Abstract
The role of intratumor heterogeneity is becoming increasingly apparent in part due to expansion in single cell technologies. Clinically, tumor heterogeneity poses several obstacles to effective cancer therapy dealing with biomarker variability and treatment responses. Matrix stiffening is known to occur during tumor progression and contribute to pathogenesis in several cancer hallmarks, including tumor angiogenesis and metastasis. However, the effects of matrix stiffening on intratumor heterogeneity have not been thoroughly studied. In this study, we applied single-cell RNA sequencing to investigate the differences in the transcriptional landscapes between stiff and compliant MMTV-PyMT mouse mammary tumors. We found similar compositions of cancer and stromal subpopulations in compliant and stiff tumors but differential intercellular communication and a significantly higher concentration of tumor-promoting, M2-like macrophages in the stiffer tumor microenvironments. Interestingly, we found that cancer cells seeded on stiffer substrates recruited more macrophages. Furthermore, elevated matrix stiffness increased Colony Stimulating Factor 1 (CSF-1) expression in breast cancer cells and reduction of CSF-1 expression on stiffer substrates reduced macrophage recruitment. Thus, our results demonstrate that tissue phenotypes were conserved between stiff and compliant tumors but matrix stiffening altered cell-cell interactions which may be responsible for shifting the phenotypic balance of macrophages residing in the tumor microenvironment towards a pro-tumor progression M2 phenotype.
Keywords: Matrix stiffness, breast cancer, macrophage, CSF-1
Graphical Abstract
1. Introduction
The extracellular matrix (ECM) contributes both structure and signaling cues to the tumor microenvironment. Over the past decade, extensive work has demonstrated how the mechanics of ECM structure itself can provide physical signals to cells. Importantly, matrix stiffness has emerged as a critical parameter of the tumor microenvironment having substantial effects on cellular behavior across many different cell types. Matrix stiffening primarily occurs through excess matrix deposition and cross-linking by either cancer or stromal cells[1]. In cancer cells, elevated matrix stiffness has been shown to regulate cell morphology and cell spreading, and promote critical cancer cell behaviors such as proliferation, migration, and epithelial-to-mesenchymal transition[2-4]. Increased matrix stiffness also affects stromal cell types including cancer-associated fibroblasts, endothelial cells, and an assortment of immune cells. Cancer associated fibroblasts are more activated on stiffer matrices which may contribute to a positive feedback loop resulting in additional matrix stiffening and fibroblast activation[5,6]. Endothelial cells are widely known to be mechanosensitive, displaying enhanced angiogenic behaviors on stiffer matrices[7]. Interestingly, matrix stiffening alone has been shown to induce tumor vasculature phenotypes in vivo[7-9]. The immune component of the tumor microenvironment, composed of numerous cell types and phenotypes, is also affected by matrix stiffness. Immune cell infiltration has been correlated with matrix stiffness and macrophages have demonstrated mechanosensitive behaviors such as cell spreading, migration, and phenotypic polarization[10-12].
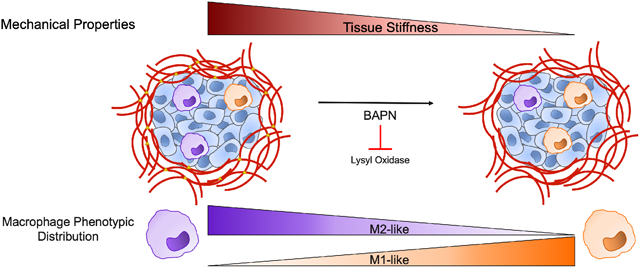
While matrix stiffening can affect cell behavior, the effect of matrix stiffening on the overall composition of the tumor microenvironment is incompletely understood. Given that matrix stiffening is known to influence the behavior of numerous cells in the tumor microenvironment, and the tumor microenvironment is complex, we implemented single cell RNA sequencing (scRNAseq) to analyze cells isolated from stiff and compliant breast tumors from the MMTV-PyMT mouse model. Our results indicate that similar cell types and phenotypes exist within both stiff and compliant tumors with a similar degree of transcriptional diversity, but stiff and compliant tumors differ in specific cell-cell signaling and altered the distribution of macrophage subsets. Specifically, we found stiffer tumors contain a higher proportion of macrophages residing in the more tumor-promoting M2-like phenotype. Additionally, we found that matrix stiffening enhances CSF-1 expression, a protein associated with M2 macrophage polarization[13], in breast cancer cells. We further demonstrated that matrix stiffness-mediated CSF-1 expression was responsible for enhanced macrophage recruitment in vitro by breast cancer cells seeded on stiffer substrates. Thus, our data indicates that stiffer tumors promote the accumulation of M2-like macrophages and this may be in part due to matrix stiffness induced secretion of the macrophage polarizing and attracting factor CSF-1 by cancer cells.
2. Methods
MMTV-PyMT Mouse Studies.
All animals experiments were conducted following a protocol approved by the Vanderbilt University Institutional Animal Care and Use Committee (IACUC). MMTV-PyMT mice of the FVB strain background were acquired from Jackson Laboratories (Stock No:002374) and housed in a facility with controlled temperature, humidity, and light (12 hr light/dark cycle). Standard rodent chow and water were provided ad libitum. Hemizygous MMTV-PyMT females began BAPN treatment (3mg/kg body weight) at the age of 4 weeks and continued treatment until 12-14 weeks of age to produce more compliant tumors as previously described[7,14-21]. Mice were euthanized with CO2 prior to tumor removal and subsequent processing.
Tumor Dissociation.
Fresh tumors were isolated in a sterile biosafety cabinet and placed in ice cold HBSS during transit from mouse facility to laboratory. Tumors were rinsed several times in ice cold HBSS and minced with sterile scalpels. Minced tumor was then enzymatically digested using the Human Tumor Dissociation Kit from Milytenyi Biotec (130-095-929). Post-enzymatic digestion, cells were passed through 100μm and 70 μm strainers (Miltenyi Biotec 130-110-916) to remove debris and undigested fragments. Cell suspensions then underwent several brief rounds of washing in 1X PBS with 3mM EDTA and an incubation in TrypLE (ThermoFisher Scientific 12604013) for 10 min to break apart cell clusters. Cells were suspended in PBS without EDTA and diluted to a concentration of 150k cells/ml for encapsulation.
Single Cell RNA-Sequencing
Single cell encapsulation and barcoding was performed as previously described[22]. Samples were sequenced in 3 batches, with 1 control and 1 BAPN tumor per batch, via Illumina NextSeq 500. Raw counts underwent quality control in Python (supplementary code) and were further analyzed in R using Seurat v3(supplementary code). Diversity scores were calculated as previously described to measure intratumoral heterogeneity[23]. Briefly, the diversity score was calculated by calculating the average distance between individual cells and the centroid within the principal component space. The centroid was calculated as the arithmetic mean of all the principal components calculated. Potential intercellular communication events were predicted using CellPhoneDB[24].
Flow Cytometry.
Cells were fixed in 4% paraformaldehyde (Electron Microscopy Sciences) in Hank’s Balanced Salt Solution (HBSS) (Gibco) for 15 min at RT, then blocked in 100 μL of FACS Buffer (HBSS without calcium, 2% FBS and 1mM EDTA) with 1% bovine serum albumin (BSA) (Sigma) for 20 min at 4°C. Cells were washed with FACS buffer between each step. Cells suspensions of 50 μL were incubated for 15 min at RT with 0.5 μL Mouse TruStain FcX (Biolegend, 101319) to prevent nonspecific Fc receptor binding. Samples were immediately stained with the following primary antibodies for 30 min at 4°C: 0.125μg/100μL eFlour 450 anti-mouse CD11b (Thermo Fisher Scientific, Clone M1/70), 0.5μg/100μL FITC anti-mouse F4/80 (Thermo Fisher Scientific, Clone BM8), 0.5μg/100μL PE anti-mouse CD86 (BD Biosciences Clone GL1), and 0.5μg/100μL APC anti-mouse CD206 (BioLegend, Clone C068C2). Cells were washed 2x with FACS buffer and analyzed using a Guava EasyCyte 12HT benchtop flow cytometer (MilliporeSigma). Flow cytometry plots were analyzed using FlowJo v10.7.1 software. Macrophages were characterized as CD11b+ F4/80+ populations. Within the gated macrophage population, M1/M2 gates were made using a control sample for each tumor, stained only for CD11b and F4/80 in the absence of M1/M2 markers to account for background fluorescence. M1 macrophages were characterized as CD86+ while M2 macrophages were CD206+.
Immunofluorescence staining.
Fresh tumors were excised and snap frozen. 8 micron sections were obtained from the VUMC Translational Pathology Shared Resource. Tumor sections were fixed with 4% (v/v) paraformaldehyde, washed with PBS, and permeabilized with 1% (v/v) triton X-100 in PBS. After permeabilization, samples were then blocked with 10% (v/v) FBS and 5% (v/v) donkey serum in PBS. Samples were stained with primary antibody (VE-Cadherin: eBioScience, eBioBV13) at 1:50 diluted in blocking solution overnight at 4°C, washed with PBS supplemented with 0.02% tween, and then incubated with secondary antibody (donkey anti-rat Alexa Fluor 594, A21209; Thermo Fisher Scientific) at 1:100 diluted in blocking solution for 1 h at room temperature in the dark. Samples were then washed, stained with DAPI, and incubated with either eFluor 660 CD68 pre-conjugated antibody (Thermo Fisher 50-0681-82) or APC CD206 (Biolegend 141708) at 1:50 diluted in blocking solution overnight at 4°C in the dark. Immunofluorescent images were taken with a Zeiss LSM800 microscope using a x40/1.1 NA water immersion objective and 488 excitation laser line.
Cell Culture.
MDA-MB-231 cells (ATCC), were cultured in DMEM media (Gibco ) supplemented with 10% Fetal Bovine Serum and 1% penicillin-streptomycin. BAC1.2F5 cells (generously provided by Dr. Richard Stanley, Albert Einstein College of Medicine) were cultured in MEM supplemented with 10% FBS, 1% penicillin-streptomycin, and 3000 U/ml of purified CSF-1 (R&D System, Minneapolis, MN, USA). Medium was replaced every 48 hours and cells were maintained in a 37°C humidified incubator of 5% (v/v) CO2. HUVECs (Lonza) between passage 3 and 5 were cultured in EBM (CC-3121; Lonza) supplemented with EGM Endothelial Cell Growth Medium SingleQuots Supplements (CC-4133; Lonza) and 1% penicillin-streptomycin.
