Abstract
Novel zinc(II) coordination compounds with imidazole (Im) and 2-methylimidazole (2-MeIm) were prepared and characterized: [ZnX2(Im)2] (X = Cl (1a), Br (1b), I (1c)) and [ZnX2(2-MeIm)2] (X = Cl (2a), Br (2b), I (2c)). Coordination compounds 1a–c were prepared mechanochemically by neat grinding while 2a–c were prepared by solution synthesis. The complexes were characterized by FT-IR and NMR spectroscopy and by powder X-ray diffraction. Crystal and molecular structures were determined by the single crystal X-ray diffraction. The characteristic of all structures is a distorted tetrahedral coordination of zinc consisting of two halide atoms and two nitrogen atoms from the imidazole (or 2-methylimidazole) ligand. Molecules in 1a–c are interconnected by hydrogen bonds into 3D structures. Structures of 1b and 1c were found to have similar unit cells and similar crystal packing and hydrogen bonding. Introduction of the 2-methylimidazole substituent introduced disorder in the crystal structures of 2a–c. Because of the very small size of the crystals data were collected by synchrotron radiation. For the disordered 2a,2b and 2c fixed geometry was used in refining of the structures. Crystal structures of 2a–c are characterized by chains of molecules connected by hydrogen bonds of the type N–H⋅⋅⋅X, with weak π⋅⋅⋅π and van der Waals interactions between the chains. The QTAIM, RDG and NCI computational analysis of 1a and 2a–c confirmed the presence of weak attractive intermolecular interactions that can be attributed to weak N–H⋅⋅⋅X and van der Waals interactions.
Keywords: Zn(II) coordination compounds, Imidazole, 2-Methylimidazole, Crystal structure, Neat grinding, Intermolecular interactions, Computational analysis
Graphical abstract

Zn(II) coordination compounds; Imidazole; 2-Methylimidazole; Crystal structure; Neat grinding; Intermolecular interactions; Computational analysis.
1. Introduction
Zinc is one of the most important essential metals as it plays an essential role in the activity of many enzymes that catalyse cellular biochemical reactions [1]. If present in high concentrations Zn(II) inhibits growth of bacterial species like Escherichia coli, Staphylococcus aureus and Pseudomonas aeruginosa [2].
Imidazole (1,3-diaza-2,4-cyclopentadiene) belongs to the N-heterocyclic group of compounds having a five-membered ring. It is the basic core of some natural products such as histidine, purine, histamine and DNA based structures. The imidazole ring contains two nitrogen atoms with different properties. One nitrogen atom can be deprotonated and in such form can coordinate transition metal atoms. The protonated nitrogen atom usually participates in hydrogen bonding. Imidazole derivatives have significant biological applications and were reported to have antifungal, antibacterial, antitumor and anti-tuberculosis activity [3, 4]. There is a great deal of interest in the synthesis and characterization of essential metal coordination compounds with heterocyclic compounds, in particular imidazole derivatives. Zinc complexes can have antibacterial and anticancer activity, some display strong photoluminescence in the blue region or can be used in green chemistry as catalysts [5, 6, 7, 8, 9, 10].
Imidazole and its derivatives have been widely used as excellent candidates for targeted metal–organic frameworks (MOFs) in the domain of coordination chemistry [11, 12]. Zeolitic imidazolate frameworks (ZIFs) belong to a subclass of MOFs that have tetrahedrally coordinated divalent cations such as zinc. ZIFs have interesting properties since they have high porosity present in MOFs, and high thermal and chemical stability of zeolites. ZIF-8 is the most well-known and studied type of ZIF. It is composed of Zn(II) interlinked with the deprotonated 2-MeIm ligand resulting in large cavities [11, 13, 14].
Imidazole and its derivatives are also often used as structural models for metalloenzymes, especially in zinc-containing enzymes [15, 16]. Our group is interested in characterization of coordination compounds of essential metals and biologically relevant ligands [17, 18, 19]. We have recently shown that mechanochemical synthesis can be very fast even by hand grinding in a mortar [20]. We are also investigating metalloproteins, especially insulin where the Zn ion is coordinated by the N atoms from the histidine imidazole ring and halide ions. By increased bromide concentration in the crystallization solution of insulin we have obtained Zn ions coordinated by two histidine and two bromide ions [21]. Interestingly, up to date the only described structure in the literature with the coordination environment similar to the above mentioned one in insulin is that of ZnCl2 with imidazole [22, 23]. This work was initiated to obtain more crystallographic data on the Zn complexes with imidazole and halides Cl, Br and I. We were further interested in the influence of the 2-methyl substitution of the imidazole ring on the intermolecular interactions. No structural data of Zn(II) coordination compounds with 2-methylimidazole and halide ions have been published till now. There is however, a deposited structure of the chloride structure (CSD ID FELQEV, J. G. Malecki, private communication). When disorder was structurally found in these complexes it was of interest to characterize the intermolecular interactions also by computational methods.
Only a limited number of other methyl-substituted zinc(II) imidazole coordination compounds with halide ligands have been structurally characterized, such as dichlorobis(1-methylimidazole)zinc(II) [24], dibromobis(1-methylimidazole)zinc(II) and diiodobis(1-methylimidazole)zinc(II) [25]. However, these complexes are N-alkylated and therefore do not have N–H hydrogen bond donors as in our structures.
In this work we report synthesis, spectroscopic characterization (FT-IR and NMR) and crystal structures of zinc(II) coordination compounds with imidazole (Im) and 2-methylimidazole (2-MeIm): [ZnX2(Im)2] (X = Cl (1a), Br (1b), I (1c)) and [ZnX2(2-MeIm)2] (X = Cl (2a), Br (2b), I (2c)). The intermolecular bonding networks for 1a, 2a, 2b and 2c were also explored with periodic DFT calculations. The wave functions and electron densities obtained for the experimentally determined atom coordinates and cell parameters in the crystals were analyzed by reduced density gradient (RDG), noncovalent interaction (NCI) plots and QTAIM analysis.
2. Experimental
2.1. Reagents and techniques
All commercially available chemicals of reagent grade were used as received without further purification. The CHN-microanalysis was carried out on a PerkinElmer 2400 Series II CHNS analyzer in the Analytical Services Laboratory at the Ruđer Bošković Institute, Zagreb. The FT-IR spectra were measured on a PerkinElmer Spectrum Two spectrometer in the spectral range 4000–450 cm−1 as KBr pellets. NMR spectra were recorded on an NMR spectrometer Bruker Avance III HD 400 mHz/54 mm Ascend in DMSO as solvent. Powder X-ray diffraction (PXRD) was performed on a Malvern Panalytical Aeris diffractometer in the Bragg-Brentano geometry with CuKα radiation (λ = 1.54184 Å) at room temperature. The samples were placed on a silicon holder and the diffractograms were measured in the 2θ range 5–40° with a step size of 0.022° and 15.0 s per step. Powder X-ray diffraction data were collected and visualized using the HighScore Plus program [26].
2.2. Synthesis
2.2.1. [ZnCl2(Im)2] (1a)
After neat grinding of zinc(II) chloride (0.340 g, 2.5 mmol) and imidazole (0.340 g, 5 mmol) with a mortar and pestle for 2 min the mixture first became pasty and solidified after 5 min. Recrystallization from methanol gave colorless crystals of good quality for X-ray diffraction. C6H8Cl2N4Zn: calcd. C 26.45; H 2.96; N 20.57%; found: C 26.83; H 2.63; N 20.60%. IR (KBr; absorption bands): ν(N–H), 3305 m; ν(C–H), 3122 s; δ(N–H), 1541 m; ν(C–N), 1424 m, 1330 m; other bands, 1258 m, 1100 m, 1072 s, 952 m, 756 s, 739 w, 704 w, 651 s, 617 w (Supplementary Figure S1, Table S1).
