Abstract
Objective:
De-centralized data analysis becomes an increasingly preferred option in the healthcare domain, as it alleviates the need for sharing primary patient data across collaborating institutions. This highlights the need for consistent harmonized data curation, pre-processing, and identification of regions of interest based on uniform criteria.
Approach:
Towards this end, this manuscript describes the Federated Tumor Segmentation (FeTS) tool, in terms of software architecture and functionality.
Main Results:
The primary aim of the FeTS tool is to facilitate this harmonized processing and the generation of gold standard reference labels for tumor sub-compartments on brain magnetic resonance imaging, and further enable federated training of a tumor sub-compartment delineation model across numerous sites distributed across the globe, without the need to share patient data.
Significance:
Building upon existing open-source tools such as the Insight Toolkit (ITK) and Qt, the FeTS tool is designed to enable training deep learning models targeting tumor delineation in either centralized or federated settings. The target audience of the FeTS tool is primarily the computational researcher interested in developing federated learning models, and interested in joining a global federation towards this effort. The tool is open sourced at https://github.com/FETS-AI/Front-End.
Keywords: federated learning, FL, OpenFL, open source, brain tumor, segmentation, machine learning
1. Introduction
Accurate delineation of solid tumors is a critical first step towards furthering downstream analyses towards predicting various clinical outcomes (D. A. Gutman et al. 2013; Mazurowski, Desjardins, and Malof 2013; Jain et al. 2014; Bonekamp et al. 2015; Macyszyn et al. 2015; Bakas, Shukla, et al. 2020), monitoring progression patterns (Akbari, Macyszyn, Da, R. L. Wolf, et al. 2014; Akbari, Macyszyn, Da, Bilello, et al. 2016; Akbari, Rathore, et al. 2020), and associating imaging patterns with cancer molecular characteristics (Bakas, Akbari, J. Pisapia, et al. 2017; Bakas, Rathore, et al. 2018; Akbari, Bakas, et al. 2018; Binder et al. 2018; Fathi Kazerooni et al. 2020). Of special note is the problem of automating delineation of glioma sub-compartments, which is a particularly challenging problem, primarily due to their inherent heterogeneity in shape, extent, location, and appearance (Inda, Bonavia, Seoane, et al. 2014). Performing this process manually is tedious, prone to misinterpretation, human error, and observer bias (Deeley et al. 2011), but importantly leads to non-reproducible annotations with intra- and inter-rater variability up to 20% and 28%, respectively (Mazzara et al. 2004; Pati, Ruchika Verma, et al. 2020). The Brain Tumor Segmentation (BraTS) challenge (Menze et al. 2014; Bakas, Akbari, Sotiras, et al. 2017; Bakas, Reyes, et al. 2018; Baid, Ghodasara, et al. 2021) has been instrumental to advance automated delineation of glioma. However, the usability of the algorithms developed as part of the challenge is an open question. To promote wider application of the methods developed for BraTS and facilitate further research, the public BraTS algorithmic repository was created to house them as containers. However, since there is no graphical interface, their results and various fusion approaches (which has been shown to perform better than expert annotators (Menze et al. 2014; Bakas, Reyes, et al. 2018)) are still out of reach for clinical researchers.
The current environment of open-source software tools targeting medical imaging is highly fragmented, with many users looking at general-purpose tools such as MevisLab (Link et al. 2004), Medical Imaging Interaction Toolkit (MITK) (I. Wolf et al. 2005), MedInria (Toussaint, Souplet, and Fillard 2007), 3D-Slicer (Kikinis, Pieper, and Vosburgh 2014), the Cancer Imaging Phenomics Toolkit (Rathore et al. 2017; Davatzikos, Rathore, et al. 2018; Pati, A. Singh, et al. 2019), and ITK-SNAP (Yushkevich et al. 2006), among many others. This issue is further exacerbated in tools that can perform deep learning (DL), are locally deployable, and provide an end-to-end solution that includes harmonized data pre-processing. Example of these DL tools are: NiftyNet (Gibson et al. 2018), DeepNeuro (Beers et al. 2020), ANTsPyNet (Tustison, Cook, et al. 2020), DLTK (Pawlowski et al. 2017), and the Generally Nuanced Deep Learning Framework (GaNDLF) (Pati, Siddhesh P. Thakur, et al. 2021). Importantly, none of these DL tools have a proper graphical interface to allow for manual quality control of pre-processed data nor allow definition of harmonized pre-processing pipelines for an entire population cohort. The ability to define and manage such pipelines is a critical first step in any cohort-based computational analysis, and particularly in complex federated learning studies (M. J. Sheller, Reina, et al. 2018; M. J. Sheller, Edwards, et al. 2020).
Increased amounts and diversity of datasets available for training machine learning (ML) models are essential to produce appropriately generalizable models (Obermeyer and Emanuel 2016). This has so far been addressed by the paradigm of sharing data to a centralized location, spearheaded by use-inspired consortia (Armato III et al. 2004; Thompson et al. 2014; Consortium 2018; Bakas, Ormond, et al. 2020; Davatzikos, Barnholtz-Sloan, et al. 2020; Habes et al. 2021). However, this approach is not scalable due to various legal and technical concerns (M. J. Sheller, Edwards, et al. 2020; Rieke et al. 2020). Federated Learning (FL) offers an alternative to this paradigm, by facilitating training of ML models by sharing ML model information among participating sites (M. J. Sheller, Reina, et al. 2018; M. J. Sheller, Edwards, et al. 2020; Rieke et al. 2020; Roth et al. 2020).
Building upon our existing efforts, in this manuscript we present the Federated Tumor Segmentation (FeTS) tool, which aims at i) enabling harmonized data curation of population cohorts for computational analyses, ii) bringing various pre-trained challenge-winning algorithms and their fusion closer to clinical experts, iii) provide a graphical interactive interface to allow for refinements of the fused outputs, and iv) empower users to perform DL training in either a localized or federated manner, the latter being done without sharing patient data. In the following sections, we will describe the methods used by the FeTS tool, the use cases it has facilitated so far, and our concluding remarks.
2. Methods
Although the FeTS tool has been designed to be agnostic to the use-case, in the rest of the manuscript we have focused on the design and application of the FeTS tool in brain tumors.