Polyacrylamide Gel Synthesis.
Polyacrylamide (PA) gels were synthesized as previously described. PA gels with stiffness of 1kPa and 10kPa were prepared by mixing 3%:0.1% or 7.5%:0.35% acrylamide [40% (w/v) stock solution] to bis-acrylamide [2% (w/v) stock solution], respectively, in Mili-Q water with HEPES and tetramethylethylenediamine (TEMED; Bio-rad) at pH 6. Ammonium persulfate was dissolved in Mili-Q water at 10% (w/v) and used to initiate polymerization. PA gels were functionalized with N-6-[(acryloyl)amido]hexanoic acid, succinimidyl ester. Type 1 rat tail collagen (Corning, Corning, NY, USA) was then covalently bound to the PA gel surfaces at 4°C in 50mM HEPES solution at pH 8. Unreacted N-6-[(acryloyl)amido]hexanoic acid, succinimidyl ester was quenched with 1:1000 ethanolamine in 50mM HEPES solution at pH8. PA gels were washed in 1X PBS and stored at 4°C in PBS with 4% penicillin-streptomycin prior to seeding. PA gels were exposed to UV light for 1 hour prior to seeding to sterilize.
Cytokine Assay.
MDA-MB-231 cells were cultured on either compliant (1kPa) or stiff (10kPa) PA gels coated with 0.1 mg/mL Type 1 rat tail collagen. After 24 hours culture on PA gels, cell culture medium was collected and utilized as directed by the Proteome Profiler Human XL Cytokine Array Kit (ARY022B; R&D Systems).
qPCR.
mRNA was isolated from cells cultured on either compliant (1kPa) or stiff (10kPa) PA gels using the RNeasy Mini Kit (Qiagen). The iScript Select cDNA Synthesis Kit (Bio-Rad) was used to generate cDNA from the isolated mRNA. Quantitative PCR was performed using SYBR green (Thermo Fisher Scientific) according to the manufacturer’s instructions. Relative expression was calculated using the 2 −ΔΔCT method using B2M as a housekeeping gene. The primers used were CSF-1: forward: 5’-CCA GTG TCA TCC TGG TCT TG-3’, reverse: 5’-CCA CCT GTC TGT CAT CCT GA-3’; B2M: forward: 5′-CAC CCC CAC TGA AAA AGA TGA G-3′, reverse: 5′-CCT CCA TGA TGC TGC TTA CAT G-3’.
Western Blot.
MDA-MB-231 cells were seeded on top of either compliant (1kPa) or stiff (10kPa) gels for 24 hours and treated with or without the FAK inhibitor PF573228 (MilliporeSigma). Cells were rinsed with 1X PBS and lysed with 4X SDS sample buffer (4X Tris-Cl/SDS, pH6.8, 30% v/v glycerol, 10% w/v SDS, 0.09% v/v 2-mercaptoethanol, and 0.012% w/v Bromophenol Blue). Standard SDS-PAGE was conducted usingBio-Rad Any kD Mini-PROTEAN (4569035; Bio-Rad gels and PVDF membranes (Bio-rad). Membrane washing steps were performed with 0.1% polyoxyethylene 20 sorbitan monolaurate (Tween; JT Baker, Phillipsburg, NJ) in Tris-buffered saline. Blocking was performed with 5% milk in the washing buffer. Primary antibodies (GAPDH Biolegend poly6314; CSF-1 Santa Cruz sc-365779) were diluted in blocking buffer at 1:1000 dilution and applied to the membranes overnight at 4°C. Horseradish-peroxidase conjugated secondary antibodies were applied to the membranes in blocking buffer at 1:2000 dilution for 1 hour at room temperature. Membranes were imaged using SuperSignal chemiluminescent substrate and a FujiFilm ImageQuant LAS-4000. Quantification of protein expression was normalized to GAPDH loading control and densitometry was performed using Fiji.
Macrophage recruitment assay.
In the macrophage recruitment assay, we utilized a modified trans-endothelial transwell migration assay. Transwells were coated with neutralized 1mg/mL collagen and allowed to polymerize before hydration and seeding. HUVECs were then seeded on top of polymerized collagen coated transwell inserts at 300,000 cells/well and cultured for 3 days to allow a monolayer to form. MDA-MB-231 cells were cultured on compliant (1kPa) or stiff (10kPa) PA gels in the bottom of the transwells below the inserts. BAC1.2F5 macrophages stained with CellTracker Green CMFDA Dye (C7025; ThermoFisher) were then seeded in the medium above the transwell insert and allowed to transmigrate through the HUVEC monolayer, collagen coating, and transwell insert pores towards the MDA-MB-231 cells cultured on PA gels in the bottom of the well. The number of recruited macrophages were measured via laser scanning confocal reflectance imaging and quantified as the number of macrophages per defined region of interest in the bottom of the transwell chamber.
Statistical Analysis
Statistical analyses were performed using GraphPad Prism 9 (GraphPad Software, La Jolla, CA, USA). Where appropriate, data were compared using unpaired t-tests with Welch’s Correction, a two-way analysis of variance (ANOVA) with Sidak multiple comparison test, or a nested t-test. Statistical significance was determined if the tested p-value was smaller than 0.05 (*), 0.01(**), 0.001 (***), or 0.0001 (****). ‘N’ represents the number of independent samples while ‘n’ represents the number of measurements taken.
3. Results
Single cell RNA sequencing reveals similar cell type composition of compliant and stiff breast tumor environments.
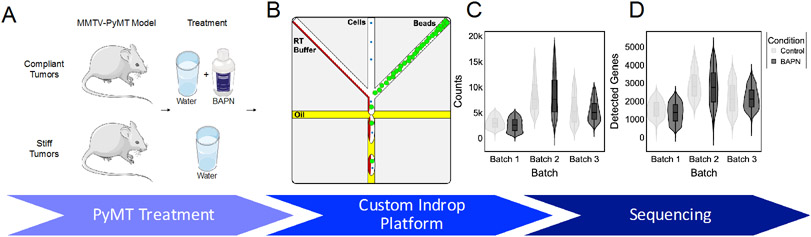
To investigate the architectural effects of matrix stiffness on the tumor microenvironment, we performed scRNAseq on stiff and compliant MMTV-PyMT mammary tumors. To obtain compliant and stiff tumors, MMTV-PyMT mice were treated with BAPN, a lysyl oxidase inhibitor, or vehicle control, respectively (Fig. 1A). Tumors were dissociated to form single cell suspensions and encapsulated using a custom inDrop platform (Fig. 1B). Tumors were excised, encapsulated, and sequenced pairwise in 3 batches on separate days and sequencing runs. All sequencing results were filtered using several quality control methods prior to analysis. Inflection point gating for total counts per cell was applied to each sample individually to remove cells with low library size and an upper threshold was applied to remove droplets that may have contained more than 1 cell[25]. Additionally, cells containing a high proportion of mitochondrial genes were removed. A total of 8,523 cells passed quality control metrics from 6 tumors with an average of ~5200 counts per cell over ~2100 genes (Fig. 1C,D). While there was batch to batch variation in preprocessed library quality, there was no difference between compliant and stiff tumors sequenced within the same batch (Fig. 1C,D).
Figure 1.
Single cell RNA-seq reveals similar transcriptional landscapes between stiff and compliant MMTV-PyMT tumors. A. Schematic of A. experimental treatment regime and B. custom Indrop platform. Violin plot of C. raw counts per cell and D. detected genes per cell across the 6 samples. N = 3.
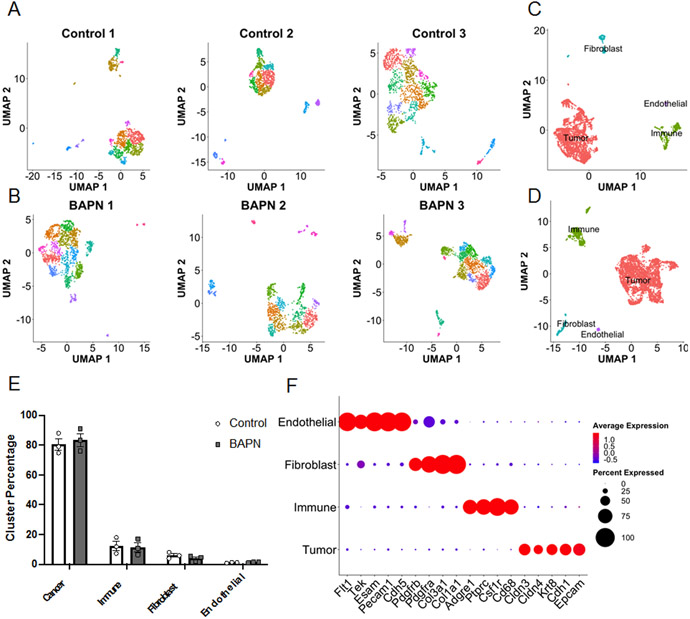
Lower dimensional embedding via UMAP revealed similar numbers of clusters in both compliant and stiff tumors detected by k-means clustering both on individual sample landscapes and samples integrated by condition using sctransform method[26](Fig. 2A,B,C,D). Using the expression of a manually curated list of marker genes, cells were assigned to 4 major cell types: cancer, immune, fibroblast, or endothelial (Fig. 2E,F,G). Cancer cells were defined as non-stromal cells that expressed epithelial markers. Both landscapes were composed of similar distributions of cell types with cancer cells being largest population of ~80% and immune cells being the next largest population at ~12% followed by fibroblasts at ~5% and endothelial cells at ~1% (Fig. 2E).
Figure 2.
Single cell RNA-seq reveals similar transcriptional landscapes between stiff and compliant MMTV-PyMT tumors. Individual UMAP projections of each individual sample from A. stiff and B. compliant tumors and clustered via Seurat. C. Stiff and D. compliant tumors integrated onto a single UMAP projection and clusters labeled by cell type determined by expression of canonical markers below. E. Distribution of libraries across the 4 main cell types. Data plotted as mean +/− SEM. N=3. F. Gene expression of the canonical cell type markers.
