Abstract
Sialodacryoadenitis virus (SDAV) is a coronavirus that is commonly found in laboratory rats and that causes sialodacryoadenitis and respiratory illness. We cloned and sequenced the 3′ terminal 9.8 kb of the genomic RNA and analyzed the structure of the viral genome. As with mouse hepatitis coronaviruses (MHVs), the SDAV genome was able to code for a spike protein, a small membrane protein, a membrane-associated protein, and a nucleocapsid protein. In addition, the hemagglutinin-esterase gene capable of encoding a protein of 439 amino acids (aa) was identified. The putative functional site for acetylesterase activity was present in the HE protein as Phe-Gly-Asp-Ser (FGDS), suggesting that the SDAV HE protein might have retained the esterase activity. Immediately upstream of the HE gene and downstream of the polymerase 1b gene, the NS2 nonstructural-protein gene was identified with a coding capacity of 274 aa. A motif of UCUAAAC was identified as a potential transcription signal for subgenomic mRNA synthesis. Large insertions of 172, 127, and 44 aa were detected in the N-terminal half of the predicted S protein of SDAV when its sequence was compared to the sequences of MHV 2, MHV JHM, and MHV A59, respectively. The sequence information on the SDAV S-protein gene was applied to a differential diagnostic PCR to detect and distinguish the rat coronavirus from mouse coronaviruses. This is the first report on the comprehensive genetic information of any rat coronavirus.
Sialodacryoadenitis virus (SDAV) is distributed worldwide in laboratory rats. SDAV infects the lacrimal and salivary glands and the upper and lower respiratory tracts of rats, causing the clinical manifestations of enlarged salivary glands, sialoadenitis, dacryoadenitis, rhinitis, tracheitis, and bronchoalveolitis (3, 9, 10). SDAV can also cause reproductive disorders and behavioral changes in the infected animals. Serologic surveys indicate that coronavirus infections are common in laboratory rats housed in research facilities (11, 16), and several outbreaks of SDAV in rat colonies have been reported (2, 6, 12, 22, 32; J. Storz, personal communication). Therefore, SDAV is an important viral pathogen in comparative laboratory medicine.
SDAV is antigenically related to the mouse hepatitis virus (MHV) serogroup of the family Coronaviridae in the order of Nidovirales (20). The MHV serogroup includes Parker's rat coronavirus (PRCV), bovine coronavirus (BCV), and human coronavirus (HCV) strain OC43. Coronavirus is an enveloped virus with a single-stranded positive-sense RNA genome of approximately 31 kb. The 5′-most 22 kb of the coronavirus genome encodes the nonstructural RNA-dependent RNA polymerase, while all the structural proteins are encoded in the 3′ terminal 9 kb of the genome (15). Although a large amount of genetic information has been accumulated for MHV and other coronaviruses, such information is not available for any rat coronavirus, mainly due to the difficulty with propagation of the virus in cell cultures. Percy and coworkers (25) reported that a subclone of L2 cells produced relatively higher titers of the virus, and subsequently, Baker et al. (1) used these cells to identify at least three structural proteins associated with the virion: spike (S) protein, membrane (M) protein, and nucleocapsid (N) protein. Antibodies specific for MHV structural proteins were able to recognize both SDAV and PRCV proteins on immunoblots. However, it has not been possible to differentiate coronaviruses from each rat coronavirus in the laboratory with antibodies (26). Defined genetic information will be of help in developing a differential diagnostic method and in providing a better understanding of this group of coronaviruses. To these ends, we have performed cDNA cloning of the entire structural-protein region of the sialodacryoadenitis rat coronavirus genome, and in this communication we report the complete sequence of the 3′ terminal 9.8 kb of the SDAV genomic RNA.
MATERIALS AND METHODS
Cells and virus.
SDAV strain 681 and L2(Percy) cells were provided by D. H. Percy (Ontario Veterinary College, Guelph, Ontario, Canada) (25). SDAV strain 681 was originally obtained from P. N. Bhatt (Yale University, New Haven, Conn.) (2). L2(Percy) cells were maintained as monolayers at 37°C with 5% CO2 in a humidified incubator. The virus was propagated in L2(Percy) cells in Dulbecco's modified Eagle's medium supplemented with 10% fetal bovine serum (CanSera, Mississauga, Ontario, Canada).
Viral RNA preparation and the first-strand cDNA synthesis.
