Abstract
Lipoprotein LppQ, a predominant 48-kDa antigen, and its corresponding gene, lppQ, were characterized in Mycoplasma mycoides subsp. mycoides SC, the etiological agent of contagious bovine pleuropneumonia. The lppQ gene is specific to M. mycoides subsp. mycoides SC and was found in the type strain and in field strains isolated in Europe, Africa, and Australia, as well as in vaccinal strains. LppQ is encoded as a precursor with a consensus sequence for prokaryotic signal peptidase II and a lipid attachment site. The leader sequence shows significant prominent transmembrane helix structure with a predicted outside-to-inside helix formation capacity. The N-terminal domain of the mature LppQ was shown to be surface exposed. It induced a strong, specific, early, and persistent immune response in naturally and experimentally infected animals. The C-terminal domain of LppQ possesses an integral membrane structure built up of repeated units, rich in hydrophobic and aromatic amino acids, which have a pore formation potential. A recombinant peptide representing the N-terminal domain of LppQ was obtained by site-directed mutagenesis of nine Mycoplasma-specific TGA (Trp) codons into universal TGG (Trp) codons and expression in Escherichia coli hosts. It was used for serodetection of cattle infected with M. mycoides subsp. mycoides SC, in which it was detected postinfection for significantly longer than conventional serological test reactions.
Mycoplasma mycoides subsp. mycoides small-colony type (SC) is the etiological agent of contagious bovine pleuropneumonia (CBPP), a highly contagious disease which represents a major threat to raising cattle, particularly in Africa. CBPP is also a problem in other parts of the world, including some European countries, where the disease suddenly reemerged 2 decades ago. CBPP is a disease of major economic concern in the affected countries, not only due to the morbidity and mortality but also due to restrictions on cattle trade imposed by international regulations. Hence, control of the disease is a priority for countries in which it is endemic, in order to eradicate the disease as quickly as possible after outbreaks and to avoid its spreading, as well as for countries which are free of CBPP, in order to keep that status. The main problem in eradication is the frequent occurrence of subacute or asymptomatic infections and the persistence of chronic carriers after the clinical phase. Serological analysis is the most important diagnostic tool for the control of CBPP, but it is significantly hampered by the relatively low sensitivity and specificity of the methods. The complement fixation test (CFT), which is currently the official and most widely used serodiagnostic test, has been shown to be relatively sensitive in the acute phase of the disease, but it levels off rather quickly and is insensitive 3 months after infection (1, 29). In contrast to CFT, immunoglobulin G (IgG) and IgA reactions to many antigens of M. mycoides subsp. mycoides SC are persistent for several months, as shown by immunoblot analysis of sera and bronchial-lavage samples which were sequentially collected from experimentally contact-infected cattle (1). In addition to problems with sensitivity, the specificity of current serological tests is reduced due to cross-reactions with other closely related members of the Mycoplasma mycoides cluster, which can lead to false-positive results (11, 14, 28, 33). By using a competitive enzyme-linked immunosorbent assay based on a monoclonal antibody which specifically recognized a yet-uncharacterized approximately 80-kDa antigen of M. mycoides subsp. mycoides SC, the specificity of serodiagnosis of CBPP could be significantly improved (21). It is therefore important to characterize specific antigens of M. mycoides subsp. mycoides SC. A few antigens of M. mycoides subsp. mycoides SC have been characterized, including the lipoproteins LppA (12) and LppB (35). The major lipoprotein, LppA, was shown to belong to a family of lipoproteins which is formed in all members of the M. mycoides cluster. LppA was recently found to contain only a few epitopes, which are specific to M. mycoides subsp. mycoides SC (15, 25). LppB is present only in strains belonging to the African cluster of M. mycoides subsp. mycoides SC and not in the European cluster (35). In order to develop more efficient serodiagnostic tools and to study the molecular mechanism of virulence which distinguishes the highly pathogenic M. mycoides subsp. mycoides SC from the other significantly less pathogenic or nonpathogenic members of the M. mycoides cluster, it is essential to acquire basic molecular knowledge about those factors which discriminate M. mycoides subsp. mycoides SC from other closely related mycoplasmas. In the present study, we have analyzed the genetic, antigenic, and biochemical properties of a newly identified lipoprotein, LppQ, which is specific to M. mycoides subsp. mycoides SC and which induces an early immune response in cattle with CBPP which persists long after other immune responses.
MATERIALS AND METHODS
Strains and growth conditions.
Mycoplasma strains used in this study are listed in Table 1. They were cultured in standard Mycoplasma medium at 37°C (5) until stationary growth phase. The cells were harvested by centrifugation at 13,000 × g for 20 min, washed three times in TES buffer (10 mM Tris-HCl, 1 mM EDTA, 0.8% NaCl, pH 8.0), and then resuspended in TES buffer to a concentration of approximately 109 ml−1. For gene cloning, Escherichia coli strains XL1-blue MRF′ {Δ(mcrA)183 Δ(mcrCB-hsdSMR-mrr)173 endA1 supE44 thi-1 recA1 gyrA96 relA1 lac[F′ proAB lacIqZΔM15 Tn10 (Tetr)]}; XLOLR {Δ(mcrA)183 Δ(mcrCB-hsdSMR-mrr)173 endA1 supE44 thi-1 recA1 gyrA96 relA1 lac [F′ proAB lacIqZΔM15 Tn10 (Tetr)] λr Su−} (Stratagene, La Jolla, Calif.); and BL21(DE3) (F− ompT hsdSB (rB− mB−) DE3 [λDE3 i21l lacI lacUV5 lacZ T7-RNA-pol (lysogenic) int]) (Novagen, Madison, Wis.) were used. All E. coli strains were grown in Luria-Bertani broth at 37°C in an orbital shaker-incubator (31). Antibiotics (ampicillin, 100 μg/ml; kanamycin, 50 μg/ml; or tetracycline, 50 μg/ml) were added when needed for the selection or stabilization of plasmids.
TABLE 1.
