Abstract
Background:
The availability of whole genome sequencing data in large studies has enabled the assessment of coding and non-coding variants across the allele frequency spectrum for their associations with blood pressure.
Methods:
We conducted a multi-ancestry whole genome sequencing analysis of blood pressure among 51,456 Trans-Omics for Precision Medicine and Centers for Common Disease Genomics program participants (stage-1). Stage-2 analyses leveraged array data from UK Biobank (N=383,145), Million Veteran Program (N= 318,891), and Reasons for Geographic and Racial Differences in Stroke (N=10,643) participants, along with whole exome sequencing data from UK Biobank (N=199,631) participants.
Results:
Two blood pressure signals achieved genome-wide significance in meta-analyses of stage-1 and stage-2 single variant findings (P<5×10−8). Among them, a rare intergenic variant at novel locus, LOC100506274, was associated with lower systolic blood pressure in stage-1 [beta (standard error)=−32.6 (6.0); P=4.99×10−8] but not stage 2 analysis (P=0.11). Furthermore, a novel common variant at the known INSR locus was suggestively associated with diastolic blood pressure in stage-1 [beta (standard error)=−0.36 (0.07); P=4.18×10−7] and attained genome-wide significance in stage-2 [beta (standard error)=-0.29 (0.03); P=7.28×10−23]. Nineteen additional signals suggestively associated with blood pressure in meta-analysis of single and aggregate rare variant findings (P<1×10−6 and P<1×10−4, respectively).
Discussion:
We report one promising but unconfirmed rare variant for blood pressure and, more importantly, contribute insights for future blood pressure sequencing studies. Our findings suggest promise of aggregate analyses to complement single variant analysis strategies and the need for larger, diverse samples and family studies to enable robust rare variant identification.
Keywords: Whole genome sequencing, blood pressure, hypertension
Graphical Abstract
INTRODUCTION
Hypertension affects nearly one third of adults and has been identified as a leading risk factor for morbidity and mortality globally.1,2 In addition to genetic influences, blood pressure (BP) is a common complex phenotype influenced by lifestyle and behavioral risk factors.3–5 Genetic factors impacting BP have been identified through multiple lines of investigation. Genome-wide association studies (GWAS) have identified over 1,000 loci influencing BP but have generally been limited to the assessment of common and low-frequency variants.6–32 Large-scale analyses of rare variants from exome sequencing and exome chip studies have also identified multiple loci influencing BP.33–36 Rare variant studies, however, have largely been restricted to coding regions of the genome. GWAS of common variants have provided empirical evidence of intergenic variants with small BP associations,6–32 while exome-based studies have identified rare coding variants with large associations.33–36 Few studies have assessed the role of non-coding rare variants in BP regulation,23,36 and no large-scale studies of high-depth whole genome sequencing (WGS) data have been conducted. Through the National Heart, Lung, and Blood Institute (NHLBI) Trans-Omics for Precision Medicine (TOPMed) and National Human Genome Research Institute (NHGRI) Center for Common Disease Genomics (CCDG) programs, WGS has now been conducted in large studies,37 providing opportunity for comprehensive exploration of common, low-frequency, and rare variants in coding and non-coding regions in relation to BP phenotypes.
The purpose of the current study was to identify novel BP signals by carrying out a WGS study of systolic BP (SBP), diastolic BP (DBP), and hypertension among a multi-ancestry sample of 51,456 participants from the TOPMed and CCDG programs. We further investigated suggestive single variant findings among up to 383,145 UK Biobank, 318,891 Million Veteran Program (MVP), and 10,643 REasons for Geographic and Racial Differences in Stroke (REGARDS) participants with genome-wide array-based genotype data. Suggestive rare variant signals were further assessed among 199,631 UK Biobank participants with whole-exome sequencing (WES) data.
METHODS
All data and materials have been made publicly available at the database of Genotypes and Phenotypes (dbGaP) and can be accessed at phs001974.
Stage-1 Studies
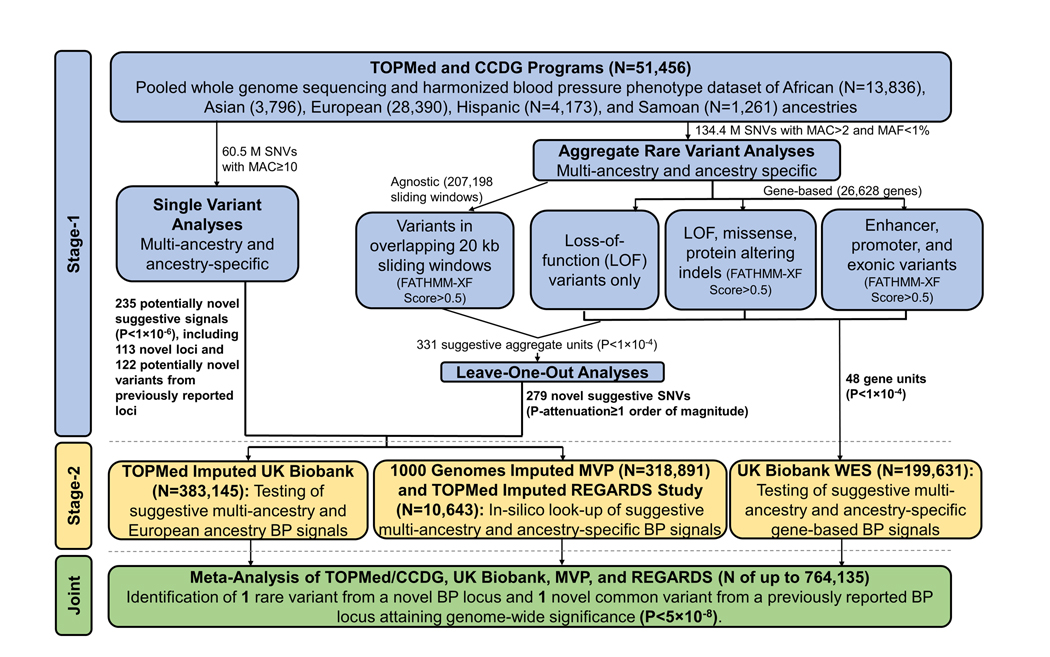
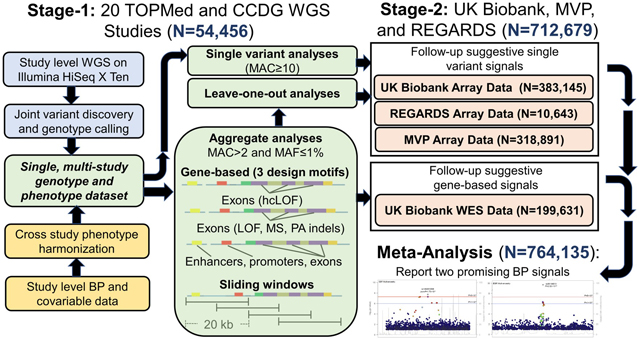
We conducted a multi-stage genomic study of BP phenotypes among up to 764,135 participants (Figure 1). The stage-1 analysis included 51,456 multi-ancestry participants from 18 TOPMed and CCDG WGS studies.37,38 BP phenotypes were harmonized across studies using a strict protocol that included adding 15 mmHg and 10 mmHg to SBP and DBP values, respectively, if a participant was taking anti-hypertension medication.39 Hypertension was defined as SBP≥140 mmHg, DBP≥90 mmHg, or use of anti-hypertension medication. WGS and BP data were pooled across studies for analyses. Stage-1 studies, WGS methods, and phenotype harmonization are detailed in the Expanded Methods, Table 1, and Table S1 (please see http://hyper.ahajournals.org).
Figure 1.
