Abstract
The Clustered Regularly Interspaced Short Palindromic Repeats/CRISPR-associated protein system (CRISPR/Cas) has recently become the most powerful tool available for genome engineering in various organisms. With efficient and proper expression of multiple guide RNAs (gRNAs), the CRISPR/Cas system is particularly suitable for multiplex genome editing. During the past several years, different CRISPR/Cas expression strategies, such as two-component transcriptional unit, single transcriptional unit, and bidirectional promoter systems, have been developed to efficiently express gRNAs as well as Cas nucleases. Significant progress has been made to optimize gRNA production using different types of promoters and RNA processing strategies such as ribozymes, endogenous RNases, and exogenous endoribonuclease (Csy4). Besides being constitutively and ubiquitously expressed, inducible and spatiotemporal regulations of gRNA expression have been demonstrated using inducible, tissue-specific, and/or synthetic promoters for specific research purposes. Most recently, the emergence of CRISPR/Cas ribonucleoprotein delivery methods, such as engineered nanoparticles, further revolutionized transgene-free and multiplex genome editing. In this review, we discuss current strategies and future perspectives for efficient expression and engineering of gRNAs with a goal to facilitate CRISPR/Cas-based multiplex genome editing.
Keywords: CRISPR/Cas, Gene expression, Genome engineering, Guide RNA, Multiplex editing
Introduction
CRISPR/Cas technology has revolutionized the field of genome engineering by dramatically improving genome editing and its applications ranging from therapeutic treatments to crop breeding. The CRISPR/Cas system is originally discovered in bacteria as an adaptive immune system, which typically comprises CRISPR loci and a Cas nuclease protein such as Cas9 (Barrangou et al. 2007). The Type II CRISPR loci consist of CRISPR RNA (crRNA) and trans-activating crRNA (tracrRNA), together forming the guide RNA (gRNA) complex. A specific gRNA is capable of directing Cas9 or other Cas nuclease to cleave a target DNA site, generating double-stranded breaks (DSBs). DSBs are primarily repaired by the non-homologous end joining (NHEJ) pathway which frequently introduces indel mutations, and by the homology-directed repair (HDR) pathway that can result in precise genome editing (Wyman and Kanaar 2006). In comparison to zinc-finger nucleases (ZFNs) and transcription activator-like effector nucleases (TALENs) (Gaj et al. 2013), CRISPR/Cas has shown clear advantages in multiplex genome editing capability by readily programing Cas nuclease to target different DNA sites with ~ 20 bp sequence of gRNA without effector nuclease modification (Cong et al. 2013; Xie et al. 2015). The intracellular levels of gRNA has been shown to correlate with Cas9 targeting and cleavage in both S. cerevisiae and mammalian cells, indicating that a sufficient amount of gRNA(s) is required at the target site for efficient genome editing (Hsu et al. 2013; Ryan et al. 2014). As a result, both gRNA specificity and expression should be optimized for CRISPR/Cas-mediated multiplex genome editing. Various studies have been conducted to improve gRNA expression efficiency by adjusting the major factors involved in the process of producing gRNAs: the type of promoter used for gRNA expression, the method for processing multiple gRNAs, and the system for CRISPR reagent delivery to targeted cells and tissues. There are diverse configurations of CRISPR/Cas expression cassettes where Cas nuclease and gRNAs are expressed by the same or different promoters. Multiple gRNAs can also be expressed with individual promoter and terminator cassettes, or with a single promoter and terminator. To improve gRNA expression efficiency for multiplex genome editing with spatiotemporal regulations, it is necessary to compare and evaluate different gRNA expression strategies. This review focuses on different types of promoters used for gRNA expression, CRISPR/Cas expression cassette configurations, and multiplex expression methods that have been demonstrated for efficient gRNA production. Molecular strategies to engineer gRNAs and their spatiotemporal expression will also be addressed in relation to their applications and future improvement for CRISPR/Cas genome editing.
Conventional approach for gRNA expression
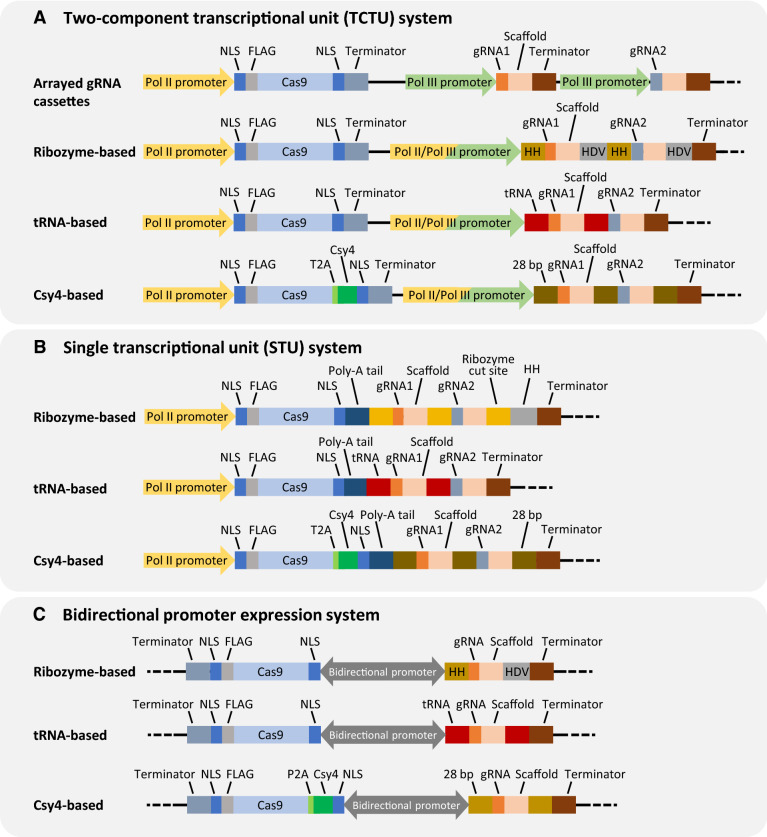
One of the main factors that impacts gRNA expression is the type of promoter used for expressing gRNA. The Cas nuclease and gRNA(s) can either be transcribed from two individual promoters or a single promoter. The conventional CRISPR/Cas-mediated genome editing system consists of a two-component transcriptional unit (TCTU), where the expression of Cas nuclease transcript and gRNA(s) is controlled separately by two promoters (Fig. 1A). The TCTU expression system can be further categorized into two types, which use either RNA polymerase II (Pol II)-based or small nuclear RNA polymerase III (Pol III)-based promoter to drive gRNA expression (Fig. 1A) (Lowder et al. 2016). In the mixed dual promoter system, Cas nuclease is typically expressed by a Pol II promoter, whereas gRNAs are expressed by a Pol III promoter, such as a U3 or U6 promoter (Fig. 1A). In eukaryotes, Pol II promoters are involved in mRNA transcription, whereas Pol III promoters function in transcription of ribosomal 5S rRNA, tRNA, and some snRNA genes. Both U3 and U6 promoters are constitutively and ubiquitously transcribed by RNA Pol III to produce small RNAs and work efficiently in plants for gRNA expression in the mixed dual promoter system (Bortesi and Fischer 2015; Jiang et al. 2013; Li et al. 2013). However, U3 and U6 promoters lack spatiotemporal control and require specific nucleotides (A or G) at the 5′ end of proto-spacer for transcription initiation (Gao and Zhao 2014; Jiang et al. 2013; Li et al. 2013; Lowder et al. 2015; Nekrasov et al. 2013; Shan et al. 2013; Tang et al. 2016; Xie et al. 2015; Yoshioka et al. 2015). Moreover, Pol III promoters are not well characterized in non-model organisms, making it difficult to find suitable heterologous U3 and U6 promoters for CRISPR/Cas editing (Gao and Zhao 2014; Sun et al. 2015; Tang et al. 2016). As for the dual Pol II promoter system, both Cas nuclease and gRNA(s) are being expressed by Pol II promoters (Fig. 1A). The dual Pol II promoter system overcomes the constitutive gRNA expression disadvantage of the mixed dual promoter system and allows spatiotemporal expression of gRNAs. Pol II promoters enhance the control of CRISPR/Cas genome editing by having a wide diversity of gRNA expression, ranging from constitutive, inducible, to tissue specific. To date, mature gRNAs processed by Pol II-driven expression have been demonstrated in many organisms (Gao et al. 2015; Gao and Zhao 2014; Nissim et al. 2014; Yoshioka et al. 2015). As discussed in the following section, the primary RNA transcript from Pol II expression can be processed by ribozymes, endogenous RNases, or Csy4 (Gao et al. 2015; Gao and Zhao 2014; Mikami et al. 2017; Nissim et al. 2014; Yoshioka et al. 2015).
