Abstract
Purpose
Carbapenem-resistant Klebsiella pneumoniae (CRKP) is one of the leading causes of healthcare-associated infections (HAIs) and is particularly pervasive in intensive care units (ICUs). This study takes ICU layout as the research object, and integrates clinical data and bacterial genome analysis to clarify the role of separate, small wards within the ICU in controlling the transmission of CRKP.
Methods
This study prospectively observed the carriage and spread of CRKP from a long-term in-hospital patient (hereafter called the Patient) colonized with CRKP in the gut and located in a separate, small ward within the ICU. The study also retrospectively investigated CRKP-HAIs in the same ICU. The relationship and transmission between CRKP isolates from the Patient and HAI events in the ICU were explored with comparative genomics.
Results
In this study, 65 CRKP-HAI cases occurred during the investigation period. Seven CRKP-HAI outbreaks were also observed. A total of 95 nonrepetitive CRKP isolates were collected, including 32 strains from the Patient in the separate small ward. Phylogenetic analysis based on core genome single-nucleotide polymorphism (cgSNP) showed that there were five possible CRKP clonal transmission events and two clonal outbreaks (A1, A2) during the study. CRKP strains from the Patient did not cause CRKP between-patient transmission or outbreaks in the ICU during the 5-year study period.
Conclusion
The presence of a long-term hospitalized patient carrying CRKP and positioned in a separate, small ward did not lead to CRKP transmission or infection outbreaks in the ICU. Combining a small-ward ICU layout with normative HAI control measures for multidrug-resistant pathogen infection was effective in reducing CRKP transmission.
Supplementary Information
The online version contains supplementary material available at 10.1007/s00134-022-06881-0.
Keywords: Carbapenem-resistant Klebsiella pneumoniae, ICU, Separate small wards, Whole-genome sequencing, Phylogenetic analysis
Introduction
The increasing prevalence of antimicrobial resistance is a serious threat to global public health [1, 2]. In 2017, the World Health Organization (WHO) listed carbapenem-resistant Enterobacteriaceae (CRE) as one of the most troubling types of resistant bacteria, and Klebsiella pneumoniae is the most notorious CRE [3]. In China, carbapenem-resistant K. pneumoniae (CRKP) accounts for about 64% of CRE infections [4]. CRKP resists carbapenems mainly via the production of carbapenemases [4, 5]. Global dissemination of CRKP has been largely attributed to the expansion of K. pneumoniae clonal group CG258, of which the main sequence types (STs) are ST258 and ST11; ST11 remains the most prevalent clone in China [6, 7]. Nosocomial infections frequently occur in intensive care units (ICUs) because of the complex ICU environment, the critical condition of the patients, the frequency of invasive operations, the heavy consumption of antibiotics, and the lack of sufficient medical staff [8, 9]. CRKP is one of the most common agents for healthcare-associated infection (HAI) and is of paramount concern in the ICU owing to its significant morbidity and mortality [10]. Infections caused by CRKP are difficult to treat because they often display resistance to multiple antibiotics, limiting therapeutic options. Therefore, it is critical to prevent the transmission of CRKP in ICUs and other healthcare settings [11, 12]. As early as 1996, the U.S. Centers for Disease Control and Prevention (CDC) recommended the implementation of single-room and contact precaution measures (SCP; single room, gown and gloves) to prevent infection by multidrug-resistant organisms (MDROs) [13].
Transmission of MDROs among patients is the main source of HAI outbreaks, especially in clinical units with crowded patients, more invasive operations, and high antibiotic consumption, such as neurosurgery and ICU settings [14]. In China, ICUs are often designed as a single large bay for the purpose of easier patient monitoring; each large room can accommodate 20–30 patients at a time, and the beds are relatively closely spaced [11, 15]. This crowded environment may contribute to the frequent occurrence of HAIs [16]. Previous studies on separate, small wards in ICUs focused on whether strict isolation measures reduced the prevalence of MDROs [17], but the impact of ward size itself on the transmission of MDROs remains poorly understood. In particular, there is a lack of studies on MDRO spread confirmed by high-resolution genomic methods [18].
In this study, a patient in a separate, small ward within the ICU (hereafter called the Patient) and the rest of the patients in the ICU were taken as the research objects. The occurrence of CRKP-HAIs in the ICU was monitored from February 2015 to December 2019. The outbreak and transmission of CRKP infections were analyzed by whole-genome sequencing, and the role of separate, small wards within the ICU in preventing transmission of CRKP was clarified.
Methods
Ward and bed setting in the ICU
There were three separate wards (Room-1, -2, -3) within the ICU, which could accommodate three, four, and two patients (Fig. 1). Each ward was equipped with a separate handwashing basin and hand disinfectant. There was a public toilet in a separate room outside the wards. There were two nurses to every three patients, with an individual nurse potentially being responsible for patients in two different wards at the same time.
Fig. 1.
