Abstract
During the last decade, there has been rapid adoption of ground and aerial platforms with multiple sensors for phenotyping various biotic and abiotic stresses throughout the developmental stages of the crop plant. High throughput phenotyping (HTP) involves the application of these tools to phenotype the plants and can vary from ground-based imaging to aerial phenotyping to remote sensing. Adoption of these HTP tools has tried to reduce the phenotyping bottleneck in breeding programs and help to increase the pace of genetic gain. More specifically, several root phenotyping tools are discussed to study the plant’s hidden half and an area long neglected. However, the use of these HTP technologies produces big data sets that impede the inference from those datasets. Machine learning and deep learning provide an alternative opportunity for the extraction of useful information for making conclusions. These are interdisciplinary approaches for data analysis using probability, statistics, classification, regression, decision theory, data visualization, and neural networks to relate information extracted with the phenotypes obtained. These techniques use feature extraction, identification, classification, and prediction criteria to identify pertinent data for use in plant breeding and pathology activities. This review focuses on the recent findings where machine learning and deep learning approaches have been used for plant stress phenotyping with data being collected using various HTP platforms. We have provided a comprehensive overview of different machine learning and deep learning tools available with their potential advantages and pitfalls. Overall, this review provides an avenue for studying various HTP platforms with particular emphasis on using the machine learning and deep learning tools for drawing legitimate conclusions. Finally, we propose the conceptual challenges being faced and provide insights on future perspectives for managing those issues.
Keywords: Biotic and abiotic stresses, Deep learning, Ground-based imaging, High throughput phenotyping, Machine learning, Unmanned aerial vehicle
Introduction
The world population is expected to reach approximately 9 to 10 billion by 2050; therefore a gain of around 25–70% above present-day production levels will be required to meet these burgeoning population demands (Hunter et al. 2017). Further, various biotic and abiotic factors cause adverse environmental conditions or stress for crop plants, resulting in a significant reduction in their yields. This significant reduction in crop yield due to stress can jeopardize global food security (Strange and Scott 2005). Enhancement of crop yields is an ever-changing global challenge for plant breeders, entomologists, pathologists, and farmers. Hence, an in-depth understanding of plant stress is pivotal for improving yield protection for sustainable production systems (Pessarakli 2019). Plant scientists rely on crop phenotyping for precise and reliable trait collection and utilization of genetic resources and tools to accomplish their research goals.
Plant phenotyping is defined as the comprehensive assessment of complex traits of plants such as development, growth, resistance, tolerance, physiology, architecture, yield, ecology, and the elementary measurement of individual quantitative parameters that form the foundation for complex trait assessment (Li et al. 2014). Breeding programs generally aim to phenotype large populations for numerous traits throughout the crop cycle (Sandhu et al. 2021b,c). This phenotyping challenge is further aggravated by the need to sample at multiple environment with replicated trials. Traditional phenotyping is very costly, laborious, destructive, and could decrease the significance or preciseness of the results. The development of automated, high throughput phenotyping (HTP) systems merged with artificial intelligence has largely overcome the problems linked with the contemporary state-of-the-art crop stress phenotyping. HTP has offered great potential for non-destructive and effective field-based plant phenotyping. Manual, semi-autonomous or autonomous platforms furnished with single or multiple sensors record temporal and spatial data, resulting in large amounts of data for storage and analysis (Kaur et al. 2021; Sandhu et al. 2021c). For the analysis and interpretation of these massive datasets, machine learning (ML) and its subtypes, i.e. deep learning (DL) approaches, are utilized (Ashourloo et al. 2014; Atieno et al. 2017; LeCun et al. 2015; Lin et al. 2019; Ramcharan et al. 2019; Sandhu et al. 2021a).
Machine learning is a multidisciplinary approach that largely relies on probability and decision theories, visualization, and optimization. Machine learning approaches can handle large amounts of data effectively and allow plant researchers to search massive datasets to discover patterns by concurrently looking at a combination of traits rather than analyzing each trait or feature separately. The capability of identifying a hierarchy of features and inferring generalized trends from given data is one of the major attributes responsible for the immense success of ML tools. Supervised and unsupervised learning are the two major ML techniques, which have been extensively used for biotic and abiotic stress phenotyping in crops (Ashourloo et al. 2014; Peña et al. 2015; Raza et al. 2015; Naik et al. 2017; Zhang et al. 2019c). Traditional ML approaches require significant efforts for feature designing, which is a laborious procedure and calls for expertise in computation and image analysis, thereby hindering traditional ML approaches for trait phenotyping applications. DL has emerged as a potential ML approach that incorporates benefits of both the advanced computing power and massive datasets and allows for hierarchical data learning (LeCun et al. 2015; Min et al. 2017). Further, DL also bypasses the need for feature designing, as the features are learned automatically from the data. The important DL models include multilayer perceptron (MLP), generative adversarial networks (GAN), convolutional neural network (CNN), and recurrent neural network (RNN) (LeCun et al. 2015). Deep CNNs primarily use DL architecture that have now attained state-of-the-art performance for crucial computer vision tasks; for instance, image classification, object recognition, and image segmentation (Pérez-Enciso and Zingaretti 2019).
In this article, we review state-of-the-art image-based HTP methods with discussion on different imaging platforms, imaging techniques, and spectral indices deployed for plant stress phenotyping. Furthermore, we provide a comprehensive overview of the different ML and DL tools available with their comparative advantages and shortcomings. We focus more on DL applications that mainly use image data because digital imaging is comparatively cheap, may be combined with the ground, manual and aerial platforms employed in a scalable manner, and do not require much technical expertise to install with off-the-shelf components for HTP of plant stress. Furthermore, we also summarize several recent studies involving ML and DL approaches for phenotyping different biotic and abiotic stresses in plants.
Phenotyping Platforms
The area of plant stress phenotyping is steadily progressing, with destructive, low throughput phenotyping protocols/methods being substituted by non-invasive high-throughput methods (Barbedo 2019). Expeditious developments in non-invasive affordable sensors and imaging techniques and tools over the decades have transformed plant phenomics. Moreover, these developments have brought harmony between the sensors, imaging techniques and analytical tools. This consonance has led to the development of one-piece compact imaging platforms for HTP studies. Several HTP platforms exist and are presently employed to phenotype different biotic and abiotic stress-associated traits in various crops (Table 1). Examples of these platforms include an automated platform i.e. “PHENOPSIS” for phenotyping plant responses to soil water stress in Arabidopsis (Granier et al. 2006); “GROWSCREEN FLUORO” to phenotype leaf growth and chlorophyll fluorescence which allowed the detection of tolerance to different abiotic stresses in Arabidopsis (Arabidopsis thaliana L.) (Jansen et al. 2009); “LemnaTec 3D Scanalyzer system” for non-invasive screening of different salinity tolerance traits in rice (Oryzae sativa L.) (Hairmansis et al. 2014); “HyperART” for non-destructive quantification of leaf traits such as leaf chlorophyll content and disease severity on leaves in four different crop species (barley, maize, tomato and rapeseed) (Bergsträsser et al. 2015); “PhenoBox” for detection of head smut and corn smut diseases on Brachypodium and maize (Zea mays L.), respectively, and salt stress response in tobacco (Nicotiana tabacum L.) (Czedik-Eysenberg et al. 2018); “PHENOVISION” for detection of drought stress and recovery in maize plants (Asaari et al. 2019); “PhénoField” for characterization of different abiotic stresses in wheat (Triticum aestivum L.) (Beauchêne et al. 2019); and the “PlantScreen™ Robotic XYZ System” for analyzing different traits associated with drought tolerance in rice (Kim et al. 2020).
Table 1.
Details of some selected imaging platforms used for trait phenotyping for biotic and abiotic stress and other morphological traits in crops
| Platform | Traits recorded | Crop | References |
|---|---|---|---|
| A. Biotic and abiotic stresses | |||
| PHENOPSIS | Plant responses to water stress | Arabidopsis (Arabidopsis thaliana) | Granier et al. (2006) |
| PHENODYN | Soil water status (drought scenarios), leaf elongation rate, and micrometeorological variable | Rice (Oryza sativa) and maize (Zea mays) | Sadok et al. (2007) |
| GROWSCREENFLUORO | Leaf growth and chlorophyll fluorescence which allows detection of stress tolerance | Arabidopsis (Arabidopsis thaliana) | Jansen et al. (2009) |
| Field monitoring support system | Occurrence of the rice bug in the field | Rice (Oryza sativa) | Fukatsu et al. ((2012) |
| BreedVision | Lodging, plant moisture content, biomass yield or tiller density | Triticale (xTriticosecale Wittmack L.) | Busemeyer et al. (2013a) |
| LemnaTec 3D scanalyzer system | Salinity tolerance traits | Rice (Oryza sativa) | Hairmansis et al. (2014) |
| HyperART | Leaf traits such as disease severity or leaf chlorophyll content | Barley (Hordeum vulgare), maize (Zea mays), tomato (Lycopersicon esculentum) and rapeseed (Brassica rapa) | Bergsträsser et al. (2015) |
| Automated video tracking platform | Resistance to aphids and other piercing-sucking insects | lettuce (Lactuca sativa) and Arabidopsis (Arabidopsis thaliana) | Kloth et al. (2015) |
| RhizoTubes | Root related traits under non-stressed and stressed conditions | Medicago (Medicago truncatula), pea (Pisum sativum), rapeseed (Brassica napus), grapes (Vitis vinifera), wheat (Triticum aestivum) | Jeudy et al. (2016) |
| RADIX | Root and shoot related traits under control and as well as stress conditions | Maize (Zea mays) | Le Marié et al. (2016) |
| PhenoBox | Detection of head smut fungus and corn smut on Brachypodium and maize, respectively, and salt stress response in Tobacco | Brachypodium (Brachypodium distachyon), maize (Zea mays), tobacco (Nicotiana benthamiana) | Czedik-Eysenberg et al. (2018) |
| PHENOVISION | Detection of drought stress and recovery | Maize (Zea mays) | Asaari et al. (2019) |
| PhénoField | Characterization of different abiotic stresses | Wheat (Triticum aestivum) | Beauchêne et al. (2019) |
| Liaphen | Leaf expansion or transpiration rate in response to water deficit | Sunflower (Helianthus annuus) | Gosseau et al. (2018) |
| PlantScreen™ robotic XYZ system | Drought tolerance traits | Rice (Oryza sativa) | Kim et al. (2020) |
| PHENOTIC | Evaluation of plant resistance to pathogens, evaluation of virulence of pathogens | Horticultural crops | Boureau 2020) |
| PhenoImage | Plant responses to water stress | Wheat (Triticum aestivum), sorghum (Sorghum bicolor) | Zhu et al. (2021) |
| B. Morphological and physiological traits (recorded under unstressed conditions) | |||
| Plant root monitoring platform (PlaRoM) | Root related traits | Arabidopsis (Arabidopsis thaliana) | Yazdanbakhsh and Fisahn (2009) |
| Phenoscope | Complex traits which involved in growth responses to the environment | Arabidopsis (Arabidopsis thaliana) | Tisné et al. (2013) |
| High-throughput rice phenotyping facility (HRPF) | Agronomic traits | Rice (Oryza sativa) | Yang et al. (2014) |
| Zeppelin NT aircraft | Leaf area index, leaf biomass, early vigour, plant height | Maize (Zea mays) | Liebisch et al. (2015) |
| Rhizoponics | Root related traits | Arabidopsis (Arabidopsis thaliana) | Mathieu et al. (2015) |
| Phenocart | Morphological traits | Wheat (Triticum aestivum) | Crain et al. (2016) |
| PhenoArch phenotyping platform | Light interception and radiation‐use efficiency | Maize (Zea mays) | Cabrera-Bosquet et al. (2016) |
| Phenovator | Growth, photosynthesis, and spectral reflectance | Arabidopsis (Arabidopsis thaliana) | Flood et al. (2016) |
| PHENOARCH | For tracking the growths of maize ear and silks | Maize (Zea mays) | Brichet et al. (2017) |
| Phenobot 1.0 | Biomass-related traits | Sorghum (Sorghum bicolor) | Salas Fernandez et al. (2017) |
| Field Scanalyzer | Morphological traits | Wheat (Triticum aestivum) | Virlet et al. (2016) |
| CropQuant | Performance-related traits | Wheat (Triticum aestivum) | Zhou et al. (2017) |
| PhenoRoots | Root related traits | Cotton (Gossypium hirsutum L.) | Martins et al. (2020) |
| Self-propelled electric HTPP platform | Plant height | Wheat (Triticum aestivum) | Pérez-Ruiz et al. (2020) |
| MVS-Pheno | Plant height and leaf traits | Maize (Zea mays) | Wu et al. (2020) |
Data has also been recorded in an automated and high throughput manner for root and shoot related traits, leaf traits, plant height, plant biomass, early vigor, radiation use efficiency, photosynthesis in different plant species such as rice, wheat, maize, sorghum (Sorghum bicolor L.), cotton (Gossypium hirsutum L.), Arabidopsis, Brachypodium (Brachypodium distachyon L.), rapeseed (Brassica napus L.), and barley (Hordeum vulgare L.) among others using different phenotyping platforms such as RootReader3D (Clark et al. 2011), GROWSCREEN-Rhizo (Nagel et al., 2012), Zeppelin NT aircraft (Liebisch et al. 2015), Phenocart (Crain et al. 2016), Phenovator (Flood et al. 2016), PHENOARCH (Brichet et al. 2017), Field Scanalyzer (Virlet et al. 2016), CropQuant (Zhou et al. 2017), and MVS-Pheno (Wu et al. 2020) (Table 1). These platforms have the potential to be utilized for HTP of traits associated with stress tolerance/resistance in different crops.
In the last decade, several state-of-the-art phenomics centers have been established across the world which utilize sensor platforms for phenotyping under controlled conditions. Major phenomics centers include ‘High-Resolution Plant Phenomics Center’ and ‘Plant Accelerator’ in Australia; ‘Leibniz Institute of Plant Genetics and Crop Plant Research’ and the ‘Julich Plant Phenotyping Center’ in Germany; the ‘National Plant Phenomics Center’ in the United Kingdom and the ‘Nanaji Deshmukh Plant Phenomics Center’ in India (Mir et al. 2019). To disseminate information about HTP, an international association of major plant phenotyping centers was also established in Germany, known as the International Plant Phenotyping Network (https://www.plant-phenotyping.org/IPPN_home).
Significant efforts are also being made to develop advanced technologies for use under field conditions at industrial and experimental scales. Some private companies, including ‘LemnaTec’, ‘PhenoSpex’, ‘Phenokey’, ‘WIWAM’, ‘Photon System Instruments’, and ‘We Provide Solutions’ provide large-scale, customized HTP platforms for both controlled and field environments (Gehan and Kellogg 2017; Mir et al. 2019). A list of major HTP platforms being utilized in different laboratories is given in Table 1. These imaging platforms can be broken down into their components to understand the evolution and scope of HTP studies. Understanding the subunits of imaging platforms that include imaging sensors, imaging techniques, and analytical tools such as spectral indices, would help justify the role of ML and DL in HTP studies and these have been summarized below.
Imaging Techniques
Phenomics has been extensively used to monitor diseases, pest infestations, drought stress, nutrient status, growth, presence of weeds and yield under stresses and normal conditions in different crop species (Barbedo 2019). Technological advancement has made novel imaging techniques available for use in HTP. Imaging techniques range from handheld mobile phones to highly flexible drone imaging using unmanned aerial vehicles (UAV). UAVs offer a platform that rapidly records data using different imaging sensors over large areas and potentially gives images with high spatial resolution. UAV can be used to cover plots or multiple fields in one flight, but their limited battery capacity reduces their utility for very large-scale HTP. Remote sensing using satellites imagery has been extensively used in assessing plant stresses since the early 1970s (Saini et al. 2022; Sishodia et al. 2020). Multispectral images obtained from satellites can be used to assess drought conditions in a particular area, crop damage due to insect pests, for example, tracking the damages caused by a swarm of locusts, or crop damage due to diseases (Kaur et al. 2021). As mentioned earlier, the field of HTP is an evolving area with consistent changes and same is true for the imaging techniques. To score minute changes in plant development with more detail and automate the imaging process, ground-based imaging platforms are also put to use. Although ground-based imaging platforms has the same limitation as drones due to limited battery capacity, but they provide more detailed and accurate images at the individual plant to individual branches, even down to the single leaf level. More detail provides more data and thus build accurate models and assessments. Moreover, the ground-based imaging platforms can be programmed to engage in a time-scheduled analysis even in the absence of the researcher, making it more convenient and efficient. All of these imaging techniques generate terabytes of data per day and thus cannot be handled manually hence they need the assistance of ML and DL programs for their management. Different machine learning methods, specifically deep learning, may efficiently deal with the millions of images quickly and with high reliability (Ashourloo et al. 2014; LeCun et al. 2015; Atieno et al. 2017). Now we are providing a brief overview of different imaging techniques used for plant stress phenotyping.
