Abstract
Objective
To detect viral load in human cytomegalovirus (HCMV) infection children after hematopoietic stem cell transplant (HSCT) by chip digital PCR (cdPCR).
Methods
The plasmid pUC57-UL83 containing the HCMV-UL83 gene and HCMV AD169 strain were used to evaluate the sensitivity of cdPCR. Either HSV-1, HSV-2, VZV, EBV, HHV-6, or HHV-7 was used to evaluate the specificity of HCMV cdPCR. The cdPCR was compared with quantitative PCR (qPCR) by detecting HCMV infection in 125 children's whole blood samples following HSCT.
Results
The limit of detection (LOD) of HCMV cdPCR was 103 copies/ml and the qPCR LOD was 297 copies/ml for plasmid pUC57-UL83. The result of HCMV cdPCR was 146 copies/ml for the HCMV AD169 strain, indicating that the sensitivity of cdPCR was higher than that of qPCR. There is no cross-reaction between HCMV cdPCR and other herpes viruses. The incidence of HCMV infection was 30.40% in 125 children following HSCT by cdPCR. The range of the HCMV viral load was from 107 copies/ml to 6600 copies/ml by cdPCR.
Conclusions
cdPCR is more sensitive than qPCR for detecting HCMV viral load. Furthermore, the cdPCR could be used to detect the viral load of HCMV infection before or after HSCT in children.
1. Introduction
Human cytomegalovirus (HCMV), a ubiquitous β-herpesvirus, has been infecting as high as 90% of the human population in developing countries [1]. Most people carry the virus in a latent form. Infection with HCMV could establish a long-lasting immunity to prevent the virus replication from latency to reactivation. The reactivation mostly occurs in immunosuppressed and immunocompromised patients [2–4]. As an important pathogen, HCMV infection causes significant morbidity and mortality in immunosuppressed individuals, especially in patients after hematopoietic stem cell transplant (HSCT) [5–7]. The reason is that the leading target cells of latent infection after primary infection are hematopoietic cells [8–10].
For high-risk children following HSCT, detection of HCMV infection should precede the appearance of clinical symptoms. For the reason that the higher or more rapidly changing viral load would correlate with both the development of HCMV disease and the higher risk of severe HCMV disease in children following HSCT [11–14]. Thus, the quantification of the viral load of HSCT recipients is crucial to the effective patient cure. The most common method is quantitative PCR (qPCR) [15]. However, the qPCR quantification relies on the standard curve, and the diversity of qPCR results in significant variability of the reported quantitative and qualitative data among different laboratories [16–18]. As a result, the divergences in qualitative data could lead to the misjudgment of the development and severity of the disease and the initiation or termination of antiviral therapies.
Our study established a chip digital PCR (cdPCR) method to detect the viral load of HCMV infection before or after HSCT in children and validate its sensitivity, specificity, and repeatability by plasmids and herpes viruses.
2. Materials and Methods
2.1. Plasmids and Virus
The UL83 gene of HCMV was cloned into the plasmid pUC57 to yield pUC57-UL83. The 3.2 × 106 copies/ml plasmid was a 10-fold serial dilution. The plasmid was double diluted 1 : 2, 1 : 4, 1 : 8, 1 : 16, 1 : 32, 1 : 64 by 3.2 × 102 copies/ml (The copy number of plasmid was obtained by the following formula: (6.02 × 1023) × plasmid concentration (g/ml)/(DNA length × 660)). HCMV AD169 strain (5.67 × 106 TCID50/ml) was 10-fold serially diluted from 5.67 × 106 TCID50/ml to 5.67 TCID50/ml. Herpes simplex virus 1 (HSV-1, KOS strain), herpes simplex virus 2 (HSV-2, G strain), varicella zoster virus (VZV, Ellen strain), Epstein–Barr virus (EBV, B95-8 strain), human herpesvirus 6 (HHV-6A, GS strain) (HHV-6 foundation), and human herpesvirus 7 (HHV-7, JI strain) (HHV-6 foundation) stored at −80°C. Each experiment has negative and positive controls. The ddH2O was used as the negative control and HCMV AD169 DNA was used as the positive control. The cdPCR and qPCR experiments were repeated three times and these experiments were completed in the second-level biosafety laboratory.