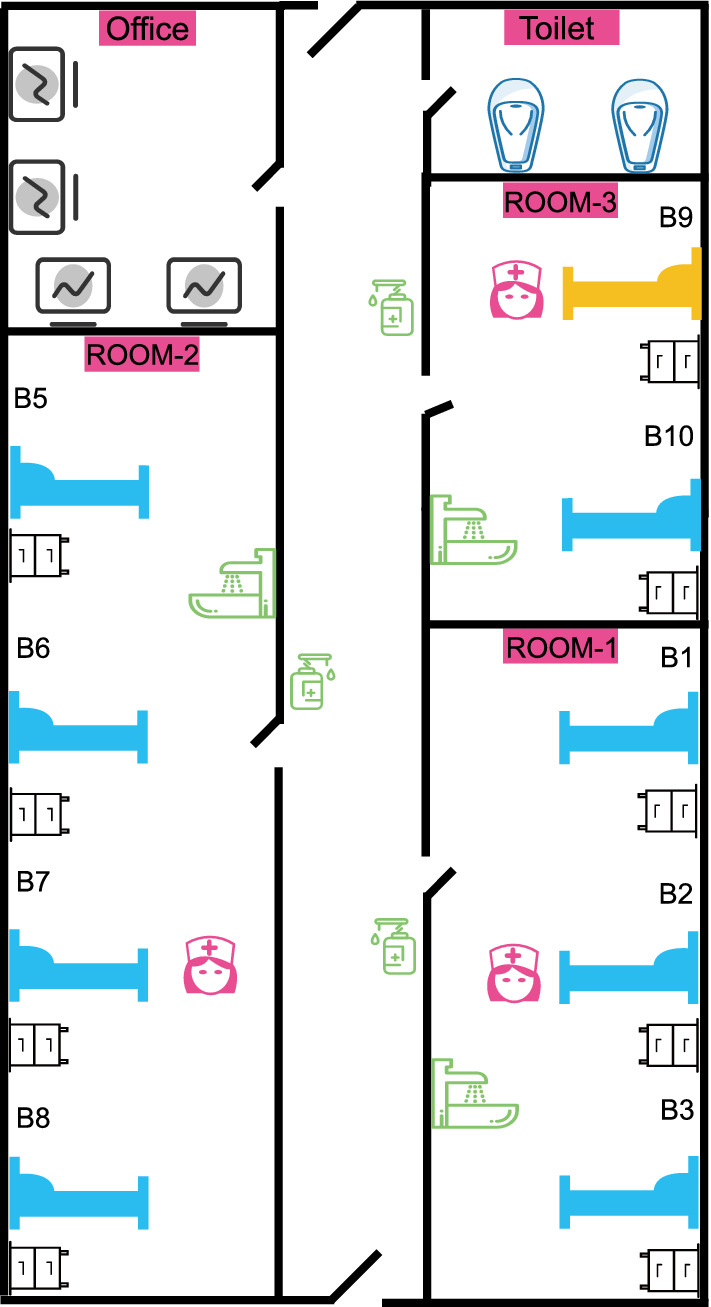
Schematic showing the ICU wards. There were three individual small wards (Room-1, Room-2, and Room-3), a doctor’s office, and a public toilet. The yellow bed shows the location of the patient with CRKP (Room-3-B9)
Prevention and control strategies for nosocomial infection in the ICU
The routine hospital infection control strategies in the ICU included personnel training, hand hygiene, environmental disinfection, medical waste management, treatment of patient excreta, surface cleaning and disinfection of furniture, monitoring and reporting of HAIs, infection prevention and control protocols for invasive procedures, occupational protection, etc. No additional infection control measures were added during the study.
Definition of nosocomial infection
CRKP-HAI was defined as an infection that occurred more than 48 h after admission, with CRKP confirmed as the cause through the culture of clinical samples [18]. A CRKP-HAI outbreak was defined as the occurrence of more than three infections in the ICU within a month. CRKP strains with less than 25 cgSNPs (core genome single-nucleotide polymorphisms) were classed as the same clone, and clonal spread was defined as ≥ 25 cgSNPs. A clonal outbreak of CRKP-HAI was defined as an HAI outbreak caused by a single CRKP clone [19, 20].
HAI diagnosis and patient management
From February 2015 to August 2019, we prospectively collected monthly fecal samples from the Patient for CRKP screening. The Patient was hospitalized for colon cancer in February 2015. Physical examination showed that the Patient had comorbidity diseases such as coronary heart disease and diabetes. The Patient stayed in Room-3, Bed 9 (Room-3-B9) from admission until the end of the study (Fig. 1). The Patient twice tested positive for bloodstream infections caused by CRKP: on April 11, 2015 and April 27, 2015. A combination of imipenem/cilastatin and tigecycline was used for therapy, and the Patient subsequently recovered. No further CRKP infections were observed in the Patient. The medical records and HAI reports for other patients admitted to the ICU were retrieved, and the occurrence of CRKP-HAI in the other patients in the ICU over the same period was retrospectively observed, including bloodstream infections (BSIs), urinary tract infections (UTIs), intra-abdominal infections (IAIs), central nervous system infections (CNS), etc.
Bacterial identification and antimicrobial susceptibility testing (AST)
A single bacterial clone was identified by matrix-assisted laser desorption/ionization time-of-flight mass spectrometry (MALDI-TOF/MS) (Bruker, Bremen, Germany) [21]. Antimicrobial susceptibility (minimum inhibitory concentrations, MICs) of 14 antimicrobial agents was determined by agar dilution methods, except for colistin and tigecycline, for which the broth microdilution method was used. The MIC results were interpreted on the basis of the breakpoints described in the Clinical and Laboratory Standards Institute document M100-S25 except for tigecycline, for which the criteria of the U.S. Food and Drug Administration (FDA) were followed.
Whole-genome sequencing (WGS) and genomic analysis
Genomic deoxyribonucleic acid (DNA) of all isolates was extracted from pure cultures of K. pneumoniae using a Gentra Puregene Yeast/Bact. Kit (QIAGEN, Hilden, Germany). Whole-genome sequencing (WGS) was performed on the extracted DNA by Novogene (Beijing, China) using the Illumina HiSeq sequencing platform. Alignment of antimicrobial-resistance (AMR) genes was performed through the ResFinder platform (https://cge.cbs.dtu.dk/services/ResFinder). Multilocus sequence typing (MLST) was performed on bacteria (https://cge.cbs.dtu.dk/services/MLST). Virulence genes were identified by blasting the Virulence Factor Database (VFDB; http://www.mgc.ac.cn/VFs).
Phylogenetic analysis
Roary v3.13.0 [22] was used to perform a pan genome analysis and create core gene alignments on 95 isolates, and SNP alignments were extracted from the core gene alignments. Using K. pneumoniae (GCF_000240185) as a reference sequence [23], Snippy v4.6.0 (https://github.com/tseemann/snippy) was used to perform reference-based mapping and identify SNPs [24]. The alignment file was filtered from variants with elevated densities of base substitutions as putative recombination events by Gubbins version 2.4.1 [25]. PHYLOViZ 2.0 (http://www.phyloviz.net/tutorials.html) was used to construct a maximum likelihood tree [26].
Constructing the putative transmission map
Genome and patient information were integrated to construct the putative map of clonal transmission of CRKP and clonal HAI outbreaks. Taking the first CRKP detected during the study as the index isolate, the minimalist transmission map was obtained by calculating the genetic distance among all strains, and the most likely transmission map was determined when the total genetic distance between strains was at a minimum (the minimum number of cgSNP). Patient information was used to distinguish transmission maps with the same possibility according to genetic distance [19].