Fig. 1.
Major expression systems for gRNA and Cas production. A Two-component transcriptional unit (TCTU) system, including the mixed dual promoter system and the dual Pol II promoter system. B Single transcriptional unit (STU) system. C Bidirectional promoter expression system. NLS nuclear localization sequence; HH hammerhead ribozyme; HDV hepatitis delta virus ribozyme; 28 bp Csy4 excision site; T2A the translational viral cleavage sequence required for Csy4 expression; P2A ribosomal skipping sequence
Diverse strategies for expressing multiple gRNAs
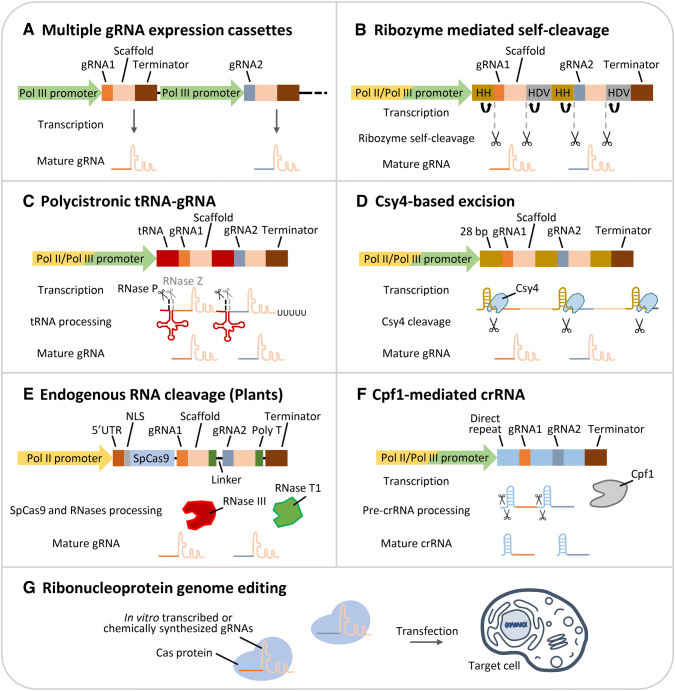
Multiple gRNAs can be produced by individual gRNA expression cassette in one plasmid or multiple plasmids, where in both cases each gRNA is expressed by its own promoter and terminator (Figs. 1A, 2A). Early reports of multiplexing gRNA expression were accomplished by assembling multiple individual gRNA expression cassettes, each transcribed from a separate Pol III promoter (e.g., U3 or U6) (Lowder et al. 2015; Ma et al. 2015; Xing et al. 2014; Zhang et al. 2016; Zhou et al. 2014). A number of cloning strategies, such as golden gate assembly, Gibson assembly, and “stackable array” method, have been used to facilitate the assembly of several individual gRNA expression cassettes (Lowder et al. 2015; Ma et al. 2015; Peterson et al. 2016). However, multiple Pol III promoters may cause undesirable “promoter cross talk effects”, where the transcriptional activity of genes driven by endogenous Pol III is reduced (Hampf and Gossen 2007; Nie et al. 2010; Wang et al. 2006). In plants, using multiple U3 or U6 promoters in gRNA expression cassettes may lead to gRNA expression variation and transgene silencing (Ma et al. 2015). Individual gRNA cassette method is also limited by plasmid cloning efficiency and insert size, especially when using the viral delivery system (Ali et al. 2015; Baltes et al. 2014; Cody et al. 2017). On the other hand, the binary vector used for the Agrobacterium-mediated transformation is less restricted to large transgene size. However, random insertion of multiple gRNA cassettes at a single locus may introduce transgene silencing (Lowder et al. 2016; Vaucheret and Fagard 2001). gRNA expression cassettes can also be delivered through multiple DNA plasmids; however, this method is often associated with poor efficiency and cytotoxicity (Kurata et al. 2018; Mans et al. 2015; Peng et al. 2016; Zhang and Matlashewski 2015). Moreover, each Agrobacterium strain only carries one type of T-DNA plasmid for transformation, making the delivery of multiple plasmids impractical in many organisms (Minkenberg et al. 2017). Therefore, a compact plasmid for multiple gRNAs or crRNAs expression is more favorable than ones with many gRNA expression cassettes.
Fig. 2.
Diverse strategies to generate multiple gRNAs for CRISPR/Cas genome editing. HH hammerhead ribozyme; HDV hepatitis delta virus ribozyme; 28 bp Csy4 excision site; 5′ UTR 5′ untranslated region; NLS nuclear localization sequence
Several different approaches have been reported to process multiple gRNAs post-transcriptionally from a single transcript using RNA-cleaving enzymes (Fig. 2B–F). Since all the gRNAs are produced from a single promoter, each gRNA likely has a similar concentration in the cell. One of the enzymes used for producing multiple gRNAs is the self-cleaving ribozyme naturally presented in living organisms (Fig. 2B). In the original study, artificial Ribozyme-gRNA-Ribozyme gene (RGR) was designed to have gRNAs flanked with hammerhead (HH) ribozyme and hepatitis delta virus (HDV) ribozyme at 5′ and 3′ end, respectively (Gao and Zhao 2014). The ribozyme method was first demonstrated for single gRNA expression in yeast, but has later been adopted for genome editing to produce multiple gRNAs in other organisms (Gao and Zhao 2014; He et al. 2017; Lee et al. 2016; Xu et al. 2017a; Zhang and Matlashewski 2015). It has been reported that multiple gRNAs can be successfully produced by ribozyme method using either Pol II or Pol III promoter in both animals and plants (He et al. 2017; Lee et al. 2016; Xu et al. 2017a; Zhang and Matlashewski 2015). In rice, primary transcript of up to two RGR units can be generated from either rice U6 (OsU6) promoter or rice Actin1 promoter (a Pol II promoter) (He et al. 2017).
The polycistronic tRNA–gRNA (PTG) is another gRNA processing strategy, which uses tRNAs and endogenous RNases for multiple gRNA expression in eukaryotes (Fig. 2C) (Dong et al. 2017; Port and Bullock 2016; Shiraki and Kawakami 2018; Tang et al. 2019; Xie et al. 2015; Zhang et al. 2019). The endogenous RNases recognize and excise the distinct tRNA structure, releasing multiple mature gRNAs from a single transcript (Fig. 2C). In comparison with ribozyme gRNA processing, tRNA processing requires shorter sequence (~ 80 nt tRNA vs. ~ 120 HH + HDV gene), and may provide sequence variability, which reduces gene silencing and instability. The PTG transcript can be expressed by both Pol II and Pol III promoters, and was first reported in rice by achieving indel mutation frequencies up to 100% under a single U3 promoter (Dong et al. 2017; Port and Bullock 2016; Shiraki and Kawakami 2018; Tang et al. 2019; Xie et al. 2015). Because the tRNA sequence contains A and B boxes acting as promoter/enhancer, it increased gRNA expression in rice by one order of magnitude (Xie et al. 2015). This gRNA processing strategy was later demonstrated in various plant, animal and microbial systems with consistently high efficiencies of multiplex genome editing (Dong et al. 2017; Port and Bullock 2016; Shiraki and Kawakami 2018; Tang et al. 2019). In addition to the tRNA–gRNA strategy, tandem repeats of gRNA–shRNA (short hairpin RNA) transcripts were also engineered for multiplex genome editing through excision of shRNAs by endogenous ribonuclease DROSHA in mammalian cells (Yan et al. 2016).
A CRISPR type III ribonuclease, Csy4, has been used to produce multiple gRNAs in various organisms (Fig. 2D) (Čermák et al. 2017; Ferreira et al. 2018; Qin et al. 2015; Tsai et al. 2014). Csy4 is a CRISPR-associated RNA endoribonuclease, which is required for crRNA biogenesis of CRISPR subtype I-F in Pseudomonas aeruginosa (Haurwitz et al. 2010; Tsai et al. 2014). Based on the characteristics of Csy4, an artificial gRNA array was made with Csy4 excision site (28 bp sequences) flanking each gRNA (Fig. 2D) (Haurwitz et al. 2010; Tsai et al. 2014). This system was applied to express multiple gRNAs or truncated gRNAs for dimeric CRISPR RNA-guided FokI nucleases (RFNs), and achieved higher editing efficiencies in mammalian system (Tsai et al. 2014; Wyvekens et al. 2015). Recently, a single Pol II promoter-driven gRNA array, which contains up to 10 gRNAs linked by optimal Csy4 ribonuclease sequences have been assembled by golden gate assembly for multiplex genome engineering in human cells (Kurata et al. 2018). The Csy4-based system using a Pol II promoter, Cestrum Yellow Leaf Curling Virus promoter (CmYLCV), was also demonstrated to be successful for plant genome editing (Čermák et al. 2017). While the Csy4-based excision method can effectively edit genome in animals and plants, a major disadvantage of this system is the requirement of transformation and expression of an exogenous Csy4 endoribonuclease.
