative transition to G (Gaudelli et al. 2017). Although deoxy-inosine:deoxy-cytidine (dI:dC) is the most stable pair, dI can also pair with dA (Alseth et al. 2014). A dI:dA pairing would give rise to a post-replicative A → T conversion. That is one of the possible explanations for the A → T conversion we observed. In another plant, we observed that an adjacent G (6th base in the protospacer) was converted to A. Accidental deamination of guanine by tadA–tadA* may give rise to xanthine (X) which can subsequently pair with T (Herraiz and Galisteo 2018). An X:T pair may result in the conversion of an original G–C base pair to A–T.
Most of the previous studies on rice have only reported evidence on genomic changes by ABE and not provided direct evidence of mutant phenotypes (Hua et al. 2018; Li et al. 2018; Yan et al. 2018). One of the reasons to select WSL5 and Z3 as the target genes in our study to demonstrate ABE effectivity was that the base editing could be translated into a detectable phenotype. We have analyzed segregation patterns of the mutation and obtained mutant phenotypes in T1 plants. White striped leaf phenotype was evident in the wsl5 mutant, whereas z3 mutant exhibited altered phenotypes like transverse dark green/green sectors in mature leaf, shortened plant height, delayed flowering, and reduced panicle size in T1 plants (Fig. 3). These results confirm the findings of the original studies on WSL5 and Z3 genes (Kim et al. 2018; Liu et al. 2018).
In comparison to wild type plants, the z3/z3 homozygous mutant in our study apparently exhibited far more reduced height than that reported by Kim et al. (2018). It is likely that this difference might be due to the use of different genotypic background in the present study (Kitaake) from that used in the earlier study (Kinmaze) (Kim et al. 2018). Our study is the first following study on both the two genes providing data on the mutant phenotypes that validate their findings. Incidentally, as described earlier in a previous paragraph, we noticed editing of 12th A in the protospacer of Z3 which causes V540A mutation. We tracked the development of those plants carrying the heterozygous and homozygous mutation for 12th A and observed similar growth retardation. Homozygous plants showed a more prominent dwarf phenotype than the heterozygous plants (supplementary Fig. 2). This unexpected finding suggests that the change in 540th amino acid can also cause a phenotype similar to the z3 which was reported to occur due to alteration at 542nd amino acids (Kim et al. 2018). From this result, it can be assumed that the V540 is as critical as the S542 residue for the natural structure and function of ZEBRA3 protein.
Base editing allows us to rapidly and precisely introduce desired single nucleotide variation in the cultivated crop genome. In the present study, we developed targeted mutants for two rice genomic loci in less than 1 year. Generation of the same kind of genetic variation would have taken much longer time through traditional breeding techniques. Besides, genetic crosses including several rounds of backcrossing generate numerous nucleotide variants, often leads to undesirable effects as a result of genotype × genotype interactions (Huang et al. 2016). By introducing SNPs, base editing can create an intended trait or attenuate an unwanted trait. Genetic modification (GM) technology, which depends on insertion, integration, and stability of DNA sequences from other species or the same species, faces tight regulation in many countries. Although base editing involves foreign DNA transformation, it does not rely on the permanent presence of that DNA sequence in the genome. After the creation of intended genetic mutation, base editing machinery is no longer needed. As edited plants without any foreign DNA remnants could address the issues of government regulation and public acceptance, generation of base editor free mutants is crucial. In rice, transgene-free mutants could be readily obtained through genetic segregation of the base editor. In the T1 generation, we obtained both homozygous and heterozygous wsl5 or z3 mutants which are Cas9–TadA–TadA* free. Earlier studies on plant ABE did not report the generation of base editor free mutants (Hua et al. 2018, 2019a; Li et al. 2018; Yan et al. 2018). Since this base editor contains Cas9 nickase (nCas9) instead of fully active Cas9 and does not create a double-strand break (DSB), it minimizes the creation of DSB associated by-products such as indel, rearrangements, and translocation (Molla and Yang 2019b). We have not detected any indel generation in the targeted loci or in the studied potential off-target loci in the mutants. Our result corroborates with other findings in ABE induced mutant rice plants (Hua et al. 2018; Li et al. 2018; Yan et al. 2018). However, ABE induced indel generation (< 0.1%) was reported in a transient assay using protoplasts (Kang et al. 2018; Li et al. 2018).