Design of the multi-stage genomic study of blood pressure phenotypes.
Table 1.
Description of the stage-1 and stage-2 studies and participants.
| Study | Sample Size | BP Measurement Method | Mean age* (s.d.) | Women, % | Ancestry, % |
Mean BMI† (s.d.) | Mean SBP‡ (s.d.) | Mean DBP‡ (s.d.) | HTN§, % | Anti-HTN medicine, % | ||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| African | Asian | European | Hispanic | Other | ||||||||||
| Stage-1 Studies | ||||||||||||||
| Amish | 1,111 | Standard sphygmomanometer | 50.7 (17.1) | 49.7 | 0.00 | 0.00 | 100 | 0.00 | 0.00 | 27.0 (4.6) | 120.7 (15.6) | 74.3 (9.7) |
16.7 | 5.2 |
| ARIC | 6545ǁ | Random zero sphygmomanometer | 54.1 (5.7) | 54.5 | 9.23 | 0.00 | 90.8 | 0.00 | 0.00 | 27.1 (5.0) | 119.1 (17.7) | 72.2 (10.5) | 34.8 | 27.2 |
| BioMe | 3,155 | Multiple sphygmomanometers | 59.1 (12.9) | 63.2 | 38.3 | 1.27 | 21.5 | 39.0 | 0.00 | 29.8 (7.3) | 133.1 (25.1) | 73.9 (13.6) | 67.5 | 54.5 |
| CARDIA | 2,930 | Random zero sphygmomanometer | 25.1 (3.6) | 57.7 | 45.1 | 0.00 | 54.9 | 0.00 | 0.00 | 24.5 (5.0) | 110.0 (10.9) | 68.5 (9.4) |
2.7 | 0.9 |
| CFS | 990 | Calibrated sphygmomanometer | 41.3 (19.5) | 53.5 | 50.6 | 0.00 | 48.9 | 0.51 | 0.00 | 31.8 (9.4) | 121.8 (17.0) | 74.1 (11.1) | 35.9 | 28.3 |
| CHS | 2,839 | Random zero sphygmomanometer | 72.5 (5.3) | 56.4 | 18.3 | 0.04 | 80.5 | 1.13 | 0.00 | 26.8 (4.7) | 136.6 (21.6) | 70.9 (11.6) | 65.9 | 46.5 |
| FHS | 3,615 | Desktop baumanometer | 38.0 (8.9) | 54.1 | 0.00 | 0.00 | 100 | 0.00 | 0.00 | 25.8 (4.8) | 119.8 (15.0) | 77.4 (10.2) | 17.9 | 4.9 |
| GeneSTAR | 1,735 | Mercury or aneroid sphygmomanometer | 41.5 (11.4) | 59.3 | 44.2 | 0.00 | 55.8 | 0.00 | 0.00 | 29.6 (7.1) | 120.3 (16.2) | 77.8 (10.8) | 28.5 | 15.9 |
| GENOA | 1,214 | Random zero sphygmomanometer | 56.3 (10.6) | 70.2 | 100 | 0.00 | 0.00 | 0.00 | 0.00 | 31.1 (6.6) | 134.5 (22.3) | 77.8 (12.2) | 69.7 | 57.7 |
| GenSalt | 1,818 | Random zero sphygmomanometer | 38.7 (9.6) | 47.3 | 0.00 | 100 | 0.00 | 0.00 | 0.00 | 23.3 (3.2) | 117.7 (15.1) | 74.3 (11.1) | 12.7 | 0.4 |
| GOLDN | 942 | Automated Dinamap | 47.9 (16.3) | 53.0 | 0.00 | 0.00 | 100.0 | 0.00 | 0.00 | 28.3 (5.7) | 115.6 (16.8) | 68.3 (9.3) |
22.9 | 17.9 |
| HCHS-SOL | 1,590 | Digital Omron | 46.9 (14.4) | 62.0 | 0.00 | 0.00 | 0.00 | 100.0 | 0.00 | 30.7 (6.9) | 123.8 (19.0) | 74.3 (11.3) | 34.3 | 21.1 |
| HyperGEN | 1,880 | Automated dinamap | 47.0 (12.8) | 63.4 | 100 | 0.00 | 0.00 | 0.00 | 0.00 | 32.0 (7.7) | 129.8 (22.5) | 74.4 (11.7) | 62.9 | 52.7 |
| JHS | 3,307 | Random zero sphygmomanometer | 55.4 (12.8) | 62.6 | 100 | 0.00 | 0.00 | 0.00 | 0.00 | 31.8 (7.3) | 127.2 (16.6) | 75.7 (8.8) |
59.4 | 52.0 |
| MESA | 4,526 | Automated dinamap | 61.0 (9.8) | 51.4 | 24.0 | 13.2 | 40.4 | 22.4 | 0.00 | 28.2 (5.3) | 124.8 (20.5) | 71.8 (10.2) | 45.6 | 35.5 |
| Samoan | 1,261 | Digital Omron | 44.6 (11.3) | 60.3 | 0.00 | 0.00 | 0.00 | 0.00 | 100 | 33.6 (6.7) | 128.4 (19.1) | 82.0 (13.0) | 32.0 | 7.2 |
| THRV | 1,138 | Automated dinamap | 49.3 (9.5) | 53.2 | 0.00 | 100 | 0.00 | 0.00 | 0.00 | 25.2 (3.5) | 130.3 (25.2) | 77.5 (13.9) | 67.6 | 50.7 |
| WHI | 10,860 | Mercury sphygmomanometer | 66.5 (6.8) | 100 | 13.2 | 1.85 | 82.2 | 2.78 | 0.00 | 28.7 (6.1) | 132.2 (18.5) | 75.7 (9.7) |
55.7 | 39.8 |
| Stage-2 Studies | ||||||||||||||
| UK Biobank (GWAS) | 383,145 | Digital Omron | 56.9 (8.0) | 54.0 | 0.00 | 0.00 | 100.0# | 0.00 | 0.00 | 27.4 (4.7) | 139.8 (19.8) | 83.3 (10.9) | 47.0 | 10.1 |
| UK Biobank (WES) | 199,631 | Digital Omron | 56.5 (8.1) | 54.4 | 0.63 | 2.50 | 93.8 | 0.00 | 3.1 | 27.4 (4.8) | 137.8 (18.7) | 82.2 (10.2) | 54.3 | 21.1 |
| MVP | 318,891 | Multiple devices | 58.6 (12.6) | 8.40 | 18.8 | 0.80 | 69.1 | 6.70 | 4.6 | 30.2 (5.8) | 138.1 (16.0) |
82.6 (11.0) |
65.6 | 48.9 |
| REGARDS | 10,643 | Aneroid sphygmomanometer | 64.4 (9.6) | 57.9 | 83.8 | 0.00 | 16.2 | 0.00 | 0.0 | 30.4 (6.6) | 130.5 (17.3) | 78.1 (10.1) | 69.1 | 62.9 |
Amish, The Amish Complex Disease Research Program; ARIC, Atherosclerosis Risk in Communities; BioMe, The IPM BioMe Biobank; BMI, Body mass index; BP, blood pressure; CARDIA, Coronary Artery Risk Development in Young Adults; CCDG, Centers for Common Disease Genomics; CFS, Cleveland Family Study; CHS, Cardiovascular Health Study; DBP, diastolic BP; FHS, Framingham Heart Study; GeneSTAR, Genetic Studies of Atherosclerosis Risk; GENOA, Genetic Epidemiology Network of Arteriopathy; GenSalt, Genetic Epidemiology Network of Salt-Sensitivity; GOLDN, Genetics of Lipid Lowering Drugs and Diet Network; HCHS-Sol, Hispanic Community Health Study-Study of Latinos; GWAS, Genome-wide association study; HTN, hypertension; HyperGen, Hypertension Genetic Epidemiology Network; JHS, Jackson Heart Study; MESA, Multi-ethnic Study of Atherosclerosis; MVP, Million Veteran Program; n, sample size; REGARDS, Reasons for Geographic and Racial Differences in Stroke; Samoan, Samoan Study; SBP, systolic BP; s.d., standard deviation; THRV, Taiwan Study of Hypertension using Rare Variants; TOPMed, Trans-omics for Precision Medicine; WES, whole-exome sequencing; WGS, whole-genome sequencing; WHI, Women’s Health Initiative.