2.2.2. [ZnBr2(Im)2] (1b)
The experimental procedure was the same as for 1a, but zinc(II) bromide (0.563 g, 2.5 mmol) was used. C6H8Br2N4Zn: calcd. C 19.94; H 2.23; N 15.51%; found: C 20.34; H 1.82; N 15.80%. IR (KBr; absorption bands): ν(N–H), 3350 m; ν(C–H), 3126 m; δ(N–H), 1541 s; ν(C–N), 1427 m, 1327 m; other bands, 1261 w, 1091 w, 1062 w, 757 m, 739 w, 706 m, 639 s (Supplementary Figure S2, Table S1).
2.2.3. [ZnI2(Im)2] (1c)
The experimental procedure was the same as for 1a, but zinc(II) iodide (0.798 g, 2.5 mmol) was used. C6H8I2N4Zn: calcd. C 15.83; H 1.77; N 12.29%; found: C 16.23; H 1.36; N 12.58%. IR (KBr; absorption bands): ν(N–H), 3342 m; ν(C–H), 3123 s; δ(N–H), 1540 m; ν(C–N), 1425 m, 1325 m; other bands, 1261 m, 1170 w, 1124 w, 1093 w, 1062 w, 759 w (Supplementary Figure S3, Table S1).
2.2.4. [ZnCl2(2-MeIm)2] (2a)
A solution of ZnCl2 (0.044 g, 0.323 mmol) in 12 mL of dry methanol and 2-methylimidazole (0.0527 g, 0.642 mmol) in 12 mL of dry methanol was maintained under reflux during 3 h. The mixture was filtered and evaporated to half of the initial volume. Slow evaporation of this solution yielded colorless crystals. C8H12Cl2N4Zn: calcd. C 31.95; H 4.02; N 18.65%; found: C 31.42; H 3.79; N 17.82%. IR (KBr; absorption bands): ν(N–H), 3216 s; ν(C–H), 3138 w, 3125 w; ν(CH3)as, 2966 w; ν(CH3)s 2925 w; ν(C–N), 1570 s; δ(N–H), 1153,1135 w; other bands, 1288 w, 1135 w, 1095 w, 749 w (Supplementary Figure S4, Table S1).
2.2.5. [ZnBr2(2-MeIm)2] (2b)
A solution of ZnBr2 (0.073 g, 0.323 mmol) in 12 mL of dry methanol and 2-methylimidazole (0.0527 g, 0.642 mmol) in 12 mL of dry methanol was maintained under reflux during 3 h. The mixture was filtered and evaporated to half of the initial volume. Slow evaporation of this solution yielded colorless crystals. C8H12Br2N4Zn: calcd. C 24.68; H 3.11; N 14.39%; found: C 25,20; H 2.85; N 14.36%. IR (KBr; absorption bands): ν(N–H), 3291s; ν(C–H), 3136 w, 3100 w; ν(CH3)as, 2965 w,ν(CH3)s 2918 w; ν(C–N), 1567 s; δ(N–H), 1152 w, 1134 w; other bands, 1426 w, 1092 w, 1016w, 686 w (Supplementary Figure S5, Table S1).
2.2.6. [ZnI2(2-MeIm)2] (2c)
A solution of ZnI2 (0.103 g, 0.323 mmol) in 12 mL of dry methanol and 2-methylimidazole (0.0527 g, 0.642 mmol) in 12 mL of dry methanol was maintained under reflux during 3 h. The mixture was filtered and evaporated to half of the initial volume. Slow evaporation of this solution yielded pale yellow crystals. C8H12I2N4Zn: calcd. C 19.88; H 2.05; N 11.59%; found: C 21.90; H 2.71; N 11.22%. IR (KBr; absorption bands): ν(N–H), 3293 s; ν(C–H), 3134 w, 3116 w; ν(CH3)as, 2962 w,ν(CH3)s 2924 w; ν(C–N), 1562 s; δ(N–H), 1152 w, 1134 w; other bands, 1424 w, 1270 w, 1092 w, 740 w (Supplementary Figure S6, Table S1).
2.3. X-ray structure determination
The single crystal X-ray diffraction data were collected by ω-scans on an Oxford Diffraction Xcalibur 3 CCD diffractometer with graphite-monochromator using MoKα radiation (λ = 0.71073 Å) at room temperature (RT) for 1bRT and 2aRT, and at 120 K for 1a. Suitable single crystal was selected and glued to a thin glass fibre. Data were also collected at the synchrotron Elettra, beamline XRD1, at 100 K by using λ of 0.7000 Å for 1b and for some very small crystals of 2a, 2b and 2c. These crystals were mounted on cryo-loops with some Paratone N oil. Data collection for 1c was performed on a XtaLAB Synergy-S diffractometer at 170 K, Data collection and reduction was performed using the CrysAlis software package [27] for all home data collection and reduction. The X-ray diffraction data were corrected for the Lorentz-polarization factor, and absorption effects by the multi-scan method. Synchrotron data were also reduced by the CrysAlis Red program. Crystal structures were solved and refined using the Olex2 program [28]. Structures 1a−c and 2a were solved by direct methods using SHELXS [29] and 2b,c by the SHELXT program [30] and refined by the full-matrix least-squares method based on F2 against all reflections using the SHELXL-97 program [31]. All non-hydrogen atoms were refined anisotropically in 1a–c and 2c. The C–H and N–H hydrogen atoms were positioned in calculated positions with Uiso(H) = 1.2Ueq(C, N) using the riding model with C–H = 0.95 Å. Most problematic were crystals of 2a–c. It was difficult to find single crystals because of twinning and they were very small in size so synchrotron data was collected. 2a and 2b were found to be severely disordered while 2c was less disordered. These structures could be refined only if the imidazole rings had fixed geometry. To obtain the most suitable geometry the Cambridge Structural Database (CSD) [32] was searched for Zn coordination compounds with N-bound 2-methylimidazole (with R < 0.05, no disorder, single-crystal X-ray structures, no errors, no polymers and no ions). There were 42 hits of such structures with 66 2-methylimidazole ligands. The mean values for interatomic distances were calculated and were used for refinement in the structures 2a–c (SHELXL DFIX command; see the corresponding cif files). Occupancies of the disordered ligands were calculated during refinement while the temperature factor was common for each ligand. Only the Zn and halide ions were refined anisotropically in 2a,b. Geometrical parameters were calculated using Olex2 and PLATON [33]. Drawings of the structures were prepared by ORTEP [34] and MERCURY [35].
Information on crystal data, data collection, and refinement are given in Table 1 (1a–c), Table 2 (2a–c) and Supplementary Table S2 for room temperature data of 1bRT and 2aRT; the data are not deposited nor discussed since the quality is not as good as the low temperature data. They are only given for comparison of the unit cells with the low temperature data.
Table 1.
Crystal data and details of crystal structure refinement for 1a–c.
| Compound | 1a | 1b | 1c |
|---|---|---|---|
| Empirical formula | C6H8Cl2N4Zn | C6H8Br2N4Zn | C6H8I2N4Zn |
| Mr | 272.43 | 361.35 | 455.33 |
| Crystal size/mm | 0.50 × 0.36 × 0.11 | 0.03 × 0.01 × 0.01 | 0.07 × 0.05 × 0.03 |
| T/K | 120(2) | 100(1) | 170(2) |
| Crystal system | monoclinic | monoclinic | monoclinic |
| Space group | P21/n | P21/c | P21/c |
| a/Å | 7.9247(3) | 7.8510(2) | 8.2052(4) |
| b/Å | 11.6605(4) | 17.3000(3) | 18.0988(8) |
| c/Å | 11.3916(5) | 8.2459(2) | 8.5262(4) |
| β/° | 105.435(4) | 98.211(2) | 99.061(5) |
| V/Å3 | 1014.69(7) | 1108.50(4) | 1250.38(10) |
| Z | 4 | 4 | 4 |
| Dcalc/g cm−3 | 1.783 | 2.165 | 2.419 |
| μ/mm−1 | 2.905 | 8.997 | 6.869 |
| F(0 0 0) | 544.0 | 688.0 | 832.0 |
| λ/Å | 0.71073 (MoKα) | 0.7000 (synchrotron) | 0.71073 (MoKα) |
| Θ range/° | 7.91–57.92 | 4.64–58.00 | 5.51–60.00 |
| h, k, l range | −10:10; −15:15; −14:12 | −10:10; −23:23; −11:10 | −11:11; −25:25; −11:11 |
| Reflections collected | 6390 | 15426 | 14007 |
| Independent reflections | 2389 | 3012 | 3571 |
| Observed reflections, I ≥ 2σ(I) | 1935 | 2800 | 2880 |
| Data/restraints/parameters | 2389/0/118 | 3012/0/118 | 3571/0/118 |
| Goodness of fit on F2, S | 0.954 | 1.015 | 0.962 |
| R, wR [I ≥ 2σ(I)] | 0.0227, 0.0494 | 0.0315, 0.0736 | 0.0292, 0.0585 |
| R, wR [all data] | 0.0321, 0.0510 | 0.0342, 0.0747 | 0.0405, 0.0619 |
| Max., min. electron density/e Å−3 | 0.38/−0.32 | 0.81/−1.06 | 0.73/−0.82 |
| CCDC no. | 2163899 | 2163904 | 2163903 |
Table 2.