Mycoplasma strains used in this study
| Species and subspecies | Strain | Collectiona | Origin | Yr isolated | Host/details | lppQc |
|---|---|---|---|---|---|---|
| M. mycoides subsp. mycoides SC | PG1 | NCTC | Unknown | 1931 | Cattle/type strain | + |
| 2059 | LPB | Spain | 1984 | Cattle/lung | + | |
| B773/125 | LNV | Portugal | 1991 | Cattle/semen | + | |
| C305 | LNV | Portugal | 1993 | Goat/lung | + | |
| O326 | LNV | Portugal | 1993 | Sheep/milk | + | |
| PO 2 | CIRAD | France | 1980 | Cattle/lung | + | |
| 2022 | LPB | France | 1984 | Cattle/lung | + | |
| L2 | IVBBE | Italy | 1993 | Cattle/lung | + | |
| 402 | LPB | Italy | 1990 | Cattle/lung | + | |
| 6479 | LPB | Italy | 1992 | Cattle/lung | + | |
| Afadé | CIRAD | Cameroonb | 1968 | Cattle | + | |
| Fatick | CIRAD | Senegal | 1968 | Cattle | + | |
| B17 | CIRAD | Chad | 1967 | Zebu | + | |
| 9050-529/1 | CIRAD | Ivory Coast | 1990 | Cattle | + | |
| 91130 | CIRAD | Ctr. Afric. Rep. | 1991 | Cattle | + | |
| 94111 | CIRAD | Rwanda | 1994 | Cattle | + | |
| 95014 | CIRAD | Tanzania | 1995 | Cattle | + | |
| T1/44 | CIRAD | Tanzania | 1952 | Cattle/vaccine strain | + | |
| T1/Sr50 | CIRAD | Tanzania | 1952 | Cattle/vaccine strain | + | |
| KH3J | CIRAD | Sudan | 1940 | Cattle/vaccine strain | + | |
| Gladysdale | AAHL | Australia | Cattle | + | ||
| V5 | AAHL | Australia | 1965 | Cattle/vaccine strain | + | |
| M. mycoides subsp. mycoides LC | Y-goat | NCTC | Australia | Goat/type strain | − | |
| 152/93 | FVLP | Gran Canaria | 1993 | Goat | − | |
| LC8065 | CIRAD | France | Cattle | − | ||
| D2482/91 | IVBBE | Switzerland | Goat | − | ||
| 8794-Inde | CIRAD | India | Goat | − | ||
| M. mycoides subsp. capri | PG3 | NCTC | Goat/type strain | − | ||
| N108 | CIRAD | Nigeria | − | |||
| capri L | CIRAD | France | Goat | − | ||
| 9139/11-91 | CIRAD | Turkey | − | |||
| Mycoplasma sp. bovine group 7 | PG50 | NCTC | Australia | Cattle/reference strain | − | |
| B5415 | NVI | Portugal | Cattle | − | ||
| CP291 | NVI | Portugal | Goat | − | ||
| PAD3186 | BVVG | India | Goat/milk | − | ||
| FRD424 | BVVG | India | Goat/milk | − | ||
| Calf 1 | NVI | Nigeria | − | |||
| D318b | NVI | Germany | − | |||
| C2306 | NVI | Portugal | − | |||
| D424 | NVI | Germany | − | |||
| QR1/92 | NVI | Australia | − | |||
| 4055 | NVI | France | − | |||
| Mycoplasma sp. serogroup L | B144P | NVI | United States | − | ||
| M. capricolum subsp. capricolum | California kid | NCTC | California | Goat/type strain | − | |
| 173/87 | CIRAD | Greece | Sheep | − | ||
| M. capricolum subsp. capripneumoniae | F38 | NCTC | Kenya | Goat/type strain | − | |
| M. capricolum subsp. capripneumoniae | 9081-487p | CIRAD | Oman | Goat | − | |
| Gabès | CIRAD | Tunisia | Goat | − | ||
| M. putrefaciens | KS1 | NCTC | Goat/type strain | − | ||
| M. agalactiae | PG2 | NCTC | Goat/type strain | − | ||
| M. bovis | PG45 | NCTC | Cattle/type strain | − |
AAHL, Australian Animal Health Laboratory, Geelong, Victoria, Australia; BVVG, BVVG, Jena, Germany; CIRAD, CIRAD-EMVT, Montpellier, France; FVLP, Faculdad de Veterinaria, Universidad de Las Palmas, Spain; IVBBE, Institute for Veterinary Bacteriology, University of Berne, Switzerland; LNV, Laboratorio Nacional de Veterinaria, Lisbon, Portugal; LPB, Laboratoire de Pathologie Bovine, Lyon, France; NCTC, National Collection of Type Cultures; Central Public Health Laboratory, London, United Kingdom; NVI, National Veterinary Institute, Uppsala, Sweden.
Isolated at Farcha Laboratory, N'Djaména, Chad, from a bovine from Afadé, Northern Cameroon.
The presence (+) or absence (−) of lppQ was determined by PCR.
DNA extraction and DNA manipulation.
Mycoplasmal DNA for the construction of a genomic library, for PCR, and for Southern blotting was extracted by the guanidium thiocyanate method (27). Ligation, subcloning, plasmid extraction, and restriction endonuclease digestion of the DNA fragments and agarose gel electrophoresis (0.7%) and photography by UV fluorescence were performed as described previously (4). Plasmid extraction was done by the alkaline lysis method with the Miniprep kit (Qiagen AG, Basel, Switzerland).
Genomic library, cloning, and DNA sequence analysis.
Genomic DNA of M. mycoides subsp. mycoides SC strain Afadé partially digested with Sau3A1 and selected for fragment sizes of 2 to 10 kb was used to construct a genomic library, using BamHI-digested λ-ZAP-express vector arms, and was packaged with the Gigapack-11 packaging system (Stratagene). The library was plated according to standard protocols using E. coli strain XL1-blue MRF′. Immunoscreening was done with sera from CBPP-infected cattle (1) by blotting phage plaques onto nitrocellulose membranes. The selected positive clones were purified and subjected to in vivo excision with the f1 helper phage and E. coli strain XLOR. Both ends of the fragment inserted in plasmid clones were sequenced by using an ampli-Taq FS dye terminator kit (Perkin-Elmer Cetus, Norwalk, Conn.) with the universal primers T3 and T7 flanking the multiple cloning site of vector pBK-CMV, pBluescript II SK(+), or pBluescript II KS(+). In order to get the complete sequence of inserts in both directions, an Exo-Ill nested-deletion library of plasmids with cloned inserts (Pharmacia Biotech, Piscataway, N.J.) was constructed according to the manufacturer's instructions. Sequencing reactions were performed with approximately 500 ng of plasmid DNA and 5 pmol of primer per reaction mixture. Sequences were determined with an ABI Prism model 310 genetic analyzer. DNA sequences were assembled and edited with the Sequencher 3.0 program (GeneCode, Ann Arbor, Mich.) to obtain contiguous sequences.