Statistical analysis
Statistical analyses were performed using the Wilcoxon rank-sum test, Chi-square test or Fisher’s exact test. Statistical significance was determined at p < 0.05. Genome-wide association analysis was performed with Scoary v1.6.16.
Data availability
The whole-genome sequences of 95 K. pneumoniae isolates have been deposited in the GenBank database under BioProject accession no. PRJNA839803.
Ethical statement
This study was conducted according to the Declaration of Helsinki, and approval was obtained from the Medical Ethics Committee at The First Affiliated Hospital of Zhejiang University. The ethics permit number is IIT20220585A.
Results
Occurrence of CRKP-HAIs and strain identification
A total of 32 CRKP isolates were prospectively collected from the Patient in the separate small ward from February 2015 to August 2019. The Patient tested positive for bloodstream infection (BSI) caused by CRKP on April 11, 2015 and April 27, 2015. Since then, the Patient did not develop a HAI caused by CRKP. A total of 30 CRKP isolates were detected in the fecal screening of the Patient.
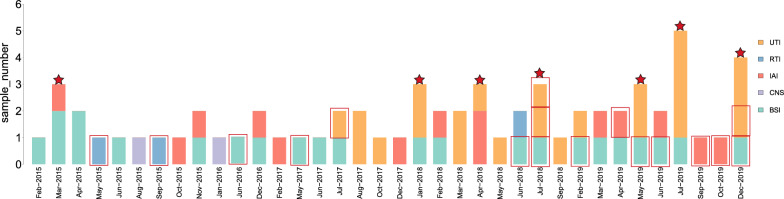
Over the same period, 63 CRKP-HAIs were diagnosed in other patients in the ICU: 11 in 2015 (17.5%), 4 in 2016 (6.3%), 9 in 2017 (14.3%), 17 in 2018 (27%), and 22 in 2019 (34.9%). HAIs were mainly comprised of UTIs (23, 36.5%), followed by BSIs (21, 33.3%), IAIs (14, 22.2%), respiratory tract infections (RTIs) (3, 4.8%), and CNS (2, 3.2%).
Seven CRKP-HAI outbreaks were observed (Fig. 2). Three outbreaks occurred in 2019 with 12 episodes of CRKP-HAI: nine UTIs (75%) and 3 BSIs (25%). Three outbreaks occurred in 2018 with nine episodes: five UTIs (55.6%), two BSIs (22.2%), and two IAIs (22.2%). One outbreak occurred in 2015 with three episodes: two BSIs (66.7%) and one IAI (33.3%).
Fig. 2.
Occurrence of CRKP-HAIs in the ICU from February 2015 to December 2019. The abscissa indicates the month of infection, the ordinate indicates the number of infection cases, the different colors indicate different infection sites, and a red border indicates that the patient had the HAI before being transferred to the ICU. The red stars indicate the months in which a CRKP outbreak occurred in the ICU. UTI urinary tract infection, RTI respiratory tract infection, IAI intra-abdominal infection, CNS central nervous system infection, BSI bloodstream infection
Transmission of CRKP in the ICU
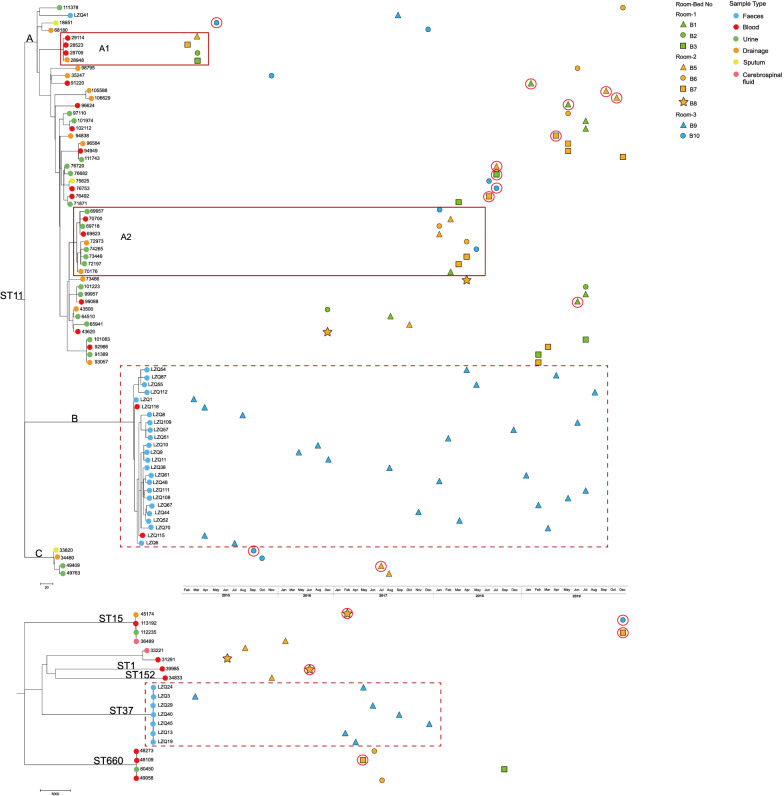
WGS of all 95 CRKP strains was performed. The predominant clone in the ICU was ST11 CRKP (76/80%), followed by ST37 (7/7.4%), ST660 (4/4.2%), ST15 (4/4.2%), ST893 (1/1.1%), ST152 (1/1.1%), ST1 (1/1.1%), and an unknown type (1/1.1%). The cgSNP-based phylogenetic analysis of the 95 CRKP strains (Fig. 3) demonstrated that there were three clades in the ST11 evolutionary tree (clade A, n = 48; clade B, n = 24; clade C, n = 4). Clade A was the most prevalent infective lineage in the ICU; all isolates in clade B were from the Patient; and clade C consisted of four CRKP isolates, which appeared only briefly and were not prevalent in the ICU.
Fig. 3.
