Abstract
NLRP3 inflammasome is suggested to contribute to the complex pathogenesis of systemic lupus erythematosus, but its role in cutaneous lupus erythematosus has not been addressed. This study investigated the expression of NLRP3 inflammasome components and levels of type I interferons in the skin of 20 patients with cutaneous lupus erythematosus. Expression of NLRP1/3, adaptor protein ASC (apoptosis-associated speck-like protein), caspase-1, interferon-α (IFN-α), myxovirus resistance protein (MxA), and interferon-induced proteins 1 and 2 (IFIT 1/2) in the skin was assessed using reverse transcription quantitative real-time PCR (RT-qPCR), western blotting and immunohistochemistry. Serum interferon-α protein levels from 12 patients were measured using digital enzyme-linked immunoassay (ELISA). Interleukin-1β expression was significantly upregulated in the lesional skin of patients with cutaneous lupus erythematosus compared with their uninvolved skin. However, NLRP1/3, ASC and caspase-1 were not significantly upregulated compared with the skin of control persons. IFN-α and IFN-induced proteins MxA and IFIT1/2 were strongly expressed in cutaneous lupus erythematosus skin. Variability in the expression of NLRP3 inflammasome components among patients suggests heterogeneity of pathological pathways in cutaneous lupus erythematosus.
Key words: cutaneous lupus erythematosus, inflammasomes, IL-1 beta, type I interferon
Lupus erythematous (LE) is an autoimmune disease with a broad spectrum of symptoms and varying organ involvement, with potentially severe clinical manifestations (1, 2). Cutaneous manifestations are the second most common symptom in patients with systemic lupus erythematosus (SLE) after articular involvement, as approximately 80% of patients have cutaneous manifestations (3, 4). The disease can also be limited to skin. Cutaneous LE (CLE) is divided into different categories, based on the clinical appearance of skin symptoms, the presence of autoantibodies, and association with systemic symptoms (3, 4).
SIGNIFICANCE
This study suggests that NLRP3 inflammasome is activated in cutaneous lupus erythematosus lesions and contributes to the ongoing chronic inflammation. The variation between patients observed in this study suggests that the NLRP3 inflammasome pathway might not be a primary determinant in all patients, but, for a subgroup of patients interleukin-1β might represent a feasible target for treatment. Upregulation of type I interferons is an established part of the pathogenesis of lupus erythematosus, but the expression of interferon-induced proteins 1 and 2 (IFIT 1/2) in lesional lupus skin represents a novel finding.
The pathophysiology of lupus involves a genetic predisposition and environmental triggers that initiate the disease process, resulting in activation of both the innate and adaptive immune systems and production of autoantibodies (1, 5). It is unclear why only some of the patients develop systemic disease. Type I interferons (IFNs) have and important role in the pathogenesis of lupus, as do T and B cell function abnormalities (1, 6–10). In CLE, keratinocytes have a pathogenic role. Ultraviolet (UV) radiation induces keratinocyte apoptosis, immune cell infiltration and cytokine production in the skin (9–11). In patients with lupus, the clearance of apoptotic bodies from the skin is diminished, leading to prolonged exposure of intra-cellular proteins to immune cells, causing the formation of autoantibodies (5, 11). The complement system is important in the removal of immune complex, and complement deficiencies are associated with the development of lupus (2, 5, 12).
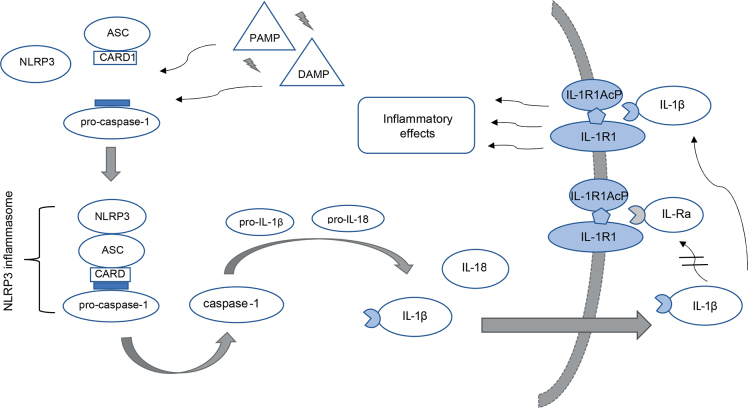
In SLE, the role of inflammasomes in chronic inflammation has been documented. Inflammasomes, a part of the innate immune response, are protein complexes found mostly in innate immune cells and in keratinocytes (13). The NLRP3 inflammasome consists of a nucleotide-binding domain-like receptor with pyrin-domain 3 (NLRP3), adaptor protein ASC (apoptosis-associated speck-like protein) and pro-caspase-1 (14). ASC has a caspase-recruitment domain (CARD), which enables self-cleavage of pro-caspase-1 into active caspase-1 (15) (Fig. 1). Caspase-1, in turn, cleaves the cytokines pro-interleukin-1β and pro-interleukin-18 to their highly proinflammatory active forms (16) (Fig. 1). NLRP3 inflammasome activation can be induced by various exogenous stimuli and has been linked to the pathogenesis of SLE and other autoimmune and autoinflammatory diseases (13, 14, 17). In lupus macrophages, neutrophil extracellular traps (NETs) activate NLRP3 inflammasome with consequent production of interleukin (IL)-1β and IL-18 (18). In the presence of anti-double-stranded (ds)DNA antibodies, which are found in patients with SLE, self-dsDNA activates the NLPR3 inflammasome and increases IL-1β secretion in monocytes (19, 20). Monocytes from the peripheral blood of patients with SLE show increased gene expression of IL-1β even in the resting state, and increased expression of NLRP1, caspase-1 and IL-1β genes when stimulated with lipopolysaccharide and adenosine triphosphate (21). The NLRP3 inflammasome has been associated with the pathogenesis of lupus nephritis in murine models and the development of cardiovascular diseases in patients with lupus (22). Belonging to the same group of NLR inflammasomes, NLRP1 has been less extensively studied in autoinflammatory diseases, but single nucleotide polymorphisms in the NLRP1 gene have been associated with SLE (23).
