Abstract
CRISPR/Cas, as a simple, versatile, robust and cost-effective system for genome manipulation, has dominated the genome editing field over the past few years. The application of CRISPR/Cas in crop improvement is particularly important in the context of global climate change, as well as diverse agricultural, environmental and ecological challenges. Various CRISPR/Cas toolboxes have been developed and allow for targeted mutagenesis at specific genome loci, transcriptome regulation and epigenome editing, base editing, and precise targeted gene/allele replacement or tagging in plants. In particular, precise replacement of an existing allele with an elite allele in a commercial variety through homology-directed repair (HDR) is a holy grail in genome editing for crop improvement as it has been very difficult, laborious and time-consuming to introgress the elite alleles into commercial varieties without any linkage drag from parental lines within a few generations in crop breeding practice. However, it still remains very challenging in crop plants. This review intends to provide an informative summary of the latest development and breakthroughs in gene replacement using CRISPR/Cas technology, with a focus on achievements, potential mechanisms and future perspectives in plant biological science as well as crop improvement.
Keywords: CRISPR/Cas, Gene targeting (GT), Gene/allele replacement, Genome editing, Homology-directed repair (HDR)
Introduction
Natural variations among crop cultivars, landraces and their wild relatives provide the genetic diversity essential for plant breeding and crop improvement. Molecular genetic studies have identified genes responsible for controlling many agriculturally important traits such as shoot branching, tiller number, flowering time, grain number, grain size, nutrient use efficiency, and resistance to both abiotic and biotic stresses (Ashikari and Matsuoka 2006; Ashkani et al. 2015; Hao and Lin 2010; Hori et al. 2016; Liu et al. 2014; Mai et al. 2014; Xu et al. 2016). Allelic differences, which have been selected during domestication and subsequent improvements, account for major differences in crop yield and other agriculturally important traits. The most valuable alleles, which are usually derived from local landraces or related species or even orthologs from other plant species, are often caused by differences of one or several single-nucleotide polymorphisms (SNPs), or defined insertion/deletions (indels) of a gene fragment in either promoter region or gene’s encoding region. Harnessing the genetic diversity and introduction of the elite alleles into commercial cultivars has been a major goal in crop breeding programs. Generally, it will take the crop breeders up to 10 years to introduce just one elite allele into commercial cultivars by crossing and back-crossing. In addition, it is very difficult to remove any undesired genes/agronomic traits derived from the parental lines by crossing if they are closely linked to the target genes. Thus, it will greatly accelerate crop improvement if we can introduce the elite alleles from landraces or the related species into the commercialized crop variety without introducing unwanted gene or DNA fragments.
The clustered regularly interspaced short palindromic repeat (CRISPR)/CRISPR-associated protein (Cas) (CRISPR/Cas) enables robust genome editing and has revolutionized both functional genomics and crop improvement. The CRISPR/Cas system which evolved as an adaptive immune response in bacteria and archaea to defend against invading viral and plasmid DNAs (Sampson et al. 2013; Sorek et al. 2013; Terns and Terns 2011), has been developed to cleave targeted specific gene locus of interest to generate double-stranded DNA breaks (DSBs) in various organisms including plants (Bogdanove and Voytas 2011; Dana 2011; Feng et al. 2013; Jinek et al. 2012; Mali et al. 2013). The so far exploited CRISPR/Cas systems were mainly classified into two classes that have been further subdivided into six types based on their characteristics (Burstein et al. 2016; Harrington et al. 2018; Liu et al. 2019; Makarova et al. 2015; Shmakov et al. 2015, 2017; Yan et al. 2019). For Class II Cas proteins, only few Cas proteins are enough to initiate the DNA cleavage. The specificity of Cas-directed DNA cleavage is strictly determined by a chimeric single guide RNA (sgRNA) and a short protospacer adjacent motif (PAM) in the genome (Cong et al. 2013; Zetsche et al. 2015). The Cas protein can induces DSBs at specific target gene sites in the genome, which are determined by the binding specificity of the DNA-binding domain and guide RNA (gRNA). DSBs are repaired by the error-prone non-homologous end joining (NHEJ) pathway or the precise homology-directed repair (HDR), or both NHEJ and HDR (Cong et al. 2013; Danner et al. 2017; Jinek et al. 2012; Puchta 1998; Zetsche et al. 2015). NHEJ is an imprecise repair pathway and frequently introduces random small deletions or insertions at the junction of the newly rejoined chromosome. Therefore, NHEJ is widely used in generating the loss-of-function knock-out mutants (Fig. 1). HDR is an alternative pathway of repairing a broken chromosome. HDR is a precise repair pathway and is stimulated by availability of the homologous DNA repair template (DRT) surrounding a DSB (Jasin and Haber 2016; Puchta 1998) (Fig. 1). HDR or homologous recombination-based gene targeting (GT) is a powerful tool for targeted allele/gene replacement or precise genome modifications such as tagging of genes of interest, and has been widely used in organisms ranging from yeast to mammalian cells (Table 1) (Chandrasegaran and Carroll 2016; Dahan-Meir et al. 2018; Hinnen et al. 1978; Thomas and Capecchi 1987). However, up to date, few successful reports have been documented in plants, especially in crops. Conducting HDR has been difficult, especially in plants, for four main reasons: (1) NHEJ is the predominant pathway for DSBs repair in somatic cells, while HDR is mainly active in the late S and G2 phase of the cell cycle (Hiom 2010; Puchta and Fauser 2014); (2) NHEJ competes with the HDR pathway. Therefore, the frequency of HDR is much lower than NHEJ (Puchta and Fauser 2014; Sun et al. 2016a; Symington and Gautier 2011); (3) timing the induction DSBs on the target gene to coincide with the delivery of sufficient donor repair template (DRT) is crucial for successful HDR events (Endo et al. 2016); (4) limited DRT delivered into plant cells for HDR (Li et al. 2019).
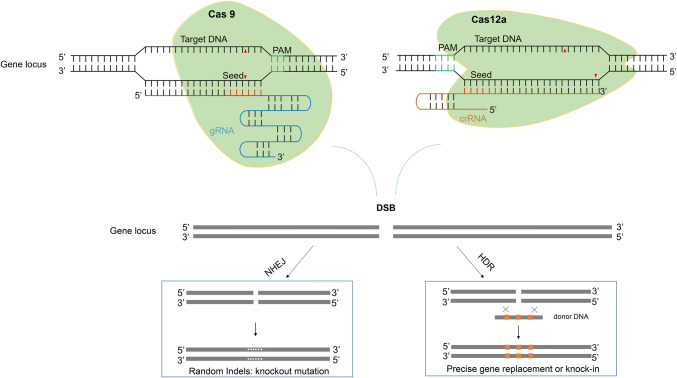
Fig. 1.
Two major pathways underlying the repair of DSB induced by CRISPR/Cas. There are two major pathways underlying the repair of double-stranded break (DSB) induced by CRISPR/Cas (Cas9 or Cas12a). One is a error-prone non-homologous end joining (NHEJ) pathway, which generally generates random indels for knock-out mutagenesis. Another one is homology-directed repair (HDR), which is a precise repair pathway and is generally used for targeted gene/allele replacement or knock-in
Table 1.
HDR in diverse plant species by CRISPR/Cas system
| Plant species | Target gene | Nucleases | Donor | Transformation | Selection | References | Note |
|---|---|---|---|---|---|---|---|
| Arabidopsis | ADH1 | Cas9 | A kanamycin (nptII) resistance cassette | Agrobaterium | Kanamycin | Schiml et al. (2014) | |
| Arabidopsis | ROS1/DME | Cas9 | GFP cassette/DME with mutant sites | Agrobaterium | Hygromycin | Miki et al. (2018) | AtDD45 promoter |
| Arabidopsis | ALS | Cas9 | Acetolactate synthase (ALS) with mutant sites | Agrobaterium | Imazapyr | Wolter et al. (2018) | EC1.1 promoter |
| Arabidopsis | ALS | LbCas12a | Acetolactate synthase (ALS) with mutant sites | Agrobaterium | Imazapyr | Wolter and Puchta (2019) | EC1.1 promoter |
| Maize | ALS2 | Cas9 | Acetolactate synthase (ALS) gene with mutant sites | Bombardment | Chlorsulfuron | Svitashev et al. (2015) | |
| Maize | ARGOS8 | Cas9 | ARGOS8 promoter sequences | Bombardment | Phosphomannose isomerase (PMI) | Shi et al. (2017) | |
| Tobacco | ALS | Cas9 | BeYDV replicons-a kanamycin resistance cassette | Agrobaterium | Kanamycin | Baltes et al. (2014) | Replicon |
| Tomato | ANT1 | Cas9 | BeYDV replicons-anthocyanin mutant 1 (ANT1) and a neomycine phosphotransferase II (NPTII) expression cassette | Agrobaterium | Kanamycin | Čermák et al. (2015) | Replicon |
| Tomato | ALC | Cas9 | ALC with mutant site | Agrobaterium | Hygromycin | Yu et al. (2017) | |
| Tomato | crtiso | Cas9 | BeYDV replicons-crtiso with mutant sites | Agrobaterium | Kanamycin | Dahan-Meir et al. (2018) | Replicon |
| Tomato | ANT1 | LbCas12a | BeYDV replicons-anthocyanin mutant 1 (ANT1) and a neomycine phosphotransferase II (NPTII) expression cassette | Agrobaterium | Kanamycin | Vu et al. (2019) | Replicon |
| Potato | ALS | Cas9 | BeYDV replicons-ALS with mutant sites | Agrobaterium | Kanamycin | Butler et al. (2016) | Replicon |
| Soybean | DD20 | Cas9 | Hygromycin phosphotransferase (HPT) gene expression cassette | Bombardment | Hygromycin | Li et al. (2015) | EFLA2 promoter |
| Soybean | DD43 | Cas9 | Hygromycin phosphotransferase (HPT) gene expression cassette | Bombardment | Hygromycin | Li et al. (2015) | EFLA2 promoter |
| Cassava | EPSPS | Cas9 | 5‐Enolpyruvylshikimate‐3‐phosphate synthase (EPSPS) with mutant sites | Agrobaterium | Glyphosate | Hummel et al. (2018) | NHEJ |
| Rice | ALS | Cas9 | Acetolactate synthase (ALS) with mutant sites | Bombardment | Bispyribac | (Sun et al. 2016b) | |
| Rice | ALS | Cas9 | Acetolactate synthase (ALS) with mutant sites | Agrobaterium | Bispyribac | Endo et al. (2016) | DNA ligase 4 knock out |
| Rice | GST | Cas9 | WDV replicons-GFP cassette | Agrobaterium | Hygromycin | Wang et al. (2017a) | replicon |
| Rice | ALS | Cas9 | Acetolactate synthase (ALS) with mutant sites | Agrobaterium | Bispyribac | (Butt et al. 2017) | RNA and DNA repair |
| Rice | CAO | FnCas12a/LbCas12a | Chlorophyllide-a oxygenase (CAO) with mutant sites | Bombardment | Hygromycin | Begemann et al. (2017) | |
| Rice | NRT1.1B | Cas9 | Nitrate transceptor NRT1.1B with mutant sites | Bombardment | Hygromycin | Li et al. (2018a) | |
| Rice | ALS | LbCas12a | Acetolactate synthase (ALS) with mutant sites | Bombardment | Bispyribac | Li et al. (2018b) | |
| Rice | ALS | LbCas12a | Acetolactate synthase (ALS) with mutant sites | Bombardment | Bispyribac | Li et al. (2019) | RNA and DNA repair |
| Wheat | Ubiquitin | Cas9 | WDV replicons-GFP cassette | Bombardment | GFP | Gil-Humanes et al. (2017) | Replicon |
As a simple, versatile, robust and cost-effective system for genome manipulation, CRISPR/Cas has dominated the genome editing field in various organisms including plants over the past few years. The application of CRISPR/Cas in crop improvement is particularly important in the context of global climate change, as well as in the face of diverse agricultural, environmental and ecological challenges. So far, various CRISPR/Cas toolboxes have been developed and allow for generation of targeted mutagenesis at specific genome loci, transcriptome regulation and epigenome editing, base editing, and precise targeted gene/allele replacement or tagging in plants. However, it is worth mentioning that if the elite allele is conferred by loss-of-function mutations such as IPA or dep-1 (Li et al. 2016), these alleles can be easily introduced in a cultivar through CRISPR/Cas9-mediated NHEJ. In fact, the majority of gene editing reports in crop plants were using NHEJ to generate loss-of-function mutations (Belhaj et al. 2013; Gao and Zhao 2014; Jiang et al. 2013b; Mao et al. 2013; Miao et al. 2013; Tang et al. 2017; Weeks et al. 2016; Xie and Yang 2013). Furthermore, single point mutations in crop plants such as C·G to T·A substitution or A·T to G·G substitution can also be generated through cytosine or adenine base editors using deaminase-dCas9 or deaminase-nCas9 fusions. However, base editing is often constrained by the distance between the targeted base and PAM sequences (Komor et al. 2016; Li et al. 2017; Lu and Zhu 2017; Shimatani et al. 2017). Besides, severe off-target effects of cytosine base editor and adenine base editor were observed both at DNA and RNA levels in cells although these may not have a major impact on crop genome editing (Jin et al. 2019; Zuo et al. 2019). Ideally, we hope to develop capacity to replace any DNA fragments in crops with a desired version, thus allowing us to quickly introduce elite alleles into commercial cultivars. However, it has been very difficult, labor-intensive and time-consuming to introgress these elite alleles into commercial varieties within a few generations in crop common breeding practice. CRISPR/Cas-mediated HDR of DSBs potentially provides us such a capability for introducing the elite alleles into commercial lines within 2–3 generations without introducing undesirable genes or traits. Thus, precise replacement of an existing allele with an elite allele in commercial variety is a holy grail in crop breeding and genetic improvement. Herein, we provide an informative summary of the latest development and breakthroughs in HDR for gene replacement or GT in plants using CRISPR/Cas technology and its potential repair underlying mechanisms. In addition, we discuss the possible factors affecting HDR as well as the challenges and future prospects in implementing precise gene replacement/modification for crop improvement through CRISPR/Cas technology.
