Abstract
Serum liver enzymes (alanine aminotransferase [ALT], aspartate aminotransferase [AST], λ-glutamyl transferase [GGT] and alkaline phosphatase [ALP]) are the leading biomarkers to measure liver injury, and they have been reported to be associated with several intrahepatic and extrahepatic diseases in observational studies. We conducted a phenome-wide association study (PheWAS) to identify disease phenotypes associated with genetically predicted liver enzymes based on the UK Biobank cohort. Univariable and multivariable Mendelian randomization (MR) analyses were performed to obtain the causal estimates of associations that detected in PheWAS. Our PheWAS identified 40 out of 1,376 pairs (16, 17, three and four pairs for ALT, AST, GGT and ALP, respectively) of genotype–phenotype associations reaching statistical significance at the 5% false discovery rate threshold. A total of 34 links were further validated in Mendelian randomization analyses. Most of the disease phenotypes that associated with genetically determined ALT level were liver-related, including primary liver cancer and alcoholic liver damage. The disease outcomes associated with genetically determined AST involved a wide range of phenotypic categories including endocrine/metabolic diseases, digestive diseases, and neurological disorder. Genetically predicted GGT level was associated with the risk of other chronic non-alcoholic liver disease, abnormal results of function study of liver, and cholelithiasis. Genetically determined ALP level was associated with pulmonary heart disease, phlebitis and thrombophlebitis of lower extremities, and hypercholesterolemia. Our findings reveal novel links between liver enzymes and disease phenotypes providing insights into the full understanding of the biological roles of liver enzymes.
Supplementary Information
The online version contains supplementary material available at 10.1007/s43657-021-00033-y.
Keywords: Liver enzyme, Disease phenotype, GWAS, Mendelian randomization
Introduction
Alanine aminotransferase (ALT), aspartate aminotransferase (AST), λ-glutamyl transferase (GGT) and alkaline phosphatase (ALP) are the leading biomarkers to measure the liver function in clinical practice, in which many physiological and pathological processes are involved. Growing epidemiological evidences showed that the variations in serum levels of liver enzymes affected both intrahepatic and extrahepatic morbidities and mortalities (Choi et al. 2018; Ghouri et al. 2010; Kaneko et al. 2019; Katzke et al. 2020; Koehler et al. 2014; Kunutsor et al. 2014, 2015; Monami et al. 2008; Siddiqui et al. 2013). However, since the levels of circulating biomarkers are easily influenced by many factors and are subject to diurnal variation (Danielsson et al. 2014; Liu et al. 2014b; Nunez et al. 2019), it was considered that the serum level of liver enzymes measuring once might bias the association estimates between liver enzymes and clinical events (Clarke et al. 1999). Moreover, previous studies were observational population-based design, and it is generally hard to disentangle the causal relationship between serum level of liver enzyme and disease phenotype, thus, the observed association might be subject to underlying confounders.
As shown in genome-wide association studies (GWAS), genetic factors are implicated in the regulation of serum levels of liver enzymes (Chambers et al. 2011; Chen et al. 2021; Stender et al. 2017; Yuan et al. 2008). Given the genotypes are generally independent of environment and the transmission of genetic information is usually unidirectional (Li et al. 2019), the genetic data from GWAS enable us to determine causal relationship between exposure and outcome via several methods including Mendelian randomization (MR) (Pingault et al. 2018). So far, MR based on single nucleotide polymorphism (SNP) or polygenetic risk scores (PRS) has been extensively used to infer the causalities (Ding et al. 2017; Ference et al. 2019; Richmond et al. 2019; Voight et al. 2012). Several MR studies have also been conducted to investigate the causal relationships between serum levels of liver enzymes and disease phenotypes (De Silva et al. 2019; Liu et al. 2016; Noordam et al. 2016; Xu et al. 2017). However, previous studies only used a few SNPs as the genetic instruments and were most limited to a single disease phenotype. To comprehensively reveal the biological roles of liver enzymes, herein, we performed a phenome-wide MR study using data from the UK Biobank cohort study.
Materials and Methods
Study Participants
This research has been conducted using the UK Biobank Resource under application numbers 58484 and 63726. The details of UK Biobank study have been previously described (Sudlow et al. 2015). Briefly, UK Biobank is a large-scale, population-based prospective cohort study, which recruited over 0.5 million participants aged 40–70 years in 2006–2010 and provided extensive measurement of baseline data and genotype data with linked national medical records for longitudinal follow-up. In this study, we excluded participants who were non-Caucasian ethnic background and with missing data on serum liver enzyme levels.
GWAS for Liver Enzymes
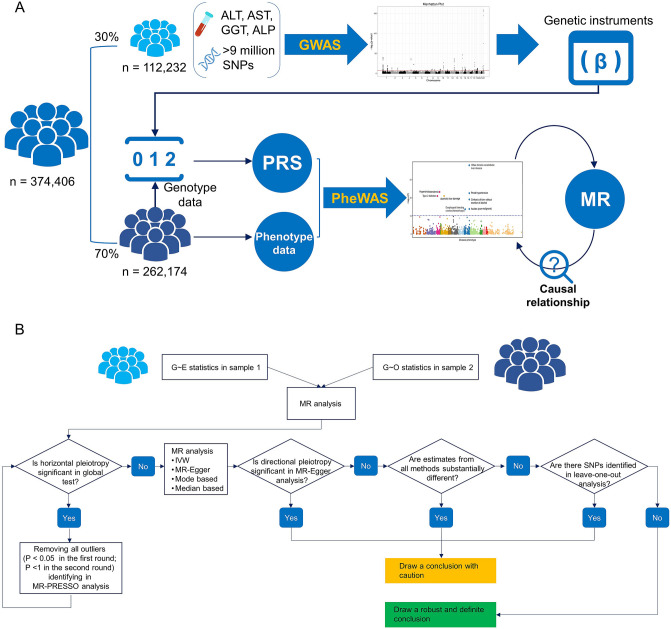
The genotype data in the UK Biobank were derived from GWAS chip (Affymetrix UK BiLEVE and UK Biobank Axiom arrays). To ensure the independence between discovery dataset and validation dataset (Chatterjee et al. 2016), we first divided the whole population into two non-overlapping groups in a ratio of 3:7 (Fig. 1A). The discovery dataset (n = 112,232) was used to perform GWAS of liver enzymes. In this dataset, participants who met any of the following conditions were excluded: (i) genetically related with at least one UK Biobank participant (kinship > 0.08); (ii) genetically inferred gender that did not match the self-reported gender; and (iii) identified as outliers in heterozygosity and missing rates. We selected the SNPs with minor allele frequency > 1%, imputation accuracy (Info) score > 0.3, and Hardy–Weinberg equilibrium p value > 10–6 to perform GWAS. The serum levels of liver enzymes were log-transformed to overcome the skewness and outliers. A total of 9,886,861 SNPs were eligible for the GWAS, where sex, age and genotyping chip were adjusted. We assigned a SNP as associated with a liver enzyme of interest using a conventional GWAS threshold (p < 5 × 10−8). SNPs were binned into loci based on pairwise linkage disequilibrium (LD; at between-SNP R2 < 0.1), with the SNP with the strongest association with the trait of interest (as defined by p value) being retained in each locus.
Fig. 1.
Study design. A UK Biobank participants were divided in a ratio of 3:7 to conduct GWAS and construct polygenic risk score (PRS) for liver enzymes. B Study flow chart of Mendelian randomization (MR) analysis
Weighted Polygenetic Risk Score
To generate a genetic proxy for serum liver enzyme, genetic variants associated with liver enzyme were retrieved from the GWAS. PRS was constructed by incorporating effect estimates of the assigned SNPs for the rest of UK Biobank participants (validation dataset; n = 262,174) (Fig. 1A). Specifically, taking ALT as an example, the PRS was created by adding up the number of ALT-related alleles for each SNP weighted for the SNP effect size on ALT level (regression beta-coefficients) and then adding up this weighted score for all assigned SNPs. As to ALT, the same processing was applied to AST, GGT and ALP.