Fig. 1.
The NLRP3 inflammasome is a cytosolic protein complex consisting of NLRP3, ASC (apoptosis-associated speck-like protein) and pro-caspase-1. The inflammasome assembly can be triggered by various pathogen-associated molecular patterns (PAMPs) and danger-associated molecular patterns (DAMPs). Upon activation, pro-caspase-1 is turned into its active form, caspase-1. Caspase-1 cleaves pro-interleukin (IL)-1β and pro-IL-18 into their mature forms, with ensuing secretion from the cell. IL-1β binds to its receptor complex consisting of IL-1R1 and IL-R1AcP and initiates a signal cascade leading to proinflammatory effects. IL-Ra is an IL-1 receptor antagonist and regulates the activity of IL-1β by competing with binding to the receptor.
In CLE, the role of the NLR inflammasomes is less studied. It is known, that UV radiation, which also aggravates the symptoms of CLE, activates NLRP3 inflammasome-dependent secretion of IL-1β and IL-18 in keratinocytes (24, 25). The current study assessed NLRP1 and NLRP3 inflammasome activity in the skin lesions of patients with discoid LE (DLE, a subtype of chronic CLE), subacute CLE (SCLE) and SLE. In addition, serum IFN-α levels, and the cutaneous expression of IFN-α, myxovirus resistance protein I (MxA) and IFN-induced protein with tetratricopeptide repeats 1 and 2 (IFIT1/2) were analysed.
MATERIALS AND METHODS
Patient samples
Twenty patients with clinically and histologically/immunohistologically confirmed CLE were recruited from patients treated at the Department of Dermatology, Helsinki University Hospital (HUH), Helsinki. Inclusion criteria were clinically confirmed CLE or SLE with active lupus skin lesion(s) at a site feasible for obtaining biopsies, and age above 18 years. The study was approved by the HUH Ethical Review Committee and all participants signed an informed consent.
Of the 20 patients, 8 had DLE, 7 had SCLE and 5 had SLE with skin manifestations. Seventeen of the patients were female and 3 were male, and their mean age was 57.8 years. The mean time elapsed from diagnosis was 8.6 years. All except 2 patients had used topical steroid and/or calcineurin inhibitor cream for treatment of skin lesions. Fourteen patients had systemic anti-inflammatory medication at the time of biopsy. Disease activity was assessed with Cutaneous Lupus Erythematosus Disease Area and Severity Index (CLASI), mean score 10.5. Characteristics of the study participants are shown in Table I.
Table I.
Characteristics of the study participants
| Subject | Diagnosis | Sex | Age, years | Time elapsed from diagnosis, years | Anti-inflammatory medication | ANAAb | ENAAb | CLASI |
|---|---|---|---|---|---|---|---|---|
| 1 | DLE | Male | 30 | 2 | HCQ | Negative | Negative | 6 |
| 2 | DLE | Female | 51 | 12 | mepacrine | Negative | RNP | 9 |
| 3 | DLE | Male | 53 | 11 | None | Negative | Negative | 10 |
| 4 | DLE | Female | 56 | >25 | None | Negative | SSB | 18 |
| 5 | DLE | Female | 59 | 0 | HCQ + MTX | Negative | SSA | 21 |
| 6 | DLE | Female | 63 | 2 | NONE | Positive | ND | 5 |
| 7 | DLE | Female | 64 | 1 | MTX | Negative | Positive | 9 |
| 8 | DLE | Female | 80 | 7 | HCQ | Negative | Negative | 5 |
| 9 | SCLE | Female | 57 | 0 | NONE | Negative | SSA | 9 |
| 10 | SCLE | Male | 59 | 1 | PRED | Negative | SSA | 12 |
| 11 | SCLE | Female | 62 | 4 | HCQ | Positive | SSA, SSB | 8 |
| 12 | SCLE | Female | 67 | 0 | PRED + etanercept | Positive | SSA | 18 |
| 13 | SCLE | Female | 67 | 0 | None | Negative | SSA | 10 |
| 14 | SCLE | Female | 74 | 3 | HCQ | Positive | SSA | 7 |
| 15 | SCLE | Female | 83 | 0 | None | Positive | SSA, SSB | 8 |
| 16 | SLE | Female | 19 | 12 | HCQ + PRED | Positive | SSA, SSB | 4 |
| 17 | SLE | Female | 40 | 26 | HCQ + MTX + PRED | Positive | SSA, SSB | 16 |
| 18 | SLE | Female | 46 | 30 | HCQ + MTX + PRED + infliximab | Positive | RNP | 14 |
| 19 | SLE | Female | 56 | 10 | HCQ + PRED + rituximab | Positive | SSA, SSB | 16 |
| 20 | SLE | Female | 69 | 25 | HCQ + PRED | Positive | SSA | 5 |
Medication dosages at the time of sampling: hydroxychloroquine 100–400 mg/day, mepacrine 100 mg/day, methotrexate 7.5–20 mg/week, prednisolone: 60 mg/day (patient 10), 20 mg/day (patient 12), 5 mg every other day (patient 16), 5 mg/day (patient 17), 5 mg/day (patient 18), 45 mg/day (patient 19) and 10 mg/day (patient 20). DLE: discoid lupus erythematosus; SCLE: subacute cutaneous lupus erythematosus; SLE: systemic lupus erythematosus; HCQ: hydroxychloroquine; MTX: methotrexate; PRED: prednisolone; ANAAb: anti-nuclear antibody; ENAAb: extractable nuclear antibodies; RNP: ribonucleoprotein; SSA: anti-Ro; SSB: anti-La.