Besides the three major gRNA processing strategies, it was reported that functional gRNA units can be processed through RNA cleavage by endogenous RNases without a specific RNA processing system in plants (Fig. 2E) (Mikami et al. 2017). SpCas9 and gRNA expression driven by a single Pol II promoter were able to achieve up to 100% editing efficiency in rice (Mikami et al. 2017). In this system, single or multiple gRNAs (with a 12 bp linker between the gRNAs) could be processed and generated by endogenous RNases (RNase III and RNase T1) (Mikami et al. 2017). This miniaturized SpCas9–gRNA expression system could benefit virus vector-mediated genome editing in plants and potentially other organisms.
CRISPR/Cpf1 is another multiplex editing system which is analogous to CRISPR/Cas9. Cas9 and Cpf1 (also known as Cas12a) are both class II CRISPR nucleases; however, Cpf1 overcomes the limitation of Cas9 for its smaller size and having the ability to process crRNAs without additional nuclease (Fig. 2F). Moreover, Cpf1 recognizes AT-rich cutting sites and produces staggered end cut differently from Cas9, enabling the CRISPR/Cpf1 system to target additional genome sites and has better performance in directional DNA insertion (Minkenberg et al. 2017; Zetsche et al. 2015). Since Cpf1 does not require tracrRNA, another advantage is the shorter gRNA length (crRNA, ~ 42 nt) needed for the CRISPR/Cpf1 system (vs. ~ 100 nt gRNA for Cas9), which helps reduce the size of expression cassette. CRISPR/Cpf1-mediated genome editing has been tested in mammalian cells and plants with different Cpf1 variants, and its multiplexing ability has been achieved by expressing a crRNA array with both Pol II and Pol III promoters (Endo et al. 2016; Tang et al. 2017b; Wang et al. 2018; Xu et al. 2017b; Zetsche et al. 2017; Zhong et al. 2017). Pol II promoter-driven crRNA production was reported to have the same or even higher editing efficiency than Pol III promoters in mammalian cells, presumably due to more efficient export of Pol II transcripts to the cytoplasm, promoting crRNAs and Cpf1 interaction (Zhong et al. 2017).
Besides intracellular transcription approaches, gRNAs can also be delivered into target cells in the form of in vitro transcribed (IVT) RNA or chemically synthesized RNA oligonucleotides (Fig. 2G) (Hendel et al. 2015; Wang et al. 2013). Unlike previously discussed methods, CRISPR/Cas ribonucleoprotein (RNP) complex allows plasmid-free and stoichiometrically controlled genome editing without foreign DNA integration (Hendel et al. 2015; Minkenberg et al. 2017; Wang et al. 2013). In this strategy, both purified Cas9 nuclease and IVT RNA or chemically synthesized sgRNAs are delivered to target cells by direct injections or synthetic nanoparticles (Fig. 2G) (Baek et al. 2016; Chen et al. 2019; Cho et al. 2013; Kim et al. 2014; Li et al. 2019; Wang et al. 2016; Woo et al. 2015). Previously, PEG-mediated transfection has been used to deliver RNP into Arabidopsis and rice protoplasts (Woo et al. 2015). While this method has constraint for plant species with a low protoplast regeneration efficiency, successful genome editing was demonstrated in wheat and maize by co-bombardment with RNPs and additional cell division promoting transcriptional factor genes (Liang et al. 2017; Svitashev et al. 2016). In addition, nanoparticles-based RNP delivery has been shown to facilitate mammalian genome editing and can be potentially applied to plant, overcoming the transformation barrier from cell walls (Cunningham et al. 2018). Most recently, various nanomaterials such as carbon dots (< 10 nm) and carbon nanotubes have been demonstrated as a fast and simple method to deliver plasmids and possibly RNPs into various mature plant cells by spraying, leaf dipping, or leaf infiltration (Demirer et al. 2019; Doyle et al. 2019).
Expression of Cas nuclease and gRNAs from a single transcript
In the single transcriptional unit (STU) system, Cas nuclease and gRNA(s) are simultaneously transcribed from a single promoter, which reduces the size of CRISPR/Cas editing cassette (Fig. 1B). This system was shown to allow successful multiplex genome editing in animals and plants, in which a single RNA Pol II promoter is used to express Cas nuclease, gRNAs and other gRNA processing elements (e.g., ribozyme or tRNA elements) (Ding et al. 2018; Tang et al. 2016, 2019; Xu et al. 2018; Yoshioka et al. 2015). The STU system is first studied in mammalian cells, where HH-gRNA-HDV cassette is linked with Cas9 by an internal ribosome entry site (IRES) (Yoshioka et al. 2015). The design allowed both Cas9 and gRNA to be expressed by a single Pol II promoter, reaching about half of the genome editing efficiency comparing to the dual Pol II promoter system (Yoshioka et al. 2015). Another STU system was produced by coexpressing sgRNA and SpCas9 mRNA separated by ribozyme cleavage sites (a short 15 nt signature) under the control of a single Pol II promoter for either inducible or constitutive genome editing in rice with mutagenesis efficiency up to 100% (Tang et al. 2016). This is the first demonstration of processing transcript containing both Cas9 and sgRNAs by cis-acting HH ribozyme (Tang et al. 2016). A more recent study developed an effective CRISPR–Cpf1 and CRISPR–Cas9 multiplex genome editing system in rice, called simplified STU (SSTU), where Cas nuclease and crRNA are co-expressed from a single Pol II promoter without additional gRNA processing machinery (Wang et al. 2018). Although editing rate is target dependent, SSTU system provides comparable mutagenesis efficiency with conventional TCTU system in general, and it is easier for construction and more efficient for viral vector-based delivery (Baltes et al. 2014; Wang et al. 2018). Moreover, enhanced genome editing efficiency as well as biallelic mutation rates could be achieved by adding poly-A linker and tRNA sequence in the STU CRISPR-LbCpf1 system, suggesting that modifications in RNA processing can greatly affect crRNA–Cas expression (Xu et al. 2018). Another STU study expressed PTG or crRNA arrays from spliced introns of Cas9 or Cpf1 genes, where the hybrid gene was able to increase editing efficiency in rice (Ding et al. 2018). Efficient multiplex genome editing and C to T base editing were also achieved by STU-Cas12a and two STU-Cas9 systems with either Csy4 ribonuclease or tRNA-driven gRNA expression (Tang et al. 2019). Recently, the dual-polymerase active human H1 promoter, with both Pol II and Pol III activity, was used to drive Cas nuclease and gRNAs expression (Gao et al. 2019). With reduced vector size, this kind of STU system has potential values in viral vector-based gene therapy applications (Gao et al. 2019). Furthermore, the Cpf1-based STU system driven by EF1a (a Pol II promoter) was able to successfully express up to 25 crRNAs by addition of a tertiary structural motif to stabilize the transcripts of Acidaminococcus Cas12a and CRISPR arrays in human cells (Campa et al. 2019).
Expression of Cas nuclease and gRNAs using bidirectional promoter systems
In addition to TCTU and STU CRISPR/Cas expression systems, bidirectional promoters (BiPs) have been recently developed to drive Cas9 and gRNA expression in opposite direction in methylotrophic yeast and rice (Liu et al. 2019; Ren et al. 2019). PHTX1, a bidirectional Pol II promoter, was designed to co-regulate the expression of Cas9 and gRNAs flanked by HH and HDV ribozymes with different terminators (Liu et al. 2019). This BiP-driven multiplexed CRISPR/Cas9 system demonstrated successful multiloci integration where multiple gene cassettes simultaneously integrated into Pichia pastoris genome (Fig. 1C) (Liu et al. 2019). Use of the constitutive rice BiP1 (OsBiP1) promoter was shown to achieve 86–93% editing efficiencies with tRNA or Csy4 gRNA processing system in rice (Fig. 1C) (Ren et al. 2019). Although the BiP-based CRISPR/Cas system was only tested in yeast and rice so far, this expression strategy can be further evaluated and modified for multiplex genome editing in various other organisms.