Disease Phenotypes and PheWAS
We identified disease phenotypes using International Classification of Diseases (ICD) code versions 10 and 9 in the electronic medical records of UK Biobank. All prevalent and incident diseases in ICD code were mapped to the corresponding phenotype via Phecode Map 1.2 (Wu et al. 2019). Self-reported diagnoses were not considered. Repeated diagnosis of an individual disease in a participant was recorded as a single event. Cases were defined as individuals having at least one documented event and controls as individuals with no record of that outcome or its related phecodes (Li et al. 2018). To ensure the statistical power, we excluded disease phenotypes that have < 20 cases. We finally included 1,376 disease phenotypes. We performed logistic regression models with adjusting for age, sex, and genotyping array to examine the associations between each disease phenotype and the PRS of ALT, AST, GGT, and ALP, respectively. To avoid the type I error, we used false discovery rate (FDR) to adjust p values from the logistic regressions. An FDR-Q value < 0.05 was deemed to be statistically significant.
Mendelian Randomization and Sensitivity Analyses
For disease phenotypes showing significant association with liver enzyme, we further performed two-sample MR study (Yavorska and Burgess 2017) to determine the potential causal relationships (Fig. 1). The statistics of genetic variant–exposure (G–E, i.e., beta-coefficients and standard errors) were derived from the GWAS of liver enzymes (Supplementary Tables S1–4). Herein, the genetic variant and exposure denote the selected SNPs and log-transformed serum level of liver enzyme, respectively. The statistics of genetic variant–outcome (G–O) were calculated from the logistic regression models that examine the associations of selected SNPs with disease phenotypes (Fig. 1B). To ensure the robustness of estimates, we established a MR framework according to previous studies (Fig. 1B) (Burgess et al. 2019; Burgess and Thompson 2017; Ference et al. 2019; Wu et al. 2020). Briefly, we first examined the horizontal pleiotropy that measured by MR-PRESSO global test and removed the outliers if the corresponding p value < 0.05. Next, we conducted the MR analysis using four methods (inverse-variance weighted [IVW], MR-Egger, median-based method and mode-based method) that are under different assumptions (Burgess et al. 2019). For the MR estimates, we could draw a robust conclusion if they fulfil all of the following conditions: (i) no significant between-SNP heterogeneity measured by Cochran Q value was detected; (ii) no significant directional pleiotropy was detected; (iii) estimates from different methods are same in terms of the direction; and (iv) no influential SNPs were detected from the “leave-one-out” analysis. Otherwise, the causal relationship is suspicious. Of note is that leaving one variant out at a time is unlikely to change the estimate substantially when there are many variants. We, therefore, left out subsets of variants by randomly chosen 20% of SNPs at a time and repeated this process for n times, where the n denotes the number of the assigned SNPs of each liver enzyme (Burgess et al. 2019; Smith et al. 2014).
Owing to the low-to-moderate correlations among serum levels of the four liver enzymes (Supplementary Fig. S1), it is reasonable to assume that the biological effects of the liver enzymes may be mutually influenced. We, therefore, conducted multivariable MR (MVMR) analyses (Burgess and Thompson 2015). Considering the underlying co-linearity and horizontal pleiotropy, we applied two robust methods (MVMR-robust and MVMR-Lasso) in the MVMR analysis (Grant and Burgess 2020).
Since the G–E statistics were derived from the GWAS for log-transformed liver enzymes, the MR results correspond to an odds ratio (OR) per 1-unit increment in log-transformed liver enzymes. This is equivalent to a 2.718 (e)-fold increase in liver enzyme on the untransformed scale, which is a substantially large change in liver enzyme levels. To better illustrate the effects of liver enzyme on disease phenotypes, we calculated G–E statistics based on associations between untransformed liver enzymes and the selected SNPs from GWAS. In this scenario, the MR results denote an OR per 1-unit increment in liver enzymes. Pearson correlation test was performed to examine the consistency between MR estimates of the two scenarios. We also performed a sensitivity analysis to examine the association between phenotypic levels of liver enzymes and risks of disease phenotypes based on the whole population. All analyses were performed using R program (version 3.6.3). The GWAS was conducted using bigsnpr and bigstatsr packages (Privé et al. 2018). The MR analysis was implemented using MendelianRandomization and MRPRESSO packages (Verbanck et al. 2018; Yavorska and Burgess 2017).
Results
GWAS and PRS
The distributions of log-transformed liver enzyme levels are shown in Supplementary Fig. S2. We identified 58,85,121 and 202 independent SNPs that were associated with levels of ALT, AST, GGT, and ALP at the GWAS-significance level, respectively (Supplementary Tables S1–4; Fig. 2). These SNPs collectively explain approximately 3.2%, 3.9%, 6.4%, and 9.2% of the variations in serum levels of ALT (F statistics = 59.5), AST (F statistics = 62.2), GGT (F statistics = 65.6), and ALP (F statistics = 75.5), respectively. The Q–Q plots of GWAS for the four liver enzymes are shown in Supplementary Fig. S3–6. The assigned SNPs identified from the GWAS are less shared among the four liver enzymes (Supplementary Fig. S7). ALT shared thirteen loci with AST. PRS was constructed and served as the genetic surrogate for liver enzyme. At the PheWAS population level, PRSs of the four liver enzymes were strictly followed normal distribution, with the mean values approximately equal zero (Supplementary Fig. S8).
Fig. 2.
Manhattan plots of GWAS for liver enzymes. A–D denotes the Manhattan plot of alanine transaminase (ALT), aspartate transaminase (AST), λ-glutamyl transferase (GGT), and alkaline phosphatase (ALP), respectively. Red dotted lines denote the cut-off of statistical GWAS-significance (p < 5 × 10−8)
PheWAS
In this analysis, we constructed a total of 1,376 genotype–phenotype pairs and identified sixteen, seventeen, three, and four pairs reaching statistical significance at the 5% FDR threshold for ALT, AST, GGT, and ALP, respectively (Fig. 3; Supplementary Tables S5–8). In the PheWAS of ALT, we found that most of the disease phenotypes reaching statistical significance are liver related, including hepatitis (not otherwise specified, NOS), alcoholic liver damage, primary liver cancer, etc. (Fig. 3A). Genetically determined ALT level was positively associated with these liver-related disease phenotypes (beta-coefficients > 0). We also observed a significantly positive association of ALT with type 2 diabetes (T2D) and disorders of iron metabolism and a significantly negative association with dementias, corneal degenerations and fracture of hand and wrist (Supplementary Table S5). The identified genotype–phenotype associations involved 25 genetic variants, of which seven loci were associated with more than one disease outcome and six loci were missense variants (Fig. 4A; Supplementary Table S9). Rs738409 (22:44,324,727:C > G), a missense variant in PNPLA3, was associated with seven disease phenotypes. Nine of the sixteen disease phenotypes shared genetic associations with ALT level at more than one locus. We did not detect significant associations of the rest of four disease phenotypes (disorders of iron metabolism, corneal degeneration, fracture of hand and wrist, and hepatomegaly) with any ALT-related genetic variant at the 5% FDR threshold.
Fig. 3.
Manhattan plots of PheWAS for liver enzymes. A–D denotes the Manhattan plot of alanine transaminase (ALT), aspartate transaminase (AST), λ-glutamyl transferase (GGT), and alkaline phosphatase (ALP), respectively. Blue dotted lines denote the cut-off of statistical PheWAS-significance (FDR-Q value < 0.05)
Fig. 4.
Associations of liver enzyme related genetic loci with disease phenotypes. The association between the genetic loci and disease phenotypes are depicted by the arcs. The area of each colored ribbon depicts the proportion of the genetic locus contributes to a particular disease phenotype. A–D denotes the circos plot of alanine transaminase (ALT), aspartate transaminase (AST), λ-glutamyl transferase (GGT), and alkaline phosphatase (ALP), respectively
In the PheWAS of AST, we detected significant correlations for 17 genotype–phenotype pairs (Fig. 3B). Genetically determined AST level was positively associated with hematuria, other chronic non-alcoholic liver disease, hallux valgus, epistaxis or throat hemorrhage, varicose veins of lower extremity, ankylosing spondylitis, disorders of iron metabolism, and other ill-defined and unknown causes of morbidity and mortality, whereas negatively associated with celiac disease, thyrotoxicosis with or without goiter, hypothyroidism (NOS), dermatitis herpetiformis, cancer of gums, multiple sclerosis, Graves’ disease, type 1 diabetes with renal manifestations, and hereditary disturbances in tooth structure (Supplementary Table S6). These genotype–phenotype correlations were distributed across 35 AST genetic loci, of which 22 loci were associated with multiple disease phenotypes. Most of the loci were either gene intron variants or variants without determined information (Fig. 4B). Sixteen of the seventeen disease phenotypes shared genetic associations with AST level at more than one locus. Gum cancer shared genetic association with AST at locus of rs56278466 (10:17,875,857:T > G), an intron variant in MRC1.