Skin biopsies (6-mm punch or excision) were obtained as follows. From 12 patients (Table I: DLE 4, 5, 6, 8; SCLE 9, 11, 12, 13, 14, 15; SLE 19, 20), 3 skin biopsies were obtained: 1 from lesional skin and 1 from anatomically corresponding healthy skin in Allprotect Tissue Reagent (Qiagen, Venlo, The Netherlands), and 1 from lesional skin in formalin. Also, serum samples were taken. From 8 patients (Table I: DLE 1, 2, 3, 7; SCLE 10; SLE 16, 17, 18), 3 skin biopsies were obtained: 1 from lesional skin and 1 from corresponding healthy skin as fresh-frozen samples, and 1 from lesional skin in formalin. Serum and fresh skin samples were stored at –80°C and Allprotect samples at –20°C.
Control skin samples (4 fresh-frozen, 4 in Allprotect, and 4 in formalin) were obtained from 12 persons not diagnosed with LE or similar disease undergoing skin graft procedures at the department’s dermatosurgery unit. All samples were processed within 18 months.
Western blot
Protein expression was analysed in fresh-frozen skin samples by western blot (WB). To achieve homogenization of skin biopsies, they were embedded in Tissue-Tek OCT Compound (Sakura Finetek Europe, Aalphen aan den Rijn, The Netherlands) and sliced using a cryo-microtome to 30 µm thickness. After washing with phosphate-buffered saline (PBS), samples were incubated in a modified RIPA lysis buffer (50 mM Tris pH 8, 150 mM NaCl, 0.5% NaDeoxycholate, 1% Triton, 1% SDS, all reagents Sigma-Aldrich/Merck, Darmstadt, Germany) over ice for 50 min. After centrifugation, the protein concentrations were determined with DC Protein Assay Kit (BioRad, Hercules, CA, USA) and absorbances measured using Multiskan EX v 2.3. spectrophotometer (Thermo Fisher Scientific, Waltham, MA, USA). The samples were run in 12% SDS-PAGE gels and transferred onto nitrocellulose membranes (Whatman, GE Healthcare Life Sciences, Chicago, IL, USA). After blocking with 5% milk in TBS/PBS-Tween 20 for 1 h at room temperature (RT), the membranes were incubated in primary antibodies diluted in 5% milk-TBS-Tween 20 overnight at +4°C. Antibody concentrations were: IL-1β (sc-7884, Santa Cruz Biotechnology, Dallas, TX, USA, 1:1000 – 1:250), caspase-1 (sc-515, Santa Cruz Biotechnology, 1:1000; 2225, Cell Signaling Technology, Danvers, MA, USA, 1:1000), ASC (AL-177, AdipoGen Life Sciences, San Diego, CA, USA, 1:1000) and β-actin (4970, Cell Signaling Technology, 1:5000). After washing with PBS-Tween 20, membranes were incubated in horse-radish peroxidase (HRP)-conjugated secondary antibody (Millipore/Merck, Darmstadt, Germany) in 5% milk-PBS-Tween 20 for 1 h at RT. BioRad ECL solution (BioRad) was used for development on film (Fuji Kodachrome).
Immunohistochemistry
Immunohistochemistry was performed with formalin-fixed paraffin-embedded (FFPE) skin samples. After deparaffinization, antigen epitope retrieval was performed using citrate buffer pH 6 and microwave heating. The sections were blocked with 2.5% normal horse blocking serum (ImmPRESS HRP Universal Antibody Polymer Detection Kit, MP-7500, Vector Laboratories, Burlingame, CA, USA) and incubated overnight at +4°C with primary antibodies diluted in 3% bovine serum albumin (BSA) as follows: NLRP3 (ab214185, Abcam, Cambridge, UK, 1:100), caspase-1 (2225, Cell Signaling Technology, 1:100), ASC (AL-177, AdipoGen Life Sciences, 1:500), IFIT1 (HPA055380, Sigma-Aldrich/Merck, 1:1000) and IFIT2 (HPA003408, Sigma-Aldrich/Merck, 1:350). After washing with PBS, the sections were incubated with secondary antibody (ImmPRESS HRP Universal Antibody Polymer Detection Kit, MP-7500, Vector Laboratories) for 30 min at RT. Bound antibodies were visualized using DAB Peroxidase (HRP) Substrate Kit (SK-4100, Vector Laboratories), except for IFIT2 with NovaRED Peroxidase (HRP) Substrate Kit (SK-4800, Vector Laboratories). Mayer’s haematoxylin was used as counterstain. Slides were mounted with Pertex mounting media (Histolab, Göteborg, Sweden). For negative control, 3% BSA without primary antibody was used. Rabbit immumnoglobulin (Ig)G isotype control (08-6199, Invitrogen, Rockford, IL, USA), NLRP1 (NBP1-97593, Novus Biologicals, Centennial, CO, USA, 1:100), MX1/MxA (D3W71, Cell Signaling Technology, 1:300) and IL-1β (3A6/12242, Cell Signaling Technology, 1:100) staining was performed with Thermo Scientific Lab Vision Autostainer 480 S (Thermo Fischer Scientific) using DAB Substrate (WellMed BV, GX Duiven, The Netherlands) and Mayer’s haematoxylin. The protocol was similar to manual staining, with the exception that primary antibody incubation was 30 min at RT. Reagents used for Autostainer protocol: Normal antibody diluent, Post-antibody blocking and Poly-HRP-Anti Mouse/Rabbit IgG (all ImmunoLogic a VWR Company, Amsterdam, The Netherlands). For IL-1β negative controls, mouse IgG Isotype Control (08-6599, Invitrogen, Rockford, IL, USA) was used.