Age in years.
Measurements in kg/m2.
Measurements in mmHg.
Defined as SBP≥140, DBP≥90 or use of anti-HTN medication.
Includes 3,934 and 2,611 participants with WGS data from TOPMed and CCDG, respectively.
All are White British.
Stage-1 Analysis
Stage-1 multi-ancestry and ancestry-specific analyses were conducted using the Analysis Commons cloud-based platform.40 Single nucleotide variants (SNVs) with minor allele count (MAC) ≥10 were individually tested for association with BP. Analyses of SBP and DBP employed a linear mixed model that accounted for familial correlations using a sparse kinship matrix,41 adjusted for age, sex, body mass index (BMI), study, and ancestry principal components (PCs), and, for the multi-ancestry analyses only, we fit separate (heterogeneous) residual variance components for each ancestry group. Single variant analyses of hypertension employed a logistic mixed model that again accounted for familial relationships and adjusted for age, sex, BMI, study, and ancestry PCs. Newly identified SNVs (r2<0.1 with any previously reported BP variant6–36) achieving suggestive significance (P<1×10−6) were moved forward for stage-2 study.
Following functional annotation using WGS Annotator software,42 rare variants [minor allele frequency (MAF)<1% and MAC>2)] were first aggregated using gene-based strategies, including: 1) Loss-of-function variants only; 2) Loss-of-function variants, missense variants, and protein altering insertion-deletions with FATHMM-XF scores>0.5; 43 and 3) Variants located in gene enhancers,44 promoters,45 and exons with FATHMM-XF scores>0.5.43 The sliding window approach was then used to aggregate variants with FATHMM-XF scores>0.5 in 20 KB chromosomal segments across the genome using a 10 KB offset.
We used a variant set mixed model association test that efficiently combines the burden and sequence kernel association tests (SMMAT-E)46 to examine associations between aggregate rare variant units and BP phenotypes employing the same generalized linear mixed model frameworks described for single variant analyses. Gene-based signals achieving suggestive significance (P<1×10−4) were moved forward for stage-2 study. Leave-one-out analyses were also conducted to identify signal driving variants among aggregate units achieving suggestive significance (P<1×10−4). Any variants whose removal attenuated the SMMAT-E p-value by one order of magnitude or more was also moved forward for stage-2 study.
Stage-2 Studies
Suggestive signals from stage-1 single variant or leave-one-out analyses were tested among up to 383,145 White British UK Biobank participants with BP and genome-wide SNP array data imputed using the TOPMed freeze 5 reference panel.37,47–49 We also carried out an in-silico look-up of individual variants using results from the previous BP GWAS of 318,891 multi-ancestry MVP23 and 10,643 multi-ancestry REGARDS participants, which utilized array data imputed to the 1KG project phase 3, version 5 and TOPMed freeze 8 reference panels, respectively. We also leveraged whole exome sequencing data from a multi-ancestry sample of 199,631 UK Biobank participants to carry-out stage-2 analyses of suggestive gene signals. Detailed descriptions of stage-2 study genotyping, phenotype harmonization, and analyses are provided in the Expanded Methods (please see http://hyper.ahajournals.org). For lead variants that were unavailable in the stage-2 samples, we assessed up to 32 proxies (r2>0.8 in the stage-1 sample), selecting the variant with the highest r2 if multiple proxies were available.
Identification and Reporting of Novel BP Signals
Single variant analysis findings with consistent associations (based on association directions and a conservative heterogeneity P>1×10−3) and a permissive inverse variance weighted fixed effects meta-analysis P<5×10−8 across stages-1 and -2 were reported in the current study. A meta-analysis P<1.88×10−6 (correcting for 26,628 independent gene tests) was used as the threshold for determining significance of aggregate gene-based signals. For variants identified through leave-one-out analyses of gene-based and sliding window units, consistency in association size and respective meta-analysis P-values of 1.88×10−6 and 2.41×10−7 (correcting for 26,628 genes and 207,198 sliding windows, respectively) was employed.
Fine-mapping of the 500 KB regions surrounding genome-wide significant signals was undertaken, sequentially adding the most significant variant in the region to the null model until no additional variants achieving P<1×10−4 were identified. Additional conditional analyses were conducted to verify the independence of suggestive signals at known loci, and expression quantitative trait locus (eQTL) analyses examined whether identified variants were associated with gene expression (Expanded Methods; please see http://hyper.ahajournals.org).
RESULTS
Characteristics of the stage-1 (TOPMed-CCDG) and stage-2 (UK Biobank, MVP, and REGARDS) study participants are shown in Table 1.
Stage-1 Single Variant Analysis Findings
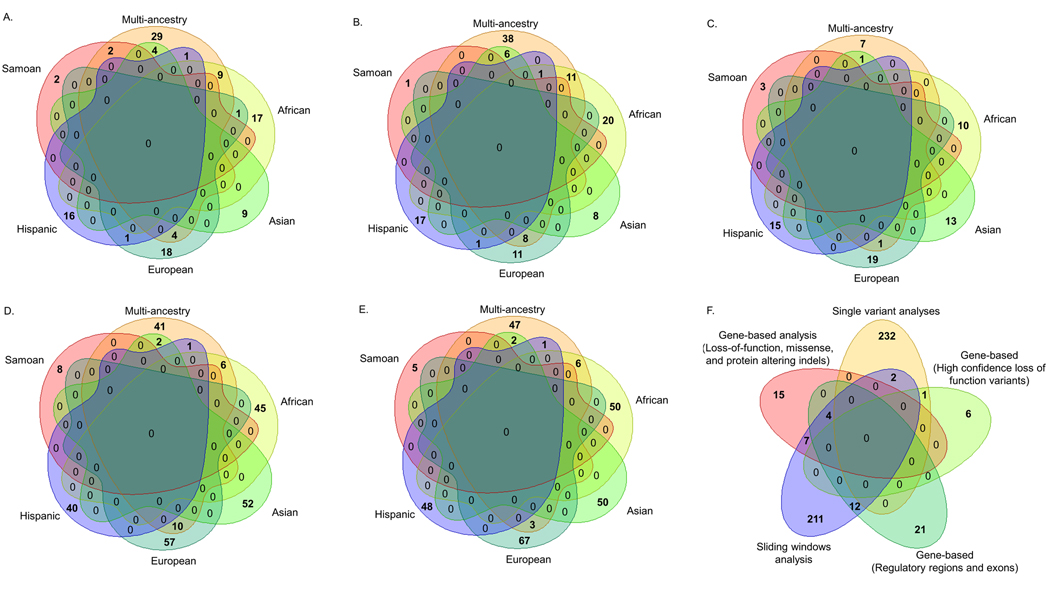
Stage-1 single variant analyses of the multi-ancestry and ancestry-specific samples identified 258 loci (113 novel plus 145 known loci) that achieved suggestive significance (P<1×10−6). Among the 113 novel loci identified, five achieved genome-wide significance [with P<5×10−8; Figure 2A; and Figure S1, Figure S2, and Tables S2-S7 (please see http://hyper.ahajournals.org)]. Novel loci were defined as those with lead variants that were neither in close proximity (>500 KB) nor correlated (r2<0.1) with a previously reported sentinel BP SNV.6–36 Suggestive associations at 145 loci were in close proximity (<500 KB) to previously reported sentinel BP SNVs (including 16 at P<5×10−8; Figure S1; please see http://hyper.ahajournals.org). Among them, 122 potentially novel variants (r2<0.1 with any previously reported sentinel BP SNVs) were identified, including two that attained genome-wide significance [Figure 2B; and Tables S2-S7 (please see http://hyper.ahajournals.org)]. Rare variants with MAC ≥10 and MAF <1% comprised 70% of novel BP signals (71% of signals at novel loci and 69% of signals from novel variants at previously reported loci). There was limited overlap of BP signals across ancestry-specific analyses (Figures 2A-B).