2.2. Patients Selection
One hundred twenty-five children following HSCT were enrolled in this study. Male/Female: 73/52, the median age is 7.5 years old. Among 125 children following HSCT, 122 children were allogeneic HSCT, and three were autologous HSCT. And in three autologous HSCTs, both the CMV status (IgG) of the donors (D) and recipient (R) patients prior to transplantation were negative. Among 122 allogeneic HSCT, there were 68 cases of both donor and recipient positive (D+ & R+), 31 cases of D+& recipient negative (R−), 18 cases of D− & R+, and 2 cases of D− & R−. There were 37 cases of acute myelocytic leukemia (AML), 17 cases of acute lymphocytic leukemia (ALL), 22 cases of aplastic anemia (AA), ten cases of myelodysplastic syndrome (MDS), three cases of lymphoma, five cases of neuroblastoma (NB), six cases of Wiskott–Aldrich syndrome (WAS), 25 cases of mucopolysaccharidosis (MPS). HSCT children were collected from May 2018 to May 2020 at the Beijing Capital Institute of Pediatrics Children's Hospital. HCMV viral load in the serum of all recipients was detected by qPCR and cdPCR before HSCT. All children were confirmed according to diagnostic criteria. All subject inclusion was approved by the Ethics Committee of the National Institute for Viral Disease Control and Prevention, China CDC.
2.3. Primers and Probe
The primers and probe of cdPCR and qPCR were designed to target the HCMV-UL83 gene.
HCMV-F 5′-GCAGCCACGGGATCGTACT-3′;
HCMV-R 5′-GGCTACCTCACACGAGCATT-3′;
HCMV-Probe 5′-CGCGAGACCGTGGAACTGCG-3′.
2.4. qPCR
140 μL diluted viral suspension or clinical whole blood samples were collected, and the DNA was extracted according to the QIAamp Viral DNA Mini Kit (Qiagen, Germany). The final elution volume was 60 μL and stored at −80°C for use. Eight μL DNA templates of each sample were added into a 25 μL qPCR reactions system [19–21], including 12.5 μL of Premix Ex Taq ™ (TaKaRa, Japan), 200 nM of each primer and probe. The cycling procedure is as follows: 95°C for 30 s; 45 cycles of 95°C for 5 s and 60°C for 30 s (CFX96, Bio-Rad, USA).
2.5. cdPCR
Eight μL DNA was added into the 25 μL cdPCR reaction system containing 5 μL ToughMix buffer (Stilla, France), 2.5 μL fluorescein (PEXBIO, China), 200 nM of each primer and probe. The cycling procedure is as follows: 95°C, 10 min; followed by 45 cycles of 94°C-5 s and 60°C-30 s. cdPCR was run in Naica™ Crystal Digital PCR system (Stilla, France). Data were analyzed using CrystalMiner.
2.6. Statistical Analyses
The data was analyzed by SPSS 20.0 software. Quantitative data were assessed by mean ± standard deviation. Count variables were assessed by the chi-square test. Relations between the expected value of diluted plasmid and those values detected by cdPCR or qPCR were assessed by Spearman's correlation. The linear relation between qPCR and cdPCR was assessed by linear regression. And each result was determined to be significantly different when P < 0.05.
3. Results
3.1. The Sensitivity, Specificity, and Repeatability of cdPCR
The linear dynamic range of the plasmid DNA containing the pUC57-UL83 gene was from 3.2 × 106 to 3.2 × 10 copies/ml. Eight μL of plasmid in each dilution was detected by qPCR and cdPCR. The plasmid with dilution from 3.2 × 106 copies/ml to 3.2 × 102 copies/ml can be detected by qPCR and cdPCR simultaneously. The 32 copies/ml diluted plasmid could not be detected by qPCR and cdPCR. To accurately understand the sensitivity of qPCR or cdPCR, plasmids 3.2 × 102 copies/ml were diluted in a 2-fold series by 1 : 2, 1 : 4, 1 : 8, 1 : 16, 1 : 32, 1 : 64. The limit of detection (LOD) of cdPCR was 103 copies/ml (2.0 copies/reaction) and the LOD of qPCR was 297 copies/ml. The results indicate that the sensitivity of cdPCR was higher than that of qPCR.
