Abstract
Bartonella species can be differentiated by microimmunofluorescence assay, sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE), and immunoblotting with murine polyclonal antisera to Bartonella henselae, B. quintana, B. elizabethae, and B. bacilliformis. A pairwise comparison on the basis of SDS-PAGE protein profiles demonstrated similarity values for proteins of different Bartonella species ranging from 28.6 to 86.4%. Antigenic relationships revealed by immunoblotting with murine antisera were equivalent to those of proteins observed by SDS-PAGE. A dendrogram obtained on the basis of protein bands of SDS-polyacrylamide gels showed that Bartonella species could be divided into three groups. B. bacilliformis was distinct from all other Bartonella species; B. grahamii, B. taylorii, B. doshiae, and B. vinsonii formed a cluster, as did B. henselae, B. quintana, B. elizabethae, and B. clarridgeiae. These relationships were consistent with those revealed by parsimony trees derived from 16S rRNA and gltA gene sequencing. SDS-PAGE analysis showed that 120-, 104-, 85-, 71-, 54-, 47-, 40-, 33-, 30-, and 19-kDa proteins were present in all species, with the 54-kDa protein being the most dominant. Proteins with a molecular mass of less than 54 kDa allow the differentiation of species and are a possible target for future species-specific antibodies and antigens.
The genus Bartonella presently contains 14 species (3, 12, 16, 17, 24): Bartonella bacilliformis, B. henselae, B. quintana, B. vinsonii, B. elizabethae, B. talpae, B. peromysci, B. grahamii, B. taylorii, B. doshiae, B. clarridgeiae, B. tribocorum, B. alsatica, and B. koehlerae. A number of studies of the genetic relationship among Bartonella species have been carried out, including determinations of guanine-plus-cytosine contents (3, 6) and whole-genome DNA-DNA hybridization analyses (9, 33). These studies have demonstrated significant levels of similarity among the Bartonella species. Comparison of 16S rRNA gene sequences also indicated a high degree of similarity, ranging from 97.9 to 99.6%, within the genus Bartonella (3). A comparison of citrate synthase gene (gltA) sequences of different Bartonella species has shown the levels of similarity between sequences to be from 83.8 to 93.5% (4). Bartonella species may, however, be differentiated by restriction fragment length polymorphism PCR of the 16S rRNA gene with a combination of the restriction endonucleases DdeI and MnlI (5) and of the citrate synthase gene (18). Few phenotypic characteristics (8, 10, 14, 26, 32) have been reported for the identification of Bartonella species. In this report, we describe the protein and antigenic characteristics of Bartonella species as determined by comparative microimmunofluorescence assay (MIF), sodium dodecyl sulfate-polyacrylamide electrophoresis (SDS-PAGE), and Western immunoblotting.
MATERIALS AND METHODS
Bartonella strains.
The sources of all strains used in this study are presented in Table 1. Bartonella isolates were cultivated on blood agar (BioMerieux, Marcy l'Etoile, France) at 37°C in a 5% carbon dioxide incubator with the exception of B. bacilliformis, for which cultivation was carried out at 32°C. After between 5 and 7 days of culture, bacteria were harvested and suspended in deionized water for use in SDS-PAGE or in phosphate-buffered saline (PBS) for use in the MIF.
TABLE 1.
Studied strains
| Species | Straina | Sourceb |
|---|---|---|
| B. bacilliformis | Monzon 812 | Bartonellosis patient, Peru |
| B. henselae | Houston-1T (ATCC 49882) | Septicemia |
| URBHLLY8 | CSD patient, Marseille, France | |
| URBHLLY9 | IE patient, Marseille, France | |
| CAL-1 | Septicemia patient, United States | |
| Cat 6 | South African cat | |
| Fizz | Swiss cat | |
| King | USA | |
| SA2 | CSD patient, United States | |
| B. quintana | FullerT (ATCC VR-358) | TF patient |
| B. elizabethae | F9251T (ATCC 49927) | IE patient |
| B. grahamii | V2T (NCTC 12860) | Blood of C. glareolus |
| B. taylorii | M6T (NCTC 12861) | Blood of Apodemus spp. |
| B. doshiae | R18T (NCTC 12862) | Blood of M. agrestis |
| B. clarridgeiae | URBCMNHC26 | French cat |
| B. vinsonii | BakerT (ATCC VR-152) | Spleen of M. pennsylvanicus |
T, reference strain.
Abbreviations: BA, bacillary angiomatosis; CSD, cat scratch disease; IE, infective endocarditis; LY, lymphadenopathy; TF, trench fever.
Antiserum production.
Murine polyclonal antisera directed against B. henselae, B. quintana, B. bacilliformis, B. elizabethae, B. taylorii, and B. vinsonii were produced in BALB/c mice. The mice were immunized intraperitoneally three times, at 7-day intervals, with 200 μg of Bartonella proteins. Sera were obtained from the mice 1 week after the final immunization. Antibody titers of all sera collected were determined by MIF.
MIF.
Antigens were deposited on 24-well microscope slides with the nib of a pen. The antigens were fixed in methanol for 10 min at room temperature. Murine antisera to Bartonella species were diluted 1:2 to 1:8,192 in PBS containing 3% milk powder and added to the wells, and the slides were incubated in a humidified chamber at 37°C for 30 min. The slides were then washed twice in PBS (5 min each) and rinsed with distilled water. After being air dried at 37°C, the slides were incubated at 37°C for 30 min with fluorescein (dechlorotriazinyl aminofluorescein)-conjugated goat anti-mouse immunoglobulin G (IgG) plus IgM (Jackson ImmunoResearch Laboratories, Inc., West Grove, Pa.) diluted 1:100 in PBS containing 0.2% Evans blue (BioMerieux). The slides were washed as described above, mounted with Fluoprep (BioMerieux), and then read on a Zeiss epifluorescence microscope (Axioskop 20; Carl Zeiss Göttingen, Germany) at 400× magnification.
