Abstract
Objective
Rates of type 2 diabetes (T2D) among adolescents are on the rise. Epigenetic changes could be associated with the metabolic alterations in adolescents with T2D.
Methods
We performed a cross sectional integrated analysis of DNA methylation data from peripheral blood mononuclear cells with serum metabolomic data from First Nation adolescents with T2D and controls participating in the Improving Renal Complications in Adolescents with type 2 diabetes through Research (iCARE) cohort study, to explore the molecular changes in adolescents with T2D.
Results
Our analysis showed that 43 serum metabolites and 36 differentially methylated regions (DMR) were associated with T2D. Several DMRs were located near the transcriptional start site of genes with established roles in metabolic disease and associated with altered serum metabolites (e.g. glucose, leucine, and gamma-glutamylisoleucine). These included the free fatty acid receptor-1 (FFAR1), upstream transcription factor-2 (USF2), and tumor necrosis factor-related protein-9 (C1QTNF9), among others.
Conclusions
We identified DMRs and metabolites that merit further investigation to determine their significance in controlling gene expression and metabolism which could define T2D risk in adolescents.
Keywords: type 2 diabetes mellitus, metabolomics, DNA methylation, integration of data, pediatrics
Introduction
Type 2 diabetes (T2D) is a global epidemic, but a major concern is the rising incidence among youth (1). In 1990 T2D accounted for only 3% of new diagnoses of diabetes among U.S. children, but by 2010 that number rose to 45% in some populations (2). In the Canadian province of Manitoba, the annual incidence of T2D in children increased from 22.8 to 35.7 cases per 100,000 children between 2007 and 2017, affecting a disproportionate number of First Nations youth (3). An understanding of the underlying pathology of T2D is paramount to improving clinical outcomes.
Adult-onset T2D progresses gradually from impaired glucose tolerance to β-cell failure, but in youth, the loss of β-cell function is accelerated (4). Single gene polymorphisms alone do not explain the rapid rise of T2D in youth observed over a single generation, suggesting the pathophysiology of T2D in youth could involve additional gene and environment interactions (5). As our study population is First Nations youth in Canada, we remain cognizant of the environmental, social and political impact of colonization which has had a powerful detrimental effect on Indigenous populations. Colonization disrupted food sovereignty and connection to the land that blocked access to traditional farming, food-gathering, hunting and fishing practices that are fundamental to the maintenance of health within First Nations populations and an associated dependence on non-traditional foods with inferior nutrient qualities. Environmental alterations such as these could have a major influence on the epigenome (changes occurring on the DNA where the DNA sequence itself is not changed), which then may affect T2D risk (6, 7).
Metabolites play a key role as both biomarkers and mediators of T2D development, and metabolic perturbations could explain the aggressive course of T2D in youth (8, 9). Though studies have characterized the circulating metabolomic profile of adults with insulin resistance and T2D, data from pediatric populations are limited. Some studies reported that aromatic and branched chain amino acids (BCAAs) were associated with insulin resistance and T2D in youth (8, 10–13). Through a combination of indirect calorimetry and mass spectrometry researchers showed that, unlike adults with T2D, changes in acylcarnitine’s and fatty acid oxidation were not observed in youth with T2D (8). Collectively these findings suggest that the metabolic perturbations of T2D are different in adolescents compared to adults. Growing evidence suggests that epigenetic modifications including DNA methylation can be affected by the nutritional and metabolic state (14). Persistent changes in the methylome could also be associated with pathogenic metabolic profiles. For example, alterations in DNA methylation have been reported in peripheral blood mononuclear cells (PBMCs) and human islets from T2D adults (7). To date, no studies have linked DNA methylation and serum metabolomic profiling of adolescents diagnosed with T2D.
Since the pathogenesis of pediatric T2D is different from T2D in adults, the objective of this study was to explore changes in the epigenetic landscape of peripheral blood mononuclear cells that are associated with an altered serum metabolome. In this study, we linked differentially methylated regions (DMRs) to five biologically important metabolites that were significantly altered in adolescents with T2D. The metabolites correlated with several DMRs in adolescents with T2D that were located near the transcriptional start sites (TSS) of several biologically relevant genes, including the free fatty acid receptor-1 (FFAR1), upstream transcription factor-2 (USF2), and the novel cytokine, tumor necrosis factor-related protein-9 (C1QTNF9). These data will help us generate new hypotheses to investigate the mechanisms that influence the metabolomic profile of adolescents diagnosed with T2D.
Material and methods
iCARE cohort
The iCARE cohort study (15) received informed consent from study participants and approval from the University of Manitoba/Health Sciences Centre Research Ethics Board (HS13255), First Nations patient and parent advisory committee and the First Nation Health and Social Secretariat of Manitoba and the iCARE participant and parent advisory committees. In this study, we performed a subgroup analysis of the iCARE cohort that consisted of 12- to 24-year-old First Nations adolescents diagnosed with T2D prior to 18 years of age (mean age of 15). The controls are normoglycemic overweight or obese adolescents at risk of developing T2D (mean age of 16). All of the patients were fasting a minimum of 8h prior to sample collection. The diagnosis of T2D was based on biochemical and clinical criteria and the absence of insulin and glutamic acid decarboxylase antibodies. All the clinical parameters are shown in Table 1 (for the whole cohort) and Supplementary Table 3 (samples selected from the cohort for SOLiD sequencing). Non-adjusted p-values were calculated using Student’s t- test and Chi-Square test.
Table 1.
Anthropometric characteristics of the primary cohort used in the study.
| Anthropometric characteristics | Control adolescents (n=42) | Adolescents with T2D (n=113) | P-value | Statistical test |
|---|---|---|---|---|
| Age (years) | 16.08 (3.11) | 15.24 (2.58) | 0.09 | t-test |
| Gender (Male/Female) | (14/28) | (35/78) | 0.78 | Chi-Square |
| Weight (kg) | 90.33 (23.56) | 86.46 (22.73) | 0.35 | t-test |
| Height (cm) | 166.00 (8.53) | 165.28 (9.64) | 0.67 | t-test |
| Waist (cm) | 104.96 (19.72) | 105.03 (18.28) | 0.98 | t-test |
| BMI (kg/m²) | 32.43 (6.79) | 31.37 (6.49) | 0.37 | t-test |
| BMI Z-score | 1.66 (0.83) | 1.79 (0.72) | 0.31 | t-test |
| Duration of diabetes (years) | / | 2.12 [3.12] | / | / |
| Albuminuria (%) | 9.5 | 39.6 | <0.001 | Chi-Square |
| Ambulatory Hypertension (%) | 22.5 | 22.8 | 0.92 | Chi-Square |
| Nocturnal Hypertension (%) | 22.5 | 32.7 | 0.23 | Chi-Square |
| Plasma glucose (mmol/L) | 3.44 (1.59) | 11.38 (5.95) | <0.001 | t-test |
| ALT (units/L) | 25.64 (23.05) | 29.28 (22.28) | 0.32 | t-test |
| AST (units/L) | 22.61 (10.35) | 22.45 (14.91) | 0.95 | t-test |
| HbA1c (mmol/mol) | 5.65 (0.24) | 9.38 (2.73) | <0.001 | t-test |
| Total Cholesterol (mmol/L) | 3.91 (0.67) | 4.45 (0.98) | <0.01 | t-test |
| Triglycerides (mmol/L) | 1.28 (0.64) | 2.19 (2.13) | <0.01 | t-test |
| HDL (mmol/L) | 1.22 (0.31) | 1.12 (0.28) | 0.06 | t-test |
| LDL (mmol/L) | 2.10 (0.51) | 2.37 (0.68) | <0.05 | t-test |
| Total Cholesterol/HDL Ratio | 3.37 (0.93) | 3.95 (1.19) | <0.05 | t-test |
| LDL/HDL Ratio | 1.83 (0.64) | 2.17 (0.79) | <0.05 | t-test |
Values are means (SD). For the variable “duration of diabetes”, values are presented as median [interquartile range]. P values ≦ 0.05 were considered significant. P-values were calculated using Student’s t-test and Chi-Square test.
T2D, type 2 diabetes mellitus; BMI, body mass index; ALT, alanine aminotransferase; AST, aspartate aminotransferase; HbA1c, glycated hemoglobin.
There are 42 controls and 113 patients with T2D included in this study.
Metabolomics analysis of T2D adolescent serum
Samples were prepared using the automated MicroLab STAR® system (Hamilton Company, Boston, U.S.A). Metabolomic methods utilized a Waters ACQUITY ultra-performance liquid chromatography (UPLC) and a Thermo Scientific Q-Exactive high resolution/accurate mass spectrometer interfaced with a heated electrospray ionization (HESI-II) source and Orbitrap mass analyzer operated at 35,000 mass resolution. [See Supplementary Methods (Supplement S1) ]. Raw data was extracted, peak-identified and QC processed using Metabolon’s hardware and software. Compounds were identified by comparison to library entries of purified standards or recurrent unknown entities. Entities with more than 20% missing values were removed, MetaboAnalyst 4.0 was used to impute values (replace the value by a small value which is half of the minimum positive value in the original data), followed by data filtering and auto scaling. Details of metabolite quantification and data scaling are found in the Supplementary Methods (Supplement S1) .