Isolation time and phylogenetic analysis of 95 CRKP isolates in the ICU. The upper figure shows the phylogenetic analysis of 76 ST11-CRKP isolates, and the lower figure shows the phylogenetic analysis of 19 other ST-type CRKP isolates. The dots on the clade indicate the sample origin of the isolate. Solid red rectangles represent clonal outbreaks in clade A, and dashed red rectangles represent CRKPs (ST11 and ST37) isolated from the Patient (Room-3-B9). Wards and beds are represented by a combination of colors and shapes. Isolates with HAIs occurring before ICU admission are indicated by red circles
The Patient in the separate small ward did not leave the ward in the 5-year study period. A total of 32 CRKP strains were collected from the Patient, including 25 for ST11 and 7 for ST37. Except for the two ST11 strains that caused the two BSI episodes, the strains were all gut-colonizing bacteria. Phylogenetic analysis showed that most of the ST11 strains from the Patient (24/25) were in clade B, and the ST37 strains were clustered in a single clade (Fig. 3). Compared with the other isolates from the Patient, one ST11 strain (LZQ41) located in clade A had a median cgSNP of 275 (range 268–278), indicating that LZQ41 might have been a temporary colonization from other patients in the ICU. ST37 CRKP was never isolated from other patients during the study period. This result shows that, from February 2015 to December 2019, neither the BSI isolates nor the gut-colonizing strains from the Patient caused transmission in the ICU, suggesting that the long duration stay by a patient with persistent gut-colonizing CRKP in a small ward was not the source of CRKP transmission in the ICU.
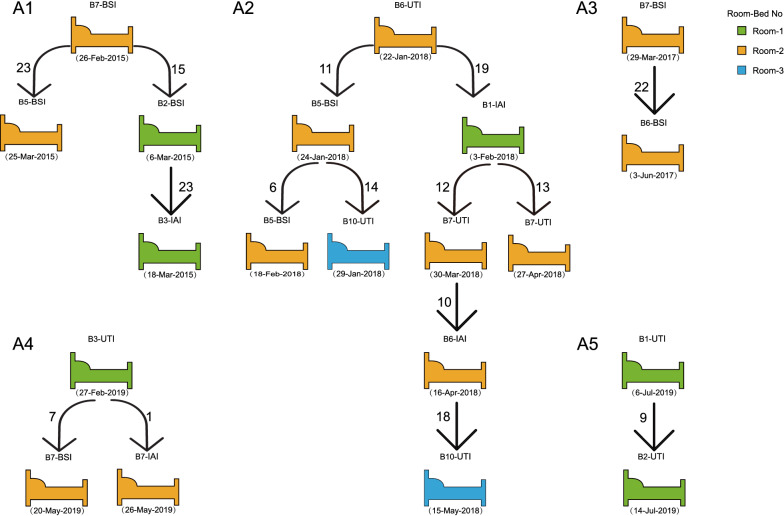
Alongside the cgSNP and phylogenetic analysis, five clonal transmission events were monitored (Fig. 4), involving 20 isolates in total from Room-1 (6, 30%), Room-2 (12, 60%), and Room-3 (2, 10%), but never connected with the Patient. The infection types of these clonal transmissions were BSI (8, 42.1%), UTI (7, 36.8%), and IAI (4, 21.2%).
Fig. 4.
From 2015 to 2019, clonal transmission and outbreaks occurred in the ICU. Green beds represent Room-1, orange beds represent Room-2, and blue beds represent Room-3. The arrows indicate different transmission maps. The bed number and infection type are marked above each hospital bed, and the date on which CRKP was detected is marked underneath each hospital bed. Numbers next to the arrows indicate the number of cgSNP between CRKP isolates when propagation events occurred. UTI urinary tract infection, IAI intra-abdominal infection, BSI bloodstream infection
Pathway of CRKP-HAI clone spread in the ICU
A phylogenetic tree of CRKP based on cgSNPs indicated that the epidemic clade A (n = 48/76, 63.16%) of ST11 caused two possible clonal outbreaks (A1, A2) (Fig. 3). Combining WGS and patient clinical information, the possible transmission maps of the CRKP clonal outbreaks were reconstructed (Fig. 4). The A1 clonal outbreak comprised four patients in total from Room-1 and Room-2, and occurred from February 26 to March 18, 2015. The A1 outbreak lasted for a total of 22 days, starting with patient Room-2-B7 (BSI) and spreading to patients Room-1-B2 (BSI) and Room-2-B5 (BSI), and then spreading within Room-1 from patient Room-1-B2 to patient Room-1-B3 (IAI). The A2 clonal outbreak comprised five patients and occurred from January 22 to May 15, 2018. This outbreak lasted a total of 113 days, starting from patient Room-2-B6 (UTI), spreading within the same ward to patient Room-2-B5 (BSI) and also to patient Room-1-B1 (IAI); it then spread from patient Room-2-B5 to patient Room-3-B10 (UTI), and from patient Room-1-B1 to patient Room-2-B7 (UTI).
Antimicrobial resistance (AMR) genetic analysis and antimicrobial susceptibility testing (AST) of epidemic clone ST11
AMR gene analysis and AST showed that clade A (except LZQ41 from the Patient) carried more AMR genes than clade B (isolates from the Patient; p < 0.05, Supplementary Material 1), and the AMR profile had a broader phenotype in clade A, with more frequent resistance to aminoglycoside. Most of the strains isolated in 2018 and 2019 carried aminoglycoside-resistance genes and showed resistance to aminoglycoside antibiotics, indicating that ST11-CRKP in the ICU was in the process of continuous dynamic evolution. Compared with clade A, clade B tended to harbor the quinolone-resistance genes oqxA and oqxB. The AMR genes and resistance phenotypes carried by clade C were similar to those of clade A (Fig. S1).
ST11-CRKP virulence genes
Virulence gene analysis revealed that clade A (except LZQ41 from the Patient) carried more acquired siderophore and hypermucoidy genes than clade B (isolates from the Patient; p < 0.05, Supplementary Material 1), including the siderophores enterobactin genes (entC, entE, entF), yersiniabactin genes (ybtA, ybtE, ybtP, ybtS, ybtT, ybtU), aerobactin genes (iucA, iucC), salmochelin gene (IroE), and hypermucoidy genes (rmpA, rmpA2). In addition, compared with clade A, clade B carried more pullulanase genes (pul), which are involved in carbohydrate metabolism. Clade C had a similar virulence gene profile to clade A but did not carry the hypervirulent genes iucA, iucC, rmpA, rmpA2, etc. These data indicate that clade A likely had higher virulence (Fig. S2).
