Abstract
Common wheat (Triticum aestivum L.) is one of the three major food crops in the world; thus, wheat breeding programs are important for world food security. Characterizing the genes that control important agronomic traits and finding new ways to alter them are necessary to improve wheat breeding. Functional genomics and breeding in polyploid wheat has been greatly accelerated by the advent of several powerful tools, especially CRISPR/Cas9 genome editing technology, which allows multiplex genome engineering. Here, we describe the development of CRISPR/Cas9, which has revolutionized the field of genome editing. In addition, we emphasize technological breakthroughs (e.g., base editing and prime editing) based on CRISPR/Cas9. We also summarize recent applications and advances in the functional annotation and breeding of wheat, and we introduce the production of CRISPR-edited DNA-free wheat. Combined with other achievements, CRISPR and CRISPR-based genome editing will speed progress in wheat biology and promote sustainable agriculture.
Keywords: Wheat, CRISPR/Cas9, Genome editing, Functional genomics, Breeding
Introduction
Common wheat is a keystone crop species. It is grown in many different environments, providing most humans with around 20% of their calories and protein (Uauy et al. 2017); thus, it occupies an important position in food security. As the global population increases, improving the yield of wheat is critical to ensure future availability. Geneticists have exploited natural or artificial wheat variations for breeding. Indeed, conventional breeding approaches have played a major role in increasing grain yields and quality based on broad genetic variations in wheat (Nadolska-Orczyk et al. 2017). However, wheat is an allohexaploid (2n = 6 × = 42, AABBDD); it harbors three closely related subgenomes inherited from three homoeologous ancestors (Petersen et al. 2006). Thus, most wheat genes have three similar but not identical copies, with functional redundancy and complementarity among the A, B, and D genomes. As a result, the probability of the simultaneous mutation of genes in the A, B, and D genomes by natural processes or induced mutagenesis is very low. Therefore, the complex polyploid nature of wheat has hindered the development of functional genomics and breeding, especially compared to other cereals, such as rice and maize.
Several genome editing technologies with the ability to change the code of life with high specificity have been developed recently (Li et al. 2019). Advances in genome editing have revolutionized life science, including plant science. The clustered regularly interspaced short palindromic repeats (CRISPR)/CRISPR-associated protein 9 (Cas9) system offers several advantages, including simplicity, versatility, high efficiency, and the ability to work with multiple targets simultaneously (multiplexing); it has surpassed other genome editing tools, becoming the most widely used gene editing technology in the world (Doudna and Charpentier 2014). Thus far, CRISPR/Cas9 has been used to create various targeted mutations in a broad range of living organisms (Char and Yang 2020; Gürel et al. 2020).
Application of the CRISPR/Cas9 system requires the DNA sequences of the target genes. Given the availability of the annotated wheat genome and the elucidation of a growing number of genes controlling important agronomic traits in other plants, it is easy to isolate orthologous genes in wheat based on homology-based cloning. In addition, CRISPR/Cas9 allows researchers to target multiple homoeoalleles simultaneously and it enables the production of targeted mutations in all copies of a gene; thus, the system holds great promise in the characterization of genes endowing important agronomic traits in polyploid wheat. Furthermore, it has been used to modify multiple genes controlling different agronomic traits in wheat. This technology will bring a new dawn to wheat biology and breeding programs. In this review, we briefly outline the utilization of the CRISPR/Cas9 system, with an emphasis on the most important breakthroughs thus far. We also summarize recent applications of genome editing in wheat. Finally, we discuss the future prospects of CRISPR/Cas9 genome editing for wheat improvement.
Genome editing with CRISPR/Cas9
Mechanism of genome editing
Genome editing technology generates site-specific double-strand breaks (DSBs) in the targeted genomic sequence using programmable sequence-specific nucleases (SSNs), and then exploits endogenous DSB repair mechanisms to generate a variety of mutations in the target region. Three types of SSNs are used to introduce DSBs at selected sites: zinc-finger nucleases, transcription activator-like effector nucleases, and CRISPR/Cas9 (Kim and Kim 2014; Zhan et al. 2020). Subsequently, the DSBs are mainly repaired via two pathways, nonhomologous end joining (NHEJ) and homologous recombination (HR) (Symington and Gautier 2011). In NHEJ, two broken ends are simply re-ligated, producing insertions and/or deletions (indels) in the target site. When homologous donor sequences are present at the DSBs, HR may be used, and desired gene modifications occur (Chen et al. 2019; Anzalone et al. 2020). Overall, SSN-induced DSBs are repaired more frequently by the NHEJ pathway than by the HR pathway (Carroll 2014; Gao 2021).
CRISPR/Cas9
CRISPR/Cas systems provide a defense against foreign plasmids or viral DNA elements in bacteria and archaea. They are divided into six types based on the assortment of cas genes and nature of the interference complex (Hille et al. 2018). Three components, mature crRNA, tracrRNA, and Cas9, are responsible for cleaving the invading elements in type II CRISPR/Cas systems. To simplify the system, a dual tracrRNA:crRNA was designed as a single guide RNA (sgRNA) to direct the production of DSBs by Cas9 in vitro (Jinek et al. 2012). Subsequently, an RNA-programmable genome editing tool, CRISPR/Cas9, was developed to create targeted mutations (Cong et al. 2013; Mali et al. 2013; Zhang et al. 2019c).