CRISPR/Cas 9-mediated HDR in plants
CRISPR/Cas9, belonging to the class II-type II CRISPR/Cas system, is a RNA-guided endonuclease that targets DNA sites through nucleotide base pairing (Fig. 1). It consists of a single gene encoding the Cas9 protein and two RNAs, a mature CRISPR RNA (crRNA) and a partially complementary trans-activating crRNA (tracrRNA). The crRNA: tracrRNA complex together with the Cas9 protein cleave the short sequences in genome DNA adjacent to PAM to produce a DSB. A crRNA and a tracrRNA are then bioengineered into one guide RNA molecule sgRNA (Fig. 1), which makes CRISPR systems easier to utilize (Jinek et al. 2012). Because of its specificity, simplicity, and versatility, CRISPR/Cas9 has been successfully applied in genome editing in many organisms including plants (Table 1) (Cong et al. 2013; Feng et al. 2013; Gao and Zhao 2014; Ma et al. 2015; Ole et al. 2014; Sun et al. 2016b; Xie and Yang 2013). However, the majority studies in reported genome editing were to generate gene knock-outs via NHEJ (Fig. 1, Table 1) (Cong et al. 2013; Endo et al. 2016; Gil-Humanes et al. 2017; Hwang et al. 2013; Jiang et al. 2013a; Miao et al. 2013; Voytas and Gao 2014; Xie et al. 2014). The CRISPR/Cas9 system has also been successfully used for gene replacement or knock-in in plants, albeit at a very lower frequency and fewer successful reports (Fig. 1, Table 1) (Baltes et al. 2014; Begemann et al. 2017; Butler et al. 2016; Butt et al. 2017; Čermák et al. 2015; Dahan-Meir et al. 2018; De Pater et al. 2018; Endo et al. 2006, 2016; Fauser et al. 2012; Gil-Humanes et al. 2017; Hahn et al. 2018; Li et al. 2015, 2018a; Miki et al. 2018; Paix et al. 2017; Sauer et al. 2016; Schiml et al. 2014; Shan et al. 2018; Shi et al. 2017; Sun et al. 2016a; Svitashev et al. 2015; Wang et al. 2017a; Wolter et al. 2018; Wolter and Puchta 2019; Yu et al. 2017). As listed in Table 1, a resistance cassette was integrated into the Alcohol Dehydrogenase (ADH1) locus in Arabidopsis via CRISPR/Cas-mediated HDR (Table 1) (Schiml et al. 2014). ALS (acetolactate synthase) gene encodes a key enzyme for the biosynthesis of branched chain amino acids including leucine, isoleucine and valine, and is a major target for a class of ALS inhibiting herbicides such as chlorsulfuron and bispyribac sodium (BS) (Mazur et al. 1987). Chlorsulfuron-resistant maize plants were generated by substitution of proline 165 with serine in the ALS2 gene using either single-stranded oligonucleotides (SSON) or double-stranded DNA vectors as DRT via CRISPR/Cas9 (Svitashev et al. 2015). To enrich the availability of DRT for HDR, an all-in-one vector which includes all components such as Cas9, two gRNAs and DRT, co-bombarded with a free DRT fragment, enabled efficiently introduction of multiple discrete point mutations in the rice ALS gene through CRISPR/Cas9-mediated HDR. Precise replacement of the wild-type ALS gene with the intended mutant version that carries two discrete point mutations at 548 and 627 sites confers rice plants higher level of herbicide resistance (Sun et al. 2016b). Precise editing of ALS gene in rice was also successful through sequential Agrobacterium-mediated transformation, albeit at a very low efficiency (Endo et al. 2016). Besides, precise modifications of the ALS gene or 5-enolpyruvylshikimate-3-phosphate synthase (EPSPS) gene in maize, soybean, and flax through CRISPR/Cas9-mediated HDR have also been reported although the HDR events occurred at a very low frequency (Sauer et al. 2016; Svitashev et al. 2015). In rice, a vector harboring the CRISPR/Cas9 components and DRT, by co-bombarding with the free DRT, was used to replace the intron region of EPSPS gene to generate the glyphosate-resistant rice plants. However, no homozygous lines were obtained even following segregation, because the authors proposed that the homozygous, precisely edited lines with modified EPSPS are lethal to the growth of rice plants (Li et al. 2016). Agrobacterium-mediated transformation method was also used for obtaining ALC (alcobaca) gene replacement in tomato, in absence and presence of the homologous repair template, and high replacement efficiency (7.69%) was achieved in T0 transgenic plants, respectively (Yu et al. 2017). In maize, the native GOS2 promoter, which confers a moderate level of constitutive expression, was inserted into the 5′-untranslated region (UTR) of the native ARGOS8 gene or was used to replace the native promoter of ARGOS8. Compared to the wild type, the developed ARGOS8 variants increased the grain yield by five bushels per acre under stress conditions and had no yield loss under well-watered conditions in field, demonstrating the utility of the CRISPR/Cas9 system in generating novel allelic variations for breeding drought tolerant crops (Shi et al. 2017). By coupling Cas9 with a chimeric single-guide RNA (cgRNA) molecule carrying both sequences for generating the double-strand breaks (sgRNA) and DRT to direct HDR, gene replacement of ALS gene in rice was achieved by CRISPR/Cas9 (Butt et al. 2017). In our previous study, we successfully replaced of an NRT1.1B allele in japonica rice with an elite allele from indica rice, which is related to improved nitrogen use efficacy, in just one generation using CRISPR/Cas9-mediated HDR (Li et al. 2018a). A sequential transformation method has been developed for GT in Arabidopsis. The parental lines expressing the Cas9 from the egg cell- and early embryo-specific DD45 gene promoter can improve the frequency of gene knock-ins and gene replacements via HDR at several endogenous sites in the Arabidopsis genome (Miki et al. 2018).
Manipulating the geminiviral replicons to enrich the DRT availability and improve HDR efficiency in plants
One of the major factors leading to the low efficiency of HDR for GT is the limited DRT delivered into plant cells for repair of DSBs (Baltes et al. 2014). Geminivirus replicons (GVR) have been used to overcome the obstacles of delivering sufficient DRTs into plant cells for CRISPR/Cas9-mediated HDR in tobacco, tomato, rice and wheat (Table 1) (Baltes et al. 2014; Butler et al. 2016; Čermák et al. 2015; Dahan-Meir et al. 2018; Gil-Humanes et al. 2017; Wang et al. 2017a). For example, in tobacco, bean yellow dwarf virus (BeYDV) replicons based HDR strategy enhanced GT frequencies one to two orders of magnitude over conventional Agrobacterium T-DNA (Baltes et al. 2014). GVR were also used to create heritable modifications to the tomato genome at frequencies tenfold higher than traditional methods of DNA delivery through Agrobacterium transformation. A strong promoter was inserted upstream of a gene anthocyanin 1 (ANT1) controlling anthocyanin biosynthesis, resulting in overexpression and ectopic accumulation of pigments in tomato tissues. In this particular case, both the pigmentation and integrated selection marker neomycine phosphotransferase II (NPTII) expression cassette facilitated the identification and analysis of the HDR events (Čermák et al. 2015). Using the wheat dwarf virus (WDV) replicons carrying CRISPR/Cas9 nucleases and DRT, HDR events were achieved at an endogenous ubiquitin locus at frequencies 12-fold greater than non-viral delivery methods. Knock-in a promoter-less T2A: GFP sequence into the third exon of the ubiquitin gene in all three of the homoeoalleles (A, B and D) of the hexaploid wheat genome was achieved within the same cell at frequencies of around 1%. However, no stable precisely edited wheat plants have been obtained (Gil-Humanes et al. 2017). Using WDV replicon and the DRTs with GFP-2A-NPTII cassette flanked by two homology arms (~ 500 bp) for targeting endogenous Actin-1 (ACT1) or glutathione S-transferase gene (GST) locus in rice, 12 out of 62 (19.4%) transgenic plants and 6 out of 78 (7.7%) plants had incorporated the GFP-2A-NPTII cassette in the predicted rice genomic loci (Wang et al. 2017a). The carotenoid isomerase (CRTISO) and phytoene synthase 1 (PSY1) genes from the carotenoid biosynthesis pathway were chosen as targets due to their easily detectable change of phenotype. By taking advantage of the geminiviral replicon amplification as a means to provide a large amount of donor template for the repair of DSBs, a fast-neutron-induced carotenoid isomerase (CRISTO) gene allele containing a 281 bp deletion was repaired with the wild-type allele through HDR at an efficiency of 25% (Dahan-Meir et al. 2018). In above reports, we noticed that the either gene replacement or knock-in events provided either additional antibiotic resistance selection and/or visible marker (such as GFP) to enrich or facilitate the identification of precise HDR events. It remains to improve the HDR efficiency in the absence of selection markers or reporters/visible markers through this replicon-based HDR technology. However, it is worth mentioning that enrichment of the homology repair template via GVR was inefficient for HDR in Arabidopsis and failed to improve GT efficiency (De Pater et al. 2018; Hahn et al. 2018; Shan et al. 2018). For example, similar numbers of GT events were obtained with or without use of bean yellow dwarf virus (BeYDV) GVR when modifying the endogenous PPO gene through HDR using a 5′ truncated PPO gene with two amino acid substitutions as a repair template (De Pater et al. 2018). In an experiment of restoring trichome formation in a glabrous Arabidopsis mutant by repairing a defective glabrous1 gene through HDR, enrichment of the homology repair template by GVR did not result in GT events (Hahn et al. 2018). While GVR increased somatic GT frequency to knock-in of GFP at the Cruciferin 3 locus in Arabidopsis, similar GT events and patterns were observed in germinal cells with or without GVR (Shan et al. 2018).