Cells were infected at a multiplicity of infection of 1 to 3 and were incubated for 3 days. The supernatant was collected and clarified with a benchtop centrifuge. Virus was pelleted through a 30% sucrose cushion with an ultracentrifuge (Beckman model XL-90) at 25,000 rpm for 2 h in an SW28 rotor. The pellets were resuspended in TE buffer (10 mM Tris-HCl, 1 mM EDTA [pH 8.0]), and viral RNA was extracted with the QIAmp viral RNA extract kit (Qiagen, Mississauga, Ontario, Canada). Specific procedures for viral RNA extraction were followed, as described in the manufacturer's instructions. For the first-strand cDNA synthesis, total cellular RNA was extracted from the virus-infected cells, and approximately 5 μg of the total cellular RNA was incubated at 70°C for 10 min with 0.5 μg of specific oligonucleotide. After chilling of the reaction mixture on ice, 200 U of SuperScript II RNase H reverse transcriptase (Gibco BRL, Mississauga, Ontario, Canada) was added. The reverse transcription (RT) reaction was carried out for 1 h at 37°C in the presence of 1 mM each dCTP, dGTP, dTTP, and dATP, 10 mM dithiothreitol, 50 mM Tris-HCl (pH 8.3), 75 mM KCl, and 3 mM MgCl2 in a reaction volume of 20 μl. The second strand was synthesized by PCR amplification.
PCR amplification.
Four microliters of the first-strand cDNA reaction was added to the PCR mixture containing final concentrations of 0.15 μg of each forward and reverse primer, 20 mM Tris-HCl (pH 8.4), 5 mM MgCl2, 50 mM KCl, 1 mM each deoxynucleoside triphosphate, and 0.5 U of Taq DNA polymerase (Gibco BRL). The PCR was performed in a Perkin-Elmer thermocycler (model PE 2400) for 30 cycles as follows: 94°C for 30 s for denaturation, 62°C for 30 s for annealing, and 72°C for 2.5 min for extension, followed by a 10-min elongation at 72°C after the final cycle. The 3′ end of the viral genome was synthesized with a 3′ Marathon rapid amplification cDNA ends kit purchased from Clontech (Palo Alto, Calif.). The PCR products were analyzed by agarose gel electrophoresis, and the residual primers were removed from the amplified products by gel filtration column chromatography (QIAquick oligonucleotide removal kit; Qiagen).
Cloning and sequencing.
The cDNA fragments were cloned into the SmaI site of the cloning vector pGEM3zf(+) (Promega, Madison, Wis.). Manipulation of DNA and general cloning procedures were followed, as described by Sambrook et al. (27). Nucleotide sequences were determined either by direct sequencing of the PCR fragments with the specific primers or by sequencing of the cloned plasmid with the universal primers. New sets of primers were designed on the basis of the sequence information obtained and were used to sequence the internal region of the clone. Nucleotide sequences were assembled and analyzed with the GeneRunner software program and the SeqWeb (version 1.1) interface connected to the GCG Wisconsin sequence analysis package provided through the Canadian Bioinformatics Resource, National Research Council (http://www.cbr.nrc.ca; Halifax, Nova Scotia, Canada).
Nucleotide sequence accession numbers.
The sequences reported in this work have been deposited in the GenBank database under accession numbers AF188191, AF188192, AF188193, AF188194, AF188195, and AF207551.
RESULTS AND DISCUSSION
Genome organization.
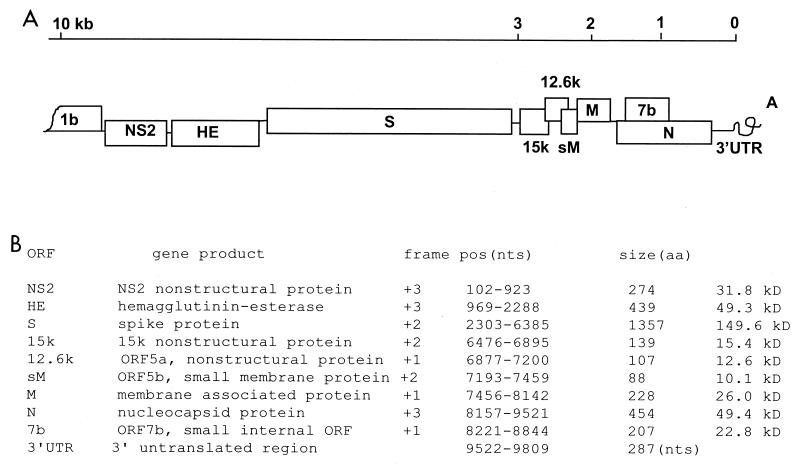
A total of seven cDNA fragments were generated by RT-PCR cDNA cloning to represent the 3′ terminal 9.8 kb of the SDAV RNA genome. The oligonucleotide primer sets used for these amplifications are listed in Table 1. Selection of the sequences used to design these primers for RT-PCR of the SDAV genome was based on the known genomic information of several strains of coronaviruses. First, the 3′ terminal 1.6-kb fragment of the SDAV genome was cloned by the 3′ rapid amplification cDNA ends method, and the nucleotide sequence of the cloned fragment was determined. On the basis of the sequence obtained, the reverse primer n1-rev-nc was designed to clone upstream fragment e. For the forward primer to be used as part of a pair with n1-rev-nc, amino acid sequences of the spike protein gene of BCV, HCV OC43, MHV A59, and MHV DVIM (GenBank accession numbers D00662, Z21849, AF029248, and AB008940, respectively) were compared, and a highly conserved region was identified. Nucleotide sequences that corresponded to the conserved region of the amino acid sequence were then examined to design the upstream primer, S2-fwd-nc. By using the primer pair of S2-fwd-nc and n1-rev-nc, the e fragment was synthesized (Table 1). Similar approaches were used for other fragment amplifications. When one or two mismatches were identified in the sequence, degenerate primers were designed, as indicated in Table 1. The contiguous overlapping sequences were assembled as a single genome. Nine major open reading frames (ORFs) were identified in the cloned 9.8-kb region of the viral genome, and these ORFs represented the coding sequences for all the structural and nonstructural proteins of the virus except the polymerase protein (Fig. 1A). The polymerase 1b coding sequence is presented in the +2 frame, and the frames for other genes are listed in Fig. 1B.
