Abstract
The circadian rhythm is generated at the cellular level by a molecular clock system that involves specific genes. Studies have revealed that circadian clock disruption is a control point in cancer progression. Colorectal cancer (CRC) is one of the cancers closely associated with circadian disruption. In the present review, the involvement of the circadian clock in CRC development was summarized. Abnormal expression of certain clock genes has been found in patients with CRC and their correlation with clinicopathological features has also been explored. The period and cryptochrome 2 (Cry2), Sirtuin1 (SIRT1) and neuronal PAS domain protein 2 (NPAS2) genes were reported to have tumour suppressor properties. Conversely, Cry1, brain and muscle ARNT-like-1, circadian locomotor output cycles kaput (CLOCK) and timeless may aggravate CRC progression, but these findings are not consistent and require to be confirmed by further research. Circadian scheduling also indicated advantages in chemotherapy treatments for patients with CRC by increasing the maximum tolerated doses and decreasing toxicities. Dysfunction of the molecular CLOCK system disrupted cellular processes to accelerate colon tumorigenesis, such as metabolism, cell cycle, DNA damage repair, proliferation and apoptosis, epithelial-mesenchymal transition and stemness. The clock gene network and how the dynamics of the system influence CRC were discussed.
Keywords: circadian clock, colorectal cancer, period, cryptochrome, CLOCK, BMAL1, TIM
1. Background
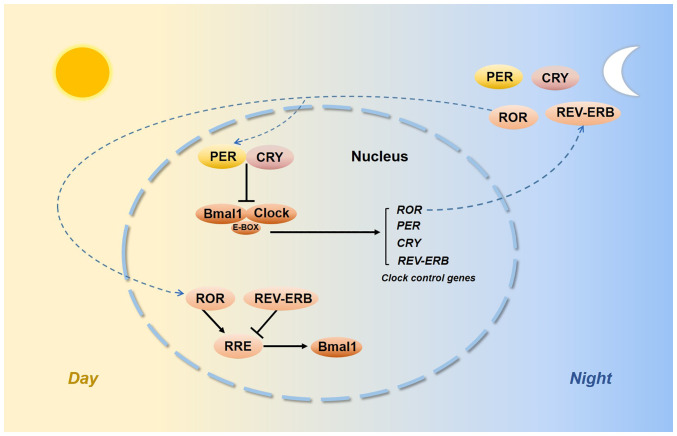
Biological rhythms exist in our internal bodies and their functions vary according to oscillations with a 24-h light/dark cycle (1). The circadian time-keeping system is controlled by both central and peripheral oscillators. The suprachiasmatic nuclei of the hypothalamus are the central pacemaker (2,3) and certain interconnected specific clock genes control the peripheral circadian rhythm. The discovery of the circadian locomotor output cycles kaput (CLOCK) and its heterodimeric partner brain and muscle ARNT-like-1 (BMAL1) led to the first site of circadian oscillations (4,5). The transactivation of E-box-containing genes by the CLOCK:BMAL1 complex was able to activate Period (Per) genes, i.e. Per1-3 and Cryptochrome (Cry) genes, i.e. Cry1 and Cry2, which then negatively repressed CLOCK:BMAL1 transcription. As auxiliary feedback loops, REV-ERB and retinoic acid receptor-related orphan receptors (RORs) bind to a common response element known as the REV-ERB/ROR response element (RRE) and their intrinsic repressive and inductive activities have been proposed to contribute to CLOCK function and BMAL1 transcription (6,7) (Fig. 1). Histone acetylation induces an open chromatin conformation that is thought to activate gene expression (8). Deacetylation would shift back to condensing chromatin and silencing gene expression. The enzymes that participate in these transitions are histone acetyltransferases (HATs) and histone deacetylases (HDACs) (9). CLOCK is an enzyme with HAT activity (10) and CLOCK specifically acetylates nonhistone targets, such as BMAL1 (11). Sirtuin1 (SIRT1) is an NAD+-dependent HDAC, and SIRT1 and CLOCK converge in a coordinated manner. SIRT1 activity depends on nicotinamide phosphoribosyl transferase, which is the rate-limiting enzyme involved in NAD+ synthesis, which is regulated by CLOCK:BMAL1 (12,13). The functions of other clock genes, such as timeless (TIM) and timeless-interacting protein, in the circadian system remain to be fully elucidated.
Figure 1.
Molecular components of the circadian clock. CLOCK, circadian locomotor output cycles kaput; BMAL1, brain and muscle ARNT-like-1; PER, period; CRY, cryptochrome; TIM, timeless; SIRT1, sirtuin1; RORs, retinoic acid receptor-related orphan receptors; RRE, REV-ERB/ROR response element.
Shifts in wake and sleep schedules or sleep deprivation desynchronize the circadian rhythm. Emerging evidence suggests that impaired circadian rhythms may be linked to causal webs of cancer initiation (14,15). Colorectal cancer (CRC) is one of the cancers that is closely associated with circadian disruption (16-18). CRC is the world's fourth most deadly cancer and approximately one in four CRC cases will be diagnosed with metastatic CRC (mCRC) (19). Multiple genetic factors contribute to the development of carcinogenesis of the large bowel, such as amplification of human epidermal growth factor receptor 2 (HER2), as well as KRAS, NRAS and BRAF mutations (20,21), chromosomal instability such as microsatellite instability (MSI) or mismatch repair (MMR) (22), which all provide a basis for optimal therapeutic intervention for individuals. Surgical excision is the first choice of treatment for CRC and chemotherapy that is given in combination with different molecular therapies (such as bevacizumab, panitumumab, cetuximab, regorafenib and aflibercept) significantly extends overall survival (OS) (23). Experimental data further indicated how the clock gene network exerts an influence on CRC progression, offers potential biomarkers for prognosis, serves as a guide for treatment decisions and provides molecular targets for treating CRC.
2. The intestine harbours a circadian clock
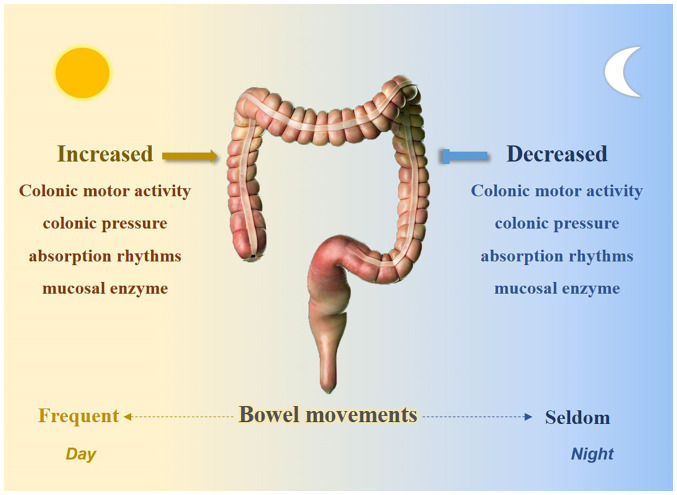
Rhythmic processes were already characterized within the intestine prior to the identification of the molecular clock. Bowel movements usually occur during the day and certain studies have suggested that a potential biological clock exists in the bowel. Research has indicated that colonic motor activity is minimal during sleep but increases significantly at morning awakening (24). Rao et al (25) determined a significant increase in colonic pressure activity after awakening. Other physiologically relevant factors, such as absorption rhythms (26) and mucosal enzymes (27), also exhibited their lowest activities at night (Fig. 2). Furthermore, the rhythmicity of circular muscle contractility and intracolonic pressure were absent in Per1/2 double-knockout mice (28). In addition, the gut microbiota exhibits circadian oscillations in both mice and humans; Per1/2 double-knockout mice lose rhythmic fluctuations in microbiota compositions and time shift-induced jet lag also disrupts diurnal oscillations in commensal bacterial abundances (29).
Figure 2.
The intestine harbors a circadian clock.
Studies have determined that clock genes have an important role in intestinal physiological activities. Per1-2, Cry1, CLOCK, BMAL1 and REV-ERBα are expressed in the epithelial cells of the colon and exhibit circadian rhythmicity (30). Per2 and BMAL1 in the myenteric plexus and epithelial cells have an important role in coordinating gastrointestinal functions, such as cell proliferation and migration (31), and rectal cell proliferation fluctuates during the day in colon cancer (32). BMAL1 was also reported to regulate intestinal drug disposition function (33).
These results clearly support that the circadian system is strongly connected with the gastrointestinal tract.
3. Associations between CRC and circadian clock genes
Clinical research indicated that disruption of the circadian rhythm accelerated CRC progression (34,35). Loss of circadian rhythms was also observed to potentiate CRC initiation in an Apcmin model (36). Abnormal expression levels of circadian clock genes were observed in CRC tissues and current studies suggested that clock genes, mainly including Per, Cry, ARNT1 (BMAL1) and CLOCK, may influence colon cancer progression.
Period genes
Period genes are identified most frequently in CRC. Per1-3 were reported to have tumour suppressor properties, but certain studies reached different conclusions (Table I). A total of seven studies determined that the expression of Per1 was significantly decreased in tumour tissue compared to healthy mucosa (37-44). Furthermore, one study indicated a decrease in Per1 in undifferentiated tumours, while no differences were obtained in differentiated colon carcinomas (45), but two studies found no significant differences in Per1 between colon tumours and healthy mucosa (46,47). Mostafaie et al (40) demonstrated a correlated decrease in Per1 and estrogen receptor-β in colorectal tumours. Furthermore, six studies reported that Per2 expression was significantly decreased in tumour tissues (39,48-52). Hasakova et al (50,53) found that Per2 was downregulated in tumour tissues of male patients with CRC, but Krugluger et al (45) determined that Per2 expression exhibited no differences in colon tumour tissues; the following six studies also confirmed this (38,40-42,47). Another six studies examined the gene expression of Per3 and all found significant decreases (37-39,41,42,54). In addition, Alexander et al (55) found a Per3 gene length polymorphism, and the 5-repeat Per3 VNTR sequence may increase the chance of colorectal adenoma formation.
