Abstract
Extracellular vesicles (EVs) have attracted enormous attention for their diagnostic and therapeutic potential. However, it has proven challenging to achieve the sensitivity to detect individual nanoscale EVs, the specificity to distinguish EV subpopulations, and a sufficient throughput to study EVs among an enormous background. To address this fundamental challenge, we developed a droplet-based optofluidic platform to quantify specific individual EV subpopulations at high throughput. The key innovation of our platform is parallelization of droplet generation, processing, and analysis to achieve a throughput (~20 million droplets/min) more than 100× greater than typical microfluidics. We demonstrate that the improvement in throughput enables EV quantification at a limit of detection = 9EVs/μL, a >100× improvement over gold standard methods. Additionally, we demonstrate the clinical potential of this system by detecting human EVs in complex media. Building on this work, we expect this technology will allow accurate quantification of rare EV subpopulations for broad biomedical applications.
Keywords: extracellular vesicles, droplet microfluidics, digital assays, diagnostics, microfluidics, parallelization
Graphical Abstract
Extracellular vesicles (EVs) are a diverse set of lipid bound nanomaterials,1 which can be found circulating in blood and carry various molecular cargo that can be representative of their cells of origin.2,3 EVs have been discovered to play an important role in intercellular communication4 and to have enormous potential as biomarkers for a wide range of biomedical applications.5–11 Because EVs are heterogeneous, there has been an effort to measure EVs with single particle resolution. This technological push is analogous to the development of single cell analysis that fueled biological discovery in the past decade.12–16 Many of the proposed techniques are based on imaging.17–20 While imaging has been successful in resolving individual EVs, it is fundamentally limited to a number of particles (<104) several orders of magnitude less than the quantity of EVs found in clinical samples (1010 EVs/mL, 108 EVs in 10 μL of plasma).21 Digital droplet techniques, where single EVs are loaded into droplets and either barcoded for downstream next generation sequencing (NGS) analysis or digital enzyme-linked immunosorbent assay (dELISA) based fluorescence detection, have been particularly successful in addressing this challenge.22–25 However, a single platform has been unable to achieve the sensitivity to detect individual EVs, the specificity to distinguish particular EV subsets based on surface protein expression, and achieve throughputs relevant to study rare EV subpopulations.
In this work, we developed a high throughput digital assay to quantify rare EV subpopulations based on their expression of surface proteins among a large background of EVs26–28 (Figure 1A). We call our approach droplet-based extracellular vesicle analysis (DEVA). DEVA’s ultrasensitivity and robustness to complex background comes primarily from two interrelated aspects of the technology. (a) The multiple proteins expressed on a targeted EV surface allows for specific capture and labeling on microbeads. Due to this specificity, <0.03% of our beads resulted in a false positive signal, 8–40× lower than typical dELISAs26–28 (Table S1). (b) At such a low false positive rate, we found that our limit of detection (LOD) was Poisson noise constrained until the bead number was increased to Nbead = O (106), which corresponded to Ndrop = O (107) to ensure a digital distribution of beads within droplets (Table S2). Accordingly, we developed a platform to measure beads at a high throughput (~20 million droplets/min) by parallelizing droplet generation, processing, and analysis, achieving a throughput >100× greater than typical in microfluidic systems using only accessible optics (<$1000) and soft-lithography fabrication. We evaluated our technology by first quantifying human neuron derived EVs spiked into PBS and achieved a LOD = 9 EVs/μL, a >100× improvement over standard single EV characterization methods.29 Moreover, we directly demonstrated the importance of the throughput of our system by showing that the LOD of DEVA improved with an increase in the number of beads up to 106 beads. Additionally, we evaluated the potential of this system for use in clinical samples by quantifying human neuron EVs spiked in complex media with a background of EVs matched to that of 2 μL of human blood and found an LOD = 11 EVs/μL.
Figure 1.
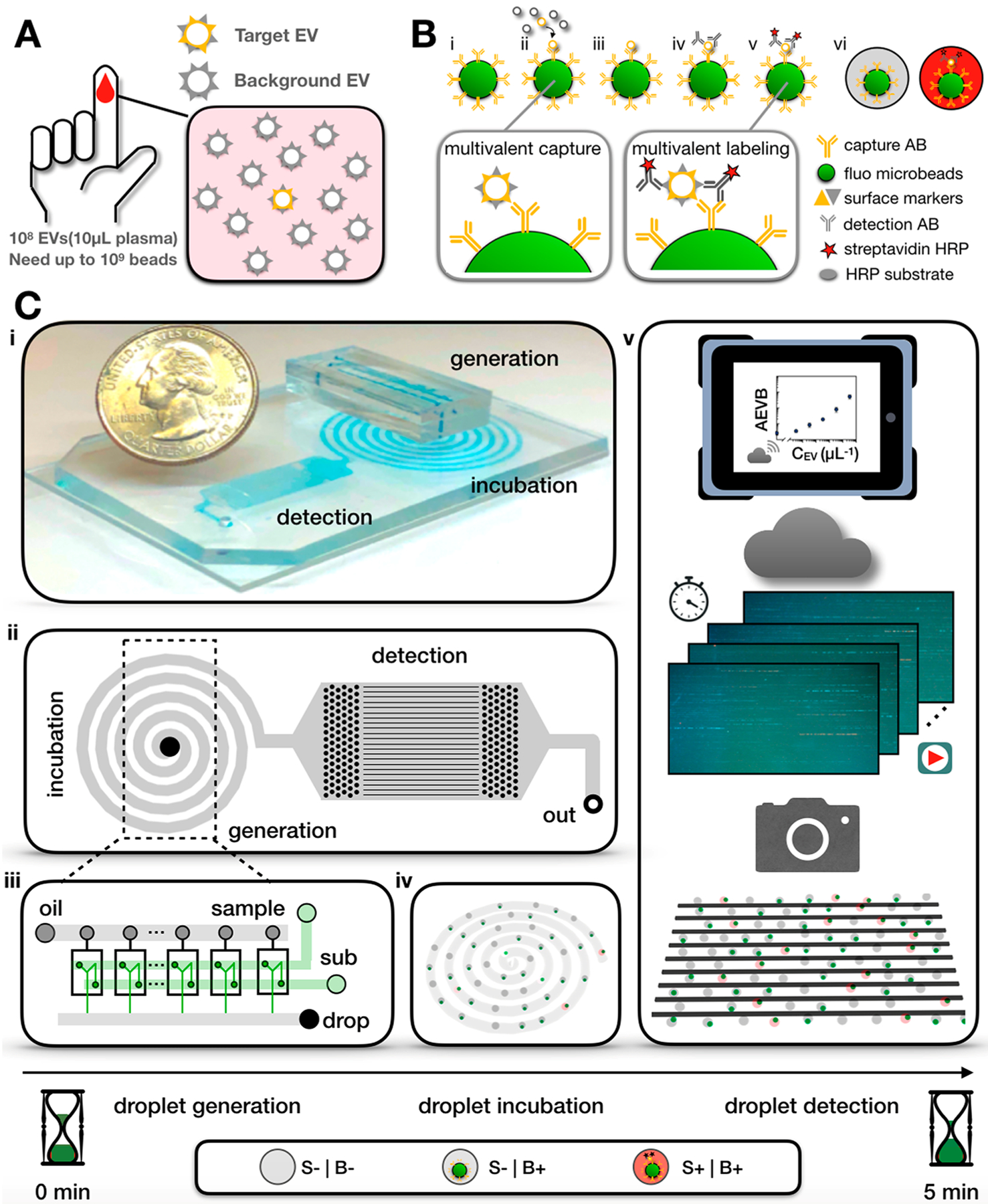
High-throughput DEVA. (A) Detecting specific EVs at single particle level is challenging, as there exists enormous EV background (108) in just 10 μL of human plasma. (B) Bead-based digital ELISA assay for single specific EV detection. (i) Paramagentic, fluorescent beads are coated with capture antibody. (ii) Antibody-coated beads are added to a sample containing target and background EVs. (iii) After bead–EV incubation, unbounded EVs are washed away. (iv) Biotinylated detection antibody is added to label the captured EVs. (v) Streptavidin–HRP is then added to bind to the biotinylated detection antibody and form an enzyme-labeled immunocomplex. (vi) The beads are mixed with HRP substrate and then partitioned into droplets. Droplets that contain an immunocomplex become fluorescent due to enzyme–substrate reaction. (C) (i) Miniaturized microfluidic platform for DEVA that integrates droplet generation, incubation, and detection. (ii) All droplets flow through a delay line chamber for incubation. Afterward, all droplets flow through a detection section consisting of 90 parallelized microfluidic channels. (iii) A parallelized flow focusing droplet generator encapsulates the beads into droplets. (iv) The EV–bead immunocomplex turns the droplet fluorescent during the incubation in the delay line. (v) A machine vision camera records the encoded fluorescent signal of 90 parallelized microfluidic channels and transfers the video to cloud computation for image analysis. S is short for substrate while B is short for beads. S+|B+: a droplet expresses signal from both substrate and bead. S−|B+: a droplet only contains bead signal. S−|B−: a droplet expresses neither a signal from substrate nor bead.