Three genotype–phenotype pairs reached the statistical significance after FDR adjustment in the PheWAS of GGT (Fig. 3C). Genetically determined GGT level was positively associated with the three disease phenotypes, which were pertained to digestive system (Supplementary Table S7). Seven GGT-related genetic variants have significant contributions to the associations of genotype–phenotype pairs at the 5% FDR threshold (Fig. 4C).
We found significantly negative association between genetically determined ALP level and four disease phenotypes, including pulmonary heart disease, phlebitis and thrombophlebitis of lower extremities, hypercholesterolemia, and circulatory disease (not elsewhere classifiable, NEC) (Fig. 3D; Supplementary Table S8). A total of 26 ALP-related genetic variants have significant contributions to the associations of genotype–phenotype pairs at the 5% FDR threshold (Fig. 4D). Eleven of the 26 loci were associated with multiple disease phenotypes. All the four disease phenotypes shared genetic associations with ALP level at more than one locus. Hypercholesterolemia was significantly associated with 18 genetic loci, including five missense variants, six gene intron variants, and one stop-gained variant in FUT2 (rs601338).
Univariable Mendelian Randomization Analysis
We performed two-sample MR analyses to identify the potential causalities based on G–E statistics (Supplementary Tables S1–4) and G–O statistics (Supplementary Tables S9–12), which were derived from two independent populations. Results from the four main methods are shown in Fig. 5 and Table 1 (Supplementary Tables S13–16). The MR estimates from liver enzymes with and without log-transformation were highly correlated (Supplementary Fig. S12). Except for dementias, type 1 diabetes with renal manifestation, and circulatory disease NEC, the MR estimates were similar across different methods and were consistent with that of PheWAS in terms of direction of the estimates (Fig. 5; Supplementary Figs. S8–11). Significant between-SNP heterogeneity was detected for epistaxis or throat hemorrhage, hypercholesterolemia, pulmonary heart disease, phlebitis and thrombophlebitis of lower extremities, and circulatory disease NEC in both IVW and MR-Egger methods (FDR-Q value of Cochran’s Q statistics < 0.05) (Table 1). Directional pleiotropy was only detected between genetically determined GGT and abnormal results of function study of liver (FDR-Q value of MR-Egger regression intercept < 0.05) (Table 1). No influential SNP was found in the “leave-one-out” analyses for the four liver enzymes (Supplementary Tables S17–20). According to the framework described in Fig. 1B, we deemed that the MR estimates were robust for most genotype–phenotype pairs.
Fig. 5.
Associations of genetically determined liver enzyme levels with disease phenotypes according to Mendelian analysis. The associations are quantified by odds ratio (OR). Rectangle in red and blue denotes a positive and negative correlation, respectively. Rectangle with black outline border denotes statistically significant, otherwise means statistically non-significant. IVW inverse-variance weighted, MBE mode-based method
Table 1.
MR estimates from the four main analyses
| Exposure | Outcome | IVW | MR-Egger | MBE | WMM | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| β (se) | FDR-Q value | Cochran's Q value | FDR-Q value | Slope (se) | FDR-Q value | Intercept (se) | FDR-Q value | Cochran's Q value | FDR-Q value | β (se) | FDR-Q value | β (se) | FDR-Q value | ||
| ALT | Hepatitis NOS | 0.221 (0.036) | 1.75E-08 | 75.9 | 1.70E-01 | 0.246 (0.066) | 5.01E-04 | − 0.013 (0.027) | 6.97E-01 | 75.6 | 1.64E-01 | 0.261 (0.053) | 4.11E-06 | 0.286 (0.050) | 1.33E-07 |
| Malignant neoplasm of liver, primary | 0.275 (0.052) | 1.01E-06 | 67.2 | 2.57E-01 | 0.358 (0.094) | 4.36E-04 | − 0.043 (0.040) | 3.69E-01 | 65.9 | 2.75E-01 | 0.332 (0.087) | 4.36E-04 | 0.309 (0.084) | 6.56E-04 | |
| Type 2 diabetes | 0.023 (0.006) | 4.36E-04 | 50.0 | 3.67E-01 | 0.004 (0.011) | 7.46E-01 | 0.009 (0.004) | 6.33E-02 | 45.2 | 4.95E-01 | 0.025 (0.009) | 9.26E-03 | 0.023 (0.009) | 1.49E-02 | |
| Disorders of iron metabolism | 0.112 (0.025) | 3.07E-05 | 44.2 | 9.42E-01 | 0.139 (0.046) | 4.94E-03 | − 0.013 (0.019) | 5.70E-01 | 43.7 | 9.33E-01 | 0.140 (0.045) | 4.57E-03 | 0.118 (0.042) | 9.26E-03 | |
| Dementias | − 0.029 (0.021) | 2.25E-01 | 46.3 | 9.06E-01 | 0.011 (0.039) | 8.01E-01 | − 0.019 (0.016) | 2.94E-01 | 44.8 | 9.26E-01 | − 0.022 (0.039) | 6.38E-01 | − 0.014 (0.035) | 7.45E-01 | |
| Alcoholic liver damage | 0.157 (0.028) | 1.67E-07 | 66.6 | 2.48E-01 | 0.124 (0.056) | 4.45E-02 | 0.014 (0.021) | 5.89E-01 | 66.1 | 2.37E-01 | 0.149 (0.056) | 1.39E-02 | 0.150 (0.039) | 3.94E-04 | |
| Corneal degenerations | − 0.240 (0.056) | 7.68E-05 | 41.4 | 9.42E-01 | − 0.306 (0.107) | 8.57E-03 | 0.030 (0.042) | 5.69E-01 | 40.9 | 9.42E-01 | − 0.280 (0.103) | 1.20E-02 | − 0.287 (0.084) | 1.47E-03 | |
| Esophageal bleeding | 0.077 (0.016) | 8.11E-06 | 62.7 | 3.89E-01 | 0.111 (0.029) | 4.85E-04 | − 0.017 (0.012) | 2.37E-01 | 60.7 | 4.43E-01 | 0.124 (0.026) | 1.28E-05 | 0.116 (0.024) | 6.13E-06 | |
| Other chronic non-alcoholic liver disease | 0.186 (0.019) | 5.13E-20 | 69.1 | 2.01E-01 | 0.209 (0.039) | 5.82E-07 | − 0.010 (0.015) | 5.82E-01 | 68.5 | 1.98E-01 | 0.114 (0.053) | 5.29E-02 | 0.181 (0.027) | 1.22E-09 | |
| Cirrhosis of liver without mention of alcohol | 0.211 (0.032) | 1.22E-09 | 35.6 | 9.42E-01 | 0.251 (0.064) | 2.88E-04 | − 0.016 (0.023) | 5.69E-01 | 35.3 | 9.33E-01 | 0.285 (0.069) | 1.40E-04 | 0.264 (0.047) | 1.98E-07 | |
| Liver abscess and sequelae of chronic liver disease | 0.110 (0.031) | 1.00E-03 | 65.3 | 2.38E-01 | 0.122 (0.057) | 5.19E-02 | − 0.006 (0.024) | 8.09E-01 | 65.1 | 2.08E-01 | 0.150 (0.047) | 2.99E-03 | 0.154 (0.045) | 1.47E-03 | |
| Portal hypertension | 0.206 (0.034) | 2.42E-08 | 61.1 | 2.38E-01 | 0.244 (0.067) | 6.74E-04 | − 0.016 (0.025) | 5.89E-01 | 61.0 | 2.07E-01 | 0.224 (0.074) | 4.94E-03 | 0.220 (0.051) | 6.44E-05 | |