Figure 2.
Venn diagrams displaying suggestive BP signals across multi-ancestry and ancestry-specific analyses for: (A.) 113 novel BP loci; (B.) 122 potentially novel variants at previously reported BP loci; (C.) 69 genes; (D.) 262 sliding windows; and (E.) 279 signal driving variants from leave-one-out analyses. (F.) This diagram displays the overlap of suggestive BP signals across 235 SNVs identified by single variant analyses, including 113 from novel loci and 122 from previously reported loci, and 279 signal driving variants from leave-one-out analyses of suggestive genes and sliding windows.
Stage-1 Aggregate Rare Variant Analysis Findings
Stage-1 aggregate rare variant analyses across the multi-ancestry and ancestry-specific samples identified 331 aggregate units (69 genes and 262 sliding windows) suggestively associated (P<1×10−4) with one or more BP phenotype [Figure 2C and 2D; and Tables S8-S13 (please see http://hyper.ahajournals.org)]. Four aggregate signals at two gene loci, GABRB3 (P=4.96×10−7) and KIF3B (minimum-P=3.23×10−8), were significant in Asian ancestry participants after Bonferroni correction for the number of aggregate units tested. Reassuringly, suggestive gene-based signals included biologically relevant BP candidates, such as: AGTRAP,50 CACNA2D3,51 ERBB4,52 PDLIM5, 22, 23 and PROCR.33 In leave-one-out analyses, removal of 279 unique rare variants attenuated SMMAT-E p-values from identified aggregate units by at least one order of magnitude [Figure 2E; and Tables S14-S19 (please see http://hyper.ahajournals.org)]. Fifty-five percent of these signal driving variants had larger p-values than those of the aggregate units from which they were derived (Tables S14-S19; please see http://hyper.ahajournals.org). Furthermore, some of the same signal-driving variants were identified across different design motifs (Figure 2F). Because aggregate analyses employed less stringent alpha thresholds and identified a high frequency of variants (38%) that fell below the MAC filtering threshold used in single variant analyses, there was limited overlap of SNVs derived from these distinct strategies (Figure 2F).
Stage-2 and Meta-Analyses of 235 BP Signals from Stage-1 Single Variant Analyses
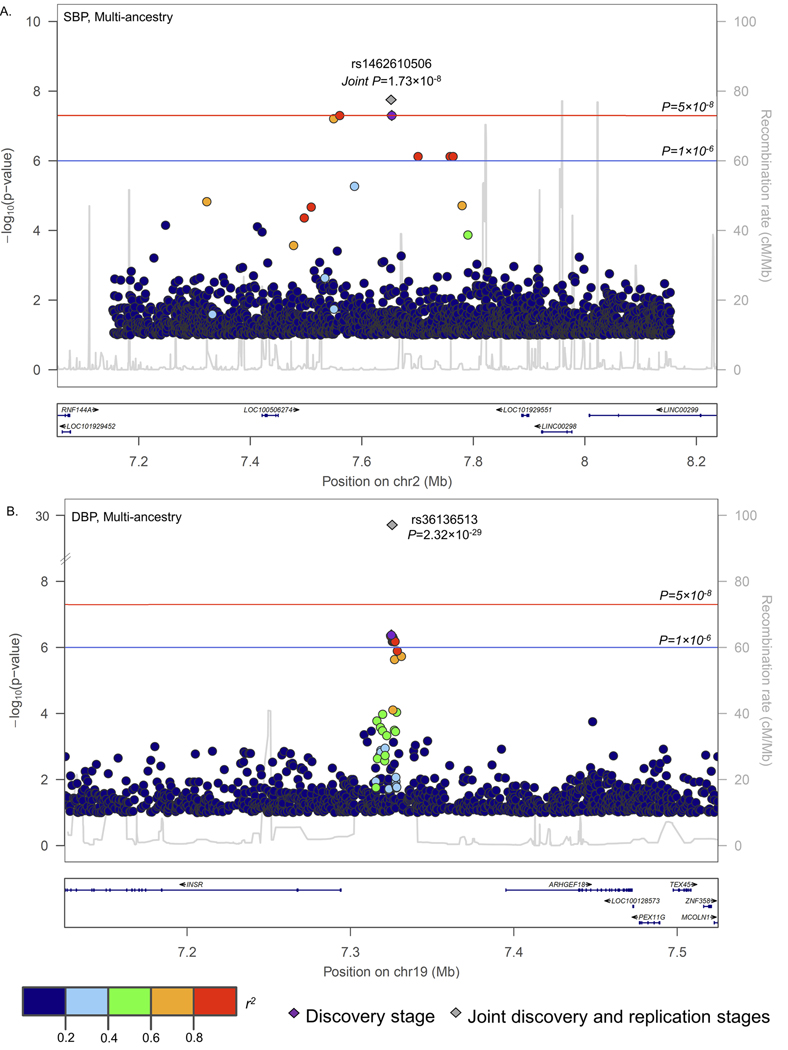
One rare SNV (rs1462610506) at the novel LOC100506274 locus and one novel SNV (rs36136513) from the previously reported INSR locus achieved consistent association directions in stage-2 analyses and genome-wide significance in our multi-ancestry meta-analyses (P=1.73×10−8 and 2.32×10−29, respectively; Table 2 and Figure 3). Twenty carriers of the rare rs1462610506 A allele had approximately 33 mmHg (95% confidence interval: 21, 44 mmHg) lower mean SBP than non-carriers in meta-analysis. This intergenic SNV displayed strong linkage disequilibrium (r2>0.8) with variants extending across a large chromosomal region (267 KB in length) harboring no known genes (Figure 3A). The novel intergenic rs36136513 variant at INSR also achieved genome-wide significance in the meta-analysis of stage-1 and stage-2 studies (Figure 3B). Each copy of the common rs36136513 G allele was associated with a modest 0.30 mmHg decrease in DBP (Table 2). Six novel rare variant loci and six novel rare variants from previously reported loci were also suggestively associated with BP in meta-analyses [P<1×10−6;; Table S20 (please see http://hyper.ahajournals.org)].
Table 2.
Genome-wide significant signals (P<5×10−8) identified in meta-analyses of stage-1 and stage-2 findings.
| Chr | Position (GRCh38) | rsID | Associated Trait | Ancestry | Nearest Gene | Classification(s) | Alleles (Ref/Alt) | Sample | AAC | Beta* | SE | P-value |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 2 | 7653714 | rs1462610506† | SBP | Multi | LOC100506274 | Intergenic | G/A | TOPMed | 13 | −32.57 | 5.97 | 4.99E-08 |
| UK Biobank | 7 | −59.79 | 37.28 | 1.09E-01 | ||||||||
| Meta-analysis | −33.25 | 5.90 | 1.73E-08 | |||||||||
| 19 | 7325055 | rs36136513° | DBP | Multi | INSR | Intergenic | A/G | TOPMed | 45,593 | −0.36 | 0.07 | 4.18E-07 |
| UKBB | 361,082 | −0.28 | 0.03 | 3.24E-22 | ||||||||
| REGARDS | 8,691 | −0.57 | 0.17 | 1.11E-03 | ||||||||
| Meta-analysis | −0.30 | 0.03 | 2.32E-29 |
AAC, alternative allele count; Alt, alternative; Chr, chromosome; DBP, diastolic blood pressure (BP); Ref, reference; REGARDS, Reasons for Geographic and Racial Differences in Stroke; SBP, systolic BP; TOPMed, Trans-Omics for Precision Medicine; UKBB, UK Biobank.
