Summary
The seven-transmembrane superfamily member 3 protein (TM7SF3) is a p53-regulated homeostatic factor that attenuates cellular stress and the unfolded protein response. Here we show that TM7SF3 localizes to nuclear speckles; eukaryotic nuclear bodies enriched in splicing factors. This unexpected location for a trans-membranal protein enables formation of stable complexes between TM7SF3 and pre-mRNA splicing factors including DHX15, LARP7, HNRNPU, RBM14, and HNRNPK. Indeed, TM7SF3 regulates alternative splicing of >330 genes, mainly at the 3′end of introns by directly modulating the activity of splicing factors such as HNRNPK. These effects are observed both in cell lines and primary human pancreatic islets. Accordingly, silencing of TM7SF3 results in differential expression of 1465 genes (about 7% of the human genome); with 844 and 621 genes being up- or down-regulated, respectively. Our findings implicate TM7SF3, as a resident protein of nuclear speckles and suggest a role for seven-transmembrane proteins as regulators of alternative splicing.
Subject areas: Biological sciences, Biochemistry, Structural biology
Graphical abstract
Highlights
-
•
TM7SF3 is the first heptahelical protein and resides mainly in nuclear speckles
-
•
TM7SF3 binds proteins involved in RNA processing and regulates alternative splicing
-
•
TM7SF3 binds splicing factors such as HNRNPK and inhibits their splicing activity
-
•
TM7SF3 knockdown affects expression of >1450 genes and promotes cellular stress
Biological sciences; Biochemistry; Structural biology.
Introduction
Protein homeostasis is regulated to a large extent by alternative splicing (AS) (Baralle and Giudice, 2017; Han et al., 2017; Heyd and Lynch, 2011; Paronetto et al., 2016). It is the process by which different combinations of splice sites in precursor mRNA (pre-mRNA) are selected to generate distinct mRNA and protein variants. It is a powerful evolutionary resource that acts widely to expand the functional and regulatory capacity of the metazoan genome (Han et al., 2017; Paronetto et al., 2016). Nearly all transcripts from mammalian multi-exon genes are alternatively spliced (Nilsen and Graveley, 2010), and a substantial fraction of these splice variants are differentially expressed in a cell- and tissue-specific manner. The spatiotemporal specificity of AS is governed by combinations of cis-regulatory elements and cognate trans-acting factors that promote or inhibit the spliceosomes assembly and activity (Baralle and Giudice, 2017; Han et al., 2017; Paronetto et al., 2016). Additional regulatory elements include transcription and epigenetic changes such as specific histone marks, DNA-methylation patterns, nucleosome occupancy, engagement of RNA binding proteins (RBP) and positioning of RNA polymerase II (Pol II) (Heyd and Lynch, 2011; Naftelberg et al., 2015). Post-translational and signaling pathways also modulate AS (Heyd and Lynch, 2011; Naftelberg et al., 2015), yet, the repertoire of elements that affect splicing remains incompletely understood.
The seven-transmembrane superfamily member 3 protein (TM7SF3) is a pro-survival factor induced by p53 that regulates protein homeostasis and attenuates cellular stress. TM7SF3 inhibits caspase 3/7 while siRNA-mediated silencing of TM7SF3 accelerates ER stress and the unfolded protein responses (UPR). This involves inhibitory phosphorylation of eIF2α activity and increased expression of ATF3, ATF4, and C/EBP, followed by induction of apoptosis (Isaac et al., 2017). TM7SF3 maintains cellular reducing power within physiological levels and reduces the content of proapoptotic proteins such as FAS, FADD, and caspase-8 (Beck et al., 2011). Accordingly, TM7SF3 inhibits cytokine-induced death and promotes insulin secretion from pancreatic β-cells (Beck et al., 2011). p53 is an upstream activator of TM7SF3 that directly binds the TM7SF3 promoter. Conversely, silencing of TM7SF3 promotes p53 activity, suggesting the existence of a negative-feedback loop, whereby p53 promotes the expression of TM7SF3 that inhibits the action of p53 (Isaac et al., 2017).
Based on these findings we wished to better understand the molecular mechanisms underlying the biological activities of TM7SF3. Here we show that TM7SF3 is not localized at the plasma membrane, like canonical seven-transmembrane proteins, but rather resides within nuclear speckles in close, proximity to the splicing machinery. Nuclear speckles, also known as interchromatin granule clusters, are nuclear domains enriched in pre-mRNA splicing factors, located in the interchromatin regions of the nucleoplasm of mammalian cells (Spector and Lamond, 2011). Nuclear speckles that serve as hubs for splicing factors also contain phosphoinositides (Balaban et al., 2021; Castano et al., 2019; Faberova et al., 2020; Mellman et al., 2008). About ∼68% of the total nuclear phosphatidylinositol-4,5-bisphosphate (PtdIns4,5P2) localizes to nuclear speckles (Sobol et al., 2018), and could facilitate binding of membranal proteins to this compartment. Yet, seven-transmembrane proteins have not been previously identified as residents of nuclear speckles or as direct regulators of alternative splicing.
Results
TM7SF3 is a nuclear protein
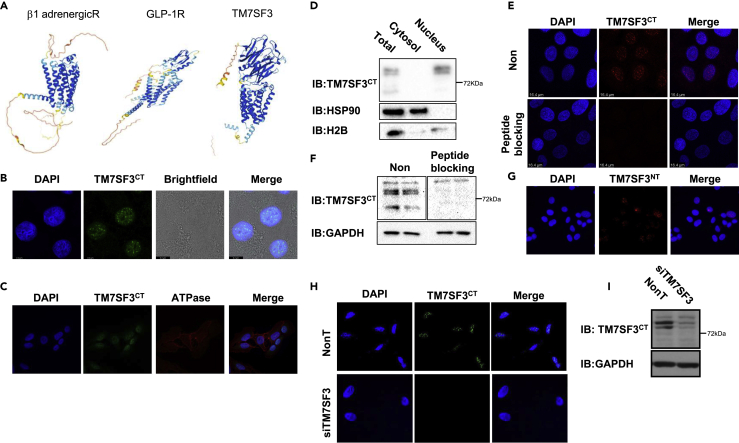
TM7SF3 is a seven transmembrane protein that regulates homeostasis and attenuates cellular stress. In silico analysis of its structure (Figure 1A), employing neural network-based model, AlphaFold2 (Jumper et al., 2021), predicts an extended N-terminal domain that somewhat resembles Class B GPCRs (e.g. GlP-1R) (Congreve et al., 2020). Yet the predicted structure of the N-terminal region is quite unique having prominent barrel-like β-pleated sheets that could serve as a ligand-binding domain. To determine its subcellular localization, polyclonal antibodies directed against a peptide (aa 551–568), located at the C-terminal domain of TM7SF3 were generated. These antibodies denoted TM7SF3CT readily interacted with the C-terminal domain of TM7SF3, but not with its N-terminal region (Figure S1). Mass spectrometry analysis of proteins immunoprecipitated by TM7SF3CT (Table S4, Figure S2) revealed a number of peptides corresponding to TM7SF3 that were identified in TM7SF3CT immunoprecipitates but not in samples precipitated by pre-immune serum, thus validating the specificity of the TM7SF3CT antibodies as being selectively directed against TM7SF3.
Figure 1.
TM7SF3 is a nuclear protein
(A) A scheme of β1 adrenergicR and GLP1R structure vs. the predicted structure of TM7SF3, based on ‘Protter’.
(B and C) U2-OS cells were fixed, stained with DAPI (blue), and immunostained with anti-TM7SF3CT alone (B) or with anti-Na+/K+ ATPase (C) antibodies.
(D) U2-OS cells were fractionated into cytosolic and nuclear-enriched fractions. Samples were immunoblotted with the indicated antibodies.
(E) U2-OS cells were stained with DAPI (blue) and immunostained with anti-TM7SF3CT (Red) antibodies that were pre-incubated with or without TM7SF3 C-terminus peptide (n = 2).
(F) Total extracts of U2-OS cells were immunoblotted with TM7SF3 antibodies pre-incubated with or without TM7SF3 C-terminus peptide (10 μg/mL). Blotting with GAPDH served as a loading control.
(G) U2-OS cells were stained with DAPI (blue) and immunostained with anti-TM7SF3NT (Green) antibodies.
(H) U2-OS cells were transfected with TM7SF3-siRNA or with a non-targeting sequence (NonT). Cells were then stained with DAPI (blue) and immunostained with anti-TM7SF3CT antibodies.
(I) U2-OS cells were transfected with TM7SF3-siRNA or with a non-targeting sequence. Total cell extracts were immunoblotted with the indicated antibodies. All experiments were carried out at least three times unless otherwise indicated.
Immunofluorescence microscopy in U2OS cells, using these antibodies, revealed that TM7SF3 localizes to the nucleus (Figure 1B), rather than the plasma membrane. Indeed, TM7SF3 failed to colocalize with a classical membrane protein, the Na+/K+ ATPase (Figure 1C). This unexpected result was verified by immunoblotting. As shown in Figure 1D, the full-length TM7SF3 migrated as ∼80 kDa protein that co-localizes with histone H2B to the nuclear fraction. To ensure that the antibodies were indeed directed against TM7SF3, we inhibited their binding to the TM7SF3 protein, using a purified TM7SF3 C-terminal synthetic peptide. Inclusion of this peptide abolished the antibodies binding to nuclear TM7SF3 both by immunofluorescence (Figure 1E) or immunoblotting (Figure 1F). To determine whether the entire TM7SF3 protein localizes to the nucleus, a different antibody that targets TM7SF3 N-terminal end was employed. This antibody also revealed the exclusive nuclear localization of TM7SF3 (Figure 1G). Given that two antibodies, directed against different epitopes localized either at the N- or at the C-terminal end of TM7SF3, bind the same nuclear protein, supports the hypothesis that TM7SF3 is indeed a nuclear resident. To further verify that the identified immunoreactive protein corresponds to TM7SF3, its expression was silenced using specific siRNAs. Such treatment abolished TM7SF3 immunoreactivity detected either by immunofluorescence (Figure 1H) or immunoblotting (Figure 1I).
TM7SF3 localizes to nuclear speckles
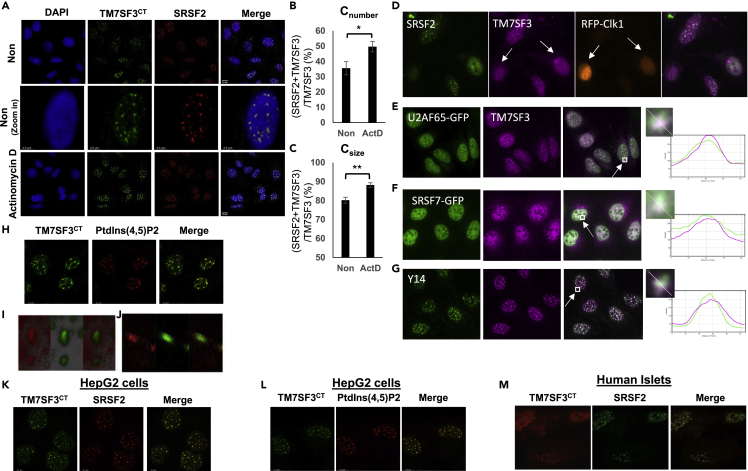
The sub-nuclear localization of TM7SF3 was evaluated next. As shown in Figure 2A, the nuclear staining of TM7SF3 in U2OS cells appeared as punctate structures that largely co-localized with immunostaining of SFRS2, a protein mainly residing within nuclear speckles (Figure 2B) (Spector et al., 1991). These structures, also known as interchromatin granule clusters, are nuclear domains enriched in pre-mRNA splicing factors, located in the interchromatin regions of the nucleoplasm of mammalian cells (Spector and Lamond, 2011). Association of splicing factors with nuclear speckles is enhanced as a result of drug-induced transcriptional inhibition (Sakashita and Endo, 2010). Indeed, we could show that when U2OS cells were treated with actinomycin D their nuclear speckles became more rounded, concomitant with a significant increased co-localization of both SFRS2 and TM7SF3 to these structures (Figures 2A–2C). Conversely, and consistent with our previous findings (Hochberg-Laufer et al., 2019) overexpression in U2OS cells of Clk1/STY kinase that disassembles nuclear speckles (Colwill et al., 1996) diminished the number of nuclear speckles concomitantly with the disappearance of TM7SF3 and of SFRS2 from these structures (Figure 2D). Other residents of nuclear speckles also co-localized with TM7SF3. These included the splicing factors U2AF65 and SRSF7 (Tang et al., 2020) and the exon junction complex factor Y14 (Chuang et al., 2015) (Figures 2E–2G). These findings suggest that TM7SF3 localizes in close proximity to a number of residents of nuclear speckles. This localization is dynamic and conditions that alter the structural integrity of nuclear speckles also affect the association of TM7SF3 with them.