2.1. Data
The starting point for FeTS users would be the identification of local multi-parametric magnetic resonance imaging (mpMRI) scans for each subject included in their study. The specific mpMRI scans considered for the case of brain gliomas, follow the convention of the BraTS challenge, and comprise i) native T1-weighted (T1), ii) Gadolinium-enhanced T1 (T1Gd), iii) native T2-weighted (T2), and iv) T2-Fluid-Attenuated-Inversion-Recovery (T2-FLAIR) sequences, characterizing the apparent anatomical tissue structure (Shukla et al. 2017). The FeTS tool deals specifically with adult-type diffuse glioma (Louis et al. 2021). In terms of radiologic appearance, glioblastomas typically comprise of 3 main sub-compartments, i) the “enhancing tumor” (ET), which represents the vascular blood-brain barrier breakdown within the tumor, ii) the “tumor core” (TC), which includes the ET and the necrotic (NCR) part, and represents the surgically relevant part of the tumor, and iii) the “whole tumor” (WT), which is defined by the union of the TC and the peritumoral edematous/infiltrated tissue (ED), and represents the complete tumor extent relevant to radiotherapy.
2.2. Modes of Operation
The functionality of the FeTS Tool can be summarized by 4 modes of operation, to facilitate an end-to-end ML-driven solid tumor study: i) data pre-processing, ii) automated segmentation using multiple pre-trained models, iii) interactive tools for manual segmentation refinements towards generating gold standard labels, and iv) ML model training.
2.2.1. Data Pre-processing for Downstream Analyses
The considered mpMRI scans are typically stored in the Digital Imaging and Communications in Medicine (DICOM) format (Pianykh 2012; Kahn et al. 2007; Mustra, Delac, and Grgic 2008). Prior to start utilizing these scans, the user needs to curate their data according to a specific folder structure, as described in the FeTS tool’s documentation ‡. Once the curated data conform to the provided definitions, the user can use this folder structure as input to the FeTS tool. The DICOM scans are then converted to the Neuroimaging Informatics Technology Initiative (NIfTI) file format (Cox et al. 2004). This process ensures that the acquired MRI scans can be easily parsed during each computational process defined in the FeTS Tool. The added benefit to this file conversion step is that all patient-identifiable metadata are removed from the DICOM scans’ header (X. Li et al. 2016; White, Blok, and Calhoun 2020). Each of these converted NIfTI scans are then registered to a common anatomical space, namely the SRI24 atlas (Rohlfing, Zahr, et al. 2010) §, to ensure a harmonized data shape ([240, 240, 155]) and voxel resolution (1mm3), thereby enabling in-tandem analysis of the mpMRI scans.
Inhomogeneity of the magnetic field is one of the most common issues observed in MRI scans (Song, Zheng, and Y. He 2017). In (Bakas, Akbari, Sotiras, et al. 2017), it has been shown that the use of non-parametric, non-uniform intensity normalization to correct for these magnetic bias fields (Sled, Zijdenbos, and Evans 1998; Tustison, Avants, et al. 2010) obliterates the MRI signal in T2-FLAIR scans, and particularly in the regions defining the abnormal T2-FLAIR signal. In this tool, we have taken advantage of this adverse effect and used the bias field-corrected scans to generate a more optimal rigid registration solution across the mpMRI scans. Specifically, the bias field-corrected scans are registered to the T1Gd scan, which itself is rigidly registered to the SRI24 atlas, resulting in 2 sets of transformation matrices per MRI scan. These matrices are then aggregated to a single matrix defining the transformation of each MRI scan from their original space to the atlas. We then apply this single aggregated matrix to the original NIfTI scans (i.e., those prior to the application of the bias field correction) to maximize the fidelity of the scans that will be used in further computational analyses. Following the co-registration of the different mpMRI scans to the template atlas, automated brain extraction (i.e., removal of all non-brain tissue from the image, including neck, fat, eyeballs, and skull) is performed. This allows further computational analyses while avoiding any potential face reconstruction/recognition (Schwarz et al. 2019). This brain extraction step is done using the Brain Mask Generator (BrainMaGe) ∥ tool (Siddhesh P Thakur et al. 2019; S. Thakur et al. 2020), which was specifically developed to address brain extraction in the presence of diffuse glioma by considering the brain shape as a prior, resulting in it being agnostic to the specific sequence/modality as input.
2.2.2. Automated Segmentation
This mode of operation provides users the ability to generate automated delineations of the tumor sub-compartments from pre-trained models. Specifically for the glioma use case, three top-performing BraTS methods are incorporated, trained on the training cohort of the BraTS challenge (Menze et al. 2014; Bakas, Akbari, Sotiras, et al. 2017; Bakas, Reyes, et al. 2018; Baid, Ghodasara, et al. 2021). These methods are i) DeepMedic (Kamnitsas et al. 2017), ii) DeepScan (McKinley, Meier, and Wiest 2018), and iii) nnU-Net (Isensee et al. 2021). Further to the generation of segmentations from these three individual pre-trained models, various label fusion strategies are also provided through the FeTS tool, to generate a single decision segmentation label from the three outputs avoiding the systematic errors of each individual pre-trained model (Pati and Bakas 2021). The exact label fusion approaches considered are: i) standard voting (Rohlfing, Russakoff, and Maurer 2004), ii) Simultaneous Truth And Performance Level Estimation (STAPLE) (Warfield, Zou, and Wells 2004; Rohlfing and Maurer Jr 2005), iii) majority voting (Huo et al. 2015), and iv) Selective and Iterative Method for Performance Level Estimation (SIMPLE) (Langerak et al. 2010). Although the fused output segmentation label is expected to provide a reasonable approximation to the actual reference standard label, manual review by expert neuroradiologists is still recommended, as further refinements might be required prior to final approval.
2.2.3. Manual Refinements Towards Reference Standard Labels
The automatically generated annotations are meant to be used as a good approximated label on which manual refinements are needed by radiology experts following a consistently communicated annotation protocol. As defined in the BraTS challenge (Menze et al. 2014; Bakas, Akbari, Sotiras, et al. 2017; Bakas, Reyes, et al. 2018; Baid, Ghodasara, et al. 2021), the reference annotations (also known as ground truth labels) comprise of the enhancing part of the tumor (ET - label ‘4’), the peritumoral edematous/infiltrated tissue (ED - label ‘2’), and the necrotic tumor core (NCR - label ‘1’). ET is described by areas with both distinctly and faintly avid enhancement on the T1Gd scan, and is considered the active portion of the tumor. NCR appears hypointense on the T1Gd scan and is the necrotic/pre-necrotic part of the tumor. ED is defined by the abnormal hyperintense signal envelope on the T2-FLAIR scans and represents the peritumoral edematous and infiltrated tissue.