CRISPR/Cas12a-mediated HDR in plants
CRISPR/Cas12a (CRISPR from Prevoltella and Francisella 1, also named as Cpf1), a new class II-type V CRISPR/Cas system, was recently exploited as an alternative to SpCas9 in genome editing (Fonfara et al. 2016; Gao et al. 2017; Kim et al. 2016, 2017; Li et al. 2018b, d, 2019; Tang et al. 2017; Wang et al. 2017b; Xu et al. 2017; Zetsche et al. 2015, 2017; Zhong et al. 2018). Cas12a recognizes the ‘TTTN’ PAM complementary with the popular SpCas9 system (‘NGG’ PAM), which enables editing AT-rich regions such as 5′ and 3′ UTR and promotor region (Zetsche et al. 2015). Cas12a is only guided by a single ~ 43 nt crRNA, which is 60 nt less than Cas9 sgRNA, making it more suitable for packaging into vectors (Zetsche et al. 2015). Thus, CRISPR/Cas12a system is emerging as an attractive tool for editing AT-rich regions (Begemann et al. 2017; Tang et al. 2017; Wang et al. 2017b). However, the gene editing efficiency of CRISPR/Cas12a is relatively low compared with CRISPR/Cas9, and many methods have been used to improve its editing efficiency including high temperature treatment (Lin et al. 2018; Malzahn et al. 2019; Moon et al. 2018). FnCas12a and LbCas12a nucleases, in the presence of a guide RNA and DRT flanked by homology DNA fragments to the target site, were demonstrated to generate precise gene insertions as well as indel mutations at the target site in the rice genome. The frequency of targeted insertion for these Cas12a nucleases reached up to 8%, higher than most other genome editing nucleases, indicative of its effective enzymatic chemistry (Table 1) (Begemann et al. 2017). Also, co-delivery of CRISPR/Cas12a ribonucleoproteins (RNP) with single-stranded DRT results in precise and targeted DNA replacement with as much as ~ 10% efficiency in C. reinhardtii. As the direct delivery of gene-editing reagents bypasses the use of transgenes, this method is potentially applicable to a wider range of species without the need to develop methods for stable transformation (Table 1) (Ferenczi et al. 2017). In addition, co-delivery of all-in-one CRISPR/Cas12a vector and free DRT through bombardment, enabled us to precisely replace the wild-type ALS gene with the intended mutant version that carries two discrete point mutations conferring rice plants herbicide resistance in our previous study albeit at a relatively lower efficiency (Table 1) (Li et al. 2018b). It is worth mentioning that although co-delivery of the free DRT fragment enriches the DRT availability and thus enhances the occurrence of HDR, and DRT itself is not harmful, random integration of this DRT fragment into the genome may raise biosafety concerns. RNA transcripts-templated homology-directed DNA repair (TT-HDR) potentially can overcome the obstacles in delivery of DRT into plant cells to repair DSBs through HDR because RNA transcripts can be produced abundantly in vivo. However, primary transcripts are often processed, modified and transported to cytosol, rendering them unavailable for HDR. TT-HDR has been previously used in yeast and human cells (Derr et al. 1991; Keskin et al. 2014; Nowacki et al. 2007; Storici et al. 2007), but not in plant cells until recently. In our previous study, we coupled LbCas12a with a single array comprising of the crRNAs flanked with ribozymes and a DRT flanked with either ribozymes or crRNA targets. The primary transcripts from the arrays underwent self-processing to release the crRNAs and DRT inside the nucleus. Using TT-HDR and DNA-free ribonucleoprotein (RNP) complexes, we achieved targeted gene replacement in rice, greatly expanding our ability to improve agriculturally important traits in crops through CRISPR/Cas12a-mediated HDR technology (Li et al. 2019). LbCas12a has recently also been reported to work well in HDR in Arabidopsis and tomato (Wolter and Puchta 2019; Vu et al. 2019). In Arabidopsis, LbCas12a successfully mediated GT of ALS gene, and the GT frequencies were higher than that of SaCas9, which greatly broadens the range of in planta gene targeting (ipGT) in AT rich regions (Wolter and Puchta 2019). In tomatoes, coupling a CRISPR/LbCas12a system with a multiple replicon system rather than a single-replicon system effectively increased HDR efficiency in approximately 3-fold, especially at higher temperatures (Vu et al. 2019).
Potential mechanisms underlying HDR
Efficient repair of DSB is important for maintaining genomic stability and survival of organisms. Compared with NHEJ, HDR is an accurate repair process. It repairs DNA DSBs using a homologous DNA sequences as template. Based on its repairing characters, HDR has been used for targeted elite allele replacement or knock-in specific genes or markers/tags to target loci. The repair mechanism of DSBs is basically conservative from yeast to plants and animals. Upon availability of a homologous template, three main potential mechanisms have been proposed for HDR: single-strand annealing (SSA), synthesis-dependent strand annealing (SDSA), and the so-called double-strand break repair (DSBR) model (Fig. 2) (Puchta 1998; Puchta and Fauser 2013).
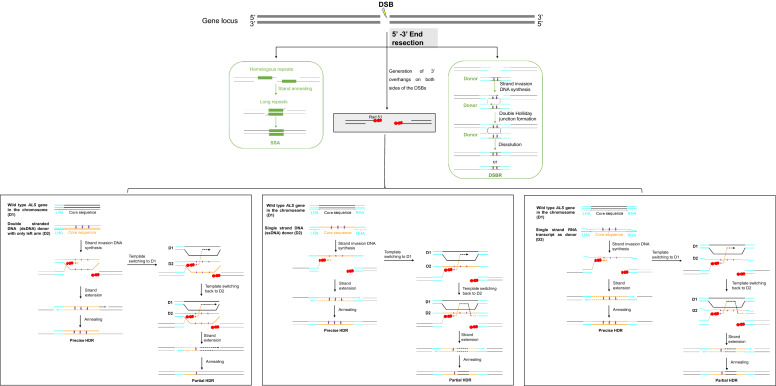
Fig. 2.
Proposed Models for HDR Pathway. Three potential mechanisms are proposed for HDR in plant cells. For SSA, complementary sequences at the two ends of a DSB anneal to the other forming a chimeric DNA molecule with the 3’-overhangs trimmed. Consequently, the sequences between the complementary sequences get lost. In DSBR, following the DSB induction, free 3’ overhangs are formed via exonuclease resection. 3’ ends interact with their homologous sequences in the donor repair templates, which leads to the formation of a Holliday junction. Resolution of the junction results in two DNA molecules that either have cross-over or gene conversion. In SDSA, the DSB is firstly resected and processed to generate 3’ overhangs on both sides of the DSB. The 3’ overhangs are then paired with the homologous arms of the donor repair template (DRT) and are extended by DNA synthesis. Finally, newly synthesized strands withdraw from the donor templates and anneal back to the locus. In addition, repair templates switch between donors and wild type during DNA synthesis, usually results in the partial HDR events. When donor fragment is dsDNA, either strand of the dsDNA can act as repair template. DSB: double strand break, SSA: single-strand annealing, DSBR: so-called double-strand break repair, SDSA: synthesis-dependent strand annealing, LHA: left homology arm, RHA: right homology arm, RT: Reverse transcription. ⋆:Nucleotides changes in DRT
During SSA, both ends of the DSB carry complementary sequences. These molecules can then anneal to one another to form a chimeric DNA molecule with the 3′-overhangs be trimmed. Consequently, the sequences between the complementary sequences will be lost. Therefore, SSA is also classified as a non-conservative HDR-mediated DSB repair mechanism (Puchta and Fauser 2013) (Fig. 2). In somatic cells, SSA can proceed as efficient as NHEJ (Knoll et al. 2014). In the cases of DSBR and SDSA, the homologous repair template can be supplied in cis or in trans. Following the DSB induction, 3′-end invasion of a single strand into a homologous double strand occurs, resulting in a D-loop. Reparative synthesis is initiated using the newly paired strand as a template. In DSBR, DNA synthesis occurs at both broken ends, respectively, so that genetic information is copied from both strands of the homologous sequences, respectively, which may lead to a crossover event (Puchta and Fauser 2013) (Fig. 2). DSBR is a prominent mechanism for meiotic recombination (Osman et al. 2011). In SDSA, both sides of the DSB is resected and processed to generate 3′ overhangs by stable complex Mre11–Rad50-Xrs2 (MRX) (Andrej and Miroslav 2004). Subsequently, Rad51 protein binds to the 3′ overhangs. The 3′ overhangs are then paired with the homologous arms of the DRT and are extended by DNA synthesis using the DRT as a template (Symington 2002). Then, the newly synthesized strands withdraw from the DRT and anneal back to the locus (Paix et al. 2017; Puchta and Fauser 2014). SDSA has been proposed as a major repair mechanism when the single-stranded oligo DNA nucleotides (ssODNs) are used as DRT (Kan et al. 2017; Paquet et al. 2016; Richardson et al. 2016). The repair process of DSBs generated by Cas nucleases is more consistent with SDSA when either ssODNs or dsDNAs is used as repair templates in human cells (Paix et al. 2017) (Fig. 2). In our previous study, we provided evidence of SDSA repair of Cas12a-induced DSBs, which enables us to precisely replace the wild-type ALS gene with the intended mutant version that carries two discrete point mutations conferring rice plants herbicide resistance. Our observation that the DRT with only left homologous arm is sufficient for precise targeted allele replacement offers a better understanding of the mechanism underlying HDR in plants (Table 1) (Li et al. 2018b).
