Abstract
The N-terminal FERM domain of focal adhesion kinase (FAK) contributes to FAK scaffolding and interacts with HER2, an oncogene and receptor tyrosine kinase. The interaction between HER2 and FAK drives resistance to FAK-kinase domain inhibitors through FAK Y397 transphosphorylation and FAK re-activation upon inhibition. As such, FAK FERM remains an attractive drug discovery target. In this report, we detail an alternative approach to targeting FAK through virtual screening-based discovery of chemical probes that target FAK FERM. We validated the binding interface between HER2 and FAK using site-directed mutagenesis and GST pull-down experiments. We assessed the ligandability of key binding residues of HER2 and FAK utilizing computational tools. We developed a virtual screening method to screen approximately 200,000 compounds against the FAK FERM domain, identifying 20 chemical probes. We performed GST pull-down screening on these compounds, discovering two hits, VS4 and VS14, with nanomolar IC50s in disrupting HER2-FAK. We performed further testing, including molecular docking, immunofluorescence, phosphorylation, and cellular invasion assays to evaluate the compounds’ biological effects. One probe, VS14, was identified with the ability to block both auto- and trans-phosphorylation of Y397. In all, these studies identify two new probes that target FAK FERM, enabling future investigation of this domain.
Keywords: Focal Adhesion Kinase, FAK FERM domain, HER2, Virtual Screening, High-throughput screening, Phosphorylation
1. INTRODUCTION
Focal adhesion kinase (FAK), a 125 kDa non-receptor tyrosine kinase overexpressed in several invasive solid tumors, is an important protein involved in tumor development with multiple implications in cancer cell signaling, proliferation, migration and metastasis (Lark et al., 2003; McLean et al., 2005; Owens et al., 1995; Weiner, Liu, Craven, & Cance, 1993). While FAK is overexpressed in most solid tumor types, it shows limited expression in its normal tissue counterpart (Cance et al., 2000; Golubovskaya, 2014; Ozkal et al., 2009). The predominance of FAK in cancerous tissue has made it a promising drug target, as inhibition of FAK has been shown to limit cancer metastasis and growth. FAK is comprised of three domains: the N-terminal 4.1 ezrin-radixin-moesin domain (FERM), the central kinase domain, and the C-terminal focal adhesion targeting (FAT) domain (T. Marlowe et al., 2019). Through FERM, FAK can function as an intracellular scaffold where it can support several protein-protein interactions to connect multiple oncogenic signaling pathways (Dunty et al., 2004; Frame, Patel, Serrels, Lietha, & Eck, 2010; T. Marlowe et al., 2019). FAK is canonically activated via autophosphorylation of its key tyrosine residue 397 (Y397) through the utilization of its own kinase domain (Fang et al., 2014; Golubovskaya et al., 2012; T. A. Marlowe, Lenzo, Figel, Grapes, & Cance, 2016).This phosphorylated tyrosine residue serves as a Src-Homology 2 (SH2)-domain docking site for SRC-family kinases, PI3K, and GRB7, leading to the full phosphorylation of FAK at effector downstream tyrosine residues Y861 and Y925 (Calalb, Zhang, Polte, & Hanks, 1996; Cobb, Schaller, Leu, & Parsons, 1994; Han & Guan, 1999; T. A. Marlowe et al., 2016).
FAK is also activated via transphosphorylation by receptor tyrosine kinases (RTKs) that directly bind to the FERM domain (T. A. Marlowe et al., 2016). We were the first to show that human epidermal growth factor receptor 2 (HER2) directly binds to the FAK-FERM domain to promote phosphorylation of key residue Y397 independently of FAK-kinase function (T. A. Marlowe et al., 2016). Furthermore, additional RTKs, including EGFR, Tie2, EphA2, and FGFR4, can directly phosphorylate FAK at Y397. Despite these data, the classical approach to targeting FAK has been by inhibiting the kinase domain, specifically the ATP binding site, to inhibit kinase enzymatic activity (Golubovskaya, 2014; Mousson et al., 2018; Roberts et al., 2008; Shi et al., 2007; Slack-Davis et al., 2007). Even though the kinase domain has been thoroughly explored as a therapeutic strategy to target FAK, partially due to advantages regarding potency, inhibiting this domain has not yet proven successful in the clinic (de Jonge et al., 2019; Infante et al., 2012; Jones et al., 2015). As a potential resistance mechanism, we have shown that cancer cells alter their kinase expression and phosphorylation levels (kinome reprogramming) upon FAK-kinase inhibition to reactivate FAK at Y397 and downstream signaling molecules (T. A. Marlowe et al., 2016). Traditional FAK-kinase inhibitors are only able to block autophosphorylation of FAK, whereby RTKs such as HER2 can trans-phosphorylate FAK at Y397 and maintain SH2 docking for Src-kinases, rendering kinase inhibitors ineffective. Furthermore, HER2-positive cancer cells are innately resistant to FAK-kinase inhibitors and do not show any viability changes upon treatment (T. A. Marlowe et al., 2016). The resistance mechanisms that evolve due to Y397 transphosphorylation could help explain why specific FAK-kinase inhibitors have showed limited efficacy in early phase I/II clinical trials (de Jonge et al., 2019; Infante et al., 2012; Jones et al., 2015).
The FERM domain remains an attractive domain for FAK inhibition, primarily due to its interactions with multiple oncogenes, receptors, integrins, and cytoskeletal proteins (Frame et al., 2010). The FAK FERM domain is required for growth factor promoted cell motility, irrespective of FAK-kinase activity (Sieg et al., 2000). Of particular interest is the protein-protein interaction between the FAK FERM F1 lobe and HER2, a known oncogene and receptor tyrosine kinase. It has been identified that the HER2-FAKinteraction drives HER2-dependent transphosphorylation of FAK at Y397 (T. A. Marlowe et al., 2016). Furthermore, the HER2-FAK interface has been characterized by molecular modeling and biological studies (T. A. Marlowe et al., 2016). GST pull-down assays, in order to determine the mechanism of this interaction, have shown that the FAK FERM domain, specifically the F1 lobe is able to bind to the HER2 kinase domain N-lobe (T.A. Marlowe et al., 2016). Molecular modeling of this interaction, as produced utilizing High Ambiguity Driven protein-protein DOCKing (HADDOCK) studies, shows that the Y397 residue, which is located in the FERM-kinase linker region, binds onto the FERM F1 lobe in order to be proximal to the HER2-FAK interaction (T.A. Marlowe et al., 2016). This interaction has been shown to be similar to a kinase-substrate interaction, as contact between the FAK FERM F1 lobe and the N-lobe of HER2 facilitates the orientation of the flexible Y397 FERM-linker, so that it would be near both the HER2 substrate-binding region and the ATP binding pocket (T.A. Marlowe et al., 2016). The HER2-FAK interaction allows significant signaling crosstalk between the two, where FAK is required for HER2 oncogenesis and HER2 allows for resistance to FAK-kinase inhibitors, including phosphorylating kinase-dead FAK (Benlimame et al., 2005; T. A. Marlowe et al., 2016). FAK activation is also involved in the acquired resistance to HER2 inhibitors, trastuzumab and lapatinib (Huang et al., 2011; Jin et al., 2017; Yang et al., 2010). By being able to disrupt this protein-protein interaction, the effects of both HER2 and FAK on cancerous cells should be drastically limited, possibly allowing for the mitigation of cancer growth/invasion and HER2-FAK crosstalk signaling. In addition, the development of FAK FERM inhibitors that block RTK-FERM interactions may be able to abrogate the resistance to FAK inhibition incurred by RTKs and promote the duration of FAK inhibitor therapy.
In this report, we describe the virtual screening-based discovery of small molecule chemical probes that inhibit the HER2- FAK interaction. We utilize site-directed mutagenesis to identify the key hotspot residues of this interaction in addition to in silico druggability software (SiteMap, FTMap) to predict possible ligand sites on the FAK FERM domain. Using structure-based virtual screening, we identify 20 putative FAK FERM chemical probes and experimentally validate 2 compounds in GST pull-down assays with IC50s in the nanomolar range. Furthermore, we show effective HER2-FAK cellular disruption, inhibition of HER2-dependent transphosphorylation of FAK, and inhibition of HER2-dependent cell invasion by these FAK FERM chemical probes. These chemical probes represent the first identified inhibitors of HER2-FAK binding and represent promising molecules to interrogate the biology of HER2-FAK signaling in cancer cells.
2. MATERIALS AND METHODS
2.1. Site-directed mutagenesis
Mutagenesis studies were performed with the QuikChange Lightning Site-Directed Mutagenesis Kit (Agilent Technologies). Mutations for the pET-chFERM construct were designed using the Agilent Primer Design program and ordered through Thermo Fisher Scientific. PCR-based amplification of nascent mutant DNA was performed as per manufacturer instructions. Subsequently, PCR product was digested with Dpn I restriction enzyme to degrade parental DNA and digested product was transformed into XL10-GOLD competent E. coli. Mini-prepped DNA was sent for DNA sequencing (University of Arizona Genetics Core) to confirm mutation of intended DNA base pair and lack of secondary mutations.