2.2.4. ML Model Training
The FeTS tool intends to utilize the data generated by the previous 3 modes of operation in 2 ways: i) to train a local collaborator model, using just the local data, or ii) participate in a federation with multiple sites to contribute the knowledge of the local collaborator data in the global consensus model, enabled by OpenFL (Reina et al. 2021). If a ML model is trained as part of a federation, no information about the actual data ever gets sent, but only each model update parameters from the collaborating sites are used to aggregate into a single global consensus model.
2.2.5. Model Architecture
Considering the impact that the potential clinical translation of the global consensus model could have, we did not follow the paradigm of training an ensemble of models (as is typically reported in related literature Menze et al. 2014; Bakas, Akbari, Sotiras, et al. 2017; Bakas, Reyes, et al. 2018; Baid, Ghodasara, et al. 2021), due to the additional computational burden the execution of multiple models introduce. We instead trained a single model architecture during the federated training, and specifically an off-the-shelf implementation of the popular 3D U-Net with residual connections (3D-ResUNet) (Ronneberger, Fischer, and Brox 2015; Çiçek et al. 2016; K. He et al. 2016; Drozdzal et al. 2016; Bhalerao and S. Thakur 2019). An illustration of the model architecture can be seen in Fig. 1. The network consists of 30 base filters, with a learning rate of lr = 5 × 10−5, optimized using the Adam optimizer (Kingma and Ba 2014). The generalized DSC score (Eq. 1) (C. H. Sudre et al. 2017; Zijdenbos et al. 1994) was calculated on the absolute complement of each tumor sub-compartment independently, as the loss function to drive the model training.
| (1) |
where RL serves as the reference standard label, PM is the predicted mask, ⊙ is the Hadamard product (Horn 1990) (i.e., component wise multiplication), and ∥x∥1 is the L1-norm (Barrodale 1968), i.e., sum of the absolute values of all components).
Figure 1.
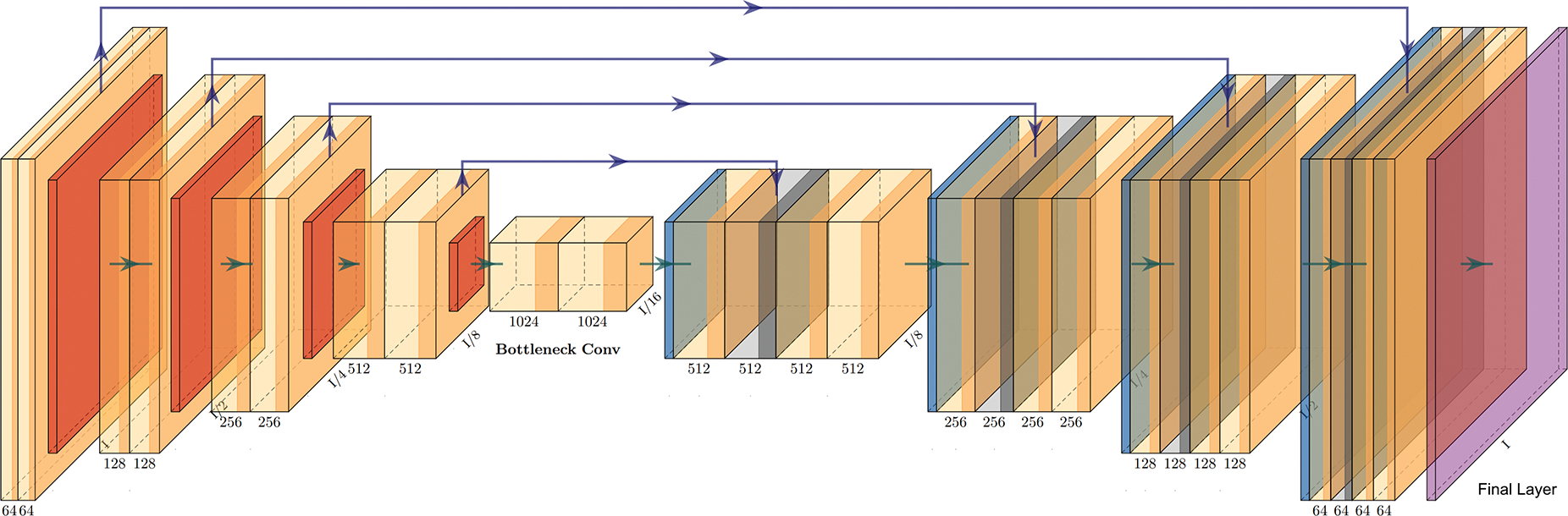
Illustration of the U-Net architecture with residual connections, plotted using PlotNeuralNet https://github.com/HarisIqbal88/PlotNeuralNet.
Previous work (Isensee et al. 2021) has shown that such mirrored DSC loss is superior in capturing variations in smaller regions. The final layer of the model is a sigmoid layer, providing three channel outputs for each voxel in the input volume, i.e., one output channel per tumor sub-compartment. While we used the floating point DSC (Shamir et al. 2019) during the training process, the generalized DSC score was calculated using a binarized version of the output (against the threshold 0.5) for the final prediction.
2.3. Open-source Software Practices
The entire source code is open-source (detailed software architecture diagram is shown in Fig. 2) and automated periodic security analysis and static code checks of the main code repository is done with public notification of alerts using Dependabot ¶ (Alfadel et al. 2021). Continuous integration is automatically performed for every pull request, with multiple automated checks and code reviews conducted prior to every code merge. All of these steps contribute in ensuring that information security teams of collaborating sites are comfortable with executing the FeTS tool on-premise.
Figure 2.