SOLiD library preparation and bioinformatics analysis
Genomic DNA was extracted from the peripheral blood mononuclear cells (PBMC) from a subset of the cohort that were included in the serum metabolomics dataset and was comprised of 21 adolescents with T2D and 10 control participants. Libraries were prepared according to the MethylMiner™ manufacturer’s protocol (ThermoFisher scientific Catalog number ME10025). MethylMiner enriches double-stranded methylated DNA based on CpG methylation density, with increased sensitivity over antibody-based methods. The methylated DNA obtained was subjected to SOLiD sequencing where 50-bp single end sequence reads were ensured by quality check (noise to signal ratio). The sequence reads were mapped to the human reference genome (hg19) using the MethylMiner™ Mapping Analysis module of the LifeScope v2.5.1 software package (Life Technologies).
Data preprocessing and normalization
Regions enriched with DNA methylation across the genome were identified using the “callpeak” function from Model-based Analysis of ChIP-seq (MACS2) (16), with model fold = [5, 30] and FDR < 0.05 on the aligned reads generating 31 peaksets. The below peak filtering was done using the DiffBind package in R (17). As we included males and females in the cohort, we removed sex chromosomes using ENCODE blacklist regions (18) and those present in at most 2 samples were excluded from downstream analyses, leaving a total of 732 984 peaks. The edgeR package (19) was used to normalize for sequencing depth and effective library size by transforming data into counts per million and performing the trimmed mean of M-value (TMM) normalization.
Linear modelling
Cell-type proportion effects were corrected for using the sva package (20) reference-free cell-type correction method. The top 2 surrogate variables were included in the regression model. A generalized linear model in edger was used to identify differentially methylated regions (DMRs) between diabetes cases and controls using the following formula: Reads ~ Diabetes status + Age + Sex + BMI + SVs. Multiple testing was corrected for using the Benjamini-Hochberg method. Statistical significance was set at FDR < 0.05.
Multivariate statistical analysis
The multiomic data sets (metabolomic and DNA methylation data) were analyzed using the multivariate statistical analysis tools found in SIMCA (version 13; Umetrics AB, Umeå, Sweden) and Metaboanalyst 5.0. Unsupervised hierarchical clustering was performed for the 43 statistically significant metabolites to identify different metabolite clusters. Based on the relevance of metabolites to the profile of T2D patients from other studies, we selected five metabolites from each of the separate clusters for data fusion (gamma-glutamylisoleucine, glucose, leucine, palmitoylcholine and sphingomyelin). We used a supervised classification method called orthogonal partial least-squares discriminant analysis (OPLS-DA) to identify the metabolites and DMRs that are most interesting for this analysis. We quantified model statistics based on the fraction of the sum of squares for the selected component (R 2), which equates to the percentage of the model variance explained, and the predictive ability (Q 2). Cross-validation was performed to predict and estimate the model performance (whether models were over fitted). For OPLS-DA models, random permutation was used whereby the class membership of individual samples are permuted randomly. In addition, ANOVA of the cross-validated residuals (CV-ANOVA) test was performed within Simca to further validate the models validated by selecting two thirds of the samples randomly and then predicting the class membership of the rest of the one third. We used variable importance in the projection scores (VIP) to prioritize the metabolites. A VIP score cutoff of >1.5 was considered in the model (21).
Peak annotation and omics data fusion and visualization
The peaks were annotated to -5000 bp to 5000 bp of the (TSS of the nearest gene) the nearest gene using the software Genomic region enrichment of Annotation s tool (GREAT v 4.04) where human genome assembly hg19 was used for annotation and for the background the whole genome was used. Omics data fusion was performed on selected metabolites and the peaks that were annotated to the closest genes. The selected metabolites and DMRs were fused based on the Pearson correlation values and visualized via R statistical software (https://www.r-project.org/) package called qgraph (22). IGV v.2.13.2 was used to make the genomic track where the BAM files for all the controls and T2D samples were used. All of the tracks were auto scaled.
Results
Patient characteristics
We used a cross sectional design to compare the serum metabolomic profiles of 113 First Nations adolescents (age range 10 - 24 years and BMI range of 19 – 48 kg/m2) with T2D to 42 normoglycemic First Nations controls. Higher levels of fasting blood glucose and HbA1c were observed in the adolescents with T2D compared to controls ( Table 1 ; p-value <0.001 using t-test). Among adolescents with T2D, the average time from diagnosis of T2D was 2.12 years. HbA1c was associated with weight (r=-0.20), waist circumference (r=-0.24) and BMI z-score (r=-0.31); however, in controls HbA1c was associated with ALT (r=0.32) ( Supplementary Figure 2 ).
Metabolites associated with T2D in adolescents
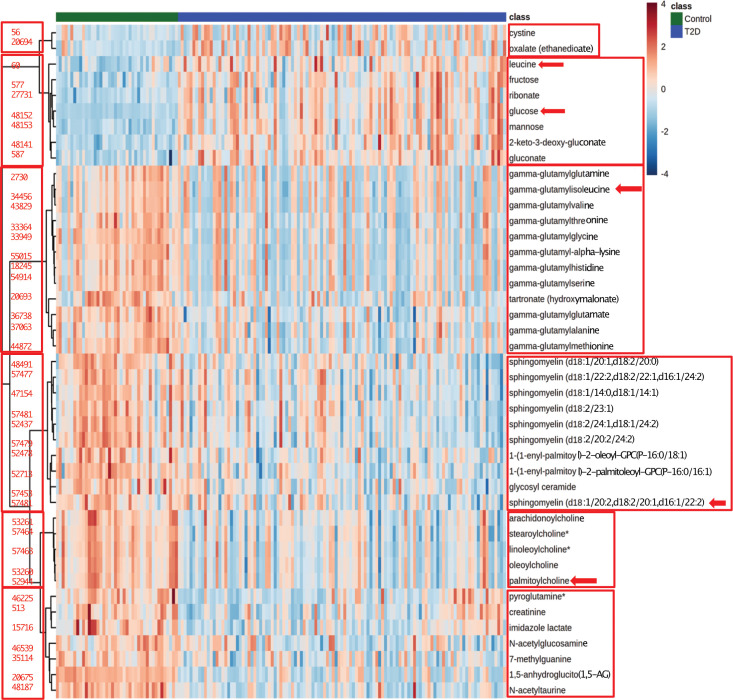
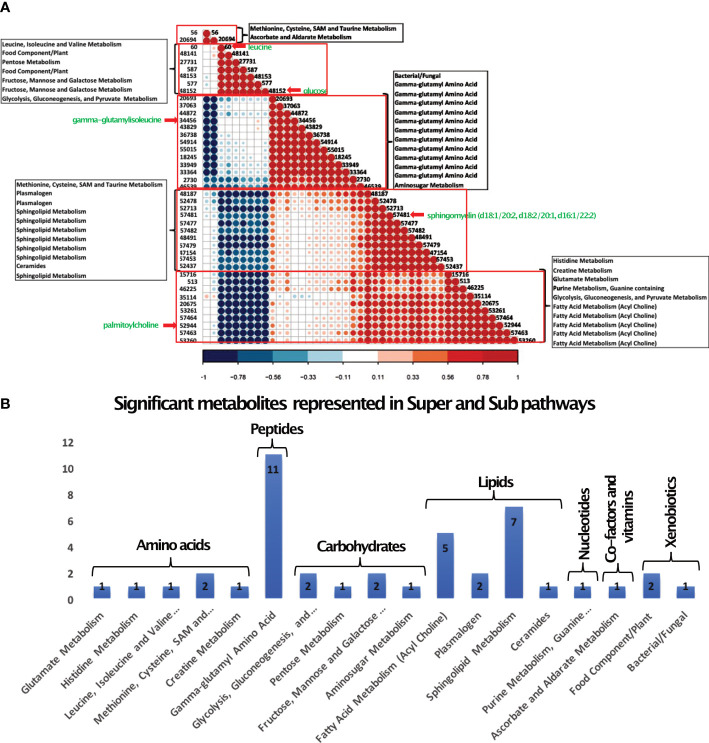
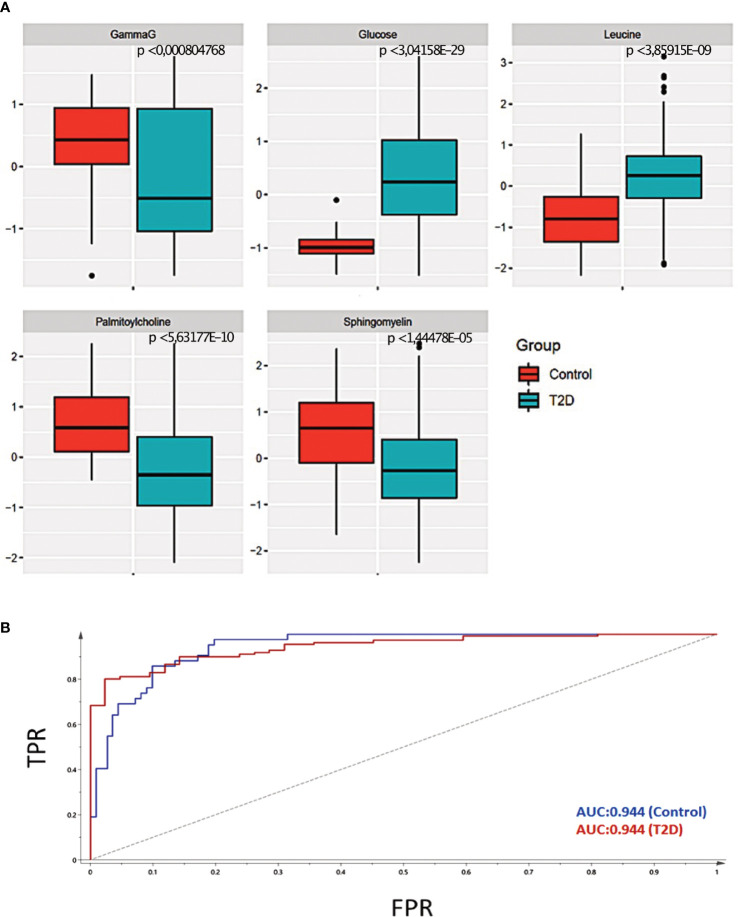
To characterize metabolic changes that are associated with T2D development in adolescents, we performed UPLC-MS/MS on the serum of fasted individuals. We initially identified a total of 820 individual metabolites. After the missing value estimation features with more than 20% missing values were removed, then after preprocessing we obtained 481 metabolites ( Supplementary Figure 1A ). A VIP score cutoff of >1.5 resulted in 43 significant metabolites ( Table 2 ). To show the most significant super pathways for the significant metabolites, we plotted a fold change vs p-value volcano plot ( Supplementary Figure 5A ) and a frequency bar plot ( Supplementary Figure 5B ). We used an unsupervised approach and performed PCA analysis where the T2D adolescents were clearly separated from the control group ( Figure 1A ). The permutation plot of the PCA ( Figure 1B ) validates the robustness of the model. Further we used an orthogonal partial least discriminant analysis (OPLS-DA) predictive model with 43 metabolites and the variation explained or goodness of fit in control vs. T2D patients (R2) of 63.8% and a predictive variation or goodness of prediction (Q 2) had a value of 59.7% ( Supplementary Figure 3A ). To verify our model, we permuted the group labels (control and T2D patients) 100 times to generate random models and observed that our model was significantly