As the integration of all cell types onto a single projection is dominated by variability in cell type marker expression, we parsed cells by cell type and re-integrated all samples together for further analysis of heterogeneity. Cells were isolated on a cell type basis and re-analyzed via Seurat to integrate the samples based on highly variable genes that exist within the specific cell type under investigation. After integration, cells were again visualized via lower dimensional embeddings and displayed thorough mixing between conditions and samples (Supplemental Fig. 1A,E,I). Several distinct subpopulations were evident from lower dimensional embeddings and clustering via Louvain algorithm with Seurat (Supplemental Fig. 1B,F,J) and were defined by distinct gene expression profiles (Supplemental Fig. 1C,G,K). Importantly, these subpopulations were composed of cells from both stiff and compliant tumors (Supplemental Fig. 1A,E,I). Interestingly, the majority of Louvain clusters detected in the cancer cells were contiguous while the clusters detected in the immune and CAF cells were more separated. Contiguous clustering suggests a spectrum of related cell states while the separation in the stromal subpopulations suggests more distinct phenotypes. To further quantify the intratumoral heterogeneity we utilized a previously published method to compute transcriptomic diversity scores based on principal component embeddings[23]. These scores were calculated for cancer cells, immune cells, and fibroblasts individually using each tumor as an independent sample. In agreement with the thorough mixing of cells between conditions and samples (Supplemental Fig. 1A,E,I), the diversity scores displayed no significant difference between cell types in stiff versus compliant tumors (Supplemental Fig. 1D,H,L). Thus, this indicates that there is significant heterogeneity that exists within the cancer and stromal cells and that this heterogeneity is conserved between stiff and compliant tumors.
Macrophages constitute the largest portion of immune cells and exhibit phenotypic heterogeneity
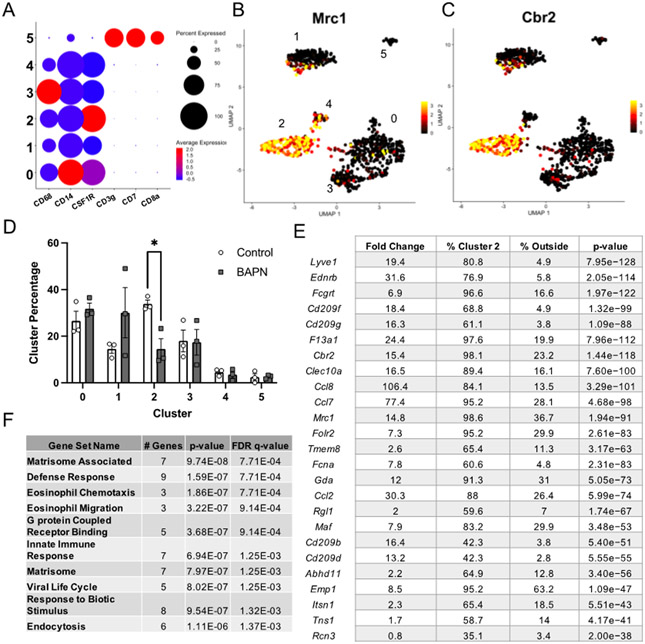
The tumor microenvironment is home to numerous types of immune cells with important pro- and anti-tumor functions. To determine the identity of immune cells captured in this study, we assessed the expression of a panel of canonical immune cell specific markers. Broad expression of macrophage markers (CD68, CD14, CSF1R) were seen in 5 of the 6 subpopulations of immune cells, approximately 97% of total immune cells, with some variation in expression levels between the clusters (Fig. 3A). The remaining small subpopulation in cluster 5, approximately ~3% of the immune cells, were identified as T-cells based on expression of CD3g, CD7, and CD8a (Fig. 3A).
Figure 3.
Immune cell annotation reveals immune cells are predominantly composed of macrophages and enrichment of M2-like macrophages in stiffer tumors. A. Expression of canonical macrophage and T-cell markers across cells in the immune category. B,C. Expression of canonical M2-like macrophage markers overlaid on UMAP projections of the cells in the immune group. D. Comparison of the distribution of immune cells to each subpopulation plotted as mean +/− SEM. N =3. E. Top 25 conserved marker genes for immune cell cluster 2. Table displays the average fold expression within cluster 2 compared to the rest of the cells, the percentage of cells expressing each transcript in cluster 2 versus the remaining clusters, and the adjusted p-value for the transcript. F. GO Term enrichment of the top 25 marker genes for the M2-like macrophage subpopulation. *p<0.05
Macrophages are a heterogeneous cell type containing complex phenotypic and functional variation[27,28]. We performed differential expression analyses between each of the macrophage subpopulations to identify marker genes for each cluster and investigate the observed heterogeneity (Supplemental Fig. 2). Examination of the top 25 marker genes in each cluster revealed heterogeneous expression of several macrophage phenotypic markers. Cluster 0 represented one of the larger clusters with approximately 30% of the total immune cells in both stiff and compliant tumor landscapes. Macrophages in this subpopulation displayed transcripts associated high expression of macrophage genes associated with both canonical polarization states, such as an important anti-inflammatory M2 polarization regulator Tlr2[29,30], the pro-inflammatory (M1-like) factor Aif1[31], and the monocyte differentiation regulator transcript Runx3[32], suggesting they may represent an intermediate polarization state (Supplemental Fig. 2). GO term analysis of the top markers revealed significant enrichment for transcripts in cell activation, cell adhesion, and secretion (Supplemental Fig. 2). Cluster 1 was composed of a subpopulation defined by high expression of transcripts traditionally involved in epithelium development and differentiation (Epcam, Cldn3, and Krt8) (Supplemental Fig. 2). Additionally, transcripts associated with both pro- and anti-inflammatory macrophage behaviors were significantly expressed in cluster 1 (Ccn1[33] and Lcn2[34], respectively). High expression of epithelial markers alongside Cd24a suggests these cells may actually represent Langerhans cells, a specialized antigen-presenting macrophage subtype[35-37] typically found in epidermal tissue but have been shown to infiltrate breast tumors[38] (Supplemental Fig. 2). Interestingly, macrophages in cluster 2 had significantly higher expression of several canonical anti-inflammatory M2 macrophage markers (Cd209, Mrc1, Cbr2, and Folr2)[39-42] and resident-like macrophage markers (F3a1, Lyve1)[41] (Fig. 3B,C,E and Supplemental Fig. 2). Due to the high expression of canonical M2 markers, we designated these macrophages as ‘M2-like’. Significant GO terms in the M2-like macrophages included categories related to the matrisome, with several C-C motif ligand chemokines (Ccl2, Ccl7, Ccl8), and eosinophil migration and chemotaxis (Fig. 3E,F). Cluster 3 was contiguous with the macrophages in cluster 0 and also significantly expressed a few pro-inflammatory transcripts associated with the M1 phenotype (Slc7a2[42], Fcgbr2[42], and Npc2[43]) and several anti-inflammatory transcripts typically associated with the M2 phenotype (Adam8[44], Spp1[45], Ctsl[42], Ctsb[42], Arg1[39,40]) (Supplemental Fig. 2), suggesting macrophages in this subpopulation may reside in an intermediate polarization state. Furthermore, cluster 3 GO terms included cell activation and secretion, similarly to cluster 0 (Supplemental Fig. 2). Cluster 4 was contiguous with cluster 2 (M2-like macrophages) and highly expressed several anti-inflammatory transcripts associated with the M2-like phenotype (Ccr2[40], Retnla[46], and Mgl2[46]) suggesting this cluster may represent a subset of M2-like macrophages, possibly M2b due to presence of Il6[47]. Altogether, these data indicate that the majority of the immune cells captured are of macrophage lineage and cluster similarly to previously defined macrophage phenotypic subsets.