Bioinformatic analysis.
Comparisons of nucleotide sequences and deduced amino acid sequences with the nonredundant GenBank, EMBL, DDBJ, and PDB databases in a search for related sequences were done using the NCBI-BLASTIN, BLASTX, and BLASTP programs (3). For the antigenicity-immunogenicity analysis of the deduced amino acid sequence, we used standard methods to locate the protein with the most antigenic determinants based on the hydrophilicity scores and the charged amino acid content in the peptide structure (20). Further investigations of secondary and tertiary protein structures were performed, including coiled-coil analysis (22) and a method for predicting transmembrane domains (19, 30) to reveal potential exposed domains of peptides.
Site-directed mutagenesis.
In order to replace the mycoplasma-specific TGA (Trp) codons with the universal TGG (Trp) codon in cloned genes, we used the overlap extension-PCR method (34). For this purpose, PCR amplifications were made in 50 μl of 10 mM Tris-HCl (pH 8.3)–2 mM MgCl2–50 mM KCl containing 2.5 nmol of each deoxynucleoside triphosphate, 2.5 U of Pwo DNA polymerase, 0.3 pmol of overlapping mutagenesis primers (containing the appropriate substitution [Table 2]), and 30 pmol of flanking primers (Table 2). The PCR products were purified and used as templates for the subsequent reactions in order to replace all TGA codons with TGG. Finally, the mutated genes were amplified by PCR using the flanking primers (Table 2) specially designed to clone the PCR fragments with the desired substitutions into the fusion expression vector pETHIS-1 (32) to obtain in-frame fusions with the vector-located polyhistidine codons.
TABLE 2.
Primers used in this study
| Namea | Sequenceb | Tempc (°C) | Used |
|---|---|---|---|
| MMMLP481 | cgccatatgCCATTAGTTGTTGTTTCATGTAAA | 54 | SDM, C |
| MMMLP482(r) | TCTTGTTTTTGCCACTCATTTTTTG | 54 | SDM |
| MMMLP483(f) | CAAAAAATGAGTGGCAAAAACAAGA | 54 | SDM |
| MMMLP484 | ccggatccCCAATTTGATAAAACTTGATTAAAGT | 54 | SDM, C |
| P48B1f | aacgaattcAGTTTTATCAAATTGGAATACAGAAAAT | 55 | SDM, C |
| P48B1r | CATTTGCTGTTTTCCAGCTCGATAAATC | 55 | SDM |
| P48B2f | GTGATTTATCGAGCTGGAAAACAGCAAATG | 55 | SDM |
| P48B2r | CATTACTTACATTCCAACTCGAAATATCTT | 55 | SDM |
| P48B3f | AAGATATTTCGAGTTGGAATGTAAGTAATG | 55 | SDM |
| P48B3r | ATATTTTTTAGATTCCAATCTGAAAGTGAT | 55 | SDM |
| P48B4f | ATCACTTTCAGATTGGAATCTAAAAAATAT | 55 | SDM |
| P48B4r | CTTTTGATACATCCCAATCTAAAGACTTAT | 55 | SDM |
| P48B5f | ATAAGTCTTTAGATTGGGATGTATCAAAAG | 55 | SDM |
| P48B5r | TTTGAAACATTCCAATTAGTTATATTTTG | 55 | SDM |
| P48B6f | TCAAAATATAACTAATTGGAATGTTTCAAAT | 55 | SDM |
| P48B6r | ACATTATCAACTCTCCAATTTTTTATATCT | 55 | SDM |
| P48B7f | AGATATAAAAAATTGGAGAGTTGATAATGTA | 55 | SDM |
| P48B7r | ccggatccTACATTTTTTACATTCCAACTTGAAATATC | 55 | SDM, C |
| P48FBF | GTTTTATCAAATTGGAATACAGAAAATGTA | 55 | SDM |
| P48FAL | TACATTTTCTGTATTCCAATTTGATAAAAC | 55 | SDM |
| P48 EcoRI | gcagaattcCCATTAGTTGTTGTTTCATGTAAA | 55 | SDM, C |
| MMMSCO5-6 | CAACCAAAGGCATCATATGA | 47.5 | P |
| MMMSCO5-7 | CTCTGAGAGGGAATAATG | 47.5 | P |
f and r, forward and reverse mutagenesis primers.
Lowercase letters indicate the nucleotides that were added to the flanking primers in order to create restriction enzyme recognition sites (italics) for cloning. Substituted nucleotides for mutagenesis are underlined.
Annealing temperature for PCR amplification.
SDM, site-directed mutagenesis; C, cloning; P, Identification of lppQ by PCR.
Expression and purification of recombinant lipoproteins.
For the expression of recombinant proteins, the host strain, E. coli BL21(DE3), harboring the respective plasmid clone was grown to mid-exponential growth phase in Luria-Bertani broth supplemented with 100 μg of ampicillin ml−1. Induction of T7 RNA polymerase in strain BL21(DE3) for the expression of the cloned genes was obtained by the addition of 1 mM isopropyl-β-d-thiogalactopyranoside (IPTG) and growth for a further 2 h. The cells were then harvested, washed with TES buffer, and resuspended in 0.1 volume of TES buffer. Total cell extracts were obtained by sonication of the cells for 1 min with a Sonifier 250 (Branson Ultrasonics, Danbury, Conn.) with the Microtip and output control 3 while the mixture was kept in ice water. Polyhistidine-tailed recombinant peptides were purified from cell extract dissolved in 6 M guanidine hydrochloride by Ni2+ chelate affinity chromatography (Qiagen AG) according to the supplier's protocol. Following elution, fractions containing the fusion proteins were dialyzed against 50 mM phosphate buffer–300 mM NaCl, pH 8.0, and analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) for the determination of purity. Protein concentrations were measured by the method of Bradford (8).