2.2. SiteMap druggability calculation
SiteMap (Schrödinger, Inc.) is a modeling program which predicts ligand/drug-binding sites on proteins and calculates the druggability of predicted binding sites (Halgren, 2009). SiteMap makes a 1 Å grid around the solvent-exposed surface of the protein (sitepoints). Sitepoints which are sufficiently enclosed and make van der Waals contacts with nearby protein residues are classified as sites. The physical properties of each site are calculated (size, exposure, enclosure, contact, hydrophobicity, hydrophilicity) and a number of scores are calculated to characterize the binding site (SiteScore, Dscore). A SiteScore >0.8 indicates a druggable site. Additionally, the Dscore, in which hydrophilicity is not capped, is a measurement used to distinguish ligand-binding sites from drug-binding sites. We ran SiteMap calculation on FAK FERM (PDB 2AL6) and ranked the predicted binding sites based on SiteScore. We subsequently visually inspected the SiteMap results and compared predicted binding sites with FTMap results.
2.3. FTMap modeling
FTMap is a fragment-docking program which identifies small organic molecule binding sites on proteins and therefore potential sites for drug discovery/fragment-based drug design (Kozakov et al., 2015). FTMap uses 16 different fragment-like molecules as probes and an energy function to search for small molecule binding sites. FTMap-predicted binding sites which bind 2 or more probes are subsequently identified as consensus binding sites. For our studies, we uploaded FAK FERM domain (PBD 2AL6) to the FTMap webserver and ran FTMap analysis. The job output file consisted of a PyMOL session file. We subsequently visually inspected the FTMap results and identified predicted binding sites which overlapped with SiteMap results.
2.4. Structure-based Virtual Screening
Schrödinger software (Small molecule drug discovery suite v2014–2) was used to perform virtual screening experiments to identify chemical probes of the HER2-FAK interaction. The virtual molecule library was designed by combining the Enamine Drug-like Set (based on Lipinski and Veber rules), Enamine 3D diversity set (based on 3-dimensional shape diversity), Enamine pharmacological diversity set (based on compounds with known pharmacological effects), and the UNC-Chapel Hill library collection. This resulted in a library of around 200,000 compounds. 2-D structures (in sdf format) were converted to 3-D structures and prepared for docking studies using LigPrep (version 3.0). Ligands were optimized using the OPLS_2005 molecular mechanics force field (Shivakumar et al., 2010). Ionization states for the compounds were generated for pH 7.0 ± 2.0, with ligands being desalted if necessary and possible tautomers generated. Specific chiralities were retained and the ring confirmation with the lowest energy was kept. Output ligands were formatted in Maestro format and utilized for subsequent docking studies. The FAK FERM crystal structure (PDB 2AL6) was downloaded into the Maestro 9.8 software and prepared for docking studies using the Protein Preparation Wizard (Sastry, Adzhigirey, Day, Annabhimoju, & Sherman, 2013). We elected to assign bond orders, add hydrogen atoms, create bonds between sulfur atoms within 3.2 Å, and removed crystallographic water molecules > 5 Å from heteroatom groups likely uninvolved with ligand binding. The protein hydrogen-bonding network was optimized using the automated optimization at pH 7.0 and the overall structure was minimized to the lowest energy state using the OPLS_2005 force field. This output structure was utilized in subsequent docking procedures. After FAK FERM was prepared for docking, the 3-D virtual grid where the virtual library would be docked was generated using the Receptor Grid Generation function of GLIDE 6.3, with the center of the grid being the F1 lobe druggable pocket predicted in previous SiteMap results. The grid was generated by using the default van der Waals radius scaling factor and partial charge cutoff of 1.0 and 0.25, respectively. Using GLIDE 6.3, we set up a virtual screening platform to identify HER2-FAK chemical probes with all docking settings set to default. To filter out hits from non-hits, three stages of GLIDE docking were performed, with each stage having more stringent requirements: HTVS (High-Throughput Virtual Screening), SP (Standard Precision), and XP (Extra Precision). Top scoring molecules were sorted based off their GLIDE docking score (kcal/mol). The top 25% of molecules from HTVS were further tested on SP docking, with the top 15% of molecules from SP docking being processed to XP docking. The top 100 molecules from XP docking were manually inspected for differences in molecular interactions, specifically proximity to Y397, erroneous poses, or similar binding modes. The top 100 molecules were subsequently clustered based on structural similarity to identify 20 structurally distinct clusters, with a single representative molecule from each cluster selected for purchase.
2.5. Structural similarity clustering
Compound similarity clustering was performed to identify structural diverse virtual hits for experimental testing. Clustering was performed with the cheminformatics package Canvas (Schrödinger Inc.) with conditions previously shown to produce the best similarity enrichment (Hu et al., 2012). A series of 2-dimensional fingerprints (molprint2D) were generated to describe chemical structure. Hierarchical clustering using molprint2D fingerprints as input and Buser similarity as the similarity metric was performed. The number of clusters (cluster stringency) was selected based on the Kelley criterion. The compounds within each cluster were then manually inspected and selected for purchase.
2.6. GST pull-down assays
Purified GST-FAK or GST constructs were incubated in NP40 buffer plus 0.1% BSA with purified HER2-ICD. GST or GST-FAK constructs were pulled down using Glutathione Sepharose 4B (GE Healthcare Life Sciences) and washed three times with NP40 buffer. Proteins were eluted off of beads in 2X Laemmli buffer (BioRad) and heated at 95 °C for 5 minutes. Samples were resolved on 4–20% gradient gels and probed for HER2-ICD using standard western blotting techniques and antibodies as described below. Secondary gels were run and stained with SimplyBlue SafeStain to confirm protein loading.
2.7. Immunofluorescence
Cells were adhered to fibronectin-coated coverslips overnight and treated with drug for 1 hour, with DMSO as a control. Alternatively, to assess effects on cell attachment, cells were also treated with drug or DMSO prior to adherence to coverslips. Subsequently, cells were rinsed once with PBS, fixed using 60% acetone + 3.7% formaldehyde at −20°C for 20 minutes and blocked using 5% FBS in PBS. Cells were then stained using anti-HER2 and anti-FAK 4.47 primary antibody at 1:100 dilution followed by staining with either Alexa Fluor-conjugated anti-rabbit (488) or anti-mouse (568) secondary antibody at 1:1000 dilution. Coverslips were mounted onto slides and analyzed using a Zeiss fluorescent microscope with AxioVision software. Images shown are representative of whole slides.
2.8. Transwell invasion assays
Transwell invasion assays were performed in 8 μm Corning BioCoat Matrigel Invasion Chambers with 8.0 μm PET Membrane and 24-well plates. For studies in FAK +/+ MEFs, cells were stably transduced with both HER2 and HER3. A total of 1 × 105 serum-starved cells were placed in the upper chamber. For drug treatment studies, cells were pretreated with drug for 30 minutes before placement into the upper chamber, with DMSO serving as the control. Drug was also added to the upper chamber for the duration of the assay. Heregulin-β1 (20 ng/mL) served as the chemotactic attractant and was added into the lower chamber. Cells were allowed to invade for 24 hours before transwell invasion assays. Invaded cells were then stained with 0.5% crystal violet for 10 minutes, washed with PBS, and then counted via microscopy. Quantitative analysis represents average results obtained from three independent experiments (6 fields of view per experiment).
2.9. Protein Purification
His-tagged avian FAK-FERM (residues 31–405) in modified pET vector (provided by Dr. Michael Eck) was expressed in BL21 (DE3) E. coli (Thermo Fisher) and purified on Ni-NTA resin (Thermo-Fisher) similarly as described (T. Marlowe et al., 2019). GST-FAK-NT (FERM) proteins in pGEX-4T1 vector was expressed in BL21 (DE3) E. coli and purified on Glutathione Sepharose 4B resin (GE Healthcare) as previously described (Golubovskaya, Finch, & Cance, 2005). Recombinant HER2-ICD (Intracellular Domain) was purchased from Thermo Fisher (cat#PV3366).
2.10. Cell Culture
Cell lines MDA-MB-453 and SkBr3 (courtesy of Katerina Gurova), FAK +/+ MEFs (courtesy of Duško Ilić) were cultured at 37°C with 5% CO2. Cells were cultured in Dulbecco’s minimal essential medium (DMEM) (GIBCO, Thermo Fisher), 10% heat-inactivated fetal bovine serum (HI FBS) (GIBCO, Thermo Fisher), 1% PenicillinStreptomycin (GIBCO, Thermo Fisher), and 0.2% Normocin (InvivoGen). Established cell lines were authenticated by the University of Arizona Genetics Core facility.
2.11. Compounds, Antibodies, & Reagents
All compounds used for testing (VS1 – VS20) are racemic mixtures and were purchased from Enamine, Ltd, with reported purities of > 90% by LC-MS. Their specific Enamine numbers are in Supplemental Table S1. HER2-ICD antibody: HER2/ErbB2 (29D8) Rabbit mAb from Cell Signaling Technologies (Cat #2165). Anti-FAK FERM antibody: Anti-FAK antibody, clone 4.47 from EDM Milliport (Cat #05–537). Anti-GAPDH antibody: GAPDH antibody (ZG003) from Invitrogen (Cat #39–8600). Heregulin beta-1 human from Sigma-Aldrich (Cat #SRP3055).
2.12. Statistical Analysis
Comparisons between groups were made using a Students t-test (GraphPad Prism 6). Data were considered significant when p<0.05. One-way ANOVA and Tukey’s multiple comparison test were used to calculate significance when comparing multiple groups within the same experiment (GraphPad Prism 6).