| Hepatomegaly | 0.145 (0.038) | 3.55E-04 | 54.8 | 6.74E-01 | 0.097 (0.069) | 2.22E-01 | 0.024 (0.029) | 4.92E-01 | 54.1 | 6.62E-01 | 0.121 (0.106) | 3.27E-01 | 0.148 (0.066) | 4.41E-02 | |
| Abnormal results of function study of liver | 0.071 (0.012) | 1.48E-07 | 63.0 | 2.48E-01 | 0.035 (0.022) | 1.60E-01 | 0.017 (0.009) | 7.78E-02 | 58.7 | 3.31E-01 | 0.050 (0.018) | 1.20E-02 | 0.048 (0.018) | 1.34E-02 | |
| Cholelithiasis | 0.041 (0.010) | 1.83E-04 | 77.5 | 9.09E-02 | 0.029 (0.021) | 2.25E-01 | 0.005 (0.008) | 5.95E-01 | 76.8 | 8.06E-02 | 0.047 (0.018) | 1.80E-02 | 0.042 (0.013) | 2.93E-03 | |
| Fracture of hand or wrist | − 0.049 (0.012) | 1.42E-04 | 41.2 | 9.42E-01 | − 0.067 (0.023) | 6.14E-03 | 0.008 (0.009) | 4.43E-01 | 40.4 | 9.42E-01 | − 0.044 (0.022) | 6.67E-02 | − 0.041 (0.020) | 6.05E-02 | |
| AST | Cancer of gums | − 0.788 (0.132) | 1.18E-07 | 87.2 | 4.97E-01 | − 0.604 (0.244) | 3.91E-02 | − 0.066 (0.073) | 4.87E-01 | 86.3 | 4.95E-01 | − 1.052 (1.535) | 6.15E-01 | − 1.034 (0.222) | 5.58E-05 |
| Thyrotoxicosis with or without goiter | − 0.087 (0.028) | 8.47E-03 | 76.2 | 2.57E-01 | − 0.090 (0.055) | 1.95E-01 | 0.001 (0.014) | 9.54E-01 | 76.1 | 2.37E-01 | − 0.092 (0.051) | 1.52E-01 | − 0.068 (0.041) | 1.83E-01 | |
| Graves' disease | 0.058 (0.046) | 3.21E-01 | 68.4 | 4.83E-01 | 0.143 (0.082) | 1.63E-01 | − 0.029 (0.023) | 3.21E-01 | 65.4 | 5.35E-01 | 0.099 (0.063) | 2.10E-01 | 0.075 (0.065) | 3.56E-01 | |
| Hypothyroidism NOS | − 0.027 (0.009) | 1.03E-02 | 59.8 | 5.15E-01 | − 0.033 (0.017) | 1.10E-01 | 0.002 (0.004) | 7.70E-01 | 59.5 | 4.95E-01 | − 0.029 (0.015) | 1.08E-01 | − 0.021 (0.014) | 2.08E-01 | |
| Type 1 diabetes | − 0.006 (0.111) | 9.58E-01 | 60.5 | 8.58E-01 | − 0.051 (0.215) | 8.65E-01 | 0.013 (0.056) | 8.65E-01 | 60.5 | 8.25E-01 | − 0.018 (0.341) | 9.59E-01 | − 0.043 (0.174) | 8.65E-01 | |
| Disorders of iron metabolism | 0.104 (0.041) | 3.62E-02 | 90.5 | 1.70E-01 | 0.091 (0.081) | 3.77E-01 | 0.004 (0.021) | 8.98E-01 | 90.4 | 1.64E-01 | 0.146 (0.082) | 1.54E-01 | 0.151 (0.061) | 3.96E-02 | |
| Multiple sclerosis | − 0.081 (0.024) | 3.48E-03 | 90.0 | 2.48E-01 | − 0.102 (0.042) | 4.23E-02 | 0.008 (0.013) | 6.64E-01 | 89.8 | 2.37E-01 | − 0.052 (0.034) | 2.21E-01 | − 0.046 (0.034) | 2.77E-01 | |
| Varicose veins of lower extremity | 0.039 (0.009) | 3.17E-04 | 85.9 | 2.10E-01 | 0.023 (0.016) | 2.45E-01 | 0.006 (0.005) | 3.26E-01 | 84.8 | 2.07E-01 | 0.038 (0.013) | 1.40E-02 | 0.035 (0.014) | 3.62E-02 | |
| Epistaxis or throat hemorrhage | 0.089 (0.020) | 1.54E-04 | 115.7 | 2.90E-02 | 0.062 (0.037) | 1.90E-01 | 0.010 (0.011) | 4.87E-01 | 115.0 | 2.59E-02 | 0.075 (0.027) | 2.24E-02 | 0.052 (0.027) | 1.23E-01 | |
| Hereditary disturbances in tooth structure | − 0.540 (0.134) | 4.46E-04 | 101.7 | 1.70E-01 | − 0.463 (0.242) | 1.19E-01 | − 0.028 (0.072) | 7.83E-01 | 101.7 | 1.64E-01 | − 0.390 (1.479) | 8.64E-01 | − 0.547 (0.215) | 3.62E-02 | |
| Celiac disease | − 0.399 (0.104) | 8.92E-04 | 28.1 | 1.36E-01 | − 0.365 (0.237) | 2.21E-01 | − 0.006 (0.036) | 9.01E-01 | 28.1 | 1.06E-01 | − 0.297 (0.138) | 7.83E-02 | − 0.345 (0.106) | 5.59E-03 | |
| Other chronic non-alcoholic liver disease | 0.125 (0.025) | 1.72E-05 | 89.9 | 2.48E-01 | 0.086 (0.057) | 2.37E-01 | 0.011 (0.014) | 5.83E-01 | 89.3 | 2.37E-01 | 0.158 (0.072) | 6.95E-02 | 0.113 (0.035) | 5.76E-03 | |
| Hematuria | 0.012 (0.008) | 2.43E-01 | 45.7 | 9.42E-01 | 0.020 (0.015) | 2.90E-01 | − 0.002 (0.004) | 6.60E-01 | 45.4 | 9.33E-01 | 0.019 (0.014) | 2.77E-01 | 0.017 (0.013) | 3.20E-01 | |
| Dermatitis herpetiformis | − 0.585 (0.162) | 1.98E-03 | 96.4 | 1.36E-01 | − 0.539 (0.332) | 1.96E-01 | − 0.014 (0.086) | 9.01E-01 | 96.4 | 1.08E-01 | − 0.564 (1.020) | 6.86E-01 | − 0.612 (0.226) | 2.56E-02 | |
| Ankylosing spondylitis | 0.104 (0.047) | 6.61E-02 | 102.9 | 8.07E-02 | 0.076 (0.081) | 4.67E-01 | 0.011 (0.025) | 7.70E-01 | 102.5 | 7.00E-02 | 0.075 (0.057) | 2.98E-01 | 0.113 (0.061) | 1.34E-01 | |
| Hallux valgus (Bunion) | 0.055 (0.012) | 3.88E-05 | 89.5 | 2.10E-01 | 0.044 (0.020) | 6.76E-02 | 0.004 (0.006) | 6.06E-01 | 88.9 | 2.01E-01 | 0.033 (0.016) | 7.91E-02 | 0.040 (0.016) | 3.91E-02 | |
| Other ill-defined and unknown causes of morbidity and mortality | 0.036 (0.007) | 2.72E-06 | 91.4 | 3.87E-01 | 0.045 (0.012) | 1.03E-03 | − 0.003 (0.004) | 4.67E-01 | 90.4 | 4.00E-01 | 0.034 (0.013) | 3.39E-02 | 0.028 (0.011) | 3.62E-02 | |
| GGT | Other chronic non-alcoholic liver disease | 0.026 (0.004) | 4.48E-09 | 146.0 | 1.70E-01 | 0.020 (0.007) | 9.02E-03 | 0.008 (0.008) | 4.09E-01 | 144.9 | 1.64E-01 | 0.022 (0.007) | 3.05E-03 | 0.021 (0.007) | 4.87E-03 |
| Abnormal results of function study of liver | 0.028 (0.003) | 6.00E-20 | 146.5 | 1.70E-01 | 0.019 (0.005) | 4.74E-04 | 0.012 (0.006) | 4.08E-02 | 141.1 | 2.01E-01 | 0.022 (0.005) | 9.79E-05 | 0.026 (0.005) | 8.63E-08 | |
| Cholelithiasis | 0.012 (0.002) | 4.96E-08 | 137.5 | 1.99E-01 | 0.011 (0.004) | 3.95E-03 | 0.001 (0.004) | 7.98E-01 | 137.4 | 1.91E-01 | 0.014 (0.003) | 3.33E-05 | 0.014 (0.003) | 1.24E-04 | |
| ALP | Hypercholesterolemia | − 0.003 (0.001) | 6.10E-04 | 248.0 | 1.30E-02 | − 0.003 (0.001) | 2.74E-02 | 0.000 (0.002) | 9.89E-01 | 247.9 | 1.12E-02 | − 0.003 (0.001) | 4.38E-02 | − 0.004 (0.001) | 1.21E-02 |
| Pulmonary heart disease | − 0.013 (0.002) | 1.64E-07 | 244.4 | 6.56E-02 | − 0.013 (0.004) | 2.51E-03 | 0.000 (0.005) | 9.93E-01 | 244.4 | 5.78E-02 | − 0.007 (0.004) | 2.15E-01 | − 0.012 (0.003) | 2.51E-03 | |
| Phlebitis and thrombophlebitis of lower extremities | − 0.009 (0.002) | 2.51E-03 | 260.5 | 1.30E-02 | − 0.007 (0.004) | 2.15E-01 | − 0.003 (0.005) | 7.19E-01 | 260.3 | 1.12E-02 | − 0.008 (0.004) | 1.84E-01 | − 0.009 (0.004) | 3.67E-02 | |
| Circulatory disease NEC | − 0.001 (0.001) | 5.79E-01 | 273.7 | 5.64E-03 | − 0.003 (0.002) | 2.66E-01 | 0.003 (0.002) | 4.47E-01 | 272.0 | 6.16E-03 | 0.002 (0.003) | 7.08E-01 | 0.000 (0.002) | 9.89E-01 | |
MR Mendelian randomization, IVW inverse-variance weighting, MBE mode-based estimate method, WMM weighted median method, FDR false discovery rate, ALT alanine aminotransferase, AST aspartate aminotransferase, ALP alkaline phosphatase, GGT λ-glutamyl transferase