SINGLE EV DETECTION ON DEVA
Our DEVA assay uses fluorescent paramagnetic microbeads (d = 5.4 μm) functionalized with a capture antibody to target EV subpopulations based on their expression of a particular surface protein (Figure 1B). We label the beads with a fluorescence signal for two key reasons. First, it allows us to calculate the ratio average EV per bead (AEVB) accurately without having to make an assumption about the number of beads that have flowed through our system. Second, it provides a convenient means to multiplex our system, wherein the color of the microbead can indicate the capture antibody.30 The microbeads are first incubated with the sample to capture target EVs. After magnetic separation and washing, captured EVs are labeled with a biotinylated labeling antibody, followed by a streptavidin-HRP enzyme, to form an enzyme-linked immunocomplex. After washing to remove unbound antibodies and enzyme, the beads are mixed with a fluorescence enzyme substrate (ThermoFisher) and loaded into d = 20 μm aqueous droplets suspended in oil (Bio-Rad) using parallelized droplet generators (Figure 1C). The droplets are incubated for 5 min on-chip to allow the immunocomplexes to generate a fluorescent signal. A fluorescent droplet corresponds to a single EV that is positive for both the protein targeted by the capture antibody and the labeling antibody. Each droplet is inspected using our microdroplet megascale detector (μMD) platform in two fluorescence channels,30 one channel to determine whether the droplet contains a fluorescent bead and the other to interrogate if the droplet is fluorescent. The ratio of beads that have captured an EV versus the total number of beads is the AEVB. To ensure the assay is in the digital regime, that is, there is one or zero EVs per bead and one or zero beads per droplet, we use >10× as many beads as EVs and >10× as many droplets as beads.
We perform high throughput fluorescence detection using a digital camera by modulating the light sources in time with a pseudorandom sequence at a rate >10× the frame rate of the camera, encoding the droplet streak with a pattern that allows it to be resolved using correlation based detection among neighboring droplets.30 Downstream of the delay line, droplets flow through a detection region that consists of 90 parallelized microfluidic channels, where their fluorescence is measured using two time-domain modulated laser diodes31 and a machine vision camera (Grasshopper) (Figures 1C, S1). In this implementation, a blue laser and a green laser module (λexBlue = 457 nm, λexGreen = 528 nm, Techhood) were each modulated by an independent pseudorandom sequence, provided by a microcontroller (Arduino), to excite the fluorophores on microbeads and HRP substrate, respectively. Each pseudorandom sequence was an algorithmically generated 63-bit maximum length sequence (MLS), chosen to be minimally autocorrelated with itself and cross correlated with one another.31 Videos from the camera are processed by either a local computer or on the cloud.
THE BENEFIT OF HIGH THROUGHPUT DIGITAL EV DETECTION
We found that increasing the number of beads until Nbead = O (106) was a key factor in achieving the LOD and the dynamic range (DR) of our DEVA platform. The LOD of a digital ELISA is a function of both the background level AEVBb, the AEVB when there are no target EVs, and the standard deviation across independent measurements of the background level, such that . We were able to achieve a background level of AEVBb = 2.6 × 10−4, compared with that in typical digital ELISA (AEBb = [2 × 10−3, 2 × 10−2]) (Table S1).27,32 With such a low false positive rate, if we had used 25 000 beads, as is typical in digital assays, we would only measure Nb = 7 false positive events. With only seven false positive events, the of the background level from shot noise alone is , which would dominate the LOD if we assume the operational variance between experiments was σexp = 5%. Given the dependence of the variance of AEVB on the number of beads Nbead, the LOD can be improved by increasing the number of beads in an experiment until Poisson noise is no longer greater than the operational variance (Figure 2A). As conventional bead-based digital ELISA detects beads within wells on a statically micromachined chip, the total number of beads in an experiment is usually between several thousands to tens of thousands,22,26,32,33 which fundamentally sets the background level AVEBb to be ~0.2%, below which it would make no further improvement to the LOD. For our platform, we found that LOD could be improved by increasing beads up to Nbead = O (106), resulting in Ndrop = 20Nbead = 2 × 107 (Figure 2B). To process Ndrop = 2 × 107 at typical throughputs of flow cytometers or microfluidic systems ϕtyp = 103 droplet/sec, it would take approximately 5 h to process a single experiment.
Figure 2.
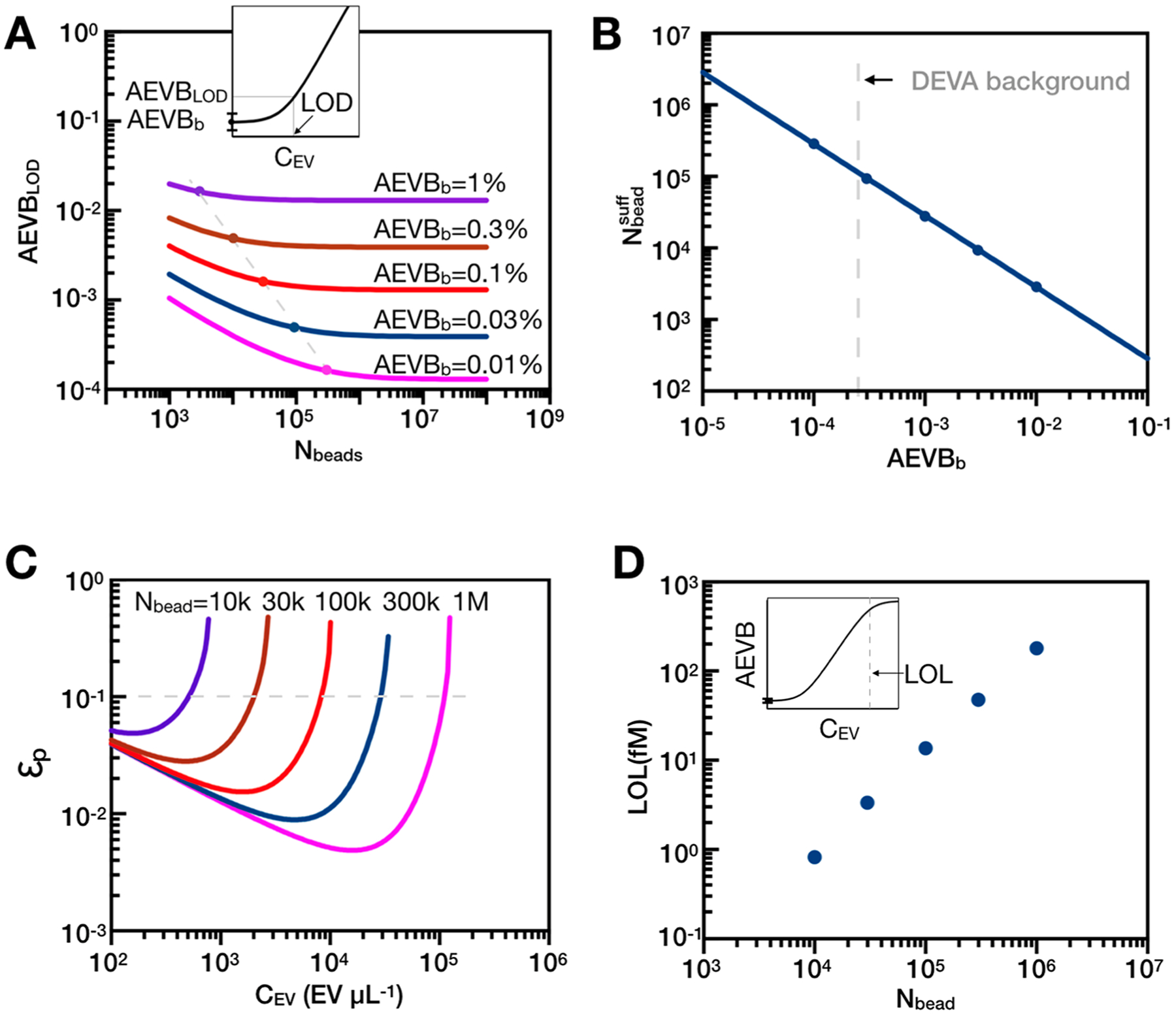
Theoretical analysis of the effect of using more beads in DEVA. (A) When background level is low, analyzing more beads is essential to minimize the imprecision coming from Poisson noise and to achieve a low limit of detection (LOD). Inset shows a schematic that defines LOD in a typical digital assay. LOD is calculated based on mean background level (AEVBb) plus three times the standard deviation. (B) Number of beads that can sufficiently minimize Poisson noise (1 dB) at different background levels. (C) Relative partition error (εp, the ratio of error over readout) will impair signal readout when there exists multiple target analytes per microbead (λ > 1). Different numbers of beads are indicated by different colors: purple, 10k; brown, 30k; red, 100k; navy, 300k; magenta, 1M. (D) More beads enable a higher limit of linearity (LOL) by keeping the partition error less than 10% of output signal. Upper left inset shows the schematic of limit of linearity at high EV concentrations.