3. RESULTS
3.1. Site-directed mutagenesis validates in-silico predictions of HER2 – FAK interface
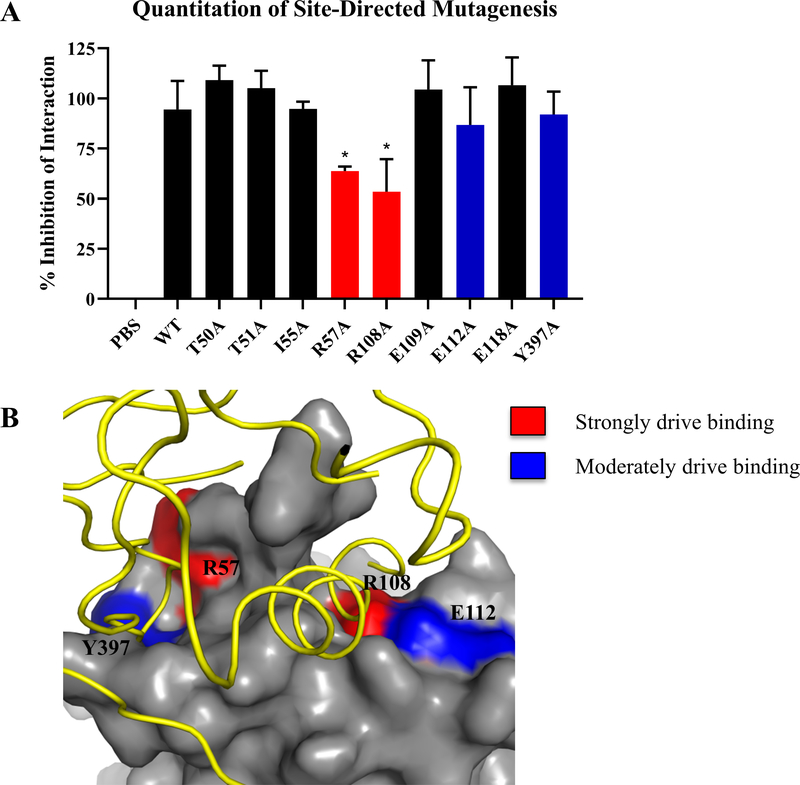
In order to determine putative small molecules that would bind to the FAK FERM F1 druggable pocket and disrupt the HER2-FAK interaction, as shown in the pipeline in Figure 1, we first started with determining the key binding residues of this interaction. Previous studies discovering the HER2-FAK direct interaction utilized HADDOCK protein-protein docking to approximate the HER2-FAK binding interface (Marlowe et al. 2019). To confirm in silico predictions of the HER2-FAK binding interface with experimental data, we proceeded with a series of site-directed mutagenesis experiments. FAK FERM F1 lobe mutations were designed based on HADDOCK-predicted surface interface residues (residues within 5 Å of cognate binding partner). Residues were mutated to alanine in order to modify the function of the original amino acid without having serious effects on protein secondary structure/folding. For the FAK FERM mutations, each mutation was made in the his-FERM construct and used for competitive pull-down assays between GST-FAK NT and HER2-ICD. Using densitometric analysis, we determined the degree of competition and therefore HER2-FAK interaction. Of the 19 mutations made for his-FERM, R57A and R108A were found to have strong decreases (statistically significant) in HER2-FAK binding while E112A and Y397A were found to have moderate decreases (Figure 2A). Representative blots of the GST pull-down competition assay are shown in Supplemental Figure S1. Interestingly, FAK mutations that disrupted binding were found to correlate well with the sites of HER2 contact in the HADDOCK model (Figure 2B). Given that the HER2-FAK interaction is a relatively strong interaction with an estimated KD of 571 nM (Supplemental Figure S2), these data suggest that residues R57 and R108 are potential hotspots that drive high-affinity binding. Also, mutation of Y397A, the site of transphosphorylation by HER2, caused a small decrease in HER2-FAK binding, implicating kinase-substrate interactions in a small proportion of the total binding process. In all, mutagenesis experiments confirmed the predicted HER2-FAK interface and therefore validated the accuracy of molecular modeling studies.
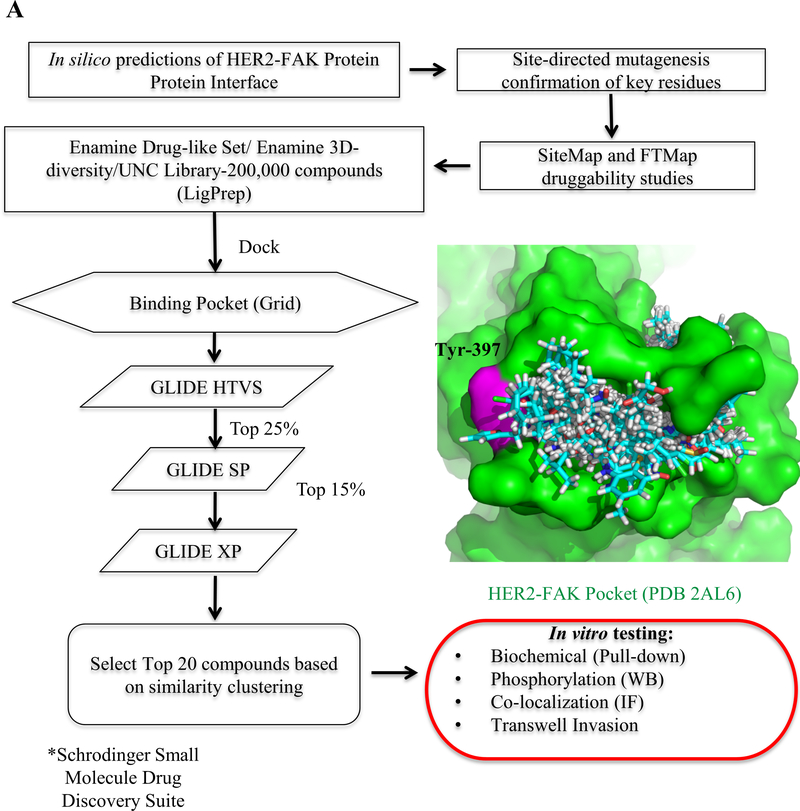
Figure 1: Structure-based virtual screening workflow to identify small molecule inhibitors of the HER2-FAK interaction.
(A) Workflow of virtual screening, starting from testing the 200,000-compound library through three increasing levels of GLIDE docking (Schrödinger, Inc.) in order to determine 20 top virtual hits for in vitro testing. The HER2-FAK pocket (PDB 2AL6) that was used for virtual screening experiments is shown in green, with key residue Tyr-397 colored in magenta. Docking poses of the top-100 ranked virtual hits are shown in cyan.
Figure 2: Validation of FAK residues predicted to be involved in HER2-FAK binding using site-directed mutagenesis.
(A) Quantitation of competitive GST pull-down assays with a series of FAK chFERM mutants. Mutations were selected based on HADDOCK molecular modeling studies. GST-FAK-NT:HER2-ICD pull-downs were competed with indicated FERM mutant and western blotting was performed to evaluate competition. Densitometry was performed to calculate % inhibition of interaction. Results are represented by three independent experiments (* denotes p < 0.05). (B) Protein-protein docking model of the HER2-FAK interaction as predicted by HADDOCK (Marlowe et al., 2016), showing the FERM regions that strongly drive binding to HER2 (R57 and R108) in red and those that moderately drive binding to HER2 (E112 and Y397) in blue. HER2 is colored in yellow and FAK is colored in gray.
3.2. HER2-FAK interface has druggable cavities near Y397
To calculate druggability of the FERM F1 lobe and the specific pockets to target for virtual-based drug discovery, we first started with the program SiteMap (Schrödinger), which utilizes a grid-based searching algorithm to search for drug-binding protein cavities and calculates the physiochemical properties of found cavities. This program is a useful tool when it comes to druggability determination of a variety of protein-protein interactions, including, but not limited to, PPIs on the cancer genome that drive cancer function (Halgren, 2010; Xu et al., 2016). For these reasons, we chose to use SiteMap to determine the key druggability of the FERM F1 lobe. SiteMap found 11 potential drug-binding sites on the FAK FERM domain (PDB 2AL6) and Site#6 was found within the F1 lobe hotspot region (Figure 3A & 3B). Site#6 was found to have a SiteScore of 0.90, where a score > 0.8 indicates a druggable site. As the SiteScore approaches 1.0, the probability of the site binding a high-affinity ligand (nanomolar) increases, suggesting the F1 lobe site as a promising site for drug discovery. Additionally, in the same F1 lobe site, we noticed a druggable cavity nearby key residue Y397 which was found to be directly phosphorylated by HER2. These data suggest that the area proximal to Y397 is druggable and that molecules targeted to this area may not only disrupt HER2-FAK interactions but also directly block Y397 phosphorylation.
Figure 3. FAK FERM druggability assessment using computational tools SiteMap and FTMap.
(A) Druggability scores of the FAK FERM F1 lobe hotspot region using SiteMap (Schrödinger, Inc.). Note: a SiteScore of 0.90 was obtained, where a score >0.80 indicates a druggable site likely to bind ligands. Other druggability metrics are indicated as well. (B) Location of the FAK FERM F1 lobe druggable site (PDB 2AL6). The white spheres represent 1Å sitepoints, which occupy the cavity of the F1 lobe druggable pocket. Y397 is labeled in yellow. (C) FTMap results for the FAK FERM domain (PDB 2AL6). Areas of the protein which are bound by fragments represent potential druggable regions and areas of focus for virtual screening/fragment-based drug discovery studies. (D) Zoomed inset of the FAK FERM F1 lobe druggable pocket. The key residues that are engaged in binding are: L37, K38, F40, E118, W120, and Y122. Note: the F1 lobe pocket was predicted to bind both benzene and ethane fragments.