Exploiting diverse strategies to improve HDR efficiency
Targeted genomic manipulation by nucleases can efficiently generate knock-out cells and organisms via NHEJ, but the efficiency of precise sequence replacement by HDR is substantially low (Doudna and Emmanuelle 2014; Jiang and Marraffini 2015). As listed in the following, diverse strategies have been exploited to improve HDR frequency.
The first strategy is to manipulate the major components involved in either NHEJ or HDR pathways in plant cells. In the process of DSBs, NHEJ is the predominant pathway. Over the past several years, various approaches have been tested by suppressing the components involved in NHEJ pathway or overexpressing the components involved HDR pathway to enhance the occurrence of HDR in plants. In Arabidopsis, a fivefold to 16-fold enhancement in HDR-based GT in a ku70 mutant and a threefold to fourfold enhancement in GT in the DNA ligase 4 (lig4) mutant compared with in wild type were achieved when using a ZFN nuclease (Qi et al. 2013). Similarly, when gRNA targeting lig4 was transformed with Cas9 prior to the GT experiment in rice, GT efficiency increased and more than one line exhibiting bi-allelic GT at the ALS locus was obtained (Table 1) (Endo et al. 2016). Moreover, HDR at a defined locus carrying an endogenous nuclear marker gene was stimulated at least tenfold in transgenic plant cells expressing nucleus-targeted RecA. The stimulation of intrachromosomal recombination demonstrates that Escherichia coli RecA protein is functional in genomic homologous recombination in plants, especially when targeted to the plant nucleus (Reiss et al. 1996). In tobacco, expression of a bacterial RecA protein in plants stimulates homologous recombination. And the number of DSBs that were repaired at both sides by homologous recombination was increased 3.3-fold (Reiss et al. 2000). In Arabidopsis, expression of the yeast RAD54 gene can enhance GT in plants (Shaked et al. 2005). RAD54 expression under the egg apparatus-specific enhancer (EASE) led to an approximately ten-fold increase in HDR for GT efficiency in Arabidopsis, when compared with wild-type plants (Even-Faitelson et al. 2011).
The second strategy is to optimizing the design of DRTs. (1) Orientation of the donor. Donor DNA complementary to the non-target strand stimulated HDR frequencies and is more effective than donor complementary to the target strand (Lin et al. 2014; Richardson et al. 2016; Yang et al. 2013). (2) Homology arm (HA) length of the donor. For editing large pieces, double stranded DNAs with relatively long homology arms are required to achieve relatively high efficiency. A 2.7 kilobase (kb) homozygous gene replacement was achieved in up to 11% of iPSC without selection when the homology arm length was around 2 kb (Byrne et al. 2015). It was found that a 600 bp homology in both arms leads to high-level genome knock-in, with 97–100% of the donor insertion events being mediated by HDR in 293 T cells (Zhang et al. 2017). (3) Cycle regulators. The combined use of CCND1, a cyclin that functions in G1/S transition, and nocodazole, a G2/M phase synchronizer, doubles HDR efficiency to up to 30% in iPSCs (Zhang et al. 2017). (4) Sufficient amount of donor repair template delivered to plant cells using RNA transcripts as templates. Limited donor repair template delivered into the nucleus is the main obstacle in GT in crop plants. Production of RNA templates in vivo by TT-HDR could potentially overcome this problem. TT-HDR has been demonstrated in yeast and human cells (Derr et al. 1991; Keskin et al. 2014; Nowacki et al. 2007; Storici et al. 2007). A chimeric sgRNA might serve as both a guide RNA and a DRT (Table 1) (Butt et al. 2017). In our previous study, by coupling Cas12a to an array of crRNAs flanked with ribozymes, and a DRT flanked with either ribozymes or crRNA sequences, we successfully obtained stable transgene-free lines with two desired mutations in the ALS gene in rice. The HDR efficiency can likely be improved if stronger promoters are used for producing more abundant RNA transcripts (Table 1) (Li et al. 2019).
Future perspectives and challenges
CRISPR/Cas provides an effective suite of molecular tools to precisely alter the genome in a user-defined manner. It has emerged as one of the foremost systems with which to edit the crop genome, with rapidly increasing agricultural applications in major cereals such as rice, wheat, and maize and other crops (Zaidi et al. 2019). Genome editing allows the alteration of endogenous genes to improve traits in crops without transferring transgenes across species boundaries. Whereas CRISPR/Cas9-mediated gene knock-out and base editing is widely used for a variety of applications in crop improvement, other important applications include precise DNA sequence editing, gene replacement, and simultaneous enhancement of multiple traits, as well as promoter and regulatory element engineering for altered gene expression patterns have been documented only in a few cases. In years to come, manipulation of diverse tools to improve HDR efficiency in crop plants, de nevo domestication and pyramiding multiple traits through HDR will become the hotspots in crop genome editing. It is worth mentioning that precisely edited crops with targeted elite allele replacement should be exempted from regulation procedure and biosafety concerns if such a variation or allele exists in crop landraces or wild relatives and occurs naturally during evolution or domestication.
Novel strategies to improve HDR efficiency in plants
Although it is now feasible to achieve precise gene/allele replacement or gene targeting in Arabidopsis and some crop species such as rice, maize and tomato (Table 1) (Begemann et al. 2017; Butt et al. 2017; Endo et al. 2016; Gil-Humanes et al. 2017; Hummel et al. 2018; Li et al. 2018a; b, 2019; Shi et al. 2017; Sun et al. 2016b; Svitashev et al. 2015; Wang et al. 2017a), it still remains challenging to perform HDR in crop plants. For example, no successful stable precisely edited wheat plants have ever been recovered (Gil-Humanes et al. 2017). Besides, even for model crop species such as rice, it is only feasible in a few laboratories due to a very lower efficiency of HDR. Moreover, manipulation of diverse tools to improve HDR efficiency in plants is particularly important for polyploidy species such as wheat because introgression and selection for wheat improvement and environmental adaptation, has taken the breeders many years to reduce deleterious allele burden (He et al. 2019). Based on the importance of HDR pathway, many approaches have been used to improve HDR efficiency in mammalian cells. To enrich the availability of DRTs, a modular RNA aptamer-streptavidin strategy increased the ratio of precisely edited to imprecisely edited alleles up to 18-fold higher than standard gene-editing methods in human cells (Carlson-Stevermer et al. 2017). Also, a 30-fold enhancement of HDR efficiency was achieved by covalently tethering a single-stranded donor oligonucleotide (ssODN) to the Cas9/guide RNA ribonucleoprotein (RNP) complex via a fused HUH endonuclease 5 (Aird et al. 2018). In human cell lines, iPS cells, and rat zygotes, Cas9 fused with a minimal N-terminal fragment of CtIP obtained twofold or more efficient transgene integration than that with only Cas9 (Charpentier et al. 2018). Also, spatial and temporal co-localization of the donor template and Cas9 via SNAP tag increases the correction HDR up to 24-fold, which is mainly caused by an increase of donor template concentration in the nucleus (Savic et al. 2018). It have been found that the HDR efficiency could be improved if the DRT is in close proximity to the DSB (Aird et al. 2018; Gu et al. 2018; Ma et al. 2017; Rolloos et al. 2015; Roy et al. 2018; Savic et al. 2018). These above strategies can be evaluated and used in crop plants to improve the HDR efficiency. Alternatively, using the egg cell-and early embryo-specific DD45 gene promoter to drive SpCas9 expression and sequential transformation in Arabidopsis has shown potential for increasing the efficiency of HDR-mediated gene editing (Miki et al. 2018), but has yet to be applied to crops.
De nevo domestication and simultaneous replacements of multiple genes for pyramiding diverse elite traits
During domestication and intensive breeding practices for higher yield and stresses tolerant crop varieties, some elite alleles may loss due to the linkage drag of deleterious genes/traits during selection. As a matter of fact, breeders need to choose the main breeding target traits and balance other multiple traits, good or bad, to breeding a new variety. Genome editing can thus be used for accurate manipulation of specific target genes for de novo domestication in a route toward ideal crops for food security. Next-generation DNA sequencing technologies have generated an immense amount of genomic data including full genome sequences for many crop species including the most important stable food species such as wheat and rice (Ling et al. 2018; Wang et al. 2018). For example, much progress has been made in understanding the genetic basis of yield- and quality-related traits in rice and maize (Chen et al. 2011; Ikeda et al. 2013; Li et al. 2018c). In wheat, exome sequencing highlights the role of wild-relative introgression in shaping the adaptive landscape of the wheat genome (He et al. 2019). The target genes/alleles for improvement are now more accessible by mining the increasing number of high-quality crop genomes and the allelic comparisons in crops and their diverse accessions including wild progenitors. It can be combined with the mining of deep-sequenced genome data to tailor crops, as well as to create a more sustainable and climate-friendly agricultural industry by precise editing the key domestication genes alongside the development of the wild progenitors into current varieties (Fernie and Yan 2019). For example, the widespread adoption of semi-dwarf green revolution varieties of cereals are conferred by mutant alleles at the wheat Rht and rice SD1 loci during breeding and domestication. However, rht and sd1 inhibit nitrogen assimilation, carbon fixation and growth, whereas GROWTH-REGULATING FACTOR 4 (GRF4) promotes these processes (Li et al. 2018c). Either the haplotype with three SNPs in GRF4 promoter region or the haplotype with two SNPs in encoding region, or the presence of both haplotypes account for increased nitrogen assimilation and grain yield (Che et al. 2015; Hu et al. 2015). Specially, the explored haplotype in promoter region only exists in selected indica cultivars, but not in modern elite varieties (Li et al. 2018a). Introgression of these haplotypes into Japonica rice varieties will take the breeder many years. Precise allele replacement will certainly accelerate this process in a shorter period of 2–3 years. Besides, along with the development of TT-HDR strategy (Table 1) (Li et al. 2019), we envision that it is also feasible to simultaneously manipulate multiple traits either by multiplexing genome editing through HDR or pyramiding the HDR events in sequential transformations or crossing.
Biosafety concerns of precisely edited crops with targeted elite allele replacement
Off-target effect has been a major concern in genome editing in animal cells, but not a barrier in applying CRISPR/Cas9 and CRISPR/Cas12a systems in precise plant breeding. Following segregation or crossing with wild-type plants, transgene-free plants could be recovered in one generation. Besides, a large-scale whole-genome sequencing analysis and CIRCLE‐seq analysis demonstrated that both CRISPR/Cas9 and CRISPR/Cas12a systems were very specific in generating targeted mutagenesis in rice and maize, respectively (Tang et al. 2018; Lee et al. 2019). These facts suggested that the off-target effect can be avoided by designing guide RNAs with high specificity and should not be a major biosafety concern. Indeed, the updated regulations across different countries allow scientists to use some genome-editing techniques in plants without government approval except for the European Union. In March 2018, the US Department of Agriculture excluded genome-edited plants from regulatory oversight (Waltz 2018). Subsequently, Canada, Brazil, Argentina and Japan announced the exclusion of the gene-edited plant products from regulation. By contrast, the Court of Justice of the European Union ruled in July 2018 that they would treat gene-edited crops as genetically modified organisms and subject to stringent regulation (Callaway 2018). However, the Australian government will not regulate the use of gene-editing techniques in plants in which proteins cut DNA at a specific target site, as long as the tools allow the host cell to repair the break naturally, rather than using a template containing genetic material to direct the repair process (https://www.nature.com/articles/d41586-019-01282-8). The Australian ruling is a “middle ground” between more lenient gene-editing rules in the United States, Canada, Brazil, Argentina and Japan, and tougher measures in the European Union. In China, where the crop genome editing research is boosting and is somehow leading the world in the last few years (https://www.sciencemag.org/news/2019/08), there are still no released regulation policies yet, even though the policy-makers show positive attitude toward gene-edited crops.