To understand the repeatability of cdPCR, a standard curve of HCMV DNA copy number was established using plasmids. Copies of serially diluted plasmids were detected by cdPCR and the coefficient of variations (CV, standard deviation/mean) of copy number detected by cdPCR was analyzed with a standard curve (Figure 1). The results showed the repeatability of cdPCR was good because the CV value was less than 15%. Good consistency was also observed between the expected value of diluted plasmid and those measured by cdPCR and qPCR (R = 0.979, P < 0.05 for cdPCR and the expected value of diluted plasmid, R = 0.939, P < 0.05, for qPCR and the expected value of diluted plasmid).
Figure 1.
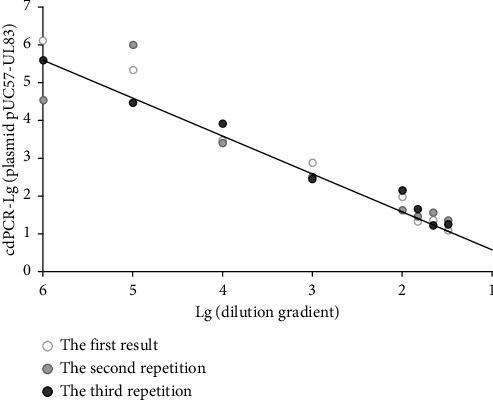
Variations of HCMV DNA copies of cdPCR compared with a standard curve. The black line shows the standard curve of the plasmid DNA. Different scatter points are the HCMV DNA copies tested by cdPCR.
To determine the specificity of cdPCR for HCMV, 7 herpes viruses, including HSV-1, HSV-2, VZV, EBV, HCMV, HHV-6A, and HHV-7, were tested by cdPCR, respectively. The results showed that cdPCR only detected the HCMV AD169 strain but not the other 6 herpes viruses, suggesting that cdPCR for HCMV has no cross-reaction with other herpes viruses.
3.2. Validation of cdPCR Using HCMV AD169 Strain
Eight μL HCMV DNA each dilution by 10-fold dilution from 5.67 × 106 TCID50/ml to 5.67 TCID50/ml was tested by qPCR and cdPCR, respectively. HCMV DNA from 5.67 × 106 TCID50/ml to 5.67 × 10 TCID50/ml virus could be detected by qPCR but not for 5.67 TCID50/ml. However, HCMV DNA of 5.67 TCID50/ml virus could be detected by cdPCR, which was 146 copies/ml (2.7 copies/reaction). The results showed that the sensitivity of cdPCR was better than that of qPCR.
3.3. HCMV Infection of Children following HSCT
To verify the sensitivity of the cdPCR method and whether it can be used for HCMV detection in the blood of HSCT patients, 125 children's whole blood samples following HSCT were tested by qPCR and cdPCR. Thirty-four samples were positive through qPCR and cdPCR, ninety-one samples were negative through qPCR. However, 4 out of 91 qPCR negative samples were positive by cdPCR. The results showed that the method of cdPCR was more sensitive than qPCR for HCMV detection (Table 1).
Table 1.