M2-like macrophages are enriched in stiffer tumors
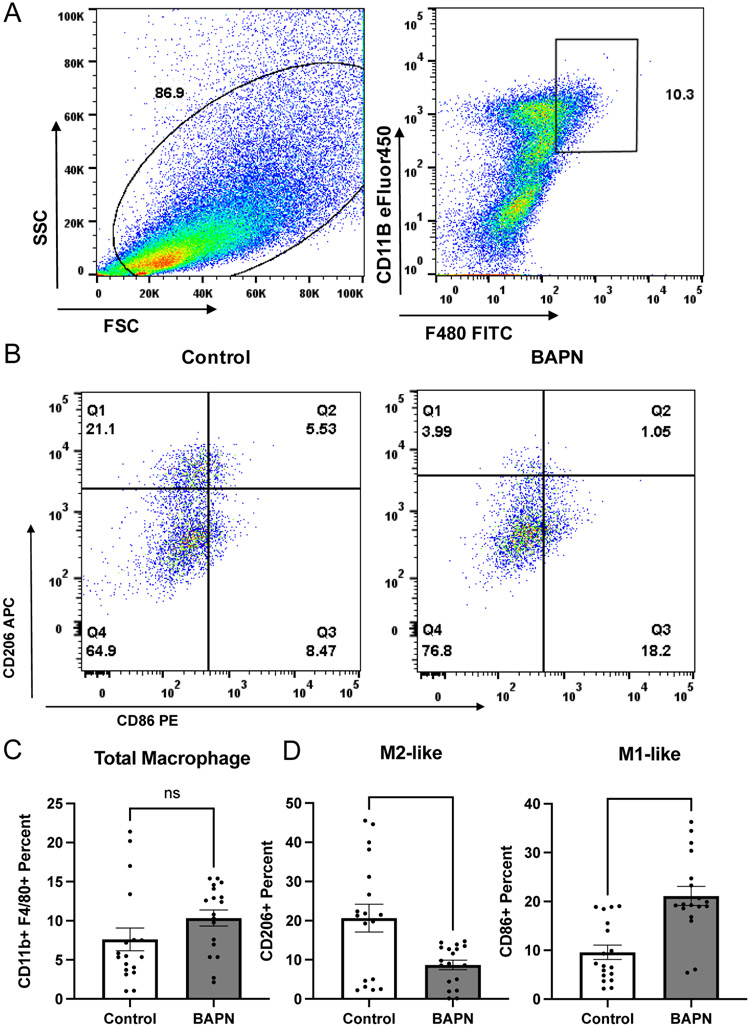
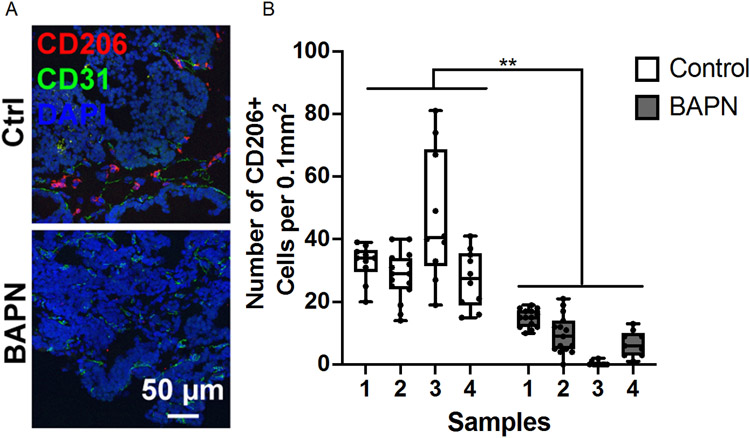
Differential expression analysis between macrophages from stiff and compliant tumors for each subpopulation yielded very few differentially expressed transcripts. However, analyzing the distribution of the macrophages in each subset identified in stiff and compliant tumors revealed significant enrichment of an M2-like macrophage subpopulation in stiffer tumors, with ~30% of macrophages in stiffer tumors mapping to the M2-like phenotype compared to ~14% in the more compliant tumors (Fig. 3D). To quantify the phenotypic distribution of macrophages in vivo and validate our scRNAseq data, we obtained stiff and compliant tumors from our MMTV-PyMT model. Tumors were dissociated and subjected to flow cytometry analysis using CD11b and F4/80 as general macrophage markers (Fig. 4A), CD86 as an M1 macrophage marker[48] (Fig. 4B), and CD206 as an M2 macrophage marker[48] (Fig. 4B). Flow cytometry revealed no significant difference between total macrophage content (Fig. 4C) but a significant increase in CD206+ macrophages in the stiffer tumors compared to compliant tumors as well as a concomitant decrease in CD86+ macrophages (Fig. 4D). Furthermore, immunofluorescence staining of tumor sections also confirmed an increase in the number of CD026+ cells per field of interest in stiff tumors compared to compliant tumors (Fig. 5A,B). This data confirms that stiff tumors contain a higher proportion of M2-like macrophages compared to compliant tumors.
Figure 4.
Quantifying macrophage polarization in the MMTV-PyMT breast tumor microenvironment via flow cytometry. A. Flow cytometry gating based on side-light vs forward light scatter intensity and double positive CD11B and F4/80 staining. B. CD206 expression (M2 marker) and CD86 expression (M1 marker) in isolated macrophages. C. Quantification of CD11b and F4/80 positive macrophages in total cell populations. D. Quantification of CD206 (left), and CD86 (right) positive macrophages in the total gated macrophage populations. Data plotted as mean +/− SEM. N=3 (number of mice), n=18 (number of data points). **p<0.01, ****p<0.0001
Figure 5.
Quantifying macrophage polarization in the MMTV-PyMT breast tumor microenvironment. A. Representative images of MMTV-PyMT tumor sections stained for DAPI (blue), CD31 (green), and CD206 (red). B. Quantification of stained tumor sections. Number of CD206+ cells per field of interest. N=4 (number of mice), n = 9-15 (number of data points). **p<0.01
Intercellular communication differs between stiff and compliant tumors
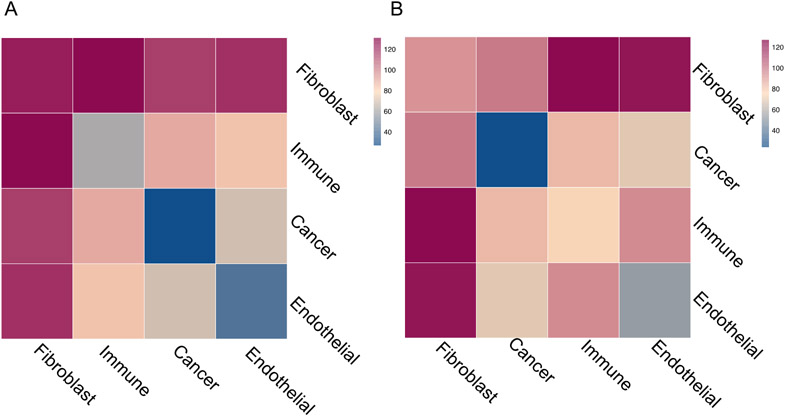
To investigate the source of M2-like macrophage enrichment in stiffer tumors, we utilized CellPhoneDB to infer cell-cell interactions in the scRNAseq data using the expression of ligands and receptors across cell types[24]. Analysis using CellPhoneDB revealed numerous potential cell-cell interactions between all the cell types in both stiff and compliant tumors (Fig. 6A,B). While many of the cell-cell interactions were shared between treatment groups, there were 45 significant interactions specific to the stiff tumors and only 7 significant interactions specific to the compliant tumors (Supplemental Fig. 3A,B). Interestingly, the network of cell-cell interactions in stiffer tumors shifted towards an increase in communication involving fibroblasts (Fig. 6A,B), with many of the ligand-receptor interactions specific to stiffer tumors involving collagen-integrin interactions with fibroblasts (Supplemental Fig. 3A,B). However, significant interactions between other cell types were present, particularly in interactions involving immune cells. Notably, the several cancer-to-immune ligand-receptor interactions were found significant only in the stiffer tumors; including TYRO3-GAS6, SPP1-PTGER4, CSF3-CSF3R, and PLXNB1-SEMA4D (Supplemental Fig. 3A,B). Furthermore, CelllPhoneDB analysis indicates that there are more immune cell-cell interactions with other cell types than cancer cell-cell interactions in stiffer tumors but the inverse is true within compliant tumors (Fig. 6A,B). Altogether, this data suggests that stiff and compliant tumors have similar degrees of heterogeneity in regards to the presence (Supplemental Fig. 1A,E,I) and diversity (Supplemental Fig. 1D,H,L) of cell states but significantly differ in the intercellular communication with stiffer tumors displaying more integrin-based fibroblast signaling and potentially more immune cell interactions with other cell types (Fig. 6A,B).
Figure 6.
Quantification of cell-cell interactions between cell-types in the MMTV-PyMT tumor microenvironment. Heat map summarizing the number of significant ligand-receptor interactions in A. control or B. BAPN treated tumors.
Matrix stiffness regulates cytokine expression in MDA-MB-231 cells.
While there were several statistically significant cell-cell interactions based on CellPhoneDB, differential expression testing revealed very few significantly expressed transcripts between stiffer and compliant tumors when comparing the same cell types. This may stem from technical limitations of our scRNAseq data resulting from the low mRNA capture efficiency of InDrop platforms as well as lower sequencing depth compared to bulk RNA sequencing. Thus, to further investigate potential cell-cell interactions responsible for M2-like macrophage enrichment, we transitioned into in vitro models using the human breast cancer cell line MDA-MB-231, a highly metastatic cell line. To determine how matrix stiffness may induce cancer-macrophage interactions to promote M2-like macrophage accumulation, we assessed how matrix stiffness regulates cytokine expression in MDA-MB-231 cells. MDA-MB-231 cells were seeded on either compliant (1kPa) or stiff (10kPa) collagen coated polyacrylamide (PA) gels. Cell lysates were collected and assayed using a human cytokine array kit which detected 105 different cytokines. 41 cytokines were found to be significantly differentially regulated by substrate stiffness (Fig. 7A). Interestingly, 3 members of the colony-stimulating factor (CSF) family, secreted glycoproteins with important roles in regulating immune cell functions and differentiation, were significantly upregulated in MDA-MB-231 cells cultured on stiff (10kPa) PA gels (Fig. 7A). This data indicates that the cancer cell cytokine secretome is affected by matrix stiffness and suggests matrix stiffening may affect intercellular signaling between cancer cells and immune cells.
Figure 7.