Table I.
Studies exploring the expression and clinical significance of period genes in patients with CRC.
| Author, year | Gene | Expression in CRC | Observations | (Refs.) |
|---|---|---|---|---|
| Mazzoccoli, 2011 | Per1 | Decreased | Low expression of Per1 may indicate a poor survival prognosis | (39) |
| Oshima, 2011 | Per1 | Decreased | Low expression of Per1 was associated with liver metastasis | (42) |
| Wu, 2016 | Per1 | Decreased | Low expression of Per1 was associated with unfavorable survival of patients with stage III/IV CRC, but with favorable OS of patients with stage II CRC | (43) |
| He, 2022 | Per1 | Decreased | Not specified | (37) |
| Karantanos, 2013 | Per1 | Decreased | Not specified | (38) |
| Orhan, 2019 | Per1 | Decreased | Not specified | (41) |
| Lu, 2015 | Per1 | No differences | Not specified | (46) |
| Nemeth, 2011 | Per1 | No differences | Not specified | (47) |
| Momma, 2017 | Per1 | Not mentioned | Positive Per1 staining was associated with larger tumor size. | (56) |
| Mostafaie, 2009 | Per1 | Not mentioned | Correlated decrease of Per1 and estrogen receptor-β in CRC | (40) |
| Krugluger, 2007 | Per1 | Not mentioned | Per1 decreased in undifferentiated tumors, while no difference was found in differentiated CRC | (45) |
| He, 2022 | Per2 | Not mentioned | High expression of Per2 was associated with immune cell infiltration and associated with unfavorable prognosis in colon adenocarcinoma | (37) |
| Karantanos, 2013 | Per2 | No differences | Not specified | (38) |
| Mostafaie, 2009 | Per2 | No differences | Not specified | (40) |
| Nemeth, 2011 | Per2 | No differences | Not specified | (47) |
| Mazzoccoli, 2011 | Per2 | Decreased | Not specified | (39) |
| Orhan, 2019 | Per2 | No differences | Not specified | (41) |
| Oshima, 2011 | Per2 | Not mentioned | High expression of Per2 associated with better outcomes | (42) |
| Krugluger, 2007 | Per2 | No differences | Not specified | (45) |
| Lu, 2015 | Per2 | Not mentioned | Per2 expression was higher in the pCR group than in the non-pCR group | (46) |
| Aroca-Siendones, 2021 | Per2 | Decreased | Low expression of Per2 was significantly associated with CRC metastasis | (49) |
| Hasakova, 2018/2019 | Per2 | Decreased | Low expression of Per2 was associated with better outcomes in male patients with CRC | (50,53) |
| Wang, 2011 | Per2 | Decreased | Not specified | (48) |
| Wang, 2016 | Per2 | Decreased | Not specified | (51) |
| Xiong, 2022 | Per2 | Decreased | Not specified | (52) |
| Momma, 2017 | Per2 | Not mentioned | Tumors with positive Per2 expression tended to exhibit a greater depth of invasion and were generally more advanced, and patients may have poorer OS | (56) |
| He, 2022 | Per3 | Decreased | Not specified | (37) |
| Karantanos, 2013 | Per3 | Decreased | Not specified | (38) |
| Mazzoccoli, 2011 | Per3 | Decreased | Lower expression of Per3 tended to be associated with poorer survival rates | (39) |
| Orhan, 2019 | Per3 | Decreased | Not specified | (41) |
| Oshima, 2011 | Per3 | Decreased | Not specified | (42) |
| Wang, 2012 | Per3 | Decreased | Incidence of death was higher in patients with Per3-negative CRC | (54) |
| Alexander, 2015 | Per3 | Not mentioned | Regarding Per3 gene length polymorphism, the 5-repeat Per3 VNTR sequence may increase the chance of colorectal adenoma formation | (55) |
CRC, colorectal cancer; Per1, period 1; OS, overall survival; pCR, pathological complete regression.
Studies have also explored the relationships among period genes and clinical-pathological features of patients with CRC. Compared to tumours that did not express period genes, Momma et al (56) indicated that positive Per1 and Per2 staining was associated with larger tumour sizes. Tumours with positivity for Per2 expression tended to have a greater depth of invasion and were generally more advanced, and patients tended to exhibit poorer OS. He et al (37) reported that Per2 was correlated with immune cell infiltration and was associated with unfavorable prognosis in colon adenocarcinoma. Hasakova et al (50,53) determined that Per2 was significantly downregulated in tumour tissues of male patients with CRC, and a negative correlation between Per2 and microRNA (miR)-34a was found. Low expression of Per2 and high expression of miR-34a were associated with significantly better outcomes in male patients. However, certain studies obtained the opposite result, claiming that high Per2 expression was associated with better survival and that low expression of the Per1 gene was related to liver metastasis (42). Low expression of Per2 was significantly associated with CRC metastasis (49). Mazzoccoli et al (39) indicated that lower expression of Per1 and Per3 in tumour tissues may be suggestive of a poorer survival prognosis. Wu et al (43) next determined that low expression levels of Per1 and miR-192 were correlated with unfavourable survival in patients with stage III/IV CRC but with better OS rates in patients with stage II CRC. In addition, Štorcelová et al (57) identified certain cell cycle regulatory genes related to the Per2 expression in human CRC tissues.
Substantial evidence indicates that the cell cycle occurs with a daily rhythm. Healthy mice exhibited rhythmicity of Per1, Per2, Wee1 and p21 in the intestine, but the circadian rhythmicity was significantly reduced in tumours (58). Knocking down Per2 subsequently inhibited p53 and caused CRC cells to acquire malignant biological features (52). Studies have explained how periodic genes regulate CRC progression by interfering with the cell cycle. DNA double-strand breaks activate the DNA damage response (DDR). The DDR is initiated by kinases such as ataxia telangiectasia mutated (ATM) (59) and the G1-S checkpoint is regulated by serine/threonine-protein kinase checkpoint kinase 2 (CHK2), whereas the G2/M checkpoint is regulated by CHK1 (60). Gery et al (61) reported that Per1 promoted DNA damage-induced apoptosis by interacting with ATM and CHK2. In addition, overexpression of c-Myc is associated with CRC (62); in Per1-knockout mice and Per2−mutant mice, c-Myc expression levels are elevated without restriction, inducing excess cyclin D1 expression that increases proliferation (63). Of note, Per2 has been indicated to be suppressed in colon cancer cell lines, which increased cyclin D and cell proliferation (64). Another study also supported that Per2 functions in tumour suppression by regulating the cell cycle (65).
Crosstalk between the circadian clock and Wnt/β-catenin signalling has been reported to be involved in CRC. β-catenin is a main signal transducer of the Wnt pathway (66) and activation of the Wnt/β-catenin pathway promotes cell proliferation and invasion (67). Wood et al (64) demonstrated that downregulation of Per2 in colon cells increased β-catenin expression. In in vivo experiments, when compared to Apc(Min/+) mice, Per2-mutant Apc(Min/+) mice developed smaller colonic polyps. A study from the same group, by Yang et al (68), discovered that increased levels of β-catenin in Apc(Min/+) mice could destabilize the Per2 protein. In addition, period genes were reported to inhibit drug resistance in CRC cells. Per2 was significantly upregulated when glucose metabolism was restricted and thus weakened the ability of cancer cells to survive chemotherapy (69). Per3 was reported to be downregulated in drug-resistant CRC cells and cancer stem-like cells (CSCs). Overexpression of Per3 strengthened the 5-fluorouracil-induced inhibitory effects on colorectal CSCs, while knockdown of Per3 decreased its inhibitory effects. In addition, Per3 overexpression decreased stemness markers, such as CD44, CD133 and SOX2. It was confirmed that Per3 reduces the chemoresistance and self-renewal capability of CRC by inhibiting both Notch and Wnt/β-catenin signalling (70). Calcium significantly prevents tumorigenesis of CRC and Per3 significantly upregulates calcium to prevent CRC tumorigenesis (71).
Cry genes
Various studies have examined the expression of Cry1 and Cry2 in CRC samples (Table II). Further evidence has indicated that Cry2 suppresses colon tumour growth, but the role of Cry1 in cancer progression remains controversial. Compared to matched normal mucosa, gene expression of Cry1 was reported to be upregulated in colon carcinoma tissues by three studies (37,72,73), but two other studies claimed that the expression of Cry1 did not differ significantly between groups (39,46). Cry2 was identified to be downregulated in CRC tissues in three studies (37,39,50,72). Another study retrospectively analysed CRC tissue samples from patients with pathological complete regression (pCR) and non-pCR and suggested that the expression of Cry2 was higher in the pCR group (46).
Table II.