Beta corresponds to the association size in mmHg and natural logarithm of the odds ratio per coded allele for the continuous and discrete blood pressure phenotypes, respectively.
rs541302407 was used as a proxy for rs1462610506 in the UK Biobank (r2=0.87).
Figure 3.
Regional association plots displaying signals at: (A.) LOC100506274; and (B.) INSR.
Stage-2 and Meta-Analyses of 331 BP Signals from Stage-1 Aggregate and Leave-One-Out Analyses
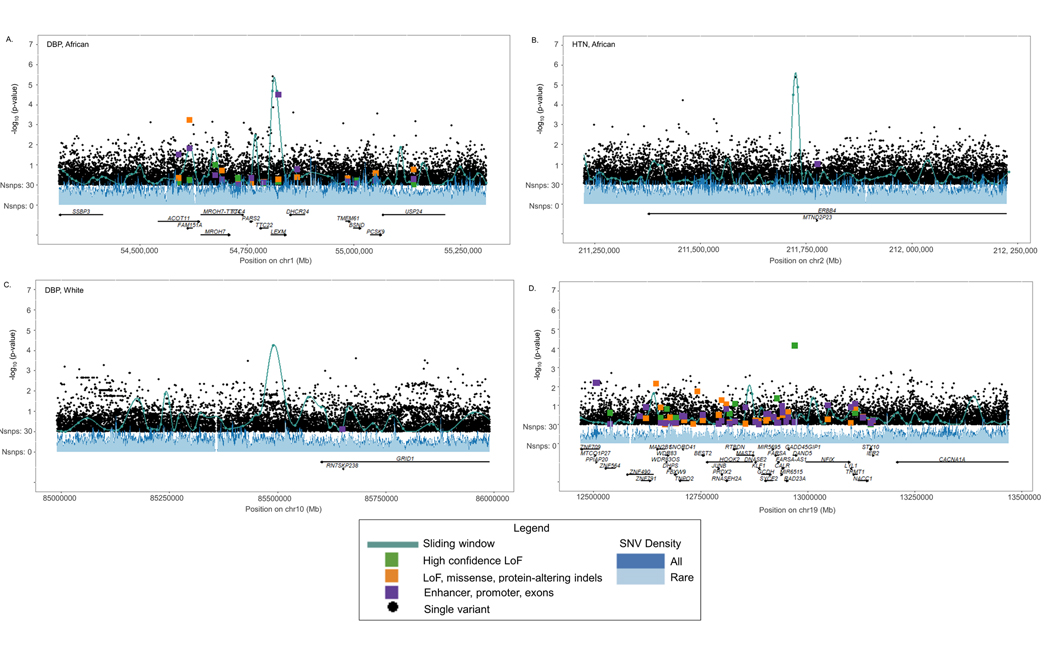
Although none of the genes identified by aggregate rare variant analyses attained genome-wide significance in the meta-analysis of stage-1 and stage-2 studies (Tables S8, S10, and S11), MZT2B, DNAJB13, and NDRGB-TPPP2 were suggestively associated with BP (P<1×10−4; Table S21). Likewise, no genome-wide significant variants, but four suggestive variants (at LEXM, ERBB4, LINC01520, and DAND5), were identified in meta-analyses of variants derived from leave-one-out analyses (P<1×10−4; Figures 4A-D; Table S22); please see http://hyper.ahajournals.org). Three of the four aggregate units harboring signal driving rare variants achieved smaller p-values compared with any of the single variants in these regions (Figures 4A-D).
Figure 4.
Lachesis plots integrating aggregate and single variant signals at: (A.) LEXM; (B.) ERBB4; (C.) LINC01520; and (D.) DAND5.
Conditional Analyses
Sequential conditional analyses did not identify any independent signals at the two newly identified loci (Figure S3). Furthermore, none of the newly identified signals (with P<1×10−6) from previously reported loci were attenuated after adjusting for previously reported variants at corresponding loci (Table S23).
DISCUSSION
In this first large-scale, multi-ancestry WGS study of BP, two novel signals achieved genome-wide significance in meta-analysis of stage-1 and stage-2 results, including one rare intergenic variant, rs1462610506, at the novel LOC100506274 locus and one new lead variant, rs36136513, at the previously reported INSR locus. The rare rs1462610506 A allele appeared to have large SBP lowering association. However, with a lack of true replication in the stage-2 sample, these findings should be interpreted with caution. As expected, we noted modest DBP lowering associations of the common rs36136513 G allele. Our analysis also identified very little overlap of BP signals across ancestries. Most of these ancestry-specific signals were untestable in other ancestry groups because they were either monomorphic or exceedingly rare. These data suggest that many rare variant associations with BP are ancestry specific. Aggregate rare variant analyses complemented our single variant approach, identifying additional novel signals that suggestively associated with BP.
Our finding of one genome-wide significant and numerous suggestive non-coding rare variant BP signals in meta-analysis of stage-1 and stage-2 findings supports the hypothesis that such variants can have large associations with complex BP phenotypes. While previous studies have suggested that rare genic variants may influence BP,23,34,35 we are among the first to identify a promising, but unconfirmed, intergenic rare variant with an association size similar to protein disrupting mutations found in monogenic BP disorders.23,36 The rare, intergenic variant at LOC100506274, a long non-coding RNA, lowered SBP by an average of 33 mmHg (95% confidence interval: 22, 45 mmHg) in carriers compared with non-carriers. The 13 rare variant carriers from our stage-1 analysis all had Asian ancestry and included individuals from six separate pedigrees across two TOPMed studies. An exceedingly rare proxy for this variant showed similar associations in white British UK Biobank participants (MAF=9.28×10−6) and, unsurprisingly, was unobserved in the 28,390 European TOPMed participants. The seven UK Biobank rare allele carriers had average BP values approximately 60 mmHg lower than that of non-carriers. Although standard errors were large and findings were non-statistically significant due to the small number of carriers in UK Biobank, the consistency in the large association sizes across ancestries and the two-staged analysis provide cautious evidence for this novel rare variant signal. Still, we cannot rule out the possibility of a false positive finding and recommend further confirmation of this signal.
Most novel signals identified by the current WGS study were derived from rare variants, which was expected given the high genome-wide coverage of common and low frequency variants by GWAS including up to one million study participants.22,23 Still, we identified one novel common variant at the previously reported INSR locus with modest DBP lowering associations. The lead variant, rs36136513, had an r2<0.1 with previously reported variants in this region,17,18,35 and its signal was unattenuated when conditioning on these variants. rs36136513 and its correlated variants were not included in GRCh37 nor were they present on the 1KG reference panel, which likely explains why this signal was discovered only through WGS.