Figure 2.
TM7SF3 localizes to nuclear speckles
(A) U2-OS cells were treated with or without Actinomycin D (5 ug/mL) for 4 h. Cells were then stained with DAPI (blue) and immunostained with anti-TM7SF3CT (Green) or anti-SFRS2 (Red) antibodies.
(B) Cnumber is the percentage of the number of objects immunostained with both TM7SF3CT and anti-SFRS2 antibodies, over the total staining with TM7SF3CT antibodies. Results are presented as means ± SEM (∗p<0.05; ∗∗p<0.01).
(C) Csize is the size-based co-localization coefficients of the objects stained with TM7SF3CT and anti-SFRS2 antibodies over the total staining with TM7SF3CT antibodies.
(D) U2OS cells overexpressing RFP-Clk1 (red), marked by arrows, were immunostained with anti-SRSF2 (green) or anti-TM7SF3CT (magenta).
(E and F) U2OS cells stably overexpressing U2AF65-GFP (E) or SRSF7-GFP (F) (green) were immunostained with anti-TM7SF3CT (magenta).
(G) U2OS cells were stained with anti-Y14 (green) or anti-TM7SF3CT (magenta). The nucleus (E-G) was stained with Hoechst (blue). The intensity of immunoreactivity of specific regions, marked by squares, is shown on the right.
(H–J) co-staining of U2-OS cells with antibodies against PtdIns(4,5)P2 (red) and TM7SF3 (green; n = 2). A z stack 3D image (I) and A z stack section (J).
(K and L) HepG2 cells were stained with DAPI (blue), and immunostained with anti-TM7SF3CT (Green), anti-SFRS2 (Red) (K), or antibodies against PtdIns(4,5)P2 (red) and TM7SF3 (green) (L).
(M) Dispersed human islets were immunostained as described in K. Experiments were carried out at least three times unless otherwise indicated.
Nuclear PtdIns4,5P2 signals for domain localization of specific proteins (Sztacho et al., 2021). These PtdIns4,5P2-binding proteins localize mainly to non-membrane bound organelles including nuclear speckles (Sztacho et al., 2021). Indeed, immunostaining of U2-OS cells showed co-localization of PIP2 and TM7SF3 (Figures 2H–2J), suggesting that TM7SF3 could be a PtdIns4,5P2-binding protein. Localization of TM7SF3 to nuclear speckles was not restricted to U2OS cells (Figure 2A), but was readily detected in HepG2 cells, where it co-localized with SRSF2 (Figure 2K), and PtdIns4,5P2 (Figure 2L). Similar staining patterns were also observed in HCT, HFF and mouse Min6 cells (Figures S3A–S3C); mouse tissues such as heart (Figure S3D) and testis (Figure S3E), as well as human pancreatic islets (Figure 2M), indicating that TM7SF3 localization to nuclear speckles is ubiquitous.
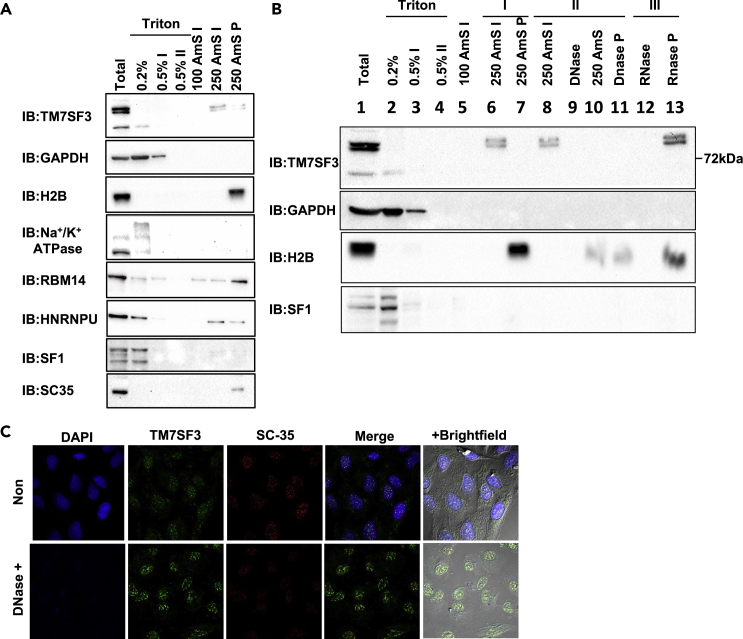
To identify the structural elements that enable the interaction of TM7SF3 with nuclear speckles, a subcellular fractionation was employed. As shown in Figure 3A TM7SF3 was resistant to extraction with 0.5% Triton X-100, unlike splicing factor 1 (SF1) that was readily solubilized by this non-ionic detergent. Similarly, TM7SF3 was resistant to solubilization following treatment with DNase or RNase, but could be eluted from cell extracts with 250 mM (NH4)2 SO4 (Figures 3A and 3B). This elution pattern was distinct from that of histone H2B or SFRS2 that were resistant to elution with 250 mM (NH4)2 SO4 (Figures 3A and 3B). This pattern was also distinct from that of membranal Na+/K+ ATPase that was eluted with 0.2% TritonX-100 (Figure 3A). The only proteins that partially co-eluted with TM7SF3 using 250 mM (NH4)2 SO4 were RBM14 [also called CoAA, RRM-containing Co-activator Activator (Auboeuf et al., 2002)] and HNRNPU (Yao et al., 2010) (Figure 3A) that are parts of the alternative splicing machinery.
Figure 3.
Subcellular fractionation and localization of TM7SF3
(A) U2-OS cells were resuspended in 0.2% Triton X-100, centrifuged and the supernatants were collected as 0.2% Triton fraction. The pellets were resuspended in 0.5% Triton X-100; centrifuged and the supernatant was collected as 0.5% Triton fractions. The pellets were resuspended with the indicated concentrations of ammonium sulfate (AMS); centrifuged and the supernatant was collected as AMS fractions. The pellets were then suspended in ‘sample buffer’ (AMS P fractions). Samples were resolved by SDS-PAGE and immunoblotted with the indicated antibodies.
(B) HFF cells were extracted in ‘sample buffer’ (1). Alternatively, the cells were sequentially incubated with the indicated solutions [0.2% (2)→ 0.5% (3)→ 0.5% (II) (4) Triton X-100→ 100 mM AMS (5)→ 250 mM AMS (6)]. The supernatants of the indicated fractions (2–6) and the 250 mM AMS pellets (7) were collected and incubated with sample buffer. Next, supernatants of the 250 mM AMS extracts (8) were treated with DNase, centrifuged and the sups were collected (9). The pellets were incubated again with AMS 250 mM, centrifuged, and supernatants were collected as AMS post-DNase fractionation (10). The pellets were incubated with ‘sample buffer’ (11) or treated with RNase and centrifuged. The supernatants (12) and pellets (13) were collected. All samples were resolved by SDS-PAGE and immunoblotted with the indicated antibodies n = 3.
(C) U2-OS cells were incubated with 0.5% Triton X-100, followed by incubation with DNase. Cells were then fixed; stained with DAPI (blue) and immunostained with anti-TM7SF3CT (Green) or anti-SFRS2 (Red) antibodies. n = 2.
Similar results were obtained when we analyzed the localization of TM7SF3 by immunofluorescence. As shown in Figure 3C, TM7SF3, unlike SFRS2, remained associated with nuclear speckles after treatment with 0.5% Triton X-100 and DNase. These findings suggest that TM7SF3 interacts with select structural elements within nuclear speckles.
TM7SF3 binding partners
To identify its binding partners TM7SF3 was immunoprecipitated from extracts of HFF or U2OS cells and the immune complexes were analyzed by mass spectrometry (MS). 28 proteins that selectively co-precipitated or were highly enriched in TM7SF3- immunoprecipitates were found (Table 1). These included 14 proteins involved in RNA binding, processing, or splicing [DHX15 (Niu et al., 2012), DDX21 (Valdez et al., 2002), NCL (Soeno et al., 2010), LMNA (Gerbino et al., 2018), HNRNPK (Liu et al., 2018), PABPC1 (Peng et al., 2017), LARP7 (Hasler et al., 2020), HNRNPU (Ye et al., 2015), HNRNPL (Fei et al., 2017), PABPC4 (Chapman et al., 2013), PTCD1 (Lightowlers and Chrzanowska-Lightowlers, 2013), DDX18 (Payne et al., 2011), RBM14 (Auboeuf et al., 2002; Zhou et al., 2017), and LARP1 (Burrows et al., 2010)]; 12 being direct RNA splicing modulators [DHX15, DDX21, NCL, LMNA, PABPC1, LARP7, HNRNPU; HNRNPL, HNRNPK, PABPC4, DDX18, and RBM14].
Table 1.
Proteins selectively immunoprecipitated by TM7SF3 antibodies
| Protein Name | Number of Repeats | Σ Coverage | Σ# Unique Peptides | |
|---|---|---|---|---|
| 1 | DHX15 | 4 | 16.6 | 12 |
| 2 | HP1BP3 | 3 | 72.69 | 65 |
| 3 | CLTC | 3 | 64.92 | 83 |
| 4 | XRCC5 | 3 | 54.64 | 25 |
| 5 | SHKBP1 | 3 | 50.00 | 21 |
| 6 | DDX21 | 3 | 44.44 | 27 |
| 7 | KCTD3 | 3 | 40.86 | 24 |
| 8 | RSL1D1 | 3 | 39.80 | 16 |
| 9 | XRCC6 | 3 | 33.33 | 16 |
| 10 | NCL | 3 | 32.39 | 23 |
| 11 | LMNA | 3 | 30.72 | 18 |
| 12 | MTPAP | 3 | 30.58 | 15 |
| 13 | HNRNPK | 3 | 26.13 | 9 |
| 14 | PABPC1 | 3 | 25.67 | 8 |
| 15 | ATAD3A | 3 | 21.16 | 11 |
| 16 | RPL4 | 3 | 20.84 | 7 |
| 17 | LARP7 | 3 | 20.36 | 5 |
| 18 | CCDC86 | 3 | 19.17 | 5 |
| 19 | HNRNPU | 3 | 18.30 | 12 |
| 20 | HNRNPL | 3 | 17.74 | 7 |
| 21 | NCOA5 | 3 | 15.54 | 7 |
| 22 | PABPC4 | 3 | 13.50 | 2 |
| 23 | GNL3 | 3 | 11.36 | 5 |
| 24 | PTCD1 | 3 | 10.57 | 7 |
| 25 | DDX18 | 3 | 8.51 | 4 |
| 26 | RBM14 | 3 | 6.28 | 3 |
| 27 | LARP1 | 3 | 5.69 | 1 |
| 28 | ATXN1L | 3 | 3.48 | 2 |
Total cell extracts of U2-OS (n = 3) or HFF cells were immunoprecipitated with pre-immune serum or anti-TM7SF3 antibodies. The beads were washed and subjected to analysis by mass spectrometry. The table summaries meta-analysis of 4 experiments of proteins absent in the pre-immune or enriched (fold≥3) in TM7SF3 IP preparations.