different from the permuted variations ( Supplementary Figure 3B ). Differential levels of the 43 significant metabolites between control and T2D patient samples are shown in ( Figure 2 ). Next, we performed unsupervised hierarchical clustering for the 43 metabolites, which formed five different clusters. Each cluster represented metabolites that mostly belong to similar sub-pathways ( Figure 3A ). Broadly, these 43 metabolites were categorized into seven super pathways, including lipids (35%), peptides (26% gamma-glutamyl amino acids), amino acids (10%), carbohydrates (17%), nucleotides (2% purine metabolism), cofactors and vitamins (2% ascorbate and aldarate metabolism), and xenobiotics (7%) ( Figure 3B ). Based on their biological relevance to metabolic health in diabetes, we selected metabolites from most of the clusters ( Figure 3A ). The differential levels of the selected metabolites (gamma-glutamyl isoleucine, glucose, leucine, palmitoyl choline and sphingomyelin) were statistically significant ( Figure 4A ; p <0.05) and were further used to integrate with the epigenetic data. To understand the robustness and accuracy of the five selected metabolites we estimated the area under the curve (AUC), which was 0.944 for both controls and T2D, demonstrating the robustness of our model ( Figure 4B ).
Table 2.
Significant Serum Metabolites in Youth with T2D.
| Biochemical | Comp ID | Super Pathway | Sub Pathway | KEGG ID | HMDB ID | PUBCHEM | Log2(FC) | FDR |
|---|---|---|---|---|---|---|---|---|
| Pyroglutamine | 46225 | Amino Acid | Glutamate Metabolism | NA | NA | 134508 | -0.95336 | 1.11E-07 |
| Imidazole Lactate | 15716 | Amino Acid | Histidine Metabolism | C05568 | HMDB02320 | 440129 | -0.54818 | 2.98E-08 |
| Leucine | 60 | Amino Acid | Leucine, Isoleucine and Valine Metabolism | C00123 | HMDB00687 | 6106 | 0.29615 | 3.06E-07 |
| Cystine | 56 | Amino Acid | Methionine, Cysteine, SAM and Taurine Metabolism | C00491 | HMDB00192 | 67678 | 0.85621 | 0.017982 |
| N-Acetyltaurine | 48187 | Amino Acid | Methionine, Cysteine, SAM and Taurine Metabolism | NA | NA | 159864 | -0.93978 | 3.07E-16 |
| Creatinine | 513 | Amino Acid | Creatine Metabolism | C00791 | HMDB00562 | 588 | -0.23991 | 0.000023 |
| Gamma-Glutamylalanine | 37063 | Peptide | Gamma-glutamyl Amino Acid | NA | HMDB29142 | 440103 | -0.66669 | 0.000081 |
| Gamma-Glutamylglutamate | 36738 | Peptide | Gamma-glutamyl Amino Acid | C05282 | HMDB11737 | 92865 | -0.63692 | 0.004894 |
| Gamma-Glutamylglutamine | 2730 | Peptide | Gamma-glutamyl Amino Acid | C05283 | HMDB11738 | 150914 | -0.68509 | 5.84E-10 |
| Gamma-Glutamylglycine | 33949 | Peptide | Gamma-glutamyl Amino Acid | NA | HMDB11667 | 165527 | -1.1881 | 2.0E-08 |
| Gamma-Glutamylhistidine | 18245 | Peptide | Gamma-glutamyl Amino Acid | NA | NA | 7017195 | -0.74326 | 0.000024 |
| Gamma-Glutamylisoleucine | 34456 | Peptide | Gamma-glutamyl Amino Acid | NA | HMDB11170 | 14253342 | -0.39281 | 0.020489 |
| Gamma-Glutamyl-Alpha-Lysine | 55015 | Peptide | Gamma-glutamyl Amino Acid | NA | NA | 65254 | -0.74009 | 0.000010 |
| Gamma-Glutamylmethionine | 44872 | Peptide | Gamma-glutamyl Amino Acid | NA | HMDB29155 | 7009567 | -1.0817 | 1.74E-09 |
| Gamma-Glutamylthreonine | 33364 | Peptide | Gamma-glutamyl Amino Acid | NA | HMDB29159 | 76078708 | -0.72121 | 0.000049 |
| Gamma-Glutamylvaline | 43829 | Peptide | Gamma-glutamyl Amino Acid | NA | HMDB11172 | 7015683 | -0.49642 | 0.008625 |
| Gamma-Glutamylserine | 54914 | Peptide | Gamma-glutamyl Amino Acid | NA | NA | 22844748 | -0.68682 | 0.000036 |
| 1,5-Anhydroglucitol (1,5-AG) | 20675 | Carbohydrate | Glycolysis, Gluconeogenesis, and Pyruvate Metabolism | C07326 | HMDB02712 | 64960 | -1.997 | 9.46E-20 |
| Glucose | 48152 | Carbohydrate | Glycolysis, Gluconeogenesis, and Pyruvate Metabolism | C00031 | HMDB00122 | 79025 | 0.95686 | 2.62E-14 |
| Ribonate | 27731 | Carbohydrate | Pentose Metabolism | C01685 | HMDB00867 | 5460677 | 1.0173 | 2.31E-09 |
| Fructose | 577 | Carbohydrate | Fructose, Mannose and Galactose Metabolism | C00095 | HMDB00660 | 5984 | 0.99634 | 3.68E-09 |
| Mannose | 48153 | Carbohydrate | Fructose, Mannose and Galactose Metabolism | C00159 | HMDB00169 | 18950 | 1.0482 | 1.41E-12 |
| N-Acetyl-glucosamine/N-Acetylgalactosamine | 46539 | Carbohydrate | Aminosugar Metabolism | NA | HMDB00215 | 24139 | -0.27191 | 0.000042 |
| Palmitoylcholine | 52944 | Lipid | Fatty Acid Metabolism (Acyl Choline) | NA | NA | 151731 | -1.0277 | 3.87E-08 |
| Oleoylcholine | 53260 | Lipid | Fatty Acid Metabolism (Acyl Choline) | NA | NA | 59040790 | -1.0407 | 8.96E-08 |
| Linoleoylcholine | 57463 | Lipid | Fatty Acid Metabolism (Acyl Choline) | NA | NA | NA | -1.1395 | 2.15E-09 |
| Stearoylcholine | 57464 | Lipid | Fatty Acid Metabolism (Acyl Choline) | NA | NA | NA | -1.1569 | 0.000000044 |
| Arachidonoylcholine | 53261 | Lipid | Fatty Acid Metabolism (Acyl Choline) | NA | NA | 122198216 | -1.2076 | 2.06E-08 |
| 1-(1-Enyl-Palmitoyl)-2-Palmitoleoyl-GPC (P-16:0/16:1) | 52713 | Lipid | Plasmalogen | NA | HMDB11207 | 52923882 | -0.50345 | 1.16E-10 |
| 1-(1-Enyl-Palmitoyl) -2-Oleoyl-GPC (P-16:0/18:1) | 52478 | Lipid | Plasmalogen | NA | NA | NA | -0.34017 | 2.49E-06 |
| Sphingomyelin (D18:2/14:0, D18:1/14:1) | 47154 | Lipid | Sphingolipid Metabolism | NA | NA | NA | -0.38072 | 0.000207 |
| Sphingomyelin (D18:1/20:1, D18:2/20:0) | 48491 | Lipid | Sphingolipid Metabolism | NA | NA | NA | -0.31732 | 1.00E-06 |
| Sphingomyelin (D18:2/24:1, D18:1/24:2) | 52437 | Lipid | Sphingolipid Metabolism | NA | NA | NA | -0.25523 | 0.000010 |
| Sphingomyelin (D18:2/23:1) | 57482 | Lipid | Sphingolipid Metabolism | NA | NA | NA | -0.34705 | 0.000085 |
| Sphingomyelin (D18:1/20:2, D18:2/20:1, D16:1/22:2) | 57481 | Lipid | Sphingolipid Metabolism | NA | NA | NA | -0.40633 | 0.000085 |
| Sphingomyelin (D18:2/20:2/24:2) | 57479 | Lipid | Sphingolipid Metabolism | NA | NA | NA | -0.51498 | 3.37E-10 |
| Sphingomyelin (D18:1/22:2, D18:2/22:1, D16:1/24:2) | 57477 | Lipid | Sphingolipid Metabolism | NA | NA | NA | -0.42945 | 2.98E-07 |
| Glycosyl Ceramide (D18:2/24:1, D18:1/24:2) | 57453 | Lipid | Ceramides | NA | NA | NA | -0.44786 | 8.52E-07 |
| 7-Methylguanine | 35114 | Nucleotide | Purine Metabolism, Guanine containing | C02242 | HMDB00897 | 11361 | -0.2625 | 0.000006 |
| Oxalate (Ethanedioate) | 20694 | Cofactors and Vitamins | Ascorbate and Aldarate Metabolism | C00209 | HMDB02329 | 971 | 1.2738 | 0.000087 |
| Gluconate | 587 | Xenobiotics | Food Component/Plant | C00257 | HMDB00625 | 10690 | 1.2306 | 1.16E-10 |
| 2-Keto-3-Deoxy-Gluconate | 48141 | Xenobiotics | Food Component/Plant | C00204 | HMDB01353 | 161227 | 1.2781 | 6.48E-10 |
| Tartronate (Hydroxymalonate) | 20693 | Xenobiotics | Bacterial/Fungal | C02287 | HMDB35227 | 45 | -0.96185 | 0.000006 |
NA, not available; FC, fold change; FDR, False discovery rate; Comp ID, Compound ID.