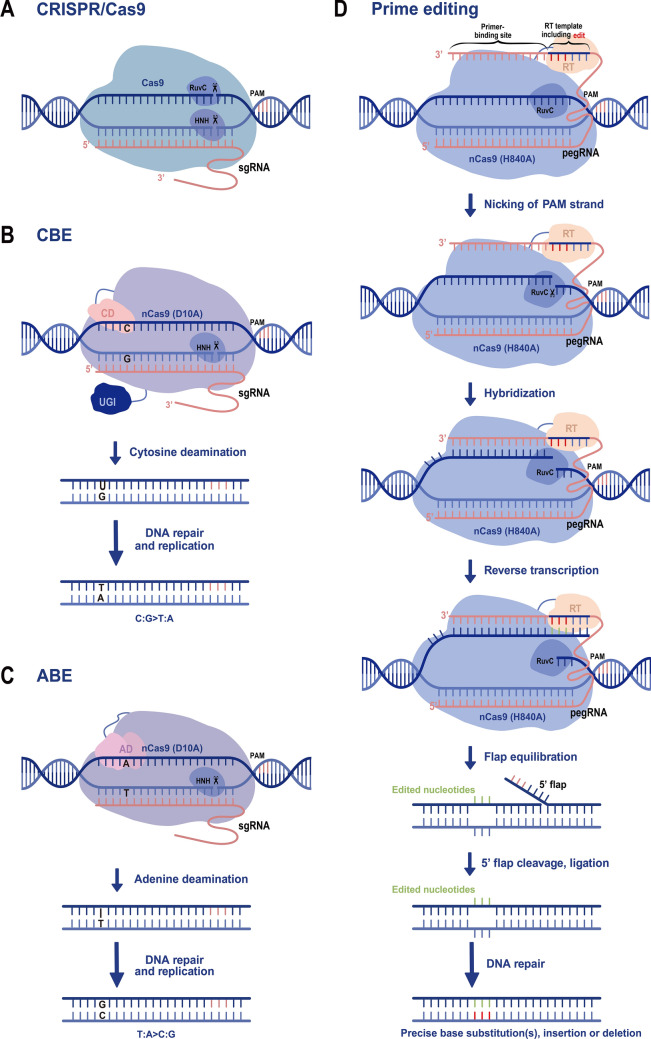
CRISPR/Cas9 contains two major components: a sgRNA, which is responsible for recognizing target DNA, and the Cas9 endonuclease, which is responsible for generating DSB at predesigned target DNA site (Fig. 1A). Cas9 from Streptococcus pyogenes (SpCas9) was the first well-characterized RNA-guided endonuclease. It is a multifunctional protein that contains two nuclease domains: the HNH domain and RuvC-like domain. Each of them cuts one DNA strand, generating blunt-end DSBs; this triggers endogenous DNA repair systems, resulting in targeted mutants. The only prerequisite for applying CRISPR/Cas9 to a given site is the presence of a protospacer-adjacent motif (PAM; NGG for SpCas9) next to the sequence of interest. For different target sites, Cas9 is constant; we can only change the guide sequence in the sgRNA.
Fig. 1.
Overview of CRISPR/Cas9 and CRISPR/Cas9-based precise genome editing. A CRISPR/Cas9-mediated genome editing. CRISPR/Cas9 contains two components: Cas9 and a sgRNA. Guided by the 20-nucleotide sequence within sgRNAs, Cas9 cleaves the DNA double-strand, generating a blunt-end DSB. B Mechanism of CBE-mediated C-to-T base editing. The CBE is comprised of nCas9 (D10A) fused with two proteins [cytosine deaminase (CD) and uracil DNA glycosylase inhibitor (UGI)] and a sgRNA. Guided by the 20-nucleotide sequence within sgRNAs, CD converts C to U within the window. With DNA repair and replication, it generates C-to-T substitutions. C Mechanism of ABE-mediated A-to-G base editing. The ABE is comprised of nCas9 (D10A) fused with adenine deaminase (AD, TadA-TadA* heterodimers), and a sgRNA. Guided by the 20-nucleotide sequence within sgRNAs, AD converts A to I within the window. With DNA repair and replication, it generates A-to-G substitutions. D Mechanism of prime editing. It contains nCas9 (H840A) fused with reverse transcriptase (RT) and pegRNA. Guided by the 20-nucleotide sequence within pegRNAs, RT primes new DNA containing the desired editing at the targeted site. After flap equilibration, cleavage, ligation, and DNA repair, the desired editing is incorporated
Base editing
Base editing, borrowed from CRISPR, is a precise genome editing approach. It generates targeted point mutations without DSBs, foreign donor templates, or HR (Komor et al. 2016; Gaudelli et al. 2017). Current base editors usually contain a sgRNA, and a catalytically impaired Cas9 nuclease [dead Cas9 (dCas9) or Cas9 nickase (Cas9n)] fused with ssDNA deaminase. The sgRNA guides the modified Cas9-deaminase to the target locus, generating ssDNA R-loop that is exposed and accessible to the deaminase (Anzalone et al. 2020). Based on the different kinds of deaminase, there are two major groups of DNA base editors: cytidine base editors (CBEs) and adenine base editors (ABEs).
With CBEs, cytidine deaminase is used to convert cytidine (C) to uridine (U) within the editing window, creating a mismatched base pair with guanine (G) on the opposite strand (Fig. 1B). However, the U intermediate is mutagenic; most organisms have evolved a uracil base excision repair (BER) pathway to excise U from genomic DNA with uracil DNA N-glycosylase. Therefore, uracil glycosylase inhibitor protein (UGI) is used to impede uracil excision, increasing the C-to-T editing efficiency of CBEs (Komor et al. 2016; Nishida et al. 2016). This approach was first used in yeast and human cells and then applied to a variety of plants, including wheat, rice, maize, tomato, and Arabidopsis (Zong et al. 2017; Shimatani et al. 2017; Chen et al. 2017).
Theoretically, dCas9/nCas9 fused with an adenosine deaminase would yield an ABE. However, there is no known natural deaminase that deaminates adenine in DNA. By extensive directed evolution and protein engineering of Escherichia coli tRNA adenine deaminase (TadA), researchers produced a deaminase variant (TadA*) that can deaminate adenine (A) in DNA. TadA-TadA* heterodimers were fused with dCas9/nCas9 to generate an ABE (Fig. 1C); adenine is deaminated to inosine (I), treated as G by the polymerase, converting AT to GC base pairs in human cells (Gaudelli et al. 2017). Several groups have shown that ABEs can be applied to plants, including wheat, rice, potato, Arabidopsis, and Brassica napus (Kang et al. 2018; Zong et al. 2018).
Many important agronomic traits involve single-nucleotide variants (Zhao et al. 2011; Hu et al. 2015). Therefore, precise editing of a single nucleotide in plants is a desirable and powerful means of accelerating crop improvement. Base editors (CBEs and ABEs) can efficiently mediate all four transition mutations (C → T, A → G, T → C, and G → A) at targeted loci; this will undoubtedly facilitate basic research and breeding in plants.
Prime editing
Prime editing, borrowed from CRISPR, is another precise genome editing method that can generate all 12 types of base substitutions, targeted small insertions, deletions, and combinations of these editing results in the target site. The prime editor mainly consists of a catalytically impaired Cas9 (Cas9n, H840A) fused with an engineered reverse transcriptase and a prime editing guide RNA (pegRNA) (Fig. 1D). The latter contains a primer-binding site at the 3′ end of the sgRNA and a reverse transcriptase template, specifying the target site and also encoding the desired sequence edit (Anzalone et al. 2019).