Satellite Imagery
Satellite imagery or remote sensing is the oldest of all HTP method. Satellites can cover a large area from 1,000 hectares to an entire county at a time. Earth observation satellites installed with multiple sensors having large apertures are used to capture ground information. These sensors vary from RGB, multispectral, hyperspectral, thermal and time of flight sensors. The multispectral sensors collect information in specific wavelengths (bands) out of the electromagnetic (EM) spectrum. With an ability to target 2–10 bands out of the EM spectrum at a time, these sensors generally target Red (R), Blue (B), and Green (G) bands which are visible to the human eye. The information collected from each specific band is then overlapped with the other bands to generate high-resolution RGB images. Other than these bands, near infra-red (NIR) bands or infrared (IR) bands are also used (Pineda et al. 2020). Multispectral sensors can have a spectral resolution as high as 0.25 m, and is continuously improving with advancements in technology.
Hyperspectral sensors can target numerous bands (up to thousands), but in a narrower spectral range (Pettorelli 2019). As they target multiple bands, their spectral resolution is higher than multispectral sensors. Studies have been conducted to use hyperspectral images for plant disease identification (Das et al. 2015; Nagasubramanian et al. 2019). Most of multispectral and hyperspectral sensors use the reflected sunlight from the earth’s surface to gather information which make these passive sensors. Conversely, active sensors include RADAR (Radio Detection and Ranging) and LiDAR (Light Detecting and Ranging), which both emit radiation to gather ground information (Teke et al. 2013). The band information gathered from multispectral and hyperspectral sensors is evaluated using different spectral indices to assess plant health and conditions.
One of the biggest limitations of satellite imaging is the high cost associated with constructing and launching satellites. A list of unique satellite sensors with their spatial resolution is provided by Pettorelli (2019). Some of the satellite involved in plant stress assessment include Resourcesat-2 and Resourcesat-2A (Indian Space Research Organization), Sentinel-2 A + B twin platform (European Space Agency) (Segarra et al. 2020), EO-1 Hyperion (National Aeronautics and Space Administration) (Apan et al. 2004), Spot-6 and Spot-7 (Centre national d'études spatiales), and KOMPSAT-3A (Korean Aerospace Industries, Ltd.).
Mobile Cameras/Imaging
Mobile phones are equipped with high-quality cameras limited to basic photography, but some manufacturers add advanced sensors like LiDAR for 3D imaging. The rapid development of smartphones with powerful computing and high-resolution cameras has also facilitated the creation of mobile applications with expanding utility. Moreover, features of smartphone technology are also being incorporated into other portable devices/instruments, thereby expanding the range of sensors and strengthening the portability and connectivity of conventional phenotyping equipment. These advancements put researchers at an advantage as they can conveniently record the phenotype with a portable handheld device. These advances, however, do not provide any practical use for research purposes, as taking pictures of each plant in the field is not practical using this method. It is equivalent to manual phenotyping as researchers would still need to go and look at every plant or its parts. Although mobile imaging can help diagnose biotic (Hallau et al. 2017) and abiotic stresses (Naik et al. 2017) using ML and DL, it doesn’t have any utility in HTP studies. Many mobile applications are available that can help a researcher or even a farmer run a quick diagnostic of symptoms before an in-depth analysis. Such mobile applications may be adapted to other important crops and their diseases, thereby contributing to better decision-making in integrated disease management but this utility is limited.
UAV Imaging
Imaging using UAV is considered the most convenient and economical for large-scale HTP studies (Ehsani and Maja 2013). UAVs take multiple photographs during their flight. The images captured include parts of the whole field which are then stitched together to form a larger image called an orthomosaic providing an overall view of the field. Open drone map, Pix4D and QGIS are some of the software applications that can be used to create these orthomosaics. Images from UAVs have high resolution as they fly closer to the ground compared to satellites. This can be particularly helpful for hyperspectral sensors as they have a low spatial resolution. UAVs can carry all the sensors that can be installed on any satellite due to increase of their payload capacity. Still, they cannot cover as much spatial area as satellites because of their limited battery capacity and flight height. Like the satellites, UAVs provide data in the form of various spectral bands that are further evaluated using spectral indices. Numerous studies have been conducted where drone imaging is used to assess biotic and abiotic stresses in plants. Some of these are provided in Table 2.
Table 2.
Different imaging techniques, plant assessment studies conducted using the imaging technique, and their advantages and limitations
| Imaging technique | Studies conducted using the technique | Advantages | Limitations |
|---|---|---|---|
| Satellite Imagery | Heat stress (Cârlan et al. 2020), wheat yield (Fieuzal et al. 2020), cotton yield (He and Mostovoy 2019), dry bean (Sankaran et al. 2019), drought assessment (Babaeian et al. 2019), hessian Fly infestation wheat (Bhattarai et al. 2019), crop water stress (Omran 2018), alien invasive species (Royimani et al. 2018), purple spot diseases asparagus (Navrozidis et al. 2018), phytophthora root rot in avocado (Salgadoe et al. 2018), heavy metal induced stress rice (Liu et al. 2018), wheat yellow rust (Zheng et al. 2018), cotton root rot (Song et al. 2017), red palm weevil attack (Bannari et al. 2017), wheat biomass (Dong et al. 2016), powdery mildew (Yuan et al. 2016), orange rust sugarcane (Apan et al. 2004) | Easily covers very large areas | High cost of satellite and their launch |
| The data can easily help predict droughts and epiphytotics as very large areas can be covered at the same time | RGB imaging is hindered by clouds and inclement weather | ||
| Temporal cycle of satellite limits use at any time | |||
| Mobile Cameras | Cercospora leaf spot(Hallau et al. 2017), iron deficiency chlorosis severity in soybean(Naik et al. 2017), salinity stress tolerance (Awlia et al. 2016) | Convenient and portable | No practical use in research as limited to only a handful of samples |
| Rapid | |||
| No operational costs | |||
| UAV/Drone imaging | Target spot and Bacterial spot Tomato (Abdulridha et al. 2020), iron deficiency soybean (Dobbels and Lorenz 2019), early stress detection (Sagan et al. 2019), dry bean (Sankaran et al. 2019), grain yield wheat (Hassan et al. 2019), plant Nitrogen content (Camino et al. 2018), wheat biomass (Yue et al. 2017), maize yield (Maresma et al. 2016), grapevine leaf stripe (Gennaro et al. 2016), bacterial Leaf Blight in rice (Das et al. 2015), low-nitrogen (low-N) stress tolerance (Zaman-Allah et al. 2015) | Very economical | Cannot cover very large areas like a satellite |
| Can cover large areas | Creating orthomosaics out of a myriad of sectional images | ||
| High resolution data | Limited battery capacity | ||
| Easy to operate with low learning curve | |||
| Imaging using robots | Nitrogen content maize (Chen et al. 2021), vegetation indices (Bai et al. 2019), Xylella fastidiosa infection Olive (Rey et al. 2019), leaf traits (Atefi et al. 2019), plant architecture (Qiu et al. 2019), heat stress and stripe rust resistance wheat (Zhang et al. 2019a), plant architecture (Young et al. 2018) | Most advanced technique | Still an evolving technique |
| Highly efficient as it provides human-like manual phenotyping results | Much work required for workflow and data management | ||
| Test images are run through models autonomously to assess plant health, no need of separate evaluation with spectral indices | High initial cost of equipment | ||
| Programmable to act like a cron job, increases efficiency | Programming skills are required |
Ground Based Imaging Platforms
Imaging using ground-based platforms is the most advanced of all of the techniques. Ground-based platforms provide very close range imaging for plant stress assessment (Mishra et al. 2020). The proximity provides human-like manual phenotyping with greater efficiency (Atefi et al. 2019). Most of the ground based platforms are autonomous systems, they can be integrated with onboard chips to evaluate the parameters of each phenotype (Vougioukas 2019). Such autonomous systems, alongside test images, add much more information to the project, such as phenotype score, metrics, parameters, generating up to terabytes of data per day. To organize all of the generated information, it becomes important to have efficient workflow managers such as PhytoOracle (Peri 2020) for improved data management and processing. Ground-based platforms are the perfect example of IoT-based intelligent systems for plant stress assessment and HTP studies (Das et al., 2019).
Spectral Indices for Plant Stress Phenotyping
Images captured by the aforementioned techniques need to be decoded, and spectral indices (SIs) are used to assess the information in these images (Hunt et al. 2013). SIs involve conducting various sets of operations on different spectral layers of an image. These sets of operations include some mathematical calculations and combination of spectral reflectance from two or more wavelengths. The result of this mathematical combination generates a number that denotes the relative abundance of the feature of interest (Jackson and Huete 1991). Various types of SIs are available to assess different types of features captured in an image. For the purposes of this review, we will discuss only vegetation indices (VIs) which are a spectral calculation conducted using different spectral bands, for decoding features and information about vegetation captured in an image. VIs can provide a plethora of information such as plant phenotype, plant architecture, stress level, and biomass (Kokhan and Vostokov 2020). The most important factor in using VIs is determining is the right kind of VIs to be incorporated for a particular application (Xue and Su 2017). Various types of VIs can be used for plant assessment studies in HTP programs. Because healthy plants reflect more IR radiation than stressed plants (Xue and Su 2017), mostly NIR or IR bands are used to monitor aspects like biotic (Mahlein et al. 2012a; Oerke et al. 2014) and abiotic stresses (Prashar and Jones 2016). Therefore, VIs involving NIR/IR data such as normalized difference vegetation index (NDVI) become of prime importance in plant stress assessment studies. A list of various types of VIs is provided in Table 3.
Table 3.
Commonly used vegetation indices for plant stress phenotyping. Rn, Rre, Rr, Rg, and Rb are the reflectance bands for NIR, Red Edge, red, green, and blue wavelengths
| Index name | Formula | Relevance | References |
|---|---|---|---|
| Normalized difference vegetation Index | (Rn − Rr)/(Rn + Rr) | Plant health monitoring, assess plant stress | Tucker (1979) |
| Triangular vegetation index | 0.5 [120(Rn − Rg) − 200 (Rr − Rg)] | Leaf chlorophyll measurements and leaf area index | Broge and Leblanc (2001) |
| Triangular greenness index | Rg − 0.39·Rr − 0.61·Rb | Leaf chlorophyll measurements | Hunt et al. (2011) |
| Normalized green red difference index | (Rg − Rr)/(Rg + Rr) | Biomass measurements | Tucker (1979) |
| Green normalized difference vegetation index | (Rn − Rg)/(Rn + Rg) | Leaf chlorophyll measurements | Gitelson et al. (1996) |
| Enhanced vegetation index | 2.5(Rn − Rr)/(Rn + 6·Rr − 7.5·Rb + 1) | Improved vegetation monitoring and biomass measurements | Huete et al. (2002) |
| Chlorophyll vegetation index | Rn·Rr/Rg2 | Leaf chlorophyll measurements | Vincini et al. (2008) |
| Chlorophyll index—green | Rn/Rg − 1 | Leaf chlorophyll measurements | Gitelson et al. (2003) |
| Chlorophyll index—red edge | Rn/Rre − 1 | Leaf chlorophyll measurements | Gitelson et al. (2003) |
| Visible atmospherically resistant index | (Rg − Rr)/(Rg + Rr − Rb) | Highlights vegetation in images | Gitelson et al. (2002) |
| Triangular chlorophyll index | 1.2(R700 − R550) − 1.5(R670 − R550)·√(R700/R670) | Leaf chlorophyll measurements | Haboudane et al. (2008) |
| Green leaf index | (2·Rg − Rr − Rb)/(2·Rg + Rr + Rb) | Differentiates vegetation from bare soil | Louhaichi et al. (2001) |
| Normalized difference red edge index | (Rn − Rre)/(Rn + Rre) | Leaf chlorophyll measurements | Gitelson and Merzlyak (1994) |
| MERIS total chlorophyll index | (R750 − R710)/(R710 − R680) | Leaf chlorophyll measurements | Dash and Curran (2004) |
Big Data and Machine Learning
With the rapid adoption of HTP platforms in agriculture, it has created enormous volume, variety, and veracity of the data with collection at multiple time points, causing big data issues. The ability to analyze and understand data is becoming critical for the development of new tools and findings. A report from the Mckinsey industry (Manyika et al. 2011) shows that the amount of data generated is increasing at approximately 50% per year, which is equivalent to a 40-fold rise in data since 2001. Even a few years ago storing such enormous data was challenging, but with better storage capacities, it is now possible for such data to be efficiently archived and potentially used in the near future. The biggest hesitation in adopting HTP for most agriculture operations at a massive scale is the challenge with analysis and interpretation of the information. Machine learning and deep learning approaches act as a valuable computer science technique, which could be adopted for increasing its utilization in agriculture, especially in monitoring plant stresses. There are numerous studies available where ML approaches have been used for processing images to identify different types and levels of stresses such as powdery mildew in cucumber (Cucumis sativus L.) (Lin et al. 2019), aflatoxin level in maize (Yao et al. 2013), leaf rust in wheat (Ashourloo et al. 2014), and salinity stress in chickpea (Cicer arietinum L.) (Atieno et al. 2017) and many more examples (Table 4). These studies provided an excellent avenue for how ML could be used for managing plant stresses.
Table 4.
Studies using various machine and deep learning tools for abiotic stress management in field crops during the last decade
| Plant | Plant trait/disease | Spectral range (nm) | Domain (field/lab) | Sensing modality | Data analysis | References |
|---|---|---|---|---|---|---|
| Barley | Drought | 400–900 | Greenhouse | Hyperspectral imaging | Simplex volume maximization | Mer et al. (2012) |
| Barley | Drought | 430–890 | Field | Hyperspectral imaging | Support vector machine | Behmann et al. (2014) |
| Barley | Drought | 400–1000 | Field | Hyperspectral imaging | Dirichlet aggregation regression | Kersting et al. (2012) |
| Chickpea | Salinity | 380–760 | Greenhouse | RGB imaging | PLS and correlation analysis | Atieno et al. (2017) |
| Chilean strawberry (Fragraria chiloensis) | Salt stress | 350–2500 | Greenhouse | Spectral reflectance imaging | ANOVA, SRI linear regression analysis, multilinear regression analysis | Garriga et al. (2014) |
| Citrus | Chlorophyll fluorescence water Stress | 400–885 | Field | Non-imaging | Regression | Zarco-Tejada et al. (2012) |
| Cotton | Water stress |
490–900 7500–13,500 |
Field | Thermal Imaging | Mapping and Correlation | Bian et al. (2019) |
| Maize | Water stress | 475–840 | Field | Non-imaging | Correlations among Indices | Zhang et al. (2019c) |
| Maize | Nitrogen, Oxygen, and Ash | 400–2500 | Field | Non-imaging | ANOVA and regression | Cabrera-Bosquet et al. (2011) |
| Maize | Weeds | 380–760 | Field | RGB imaging | Image processing and color segmentation | Burgos-Artizzu et al. (2011) |
| Maize | Weeds | 380–760 | Field | RGB imaging | Support vector machine for separating plants based on spectral components | Guerrero et al. (2012) |
| Oilseed rape | Proteins | 500–900 | Laboratory | Near-infrared and hyperspectral imaging | PLS | Zhang et al. (2015) |
| Pepper | Nitrogen | 380–1030 | Laboratory | Hyperspectral imaging | PLSR | Yu et al. (2014) |
| Rice | Nitrogen | 400–1000 | Field | Hyperspectral imaging | PLSR | Onoyama et al. (2013) |
| Rice | Salinity | 380–760 | Field | RGB imaging | Smoothing splines curves and gene mapping | Al-Tamimi et al. (2016) |
| Rice | Salt stress | 400–500 | Greenhouse | Fluorescence imaging | Hierarchical clustering, Pearson correlation analysis, non-linear mixed model | Campbell et al. (2015) |
| Soybean | Iron | 380–760 | Field | RGB imaging | Decision trees, random forests, KNN, LDA and QDA | Naik et al. (2017) |
| Soybean | Drought | 400–780 | Greenhouse | Hyperspectral fluorescence imaging | PLSR | Mo et al. (2015) |
| Spinach | Quality | 400–1000 | Lab | Hyperspectral imaging | PLS-DA | Diezma et al. (2013) |
| Spinach | Drought | 380–760, and 8000–14,000 | Field | Visible and thermal imaging | Support vector machine and Gaussian processes classifier | Raza et al. (2014) |
| Spring Wheat | Water Stress | 350–2500 | Field | Non-imaging | Vegetation indices and regression | Wang et al. (2015) |
| Sunflower | Weed | 450–780 | Field | Visible light and multispectral imaging | Classification | Peña et al. (2015) |
| Vineyard | Water stress variability | 530–800 | Field | Thermal and multispectral Imaging | Regression | Baluja et al. (2012) |
| Wheat | Water stress | 841–1652 | Landsat data | Multispectral imaging | Indices and regression | Dangwal et al. (2016) |
| Wheat | Nitrogen | 400–1000 | Both | Hyperspectral imaging | PLSR | Vigneau et al. (2011) |
| Wheat | Nitrogen, phosphorus, potassium, sulfur | 350–2500 | Field | Hyperspectral imaging | Regression | Mahajan et al. (2014) |
| Wheat | Ozone | 300–1100 | Field | Non-imaging | Correlation analysis | Chi et al. (2016) |
ML provides an alternative opportunity to extract valuable information for making conclusions, which were previously difficult because of issues discovering patterns from the datasets (Sandhu et al. 2020, 2021b). ML is an interdisciplinary approach for data analysis using probability, statistics, classification, regression, decision theory, data visualization, and neural networks to relate information extracted with the phenotype obtained (Samuel 1959). ML provides a significant advantage to plant breeders, pathologists, physiologists and agronomists for the extraction of multiple parameters to analyze traits together instead of traditional methods that focused on a single feature at a time. The other important advantage with ML is directly linking the variables extracted from HTP data to plant stresses (Zhang et al. 2019c), biomass accumulation (Busemeyer et al. 2013b), grain yield (Crain et al. 2018), and soil characteristics. ML’s greatest success involves inferring trends from the data and generalizing the results by training the model. There has been a recent adoption of ML in bioinformatics (Min et al. 2017), cell biology, epigenetics (Samantara et al. 2021), plant breeding (Sandhu et al. 2021a), pathology (Ashourloo et al. 2014), computer vision, image processing (Rousseau et al. 2013), voice recognition, and disease classification (Fuentes et al. 2017). The main driving forces behind applying these techniques in agriculture involve their use by commercial companies and a reduction in the cost of sensors and imaging platforms (Araus and Cairns 2014).