In contrast to previous GWAS, we identified minimal overlap of genome-wide significant and suggestive BP signals across our ancestry-specific analyses.10,15,27 Compared with GWAS, which primarily target common and low frequency SNPs, most variants identified in the current analyses were rare and demonstrated large associations with BP. Approximately 72% of the identified ancestry-specific variants were either monomorphic or below our MAC filter in the other ancestral groups. Consistent with these findings, population genetics suggests that deleterious rare variants are more likely to have arisen recently in human history and show geographic clustering compared with more neutral variants typically identified by GWAS. While we cannot rule out the possibility of false positive findings, this latter phenomenon could also explain the observed lack of overlap across populations.53,54 Only 38% of the rare variants identified in non-European ancestry groups were available for stage-2 study. Because the most deleterious rare variants tend to be private to single populations,54 ancestry-specific rare variant signals identified here are promising for verification in future studies providing more comprehensive rare variant coverage in diverse populations.
Three of four suggestive aggregate rare variant signals for BP attained smaller p-values than any one of their individual signal driving variants. Likewise, at aggregate unit sites where suggestive signal driving variants were discovered in the stage-1 study, the identified aggregate units often outperformed SNVs included in single variant analyses across the surrounding 1 MB region. These data suggest that aggregate rare variant analyses may discover BP loci missed by traditional single variant analyses. In contrast to the unique information derived from aggregate and single variant analysis strategies, leave-one-out analyses across certain aggregation design motifs provided similar information. Future WGS studies may be able to reduce the number of statistical tests conducted by minimizing designs that provide redundant information.
As the first large-scale WGS analysis of BP phenotypes, the current study has important strengths. The large and ancestrally diverse stage-1 sample allowed for the identification of promising, rare variants with large BP associations in the multi-ancestry and ancestry-specific analyses. In addition, harmonization of BP phenotypes combined with joint calling of genotypes from WGS across studies allowed for pooling of individual level stage-1 data. Using one large multi-study dataset, we were able to conduct multi-ancestry and ancestry specific mega-analyses, which have power advantages for rare variant study compared with traditional meta-analysis techniques.55 Certain limitations should also be acknowledged. Although this represents the largest WGS study of BP, our sample size was modest compared with recent BP GWAS and power was limited. For this reason, we employed a somewhat permissive p-value (5×10−8) for determining statistical significance. In addition, because the UK Biobank sample was comprised predominantly of white British participants and many rare variants were unavailable for look-up in the multi-ancestry MVP program and REGARDS study, we did not have the opportunity to verify 62% of non-European ancestry-specific signals. In addition, the inclusion of non-functional variants in aggregate analyses could have also reduced statistical power of these tests, despite filtering on predicted functional relevance. We were also unable to verify aggregate signals in non-coding regions since only WES data were available for stage-2 study. Furthermore, while eQTL analyses were performed, only 838 samples with WGS data were available in the GTEx database. Hence, many of rare variant signals were unavailable or had minor allele counts too low for suitable functional analyses. Future sequencing studies, either in large ancestry-specific samples or families enriched for the identified rare variants, will be needed to confirm and infer the function of novel findings discovered in our WGS study.
Supplementary Material
NOVELTY AND RELEVANCE.
What is new?
The stage-1 study represents the first large-scale whole genome sequencing analysis of blood pressure phenotypes.
Variants across the allele frequency spectrum were investigated for associations with blood pressure and replication in large, independent stage-2 samples with imputed array or exome sequencing data.
What is relevant?
We identified two promising loci for BP, including a rare variant with large association size.
Aggregate analyses identified suggestive BP signals.
Clinical/Pathophysiological implications?
Rare, intergenic variants may have large associations with BP but require larger, diverse samples or family studies for confirmation.
Aggregate analyses may complement single variant analyses for rare variant discovery.
PERSPECTIVES.
Using a large-scale WGS discovery approach, the current study reported two novel variants for BP and highlighted the potential utility of aggregate analysis techniques as a complement to traditional single variant analyses for novel rare variant discovery. Reported signals included one promising but unconfirmed rare variant finding from the LOC100506274 locus, which achieved genome-wide significance in meta-analysis and stage-1 analysis but did not replicate at nominal significance in stage-2 analysis, and one common variant from the previously reported INSR locus. Aside from these signals, our findings were largely null and provide a similar lesson to that of early BP GWAS studies. Namely, even larger sample sizes will be needed to identify many rare variant signals for BP. With the forthcoming emergence of WGS data in all 500,000 UK Biobank participants, comprised predominantly of white British individuals, we speculate that new rare variant loci for BP will be discovered. WGS studies in much larger non-European populations will also be needed to identify ancestry-specific rare variants and avoid the Eurocentric biases long observed in GWAS.
ACKNOWLEDGEMENTS
Please see data supplement: http://hyper.ahajournals.org.
SOURCES OF FUNDING
Please see data supplement: http://hyper.ahajournals.org.
Non-standard abbreviations and acronyms
- Amish
The Amish Complex Disease Research Program
- ARIC
Atherosclerosis Risk in Communities
- BioMe
The IPM BioMe Biobank
- BMI
Body mass index
- BP
Blood pressure
- CARDIA
Coronary Artery Risk Development in Young Adults
- CCDG
Center for Common Disease Genomics
- CFS
Cleveland Family Study
- CHS
Cardiovascular Health Study
- dbGaP
Database of Genotypes and Phenotypes
- DBP
Diastolic BP
- eQTL
expression quantitative trait loci
- FHS
Framingham Heart Study
- GeneSTAR
Genetic Studies of Atherosclerosis Risk
- GENOA
Genetic Epidemiology Network of Arteriopathy
- GenSalt
Genetic Epidemiology Network of Salt-Sensitivity
- GOLDN
Genetics of Lipid Lowering Drugs and Diet Network
- GWAS
Genome-wide association studies
- HCHS-Sol
Hispanic Community Health Study-Study of Latinos
- HTN
Hypertension
- HyperGen
Hypertension Genetic Epidemiology Network
- JHS
Jackson Heart Study
- MAC
Minor allele count
- MAF
Minor allele frequency
- MESA
Multi-ethnic Study of Atherosclerosis
- MVP
Million Veteran Program
- NHLBI
National Heart, Lung, and Blood Institute
- NHGRI
National Human Genome Research Institute
- PCs
Principal components
- REGARDS
Reasons for Geographic and Racial Differences in Stroke
- Samoan
Samoan Study
- SBP
Systolic BP
- SMMAT-E
Set mixed model association test that efficiently combines the burden and sequence kernel association tests
- SNV
Single nucleotide variant
- THRV
Taiwan Study of Hypertension using Rare Variants
- TOPMed
Trans-Omics for Precision Medicine
- WES
Whole exome sequencing
- WGS
Whole genome sequencing
- WHI
Women’s Health Initiative
Footnotes
DISCLOSURES
Please see data supplement: http://hyper.ahajournals.org.
DATA AVAILABILITY
Genomic summary results for the TOPMed stage-1 analyses are available for controlled access through the database of Genotypes and Phenotypes (dbGaP) accession phs001974.