The estimates were derived from MR analyses based on untransformed liver enzyme levels, and the outliers identified by MR-PRESSO method have been removed
The P value in bold denotes statistical significance
Results from IVW method suggest that a genetically determined high serum ALT level was associated with increased risk of hepatitis NOS (OR = 1.25, 95%CI 1.16–1.34), primary liver cancer (1.32, 1.19–1.46), T2D (1.02, 1.01–1.03), disorders of iron metabolism (1.12, 1.07–1.17), alcoholic liver damage (1.17, 1.11–1.23), esophageal bleeding (1.08, 1.05–1.12), liver abscess and sequelae of chronic liver disease (1.12, 1.05–1.19), non-alcoholic cirrhosis (1.24, 1.16–1.32), choleithiasis (1.04, 1.02–1.06), hepatomegaly (1.16, 1.07–1.24), portal hypertension (1.16, 1.07–1.24), other chronic non-alcoholic liver disease (1.20, 1.16–1.25), and abnormal results of function study of liver (1.07, 1.05–1.10) (Fig. 5; Table 1). These correlations have also been demonstrated in most of other methods, although the nuances of the MR estimates. By contrast, genetically determined serum ALT level was negatively associated with risk of corneal degeneration (OR = 0.79, 95%CI 0.70–0.88) and fracture of hand or wrist (0.95, 0.93–0.97).
According to the estimates of IVW methods, genetically predicted AST level was positively correlated with other chronic non-alcoholic liver disease (OR = 1.13, 95%CI 1.08–1.19), disorder of iron metabolism (1.11, 1.02–1.20), other ill-defined and unknown causes of morbidity and mortality (1.04, 1.02–1.05), varicose veins of lower extremity (1.04, 1.02–1.06), hallux valgus (1.06, 1.03–1.08), and ankylosing spondylitis (1.11, 1.01–1.22). On the contrary, genetically determined AST level was negatively associated with multiple sclerosis (OR = 0.92, 95%CI 0.88–0.97), thyrotoxicosis with or without goiter (0.92, 0.87–0.97), celiac disease (0.67, 0.55–0.82), hereditary disturbances in tooth structure (0.58, 0.45–0.76), dermatitis herpetiformis (0.56, 0.41–0.77), and cancer of the gums (0.45, 0.34–0.59) (Fig. 5).
Genetically predicted GGT level was positively associated with the risk of other chronic non-alcoholic liver disease (OR = 1.03, 95%CI 1.02–1.03), abnormal results of function study of liver (1.03, 1.02–1.03), and cholelithiasis (1.01, 1.01–1.02). Genetically determined ALP level was negatively associated with pulmonary heart disease (OR = 0.987, 95%CI 0.983–0.991), phlebitis and thrombophlebitis of lower extremities (0.991, 0.987–0.996) and hypercholesterolemia (0.997, 0.995–0.998). These associations have also been demonstrated in MR methods other than IVW.
Multivariable Mendelian Randomization Analysis
In the analysis of MVMR, we further validated the causal relationships of liver enzymes with disease phenotypes based on 432 genetic variants (Supplementary Table S21). The results of MVMR-Lasso and MVMR-robust methods were not only concordant with each other but were also largely consistent with the results from univariable MR analysis (Supplementary Table S22). The significant associations of genetically determined ALT level with corneal degenerations, hepatomegaly, and fracture of hand or wrist that detected in univariable MR analysis were disappeared according to both MVMR-Lasso and MVMR-robust analyses. The associations of genetically determined AST with hypothyroidism, disorders of iron metabolism and other chronic non-alcoholic liver disease were become statistically non-significant. Additionally, we did not detect the association between genetically determined GGT level and other chronic non-alcoholic liver disease in the MVMR analyses. The results of sensitivity analysis were shown in Supplementary Table S23. In contrast to associations detected from PheWAS and MR analysis, we identified much more associations that reach a statistical significance (353 for ALT, 485 for AST, 729 for GGT, and 819 for ALP).
Discussion
In this MR-PheWAS analysis, we reported associations of liver enzymes with a set of intrahepatic and extrahepatic disease phenotypes. Genetically determined ALT level was not only associated with several intrahepatic disease phenotypes but also associated with extrahepatic outcomes such as T2D and corneal degeneration. The AST-related disease phenotypes were not limited to liver or digestive system and were more diverse than that of ALT in terms of disease categories. In contrast, genetically predicted GGT level was only found to be positively associated with three hepatic outcomes. Genetically determined ALP level was found to be negatively associated with hypercholesterolemia and two circulatory system diseases, that are pulmonary heart disease and phlebitis and thrombophlebitis of lower extremities.
ALT and AST are found abundantly within hepatocytes, and they catalyse the transfer of amino groups to generate products of gluconeogenesis and amino acid metabolism (Kunutsor et al. 2014). Aminotransferase in blood is deemed to be a consequence of the liver cell membrane damage, which leads the subsequent leakage of intracellular enzymes into the circulation (Sookoian and Pirola 2015). Serum level of ALT has been regarded as the leading biomarker and even the gold standard of liver injury in clinical practice for decades (Pratt and Kaplan 2000). AST is also found in many extrahepatic tissues, and thus it is not a specific marker for liver injury. ALP and GGT are other two membrane-bound enzymes that mainly found in biliary epithelial cells and hepatocytes. ALP is considered to be a sensitive marker for cholestasis with a reported sensitivity of 85% (Lawrence and Steiner 2017). Elevations of serum levels of GGT are attributed to cholestasis or biliary hyperplasia resulting in enzyme induction (Lawrence and Steiner 2017). Previous observational studies indicated a wide spectrum of effects of liver enzymes on diseases phenotypes, although the underlying mechanisms are not completely clear (Kunutsor et al. 2015; Rahmani et al. 2019).