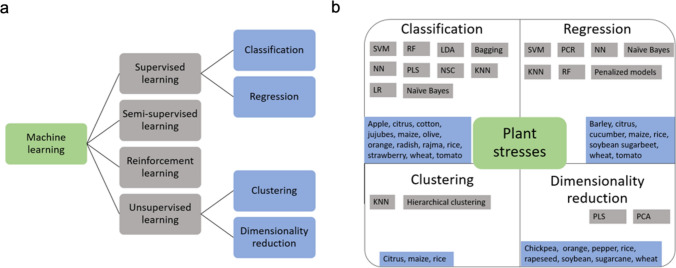
ML involves learning the patterns from the dataset using computerized models to make reliable conclusions without being explicitly told (Sandhu et al. 2022a, 2022b). ML efficiently uses its experience for identifying the underlying structure, pattern, similarities, or dissimilarities in the provided dataset for classification and prediction problems (LeCun et al. 2015). Typically, an ML model consists of a calibration process where a model is trained on a given large data set called a training set. The remaining dataset on which the model’s performance is validated is called the testing set. The accuracy and precision of the calibrated model classify its use for future applications. Generally, during the model training process, two approaches, namely, supervised and unsupervised machine learning are used. Supervised ML models involve providing a label for the data during the training process; for example, differentiating wheat and rice images, labels are provided for the two crops while training the model. On the other hand, the unsupervised model does not involve the use of labels during the training process, while the model attempts to differentiate both crops on its own by learning similarities and dissimilarities. Numerous studies have used ML for managing biotic and abiotic stresses in HTP and involve applications like support vector machine, discriminant analysis, k-means clustering, neural networks, clustering, dimensional reduction, and least discriminant analysis (Table 4). All of these models help to identify, classify, quantify, and predict different phenotyping components in plants. The overview of all these ML learning models can be found in Fig. 1 and the section below.
Fig. 1.
Classification of machine and deep learning models is provided which are mostly used in high throughput phenotyping (a, b)
Supervised Learning
Supervised learning provides the power to process data using machine language. The main goal of supervised learning is to use the input and output variables for mapping the relationship. The main interest is to build a mapping function such that if you used new input data, predictions could be made about the output. Training supervised learning models involves learning its parameters and is focused on lowering the training data’s loss function. Supervised learning can be further classified into classification and regression problems (Fig. 1). A classification problem involves categorizing the output variables by building a relationship within them. For example, grouping plants based on disease severity rating. Here, we review the studies using classification algorithms for plant stress detection. While in case of regression problems, the output consists of real values, and the model aims to predict the data using a trained model on the training data set. For example, the prediction of a plant’s grain yield can be done by training the model on the previous year’s data set. There are various supervised machine learning models which have been used for plant stress phenotyping and are described below.
Linear Discriminant Analysis
Linear discriminant analysis (LDA) uses a linear combination of features for categorizing the output into two or more classes. LDA has continuous independent variables with a categorical dependent variable (Kim et al. 2011). Kim et al. (2011) applied the discriminant analysis to classify 14 different Arabidopsis seeds into two groups using direct analysis in real-time and mass spectrometry. LDA and support vector machine (SVM) were used for the early detection of almond (Prunus dulcis) red leaf blotch using high-resolution hyperspectral and thermal imagery, and it was helpful for the differentiation between the trees that did not have symptoms and those with high infestation (Peña et al. 2015). In another study, various classification algorithms such as LDA, quadratic discriminant analysis (QDA), KNN and soft independent modelling of classification analogies were used to detect Huanglongbing in citrus orchards using the images acquired through visible-near infrared spectroscopy. The classification accuracies of QDA and SIMCA were 95% and 92%, respectively (Sankaran et al. 2011).
Support Vector Machine
The support vector machine (SVM) develops a hyperplane to have a maximum distance from the nearest example in the training data set. This hyperplane helps in the clear separation between different classes with maximization of the difference between the classes (Eichner et al. 2011). Eichner et al. (2011) utilized the support vector machines for detecting the intron retentions and exon skipping in tiling arrays. SVM was used for the segmentation of images, which were helpful in the analysis of Salmonella typhimurium (human pathogen) attack on Arabidopsis spp (Schikora et al. 2012). LDA and SVM classification methods were used to detect verticillium wilt in olive (Olea europaea) by using thermal and hyperspectral images. SVM performed better, having a classification accuracy of 79.2% as compared to LDA, which only had an accuracy of 59.0% (Calderón et al. 2015). This method was used to identify regions of canopy showing response to soil water deficit in spinach (Spinacia oleracea) using infrared thermal images. Results indicated that an average accuracy of 96.3% was obtained for SVM (Raza et al. 2014). In another study, SVM was used to detect drought stress in barley (Hordeum vulgare) plants from a series of hyperspectral images. The final data set contained 211,500 tests and 211,500 training instances. Five supervised prediction methods for deriving local stress levels were evaluated: one-vs.-one Support Vector Machine (SVM), one-vs.-all SVM, Support Vector Regression (SVR), Support Vector Ordinal Regression (SVORIM) and Linear Ordinal SVM classification. The highest accuracy was achieved by the one-vs.-one SVM (83%) (Behmann et al. 2014). Furthermore, SVM was used to identify weeds with green spectral components masked and unmasked. The masked plants were detected by identifying support vectors (Guerrero et al. 2012).
Logistic Regression
Logistic regression uses a logistic function for classifying binary variables. A logistic function usually classifies the dependent variable into two classes using all of the predictors with the odds ratio. Output with more than two values can be applied using multinomial logistic regression. Hyperspectral imaging was used for the early detection of apple scab for managing the application of pesticides and crop management strategies in the orchard. Logistic regression was efficiently used in this study for separating the infected and non-infected plants by selecting hyperspectral bands based on classification algorithms (Delalieux et al. 2007).
Random Forest
The working principle of random forests (RF) relies on the ensemble learning algorithm. This uses the tree-building process for classifying individuals into separate nodes of the tree. Random forests have several advantages over other tree-based classifications because of its ability to handle noise, control model overfitting, and a capacity to handle many variables. Random forest was used for feature selection from spectroradiometer data for detecting phaeosphaeria leaf spot in maize (Adam et al. 2017).
Linear Regression
Linear regression is used in most phenomics studies because of its simplicity and interpretation of data. The focus is how much variation in the target is explained by a particular feature in the dataset. A regression model was developed between crop water stress index (CWSI) and vegetation indices (VI) in maize to measure water stress by using multispectral images and regression models between VI and CWSI, which successfully mapped water stress (Zhang et al. 2019c). In another study, Pearson correlations and linear regression were calculated between thermal indices and leaf stomatal conductance (gs) and stem water potential (ѱSTEM) to measure water status variability in a vineyard using multispectral and thermal imagery (Baluja et al. 2012). Also, linear regression models were developed based on correlation coefficients between nitrogen absorption and different radiometric variables to determine nitrogen uptake in rice paddy by using two-band imaging systems, namely the visible red band (650–670 nm) and the near-infrared band (820–900 nm). The equation developed for nitrogen absorption per unit area based on the two band reflectance values indicated a high r-value (0.96) (Shibayama et al. 2009). In addition, the correlation and linear relationship between projected shoot area and shoot fresh weight were calculated to study salinity tolerance in rice by high throughput phenotyping technologies (Hairmansis et al. 2014), and a mixed linear model with the use of REML procedure was used for the detection of salinity tolerance in barley using infrared thermography (Sirault et al. 2009).
Multiple Linear Regression
Multiple linear regression (MLR), also known as multiple regression, predicts the outcome using several explanatory variables. MLR models the linear relationship between the predictors for predicting the outcome. Hyperspectral images were used to measure powdery mildew using multivariate linear regression (MLR), PLSR and Fisher linear discriminant analysis (FLDA) as data analysis techniques. The PLSR model's performance performed better than the MLR model, whereas FLDA had the highest accuracy (Zhang et al. 2012a). Bacterial spot in tomato (Lycopersicon esculentum) was measured using the spectral images and data analyzed using partial least squares (PLS) regression, analysis of correlation coefficient spectrum, and stepwise multiple linear regression (SMLR). Various predictive models were developed for the prediction of bacterial spot (Jones et al. 2010).
Partial Least Square Regression
Partial least square regression (PLSR) is a very beneficial tool for modelling and multivariate analysis since it can handle a large number of variables and collinearity among the variables (Yu et al. 2014). High value of correlation coefficient (r), and low values of RMSE are selected for developing the best model (Zhang et al. 2015). The PLSR model was used to estimate nitrogen content in rice (Oryza sativa) using ground-based hyperspectral imaging, and the relationship between the reflectance of the rice plant and nitrogen content was used to develop the PLSR model. The best model for the estimation of rice nitrogen content was constructed by combining the reflectance and the temperature data, which had low RMSE (0.95 g/m2) and RE (13%) (Onoyama et al. 2013). In another study, nitrogen content in citrus leaves was measured by hyperspectral imaging at 719 nm, and analysis was done using PLS. Stepwise multiple linear regression (SMLR) calibration models and the MLR calibration model performed better at 70% accuracy (Min et al. 2008). Also, the PLSR model successfully predicted drought stress in soybean plants, and its accuracy for two cultivars in an 8-day treatment group and a 6-day treatment group was 0.973 and 0.969, respectively (Mo et al. 2015).
Unsupervised Learning
Unsupervised learning finds the structure present in the unlabeled data set where output variables are not provided. The main focus in unsupervised learning is to identify the underlying structure present in the data to get more insight into the data. Unsupervised learning can be grouped into clustering of the dataset or extraction of latent factors using dimensionality reduction approaches (Fig. 1a). Clustering involves finding similarities in the dataset and later grouping those individuals. The dissimilar individuals are grouped into separate clusters. There are various clustering approaches including K-mean clustering and hierarchical clustering.
K-means Clustering
K-nearest neighbor (K-NN) is a nonparametric supervised and unsupervised classification method that assigns the object to its nearest neighbor using the most popular vote for classification. The number of classes depends upon the value of K specified before classification. These methods were used for classification and K-NN gave better results than Bayes rule for weed detection in cereal crops. The K-NN decision rule has high storage demand and is used for pattern recognition (Pérez et al. 2000). In another study, K-means clustering was used to extract values for the plant surface in the RGB channels for salinity tolerance in Arabidopsis thaliana (Awlia et al. 2016).
Dimensionality Reduction
Dimensionality reduction approaches try to explain the whole dataset with a few numbers of variables with the extraction of useful or latent variables. The most common dimensionality reduction techniques is principal component analysis (PCA) and it involves reducing the data’s dimensionality with the extraction of completely independent variables while minimizing information loss. PCA aims to explain the majority of variance present in the dataset using a few principal components. Stepwise discriminant analysis was performed to identify insect infestation in jujubes (Hovenia acerba Lindl.) by using visible and NIR spectroscopy. PCA was used for analyzing the spectra (Wang et al. 2011). In another study, aflatoxin B1 in maize (Zea mays) was detected by hyperspectral imaging and the data was analyzed using PCA. PCA was used for the reduction of dimensionality of the data. Stepwise factorial discriminant analysis was carried out on the latent variables provided by the PCAs (Wang et al. 2011). PCA analysis was also done to predict toxigenic fungi on maize with hyperspectral imaging (400–1000 nm), and discriminant analysis was done to create a model for fungal growth identification (Del Fiore et al. 2010). Furthermore, Fusarium infestation in wheat (Triticum aestivum) was identified with hyperspectral imaging and diseased and healthy plants were identified using PCA (Bauriegel et al. 2011).
Deep Learning
In the ML section, we provided information about various tools which could potentially be used for extracting valuable information from plants. However, there is rapid adoption of DL tools for easier data analysis of large numbers of images with high accuracy. DL is a branch of ML which uses a deep network of neurons and layers for processing information (LeCun et al. 2015). DL has provided remarkable achievements in other disciplines such as fraud detection, automated financial management, autonomous vehicles, consumer analytics, and automated medical diagnostics (Min et al. 2017). DL is a type of ML that does not require the prior feature selection, which is the characteristic of traditional ML models and the most time-consuming and error-prone step (Sandhu et al. 2021b). DL models involve automatically learning the pattern from a large data set using non-linear activation functions for making conclusions such as classification or predictions (Sandhu et al. 2021d).
The important DL models used for phenomics include, but are not limited to, a multilayer perceptron (MLP), generative adversarial networks (GAN), convolutional neural network (CNN), and recurrent neural network (RNN). It has been observed that CNN is superior for image analysis, and different CNN image recognition architecture is utilized for plants, namely ResNet, ZFNet, VGGNet, GoogleNet, and AlexNet (Fuentes et al. 2017). The general outline of deep learning consists of multiple connected neurons across the layers. The concept of DL was proposed in the 1950s (Samuel 1959) and is designed to work using a logic structure as a human would in completing a particular task. There has been rapid advancement in the adoption of DL models because of the development of efficient algorithms for estimation of complex hyperparameters for training the models (Sandhu et al. 2020). Deep learning consists of multiple terms, some of which can be found in Table 5.
Table 5.
Terms used in deep learning models for the model’s optimization and prediction
| Term | Definition |
|---|---|
| Activation function | The function that produces the neuron’s output |
| Batch | Partition of the data into different sets within a given epoch |
| Dropout | Removal of a fixed number of neurons during each training set for controlling overfitting |
| Early stopping | A strategy to control overfitting by the early stopping of the model |
| Epoch | Shifting the training set into different batches |
| Neuron | A primary entity of the DL model which learns the information and provides the output to the next layer using different activation functions |
| Regularization | Works as a penalty for neurons’ weight during model training |
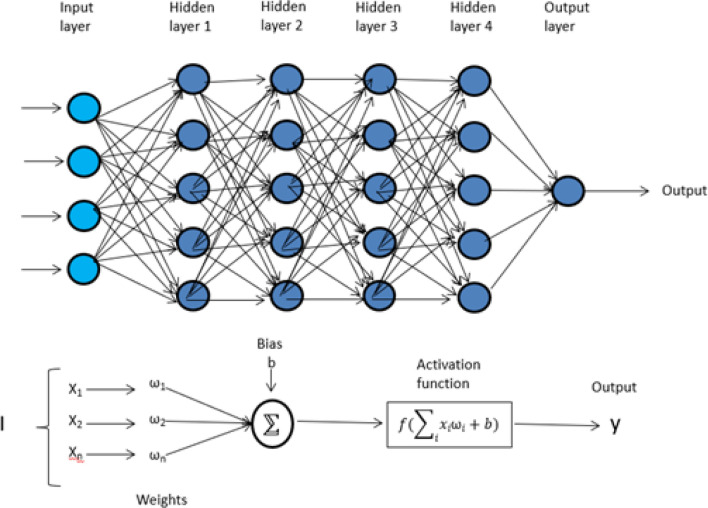
MLP is the most commonly used neural network in genomics which consists of multiple fully connected layers, namely input, hidden, and an output layer which is connected by a dense network of neurons. The input layer consists of all of the input features. The first hidden layer uses a different number of neurons to learn a weight parameter with a constant bias while training the model. The output of the first hidden layer acts as input for the second hidden layer and continues in this sequence. The final layer is known as an output layer where input from the last hidden layer converges to a single value. The output from the first hidden layer can be represented as:
Where Z1 is the output of the first hidden layer, b0 is the bias estimated with the rest of the weights W0, and 0(x) is the particular activation function used in the model (Gulli and Pal 2017). The detailed working and model of the MLP is presented in Fig. 2.
Fig. 2.