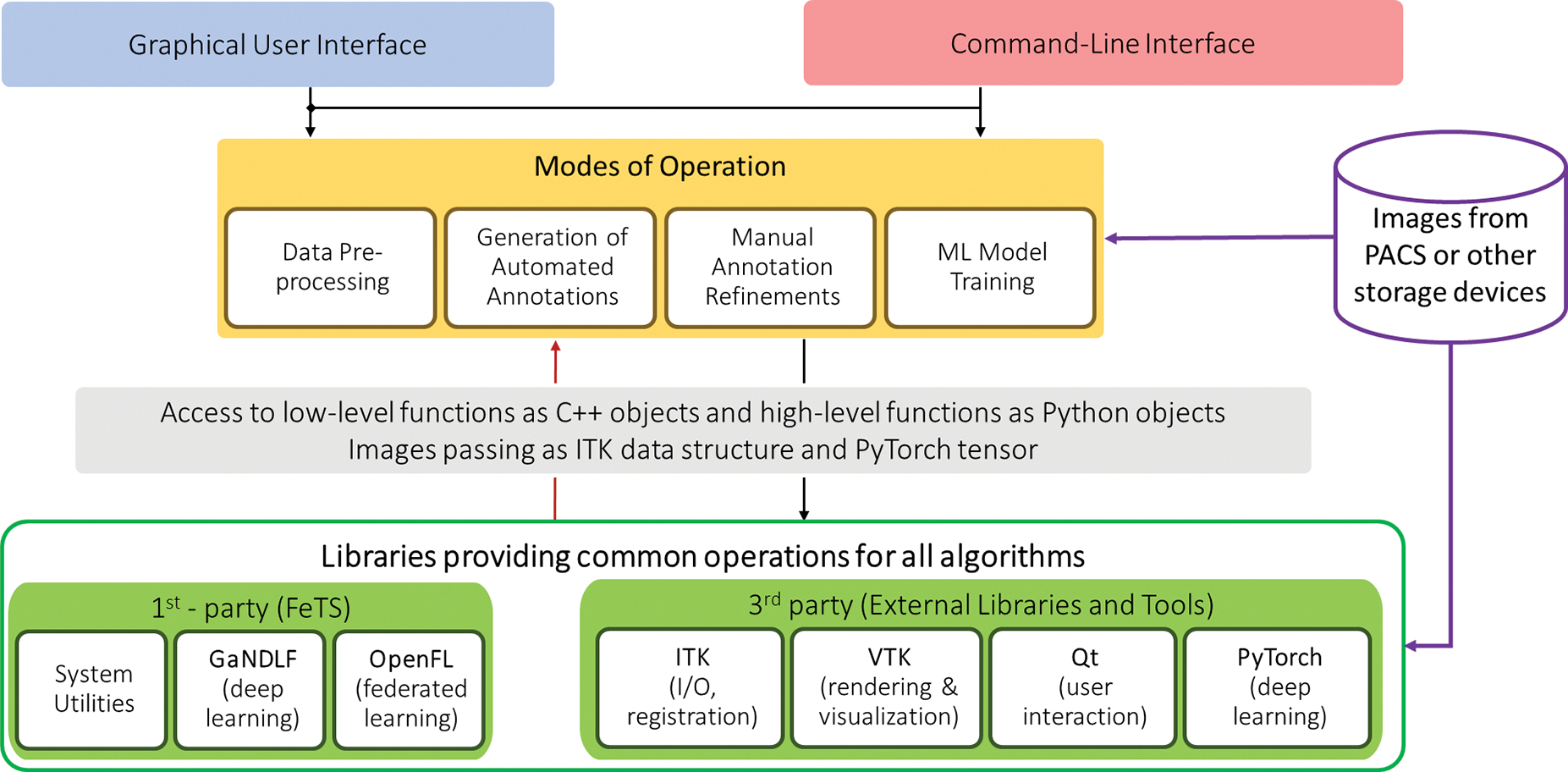
Detailed diagram of the FeTS Tool’s software architecture showing the various inter-dependencies between the components and the 4 main modes of operation.
3. Use Cases
3.1. The Largest to-date Real-World Federated Learning Initiative
The FeTS tool has enabled the largest to-date federation, utilizing data across 71 sites (Fig. 3), focusing on training a single consensus model to automatically delineate tumor sub-regions, without sharing patient data, and thereby overcoming legal, privacy, and data-ownership challenges. Specifically, the FeTS tool aimed at i) enabling clinical experts and researchers to work with the state-of-the-art segmentation algorithms (Kamnitsas et al. 2017; McKinley, Meier, and Wiest 2018; Isensee et al. 2021) and label fusion approaches (Pati and Bakas 2021), which directly resulted in easier quantification of tumor sub-compartments and the generation of reference standard labels in unseen radiologic scans.
Figure 3.
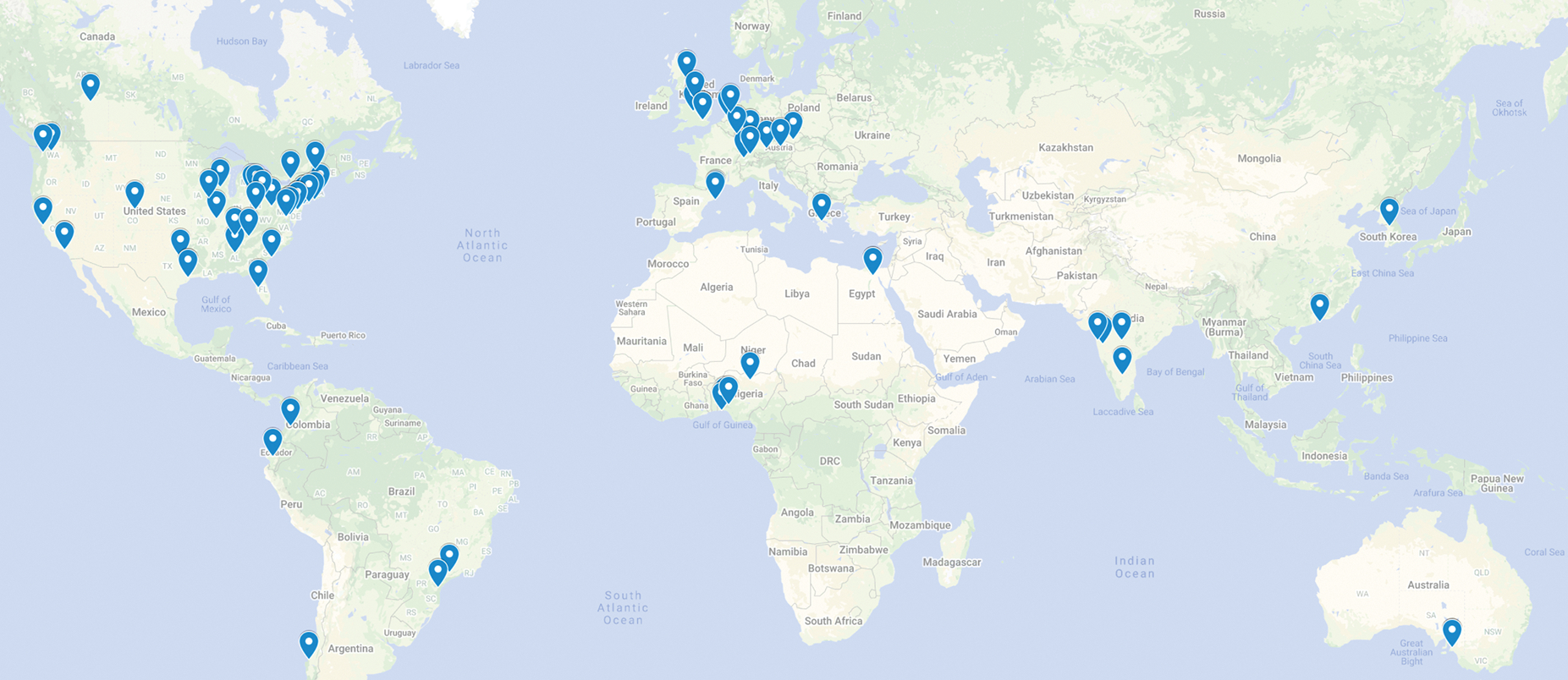
Illustration of the sites where the FeTS Tool was deployed.