Studies exploring the expression and clinical significance of CRY genes in patients with CRC.
| Author, year | Gene | Expression in CRC | Observations | (Refs.) |
|---|---|---|---|---|
| Hasakova, 2018 | Cry1 | Upregulated | High expression of Cry1 was associated with unfavorable survival in females. Higher expression of Cry1 in the right-sided colon tumor tissue tended to be associated with unfavorable survival in females | (50) |
| He, 2022 | Cry1 | Upregulated | Not mentioned | (37) |
| Yu, 2013 | Cry1 | Upregulated | High Cry1 expression was associated with lymph node metastasis and TNM stage III-IV and tended to be associated with poor OS in patients with CRC | (73) |
| Aroca-Siendones, 2021 | Cry1 | Not mentioned | High expression of Cry1 was associated with lower OS and DFS at five years in patients with CRC | (49) |
| Lu, 2015 | Cry1 | No differences | Not specified | (46) |
| Mazzoccoli, 2011 | Cry1 | No differences | In patients with CRC, elevated Cry1 expression was associated with poorer survival. Cry1 was particularly decreased in elderly female patients with CRC and in tumors located in the transverse colon tend to exhibit lower Cry expression level. | (39) |
| Lu, 2015 | Cry2 | Not mentioned | Cry2 expression was higher in tissue with pathological complete regression | (46) |
| Hasakova, 2018 | Cry2 | Decreased | Not specified | (50) |
| He, 2022 | Cry2 | Decreased | Patients with CRC with high Cry2 expression had a tendency of unfavorable survival prognosis | (37) |
| Mazzoccoli, 2011 | Cry2 | Decreased | Patients with CRC with high Cry2 expression had an unfavorable survival prognosis | (39) |
CRC, colorectal cancer; Cry1, cryptochrome 1; OS, overall survival; DFS, disease-free survival.
The Cry1 and Cry2 genes were also associated with adverse clinical-pathological features in patients with CRC. It was determined that high Cry1 expression was correlated with poor OS rates in patients with CRC (73). Another study also indicated that high expression of Cry1 was associated with lower OS and disease-free survival at five years in patients with CRC (49). Hasakova et al (50,72) indicated that the expression of Cry1 and Cry2 was correlated differently with sex or tumour location in colon carcinoma tissues. Cry1 expression increased significantly in females with distant metastases, while females without distant metastases frequently exhibited downregulation of Cry2 expression. Better survival rates were associated with low expression of Cry2, high expression of Cry1 in tumour tissue was associated with an unfavorable survival prognosis, but this was not observed in males. In addition, higher Cry1 expression in right-sided colon tumour tissues was associated with worse survival rates in females, and the expression of Cry1 in the left-sided colon tumour tissues was higher than in the adjacent tissue in males. Furthermore, Cry2 expression was associated with tumour location in males with grade 2 cancer. However, Mazzoccoli et al (74) indicated that patients with CRC with elevated Cry1 or Cry2 expression levels had poorer survival rates. The Cry1 gene was particularly decreased in elderly female patients with CRC and tumours located in the transverse colon tended to exhibit lower Cry expression levels.
WEE1 was reported to affect the timing of the M phase (75) and downregulation of WEE1 was identified in CRC (76). Studies have indicated that both Cry1 and Cry2 deletions lead to elevated WEE1 levels (63,77) and thus affected the cell cycle and proliferation of CRC. However, this requires to be confirmed by further research.
BMAL1 and CLOCK genes
CLOCK and its heterodimeric partner, ARNTL1 (BMAL1), are the core components of the circadian clock system, and modified expression of these two genes in CRC tumour tissues is related to clinico-pathological features (Table III).
Table III.
Studies exploring the expression and clinical significance of BMAL1/CLOCK in patients with CRC.
| Author, year | Gene | Expression in CRC | Observations | (Refs.) |
|---|---|---|---|---|
| Karantanos, 2013 | BMAL1 | Upregulated | No correlation was found between BMAL1 gene expression and clinical significance | (38) |
| Burgermeister, 2019 | BMAL1 | Decreased | Patients with CRC with low expression of BMAL1 tended to have a good response to bevacizumab | (78) |
| Oshima, 2011 | BMAL1 | Not mentioned | High expression of BMAL1 was associated with liver metastasis in patients with CRC | (42) |
| Zhang, 2021 | BMAL1 | Not mentioned | Patients with CRC with high expression of BMAL1 had enrichment of EMT and invasive gene signatures | (79) |
| He, 2022 | BMAL1 | Decreased | Not specified | (37) |
| Aroca-Siendones, 2021 | BMAL1 | Not mentioned | Low expression of BMAL1 was associated with metastasis | (49) |
| Karantanos, 2013 | CLOCK | Upregulated | No correlation was found between CLOCK gene expression and clinical significance | (38) |
| Momma, 2017 | CLOCK | Not mentioned | Colon tumors with positive CLOCK expression were generally more advanced and invasive | (56) |
| Mostafaie, 2009 | CLOCK | Not mentioned | CLOCK was significantly higher in G(2) tumors of male patients | (40) |
| He, 2022 | CLOCK | Upregulated | Patients with CRC with higher expression of CLOCK tended to have lymph node metastasis and tended to be more advanced | (37) |
| Lu, 2015 | CLOCK | Not mentioned | CLOCK expression levels were elevated in CRC tissue samples from patients with pCR vs. non-pCR | (46) |
| Karantanos, 2013 | CLOCK | Upregulated | Polymorphism in the CLOCK1 gene significantly increased the risk of CRC susceptibility | (38) |
| Alhopuro, 2010 | CLOCK | Not mentioned | CLOCK mutations occurred in nearly half of MSI CRCs | (84) |
CRC, colorectal cancer; CLOCK, circadian locomotor output cycles kaput; BMAL1, brain and muscle ARNT-like-1; EMT, epithelial to mesenchymal transition; MSI, microsatellite instability; pCR, pathological complete regression.
BMAL1 may be a biomarker for CRC treatment prognoses. Studies indicated that the expression levels of BMAL1 decreased by half in mCRC tumour tissues, high expression levels of BMAL1 were associated with reduced efficacy of the anti-angiogenic drug bevacizumab (Beva) and patients who had low expression levels of BMAL1 tended to have good responses to Beva therapy (78); furthermore, upregulation of the BMAL1 gene was correlated with liver metastasis in patients with CRC (42). Patients with CRC with high BMAL1 expression exhibited a significant enrichment of epithelial-mesenchymal transition (EMT) and invasion-associated gene signatures (79). Another study suggested that BMAL1 expression decreased in CRC (37), and low BMAL1 expression was significantly associated with metastasis (49). However, Karantanos et al (38) determined that the BMAL1 gene expression levels were higher in cancerous tissues, but no correlation was found with the clinical significance.
Momma et al (56) indicated that colon tumours with positive CLOCK expression were generally more advanced and invasive. In addition, Mostafaie et al (40) reported that CLOCK expression was significantly higher in G(2) tumours of male patients than that in female patients. Another study suggested that the CLOCK gene was expressed in all CRC tissue samples and colon tumours with higher expression levels of CLOCK were associated with lymph node metastasis and tended to be more advanced (37). Accordingly, higher CLOCK expression was found in CRC cell lines with greater metastatic potential (80,81). However, the CLOCK expression levels were significantly elevated in CRC tissue samples from patients who achieved a pCR compared to those from non-pCR patients (46). Karantanos et al (38) determined that CLOCK gene expression levels were higher in colorectal cancerous tissues. They also performed a case-control study (82) and found that a polymorphism in the CLOCK1 gene significantly increased CRC susceptibility. Mutations in DNA MMR system genes, such as MutL homolog1, MutS homolog 2 (MSH2) and MSH6 result in dysfunctional DNA repair, leading to MSI. MSI accounts for ~15% of CRCs and is thus the main hallmark of CRC (83). Alhopuro et al (84) demonstrated that CLOCK mutations occurred in nearly half of MSI CRCs (total of 101 MSI CRCs), and restoring CLOCK expression in cells enhanced the protective activity against UV-induced apoptosis.
Clockwork genes drive the metabolism and the cell cycle. Fuhr et al (85) identified a reciprocal connection between the circadian clock and glycolysis. Disruption of BMAL1 impacts hexokinase domain containing 1, a glycolysis gene, and leads to increased glycolytic activity. A Japanese study implanted colon tumours in mice and found that the growth of CLOCK-mutant colon tumours was slower than that of wild-type colon tumours. They further indicated that the BMAL1 and CLOCK genes control the time-dependent variations in cellular iron levels by promoting the mRNA expression of iron regulatory protein 2 and transferrin receptor 1. High levels of iron uptake may be associated with an increased risk of CRC (86,87) and thus promote tumour proliferation (88). However, certain studies indicated that both the BMAL1 and CLOCK genes may act as tumour suppressors. CLOCK-binding elements were identified near p21 and BRCA1, which were indicated to interfere with the DNA repair response and cell cycle (84). Sakamoto and Takenoshita (89) observed that overexpression of CLOCK and BMAL1 prevented CRC cells from entering the G1 to S phase by suppressing CyclinD1 to inhibit cell growth. In CLOCK knockout mice, the mRNA levels of WEE1 decreased significantly. Another study demonstrated that inhibiting BMAL1 expressions in murine colon cancer cells decreased DNA damage (90). Zhang et al (91) found that under different P53 backgrounds, BMAL1 knockdown in CRC cells induced a relatively equal activation of AKT/mTOR but ultimately resulted in different cell fates. Under a wild-type P53 background, BMAL1 knockdown cells rapidly underwent apoptosis, while the surviving cells contained less P53.