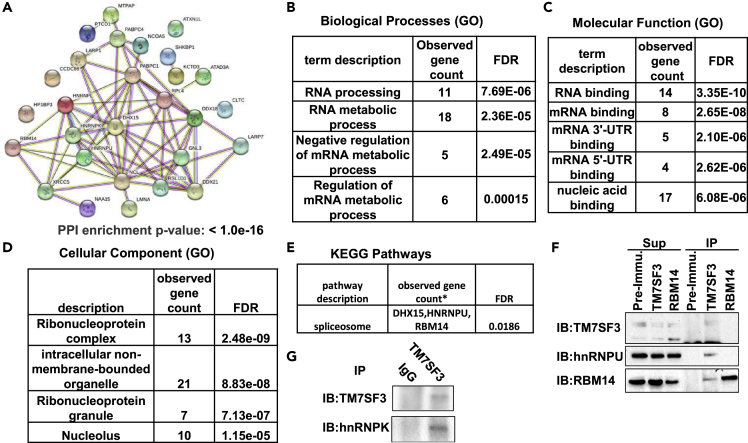
Gene ontology analysis revealed that many of the TM7SF3 binding partners form networks (Figure 4A) involved in RNA binding (11 out of 28) or RNA metabolic processes (18 out of 28) (Figure 4B). The molecular function ascribed to most (14 out 28) is RNA binding, or nucleic acid binding (17 out 28 (Figure 4C). Most of them (21 out of 28) localize to intracellular membrane-bound organelles and 13 out of the 28 are part of ribonucleoprotein complexes (Figure 4D). KEGG pathways analysis revealed that three of the proteins are part of the spliceosome (DHX15, HNRNPU, RBM14) (Figure 4E). This functional analysis implicates a role for TM7SF3 in RNA splicing. Complex formation between TM7SF3 and elements/splicing factors that regulate the spliceosome machinery could be further illustrated by co-immunoprecipitation experiments, followed by Western blotting. As shown in Figures 4F and 4G, TM7SF3, RBM14, HNRNPU, and HNRNPK co-precipitated by the TM7SF3CT antibodies from RIPA extracts of U2-OS cells, but not by pre-immune serum. These findings support the MS analysis that identified RBM14, HNRNPU and HNRNPK as TM7SF3 binding partners.
Figure 4.
Analysis of proteins that co-IP with TM7SF3
(A) STRING analysis was performed to identify the network of proteins that co-IP with TM7SF3 (cf. Table 1).
(B–E) Tables of proteins enrichment Gene Onthology (GO).
(F and G) Validation of the co-IP and Mass spectrometry data detailed in Table 1 by Western blotting with antibodies against HNRNPU, RBM14 (F), and HNRNPK (G) (n = 3).
TM7SF3 regulates alternative splicing
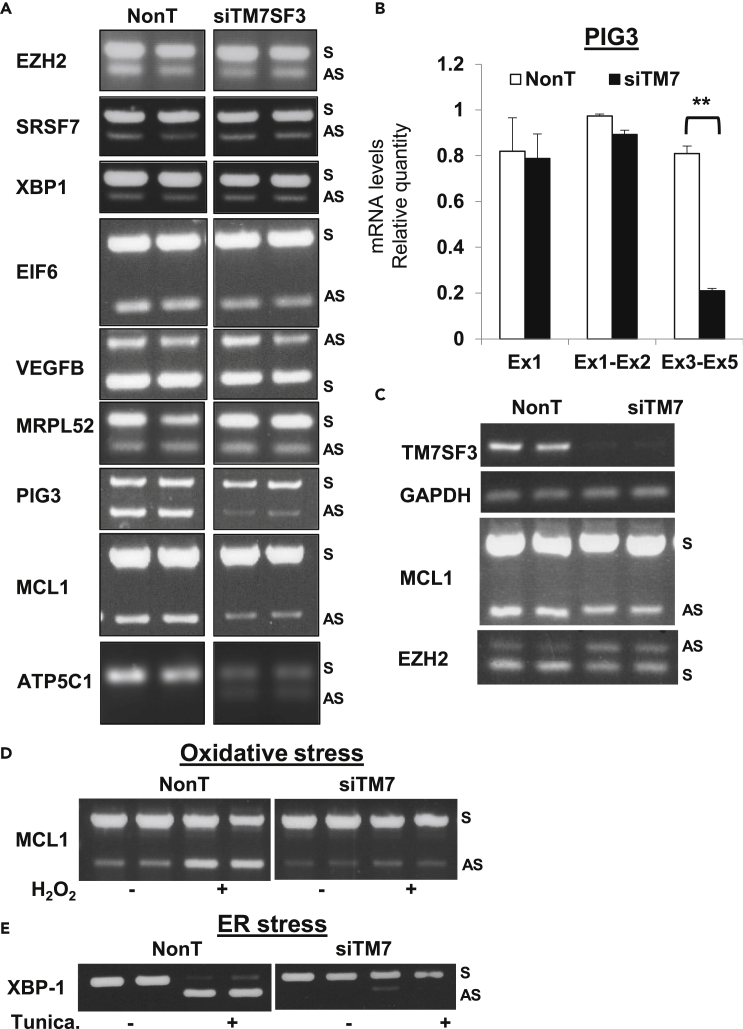
Nuclear speckles serve as hubs for factors involved in pre-mRNA splicing and alternative splicing (AS). Given that TM7SF3 directly interacts with a number of splicing factors, we wished to determine whether TM7SF3 plays a role in this process. AS is a ubiquitous phenomenon that affects almost any gene, therefore the pre-mRNAs chosen for analysis were randomly selected. As shown in Figure 5A, silencing of TM7SF3 in U2OS cells selectively inhibited AS of a number of pre-mRNAs, including PIG3 and ATP5C1 but it did not affect AS of other pre-mRNAs (e.g. EZH2, SRSF7, XBP1, EIF6, VEDFB, and MRPL52). In certain instances, silencing of TM7SF3 affected both basal and alternative splicing (cf. MCL1, Figure 5A). The affected exons were Ex4 of PIG3 (Figure 5B) and Ex2 of MCL-1 (Figure 5C). In contrast, silencing of TM7SF3 in HFF cells promoted AS of EZH2 (Figure 5C), suggesting that depending on the cell type, TM7SF3 can either promote or inhibit AS of a given pre-mRNA. Based on the above results, we wished to determine how general are the effects of TM7SF3 on alternative splicing and whether it affects specific splice sites. To this end, TM7SF3 was silenced in either human U2OS or mouse MIN6 cells, and total RNAseq analysis was performed. The results were analyzed for changes in local splicing variations (LSVs) between cells expressing siTM7SF3 and control cells, using the MAJIQ/VOILA algorithm (Vaquero-Garcia et al., 2016). Such analysis revealed significant alterations in splicing of mRNAs derived from >330 genes, some having multiple LSVs. These alterations encompassed differential splicing of cassette exons and retained introns and increased usage of cryptic splice sites (Table S5).
Figure 5.
TM7SF3 affects alternative splicing both under basal conditions and under stress
(A) U2-OS cells were transfected with TM7SF3-siRNA or with a non-targeting sequence. RNA was extracted, and splicing variants of the indicated genes were detected by PCR.
(B) Bar graphs represent the mean expression of PIG3 exons that were determined by real-time PCR with primers targeting Ex1-2 junction and Ex3-5 junction, n = 3. Results are presented as means ± SEM (∗∗p<0.01).
(C) HFF cells were transfected with TM7SF3-siRNA or with a non-targeting sequence. Splicing variants of the indicated genes were detected by PCR.
(D and E) U2-OS were transfected with TM7SF3-siRNA or with a non-targeting sequence. Cells were then treated with H2O2 (500 μM; 3 h) (D) or tunicamycin (5ug/mL; 8 h) (E). Splicing variants of MCL1 (D) or XBP-1 (E) were detected by PCR.
TM7SF3 regulates alternative splicing under stress
Induction of cellular stress alters both the structure of nuclear speckles (Figure 2) as well as the profile of AS (Paronetto et al., 2016), therefore we checked whether TM7SF3 affects AS under stress. Treatment of U2OS cells with the stress inducers H2O2 (Figure 5D) or tunicamycin (Figure 5E) markedly induced AS of MCL1 and XBP-1, respectively, whereas prior treatment of these cells with TM7SF3 siRNA significantly inhibited this process. These findings suggest that TM7SF3 regulates AS both under basal conditions and under stress. Of interest, whereas silencing of TM7SF3 failed to alter AS of XBP-1 in naive U2OS cells, it effectively stimulated its AS under stress conditions, indicating that TM7SF3 engages with a different set of splicing factors, depending on the state of cellular stress. Given that AS of XBP-1 takes place in the cytoplasm (Hassler et al., 2015) whereas TM7SF3 resides in nuclear speckles, these findings further suggest that TM7SF3 presumably regulates the availability of splicing factors that act in the cytoplasm, outside the nuclear splicesome machinery (Jurica, 2008).
TM7SF3 regulates alternative splicing preferentially at the 3′end of introns
To gain a deeper insight into the splicing machinery regulated by TM7SF3, we tested a compendium of motifs that serve as binding sites for RNA binding proteins (Giudice et al., 2016), and are enriched around splice sites. In particular, we were interested in those motifs that either increased or decreased on TM7SF3 knockdown. Because sequence elements that regulate splicing have strong location-specific effects, we performed this analysis separately for different regions relative to the splice sites, namely the last 50 nt in the exon that precedes the splice junction, the first and last100 nt of the intron, and the first 50 nt of the downstream exon. Using ATtRACT (Giudice et al., 2016) and HOMER (Hypergeometric Optimization of Motif Enrichment (Heinz et al., 2010)) as databases we looked for significant enrichment of motifs in these regions over the corresponding regions around all other splice sites in the human genome. We could identify many motifs to be significantly (FDR-adjusted pvalue<0.05) enriched in the last 100 nt before splice acceptors that exhibited increased usage upon TM7SF3 knockdown (58 out of 1583 motifs, as opposed to 3–18 motifs enriched in the other regions tested, Table S6, Figure S6). The motifs at the start/end of introns, having the highest confidence of enrichment in siTM7SF3 cells, served as binding sites for splicing regulators (SRSF1, HNRNPC, PCBP1, PCBP2, HEN1, SRSF5, SFPQ, and SF1, Table S6) that formed a highly significant network with the RNA binding proteins that co-immunoprecipitated with TM7SF3 (Figure S4). These findings suggest a functional interaction between these splicing regulators and TM7SF3 at the splice sites.