Comparisons between qPCR and cdPCR.
| qPCR | cdPCR | Total | P | |
|---|---|---|---|---|
| Positive | Negative | |||
| Positive | 34 | 0 | 34 | <0.05 |
| Negative | 4 | 87 | 91 | |
| Total | 38 | 87 | 125 | |
Subsequently, one hundred twenty-five children's whole blood samples following HSCT were tested by cdPCR to investigate the viral load of HCMV infection. The HCMV viral load was from 107 copies/ml to 6600 copies/ml by cdPCR. In 68 cases of D+ & R+, HCMV was detected in 28 cases (41.18%), and the viral load was 107–6600 copies/ml. HCMV was detected in 7 cases (22.58%) with a viral load of 437 to 5314 copies/ml in 31 D+ & R− cases. There were 18 cases of D− & R+, and three cases (16.67%) of HCMV were detected, and the viral load was 463–883 copies/ml. There were 5 cases of D− & R−, and HCMV was not detected. The HCMV infection rate was 40.54% (15/37) among AML cases, and the HCMV viral load was from 107 copies/ml to 5314 copies/ml. The HCMV infection rate was 41.18% (7/17) in ALL cases, and the HCMV viral load was from 137 copies/ml to 1779 copies/ml. The HCMV infection rate was 22.73% (5/22) among AA cases, and the HCMV viral load was from 154 copies/ml to 6600 copies/ml. The HCMV infection rate was 40% (4/10) among MDS cases, and the HCMV viral load was from 999 copies/ml to 5957 copies/ml. The HCMV infection rate was 33.33% (1/3) among lymphoma cases, and the HCMV viral load was 2494 copies/ml. There was no HCMV infection in NB cases. One case (16.67%) was infected with HCMV in WAS cases, and the HCMV viral load was 801 copies/ml. The HCMV infection rate was 20% (5/25) among MPS cases, and the HCMV viral load was from 351 copies/ml to 5100 copies/ml (Table 2). Due to the small number of cases, the HCMV infection rate of patients with different primary diseases is not statistically different, and the viral load of HCMV infection varies among different diseases group without significant variation.
Table 2.
Characteristics of 125 children following HSCT.
| Children patient characteristics | No. of children patients (no. of HCMV Positive) | |
|---|---|---|
| Age (years) | 0–6 | 70 (17) |
| 7–12 | 43 (17) | |
| ≥12 | 12 (4) | |
|
| ||
| Sex | Male | 73 (22) |
| Female | 52 (16) | |
|
| ||
| HCMV IgG | D+ & R+ | 68 (28) |
| D+ & R− | 31 (7) | |
| D− & R+ | 18 (3) | |
| D− & R− | 5 (0) | |
|
| ||
| Disease | AML | 37 (15) |
| ALL | 17 (7) | |
| AA | 22 (5) | |
| MDS | 10 (4) | |
| Lymphoma | 3 (1) | |
| NB | 5 (0) | |
| WAS | 6 (1) | |
| MPS | 25 (5) | |
|
| ||
| HSCT | Allogeneic | 122 (38) |
| Autologous | 3 (0) | |
|
| ||
| Source of HSCT | BM + PBSCT | 98 (32) |
| PBSCT | 18 (3) | |
| UCB-HSCT | 9 (3) | |
|
| ||
| GVHD (grade) | 0 | 83 (23) |
| 1-2 | 30 (12) | |
| 3-4 | 12 (3) | |
Abbreviations: HSCT, hematopoietic stem cell transplantation; HCMV, human cytomegalovirus; D, donor patients; R, recipient patients; AML, acute myelocytic leukemia; ALL, acute lymphocytic leukemia; AA, aplastic anemia; MDS, myelodysplastic syndrome; NB, neuroblastoma; WAS, Wiskott–Aldrich syndrome; MPS, mucopolysaccharidosis; BM, bone marrow; PBHSCT, peripheral blood stem cell transplantation; GVHD, graft vs. host disease.
The detection rate of HCMV was 30.40% (38/125) in 125 children following HSCT, and the range of HCMV viral load was from 107 copies/ml to 6600 copies/ml. The detection rate in the male group was 30.14% (22/73) and in the female group was 30.77% (16/52). The detection rate of HCMV was 89.47% (34/38) in the HCMV-positive children following HSCT aged 0–12. In the aged 0–6 group, the detection rate in males was 25.64% (10/39) and in females was 22.58% (7/31). In the aged 7–12 group, the detection rate in males was 39.29% (11/28) and in females was 40% (6/15). For over 12 years old children following HSCT, the HCMV detection rate was 33.33% (4/12), the detection rate in males was16.67% (1/6), and the detection rate in females was 50% (3/6) (Table 3), suggesting that HCMV infection is mainly found in patients under 12 years of age following HSCT, and the gender does not affect HCMV infection rate.