Discussion
Because of the limited therapeutic options and extremely high mortality, CRKP infection has become one of the most troubling problems in healthcare [27, 28]. CRKP infections account for 64% of CRE infection in China [4]. In Europe, the incidence of CRKP infection increased sharply from 1% in 2009 to 15% in 2010 and 34% in 2016 [29]. CRKP can lead to different types of infection, including BSIs, UTIs, and CNS, with a mortality rate of up to 42.1% [30]. Both the WHO and the CDC list CREs, especially CRKP, as the most urgent pandrug-resistant bacteria to be controlled, and have formulated prevention and control guidelines [31]. The prevention and control of CRKP require timely identification of infection or colonization, patient isolation, minimization of physical contact, disinfection of the environment, hand hygiene, and rational management of antibiotics use [32]. Bernard et al. evaluated the application of CRKP infection–prevention measures by comparing 34 related studies. Active screening, rational use of antibiotics, and exposure prevention effectively reduced the spread of CRKP, but staff behavioral changes were a greater barrier [33, 34].
A CRKP outbreak in Israel that was successfully controlled demonstrated that admission screening for CRKP, strict isolation of patients, and a separate medical cohort are of great value for timely control of a CRKP outbreak [35]. Severe patient illness and substandard healthcare procedures, such as substantial invasive operations, heavy consumption of antibiotics, and an insufficient number of medical staff, often hold back the implementation of infection–prevention measures, and make ICUs particularly likely to have a high incidence and prevalence of CRKP infections [8, 9]. Surveillance data show that the prevalence of CRKP in ICUs can be as high as 20.8–48.1% [36, 37], which is significantly higher than that in other clinical departments. A major factor in this is the high incidence of CRKP clone transmission in ICUs. Molecular epidemiological studies showed that ST11 was the predominant clone in ICUs in China [38], which may be because of excessive patient crowding in large wards [11, 14]. Therefore, changing the ward layout may help control the prevalence of MDROs, including CRKP. Halaby et al. showed that in large-room ICUs where patients were concentrated, extended-spectrum β-lactamase–producing K. pneumoniae (ESBL-Kp) was continuously detected even after the implementation of contact isolation, but detection of ESBL-Kp decreased by 83.3% after the implementation of single-room isolation for patients [18]. Biehl et al. found similar benefits to single-room isolation in a multicenter prospective study on HAIs [17]. However, these studies focused on observing the changes in the prevalence of antibiotic-resistant bacteria, without analyzing the specific mechanisms through which single-room wards can reduce MDRO transmission. Thus, whether the decrease in MDROs was caused by blocked transmission or improved patient care remained unknown.
In this study, the Patient in the separate, small ward in the ICU and the other patients in the ICU were selected as the research objects (Fig. 1), and CRKP-HAIs in the ICU were observed for 5 years. A total of 65 CRKP-HAI cases were monitored in the ICU during the 5 years, including two BSI episodes in the Patient. HAIs in the ICU mainly comprised BSIs and UTIs, which accounted for 70.8% of all HAIs (Fig. 2). WGS analysis of 95 CRKP isolates collected in the past 5 years showed that ST11 was the main type of CRKP. However, phylogenetic analysis showed that the strains isolated from the Patient and from other patients in the ICU belonged to different clades. Intestinal colonized strains from the Patient did not spread outward and cause transmission in the ICU (Fig. 3). CRKP-HAIs in other patients in the ICU were caused by strains of a new subclone. SY37 CRKP, a specific ST type isolated only from the Patient, also did not cause any HAIs.
The five clonal transmission events and two clonal outbreaks that occurred in the ICU were studied by integrating patient information, HAI data, microbiological examination, and WGS analysis (Fig. 4). The results showed that all clonal transmission and outbreaks were caused by a new subclone that spread in the ICU within a short period of time. None of the clonal outbreaks in the ICU involved the Patient. These results clearly demonstrate that establishing a separate, small ward in the ICU can effectively control CRKP outbreaks and epidemics without additional infection control measures. Our results also indicate that blocking the transmission of CRKP from outside the ICU greatly helps control the prevalence of MDROs in the ICU. Bracco et al. also explored the role of ICU building layout in the control of HAIs. Their study compared the incidence of HAIs in patients staying in large rooms or separate small wards in a ICU, and found that HAI incidence for patients in the small wards was reduced by 68% compared with that for patients in large rooms [39]. However, the study was limited to bacterial detection rates and lacked genomic evidence to explain the mechanism. Our study showed that small ICU wards are effective for CRKP prevention because they reduce or avoid transmission among patients.
In addition to the physical barrier provided by the small ward, reduced CRKP transmission may have been related to bacterial virulence and antibiotic resistance. Virulence and AMR genetic analyses of CRKP showed that ST11 clade A, which led to the CRKP outbreak, carried more acquired siderophore and hypermucoidy genes (such as entC, entE, entF, ybtA, ybtE, ybtP, ybtS, ybtT, ybtU, rmpA, rmpA2, etc.), whereas clade B carried more pullulanase genes (pul), which are involved in carbohydrate metabolism (Figs. S1, S2) [2]. Clade A had more aminoglycoside-resistance genes, and clade B harbored more quinolone-resistance genes (oqxA and oqxB). The AMR genes and resistance phenotypes of clade C were similar to those of clade A (Fig. S1). However, the differences in genes related to virulence and resistance cannot explain the prevalence of clade A in the ICU. Although the virulent genes in CRKP are beneficial to its pathogenesis [40, 41], the oqx genes in clade B can promote its survival in the environment and thus spread in hospitals. It will be necessary to study the genetic factors related to transmission advantages for different clades in the future.
Our study has some limitations. We analyzed the ICU CRKP-HAIs retrospectively, and it is impossible to determine the impact of other infection control measures. In the future, a prospective study should be carried out to continuously track the patients admitted to the ICU, along with the implementation of various infection control measures and the collection of epidemiological evidence and relevant environmental microbial strains. Genomic comparison of outbreak strains in a prospective study would add to the findings in the present study.
Conclusion
In conclusion, by integrating epidemiological data with WGS analysis, we described CRKP outbreaks and transmission in an ICU with separate small wards during a 5-year period. We found that a patient carrying CRKP who was hospitalized in a separate, small ward for the entire study period did not spread CRKP to the other ICU patients. Our study clarifies the value of small-ward layouts in the control of CRKP-HAIs in ICUs; whole genomic analysis indicated that the transmission block of CRKP is likely the major explanation for the lack of spread.