Sera, polyclonal antibodies, and immunoblot analysis.
Sera taken sequentially from cattle which had undergone a controlled experimental infection with M. mycoides subsp. mycoides SC, European strain L2 (animal 502) and African strain Afadé (animal 511), were described in detail (1). Field serum from a cow which was naturally infected with M. mycoides subsp. mycoides SC during an outbreak of CBPP in Italy in 1992, with a CFT titer of 1:1,280, was obtained from F. Santini, Teramo, Italy. In order to obtain polyclonal serum directed against recombinant proteins, rabbits were immunized subcutaneously with 200 μg of the appropriate purified protein in a volume of 500 μl of TES buffer emulsified with 500 μl of Freund's complete adjuvant (Difco Laboratories, Detroit, Mich.) as described previously (16). After 3 weeks, the mice were booster immunized with the same amount of protein emulsified in Freund's incomplete adjuvant. Blood was taken 2 weeks later. Animal experimentation was approved and supervised by the local ethics committee.
Immunoblotting was performed according to standard protocols (4). Rabbit hyperimmune serum was diluted 1:1,000, and bovine serum from experimentally infected animals (1) was used at a dilution of 1:100. For the detection of bound antibodies, affinity-purified goat phosphatase-labeled anti-rabbit IgG (H + L) (catalog no. 075-1506; Kirkegaard & Perry Laboratories, Gaithersburg, Md.) was diluted 1:2,000 and the monoclonal antibody anti-bovine IgG (catalog no. A7554; Sigma Aldrich Fluka Chemie, Buchs, Switzerland) was diluted 1:5,000.
Metabolic labeling with [14C]palmitic acid.
Ten microcuries (370 kBq) of [U-14C]palmitic acid (840 mCi mmol−1; Amersham) was dried, resuspended in 150 μl of ethanol, and added to a 50-ml culture of M. mycoides subsp. mycoides SC strain Afadé in the early exponential growth phase. The culture was grown for a further 16 h until late exponential phase and was then harvested by centrifugation, washed three times, and resuspended in 1 ml of TES buffer. A 20-μl sample was withdrawn for testing by SDS-PAGE for further use in membrane fractionation by Triton X-114 phase partitioning.
Triton X-114 phase partitioning.
M. mycoides subsp. mycoides SC cell components were separated into hydrophobic and hydrophilic fractions by the Triton X-114 (Fluka Chemicals, Buchs, Switzerland) partitioning method (7). Prewashed condensed Triton X-114 was added to 1 ml of [14C]palmitic acid-labeled M. mycoides subsp. mycoides SC to a final concentration of 1% (wt/vol) in a 1.5-ml conical tube, and the mixture was incubated for 30 min at 4°C with gentle rocking. The insoluble components were then removed by centrifugation at 4°C for 5 min at 13,000 × g. The Triton X-114-soluble fraction was incubated at 37°C for 15 min to allow condensation of the detergent phase, which was then separated by centrifugation at 37°C for 5 min at 13,000 × g. The upper aqueous phase was transferred to a new tube and chilled at 4°C, and Triton X-114 was added to a final concentration of 1%. The lower (detergent) phase was adjusted to its original volume with buffer. Both vials were rocked at 4°C for 15 min and then incubated for 30 min at 37°C, followed by centrifugation at 37°C for 5 min at 13,000 × g. This cycle was repeated three times to ensure complete partitioning. Both phases were adjusted to 1 ml. Unincorporated palmitic acid was extracted by chloroform. Samples from the detergent phase, the chloroform-extracted detergent phase, and the aqueous phase and whole labeled M. mycoides subsp. mycoides SC were mixed with equal volumes of SDS sample buffer, run on 5 to 15% gradient SDS-PAGE, and blotted onto a nitrocellulose membrane. The membrane was exposed to molecular screening (screen type, CS Molecular Imager GS 363 [BioRad, Hercules, Calif.]) for 2 weeks. The membranes were subsequently used for Western blotting with anti-LppQ serum.
Nucleotide sequence accession number.
The GenBank-EMBL DNA sequence accession number of the cloned fragment encoding the LppQ peptide is AF072716.
RESULTS
Cloning and sequence analysis of lppQ from M. mycoides subsp. mycoides SC.
An expression library based on bacteriophage vector λ-ZAP express was established from genomic DNA of M. mycoides subsp. mycoides SC strain Afadé (Table 1). Approximately 50,000 recombinant phage clones were screened using sera from cattle experimentally infected with the homologous strain (1), and the positive clones were converted to phagemid by in vivo excision with the f1 helper phage. DNA sequence analysis revealed the presence of a gene encoding a potential lipoprotein in one clone, plasmid pJFFmaO5. The entire 3.6-kb insert of plasmid pJFFmaO5 was sequenced in both directions, and DNA primers derived from it were used in a PCR with genomic DNA of strain Afadé to analyze the integrity of the clone. The analysis revealed that it was composed of two noncontiguous fragments, with the lipoprotein gene located on a contiguous segment of 1,764 bp. This segment contained an open reading frame (ORF) of 1,335 bp encoding a protein of 445 amino acids with a calculated molecular mass of 52.08 kDa which showed characteristics of a lipoprotein. It was named LppQ, and its corresponding gene was named lppQ. The coding sequence of lppQ starts with ATG and ends with TAG. It contains 10 mycoplasma-specific TGA (Trp) codons, which are utilized as stop codons in most other organisms. The ORF of lppQ is preceded by a consensus sequence for a ribosomal binding site located 6 nucleotides upstream of the start codon and a stem-loop structure (ΔG, −12.8 kcal) representing a potential rho-independent transcription termination signal (Fig. 1).
FIG. 1.