SDS-PAGE and Western immunoblotting.
SDS-PAGE was performed by a modification of the method described by Laemmli (22), using a 12% polyacrylamide separating gel and a 5% stacking gel. Aliquots (10 μl) of antigens preparations, previously adjusted to contain 4 mg of protein/ml, were suspended in an equal volume of sample buffer (0.0625 M Tris hydrochloride [pH 8.0], 2% SDS, 5% 2-mercaptoethanol, 10% glycerol, 0.02% bromophenol blue) and heated for 5 min at 100°C. The dissolved antigens were separated in an electrophoretic cell (Mini Protein II; Bio-Rad, Richmond, Calif.) at a constant current (8 to 10 mA per gel) for 3 to 4 h in running buffer (25 mM Tris, 192 mM glycine, 0.1% SDS). Prestained SDS-PAGE standards (Low-Range; Bio-Rad) were used on each gel as a reference.
The separated antigens were transferred to 0.45-μm-pore-size nitrocellulose membranes (Hybond-C; Amersham, Little Chalfont, United Kingdom) in an electrophoretic transfer cell (Mini Trans-Blot; Bio-Rad) with transfer buffer (2.5 mM Tris base, 192 mM glycine, 20% methanol) at 50 V and 4°C for 1 h. After transfer was completed, the nitrocellulose membranes were incubated overnight in PBS with 5% nonfat dry milk to block nonspecific binding sites, washed three times with PBS, and air dried. The membranes were then incubated at room temperature for 1 h with murine antisera diluted 1:100 in PBS containing 3% nonfat dry milk and washed as described above. After being incubated at room temperature for 1 h with peroxidase-conjugated F(ab′)2 fragment goat anti-mouse IgG (heavy and light chains; AffiniPure; Jackson ImmunoResearch) diluted 1:500 in PBS containing 3% nonfat dry milk, the blots were washed in PBS and bound peroxidase enzyme was detected with 4-chloro-1-naphthol as the substrate.
Calculation of protein molecular sizes.
A standard curve was generated by using molecular mass standards on each Western blot so that the sizes of the Bartonella proteins could be determined.
Numerical taxonomic analysis.
A score of 1 was given for each band in the SDS-PAGE profile or Western blot profile. These scores were used to construct a dendrogram from the matrix by the unweighted-pair group method with an arithmetic mean (UPGMA), available in the PC-TAXAN software package (Sea Grant College, University of Maryland, College Park), according to the manufacturer's instructions. The Jaccard coefficients (SJ) derived from matrix analysis were used as measures of similarity of antigenicity among different Bartonella species. Parsimony trees inferred from alignment of Bartonella 16S rRNA and gltA gene sequences obtained from GenBank were compared with the obtained dendrograms.
RESULTS
While MIF titers of the polyclonal mouse antisera were highest against the homologous antigens (Table 2), cross-reactive antibodies against the other Bartonella species could generally be detected. The highest titers of cross-reacting antibodies were in antisera to B. henselae when tested against B. quintana.
TABLE 2.
MIF titers of Bartonella antigens, obtained with murine antisera
| Species no. | Speciesa | MIF titer determined using antiserum to species:
|
|||||
|---|---|---|---|---|---|---|---|
| 1 | 2 | 4 | 5 | 6 | 9 | ||
| 1 | B. bacilliformis | 4,096 | 16 | 16 | 4 | 128 | |
| 2 | B. henselae H | 128 | 4,096 | 64 | 32 | 16 | 16 |
| 3 | B. henselae M | 128 | 2,048 | 32 | 32 | 16 | 16 |
| 4 | B. quintana | 128 | 512 | 4,096 | 8 | 64 | 4 |
| 5 | B. elizabethae | 128 | 64 | 16 | 1,024 | 32 | |
| 6 | B. grahamii | 64 | 16 | 16 | 4 | 1,024 | |
| 7 | B. taylorii | 128 | 16 | 16 | 8 | 512 | |
| 8 | B. doshiae | 128 | 16 | 16 | 8 | 64 | 32 |
| 9 | B. vinsonii | 128 | 16 | 16 | 16 | 64 | 1,024 |
| 10 | B. clarridgeiae | 128 | 16 | 16 | 8 | 32 | 16 |
B. henselae H and B. henselae M are B. henselae subspecies hountonae and B. henselae massiliae, respectively.
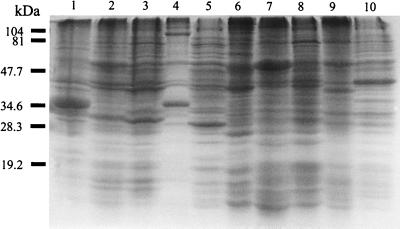
SDS-PAGE analysis of Bartonella species showed distinct protein profiles (Fig. 1), with differences being evident mainly among proteins with molecular masses smaller than 54 kDa. Although minor differences between B. grahamii and B. taylorii were detected, their SDS-PAGE profiles were almost identical. B. bacilliformis was clearly distinct from all other Bartonella species. Different strains belonging to the same species had identical protein profiles, with the exception of the two B. henselae serotypes, which showed only minor differences (data not shown). SDS-PAGE of Bartonella species antigens showed that several proteins, including those of 120, 104, 85, 71, 54, 47, 40, 33, 30, and 19 kDa, were common to all (Fig. 1; Table 3). The 54-kDa protein band gave the most prominent staining reaction in each species (Fig. 1; Table 3), and dominant and species-specific proteins of less than 54 kDa were also observed (see Fig. 4).