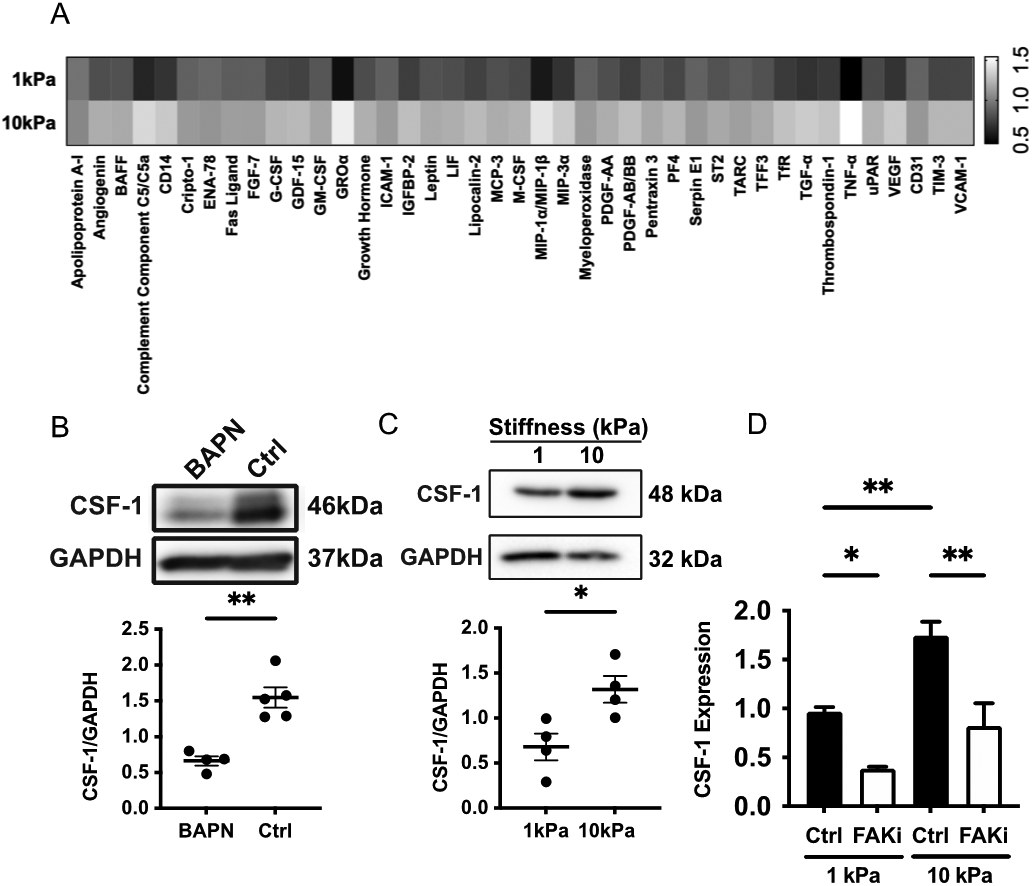
Matrix stiffness mediates cytokine expression in MDA-MB-231 cells. A. Heat map displaying significantly differentially expressed cytokines between MDA-MB-231 cells on compliant (1kPa) or stiff (10kPa) gels. B. Western blot image and quantification of CSF-1 in compliant (BAPN) or stiff (ctrl) PyMT tumors. C. Western blot image and quantification of CSF-1 in MDA-MB-231 cells cultured on compliant or stiff PA gels. D. qPCR of CSF-1 expression in MDA-MB-231 cells cultured on compliant or stiff PA gels and treated with a FAK inhibitor (FAKi). All data represented as mean +/− sem. *p<0.05, **p<0.01
Increased matrix stiffness upregulates CSF-1 in MDA-MB-231 cells and is dependent on FAK-mediated mechanotransduction
To further investigate how matrix stiffness may mediate intercellular communication between cancer and immune cells, we focused on the CSF family of cytokines as CSF1, CSF2, and CSF3 were upregulated in MDA-MB-231 cells cultured on stiff (10kPa) PA gels (Fig. 7A), and they are known to regulate macrophage function and polarization[49]. As our scRNAseq data only revealed appreciable expression of the CSF-1 receptor on our macrophage populations, we hypothesized that mechanical regulation of CSF-1 in the MDA-MB-231 cells may regulate macrophage recruitment. To determine if CSF-1 protein expression is higher in stiffer MMTV-PyMT tumors, we performed western blotting on lysates derived from compliant (BAPN) and stiff (control) MMTV-PyMT tumors and found that CSF-1 expression was significantly higher in stiffer tumors (Fig. 7B). To confirm that increased substrate stiffness upregulates CSF-1, we cultured MDA-MB-231 cells on compliant (1kPa) and stiff (10kPa) PA gels and performed western blotting on cell lysates. As expected, western blotting revealed protein expression of CSF-1 on stiff PA gels compared to compliant PA gels (Fig. 7C). Prior work shows that the focal adhesion kinase (FAK) is an important protein in the mechanotransduction of substrate stiffness in cancer cells[50]. To determine if mechanical regulation of CSF-1 in MDA-MB-231 cells is regulated by FAK, we treated MDA-MB-231 cells cultured on compliant (1kPa) and stiff (10kPa) gels with PF573228, a small molecule FAK inhibitor. Western blotting revealed that inhibition of FAK via PF573228 significantly reduced the expression of CSF-1 in MDA-MB-231 cells (Fig. 7D). Together, this data indicates that matrix stiffness regulates CSF-1 expression via FAK in MDA-MB-231 cells.
Matrix stiffness regulates macrophage recruitment through CSF-1
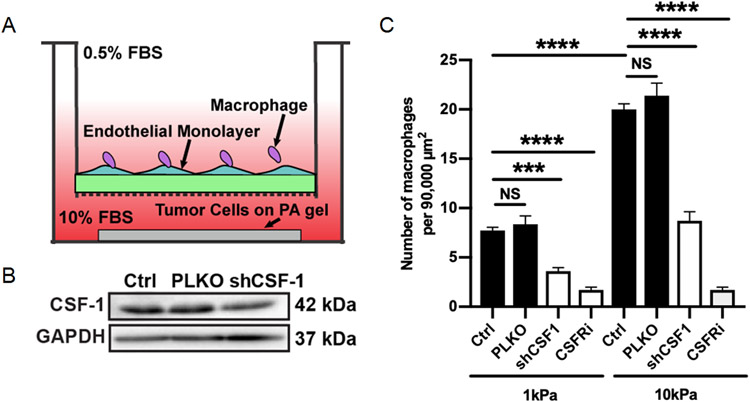
To confirm the functional importance of tumor derived CSF-1 in cancer-macrophage intercellular communication, we utilized an in vitro transwell-based assay to determine how stiffness mediated CSF-1 expression effects macrophage recruitment. In brief, BAC1.2F5 macrophages were seeded on top of a transwell insert with a Human Umbilical Vein Endothelial Cell (HUVEC) monolayer and MDA-MB-231 cells were seeded in the bottom of the well on compliant (1kPa) or stiff (10kPa) PA gels (Fig. 8B). Macrophage recruitment was quantified as the number of macrophages that migrated through the HUVEC monolayer and transwell insert membrane towards the MDA-MB-231 cells that were imaged 24 hours after seeding. As expected, MDA-MB-231 cells cultured on stiffer PA gels recruited significantly more macrophages than those on compliant PA gels (Fig. 8C). Furthermore, reduction of CSF-1 expression in MDA-MB-231 cells via shRNA knockdown resulted in significantly less macrophage recruitment (Fig. 8A,C). Similarly, inhibition of CSF-1 receptor on macrophages using a CSF-1 receptor inhibitor resulted in significantly less macrophage recruitment (Fig. 8C). Thus, our data suggests matrix stiffness facilitates a cancer-macrophage intercellular interaction by increasing CSF-1 expression in cancer cells.
Figure 8.
Stiffness mediated CSF-1 expression promotes macrophage recruitment. A. Representative western blot confirming CSF-1 knockdown via shCSF-1. B. Schematic diagram of modified transwell assay used to measure macrophage recruitment. C. Quantification of the number of macrophages that migrated through the transwell towards MDA-MB-231 cells cultured on compliant or stiff PA gels with or without CSF-1 knockdown or with or without a CSF-1R inhibitor (CSF-1R). Data plotted as mean +/− SEM. N=3, n=25-30. ***p<0.001, ****p<0.0001
4. Discussion
To profile the transcriptional landscapes and investigate phenotypic differences caused by tumor stiffness, we performed scRNAseq on all cells isolated from stiff and compliant PyMT mammary tumors. Both stiff and compliant tumors exhibit significant intratumor heterogeneity in the cancer and stromal cells (Supplemental Fig. 1A,E,I). Interestingly, much of the heterogeneity was conserved between conditions with both stiff and compliant tumors containing roughly the same subpopulations of cells with similar diversity of transcriptional profiles (Supplemental Fig. 1D,H,L). However, there were differences in cell-cell interactions between stiff and compliant tumors with stiffer tumor interaction networks increasing the number of ECM-component and integrin-based fibroblast receptor-ligand interactions (Fig. 6) as expected in stiffer more fibrotic tumors[18]. Furthermore, a significantly higher percentage of M2-like macrophages reside in the stiffer tumor microenvironment. Thus, while matrix stiffness does not induce novel cell phenotypes, it may affect intercellular signaling and adjust the phenotypic balances within the tumor microenvironment.
Our findings synergize well with recent reports using scRNAseq showing stromal subpopulations from different patients were highly similar in their expression states but varied in their proportions[51] and CAF subsets were highly similar between primary tumor and lymph node metastases[52]. Together, these studies suggest stromal subpopulations may be highly conserved between tumors, and the intertumoral heterogeneity may predominantly come in the form of intercellular communication and varying tumor composition.
Our scRNAseq (Fig. 3D), flow cytometry (Fig. 4), and immunostaining (Fig. 5) data indicate an elevation in M2-like macrophage presence in stiffer tumors. Additionally, while not evident in the scRNAseq data (Fig 3D), our flow cytometry data indicate a significant decrease in M1-like macrophages in stiffer tumors (Fig. 4). This discrepancy is likely due to the fact that our scRNAseq data did not resolve any specific M1-like clusters according to canonical markers (such as CD86) which could arise from technical aberrations or the actual complexity of macrophage polarization phenotypes[28,53]. It is known that tumor associated macrophages specifically contribute to tumor progression by promoting angiogenesis, facilitating cancer cell invasion, and repressing anti-tumor immunity[54-58]. The presence of macrophages within the tumor microenvironment has prognostic value in several cancers, with higher macrophage density being correlated with worse outcomes[59-61]. Traditionally, tumor associated macrophages exhibiting an alternatively activated M2 phenotype exert pro-tumoral effects while the classically activated M1 phenotype may exert tumor suppressing effects[62-64]. Furthermore, previous studies have revealed that BAPN treatment in the MMTV-PyMT model delays primary tumor development and metastatic lung burden[21]. As the M2 phenotype is associated with tumor progression and elevated matrix stiffening is associated with delayed primary tumor development and metastasis, this finding suggests another mechanism by which matrix stiffening may reshape the tumor microenvironment to further cancer progression. However, the mechanism by which matrix stiffness drives M2-like macrophage enrichment remains unknown.
Macrophage accumulation could occur through several mechanisms. Stiffer matrices may 1.) preferentially recruit M2-like macrophages, 2.) promote proliferation and survival of M2-like macrophages, 3.) shift macrophages towards an M2-like phenotype, or 4.) decrease infiltration, proliferation, or survival of M1-like macrophages. Extracellular matrix stiffness could induce expression of chemokines or other attractants by either cancer or stromal cells that lead to infiltration of M2-like macrophages. For example, previous work has demonstrated that hypoxia in the breast cancer microenvironment may induce intercellular signaling between that ultimately leads to increased macrophage recruitment via cancer secreted CSF-1[65]. Interestingly, while a large portion of significant cell-cell interactions detected specifically in stiffer tumors were focused between fibroblasts to fibroblasts, we detected cancer-to-immune cell ligand-receptor interactions which could contribute to M2-like accumulation. Another interesting possibility could be differential macrophage infiltration due to changes in the tumor endothelium. We have previously shown that matrix stiffening leads to significant permeability in the tumor endothelium[7]. Thus, it may be possible for more macrophages to enter the stiffer tumor microenvironment, bypassing a more permissive vasculature than in compliant tumors. However, this does not completely explain the enrichment for M2-like macrophages as similar amount of total macrophages were observed in the stiff and compliant tumors.