In addition to improving LOD, increasing the bead number also improves the dynamic range (DR) of DEVA. The upper value of the DR of a digital assay is limited by partitioning error εp, which becomes significant when the number of EVs becomes similar to the number of beads Nbeads ~ NEVs, making it increasingly probable that there will be more than one EV bound to each positive bead (λ > 1, λ is the average number of EVs per bead). This partitioning error (εp) is modeled based on the binomial process34 and is expressed as , where Zc = 1.96 for 95% confidence interval and (Figure 2C). Incorporating more beads into our assay thus improves the LOL by keeping partitioning error low (εp < 10% of final readout) for increasing quantities of target EVs (Figure 2D). Leveraging our system’s high throughput to measure large numbers of beads, both the LOD and the LOL can be improved, resulting in an expanded dynamic range for target EVs. From a user’s perspective, if the number of EVs falls below the LOD or over the LOL, the system would report accordingly.
MICROFLUIDIC INTEGRATION OF OUR HIGH THROUGHPUT BEAD ANALYSIS
Building on our previous work on high throughput detection of droplets using time-domain-encoded optofluidics,30 we developed a high throughput single EV analysis platform by parallelizing droplet generation, processing, and analysis to achieve a throughput >100× greater than typical in microfluidic systems (Figure 1C). We updated our previous platform to be applied to DEVA in two main aspects. First, we used a digital camera, which uses a global shutter instead of rolling shutter,35 to prevent uncertainty in the phase of the time domain modulated fluorescent streaks across the image’s 90 parallelized microfluidic channels. Second, we optimized the image processing algorithms and applied GPU acceleration for image analysis. The resulting calculations for each experiment decreased from 1000 to 5 min.
The workflow of our detection approach is described below. A machine vision camera (Grasshopper3) with a macro lens (Computar) records the fluorescence signal from all 90 microfluidic channels for image analysis (Figure 3A,C). Each detection channel has a width of 40 μm and a height of 45 μm, and the entire detection region occupies a space of 15 mm× 9 mm. A multiband pass filter (λcwl = 485 ± 10 nm, 559.5 ± 12.5 nm; Semrock) is incorporated between the camera and the device to block scattered excitation light. The fluorescence signal from bead and substrate are chosen to have spectrally separated absorption spectra so they can be independently excited by our blue laser (λexBlue = 457 nm) and the green laser (λexGreen = 528 nm) (Figure S2), respectively. Each laser is modulated by a unique 63-bit MLS sequence (Figure 3B) so they can be resolved via correlation detection (Figure 3D).31,36 The velocity of the droplets and exposure time of our camera (45 ms) is chosen to generate a 63-bit MLS pattern in each droplet’s imaged streak length of 5 mm, which can be resolved by our optical system that has a resolution of 30 μm. A threshold is chosen to identify beads and positive droplets from the correlation signal of both the bead and the substrate fluorescence channel, by choosing a threshold that equals three standard deviations above the noise floor in the correlation signal. Additionally, we analyze the fluorescence signal of droplets with beads versus those without, and droplets with an enzyme versus those without, in Figure S3. We find that we can separate droplets with beads versus those without with an AUC = 1 (N = 41 droplets) and droplets with and without enzyme with an AUC = 1 (N = 118 droplets). Further details of our multidimensional correlation analysis were reported previously.30 An example of raw imaging data, and the corresponding correlation analysis, of a typical signal of a droplet containing a microbead that has and has not captured a single EV appear in Figure 3E. To characterize this approach, we spiked known quantities of fluorescent microbeads into our detection platform and quantified the number of measured beads (Figure S3) (R2 = 0.989). To match the throughput of our fluorescence detection, we integrated high throughput droplet generation (20 million droplets/min, d = 19.5 μm) by parallelizing 10 flow focusing droplet generators onto a single chip (Figure S4). We used a three-dimensional ladder geometry to uniformly distribute fluids to each droplet generator and collect each generator’s output.35
Figure 3.
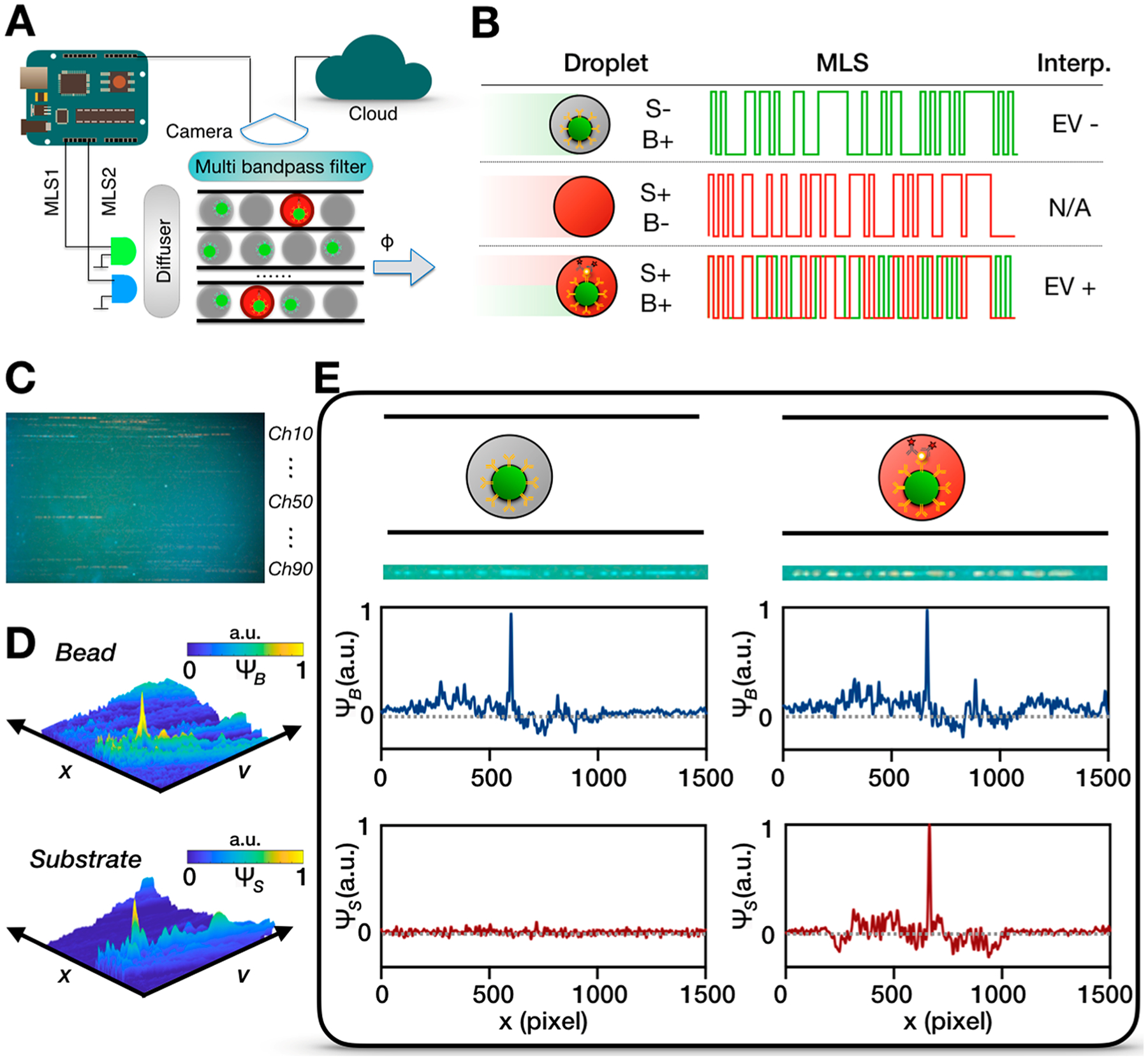
High throughput detection schematic for DEVA. (A) A microcontroller synchronizes the laser excitation and the camera recording. A diffuser is used to ensure uniform excitation across the chip. Multipass filters between the detection chip and the camera reduces background signal. The video is transmitted to the cloud that uses parallelized computation for image processing. (B) The fluorescent signal from the substrate and bead are encoded using different lasers. Truth table showing the interpretation on the fluorescent signal. S, substrate. B, bead. (C) Example of a frame recorded by the camera. (D) Decoding fluorescent signal of the bead and substrate channels, respectively. A plot of the correlation signal for bead or substrate as a function of position in the channel x and the velocity of the droplet v. (E) Example figures demonstrating that we can identify signal from a droplet that contains a bead but no EV (left) and a bead that has captured a single EV (right). Schematics and raw camera frames are shown respectively, followed by the correlation signal.