TABLE 1.
Oligonucleotides used to synthesize the cDNA fragments of SDAV genomic RNA
| cDNA fragment | Primer | Primer sequence | Nucleotide positions |
|---|---|---|---|
| f | 1b-fwd-nc | 5′-GCTGGTACGGCTGTTGTTAG-3′ | 1–10 |
| he-rev-nc | 5′-TCWGAACGACTGTCACC-3′ | 1095–1111 | |
| c | he-fwd-nc | 5′-GGTGACAGTCGTTCWGA-3′ | 1095–1111 |
| s1-rev-nc | 5′-GTAAGCAATAAYGTGG-3′ | 2484–2499 | |
| b | s1-fwd-nc | 5′-CYACRTTATTGCTTAMTGGG-3′ | 2484–2499 |
| hypvar-rev | 5′-AGAGTGACTCAGCCTGAACA-3′ | 3394–3413 | |
| a | 1stf | 5′-CCWCTMAAYTGGGAGCGTA-3′ | 3329–3347 |
| 1str | 5′-MGCWGAAACACGGCCRCTAT-3′ | 4320–4339 | |
| d | hypvar-fwd | 5′-GCAATTGTTCGGCTGATG-3′ | 3996–4013 |
| s2-rev-nc | 5′-TGTRCCAACKTCCTTGAG-3′ | 6164–6181 | |
| e | s2-fwd-nc | 5′-CTCAAGGAMGTTGGYACA-3′ | 6164–6181 |
| n1-rev-nc | 5′-TTGGGCTGAGTAGTTGCAGT-3′ | 8310–8329 | |
| g | nf1 | 5′-GCAGAAGCTCCTCTGGAAAC-3′ | 8191–8210 |
| ap1 | 5′-CCATCCTAATACGACCACTATAGGGC-3′ |
R = A + G; Y = C + T; M = A + C; K = G + T; W = A + T.
FIG. 1.
Genome structure and the coding assignments of SDAV. (A) ORFs identified in the 3′ terminal 9.8 kb of the viral genome. (B) The gene products of the coding sequences and their structural characteristics. nts, nucleotides; aa, amino acid.
Consistent with a previous report on SDAV structural proteins (1), the coding sequences for the S, M, and N proteins were identified. Downstream of the S-protein gene, two overlapping potentially nonstructural-protein genes were located, and these were predicted to encode 15- and 12.6-kDa nonstructural proteins. A small internal ORF (ORF7b) was also found in the +2 frame within the N-protein-coding sequence. Following the N-protein gene, the 3′ untranslated region (UTR) was identified to be 298 nucleotides, followed by a stretch of polyadenylation tail.
The nonstructural NS2 gene, located immediately downstream of RNA polymerase gene 1b, has been shown to be heterogeneous in coronavirus. In MHV JHM variant Wb1, a large portion of the 5′ end of the NS2 gene is deleted, and as a result, the NS2 protein is not expressed (29). However, in SDAV the NS2 nonstructural-protein gene was identified to code for a polypeptide of 274 amino acids.
Similar to the NS2 gene, the hemagglutinin-esterase (HE) gene is also known to be optional in coronaviruses. In MHV A59 and MHV 2, the HE gene lacks the 5′ end sequence, and therefore, the HE gene is not expressed (18, 30). MHV JHM variants 1, 2, and 3 also showed strain variations within the HE gene, with truncations that occurred to different extents (34). In contrast, MHV JHM, MHV 4, and MHV DIVM, as well as BCV and HCV OC43, carry the functional HE protein as a major structural component (5, 19). We found that in SDAV the HE gene downstream of the NS2 gene has an intact coding sequence capable of producing a protein of 439 amino acids. The SDAV HE protein was highly conserved compared with those of MHVs, with an amino acid identity of 91%, but the identity between the SDAV HE protein and the BCV or OC43 HE protein was lower, with only 58% homology. Nevertheless, the functional site for acetylesterase activity, known as Phe-Gly-Asp-Ser (FGDS) of the HE protein in BCV, HCV OC43, and MHVs, including the HE protein in human influenza type C virus, was well maintained in SDAV at positions 42 to 45. It has been reported that SDAV did not exhibit acetylesterase activity, and the antibody generated against the BCV HE protein failed to identify the HE protein from SDAV virions (7). However, our sequence information demonstrates the presence of a complete HE-coding sequence in the SDAV genome, and therefore, it is conceivable that the HE protein is expressed (Fig. 1). The reason for the failure to detect the HE protein from SDAV needs to be further investigated.