FIG. 1.
Coomassie brilliant blue staining profiles of Bartonella strains after SDS-PAGE. Lanes: 1, B. bacilliformis; 2, B. henselae serotype Houston; 3, B. henselae serotype Marseille; 4, B. clarridgeiae; 5, B. quintana; 6, B. elizabethae; 7, B. grahamii; 8, B. taylorii; 9, B. doshiae; 10, B. vinsonii. The positions of molecular mass markers of 104, 81, 47.7, 34.6, 28.3, and 19.2 kDa are noted on the left.
TABLE 3.
Bartonella genus-specific antigens detected by SDS-PAGE and Western blotting with antibodies against various Bartonella species
| Protein size (kDa) | SDS- PAGE | Results of immunoblot with murine serum toa:
|
|||||
|---|---|---|---|---|---|---|---|
| bac | hen | qui | eli | gra | vin | ||
| 120 | + | ++ | + | ++ | ++ | + | + |
| 104 | + | + | ++ | ++ | ++ | ++ | + |
| 85 | + | + | ++ | + | + | + | − |
| 71 | + | + | + | − | +++ | − | − |
| 54 | ++++ | ++++ | ++++ | +++ | ++++ | ++++ | +++ |
| 47 | + | +++ | + | ++ | ++ | − | +++ |
| 40 | + | +++ | ++ | − | + | + | − |
| 33 | + | + | + | − | − | − | − |
| 30 | + | +++ | + | +++ | ++ | − | − |
| 19 | + | − | ++ | +++ | − | − | − |
Antigens were B. bacilliformis (bac), B. henselae (hen), B. quintana (qui), B. elizabethae (eli), B. grahamii (gra), and B. vinsonii (vin). −, no band evident; +, weak band; ++, moderate-intensity band; +++, intense band; ++++, very intense band.
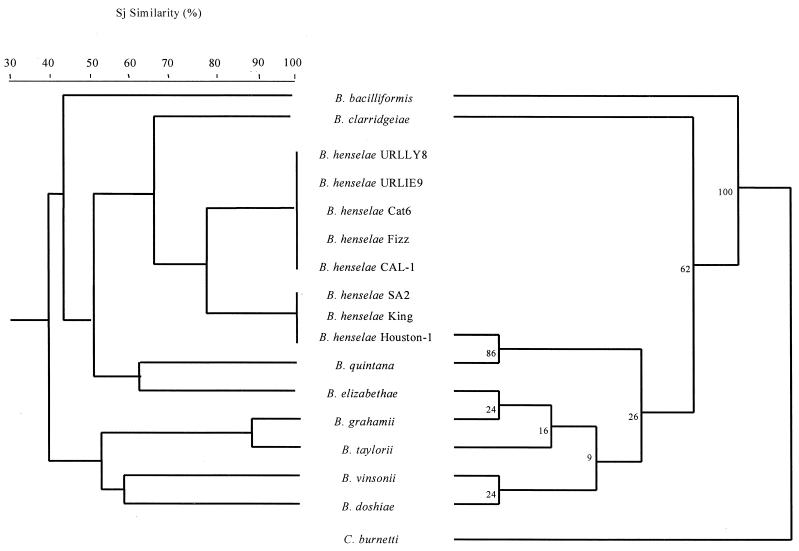
FIG. 4.
Dendrogram of Bartonella strains representative of Coxiella inferring Sj similarities obtained by UPGMA on the basis of immunoblotting with murine antiserum to B. henselae (left), and parsimony tree of Bartonella strains and Coxiella burnetii derived from gltA gene comparison (right). The support for each branch, as determined from bootstrap samples, is indicated by the value at the node. This analysis provided tree topology only, and lengths of both the vertical lines and the horizontal lines are not significant. The GenBank accession numbers for gltA gene sequences included are as follows: B. bacilliformis, Z70021; B. henselae, L38987; B. quintana, Z70014; B. elizabethae, Z70009; B. grahamii, Z70016; B. taylorii, Z70013; B. doshiae, Z70017; B. vinsonii, Z70015; and C. burnetii, M36338.