Spatiotemporal and inducible expression of gRNAs
Most CRISPR/Cas-based expression systems use RNA Pol III promoters, such as U3 and U6, to constitutively express gRNAs; however, Pol II promoter-driven gRNA expression is preferable for applications that require strict control over spatiotemporal expression. Gao and Zhao first demonstrated the use of Pol II promoter instead of Pol III promoter for gRNA expression in yeast (Gao and Zhao 2014). The Pol II promoter which transcribed alcohol dehydrogenase 1 (ADH1) was used for targeted DNA cleavage, and the success had brought others to select different promoters suitable for experiments that need precise spatial and temporal gRNA expression (Gao and Zhao 2014). Several Pol II promoters have been used in multiplex genome editing in mammalian cells, such as CAG, CMV, MHCK7, human ubiquitin C, and human histone H2A1 promoters (Nissim et al. 2014; Yoshioka et al. 2015). MHCK7 promoter is a muscle/heart-specific promoter which has been demonstrated to drive gRNA expression in mice muscle, potentially helpful for neuromuscular disorder treatment (Xu et al. 2017a). Plant Pol II promoters, such as maize ubiquitin, CmYLCV, and pathogenesis-related protein gene promoters (PR1 and PR5) have also been demonstrated for gRNA/Cas9 expression (Čermák et al. 2017; Ding et al. 2018; Tang et al. 2016, 2019; Wang et al. 2018). PR1 and PR5 expression are induced under stress conditions and their promoters have been used for expressing multiple gRNAs and crRNAs in Cas9- and Cpf1-mediated genome editing in rice (Ding et al. 2018). Tissue-specific promoters, such as SMB promoter (SOMBRERO/ANAC033, root cap-specific) and TMM promoter (TOO MANY MOUTHS, expressed early in the stomatal cell lineage), have been utilized to develop the CRISPR-TSKO (tissue-specific knockout) system for efficient somatic mutagenesis in particular plant cell types, tissues, and organs (Decaestecker et al. 2019). This system greatly benefits functional characterization of fundamentally important genes without loss of gene function in a system-wide manner. Besides native promoters, synthetic promoters can be made to have enhanced specificity and/or strength comparing to their natural counterparts in plants (Liu and Stewart 2016). Previously, transgenic tobacco and Arabidopsis plants which contain disease-inducible synthetic promoters have been developed to have potential use as phytosensors (Liu et al. 2013). Such promoter could also be applied for producing gRNA which will only be expressed in response to defense signal molecules or pathogen infection. Inducible Cas9 gRNA expression can also be accomplished using Pol II with decoupled human tRNA promoter, where the engineered tRNA variants allow sufficient gRNA processing with no detectable promoter activity in mammalian cells (Knapp et al. 2019). Together, the use of tissue-specific and inducible promoters for gRNA expression can greatly increase the flexibility for the CRISPR/Cas-based multiplex genome editing.
Engineering gRNAs to improve stability as well as editing specificity and efficiency
Besides gRNA expression optimization, gRNA can be engineered and synthesized to improve both the specificity and the efficiency of genome editing. Synthesized gRNAs with high purity and structural modifications can form RNP complex with Cas nuclease, enabling more efficient editing. Such alterations include spacer length changes, sequence modifications, RNA–DNA hybrid gRNA, covalent chemical modifications, and independent components (RNA or DNA) incorporations (Moon et al. 2019). Changes in spacer length alter gRNA targeting specificity and can either promote orthogonal or base editing applications (Dahlman et al. 2015; Kiani et al. 2015; Ryu et al. 2018). Modified gRNA sequence was shown to improve specificity through efficient T7 in vitro transcription with additional guanidines at the 5′ end of the spacer (Cho et al. 2014). In addition, gRNA stability increases with structure mimicking mRNA (adding 5′ cap and 3′ poly-A tail), optimizing CRISPR/Cas genome editing efficiency in human cells (Mu et al. 2019). Partial replacement of RNA with DNA in both spacer and scaffold was also shown to increase gRNA stability in human cells (Yin et al. 2018). Moreover, improved editing efficiency in human cells was reported by co-delivery of Cas9 mRNA or protein with chemically modified gRNAs comprising 2′-O-methyl 3′phosphorothioate (MS) or 2′-O-methyl 3′thioPACE (MSP) functional groups at terminal nucleotides (Hendel et al. 2015). Besides chemical modification with functional groups, RNA and DNA can be incorporated with gRNA to recruit transcription-related proteins, serve as enzyme or enzyme substrate, or act as donor for HDR-mediated repair (Jing et al. 2018; Lee et al. 2017; Lowder et al. 2018; Moon et al. 2019; Shechner et al. 2015; Tang et al. 2017a; Zalatan et al. 2015). For example, engineered gRNA scaffolds with bacteriophage coat protein MS2-binding RNA aptamers were shown to successfully recruit transcriptional activators and achieve multiplexed CRISPR/Cas9-based transcriptional activation in Arabidopsis and rice (Lowder et al. 2018).
Conclusion and perspectives
Efficient expression of multiple gRNAs plays a key role in achieving optimal and/or multiplex genome editing. Diverse CRISPR/Cas expression cassette configurations, transcript processing processes, and delivery strategies are available nowadays to optimize gRNA expression. Typically, expression of Cas nuclease and gRNAs has been achieved using TCTU systems. With the need to perform coordinated expression and minimize vector size, a more compact STU system encoding both Cas nuclease and gRNAs in a single transcript has been demonstrated to be effective for genome editing in various organisms. It should be noted that the expression of gRNA and Cas nuclease by separate Pol II promoters in the TCTU system provides more flexibility towards spatiotemporal induction of genome editing comparing to the STU system. Moreover, gRNAs can be expressed by individual gRNA expression cassette through TCTU approach, which favors application where different gRNAs are to be expressed in different tissues or different time periods. Other important factors for selecting strategies for multiple gRNA expression include the number of gRNAs to be expressed, the cloning efficiency, and the characteristics of target organisms. Ribozyme-based methods have disadvantage in applications with increased number of gRNAs, since the production of gRNA decreases with multiple gRNAs (Xu et al. 2017a). In addition, tRNAs are relatively shorter and more variable than ribozymes, which reduces vector size and avoids gene silencing caused by repeated gRNA arrays (Shiraki and Kawakami 2018). When conducting the zebrafish CRISPR/Cas genome editing, the endogenous tRNA-based gRNA processing also showed advantages over Csy4-based gRNA processing due to Csy4 toxicity to zebrafish (Qin et al. 2015).
Tissue-specific mutagenesis can be accomplished by CRISPR/Cas system, where Cas nuclease or gRNAs are expressed by a tissue-specific promoter. Pol III promoters usually allows constitutive expression of gRNAs, whereas Pol II promoters enable constitutive, spatiotemporal and inducible gRNA expression. When selecting reliable gRNA expression promoter for individual organisms, it should be considered that the efficiency of the promoter may vary even among closely related species. For example, the performance of gRNA promoters, especially U6 promoters, are not consistent among Aspergilli fungi. AoU6 (U6 promoter of A. oryzae) and AfU3 (U3 promoter of A. fumigatus) promoter can successfully produce gRNA in A. oryzae and A. nidulans, respectively, while AnU6 (U6 promoter A. nidulans) and AfU6 (U6 promoter of A. fumigatus) promoters failed to drive gRNA expression in A. nidulans (Katayama et al. 2016; Nødvig et al. 2018; Song et al. 2018). As for A. niger, endogenous tRNA promoter and 5S rRNA gene (both Pol III promoter) have been demonstrated to drive gRNA expression (Song et al. 2018; Zheng et al. 2018). In addition, it has been reported that the gene editing efficiencies of tRNA promoters vary between different yeast strains; on the other hand, 5S rRNA gene is highly conserved and abundant in cells, indicating its broader application for gRNA expression in eukaryotes (Ryan et al. 2014; Zheng et al. 2018). In Drosophila, CRISPR/Cas technology has been combined with Gal4/UAS (upstream activating system) system for tissue-specific loss-of-function analysis, where Cas nuclease is tissue specifically expressed by Gal4, while gRNAs are ubiquitously expressed or UAS-driven (Port et al. 2014; Port and Bullock 2016; Xue et al. 2014). A more recent study achieved efficient tissue-specific gene knockouts with Cas9 driven by a tissue-specific enhancer, and ubiquitously expressed multi-gRNAs driven by U6 promoter (Poe et al. 2019). This method is more effective in loss-of-function analysis and in reducing the potential cell toxicity caused by excessive Cas nuclease derived from Gal4/UAS system in Drosophila neurons (Jiang et al. 2014; Poe et al. 2019). It is possible that the tissue-specific enhancer can also be used for gRNA expression, further expanding the utility of CRISPR/Cas toolkits.