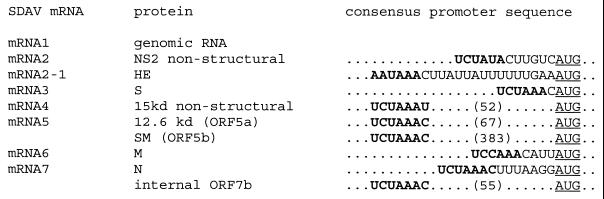
In coronaviruses, a short intergenic consensus sequence is located immediately upstream of the individual ORFs, and this sequence is a signal for discontinuous transcription. We examined the intergenic sequences of the SDAV genome, and the identified intergenic sequences for individual genes are listed in Fig. 2. A motif of UCUAAAC was identified immediately upstream of each ORF as a likely intergenic transcription signal for SDAV. The consensus intergenic promoter sequence appeared to be slightly diverse, with single or double nucleotide variations. In the NS2 nonstructural-protein gene, A at position +5 was mutated to U to result in UCUAUAC, where the first U of the consensus sequence is +1, whereas C at position +7 was changed to U to result in UCUAAAU for the 15-kDa nonstructural-protein gene. In the M-protein gene, U at position +3 was changed to C to result in UCCAAAC. Double mutations were noticed for the HE gene, in which both U at position +1 and C at position +2 were altered to A, resulting in AAUAAAC. No intergenic promoter sequence was identified upstream of the small membrane (sM)-protein-coding sequence except the one located 67 nucleotides upstream of the 12.6-kDa nonstructural-protein gene. Thus, it is likely that the sM protein is translated from mRNA5, the same mRNA which is used for translation of the 12.6-kDa nonstructural protein (Fig. 2). Therefore, mRNA5 of SDAV is likely a bicistronic mRNA, unlike that of BCV and HCV OC43 in which the mRNA5 is monocistronic (15).
FIG. 2.
Intergenic consensus transcription signals for SDAV. The putative intergenic consensus sequences are indicated as boldface characters, and the first AUG methionine codon for the ORF following the intergenic consensus promoter sequence is underlined. Numbers in parentheses indicate the number of nucleotides between the consensus sequence and the start of the gene.
Sequence comparisons with other coronaviruses.
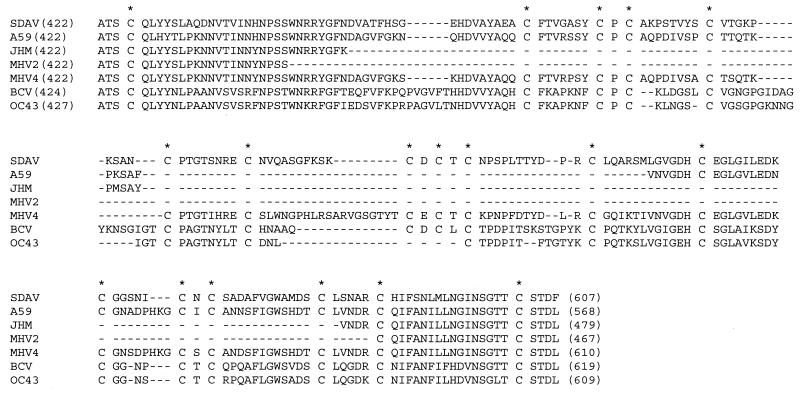
The overall sequence similarities of the SDAV genes to the sequences of the genes of other coronaviruses within the same serogroup are presented in Table 2. The SDAV genome was more similar to MHV A59 than BCV, with overall homologies of 86.1 and 66.8%, respectively. More specifically, the NS2, HE, M-protein and N-protein genes including the 3′ UTR were highly conserved between the rat coronavirus and mouse coronavirus, with similarities of 91 to 99%. The sequence homologies of the sM-protein genes were also high between SDAV and MHV A59, with an amino acid identity of 85.2%, while the similarities of the S-protein genes were 76.2%. The relatively low degrees of similarity of the S-protein genes were largely due to the addition of sequences to the N-terminal half of the S-protein gene in SDAV. In MHV, the sequence of the N-terminal half of the S-protein gene is known to be heterogeneous, and this heterogeneous region has been suggested to be associated with viral pathogenesis (8, 23, 24, 31). When the SDAV S-protein sequence was compared to the corresponding sequences of MHV 2, MHV JHM, and MHV A59, a hypervariable region was identified that included a large addition of 172, 127, and 44 amino acid residues, respectively (Fig. 3). In contrast, a relatively small but distinct insertion (amino acid positions 474 to 478) and deletions (positions 497 to 498 and 543 to 545) were identified within the S-protein gene sequence in the SDAV genome when the sequence was compared to that of the S-protein gene sequence of the MHV 4 genome.