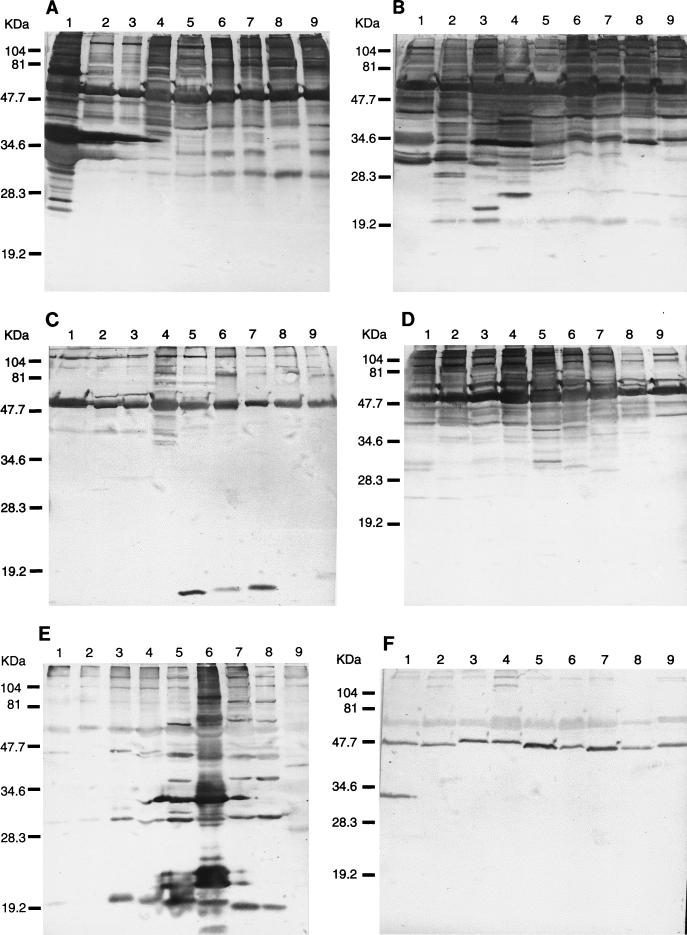
Western blots revealed unique profiles for each species (Fig. 2A to D), with the exception of antisera to B. grahamii and B. vinsonii, which could not differentiate Bartonella species (Fig. 2E and F). Although there were visible differences among B. henselae serotypes and between B. grahamii and B. taylorii, these organisms generally had very similar profiles (Fig. 2A to D). Species-specific antigens were clearly identified in a low-molecular-mass range. Each polyclonal murine antiserum reacted with the 120-, 104-, and 54-kDa antigen bands of each Bartonella species (Fig. 2; Table 3), and in addition, antisera to B. henselae and B. quintana (Fig. 2; Table 3) reacted with a 19-kDa protein in each species. Homologous Western blots identified several antigens, and the most immunoreactive bands were detected with anti-B. bacilliformis serum. Eleven of these bands had estimated molecular sizes of 120, 104, 71, 54, 47, 41, 37, 33, 30, 27, and 25 kDa and were highly reactive. The less-reactive antisera were mostly against B. grahamii and B. vinsonii (Fig. 2E and F). Species-specific immunoreactive bands of 41, 23, 21, 32, and 26 kDa were prominently observed for B. bacilliformis, B. henselae subtype Houston, B. henselae subtype Marseille, B. quintana, and B. elizabethae, respectively (Fig. 2; Table 4).
FIG. 2.
Immunoblotting of Bartonella antigens with murine antisera to B. bacilliformis (A), B. henselae (B), B. grahamii (C), B. elizabethae (D), B. quintana (E), and B. vinsonii (F). (A to C) Lanes: 1, B. bacilliformis; 2, B. henselae Houston; 3, B. henselae Marseille; 4, B. quintana; 5, B. elizabethae; 6, B. grahamii; 7, B. taylorii; 8, B. doshiae; 9, B. vinsonii. (D to F) Lanes: 1, B. vinsonii; 2, B. doshiae; 3, B. taylorii; 4, B. grahamii; 5, B. elizabethae; 6, B. quintana; 7, B. henselae Marseille; 8, B. henselae Houston; 9, B. bacilliformis. The positions of molecular mass standards are noted on the left of each panel.
TABLE 4.
Dominant and species-specific proteins of less than 54 kDa in Bartonella species
| Method used | Protein (kDa) | Results fora:
|
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| B. bacilliformis | B. henselae Houston | B. henselae Marseille | B. quintana | B. elizabethae | B. grahamii | B. taylorii | B. doshiae | B. vinsonii | B. clarridgeiae | ||
| SDS-PAGE | 41 | +++ | +++ | ++ | ++ | ++ | +++ | +++ | |||
| 40.5 | ++ | ++ | |||||||||
| 40 | +++ | +++ | +++ | ++ | ++ | ++ | +++ | ||||
| 37 | +++ | ++ | |||||||||
| 34 | +++++ | ++++ | |||||||||
| 32 | +++ | ||||||||||
| 31 | ++ | ++ | ++ | ++ | |||||||
| 30 | +++ | +++ | ++ | ++ | ++ | ++ | |||||
| 29 | ++++ | +++ | ++ | ||||||||
| 27 | +++ | ++ | ++++ | ||||||||
| 26 | +++ | ||||||||||
| 23 | ++ | ++ | |||||||||
| Immunoblotting with antiserum to B. henselae | 41 | +++ | ++ | ++ | ++ | +++ | NT | ||||
| 38 | ++ | NT | |||||||||
| 35 | + | + | ++ | NT | |||||||
| 34 | ++++ | ++ | ++++ | ++++ | ++ | ++ | ++ | ++++ | NT | ||
| 31 | ++ | NT | |||||||||
| 30 | ++ | +++ | ++ | NT | |||||||
| 29 | ++ | ++ | ++ | ++ | ++ | ++ | NT | ||||
| 28 | ++++ | ++ | NT | ||||||||
| 24 | +++ | ++ | ++ | ++ | ++ | NT | |||||
| 23 | ++ | NT | |||||||||
| 21 | +++ | NT | |||||||||
| 19 | ++ | +++ | ++ | ++ | ++ | ++ | NT | ||||
| Immunoblotting with antiserum to B. quintana | 45 | ++ | ++ | ++ | NT | ||||||
| 44 | +++ | +++ | +++ | +++ | ++ | NT | |||||
| 41 | ++ | NT | |||||||||
| 37 | +++ | +++ | +++ | NT | |||||||
| 34 | +++ | ++++ | +++ | NT | |||||||
| 32 | ++ | NT | |||||||||
| 31 | ++ | NT | |||||||||
| 30 | +++ | +++ | +++ | ++ | +++ | ++ | ++ | NT | |||
| 29 | ++ | NT | |||||||||
| 28 | ++ | NT | |||||||||
| 23 | ++ | NT | |||||||||
+, weak; ++, moderate; +++, intense; ++++, very intense; NT, not tested.