RNA extraction and RT-qPCR analysis
RNA was extracted from biopsies stored in Allprotect Tissue Reagent with RNeasy Lipid Tissue Mini Kit (Qiagen). Over ice, the samples were cut to small pieces and incubated in QIAzol Lysis Reagent (RNeasy Lipid Tissue Mini Kit) for 30–60 min. Extraction was completed following the manufacturer’s instructions. RNAs were stored at –80°C until RNA concentration and purity were quantified with NanoDrop (Thermo Fisher Scientific). Complementary DNA synthesis was performed with High-Capacity cDNA Reverse Transcription Kit (4368814, Applied Biosystems/Thermo Fisher Scientific) according to manufacturer’s instructions, in a Finnzymes DNA Engine gradient cycler (Thermo Fisher Scientific). Quantitative PCR was run with iQ SYBR Green Supermix (BioRad) using the LightCycler 480 machine and software set (Roche, Basel, Switzerland). Primers were for caspase-1 (Microsynth, Balgach, Switzerland), IL-1β (Microsynth) and IFN-α1 (Oligomer, Helsinki, Finland). Glyceraldehyde 3-phosphate dehydrogenase (GAPDH) (Oligomer) was used as a reference gene. Primer sequences and additional information are presented in Tables SI and SII. Melting curve analysis was performed to assess the specificity of the amplified product. Detailed information on reverse transcription quantitative real-time PCR (RT-qPCR) reaction protocol is given in Table SIII. qPCR data analysis (mRNA relative fold-change) was performed using the –2–ΔΔCt method (26). For the relative expression calculations, the sample for comparison was chosen in random from the control person group.
Serum IFN-α protein levels were quantified from serum samples (12 patients) using a digital-ELISA assay (27) (Simoa assay, Quanterix, Lexington, MA, USA), in accordance with the manufacturer’s Homebrew Simoa Assay Kit instructions, utilizing 2 autoantibodies specific for IFN-α isolated and cloned from 2 patients with autoimmune polyendocrinopathy syndrome type 1 (APS1) (28). The 8H1 antibody clone was used as a capture antibody, after coating on paramagnetic beads (0.3 mg/ml), and the 12H5 antibody was biotinylated (biotin-to-antibody ratio 30:1) and used as the detector. Recombinant IFNa17/aI (PBL Assay Science, Piscataway, NJ, USA) was used to generate a standard curve, after cross-reactivity testing. Each serum sample was analysed in duplicate and in dilution 1:10. The limit of detection (LOD) was calculated as the mean value +3SD of reactivity from all blank runs, and found to be 0.63 fg/mL, including the dilution factor.
Statistical analyses were performed with IBM SPSS Statistics 24.
RESULTS
NLRP1, NLRP3 and ASC are expressed in the skin of both patients with cutaneous lupus erythematosus and control persons
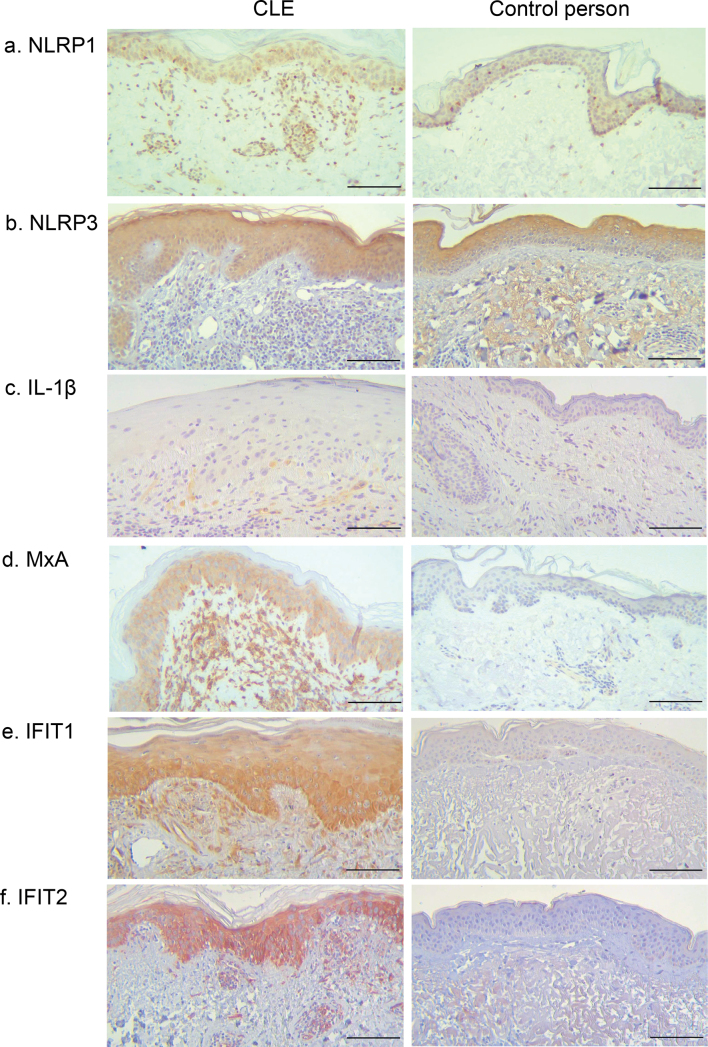
Immunohistochemistry revealed NLRP1 protein expression in all CLE skin samples (19/19, 1 sample was excluded due to poor quality of tissue slide) and in control person samples (Fig. 2a). Expression was found both in keratinocytes and inflammatory cells (lymphocytes) in all but 5 patients with CLE, who only showed expression in lymphocytes. Control person samples showed similarly positivity in both keratinocytes and inflammatory cells (lymphocytes); however, the number of inflammatory cells was significantly lower in control skin compared with CLE. NLRP3 protein was also detected by immunohistochemistry in all CLE (20/20) and control person skin samples (Fig. 2b). NLRP3 was expressed mainly in keratinocytes, but also in lymphocytes and monocytes in 50% of the lupus samples. Expression was comparable in all lupus subtypes. Control person sample keratinocytes showed NLRP3 expression, while the low number of immune cells did not show any positivity.