REFERENCES
- 1.Mills KT, Bundy JD, Kelly TN, Reed JE, Kearney PM, Reynolds K, Chen J, He J. Global Disparities of Hypertension Prevalence and Control. Circulation. 2016;134:441–450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lim SS, Vos T, Flaxman AD, Danaei G, Shibuya K, Adair-Rohani H, Amann M, Anderson HR, Andrews KG, Aryee M, et al. A comparative risk assessment of burden of disease and injury attributable to 67 risk factors and risk factor clusters in 21 regions, 1990–2010: a systematic analysis for the Global Burden of Disease Study 2010. Lancet. 2013;380:2224–2260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cutler JA, Follmann D, Allender PS. Randomized trials of sodium reduction: an overview. Am J Clin Nutr. 1997;65:643S–651S. [DOI] [PubMed] [Google Scholar]
- 4.Neter JE, Stam BE, Kok FJ, Grobbee DE, Geleijnse JM. Influence of Weight Reduction on Blood Pressure: A Meta-Analysis of Randomized Controlled Trials. Hypertension. 2003;42:878–884. [DOI] [PubMed] [Google Scholar]
- 5.Xin X, He J, Frontini MG, Ogden LG, Motsamai OI, Whelton PK. Effects of Alcohol Reduction on Blood Pressure. Hypertension. 2001;38:1112–1117. [DOI] [PubMed] [Google Scholar]
- 6.Newton-Cheh C, Larson MG, Vasan RS, Levy D, Bloch KD, Surti A, Guiducci C, Kathiresan S, Benjamin EJ, Struck J, et al. Association of common variants in NPPA and NPPB with circulating natriuretic peptides and blood pressure. Nat Genet. 2009;41:348–353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cho YS, Go MJ, Kim YJ, Heo JY, Oh JH, Ban H-J, Yoon D, Lee MH, Kim DJ, Park M, et al. A large-scale genome-wide association study of Asian populations uncovers genetic factors influencing eight quantitative traits. Nat Genet. 2009;41:527–534. [DOI] [PubMed] [Google Scholar]
- 8.Johnson T, Gaunt TR, Newhouse SJ, Padmanabhan S, Tomaszewski M, Kumari M, Morris RW, Tzoulaki I, O’Brien ET, Poulter NR, et al. Blood pressure loci identified with a gene-centric array. Am J Hum Genet. 2011;89:688–700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ganesh SK, Tragante V, Guo W, Guo Y, Lanktree MB, Smith EN, Johnson T, Castilly BA, Barnard J, Baumert J, et al. Loci influencing blood pressure identified using a cardiovascular gene-centric array. Hum Mol Genet 2013;22:1663–1678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Franceschini N, Fox E, Zhang Z, Edwards TL, Nalls MA, Sung YJ, Tayo BO, Sun YV, Gottesman O, Adeyemo A, et al. Genome-wide Association Analysis of Blood-Pressure Traits in African-Ancestry Individuals Reveals Common Associated Genes in African and Non-African Populations. Am J Hum Genet. 2013;93:545–554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tragante V, Barnes MR, Ganesh SK, Lanktree MB, Guo W, Franceschini N, Smith EN, Johnson T, Holmes MV, Padmanabhan S, et al. Gene-centric meta-analysis in 87,736 individuals of European ancestry identifies multiple blood-pressure-related loci. Am J Hum Genet. 2014;94:349–360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Simino J, Shi G, Bis JC, Chasman DI, Ehret GB, Gu X, Guo X, Hwang S-J, Sijbrands E, Smith AV, et al. Gene-age interactions in blood pressure regulation: a large-scale investigation with the CHARGE, Global BPgen, and ICBP Consortia. Am J Hum Genet. 2014;95:24–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ganesh SK, Chasman DI, Larson MG, Guo X, Verwoert G, Bis JC, Gu X, Smith AV, Yang M-L, Zhang Y, et al. Effects of long-term averaging of quantitative blood pressure traits on the detection of genetic associations. Am J Hum Genet. 2014;95:49–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhu X, Feng T, Tayo BO, Liang J, Young JH, Franceschini N, Smith JA, Yanek LR, Sun YV, Edwards TL, et al. Meta-analysis of correlated traits via summary statistics from GWASs with an application in hypertension. Am J Hum Genet. 2015;96:21–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kato N, Takeuchi F, Tabara Y, Kelly TN, Go MJ, Sim X, Tay WT, Chen C-H, Zhang Y, Yamamoto K, et al. Meta-analysis of genome-wide association studies identifies common variants associated with blood pressure variation in east Asians. Nat Genet. 2011;43:531–538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Deo R, Nalls MA, Avery CL, Smith JG, Evans DS, Keller MF, Butler AM, Buxbaum SG, Li GMiguel Quibrera P, et al. Common genetic variation near the connexin-43 gene is associated with resting heart rate in African Americans: a genome-wide association study of 13,372 participants. Hear Rhythm. 2013;10:401–408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ehret GB, Ferreira T, Chasman DI, Jackson AU, Schmidt EM, Johnson T, Thorleifsson G, Luan J, Donnelly LA, Kanoni S, et al. The genetics of blood pressure regulation and its target organs from association studies in 342,415 individuals. Nat Genet. 2016;48:1171–1184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hoffmann TJ, Ehret GB, Nandakumar P, Ranatunga D, Schaefer C, Kwok PY, Iribarren C, Chakravarti A, Risch N. Genome-wide association analyses using electronic health records identify new loci influencing blood pressure variation. Nat Genet. 2017;49:54–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Liang J, Le TH, Edwards DRV, Tayo BO, Gaulton KJ, Smith JA, Lu Y, Jensen RA, Chen G, Yanek LR, et al. Single-trait and multi-trait genome-wide association analyses identify novel loci for blood pressure in African-ancestry populations. PLoS Genet. 2017;13: e1006728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wain LV, Vaez A, Jansen R, Joehanes R, van der Most PJ, Erzurumluoglu AM, O’Reilly PF, Cabrera CP, Warren HR, Rose LM, et al. Novel Blood Pressure Locus and Gene Discovery Using Genome-Wide Association Study and Expression Data Sets From Blood and the Kidney. Hypertenion 2017;70:e4–e19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Warren HR, Evangelou E, Cabrera CP, Gao H, Ren M, Mifsud B, Ntalla I, Surendran P, Liu C, Cook JP, et al. Genome-wide association analysis identifies novel blood pressure loci and offers biological insights into cardiovascular risk. Nat Genet. 2017;49;403–415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Evangelou E, Warren HR, Mosen-Ansorena D, Mifsud B, Pazoki R, Gao H, Ntritsos G, Dimou N, Cabrera CP, Karaman I, et al. Genetic analysis of over 1 million people identifies 535 new loci associated with blood pressure traits. Nat Genet. 2018;50:1412–1425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Giri A, Hellwege JN, Keaton JM, Park J, Qiu C, Warren HR, Torstenson ES, Kovesdy CP, Sun YV, Wilson OD, et al. Trans-ethnic association study of blood pressure determinants in over 750,000 individuals. Nat Genet. 2019;51:51–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Takeuchi F, Akiyama M, Matoba N, Katsuya T, Nakatochi M, Tabara Y, Narita A, Saw W-Y, Moon S, Spracklen CN, et al. Interethnic analyses of blood pressure loci in populations of East Asian and European descent. Nat Commun. 2018;9:5052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Takeuchi F, Isono M, Katsuya T, Yamamoto K, Yokota M, Sugiyama T, Nabika T, Fujioka A, Ohnaka K, Asano H, et al. Blood pressure and hypertension are associated with 7 loci in the Japanese population. Circulation. 2010;121:2302–2309. [DOI] [PubMed] [Google Scholar]