TM7SF3 regulates the activity of splicing factors
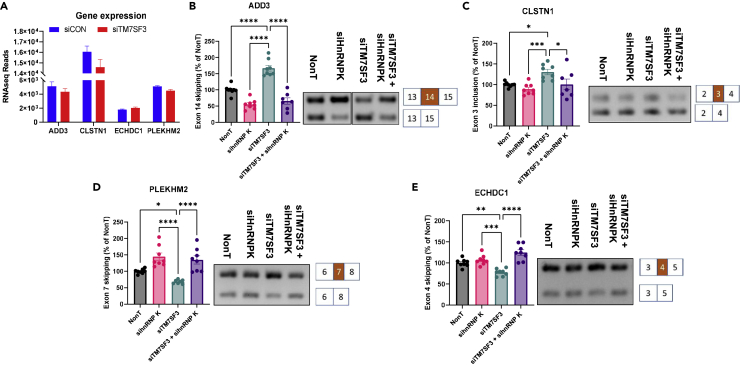
To gain a deeper mechanistic insight into the mode of action of TM7SF3, its expression was silenced independent of, or concomitantly with silencing of HNRNPK that forms stable complexes with TM7SF3 (Figure 4G and, Table 1). HNRNPK binding motifs were selected by comparing previous findings (Selvanathan et al., 2015) with the LSVs data (Figure S5). Only genes, whose global expression was not affected upon silencing of TM7SF3 (Figure S5) were selected for further analysis. Furthermore, alternative-splicing events having functional consequences were selected for analysis. Indeed, as shown in Figure 6A silencing of TM7SF3 did not affect the expression of ADD3, GLSTN1, PLEKHM2, or ECHDC1. Exon skipping in these genes was analyzed next. Silencing of TM7SF3 promoted significant skipping of Ex14 of cytoskeletal gene Adducin 3 (ADD3) whereas silencing of HNRNPK exerted a significant opposite effect (Figure 6B). Silencing of both TM7SF3 and HNRNPK eliminated the effects of siTM7SF3 on ADD3-Ex14. Of interest, upregulation of Ex14 inclusion significantly correlates with poor prognosis in lung cancer (Wang et al., 2021), suggesting that TM7SF3 might act as a promoter of this process. Similar, albeit less dramatic effects were observed when inclusion of Ex3 of CLSTN1 was analyzed (Figure 6C). Aberrant AS of this exon was linked to the development of gastric cancer (Feng et al., 2020). Opposing effects of siTM7SF3 and siHNRNPK were also observed when analyzing the skipping of Ex7 of PLEKHM2 (Figure 6D) or Ex4 of ECHDC1 (Figure 6E). Here silencing of TM7SF3 inhibited significant skipping of these exons, whereas siHNRNPK exerted an opposite effect. These results suggest that TM7SF3, by acting as an upstream inhibitor of HNRNPK, regulates exon inclusion or skipping in a process that requires an active HNRNPK.
Figure 6.
TM7SF3 modulates the activity of HNRNPK
(A) Voila analysis of RNAseq data of genes whose expression is not affected by siTM7SF3.
(B–E) U2-OS cells were transfected with indicated siRNAs or with a non-targeting sequence. RNA was extracted, and splicing variants of the indicated genes ADD3 (B), CLSTN1(C), PLEKHM2 (D), and ECHDC1 (E) were determined by PCR. Bar graphs represent the mean percentage of the spliced-out exons shown in each scheme (NonT = 100%) as measured by PCR products densitometry in 4 independent experiments Results are presented as means ± SEM (∗p < 0.05; ∗∗p < 0.01; ∗∗∗p < 0.001).
TM7SF3 induces differential gene expression
Given that AS affects gene expression, we utilized RNAseq analysis to determine the effects of TM7SF3 on the gene expression profile of U2OS cells. PCA analysis of the transcriptome and hierarchical clustering revealed significant differential expression of 1465 genes between control cells and cells in which TM7SF3 has been silenced. Silencing upregulated the expression of 844 genes, whereas the expression of 621 genes was downregulated (Figure S5). Gene ontology analysis revealed that silencing induced a number of pathways and biological processes, such as DNA damage responses (17 out of 120 genes) and telomere stress induced senescence (15 out of 80 genes) (Table S7). Conversely, silencing of TM7SF3 inhibited extracellular matrix degradation (29 out of 298 genes) and cell adhesion (42 out of 625 genes) (Table S8). Complementary analysis using a different set of algorithms (Table S9) showed additional pathways being affected by silencing of TM7SF3, that included e2F signaling, nucleosome assembly, DNA methylation, and histone modifications, all involved in RNA processing and splicing (Lev Maor et al., 2015; Warns et al., 2016). Collectively, these findings suggest that TM7SF3, being a regulator of alternative splicing, affects the expression of genes covering a large spectrum of biological responses.
Discussion
Alternative splicing (AS) is a critical mechanism for the generation of proteome diversity. Expression of nearly 95% of human multi-exon genes involves alternative splicing (Chen and Manley, 2009), yet the underlying molecular elements that regulate this process remain incompletely understood. Here we show that a seven transmembrane protein TM7SF3 resides in nuclear speckles where it regulates AS both under basal and stress conditions. Hence, TM7SF3 appears to be the first identified seven transmembrane protein having such unique nuclear localization and function as a modulator of the splicing machinery. We further show that TM7SF3 forms complexes with a number of RNA-binding proteins directly involved in AS such as HNRNPU, HNRNPK, and RBM14. The fact that TM7SF3 remains associated with these proteins even following extraction with a combination of detergents (deoxycholate and SDS) supports the notion that TM7SF3 forms relatively tight contact with the AS machinery. At the molecular level, we show that TM7SF3 binds splicing factors and RNA-binding proteins that regulate alternative splicing mainly at the 3′-end of introns. Of interest, TM7SF3 alters the pattern of AS in a cell-specific manner, because a different repertoire of AS sites are affected by silencing of TM7SF3 in U2OS cells (present study) and MIN6 cell line (not shown).
Changes in AS affect patterns of gene expression (Fiszbein and Kornblihtt, 2017). Transcriptome analysis revealed that silencing of TM7SF3 alters the expression of >1,400 proteins, involved in a large number of pathways and biological processes, such as DNA damage responses and telomere stress induced senescence. siTM7SF3 also inhibited extracellular matrix disassembly and cell adhesion, presumably through interference with expression of genes such as MMP1, MMP7, CD44, FN1, and COL8A1. These findings are in accordance with the role of TM7SF3 as a regulator of protein homeostasis (Isaac et al., 2017). Most relevant, silencing of TM7SF3 affected nucleosome assembly, DNA methylation, and histone modifications. These processes regulate RNA proccing and splicing (Lev Maor et al., 2015; Warns et al., 2016) in part by controlling the rate of Pol II-mediated RNA elongation that governs the extent of alternative splicing (Lev Maor et al., 2015; Warns et al., 2016).
One of the exciting and unexpected finding of our study stems from that fact that TM7SF3, a seven transmembrane protein, is found localized to nuclear speckles that are not membranal structures. It raises the interesting question, of how TM7SF3 is spatially localized within speckles and what replaces the conventional membranous structures needed to maintain its proper topology. In that respect, G protein-coupled receptors are localized at the nuclear membrane and nucleoplasm (Branco and Allen, 2015; Campden et al., 2015; Gobeil et al., 2006; Lee et al., 2004) and their intracellular signaling is ascribed to nuclear translocation of peripheral ligands or to endogenously produced non-secreted ligands (Boivin et al., 2008; Gobeil et al., 2006). Nuclei of many mammalian cells contain numerous channels arising from invagination of nuclear envelopes (Lagace and Ridgway, 2005) or inner nuclear membrane (Isaac et al., 2001). Thus, the existence of distinct, dynamic tubular membranes within the nucleus, termed the nucleoplasmic reticulum and R-rings/nucleolar channel systems, may reconcile the apparent nucleoplasmic localization of GPCRs with their integral membranous association characteristics (Boivin et al., 2008; Gobeil et al., 2006). However, none of the heptahelical receptors described above is selectively localized to nuclear speckles, and none was assigned a specific function related to alternative splicing. The fact that both antibodies directed against the N- or C-terminal end of TM7SF3 interact with this protein in nuclear speckles, combined with the fact that silencing of TM7SF3 by specific siRNAs abolishes this immunoreactivity, supports the idea that the entire TM7SF3 protein, rather than its fragment thereof, resides in nuclear speckles.
Our findings indicate that TM7SF3 co-localizes with PtdIns4,5P2 within nuclear speckles. These observations fit well with the findings that more than 300 nuclear PtdIns4,5P2-interacting proteins involved in chromatin remodeling or RNA processing were already identified (Lewis et al., 2011). These proteins localize mainly to non-membrane bound organelles—nuclear speckles and nucleolus and are connected to the actin nucleoskeleton (Gonzales and Anderson, 2006; Sztacho et al., 2021). The co-localization of TM7SF3 with PtdIns4,5P2 within nuclear speckles might indicate that these structures serve as binding partners for TM7SF3, and stabilize its anchorage to nuclear speckles. Furthermore, both PtdIns4,5P2 (Gonzales and Anderson, 2006) and TM7SF3 are found in the detergent-insoluble nuclear fraction, suggesting that both are linked to the nuclear matrix. Given that RBM14, HNRNPU and HNRNPK, are TM7SF3-binding partner, and are also constituents of paraspeckles (Hennig et al., 2015; Hirose et al., 2019), TM7SF3 might be peripherally associated with these structures as well.
Of interest, the residency of TM7SF3 to nuclear speckles and its effects on AS are dynamic processes. Conditions that induce cellular stress such as inhibition of transcription by actinomycin D, increase the cellular content of TM7SF3 in nuclear speckles, similar to the increase in other splicing factors such as SFRS2. In that respect, TM7SF3 resembles other residents of nuclear speckles that are PtdIns4,5P2- interacting proteins, such as the non-canonical poly(A) polymerase, termed Star-PAP (nuclear speckle targeted PIPKIα regulated-poly(A) polymerase) that responds to oxidative stress by regulating 3′-end formation and expression of its target mRNAs (Mellman et al., 2008). Conversely, overexpression of Clk1/STY kinase, responsible for serine phosphorylations on RS domains in SR proteins, causes the disassembly of nuclear speckles (Chuang et al., 2015); nucleoplasmic dispersion of endogenous splicing factors such as SFRS2, and disappearance of TM7SF3 from these structures. These findings indicate that similar to other splicing factors and other residents of nuclear speckles, TM7SF3, shuttles in and out of nuclear speckles, depending on the homeostatic conditions and the cellular demand for elements of the splicing machinery.
At the molecular level, we could show that TM7SF3 acts as an upstream regulator of splicing factors such as HNRNPK. TM7SF3 inhibits the action of HNRNPK, whereas silencing of both proteins results in similar effects to those of silencing of HNRNPK alone. Given that TM7SF3 forms tight complexes with HNRNPK and other splicing factors, it is reasonable to assume that TM7SF3 alters the function of HNRNPK through direct interactions with this protein and other splicing factors as well. The inhibitory effects of TM7SF3 on HNRNPK activity might have functional consequences. Aberrant AS of Ex 3 of CLSTN1, that is regulated by TM7SF3 (Figure 6), is linked to development of gastric cancer (Feng et al., 2020). Similarly, AS of ADD3 Ex14 is a key lung cancer-related splicing event because inclusion of ADD3-Ex14 promotes lung cancer cell proliferation and migration and positively correlates with the development of lung adenocarcinoma and squamous cell carcinoma (Wang et al., 2021). By promoting inclusion of Ex14 (Figure 6), TM7SF3 might support cancer cell growth, which is in line with its inhibitory effects on p53 activity (vide infra).
As we have previously shown, TM7SF3 is a p53-regulated homeostatic factor that attenuates cellular stress and induction of the unfolded protein response (UPR) (Isaac et al., 2017). Our current findings are well in line with this concept as they provide the underlying mechanism to the mode of action of TM7SF3. Being a regulator of AS it is easy to rationalize why silencing of TM7SF3 deranges AS, promotes cellular stress, and induces UPR. The current findings also fit well with the observation that TM7SF3 inhibits expression of p53 (Isaac et al., 2017). Given that p53 attenuates the UPR as evidenced by augmented alternative splicing of XBP1 in p53-deficient mice (Dioufa et al., 2012), and given that silencing of TM7SF3 under stress inhibits AS of XBP1 (current study), inhibition of p53 expression, induced by TM7SF3, is expected to relief the inhibitory action of p53 on AS of XBP1. Of interest, HNRNPK, that serves as transcriptional co-activator of p53 (Lee et al., 2012), is a downstream effector of TM7SF3. We have previously shown that p53 promotes transcription of TM7SF3 (Isaac et al., 2017), hence, HNRNPK could indirectly promote expression of TM7SF3 that then inhibits the function of HNRNPK, thus generating a negative-feed-back loop. This conclusion is supported by our previous findings that TM7SF3 inhibits the action of p53 (Isaac et al., 2017).