We then proceeded with the program FTMap, which is a fragment docking/solvent mapping program to guide in structure-based virtual screening/fragment-based drug discovery applications (Kozakov et al., 2015). FTMap provides additional information in that identified sites are not only protein cavities but are also pockets likely to bind organic fragments, which is especially useful for virtual screening and fragment discovery studies. FT Map results using FAK FERM (2AL6) showed multiple sites likely to bind organic fragments (Figure 3C). When analyzing potential fragment-binding sites on the F1 lobe, we found one site comprised of residues L37, K38, F40, R108, E118, W120, and Y122 (Figure 3D). Interestingly, benzene and ethane fragments were found to bind within the F1 lobe site, indicating potential starting points for drug discovery. In all, these data confirmed the F1 lobe “hotspot” as druggable and indicated surface areas which may be particularly important for high-affinity ligand binding.
3.3. Structure-based virtual screening to identify HER2-FAK inhibitors
To identify possible inhibitors to the HER2-FAK interaction, we virtually screened a library (around 200,000 compounds), whichwas designed by combining virtual structures of the Enamine Drug-like Set (based on Lipinski and Veber rules), Enamine 3D Diversity Set, and UNC library collection. The crystal structure of the FAK FERM domain (PBD 2AL6) was used and the GLIDE docking grid (Schrödinger) was set centroid of the HER2-binding pocket as determined by molecular modeling. The small molecule library was subsequently docked and scored against the GLIDE grid using three types of GLIDE docking algorithms: HTVS (High-Throughput Virtual Screening), SP (Standard Precision), and XP (Extra Precision). The top 100 scored molecules after XP docking were then clustered based on hierarchical structure similarity and one representative molecule from each cluster (20 total) was selected in order to test a diverse set of compounds, as whown in Figure 3B (Banks et al., 2005). As shown in Table 1 and Supplemental Table S1, each compound represented a different structural class with different predicted binding affinities (Glide gscore). These 20 compounds were purchased and tested, with GST pull-down as our biochemical assay.
Table 1:
GLIDE gscores of virtual screening hits from XP assay
| Name | Cluster | Docking Rank |
Glide gscore |
|---|---|---|---|
|
| |||
| VS1 | 1 | 1 | −8.056654 |
| VS2 | 2 | 100 | −5.074416 |
| VS3 | 3 | 48 | −5.219695 |
| VS4* | 4 | 3 | −8.128526 |
| VS5 | 5 | 55 | −5.208667 |
| VS6 | 6 | 23 | −5.528736 |
| VS7 | 7 | 8 | −6.120372 |
| VS8 | 8 | 14 | −5.626886 |
| VS9 | 9 | 9 | −5.855779 |
| VS10 | 10 | 11 | −5.780904 |
| VS11 | 11 | 20 | −5.58567 |
| VS12 | 12 | 40 | −5.326291 |
| VS13 | 13 | 22 | −5.810899 |
| VS14* | 14 | 12 | −5.879422 |
| VS15 | 15 | 10 | −6.037577 |
| VS16 | 16 | 39 | −5.331381 |
| VS17 | 17 | 97 | −4.893491 |
| VS18 | 18 | 66 | −5.267777 |
| VS19 | 19 | 36 | −5.386051 |
| VS20 | 20 | 71 | −5.019572 |
Notes: Binding data of compounds derived from virtual screening. The docking rank is based on the docking score, a more inclusive measurement of compound binding. Those labeled with asterisks (VS4 and VS14) were determined to have the best binding through GST pull-down assays.
3.4. Biochemical testing of compounds from virtual screening identifies VS4 and VS14 as FERM chemical probes
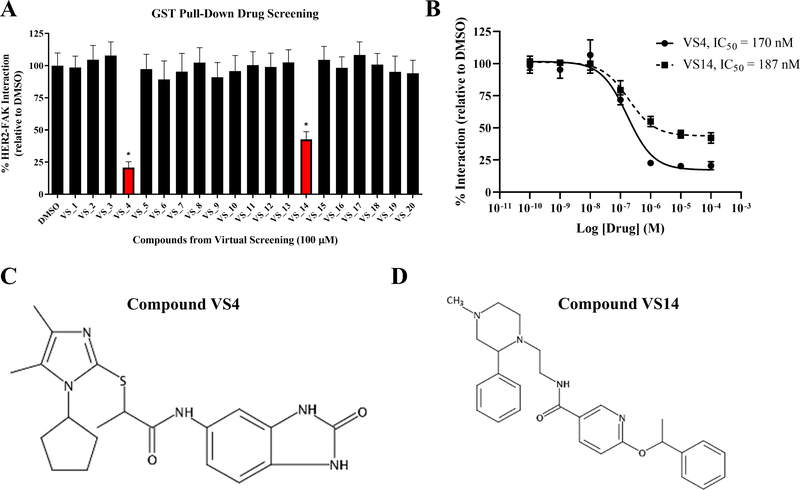
The 20 compounds identified from virtual screening experiments were processed for activity testing using competitive GST pull-down assays to measure the effect on binding between GST-FAK-NT (FERM) and His-HER2-ICD. For the GST pull-down screening assay, an initial compound concentration of 100 μM was selected in order to detect even small inhibition of HER2-FAK binding. As observed in Figure 4A, 2 of 20 compounds (VS4 and VS14) caused significant inhibition of the HER2-FAK interaction. All other compounds caused minimal inhibition of the HER2-FAK interaction. Next, we performed dose-response experiments in order to better evaluate the activity of compounds VS4 and VS14 on a quantitative level. IC50 and Imax analyses provided an estimate of drug potency and efficacy, which could inform and direct future comparative and optimization studies. For quantitative GST pull-down assays, the dose of VS4 and VS14 was titrated from 10−10 to 10−4 M and effect on the HER2-FAK interaction was measured by densitometric analysis. As shown in Figure 4B, compound VS4 had greater efficacy than VS14 in the ability to inhibit the HER2-FAK interaction (VS4 Imax = 81.7%, VS14 Imax = 57.8%). However, compounds VS4 and VS14 were equipotent, as both compounds had similar IC50 values (VS4 IC50 = 170 nM, VS14 IC50 = 187 nM). The structures of VS4 ((rac)-2-((1-cyclopentyl-4,5-dimethyl-1H-imidazol-2-yl)thio)-N-(2-oxo-2,3-dihydro-1H-benzo[d]imidazol-5-yl)propanamide) and VS14 ((rac)-N-(2-(4-methyl-2-phenylpiperazin-1-yl)ethyl)-6-(1-phenylethoxy)nicotinamide) are shown in Figure 4C and 4D, respectively.
Figure 4: Evaluation of HER2-FAK virtual compounds in GST pull-down assays.
(A) Results from GST pull-down drug screening with 20 virtual hits from virtual screening studies. GST-FAK-NT: HER2-ICD pull-downs were competed with 100 μM compound and the remaining bound HER2 was evaluated via western blotting. Results from three independent experiments were quantified via ImageJ densitometry. Error bars represent standard deviations of the mean (* denotes p < 0.05). Compounds VS4 and VS14 significantly reduced the HER2-FAK interaction. (B) Quantitative GST pull-down assays performed with compounds VS4 and VS14. Titrating concentrations (10−10 to 10−4 M) were added to compete off the GST-FAK-NT: HER2-ICD interaction. IC50s were calculated using Dose-response – Inhibition nonlinear regression algorithm (log(inhibitor) vs. response) in Graphpad Prism 6. (C) Structure of VS4. (D) Structure of VS14.
3.5. VS4 is predicted to bind proximal to the HER2-FAK interaction, while VS14 binds proximal to Y397
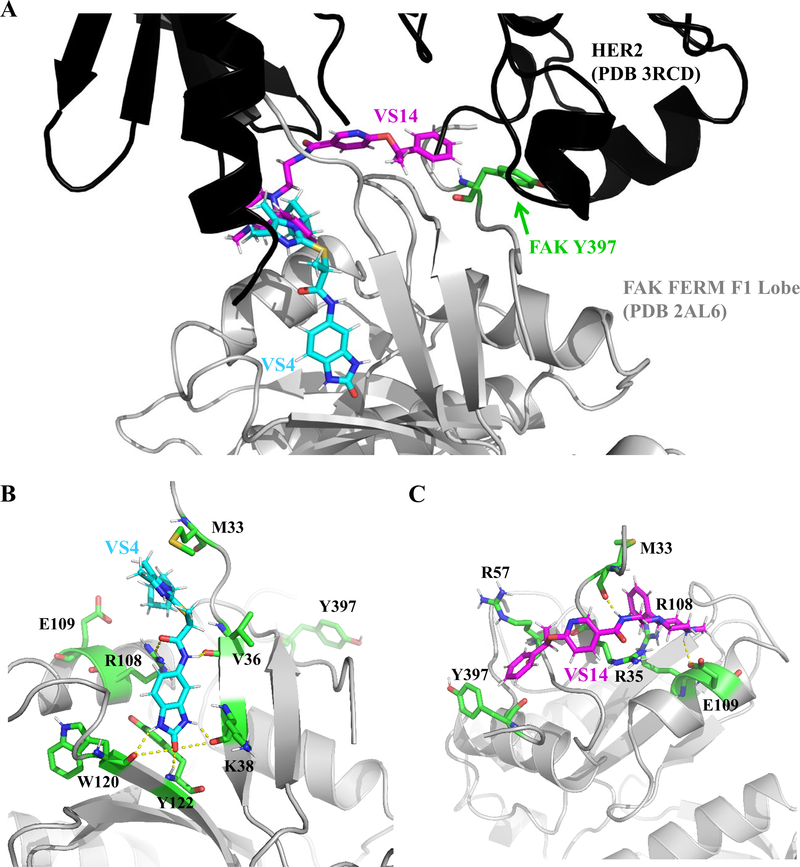
Because compounds VS4 and VS14 showed different activities in the ability to displace HER2-FAK interactions, we performed molecular docking experiments to explain such differences. Using GLIDE XP docking, we docked compounds VS4 and VS14 to the FAK FERM F1 lobe hotspot (HER2-binding site) and selected the top scoring pose for each molecule. As shown in Figure 5A, compounds VS4 and VS14 occupied different pockets within the entire HER2-FAK interface. Interestingly, compound VS4 bound to the F1 lobe near residues M33, E34, V36, K38, E112, W120, K121, and Y122 while compound VS14 bound more proximal to key FAK residue Y397 (Figure 5B and 5C).