This FeTS initiative followed a staged approach. The initial stage was conducted over a smaller consortium of 23 sites with data of 2, 200 patients (Baid, Pati, et al. 2021; Pati, Baid, Edwards, et al. 2022). The consensus model developed from this initial federation was evaluated on unseen validation data from each collaborator and showed an average model performance improvement of 11.1%, when compared to a model trained on the publicly available BraTS data (n = 231) (Baid, Pati, et al. 2021). Comparing against the model trained on publicly available data, the final consensus model (developed using 6, 314 patients) produced an improvement of 33% to delineate the surgically targetable tumor, and 23% over the tumor’s entire extent (Pati, Baid, Edwards, et al. 2022).
3.2. The First Challenge on Federated Learning
International computational competition (i.e., challenges) have become the accepted community standard for validating medical image analysis methods. However, the actual performance of submitted algorithms on “real-world” clinical data is often unclear, since the data included in these challenges usually follow a strict acquisition protocol from few sites and they do not represent the extent of patient populations across the globe. The obvious solution of simply collecting more data from more sites does not scale well, due to privacy and ownership hurdles.
Towards addressing these hurdles, the FeTS tool enabled the first challenge ever proposed for federated learning, i.e., the FeTS challenge 2021+ (Pati, Baid, Zenk, et al. 2021). Specifically, the FeTS challenge focused on two tasks: 1) federated training, which aimed at effective weight aggregation methods for the creation of a consensus model given a pre-defined segmentation algorithm; and 2) federated evaluation, which aimed to evaluate algorithms that accurately and robustly produce brain tumor segmentations “in-the-wild”, i.e., across different medical sites, demographic distributions, MRI scanners, and image acquisition parameters. The FeTS tool enabled both these tasks. In both the tasks, the FeTS tool enabled the processing of all clinically acquired multi-institutional MRI scans used by the remote sites used to validate the participating algorithms. Specifically for task 1, the federated learning component of the FeTS tool Reina et al. 2021 was used as the backbone to develop the weight aggregation methods.
4. Discussion
In this manuscript, we have described the Federated Tumor Segmentation (FeTS) tool, which has successfully facilitated 1) the first ever challenge on federated learning (Pati, Baid, Zenk, et al. 2021), and 2) the largest to-date real-world federation including data from 71 sites across the globe (Baid, Pati, et al. 2021). The FeTS tool has so far established an end-to-end solution for training ML models for neuroradiological mpMRI scans, while also incorporating a well-accepted robust harmonized pre-processing pipeline.
The FeTS tool was able to successfully process all the collaborators’ routine clinical data during the real-world federation, with sites spanning across 6 continents (Fig 3). This proved its out-of-the-box ability to handle processing of data “in the wild”, i.e., from a wide array of unseen scanner types and acquisition protocols, and importantly without any additional considerations. Furthermore, during its application in the first federated learning challenge (Pati, Baid, Zenk, et al. 2021), the FeTS tool has enabled i) the challenge participants to develop novel FL aggregation strategies, and ii) the “in-the-wild” evaluation of trained tumor segmentation ML models, across international collaborating healthcare sites.
One of its current limitations lie in the fact that even though its software stack has been designed and developed to be agnostic to the use-case, it has currently been tuned specifically for application in neuroradiological mpMRI scans. This is primarily due to the maturity and the widely evaluated specific pipelines and pre-trained segmentation models. Nonetheless, all incorporated algorithms can be trained and applied on data from different anatomies and modalities, notwithstanding their performance evaluation. Moreover, extension of the current pre-processing pipeline to include different imaging modalities would also be useful. Another potentially considered limitation, is the focus of the tool capabilities only defined for delineation tasks, and not offering the basis for classification problems. We are actively working towards exploring data augmentation techniques, as well as application of the tool to other anatomies and pathologies in future research projects.
In conclusion, we have developed the FeTS tool following numerous years of research and development on the BraTS challenge, and evaluated its performance and robustness during the largest to-date real-world federation. The FeTS tool can be currently used for any computational processing that would relate to brain tumor mpMRI scans. Particularly important for addressing the need to perform harmonized processing at remote healthcare sites prior to sharing data for varying consortia, as well as for the generation of centralized datasets hosted by authoritative organizations for de-centralized evaluation of computational algorithms. Towards this end, we expect the FeTS tool to pave the way for the generation of uniform datasets, which could contribute in improving our mechanistic understanding of disease.
Acknowledgments
Research reported in this publication was partly supported by the Informatics Technology for Cancer Research (ITCR) program of the National Cancer Institute (NCI) of the National Institutes of Health (NIH), under award number U01CA242871. The content of this publication is solely the responsibility of the authors and does not represent the official views of the NIH.