Crystal data and details of crystal structure refinement for 2a–c.
| Compound | 2a | 2b | 2c |
|---|---|---|---|
| Empirical formula | C8H12Cl2N4Zn | C8H12Br2N4Zn | C8H12I2N4Zn |
| Mr | 300.50 | 389.41 | 483.39 |
| Crystal size/mm | 0.10 × 0.05 × 0.04 | 0.03 × 0.02 × 0.02 | 0.05 × 0.03 × 0.02 |
| T/K | 100(1) | 100(1) | 100(1) |
| Crystal system | monoclinic | monoclinic | monoclinic |
| Space group | P21/n | P21/c | P21/c |
| a/Å | 7.98540(10) | 22.6408(4) | 14.2210(4) |
| b/Å | 22.0564(2) | 8.14598(13) | 8.4840(2) |
| c/Å | 13.8626(2) | 22.5579(4) | 23.1938(6) |
| β/° | 101.0510(10) | 113.8298(19) | 94.674(3) |
| V/Å3 | 2396.33(5) | 3805.70(12) | 2789.05(14) |
| Z | 8 | 12 | 8 |
| Dcalc/g cm−3 | 1.666 | 2.039 | 2.302 |
| μ/mm−1 | 2.374 | 7.869 | 5.940 |
| F(0 0 0) | 1216.0 | 2256.0 | 1792.0 |
| λ/Å | 0.7000 (synchrotron) | 0.7000 (synchrotron) | 0.7000 (synchrotron) |
| Θ range/° | 3.464–52.998 | 3.576–53.998 | 3.47–55.998 |
| h, k, l range | −10:10; −28:28; −17:17 | −29:29: −10:10; −29:29 | −18:19, −11:11; −30:30 |
| Reflections collected | 32304 | 50409 | 11616 |
| Independent reflections | 5180 | 8406 | 11616 |
| Observed reflections, I ≥ 2σ(I) | 4949 | 7378 | 10113 |
| Data/restraints/parameters | 5180/84/226 | 8406/152/401 | 11616/50/272 |
| Goodness of fit on F2, S | 1.055 | 1.086 | 1.046 |
| R, wR [I ≥ 2σ(I)] | 0.0688, 0.1690 | 0.0826, 0.2090 | 0.0764, 0.2164 |
| R, wR [all data] | 0.0706, 0.1705 | 0.0910, 0.2171 | 0.0834, 0.2227 |
| Max., min. electron density/e Å−3 | 2.54/−1.80 | 3.58/−1.79 | 3.84/−2.23 |
| CCDC no. | 2163901 | 2163902 | 2163900 |
2.4. Computational methods
The initial atomic coordinates and crystal cell parameters for the periodic DFT calculations of 1a, 2a, 2b and 2c crystal structures were taken from our X-ray diffraction experiments. For the disordered structures, 2a, 2b and 2c, calculations were performed for several proposed structures. Calculations started with geometry optimization for all hydrogen atoms within the entire crystal cell of the crystal structures. The coordinates of all atoms heavier than hydrogen were fixed and all of the hydrogen atoms were geometry-optimized under periodic conditions using the CP2K software package [36, 37]. Periodic H atom geometry optimization of the crystal cell was performed with the PBO functional [38], GTH pseudopotential [39, 40], optimized DZVP-MOLOPT-GTH basis sets [41], 400 Ry cutoff for the plane wave grid and the Grimme D3 dispersion correction [42]. Single point calculation was performed with the same level of theory with 800 Ry cutoff for the plane wave grid and the Grimme D3 dispersion correction to obtain the wave functions and electron density of the periodic cells. The QTAIM analysis, reduced density gradient (RDG) and noncovalent interaction (NCI) plots [43] from the obtained periodic electron densities for the crystal structures were performed with CRITIC2 [44] and VMD [45] software packages. Hirshfeld surfaces were calculated by CrystalExplorer [46].
3. Results and discussion
3.1. Synthesis
Coordination compounds 1a–c were obtained by the reaction of ZnX2 and imidazole in the 1:2 ratio mechanochemically by neat grinding (NG) and liquid-assisted grinding (LAG; with methanol). NG and LAG gave the same diffraction patterns so the liquid was not needed in these reactions (Scheme 1). Single crystals were obtained by recrystallization from methanol solutions. From the powder diffraction patterns we saw only a few maxima in 1b and 1c that could not be interpreted by comparison with the calculated diffractograms i.e. only a very small amount of impurities that could not be analysed. M. Takaya [47] has recently published a new method of obtaining zinc(II) complexes at high yields by direct mixing of solid ligands and solid Zn(OH)2 or ZnO with or without the addition of small amount water. The author described the synthesis of the bis(imidazolato)zinc(II) complex, and its characterization by elemental analysis and FT-IR spectroscopy but not by the single-crystal X-ray diffraction. Here, zinc halides were used with success.
Scheme 1.
Synthesis of 1a–c and 2a–c.
Coordination compounds 2a–c were prepared by the solution-based method, by heating the reaction mixture under reflux. This method gave single-crystals suitable for the crystal structure determination of 2a and 2c after evaporation. However, 2b is hygroscopic so the solution had to be kept in a desiccator above CaCl2 to obtain crystals. NG and LAG methods were also tried but resulted in sticky, gummy products.
3.2. IR and NMR spectroscopy
The most important IR bands are given in Table S1 in the Supplementary. The N–H stretching vibrations in heterocyclic compounds occur in the region 3500−3000 cm−1 [48]. The IR spectra of complexes 1a–c show a medium-intensity and broad bands at 3305 and 3350 and 3342 cm−1, respectively. These bands can be assigned to ν(N–H), while the broadness is indicative of hydrogen bonding, being in accord with the crystal structures. The IR spectra of the complexes 2a–c exhibit broad bands at 3216, 3291 and 3293 cm−1, respectively which is attributed to the N–H stretching vibration from the 2-MeIm ligands [49, 50]. The other frequencies 3138, 3136 and 3134 cm−1 are attributed to C–H stretching of the heteroatomic ring. The C–N stretching modes shift to lower frequencies which is indication of the weakening of the C–N bond caused by coordination of the nitrogen atom of the imidazole ring to Zn.
In the Zn(II) coordination compounds where imidazole or 2-methylimidazole are involved in coordination of Zn the chemical shifts for all hydrogen atoms are shifted upfield in comparison with their free ligands. The increase of the π-density of the ring carbons is caused by the π-back-bonding from the metal d-orbitals into the ligand π-system, which results in an upfield change in the chemical shift of the ring protons. Table S3 and Supplementary Figures S7-S12 show 1H NMR chemical shifts of the free imidazole ligand [50, 51] and their coordinated zinc(II) complexes.
3.3. Crystal structures
The characteristic of all molecular structures is a distorted tetrahedron N2X2 (X = Cl, Br, I) consisting of two halide atoms and two nitrogen atoms from the imidazole (or 2-methylimidazole) ligand. Since our structure of 1a is at low temperature it will be presented here although the room temperature structure has been published previously [22, 23]. Comparison of the calculated and observed PXRD data is given in the Supplementary Figures S13 (1a–c) and S14 (2a–c).