TABLE 2.
Sequence identities between SDAV and other coronaviruses of the same serogroupa
| Protein | % Sequence identity
|
|||
|---|---|---|---|---|
| BCV | MHV JHM | MHV A59 | HCV OC43 | |
| NS2 nonstructural | 47.8 | 91.3 | 90.8 | NAb |
| HE protein | 57.9 | 91.1 | NA | 58.6 |
| S protein | 64.5 | 76.2 | 79.2 | 64.4 |
| sM protein | 64.4 | 85.2 | 85.2 | 66.7 |
| M protein | 84.5 | 93.9 | 95.6 | 82.3 |
| N protein | 70.8 | 93.6 | 93.4 | NA |
| 3′ noncoding (nucleotides) | 73.3 | 98.3 | 98.7 | NA |
| Overallc | 66.8 | NA | 86.1 | NA |
FIG. 3.
Sequence heterogeneity in the N-terminal half of the S protein of SDAV. Numbers in parentheses indicate amino acid positions of the spike protein. Asterisks indicate conservation of the cysteine residues, and hyphens indicate amino acid deletions. GenBank accession numbers are as follows: for SDAV, AF188193; for MHV A59, M18379; for MHV JHM, D00093; for MHV 2, AF107212; for MHV-4, S51114; for BCV, D00662; for HCV OC43 S62886.
Interestingly, within the stretch of 185 amino acids that represents the hypervariable region of the S protein, a total of 18 cysteine residues were identified (Fig. 3). Furthermore, all of the 18 cysteines present in this region were highly conserved among coronaviruses of the same serogroup regardless of the sequence heterogeneity. The abundance and the perfect conservation of cysteine residues within this region may reflect the native conformational structure of the spike protein in which the S1 portion forms the bulbous part of the peplomeric structure of the S protein protruding on the viral envelope, and thus, the conservation of the cysteine residues may be required to maintain the biologically active structure of the bulbous portion.
The S proteins of MHV A59, MHV JHM, and BCV are proteolytically cleaved into two subunits to yield the S1 and S2 subunit proteins, and the cleavage of S protein has been suggested to be related to the fusogenicity of coronavirus (4, 35). The cleavage sequences are RRAHR (Arg-Arg-Ala-His-Arg), RRARR, and RRSRR for MHV A59, MHV JHM, and both BCV and HCV OC43, respectively (14, 17, 23, 28). In contrast, the counterpart sequences of the S proteins of MHV DVIM, MHV Y, and MHV 2 are HRARS, HRARR, and RRARS, respectively, and none of these are cleaved (13, 19, 21, 33). In SDAV, the putative cleavage sequence of HRARR was identified at positions 752 to 756. This cleavage sequence was identical to the sequence of MHV Y; however, unlike the MHV Y S protein, the SDAV S protein was reported to undergo an efficient cleavage to yield two subunit proteins (7). Therefore, the cleavage recognition sequence does not seem to be a sole determinant for the proteolytic cleavage of the S protein, as was previously thought. The protein conformation adjacent to the cleavage site and/or the degree of glycosylation may influence the exposure of the cleavage sequence to the proteolytic enzyme, thereby affecting the extent of proteolytic cleavage.
Differential PCR for SDAV and MHV.