Promoters can also be modified or synthesized to achieve efficient gRNA expression in both model species and diverse organisms that do not have well-characterized Pol III promoters. Synthetic hybrid RNA Pol III promoters which combined native Pol III promoters with tRNA were used for producing gRNAs to disrupt and integrate gene in oleaginous yeast, Yarrowia lipolytica (Schwartz et al. 2016). The three different Pol III–tRNA hybrid promoters tested all performed higher gene disruption rates compared to native SNR52 Pol III promoter (Schwartz et al. 2016). Plant synthetic promoters with strengthened spatiotemporal and inducible regulations can also be used for gRNA expression (Liu and Stewart 2016). While most of these spatiotemporal and inducible promoters were not originally developed for improving CRISPR/Cas genome editing, it is worthwhile to test their efficiency for gRNA expression (Liu and Stewart 2016). A more compact vector size can also be achieved by synthetic and minimal promoters, which should greatly facilitate the viral-based delivery methods (Baltes et al. 2014). Together, appropriate selection of promoters and adequate arrangement of Cas and gRNA expression units should facilitate the optimal expression of single and multiple gRNAs which is essential for efficient genome editing in various organisms.
Acknowledgements
This work was supported by NSF Plant Genome Research Project Grant (1740874) and the USDA National Institute of Food and Agriculture and Hatch Appropriations under Project PEN04659 and Accession #1016432.
Compliance with ethical standards
Conflict of interest
On behalf of all authors, the corresponding author states that there is no conflict of interest.
References
- Ali Z, Abul-faraj A, Li L, Ghosh N, Piatek M, Mahjoub A, Aouida M, Piatek A, Baltes NJ, Voytas DF, Dinesh-Kumar S, Mahfouz MM. Efficient virus-mediated genome editing in plants using the CRISPR/Cas9 system. Mol Plant. 2015;8:1288–1291. doi: 10.1016/j.molp.2015.02.011. [DOI] [PubMed] [Google Scholar]
- Baek K, Kim DH, Jeong J, Sim SJ, Melis A, Kim J-S, Jin E, Bae S. DNA-free two-gene knockout in Chlamydomonas reinhardtii via CRISPR-Cas9 ribonucleoproteins. Sci Rep. 2016;6:30620. doi: 10.1038/srep30620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baltes NJ, Gil-Humanes J, Cermak T, Atkins PA, Voytas DF. DNA replicons for plant genome engineering. Plant Cell. 2014;26:151–163. doi: 10.1105/tpc.113.119792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrangou R, Fremaux C, Deveau H, Richards M, Boyaval P, Moineau S, Romero DA, Horvath P. CRISPR provides acquired resistance against viruses in prokaryotes. Science. 2007;315:1709–1712. doi: 10.1126/science.1138140. [DOI] [PubMed] [Google Scholar]
- Bortesi L, Fischer R. The CRISPR/Cas9 system for plant genome editing and beyond. Biotechnol Adv. 2015;33:41–52. doi: 10.1016/j.biotechadv.2014.12.006. [DOI] [PubMed] [Google Scholar]
- Campa CC, Weisbach NR, Santinha AJ, Incarnato D, Platt RJ. Multiplexed genome engineering by Cas12a and CRISPR arrays encoded on single transcripts. Nat Methods. 2019;16:887–893. doi: 10.1038/s41592-019-0508-6. [DOI] [PubMed] [Google Scholar]
- Čermák T, Curtin SJ, Gil-Humanes J, Čegan R, Kono TJY, Konečná E, Belanto JJ, Starker CG, Mathre JW, Greenstein RL, Voytas DF. A multipurpose toolkit to enable advanced genome engineering in plants. Plant Cell. 2017;29:1196–1217. doi: 10.1105/tpc.16.00922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen G, Abdeen AA, Wang Y, Shahi PK, Robertson S, Xie R, Suzuki M, Pattnaik BR, Saha K, Gong S. A biodegradable nanocapsule delivers a Cas9 ribonucleoprotein complex for in vivo genome editing. Nat Nanotechnol. 2019;14:974–980. doi: 10.1038/s41565-019-0539-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho SW, Lee J, Carroll D, Kim J-S, Lee J. Heritable gene knockout in Caenorhabditis elegans by direct injection of Cas9-sgRNA ribonucleoproteins. Genetics. 2013;195:1177–1180. doi: 10.1534/genetics.113.155853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho SW, Kim S, Kim Y, Kweon J, Kim HS, Bae S, Kim J-S. Analysis of off-target effects of CRISPR/Cas-derived RNA-guided endonucleases and nickases. Genome Res. 2014;24:132–141. doi: 10.1101/gr.162339.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cody WB, Scholthof HB, Mirkov TE. Multiplexed gene editing and protein overexpression using a tobacco mosaic virus viral vector. Plant Physiol. 2017;175:23–35. doi: 10.1104/pp.17.00411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cong L, Ran FA, Cox D, Lin S, Barretto R, Habib N, Hsu PD, Wu X, Jiang W, Marraffini LA, Zhang F. Multiplex genome engineering using CRISPR/Cas systems. Science. 2013;339:819–823. doi: 10.1126/science.1231143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cunningham FJ, Goh NS, Demirer GS, Matos JL, Landry MP. Nanoparticle-mediated delivery towards advancing plant genetic engineering. Trends Biotechnol. 2018;36:882–897. doi: 10.1016/j.tibtech.2018.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dahlman JE, Abudayyeh OO, Joung J, Gootenberg JS, Zhang F, Konermann S. Orthogonal gene knockout and activation with a catalytically active Cas9 nuclease. Nat Biotechnol. 2015;33:1159–1161. doi: 10.1038/nbt.3390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Decaestecker W, Andrade Buono R, Pfeiffer M, Vangheluwe N, Jourquin J, Karimi M, van Isterdael G, Beeckman T, Nowack MK, Jacobs TB. CRISPR-TSKO: a technique for efficient mutagenesis in specific cell types, tissues, or organs in Arabidopsis. Plant Cell. 2019 doi: 10.1105/tpc.19.00454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demirer GS, Zhang H, Matos JL, Goh NS, Cunningham FJ, Sung Y, Chang R, Aditham AJ, Chio L, Cho M-J, Staskawicz B, Landry MP. High aspect ratio nanomaterials enable delivery of functional genetic material without DNA integration in mature plants. Nat Nanotechnol. 2019;14:456–464. doi: 10.1038/s41565-019-0382-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding D, Chen K, Chen Y, Li H, Xie K. Engineering introns to express RNA guides for Cas9- and Cpf1-mediated multiplex genome editing. Mol Plant. 2018;11:542–552. doi: 10.1016/j.molp.2018.02.005. [DOI] [PubMed] [Google Scholar]
- Dong F, Xie K, Chen Y, Yang Y, Mao Y. Polycistronic tRNA and CRISPR guide-RNA enables highly efficient multiplexed genome engineering in human cells. Biochem Biophys Res Commun. 2017;482:889–895. doi: 10.1016/j.bbrc.2016.11.129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doyle C, Higginbottom K, Swift TA, Winfield M, Bellas C, Benito-Alifonso D, Fletcher T, Galan MC, Edwards K, Whitney HM. A simple method for spray-on gene editing in planta. bioRxiv. 2019 doi: 10.1101/805036. [DOI] [Google Scholar]
- Endo A, Masafumi M, Kaya H, Toki S. Efficient targeted mutagenesis of rice and tobacco genomes using Cpf1 from Francisella novicida. Sci Rep. 2016;6:38169. doi: 10.1038/srep38169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferreira R, Skrekas C, Nielsen J, David F. Multiplexed CRISPR/Cas9 genome editing and gene regulation using Csy4 in Saccharomyces cerevisiae. ACS Synth Biol. 2018;7:10–15. doi: 10.1021/acssynbio.7b00259. [DOI] [PubMed] [Google Scholar]