3.3.1. Crystal structures of 1a, 1b and 1c
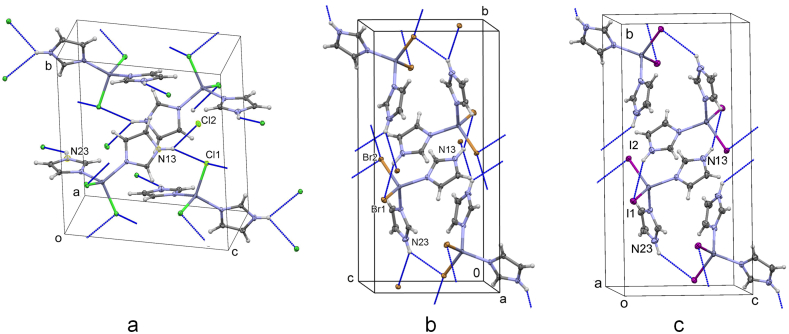
The major difference in the structures of 1a, 1b and 1c is in the orientation of the imidazole ligand, Figure 1. Overlay of the molecules shows an almost identical orientation of the ligands in 1b and 1c, being different from that in 1a. The unit cell parameters of 1b and 1c are very similar (Table 1) resulting in a very similar crystal packing and orientation of the hydrogen bonds. Selected bond distances are given in Table 3. The Zn–N distances are similar in all three compounds, whereas those of Zn–X increase with the size of the ligand. They are consistent with those in the related compounds [22, 23, 24, 25] and fall within the range of bond lengths in complexes of tetrahedrally coordinated Zn with two N-coordinated ligands and two halide atoms, found in the CSD [32]. There are 932 entries of such complexes having the Zn–N bond with a mean value of 2.09(8) Å (range 1.90–2.50 Å), 670 entries with the Zn–Cl bond (mean value 2.23(3) Å; range 2.13–2.44 Å), 165 entries with compounds having the Zn–Br bond (mean value 2.36(3) Å; range 2.28–2.48 Å) and 97 entries with the Zn–I bond (mean value 2.56(3) Å; range 2.50–2.71 Å). The two imidazole and two halide ligands form a distorted octahedron around zinc (Figure 1a,b). The angles N–Zn–N range from 105.66(6)° (in 1a) to 110.96(11)° (in 1c), N–Zn−X angles range from 105.28(4)° to 115.08(4)° (both in 1a), and X–Zn−X angles range from 110.87(2)° (in 1a) to 115.34(2)° (in 1c). Differences in the angles X–Zn−X are also consequence of the increasing ligand size from Cl to I.
Figure 1.
(a) Molecular structure of 1a with the atom labelling scheme. The ellipsoids are at the 40 % probability level. Hydrogen atoms are depicted as spheres of arbitrary radii. 1b and 1c have an equivalent atom labelling scheme as 1a. (b) Overlayed structures of 1a (green), 1b (brown) and 1c (violet).
Table 3.
Bond distances (Å) for the tetrahedral arrangement around the zinc atom in the coordination compounds 1a–c.
| 1a (X = Cl) | 1b (X = Br) | 1c (X = I) | |
|---|---|---|---|
| Zn–N11 | 2.0055(14) | 2.0038(17) | 2.003(3) |
| Zn–N21 | 1.9998(15) | 2.0070(16) | 2.013(2) |
| Zn–X1 | 2.2642(5) | 2.3985(3) | 2.5898(5) |
| Zn–X2 | 2.2455(4) | 2.4033(3) | 2.5956(4) |
1a was previously prepared by the solution-based method and structurally defined at room temperature [22, 23]. Molecules in 1a–c are interconnected by hydrogen bonds of the N–H⋅⋅⋅X type into a three-dimensional network (Figure 2). In 1a atom N13 is involved in a bifurcated hydrogen bond to Cl1 and Cl2 of 3.359(2) and 3.324(2) Å, respectively. N23 forms a hydrogen bond to Cl1 of 3.240(2) Å (Figure 2a). In 1b N13 is hydrogen bonded to Br1 at 3.478(2) Å, N23 to Br2 at 3.519(2) Å, while the hydrogen bond to another Br2 (3.634(2) Å) is of border value (Figure 2b). In 1c both I atoms accept only one hydrogen bond, N13–H⋅⋅⋅I1 of 3.703(3) and N23–H⋅⋅⋅I2 of 3.732(4) (Figure 2c). Hydrogen bond parameters are given in Table S4 in the Supplementary.
Figure 2.
Packing of (a) 1a, (b) 1b and (c) 1c in the unit cell. Hydrogen bonds are shown by blue lines.
3.3.2. Crystal structures of 2a, 2b and 2c
The deposited structure of 2a (J. G. Malecki, private communication to CSD) was never published. The unit cell differs from our 2a (CSD ID FELQEV, P21/c, a = 7.5332(7), b = 21.9542(14), c = 8.1101(6) Å), and the powder patterns are similar but not in agreement, Supplementary Figure S14.
The 2-MeIm ligands had fixed geometry during refinement of crystal structures. Therefore, for the structures of 2a–c the bonds and angles are not given and only the intermolecular interactions are discussed. The overall structures are very similar. The molecules are connected into chains by hydrogen bonds of the N–H⋅⋅⋅X type. The chains are further connected by π⋅⋅⋅π interactions between pairs of 2-MeIm rings from adjacent chains into layers, and finally, the weakest intermolecular interactions are between the layers. All intermolecular contacts that are given are between the ligands with major occupancies and are rounded to 2 decimals (there are no e.s.d.’s because the ligands had fixed geometry). There exist also contacts between minor components and also mixed ones since their appearance in the crystal is random.
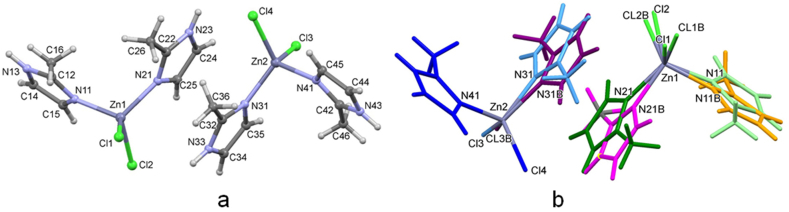
In 2a there are two independent molecules in the asymmetric unit (Figure 3a,b). Molecule 1 (Zn1) has both 2-MeIm ligands disordered, while in molecule 2 only one is disordered. The disorder is such that in each pair of disordered 2-MeIm ligands they are rotated approx. by 180° and slightly shifted. This type of disorder is found also in 2b. Halogen atoms are disordered as well.
Figure 3.
(a) Molecular structure of 2a with the atom labelling scheme. Only the major part of the two independent disordered molecules is given. (b) Disordered parts in the two independent molecules in 2a are shown in different colors. The major parts in molecule 1 (Zn1) are colored light green (2-MeIm ring N11) and dark green (2-MeIm ring N21), and in molecule 2 (Zn2) light blue (2-MeIm ring N31) and dark blue (2-MeIm ring N41). The minor parts are labelled B. The minor parts in molecule 1 are colored orange (2-MeIm ring N11B) and purple (2-MeIm ring N21B), and in molecule 2 violet (2-MeIm ring N31B).
Molecules 1 and 2 are interconnected by hydrogen bonds N–H⋅⋅⋅Cl type into chains parallel to [100], Figure 4a. Five hydrogen bonds are in the range 3.15–3.52 Å. There are only weak π⋅⋅⋅π interactions between the 2-MeIm ligands (centroid distance (ring N21)⋅⋅⋅(ring N31 from the neighbouring chain) of 3.61 Å) connecting neighbouring chains into layers parallel to (001). The two orientations of the 2-MeIm ligands (Figure 4b) have only a small influence on the size of the molecule along [001] where the chains are linked only by weak van der Waals intermolecular interactions thus contributing to the disorder.