Supplementary Information
Below is the link to the electronic supplementary material.
Fig. S1 Antibiotic resistance genetic analysis and antibiotic susceptibility test results for ST11-CRKP. Green squares indicate the presence of antibiotic-resistance genes, and white indicates the absence of antibiotic-resistance genes. Orange squares indicate the presence of relevant antibiotic-resistant phenotypes, and white indicates antibiotic sensitivity. (PDF 533 KB)
Fig. S2 The virulence gene alignment results for ST11-CRKP. Blue squares represent the presence of virulence genes, and white represent their absence. (PDF 739 KB)
Funding
This project was supported by the National Key Research and Development Program of China (2021YFC2300300), the National Natural Science Foundation of China (82072314), the Research Project of Jinan Microecological Biomedicine Shandong Laboratory (JNL-2022006B, JNL-2022011B), and the CAMS Innovation Fund for Medical Sciences (2019-I2M-5-045).
Declarations
Conflicts of interest
The authors declare that they have no conflicts of interest.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Xiaohui Chi, Xiaohua Meng and Luying Xiong contributed equally to this work.
Contributor Information
Beiwen Zheng, Email: zhengbw@zju.edu.cn.
Yonghong Xiao, Email: xiaoyonghong@zju.edu.cn.
References
- 1.Bassetti M, Giacobbe DR, Giamarellou H, Viscoli C, Daikos GL, Dimopoulos G, De Rosa FG, Giamarellos-Bourboulis EJ, Rossolini GM, Righi E, Karaiskos I, Tumbarello M, Nicolau DP, Viale PL, Poulakou G, M. Critically Ill Patients Study Group of the European Society of Clinical, D. Infectious, C. Hellenic Society of, A. Societa Italiana di Terapia Management of KPC-producing Klebsiella pneumoniae infections. Clin Microbiol Infect. 2018;24(2):133–144. doi: 10.1016/j.cmi.2017.08.030. [DOI] [PubMed] [Google Scholar]
- 2.Holt KE, Wertheim H, Zadoks RN, Baker S, Whitehouse CA, Dance D, Jenney A, Connor TR, Hsu LY, Severin J, Brisse S, Cao H, Wilksch J, Gorrie C, Schultz MB, Edwards DJ, Nguyen KV, Nguyen TV, Dao TT, Mensink M, Minh VL, Nhu NT, Schultsz C, Kuntaman K, Newton PN, Moore CE, Strugnell RA, Thomson NR. Genomic analysis of diversity, population structure, virulence, and antimicrobial resistance in Klebsiella pneumoniae, an urgent threat to public health. Proc Natl Acad Sci USA. 2015;112(27):E3574–E3581. doi: 10.1073/pnas.1501049112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wang M, Earley M, Chen L, Hanson BM, Yu Y, Liu Z, Salcedo S, Cober E, Li L, Kanj SS, Gao H, Munita JM, Ordonez K, Weston G, Satlin MJ, Valderrama-Beltran SL, Marimuthu K, Stryjewski ME, Komarow L, Luterbach C, Marshall SH, Rudin SD, Manca C, Paterson DL, Reyes J, Villegas MV, Evans S, Hill C, Arias R, Baum K, Fries BC, Doi Y, Patel R, Kreiswirth BN, Bonomo RA, Chambers HF, Fowler VG, Jr, Arias CA, van Duin D, I. Multi-Drug Resistant Organism Network Clinical outcomes and bacterial characteristics of carbapenem-resistant Klebsiella pneumoniae complex among patients from different global regions (CRACKLE-2): a prospective, multicentre, cohort study. Lancet Infect Dis. 2022;22(3):401–412. doi: 10.1016/S1473-3099(21)00399-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zhang R, Liu L, Zhou H, Chan EW, Li J, Fang Y, Li Y, Liao K, Chen S. Nationwide surveillance of clinical carbapenem-resistant Enterobacteriaceae (CRE) strains in China. EBioMedicine. 2017;19:98–106. doi: 10.1016/j.ebiom.2017.04.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bi S, Yao X, Huang C, Zheng X, Xuan T, Sheng J, Xu K, Zheng B, Yang Q. Antagonistic effect between tigecycline and meropenem: successful management of KPC-producing Klebsiella pneumoniae infection. Infection. 2019;47(3):497–500. doi: 10.1007/s15010-019-01274-w. [DOI] [PubMed] [Google Scholar]
- 6.Chen L, Mathema B, Chavda KD, DeLeo FR, Bonomo RA, Kreiswirth BN. Carbapenemase-producing Klebsiella pneumoniae: molecular and genetic decoding. Trends Microbiol. 2014;22(12):686–696. doi: 10.1016/j.tim.2014.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wu X, Han H, Chen C, Zheng B. Genomic characterisation of a colistin-resistant Klebsiella pneumoniae ST11 strain co-producing KPC-2, FloR, CTX-M-55, SHV-12, FosA and RmtB causing a lethal infection. J Glob Antimicrob Resist. 2019;19:78–80. doi: 10.1016/j.jgar.2019.08.023. [DOI] [PubMed] [Google Scholar]
- 8.Weist K, Pollege K, Schulz I, Ruden H, Gastmeier P. How many nosocomial infections are associated with cross-transmission? A prospective cohort study in a surgical intensive care unit. Infect Control Hosp Epidemiol. 2002;23(3):127–132. doi: 10.1086/502021. [DOI] [PubMed] [Google Scholar]
- 9.Crnich CJ, Safdar N, Maki DG. The role of the intensive care unit environment in the pathogenesis and prevention of ventilator-associated pneumonia. Respir Care. 2005;50(6):813–836. [PubMed] [Google Scholar]
- 10.Papagiannitsis CC, Izdebski R, Baraniak A, Fiett J, Herda M, Hrabak J, Derde LP, Bonten MJ, Carmeli Y, Goossens H, Hryniewicz W, Brun-Buisson C, Gniadkowski M, W.P. Mosar Wp, W.P.s. groups Survey of metallo-beta-lactamase-producing Enterobacteriaceae colonizing patients in European ICUs and rehabilitation units, 2008–2011. J Antimicrob Chemother. 2015;70(7):1981–1988. doi: 10.1093/jac/dkv055. [DOI] [PubMed] [Google Scholar]