A pairwise comparison of SDS-PAGE protein profiles demonstrated protein similarity values ranging from 28.6% (for B. clarridgeiae and B. doshiae) to 86.4% (for B. grahamii and B. taylorii). The protein similarity value for B. henselae strains Houston and Marseille was 76.3%. However, the similarity between different strains within each serotype was 100% (Table 5). Antigenic relationships identified by Western blotting with murine antiserum to B. henselae were similar to those found in protein profiles obtained by SDS-PAGE (Table 5).
TABLE 5.
Antigenic Sj similarity among Bartonella isolate pairs as revealed by SDS-PAGE and immunoblotting with antiserum to B. henselae
| Isolate no. | Bartonella isolate | Antigenic similarity (Sj; %) among Bartonella isolatea:
|
|||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | 12 | 13 | 14 | 15 | 16 | ||
| 1 | B. bacilliformis | 56.0 | 56.0 | 56.0 | 48.1 | 48.1 | 48.1 | 48.1 | 48.1 | NT | 60.9 | 50.0 | 22.2 | 25.9 | 30.4 | 33.3 | |
| 2 | B. henselae | 34.7 | 100 | 100 | 75.0 | 75.0 | 75.0 | 75.0 | 75.0 | NT | 64.3 | 60.7 | 23.5 | 26.5 | 30.0 | 32.1 | |
| 3 | B. henselae | 34.7 | 100 | 100 | 75.0 | 75.0 | 75.0 | 75.0 | 75.0 | NT | 64.3 | 60.7 | 23.5 | 26.5 | 30.0 | 32.1 | |
| 4 | B. henselae | 34.7 | 100 | 100 | 75.0 | 75.0 | 75.0 | 75.0 | 75.0 | NT | 64.3 | 60.7 | 23.5 | 26.5 | 30.0 | 32.1 | |
| 5 | B. henselae | 45.7 | 76.3 | 76.3 | 76.3 | 100 | 100 | 100 | 100 | NT | 56.7 | 48.4 | 26.5 | 25.7 | 29.0 | 26.7 | |
| 6 | B. henselae | 45.7 | 76.3 | 76.3 | 76.3 | 100 | 100 | 100 | 100 | NT | 56.7 | 48.4 | 26.5 | 25.7 | 29.0 | 26.7 | |
| 7 | B. henselae | 45.7 | 76.3 | 76.3 | 76.3 | 100 | 100 | 100 | 100 | NT | 56.7 | 48.4 | 26.5 | 25.7 | 29.0 | 26.7 | |
| 8 | B. henselae | 45.7 | 76.3 | 76.3 | 76.3 | 100 | 100 | 100 | 100 | NT | 56.7 | 48.4 | 26.5 | 25.7 | 29.0 | 26.7 | |
| 9 | B. henselae | 45.7 | 76.3 | 76.3 | 76.3 | 100 | 100 | 100 | 100 | NT | 56.7 | 48.4 | 26.5 | 25.7 | 29.0 | 26.7 | |
| 10 | B. clarridgeiae | 40.0 | 61.5 | 61.5 | 61.5 | 64.1 | 64.1 | 64.1 | 64.1 | 64.1 | NT | NT | NT | NT | NT | NT | |
| 11 | B. quintana | 43.5 | 50.0 | 50.0 | 50.0 | 59.5 | 59.5 | 59.5 | 59.5 | 59.5 | 61.5 | 65.4 | 33.3 | 36.7 | 42.3 | 45.8 | |
| 12 | B. elizabethae | 42.9 | 48.9 | 48.9 | 48.9 | 54.3 | 54.3 | 54.3 | 54.3 | 54.3 | 55.8 | 59.1 | 34.5 | 37.9 | 38.5 | 47.8 | |
| 13 | B. grahamii | 35.9 | 32.5 | 32.5 | 32.5 | 31.7 | 31.7 | 31.7 | 31.7 | 31.7 | 38.9 | 39.5 | 39.0 | 94.7 | 65.0 | 63.2 | |
| 14 | B. taylorii | 42.1 | 31.7 | 31.7 | 31.7 | 31.0 | 31.0 | 31.0 | 31.0 | 31.0 | 37.8 | 38.5 | 41.5 | 86.4 | 61.9 | 68.4 | |
| 15 | B. doshiae | 37.1 | 33.3 | 33.3 | 33.3 | 32.4 | 32.4 | 32.4 | 32.4 | 32.4 | 28.6 | 33.3 | 30.0 | 52.2 | 50.0 | 64.7 | |
| 16 | B. vinsonii | 36.8 | 33.3 | 33.3 | 33.3 | 32.5 | 32.5 | 32.5 | 32.5 | 32.5 | 40.0 | 44.4 | 36.6 | 50.0 | 48.1 | 54.5 | |
The values on the upper right are the antigenic similarities of Bartonella spp. as determined by SDS-PAGE, and the values on the lower left are the antigenic similarities of Bartonella spp. as determined by immunoblotting with antiserum to B. henselae.
NT, not tested.