Fig. 2.
Representative immunostaining images of cutaneous lupus erythematosus (CLE) patients and control persons. Scale bar: 100 μm. (a) Patient 12 (subacute CLE), (b) patient 1 (discoid LE; DLE), (c) patient 7 (DLE), (d) patient 16 (systemic lupus erythematosus), (e) patient 5 (DLE), (f) patient 12 (subacute CLE).
ASC was detected by WB in all CLE skin biopsies, both in healthy and lesional skin of the patients (Fig. 3). ASC expression was most prominent in DLE and SLE samples, but low expression was observed in the skin of control persons (Fig. 3). Immunohistochemistry confirmed ASC positivity in all CLE and control person skin samples and was detected both in keratinocytes and immune cells (mostly lymphocytes).
Fig. 3.
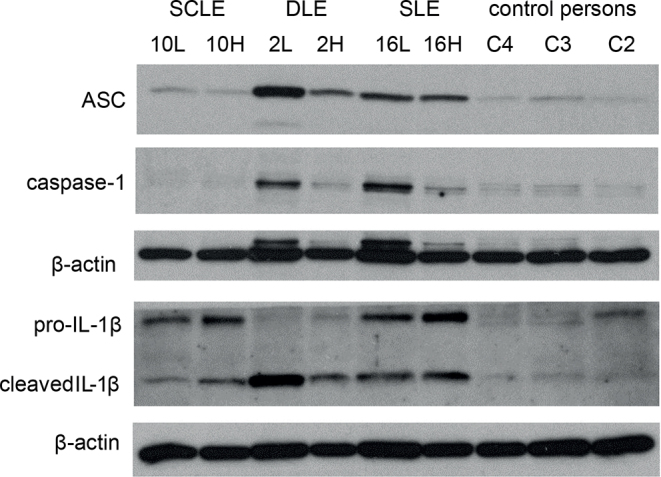
ASC (apoptosis-associated speck-like protein), caspase-1 and interleukin (IL)-1β protein expression in the skin of 3 patients with cutaneous lupus erythematosus (CLE) and 3 control persons, as detected by western blot. From patients with CLE there were 2 biopsies: 1 from lesional skin (L) and 1 from corresponding healthy skin (H). DLE: discoid lupus erythematosus; SCLE: subacute cutaneous lupus erythematosus.
Caspase-1 is expressed in lesional cutaneous lupus erythematosus skin
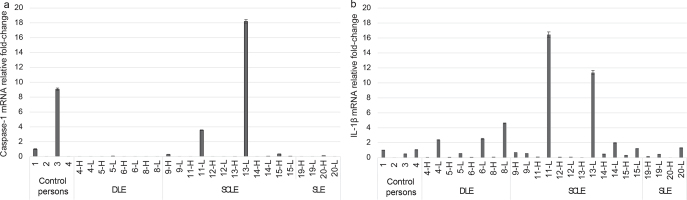
Caspase-1 mRNA was clearly increased in the lesional skin of 2 patients with CLE, both of whom had SCLE (Fig. 4a). The patient with the highest relative fold-change increase in caspase-1 mRNA (case 13) had been recently diagnosed with the disease and, at the time of sampling, was not yet using any systemic anti-inflammatory medication. Caspase-1 protein could be detected by WB in 4/8 lesional lupus skin samples (2 SLE and 2 DLE) and only minutely in control person skin (Fig. 3). Of note, due to the availability of patient samples and restrictions in sample size, 12 samples were analysed with RT-qPCR and another 8 samples with WB (see Materials and Methods section). Immunohistochemistry showed caspase-1 positivity in all CLE and control person samples.
Fig. 4.
(a) Caspase-1 mRNA relative fold-change and (b) interleukin (IL)-1β mRNA relative fold-change in the skin of discoid lupus erythematosus (DLE), subacute cutaneous lupus erythematosus (SCLE) and systemic lupus erythematosus (SLE) patients and control persons. From the study subjects there were 2 biopsies: 1 from cutaneous lupus erythematosus (CLE) lesion (L) and 1 from corresponding healthy skin (H).
The specificity of the polyclonal antibodies (NLRP3, ASC, caspase-1) was confirmed by immunostaining with polyclonal rabbit IgG isotype control antibody, yielding negative staining. In addition, staining with BSA without primary antibody yielded negative results.
Increased expression of interleukin-1β in the skin of patients with cutaneous lupus erythematosus
Pro-IL-1β was detected by WB in 7/8 lupus skin samples, and with mature (cleaved) form of IL-1β in 5 of those samples (Fig. 3). IL-1β was detected in both lesional and healthy skin of the patients with CLE. Control person skin samples showed no, or very low, amount, of IL-1β. IL-1β mRNA was increased more than 2-fold in 5/12 patients with CLE, especially in DLE and SCLE subtypes (Fig. 4b). IL-1β mRNA increase was observed in the lesional, but not in the healthy, skin sample of each patient, and the difference was statistically significant (p < 0.01, Mann–Whitney U test). By immunohistochemistry, IL-1β positivity was seen in 7/20 of CLE skin samples (3 DLE, 3 SCLE, 1 SLE) with expression in immune cells (macrophages, 5 patients) and in keratinocytes (2 patients) (Fig. 2c). Expression was not found in control person samples.
The highest fold-change increases in IL-1β correlated with highest caspase-1 mRNA fold-changes (Fig. 4). The patients with increased skin IL-1β mRNA were using either no anti-inflammatory medications or only hydroxychloroquine; in the skin of the patients who used combination of 2 or more drugs there was no IL-1β mRNA observed, even though they had an active disease measured by CLASI score (Table I).