TM7SF3 has also been implicated as a potential player in the inhibition of cytokine-induced death and in the promotion of insulin secretion from pancreatic beta cells (Beck et al., 2011). Here we show that TM7SF3 resides in nuclear speckles of human pancreatic islets where it can alter AS. Alternative splicing events occurring in pancreatic β cells regulate β cell functionality (Juan-Mateu et al., 2016). Given that TM7SF3 is one of ten genes found in the 700 kb murine type 1 diabetes locus, Idd6.3 (Hung et al., 2006), our findings support a special role for TM7SF3 in the regulation of β cell function.
In summary, identification of TM7SF3 as a regulator of AS places this seven transmembrane protein at a key position in one of the most complex posttranscriptional regulatory mechanism affecting gene expression. Its essential role in this process is exemplified by the deranged AS that occurs once TM7SF3 is silenced. This exerts a major impact on cellular functionality, exemplified by the inhibition of GSIS in pancreatic β cells (Beck et al., 2011). Identifying the seven transmembrane TM7SF3 as a resident of nuclear speckles implicates seven-transmembrane proteins in the modulation of nuclear functions including alternative splicing.
Limitations of the study
-
-
The assignment of TM7SF3 as a seven-transmembrane protein is solely based on computerized prediction employing neural network-based model, AlphaFold2. Actual X-ray crystallography is required to validate this assignment.
-
-
Co-immunoprecipitation of TM7SF3 with its binding partners was limited due to the need to use a strong detergent such as SDS to dissolve TM7SF3. This resulted in detection only of high-affinity binding partners of TM7SF3.
-
-
Overexpression of TM7SF3 resulted in its perinuclear localization, suggesting that a missing factor is required to localize TM7SF3 to nuclear speckles.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Mouse monoclonal anti-hnRNPK | Santa Cruz Biotechnology | Cat#: sc-28380 RRID:AB_627734 |
| Mouse anti-IgG2B | Santa Cruz Biotechnology | Cat#: sc-3879 RRID:AB_737262 |
| Mouse monoclonal anti-hnRNPU | Millipore | Cat#: 05-1516 RRID:AB_10563506 |
| Mouse monoclonal anti-GAPDH | Millipore | Cat# MAB374 RRID:AB_2107445 |
| Rabbit polyclonal anti-H2B | Millipore | Cat#07371 RRID:AB_310561 |
| Rabbit polyclonal anti-HSP90 | Cell Signaling Technology | Cat#: 4874 RRID: AB_2121214 |
| Mouse monoclonal anti-Y14 | Abcam | Cat# ab5828 RRID:AB_2269494 |
| Rabbit polyclonal anti-U2AF65 | Abcam | Cat# ab37530 RRID:AB_883336 |
| Mouse monoclonal anti-PI(4,5)P2 | Echelon biosciences | Cat#Z-P045 RRID:AB_427225 |
| Rabbit polyclonal anti-SF1 | Antibody Verify | N/A |
| Rabbit polyclonal anti-RBM14 | Antibody Verify | N/A |
| Guinea Pig polyclonal anti-Insulin | DAKO | Cat#: A0564 N/A |
| Mouse monoclonal anti-SRSF2 | Sigma | Cat# S4045 RRID:AB_477511 |
| Rabbit polyclonal anti-TM7SF3 N-terminus | Sigma | Cat# HPA057640 N/A |
| Rabbit polyclonal anti-TM7SF3 C-Terminus | Sigma, This paper | N/A |
| Mouse monoclonal anti-α1-subunit of Na+/K+-ATPase (6H) | Dr. M. J. Caplan, Yale |
N/A |
| Oligonucleotides | ||
| Primers for qPCR and PCR, see Tables S2 and S3 | This paper | N/A |
| siRNA-human TM7SF3 | Horizon Discovery | Cat# M-005745-00-0010 |
| Human siCON | Horizon Discovery | Cat# D-OO1206-13-20 |
| siRNA-Human hnRNPK | Horizon Discovery | Cat# M-011692-00-0005 |
| Chemicals, peptides, and recombinant proteins | ||
| Protein A-agarose beads | Santa Cruz | Cat# sc-2001 |
| Blocking peptide - IKELFQKEQPAGERTPL | Sigma | Customized N/A |
| Actinomycin D | Sigma | Cat# A1410 |
| High-capacity cDNA reverse transcription kit | ThermoFisher Scientific | Cat# 4368813 |
| Lipofectamine 2000 reagent | ThermoFisher Scientific | Cat# 13778-075 |
| SYBR Green Master Mix | ThermoFisher Scientific | Cat# A25743 |
| Experimental models: Cell line | ||
| U2-OS | ATCC | Cat# HTB-96 |
| HepG2 | ATCC | Cat# HB-8065 |
| HFF | ATCC | Cat# SCRC-1041 |
| Software and algorithms | ||
| ImageJ | NIH | https://imagej.nih.gov/ij/ |
| Prism | Graphpad | https://www.graphpad.com/scientificsoftware/prism/ |
| Image Lab | BioRad | https://www.bio-rad.com/en-us/product/chemidoc-xrs-system?ID=NINJHRKG4 |
| STRING analysis | Szklarczyk et al., 2015) PMID:25352553 | https://string-db.org/ |
| AlphaFold2 |
Jumper et al. (2021) {PMID: 34265844} |
AlphaFold2.ipynb - Colaboratory (google.com) |
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact Yehiel Zick (Weizmann Institute of Science, Rehovot, Israel; Yehiel.Zick@weizmann.ac.il).
Material availability
Plasmids generated are available by the lead contact Yehiel Zick (Weizmann Institute of Science, Rehovot, Israel; Yehiel.Zick@weizmann.ac.il).
Experimental model and subject details
Cell lines
Cell lines that have been used for this study and media are described in Table S1.
Method details
Cloning of mouse TM7SF3 (GI:7288142) expressed in bacterial expression plasmids
Recombinant TM7SF3 protein fragments were used for antibody validation. An N-terminus fragment (aa 24-290) and a C-terminus fragment (aa 496-564) were both tagged at their amino terminus with 6xHis and MBP (Maltose binding protein)-TEVH. The plasmids were generated by Transfer-PCR (TPCR) (Ben-Ari Fuchs, 2016), which combines PCR amplification from the mouse TM7SF3-Myc vectors (Targets: N- or C- terminus) (Isaac et al., 2017) and subsequent integration of the PCR product into a recipient vector (pETMBPH-TEVH). For all constructs, primers were designed as follows: every two primers included a target-gene specific sequence at the 3′ end and sequences corresponding to the integration site in the recipient vector at the 5′ end (underlined sequence for the specific target of TM7SF3 fragments). Primers for N-terminus of TM7SF3 (24-290) 5′-TCCGCGGGTGAAAACCTGTACTTCCAGGGTAGCGACGGTTTCCTTGAG-3′; 5′-GTGGTGGTGCTCGAGTGCGGCCGCAAGCTTTCATTTCGTAGATACTCTTC-3′. Primers for C-terminus of TM7SF3 (496-564) 5′-TCCGCGGGTGAAAACCTGTACTTCCAGGGTCAGATTCGAAGAGAAAGAGG-3′; 5′-GTGGTGGTGCTCGAGTGCGGCCGCAAGCTTTCACAGAAGCAAGGGTGT-3′. Following completion of the PCR reaction, 1 μL of restriction enzyme Dpn1 (20 U/μl) was added to 10 μL of PCR reaction, followed by incubation for 2h at 37°C, to remove the parental methylated DNA (TM7SF3-Myc). The DNA was then directly transformed into competent E. coli XL-1Blue.
Cell cultures, transfection, and preparation of cell extracts
Cells were grown at 37°C, 5% CO2 in the indicated medium (Table S1) that was supplemented with Glutamine (2 mM) and Pen/Strep/Ampho. (100 units/ml;100 μg/mL; 250 ng/ml). A stably integrated Tet-inducible β-globin mini-gene termed E3 in U2OS cells (U2OS-E3 cells) that stably express RFP-Clk1 or GFP-tagged splicing factors from a BAC construct: U2AF65-GFP, SRSF2-GFP, and SRSF7-GFP that mark nuclear speckles, were generated as previously described (Alkalay, 2020; Hochberg-Laufer et al., 2019). Induction of expression was carried out using 1 μg/mL doxycycline (Sigma). Cells were seeded in 24-well/12-well plates (15 × 104/50 × 104 cells/well, respectively) and were transfected with siRNA SMARTpools using Dharmafect-4 transfection reagent (Dharmacon), according to the manufacturer’s instructions. Cells were then treated as indicated, washed three times with PBS and harvested in RIPA buffer (50 mM Tris/HCl, 150 mM NaCl, 0.5 mM EGTA, 2 mM sodium orthovanadate, 10 mM NaF, 10 mM sodium pyrophosphate, 80 mM β-glycerophosphate, 1% NP-40, 0.5% Deoxycholate, 0.05% SDS, 1 mM PMSF and protease inhibitors 1:100, pH 8). Cell extracts were centrifuged at 20,000xg for 15min at 4°C, and the supernatants were collected. Samples (10–40 μg) were mixed with 5x Laemmle sample-buffer, boiled, and resolved by 8–12% SDS-PAGE under reducing conditions. The proteins were transferred to nitrocellulose membrane for Western blotting with the indicated antibodies.
Subcellular fractionation by differential detergent and salt extraction
Subcellular fractionation of U2-OS or HFF cells was carried out as described (Fey et al., 1986). Cells were washed three times in ice cold PBS (with 1 mM EDTA). Pellets were resuspended in cytoskeleton (CSK) buffer (100 mM NaCl, 300 mM sucrose, 10 mM Pipes [pH 6.8], 3 mM MgCl2, 1 mM EGTA, and 0.2% Triton X-100) and were incubated at 4°C for 10 min. After centrifugation at 1000g for 3 min, the supernatants, which contained soluble proteins, were collected. The remaining pellets were resuspended again in CSK buffer containing 0.5% Triton X-100, re-centrifuged, and the supernatants collected. The remaining nuclear pellets were further incubated for 7 min at 4°C in ammonium sulfate buffer (100 mM ammonium sulfate, 300 mM sucrose, 10 mM Pipes [pH 6.8], 3 mM MgCl2, 1 mM EGTA and 0.5% Triton X-100). The eluted proteins were collected following centrifugation as above. This step was repeated with a higher concentration of ammonium sulfate buffer (250 mM) to elute TM7SF3 and other proteins. The pellets were then sonicated with 1xLaemmli sample buffer for 10 min (30 s on; 30 s off). Where indicated, the chromatin fraction was extracted from the pellets, prior to sonication, following digestion for 1h at 22°C in Triton X-100-free CSK buffer, containing 500 U/ml DNAase I (5 PRIME, Hamburg, Germany). Digestion in DNAase I was terminated by addition of ammonium sulfate to a final concentration of 0.25 M. The nuclear matrix was pelleted at 1,000xg for 5 min, and the chromatin fraction was collected in the supernatant.
Culture of dispersed human islets
Isolated human islets (∼90% purity) were provided by the European Consortium for Islets Transplantation (Islet for Basic Research program) through a Juvenile Diabetes Research Foundation Award 31-2008-413. Islets were cultured at 37°C in a 5% CO2 humidified atmosphere in CMRL 1066 medium containing 10% (v/v) FBS, 2mM L-glutamine, 100 units/ml penicillin, 100 μg/mL streptomycin, 0.25 μg/mL amphotericin, and 40 μg/mL gentamycin. The medium was changed every other day. Human islets studies received the WIS Ethics Committee approval. Intact human islets were dispersed by a 4 min incubation at 37°C with 1 mg/ml Trypsin/EDTA and by passing the cells twice through a 21G needle. Trypsinized islets were washed with CMRL 1066 medium containing 10% FBS and were resuspended in CMRL 1066 containing 10% FBS.