Figure 5: Molecular docking analysis of compounds VS4 and VS14.
(A) Molecular docking studies of compounds VS4 and VS14 to the FAK FERM F1 lobe site (HER2-FAK binding pocket) using GLIDE XP docking software (Schrödinger, Inc.). Note: compounds VS4 and VS14 occupy different regions of the HER2-FAK interface. (B) Residues (colored green) implicated in the binding of VS4 to FAK FERM F1 lobe. Note: VS4 is predicted to form a hydrogen-bond with the side chain of R108, potentially explaining greater efficacy in displacing HER2-FAK interactions. (C) Residues (colored green) implicated in binding between VS14 and FAK FERM F1 lobe. Note: VS14 binds to a pocket adjacent to key tyrosine 397, potentially promoting direct inhibitory effects on Y397 phosphorylation.
VS4 was predicted to bind to the FTMap-predicted F1 lobe binding pocket. Specifically, the benzimidazole ring of VS4, was predicted to make multiple direct hydrogen bonding contacts with residues K38, W120, and Y122, strengthening the interaction that VS4 has for the interior of the FAK FERM F1 lobe. VS4 also forms a hydrogen bond with the side chain of R108, which was previously determined to be a hotspot residue for HER2-FAK binding. This potentially explains the increased HER2-FAK disruption observed with compound VS4 as compared to VS14. Molecular modeling predicted similar binding energies for the two stereoisomers of VS4.
The binding of compound VS14 may promote more direct effects on Y397, as the benzene ring from the phenethyl group of VS14 is predicted to form van der Waals interactions and π-π stacking with the side chain of Y397. Furthermore, the amide of VS14 forms a hydrogen bond with the backbone carbonyl of M33 and the 4-methyl ammonium moiety forms a salt bridge with the side chain of E109. Molecular modeling predicted the optimal stereoisomer of VS14 to be N-(2-((S)-4-methyl-2-phenylpiperazin-1-yl)ethyl)-6-((R)-1-phenylethoxy)nicotinamide. In all, molecular docking studies identified two distinct binding modes for compounds VS4 and VS14, suggesting separate mechanisms of activity.
3.6. VS14 is able to block both auto- and trans-phosphorylation of FAK at Y397
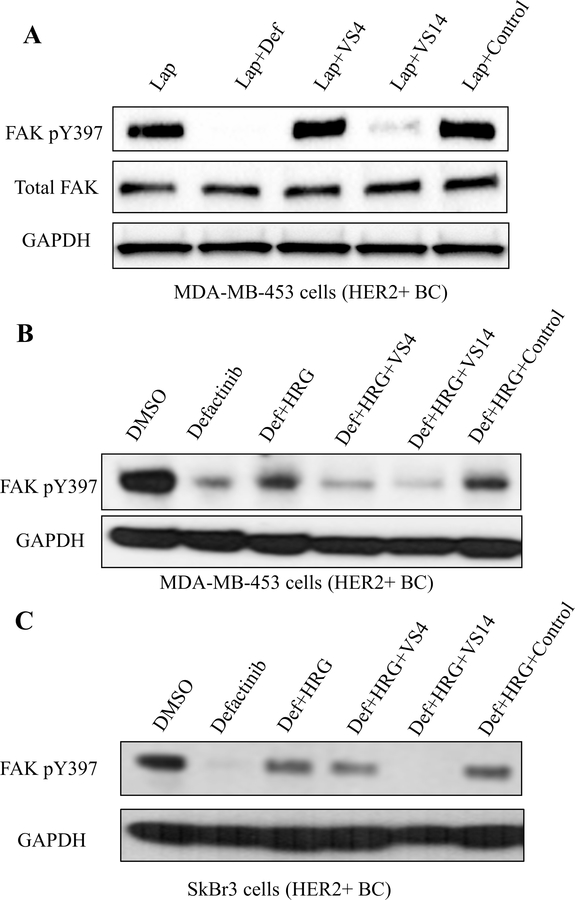
As the HER2-FAK interaction site was found structurally to be proximal to the key Y397 region and compound VS14 was predicted to bind to the FAK FERM F1 lobe pocket adjacent to Y397, we decided to test VS4 and VS14 in FAK autophosphorylation assays. We hypothesized that compound VS14, which binds near FAK Y397, may directly inhibit Y397 and prevent both auto- and trans-phosphorylation mechanisms. To test this, we designed an immunoblot experiment which only measured FAK Y397 autophosphorylation levels. MDA-MB-453 cells were serum-starved overnight, pretreated with lapatinib (HER2-kinase inhibitor) for 1 hour to prevent transphosphorylation, and then treated with defactinib (positive control), VS4, VS14, or the negative control for 1 hour to measure effects on autophosphorylation. As shown in Figure 6A, defactinib completely inhibited levels of FAK Y397 autophosphorylation, as expected. However, compound VS14, also inhibited FAK Y397 autophosphorylation to near complete levels, indicating direct Y397 effects of compound VS14. Compound VS4, which binds to the FAK FERM F1 lobe more distal to Y397, and negative control compound had no effects on Y397 autophosphorylation. In all, this data indicate that compound VS14 can inhibit FAK phosphorylation by both auto- and trans-phosphorylation mechanisms, providing potential biological utility by evading drug resistance and prolonging anti-FAK therapy.
Figure 6: Western blotting studies evaluating the effect of VS4/VS14 on phosphorylation of FAK Y397.
(A) Immunoblot-based auto-phosphorylation assay in HER2+ MDA-MB-453 breast cancer cells. Cells were serum starved overnight, pre-treated with lapatinib (1 μM) for 1h to prevent transphosphorylation, and then treated with additional drug (defactinib – 1μM, VS4/VS14/control – 10 μM) for 1 hour to test effects on auto-phosphorylation. As expected, defactinib fully reduced auto-phosphorylation at Y397. However, compound VS14 also significantly inhibited auto-phosphorylation at Y397 while compounds VS4 and the negative control had no effect. (B) Immunoblot-based transphosphorylation assay in HER2+ MDA-MB-453 breast cancer cells. Cells were serum-starved overnight, treated with drug (defactinib – 1μM, VS4/VS14/control – 10 μM) for 1h, then stimulated with HRG (20 ng/mL) for 30 min. Both compounds VS4 and VS14 inhibited transphosphorylation at Y397 by HER2. (C) Immunoblot-based transphosphorylation assay in HER2+ SkBr3 breast cancer cells similarly performed as (A). Both compounds VS4 and VS14 inhibited transphosphorylation at Y397 by HER2. Compound VS14 is a dual inhibitor of both auto- and transphosphorylation.
To test the effects of compounds VS4 and VS14 on FAK transphosphorylation by HER2 in cells, we designed an immunoblot experiment which only measured FAK Y397 transphosphorylation levels. We used MDA-MB-453 and SkBr3 HER2+ breast cancer cells, serum-starved overnight, pre-treated with defactinib (FAK-kinase inhibitor) for 1 hour to inhibit auto-phosphorylation, and then stimulated with Heregulin β1 (HRG) for 30 min to induce transphosphorylation. As shown in Figure 6B and 6C, the addition of HRG increased the levels of pY397 compared to defactinib-only control, serving as the maximum amount of transphosphorylation signal. We then co-treated with both defactinib and VS4/VS14/control compound to test the effects of HER2-FAK inhibitors on Y397 transphosphorylation. The negative control was shown not to inhibit HER2-FAK interactions, the identity of which is shown in Supplemental Figure S3. In MDA-MB-453 and SkBr3 cells, both compound VS4 and VS14 at 10 μM prevented Y397 transphosphorylation by HER2 while control compound had no effects on transphosphorylation. These data indicate that HER2-FAK inhibitors, which inhibit HER2-FAK protein-protein interactions, can also inhibit phosphorylation of FAK by HER2.