Figure 4.
(a) Crystal packing of 2a along the c-axis; chains of hydrogen bonded molecules are parallel to [100]. (b) Packing along the a-axis. Disordered molecules of 2a are colored as in Figure 3. Hydrogen bonds are shown by light blue lines.
Crystal structure of 2b was the most complicated one to solve since there are three independent molecules in the asymmetric unit (Figure 5a–c). Molecules Zn1 and Zn3 have both ligands disordered while Zn2 has only one disordered Br atom.
Figure 5.
Three independent molecules in 2b: (a) molecule 1 (Zn1); (b) molecule 2 (Zn2) is shown with the atom labelling scheme; (c) molecule 3 (Zn3). Major disordered ligands are labelled A, and the minor ones B. Molecules 1 and 3 have similar numbering scheme as molecule 2 as indicated by the labelled N atoms (e.g. in molecule 1: N11A, C12A, N13A…; N21A, C22A, N23A…).
Two types of chains are formed by hydrogen bonds of the type N–H⋅⋅⋅Br, in one chain molecules 1 (Zn1) and 2 (Zn2) are linked (Figure 6a), while in the other only molecules 3 are hydrogen bonded (Figure 6b). Both chains are parallel to [010]. Four hydrogen bonds within molecules labelled A are in the range 3.50–3.62 Å. There are also two types of layers parallel to (100) formed by π⋅⋅⋅π interactions, one is built up of molecules 1 and 2, and the other of molecules 3 (Figure 6c; distance between centroids: (ring N51A)⋅⋅⋅(ring N51A from the neighbouring chain) 3.66 Å, (ring N11A)⋅⋅⋅(ring N41A from the neighbouring chain) 3.60 Å). They alternate along [100] and are connected only by weak van der Waals interactions.
Figure 6.
The major parts in the molecules of 2b. (a) Molecules 1 (labelled Zn1, in purple color) and 2 (labelled Zn2, in cyan) connected by hydrogen bonds into chains; (b) molecules 3 (Zn3, red-brown) connected by hydrogen bonding into chains. The chains are parallel to [010]. (c) Packing of the molecules in the unit cell along the b-axis.
In 2c there are two independent molecules in the asymmetric unit (Figure 7). This crystal structure is the least disordered. There was some small leftover electron density in the difference Fourier map but it could not be modelled.
Figure 7.
Molecular structure of 2c with the atom labelling scheme.
Molecules 1 and 2 are linked by hydrogen bonds of the type N–H⋅⋅⋅I into chains along [010], Figure 8a. Τhree hydrogen bonds are in the range 3.75–3.83 Å. Layers parallel to (100) are formed by π⋅⋅⋅π interactions between 2-MeIm ligands of the two molecules (Figure 8b, centroid distance (ring N21)⋅⋅⋅(ring N31 from the neighbouring chain) 3.83 Å). They alternate along [010] and are connected only by weak van der Waals interactions. In this structure the layers are shifted by approx. half of the molecule size along the c-axis. This is similar as in 2a but quite different from 2b, so there are no preferred interactions in this direction being another indicator of the weakness of intermolecular contacts between the layers.
Figure 8.
(a) Two chains (formed by hydrogen bonding) in the structure of 2c parallel to [010]. (b) Packing of the molecules in the unit cell along the b-axis. Interactions between the chains are shown. Hydrogen bonds are shown by blue lines.
3.4. Computation analysis
Intermolecular interactions for the crystal structure of 1a (Table 4 and Figure 9) and representative structures for disordered 2a–c were also explored with reduced gradient analysis (RDG), non-covalent interaction (NCI) plots and QTAIM descriptors. We were interested in computational analysis of the disordered structures 2a–c and 1a was calculated only for comparison. The obtained results for 2c are presented in Figures 10 and 11, and Table 5, and for 2a and 2b in Supplementary Figures S15 and S16, respectively. The RDG and NCI plots, developed by Johnson et al. [43], based on electron density and its derivative analysis, enable visualization of the domains involved in either attractive or repulsive intermolecular interactions. The “spikes” in the RDG plot and color-coded red-green-blue domains of the NCI plots are interpreted as the descriptors for non-bonding, weak attractive and strong attractive interactions between the molecules in solids, respectively [43, 52, 53]. The RDG and NCI plots were successfully applied regarding hydrogen bonds, electrostatic, stacking and van der Waals interactions, molecular aggregations and crystal packing [43, 52]. The RDG and NCI plots obtained from the periodic cell electron density for 1a unit are shown in Figure 9. The presence of the “spikes” above the value of −0.01 a.u. in RDG plot (Figure 9a) indicate the presence of only weak attractive intermolecular interactions. Attractive domains, represented in the NCI plot with green ellipsoids (Figure 9b), can be attributed to weak N–H⋯Cl contacts. The RDG and NCI plots obtained for the structure of 2c unit (which is the least disordered of complexes 2) are shown in Figure 10. The presence of the “spikes” above the value of −0.01 a.u. in the RDG plot (Figure 10a) indicate presence of only weak attractive intermolecular interactions. Attractive domains represented in the NCI plot with green ellipsoids (Figure 10b) can be attributed to weak N–H⋅⋅⋅I contacts. Broad volume between the 2-MeIm units (Figure 10b) suggests that the electron distribution in the bonding region involves several atoms and can be attributed to weak van der Waals interactions. Similar results were also obtained for the representative structures of disordered 2a and 2b, Supplementary Figures S15 and S16, respectively. Besides strong interactions between the ions, weak N–H⋅⋅⋅X and van der Waals interactions can be observed in these cases as well. The interactions between the chains are also of the same kind, see for example RDG and NCI plots obtained for 2c, Figure 11a,b.
Table 4.
QTAIM descriptorsa of the selected bond path (3, −1) critical points (BCP) for intermolecular interactions in the crystal structure of 1a. All values are in atomic units (a.u.).
| BCP | ρ(r) | ∇2ρ(r) | G(r) | V(r) | H(r) | |V(r)|/G(r) | r/Åb | |
|---|---|---|---|---|---|---|---|---|
| 1 | Zn⋯Cl1 | 0.0721 | 0.1719 | 0.0359 | −0.0288 | 0.0071 | 0.802 | – |
| 2 | Zn⋯Cl2 | 0.0748 | 0.2243 | 0.0381 | −0.0201 | 0.0180 | 0.528 | – |
| 3 | Zn⋯N11 | 0.0865 | 0.2412 | 0.0486 | −0.0369 | 0.0117 | 0.759 | – |
| 4 | Zn⋯N21 | 0.0875 | 0.1804 | 0.0495 | −0.0540 | −0.0044 | 1.091 | – |
| 5 | Cl1⋯H23 | 0.0188 | 0.0581 | 0.0038 | 0.0069 | 0.0107 | 1.816 | 2.342 |
| 6 | Cl1⋯H13 | 0.0131 | 0.0419 | 0.0021 | 0.0062 | 0.0084 | 2.952 | 2.521 |
| 7 | Cl2⋯H13 | 0.0117 | 0.0380 | 0.0017 | 0.0060 | 0.0078 | 3.529 | 2.592 |
ρ(r) - electron density, ∇2ρ(r) - Laplacian of the electron density, G(r) - kinetic energy density, V(r) - potential energy density, H(r) - energy density and |V(r)|/G(r) - ratio of the potential energy to the kinetic energy density.
The distance obtained with a geometry optimization of all of the hydrogen atoms within the entire crystal cell.
Figure 9.
(a) Reduced Density Gradient (RDG) plot showing the presence of noncovalent interactions and (b) selected non-covalent interaction (NCI) domains plot in structure of 1a. The reduced gradient isosurfaces at s = 0.5 a.u. are colored on a red-green-blue color scale according to the sign(λ2)ρ values ranging from −0.04 to 0.04 a.u. indicating non-bonding, weak attractive and strong attractive interactions, respectively.
Figure 10.