In this study, we first conducted a large-scale GWAS for the liver enzymes, identifying a set of genetic variants associated with serum levels of liver enzyme, of which most are novel associations and some are concordant with previous GWAS (Chambers et al. 2011). Leveraging the genetic variants, we constructed PRS of liver enzymes and detected numerous links between genetically determined liver enzyme and disease phenotypes. Most of the links were validated in the subsequent MR analysis, which might indicate a causal relationship between liver enzyme and disease phenotype. However, the detected causal relationships still need further validation. For ALT, twelve out of the sixteen identified disease phenotypes were closely link to the liver, including hepatitis, primary liver cancer, and non-alcoholic cirrhosis. Genetically predicted level of ALT was positively associated with the risk of these hepatic outcomes. In other words, ALT might have its own role in the onset of these diseases rather than only serving as an indicator. This result might be contradictory with the conventional perception that elevated ALT level is a consequence instead of a cause of liver diseases. However, we found that the hepatic disease phenotypes shared many genetic variants with ALT, of which the most noticeable variant is rs738409 of PNPLA3. Growing evidences have shown that rs738409 of PNPLA3 was significantly associated with a wide range of liver diseases from non-alcoholic fatty liver disease to hepatocellular carcinoma (Liu et al. 2014a; Romeo et al. 2008). The similar phenomenon was also observed for rs58542926 of TM6SF2 and rs28929474 of SERPINA1 (Mandorfer et al. 2018; Tang et al. 2019). These genetic variants including those have not been extensively investigated (e.g., rs41318029 of ABCC2) might mediate the biological effect of ALT on liver diseases. Since the elevation of ALT level usually precedes the genesis of liver diseases, ALT can be termed as a cause as well as a strong indicator for these diseases. Consistent to a previous MR study and an observational study (Goessling et al. 2008; Liu et al. 2016), we found that genetically determined ALT level was positively associated with T2D. T2D shared genetic associations with ALT level at 10 loci, including rs1801282 of PPARγ that has been demonstrated to be associated with risk of T2D (Rehman et al. 2020). Moreover, ALT is thought to cause diabetes via insulin resistance with hepatic steatosis aggravating insulin resistance and creating a vicious cycle (Jacobs et al. 2011). We also observed a positive correlation between genetically determined ALT level and iron metabolism disorder, which has been reported in an observational study (Ruhl and Everhart 2003). Hepatic iron could play a role in liver injury by promoting oxidative stress either as a catalyst in the formation of free radicals or as a direct cause of lipid peroxidation (Bloomer and Brown 2019). However, the mechanism under the correlation of ALT with iron homeostasis remains elusive. Interestingly, we found negative associations of genetically determined ALT level with corneal degeneration and fracture of hand or wrist, although the association became non-significant in the MVMR analysis. No shared genetic variant was detected between ALT and these two disease phenotypes. These associations should be further validated in future researches.
For AST, we identified 17 pairs of genotype–phenotype associations, which covered a wide range of phenotypic categories including endocrine/metabolic diseases, digestive diseases, and neurological disorder. The results of MR-IVW analysis suggested a potential causal effect of AST level on 14 of the seventeen phenotypes. Moreover, these associations were rarely reported in observational studies, except for multiple sclerosis, iron metabolism disorder, chronic non-alcoholic liver disease, and thyrotoxicosis (Jensen et al. 2003; Tremlett et al. 2006; Wu et al. 2010). Most of the shared genetic variants between AST and disease phenotypes have not been previously investigated but showed significant association with several diseases in the current study. For instance, rs1033499 A allele of TSBP1 was associated with celiac disease and thyrotoxicosis with a FDR-Q value of 5.00 × 10–255 and 5.41 × 10–11, respectively. By contrast, a few of the shared genetic variants were also related to diseases that not reported in our study. For example, the MHC rs9257809 G allele was associated with Barrett’s oesophagus according to a previous study (Su et al. 2012). In our study, we found that this variant was significantly associated with celiac disease with a FDR-Q value as low as 8.92 × 10–127. Our findings not only suggested multiple function of AST but also indicated a potential biological effect of the selected genetic variants. Genetically determined GGT and ALP levels were associated with limited disease phenotypes. For GGT, the genotype–phenotype correlation remained significant according to all MR methods, suggesting a robust causal relationship between GGT level and the three hepatic diseases. Genetically determined ALP level was negatively associated with hypercholesterolemia, although the effect was weak. This result was contradictory with previously observational study that reported a positive association between ALP level and hypercholesterolemia (Webber et al. 2010). Hypercholesterolemia shared genetic associations with ALP level at 18 loci, of which 12 loci showed negative association with hypercholesterolemia. In this regard, the association between ALP level and hypercholesterolemia might be confounded in observational studies and warrants further research. We also observed a significant negative association between genetically ALP level and pulmonary heart disease in terms of most MR analyses. This link has not been reported elsewhere. Of note is that between-SNP heterogeneity was significant for the four genotype–phenotype pairs of ALP, although we have aforehand removed outliers that identified in MR-PRESSO analysis. The MR estimates of ALP should be interpreted with cautions.
Our study has limitations. First, although the discovery and validation cohort of liver enzyme PRS were independent, they were both derived from UK Biobank cohort. Our results need further external validation. Second, the disease phenotypes were retrieved from electronic health records, which might be subject to misclassification and underreporting. Third, it is hard to interpret most links identified in our study based on the current evidences, which is a common defect for PheWAS. However, our findings can serve as hypothesis-generators for future studies into the biological mechanisms underpinning these links. Finally, MR analysis lacks the ability to assess the effect of an acute increase of exposure.
In summary, leveraging a large-scale population data, this MR-PheWAS study revealed several novel links between liver enzyme and disease phenotypes and provided insights into the full understanding of the roles of liver enzymes in biological processes.
Supplementary Information
Below is the link to the electronic supplementary material.
Acknowledgements
We sincerely appreciate the great works of UK Biobank collaborators.
Authors' contributions
Conceptualization: ZL and XC; data curation: ZL and RZ; formal analysis: ZL, CS, and YJ; methodology: ZL, LJ, and XC; supervision: LJ and XC; visualization: ZL and CS; writing-original draft: ZL, CS, and YJ; writing-review and editing: all authors.
Funding
This work was supported by the National Postdoctoral Program for Innovative Talents (grant number: BX2021077), National Natural Science Foundation of China (grant numbers: 91846302, 82073637), the National Key Research and Development Program of China (grant numbers: 2017YFC0907000, 2017YFC0907500, 2019YFC1315804, 2019FY101103), key basic research grants from the Science and Technology Commission of Shanghai Municipality (grant number: 16JC1400500), the Shanghai Municipal Science and Technology Major Project (grant number: 2017SHZDZX01), Three-Year Action Plan for Strengthening Public Health System in Shanghai (grant number: GWV-10.2-YQ32), and Innovation Grant from Science and Technology Commission of Shanghai Municipality, China (grant number: 20ZR1405600).
Data availability
The UK Biobank data are available from the UK Biobank on request (www.ukbiobank.ac.uk/).
Declarations
Conflicts of interests
The authors declared none conflicts of interests. LJ is the Editor-in-Chief of Phenomics, and he was not involved in reviewing this paper.
Ethical approval and consent to participate
The UK Biobank received ethical approval from the research ethics committee (REC reference for UK Biobank 11/NW/0382), and participants provided written informed consent.
Consent for publication
Not applicable.
Footnotes
Zhenqiu Liu, Chen Suo and Yanfeng Jiang contributed equally.