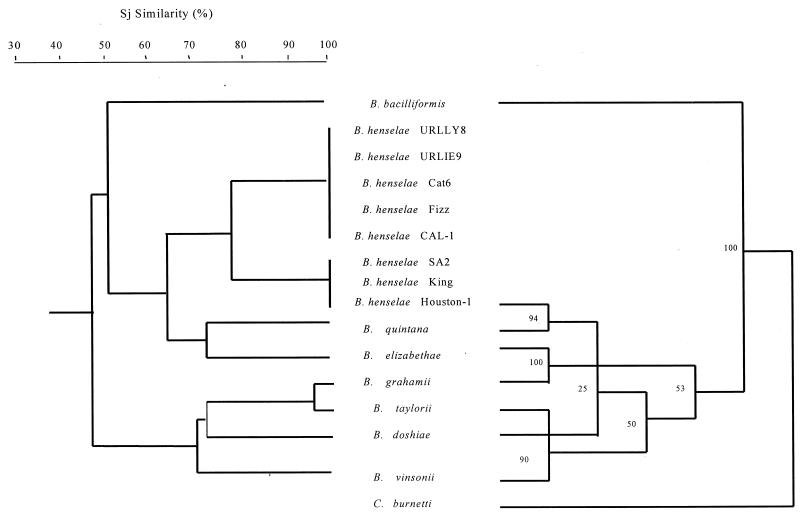
If an SJ similarity value of 50.1% was used as the cutoff for the dendrogram obtained from SDS-PAGE, the Bartonella species could be divided into three groups (Fig. 3). B. bacilliformis was the single member of the first group and was most distant; B. grahamii, B. taylorii, B. doshiae, and B. vinsonii formed a second cluster, while B. henselae, B. quintana, B. elizabethae, and B. clarridgeiae formed a third (Fig. 3). The dendrogram based on immunoreactive bands observed with murine antiserum to B. henselae also identified these three groups (Fig. 4).
FIG. 3.
Dendrogram of Bartonella strains inferring Sj similarities obtained by UPGMA on the basis of the molecular masses of different protein bands on SDS-PAGE (left), and parsimony tree of Bartonella strains and Coxiella burnetii derived from 16S rRNA gene comparison (right). The numbers at the nodes are the proportions of 100 bootstrap resamplings that support the topology shown. This analysis provided tree topology only, and the lengths of both the vertical lines and the horizontal lines are not significant. The GenBank accession numbers for the 16S rRNA gene sequences included are as follows: B. bacilliformis, Z70003; B. henselae, Z11684; B. quintana, M11927; B. clarridgeiae, X97822; B. elizabethae, L01260; B. grahamii, Z31349; B. taylorii, Z31350; B. doshiae, Z31351; B. vinsonii, Z31352; and C. burnetii, D89792.
As for comparison of these dendrograms with phylogenic trees, SDS-PAGE and Western blot findings were consistent with the analysis of the 16S rRNA and gltA gene data (100% bootstrap) (Fig. 3 and 4) obtained by parsimony analysis, showing the relatively deeply rooted divergence of B. bacilliformis from other Bartonella species. However, the two other groups were not identified, and none of the significant bootstrap values was found in a phylogenic tree inferred from the 16S rRNA gene sequence data. However, three significant nodes were determined from the gltA-derived data. Two groups were also found with the Western blot data based on UPGMA; one contained B. taylorii and B. vinsonii (90% bootstrap value) (Fig. 4), and the second included B. henselae and B. quintana (94% bootstrap value) (Fig. 4). The third node, clustering B. elizabethae and B. grahamii (100% bootstrap) (Fig. 4), was not found when Western blot data were used.
DISCUSSION
In this study, nine Bartonella species were clearly differentiated by their distinct SDS-PAGE protein profiles, with bands of molecular mass less than 54 kDa being responsible for most of the observed differences. Western blotting with antisera to B. quintana, B. henselae, B. bacilliformis, and B. elizabethae also enabled the differentiation of all species and subspecies of Bartonella studied. These results support the earlier suggestion that phenotypic criteria may be used to characterize Bartonella spp. (26). They also show that SDS-PAGE and Western blotting are effective methods for phenotypic identification of the Bartonella group to the species level, in accord with the observed specific reactivity between B. quintana and B. henselae. Moreover, the identification of genus- and species-specific bands may allow the identification of genus- and species-specific polyclonal and monoclonal antibodies. The species-specific antigen could be the source of species-specific serology.
Data obtained in SDS-PAGE and Western blot analyses enabled us to determine that there are three groups within the species Bartonella. The first contains only B. bacilliformis, the etiological agent of Carrion's disease, whose distribution is limited to the inter-Andean valleys of South America. The trees derived from the similarity matrix based on our SDS-PAGE and Western blotting results indicate that B. bacilliformis is the most distant of the Bartonella species, which is consistent with the results of previous DNA hybridization studies (9, 33) and with data obtained in a series of studies of both 16S rRNA and gltA gene sequences (4). The second cluster contains B. henselae, B. quintana, B. elizabethae, and B. clarridgeiae. B. henselae, a new species first described in 1992 (29), contains two serotypes and two genotypes (2, 11, 25, 30, 31), and we believe that they are subspecies (Z. Liang, V. Roux, B. La Scola, and D. Raoult, unpublished data). The dendrogram confirms that B. henselae is very closely related to B. quintana. These species can cause a number of clinical manifestations, some of which, such as bacillary angiomatosis and endocarditis, are identical (7, 15, 19, 21, 27, 28). Both exhibit a high degree of serological cross-reactivity (1, 13, 23). DNA-DNA hybridization experiments (9, 33) and comparisons of the sequences of the 16S rRNA and gltA genes revealed a high level of genetic homology (3, 4). B. clarridgeiae was isolated from an American cat in 1996 (24), and it has subsequently been suggested that this organism may be one of the etiological agents of cat scratch disease (20). The dendrogram produced in the present study demonstrates that this species is very closely related to B. henselae. To date, only a single isolate of B. elizabethae has been obtained from a human patient with endocarditis (9). This isolate clustered differently in our study and was found to be closely related to B. quintana and B. henselae; 16S rRNA and gltA gene sequence analysis showed it to cluster with B. grahamii.
The third cluster contains B. grahamii, B. taylorii, B. doshiae, and B. vinsonii. These organisms were isolated from rodents; B. vinsonii was also isolated from dogs.