High expression of type I interferon and interferon-induced proteins in patients with cutaneous lupus erythematosus
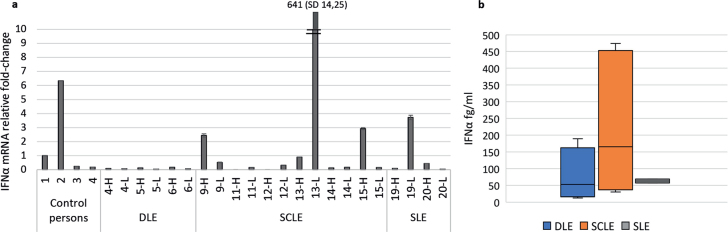
Expression of IFN-α mRNA was increased in the skin of 4/11 patients (3 patients with SCLE and 1 with SLE) and 1/4 control persons (Fig. 5a). One patient sample was excluded from the analysis due to high variability between reaction replicates. Very high IFN-α fold-change (more than 600-fold) was observed in the skin of the same patient with SCLE who also had high caspase-1 and IL-1β mRNA fold-change (case 13). The other patients with IFN-α mRNA fold-change increase did not have increased caspase-1 or IL-1β mRNA in their skin. Serum IFN-α levels were highest among patients with SCLE (Fig. 5b).
Fig. 5.
Interferon (IFN)-α1 mRNA in the skin and IFN-α levels in the serum of patients with cutaneous lupus erythematosus (CLE). (a) IFN-α1 mRNA fold-change in the skin of lupus patients and control persons. From lupus patients, skin biopsies were both form lesional skin (L) and corresponding healthy skin (H). (b) Serum IFN-α levels from lupus patients quantified with Simoa digital enzyme-linked immunoassay (ELISA). In discoid LE (DLE) patients range 12.7–189.5 fg/ml, mean 77.1 fg/ml, in subacute CLE (SCLE patients range 30.3–474.2 fg/ml, mean 220.2 fg/ml, and in systemic LE (SLE) patients range 57.6–68.9 fg/ml, mean 63.3 fg/ml.
Expression of MxA protein, which is considered a marker for type I IFN activity (29, 30), was found by immunohistochemistry in 19/20 patients with CLE with moderate to strong expression in both keratinocytes and inflammatory cells (lymphocytes) (Fig. 2d). Expression of MxA protein was not found in control person samples. We also studied the expression of IFN-induced protein with tetratricopeptide repeats (IFIT) 1 and 2 by immunohistochemistry (in 12 patient samples from which RT-qPCR was also performed), since we recently showed that these proteins are upregulated on UV photoprovocation in CLE skin (31). Strong IFIT1 and IFIT2 expression was observed in the keratinocytes of 10/12 CLE skin samples and the expression pattern was similar across the 3 subtypes (Fig. 2e–f and Table II). No expression was observed in the skin samples of control persons.
Table II.
Immunohistochemistry analysis for interferon-induced protein with tetratricopeptide repeats 1 and 2 (IFIT1 and IFIT2)
| Patients | IFIT1 keratinocytes/inflammatory cells | IFIT2 keratinocytes/inflammatory cells |
|---|---|---|
| 4 | +++/− | ++/+ |
| 5 | +++/+ | +++/+ |
| 6 | ++/+ | ++/+ |
| 8 | −/ − | −/ − |
| 9 | ++/− | ++/− |
| 11 | +/+ | +/+ |
| 12 | +++/− | +++/− |
| 13 | +/− | ++/− |
| 14 | +++/+ | +++/+ |
| 15 | +/− | ++/− |
| 19 | +/− | ++/− |
| 20 | −/ − | −/ − |
Keratinocyte immunostaining is graded as follows: − negative; + < 30% cells positive; ++ 30– 70% cells positive, +++ > 70% cells positive. Inflammatory cell immunostaining is graded as follows: − negative, + > 10% positive.
DISCUSSION
NLRP3 inflammasome is known to be expressed in epithelial tissues, including skin, and previous studies have shown expression of pro-IL β and NLRP3 inflammasome components (ASC, caspase-1) in cultured keratinocytes (24, 32, 33). NLRP1 is expressed at high levels in keratinocytes (34). Our results are consistent with these studies as NLRP1/3 and ASC were expressed both in CLE and in control person skin. Caspase-1 was expressed also in the skin of control persons, but marked expression was observed in lesional skin of patients with CLE. As judged by immunohistochemistry, IL1β positivity was found only in lupus skin samples and in none of the controls. Moreover, mature IL-1β detected by WB and increase in IL-1β mRNA in the skin of patients with CLE suggests that NLRP3 inflammasome is activated in patients with CLE. As the differences in the expression of ASC and NLRP3 do not explain the increased IL-1β production, this suggests that, in CLE, there are factors that directly activate the assembly of NLRP3 inflammasome. IL-1β has several effects on promoting inflammation, including recruitment of immune cells from circulation by inducing chemokines and expression of adhesion molecules in endothelial cells, and promoting the synthesis of IL-6, IL-23 and IL-17 (35, 36).
IL-1β could represent a potential target for treatment for a selected group of patients with lupus. After its release, IL-1β binds to the IL-1 receptor type I (IL-1RI) initiating intracellular signalling and transcription of proinflammatory genes (37) (Fig. 1). IL-1Ra regulates the activity of IL-1β by competitively binding to the IL-1R1 (36, 37) (Fig. 1). Anakinra is a recombinant form of the naturally occurring IL-1Ra (37, 38). Studies of the efficacy of anakinra and other drugs targeting IL-1 (canakinumab and rilonacept) in lupus are very limited.