DEVELOPMENT AND CHARACTERIZATION OF ASSAY CONDITIONS FOR ULTRASENSITIVE SINGLE EV DETECTION
As a model system to evaluate DEVA, we used EVs isolated from a human neuronal cell line37 (SI). From this cell culture media, we isolated EVs using a commercial total-exosome isolation kit (ThermoFisher). We chose human neuron-derived EVs because neuronal EVs participate in neuron-glial communication, neuroinflammation, and propagation of pathogenic proteins such as amyloid-beta.38–40 These processes help create the neurodegenerative microenvironment of evolving traumatic brain injury pathology, along with other chronic neurologic disease pathologies.9 Moreover, as EVs can cross the blood-brain-barrier while remaining intact, they have great potential as biomarkers to monitor central nervous system injury and recovery.41
We characterized the EVs from this model system using scanning electron microscopy (SEM), nanoparticle tracking analysis (NTA), and Nanoview, which is a commercial platform for EV concentration, phenotype, and biomarker colocalization analysis. SEM demonstrated that, qualitatively, our antibody functionalized magnetic microbeads were capturing single EVs (Figures 4A, S5). NTA analysis was used to quantify the concentration of our cell culture derived EVs(1.4 × 109EV/mL) and their size distribution (d = 155 nm) (Figure 4B). We used Nanoview to determine which antibody to use to capture and to label EVs in our model system. With Nanoview, we quantified the heterogeneous protein expression on the EVs’ surface. In this assay, EVs are first captured on antibody coated chips, then labeled with antibodies (CD9/CD63/CD81) conjugated with different fluorophores (Figure 4C). By quantifying the fluorescent events in each channel, we found that CD81 showed significant higher expression than CD63 or CD9 on our human neuron derived EVs (p < 0.05, p < 0.0001), which was also significantly higher than the MIgG negative control (p < 0.0001). Hence, we chose to functionalize our beads with anti-CD81 as capture antibody in DEVA. After EV capture, we used 1% SDS to lyse the captured CD81+ EVs and compared their protein cargo to the total EV input. The result showed 11% (9.6–12.4%) of all EVs expressed at least one CD81 marker on their surface (Figure S6). Among CD81+ EVs, we found that 67% (64.3%–69.8%) of EVs coexpressed another copy of CD81 that could be labeled with a fluorophore-conjugated antibody. Consistent with the observation of others, we found heterogeneity among EVs’ surface protein “pan-EV” markers even though they were derived from the same cell line.18,42,43 We chose to use CD81 for capture and labeling, rather than neuron specific surface markers, because this protein could be characterized using established Nanoview assay kits, allowing us to quantify and benchmark the performance of DEVA. Improving the performance of DEVA assay required optimization in a multidimensional parameter space, including the concentration of several reagents and blocking conditions. For instance, we evaluated the concentration of the labeling antibody and HRP enzyme to balance trade-off between nonspecific and specific labeling (Figure S7). The concentration of labeling antibody (6.6 nM) and HRP (1 nM) achieved the current performance that allowed us to validate our technology, and we believe that even better performance may be possible by further optimization.
Figure 4.
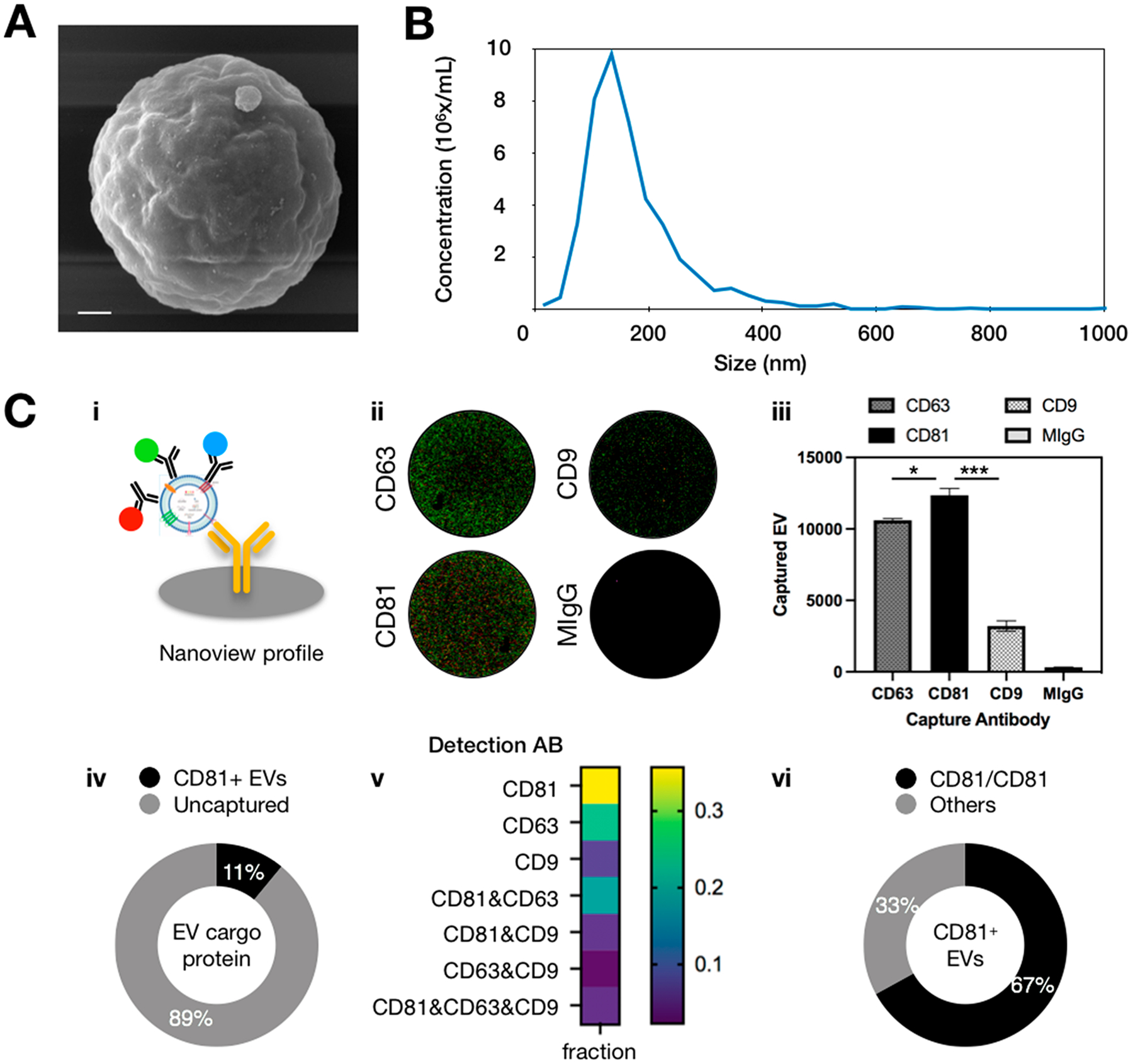
Characterization of human neuron EVs for DEVA. (A) SEM of an EV captured on an antibody-coated microbead. Scale bar: 300 nm. (B) NTA analysis of the size distribution and the concentration of human neuron derived EVs. (C) (i) Schematic of Nanoview analysis of EV surface proteins by immunocapture and immunolabeling. Red stands for CD63, green stands for CD81, blue stands for CD9. (ii) Fluorescent image recorded by Nanoview showing the surface protein profiling on captured EVs. (iii) Nanoview chip captured most human neuron EVs on the anti-CD81-coated chip. * indicates p < 0.05, *** indicates p < 0.001. (iv) Eleven percent of human neuron EVs express at least one CD81 protein based on immunoisolation and protein calibration. (v) Surface protein profiling of CD81+ EVs revealed by Nanoview. Each marker group represents a distinct EV subpopulation with no overlap. (vi) Among the CD81+ EVs, 67% of them expressed at least one other CD81 protein.