In the prime editing system, the complex binds to the target DNA site and nicks the PAM-containing strand, generating a 3′ end. Then, it hybridizes to the primer-binding site of the pegRNA and primes reverse transcription of the template containing the desired edits. After equilibration between the edited 3′ flap and the unedited 5′ flap, 5′ flap excision, ligation, and DNA repair, DNAs are stably edited in the desired manner (Anzalone et al. 2019). It was first used to correct mutations in human cells and has since been successfully applied to rice plants and wheat protoplasts (Li et al. 2020a; Lin et al. 2020; Xu et al. 2020; Hua et al. 2020). This search-and-replace method is a versatile and precise genome editing tool that does not require DSBs, or donor DNA templates.
Recent applications of genome editing in wheat
When just one homolog mutates, no mutant phenotype might be observed due to masking by other homologs (Borrill et al. 2015). This genetic redundancy and complementarity has hindered the development of wheat biology. Now that CRISPR/Cas9 has been used to target multiple homoeoalleles simultaneously (Table 1), it will accelerate progress in functional genomics and molecular breeding in wheat.
Table 1.
List of wheat endogenous gene(s) targeted using CRISPR in the literature
| Targeted gene(s) | Genome editing method | PAM type(s) | Repair result | Delivery | Efficiency | Reference |
|---|---|---|---|---|---|---|
| MLO | CRISPR/Cas9 | CCN | Indels | Particle bombardment | 5.6% | Wang et al. (2014) |
| GASR7, GW2, DEP1, NAC2, PIN1, LOX2 | CRISPR/Cas9 | CCN, NGG | Indels | Particle bombardment | 1.0–9.5% | Zhang et al. (2016) |
| EDR1 | CRISPR/Cas9 | CCN | Indels | Particle bombardment | N.A | Zhang et al. (2017) |
| gliadin | CRISPR/Cas9 | CCN | Indels | Particle bombardment | 62.3–75.1% | Sanchez-Leon et al. (2018) |
| PDS | CRISPR/Cas9 | CCN, NGG | Indels | Agrobacterium | 11–17% | Howells et al. (2018) |
| GW2, Lpx-1, MLO | CRISPR/Cas9 | CCN, NGG | Indels | Particle bombardment | 3–32% | Wang et al. (2018a) |
| CKX2-1, GLW7, GW2, GW8 | CRISPR/Cas9 | CCN, NGG | Indels | Agrobacterium | 10% | Zhang et al. (2019d) |
| Qsd1 | CRISPR/Cas9 | NGG | Indels | Agrobacterium | 37.5% | Abe et al. (2019) |
| DA1, DA2, NCED1, LPR2 | CRISPR/Cas9 | CCN, NGG | Indels | Agrobacterium | 20.8–54.2% | Zhang et al. (2019b) |
| GW7 | CRISPR/Cas9 | NGG | Indels | Particle bombardment | 1.1–8.3% | Wang et al. (2019) |
| Ms45 | CRISPR/Cas9 | NGG | Indels | Agrobacterium | N.A | Singh et al. (2018) |
| Ms1 | CRISPR/Cas9 | CCN, NGG | Indels | Agrobacterium | 5% | Okada et al. (2019) |
| NP1 | CRISPR/Cas9 | NGG | Indels | Particle bombardment | 14.0–54.2% | Li et al. (2020b) |
| Ms2 | CRISPR/Cas9 | CCN, NGG | Indels | Agrobacterium | 2.3–8.27% | Tang et al. (2020) |
| MTL, Waxy | CRISPR/Cas9 | CCN, NGG | Indels | Agrobacterium | 8.9–71.3% | Liu et al. (2020a) |
| Q | CRISPR/Cas9 | NGG | Indels | Agrobacterium | 44.0–45.6% | Liu et al. (2020b) |
| CENH3 | CRISPR/Cas9 | CCN, NGG | Indels, restored frameshift | Particle bombardment | N.A | Lv et al. (2020) |
| SBEIIa | CRISPR/Cas9 | CCN, NGG | Indels | Particle bombardment | 38.5–66.7% | Li et al. (2020c) |
| LOX2 | BE3 | NGG | C-to-T conversion | Particle bombardment | 1.25% | Zong et al. (2017) |
| ALS, ACCase | BE3 | CCN, NGG | C-to-T conversion | Particle bombardment | 2.5% | Zhang et al. (2019a) |
| ALS, MTL | A3A-PBE | NGG | C-to-T conversion | Particle bombardment | 16.7–22.5% | Zong et al. (2018) |
| DEP1, GW2 | ABE7.10 | NGG | A-to-G conversion | Particle bombardment | 0.4–1.1% | Li et al. (2018) |
| GW2, LOX2, MLO, GASR7, DME | Prime editing | NGG | Point mutations, targeted insertions and deletions | Particle bombardment | N.A | Lin et al. (2020) |
NA not available
Improving wheat yields and grain quality
As the world’s population grows, wheat yields will need to substantially increase to ensure global food security. This fact has pushed scientists to investigate and breed innovative wheat varieties. Using CRISPR/Cas9, many negative regulatory genes have been knocked out to improve wheat yields and quality. For example, GASR7 is a gibberellin-regulated gene that controls grain length in rice. Simultaneous targeting of all three TaGASR7 homoeologs significantly elevated the thousand kernel weight, irrespective of the varietal background (Zhang et al. 2016). Similarly, GW2, encoding a RING-type E3 ligase that controls rice grain weight, was knocked out to increase the length and width of wheat grains and, hence, grain yields (Wang et al. 2018b; Zhang et al. 2018).
To meet different customers’ needs, grain quality is an important trait that should be improved. For example, gluten proteins, encoded by gliadin genes in wheat, are major factors triggering celiac disease in genetically predisposed individuals. Researchers designed two sgRNAs to target the conserved region of a-gliadin genes; they generated low-gluten wheat, for which the immunoreactivity was reduced by 85% (Sanchez-Leon et al. 2018). Moreover, targeted mutagenesis of TaSBEIIa by CRISPR/Cas9 successfully generated high-amylose wheat with a significantly increased resistant starch content (Li et al. 2020d). Therefore, wheat yields and quality traits can be successfully improved using CRISPR/Cas9.