- 11.Wei L, Wu L, Wen H, Feng Y, Zhu S, Liu Y, Tang L, Doughty E, van Schaik W, McNally A, Zong Z. Spread of Carbapenem-Resistant Klebsiella pneumoniae in an Intensive Care Unit: a whole-genome sequence-based prospective observational study. Microbiol Spectr. 2021;9(1):e0005821. doi: 10.1128/Spectrum.00058-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Debby BD, Ganor O, Yasmin M, David L, Nathan K, Ilana T, Dalit S, Smollan G, Galia R. Epidemiology of carbapenem resistant Klebsiella pneumoniae colonization in an intensive care unit. Eur J Clin Microbiol Infect Dis. 2012;31(8):1811–1817. doi: 10.1007/s10096-011-1506-5. [DOI] [PubMed] [Google Scholar]
- 13.Abad C, Fearday A, Safdar N. Adverse effects of isolation in hospitalised patients: a systematic review. J Hosp Infect. 2010;76(2):97–102. doi: 10.1016/j.jhin.2010.04.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gu B, Bi R, Cao X, Qian H, Hu R, Ma P. Clonal dissemination of KPC-2-producing Klebsiella pneumoniae ST11 and ST48 clone among multiple departments in a tertiary teaching hospital in Jiangsu Province, China. Ann Transl Med. 2019;7(23):716. doi: 10.21037/atm.2019.12.01. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Qiao F, Huang W, Gao S, Cai L, Zhu S, Wei L, Kang Y, Tao C, Zong Z. Risk factor for intestinal carriage of carbapenem-resistant Acinetobacter baumannii and the impact on subsequent infection among patients in an intensive care unit: an observational study. BMJ Open. 2020;10(9):e035893. doi: 10.1136/bmjopen-2019-035893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Suleyman G, Alangaden GJ. Nosocomial fungal infections: epidemiology, infection control, and prevention. Infect Dis Clin North Am. 2021;35(4):1027–1053. doi: 10.1016/j.idc.2021.08.002. [DOI] [PubMed] [Google Scholar]
- 17.Biehl LM, Higgins P, Wille T, Peter K, Hamprecht A, Peter S, Dorfel D, Vogel W, Hafner H, Lemmen S, Panse J, Rohde H, Klupp EM, Schafhausen P, Imirzalioglu C, Falgenhauer L, Salmanton-Garcia J, Stecher M, Vehreschild JJ, Seifert H, Vehreschild M. Impact of single-room contact precautions on hospital-acquisition and transmission of multidrug-resistant Escherichia coli: a prospective multicentre cohort study in haematological and oncological wards. Clin Microbiol Infect. 2019;25(8):1013–1020. doi: 10.1016/j.cmi.2018.12.029. [DOI] [PubMed] [Google Scholar]
- 18.Halaby T, Al Naiemi N, Beishuizen B, Verkooijen R, Ferreira JA, Klont R, Vandenbroucke-Grauls C. Impact of single room design on the spread of multi-drug resistant bacteria in an intensive care unit. Antimicrob Resist Infect Control. 2017;6:117. doi: 10.1186/s13756-017-0275-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Snitkin ES, Zelazny AM, Thomas PJ, Stock F, Henderson DK, Palmore TN, Segre JA, N.C.S.P. Group Tracking a hospital outbreak of carbapenem-resistant Klebsiella pneumoniae with whole-genome sequencing. Sci Transl Med. 2012;4(148):148ra116. doi: 10.1126/scitranslmed.3004129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.David S, Reuter S, Harris SR, Glasner C, Feltwell T, Argimon S, Abudahab K, Goater R, Giani T, Errico G, Aspbury M, Sjunnebo S, Eu SWG, Group ES, Feil EJ, Rossolini GM, Aanensen DM, Grundmann H. Epidemic of carbapenem-resistant Klebsiella pneumoniae in Europe is driven by nosocomial spread. Nat Microbiol. 2019;4(11):1919–1929. doi: 10.1038/s41564-019-0492-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zheng B, Chen Y, Violetta L, Xiao Y, Li L. Bloodstream infections caused by Entero-bacteriaceae in China. Lancet Infect Dis. 2019;19(8):810–811. doi: 10.1016/S1473-3099(19)30352-4. [DOI] [PubMed] [Google Scholar]
- 22.Page AJ, Cummins CA, Hunt M, Wong VK, Reuter S, Holden MT, Fookes M, Falush D, Keane JA, Parkhill J. Roary: rapid large-scale prokaryote pan genome analysis. Bioinformatics. 2015;31(22):3691–3693. doi: 10.1093/bioinformatics/btv421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Liu P, Li P, Jiang X, Bi D, Xie Y, Tai C, Deng Z, Rajakumar K, Ou HY. Complete genome sequence of Klebsiella pneumoniae subsp. pneumoniae HS11286, a multidrug-resistant strain isolated from human sputum. J Bacteriol. 2012;194(7):1841–1842. doi: 10.1128/JB.00043-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Stohr J, Kluytmans-van den Bergh MFQ, Weterings V, Rossen JWA, Kluytmans J. Distinguishing bla KPC Gene-Containing IncF Plasmids from Epidemiologically Related and Unrelated Enterobacteriaceae Based on Short- and Long-Read Sequence Data. Antimicrob Agents Chemother. 2021 doi: 10.1128/AAC.00147-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Croucher NJ, Page AJ, Connor TR, Delaney AJ, Keane JA, Bentley SD, Parkhill J, Harris SR. Rapid phylogenetic analysis of large samples of recombinant bacterial whole genome sequences using Gubbins. Nucleic Acids Res. 2015;43(3):e15. doi: 10.1093/nar/gku1196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lieberman TD, Michel JB, Aingaran M, Potter-Bynoe G, Roux D, Davis MR, Jr, Skurnik D, Leiby N, LiPuma JJ, Goldberg JB, McAdam AJ, Priebe GP, Kishony R. Parallel bacterial evolution within multiple patients identifies candidate pathogenicity genes. Nat Genet. 2011;43(12):1275–1280. doi: 10.1038/ng.997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Acevedo Ugarriza LE, Michalik-Provasek J, Newkirk H, Liu M, Gill JJ, Ramsey J. Complete Genome Sequence of Klebsiella pneumoniae Myophage Magnus. Microbiol Resour Announc. 2019 doi: 10.1128/MRA.01049-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Asensio A, Oliver A, Gonzalez-Diego P, Baquero F, Perez-Diaz JC, Ros P, Cobo J, Palacios M, Lasheras D, Canton R. Outbreak of a multiresistant Klebsiella pneumoniae strain in an intensive care unit: antibiotic use as risk factor for colonization and infection. Clin Infect Dis. 2000;30(1):55–60. doi: 10.1086/313590. [DOI] [PubMed] [Google Scholar]