EVALUATION OF THE PERFORMANCE OF DEVA FOR SINGLE EV ANALYSIS
We initially evaluated the performance of DEVA by quantifying human neuron EVs spiked into PBS at known concentrations. We first evaluated the assay qualitatively by examining the droplets under a microscope (Leica) (Figures 5A, S8). Assays run with increasing quantities of EVs showed an increase in the number of red fluorescent droplets that contain beads, each of these instances indicating a single detected targeted EV. Subsequently, we quantitatively analyzed the performance of DEVA by measuring the response of our μMD system to a serial dilution of human neuron EVs from 0–105 EV/μL spiked into 100 μL PBS (Figure 5B). The background level, when no EVs were spiked, was AEVBb = 0.026%, which is 8–40 times lower compared with typical dELISA.28,33 We quantified the LOD of DEVA for detecting targeted EVs in PBS as LOD = 9EVs/μL. Within the dynamic range (DR, 9–5 × 105 EVs/μL), DEVA showed good linearity (Figure 5C, R2 = 0.9976).
Figure 5.
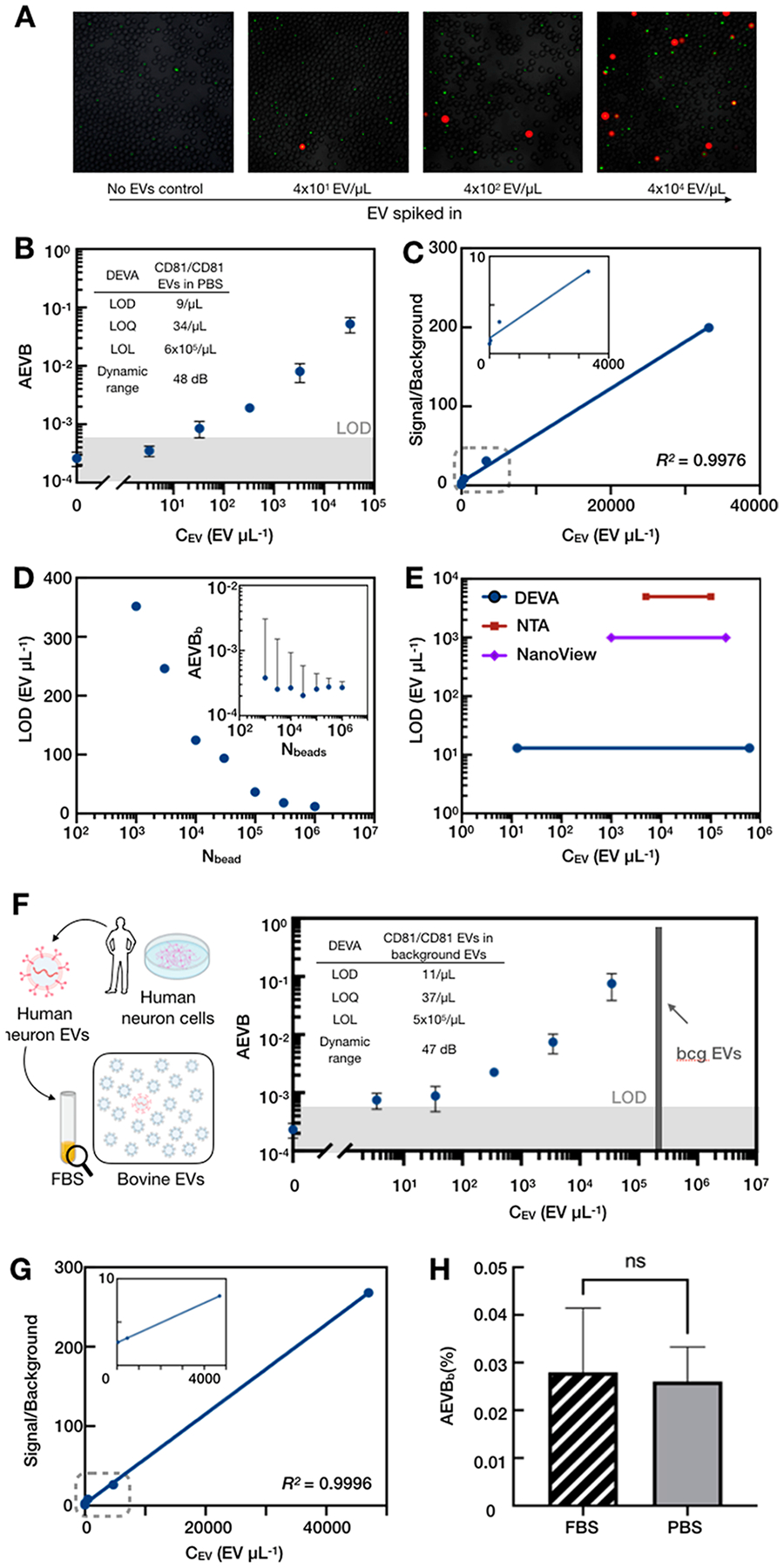
Benchmarking and characterization of DEVA. (A) Fluorescent microscopic images for DEVA assay on serial diluted human neuron EVs. (B) Calibration curve for DEVA detecting CD81/CD81 human neuron EVs spiked into PBS. Inset shows the detailed characterization. Error bars were calculated from experimental replicates (N = 3). (C) Signal over background for the calibration curve in A. Inset shows the zoomed-in view of signal-overbackground (y-axis) from 0 to 4000 EVs (x-axis). (D) Analyzing more beads improves DEVA’s LOD by minimizing the background imprecision coming from Poisson noise. Inset shows that increasing the number of beads analyzed decreases the variance of the AEVBb measurement. (E) Comparing the sensitivity and the dynamic range of DEVA, NTA, and Nanoview. The LOL of DEVA was calculated based on the number of beads quantified. (F) Calibration curve for DEVA detecting CD81/CD81 human neuron EVs with background (bcg) EVs (background EV concentration in final sample volume is indicated by gray bar). Inset shows detailed characterization. Error bars were calculated from experimental replicates (N = 3). (G) Signal over background for the calibration curve for DEVA in complex media. Inset shows the zoomed-in view from 0 to 5000 EVs/μL. (H) DEVA shows no significant difference (student t test) on the background level (AEVBb) in PBS or complex media.
We directly evaluated the role of the number of beads used in a DEVA assay to reduce the Poisson noise in the measurement and thus improve the LOD. Using the same assay conditions, we calculated the AEVBb by analyzing 103 to 106 beads. As expected, analyzing more beads decreased the standard deviation of the measurement of the response to a blank input AEVBb, which decreased the LOD of DEVA (Figure 5D). We also compared the performance of DEVA with conventional EV characterization platforms NTA and Nanoview, using aliquots of the same samples processed by our μMD (Figure S9). NTA is a common tool for EV quantification and surface protein analysis.29 We found, consistent with the literature, the LOD for NTA is ~O (104 EV/μL) with DR (104–2 × 105 EV/μL). Nanoview provides a multiplexed surface protein profiling of individual EVs. We quantified the LOD of Nanoview to be ~O (103 EV/μL) and the DR to be 2.3 log range, similar to their commercial characterization(2.8 log). In comparison to these technologies, DEVA showed a 100× better LOD and a 200× better DR for single EV characterization (Figure 5E).