3.7. VS14 treatment reduces HER2-FAK co-localization in breast cancer cells
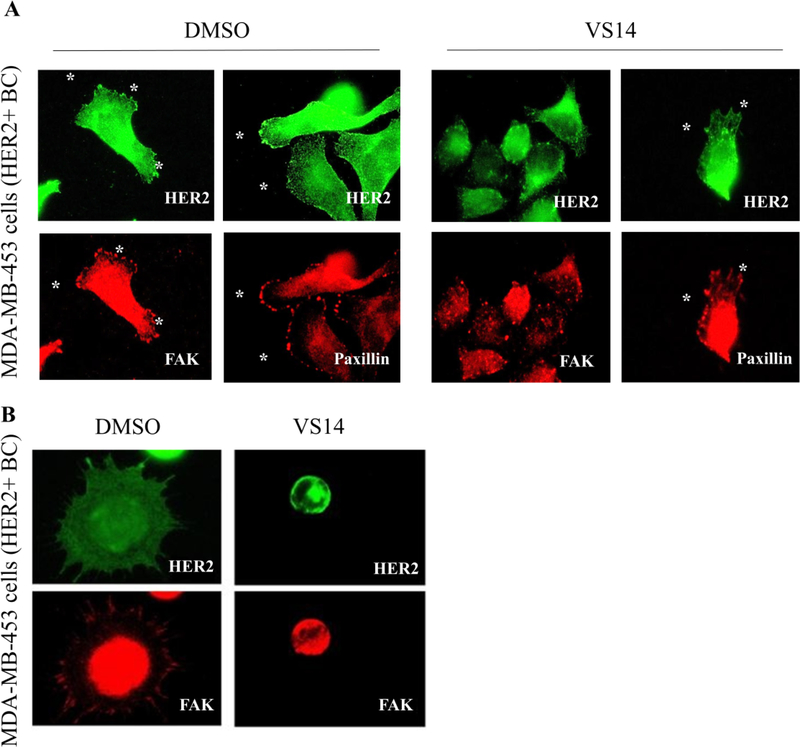
As in vitro assays showed that compound VS14 was able to disrupt HER2-FAK interactions and inhibit both auto- and trans-phosphorylation of FAK Y397 by HER2, we hypothesized that VS14 was causing changes in FAK localization. To test this, we designed and performed immunofluorescence assays of HER2+ MDA-MB-453 cells treated with DMSO or VS14 and stained for HER2 and FAK (Figure 7A). In DMSO-treated cells, both HER2 and FAK were prominently found at focal adhesion sites. However, in VS14-treated cells, FAK was clustered within cytoplasm and de-localized from focal adhesions. VS14-induced protein delocalization was specific for FAK, as HER2 localization remained unaffected by VS14 treatment. From a cell morphology perspective, the VS14 treated cells displayed cell rounding and lacked elongation as compared to the DMSO-treated cells. Suggestive of changes in cell adhesion, highly motile HER2/3+ FAK+/+ MEFs were pre-treated with DMSO or VS14 and attached to fibronectin-coated coverslips. As shown in Figure 7B, the majority of VS14-treated cells did not attach while those that did were highly rounded and lacked focal adhesions. Overall, this data shows that compound VS14 specifically causes FAK delocalization and connects VS14-induced reduction of FAK Y397 phosphorylation with FAK-dependent cellular adhesion.
Figure 7: Compound VS14 promotes FAK delocalization and prevents cellular adhesion.
(A) Immunofluorescent-staining of FAK in HER2+ MDA-MB-453 breast cancer cells treated with DMSO or 10 μM compound VS14. In DMSO treated cells, FAK was localized to focal adhesions (white asterisks). In VS14 treated cells, FAK was de-localized from focal adhesions and clustered in the cytoplasm. HER2 and Paxillin localization was unchanged by VS14 treatment, supporting the FAK specificity of VS14. (B) In MDA-MB-453 breast cancer cells, VS14 pre-treated cells did not attach to fibronectin-coated coverslips, unlike DMSO treated cells. The VS14 treated cells were round and lacked focal adhesions.
3.8. VS14 treatment reduces HER2 mediated invasion
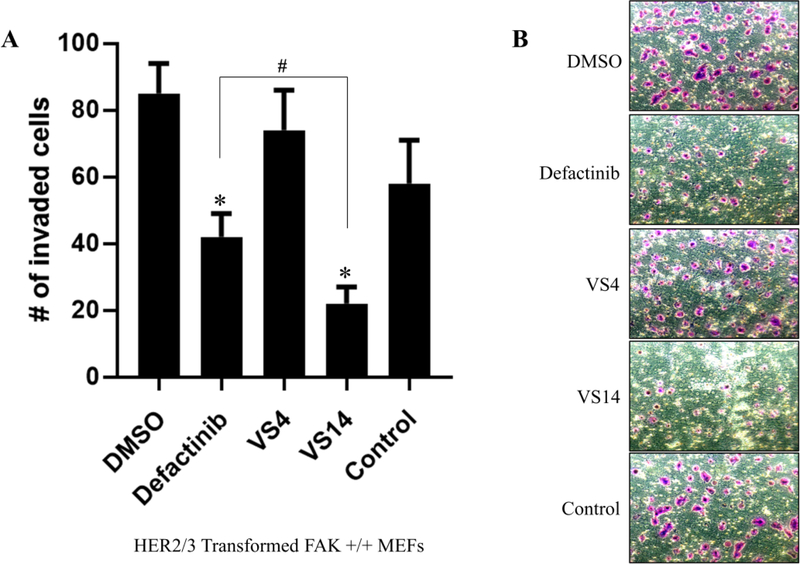
To evaluate the effects of compounds VS4 and VS14 on FAK-regulated biological functions compared to FAK-kinase inhibitors, cellular invasion assays were performed. We started with HER2-directed invasion in HER2/HER3+ FAK+/+ MEFs, as previous studies showed that HER2-directed invasion occurs independently of FAK-kinase activity (Marlowe et al. 2016. MCT). We pretreated cells with DMSO, defactinib, VS4, VS14, or control compound for 30 min and then subjected cells to transwell invasion assays. As shown in Figure 8, defactinib treatment reduced HER2-directed invasion by approximately 50%; however, it did not completely inhibit total cell invasion. In comparison, compound VS14, which inhibited FAK auto- and trans-phosphorylation in previous experiments, reduced HER2-directed invasion by approximately 75%, which was better than defactinib treatment and statistically significant. Compounds VS4 and control, however, had limited effect on HER2-directed migration/invasion. This data indicates that direct FAK Y397 inhibition is required to maximally inhibit HER2-directed invasion.
Figure 8: Effects of compounds VS4 and VS14 on cell invasion.
(A) Transwell invasion assays of HER2/3-transformed FAK +/+ MEFs treated with 10 μM defactinib, VS4, VS14, or negative control. HRG (20 ng/mL) was added in the lower chamber for chemotaxis and invaded cells were stained with crystal violet. Defactinib treatment reduced invasion however VS14 was more effective. Compound VS4 and the negative control had limited effect on invasion. Quantitation represents averaged results from six fields of view in triplicate. Error bars represent standard deviations from the mean. One-way ANOVA statistics were performed (* denotes p < 0.05 compared to DMSO, and # denotes p < 0.05 compared to defactinib). (B) Representative images from experiment in (A).
4. DISCUSSION
FAK has emerged as a promising drug target due to its function as a critical component in multiple oncogenic signaling pathways, cancer invasion, and metastasis (Sulzmaier, Jean, & Schlaepfer, 2014). In our efforts to target the FAK FERM domain, specifically the HER2-FAK interaction, we identified the hotspot residues that drive the HER2-FAK interaction using molecular modeling and validated those predictions utilizing site directed mutagenesis. Subsequently, we virtually screened a 200,000-compound library against this pocket and identified 20 putative HER2-FAK probes using GST pull-down assays. Two compounds, VS4 and VS14, were found to decrease the interaction between FAK FERM and HER2. These chemical probes, discovered using molecular modeling and in vitro assays, were found to have different mechanisms of action, where VS4 binds directly to the FERM F1 lobe, specifically the interface with HER2, while VS14 binds proximal to the major phosphorylation site Y397. Compound VS14 was found to also inhibit HER2-FAK co-localization in breast cancer cells and HER2-directed cellular invasion, identifying this compound as a potential FAK FERM chemical probe for future biological investigation.
Site-directed mutagenesis is a powerful method to validate virtual models of protein-protein interactions (DeLano, 2002). By mutating integral residues to alanine and measuring the change in interaction between the proteins of interest, the importance of each residue is inferred, as a decrease in affinity occurs when an important residue is knocked out. From there, compounds can be devised and tested to bind to these critical residues In the HER2-FAKinteraction, two residues (R57 and R108) were shown to be critical for binding due to a decrease in interaction after mutation to alanine. That information was used to develop a virtual screening pipeline to identify compounds that were predicted to bind proximal to R57/R108 and contain functional groups capable of forming hydrogen bonds with the side chain(s) of R57/R108. Intriguingly, hit probe VS4, is predicted to form hydrogen bonds with R108, which would effectively compete with interactions between HER2 and FAK FERM. Future X-ray co-crystallography and NMR experiments are in development to fully validate the predicted HER2-FAK protein-protein interaction model.