Stokes et al (36) indicated that CRC tumours only have a weak CLOCK function in vivo. Loss of BMAL1 increases tumour initiation. Intestinal organoid assays indicated that loss of BMAL1 increases self-renewal in a Yes-associated protein 1-dependent manner. BMAL1 was also reported to affect the invasion and metastasis of CRC by promoting EMT. BMAL1 is involved in maintaining the epithelial-mesenchymal equilibrium of CRC cells and influences migration, invasion, adhesion and chemoresistance. BMAL1 knockdown increased the expression of epithelial markers, such as E-cadherin, epithelial cell adhesion molecule and cytokeratin 20, but decreased the expression of Twist and mesenchymal markers (e.g., vimentin and N-cadherin) in CRC cells. In addition, the invasion and drug resistance capacities also decreased in BMAL1-knockdown CRC cells (79). In addition, BMAL1 was reported to promote CRC metastasis by stimulating exosome secretion (92).
Although the BMAL1 and CLOCK expression levels were observed to be related to advanced pathological clinical features, certain experiments have indicated that these two genes may inhibit CRC cell growth. This inconsistency requires to be clarified by further research.
Other clock genes
Altered expression levels of other circadian clock genes were also detected in CRC tissue samples (Table IV). Compared to adjacent normal colon tissues, the SIRT1 and RORA expression levels decreased in CRC tumour tissues, while TIM was reported to be increased (37,39,93,94). High TIM levels were an unfavourable prognostic factor significantly associated with microsatellite instability and proximal lymph nodes and were prevalent in TNM stages III-IV (39). However, the most recent study indicated that loss of TIM expression is associated with advanced tumour stage, metastasis and microsatellite stability status, and TIM expression levels are inversely correlated with a set of gene signatures of EMT markers (95). In addition, NPAS2 expression was significantly downregulated in CRC tumour tissues. Low NPAS2 expression was associated with tumour size, TNM stage and metastases (96).
Table IV.
Studies exploring the expression and clinical significance of SIRT1, RORA and TIM in patients with CRC.
| Author, year | Gene | Expression in CRC | Observations | (Refs.) |
|---|---|---|---|---|
| Pazienza, 2012 | SIRT1 | Decreased | Not specified | (94) |
| He, 2022 | RORA | Decreased | Not specified | (37) |
| Gu, 2018 | RORA | Decreased | Not specified | (93) |
| Mazzoccoli, 2011 | TIM | Upregulated | High TIM levels were associated with MSI and proximal lymph nodes, and were prevalent in TNM stages III-IV | (39) |
| Colangelo T, 2022 | TIM | Not mentioned | Loss of TIM expression is associated with advanced tumor stage, metastasis and microsatellite stability status; TIM expression was inversely correlated with a set of EMT marker genes | (95) |
| Xue, 2014 | NPAS2 | Decreased | Low expression of NPAS2 was associated with tumor size, TNM stage and metastasis | (96) |
CRC, colorectal cancer; MSI, microsatellite instability; EMT, epithelial to mesenchymal transition; NPAS2, neuronal PAS domain protein 2; TIM, timeless; SIRT1, sirtuin1; ROR, retinoic acid receptor-related orphan receptor.
TIM has no apparent circadian clock function in mammals, but research has identified it as being important for CRC progression (97). Recent data revealed that TIM depletion increases γH2AX, a marker of DNA damage, and increases CHK1 and CDK1 phosphorylation (98). A report also demonstrated that overexpression of TIM in colon cancer cells suppressed G2/M arrest, while TIM depletion increased CHK1 and CDK1 phosphorylation and triggered G2/M arrest (98). Another study suggested that TIM inhibited EMT, and its ectopic silencing promoted invasion, migration and stemness in CRC cells (95). Overexpression of SIRT1 in Apc(Min/+) mice resulted in reduced neoplasia, while SIRT1-knockout mice had an increased tumour incidence (99). NPAS2 increased the G0/G1 phase population in CRC cells and inhibited proliferation, invasion and migration in vitro (96).
4. Influence of the circadian clock in CRC treatment
Circadian scheduling produced advantages in chemotherapy treatments of patients with CRC. A 24-h programmable time ambulatory pump was used to administer oxaliplatin by venous infusion against mCRC. This type of treatment increased the maximum tolerated doses and decreased the toxicities of oxaliplatin (100). Another previous phase II trial also proved that if chemo-drug delivery was chronomodulated rather than constant over time, chemotherapy was more effective and less toxic (101). The addition of cetuximab to chronotherapy in patients with mCRC increased the chance of complete resection compared to conventional chemotherapy (102). However, an international randomized trial reported that chronomodulated FLO4 provided no survival benefit compared with ordinary FOLFOX2 in patients with mCRC (103). Of note, Innominato et al (104) chronomodulated irinotecan, oxaliplatin, 5-fluorouracil and leucovorin dosing times against mCRC and found encouraging activity in second-line treatments, with limited haematological toxicity. Another study indicated sex differences in the advantages of chronomodulated irinotecan delivery to minimize adverse events; it is better to administer irinotecan in the morning for males and in the afternoon for females (105).
Fluoropyrimidine-related toxicity is strongly affected by the activity of dihydropyrimidine dehydrogenase (DPD), a 5-fluorouracil-metabolizing enzyme (106). Krugluger et al (45) revealed that disturbed transcription of Per1 may be a cause of disrupted daily DPD oscillation in CRC cells. Fang et al (107) demonstrated that the expression levels of Cry2 are elevated in chemoresistant CRC samples and knockdown of Cry2 increased oxaliplatin sensitivity in CRC cells. Circadian delivery of irinotecan to Caco-2 colon cancer cells indicated that chronomodulated chemotherapy may be an optimized option (108). Hesse et al (109) established a mathematical model to predict the impact of various parameters (e.g., BMAL1 degradation rates, cytosolic BMAL1 degradation rates and CLOCK activation rates) on irinotecan toxicity, but clinical evidence is required to prove this hypothesis.
These findings indicated that the circadian clock system may guide oncological treatments of CRC.
5. Conclusion
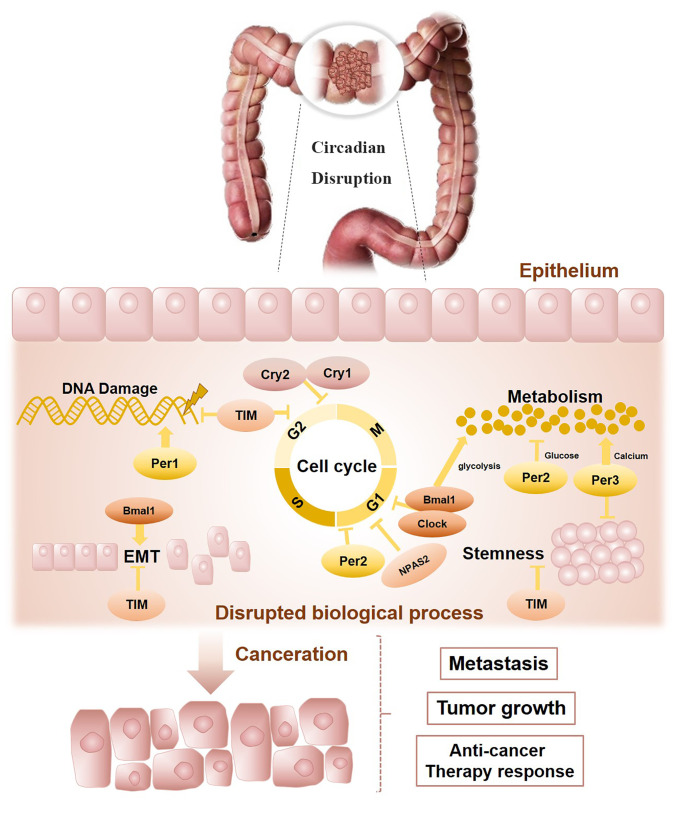
The present review outlines the connections between the circadian clock and CRC progression (Fig. 3). Bowel physiology indicates a circadian rhythm and molecular clockwork dysregulation may host the colorectal carcinogenesis process. According to previously published reports, circadian clock genes are related to CRC progression, patient survival and response to chemotherapy. Although the findings are not all consistent, it may be hypothesized that this largely resulted from individual differences, such as different ethnicities, gender and age. Furthermore, multiple detection methods may also lead to different findings. Based on the circadian rhythm system, chronotherapy may be an optimal method for effective treatment, particularly to increase the maximum tolerated doses and alleviate the toxicity of chemotherapy agents. In addition, the oscillating circadian clock regulates multiple cellular activities related to colorectal carcinogenesis, such as metabolism, the cell cycle, DNA damage response, EMT processes and stemness. Finally, it may be concluded that the circadian clock is tightly correlated with CRC prognosis and allows further advances in modern therapeutic approaches. The relationship between the circadian clock and CRC warrants further investigation.
Figure 3.
Circadian disruption promotes the progression of colorectal cancer. CLOCK, circadian locomotor output cycles kaput; BMAL1, brain and muscle ARNT-like-1; Per1, 2, 3 period1; 2, 3; Cry1, 2, cryptochrome 1, 2; TIM, timeless; EMT, epithelial-mesenchymal transition; NPAS2, neuronal PAS domain protein 2.
Acknowledgments
Not applicable.