CHARACTERIZATION OF BACKGROUND INVARIANCE OF DEVA
We evaluated the performance of DEVA to quantify rare EVs in the presence of complex media. To this end, we spiked human neuron EVs into fetal bovine serum (FBS) that contained 2 × 107 bovine EVs as background (Figure S10), similar to the number of human EVs present in a 2 μL blood from a typical finger prick. We chose FBS because it models a complex background but does not contain any EVs positive for hum-CD81.24,30 We analyzed the performance of DEVA by measuring the response of our μMD system to a serial dilution of human neuron EVs from 0–105 EV/μL spiked into 100 μL FBS (Figure 5F). The LOD of DEVA in complex media is 11EVs/μL. DEVA demonstrated good linearity within the dynamic range (Figure 5G, R2 = 0.9996). We found that the bovine EV background did not have a significant effect on the LOD of DEVA compared to measurements in PBS (Figure 5H), demonstrating the potential of this technology to be applied to detecting rare EVs in clinical samples. Additionally, we demonstrated the feasibility of quantifying endogenous EVs in human plasma, using isotope antibodies for capture and labeling as a negative control (Figure S11).
DISCUSSION
Our DEVA platform with its high sensitivity (LOD = 9 EVs/μL) and its high droplet throughput (20 M droplets/min) makes it possible to quantify sparse EV subpopulations in complex media. The key to our device’s sensitivity is its processing of tens of millions of droplets, and its capability to scale its processing rate only at a cost in computation. While in this first demonstration we performed a single-plex assay to quantify EVs based on CD81/CD81 expression, we can combine our droplet throughput and a multicolor detection approach to analyze multiple EV subpopulations simultaneously. Multiplexing can be increased by running assays in parallel, using microbeads barcoded with distinct ratios of concentrations of multiple dyes, as has been done by Luminex for nondigital ELISA.44 Alternatively, or in combination, the sample can be divided to be mixed with different panels of microbeads in individual sets of channels of the n = 90 detection channels. By combining these two approaches, it is possible for DEVA to achieve >100 multiplexed EV assays. Recently, a growing number of studies has shown the clinical value of measuring <8 of EV subpopulations.5,7,45–49 In prior work, we describe a duplex protein assay as a starting point.30 In addition to EVs, our approach can be applied to quantify other nanoscale objects, such as mitochondria to study their dysfunction in Alzheimer’s disease50 or HIV virus to study transfection mechanisms.51 Additionally, we are encouraged by recent advances in digital assays, which either obviate the need for droplets by replacing enzymatic amplification with rolling circle amplification directly on beads or obviate the need for microfluidics by using beads that template droplets.28,52 These techniques, incorporated with DEVA, could provide a further 10× improvement in throughput and allow a simpler implementation, which can be leveraged to increase multiplexing, further improve sensitivity, and allow point-of-care use. In addition to improving the LOD, the throughput of our system can be leveraged to increase the LOL by reducing partitioning error at high concentrations of EVs, resulting in an increased dynamic range. By applying high throughput digital droplet detection to single EV analysis, the μMD allows ultrasensitive, multiplexed EV detection in complex media, opening a broad range of new possibilities for clinical diagnostics and biological inquiry.
Supplementary Material
Funding
This work was supported by a grant from the Paul G. Allen Frontiers group (Grant 12347), the New Jersey Commission for Brain Injury Research, Grant W81XWH1920002 from the Department of Defense, grant RM1 HG010023 from the National Institutes of Health, and Grant 5R21CA236653 from the National Cancer Institute. Y.A. is supported by a National Defense Science and Engineering Graduate Fellowship from the Department of Defense.
Footnotes
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/acs.nanolett.2c00274.
Materials and methods for microfluidic device fabrication, optical setup, human neuron cell culture, preparation of antibody coated magnetic microbeads, detection antibody biotinylation, video processing and analysis, fabrication of 10-channel droplet microfluidics device, calculation of partitioning error; comparison between DEVA and typical commercial or research-based digital ELISA platforms, comparison between DEVA and typical commercial or research-based single EV analysis platforms, photograph of experimental setup, laser excitation characterization, DEVA fluorescence characterization, demonstration of parallelized droplet generators, SEM images of beads capturing individual EVs, protein quantification from isolated EVs, optimization of reagent concentrations for DEVA, microscopic images of fluorescent droplets, human neuron EV characterization using conventional tools, NTA analysis on isolated EVs, characterization of human plasma EVs on DEVA (PDF)
Complete contact information is available at: https://pubs.acs.org/10.1021/acs.nanolett.2c00274
The authors declare the following competing financial interest(s): David Issadore is a founder and holds shares in the startup company Chip Diagnostics.
Contributor Information
Zijian Yang, Department of Mechanical Engineering and Applied Mechanics, School of Engineering and Applied Science, University of Pennsylvania, Philadelphia, Pennsylvania 19104, United States.
Yasemin Atiyas, Department of Bioengineering, School of Engineering and Applied Science, University of Pennsylvania, Philadelphia, Pennsylvania 19104, United States.
Hanfei Shen, Department of Bioengineering, School of Engineering and Applied Science, University of Pennsylvania, Philadelphia, Pennsylvania 19104, United States.
Michael J. Siedlik, Department of Bioengineering, School of Engineering and Applied Science, University of Pennsylvania, Philadelphia, Pennsylvania 19104, United States
Jingyu Wu, Department of Chemical and Biomolecular Engineering, University of Pennsylvania, Philadelphia, Pennsylvania 19104, United States.
Kryshawna Beard, Department of Pharmacology, Perelman School of Medicine, University of Pennsylvania, Philadelphia, Pennsylvania 19104, United States.
Gennadiy Fonar, Center for Brain Injury and Repair, Department of Neurosurgery, University of Pennsylvania, Philadelphia, Pennsylvania 19104, United States.
Jean Pierre Dolle, Center for Brain Injury and Repair, Department of Neurosurgery, University of Pennsylvania, Philadelphia, Pennsylvania 19104, United States.
Douglas H. Smith, Center for Brain Injury and Repair, Department of Neurosurgery, University of Pennsylvania, Philadelphia, Pennsylvania 19104, United States
James H. Eberwine, Department of Pharmacology, Perelman School of Medicine, University of Pennsylvania, Philadelphia, Pennsylvania 19104, United States
David F. Meaney, Department of Bioengineering, School of Engineering and Applied Science, University of Pennsylvania, Philadelphia, Pennsylvania 19104, United States
David A. Issadore, Department of Bioengineering, School of Engineering and Applied Science and Department of Electrical and Systems Engineering, School of Engineering and Applied Science, University of Pennsylvania, Philadelphia, Pennsylvania 19104, United States.