The super and the sub pathways of metabolites determined by their KEGG, HMDB and, PUBCHEM ids. The folds changes are converted to log2 values. FDR calculated unpaired t-test. The negative sign indicates the lower levels and positive value is higher levels as compared to the controls.
Figure 1.
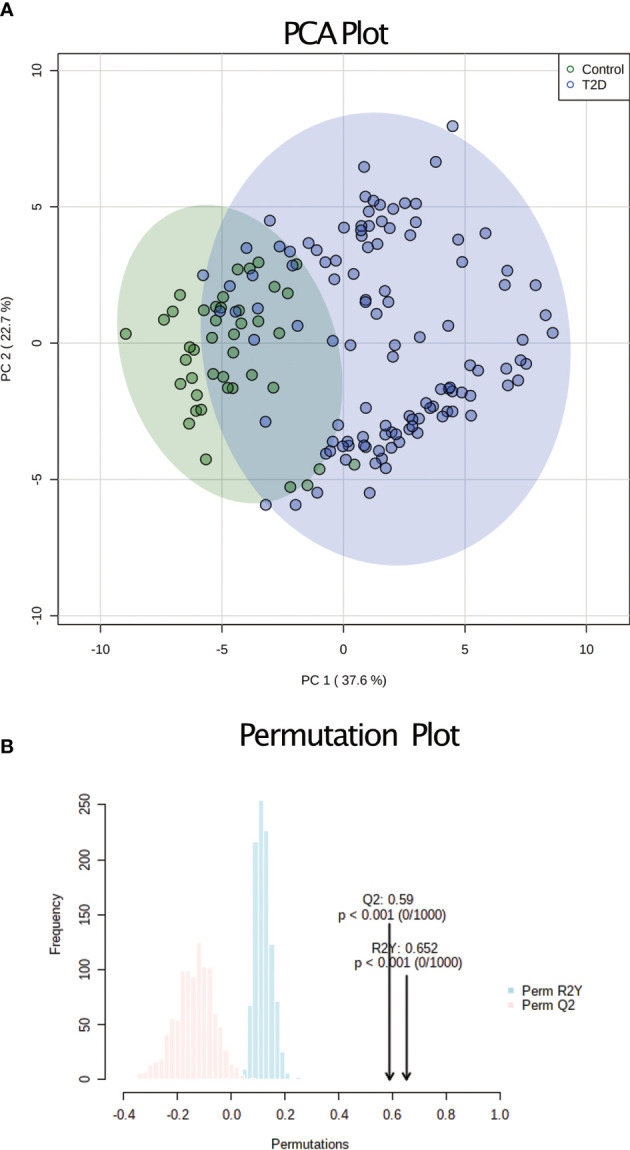
Unsupervised analysis of serum metabolomics. (A) PCA analysis based on the 43 metabolites and all 155 samples. The controls are shown in green and the T2D samples in blue (B) Permutation conducted to validate the variation obtained during the PCA. The R2 (shown in blue) and Q2 (shown in red) values indicate the robustness of the PCA model.
Figure 2.
Heatmap of differential levels of 43 serum metabolites. 43 metabolites that were significantly different between T2D adolescents (blue color class) and control adolescents (green color class). Red color indicates the increased and the blue indicates reduced levels. The red boxes show the six major clusters formed. On the left of cluster is shown the compound identification for the respective metabolite. The red arrows show the five metabolites that were used for data integration.
Figure 3.
Pathway analysis of Serum Metabolites. (A) Pearson correlation-based clustering of the significant metabolites. Five major clusters obtained are shown in red boxes. Each metabolite is represented by their compound identification ( Table 2 shows the respective metabolites). The sub pathways of the metabolites are represented in the boxes. The five metabolites chosen for data integration are indicated by a red arrow. (B) 43 metabolites were categorized into seven super pathways, including lipids (35%), peptides (26% gamma-glutamyl amino acids), amino acids (10%), carbohydrates (17%), nucleotide (2% purine metabolism), cofactors and vitamins (2% ascorbate and aldarate metabolism), and xenobiotics (7%).
Figure 4.
Selection of representative metabolites for data integration. (A) Statistical significance of the five selected metabolites for data integration. The p-value is estimated using an unpaired t-test. The p-value < 0.05 is considered to the significant. (B) The Area Under the Curve (AUC) of the five selected metabolites was 0.94, demonstrating a high accuracy of prediction.
Differential methylation of DNA in adolescents with T2D
To describe the epigenetic changes associated with adolescents with T2D in First Nations youth, we performed DNA methylation profiling on PBMC genomic DNA from the iCARE cohort study participants. The bioinformatic pipeline used to obtain the significant DMRs is shown in ( Supplementary Figure 1B ). After peak calling and peak filtering, we obtained 732984 peaks. Using linear regression comparing adolescents with and without T2D, we controlled for age, sex, BMI and surrogate variables to correct for cell type differences, we obtained 459 significant peaks ( Supplementary Table 1 ). Peaks were annotated to cis-regulatory regions of the TSS of the nearest genes (-5000 kb upstream to 5000 kb downstream). Using the above criteria, we obtained 42 regions out of which 36 significant DMRs were within the 5kb upstream or downstream window of the TSS and six were close to more than one gene ( Table 3 ). Among these 36 DMRs seven were located near the TSS. Some of these genes have biological relevance to T2D, such as FFAR1, USF2, C1QTNF9, Arylsulfatase A (ARSA), Chromodomain Helicase DNA Binding Protein 8 (CHD8), Protocadherin Alpha 1 (PCDHA1) and Natriuretic Peptide B (NPPB) ( Table 3 ). The methylation peaks of the nearest DMRs to the FFAR1, C1QTNF9 and USF2 genes are represented in Supplementary Figures 6A–C ).
Table 3.
Significant peaks that are differentially methylated between T2D and controls.