Structure of LppQ. (Top) Genetic structure of the 1,764-bp segment of M. mycoides subsp. mycoides SC strain Afadé, cloned in plasmid pJFFmaO5. The box represents the ORF of lppQ. Dotted segment, precursor signal sequence; hatched segment, antigenic, surface-exposed N-terminal half; open segment, integral membrane C-terminal half. The circle with a stem represents the transcriptional stop signal. (Middle) Transmembrane helix prediction diagram; aa, amino acids. (Bottom) The solid line and left-hand scale represent the hydrophilicity diagram calculated according to the method of Hopp and Woods (20); the dotted line and right-hand scale show the predicted value of the coiled-coil tertiary structure calculated using a window size of 14 aa on the Lupas scale (22).
Sequence similarity analysis of the amino acid sequence deduced from lppQ, using the mycoplasmal gene code with the Swiss-PROT and GenBank sequence databases (using the programs BLASTP and BLASTX, respectively) revealed no significant similarity to any other known proteins or ORFs of any member of the class Mollicutes. It revealed similarity only to the surface-located membrane protein Lmp1 of Borrelia burgdorferi (GenBank-EMBL accession no. AE001131), showing 26% identity and 44% similarity on the amino acid level. Analysis of the amino acid sequence of LppQ revealed a typical prokaryotic signal peptidase cleavage site in the N-terminal portion containing two Lys residues in the first 7 amino acids and ending with Val-Val-Val-Ser-Cys, with a cysteine residue at position 28 (17). The leader sequence shows a typical transmembrane helix structure with the significant outside-to-inside helix formation score of 1,041 on the TM prediction scale (Fig. 1) (2), indicating that LppQ is a surface-located lipoprotein. Analysis of the amino acid sequence of the mature LppQ protein for hydrophilicity by the method of Hopp and Woods (20) revealed the N-terminal portion represented by the first 168 amino acids to be particularly hydrophilic while the C-terminal domain, represented by the last 250 amino acids, showed more hydrophobic patterns (Fig. 1). The three most hydrophilic peaks of the N-terminal domain were associated in the same locations with significant scores for coiled-coil tertiary structure, while the C-terminal domain was devoid of such tertiary structures (Fig. 1). In addition, the N-terminal domain showed a higher score (36%) for the most-charged amino acids than the C-terminal domain (23.6%). Moreover, the C-terminal domain of LppQ was shown to be built up of nine repeated units composed of 25 amino acids, rich in hydrophobic and aromatic residues (W-X[4]-[V/I]-X[2]-[M/L]-X[2]-M-F-X[5]-F-N-X[2]-[I/L]-X[2]), which gives further support to its integral membrane localization (30).
PCR amplifications using the primers MMMSC05-6 and MMMSC05-7 flanking lppQ (Table 2) and genomic DNA from a large number of Mycoplasma strains (Table 1) as a template revealed that lppQ is present in all strains of M. mycoides subsp. mycoides SC tested, which were isolated in many different countries and continents. However, lppQ was not amplified from any other, even closely related, Mycoplasma strain (Table 1).
Expression of recombinant LppQ.
Since the lppQ gene contained several mycoplasma-specific TGA (Trp) codons, which are recognized in E. coli as stop codons, they had to be changed to TGG (Trp) by site-directed mutagenesis in order to express the gene in heterologous hosts and to produce recombinant proteins. The modified gene was designated lppQm. Sequence analysis revealed that it contained the desired mutations. The mutated gene was subsequently amplified by PCR using the oligonucleotide primers P48B7r and P48EcoRI (Table 2) and cloned into EcoRI/BamHI-digested pETHIS-1 to obtain plasmid pJFFmaLP48-MuHis1, which expressed in E. coli a polyhistidine-tailed molecule designated LppQ′. The recombinant polyhistidine-tailed C-terminal domain of LppQ (LppQ-C′) was obtained from plasmid pJFFmaLppQ-C, which was produced by PCR amplification with lppQm as a template and primers P48B1f and P48B7r (Table 2), followed by cloning the amplification product into EcoRI/BamHI-digested pETHIS-1. Finally, plasmid pJFFLP48-11 was constructed by cloning PCR amplification products obtained with primers MMMLP481 and MMMLP484 (Table 2) and lppQm as a template into NdeI/BamHI-digested pETHIS-1, in order to obtain the polyhistidine-tailed N-terminal domain of LppQ, named LppQ-N′. Inserts of all three recombinant plasmids were sequenced and confirmed the correct reading frames and fusion with the codons for polyhistidine residues. The three recombinant peptides were produced in E. coli strain BL21(DE3) harboring the respective plasmids. Purification of the peptides was performed by Ni2+ chelation chromatography (see Materials and Methods), and the purified peptides were analyzed by SDS-PAGE. Typically, 50-ml cultures yielded 2 to 4 mg of purified peptide.
Membrane location and antigenic structure of LppQ.
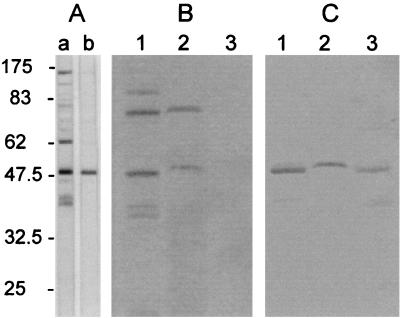
In order to identify LppQ and its two major domains, the C-terminal and N-terminal domains, monospecific polyclonal antibodies were made against the peptides LppQ′, LppQ-C′, and LppQ-N′. Total-cell antigens of M. mycoides subsp. mycoides SC were reacted on immunoblots with monospecific anti-LppQ′ antibodies and compared to the reaction with sera from experimentally infected cattle (1). This analysis identified LppQ as the predominant antigenic protein, with an apparent molecular mass of 48 kDa (Fig. 2), which was previously recognized as one of the important antigenic proteins in experimentally infected cattle (1). The predicted membrane location of LppQ was confirmed experimentally by Triton X-114 phase partitioning of M. mycoides subsp. mycoides SC cells labeled with [14C]palmitate during growth (see Materials and Methods). Autoradiography of the different fractions which were separated by SDS-PAGE and transferred onto nitrocellulose membranes revealed the 48-kDa lipoprotein band for LppQ in the Triton X-114 micelle phase containing the integral hydrophobic membrane proteins (Fig. 2). The identity of LppQ on the nitrocellulose membrane was confirmed by immunoreaction with anti-LppQ′. We interpret the weaker reaction of anti-LppQ′ with the 48-kDa protein which is seen in the aqueous phase (Fig. 2) to be due to a precursor of LppQ, which is thus not labeled with [14C]palmitate.