Table 3.
HCMV infection rate by cdPCR in children following HSCT.
| Age | No. | HCMV positive | Positive rate (%) | Male | Female | ||||
|---|---|---|---|---|---|---|---|---|---|
| Positive | Negative | Positive rate (%) | Positive | Negative | Positive rate (%) | ||||
| 0–6 | 70 | 17 | 24.28 | 10 | 29 | 25.64 | 7 | 24 | 22.58 |
| 7–12 | 43 | 17 | 39.53 | 11 | 17 | 39.29 | 6 | 9 | 40 |
| ≥12 | 12 | 4 | 33.33 | 1 | 5 | 16.67 | 3 | 3 | 50 |
| Total | 125 | 38 | 30.40 | 22 | 51 | 30.14 | 16 | 36 | 30.77 |
The prognosis of 125 children following HSCT was analyzed retrospectively. GVHD was found in 13 children (the range of the HCMV viral load was from 437 copies/ml to 4457 copies/ml, and the median of viral load was 693 copies/ml) (34.21%) of 38 HCMV-positive children and 25 children (28.74%) of the 87 HCMV-negative children (P > 0.05). HCMV-positive children were slightly more likely to develop GVHD than HCMV-negative children. Among the 38 positive children, five children died following HSCT. The coinfection with EBV occurred in 2 of the 5 deaths.
4. Discussion
The increase of HCMV viral load in clinical samples can predict disease progression and outcome in patients [11]. However, the lack of well-established viral load thresholds has limited HCMV qPCR in clinical applications. The results of qPCR cannot be directly compared between different hospitals without consensus standardization. Thus, it is hard to use the viral loads' value to initiate preemptive therapy for patients infected with HCMV [14, 16, 22]. A variety of factors can lead to large changes in viral load, such as detection methods, disease severity (course), and sample quality [23–25].
Digital PCR solves the shortcomings of qPCR. Droplet digital PCR (ddPCR) and cdPCR are two types of commercial digital PCR technic. As our results, many studies show that the sensitivity of digital PCR is significantly higher than that of qPCR [20, 26, 27]. Furthermore, the sensitivity of ddPCR for HCMV is 100 copies/ml [28]. Our results also showed that the lowest viral load of cdPCR in detecting whole blood samples was 107 copies/ml, lower than qPCR. In the clinical context, qPCR is a common method for detecting HCMV viral load. But the sensitivity of qPCR was less than that of cdPCR. The cdPCR can detect a lower viral load of HCMV infection than qPCR under its threshold. The cdPCR is conducted through an advanced cutting-edge microfluidic chip (Sapphire chip) 2D array of microchamber to complete PCR reaction, and cdPCR can conduct three-color multiplexing amplification [29, 30]. Due to simplified steps, cdPCR effectively reduces the risk of contamination. Thus, in this study, an HCMV cdPCR method was established to detect the viral load of HCMV infection.
It is well known that HCMV is the most common transmissible virus in children following HSCT and is considered to be the major risk factor for transplantation. Studies have shown that almost all HCMV viremia after bone marrow transplantation occurs in HCMV-positive recipients, and only a few patients can be transmitted from the donor [31]. For patients following HSCT, myeloablation may reduce immunity and lead to HCMV reinfection [32]. Our results also showed that the HCMV detection rate was 57.85% in HCMV-positive recipients prior to transplantation, which was higher than that in HCMV-negative recipients prior to transplantation (22.58%). Unlike those who have undergone solid organ transplantation, the HCMV-positive donor is the high-risk group [32].