- Gaj T, Gersbach CA, Barbas CF. ZFN, TALEN, and CRISPR/Cas-based methods for genome engineering. Trends Biotechnol. 2013;31:397–405. doi: 10.1016/j.tibtech.2013.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao Y, Zhao Y. Self-processing of ribozyme-flanked RNAs into guide RNAs in vitro and in vivo for CRISPR-mediated genome editing. J Integr Plant Biol. 2014;56:343–349. doi: 10.1111/jipb.12152. [DOI] [PubMed] [Google Scholar]
- Gao Y, Zhang Y, Zhang D, Dai X, Estelle M, Zhao Y. Auxin binding protein 1 (ABP1) is not required for either auxin signaling or Arabidopsis development. Proc Natl Acad Sci USA. 2015;112:2275–2280. doi: 10.1073/pnas.1500365112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao Z, Herrera-Carrillo E, Berkhout B. A single H1 promoter can drive both guide RNA and endonuclease expression in the CRISPR-Cas9 system. Mol Ther Nucleic Acids. 2019;14:32–40. doi: 10.1016/j.omtn.2018.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hampf M, Gossen M. Promoter crosstalk effects on gene expression. J Mol Biol. 2007;365:911–920. doi: 10.1016/j.jmb.2006.10.009. [DOI] [PubMed] [Google Scholar]
- Haurwitz RE, Jinek M, Wiedenheft B, Zhou K, Doudna JA. Sequence- and structure-specific RNA processing by a CRISPR endonuclease. Science. 2010;329:1355–1358. doi: 10.1126/science.1192272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He Y, Zhang T, Yang N, Xu M, Yan L, Wang L, Wang R, Zhao Y. Self-cleaving ribozymes enable the production of guide RNAs from unlimited choices of promoters for CRISPR/Cas9 mediated genome editing. J Genet Genomics. 2017;44:469–472. doi: 10.1016/j.jgg.2017.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hendel A, Bak RO, Clark JT, Kennedy AB, Ryan DE, Roy S, Steinfeld I, Lunstad BD, Kaiser RJ, Wilkens AB, Bacchetta R, Tsalenko A, Dellinger D, Bruhn L, Porteus MH. Chemically modified guide RNAs enhance CRISPR-Cas genome editing in human primary cells. Nat Biotechnol. 2015;33:985–989. doi: 10.1038/nbt.3290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu PD, Scott DA, Weinstein JA, Ran FA, Konermann S, Agarwala V, Li Y, Fine EJ, Wu X, Shalem O, Cradick TJ, Marraffini LA, Bao G, Zhang F. DNA targeting specificity of RNA-guided Cas9 nucleases. Nat Biotechnol. 2013;31:827. doi: 10.1038/nbt.2647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang W, Zhou H, Bi H, Fromm M, Yang B, Weeks DP. Demonstration of CRISPR/Cas9/sgRNA-mediated targeted gene modification in Arabidopsis, tobacco, sorghum and rice. Nucleic Acids Res. 2013;41:e188–e188. doi: 10.1093/nar/gkt780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang W, Brueggeman AJ, Horken KM, Plucinak TM, Weeks DP. Successful transient expression of Cas9 and single guide RNA genes in Chlamydomonas reinhardtii. Eukaryot Cell. 2014;13:1465–1469. doi: 10.1128/EC.00213-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jing X, Xie B, Chen L, Zhang N, Jiang Y, Qin H, Wang H, Hao P, Yang S, Li X. Implementation of the CRISPR-Cas13a system in fission yeast and its repurposing for precise RNA editing. Nucleic Acids Res. 2018;46:e90–e90. doi: 10.1093/nar/gky433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katayama T, Tanaka Y, Okabe T, Nakamura H, Fujii W, Kitamoto K, Maruyama J. Development of a genome editing technique using the CRISPR/Cas9 system in the industrial filamentous fungus Aspergillus oryzae. Biotechnol Lett. 2016;38:637–642. doi: 10.1007/s10529-015-2015-x. [DOI] [PubMed] [Google Scholar]
- Kiani S, Chavez A, Tuttle M, Hall RN, Chari R, Ter-Ovanesyan D, Qian J, Pruitt BW, Beal J, Vora S, Buchthal J, Kowal EJK, Ebrahimkhani MR, Collins JJ, Weiss R, Church G. Cas9 gRNA engineering for genome editing, activation and repression. Nat Methods. 2015;12:1051–1054. doi: 10.1038/nmeth.3580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S, Kim D, Cho SW, Kim J, Kim J-S. Highly efficient RNA-guided genome editing in human cells via delivery of purified Cas9 ribonucleoproteins. Genome Res. 2014;24:1012–1019. doi: 10.1101/gr.171322.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knapp DJHF, Michaels YS, Jamilly M, Ferry QRV, Barbosa H, Milne TA, Fulga TA. Decoupling tRNA promoter and processing activities enables specific Pol-II Cas9 guide RNA expression. Nat Commun. 2019;10:1490. doi: 10.1038/s41467-019-09148-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurata M, Wolf NK, Lahr WS, Weg MT, Kluesner MG, Lee S, Hui K, Shiraiwa M, Webber BR, Moriarity BS. Highly multiplexed genome engineering using CRISPR/Cas9 gRNA arrays. PLoS ONE. 2018;13:e0198714–e0198714. doi: 10.1371/journal.pone.0198714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee RTH, Ng ASM, Ingham PW. Ribozyme mediated gRNA generation for in vitro and in vivo CRISPR/Cas9 mutagenesis. PLoS ONE. 2016;11:e0166020–e0166020. doi: 10.1371/journal.pone.0166020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee K, Mackley VA, Rao A, Chong AT, Dewitt MA, Corn JE, Murthy N. Synthetically modified guide RNA and donor DNA are a versatile platform for CRISPR-Cas9 engineering. Elife. 2017;6:e25312. doi: 10.7554/eLife.25312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Aach J, Norville JE, Mccormack M, Bush J, Church GM, Sheen J. Multiplex and homologous recombination-mediated plant genome editing via guide RNA/Cas9. Nat Biotechnol. 2013;31:688–691. doi: 10.1038/nbt.2654.Multiplex. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Bolinger J, Yu Y, Glass Z, Shi N, Yang L, Wang M, Xu Q. Intracellular delivery and biodistribution study of CRISPR/Cas9 ribonucleoprotein loaded bioreducible lipidoid nanoparticles. Biomater Sci. 2019;7:596–606. doi: 10.1039/C8BM00637G. [DOI] [PubMed] [Google Scholar]
- Liang Z, Chen K, Li T, Zhang Y, Wang Y, Zhao Q, Liu J, Zhang H, Liu C, Ran Y, Gao C. Efficient DNA-free genome editing of bread wheat using CRISPR/Cas9 ribonucleoprotein complexes. Nat Commun. 2017;8:14261. doi: 10.1038/ncomms14261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu W, Stewart CN. Plant synthetic promoters and transcription factors. Curr Opin Biotechnol. 2016;37:36–44. doi: 10.1016/j.copbio.2015.10.001. [DOI] [PubMed] [Google Scholar]
- Liu W, Mazarei M, Rudis MR, Fethe MH, Peng Y, Millwood RJ, Schoene G, Burris JN, Stewart CN., Jr Bacterial pathogen phytosensing in transgenic tobacco and Arabidopsis plants. Plant Biotechnol J. 2013;11:43–52. doi: 10.1111/pbi.12005. [DOI] [PubMed] [Google Scholar]
- Liu Q, Shi X, Song L, Liu H, Zhou X, Wang Q, Zhang Y, Cai M. CRISPR–Cas9-mediated genomic multiloci integration in Pichia pastoris. Microb Cell Fact. 2019;18:144. doi: 10.1186/s12934-019-1194-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowder L, Zhang D, Baltes NJ, Paul JW, Tang X, Zheng X, Voytas DF, Hsieh T-F, Zhang Y, Qi Y. A CRISPR/Cas9 toolbox for multiplexed plant genome editing and transcriptional regulation. Plant Physiol. 2015;169:971–985. doi: 10.1104/pp.15.00636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowder L, Malzahn A, Qi Y. Rapid evolution of manifold CRISPR systems for plant genome editing. Front Plant Sci. 2016;7:1683. doi: 10.3389/fpls.2016.01683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowder LG, Zhou J, Zhang Y, Malzahn A, Zhong Z, Hsieh T, Voytas DF, Zhang Y, Qi Y. Robust transcriptional activation in plants using systems. Mol Plant. 2018;11:245–256. doi: 10.1016/j.molp.2017.11.010. [DOI] [PubMed] [Google Scholar]