| Chromosome | Start | End | Peak number | Nearest gene | Distance from TSS | Methylation Status |
|---|---|---|---|---|---|---|
| chr22 | 51064477 | 51064877 | Peak117 | ARSA | 1923 | Decreased |
| chr8 | 22412013 | 22412413 | Peak135 | SORBS3 | 3005 | Decreased |
| chr11 | 67810920 | 67811320 | Peak137 | TCIRG1 | 4637 | Decreased |
| chr1 | 11919744 | 11920144 | Peak16 | NPPB | -956 | Increased |
| chr19 | 38016396 | 38016796 | Peak166 | ZNF793 | 2289 | Increased |
| chr1 | 27189779 | 27190179 | Peak176 | SFN | 346 | Decreased |
| chr4 | 190943560 | 190943960 | Peak18 | FRG2 | 4652 | Decreased |
| chr19 | 3767151 | 3767551 | Peak184 | MRPL54 | 4689 | Decreased |
| chr19 | 3767151 | 3767551 | Peak184 | RAX2 | 4882 | Decreased |
| chr19 | 4557053 | 4557766 | Peak185 | SEMA6B | 2410 | Decreased |
| chr16 | 68269125 | 68269525 | Peak190 | ESRP2 | 1162 | Decreased |
| chr20 | 61923486 | 61923886 | Peak205 | COL20A1 | -852 | Decreased |
| chr11 | 823636 | 824345 | Peak21 | EFCAB4A | -3594 | Decreased |
| chr19 | 35843080 | 35843480 | Peak271 | FFAR1 | 835 | Decreased |
| chr11 | 56057882 | 56058282 | Peak272 | OR8H1 | 484 | Decreased |
| chr14 | 21899880 | 21900280 | Peak275 | CHD8 | -213 | Decreased |
| chr11 | 840258 | 840658 | Peak289 | POLR2L | 2087 | Decreased |
| chr11 | 840258 | 840658 | Peak289 | TSPAN4 | -3988 | Decreased |
| chr12 | 108083057 | 108083457 | Peak310 | PWP1 | 3748 | Increased |
| chr12 | 51478875 | 51479275 | Peak320 | CSRNP2 | -1742 | Increased |
| chr22 | 30727674 | 30728074 | Peak338 | TBC1D10A | -4984 | Increased |
| chr11 | 67191345 | 67191745 | Peak34 | RPS6KB2 | -4427 | Decreased |
| chr6 | 24723903 | 24724303 | Peak380 | C6ORF62 | -3039 | Decreased |
| chr11 | 46726740 | 46727140 | Peak381 | ARHGAP1 | -4791 | Decreased |
| chr11 | 46726740 | 46727140 | Peak381 | ZNF408 | 4572 | Decreased |
| chr19 | 1065608 | 1066008 | Peak404 | HMHA1 | -114 | Decreased |
| chr17 | 79782003 | 79782403 | Peak42 | FAM195B | 2335 | Decreased |
| chr13 | 24882421 | 24882821 | Peak428 | C1QTNF9 | 1317 | Decreased |
| chr16 | 58030446 | 58031214 | Peak432 | USB1 | -4447 | Decreased |
| chr16 | 58030446 | 58031214 | Peak432 | ZNF319 | 2932 | Decreased |
| chr6 | 30587385 | 30587785 | Peak435 | MRPS18B | 2099 | Increased |
| chr6 | 30587385 | 30587785 | Peak435 | PPP1R10 | -2564 | Increased |
| chr19 | 35757516 | 35757916 | Peak44 | USF2 | -2252 | Decreased |
| chr2 | 18765729 | 18766129 | Peak458 | NT5C1B | 4883 | Decreased |
| chr9 | 139835090 | 139835490 | Peak51 | C8G | -4423 | Decreased |
| chr9 | 139835090 | 139835490 | Peak51 | FBXW5 | 3812 | Decreased |
| chr5 | 140167607 | 140168007 | Peak64 | PCDHA1 | 1931 | Decreased |
| chr20 | 39990179 | 39990942 | Peak75 | EMILIN3 | 4906 | Decreased |
| chr2 | 241512174 | 241512574 | Peak76 | RNPEPL1 | 4270 | Decreased |
| chr19 | 11642058 | 11642458 | Peak77 | ECSIT | -2269 | Decreased |
| chr13 | 21349929 | 21350329 | Peak81 | N6AMT2 | -2041 | Increased |
| chr17 | 48613911 | 48614311 | Peak87 | EPN3 | 4207 | Decreased |
All the significant peaks that were annotated using GREAT -5000 to 5000 bp of the TSS. Chromosome indicates the chromosome number of the peak, start indicates the start of peak, end indicates the end of peak, peak number is the unique number of a peak, nearest gene indicates the closest gene to the center of the peak, Distance from TSS indicates the distance of the peak from the TSS of the gene. The methylation status is indicated as increased or decreased.
Correlation between metabolites and DNA methylation in youth with T2D
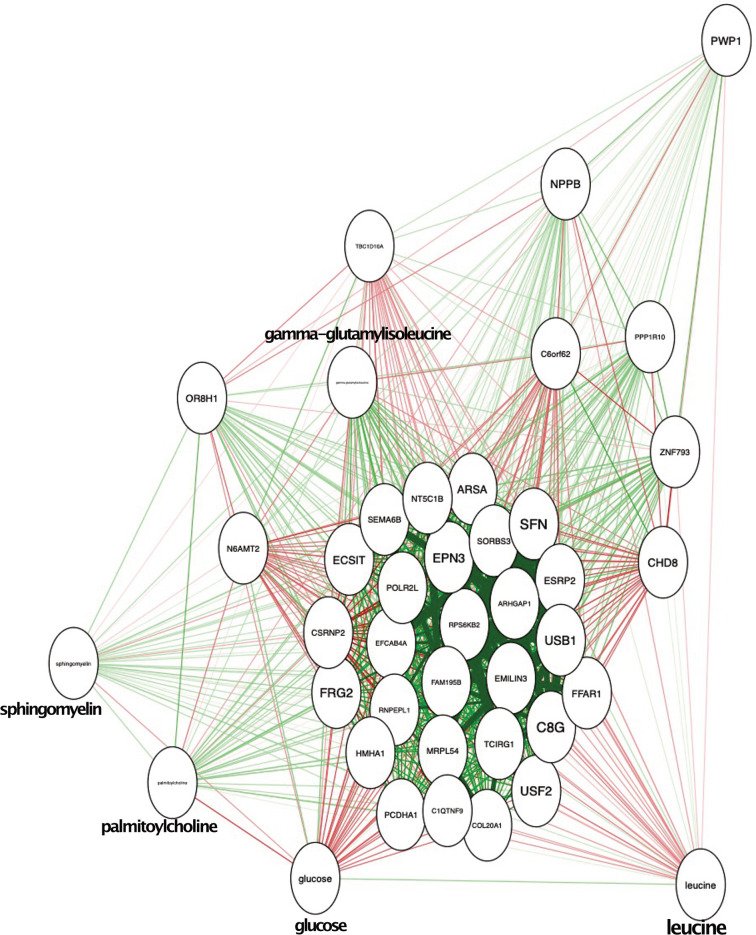
To improve our understanding of mechanisms involved in metabolic perturbations in youth-onset T2D, we investigated the link between DNA methylation and altered levels of metabolites. To predict the T2D status using the selected 5 metabolites the value of the Area Under the Curve (AUC) was 0.94, demonstrating a high accuracy of prediction ( Figure 4B ). We used a graph-based correlation method to find the significant correlations between the five representative metabolites and the 36 DMRs that were located within the -5kb and 5kb window of the TSS of the nearest genes. Figure 5 shows that upon data integration, the metabolites were negatively (green lines) as well as positively correlated (red lines) to several genes. Glucose was correlated with 31 DMRs, leucine correlated to 7 DMRs, gamma-glutamylisoleucine correlated with 30 DMRs, palmitoylcholine was correlated with 10 DMRs and sphingomyelin was correlated to a single DMR ( Supplementary Table 2 ). Interestingly, FFAR1 was negatively correlated to glucose and leucine, but was positively correlated to gamma-glutamylisoleucine. USF2 was also positively correlated with gamma-glutamylisoleucine and negatively correlated with leucine and glucose. C1QTNF9 was positively correlated to gamma-glutamylisoleucine and negatively correlated to glucose. We further correlated all the metabolites and 36 DMRs and found several of them to be highly correlated ( Supplementary Figure 4 ). The number of DMRs that correlated to metabolites are shown in Supplementary Table 4 .
Figure 5.
Integration of metabolic and epigenomic data. The red lines indicate the positive correlation and green lines indicate the negative correlation. The five metabolites and the genes are shown in the circles.
Discussion
T2D in adolescents is aggressive and phenotypically different from T2D in adults (4, 23). The mechanisms underlying these differences are poorly understood although detrimental environmental exposures related to poverty, food insecurity, and poor housing related to the impact of colonization likely have an important role (24). To our knowledge this is the first study to link differential DNA methylation in PBMCs with the serum metabolome in youth-onset T2D. Using stringent VIP scores, we identified 43 metabolites associated with T2D and by peak calling for adolescents with T2D vs the controls, we obtained 459 significant peaks. Among these 459 DMRs, 36 were located near the TSS of genes. Some of these DMRs correlated with the selected metabolites, including 31 that were associated with fasting glucose levels, 7 correlated with leucine, 30 with gamma-glutamyl isoleucine and 10 with palmitoyl choline. DMRs that strongly associated with several of the metabolites in T2D patients included DMRs near the TSS in FFAR1, USF2, and C1QTNF9. Interestingly, these three genes have biological relevance in T2D. This data highlights that complementary information provided by epigenetic marks provide new insight into the metabolic perturbations occurring in adolescents with T2D. Future research will examine the novel role for the DMRs near these genes in regulating gene expression and serum metabolite levels in T2D.
We found that amino acids were the most commonly altered metabolite in the circulation of adolescents with T2D. In adults, high levels of aromatic and BCAAs are predictive of future T2D development (25) and a strong negative association exists between these levels and insulin sensitivity (26, 27). Altered amino acid catabolism in adipose tissue is believed to be the underlying reason that amino acid levels are altered in obese and insulin resistant adults (28). In adolescents, elevated levels of BCAAs have been reported to be associated with obesity (11) and impaired diastolic cardiac function (13). Unlike adults, increased levels of BCAAs were positively associated with beta-cell function, relative to insulin sensitivity in adolescents (8, 10). Consistent with these findings we observed that the BCAA leucine was increased in the serum of adolescents with T2D. Given that seven DMRs correlated with serum leucine levels in T2D, this finding sets the stage to examine whether these DMRs are involved in regulating leucine levels in adolescents that could underlie the differential effects of BCAAs on beta-cell function in adolescents and adults with T2D. Notably, levels of a broad range of gamma-glutamyl dipeptides were also reduced in adolescents with T2D. Gamma-glutamyl amino acids are considered to be involved in regulating oxidative stress through their involvement in glutathione production.