Multi-layer perceptron showing the working of the neural network using input, hidden, and output layer. The bottom half of the figure shows the weight associated with each neuron and transformation using an activation function. Y represents the final output from the model and is achieved by optimizing other hyperparameters
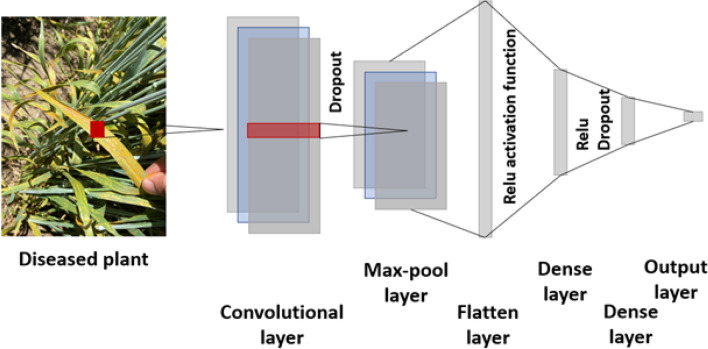
Convolutional neural network (CNN) is used when variables are distributed in one or two dimensions and is mostly used for plant image analysis. CNN is a special case of DL which uses fully connected layers known as convolutional layers. In each of the convolutional layers, the convolution operation is performed along with the inputs of predefined strides and width (Pérez-Enciso and Zingaretti 2019). Finally, smoothing the results is performed along the pooling layer. The detailed layout and working of CNN are provided in Fig. 3. CNN is the most commonly used classification model used in plant studies, especially for classifying different disease types. The biggest advantage of CNN involves utilization of the raw image as input without any prior pre-processing of the image. CNN has demonstrated superiority in plant detection and diagnosis, classification of fruits and flowers, and detection of disease severity in infected plants (Sandhu et al. 2020, 2021b).
Fig. 3.
The most straightforward design of convolutional neural networks with convolutional, max pool, flatten, dense and output layer. Each region of interest is processed separately in the CNN for predicting the output
Ramcharan et al. (2019) showed that CNN could be used in mobile-based apps for detecting foliar disease symptoms in cassava (Manihot esculenta Crantz). They also suggested that model performances differed when trained on real-world conditions and existing images, demonstrating the importance of lighting and orientation for efficient model performance. Powdery mildew is a devastating disease in cucumber during the middle and final stages of growth. Quantitative assessment of disease over the cucumber (Cucumis sativus) leaves is especially important for plant breeders for making selections. Lin et al. (2019) segmented the diseased leaf images at pixel level using a segmentation model on a CNN and achieved a pixel accuracy up to 96%. The CNN model employed in their study outperformed the existing segmentation models, namely, random forest, K-means, and support vector machines. Similarly, deep convolutional neural networks were used to detect 10 different diseases in rice and differentiate them from the healthy plants on a dataset of 500 images. That study showed that using ten-fold cross-validation, deep CNN models gave an accuracy of 95.48%, which was significantly higher than the conventional machine learning models (Lu et al. 2017) (Table 6).
Table 6.
Studies using various machine and deep learning tools for biotic stress management in field crops during the last decade
| Plant | Plant trait/disease | Pathogen | Spectral range (nm) | Domain (field/lab) | Sensing modality | Data analysis | References |
|---|---|---|---|---|---|---|---|
| Almond | Red leaf blotch | Polystigma amygdalinum | 400–790 | Field | RGB, hyperspectral and thermal imaging | ANOVA, LDA, SVM | López-López et al. (2016) |
| Apple | Scab | Venturia inaequalis | 400–800 | Field | RGB and 3D imaging | Image processing and segmentation | Chéné et al. (2012) |
| Arabidopsis | Salmonella bacteria | Salmonella typhimurium | 380–760 | Laboratory | RGB imaging | Support vector machine | Schikora et al. (2012) |
| Banana | Black Sigatoka and fusarium wilt | Mycosphaerella fijiensis | 380–760 | Field | RGB imaging | Deep learning and image processing | Sanga et al. (2020) |
| Banana | Black Sigatoka and speckle | Mycosphaerella fijiensis | 380–760 | Field | RGB imaging | Deep learning | Amara et al. (2017) |
| Banana | Black Sigatoka | Mycosphaerella fijiensis | 380–1024 | Laboratory | Hyperspectral imaging | Visual inspection | Ochoa et al. (2016) |
| Blackgram | Yellow mosaic disease | Yellow mosaic virus | 350–2500 | Field | Non-imaging | Multinomial logistic regression | Prabhakar et al. (2013a) |
| Barley | Powdery mildew, and rust | Blumeria graminis hordei, and Puccinia hordei | 400–2500 | Greenhouse | Hyperspectral imaging | Simplex volume maximization | Wahabzada et al. (2015) |
| Cassava | Cassava mosaic, cassava brown streak, and green might | Bemisia tabaci | 380–750 | Field | RGB imaging | Deep learning | Ramcharan et al. (2019) |
| Cassava | Brown leaf spot, cassava brown streak, and red mite | Bemisia tabaci | 380–750 | Field | RGB imaging | Deep learning | Ramcharan et al. (2017) |
| Chili Pepper | Aflatoxins | Aspergillus flavus, Aspergillus parasiticus | 400–720 | Laboratory | Hyperspectral imaging | Fisher discrimination power | Ataş et al. 2012) |
| Citrus | Huanglongbing | Candidatus spp | 530–690 | Laboratory | Fluorescence imaging | Support vector machine and artificial neural network | Wetterich et al. (2017) |
| Citrus | Huanglongbing | Candidatus spp | 530–1000 | Field | NIR spectroscopy | Support vector machine and vegetation indices | Garcia-Ruiz et al. (2013) |
| Citrus | Huanglongbing | Candidatus spp | 350–2500 | Field | Infra-red spectroscopy | Linear and quadratic discriminant analysis, k nearest neighbor and support vector machine | Sankaran et al. (2011) |
| Cotton | Mealybug | Phenacoccus solenopsis | 350–2500 | Field | Spectro-radiometry | Linear discriminant analysis, and multivariate logistic regression | Prabhakar et al. (2013b) |
| Cucumber | Cucumber mosaic virus (CMV), Cucumber green mottle mosaic virus (CGMMV), Powdery mildew | Sphaerotheca fuliginea (Powdery mildew) | 400–1000 nm | Greenhouse | Hyperspectral and fluorescence Imaging | Linear regression | Berdugo et al. (2014) |
| Cucumber | Powdery mildew | Podosphaera xanthii | 400–700 | Laboratory | RGB imaging | CNN | Lin et al. (2019) |
| Jujubes | Internal insect infestation | Ancylis sativa | 400–2000 | Laboratory | NIR spectroscopy | Classification | Wang et al. (2011) |
| Maize | Northern leaf blight | Setosphaeria turcica | 380–760 | Field | RGB Imaging | Deep learning | DeChant et al. (2017) |
| Maize | Phaeosphaeria Leaf Spot | Phaeosphaeria maydis | 350–2500 | Field | Hyperspectral Imaging | Random forest classifier | Adam et al. (2017) |
| Maize | Aflatoxin | Aspergillus flavus, Aspergillus parasiticus | 1100–1700 | Lab | Hyperspectral Imaging | Deep learning | Kandpal et al. (2015) |
| Maize | Aflatoxin | Aspergillus flavus | 1000–2500 | Laboratory | Hyperspectral Imaging | PCA and stepwise factorial discriminant analysis | Wang et al. (2014) |
| Maize | Aflatoxin | Aspergillus flavus | 400–700 | Field and laboratory | Hyperspectral Imaging | Discriminant analysis | Yao et al. (2013) |
| Maize | |||||||
| Oilseed rape | Alternaria spots | Alternaria alternata | 400–2500 | Laboratory | Hyperspectral and thermal Imaging | Neural networks | Baranowski et al. (2015) |
| Olive | Wilt | Verticillium dahliae | 400–885 and 7500–13,000 | Field | Hyperspectral and thermal Imaging | Support vector machine and linear discriminant analysis | Calderón et al. (2015) |
| Orange | |||||||
| Pearl millet | Mildew diseases | Sclerospora graminicola | 380–760 | Field | RGB Imaging | Deep learning | Coulibaly et al. (2019) |
| Rice | Aflatoxin | Aspergillus flavus | 950–1650 | Laboratory | Non-imaging | PLSR | Dachoupakan Sirisomboon et al. (2013) |
| Rice | Rice blast, false smut, brown spot, bakanae disease, sheath blight, bacterial leaf blight, and bacterial wilt | Magnaporthe grisea | 380–760 | Field | RGB Imaging | Deep learning | Lu et al. (2017) |
| Strawberry | Anthracnose | Colletotrichum gloeosporioides | 400–1000 | Greenhouse | Hyperspectral Imaging | Discriminant analysis | Yeh et al. (2016) |
| Sugarbeet | |||||||
| Sugarbeet | Cercospora leaf spot | Cercospora beticola | 400–1000 | Greenhouse | Hyperspectral Imaging | SAM | Mahlein et al. (2012b) |
| Sugarbeet | Cercospora leaf spot | Cercospora beticola | 380–760 | Greenhouse and field | RGB Imaging | Support vector machine with radial basis kernel function | Hallau et al. (2017) |
| Sugarbeet | Nematode and root rot | Heterodera schachtii and Rhizoctonia solani | 400–2500 | Field | Hyperspectral Imaging | Vegetation indices and classification approaches | Hillnhütter et al. (2011) |
| Sugarcane | African stalk borer, Smut, Sugarcane thrips, Brown rust |
Eldana saccharina (African stalk borer), Sporisorium scitamineum (Smut), Fulmekiola serrata (Sugarcane thrips), Puccinia melanocephala (Brown rust ) |
1100–2300 | Field | NIR spectroscopy | PCA, PLS | Mahlein et al. (2012b) |
| Wheat | Head blight | Fusarium sp. | 400–1000 | Field | RGB and multispectral imaging | Linear and quadratic regression | Dammer et al. (2011) |
| Wheat | Head blight | Fusarium sp. | 400–1000 | Laboratory | Visible/near-infrared hyperspectral imaging | PCA and LDA | Shahin and Symons (2011) |
| Wheat | Head blight | Fusarium sp. | 400–1000 | Laboratory | Hyperspectral imaging | PCA | Bauriegel et al. (2011) |
| Wheat | Leaf rust and powdery mildew | Puccinia triticina, and Blumeria graminis | 370–800 | Greenhouse | Hyperspectral imaging | Linear regression | Bürling et al. (2011) |
| Wheat | Leaf rust | Puccinia triticina | 350–2500 | Field | Hyperspectral imaging | Vegetation indices | Zhang et al. (2012a) |
| Wheat | Powdery mildew | Blumeria graminis | 350–2500 | Field | Hyperspectral imaging | Vegetation indices | Cao et al. 2013) |
| Wheat | Powdery mildew | Blumeria graminis | 450–950 | Field and laboratory | Hyperspectral imaging | Multivariate linear regression and PLSR | Zhang et al. 2012b) |
| Wheat | |||||||
| Wheat | Wheat streak mosaic virus | Tritimovirus spp. | 380–760 | Field | RGB imaging | Expectation maximization for analysing hue values differences | Casanova et al. (2014) |
| Wheat | Powdery mildew | Blumeria graminis | 400–2450 | Field | Hyperspectral imaging | Classification | Mewes et al. (2011) |
| Wheat | Mycotoxins | Fusarium spp. | 400–1000 | Greenhouse, Field | Hyperspectral imaging | Classification | Bauriegel et al. (2011) |
| Wheat | Leaf Rust | Puccinia triticina | 350–2500 | Greenhouse | Hyperspectral imaging | Classification | Ashourloo et al. (2014) |
| Wheat | Stripe rust | Puccinia striiformis | 400–900 | Field | Hyperspectral imaging | Correlation | Devadas et al. (2015) |
| Tomato | Gray mold, leaf mold, and canker | Botrytis cinerea | 380–780 | Field | Fluorescence and hyperspectral imaging | CNN | Fuentes et al. (2017) |
| Tomato | |||||||
| Tomato | Gray mold | Botrytis cinerea | 380–1050 | Greenhouse | Hyperspectral imaging | GA-PLS | Kong et al. (2014) |
| Tomato | Early blight | Alternaria solani | 380–1023 | Greenhouse | Hyperspectral imaging | ELM | Xie et al. (2015) |
| Tomato | Powdery mildew | Oidium neolycopersici | 380–760 and 7000–13,000 | Greenhouse | Thermal and visible light imaging | Support vector machine | Raza et al. (2015) |
| Tomato | Powdery mildew | Oidium neolycopersici | 380–760 | Greenhouse | Color image processing (using self-organizing map) | Bayesian classifier | Hernández-Rabadán et al. (2014) |
| Tomato | Leaf curl virus | Begomovirus spp. | 380–760 | Laboratory | Gray level co occurrence matrix detection | Multilayer perceptron, quadratic kernel, and support vector machine | Mokhtar et al. (2015) |
| Zucchini | Soft rot | Dickeya dadantii | 7500–13,000 | Greenhouse | Fluorescence imaging | Logistic regression analysis, and artificial neural networks | Pérez-Bueno et al. (2016) |
The main bottleneck in DL involves optimization of hyperparameters that involve the number of neurons, activation functions, number of epochs, total number of convolutional layers, number of filters and hidden layers, solver, dropout, and regularization. There are many different combinations which could be used for these hyperparameters resulting in a huge computational burden and requires strong programming skills. There are four most common approaches used for selecting these hyperparameters, namely, Latin hypercube sampling, grid search, random search, and optimization (Cho and Hegde 2019). Furthermore, there is a problem of overfitting in DL models, if it is not accounted for (Sandhu et al. 2021b). Tools such as early stopping, L1 and L2 regularization, and dropout are available for reducing the chances of overfitting in the trained model. The detailed information about the optimization of hyperparameters and tools for controlling overfitting is referred to in other texts (Cho and Hegde 2019).
Conclusion
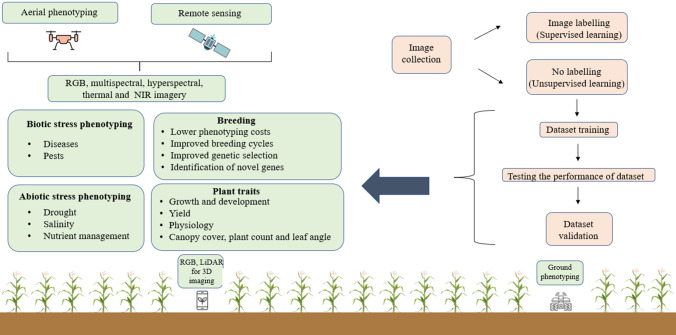
Plant stress phenotyping is an important parameter for predicting crop losses caused by various biotic and abiotic stresses. It can be used to identify superior disease resistant and stress-tolerant genotypes as well as to assess disease management decisions (Fig. 4). The phenotypic parameters include not only morphological data, but also a large number of physiological and biochemical data, as well as deeper mechanistic data, allowing scientists to identify and predict heritable traits through controlled phenotypic and genotypic studies. Current methods for stress severity phenotyping are used at various scales, such as the number of plants affected or exact counts of lesion numbers, or estimates of the severity or surface area affected by a particular biotic/abiotic stress at the canopy of single plant and field levels (Table 4). Stationary HTP platforms can carry a variety of sensors to monitor both pot crops and crops in specific field areas at the same time. Their operation, however, is limited to a small area, and their construction is costly. More research is needed in the future to improve UAV-based sensing for plant phenotyping. High-performance and low cost UAVs should be introduced in future studies. For long-term and large-field plant phenotyping, high-performance UAVs with high flight stability, precision, long flight duration, and heavy payload are required. Unlike ground-based phenotyping, UAV-based phenotyping is afflicted by a serious issue: the safety of the UAV and its sensors.
Fig. 4.
The outline of different phenotyping tools and platforms for plant stress detection in the field
Presently, RGB and multispectral sensors are primarily used to estimate crop height, biomass and other agronomic traits under normal and stressed conditions. However, the utilization of hyperspectral sensors is still in its infancy. Hyperspectral imaging has emerged as a cutting-edge/emerging method to assess crop attributes such as water content, leaf nitrogen concentration, chlorophyll content, leaf area index, and other physiology and biochemistry parameters. The interference of soil signals, ambient air, and canopy temperature on thermal infrared cameras' imaging is the main limitation. The collected data information is heavily reliant on these sensors and has an impact on the final phenotyping results. Simultaneous acquisition and analysis of the same crop phenotypes by multiple sensors can provide a more comprehensive and accurate assessment of crop traits. For instance, in the process of screening crop biotic and abiotic stress tolerance, a variety of phenotypic data is typically combined for effective and accurate analysis. Further, the data types and formats collected by different phenotypic sensors varies, combining and managing the data collected by multiple sensors, as well as the development of novel sensors such as laser-induced breakdown spectroscopy and electrochemical micro-sensing (Li et al. 2019) for detection of plant signal molecules is another challenge for future phenotypic data analysis, necessitating collaboration between multidisciplinary laboratories.
In addition, environmental factors are also critical and should be given at least as much attention as the traits that are measured, which leads to the next question: how can all of the environmental impacts be measured? Envirotyping, defined as a comprehensive set of next-generation high-throughput accurate envirotyping technologies, could aid in addressing this issue (Xu 2016). Furthermore, by integrating multi-typing data, the genotyping environment management interaction could be investigated, and predictive phenomics would be possible (Xu 2016; Araus et al. 2018). High-throughput and accurate phenotyping, when combined with optimized experimental design, high quality field trials, a robust crop model, envirotyping, and other strategies, will improve heritability and genetic gain potential (Araus et al. 2018).