Expression of IL-1β was most prominently observed in SCLE and DLE patients. Many of the patients were on anti-inflammatory drugs at the time of tissue sampling, which could have influenced inflammasome activation, especially in the SLE group, where all the patients were using 2 or more systemic anti-inflammatory drugs. It was not possible to stop the medications before participation, due to the severe nature of the disease.
Type I interferons have previously been shown to play important part in the pathogenesis of CLE (39). The current results are consistent with this, showing increased IFN-α mRNA levels and expression of MxA protein in the skin of patients with CLE. A previous study indicated that, in SLE, long-term type I IFN exposure causes inflammasome activation in monocytes with increased release of IL-1β, mediated by interferon regulatory factor I (IFR-1) (40). In the same study, expression of caspase-1 and IRF-1 mRNA was also investigated from skin biopsies from DLE and SCLE patients, showing a correlation between them (40). In the current study, although a similar increased expression of both IL-1β and IFN-α was observed in the skin of 1 patient with CLE, no statistically significant correlations were found.
Type I IFNs stimulate the expression of IFIT family proteins, for example, during viral infections (41, 42). IFIT proteins have antiviral activity as well as effects on immune responses, and IFIT2 is involved in IFN-induced apoptosis (41, 42). IFIT1 and IFIT2 are usually not expressed under normal conditions. We have previously shown that IFIT1 and IFIT2 proteins are upregulated in the skin of patients with CLE after UV photoprovocation (31). In this study, we found expression of both IFIT1 and IFIT2 in lesional lupus skin that had not undergone photoprovocation, indicating that IFIT proteins are upregulated in CLE skin even by environmental UV exposure.
In summary, NLRP3 inflammasome activity and IL-1β expression are increased in the skin of patients with CLE. This does not seem to correlate with expression of MxA, IFIT1/2, serum IFN-α or skin IFN-α mRNA levels, however. There seems to be significant variation in the expression pattern of NLRP3 inflammasome components among patients with CLE, suggesting that CLE is pathogenetically a heterogeneous disease with distinct subtypes of inflammation. Individualization of treatment according to the disease subtype could be beneficial to the patients.
ACKNOWLEDGEMENTS
This study has been supported by a grant of The Finnish Dermatological Society (KM) and Finska Läkaresällskapet (JP, KKE), and by Helsinki University Hospital Research Funds. DD acknowledges support from the ANR (CE17001002).
Footnotes
The authors have no conflicts of interest to declare.
REFERENCES
- 1.Liu Z, Davidson A. Taming lupus – a new understanding of pathogenesis is leading to clinical advances. Nat Med 2012; 18: 871–882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lisnevskaia L, Murphy G, Isenberg D. Systemic lupus erythematosus. Lancet 2014; 384: 1878–1888. [DOI] [PubMed] [Google Scholar]
- 3.Stannard JN, Kahlenberg JM. Cutaneous lupus erythematosus: updates on pathogenesis and associations with systemic lupus. Curr Opin Rheumatol 2016; 28: 453–459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rothfield N, Sontheimer RD, Bernstein M. Lupus erythematosus: systemic and cutaneous manifestations. Clin Dermatol 2006; 24: 348–362. [DOI] [PubMed] [Google Scholar]
- 5.Kirchhof MG, Dutz JP. The immunopathology of cutaneous lupus erythematosus. Rheum Dis Clin North Am 2014; 40: 455–474, viii. [DOI] [PubMed] [Google Scholar]
- 6.Hagberg N, Rönnblom L. Systemic lupus erythematosus – a disease with a dysregulated type I interferon System. Scand J Immunol 2015; 82: 199–207. [DOI] [PubMed] [Google Scholar]
- 7.Bengtsson AA, Rönnblom L. Role of interferons in SLE. Best Pract Res Clin Rheumatol 2017; 31: 415–428. [DOI] [PubMed] [Google Scholar]
- 8.Bezalel S, Guri KM, Elbirt D, Asher I, Sthoeger ZM. Type I interferon signature in systemic lupus erythematosus. Isr Med Assoc J 2014; 16: 246–249. [PubMed] [Google Scholar]
- 9.Robinson ES, Werth VP. The role of cytokines in the pathogenesis of cutaneous lupus erythematosus. Cytokine 2015; 73: 326–334. [DOI] [PubMed] [Google Scholar]
- 10.Ribero S, Sciascia S, Borradori L, Lipsker D. The cutaneous spectrum of lupus erythematosus. Clin Rev Allergy Immunol 2017; 53: 291–305. [DOI] [PubMed] [Google Scholar]
- 11.Achtman JC, Werth VP. Pathophysiology of cutaneous lupus erythematosus. Arthritis Res Ther 2015; 17: 182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Weidenbusch M, Kulkarni OP, Anders H. The innate immune system in human systemic lupus erythematosus. Clin Sci 2017; 131: 625–634. [DOI] [PubMed] [Google Scholar]
- 13.Shen HH, Yang YX, Meng X, Luo XY, Li XM, Shuai ZW, et al. NLRP3: a promising therapeutic target for autoimmune diseases. Autoimmun Rev 2018; 17: 694–702. [DOI] [PubMed] [Google Scholar]
- 14.Deuteraiou K, Kitas G, Garyfallos A, Dimitroulas T. Novel insights into the role of inflammasomes in autoimmune and metabolic rheumatic diseases. Rheumatol Int 2018; 38: 1345–1354. [DOI] [PubMed] [Google Scholar]