It is highly possible that matrix stiffness in the tumor microenvironment polarizes macrophages towards the M2-like phenotype. Macrophages are mechanosensitivity to substrate stiffness[11,66] and the effect of matrix stiffness on macrophage polarization has been studied numerous times, with some mixed findings[67-72]. There is evidence for increased M2 polarization on both stiff[67,70,71] or soft[69] matrices. To further complicate these conflicting findings, the studies employed different macrophage sources coupled with systems possessing different dimensionality (2D vs 3D), ligand availability, and stiffness ranges. Additionally, these studies were completed on macrophages cultured in vitro using methods developed to polarize macrophages with a chemical stimulus. Importantly, our data provides indirect evidence for macrophage polarization towards an M2 phenotype under stiffer conditions and, to our knowledge, is the only study to use an in vivo model of matrix stiffening.
Importantly, we have shown that MDA-MB-231 cells alter their cytokine secretome in response to increased matrix stiffness. Notably, CSF-1 is upregulated in stiffer MMTV-PyMT tumors and MDA-MB-231 cells cultured on stiffer substrates (Fig. 7). CSF-1, also known as macrophage CSF (M-CSF), is a member of the family of the colony stimulating factors[49]. CSF members are regulatory cytokines that facilitate intercellular communication, paracrine, autocrine, or endocrine, via binding to extracellular CSF receptors [49]. In particular, CSF-1 promotes macrophage polarization towards the M2 phenotype and CSF1-R inhibition has been shown to reduce M2 gene expression in vivo[13]. In cancer, CSF-1 has been correlated with worse prognosis[73]. As such, there has been a recent focus on targeting CSF-1 in cancer patients as a therapeutic strategy and there have been two clinical trials completed utilizing an anti-CSF-1 antibody in combination with additional chemotherapy agents in patients with various types of breast cancer[74,75]. Our findings suggest that matrix stiffness may induce M2-like macrophage accumulation via a cancer-macrophage intercellular communication through CSF-1. Thus anti-CSF-1 drugs may also be effective in inhibiting the accumulation of tumor promoting M2-like macrophages in stiffer tumors.
5. Conclusion
Therapies targeting extracellular matrix stiffness have become increasingly popular due to the known effects of matrix stiffness on cellular behavior, however, these therapies are unlikely to work as standalone treatments and it will be important to understand what additional therapies will be viable if matrix stiffening can be attenuated[76]. Our results indicate that while the overall cell populations close resemble each other in stiff and compliant tumor microenvironments, the cell-cell interactions between cell types and the distribution of phenotypic cell subtypes are different. Specifically, more integrin-based fibroblast cell-cell interactions exist in stiffer tumors and a higher proportion of the tumor promoting M2-like macrophages reside within stiffer tumors. Furthermore, our data suggests that matrix stiffening in the tumor microenvironment may drive M2-like macrophage accumulation through intercellular cross-talk between cancer cells and macrophages via cancer secreted CSF-1. Given that tumor angiogenesis and metastasis are affected by both matrix stiffening[3,7,77] and M2-like macrophage interactions[58,78], accumulation of M2-like macrophages may represent an alternative or reinforcing mechanism by which matrix stiffness alters tumor angiogenesis and metastasis.
Supplementary Material
Acknowledgments
This work was funded by the NIH NHLBI (Award number HL127499 and GM131178) and the W.M. Keck Foundation to CAR. KSL, ANS, BC, and AJS were funded by R01DK103831 and P50CA236733. We would like to thank the Vanderbilt Technologies for Advanced Genomics (VANTAGE) core facility for their help with this work. The BAC1.2F5 cells were graciously donated by Dr. Richard Stanley, Albert Einstein College of Medicine.
Footnotes
Disclosures
The author(s) declare no competing interests.
References
- [1].Lu P, Weaver VM, Werb Z, The extracellular matrix: A dynamic niche in cancer progression, J Cell Biol. 196 (2012) 395–406. 10.1083/jcb.201102147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Ulrich TA, de Juan Pardo EM, Kumar S, The mechanical rigidity of the extracellular matrix regulates the structure, motility, and proliferation of glioma cells, Cancer Res. 69 (2009) 4167–4174. 10.1158/0008-5472.CAN-08-4859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Wei SC, Fattet L, Tsai JH, Guo Y, Pai VH, Majeski HE, Chen AC, Sah RL, Taylor SS, Engler AJ, Yang J, Matrix stiffness drives epithelial–mesenchymal transition and tumour metastasis through a TWIST1–G3BP2 mechanotransduction pathway, Nat. Cell Biol 17 (2015) 678–688. 10.1038/ncb3157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Rice AJ, Cortes E, Lachowski D, Cheung BCH, Karim SA, Morton JP, del Río Hernández A, Matrix stiffness induces epithelial–mesenchymal transition and promotes chemoresistance in pancreatic cancer cells, Oncogenesis. 6 (2017) e352–e352. 10.1038/oncsis.2017.54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Schwager SC, Bordeleau F, Zhang J, Antonyak MA, Cerione RA, Reinhart-King CA, Matrix stiffness regulates microvesicle-induced fibroblast activation, Am. J. Physiol.-Cell Physiol 317 (2019) C82–C92. 10.1152/ajpcell.00418.2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Calvo F, Ege N, Grande-Garcia A, Hooper S, Jenkins RP, Chaudhry SI, Harrington K, Williamson P, Moeendarbary E, Charras G, Sahai E, Mechanotransduction and YAP-dependent matrix remodelling is required for the generation and maintenance of cancer-associated fibroblasts, Nat. Cell Biol 15 (2013) 637–646. 10.1038/ncb2756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Bordeleau F, Mason BN, Lollis EM, Mazzola M, Zanotelli MR, Somasegar S, Califano JP, Montague C, LaValley DJ, Huynh J, Mencia-Trinchant N, Abril YLN, Hassane DC, Bonassar LJ, Butcher JT, Weiss RS, Reinhart-King CA, Matrix stiffening promotes a tumor vasculature phenotype, Proc. Natl. Acad. Sci 114 (2017) 492–497. 10.1073/pnas.1613855114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Mason BN, Starchenko A, Williams RM, Bonassar LJ, Reinhart-King CA, Tuning three-dimensional collagen matrix stiffness independently of collagen concentration modulates endothelial cell behavior, Acta Biomater. 9 (2013) 4635–4644. 10.1016/j.actbio.2012.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Yeh Y-T, Hur SS, Chang J, Wang K-C, Chiu J-J, Li Y-S, Chien S, Matrix Stiffness Regulates Endothelial Cell Proliferation through Septin 9, PLoS ONE. 7 (2012) e46889. 10.1371/journal.pone.0046889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Acerbi I, Cassereau L, Dean I, Shi Q, Au A, Park C, Chen YY, Liphardt J, Hwang ES, Weaver VM, Human breast cancer invasion and aggression correlates with ECM stiffening and immune cell infiltration, Integr. Biol 7 (2015) 1120–1134. 10.1039/c5ib00040h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Adlerz KM, Aranda-Espinoza H, Hayenga HN, Substrate elasticity regulates the behavior of human monocyte-derived macrophages, Eur. Biophys. J 45 (2016) 301–309. 10.1007/s00249-015-1096-8. [DOI] [PubMed] [Google Scholar]
- [12].McWhorter FY, Davis CT, Liu WF, Physical and mechanical regulation of macrophage phenotype and function, Cell. Mol. Life Sci 72 (2015) 1303–1316. 10.1007/s00018-014-1796-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Pyonteck SM, Akkari L, Schuhmacher AJ, Bowman RL, Sevenich L, Quail DF, Olson OC, Quick ML, Huse JT, Teijeiro V, Setty M, Leslie CS, Oei Y, Pedraza A, Zhang J, Brennan CW, Sutton JC, Holland EC, Daniel D, Joyce JA, CSF-1R inhibition alters macrophage polarization and blocks glioma progression, Nat. Med 19 (2013) 1264–1272. 10.1038/nm.3337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Bordeleau F, Califano JP, Negrón Abril YL, Mason BN, LaValley DJ, Shin SJ, Weiss RS, Reinhart-King CA, Tissue stiffness regulates serine/arginine-rich protein-mediated splicing of the extra domain B-fibronectin isoform in tumors, Proc. Natl. Acad. Sci 112 (2015) 8314–8319. 10.1073/pnas.1505421112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Miroshnikova YA, Rozenberg GI, Cassereau L, Pickup M, Mouw JK, Ou G, Templeman KL, Hannachi E-I, Gooch KJ, Sarang-Sieminski AL, García AJ, Weaver VM, α5β1-Integrin promotes tension-dependent mammary epithelial cell invasion by engaging the fibronectin synergy site, Mol. Biol. Cell 28 (2017) 2958–2977. 10.1091/mbc.e17-02-0126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Wang W, Lollis EM, Bordeleau F, Reinhart-King CA, Matrix stiffness regulates vascular integrity through focal adhesion kinase activity, FASEB J. 33 (2018) 1199–1208. 