In this study, up to 30.40% of samples were positive for HCMV by cdPCR. About 90% of HCMV infections occurred before the age of 12 in these children following HSCT. Among 125 children following HSCT, neoplastic diseases (AML, ALL, and lymphoma) have a higher HCMV infection rate than non-neoplastic diseases (AA, WAS, and MPS). It is reasonable that the immune status of neoplastic children is weaker than that of non-neoplastic children, which makes HCMV infection more likely to occur. Our results suggest that HCMV-positive children were more likely to develop GVHD than HCMV-negative children, consistent with previous studies. In other words, it is necessary to pay attention to children following HSCT with HCMV and HCMV-infected children after bone marrow transplant. Therefore, timely monitoring and accurate quantification of HCMV viral loads in children following HSCT can provide an important basis for clinical antiviral treatment.
5. Conclusion
In conclusion, the cdPCR method of HCMV DNA was established and confirmed for detecting HCMV viral load in this study.
Acknowledgments
The authors would like to thank HHV-6 Foundation for providing human herpesvirus 6A (GS strain) and human herpesvirus 7 (JI strain). This work was supported by the Research Project Supported by the China Mega-Project for Infectious Disease (2018ZX10102001, 2018ZX10711001, and 2018ZX10734404), National Pathogen Resource Collection Center (NPRC-32), and the SKLID Development Grant (2011SKLID104).
Data Availability
The data used to support the findings of this study are available from the corresponding author upon request.
Ethical Approval
All children were confirmed according to diagnostic criteria. All subjects were approved by the Ethics Committee of the National Institute for Viral Disease Control and Prevention. The subject was approved by the Ethics Committee of the National Institute for Viral Disease Control and Prevention (Reference code: IVDC2020-014).
Consent
The authors affirm that human research participants provided informed consent for the publication of the data.
Conflicts of Interest
The authors declare that they have no conflicts of interest.
Authors' Contributions
Jun Han, Hai-Lan Yao, and Wen-Jun Wang conceived and designed the experiments. Wen-Jun Wang, Miao-Feng, and Feng-He performed the experiments. Wen-Jun Wang analyzed the data. Jun Han, Juan Song, Qin-Qin Song, and Dong Xia contributed reagents, materials, and analysis tools. Wen-Jun Wang and Jun Han wrote the paper. All authors read and approved the final manuscript.
References
- 1.Griffiths P., Baraniak I., Reeves M. The pathogenesis of human cytomegalovirus. Journal of Pathology . 2015;235(2):288–297. doi: 10.1002/path.4437. [DOI] [PubMed] [Google Scholar]
- 2.Dupont L., Reeves M. B. Cytomegalovirus latency and reactivation: recent insights into an age old problem. Reviews in Medical Virology . 2016;26(2):75–89. doi: 10.1002/rmv.1862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Goodrum F., Caviness K., Zagallo P. Human cytomegalovirus persistence. Cellular Microbiology . 2012;14(5):644–655. doi: 10.1111/j.1462-5822.2012.01774.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Reeves M., Sinclair J. Aspects of human cytomegalovirus latency and reactivation. Current Topics in Microbiology and Immunology . 2008;325:297–313. doi: 10.1007/978-3-540-77349-8_17. [DOI] [PubMed] [Google Scholar]
- 5.Bissinger A. L., Sinzger C., Kaiserling E., Jahn G. Human cytomegalovirus as a direct pathogen: correlation of multiorgan involvement and cell distribution with clinical and pathological findings in a case of congenital inclusion disease. Journal of Medical Virology . 2002;67(2):200–206. doi: 10.1002/jmv.2208. [DOI] [PubMed] [Google Scholar]
- 6.Martin D. F., Sierra-Madero J., Walmsley S., et al. A controlled trial of valganciclovir as induction therapy for cytomegalovirus retinitis. New England Journal of Medicine . 2002;346(15):1119–1126. doi: 10.1056/nejmoa011759. [DOI] [PubMed] [Google Scholar]