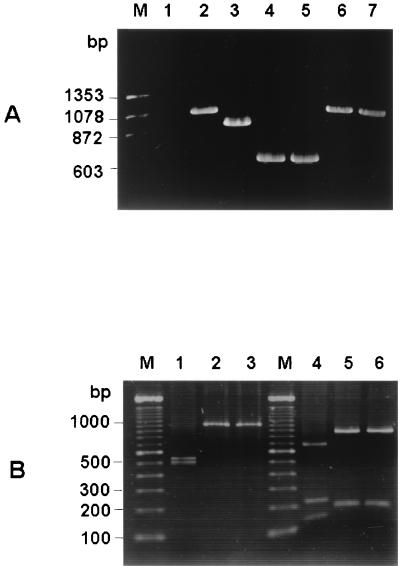
Rat coronavirus and mouse coronavirus share antigenic similarities, and the antisera raised against SDAV cross-react with MHV strains. The sensitive indirect fluorescent-antibody test and the enzyme immunoassay were not able to differentiate antibodies to MHV or SDAV. Thus, at present no diagnostic method is available for differentiation of rat coronavirus and mouse coronavirus. The hypervariable region identified in the SDAV S sequence may be used as a genetic marker to develop a reliable differential diagnostic PCR. On the basis of the sequence divergence observed in the S-protein gene of the rat coronavirus SDAV and mouse coronavirus MHV strains, we attempted to demonstrate a principle of PCR differentiation for rodent coronaviruses. Using a primer pair that encompasses the hypervariable region (1stf and 1str in Table 1), we amplified part of the S-protein genes of SDAV and MHV strains (Fig. 4A). SDAV was amplified as a 1,010-bp fragment (lane 2), while MHV A59, MHV JHM, and MHV 2 were amplified as 893-, 626-, and 590-bp fragments, respectively (lanes 3, 4, and 5, respectively). BCV and HCV OC43 were amplified as fragments of 1,046 and 1,016 bp, respectively (lanes 6 and 7, respectively). The BCV and HCV OC43 fragments were similar in size to the SDAV S-protein fragment. Since rodents are not naturally susceptible to BCV and HCV OC43, the similar sizes of the BCV and HCV OC43 fragments that were amplified may not be a practical matter of concern in the development of a differential diagnostic PCR for rodent coronaviruses. However, it may still be possible that BCV and HCV OC43 may cross the species barrier and infect rodent species. To further differentiate SDAV infection from BCV or HCV OC43 infection, we used a restriction analysis of the PCR products. By digestion with KpnI, the SDAV product generated 485- and 526-bp fragments (Fig. 4B, lane 1), whereas both the BCV and HCV OC43 products remained as uncleaved forms of 1,010 and 1,045 bp, respectively (Fig. 4B, lanes 2 and 3, respectively). Furthermore, the MfeI digestion yielded four cleaved fragments of 663, 215, 127, and 6 bp for SDAV (Fig. 4B, lane 4), while only two fragments were generated by the same enzyme for both BCV (821 and 189 bp) and HCV OC43 (856 and 189 bp) (Fig. 4B, lanes 5 and 6, respectively). The 6-bp fragment of SDAV generated by MfeI digestion is not seen on the gel in Fig. 4B. Therefore, the amplification of the hypervariable region of the S-protein gene by PCR and the subsequent digestion with KpnI or/and MfeI allows us to differentiate SDAV from the other coronaviruses.
FIG. 4.
(A) Differential amplifications of the hypervariable region of rodent coronaviruses. Total RNA was extracted from virus-infected cells 24 h postinfection. RT-PCR was performed with the primer pair which was used for amplification of fragment a (1stf and 1str in Table 1), and the PCR products were electrophoresed on a 1% agarose gel. Lanes: M, molecular weight marker; 1, uninfected cells; 2, SDAV-infected cells; 3, MHV A59-infected cells; 4, MHV JHM-infected cells; 5, MHV 2-infected cells; 6, BCV-infected cells; 7, HCV OC43 infected cells. (B) Restriction patterns of PCR fragment a from SDAV (lanes 1 and 4), BCV (lanes 2 and 5), and HCV OC43 (lanes 3 and 6). Lanes 1, 2, and 3, digestion patterns obtained with restriction endonuclease KpnI; lanes 4, 5, and 6, digestion patterns obtained with restriction endonuclease MfeI; lane M, 100-bp ladder molecular weight marker. For the sizes of the digested fragments, see the text.
Two antigenically related coronaviruses, SDAV and PRCV, have been isolated from laboratory rats. These viruses have slightly dissimilar tissue tropisms and therefore cause distinguishable diseases in the infected animals (Rat coronaviruses, technical bulletin, vol. 2, The Charles River Breeding Laboratories, Inc., Wilmington, Mass., 1983). Diagnosis of virus infection has been based on clinical signs, histopathologic lesions, and antibody detection, but it has not been possible to differentiate the two rat coronaviruses biochemically in the laboratory. Thus, the development of a differential diagnostic method is required. In the current study, we have determined the entire genomic sequence of SDAV except for that of the replicase gene of the virus. The data on the rat coronavirus genome sequence presented in this study are the first to be presented in a comprehensive report. On the basis of the sequence information, we have developed a PCR-based diagnostic test to differentiate the SDAV genome from mouse coronavirus MHV JHM, MHV A59, bovine coronavirus, and human coronavirus OC43. The genomic sequence of other rat coronavirus including the PRCV is not yet available. Thus, at present it is still not possible to distinguish the two rat coronaviruses, SDAV and PRCV. We are working to complete the sequencing of PRCV, and we may be able to devise an improved diagnostic test to further differentiate the two rat coronaviruses from one another. The genetic information reported in this communication will allow a better understanding of the molecular biology of rat coronavirus in general.