Phenotypic researchers prefer mobile high-throughput plant phenotypings that can be transported to the experimental sites (Roitsch et al. 2019). This indicates that the development of mobile high-throughput plant phenotyping must focus on flexibility and portability, and that modular and customizable designs will be appreciated by the phenotyping community. With recent advances in artificial intelligence analysis techniques, 5th generation mobile networks, and cloud-based innovations, more smart "pocket" phenotyping tools are likely to be introduced, potentially replacing manual field phenotyping, which has been practiced for hundreds of years.
Overall, whether phenotyping in the greenhouse or in the field, ground-based proximal phenotyping or aerial large-scale remote sensing, the future of high-throughput plant phenotyping lies in enhancing spatial–temporal resolution, turnaround time in data analysis, sensor integration, human–machine interaction, throughput, operational stability, operability, automation, and accessibility. It is worth mentioning that the selection, development, and application of high-throughput plant phenotypings should be directed by concrete project requirements, and practical application scenarios, specific phenotypic tasks, such as field coverage (Kim, 2020), instead of supposing that the more devices, innovations, and funds with which the high-throughput plant phenotyping is furnished, the better; partly because collecting a large amount of data does not imply that all of it is useful (Haagsma et al. 2020). Even in some cases, the experimental implications of using a single sensor and multiple sensors are similar (Meacham-Hensold et al. 2020), and data acquired from different devices is redundant.
Challenges and Future Prospects Nevertheless, combining various high-throughput plant phenotypings for comparative verification and detailed evaluation could open up new avenues for inspection, extraction, and quantification of complex physiological functional phenotypes. However, the technical issues of developing standards and coordinating calibrations for these numerous combinations are challenging tasks. Furthermore, to achieve a true sense of “cost-effective phenotyping”, the trade-off between phenotyping technique investment and manpower cost should be noted, which is primarily dependent on the different objectives (Reynolds et al. 2019).
After collecting massive amounts of data, the next concern is how to handle “Big Data”? Wilkinson et al. proposed the FAIR (findable, available, identifiable, and reusable) principle to enable the finding and reuse of data all over different individuals or groups (Wilkinson et al. 2016), which means all essential metadata, such as resource and data acquisition information, measurement protocols, and environmental conditions, data description should be clearly addressable. Several efforts for data management and analysis have also been made in accordance with the FAIR principle, such as PHENOPSIS DB (Fabre et al. 2011) and CropSight (Reynolds et al. 2019). Data sharing and standardization across communities continues to be a major challenge. Another issue is a lack of funding and data infrastructure to manage these data sets (Bolger et al. 2019). We support the creation of OPEN data infrastructures and the publication of primary data with DOIs. This would make data reuse easier, as well as the development, testing, and comparison of technologies.
Online databases, such as http://www.plant-image-analysis.org, can effectively connect developers and users. The creation of the PlantVillage database, which comprised 54,306 images of 14 crop species and 26 diseases, opened up new avenues for meeting some research needs (crops, number of diseases, severity of expression, stages of disease infection, and so on), along with the need for a publicly accessible, open-source, shared database of annotated plant stresses at the individual leaf scale. PlantVillage data has also been overexplored, and new data sources will be required for the development of robust models. Some other open-source databases are also available, including the maize northern leaf blight disease database (Wiesner-Hanks et al. 2018), which contains 18,222 digital images of maize leaves in the field, either captured manually, mounted on a boom, or taken by UAVs. Human experts annotated approximately 105,705 NLB lesions, making this the largest publicly accessible annotated image set for a plant disease. However, comprehensive management platforms covering software, web-based tools, toolkits, pipelines, ML and DL tools, and other phenotypic solutions are still lacking, which will be a significant milestone as well as a significant challenge.
Data that is poorly annotated and in a disorganized format, on the other hand, could generate noise or disordered waves. Certainly, standard constraints are being proposed that are relevant. Krajewski et al. (2015), for instance, published a technical paper with effective recommendations (at http://cropnet.pl/phenotypes) and initiatives (such as http://wheatis.org), taking another step toward establishing globally practical solutions. Furthermore, initiating relevant phenotyping standardization can improve comprehension and explanation of biological phenomena, contributing to the transformation of biological knowledge and the creation of a true coherent semantic network.
As technologies for generating and storing high-throughput measurement data become more accessible to researchers, the role of ML is expected to grow in importance in the coming years. Nonetheless, several challenges still remain. The success of ML is critically dependent on the availability of large collections of samples with sufficient common features. The accumulation of such collections has been useful in other areas for complex analyses (ENCODE Project Consortium, 2012), but for plant research, the required investment is very massive and appears to be feasible only for commercially relevant crops such as rice, wheat, and maize. A variety of efforts, from local to international, are underway to build phenotyping centers that automate and standardize high-throughput measurements of plant phenotypes at all stages—a process known as phenomics (Yang et al. 2020). These will provide more data in a more objective and accurate manner, but it will take some time before sufficiently large and rich data sets are available.
The most interesting scientific challenge is to create (novel) ML approaches for problems with data that are too complex, heterogeneous, and variable for current techniques to handle. Current ML-based techniques focus on a single stress or disease on a leaf or canopy, but in real-world situations, numerous diseases and stresses may appear on a single leaf or canopy. ML platforms must be flexible and robust, with the ability to distinguish multiple disease symptoms on a single leaf or within the same plant canopy. The training dataset should include multi-location, multi-year, and diverse plant stress symptom images. The quantity of data needed to train ML models is determined by the complexity of the problem and the complexity of the learning algorithm; thus, for wider applicability, the training data should really be continuously updated using techniques such as artificial learning to reflect the complexity of stress symptoms for the targeted crop. The plant community requires universal datasets for various stresses in order to train robust ML models for practical application in decision support. To ensure robust in-field performance of ML, the datasets should include realistic and potentially degraded sensing environments (e.g., cloudy, fog, low light, and saturated lighting).
A future opportunity for ML is to support decision-making in plant research, from predicting which region of the genome should be modified to achieve a desired phenotype (in genetic modification strategies) to make sure optimal local growth conditions by assessing crop performance in vivo in the greenhouse or on the field. While these are mainly engineering challenges, effective ML methods will provide powerful tools to researchers, especially as they improve their ability to allow interpretation of their decisions. In this way, ML can aid in addressing the challenges of ensuring food security for growing populations in rapidly changing environments.
DL is a flexible tool for making sense of the vast datasets in plant stress ICQP (identification, classification, quantification, and prediction) (Singh et al. 2018). Most DL models, however, have been applied in biotic stress and disease detection due to their excellent disease identification. In the coming years, DL could be used to quantify abiotic stress and predict the loci that control stress tolerance by integrating genetic and other omics data with different sensor technologies (thermal, hyperspectral, CT, terahertz, radar, MRI etc.) and large multiscale (lab-to-field) phenotypic datasets that include weather and other environmental parameters. This approach is most likely to work best with very large and well-annotated datasets, emphasizing the importance of open data. The most difficult issue with deep learning tasks is a lack of adequate training samples with labels (Li et al. 2019). Every day, petabytes of data are added, excluding the zettabytes that already exist. This tremendous progress is amassing data that cannot be labelled without the assistance of humans. The current success of supervised learning techniques is typically due to the availability of large datasets and readily available labels (Gregor and LeCun 2010). Unsupervised learning techniques, on the other hand, will become the primary considered solution as data complexity and size increase rapidly (Ranzato et al. 2013).
Finally, advances in artificial learning and transfer learning can be used to improve ML models for new goals and plant stresses (e.g., disease, pest, flooding, drought, salinity, nutrient, temperature, weed stresses). We encourage the creation and widespread distribution of community-wide curated, labelled datasets, which would significantly accelerate deployment while also improving interactions with ML experts who would use these curated datasets as benchmark data, similar to MNIST, ImageNet, or CIFAR. The development of such open, labelled, broad-spectrum plant stress datasets across various plant species will eliminate data duplication, allowing plant scientists with smaller datasets to use DL through transfer learning. We also strongly recommend using imaging data from field conditions to create training datasets, as this will result in robust models that can be embedded on high-throughput systems such as UAVs and other autonomous systems. The convergence of imaging platforms advances in DL approaches, as well as the availability of storage and computing resources, makes this an interesting time to be pursuing plant phenotyping, including stress-related traits.
Authors' contributions
All authors significantly contributed to planning, writing, and editing of the manuscript.
Funding
The authors have not received funding for conducting this study.
Availability of Data and Materials
Not applicable.
Code Availability
Not applicable.
Declarations
Conflicts of Interest
The authors declare that they have no conflict of interest.
Ethics Approval
Not applicable.
Consent to Publish
Not applicable.
Consent to Participate
Not applicable.
References
- Abdulridha J, Ampatzidis Y, Kakarla SC, Roberts P. Detection of target spot and bacterial spot diseases in tomato using UAV-based and benchtop-based hyperspectral imaging techniques. Precision Agric. 2020;21:955–978. doi: 10.1007/s11119-019-09703-4. [DOI] [Google Scholar]
- Adam E, Deng H, Odindi J, et al. Detecting the early stage of phaeosphaeria leaf spot infestations in maize crop using in situ hyperspectral data and guided regularized random forest algorithm. J Spectrosc. 2017;2017:1–8. doi: 10.1155/2017/6961387. [DOI] [Google Scholar]
- Al-Tamimi N, Brien C, Oakey H, et al. Salinity tolerance loci revealed in rice using high-throughput non-invasive phenotyping. Nat Commun. 2016;7:13342. doi: 10.1038/ncomms13342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amara J, Bouaziz B, Algergawy A (2017) A deep learning-based approach for banana leaf diseases.
- Apan A, Held A, Phinn S, Markley J. Detecting sugarcane ‘orange rust’ disease using EO-1 hyperion hyperspectral imagery. Int J Remote Sens. 2004;25:489–498. doi: 10.1080/01431160310001618031. [DOI] [Google Scholar]
- Araus JL, Cairns JE. Field high-throughput phenotyping: the new crop breeding frontier. Trends Plant Sci. 2014;19(1):52–61. doi: 10.1016/j.tplants.2013.09.008. [DOI] [PubMed] [Google Scholar]
- Araus JL, Kefauver SC, Zaman-Allah M, Olsen MS, Cairns JE. Translating high-throughput phenotyping into genetic gain. Trends Plant Sci. 2018;23(5):451–466. doi: 10.1016/j.tplants.2018.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asaari MSM, Mertens S, Dhondt S, et al. Analysis of hyperspectral images for detection of drought stress and recovery in maize plants in a high-throughput phenotyping platform. Comput Electron Agric. 2019;162:749–758. doi: 10.1016/j.compag.2019.05.018. [DOI] [Google Scholar]
- Ashourloo D, Mobasheri M, Huete A. Evaluating the effect of different wheat rust disease symptoms on vegetation indices using hyperspectral measurements. Remote Sens (basel) 2014;6:5107–5123. doi: 10.3390/rs6065107. [DOI] [Google Scholar]
- Ataş M, Yardimci Y, Temizel A. A new approach to aflatoxin detection in chili pepper by machine vision. Comput Electron Agric. 2012;87:129–141. doi: 10.1016/j.compag.2012.06.001. [DOI] [Google Scholar]
- Atefi A, Ge Y, Pitla S, Schnable J. In vivo human-like robotic phenotyping of leaf traits in maize and sorghum in greenhouse. Comput Electron Agric. 2019;163:104854. doi: 10.1016/j.compag.2019.104854. [DOI] [Google Scholar]
- Atieno J, Li Y, Langridge P, et al. Exploring genetic variation for salinity tolerance in chickpea using image-based phenotyping. Sci Rep. 2017;7:1300. doi: 10.1038/s41598-017-01211-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Awlia M, Nigro A, Fajkus J, et al. High-throughput non-destructive phenotyping of traits that contribute to salinity tolerance in Arabidopsis thaliana. Front Plant Sci. 2016;7:1414. doi: 10.3389/fpls.2016.01414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Babaeian E, Sadeghi M, Jones SB, et al. Ground, proximal and satellite remote sensing of soil moisture. Rev Geophys. 2019 doi: 10.1029/2018RG000618. [DOI] [Google Scholar]
- Bai G, Ge Y, Scoby D, et al. NU-Spidercam: a large-scale, cable-driven, integrated sensing and robotic system for advanced phenotyping, remote sensing, and agronomic research. Comput Electron Agric. 2019;160:71–81. doi: 10.1016/j.compag.2019.03.009. [DOI] [Google Scholar]
- Baluja J, Diago MP, Balda P, et al. Assessment of vineyard water status variability by thermal and multispectral imagery using an unmanned aerial vehicle (UAV) Irrig Sci. 2012;30:511–522. doi: 10.1007/s00271-012-0382-9. [DOI] [Google Scholar]
- Bannari A, Mohamed AMA, El-Battay A (2017) Water stress detection as an indicator of red palm weevil attack using worldview-3 data. In: 2017 IEEE International Geoscience and Remote Sensing Symposium (IGARSS). IEEE, p 4000–4003
- Baranowski P, Jedryczka M, Mazurek W, et al. Hyperspectral and thermal imaging of oilseed rape (Brassica napus) response to fungal species of the genus Alternaria. PLoS One. 2015;10:e0122913. doi: 10.1371/journal.pone.0122913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barbedo JGA. A review on the use of unmanned aerial vehicles and imaging sensors for monitoring and assessing plant stresses. Drones. 2019;3:40. doi: 10.3390/drones3020040. [DOI] [Google Scholar]
- Bauriegel E, Giebel A, Geyer M, et al. Early detection of Fusarium infection in wheat using hyper-spectral imaging. Comput Electron Agric. 2011;75:304–312. doi: 10.1016/j.compag.2010.12.006. [DOI] [Google Scholar]
- Beauchêne K, Leroy F, Fournier A, et al. Management and characterization of abiotic stress via PhénoField®, a high-throughput field phenotyping platform. Front Plant Sci. 2019;10:904. doi: 10.3389/fpls.2019.00904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Behmann J, Schmitter P, Steinrücken J, Plümer L. Ordinal classification for efficient plant stress prediction in hyperspectral data. Int Arch Photogramm Remote Sens Spatial Inf Sci. 2014;XL-7:29–36. doi: 10.5194/isprsarchives-XL-7-29-2014. [DOI] [Google Scholar]