10.1096/fj.201800841R. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Wang W, Miller JP, Pannullo SC, Reinhart-King CA, Bordeleau F, Quantitative assessment of cell contractility using polarized light microscopy, J. Biophotonics 11 (2018) e201800008. 10.1002/jbio.201800008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Levental KR, Yu H, Kass L, Lakins JN, Egeblad M, Erler JT, Fong SFT, Csiszar K, Giaccia A, Weninger W, Yamauchi M, Gasser DL, Weaver VM, Matrix crosslinking forces tumor progression by enhancing integrin signaling, Cell. 139 (2009) 891–906. 10.1016/j.cell.2009.10.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Lopez JI, Kang I, You W-K, McDonald DM, Weaver VM, In situ force mapping of mammary gland transformation, Integr. Biol. Quant. Biosci. Nano Macro 3 (2011) 910–921. 10.1039/c1ib00043h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Nicolas-Boluda A, Vaquero J, Vimeux L, Guilbert T, Barrin S, Kantari-Mimoun C, Ponzo M, Renault G, Deptula P, Pogoda K, Bucki R, Cascone I, Courty J, Fouassier L, Gazeau F, Donnadieu E, Tumor stiffening reversion through collagen crosslinking inhibition improves T cell migration and anti-PD-1 treatment, ELife. 10 (2021) e58688. 10.7554/eLife.58688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Tang H, Leung L, Saturno G, Viros A, Smith D, Di Leva G, Morrison E, Niculescu-Duvaz D, Lopes F, Johnson L, Dhomen N, Springer C, Marais R, Lysyl oxidase drives tumour progression by trapping EGF receptors at the cell surface, Nat. Commun 8 (2017) 14909. 10.1038/ncomms14909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Herring CA, Banerjee A, McKinley ET, Simmons AJ, Ping J, Roland JT, Franklin JL, Liu Q, Gerdes MJ, Coffey RJ, Lau KS, Unsupervised Trajectory Analysis of Single-Cell RNA-Seq and Imaging Data Reveals Alternative Tuft Cell Origins in the Gut, Cell Syst. 6 (2018) 37–51.e9. 10.1016/j.cels.2017.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Ma L, Hernandez MO, Zhao Y, Mehta M, Tran B, Kelly M, Rae Z, Hernandez JM, Davis JL, Martin SP, Kleiner DE, Hewitt SM, Ylaya K, Wood BJ, Greten TF, Wang XW, Tumor cell biodiversity drives microenvironmental reprogramming in liver cancer, Cancer Cell. 36 (2019) 418–430.e6. 10.1016/j.ccell.2019.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Efremova M, Vento-Tormo M, Teichmann SA, Vento-Tormo R, CellPhoneDB: inferring cell–cell communication from combined expression of multi-subunit ligand–receptor complexes, Nat. Protoc. 15 (2020) 1484–1506. 10.1038/s41596-020-0292-x. [DOI] [PubMed] [Google Scholar]
- [25].Chen B, Ramirez-Solano MA, Heiser CN, Liu Q, Lau KS, Processing single-cell RNA-seq data for dimension reduction-based analyses using open-source tools, STAR Protoc. 2 (2021) 100450. 10.1016/j.xpro.2021.100450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Hafemeister C, Satija R, Normalization and variance stabilization of single-cell RNA-seq data using regularized negative binomial regression, Genome Biol. 20 (2019) 296. 10.1186/s13059-019-1874-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Lawrence T, Natoli G, Transcriptional regulation of macrophage polarization: enabling diversity with identity, Nat. Rev. Immunol 11 (2011) 750–761. 10.1038/nri3088. [DOI] [PubMed] [Google Scholar]
- [28].Murray PJ, Macrophage Polarization, Annu. Rev. Physiol 79 (2017) 541–566. 10.1146/annurev-physiol-022516-034339. [DOI] [PubMed] [Google Scholar]
- [29].Shiau D-J, Kuo W-T, Davuluri GVN, Shieh C-C, Tsai P-J, Chen C-C, Lin Y-S, Wu Y-Z, Hsiao Y-P, Chang C-P, Hepatocellular carcinoma-derived high mobility group box 1 triggers M2 macrophage polarization via a TLR2/NOX2/autophagy axis, Sci. Rep 10 (2020) 13582. 10.1038/s41598-020-70137-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Chang C-P, Su Y-C, Hu C-W, Lei H-Y, TLR2-dependent selective autophagy regulates NF- κ B lysosomal degradation in hepatoma-derived M2 macrophage differentiation, Cell Death Differ. 20 (2013) 515–523. 10.1038/cdd.2012.146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Yang ZF, Ho DW, Lau CK, Lam CT, Lum CT, Poon RTP, Fan ST, Allograft inflammatory factor-1 (AIF-1) is crucial for the survival and pro-inflammatory activity of macrophages, Int. Immunol 17 (2005) 1391–1397. 10.1093/intimm/dxh316. [DOI] [PubMed] [Google Scholar]
- [32].Sánchez-Martín L, Estecha A, Samaniego R, Sánchez-Ramón S, Vega MÁ, Sánchez-Mateos P, The chemokine CXCL12 regulates monocyte-macrophage differentiation and RUNX3 expression, Blood. 117 (2011) 88–97. 10.1182/blood-2009-12-258186. [DOI] [PubMed] [Google Scholar]
- [33].Bai T, Chen C-C, Lau LF, Matricellular Protein CCN1 Activates a Proinflammatory Genetic Program in Murine Macrophages, J. Immunol 184 (2010) 3223–3232. 10.4049/jimmunol.0902792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Guo H, Jin D, Chen X, Lipocalin 2 is a Regulator Of Macrophage Polarization and NF-κB/STAT3 Pathway Activation, Mol. Endocrinol 28 (2014) 1616–1628. 10.1210/me.2014-1092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Doebel T, Voisin B, Nagao K, Langerhans Cells - The Macrophage in Dendritic Cell Clothing, Trends Immunol. 38 (2017) 817–828. 10.1016/j.it.2017.06.008. [DOI] [PubMed] [Google Scholar]
- [36].Ouchi T, Nakato G, Udey MC, EpCAM Expressed by Murine Epidermal Langerhans Cells Modulates Immunization to an Epicutaneously Applied Protein Antigen, J. Invest. Dermatol 136 (2016) 1627–1635. 10.1016/j.jid.2016.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Stutte S, Jux B, Esser C, Förster I, CD24a Expression Levels Discriminate Langerhans Cells from Dermal Dendritic Cells in Murine Skin and Lymph Nodes, J. Invest. Dermatol 128 (2008) 1470–1475. 10.1038/sj.jid.5701228. [DOI] [PubMed] [Google Scholar]
- [38].Tsuge T, Yamakawa M, Tsukamoto M, Infiltrating dendritic/Langerhans cells in primary breast cancer, Breast Cancer Res. Treat 59 (2000) 141–152. 10.1023/A:1006396216933. [DOI] [PubMed] [Google Scholar]
- [39].Zhang Y-H, He M, Wang Y, Liao A-H, Modulators of the Balance between M1 and M2 Macrophages during Pregnancy, Front. Immunol 8 (2017). 10.3389/fimmu.2017.00120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Mantovani A, Sozzani S, Locati M, Allavena P, Sica A, Macrophage polarization: tumor-associated macrophages as a paradigm for polarized M2 mononuclear phagocytes, Trends Immunol. 23 (2002) 549–555. 10.1016/S1471-4906(02)02302-5. [DOI] [PubMed] [Google Scholar]
- [41].Cochain C, Vafadarnejad E, Arampatzi P, Pelisek J, Winkels H, Ley K, Wolf D, Saliba A-E, Zernecke A, Single-Cell RNA-Seq Reveals the Transcriptional Landscape and Heterogeneity of Aortic Macrophages in Murine Atherosclerosis, Circ. Res 122 (2018) 1661–1674. 10.1161/CIRCRESAHA.117.312509. [DOI] [PubMed] [Google Scholar]
- [42].Orecchioni M, Ghosheh Y, Pramod AB, Ley K, Macrophage Polarization: Different Gene Signatures in M1(LPS+) vs. Classically and M2(LPS–) vs. Alternatively Activated Macrophages, Front. Immunol 10 (2019). 10.3389/fimmu.2019.01084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Kamata T, Jin H, Giblett S, Patel B, Patel F, Foster C, Pritchard C, The cholesterol-binding protein NPC2 restrains recruitment of stromal macrophage-lineage cells to early-stage lung tumours, EMBO Mol. Med 7 (2015) 1119–1137. 10.15252/emmm.201404838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Puolakkainen P, Koski A, Vainionpää S, Shen Z, Repo H, Kemppainen E, Mustonen H, Seppänen H, Anti-inflammatory macrophages activate invasion in pancreatic adenocarcinoma by increasing the MMP9 and ADAM8 expression, Med. Oncol 31 (2014) 884. 10.1007/s12032-014-0884-9. [DOI] [PubMed] [Google Scholar]
- [45].Zhang Y, Du W, Chen Z, Xiang C, Upregulation of PD-L1 by SPP1 mediates macrophage polarization and facilitates immune escape in lung adenocarcinoma, Exp. Cell Res 359 (2017) 449–457. 10.1016/j.yexcr.2017.08.028. [DOI] [PubMed] [Google Scholar]
- [46].Liu K, Zhao E, Ilyas G, Lalazar G, Lin Y, Haseeb M, Tanaka KE, Czaja MJ, Impaired macrophage autophagy increases the immune response in obese mice by promoting proinflammatory macrophage polarization, Autophagy. 11 (2015) 271–284. 10.1080/15548627.2015.1009787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Wang L, Zhang S, Wu H, Rong X, Guo J, M2b macrophage polarization and its roles in diseases, J. Leukoc. Biol 106 (2019) 345–358. 10.1002/JLB.3RU1018-378RR. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Moradi-Chaleshtori M, Shojaei S, Mohammadi-Yeganeh S, Hashemi SM, Transfer of miRNA in tumor-derived exosomes suppresses breast tumor cell invasion and migration by inducing M1 polarization in macrophages, Life Sci. 282 (2021) 119800. 10.1016/j.lfs.2021.119800. [DOI] [PubMed] [Google Scholar]