ACKNOWLEDGMENTS
This study was supported by a grant from the Medical Research Council of Canada awarded to D.Y.
We thank D. H. Percy for providing SDAV and L2(Percy) cells and for valuable advice throughout the study.
The authors acknowledge the use of the Canadian Bioinformatics Resource (http://www.cbr.nrc.ca/) in this research.
REFERENCES
- 1.Baker M G, Percy D H, Hovland D J, MacInnes J I. Preliminary characterization of the structural proteins of the coronaviruses, sialodacryoadenitis virus and Parker's rat coronavirus. Can J Vet Res. 1994;58:99–103. [PMC free article] [PubMed] [Google Scholar]
- 2.Bhatt P N, Percy D H, Jonas A M. Characterization of the virus of sialodacryoadenitis of rats: a member of the coronavirus group. J Infect Dis. 1972;126:123–130. doi: 10.1093/infdis/126.2.123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bhatt P N, Jacoby R O, Jonas A M. Respiratory infection in mice with sialodacryoadenitis virus, a coronavirus of rats. Infect Immun. 1977;18:823–827. doi: 10.1128/iai.18.3.823-827.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bos E C W, Heunen L, Luytjes W, Spaan W J M. Mutational analysis of the murine coronavirus spike protein: effect on cell-to-cell fusion. Virology. 1995;214:453–463. doi: 10.1006/viro.1995.0056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brian D, Hogue B, Kienzle T E. The coronavirus hemagglutinin-esterase glycoprotein. In: Siddell S G, editor. The Coronaviridae. New York, N.Y: Plenum Press; 1995. pp. 165–179. [Google Scholar]
- 6.Compton S R, Vivas-Genzales B E, Macy J D. Reverse transcriptase polymerase chain reaction based diagnosis and molecular characterization of a new rat coronavirus strain. Lab Anim Sci. 1999;49:506–513. [PubMed] [Google Scholar]
- 7.Gagneten S, Scanga C A, Dveksler G S, Beauchemin N, Percy D, Holmes K V. Attachment glycoproteins and receptor specificity of rat coronaviruses. Lab Anim Sci. 1996;46:159–166. [PubMed] [Google Scholar]
- 8.Gallagher T M, Parker S E, Buchmeier M J. Neutralization resistant variants of a neurotropic coronavirus are generated by deletions within the amino-terminal half of the spike glycoprotein. J Virol. 1990;64:731–741. doi: 10.1128/jvi.64.2.731-741.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jacoby R O, Bhatt P N, Jonas A M. Pathogenesis of sialodacryoadenitis virus in gnotobiotic rats. Vet Pathol. 1975;12:196–209. doi: 10.1177/030098587501200305. [DOI] [PubMed] [Google Scholar]
- 10.Jacoby R O, Bhatt P N, Jonas A M. The laboratory rat. In: Baker J H, editor. Biology and diseases. Vol. 1. New York, N.Y: Academic Press, Inc.; 1979. [Google Scholar]
- 11.Kraft V, Meyer B. Seromonitoring in small laboratory animal colonies. A five year study: 1984–1988. Z Versuchstierkd. 1990;33:29–35. [PubMed] [Google Scholar]
- 12.Kunita S, Mori M, Terada E. Sequence analysis of the nucleocapsid protein gene of rat coronavirus SDAV-681. Virology. 1993;193:520–523. doi: 10.1006/viro.1993.1158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kunita S, Zhang L, Homberger F R, Compton S R. Molecular characterization of the S proteins of two enterotropic murine coronavirus strains. Virus Res. 1995;35:277–289. doi: 10.1016/0168-1702(94)00089-U. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kunkel F, Herrler G. Structural and functional analysis of the surface protein of human coronavirus OC43. Virology. 1993;195:195–202. doi: 10.1006/viro.1993.1360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lai M M C, Cavanagh D. The molecular biology of coronaviruses. Adv Virus Res. 1997;48:1–100. doi: 10.1016/S0065-3527(08)60286-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lussier G, Descoteaux J P. Prevalence of natural virus infections in laboratory mice and rats used in Canada. Lab Anim Sci. 1986;36:145–160. [PubMed] [Google Scholar]