Considering the nature of precise gene/allele replacement, it simply edits the existing genes within crops using this technology to speed up a process that otherwise elite alleles have occurred in landraces or wild relatives in nature. Through this process, we’re able to improve crops in a user-defined manner, quickly, efficiently and cost-effectively without compromising other elite agronomy traits and without linkage drag of deleterious genes. Besides, no foreign DNA/genes exist in the precisely edited lines following segregation in the progeny or crossing with wild types, or using transgene-killer CRISPR technology (He et al. 2018). Thus, we here would like to propose that it is unnecessary to regulate the crops with precisely gene/allele replacement, if the resulting alleles exist in local landraces or wild relatives, or such alleles can be obtained through other conventional mutagenesis breeding methods.
Concluding remarks
Precise replacement of an existing allele with an elite allele in commercial variety through HDR is a holy grail in crop improvement. Although the impressive progresses have been achieved in the past few years, it still remains very challenging in crop plants. Understanding the potential mechanisms underlying HDR and manipulating the DNA repair pathways will benefit the improvement of HDR efficiency in crop plants. Furthermore, diverse strategies used for HDR in mammalian cells may facilitate us to achieve a better efficacy in crop plants. Through CRISPR/Cas-mediated gene replacement, we’re able to improve crops in a user-defined manner, quickly, efficiently and cost-effectively without compromising other elite agronomy traits and without linkage drag of deleterious genes. Following the technology improvement by different modulations for HDR, it is reasonable to expect that in the long run, precise gene/allele replacement through CRISPR/Cas system, in combination with conventional breeding methods, will be widely used for breeding of diverse crop elite varieties for sustainable agriculture and better environment to ensure global food security.
Acknowledgements
Some mentioned works in this review are partly funded by the Ministry of Agriculture of China (Grant nos. 2019ZX08010001 and 2019ZX08010003), the Central Non-Profit Fundamental Research Funding supported by Institute of Crop Sciences, Chinese Academy of Agricultural Sciences (S2018QY05).
Author contributions
SYL and LQX wrote the manuscript.
Compliance with ethical standards
Conflict of interest
There are no conflicts of interest.
References
- Aird EJ, Lovendahl KN, St. Martin A, Harris RS, Gordon WR. Increasing Cas9-mediated homology-directed repair efficiency through covalent tethering of DNA repair template. Commun Biol. 2018;1:54. doi: 10.1038/s42003-018-0054-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andrej D, Miroslav C. DNA double-strand break repair by homologous recombination. Mutat Res. 2004;566:131–167. doi: 10.1016/j.mrrev.2003.07.001. [DOI] [PubMed] [Google Scholar]
- Ashikari M, Matsuoka M. Identification, isolation and pyramiding of quantitative trait loci for rice breeding. Trends Plant Sci. 2006;11:344–350. doi: 10.1016/j.tplants.2006.05.008. [DOI] [PubMed] [Google Scholar]
- Ashkani S, Rafii MY, Shabanimofrad M, Miah G, Sahebi M, Azizi P, Tanweer FA, Akhtar MS, Nasehi A. Molecular breeding strategy and challenges towards improvement of blast disease resistance in rice crop. Front Plant Sci. 2015;6:1–14. doi: 10.3389/fpls.2015.00886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baltes NJ, Gil-Humanes J, Cermak T, Atkins PA, Voytas DF. DNA replicons for plant genome engineering. Plant Cell. 2014;26:151–163. doi: 10.1105/tpc.113.119792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Begemann MB, Gray BN, January E, Gordon GC, He Y, Liu H, Wu X, Brutnell TP, Mockler TC, Oufattole M. Precise insertion and guided editing of higher plant genomes using Cpf1 CRISPR nucleases. Sci Rep. 2017;7:11606. doi: 10.1038/s41598-017-11760-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belhaj K, Chaparro-Garcia A, Kamoun S, Nekrasov V. Plant genome editing made easy: targeted mutagenesis in model and crop plants using the CRISPR/Cas system. Plant Methods. 2013;9:39. doi: 10.1186/1746-4811-9-39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bogdanove AJ, Voytas DF. TAL effectors: customizable proteins for DNA targeting. Science. 2011;333:1843–1846. doi: 10.1126/science.1204094. [DOI] [PubMed] [Google Scholar]
- Burstein D, Harrington LB, Strutt SC, Probst AJ, Anantharaman K, Thomas BC, Doudna JA, Banfield JF. New CRISPR–Cas systems from uncultivated microbes. Nature. 2016;542:237. doi: 10.1038/nature21059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butler NM, Baltes NJ, Voytas DF, Douches DS. Geminivirus-mediated genome editing in potato (Solanum tuberosum L.) using sequence-specific nucleases. Front Plant Sci. 2016;7:1–13. doi: 10.3389/fpls.2016.01045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butt H, Eid A, Ali Z, Atia MAM, Mokhtar MM, Hassan N, Lee CM, Bao G, Mahfouz MM. Efficient CRISPR/Cas9-mediated genome editing using a chimeric single-guide RNA molecule. Front Plant Sci. 2017;8:1441. doi: 10.3389/fpls.2017.01441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Byrne SM, Luis O, Prashant M, John A, Church GM. Multi-kilobase homozygous targeted gene replacement in human induced pluripotent stem cells. Nucleic Acids Res. 2015;43:e21. doi: 10.1093/nar/gku1246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Callaway E. CRISPR plants now subject to tough GM laws in Europe. Nature. 2018;560:16. doi: 10.1038/d41586-018-05814-6. [DOI] [PubMed] [Google Scholar]
- Carlson-Stevermer J, Abdeen AA, Kohlenberg L, Goedland M, Molugu K, Lou M, Saha K. Assembly of CRISPR ribonucleoproteins with biotinylated oligonucleotides via an RNA aptamer for precise gene editing. Nat Commun. 2017;8:1711. doi: 10.1038/s41467-017-01875-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Čermák T, Baltes NJ, Čegan R, Zhang Y, Voytas DF. High-frequency, precise modification of the tomato genome. Genome Biol. 2015;16:232. doi: 10.1186/s13059-015-0796-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chandrasegaran S, Carroll D. Origins of programmable nucleases for genome engineering. J Mol Biol. 2016;428:963–989. doi: 10.1016/j.jmb.2015.10.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charpentier M, Ahy K, Menoret S, Brion A, Lamribet K, Dardillac E, Boix C, Perrouault L, Tesson L, Geny S. CtIP fusion to Cas9 enhances transgene integration by homology-dependent repair. Nat Commun. 2018;9:1133. doi: 10.1038/s41467-018-03475-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Che R, Tong H, Shi B, Liu Y, Fang S, Liu D, Xiao Y, Hu B, Liu L, Wang H, et al. Control of grain size and rice yield by GL2-mediated brassinosteroid responses. Nat Plants. 2015;2:15195. doi: 10.1038/nplants.2015.195. [DOI] [PubMed] [Google Scholar]
- Chen J, Liu Y, Ni J, Wang Y, Bai Y, Shi J, Gan J, Wu Z, Wu P. OsPHF1 regulates the plasma membrane localization of low-and high-affinity inorganic phosphate transporters and determines inorganic phosphate uptake and translocation in rice. Plant Physiol. 2011;157:269–278. doi: 10.1104/pp.111.181669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cong L, Ran FA, Cox D, Lin S, Barretto R, Habib N, Hsu PD, Wu X, Jiang W, Marraffini LA, et al. Multiplex genome engineering using CRISPR/Cas systems. Science. 2013;339:819–823. doi: 10.1126/science.1231143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dahan-Meir T, Filler-Hayut S, Melamed-Bessudo C, Bocobza S, Czosnek H, Aharoni A, Levy AA. Efficient in planta gene targeting in tomato using geminiviral replicons and the CRISPR/Cas9 system. Plant J. 2018;95:5–16. doi: 10.1111/tpj.13932. [DOI] [PubMed] [Google Scholar]
- Dana C. Genome engineering with zinc-finger nucleases. Genetics. 2011;188:773–782. doi: 10.1534/genetics.111.131433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danner E, Bashir S, Yumlu S, Wurst W, Wefers B, Kühn R. Control of gene editing by manipulation of DNA repair mechanisms. Mamm Genome. 2017;28:1–13. doi: 10.1007/s00335-017-9688-5. [DOI] [PubMed] [Google Scholar]
- De Pater S, Klemann B, Hooykaas PJJ. True gene-targeting events by CRISPR/Cas-induced DSB repair of the PPO locus with an ectopically integrated repair template. Sci Rep. 2018;8:3338. doi: 10.1038/s41598-018-21697-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Derr LK, Strathern JN, Garfinkel DJ. RNA-mediated recombination in S. cerevisiae. Cell. 1991;67:355–364. doi: 10.1016/0092-8674(91)90187-4. [DOI] [PubMed] [Google Scholar]
- Doudna JA, Emmanuelle C. The new frontier of genome engineering with CRISPR-Cas9. Science. 2014;346:1258096. doi: 10.1126/science.1258096. [DOI] [PubMed] [Google Scholar]
- Endo M, Ishikawa Y, Osakabe K, Nakayama S, Kaya H, Araki T, Shibahara K-I, Abe K, Ichikawa H, Valentine L, et al. Increased frequency of homologous recombination and T-DNA integration in Arabidopsis CAF-1 mutants. EMBO J. 2006;25:5579–5590. doi: 10.1038/sj.emboj.7601434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Endo M, Mikami M, Toki S. Biallelic gene targeting in rice. Plant Physiol. 2016;170:667–677. doi: 10.1104/pp.15.01663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Even-Faitelson L, Samach A, Melamed-Bessudo C, Avivi-Ragolsky N, Levy AA. Localized egg-cell expression of effector proteins for targeted modification of the Arabidopsis genome. Plant J. 2011;68:929–937. doi: 10.1111/j.1365-313X.2011.04741.x. [DOI] [PubMed] [Google Scholar]