Speeding wheat hybrid seed production
Heterosis is widely exploited to improve crop productivity and other agronomic traits. However, due to wheat’s strong inbreeding habit, it has been a great challenge to develop male-sterile wheat lines for the production of hybrid seed (Singh et al. 2018). The identification and manipulation of male sterility genes is the first step to generate novel male-sterile wheat mutants. Given the recent molecular identification of male fertility genes in plants, it is possible to clone the homoeoalleles using homology-based cloning, and then create male-sterile hexaploid wheat lines using CRISPR/Cas9. Indeed, great progress has been achieved recently. For instance, NP1 encodes a putative glucose-methanol-choline oxidoreductase that is required for male sterility in rice (Chang et al. 2016). Using an optimized CRISPR/Cas9 system, our group simultaneously disrupted three TaNP1 homoeoalleles in wheat. The resulting Tanp1 triple mutants showed complete male sterility (i.e., produced no pollen) (Li et al. 2020b). Similarly, the targeted knockout of Ms1, which is responsible for pollen exine development and male fertility (Tucker et al. 2017; Wang et al. 2017b), produced complete male sterility in commercial wheat cultivars (Okada et al. 2019). These male sterility mutants could accelerate hybrid breeding of wheat. Very recently, TaCENH3α was edited using CRISPR/Cas9; paternal haploid inducer wheat lines were generated with an induction rate of ~ 7% (Lv et al. 2020). This could be used for additional new breeding technologies and paves the way for reducing the cost of goods in wheat seed production.
Increasing disease resistance in wheat
Diseases induced by fungi, bacteria, and viruses could reduce wheat yields and quality dramatically. CRISPR/Cas9 has been used to knock out disease-susceptibility genes to generate disease-resistant wheat. In plants, a loss of function of MILDEW-RESISTANCE LOCUS (MLO) confers broad-spectrum resistance to powdery mildew (Gil-Humanes and Voytas 2014). This makes MLO an ideal target for CRISPR/Cas9 to enhance resistance to powdery mildew. Researchers knocked out all six MLO alleles in wheat; they produced a Tamlo triple mutant showing increased resistance to powdery mildew disease (Wang et al. 2014). Similarly, the gene encoding enhanced disease resistance1 (EDR1), a negative factor against powdery mildew defenses, has been simultaneously modified using CRISPR/Cas9, generating wheat with improved powdery mildew resistance (Zhang et al. 2017). Thus, CRISPR/Cas9 is an important means to enhance disease resistance in wheat.
Generating CRISPR-edited DNA-free wheat
CRISPR/Cas9 is widely used to improve agricultural traits by knocking out unwanted genes or genes conferring undesirable phenotypes. However, this process usually involves transgenic intermediates, which causes regulatory concerns and is not accepted worldwide (Zhang et al. 2020). For public acceptance, gene removal or bypassing foreign elements to edit endogenous genes is a good choice (He and Zhao 2020). Based on the reagents needed for CRISPR-mediated editing, there are two main ways to produce CRISPR-edited DNA-free plants.
In the vector-based method, a vector is delivered into wheat callus using Agrobacterium or particle bombardment. It then integrates into the genome and the encoded genome editing elements are expressed, enabling targeted gene knockout. Targeted knockout wheat with foreign DNA is generated in the T0 generation. Ultimately, the foreign DNA can be segregated by selfing and crossing. For example, researchers created a triple-knockout mutant of TaQsd1 via Agrobacterium-delivered CRISPR/Cas9. The mutant was then crossed with wild-type wheat plants, producing transgene-free triple-recessive TaQsd1 mutants that exhibited longer seed dormancy (Abe et al. 2019). Similarly, a marker-free wheat mutant was obtained among the offspring of T0 plants (Wang et al. 2017a).
Sometimes, vectors are not integrated into the genome; instead, they may transiently express their encoded genome editing elements to knock out genes. A targeted gene-modified plantlet without foreign DNA is generated in the T0 generation. This approach has been reported in wheat for the first time. Researchers delivered vectors containing CRISPR/Cas9 elements into wheat callus through particle bombardment; the plantlet was subsequently regenerated without antibiotic selection. This transient expression-based CRISPR/Cas9 system produced transgene-free, homozygous mutants (Zhang et al. 2016). In addition, transgene-free wheat carrying nucleotide substitutions have been generated by transiently expressing CBEs or ABEs (Zong et al. 2017; Li et al. 2018).
In the non-vector method, Cas9 and sgRNAs are transcribed in vitro and then delivered into immature wheat embryos through particle bombardment. DNA-free genome-edited wheat plants have been generated. Though the editing efficiency was lower, the specificity was higher than with a vector-based system (Zhang et al. 2016). Moreover, nCas9-PBE mRNA and sgRNA were transcribed in vitro and delivered into immature wheat embryos. DNA-free base editing at TaALS-P174 was obtained, endowing wheat with resistance to the herbicide nicosulfuron (Zhang et al. 2019a). In addition, Cas9 can be expressed in vitro and assembled with the sgRNA into a Cas9/sgRNA ribonucleoprotein, which is delivered into immature wheat embryos by particle bombardment. The ribonucleoprotein cleaves the target site immediately and is quickly degraded, generating DNA-free edited wheat (Liang et al. 2017).
The final CRISPR-edited DNA-free products are similar to natural and artificial mutants, which are not subject to GMO regulations. We believe that this is the direction of future breeding, and it will play a vital role in realizing sustainable agriculture in the future.
Concluding remarks and future perspectives
Though large genome and complex polyploid nature have hindered the development of wheat genetic engineering and breeding in the past, several powerful tools are now available to advance wheat biology (Li et al. 2020c). In particular, the development of CRISPR/Cas9 technology has been widely used in wheat genome editing. This technology allows multiplex genome engineering, which has enabled the production of loss-of-function triple wheat mutants; thus, it is a powerful tool for introducing desired traits conferred by a loss-of-function mutation into commercial cultivars via NHEJ. For example, a recessive genic male-sterile (GMS) mutant has several advantages for hybrid wheat production (Li et al. 2020b). As additional genes required for genic male sterility are identified, CRISPR/Cas9-mediated disruption of these genes will enable the rapid production of male-sterile wheat. This represents a promising method for manipulating recessive sterility genes to capture heterosis in wheat.