We identified an association between lipid metabolism and adolescents with T2D. Reductions in circulating levels of several sphingomyelins, lysophosphatidylcholines and acyl-alkyl phosphatidylcholine (plasmalogens) were observed. These acyl-alkyl phosphatidylcholines belong to a class of antioxidant plasmalogens and could reflect the state of oxidative stress. On the other hand, since lysophosphatidylcholines in the bloodstream are derived from oxidation of phosphatidylcholine in low density lipoproteins, could suggests a reduction in its oxidation. These results are consistent with a previous study showing that serum levels of acyl-alkyl phosphatidylcholines and lysophosphatidylcholines are reduced in obese children (29). In adult populations, elevated sphingolipids are generally associated with obesity and greater insulin resistance (30). We observed reductions in a number of sphingolipids in First Nations adolescents with T2D. This finding is consistent with another study of normoglycemic North American Indigenous adolescents and young adults (31) that identified an association between obesity and lowered sphingolipid. Thus, there appears to be a role for altered lipid metabolism in the natural history of T2D. In light of this, nutritional strategies developed by and for First Nations people should be crucial to improving their health status as a whole (32).
We identified a DMR near FFAR1 that was positively correlated with gamma-glutamylisoleucine and negatively correlated with glucose and leucine. FFAR1 (also known as GPR40) induces the Gαq signaling cascade which activates phospholipase C and inositol 1,4,5-triphosphate (IP3) formation, stimulating Ca2+ mobilization from the endoplasmic reticulum and triggering insulin secretion (33). Free fatty acids are proposed to potentiate glucose-stimulated insulin secretion through FFAR1 activation, evidenced by a reduction in Ca2+ oscillations following the inactivation of FFAR1 using GW1100 (a GPR40 inhibitor) (34). Given that FFAR1 mRNA and protein expression are reduced in the islets of diabetic mice (35) and an FFAR1 agonist improved glucose and lipid metabolism in obese mice (36), it is conceivable that altered DNA methylation near the TSS of the FFAR1 gene could be associated with alterations in insulin secretion and glucose homeostasis in adolescents with T2D, although this hypothesis requires further investigation.
Another relevant discovery was the identification of a DMR near the TSS of the USF2 gene that correlated with glucose, leucine and gamma-glutamylisoleucine. USF2 is a ubiquitous basic helix-loop-helix transcription factor that binds to E-box elements. High glucose levels upregulate USF2 expression in the liver and USF2 regulates SREBP-1c and stimulates fatty acid synthesis in the liver that leads to lipid accumulation (37, 38). This is consistent with our previous finding that hepatic steatosis was 3-fold higher in First Nation adolescents with T2D compared to normoglycemic controls (39), although whether methylation of the USF2 promoter is a contributing factor remains to be investigated.
We also identified a DMR near the TSS of the C1QTNF9 gene, that was correlated with serum glucose and gamma-glutamylisoleucine levels in adolescents with T2D. C1QTNF9 encodes a novel cytokine, termed CTRP9, that is a paralog of adiponectin and is expressed by adipose tissue, heart and endothelium. CTRP9 protects cells against high glucose and palmitate-induced oxidative stress (40, 41). CTRP9 has been reported to attenuate diabetic nephropathy and improve cardiac function in obese and diabetic mice (42, 43). However, increased CTRP9 in the circulation correlated with insulin resistance in humans (44, 45), suggesting that more studies investigating CTRP9 actions in T2D are necessary. Given that Dart et al. (46, 47) reported a higher incidence and earlier onset of major diabetes-related complications in a cohort of adolescents with T2D compared to a cohort of adolescents with type 1 diabetes, follow-up studies will examine the association between alterations in DNA methylation and the risk for complications of diabetes in youth. These findings correspond with a growing body of literature linking genome-wide alterations in DNA methylation to complications of diabetes (48, 49).
In Canada, First Nation youth account for a disproportionate number of T2D diagnoses (50). Environmental influences, including nutrition, have a major role in defining T2D risk, but T2D in youth is also associated with poverty and lower socioeconomic status (47, 48, 51). Epigenetic changes mediate environmental influences on the genetic architecture. We uncovered several DNA methylation and metabolite alterations that provide important new knowledge about the cellular changes occurring in First Nations adolescents associated with T2D. This supports the theory that social inequities, purposeful starvation, dispossession of land and traditional ways of living in First Nations youth induced molecular and biological manifestations of chronic disease risk (49). However, we also acknowledge that our study cannot separate whether these changes in metabolites and DNA methylation are a consequence of T2D development or contribute to the development of T2D in youth. One of the major limitations of our analysis is the small sample size. Since our study only captures a snapshot of the changes following a diagnosis of T2D, further studies are warranted in a larger sample size in the iCARE cohort, as well as replication in other populations to determine whether these findings are generalizable to the wider population of adolescents with T2D or whether these changes are unique to First Nation adolescents. Future work in longitudinal settings would also provide a clearer picture of the mechanisms involved in the development of T2D and its associated complications in adolescents. Finally, we recognize that the serum metabolomic profile may not be reflective of the metabolic changes in all tissues. Moreover, the DNA methylation patterns in PBMCs may not reflect methylation and gene expression changes in all tissues that generate serum metabolites. However, some of the genes and metabolites have been separately linked to altered metabolism in T2D by previous studies, suggesting that our integrative approach provides relevant information about youth-onset T2D. Nonetheless, future functional studies determining mechanisms of how DNA methylation induces changes in gene expression and the observed metabolites are warranted.
In summary, we integrated serum metabolomic and genome-wide DNA methylation data in a systems medicine-based approach that is well suited to generate new hypotheses about the complex and variable factors that contribute to the development of T2D in pediatric populations. We identified several robust candidate DMRs located near the TSS of genes such as USF2, FFAR1 and C1QTNF9 that are correlated to a collection of metabolites that are relevant to metabolic homeostasis in T2D. We will test the hypothesis that T2D induced DNA methylation of USF2, FFAR1 and C1QTNF9, among others, affect gene expression and metabolite levels. This study lays the groundwork for future research about how epigenetic and metabolite alterations relate to T2D pathogenesis in adolescents and evaluate their predictive power in the development of T2D and its associated complications. Our findings generate new hypotheses that will test whether the identified DMRs regulate the expression of the nearby genes in metabolic cell types and whether altered expression of these genes influences metabolite levels. Elucidating the linkages between DNA methylation patterns in PBMCs and circulating metabolites would alsoserum highlight the advantage of integrating data from multiple sources.
Data availability statement
The DNA methylation and metabolomic data presented in the study are deposited in the European Genome-phenome Archive database (EGAS00001003816). The remaining data are available from the corresponding author upon reasonable request and approval by the iCARE data access committee.
Ethics statement
The studies involving human participants were reviewed and approved by the University of Manitoba/Health Sciences Research ethics board. The iCARE cohort study (15) received informed consent from study participants and approval from the University of Manitoba/Health Sciences Centre Research Ethics Board (HS13255), First Nations patient and parent advisory committee and the First Nation Health and Social Secretariat of Manitoba and the iCARE participant and parent advisory committees. Written informed consent to participate in this study was provided by the participants’ legal guardian/next of kin.
Author contributions
PA, MJ and AA conceived and designed the data analysis strategy and integration. MF prepared clinical samples for analysis. PA, NH, CT and AA performed data analysis. VWD, MJ and JD provided oversight, analysis and coordination of all aspects listed above. BW, AD, ES and JM developed the iCARE cohort study, sample collection and clinical analyses. PA, BW, MJ, AA and VWD wrote the manuscript. All authors edited and reviewed the manuscript. VWD is the guarantor of this work and, as such, had full access to all data in the study and takes responsibility for the integrity of the data and the accuracy of the data analysis. All authors contributed to the article and approved the submitted version.
Funding
This research is supported by a CIHR grant (MOP#142309) to AD and BW and an Environments, Genes and Chronic Disease CIHR Team Grant #144626 to VWD, BW, JD et al. Funding sources were not involved in the study design, collection, interpretation of the data or preparation of the manuscript.
Acknowledgments
The authors are grateful to Dr. Wanda Phillips-Beck of Nanaandawewigamig First Nations Health and Social Secretariat of Manitoba for providing comments about this paper from an Indigenous perspective. PA was the recipient of a postdoctoral fellowship from Research Manitoba. JMM holds an Applied Public Health Chair awarded by the Canadian Institutes for Health Research (CIHR) #CPP-137910. JRD was a Canada Research Chair (Tier 1) in Chromatin Dynamics. AA is supported by the National Institute for Health Research (NIHR) Surgical Reconstruction and Microbiology Research Centre (SRMRC), Birmingham, UK. VWD was the Allen Rouse-Manitoba Medical Services Foundation Basic Scientist. The views expressed in this publication are those of the authors and not necessarily those of the National Health Service (NHS), the National Institute for Health Research (NIHR), UK.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fendo.2022.934706/full#supplementary-material
Schematic workflow of the analysis for epigenetic and metabolomic datasets. (A) Shows the workflow of the data processing steps, normalization and, data analysis steps followed for obtaining the significant metabolites. (B) Shows the workflow of data processing, normalization and, analysis to obtain the significant DMRs.
Pearson correlation between all the clinical variables of primary cohort. (A) Clinical variables of controls that include age, height, weight, waist size, BMI, glucose levels, AST, ALT and HbA1c levels. Similarly, (B) Clinical variables of T2D patients.
Serum metabolomic profile of youth with T2D. (A) OPLSDA model based on the significant metabolites (i.e. 43 metabolites). The controls are shown in green and the T2D samples are in blue. (B) Permutation conducted to validate the variation obtained during the OPLSDA. The R2 (shown in green) and Q2 (shown in blue) values indicate the robustness of the OPLSDA model.