(a) Reduced Density Gradient (RDG) plot showing the presence of noncovalent interactions and (b) selected non-covalent interaction (NCI) domains plot in a representative structure for disordered 2c. The reduced gradient isosurfaces at s = 0.5 a.u. are colored on a red-green-blue color scale according to the sign(λ2)ρ values ranging from −0.04 to 0.04 a.u. indicating non-bonding, weak attractive and strong attractive interactions, respectively.
Figure 11.
(a) Reduced Density Gradient (RDG) plot showing the presence of selected noncovalent interactions and (b) selected non-covalent interaction (NCI) domains plot between the chains in a representative structure for disordered 2c. The reduced gradient isosurfaces at s = 0.5 a.u. are colored on a red-green-blue color scale according to the sign(λ2)ρ values ranging from −0.04 to 0.04 a.u. indicating non-bonding, weak attractive and strong attractive interactions, respectively.
Table 5.
QTAIM descriptorsa of the selected bond path (3, −1) critical points (BCP) for intermolecular interactions in a representative structure for 2c. All values are in atomic units (a.u.).
| BCP | ρ(r) | ∇2ρ(r) | G(r) | V(r) | H(r) | |V(r)|/G(r) | r/Å b | |
|---|---|---|---|---|---|---|---|---|
| 1 | Zn1⋯I1 | 0.0567 | 0.0735 | 0.0240 | −0.0297 | −0.0057 | 1.237 | – |
| 2 | Zn1⋯I2 | 0.0558 | 0.0435 | 0.0234 | −0.0359 | −0.0102 | 1.534 | – |
| 3 | Zn1⋯N11 | 0.0820 | 0.2957 | 0.0444 | −0.0149 | 0.0195 | 0.336 | – |
| 4 | Zn1⋯N21 | 0.0886 | 0.3054 | 0.0505 | −0.0247 | 0.0258 | 0.489 | – |
| 5 | Zn2⋯I3 | 0.0572 | 0.0545 | 0.0244 | −0.0352 | −0.0108 | 1.443 | – |
| 6 | Zn2⋯I4 | 0.0568 | 0.0815 | 0.0241 | −0.0278 | −0.0037 | 1.154 | – |
| 7 | Zn2⋯N31 | 0.0858 | 0.2212 | 0.0479 | −0.0406 | 0.0074 | 0.848 | – |
| 8 | Zn2⋯N41 | 0.0823 | 0.2969 | 0.0447 | −0.0152 | 0.0295 | 0.340 | – |
| 9 | I2⋯H43 | 0.0135 | 0.0286 | 0.0022 | 0.0027 | 0.0049 | 1.227 | 2.7722 |
| 10 | I3⋯H33 | 0.0116 | 0.0280 | 0.0017 | 0.0036 | 0.0053 | 2.118 | 2.8388 |
| 11 | I4⋯H13 | 0.0120 | 0.0248 | 0.0018 | 0.0026 | 0.0044 | 1.444 | 2.8302 |
ρ(r) - electron density, ∇2ρ(r) - Laplacian of the electron density, G(r) - kinetic energy density, V(r) - potential energy density, H(r) - energy density and |V(r)|/G(r) - ratio of the potential energy to the kinetic energy density.
The distance obtained with a geometry optimization of all of the hydrogen atoms within the entire crystal cell.
Characterization of the intermolecular interactions was also done by topological analysis of the electron density within Bader’s QTAIM analysis [54, 55] using the bond path (3, −1) critical point (BCP) properties. Within QTAIM, the bond strength can be estimated from the electron density value (ρ(r)) and also from the magnitude and sign of the energy density (H(r)). The difference between closed-shell and shared-shell bonding interactions can be determined from the Laplacian of the electron density (∇2ρ(r)) value, which reflects the degree of density concentration or depletion. The hydrogen-bond energy is correlated with the value of the potential energy density (V(r)) at the BCP [56]. The potential energy density to kinetic energy density ratio, (|V(r)|/G(r)), has a value lower than 1 for ionic, hydrogen-bond and van der Waals interactions, a value 1–2 for mixed character interactions and a value greater than 2 for covalent bonds [55, 56, 57]. The selected bond path (3, −1) critical points and their properties, associated with the bonding interactions in the 1a unit are given in Table 4. For BCPs 1 to 4, associated with the Zn⋅⋅⋅Cl and Zn⋅⋅⋅N interactions, the large electron density values, positive Laplacian and energy density values and negative potential energy density values suggest a strong closed-shell ionic bonded interaction. The BCPs 5 to 7, the N–H⋅⋅⋅Cl interactions, with small electron density and Laplacian values, and large potential energy density and kinetic energy density ratio can be classified as weak hydrogen bonds with mixed character interactions. Similar results were also obtained for the representative structure of disordered 2a, Table S6. The potential energy density and the kinetic energy density ratio obtained for BCPs associated with the Zn⋅⋅⋅Ι interactions for 2c, Table 5, suggests somewhat mixed character that can be attributed to larger polarizability of I− ions.
Hirshfeld surfaces were calculated for complexes 1a-c (Figure S17) and 2c (Figure S18). Histograms of the di - de distances, given in the Supplementary Figures S17 and S18, show similarity of 1b and 1c, as well as similarity of two symmetrically independent complexes in 2c.
4. Conclusion
Coordination compounds 1a–c were prepared by an easy and fast mechanochemical synthesis without use of solvents. The Zn atom is coordinated tetrahedrally by two Cl−, Br− or I− anions and two imidazole or 2-methylimidazole ligands. The ZnX2L2 tetrahedron is slightly distorted. In 1a–c the molecules are hydrogen bonded into 3D networks. Coordination compounds 2a,b have significantly disordered 2-MeIm ligands. The molecules in 2a–c are connected into 1D chains by hydrogen bonds of the N–H⋅⋅⋅X type. The chains are connected by π⋅⋅⋅π interactions between pairs of 2-MeIm rings into 2D layers. Weak van der Waals interactions connect the layers into 3D structures. The RDG and NCI analysis for 1a and 2a–c structures shows presence of only weak attractive N–H⋅⋅⋅X and van der Waals intermolecular interactions between the units and the chains. The QTAIM analysis for BCPs associated with N–H⋅⋅⋅X interactions suggests that these interactions can be classified as weak hydrogen bonds with mixed character.
Declarations
Author contribution statement
M. Tašner, D. Vušak, I. Kekez: Performed the experiments; Analyzed and interpreted the data; Wrote the paper.
A. Gabud: Performed the experiments; Analyzed and interpreted the data.
V. Pilepić, D. Mrvoš-Sermek, D. Matković-Čalogović: Conceived and designed the experiments; Performed the experiments; Analyzed and interpreted the data; Wrote the paper.
Funding statement
This work was supported by Croatian Science Foundation [IP-2014-09-4274], and by the University of Zagreb, Croatia [for years 2020, 2021].
Data availability statement
Data associated with this study has been deposited at Cambridge Structural Database under the accession number CCDC 2163899-2163904.
Declaration of interests statement
The authors declare no conflict of interest.
Additional information
CCDC 2163899-2163904 contain the supplementary crystallographic data for this paper. These data can be obtained free of charge via www.ccdc.cam.ac.uk/data_request/cif (or from the Cambridge Crystallographic Data Centre (CCDC), 12 Union Road, Cambridge CB2 1EZ, UK; fax: +44(0)1223-336033; email: deposit@ccdc.cam.ac.uk).
Supplementary content related to this article has been published online at https://doi.org/10.1016/j.heliyon.2022.e11100.
Acknowledgements
We are grateful to Maurizio Polentarutti, Alberto Cassetta and Nicola Demitri for valuable support during beam time at synchrotron Elettra, Italy and to the University of Zagreb Computing Centre (SRCE) for resources made available.
Contributor Information
Draginja Mrvoš-Sermek, Email: mrvos@chem.pmf.hr.
Dubravka Matković-Čalogović, Email: dubravka@chem.pmf.hr.