Some valuable alleles are often caused by one or several SNPs or defined insertion/deletions. The introduction of such valuable alleles into commercial cultivars requires 8–10 years by crossing and back-crossing to eliminate unexpected linked traits (Chen et al. 2019). Moreover, breeding is a complicated matter. Sometimes it is not enough to create a good variety by simply modifying one or two genes. For example, five Puccinia graminis f. sp. Tritici (Pgt) resistance genes have been introduced into bread wheat, conferring wheat with broad-spectrum resistance in the field (Luo et al. 2021). Nowadays, CRISPR-mediated precise genome editing is a useful means to achieve these targeted substitutions and replacements by modifying endogenous genes without introducing linkage drag; it can also introduce new alleles (segregating as a single locus) into a predetermined genomic site. Thus, this approach could accelerate the breeding process.
Though precise gene modification has been achieved in Arabidopsis, rice, maize, and tomato, it is only feasible in a few laboratories with low efficiency (Li and Xia 2020). To date, except for base editing, precise editing in wheat has not been achieved (Gil-Humanes et al. 2017; Lin et al. 2020). Therefore, precise gene modification in wheat remains a challenge. Given the increased focus of researchers on the mechanism of HR, we believe that precise gene editing via HR will be used for wheat breeding in the near future. The alternative is the dominant repair pathway—NHEJ, which has been exploited to generate gene replacements and gene knock-ins in rice (Li et al. 2016; Dong et al. 2020). It is a promising method to achieve precise genome modification in wheat, and it may facilitate wheat breeding by modifying gene functions or introducing new alleles into a predetermined genomic safe harbor.
Transgenerational CRISPR/Cas9 activity has been used to modify multiple target sites in tomato and wheat (Rodriguez-Leal et al. 2017; Wang et al. 2018a). This suggests that valuable, desired phenotypes in elite wheat germplasms, which are recalcitrant to transformation, could be induced by crossing with lines carrying CRISPR/Cas9 elements. In addition, wheat genes have been successfully edited via pollination using CRISPR/Cas9-transgenic maize as a haploid inducer (Kelliher et al. 2020; Budhagatapalli et al. 2020). Such haploid induction-mediated genome editing would not only reduce the genotype dependence on site-specific mutagenesis in wheat, but also provide a path to produce transgene-free gene-edited inbred wheat lines. Collectively, these technologies will accelerate wheat breeding.
Some studies have reported that although CRISPR/Cas9 can cleave a target site, sometimes it also cleaves sites with a few mismatches to the target site. This off-target effect is a major concern in gene therapy, but this issue might not be a barrier in plant biotechnology. The putative off-target mutation could be eliminated through back-crossing or crossing with wild-type plants. Moreover, it is advisable to design target sites using web-based tools to reduce off-target mutations by leveraging computation.
The fields of genome editing and wheat biology are attracting more and more excellent scientists, and as the number of available CRISPR/Cas platforms increases, additional tools for precisely fine-tuning gene expression will become available in wheat. Combined with other achievements, including the production of high-quality genome sequences and improved transgenic methods, CRISPR and CRISPR-based genome editing will bring functional genomics and rational design-based molecular breeding of polyploid wheat to the forefront of wheat biology. We believe that transgene-free, gene-edited wheat will play a critical role in addressing environmental issues while promoting sustainable agriculture. Significantly, it is not a replacement for traditional breeding; it is just one of the methods advancing wheat breeding programs and accelerating wheat biology.
Acknowledgements
We thank Dr. Jessica Habashi for critical reading of the manuscript. This work was financially supported by the National Key R&D Program of China (2017YFD0101001), Beijing Municipal Government Science Foundation (IDHT20170513), Provincial Natural Science Foundation of Hebei for Excellent Young Scholar (C2020204062), and Starting Grant from Hebei Agricultural University (YJ201958).
Declarations
Conflict of interest
The authors declare no conflicts of interest.
References
- Abe F, Haque E, Hisano H, Tanaka T, Kamiya Y, Mikami M, Kawaura K, Endo M, Onishi K, Hayashi T, et al. Genome-edited triple-recessive mutation alters seed dormancy in wheat. Cell Rep. 2019;28(5):1362–1369. doi: 10.1016/j.celrep.2019.06.090. [DOI] [PubMed] [Google Scholar]
- Anzalone A, Randolph P, Davis J, Sousa A, Koblan L, Levy J, Chen P, Wilson C, Newby G, Raguram A, et al. Search-and-replace genome editing without double-strand breaks or donor DNA. Nature. 2019;576:149–157. doi: 10.1038/s41586-019-1711-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anzalone A, Koblan L, David L. Genome editing with CRISPR/Cas nucleases, base editors, transposases and prime editors. Nat Biotechnol. 2020;38(7):824–844. doi: 10.1038/s41587-020-0561-9. [DOI] [PubMed] [Google Scholar]
- Borrill P, Adamski N, Uauy C. Genomics as the key to unlocking the polyploid potential of wheat. New Phytol. 2015;208(4):1008–1022. doi: 10.1111/nph.13533. [DOI] [PubMed] [Google Scholar]
- Budhagatapalli N, Halbach T, Hiekel S, Büchner H, Müuller A, Kumlehn J. Site-directed mutagenesis in bread and durum wheat via pollination by cas9/guide RNA-transgenic maize used as haploidy inducer. Plant Biotechnol J. 2020;18:2376–2378. doi: 10.1111/pbi.13415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carroll D. Genome engineering with targetable nucleases. Annu Rev Biochem. 2014;83:409–439. doi: 10.1146/annurev-biochem-060713-035418. [DOI] [PubMed] [Google Scholar]