Footnotes
References
- Akbari Hamed, Bakas Spyridon, et al. (2018). “In vivo evaluation of EGFRvIII mutation in primary glioblastoma patients via complex multiparametric MRI signature”. In: Neuro-oncology 20.8, pp. 1068–1079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akbari Hamed, Macyszyn Luke, Da Xiao, Bilello Michel, et al. (2016). “Imaging surrogates of infiltration obtained via multiparametric imaging pattern analysis predict subsequent location of recurrence of glioblastoma”. In: Neurosurgery 78.4, pp. 572–580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akbari Hamed, Macyszyn Luke, Da Xiao, Wolf Ronald L, et al. (2014). “Pattern analysis of dynamic susceptibility contrast-enhanced MR imaging demonstrates peritumoral tissue heterogeneity”. In: Radiology 273.2, pp. 502–510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akbari Hamed, Rathore Saima, et al. (2020). “Histopathology-validated machine learning radiographic biomarker for noninvasive discrimination between true progression and pseudo-progression in glioblastoma”. In: Cancer 126.11, pp. 2625–2636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alfadel Mahmoud et al. (2021). “On the Use of Dependabot Security Pull Requests”. In: 2021 IEEE/ACM 18th International Conference on Mining Software Repositories (MSR). IEEE, pp. 254–265. [Google Scholar]
- Armato III, Samuel G et al. (2004). “Lung image database consortium: developing a resource for the medical imaging research community”. In: Radiology 232.3, pp. 739–748. [DOI] [PubMed] [Google Scholar]
- Baid Ujjwal, Ghodasara Satyam, et al. (2021). “The rsna-asnr-miccai brats 2021 benchmark on brain tumor segmentation and radiogenomic classification”. In: arXiv preprint arXiv:2107.02314. [Google Scholar]
- Baid Ujjwal, Pati Sarthak, et al. (Nov. 2021). “NIMG-32. The Federated Tumor Segmentation (Fets) Initiative: The First Real-World Large-Scale Data-Private Collaboration Focusing On Neuro-Oncology”. In: Neuro-oncology 23.Supplement 6, pp. vi135–vi136. issn: 1522–8517. doi: 10.1093/neuonc/noab196.532. [DOI] [Google Scholar]
- Bakas Spyridon, Akbari Hamed, Pisapia Jared, et al. (2017). “In vivo detection of EGFRvIII in glioblastoma via perfusion magnetic resonance imaging signature consistent with deep peritumoral infiltration: the φ-index”. In: Clinical Cancer Research 23.16, pp. 4724–4734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bakas Spyridon, Akbari Hamed, Sotiras Aristeidis, et al. (2017). “Advancing the cancer genome atlas glioma MRI collections with expert segmentation labels and radiomic features”. In: Scientific data 4.1, pp. 1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bakas Spyridon, Ormond David Ryan, et al. (2020). “iGLASS: imaging integration into the Glioma Longitudinal Analysis Consortium”. In: Neuro-oncology 22.10, pp. 1545–1546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bakas Spyridon, Rathore Saima, et al. (2018). “NIMG-40. Non-invasive in vivo signature of idh1 mutational status in high grade glioma, from clinically-acquired multi-parametric magnetic resonance imaging, using multivariate machine learning”. In: Neuro-Oncology 20.suppl 6, pp. vi184–vi185. [Google Scholar]
- Bakas Spyridon, Reyes Mauricio, et al. (2018). “Identifying the best machine learning algorithms for brain tumor segmentation, progression assessment, and overall survival prediction in the BRATS challenge”. In: arXiv preprint arXiv:1811.02629. [Google Scholar]
- Bakas Spyridon, Shukla Gaurav, et al. (2020). “Overall survival prediction in glioblastoma patients using structural magnetic resonance imaging (MRI): advanced radiomic features may compensate for lack of advanced MRI modalities”. In: Journal of Medical Imaging 7.3, p. 031505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrodale Ian (1968). “L1 approximation and the analysis of data”. In: Journal of the Royal Statistical Society: Series C (Applied Statistics) 17.1, pp. 51–57. [Google Scholar]
- Beers Andrew et al. (2020). “DeepNeuro: an open-source deep learning toolbox for neuroimaging”. In: Neuroinformatics, pp. 1–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhalerao Megh and Thakur Siddhesh (2019). “Brain tumor segmentation based on 3D residual U-Net”. In: International MICCAI Brainlesion Workshop. Springer, pp. 218–225. [Google Scholar]
- Binder Zev A et al. (2018). “Epidermal growth factor receptor extracellular domain mutations in glioblastoma present opportunities for clinical imaging and therapeutic development”. In: Cancer cell 34.1, pp. 163–177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonekamp David et al. (2015). “Association of overall survival in patients with newly diagnosed glioblastoma with contrast-enhanced perfusion MRI: Comparison of intraindividually matched T1-and T2*-based bolus techniques”. In: Journal of Magnetic Resonance Imaging 42.1, pp. 87–96. [DOI] [PubMed] [Google Scholar]
- Çiçek Özgün et al. (2016). “3D U-Net: learning dense volumetric segmentation from sparse annotation”. In: International conference on medical image computing and computer-assisted intervention. Springer, pp. 424–432. [Google Scholar]
- Consortium, The GLASS (Feb. 2018). “Glioma through the looking GLASS: molecular evolution of diffuse gliomas and the Glioma Longitudinal Analysis Consortium”. In: Neuro-Oncology 20.7, pp. 873–884. issn: 1522–8517. doi: 10.1093/neuonc/noy020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox RW et al. (Jan. 2004). “A (sort of) new image data format standard: NiFTI-1”. In: 10th Annual Meeting of the Organization for Human Brain Mapping 22. [Google Scholar]
- Davatzikos Christos, Barnholtz-Sloan Jill S, et al. (2020). “AI-based prognostic imaging biomarkers for precision neuro-oncology: the ReSPOND consortium”. In: Neuro-oncology 22.6, pp. 886–888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davatzikos Christos, Rathore Saima, et al. (2018). “Cancer imaging phenomics toolkit: quantitative imaging analytics for precision diagnostics and predictive modeling of clinical outcome”. In: Journal of medical imaging 5.1, p. 011018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deeley MA et al. (2011). “Comparison of manual and automatic segmentation methods for brain structures in the presence of space-occupying lesions: a multi-expert study”. In: Physics in Medicine & Biology 56.14, p. 4557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drozdzal Michal et al. (2016). “The importance of skip connections in biomedical image segmentation”. In: Deep Learning and Data Labeling for Medical Applications. Springer, pp. 179–187. [Google Scholar]