References
- Bloomer SA, Brown KE. Iron-induced liver injury: a critical reappraisal. Int J Mol Sci. 2019;20(9):2132. doi: 10.3390/ijms20092132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burgess S, Thompson SG. Multivariable mendelian randomization: the use of pleiotropic genetic variants to estimate causal effects. Am J Epidemiol. 2015;181(4):251–260. doi: 10.1093/aje/kwu283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burgess S, Thompson SG. Interpreting findings from Mendelian randomization using the MR-egger method. Eur J Epidemiol. 2017;32(5):377–389. doi: 10.1007/s10654-017-0255-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burgess S, Davey Smith G, Davies NM, Dudbridge F, Gill D, Glymour MM, et al. Guidelines for performing Mendelian randomization investigations. Wellcome Open Res. 2019;4:186. doi: 10.12688/wellcomeopenres.15555.2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chambers JC, Zhang W, Sehmi J, Li X, Wass MN, Van der Harst P, et al. Genome-wide association study identifies loci influencing concentrations of liver enzymes in plasma. Nat Genet. 2011;43(11):1131–1138. doi: 10.1038/ng.970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chatterjee N, Shi J, García-Closas M. Developing and evaluating polygenic risk prediction models for stratified disease prevention. Nat Rev Genet. 2016;17(7):392–406. doi: 10.1038/nrg.2016.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen VL, Du X, Chen Y, Kuppa A, Handelman SK, Vohnoutka RB, et al. Genome-wide association study of serum liver enzymes implicates diverse metabolic and liver pathology. Nat Commun. 2021;12(1):816. doi: 10.1038/s41467-020-20870-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi KM, Han K, Park S, Chung HS, Kim NH, Yoo HJ, et al. Implication of liver enzymes on incident cardiovascular diseases and mortality: a nationwide population-based cohort study. Sci Rep. 2018;8(1):3764. doi: 10.1038/s41598-018-19700-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clarke R, Shipley M, Lewington S, Youngman L, Collins R, Marmot M, et al. Underestimation of risk associations due to regression dilution in long-term follow-up of prospective studies. Am J Epidemiol. 1999;150(4):341–353. doi: 10.1093/oxfordjournals.aje.a010013. [DOI] [PubMed] [Google Scholar]
- Danielsson J, Kangastupa P, Laatikainen T, Aalto M, Niemelä O. Impacts of common factors of life style on serum liver enzymes. World J Gastroenterol. 2014;20(33):11743–11752. doi: 10.3748/wjg.v20.i33.11743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Silva NMG, Borges MC, Hingorani AD, Engmann J, Shah T, Zhang X, et al. Liver function and risk of type 2 diabetes: bidirectional mendelian randomization study. Diabetes. 2019;68(8):1681–1691. doi: 10.2337/db18-1048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding M, Huang T, Bergholdt HK, Nordestgaard BG, Ellervik C, Qi L. Dairy consumption, systolic blood pressure, and risk of hypertension: Mendelian randomization study. BMJ. 2017;356:j1000. doi: 10.1136/bmj.j1000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ference BA, Ray KK, Catapano AL, Ference TB, Burgess S, Neff DR, et al. Mendelian randomization study of ACLY and cardiovascular disease. N Engl J Med. 2019;380(11):1033–1042. doi: 10.1056/NEJMoa1806747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghouri N, Preiss D, Sattar N. Liver enzymes, nonalcoholic fatty liver disease, and incident cardiovascular disease: a narrative review and clinical perspective of prospective data. Hepatology. 2010;52(3):1156–1161. doi: 10.1002/hep.23789. [DOI] [PubMed] [Google Scholar]
- Goessling W, Massaro JM, Vasan RS, D'Agostino RB, Sr, Ellison RC, Fox CS. Aminotransferase levels and 20-year risk of metabolic syndrome, diabetes, and cardiovascular disease. Gastroenterology. 2008;135(6):1935–1944. doi: 10.1053/j.gastro.2008.09.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grant A, Burgess S (2020) Pleiotropy robust methods for multivariable Mendelian randomization. arXiv 2020, 11997 [DOI] [PMC free article] [PubMed]
- Jacobs M, van Greevenbroek MM, van der Kallen CJ, Ferreira I, Feskens EJ, Jansen EH, et al. The association between the metabolic syndrome and alanine amino transferase is mediated by insulin resistance via related metabolic intermediates (the Cohort on Diabetes and Atherosclerosis Maastricht [CODAM] study) Metabolism. 2011;60(7):969–975. doi: 10.1016/j.metabol.2010.09.006. [DOI] [PubMed] [Google Scholar]
- Jensen PD, Jensen FT, Christensen T, Nielsen JL, Ellegaard J. Relationship between hepatocellular injury and transfusional iron overload prior to and during iron chelation with desferrioxamine: a study in adult patients with acquired anemias. Blood. 2003;101(1):91–96. doi: 10.1182/blood-2002-06-1704. [DOI] [PubMed] [Google Scholar]
- Kaneko K, Yatsuya H, Li Y, Uemura M, Chiang C, Hirakawa Y, et al. Association of gamma-glutamyl transferase and alanine aminotransferase with type 2 diabetes mellitus incidence in middle-aged Japanese men: 12-year follow up. J Diabetes Investig. 2019;10(3):837–845. doi: 10.1111/jdi.12930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katzke V, Johnson T, Sookthai D, Hüsing A, Kühn T, Kaaks R. Circulating liver enzymes and risks of chronic diseases and mortality in the prospective EPIC-Heidelberg case-cohort study. BMJ Open. 2020;10(3):e033532. doi: 10.1136/bmjopen-2019-033532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koehler EM, Sanna D, Hansen BE, van Rooij FJ, Heeringa J, Hofman A, et al. Serum liver enzymes are associated with all-cause mortality in an elderly population. Liver Int. 2014;34(2):296–304. doi: 10.1111/liv.12311. [DOI] [PubMed] [Google Scholar]
- Kunutsor SK, Apekey TA, Seddoh D, Walley J. Liver enzymes and risk of all-cause mortality in general populations: a systematic review and meta-analysis. Int J Epidemiol. 2014;43(1):187–201. doi: 10.1093/ije/dyt192. [DOI] [PubMed] [Google Scholar]
- Kunutsor SK, Apekey TA, Van Hemelrijck M, Calori G, Perseghin G. Gamma glutamyltransferase, alanine aminotransferase and risk of cancer: systematic review and meta-analysis. Int J Cancer. 2015;136(5):1162–1170. doi: 10.1002/ijc.29084. [DOI] [PubMed] [Google Scholar]
- Lawrence YA, Steiner JM. Laboratory evaluation of the liver. Vet Clin N Am Small Anim Pract. 2017;47(3):539–553. doi: 10.1016/j.cvsm.2016.11.005. [DOI] [PubMed] [Google Scholar]
- Li X, Meng X, Spiliopoulou A, Timofeeva M, Wei WQ, Gifford A, et al. MR-PheWAS: exploring the causal effect of SUA level on multiple disease outcomes by using genetic instruments in UK Biobank. Ann Rheum Dis. 2018;77(7):1039–1047. doi: 10.1136/annrheumdis-2017-212534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Meng X, He Y, Spiliopoulou A, Timofeeva M, Wei WQ, et al. Genetically determined serum urate levels and cardiovascular and other diseases in UK Biobank cohort: a phenome-wide Mendelian randomization study. PLoS Med. 2019;16(10):e1002937. doi: 10.1371/journal.pmed.1002937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu YL, Patman GL, Leathart JB, Piguet AC, Burt AD, Dufour JF, et al. Carriage of the PNPLA3 rs738409 C >G polymorphism confers an increased risk of non-alcoholic fatty liver disease associated hepatocellular carcinoma. J Hepatol. 2014;61(1):75–81. doi: 10.1016/j.jhep.2014.02.030. [DOI] [PubMed] [Google Scholar]
- Liu Z, Que S, Xu J, Peng T. Alanine aminotransferase-old biomarker and new concept: a review. Int J Med Sci. 2014;11(9):925–935. doi: 10.7150/ijms.8951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Au Yeung SL, Lin SL, Leung GM, Schooling CM. Liver enzymes and risk of ischemic heart disease and type 2 diabetes mellitus: a mendelian randomization study. Sci Rep. 2016;6:38813. doi: 10.1038/srep38813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mandorfer M, Bucsics T, Hutya V, Schmid-Scherzer K, Schaefer B, Zoller H, et al. Liver disease in adults with α1-antitrypsin deficiency. United Eur Gastroenterol J. 2018;6(5):710–718. doi: 10.1177/2050640618764057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monami M, Bardini G, Lamanna C, Pala L, Cresci B, Francesconi P, et al. Liver enzymes and risk of diabetes and cardiovascular disease: results of the Firenze Bagno a Ripoli (FIBAR) study. Metabolism. 2008;57(3):387–392. doi: 10.1016/j.metabol.2007.10.015. [DOI] [PubMed] [Google Scholar]