REFERENCES
- (1).Kalluri R; LeBleu VS The biology, function, and biomedical applications of exosomes. Science (80-.). 2020, DOI: 10.1126/science.aau6977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (2).Costa-Silva B; Aiello NM; Ocean AJ; Singh S; Zhang H; Thakur BK; Becker A; Hoshino A; Mark MT; Molina H; Xiang J; Zhang T; Theilen TM; García-Santos G; Williams C; Ararso Y; Huang Y; Rodrigues G; Shen TL; Labori KJ; Lothe IMB; Kure EH; Hernandez J; Doussot A; Ebbesen SH; Grandgenett PM; Hollingsworth MA; Jain M; Mallya K; Batra SK; Jarnagin WR; Schwartz RE; Matei I; Peinado H; Stanger BZ; Bromberg J; Lyden D Pancreatic cancer exosomes initiate pre-metastatic niche formation in the liver. Nat. Cell Biol 2015, 17, 816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (3).Shao H; Chung J; Balaj L; Charest A; Bigner DD; Carter BS; Hochberg FH; Breakefield XO; Weissleder R; Lee H Protein typing of circulating microvesicles allows real-time monitoring of glioblastoma therapy. Nat. Med 2012, 18, 1835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (4).Mathieu M; Martin-Jaular L; Lavieu G; Théry C Specificities of secretion and uptake of exosomes and other extracellular vesicles for cell-to-cell communication. Nat. Cell Biol 2019, 21, 9. [DOI] [PubMed] [Google Scholar]
- (5).Chen G; Huang AC; Zhang W; Zhang G; Wu M; Xu W; Yu Z; Yang J; Wang B; Sun H; Xia H; Man Q; Zhong W; Antelo LF; Wu B; Xiong X; Liu X; Guan L; Li T; Liu S; Yang R; Lu Y; Dong L; McGettigan S; Somasundaram R; Radhakrishnan R; Mills G; Lu Y; Kim J; Chen YH; Dong H; Zhao Y; Karakousis GC; Mitchell TC; Schuchter LM; Herlyn M; Wherry EJ; Xu X; Guo W Exosomal PD-L1 contributes to immunosuppression and is associated with anti-PD-1 response. Nature 2018, 560, 382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (6).Yang Z; LaRiviere MJ; Ko J; Till JE; Christensen T; Yee SS; Black TA; Tien K; Lin A; Shen H; Bhagwat N; Herman D; Adallah A; Vollmer CM; Katona BW; Stanger BZ; Issadore D; Carpenter EL Carpenter, A multianalyte panel consisting of extracellular vesicle miRNAs and mRNAs, cfDNA, and CA19–9 shows utility for diagnosis and staging of pancreatic ductal adenocarcinoma. Clin. Cancer Res 2020, 26, 3248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (7).Yang KS; Im H; Hong S; Pergolini I; Del Castillo AF; Wang R; Clardy S; Huang CH; Pille C; Ferrone S; Yang R; Castro CM; Lee H; Del Castillo CF; Weissleder R Multiparametric plasma EV profiling facilitates diagnosis of pancreatic malignancy. Sci. Transl. Med 2017, DOI: 10.1126/scitranslmed.aal3226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (8).Hoshino A; Kim HS; Bojmar L; Gyan KE; Cioffi M; Hernandez J; Zambirinis CP; Rodrigues G; Molina H; Heissel S; Mark MT; Steiner L; Benito-Martin A; Lucotti S; Di Giannatale A; Offer K; Nakajima M; Williams C; Nogues L; Pelissier Vatter FA; Hashimoto A; Davies AE; Freitas D; Kenific CM; Ararso Y; Buehring W; Lauritzen P; Ogitani Y; Sugiura K; Takahashi N; Aleckovic M; Bailey KA; Jolissant JS; Wang H; Harris A; Schaeffer LM; Garcia-Santos G; Posner Z; Balachandran VP; Khakoo Y; Raju GP; Scherz A; Sagi I; Scherz-Shouval R; Yarden Y; Oren M; Malladi M; Petriccione M; De Braganca KC; Donzelli M; Fischer C; Vitolano S; Wright GP; Ganshaw L; Marrano M; Ahmed A; DeStefano J; Danzer E; Roehrl MHA; Lacayo NJ; Vincent TC; Weiser MR; Brady MS; Meyers PA; Wexler LH; Ambati SR; Chou AJ; Slotkin EK; Modak S; Roberts SS; Basu EM; Diolaiti D; Krantz BA; Cardoso F; Simpson AL; Berger M; Rudin CM; Simeone DM; Jain M; Ghajar CM; Batra SK; Stanger BZ; Bui J; Brown KA; Rajasekhar VK; Healey JH; de Sousa M; Kramer K; Sheth S; Baisch J; Pascual V; Heaton TE; La Quaglia MP; Pisapia DJ; Schwartz R; Zhang H; Liu Y; Shukla A; Blavier L; DeClerck YA; LaBarge M; Bissell MJ; Caffrey TC; Grandgenett PM; Hollingsworth MA; Bromberg J; Costa-Silva B; Peinado H; Kang Y; Garcia BA; O’Reilly EM; Kelsen D; Trippett TM; Jones DR; Matei IR; Jarnagin WR; Lyden D Extracellular Vesicle and Particle Biomarkers Define Multiple Human Cancers. Cell. 2020, 182, 1044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (9).Beard K; Meaney DF; Issadore D Clinical Applications of Extracellular Vesicles in the Diagnosis and Treatment of Traumatic Brain Injury. J. Neurotrauma 2020, 37, 2045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (10).Muraoka S; DeLeo AM; Sethi MK; Yukawa-Takamatsu K; Yang Z; Ko J; Hogan JD; Ruan Z; You Y; Wang Y; Medalla M; Ikezu S; Chen M; Xia W; Gorantla S; Gendelman HE; Issadore D; Zaia J; Ikezu T Proteomic and biological profiling of extracellular vesicles from Alzheimer’s disease human brain tissues. Alzheimer’s Dementia 2020, 16, 896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (11).Beard K; Yang Z; Haber M; Flamholz M; Diaz-Arrastia R; Sandsmark D; Meaney DF; Issadore D Extracellular vesicles as distinct biomarker reservoirs for mild traumatic brain injury diagnosis. Brain Commun. 2021, DOI: 10.1093/braincomms/fcab151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (12).Habib N; Avraham-Davidi I; Basu A; Burks T; Shekhar K; Hofree M; Choudhury SR; Aguet F; Gelfand E; Ardlie K; Weitz DA; Rozenblatt-Rosen O; Zhang F; Regev A Massively parallel single-nucleus RNA-seq with DroNc-seq. Nat. Methods 2017, 14, 955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (13).Klein AM; Mazutis L; Akartuna I; Tallapragada N; Veres A; Li V; Peshkin L; Weitz DA; Kirschner MW Kirschner, Droplet barcoding for single-cell transcriptomics applied to embryonic stem cells. Cell. 2015, 161, 1187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (14).Macosko EZ; Basu A; Satija R; Nemesh J; Shekhar K; Goldman M; Tirosh I; Bialas AR; Kamitaki N; Martersteck EM; Trombetta JJ; Weitz DA; Sanes JR; Shalek AK; Regev A; McCarroll SA Highly parallel genome-wide expression profiling of individual cells using nanoliter droplets. Cell. 2015, 161, 1202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (15).Grosselin K; Durand A; Marsolier J; Poitou A; Marangoni E; Nemati F; Dahmani A; Lameiras S; Reyal F; Frenoy O; Pousse Y; Reichen M; Woolfe A; Brenan C; Griffiths AD; Vallot C; Gérard A High-throughput single-cell ChIP-seq identifies heterogeneity of chromatin states in breast cancer. Nat. Genet 2019, 51, 1060. [DOI] [PubMed] [Google Scholar]
- (16).Saliba AE; Westermann AJ; Gorski SA; Vogel J Single-cell RNA-seq: Advances and future challenges. Nucleic Acids Res. 2014, 42, 8845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (17).Beekman P; Enciso-Martinez A; Rho HS; Pujari SP; Lenferink A; Zuilhof H; Terstappen LWMM; Otto C; Le Gac S Immuno-capture of extracellular vesicles for individual multi-modal characterization using AFM, SEM and Raman spectroscopy. Lab Chip. 2019, 19, 2526. [DOI] [PubMed] [Google Scholar]
- (18).Lee K; Fraser K; Ghaddar B; Yang K; Kim E; Balaj L; Chiocca EA; Breakefield XO; Lee H; Weissleder R Multiplexed Profiling of Single Extracellular Vesicles. ACS Nano 2018, 12, 494–503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (19).Carney RP; Hazari S; Colquhoun M; Tran D; Hwang B; Mulligan MS; Bryers JD; Girda E; Leiserowitz GS; Smith ZJ; Lam KS Multispectral Optical Tweezers for Biochemical Fingerprinting of CD9-Positive Exosome Subpopulations. Anal. Chem 2017, 89, 5357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (20).Smith ZJ; Lee C; Rojalin T; Carney RP; Hazari S; Knudson A; Lam K; Saari H; Ibañez EL; Viitala T; Laaksonen T; Yliperttula M; Wachsmann-Hogiu S Single exosome study reveals subpopulations distributed among cell lines with variability related to membrane content. J. Extracell. Vesicles 2015, 4, 28533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (21).Johnsen KB; Gudbergsson JM; Andresen TL; Simonsen JB What is the blood concentration of extracellular vesicles? Implications for the use of extracellular vesicles as blood-borne biomarkers of cancer. Biochim. Biophys. Acta - Rev. Cancer 2019, 1871, 109. [DOI] [PubMed] [Google Scholar]