Co-immunoprecipitation
Protein A-agarose beads (20 μL packed per point) were washed with ice-cold 0.1 M Tris-HCl (pH 8.5) followed by centrifugation at 1000xg for 2 min at 4°C. The beads were washed twice with buffer-I containing 10 mM Tris, 150 mM NaCl, 1 mM EGTA, 1 mM EDTA, 1% Triton X-100, 0.2 mM Sodium orthovanadate, and protease inhibitor cocktail 1:100, pH 7.4. The washed beads were then incubated with polyclonal TM7SF3CT antibodies for 4 h at 4°C. 12,000 xg supernatants of cell extracts in RIPA buffer, containing 0.6–1 mg of protein, were diluted 1:5 in buffer-I and underwent pre-clearing by incubation with protein A-agarose beads for 2 h at 4°C. The supernatants were incubated overnight at 4°C with the immobilized TM7SF3CT antibodies. Immunocomplexes were washed three times with buffer-I, and were analyzed by SDS-PAGE and Western blotting using a BioRad Mini-PROTEAN system (BioRad, Hercules, CA). For mass-spectrometry, protein bands were excised from SDS gels that were stained with GelCode and subjected to mass-spectrometry analysis.
Immunofluorescence
For subcellular localization of TM7SF3, cells (5 × 104 cells/well; 24 wells/plate) were grown on glass coverslips; washed with PBS, and fixed with 4% paraformaldehyde for 17 min at 22°C. Alternatively, cells were fixed with MeOH for 5 min followed by incubation with acetone for 2 min at −20°C as indicated. Cells were then permeabilized with 0.5% Triton-X100 in PBS for 4 min, and thereafter blocked for 30 min in a blocking solution (Tris 10 mM, NaCl 150 mM, 0.5% Triton X-100, 10% normal Goat serum, 2% BSA, 1% Glycine, pH 7.4). Cells were subjected to indirect immunofluorescence with the indicated antibodies diluted in a blocking solution for 1 h at 22°C. Cells were then washed with PBS, and primary antibodies were detected with Alexa594 Goat anti-mouse and/or Alexa488 Goat anti-rabbit secondary antibodies (ThermoFisher, diluted 1:200 in PBS with 20% Normal horse serum) for 1 h at 22°C. Cells were then washed with PBS. Staining of nuclei was performed using DAPI (0.5 μg/mL) or Hoechst 33342 as indicated. The specimens were washed several times with PBS, and were mounted with Immu-mount (Thermo Scientific; Cheshire, UK) overnight on glass microscope slides. Images were obtained as single optical slides using LSM 710 Confocal microscope (for Figures 1C, 1G, 1H, and 2M) (Zeiss, Jena, Germany), Leica TCS SP8 confocal microscope (for Figures 1B, 1E, 2A, and 2H–2L), or Olympus IX83 inverted microscope (for Figures 2D–2G). Quantitative analysis of co-localization of TM7SF3 and SC-35 (SRSF2) proteins was carried out as described previously (Rizk et al., 2014). In brief, 8–9 frames of U2-OS cells that were immunostained with TM7SF3 and SC-35 antibodies as described above, were processed by Fiji software. Nuclei in each frame were chosen as the region of interest by DAPI staining. Then, segmentation and quantification were done using the ‘Squassh’ plugin, and the statistical analysis was performed with the open code source R-statistics.
RNA extraction, RT-PCR and qRT-PCR
Total RNA was extracted using PerfectPure RNA kit (5-prime, MD). RNA was quantified using nano-drop, and cDNA was generated by cDNA Reverse Transcription kit (Applied Biosystems, CA). Quantitative detection of mRNA transcripts was carried out by real-time PCR using ABI-Prism 7300 instrument (Applied Biosystems, CA); SYBR Green PCR mix (Invitrogen), and specific primers (400 nM final concentration). Results were normalized to mRNA levels of Actin or HPRT. Splice variants were examined by conventional PCR. Primers used for qRT- and RT-PCR are listed in Tables S2 and S3.
Mass spectrometry
Analysis by mass spectrometry was carried out essentially as described (Isaac et al., 2013). In brief, protein extracts were subjected to immunoprecipitation with TM7SF3CT antibodies or with pre-immune serum (control). The immunoprecipitated proteins were resolved by SDS-PAGE and the gel bands were subjected to in-gel trypsin digestion followed by a desalting step. The resulting peptides were analyzed using nanoflow liquid chromatography (nanoAcquity) coupled to high resolution, high mass accuracy mass spectrometry (Q Exactive Plus). Samples were analyzed in a random order on the instrument in discovery mode. Data were analyzed using Proteome Discoverer version 1.4 and searched against the Uniprot database, to which a list of common lab contaminants was added. Two search algorithms-SequestHT and Mascot, were employed. Relative quantitation was conducted with the Scaffold PTM software (Proteome Software Inc., USA) and the A-score algorithm (Zhai et al., 2008).
RNA-seq and data analysis
TM7S3 was silenced using specific siRNAs, in human U2OS cells. Cells treated with non-targeting siRNAs served as controls. RNA was extracted from three replicates of siTM7-treated and three siCON-treated U2OS cells. Six samples altogether were used. Total RNA was isolated using RNeasy Kit (Qiagen, Hilden, Germany) according to the manufacturer’s protocol. Total RNA was fragmented followed by reverse transcription and second strand cDNA synthesis. The double strand cDNA was subjected to end repair, A base addition, adapter ligation and PCR amplification to create libraries. Libraries were evaluated by Qubit and TapeStation. 75bp single reads were sequenced on 4 lanes of an Illumina NextSeq High output 75 cycles. The output was ∼56 million reads per sample. Poly-A/T stretches and Illumina adapters were trimmed from the reads using cutadapt (Martin, 2011); resulting reads shorter than 30bp were discarded. Data analysis was carried our using GeneAnalytics software (URL geneanalytics.genecards.org) as described (Ben-Ari Fuchs et al., 2016). Reads were mapped to the Homo Sapiens GRCm38 reference genome using STAR (Dobin et al., 2013), supplied with gene annotations downloaded from Ensembl, build 88 (and with EndToEnd option and out Filter Mismatch Nover Lmax was set to 0.04). Mapping of reads to the genome was good, with ∼94% reads being mapped. Counting was also good with ∼85% of the uniquely mapped reads being counted. The highest fraction of counts from a single gene was 1.8%. Expression levels for each gene were quantified using htseq-count (Anders et al., 2015), using the gtf above. Differentially expressed genes were identified and analysis was performed using DESeq2 (Love et al., 2014) with the beta Prior, cooks Cutoff and independent Filtering parameters set to False. Raw p values were adjusted for multiple testing using the procedure of Benjamini and Hochberg. In an alternative procedure DESeq2 (R v3.5.1, package v1.22.2 (Love et al., 2014) was used on the raw count values for each sample to identify differential expressed genes between the two conditions. The Wald test was used within DESeq2, and all genes were subsequently ranked according to their Wald test statistic. To identify the biological processes associated with this ranked gene set, gene set enrichment analysis (GSEA) was employed using the fgsea package with 10000 permutations (package v1.8.0 (Korotkevich et al., 2019)), along with the following MSigDB 7.0 collections: KEGG and REACTOME curated gene sets, and Gene Ontology (GO) gene sets. The default two-sided enrichment p-value with Benjamini–Hochberg correction from the fgsea package was utilized.
Differential alternative splicing analysis
Alternative splicing events were analyzed using MAJIQ and VOILA with the default parameters (Vaquero-Garcia et al., 2016). Briefly, uniquely mapped, junction-spanning reads were used by MAJIQ to construct splice graphs for transcripts by using the Ensembl transcriptome annotation supplemented with de-novo detected junctions. Here, de-novo refers to junctions that were not in the Ensembl transcriptome database but had sufficient evidence in the RNA-Seq data (default: at least three reads mapping to at least two different start positions). The resulting gene splice graphs were analyzed for all identified local splice variations (LSVs), defined as splits in a splice graph to or from a given exon. Redundant LSVs were removed, and each junction in the remaining LSVs was quantified for its expected percent spliced in (PSI) value in the control and siTM7SF3 samples and expected change in PSI (ΔPSI) between WT and siTM7SF3 samples. PSI and ΔPSI correspond to the percent of isoforms and change in percent of isoforms, respectively, that use a specific junction compared to the other junctions in the given LSV. Junctions with P(ΔΨ >0.2) of >0.95, where ΔΨ is the relative change in the percent selected index (PSI, Ψ) for each junction involved in an LSV were selected and were utilized by VOILA to produce gene and LSV splice graphs as well as violin plots representing PSI and ΔPSI quantifications.
Analysis of motif enrichment around splice sites
To test for enrichment of sequence motifs around splice sites affected by siTM7SF3 we extracted sequences surrounding the 425 splice sites with significant differences between the RNAi and control condition as determined using MAJIQ/Voila from the human genome (hg19). Specifically, we selected the 50 nt in the exon that precedes the splice junction, the first 100 nt of the intron, the last 100 nt of the intron, and the first 50 nt of the downstream exon, separately for the alternative splice sites showing increased or decreased usage upon siTM7SF3. As background set for determining enrichment, we used the corresponding regions surrounding all exons with robust evidence for alternative splicing in RNAseq data. We used the findMotifs.pl tool of HOMER (Hypergeometric Optimization of Motif EnRichment (Heinz et al., 2010)) to test for enrichment of either the RNA binding protein motifs provided with HOMER (all.rna.motifs) or a separate compendium of motifs of RNA binding proteins (Giudice et al., 2016).
Quantification and statistical analysis
The intensity of bands in autoradiograms was determined by densitometry that was carried out on exposures within the linear range. Graphic analysis was performed with NIH image software. Results are presented as means ± SEM. For comparison between the two groups, a two-sample equal variance student t-test (two-tailed) was used when appropriate; p values less than 0.05 were considered significant. ∗, ∗∗, ∗∗∗, ∗∗∗∗ correspond to p < 0.05, 0.01, 0.001, and 0.0001 respectively.
Acknowledgments
Insightful discussions with Dr. Sanford Sampson (Bar Ilan University, Ramat Gan, Israel) are greatly acknowledged. YZ was supported by The Israel Science Foundation (776/13) and by the Samuil and Petr Polsky Prostate Cancer Research Fund (7218400101). ARM is supported by a UCSD/UCLA Diabetes Center Grant (P30 DK063491) and by the NIH/NIDDK (1R01DK123422-01). N.D. was supported in part by a Ruth L. Kirschstein Institutional National Research Service Award T32 GM008666 from the National Institute of General Medical Sciences.
Author contributions
R.I., Y.V., H.S-A., S.N-E., and A.Y. performed studies and analyzed data. M.M., N.D., A.R.M., and N.J.G.W. analyzed RNA-seq and splicing data. R.I., Y. S-T., E.E., and Y.Z. designed the studies. R.I. and Y.Z. wrote the manuscript, with input from all other authors.
Declaration of interests
All authors declare no conflicts of interest.
Inclusion and diversity
We support inclusive, diverse, and equitable conduct of research.
Published: November 18, 2022
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.isci.2022.105270.
Supplemental information
425 LSVs that occur on silencing of TM7SF3 in U2-OS are listed in this file.
Enrichment by location: Overview of motif enrichment by location and effects of siTM7SF3 on usage of the adjacent splice site. The number of motifs with significant enrichment after multiple testing correction (FDR adjusted p-value <0.05; Benjamini-Hochberg) is reported for each location relative to differentially spliced regions and separately for up- or downregulation of splice site usage on knock-down of TM7SF3. Homer enrichment sigFDR: Sequence motifs showing significant enrichment around splice sites affected by siTM7SF3 compared to the background set (cf. STAR Methods). The percent of test and background sequences containing the motif, the location relative to the splice site, the effect of siTM7SF3 on splicing behavior, the motif database used and the RNA binding protein predicted to bind the motif are reported.
Data lists of the biological processes involved is detailed in this Excel file.
Data lists of the biological processes involved is detailed in this Excel file.
Data and code availability
-
•
Data reported in this paper will be shared by the lead contact upon request.