The two chemical probes that were discovered to inhibit the HER2-FAK interaction, VS4 and VS14, are able to do so through separate predicted binding pockets at the protein-protein interaction interface. Although both inhibitors are equipotent at HER2-FAK inhibition, VS4 does have a greater Imax than VS14, possibly explained by the molecular modeling-determined binding mode of each inhibitor to the surrounding residues in the binding pocket. Specifically, VS4 binds to residues K38, W120, and Y122 key residues in the FERM F1 lobe binding pocket as determined through docking studies. Intriguingly, this is the same F1 lobe pocket identified by FTMap studies and we speculate that this pocket may be more essential for HER2-FAK binding compared to the VS14 binding pocket. VS14, on the other hand, binds more proximal to R57, R108, and Y397, a region that is perhaps more critical for Y397 phosphorylation rather that direct HER2-FAK binding. This difference in binding mode could explain how VS14 is able to affect both auto- and trans-phosphorylation while VS4 is only able to affect transphosphorylation. In fact, molecular docking studies suggest potential van der Waals forces and π-π stacking between the side chain of Y397 and the benzene ring of VS14. Furthermore, previous structural studies have shown that the Y397 FERM-kinase linker region forms an auto-inhibitory interaction with the FERM F1-lobe to regulate Y397 phosphorylation (Lietha et al., 2007). We speculate that compound VS14 could be stabilizing linker-F1 lobe interactions as a mechanism of inhibiting both auto- and trans-phosphorylation of FAK Y397. Ultimately, co-crystallization and/or NMR data of the VS14-FERM complex would provide additional structural data to confirm the compound mechanism of action.
In vitro evaluation of both chemical probes that tested their ability to decrease FAK phosphorylation, cellular invasion, and FAK localization, showed that VS14 was a superior probe at inhibiting the biological functions of FAK compared to VS4. VS14 inhibited both types of phosphorylation, auto- and trans-, while VS4 was only able to interrupt Y397 transphosphorylation. In addition, VS14 showed the ability to delocalize FAK from focal adhesions and prevent HER2-mediated invasion more so than DMSO, negative control compound, and defactinib, a FAK-kinase domain inhibitor. These unique properties of VS14 highlight the importance of inhibiting both auto- and trans-phosphorylation of FAK Y397 in order to produce a maximum anti-FAK pharmacological effect. In further support, multiple genetic studies show that FAK Y397F mutation or FAK FERM deletion is more effective at blocking FAK-driven migration than FAK-kinase inhibition alone (K454R) (Sieg et al., 2000; Sieg, Hauck, & Schlaepfer, 1999). These mechanistic observations may explain why phase I/II clinical trials using single-agent FAK-kinase inhibitor treatment in solid tumors have failed due to limited efficacy (de Jonge et al., 2019; Infante et al., 2012; Jones et al., 2015). Further biological investigation of our discovered FAK FERM chemical probes will allow us to determine the optimal strategy (kinase vs. scaffold) to target FAK in cancer and under which molecular context. A future area of research will be the comparison of resistance mechanisms between FAK-kinase inhibitors and FAK-FERM inhibitors, specifically in relation to kinome reprogramming. An interesting question will be whether FAK FERM chemical probes can inhibit other RTK-FAK interactions (e.g. EGFR-FAK, cMet-FAK, RET-FAK) and how disruption of these interactions affects FAK phosphorylation. Also, we will explore combinational studies using both FAK-kinase and FAK-FERM inhibitors in an attempt to maximally block both kinase and scaffold signaling pathways. Finally, further in-depth mechanistic investigation of our FAK-FERM chemical probes using technologies such as mass-spectrometry may identify novel FERM interactions that regulate FAK biology.
While these FAK FERM chemical probes show importance as biological tools, additional work is necessary to develop these individual chemical probes into drug-like candidates. Further biochemical, biophysical, and structural characterization of VS4/VS14 is necessary to evaluate compound mechanism of action and enable structure-based optimization. These studies include direct binding assays such as surface plasmon resonance (SPR) and isothermal titration calorimetry (ITC) to identify the direct binding partner and affinity constants (KD). Co-crystallization of the VS14-FERM complex would provide valuable structural information that would aid in the design of VS14 analogs that have additional interactions with Y397 and enhanced binding affinity by linking/fusing compound VS14 to VS4. Significant medicinal chemistry would have to be performed to optimize both the biochemical and cellular properties of VS4/VS14. Selectivity assays that measure FERM-domain specificity and proteome-wide specificity would be helpful in the development of selective FERM compounds. Finally, extensive in vivo studies, including efficacy, PK/PD, and toxicity, as well as studying FAK-kinase inhibitor resistance models, would be required to test VS14 derivatives, verify their mechanism of action, and assess their safety. Future studies in HER2 isogenic mouse models and FAK-kinase inhibitor acquired-resistance mouse models would be useful in testing VS14 and its derivatives in vivo. Nonetheless, the identified FAK FERM chemical probes have promising biological activity for immediate cellular investigation in a variety of cancer cell models.
5. CONCLUSIONS
In this study, we utilized a virtual screening pipeline, followed by experimental testing, to identify two compounds with nanomolar potency that are capable of inhibiting the protein-protein interaction between HER2 and FAK FERM. Further molecular modeling on these compounds (VS4 and VS14) showed that VS4 binds closer to the HER2-FAK interaction while VS14 binds more proximal to the phosphorylation site Y397. Biological studies, including phosphorylation, cellular localization, and cellular invasion assays determined the effects of these compounds. Chemical probe VS14 successfully blocked both auto- and trans-phosphorylation of FAK, disrupted HER2-FAK co-localization in cells, and inhibited HER2-dependent cellular invasion. In all, this work has identified new chemical probes of the FAK FERM domain, allowing future investigation of the biological significance of this critical domain in cancer.
Supplementary Material
ACKNOWLEDGMENTS
We would like to thank Dr. Warren S. Weiner for his valuable insight and assistance in the preparation and revision of the manuscript.
Footnotes
CONFLICTS OF INTEREST
The authors declare no conflict of interest.
Publisher's Disclaimer: This article has been accepted for publication and undergone full peer review but has not been through the copyediting, typesetting, pagination and proofreading process, which may lead to differences between this version and the Version of Record. Please cite this article as doi: 10.1111/CBDD.13671
DATA AVAILABILITY STATEMENT
The data that support the findings of this study are available from the corresponding author upon reasonable request.
REFERENCES
- Banks JL, Beard HS, Cao Y, Cho AE, Damm W, Farid R, … Levy RM (2005). Integrated Modeling Program, Applied Chemical Theory (IMPACT). J Comput Chem, 26(16), 1752–1780. doi: 10.1002/jcc.20292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benlimame N, He Q, Jie S, Xiao D, Xu YJ, & Loignon M (2005). FAK signaling is critical for ErbB-2/ErbB-3 receptor cooperation for oncogenic transformation and invasion. J Cell Biol, 171, 505–516. doi: 10.1083/jcb.200504124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calalb MB, Zhang X, Polte TR, & Hanks SK (1996). Focal adhesion kinase tyrosine-861 is a major site of phosphorylation by Src. Biochem Biophys Res Commun, 228(3), 662–668. doi: 10.1006/bbrc.1996.1714 [DOI] [PubMed] [Google Scholar]
- Cance WG, Harris JE, Iacocca MV, Roche E, Yang XH, Chang JL, … Xu LH (2000). Immunohistochemical analyses of focal adhesion kinase expression in benign and malignant human breast and colon tissues: Correlation with preinvasive and invasive phenotypes. Clinical Cancer Research, 6(6), 2417–2423. [PubMed] [Google Scholar]
- Cobb BS, Schaller MD, Leu TH, & Parsons JT (1994). Stable association of pp60src and pp59fyn with the focal adhesion-associated protein tyrosine kinase, pp125FAK. Molecular & Cellular Biology, 14(1), 147–155. doi: 10.1128/mcb.14.1.147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Jonge MJA, Steeghs N, Lolkema MP, Hotte SJ, Hirte HW, van der Biessen DAJ, … Siu LL (2019). Phase I Study of BI 853520, an Inhibitor of Focal Adhesion Kinase, in Patients with Advanced or Metastatic Nonhematologic Malignancies. Target Oncol, 14(1), 43–55. doi: 10.1007/s11523-018-00617-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeLano WL (2002). Unraveling hot spots in binding interfaces: progress and challenges. Curr Opin Struct Biol, 12(1), 14–20. doi: 10.1016/s0959-440x(02)00283-x [DOI] [PubMed] [Google Scholar]