- Chang Z, Chen Z, Wang N, Xie G, Lu J, Yan W, Zhou J, Tang X, Deng X. Construction of a male sterility system for hybrid rice breeding and seed production using a nuclear male sterility gene. Proc Natl Acad Sci USA. 2016;113(49):14145–14150. doi: 10.1073/pnas.1613792113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Char S, Yang B. Genome editing in grass plants. aBIOTECH. 2020;1:41–57. doi: 10.1007/s42994-019-00005-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y, Wang Z, Ni H, Xu Y, Chen Q, Jiang L. CRISPR/Cas9-mediated base-editing system efficiently generates gain-of-function mutations in Arabidopsis. Sci China Life Sci. 2017;60(5):520–523. doi: 10.1007/s11427-017-9021-5. [DOI] [PubMed] [Google Scholar]
- Chen K, Wang Y, Zhang R, Zhang H, Gao C. CRISPR/Cas genome editing and precision plant breeding in agriculture. Annu Rev Plant Biol. 2019;70:667–697. doi: 10.1146/annurev-arplant-050718-100049. [DOI] [PubMed] [Google Scholar]
- Cong L, Ran FA, Cox D, Lin S, Barretto R, Habib N, Hsu P, Wu X, Jiang W, Marraffifini LA, et al. Multiplex genome engineering using CRISPR/Cas systems. Science. 2013;339:819–823. doi: 10.1126/science.1231143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong O, Yu S, Jain R, Zhang N, Duong PQ, Butler C, Li Y, Lipzen A, Martin JA, Barry KW, et al. Marker-free carotenoid-enriched rice generated through targeted gene insertion using CRISPR-Cas9. Nat Commun. 2020;11:1178. doi: 10.1038/s41467-020-14981-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doudna JA, Emmanuelle C. The new frontier of genome engineering with CRISPR-Cas9. Science. 2014;346:1258096. doi: 10.1126/science.1258096. [DOI] [PubMed] [Google Scholar]
- Gao C. Genome engineering for crop improvement and future agriculture. Cell. 2021 doi: 10.1016/j.cell.2021.01.005. [DOI] [PubMed] [Google Scholar]
- Gaudelli NM, Komor AC, Rees HA, Packer MS, Badran AH, Bryson DI, Liu DR. Programmable base editing of A•T to G•C in genomic DNA without DNA cleavage. Nature. 2017;551:464–471. doi: 10.1038/nature24644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gil-Humanes J, Voytas DF. Wheat rescued from fungal disease. Nat Biotechnol. 2014;32(9):886–887. doi: 10.1038/nbt.3013. [DOI] [PubMed] [Google Scholar]
- Gil-Humanes J, Wang Y, Liang Z, Shan Q, Ozuna CV, Sanchez-Leon S, Baltes NJ, Starker C, Barro F, Gao C, et al. High efficiency gene targeting in hexaploid wheat using DNA replicons and CRISPR/Cas9. Plant J. 2017;89:1251–1262. doi: 10.1111/tpj.13446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gürel F, Zhang Y, Sretenovic S, Qi Y. CRISPR-Cas nucleases and base editors for plant genome editing. aBIOTECH. 2020;1:74–87. doi: 10.1007/s42994-019-00010-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He Y, Zhao Y. Technological breakthroughs in generating transgene-free and genetically stable CRISPR-edited plants. aBIOTECH. 2020;1:88–96. doi: 10.1007/s42994-019-00013-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He F, Pasam R, Shi F, Kant S, Keeble-Gagnere G, Kay P, Forrest K, Fritz A, Hucl P, Wiebe K, et al. Exome sequencing highlights the role of wild-relative introgression in shaping the adaptive landscape of the wheat genome. Nat Genet. 2019;51:896–904. doi: 10.1038/s41588-019-0463-2. [DOI] [PubMed] [Google Scholar]
- Hille F, Richter H, Wong SP, BratoviM RS, Charpentier E. The biology of CRISPR-Cas: backward and forward. Cell. 2017;172:1239–1259. doi: 10.1016/j.cell.2017.11.032. [DOI] [PubMed] [Google Scholar]
- Howells RM, Craze M, Bowden S, Wallington EJ. Efficient generation of stable, heritable gene edits in wheat using CRISPR/Cas9. BMC Plant Biol. 2018;18(1):215. doi: 10.1186/s12870-018-1433-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu B, Wang W, Ou S, Tang J, Li H, Che R, Zhang Z, Chai X, Wang H, Wang Y, et al. Variation in NRT1.1B contributes to nitrate-use divergence between rice subspecies. Nat Genet. 2015;47(7):834–838. doi: 10.1038/ng.3337. [DOI] [PubMed] [Google Scholar]
- Hua K, Jiang Y, Tao X, Zhu J. Precision genome engineering in rice using prime editing system. Plant Biotechnol J. 2020;18(11):2167–2169. doi: 10.1111/pbi.13395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jinek M, Chylinski K, Fonfara I, Hauer M, Doudna JA, Charpentier E. A programmable dual RNA-guided DNA endonuclease in adaptive bacterial immunity. Science. 2012;337(6096):816–821. doi: 10.1126/science.1225829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang B, Yun J, Kim S, Shin Y, Ryu J, Choi M, Woo J, Kim J. Precision genome engineering through adenine base editing in plants. Nat Plants. 2018;4:427–431. doi: 10.1038/s41477-018-0178-x. [DOI] [PubMed] [Google Scholar]
- Kelliher T, Starr D, Su X, Tang G, Chen Z, Carter J, Wittich PE, Dong S, Green J, Burch E, et al. One-step genome editing of elite crop germplasm during haploid induction. Nat Biotechnol. 2019;37:287–292. doi: 10.1038/s41587-019-0038-x. [DOI] [PubMed] [Google Scholar]
- Kim H, Kim J. A guide to genome engineering with programmable nucleases. Nat Rev Genet. 2014;15(5):321–334. doi: 10.1038/nrg3686. [DOI] [PubMed] [Google Scholar]
- Komor AC, Kim YB, Packer MS, Zuris JA, Liu DR. Programmable editing of a target base in genomic DNA without double-stranded DNA cleavage. Nature. 2016;533:420–424. doi: 10.1038/nature17946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li SY, Xia LQ. Precise gene replacement in plants through CRISPR/Cas genome editing technology: current status and future perspectives. aBIOTECH. 2020;1:58–73. doi: 10.1007/s42994-019-00009-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Meng X, Zong Y, Chen K, Zhang H, Liu J, Li J, Gao C. Gene replacements and insertions in rice by intron targeting using CRISPR-Cas9. Nat Plants. 2016;2:16139. doi: 10.1038/nplants.2016.139. [DOI] [PubMed] [Google Scholar]