- 26.Ho JE, Levy D, Rose L, Johnson AD, Ridker PM, Chasman DI. Discovery and replication of novel blood pressure genetic loci in the Women’s Genome Health Study. J Hypertens. 2011;29:62–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kato N, Loh M, Takeuchi F, Verweij N, Wang X, Zhang W, Kelly TN, Saleheen D, Lehne B, Leach IM, et al. Trans-ancestry genome-wide association study identifies 12 genetic loci influencing blood pressure and implicates a role for DNA methylation. Nat Genet. 2015;47:1282–1293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Padmanabhan S, Melander O, Johnson T, Di Blasio AM, Lee WK, Gentilini D, Hastie CE, Menni C, Monti MC, Delles C, et al. Genome-wide association study of blood pressure extremes identifies variant near UMOD associated with hypertension. PLoS Genet. 2010;6:e1001177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Johnson AD, Newton-Cheh C, Chasman DI, Ehret GB, Johnson T, Rose L, Rice K, Verwoert GC, Launer LJ, Gudnason V, et al. Association of hypertension drug target genes with blood pressure and hypertension in 86 588 individuals. Hypertension. 2011;57:903–910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wain LV, Verwoert GC, O’Reilly PF, Shi G, Johnson T, Johnson AD, Bochud M, Rice KM, Henneman P, Smith AV, et al. Genome-wide association study identifies six new loci influencing pulse pressure and mean arterial pressure. Nat Genet. 2011;43:1005–1011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ehret GB, Munroe PB, Rice KM, Bochud M, Johnson AD, Chasman DI, Smith AV, Tobin M, Verwoert GC, Hwang SJ, et al. Genetic variants in novel pathways influence blood pressure and cardiovascular disease risk. Nature. 2011;478:103–109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Newton-Cheh C, Johnson T, Gateva V, Tobin MD, Bochud M, Coin L, Najjar SS, Zhao JH, Heath SC, Eyheramendy S, et al. Genome-wide association study identifies eight loci associated with blood pressure. Nat Genet. 2009;41:666–676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Surendran P, Drenos F, Young R, Warren H, Cook JP, Manning AK, Grarup N, Sim X, Barnes DR, Witkowska K, et al. Trans-ancestry meta-analyses identify rare and common variants associated with blood pressure and hypertension. Nat Genet. 2016;48:1151–1161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yu B, Pulit SL, Hwang S-J, Brody JA, Amin N, Auer PL, Bis JC, Boerwinkle E, Burke GL, Chakravarti A, et al. Rare Exome Sequence Variants in CLCN6 Reduce Blood Pressure Levels and Hypertension Risk. Circ Cardiovasc Genet. 2016;9:64–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Liu C, Kraja AT, Smith JA, Brody JA, Franceschini N, Bis JC, Rice K, Morrison AC, Lu Y, Weiss S, et al. Meta-analysis identifies common and rare variants influencing blood pressure and overlapping with metabolic trait loci. Nat Genet. 2016;48:1162–1170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Surendran P, Feofanova EV, Lahrouchi N, Ntalla I, Karthideyan S, Cook J, Chen L, Mifsud B, Yao C, Kraja AT, et al. Discovery of rare variants associated with blood pressure regulation through meta-analysis of 1.3 million individuals. Nat Genet. 2020; 52:1314–1332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Taliun D, Harris DN, Kessler MD, Carlson J, Szpiech ZA, Torres R, Gagliano Taliun SA, Corvelo A, Gogarten SM, Kang HM, et al. Sequencing of 53,831 diverse genomes from the NHLBI TOPMed Program. Nature 2021; 590:290–299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Jun G, Wing MK, Abecasis GR, Kang HM. An efficient and scalable analysis framework for variant extraction and refinement from population-scale DNA sequence data. Genome Res. 2015;25:918–925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tobin MD, Sheehan NA, Scurrah KJ, Burton PR. Adjusting for treatment effects in studies of quantitative traits: Antihypertensive therapy and systolic blood pressure. Stat Med. 2005;24:2911–2935. [DOI] [PubMed] [Google Scholar]
- 40.Brody JA, Morrison AC, Bis JC, O’Connell JR, Brown MR, Huffman JE, Ames DC, Carroll A, Conomos MP, Gabriel S, et al. Analysis commons, a team approach to discovery in a big-data environment for genetic epidemiology. Nat Genet. 2017;49:1560–1563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Conomos MP, Reiner AP, Weir BS, Thornton TA. Model-free Estimation of Recent Genetic Relatedness. Am J Hum Genet. 2016;98:127–148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Liu X, White S, Peng B, Johnson AD, Brody JA, Li AH, Huang Z, Carroll A, Wei P, Gibbs R, Klein RJ, Boerwinkle E. WGSA: an annotation pipeline for human genome sequencing studies. J Med Genet. 2016;53:111–112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rogers MF, Shihab HA, Mort M, Cooper DN, Gaunt TR, Campbell C. FATHMM-XF: accurate prediction of pathogenic point mutations via extended features. Bioinformatics. 2018;34:511–513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Fishilevich S, Nudel R, Rappaport N, Hadar R, Plaschkes I, Iny Stein T, Rosen N, Kohn A, Twik M, Safran M, Lancet D, Cohen D. GeneHancer: genome-wide integration of enhancers and target genes in GeneCards. Database. 2017;bax028. [DOI] [PMC free article] [PubMed]
- 45.Lizio M, Harshbarger J, Shimoji H, Severin J, Kasukawa T, Sahin S, Abugessaisa I, Fukada S, Fumi H, Ishikawa-Kato S, et al. Gateways to the FANTOM5 promoter level mammalian expression atlas. Genome Biol. 2015;16:22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Chen H, Huffman JE, Brody JA, Wang C, Lee S, Li Z, Gogarten SM, Sofer T, Bielak LF, Bis JC, et al. Efficient Variant Set Mixed Model Association Tests for Continuous and Binary Traits in Large-Scale Whole-Genome Sequencing Studies. Am J Hum Genet. 2019;104:260–274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Das S, Forer L, Schönherr S, Sidore C, Locke AE, Kwong A, Vrieze SI, Chew EY, Levy S, McGue M, et al. Next-generation genotype imputation service and methods. Nat Genet. 2016;48:1284–1287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Loh PR, Danecek P, Palamara PF, Fuchsberger C, Reshef YA, Finucane HK, Schoenherr S, Forer L, McCarthy S, Abecasis GR, Drubin R, Price AL. A. Reference-based phasing using the Haplotype Reference Consortium panel. Nat Genet. 2016;48:1443–1448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Casper J, Zweig AS, Villarreal C, Tyner C, Speir ML, Rosenbloom KR, Raney BJ, Lee CM, Lee BT, Karolchik D, et al. The UCSC Genome Browser database: 2018 update. Nucleic Acids Res. 2017;46:D762–D769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zhou W, Nielsen JB, Fritsche LG, Dey R, Gabrielsen ME, Wolford BN, LeFaive J, VandeHaar P, Gagliano SA, Gifford A, et al. Efficiently controlling for case-control imbalance and sample relatedness in large-scale genetic association studies. Nat Genet. 2018;50:1335–1341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.1000 Genomes Project Consortium, Auton A, Brooks LD, Durbin RM, Garrison EP, Kang HM, Korbel JO, Marchini JL, McCarthy S, McVean GA, Abecasis GR. A global reference for human genetic variation. Nature. 2015;526:68–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Fagerberg L, Hallstrom BM, Oksvold P, Kampf C, Djureinovic D, Odeberg J, Havuka M, Tahmasebpoor S, Danielsson A, Edlund K, et al. Analysis of the human tissue-specific expression by genome-wide integration of transcriptomics and antibody-based proteomics. Mol Cell Proteomics. 2014;13:397–406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kiezun A, Pulit SL, Francioli LC, van Dijk F, Swertz M, Boosmsma DI, van Duijn CM, Slagboom PE, van Ommen GJB, Wijmenga C, Genome of the Netherlands Consortium, de Bakker PIW, Sunyaev SR. Deleterious Alleles in the Human Genome Are on Average Younger Than Neutral Alleles of the Same Frequency. PLoS Genet. 2013;9:e1003301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Mathieson I, McVean G. Demography and the Age of Rare Variants. PLoS Genet. 2014;10:e1004528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Liu DJ, Peloso GM, Zhan X, Holmen OL, Zawistowski M, Feng S, Nikpay M, Auer PL, Goel A, Zhang H, et al. Meta-analysis of gene-level tests for rare variant association. Nat. Genet. 2014;46:200–204. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Genomic summary results for the TOPMed stage-1 analyses are available for controlled access through the database of Genotypes and Phenotypes (dbGaP) accession phs001974.