- Ma X, Zhang Q, Zhu Q, Liu W, Chen Y, Qiu R, Wang B, Yang Z, Li H, Lin Y, Xie Y, Shen R, Chen S, Wang Z, Chen Y, Guo J, Chen L, Zhao X, Dong Z, Liu Y-G. A robust CRISPR/Cas9 system for convenient, high-efficiency multiplex genome editing in monocot and dicot plants. Mol Plant. 2015;8:1274–1284. doi: 10.1016/j.molp.2015.04.007. [DOI] [PubMed] [Google Scholar]
- Mans R, van Rossum HM, Wijsman M, Backx A, Kuijpers NGA, van den Broek M, Daran-Lapujade P, Pronk JT, van Maris AJA, Daran J-MG. CRISPR/Cas9: a molecular Swiss army knife for simultaneous introduction of multiple genetic modifications in Saccharomyces cerevisiae. FEMS Yeast Res. 2015 doi: 10.1093/femsyr/fov004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mikami M, Toki S, Endo M. In planta processing of the SpCas9-gRNA complex. Plant Cell Physiol. 2017;58:1857–1867. doi: 10.1093/pcp/pcx154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minkenberg B, Wheatley M, Yang Y. CRISPR/Cas9-enabled multiplex genome editing and its application. In: Weeks DP, Yang B, editors. Gene editing in plants. UK: Academic Press; 2017. pp. 111–132. [DOI] [PubMed] [Google Scholar]
- Moon SB, Kim DY, Ko J-H, Kim J-S, Kim Y-S. Improving CRISPR genome editing by engineering guide RNAs. Trends Biotechnol. 2019;37:870–881. doi: 10.1016/j.tibtech.2019.01.009. [DOI] [PubMed] [Google Scholar]
- Mu W, Zhang Y, Xue X, Liu L, Wei X, Wang H. 5′ capped and 3′ polyA-tailed sgRNAs enhance the efficiency of CRISPR-Cas9 system. Protein Cell. 2019;10:223–228. doi: 10.1007/s13238-018-0552-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nekrasov V, Staskawicz B, Weigel D, Jones JDG, Kamoun S. Targeted mutagenesis in the model plant Nicotiana benthamiana using Cas9 RNA-guided endonuclease. Nat Biotechnol. 2013;31:691. doi: 10.1038/nbt.2655. [DOI] [PubMed] [Google Scholar]
- Nie L, Das TM, Wang Y, Su Q, Zhao Y, Feng Y. Regulation of U6 promoter activity by transcriptional interference in viral vector-based RNAi. Genomics Proteomics Bioinformatics. 2010;8:170–179. doi: 10.1016/S1672-0229(10)60019-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nissim L, Perli SD, Fridkin A, Perez-Pinera P, Lu TK. Multiplexed and programmable regulation of gene networks with an integrated RNA and CRISPR/Cas toolkit in human cells. Mol Cell. 2014;54:698–710. doi: 10.1016/j.molcel.2014.04.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nødvig CS, Hoof JB, Kogle ME, Jarczynska ZD, Lehmbeck J, Klitgaard DK, Mortensen UH. Efficient oligo nucleotide mediated CRISPR-Cas9 gene editing in Aspergilli. Fungal Genet Biol. 2018;115:78–89. doi: 10.1016/j.fgb.2018.01.004. [DOI] [PubMed] [Google Scholar]
- Peng R, Lin G, Li J. Potential pitfalls of CRISPR/Cas9-mediated genome editing. FEBS J. 2016;283:1218–1231. doi: 10.1111/febs.13586. [DOI] [PubMed] [Google Scholar]
- Peterson BA, Haak DC, Nishimura MT, Teixeira PJPL, James SR, Dangl JL, Nimchuk ZL. Genome-wide assessment of efficiency and specificity in CRISPR/Cas9 mediated multiple site targeting in arabidopsis. PLoS ONE. 2016;11:e0162169–e0162169. doi: 10.1371/journal.pone.0162169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poe AR, Wang B, Sapar ML, Ji H, Li K, Onabajo T, Fazliyeva R, Gibbs M, Qiu Y, Hu Y, Han C. Robust CRISPR/Cas9-mediated tissue-specific mutagenesis reveals gene redundancy and perdurance in Drosophila. Genetics. 2019;211:459–472. doi: 10.1534/genetics.118.301736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Port F, Bullock SL. Augmenting CRISPR applications in Drosophila with tRNA-flanked sgRNAs. Nat Methods. 2016;13:852–854. doi: 10.1038/nmeth.3972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Port F, Chen H-M, Lee T, Bullock SL. Optimized CRISPR/Cas tools for efficient germline and somatic genome engineering in Drosophila. Proc Natl Acad Sci USA. 2014;111:E2967–E2976. doi: 10.1073/pnas.1405500111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin W, Liang F, Feng Y, Bai H, Yan R, Li S, Lin S. Expansion of CRISPR/Cas9 genome targeting sites in zebrafish by Csy4-based RNA processing. Cell Res. 2015;25:1074–1077. doi: 10.1038/cr.2015.95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren Q, Zhong Z, Wang Y, You Q, Li Q, Yuan M, He Y, Qi C, Tang X, Zheng X, Zhang T, Qi Y, Zhang Y. Bidirectional promoter-based CRISPR-Cas9 systems for plant genome editing. Front Plant Sci. 2019;10:1173. doi: 10.3389/fpls.2019.01173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryan OW, Skerker JM, Maurer MJ, Li X, Tsai JC, Poddar S, Lee ME, DeLoache W, Dueber JE, Arkin AP, Cate JHD. Selection of chromosomal DNA libraries using a multiplex CRISPR system. Elife. 2014;3:e03703. doi: 10.7554/eLife.03703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryu S-M, Koo T, Kim K, Lim K, Baek G, Kim S-T, Kim HS, Kim D, Lee H, Chung E, Kim J-S. Adenine base editing in mouse embryos and an adult mouse model of Duchenne muscular dystrophy. Nat Biotechnol. 2018;36:536. doi: 10.1038/nbt.4148. [DOI] [PubMed] [Google Scholar]
- Schwartz CM, Hussain MS, Blenner M, Wheeldon I. Synthetic RNA polymerase III promoters facilitate high-efficiency CRISPR−Cas9 mediated genome editing in Yarrowia lipolytica. ACS Synth Biol. 2016;5:356–359. doi: 10.1021/acssynbio.5b00162. [DOI] [PubMed] [Google Scholar]
- Shan Q, Wang Y, Li J, Zhang Y, Chen K, Liang Z, Zhang K, Liu J, Xi JJ, Qiu J-L, Gao C. Targeted genome modification of crop plants using a CRISPR-Cas system. Nat Biotechnol. 2013;31:686. doi: 10.1038/nbt.2650. [DOI] [PubMed] [Google Scholar]
- Shechner DM, Hacisuleyman E, Younger ST, Rinn JL. Multiplexable, locus-specific targeting of long RNAs with CRISPR-Display. Nat Methods. 2015;12:664–670. doi: 10.1038/nmeth.3433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiraki T, Kawakami K. A tRNA-based multiplex sgRNA expression system in zebrafish and its application to generation of transgenic albino fish. Sci Rep. 2018;8:13366. doi: 10.1038/s41598-018-31476-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song L, Ouedraogo J-P, Kolbusz M, Nguyen TTM, Tsang A. Efficient genome editing using tRNA promoter-driven CRISPR/Cas9 gRNA in Aspergillus niger. PLoS ONE. 2018;13:e0202868–e0202868. doi: 10.1371/journal.pone.0202868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun X, Hu Z, Chen R, Jiang Q, Song G, Zhang H, Xi Y. Targeted mutagenesis in soybean using the CRISPR-Cas9 system. Sci Rep. 2015;5:10342. doi: 10.1038/srep10342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Svitashev S, Schwartz C, Lenderts B, Young JK, Mark Cigan A. Genome editing in maize directed by CRISPR-Cas9 ribonucleoprotein complexes. Nat Commun. 2016;7:13274. doi: 10.1038/ncomms13274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang X, Zheng X, Qi Y, Zhang D, Cheng Y, Tang A, Voytas DF, Zhang Y. A single transcript CRISPR-Cas9 system for efficient genome editing in plants. Mol Plant. 2016;9:1088–1091. doi: 10.1016/j.molp.2016.05.001. [DOI] [PubMed] [Google Scholar]
- Tang W, Hu JH, Liu DR. Aptazyme-embedded guide RNAs enable ligand-responsive genome editing and transcriptional activation. Nat Commun. 2017;8:15939. doi: 10.1038/ncomms15939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang X, Lowder LG, Zhang T, Malzahn AA, Zheng X, Voytas DF, Zhong Z, Chen Y, Ren Q, Li Q, Kirkland ER, Zhang Y, Qi Y. A CRISPR—Cpf1 system for efficient genome editing and transcriptional repression in plants. Nat Plants. 2017;3:1–5. doi: 10.1038/nplants.2017.18. [DOI] [PubMed] [Google Scholar]