FIG. 2.
Identification and characterization of the membrane lipoprotein LppQ. (A) Immunoblots containing total antigens of M. mycoides subsp. mycoides SC strain Afadé were reacted with serum from a cow experimentally infected with the homologous strain (1) (lane a) or with monospecific polyclonal rabbit anti-LppQ′ antibodies (lane b). (B) Autoradiography of [14C]palmitate-labeled M. mycoides subsp. mycoides SC strain Afadé. Lane 1, total antigens; lane 2, Triton X-114 detergent phase; lane 3, aqueous phase. (C) Filter containing the same samples as in panel B reacted with anti-LppQ′ antibodies. The scale to the left of panel A is in kilodaltons.
In order to analyze the antigenic structure of LppQ, which is predicted from theoretical considerations to be localized on the N-terminal domain of the molecule, whole recombinant LppQ′, as well as recombinant C-terminal and N-terminal peptides (LppQ-C′ and LppQ-N′, respectively), have been analyzed on immunoblots using monospecific anti-LppQ antiserum and field sera from cattle naturally infected with M. mycoides subsp. mycoides SC. Polyclonal antibody raised in rabbits against purified LppQ′ reacted against purified recombinant LppQ-C′, LppQ-N′, and whole LppQ′, while serum derived from cattle naturally infected with M. mycoides subsp. mycoides SC reacted against only the LppQ-N′ and whole LppQ′ but not against the C-terminal LppQ-C′ (Fig. 3). This confirmed the structural predictions for LppQ, based on theoretical considerations, which showed that the N-terminal domain of LppQ contained in the recombinant peptide LppQ-N′ is surface exposed and shows strong antigenic characteristics, while the C-terminal domain possesses no particular immunogenicity and seems to be an integral membrane structure.
FIG. 3.
Antigenic domains of LppQ. The immunoblots contain recombinant purified LppQ-C′ (lanes 1), LppQ-N′ (lanes 2), and whole LppQ′ (lanes 3). The filter in panel A was reacted with monospecific polyclonal rabbit anti-LppQ′. The filter in panel B was reacted with a field serum of a naturally infected cow suffering from CBPP. St, prestained molecular mass standard (in kilodaltons).
Antigenic specificity of LppQ.
In order to study the antigenic specificities of the different domains of LppQ for M. mycoides subsp. mycoides SC, polyclonal monospecific antibodies directed against LppQ-C′ and LppQ-N′ were reacted on immunoblots containing total antigens of different members of the M. mycoides cluster. Anti-LppQ-N′ antibodies strongly reacted with the 48-kDa band of LppQ in M. mycoides subsp. mycoides SC strains (PG1, Afadé, and L2), but not with any of the other tested mycoplasmas of the cluster (Fig. 4), showing the high specificity of the surface-exposed domain of LppQ for M. mycoides subsp. mycoides SC. Anti-LppQ-C′ antiserum reacted not only with the 48-kDa LppQ of M. mycoides subsp. mycoides SC but also with proteins of different molecular masses from Mycoplasma sp. bovine group 7, M. mycoides subsp. capri, Mycoplasma capricolum subsp. capripneumoniae, and also to some extent Mycoplasma putrefaciens (Fig. 4). As a control, total antigens of the same strains were reacted on immunoblots with sera of cows infected with M. mycoides subsp. mycoides SC, which demonstrated the vast number of cross-reacting antigens of the different members of the M. mycoides cluster (Fig. 4). The immunogenic behavior of LppQ was studied with sera sequentially collected before and after infection of cattle with an African and a European strain of M. mycoides subsp. mycoides SC (1). Immunoblots were loaded with approximately 5 μg of LppQ-N′ and reacted with the sera as described previously (1). Sera from cattle infected with the African strain Afadé of M. mycoides subsp. mycoides SC showed a strong reaction to LppQ-N′ starting 28 days postinfection (p.i.) which continued until day 134 p.i., when the animal was slaughtered (Fig. 5). Day 28 corresponded to the appearance of the first positive CFT titers. No reaction was detected prior to day 28 p.i. In contrast to anti-LppQ-N′ reactions, which remained strong until the end of the observation period, CFT titers strongly declined at day 84 p.i. (1). Similarly, sera from cattle infected with the European strain L2 showed strong reactions with LppQ-N′ starting on day 92 p.i., when the first positive CFT was obtained. The LppQ-N′ reactions remained very strong until the end of the experimental infection 224 days p.i. (Fig. 5), when CFT had been negative for over a month (1).
FIG. 4.
Immunogenic specificity of LppQ. The immunoblots contain total-cell antigens. Lane 1, M. mycoides subsp. mycoides SC strain Afadé; lane 2, M. mycoides subsp. mycoides SC strain L2; lane 3, M. mycoides subsp. mycoides SC strain PG1; lane 4, M. mycoides subsp. mycoides LC strain Y-goat; lane 5, Mycoplasma sp. bovine group 7 strain PG50; lane 6, M. mycoides subsp. capri strain PG3; lane 7, M. capricolum subsp. capricolum strain California Kid; lane 8, M. capricolum subsp. capripneumoniae strain F38; lane 9, M. putrefaciens strain KS1. The blot in panel A was reacted with anti-LppQ-N′ antibodies. The blot in panel B was reacted with anti-LppQ-C′ antibodies. The blot in panel C (control) was reacted with a field serum from a cow which was infected experimentally with M. mycoides subsp. mycoides SC strain Afadé. St, prestained molecular mass standard (in kilodaltons).
FIG. 5.
Immunogenicity of LppQ. The immunoblots contain purified recombinant LppQ-N′ reacted with cow sera taken sequentially before and after experimental infection with M. mycoides subsp. mycoides SC. (A) African strain Afadé; (B) European strain L2. The numbers indicate days before (−) or after infection.