- Fauser F, Roth N, Pacher M, Ilg G, Sánchez-Fernández R, Biesgen C, Puchta H. In planta gene targeting. Proc Natl Acad Sci. 2012;109:7535–7540. doi: 10.1073/pnas.1202191109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng Z, Zhang B, Ding W, Liu X, Yang DL, Wei P, Cao F, Zhu S, Zhang F, Mao Y, et al. Efficient genome editing in plants using a CRISPR/Cas system. Cell Res. 2013;23:1229–1232. doi: 10.1038/cr.2013.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferenczi A, Pyott DE, Xipnitou A, Molnar A. Efficient targeted DNA editing and replacement in Chlamydomonas reinhardtii using Cpf1 ribonucleoproteins and single-stranded DNA. Proc Natl Acad Sci. 2017;114:13567–13572. doi: 10.1073/pnas.1710597114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernie AR, Yan J. De novo domestication: an alternative route toward new crops for the future. Molecular Plant. 2019;12:615–631. doi: 10.1016/j.molp.2019.03.016. [DOI] [PubMed] [Google Scholar]
- Fonfara I, Richter H, Bratovič M, Le Rhun A, Charpentier E. The CRISPR-associated DNA-cleaving enzyme Cpf1 also processes precursor CRISPR RNA. Nature. 2016;532:517. doi: 10.1038/nature17945. [DOI] [PubMed] [Google Scholar]
- Gao Y, Zhao Y. Self-processing of ribozyme-flanked RNAs into guide RNAs in vitro and in vivo for CRISPR-mediated genome editing. J Integr Plant Biol. 2014;56:343–349. doi: 10.1111/jipb.12152. [DOI] [PubMed] [Google Scholar]
- Gao L, Cox DBT, Yan WX, Manteiga JC, Schneider MW, Yamano T, Nishimasu H, Nureki O, Crosetto N, Zhang F. Engineered Cpf1 variants with altered PAM specificities. Nat Biotechnol. 2017;35:789. doi: 10.1038/nbt.3900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gil-Humanes J, Wang Y, Liang Z, Shan Q, Ozuna CV, Sánchez-León S, Baltes NJ, Starker C, Barro F, Gao C, et al. High-efficiency gene targeting in hexaploid wheat using DNA replicons and CRISPR/Cas9. Plant J. 2017;89:1251–1262. doi: 10.1111/tpj.13446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu B, Posfai E, Rossant J. Efficient generation of targeted large insertions by microinjection into two-cell-stage mouse embryos. Nat Biotechnol. 2018;36:632–637. doi: 10.1038/nbt.4166. [DOI] [PubMed] [Google Scholar]
- Hahn F, Eisenhut M, Mantegazza O, Weber APM. Homology-directed repair of a defective glabrous gene in Arabidopsis with Cas9-based gene targeting. Front Plant Sci. 2018;9:1–13. doi: 10.3389/fpls.2018.00001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hao W, Lin HX. Toward understanding genetic mechanisms of complex traits in rice. J Genet Genomics. 2010;37:653–666. doi: 10.1016/S1673-8527(09)60084-9. [DOI] [PubMed] [Google Scholar]
- Harrington LB, Burstein D, Chen JS, Paez-Espino D, Ma E, Witte IP, Cofsky JC, Kyrpides NC, Banfield JF, Doudna JA. Programmed DNA destruction by miniature CRISPR-Cas14 enzymes. Science. 2018;362:839–842. doi: 10.1126/science.aav4294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He Y, Zhu M, Wang L, Wu J, Wang Q, Wang R, Zhao Y. Programmed self-elimination of the CRISPR/Cas9 construct greatly accelerates the Isolation of edited and transgene-free rice plants. Mol Plant. 2018;11:1210–1213. doi: 10.1016/j.molp.2018.05.005. [DOI] [PubMed] [Google Scholar]
- He F, Pasam R, Shi F, Kant S, Keeble-Gagnere G, Kay P, Forrest K, Fritz A, Hucl P, Wiebe K, et al. Exome sequencing highlights the role of wild-relative introgression in shaping the adaptive landscape of the wheat genome. Nat Genet. 2019;51:896–904. doi: 10.1038/s41588-019-0382-2. [DOI] [PubMed] [Google Scholar]
- Hinnen A, Hicks JB, Fink GR. Transformation of yeast. Proc Natl Acad Sci. 1978;75:1929–1933. doi: 10.1073/pnas.75.4.1929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiom K. Coping with DNA double strand breaks. DNA Repair. 2010;9:1256–1263. doi: 10.1016/j.dnarep.2010.09.018. [DOI] [PubMed] [Google Scholar]
- Hori K, Matsubara K, Yano M. Genetic control of flowering time in rice: integration of Mendelian genetics and genomics. Theor Appl Genet. 2016;129:2241–2252. doi: 10.1007/s00122-016-2773-4. [DOI] [PubMed] [Google Scholar]
- Hu J, Wang Y, Fang Y, Zeng L, Xu J, Yu H, Shi Z, Pan J, Zhang D, Kang S, et al. A rare allele of GS2 enhances grain size and grain yield in rice. Mol Plant. 2015;8:1455–1465. doi: 10.1016/j.molp.2015.07.002. [DOI] [PubMed] [Google Scholar]
- Hummel AW, Chauhan RD, Cermak T, Mutka AM, Vijayaraghavan A, Boyher A, Starker CG, Bart R, Voytas DF, Taylor NJ. Allele exchange at the EPSPS locus confers glyphosate tolerance in cassava. Plant Biotechnol J. 2018;16:1275–1282. doi: 10.1111/pbi.12868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang WY, Yanfang F, Deepak R, Maeder ML, Tsai SQ, Sander JD, Peterson RT, Joanna J-R, Keith J. Efficient genome editing in zebrafish using a CRISPR-Cas system. Nat Biotechnol. 2013;31:227–229. doi: 10.1038/nbt.2501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikeda M, Miura K, Aya K, Kitano H, Matsuoka M. Genes offering the potential for designing yield-related traits in rice. Curr Opin Plant Biol. 2013;16:213–220. doi: 10.1016/j.pbi.2013.02.002. [DOI] [PubMed] [Google Scholar]
- Jasin M, Haber JE. The democratization of gene editing: insights from site-specific cleavage and double-strand break repair. DNA Repair. 2016;44:6–16. doi: 10.1016/j.dnarep.2016.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang W, Marraffini LA. CRISPR-Cas: new tools for genetic manipulations from bacterial immunity systems. Annu Rev Microbiol. 2015;69:209–228. doi: 10.1146/annurev-micro-091014-104441. [DOI] [PubMed] [Google Scholar]
- Jiang W, Bikard D, Cox D, Zhang F, Marraffini LA. RNA-guided editing of bacterial genomes using CRISPR-Cas systems. Nat Biotechnol. 2013;31:233. doi: 10.1038/nbt.2508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang W, Zhou H, Bi H, Fromm M, Yang B, Weeks DP. Demonstration of CRISPR/Cas9/sgRNA-mediated targeted gene modification in Arabidopsis, tobacco, sorghum and rice. Nucleic Acids Res. 2013;41:e188. doi: 10.1093/nar/gkt780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin S, Zong Y, Gao Q, Zhu Z, Wang Y, Qin P, Liang C, Wang D, Qiu JL, Zhang F, et al. Cytosine, but not adenine, base editors induce genome-wide off-target mutations in rice. Science. 2019;364:292–295. doi: 10.1126/science.aaw7166. [DOI] [PubMed] [Google Scholar]
- Jinek M, Chylinski K, Fonfara I, Hauer M, Doudna JA, Charpentier E. A programmable dual-RNA–guided DNA endonuclease in adaptive bacterial immunity. Science. 2012;337:816–821. doi: 10.1126/science.1225829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kan Y, Ruis B, Takasugi T, Hendrickson EA. Mechanisms of precise genome editing using oligonucleotide donors. Genome Res. 2017;27:1099–1111. doi: 10.1101/gr.214775.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keskin H, Shen Y, Huang F, Patel M, Yang T, Ashley K, Mazin AV, Storici F. Transcript-RNA-templated DNA recombination and repair. Nature. 2014;515:436. doi: 10.1038/nature13682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim D, Kim J, Hur JK, Been KW, Yoon S, Kim JS. Genome-wide analysis reveals specificities of Cpf1 endonucleases in human cells. Nat Biotechnol. 2016;34:863. doi: 10.1038/nbt.3609. [DOI] [PubMed] [Google Scholar]
- Kim H, Kim ST, Ryu J, Kang BC, Kim JS, Kim SG. CRISPR/Cpf1-mediated DNA-free plant genome editing. Nat Commun. 2017;8:14406. doi: 10.1038/ncomms14406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knoll A, Fauser F, Puchta H. DNA recombination in somatic plant cells: mechanisms and evolutionary consequences. Chromosome Res. 2014;22:191–201. doi: 10.1007/s10577-014-9415-y. [DOI] [PubMed] [Google Scholar]
- Komor AC, Kim YB, Packer MS, Zuris JA, Liu DR. Programmable editing of a target base in genomic DNA without double-stranded DNA cleavage. Nature. 2016;533:420–424. doi: 10.1038/nature17946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee K, Zhang Y, Kleinstiver BP, Guo JA, Aryee MJ, Miller J, Malzahn A, Zarecor S, Lawrence-Dill CJ, Joung JK, et al. Activities and specificities of CRISPR/Cas9 and Cas12a nucleases for targeted mutagenesis in maize. Plant Biotechnol J. 2019;17:362–372. doi: 10.1111/pbi.12982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z, Liu ZB, Xing A, Moon BP, Koellhoffer JP, Huang L, Ward RT, Clifton E, Falco SC, Cigan AM. Cas9-guide RNA directed genome editing in soybean. Plant Physiol. 2015;169:960–970. doi: 10.1104/pp.15.00783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Meng X, Zong Y, Chen K, Zhang H, Liu J, Li J, Gao C. Gene replacements and insertions in rice by intron targeting using CRISPR-Cas9. Nat Plants. 2016;2:16139. doi: 10.1038/nplants.2016.139. [DOI] [PubMed] [Google Scholar]
- Li J, Sun Y, Du J, Zhao Y, Xia L. Generation of targeted point mutations in rice by a modified CRISPR/Cas9 system. Mol Plant. 2017;10:526–529. doi: 10.1016/j.molp.2016.12.001. [DOI] [PubMed] [Google Scholar]
- Li J, Zhang X, Sun Y, Zhang J, Du W, Guo X, Li S, Zhao Y, Xia L. Efficient allelic replacement in rice by gene editing: a case study of the NRT1.1B gene. J Integr Plant Biol. 2018;60:536–540. doi: 10.1111/jipb.12650. [DOI] [PubMed] [Google Scholar]
- Li S, Li J, Zhang J, Du W, Fu J, Sutar S, Zhao Y, Xia L. Synthesis-dependent repair of Cpf1-induced double strand DNA breaks enables targeted gene replacement in rice. J Exp Bot. 2018;69:4715–4721. doi: 10.1093/jxb/ery245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S, Tian Y, Wu K, Ye Y, Yu J, Zhang J, Liu Q, Hu M, Li H, Tong Y, et al. Modulating plant growth–metabolism coordination for sustainable agriculture. Nature. 2018;560:595–600. doi: 10.1038/s41586-018-0415-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S, Zhang X, Wang W, Guo X, Wu Z, Du W, Zhao Y, Xia L. Expanding the scope of CRISPR/Cpf1-mediated genome editing in rice. Mol Plant. 2018;11:995–998. doi: 10.1016/j.molp.2018.03.009. [DOI] [PubMed] [Google Scholar]