Abbreviations
- CRC
colorectal cancer
- mCRC
metastatic CRC
- CLOCK
circadian locomotor output cycles kaput
- BMAL1
brain and muscle ARNT-like-1
- Per1
period1
- Cry1
cryptochrome 1
- TIM
timeless
- SIRT1
sirtuin1
- RORs
retinoic acid receptor-related orphan receptors
- RRE
REV-ERB/ROR response element
- HER2
human epidermal growth factor receptor 2
- MSI
microsatellite instability
- MMR
mismatch repair
- HAT
histone acetyltransferases
- HDAC
histone deacetylases
- DDR
DNA damage response
- ATM
ataxia telangiectasia mutated
- pCR
pathological complete regression
- Beva
bevacizumab
- CSCs
cancer stem-like cells
- EMT
epithelial-mesenchymal transition
- DPD
dihydropyrimidine dehydrogenase
- NPAS2
neuronal PAS domain protein 2
- OS
overall survival
- miR
microRNA
- MSH2
MutS homolog 2
Funding Statement
This work was supported by grants from the National Natural Science Foundation of China (grant no. 82104647), the Chinese Postdoctoral Science Foundation (grant no. 2021M700964) and Guangzhou Science and Technology Bureau (grant no. 201904010396).
Availability of data and materials
Not applicable.
Authors' contributions
LL was involved in the conception and design of the structure of the manuscript. XR then performed the literature search, generated the figures and wrote the manuscript.
Ethics approval and consent to participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Shafer OT, Levine JD, Truman JW, Hall JC. Flies by night: Effects of changing day length on Drosophila's circadian clock. Curr Biol. 2004;14:424–432. doi: 10.1016/j.cub.2004.02.038. [DOI] [PubMed] [Google Scholar]
- 2.Rusak B, Zucker I. Neural regulation of circadian rhythms. Physiol Rev. 1979;59:449–526. doi: 10.1152/physrev.1979.59.3.449. [DOI] [PubMed] [Google Scholar]
- 3.Dibner C, Schibler U, Albrecht U. The mammalian circadian timing system: Organization and coordination of central and peripheral clocks. Annu Rev Physiol. 2010;72:517–549. doi: 10.1146/annurev-physiol-021909-135821. [DOI] [PubMed] [Google Scholar]
- 4.Gekakis N, Staknis D, Nguyen HB, Davis FC, Wilsbacher LD, King DP, Takahashi JS, Weitz CJ. Role of the CLOCK protein in the mammalian circadian mechanism. Science. 1998;280:1564–1569. doi: 10.1126/science.280.5369.1564. [DOI] [PubMed] [Google Scholar]
- 5.Antoch MP, Song EJ, Chang AM, Vitaterna MH, Zhao Y, Wilsbacher LD, Sangoram AM, King DP, Pinto LH, Takahashi JS. Functional identification of the mouse circadian Clock gene by transgenic BAC rescue. Cell. 1997;89:655–667. doi: 10.1016/S0092-8674(00)80246-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Preitner N, Damiola F, Lopez-Molina L, Zakany J, Duboule D, Albrecht U, Schibler U. The orphan nuclear receptor REV-ERBalpha controls circadian transcription within the positive limb of the mammalian circadian oscillator. Cell. 2002;110:251–260. doi: 10.1016/S0092-8674(02)00825-5. [DOI] [PubMed] [Google Scholar]
- 7.Cho H, Zhao X, Hatori M, Yu RT, Barish GD, Lam MT, Chong LW, DiTacchio L, Atkins AR, Glass CK, et al. Regulation of circadian behaviour and metabolism by REV-ERB-α and REV-ERB-β. Nature. 2012;485:123–127. doi: 10.1038/nature11048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Strahl BD, Allis CD. The language of covalent histone modifications. Nature. 2000;403:41–45. doi: 10.1038/47412. [DOI] [PubMed] [Google Scholar]
- 9.Grunstein M. Histone acetylation in chromatin structure and transcription. Nature. 1997;389:349–352. doi: 10.1038/38664. [DOI] [PubMed] [Google Scholar]
- 10.Doi M, Hirayama J, Sassone-Corsi P. Circadian regulator CLOCK is a histone acetyltransferase. Cell. 2006;125:497–508. doi: 10.1016/j.cell.2006.03.033. [DOI] [PubMed] [Google Scholar]
- 11.Hirayama J, Sahar S, Grimaldi B, Tamaru T, Takamatsu K, Nakahata Y, Sassone-Corsi P. CLOCK-mediated acetylation of BMAL1 controls circadian function. Nature. 2007;450:1086–1090. doi: 10.1038/nature06394. [DOI] [PubMed] [Google Scholar]
- 12.Nakahata Y, Kaluzova M, Grimaldi B, Sahar S, Hirayama J, Chen D, Guarente LP, Sassone-Corsi P. The NAD+-dependent deacetylase SIRT1 modulates CLOCK-mediated chromatin remodeling and circadian control. Cell. 2008;134:329–340. doi: 10.1016/j.cell.2008.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Asher G, Gatfield D, Stratmann M, Reinke H, Dibner C, Kreppel F, Mostoslavsky R, Alt FW, Schibler U. SIRT1 regulates circadian clock gene expression through PER2 deacetylation. Cell. 2008;134:317–328. doi: 10.1016/j.cell.2008.06.050. [DOI] [PubMed] [Google Scholar]
- 14.Sulli G, Lam MTY, Panda S. Interplay between circadian clock and cancer: New frontiers for cancer treatment. Trends Cancer. 2019;5:475–494. doi: 10.1016/j.trecan.2019.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Erren TC, Morfeld P, Foster RG, Reiter RJ, Groß JV, Westermann IK. Sleep and cancer: Synthesis of experimental data and meta-analyses of cancer incidence among some 1,500,000 study individuals in 13 countries. Chronobiol Int. 2016;33:325–350. doi: 10.3109/07420528.2016.1149486. [DOI] [PubMed] [Google Scholar]
- 16.Papantoniou K, Devore EE, Massa J, Strohmaier S, Vetter C, Yang L, Shi Y, Giovannucci E, Speizer F, Schernhammer ES. Rotating night shift work and colorectal cancer risk in the nurses' health studies. Int J Cancer. 2018;143:2709–2717. doi: 10.1002/ijc.31655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Shi Y, Liu L, Hamada T, Nowak JA, Giannakis M, Ma Y, Song M, Nevo D, Kosumi K, Gu M, et al. Night-shift work duration and risk of colorectal cancer according to IRS1 and IRS2 expression. Cancer Epidemiol Biomarkers Prev. 2020;29:133–140. doi: 10.1158/1055-9965.EPI-19-0325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bishehsari F, Engen PA, Voigt RM, Swanson G, Shaikh M, Wilber S, Naqib A, Green SJ, Shetuni B, Forsyth CB, et al. Abnormal eating patterns cause circadian disruption and promote alcohol-associated colon carcinogenesis. Cell Mol Gastroenterol Hepatol. 2020;9:219–237. doi: 10.1016/j.jcmgh.2019.10.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68:394–424. doi: 10.3322/caac.21492. [DOI] [PubMed] [Google Scholar]
- 20.Pelullo M, Nardozza F, Zema S, Quaranta R, Nicoletti C, Besharat ZM, Felli MP, Cerbelli B, d'Amati G, Palermo R, et al. Kras/ADAM17-dependent Jag1-ICD reverse signaling sustains colorectal cancer progression and chemoresistance. Cancer Res. 2019;79:5575–5586. doi: 10.1158/0008-5472.CAN-19-0145. [DOI] [PubMed] [Google Scholar]
- 21.Afrăsânie VA, Marinca MV, Alexa-Stratulat T, Gafton B, Păduraru M, Adavidoaiei AM, Miron L, Rusu C. KRAS, NRAS, BRAF, HER2 and microsatellite instability in metastatic colorectal cancer-practical implications for the clinician. Radiol Oncol. 2019;53:265–274. doi: 10.2478/raon-2019-0033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Slik K, Turkki R, Carpén O, Kurki S, Korkeila E, Sundström J, Pellinen T. CDX2 loss with microsatellite stable phenotype predicts poor clinical outcome in stage II colorectal carcinoma. Am J Surg Pathol. 2019;43:1473–1482. doi: 10.1097/PAS.0000000000001356. [DOI] [PubMed] [Google Scholar]
- 23.Wolpin BM, Mayer RJ. Systemic treatment of colorectal cancer. Gastroenterology. 2008;134:1296–1310. doi: 10.1053/j.gastro.2008.02.098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Narducci F, Bassotti G, Gaburri M, Morelli A. Twenty four hour manometric recording of colonic motor activity in healthy man. Gut. 1987;28:17–25. doi: 10.1136/gut.28.1.17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rao SS, Sadeghi P, Beaty J, Kavlock R, Ackerson K. Ambulatory 24-h colonic manometry in healthy humans. Am J Physiol Gastrointest Liver Physiol. 2001;280:G629–G639. doi: 10.1152/ajpgi.2001.280.4.G629. [DOI] [PubMed] [Google Scholar]
- 26.Clench J, Reinberg A, Dziewanowska Z, Ghata J, Smolensky M. Circadian changes in the bioavailability and effects of indomethacin in healthy subjects. Eur J Clin Pharmacol. 1981;20:359–369. doi: 10.1007/BF00615406. [DOI] [PubMed] [Google Scholar]
- 27.Markiewicz A, Kamiński M, Chocilowski W, Gomoluch T, Bołdys H, Skrzypek B. Circadian rhythms of four marker enzymes activity of the jejunal villi in man. Acta Histochem. 1983;72:91–99. doi: 10.1016/S0065-1281(83)80015-4. [DOI] [PubMed] [Google Scholar]