- Berdugo CA, Zito R, Paulus S, Mahlein AK. Fusion of sensor data for the detection and differentiation of plant diseases in cucumber. Plant Pathol. 2014;63(6):1344–1356. doi: 10.1111/ppa.12219. [DOI] [Google Scholar]
- Bergsträsser S, Fanourakis D, Schmittgen S, et al. HyperART: non-invasive quantification of leaf traits using hyperspectral absorption-reflectance-transmittance imaging. Plant Methods. 2015;11:1. doi: 10.1186/s13007-015-0043-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhattarai GP, Schmid RB, McCornack BP. Remote sensing data to detect hessian fly infestation in commercial wheat fields. Sci Rep. 2019;9:6109. doi: 10.1038/s41598-019-42620-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bian J, Zhang Z, Chen J, et al. Simplified evaluation of cotton water stress using high resolution unmanned aerial vehicle thermal imagery. Remote Sens (basel) 2019;11:267. doi: 10.3390/rs11030267. [DOI] [Google Scholar]
- Bolger AM, Poorter H, Dumschott K, Bolger ME, Arend D, Osorio S, Gundlach H, Mayer KF, Lange M, Scholz U, Usadel B. Computational aspects underlying genome to phenome analysis in plants. Plant J. 2019;97(1):182–198. doi: 10.1111/tpj.14179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boureau T. PHENOTIC platform. Portail Data INRAE. 2020 doi: 10.15454/u2bwfj. [DOI] [Google Scholar]
- Brichet N, Fournier C, Turc O, et al. A robot-assisted imaging pipeline for tracking the growths of maize ear and silks in a high-throughput phenotyping platform. Plant Methods. 2017;13:96. doi: 10.1186/s13007-017-0246-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broge NH, Leblanc E. Comparing prediction power and stability of broadband and hyperspectral vegetation indices for estimation of green leaf area index and canopy chlorophyll density. Remote Sens Environ. 2001;76:156–172. doi: 10.1016/S0034-4257(00)00197-8. [DOI] [Google Scholar]
- Burgos-Artizzu XP, Ribeiro A, Guijarro M, Pajares G. Real-time image processing for crop/weed discrimination in maize fields. Comput Electron Agric. 2011;75:337–346. doi: 10.1016/j.compag.2010.12.011. [DOI] [Google Scholar]
- Bürling K, Hunsche M, Noga G. Use of blue-green and chlorophyll fluorescence measurements for differentiation between nitrogen deficiency and pathogen infection in winter wheat. J Plant Physiol. 2011;168:1641–1648. doi: 10.1016/j.jplph.2011.03.016. [DOI] [PubMed] [Google Scholar]
- Busemeyer L, Mentrup D, Möller K, et al. BreedVision–a multi-sensor platform for non-destructive field-based phenotyping in plant breeding. Sensors. 2013;13:2830–2847. doi: 10.3390/s130302830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Busemeyer L, Ruckelshausen A, Möller K, et al. Precision phenotyping of biomass accumulation in triticale reveals temporal genetic patterns of regulation. Sci Rep. 2013;3:2442. doi: 10.1038/srep02442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cabrera-Bosquet L, Sánchez C, Rosales A, et al. Near-infrared reflectance spectroscopy (NIRS) assessment of δ(18)O and nitrogen and ash contents for improved yield potential and drought adaptation in maize. J Agric Food Chem. 2011;59:467–474. doi: 10.1021/jf103395z. [DOI] [PubMed] [Google Scholar]
- Cabrera-Bosquet L, Fournier C, Brichet N, et al. High-throughput estimation of incident light, light interception and radiation-use efficiency of thousands of plants in a phenotyping platform. New Phytol. 2016;212:269–281. doi: 10.1111/nph.14027. [DOI] [PubMed] [Google Scholar]
- Calderón R, Navas-Cortés J, Zarco-Tejada P. Early detection and quantification of verticillium wilt in olive using hyperspectral and thermal imagery over large areas. Remote Sens (basel) 2015;7:5584–5610. doi: 10.3390/rs70505584. [DOI] [Google Scholar]
- Camino C, González-Dugo V, Hernández P, et al. Improved nitrogen retrievals with airborne-derived fluorescence and plant traits quantified from VNIR-SWIR hyperspectral imagery in the context of precision agriculture. Int J Appl Earth Obs Geoinf. 2018;70:105–117. doi: 10.1016/j.jag.2018.04.013. [DOI] [Google Scholar]
- Campbell MT, Knecht AC, Berger B, Brien CJ, Wang D, Walia H. Integrating image-based phenomics and association analysis to dissect the genetic architecture of temporal salinity responses in rice. Plant Physiol. 2015;168(4):1476–1489. doi: 10.1104/pp.15.00450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao X, Luo Y, Zhou Y, et al. Detection of powdery mildew in two winter wheat cultivars using canopy hyperspectral reflectance. Crop Prot. 2013;45:124–131. doi: 10.1016/j.cropro.2012.12.002. [DOI] [Google Scholar]
- Cârlan I, Haase D, Große-Stoltenberg A, Sandric I. Mapping heat and traffic stress of urban park vegetation based on satellite imagery—a comparison of Bucharest, Romania and Leipzig, Germany. Urban Ecosyst. 2020;23:363–377. doi: 10.1007/s11252-019-00916-z. [DOI] [Google Scholar]
- Casanova JJ, O’Shaughnessy SA, Evett SR, Rush CM. Development of a wireless computer vision instrument to detect biotic stress in wheat. Sensors. 2014;14:17753–17769. doi: 10.3390/s140917753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Z, Wang J, Wang T, et al. Automated in-field leaf-level hyperspectral imaging of corn plants using a Cartesian robotic platform. Comput Electron Agric. 2021;183:105996. doi: 10.1016/j.compag.2021.105996. [DOI] [Google Scholar]
- Chéné Y, Rousseau D, Lucidarme P, et al. On the use of depth camera for 3D phenotyping of entire plants. Comput Electron Agric. 2012;82:122–127. doi: 10.1016/j.compag.2011.12.007. [DOI] [Google Scholar]
- Chi G, Huang B, Shi Y, et al. Detecting ozone effects in four wheat cultivars using hyperspectral measurements under fully open-air field conditions. Remote Sens Environ. 2016;184:329–336. doi: 10.1016/j.rse.2016.07.020. [DOI] [Google Scholar]
- Cho M, Hegde C (2019) Reducing the search space for hyperparameter optimization using group sparsity. In: ICASSP 2019–2019 IEEE International Conference on Acoustics, Speech and Signal Processing (ICASSP), IEEE, p 3627–3631
- Clark RT, MacCurdy RB, Jung JK, et al. Three-dimensional root phenotyping with a novel imaging and software platform. Plant Physiol. 2011;156:455–465. doi: 10.1104/pp.110.169102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coulibaly S, Kamsu-Foguem B, Kamissoko D, Traore D. Deep neural networks with transfer learning in millet crop images. Comput Ind. 2019;108:115–120. doi: 10.1016/j.compind.2019.02.003. [DOI] [Google Scholar]
- Crain JL, Wei Y, Barker J, et al. Development and deployment of a portable field phenotyping platform. Crop Sci. 2016;56:965–975. doi: 10.2135/cropsci2015.05.0290. [DOI] [Google Scholar]
- Crain J, Mondal S, Rutkoski J, et al. Combining high-throughput phenotyping and genomic information to increase prediction and selection accuracy in wheat breeding. Plant Genome. 2018 doi: 10.3835/plantgenome2017.05.0043. [DOI] [PubMed] [Google Scholar]
- Czedik-Eysenberg A, Seitner S, Güldener U, et al. The “PhenoBox”, a flexible, automated, open-source plant phenotyping solution. New Phytol. 2018;219:808–823. doi: 10.1111/nph.15129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dachoupakan Sirisomboon C, Putthang R, Sirisomboon P. Application of near infrared spectroscopy to detect aflatoxigenic fungal contamination in rice. Food Control. 2013;33:207–214. doi: 10.1016/j.foodcont.2013.02.034. [DOI] [Google Scholar]
- Dammer K-H, Möller B, Rodemann B, Heppner D. Detection of head blight (Fusarium ssp.) in winter wheat by color and multispectral image analyses. Crop Prot. 2011;30:420–428. doi: 10.1016/j.cropro.2010.12.015. [DOI] [Google Scholar]
- Dangwal N, Patel NR, Kumari M, Saha SK. Monitoring of water stress in wheat using multispectral indices derived from Landsat-TM. Geocarto Int. 2016;31:682–693. doi: 10.1080/10106049.2015.1073369. [DOI] [Google Scholar]
- Das PK, Laxman B, Rao SVCK, et al. Monitoring of bacterial leaf blight in rice using ground-based hyperspectral and LISS IV satellite data in Kurnool, Andhra Pradesh, India. Int J Pest Manag. 2015;61:359–368. doi: 10.1080/09670874.2015.1072652. [DOI] [Google Scholar]
- Das R, Banerjee M, De S, editors. Emerging trends in disruptive technology management for sustainable development, illustrated edition. Boca Raton: CRC Press; 2019. [Google Scholar]
- Dash J, Curran PJ. The MERIS terrestrial chlorophyll index. Int J Remote Sens. 2004;25:5403–5413. doi: 10.1080/0143116042000274015. [DOI] [Google Scholar]
- DeChant C, Wiesner-Hanks T, Chen S, et al. Automated identification of northern leaf blight-infected maize plants from field imagery using deep learning. Phytopathology. 2017;107:1426–1432. doi: 10.1094/PHYTO-11-16-0417-R. [DOI] [PubMed] [Google Scholar]
- Del Fiore A, Reverberi M, Ricelli A, et al. Early detection of toxigenic fungi on maize by hyperspectral imaging analysis. Int J Food Microbiol. 2010;144:64–71. doi: 10.1016/j.ijfoodmicro.2010.08.001. [DOI] [PubMed] [Google Scholar]
- Delalieux S, van Aardt J, Keulemans W, et al. Detection of biotic stress (Venturia inaequalis) in apple trees using hyperspectral data: non-parametric statistical approaches and physiological implications. Eur J Agron. 2007;27:130–143. doi: 10.1016/j.eja.2007.02.005. [DOI] [Google Scholar]
- Devadas R, Lamb DW, Backhouse D, Simpfendorfer S. Sequential application of hyperspectral indices for delineation of stripe rust infection and nitrogen deficiency in wheat. Precis Agric. 2015;16(5):477–491. doi: 10.1007/s11119-015-9390-0. [DOI] [Google Scholar]
- Diezma B, Lleó L, Roger JM, et al. Examination of the quality of spinach leaves using hyperspectral imaging. Postharvest Biol Technol. 2013;85:8–17. doi: 10.1016/j.postharvbio.2013.04.017. [DOI] [Google Scholar]
- Dobbels AA, Lorenz AJ. Soybean iron deficiency chlorosis high throughput phenotyping using an unmanned aircraft system. Plant Methods. 2019;15:97. doi: 10.1186/s13007-019-0478-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong T, Liu J, Qian B, et al. Estimating winter wheat biomass by assimilating leaf area index derived from fusion of Landsat-8 and MODIS data. Int J Appl Earth Obs Geoinf. 2016;49:63–74. doi: 10.1016/j.jag.2016.02.001. [DOI] [Google Scholar]
- Ehsani R, Maja JM. The rise of small UAVs in precision agriculture. Resour Mag. 2013;20:18–19. [Google Scholar]
- Eichner J, Zeller G, Laubinger S, Rätsch G. Support vector machines-based identification of alternative splicing in Arabidopsis thaliana from whole-genome tiling arrays. BMC Bioinformatics. 2011;12:55. doi: 10.1186/1471-2105-12-55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ENCODE Project Consortium An integrated encyclopedia of DNA elements in the human genome. Nature. 2012;489(7414):57–74. doi: 10.1038/nature11247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fabre J, Dauzat M, Nègre V, Wuyts N, Tireau A, Gennari E, Neveu P, Tisné S, Massonnet C, Hummel I, Granier C. PHENOPSIS DB: an information system for Arabidopsis thaliana phenotypic data in an environmental context. BMC Plant Biol. 2011;11(1):1–7. doi: 10.1186/1471-2229-11-77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fieuzal R, Bustillo V, Collado D, Dedieu G. Combined use of multi-temporal landsat-8 and sentinel-2 images for wheat yield estimates at the intra-plot spatial scale. Agronomy. 2020;10:327. doi: 10.3390/agronomy10030327. [DOI] [Google Scholar]
- Flood PJ, Kruijer W, Schnabel SK, et al. Phenomics for photosynthesis, growth and reflectance in Arabidopsis thaliana reveals circadian and long-term fluctuations in heritability. Plant Methods. 2016;12:14. doi: 10.1186/s13007-016-0113-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuentes A, Yoon S, Kim SC, Park DS. A robust deep-learning-based detector for real-time tomato plant diseases and pests recognition. Sensors. 2017 doi: 10.3390/s17092022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukatsu T, Watanabe T, Hu H, et al. Field monitoring support system for the occurrence of Leptocorisa chinensis Dallas (Hemiptera: Alydidae) using synthetic attractants, field servers, and image analysis. Comput Electron Agric. 2012;80:8–16. doi: 10.1016/j.compag.2011.10.005. [DOI] [Google Scholar]
- Garcia-Ruiz F, Sankaran S, Maja JM, et al. Comparison of two aerial imaging platforms for identification of Huanglongbing-infected citrus trees. Comput Electron Agric. 2013;91:106–115. doi: 10.1016/j.compag.2012.12.002. [DOI] [Google Scholar]
- Garriga M, Retamales JB, Romero-Bravo S, Caligari PD, Lobos GA. Chlorophyll, anthocyanin, and gas exchange changes assessed by spectroradiometry in Fragaria chiloensis under salt stress. J Integr Plant Biol. 2014;56(5):505–515. doi: 10.1111/jipb.12193. [DOI] [PubMed] [Google Scholar]
- Gehan MA, Kellogg EA. High-throughput phenotyping. Am J Bot. 2017;104:505–508. doi: 10.3732/ajb.1700044. [DOI] [PubMed] [Google Scholar]
- Gennaro SD, Battiston E, Marco SD, et al. Unmanned Aerial Vehicle (UAV)-based remote sensing to monitor grapevine leaf stripe disease within a vineyard affected by esca complex. Phytopathol Mediterr. 2016;55:262–275. [Google Scholar]
- Gitelson A, Merzlyak MN. Quantitative estimation of chlorophyll-a using reflectance spectra: experiments with autumn chestnut and maple leaves. J Photochem Photobiol Biol. 1994;22:247–252. doi: 10.1016/1011-1344(93)06963-4. [DOI] [Google Scholar]
- Gitelson AA, Kaufman YJ, Merzlyak MN. Use of a green channel in remote sensing of global vegetation from EOS-MODIS. Remote Sens Environ. 1996;58:289–298. doi: 10.1016/S0034-4257(96)00072-7. [DOI] [Google Scholar]
- Gitelson AA, Kaufman YJ, Stark R, Rundquist D. Novel algorithms for remote estimation of vegetation fraction. Remote Sens Environ. 2002;80:76–87. doi: 10.1016/S0034-4257(01)00289-9. [DOI] [Google Scholar]
- Gitelson AA, Gritz Y, Merzlyak MN. Relationships between leaf chlorophyll content and spectral reflectance and algorithms for non-destructive chlorophyll assessment in higher plant leaves. J Plant Physiol. 2003;160:271–282. doi: 10.1078/0176-1617-00887. [DOI] [PubMed] [Google Scholar]
- Gosseau F, Blanchet N, Varès D, et al. Heliaphen, an outdoor high-throughput phenotyping platform for genetic studies and crop modeling. Front Plant Sci. 2018;9:1908. doi: 10.3389/fpls.2018.01908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Granier C, Aguirrezabal L, Chenu K, et al. PHENOPSIS, an automated platform for reproducible phenotyping of plant responses to soil water deficit in Arabidopsis thaliana permitted the identification of an accession with low sensitivity to soil water deficit. New Phytol. 2006;169:623–635. doi: 10.1111/j.1469-8137.2005.01609.x. [DOI] [PubMed] [Google Scholar]
- Gregor K, LeCun Y (2010) Learning fast approximations of sparse coding. In: Proceedings of the 27th international conference on international conference on machine learning, p 399–406
- Guerrero JM, Pajares G, Montalvo M, et al. Support vector machines for crop/weeds identification in maize fields. Expert Syst Appl. 2012;39:11149–11155. doi: 10.1016/j.eswa.2012.03.040. [DOI] [Google Scholar]
- Gulli A, Pal S. Deep learning with Keras: implementing deep learning models and neural networks with the power of Python. Birmingham: Packt Publishing; 2017. [Google Scholar]
- Haagsma M, Page G, Johnson J, Still C, Waring K, Sniezko R, Selker J, (2020). Is more data better? A comparison of multi-and hyperspectral imaging in phenotyping. In: EGU General Assembly Conference Abstracts, p 10673
- Haboudane D, Tremblay N, Miller JR, Vigneault P. Remote estimation of crop chlorophyll content using spectral indices derived from hyperspectral data. IEEE Trans Geosci Remote Sensing. 2008;46:423–437. doi: 10.1109/TGRS.2007.904836. [DOI] [Google Scholar]
- Hairmansis A, Berger B, Tester M, Roy SJ. Image-based phenotyping for non-destructive screening of different salinity tolerance traits in rice. Rice (NY) 2014;7:16. doi: 10.1186/s12284-014-0016-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hallau L, Neumann M, Klatt B, et al. Automated identification of sugar beet diseases using smartphones. Plant Pathol. 2017;67:399–410. doi: 10.1111/ppa.12741. [DOI] [Google Scholar]
- Hassan MA, Yang M, Rasheed A, et al. A rapid monitoring of NDVI across the wheat growth cycle for grain yield prediction using a multi-spectral UAV platform. Plant Sci. 2019;282:95–103. doi: 10.1016/j.plantsci.2018.10.022. [DOI] [PubMed] [Google Scholar]