- 7.Vanegas F., Montalvo R. D., Alvarez O. A., Donelson S. S., Lee M. Massive upper gastrointestinal hemorrhage due to cytomegalovirus infection in two patients with acquired immunodeficiency syndrome. Southern Medical Journal . 2000;93(2):235–237. doi: 10.1097/00007611-200002000-00020. [DOI] [PubMed] [Google Scholar]
- 8.Kotton C. N. Management of cytomegalovirus infection in solid organ transplantation. Nature Reviews Nephrology . 2010;6:711–721. doi: 10.1038/nrneph.2010.141. [DOI] [PubMed] [Google Scholar]
- 9.Snydman D. R. Persistent clinical impact of cytomegalovirus in organ transplantation. Clinical Infectious Diseases: An Official Publication of the Infectious Diseases Society of America . 2008;47:883–8841. doi: 10.1086/591533. [DOI] [PubMed] [Google Scholar]
- 10.Zheng L. I. H., Wang Q., Zhang Yi Y., et al. Onset of coronary heart disease is associated with HCMV infection and increased CD14+CD16+ monocytes in a population of weifang, China. Biomedical and Environmental Sciences . 2020;33(8):573–582. doi: 10.3967/bes2020.076. [DOI] [PubMed] [Google Scholar]
- 11.Emery V. C., Sabin C. A., Cope A. V., Gor D., Hassan-Walker A. F., Griffiths P. D. Application of viral-load kinetics to identify patients who develop cytomegalovirus disease after transplantation. Lancet . 2000;355(9220):2032–2036. doi: 10.1016/s0140-6736(00)02350-3. [DOI] [PubMed] [Google Scholar]
- 12.Griffiths P. D., Rothwell E., Raza M., et al. Randomized controlled trials to define viral load thresholds for cytomegalovirus pre-emptive therapy. PLoS One . 2016;11(9) doi: 10.1371/journal.pone.0163722.e0163722 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Natori Y., Alghamdi A., Tazari M., et al. Use of Viral load as a surrogate marker in clinical studies of cytomegalovirus in solid organ transplantation: a systematic review and meta-analysis. Clinical Infectious Diseases: An Official Publication of the Infectious Diseases Society of America . 2018;66:617–631. doi: 10.1093/cid/cix793. [DOI] [PubMed] [Google Scholar]
- 14.Ross S. A., Pati P., Jensen T. L., et al. Cytomegalovirus genetic diversity following primary infection. Journal of Infectious Diseases . 2020;221(5):715–720. doi: 10.1093/infdis/jiz507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gao Y., Tie Y. Q., Zhao L. Q., et al. Rapid internal control reference recombinase-aided amplification assays for EBV and CMV detection. Biomedical and Environmental Sciences . 2021;34(8):650–655. doi: 10.3967/bes2021.091. [DOI] [PubMed] [Google Scholar]
- 16.Hayden R. T., Yan X., Wick M. T., et al. Factors contributing to variability of quantitative viral PCR results in proficiency testing samples: a multivariate analysis. Journal of Clinical Microbiology . 2012;50(2):337–345. doi: 10.1128/jcm.01287-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hayden R. T., Preiksaitis J., Tong Y., et al. Commutability of the first world health organization international standard for human cytomegalovirus. Journal of Clinical Microbiology . 2015;53(10):3325–3333. doi: 10.1128/jcm.01495-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Razonable R. R., Hayden R. T. Clinical utility of viral load in management of cytomegalovirus infection after solid organ transplantation. Clinical Microbiology Reviews . 2013;26(4):703–727. doi: 10.1128/cmr.00015-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pavšič J., Devonshire A., Blejec A., et al. Inter-laboratory assessment of different digital PCR platforms for quantification of human cytomegalovirus DNA. Analytical and Bioanalytical Chemistry . 2017;409(10):2601–2614. doi: 10.1007/s00216-017-0206-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sedlak R. H., Cook L., Cheng A., Magaret A., Jerome K. R. Clinical utility of droplet digital PCR for human cytomegalovirus. Journal of Clinical Microbiology . 