- 28.Hoogerwerf WA, Shahinian VB, Cornélissen G, Halberg F, Bostwick J, Timm J, Bartell PA, Cassone VM. Rhythmic changes in colonic motility are regulated by period genes. Am J Physiol Gastrointest Liver Physiol. 2010;298:G143–G150. doi: 10.1152/ajpgi.00402.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Thaiss CA, Zeevi D, Levy M, Zilberman-Schapira G, Suez J, Tengeler AC, Abramson L, Katz MN, Korem T, Zmora N, et al. Transkingdom control of microbiota diurnal oscillations promotes metabolic homeostasis. Cell. 2014;159:514–529. doi: 10.1016/j.cell.2014.09.048. [DOI] [PubMed] [Google Scholar]
- 30.Sládek M, Rybová M, Jindráková Z, Zemanová Z, Polidarová L, Mrnka L, O'Neill J, Pácha J, Sumová A. Insight into the circadian clock within rat colonic epithelial cells. Gastroenterology. 2007;133:1240–1249. doi: 10.1053/j.gastro.2007.05.053. [DOI] [PubMed] [Google Scholar]
- 31.Hoogerwerf WA, Hellmich HL, Cornélissen G, Halberg F, Shahinian VB, Bostwick J, Savidge TC, Cassone VM. Clock gene expression in the murine gastrointestinal tract: Endogenous rhythmicity and effects of a feeding regimen. Gastroenterology. 2007;133:1250–1260. doi: 10.1053/j.gastro.2007.07.009. [DOI] [PubMed] [Google Scholar]
- 32.Brandi G, Calabrese C, Pantaleo MA, Morselli Labate A, Di Febo G, Hakim R, De Vivo A, Di Marco MC, Biasco G. Circadian variations of rectal cell proliferation in patients affected by advanced colorectal cancer. Cancer Lett. 2004;208:193–196. doi: 10.1016/j.canlet.2003.11.015. [DOI] [PubMed] [Google Scholar]
- 33.Yu F, Zhang T, Zhou C, Xu H, Guo L, Chen M, Wu B. The circadian clock gene Bmal1 controls intestinal exporter MRP2 and drug disposition. Theranostics. 2019;9:2754–2767. doi: 10.7150/thno.33395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lévi F, Dugué PA, Innominato P, Karaboué A, Dispersyn G, Parganiha A, Giacchetti S, Moreau T, Focan C, Waterhouse J, et al. Wrist actimetry circadian rhythm as a robust predictor of colorectal cancer patients survival. Chronobiol Int. 2014;31:891–900. doi: 10.3109/07420528.2014.924523. [DOI] [PubMed] [Google Scholar]
- 35.Innominato PF, Focan C, Gorlia T, Moreau T, Garufi C, Waterhouse J, Giacchetti S, Coudert B, Iacobelli S, Genet D, et al. Circadian rhythm in rest and activity: A biological correlate of quality of life and a predictor of survival in patients with metastatic colorectal cancer. Cancer Res. 2009;69:4700–4707. doi: 10.1158/0008-5472.CAN-08-4747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Stokes K, Nunes M, Trombley C, Flôres DEFL, Wu G, Taleb Z, Alkhateeb A, Banskota S, Harris C, Love OP, et al. The circadian clock gene, Bmal1, regulates intestinal stem cell signaling and represses tumor initiation. Cell Mol Gastroenterol Hepatol. 2021;12:1847–1872.e0. doi: 10.1016/j.jcmgh.2021.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.He A, Huang Z, Zhang R, Lu H, Wang J, Cao J, Feng Q. Circadian clock genes are correlated with prognosis and immune cell infiltration in colon adenocarcinoma. Comput Math Methods Med. 2022;2022:1709918. doi: 10.1155/2022/4957996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Karantanos T, Theodoropoulos G, Gazouli M, Vaiopoulou A, Karantanou C, Lymberi M, Pektasides D. Expression of clock genes in patients with colorectal cancer. Int J Biol Markers. 2013;28:280–285. doi: 10.5301/JBM.5000033. [DOI] [PubMed] [Google Scholar]
- 39.Mazzoccoli G, Panza A, Valvano MR, Palumbo O, Carella M, Pazienza V, Biscaglia G, Tavano F, Di Sebastiano P, Andriulli A, Piepoli A. Clock gene expression levels and relationship with clinical and pathological features in colorectal cancer patients. Chronobiol Int. 2011;28:841–851. doi: 10.3109/07420528.2011.615182. [DOI] [PubMed] [Google Scholar]
- 40.Mostafaie N, Kállay E, Sauerzapf E, Bonner E, Kriwanek S, Cross HS, Huber KR, Krugluger W. Correlated downregulation of estrogen receptor beta and the circadian clock gene Per1 in human colorectal cancer. Mol Carcinog. 2009;48:642–647. doi: 10.1002/mc.20510. [DOI] [PubMed] [Google Scholar]
- 41.Orhan T, Nielsen PB, Hviid TVF, Rosen AW, Gögenür I. Expression of circadian clock genes in human colorectal cancer tissues using droplet digital PCR. Cancer Invest. 2019;37:90–98. doi: 10.1080/07357907.2019.1571079. [DOI] [PubMed] [Google Scholar]
- 42.Oshima T, Takenoshita S, Akaike M, Kunisaki C, Fujii S, Nozaki A, Numata K, Shiozawa M, Rino Y, Tanaka K, et al. Expression of circadian genes correlates with liver metastasis and outcomes in colorectal cancer. Oncol Rep. 2011;25:1439–1446. doi: 10.3892/or.2011.1207. [DOI] [PubMed] [Google Scholar]
- 43.Wu S, Fesler A, Ju J. Implications of circadian rhythm regulation by microRNAs in colorectal cancer. Cancer Transl Med. 2016;2:1–6. doi: 10.4103/2395-3977.177555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Expression of PER, CRY, and TIM genes for the pathological features of colorectal cancer patients [Retraction] Onco Targets Ther. 2016;9:5699. doi: 10.2147/OTT.S121862. No authors listed. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Krugluger W, Brandstaetter A, Kállay E, Schueller J, Krexner E, Kriwanek S, Bonner E, Cross HS. Regulation of genes of the circadian clock in human colon cancer: Reduced period-1 and dihydropyrimidine dehydrogenase transcription correlates in high-grade tumors. Cancer Res. 2007;67:7917–7922. doi: 10.1158/0008-5472.CAN-07-0133. [DOI] [PubMed] [Google Scholar]
- 46.Lu H, Chu Q, Xie G, Han H, Chen Z, Xu B, Yue Z. Circadian gene expression predicts patient response to neoadjuvant chemo-radiation therapy for rectal cancer. Int J Clin Exp Pathol. 2015;8:10985–10994. [PMC free article] [PubMed] [Google Scholar]
- 47.Nemeth C, Humpeler S, Kallay E, Mesteri I, Svoboda M, Rögelsperger O, Klammer N, Thalhammer T, Ekmekcioglu C. Decreased expression of the melatonin receptor 1 in human colorectal adenocarcinomas. J Biol Regul Homeost Agents. 2011;25:531–542. [PubMed] [Google Scholar]
- 48.Wang Y, Hua L, Lu C, Chen Z. Expression of circadian clock gene human Period2 (hPer2) in human colorectal carcinoma. World J Surg Oncol. 2011;9:166. doi: 10.1186/1477-7819-9-166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Aroca-Siendones MI, Moreno-SanJuan S, Puentes-Pardo JD, Verbeni M, Arnedo J, Escudero-Feliu J, García-Costela M, García-Robles A, Carazo Á, León J. Core circadian clock proteins as biomarkers of progression in colorectal cancer. Biomedicines. 2021;9:967. doi: 10.3390/biomedicines9080967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hasakova K, Vician M, Reis R, Zeman M, Herichova I. Sex-dependent correlation between survival and expression of genes related to the circadian oscillator in patients with colorectal cancer. Chronobiol Int. 2018;35:1423–1434. doi: 10.1080/07420528.2018.1488722. [DOI] [PubMed] [Google Scholar]
- 51.Wang Y, Cheng Y, Yu G, Jia B, Hu Z, Zhang L. Expression of PER, CRY, and TIM genes for the pathological features of colorectal cancer patients. Onco Targets Ther. 2016;9:1997–2005. doi: 10.2147/OTT.S96925. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 52.Xiong Y, Zhuang Y, Zhong M, Qin W, Huang B, Zhao J, Gao Z, Ma J, Wu Z, Hong X, et al. Period 2 suppresses the malignant cellular behaviors of colorectal cancer through the epithelial-mesenchymal transformation process. Cancer Control. 2022;29:10732748221081369. doi: 10.1177/10732748221081369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Hasakova K, Reis R, Vician M, Zeman M, Herichova I. Expression of miR-34a-5p is up-regulated in human colorectal cancer and correlates with survival and clock gene PER2 expression. PLoS One. 2019;14:e0224396. doi: 10.1371/journal.pone.0224396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wang X, Yan D, Teng M, Fan J, Zhou C, Li D, Qiu G, Sun X, Li T, Xing T, et al. Reduced expression of PER3 is associated with incidence and development of colon cancer. Ann Surg Oncol. 2012;19:3081–3088. doi: 10.1245/s10434-012-2279-5. [DOI] [PubMed] [Google Scholar]