- Li C, Zong Y, Wang Y, Jin S, Zhang D, Song Q, Zhang R, Gao C. Expanded base editing in rice and wheat using a Cas9-adenosine deaminase fusion. Genome Biol. 2018;19:59. doi: 10.1186/s13059-018-1443-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Li Y, Ma L. CRISPR/Cas9-Based genome editing and its applications for functional genomic analyses in plants. Small Methods. 2019;3(3):1800473–1800493. doi: 10.1002/smtd.201800473. [DOI] [Google Scholar]
- Li H, Li J, Chen J, Yan L, Xia L. Precise modifications of both exogenous and endogenous genes in rice by prime editing. Mol Plant. 2020;13(5):671–674. doi: 10.1016/j.molp.2020.03.011. [DOI] [PubMed] [Google Scholar]
- Li J, Wang Z, He G, Ma L, Deng X. CRISPR/Cas9-mediated disruption of TaNP1 genes results in complete male sterility in bread wheat. J Genet Genomics. 2020;47(5):263–272. doi: 10.1016/j.jgg.2020.05.004. [DOI] [PubMed] [Google Scholar]
- Li J, Yang J, Li Y, Ma L. Current strategies and advances in wheat biology. Crop J. 2020;8(6):879–891. doi: 10.1016/j.cj.2020.03.004. [DOI] [Google Scholar]
- Li JY, Jiao G, Sun Y, Chen J, Zhong Y, Yan L, Jiang D, Ma Y, Xia L. Modification of starch composition, structure and properties through editing of TaSBEIIa in both winter and spring wheat varieties by CRISPR/Cas9. Plant Biotechnol J. 2020 doi: 10.1111/pbi.13519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang Z, Chen K, Li T, Zhang Y, Wang Y, Zhao Q, Liu J, Zhang H, Liu C, Ran Y, et al. Efficient DNA-free genome editing of bread wheat using CRISPR/Cas9 ribonucleoprotein complexes. Nat Commun. 2017;8:14261. doi: 10.1038/ncomms14261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin Q, Zong Y, Xue C, Wang S, Jin S, Zhu Z, Wang Y, Anzalone A, Raguram A, Doman J, et al. Prime genome editing in rice and wheat. Nat Biotechnol. 2020;38:582–585. doi: 10.1038/s41587-020-0455-x. [DOI] [PubMed] [Google Scholar]
- Liu H, Wang K, Jia Z, Gong Q, Lin Z, Du L, Pei X, Ye X. Efficient induction of haploid plants in wheat by editing of TaMTL using an optimized Agrobacterium-mediated CRISPR system. J Exp Bot. 2020;71(4):1337–1349. doi: 10.1093/jxb/erz529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H, Wang K, Tang H, Gong Q, Du L, Pei X, Ye X. CRISPR/Cas9 editing of wheat TaQ genes alters spike morphogenesis and grain threshability. J Genet Genomics. 2020 doi: 10.1016/j.jgg.2020.08.004. [DOI] [PubMed] [Google Scholar]
- Lv J, Yu K, Wei J, Gui H, Liu C, Liang D, Wang Y, Zhou H, Carlin R, Rich R, et al. Generation of paternal haploids in wheat by genome editing of the centromeric histone CENH3. Nat Biotechnol. 2020;38(12):1397–1401. doi: 10.1038/s41587-020-0728-4. [DOI] [PubMed] [Google Scholar]
- Mali P, Yang L, Esvelt KM, Aach J, Guell M, DiCarlo JE, Norville JE, Church GM. RNA-guided human genome engineering via Cas9. Science. 2013;339:823–826. doi: 10.1126/science.1232033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nadolska-Orczyk A, Rajchel I, Orczyk W, Gasparis S. Major genes determining yield-related traits in wheat and barley. Theor Appl Genet. 2017;130:1081–1098. doi: 10.1007/s00122-017-2880-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishida K, Arazoe T, Yachie N, Banno S, Kakimoto M, Tabata M, Mochizuki M, Miyabe A, Araki M, Hara K, et al. Targeted nucleotide editing using hybrid prokaryotic and vertebrate adaptive immune systems. Science. 2016;353(6305):aaf8729. doi: 10.1126/science.aaf8729. [DOI] [PubMed] [Google Scholar]
- Okada A, Arndell T, Borisjuk N, Sharma N, Watson-Haigh N, Tucker E, Baumann U, Langridge P, Whitford R. CRISPR/Cas9-mediated knockout of Ms1 enables the rapid generation of male-sterile hexaploid wheat lines for use in hybrid seed production. Plant Biotechnol J. 2019;17(10):1905–1913. doi: 10.1111/pbi.13106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petersen G, Seberg O, Yde M, Berthelsen K. Phylogenetic relationships of Triticum and Aegilops and evidence for the origin of the A, B, and D genomes of common wheat (Triticum aestivum) Mol Phylogenet Evol. 2006;39(1):70–82. doi: 10.1016/j.ympev.2006.01.023. [DOI] [PubMed] [Google Scholar]
- Rodriguez-Leal D, Lemmon ZH, Man J, Bartlett ME, Lippman ZB. Engineering quantitative trait variation for crop improvement by genome editing. Cell. 2017;171(2):470–480. doi: 10.1016/j.cell.2017.08.030. [DOI] [PubMed] [Google Scholar]
- Sanchez-Leon S, Gil-Humanes J, Ozuna CV, Gimenez MJ, Sousa C, Voytas DF, Barro F. Low-gluten, nontransgenic wheat engineered with CRISPR/Cas9. Plant Biotechnol J. 2018;16(4):902–910. doi: 10.1111/pbi.12837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimatani Z, Kashojiya S, Takayama M, Terada R, Arazoe T, Ishii H, Teramura H, Yamamoto T, Komatsu H, Miura K, et al. Targeted base editing in rice and tomato using a CRISPR-Cas9 cytidine deaminase fusion. Nat Biotechnol. 2017;35(5):441–443. doi: 10.1038/nbt.3833. [DOI] [PubMed] [Google Scholar]
- Singh M, Kumar M, Albertsen MC, Young JK, Cigan AM. Concurrent modifications in the three homeologs of Ms45 gene with CRISPR-Cas9 lead to rapid generation of male sterile bread wheat (Triticum aestivum L.) Plant Mol Biol. 2018;97:371–383. doi: 10.1007/s11103-018-0749-2. [DOI] [PubMed] [Google Scholar]
- Symington L, Gautier J. Double-strand break end resection and repair pathway choice. Annu Rev Genet. 2011;45:247–271. doi: 10.1146/annurev-genet-110410-132435. [DOI] [PubMed] [Google Scholar]