- Kazerooni Fathi, Anahita et al. (2020). “Imaging signatures of glioblastoma molecular characteristics: a radiogenomics review”. In: Journal of Magnetic Resonance Imaging 52.1, pp. 54–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibson Eli et al. (2018). “NiftyNet: a deep-learning platform for medical imaging”. In: Computer methods and programs in biomedicine 158, pp. 113–122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutman David A et al. (2013). “MR imaging predictors of molecular profile and survival: multi-institutional study of the TCGA glioblastoma data set”. In: Radiology 267.2, pp. 560–569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Habes Mohamad et al. (2021). “The Brain Chart of Aging: Machine-learning analytics reveals links between brain aging, white matter disease, amyloid burden, and cognition in the iSTAGING consortium of 10,216 harmonized MR scans”. In: Alzheimer’s & Dementia 17.1, pp. 89–102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He Kaiming et al. (2016). “Deep residual learning for image recognition”. In: Proceedings of the IEEE conference on computer vision and pattern recognition, pp. 770–778. [Google Scholar]
- Horn Roger A (1990). “The hadamard product”. In: Proc. Symp. Appl. Math. Vol. 40, pp. 87–169. [Google Scholar]
- Huo Jie et al. (2015). “Label fusion for multi-atlas segmentation based on majority voting”. In: International Conference Image Analysis and Recognition. Springer, pp. 100–106. [Google Scholar]
- Inda Maria-del-Mar, Bonavia Rudy, Seoane Joan, et al. (2014). “Glioblastoma multiforme: a look inside its heterogeneous nature”. In: Cancers 6.1, pp. 226–239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Isensee Fabian et al. (2021). “nnU-Net: a self-configuring method for deep learning-based biomedical image segmentation”. In: Nature Methods 18.2, pp. 203–211. [DOI] [PubMed] [Google Scholar]
- Jain Rajan et al. (2014). “Outcome prediction in patients with glioblastoma by using imaging, clinical, and genomic biomarkers: focus on the nonenhancing component of the tumor”. In: Radiology 272.2, pp. 484–493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kahn Charles E et al. (2007). “DICOM and radiology: past, present, and future”. In: Journal of the American College of Radiology 4.9, pp. 652–657. [DOI] [PubMed] [Google Scholar]
- Kamnitsas Konstantinos et al. (2017). “Efficient multi-scale 3D CNN with fully connected CRF for accurate brain lesion segmentation”. In: Medical image analysis 36, pp. 61–78. [DOI] [PubMed] [Google Scholar]
- Kikinis Ron, Pieper Steve D, and Vosburgh Kirby G (2014). “3D Slicer: a platform for subject-specific image analysis, visualization, and clinical support”. In: Intraoperative imaging and image-guided therapy. Springer, pp. 277–289. [Google Scholar]
- Kingma Diederik P and Ba Jimmy (2014). “Adam: A method for stochastic optimization”. In: arXiv preprint arXiv:1412.6980. [Google Scholar]
- Langerak Thomas Robin et al. (2010). “Label fusion in atlas-based segmentation using a selective and iterative method for performance level estimation (SIMPLE)”. In: IEEE transactions on medical imaging 29.12, pp. 2000–2008. [DOI] [PubMed] [Google Scholar]
- Li Xiangrui et al. (2016). “The first step for neuroimaging data analysis: DICOM to NIfTI conversion”. In: Journal of neuroscience methods 264, pp. 47–56. [DOI] [PubMed] [Google Scholar]
- Link F et al. (2004). “A Flexible Research and Development Platform for Medical Image Processing and Visualization”. In: Proceeding Radiology Society of North America (RSNA), Chicago. [Google Scholar]
- Louis David N et al. (2021). “The 2021 WHO classification of tumors of the central nervous system: a summary”. In: Neuro-oncology 23.8, pp. 1231–1251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macyszyn Luke et al. (2015). “Imaging patterns predict patient survival and molecular subtype in glioblastoma via machine learning techniques”. In: Neuro-oncology 18.3, pp. 417–425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mazurowski Maciej Andrzej, Desjardins Annick, and Malof Jordan Milton (2013). “Imaging descriptors improve the predictive power of survival models for glioblastoma patients”. In: Neuro-oncology 15.10, pp. 1389–1394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mazzara Gloria P et al. (2004). “Brain tumor target volume determination for radiation treatment planning through automated MRI segmentation”. In: International Journal of Radiation Oncology* Biology* Physics 59.1, pp. 300–312. [DOI] [PubMed] [Google Scholar]
- McKinley Richard, Meier Raphael, and Wiest Roland (2018). “Ensembles of densely-connected CNNs with label-uncertainty for brain tumor segmentation”. In: International MICCAI Brainlesion Workshop. Springer, pp. 456–465. [Google Scholar]
- Menze Bjoern H et al. (2014). “The multimodal brain tumor image segmentation benchmark (BRATS)”. In: IEEE transactions on medical imaging 34.10, pp. 1993–2024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mustra Mario, Delac Kresimir, and Grgic Mislav (2008). “Overview of the DICOM standard”. In: 2008 50th International Symposium ELMAR. Vol. 1. IEEE, pp. 39–44. [Google Scholar]
- Obermeyer Ziad and Emanuel Ezekiel J (2016). “Predicting the future—big data, machine learning, and clinical medicine”. In: The New England journal of medicine 375.13, p. 1216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pati Sarthak, Baid Ujjwal, Edwards Brandon, et al. (2022). Federated Learning Enables Big Data for Rare Cancer Boundary Detection. arXiv: 2204.10836 [cs.LG]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pati Sarthak, Baid Ujjwal, Zenk Maximilian, et al. (2021). “The federated tumor segmentation (fets) challenge”. In: arXiv preprint arXiv:2105.05874. [Google Scholar]
- Pati Sarthak and Bakas Spyridon (Mar. 2021). LabelFusion: Medical Image label fusion of segmentations. Version 1.0.10. [Google Scholar]
- Pati Sarthak, Singh Ashish, et al. (2019). “The cancer imaging phenomics toolkit (captk): Technical overview”. In: International MICCAI Brainlesion Workshop. Springer, pp. 380–394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pati Sarthak, Thakur Siddhesh P., et al. (2021). GaNDLF: A Generally Nuanced Deep Learning Framework for Scalable End-to-End Clinical Workflows in Medical Imaging. arXiv: 2103.01006 [cs.LG]. [Google Scholar]