- Li S, Li J, He Y, Xu M, Zhang J, Du W, Zhao Y, Xia L. Precise gene replacement in rice by RNA transcript-templated homologous recombination. Nat Biotechnol. 2019;37:445–450. doi: 10.1038/s41587-019-0065-7. [DOI] [PubMed] [Google Scholar]
- Lin S, Staahl BT, Alla RK, Doudna JA (2014) Enhanced homology-directed human genome engineering by controlled timing of CRISPR/Cas9 delivery. eLife 3:e04766 [DOI] [PMC free article] [PubMed]
- Lin L, He X, Zhao T, Gu L, Liu Y, Liu X, Liu H, Yang F, Tu M, Tang L, et al. Engineering the direct repeat sequence of crRNA for optimization of FnCpf1-mediated genome editing in human cells. Mol Ther. 2018;26:2650–2657. doi: 10.1016/j.ymthe.2018.08.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ling HQ, Ma B, Shi X, Liu H, Dong L, Sun H, Cao Y, Gao Q, Zheng S, Li Y, et al. Genome sequence of the progenitor of wheat A subgenome Triticum urartu. Nature. 2018;557:424–428. doi: 10.1038/s41586-018-0108-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Wu H, Chen H, Liu Y, He J, Kang H, Sun Z, Pan G, Wang Q, Hu J, et al. A gene cluster encoding lectin receptor kinases confers broad-spectrum and durable insect resistance in rice. Nat Biotechnol. 2014;33:301. doi: 10.1038/nbt.3069. [DOI] [PubMed] [Google Scholar]
- Liu JJ, Orlova N, Oakes BL, Ma E, Spinner HB, Baney KLM, Chuck J, Tan D, Knott GJ, Harrington LB, et al. CasX enzymes comprise a distinct family of RNA-guided genome editors. Nature. 2019;566:218–223. doi: 10.1038/s41586-019-0908-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu Y, Zhu JK. Precise editing of a target base in the rice genome using a modified CRISPR/Cas9 system. Mol Plant. 2017;10:523–525. doi: 10.1016/j.molp.2016.11.013. [DOI] [PubMed] [Google Scholar]
- Ma X, Zhang Q, Zhu Q, Liu W, Chen Y, Qiu R, Wang B, Yang Z, Li H, Lin Y, et al. A robust CRISPR/Cas9 system for convenient, high-efficiency multiplex genome editing in monocot and dicot plants. Mol plant. 2015;8:1274–1284. doi: 10.1016/j.molp.2015.04.007. [DOI] [PubMed] [Google Scholar]
- Ma M, Zhuang F, Hu X, Wang B, Wen XZ, Ji JF, Xi JJ. Efficient generation of mice carrying homozygous double-floxp alleles using the Cas9-Avidin/Biotin-donor DNA system. Cell Res. 2017;27:578. doi: 10.1038/cr.2017.29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mai CD, Phung NT, To HT, Gonin M, Hoang GT, Nguyen KL, Do VN, Courtois B, Gantet P. Genes controlling root development in rice. Rice. 2014;7:30. doi: 10.1186/s12284-014-0030-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makarova KS, Wolf YI, Alkhnbashi OS, Costa F, Shah SA, Saunders SJ, Barrangou R, Brouns SJJ, Charpentier E, Haft DH, et al. An updated evolutionary classification of CRISPR-Cas systems. Nat Rev Microbiol. 2015;13:722. doi: 10.1038/nrmicro3569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mali P, Yang L, Esvelt KM, Aach J, Guell M, DiCarlo JE, Norville JE, Church GM. RNA-guided human genome engineering via Cas9. Science. 2013;339:823–826. doi: 10.1126/science.1232033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malzahn AA, Tang X, Lee K, Ren Q, Sretenovic S, Zhang Y, Chen H, Kang M, Bao Y, Zheng X, et al. Application of CRISPR-Cas12a temperature sensitivity for improved genome editing in rice, maize, and Arabidopsis. BMC Biol. 2019;17:9. doi: 10.1186/s12915-019-0629-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao Y, Zhang H, Xu N, Zhang B, Gou F, Zhu J-K. Application of the CRISPR-Cas system for efficient genome engineering in plants. Mol Plant. 2013;6:2008–2011. doi: 10.1093/mp/sst121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mazur BJ, Chui CF, Smith JK. Isolation and characterization of plant genes coding for Acetolactate Synthase, the target enzyme for two classes of herbicides. Plant Physiol. 1987;85:1110–1117. doi: 10.1104/pp.85.4.1110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miao J, Guo D, Zhang J, Huang Q, Qin G, Zhang X, Wan J, Gu H, Qu LJ. Targeted mutagenesis in rice using CRISPR-Cas system. Cell Res. 2013;23:1233–1236. doi: 10.1038/cr.2013.123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miki D, Zhang W, Zeng W, Feng Z, Zhu J-K. CRISPR/Cas9-mediated gene targeting in Arabidopsis using sequential transformation. Nat Commun. 2018;9:1967. doi: 10.1038/s41467-018-04416-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moon SB, Lee JM, Kang JG, Lee NE, Ha DI, Kim DY, Kim SH, Yoo K, Kim D, Ko JH, et al. Highly efficient genome editing by CRISPR-Cpf1 using CRISPR RNA with a uridinylate-rich 3′-overhang. Nat Commun. 2018;9:3651. doi: 10.1038/s41467-018-06129-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nowacki M, Vijayan V, Zhou Y, Schotanus K, Doak TG, Landweber LF. RNA-mediated epigenetic programming of a genome-rearrangement pathway. Nature. 2007;451:153. doi: 10.1038/nature06452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ole N, Martin J, Doudna JA. Evolution of CRISPR RNA recognition and processing by Cas9 endonucleases. Nucleic Acids Res. 2014;42:1341–1353. doi: 10.1093/nar/gkt922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osman K, Higgins JD, Sanchez-Moran E, Armstrong SJ, Franklin FCH. Pathways to meiotic recombination in Arabidopsis thaliana. New Phytol. 2011;190:523–544. doi: 10.1111/j.1469-8137.2011.03665.x. [DOI] [PubMed] [Google Scholar]
- Paix A, Folkmann A, Goldman DH, Kulaga H, Grzelak MJ, Rasoloson D, Paidemarry S, Green R, Reed RR, Seydoux G. Precision genome editing using synthesis-dependent repair of Cas9-induced DNA breaks. Proc Natl Acad Sci. 2017;114:E10745–E10754. doi: 10.1073/pnas.1711979114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paquet D, Kwart D, Chen A, Sproul A, Jacob S, Teo S, Olsen KM, Gregg A, Noggle S, Tessier-Lavigne M. Efficient introduction of specific homozygous and heterozygous mutations using CRISPR/Cas9. Nature. 2016;533:125–129. doi: 10.1038/nature17664. [DOI] [PubMed] [Google Scholar]
- Puchta H. Repair of genomic double-strand breaks in somatic plant cells by one-sided invasion of homologous sequences. Plant J. 1998;13:331–339. doi: 10.1046/j.1365-313X.1998.00035.x. [DOI] [Google Scholar]
- Puchta H, Fauser F. Gene targeting in plants: 25 years later. Int J Dev Biol. 2013;57:629–637. doi: 10.1387/ijdb.130194hp. [DOI] [PubMed] [Google Scholar]
- Puchta H, Fauser F. Synthetic nucleases for genome engineering in plants: prospects for a bright future. Plant J. 2014;78:727–741. doi: 10.1111/tpj.12338. [DOI] [PubMed] [Google Scholar]
- Qi Y, Zhang Y, Zhang F, Baller JA, Cleland SC, Ryu Y, Starker CG, Voytas DF. Increasing frequencies of site-specific mutagenesis and gene targeting in Arabidopsis by manipulating DNA repair pathways. Genome Res. 2013;23:547–554. doi: 10.1101/gr.145557.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reiss B, Klemm M, Kosak H, Schell J. RecA protein stimulates homologous recombination in plants. Proc Natl Acad Sci USA. 1996;93:3094–3098. doi: 10.1073/pnas.93.7.3094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reiss B, Schubert I, Köpchen K, Wendeler E, Schell J, Puchta H. RecA stimulates sister chromatid exchange and the fidelity of double-strand break repair, but not gene targeting, in plants transformed by Agrobacterium. Proc Natl Acad Sci USA. 2000;97:3358–3363. doi: 10.1073/pnas.97.7.3358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richardson CD, Ray GJ, DeWitt MA, Curie GL, Corn JE. Enhancing homology-directed genome editing by catalytically active and inactive CRISPR-Cas9 using asymmetric donor DNA. Nat Biotechnol. 2016;34:339–344. doi: 10.1038/nbt.3481. [DOI] [PubMed] [Google Scholar]
- Rolloos M, Hooykaas PJJ, Van Der Zaal BJ. Enhanced targeted integration mediated by translocated I-SceI during the Agrobacterium mediated transformation of yeast. Sci Rep. 2015;5:8345. doi: 10.1038/srep08345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roy KR, Smith JD, Vonesch SC, Lin G, Tu CS, Lederer AR, Chu A, Suresh S, Nguyen M, Horecka J, et al. Multiplexed precision genome editing with trackable genomic barcodes in yeast. Nat Biotechnol. 2018;36:512. doi: 10.1038/nbt.4137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sampson TR, Saroj SD, Llewellyn AC, Tzeng YL, Weiss DS. A CRISPR/Cas system mediates bacterial innate immune evasion and virulence. Nature. 2013;497:254. doi: 10.1038/nature12048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sauer NJ, Narváez-Vásquez J, Mozoruk J, Miller RB, Warburg ZJ, Woodward MJ, Mihiret YA, Lincoln TA, Segami RE, Sanders SL, et al. Oligonucleotide-mediated genome editing provides precision and function to engineered nucleases and antibiotics in plants. Plant Physiol. 2016;170:1917–1928. doi: 10.1104/pp.15.01696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Savic N, Ringnalda FC, Lindsay H, Berk C, Bargsten K, Li Y, Neri D, Robinson MD, Ciaudo C, Hall J (2018) Covalent linkage of the DNA repair template to the CRISPR-Cas9 nuclease enhances homology-directed repair. Elife 7 [DOI] [PMC free article] [PubMed]
- Schiml S, Fauser F, Puchta H. The CRISPR/Cas system can be used as nuclease for in planta gene targeting and as paired nickases for directed mutagenesis in Arabidopsis resulting in heritable progeny. Plant J. 2014;80:1139–1150. doi: 10.1111/tpj.12704. [DOI] [PubMed] [Google Scholar]