Appendix A. Supplementary data
The following is the supplementary data related to this article:
References
- 1.Krezel A., Maret W. The biological inorganic chemistry of zinc ions. Arch. Biochem. Biophys. 2016;611:3–19. doi: 10.1016/j.abb.2016.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Khan M.S., Siddiqui S.A., Siddiqui M.S.R.A., Goswami U., Srinivasan K.V., Khan M.I. Antibacterial activity of synthesized 2,4,5-trisubstituted imidazole derivatives. Chem. Biol. Drug Des. 2008;72:197–204. doi: 10.1111/j.1747-0285.2008.00691.x. [DOI] [PubMed] [Google Scholar]
- 3.Kerru N., Gummidi L., Maddila S., Gangu K.K., Jonnalagadda S.B. A review on recent advances in nitrogen-containing molecules and their biological applications. Molecules. 2020;25:1909. doi: 10.3390/molecules25081909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Serdaliyeva D., Nurgozhin T., Satbayeva E., Khayitova M., Seitaliyeva A., Ananyeva L. Review of pharmacological effects of imidazole derivatives. J. Clin. Med. Kaz. 2022;19:11–15. [Google Scholar]
- 5.Rehman S. Synthesis, characterization and antimicrobial studies of transition metal complexes of imidazole derivative. Bull. Chem. Soc. Ethiop. 2010;24(2):201–207. [Google Scholar]
- 6.Zhoo G., Lin H. Metal complexes with aromatic N-containing ligands as potential agents in cancer treatment. Curr. Med. Chem. 2005;5:137–147. doi: 10.2174/1568011053174873. [DOI] [PubMed] [Google Scholar]
- 7.Zhang H.-B., Zhang X.-F., Chai L.-Q., Tang L.-J., Zhang H.-S. Zinc(II) and Cadmium(II) complexes containing imidazole ring: structural, spectroscopic, antibacterial, DFT calculations and Hirshfeld surface analysis. Inorg. Chim. Acta. 2020;507 [Google Scholar]
- 8.Li C.-G., Chai Y.-M., Chai L.-Q., Xu L.-Y. Novel zinc (II) and nickel (II) complexes of a quinazoline-based ligand with an imidazole ring: synthesis, spectroscopic property, antibacterial activities, time-dependent density functional theory calculations and Hirshfeld surface analysis. Appl. Organomet. Chem. 2022;36 [Google Scholar]
- 9.Tong Y.P., Zheng S.L., Chen X.M. Structures, photoluminescence and theoretical studies of two ZnII complexes with substituted 2-(2-hydroxyphenyl)benzimidazoles. Eur. J. Inorg. Chem. 2005;18:3734–3741. [Google Scholar]
- 10.Nakatake D., Yokote Y., Matsushima Y., Yazaki R., Ohshima T. A highly stable but highly reactive zinc catalyst for transesterification supported by a bis(imidazole) ligand. Green Chem. 2016;18:1524–1530. [Google Scholar]
- 11.Chen S.S. The roles of imidazole ligands in coordination supramolecular systems. CrystEngComm. 2016;18:6543–6565. [Google Scholar]
- 12.Katsoulidis A.P., Antypov D., Whitehead G.F.S., Carrington E.J., Adams D.J., Berry N.G., Darling G.R., Dyer M.S., Rosseinsky M.J. Chemical control of structure and guest uptake by a conformationally mobile porous material. Nature. 2019;565:213–217. doi: 10.1038/s41586-018-0820-9. [DOI] [PubMed] [Google Scholar]
- 13.Huang X.C., Lin Y.Y., Zhang J.P., Chen X.M. Ligand-directed strategy for zeolite-type metal-organic frameworks: zinc(II) imidazolates with unusual zeolitic topologies. Angew. Chem. Int. Ed. 2006;45:1557–1559. doi: 10.1002/anie.200503778. [DOI] [PubMed] [Google Scholar]
- 14.Brekalo I., Yuan W., Mottillo C., Lu Y., Zhang Y., Casaban J., Holman K.T., James S.L., Duarte F., Williams P.A., Harris K.D.M., Friščić T. Manometric real-time studies of the mechanochemical synthesis of zeolitic imidazolate frameworks. Chem. Sci. 2020;11:2141–2147. doi: 10.1039/c9sc05514b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yang S.P., Tong Y.X., Zhu H.L., Cao H., Chen X.M., Ji L.N. Three transition metal complexes formed with tripodal polyimidazole ligands: synthesis, crystal structures and reactivity toward superoxide. Polyhedron. 2001;20:223–229. [Google Scholar]
- 16.Soayed A.A., Refaat H.M., Noor El-Din D.A. Metal complexes of moxifloxacin-imidazole mixed ligands: characterization and biological studies. Inorg. Chim. Acta. 2013;406:230–240. [Google Scholar]
- 17.Vušak D., Smrečki N., Prugovečki B., Đilović I., Kirasić I., Žilić D., Muratović S., Matković-Čalogović D. Cobalt, nickel and copper complexes with glycinamide: structural insights and magnetic properties. RSC Adv. 2019;9:21637–21645. doi: 10.1039/c9ra03693h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pejić J., Vušak D., Szalontai G., Prugovečki B., Mrvoš-Sermek D., Matković-Čalogović D., Sabolović J. Disorder at the Chiral Cα Center and room-temperature solid-state cis–trans isomerization; synthesis and structural characterization of copper(II) complexes with D-allo, L-isoleucine. Cryst. Growth Des. 2018;18:5138–5154. [Google Scholar]
- 19.Vušak D., Smrečki N., Muratović S., Žilić D., Prugovečki B., Matković-Čalogović D. Structural diversity in the coordination compounds of cobalt, nickel and copper with N-alkylglycinates: crystallographic and ESR study in the solid state. RSC Adv. 2021;11:23779–23790. doi: 10.1039/d1ra04219j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tašner M., Mrvoš-Sermek D., Hajdarpašić E., Matković-Čalogović D. Dinuclear copper(II) acetate complex with caffeine, a fast mechanochemical synthesis. Contrib., Sect. Nat., Math. Biotech. Sci. 2018;39 91‒101. [Google Scholar]
- 21.Matković-Čalogović D., Prugovečki B., Đilović I. Zn(II) coordination in human and bovine bromo- and iodo-insulin hexamers. Acta Crystallogr. 2012;A68:s249. [Google Scholar]
- 22.Lundberg B.K.S. The crystal structure of di-imidazole-zinc(II) dichloride. Acta Crystallogr. 1966;21:901–909. doi: 10.1107/s0365110x66004171. [DOI] [PubMed] [Google Scholar]
- 23.Mroué K.H., Power William P. High-field solid-state 67Zn NMR spectroscopy of several zinc-amino acid complexes. J. Phys. Chem. 2010;A114:324–335. doi: 10.1021/jp908325n. [DOI] [PubMed] [Google Scholar]
- 24.Musie G.T., Li X., Powell D.R. Dichlorobis(1-methylimidazole)zinc(II) Acta Crystallogr. 2004;E60:m471–m472. [Google Scholar]
- 25.Hobbollahi E., Veselkova B., List M., Redhammer G., Monkowius U. Synthesis, crystal structures and blue emission of zinc(II) halide complexes of 1-alkyl-imidazole and (–)-nicotine. Z. Naturforsch., B: Chem. Sci. 2016;71:1269–1277. [Google Scholar]
- 26.Degen T., Sadki M., Bron E., König U., Nénert G. The HighScore suite. Powder Diffr. 2014;29(S2):S13–S18. [Google Scholar]
- 27.CrysAlisPro Software System . Rigaku Oxford Diffraction; 2020. Version 1.171.41.92a. [Google Scholar]
- 28.Dolomanov O.V., Bourhis L.J., Gildea R.J., Howard J.A.K., Puschmann H. OLEX2: a complete structure solution, refinement and analysis program. J. Appl. Crystallogr. 2009;42:339–341. [Google Scholar]
- 29.Sheldrick G.M. A short history of SHELX. Acta Crystallogr., Sect. A: Found. Crystallogr. 2008;64:112–122. doi: 10.1107/S0108767307043930. [DOI] [PubMed] [Google Scholar]