- Tang H, Liu H, Zhou Y, Liu H, Du L, Wang K, Ye X. Fertility recovery of wheat male sterility controlled by Ms2 using CRISPR/Cas9. Plant Biotechnol J. 2020 doi: 10.1111/pbi.13482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tucker E, Baumann U, Kouidri A, Suchecki R, Baes M, Garcia M, Okada T, Dong C, Wu Y, Sandhu A, et al. Molecular identification of the wheat male fertility gene Ms1 and its prospects for hybrid breeding. Nat Commun. 2017;8:869. doi: 10.1038/s41467-017-00945-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uauy C, Wulff B, Dubcovsky J. Combining traditional mutagenesis with new high-throughput sequencing and genome editing to reveal hidden variation in polyploid wheat. Annu Rev Genet. 2017;51:435–454. doi: 10.1146/annurev-genet-120116-024533. [DOI] [PubMed] [Google Scholar]
- Wang Y, Cheng X, Shan Q, Zhang Y, Liu J, Gao C, Qiu J. Simultaneous editing of three homoeoalleles in hexaploid bread confers heritable resistance to powdery mildew. Nat Biotechnol. 2014;32(9):947–951. doi: 10.1038/nbt.2969. [DOI] [PubMed] [Google Scholar]
- Wang K, Liu H, Du L, Ye X. Generation of marker-free transgenic hexaploid wheat via an Agrobacterium-mediated co-transformation strategy in commercial Chinese wheat varieties. Plant Biotechnol J. 2017;15(5):614–623. doi: 10.1111/pbi.12660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z, Li J, Chen S, Heng Y, Chen Z, Yang J, Zhou K, Pei J, He H, Deng X, et al. Poaceae-specific MS1 encodes a phospholipid-binding protein for male fertility in bread wheat. Proc Natl Acad Sci USA. 2017;114(47):12614–12619. doi: 10.1073/pnas.1715570114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W, Pan Q, He F, Akhunova A, Chao S, Trick H, Akhunov E. Transgenerational CRISPR-Cas9 activity facilitates multiplex gene editing in allopolyploid wheat. Crispr J. 2018;1(1):65–74. doi: 10.1089/crispr.2017.0010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W, Simmonds J, Pan Q, Davidson D, He F, Battal A, Akhunova A, Trick H, Uauy C, Akhunov E. Gene editing and mutagenesis reveal inter-cultivar differences and additivity in the contribution of TaGW2 homoeologues to grain size and weight in wheat. Theor Appl Genet. 2018;131:2463–2475. doi: 10.1007/s00122-018-3166-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W, Pan Q, Tian B, He F, Chen Y, Bai G, Akhunova A, Trick H, Akhunov E. Gene editing of the wheat homologs of TONNEAU1-recruiting motif encoding gene affects grain shape and weight in wheat. Plant J. 2019;100(2):251–264. doi: 10.1111/tpj.14440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu W, Zhang C, Yang Y, Zhao S, Kang G, He X, Song J, Yang J. Versatile nucleotides substitution in plant using an improved prime editing system. Mol Plant. 2020;13(5):675–678. doi: 10.1016/j.molp.2020.03.012. [DOI] [PubMed] [Google Scholar]
- Zhan X, Lu Y, Zhu J, Botella J. Genome editing for plant research and crop improvement. J Integr Plant Biol. 2020 doi: 10.1111/jipb.13063. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Liang Z, Zong Y, Wang Y, Liu J, Chen K, Qiu J, Gao C. Efficient and transgene-free genome editing in wheat through transient expression of CRISPR/Cas9 DNA or RNA. Nat Commun. 2016;7:12617. doi: 10.1038/ncomms12617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Bai Y, Wu G, Zou S, Chen Y, Gao C, Tang D. Simultaneous modification of three homoeologs of TaEDR1 by genome editing enhances powdery mildew resistance in wheat. Plant J. 2017;91(4):714–724. doi: 10.1111/tpj.13599. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Li D, Zhang D, Zhao X, Cao X, Dong L, Liu J, Chen K, Zhang H, Gao C, et al. Analysis of the functions of TaGW2 homoeologs in wheat grain weight and protein content traits. Plant J. 2018;94(5):857–866. doi: 10.1111/tpj.13903. [DOI] [PubMed] [Google Scholar]
- Zhang R, Liu J, Chai Z, Chen S, Bai Y, Zong Y, Chen K, Li J, Jiang L, Gao C. Generation of herbicide tolerance traits and a new selectable marker in wheat using base editing. Nat Plants. 2019;5:480–485. doi: 10.1038/s41477-019-0405-0. [DOI] [PubMed] [Google Scholar]
- Zhang S, Zhang R, Gao J, Gu T, Song G, Li W, Li D, Li Y, Li G. Highly efficient and heritable targeted mutagenesis in wheat via the Agrobacterium tumefaciens-mediated CRISPR/Cas9 system. Int J Mol Sci. 2019;20(17):4257. doi: 10.3390/ijms20174257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Malzahn AA, Sretenovic S, Qi Y. The emerging and uncultivated potential of CRISPR technology in plant science. Nat Plants. 2019;5:778–794. doi: 10.1038/s41477-019-0461-5. [DOI] [PubMed] [Google Scholar]
- Zhang Z, Hua L, Gupta A, Tricoli D, Edwards KJ, Yang B, Li W. Development of an Agrobacterium-delivered CRISPR/Cas9 system for wheat genome editing. Plant Biotechnol J. 2019;17(8):1623–1635. doi: 10.1111/pbi.13088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Pribil M, Palmgren M, Gao C. A CRISPR way for accelerating improvement of food crops. Nat food. 2020;1:200–205. doi: 10.1038/s43016-020-0051-8. [DOI] [Google Scholar]
- Zhao K, Tung C, Eizenga G, Wright M, Ali M, Price A, Norton G, Islam M, Reynolds A, Mezey J, et al. Genome-wide association mapping reveals a rich genetic architecture of complex traits in Oryza sativa. Nat Commun. 2011;2(467):1–10. doi: 10.1038/ncomms1467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zong Y, Wang Y, Li C, Zhang R, Chen K, Ran Y, Qiu J, Wang D, Gao C. Precise base editing in rice, wheat and maize with a Cas9-cytidine deaminase fusion. Nat Biotechnol. 2017;35(5):438–440. doi: 10.1038/nbt.3811. [DOI] [PubMed] [Google Scholar]
- Zong Y, Song Q, Li C, Jin S, Zhang D, Wang Y, Qiu J, Gao C. Efficient C-to-T base editing in plants using a fusion of nCas9 and human APOBEC3A. Nat Biotechnol. 2018;36:950–953. doi: 10.1038/nbt.4261. [DOI] [PubMed] [Google Scholar]