- Noordam R, Smit RA, Postmus I, Trompet S, van Heemst D. Assessment of causality between serum gamma-glutamyltransferase and type 2 diabetes mellitus using publicly available data: a Mendelian randomization study. Int J Epidemiol. 2016;45(6):1953–1960. doi: 10.1093/ije/dyw306. [DOI] [PubMed] [Google Scholar]
- Nunez DJ, Alexander M, Yerges-Armstrong L, Singh G, Byttebier G, Fabbrini E, et al. Factors influencing longitudinal changes of circulating liver enzyme concentrations in subjects randomized to placebo in four clinical trials. Am J Physiol Gastrointest Liver Physiol. 2019;316(3):G372–g386. doi: 10.1152/ajpgi.00051.2018. [DOI] [PubMed] [Google Scholar]
- Pingault JB, O'Reilly PF, Schoeler T, Ploubidis GB, Rijsdijk F, Dudbridge F. Using genetic data to strengthen causal inference in observational research. Nat Rev Genet. 2018;19(9):566–580. doi: 10.1038/s41576-018-0020-3. [DOI] [PubMed] [Google Scholar]
- Pratt DS, Kaplan MM. Evaluation of abnormal liver-enzyme results in asymptomatic patients. N Engl J Med. 2000;342(17):1266–1271. doi: 10.1056/nejm200004273421707. [DOI] [PubMed] [Google Scholar]
- Privé F, Aschard H, Ziyatdinov A, Blum MGB. Efficient analysis of large-scale genome-wide data with two R packages: bigstatsr and bigsnpr. Bioinformatics. 2018;34(16):2781–2787. doi: 10.1093/bioinformatics/bty185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rahmani J, Miri A, Namjoo I, Zamaninour N, Maljaei MB, Zhou K, et al. Elevated liver enzymes and cardiovascular mortality: a systematic review and dose-response meta-analysis of more than one million participants. Eur J Gastroenterol Hepatol. 2019;31(5):555–562. doi: 10.1097/meg.0000000000001353. [DOI] [PubMed] [Google Scholar]
- Rehman K, Jabeen K, Awan FR, Hussain M, Saddique MA, Akash MSH. Biochemical investigation of rs1801282 variations in PPAR-γ gene and its correlation with risk factors of diabetes mellitus in coronary artery disease. Clin Exp Pharmacol Physiol. 2020;47(9):1517–1529. doi: 10.1111/1440-1681.13339. [DOI] [PubMed] [Google Scholar]
- Richmond RC, Anderson EL, Dashti HS, Jones SE, Lane JM, Strand LB, et al. Investigating causal relations between sleep traits and risk of breast cancer in women: Mendelian randomisation study. BMJ. 2019;365:l2327. doi: 10.1136/bmj.l2327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romeo S, Kozlitina J, Xing C, Pertsemlidis A, Cox D, Pennacchio LA, et al. Genetic variation in PNPLA3 confers susceptibility to nonalcoholic fatty liver disease. Nat Genet. 2008;40(12):1461–1465. doi: 10.1038/ng.257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruhl CE, Everhart JE. Relation of elevated serum alanine aminotransferase activity with iron and antioxidant levels in the United States. Gastroenterology. 2003;124(7):1821–1829. doi: 10.1016/s0016-5085(03)00395-0. [DOI] [PubMed] [Google Scholar]
- Siddiqui MS, Sterling RK, Luketic VA, Puri P, Stravitz RT, Bouneva I, et al. Association between high-normal levels of alanine aminotransferase and risk factors for atherogenesis. Gastroenterology. 2013;145(6):1271–1279.e1271-1273. doi: 10.1053/j.gastro.2013.08.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith JG, Luk K, Schulz CA, Engert JC, Do R, Hindy G, et al. Association of low-density lipoprotein cholesterol-related genetic variants with aortic valve calcium and incident aortic stenosis. JAMA. 2014;312(17):1764–1771. doi: 10.1001/jama.2014.13959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sookoian S, Pirola CJ. Liver enzymes, metabolomics and genome-wide association studies: from systems biology to the personalized medicine. World J Gastroenterol. 2015;21(3):711–725. doi: 10.3748/wjg.v21.i3.711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stender S, Kozlitina J, Nordestgaard BG, Tybjærg-Hansen A, Hobbs HH, Cohen JC. Adiposity amplifies the genetic risk of fatty liver disease conferred by multiple loci. Nat Genet. 2017;49(6):842–847. doi: 10.1038/ng.3855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su Z, Gay LJ, Strange A, Palles C, Band G, Whiteman DC, et al. Common variants at the MHC locus and at chromosome 16q24.1 predispose to Barrett's esophagus. Nat Genet. 2012;44(10):1131–1136. doi: 10.1038/ng.2408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sudlow C, Gallacher J, Allen N, Beral V, Burton P, Danesh J, et al. UK biobank: an open access resource for identifying the causes of a wide range of complex diseases of middle and old age. PLoS Med. 2015;12(3):e1001779. doi: 10.1371/journal.pmed.1001779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang S, Zhang J, Mei TT, Guo HQ, Wei XH, Zhang WY, et al. Association of TM6SF2 rs58542926 T/C gene polymorphism with hepatocellular carcinoma: a meta-analysis. BMC Cancer. 2019;19(1):1128. doi: 10.1186/s12885-019-6173-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tremlett H, Seemüller S, Zhao Y, Yoshida EM, Oger JD, Petkau J. Liver test abnormalities in multiple sclerosis: findings from placebo-treated patients. Neurology. 2006;67(7):1291–1293. doi: 10.1212/01.wnl.0000238515.27055.62. [DOI] [PubMed] [Google Scholar]
- Verbanck M, Chen CY, Neale B, Do R. Detection of widespread horizontal pleiotropy in causal relationships inferred from Mendelian randomization between complex traits and diseases. Nat Genet. 2018;50(5):693–698. doi: 10.1038/s41588-018-0099-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voight BF, Peloso GM, Orho-Melander M, Frikke-Schmidt R, Barbalic M, Jensen MK, et al. Plasma HDL cholesterol and risk of myocardial infarction: a Mendelian randomisation study. Lancet. 2012;380(9841):572–580. doi: 10.1016/s0140-6736(12)60312-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Webber M, Krishnan A, Thomas NG, Cheung BM. Association between serum alkaline phosphatase and C-reactive protein in the United States National Health and Nutrition Examination Survey 2005–2006. Clin Chem Lab Med. 2010;48(2):167–173. doi: 10.1515/cclm.2010.052. [DOI] [PubMed] [Google Scholar]
- Wu W, Sun Z, Yu J, Meng Q, Wang M, Miao J, et al. A clinical retrospective analysis of factors associated with apathetic hyperthyroidism. Pathobiology. 2010;77(1):46–51. doi: 10.1159/000272954. [DOI] [PubMed] [Google Scholar]
- Wu P, Gifford A, Meng X, Li X, Campbell H, Varley T, et al. Mapping ICD-10 and ICD-10-CM codes to phecodes: workflow development and initial evaluation. JMIR Med Inform. 2019;7(4):e14325. doi: 10.2196/14325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu F, Huang Y, Hu J, Shao Z. Mendelian randomization study of inflammatory bowel disease and bone mineral density. BMC Med. 2020;18(1):312. doi: 10.1186/s12916-020-01778-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu L, Jiang CQ, Lam TH, Zhang WS, Zhu F, Jin YL, et al. Mendelian randomization estimates of alanine aminotransferase with cardiovascular disease: Guangzhou Biobank Cohort study. Hum Mol Genet. 2017;26(2):430–437. doi: 10.1093/hmg/ddw396. [DOI] [PubMed] [Google Scholar]
- Yavorska OO, Burgess S. MendelianRandomization: an R package for performing Mendelian randomization analyses using summarized data. Int J Epidemiol. 2017;46(6):1734–1739. doi: 10.1093/ije/dyx034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan X, Waterworth D, Perry JR, Lim N, Song K, Chambers JC, et al. Population-based genome-wide association studies reveal six loci influencing plasma levels of liver enzymes. Am J Hum Genet. 2008;83(4):520–528. doi: 10.1016/j.ajhg.2008.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The UK Biobank data are available from the UK Biobank on request (www.ukbiobank.ac.uk/).