In contrast to ABE, cytosine base editors (CBEs) have been reported to generate indels up to 9.61% in rice (Li et al. 2017) and up to 10–69% in tomato and rice (Shimatani et al. 2017). The superior performance of ABEs in terms of generating very low indel mutation than CBEs might be due to the less active cellular inosine excision repair system than the uracil excision repair system (Gaudelli et al. 2017). Off-target editing by Cas9 or any other nucleases is one of the major concerns in genome editing experiments. Similarly, base editors can also generate off-target editing due to the nonspecific interaction of nCas9 with partially homologous loci. When we assayed editing in a total of six potential off-target loci, no off-target mutations were detected demonstrating the precise nature of ABE in rice. These results are consistent with the data obtained in two earlier studies in rice (Hua et al. 2018; Yan et al. 2018). However, off-target editing may vary case-by-case depending on the unintended interaction of sgRNA, deaminase and Cas9.
In conclusion, we have developed an adenine base editing system for plants and generated precise rice mutants for two distinct loci within a very short period of time by employing CRISPR/Cas-mediated adenine base editors. Our study demonstrates that ABEs can be used to generate precise and heritable base change mutations in plants and to rapidly develop edited, yet transgene-free, crops. This work will help pave the way to functionally validate many SNPs or achieve desired agronomic traits by targeted A → G or T → C single base substitution in plant genome for natural and induced variants of genes.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Acknowledgements
Kutubuddin Molla would like to acknowledge the United States-India Educational Foundation (USIEF), New Delhi and the US Department of State for the Fulbright-Nehru Postdoctoral Fellowship. This work was supported by National Science Foundation Plant Genome Research Program Grant No. 1740874 and the USDA National Institute of Food and Agriculture and Hatch Appropriations under Project #PEN04659 and Accession #1016432 to Yinong Yang.
Compliance with ethical standards
Conflict of interest
On behalf of all authors, the corresponding author states that there is no conflict of interest.
Contributor Information
Kutubuddin A. Molla, Email: kutubuddin.molla@icar.gov.in
Justin Shih, Email: jws52@psu.edu.
Yinong Yang, Email: yuy3@psu.edu.
References
- Alseth I, Dalhus B, Bjørås M. Inosine in DNA and RNA. Curr Opin Genet Dev. 2014;26:116–123. doi: 10.1016/j.gde.2014.07.008. [DOI] [PubMed] [Google Scholar]
- Doudna JA, Charpentier E. The new frontier of genome engineering with CRISPR-Cas9. Science. 2014;346:1258096. doi: 10.1126/science.1258096. [DOI] [PubMed] [Google Scholar]
- Fauser F, Schiml S, Puchta H. Both CRISPR/Cas-based nucleases and nickases can be used efficiently for genome engineering in Arabidopsis thaliana. Plant J. 2014;79:348–359. doi: 10.1111/tpj.12554. [DOI] [PubMed] [Google Scholar]
- Gaudelli NM, Packer MS, Liu DR, Komor AC, Bryson DI, Badran AH, Rees HA, Gaudelli NM. Programmable base editing of A•T to G•C in genomic DNA without DNA cleavage. Nature. 2017;551:464–471. doi: 10.1038/nature24644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herraiz T, Galisteo J. Nitrosative deamination of 2′-deoxyguanosine and DNA by nitrite, and antinitrosating activity of β-carboline alkaloids and antioxidants. Food Chem Toxicol. 2018;112:282–289. doi: 10.1016/j.fct.2017.12.042. [DOI] [PubMed] [Google Scholar]