- 15.Jo E, Kim JK, Shin D, Sasakawa C. Molecular mechanisms regulating NLRP3 inflammasome activation. Cell Mol Immunol 2016; 13: 148–159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.He Y, Hara H, Núñez G. Mechanism and regulation of NLRP3 inflammasome activation. Trends Biochem Sci 2016; 41: 1012–1021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kahlenberg JM, Kang I. Advances in disease mechanisms and translational technologies: clinicopathologic significance of inflammasome activation in autoimmune diseases. Arthritis Rheumatol 2020; 72: 386–395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kahlenberg JM, Carmona-Rivera C, Smith CK, Kaplan MJ. Neutrophil extracellular trap-associated protein activation of the NLRP3 inflammasome is enhanced in lupus macrophages. J Immunol 2013; 190: 1217–1226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Shin MS, Kang Y, Lee N, Wahl ER, Kim SH, Kang KS, et al. Self double-stranded (ds)DNA induces IL-1β production from human monocytes by activating NLRP3 inflammasome in the presence of anti-dsDNA antibodies. J Immunol 2013; 190: 1407–1415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhang H, Fu R, Guo C, Huang Y, Wang H, Wang S, et al. Anti-dsDNA antibodies bind to TLR4 and activate NLRP3 inflammasome in lupus monocytes/macrophages. J Transl Med 2016; 14: 156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.da Cruz, HLA, Cavalcanti CAJ, de Azêvedo Silva J, de Lima, CAD, Fragoso TS, Barbosa AD, et al. Differential expression of the inflammasome complex genes in systemic lupus erythematosus. Immunogenetics 2020; 72: 217–224. [DOI] [PubMed] [Google Scholar]
- 22.Kahlenberg JM, Kaplan MJ. The inflammasome and lupus: another innate immune mechanism contributing to disease pathogenesis? Curr Opin Rheumatol 2014; 26: 475–481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pontillo A, Girardelli M, Kamada AJ, Pancotto JA, Donadi EA, Crovella S, et al. Polimorphisms in inflammasome genes are involved in the predisposition to systemic lupus erythematosus. Autoimmunity 2012; 45: 271–278. [DOI] [PubMed] [Google Scholar]
- 24.Feldmeyer L, Keller M, Niklaus G, Hohl D, Werner S, Beer H. The inflammasome mediates UVB-induced activation and secretion of interleukin-1beta by keratinocytes. Curr Biol 2007; 17: 1140–1145. [DOI] [PubMed] [Google Scholar]
- 25.Sand J, Haertel E, Biedermann T, Contassot E, Reichmann E, French LE, et al. Expression of inflammasome proteins and inflammasome activation occurs in human, but not in murine keratinocytes. Cell Death Dis 2018; 9: 24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods 2001; 25: 402–408. [DOI] [PubMed] [Google Scholar]
- 27.Rodero MP, Decalf J, Bondet V, Hunt D, Rice GI, Werneke S, et al. Detection of interferon alpha protein reveals differential levels and cellular sources in disease. J Exp Med 2017; 214: 1547–1555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Meyer S, Woodward M, Hertel C, Vlaicu P, Haque Y, Kärner J, et al. AIRE-deficient patients harbor unique high-affinity disease-ameliorating autoantibodies. Cell 2016; 166: 582–595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Haller O, Kochs G. Human MxA protein: an interferon-induced dynamin-like GTPase with broad antiviral activity. J Interferon Cytokine Res 2011; 31: 79–87. [DOI] [PubMed] [Google Scholar]
- 30.Wenzel J, Zahn S, Bieber T, Tüting T. Type I interferon-associated cytotoxic inflammation in cutaneous lupus erythematosus. Arch Dermatol Res 2009; 301: 83–86. [DOI] [PubMed] [Google Scholar]
- 31.Katayama S, Panelius J, Koskenmies S, Skoog T, Mahonen K, Kisand K, et al. Delineating the healthy human skin UV Response and early induction of interferon pathway in cutaneous lupus erythematosus. J Invest Dermatol 2019; 139: 2058–2061.e4. [DOI] [PubMed] [Google Scholar]
- 32.Peeters PM, Wouters EF, Reynaert NL. Immune homeostasis in epithelial cells: evidence and role of inflammasome signaling reviewed. J Immunol Res 2015; 2015: 828264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Watanabe H, Gaide O, Pétrilli V, Martinon F, Contassot E, Roques S, et al. Activation of the IL-1beta-processing inflammasome is involved in contact hypersensitivity. J Invest Dermatol 2007; 127: 1956–1963. [DOI] [PubMed] [Google Scholar]
- 34.Fenini G, Karakaya T, Hennig P, Di Filippo M, Beer HD. The NLRP1 inflammasome in human skin and beyond. Int J Mol Sci 2020; 21: 4788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Dinarello CA. Immunological and inflammatory functions of the interleukin-1 family. Annu Rev Immunol 2009; 27: 519–550. [DOI] [PubMed] [Google Scholar]
- 36.Dinarello CA. Interleukin-1 in the pathogenesis and treatment of inflammatory diseases. Blood 2011; 117: 3720–3732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lane T, Lachmann HJ. The emerging role of interleukin-1β in autoinflammatory diseases. Curr Allergy Asthma Rep 2011; 11: 361–368. [DOI] [PubMed] [Google Scholar]
- 38.Dinarello CA. Blocking interleukin-1β in acute and chronic autoinflammatory diseases. J Intern Med 2011; 269: 16–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wenzel J, Tüting T. Identification of type I interferon-associated inflammation in the pathogenesis of cutaneous lupus erythematosus opens up options for novel therapeutic approaches. Exp Dermatol 2007; 16: 454–463. [DOI] [PubMed] [Google Scholar]
- 40.Liu J, Berthier CC, Kahlenberg JM. Enhanced inflammasome activity in systemic lupus erythematosus is mediated via type I interferon-induced up-regulation of interferon regulatory factor 1. Arthritis Rheumatol 2017; 69: 1840–1849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Diamond MS, Farzan M. The broad-spectrum antiviral functions of IFIT and IFITM proteins. Nat Rev Immunol 2013; 13: 46–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Reich NC. A death-promoting role for ISG54/IFIT2. J Interferon Cytokine Res 2013; 33: 199–205. [DOI] [PMC free article] [PubMed] [Google Scholar]