A previous investigation (4) has shown that analysis of the gltA gene is more suitable for phylogenic studies of Bartonella species than is analysis of the 16S rRNA gene. The results of our study support this finding, as the taxonomic relationships among the Bartonella species we inferred were generally consistent with those derived from phylogenetic analysis of the gltA gene data.
ACKNOWLEDGMENTS
We are grateful to Hervé Tissot-Dupont and Veronique Roux for making the dendrogram and to P. Kelly for reviewing the manuscript.
REFERENCES
- 1.Baneth G, Kordick D L, Hegarty B C, Breitschwerdt E B. Comparative seroreactivity to Bartonella henselae and Bartonella quintana among cats from Israel and North Carolina. Vet Microbiol. 1996;50:95–103. doi: 10.1016/0378-1135(96)00006-5. [DOI] [PubMed] [Google Scholar]
- 2.Bergmans A M C, Schellekens J F P, Embden J A V, Schouls L M. Predominance of two Bartonella henselae variants among cat-scratch disease patients in The Netherlands. J Clin Microbiol. 1996;34:254–260. doi: 10.1128/jcm.34.2.254-260.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Birtles R J, Harrison T G, Saunders N A, Molyneux D H. Proposals to unify the genera Grahamella and Bartonella, with descriptions of Bartonella talpae comb. nov., Bartonella peromysci comb. nov., and three new species, Bartonella grahamii sp. nov., Bartonella taylorii sp. nov., and Bartonella doshiae sp. nov. Int J Syst Bacteriol. 1995;45:1–8. doi: 10.1099/00207713-45-1-1. [DOI] [PubMed] [Google Scholar]
- 4.Birtles R J, Raoult D. Comparison of partial citrate synthase gene (gltA) sequences for phylogenetic analysis of Bartonella species. Int J Syst Bacteriol. 1996;46:891–897. doi: 10.1099/00207713-46-4-891. [DOI] [PubMed] [Google Scholar]
- 5.Birtles R J. Differentiation of Bartonella species using restriction endonuclease analysis of PCR-amplified 16S rRNA genes. FEMS Microbiol Lett. 1995;129:261–265. doi: 10.1111/j.1574-6968.1995.tb07590.x. [DOI] [PubMed] [Google Scholar]
- 6.Brenner D J, O'Connor S P, Winkler H H, Steigerwalt A G. Proposals to unify the genera Bartonella and Rochalimaea, with descriptions of Bartonella quintana comb. nov., Bartonella vinsonii comb. nov., Bartonella henselae comb. nov., and Bartonella elizabethae comb. nov., and to remove the family Bartonellaceae from the order Rickettsiales. Int J Syst Bacteriol. 1993;43:777–786. doi: 10.1099/00207713-43-4-777. [DOI] [PubMed] [Google Scholar]
- 7.Brouqui P, Lascola B, Roux V, Raoult D. Chronic Bartonella quintana bacteremia in homeless patients. N Engl J Med. 1999;340:184–189. doi: 10.1056/NEJM199901213400303. [DOI] [PubMed] [Google Scholar]
- 8.Burgess A W, Anderson B E. Outer membrane proteins of Bartonella henselae and their interaction with human endothelial cells. Microb Pathog. 1998;25:157–164. doi: 10.1006/mpat.1998.0223. [DOI] [PubMed] [Google Scholar]
- 9.Daly J S, Worthington M G, Brenner D J, Moss C W, Hollis D G, Weyant R S, Steigerwalt A G, Weaver R E, Daneshvar M I, O'Connor S P. Rochalimaea elizabethae sp. nov. isolated from a patient with endocarditis. J Clin Microbiol. 1993;31:872–881. doi: 10.1128/jcm.31.4.872-881.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Drancourt M, Raoult D. Proposed tests for the routine identification of Rochalimaea species. Eur J Clin Microbiol Infect Dis. 1993;12:710–713. doi: 10.1007/BF02009387. [DOI] [PubMed] [Google Scholar]
- 11.Drancourt M, Birtles R, Chaumentin G, Vandenesch F, Etienne J, Raoult D. A new serotype of Bartonella henselae causes endocarditis and cat-scratch disease. Lancet. 1996;347:441–443. doi: 10.1016/s0140-6736(96)90012-4. [DOI] [PubMed] [Google Scholar]
- 12.Droz S, Chi B, Horn E, Steigerwalt A G, Whitney A M, Brenner D J. Bartonella koehlerae sp. nov., isolated from cats. J Clin Microbiol. 1999;37:1117–1122. doi: 10.1128/jcm.37.4.1117-1122.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dupont M, Larclause A S D, Brouqui P, Drancourt M, Raoult D, Mascarel A D, Lacut J. Evaluation of serological response to Bartonella henselae, Bartonella quintana, and Afipia felis antigens in 64 patients with suspected cat-scratch disease. Scand J Infect Dis. 1996;28:361–366. doi: 10.3109/00365549609037920. [DOI] [PubMed] [Google Scholar]
- 14.Freeland R L, Scholl D T, Rohde K R, Shelton L J, O'Reilly K L. Identification of Bartonella-specific immunodominant antigens recognized by the feline humoral immune system. Clin Diagn Lab Immunol. 1999;6:558–566. doi: 10.1128/cdli.6.4.558-566.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hadfield T L, Warren R, Kass M, Brun E, Levy C. Endocarditis caused by Rochalimaea henselae. Hum Pathol. 1993;24:1140–1141. doi: 10.1016/0046-8177(93)90196-n. [DOI] [PubMed] [Google Scholar]
- 16.Heller R, Riegel P, Hansmann Y, Delacour G, Bermond D, Dehio C, Lamarque F, Monteil H, Chomel B, Piemont Y. Bartonella tribocorum sp. nov., a new Bartonella species isolated from the blood of wild rats. Int J Syst Bacteriol. 1998;48:1333–1339. doi: 10.1099/00207713-48-4-1333. [DOI] [PubMed] [Google Scholar]