- Dunty JM, Gabarra-Niecko V, King ML, Ceccarelli DF, Eck MJ, & Schaller MD (2004). FERM domain interaction promotes FAK signaling. Mol Cell Biol, 24(12), 5353–5368. doi: 10.1128/MCB.24.12.5353-5368.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang X, Liu X, Yao L, Chen C, Lin J, Ni P, … Fan Q (2014). New insights into FAK phosphorylation based on a FAT domain-defective mutation. PLoS One, 9(9), e107134. doi: 10.1371/journal.pone.0107134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frame MC, Patel H, Serrels B, Lietha D, & Eck MJ (2010). The FERM domain: organizing the structure and function of FAK. Nat Rev Mol Cell Biol, 11(11), 802–814. doi: 10.1038/nrm2996 [DOI] [PubMed] [Google Scholar]
- Golubovskaya VM (2014). Targeting FAK in human cancer: from finding to first clinical trials. Front Biosci (Landmark Ed), 19, 687–706. doi: 10.2741/4236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Golubovskaya VM, Figel S, Ho BT, Johnson CP, Yemma M, Huang G, … Cance WG (2012). A small molecule focal adhesion kinase (FAK) inhibitor, targeting Y397 site: 1-(2-hydroxyethyl)-3, 5, 7-triaza-1-azoniatricyclo [3.3.1.1(3,7)]decane; bromide effectively inhibits FAK autophosphorylation activity and decreases cancer cell viability, clonogenicity and tumor growth in vivo. Carcinogenesis, 33(5), 1004–1013. doi: 10.1093/carcin/bgs120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Golubovskaya VM, Finch R, & Cance WG (2005). Direct interaction of the N-terminal domain of focal adhesion kinase with the N-terminal transactivation domain of p53. J Biol Chem, 280(26), 25008–25021. doi: 10.1074/jbc.M414172200 [DOI] [PubMed] [Google Scholar]
- Halgren TA (2009). Identifying and characterizing binding sites and assessing druggability. J Chem Inf Model, 49(2), 377–389. doi: 10.1021/ci800324m [DOI] [PubMed] [Google Scholar]
- Han DC, & Guan JL (1999). Association of focal adhesion kinase with Grb7 and its role in cell migration. J Biol Chem, 274(34), 24425–24430. doi: 10.1074/jbc.274.34.24425 [DOI] [PubMed] [Google Scholar]
- Hu G, Kuang G, Xiao W, Li W, Liu G, & Tang Y (2012). Performance evaluation of 2D fingerprint and 3D shape similarity methods in virtual screening. J Chem Inf Model, 52(5), 1103–1113. doi: 10.1021/ci300030u [DOI] [PubMed] [Google Scholar]
- Huang C, Park CC, Hilsenbeck SG, Ward R, Rimawi MF, Wang YC, … Schiff R (2011). beta1 integrin mediates an alternative survival pathway in breast cancer cells resistant to lapatinib. Breast Cancer Res, 13(4), R84. doi: 10.1186/bcr2936 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Infante JR, Camidge DR, Mileshkin LR, Chen EX, Hicks RJ, Rischin D, … Siu LL (2012). Safety, pharmacokinetic, and pharmacodynamic phase I dose-escalation trial of PF-00562271, an inhibitor of focal adhesion kinase, in advanced solid tumors. J Clin Oncol, 30(13), 1527–1533. doi: 10.1200/JCO.2011.38.9346 [DOI] [PubMed] [Google Scholar]
- Jin MH, Nam AR, Park JE, Bang JH, Bang YJ, & Oh DY (2017). Resistance Mechanism against Trastuzumab in HER2-Positive Cancer Cells and Its Negation by Src Inhibition. Mol Cancer Ther, 16(6), 1145–1154. doi: 10.1158/1535-7163.MCT-16-0669 [DOI] [PubMed] [Google Scholar]
- Jones SF, Siu LL, Bendell JC, Cleary JM, Razak AR, Infante JR, … Shapiro GI (2015). A phase I study of VS-6063, a second-generation focal adhesion kinase inhibitor, in patients with advanced solid tumors. Invest New Drugs, 33(5), 1100–1107. doi: 10.1007/s10637-015-0282-y [DOI] [PubMed] [Google Scholar]
- Kozakov D, Grove LE, Hall DR, Bohnuud T, Mottarella SE, Luo L, … Vajda S (2015). The FTMap family of web servers for determining and characterizing ligand-binding hot spots of proteins. Nat Protoc, 10(5), 733–755. doi: 10.1038/nprot.2015.043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lark AL, Livasy CA, Calvo B, Caskey L, Moore DT, Yang XH, & Cance WG (2003). Overexpression of focal adhesion kinase in primary colorectal carcinomas and colorectal liver metastases: Immunohistochemistry and real-time PCR analyses. Clinical Cancer Research, 9(1), 215–222. [PubMed] [Google Scholar]
- Lietha D, Cai X, Ceccarelli DF, Li Y, Schaller MD, & Eck MJ (2007). Structural basis for the autoinhibition of focal adhesion kinase. Cell, 129(6), 1177–1187. doi: 10.1016/j.cell.2007.05.041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marlowe T, Dementiev A, Figel S, Rivera A, Flavin M, & Cance W (2019). High resolution crystal structure of the FAK FERM domain reveals new insights on the Druggability of tyrosine 397 and the Src SH3 binding site. BMC Mol Cell Biol, 20(1), 10. doi: 10.1186/s12860-019-0193-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marlowe TA, Lenzo FL, Figel SA, Grapes AT, & Cance WG (2016). Oncogenic Receptor Tyrosine Kinases Directly Phosphorylate Focal Adhesion Kinase (FAK) as a Resistance Mechanism to FAK-Kinase Inhibitors. Mol Cancer Ther, 15(12), 3028–3039. doi: 10.1158/1535-7163.MCT-16-0366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLean GW, Carragher NO, Avizienyte E, Evans J, Brunton VG, & Frame MC (2005). The role of focal-adhesion kinase in cancer. A ew therapeutic opportunity. Nature Reviews Cancer, 5(7), 505–515. doi: 10.1038/nrc1647 [DOI] [PubMed] [Google Scholar]
- Mousson A, Sick E, Carl P, Dujardin D, De Mey J, & Ronde P (2018). Targeting Focal Adhesion Kinase Using Inhibitors of Protein-Protein Interactions. Cancers (Basel), 10(9). doi: 10.3390/cancers10090278 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Owens LV, Xu LH, Craven RJ, Dent GA, Weiner TM, Kornberg L, … Cance WG (1995). Overexpression of the Focal Adhesion Kinase (P125(Fak)) in Invasive Human Tumors. Cancer Research, 55(13), 2752–2755. [PubMed] [Google Scholar]
- Ozkal S, Paterson JC, Tedoldi S, Hansmann ML, Kargi A, Manek S, … Marafioti T (2009). Focal adhesion kinase (FAK) expression in normal and neoplastic lymphoid tissues. Pathol Res Pract, 205(11), 781–788. doi: 10.1016/j.prp.2009.07.002 [DOI] [PubMed] [Google Scholar]
- Roberts WG, Ung E, Whalen P, Cooper B, Hulford C, Autry C, … Vajdos F (2008). Antitumor activity and pharmacology of a selective focal adhesion kinase inhibitor, PF-562,271. Cancer Res, 68(6), 1935–1944. doi: 10.1158/0008-5472.CAN-07-5155 [DOI] [PubMed] [Google Scholar]
- Sastry GM, Adzhigirey M, Day T, Annabhimoju R, & Sherman W (2013). Protein and ligand preparation: parameters, protocols, and influence on virtual screening enrichments. J Comput Aided Mol Des, 27(3), 221–234. doi: 10.1007/s10822-013-9644-8 [DOI] [PubMed] [Google Scholar]
- Shi Q, Hjelmeland AB, Keir ST, Song L, Wickman S, Jackson D, … Rich JN (2007). A novel low-molecular weight inhibitor of focal adhesion kinase, TAE226, inhibits glioma growth. Mol Carcinog, 46(6), 488–496. doi: 10.1002/mc.20297 [DOI] [PubMed] [Google Scholar]
- Shivakumar D, Williams J, Wu Y, Damm W, Shelley J, & Sherman W (2010). Prediction of Absolute Solvation Free Energies using Molecular Dynamics Free Energy Perturbation and the OPLS Force Field. J Chem Theory Comput, 6(5), 1509–1519. doi: 10.1021/ct900587b [DOI] [PubMed] [Google Scholar]
- Sieg DJ, Hauck CR, Ilic D, Klingbeil CK, Schaefer E, Damsky CH, & Schlaepfer DD (2000). FAK integrates growth-factor and integrin signals to promote cell migration. Nat Cell Biol, 2(5), 249–256. doi: 10.1038/35010517 [DOI] [PubMed] [Google Scholar]
- Sieg DJ, Hauck CR, & Schlaepfer DD (1999). Required role of focal adhesion kinase (FAK) for integrin-stimulated cell migration. J Cell Sci, 112(Pt), 2677–2691. [DOI] [PubMed] [Google Scholar]
- Slack-Davis JK, Martin KH, Tilghman RW, Iwanicki M, Ung EJ, Autry C, … Parsons JT (2007). Cellular characterization of a novel focal adhesion kinase inhibitor. J Biol Chem, 282(20), 14845–14852. doi: 10.1074/jbc.M606695200 [DOI] [PubMed] [Google Scholar]
- Sulzmaier FJ, Jean C, & Schlaepfer DD (2014). FAK in cancer: mechanistic findings and clinical applications. Nat Rev Cancer, 14(9), 598–610. doi: 10.1038/nrc3792 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiner TM, Liu ET, Craven RJ, & Cance WG (1993). Expression of focal adhesion kinase gene and invasive cancer. Lancet, 342(8878), 1024–1025. [DOI] [PubMed] [Google Scholar]
- Xu D, Jalal SI, Sledge GW, & Meroueh SO (2016). Small-molecule binding sites to explore protein–protein interactions in the cancer proteome. Molecular BioSystems, 12(10), 3067–3087. doi: 10.1039/c6mb00231e [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang XH, Flores LM, Li Q, Zhou P, Xu F, Krop IE, & Hemler ME (2010). Disruption of laminin-integrin-CD151-focal adhesion kinase axis sensitizes breast cancer cells to ErbB2 antagonists. Cancer Res, 70(6), 2256–2263. doi: 10.1158/0008-5472.CAN-09-4032 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data that support the findings of this study are available from the corresponding author upon reasonable request.