2014;52(8):2844–2848. doi: 10.1128/jcm.00803-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.From the American Association of Neurological Surgeons (AANS), American Society of Neuroradiology (ASNR), Cardiovascular and Interventional Radiology Society of Europe (CIRSE), et al. Multisociety consensus quality improvement revised consensus statement for endovascular therapy of acute ischemic stroke. International Journal of Stroke . 2018;13(6):612–632. doi: 10.1177/1747493018778713. [DOI] [PubMed] [Google Scholar]
- 22.Pang X. L., Fox J. D., Fenton J. M., Miller G. G., Caliendo A. M., Preiksaitis J. K. Interlaboratory comparison of cytomegalovirus viral load assays. American Journal of Transplantation . 2009;9(2):258–268. doi: 10.1111/j.1600-6143.2008.02513.x. [DOI] [PubMed] [Google Scholar]
- 23.Dioverti M. V., Lahr B. D., Germer J. J., Yao J. D., Gartner M. L., Razonable R. R. Comparison of standardized cytomegalovirus (CMV) viral load thresholds in whole blood and plasma of solid organ and hematopoietic stem cell transplant recipients with CMV infection and disease. Open Forum Infectious Diseases . 2017;4(3) doi: 10.1093/ofid/ofx143.ofx143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Naegele K., Lautenschlager I., Gosert R., et al. Cytomegalovirus sequence variability, amplicon length, and DNase-sensitive non-encapsidated genomes are obstacles to standardization and commutability of plasma viral load results. Journal of Clinical Virology . 2018;104:39–47. doi: 10.1016/j.jcv.2018.04.013. [DOI] [PubMed] [Google Scholar]
- 25.Preiksaitis J. K., Hayden R. T., Tong Y., et al. Are we there yet? Impact of the first international standard for cytomegalovirus DNA on the harmonization of results reported on plasma samples. Clinical Infectious Diseases: An Official Publication of the Infectious Diseases Society of America . 2016;63:583–589. doi: 10.1093/cid/ciw370. [DOI] [PubMed] [Google Scholar]
- 26.Pavšič J., Žel J., Milavec M. Assessment of the real-time PCR and different digital PCR platforms for DNA quantification. Analytical and Bioanalytical Chemistry . 2016;408(1):107–121. doi: 10.1007/s00216-015-9107-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wang W. J., Feng S. Q., He F., et al. The viral load of epstein-barr virus in blood of children after hematopoietic stem cell transplantation. Biomedical and Environmental Sciences . 2022;35(9):804–810. doi: 10.3967/bes2022.052. [DOI] [PubMed] [Google Scholar]
- 28.Cao G., Tan C., Zhang Y., et al. Digital droplet polymerase chain reaction analysis of common viruses in the aqueous humour of patients with posner-schlossman syndrome in Chinese population. Clinical and Experimental Ophthalmology . 2019;47(4):513–520. doi: 10.1111/ceo.13440. [DOI] [PubMed] [Google Scholar]
- 29.Madic J., Zocevic A., Senlis V., et al. Three-color crystal digital PCR. Biomol detect quantif 10. 2016. pp. 34–461. [DOI] [PMC free article] [PubMed]
- 30.Pomari E., Piubelli C., Perandin F., Bisoffi Z. Digital PCR: a new technology for diagnosis of parasitic infections. Clinical Microbiology and Infections . 2019;25(12):1510–1516. doi: 10.1016/j.cmi.2019.06.009. [DOI] [PubMed] [Google Scholar]
- 31.Boeckh M., Nichols W. G. The impact of cytomegalovirus serostatus of donor and recipient before hematopoietic stem cell transplantation in the era of antiviral prophylaxis and preemptive therapy. Blood . 2004;103(6):2003–2008. doi: 10.1182/blood-2003-10-3616. [DOI] [PubMed] [Google Scholar]
- 32.Griffiths P., Reeves M. Pathogenesis of human cytomegalovirus in the immunocompromised host. Nature Reviews Microbiology . 2021;19(12):759–773. doi: 10.1038/s41579-021-00582-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The data used to support the findings of this study are available from the corresponding author upon request.