- 17.Heller R, Kubina M, Mariet P, Riegel P, Delacour G, Dehio C, Lamarque F, Kasten R, Boulouis H J, Monteil H, Chomel B, Piemont Y. Bartonella alsatica sp. nov., a new Bartonella species isolated from the blood of wild rabbits. Int J Syst Bacteriol. 1999;49:283–288. doi: 10.1099/00207713-49-1-283. [DOI] [PubMed] [Google Scholar]
- 18.Joblet C, Roux V, Drancourt M, Gouvernet J, Raoult D. Identification of Bartonella (Rochalimaea) species among fastidious gram-negative bacteria on the basis of the partial sequence of the citrate-synthase gene. J Clin Microbiol. 1995;33:1879–1883. doi: 10.1128/jcm.33.7.1879-1883.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Koehler J E, Quinn F D, Berger T G, LeBoit P E, Tappero J W. Isolation of Rochalimaea species from cutaneous and osseous lesions of bacillary angiomatosis. N Engl J Med. 1992;327:1625–1631. doi: 10.1056/NEJM199212033272303. [DOI] [PubMed] [Google Scholar]
- 20.Kordick D L, Hilyard E J, Hadfield T L, Wilson K H, Steigerwalt A G, Brenner D J, Breitschwerdt E B. Bartonella clarridgeiae, a newly recognized zoonotic pathogen causing inoculation papules, fever, and lymphadenopathy (cat scratch disease) J Clin Microbiol. 1997;35:1813–1818. doi: 10.1128/jcm.35.7.1813-1818.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kordick D L, Wilson K H, Sexton D J, Hadfield T L, Berkhoff H A, Breitschwerdt E B. Prolonged Bartonella bacteremia in cats associated with cat-scratch disease patients. J Clin Microbiol. 1995;33:3245–3251. doi: 10.1128/jcm.33.12.3245-3251.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Laemmli U K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature (London) 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 23.La Scola B, Raoult D. Serological cross-reactions between Bartonella quintana, Bartonella henselae, and Coxiella burnetii. J Clin Microbiol. 1996;34:2270–2274. doi: 10.1128/jcm.34.9.2270-2274.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lawson P A, Collins M D. Description of Bartonella clarridgeiae sp. nov. isolated from the cat of a patient with Bartonella henselae septicemia. Med Microbiol Lett. 1996;5:64–73. [Google Scholar]
- 25.Mainardi J L, Figliolini C, Goldstein F W, Blanche P, Baret-Rigoulet M, Galezowski N, Fournier P E, Raoult D. Cat scratch disease due to Bartonella henselae serotype Marseille (Swiss cat) in a seronegative patient. J Clin Microbiol. 1998;36:2800. doi: 10.1128/jcm.36.9.2800-2800.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Maurin M, Roux V, Stein A, Ferrier F, Viraben R, Raoult D. Isolation and characterization by immunofluorescence, sodium dodecyl sulfate-polyacrylamide gel electrophoresis, Western blot, restriction fragment length polymorphism-PCR, 16S rRNA gene sequencing, and pulsed-field gel electrophoresis of Rochalimaea quintana from a patient with bacillary angiomatosis. J Clin Microbiol. 1994;32:1166–1171. doi: 10.1128/jcm.32.5.1166-1171.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Raoult D, Fournier P E, Drancourt M, Marrie T J, Etienne J, Cosserat J, Cacoub P, Poinsingnon Y, Leclercq P, Sefton A M. Diagnosis of 22 new cases of Bartonella endocarditis. Ann Intern Med. 1996;125:646–652. doi: 10.7326/0003-4819-125-8-199610150-00004. [DOI] [PubMed] [Google Scholar]
- 28.Raoult D, Drancourt M, Carta A, Gastaut J A. Bartonella (Rochalimaea) quintana isolation in patient with chronic adenopathy, lymphopenia, and a cat. Lancet. 1994;343:977. doi: 10.1016/s0140-6736(94)90102-3. [DOI] [PubMed] [Google Scholar]
- 29.Regnery R L, Anderson B E, Clarridge III J E, Rodriguez-Barradas M C, Jones D C, Carr J H. Characterization of a novel Rochalimaea species, R. henselae sp. nov., isolated from blood of a febrile, human immunodeficiency virus-positive patient. J Clin Microbiol. 1992;30:265–274. doi: 10.1128/jcm.30.2.265-274.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sander A, Ruess M, Bereswill S, Schuppler M, Steinbrueckner B. Comparison of different DNA fingerprinting techniques for molecular typing of Bartonella henselae isolates. J Clin Microbiol. 1998;36:2973–2981. doi: 10.1128/jcm.36.10.2973-2981.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sander A, Ruess M, Deichmann K, Bohm N, Bredt W. Two different genotypes of Bartonella henselae in children with cat-scratch disease and their pet cats. Scand J Infect Dis. 1998;30:387–391. doi: 10.1080/00365549850160693. [DOI] [PubMed] [Google Scholar]
- 32.Slater L N, Welch D F, Min K W. Rochalimaea henselae causes bacillary angiomatosis and peliosis hepatis. Arch Intern Med. 1992;152:602–606. [PubMed] [Google Scholar]
- 33.Welch D F, Pickett D A, Slater L N, Steigerwalt A G, Brenner D J. Rochalimaea henselae sp. nov., a cause of septicemia, bacillary angiomatosis, and parenchymal bacillary peliosis. J Clin Microbiol. 1992;30:275–280. doi: 10.1128/jcm.30.2.275-280.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]