- Shaked H, Melamed-Bessudo C, Levy AA. High-frequency gene targeting in Arabidopsis plants expressing the yeast RAD54 gene. Proc Natl Acad Sci USA. 2005;102:12265–12269. doi: 10.1073/pnas.0502601102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shan Q, Baltes NJ, Atkins P, Kirkland ER, Zhang Y, Baller JA, Lowder LG, Malzahn AA, Haugner JC, Seelig B, et al. ZFN, TALEN and CRISPR-Cas9 mediated homology directed gene insertion in Arabidopsis: a disconnect between somatic and germinal cells. J Genet Genom. 2018;45:681–684. doi: 10.1016/j.jgg.2018.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi J, Gao H, Wang H, Lafitte HR, Archibald RL, Yang M, Hakimi SM, Mo H, Habben JE. ARGOS8 variants generated by CRISPR-Cas9 improve maize grain yield under field drought stress conditions. Plant Biotechnol J. 2017;15:207–216. doi: 10.1111/pbi.12603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimatani Z, Kashojiya S, Takayama M, Terada R, Arazoe T, Ishii H, Teramura H, Yamamoto T, Komatsu H, Miura K, et al. Targeted base editing in rice and tomato using a CRISPR-Cas9 cytidine deaminase fusion. Nat Biotechnol. 2017;35:441. doi: 10.1038/nbt.3833. [DOI] [PubMed] [Google Scholar]
- Shmakov S, Abudayyeh Omar O, Makarova Kira S, Wolf Yuri I, Gootenberg Jonathan S, Semenova E, Minakhin L, Joung J, Konermann S, Severinov K, et al. Discovery and functional characterization of diverse class 2 CRISPR-Cas systems. Mol Cell. 2015;60:385–397. doi: 10.1016/j.molcel.2015.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shmakov S, Smargon A, Scott D, Cox D, Pyzocha N, Yan W, Abudayyeh OO, Gootenberg JS, Makarova KS, Wolf YI, et al. Diversity and evolution of class 2 CRISPR-Cas systems. Nat Rev Microbiol. 2017;15:169. doi: 10.1038/nrmicro.2016.184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sorek R, Lawrence CM, Wiedenheft B. CRISPR-mediated adaptive immune systems in bacteria and archaea. Annu Rev Biochem. 2013;82:237–266. doi: 10.1146/annurev-biochem-072911-172315. [DOI] [PubMed] [Google Scholar]
- Storici F, Bebenek K, Kunkel TA, Gordenin DA, Resnick MA. RNA-templated DNA repair. Nature. 2007;447:338–341. doi: 10.1038/nature05720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Y, Li J, Xia L (2016a) Precise genome modification via sequence-specific nucleases-mediated gene targeting for crop improvement. Front Plant Sci 7 [DOI] [PMC free article] [PubMed]
- Sun Y, Zhang X, Wu C, He Y, Ma Y, Hou H, Guo X, Du W, Zhao Y, Xia L. Engineering herbicide-resistant rice plants through CRISPR/Cas9-mediated homologous recombination of Acetolactate synthase. Mol Plant. 2016;9:628–631. doi: 10.1016/j.molp.2016.01.001. [DOI] [PubMed] [Google Scholar]
- Svitashev S, Young JK, Schwartz C, Gao H, Falco SC, Cigan AM. Targeted mutagenesis, precise gene editing, and site-specific gene insertion in maize using Cas9 and guide RNA. Plant Physiol. 2015;169:931–945. doi: 10.1104/pp.15.00793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Symington LS. Role of RAD52 epistasis group genes in homologous recombination and double-strand break repair. Microbiol Mol Biol Rev. 2002;66:630–670. doi: 10.1128/MMBR.66.4.630-670.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Symington LS, Gautier J. Double-strand break end resection and repair pathway choice. Annu Rev Genet. 2011;45:247. doi: 10.1146/annurev-genet-110410-132435. [DOI] [PubMed] [Google Scholar]
- Tang X, Lowder LG, Zhang T, Malzahn AA, Zheng X, Voytas DF, Zhong Z, Chen Y, Ren Q, Li Q, et al. A CRISPR-Cpf1 system for efficient genome editing and transcriptional repression in plants. Nat Plants. 2017;3:17018. doi: 10.1038/nplants.2017.18. [DOI] [PubMed] [Google Scholar]
- Tang X, Liu G, Zhou J, Ren Q, You Q, Tian L, Xin X, Zhong Z, Liu B, Zheng X, et al. A large-scale whole-genome sequencing analysis reveals highly specific genome editing by both Cas9 and Cpf1 (Cas12a) nucleases in rice. Genome Biol. 2018;19:84. doi: 10.1186/s13059-018-1458-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terns MP, Terns RM. CRISPR-based adaptive immune systems. Curr Opin Microbiol. 2011;14:321–327. doi: 10.1016/j.mib.2011.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas KR, Capecchi MR. Site-directed mutagenesis by gene targeting in mouse embryo-derived stem cells. Cell. 1987;51:503–512. doi: 10.1016/0092-8674(87)90646-5. [DOI] [PubMed] [Google Scholar]
- Voytas DF, Gao C. Precision genome engineering and agriculture: opportunities and regulatory challenges. PLoS Biol. 2014;12:e1001877. doi: 10.1371/journal.pbio.1001877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vu TV, Sivankalyani V, Kim EJ, Tran MT, Kim J, Sung YW, Doan DTH, and Kim JY (2019) Highly efficient homology-directed repair using transient CRISPR/Cpf1-geminiviral replicon in tomato. bioRxiv:521419 [DOI] [PMC free article] [PubMed]
- Waltz E. With a free pass, CRISPR-edited plants reach market in record time. Nat Biotechnol. 2018;36:6. doi: 10.1038/nbt0118-6b. [DOI] [PubMed] [Google Scholar]
- Wang M, Lu Y, Botella JR, Mao Y, Hua K, Zhu JK. Gene targeting by homology-directed repair in rice using a geminivirus-based CRISPR/Cas9 system. Mol Plant. 2017;10:1007–1010. doi: 10.1016/j.molp.2017.03.002. [DOI] [PubMed] [Google Scholar]
- Wang M, Mao Y, Lu Y, Tao X, Zhu JK. Multiplex gene editing in rice using the CRISPR-Cpf1 system. Mol Plant. 2017;10:1011–1013. doi: 10.1016/j.molp.2017.03.001. [DOI] [PubMed] [Google Scholar]
- Wang W, Mauleon R, Hu Z, Chebotarov D, Tai S, Wu Z, Li M, Zheng T, Fuentes RR, Zhang F, et al. Genomic variation in 3,010 diverse accessions of Asian cultivated rice. Nature. 2018;557:43–49. doi: 10.1038/s41586-018-0063-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weeks DP, Spalding MH, Yang B. Use of designer nucleases for targeted gene and genome editing in plants. Plant Biotechnol J. 2016;14:483–495. doi: 10.1111/pbi.12448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolter F, Puchta H (2019) In planta gene targeting can be enhanced by the use of CRISPR/Cas12a. Plant J [DOI] [PubMed]
- Wolter F, Klemm J, Puchta H. Efficient in planta gene targeting in Arabidopsis using egg cell-specific expression of the Cas9 nuclease of Staphylococcus aureus. Plant J. 2018;94:735–746. doi: 10.1111/tpj.13893. [DOI] [PubMed] [Google Scholar]
- Xie K, Yang Y. RNA-guided genome editing in plants using a CRISPR–Cas system. Mol Plant. 2013;6:1975–1983. doi: 10.1093/mp/sst119. [DOI] [PubMed] [Google Scholar]
- Xie K, Zhang J, Yang Y. Genome-wide prediction of highly specific guide RNA spacers for CRISPR-Cas9-mediated genome editing in model plants and major crops. Mol Plant. 2014;7:923–926. doi: 10.1093/mp/ssu009. [DOI] [PubMed] [Google Scholar]
- Xu Q, Zhao M, Wu K, Fu X, Liu Q. Emerging insights into heterotrimeric G protein signaling in plants. J Genet Genom. 2016;43:495–502. doi: 10.1016/j.jgg.2016.06.004. [DOI] [PubMed] [Google Scholar]
- Xu R, Qin R, Li H, Li D, Li L, Wei P, Yang J. Generation of targeted mutant rice using a CRISPR-Cpf1 system. Plant Biotechnol J. 2017;15:713–717. doi: 10.1111/pbi.12669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan WX, Hunnewell P, Alfonse LE, Carte JM, Keston-Smith E, Sothiselvam S, Garrity AJ, Chong S, Makarova KS, Koonin EV, et al. Functionally diverse type V CRISPR-Cas systems. Science. 2019;363:88–91. doi: 10.1126/science.aav7271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang L, Guell M, Byrne S, Yang JL, De Los Angeles A, Mali P, Aach J, Kim-Kiselak C, Briggs AW, Rios X, et al. Optimization of scarless human stem cell genome editing. Nucleic Acids Res. 2013;41:9049–9061. doi: 10.1093/nar/gkt555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu QH, Wang B, Li N, Tang Y, Yang S, Yang T, Xu J, Guo C, Yan P, Wang Q, et al. CRISPR/Cas9-induced targeted mutagenesis and gene replacement to generate long-shelf life tomato lines. Sci Rep. 2017;7:11874. doi: 10.1038/s41598-017-12262-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaidi SSA, Vanderschuren H, Qaim M, Mahfouz MM, Kohli A, Mansoor S, Tester M. New plant breeding technologies for food security. Science. 2019;363:1390–1391. doi: 10.1126/science.aav6316. [DOI] [PubMed] [Google Scholar]
- Zetsche B, Gootenberg JS, Abudayyeh OO, Slaymaker IM, Makarova KS, Essletzbichler P, Volz SE, Joung J, van der Oost J, Regev A, et al. Cpf1 is a single RNA-guided endonuclease of a class 2 CRISPR-Cas system. Cell. 2015;163:759–771. doi: 10.1016/j.cell.2015.09.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zetsche B, Heidenreich M, Mohanraju P, Fedorova I, Kneppers J, DeGennaro EM, Winblad N, Choudhury SR, Abudayyeh OO, Gootenberg JS, et al. Multiplex gene editing by CRISPR-Cpf1 using a single crRNA array. Nat Biotechnol. 2017;35:178. doi: 10.1038/nbt0217-178b. [DOI] [PubMed] [Google Scholar]
- Zhang JP, Li XL, Li GH, Chen W, Arakaki C, Botimer GD, Baylink D, Zhang L, Wen W, Fu YW, et al. Efficient precise knockin with a double cut HDR donor after CRISPR/Cas9-mediated double-stranded DNA cleavage. Genome Biol. 2017;18:35. doi: 10.1186/s13059-017-1164-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong Z, Zhang Y, You Q, Tang X, Ren Q, Liu S, Yang L, Wang Y, Liu X, Liu B, et al. Plant genome editing using FnCpf1 and LbCpf1 nucleases at redefined and altered PAM sites. Mol Plant. 2018;11:999–1002. doi: 10.1016/j.molp.2018.03.008. [DOI] [PubMed] [Google Scholar]
- Zuo E, Sun Y, Wei W, Yuan T, Ying W, Sun H, Yuan L, Steinmetz LM, Li Y, Yang H. Cytosine base editor generates substantial off-target single-nucleotide variants in mouse embryos. Science. 2019;364:289–292. doi: 10.1126/science.aav9973. [DOI] [PMC free article] [PubMed] [Google Scholar]