- [49].Metcalf D, The Colony-Stimulating Factors and Cancer, Cancer Immunol. Res 1 (2013) 351–356. 10.1158/2326-6066.CIR-13-0151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Cooper J, Giancotti FG, Integrin Signaling in Cancer: Mechanotransduction, Stemness, Epithelial Plasticity, and Therapeutic Resistance, Cancer Cell. 35 (2019) 347–367. 10.1016/j.ccell.2019.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Puram SV, Tirosh I, Parikh AS, Patel AP, Yizhak K, Gillespie S, Rodman C, Luo CL, Mroz EA, Emerick KS, Deschler DG, Varvares MA, Mylvaganam R, Rozenblatt-Rosen O, Rocco JW, Faquin WC, Lin DT, Regev A, Bernstein BE, Single-Cell Transcriptomic Analysis of Primary and Metastatic Tumor Ecosystems in Head and Neck Cancer, Cell. 171 (2017) 1611–1624.e24. 10.1016/j.cell.2017.10.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Pelon F, Bourachot B, Kieffer Y, Magagna I, Mermet-Meillon F, Bonnet I, Costa A, Givel A-M, Attieh Y, Barbazan J, Bonneau C, Fuhrmann L, Descroix S, Vignjevic D, Silberzan P, Parrini MC, Vincent-Salomon A, Mechta-Grigoriou F, Cancer-associated fibroblast heterogeneity in axillary lymph nodes drives metastases in breast cancer through complementary mechanisms, Nat. Commun 11 (2020) 1–20. 10.1038/s41467-019-14134-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Martinez FO, Gordon S, The M1 and M2 paradigm of macrophage activation: time for reassessment, F1000Prime Rep. 6 (2014). 10.12703/P6-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Solinas G, Schiarea S, Liguori M, Fabbri M, Pesce S, Zammataro L, Pasqualini F, Nebuloni M, Chiabrando C, Mantovani A, Allavena P, Tumor-Conditioned Macrophages Secrete Migration-Stimulating Factor: A New Marker for M2-Polarization, Influencing Tumor Cell Motility, J. Immunol 185 (2010) 642–652. 10.4049/jimmunol.1000413. [DOI] [PubMed] [Google Scholar]
- [55].Kurahara H, Shinchi H, Mataki Y, Maemura K, Noma H, Kubo F, Sakoda M, Ueno S, Natsugoe S, Takao S, Significance of M2-Polarized Tumor-Associated Macrophage in Pancreatic Cancer, J. Surg. Res 167 (2011) e211–e219. 10.1016/j.jss.2009.05.026. [DOI] [PubMed] [Google Scholar]
- [56].Sica A, Schioppa T, Mantovani A, Allavena P, Tumour-associated macrophages are a distinct M2 polarised population promoting tumour progression: Potential targets of anti-cancer therapy, Eur. J. Cancer 42 (2006) 717–727. 10.1016/j.ejca.2006.01.003. [DOI] [PubMed] [Google Scholar]
- [57].Tripathi C, Tewari BN, Kanchan RK, Baghel KS, Nautiyal N, Shrivastava R, Kaur H, Bhatt MLB, Bhadauria S, Macrophages are recruited to hypoxic tumor areas and acquire a Pro-Angiogenic M2-Polarized phenotype via hypoxic cancer cell derived cytokines Oncostatin M and Eotaxin, Oncotarget. 5 (2014) 5350–5368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [58].Chen Y, Zhang S, Wang Q, Zhang X, Tumor-recruited M2 macrophages promote gastric and breast cancer metastasis via M2 macrophage-secreted CHI3L1 protein, J. Hematol. Oncol.J Hematol Oncol 10 (2017) 36. 10.1186/s13045-017-0408-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [59].Ding T, Xu J, Wang F, Shi M, Zhang Y, Li S-P, Zheng L, High tumor-infiltrating macrophage density predicts poor prognosis in patients with primary hepatocellular carcinoma after resection, Hum. Pathol 40 (2009) 381–389. 10.1016/j.humpath.2008.08.011. [DOI] [PubMed] [Google Scholar]
- [60].Takanami I, Takeuchi K, Kodaira S, Tumor-Associated Macrophage Infiltration in Pulmonary Adenocarcinoma: Association with Angiogenesis and Poor Prognosis, Oncology. 57 (1999) 138–142. 10.1159/000012021. [DOI] [PubMed] [Google Scholar]
- [61].Subimerb C, Pinlaor S, Khuntikeo N, Leelayuwat C, Morris A, McGrath MS, Wongkham S, Tissue invasive macrophage density is correlated with prognosis in cholangiocarcinoma, Mol. Med. Rep 3 (2010) 597–605. 10.3892/mmr_00000303. [DOI] [PubMed] [Google Scholar]
- [62].Najafi M, Goradel NH, Farhood B, Salehi E, Nashtaei MS, Khanlarkhani N, Khezri Z, Majidpoor J, Abouzaripour M, Habibi M, Kashani IR, Mortezaee K, Macrophage polarity in cancer: A review, J. Cell. Biochem 120 (2019) 2756–2765. 10.1002/jcb.27646. [DOI] [PubMed] [Google Scholar]
- [63].Ma J, Liu L, Che G, Yu N, Dai F, You Z, The M1 form of tumor-associated macrophages in non-small cell lung cancer is positively associated with survival time, BMC Cancer. 10 (2010) 112. 10.1186/1471-2407-10-112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [64].Ohri CM, Shikotra A, Green RH, Waller DA, Bradding P, Macrophages within NSCLC tumour islets are predominantly of a cytotoxic M1 phenotype associated with extended survival, Eur. Respir. J 33 (2009) 118–126. 10.1183/09031936.00065708. [DOI] [PubMed] [Google Scholar]
- [65].Chaturvedi P, Gilkes DM, Takano N, Semenza GL, Hypoxia-inducible factor-dependent signaling between triple-negative breast cancer cells and mesenchymal stem cells promotes macrophage recruitment, Proc. Natl. Acad. Sci 111 (2014) E2120–E2129. 10.1073/pnas.1406655111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [66].Hind LE, Dembo M, Hammer DA, Macrophage motility is driven by frontal-towing with a force magnitude dependent on substrate stiffness, Integr. Biol 7 (2015) 447–453. 10.1039/c4ib00260a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [67].Friedemann M, Kalbitzer L, Franz S, Moeller S, Schnabelrauch M, Simon J-C, Pompe T, Franke K, Instructing Human Macrophage Polarization by Stiffness and Glycosaminoglycan Functionalization in 3D Collagen Networks, Adv. Healthc. Mater 6 (2017) 1600967. 10.1002/adhm.201600967. [DOI] [PubMed] [Google Scholar]
- [68].Sridharan R, Ryan EJ, Kearney CJ, Kelly DJ, O’Brien FJ, Macrophage Polarization in Response to Collagen Scaffold Stiffness Is Dependent on Cross-Linking Agent Used To Modulate the Stiffness, ACS Biomater. Sci. Eng 5 (2019) 544–552. 10.1021/acsbiomaterials.8b00910. [DOI] [PubMed] [Google Scholar]
- [69].Sridharan R, Cavanagh B, Cameron AR, Kelly DJ, O’Brien FJ, Material stiffness influences the polarization state, function and migration mode of macrophages, Acta Biomater. 89 (2019) 47–59. 10.1016/j.actbio.2019.02.048. [DOI] [PubMed] [Google Scholar]
- [70].Okamoto T, Takagi Y, Kawamoto E, Park EJ, Usuda H, Wada K, Shimaoka M, Reduced substrate stiffness promotes M2-like macrophage activation and enhances peroxisome proliferator-activated receptor γ expression, Exp. Cell Res 367 (2018) 264–273. 10.1016/j.yexcr.2018.04.005. [DOI] [PubMed] [Google Scholar]
- [71].Wu S, Yue H, Wu J, Zhang W, Jiang M, Ma G, The interacting role of physical stiffness and tumor cells on the macrophages polarization, Colloids Surf. Physicochem. Eng. Asp 552 (2018) 81–88. 10.1016/j.colsurfa.2018.04.026. [DOI] [Google Scholar]
- [72].Xue YZB, Niu YM, Tang B, Wang CM, PCL/EUG scaffolds with tunable stiffness can regulate macrophage secretion behavior, Prog. Biophys. Mol. Biol 148 (2019) 4–11. 10.1016/j.pbiomolbio.2019.05.006. [DOI] [PubMed] [Google Scholar]
- [73].Sapi E, The Role of CSF-1 in Normal Physiology of Mammary Gland and Breast Cancer: An Update, Exp. Biol. Med 229 (2004) 1–11. 10.1177/153537020422900101. [DOI] [PubMed] [Google Scholar]
- [74].Kuemmel S, Campone M, Loirat D, López RL, Beck JT, Laurentiis MD, Im S-A, Kim S-B, Kwong A, Steger GG, Adelantado EZ, Duhoux FP, Greil R, Kuter I, Lu Y-S, Tibau A, Özgüroğlu M, Scholz C, Singer CF, Vega E, Wimberger P, Zamagni C, Couillebault X-M, Fan L, Guerreiro N, Mataraza J, Sand-Dejmek J, Chan A, A Randomized Phase II Study of Anti-CSF-1 Monoclonal Antibody Lacnotuzumab (MCS110) Combined with Gemcitabine and Carboplatin in Advanced Triple Negative Breast Cancer, Clin. Cancer Res (2021). 10.1158/1078-0432.CCR-20-3955. [DOI] [PubMed] [Google Scholar]
- [75].Calvo A, Joensuu H, Sebastian M, Naing A, Bang Y-J, Martin M, Roda D, Hodi FS, Veloso A, Mataraza J, Baneyx G, Guerreiro N, Liao S, Rabault B, Fjaellskog M-L, Cervantes A, Phase Ib/II study of lacnotuzumab (MCS110) combined with spartalizumab (PDR001) in patients (pts) with advanced tumors., J. Clin. Oncol 36 (2018) 3014–3014. 10.1200/JCO.2018.36.15_suppl.3014. [DOI] [Google Scholar]
- [76].Lampi MC, Reinhart-King CA, Targeting extracellular matrix stiffness to attenuate disease: From molecular mechanisms to clinical trials, Sci. Transl. Med 10 (2018) eaao0475. 10.1126/scitranslmed.aao0475. [DOI] [PubMed] [Google Scholar]
- [77].Reid SE, Kay EJ, Neilson LJ, Henze A-T, Serneels J, McGhee EJ, Dhayade S, Nixon C, Mackey JB, Santi A, Swaminathan K, Athineos D, Papalazarou V, Patella F, Román-Fernández Á, ElMaghloob Y, Hernandez-Fernaud JR, Adams RH, Ismail S, Bryant DM, Salmeron-Sanchez M, Machesky LM, Carlin LM, Blyth K, Mazzone M, Zanivan S, Tumor matrix stiffness promotes metastatic cancer cell interaction with the endothelium, EMBO J. (2017) e201694912. 10.15252/embj.201694912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [78].Solinas G, Germano G, Mantovani A, Allavena P, Tumor-associated macrophages (TAM) as major players of the cancer-related inflammation, J. Leukoc. Biol 86 (2009) 1065–1073. 10.1189/jlb.0609385. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.