- 55.Alexander M, Burch JB, Steck SE, Chen CF, Hurley TG, Cavicchia P, Ray M, Shivappa N, Guess J, Zhang H, et al. Case-control study of the PERIOD3 clock gene length polymorphism and colorectal adenoma formation. Oncol Rep. 2015;33:935–941. doi: 10.3892/or.2014.3667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Momma T, Okayama H, Saitou M, Sugeno H, Yoshimoto N, Takebayashi Y, Ohki S, Takenoshita S. Expression of circadian clock genes in human colorectal adenoma and carcinoma. Oncol Lett. 2017;14:5319–5325. doi: 10.3892/ol.2017.6876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Štorcelová M, Vicián M, Reis R, Zeman M, Herichová I. Expression of cell cycle regulatory factors hus1, gadd45a, rb1, cdkn2a and mre11a correlates with expression of clock gene per2 in human colorectal carcinoma tissue. Mol Biol Rep. 2013;40:6351–6361. doi: 10.1007/s11033-013-2749-2. [DOI] [PubMed] [Google Scholar]
- 58.Soták M, Polidarová L, Ergang P, Sumová A, Pácha J. An association between clock genes and clock-controlled cell cycle genes in murine colorectal tumors. Int J Cancer. 2013;132:1032–1041. doi: 10.1002/ijc.27760. [DOI] [PubMed] [Google Scholar]
- 59.Bednarski JJ, Sleckman BP. At the intersection of DNA damage and immune responses. Nat Rev Immunol. 2019;19:231–242. doi: 10.1038/s41577-019-0135-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Ciccia A, Elledge SJ. The DNA damage response: Making it safe to play with knives. Mol Cell. 2010;40:179–204. doi: 10.1016/j.molcel.2010.09.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Gery S, Komatsu N, Baldjyan L, Yu A, Koo D, Koeffler HP. The circadian gene per1 plays an important role in cell growth and DNA damage control in human cancer cells. Mol Cell. 2006;22:375–382. doi: 10.1016/j.molcel.2006.03.038. [DOI] [PubMed] [Google Scholar]
- 62.Arango D, Mariadason JM, Wilson AJ, Yang W, Corner GA, Nicholas C, Aranes MJ, Augenlicht LH. c-Myc overexpression sensitises colon cancer cells to camptothecin-induced apoptosis. Br J Cancer. 2003;89:1757–1765. doi: 10.1038/sj.bjc.6601338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Borgs L, Beukelaers P, Vandenbosch R, Belachew S, Nguyen L, Malgrange B. Cell 'circadian' cycle: New role for mammalian core clock genes. Cell Cycle. 2009;8:832–837. doi: 10.4161/cc.8.6.7869. [DOI] [PubMed] [Google Scholar]
- 64.Wood PA, Yang X, Taber A, Oh EY, Ansell C, Ayers SE, Al-Assaad Z, Carnevale K, Berger FG, Peña MM, Hrushesky WJ. Period 2 mutation accelerates ApcMin/+ tumorigenesis. Mol Cancer Res. 2008;6:1786–1793. doi: 10.1158/1541-7786.MCR-08-0196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Fu L, Pelicano H, Liu J, Huang P, Lee C. The circadian gene Period2 plays an important role in tumor suppression and DNA damage response in vivo. Cell. 2002;111:41–50. doi: 10.1016/S0092-8674(02)00961-3. [DOI] [PubMed] [Google Scholar]
- 66.Shen P, Pichler M, Chen M, Calin GA, Ling H. To Wnt or lose: The missing non-coding linc in colorectal cancer. Int J Mol Sci. 2017;18:2003. doi: 10.3390/ijms18092003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Filipovich A, Gehrke I, Poll-Wolbeck SJ, Kreuzer KA. Physiological inhibitors of Wnt signaling. Eur J Haematol. 2011;86:453–465. doi: 10.1111/j.1600-0609.2011.01592.x. [DOI] [PubMed] [Google Scholar]
- 68.Yang X, Wood PA, Ansell CM, Ohmori M, Oh EY, Xiong Y, Berger FG, Peña MM, Hrushesky WJ. Beta-catenin induces beta-TrCP-mediated PER2 degradation altering circadian clock gene expression in intestinal mucosa of ApcMin/+ mice. J Biochem. 2009;145:289–297. doi: 10.1093/jb/mvn167. [DOI] [PubMed] [Google Scholar]
- 69.Schroll MM, LaBonia GJ, Ludwig KR, Hummon AB. Glucose restriction combined with autophagy inhibition and chemotherapy in HCT 116 spheroids decreases cell clonogenicity and viability regulated by tumor suppressor genes. J Proteome Res. 2017;16:3009–3018. doi: 10.1021/acs.jproteome.7b00293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Zhang F, Sun H, Zhang S, Yang X, Zhang G, Su T. Overexpression of PER3 inhibits self-renewal capability and chemoresistance of colorectal cancer stem-like cells via inhibition of notch and β-catenin signaling. Oncol Res. 2017;25:709–719. doi: 10.3727/096504016X14772331883976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Wang JL, Lin YW, Chen HM, Kong X, Xiong H, Shen N, Hong J, Fang JY. Calcium prevents tumorigenesis in a mouse model of colorectal cancer. PLoS One. 2011;6:e22566. doi: 10.1371/journal.pone.0022566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Hasakova K, Vician M, Reis R, Zeman M, Herichova I. The expression of clock genes cry1 and cry2 in human colorectal cancer and tumor adjacent tissues correlates differently dependent on tumor location. Neoplasma. 2018;65:986–992. doi: 10.4149/neo_2018_180122N47. [DOI] [PubMed] [Google Scholar]
- 73.Yu H, Meng X, Wu J, Pan C, Ying X, Zhou Y, Liu R, Huang W. Cryptochrome 1 overexpression correlates with tumor progression and poor prognosis in patients with colorectal cancer. PLoS One. 2013;8:e61679. doi: 10.1371/journal.pone.0061679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Mazzoccoli G, Colangelo T, Panza A, Rubino R, De Cata A, Tiberio C, Valvano MR, Pazienza V, Merla G, Augello B, et al. Deregulated expression of cryptochrome genes in human colorectal cancer. Mol Cancer. 2016;15:6. doi: 10.1186/s12943-016-0492-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Heald R, McLoughlin M, McKeon F. Human wee1 maintains mitotic timing by protecting the nucleus from cytoplasmically activated Cdc2 kinase. Cell. 1993;74:463–474. doi: 10.1016/0092-8674(93)80048-J. [DOI] [PubMed] [Google Scholar]
- 76.Backert S, Gelos M, Kobalz U, Hanski ML, Böhm C, Mann B, Lövin N, Gratchev A, Mansmann U, Moyer MP, et al. Differential gene expression in colon carcinoma cells and tissues detected with a cDNA array. Int J Cancer. 1999;82:868–874. doi: 10.1002/(SICI)1097-0215(19990909)82:6<868::AID-IJC16>3.0.CO;2-W. [DOI] [PubMed] [Google Scholar]
- 77.van der Horst GT, Muijtjens M, Kobayashi K, Takano R, Kanno S, Takao M, de Wit J, Verkerk A, Eker AP, van Leenen D, et al. Mammalian Cry1 and Cry2 are essential for maintenance of circadian rhythms. Nature. 1999;398:627–630. doi: 10.1038/19323. [DOI] [PubMed] [Google Scholar]
- 78.Burgermeister E, Battaglin F, Eladly F, Wu W, Herweck F, Schulte N, Betge J, Härtel N, Kather JN, Weis CA, et al. Aryl hydrocarbon receptor nuclear translocator-like (ARNTL/BMAL1) is associated with bevacizumab resistance in colorectal cancer via regulation of vascular endothelial growth factor A. EBioMedicine. 2019;45:139–154. doi: 10.1016/j.ebiom.2019.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Zhang Y, Devocelle A, Desterke C, de Souza LEB, Hadadi É, Acloque H, Foudi A, Xiang Y, Ballesta A, Chang Y, Giron-Michel J. BMAL1 knockdown leans epithelial-mesenchymal balance toward epithelial properties and decreases the chemoresistance of colon carcinoma cells. Int J Mol Sci. 2021;22:5247. doi: 10.3390/ijms22105247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Wang L, Chen B, Wang Y, Sun N, Lu C, Qian R, Hua L. hClock gene expression in human colorectal carcinoma. Mol Med Rep. 2013;8:1017–1022. doi: 10.3892/mmr.2013.1643. [DOI] [PubMed] [Google Scholar]
- 81.Wang Y, Sun N, Lu C, Bei Y, Qian R, Hua L. Upregulation of circadian gene 'hClock' contribution to metastasis of colorectal cancer. Int J Oncol. 2017;50:2191–2199. doi: 10.3892/ijo.2017.3987. [DOI] [PubMed] [Google Scholar]
- 82.Karantanos T, Theodoropoulos G, Gazouli M, Vaiopoulou A, Karantanou C, Stravopodis DJ, Bramis K, Lymperi M, Pektasidis D. Association of the clock genes polymorphisms with colorectal cancer susceptibility. J Surg Oncol. 2013;108:563–567. doi: 10.1002/jso.23434. [DOI] [PubMed] [Google Scholar]
- 83.Kurzawski G, Suchy J, Debniak T, Kładny J, Lubiński J. Importance of microsatellite instability (MSI) in colorectal cancer: MSI as a diagnostic tool. Ann Oncol. 2004;15(Suppl 4):iv283–iv284. doi: 10.1093/annonc/mdh940. [DOI] [PubMed] [Google Scholar]
- 84.Alhopuro P, Björklund M, Sammalkorpi H, Turunen M, Tuupanen S, Biström M, Niittymäki I, Lehtonen HJ, Kivioja T, Launonen V, et al. Mutations in the circadian gene CLOCK in colorectal cancer. Mol Cancer Res. 2010;8:952–960. doi: 10.1158/1541-7786.MCR-10-0086. [DOI] [PubMed] [Google Scholar]