- 17.Luytjes W, Sturman L S, Bredenbeek P J, Charite J, van der Zeijst B A M, Horzinek M C, Spaan W J M. Primary structure of the glycoprotein E2 of coronavirus MHV-A59 and identification of the trypsin cleavage site. J Virol. 1987;161:479–487. doi: 10.1016/0042-6822(87)90142-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Luytjes W, Bredenbeek P J, Noten A F H, Horzinek M C, Spaan W J M. Sequence of mouse hepatitis virus A59 mRNA2: indications for RNA-recombination between coronaviruses and influenza C virus. Virology. 1988;166:415–422. doi: 10.1016/0042-6822(88)90512-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Morita E, Ebina H, Muto A, Himeno H, Hatakeyama K, Sugiyama K. Primary structure of hemagglutinin-esterase and spike glycoproteins of murine coronavirus DVIM (diarrhea virus in mice) Virus Genes. 1998;17:123–128. doi: 10.1023/A:1008060522426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Murphy F A, Fauquet C M, Bishop D H L, Ghabrial S L, Jarvis A W, Martelli G P, Mayo M A, Summers M D. Virus taxonomy: the classification and nomenclature of viruses. The sixth report of the International Committee on Taxonomy of Viruses. Vienna, Austria: Springer-Verlag; 1995. [Google Scholar]
- 21.Nakagawa M, Saito M, Suzuki E, Nakayama K, Matsubara J, Muto T. Ten years-long survey on pathogen status of mouse and rat breeding colonies. Exp Anim. 1984;33:115–120. doi: 10.1538/expanim1978.33.1_115. [DOI] [PubMed] [Google Scholar]
- 22.Nunoya T, Itabashi M, Kudow S, Hayashi K, Tajima M. An epizootic outbreak of sialodacryoadenitis in rats. Jpn J Vet Sci. 1977;39:445–450. doi: 10.1292/jvms1939.39.445. [DOI] [PubMed] [Google Scholar]
- 23.Parker M D, Yoo D, Cox G J, Babiuk L A. Primary structure of the S peplomer gene of bovine coronavirus and surface expression in insect cells. J Gen Virol. 1990;71:263–270. doi: 10.1099/0022-1317-71-2-263. [DOI] [PubMed] [Google Scholar]
- 24.Parker S E, Gallagher T M, Buchmeier M J. Sequence analysis reveals extensive polymorphism and evidence of deletions within the E2 glycoprotein gene of several strains of murine hepatitis virus. Virology. 1989;173:664–673. doi: 10.1016/0042-6822(89)90579-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Percy D H, Bond S, MacInnes J. Replication of sialodacryoadenitis virus in mouse L-2 cells. Arch Virol. 1989;104:323–333. doi: 10.1007/BF01315553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Percy D H, Williams K L, Paturzo F X. A comparison of the sensitivity and specificity of sialodacryoadenitis virus, Parker's rat coronavirus, and mouse hepatitis virus-infected cells as a source of antigen for the detection of antibody to rat coronaviruses. Arch Virol. 1991;119:175–180. doi: 10.1007/BF01310668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 28.Schmidt I, Skinner M, Siddell S G. Nucleotide sequence of the gene encoding the surface projection glycoprotein of coronavirus MHV-JHM. J Gen Virol. 1987;68:47–56. doi: 10.1099/0022-1317-68-1-47. [DOI] [PubMed] [Google Scholar]
- 29.Schwarz B, Routledge E, Siddell S G. Murine coronavirus nonstructural protein NS2 is not essential for virus replication in transformed cells. J Virol. 1990;64:4784–4791. doi: 10.1128/jvi.64.10.4784-4791.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Spaan W, Cavanagh D, Horzinek M C. Coronaviruses: structure and genome expression. J Gen Virol. 1988;69:2939–2952. doi: 10.1099/0022-1317-69-12-2939. [DOI] [PubMed] [Google Scholar]
- 31.Tsai C W, Chang S C, Chang M F. A 12-amino acid stretch in the hypervariable region of the spike protein S1 subunit is critical for cell fusion activity of mouse hepatitis virus. J Biol Chem. 1999;274:26085–26090. doi: 10.1074/jbc.274.37.26085. [DOI] [PubMed] [Google Scholar]
- 32.Utsumi K, Maeda T, Tatsumi H, Fujiwara K. Some clinical and epizootiological observations of infectious sialodacryoadenitis in rats. Exp Anim. 1978;27:283–287. doi: 10.1538/expanim1978.27.3_283. [DOI] [PubMed] [Google Scholar]
- 33.Yamada Y K, Takimoto K, Yabe M, Taguchi F. Acquired fusion activity of a murine coronavirus MHV-2 variant with mutations in the proteolytic cleavage site and the signal sequence of the S protein. Virology. 1997;227:215–219. doi: 10.1006/viro.1996.8313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yokomori K, Stohlman S A, Lai M M C. The detection and characterization of multiple hemagglutinin-esterase (HE)-defective viruses in the mouse brain during subacute dymyelination induced by mouse hepatitis virus. Virology. 1993;192:170–180. doi: 10.1006/viro.1993.1019. [DOI] [PubMed] [Google Scholar]
- 35.Yoo D, Parker M D, Babiuk L A. The S2 subunit of the spike glycoprotein of bovine coronavirus mediates membrane fusion in insect cells. Virology. 1991;180:395–399. doi: 10.1016/0042-6822(91)90045-D. [DOI] [PMC free article] [PubMed] [Google Scholar]