Correlation plot for all the significant metabolites and 36 DMRs. Red color shows the positively correlated and blue shows negatively correlated. Non correlated ones are shown in white. The five metabolites chosen for data integration are indicated by a red arrow.
Clusters of super pathways for metabolites. a) x-axis of the volcano plot for the super pathways shows the log2 fold change and y-axis shows the p-value. b) x-axis of the bar plot shows the super pathways, and the y-axis shows the frequency of each pathway.
Visualization of DMRs genomic location using IGV genome viewer. Red color is for the 10 control samples and blue color for 21 T2D samples. All the shown regions have decreased methylation as compared to controls. The two vertical lines in each figure shows the approx. peak region. (A) shows genomic location of DMR nearest to TSS of the gene FFAR1. (B) shows genomic location of the DMR nearest to TSS of gene C1QTNF9. (C) shows genomic location of the DMR nearest to TSS of the gene USF2.
Significant DMRs/peaks file. The table contains all the significant DMRs/peaks obtained in the DNA methylation analysis. The file contains the genomic location of each peak followed by the significance level.
Data integration correlation values. The table contains the correlation values obtained from the DMR and five metabolites integration. The green colored values show the negative correlation and red values show the positive correlation.
Information of the DNA methylation sequencing samples. The table provides the information of the samples that were sequenced and submitted in the European Genome-phenome Archive.
All the correlated metabolites and DMRs
References
- 1. Bullock A, Sheff K. Incidence trends of type 1 and type 2 diabetes among youth-2012. N Engl J Med (2017) 377:301. doi: 10.1056/NEJMc1706291 [DOI] [PubMed] [Google Scholar]
- 2. Pinhas-Hamiel O, Zeitler P. Acute and chronic complications of type 2 diabetes mellitus in children and adolescents. Lancet (2007) 369:1823–31. doi: 10.1016/S0140-6736(07)60821-6 [DOI] [PubMed] [Google Scholar]
- 3. Ruth C, Sellers E, Chartrand C, Mcleod L, Prior H, Sirski M, et al. Type 2 diabetes in manitoba. Manitoba centre for health policy autumn (2020). Available at: http://mchp-appserv.cpe.umanitoba.ca/reference/T2DM_Report_web.pdf.
- 4. Bacha F, Gungor N, Lee S, Arslanian SA. Progressive deterioration of beta-cell function in obese youth with type 2 diabetes. Pediatr Diabetes (2013) 14:106–11. doi: 10.1111/j.1399-5448.2012.00915.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Manolio TA, Collins FS, Cox NJ, Goldstein DB, Hindorff LA, Hunter DJ, et al. Finding the missing heritability of complex diseases. Nature (2009) 461:747–53. doi: 10.1038/nature08494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Agarwal P, Morriseau TS, Kereliuk SM, Doucette CA, Wicklow BA, Dolinsky VW. Maternal obesity, diabetes during pregnancy and epigenetic mechanisms that influence the developmental origins of cardiometabolic disease in the offspring. Crit Rev Clin Lab Sci (2018) 55:71–101. doi: 10.1080/10408363.2017.1422109 [DOI] [PubMed] [Google Scholar]
- 7. Ling C, Ronn T. Epigenetics in human obesity and type 2 diabetes. Cell Metab (2019) 29:1028–44. doi: 10.1016/j.cmet.2019.03.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Mihalik SJ, Michaliszyn SF, De Las Heras J, Bacha F, Lee S, Chace DH, et al. Metabolomic profiling of fatty acid and amino acid metabolism in youth with obesity and type 2 diabetes: evidence for enhanced mitochondrial oxidation. Diabetes Care (2012) 35:605–11. doi: 10.2337/DC11-1577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Frohnert BI, Rewers MJ. Metabolomics in childhood diabetes. Pediatr Diabetes (2016) 17:3–14. doi: 10.1111/pedi.12323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Michaliszyn SF, Sjaarda LA, Mihalik SJ, Lee S, Bacha F, Chace DH, et al. Metabolomic profiling of amino acids and beta-cell function relative to insulin sensitivity in youth. J Clin Endocrinol Metab (2012) 97:E2119–2124. doi: 10.1210/jc.2012-2170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Mccormack SE, Shaham O, Mccarthy MA, Deik AA, Wang TJ, Gerszten RE, et al. Circulating branched-chain amino acid concentrations are associated with obesity and future insulin resistance in children and adolescents. Pediatr Obes (2013) 8:52–61. doi: 10.1111/j.2047-6310.2012.00087.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Newbern D, Gumus Balikcioglu P, Balikcioglu M, Bain J, Muehlbauer M, Stevens R, et al. Sex differences in biomarkers associated with insulin resistance in obese adolescents: metabolomic profiling and principal components analysis. J Clin Endocrinol Metab (2014) 99:4730–9. doi: 10.1210/jc.2014-2080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Guillemette L, Dart A, Wicklow B, Dolinsky VW, Cheung D, Jassal DS, et al. Cardiac structure and function in youth with type 2 diabetes in the iCARE cohort study: Cross-sectional associations with prenatal exposure to diabetes and metabolomic profiles. Pediatr Diabetes (2020) 21:233–42. doi: 10.1111/pedi.12954 [DOI] [PubMed] [Google Scholar]
- 14. Wang Z, Long H, Chang C, Zhao M, Lu Q. Crosstalk between metabolism and epigenetic modifications in autoimmune diseases: A comprehensive overview. Cell Mol Life Sci (2018) 75:3353–69. doi: 10.1007/s00018-018-2864-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Dart AB, Wicklow BA, Sellers EA, Dean HJ, Malik S, Walker J, et al. The improving renal complications in adolescents with type 2 diabetes through the REsearch (iCARE) cohort study: rationale and protocol. Can J Diabetes (2014) 38:349–55. doi: 10.1016/j.jcjd.2014.07.224 [DOI] [PubMed] [Google Scholar]
- 16. Zhang Y, Liu T, Meyer CA, Eeckhoute J, Johnson DS, Bernstein BE, et al. Model-based analysis of ChIP-seq (MACS). Genome Biol (2008) 9:R137. doi: 10.1186/gb-2008-9-9-r137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Ross-Innes CS, Stark R, Teschendorff AE, Holmes KA, Ali HR, Dunning MJ, Brown GD, et al. Differential oestrogen receptor binding is associated with clinical outcome in breast cancer. Nature (2012) 481:389–93. doi: 10.1038/nature10730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Amemiya HM, Kundaje A, Boyle AP. The ENCODE blacklist: Identification of problematic regions of the genome. Sci Rep (2019) 9:9354 doi: 10.1038/s41598-019-45839-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Robinson MD, Mccarthy DJ, Smyth GK. edgeR: a bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics (2010) 26:139–40. doi: 10.1093/bioinformatics/btp616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Leek JT, Johnson WE, Parker HS, Jaffe A, Storey JD. Bioinformatics (2012) 28:882–3. doi: 10.1093/bioinformatics/bts034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Chiu CY, Yeh KW, Lin G, Chiang MH, Yang SC, Chao WJ, et al. Metabolomics reveals dynamic metabolic changes associated with age in early childhood. PloS One (2016) 11:e0149823. doi: 10.1371/journal.pone.0149823 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Epskamp SC, Ang´Elique OJ, Waldor LJ, Schmittmann VD, Borsboom D. Qgraph: Network visualizations of relationships in psychometric data. J Stat Software (2012) 48:1–18. doi: 10.18637/jss.v048.i04 [DOI] [Google Scholar]
- 23. Rise Consortium Investigators . Effects of treatment of impaired glucose tolerance or recently diagnosed type 2 diabetes with metformin alone or in combination with insulin glargine on beta-cell function: Comparison of responses in youth and adults. Diabetes (2019) 68:1670–80. doi: 10.2337/db19-0299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. McGavock J, Wicklow B, Dart AB. Type 2 diabetes in youth is a disease of poverty. Lancet (2017) 390:1829. doi: 10.1016/S0140-6736(17)32461-3 [DOI] [PubMed] [Google Scholar]
- 25. Wang TJ, Larson MG, Vasan RS, Cheng S, Rhee EP, Mccabe E, et al. Metabolite profiles and the risk of developing diabetes. Nat Med (2011) 17:448–53. doi: 10.1038/nm.2307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Newgard CB, An J, Bain JR, Muehlbauer MJ, Stevens RD, Lien LF, et al. A branched-chain amino acid-related metabolic signature that differentiates obese and lean humans and contributes to insulin resistance. Cell Metab (2009) 9:311–26. doi: 10.1016/j.cmet.2009.02.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Tai ES, Tan ML, Stevens RD, Low YL, Muehlbauer MJ, Goh DL, et al. Insulin resistance is associated with a metabolic profile of altered protein metabolism in Chinese and Asian-Indian men. Diabetologia (2010) 53:757–67. doi: 10.1007/s00125-009-1637-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Newgard CB. Interplay between lipids and branched-chain amino acids in development of insulin resistance. Cell Metab (2012) 15:606–14. doi: 10.1016/j.cmet.2012.01.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Mihalik SJ, Goodpaster BH, Kelley DE, Chace DH, Vockley J, Toledo FG, et al. Increased levels of plasma acylcarnitines in obesity and type 2 diabetes and identification of a marker of glucolipotoxicity. Obes (Silver Spring) (2010) 18:1695–700. doi: 10.1038/oby.2009.510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Holland WL, Summers SA. Sphingolipids, insulin resistance, and metabolic disease: new insights from in vivo manipulation of sphingolipid metabolism. Endocr Rev (2008) 29:381–402. doi: 10.1210/er.2007-0025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Zhao Q, Zhu Y, Best LG, Umans JG, Uppal K, Tran VT, et al. Metabolic profiles of obesity in American indians: The strong heart family study. PloS One (2016) 11:e0159548. doi: 10.1371/journal.pone.0159548 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Willows N, Dyck Fehderau D, Raine KD. Analysis grid for environments linked to obesity (ANGELO) framework to develop community-driven health programmes in an indigenous community in Canada. Health Soc Care Community (2016) 24:567–75. doi: 10.1111/hsc.12229 [DOI] [PubMed] [Google Scholar]