- (22).Ko J; Wang Y; Sheng K; Weitz DA; Weissleder R Sequencing-Based Protein Analysis of Single Extracellular Vesicles. ACS Nano 2021, 15, 5631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (23).Liu C; Xu X; Li B; Situ B; Pan W; Hu Y; An T; Yao S; Zheng L Single-Exosome-Counting Immunoassays for Cancer Diagnostics. Nano Lett. 2018, 18, 4226–4232. [DOI] [PubMed] [Google Scholar]
- (24).Wei P; Wu F; Kang B; Sun X; Heskia F; Pachot A; Liang J; Li D Plasma extracellular vesicles detected by Single Molecule array technology as a liquid biopsy for colorectal cancer. J. Extracell. Vesicles 2020, 9, 1809765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (25).Wu F; Gu Y; Kang B; Heskia F; Pachot A; Bonneville M; Wei P; Liang J PD-L1 detection on circulating tumor-derived extracellular vesicles (T-EVs) from patients with lung cancer. Transl. Lung Cancer Res 2021, 10, 2441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (26).Rissin DM; Kan CW; Campbell TG; Howes SC; Fournier DR; Song L; Piech T; Patel PP; Chang L; Rivnak AJ; Ferrell EP; Randall JD; Provuncher GK; Walt DR; Duffy DC Single-molecule enzyme-linked immunosorbent assay detects serum proteins at subfemtomolar concentrations. Nat. Biotechnol 2010, 28, 595–599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (27).Cohen L; Cui N; Cai Y; Garden PM; Li X; Weitz DA; Walt DR Single Molecule Protein Detection with Attomolar Sensitivity Using Droplet Digital Enzyme-Linked Immunosorbent Assay. ACS Nano 2020, 14, 9491. [DOI] [PubMed] [Google Scholar]
- (28).Wu C; Garden PM; Walt DR Ultrasensitive Detection of Attomolar Protein Concentrations by Dropcast Single Molecule Assays. J. Am. Chem. Soc 2020, 142, 12314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (29).Cho S; Yi J; Kwon Y; Kang H; Han C; Park J Multifluorescence Single Extracellular Vesicle Analysis by Time-Sequential Illumination and Tracking. ACS Nano 2021, 15, 11753. [DOI] [PubMed] [Google Scholar]
- (30).Yelleswarapu V; Buser JR; Haber M; Baron J; Inapuri E; Issadore D Mobile platform for rapid sub–picogram-per-milliliter, multiplexed, digital droplet detection of proteins. Proc. Natl. Acad. Sci. U. S. A 2019, 116, 4489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (31).Yelleswarapu VR; Jeong H-H; Yadavali S; Issadore D Ultra-high throughput detection (1 million droplets per second) of fluorescent droplets using a cell phone camera and time domain encoded optofluidics. Lab Chip. 2017, 17, 1083–1094. [DOI] [PubMed] [Google Scholar]
- (32).Kan CW; Tobos CI; Rissin DM; Wiener AD; Meyer RE; Svancara DM; Comperchio A; Warwick C; Millington R; Collier N; Duffy DC Digital enzyme-linked immunosorbent assays with sub-attomolar detection limits based on low numbers of capture beads combined with high efficiency bead analysis. Lab Chip. 2020, 20, 2122. [DOI] [PubMed] [Google Scholar]
- (33).Chang L; Rissin DM; Fournier DR; Piech T; Patel PP; Wilson DH; Duffy DC Single molecule enzyme-linked immunosorbent assays: theoretical considerations. J. Immunol. Methods 2012, 378, 102–115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (34).Dube S; Qin J; Ramakrishnan R Mathematical analysis of copy number variation in a DNA sample using digital PCR on a nanofluidic device. PLoS One. 2008, 3, e2876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (35).Jeong H-H; Yelleswarapu VR; Yadavali S; Issadore D; Lee D Kilo-scale droplet generation in three-dimensional monolithic elastomer device (3D MED). Lab Chip. 2015, 15, 4387–4392. [DOI] [PubMed] [Google Scholar]
- (36).MacWilliams FJ; Sloane NJA Sloane, Pseudo-random sequences and arrays. Proc. IEEE 1976, 64, 1715–1729. [Google Scholar]
- (37).Zhang Y; Pak C; Han Y; Ahlenius H; Zhang Z; Chanda S; Marro S; Patzke C; Acuna C; Covy J; Xu W; Yang N; Danko T; Chen L; Wernig M; Sudhof TC Rapid single-step induction of functional neurons from human pluripotent stem cells. Neuron. 2013, 78, 785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (38).Sardar Sinha M; Ansell-Schultz A; Civitelli L; Hildesjö C; Larsson M; Lannfelt L; Ingelsson M; Hallbeck M Alzheimer’s disease pathology propagation by exosomes containing toxic amyloid-beta oligomers. Acta Neuropathol. 2018, 136, 41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (39).Fruhbeis C; Frohlich D; Kuo WP; Kramer-Albers E-M Extracellular vesicles as mediators of neuron-glia communication. Front. Cell. Neurosci 2013, DOI: 10.3389/fncel.2013.00182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (40).Yang Y; Boza-Serrano A; Dunning CJR; Clausen BH; Lambertsen KL; Deierborg T Inflammation leads to distinct populations of extracellular vesicles from microglia. J. Neuro-inflammation 2018, DOI: 10.1186/s12974-018-1204-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (41).Chen CC; Liu L; Ma F; Wong CW; Guo XE; Chacko JV; Farhoodi HP; Zhang SX; Zimak J; Ségaliny A; Riazifar M; Pham V; Digman MA; Pone EJ; Zhao W Elucidation of Exosome Migration Across the Blood–Brain Barrier Model In Vitro. Cell. Mol. Bioeng 2016, 9, 509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (42).Koliha N; Wiencek Y; Heider U; Jüngst C; Kladt N; Krauthäuser S; Johnston ICD; Bosio A; Schauss A; Wild S A novel multiplex bead-based platform highlights the diversity of extracellular vesicles. J. Extracell. Vesicles 2016, 5, 29975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (43).Kuypers S; Smisdom N; Pintelon I; Timmermans JP; Ameloot M; Michiels L; Hendrix J; Hosseinkhani B Unsupervised Machine Learning-Based Clustering of Nanosized Fluorescent Extracellular Vesicles. Small. 2021, 17, 2006786. [DOI] [PubMed] [Google Scholar]
- (44).Fulton RJ; McDade RL; Smith PL; Kienker LJ; Kettman JR Clinical Chemistry 1997, 43, 1749. [PubMed] [Google Scholar]
- (45).Wang X; Zhong W; Bu J; Li Y; Li R; Nie R; Xiao C; Ma K; Huang X; Li Y Exosomal protein CD82 as a diagnostic biomarker for precision medicine for breast cancer. Mol. Carcinog 2019, 58, 674. [DOI] [PubMed] [Google Scholar]
- (46).Yoshioka Y; Kosaka N; Konishi Y; Ohta H; Okamoto H; Sonoda H; Nonaka R; Yamamoto H; Ishii H; Mori M; Furuta K; Nakajima T; Hayashi H; Sugisaki H; Higashimoto H; Kato T; Takeshita F; Ochiya T Ultra-sensitive liquid biopsy of circulating extracellular vesicles using ExoScreen. Nat. Commun 2014, 5, 3591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (47).Chen Z; Liang Q; Zeng H; Zhao Q; Guo Z; Zhong R; Xie M; Cai X; Su J; He Z; Zheng L; Zhao K Exosomal CA125 as A promising biomarker for ovarian cancer diagnosis. J. Cancer 2020, 11, 6445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (48).Zhang P; Wu X; Gardashova G; Yang Y; Zhang Y; Xu L; Zeng Y Molecular and functional extracellular vesicle analysis using nanopatterned microchips monitors tumor progression and meta-stasis. Sci. Transl. Med 2020, DOI: 10.1126/scitranslmed.aaz2878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (49).Zhang P; Zhou X; Zeng Y Multiplexed immunophenotyping of circulating exosomes on nano-engineered ExoProfile chip towards early diagnosis of cancer. Chem. Sci 2019, DOI: 10.1039/c9sc00961b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (50).Adav SS; Park JE; Sze SK Quantitative profiling brain proteomes revealed mitochondrial dysfunction in Alzheimer’s disease. Mol. Brain 2019, x DOI: 10.1186/s13041-019-0430-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (51).Imbeault M; Lodge R; Ouellet M; Tremblay MJ Efficient magnetic bead-based separation of HIV-1-infected cells using an improved reporter virus system reveals that p53 up-regulation occurs exclusively in the virus-expressing cell population. Virology. 2009, 393, 160. [DOI] [PubMed] [Google Scholar]
- (52).Wang Y; Shah V; Lu A; Pachler E; Cheng B; Di Carlo D Counting of enzymatically amplified affinity reactions in hydrogel particle-templated drops. Lab Chip. 2021, 21, 3438. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