- Hua K, Tao X, Yuan F, Wang D, Zhu JK. Precise A•T to G•C base editing in the rice genome. Mol Plant. 2018;11:627–630. doi: 10.1016/j.molp.2018.02.007. [DOI] [PubMed] [Google Scholar]
- Hua K, Tao X, Zhu JK. Expanding the base editing scope in rice by using Cas9 variants. Plant Biotechnol J. 2019;17(2):499–504. doi: 10.1111/pbi.12993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hua K, Tao X, Han P, Wang R, Zhu JK. Genome engineering in rice using Cas9 variants that recognize NG PAM sequences. Mol Plant. 2019;12:1003–1014. doi: 10.1016/j.molp.2019.03.009. [DOI] [PubMed] [Google Scholar]
- Huang TK, Puchta H. CRISPR/Cas-mediated gene targeting in plants: finally a turn for the better for homologous recombination. Plant Cell Rep. 2019;38:443–453. doi: 10.1007/s00299-019-02379-0. [DOI] [PubMed] [Google Scholar]
- Huang S, Weigel D, Beachy RN, Li J. A proposed regulatory framework for genome-edited crops. Nat Genet. 2016;48:109–111. doi: 10.1038/ng.3484. [DOI] [PubMed] [Google Scholar]
- Ishizaki T. CRISPR/Cas9 in rice can induce new mutations in later generations, leading to chimerism and unpredicted segregation of the targeted mutation. Mol Breed. 2016;36:165. doi: 10.1007/s11032-016-0591-7. [DOI] [Google Scholar]
- Kang B-C, Choi M, Ryu J, Woo JW, Yun J-Y, Kim J-S, Shin Y, Kim S-T. Precision genome engineering through adenine base editing in plants. Nat Plants. 2018;4:427–431. doi: 10.1038/s41477-018-0178-x. [DOI] [PubMed] [Google Scholar]
- Kim SH, Kwon CT, Song G, Koh HJ, An G, Paek NC. The rice zebra3 (z3) mutation disrupts citrate distribution and produces transverse dark-green/green variegation in mature leaves. Rice. 2018;11:1–15. doi: 10.1186/s12284-017-0196-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim HS, Jeong YK, Hur JK, Kim JS, Bae S. Adenine base editors catalyze cytosine conversions in human cells. Nat Biotechnol. 2019;37(10):1145–1148. doi: 10.1038/s41587-019-0254-4. [DOI] [PubMed] [Google Scholar]
- Komor AC, Kim YB, Packer MS, Zuris JA, David R. Programmable editing of a target base in genomic DNA without double-stranded DNA cleavage. Nature. 2016;533(7603):420–424. doi: 10.1038/nature17946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Sun Y, Du J, Zhao Y, Xia L. Generation of targeted point mutations in rice by a modified CRISPR/Cas9 system. Mol Plant. 2017;10:526–529. doi: 10.1016/j.molp.2016.12.001. [DOI] [PubMed] [Google Scholar]
- Li C, Zong Y, Wang Y, Jin S, Zhang D, Song Q, Zhang R, Gao C. Expanded base editing in rice and wheat using a Cas9-adenosine deaminase fusion. Genome Biol. 2018;19:1–9. doi: 10.1186/s13059-018-1443-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X, Lan J, Huang Y, Cao P, Zhou C, Ren Y, He N, Liu S, Tian Y, Nguyen T, Jiang L, Wan J. WSL5, a pentatricopeptide repeat protein, is essential for chloroplast biogenesis in rice under cold stress. J Exp Bot. 2018;69:3949–3961. doi: 10.1093/jxb/ery214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malina A, Cameron CJF, Robert F, Blanchette M, Dostie J, Pelletier J. PAM multiplicity marks genomic target sites as inhibitory to CRISPR-Cas9 editing. Nat Commun. 2015;6:4–9. doi: 10.1038/ncomms10124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mei C, Qi M, Sheng G, Yang Y. Inducible overexpression of a rice allene oxide synthase gene increases the endogenous jasmonic acid level, PR gene expression, and host resistance to fungal infection. Mol Plant-Microb Interact. 2007;19:1127–1137. doi: 10.1094/MPMI-19-1127. [DOI] [PubMed] [Google Scholar]