DISCUSSION
We have cloned and expressed the gene lppQ encoding a predominant immunogenic 48-kDa lipoprotein of M. mycoides subsp. mycoides SC. The lppQ gene is specific to M. mycoides subsp. mycoides SC and was found in all strains tested, including the type strain and European, African, and historic Australian isolates, as well as vaccine strains, but it was absent in all other closely related members of the M. mycoides cluster and in other mycoplasmas of ruminants. The oligonucleotide primer pair MMMSCO5-7 and MMMSCO5-6 can therefore be used in a confirmatory PCR method for the genetic identification of M. mycoides subsp. mycoides SC, together with previously established methods (6, 13, 24). It must be noted that no DNA sequence with similarity to that of lppQ has been found in other species of the class Mollicutes, based on currently accessible DNA sequences in databases.
Theoretical considerations based on extensive analysis of the amino acid sequence of LppQ, which were experimentally confirmed by immunological and biochemical methods, and recombinant peptides derived from LppQ showed that LppQ is a surface-exposed and membrane-associated lipoprotein. It is composed of two distinct domains, a surface-exposed, strongly immunogenic, and highly specific hydrophilic N-terminal domain and a C-terminal integral membrane domain which is composed of repeated units rich in hydrophobic and aromatic amino acids. The repeated units might be involved in pore formation, but their function is still unknown. Lipoproteins, in particular those of mycoplasmas, are expected to play a role in mechanisms of pathogenicity, since they are known to induce proinflammatory cytokines and might adopt the function of lipopolysaccharides, which are missing in mycoplasmas (9, 10, 18, 23, 26). How far LppQ is directly involved in the pathogenicity of M. mycoides subsp. mycoides SC remains to be determined. However, it is interesting to note that the only mycoplasma species that showed a relatively marked immunological cross-reaction with the C-terminal domain of LppQ, which is the part that has pore formation potential, was M. capricolum subsp. capripneumoniae, which is the only other severe pathogen of the M. mycoides cluster and which might possess a protein that is structurally, but not antigenically, similar to LppQ. The surface-exposed N-terminal domain of LppQ was shown to be antigenically specific to M. mycoides subsp. mycoides SC. It was shown to induce a strong, early, and persistent immune response in cattle infected experimentally with either an African or a European strain of M. mycoides subsp. mycoides SC. Strong serological reactions to recombinant LppQ-N′ were also detected with sera from cattle of herds in which there had been natural outbreaks of CBPP and that had either high or low CFT titers. This immunogenic characterization of the N-terminal domain of LppQ makes this molecule a most valuable candidate for development of specific and sensitive serological test methods for the control of CBPP.
ACKNOWLEDGMENTS
We are particularly grateful to Margrit Krawinkler and Yvonne Schlatter for expert help with cultivation of mycoplasmas, DNA sequencing, and PCR. We thank Fedrigo Santini, Instituto Zooprofilattico Spermintale dell'Abruzzo e del Molise, Teramo, Italy, for the gift of sera.
This study is part of the European COST Action 826 on ruminant mycoplasmoses and was supported by grant no. C96.0073 of the Swiss Ministry of Education and Science and by the Swiss Federal Veterinary Office.
REFERENCES
- 1.Abdo E-M, Nicolet J, Miserez R, Gonçalves R, Regalla J, Griot C, Bensaide A, Krampe M, Frey J. Humoral and bronchial immune responses in cattle experimentally infected with Mycoplasma mycoides subsp. mycoides small colony type. Vet Microbiol. 1998;59:109–122. doi: 10.1016/s0378-1135(97)00184-3. [DOI] [PubMed] [Google Scholar]
- 2.Adams G A, Rose J K. Structural requirements of a membrane-spanning domain for protein anchoring and cell surface transport. Cell. 1985;41:1007–1015. doi: 10.1016/s0092-8674(85)80081-7. [DOI] [PubMed] [Google Scholar]
- 3.Altschul S F, Gish W, Miller W, Myers E W, Lipman D J. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 4.Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K. Current protocols in molecular biology. New York, N.Y: John Wiley & Sons; 1999. [Google Scholar]
- 5.Bannerman E S, Nicolet J. Isolation and identification of porcine Mycoplasma in Switzerland. Schweiz Arch Tierheilkd. 1971;113:697–710. [PubMed] [Google Scholar]
- 6.Bashiruddin J B, Taylor T K, Gould A R. A PCR-based test for the specific identification of Mycoplasma mycoides subspecies mycoides SC. J Vet Diagn Investig. 1994;6:428–434. doi: 10.1177/104063879400600405. [DOI] [PubMed] [Google Scholar]
- 7.Bordier C. Phase separation of integral membrane proteins in Triton X-114 solution. J Biol Chem. 1981;256:1604–1607. [PubMed] [Google Scholar]
- 8.Bradford M M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- 9.Brenner C, Wroblewski H, Le Henaff M, Montagnier L, Blanchard A. Spiralin, a mycoplasmal membrane lipoprotein, induces T-cell-independent B-cell blastogenesis and secretion of proinflammatory cytokines. Infect Immun. 1997;65:4322–4329. doi: 10.1128/iai.65.10.4322-4329.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Calcutt M J, Kim M F, Karpas A B, Mühlradt P F, Wise K S. Differential posttranslational processing confers intraspecies variation of a major surface lipoprotein and a macrophage-activating lipopeptide of Mycoplasma fermentans. Infect Immun. 1999;67:760–771. doi: 10.1128/iai.67.2.760-771.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cheng X, Frey J, Krawinkler M, Nicolet J. Immunological cross-reactions within the Mycoplasma mycoides cluster with field sera reacting for contagious bovine pleuropneumonia (CBPP) IOM Lett. 1994;3:33. . (Abstract.) [Google Scholar]