- 30.Sheldrick G.M. SHELXT - integrated space-group and crystal-structure determination. Acta Crystallogr. C: Struct. Chem. 2015;71:3–8. doi: 10.1107/S2053273314026370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sheldrick G.M. Crystal structure refinement with SHELXL. Acta Crystallogr. C: Struct. Chem. 2015;71:3–8. doi: 10.1107/S2053229614024218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Groom C.R., Bruno I.J., Lightfoot M.P., Ward S.C. The Cambridge Structural Database. Acta Crystallogr. 2016;B72:171–179. doi: 10.1107/S2052520616003954. Version 2020.2.0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Spek A. Structure validation in chemical crystallography. Acta Crystallogr. Sect. D Biol. Crystallogr. 2009;65:148–155. doi: 10.1107/S090744490804362X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Farrugia L.J. ORTEP-3 for Windows - a version of ORTEP-III with a Graphical User Interface (GUI) J. Appl. Crystallogr. 1997;30:565–566. [Google Scholar]
- 35.Macrae C.F., Bruno I.J., Chisholm J.A., Edgington P.R., McCabe P., Pidcock E., Rodriguez-Monge L., Taylor R., van de Streek J., Wood P.A. Mercury CSD 2.0 - new features for the visualization and investigation of crystal structures. J. Appl. Crystallogr. 2008;41:466–470. [Google Scholar]
- 36.Kühne T.D., Iannuzzi M., Del Ben M., Rybkin V.V., Seewald P., Stein F., Laino T., Khaliullin R.Z., Schütt O., Schiffmann F., Golze D., Wilhelm J., Chulkov S., Bani-Hashemian M.H., Weber V., Borštnik U., Taillefumier M., Jakobovits A.S., Lazzaro A., Pabst H., Müller T., Schade R., Guidon M., Andermatt S., Holmberg N., Schenter G.K., Hehn A., Bussy A., Belleflamme F., Tabacchi G., Glöß A., Lass M., Bethune I., Mundy C.J., Plessl C., Watkins M., VandeVondele J., Krack M., Hutter J. CP2K: an electronic structure and molecular dynamics software package - Quickstep: efficient and accurate electronic structure calculations. J. Chem. Phys. 2020;152 doi: 10.1063/5.0007045. https://www.cp2k.org/ CP2K version 8.2. The CP2K developers group. 2021. [DOI] [PubMed] [Google Scholar]
- 37.Hutter J., Iannuzzi M., Schiffmann F., VandeVondele J. CP2K: atomistic simulations of condensed matter systems: cp2k Simulation Software. WIREs Comput. Mol. Sci. 2014;4:15–25. [Google Scholar]
- 38.Perdew J.P., Burke K., Ernzerhof M. Generalized gradient approximation made simple. Phys. Rev. Lett. 1996;77:3865–3868. doi: 10.1103/PhysRevLett.77.3865. [DOI] [PubMed] [Google Scholar]
- 39.Goedecker S., Teter M., Hutter J. Separable dual-space Gaussian pseudopotentials. Phys. Rev. B. 1996;54:1703–1710. doi: 10.1103/physrevb.54.1703. [DOI] [PubMed] [Google Scholar]
- 40.Hartwigsen C., Goedecker S., Hutter J. Relativistic separable dual-space Gaussian pseudopotentials from H to Rn. Phys. Rev. B. 1998;58:3641–3662. doi: 10.1103/physrevb.54.1703. [DOI] [PubMed] [Google Scholar]
- 41.VandeVondele J., Hutter J. Gaussian basis sets for accurate calculations on molecular systems in gas and condensed phases. J. Chem. Phys. 2007;127 doi: 10.1063/1.2770708. [DOI] [PubMed] [Google Scholar]
- 42.Grimme S., Antony J., Ehrlich S., Krieg H. A consistent and accurate ab initio parametrization of density functional dispersion correction (DFT-D) for the 94 elements H-Pu. J. Chem. Phys. 2010;132 doi: 10.1063/1.3382344. [DOI] [PubMed] [Google Scholar]
- 43.Johnson E.R., Keinan S., Mori-Sanchez P., Contreras-Garcia J., Cohen A.J., Yang W. Revealing noncovalent interactions. J. Am. Chem. Soc. 2010;132:6498–6506. doi: 10.1021/ja100936w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Otero-De-La-Roza A., Johnson E.R., Luaña V. Critic2: a program for real-space analysis of quantum chemical interactions in solids. Comput. Phys. Commun. 2014;185:1007–1018. [Google Scholar]
- 45.Humphrey W., Dalke A., Schulten K. VMD: visual molecular dynamics. J. Mol. Graph. 1996;14:33–38. doi: 10.1016/0263-7855(96)00018-5. VMD version 1.9.4 (2022) [DOI] [PubMed] [Google Scholar]
- 46.Spackman P.R., Turner M.J., McKinnon J.J., Wolff S.K., Grimwood D.J., Jayatilaka D., Spackman M.A. CrystalExplorer: a program for Hirshfeld surface analysis, visualization and quantitative analysis of molecular crystals. J. Appl. Crystallogr. 2021;54(3):1006–1011. doi: 10.1107/S1600576721002910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Takaya M. A New synthetic method of zinc(II) complexes based on mixing. Yakugaku Zasshi. 2005;125(10):829–832. doi: 10.1248/yakushi.125.829. [DOI] [PubMed] [Google Scholar]
- 48.Ramasamy R. Vibrational spectroscopic studies of imidazole. Armen. J. Phys. 2015;8(1):51–55. [Google Scholar]
- 49.Semerci F., Yesilel O.Z., Sahin E. Cu(II) and Zn(II)-pyridine-2,3-dicarboxylate complexes with 2-methylimidazole: syntheses, crystal structures, spectroscopic and thermal analyses. J. Inorg. Organomet. Polym. 2010;20:334–342. [Google Scholar]
- 50.Di Santo A., Pérez H., Echevernia G.A., Piro O.E., Iglesias R.A., Carbonio R.E., Altabef A.B., Gil D.M. Exploring weak intermolecular interactions in thiocyanate-bonded Zn(II) and Cd(II) complexes with methylimidazole: crystal structures, Hirshfeld surface analysis and luminescence properties. RCS Adv. 2018;8:23891–23902. doi: 10.1039/c8ra04452j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Pinto D.C.G.A., Santos C.M.M., Silva A.M.S. In: Recent Research Developments in Heterocyclic Chemistry. Pinho e Melo T.M.V.D., editor. Research Signpost; Kerala, India: 2007. Advanced NMR techniques for structural characterization of heterocyclic structures; pp. 397–475. [Google Scholar]
- 52.Otero-de-la-Roza A., Johnson E.R., Contreras-García J. Revealing non-covalent interactions in solids: NCI plots revisited. Phys. Chem. Chem. Phys. 2012;14:12165–12172. doi: 10.1039/c2cp41395g. [DOI] [PubMed] [Google Scholar]
- 53.Saleh G., Gatti C., Lo Presti L. Non-covalent interaction via the reduced density gradient: independent atom model vs experimental multipolar electron densities. Computational and Theoretical Chemistry. 2012;998:148–163. [Google Scholar]
- 54.Bader R.F.W. Oxford University Press; Oxford, New York: 1994. Atoms in Molecules: A Quantum Theory. [Google Scholar]
- 55.Popelier P.L.A. On the full topology of the Laplacian of the electron density. Coord. Chem. Rev. 2000;197:169–189. [Google Scholar]
- 56.Grabowski S.J. What is the covalency of hydrogen bonding? Chem. Rev. 2011;111:2597–2625. doi: 10.1021/cr800346f. [DOI] [PubMed] [Google Scholar]
- 57.Espinosa E., Alkorta I., Elguero J., Molins E. From weak to strong interactions: A comprehensive analysis of the topological and energetic properties of the electron density distribution involving X-H⋅⋅⋅F-Y systems. J. Chem. Phys. 2002;117:5529–5542. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data associated with this study has been deposited at Cambridge Structural Database under the accession number CCDC 2163899-2163904.