- 33. Prentki M, Corkey BE, Madiraju SRM. Lipid-associated metabolic signalling networks in pancreatic beta cell function. Diabetologia (2020) 63:10–20. doi: 10.1007/s00125-019-04976-w [DOI] [PubMed] [Google Scholar]
- 34. Briscoe CP, Peat AJ, Mckeown SC, Corbett DF, Goetz AS, Littleton TR, et al. Pharmacological regulation of insulin secretion in MIN6 cells through the fatty acid receptor GPR40: identification of agonist and antagonist small molecules. Br J Pharmacol (2006) 148:619–28. doi: 10.1038/sj.bjp.0706770 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Kohara K, Obata A, Kimura T, Shimoda M, Moriuchi S, Okauchi S, et al. Suppression of free fatty acid receptor 1 expression in pancreatic beta-cells in obese type 2 diabetic db/db mice: A potential role of pancreatic and duodenal homeobox factor 1. Endocr J (2019) 66:43–50. doi: 10.1507/endocrj.EJ18-0203 [DOI] [PubMed] [Google Scholar]
- 36. Chen Y, Ren Q, Zhou Z, Deng L, Hu L, Zhang L, et al. HWL-088, a new potent free fatty acid receptor 1 (FFAR1) agonist, improves glucolipid metabolism and acts additively with metformin in ob/ob diabetic mice. Br J Pharmacol (2020) 177:2286–302. doi: 10.1111/bph.14980 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Van Deursen D, Jansen H, Verhoeven AJ. Glucose increases hepatic lipase expression in HepG2 liver cells through upregulation of upstream stimulatory factors 1 and 2. Diabetologia (2008) 51:2078–87. doi: 10.1007/s00125-008-1125-6 [DOI] [PubMed] [Google Scholar]
- 38. Czech MP, Tencerova M, Pedersen DJ, Aouadi M. Insulin signalling mechanisms for triacylglycerol storage. Diabetologia (2013) 56:949–64. doi: 10.1007/s00125-013-2869-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Wittmeier KD, Wicklow BA, Macintosh AC, Sellers EA, Ryner LN, Serrai H, et al. Hepatic steatosis and low cardiorespiratory fitness in youth with type 2 diabetes. Obes (Silver Spring) (2012) 20:1034–40. doi: 10.1038/oby.2011.379 [DOI] [PubMed] [Google Scholar]
- 40. Cheng Y, Qi Y, Liu S, Di R, Shi Q, Li J, et al. C1q/TNF-related protein 9 inhibits high glucose-induced oxidative stress and apoptosis in retinal pigment epithelial cells through the activation of AMPK/Nrf2 signaling pathway. Cell Transplant (2020) 29:963689720962052. doi: 10.1177/0963689720962052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Zuo A, Li J, Zhao X, Li T, Lei S, Chen J, et al. Globular CTRP9 protects cardiomyocytes from palmitic acid-induced oxidative stress by enhancing autophagic flux. Chem Biol Interact (2020) 329:109094. doi: 10.1016/j.cbi.2020.109094 [DOI] [PubMed] [Google Scholar]
- 42. Su H, Yuan Y, Wang XM, Lau WB, Wang Y, Wang X, et al. Inhibition of CTRP9, a novel and cardiac-abundantly expressed cell survival molecule, by TNFalpha-initiated oxidative signaling contributes to exacerbated cardiac injury in diabetic mice. Basic Res Cardiol (2013) 108:315. doi: 10.1007/s00395-012-0315-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Hu H, Li W, Liu M, Xiong J, Li Y, Wei Y, et al. C1q/Tumor necrosis factor-related protein-9 attenuates diabetic nephropathy and kidney fibrosis in db/db mice. DNA Cell Biol (2020) 39:938–48. doi: 10.1089/dna.2019.5302 [DOI] [PubMed] [Google Scholar]
- 44. Jia Y, Luo X, Ji Y, Xie J, Jiang H, Fu M, et al. Circulating CTRP9 levels are increased in patients with newly diagnosed type 2 diabetes and correlated with insulin resistance. Diabetes Res Clin Pract (2017) 131:116–23. doi: 10.1016/j.diabres.2017.07.003 [DOI] [PubMed] [Google Scholar]
- 45. Moradi N, Fadaei R, Emamgholipour S, Kazemian E, Panahi G, Vahedi S, et al. Association of circulating CTRP9 with soluble adhesion molecules and inflammatory markers in patients with type 2 diabetes mellitus and coronary artery disease. PloS One (2018) 13:e0192159. doi: 10.1371/journal.pone.0192159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Dart AB, Sellers EA, Martens PJ, Rigatto C, Brownell MD, Dean HJ. High burden of kidney disease in youth-onset type 2 diabetes. Diabetes Care (2012) 35:1265–71. doi: 10.2337/dc11-2312 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Dart AB, Martens PJ, Rigatto C, Brownell MD, Dean HJ, Sellers EA. Earlier onset of complications in youth with type 2 diabetes. Diabetes Care (2014) 37:436–43. doi: 10.2337/dc13-0954 [DOI] [PubMed] [Google Scholar]
- 48. Dabelea D, Stafford JM, Mayer-Davis EJ, D’agostino R, Jr., Dolan L, Imperatore G, et al. Association of type 1 diabetes vs type 2 diabetes diagnosed during childhood and adolescence with complications during teenage years and young adulthood. JAMA (2017) 317:825–35. doi: 10.1001/jama.2017.0686 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Mosby I, Galloway T. “Hunger was never absent”: How residential school diets shaped current patterns of diabetes among indigenous peoples in Canada. CMAJ (2017) 189:E1043–5. doi: 10.1503/cmaj.170448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Amed S, Dean HJ, Panagiotopoulos C, Sellers EA, Hadjiyannakis S, Laubscher TA, et al. Type 2 diabetes, medication-induced diabetes, and monogenic diabetes in Canadian children: a prospective national surveillance study. Diabetes Care (2010) 33:786–91. doi: 10.2337/dc09-1013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Copeland KC, Zeitler P, Geffner M, Guandalini C, Higgins J, Hirst K, et al. Characteristics of adolescents and youth with recent-onset type 2 diabetes: the TODAY cohort at baseline. J Clin Endocrinol Metab (2011) 96:159–67. doi: 10.1210/jc.2010-1642 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Schematic workflow of the analysis for epigenetic and metabolomic datasets. (A) Shows the workflow of the data processing steps, normalization and, data analysis steps followed for obtaining the significant metabolites. (B) Shows the workflow of data processing, normalization and, analysis to obtain the significant DMRs.
Pearson correlation between all the clinical variables of primary cohort. (A) Clinical variables of controls that include age, height, weight, waist size, BMI, glucose levels, AST, ALT and HbA1c levels. Similarly, (B) Clinical variables of T2D patients.
Serum metabolomic profile of youth with T2D. (A) OPLSDA model based on the significant metabolites (i.e. 43 metabolites). The controls are shown in green and the T2D samples are in blue. (B) Permutation conducted to validate the variation obtained during the OPLSDA. The R2 (shown in green) and Q2 (shown in blue) values indicate the robustness of the OPLSDA model.
Correlation plot for all the significant metabolites and 36 DMRs. Red color shows the positively correlated and blue shows negatively correlated. Non correlated ones are shown in white. The five metabolites chosen for data integration are indicated by a red arrow.
Clusters of super pathways for metabolites. a) x-axis of the volcano plot for the super pathways shows the log2 fold change and y-axis shows the p-value. b) x-axis of the bar plot shows the super pathways, and the y-axis shows the frequency of each pathway.
Visualization of DMRs genomic location using IGV genome viewer. Red color is for the 10 control samples and blue color for 21 T2D samples. All the shown regions have decreased methylation as compared to controls. The two vertical lines in each figure shows the approx. peak region. (A) shows genomic location of DMR nearest to TSS of the gene FFAR1. (B) shows genomic location of the DMR nearest to TSS of gene C1QTNF9. (C) shows genomic location of the DMR nearest to TSS of the gene USF2.
Significant DMRs/peaks file. The table contains all the significant DMRs/peaks obtained in the DNA methylation analysis. The file contains the genomic location of each peak followed by the significance level.
Data integration correlation values. The table contains the correlation values obtained from the DMR and five metabolites integration. The green colored values show the negative correlation and red values show the positive correlation.
Information of the DNA methylation sequencing samples. The table provides the information of the samples that were sequenced and submitted in the European Genome-phenome Archive.
All the correlated metabolites and DMRs
Data Availability Statement
The DNA methylation and metabolomic data presented in the study are deposited in the European Genome-phenome Archive database (EGAS00001003816). The remaining data are available from the corresponding author upon reasonable request and approval by the iCARE data access committee.