- 29.Di Pilato V, Errico G, Monaco M, Giani T, Del Grosso M, Antonelli A, David S, Lindh E, Camilli R, Aanensen DM, Rossolini GM, Pantosti A, A.-I.L.S.G.o.c.-p.K. pneumoniae The changing epidemiology of carbapenemase-producing Klebsiella pneumoniae in Italy: toward polyclonal evolution with emergence of high-risk lineages. J Antimicrob Chemother. 2021;76(2):355–361. doi: 10.1093/jac/dkaa431. [DOI] [PubMed] [Google Scholar]
- 30.Zhang H, Wang J, Zhou W, Yang M, Wang R, Yan X, Cai Y. Risk factors and prognosis of carbapenem-resistant Klebsiella pneumoniae infections in respiratory intensive care unit: a retrospective study. Infect Drug Resist. 2021;14:3297–3305. doi: 10.2147/IDR.S317233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.WHO . Prevention of hospital-acquired infections. WHO; 2002. [Google Scholar]
- 32.Kim NH, Han WD, Song KH, Seo HK, Shin MJ, Kim TS, Park KU, Ahn S, Yoo JS, Kim ES, Kim HB. Successful containment of carbapenem-resistant Enterobacteriaceae by strict contact precautions without active surveillance. Am J Infect Control. 2014;42(12):1270–1273. doi: 10.1016/j.ajic.2014.09.004. [DOI] [PubMed] [Google Scholar]
- 33.Borer A, Eskira S, Nativ R, Saidel-Odes L, Riesenberg K, Livshiz-Riven I, Schlaeffer F, Sherf M, Peled N. A multifaceted intervention strategy for eradication of a hospital-wide outbreak caused by carbapenem-resistant Klebsiella pneumoniae in Southern Israel. Infect Control Hosp Epidemiol. 2011;32(12):1158–1165. doi: 10.1086/662620. [DOI] [PubMed] [Google Scholar]
- 34.Okeah BO, Morrison V, Huws JC. Antimicrobial stewardship and infection prevention interventions targeting healthcare-associated Clostridioides difficile and carbapenem-resistant Klebsiella pneumoniae infections: a scoping review. BMJ Open. 2021;11(8):e051983. doi: 10.1136/bmjopen-2021-051983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cohen MJ, Block C, Levin PD, Schwartz C, Gross I, Weiss Y, Moses AE, Benenson S. Institutional control measures to curtail the epidemic spread of carbapenem-resistant Klebsiella pneumoniae: a 4-year perspective. Infect Control Hosp Epidemiol. 2011;32(7):673–678. doi: 10.1086/660358. [DOI] [PubMed] [Google Scholar]
- 36.Qin X, Wu S, Hao M, Zhu J, Ding B, Yang Y, Xu X, Wang M, Yang F, Hu F. The colonization of carbapenem-resistant Klebsiella pneumoniae: epidemiology, resistance mechanisms, and risk factors in patients admitted to intensive care units in China. J Infect Dis. 2020;221(Suppl 2):S206–S214. doi: 10.1093/infdis/jiz622. [DOI] [PubMed] [Google Scholar]
- 37.Li Y, Shen H, Zhu C, Yu Y. Carbapenem-resistant Klebsiella pneumoniae infections among ICU admission patients in central china: prevalence and prediction model. Biomed Res Int. 2019;2019:9767313. doi: 10.1155/2019/9767313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Liao W, Liu Y, Zhang W. Virulence evolution, molecular mechanisms of resistance and prevalence of ST11 carbapenem-resistant Klebsiella pneumoniae in China: a review over the last 10 years. J Glob Antimicrob Resist. 2020;23:174–180. doi: 10.1016/j.jgar.2020.09.004. [DOI] [PubMed] [Google Scholar]
- 39.Bracco D, Dubois MJ, Bouali R, Eggimann P. Single rooms may help to prevent nosocomial bloodstream infection and cross-transmission of methicillin-resistant Staphylococcus aureus in intensive care units. Intensive Care Med. 2007;33(5):836–840. doi: 10.1007/s00134-007-0559-5. [DOI] [PubMed] [Google Scholar]
- 40.Zhou K, Xiao T, David S, Wang Q, Zhou Y, Guo L, Aanensen D, Holt KE, Thomson NR, Grundmann H, Shen P, Xiao Y. Novel subclone of carbapenem-resistant Klebsiella pneumoniae sequence type 11 with enhanced virulence and transmissibility, China. Emerg Infect Dis. 2020;26(2):289–297. doi: 10.3201/eid2602.190594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gu D, Dong N, Zheng Z, Lin D, Huang M, Wang L, Chan EW, Shu L, Yu J, Zhang R, Chen S. A fatal outbreak of ST11 carbapenem-resistant hypervirulent Klebsiella pneumoniae in a Chinese hospital: a molecular epidemiological study. Lancet Infect Dis. 2018;18(1):37–46. doi: 10.1016/S1473-3099(17)30489-9. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1 Antibiotic resistance genetic analysis and antibiotic susceptibility test results for ST11-CRKP. Green squares indicate the presence of antibiotic-resistance genes, and white indicates the absence of antibiotic-resistance genes. Orange squares indicate the presence of relevant antibiotic-resistant phenotypes, and white indicates antibiotic sensitivity. (PDF 533 KB)
Fig. S2 The virulence gene alignment results for ST11-CRKP. Blue squares represent the presence of virulence genes, and white represent their absence. (PDF 739 KB)
Data Availability Statement
The whole-genome sequences of 95 K. pneumoniae isolates have been deposited in the GenBank database under BioProject accession no. PRJNA839803.