-
•
This paper does not report original code.
-
•
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
References
- Alkalay E. The Sub-Nuclear Localization of RNA-Binding Proteins in KSHV-Infected Cells. Cells. 2020;9:1958. doi: 10.3390/cells9091958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anders S., Pyl P.T., Huber W. HTSeq--a Python framework to work with high-throughput sequencing data. Bioinformatics. 2015;31:166–169. doi: 10.1093/bioinformatics/btu638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Auboeuf D., Honig A., Berget S.M., O'Malley B.W. Coordinate regulation of transcription and splicing by steroid receptor coregulators. Science. 2002;298:416–419. doi: 10.1126/science.1073734. [DOI] [PubMed] [Google Scholar]
- Balaban C., Sztacho M., Blazikova M., Hozak P. The F-Actin-Binding MPRIP forms phase-separated condensates and associates with PI(4,5)P2 and active RNA polymerase II in the cell nucleus. Cells. 2021;10 doi: 10.3390/cells10040848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baralle F.E., Giudice J. Alternative splicing as a regulator of development and tissue identity. Nat. Rev. Mol. Cell Biol. 2017;18:437–451. doi: 10.1038/nrm.2017.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beck A., Isaac R., Lavelin I., Hart Y., Volberg T., Shatz-Azoulay H., Geiger B., Zick Y. An siRNA screen identifies transmembrane 7 superfamily member 3 (TM7SF3), a seven transmembrane orphan receptor, as an inhibitor of cytokine-induced death of pancreatic beta cells. Diabetologia. 2011;54:2845–2855. doi: 10.1007/s00125-011-2277-3. [DOI] [PubMed] [Google Scholar]
- Ben-Ari Fuchs S. GeneAnalytics: An integrative gene set analysis tool for next generation sequencing, RNAseq and microarray data. OMICS. 2016;20:139–151. doi: 10.1089/omi.2015.0168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boivin B., Vaniotis G., Allen B.G., Hébert T.E. G protein-coupled receptors in and on the cell nucleus: a new signaling paradigm? J. Recept. Signal Transduct. Res. 2008;28:15–28. doi: 10.1080/10799890801941889. [DOI] [PubMed] [Google Scholar]
- Branco A.F., Allen B.G. G protein-coupled receptor signaling in cardiac nuclear membranes. J. Cardiovasc. Pharmacol. 2015;65:101–109. doi: 10.1097/FJC.0000000000000196. [DOI] [PubMed] [Google Scholar]
- Burrows C., Abd Latip N., Lam S.J., Carpenter L., Sawicka K., Tzolovsky G., Gabra H., Bushell M., Glover D.M., Willis A.E., Blagden S.P. The RNA binding protein Larp1 regulates cell division, apoptosis and cell migration. Nucleic Acids Res. 2010;38:5542–5553. doi: 10.1093/nar/gkq294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campden R., Audet N., Hébert T.E. Nuclear G protein signaling: new tricks for old dogs. J. Cardiovasc. Pharmacol. 2015;65:110–122. doi: 10.1097/FJC.0000000000000198. [DOI] [PubMed] [Google Scholar]
- Castano E., Yildirim S., Fáberová V., Krausová A., Uličná L., Paprčková D., Sztacho M., Hozák P. Nuclear phosphoinositides-versatile regulators of genome functions. Cells. 2019;8:649. doi: 10.3390/cells8070649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chapman K.M., Powell H.M., Chaudhary J., Shelton J.M., Richardson J.A., Richardson T.E., Hamra F.K. Linking spermatid ribonucleic acid (RNA) binding protein and retrogene diversity to reproductive success. Mol. Cell. Proteomics. 2013;12:3221–3236. doi: 10.1074/mcp.M113.030585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen M., Manley J.L. Mechanisms of alternative splicing regulation: insights from molecular and genomics approaches. Nat. Rev. Mol. Cell Biol. 2009;10:741–754. doi: 10.1038/nrm2777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chuang T.W., Lee K.M., Tarn W.Y. Function and pathological implications of exon junction complex factor Y14. Biomolecules. 2015;5:343–355. doi: 10.3390/biom5020343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colwill K., Pawson T., Andrews B., Prasad J., Manley J.L., Bell J.C., Duncan P.I. The Clk/Sty protein kinase phosphorylates SR splicing factors and regulates their intranuclear distribution. EMBO J. 1996;15:265–275. [PMC free article] [PubMed] [Google Scholar]
- Congreve M., de Graaf C., Swain N.A., Tate C.G. Impact of GPCR structures on drug discovery. Cell. 2020;181:81–91. doi: 10.1016/j.cell.2020.03.003. [DOI] [PubMed] [Google Scholar]
- Dioufa N., Chatzistamou I., Farmaki E., Papavassiliou A.G., Kiaris H. p53 antagonizes the unfolded protein response and inhibits ground glass hepatocyte development during endoplasmic reticulum stress. Exp. Biol. Med. 2012;237:1173–1180. doi: 10.1258/ebm.2012.012140. [DOI] [PubMed] [Google Scholar]
- Dobin A., Davis C.A., Schlesinger F., Drenkow J., Zaleski C., Jha S., Batut P., Chaisson M., Gingeras T.R. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29:15–21. doi: 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fáberová V., Kalasová I., Krausová A., Hozák P. Super-resolution localisation of nuclear PI(4)P and identification of its interacting proteome. Cells. 2020;9:1191. doi: 10.3390/cells9051191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fei T., Chen Y., Xiao T., Li W., Cato L., Zhang P., Cotter M.B., Bowden M., Lis R.T., Zhao S.G., et al. Genome-wide CRISPR screen identifies HNRNPL as a prostate cancer dependency regulating RNA splicing. Proc. Natl. Acad. Sci. USA. 2017;114:E5207–E5215. doi: 10.1073/pnas.1617467114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng H., Jin Z., Liu K., Peng Y., Jiang S., Wang C., Hu J., Shen X., Qiu W., Cheng X., Zhao R. Identification and validation of critical alternative splicing events and splicing factors in gastric cancer progression. J. Cell Mol. Med. 2020;24:12667–12680. doi: 10.1111/jcmm.15835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fey E.G., Krochmalnic G., Penman S. The nonchromatin substructures of the nucleus: the ribonucleoprotein (RNP)-containing and RNP-depleted matrices analyzed by sequential fractionation and resinless section electron. J. Cell Biol. 1986;102:1654–1665. doi: 10.1083/jcb.102.5.1654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fiszbein A., Kornblihtt A.R. Alternative splicing switches: important players in cell differentiation. Bioessays. 2017;39:1600157. doi: 10.1002/bies.201600157. [DOI] [PubMed] [Google Scholar]
- Gerbino A., Procino G., Svelto M., Carmosino M. Role of lamin A/C gene mutations in the signaling defects leading to cardiomyopathies. Front. Physiol. 2018;9:1356. doi: 10.3389/fphys.2018.01356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giudice G., Sanchez-Cabo F., Torroja C., Lara-Pezzi E. ATtRACT-a database of RNA-binding proteins and associated motifs. Database. 2016;2016 doi: 10.1093/database/baw035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gobeil F., Fortier A., Zhu T., Bossolasco M., Leduc M., Grandbois M., Heveker N., Bkaily G., Chemtob S., Barbaz D. G-protein-coupled receptors signalling at the cell nucleus: an emerging paradigm. Can. J. Physiol. Pharmacol. 2006;84:287–297. doi: 10.1139/y05-127. [DOI] [PubMed] [Google Scholar]
- Gonzales M.L., Anderson R.A. Nuclear phosphoinositide kinases and inositol phospholipids. J. Cell. Biochem. 2006;97:252–260. doi: 10.1002/jcb.20655. [DOI] [PubMed] [Google Scholar]
- Han H., Braunschweig U., Gonatopoulos-Pournatzis T., Weatheritt R.J., Hirsch C.L., Ha K.C.H., Radovani E., Nabeel-Shah S., Sterne-Weiler T., Wang J., et al. Multilayered control of alternative splicing regulatory networks by transcription factors. Mol. Cell. 2017;65:539–553.e7. doi: 10.1016/j.molcel.2017.01.011. [DOI] [PubMed] [Google Scholar]
- Hasler D., Meduri R., Bąk M., Lehmann G., Heizinger L., Wang X., Li Z.T., Sement F.M., Bruckmann A., Dock-Bregeon A.C., et al. The alazami syndrome-associated protein LARP7 guides U6 small nuclear RNA modification and contributes to splicing robustness. Mol. Cell. 2020;77:1014–1031.e13. doi: 10.1016/j.molcel.2020.01.001. [DOI] [PubMed] [Google Scholar]
- Hassler J.R., Scheuner D.L., Wang S., Han J., Kodali V.K., Li P., Nguyen J., George J.S., Davis C., Wu S.P., et al. The IRE1alpha/XBP1s pathway is essential for the glucose response and protection of beta cells. PLoS Biol. 2015;13:e1002277. doi: 10.1371/journal.pbio.1002277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heinz S., Benner C., Spann N., Bertolino E., Lin Y.C., Laslo P., Cheng J.X., Murre C., Singh H., Glass C.K. Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol. Cell. 2010;38:576–589. doi: 10.1016/j.molcel.2010.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hennig S., Kong G., Mannen T., Sadowska A., Kobelke S., Blythe A., Knott G.J., Iyer K.S., Ho D., Newcombe E.A., et al. Prion-like domains in RNA binding proteins are essential for building subnuclear paraspeckles. J. Cell Biol. 2015;210:529–539. doi: 10.1083/jcb.201504117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heyd F., Lynch K.W. Degrade, move, regroup: signaling control of splicing proteins. Trends Biochem. Sci. 2011;36:397–404. doi: 10.1016/j.tibs.2011.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirose T., Yamazaki T., Nakagawa S. Molecular anatomy of the architectural NEAT1 noncoding RNA: the domains, interactors, and biogenesis pathway required to build phase-separated nuclear paraspeckles. Wiley Interdiscip. Rev. RNA. 2019;10:e1545. doi: 10.1002/wrna.1545. [DOI] [PubMed] [Google Scholar]
- Hochberg-Laufer H., Neufeld N., Brody Y., Nadav-Eliyahu S., Ben-Yishay R., Shav-Tal Y. Availability of splicing factors in the nucleoplasm can regulate the release of mRNA from the gene after transcription. PLoS Genet. 2019;15:e1008459. doi: 10.1371/journal.pgen.1008459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hung M.S., Avner P., Rogner U.C. Identification of the transcription factor ARNTL2 as a candidate gene for the type 1 diabetes locus Idd6. Hum. Mol. Genet. 2006;15:2732–2742. doi: 10.1093/hmg/ddl209. [DOI] [PubMed] [Google Scholar]
- Isaac C., Pollard J.W., Meier U.T. Intranuclear endoplasmic reticulum induced by Nopp140 mimics the nucleolar channel system of human endometrium. J. Cell Sci. 2001;114:4253–4264. doi: 10.1242/jcs.114.23.4253. [DOI] [PubMed] [Google Scholar]
- Isaac R., Boura-Halfon S., Gurevitch D., Shainskaya A., Levkovitz Y., Zick Y. Selective serotonin reuptake inhibitors (SSRIs) inhibit insulin secretion and action in pancreatic beta cells. J. Biol. Chem. 2013;288:5682–5693. doi: 10.1074/jbc.M112.408641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Isaac R., Goldstein I., Furth N., Zilber N., Streim S., Boura-Halfon S., Elhanany E., Rotter V., Oren M., Zick Y. TM7SF3, a novel p53-regulated homeostatic factor, attenuates cellular stress and the subsequent induction of the unfolded protein response. Cell Death Differ. 2017;24:132–143. doi: 10.1038/cdd.2016.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Juan-Mateu J., Villate O., Eizirik D.L. Mechanisms in endocrinology: alternative splicing: the new frontier in diabetes research. Eur. J. Endocrinol. 2016;174:R225–R238. doi: 10.1530/EJE-15-0916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jumper J., Evans R., Pritzel A., Green T., Figurnov M., Ronneberger O., Tunyasuvunakool K., Bates R., Žídek A., Potapenko A., et al. Highly accurate protein structure prediction with AlphaFold. Nature. 2021;596:583–589. doi: 10.1038/s41586-021-03819-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jurica M.S. Detailed close-ups and the big picture of spliceosomes. Curr. Opin. Struct. Biol. 2008;18:315–320. doi: 10.1016/j.sbi.2008.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korotkevich G., Sukhov V., Sergushichev A. Fast gene set enrichment analysis. bioRxiv. 2019 doi: 10.1101/060012. Preprint at. [DOI] [Google Scholar]