- 85.Fuhr L, El-Athman R, Scrima R, Cela O, Carbone A, Knoop H, Li Y, Hoffmann K, Laukkanen MO, Corcione F, et al. The circadian clock regulates metabolic phenotype rewiring via HKDC1 and modulates tumor progression and drug response in colorectal cancer. EBioMedicine. 2018;33:105–121. doi: 10.1016/j.ebiom.2018.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Nelson RL. Iron and colorectal cancer risk: Human studies. Nutr Rev. 2001;59:140–148. doi: 10.1111/j.1753-4887.2001.tb07002.x. [DOI] [PubMed] [Google Scholar]
- 87.Osborne NJ, Gurrin LC, Allen KJ, Constantine CC, Delatycki MB, McLaren CE, Gertig DM, Anderson GJ, Southey MC, Olynyk JK, et al. HFE C282Y homozygotes are at increased risk of breast and colorectal cancer. Hepatology. 2010;51:1311–1318. doi: 10.1002/hep.23448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Okazaki F, Matsunaga N, Okazaki H, Azuma H, Hamamura K, Tsuruta A, Tsurudome Y, Ogino T, Hara Y, Suzuki T, et al. Circadian clock in a mouse colon tumor regulates intracellular iron levels to promote tumor progression. J Biol Chem. 2016;291:7017–7028. doi: 10.1074/jbc.M115.713412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Sakamoto W, Takenoshita S. Overexpression of both clock and BMAL1 inhibits entry to S phase in human colon cancer cells. Fukushima J Med Sci. 2015;61:111–124. doi: 10.5387/fms.2015-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Zeng ZL, Wu MW, Sun J, Sun YL, Cai YC, Huang YJ, Xian LJ. Effects of the biological clock gene Bmal1 on tumour growth and anti-cancer drug activity. J Biochem. 2010;148:319–326. doi: 10.1093/jb/mvq069. [DOI] [PubMed] [Google Scholar]
- 91.Zhang Y, Devocelle A, Souza L, Foudi A, Tenreira Bento S, Desterke C, Sherrard R, Ballesta A, Adam R, Giron-Michel J, Chang Y. BMAL1 knockdown triggers different colon carcinoma cell fates by altering the delicate equilibrium between AKT/mTOR and P53/P21 pathways. Aging (Albany NY) 2020;12:8067–8083. doi: 10.18632/aging.103124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Dong P, Wang Y, Liu Y, Zhu C, Lin J, Qian R, Hua L, Lu C. BMAL1 induces colorectal cancer metastasis by stimulating exosome secretion. Mol Biol Rep. 2022;49:373–384. doi: 10.1007/s11033-021-06883-z. [DOI] [PubMed] [Google Scholar]
- 93.Gu D, Li S, Ben S, Du M, Chu H, Zhang Z, Wang M, Zhang ZF, Chen J. Circadian clock pathway genes associated with colorectal cancer risk and prognosis. Arch Toxicol. 2018;92:2681–2689. doi: 10.1007/s00204-018-2251-7. [DOI] [PubMed] [Google Scholar]
- 94.Pazienza V, Piepoli A, Panza A, Valvano MR, Benegiamo G, Vinciguerra M, Andriulli A, Mazzoccoli G. SIRT1 and the clock gene machinery in colorectal cancer. Cancer Invest. 2012;30:98–105. doi: 10.3109/07357907.2011.640650. [DOI] [PubMed] [Google Scholar]
- 95.Colangelo T, Carbone A, Mazzarelli F, Cuttano R, Dama E, Nittoli T, Albanesi J, Barisciano G, Forte N, Palumbo O, et al. Loss of circadian gene timeless induces EMT and tumor progression in colorectal cancer via Zeb1-dependent mechanism. Cell Death Differ. 2022;29:1552–1568. doi: 10.1038/s41418-022-00935-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Xue X, Liu F, Han Y, Li P, Yuan B, Wang X, Chen Y, Kuang Y, Zhi Q, Zhao H. Silencing NPAS2 promotes cell growth and invasion in DLD-1 cells and correlated with poor prognosis of colorectal cancer. Biochem Biophys Res Commun. 2014;450:1058–1062. doi: 10.1016/j.bbrc.2014.06.104. [DOI] [PubMed] [Google Scholar]
- 97.Yang X, Wood PA, Hrushesky WJ. Mammalian TIMELESS is required for ATM-dependent CHK2 activation and G2/M checkpoint control. J Biol Chem. 2010;285:3030–3034. doi: 10.1074/jbc.M109.050237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Neilsen BK, Frodyma DE, McCall JL, Fisher KW, Lewis RE. ERK-mediated TIMELESS expression suppresses G2/M arrest in colon cancer cells. PLoS One. 2019;14:e209224. doi: 10.1371/journal.pone.0209224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Pruitt K, Zinn RL, Ohm JE, McGarvey KM, Kang SH, Watkins DN, Herman JG, Baylin SB. Inhibition of SIRT1 reactivates silenced cancer genes without loss of promoter DNA hypermethylation. PLoS Genet. 2006;2:e40. doi: 10.1371/journal.pgen.0020040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Levi F, Perpoint B, Garufi C, Focan C, Chollet P, Depres-Brummer P, Zidani R, ienza S, Itzhaki M, Iacobelli S, et al. Oxaliplatin activity against metastatic colorectal cancer. A phase II study of 5-day continuous venous infusion at circadian rhythm modulated rate. Eur J Cancer. 1993;29A:1280–1284. doi: 10.1016/0959-8049(93)90073-O. [DOI] [PubMed] [Google Scholar]
- 101.Lévi FA, Zidani R, Vannetzel JM, Perpoint B, Focan C, Faggiuolo R, Chollet P, Garufi C, Itzhaki M, Dogliotti L, et al. Chronomodulated versus fixed-infusion-rate delivery of ambulatory chemotherapy with oxaliplatin, fluorouracil, and folinic acid (leucovorin) in patients with colorectal cancer metastases: A randomized multi-institutional trial. J Natl Cancer Inst. 1994;86:1608–1617. doi: 10.1093/jnci/86.21.1608. [DOI] [PubMed] [Google Scholar]
- 102.Lévi F, Karaboué A, Gorden L, Innominato PF, Saffroy R, Giacchetti S, Hauteville D, Guettier C, Adam R, Bouchahda M. Cetuximab and circadian chronomodulated chemotherapy as salvage treatment for metastatic colorectal cancer (mCRC): Safety, efficacy and improved secondary surgical resectability. Cancer Chemother Pharmacol. 2011;67:339–348. doi: 10.1007/s00280-010-1327-8. [DOI] [PubMed] [Google Scholar]
- 103.Innominato PF, Giacchetti S, Moreau T, Smaaland R, Focan C, Bjarnason GA, Garufi C, Iacobelli S, Tampellini M, Tumolo S, et al. Prediction of survival by neutropenia according to delivery schedule of oxaliplatin-5-fluorouracil-leucovorin for metastatic colorectal cancer in a randomized international trial (EORTC 05963) Chronobiol Int. 2011;28:586–600. doi: 10.3109/07420528.2011.597532. [DOI] [PubMed] [Google Scholar]
- 104.Innominato PF, Karaboué A, Focan C, Chollet P, Giacchetti S, Bouchahda M, Ulusakarya A, Torsello A, Adam R, Lévi FA, Garufi C. Efficacy and safety of chronomodulated irinotecan, oxaliplatin, 5-fluorouracil and leucovorin combination as first- or second-line treatment against metastatic colorectal cancer: Results from the international EORTC 05011 trial. Int J Cancer. 2020;148:2512–2521. doi: 10.1002/ijc.33422. Epub ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Innominato PF, Ballesta A, Huang Q, Focan C, Chollet P, Karaboué A, Giacchetti S, Bouchahda M, Adam R, Garufi C, Lévi FA. Sex-dependent least toxic timing of irinotecan combined with chronomodulated chemotherapy for metastatic colorectal cancer: Randomized multicenter EORTC 05011 trial. Cancer Med. 2020;9:4148–4159. doi: 10.1002/cam4.3056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Henricks LM, Opdam FL, Beijnen JH, Cats A, Schellens JHM. DPYD genotype-guided dose individualization to improve patient safety of fluoropyrimidine therapy: Call for a drug label update. Ann Oncol. 2017;28:2915–2922. doi: 10.1093/annonc/mdx411. [DOI] [PubMed] [Google Scholar]
- 107.Fang L, Yang Z, Zhou J, Tung JY, Hsiao CD, Wang L, Deng Y, Wang P, Wang J, Lee MH. Circadian clock gene CRY2 degradation is involved in chemoresistance of colorectal cancer. Mol Cancer Ther. 2015;14:1476–1487. doi: 10.1158/1535-7163.MCT-15-0030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Ballesta A, Dulong S, Abbara C, Cohen B, Okyar A, Clairambault J, Levi F. A combined experimental and mathematical approach for molecular-based optimization of irinotecan circadian delivery. PLoS Comput Biol. 2011;7:e1002143. doi: 10.1371/journal.pcbi.1002143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Hesse J, Martinelli J, Aboumanify O, Ballesta A, Relógio A. A mathematical model of the circadian clock and drug pharmacology to optimize irinotecan administration timing in colorectal cancer. Comput Struct Biotechnol J. 2021;19:5170–5183. doi: 10.1016/j.csbj.2021.08.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Not applicable.