- 12.Cheng X, Nicolet J, Miserez R, Kuhnert P, Krampe M, Pilloud T, Abdo E-M, Griot C, Frey J. Characterization of the gene for an immunodominant 72 kDa lipoprotein of Mycoplasma mycoides subsp. mycoides small colony type. Microbiology. 1996;142:3515–3524. doi: 10.1099/13500872-142-12-3515. [DOI] [PubMed] [Google Scholar]
- 13.Dedieu L, Mady V, Lefevre P C. Development of a selective polymerase chain reaction assay for the detection of Mycoplasma mycoides subsp. mycoides SC (contagious bovine pleuropneumonia agent) Vet Microbiol. 1994;42:327–339. doi: 10.1016/0378-1135(94)90064-7. [DOI] [PubMed] [Google Scholar]
- 14.Etheridge J R, Buttery S H. Improving the specificity and yield of the contagious bovine pleuropneumonia complement fixation test antigen. Res Vet Sci. 1976;20:201–206. [PubMed] [Google Scholar]
- 15.Frey J, Cheng X, Monnerat M P, Abdo E-M, Krawinkler M, Bolske G, Nicolet J. Genetic and serological analysis of the immunogenic 67-kDa lipoprotein of Mycoplasma sp. bovine group 7. Res Microbiol. 1998;149:55–64. doi: 10.1016/s0923-2508(97)83624-8. [DOI] [PubMed] [Google Scholar]
- 16.Harlow E, Lane D. Antibodies. A laboratory manual. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1988. [Google Scholar]
- 17.Hayashi S, Wu H C. Lipoproteins in bacteria. J Bioenerg Biomembr. 1990;22:451–471. doi: 10.1007/BF00763177. [DOI] [PubMed] [Google Scholar]
- 18.Herbelin A, Ruuth E, Delorme D, Michel Herbelin C, Praz F. Mycoplasma arginini TUH-14 membrane lipoproteins induce production of interleukin-1, interleukin-6, and tumor necrosis factor alpha by human monocytes. Infect Immun. 1994;62:4690–4694. doi: 10.1128/iai.62.10.4690-4694.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hofmann K, Stoffel W. TMbase—a database of membrane spanning protein segments. Biol Chem Hoppe-Seyler. 1993;347:166. [Google Scholar]
- 20.Hopp T P, Woods K R. Prediction of protein antigenic determinants from amino acid sequences. Proc Natl Acad Sci USA. 1981;78:3824–3828. doi: 10.1073/pnas.78.6.3824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.LeGoff C, Thiaucourt F. A competitive ELISA for the specific diagnosis of contagious bovine pleuropneumonia (CBPP) Vet Microbiol. 1998;60:179–191. doi: 10.1016/s0378-1135(98)00156-4. [DOI] [PubMed] [Google Scholar]
- 22.Lupas A, Van D M, Stock J. Predicting coiled coils from protein sequences. Science. 1991;252:1162–1164. doi: 10.1126/science.252.5009.1162. [DOI] [PubMed] [Google Scholar]
- 23.Marie C, Roman-Roman S, Rawadi G. Involvement of mitogen-activated protein kinase pathways in interleukin-8 production by human monocytes and polymorphonuclear cells stimulated with lipopolysaccharide or Mycoplasma fermentans membrane lipoproteins. Infect Immun. 1999;67:688–693. doi: 10.1128/iai.67.2.688-693.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Miserez R, Pilloud T, Cheng X, Nicolet J, Griot C, Frey J. Development of a sensitive nested PCR method for the specific detection of Mycoplasma mycoides subsp. mycoides SC. Mol Cell Probes. 1997;11:103–111. doi: 10.1006/mcpr.1996.0088. [DOI] [PubMed] [Google Scholar]
- 25.Monnerat M P, Thiaucourt F, Poveda J B, Nicolet J, Frey J. Genetic and serological analysis of lipoprotein LppA in Mycoplasma mycoides subsp. mycoides LC and Mycoplasma mycoides subsp. capri. Clin Diagn Lab Immunol. 1999;6:224–230. doi: 10.1128/cdli.6.2.224-230.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mühlradt P F, Frisch M. Purification and partial biochemical characterization of a Mycoplasma fermentans-derived substance that activates macrophages to release nitric oxide, tumor necrosis factor, and interleukin-6. Infect Immun. 1994;62:3801–3807. doi: 10.1128/iai.62.9.3801-3807.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pitcher D G, Saunders N A, Owen R J. Rapid extraction of bacterial genomic DNA with guanidium thiocyanate. Lett Appl Microbiol. 1989;8:151–156. [Google Scholar]
- 28.Poumarat F, Perrin M, Belli P, Longchambon D, Le Goff C, Martel J L. Studies of the origin of false positive reactions in the serodiagnosis of contagious bovine pleuropneumonia. Rev Elev Med Vet Pays Trop. 1989;42:371–378. [PubMed] [Google Scholar]
- 29.Poumarat F, Perrin M, Belli P, Martel J L. Correlation of the excretion of mycoplasma and kinetics of antibodies detected by complement fixation, passive hemagglutination and rapid seroagglutination in Mycoplasma mycoides subsp. mycoides SC experimental infection in cattle. Rev Elev Med Vet Pays Trop. 1989;42:357–364. [PubMed] [Google Scholar]
- 30.Reithmeier R A. Characterization and modeling of membrane proteins using sequence analysis. Curr Opin Struct Biol. 1995;5:491–500. doi: 10.1016/0959-440x(95)80034-4. [DOI] [PubMed] [Google Scholar]
- 31.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 32.Schaller A, Kuhn R, Kuhnert P, Nicolet J, Anderson T J, MacInnes J I, Segers R P A M, Frey J. Characterization of apxIVA, a new RTX determinant of Actinobacillus pleuropneumoniae. Microbiology. 1999;145:2105–2116. doi: 10.1099/13500872-145-8-2105. [DOI] [PubMed] [Google Scholar]
- 33.Stärk K D C, Vicari A, Kihm U, Nicolet J. Surveillance of contagious bovine pleuropneumonia in Switzerland. Rev Sci Tech. 1995;14:621–629. doi: 10.20506/rst.14.3.868. [DOI] [PubMed] [Google Scholar]
- 34.Urban A, Neukirchen S, Jaeger K E. A rapid and efficient method for site-directed mutagenesis using one-step overlap extension PCR. Nucleic Acids Res. 1997;25:2227–2228. doi: 10.1093/nar/25.11.2227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Vilei E M, Abdo E-M, Nicolet J, Botelho A, Gonçalves R, Frey J. Genomic and antigenic differences between the European and African/Australian clusters of Mycoplasma mycoides subsp. mycoides SC. Microbiology. 2000;146:477–486. doi: 10.1099/00221287-146-2-477. [DOI] [PubMed] [Google Scholar]