- He L, Mostovoy G. Cotton yield estimate using sentinel-2 data and an ecosystem model over the southern US. Remote Sens (basel) 2019;11:2000. doi: 10.3390/rs11172000. [DOI] [Google Scholar]
- Hernández-Rabadán DL, Ramos-Quintana F, Guerrero Juk J. Integrating SOMs and a Bayesian classifier for segmenting diseased plants in uncontrolled environments. Sci World J. 2014;2014:214674. doi: 10.1155/2014/214674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hillnhütter C, Mahlein AK, Sikora RA, Oerke EC. Remote sensing to detect plant stress induced by Heterodera schachtii and Rhizoctonia solani in sugar beet fields. Field Crops Res. 2011;122:70–77. doi: 10.1016/j.fcr.2011.02.007. [DOI] [Google Scholar]
- Huete A, Didan K, Miura T, et al. Overview of the radiometric and biophysical performance of the MODIS vegetation indices. Remote Sens Environ. 2002;83:195–213. doi: 10.1016/S0034-4257(02)00096-2. [DOI] [Google Scholar]
- Hunt ER, Daughtry CST, Eitel JUH, Long DS. Remote sensing leaf chlorophyll content using a visible band index. Agron J. 2011;103:1090. doi: 10.2134/agronj2010.0395. [DOI] [Google Scholar]
- Hunt ER, Doraiswamy PC, McMurtrey JE, et al. A visible band index for remote sensing leaf chlorophyll content at the canopy scale. Int J Appl Earth Obs Geoinf. 2013;21:103–112. doi: 10.1016/j.jag.2012.07.020. [DOI] [Google Scholar]
- Hunter MC, Smith RG, Schipanski ME, et al. Agriculture in 2050: recalibrating targets for sustainable intensification. Bioscience. 2017;67:386–391. doi: 10.1093/biosci/bix010. [DOI] [Google Scholar]
- Jackson RD, Huete AR. Interpreting vegetation indices. Prev Vet Med. 1991;11:185–200. doi: 10.1016/S0167-5877(05)80004-2. [DOI] [Google Scholar]
- Jansen M, Gilmer F, Biskup B, et al. Simultaneous phenotyping of leaf growth and chlorophyll fluorescence via GROWSCREEN FLUORO allows detection of stress tolerance in Arabidopsis thaliana and other rosette plants. Funct Plant Biol. 2009;36:902–914. doi: 10.1071/FP09095. [DOI] [PubMed] [Google Scholar]
- Jeudy C, Adrian M, Baussard C, et al. RhizoTubes as a new tool for high throughput imaging of plant root development and architecture: test, comparison with pot grown plants and validation. Plant Methods. 2016;12:31. doi: 10.1186/s13007-016-0131-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones CD, Jones JB, Lee WS. Diagnosis of bacterial spot of tomato using spectral signatures. Comput Electron Agric. 2010;74:329–335. doi: 10.1016/j.compag.2010.09.008. [DOI] [Google Scholar]
- Kandpal LM, Lee S, Kim MS, et al. Short wave infrared (SWIR) hyperspectral imaging technique for examination of aflatoxin B1 (AFB1) on corn kernels. Food Control. 2015;51:171–176. doi: 10.1016/j.foodcont.2014.11.020. [DOI] [Google Scholar]
- Kaur B, Sandhu KS, Kamal R, et al. Omics for the improvement of abiotic, biotic, and agronomic traits in major cereal crops: applications, challenges, and prospects. Plants. 2021;10:1989. doi: 10.3390/PLANTS10101989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kersting K, Xu Z, Wahabzada M, et al (2012) Pre-symptomatic prediction of plant drought stress using dirichlet-aggregation regression on hyperspectral images
- Kim JY. Roadmap to high throughput phenotyping for plant breeding. J Biosyst Eng. 2020;45(1):43–55. doi: 10.1007/s42853-020-00043-0. [DOI] [Google Scholar]
- Kim SW, Kim HJ, Kim JH, et al. A rapid, simple method for the genetic discrimination of intact Arabidopsis thaliana mutant seeds using metabolic profiling by direct analysis in real-time mass spectrometry. Plant Methods. 2011;7:14. doi: 10.1186/1746-4811-7-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim SL, Kim N, Lee H, et al. High-throughput phenotyping platform for analyzing drought tolerance in rice. Planta. 2020;252:38. doi: 10.1007/s00425-020-03436-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kloth KJ, Ten Broeke CJ, Thoen MP, et al. High-throughput phenotyping of plant resistance to aphids by automated video tracking. Plant Methods. 2015;11:4. doi: 10.1186/s13007-015-0044-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kokhan S, Vostokov A. Using vegetative indices to quantify agricultural crop characteristics. J Ecol Eng. 2020;21:120–127. doi: 10.12911/22998993/119808. [DOI] [Google Scholar]
- Kong W, Liu F, Zhang C, et al. Fast detection of peroxidase (POD) activity in tomato leaves which infected with Botrytis cinerea using hyperspectral imaging. Spectrochim Acta A Mol Biomol Spectrosc. 2014;118:498–502. doi: 10.1016/j.saa.2013.09.009. [DOI] [PubMed] [Google Scholar]
- Krajewski P, Chen D, Ćwiek H, van Dijk AD, Fiorani F, Kersey P, Klukas C, Lange M, Markiewicz A, Nap JP, van Oeveren J. Towards recommendations for metadata and data handling in plant phenotyping. J Exp Bot. 2015;66(18):5417–5427. doi: 10.1093/jxb/erv271. [DOI] [PubMed] [Google Scholar]
- Le Marié C, Kirchgessner N, Flütsch P, et al. RADIX: rhizoslide platform allowing high throughput digital image analysis of root system expansion. Plant Methods. 2016;12:40. doi: 10.1186/s13007-016-0140-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LeCun Y, Bengio Y, Hinton G. Deep learning. Nature. 2015;521:436–444. doi: 10.1038/nature14539. [DOI] [PubMed] [Google Scholar]
- Li L, Zhang Q, Huang D. A review of imaging techniques for plant phenotyping. Sensors. 2014;14:20078–20111. doi: 10.3390/s141120078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Wang C, Wang X, Hou P, Luo B, Song P, Pan D, Li A, Chen L. Disposable stainless steel-based electrochemical microsensor for in vivo determination of indole-3-acetic acid in soybean seedlings. Biosens Bioelectron. 2019;126:193–199. doi: 10.1016/j.bios.2018.10.041. [DOI] [PubMed] [Google Scholar]
- Liebisch F, Kirchgessner N, Schneider D, et al. Remote, aerial phenotyping of maize traits with a mobile multi-sensor approach. Plant Methods. 2015;11:9. doi: 10.1186/s13007-015-0048-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin K, Gong L, Huang Y, et al. Deep learning-based segmentation and quantification of cucumber powdery mildew using convolutional neural network. Front Plant Sci. 2019;10:155. doi: 10.3389/fpls.2019.00155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu M, Wang T, Skidmore AK, Liu X. Heavy metal-induced stress in rice crops detected using multi-temporal Sentinel-2 satellite images. Sci Total Environ. 2018;637–638:18–29. doi: 10.1016/j.scitotenv.2018.04.415. [DOI] [PubMed] [Google Scholar]
- López-López M, Calderón R, González-Dugo V, Zarco-Tejada PJ, Fereres E. Early detection and quantification of almond red leaf blotch using high-resolution hyperspectral and thermal imagery. Remote Sensing. 2016;8(4):276. doi: 10.3390/rs8040276. [DOI] [Google Scholar]
- Louhaichi M, Borman MM, Johnson DE. Spatially located platform and aerial photography for documentation of grazing impacts on wheat. Geocarto Int. 2001;16:65–70. doi: 10.1080/10106040108542184. [DOI] [Google Scholar]
- Lu Y, Yi S, Zeng N, et al. Identification of rice diseases using deep convolutional neural networks. Neurocomputing. 2017;267:378–384. doi: 10.1016/j.neucom.2017.06.023. [DOI] [Google Scholar]
- Mahajan GR, Sahoo RN, Pandey RN, et al. Using hyperspectral remote sensing techniques to monitor nitrogen, phosphorus, sulphur and potassium in wheat (Triticum aestivum L.) Precis Agric. 2014;15:499–522. doi: 10.1007/s11119-014-9348-7. [DOI] [Google Scholar]
- Mahlein A-K, Oerke E-C, Steiner U, Dehne H-W. Recent advances in sensing plant diseases for precision crop protection. Eur J Plant Pathol. 2012;133:197–209. doi: 10.1007/s10658-011-9878-z. [DOI] [Google Scholar]
- Mahlein A-K, Steiner U, Hillnhütter C, et al. Hyperspectral imaging for small-scale analysis of symptoms caused by different sugar beet diseases. Plant Methods. 2012;8:3. doi: 10.1186/1746-4811-8-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manyika J, Chui M, Brown B, Bughin J, Dobbs R, Roxburgh C, Byers A.H (2011) Big data: the next frontier for innovation, competition, and productivity. https://www.mckinsey.com/business-functions/mckinsey-digital/our-insights/big-data-the-next-frontier-for-innovation
- Maresma Á, Ariza M, Martínez E, et al. Analysis of vegetation indices to determine nitrogen application and yield prediction in maize (Zea mays L.) from a standard UAV service. Remote Sens (basel) 2016;8:973. doi: 10.3390/rs8120973. [DOI] [Google Scholar]
- Martins SM, de Brito GG, da Gonçalves WC, et al. PhenoRoots: an inexpensive non-invasive phenotyping system to assess the variability of the root system architecture. Sci Agric. 2020 doi: 10.1590/1678-992x-2018-0420. [DOI] [Google Scholar]
- Mathieu L, Lobet G, Tocquin P, Périlleux C. “Rhizoponics”: a novel hydroponic rhizotron for root system analyses on mature Arabidopsis thaliana plants. Plant Methods. 2015;11:3. doi: 10.1186/s13007-015-0046-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meacham-Hensold K, Fu P, Wu J, Serbin S, Montes CM, Ainsworth E, Guan K, Dracup E, Pederson T, Driever S, Bernacchi C. Plot-level rapid screening for photosynthetic parameters using proximal hyperspectral imaging. J Exp Bot. 2020;71(7):2312–2328. doi: 10.1093/jxb/eraa068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mer CR, Wahabzada M, Ballvora A, et al. Early drought stress detection in cereals: simplex volume maximisation for hyperspectral image analysis. Funct Plant Biol. 2012;39:878–890. doi: 10.1071/FP12060. [DOI] [PubMed] [Google Scholar]
- Mewes T, Franke J, Menz G. Spectral requirements on airborne hyperspectral remote sensing data for wheat disease detection. Precision Agric. 2011;12(6):795–812. doi: 10.1007/s11119-011-9222-9. [DOI] [Google Scholar]
- Min M, Lee WS, Burks TF, Jordan JD, Schumann AW, Schueller JK, Xie H. Design of a hyperspectral nitrogen sensing system for orange leaves. Comput Electron Agric. 2008;63(2):215–226. doi: 10.1016/j.compag.2008.03.004. [DOI] [Google Scholar]
- Min S, Lee B, Yoon S. Deep learning in bioinformatics. Brief Bioinformatics. 2017;18:851–869. doi: 10.1093/bib/bbw068. [DOI] [PubMed] [Google Scholar]
- Mir RR, Reynolds M, Pinto F, et al. High-throughput phenotyping for crop improvement in the genomics era. Plant Sci. 2019;282:60–72. doi: 10.1016/j.plantsci.2019.01.007. [DOI] [PubMed] [Google Scholar]
- Mishra P, Polder G, Vilfan N. Close range spectral imaging for disease detection in plants using autonomous platforms: a review on recent studies. Curr Robot Rep. 2020;1:43–48. doi: 10.1007/s43154-020-00004-7. [DOI] [Google Scholar]
- Mo C, Kim MS, Kim G, et al. Detecting drought stress in soybean plants using hyperspectral fluorescence imaging. J Biosyst Eng. 2015;40:335–344. doi: 10.5307/JBE.2015.40.4.335. [DOI] [Google Scholar]
- Mokhtar U, Ali MAS, Hassanien AE, Hefny H, et al. Identifying two of tomatoes leaf viruses using support vector machine. In: Mandal JK, Satapathy SC, Kumar Sanyal M, et al., editors. Information systems design and intelligent applications. New Delhi: Springer India; 2015. pp. 771–782. [Google Scholar]
- Nagasubramanian K, Jones S, Singh AK, et al. Plant disease identification using explainable 3D deep learning on hyperspectral images. Plant Methods. 2019;15:98. doi: 10.1186/s13007-019-0479-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagel KA, Putz A, Gilmer F, et al. GROWSCREEN-Rhizo is a novel phenotyping robot enabling simultaneous measurements of root and shoot growth for plants grown in soil-filled rhizotrons. Funct Plant Biol. 2012;39:891. doi: 10.1071/FP12023. [DOI] [PubMed] [Google Scholar]
- Naik HS, Zhang J, Lofquist A, et al. A real-time phenotyping framework using machine learning for plant stress severity rating in soybean. Plant Methods. 2017;13:23. doi: 10.1186/s13007-017-0173-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Navrozidis I, Alexandridis TK, Dimitrakos A, et al. Identification of purple spot disease on asparagus crops across spatial and spectral scales. Comput Electron Agric. 2018;148:322–329. doi: 10.1016/j.compag.2018.03.035. [DOI] [Google Scholar]
- Ochoa D, Cevallos J, Vargas G, et al. Hyperspectral imaging system for disease scanning on banana plants. In: Kim MS, Chao K, Chin BA, et al., editors. Sensing for agriculture and food quality and safety VIII. Bellingham: SPIE; 2016. p. 98640M. [Google Scholar]
- Oerke E-C, Mahlein A-K, Steiner U. Proximal sensing of plant diseases. In: Gullino ML, Bonants PJM, editors. Detection and diagnostics of plant pathogens. Dordrecht: Springer Netherlands; 2014. pp. 55–68. [Google Scholar]
- Omran E-S. Remote estimation of vegetation parameters using narrowband sensor for precision agriculture in arid environment. Egypt J Soil Sci. 2018;58:73–92. doi: 10.21608/ejss.2018.5614. [DOI] [Google Scholar]
- Onoyama H, Ryu C, Suguri M, Iida M. Potential of hyperspectral imaging for constructing a year-invariant model to estimate the nitrogen content of rice plants at the panicle initiation stage. IFAC Proc Vol. 2013;46:219–224. doi: 10.3182/20130828-2-SF-3019.00054. [DOI] [Google Scholar]
- Peña JM, Torres-Sánchez J, Serrano-Pérez A, et al. Quantifying efficacy and limits of unmanned aerial vehicle (UAV) technology for weed seedling detection as affected by sensor resolution. Sensors. 2015;15:5609–5626. doi: 10.3390/s150305609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pérez AJ, López F, Benlloch JV, Christensen S. Colour and shape analysis techniques for weed detection in cereal fields. Comput Electron Agric. 2000;25:197–212. doi: 10.1016/S0168-1699(99)00068-X. [DOI] [Google Scholar]
- Pérez-Bueno ML, Pineda M, Cabeza FM, Barón M. Multicolor fluorescence imaging as a candidate for disease detection in plant phenotyping. Front Plant Sci. 2016;7:1790. doi: 10.3389/fpls.2016.01790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pérez-Enciso M, Zingaretti LM. A guide for using deep learning for complex trait genomic prediction. Genes (basel) 2019 doi: 10.3390/genes10070553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pérez-Ruiz M, Prior A, Martinez-Guanter J, et al. Development and evaluation of a self-propelled electric platform for high-throughput field phenotyping in wheat breeding trials. Comput Electron Agric. 2020;169:105237. doi: 10.1016/j.compag.2020.105237. [DOI] [Google Scholar]
- Peri S (2020) PhytoOracle: a scalable, modular framework for phenomics data processing and trait extraction. In: Plant and Animal Genome XXVIII Conference (January 11–15, 2020)
- Pessarakli M. Handbook of plant and crop stress. 4. Boca Raton, FL: CRC Press, Taylor & Francis Group; 2019. [Google Scholar]
- Pettorelli N. Satellite remote sensing and the management of natural resources. Oxford: Oxford University Press; 2019. [Google Scholar]
- Pineda M, Barón M, Pérez-Bueno M-L. Thermal imaging for plant stress detection and phenotyping. Remote Sens (basel) 2020;13:68. doi: 10.3390/rs13010068. [DOI] [Google Scholar]
- Prabhakar M, Prasad YG, Desai S, et al. Hyperspectral remote sensing of yellow mosaic severity and associated pigment losses in Vigna mungo using multinomial logistic regression models. Crop Prot. 2013;45:132–140. doi: 10.1016/j.cropro.2012.12.003. [DOI] [Google Scholar]
- Prabhakar M, Prasad YG, Vennila S, et al. Hyperspectral indices for assessing damage by the solenopsis mealybug (Hemiptera: Pseudococcidae) in cotton. Comput Electron Agric. 2013;97:61–70. doi: 10.1016/j.compag.2013.07.004. [DOI] [Google Scholar]
- Prashar A, Jones HG. Assessing drought responses using thermal infrared imaging. Methods Mol Biol. 2016;1398:209–219. doi: 10.1007/978-1-4939-3356-3_17. [DOI] [PubMed] [Google Scholar]
- Qiu Q, Sun N, Bai H, et al. Field-based high-throughput phenotyping for maize plant using 3D LiDAR point cloud generated with a “Phenomobile”. Front Plant Sci. 2019;10:554. doi: 10.3389/fpls.2019.00554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramcharan A, Baranowski K, McCloskey P, et al. Deep learning for image-based cassava disease detection. Front Plant Sci. 2017;8:1852. doi: 10.3389/fpls.2017.01852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramcharan A, McCloskey P, Baranowski K, et al. A mobile-based deep learning model for cassava disease diagnosis. Front Plant Sci. 2019;10:272. doi: 10.3389/fpls.2019.00272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ranzato MA, Mnih V, Susskind JM, Hinton GE. Modeling natural images using gated MRFs. IEEE Trans Pattern Anal Mach Intell. 2013;35(9):2206–2222. doi: 10.1109/TPAMI.2013.29. [DOI] [PubMed] [Google Scholar]