- Molla KA, Yang Y. Predicting CRISPR/Cas-induced mutations for precise genome editing. Trends Biotechnol. 2019 doi: 10.1016/j.tibtech.2019.08.002. [DOI] [PubMed] [Google Scholar]
- Molla KA, Yang Y. CRISPR/Cas-mediated base editing: technical considerations and practical applications. Trends Biotechnol. 2019;37:1121–1142. doi: 10.1016/j.tibtech.2019.03.008. [DOI] [PubMed] [Google Scholar]
- Molla KA, Debnath AB, Ganie SA, Mondal TK. Identification and analysis of novel salt responsive candidate gene based SSRs (cgSSRs) from rice (Oryza sativa L) BMC Plant Biol. 2015 doi: 10.1186/s12870-015-0498-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishida K, Arazoe T, Yachie N, Banno S, Kakimoto M, Tabata M, Mochizuki M, Miyabe A, Araki M, Hara KY, Shimatani Z, Kondo A. Targeted nucleotide editing using hybrid prokaryotic and vertebrate adaptive immune systems. Science. 2016;353(6305):729. doi: 10.1126/science.aaf8729. [DOI] [PubMed] [Google Scholar]
- Ren B, Liu L, Li S, Kuang Y, Wang J, Zhang D, Zhou X, Lin H, Zhou H. Cas9-NG greatly expands the targeting scope of the genome-editing toolkit by recognizing NG and other atypical PAMs in rice. Mol Plant. 2019;12:1015–1026. doi: 10.1016/j.molp.2019.03.010. [DOI] [PubMed] [Google Scholar]
- Shimatani Z, Kashojiya S, Takayama M, Terada R, Arazoe T, Ishii H, Teramura H, Yamamoto T, Komatsu H, Miura K, Ezura H, Nishida K, Ariizumi T, Kondo A. Targeted base editing in rice and tomato using a CRISPR-Cas9 cytidine deaminase fusion. Nat Biotechnol. 2017;35:441–443. doi: 10.1038/nbt.3833. [DOI] [PubMed] [Google Scholar]
- Untergasser A, Cutcutache I, Koressaar T, Ye J, Faircloth BC, Remm M, Rozen SG. Primer3-new capabilities and interfaces. Nucleic Acids Res. 2012;40:1–12. doi: 10.1093/nar/gks596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie K, Yang Y. RNA-Guided genome editing in plants using a CRISPR-Cas system. Mol Plant. 2013;6:1975–1983. doi: 10.1093/mp/sst119. [DOI] [PubMed] [Google Scholar]
- Xie K, Minkenberg B, Yang Y. Boosting CRISPR/Cas9 multiplex editing capability with the endogenous tRNA-processing system. Proc Natl Acad Sci. 2015;112:3570–3575. doi: 10.1073/pnas.1420294112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie X, Zeng D, Liu Y-G, Li G, Ma X, Zhu Q. CRISPR-GE: a convenient software toolkit for CRISPR-based genome editing. Mol Plant. 2017;10:1246–1249. doi: 10.1016/j.molp.2017.06.004. [DOI] [PubMed] [Google Scholar]
- Xu RF, Li H, Qin RY, Li J, Qiu CH, Yang YC, Ma H, Li L, Wei PC, Yang JB. Generation of inheritable and “transgene clean” targeted genome-modified rice in later generations using the CRISPR/Cas9 system. Sci Rep. 2015;5:1–10. doi: 10.1038/srep11491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan F, Kuang Y, Ren B, Wang J, Zhang D, Lin H, Yang B, Zhou X, Zhou H. Highly efficient A·T to G·C base editing by Cas9n-guided tRNA adenosine deaminase in rice. Mol Plant. 2018;11:631–634. doi: 10.1016/j.molp.2018.02.008. [DOI] [PubMed] [Google Scholar]
- Zhong Z, Sretenovic S, Ren Q, Yang L, Bao Y, Qi C, Yuan M, He Y, Liu S, Liu X, Wang J, Huang L, Wang Y, Baby D, Wang D, Zhang T, Qi Y, Zhang Y. Improving plant genome editing with high-fidelity xCas9 and non-canonical PAM-targeting Cas9-NG. Mol Plant. 2019;12:1027–1036. doi: 10.1016/j.molp.2019.03.011. [DOI] [PubMed] [Google Scholar]
- Zong Y, Wang Y, Li C, Zhang R, Chen K, Ran Y, Qiu JL, Wang D, Gao C. Precise base editing in rice, wheat and maize with a Cas9-cytidine deaminase fusion. Nat Biotechnol. 2017;35:438–440. doi: 10.1038/nbt.3811. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.