- Pati Sarthak, Verma Ruchika, et al. (2020). “Reproducibility analysis of multi-institutional paired expert annotations and radiomic features of the Ivy Glioblastoma Atlas Project (Ivy GAP) dataset”. In: Medical physics 47.12, pp. 6039–6052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pawlowski Nick et al. (2017). “DLTK: State of the Art Reference Implementations for Deep Learning on Medical Images”. In: arXiv preprint arXiv:1711.06853. [Google Scholar]
- Pianykh Oleg S (2012). Digital imaging and communications in medicine (DICOM): a practical introduction and survival guide. Springer. [Google Scholar]
- Rathore Saima et al. (2017). “Brain cancer imaging phenomics toolkit (brain-CaPTk): an interactive platform for quantitative analysis of glioblastoma”. In: International MICCAI Brainlesion Workshop. Springer, pp. 133–145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reina G Anthony et al. (2021). “OpenFL: An open-source framework for Federated Learning”. In: arXiv preprint arXiv:2105.06413. [Google Scholar]
- Rieke Nicola et al. (2020). “The future of digital health with federated learning”. In: NPJ digital medicine 3.1, pp. 1–7. doi: 10.1038/s41746-020-00323-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rohlfing Torsten and Maurer Calvin R Jr (2005). “Multi-classifier framework for atlas-based image segmentation”. In: Pattern Recognition Letters 26.13, pp. 2070–2079. [Google Scholar]
- Rohlfing Torsten, Russakoff Daniel B, and Maurer Calvin R (2004). “Performance-based classifier combination in atlas-based image segmentation using expectation-maximization parameter estimation”. In: IEEE transactions on medical imaging 23.8, pp. 983–994. [DOI] [PubMed] [Google Scholar]
- Rohlfing Torsten, Zahr Natalie M, et al. (2010). “The SRI24 multichannel atlas of normal adult human brain structure”. In: Human brain mapping 31.5, pp. 798–819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ronneberger Olaf, Fischer Philipp, and Brox Thomas (2015). “U-net: Convolutional networks for biomedical image segmentation”. In: International Conference on Medical image computing and computer-assisted intervention. Springer, pp. 234–241. [Google Scholar]
- Roth Holger R et al. (2020). “Federated learning for breast density classification: A real-world implementation”. In: Domain Adaptation and Representation Transfer, and Distributed and Collaborative Learning. Springer, pp. 181–191. [Google Scholar]
- Schwarz Christopher G et al. (2019). “Identification of anonymous MRI research participants with face-recognition software”. In: New England Journal of Medicine 381.17, pp. 1684–1686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shamir Reuben R et al. (2019). “Continuous dice coefficient: a method for evaluating probabilistic segmentations”. In: arXiv preprint arXiv:1906.11031. [Google Scholar]
- Sheller Micah J, Edwards Brandon, et al. (2020). “Federated learning in medicine: facilitating multi-institutional collaborations without sharing patient data”. In: Scientific reports 10.1, pp. 1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheller Micah J, Reina G Anthony, et al. (2018). “Multi-institutional deep learning modeling without sharing patient data: A feasibility study on brain tumor segmentation”. In: International MICCAI Brainlesion Workshop. Springer, pp. 92–104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shukla Gaurav et al. (2017). “Advanced magnetic resonance imaging in glioblastoma: a review”. In: Chin Clin Oncol 6.4, p. 40. [DOI] [PubMed] [Google Scholar]
- Sled John G, Zijdenbos Alex P, and Evans Alan C (1998). “A nonparametric method for automatic correction of intensity nonuniformity in MRI data”. In: IEEE transactions on medical imaging 17.1, pp. 87–97. [DOI] [PubMed] [Google Scholar]
- Song Shuang, Zheng Yuanjie, and He Yunlong (2017). “A review of methods for bias correction in medical images”. In: Biomedical Engineering Review 1.1. [Google Scholar]
- Sudre Carole H et al. (2017). “Generalised dice overlap as a deep learning loss function for highly unbalanced segmentations”. In: Deep learning in medical image analysis and multimodal learning for clinical decision support. Springer, pp. 240–248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thakur Siddhesh et al. (2020). “Brain extraction on MRI scans in presence of diffuse glioma: Multi-institutional performance evaluation of deep learning methods and robust modality-agnostic training”. In: NeuroImage 220, p. 117081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thakur Siddhesh P et al. (2019). “Skull-stripping of glioblastoma MRI scans using 3D deep learning”. In: International MICCAI Brainlesion Workshop. Springer, pp. 57–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson Paul M et al. (2014). “The ENIGMA Consortium: large-scale collaborative analyses of neuroimaging and genetic data”. In: Brain imaging and behavior 8.2, pp. 153–182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toussaint Nicolas, Souplet Jean-Christophe, and Fillard Pierre (2007). “MedINRIA: medical image navigation and research tool by INRIA”. In: Proc. of MICCAI’07 Workshop on Interaction in medical image analysis and visualization. [Google Scholar]
- Tustison Nicholas J, Avants Brian B, et al. (2010). “N4ITK: improved N3 bias correction”. In: IEEE transactions on medical imaging 29.6, pp. 1310–1320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tustison Nicholas J, Cook Philip A, et al. (2020). “ANTsX: A dynamic ecosystem for quantitative biological and medical imaging”. In: medRxiv. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warfield Simon K, Zou Kelly H, and Wells William M (2004). “Simultaneous truth and performance level estimation (STAPLE): an algorithm for the validation of image segmentation”. In: IEEE transactions on medical imaging 23.7, pp. 903–921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White Tonya, Blok Elisabet, and Calhoun Vince D (2020). “Data sharing and privacy issues in neuroimaging research: Opportunities, obstacles, challenges, and monsters under the bed”. In: Human Brain Mapping. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolf Ivo et al. (2005). “The medical imaging interaction toolkit”. In: Medical image analysis 9.6, pp. 594–604. [DOI] [PubMed] [Google Scholar]
- Yushkevich Paul A. et al. (2006). “User-Guided 3D Active Contour Segmentation of Anatomical Structures: Significantly Improved Efficiency and Reliability”. In: Neuroimage 31.3, pp. 1116–1128. [DOI] [PubMed] [Google Scholar]
- Zijdenbos Alex P et al. (1994). “Morphometric analysis of white matter lesions in MR images: method and validation”. In: IEEE transactions on medical imaging 13.4, pp. 716–724. [DOI] [PubMed] [Google Scholar]