- Tang X, Ren Q, Yang L, Bao Y, Zhong Z, He Y, Liu S, Qi C, Liu B, Wang Y, Sretenovic S, Zhang Y, Zheng X, Zhang T, Qi Y, Zhang Y. Single transcript unit CRISPR 2.0 systems for robust Cas9 and Cas12a mediated plant genome editing. Plant Biotechnol J. 2019;17:1431–1445. doi: 10.1111/pbi.13068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai SQ, Wyvekens N, Khayter C, Foden JA, Thapar V, Reyon D, Goodwin MJ, Aryee MJ, Joung JK. Dimeric CRISPR RNA-guided FokI nucleases for highly specific genome editing. Nat Biotechnol. 2014;32:569–576. doi: 10.1038/nbt.2908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaucheret H, Fagard M. Transcriptional gene silencing in plants: targets, inducers and regulators. Trends Genet. 2001;17:29–35. doi: 10.1016/S0168-9525(00)02166-1. [DOI] [PubMed] [Google Scholar]
- Wang S, Shi Z, Liu W, Jules J, Feng X. Development and validation of vectors containing multiple siRNA expression cassettes for maximizing the efficiency of gene silencing. BMC Biotechnol. 2006;6:50. doi: 10.1186/1472-6750-6-50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, Yang H, Shivalila CS, Dawlaty MM, Cheng AW, Zhang F, Jaenisch R. One-step generation of mice carrying mutations in multiple genes by CRISPR/Cas-mediated genome engineering. Cell. 2013;153:910–918. doi: 10.1016/j.cell.2013.04.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang M, Zuris JA, Meng F, Rees H, Sun S, Deng P, Han Y, Gao X, Pouli D, Wu Q, Georgakoudi I, Liu DR, Xu Q. Efficient delivery of genome-editing proteins using bioreducible lipid nanoparticles. Proc Natl Acad Sci. 2016;113:2868–2873. doi: 10.1073/pnas.1520244113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang M, Mao Y, Lu Y, Wang Z, Tao X, Zhu J-K. Multiplex gene editing in rice with simplified CRISPR-Cpf1 and CRISPR-Cas9 systems. J Integr Plant Biol. 2018;60:626–631. doi: 10.1111/jipb.12667. [DOI] [PubMed] [Google Scholar]
- Woo JW, Kim J, Il KS, Corvalán C, Cho SW, Kim H, Kim S-G, Kim S-T, Choe S, Kim J-S. DNA-free genome editing in plants with preassembled CRISPR-Cas9 ribonucleoproteins. Nat Biotechnol. 2015;33:1162–1164. doi: 10.1038/nbt.3389. [DOI] [PubMed] [Google Scholar]
- Wyman C, Kanaar R. DNA double-strand break repair: all’s well that ends well. Annu Rev Genet. 2006;40:363–383. doi: 10.1146/annurev.genet.40.110405.090451. [DOI] [PubMed] [Google Scholar]
- Wyvekens N, Topkar VV, Khayter C, Joung JK, Tsai SQ. Dimeric CRISPR RNA-guided FokI-dCas9 nucleases directed by truncated gRNAs for highly specific genome editing. Hum Gene Ther. 2015;26:425–431. doi: 10.1089/hum.2015.084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie K, Minkenberg B, Yang Y. Boosting CRISPR/Cas9 multiplex editing capability with the endogenous tRNA-processing system. Proc Natl Acad Sci USA. 2015;112:3570–3575. doi: 10.1073/pnas.1420294112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xing H-L, Dong L, Wang Z-P, Zhang H-Y, Han C-Y, Liu B, Wang X-C, Chen Q-J. A CRISPR/Cas9 toolkit for multiplex genome editing in plants. BMC Plant Biol. 2014;14:327. doi: 10.1186/s12870-014-0327-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu L, Zhao L, Gao Y, Xu J, Han R. Empower multiplex cell and tissue-specific CRISPR-mediated gene manipulation with self-cleaving ribozymes and tRNA. Nucleic Acids Res. 2017;45:e28–e28. doi: 10.1093/nar/gkw1048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu R, Qin R, Li H, Li D, Li L, Wei P, Yang J. Generation of targeted mutant rice using a CRISPR-Cpf1 system. Plant Biotechnol J. 2017;15:713–717. doi: 10.1111/pbi.12669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu R, Qin R, Li H, Li J, Yang J, Wei P. Enhanced genome editing in rice using single transcript unit CRISPR- Lb Cpf1 systems. Plant Biotechnol J. 2018 doi: 10.1111/pbi.13028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue Z, Wu M, Wen K, Ren M, Long L, Zhang X, Gao G. CRISPR/Cas9 mediates efficient conditional mutagenesis in Drosophila. G3 (Bethesda) 2014;4:2167–2173. doi: 10.1534/g3.114.014159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan Q, Xu K, Xing J, Zhang T, Wang X, Wei Z, Ren C, Liu Z, Shao S, Zhang Z. Multiplex CRISPR/Cas9-based genome engineering enhanced by Drosha-mediated sgRNA-shRNA structure. Sci Rep. 2016;6:38970. doi: 10.1038/srep38970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin H, Song C-Q, Suresh S, Kwan S-Y, Wu Q, Walsh S, Ding J, Bogorad RL, Zhu LJ, Wolfe SA, Koteliansky V, Xue W, Langer R, Anderson DG. Partial DNA-guided Cas9 enables genome editing with reduced off-target activity. Nat Chem Biol. 2018;14:311. doi: 10.1038/nchembio.2559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshioka S, Fujii W, Ogawa T, Sugiura K, Naito K. Development of a mono-promoter-driven CRISPR/Cas9 system in mammalian cells. Sci Rep. 2015;5:18341. doi: 10.1038/srep18341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zalatan JG, Lee ME, Almeida R, Gilbert LA, Whitehead EH, La RM, Tsai JC, Weissman JS, Dueber JE, Qi LS. Resource engineering complex synthetic transcriptional programs with CRISPR RNA scaffolds. Cell. 2015;160:339–350. doi: 10.1016/j.cell.2014.11.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zetsche B, Gootenberg JS, Abudayyeh OO, Slaymaker IM, Makarova KS, Essletzbichler P, Volz SE, Joung J, van der Oost J, Regev A, Koonin EV, Zhang F. Cpf1 is a single RNA-guided endonuclease of a class 2 CRISPR-Cas system. Cell. 2015;163:759–771. doi: 10.1016/j.cell.2015.09.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zetsche B, Heidenreich M, Mohanraju P, Kneppers J, Degennaro EM, Winblad N, Choudhury SR, Abudayyeh OO, Gootenberg JS, Wu WY, Scott DA, Severinov K, Van Der OJ, Sciences C, Sciences F, Academy R. Multiplex gene editing by CRISPR-Cpf1 through autonomous processing of a single crRNA array. Nat Biotechnol. 2017;35:31–34. doi: 10.1038/nbt.3737.Multiplex. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang W-W, Matlashewski G. CRISPR-Cas9-mediated genome editing in Leishmania donovani. MBio. 2015;6:e00861–e00861. doi: 10.1128/mBio.00861-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z, Mao Y, Ha S, Liu W, Botella JR, Zhu J-K. A multiplex CRISPR/Cas9 platform for fast and efficient editing of multiple genes in Arabidopsis. Plant Cell Rep. 2016;35:1519–1533. doi: 10.1007/s00299-015-1900-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Wang J, Wang Z, Zhang Y, Shi S, Nielsen J, Liu Z. A gRNA-tRNA array for CRISPR-Cas9 based rapid multiplexed genome editing in Saccharomyces cerevisiae. Nat Commun. 2019;10:1053. doi: 10.1038/s41467-019-09005-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng X, Zheng P, Zhang K, Cairns TC, Meyer V, Sun J, Ma y. 5S rRNA promoter for guide RNA expression enabled highly efficient CRISPR/Cas9 genome editing in Aspergillus niger. ACS Synth Biol. 2018;8:1568–1574. doi: 10.1021/acssynbio.7b00456. [DOI] [PubMed] [Google Scholar]
- Zhong G, Wang H, Li Y, Tran MH, Farzan M. Cpf1 proteins excise CRISPR RNAs from mRNA transcripts in mammalian cells. Nat Chem Biol. 2017;13:839–841. doi: 10.1038/nchembio.2410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou H, Liu B, Weeks DP, Spalding MH, Yang B. Large chromosomal deletions and heritable small genetic changes induced by CRISPR/Cas9 in rice. Nucleic Acids Res. 2014;42:10903–10914. doi: 10.1093/nar/gku806. [DOI] [PMC free article] [PubMed] [Google Scholar]