- Lagace T.A., Ridgway N.D. The rate-limiting enzyme in phosphatidylcholine synthesis regulates proliferation of the nucleoplasmic reticulum. Mol. Biol. Cell. 2005;16:1120–1130. doi: 10.1091/mbc.E04-10-0874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee D.K., Lança A.J., Cheng R., Nguyen T., Ji X.D., Gobeil F., Jr., Chemtob S., George S.R., O'Dowd B.F. Agonist-independent nuclear localization of the Apelin, angiotensin AT1, and bradykinin B2 receptors. J. Biol. Chem. 2004;279:7901–7908. doi: 10.1074/jbc.M306377200. [DOI] [PubMed] [Google Scholar]
- Lee S.W., Lee M.H., Park J.H., Kang S.H., Yoo H.M., Ka S.H., Oh Y.M., Jeon Y.J., Chung C.H. SUMOylation of hnRNP-K is required for p53-mediated cell-cycle arrest in response to DNA damage. EMBO J. 2012;31:4441–4452. doi: 10.1038/emboj.2012.293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lev Maor G., Yearim A., Ast G. The alternative role of DNA methylation in splicing regulation. Trends Genet. 2015;31:274–280. doi: 10.1016/j.tig.2015.03.002. [DOI] [PubMed] [Google Scholar]
- Lewis A.E., Sommer L., Arntzen M.Ø., Strahm Y., Morrice N.A., Divecha N., D'Santos C.S. Identification of nuclear phosphatidylinositol 4, 5-bisphosphate-interacting proteins by neomycin extraction. Mol. Cell. Proteomics. 2011;10 doi: 10.1074/mcp.M110.003376. M110.003376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lightowlers R.N., Chrzanowska-Lightowlers Z.M.A. Human pentatricopeptide proteins: only a few and what do they do? RNA Biol. 2013;10:1433–1438. doi: 10.4161/rna.24770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu L., Luo C., Luo Y., Chen L., Liu Y., Wang Y., Han J., Zhang Y., Wei N., Xie Z., et al. MRPL33 and its splicing regulator hnRNPK are required for mitochondria function and implicated in tumor progression. Oncogene. 2018;37:86–94. doi: 10.1038/onc.2017.314. [DOI] [PubMed] [Google Scholar]
- Love M.I., Huber W., Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15:550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet. Journal. 2011;17 doi: 10.14806/ej.17.1.200. [DOI] [Google Scholar]
- Mellman D.L., Gonzales M.L., Song C., Barlow C.A., Wang P., Kendziorski C., Anderson R.A. A PtdIns4, 5P2-regulated nuclear poly(A) polymerase controls expression of select mRNAs. Nature. 2008;451:1013–1017. doi: 10.1038/nature06666. [DOI] [PubMed] [Google Scholar]
- Naftelberg S., Schor I.E., Ast G., Kornblihtt A.R. Regulation of alternative splicing through coupling with transcription and chromatin structure. Annu. Rev. Biochem. 2015;84:165–198. doi: 10.1146/annurev-biochem-060614-034242. [DOI] [PubMed] [Google Scholar]
- Nilsen T.W., Graveley B.R. Expansion of the eukaryotic proteome by alternative splicing. Nature. 2010;463:457–463. doi: 10.1038/nature08909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niu Z., Jin W., Zhang L., Li X. Tumor suppressor RBM5 directly interacts with the DExD/H-box protein DHX15 and stimulates its helicase activity. FEBS Lett. 2012;586:977–983. doi: 10.1016/j.febslet.2012.02.052. [DOI] [PubMed] [Google Scholar]
- Paronetto M.P., Passacantilli I., Sette C. Alternative splicing and cell survival: from tissue homeostasis to disease. Cell Death Differ. 2016;23:1919–1929. doi: 10.1038/cdd.2016.91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Payne E.M., Bolli N., Rhodes J., Abdel-Wahab O.I., Levine R., Hedvat C.V., Stone R., Khanna-Gupta A., Sun H., Kanki J.P., et al. Ddx18 is essential for cell-cycle progression in zebrafish hematopoietic cells and is mutated in human AML. Blood. 2011;118:903–915. doi: 10.1182/blood-2010-11-318022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng Y., Yuan J., Zhang Z., Chang X. Cytoplasmic poly(A)-binding protein 1 (PABPC1) interacts with the RNA-binding protein hnRNPLL and thereby regulates immunoglobulin secretion in plasma cells. J. Biol. Chem. 2017;292:12285–12295. doi: 10.1074/jbc.M117.794834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rizk A., Paul G., Incardona P., Bugarski M., Mansouri M., Niemann A., Ziegler U., Berger P., Sbalzarini I.F. Segmentation and quantification of subcellular structures in fluorescence microscopy images using Squassh. Nat. Protoc. 2014;9:586–596. doi: 10.1038/nprot.2014.037. [DOI] [PubMed] [Google Scholar]
- Sakashita E., Endo H. SR and SR-related proteins redistribute to segregated fibrillar components of nucleoli in a response to DNA damage. Nucleus. 2010;1:367–380. doi: 10.4161/nucl.1.4.12683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selvanathan S.P., Graham G.T., Erkizan H.V., Dirksen U., Natarajan T.G., Dakic A., Yu S., Liu X., Paulsen M.T., Ljungman M.E., et al. Oncogenic fusion protein EWS-FLI1 is a network hub that regulates alternative splicing. Proc. Natl. Acad. Sci. USA. 2015;112:E1307–E1316. doi: 10.1073/pnas.1500536112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sobol M., Krausová A., Yildirim S., Kalasová I., Fáberová V., Vrkoslav V., Philimonenko V., Marášek P., Pastorek L., Čapek M., et al. Nuclear phosphatidylinositol 4, 5-bisphosphate islets contribute to efficient RNA polymerase II-dependent transcription. J. Cell Sci. 2018;131:jcs211094. doi: 10.1242/jcs.211094. [DOI] [PubMed] [Google Scholar]
- Soeno Y., Taya Y., Stasyk T., Huber L.A., Aoba T., Hüttenhofer A. Identification of novel ribonucleo-protein complexes from the brain-specific snoRNA MBII-52. RNA. 2010;16:1293–1300. doi: 10.1261/rna.2109710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spector D.L., Fu X.D., Maniatis T. Associations between distinct pre-mRNA splicing components and the cell nucleus. EMBO J. 1991;10:3467–3481. doi: 10.1002/j.1460-2075.1991.tb04911.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spector D.L., Lamond A.I. Nuclear speckles. Cold Spring Harb. Perspect. Biol. 2011;3:a000646. doi: 10.1101/cshperspect.a000646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szklarczyk D., Franceschini A., Wyder S., Forslund K., Heller D., Huerta-Cepas J., Simonovic M., Roth A., Santos A., Tsafou K.P., et al. STRING v10: protein-protein interaction networks, integrated over the tree of life. Nucleic Acids Res. 2015;43:D447–D452. doi: 10.1093/nar/gku1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sztacho M., Salovska B., Cervenka J., Balaban C., Hoboth P., Hozak P. Limited proteolysis-coupled mass spectrometry identifies phosphatidylinositol 4,5-bisphosphate effectors in human nuclear proteome. Cells. 2021;10:68. doi: 10.3390/cells10010068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang S.J., Shen H., An O., Hong H., Li J., Song Y., Han J., Tay D.J.T., Ng V.H.E., Bellido Molias F., et al. Cis- and trans-regulations of pre-mRNA splicing by RNA editing enzymes influence cancer development. Nat. Commun. 2020;11:799. doi: 10.1038/s41467-020-14621-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valdez B.C., Yang H., Hong E., Sequitin A.M. Genomic structure of newly identified paralogue of RNA helicase II/Gu: detection of pseudogenes and multiple alternatively spliced mRNAs. Gene. 2002;284:53–61. doi: 10.1016/s0378-1119(01)00888-5. [DOI] [PubMed] [Google Scholar]
- Vaquero-Garcia J., Barrera A., Gazzara M.R., González-Vallinas J., Lahens N.F., Hogenesch J.B., Lynch K.W., Barash Y. A new view of transcriptome complexity and regulation through the lens of local splicing variations. Elife. 2016;5:e11752. doi: 10.7554/eLife.11752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J.Z., Fu X., Fang Z., Liu H., Zong F.Y., Zhu H., Yu Y.F., Zhang X.Y., Wang S.F., Huang Y., Hui J. QKI-5 regulates the alternative splicing of cytoskeletal gene ADD3 in lung cancer. J. Mol. Cell Biol. 2021;13:347–360. doi: 10.1093/jmcb/mjaa063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warns J.A., Davie J.R., Dhasarathy A. Connecting the dots: chromatin and alternative splicing in EMT. Biochem. Cell. Biol. 2016;94:12–25. doi: 10.1139/bcb-2015-0053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao Z., Duan S., Hou D., Wang W., Wang G., Liu Y., Wen L., Wu M. B23 acts as a nucleolar stress sensor and promotes cell survival through its dynamic interaction with hnRNPU and hnRNPA1. Oncogene. 2010;29:1821–1834. doi: 10.1038/onc.2009.473. [DOI] [PubMed] [Google Scholar]
- Ye J., Beetz N., O'Keeffe S., Tapia J.C., Macpherson L., Chen W.V., Bassel-Duby R., Olson E.N., Maniatis T. hnRNP U protein is required for normal pre-mRNA splicing and postnatal heart development and function. Proc. Natl. Acad. Sci. USA. 2015;112:E3020–E3029. doi: 10.1073/pnas.1508461112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhai B., Villén J., Beausoleil S.A., Mintseris J., Gygi J.P. Phosphoproteome analysis of Drosophila melanogaster embryos. J. Proteome Res. 2008;7:1675–1682. doi: 10.1021/pr700696a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou L.T., Ye S.H., Yang H.X., Zhou Y.T., Zhao Q.H., Sun W.W., Gao M.M., Yi Y.H., Long Y.S. A novel role of fragile X mental retardation protein in pre-mRNA alternative splicing through RNA-binding protein 14. Neuroscience. 2017;349:64–75. doi: 10.1016/j.neuroscience.2017.02.044. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
425 LSVs that occur on silencing of TM7SF3 in U2-OS are listed in this file.
Enrichment by location: Overview of motif enrichment by location and effects of siTM7SF3 on usage of the adjacent splice site. The number of motifs with significant enrichment after multiple testing correction (FDR adjusted p-value <0.05; Benjamini-Hochberg) is reported for each location relative to differentially spliced regions and separately for up- or downregulation of splice site usage on knock-down of TM7SF3. Homer enrichment sigFDR: Sequence motifs showing significant enrichment around splice sites affected by siTM7SF3 compared to the background set (cf. STAR Methods). The percent of test and background sequences containing the motif, the location relative to the splice site, the effect of siTM7SF3 on splicing behavior, the motif database used and the RNA binding protein predicted to bind the motif are reported.
Data lists of the biological processes involved is detailed in this Excel file.
Data lists of the biological processes involved is detailed in this Excel file.
Data Availability Statement
-
•
Data reported in this paper will be shared by the lead contact upon request.
-
•
This paper does not report original code.
-
•
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.