- Raza S-A, Smith HK, Clarkson GJJ, et al. Automatic detection of regions in spinach canopies responding to soil moisture deficit using combined visible and thermal imagery. PLoS ONE. 2014;9:e97612. doi: 10.1371/journal.pone.0097612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raza S-A, Prince G, Clarkson JP, Rajpoot NM. Automatic detection of diseased tomato plants using thermal and stereo visible light images. PLoS One. 2015;10:e0123262. doi: 10.1371/journal.pone.0123262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rey B, Aleixos N, Cubero S, Blasco J. Xf-Rovim. A field robot to detect olive trees infected by Xylella fastidiosa using proximal sensing. Remote Sens (basel) 2019;11:221. doi: 10.3390/rs11030221. [DOI] [Google Scholar]
- Reynolds D, Ball J, Bauer A, Davey R, Griffiths S, Zhou J. CropSight: a scalable and open-source information management system for distributed plant phenotyping and IoT-based crop management. Gigascience. 2019;8(3):giz009. doi: 10.1093/gigascience/giz009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roitsch T, Cabrera-Bosquet L, Fournier A, Ghamkhar K, Jiménez-Berni J, Pinto F, Ober ES. New sensors and data-driven approaches—a path to next generation phenomics. Plant Sci. 2019;282:2–10. doi: 10.1016/j.plantsci.2019.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rousseau C, Belin E, Bove E, et al. High throughput quantitative phenotyping of plant resistance using chlorophyll fluorescence image analysis. Plant Methods. 2013;9:17. doi: 10.1186/1746-4811-9-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Royimani L, Mutanga O, Odindi J, et al. Advancements in satellite remote sensing for mapping and monitoring of alien invasive plant species (AIPs) Phys Chem Earth, Parts a/b/c. 2018 doi: 10.1016/j.pce.2018.12.004. [DOI] [Google Scholar]
- Sadok W, Naudin P, Boussuge B, et al. Leaf growth rate per unit thermal time follows QTL-dependent daily patterns in hundreds of maize lines under naturally fluctuating conditions. Plant Cell Environ. 2007;30:135–146. doi: 10.1111/j.1365-3040.2006.01611.x. [DOI] [PubMed] [Google Scholar]
- Sagan V, Maimaitijiang M, Sidike P, et al. Uav/satellite multiscale data fusion for crop monitoring and early stress detection. Int Arch Photogramm Remote Sens Spatial Inf Sci. 2019;XLII-2/W13:715–722. doi: 10.5194/isprs-archives-XLII-2-W13-715-2019. [DOI] [Google Scholar]
- Saini DK, Chopra Y, Singh J, et al. Comprehensive evaluation of mapping complex traits in wheat using genome-wide association studies. Mol Breed. 2022;42:1. doi: 10.1007/s11032-021-01272-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salas Fernandez MG, Bao Y, Tang L, Schnable PS. A high-throughput, field-based phenotyping technology for tall biomass crops. Plant Physiol. 2017;174:2008–2022. doi: 10.1104/pp.17.00707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salgadoe A, Robson A, Lamb D, et al. Quantifying the severity of phytophthora root rot disease in avocado trees using image analysis. Remote Sens (basel) 2018;10:226. doi: 10.3390/rs10020226. [DOI] [Google Scholar]
- Samantara K, Shiv A, de Sousa LL, Sandhu KS, Priyadarshini P, Mohapatra SR. A comprehensive review on epigenetic mechanisms and application of epigenetic modifications for crop improvement. Environ Exp Bot. 2021;188:104479. doi: 10.1016/j.envexpbot.2021.104479. [DOI] [Google Scholar]
- Samuel AL. Some studies in machine learning using the game of checkers. IBM J Res Dev. 1959;3:210–229. doi: 10.1147/rd.33.0210. [DOI] [Google Scholar]
- Sandhu KS, Lozada DN, Zhang Z, et al. Deep learning for predicting complex traits in spring wheat breeding program. Front Plant Sci. 2020;11:613325. doi: 10.3389/fpls.2020.613325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandhu KS, Mihalyov PD, Lewien MJ, et al. Combining genomic and phenomic information for predicting grain protein content and grain yield in spring wheat. Front Plant Sci. 2021;12:613300. doi: 10.3389/fpls.2021.613300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandhu KS, Patil SS, Pumphrey MO, Carter AH. Multi-trait machine and deep learning models for genomic selection using spectral information in a wheat breeding program. The Plant Genome. 2021 doi: 10.1002/TPG2.20119. [DOI] [PubMed] [Google Scholar]
- Sandhu KS, Mihalyov PD, Lewien MJ, Pumphrey MO, Carter AH. Genomic selection and genome-wide association studies for grain protein content stability in a nested association mapping population of wheat. Agronomy. 2021;11(12):2528. doi: 10.3390/agronomy11122528. [DOI] [Google Scholar]
- Sandhu KS, Aoun M, Morris CF, Carter AH. Genomic selection for end-use quality and processing traits in soft white winter wheat breeding program with machine and deep learning models. Biol. 2021;10:689. doi: 10.3390/BIOLOGY10070689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandhu KS, Merrick L, Lewien MJ, et al. Prospectus of genomic selection and phenomics in cereal, legume and oilseed breeding programs. Front Genet. 2022;12:829131. doi: 10.3389/fgene.2021.829131. [DOI] [Google Scholar]
- Sandhu KS, Patil SS, Aoun M, Carter AH. Multi-trait multi-environment genomic prediction for end-use quality traits in winter wheat. Front Gen. 2022;13:831020. doi: 10.3389/fgene.2022.831020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanga S, Mero V, Machuve D, Mwanganda D. Mobile-based deep learning models for banana diseases detection. Eng Technol Appl Sci Res. 2020;10:5674. doi: 10.48084/etasr.3452. [DOI] [Google Scholar]
- Sankaran S, Mishra A, Maja JM, Ehsani R. Visible-near infrared spectroscopy for detection of Huanglongbing in citrus orchards. Comput Electron Agric. 2011;77:127–134. doi: 10.1016/j.compag.2011.03.004. [DOI] [Google Scholar]
- Sankaran S, Quirós JJ, Miklas PN. Unmanned aerial system and satellite-based high resolution imagery for high-throughput phenotyping in dry bean. Comput Electron Agric. 2019;165:104965. doi: 10.1016/j.compag.2019.104965. [DOI] [Google Scholar]
- Schikora M, Neupane B, Madhogaria S, et al. An image classification approach to analyze the suppression of plant immunity by the human pathogen Salmonella Typhimurium. BMC Bioinformatics. 2012;13:171. doi: 10.1186/1471-2105-13-171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Segarra J, Buchaillot ML, Araus JL, Kefauver SC. Remote sensing for precision agriculture: sentinel-2 improved features and applications. Agronomy. 2020;10:641. doi: 10.3390/agronomy10050641. [DOI] [Google Scholar]
- Shahin MA, Symons SJ. Detection of Fusarium damaged kernels in Canada Western Red Spring wheat using visible/near-infrared hyperspectral imaging and principal component analysis. Comput Electron Agric. 2011;75:107–112. doi: 10.1016/j.compag.2010.10.004. [DOI] [Google Scholar]
- Shibayama M, Sakamoto T, Takada E, et al. Continuous monitoring of visible and near-infrared band reflectance from a rice paddy for determining nitrogen uptake using digital cameras. Plant Prod Sci. 2009;12:293–306. doi: 10.1626/pps.12.293. [DOI] [Google Scholar]
- Singh AK, Ganapathysubramanian B, Sarkar S, Singh A. Deep learning for plant stress phenotyping: trends and future perspectives. Trends Plant Sci. 2018;23(10):883–898. doi: 10.1016/j.tplants.2018.07.004. [DOI] [PubMed] [Google Scholar]
- Sirault XRR, James RA, Furbank RT. A new screening method for osmotic component of salinity tolerance in cereals using infrared thermography. Functional Plant Biol. 2009;36:970. doi: 10.1071/FP09182. [DOI] [PubMed] [Google Scholar]
- Sishodia RP, Ray RL, Singh SK. Applications of remote sensing in precision agriculture: a review. Remote Sensing. 2020;12(19):3136. doi: 10.3390/rs12193136. [DOI] [Google Scholar]
- Song X, Yang C, Wu M, et al. Evaluation of sentinel-2A satellite imagery for mapping cotton root rot. Remote Sens (basel) 2017;9:906. doi: 10.3390/rs9090906. [DOI] [Google Scholar]
- Strange RN, Scott PR. Plant disease: a threat to global food security. Annu Rev Phytopathol. 2005;43:83–116. doi: 10.1146/annurev.phyto.43.113004.133839. [DOI] [PubMed] [Google Scholar]
- Teke M, Deveci HS, Haliloglu O, et al (2013) A short survey of hyperspectral remote sensing applications in agriculture. In: 2013 6th International Conference on Recent Advances in Space Technologies (RAST). IEEE, p 171–176
- Tisné S, Serrand Y, Bach L, et al. Phenoscope: an automated large-scale phenotyping platform offering high spatial homogeneity. Plant J. 2013;74:534–544. doi: 10.1111/tpj.12131. [DOI] [PubMed] [Google Scholar]
- Tucker CJ. Red and photographic infrared linear combinations for monitoring vegetation. Remote Sens Environ. 1979;8:127–150. doi: 10.1016/0034-4257(79)90013-0. [DOI] [Google Scholar]
- Vigneau N, Ecarnot M, Rabatel G, Roumet P. Potential of field hyperspectral imaging as a non destructive method to assess leaf nitrogen content in wheat. Field Crops Res. 2011;122:25–31. doi: 10.1016/j.fcr.2011.02.003. [DOI] [Google Scholar]
- Vincini M, Frazzi E, D’Alessio P. A broad-band leaf chlorophyll vegetation index at the canopy scale. Precis Agric. 2008;9:303–319. doi: 10.1007/s11119-008-9075-z. [DOI] [Google Scholar]
- Virlet N, Sabermanesh K, Sadeghi-Tehran P, Hawkesford MJ. Field Scanalyzer: an automated robotic field phenotyping platform for detailed crop monitoring. Funct Plant Biol. 2016;44:143–153. doi: 10.1071/FP16163. [DOI] [PubMed] [Google Scholar]
- Vougioukas SG. Agricultural robotics. Annu Rev Control Robot Auton Syst. 2019;2:365–392. doi: 10.1146/annurev-control-053018-023617. [DOI] [Google Scholar]
- Wahabzada M, Mahlein A-K, Bauckhage C, et al. Metro maps of plant disease dynamics–automated mining of differences using hyperspectral images. PLoS One. 2015;10:e0116902. doi: 10.1371/journal.pone.0116902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Nakano K, Ohashi S. Nondestructive detection of internal insect infestation in jujubes using visible and near-infrared spectroscopy. Postharvest Biol Technol. 2011;59:272–279. doi: 10.1016/j.postharvbio.2010.09.017. [DOI] [Google Scholar]
- Wang W, Heitschmidt GW, Ni X, et al. Identification of aflatoxin B1 on maize kernel surfaces using hyperspectral imaging. Food Control. 2014;42:78–86. doi: 10.1016/j.foodcont.2014.01.038. [DOI] [Google Scholar]
- Wang X, Zhao C, Guo N, et al. Determining the canopy water stress for spring wheat using canopy hyperspectral reflectance data in loess plateau semiarid regions. Spectrosc Lett. 2015;48:492–498. doi: 10.1080/00387010.2014.909495. [DOI] [Google Scholar]
- Wetterich CB, de Oliveira F, Neves R, Belasque J, et al. Detection of Huanglongbing in Florida using fluorescence imaging spectroscopy and machine-learning methods. Appl Opt. 2017;56:15. doi: 10.1364/AO.56.000015. [DOI] [Google Scholar]
- Wiesner-Hanks T, Stewart EL, Kaczmar N, DeChant C, Wu H, Nelson RJ, Lipson H, Gore MA. Image set for deep learning: field images of maize annotated with disease symptoms. BMC Res Notes. 2018;11(1):1–3. doi: 10.1186/s13104-018-3548-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilkinson MD, Dumontier M, Aalbersberg IJJ, et al. The FAIR Guiding Principles for scientific data management and stewardship. Sci Data. 2016;3:160018. doi: 10.1038/sdata.2016.18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu S, Wen W, Wang Y, et al. MVS-pheno: a portable and low-cost phenotyping platform for maize shoots using multiview stereo 3D reconstruction. Plant Phenomics. 2020;2020:1848437. doi: 10.34133/2020/1848437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie C, Shao Y, Li X, He Y. Detection of early blight and late blight diseases on tomato leaves using hyperspectral imaging. Sci Rep. 2015;5:16564. doi: 10.1038/srep16564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Y. Envirotyping for deciphering environmental impacts on crop plants. Theor Appl Genet. 2016;129(4):653–673. doi: 10.1007/s00122-016-2691-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue J, Su B. Significant remote sensing vegetation indices: a review of developments and applications. J Sensors. 2017;2017:1–17. doi: 10.1155/2017/1353691. [DOI] [Google Scholar]
- Yang W, Guo Z, Huang C, et al. Combining high-throughput phenotyping and genome-wide association studies to reveal natural genetic variation in rice. Nat Commun. 2014;5:5087. doi: 10.1038/ncomms6087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang W, Feng H, Zhang X, et al. Crop phenomics and high-throughput phenotyping: past decades, current challenges, and future perspectives. Mol Plant. 2020;13:187–214. doi: 10.1016/j.molp.2020.01.008. [DOI] [PubMed] [Google Scholar]
- Yao H, Hruska Z, Kincaid R, et al. Detecting maize inoculated with toxigenic and atoxigenic fungal strains with fluorescence hyperspectral imagery. Biosys Eng. 2013;115:125–135. doi: 10.1016/j.biosystemseng.2013.03.006. [DOI] [Google Scholar]
- Yazdanbakhsh N, Fisahn J. High throughput phenotyping of root growth dynamics, lateral root formation, root architecture and root hair development enabled by PlaRoM. Funct Plant Biol. 2009;36:938–946. doi: 10.1071/FP09167. [DOI] [PubMed] [Google Scholar]
- Yeh Y-H, Chung W-C, Liao J-Y, et al. Strawberry foliar anthracnose assessment by hyperspectral imaging. Comput Electron Agric. 2016;122:1–9. doi: 10.1016/j.compag.2016.01.012. [DOI] [Google Scholar]
- Young SN, Kayacan E, Peschel JM. Design and field evaluation of a ground robot for high-throughput phenotyping of energy sorghum. Precis Agric. 2018 doi: 10.1007/s11119-018-9601-6. [DOI] [Google Scholar]
- Yu K-Q, Zhao Y-R, Li X-L, et al. Hyperspectral imaging for mapping of total nitrogen spatial distribution in pepper plant. PLoS One. 2014;9:e116205. doi: 10.1371/journal.pone.0116205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan L, Pu R, Zhang J, et al. Using high spatial resolution satellite imagery for mapping powdery mildew at a regional scale. Precision Agric. 2016;17:332–348. doi: 10.1007/s11119-015-9421-x. [DOI] [Google Scholar]
- Yue J, Yang G, Li C, et al. Estimation of winter wheat above-ground biomass using unmanned aerial vehicle-based snapshot hyperspectral sensor and crop height improved models. Remote Sens (basel) 2017;9:708. doi: 10.3390/rs9070708. [DOI] [Google Scholar]
- Zaman-Allah M, Vergara O, Araus JL, et al. Unmanned aerial platform-based multi-spectral imaging for field phenotyping of maize. Plant Methods. 2015;11:35. doi: 10.1186/s13007-015-0078-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zarco-Tejada PJ, González-Dugo V, Berni JAJ. Fluorescence, temperature and narrow-band indices acquired from a UAV platform for water stress detection using a micro-hyperspectral imager and a thermal camera. Remote Sens Environ. 2012;117:322–337. doi: 10.1016/j.rse.2011.10.007. [DOI] [Google Scholar]
- Zhang J, Pu R, Huang W, et al. Using in-situ hyperspectral data for detecting and discriminating yellow rust disease from nutrient stresses. Field Crops Res. 2012;134:165–174. doi: 10.1016/j.fcr.2012.05.011. [DOI] [Google Scholar]
- Zhang J-C, Pu R, Wang J, et al. Detecting powdery mildew of winter wheat using leaf level hyperspectral measurements. Comput Electron Agric. 2012;85:13–23. doi: 10.1016/j.compag.2012.03.006. [DOI] [Google Scholar]
- Zhang C, Liu F, Kong W, He Y. Application of visible and near-infrared hyperspectral imaging to determine soluble protein content in oilseed rape leaves. Sensors. 2015;15:16576–16588. doi: 10.3390/s150716576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang C, Pumphrey MO, Zhou J, et al. Development of an automated high- throughput phenotyping system for wheat evaluation in a controlled environment. Trans ASABE. 2019;62:61–74. doi: 10.13031/trans.12856. [DOI] [Google Scholar]
- Zhang L, Zhang H, Niu Y, Han W. Mapping maize water stress based on UAV multispectral remote sensing. Remote Sens (basel) 2019;11:605. doi: 10.3390/rs11060605. [DOI] [Google Scholar]
- Zheng Q, Huang W, Cui X, et al. New spectral index for detecting wheat yellow rust using sentinel-2 multispectral imagery. Sensors. 2018 doi: 10.3390/s18030868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou J, Reynolds D, Websdale D, et al. CropQuant: an automated and scalable field phenotyping platform for crop monitoring and trait measurements to facilitate breeding and digital agriculture. BioRxiv. 2017 doi: 10.1101/161547. [DOI] [Google Scholar]
- Zhu F, Saluja M, Dharni JS, et al. PhenoImage : An open-source graphical user interface for plant image analysis. Plant Phenome J. 2021 doi: 10.1002/ppj2.20015. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Not applicable.
Not applicable.